ABSTRACT
The human intestine hosts diverse microbial communities that play a significant role in maintaining gut-skin homeostasis. When the relationship between gut microbiome and the immune system is impaired, subsequent effects can be triggered on the skin, potentially promoting the development of skin diseases. The mechanisms through which the gut microbiome affects skin health are still unclear. Enhancing our understanding on the connection between skin and gut microbiome is needed to find novel ways to treat human skin disorders. In this review, we systematically evaluate current data regarding microbial ecology of healthy skin and gut, diet, pre- and probiotics, and antibiotics, on gut microbiome and their effects on skin health. We discuss potential mechanisms of the gut-skin axis and the link between the gut and skin-associated diseases, such as psoriasis, atopic dermatitis, acne vulgaris, rosacea, alopecia areata, and hidradenitis suppurativa. This review will increase our understanding of the impacts of gut microbiome on skin conditions to aid in finding new medications for skin-associated diseases.
KEYWORDS: Gut microbiome, gastrointestinal health, gut dysbiosis, skin disease, probiotics, prebiotics, skin-gut axis, dietary components
Introduction
Skin and gut both are active, complex immunological and neuroendocrine organs that are exposed to the outside environment on a frequent basis and host a wide range of microbiomes.1,2 The skin and gut must operate appropriately in order to enable organisms to maintain homeostasis and survive.3 Notably, the skin is the body’s largest organ and serves as a defensive obstruction against injuries and microbial assault.4 The gut, on the other hand, consists of trillions of microbial communities, being recognized as a virtual organ closely associated with health and longevity of the host. The gut microbiome has both beneficial and adverse impacts on normal physiology and homeostasis of both gut and skin tissues.3,5
The three most essential roles that the gut microbiome plays from birth are protection, providing metabolic activities and immune system development and regulation.6,7 At the beginning of life, gut microbial communities have a role in defending the host against pathogenic organisms. Throughout the life, they provide metabolic services, such as digestion of breast milk and other food. Members of microbiome help in degradation of toxins and drugs and in the biosynthesis of vitamins.6 The gut is considered to be in a state of symbiosis, because it is populated by a diverse group of microorganisms, and the host tolerates these commensal bacteria and associated benign antigens.8–10 The ability of the immune system to build a tolerance to benign antigens is built through ontogeny and the reduction of microbiome-dependent inflammatory responses.10,11 For example, in mice, long-term colonization by benign Hymenolepis diminuta results in modification of the immune system without causing dysbiosis and give protection against immune-mediated inflammatory diseases (IMIDs).12
However, any alteration among gut microbial diversity (dysbiosis) can increase host vulnerability and disrupt mucosal immunological tolerance,7 which can subsequently influence skin health.13 Several dermatologic conditions, such as acne, atopic dermatitis, psoriasis, and rosacea are linked with intestinal dysbiosis.223 Many studies have associated gastrointestinal health with skin homeostasis and allostasis, and there is evidence of a bidirectional interaction between the gut and the skin.2 Members of the gut microbiome can influence skin conditions through their metabolic activity and immunological impact.3 For example, commensal gut microbes can foster skin allostasis by controlling T-cell differentiation.2
Based on existing studies, we hypothesize that the gut microbiomes are vital to the gut-skin axis.3 Thus, this review is intended to evaluate the interlinks between gut and skin microbiome and following skin conditions. Our primary objective is to review and analyze microbial ecology of skin and gut tissues and to study the characteristics and mechanisms of microbe-transmitted interactions along the gut-skin axis. Diet and probiotics have an enormous impact on the composition and metabolic activities of gut microbiome, which subsequently impacts the skin.14 Earlier, Polkowska-Pruszyska et al.,15 have discussed how changes in gut microbial communities could trigger an immunological response, resulting in allergies, acne vulgaris, atopic dermatitis (AD), alopecia areata (AA), rosacea, hidradenitis suppurativa (HS) and other skin diseases. De Pessemier et al.,16 recently demonstrated an association between dietary products and skin dysbiosis in their research. The present review paper, on the contrary, will give an integrated and structured view of the therapeutic effects of drugs, prebiotics, and probiotics on the gut microbiome and skin, as well as dietary effects on skin conditions. The secondary objective of this paper is to discuss the connection between microbial communities of the gut and skin, and their significance in dermatologic conditions, such as psoriasis, atopic dermatitis (AD), acne vulgaris, rosacea, alopecia areata (AA), and hidradenitis suppurativa (HS).
Methodology
Criteria for literature search, study selection, and exclusion
The present review paper followed the PRISMA guideline. Figure 1. depicts a flow diagram. Articles published between 2000 and 2021 were searched to find related information on the association between the gut microbiome and skin health. Articles were searched on Google Scholar, ResearchGate and PubMed using keywords like gut microbiome, skin microbiome, intestinal dysbiosis, skin–gut interactions, gut–skin microbiome, skin homeostasis, skin–gut allostasis, commensal bacteria, alteration in the gut microbiome, skin and gut microbial ecology, the role of the gut microbiome in the pathogenesis of skin diseases, gut microbes in skin disease prevention and treatment, gut–skin axis, diet influencing skin inflammation through gut microbes, drugs affecting gut-skin axis, biologics influencing skin-gut axis. The selection of such keywords ensured that all papers relating to gut microbiota and skin diseases were included. The current analysis only included peer-reviewed, high-quality research data, omitting preprints, non-English papers, and duplicate databases.
Figure 1.
Prisma flowchart. This diagram represents the Prisma flowchart and demonstrates database searching, screening, excluding, retrieval, and eligibility findings for the final full-text studies used in this review. This illustration is based on17 and created with BioRender.com (2022).
The skin and gut microbial ecology
As the composition of human microbiome varies significantly between individuals and contributes both positively and negatively to host immunity, a broad understanding on the microbial ecology of humans is necessary. Humans acquire a maternal microbiome mainly from the birth canal, and the microbial community changes over time.18,19 Skin, oral tissues, airways, gut and vagina generally harbor hundreds of microbial genera and species unlike other tissues and organs.18,20
The skin is the largest organ of the human body and provides multiple niches for colonization by diverse microbial communities, such as the stratum corneum, hair follicles, and sebaceous glands.21 Skin acts as the boundary between the external and internal environment of the human body, and the skin microbiome is an interface that contributes to human immunity.22 Culture-independent methods have revealed that healthy human skin harbors >1,000 bacterial species, mainly within the genera Brevibacterium, Propionibacterium, Micrococcus, Staphylococcus, Streptococcus, and Corynebacterium, and Malassezia is the main fungal genus.21,23 The normal microbial communities of the skin can interact with the host in both commensal and parasitic ways.24
A number of studies have highlighted the predominant factors contributing to the ecosystem of the human skin microbiome. Several skin regions with versatile characteristics, such as dry sites (volar forearm, buttock, hypothenar palm), moist sites (axillary vault, antecubital fossa, inguinal crease, umbilicus), and oily sites (glabella, alar crease, occiput, manubrium) differ from each other based on microbial composition (Figure 2).29
Figure 2.
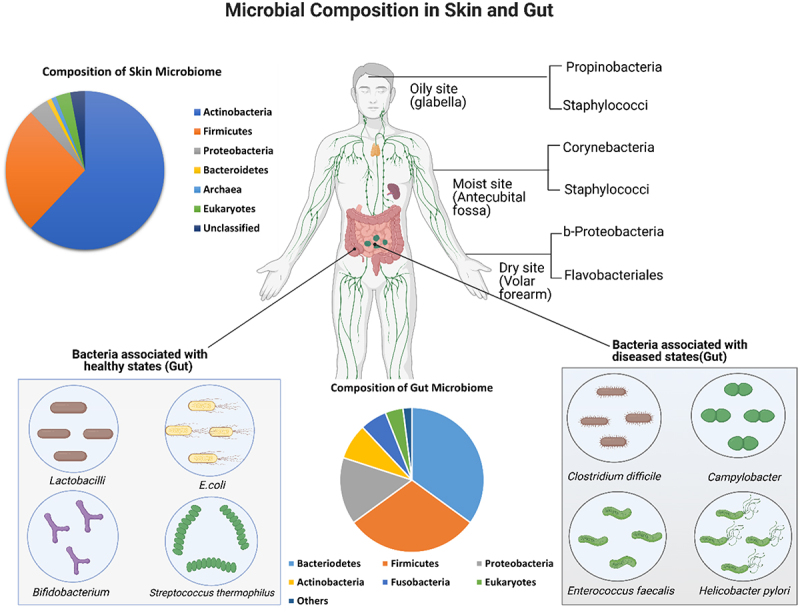
Microbial composition of gut and skin. Skin, the largest organ of the human body, shelters numerous commensal microbes (bacteria, fungi and viruses) and prevents entry by foreign pathogens by acting as a physical barrier. Skin can be broadly categorized as sebaceous or oily (glabella), moist (antecubital fossa) or dry (volar forearm), according to the physiological characteristics of each skin site. Specific microbial groups dominate different skin sites. Like skin, human gut is a home to innumerable amounts of microbes. A number of gut bacteria (e.g, Lactobacilli, E. coli, Bifidobacterium, Streptococcus thermophilus) contribute to maintenance of human health state, whereas others (e.g., Clostridium difficile, Campylobacter, Enterococcus faecalis, Helicobacter pylori) are more prevalent in disease states. The illustration was adapted from “Immune Organs in the Human Body”, by BioRender.com (2022). Retrieved from https://app.biorender.com/biorender-templates, associated information based on .25–27,28
Ecology of gut microbes has been extensively studied for decades, as the human gastrointestinal (GI) tract is one of the most complex systems in the human body. The gastrointestinal system starts from the oral cavity and finishes in the anus after passing through the stomach and intestines.30 The GI tract is considered more microbially enriched than any other organ in the human body and has the highest diversity of microorganisms. Up to 1013 microbial cells, belonging to all three domains of life (Bacteria, Archaea and Eukarya), and viruses, comprise the microbial community of the GI tract. Microbial diversity in the gut varies between individuals.18,31 Based on chemical composition and physical state, the GI tract can be divided into distinct sections. The upper portion of the GI tract, stomach, and small intestine have comparatively low numbers of bacteria (103 to 104 cells in total) due to low pH and shorter transition period. The colon is the most colonized region of the GI tract, where an estimate of 1010 to 1011 bacterial cells resides.30 The gut can harbor 1000 different bacterial species belonging to phyla such as Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria, Verrucomicrobia, Fusobacteria, Tenericutes, Spirochaetes , Cyanobacteria, and Saccharibacteria (Figure 2).30–32
Fungal diversity, in contrast, is relatively limited in the human gut. Pyrosequencing has revealed nearly 66 genera of fungi from 98 human individuals; the prevalent fungal genera being Saccharomyces, Candida, and Cladosporium.33 Abundance of fungi such as Candida, Aspergillus, Fusarium, and Cryptococcus in the gut may have a pathogenic effect on the host.30 For example, children with type I diabetes have high numbers of Saccharomyces and Candida yeasts in their gut.34 Among Archaea, the predominant genera in the human gut are Methanobrevibacter, Methanosphaera, Nitrososphaera, Thermogynomonas, and Thermoplasma.33,35 Viruses and phages, on the other hand, can act as reservoirs of genetic material in the gut and destroy microbial cells.36
Characteristics of a healthy gut
Microbes colonize the human gut at birth.37–40 The initial colonization plays an essential role in determining the composition of the gut microbiome as the person grows older.41 Progression of the intestinal microbiome is influenced by contact between host and microbes. For instance, analogous microbiomes are present in the intestine of infants and the vagina of their mothers.42 During the early life (above the age of 3), the composition of gut microbiome changes with time until it becomes relatively stable.5 A normal gut harbors bacterial genus such as Bacillus, Lactobacillus, Enterococcus, Clostridium, and Ruminococcus.43 One of the most notable changes in the gut microbiome is the ratio between the phyla Firmicutes and Bacteroidetes, as higher Firmicutes level is reported in obesity.44 The gut microbiome has a number of functions, including gathering of indigestible food particles in feces and assimilating nutritious particles, such as vitamins and minerals,45 while collaborating with the liver to detoxify and eliminate xenobiotics, which are toxic foreign compounds that are omnipresent in the environment.46 Millions of microbial genes have been identified in the gut that support essential and necessary human functions.47,48 The gut is the primary site of intricate interactions between the genes and the extrinsic immunological influence on the body, making it one of the most important organs for communicating with the environment.49 Several microbial ecosystems consisting of the entire mucosal lining are maintained in the human gastrointestinal tract.50 When villi, microvilli, crypts, and folds are considered, the gut epithelium of an adult spans a surface area of around 300 m2; this barrier typically contains just one cell layer and is therefore vulnerable. Bacteria that enter through the gut wall and into the bloodstream can cause a systemic inflammation in the absence of a healthy intestinal wall.51 The gut wall, on the other hand, is protected by a variety of chemical and physical innate defense mechanisms that function in tandem with a local adaptive immune system.52
The role of gut microbiome in epithelial cell renewal and intestinal integrity regulation is crucial to a person’s overall and intestinal immune systems.51 The local adaptive immune system is dominated by an immunoglobulin A (IgA)-producing B-cell population, which primarily offers an anti-inflammatory first-line response by producing secretory IgA (sIgA) antibodies that conduct immune exclusion.52,53 Immune tolerance toward dietary and environmental antigens is modified by gut microorganisms, which also protect against potential pathogens.2 Such protection is achieved, besides directly triggering the immune protective responses, indirectly by attachment of microbes to epithelial cells.48 Their attachment to gut epithelium can provide colonization resistance in the gut. By such process, resident microorganisms can block colonization of the gut by exogenous pathogenic microbes, such as Clostridium difficile and Helicobacter pylori.54
Gut-skin communication through immuno-cross-linking
Gut-skin communication occurs through the activities of immunological components which are present between the gut and the skin. Regulating host’s interaction with the microbiota is a fundamental function of immune system, thereby the regions colonized by commensals, such as the skin and the GI tract, encase the substantial volume of immune cells in the body. Through the dominant activity on the immune system, the commensal microbial communities play an important role in boosting barrier immunity along with their own confinement to safeguard their ecological niche. Curtailing the contact between microorganisms and the gut epithelial membrane to minimize the inflammatory responses and microbial translocation is crucial in preserving the host’s homeostatic balance. To achieve this segregation, gut epithelial cell barrier, mucus layer, T cells, IgA, and dendritic cells (DCs) collectively forge the shield named ‘mucosal firewall’ (Figure 3). The mucosal firewall limits the translocation of commensal bacteria to the lymphoid tissues, preventing the inflammation of the gut and skin.59 These lymphoid tissues are usually known as gut-associated lymphoid tissues (GALTs) (Figure 3). GALTs are typically mucosa-associated lymphoid tissues (MALTs) that work as a barrier between the host and the environment.55 GALTs are comprised of microfold cells (M cells), which have evolved into phagocytose and transcytose (a process through which particles are transported across the mucosal barrier to the lamina propria via unique cellular pathways) gut lumen macromolecules, particulate antigens, and harmful or commensal bacteria through epithelium (Figure 3).60,61 Along with M cells, conventional lymphocytes (regulatory T cells (Tregs), helper T cells (Th cells), cytotoxic T lymphocytes, IgA producing B cells), and professional phagocytes (DCs, mast cells, neutrophils, and macrophages) and unconventional lymphocytes, such as innate lymphoid cells (ILCs) and mucosal-associated invariant T (MAIT) cells, are also the constituents of GALTs (Figure 3).55,62 In addition, reports indicate that gut microbiota is behind the fundamental development mechanism of GALTs (Figure 3).63,64 Peyer’s patches, crypt cells of the intestinal epithelium, isolated lymphoid follicles (ILFs) of the intestine, appendix, and mesenteric lymph nodes (mLNs) are the most common histologic components of GALTs.63,64 The hematopoietic cell type lymphoid tissue inducer (LTi) cell and its interplay with gut microbial colonization control the formation of gut secondary lymphoid organs.55
Figure 3.
