Abstract
The visual sensory system is essential for animals to perceive their environment and is thus under strong selection. In aquatic environments, light intensity and spectrum differ primarily along a depth gradient. Rhodopsin (RH1) is the only opsin responsible for dim‐light vision in vertebrates and has been shown to evolve in response to the respective light conditions, including along a water depth gradient in fishes. In this study, we examined the diversity and sequence evolution of RH1 in virtually the entire adaptive radiation of cichlid fishes in Lake Tanganyika, focusing on adaptations to the environmental light with respect to depth. We show that Tanganyikan cichlid genomes contain a single copy of RH1. The 76 variable amino acid sites detected in RH1 across the radiation were not uniformly distributed along the protein sequence, and 31 of these variable sites show signals of positive selection. Moreover, the amino acid substitutions at 15 positively selected sites appeared to be depth‐related, including three key tuning sites that directly mediate shifts in the peak spectral sensitivity, one site involved in protein stability and 11 sites that may be functionally important on the basis of their physicochemical properties. Among the strongest candidate sites for deep‐water adaptations are two known key tuning sites (positions 292 and 299) and three newly identified variable sites (37, 104 and 290). Our study, which is the first comprehensive analysis of RH1 evolution in a massive adaptive radiation of cichlid fishes, provides novel insights into the evolution of RH1 in a freshwater environment.
Keywords: freshwater fish, opsin, photic environment, rod photoreceptor, spectral tuning, vision
1. INTRODUCTION
The survival of animals depends heavily on their ability to perceive their surroundings (Escobar‐Camacho & Carleton, 2015). Among the different sensory systems, vision allows animals to orientate themselves, to escape from predators, to find shelter, to sense potential food items, to communicate amongst each other, to find mates, to choose a habitat and/or to explore new environments (Escobar‐Camacho & Carleton, 2015; Musilova et al., 2021). The visual sensory system is thus under strong selection (e.g., Schott et al., 2014; Terai et al., 2002; Torres‐Dowdall et al., 2015) and visual adaptations have been implicated with diversification in different terrestrial and aquatic taxa (Boughman, 2002; Maan et al., 2006; Seehausen et al., 2008; Terai et al., 2006; Wright et al., 2020).
In vertebrates, two types of photoreceptor cells located in the retina—cones and rods—are typically responsible for bright‐light (photopic) and dim‐light (scotopic) vision, respectively (Bowmaker, 2008; but see Musilova et al., 2021). Cones and rods harbour visual pigments, which are composed of an opsin protein bound to a light‐sensitive vitamin A‐derived, nonprotein chromophore (Bowmaker, 2008). Each visual pigment absorbs a specific waveband in the range of the ultraviolet (UV) to the red light spectrum, and can be characterized by its maximal spectral sensitivity ( ) (Carleton et al., 2020). Vertebrates possess up to four main types of cone opsins, the short‐wavelength sensitive SWS1 and SWS2, the middle‐wavelength sensitive RH2, and the long‐wavelength sensitive LWS, plus one rod opsin (rhodopsin, RH1; Bowmaker, 2008; Musilova et al., 2021; Musilova, Cortesi, et al., 2019). These opsins belong to a larger gene family, the transmembrane G protein‐coupled receptors (GPCRs), which initiate signalling pathways through the activation of G proteins. GPCRs feature largely conserved structural motifs including seven transmembrane alpha‐helices (TM) that form a ligand‐binding pocket (Bowmaker, 2008; Palczewski et al., 2000; Terakita, 2005). In visual opsin proteins, the ligand‐binding pocket is called retinal binding pocket and binds to the chromophore via a retinal‐binding site (located in TM VII) and a Schiff base counterion (in TM III; Bowmaker, 2008; Palczewski et al., 2000; Terakita, 2005). In addition, a disulphide bridge between two amino acid sites maintains the organization of the seven‐helix transmembrane motif by bringing together TM III and the extracellular loop between TM IV and V. Visual opsin proteins are hence characterized by conserved amino acid sites (including those responsible for structural stability and chromophore binding) and highly variable sites, whereby a number of amino acid changes at these latter sites have been suggested to be adaptive by fine‐tuning the visual sensitivity to match the local light environment (Bowmaker, 2008; Carleton et al., 2020; Hauser & Chang, 2017; Musilova et al., 2021). Next to amino acid substitutions, adaptive changes in opsins may also occur through gene duplication or loss, or the expression of different opsin palettes (e.g., Carleton et al., 2020; Hauser & Chang, 2017; Lin et al., 2017; Musilova, Indermaur, et al., 2019). Thus, different strategies can be applied to assess whether changes in visual opsin proteins are adaptive and correlate with ecology, including positive selection analyses and photoreceptor sensitivity (
) (Carleton et al., 2020). Vertebrates possess up to four main types of cone opsins, the short‐wavelength sensitive SWS1 and SWS2, the middle‐wavelength sensitive RH2, and the long‐wavelength sensitive LWS, plus one rod opsin (rhodopsin, RH1; Bowmaker, 2008; Musilova et al., 2021; Musilova, Cortesi, et al., 2019). These opsins belong to a larger gene family, the transmembrane G protein‐coupled receptors (GPCRs), which initiate signalling pathways through the activation of G proteins. GPCRs feature largely conserved structural motifs including seven transmembrane alpha‐helices (TM) that form a ligand‐binding pocket (Bowmaker, 2008; Palczewski et al., 2000; Terakita, 2005). In visual opsin proteins, the ligand‐binding pocket is called retinal binding pocket and binds to the chromophore via a retinal‐binding site (located in TM VII) and a Schiff base counterion (in TM III; Bowmaker, 2008; Palczewski et al., 2000; Terakita, 2005). In addition, a disulphide bridge between two amino acid sites maintains the organization of the seven‐helix transmembrane motif by bringing together TM III and the extracellular loop between TM IV and V. Visual opsin proteins are hence characterized by conserved amino acid sites (including those responsible for structural stability and chromophore binding) and highly variable sites, whereby a number of amino acid changes at these latter sites have been suggested to be adaptive by fine‐tuning the visual sensitivity to match the local light environment (Bowmaker, 2008; Carleton et al., 2020; Hauser & Chang, 2017; Musilova et al., 2021). Next to amino acid substitutions, adaptive changes in opsins may also occur through gene duplication or loss, or the expression of different opsin palettes (e.g., Carleton et al., 2020; Hauser & Chang, 2017; Lin et al., 2017; Musilova, Indermaur, et al., 2019). Thus, different strategies can be applied to assess whether changes in visual opsin proteins are adaptive and correlate with ecology, including positive selection analyses and photoreceptor sensitivity ( ) estimations (reviewed by Carleton et al., 2020).
) estimations (reviewed by Carleton et al., 2020).
In aquatic environments, the visual scenes vary dramatically depending on several different biotic and abiotic factors (Munz & McFarland, 1977; Warrant & Locket, 2004; Yokoyama & Yokoyama, 1996). As light (mostly provided by the sun) passes through the water column, it is continuously absorbed and scattered, resulting in an increasingly narrowed light spectrum with depth. As a consequence, shallow‐water species are exposed to UV and a broader range of the visible light spectrum compared to those living in deep waters where blue and blue–green light dominates (Yokoyama & Yokoyama, 1996). In addition, rivers, lakes and oceans have different optical properties that affect light transmission and absorption in the water column (Yokoyama & Yokoyama, 1996). For example, the presence of phytoplankton and dissolved organic material as well as turbidity reduce light penetration, most notably in inshore and inland water bodies. Accordingly, the visual system of fish has been shown to evolve and adapt both anatomically and physiologically to divergent light environments (Carleton et al., 2020; Davies et al., 2012; Musilova, Cortesi, et al., 2019; Seehausen et al., 2008). For instance, deep‐sea fishes have evolved different phenotypic adaptations to catch more photons, such as big eyes, tubular eye structures, reflective tapeta that are wavelength‐selective, multilayer retinae and lenses containing yellow pigment (Musilova, Cortesi, et al., 2019; Warrant & Locket, 2004).
RH1 is the single opsin in rods and is responsible for perceiving the achromatic visual information from the environment, typically featuring a  ‐value around 500 nm (i.e., in the blue–green part of the light spectrum; Bowmaker, 2008; Hauser & Chang, 2017; Munz & McFarland, 1977). RH1 is the best studied opsin gene and the first GPCR that had its crystal structure determined (Palczewski et al., 2000). Structural conformation studies found that RH1 exists as dimers and that the oligomeric state of RH1 is important for the activation of G proteins (Fotiadis et al., 2003; Guo et al., 2005). Further, through phylogenetic comparative analyses and in vitro protein reconstructions, specific amino acid substitutions at so‐called “key tuning” sites have been identified that change
‐value around 500 nm (i.e., in the blue–green part of the light spectrum; Bowmaker, 2008; Hauser & Chang, 2017; Munz & McFarland, 1977). RH1 is the best studied opsin gene and the first GPCR that had its crystal structure determined (Palczewski et al., 2000). Structural conformation studies found that RH1 exists as dimers and that the oligomeric state of RH1 is important for the activation of G proteins (Fotiadis et al., 2003; Guo et al., 2005). Further, through phylogenetic comparative analyses and in vitro protein reconstructions, specific amino acid substitutions at so‐called “key tuning” sites have been identified that change  and, hence, the function of RH1 (Hunt et al., 2014; Sugawara et al., 2005; Yokoyama et al., 2008). Previous studies also found that deep‐water living fishes show specific molecular adaptations and/or have expanded their RH1 repertoire to efficiently adjust their visual system to the light environment at depth (Hunt et al., 1996; Musilova, Cortesi, et al., 2019; Nagai et al., 2011; Sugawara et al., 2005). For example, more than 20 amino acid substitutions at key tuning sites have been identified that mediate a shift in
and, hence, the function of RH1 (Hunt et al., 2014; Sugawara et al., 2005; Yokoyama et al., 2008). Previous studies also found that deep‐water living fishes show specific molecular adaptations and/or have expanded their RH1 repertoire to efficiently adjust their visual system to the light environment at depth (Hunt et al., 1996; Musilova, Cortesi, et al., 2019; Nagai et al., 2011; Sugawara et al., 2005). For example, more than 20 amino acid substitutions at key tuning sites have been identified that mediate a shift in  towards shorter wavelengths (blue‐shift), and the emergence of differently tuned RH1 copies has been suggested to increase visual sensitivity in deep waters (Hofmann et al., 2009; Musilova, Cortesi, et al., 2019; Sugawara et al., 2005). Such adaptive changes generally optimize visual sensitivity by matching the peak absorbance of RH1 with the peak wavelength of the environmental light (blue and blue–green light in deep waters, as compared to blue‐to‐red light in shallow waters; Carleton et al., 2020; Yokoyama, 2008). Finally, some fish species living in deep waters, where hydrostatic pressure is elevated, were shown to feature particular amino acid substitutions in RH1 associated with protein stability (Porter et al., 2016). More precisely, the steadiness of the protein structural conformation (opsin dimers) and, hence, the function of RH1 is maintained in deep‐water fishes through a reduction of the protein's adiabatic compressibility (Porter et al., 2016).
towards shorter wavelengths (blue‐shift), and the emergence of differently tuned RH1 copies has been suggested to increase visual sensitivity in deep waters (Hofmann et al., 2009; Musilova, Cortesi, et al., 2019; Sugawara et al., 2005). Such adaptive changes generally optimize visual sensitivity by matching the peak absorbance of RH1 with the peak wavelength of the environmental light (blue and blue–green light in deep waters, as compared to blue‐to‐red light in shallow waters; Carleton et al., 2020; Yokoyama, 2008). Finally, some fish species living in deep waters, where hydrostatic pressure is elevated, were shown to feature particular amino acid substitutions in RH1 associated with protein stability (Porter et al., 2016). More precisely, the steadiness of the protein structural conformation (opsin dimers) and, hence, the function of RH1 is maintained in deep‐water fishes through a reduction of the protein's adiabatic compressibility (Porter et al., 2016).
