ABSTRACT
Rickettsia spp. are obligate intracellular bacterial pathogens that have evolved a variety of strategies to exploit their host cell niche. However, the bacterial factors that contribute to this intracellular lifestyle are poorly understood. Here, we show that the conserved ankyrin repeat protein RARP-1 supports Rickettsia parkeri infection. Specifically, RARP-1 promotes efficient host cell entry and growth within the host cytoplasm, but it is not necessary for cell-to-cell spread or evasion of host autophagy. We further demonstrate that RARP-1 is not secreted into the host cytoplasm by R. parkeri. Instead, RARP-1 resides in the periplasm, and we identify several binding partners that are predicted to work in concert with RARP-1 during infection. Altogether, our data reveal that RARP-1 plays a critical role in the rickettsial life cycle.
IMPORTANCE Rickettsia spp. are obligate intracellular bacterial pathogens that pose a growing threat to human health. Nevertheless, their strict reliance on a host cell niche has hindered investigation of the molecular mechanisms driving rickettsial infection. This study yields much-needed insight into the Rickettsia ankyrin repeat protein RARP-1, which is conserved across the genus but has not yet been functionally characterized. Earlier work had suggested that RARP-1 is secreted into the host cytoplasm. However, the results from this work demonstrate that R. parkeri RARP-1 resides in the periplasm and is important both for invasion of host cells and for growth in the host cell cytoplasm. These results reveal RARP-1 as a novel regulator of the rickettsial life cycle.
KEYWORDS: RARP-1, Rickettsia parkeri, intracellular pathogen
INTRODUCTION
Intracellular bacterial pathogens face considerable challenges and opportunities when invading and occupying their host cell niche. The host cell membrane physically occludes entry and the endolysosomal pathway imperils invading microbes. Moreover, host cell defenses like autophagy create a hostile environment for internalized bacteria. If a bacterium successfully navigates these obstacles, however, it can conceal itself from humoral immunity, commandeer host metabolites, and exploit host cell biology to support infection. Not surprisingly, the host cell niche has provided fertile ground for the evolution of diverse lifestyles across many well-studied bacterial pathogens, such as Shigella, Listeria, Salmonella, and Legionella (1, 2). The prospect of uncovering unique infection strategies invites a thorough investigation of these adaptations in more enigmatic pathogens.
Members of the genus Rickettsia include emerging global health threats that can cause mild to severe diseases, such as typhus and Rocky Mountain spotted fever (3). These Gram-negative bacterial pathogens are transmitted from arthropod vectors to vertebrate hosts, where they primarily target the vascular endothelium. As obligate intracellular pathogens, Rickettsia spp. define the extreme end of adaptation to intracellular life and are completely dependent on their hosts for survival (4). Consequently, they have evolved a complex life cycle to invade, grow, and disseminate across host tissues.
As the first step of their life cycle, Rickettsia spp. adhere to and invade host cells by inducing phagocytosis (5–7). Once inside, these bacteria rapidly escape the phagocytic vacuole to access the host cytoplasm (8, 9). To establish a hospitable niche for proliferation, Rickettsia spp. scavenge host nutrients, modulate apoptosis, and thwart antimicrobial autophagy (10–13). Successful colonization of the host cytoplasm allows Rickettsia spp. to spread to neighboring cells. Members of the spotted fever group (SFG) Rickettsia hijack the host actin cytoskeleton, forming tails that propel the bacteria around the cytoplasm, and then protrude through cell-cell junctions to repeat the infection cycle (14, 15).
Recent work using the model SFG member Rickettsia parkeri has highlighted a short list of surface-exposed proteins and secreted effectors that manipulate host cell processes during infection (4). For example, the surface protein Sca2 nucleates actin at the bacterial pole and promotes motility by mimicking host formins (14). Sca4, a secreted effector, interacts with host vinculin to reduce intercellular tension and facilitate protrusion engulfment (15). Additionally, methylation of outer membrane proteins, like OmpB, protects R. parkeri from ubiquitination and autophagy (13, 16). Despite these advances, our knowledge of the factors that govern the multistep rickettsial life cycle is still limited. Indeed, Rickettsia spp. genomes are replete with hypothetical proteins that are conserved even among less virulent members of the genus (17), but a paucity of genetic tools has stunted investigation of these proteins. Such factors could support infection directly, by targeting host processes, or indirectly, by controlling the bacterial mediators at the host-pathogen interface. Thus, it is critical to reveal how these uncharacterized proteins contribute to infection.
In a recent transposon mutagenesis screen of R. parkeri (18), we identified over 100 mutants that exhibited defects in infection. Although several hits from this screen have been functionally characterized (13–16), many play unknown roles during infection. One such unexplored hit is the Rickettsia ankyrin repeat protein 1 (RARP-1), which is conserved across the genus and predicted to be secreted into the host cytoplasm (19). To better understand the factors that influence the rickettsial life cycle, we investigated the function of RARP-1 during R. parkeri infection. We demonstrated that RARP-1 promotes both efficient host cell invasion and growth in the host cytoplasm, but it is otherwise dispensable for cell-to-cell spread and avoidance of host autophagy. Although prior work indicated that RARP-1 is secreted into the host cytoplasm (19), we found instead that it localizes to the R. parkeri periplasm. Furthermore, we showed that RARP-1 interacts with a variety of factors that are predicted to support bacterial fitness. Our results suggest that RARP-1 is a Rickettsia-specific tool that promotes the obligate intracellular life cycle.
RESULTS
Transposon mutagenesis of rarp-1 impairs R. parkeri infection.
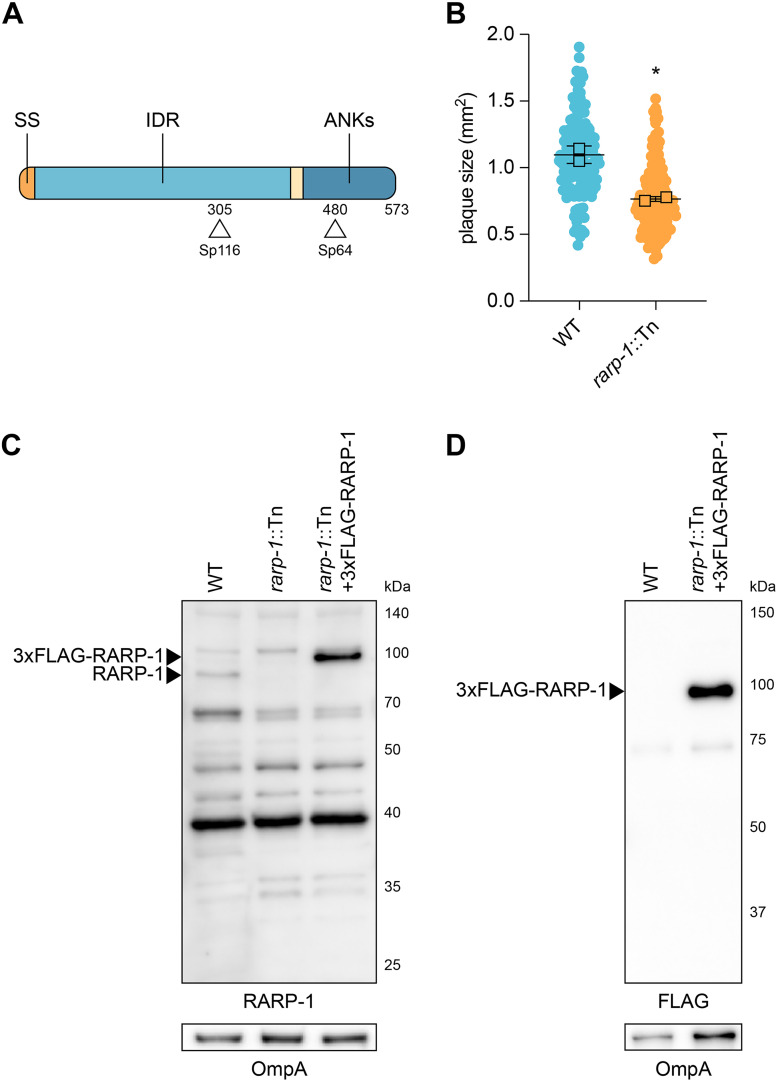
In a previous mariner-based transposon mutagenesis screen (18), we identified a number of R. parkeri mutants that displayed abnormal plaque sizes after infection of Vero host cell monolayers. We hypothesized that the plaque phenotypes for these mutants were due to defects in growth, cell-to-cell spread, or other steps of the rickettsial life cycle. Two such small plaque (Sp) mutants contained a transposon (Tn) insertion within the rarp-1 gene, giving a predicted truncation of RARP-1 at residues 305 (Sp116) and 480 (Sp64) (Fig. 1A). RARP-1 is a 573-amino-acid protein conserved across the Rickettsia genus, but the lack of loss-of-function mutants has thus far prevented characterization of RARP-1 function. Due to the upstream position of its Tn insertion within the rarp-1 coding DNA sequence, we focused on Sp116 (herein referred to as rarp-1::Tn) for all subsequent studies and confirmed that it formed smaller plaques than green fluorescent protein (GFP)-expressing wild-type bacteria (WT) (Fig. 1B). We generated polyclonal antibodies against a RARP-1 peptide upstream of the Tn insertion site to assess RARP-1 expression in the mutant. As expected, the rarp-1::Tn mutant did not express the full-length protein, as shown by immunoblotting (Fig. 1C). Furthermore, we were unable to detect an obvious band consistent with the expected 30-kDa product resulting from Tn insertion. Altogether, these results suggest that the loss of RARP-1 expression in the rarp-1::Tn mutant leads to a small plaque phenotype.
FIG 1.
Transposon mutagenesis of rarp-1 impairs R. parkeri infection. (A) R. parkeri RARP-1 contains an N-terminal Sec secretion signal (SS; orange), a central intrinsically disordered region (IDR; light blue), and C-terminal ankyrin repeats (ANKs; dark blue). Tn insertions at residues 305 (Sp116) and 480 (Sp64) are indicated (arrowheads). (B) Plaque areas in infected Vero cell monolayers. Means from two independent experiments (squares) are superimposed over the raw data (circles) and were used to calculate means ± standard deviations (SD) and P values (unpaired two-tailed t test). *, P < 0.05 relative to WT. (C) Western blot for RARP-1 using purified R. parkeri strains. 3×FLAG-tagged and endogenous RARP-1 are indicated (arrowheads). OmpA, loading control. (D) Western blot for FLAG using purified R. parkeri strains. 3×FLAG-tagged RARP-1 is indicated (arrowhead). OmpA, loading control.
RARP-1 supports bacterial growth and is dispensable for cell-to-cell spread.