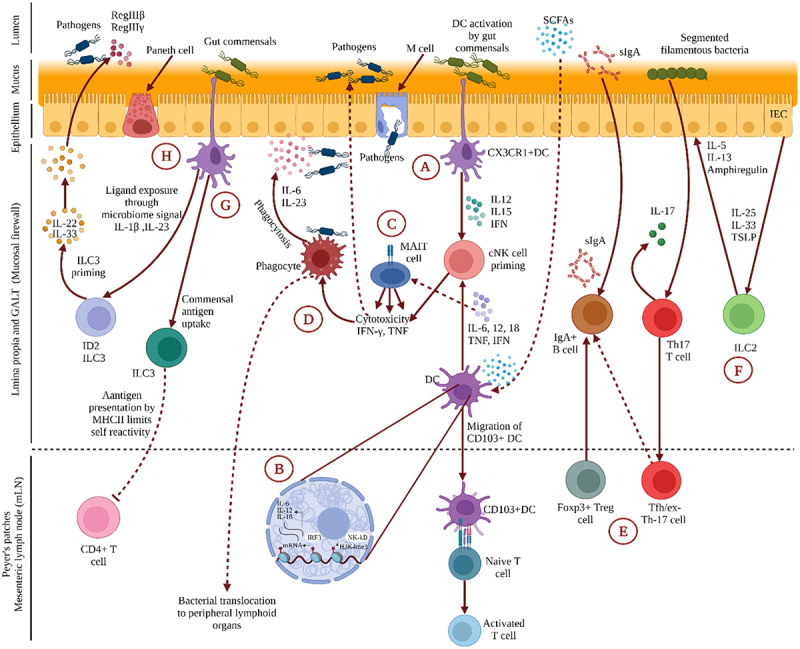
Gut-Skin communication through immuno-cross-linking. This illustration represents the immunological crosstalk between the gut and skin. (A) CX3CR1+ DCs generate dendrites for phagocytosis at homeostatic condition, whereas CD103+ DCs relocate to Peyer’s patches or mesenteric lymph nodes to deliver antigens to naive T lymphocytes. DC secretes interleukin (IL)-12, IL-15, and interferon (IFN) in response to commensal activation to stimulate conventional NK (cNK) cells. (B) As metabolic by-products, short-chain fatty acids (SCFAs) upregulate H3K4me3 in DC and enhance the production of IL-6, IL-12, IFN, and tumor necrosis factor (TNF), which is an alternative way to train cNK cells. Trained cNK cells have the necessary cytotoxicity and cytokine production capacity to fight bacteria and viruses. (C) MAIT cells can be directly stimulated to create IFN-γ by IL-12 or IL-15 in combination with IL-18 produced by APCs in response to TLR ligands. (D) TNF-like protein, a gut-associated pro-inflammatory cytokine, activates MAIT cells when coupled with IL-12 and IL-18. Phagocytes help the body defend itself by phagocytosing and producing cytokines like IL-6 and IL-23. (E) Foxp3+ Treg cells and Tfh/ex-Th17 cells cluster in Peyer’s patches, promoting B cell class switching and secretory (s)IgA production. These help to compartmentalize the commensal microbiome and modulate the diversity of the homeostatic microbiome. (F) ILC2 is activated by IL-25, IL-33, and thymic stromal lymphopoietin (TSLP) produced by intestinal epithelial cells (IEC) in response to commensal bacteria. (G) ILC3 expressing MHC II is capable of delivering commensal antigens to CD4 + T cells, reducing their self-reactivity. (H) In an ID2-dependent manner, microbial signals are also used to prime ILC3. ILC3, which has been primed secretes IL-22 and participates in the pathogen defense by stimulating the synthesis of antimicrobial peptides, such as REGIIIβ and REGIIIγ. This illustration was based on 55–58 and created in BioRender.com (2022).
Both conventional lymphocytes and professional phagocytes secrete antimicrobial peptides (AMPs) for the maintenance of homeostasis. AMPs are convoluted evolving compounds produced by gut epithelial cells, paneth cells, and immunological cells in the digestive tract (Figure 3).65 AMPs, such as α- and β- defensins, are secreted by localized immune cell types, namely macrophages, T cells, B cells and mast cells (MC).66 Furthermore, MCs can produce the AMP cathelicidin and contribute to microbiome-tissue homeostasis in the dermis. Pathogens or their components can directly bind to the TLRs, (NOD)-like receptors (NLRs), and (RIG-I)-like receptors (RLRs) and activate complement receptors of MCs, subsequently releasing inflammatory mediators, which aid in antimicrobial immune responses.67 By expression of co-stimulatory molecules and secretion of inflammatory cytokines, TLR4 elicits innate responses. An earlier study in a mouse model substantiated that TLRs generally function as pattern-recognition receptors (PRRs) with the ability to recognize a wide range of pathogen-associated molecular patterns (PAMPs), including proteins, lipoproteins, lipids, nucleic acids, and glycans, and aid the initiation of innate immunity.68 Development of ILFs occurs as soon as PRRs recognize PAMPs of enteric bacteria and activate the downstream signaling pathways. Several AMPs, such as REGIIIβ and REGIIIγ can be produced as the result of Peyer’s patch priming by TLR pathways. On the other hand, enteric bacterial infection can occur due to the inhibition of TLR pathway. For example, TLR2 deficiency impairs the integrity of the intestinal epithelial barrier and disrupts the balance between commensal bacteria and host defense, exacerbating colitis.55,69
As antigen presenting cells, DCs play an important role in producing tight junction proteins and extending dendrites into the lumen through the junctions.70 Through the adherence to C-X3-C Motif Chemokine Receptor 1 (CX3CR1), DCs give rise to the formation of trans-epithelial dendrites and the delivery of antigens for sampling (Figure 3). DCs are also capable of forming trans-epithelial dendrites and phagocytosing invasive enteric pathogens.71
ILCs play imperative parts in immunity, homeostasis, and inflammation in various tissues, including the intestine, lungs, skin, liver, adipose tissue, and mesenteric lymph nodes.72,73 ILCs are classified in three groups by the developmental process, transcription factor expression, and secretion profiles of interleukins (IL)-17A, IL-17 F, IL-22, and GM-CSF, namely, ILC1, ILC2, and ILC3, which are the innate counterparts of T helper cells, Th1, Th2, and Th17, respectively.73–75 Among the helper ILC subsets, ILC1 has a comparatively low frequency in the fetal intestine, as the gut microbiome is not established. This indicates that ILC1 development depends on commensal bacteria.55,71 Th1 cells and ILC1s eradicate viruses, bacteria, or protozoa from the body through production of IFN-γ.75 Th2 and ILC2 cells, which produce IL-5 and IL-13, respectively, are behind development of allergies and assist in the elimination of helminths. ILC3s and Th17 cells also contribute to autoimmunity by the secretion of IL-17 and IL-22, respectively, which provide protection against fungal and extracellular bacterial infections (Figure 3).75
Meanwhile, the most common subgroup of T-cells that can identify bacterial particles is mucosal-associated invariant T (MAIT) cells, also known as evolutionarily conserved T-cells.76 MAIT cells play a crucial role in elimination of bacterial infections from the body, and their role in defense against viral infection has also been reported.67 MAIT cells detect antigens via Major histocompatibility complex class I-related gene protein (MR1), a protein predominantly expressed by B cells.77 When MAIT cells come into contact with a variety of bacteria, they respond by detecting vitamin B2 biosynthesis pathway products via T-cell receptor (TCR) recognition.67
Mechanisms of the interaction at the gut-skin axis
Disruption of gut integrity, and an imbalance within microbial communities can have a significant impact on the overall homeostasis of skin.2 Gut-skin axis is a term used for the intricate interaction between the gut and the skin.16 The gut microbiome interacts with the skin largely to manage systemic and local inflammation through engaging with the immune system (Figure 4).15,78 The microbial communities maintain the gut barrier integrity mainly by converting undigestible complex polysaccharides into vitamins (specifically K and B12), and SCFAs (specifically butyrate and propionate).79,80 For example, butyrate wanes the intestinal barrier permeability and enhances epithelial barrier integrity.16 The mucus layer of the gut acts as the primary barrier and prevents microbial relocation to other host tissues (Figure 4).81,82 The gut mucosal defense is provided by innate immune cells of GALT. They recognize nonspecific infections and activate both the innate and adaptive immune systems by presenting those antigens.63,83 AMPs, macrophages, and CD103+ CD11b+ DCs mainly limit the translocation of pathogenic microbes by eliminating them.81,84,85 Defensins act against bacteria by generating pores in their membranes. This results in cell death if appropriate thresholds are exceeded. Cathelicidins (LL-37 in humans) help to keep the epithelial barrier intact. While their principal mechanism of action is to break bacterial membranes, they also possess immunomodulatory effects. Enhanced tight junction protein production, as well as post-translational effects, such as tight junction repositioning, are principally responsible for the gut epithelial barrier integrity maintenance. As a result, cathelicidins predominantly are employed when the epithelial barrier is breached.65 Differentiation of gut commensal bacteria-specific Tregs, IgA-producing B cells, and Th17 cells are the outcomes of commensal antigens presentation by DCs.84 DCs establish the specificity of CD4+ Th17 cells toward commensal microbes as the result of Major histocompatibility complex II (MHCII) antigen presentation. The CD4+ Th17 cells produce the cytokine Interleukin 22 (IL-22), which enhances secretion of host AMPs.86,87 The integrity of the intestinal barrier along with the action of mucus, immune cells, IgA, and antimicrobial peptides (AMPs) produced by epithelial cells prevents the entrance of gut bacteria into the bloodstream, ultimately maintaining skin homeostasis.59 Especially, secretory IgA controls the inflammatory responses against the gut microbes by spatially dissociating the host tissue and gut microbes.11,88,89 The commensal bacteria-specific lymphocytes accumulate in Peyer’s patches and lamina propria of the gut, which shapes the gut microbial profile toward homeostatic balance. Promotion of class switching by Tregs and IgA production against the commensal bacteria takes place in the Peyer’s patches of the gut.90,91 However, the link between skin health and immunological responses caused by the gut microbiome is still largely unclear and requires further research.92,93
Figure 4.
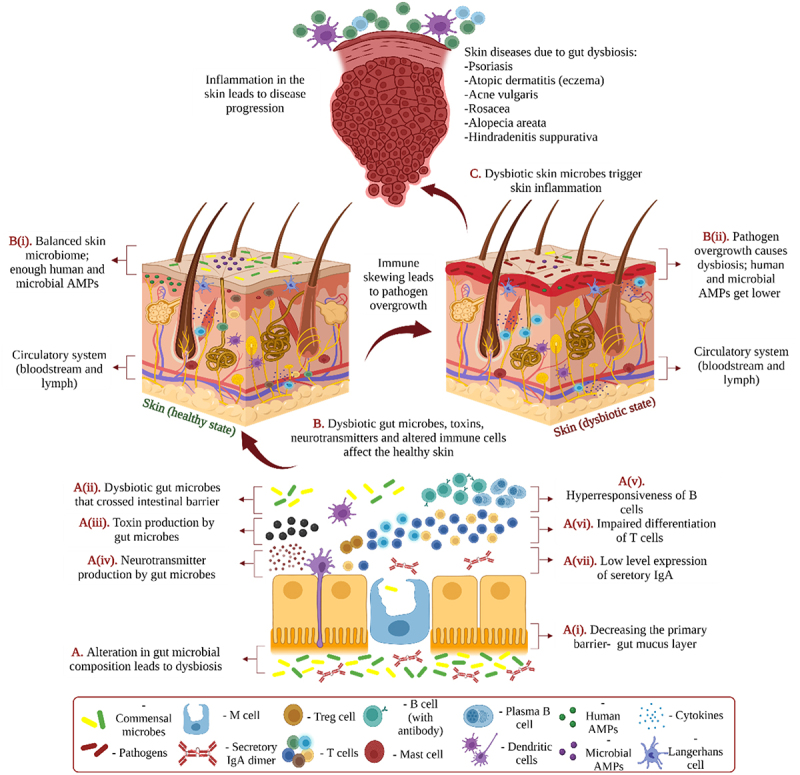
Mechanisms of the interaction at the gut-skin axis. This illustration represents the underlying mechanisms of gut-skin interaction. Various dietary components, illnesses, lifestyles, prebiotics, antibiotics, probiotics, and novel biological drugs can alter gut microbial communities. A. The alteration can lead to dysbiosis, which can further (i) decrease the gut mucus layer, (ii) results in the passage of microbes through the intestinal barrier, (iii) cause the production of toxic products, (iv) induce harmful effects by neurotransmitters of the gut microbes or the host, (v) produce B cell hyperresponsiveness, (vi) impair T cell differentiation, (vii) create low levels of IgA secretion. B. Dysbiotic gut microbes, toxic products, neurotransmitters, and altered immune cells pass through the circulatory system turning the skin condition from healthy (left) to dysbiotic (right). (i) Healthy skin possesses a balanced composition of microbes and proper quantities of human and microbial AMPs. (ii) A dysbiotic skin condition is induced by the pathogen due to improper immune system functioning and low quantities of human and microbial AMPs. C. Dysbiotic skin microbes trigger skin inflammation and can be involved in the onset of a variety of skin illnesses. This illustration is based on 15and 16and created with BioRender.com (2022).
In the first part of the twentieth century, dermatologists Stokes and Pillsbury were the first to suggest that the gut and skin communicate with the brain.3,94 GABA, acetylcholine, dopamine, and serotonin are among the neurotransmitters produced by gut microbes. These neurotransmitters can modulate the function of the skin through the nervous system. They can also create systemic effects by entering the bloodstream through the intestinal epithelium (Figure 4).95 For example, an experiment conducted on NC/Nga mice (an inbred mouse line employed as a human AD model) demonstrated the role of GABA in AD-like skin lesion mitigation. This experiment led to the conclusion that by increasing the production of serum immunoglobulin E (IgE) and splenocyte IL-4, GABA balances the T helper cell type 1 (Th1) and T helper cell type 2 (Th2) levels, keeping the Th1 predominant, which effectively wanes AD-like skin lesions in human.96 By suppressing the type I collagen degrading enzyme matrix metalloproteinase-I (MMP-I), GABA also increases the expression of human type I collagen (COL1A1 and COL1A2) and maintains skin elasticity.97 Neurotransmitters can also create negative effects, e.g., dopamine can inhibit hair growth by stimulating catagen induction.98
Antibiotics, prebiotics, probiotics, lifestyles, long-term diets, and illnesses can influence the gut microbiome. Furthermore, changes in the major strains of the gut microbiome can occur as people age.99–102 Skin inflammation may also result from minute changes in a single bacterial species of the intestinal microbiome.92,93 These, in turn, may lead to diseases e.g., acne, alopecia areata, atopic dermatitis, psoriasis, rosacea, and hidradenitis suppurativa (Figure 4).16,103,104 Table 1 lists the microbial species and metabolites associated with various skin effects.
Table 1.
Microbial species and metabolites from the gut that have been associated with skin effects.
|
A. Microbial species | |||||
|---|---|---|---|---|---|
| Organism | Effects on skin | Mechanism | References | ||
| Faecalibacterium prausnitzii, Akkermansia muciniphila and Ruminoccocus | Protection against psoriasis | Prevention of colonization of pathogenic flora on skin by competitive inhibition and the SCFAs production | 158,159,169,213 | ||
| Helicobacter pylori | Rosacea-related signs and symptoms | Production of cytotoxin and by proliferating the production of reactive oxygen species-nitric oxide [NO], which causes gut mucosal inflammation and changes physiological processes in the skin including vasodilation, inflammation and immunomodulation. | 194 | ||
| Faecalibacterium prausnitzii | Chronic atopic dermatitis progression resulting in gut epithelial barrier impairment | Dysregulation of gut epithelial inflammation | 180 | ||
| Lactobacillus casei | Decrease skin inflammation | Alteration of the number of cytotoxic CD8 + T cells | 171 | ||
| Lactobacillus paracasei | Reduce the size of acne lesions as well as inflammation | Inhibition of mast cell degranulation, TNF-α release, edema and vasodilation, and thereby speeding up the restoration of barrier function |
145,171 136 |
||
| Bifidobacterium animalis subsp. lactis [LKM512] | Reduce the scratching behavior in atopic dermatitis | Increase of levels of the kynurenic acid metabolite | 223 | ||
| Bacteroides thetaiotaomicron | Alleviate the allergic symptoms of atopic dermatitis as well as Crohn’s disease like other chronic inflammatory diseases | Anti-inflammatory action | 228 | ||
| Larger number of Clostridium difficile and Escherichia coli |
Onset of atopic dermatitis symptoms in childhood | Immune dysregulation as a result of decreased Treg cell inducing beneficial bacteria | 16,224 | ||
| Decrease in Firmicutes and increase in Bacteroides |
Development of acne vulgaris |
Dysbiosis by altering the serological cytokine levels promoting inflammation |
15,188 |
||
|
B. Metabolites | |||||
| Metabolites |
Effects on skin |
Mechanism |
References |
||
| SCFAs | Increase the epithelial barrier function and skin-inflammation | Development of Tregs within the colon, DCs precursors, and IL-10 production |
120,229 13,106 |
||
| GABA | Itch restriction | Inhibition of neurons which are responsible for itch-signaling in the spinal cord | 230,223 | ||
| Tryptophan | Regulate skin inflammation | Activation of AhR and inhibition of TSLP production in keratinocytes | 231 | ||
| Dopamine | Inhibition of hair growth | Through the stimulation of catagen induction | 98 | ||
| Serotonin | Involved in skin pigmentation | Modulation of melatonin | 16,223 | ||
| Acetylcholine | Barrier function | Not reported | 16 | ||
| Phenol & p-cresol | Impaired epidermal barrier function | Skin hydration reduction and disruption of keratinization | 171,232 | ||
| Propionic acid | Promote skin homeostasis by reducing inflammation | Antimicrobial effects | 2 | ||
| Sodium butyrate | Treat psoriasis and other hyperproliferative skin diseases | Modulation of several key cellular processes including differentiation, proliferation, and apoptosis. | 137 | ||
| Galactoligosaccharides and fructooligosaccharides |
Reduction of infant eczema and allergy | Through the stimulation of Tregs | 13,107 | ||
| Polysaccharide A and retinoic acid | Suppress inflammation | Induction of accumulation of Tregs | 153 | ||
| Saturated fats and higher amount of glycemic load |
Development of acne | Impairment in nutrient signaling. SREBP-1 overexpression and increased sebum synthesis of fatty acids (e.g., free oleic acid) and triglycerides which promotes flourishing. P. acnes growth | 107 | ||
| High-peptides and unsaturated omega-3 fatty acids | Act against hypersensitivity (allergies) and asthma | Through the development of Tregs | 13 | ||
| High-fat and alcohol | Promote skin inflammation and oxidative stress. Impairment of colonic epithelial integrity and barrier function. |
Increase of pro-inflammatory cytokines secretion | 122,123 | ||
Diet, drugs and other consumed substances affect skin through gut microbiome
Several studies have related the diversity and pathogenicity of the gut microbiome to skin disorders, which can be significantly altered by long-term dietary patterns.43,105–107 Diet can affect the skin condition both positively and negatively through alteration of the gut microbiome, indicating that there is a relationship between the skin and the gut.16 Not only diet, but also many synthetic and natural products consumed by humans as drugs can provide direct and indirect evidence on the connection between gut microbiome and skin. Direct effects of antibiotics are perhaps the finest example of the link between gut and skin microbiota.108 Although the prime target of antibiotics use is the elimination of infectious pathogens, they can also be useful in treating noninfectious cutaneous diseases because of their anti-inflammatory and immunomodulatory properties.109,110 In contrast to antibiotics, prebiotics and probiotics invigorate the gut mirobiome.111 Prebiotics are typically known as nondigestible carbohydrates, which promote the flourishment of probiotic bacteria in the gut.112,113 On the other hand, probiotics influence the gut and skin according to nutritional status and medical conditions.113 They are the key players in balancing the gut microbiome, which eventually also regulates human health.223 The effects of diet, antibiotics, prebiotics, probiotics, and novel biologic drugs on the relationship of the gut microbiome with skin health are discussed below.
