Abstract
Chemo-radiotherapy (CRT) remains the main treatment modality for non-small-cell lung cancer (NSCLC). However, its clinical efficacy is largely limited by individual variations in radio-sensitivity and radiotherapy-associated toxicity. There is an urgent need to identify genetic determinants that can explain patients’ likelihood to develop recurrence and radiotherapy-associated toxicity following CRT. In this study, we performed comprehensive genomic profiling, using a 474-cancer- and radiotherapy-related gene panel, on pretreatment biopsy samples from patients with unresectable stage III NSCLCs who underwent definitive CRT. Patients’ baseline clinical characteristics and genomic features, including tumor genetic, genomic and molecular pathway alterations, as well as single nucleotide polymorphisms (SNPs), were correlated with progression-free survival (PFS), overall survival (OS), and radiotherapy-associated pneumonitis and/or esophagitis development after CRT. A total of 122 patients were enrolled between 2014 and 2019, with 84 (69%) squamous cell carcinomas and 38 (31%) adenocarcinomas. Genetic analysis confirmed the association between the KEAP1-NRF2 pathway gene alterations and unfavorable survival outcome, and revealed alterations in FGFR family genes, MET, PTEN, and NOTCH2 as potential novel and independent risk factors of poor post-CRT survival. Combined analysis of such alterations led to improved stratification of the risk populations. In addition, patients with EGFR activating mutations or any oncogenic driver mutations exhibited improved OS. On the other hand, we also identified genetic markers in relation to radiotherapy-associated thoracic toxicity. SNPs in the DNA repair-associated XRCC5 (rs3835) and XRCC1 (rs25487) were associated with an increased risk of high-grade esophagitis and pneumonitis respectively. MTHFR (rs1801133) and NQO1 (rs1800566) were additional risk alleles related to higher susceptibility to pneumonitis and esophagitis overall. Moreover, through their roles in genome integrity and replicative fidelity, somatic alterations in ZNF217 and POLD1 might also serve as risk predictors of high-grade pneumonitis and esophagitis. Taken together, leveraging targeted next-generating sequencing, we identified a set of novel clinically applicable biomarkers that might enable prediction of survival outcomes and risk of radiotherapy-associated thoracic toxicities. Our findings highlight the value of pre-treatment genetic testing to better inform CRT outcomes and clinical actions in stage III unresectable NSCLCs.
Keywords: non-small cell lung cancer, radiotherapy, radiation sensitivity, biomarker, genetic variation, prognosis, chemo-radiotherapy, radiotherapy-associated toxicity
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide and in China, among which approximately 85% of patients have non-small cell lung cancers (NSCLCs) (1, 2). NSCLC is sub-categorized based on histological features into mainly adenocarcinoma (ADC) and squamous cell carcinoma (SCC) (1). Over the past two decades, many therapeutic advances have been made given our deepened understanding of lung cancer etiology. The identification of actionable molecular targets has revolutionized the management of NSCLC, with targeted therapies demonstrating remarkable clinical benefits in patients carrying specific driver mutations (1). Nevertheless, the majority of lung cancer patients still require radiotherapy for cure or palliative care. In particular, for NSCLC patients with unresectable locally advanced tumors, especially SCC, the combination of chemotherapy and thoracic radiation, given either concurrently or sequentially, remains the standard of care (3).
Radiotherapy, together with the radio-sensitizing effect of chemotherapy, results in enhanced anti-tumor efficacy, although at the expense of significant normal tissue toxicity. Radiotherapy-induced lung injury (known as radiation pneumonitis in an early phase and pulmonary fibrosis in the late phase), as well as esophagitis are common adverse events following thoracic radiation (4, 5). There are considerable variations between patients in their likelihood to develop severe adverse events following a given dose of radiation, which consequently, limits the maximum dose that can be administered to the majority (6). Similarly, there are substantial differences in individual response to chemo-radiotherapy (CRT) and the risk of resistance development. It has been long recognized that genetic variations between individuals or tumors are major contributors to the differences in radio-sensitivity and risks of developing radiotherapy-associated toxicity, and thus, must be taken into consideration for personalized radiotherapy dose-prescription.
Our understanding of differential response to radiotherapy begins with the discovery of several genetic syndromes caused by mutations in the DNA repair pathways, which can lead to life-threatening radiotherapy toxicity (7, 8). Subsequently, a number of other molecular processes, such as scavenging of reactive oxygen species (ROS), apoptosis, proliferation and inflammatory response, have been implicated in the development of radiation-induced toxicity (9–12). While multiple approaches, including candidate gene approach and genome-wide association studies, were undertaken to identify genetic variants that might explain the differences in individual response to radiotherapy, no robust biomarkers with convincing clinical applicability have been identified (13). In addition, a lung tissue-specific, comprehensive analysis for personalized radiotherapy is still lacking. Here, taking advantage of next-generation sequencing (NGS) technology, we performed comprehensive genomic profiling on 474 cancer- and radio-sensitivity-related genes of the tumor biopsies from 122 unresectable stage III NSCLC patients prior to radiation therapy, and identified a set of promising biomarkers for predicting radiation survival and toxicity, which may prove beneficial for guiding clinical treatment decision-making.
Material and Methods
Patient Enrollment
The patients with NSCLC in the study were treated with CRT at the multiple centers between October 2014 and March 2019. Eligible patients for this study were determined based on the following criteria: histological diagnosis of unresectable stage IIIA-C NSCLC based on the tumor, node and metastasis (TNM) staging system without severe pleural or pericardial effusion, age older than 18 years, adequate lung, bone marrow, renal, hepatic, and cardiac function, and no history of systemic treatment or radiotherapy for thoracic cancers. The study was approved by the Ethical Review Board of the Oncology Center of Shandong Provincial Hospital, and all patients provided written informed consent.
Treatment and Assessments
All patients in this study received standard definitive CRT (dCRT). A median of five cycles of cisplatin- or paclitaxel-based chemotherapy were given concurrently or sequentially with radiotherapy. The choice of chemotherapy regimen was left to the investigator’s discretion. Three-dimensional conformal radiation therapy (3D-CRT) or intensity-modulated radiation therapy (IMRT) was administered at a total dose of 50-70 Gy.
