Abstract
The human microbiota acts as a diverse source of molecular cues that influence the development and homeostasis of the immune system. Beyond endogenous roles in the human holobiont, host-microbial interactions also alter outcomes for immune-related diseases and treatment regimens. Over the past decade, sequencing analyses of cancer patients have revealed correlations between microbiota composition and the efficacy of cancer immunotherapies such as checkpoint inhibitors. However, very little is known about the exact mechanisms that link specific microbiota with patient responses, limiting our ability to exploit these microbial agents for improved oncology care. Here, we summarize current progress towards a molecular understanding of host-microbial interactions in the context of checkpoint inhibitor immunotherapies. By highlighting the successes of a limited number of studies focused on identifying specific, causal molecules, we underscore how the exploration of specific microbial features such as proteins, enzymes, and metabolites may translate into precise and actionable therapies for personalized patient care in the clinic.
Keywords: Cancer immunotherapy, Checkpoint inhibitor, Gut microbiota, Microbial metabolites
Abbreviations: CAR, chimeric antigen receptor; SCFA, short-chain fatty acid; GPCR, G protein-coupled receptor; FMT, fecal microbiota transplant; ABX, antibiotic; GF, germ-free; SPF, specific pathogen-free; OTU, operational taxonomic unit; ASV, amplicon sequence variant; MM, metastatic melanoma; RCC, renal cell carcinoma; NSCLC, non-small cell lung cancer; HCC, hepatocellular carcinoma; Th1, T helper 1; Treg, regulatory T; Th17, T helper 17; Tc1, type 1 cytotoxic T; Tmp, memory precursor T; BMDC, bone marrow-derived dendritic cell; MNP, mononuclear phagocyte; NK, natural killer; cdAMP, cyclic di-adenosine monophosphate; MAMP, microbe-associated molecular pattern; MDP, muramyl dipeptide; EcN, Escherichia coli Nissle 1917
Introduction
Precision cancer treatment is built upon biological differences that govern tumor growth and development within individual patients. For example, tumor-intrinsic factors such as key oncogenic driver mutations, the presence or absence of specific tumor cell-surface receptors, and dysregulated signaling pathways can guide therapeutic selection [1], [2], [3]. Beyond tumor cell autonomous features, the interplay between the self and the “altered self” tumor via host immunity has proven an incredibly fruitful area for clinical intervention [4], [5], [6]. Grouped under the broad umbrella of cancer immunotherapies, these immune cell-targeted approaches utilize diverse immune responses to treat solid and hematologic malignancies [5,6].
The two most prevalent types of cancer immunotherapies in the clinic both rely on T cell-driven cytotoxicity. Chimeric antigen receptor (CAR) T cells are artificially produced using a patient's own T cells that are genetically reprogrammed to recognize and kill cancer cells through viral transduction of a transgene CAR specific for a tumor antigen [5]. A second approach, immune checkpoint blockade, boosts naturally occurring or therapeutically induced antitumor adaptive responses through targeting regulatory pathways known as immune checkpoints [6]. Currently, numerous antibodies against the CTLA4 and PD-1/PD-L1 inhibitory pathways have been clinically approved to treat a wide variety of tumor types including melanoma, non-small cell lung cancers, and renal cell carcinoma [7]. Because these drugs act through the potentiation of an adaptive immune response, the clinical success of checkpoint inhibitors relies on pre-existing tumor immunity [8]. Therefore, their activity is often lower in “cold” tumors that exhibit fewer accessible neoantigens and lower levels of prior T cell infiltration [9]. Even within patient populations suffering from malignancies approved for treatment, response is variable and likely results from a combination of host-intrinsic and host-associated factors [10].
In recent years, the human microbiota has been significantly correlated with various host phenotypes including immune system development and homeostasis [11]. These microorganisms, which include bacteria, viruses, archaea, and fungi, inhabit every mucosal and barrier surface of the body as well as some internal organs to form complex, individual- and location-specific ecosystems [12]. Cohabitation between the host and microbes is quite prominent within the gut, which contains a roughly equivalent number of microbial cells as there are human cells throughout the body [13]. The genetic information from these microorganisms – the gut microbiome – greatly outnumbers human genes and provides a rich source for metabolic and signaling activity that can vary from person to person. Microbially generated biomolecules are sensed by the host to elicit immune cell differentiation and activation. These molecular connections are highlighted by the activities of short-chain fatty acids (SCFAs) and secondary bile acids, which directly bind to G-protein coupled receptors (GPCRs) [14,15] and nuclear receptors such as RORɣt [16], [17], [18], respectively, to generate specific helper T subsets among other immune cell phenotypes.
Together, the genetic and metabolic output of the microbiota can shape many aspects of host immunity. These activities can affect both initial responses to “altered self” entities like tumors and the clinical efficacy of immune-targeted therapies like checkpoint blockade. Nevertheless, very little is known about the exact molecular mechanisms by which the microbiota can alter therapeutic responses to checkpoint inhibitors. In this review, we summarize our current understanding of the relationship between microbiota and checkpoint blockade efficacy with an emphasis on the exact microbial agents that underlie these correlations. By concentrating on the molecular basis of host-microbial interactions during cancer treatment, this reductive approach may circumvent the inherent difficulties in modulating microbiota output through bulk, ecological means such as fecal microbiota transplants (FMTs), antibiotics (ABX), or colonization with defined communities. In turn, this molecular focus may enable us to predict and augment immunotherapy activity as a new form of precision medicine using synthetic molecules.
Establishing correlations
Landmark experiments in mouse models have demonstrated that the microbiota alters the efficacy of immune checkpoint inhibitors [19,20]. To first show this dependency, bulk methods to alter the host microbiota were employed. In one study, the growth of B16 melanoma ectopic tumors in response to anti-PD-L1 was found to depend on the vendor origin of the animals [19]. The importance of the microbiota itself on this phenotype was then elucidated through cohousing of animals or direct cross-colonization using FMT. In a related study, treatment of MCA205 fibrosarcoma ectopic tumors with anti-CTLA4 was found to require the microbiota, with a loss of antitumor activity observed in both germ-free (GF) or ABX-treated models compared to specific pathogen-free (SPF) mice [20].
Beyond broad methods to alter entire microbiota communities, genomic sequencing has been employed to identify specific microbes that correlate with activity [21,22]. These techniques are often performed using bacterial 16S rRNA sequencing (Fig. 1A). Here, a genomic locus encoding the 16S ribosomal RNA subunit is targeted using primers that recognize sequences highly conserved across all bacteria. The resulting amplicon contains sequences from hypervariable regions – usually V3 to V5 – that can be used to identify bacteria within the mixtures. Taxonomic assignment can be carried out on the sequences using two common methods. After pairwise sequence alignments, sequences that share a high percentage similarity within the hypervariable regions can be clustered into operational taxonomic units (OTUs). A single, representative amplicon within the OTU is then aligned to a reference database to assign a single taxon to the OTU. Depending on the clustering stringency, the reference-based assignment of OTU clusters can underestimate the actual diversity within the sequenced community by combining highly similar but nonequivalent amplicons [23,24]. OTU clustering can generally yield bacterial identifications down to the taxonomic level of genus, whereas species-level discrimination requires additional sequencing techniques such as quantitative PCR using species-specific primers or shotgun metagenomic sequencing [25].
Fig. 1.
Methods to establish and verify correlations between microbiota and immunotherapy response. [A] Bacterial community composition can be measured by operational taxonomic unit (OTU) clustering or amplicon sequence variant (ASV) assignment of 16S rRNA sequencing libraries that reflect the abundance of different bacterial taxa. [B] Avatar mice that are colonized with microbiota from responding or non-responding patients can be used to phenotype tumor growth and immune checkpoint treatment, which may phenocopy drug efficacy in the original fecal microbiota transplant (FMT) sources. [C] Microbiota that elicit specific immune cell types can be rationally selected through iterative in vivo passaging along with correlation analysis between microbiota composition and immune cell levels. Figure created with Biorender.com.