The small plaques formed by the rarp-1::Tn mutant could be the result of defects in one or more steps of the rickettsial life cycle, and determining when RARP-1 acts during infection would support characterization of its function. We first performed infectious focus assays in A549 host cell monolayers to assess the growth and cell-to-cell spread of the rarp-1::Tn mutant on a shorter timescale than is required for plaque formation (28 h versus 5 days postinfection). A549 cells support all known aspects of the R. parkeri life cycle, and the use of gentamicin prevents asynchronous invasion events (15). Consistent with the small plaque phenotype, the rarp-1::Tn mutant generated smaller foci than WT bacteria (Fig. 2A). To confirm that this phenotype was due specifically to the disruption of rarp-1, we complemented the rarp-1::Tn mutant with a plasmid expressing 3×FLAG-tagged RARP-1 (rarp-1::Tn + 3×FLAG-RARP-1) (see Fig. S1A in the supplemental material). Since rarp-1 is predicted to be part of an operon (19), we selected a 247-bp region immediately upstream of the first gene in the operon (encoding the outer membrane channel TolC) as a putative promoter to drive rarp-1 expression. This construct was sufficient for expression of epitope-tagged RARP-1 in the rarp-1::Tn mutant (Fig. 1C and D). Importantly, the complemented strain exhibited infectious focus sizes comparable to those of the WT (Fig. 2A), indicating that the putative promoter and epitope-tagged RARP-1 are functionally relevant. Thus, RARP-1 specifically supports the size of R. parkeri infectious foci.
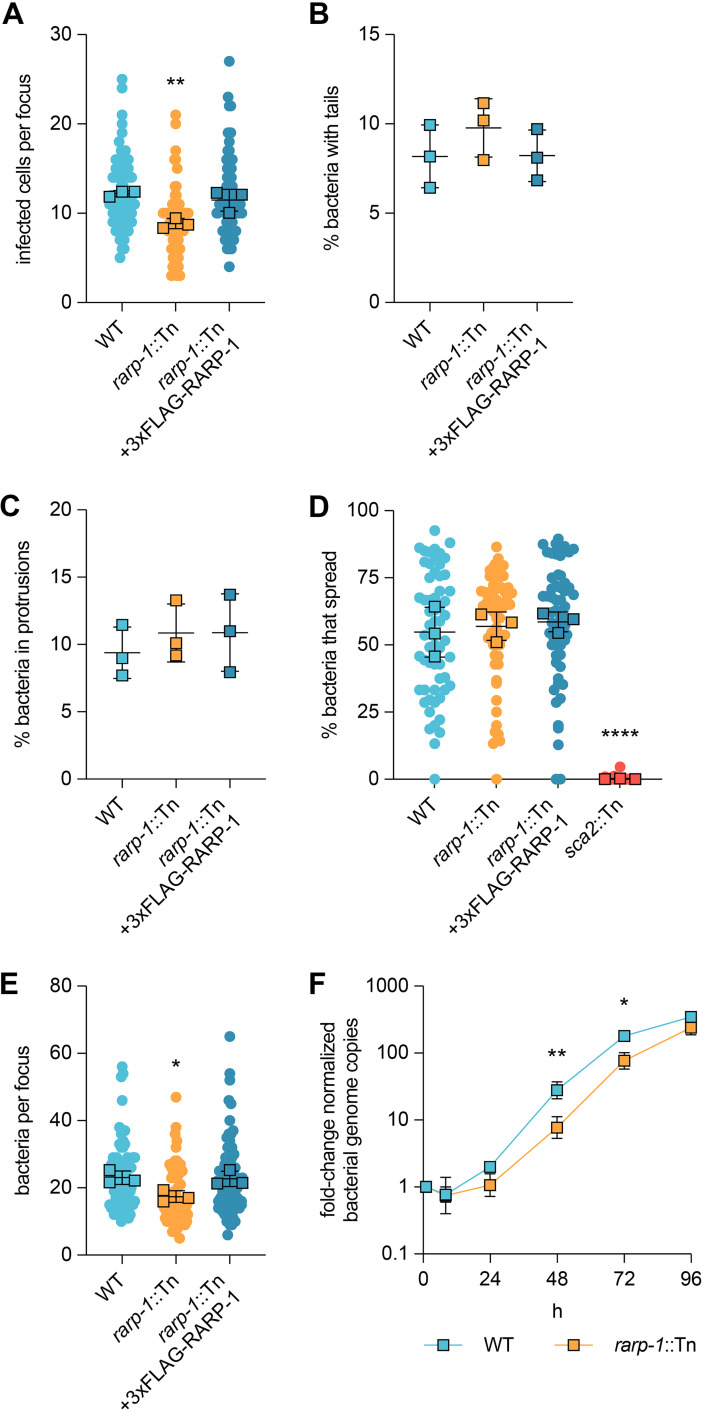
FIG 2.
RARP-1 supports bacterial growth and is dispensable for cell-to-cell spread. (A) Infected cells per focus during infection of A549 cells. The means from three independent experiments (squares) are superimposed over the raw data (circles) and were used to calculate the means ± SD and P values (one-way ANOVA with post hoc Dunnett’s test). **, P < 0.01 relative to WT. (B) Percentage of bacteria with actin tails during infection of A549 cells. (C) Percentage of bacteria within a protrusion during infection of A549 cells. In panels B and C, the percentages were determined from three independent experiments (≥380 bacteria were counted for each infection) and were used to calculate the means ± SD and P values (one-way ANOVA with post hoc Dunnett’s test). n.s., not significant relative to WT. (D) Percentage of bacteria per focus that spread from infected donor cells to uninfected recipient cells by mixed-cell assay in A549 cells. The means from three independent experiments (squares) are superimposed over the raw data (circles) and were used to calculate the means ± SD and P values (one-way ANOVA with post hoc Dunnett’s test). ****, P < 0.0001 relative to WT. The sca2::Tn mutant was used as a positive control. (E) Bacteria per focus during infection of A549 cells. The means from three independent experiments (squares) are superimposed over the raw data (circles) and were used to calculate the means ± SD and P values (one-way ANOVA with post hoc Dunnett’s test). *, P < 0.05). These data correspond to the same set of infectious focus assays displayed in panel A. (F) Growth curves as measured by R. parkeri (17-kDa surface antigen) genome equivalents per Vero host cell (GAPDH) genome equivalent normalized to 1 h postinfection. The means ± SD for triplicate samples from a representative experiment were compared at each time point after log2 transformation (unpaired two-tailed t test). *, P < 0.05; **, P < 0.01 relative to WT.
A reduction in infectious focus size could be caused by defects in cell-to-cell spread. For example, Tn mutagenesis of sca2 and sca4 specifically disrupts spread by limiting actin tail formation and protrusion resolution, respectively, leading to smaller infectious foci (14, 15). Loss of RARP-1 did not alter the frequency of actin tails or protrusions (Fig. 2B and C), suggesting that spread may not be regulated by RARP-1. As an orthogonal approach, we also evaluated the efficiency of spread by performing a mixed-cell infectious focus assay (15). In this assay, donor host cells stably expressing a cytoplasmic marker are infected, mixed with unlabeled recipient host cells, and then infection of the mixed monolayer is allowed to progress. Bacteria that spread to unlabeled recipient cells can thus be distinguished from bacteria that remain in the labeled donor cell for each focus. As expected, a sca2::Tn mutant failed to spread from infected donor cells (Fig. 2D). In contrast, the rarp-1::Tn mutant exhibited similar efficiency of spread from donors to recipients as WT bacteria. Altogether, these results indicate that RARP-1 is dispensable for cell-to-cell spread.
Alternatively, a reduction in infectious focus size could be caused by defects in bacterial growth. When performing the infectious focus assays, we noted that the number of rarp-1::Tn mutant bacteria within the infectious foci was reduced compared to WT (Fig. 2E). This was in contrast to Tn mutants of sca2 and sca4, which do not exhibit reduced bacterial loads despite forming smaller foci (14, 15). Restoring RARP-1 expression in the complemented strain rescued the bacterial load defect (Fig. 2E), suggesting that RARP-1 regulates bacterial growth. To determine if the rarp-1:Tn mutant displayed altered growth behavior over the course of infection, we used quantitative PCR (qPCR) to monitor bacterial genome equivalents during infection of Vero host cell monolayers. In agreement with the bacterial load defect observed in the infectious focus assay, the rarp-1::Tn mutant exhibited a growth defect compared to WT (Fig. 2F), with a doubling time of 8.4 h versus 6.3 h approximated from exponential-phase growth. Furthermore, the viability of rarp-1::Tn mutant bacteria during infection was identical to that of the WT (see Fig. S2 in the supplemental material); thus, the observed growth defects for the rarp-1::Tn mutant are not due to the generation of nonviable progeny. Together, our data support a role for RARP-1 during bacterial growth in multiple cell types.
RARP-1 is dispensable for evasion of autophagy.
Given the rarp-1::Tn mutant growth defect, we hypothesized that RARP-1 promotes bacterial growth by preventing clearance from the host cell. R. parkeri avoids recognition and destruction by the host cell autophagy machinery using the abundant outer membrane protein OmpB (13). Bacteria lacking OmpB are readily polyubiquitinated by the host cell and associate with LC3-positive autophagic membranes. We tested whether the rarp-1::Tn mutant likewise associates with LC3 during infection of A549 cells. In contrast to an ompB::Tn mutant, the rarp-1::Tn mutant failed to mobilize host LC3 (Fig. 3A). Thus, loss of RARP-1 expression does not render this mutant more susceptible to autophagic clearance, indicating that RARP-1 supports growth through a different mechanism.
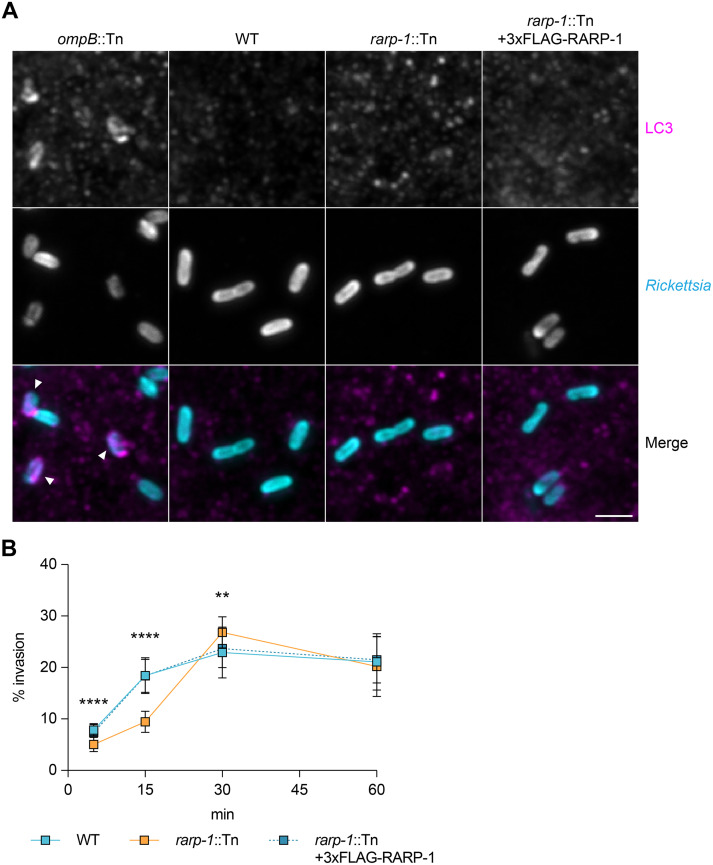
FIG 3.