The follow-up of all patients was conducted 1 month after radiotherapy, and then every 3 months during the first year. After that, the patients were followed up every 3-6 months. Radiotherapy-associated thoracic toxicities were graded according to the toxicity criteria of the Radiation Therapy Oncology Group (RTOG) and the European Organization for Research and Treatment of Cancer (EORTC) (14). For toxicity criteria of pneumonitis, grade 1 includes mild symptoms of dry cough or dyspnea on exertion; grade 2 includes persistent cough requiring narcotic or antitussive agents, or dyspnea with minimal effort but not at rest; grade 3 includes severe cough unresponsive to narcotic antitussive agent or dyspnea at rest, clinical or radiological evidence of acute pneumonitis, or requirement of intermittent oxygen or steroids; and grade 4 includes severe respiratory insufficiency or continuous oxygen or assisted ventilation. For esophagitis, grade 1 includes mild dysphagia or odynophagia, requirement of topical anesthetic, non-narcotic analgesics, or soft diet; grade 2 includes moderate dysphagia or odynophagia, requirement of narcotic analgesics, puree or liquid diet; grade 3 includes severe dysphagia or odynophagia with dehydration or weight loss >15% from pretreatment baseline, requirement of nasogastric feeding tube, intravenous fluids, or hyperalimentation; grade 4 includes complete obstruction, ulceration, perforation, or fistula. Treatment responses were assessed using CT imaging at each follow-up and compared to the images at baseline or from the last follow-up and were evaluated according to the Response Evaluation Criteria in Solid Tumors (RECIST), version 1.1. Progression-free survival (PFS) was defined as the time from the beginning of treatment to disease progression. Patients who had not progressed were censored at the date of their last scan. Overall survival (OS) was defined from the beginning of treatment to the time of death from any cause or the last follow-up.
DNA Extraction and Library Preparation
All tumor samples were formalin-fixed paraffin-embedded (FFPE, 10 µm) and were obtained from original biopsies prior to any treatment. At least 10% tumor content of all samples, as determined by pathologists, was required. NGS was performed in a CLIA-certified and CAP-accredited laboratory (Nanjing Geneseeq Technology Inc., Nanjing, China). Genomic DNA was extracted from de-paraffinized FFPE sections using QIAamp DNA FFPE Tissue Kit (Qiagen) according to the manufacturer’s instructions. Quantity and quality of DNA were assessed using Qubit 3.0 fluorometer and Nanodrop 2000 (ThermoFisher), respectively. DNA was fragmented into 350 bp using the Covaris M220 sonication system and purified with Agencourt AMPure XP beads (Beckman Coulter).
DNA (50 ng) libraries were prepared with KAPA hyper library preparation kit (KAPA Biosystems). Libraries with different indices were pooled for targeted enrichment with IDT xGen Lockdown Reagents and a customized enrichment panel (IDT) covering 474 cancer-related genes with whole-exon coverage, including those that have been implicated in radiotherapy response and/or radiotherapy-associated adverse effects (Radiotron®, Nanjing Geneseeq Technology Inc., Nanjing; Supplementary Table 1 ). Libraries were captured with Dynabeads M-270 (Life Technologies) and xGen Lockdown hybridization and wash kit (IDT). The captured library was further on-beads PCR amplified with Illumina p5 (5’ AAT GAT ACG GCG ACC GA 3’) and p7 (5’ CAA GCA GAA GAC GGC ATA CGA GAT 3’) primers in KAPA Hifi HotStart ReadyMix (KAPA Biosystems) and purified with Agencourt AMPure XP beads. Sequencing libraries were sized on the Agilent Bioanalyzer 2100 (Agilent Technologies) and their concentrations analyzed by qPCR with KAPA Library Quantification kit (KAPA Biosystems). The final libraries were sequenced on an Illumina Hiseq 4000 platform to a mean coverage depth of ~350.
Sequencing Data Analysis
NGS read preprocessing, including quality control of FASTQ files and removing leading/trailing low quality (quality reading below 15) or N bases, was conducted with Trimmomatic (15). Qualified pair-end reads were aligned to the reference human genome hg19 with Burrows-Wheeler Aligner (v0.7.12) (16). PCR deduplication was performed using Picard and local realignment around indels and base quality score recalibration was performed using GATK3. Samples with mean dedup depth of less than 30X were also removed. Cross-sample contamination was quantified by using ContEst (Broad Institute). Single nucleotide polymorphisms (SNPs) were identified if present in >1% population frequency in the 1000g, genomAD, or ExAC databases. Somatic single nucleotide variants (SNVs) and indels were identified using VarScan2 (17) with the following parameters: i) minimum read depth=20; ii) minimum variant supporting reads=5, mapped to both strands; iii) minimum base quality=15; iv) strand bias no greater than 10%. Somatic variants were further filtered through an internally collected list of recurrent sequencing errors and if present in >1% population frequency in the 1000g, genomAD or ExAC database. Copy number variations (CNVs) were detected using CNVkit (18). CNV gain and loss were identified if depth ratio were above 1.6 or below 0.6, respectively. Final list of mutations was annotated using vcf2maf (call VEP for annotation). Panel tumor mutational burden (TMB) was counted by summing all base substitutions and indels in the coding region of targeted genes, including synonymous alterations to reduce sampling noise and excluding known driver mutations as they are over-represented in the Panel, as previously described (19).
Statistical Analysis
For comparisons of proportion between groups, Fisher’s exact tests were performed. For non-normally distributed data, such as TMB, differences between two groups were evaluated with the non-parametric Mann-Whitney/Wilcoxon rank-sum test. For survival analyses, Kaplan-Meier curves were estimated using the log-rank test, and hazard ratios (HRs) for PFS and OS were calculated by Cox proportional hazards model. Multivariable survival analysis was performed using the Cox regression model. A two-sided P value of less than 0.05 was considered to be statistically significant unless otherwise indicated. All statistical analyses were done in R (v.3.5.2).
Results
Patient Overview
We retrospectively performed analyses on 122 patients with unresectable stage III NSCLC, who underwent dCRT. Patients’ baseline characteristics were summarized in Table 1 . Median age of the study cohort was 62 years. Histological subtypes included 84 SCC (68.9%) and 38 ADC (31.1%). Consistent with a higher proportion of SCC patients, there were more male patients (87.7%) and former smokers (74.6%) in the study cohort. 51.6% (63/122) patients received concurrent dCRT and the remaining (48.4%) received sequential dCRT. At data cutoff, the median follow-up time was 30.1 months. Median PFS and OS of the study cohort were 11.4 and 34.6 months, respectively ( Supplementary Figure 1 ). A total of 51 (41.8%) patients developed grade 2 or higher toxicity, with 39 patients developed grade 2 or higher pneumonitis and 16 cases of esophagitis.
Table 1.