Alternatively, 16S rRNA sequencing results can be taxonomically assigned by exact sequences known as amplicon sequence variants (ASVs) [26,27]. Here, different sequences are treated as separate entities rather than clustered into high similarity OTUs. To avoid the inclusion of artificial ASVs caused by inherent sequencing errors, the number of reads for each ASV is used to derive a confidence score for its presence in the mixture, which can be filtered to remove potential artifacts. Because ASV assignments can discriminate single nucleotide differences, this method has been proposed as a higher resolution approach to catalog sample diversity compared to OTU clustering [27]. Moreover, the use of ASVs also allows for the direct evaluation of multiple datasets without the need to recluster OTUs. Nevertheless, both OTU clustering and ASV assignment of 16S rRNA sequences provide cost-effective means to rapidly measure community composition across human microbiota samples. For example, in the initial studies described above, changes in anti-PD-L1 and anti-CTLA4 efficacy were correlated with the presence of Bifidobacterium and Bacteroides species, respectively [19,20]. The identification of these correlated genera enabled their direct testing in the same animal models through gut colonization, proving that these bacteria were sufficient to induce changes in checkpoint blockade response.
Genomic sequencing has also enabled correlations between microbiota composition and therapeutic efficacy in humans. To date, more than 20 studies have used a combination of 16S rRNA and/or shotgun metagenomic sequencing to identify bacteria in patient populations treated with immune checkpoint inhibitors [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50] (Table 1). Although many early studies were performed using samples from melanoma patients [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], these analyses have also been extended to other tumor types including non-small cell lung cancer, renal cell carcinoma, hepatocellular carcinoma, and gastrointestinal cancers [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50]. In all cases, analyses revealed unique bacterial OTUs or species that positively correlated with patient outcomes. However, the identified bacteria were largely study-specific, with only a few species like Faecalibacterium prausnitzii [[28], [29], [30],32] and Akkermansia muciniphila [36,38,41,46] enriched across multiple analyses. These differences may be attributed to the relatively small cohort sizes of some studies along with other sources of variability that influence microbiota composition including geography, age, diet, and other patient characteristics as well as distinct methods used for sampling, sequence clustering, and correlation analyses [51,52]. Nevertheless, these foundational studies established that the microbiota differed between responding and nonresponding patients.
Table 1.
Summary of large-scale, untargeted studies correlating microbiota taxa with immunotherapy efficacy. MM = metastatic melanoma, NSCLC = non-small cell lung cancer, RCC = renal cell carcinoma, HCC = hepatocellular carcinoma, GI = gastrointestinal.
| Cancer | Therapy | Sample Size | Major Positively Correlated Taxa | Year | Reference |
|---|---|---|---|---|---|
| Metastatic melanoma | |||||
| MM | ɑPD-1 +/- ɑCTLA-4 | 39 | Bacteroides caccae, Faecalibacterium prausnitzii, Bacteroides thetaiotaomicron, Holdemania filiformis, Dorea formicogenerans | 2017 | [28] |
| MM | ɑCTLA-4 | 26 | Faecalibacterium prausnitzii, Gemmiger formicilis, Clostridium XIVa | 2017 | [29] |
| MM | ɑPD-1 | 43 | Clostridiales, Ruminococcaceae, Ruminococcus bromii, Faecalibacterium, Phascolarctobacterium | 2018 | [30] |
| MM | ɑPD-1 | 42 | Bifidobacterium spp., Enterococcus faecium, Collinsella aerofaciens, Klebsiella pneumoniae, Veillonella parvula, Parabacteroides merdae | 2018 | [31] |
| MM | ɑCTLA-4 and/or ɑPD-1 | 27 | Faecalibacterium prausnitzii, Coprococcus eutactus, Prevotella stercorea, Streptococcus spp., Lachnospiraceae | 2019 | [32] |
| MM | ɑCTLA-4 | 38 | Faecalibacterium, Gemmiger | 2020 | [33] |
| MM | ɑPD-1 +/- ɑCTLA-4 | 25 | Streptococcus parasanguinis, Bacteroides massiliensis | 2020 | [34] |
| MM | ɑCTLA-4 + ɑPD-1 | 54 | Bacteroides stercoris, Parabacteroides distasonis, Fournierella massiliensis | 2021 | [35] |
| MM | ɑCTLA-4 and/or ɑPD-1 | 165 | Akkermansia muciniphila, Doria formicigenerans, Bifidobacterium pseudocatenulatum, Roseburia spp. | 2022 | [36] |
| MM | ɑPD-1 | 94 | Ruminococcus spp., Blautia spp., Eubacteria rectale, Anaerostipes hadrus | 2022 | [37] |
| Other cancers | |||||
| NSCLC, RCC | ɑPD-1 | 100 | Akkermansia muciniphila, Alistipes indistinctus, Enterococcus spp. | 2018 | [38] |
| NSCLC, gastric | ɑPD-1 | 38 | Ruminococcaceae | 2018 | [39] |
| NSCLC | ɑPD-1 | 25 | Alistipes putredinis, Prevotella copri, Bifidobacterium longum, Lachnobacterium, Lachnospiraceae, Shigella | 2019 | [40] |
| HCC | ɑPD-1 | 8 | Bifidobacterium dentium, Akkermansia muciniphila, Lactobacillus oris, Dialister invisus, Coprococcus comes | 2019 | [41] |
| NSCLC | ɑPD-1 | 17 | Lactobacillus, Clostridium, Syntrophococcus | 2019 | [42] |
| NSCLC | ɑPD-1 | 63 | Alistipes spp., Desulfovibrio desulfuricans, Bacteroides nordii, Methanobrevibacter smithii, Parabacteroides merdae | 2020 | [43] |
| NSCLC | ɑPD-1 or ɑPD-L1 | 54 | Ruminococcaceae UCG 13, Agathobacter | 2020 | [44] |
| GI cancers | ɑPD-1 +/- ɑCTLA-4 or ɑPD-L1 | 74 | Ruminococcaceae, Clostridium sp. CAG:352, Prevotella, Dialister, Lachnospiraceae | 2020 | [45] |
| RCC | ɑPD-1 | 58 | Akkermansia muciniphila, Bacteroides salyersiae | 2020 | [46] |
| RCC | ɑPD-1 +/- ɑCTLA-4 | 31 | Prevotella copri, Bifidobacterium adolescentis, Barnesiella intestinihominis, Odoribacter splanchicus, Bacteroides eggerthii | 2020 | [47] |
| NSCLC | ɑPD-1 | 75 | Desulfovibrio, Bifidobacterium, Odoribacteriaceae, Anaerostipes, Rikenellaceae, Faecalibacterium, Alistipes | 2021 | [48] |
| Thoracic carcinoma | ɑPD-1 | 42 | Akkermansiaceae, Enterococcaceae, Enterobacteriaceae, Carnobacteriaceae, Clostridiales family XI | 2021 | [49] |
| NSCLC | ɑPD-1 +/- ɑCTLA-4 or ɑPD-L1 | 65 | Ruminococcus, Akkermansia, Faecalibacterium | 2022 | [50] |
Although an exciting starting point, deciphering these individual correlations remains challenging. Because the microbiota is a co-existing community of multiple microorganisms, it is unclear if each correlated microbe is a causal agent or a result of other influences on the community ecology such as keystone species [53]. Moreover, a mixed microbial population may yield emergent properties through collective interactions that cannot be recapitulated by individual correlated taxa [54]. Even in the case of an individual, causal species, strains of the same bacterial species can possess significantly different metabolic outputs [55], further complicating monocolonization experiments to establish if a species is a causal agent. Finally, while phylogeny has helped to organize the correlated species, these associations do not fully predict the activity of related taxa [56], necessitating validation experiments to prove causality. These difficulties underscore a key question that arises from current correlations – what are the exact molecular features that underlie microbial modulation of checkpoint immunotherapy? A focus on shared function rather than phylogeny may explain the activities of multiple, unrelated species and establish defining traits to predict phenotypes resulting from individual species or communities.