RARP-1 is dispensable for evasion of host cell autophagy and supports host cell invasion. (A) Recruitment of LC3 during infection of A549 cells. Samples were stained for LC3 (magenta) and bacteria (cyan). The ompB::Tn mutant was used as a positive control, and bacteria associated with LC3-positive membranes are indicated (arrowheads). Scale bar, 2 μm. (B) Efficiency of invasion into A549 cells. The means ± SD from a representative experiment (n = 20 fields of view each with ≥45 bacteria) were compared at each time point (one-way ANOVA with post hoc Dunnett’s test). **, P < 0.01; ****, P < 0.0001 relative to WT.
RARP-1 supports host cell invasion.
We next wanted to determine if RARP-1 plays other roles in the infection cycle upstream of growth inside the host cytoplasm. We tested whether the rarp-1::Tn mutant exhibited defects during invasion of A549 host cells by using differential immunofluorescent staining (6). In this assay, bacteria are stained both before and after host cell permeabilization to distinguish external and internal bacteria, respectively. Invasion of the rarp-1::Tn mutant was delayed compared to WT but otherwise recovered within 30 min postinfection (Fig. 3B). We observed similar invasion kinetics for WT bacteria and the complemented strain, indicating that the delayed invasion of the rarp-1::Tn mutant is due to loss of RARP-1 expression. Thus, RARP-1 supports efficient host cell invasion. We therefore turned our investigation to the localization and binding partners of RARP-1 so that we could reveal how this factor contributes to infection.
RARP-1 is not secreted into the host cytoplasm by R. parkeri.
RARP-1 contains an N-terminal Sec secretion signal and several C-terminal ankyrin repeats. Ankyrin repeats are often involved in protein-protein interactions (20), and various intracellular pathogens secrete ankyrin repeat-containing proteins to target an array of host cell processes (21, 22). Previous work with the typhus group Rickettsia species R. typhi suggested that RARP-1 is delivered into host cells through a noncanonical mechanism mediated by the Sec translocon and TolC (19). We originally hypothesized that R. parkeri also secretes RARP-1 to target host cell functions and ultimately promote bacterial growth and invasion. To monitor secretion of RARP-1 during infection of A549 cells, we used selective lysis to separate supernatants containing the infected host cytoplasm from pellets containing intact bacteria. A protein that is secreted during infection should be detected in both the supernatant and pellet fractions by immunoblotting, as was observed for the secreted effector Sca4 (Fig. 4A, middle panel). The absence of the bacterial RNA polymerase subunit RpoA in the supernatant fraction confirmed that our lysis conditions did not cause bacterial lysis and release of nonsecreted bacterial proteins (Fig. 4A, bottom panel). Unexpectedly, we detected 3xFLAG-RARP-1 in the bacterial pellet but not in the supernatant fraction of cells infected with the rarp-1::Tn + 3×FLAG-RARP-1 complemented strain (Fig. 4A, top). Similar results were observed for a 3×FLAG-RARP-1 construct containing an additional Ty1 epitope tag inserted proximal to the C terminus (see Fig. S1A in the supplemental material), suggesting that the lack of detection was not due to proteolytic processing of the RARP-1 protein. As with the 3×FLAG-RARP-1 construct, this dual-tagged variant rescued the rarp-1::Tn mutant infectious focus defects (see Fig. S1B and C), demonstrating the functional relevance of the tagged RARP-1 construct. Moreover, endogenous RARP-1 protein was detectable in the WT bacterial pellet but not in the supernatant fraction with our polyclonal antibody (see Fig. S1D), confirming that the epitope-tagged constructs recapitulate the behavior of the endogenous protein. Together, these results suggest that RARP-1 is not secreted by R. parkeri into the host cytoplasm.
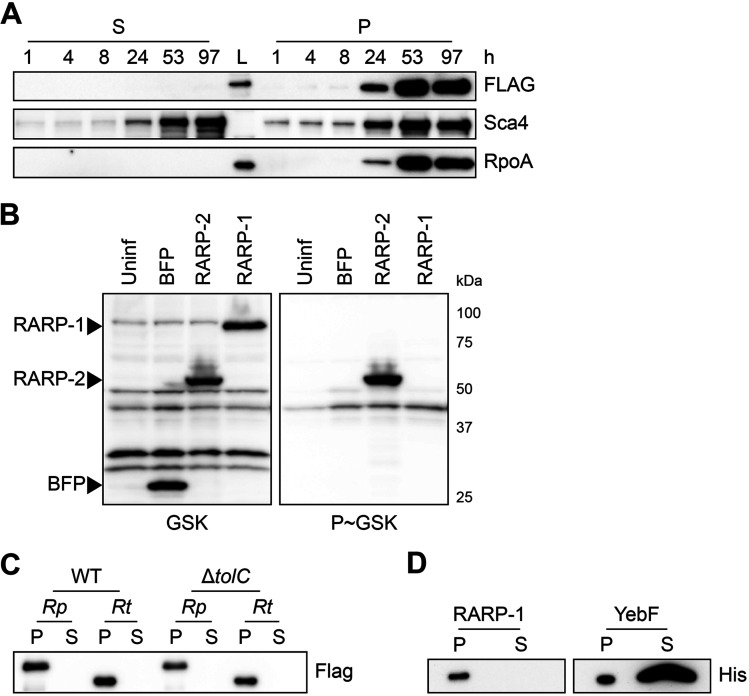
FIG 4.
RARP-1 is not secreted. (A) Western blots for FLAG (top) and Sca4 (middle) during infection of A549 cells with rarp-1::Tn + 3×FLAG-RARP-1 bacteria. Infected host cells were selectively lysed at various time points to separate supernatants (S) containing the infected host cytoplasm from pellets (P) containing intact bacteria. RpoA (bottom) served as a control for bacterial lysis or contamination of the infected cytoplasmic fraction. L, ladder. (B) Western blot for GSK-tagged constructs during infection of Vero cells. Whole-cell infected lysates were probed with antibodies against the GSK tag (left) or its phosphorylated form (P~GSK, right) to detect exposure to the host cytoplasm. BFP (nonsecreted) and RARP-2 (secreted) were used as controls. Uninf, uninfected whole-cell lysate. (C) Western blot for FLAG using N-terminal FLAG-tagged R. parkeri (Rp) or R. typhi (Rt) RARP-1 expressed by WT or ΔtolC E. coli. (D) Western blot for His using C-terminal Myc-6×His-tagged R. typhi RARP-1 or C-terminal 6×His-tagged E. coli YebF expressed by WT E. coli. For panels C and D, cultures were pelleted (P) and the culture supernatant (S) was filtered and precipitated to concentrate proteins released into the medium.
As an alternative strategy to evaluate RARP-1 secretion, we introduced glycogen synthase kinase (GSK)-tagged constructs into R. parkeri. This system has been used to assess secretion of effector proteins by Rickettsia spp. and other bacteria, and it does not rely on the selective lysis of infected samples (23, 24). GSK-tagged proteins become phosphorylated by host kinases upon entering the host cytoplasm, and secretion of the tagged protein can be validated by phospho-specific antibodies (25). Although GSK-tagged RARP-2, a known secreted effector (23), was phosphorylated, GSK-tagged RARP-1 and a nonsecreted control (BFP) were not phosphorylated during infection (Fig. 4B). These results provide further evidence that RARP-1 is not secreted into the host cytoplasm by R. parkeri.
Heterologously expressed RARP-1 is not secreted by E. coli.
We were surprised by the results above since previous work suggested that RARP-1 is delivered into host cells by R. typhi. Heterologous expression in Escherichia coli provided evidence that R. typhi RARP-1 is secreted in a Sec- and TolC-dependent manner (19). Following the methodology described for that work, we assessed secretion of R. parkeri and R. typhi RARP-1 by WT and ΔtolC E. coli. In this assay, E. coli cultures expressing RARP-1 are pelleted and the culture supernatant is then filtered and precipitated to concentrate proteins released into the extracellular milieu. Although R. parkeri RARP-1 was clearly detectable in the bacterial pellets of both strains, it was not observed in the supernatants for either strain (Fig. 4C). Likewise, we were unable to detect secretion of R. typhi RARP-1 by either strain, in contrast to the previously described secretion pattern for this protein. To confirm that our use of an N-terminal 3×FLAG tag did not disrupt secretion by E. coli, we generated an R. typhi RARP-1 construct with a C-terminal Myc-6×His tag, as described in the previous work. Again, we were unable to detect secretion of R. typhi RARP-1 (Fig. 4D). To validate our ability to detect secreted proteins in the culture supernatant, we assessed secretion of 6×His-tagged YebF, a protein known to be exported into the medium by E. coli (26). As expected, YebF was observed in both the bacterial pellet and culture supernatant. The lack of RARP-1 secretion by E. coli is consistent with our immunoblotting results for infection with R. parkeri, suggesting that RARP-1 is not a secreted effector.
RARP-1 resides within R. parkeri.
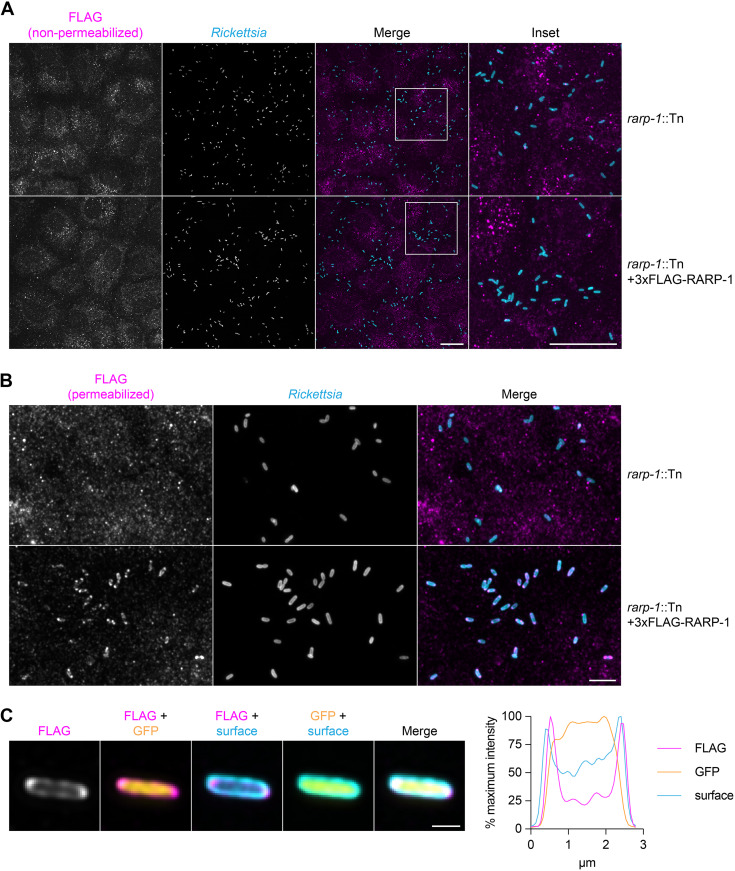
Given that RARP-1 is not secreted by R. parkeri, we next investigated where it localized during infection by using differential immunofluorescent staining (15). In this assay, infected A549 host cells are first selectively permeabilized such that only the host cell contents and bacterial surface are accessible for staining. Then, the bacteria are permeabilized with lysozyme and detergent to permit immunostaining of proteins inside the bacteria. By staining with a FLAG tag-specific antibody either with or without this second permeabilization step, we can distinguish the localization of tagged proteins inside or outside the bacteria, respectively. We predicted that epitope-tagged RARP-1 expressed by the rarp-1::Tn + 3×FLAG-RARP-1-complemented strain would be absent from the host cytoplasm but present inside permeabilized bacteria. In agreement with our immunoblotting results above, we did not detect specific FLAG staining in the host cytoplasm after infection with the complemented strain, similar to results with the rarp-1::Tn mutant (Fig. 5A). We also did not detect the protein on the bacterial surface. Instead, 3×FLAG-RARP-1 was only detectable after permeabilizing bacteria with lysozyme and detergent. Under these conditions, the 3×FLAG-RARP-1 signal surrounded the bacteria with variable localization patterns and often formed bipolar puncta (Fig. 5B). Line scan analysis of permeabilized bacteria confirmed that 3×FLAG-RARP-1 localized adjacent to the bacterial cytoplasm (Fig. 5C). These localization patterns suggest that RARP-1 is not secreted into the host cytoplasm but instead localizes within R. parkeri. The presence of an N-terminal Sec secretion signal and the lack of predicted transmembrane domains suggest that RARP-1 localizes to the R. parkeri periplasmic space or is otherwise associated with the inner or outer membrane leaflets facing the periplasm.