Breastmilk and formula
The gut microbiome of a newborn is highly dependent on diet. Breastfed infants have much higher levels of bacteria belonging to the class Actinobacteria.114,115 Breastfeeding influences gut microbiome diversity by increasing especially colonization of genera belonging to Lactobacillus and Bifidobacterium.114–118 Colonization of the gut by the bacterial class γ-Proteobacteria, which contains many proinflammatory species118 is often observed in formula-fed infants.119 Furthermore, babies fed with formula are more likely to be colonized by members of the phylum Bacteroides, and the opportunists Escherichia coli and Clostridium difficile.13,120 High quantities of oligosaccharides and various fatty acids present in breast milk positively influence the gut microbiome and its metabolites that can act against hypersensitivity (allergy) and asthma through stimulation of Tregs.13
High and low-fat diet
In the gut, a diet high in industrial trans-fatty acids increases the number of harmful microbes (such as Desulfovibrionaceae and Proteobacteria) while suppressing populations of advantageous microorganisms (e.g. members of Bacteroidetes, Lachnospiraceae, and Bacteroidales).121 Refined and hydrogenated oils (e.g., soybean, sunflower, safflower, canola, corn, and vegetable oils) can cause inflammation in the gut, which then manifests on the skin. Skin wound healing may also be delayed by a high-fat diet and alcohol, which exacerbate the inflammation of the skin and oxidative stress.122 High-fat diets are responsible for reducing gut microbial diversity and inducing the production of higher concentrations of lipopolysaccharides. This leads to a loss of colonic epithelial integrity and barrier function, a reduction in mucus layer thickness, and an increase in the release of pro-inflammatory cytokines, all of which further lead to systemic inflammation.123
Protein-rich diet
The gut microbiome can synthesize and subsequently export excessive dietary proteins and amino acids, resulting in the production of indoxyl sulfate (IS), trimethylamine N-oxide (TMAO), and p-cresyl sulfate-like toxins.5 These toxins are involved in several skin diseases, e.g., peripheral arthritis and psoriasis.106,124 In contrast, a high-collagen peptide diet, which contains high quantities of microbes, can protect the skin from aging and promote wound healing.125 The abundance of gut-commensal genera, such as Lactobacillus and Bifidobacterium, is increased with the ingestion of a diet containing whey and pea protein extracts, and the presence of pathogenic Bacteroides fragilis and Clostridium perfringens is simultaneously decreased. SCFA levels in the intestinal mucosa are increased with the consumption of pea proteins, which are critical for keeping the mucosal barrier intact.106
Dietary fiber
In general, a fiber-rich diet has a remarkably beneficial effect on the gut microbiome and is therefore well studied. Dietary fiber cannot be digested by the human body and is mainly metabolized by the microbiome in the colon.105 The metabolic process induces the growth of many bacterial groups. Eating foods rich in dietary fiber, especially whole grains, dramatically increases the populations of Bifidobacteria and the Lactobacillus/Enterococcus group.105 Foods containing complex dietary carbohydrates can be converted into SCFAs, including propionate, acetate, and butyrate through fermentation by the gut microbiome.2,5,105,120 Gut commensal microorganisms influence mucosal immunity through the development of Tregs within the colon, which is mediated through SCFAs.120 By increasing epithelial barrier function and inducing various anti-inflammatory effects, the SCFAs strengthen the gut’s function and integrity,5 modulate respiratory diseases,13 prevent the development of inflammatory disorders, e.g., allergy, arthritis, and colitis,2 regulate the metabolism of lipids and glucose,5 and inhibit the buildup of potentially harmful metabolic by-products, such as D-lactate.41 SCFAs play a role in the expression of Foxp3 (Forkhead box protein P3) that helps in regulating the development and function of Tregs, thus improving the regulatory T cell function.126,127 The anti-inflammatory effects by SCFAs are aided by G-protein coupled receptor 43, TGF-β, and/or IL-10, in addition to Tregs.128 On the skin, the anti-inflammatory actions could be mediated through resident Tregs, which become less abundant in certain inflammatory dermatoses.120 SCFAs derived from fiber through the gut can also influence the prevalence of certain skin microbial groups, which subsequently affect cutaneous immune defense mechanisms. By preventing the growth of harmful bacteria on the skin and lowering inflammation, the skin microbiome can collaborate with the immune system to promote skin homeostasis.26 For example, propionic acid, which is formed during the fermentation of dietary fiber by the Propionibacterium genus, is antibacterial and can kill the most prevalent community-acquired methicillin-resistant Staphylococcus aureus strains USA300.2
Antibiotics
One of the most well-studied effects on the gut microbiome is that of antibiotics. They have the potential to change the composition and function of the gut microbiome by killing or inhibiting the growth of specific microbial groups and by changing the molecular patterns associated with the microbiome. The effect of antimicrobials on the gut microbiome leads to dysbiosis,108 and consequently, skin abnormalities may occur. For example, oral administration of vancomycin for the treatment of skin wounds reduced bacterial diversity and regenerating gene III gamma (RegIIIγ) expression, potentially delaying wound-healing repair mechanisms.226 Antibiotic use, over time, leads to spread of antibacterial resistance and a decline in non-target bacterial populations, which can result in flourishment of pathogenic bacteria or fungi.111,129 For example, the use of a set of antimicrobials, levofloxacin and moxifloxacin, is associated with significantly increased numbers of Candida species in the human gut.130 The yeast Candida can colonize the small intestine and manifest as skin redness, which in turn accelerates aging.131
Prebiotics
International Scientific Association for Probiotics and Prebiotics (ISAPP) declares that the substrates that upon utilization by microorganisms provide useful properties to the host are prebiotics.132 They play a significant role in enhancing the number of gut microbes and their function.133,134 Prebiotics, such as fructooligosaccharides, galactooligosaccharides, inulin, polydextrose, lactulose, sorbitol, and xylitol are a promising group of compounds that modulate the gut microbiome and can also provide skin benefits.135 The prebiotics, galactooligosaccharides (GOS), are one of the major components of breast milk that can be fermented into SCFAs by members in the genus Bifidobacterium. This was shown by the number of human milk oligosaccharides (HMOs) decreasing in feces along with the increased presence of Bifidobacterium genera. HMO utilization by Bifidobacterium spp. can be considered as a prebiotic impact on the infant gut microbiome. Similarly, the human milk fatty acid palmitate plays a prebiotic effect on the infant gut microbiome by positively influencing the abundance of Bifidobacterium spp. and Lactobacillus spp.43
The effect of prebiotics on the skin condition is also obvious. For example, a Lactobacillus extract helps to reduce the size of acne lesions as well as inflammation by reducing skin erythema, improving skin barrier function and lowering the microbial counts on skin.136 Furthermore, GOSs have been used in the treatment of photoaging diseases for decades.137 In a study, the combination of GOS and Bifidobacterium reduced trans-epidermal water loss (TEWL or TWL) and prevented skin erythema.137,138 Atopic dermatitis and eczema have also been treated with GOS.139 Another prebiotic (carbohydrate) derived metabolite, sodium butyrate,140 is commonly used to treat hyperproliferative skin diseases (including psoriasis) through its modulation of key cellular processes, such as differentiation, proliferation, and apoptosis.137 Sodium butyrate is a compound derived from the gut microbiome that commonly affects the cell cycle, protease enzymes, and tumor growth factors (TGF-β). Apoptosis in keratinocyte (HaCaT) cells can be induced by sodium butyrate through up-regulation of the Fas receptor (death receptor) with the concomitant triggering of caspases 8 and 3. Another effect is an induced expression of p52 and TGF-β, suggesting a connection with cell proliferation and terminal differentiation.137 Differentiation of keratinocytes can also be enhanced by the joint action of sodium butyrate and PD153035 (an epidermal growth factor receptor inhibitor).141
Probiotics
Probiotics are living organisms that, when consumed in sufficient proportions, confer health benefits. They can be formulated, for example, as food, drugs, and dietary supplements.6,113,142 Probiotics can prevent gut colonization by pathogens and support anti-inflammatory responses by producing metabolites with anti-inflammatory properties. The most common probiotic microbes currently in use belong to the genera Bacillus, Bifidobacterium, Enterococcus, Escherichia, Lactobacillus, Saccharomyces, and Streptococcus.143,144 Several beneficial effects of probiotic consumption have been demonstrated on many dermatological conditions, thus proving the existence of the gut-skin axis. For example, skin sensitivity and significant restoration of skin barrier function were reported in individuals after daily oral administration of Lactobacillus paracasei.145 In a study conducted on mice, addition of Lactobacillus reuteri to drinking water resulted in improved epidermal thickness, increased folliculogenesis, lower skin pH, and enhanced production of sebum-producing epithelial cells. Accordingly, mice provided with the supplemented water had shinier and thicker fur than those that did not receive the probiotic.2,3 In another experiment, administration of Lactobacillus johnsonii in mice exhibited a therapeutic effect in restoring skin damage after exposure to UV radiation.15 The beneficial role of probiotics in preventing dermatological diseases has also been shown. For example, administration of E. coli strain Nissle improved skin health in patients with acne vulgaris.15,146 Orally administered doxycycline and probiotics were effective in a study in patients suffering from rosacea.135 In another study, the probability of developing atopic dermatitis was decreased throughout the postnatal period in mice by Lactobacillus sakei WIKIM30 consumption. The mice that had probiotics exhibited altered gut microbial composition along with improved skin lesions due to induction of Tregs. The risk of atopic dermatitis development was also found considerably lower in children who took probiotic supplements in the post-neonatal period of life.15
Novel biologic drugs
‘Novel biologics’ refers to substances generated from products of biological entities and may become therapeutic for dermatological conditions.147 A few immune-based diseases are now being treated with novel biologic drugs by therapeutic manipulation. Some therapeutic biologicals authorized by the European Medicines Agency are Adalimumab (Humira), Infliximab (Remicade), Rituximab (MabThera), and Etanercept (Enbrel). All of these drugs have a direct involvement with skin and gastrointestinal tract.148,149 Among all intrinsic and extrinsic factors, drugs are indicated as the major one that can remarkably interrupt gut microbial ecology. Gut microbial composition may be affected by drugs in two common ways. First, drugs can influence the translocation of microbes from other body parts to the intestine, and secondly, they can affect the local bacterial growth directly.150 Biologics such as Etanercept and Infliximab, which are used to treat psoriasis, may affect some microbial species, e.g. Clostridium citroniae, Collinsella aerofaciens, Dorea formicigenerans, Parabacteroides distasonis, Prevotella copri, and Ruminococcus gnavus.149,151
Technological advancements have opened up new possibilities for the selection and manufacture of monoclonal antibodies that target specific diseases.149 For example, the only monoclonal antibody authorized for individuals suffering from chronic spontaneous urticaria is omalizumab. These novel biologics are regarded as an efficient and safe treatment, but their availability and high cost severely impair their usefulness.152
The link between gut and skin disease
Although a healthy gut is essential for host health, gut microbiome overgrowth and changes in diversity can result in a skin disease. Specific metabolic byproducts of gut microbes can directly influence normal physiology and disease processes,5 as discussed in previous paragraphs. Factors that influence the diversity and composition of the gut microbial community can be grouped into categories, namely non-host factors (such as environmental determinants), host factors (such as pancreatic enzymes, bile acids, pH), and bacterial factors (such as microbial enzymes, adhesive ability).225 Gut dysbiosis153 negatively impacts skin health and is considered to contain biomarkers such as free phenol, p-cresol, and aromatic amino acid derivatives produced by a disturbed gut.3 Dysbiosis in the gut contributes to the three common skin disorders: psoriasis, atopic dermatitis, and acne.2 There are also reports on the association of gut dysbiosis with some less common but potentially more serious diseases, such as rosacea, alopecia areata, hidradenitis suppurativa,15 erythema nodosum, and pyoderma gangrenosum.135 Furthermore, epidemiological studies and clinical trials suggest that modulation of the gut microbiome may influence susceptibility to allergic diseases and asthma.154
The skin plays a crucial part in maintaining homeostasis of the body by performing several important functions, such as regulation of water balance and temperature control. For these functions to be performed, the skin must undergo a renewal and turnover process, known as skin regeneration. Once epidermal cells differentiate from stem cells, epidermal cells undergo a differentiation process called keratinization, which is regulated by specific transcriptional processes. The gut microbiome influences the signaling processes that maintain epidermal differentiation and thus affect skin homeostasis.155 Several studies provided more information regarding a correlation between the gut microbiome and skin health. For example, the genetic material of gut bacteria has been identified in plasma samples of patients with psoriasis. In the study of 54 patients and 27 healthy controls, bacterial DNA was found in 16 out of 54 plaque psoriasis patients and was not detected in any controls. Furthermore, those patients with psoriasis were also detected with an increase in systemic inflammatory response markers (SIRs) such as IFN-γ, IL-1β, IL-6, IL-12, and TNF compared with the healthy control. Sequencing of the bacterial DNA identified the same type of organisms commonly found in the gut flora, pointing toward a possible link between the gut microbiome with psoriasis.3,5,156 Patients with Crohn’s disease are also mostly encountered with psoriasis as a comorbidity.107,155 Such findings suggest a correlation between altered gut microbiome and skin condition.2 Table 2 represents several human clinical studies that have used gut microbial interventions for human skin diseases. Next, we will take a closer look at the specific diseases where gut microbiome affects the development of the condition.
Table 2.