Clinical characteristics of the patients.
| Characteristics | No. of patients (%) |
|---|---|
| Sex | |
| Male | 107 (87.7%) |
| Female | 15 (12.3%) |
| Age | |
| <65 | 73 (59.8%) |
| ≥65 | 49 (40.2%) |
| Median age (range) | 62 (33–84) |
| Histological subtypes | |
| Adenocarcinoma | 38 (31.1%) |
| Squamous cell carcinoma | 84 (68.9%) |
| Smoking history | |
| Former smokers | 91 (74.6%) |
| Never smokers | 31 (25.4%) |
| Treatment regimen | |
| SCRT | 59 (48.4%) |
| CCRT | 63 (51.6%) |
| Radiation type | |
| 3DCRT | 30 (24.6%) |
| IMRT | 92 (75.4%) |
| Radiation dose | |
| <60 Gy | 28 (23.0%) |
| =60 Gy | 66 (54.0%) |
| >60 Gy | 28 (23.0%) |
| RT-related pneumonitis | |
| Grade 0-1 | 83 (68.0%) |
| Grade 2 | 18 (14.8%) |
| Grade 3 | 19 (15.6%) |
| Grade 4 | 2 (1.6%) |
| RT-related esophagitis | |
| Grade 0-1 | 106 (86.9%) |
| Grade 2 | 8 (6.55%) |
| Grade 3 | 8 (6.55%) |
| Grade 4 | 0 (0%) |
SCRT, sequential chemoradiotherapy; CCRT, concurrent chemoradiotherapy
Genomic profiling on baseline tissues using a targeted NGS panel covering 474 cancer- and radiotherapy-associated genes ( Supplementary Table 1 ) revealed the mutational landscape of the cohort ( Figure 1A ). The most frequently altered genes were TP53 (ADC, 78.9%; SCC, 94.0%), MCL1 (ADC, 52.6%; SCC, 62.7%), MYC (ADC, 23.7%; SCC, 38.5%), NOS2 (ADC, 26.3%; SCC, 28.9%) and EGFR (ADC, 31.6%; SCC 24.1%). Notably, we observed a high frequency of TP53 and a low frequency of EGFR mutations in our ADC patients, which was consistent with an enrichment of former smokers in our study. The median TMB of the cohort is 13.4 mutations/Mb, with no significant difference observed between ADC (12.4 mutations/Mb) and SCC (13.9 mutations/Mb; P=0.68).
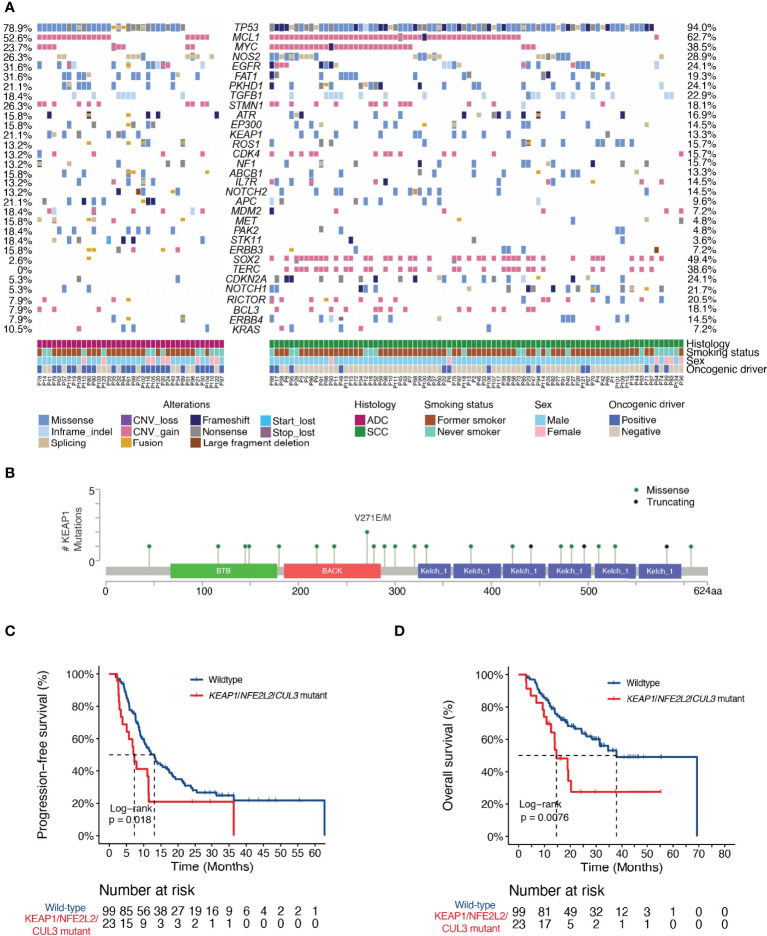
Figure 1.
Landscape of genetic variations and the associations of dCRT survival outcomes with the KEAP1-NRF2 pathway. (A) The distribution of various genetic variations in each patient was shown. Clinical characteristics of each patient were shown at the bottom. ADC, adenocarcinoma; SCC, squamous cell carcinoma. (B) Lollipop plot showing the distribution of KEAP1 mutations in the study cohort. (C, D) Kaplan-Meier estimates of (C) PFS and (D) OS in the full analysis set comparing patients with and without KEAP1-NRF2 pathway gene mutations. HR denotes hazard ratio; CI denotes confidence interval. Tick marks indicate censored data.
Predictive Markers for Survival Outcome Following CRT
First, we examined potential associations between clinical characteristics and survival outcome following dCRT. No significant difference in survival outcomes was observed comparing patients with different histological subtypes (PFS, P=0.27; OS, P=0.76). Patients treated with concurrent and sequential dCRT also had similar PFS outcome (P=0.56, Supplementary Figure 2A ), albeit a small trend towards increased OS in patients treated with concurrent dCRT (HR [95% CI] =0.63 [0.37-1.1], P=0.10, Supplementary Figure 2B ). Patients treated with 3D-conformal RT had lower risk of progression compared with those treated with intensity-modulated RT (HR [95% CI] =0.53 [0.31-0.90], P=0.01, Supplementary Figure 2C ), which did not translate into an OS difference (P=0.92, Supplementary Figure 2D ). Patients with smoking histories had worse outcome compared with never smokers (PFS, HR [95% CI] =1.95 [1.15-3.32], P=0.01; OS, HR [95% CI] =1.58 [0.79-3.16], P=0.19, Supplementary Figures 2E, F ). A higher overall dose was associated with a trend towards improved PFS and prolonged OS (PFS, HR [95% CI] =0.73 [0.45-1.18], P=0.2; OS, HR [95% CI] =0.56 [0.31-1.0], P=0.05, Supplementary Figures 2G, H ).