FIG 5.
RARP-1 resides within R. parkeri. (A) Images of rarp-1::Tn (top) and rarp-1::Tn + 3×FLAG-RARP-1 (bottom) bacteria during infection of A549 cells. Samples were stained for FLAG (magenta) and the bacterial surface (cyan) without permeabilization of bacteria. Scale bars, 20 μm. (B) Images of rarp-1::Tn (top) and rarp-1::Tn + 3×FLAG-RARP-1 (bottom) bacteria during infection of A549 cells. The bacterial surface (cyan) was stained prior to permeabilization by lysozyme and detergent and subsequent staining for FLAG (magenta). Scale bar, 5 μm. (C) Subcellular localization of 3×FLAG-RARP-1 in a representative rarp-1::Tn + 3×FLAG-RARP-1 bacterium during infection of A549 cells. The bacterial surface (cyan) was stained prior to permeabilization by lysozyme and detergent and subsequent staining for FLAG (magenta). GFP (yellow) demarcates the bacterial cytoplasm. Scale bar, 1 μm. A pole-to-pole 0.26-μm-width line scan (right) was generated for FLAG, GFP, and the bacterial surface.
RARP-1 interacts with other bacterial factors that access the periplasm.
Based on the 3×FLAG-RARP-1 localization pattern, we hypothesized that RARP-1 might interact with other factors in the R. parkeri periplasm to support growth and host cell invasion. To test this hypothesis, we isolated rarp-1::Tn + 3×FLAG-Ty1-RARP-1 bacteria and treated them with lysozyme-containing lysis buffer to release nonsecreted proteins for pulldown. As a control, we also prepared lysates from WT bacteria that do not express tagged RARP-1. We then immunoprecipitated the lysates with a FLAG tag-specific antibody, performed an acid elution to release bound proteins, and analyzed the eluates by mass spectrometry to identify putative RARP-1 binding partners (see Fig. S3A and B in the supplemental material). Proteins that were present in the tagged lysate pulldown but absent from the untagged lysate pulldown were called hits (Table 1; see also Data Set S1 in the supplemental material).
TABLE 1.
Coimmunoprecipitation of lysozyme-permeabilized bacteria reveals that RARP-1 interacts with other bacterial factors that access the periplasma
| Gene ID | Description |
|---|---|
| MC1_RS01995 | RvhB10; T4SS outer membrane core complex |
| MC1_RS00605 | Sca2; autotransporter; surface actin nucleation |
| MC1_RS00420 | Hypothetical lipoprotein |
| MC1_RS06520 | Hypothetical porin |
| MC1_RS02895 | Hypothetical lipoprotein |
| MC1_RS00535 | Hypothetical porin |
| MC1_RS00570 | OmpW family protein; porin |
| MC1_RS01970 | RvhB9a; T4SS outer membrane core complex |
| MC1_RS01990 | RvhB9b; T4SS outer membrane core complex |
| MC1_RS06075 | Pal; peptidoglycan-associated lipoprotein |
| MC1_RS05020 | 50S ribosomal protein L17 |
| MC1_RS06525 | Hypothetical porin |
| MC1_RS02795 | PcaH; protocatechuate-3,4-dioxgenase |
| MC1_RS00865 | HflC; protease modulator |
| MC1_RS06550 | 17-kDa lipoprotein surface antigen |
Putative RARP-1 binding partners are ordered by decreasing spectral count. MC1_RS05020 is the only hit not predicted to access the periplasm.
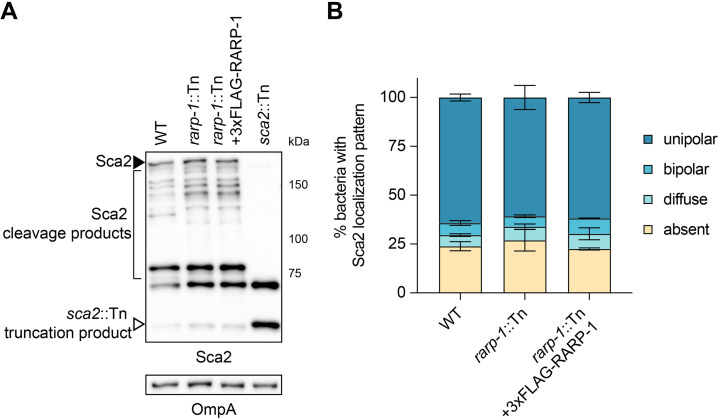
Of the hits identified, only Sca2 has been functionally characterized in R. parkeri (14). Although Sca2 promotes late-stage actin-based motility, the rarp-1::Tn mutant formed actin tails at frequencies comparable to WT (Fig. 2A), indicating that the loss of RARP-1 does not dramatically impair Sca2 function. However, it is possible that RARP-1 functions in a more subtle way to influence Sca2 activity. To test this hypothesis, we used immunoblotting to assess Sca2 expression in the rarp-1::Tn mutant (Fig. 6A). The abundance of full-length Sca2 and its processed products was comparable between the rarp-1::Tn mutant and the complemented strain, suggesting that RARP-1 does not grossly impact Sca2 levels. Likewise, we observed similar patterns of Sca2 localization between strains (Fig. 6B), suggesting that RARP-1 does not play a role in the polar positioning of Sca2. Taken together, these results suggest that RARP-1 does not regulate the activity of its putative binding partner Sca2.
FIG 6.
RARP-1 does not regulate the abundance or localization of Sca2. (A) Western blot for Sca2 from purified R. parkeri strains. Full-length Sca2 (arrowhead), Sca2 cleavage products (bracket), and the truncation product in the sca2::Tn mutant (open arrowhead) are indicated. (B) Percentage of bacteria with the indicated Sca2 localization pattern during infection of A549 cells. Percentages were determined from two independent experiments (≥350 bacteria were counted for each infection) and were used to calculate the means ± SD and P values (one-way ANOVA with post hoc Dunnett’s test). n.s., not significant relative to WT.
Additional hits identified in our analysis include the type IV secretion system outer membrane components RvhB9 and RvhB10, as well as several hypothetical lipoproteins and porins (Table 1). At this time, none of these proteins has been functionally characterized in R. parkeri. Consistent with RARP-1 localization to the periplasm, however, nearly all of these hits are predicted to reside in the periplasm or otherwise access and transit the periplasm en route to the bacterial surface. Thus, it remains possible that RARP-1 acts with one or more of these binding partners to support growth and host cell invasion.
DISCUSSION
After host cell invasion, obligate intracellular bacteria must scavenge host nutrients, proliferate, and avoid destruction by their hosts (2). Disruption of one or all of these activities will diminish intracellular bacterial loads and ultimately reduce pathogenicity. While many studies have revealed important regulators of invasion, nutrient acquisition, and bacterial growth for other species, little is known about the factors that support rickettsial physiology during infection, and only recently have we begun to uncover the protective strategies Rickettsia spp. employ to ward off host cell defenses (13, 16). Consequently, we sought to better understand the genetic determinants of rickettsial infection using our functional genetic approaches in R. parkeri. We found that RARP-1 likely resides in the periplasm, where it interacts with proteins predicted or known to drive bacterial fitness or interactions with the host. Furthermore, our results suggest that RARP-1 supports the R. parkeri life cycle by promoting bacterial growth as well as efficient host cell invasion.
Several studies have identified rickettsial surface proteins and candidate secreted effectors that facilitate invasion. For example, the outer membrane proteins OmpA and OmpB, respectively, interact with ɑ2β1 integrin and Ku70 at the host cell surface to support receptor-mediated invasion (5, 7), while the effectors RalF and Risk1 modulate host membrane phosphoinositides during entry (27, 28). Loss of RARP-1 expression led to a transient invasion delay, indicating that RARP-1 also plays a role in host cell entry. A similar invasion delay was reported for an ompB::Tn mutant (13), suggesting that Rickettsia spp. use several redundant strategies to enter their hosts. Although RARP-1 itself is not exported from the bacterium, one or more of the RARP-1 interaction partners may contribute to efficient internalization. Loss of RARP-1 expression would therefore have pleiotropic effects on infection by hindering both invasion and growth. Alternatively, it is possible that the rarp-1::Tn mutant invasion delay is the result of defective growth in the preceding infection cycles when the bacteria were harvested. Indeed, invasion competency of the intracellular bacterial pathogen Brucella abortus is linked to cell cycle progression (29). Perhaps rickettsial invasion efficiency relies on robust growth, without which the invasion program is impaired.
Loss of RARP-1 expression also reduced bacterial loads, persisting long after the initial invasion delay was overcome. This defect suggests that RARP-1 plays a role in bacterial growth through the regulation of bacterial physiology or avoidance of host defenses. Normally, R. parkeri shields itself from autophagy receptors by methylating outer membrane proteins such as OmpB (13). Loss of OmpB or the methyltransferases PKMT1 and PKMT2 promotes autophagy of R. parkeri and reduction of intracellular bacterial burdens (16). Since the rarp-1::Tn mutant did not display enhanced recruitment of the autophagy marker LC3, we concluded that the loss of RARP-1 does not render this mutant more susceptible to autophagy. Nevertheless, we cannot rule out that growth of the rarp-1::Tn mutant is restricted by other host defense strategies employed by the cell lines used in this study.