Verifying correlations with host phenotypes
As a first and critical step, murine tumor models are often used to establish the effects of a single microbe or defined community on checkpoint blockade. As mentioned above, early studies examining murine microbiota compositions found that Bifidobacterium and Bacteroides species correlated with responsiveness to checkpoint inhibitor treatment [19,20]. Correlations were then directly validated in tumor models by supplementation onto an intact, nonresponsive microbiota as well as monocolonization with ABX-pretreated or GF animals . This reconstitution-based approach has also been extended to human-relevant microbiota. Three studies used FMTs to colonize ABX-pretreated or GF animals with fecal material from responding or nonresponding patients (Fig. 1B), and each study independently found that reconstitution could phenocopy the antitumor effects seen in patients [30,31,38]. These “avatar” mouse models establish both causality and sufficiency of the reconstituted microbes and provide opportunities to discover other host immune phenotypes that correlate with response. In all three studies, mice colonized with responding microbiota showed evidence of increased type 1 immunity, with each study demonstrating related cellular phenotypes including increased CD8+ T cell infiltration into tumors [30] as well as increased amounts of antigen-specific CD8+ [31] and CXCR3+CD4+ T cells [38]. However, the avatar system alone does not directly explain how these different and incompletely defined communities converge on the observed adaptive immune activity.
Despite these unknowns, reconstitution of a functional microbiota using fecal material from responding donors has progressed to clinical trials. Two recent reports have outlined early-stage clinical data using ABX pretreatment and FMT to treat patients refractory to prior anti-PD-1 therapy [57,58]. In one trial (NCT03353402), patients received FMTs from one of two donors that were previously treated for metastatic melanoma with anti-PD-1 and achieved complete response for over one year [57]. Three of five patients receiving FMTs from the same donor showed clinical responses, whereas the five patients receiving FMTs from the second donor showed no clinical benefit. Principal component analysis of pre- and post-FMT microbiota populations by 16S rRNA showed that both groups shifted towards the composition of the original donor. FMTs elicited increased CD68+ antigen-presenting cell infiltration into the gut lamina propria, and responding patients showed increased CD8+ T cell infiltration into tumors.
In the second trial (NCT03341143), seven donors successfully treated with anti-PD-1 therapy were used to treat 16 patients, and six of the recipients showed partial or complete responses [58]. Although different patients receiving FMTs from the same donor did not always show similar responses, multidimensional Euclidean analysis found that the microbiota of responding patients shifted significantly towards the donor composition, which was not consistently observed in the non-responding patients. Longitudinal single cell analyses of peripheral mononuclear blood cells and tumor-infiltrating immune cells revealed significant increases in cytolytic CD56+ CD8+ T cells and terminally differentiated effector memory CD8+ T cells (CCR7−CD45RA+). Decreases were also observed in overall and CXCL8-expressing myeloid cells as well as regulatory T cells (Tregs). These studies show that FMTs may be able to recapitulate the responding phenotype of donors in a subset of patients.
In a complementary approach, the host phenotype can be used to guide the selection and rational design of an active microbial consortium [Fig. 1C]. Defined consortia have been developed to elicit the development of multiple T cell subtypes including Tregs [59], T helper 17 (Th17) [60], and most recently IFNɣ-expressing CD8+ T cells [61]. To achieve this, GF animals were first colonized by FMT from human donors, and microbiota from animals demonstrating the highest level of the desired cell type were passaged in vivo into new recipients. The consortium was further modulated through selective ABX administration, and OTUs identified by genomic sequencing were correlated with the prevalence of IFNɣ+CD8+ T cells across the different conditions, leading to a final community of 11 strains. Reconstitution of GF mice or supplementation of SPF mice with the 11-strain mixture improved the activity of checkpoint blockade in multiple tumor models, and this activity was dependent on CD8+ T cells as demonstrated by antibody depletion studies. The development of IFNɣ+CD8+ T cells required CD103+ lamina propria dendritic cells (DCs) through an MHC class Ia-dependent mechanism; however, the exact molecular requirements from the microbes are unknown. Nevertheless, this consortium as well as others are currently being explored in clinical trials (NCT04208958).
Although potentially feasible in clinical settings, reconstitution methods face several technical challenges. For donor derived samples used for FMTs, the selection of the donor and the successful engraftment of the FMT microbiota appears to be crucial in preliminary clinical data. Moreover, the material supply may be limiting, its composition and efficacy may change over time as new samples are acquired, and standardization of FMT harvesting and administration techniques are needed. For defined consortia, colonization levels of each member microbe may differ between individuals and affect efficacy, which may be resolved through the optimization of dosing and long-term monitoring of microbiota composition in patients. To address these concerns, a better understanding of the molecular basis of these bioactive microbes may provide an alternative route for treatment that avoids the ecological and practical variables of reconstitution.
Molecular precision via microbial mechanisms
The discovery of microbial mechanisms and molecules that elicit complex host responses like antitumor immunity is unsurprisingly quite challenging. Shotgun metagenomic sequencing of responding or nonresponding microbiota often shows enrichment of numerous metabolic and signaling processes [32], from which it remains difficult to directly implicate a specific pathway as a causative agent. Similarly, characterization of active and inactive isolates from a single species via whole genome sequencing can yield hundreds of unique genes between strain genomes [55], and further bioinformatic analyses require sufficient functional annotation of the gene products. Finally, metabolomic analyses of cecal or serum contents can generate hundreds of potential metabolite leads [61] and often relies on spectral matching to established reference libraries that may underrepresent microbial products [62]. Despite these significant challenges, a small number of studies have highlighted how a molecular understanding of microbial activity may provide new paths towards improved and personalized cancer care.
T cell stimulation
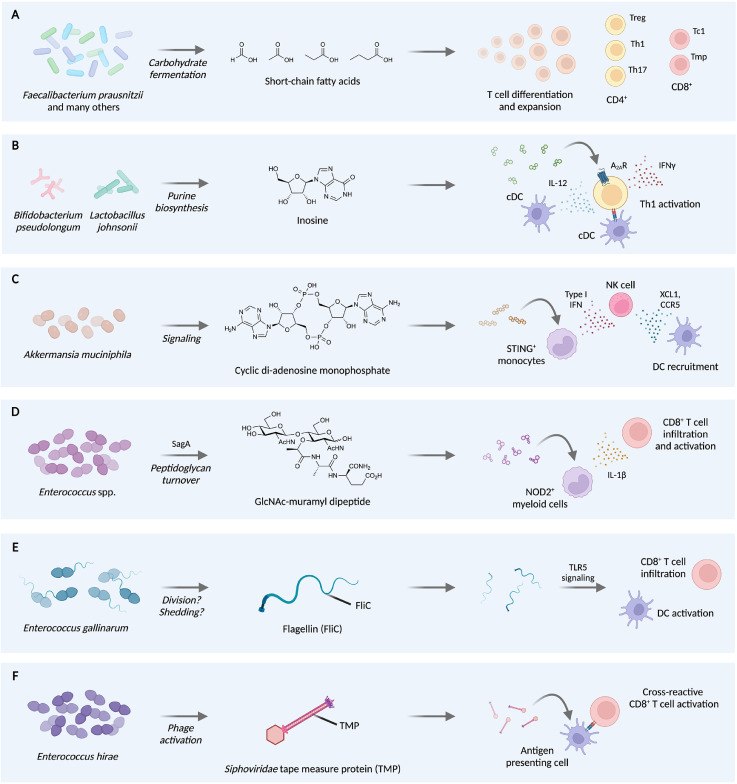
One of the best studied microbial metabolites is a class of aliphatic carboxylates known as short-chain fatty acids (SCFAs) [63,64] (Fig. 2A). SCFAs most commonly consist of a mixture of formate, acetate, propionate, and butyrate and are produced by the bacterial fermentation of carbohydrates not digested or absorbed by the host like dietary fiber [65]. These molecules have been shown to have profound effects on host immunity, including the expansion of Treg cells [59,66,67] and the development of effector CD4+ and CD8+ T cells [68], [69], [70], [71]. Gut microbes that generate SCFAs, including the major butyrate producer Faecalibacterium prausnitzii [72], as well as fecal SCFA concentrations in patients have been correlated with clinical efficacy of PD-1/PD-L1-targeting checkpoint inhibitors [[28], [29], [30],73]. Moreover, fiber intake in both mouse models and humans have been linked with improved activity of anti-PD-1/PD-L1 [74,75]; however, fecal and serum SCFA levels did not broadly correlate with fiber intake in mouse experiments [75].