Cichlid fishes (Cichliformes, Cichlidae), which are currently undergoing dramatic adaptive radiations, most notably in the African Great Lakes, are an important model system in evolutionary biology (Kocher, 2004; McGee et al., 2020; Salzburger, 2018). These radiations have resulted in arrays of closely related species living in various habitats, including at different depths. Cichlids thus also emerge as a great model system to investigate the visual sensory system in general and visual opsin genes in particular (e.g., Escobar‐Camacho & Carleton, 2015; Musilova, Indermaur, et al., 2019; Musilova et al., 2021; Schneider et al., 2020; Torres‐Dowdall et al., 2015; Wright et al., 2020). Previous studies investigated the achromatic visual system of a handful of East African cichlid species (Malinsky et al., 2015; Miyagi et al., 2012; Nagai et al., 2011; Schott et al., 2014; Sugawara et al., 2005; Terai et al., 2017; Torres‐Dowdall et al., 2015). A common finding so far is that cichlid species living at depth have evolved similar molecular changes at key tuning sites that may shift  towards shorter wavelengths (blue‐shift; Sugawara et al., 2005) to better match the peak wavelength of the environmental light. On a small subset of Lake Tanganyika (LT) cichlid species, Nagai et al. (2011) found that most deep‐water species have a serine (S) at amino acid position 292, while the majority of shallow‐water species feature an alanine (A). Note that the substitution A292S is predicted to shift
towards shorter wavelengths (blue‐shift; Sugawara et al., 2005) to better match the peak wavelength of the environmental light. On a small subset of Lake Tanganyika (LT) cichlid species, Nagai et al. (2011) found that most deep‐water species have a serine (S) at amino acid position 292, while the majority of shallow‐water species feature an alanine (A). Note that the substitution A292S is predicted to shift  by about −9 nm and has therefore been suggested to mediate a depth‐related adaptation (Musilova, Cortesi, et al., 2019; Yokoyama et al., 2008).
by about −9 nm and has therefore been suggested to mediate a depth‐related adaptation (Musilova, Cortesi, et al., 2019; Yokoyama et al., 2008).
In this study, we investigated the molecular evolution of RH1 in virtually the entire adaptive radiation of cichlid fishes from LT, which represents the ecologically, morphologically and behaviourally most diverse cichlid radiation (Ronco et al., 2021; Salzburger, 2018; Salzburger et al., 2014). Making use of available whole genome sequences of nearly all cichlid species occurring in the lake, we (i) identified and newly assembled RH1 gene sequences and screened for possible gene duplications; (ii) quantified the diversity of both nucleotide and amino acid sequences of RH1; (iii) tested if environmentally selective pressures have shaped RH1 protein sequence evolution; and (iv) screened for candidate amino acid substitutions that are associated with depth and hence potentially represent depth‐related adaptations. Our investigation of about 250 closely related taxa provides comprehensive insights into the evolution of RH1 in a freshwater environment.
2. MATERIALS AND METHODS
2.1. Data set
For this study, we revisited the Illumina sequence data from our previous work, in which we had produced whole genome sequences for a nearly taxonomically complete sample of the cichlid fish fauna of LT (raw sequencing data are available on NCBI under the BioProject accession no. PRJNA550295, https://www.ncbi.nlm.nih.gov/bioproject/; Ronco et al., 2021). Making use of the available raw DNA reads, we newly assembled the intron‐less RH1 coding sequence (CDS). Our sample included sequence data for 245 taxa belonging to 12 tribes that are part of the adaptive radiation of cichlid fishes in LT, 16 riverine species nested within the Tanganyikan cichlid radiation (from the tribes Haplochromini, Serranochromini and Lamprologini), four Haplochromini species occurring in LT and adjacent rivers, one Oreochromini species that is a secondary colonizer to LT, and five riverine outgroup species (from the tribes Gobiocichlini, Heterotilapini, Tilapiini and Steatocranini; Table S1). In total, we thus used raw read data from 517 specimens, representing one to four male and female individuals of 271 cichlid species in 19 tribes. For depth‐related analyses, we refined the species‐specific habitat categories from Ronco et al. (2021) into four categories of depth of occurrence, resulting in 61 shallow‐water living species (primarily occurring at a depth of 0–10 m); 119 intermediate‐water living species (10–20 m); 42 deep‐water species (>20 m); and 50 species living at unknown depth or for which the above categories would not capture the ecology of the species adequately (e.g., species occurring in highly variable depth ranges). For analytical reasons, only the categories shallow‐ and deep‐water living species were used in the positive selection tests with the branch‐site model (see Section 2.6) and in the depth‐related substitutions analysis (see Section 2.7).
2.2. Identification and assembly of rhodopsin
In a first step, we used bwa‐mem (version 0.7.17, Li & Durbin, 2009) to map all raw reads of each individual to the phylogenetically equidistant Nile tilapia reference genome (Oreochromis niloticus; RefSeq accession GCF_001858045.2, female) and extracted all reads that mapped to the Nile tilapia RH1 CDS (1065 bp). To improve the mapping, we removed the existing trim annotations and performed a second, individual‐based, assembly with geneious (version 2020.1.2, www.geneious.com, parameters: dissolve contigs and re‐assemble, geneious as mapper, Medium Sensitivity/Fast as Sensitivity, Iterate up to 5 times as Fine Tuning). We then obtained the consensus CDS from the individual sequence reads per specimen with geneious (parameters: 65%—strict as Threshold, Total as Assign quality) and performed a multiple sequence alignment of all consensus CDS plus the reference CDS of Nile tilapia with mafft (version 7.310, Katoh & Standley, 2013). One individual of Neolamprologus multifasciatus (Neomul, IRF8, Lamprologini) had an additional base in the form of an “N” at nucleotide position 843. However, since the other specimen of this species did not feature this “N” (nor did any other sequence), we deleted this position in specimen IRF8. We then translated every CDS into amino acids (AA) and checked for the presence of start/stop codons, early stop codons or sequence lengths not dividable by 3. Five sequences had early stop codons and were consequently excluded from the downstream analyses (no species had to be removed from the original data set). The resulting 512 RH1 AA sequences were intact with a length of 355 AA.
2.3. Rhodopsin copy number
To screen for potential duplications of RH1 (see Musilova, Cortesi, et al., 2019), we performed blastn searches (E‐value cut‐off of 1e‐5; version 2.2.28, https://blast.ncbi.nlm.nih.gov) in all available Tanganyikan cichlid draft genome assemblies (n = 509, see Ronco et al., 2021), using the newly assembled RH1 CDS as queries. As the extra‐ocular rhodopsin 1 (exoRH1) is highly similar to RH1 (72.39% CDS sequence identity and 74.37% AA sequence identity in the reference genome, see Mano et al., 1999), we used the same workflow to confirm the presence of both genes, RH1 and exoRH1, in the available draft assemblies. As an additional strategy to test for possible RH1 duplicates, we determined, for each cichlid specimen, the ratio of the median coverage (of mapped reads) in the reference's RH1 coding region vs. the median coverage of the entire genome. The coverage of the RH1 coding region was calculated using samtools (version 1.7, Li et al., 2009). To obtain an estimate of the genome‐wide coverage, we first calculated, for each genome, the coverage distribution of mapped reads to the reference genome using bedtools (version 2.27.1, Quinlan & Hall, 2010). We then only retained positions with (i) a coverage of at least 1, to not deflate the estimate by regions that are not present in the reference, and (ii) below 50, to exclude sites that are highly duplicated or where mapping is ambiguous (note that this is an arbitrary threshold set after visual inspection of the mapping distributions). Finally, we also manually inspected the raw read alignments to search for an excess of heterozygous sites, which could indicate functionally different RH1 copies.
2.4. Rhodopsin gene trees
To decrease the computational complexity of the phylogenetic analyses, we first reduced the multiple alignments for both CDS and AA to unique sequences. This resulted in 238 unique CDS and 158 unique AA sequences (including the reference sequence). Ambiguous characters in the AA alignment (due to heterozygous nucleotide sites) were masked by “X.” We then used jmodeltest (version 2.1.10, Darriba et al., 2012) and paup* (version 4.0a, Wilgenbusch & Swofford, 2003) to identify the most appropriate nucleotide substitution models, which turned out to be the GTR+G+I model in both cases (based on Akaike Information Criterion [AIC] values). prottest (version 4.3.2, Darriba et al., 2011) was used to determine the best protein substitution model (JTT+I+G+F, according to AIC). CDS and AA phylogenies were generated using mrbayes (version 3.2.7, Ronquist & Huelsenbeck, 2003) and iq‐tree (version 2.0, Nguyen et al., 2014). Bayesian inference calculations with mrbayes were performed with four chains, 107 generations with a sample frequency of 1000 and a burn‐in of 25%. The maximum‐likelihood (ML) analyses with iq‐tree were performed with 1000 bootstrap replicates and 1000 iterations for the UFBoot stopping rule, with/without a more thorough NNI (Nearest Neighbour Interchange) search. We ran both analyses four times for CDS and AA, respectively, specifying the Nile tilapia as outgroup. The best topology of these four replicates for Bayesian inference and ML was selected on the basis of topology tests (khtest, shtest and autest) in paup*. The topology distance between trees (Robinson–Foulds distance, Robinson & Foulds, 1981) was calculated using the dist.topo function from the R package ape (version 4.0.3 and version 5.4‐1, Paradis et al., 2004; Team R Development Core, 2018). Finally, a haplotype genealogy based on the CDS multiple alignment and the best gene tree (ML) was built with haplotype viewer (http://www.cibiv.at/~greg/haploviewer, Salzburger et al., 2011) and colour‐coded according to the depth categories.
2.5. Rhodopsin amino acid substitutions
To visualize the AA changes in RH1 on the gene trees, we modified the Bayesian and ML trees built with unique CDS sequences (see above), to recover all initial CDS tip labels (reversing the merging of unique CDS sequences). A length of 10−4 was assigned to each branch for visualization purposes. We then again reduced each gene tree by pruning tip labels of individuals with identical CDS, but this time only within species (resulting in a total of 343 tip labels including the reference). Using paup* (command: describe; parameters: apoList=yes chgList=yes diagnose=yes brlens=yes), we then mapped the AA changes (ambiguous characters as “X”) on the gene trees and reported the Consistency Index (CI) and Retention Index (RI) of the respective substitutions. Gene trees were visualized with changes on branches using the R packages ape, phytools (version 0.7‐47; Revell, 2012), phangorn (version 2.5.5; Schliep, 2011), and seqinr (version 4.2‐4; Charif & Lobry, 2007). Moreover, we mapped the substitutions of variable known key tuning sites in the data set on the gene trees and investigated the potential effect of specific changes on the maximal spectral sensitivity value using the reconstructions performed in Yokoyama et al. (2008) and Musilova, Cortesi, et al. (2019). We also mapped the substitutions of variable sites known to be involved in the maintenance of RH1 protein stability along the depth gradient (Porter et al., 2016).