Next, we explored the associations between individual genomic alterations (non-synonymous alterations that occur in at least 5% of the cohort) and dCRT survival outcome. We identified 19 patients with mutations in Kelch Like ECH Associated Protein 1 (KEAP1, Figures 1A, B ). KEAP1 is an E3 ubiquitin ligase that functions as a sensor for oxidative stress and negatively regulates NRF2, a transcription factor upstream of cytoprotective and antioxidant genes, in the absence of stress (20). Of the 19 KEAP1-mutant patients, three carried nonsense mutations and the rest carried missense mutations that were all except three predicted to be deleterious or potentially damaging to protein function by SIFT or PolyPhen. Specifically, we detected three mutations in the BTB domain and four in the intervening BACK (BTB and C-terminal Kelch) domain, both of which mediate its interaction with Cullin 3 (Cul3) for protein ubiquitination, as well as ten mutations located in the Kelch repeat domains that mediates interaction with NRF2 ( Figure 1B ). KEAP1 mutations have been reported to correlate with an increased rate of local recurrence in NSCLC patients treated with radiotherapy (21). Indeed, our data independently showed that patients with KEAP1 mutations had shorter median PFS (6.7 months vs. 12.2 months; HR [95% CI] = 2.17 [1.24-3.81], P = 0.006, Supplementary Figure 3A ) and OS (18.8 vs. 37.8 months; HR [95% CI] =2.37 [1.23-4.55], P=0.008, Supplementary Figure 3B ) compared with those with the wild-type gene. Considering patients with deleterious mutations in genes in the KEAP1-NRF2 pathway (KEAP1, NFE2L2 or CUL3) showed a consistent increase in the risk of disease progression (HR [95% CI] =1.86 [1.1-3.15], P=0.02, Figure 1C ), as well as decreased OS (HR=2.27 [1.22-4.23], P=0.008, Figure 1D ).
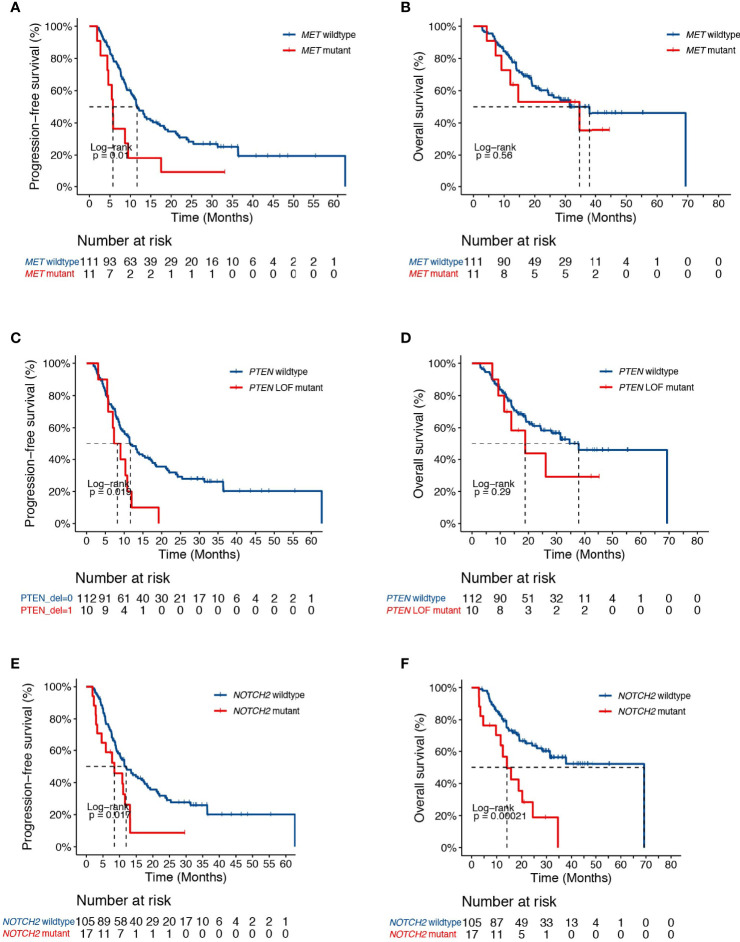
Univariate analysis revealed additional associations of survival outcome following dCRT with variations in several key genes that play important roles in lung cancer initiation and progression. The MET oncogene, which encodes a receptor tyrosine kinase, has become an important target for the treatment of NSCLC. We identified 11 patients with MET alterations, of which one had MET amplification, four carried exon 14 skipping mutations, three had MET fusions, and one patient with both amplification and an exon 14 skipping mutation ( Figure 1A ). The presence of MET alterations had a negative impact on disease progression, with shorter PFS than those with the wild-type gene (HR [95% CI] =2.33 [1.2-4.52], P=0.01, Figure 2A ). No significant difference in OS was found comparing patients with MET alterations and those with the wild-type gene (HR [95% CI] =1.29 [0.55-3.03], P=0.56, Figure 2B ). PTEN is an important tumor suppressor gene in lung cancers. Patients with deleterious mutations in PTEN, including nonsense, frameshift and splicing alterations exhibited a higher progression risk than those without (PFS, HR [95% CI] =2.19 [1.12-4.27], P=0.02, Figure 2C ). Similarly, no OS difference was found comparing patients with and without PTEN alterations (HR [95% CI] =1.57 [0.67-3.68], P=0.29, Figure 2D ), suggesting that both MET and PTEN alterations might serve as predictive markers of dCRT response. Preclinical studies have suggested that the NOTCH signaling pathway might also promote radiation resistance (22). Mutations in NOTCH2 were found to correlate with unfavorable survival outcome (PFS, HR [95% CI] = 2.0 [1.12-3.57], P=0.02; OS, HR [95% CI] = 3.12 [1.65-5.89], P=0.0002, Figures 2E, F ).
Figure 2.
Clinical associates of dCRT survival outcomes in NSCLC. (A, B) Kaplan-Meier estimates of (A) PFS and (B) OS in the full analysis set comparing patients with and without MET alterations. (C, D) Kaplan-Meier estimates of (C) PFS and (D) OS in the full analysis set comparing patients with and without PTEN deleterious mutations. (E, F) Kaplan-Meier estimates of (E) PFS or (F) OS in the full analysis set comparing patients with and without NOTCH2 alterations.
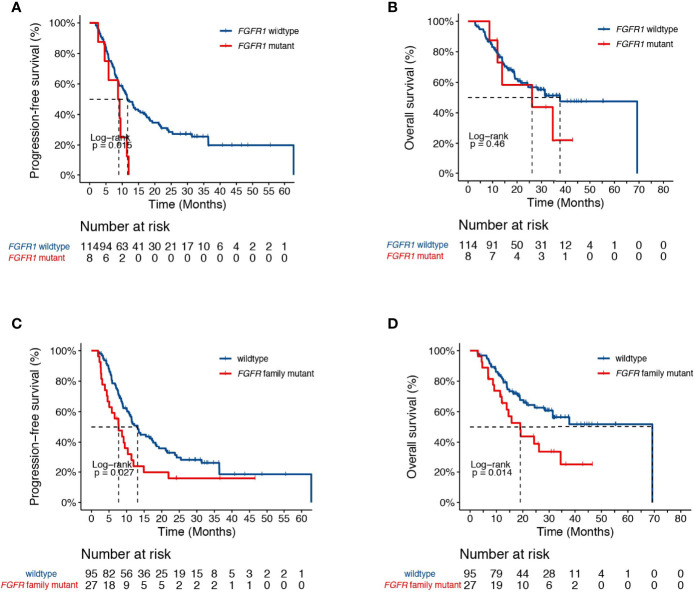
Genetic analysis also revealed alterations in FGFR1 in association with higher risk of progression in our study cohort (HR [95% CI] = 2.44 [1.16-5.14], P=0.015, Figure 3A ). No difference in survival was observed between FGFR1 wildtype and mutant patients (HR [95% CI] = 1.41 [0.56-3.56], P=0.46, Figure 3B ). Given that FGFR signaling is often dysregulated in NSCLC and have been implicated in radiation resistance in preclinical studies (23, 24), we sought to further test the association between FGFR family genes and patient survival. Indeed, we found that genetic alterations in the FGFR family receptors, including FGFR1-4, were associated with earlier progression (PFS, HR [95% CI] = 1.72 [1.06-2.79], P=0.03; OS, HR [95% CI] = 2.04 [1.14-3.65], P=0.01, Figures 3C, D ).