Human clinical studies that have used gut microbial interventions for human skin diseases.
| Participants | Intervention | Key findings | References |
|---|---|---|---|
| 43 children (aged 4–17 years) | Bifidobacterium animalis subsp. lactis CECT 8145, Lactobacillus casei CECT 9104, and Bifidobacterium longum CECT 7347 (mixture) for 12 weeks. | Reducing the severity of AD according to the SCORAD index | 233 |
| 50 children (aged 4 to 17 years) | Bifidobacterium lactis CECT8145, Bifidobacterium longum CECT 7347, and Lactobacillus casei CECT 9104 (mixture) for 12 weeks | Reducing the severity of AD according to the SCORAD index | 234 |
| 31 patients (aged 18–75 years) | Lactobacillus johnsonii NCC 533, twice daily for 3 weeks | Reducing the severity of AD according to the SCORAD index | 235 |
| 20 Caucasian adult | Lactobacillus rhamnosus for 12 weeks | Reducing the acne severity | 236 |
| 101 Japanese female students (aged 18–23 years) | B. breve strain Yakult (YIT 12272), Lactococcus lactis YIT 2027, and Streptococcus thermophilus YIT 2021 (mixture) for 4 weeks | Enhancing stool consistency, defecation frequency, and feces quantity to maintain healthy skin | 237 |
| 110 volunteers (aged 41 and 59 years) | Lactobacillus plantarum HY7714 for 12 weeks | Enhancing skin hydration, reduction of wrinkle depth, and overall skin shine and elasticity | 238 |
| 166 pregnant women | Bifidobacterium breve M-16 V and Bifidobacterium longum BB536 for 4 weeks | Reducing the risk of newborns eczema | 239 |
| 45 females (aged 18 to 35 years) |
Lactobacillus acidophilus, Lactobacillus delbrueckii, and Bifidobacterium bifidum (mixture) for 12 weeks |
Improving the total lesion for acne patients | 240 |
| 26 male and female (aged 18–60 years) | Bifidobacterium infantis 35624 for 6‒8 weeks | Improving the CRP levels in psoriasis patients | 170 |
| 30 women (aged 30–48 years) | L. lactis strain H61 (60 mg), daily for 8 weeks | Improving hair follicles and skin hydration | 241 |
| A woman (aged 47 years) | Lactobacillus sporogenes and biotin (mixture) | Reducing the risk of psoriasis | 242 |
| 20 children (aged 4–15 years) | L. acidophilus L-92 strain for 8 weeks | Effective as Serum Markers of Atopic Dermatitis in Children | 243 |
| 56 Patients (aged 18 to 30 years) |
L. bulgaricus and Streptococcus thermophilus for 12 weeks |
Reducing the severity of acne vulgaris by selectively lowering TGs in skin surface lipids. | 244 |
| 66 female volunteers (aged 35–45 years) | Bifidobacterium longum, twice daily for 2 months | Improving skin hydration and physical aggression | 245 |
| 70 patients (12 years of age or older) | Enterococcus faecalis SL-5 for 8 weeks | Improving the severity of acne | 246 |
| 474 patients | Lactobacillus rhamnosus for 35 weeks | Reducing the risk of eczema | 247 |
| 59 patients |
Lactobacillus rhamnosus and Bifidobacterium lactis for 12 weeks |
Reducing the severity of AD according to the SCORAD index | 248 |
| 56 children (aged 6–18 months) | Lactobacillus fermentum VRI-033 PCC for 8 weeks | Reducing the severity of AD according to the SCORAD index | 249 |
| 41 children (aged 1–13 years) | Lactobacillus rhamnosus 19070–2 and Lactobacillus reuteri DSM 12246 for 6 weeks | Reducing the severity of atopic eczema | 250 |
| 159 pregnant women |
Lactobacillus rhamnosus strain GG for 4 weeks | Reducing the severity of atopic eczema | 251 |
| 159 patients | Lactobacillus GG for 4 weeks | Reducing the severity of atopic eczema in children | 252 |
| 27 infants | Bifidobacterium lactis Bb-12 or Lactobacillus strain GG (ATCC 53103) | Reducing the severity of atopic eczema | 253 |
Psoriasis
Psoriasis is a systemic autoimmune condition associated with improper activation of immune-mediated pathways that drive the immune cells to attack self-skin cells, subsequently resulting in increased levels of pro-inflammatory cytokines. Inflammatory bowel disease (IBD), a second known comorbidity of psoriasis, is associated with the gut microbiome through development of pro-inflammatory Th17 cells.5,157 A reduction in the abundance of potentially beneficial microbes in psoriasis patients could alter balance of immune system, thus affecting skin health.158,159 Specifically, psoriasis patients have reduced numbers of Bacteroidetes (including the Bacteroides genus), Proteobacteria, and Actinobacteria along with relatively low amounts of Akkermansia muciniphila15,160,161 and a lower prevalence of the phylum Firmicutes.162 A reduced abundance of Akkermansia and Ruminoccocus was also observed in patients with psoriasis, psoriatic arthritis (PsA), and IBD.159,160,163
Biomarkers of a damaged intestinal barrier, such as elevated levels of claudin-3 and intestinal fatty acid-binding protein, have been observed among psoriasis patients.15 Furthermore, microbial production of TMAO upon utilization of choline can lead to psoriasis.106,124 Several studies have shown that Bacteroides produces polysaccharide A, activates Tregs and promotes the anti-inflammatory response.158,164–166 Thus, a remarkable reduction in Bacteroides count in psoriasis could lead to immune alteration and pro-inflammatory response.158 As the natural colonizers of the mucin layer in the human gut, Akkermansia muciniphila and Ruminoccocus prevent pathogen colonization by competitive inhibition.157,159,167,168 Moreover, Bacteroides, Akkermansia muciniphila and Ruminoccocus produce SCFAs.126 Therefore, the reduction of these beneficial microbes results in the impairment of the gut barrier function, promoting bacterial translocation from the gut into the systemic circulation in psoriasis patients.126,156,166
Oral administration of antibiotics, prebiotics, probiotics, and newly developed fecal transplantation are promising therapeutic methods for psoriasis.169 Probiotics have been shown to improve the disease course of psoriasis by increasing TNF-α (tumor necrosis factor alpha) production by epithelial cells, by improving the barrier function, and by regulating the NF-kβ (Nuclear factor kappa B) pathway that affects the development of psoriasis pathogenesis.169 A study by Groeger and colleagues in 2013 found that Bifidobacterium infantis 35624 supplementation significantly decreased plasma levels of TNF-α, IL-6, and C-reactive protein (CRP) (the markers of inflammation in the body) among psoriasis sufferers.170 Although studies have shown that probiotic treatments lead to a significant change in skin inflammation, there is still no evidence to support their use in the treatment of psoriasis. Therefore, further research is needed in this field.171
Atopic dermatitis (eczema)
Several investigations on the role of gut microbiome in the pathogenesis of atopic dermatitis (AD) in infants and children suggest that infants with limited heterogeneity in the gut microbiome develop AD later in life.5,172–175 This is contradicting to other results, which suggest that gut microbial diversity facilitates AD. For example, a lower abundance of Bifidobacterium in the gut was observed in patients with AD compared to healthy controls.15,176,177 However, alteration of gut microbial composition alone cannot promote the development of AD. Interaction of specific microbes with the immune system, together with other external factors, such as diet, could better explain the pathogenesis of AD. The immunopathology behind AD prognosis is linked with an imbalance of Th1 and Th2. The effector skin-resident dendritic cells of AD patients travel to the local lymph node where they activate the naïve T lymphocytes and induce their differentiation into effector Th2 subtype of effector T lymphocytes.223 Once activated and recruited back to the skin, Th2s substantially produce inflammatory cytokines, such as IL-4, IL-5, and IL-13, which ultimately causes enhanced production of IgE, commonly found in AD patient.178,223
Two metagenomic studies conducted in South Korea analyzed fecal samples of patients with AD and revealed a decreased abundance in Faecalibacterium prausnitzii along with a significant reduction in SCFA production when compared with healthy controls.2,179,180 According to a study, dysbiosis of F. prausnitzii in patients with AD was associated with an increased expression of a variety of nutrients. These nutrients, such as the mucin components GalNAc and L-fucose, are released from damaged gut epithelium, indicating a leaky gut and dysregulation of inflammation of the gut epithelium. Increased gut permeability was identified due to damaged gut epithelium, allowing various toxins, food residues, and pathogens to access blood circulation. The metabolites and toxins ultimately paved their way to the skin and induced Th2-type immune responses by releasing pro-inflammatory cytokines, which underlies the prognosis of AD.180
Zheng et al.181 demonstrated that an increased abundance of Akkermansia muciniphila in infants with AD is associated with intestinal barrier dysfunction and skin lesion deterioration.15,181 In addition to, or potentially mediated by the gut microbiome, diet may also impact AD. For example, a gluten-containing diet can impair the intestinal barrier, causing leaky gut, and gluten sensitivity is associated with gut dysbiosis.135,182,183 According to a recent study, probiotic lactobacilli and enterococci of human origin can help to improve gut microbiome dysbiosis, and hence could be used as a treatment for disorders involving low SCFA synthesis in the gut.184 Furthermore, Fecal microbiota transplant (FMT) could be a potential therapeutic option for AD.185 However, research-oriented studies in this section are required to gain a better insight for future therapeutic interventions.
Acne vulgaris
Acne vulgaris is identified by inflammatory skin lesions (papules and pustules), non‐inflammatory comedones, or a combination of the two.15,135 Several studies have indicated a correlation of gut dysbiosis with acne vulgaris. A recent investigation revealed a significant reduction in the prevalence of Actinobacteria, Bifidobacterium, Butyricicoccus, Coprobacillus, and Lactobacillus species along with an increased abundance of Proteobacteria in persons with acne vulgaris.15 According to a hypothesis, sterol regulatory element-binding protein 1 (SREBP-1), sebum fatty acid, and sebum triglyceride become stimulated by nutrient signaling disruption and lead to flourishment of Propionibacterium acnes.186 In addition to the gut microbiome, various metabolic pathways also influence the pathophysiology of acne vulgaris, such as the mTOR pathway, which becomes activated by high glycemic load. The high glycemic load is the sole contributor to increased insulin/insulin-like growth factor (IGF-1) signaling, enhancing the cytoplasmic expression of FoxO1 (Forkhead box transcription factor O1). FoxO1 then stimulates the mammalian target of rapamycin complex 1 (mTORC1), which ultimately leads to acne development.2
Besides the mTOR pathway, high-fat diet has an influence on acne development.187 Guo et al. found that there is a lower release rate of AMPs in the small intestine of mice due to high-fat diet, which promotes growth of Firmicutes than Bacteroidetes, resulting in dysbiosis and alteration of serological cytokine levels that promote inflammation.188 Several acne patients have also been found to develop Hyperchlorhydria (low level of stomach acid). Low acidity levels allow colonic bacteria to migrate to the distal region of the small intestine, creating conditions for intestinal dysbiosis and growth of small intestinal bacteria, which can increase intestinal permeability and promote inflammation of the skin.189
Noureldein & Eid190 demonstrated that the inhibition of mTORC1 together with changes in the gut microbiome improve the level of glucose tolerance and reduce hyperinsulinemia. Supporting this hypothesis, a reduced level of acne lesion was found in patients supplied with a low glycemic load diet for 12 weeks.191 Also, probiotics may reduce acne inflammation by reducing levels of inflammatory cytokines, by increasing CD8 cell recruitment, by suppressing IL−1α, and by activating Tregs.112,192
Rosacea
Rosacea is a common dermatological condition that is characterized by pustules, persistent erythema, excessive fibrous tissue proliferation, papules, telangiectasia, and periocular changes. The coexistence of several gastrointestinal comorbidities with rosacea has been observed, which connects the condition with altered gut microbiome.15,107,161 Moreover, H. pylori infection, IBD, and small intestinal bacterial overgrowth (SIBO) are potentially associated with the pathogenesis of rosacea.15,135,161 The crosstalk between the immune system and the gut-associated diseases further demonstrates the link between gut and rosacea. According to reports, the role of H. pylori in rosacea pathogenesis is associated with several immune-pathological and inflammatory mediators and toxic factors.193–195 For example, H. pylori can substantially increase ROS generation (Reactive oxygen species), which has an inflammatory effect on the gut. Among the ROS, NO (nitric oxide) causes inflammation of the gut mucosa and alters the skin physiological processes, including vasodilation, inflammation and immunomodulation, ultimately resulting in the clinical manifestations associated with rosacea.194 Another mechanism justifying the role of H. pylori in rosacea development is the production of a cytotoxin, which induces the production of pro-inflammatory cytokines such as TNF-α and IL-8. As a result of these inflammatory cytokine releases, inflammation of gastric mucosa and clinical manifestation of rosacea are seen.193,194
The eradication of H. pylori reduces rosacea symptoms as well as associated gastrointestinal issues.196 Bacillus subtilis-3 is effective against H. pylori, allowing spore colonization of the gastrointestinal system and subsequent microbiome modification by permitting passage over the gastric barrier.197 Many studies have shown that probiotics can be used to treat chronic inflammatory rosacea, which could be used as an additional therapy in patients.198,199
Alopecia areata (AA)
AA, or spot baldness, is an autoimmune disorder in which the hair-loss pattern is in either a limited or all areas of the body. AA has no age restriction.15 AA is an auto-immune disease where the auto-reactive T lymphocyte interacts with a follicular auto-antigen presented by perifollicular or follicular cell. Their interaction gives rise to activation and induction of the T Lymphocytes, especially Th1, to produce pro-inflammatory cytokines (such as IFN-γ). IFN-γ disrupts the anagen growth phase, ultimately resulting in hair loss and other manifestations of AA.200,201
When compared with a healthy population, a high proportion of persons with AA also exhibit ulcerative colitis, indicating an association of the gut with AA.15,200,202 The driving force behind this association is accumulation of auto-reactive T lymphocytes that gain tolerance against apoptotic cell death, causing prolonged chronic inflammation and hair loss due to inflammatory cytokine (IFN-γ and IL-2) production by the autoreactive Th1 cells.200
FMT is thought to be a safe and effective way to reestablish a healthy gut microbial ecosystem by delivering beneficial bacteria and nutrients directly or indirectly.203 FMT could possibly be used to treat alopecia areata by restoring the homeostasis of the gut flora.204
Hidradenitis suppurativa
HS is a chronic disorder of the skin characterized by inflammation of hair follicles in intertriginous areas.15 The mechanism underlying this chronic disorder needs to be deeply researched, as very little evidence and studies are available for inferring correlation with the disease pathogenesis and gut microbiome. Although the pathogenesis of HS is not yet clear, follicular occlusion syndrome, disturbance of the inflammatory cytokines, such as TNF-α, IL-1ß and IL-17, and fluctuation in the microbial composition are known to be crucial.205–210 The possible linkage between HS pathology and altered gut microbiome is supported by some features that are found in both HS and acne vulgaris, such as the coexistence of IBD and metabolic syndrome, and a negative association with a high-sugar diet.15,211,212 IBD was observed at high frequencies in patients with HS when compared with healthy controls. Persons with HS also had a significant reduction in Faecalibacterium prausnitzii and increased abundance of E. coli in their gut.15,213,214
Gut microbial dysbiosis can potentially induce HS by influencing skin microbes through mediating systemic inflammatory pathways.2,215 According to an interesting hypothesis, colonic dysbiosis resulting from high-fat diet results in inflammatory cytokine (e.g., TNF-α, IL-1β, and IL-17) elevation, and ultimately in formation of HS lesion by enhancing matrix metalloproteinase levels.215 Probiotics are considered an effective option in treating HS for restoring a healthy cutaneous microbiome, as they are capable of replenishing the abundance of Cutibacterium spp., Corynebacterium, and Staphylococcus within the communities.216 Avoiding a high-fat diet can be a great secondary preventive measure for HS.217 However, finding the potential primary preventive measure (e.g., microbial marker) for HS is still under rigorous research.
Conclusion and future prospective
Understanding the relationship between gut and skin microbial communities is an emerging area of research. A number of findings we have summarized here provides sufficient evidence for the existence of a skin–gut axis. Although limited research is available on the pathobiological drivers on the gut-skin axis in disease, our review has provided a brief understanding on the linkage between the gut microbiome with skin diseases and indicated direction of potential future studies. Whereas specific skin diseases have been connected with gut health and the balance of microbiome within the gut, the exact mechanisms of influencing skin by intestinal microorganisms are to be elucidated. Besides acting through the immune system, catabolic products of diet and microbial compounds can impact the gut epithelium by altering gut physiology, leading to a variety of secretory products that circulate throughout the body and enter the skin. Skin commensal microbiome can be affected by the bioactive compounds, such as neurotransmitters, hormones, and SCFAs, which are the end-products of gut microbial metabolism. Ingested compounds and chemicals can thus have an immediate impact on the skin appearance and activity. However, the mechanisms behind this interaction are multifactorial and currently mainly based on theory. Further studies that can detail the fate of specific compounds (e.g., by labeling), being transferred from the intestine to the skin and their mechanism of action on skin cells and/or microbiome would be needed to positively prove such connection. For example, metabolites, such as ceramide,218–227 have been radioactively labeled prior to oral consumption and tracked on the skin of mice. Similar labeling studies on microbial products, such as SCFAs, would provide strong evidence toward direct effects of gut microbiome on the skin.
Another line of research having many promising prospects on the gut-skin axis are dietary supplements promoting health of gut microbiome, including pre- and probiotics. This research area has been massively studied in the past due to many beneficial health effects of fermented foods. In our review, we have listed several connections of probiotics with the skin condition, for example, Lactobacillus reuteri, which improves epidermal thickness and increased folliculogenesis after ingestion by mice.3 Further studies linking probiotics with skin would likely provide new significant knowledge on the importance of specific members of the gut microbial community in skin health. Linking together studies on probiotics and tracking of specific microbial products from gut to the skin could reveal exciting new information on the gut-skin axis in the future.