2.6. Positive selection analysis
Positively selected sites were identified using CodeML from the paml software package (version 4.9; Yang, 2007). To assess the impact of different tree topologies, we tested random site models (M1a vs. M2a and M7 vs. M8) and branch‐site model (H0 vs. HA) on both the Bayesian and ML gene tree. Each pair of models was compared using a Likelihood Ratio Test (LRT) with a χ2 distribution. For the branch‐site model, the background branches included species living in shallow waters (77 CDS tip labels) as opposed to those living in deep waters (58 CDS tip labels). To do so, we used a reduced version of the gene trees that included only species belonging to these two most extreme depth categories. We then filtered the CDS multiple alignment to retain only sequences that are represented in the pruned gene trees. We performed the same analyses with hyphy (version 2.3.13.20180525beta(MP); Kosakovsky Pond et al., 2005) to confirm our findings using the ML gene tree. FEL (Fixed Effects Likelihood) and FUBAR (Fast Unconstrained Bayesian AppRoximation for inferring selection) were performed to test for pervasive site‐level selection, which is equivalent to random site models in CodeML. We also tested for lineage‐specific evolution using the branch‐site method aBS‐REL (Adaptive Branch‐Site Random Effects Likelihood), which is equivalent to branch‐site models in CodeML, with shallow‐ and deep‐water living species as categories.
2.7. Depth‐related substitution analysis
To investigate putative depth‐related substitutions, we retrieved AAs at variable sites for each species living in the shallow and deep waters included in the species tree from Ronco et al. (2021) and recorded each species as a binary AA score (absence or presence of a particular AA) at each position and the binary depth score (shallow vs. deep). For each site and AA, we then fitted models of trait evolution for discrete characters based on the species tree using bayestraits (version 3.0.2; http://www.evolution.rdg.ac.uk/). More specifically, for each site we compared the likelihood of an independent model (assuming two binary traits that evolve independently along the phylogeny) and a dependent model (assuming two binary traits that evolve dependently along the phylogeny). We applied the Markov Chain Monte Carlo method with 108 iterations and a sample frequency of 2000. We then retained the last 25,000 iterations and assessed the convergence of each chain by calculating the effective sample size (ESS) for each parameter. As all chains converged (ESS > 200), we summarized each model by calculating the mean log‐likelihood of the posterior distribution and its derived AIC. Significance was assessed by comparing the difference in AIC of the independent model minus the dependent model, so that positive values indicate that the two traits probably evolved dependently along the phylogeny.
3. RESULTS
3.1. Rhodopsin diversity in Tanganyikan cichlids
We identified and newly assembled the intron‐less RH1 gene in 271 cichlid species covering the entire cichlid species flock in LT plus some outgroup taxa and species nested within the radiation. In total, 32 species were represented by one individual, 164 species by two or more individuals that had identical CDS, and 75 species were represented by two or more individuals with different CDS due to incompleteness, homozygous single nucleotide polymorphisms (SNPs) and/or individuals with heterozygous sites (Table S1). Five incomplete CDS with early stop codons were removed from the analysis: JWA8 (Lamprologus meleagris, Lamprologini), IZI8 (Neolamprologus christyi, Lamprologini), ISA1 (Trematocara marginatum, Trematocarini), JXH4 (Petrochromis orthognathus, Tropheini) and LCF3 (Tropheus sp. “kirschfleck,” Tropheini). A visual inspection revealed that all these incomplete CDS were due to regions with no coverage (i.e., lack of raw read data) leading to apparent frame shift mutations (i.e., the lack of nucleotides shifts the reading frame and thereby creates early stop codons). However, because the respective other individual of these five species had a complete CDS and could hence be used for downstream analyses, all species present in the original data set were also included in our analyses. Individuals of 18 species featured differences in the CDS between individuals due to homozygous SNPs, 44 species showed intraspecific differences due to heterozygous sites, and eight species showed differences between individuals due to both homozygous SNPs and heterozygous sites (Figure S1). The minimum CDS and AA sequence identity was 95.3% and 92.5%, respectively, among all pairs of individuals (AA heterozygous sites masked; the CDS multiple alignment is available on Dryad: https://doi.org/10.5061/dryad.4mw6m90c7). No evidence of sex‐specific differences was found across the data set. Intraspecific sequence variation was generally low or absent, except for three species that had more than one nucleotide/AA difference due to homozygous SNPs: XenniS (16/12, Xenotilapia nigrolabiata, Ectodini), Lamorn (7/6, Lamprologus ornatipinnis, Lamprologini) and TelteS (11/8, Telmatochromis temporalis, Lamprologini).
Among the 1065 nucleotide positions of RH1, 212 positions were variable across the data set (including the reference), of which 57 involved first codon positions, 37 second codon positions and 118 third codon positions (Figure S2A). This resulted in 154 variable codons and 76 variable AA sites (13 variable AA sites due to heterozygosity). Codons were represented by one to 13 codon variants across the data set (median = 1, mean = 1.814) and by one to six different AAs (median = 1, mean = 1.322, Figure S2A). Positions 162 (TM IV), 213 (TM V) and 217 (TM V) showed the greatest AA diversity with six AA variants each. The N‐terminal (extracellular side) and the seven transmembrane alpha‐helices (TM I–VII), which make up 65.35% (232/355 AA sites) of the whole protein sequence, contained most of the variable AA sites (60/76; 78.95%). The variable AA sites were not uniformly distributed along the alignment and most of them were found in the N‐terminal and TM I, IV, V, VI and VII with 10, 6, 7, 8, 10 and 9 changes, respectively (in addition to variable AA sites due to heterozygosity: one in TM IV, two in TM V and one in TM VII; Figure S2A). More than 25% of the AA sites in each of these six regions were variable (with and without positions variable due to heterozygosity; Figure S2B). For all individuals including the reference, the retinal‐binding site was fixed at K296 (TM VII), the Schiff base counterion at E113 (TM III), and the disulphide bond sites at C110 (TM III) and C187 (extracellular loop) (Bowmaker, 2008; Terakita, 2005). The conserved tripeptide sequence in TM III involved in G protein interactions was fixed at E134/R135/W136 (Menon et al., 2001). Moreover, of the 27 known key tuning sites in RH1 (Musilova, Cortesi, et al., 2019; Yokoyama et al., 2008), six positions were variable (TM II: site 83, extracellular loop: site 183, TM V: site 214, TM VII: sites 292, 299 and 300). From the AA sites known to be involved in protein stability at depth (Porter et al., 2016), two were fixed (E196 and I275), one was fixed for the majority of species (F159; except for one shallow‐water living species with a heterozygous site {FV} and one species living at unknown depth with V), while one site was highly variable (six possible AAs at position 213: A, I, L, M, S, T or {LT}).
3.2. Rhodopsin copy number
We did not find evidence for multiple RH1 copies in the Tanganyikan cichlids’ draft genome assemblies (the full blastn report is available on Dryad: https://doi.org/10.5061/dryad.4mw6m90c7). The blastn analysis with the respective RH1 CDS recovered from the raw reads as query resulted in a highest‐scoring hit to RH1 and a second highest‐scoring hit to exoRH1 for all cichlid genomes. The ratio of the median coverage on the reference's RH1 CDS vs. the median coverage on the reference's overall genome did not reveal any evidence for a duplication of RH1, except for the two Neolamprologus splendens individuals that showed a coverage in RH1 of more than twice the genome‐wide median coverage (Figure 1). However, a close inspection of the read data of the two N. splendens genomes revealed a coverage distribution with high variance when mapped to the Nile tilapia reference genome. Although many sites showed comparatively high coverage, the highest proportion of positions was covered by only four or fewer reads, pushing down the genome‐wide median coverage in these two specimens compared to the other genomes where the coverage was normally distributed with much less variance. We can attribute the much greater variation in coverage in the two N. splendens individuals to the comparatively high level of fragmentation of the extracted genomic DNA that was initially recovered from them (the original electropherograms indicated a fragment size distributed around 500–700 bp for the two N. splendens individuals, while the other samples’ fragments were typically centred around >10 kb). This, in turn, is probably a consequence of these two specimens having been collected more than 30 years ago, whereas the vast majority of the remaining Tanganyikan cichlid genomes were sequenced from recent material producing high‐quality DNA extracts (for details see Ronco et al., 2021). Furthermore, since the RH1 CDS of the two N. splendens individuals did not show any excess of heterozygous sites, as would be expected in the presence of a second, functionally different RH1 copy in a genome, we tentatively assume that the observed signals of an elevated median coverage in RH1 in N. splendens are artefacts.
FIGURE 1.
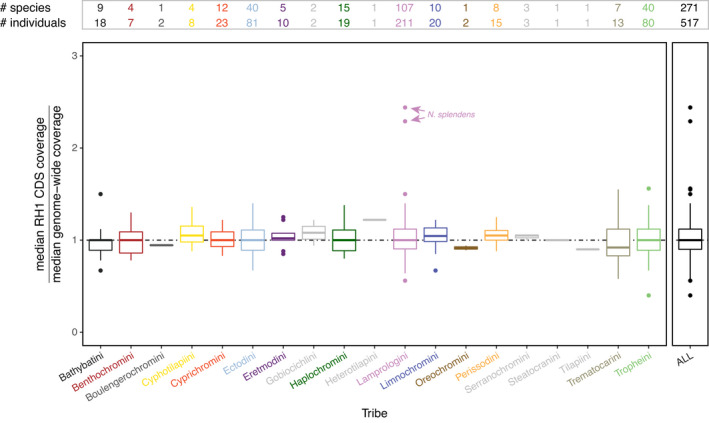
Coverage of mapped read estimates. Boxplots showing the median coverage in the CDS (coding sequence) of the rhodopsin (RH1) gene over an estimate of the median genome‐wide coverage in the cichlid fauna of Lake Tanganyika (LT) as well as in outgroup taxa. Boxplots are shown for each cichlid tribe separately and for the entire data set (ALL); the cichlid tribes occurring in LT are colour‐coded as in Ronco et al. (2021). The boxes’ centre lines show the median, box limits show first and third quartiles, and whiskers show 1.5× the interquartile ranges. Outliers are represented by dots. The horizontal dashed line in black is set to 1 (equal coverage across the whole genome). The number of species and individuals for each tribe is reported at the top. Note that the two genomes of Neolamprologus splendens (marked with arrows) show elevated ratios, which we interpret as artefacts (see Section 3.2 Rhodopsin copy number)
3.3. Rhodopsin gene trees
The phylogenetic analyses based on Bayesian and ML of the RH1 CDS and AA sequences resulted in similar tree topologies, with a Robinson–Foulds distance of 125 between gene trees and of 106 between AA trees (Figures S3–S6; the tree files are available on Dryad: https://doi.org/10.5061/dryad.4mw6m90c7). Overall, the gene trees resembled the species tree topology at the tribal and genus level (see Ronco et al., 2021), whereas species‐level relationships often remained unresolved. This is largely due to the overall short length of the RH1 CDS and AA sequences, and the relatively small number of variable positions (1/5 of the sequences) in proportion to the number of taxa analysed. For the same reasons, the Bayesian posterior probability and ML bootstrap values were low in parts. The topology tests revealed that the ML CDS and AA trees were more likely than the Bayesian trees. Therefore, we mainly present results using the ML trees in the following. The haplotype genealogy based on the ML gene tree (Figure 2) showed that, if species shared the same RH1 gene sequence (i.e., haplotype), this only occurred between species within but not between tribes (except for Tropheini and Haplochromini, but which should be considered as one clade because Tropheini is phylogenetically nested with Haplochromini; Salzburger et al., 2005). It further appears that within tribes—specifically in the Ectodini, Lamprologini and Tropheini/Haplochromini—shallow‐, intermediate‐, or deep‐water living species can share the same RH1 haplotype (Figure 2).