Figure 3.
Associations of dCRT survival outcomes with the FGFR pathway. (A, B) Kaplan-Meier estimates of (A) PFS and (B) OS in the full analysis set comparing patients with and without FGFR1 alterations. (C, D) Kaplan-Meier estimates of (C) PFS and (D) OS in the full analysis set comparing patients with and without alterations in the FGFR family genes.
Based on these findings, we also assessed if other oncogenic mutations might influence patient outcome. Overall, the presence of oncogenic driver mutations in key lung cancer targets, such as activating mutations in EGFR, ERBB2, KRAS, MET, as well as ALK, RET, and ROS1 fusions, had no impact on patient progression (HR [95% CI] =0.87 [0.55-1.38], P=0.56, Supplementary Figure 4A ). Interestingly, carriers of oncogenic mutations had significantly improved OS compared with those without (HR [95% CI] =0.52 [0.27-0.99], P=0.04, Supplementary Figure 4B ). Similarly, patients with activating EGFR mutations alone had no significant impact on PFS but demonstrated increased OS compared with EGFR wild-type patients (PFS, HR [95% CI] =0.77 [0.46-1.26], P=0.3; OS, HR [95% CI] =0.42 [0.20-0.89], P=0.02, Supplementary Figures 4C, D ), which is likely explained by the potential use of subsequent targeted therapies in these patients. We also examined the effect of TMB on patient recurrence, and no significant association was identified.
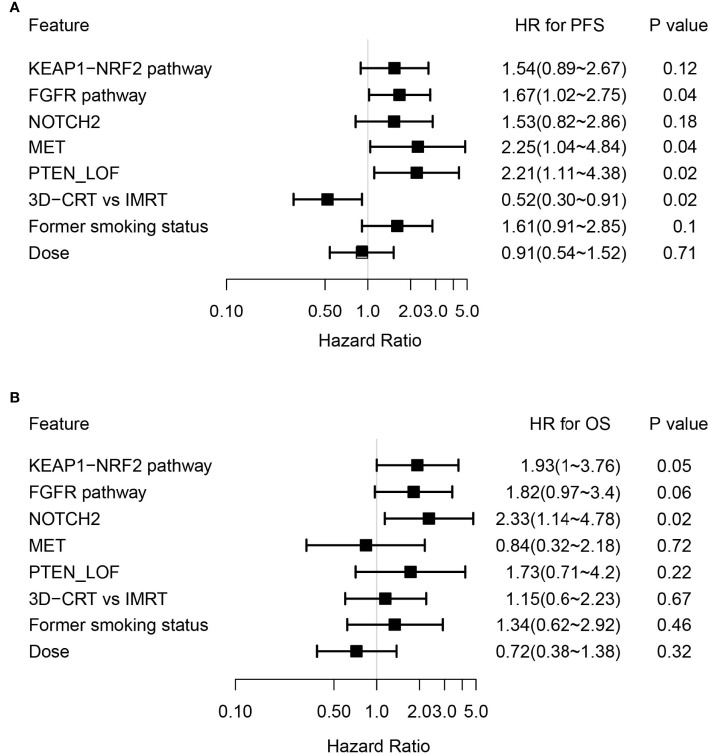
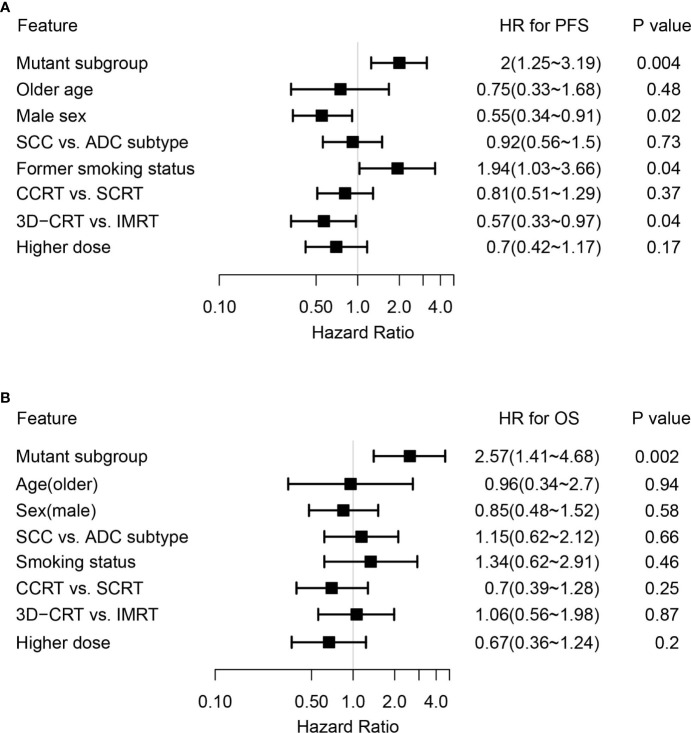
By adjusting for differences in clinical characteristics, including types of RT, smoking histories and overall dose, multivariate Cox analysis showed that alterations in MET and deleterious mutations in PTEN, as well as the FGFR pathway gene alterations remained independent predictive factors for reduced PFS following CRT ( Figure 4A ). On the other hand, the associations of OS with alterations in NOTCH2 and those in genes in the FGFR and the KEAP1-NRF2 pathways remained independent ( Figure 4B ). Subgroup analysis considering all patients who had received the recommended doses of 60-66Gy was performed and we found that deleterious mutations in PTEN and FGFR1 mutations remained independently associated with poorer PFS ( Supplementary Table 2 ). In addition, mutations in NOTCH2 remained independent predictors of unfavorable OS outcome ( Supplementary Table 2 ). Building on these associations and the largely exclusive nature of KEAP1, MET, PTEN, NOTCH2 and FGFR family gene alterations, we next sought to improve the stratification of NSCLC patients with differential survival outcome. Combined analysis of the mutant subgroup showed markedly improved risk stratification of our patients with significant PFS (HR=2.09, 95% CI=1.36-3.2, P=0.0006) and OS (HR= 2.73, 95% CI=1.54-4.85, P=0.0003) differences ( Figures 5A, B ). In addition, multivariate analysis incorporating all relevant clinical characteristics, including histology, modes of CRT, smoking status and overall dose, showed that the mutant subgroup remained a strong independent predictor of survival outcomes ( Figures 6A, B ).