Prior work reported that RARP-1 was robustly secreted into the host cytoplasm by R. typhi, and experiments in E. coli suggested that RARP-1 relied on a noncanonical Sec- and TolC-dependent pathway for export (19). We were unable to detect secretion of endogenous or epitope-tagged RARP-1 into the host cytoplasm by R. parkeri, even though the tagged constructs functionally complemented the rarp-1::Tn mutant phenotype. Similarly, we were unable to detect phosphorylation of GSK-tagged RARP-1 in infected cell lysates as an orthogonal secretion assay. Notably, this lack of secretion was observed during infection of multiple host cell types and for both WT and rarp-1::Tn backgrounds. We also could not detect secretion of RARP-1 by E. coli, despite testing both R. parkeri and R. typhi homologs under the same conditions previously published (19). It is formally possible that our use of a different E. coli K-12 strain (BW25513 rather than C600) prevented release of RARP-1 due to incompatibility with the endogenous secretion system. Since R. typhi is a biosafety level 3 pathogen, we are not able to assess secretion of R. typhi RARP-1 by R. parkeri, and a loss-of-function rarp-1 mutant does not exist in R. typhi. Altogether, our data suggest that RARP-1 is not secreted into the host cytoplasm by R. parkeri; instead, it is likely targeted by its Sec secretion signal to the periplasm, where it stays to support bacterial growth and invasion.
RARP-1 is not predicted to possess enzymatic activity, but it does contain a large central intrinsically disordered region (IDR). The structural plasticity of IDRs affords them diverse biological functions, such as chaperone recruitment, passage through narrow protein channels, and binding of multiple protein partners (30–33). RARP-1 also possesses several C-terminal ankyrin repeats (ANKs), and many intracellular bacterial pathogens secrete ANK-containing effectors to target host cell functions (21, 22). Nevertheless, ANKs have been shown to support the activity of bacterial proteins that are not secreted into the extracellular milieu. For example, AnkB localizes to the periplasm of Pseudomonas aeruginosa, where it protects against oxidative stress (34), and Bd3460 of Bdellovibrio bacteriovorus forms complexes with endopeptidases in the periplasm to prevent degradation of its own cell wall (35). Although ANKs are best known for mediating protein-protein interactions, recent work has demonstrated that ANKs can also bind sugars and lipids (36, 37). Future mutational and biochemical analyses may reveal if the RARP-1 IDR and ANKs are necessary for interactions with its putative binding partners or if these domains otherwise support RARP-1 activity.
Our data suggest that RARP-1 interacts with several classes of proteins to support growth and invasion. Because many of these binding partners have not been functionally characterized, we focused our attention on the interaction between RARP-1 and Sca2. Sca2 is required for late-stage actin-based motility in mammalian and tick cells (14, 38), and it is necessary for virulence in animal models of SFG rickettsial infection (39). Tn mutagenesis of rarp-1 did not reduce actin tail frequency or Sca2 localization to the cell poles, suggesting that RARP-1 does not govern Sca2 function. Nevertheless, it is possible that Sca2 supports the localization or function of RARP-1 in the periplasm as it acts on other factors to regulate invasion and growth.
We also detected interactions between RARP-1 and components of the Rickettsiales vir homolog type IV secretion system (rvh T4SS). In the canonical vir T4SS of Agrobacterium tumefaciens, VirB9 and VirB10, together with VirB7, form a core complex positioned in the periplasm and outer membrane (40). It is unknown to what extent the rvh subunits play similar roles as their vir counterparts and how RARP-1 might be involved. Furthermore, few rvh T4SS effectors are known, and none of them have been shown to modulate growth (23, 27, 28). Recent work suggested that the putative rvh T4SS effector Risk1 promotes host cell invasion by R. typhi (28); whether Risk1 plays a similar role in R. parkeri or if its secretion is impacted in the rarp-1::Tn mutant is unknown. As new effectors are characterized, it will be important to determine if their secretion depends on the interaction between RARP-1 and the rvh T4SS.
Interestingly, many of the RARP-1 binding partners we identified include predicted porins and lipoproteins of unknown function. Porins are major components of the outer membrane and regulate the transport of hydrophilic compounds such as nutrients, toxins, and antibiotics (41). Homologs of MC1_RS00535 and MC1_RS00570 have been identified on the surface of the related SFG member R. rickettsii (42), but the substrates for these and other rickettsial porins have yet to be characterized. Lipoproteins are lipid-modified proteins that anchor to the membrane and support many aspects of bacterial physiology, including nutrient uptake, protein folding, signal transduction, and cell division (43). Based on remote homology predictions (via HHpred [44]), the lipoproteins identified in this study appear to be unique to the Rickettsia genus and remain largely uncharacterized. Indeed, although Tn mutagenesis of the 17-kDa lipoprotein surface antigen reduces R. parkeri plaque size, its function is unknown (18, 45). In future studies, it will be important to investigate how disruption of one or more of these factors contributes to the invasion and growth defects we observed for the rarp-1::Tn mutant.
The remaining RARP-1 interaction partners include homologs of proteins with known roles in bacterial physiology. These include factors that regulate cell division (Pal [46]), metabolism (PcaH [47]), and protein stability (HflC [48]), but none of these functions has been experimentally validated in Rickettsia spp. Although we did not observe any obvious morphological defects for the rarp-1::Tn mutant, the interaction between RARP-1 and a Pal homolog could influence rickettsial growth in a more subtle manner. It is also possible that disruption of metabolism or membrane protein quality control underlies the rarp-1::Tn mutant invasion and growth defects.
Our work uncovers an important role for RARP-1 in supporting the R. parkeri life cycle. Through its targeting to the periplasm, we propose that RARP-1 regulates invasion and growth by acting in concert with one or more of the factors revealed in our study. Further work is needed to characterize these interactions, since many of the RARP-1 binding partners we identified have unknown functions in the Rickettsia genus. Expansion of the rickettsial toolkit could facilitate these efforts as well as help determine if there is temporal or spatial control of RARP-1 activity during the R. parkeri life cycle. Moreover, structure-function analyses of RARP-1 will provide valuable insights into its mechanism of action in particular and the function of ANK- and IDR-containing proteins in general. Homologs of RARP-1 are notably absent outside the genus, despite conservation of the protein across Rickettsia spp. (19). We therefore speculate that RARP-1 represents a core and unique adaptation to the demands of the host cell niche, and future studies may extend its relevance to infection of arthropod vectors. The success of Rickettsia spp. hinges on their ability to access and thrive within the complex environment of the host cytoplasm. Continued investigation into the factors that support these fundamental processes will not only improve our understanding of rickettsial biology, but also will highlight the diverse strategies underpinning obligate intracellular bacterial life.
MATERIALS AND METHODS
Cell culture.
A549 human lung epithelial and Vero monkey kidney epithelial cell lines were obtained from the University of California, Berkeley Cell Culture Facility (Berkeley, CA). A549 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco catalog number 11965118) containing 10% fetal bovine serum (FBS). Vero cells were maintained in DMEM containing 5% FBS. A549 cells stably expressing cytoplasmic TagRFP-T (A549-TRT) were generated by retroviral transduction as previously described (15). Cell lines were confirmed to be mycoplasma-negative in a MycoAlert PLUS assay (Lonza catalog number LT07-710) performed by the Koch Institute High-Throughput Sciences Facility (Cambridge, MA).
Plasmid construction.
pRAM18dSGA-3×FLAG-RARP-1 was generated from pRAM18dSGA[MCS] (kindly provided by Ulrike Munderloh) and contains the 247 bp immediately upstream of the tolC start codon (MC1_RS01570), the first 23 amino acids (aa) of R. parkeri RARP-1 (MC1_RS01585) containing the Sec SS, a HVDYKDHDGDYKDHDIDYKDDDDKHV sequence (3×FLAG epitope tag underlined), the remaining 550 aa of RARP-1, and the R. parkeri ompA terminator (MC1_RS06480). pRL0079 is identical to pRAM18dSGA-3×FLAG-RARP-1 but contains GSGGEVHTNQDPLDGGT (Ty1 epitope tag underlined) between residues 396 and 397.
pRL0284 was generated from pRAM18dSGA[MCS] and contains the R. parkeri ompA promoter, an N-terminal MSGRPRTTSFAESGS sequence (GSK epitope tag underlined), TagBFP from pRAM18dRA-2×TagBFP (15), and the ompA terminator. pRL0285 is identical to pRL0284 but contains R. parkeri RARP-2 (MC1_RS04780) in place of TagBFP. Similarly, pRL0286 contains R. parkeri RARP-1 in place of TagBFP, but GSMSGRPRTTSFAESGS was inserted after the Sec SS (as in pRAM18dSGA-3×FLAG-RARP-1) instead of at the N terminus.
pRL0287 was generated from pEXT20 (kindly provided by Michael Laub) and contains the R. parkeri RARP-1 insert with intervening 3×FLAG epitope tag from pRAM18dSGA-3xFLAG-RARP-1. pRL0288 is identical to pRL0287, except the 23-aa Sec SS of R. typhi RARP-1 (RT0218) and the remaining 563 aa of R. typhi RARP-1 were used. In contrast, pRL0289 contains the full 586 aa of R. typhi RARP-1 with a C-terminal KGEFEAYVEQKLISEEDLNSAVDHHHHHH sequence (Myc and 6×His epitope tags underlined) as previously described (19). For pRL0290, a C-terminal VDHHHHHH sequence (6×His epitope tag underlined) was added to E. coli YebF (NCBI b1847).
Generation of R. parkeri strains.
Parental R. parkeri strain Portsmouth (kindly provided by Chris Paddock) and all derived strains were propagated by infection and mechanical disruption of Vero cells grown in DMEM containing 2% FBS at 33°C as previously described (15, 18). Bacteria were clonally isolated and expanded from plaques formed after overlaying infected Vero cell monolayers with agarose as previously described (18). When appropriate, bacteria were further purified by centrifugation through a 30% MD-76R gradient (Mallinckrodt Inc. catalog number 1317-07) as previously described (15). Bacterial stocks were stored as aliquots at −80°C to minimize variability due to freeze-thaws. Titers were determined for bacterial stocks by plaque assay (15), and plaque sizes (Fig. 1B) were measured with ImageJ after 5 days of infection.
Bacteria were transformed with plasmids by small-scale electroporation as previously described (18), except infections were scaled down to a T25 flask, and bacteria were electroporated with 1 μg dialyzed plasmid DNA. When appropriate, rifampicin (200 ng/mL) or spectinomycin (50 μg/mL) was included to select for transformants. GFP-expressing WT bacteria were generated as previously described (15); this control strain behaves similarly to the parental WT strain in a variety of assays, such as actin tail assays (14), mixed-cell assays (9), and infectious focus assays (data not shown). The rarp-1::Tn and sca2::Tn mutants were generated as previously described (18), and the genomic locations of the Tn insertion sites were determined by semirandom nested PCR and Sanger sequencing. The expanded strains were verified by PCR amplification of the Tn insertion site using primers flanking the region. The ompBSTOP::Tn mutant (referred to as ompB::Tn in this work; kindly provided by Matthew Welch) was generated as previously described (13).
Infectious focus assays.