FIGURE 2.
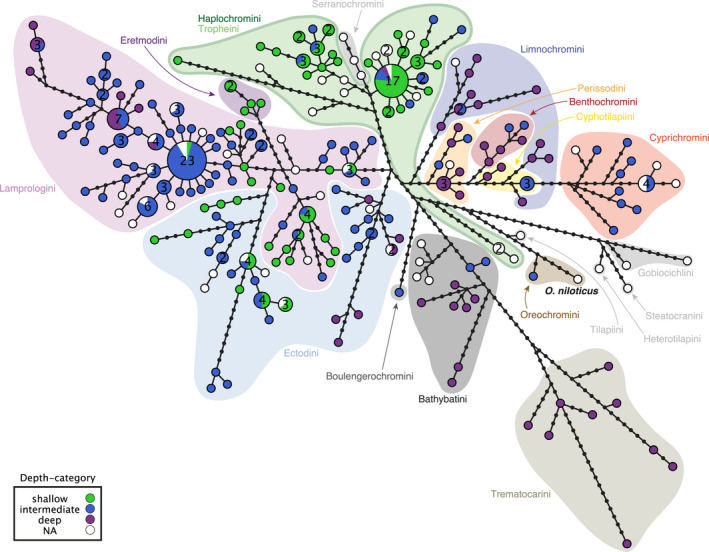
Rhodopsin nucleotide haplotype genealogy based on the maximum‐likelihood gene tree. Each pie chart represents a unique CDS (coding sequence) of the rhodopsin gene, and its size corresponds to the number of individuals that share this haplotype (whenever two or more individuals share a haplotype, the number of individuals is reported inside the pie charts). Pie charts are colour‐coded according to depth category (see box), and cichlid tribes are indicated with coloured background shadings (according to Ronco et al., 2021); the reference sequence (Oreochromis niloticus) is indicated in bold. The black dots represent hypothetical (unsampled) haplotypes. Note that Haplochromini and Tropheini have the same green background shading, because they form one clade, with Tropheini being phylogenetically nested within Haplochromini (Salzburger et al., 2005)
3.4. Rhodopsin amino acid substitutions
RH1 AA changes occurred on both internal (including close to the root) and terminal branches (Figures S3–S6). No major difference in the number and location of AA changes was found between the Bayesian and ML gene trees. Changes mapped on the ML gene tree occurred at different frequency depending on the AA sites and varied from one to 18 changes (median = 3, mean = 5, Table S2). Position 162 featured the highest number of AA changes mapped on the gene tree.
The mapping of changes at (variable) known key tuning sites (positions 83, 183, 214, 292, 299 and 300) revealed that all Tanganyikan cichlids featured at least one change in RH1 that may fine‐tune  (absent in Oreochromini species, riverine outgroup species, and three riverine Haplochromini species; Figure 3a). Of 28 changes in total, 12 substitutions mapped on the ML gene tree are likely to shift
(absent in Oreochromini species, riverine outgroup species, and three riverine Haplochromini species; Figure 3a). Of 28 changes in total, 12 substitutions mapped on the ML gene tree are likely to shift  towards shorter (blue‐shift) and longer (red‐shift) wavelength (nine and three changes, respectively, Figure S7; see Yokoyama et al., 2008 and Musilova, Cortesi, et al., 2019). Interestingly, the mapped changes at the two variable sites involved in protein stability at depth (see Porter et al., 2016) revealed a single change for AA position 159, while 15 changes were mapped at AA position 213 (Figure 3a).
towards shorter (blue‐shift) and longer (red‐shift) wavelength (nine and three changes, respectively, Figure S7; see Yokoyama et al., 2008 and Musilova, Cortesi, et al., 2019). Interestingly, the mapped changes at the two variable sites involved in protein stability at depth (see Porter et al., 2016) revealed a single change for AA position 159, while 15 changes were mapped at AA position 213 (Figure 3a).
FIGURE 3.
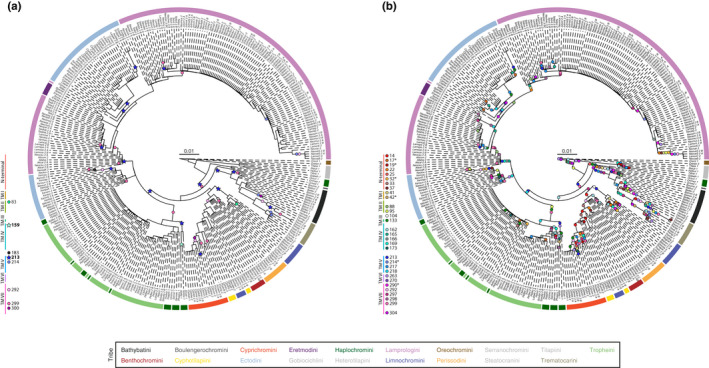
Amino acid substitutions in the rhodopsin protein mapped on the maximum‐likelihood (ML) gene tree. The coloured arches around the ML gene trees indicate the tribe to which a species belongs (see Figure S3 and Table S1 for full species names). The number and the letter(s) next to each abbreviated species name represent the number of individuals and their IDs (see Table S1 [CDS tip label]). Each coloured circle corresponds to a change occurring at a specific amino acid position along the rhodopsin protein sequence. TM: transmembrane alpha‐helix. (a) Amino acid substitutions in known key tuning sites (circles) and in sites involved in protein stability along the depth gradient (in bold, star‐shaped symbol). (b) Amino acid substitutions in positively selected sites (CodeML random‐site models M2a and M8). Positively selected sites present in M8 but not in M2a are marked with an asterisk (*)
3.5. Positive selection analysis
Variation in dN/dS (the ratio of nonsynonymous to synonymous substitutions) was found when comparing the site models M1a vs. M2a and M7 vs. M8 using both the Bayesian and ML gene trees (p < 10−4, Table 1; Table S3). The models M2a and M8 using the ML gene tree revealed similar results with 25 and 31 positively selected sites, respectively (BEB analysis, p > 95%, Figures 3b and 4; Table S4). Hence, 7.04/8.73% of sites showed signals of positive selection with an average dN/dS of 10.17/9.68 (under M2a and M8, respectively). The majority of positively selected sites were found in the N‐terminal and TM IV, V and VII (Figure 4). Interestingly, no positively selected site was detected in the intracellular loops of RH1. Positive selection results using the Bayesian tree are available in the Supporting Information (including hyphy results using the ML gene tree, Tables S3–S7).
TABLE 1.
Rhodopsin positive selection results based on a CodeML random‐site model comparison (M1a vs. M2a and M7 vs. M8a) and the maximum‐likelihood gene tree. The pairs of models were tested using a Likelihood Ratio Test (LRT) following a χ2 distribution. Values of each site class ω0 , ω1 and ω2 are specified for models M1a and M2a. The shape parameters p and q are specified for models M7 and M8. The value ωp corresponds to the positively selected site class for M8. The proportion of each site class is given in parentheses
| Parameter | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Model | np | lnL | κ | ω0 /p | ω1 /q | ω2 /ωp | Null | LRT | df | p |
| M1a | 3 | −5266 | 2.144 | 0.001 (91.7%) | 1 (8.3%) | |||||
| M2a | 5 | −4999 | 2.702 | 0.017 (89.9%) | 1 (83%) | 9.797 (1.7%) | M1a | 534 | 2 | <.0001 |
| M7 | 3 | −5314 | 2.377 | 0.007 | 0.018 | |||||
| M8 | 5 | −5005 | 2.807 | 0.028 | 0.205 | 9.548 (2%) | M7 | 618 | 2 | <.0001 |
Abbreviations: np, number of parameters; lnL, ln likelihood; κ, transition/transversion ratio; Null, null model; df, degrees of freedom.
FIGURE 4.
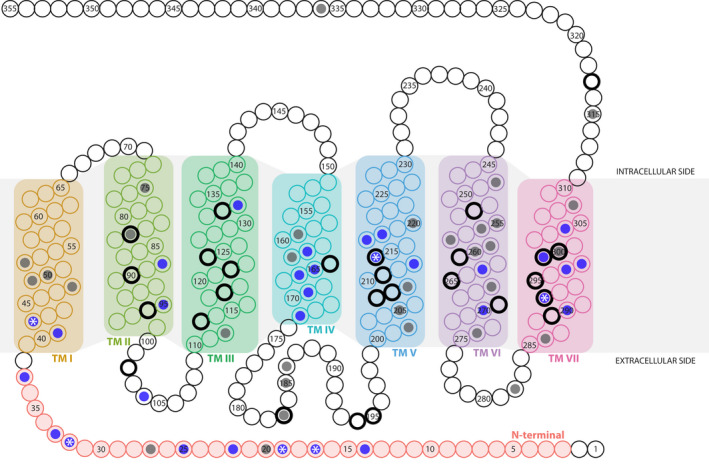
Schematic of the rhodopsin (RH1) protein (based on Oreochromis niloticus) showing the variable amino acid sites and the positively selected amino acid sites (based on CodeML random site models M1a vs. M2a and M7 vs. M8). Each circle represents an amino acid position in RH1. Variable sites in our data set are colour‐coded with a dark grey dot, and positively selected sites among those are highlighted with a blue dot. Positively selected sites present in M8 but not in M2a results are indicated with a white asterisk (*). The 27 key tuning sites in RH1 (see Musilova, Cortesi, et al., 2019; Sugawara et al., 2005) are marked with a black outer circle. The transmembrane alpha‐helices (TM) of rhodopsin are highlighted with different background colours (from left to right): TM I, II, III, IV, V, VI, VII. Sites that are part of extra‐ and intracellular loops are shown in white, except for the N‐terminal site that is colour‐coded in red
The majority of changes at positively selected sites mapped on both internal and terminal branches on the ML gene tree (Figure 3b). Moreover, substitutions in positively selected sites belonging to the same region of the protein did not appear to co‐occur on specific branches. Among the six key tuning sites variable in our data set, a single site (299) was found to be under positive selection according to M2a, and three sites (214, 292 and 299) according to M8. Moreover, among sites preserving protein stability at depth, a single site (213) was found to be under positive selection according to M2a and M8. The positive selection test using the branch‐site models H0 and HA (with both the Bayesian and ML gene trees) and the category “shallow‐water living species” as background branches and the category “deep‐water living species” as foreground branches did not reveal a difference in RH1 sequence evolution between these groups (Table S6; aBS‐REL results using hyphy were congruent with CodeML results).