Funding Statement
Open access to this publication is funded by Helsinki University Library.
Abbreviations
- AA
Alopecia areata
- AD
Atopic dermatitis
- AhR
Aryl hydrocarbon receptor
- AMP
Antimicrobial peptides
- COL1A1
Collagen type 1 alpha 1
- COL1A2
Collagen type 1 alpha 2
- CRP
C-reactive protein
- CX3CR1
C-X3-C Motif Chemokine Receptor 1
- DC
Dendritic cell
- FoxO1
Forkhead box transcription factor O1
- Foxp3
Forkhead box protein P3
- FMT
Fecal microbiota transplant
- GalNAc
N-Acetylgalactosamine
- GABA
Gamma-aminobutyric acid
- GALTs
Gut-associated lymphoid tissues
- GI
Gastrointestinal
- GM-CSF
Granulocyte-macrophage colony-stimulating factor
- GOS
Galacto-oligosaccharides
- HS
Hidradenitis suppurativa
- HMO
Human milk oligosaccharide
- IFN
Interferon
- IBD
Inflammatory bowel disease
- IEC
Intestinal epithelial cell
- IgA
Immunoglobulin A
- IgE
Immunoglobulin E
- IGF
Insulin-like growth factor
- IMID
Immune-mediated inflammatory diseases
- IL-10
Interleukin 10
- ILCs
Innate lymphoid cells
- ILFs
Isolated lymphoid follicles
- IL
Interleukin
- IS
Indoxyl sulfate
- ISAPP
International Scientific Association for Probiotics and Prebiotics
- MAIT
Mucosal-associated invariant T cells
- MALTs
Mucosa-associated lymphoid tissues
- LTi
Lymphoid tissue inducer
- MHC II
Major histocompatibility complex II
- mLN
Mediastinal lymph node
- MMP
Matrix metalloproteinase
- MR1
Major histocompatibility complex class I-related gene protein
- mTOR
Mammalian target of rapamycin
- mTORC1
Mammalian target of rapamycin complex 1
- NF-kβ
Nuclear factor kappa B
- NK
Natural Killer cells
- NO
Nitric oxide
- NLR
NOD like receptor
- NOD
Nucleotide oligomerization domain
- PRRs
Pattern-recognition receptors
- PAMPs
Pathogen-associated molecular patterns
- RegIII
Regenerating gene III
- RIG
Retinoic acid-inducible gene I
- RLR
RIG-like receptor
- ROS
Reactive oxygen species
- SIBO
Small intestinal bacterial overgrowth
- SCFA
Short chain fatty acid
- SREBP-1
Sterol regulatory element-binding protein 1
- SIRs
Systemic inflammatory response markers
- Tregs
T regulatory cells
- TEWL or TWL
Trans-epidermal water loss
- TLR
Toll like receptor
- TMAO
Trimethylamine N-oxide
- TGF-β
Tumor growth factor beta
- TNF-α
Tumor necrosis factor alpha
- TSLP
Thymic stromal lymphopoietin
- Th1
T helper cell type 1
- Th2
T helper cell type 2
Disclosure statement
No potential conflict of interest was reported by the authors.
Author contributions
M.R.M designed and supervised the study. M.R.M, S.A, S.K.T, I.Z.E, M.S.H, S.B, L.M, S.A and M.R.H collected necessary data and participated in the writing manuscript. A.M.P & M.A reviewed the draft thoroughly and A.M.P also revised the final manuscript for further changes in format. M.R.M also acted as corresponding author. All authors declared no objection for submitting the final manuscript.
References
- 1.Coates M, Lee MJ, Norton D, MacLeod AS, Norton D, MacLeod AS, MacLeod AS.. The skin and intestinal microbiota and their specific innate immune systems. Front Immunol. 2019;10:10. doi: 10.3389/fimmu.2019.02950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salem I, Ramser A, Isham N, Ghannoum MA. The gut microbiome as a major regulator of the gut-skin axis. Front Microbiol. 2018;9:1459. doi: 10.3389/fmicb.2018.01459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Neill CA, Monteleone G, McLaughlin JT, Paus R. The gut-skin axis in health and disease: a paradigm with therapeutic implications. BioEssays. 2016;38(11):1167–29. doi: 10.1002/bies.201600008. [DOI] [PubMed] [Google Scholar]
- 4.Hsu DK, Fung MA, Chen HL. Role of skin and gut microbiota in the pathogenesis of psoriasis, an inflammatory skin disease. Med Microecol. 2020;4:100016. doi: 10.1016/j.medmic.2020.100016. [DOI] [Google Scholar]
- 5.Ellis SR, Nguyen M, Vaughn AR, Notay M, Burney WA, Sandhu S, Sivamani RK. The skin and gut microbiome and its role in common dermatologic conditions. Microorganisms. 2019;7(11):550. doi: 10.3390/microorganisms7110550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guarner F, Khan AG, Garisch J, Eliakim R, Gangl A, Thomson A, Krabshuis J, Lemair T, Kaufmann P, de Paula JA, et al. World gastroenterology organisation global guidelines. J Clin Gastroenterol. 2012;46(6):468–481. doi: 10.1097/mcg.0b013e3182549092. [DOI] [PubMed] [Google Scholar]
- 7.Renz H, Brandtzaeg P, Hornef M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat Rev Immunol. 2011;12(1):9–23. doi: 10.1038/nri3112. [DOI] [PubMed] [Google Scholar]
- 8.Malard F, Dore J, Gaugler B, Mohty M. Introduction to host microbiome symbiosis in health and disease. Mucosal Immunol. 2020;14(3):547–554. doi: 10.1038/s41385-020-00365-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen J, Obin MS, Zhao L. The gut microbiota, obesity and insulin resistance. Mol Aspects Med. 2013;34(1):39–58. doi: 10.1016/j.mam.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Weng M, Walker WA. The role of gut microbiota in programming the immune phenotype. J Dev Orig Health Dis. 2013;4(3):203–214. doi: 10.1017/s2040174412000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roland MM, Mohammed AD, Kubinak JL. How MHCII signaling promotes benign host-microbiota interactions. PLoS Pathog. 2020;16(6):e1008558. Public Library of Science. doi: 10.1371/journal.ppat.1008558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parfrey LW, Jirků M, Šíma R, Jalovecká M, Sak B, Grigore K, Pomajbíková KJ. A benign helminth alters the host immune system and the gut microbiota in a rat model system. PLoS ONE . 2017:12(8). doi: 10.1371/journal.pone.0182205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pascal M, Perez-Gordo M, Caballero T, Escribese MM, Lopez Longo MN, Luengo O, Manso L, Matheu V, Seoane E, Zamorano M, et al. Microbiome and allergic diseases. Front Immunol. 2018;9:1584. doi: 10.3389/fimmu.2018.01584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holscher HD. Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes. 2017;8(2):172–184. doi: 10.1080/19490976.2017.1290756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polkowska‐Pruszyńska B, Gerkowicz A, Krasowska D. The gut microbiome alterations in allergic and inflammatory skin diseases – an update. J EurAcad Dermatol Venereol. 2019;34(3):455–464. doi: 10.1111/jdv.15951. [DOI] [PubMed] [Google Scholar]
- 16.De Pessemier B, Grine L, Debaere M, Maes A, Paetzold B, Callewaert C. Gut–skin axis: current knowledge of the interrelationship between microbial dysbiosis and skin conditions. Microorganisms. 2021;9(2):353. doi: 10.3390/microorganisms9020353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan SE, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021:71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godoy-Vitorino F. Human microbial ecology and the rising new medicine. Ann Transll Med. 2019;7(14):342. doi: 10.21037/atm.2019.06.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uhr GT, Dohnalová L, Thaiss CA. The dimension of time in host-microbiome interactions. MSystems. 2019;4(1). doi: 10.1128/msystems.00216-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson CL, Versalovic J. The human microbiome and its potential importance to pediatrics. Pediatrics. 2012;129(5):950–960. doi: 10.1542/peds.2011-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fredricks DN. Microbial ecology of human skin in health and disease. J Invest Dermatol Symp Proc. 2001;6(3):167–169. doi: 10.1046/j.0022-202x.2001.00039.x. [DOI] [PubMed] [Google Scholar]
- 22.Gallo RL. Human skin is the largest epithelial surface for interaction with microbes. J Invest Dermatol. 2017;137(6):1213–1214. doi: 10.1016/j.jid.2016.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cundell AM. Microbial ecology of the human skin. Microb Ecol. 2016;76(1):113–120. doi: 10.1007/s00248-016-0789-6. [DOI] [PubMed] [Google Scholar]
- 24.Chiller K, Selkin BA, Murakawa GJ. Skin microflora and bacterial infections of the skin. J Invest Dermatol Symp Proc. 2001;6(3):170–174. doi: 10.1046/j.0022-202x.2001.00043.x. [DOI] [PubMed] [Google Scholar]
- 25.Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol. 2018;16(3):143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 26.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Z, Knight R. Dietary effects on human gut microbiome diversity. Br J Nutr. 2014;113(S1):S1–S5. doi: 10.1017/s0007114514004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. BMJ. 2018:k2179. doi: 10.1136/bmj.k2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hillman ET, Lu H, Yao T, Nakatsu CH. Microbial ecology along the gastrointestinal tract. Microbes and Environ. 2017;32(4):300–313. doi: 10.1264/jsme2.me17017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lau K, Srivatsav V, Rizwan A, Nashed A, Liu R, Shen R, Akhtar M. Bridging the Gap between gut microbial dysbiosis and cardiovascular diseases. Nutrients. 2017;9(8):859. doi: 10.3390/nu9080859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bor B, Bedree J, Shi W, McLean J, He X. Saccharibacteria (TM7) in the human oral microbiome. J Dent Res. 2019;98(5):500–509. doi: 10.1177/0022034519831671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, Lewis JD, Bushman FD. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS ONE. 2013;8(6):e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Honkanen J, Vuorela A, Muthas D, Orivuori L, Luopajärvi K, Tejesvi MVG, Lavrinienko A, Pirttilä AM, Fogarty CL, Härkönen T, et al. Fungal dysbiosis and intestinal inflammation in children with beta-cell autoimmunity. Front Immunol. 2020;11:11. doi: 10.3389/fimmu.2020.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dridi B, Raoult D, Drancourt M. Archaea as emerging organisms in complex human microbiomes. Anaerobe. 2011;17(2):56–63. doi: 10.1016/j.anaerobe.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Camp JG, Kanther M, Semova I, Rawls JF. Patterns and scales in gastrointestinal microbial ecology. Gastroenterology. 2009;136(6):1989–2002. doi: 10.1053/j.gastro.2009.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen R, Scott KP, Khan S, Martin JC, Berry SH, Stevenson M, Okpapi A, Munro MJ, Hold GL. First-pass meconium samples from healthy term vaginally-delivered neonates: an analysis of the microbiota. PLOS ONE. 2015;10(7):e0133320. doi: 10.1371/journal.pone.0133320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiménez E, Marín ML, Martín R, Odriozola JM, Olivares M, Xaus J, Fernández L, Rodríguez JM. Is meconium from healthy newborns actually sterile? Res Microbiol. 2008;159(3):187–193. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 39.Tapiainen T, Paalanne N, Tejesvi MV, Koivusaari P, Korpela K, Pokka T, Salo J, Kaukola T, Pirttilä AM, Uhari M, et al. Maternal influence on the fetal microbiome in a population-based study of the first-pass meconium. Pediatr Res. 2018;84(3):371–379. doi: 10.1038/pr.2018.29. [DOI] [PubMed] [Google Scholar]
- 40.Yang J, Yao S, Cheng K, Xu L, Hou L, Wei Y, Feng H, Yu X, Zhang X, Tong X, et al. Comparison of meconium microbiome in dizygotic and monozygotic twins born by Caesarean section (CS). Front Microbiol. 2020;11:1139. doi: 10.3389/fmicb.2020.01139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sekirov I, Russell SL, Caetano M, Antunes L, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90(3):859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 42.Mueller NT, Bakacs E, Combellick J, Grigoryan Z, Dominguez-Bello MG. The infant microbiome development: mom matters. Trends Mol Med. 2015;21(2):109–117. doi: 10.1016/j.molmed.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano G, Gasbarrini A, Mele MC. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 2019;7(1):14. doi: 10.3390/microorganisms7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santacruz A, Collado MC, García-Valdés L, Segura MT, Martín-Lagos JA, Anjos T, Martí-Romero M, Lopez RM, Florido J, Campoy C, et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br J Nutr. 2010;104(1):83–92. doi: 10.1017/s0007114510000176. [DOI] [PubMed] [Google Scholar]
- 45.Edwards SM, Cunningham SA, Dunlop AL, Corwin EJ. The Maternal gut microbiome during pregnancy. MCN: Ame J Matern Child Nurs. 2017;42(6):310–317. doi: 10.1097/nmc.0000000000000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayer EA, Savidge T, Shulman RJ. Brain–gut microbiome interactions and functional bowel disorders. Gastroenterology. 2014;146(6):1500–1512. doi: 10.1053/j.gastro.2014.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boyle RJ, Lahtinen SJ, Tang MLK. Probiotics and Skin. Nutr Skin. 2011:111–127. doi: 10.1007/978-1-4419-7967-4_8. [DOI] [Google Scholar]
- 48.Kosiewicz MM, Dryden GW, Chhabra A, Alard P. Relationship between gut microbiota and development of T cell associated disease. FEBS Lett. 2014;588(22):4195–4206. doi: 10.1016/j.febslet.2014.03.019. [DOI] [PubMed] [Google Scholar]
- 49.Brandtzaeg P. The gut as communicator between environment and host: immunological consequences. Eur J Pharmacol. 2011;668:S16–S32. doi: 10.1016/j.ejphar.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 50.Thursby E, Juge N. Introduction to the human gut microbiota. Biochem Jl. 2017;474(11):1823–1836. doi: 10.1042/bcj20160510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Power SE, O’Toole PW, Stanton C, Ross RP, Fitzgerald GF. Intestinal microbiota, diet and health. Br J Nutr. 2013;111(3):387–402. doi: 10.1017/s0007114513002560. [DOI] [PubMed] [Google Scholar]
- 52.Brandtzaeg P. Mucosal immunity: induction, dissemination, and effector functions. Scand J Immunol. 2009;70(6):505–515. doi: 10.1111/j.1365-3083.2009.02319.x. [DOI] [PubMed] [Google Scholar]
- 53.Corthésy B. Role of secretory immunoglobulin A and secretory component in the protection of mucosal surfaces. Future Microbiol. 2010;5(5):817–829. doi: 10.2217/fmb.10.39. [DOI] [PubMed] [Google Scholar]
- 54.Canny GO, McCormick BA. Bacteria in the intestine, helpful residents or enemies from within? Infect Immun. 2008;76(8):3360–3373. doi: 10.1128/iai.00187-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiao Y, Wu L, Huntington ND, and Zhang X. Crosstalk between gut microbiota and innate immunity and its implication in autoimmune diseases. Front Immunol. 2020:11. doi: 10.3389/fimmu.2020.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30(6):492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Belkaid Y, Hand T. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hinks TSC, Zhang XW. MAIT cell activation and functions. Front Immunol. 2020;11:11. doi: 10.3389/fimmu.2020.01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Macpherson AJ, Slack E, Geuking MB, McCoy KD. The mucosal firewalls against commensal intestinal microbes. Semin Immunopathol. 2009;31(2):145–149. doi: 10.1007/s00281-009-0174-3. [DOI] [PubMed] [Google Scholar]
- 60.Dillon A, Lo DD. M cells: intelligent engineering of mucosal immune surveillance. Front Immunol. 2019;10:10. doi: 10.3389/fimmu.2019.01499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mabbott NA, Donaldson DS, Ohno H, Williams IR, Mahajan A. Microfold (M) cells: important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol. 2013;6(4):666–677. doi: 10.1038/mi.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mörbe UM, Jørgensen PB, Fenton TM, von Burg N, Riis LB, Spencer J, Agace WW. Human gut-associated lymphoid tissues (GALT); diversity, structure, and function. Mucosal Immunol. 2021;14(4):793–802. doi: 10.1038/s41385-021-00389-4. [DOI] [PubMed] [Google Scholar]