Confluent A549 cells (approximately 3.5 × 105 cells/cm2) were grown on 12-mm coverslips in 24-well plates and were infected at a multiplicity of infection (MOI) of 0.005 to 0.025, centrifuged at 200 × g for 5 min at room temperature (RT), and incubated at 33°C for 1 h. Infected cells were washed three times with phosphate-buffered saline (PBS) before adding complete medium with 10 μg/mL gentamicin. Infections progressed for 28 h at 33°C until fixation with 4% paraformaldehyde (PFA) in PBS for 10 min at RT. Fixed samples were incubated with 0.1 M glycine in PBS for 10 min at RT to quench residual PFA. Samples were then washed three times with PBS, permeabilized with 0.5% Triton X-100 in PBS for 5 min at RT, and washed another three times with PBS. Samples were then incubated with blocking buffer (2% bovine serum albumin [BSA] in PBS) for 30 min at RT. Primary and secondary antibodies were diluted in blocking buffer and incubated for 1 h each at RT with three 5-min PBS washes after each incubation step. The following antibodies and stains were used: mouse anti-β-catenin (Cell Signaling Technology catalog number 2677) to detect host membrane, rabbit anti-Rickettsia I7205 (kindly provided by Ted Hackstadt), goat anti-mouse conjugated to Alexa Fluor 568 (Invitrogen catalog number A-11004), goat anti-rabbit conjugated to Alexa Fluor 488 (Invitrogen catalog number A-11008), and Hoechst stain (Invitrogen catalog number H3570) to detect host nuclei. Coverslips were mounted using ProLong Gold Antifade mountant (Invitrogen catalog number P36934) mages were acquired using a 60× UPlanSApo (1.30 numerical aperture [NA]) objective on an Olympus IXplore Spin microscope system. Image analysis was performed with ImageJ. For each strain, 20 to 35 foci were imaged, and the number of infected cells and bacteria per focus was calculated.
Actin tail and protrusion assays.
Confluent A549 cells were infected and processed as above, but an MOI of 0.3 to 0.6 was used and phalloidin conjugated to Alexa Fluor 647 (Invitrogen catalog number A22287) was included to detect actin. For each strain, ≥380 bacteria were imaged using a 100× UPlanSApo (1.35 NA) objective, and the percentage of bacteria with tails (>1 bacterial length) and the percentage of bacteria within protrusions were calculated.
Mixed-cell assays.
A549-TRT donor cells were plated in 96-well plates, and unlabeled A549 recipient cells were plated in 6-well plates and grown to confluence. Donors were infected at an MOI of 9 to 10, centrifuged at 200 × g for 5 min at RT, and incubated at 33°C for 1 h. Infected donors and uninfected recipients were washed with PBS, lifted with citric saline (135 mM KCl, 15 mM sodium citrate) at 37°C to preserve cell surface receptors, recovered in complete medium, washed twice with complete medium to remove residual citric saline, and resuspended in complete medium with 10 μg/mL gentamicin (6 × 105 cells/mL donors and 8 × 105 cells/mL recipients). Cells were then mixed at a 1:125 ratio (5.3 μL donors and 500 μL recipients) and plated on 12-mm coverslips in 24-well plates. Infections progressed for 31 h at 33°C until fixation with 4% PFA in PBS for 1 h at RT. Fixed samples were processed as above, except the following antibodies and stains were used: mouse anti-Rickettsia 14-13 (kindly provided by Ted Hackstadt), goat anti-mouse conjugated to Alexa Fluor 647 (Invitrogen catalog number A-21235), and phalloidin-iFluor 405 reagent (Abcam catalog number ab176752). For each strain, 20 foci were imaged using a 60× objective, and the percentage of bacteria per focus that had spread to recipient cells was calculated.
Growth curves.
Confluent Vero cells (approximately 4 × 105 cells/cm2) were grown in 24-well plates and infected in triplicate at an MOI of 0.025, centrifuged at 200 × g for 5 min at RT, and incubated at 33°C for 1 h. Infected cells were washed three times with serum-free DMEM before adding complete medium and allowing infections to progress at 33°C. To harvest samples at the indicated time point, infected cells were scraped into the medium and centrifuged at 20,000 × g for 5 min. The resulting pellets were resuspended in 600 μL nuclei lysis solution (Promega catalog number A7941), boiled for 10 min to release genomic DNA, and processed with a Wizard genomic DNA purification kit (Promega catalog number A1125) according to the manufacturer’s instructions. After air-drying, the DNA pellets were resuspended in 100 μL H2O, incubated at 65°C for 1 h, and allowed to completely rehydrate overnight at RT. For qPCR, runs were carried out on a LightCycler 480 (Roche) at the MIT BioMicro Center (Cambridge, MA). Primers to the R. parkeri 17-kDa surface antigen gene (MC1_RS06550; 5′-TTCGGTAAGGGCAAAGGACA-3′ and 5′-GCACCGATTTGTCCACCAAG-3′) and to Chlorocebus sabaeus GAPDH (5′-AATGGGACTGAAGCTCCTGC-3′ and 5′-ATCACCACCCCTCTACCTCC-3′) were used to determine bacterial and host genome equivalents, respectively, relative to a standard curve prepared from a pooled mixture of the 96-h time point WT infection samples. Results from each biological replicate were normalized to the 1-h time point, and fold change was calculated. Doubling times were computed from the 24 to 48 h of exponential growth for each strain.
Viability assays.
Confluent A549 cells grown in 24-well plates were infected at an MOI of 0.2 to 0.8, centrifuged at 200 × g for 5 min at RT, and incubated at 33°C for 48 h. Bacteria were released by incubating infected cells with ice-cold H2O for 2.5 min, immediately recovered in ice-cold 250 mM sucrose, and stained for 15 min at RT using a LIVE/DEAD BacLight bacterial viability kit (Thermo Scientific catalog number L7012) according to the manufacturer’s instructions. R. parkeri cells were heat-killed by incubating at 65°C for 20 min prior to staining. For each condition, ≥160 bacteria were imaged using a 60× objective, and the percentage of viable bacteria was calculated.
LC3 recruitment assays.
Confluent A549 cells grown on 12-mm coverslips in 24-well plates were infected at an MOI of 1.8 to 3.6, centrifuged at 200 × g for 5 min at RT, and incubated at 33°C for 2 h until fixation with 4% PFA in PBS for 10 min. Fixed samples were processed as above, except cells were permeabilized with 100% methanol for 5 min at RT instead of Triton X-100, and the following antibodies and stains were used: rabbit anti-LC3B (ABclonal catalog number A7198), mouse anti-Rickettsia 14-13, goat anti-rabbit conjugated to Alexa Fluor 568 (Invitrogen catalog number A-11011), goat anti-mouse conjugated to Alexa Fluor 488 (Invitrogen catalog number A-11001), and Hoechst. Representative images were acquired using a 100× objective.
Invasion assays.
Confluent A549 cells grown on 12-mm coverslips in 24-well plates were placed on ice and the medium was replaced with 500 μL ice-cold complete medium. The cells were then infected at an MOI of 0.7 to 1.2, centrifuged at 200 × g for 5 min at 4°C, 500 μL of 37°C complete medium was added, and the plates were immediately moved to 37°C until fixation with 4% PFA in PBS for 10 min. Fixed samples were incubated with 0.1 M glycine in PBS for 10 min at RT to quench residual PFA. Samples were then washed three times with PBS and incubated with blocking buffer for 30 min at RT. To stain external bacteria, primary and secondary antibodies were diluted in blocking buffer and incubated for 30 min each at RT with three 5-min PBS washes after each incubation step. The following antibodies and stains were used: mouse anti-Rickettsia 14-13 and goat anti-mouse conjugated to Alexa Fluor 647. The samples were then fixed with 4% PFA in PBS for 5 min at RT, washed three times with PBS, and quenched with 0.1 M glycine in PBS for 10 min at RT. Samples were then washed three times with PBS, permeabilized with 0.5% Triton X-100 in PBS for 5 min at RT, and washed another three times with PBS. To stain both external and internal bacteria, primary and secondary antibodies were diluted in blocking buffer and incubated for 30 min each at RT with three 5-min PBS washes after each incubation step. The following antibodies and stains were used: mouse anti-Rickettsia 14-13 and goat anti-mouse conjugated to Alexa Fluor 488. For each strain, 20 fields of view each containing ≥45 bacteria were imaged using a 60× objective. To facilitate analysis, internal and external bacteria were quantified using ilastik (49); the pixel classifier was trained to distinguish bacteria from background, and then the object classifier was trained to distinguish between internal (single-stained) and external (double-stained) bacteria.
RARP-1 antibody production.
The RARP-1 peptide antigen (SNEMHEAQVASNEHND, corresponding to residues 159 to 174), was selected and synthesized by New England Peptide (Gardner, MA). The peptide antigen was conjugated to keyhole limpet hemocyanin and used for immunization by Pocono Rabbit Farm and Laboratory (Canadensis, PA) according to their 70-day rabbit polyclonal antibody protocol.
R. parkeri RARP-1 secretion assays.
Confluent A549 cells grown in 24-well plates were infected at an MOI of 0.5 to 1.0, centrifuged at 200 × g for 5 min at RT, and incubated at 33°C until the indicated harvest time point. Infected cells were washed three times with PBS, lifted with trypsin-EDTA, and centrifuged at 2,400 × g for 5 min at RT. The resulting pellets were resuspended in selective lysis buffer (50 mM HEPES [pH 7.9], 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% IGEPAL) containing protease inhibitors (Sigma-Aldrich catalog number P1860), incubated on ice for 15 min, and centrifuged at 11,300 × g for 10 min at 4°C. The resulting pellets were washed with PBS and boiled in loading buffer (50 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 10% glycerol, 0.1% bromophenol blue, 5% β-mercaptoethanol). The resulting supernatants were passed through a 0.22-μm cellulose acetate filter (Thermo Scientific catalog number F2517-1) by centrifugation at 6,700 × g for 10 min at 4°C, combined with loading buffer (to a final volume equal to the final pellet volume), and boiled. Lysates were analyzed by Western blotting using rabbit anti-FLAG (Cell Signaling Technology catalog number 2368), rabbit anti-Sca4 (15), mouse anti-Ty1 (kindly provided by Sebastian Lourido), rabbit RARP-1 peptide antisera, and mouse anti-RpoA (BioLegend catalog number 663104).
R. parkeri GSK secretion assays.
Confluent Vero cells grown in 24-well plates were infected with the indicated strains, centrifuged at 200 × g for 5 min at RT, and incubated at 33°C with spectinomycin for 72 h (when infected cells were approximately 90% rounded) before harvesting. Infected cells were washed with ice-cold serum-free DMEM, directly lysed in loading buffer, and boiled. Lysates were analyzed by Western blotting using rabbit anti-GSK-3β-Tag (Cell Signaling Technology catalog number 9325) and rabbit anti-phospho-GSK-3β (Cell Signaling Technology catalog number 9336).
E. coli secretion assays.
E. coli K-12 BW25113 (WT) and JW5503-1 (ΔtolC) from the Keio Knockout Collection (50) were obtained from Horizon Discovery. SDS sensitivity and the Kanr cassette insertion site were confirmed for the ΔtolC strain. Secretion assay samples were collected and processed as previously described (19). Bacterial pellets and precipitated proteins were boiled in loading buffer. Bacterial pellet lysates (equivalent to 0.025 optical density at 600 nm [OD600]/mL of cultured cells), and precipitated culture supernatants (equivalent to 2 mL of culture supernatant prior to precipitation) were analyzed by Western blotting using rabbit anti-FLAG and horseradish peroxidase-conjugated mouse anti-His (ABclonal catalog number AE028).