Fig. 2.
Molecular mechanisms of microbiota-mediated potentiation of checkpoint blockade. [A] F. prausnitzii and other microbes produce short-chain fatty acids (SCFAs), which stimulate differentiation and expansion of multiple T cell subsets, including regulatory T (Treg), T helper 1 (Th1), T helper 17, cytotoxic T (Tc1), and memory precursor T (Tmp) cells. [B] B. pseudolongum and L. johnsonii synthesize the purine inosine, which engages A2AR on Th1 cells to elicit activation in an IL-12- and antigen-dependent manner. [C] A. muciniphilia generates cyclic di-AMP, which activates STING+ monocytes to activate natural killer (NK) cells that in turn recruit dendritic cells (DCs). [D] Enterococcus species express and secrete the peptidoglycan hydrolase SagA to produce GlcNAc-MDP and other muropeptides, which activates NOD2+ myeloid cells and leads to increased CD8+ T cell infiltration and activation. [E] E. gallinarum produces flagellin, which activates DCs and increases CD8+ T cell infiltration in a TLR5-dependent manner. [F] E. hirae hosts a phage, in which the tape measure protein (TMP) contains an MHC-I-restricted antigen that can activate CD8+ T cells to cross-react with PSMB4-overexpressing tumors. Figure created with Biorender.com.
Conversely, recent data has found that butyrate may negatively affect CTLA4 targeting [33]. Serum levels of butyrate were found to inversely correlate with progression-free and overall survival in a small cohort of metastatic melanoma patients treated with ipilimumab. In mice, direct administration of sodium butyrate in drinking water also led to a partial blunting of anti-CTLA4 efficacy against tumor growth as well as decreased DC maturation and T cell priming. Together, these data suggest that the effects of SCFAs on checkpoint blockade are not monolithic and may depend on the therapeutic target of the checkpoint inhibitor.
Direct stimulation of T cells by microbial metabolites may be a general feature of synergistic microbiota. Using a phenotype-driven approach, three strains of Bifidobacterium psuedolongum, Lactobacillus johnsonii, and Olsenella spp. were isolated from mice that responded to anti-CTLA4 treatment of the azoxymethane and dextran sodium sulfate mouse model of colorectal cancer [76]. Untargeted serum metabolomics of GF animals colonized with B. pseudolongum or a non-responsive microbe showed elevated levels of the purine nucleoside inosine and related degradation products including hypoxanthine (Fig. 2B). Inosine has been ascribed both immunosuppressive and immunostimulatory roles by altering T helper 1 (Th1) differentiation. Here, inosine treatment of CD4+ T cell and bone marrow-derived dendritic cell (BMDC) co-cultures led to increased expression of the Th1 master transcription factor T-bet in the presence of anti-CD3/CD28 or IFNɣ co-stimulation. This increase in T-bet-expressing CD4+ T cells was also observed in the small intestine lamina propria colonized by B. pseudolongum.
Inosine is a known ligand for adenosine 2A receptor (A2AR) on T cells [77]. Adoptive transfer of wild-type or A2AR-deficient T cells revealed that rejection of MC38 colorectal cancer ectopic tumors in B. pseudolongum-colonized mice via anti-CTLA4 required this receptor. Improvement of anti-CTLA4 efficacy via monocolonization with Akkermansia muciniphila and L. johnsonii was also shown to depend on A2AR using the adoptive transfer model. Interestingly, inosine alone was not sufficient to augment anti-CTLA4 activity. However, co-administration of inosine and the potent TLR9 agonist CpG in GF mice could recapitulate the enhanced anti-tumor activity of B. pseudolongum colonization and anti-CTLA4 therapy in multiple tumor models. Thus, unrelated microbes can possess overlapping immune modulatory activities through the production of identical or related metabolites and the engagement of a conserved host signaling pathway. Nevertheless, the inability of the metabolite alone to recapitulate the antitumor phenotype in GF mice suggests the involvement of additional, integral mechanisms caused by gut colonization.
Myeloid cell stimulation
Changes in myeloid cell populations and activation may also underlie the effects of gut microbiota on therapeutic response. For example, EL4 lymphoma ectopic tumors in SPF mice showed significant differences in mononuclear phagocyte (MNP) populations compared to tumors in GF mice, with broad increases in anti-tumorigenic monocytes and macrophages as well as dendritic cells in the presence of a gut microbiota [74]. Tumor lysates from SPF mice were enriched in type I interferon, which led to IFN-dependent recruitment of natural killer (NK) cells into the tumor microenvironment. In turn, NK cells in SPF mice produced higher levels of the DC chemoattractants XCL1 and CCL5. A similar intratumoral expression profile could be stimulated using the bacterial metabolite, cyclic di-adenosine monophosphate (cdAMP) (Fig. 2C). This immune activating molecule initiates host STING signaling upstream of type I interferon production [78], providing a potential mechanism for the observed SPF-mediated shifts in MNP populations. Supplementation of MC38 tumor-bearing mice with a high-fiber diet also led to similar changes in intratumoral MNP amounts and improved responses to PD-1/PD-L1 targeting.
To identify correlations between differentially abundant microbes caused by fiber treatment and cell type or phenotypic changes, transkingdom network analysis [79] was employed, revealing that Akkermansia muciniphila strongly influenced network structure. As validation, colonization of GF mice with A. muciniphila was found to elicit comparable MNP population changes. A. muciniphila was also shown to produce cdAMP both in vitro and in vivo, providing a potential microbial source for STING agonism. Host phenotypes within this STING-type I interferon-NK-dendritic cell signaling axis correlated with response to checkpoint blockade in melanoma patients, providing microbe-elicited biomarkers that may potentially predict therapeutic response. Moreover, these data along with other previous work on A. muciniphila [38,65] suggest that bacteria may generate multiple immune active metabolites, which can each independently contribute to complementary antitumor signaling pathways.
Immune activation by the recognition of other conserved microbe-associated molecular patterns (MAMPs) has also been implicated in improved checkpoint inhibitor efficacy. For example, colonization of B16 melanoma tumor-bearing mice with species of Enterococcus were shown to synergize with anti-PD-L1 treatment [80]. Although this activity extended to numerous Enterococcus species including E. faecium, E. durans, E. hirae, and E. mundtii, the synergistic effects of these microbes were not observed in the related species E. faecalis. Using targeted bioinformatic analyses, the active Enterococcus species were found to possess highly conserved orthologs of the E. faecium protein SagA. SagA is a D,L-endopeptidase that can degrade bacterial peptidoglycan into immune active muropeptide fragments [81] (Fig. 2D). Previous work has demonstrated that SagA can broadly improve tolerance towards enteric infection through innate immune signaling and improved barrier functions within the gut [82,83]. Here, transgenic expression of SagA was sufficient to improve the antitumor effects of E. faecalis and to increase intratumoral levels of effector and antigen-specific CD8+ T cells. SagA expression within the gut also improved the efficacy of anti-PD-1 and anti-CTLA4 against two other tumor models, suggesting the involvement of a conserved mechanism upstream of effector T cell activity.