Figure 4.
Multivariate Cox analysis of genetic features associated with survival outcomes. (A, B) Forest plots showing key genetic and clinical features in association with (A) PFS and (B) OS following dCRT treatment by multivariate analysis.
Figure 5.
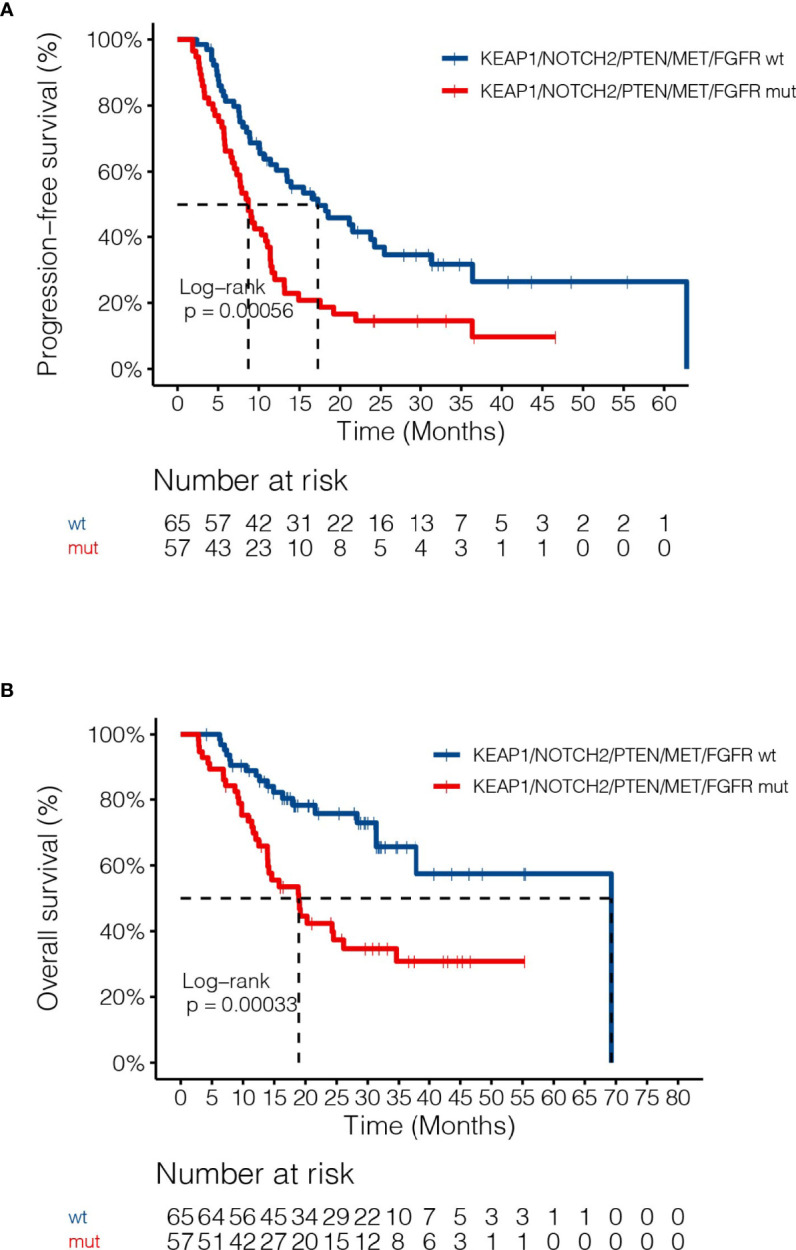
Associations of dCRT survival outcomes with the mutant subgroup. (A, B) Kaplan-Meier estimates of (A) PFS and (B) OS in the full analysis set comparing patients harboring any of the MET, NOTCH2 and PTEN loss of function, as well as FGFR and KEAP1-NRF2 pathway gene alterations and those without.
Figure 6.
Multivariate Cox analysis of clinical features associated with survival outcomes in the genetically altered subgroup. (A, B) Forest plots showing the mutant subgroup and all relevant clinical features in association with (A) PFS and (B) OS following dCRT treatment by multivariate analysis. Mutant subgroup: patients with any of the MET, NOTCH2 and PTEN loss of function, as well as FGFR and KEAP1-NRF2 pathway gene alterations; ADC: adenocarcinoma; SCC: squamous cell carcinoma.
SNPs and Somatic Mutations Predictive of Radiation Toxicity
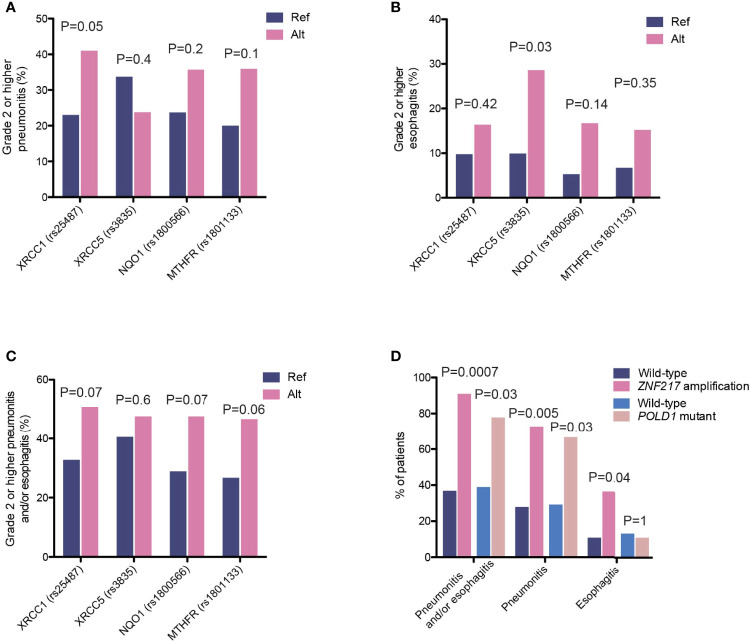
To identify potential risk factors that could explain individual variations in their likelihood to develop radiotherapy-associated toxicity, we first examined the potential effects of various clinical characteristics and treatment regimens. None of the clinical features, including age, smoking status, dose, sequential or concurrent combinations of chemo- and radiotherapy, or delivery methods, had a significant influence on the development of radiation-induced thoracic toxicity, including grade 2 or higher pneumonitis and esophagitis. In recognition that radiotherapy-associated toxicity are manifested as damages to the normal tissue surrounding the site of lesion, we analyzed the association between SNPs and the incidence of radiation-induced thoracic toxicity. Consistent with existing studies demonstrating the associations of radiotherapy-associated toxicity with SNPs in genes in the DNA damage repair, oxidative reduction and metabolic pathways, we identified SNPs in X-ray repair cross-complementing 1 (XRCC1, rs25487, c.1196A>G; OR=2.31 [95%CI, 1.0-5.56]; P=0.05) and XRCC5 (rs3835, c.2110-2408G>A; OR=3.59 [95% CI, 0.93-12.96]; P=0.03), which conferred increased risks of radiation-induced pneumonitis and esophagitis, respectively ( Figures 7A, B ). Further analysis revealed a stronger association between the XRCC5 allele with severe (grade 3 or higher) esophagitis (OR=5.71 [95% CI, 1.30-25.0]; P=0.03). In addition to these two SNPs, MTHFR (rs1801133, c.665C>T) and NAD(P)H Quinone Dehydrogenase 1 (NQO1, rs1800566, c.559C>T) were associated with trends towards higher incidence of radiotherapy-associated toxicity overall ( Figure 7C ).