- 63.Brandtzaeg P, Kiyono H, Pabst R, Russell MW. Terminology: nomenclature of mucosa-associated lymphoid tissue. Mucosal Immunol. 2007;1(1):31–37. doi: 10.1038/mi.2007.9. [DOI] [PubMed] [Google Scholar]
- 64.Layhadi JA, Shamji MH. Uncovering the immunological properties of isolated lymphoid follicles. Allergy. 2020;76(4):1292–1293. doi: 10.1111/all.14598. [DOI] [PubMed] [Google Scholar]
- 65.Gubatan J, Holman DR, Puntasecca CJ, Polevoi D, Rubin SJ, Rogalla S. Antimicrobial peptides and the gut microbiome in inflammatory bowel disease. World J Gastroenterol. 2021;27(43):7402–7422. doi: 10.3748/wjg.v27.i43.7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grigat J, Soruri A, Forssmann U, Riggert J, Zwirner J. Chemoattraction of macrophages, T Lymphocytes, and mast cells is evolutionarily conserved within the human α-defensin family. J Immunol. 2007;179(6):3958–3965. doi: 10.4049/jimmunol.179.6.3958. [DOI] [PubMed] [Google Scholar]
- 67.Lunjani N, Ahearn-Ford S, Dube FS, Hlela C, O’Mahony L. Mechanisms of microbe-immune system dialogue within the skin. Genes Immun. 2021;22(5–6):276–288. doi: 10.1038/s41435-021-00133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol. 2009;21(4):317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Asquith MJ, Boulard O, Powrie F, Maloy KJ. Pathogenic and protective roles of MyD88 in Leukocytes and epithelial cells in mouse models of inflammatory bowel disease. Gastroenterology. 2010;139(2):519–529.e2. doi: 10.1053/j.gastro.2010.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang YJ, Marsland BJ, Bunyavanich S, O’Mahony L, Leung DY, Muraro A, Fleisher TA. The microbiome in allergic disease: current understanding and future opportunities—2017 PRACTALL document of the American academy of allergy, Asthma & immunology and the European academy of allergy and clinical immunology. J Allergy Clin Immunol. 2017;139(4):1099–1110. doi: 10.1016/j.jaci.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rizzello V, Bonaccorsi I, Dongarrà ML, Fink LN, Ferlazzo G. Role of Natural Killer and Dendritic Cell Crosstalk in Immunomodulation by Commensal Bacteria Probiotics. J Biomed Biotechnol. 2011;2011:1–10. doi: 10.1155/2011/473097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Klose CSN, Artis D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat Immunol. 2016;17(7):765–774. doi: 10.1038/ni.3489. [DOI] [PubMed] [Google Scholar]
- 73.Withers DR, Hepworth MR. Group 3 innate lymphoid cells: communications hubs of the intestinal immune system. Front Immunol. 2017;8:1298. doi: 10.3389/fimmu.2017.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kobayashi T, Ricardo-Gonzalez RR, Moro K. Skin-resident innate lymphoid cells – cutaneous innate guardians and regulators. Trends Immunol. 2020;41(2):100–112. doi: 10.1016/j.it.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng M, Zhu J. Innate lymphoid cells and intestinal inflammatory disorders. Int J Mol Sci. 2022;23(3):1856. doi: 10.3390/ijms23031856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Legoux F, Salou M, Lantz O. MAIT cell development and functions: the microbial connection. Immunity ScienceDirect. 2020. October 13;53(4):710–723. doi: 10.1016/j.immuni.2020.09.009. [DOI] [PubMed] [Google Scholar]
- 77.Gebru YA, Choi MR, Raja G, Gupta H, Sharma SP, Choi YR, Kim HS, Yoon SJ, Kim DJ, Suk KT. Pathophysiological roles of mucosal-associated invariant t cells in the context of gut microbiota-liver axis. Microorganisms. 2021;9(2):296. doi: 10.3390/microorganisms9020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vojvodic A, Peric-Hajzler Z, Matovic D, Vojvodic P, Vlaskovic-Jovicevic T, Sijan G, Dimitrijevic S, Stepic N, Wollina U, Badr BAE, et al. Gut microbiota and the alteration of immune balance in skin diseases: from nutraceuticals to fecal transplantation. Open Access Macedonian J Med Sci. 2019;7(18):3034–3038. doi: 10.3889/oamjms.2019.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. 2013;24(2):160–168. doi: 10.1016/j.copbio.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 80.Scott KP, Gratz SW, Sheridan PO, Flint HJ, Duncan SH. The influence of diet on the gut microbiota. Pharmacol Res. 2013;69(1):52–60. doi: 10.1016/j.phrs.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 81.Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity. 2017;46(4):562–576. doi: 10.1016/j.immuni.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Konrad A, Cong Y, Duck W, Borlaza R, Elson CO. Tight mucosal compartmentation of the murine immune response to antigens of the enteric microbiota. Gastroenterology. 2006;130(7):2050–2059. doi: 10.1053/j.gastro.2006.02.055. [DOI] [PubMed] [Google Scholar]
- 83.Mowat AM. Anatomical basis of tolerance and immunity to intestinal antigens. Nat Rev Immunol. 2003;3(4):331–341. doi: 10.1038/nri1057. [DOI] [PubMed] [Google Scholar]
- 84.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Sci (New York, N Y). 2004;303(5664):1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 85.Naik S, Bouladoux N, Linehan JL, Han SJ, Harrison OJ, Wilhelm C, Conlan S, Himmelfarb S, Byrd AL, Deming C, et al. Commensal–dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520(7545):104–108. doi: 10.1038/nature14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez M, Laufer T, Ignatowicz L, Ivanov I. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity. 2014;40(4):594–607. doi: 10.1016/j.immuni.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203(10):2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaetzel CS. Cooperativity among secretory IgA, the polymeric immunoglobulin receptor, and the gut microbiota promotes host–microbial mutualism. Immunol Lett. 2014;162(2):10–21. doi: 10.1016/j.imlet.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mantis NJ, Rol N, Corthésy B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4(6):603–611. doi: 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Suzuki K, Maruya M, Kawamoto S, Sitnik K, Kitamura H, Agace WW, Fagarasan S. The sensing of environmental stimuli by follicular dendritic cells promotes immunoglobulin A generation in the gut. Immunity. 2010;33(1):71–83. doi: 10.1016/j.immuni.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 91.Wang X, Cho B, Suzuki K, Xu Y, Green JA, An J, Cyster JG. Follicular dendritic cells help establish follicle identity and promote B cell retention in germinal centers. J Exp Med. 2011;208(12):2497–2510. doi: 10.1084/jem.20111449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lazar V, Ditu LM, Pircalabioru GG, Gheorghe I, Curutiu C, Holban AM, Picu A, Petcu L, Chifiriuc MC. Aspects of gut microbiota and immune system interactions in infectious diseases, immunopathology, and cancer. Front Immunol. 2018;9:1830. doi: 10.3389/fimmu.2018.01830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu HJ, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. 2012;3(1):4–14. doi: 10.4161/gmic.19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arck P, Handjiski B, Hagen E, Pincus M, Bruenahl C, Bienenstock J, Paus R. Is there a ‘gut-brain-skin axis’? Exp Dermatol. 2010;19(5):401–405. doi: 10.1111/j.1600-0625.2009.01060.x. [DOI] [PubMed] [Google Scholar]
- 95.Lyte M. Microbial endocrinology and the microbiota-gut-brain axis. Adv Exp Med Biol. 2014:3–24. doi: 10.1007/978-1-4939-0897-4_1. [DOI] [PubMed] [Google Scholar]
- 96.Hokazono H, Omori T, Ono K. Effects of single and combined administration of fermented barley extract and γ-aminobutyric acid on the development of atopic dermatitis in NC/Nga mice. Biosci Biotechnol Biochem. 2010;74(1):135–139. doi: 10.1271/bbb.90653. [DOI] [PubMed] [Google Scholar]
- 97.Uehara E, Hokazono H, Sasaki T, Yoshioka H, Matsuo N. Effects of GABA on the expression of type I collagen gene in normal human dermal fibroblasts. Biosci Biotechnol Biochem. 2017;81(2):376–379. doi: 10.1080/09168451.2016.1238296. [DOI] [PubMed] [Google Scholar]
- 98.Langan E, Lisztes E, Bíró T, Funk W, Kloepper J, Griffiths C, Paus R. Dopamine is a novel, direct inducer of catagen in human scalp hair folliclesin vitro. Bri J Dermatol. 2012;168(3):520–525. doi: 10.1111/bjd.12113. [DOI] [PubMed] [Google Scholar]
- 99.Ainonen S, Tejesvi MV, Mahmud MR, Paalanne N, Pokka T, Li W, Nelson KE, Salo J, Renko M, Vänni P, et al. Antibiotics at birth and later antibiotic courses: effects on gut microbiota. Pediatr Res. 2021;91(1):154–162. doi: 10.1038/s41390-021-01494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Conlon M, Bird A. The impact of diet and lifestyle on gut microbiota and human health. Nutrients. 2014;7(1):17–44. doi: 10.3390/nu7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rodríguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N, Avershina E, Rudi K, Narbad A, Jenmalm MC, et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis. 2015;26. doi: 10.3402/mehd.v26.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tojo R. Intestinal microbiota in health and disease: role of bifidobacteria in gut homeostasis. World J Gastroenterol. 2014;20(41):15163. doi: 10.3748/wjg.v20.i41.15163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Drake D. The gut microbiome‐skin axis. Skin Microbiome Handb. 2020:33–44. doi: 10.1002/9781119593058.ch2. [DOI] [Google Scholar]
- 104.Ramasamy S, Barnard E, Dawson TL Jr, Li H. The role of the skin microbiota in acne pathophysiology. Br J Dermatol. 2019;181(4):691–699. doi: 10.1111/bjd.18230. [DOI] [PubMed] [Google Scholar]
- 105.Graf D, Di Cagno R, Fåk F, Flint HJ, Nyman M, Saarela M, Watzl B. Contribution of diet to the composition of the human gut microbiota. Microb Ecol Health Dis. 2015;26:26164. doi: 10.3402/mehd.v26.26164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, Abrouk M, Farahnik B, Nakamura M, Zhu TH, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15(1). doi: 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vaughn AR, Notay M, Clark AK, Sivamani RK. Skin-gut axis: the relationship between intestinal bacteria and skin health. World J Dermatol. 2017;6(4):52–58. doi: 10.5314/wjd.v6.i4.52. [DOI] [Google Scholar]
- 108.Purchiaroni F, Tortora A, Gabrielli M, Bertucci F, Gigante G, Ianiro G, Ojetti V, Scarpellini E, Gasbarrini A. The role of intestinal microbiota and the immune system. Eur Rev Med Pharmacol Sci. 2013;17:323–333. [PubMed] [Google Scholar]
- 109.Madke B, Pradhan S, Kabra P, Singh A. Anti-inflammatory and immunomodulatory effects of antibiotics and their use in dermatology. Indian J Dermatol. 2016;61(5):469. doi: 10.4103/0019-5154.190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ribeiro CFA, Silveira GGDOS, Cândido EDS, Cardoso MH, Espínola Carvalho CM, Franco OL. Effects of antibiotic treatment on gut microbiota and how to overcome its negative impacts on human health. ACS Infect Dis. 2020;6(10):2544–2559. doi: 10.1021/acsinfecdis.0c00036. [DOI] [PubMed] [Google Scholar]
- 111.Sinha S, Lin G, Ferenczi K. The skin microbiome and the gut-skin axis. Clin Dermatol. 2021;39(5):829–839. doi: 10.1016/j.clindermatol.2021.08.021. [DOI] [PubMed] [Google Scholar]
- 112.Baquerizo Nole KL, Yim E, Keri JE. Probiotics and prebiotics in dermatology. J Am Acad Dermatol. 2014;71(4):814–821. doi: 10.1016/j.jaad.2014.04.050. [DOI] [PubMed] [Google Scholar]
- 113.Linares DM, Ross P, Stanton C. Beneficial microbes: the pharmacy in the gut. Bioengineered. 2015;7(1):11–20. doi: 10.1080/21655979.2015.1126015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kordy K, Gaufin T, Mwangi M, Li F, Cerini C, Lee DJ, Adisetiyo H, Woodward C, Pannaraj PS, Tobin NH, et al. Contributions to human breast milk microbiome and enteromammary transfer of Bifidobacterium breve. PLOS ONE. 2020;15(1):e0219633. doi: 10.1371/journal.pone.0219633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shamash M, Maurice CF. Phages in the infant gut: a framework for virome development during early life. ISME J Published. 2021. doi: 10.1038/s41396-021-01090-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Duranti S, Lugli GA, Mancabelli L, Armanini F, Turroni F, James K, Ferretti P, Gorfer V, Ferrario C, Milani C, et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome. 2017:5(1). doi: 10.1186/s40168-017-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tanaka M, Nakayama J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol Int. 2017;66(4):515–522. doi: 10.1016/j.alit.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 118.Zeng MY, Inohara N, Nuñez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2016;10(1):18–26. doi: 10.1038/mi.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.O’Sullivan A, Farver M, Smilowitz JT. The influence of early infant-feeding practices on the intestinal microbiome and body composition in infants. Nutr Metab Insights. 2015;8(Suppl 1):1–9. doi: 10.4137/NMI.S29530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Egawa G, Honda T, Kabashima K. SCFAs control skin immune responses via increasing tregs. J Invest Dermatol. 2017;137(4):800–801. doi: 10.1016/j.jid.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 121.Ge Y, Liu W, Tao H, Zhang Y, Liu L, Liu Z, Qiu B, Xu T. Effect of industrial trans-fatty acids-enriched diet on gut microbiota of C57BL/6 mice. Eur J Nutr. 2019;58(7):2625–2638. doi: 10.1007/s00394-018-1810-2. [DOI] [PubMed] [Google Scholar]
- 122.Rosa DF, Sarandy MM, Novaes RD, Freitas MB, Do Carmo Gouveia Pelúzio M, Gonçalves RV. High-fat diet and alcohol intake promotes inflammation and impairs skin wound healing in wistar rats. Mediators Inflamm. 2018;2018:4658583. doi: 10.1155/2018/4658583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Morales P, Fujio S, Navarrete P, Ugalde JA, Magne F, Carrasco-Pozo C, Tralma K, Quezada M, Hurtado C, Covarrubias N, et al. Impact of dietary lipids on colonic function and microbiota: an experimental approach involving orlistat-induced fat malabsorption in human volunteers. Clin Transl Gastroenterol. 2016;7(4):e161. doi: 10.1038/ctg.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Coras R, Kavanaugh A, Boyd T, Huynh D, Lagerborg KA, Xu YJ, Rosenthal SB, Jain M, Guma M. Choline metabolite, trimethylamine N-oxide (TMAO), is associated with inflammation in psoriatic arthritis. Clin Exp Rheumatol. 2019;37:481–484. [PMC free article] [PubMed] [Google Scholar]
- 125.Mei F, Duan Z, Chen M, Lu J, Zhao M, Li L, Shen X, Xia G, Chen S. Effect of a high-collagen peptide diet on the gut microbiota and short-chain fatty acid metabolism. J Funct Foods. 2020;75:104278. doi: 10.1016/j.jff.2020.104278. [DOI] [Google Scholar]
- 126.Komine M. Recent advances in psoriasis research; the clue to mysterious relation to gut microbiome. Int J Mol Sci. 2020;21(7):2582. doi: 10.3390/ijms21072582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic t reg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ramirez J, Guarner F, Bustos Fernandez L, Maruy A, Sdepanian VL, Cohen H. Antibiotics as major disruptors of gut microbiota. Front Cell Infect Microbiol. 2020;10:572912. doi: 10.3389/fcimb.2020.572912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Samonis G, Kofteridis DP, Maraki S, Alegakis D, Mantadakis E, Papadakis JA, Gikas AH, Falagas M. Levofloxacin and moxifloxacin increase human gut colonization by candida species. Antimicrob Agents Chemother. 2005;49(12):5189. doi: 10.1128/aac.49.12.5189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gu ZQ, Tseng KY, Tsai YH. Candida gut commensalism and inflammatory disease. Med Microecol. 2020;3:100008. doi: 10.1016/j.medmic.2020.100008. [DOI] [Google Scholar]