To better define the molecular basis of this pathway, anti-PD-L1 was co-administered with muramyl dipeptide (MDP), a synthetic analog of the SagA-produced muropeptides. The small molecule combination therapy also inhibited tumor growth, and the activity of SagA required the host muropeptide receptor NOD2 [84,85]. Single cell RNA sequencing of intratumoral CD45+ cells from the MDP co-treatment model uncovered significant population changes across myeloid cell clusters, including increased amounts of monocytes and decreased amounts of tumor-associated macrophages. In addition, broad increases in NOD2-dependent signaling including NF-κB and MAPK pathways were observed, leading to a pro-inflammatory environment within the tumor.
Beyond small molecules, proteinaceous MAMPs have also been exploited to improve checkpoint blockade. Enterococcus gallinarum is one of only two motile species of Enterococcus via the production of flagellae [86], thread-like projections that are multiple microns in length and consist of repeating units of the protein flagellin (Fig. 2E). Flagellin is recognized by the host receptor TLR5 [87], and this protein has been extensively examined for its ability to activate host immunity as an antitumor agent [88]. The E. gallinarum strain MRx0518 isolated from a healthy human fecal sample was shown to be a potent activator of NF-κB- and TLR5-dependent signaling in vitro compared to the highly motile, murine-derived E. gallinarum strain DSM100110 [89]. This activity was also observable within the bacterial supernatant and was depleted upon trypsin treatment, suggesting that MRx0518 likely shed highly potent flagellin protein. Mutation within the flagellin fliC gene of MRx0518 abrogated activity, and transfer of fliC to DSM100110 led to increased TLR5 activation, showing that the flagellin protein was sufficient for the observed in vitro response. Phylogenetic analysis of the FliC protein sequence across E. gallinarum genomes showed two distinct clades, suggesting that the potent FliC isoform may be shared by a subset of strains.
As a monotherapy, preliminary data from a small cohort of breast cancer patients have shown that oral supplementation with MRx0518 increases gene signatures for myeloid inflammation and inflammatory cytokines as well as increased activated DCs and CD8+ T cell infiltration (NCT03934827). MRx0518 is also under investigation as a co-therapy with anti-PD-1 and anti-PD-L1 in phase II studies for the treatment for multiple cancer types (NCT03637803, NCT05107427). Although well-tolerated in the current trial, a separate study has found that E. gallinarum can translocate from the gut upon barrier disruption and promote autoimmunity [90], underscoring how strain-level variability and other microbial activities are important considerations in the selection of new probiotics.
Molecular mimicry
Finally, molecular mimicry between microbial and host antigens may yield antitumor immune activity. T cells that react strongly to self-antigens are eliminated during their development in the thymus [91]; however, some self-reactive conventional T cells can escape clonal deletion and are controlled by anergy, ignorance, or active Treg-mediated suppression [92]. In some cases, these self-reactive T cells may be aberrantly activated by similar immunogenic microbial antigens [93]. Although this activation can lead to autoimmunity, microbe-specific T cells have been associated with improved outcomes for cancer patients [20,38,94,95]. Thus, cross-reactivity between microbial and host antigens may lead to T cell-mediated killing of tumor cells expressing self- or neoantigens that are recognized by microbe-specific T cells. Treatment of MCA205 tumor-bearing mice with cyclophosphamide, a chemotherapy that enables bacterial translocation from the gut, led to decreased tumor growth upon colonization with the Enterococcus hirae 13144 strain but not other tested strains [96]. Stimulation of CD8+ T cells from colonized mice with DCs preincubated with different E. hirae strains showed recall responses specific for 13144.
To identify the potential antigen, bioinformatic mining of the genome for 13144-specific proteins was used, followed by in silico prediction of MHC-I binding peptides and ex vivo recall response using nonapeptide libraries. These screening efforts led to the discovery of an epitope derived from a tail length tape measure protein (TMP) within a prophage sequence from 13144 (Fig. 2F). Vaccination of mice with DCs loaded with TMP peptide both before and after MCA205 tumor inoculation led to decreases in tumor size. The TMP antigen aligned with a sequence within the host protein PSMB4, which is over-expressed in MCA205 cells. Point mutations within the putative PSMB4 antigen ablated the effects of 13144 colonization during CTX treatment, suggesting the synergistic effects of 13144 and CTX were due to antigen-specific T cells. Treatment of MCA205 tumors with anti-PD-1 were also enhanced by 13144 colonization in an antigen-specific manner, suggesting that antigen mimicry may function across multiple therapeutic interventions.
Exploiting molecular mechanisms to engineer next-generation probiotics
The identification of specific microbial mechanisms that improve checkpoint blockade provides the unique opportunity to engineer therapeutic probiotics with defined functions (Table 2). Although probiotic bacteria such as Lactobacillus, Lactococcus, Bifidobacterium, and Escherichia strains have been heavily characterized as human supplements [97], [98], [99], strains of these bacteria can exhibit conflicting activity during checkpoint blockade [55]. In fact, recent clinical observations have found that use of standard probiotics in general do not correlate with improved efficacy of checkpoint inhibitors [75]. By incorporating exact molecular features into a probiotic chassis, engineered strains can take advantage of both their long history of human use as well as the defined immunomodulatory activities of the newly incorporated pathway. Moreover, the transfer of microbial mechanisms away from natural sources can also obviate other confounding issues such as ABX resistance. For example, some Enterococcus species can acquire vancomycin resistance and lead to nosocomial infections [100]. To avoid these issues, SagA from E. faecium was stably incorporated into the genome of probiotic Lactococcus lactis as a thymidine auxotroph [80], providing a further means to control dissemination of the engineered probiotic. SagA-expressing L. lactis was able to efficiently improve anti-PD-L1 treatment of B16 tumors similar to the E. faecium source strain. Moreover, expression of the catalytically dead SagA mutant in L. lactis demonstrated that this antitumor phenotype was due to SagA enzymatic activity, providing a defined mechanism of action for the probiotic.
Table 2.
Engineered probiotics with defined antitumor functions. Δ indicates a gene deletion. fbr = feedback resistant mutant, fnrS = fumarate and nitrate reductase, thyA = thymidylate synthase, dapA = 4-hydroxy-tetrahydrodipicolinate synthase.
| Probiotic Host | Genetic Modifications | Method of Incorporation | Expression Promoter | Safety Measure(s] | Molecule(s] Produced | Delivery Method | Tumor Model(s] |
|---|---|---|---|---|---|---|---|
| Lactococcus lactis | sagA (E. faecium) | Genomic | PsagA | ΔthyA | Muropeptides | Oral | B16 |
| Escherichia coli Nissle 1917 | dacA (L. monocytogenes) | Genomic | PfnrS | ΔthyA, ΔdapA | Cyclic-di-AMP | Intratumoral | CT26, B16, A20 |
| Escherichia coli Nissle 1917 | ΔargR, argAfbr (E. coli) | Genomic (endogenous arg operon) | Parg (endogenous) | n/a | Arginine | Intratumoral | MC38, B16-OVA + OT-I adoptive transfer |
In addition to immunomodulatory activities, other probiotic features can be leveraged to further improve therapeutic outcomes. The Escherichia coli strain Nissle 1917 (EcN) has been extensively studied for its use in vivo [99]. Interestingly, EcN has a propensity to colonize solid tumors when injected intraveneously [101], which can allow for intratumoral delivery of gene products or metabolites. For instance, EcN has been explored as a biotherapeutic agent to activate STING within the tumor microenvironment through the production of cyclic dinucleotides [102]. Inducible expression of the diadenylate cyclase dacA from Listeria monocytogenes led to significant production of cdAMP in vitro as well as type I interferon production by BMDCs in a STING-dependent manner. Therapeutic administration of engineered EcN in vivo led to decreased tumor growth and prolonged survival in multiple model systems and also elicited immunological memory in a tumor cell rechallenge experiment. The microbe SYNB1891 is currently under investigation in a phase I clinical trial in combination with anti-PD-L1 (NCT04167137).