Figure 7.
Genetic variants associated with incidence of high-grade radiation toxicity. (A–C) The proportions of patients carrying the indicated polymorphisms that developed high-grade (A) radiation-induced pneumonitis, (B) radiation-induced esophagitis, and (C) overall pneumonitis and esophagitis. (D) The proportions of patients carrying ZNF217 amplification or POLD1 mutations that developed high-grade radiation-induced toxicity events as indicated.
We also observed enrichments of several somatic aberrations in patients who developed grade 2 or higher pneumonitis and/or esophagitis ( Figure 7D ). The zinc-finger protein 217 (ZNF217) gene is frequently amplified in human cancers (25, 26). It encodes a Kruppel-like transcription factor that mediates complex molecular processes through the regulation of gene expression. A total of 11 patients carried ZNF217 amplifications, ten of which developed grade 2 or higher pneumonitis and esophagitis (90.9% vs. 36.9%, OR=16.7 [95% CI, 2.24-748.4], P=0.0007, Figure 7D ). Of these, four developed radiation-induced esophagitis (36.4% vs. 10.8%, OR=4.6 [95% CI, 0.86-21.7], P=0.04) and eight developed pneumonitis (72.7% vs. 27.9%, OR=6.8 [95% CI, 1.5-42.1], P=0.005). Similarly, ZNF217 amplification was associated with severe pneumonitis and esophagitis (grade 3 or higher; OR=7.5 [95% CI, 2.0-28.0], P=0.003), as well as severe radiation-induced pneumonitis (OR=4.9 [95% CI, 1.3-18.2], P=0.02). In addition, we found that mutations in POLD1, encoding the DNA polymerase delta 1 that is a key protein for ensuring the replicative fidelity of DNA, were also associated with an increase in overall toxicity risk (pneumonitis and/or esophagitis, 77.8% vs. 38.9%, OR=5.4 [95% CI, 0.97-55.7], P=0.03, Figure 7D ). Six (out of nine) POLD1 mutations were predicted to have functional consequences (SIFT score ≤ 0.01). Of the seven patients who developed toxicity, six had pneumonitis and one had esophagitis. It is worth noting that somatic alterations in both genes showed stronger associations with radiation pneumonitis, as compared to esophagitis, likely reflecting interactions of these tumor cells with the local microenvironment.
Discussion
As we enter the era of personalized medicine, there is an area of unmet needs for identifying genetic determinants that explain individual differences in dCRT response. The development of disease recurrence and severe radiation-induced toxicity following dCRT could negatively impact patients’ survival outcome and quality of life. In this study, by comprehensive profiling of 122 unresectable stage IIIA-C NSCLC patients who underwent dCRT, we identified a number of highly relevant and novel genetic and pathway-level features that might serve as potential biomarkers for predicting response to CRT.
The KEAP1-NRF2 pathway is often altered in NSCLC. In line with previous studies that have demonstrated the potential prognostic or predictive value of the dysregulation in the KEAP1-NRF2 pathway in NSCLC patients following chemotherapy and/or radiotherapy (20, 21, 27, 28), our findings independently confirmed its association with poor outcome in NSCLC patients following dCRT. Multiple others signaling pathways, including the MAPK, PI3K/AKT, FGFR and NOTCH pathways, have been implicated in radio-resistance, through their regulation of cellular proliferation, differentiation, apoptosis, invasion and maintenance of cancer stem cells (22, 23, 29–32). However, most findings were based on preclinical studies using in vitro or animal models. Here, we provided the first clinical evidence showing the associations of dCRT recurrence with several highly functionally relevant lung cancer genes, including MET, PTEN, FGFR1-4, and NOTCH2. Of these, MET, PTEN and FGFRs remained independent predictors of PFS by multivariate analysis. In particular, MET is commonly activated in NSCLC, which can be a result of gene amplification and exon 14 skipping mutations. In our study, the majority of MET-altered cases could lead to activation of the protein, which included MET exon 14 skipping mutations, gene rearrangement and amplification. While the presence of MET alterations was associated with worse outcome when treated with dCRT, no difference in OS was detected. Despite insufficient clinical information on the subsequent lines of treatment, these MET-altered cases might have derived long-term benefit from MET-targeted therapies. Similarly, while the presence of other potentially targetable driver mutations, such as EGFR activating mutations, had no impact on disease progression, it was associated with improved OS, which is also likely explained by the later use of targeted therapies in these patients. Thus, our data suggest that MET alteration is likely a negative predictive, rather than prognostic, factor of dCRT recurrence.
The FGFR family receptors activate multiple signaling pathways, including the RAS/MAPK, PI3K, and STAT pathways, which play important roles in cancer initiation and development (33). Emerging evidence suggest that FGFR may be implicated in variable response to radiotherapy. Pre-clinical studies in NSCLC cell lines, xenograft models and genetically engineered mouse models have shown that FGFR inhibition can enhance radiation response, which may be through the upregulation of cellular apoptosis and autophagy (23) and/or polarization of tumor-associated macrophages towards the M1 phenotype (24). In our study, we provide the first clinical evidence for the role of FGFR in mediating dCRT response. Patients with FGFR family gene alterations demonstrated reduced PFS outcome. Furthermore, the negative association of FGFRs with PFS also translated into poor OS outcome.