RARP-1 localization assays.
Confluent A549 cells grown in 24-well plates were infected at an MOI of 0.3 to 0.6, centrifuged at 200 × g for 5 min at RT, and incubated at 33°C for 27 h until fixation with 4% PFA in PBS for 1 h. Fixed samples were incubated with 0.1 M glycine in PBS for 10 min at RT to quench residual PFA. Samples were then washed three times with PBS, permeabilized with 0.5% Triton X-100 in PBS for 5 min at RT, and washed another three times with PBS. Samples were then incubated with goat serum blocking buffer (2% BSA and 10% normal goat serum in PBS) for 30 min at RT. To stain host cell contents and bacterial surface proteins, primary and secondary antibodies were diluted in goat serum blocking buffer and incubated for 3 h at 37°C and 1 h at RT, respectively, with three 5-min PBS washes after each incubation step. To stain nonpermeabilized bacteria, rabbit anti-FLAG, mouse anti-Rickettsia 14-13, goat anti-rabbit conjugated to Alexa Fluor 647 (Invitrogen catalog number A-21245), and goat anti-mouse conjugated to Alexa Fluor 488 were used, and coverslips were mounted after washing. To stain permeabilized bacteria, only mouse anti-Rickettsia 14-13 and goat anti-mouse conjugated to Alexa Fluor 488 were used in the first round of staining, and coverslips were instead fixed with 4% PFA in PBS for 5 min at RT after washing. These samples were then incubated with 0.1 M glycine in PBS for 10 min at RT to quench residual PFA and washed three times with PBS. To expose proteins inside the bacteria for staining, these samples were incubated with lysozyme reaction buffer (0.8× PBS, 50 mM glucose, 5 mM EDTA, 0.1% Triton X-100, 5 mg/mL lysozyme [Sigma catalog number L6876]) for 20 min at 37°C, and then washed three times with PBS. Rabbit anti-FLAG and goat anti-rabbit conjugated to Alexa Fluor 647 were diluted in goat serum blocking buffer and incubated for 3 h at 37°C and 1 h at RT, respectively, with three 5-min PBS washes after each incubation step. Coverslips were mounted after the second round of staining. For subcellular localization images, goat anti-mouse conjugated to Alexa Fluor 488 was replaced with Alexa Fluor 405 (Invitrogen catalog number A-31553) to permit imaging of bacterial GFP. Representative images were acquired using a 100× objective, deconvolved by performing five iterations of the cellSens (Olympus) advanced maximum likelihood estimation algorithm, and a 0.26-μm-width pole-to-pole line scan was performed with ImageJ.
Sca2 localization assays.
Confluent A549 cells grown in 24-well plates were infected at an MOI of 0.3 to 0.6, centrifuged at 200 × g for 5 min at RT, and incubated at 33°C for 28 h until fixation with 4% PFA in PBS for 10 min. Fixed samples were processed as in the infectious focus assays, except the following antibodies and stains were used: rabbit anti-Sca2 (kindly provided by Matthew Welch), mouse anti-Rickettsia 14-13, goat anti-rabbit conjugated to Alexa Fluor 568, goat anti-mouse conjugated to Alexa Fluor 488, phalloidin conjugated to Alexa Fluor 647, and Hoechst. For each strain, ≥350 bacteria were imaged using a 100× objective, and the Sca2 localization pattern was determined (following the classification scheme described in reference 14).
Immunoblotting of RARP-1 and Sca2 from purified bacteria.
Purified bacteria were boiled in loading buffer and analyzed by Western blotting using rabbit RARP-1 peptide antisera, rabbit anti-FLAG, rabbit anti-Sca2, and mouse anti-OmpA 13-3 (kindly provided by Ted Hackstadt). For Fig. 1C and 6A, the parental WT R. parkeri strain lacking pRAM18dRGA+OmpApr-GFPuv was used. In Fig. 1C and D, the apparent molecular weight (MW) of RARP-1 is greater than its predicted MW (60 kDa). This aberrant migration by SDS-PAGE is typical of proteins with IDRs (51).
Coimmunoprecipitation assays.
Two replicate samples each of WT and rarp-1::Tn + 3×FLAG-Ty1-RARP-1 bacteria were processed in parallel for FLAG coimmunoprecipitation. For each sample, bacteria purified from a fully infected T175 flask were centrifuged at 16,200 × g for 2 min at RT, resuspended in 1 mL immunoprecipitation lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) containing 50 U/μL Ready-Lyse lysozyme (Lucigen catalog number R1804M) and protease inhibitors, incubated for 25 min at RT, and centrifuged at 11,300 × g for 15 min at 4°C. The resulting supernatants were precleared twice by incubation with 28 μL of 50% mouse IgG agarose slurry (Sigma catalog number A0919) for 30 min at 4°C. The precleared input lysates were then incubated with 28 μL 50% anti-FLAG M2 magnetic bead slurry (Sigma catalog number M8823) overnight at 4°C. The bound complexes were washed four times with 500 μL of ice-cold immunoprecipitation wash buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl) containing protease inhibitors, eluted by incubation with 65.2 μL 0.1 M glycine (pH 2.8) for 20 min at RT, and neutralized with 9.8 μL 1 M Tris-HCl (pH 8.5). The neutralized eluates were then combined with loading buffer and submitted to the Whitehead Institute Proteomics Core Facility (Cambridge, MA) for sample workup and mass spectrometry analysis. Bacterial inputs were evaluated by immunoblotting for Sca4 and FLAG.
Mass spectrometry.
Samples were run 1 cm into an SDS-PAGE gel, excised, and then reduced, alkylated, and digested with trypsin overnight at 37°C. The resulting peptides were extracted, concentrated, and injected onto a nanoACQUITY ultraperformance liquid chromatograph (Waters) equipped with a self-packed Aeris 3.6-μm C18 analytical column (20 cm by 75 μm; Phenomenex). Peptides were eluted using standard reverse-phase gradients. The effluent from the column was analyzed using an Orbitrap Elite mass spectrometer (nanospray configuration; Thermo Scientific) operated in a data-dependent manner. Peptides were identified using SEQUEST (Thermo Scientific), and the results were compiled in Scaffold (Proteome Software). RefSeq entries for R. parkeri strain Portsmouth (taxonomy ID 1105108) and Homo sapiens (taxonomy ID 9606) were downloaded from NCBI and concatenated with a database of common contaminants. Peptide identifications were accepted at a threshold of 95%. Protein identifications were accepted with a threshold of 99% and two unique peptides. Rickettsial proteins that were present in both replicates of the tagged (rarp-1::Tn + 3×FLAG-Ty1-RARP-1) lysate pulldown but absent from both replicates of the untagged (WT) lysate pulldown were called hits.
Statistical analyses.
Statistical analysis was performed using Prism 9 (GraphPad Software). Graphical representations, statistical parameters, and significance are reported in the figure legends. Data were considered statistically significant when P was <0.05, as determined by an unpaired Student’s t test or one-way analysis of variance (ANOVA) with post hoc Dunnett’s test.
Data availability.
Mass spectral data and the protein sequence database used for searches have been deposited in the public proteomics repository MassIVE (https://massive.ucsd.edu, MSV000088867).
ACKNOWLEDGMENTS
We are grateful to Michael Laub, Jon McGinn, and Brandon Sit for critical review of the manuscript. We thank Ted Hackstadt, Michael Laub, Sebastian Lourido, Ulrike Munderloh, Chris Paddock, and Matthew Welch for reagents and Sebastian Lourido, Elizabeth Boydston, Natasha Kafai, and Adam Nock for technical help. We also thank Whitehead Institute Proteomics Core Facility members Eric Spooner and Edward Dudek for experimental support. This work was supported by NIH/NIGMS T32GM007287 and T32GM136540 (A.G.S., R.E.H.), NIH/NIGMS R00GM115765 (R.L.L.), and NIH/NIAID R01AI155489 (R.L.L.).
Footnotes
Supplemental material is available online only.
Contributor Information
Rebecca L. Lamason, Email: rlamason@mit.edu.