Similarly, natural probiotic isolates of Bifidobacterium also present tumor-homing properties. Administration of four Bifidobacterium species either by oral gavage or intravenous injection led to a significant increase in live bacteria within the tumor microenvironment [103]. These bacteria improved the efficacy of anti-CD47 treatment, which targets the CD47-SIRPɑ phagocytosis checkpoint to increase direct tumor cell clearance [104]. This antitumor activity was recapitulated by intratumoral injection of bacteria, suggesting that tumor-resident microbes directly contributed to the phenotype. Bifidobacterium required STING signaling to increase anti-CD47 activity; however, it remains unclear if this effect was due to microbially-produced STING agonists or indirect activation of cGAS. Notably, recent evidence has shown that only certain strains of Bifidobacterium bifidum augment anti-PD-1 blockade [55]. Thus, the overall activity of individual isolates may be dictated by multiple strain-specific properties including both metabolic output and tumor localization.
Intratumoral colonization has also been employed to produce immune active metabolites that are not microbial-specific. Various amino acids including serine [105], asparagine [106], and arginine [107] have been demonstrated to modulate T cell proliferation and function. Previous work has demonstrated that EcN metabolic pathways can be engineered to convert harmful metabolites into non-toxic products to treat metabolic disorders [108]. Using a similar approach, EcN was used to convert intratumoral ammonia to arginine [109]. Systemic administration of arginine-producing EcN along with anti-PD-L1 inhibited growth of MC38 tumors and protected mice from tumor cell rechallenge. Although oral administration of arginine could also improve anti-PD-L1 activity, this approach required high daily doses of arginine, which would be impractical in clinical settings. Moreover, direct injection of the amino acid into tumors could not recapitulate this effect, highlighting how the persistent production of arginine by colonizing EcN was crucial to stably increase arginine intratumoral concentrations.
Conclusions and future directions
The clinical revolution enabled by checkpoint blockade has revealed intricate relationships between host, malignancy, and microbiota. Fundamental studies from patients have established that responding and non-responding patients possess distinct microbiota compositions. In parallel, colonization experiments in murine models have confirmed that specific microbes causally alter checkpoint inhibitor efficacy. These studies have inspired ecological approaches including FMT, defined colonization, and dietary modifications to alter the gut microbiota and improve treatment response. In the future, the expansion of these reconstitution studies to larger patient cohorts and other indications will reveal how broadly these ecological methods may work. Follow-up studies to compare microbial populations within a treatment cohort will be necessary to confirm whether these treatments change the microbiota in a general and predictable fashion. Similarly, the rational use of these methods may benefit from further precision. For example, matching each patient with the necessary ABX pretreatments, specific FMT donors, individualized dosing regimens of defined consortia, or chemically defined dietary fibers may broaden the applicability of these therapies.
Conversely, a reductive, molecular view of microbial output may simplify microbiota-mediated therapies. Although challenging, these approaches yield an enhanced level of precision through the identification of defined molecular factors that enhance checkpoint blockade. In turn, the identified proteins, enzymes, and metabolites can provide a straightforward path towards combination therapies, both through small molecules and biotherapeutic agents. Beyond translational benefits, these studies have revealed basic mechanisms of immune-microbial interactions that alter host health. Somewhat unsurprisingly, many synergistic effects with checkpoint blockade have coalesced around improved type 1 immunity. However, microbes and their output act through multiple pathways including stimulation of innate signaling pathways as well as direct activity on T cells, offering multiple routes to overcome resistance.
Looking forward, we have likely only scratched the surface of molecular interactions between the host and its microbes that affect oncology care. For example, most studies have centered on bacteria rather than other microbes like fungi and archaea. In addition, the field has largely ignored microbes that are anti-correlated with clinical responses and may possess immune-suppressive factors to counteract therapeutic efficacy. Beyond the gut, microbiota found within distal organs or solid tumors [103,110,111] may generate molecules that can directly influence the tumor microenvironment and its responsiveness to checkpoint inhibitors. Finally, recent evidence has suggested that microbes can alter the efficacy of new therapeutic antibodies [103,112] as well as CAR T cell approaches [113], which underscore the broad scope of microbiota-mediated effects on clinical interventions. Thus, further molecular studies at the host-microbial interface will enable us to better explain, predict, and augment responses to current and next-generation cancer immunotherapies.
Author contribution statement
M.E.G. and H.C.H. conceived of and drafted the manuscript.
Funding
This work was supported by the National Cancer Institute (5R01CA245292, H.C.H.). Support was also provided from the Hope Funds for Cancer Research (HCFR-19-03-02, M.E.G.) and the Melanoma Research Foundation (career development award, M.E.G.).
Declaration of Competing Interest
M.E.G. and H.C.H. have filed a patent application (PCT/US2020/019038) to use SagA-expressing microbes as a therapeutic agent for cancer and infection.
Contributor Information
Matthew E. Griffin, Email: griffin@uci.edu.
Howard C. Hang, Email: hhang@scripps.edu.
References
- 1.Kim D., Xue J.Y., Lito P. Targeting KRAS(G12C): from inhibitory mechanism to modulation of antitumor effects in patients. Cell. 2020;183:850–859. doi: 10.1016/j.cell.2020.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oh D.Y., Bang Y.J. HER2-targeted therapies - a role beyond breast cancer. Nat Rev Clin Oncol. 2020;17:33–48. doi: 10.1038/s41571-019-0268-3. [DOI] [PubMed] [Google Scholar]
- 3.Spranger S., Bao R., Gajewski T.F. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 4.Sahin U., Türeci Ö. Personalized vaccines for cancer immunotherapy. Science. 2018;359:1355–1360. doi: 10.1126/science.aar7112. [DOI] [PubMed] [Google Scholar]
- 5.June C.H., O'Connor R.S., Kawalekar O.U., Ghassemi S., Milone M.C. CAR T cell immunotherapy for human cancer. Science. 2018;359:1361–1365. doi: 10.1126/science.aar6711. [DOI] [PubMed] [Google Scholar]
- 6.Ribas A., Wolchok J.D. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaddepally R.K., Kharel P., Pandey R., Garje R., Chandra A.B. Review of Indications of FDA-approved immune checkpoint inhibitors per NCCN guidelines with the level of evidence. Cancers. 2020;12 doi: 10.3390/cancers12030738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Appleton E., et al. Kickstarting immunity in cold tumours: localised tumour therapy combinations with immune checkpoint blockade. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.754436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonaventura P., et al. Cold tumors: a therapeutic challenge for immunotherapy. Front Immunol. 2019;10:168. doi: 10.3389/fimmu.2019.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma P., Hu-Lieskovan S., Wargo J.A. A. Ribas, Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ansaldo E., Farley T.K., Belkaid Y. Control of immunity by the microbiota. Annu Rev Immunol. 2021;39:449–479. doi: 10.1146/annurev-immunol-093019-112348. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert J.A., et al. Current understanding of the human microbiome. Nat Med. 2018;24:392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sender R., Fuchs S., Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14 doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colosimo D.A., et al. Mapping interactions of microbial metabolites with human G-protein-coupled receptors. Cell Host Microbe. 2019;26:273–282. doi: 10.1016/j.chom.2019.07.002. e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H., et al. A forward chemical genetic screen reveals gut microbiota metabolites that modulate host physiology. Cell. 2019;177:1217–1231. doi: 10.1016/j.cell.2019.03.036. e1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hang S., et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature. 2019;576:143–148. doi: 10.1038/s41586-019-1785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song X., et al. Microbial bile acid metabolites modulate gut RORgamma(+) regulatory T cell homeostasis. Nature. 2020;577:410–415. doi: 10.1038/s41586-019-1865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paik D., et al. Human gut bacteria produce Th17-modulating bile acid metabolites. Nature. 2022 doi: 10.1038/s41586-022-04480-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sivan A., et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vetizou M., et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinstock G.M. Genomic approaches to studying the human microbiota. Nature. 2012;489:250–256. doi: 10.1038/nature11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bharti R., Grimm D.G. Current challenges and best-practice protocols for microbiome analysis. Brief Bioinform. 2021;22:178–193. doi: 10.1093/bib/bbz155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar R.C. Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics. 2018;34:2371–2375. doi: 10.1093/bioinformatics/bty113. [DOI] [PubMed] [Google Scholar]