3.6. Depth‐related substitution analysis
We found evidence for 23 depth‐related substitutions in our data set; that is, AAs at 15 specific RH1 positions are likely to have evolved in dependence on the depth at which a species occurs (Figure 5; Figures S8 and S9). Three of these AA positions were found in the N‐terminal (sites 17, 33 and 37), one in TM I (42) and TM II (95), one in the extracellular loop (104), two each in TM IV (162 and 165) and TM V (213 and 214), and five in TM VII (290, 292, 297, 298 and 299). In contrast, no depth‐related substitution was found in the intracellular loops and in TM III and TM VI. All detected depth‐related substitutions occurred at sites showing signals of positive selection (under M8 using the ML gene tree), including at three known key tuning sites (214, 292 and 299) and one site (213) involved in the maintenance of protein stability.
FIGURE 5.
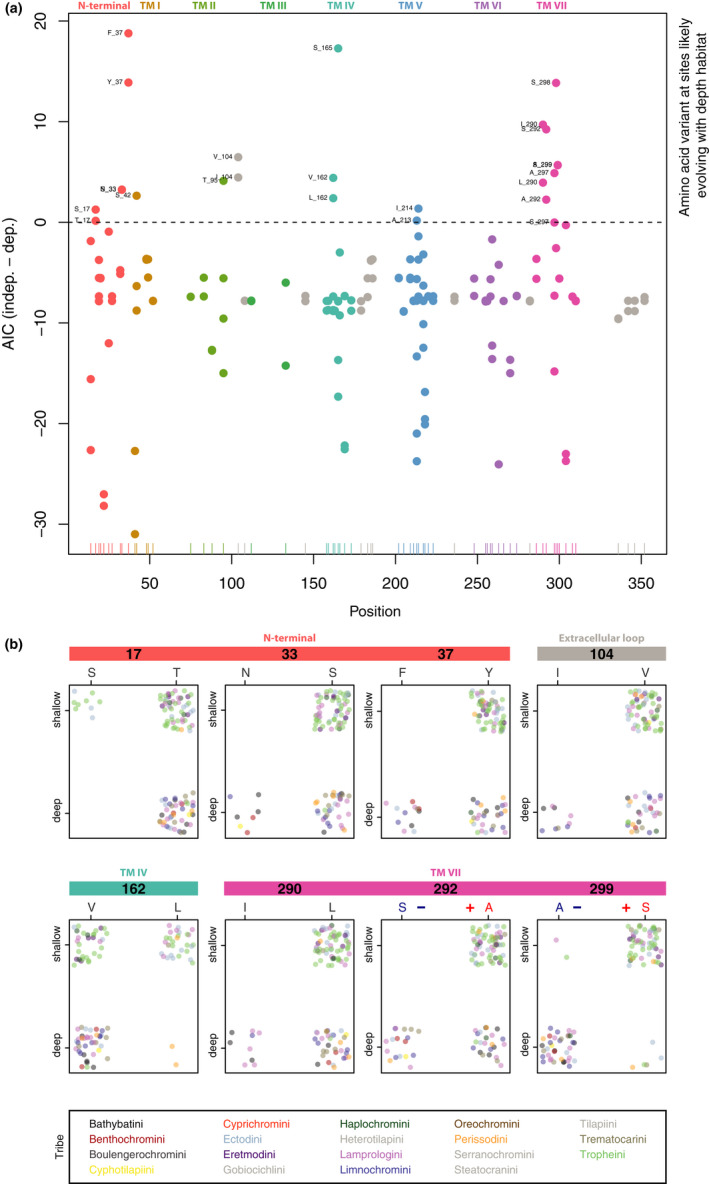
Depth‐related substitution analysis of rhodopsin (RH1) amino acid sites. (a) Dotplots of bayestraits results. Each dot corresponds to the difference in AICs of the independent model and the dependent model (absence/presence of an amino acid at a specific site in RH1 and depth at which species occur) and is colour‐coded according to the RH1 regions (TM: transmembrane alpha‐helix; see Figure 4). The x‐axis shows the positions along the RH1 protein sequence, and the y‐axis shows the difference in AICs between the two models. The horizontal dashed line is fixed at zero, meaning that dots above this threshold indicate those amino acids that are associated with the water depth at which a species occurs (shallow‐ vs. deep‐water living species). (b) Dotplots of sites with exactly two amino acid variants associated with water depth (colour‐coded as in (a)). Each individual is represented by a single dot, colour‐coded according to tribe. The x‐axis represents the amino acid variants, and the y‐axis represents the shallow‐ and deep‐water living species (plotted with jitter points and without ambiguous amino acid sites for better visualization). Note that sites 292 and 299 are among the known key tuning sites in RH1, with substitutions predicted to shift the peak spectral sensitivity (the blue “–“ symbol indicates a predicted shift towards shorter wavelengths, and the red “+” indicates a predicted shift towards longer wavelengths). An extended version of this figure showing all identified sites is provided as Figure S8
We found that, among the AA variants at sites probably evolving in dependence on the depth, the commonly observed pattern is that certain AA variants (F37, I104, I290, S292, A297 and A299, and with weaker effect also N33, S42, T95 and A213, Figure 5b; Figures S8 and S9) are found only (or predominantly) in deep‐water living species, whereas the alternative variant is found both in shallow‐ and deep‐water living species. The opposite case with AAs exclusively found in the shallow‐water living species is rarer (S17, L162, S165 and S298), and similarly, the alternative variants at these positions are then found in both deep‐ and some shallow‐water living species. For eight AA sites, we detected two AA variants associated with depth (sites 17, 33, 37, 104, 162, 290, 292 and 299; Figure 5b), of which a single AA site revealed an almost perfect bidirectional association: S299 is found in most shallow‐water living species, while A299 is found in most deep‐water living species. Our list of AA sites putatively pertinent for depth‐related adaptations thus contains positions previously suggested to be relevant in deep waters (e.g., 292 and 299) as well as candidate positions that have not previously been identified in the context of deep‐water adaptations in fishes (e.g., 37, 104 and 290).
4. DISCUSSION
Rhodopsin (RH1) is the only visual opsin type present in the rod cells of the retina, and is responsible for dim‐light vision in vertebrates (Bowmaker, 2008; but see Musilova et al., 2021). While in terrestrial vertebrates adaptive changes in their single copy of RH1 are commonly found in association with a nocturnal lifestyle or crepuscular activity patterns (e.g., Castiglione & Chang, 2018; Hauzman et al., 2017), molecular adaptations in RH1 of fishes—as the main group of vertebrates living in the aquatic realm—are most often associated with the water depth at which a species occurs, and in some cases also with water turbidity (Musilova et al., 2021). In addition to acquiring specific changes in the protein‐coding sequence of RH1 in response to the light environment in deeper waters (Hofmann et al., 2009; Hope et al., 1997; Hunt et al., 1996; Malinsky et al., 2015; Musilova, Cortesi, et al., 2019; Nagai et al., 2011; Sugawara et al., 2005), some fishes were shown to have expanded their RH1 repertoire via gene duplication and subsequent functional diversification, resulting in intraspecific arrays of differently tuned copies of RH1 (Musilova, Cortesi, et al., 2019). In the present study, we investigated the evolution of RH1 in the massive adaptive radiation of cichlids in LT, where these fishes have diversified into a multitude of ecological niches in the shallow, intermediate and deep (up to the oxycline at a depth of about 200 m; Talling, 1991) waters (Table S1), starting from a common ancestor that colonized the emerging lake about 10 million years ago (Ronco et al., 2021).
Our in‐depth analyses of RH1 in the endemic cichlid fauna of the LT basin based on whole‐genome raw sequence data of 517 specimens from 271 species revealed the presence of a single copy of RH1 per genome across the radiation (Figure 1). We note that for two genomes—namely those of the two representatives of Neolamprologus splendens—a coverage pattern in the CDS of RH1 was retrieved that, when compared to the genome‐wide median, would be compatible with a gene duplication scenario for RH1 (Figure 1). However, upon closer inspection, we tentatively argue that the seemingly higher coverage in the CDS of RH1 in N. splendens is an artefact. Thus, unlike what has been found in deep‐sea fishes and in some species of freshwater fishes living in murky waters (Musilova, Cortesi, et al., 2019; Musilova et al., 2021), the adaptation to scotopic light conditions does not seem to have involved lineage‐ or species‐specific duplications of RH1 in LT cichlids.
When focusing on RH1 sequence evolution in the adaptive radiation of cichlid fishes in LT, we identified a total of 237 unique CDS (haplotypes) and 157 unique AA sequences in the data set (excluding the reference sequence in both cases). In the Bayesian and ML gene trees reconstructed from these data, haplotypes and AA sequences clustered according to tribes and genera—thus reflecting phylogenetic relationships as established in a species‐tree analysis from genome‐wide SNPs (Ronco et al., 2021)—but not according to depth at which a species occurs, nor to feeding ecology (Figure 2; Figures S3 and S4). This is best illustrated by the haplotype genealogy based on unique CDS of the RH1 gene in LT cichlids (Figure 2), highlighting that no single RH1 haplotype is shared between tribes (except between Haplochromini and Tropheini, which belong to the same clade with Tropheini being phylogenetically nested in Haplochromini; Salzburger et al., 2005), whereas within tribes, RH1 haplotypes are occasionally shared between depth categories. When individually mapped on the species tree (Ronco et al., 2021), we found that 56.7% (89 out of 157) of the AA substitutions occurred more than once (up to 20 times; the species tree with mapped AA changes is available on Dryad: https://doi.org/10.5061/dryad.4mw6m90c7), again highlighting that convergent evolution is common in cichlid adaptive radiations (Muschick et al., 2012), in this case on the molecular level. Some of these convergent cases, especially among more closely related species, could also be the result of introgression and/or incomplete lineage sorting (Salzburger, 2018).
Intraspecific sequence variation in RH1 was very low or absent for the vast majority of species, except for three species that featured noticeable degrees of intraspecific sequence variation between the two specimens examined (Figure S1). There is no obvious reason—for example, with respect to phylogeny, ecology, demography, behaviour or morphology—that could explain why these three species are more diverse in RH1 than others. To some extent, however, intraspecific variation should be expected in instances of adaptive radiation, which are characterized by incomplete lineage sorting and occasional hybridization (see Salzburger, 2018). Lastly, there was no indication for sex‐specific nucleotide differences across the data set.