Conrad W. Mullineaux, Queen Mary University of London
REFERENCES
- 1.Kumar Y, Valdivia RH. 2009. Leading a sheltered life: intracellular pathogens and maintenance of vacuolar compartments. Cell Host Microbe 5:593–601. 10.1016/j.chom.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ray K, Marteyn B, Sansonetti PJ, Tang CM. 2009. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol 7:333–340. 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- 3.Walker DH, Ismail N. 2008. Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat Rev Microbiol 6:375–386. 10.1038/nrmicro1866. [DOI] [PubMed] [Google Scholar]
- 4.McGinn J, Lamason RL. 2021. The enigmatic biology of rickettsiae: recent advances, open questions and outlook. Pathog Dis 79:ftab019. 10.1093/femspd/ftab019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan YGY, Cardwell MM, Hermanas TM, Uchiyama T, Martinez JJ. 2009. Rickettsial outer-membrane protein B (rOmpB) mediates bacterial invasion through Ku70 in an actin, c-Cbl, clathrin and caveolin 2-dependent manner. Cell Microbiol 11:629–644. 10.1111/j.1462-5822.2008.01279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reed SCO, Serio AW, Welch MD. 2012. Rickettsia parkeri invasion of diverse host cells involves an Arp2/3 complex, WAVE complex and Rho-family GTPase-dependent pathway. Cell Microbiol 14:529–545. 10.1111/j.1462-5822.2011.01739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hillman RD, Baktash YM, Martinez JJ. 2013. OmpA-mediated rickettsial adherence to and invasion of human endothelial cells is dependent upon interaction with α2β1 integrin. Cell Microbiol 15:727–741. 10.1111/cmi.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teysseire N, Boudier JA, Raoult D. 1995. Rickettsia conorii entry into Vero cells. Infect Immun 63:366–374. 10.1128/iai.63.1.366-374.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borgo GM, Burke TP, Tran CJ, Lo NTN, Engström P, Welch MD. 2021. A patatin-like phospholipase mediates Rickettsia parkeri escape from host membranes. bioRxiv 10.1101/2021.10.21.465009. [DOI] [PMC free article] [PubMed]
- 10.Driscoll TP, Verhoeve VI, Guillotte ML, Lehman SS, Rennoll SA, Beier-Sexton M, Rahman MS, Azad AF, Gillespie JJ. 2017. Wholly Rickettsia! Reconstructed metabolic profile of the quintessential bacterial parasite of eukaryotic cells. mBio 8:e00859-17. 10.1128/mBio.00859-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahyong V, Berdan CA, Burke TP, Nomura DK, Welch MD. 2019. A metabolic dependency for host isoprenoids in the obligate intracellular pathogen Rickettsia parkeri underlies a sensitivity to the statin class of host-targeted therapeutics. mSphere 4:e00536-19. 10.1128/mSphere.00848-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clifton DR, Goss RA, Sahni SK, van Antwerp D, Baggs RB, Marder VJ, Silverman DJ, Sporn LA. 1998. NF-κB-dependent inhibition of apoptosis is essential for host cell survival during Rickettsia rickettsii infection. Proc Natl Acad Sci U S A 95:4646–4651. 10.1073/pnas.95.8.4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engström P, Burke TP, Mitchell G, Ingabire N, Mark KG, Golovkine G, Iavarone AT, Rape M, Cox JS, Welch MD. 2019. Evasion of autophagy mediated by Rickettsia surface protein OmpB is critical for virulence. Nat Microbiol 4:2538–2551. 10.1038/s41564-019-0583-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reed SCO, Lamason RL, Risca VI, Abernathy E, Welch MD. 2014. Rickettsia actin-based motility occurs in distinct phases mediated by different actin nucleators. Curr Biol 24:98–103. 10.1016/j.cub.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamason RL, Bastounis E, Kafai NM, Serrano R, del Álamo JC, Theriot JA, Welch MD. 2016. Rickettsia Sca4 reduces vinculin-mediated intercellular tension to promote spread. Cell 167:670–683.e10. 10.1016/j.cell.2016.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engström P, Burke TP, Tran CJ, Iavarone AT, Welch MD. 2021. Lysine methylation shields an intracellular pathogen from ubiquitylation and autophagy. Sci Adv 7:eabg2517. 10.1126/sciadv.abg2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillespie JJ, Williams K, Shukla M, Snyder EE, Nordberg EK, Ceraul SM, Dharmanolla C, Rainey D, Soneja J, Shallom JM, Vishnubhat ND, Wattam R, Purkayastha A, Czar M, Crasta O, Setubal JC, Azad AF, Sobral BS. 2008. Rickettsia phylogenomics: unwinding the intricacies of obligate intracellular life. PLoS One 3:e2018. 10.1371/journal.pone.0002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamason RL, Kafai NM, Welch MD. 2018. A streamlined method for transposon mutagenesis of Rickettsia parkeri yields numerous mutations that impact infection. PLoS One 13:e0197012. 10.1371/journal.pone.0197012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaur SJ, Rahman MS, Ammerman NC, Beier-Sexton M, Ceraul SM, Gillespie JJ, Azad AF. 2012. TolC-dependent secretion of an ankyrin repeat-containing protein of Rickettsia typhi. J Bacteriol 194:4920–4932. 10.1128/JB.00793-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mosavi LK, Cammett TJ, Desrosiers DC, Peng Z. 2004. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci 13:1435–1448. 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Khodor S, Price CT, Kalia A, Kwaik YA. 2010. Ankyrin-repeat containing proteins of microbes: a conserved structure with functional diversity. Trends Microbiol 18:132–139. 10.1016/j.tim.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aistleitner K, Clark T, Dooley C, Hackstadt T. 2020. Selective fragmentation of the trans-Golgi apparatus by Rickettsia rickettsii. PLoS Pathog 16:e1008582. 10.1371/journal.ppat.1008582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehman SS, Noriea NF, Aistleitner K, Clark TR, Dooley CA, Nair V, Kaur SJ, Rahman MS, Gillespie JJ, Azad AF, Hackstadt T. 2018. The rickettsial ankyrin repeat protein 2 is a type IV secreted effector that associates with the endoplasmic reticulum. mBio 9:e00975-18. 10.1128/mBio.00975-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bauler LD, Hackstadt T. 2014. Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol 196:1325–1334. 10.1128/JB.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torruellas GJ, Ferracci F, Jackson MW, Joseph SS, Pattis I, Plano LRW, Fischer W, Plano GV. 2006. Measurement of effector protein injection by type III and type IV secretion systems by using a 13-residue phosphorylatable glycogen synthase kinase tag. Infect Immun 74:5645–5657. 10.1128/IAI.00690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang G, Brokx S, Weiner JH. 2006. Extracellular accumulation of recombinant proteins fused to the carrier protein YebF in Escherichia coli. Nat Biotechnol 24:100–104. 10.1038/nbt1174. [DOI] [PubMed] [Google Scholar]
- 27.Rennoll-Bankert KE, Rahman MS, Gillespie JJ, Guillotte ML, Kaur SJ, Lehman SS, Beier-Sexton M, Azad AF. 2015. Which way in? The RalF Arf-GEF orchestrates Rickettsia host cell invasion. PLoS Pathog 11:e1005115. 10.1371/journal.ppat.1005115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voss OH, Gillespie JJ, Lehman SS, Rennoll SA, Beier-Sexton M, Rahman MS, Azad AF. 2020. Risk1, a phosphatidylinositol 3-kinase effector, promotes Rickettsia typhi intracellular survival. mBio 11:e00820-20. 10.1128/mBio.00820-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deghelt M, Mullier C, Sternon J-F, Francis N, Laloux G, Dotreppe D, Van der Henst C, Jacobs-Wagner C, Letesson J-J, De Bolle X. 2014. G1-arrested newborn cells are the predominant infectious form of the pathogen Brucella abortus. Nat Commun 5:4366. 10.1038/ncomms5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dyson HJ, Wright PE. 2005. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6:197–208. 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 31.Rodgers L, Gamez A, Riek R, Ghosh P. 2008. The type III secretion chaperone SycE promotes a localized disorder-to-order transition in the natively unfolded effector YopE. J Biol Chem 283:20857–20863. 10.1074/jbc.M802339200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Housden NG, Hopper JTS, Lukoyanova N, Rodriguez-Larrea D, Wojdyla JA, Klein A, Kaminska R, Bayley H, Saibil HR, Robinson CV, Kleanthous C. 2013. Intrinsically disordered protein threads through the bacterial outer membrane porin OmpF. Science 340:1570–1574. 10.1126/science.1237864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holmes JA, Follett SE, Wang H, Meadows CP, Varga K, Bowman GR. 2016. Caulobacter PopZ forms an intrinsically disordered hub in organizing bacterial cell poles. Proc Natl Acad Sci U S A 113:12490–12495. 10.1073/pnas.1602380113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howell ML, Alsabbagh E, Ma J-F, Ochsner UA, Klotz MG, Beveridge TJ, Blumenthal KM, Niederhoffer EC, Morris RE, Needham D, Dean GE, Wani MA, Hassett DJ. 2000. AnkB, a periplasmic ankyrin-like protein in Pseudomonas aeruginosa, is required for optimal catalase B (KatB) activity and resistance to hydrogen peroxide. J Bacteriol 182:4545–4556. 10.1128/JB.182.16.4545-4556.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lambert C, Cadby IT, Till R, Bui NK, Lerner TR, Hughes WS, Lee DJ, Alderwick LJ, Vollmer W, Sockett RE, Sockett ER, Lovering AL. 2015. Ankyrin-mediated self-protection during cell invasion by the bacterial predator Bdellovibrio bacteriovorus. Nat Commun 6:8884. 10.1038/ncomms9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woodford CR, Thoden JB, Holden HM. 2015. A new role for the ankyrin repeat revealed by the study of the N-formyltransferase from Providencia alcalifaciens. Biochemistry 54:631–638. 10.1021/bi501539a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim DH, Park M-J, Gwon GH, Silkov A, Xu Z-Y, Yang EC, Song S, Song K, Kim Y, Yoon HS, Honig B, Cho W, Cho Y, Hwang I. 2014. Chloroplast targeting factor AKR2 evolved from an ankyrin repeat domain coincidentally binds two chloroplast lipids. Dev Cell 30:598–609. 10.1016/j.devcel.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris EK, Jirakanwisal K, Verhoeve VI, Fongsaran C, Suwanbongkot C, Welch MD, Macaluso KR. 2018. Role of Sca2 and RickA in the dissemination of Rickettsia parkeri in Amblyomma maculatum. Infect Immun 86:e00123-18. 10.1128/IAI.00123-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleba B, Clark TR, Lutter EI, Ellison DW, Hackstadt T. 2010. Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect Immun 78:2240–2247. 10.1128/IAI.00100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costa TRD, Harb L, Khara P, Zeng L, Hu B, Christie PJ. 2021. Type IV secretion systems: advances in structure, function, and activation. Mol Microbiol 115:436–452. 10.1111/mmi.14670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vergalli J, Bodrenko IV, Masi M, Moynié L, Acosta-Gutiérrez S, Naismith JH, Davin-Regli A, Ceccarelli M, van den Berg B, Winterhalter M, Pagès J-M. 2020. Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria. Nat Rev Microbiol 18:164–176. 10.1038/s41579-019-0294-2. [DOI] [PubMed] [Google Scholar]
- 42.Gong W, Xiong X, Qi Y, Jiao J, Duan C, Wen B. 2014. Identification of novel surface-exposed proteins of Rickettsia rickettsii by affinity purification and proteomics. PLoS One 9:e100253. 10.1371/journal.pone.0100253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kovacs-Simon A, Titball RW, Michell SL. 2011. Lipoproteins of bacterial pathogens. Infect Immun 79:548–561. 10.1128/IAI.00682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gabler F, Nam S-Z, Till S, Mirdita M, Steinegger M, Söding J, Lupas AN, Alva V. 2020. Protein sequence analysis using the MPI bioinformatics toolkit. Curr Protoc Bioinforma 72:e108. 10.1002/cpbi.108. [DOI] [PubMed] [Google Scholar]
- 45.Anderson BE. 1990. The 17-kilodalton protein antigens of spotted fever and typhus group rickettsiae. Ann N Y Acad Sci 590:326–333. 10.1111/j.1749-6632.1990.tb42240.x. [DOI] [PubMed] [Google Scholar]
- 46.Yakhnina AA, Bernhardt TG. 2020. The Tol-Pal system is required for peptidoglycan-cleaving enzymes to complete bacterial cell division. Proc Natl Acad Sci U S A 117:6777–6783. 10.1073/pnas.1919267117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harwood CS, Parales RE. 1996. The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol 50:553–590. 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 48.Bittner L-M, Arends J, Narberhaus F. 2017. When, how and why? Regulated proteolysis by the essential FtsH protease in Escherichia coli. Biol Chem 398:625–635. 10.1515/hsz-2016-0302. [DOI] [PubMed] [Google Scholar]
- 49.Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, Schiegg M, Ales J, Beier T, Rudy M, Eren K, Cervantes JI, Xu B, Beuttenmueller F, Wolny A, Zhang C, Koethe U, Hamprecht FA, Kreshuk A. 2019. ilastik: interactive machine learning for (bio)image analysis. Nat Methods 16:1226–1232. 10.1038/s41592-019-0582-9. [DOI] [PubMed] [Google Scholar]
- 50.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio Collection. Mol Syst Biol 2:2006.0008. 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tompa P. 2002. Intrinsically unstructured proteins. Trends Biochem Sci 27:527–533. 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S3 and Table S1. Download jb.00182-22-s0001.pdf, PDF file, 0.3 MB (287.5KB, pdf)
Data Set S1. Download jb.00182-22-s0002.xlsx, XLSX file, 0.02 MB (16.7KB, xlsx)
Data Availability Statement
Mass spectral data and the protein sequence database used for searches have been deposited in the public proteomics repository MassIVE (https://massive.ucsd.edu, MSV000088867).