- 24.Wei Z.G., et al. Comparison of methods for picking the operational taxonomic units from amplicon sequences. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.644012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson J.S., et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019;10:5029. doi: 10.1038/s41467-019-13036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eren A.M., et al. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol. 2013;4 doi: 10.1111/2041-210X.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callahan B.J., McMurdie P.J., Holmes S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017;11:2639–2643. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frankel A.E., et al. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia. 2017;19:848–855. doi: 10.1016/j.neo.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaput N., et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28:1368–1379. doi: 10.1093/annonc/mdx108. [DOI] [PubMed] [Google Scholar]
- 30.Gopalakrishnan V., et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matson V., et al. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters B.A., et al. Relating the gut metagenome and metatranscriptome to immunotherapy responses in melanoma patients. Genome Med. 2019;11:61. doi: 10.1186/s13073-019-0672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coutzac C., et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat Commun. 2020;11:2168. doi: 10.1038/s41467-020-16079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wind T.T., et al. Gut microbial species and metabolic pathways associated with response to treatment with immune checkpoint inhibitors in metastatic melanoma. Melanoma Res. 2020;30:235–246. doi: 10.1097/CMR.0000000000000656. [DOI] [PubMed] [Google Scholar]
- 35.Andrews M.C., et al. Gut microbiota signatures are associated with toxicity to combined CTLA-4 and PD-1 blockade. Nat Med. 2021;27:1432–1441. doi: 10.1038/s41591-021-01406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee K.A., et al. Cross-cohort gut microbiome associations with immune checkpoint inhibitor response in advanced melanoma. Nat Med. 2022;28:535–544. doi: 10.1038/s41591-022-01695-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCulloch J.A., et al. Intestinal microbiota signatures of clinical response and immune-related adverse events in melanoma patients treated with anti-PD-1. Nat Med. 2022;28:545–556. doi: 10.1038/s41591-022-01698-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Routy B., et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 39.Fukuoka S., et al. Association of gut microbiome with immune status and clinical response in solid tumor patients who received on anti-PD-1 therapies. Journal of Clinical Oncology. 2018;36:3011. [Google Scholar]
- 40.Jin Y., et al. The diversity of gut microbiome is associated with favorable responses to anti-programmed death 1 immunotherapy in chinese patients with NSCLC. J Thorac Oncol. 2019;14:1378–1389. doi: 10.1016/j.jtho.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Zheng Y., et al. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J Immunother Cancer. 2019;7:193. doi: 10.1186/s40425-019-0650-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katayama Y., et al. The role of the gut microbiome on the efficacy of immune checkpoint inhibitors in Japanese responder patients with advanced non-small cell lung cancer. Transl Lung Cancer Res. 2019;8:847–853. doi: 10.21037/tlcr.2019.10.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song P., et al. Relationship between intestinal flora structure and metabolite analysis and immunotherapy efficacy in Chinese NSCLC patients. Thorac Cancer. 2020;11:1621–1632. doi: 10.1111/1759-7714.13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hakozaki T., et al. The gut microbiome associates with immune checkpoint inhibition outcomes in patients with advanced non-small cell lung cancer. Cancer Immunol Res. 2020;8:1243–1250. doi: 10.1158/2326-6066.CIR-20-0196. [DOI] [PubMed] [Google Scholar]
- 45.Peng Z., et al. The gut microbiome is associated with clinical response to anti-PD-1/PD-L1 immunotherapy in gastrointestinal cancer. Cancer Immunol Res. 2020;8:1251–1261. doi: 10.1158/2326-6066.CIR-19-1014. [DOI] [PubMed] [Google Scholar]
- 46.Derosa L., et al. Gut bacteria composition drives primary resistance to cancer immunotherapy in renal cell carcinoma patients. Eur Urol. 2020;78:195–206. doi: 10.1016/j.eururo.2020.04.044. [DOI] [PubMed] [Google Scholar]
- 47.Salgia N.J., et al. Stool microbiome profiling of patients with metastatic renal cell carcinoma receiving anti-PD-1 immune checkpoint inhibitors. Eur Urol. 2020;78:498–502. doi: 10.1016/j.eururo.2020.07.011. [DOI] [PubMed] [Google Scholar]
- 48.Zhang C., et al. Commensal microbiota contributes to predicting the response to immune checkpoint inhibitors in non-small-cell lung cancer patients. Cancer Sci. 2021;112:3005–3017. doi: 10.1111/cas.14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin H., et al. The commensal consortium of the gut microbiome is associated with favorable responses to anti-programmed death protein 1 (PD-1) therapy in thoracic neoplasms. Cancer Biol Med. 2021 doi: 10.20892/j.issn.2095-3941.2020.0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newsome R.C., et al. Interaction of bacterial genera associated with therapeutic response to immune checkpoint PD-1 blockade in a United States cohort. Genome Med. 2022;14:35. doi: 10.1186/s13073-022-01037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conlon M.A., Bird A.R. The impact of diet and lifestyle on gut microbiota and human health. Nutrients. 2014;7:17–44. doi: 10.3390/nu7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Panek M., et al. Methodology challenges in studying human gut microbiota - effects of collection, storage, DNA extraction and next generation sequencing technologies. Sci Rep. 2018;8:5143. doi: 10.1038/s41598-018-23296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trosvik P., de Muinck E.J. Ecology of bacteria in the human gastrointestinal tract–identification of keystone and foundation taxa. Microbiome. 2015;3:44. doi: 10.1186/s40168-015-0107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McDonald J.E., Marchesi J.R., Koskella B. Application of ecological and evolutionary theory to microbiome community dynamics across systems. Proc Biol Sci. 2020;287 doi: 10.1098/rspb.2020.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee S.H., et al. Bifidobacterium bifidum strains synergize with immune checkpoint inhibitors to reduce tumour burden in mice. Nat Microbiol. 2021;6:277–288. doi: 10.1038/s41564-020-00831-6. [DOI] [PubMed] [Google Scholar]
- 56.Gopalakrishnan V., Helmink B.A., Spencer C.N., Reuben A., Wargo J.A. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 2018;33:570–580. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baruch E.N., et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 2021;371:602–609. doi: 10.1126/science.abb5920. [DOI] [PubMed] [Google Scholar]
- 58.Davar D., et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science. 2021;371:595–602. doi: 10.1126/science.abf3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Atarashi K., et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 60.Atarashi K., et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tanoue T., et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. 2019;565:600–605. doi: 10.1038/s41586-019-0878-z. [DOI] [PubMed] [Google Scholar]
- 62.Han S., et al. A metabolomics pipeline for the mechanistic interrogation of the gut microbiome. Nature. 2021;595:415–420. doi: 10.1038/s41586-021-03707-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.den Besten G., et al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blaak E.E., et al. Short chain fatty acids in human gut and metabolic health. Benef Microbes. 2020;11:411–455. doi: 10.3920/BM2020.0057. [DOI] [PubMed] [Google Scholar]