At the level of the RH1 protein, we found that, out of a total of 355 AA positions, 76 sites were variable across the data set (Figure 4; Figure S2). These included six out of the 27 known key tuning sites (Musilova, Cortesi, et al., 2019; Yokoyama et al., 2008)—namely sites 83, 183, 214, 292, 299 and 300 (Figure S2A)—and two AA sites (159 and 213) implicated in protein stability (Porter et al., 2016). Close to half of the variable AA sites (31) were additionally identified as having evolved under positive selection according to the random site model (M8) in paml (Figure 3b; Table S4). Among these were three key tuning sites (214, 292 and 299; Musilova, Cortesi, et al., 2019; Yokoyama et al., 2008) and one site (213) involved in the maintenance of protein stability at depth (Figure 3a; Porter et al., 2016). The proportion of sites showing a signal of positive selection in RH1 of Tanganyikan cichlids (8.73%; Table 1), as well as the average dN/dS (9.68), is in line with what has previously been reported for this group of fishes: examining RH1 in 16 cichlid species from lakes Victoria, Malawi and Tanganyika (three from LT plus Oreochromis niloticus in our data set) and using a neighbour‐joining tree as a backbone, Spady et al. (2005) found a proportion of positively selected sites of 6.9% and an average dN/dS of 14.07; and Schott et al. (2014) found that 7.1% of the sites were under positive selection with an average dN/dS of 13.69 in a set of 32 African Great Lake cichlids (20 from LT in our data set) and a Bayesian gene tree as backbone (and 6.6% positively selected sites and a dN/dS of 14.37 when using an ML gene tree). A comparison between their (Schott et al., 2014; Spady et al., 2005) and our new findings (Table S4) reveals that 15 positively selected sites (41, 42, 95, 133, 162, 165, 166, 169, 213, 217, 218, 263, 297, 298 and 299) are common to the three studies.
The distribution of the variable (and also of the positively selected) AA sites along the protein was not random in our data set, with the majority of variable sites (60/76) being located in the N‐terminal (extracellular side) and the transmembrane alpha‐helices I, IV, V, VI and VII (Figure 4; Figure S2A and B), and most (23/31) of the positively selected sites in the N‐terminal and TM IV, V and VII (Figure 4). On the other hand, TM II and III, and the intracellular side of RH1 were found to be rather conserved and to have primarily evolved under purifying selection in LT cichlids. Also, the AAs at the retinal‐binding site 296, the Schiff base counterion at position 113, and at the disulphide bond sites 110 and 187 were conserved in all cichlid species included in this study, as well as in Nile tilapia, and congruent with what has been reported previously (Bowmaker, 2008; Menon et al., 2001; Terakita, 2005). This suggests that—at least over short evolutionary timescales—some AA sites of RH1 are, more than others, involved in rapid adaptive evolution, which may in part be explained by functional constraints on the respective other regions. For example, the conserved TM III not only contains the disulphide bond site 110 and a fixed tripeptide motif (sites 134, 135 and 136; Menon et al., 2001), but also seven AA sites involved in the formation of the retinal binding pocket (positions 114, 117, 120 and 121, and key tuning sites 113, 118 and 122; Musilova, Cortesi, et al., 2019; Ou et al., 2011; Yokoyama, 2008). By contrast, the regions of RH1 around the dimerization interface (TM IV and V; Fotiadis et al., 2003; Guo et al., 2005) and adjacent to the retinal binding site (position in TM VII) as well as the N‐terminal region emerge as mutational hotspots and main targets of positive selection in LT cichlids (Figure 4).
Because, in fishes, adaptive evolution in RH1 is known to be impacted by the ambient light environment along the depth gradient (Hunt et al., 1996; Musilova, Cortesi, et al., 2019; Nagai et al., 2011; Sugawara et al., 2005), and because the roughly 250 cichlid species in LT cover a vast range of niches including from shallow to deep waters, we were particularly interested in depth‐related adaptations in RH1 in this spectacular example of adaptive radiation. We thus applied phylogenetic comparative methods to test for potential associations between the variable AAs and depth, and, in doing so, identified 23 substitutions at 15 AA sites that are putatively involved in deep‐water visual adaptations in LT cichlids (Figure 5a; Figure S8). This list contains AA sites previously suggested to be involved in deep‐water adaptations, including three previously known key tuning sites (214, 292 and 299) and one site (213) likely to be involved in the maintenance of protein stability (Hope et al., 1997; Hunt et al., 1996, 2001; Malinsky et al., 2015; Musilova, Cortesi, et al., 2019; Nagai et al., 2011; Porter et al., 2016; Sugawara et al., 2005; Varela & Ritchie, 2015), as well as 11 novel candidate sites that have not yet been implicated with living at depth and for which the exact functions are currently unknown (sites 17, 33, 37, 42, 95, 104, 162, 165, 290, 297 and 298) (Figure 5a; Figures S8 and S9). Our results may thus serve as a starting point for future functional tests to determine the effect of these particular AA substitutions on  of RH1.
of RH1.
Importantly, all AA substitutions in RH1 that we identified as candidates for deep‐water adaptations in the cichlid adaptive radiation of LT also show signals of positive selection (Figure 3; Table S4) and occurred repeatedly over the course of the radiation (https://doi.org/10.5061/dryad.4mw6m90c7). Some of these sites, and specific AA substitutions associated with them, emerge as particularly strong candidates for deep‐water adaptations, based on what is already known about their function (or their occurrence) in other deep‐living species of fish. For example, previous work in both deep‐sea and deep‐living freshwater fishes revealed specific depth‐related molecular adaptations at key tuning sites in RH1 (Hope et al., 1997; Hunt et al., 1996, 2001; Malinsky et al., 2015; Nagai et al., 2011; Varela & Ritchie, 2015), including N83, S292 and A299, which all mediate a blue‐shift in  that is considered adaptive at depth. In our data set, all species featured D83, except for the deep‐water living Baileychromis centropomoides (N83) and two out of three Bathybates vittatus individuals (deep‐water living; heterozygous site with {DN}); all shallow‐water living species had A292, while deep‐water living species had either A292 or S292; and most shallow‐water living species had S299, while most deep‐water living species had A299 (Figure 5b; Figures S8 and S9). This suggests strongly that also in deep‐water living LT cichlids the key tuning sites 292 and 299 were involved in adjusting the visual system to the light environment at depth. AA substitutions at sites other than key tuning sites have also been implicated in depth‐related adaptations in fishes. Malinsky et al. (2015), for example, identified AAs at four positions to be associated with the deep‐water benthic cichlid ecomorph in crater lake Massoko. We found that three of them (V162, S166, A298) are also present in the majority of deep‐water living cichlid species in LT, while there is a difference in the fourth one (G297 in the benthic ectomorph in Lake Massoko vs. predominantly A297 in deep‐water living species in LT; Figure 5B; Figures S8 and S9). It is of note that V162, which was also among the three sites with the largest number of different AAs across the data set (the others being 213 and 217; Figures S2 and S9), was also retrieved with the depth‐related substitution analysis (just as A297 and S298 were). Without doubt, however, it would be necessary to determine the spectral sensitivity properties of the newly identified variants of RH1 by measuring the absorption spectrum of the RH1 pigments (as, e.g., reported in Sugawara et al., 2005), to confirm whether these indeed mediate the hypothesized blue‐shift of
that is considered adaptive at depth. In our data set, all species featured D83, except for the deep‐water living Baileychromis centropomoides (N83) and two out of three Bathybates vittatus individuals (deep‐water living; heterozygous site with {DN}); all shallow‐water living species had A292, while deep‐water living species had either A292 or S292; and most shallow‐water living species had S299, while most deep‐water living species had A299 (Figure 5b; Figures S8 and S9). This suggests strongly that also in deep‐water living LT cichlids the key tuning sites 292 and 299 were involved in adjusting the visual system to the light environment at depth. AA substitutions at sites other than key tuning sites have also been implicated in depth‐related adaptations in fishes. Malinsky et al. (2015), for example, identified AAs at four positions to be associated with the deep‐water benthic cichlid ecomorph in crater lake Massoko. We found that three of them (V162, S166, A298) are also present in the majority of deep‐water living cichlid species in LT, while there is a difference in the fourth one (G297 in the benthic ectomorph in Lake Massoko vs. predominantly A297 in deep‐water living species in LT; Figure 5B; Figures S8 and S9). It is of note that V162, which was also among the three sites with the largest number of different AAs across the data set (the others being 213 and 217; Figures S2 and S9), was also retrieved with the depth‐related substitution analysis (just as A297 and S298 were). Without doubt, however, it would be necessary to determine the spectral sensitivity properties of the newly identified variants of RH1 by measuring the absorption spectrum of the RH1 pigments (as, e.g., reported in Sugawara et al., 2005), to confirm whether these indeed mediate the hypothesized blue‐shift of  in deep‐water living Tanganyikan cichlid species or contribute to protein stability at depth.
in deep‐water living Tanganyikan cichlid species or contribute to protein stability at depth.
5. CONCLUSION
Cichlid fishes are an important model group to investigate the visual sensory system in general and visual opsin genes in particular in the aquatic environment. In this study, we present the first all‐inclusive analysis of RH1 molecular evolution in an entire massive adaptive radiation, that of cichlid fishes in LT. Our in‐depth genomic investigations revealed the presence of a single copy of RH1 per genome across the cichlid fauna of LT. The AA differences identified across the radiation were not uniformly distributed along the protein, and 31 out of the 76 variable AA sites showed signatures of positive selection. Six out of the 27 known key tuning sites in RH1 are variable in LT cichlids, of which three are likely to have evolved under positive selection. Through phylogenetic comparative analyses, we identified 23 AA substitutions at 15 sites that are associated with water depth. These include three known key‐tuning sites, one site with a putative function in protein stability with respect to water depth, as well as 11 novel candidate sites for deep‐water adaptations in (cichlid) fishes. Importantly, all the candidates we identified based on the depth‐related substitution analyses also emerged as potentially important sites based on our molecular evolutionary inferences, phylogenetic comparisons and positive selection tests. Together, our integrative study on the molecular evolution of the visual system of cichlid fishes in LT provides a comprehensive view on the patterns of RH1 evolution in a freshwater environment and more generally on the evolutionary dynamics of environmentally driven adaptations.
AUTHOR CONTRIBUTIONS
V.R., W.S. and Z.M. designed the project. V.R. performed the rhodopsin analyses under the supervision of F.R., Z.M. and W.S. V.R. and F.R. performed the coverage calculations and the depth‐related substitution analysis. V.R. and W.S. wrote the initial manuscript draft. All authors commented on the manuscript and approved the final version.
Supporting information
Supplementary Material
Fig S2
Fig S8
ACKNOWLEDGEMENTS
We thank N. Boileau, F. Cortesi, A. Fages and P. Tschopp for valuable feedback on the project and the analyses; N. Boileau and B. Stelbrink for their help with mrbayes; R. A. W. Wiberg for advice with CodeML; A. Indermaur for his help categorizing shallow‐, intermediate‐ and deep‐water living species; and the Subject Editor as well as two anonymous reviewers for valuable comments. Calculations were performed at sciCORE (https://scicore.unibas.ch/), the Center for Scientific Computing at the University of Basel with support from the Swiss Institute of Bioinformatics (SIB). This work was supported by the European Research Council (ERC, Consolidator Grant No. 617585 “CICHLID~X” to W.S.), the Swiss National Science Foundation (SNF; grant 176039 to W.S.; grant PROMYS 166550 to Z.M.), Charles University (Primus to Z.M.) and the Czech Science Foundation (21‐31712S to Z.M.). Open access funding provided by Universitat Basel.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
Ricci, V. , Ronco, F. , Musilova, Z. , & Salzburger, W. (2022). Molecular evolution and depth‐related adaptations of rhodopsin in the adaptive radiation of cichlid fishes in Lake Tanganyika. Molecular Ecology, 31, 2882–2897. 10.1111/mec.16429
Handling Editor: Philine G. D. Feulner
Contributor Information
Virginie Ricci, Email: virginie.ricci@unibas.ch.