- 132.Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, Scott K, Stanton C, Swanson KS, Cani PD, et al. Expert consensus document: the International scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017;14(8):491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 133.Azad M, Sarker M, Li T, Yin J. Probiotic species in the modulation of gut microbiota: an overview. Biomed Res Int. 2018;2018:9478630. doi: 10.1155/2018/9478630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang S, Xiao Y, Tian F, Zhao J, Zhang H, Zhai Q, Chen W. Rational use of prebiotics for gut microbiota alterations: specific bacterial phylotypes and related mechanisms. J Funct Foods. 2020;66:103838. doi: 10.1016/j.jff.2020.103838. [DOI] [Google Scholar]
- 135.Szántó M, Dózsa A, Antal D, Szabó K, Kemény L, Bai P. Targeting the gut-skin axis-probiotics as new tools for skin disorder management? Exp Dermatol. 2019;28(11):1210–1218. doi: 10.1111/exd.14016. [DOI] [PubMed] [Google Scholar]
- 136.Muizzuddin N, Maher W, Sullivan M, Schnittger S, Mammone T. Physiological effect of a probiotic on skin. J Cosmet Sci. 2012;63:385–395. [PubMed] [Google Scholar]
- 137.Lolou V, Panayiotidis MI. Functional role of probiotics and prebiotics on skin health and disease. Fermentation. 2019;5(2):41. doi: 10.3390/fermentation5020041. [DOI] [Google Scholar]
- 138.Hong KB, Jeong M, Han KS, Hwan Kim J, Park Y, Suh HJ. Photoprotective effects of galacto-oligosaccharide and/or Bifidobacterium longum supplementation against skin damage induced by ultraviolet irradiation in hairless mice. Int J Food Sci Nutr. 2015;66(8):923–930. doi: 10.3109/09637486.2015.1088823. [DOI] [PubMed] [Google Scholar]
- 139.Davani-Davari D, Negahdaripour M, Karimzadeh I, Seifan M, Mohkam M, Masoumi S, Berenjian A, Ghasemi Y. Prebiotics: definition, types, sources, mechanisms, and clinical applications. Foods. 2019;8(3):92. doi: 10.3390/foods8030092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Manning TS, Gibson GR. Prebiotics. Best Pract Res Clin Gastroenterol. 2004;18(2):287–298. doi: 10.1016/j.bpg.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 141.Leon Carrion S, Sutter CH, Sutter TR. Combined treatment with sodium butyrate and PD153035 enhances keratinocyte differentiation. Exp Dermatol. 2014;23(3):211–214. doi: 10.1111/exd.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vollmer D, West V, Lephart E. Enhancing skin health: by oral administration of natural compounds and minerals with implications to the dermal microbiome. Int J Mol Sci. 2018;19(10):3059. doi: 10.3390/ijms19103059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Al-Ghazzewi F, Tester R. Impact of prebiotics and probiotics on skin health. Benef Microbes. 2014;5(2):99–107. doi: 10.3920/bm2013.0040. [DOI] [PubMed] [Google Scholar]
- 144.Fijan S. Microorganisms with claimed probiotic properties: an overview of recent literature. Int J Environ Res Public Health. 2014;11(5):4745–4767. doi: 10.3390/ijerph110504745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Benhadou F, Mintoff D, Schnebert B, Thio H. Psoriasis and microbiota: a systematic review. Diseases. 2018;6(2):47. doi: 10.3390/diseases6020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Manzhalii E, Hornuss D, and Stremmel W. Intestinal-borne dermatoses significantly improved by oral application ofEscherichia coliNissle 1917. World J Gastroenterol. 2016;22(23):5415. doi: 10.3748/wjg.v22.i23.5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sehgal V, Khurana A, Pandhi D. Biologics in dermatology: an integrated review. Indian J Dermatol. 2014;59(5):425–441. doi: 10.4103/0019-5154.139859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Conticini E, Sota J, Falsetti P, Lamberti A, Miracco C, Guarnieri A, Frediani B, Cantarini L. Biologic drugs in the treatment of polyarteritis nodosa and deficit of adenosine deaminase 2: a narrative review. Autoimmun Rev. 2021;20(4):102784. doi: 10.1016/j.autrev.2021.102784. [DOI] [PubMed] [Google Scholar]
- 149.Johnston SL. Biologic therapies: what and when? J Clin Pathol. 2007;60(1):8–17. doi: 10.1136/jcp.2005.032300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Weersma RK, Zhernakova A, Fu J. Interaction between drugs and the gut microbiome. Gut. 2020;69(8):1510–1519. doi: 10.1136/gutjnl-2019-320204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Myers B, Brownstone N, Reddy V, Chan S, Thibodeaux Q, Truong A, Bhutani T, Chang HW, Liao W. The gut microbiome in psoriasis and psoriatic arthritis. Best Pract Res Clin Rheumatol. 2019;33(6):101494. doi: 10.1016/j.berh.2020.101494. [DOI] [PubMed] [Google Scholar]
- 152.Di Filippo P, Russo D, Attanasi M, Di Pillo S, Chiarelli F. Immunological targets of biologic drugs in allergic skin diseases in children. Biomedicines. 2021;9(11):1615. doi: 10.3390/biomedicines9111615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Forbes JD, Van Domselaar G, Bernstein CN. The gut microbiota in immune-mediated inflammatory diseases. Front Microbiol. 2016;7:1081. doi: 10.3389/fmicb.2016.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Simonyte Sjödin K, Vidman L, Rydén P, West CE. Emerging evidence of the role of gut microbiota in the development of allergic diseases. Curr Opin Allergy Clin Immunol. 2016;16(4):390–395. doi: 10.1097/aci.0000000000000277. [DOI] [PubMed] [Google Scholar]
- 155.Alzahrani Y, Alesa D, Alshamrani H, Alamssi D, Alzahrani N, Almohammadi M. The role of gut microbiome in the pathogenesis of psoriasis and the therapeutic effects of probiotics. J Fami Med Primary Care. 2019;8(11):3496. doi: 10.4103/jfmpc.jfmpc_709_19. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 156.Ramírez-Boscá A, Navarro-López V, Martínez-Andrés A, Such J, Francés R, Horga De La Parte J, Asín-Llorca M. Identification of bacterial DNA in the ––peripheral blood of patients with active Psoriasis. JAMA Dermatol. 2015;151(6):670. doi: 10.1001/jamadermatol.2014.5585. [DOI] [PubMed] [Google Scholar]
- 157.Tan L, Zhao S, Zhu W, Wu L, Li J, Shen M, Lei L, Chen X, Peng C. The Akkermansia muciniphila is a gut microbiota signature in psoriasis. Exp Dermatol. 2018;27(2):144–149. doi: 10.1111/exd.13463. [DOI] [PubMed] [Google Scholar]
- 158.de Francesco MA, Caruso A. The gut microbiome in psoriasis and crohn’s disease: is its perturbation a common denominator for their pathogenesis? Vaccines. 2022;10(2):244. doi: 10.3390/vaccines10020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Scher JU, Ubeda C, Artacho A, Attur M, Isaac S, Reddy SM, Marmon S, Neimann A, Brusca S, Patel T, et al. Decreased Bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015;67(1):128–139. doi: 10.1002/art.38892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Huang L, Gao R, Yu N, Zhu Y, Ding Y, Qin H. Dysbiosis of gut microbiota was closely associated with psoriasis. Sci China Life Sci. 2018;62(6):807–815. doi: 10.1007/s11427-018-9376-6. [DOI] [PubMed] [Google Scholar]
- 161.Mann EA, Bae E, Kostyuchek D, Chung HJ, McGee JS. The Gut microbiome: human health and inflammatory skin diseases. Ann Dermatol. 2020;32(4):265. doi: 10.5021/ad.2020.32.4.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chen Y, Ho HJ, Tseng C, Lai Z, Shieh J, Wu C. Intestinal microbiota profiling and predicted metabolic dysregulation in psoriasis patients. Exp Dermatol. 2018;27(12):1336–1343. doi: 10.1111/exd.13786. [DOI] [PubMed] [Google Scholar]
- 163.Yan D, Issa N, Afifi L, Jeon C, Chang HW, Liao W. The role of the skin and gut microbiome in psoriatic disease. Curr Dermatol Rep. 2017;6(2):94–103. doi: 10.1007/s13671-017-0178-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kierasińska M, Donskow-Łysoniewska K. Both the microbiome and the macrobiome can influence immune responsiveness in psoriasis. Cent Eur J Immunol. 2021;46(4):502–508. doi: 10.5114/ceji.2021.110314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mosca A, Leclerc M, Hugot JP. Gut microbiota diversity and human diseases: should we reintroduce key predators in our ecosystem? Front Microbiol. 2016:7. doi: 10.3389/fmicb.2016.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Visser MJE, Kell DB, Pretorius E. Bacterial dysbiosis and translocation in psoriasis vulgaris. Front Cell Infect Microbiol. 2019;9:9. doi: 10.3389/fcimb.2019.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bland J. Intestinal Microbiome, akkermansia muciniphila, and medical nutrition therapy. Integr Med (Encinitas, Calif). 2016;15:14–16. [PMC free article] [PubMed] [Google Scholar]
- 168.Zhang T, Li Q, Cheng L, Buch H, Zhang F. Akkermansia muciniphila is a promising probiotic. Microb Biotechnol. 2019;12(6):1109–1125. doi: 10.1111/1751-7915.13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rungjang A, Meephansan J, Bing Thio H. Skin and gut microbiota in psoriasis: a systematic review. Hum Microbiome. 2021. doi: 10.5772/intechopen.92686. [DOI] [Google Scholar]
- 170.Groeger D, O’Mahony L, Murphy EF, Bourke JF, Dinan TG, Kiely B, Shanahan F, Quigley EM. Bifidobacterium infantis35624 modulates host inflammatory processes beyond the gut. Gut Microbes. 2013;4(4):325–339. doi: 10.4161/gmic.25487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chen L, Li J, Zhu W, Kuang Y, Liu T, Zhang W, Chen X, Peng C. Skin and gut microbiome in psoriasis: gaining insight into the pathophysiology of it and finding novel therapeutic strategies. Front Microbiol. 2020:11. doi: 10.3389/fmicb.2020.589726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. 2012;129(2):434–440.e2. doi: 10.1016/j.jaci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 173.Forno E, Onderdonk AB, McCracken J, Litonjua AA, Laskey D, Delaney ML, DuBois AM, Gold DR, Ryan LM, Weiss ST, et al. Diversity of the gut microbiota and eczema in early life. Clin Mol Allergy. 2008;6(1). doi: 10.1186/1476-7961-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ismail IH, Oppedisano F, Joseph SJ, Boyle RJ, Licciardi PV, Robins-Browne RM, Tang ML. Reduced gut microbial diversity in early life is associated with later development of eczema but not atopy in high-risk infants. Pediatr Allergy and Immunol. 2012;23(7):674–681. doi: 10.1111/j.1399-3038.2012.01328.x. [DOI] [PubMed] [Google Scholar]
- 175.Wang M, Karlsson C, Olsson C, Adlerberth I, Wold AE, Strachan DP, Martricardi PM, Åberg N, Perkin MR, Tripodi S, et al. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J Allergy Clin Immunol. 2008;121(1):129–134. doi: 10.1016/j.jaci.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 176.Lunjani N, Satitsuksanoa P, Lukasik Z, Sokolowska M, Eiwegger T, O’Mahony L. Recent developments and highlights in mechanisms of allergic diseases: microbiome. Allergy. 2018;73(12):2314–2327. doi: 10.1111/all.13634. [DOI] [PubMed] [Google Scholar]
- 177.Watanabe S, Narisawa Y, Arase S, Okamatsu H, Ikenaga T, Tajiri Y, Kumemura M. Differences in fecal microflora between patients with atopic dermatitis and healthy control subjects. J Allergy Clin Immunol. 2003;111(3):587–591. doi: 10.1067/mai.2003.105. [DOI] [PubMed] [Google Scholar]
- 178.Biedermann T, Skabytska Y, Kaesler S, Volz T. Regulation of T cell immunity in atopic dermatitis by microbes. The Yin and Yang of Cutaneous Inflammation Frontiers Immunol. 2015:6. doi: 10.3389/fimmu.2015.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kim HJ, Kim HY, Lee SY, Seo JH, Lee E, Hong SJ. Clinical efficacy and mechanism of probiotics in allergic diseases. Korean J Pediatr. 2013;56(9):369. doi: 10.3345/kjp.2013.56.9.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Song H, Yoo Y, Hwang J, Na YC, Kim HS. Faecalibacterium prausnitzii subspecies–level dysbiosis in the human gut microbiome underlying atopic dermatitis. J Allergy Clin Immunol. 2016;137(3):852–860. doi: 10.1016/j.jaci.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 181.Zheng H, Liang H, Wang Y, Miao M, Shi T, Yang F, Liu E, Yuan W, Ji ZS, Li DK. Altered gut microbiota composition associated with eczema in infants. PLOS ONE. 2016;11(11):e0166026. doi: 10.1371/journal.pone.0166026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.de Sousa Moraes LF, Grzeskowiak LM, de Sales Teixeira TF, Gouveia Peluzio MDC. Intestinal microbiota and probiotics in celiac disease. Clin Microbiol Rev. 2014;27(3):482–489. doi: 10.1128/cmr.00106-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Uhde M, Ajamian M, Caio G, de Giorgio R, Indart A, Green PH, Verna EC, Volta U, Alaedini A. Intestinal cell damage and systemic immune activation in individuals reporting sensitivity to wheat in the absence of coeliac disease. Gut. 2016;65(12):1930–1937. doi: 10.1136/gutjnl-2016-311964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nagpal R, Wang S, Ahmadi S, Hayes J, Gagliano J, Subashchandrabose S, Kitzman DW, Becton T, Read R, Yadav H. Human-origin probiotic cocktail increases short-chain fatty acid production via modulation of mice and human gut microbiome. Sci Rep. 2018;8(1). doi: 10.1038/s41598-018-30114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kim JH, Kim K, Kim W. Gut microbiota restoration through fecal microbiota transplantation: a new atopic dermatitis therapy. Exp Mol Med. 2021;53(5):907–916. doi: 10.1038/s12276-021-00627-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zouboulis C, Jourdan E, Picardo M. Acne is an inflammatory disease and alterations of sebum composition initiate acne lesions. J EurAcad Dermatol Venereol. 2013;28(5):527–532. doi: 10.1111/jdv.12298. [DOI] [PubMed] [Google Scholar]
- 187.Penso L, Touvier M, Deschasaux M, Szabo De Edelenyi F, Hercberg S, Ezzedine K, Sbidian E. Association between adult acne and dietary behaviors. JAMA Dermatol. 2020;156(8):854. doi: 10.1001/jamadermatol.2020.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Guo X, Li J, Tang R, Zhang G, Zeng H, Wood RJ, Liu Z. High fat diet alters gut microbiota and the expression of paneth cell-antimicrobial peptides preceding changes of circulating inflammatory cytokines. Mediators Inflamm. 2017;2017:1–9. doi: 10.1155/2017/9474896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lee YB, Byun EJ, Kim HS. Potential role of the microbiome in acne: a comprehensive review. J clini med. 2019;8(7):987. doi: 10.3390/jcm8070987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Noureldein MH, Eid AA. Gut microbiota and mTOR signaling: insight on a new pathophysiological interaction. Microb Pathog. 2018;118:98–104. doi: 10.1016/j.micpath.2018.03.021. [DOI] [PubMed] [Google Scholar]
- 191.Smith RN, Braue A, Varigos GA, Mann NJ. The effect of a low glycemic load diet on acne vulgaris and the fatty acid composition of skin surface triglycerides. J Dermatol Sci. 2008;50(1):41–52. doi: 10.1016/j.jdermsci.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 192.Volkova LA, Khalif IL, Kabanova IN. Vliiane disbakterioza kishechnika na techenie vul’garnykh ugreĭ [Impact of the impaired intestinal microflora on the course of acne vulgaris]. Klin Med (Mosk). 2001;79(6):39–41. Russian. PMID: 11525176 [PubMed] [Google Scholar]
- 193.Holmes AD. Potential role of microorganisms in the pathogenesis of rosacea. J Am Acad Dermatol. 2013;69(6):1025–1032. doi: 10.1016/j.jaad.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 194.Joura MI, Brunner A, Nemes-Nikodém V, Sárdy M, Ostorházi E. Interactions between immune system and the microbiome of skin, blood and gut in pathogenesis of rosacea. Acta Microbiol Immunol Hung. 2021;68(1):1–6. doi: 10.1556/030.2021.01366. [DOI] [PubMed] [Google Scholar]