- 65.Parada Venegas D., et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. 2019;10:277. doi: 10.3389/fimmu.2019.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith P.M., et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arpaia N., et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park J., et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2015;8:80–93. doi: 10.1038/mi.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun M., et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat Commun. 2018;9:3555. doi: 10.1038/s41467-018-05901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bachem A., et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8(+) T cells. Immunity. 2019;51:e285. doi: 10.1016/j.immuni.2019.06.002. 285-297. [DOI] [PubMed] [Google Scholar]
- 71.He Y., et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8(+) T cell immunity. Cell Metab. 2021;33:988–1000. doi: 10.1016/j.cmet.2021.03.002. e1007. [DOI] [PubMed] [Google Scholar]
- 72.Lopez-Siles M., Duncan S.H., Garcia-Gil L.J., Martinez-Medina M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 2017;11:841–852. doi: 10.1038/ismej.2016.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nomura M., et al. Association of short-chain fatty acids in the gut microbiome with clinical response to treatment with nivolumab or pembrolizumab in patients With solid cancer tumors. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lam K.C., et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell. 2021;184:5338–5356.e5321. doi: 10.1016/j.cell.2021.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spencer C.N., et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science. 2021;374:1632–1640. doi: 10.1126/science.aaz7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mager L.F., et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science. 2020;369:1481–1489. doi: 10.1126/science.abc3421. [DOI] [PubMed] [Google Scholar]
- 77.Welihinda A.A., Kaur M., Greene K., Zhai Y., Amento E.P. The adenosine metabolite inosine is a functional agonist of the adenosine A2A receptor with a unique signaling bias. Cell Signal. 2016;28:552–560. doi: 10.1016/j.cellsig.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hopfner K.P., Hornung V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat Rev Mol Cell Biol. 2020;21:501–521. doi: 10.1038/s41580-020-0244-x. [DOI] [PubMed] [Google Scholar]
- 79.Rodrigues R.R., Shulzhenko N., Morgun A. Transkingdom networks: a systems biology approach to identify causal members of host-microbiota interactions. Methods Mol Biol. 2018;1849:227–242. doi: 10.1007/978-1-4939-8728-3_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Griffin M.E., et al. Enterococcus peptidoglycan remodeling promotes checkpoint inhibitor cancer immunotherapy. Science. 2021;373:1040–1046. doi: 10.1126/science.abc9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim B., et al. Enterococcus faecium secreted antigen A generates muropeptides to enhance host immunity and limit bacterial pathogenesis. eLife. 2019;8:e45343. doi: 10.7554/eLife.45343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rangan K.J., et al. A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens. Science. 2016;353:1434–1437. doi: 10.1126/science.aaf3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pedicord V.A., et al. Exploiting a host-commensal interaction to promote intestinal barrier function and enteric pathogen tolerance. Sci Immunol. 2016;1:eaai7732. doi: 10.1126/sciimmunol.aai7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Girardin S.E., et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 85.Inohara N., et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 86.Palmer K.L., et al. Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio. 2012;3:e00311–e00318. doi: 10.1128/mBio.00318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hayashi F., et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 88.Hajam I.A., Dar P.A., Shahnawaz I., Jaume J.C., Lee J.H. Bacterial flagellin-a potent immunomodulatory agent. Exp Mol Med. 2017;49:e373. doi: 10.1038/emm.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Laute-Caly D.L., et al. The flagellin of candidate live biotherapeutic Enterococcus gallinarum MRx0518 is a potent immunostimulant. Sci Rep. 2019;9:801. doi: 10.1038/s41598-018-36926-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vieira S.Manfredo, et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science. 2018;359:1156–1161. doi: 10.1126/science.aar7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stritesky G.L., Jameson S.C., Hogquist K.A. Selection of self-reactive T cells in the thymus. Annu Rev Immunol. 2012;30:95–114. doi: 10.1146/annurev-immunol-020711-075035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Walker L.S., Abbas A.K. The enemy within: keeping self-reactive T cells at bay in the periphery. Nat Rev Immunol. 2002;2:11–19. doi: 10.1038/nri701. [DOI] [PubMed] [Google Scholar]
- 93.Rojas M., et al. Molecular mimicry and autoimmunity. J Autoimmun. 2018;95:100–123. doi: 10.1016/j.jaut.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 94.Daillère R., et al. Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity. 2016;45:931–943. doi: 10.1016/j.immuni.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 95.Rong Y., et al. Reactivity toward Bifidobacterium longum and Enterococcus hirae demonstrate robust CD8(+) T cell response and better prognosis in HBV-related hepatocellular carcinoma. Exp Cell Res. 2017;358:352–359. doi: 10.1016/j.yexcr.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 96.Fluckiger A., et al. Cross-reactivity between tumor MHC class I-restricted antigens and an enterococcal bacteriophage. Science. 2020;369:936–942. doi: 10.1126/science.aax0701. [DOI] [PubMed] [Google Scholar]
- 97.Sanders M.E., et al. Safety assessment of probiotics for human use. Gut Microbes. 2010;1:164–185. doi: 10.4161/gmic.1.3.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sanders M.E., Merenstein D., Merrifield C.A., Hutkins R. Probiotics for human use. Nutrition Bulletin. 2018;43:212–225. [Google Scholar]
- 99.Sonnenborn U. Escherichia coli strain Nissle 1917-from bench to bedside and back: history of a special Escherichia coli strain with probiotic properties. FEMS Microbiol Lett. 2016;363 doi: 10.1093/femsle/fnw212. [DOI] [PubMed] [Google Scholar]
- 100.Lebreton F., et al. Tracing the enterococci from paleozoic origins to the hospital. Cell. 2017;169:849–861. doi: 10.1016/j.cell.2017.04.027. e813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stritzker J., et al. Tumor-specific colonization, tissue distribution, and gene induction by probiotic Escherichia coli Nissle 1917 in live mice. Int J Med Microbiol. 2007;297:151–162. doi: 10.1016/j.ijmm.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 102.Leventhal D.S., et al. Immunotherapy with engineered bacteria by targeting the STING pathway for anti-tumor immunity. Nat Commun. 2020;11:2739. doi: 10.1038/s41467-020-16602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shi Y., et al. Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via STING signaling. J Exp Med. 2020;217 doi: 10.1084/jem.20192282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Feng M., et al. Phagocytosis checkpoints as new targets for cancer immunotherapy. Nat Rev Cancer. 2019;19:568–586. doi: 10.1038/s41568-019-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ma E.H., et al. Serine Is an essential metabolite for effector T cell expansion. Cell Metab. 2017;25:345–357. doi: 10.1016/j.cmet.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 106.Wu J., et al. Asparagine enhances LCK signalling to potentiate CD8(+) T-cell activation and anti-tumour responses. Nat Cell Biol. 2021;23:75–86. doi: 10.1038/s41556-020-00615-4. [DOI] [PubMed] [Google Scholar]
- 107.Geiger R., et al. L-Arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell. 2016;167:829–842. doi: 10.1016/j.cell.2016.09.031. e813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kurtz C.B., et al. An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci Transl Med. 2019;11:eaau7975. doi: 10.1126/scitranslmed.aau7975. [DOI] [PubMed] [Google Scholar]
- 109.Canale F.P., et al. Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature. 2021;598:662–666. doi: 10.1038/s41586-021-04003-2. [DOI] [PubMed] [Google Scholar]
- 110.Riquelme E., et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178:795–806. doi: 10.1016/j.cell.2019.07.008. e712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nejman D., et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368:973–980. doi: 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee B., et al. Modulation of the gut microbiota alters the tumour-suppressive efficacy of Tim-3 pathway blockade in a bacterial species- and host factor-dependent manner. Microorganisms. 2020;8 doi: 10.3390/microorganisms8091395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Smith M., et al. Gut microbiome correlates of response and toxicity following anti-CD19 CAR T cell therapy. Nat Med. 2022;28:713–723. doi: 10.1038/s41591-022-01702-9. [DOI] [PMC free article] [PubMed] [Google Scholar]