Walter Salzburger, Email: walter.salzburger@unibas.ch.
DATA AVAILABILITY STATEMENT
The sequenced genomes and their raw reads are available from NCBI under the BioProject accession no. PRJNA550295 (https://www.ncbi.nlm.nih.gov/bioproject/; Ronco et al., 2021). blastn results, gene multiple alignments and phylogenetic trees have been deposited on Dryad (https://doi.org/10.5061/dryad.4mw6m90c7). Scripts as well as data have been deposited on GitHub (https://github.com/Ninet93/RH1_Ricci_et_al.git).
REFERENCES
- Boughman, J. W. (2002). How sensory drive can promote speciation. Trends in Ecology and Evolution, 17(12), 571–577. 10.1016/S0169-5347(02)02595-8 [DOI] [Google Scholar]
- Bowmaker, J. K. (2008). Evolution of vertebrate visual pigments. Vision Research, 48, 2022–2041. 10.1016/j.visres.2008.03.025 [DOI] [PubMed] [Google Scholar]
- Carleton, K. L. , Escobar‐Camacho, D. , Stieb, S. M. , Cortesi, F. , & Marshall, N. J. (2020). Seeing the rainbow: Mechanisms underlying spectral sensitivity in teleost fishes. Journal of Experimental Biology, 223(8), jeb193334. 10.1242/jeb.193334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglione, G. M. , & Chang, B. S. W. (2018). Functional trade‐offs and environmental variation shaped ancient trajectories in the evolution of dim‐light vision. eLife, 7, 1–30. 10.7554/eLife.35957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charif, D. , & Lobry, J. R. (2007). SeqinR 1.0‐2: A contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. In Bastolla U., Porto M., Roman H. E., & Vendruscolo M. (Eds.), Structural approaches to sequence evolution: Molecules, networks, populations, series Biological and Medical Physics, Biomedical Engineering (pp. 207–232). Springer Verlag. [Google Scholar]
- Darriba, D. , Taboada, G. L. , Doallo, R. , & Posada, D. (2011). ProtTest 3: Fast selection of best‐fit models of protein evolution. Bioinformatics, 27(8), 1164–1165. 10.1093/bioinformatics/btr088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba, D. , Taboada, G. L. , Doallo, R. , & Posada, D. (2012). JModelTest 2: More models, new heuristics and parallel computing. Nature Methods, 9(8), 772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, W. I. L. , Collin, S. P. , & Hunt, D. M. (2012). Molecular ecology and adaptation of visual photopigments in craniates. Molecular Ecology, 21(13), 3121–3158. 10.1111/j.1365-294X.2012.05617.x [DOI] [PubMed] [Google Scholar]
- Escobar‐Camacho, D. , & Carleton, K. L. (2015). Sensory modalities in cichlid fish behavior. Current Opinion in Behavioral Sciences, 6, 115–124. 10.1016/j.cobeha.2015.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotiadis, D. , Liang, Y. , Filipek, S. , Saperstein, D. A. , Engel, A. , & Palczewski, K. (2003). Rhodopsin dimers in native disc membranes. Nature, 421(6919), 127–128. [DOI] [PubMed] [Google Scholar]
- Guo, W. , Shi, L. , Filizola, M. , Weinstein, H. , & Javitch, J. A. (2005). Crosstalk in G protein‐coupled receptors: Changes at the transmembrane homodimer interface determine activation. Proceedings of the National Academy of Sciences of the United States of America, 102(48), 17495–17500. 10.1073/pnas.0508950102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser, F. E. , & Chang, B. S. (2017). Insights into visual pigment adaptation and diversity from model ecological and evolutionary systems. Current Opinion in Genetics and Development, 47(November), 110–120. 10.1016/j.gde.2017.09.005 [DOI] [PubMed] [Google Scholar]
- Hauzman, E. , Bonci, D. M. O. , Suárez‐Villota, E. Y. , Neitz, M. , & Ventura, D. F. (2017). Daily activity patterns influence retinal morphology, signatures of selection, and spectral tuning of opsin genes in colubrid snakes. BMC Evolutionary Biology, 17(1), 1–14. 10.1186/s12862-017-1110-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, C. M. , O’Quin, K. E. , Marshall, N. J. , Cronin, T. W. , Seehausen, O. , & Carleton, K. L. (2009). The eyes have it: Regulatory and structural changes both underlie cichlid visual pigment diversity. PLoS Biology, 7(12), e1000266. 10.1371/journal.pbio.1000266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope, A. J. , Partridge, J. C. , Dulai, K. S. , & Hunt, D. M. (1997). Mechanisms of wavelength tuning in the rod opsins of deep‐sea fishes. Proceedings of the Royal Society B: Biological Sciences, 264(1379), 155–163. 10.1098/rspb.1997.0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, D. M. , & Collin, S. P. (2014). The evolution of photoreceptors and visual photopigments in vertebrates. In Hunt D. M., Hankins M. W., Collin S. P. & Marshall N. J. (Eds.), Evolution of visual and non‐visual pigments (pp. 163–217). 10.1007/978-1-4614-4355-1 [DOI] [Google Scholar]
- Hunt, D. M. , Dulai, K. S. , Partridge, J. C. , Cottrill, P. , & Bowmaker, J. K. (2001). The molecular basis for spectral tuning of rod visual pigments in deep‐sea fish. Journal of Experimental Biology, 204(19), 3333–3344. 10.1242/jeb.204.19.3333 [DOI] [PubMed] [Google Scholar]
- Hunt, D. M. , Fitzgibbon, J. , Slobodyanyuk, S. J. , & Bowmaker, J. K. (1996). Spectral tuning and molecular evolution of rod visual pigments in the species flock of cottoid fish in Lake Baikal. Vision Research, 36(9), 1217–1224. 10.1016/0042-6989(95)00228-6 [DOI] [PubMed] [Google Scholar]
- Katoh, K. , & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30(4), 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher, T. D. (2004). Adaptive evolution and explosive speciation: The cichlid fish model. Nature Reviews Genetics, 5(4), 288–298. 10.1038/nrg1316 [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond, S. L. , Frost, S. D. W. , & Muse, S. V. (2005). HyPhy: Hypothesis testing using phylogenies. Bioinformatics, 21(5), 676–679. 10.1093/bioinformatics/bti079 [DOI] [PubMed] [Google Scholar]
- Li, H. , & Durbin, R. (2009). Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics, 25(14), 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. , Marth, G. , Abecasis, G. , & Durbin, R. (2009). The sequence alignment/map format and SAMtools. Bioinformatics, 25(16), 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J. J. , Wang, F. Y. , Li, W. H. , & Wang, T. Y. (2017). The rises and falls of opsin genes in 59 ray‐finned fish genomes and their implications for environmental adaptation. Scientific Reports, 7(1), 1–13. 10.1038/s41598-017-15868-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maan, M. E. , Hofker, K. D. , Van Alphen, J. J. M. , & Seehausen, O. (2006). Sensory drive in cichlid speciation. The American Naturalist, 167(6), 947–954. 10.1086/503532 [DOI] [PubMed] [Google Scholar]
- Malinsky, M. , Challis, R. J. , Tyers, A. M. , Schiffels, S. , Terai, Y. , Ngatunga, B. P. , Miska, E. A. , Durbin, R. , Genner, M. J. , & Turner, G. F. (2015). Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science, 350(6267), 1493–1498. 10.1126/science.aac9927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano, H. , Kojima, D. , & Fukada, Y. (1999). Exo‐rhodopsin: A novel rhodopsin expressed in the zebrafish pineal gland. Molecular Brain Research, 73(1–2), 110–118. 10.1016/S0169-328X(99)00242-9 [DOI] [PubMed] [Google Scholar]
- McGee, M. D. , Borstein, S. R. , Meier, J. I. , Marques, D. A. , Mwaiko, S. , Taabu, A. , Kishe, M. A. , O’Meara, B. , Bruggmann, R. , Excoffier, L. , & Seehausen, O. (2020). The ecological and genomic basis of explosive adaptive radiation. Nature, 586, 75–79. 10.1038/s41586-020-2652-7 [DOI] [PubMed] [Google Scholar]
- Menon, S. T. , Han, M. , & Sakmar, T. P. (2001). Rhodopsin: Structural basis of molecular physiology. Physiological Reviews, 81(4), 1659–1688. 10.1152/physrev.2001.81.4.1659 [DOI] [PubMed] [Google Scholar]
- Miyagi, R. , Terai, Y. , Aibara, M. , Sugawara, T. , Imai, H. , Tachida, H. , Mzighani, S. I. , Okitsu, T. , Wada, A. , & Okada, N. (2012). Correlation between nuptial colors and visual sensitivities tuned by opsins leads to species richness in sympatric Lake Victoria cichlid fishes. Molecular Biology and Evolution, 29(11), 3281–3296. 10.1093/molbev/mss139 [DOI] [PubMed] [Google Scholar]
- Munz, F. W. , & McFarland, W. N. (1977). Evolutionary adaptations of fishes to the photic environment. In Crescitelli F. (Ed.), The visual system in vertebrates (pp. 193–274). Springer Verlag. 10.1007/978-3-642-66468-7_4 [DOI] [Google Scholar]
- Muschick, M. , Indermaur, A. , & Salzburger, W. (2012). Convergent evolution within an adaptive radiation of cichlid fishes. Current Biology, 22(24), 10.1016/j.cub.2012.10.048 [DOI] [PubMed] [Google Scholar]
- Musilova, Z. , Cortesi, F. , Matschiner, M. , Davies, W. I. L. , Patel, J. S. , Stieb, S. M. , De Busserolles, F. , Malmstrøm, M. , Tørresen, O. K. , Brown, C. J. , Mountford, J. K. , Hanel, R. , Stenkamp, D. L. , Jakobsen, K. S. , Carleton, K. L. , Jentoft, S. , Marshall, J. , & Salzburger, W. (2019). Vision using multiple distinct rod opsins in deep‐sea fishes. Science, 364(6440), 588–592. 10.1126/science.aav4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musilova, Z. , Indermaur, A. , Bitja‐Nyom, A. R. , Omelchenko, D. , Kłodawska, M. , Albergati, L. , Remišová, K. , & Salzburger, W. (2019). Evolution of the visual sensory system in cichlid fishes from crater lake Barombi Mbo in Cameroon. Molecular Ecology, 28, 5010–5031. 10.1111/mec.15217 [DOI] [PubMed] [Google Scholar]
- Musilova, Z. , Salzburger, W. , & Cortesi, F. (2021). The visual opsin gene repertoires of teleost fishes: Evolution, ecology, and function. Annual Review of Cell and Developmental Biology, 37(1), 441–468. 10.1146/annurev-cellbio-120219-024915 [DOI] [PubMed] [Google Scholar]