- 195.Yang X. Relationship between Helicobacter pylori and Rosacea: review and discussion. BMC Infect Dis. 2018;18(1). doi: 10.1186/s12879-018-3232-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Szlachcic A. The link between Helicobacter pylori infection and rosacea. J EurAcad Dermatol Venereol. 2002;16(4):328–333. doi: 10.1046/j.1468-3083.2002.00497.x. [DOI] [PubMed] [Google Scholar]
- 197.Pinchuk IV, Bressollier P, Verneuil B, Fenet B, Sorokulova IB, Mégraud F, Urdaci MC. In vitro anti- Helicobacter pylori activity of the probiotic strain Bacillus subtilis 3 is due to secretion of antibiotics. Antimicrob Agents Chemother. 2001;45(11):3156–3161. doi: 10.1128/aac.45.11.3156-3161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Buianova I, Girnyk G, Senyshyn N, Hepburn IS. Gut-skin connection: role of intestinal biome in rosacea. Ame J Gastroenterol. 2018;113(Supplement):S126. doi: 10.14309/00000434-201810001-00216. [DOI] [Google Scholar]
- 199.Kober MM, Bowe WP. The effect of probiotics on immune regulation, acne, and photoaging. Int J Women’s Dermatol. 2015;1(2):85–89. doi: 10.1016/j.ijwd.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Katsinelos P, Kountouras J, Paroutoglou G, Zavos C. Alopecia areata, primary sclerosing cholangitis, and ulcerative colitis: autoimmunity and apoptosis as common links? Dig Dis Sci. 2007;52(5):1288–1292. doi: 10.1007/s10620-006-9265-3. [DOI] [PubMed] [Google Scholar]
- 201.Simakou T, Butcher JP, Reid S, Henriquez FL. Alopecia areata: a multifactorial autoimmune condition. J Autoimmun. 2019;98:74–85. doi: 10.1016/j.jaut.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 202.Borde A, Åstrand A. Alopecia areata and the gut—the link opens up for novel therapeutic interventions. Expert Opin Ther Targets. 2018;22(6):503–511. doi: 10.1080/14728222.2018.1481504. [DOI] [PubMed] [Google Scholar]
- 203.Ooijevaar R, Terveer E, Verspaget H, Kuijper E, Keller J. Clinical application and potential of fecal microbiota transplantation. Annu Rev Med. 2019;70(1):335–351. doi: 10.1146/annurev-med-111717-122956. [DOI] [PubMed] [Google Scholar]
- 204.Xie WR, Yang XY, Xia HHX, Wu LH, He XX. Hair regrowth following fecal microbiota transplantation in an elderly patient with alopecia areata: a case report and review of the literature. World J Clin Cases. 2019;7(19):3074–3081. doi: 10.12998/wjcc.v7.i19.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Balato A, Cacciapuoti S, Di Caprio R, Marasca C, Masarà A, Raimondo A, Fabbrocini G. Human microbiome: composition and role in inflammatory skin diseases. Arch Immunol Ther Exp (Warsz). 2018;67(1):1–18. doi: 10.1007/s00005-018-0528-4. [DOI] [PubMed] [Google Scholar]
- 206.Kelly G, Hughes R, McGarry T, Born M, Adamzik K, Fitzgerald R, Lawlor C, Tobin A, Sweeney C, Kirby B. Dysregulated cytokine expression in lesional and non lesional skin in hidradenitis suppurativa. Bri J Dermatol. 2015;173(6):1431–1439. doi: 10.1111/bjd.14075. [DOI] [PubMed] [Google Scholar]
- 207.Laffert MV, Helmbold P, Wohlrab J, Fiedler E, Stadie V, Marsch WC. Hidradenitis suppurativa (acne inversa): early inflammatory events at terminal follicles and at interfollicular epidermis*. Exp Dermatol. 2009;19(6):533–537. doi: 10.1111/j.1600-0625.2009.00915.x. [DOI] [PubMed] [Google Scholar]
- 208.McCarthy S, Barrett M, Kirthi S, Pellanda P, Vlckova K, Tobin AM, Murphy M, Shanahan F, O’Toole PW. Altered skin and gut microbiome in hidradenitis suppurativa. J Invest Dermatol. 2022;142(2):459–468.e15. doi: 10.1016/j.jid.2021.05.036. [DOI] [PubMed] [Google Scholar]
- 209.Nikolakis G, Join-Lambert O, Karagiannidis I, Guet-Revillet H, Zouboulis CC, Nassif A. Bacteriology of hidradenitis suppurativa/acne inversa: a review. J Am Acad Dermatol. 2015;73(5):S12–S18. doi: 10.1016/j.jaad.2015.07.041. [DOI] [PubMed] [Google Scholar]
- 210.Sabat R, Jemec GBE, Matusiak U, Kimball AB, Prens E, Wolk K. Hidradenitis suppurativa. Nat Revi Dis Primers . 2020:6(1). doi: 10.1038/s41572-020-0149-1. [DOI] [PubMed] [Google Scholar]
- 211.Maarouf M, Platto JF, Shi VY. The role of nutrition in inflammatory pilosebaceous disorders: implication of the skin‐gut axis. Australas J Dermatol. 2018:60(2. doi: 10.1111/ajd.12909. [DOI] [PubMed] [Google Scholar]
- 212.Principi M, Cassano N, Contaldo A, Iannone A, Losurdo G, Barone M, Mastrolonardo M, Vena GA, Ierardi E, Di Leo A. Hydradenitis suppurativa and inflammatory bowel disease: an unusual, but existing association. World J Gastroenterol. 2016;22(20):4802. doi: 10.3748/wjg.v22.i20.4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Eppinga H, Sperna Weiland CJ, Thio HB, van der Woude CJ, Nijsten TEC, Peppelenbosch MP, Konstantinov SR. Similar depletion of protective faecalibacterium prausnitzii in psoriasis and inflammatory bowel disease, but not in hidradenitis suppurativa. J Crohn’s Colitis. 2016;10(9):1067–1075. doi: 10.1093/ecco-jcc/jjw070. [DOI] [PubMed] [Google Scholar]
- 214.Mintoff D, Borg I, Pace NP. The clinical relevance of the microbiome in hidradenitis suppurativa: a systematic review. Vaccines. 2021;9(10):1076. doi: 10.3390/vaccines9101076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Molnar J, Mallonee CJ, Stanisic D, Homme RP, George AK, Singh M, Tyagi SC. Hidradenitis Suppurativa and 1-carbon metabolism: role of gut microbiome, matrix metalloproteinases, and hyperhomocysteinemia. Front Immunol. 2020;11:11. doi: 10.3389/fimmu.2020.01730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ring HC, Thorsen J, Fuursted K, Bjarnsholt T, Bay L, Saunte DM, Thomsen SF, Jemec GB. Probiotics in hidradenitis suppurativa: a potential treatment option? Clin Exp Dermatol. 2021;47(1):139–141. doi: 10.1111/ced.14838. [DOI] [PubMed] [Google Scholar]
- 217.Kurzen H, Kurzen M. Secondary prevention of hidradenitis suppurativa. Dermatol Rep. 2019;11(2). doi: 10.4081/dr.2019.8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ueda O, Hasegawa M, Kitamura S. Distribution in skin of ceramide after oral administration to rats. Drug Metab Pharmacokinet. 2009;24(2):180–184. doi: 10.2133/dmpk.24.180. [DOI] [PubMed] [Google Scholar]
- 219.Clapp M, Aurora N, Herrera L, Bhatia M, Wilen E, Wakefield S. Gut microbiota’s effect on mental health: the gut-brain axis. Clini Pract. 2017;7(4):131–136. doi: 10.4081/cp.2017.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Edwards RA, Vega AA, Norman HM, Ohaeri M, Levi K, Dinsdale EA, Cinek O, Aziz RK, McNair K, Barr JJ, et al. Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat Microbiol. 2019;4(10):1727–1736. doi: 10.1038/s41564-019-0494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Hasan N, Yang H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ. 2019;7:e7502. doi: 10.7717/peerj.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Lee ES, Song EJ, Nam YD, Lee SY. Probiotics in human health and disease: from nutribiotics to pharmabiotics. J Microbiol. 2018;56(11):773–782. doi: 10.1007/s12275-018-8293-y. [DOI] [PubMed] [Google Scholar]
- 223.Lee SY, Lee E, Park YM, Hong SJ. Microbiome in the gut-skin axis in atopic dermatitis. Allergy Asthma Immunol Res. 2018;10(4):354. doi: 10.4168/aair.2018.10.4.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, Adams H, van Ree R, Stobberingh EE. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA birth cohort study. Gut. 2007;56(5):661–667. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Penders J, Stobberingh EE, Brandt PAVD, Thijs C. The role of the intestinal microbiota in the development of atopic disorders. Allergy. 2007;62(11):1223–1236. doi: 10.1111/j.1398-9995.2007.01462.x. [DOI] [PubMed] [Google Scholar]
- 226.Zhang M, Jiang Z, Li D, Jiang D, Wu Y, Ren H, Peng H, Lai Y. Oral antibiotic treatment induces skin microbiota dysbiosis and influences wound healing. Microb Ecol. 2015;69(2):415–421. doi: 10.1007/s00248-014-0504-4. [DOI] [PubMed] [Google Scholar]
- 227.Zhang YJ, Li S, Gan RY, Zhou T, Xu DP, Li HB. Impacts of gut bacteria on human health and diseases. Int J Mol Sci. 2015;16(12):7493–7519. doi: 10.3390/ijms16047493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Delday M, Mulder I, Logan ET, Grant G. Bacteroides thetaiotaomicron ameliorates colon inflammation in preclinical models of Crohn’s disease. Inflamm Bowel Dis. 2018;25(1):85–96. doi: 10.1093/ibd/izy281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Silva YP, Bernardi A, Frozza RL. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front Endocrinol (Lausanne). 2020;11:11. doi: 10.3389/fendo.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Akiyama T, Iodi Carstens M, Carstens E. Transmitters and pathways mediating inhibition of spinal itch-signaling neurons by scratching and other counterstimuli. PLoS ONE. 2011;6(7):e22665. doi: 10.1371/journal.pone.0022665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Li W, Yosipovitch G. The role of the microbiome and microbiome-derived metabolites in atopic dermatitis and non-histaminergic itch. Am J Clin Dermatol. 2020;21(S1):44–50. doi: 10.1007/s40257-020-00538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Miyazaki K, Masuoka N, Kano M, Iizuka R. Bifidobacterium fermented milk and galacto-oligosaccharides lead to improved skin health by decreasing phenols production by gut microbiota. Benef Microbes. 2014;5(2):121–128. doi: 10.3920/bm2012.0066. [DOI] [PubMed] [Google Scholar]
- 233.Climent E, Martinez-Blanch JF, Llobregat L, Ruzafa-Costas B, Carrión-Gutiérrez MN, Ramírez-Boscá A, Prieto-Merino D, Genovés S, Codoñer FM, Ramón D, et al. Changes in gut microbiota correlates with response to treatment with probiotics in patients with atopic dermatitis. a post hoc analysis of a clinical trial. Microorganisms. 2021;9(4):854. doi: 10.3390/microorganisms9040854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Navarro-López V, Ramírez-Boscá A, Ramón-Vidal D, Ruzafa-Costas B, Genovés-Martínez S, Chenoll-Cuadros E, Carrión-Gutiérrez M, Horga De La Parte J, Prieto-Merino D, Codoñer-Cortés FM. Effect of oral administration of a mixture of probiotic strains on scorad index and use of topical steroids in young patients with moderate atopic dermatitis. JAMA Dermatol. 2018;154(1):37. doi: 10.1001/jamadermatol.2017.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Blanchet-Réthoré S, Bourdès V, Mercenier A, Haddar CH, Verhoeven PO, Andres P. Effect of a lotion containing the heat-treated probiotic strain Lactobacillus johnsonii NCC 533 on Staphylococcus aureus colonization in atopic dermatitis. Clin Cosmet Investig Dermatol. 2017;10:249–257. doi: 10.2147/ccid.s135529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Fabbrocini G, Bertona M, Picazo Ó, Pareja-Galeano H, Monfrecola G, Emanuele E. Supplementation with Lactobacillus rhamnosus SP1 normalises skin expression of genes implicated in insulin signalling and improves adult acne. Benef Microbes. 2016;7(5):625–630. doi: 10.3920/bm2016.0089. [DOI] [PubMed] [Google Scholar]
- 237.Mori N, Kano M, Masuoka N, Konno T, Suzuki Y, Miyazaki K, Ueki Y. Effect of probiotic and prebiotic fermented milk on skin and intestinal conditions in healthy young female students. Biosci Microbiota, Food and Health. 2016;35(3):105–112. doi: 10.12938/bmfh.2015-022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Lee DE, Huh CS, Ra J, Choi ID, Jeong JW, Kim SH, Ryu JH, Seo YK, Koh JS, Lee JH, et al. Clinical evidence of effects of Lactobacillus plantarum HY7714 on skin aging: a randomized, double blind, placebo-controlled study. J Microbiol Biotechnol. 2015;25(12):2160–2168. doi: 10.4014/jmb.1509.09021. [DOI] [PubMed] [Google Scholar]
- 239.Enomoto T, Sowa M, Nishimori K, Shimazu S, Yoshida A, Yamada K, Furukawa F, Nakagawa T, Yanagisawa N, Iwabuchi N, et al. Effects of bifidobacterial supplementation to pregnant women and infants in the prevention of allergy development in infants and on fecal microbiota. Allergol Int. 2014;63(4):575–585. doi: 10.2332/allergolint.13-oa-0683. [DOI] [PubMed] [Google Scholar]
- 240.Jung GW, Tse JE, Guiha I, Rao J. Prospective, randomized, open-label trial comparing the safety, efficacy, and tolerability of an acne treatment regimen with and without a probiotic supplement and minocycline in subjects with mild to moderate acne. J Cutan Med Surg. 2013;17(2):114–122. doi: 10.2310/7750.2012.12026. [DOI] [PubMed] [Google Scholar]
- 241.Kimoto-Nira H, Aoki R, Sasaki K, Suzuki C, Mizumachi K. Oral intake of heat-killed cells of Lactococcus lactis strain H61 promotes skin health in women. J Nutr Sci. 2012:1. doi: 10.1017/jns.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Vijayashankar M, Raghunath N. Pustular psoriasis responding to probiotics – a new insight. Our Dermatol Online. 2012;3(4):326–329. doi: 10.7241/ourd.20124.71. [DOI] [Google Scholar]
- 243.Torii S, Torii A, Itoh K, Urisu A, Terada A, Fujisawa T, Yamada K, Suzuki H, Ishida Y, Nakamura F, et al. Effects of oral administration of Lactobacillus acidophilus L-92 on the symptoms and serum markers of atopic dermatitis in children. Int Arch Allergy Immunol. 2011;154(3):236–245. doi: 10.1159/000321110. [DOI] [PubMed] [Google Scholar]
- 244.Kim J, Ko Y, Park YK, Kim NI, Ha WK, Cho Y. Dietary effect of lactoferrin-enriched fermented milk on skin surface lipid and clinical improvement of acne vulgaris. Nutrition. 2010;26(9):902–909. doi: 10.1016/j.nut.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 245.Guéniche A, Bastien P, Ovigne JM, Kermici M, Courchay G, Chevalier V, Breton L, Castiel-Higounenc I. Bifidobacterium longum lysate, a new ingredient for reactive skin. Exp Dermatol. 2010;19(8):1–8. doi: 10.1111/j.1600-0625.2009.00932.x. [DOI] [PubMed] [Google Scholar]
- 246.Kang BS, Seo JG, Lee GS, Kim JH, Kim SY, Han YW, Kang H, Kim HO, Rhee JH, Chung MJ, et al. Antimicrobial activity of enterocins from Enterococcus faecalis SL-5 against propionibacterium acnes, the causative agent in acne vulgaris, and its therapeutic effect. J Microbiol. 2009;47(1):101–109. doi: 10.1007/s12275-008-0179-y. [DOI] [PubMed] [Google Scholar]
- 247.Wickens K, Black PN, Stanley TV, Mitchell E, Fitzharris P, Tannock GW, Purdie G, Crane J. A differential effect of 2 probiotics in the prevention of eczema and atopy: a double-blind, randomized, placebo-controlled trial. J Allergy Clin Immunol. 2008;122(4):788–794. doi: 10.1016/j.jaci.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 248.Sistek D, Kelly R, Wickens K, Stanley T, Fitzharris P, Crane J. Is the effect of probiotics on atopic dermatitis confined to food sensitized children? Clin Exp Allergy. 2006;36(5):629–633. doi: 10.1111/j.1365-2222.2006.02485.x. [DOI] [PubMed] [Google Scholar]
- 249.Weston S. Effects of probiotics on atopic dermatitis: a randomised controlled trial. Arch Dis Child. 2005;90(9):892–897. doi: 10.1136/adc.2004.060673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Rosenfeldt V, Benfeldt E, Valerius NH, Pærregaard A, and Michaelsen KF. Effect of probiotics on gastrointestinal symptoms and small intestinal permeability in children with atopic dermatitis. J Pediatr. 2004;145(5):612–616. doi: 10.1016/j.jpeds.2004.06.068. [DOI] [PubMed] [Google Scholar]
- 251.Rautava S, Kalliomäki M, Isolauri E. Probiotics during pregnancy and breast-feeding might confer immunomodulatory protection against atopic disease in the infant. J Allergy Clin Immunol. 2002;109(1):119–121. doi: 10.1067/mai.2002.120273. [DOI] [PubMed] [Google Scholar]
- 252.Kalliomäki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. The Lancet. 2001;357(9262):1076–1079. doi: 10.1016/s0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 253.Isolauri E, Arvola T, Sütas Y, Moilanen E, Salminen S. Probiotics in the management of atopic eczema. Clini Exp Allergy. 2000;30(11):1605–1610. doi: 10.1046/j.1365-2222.2000.00943.x. [DOI] [PubMed] [Google Scholar]