- Nagai, H. , Terai, Y. , Sugawara, T. , Imai, H. , Nishihara, H. , Hori, M. , & Okada, N. (2011). Reverse evolution in RH1 for adaptation of cichlids to water depth in Lake Tanganyika. Molecular Biology and Evolution, 28(6), 1769–1776. 10.1093/molbev/msq344 [DOI] [PubMed] [Google Scholar]
- Nguyen, L. T. , Schmidt, H. A. , Von Haeseler, A. , & Minh, B. Q. (2014). IQ‐TREE: A fast and effective stochastic algorithm for estimating maximum‐likelihood phylogenies. Molecular Biology and Evolution, 32(1), 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, W.‐b. , Yi, T. , Kim, J. M. , & Khorana, H. G . (2011). The roles of transmembrane domain helix‐III during rhodopsin photoactivation. PLoS One, 6(2), 1–13. 10.1371/journal.pone.0017398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski, K. , Kumasaka, T. , Hori, T. , Behnke, C. A. , Motoshima, H. , Fox, B. A. , Le Trong, I. , Teller, D. C. , Okada, T. , Stenkamp, R. E. , Yamamoto, M. , & Miyano, M. (2000). Crystal structure of rhodopsin: A G protein‐coupled receptor. Science, 289(5480), 739–745. 10.1126/science.289.5480.739 [DOI] [PubMed] [Google Scholar]
- Paradis, E. , Claude, J. , & Strimmer, K. (2004). APE: Analyses of phylogenetics and evolution in R language. Bioinformatics, 20(2), 289–290. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- Porter, M. L. , Roberts, N. W. , & Partridge, J. C. (2016). Evolution under pressure and the adaptation of visual pigment compressibility in deep‐sea environments. Molecular Phylogenetics and Evolution, 105, 160–165. 10.1016/j.ympev.2016.08.007 [DOI] [PubMed] [Google Scholar]
- Quinlan, A. R. , & Hall, I. M. (2010). BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics, 26(6), 841–2. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell, L. J. (2012). phytools: An R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution, 3(2), 217–223. 10.1111/j.2041-210X.2011.00169.x [DOI] [Google Scholar]
- Robinson, D. F. , & Foulds, L. R. (1981). Comparison of phylogenetic trees. Mathematical Biosciences, 53(1–2), 10.1016/0025-5564(81)90043-2 [DOI] [Google Scholar]
- Ronco, F. , Matschiner, M. , Böhne, A. , Boila, A. , Büscher, H. H. , El Taher, A. , Indermaur, A. , Malinsky, M. , Ricci, V. , Kahmen, A. , Jentoft, S. , & Salzburger, W. (2021). Drivers and dynamics of a massive adaptive radiation in cichlid fishes. Nature, 589(February), 76–81. 10.1038/s41586-020-2930-4 [DOI] [PubMed] [Google Scholar]
- Ronquist, F. , & Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19(12), 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Salzburger, W. (2018). Understanding explosive diversification through cichlid fish genomics. Nature Reviews Genetics, 19(11), 705–717. 10.1038/s41576-018-0043-9 [DOI] [PubMed] [Google Scholar]
- Salzburger, W. , Bocxlaer, B. V. , & Cohen, A. S. (2014). Ecology and evolution of the African great lakes and their faunas. Annual Review of Ecology, Evolution, and Systematics, 45, 519–545. 10.1146/annurev-ecolsys-120213-091804 [DOI] [Google Scholar]
- Salzburger, W. , Ewing, G. B. , & Von Haeseler, A. (2011). The performance of phylogenetic algorithms in estimating haplotype genealogies with migration. Molecular Ecology, 20(9), 1952–1963. 10.1111/j.1365-294X.2011.05066.x [DOI] [PubMed] [Google Scholar]
- Salzburger, W. , Mack, T. , Verheyen, E. , & Meyer, A. (2005). Out of Tanganyika: Genesis, explosive speciation, key‐innovations and phylogeography of the haplochromine cichlid fishes. BMC Evolutionary Biology, 5(1983), 1–15. 10.1186/1471-2148-5-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep, K. P. (2011). phangorn: Phylogenetic analysis in R. Bioinformatics, 27(4), 592–593. 10.1093/bioinformatics/btq706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, R. F. , Rometsch, S. J. , Torres‐Dowdall, J. , & Meyer, A. (2020). Habitat light sets the boundaries for the rapid evolution of cichlid fish vision, while sexual selection can tune it within those limits. Molecular Ecology, 29(8), 1476–1493. 10.1111/mec.15416 [DOI] [PubMed] [Google Scholar]
- Schott, R. K. , Refvik, S. P. , Hauser, F. E. , López‐Fernández, H. , & Chang, B. S. W. (2014). Divergent positive selection in rhodopsin from lake and riverine cichlid fishes. Molecular Biology and Evolution, 31(5), 1149–1165. 10.1093/molbev/msu064 [DOI] [PubMed] [Google Scholar]
- Seehausen, O. , Terai, Y. , Magalhaes, I. S. , Carleton, K. L. , Mrosso, H. D. J. , Miyagi, R. , Van Der Sluijs, I. , Schneider, M. V. , Maan, M. E. , Tachida, H. , Imai, H. , & Okada, N. (2008). Speciation through sensory drive in cichlid fish. Nature, 455(7213), 620–626. 10.1038/nature07285 [DOI] [PubMed] [Google Scholar]
- Spady, T. C. , Seehausen, O. , Loew, E. R. , Jordan, R. C. , Kocher, T. D. , & Carleton, K. L. (2005). Adaptive molecular evolution in the opsin genes of rapidly speciating cichlid species. Molecular Biology and Evolution, 22(6), 1412–1422. 10.1093/molbev/msi137 [DOI] [PubMed] [Google Scholar]
- Sugawara, T. , Terai, Y. , Imai, H. , Turner, G. F. , Koblmüller, S. , Sturmbauer, C. , Shichida, Y. , & Okada, N. (2005). Parallelism of amino acid changes at the RH1 affecting spectral sensitivity among deep‐water cichlids from Lakes Tanganyika and Malawi. Proceedings of the National Academy of Sciences of the United States of America, 102(15), 5448–5453. 10.1073/pnas.0405302102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talling, J. F. (1991). Lake Tanganyika and its life. Coulter G. W. (Ed.) with contributions from J.‐J. Tiercelin, A. Mondegver, R.E. Hecky and R.H. Spigel. Aquatic conservation: Marine and freshwater ecosystems (pp. 190‐191). British Museum (Natural History) Publications — Oxford University Press. 10.1016/0169-5347(91)90231-l [DOI] [Google Scholar]
- Team R Development Core (2018). A language and environment for statistical computing. R Foundation for Statistical Computing; (p. 2). https://www.R‐project.org [Google Scholar]
- Terai, Y. , Mayer, W. E. , Klein, J. , Tichy, H. , & Okada, N. (2002). The effect of selection on a long wavelength‐sensitive (LWS) opsin gene of Lake Victoria cichlid fishes. Proceedings of the National Academy of Sciences of the United States of America, 99(24), 15501–15506. 10.1073/pnas.232561099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai, Y. , Miyagi, R. , Aibara, M. , Mizoiri, S. , Imai, H. , Okitsu, T. , Wada, A. , Takahashi‐Kariyazono, S. , Sato, A. , Tichy, H. , Mrosso, H. D. J. , Mzighani, S. I. , & Okada, N. (2017). Visual adaptation in Lake Victoria cichlid fishes: Depth‐related variation of color and scotopic opsins in species from sand/mud bottoms. BMC Evolutionary Biology, 17(1), 1–12. 10.1186/s12862-017-1040-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai, Y. , Seehausen, O. , Sasaki, T. , Takahashi, K. , Mizoiri, S. , Sugawara, T. , Sato, T. , Watanabe, M. , Konijnendijk, N. , Mrosso, H. D. J. , Tachida, H. , Imai, H. , Shichida, Y. , & Okada, N. (2006). Divergent selection on opsins drives incipient speciation in Lake Victoria cichlids. PLoS Biology, 4(12), 2244–2251. 10.1371/journal.pbio.0040433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakita, A. (2005). The opsins. Genome Biology, 6(3), 1–9. 10.1186/gb-2005-6-3-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres‐Dowdall, J. , Henning, F. , Elmer, K. R. , & Meyer, A. (2015). Ecological and lineage‐specific factors drive the molecular evolution of rhodopsin in cichlid fishes. Molecular Biology and Evolution, 32(11), 2876–2882. 10.1093/molbev/msv159 [DOI] [PubMed] [Google Scholar]
- Varela, A. I. , & Ritchie, P. A. (2015). Critical amino acid replacements in the rhodopsin gene of 19 teleost species occupying different light environments from shallow‐waters to the deep‐sea. Environmental Biology of Fishes, 98, 193–200. 10.1007/s10641-014-0249-4 [DOI] [Google Scholar]
- Warrant, E. J. , & Locket, N. A. (2004). Vision in the deep sea. Biological Reviews of the Cambridge Philosophical Society, 79, 671–712. 10.1017/S1464793103006420 [DOI] [PubMed] [Google Scholar]
- Wilgenbusch, J. C. , & Swofford, D. (2003). Inferring evolutionary trees with PAUP *. Current Protocols in Bioinformatics, 00(1), 6.4.1‐6.4.28. 10.1002/0471250953.bi0604s00 [DOI] [PubMed] [Google Scholar]
- Wright, D. S. , van Eijk, R. , Schuart, L. , Seehausen, O. , Groothuis, T. G. G. , & Maan, M. E. (2020). Testing sensory drive speciation in cichlid fish: Linking light conditions to opsin expression, opsin genotype and female mate preference. Journal of Evolutionary Biology, 33, 422–434. 10.1111/jeb.13577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution, 24(8), 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Yokoyama, S. (2008). Evolution of dim‐light and color vision pigments. Annual Review of Genomics and Human Genetics, 9, 259–282. 10.1146/annurev.genom.9.081307.164228 [DOI] [PubMed] [Google Scholar]
- Yokoyama, S. , Tada, T. , Zhang, H. , & Britt, L. (2008). Elucidation of phenotypic adaptations: Molecular analyses of dim‐light vision proteins in vertebrates. Proceedings of the National Academy of Sciences of the United States of America, 105(36), 13480–13485. 10.1073/pnas.0802426105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama, S. , & Yokoyama, R. (1996). Adaptive evolution of photoreceptors and visual pigments in vertebrates. Annual Review of Ecology and Systematics, 27, 543–567. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Fig S2
Fig S8
Data Availability Statement
The sequenced genomes and their raw reads are available from NCBI under the BioProject accession no. PRJNA550295 (https://www.ncbi.nlm.nih.gov/bioproject/; Ronco et al., 2021). blastn results, gene multiple alignments and phylogenetic trees have been deposited on Dryad (https://doi.org/10.5061/dryad.4mw6m90c7). Scripts as well as data have been deposited on GitHub (https://github.com/Ninet93/RH1_Ricci_et_al.git).