In addition to disease recurrence, a subset of patients would also develop severe, and often long-term, radiation-induced toxicities, which can negatively impact their quality of life. Understanding the genetic basis underlying individual differences in the development of dCRT-associated adverse events would allow for the risk stratification of patients and consequently personalized dCRT regimens, which would maximize tumor control while minimizing damage to the local tissue. The involvement of SNPs in the various damage and stress-response genes in mediating radiation-induced toxicities has been extensively studied in multiple cancer types (9–12, 34–37). Early association studies have employed a candidate gene approach, which has led to the identification of several key genes that may serve as potential predictors of radiation-induced toxicities, including ATM, base excision repair genes (XRCC1-5), mismatch repair genes (MSH2, MLH1), and oxidative damage-detoxification genes (GSTM1, GSTT1) (38–45). As we shift from candidate gene approach to genome-wide association studies, such as the multi-centered RAPPER (Radiogenomics: Assessment of Polymorphisms for Predicting the Effects of Radiotherapy) study (46), more genetic polymorphisms associated with radio-toxicity have been identified. However, these studies are often underpowered and difficult to replicate due to the small effect size of individual SNPs on radiotoxicity, and rarely lead to clinically useful biomarkers. A gene-expression-based radio-sensitivity model has been developed (47–50) and clinically validated in multiple cancer types (51–55). These data provide the prescription framework for genomic-based radiotherapy and emphasize the importance of multi-gene testing as response to CRT is dependent on the combined influence of genetic variations at multiple loci. However, it remains challenging to adopt RNA-sequencing routinely into clinical practice, particularly on formalin-fixed paraffin-embedded samples.
Taking advantage of the NGS technology, our study independently verified the predictive potential of the DNA damage repair and oxidative stress pathway gene variants for radiotherapy-associated toxicity. Specifically, we identified polymorphisms in XRCC1/XRCC5 (x-ray repair cross-complementing 1/5), encoding two key genes responsible for base excision repair, that were associated with differential risks of high-grade toxicity. The XRCC1 rs25487 allele has been associated with severe oral mucositis in oropharyngeal carcinoma patients treated with radiotherapy (45). The XRCC5 rs3835 allele has been implicated in the development of severe radiation pneumonitis in NSCLC patients (56). In our study, no significant association between this particular allele with radiation pneumonitis was observed. Instead, we showed that XRCC5 might increase the risk of developing severe radiation esophagitis. In addition, we identified two risk alleles in MTHFR and NQO1 that were associated with radiotoxicity. MTHFR encodes a methylenetetrahydrofolate reductase, which participates in folate metabolism and the regulation of DNA methylation and repair (57, 58). On the other hand, NQO1 is involved in the regulation of reactive oxygen species and continued oxidative stress can also induce DNA damage and chronic inflammation (59, 60). Combinatorial testing for these genetic variations might be useful for identifying patients who are susceptible to radiation toxicity. However, large-scale studies are needed to fully assess the predictive potential of these particular polymorphisms or variations in DNA damage repair and oxidative stress pathways.
Solid tumors often exhibit complex interactions with their surrounding tissues via stromal components, the vasculature and immune cells, among others (61). However, it has never been reported that somatic mutations could influence a patient’s likelihood of developing radiation toxicity. Here, we report the associations of ZNF217 amplification and POLD1 mutations with increased likelihoods of developing radiation toxicity. In particular, ZNF217 amplification was associated increased risks of developing severe (grade 3 or higher) pneumonitis and esophagitis. ZNF217 is commonly amplified in human cancers (25, 26). While the presence of ZNF217 amplification itself may indicate loss of genome integrity, there may also be functional consequences given that ZNF217 encodes a transcription factor that mediates a diverse array of cellular processes through the regulation of various target gene expressions (62). Importantly, ZNF217 may have a role in DNA damage repair as it has been shown to repress the levels of BRCA1 (63). Likewise, the role of POLD1 in controlling replicative fidelity has been firmly established (64, 65). Thus, we speculate that ZNF217 amplification and/or POLD1 mutations in the tumor may affect overall genome stability and lead to the generation of tumor-specific neoantigens and consequently extensive lymphocyte infiltration (66). Conceivably, this could also exacerbate inflammation in the surrounding normal tissue following radiation. Interestingly, similar to our work in small-cell lung cancer (67), somatic alterations are more likely to affect radiotherapy-associated toxicity at the site of the lesion, as they were more commonly associated with pneumonitis than with esophagitis. Due to the retrospective nature of this current study, our cohort consisted of relatively high proportions of SCC and former smokers, as well as widely varied radiation doses which is associated with radiotherapy toxicity (68). Thus, the predictive value of these novel variations’ merits further investigation. Nevertheless, our data indicate that extra caution should be exercised when giving radiotherapy to NSCLC patients carrying such mutations.
Our observations from clinical data of genetic associations with CRT survival outcome and toxicity provide a set of candidate predictive biomarkers present in normal and also tumor tissues. The mechanisms by which some of these genetic variants act to promote development of adverse response or cancer recurrence remain to be elucidated, although it is likely through their combined influence on important oncogenic signaling pathways, as well as DNA damage repair, oxidative and inflammatory response pathways. Due to the lack of sufficient data from published work or public databases such as TCGA, future work should involve validation of these potential biomarkers in a larger set of cohorts and generation of multifactorial prediction models of the expected treatment outcome. Taken together, our results demonstrate the clinical utility of NGS panels in identifying predictive biomarkers for response to CRT and suggest that testing for these susceptibility loci would prove beneficial in improving personalized CRT in NSCLC patients.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: Genome Sequence Archive for Human (GSA- Human); HRA002626.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethical Review Board of the Oncology Center of Shandong Provincial Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Conceptualization: JY, SY, YS, XW, and XNW. Investigation: JY, SY, YS. Resources: DM, HG, JW, JM, ZY, XZ, LD, PC. Data curation: KH, ST, LL. Data analysis and interpretation: JP, JCY, JZ, DM and JM. Manuscript writing: All authors. Manuscript review & editing: All authors. Funding acquisition: JY. Supervision: JY, SY, YS. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Natural Science Foundation of China (grant No. NSFC81872475 and NSFC82073345 to SY; NSFC81874254 to PC); and Research Unit of Radiation Oncology, Chinese Academy of Medical Sciences (2019RU071), the foundation of National Natural Science Foundation of China (NSFC81627901, NSFC81972863 and NSFC82030082), the foundation of Natural Science Foundation of Shandong (ZR201911040452) and the Academic Promotion Program of Shandong First Medical University (2019ZL002) to JY.
Conflict of Interest
JP, JY, JZ, HB, XW, XNW and YS are staff of Nanjing Geneseeq Technology Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgment
The authors thank all investigators and research staff at the following hospitals and research sites: Fujian Zhangzhou Hospital, Nanjing Gulou Hospital, Cancer Hospital of The University of Chinese Academy of Science (Zhejiang Cancer Hospital), Wuhan University Renmin Hospital, Yantai Yuhuangding Hospital, Liuzhou Gongren Hospital, Huazhong University of Science and Technology for providing cases and samples.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.928605/full#supplementary-material
References
- 1. Herbst RS, Morgensztern D, Boshoff C. The Biology and Management of non-Small Cell Lung Cancer. Nature (2018) 553(7689):446–54. doi: 10.1038/nature25183 [DOI] [PubMed] [Google Scholar]
- 2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer Statistics in China, 2015. CA Cancer J Clin (2016) 66(2):115–32. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 3. Dillman RO, Seagren SL, Propert KJ, Guerra J, Eaton WL, Perry MC, et al. A Randomized Trial of Induction Chemotherapy Plus High-Dose Radiation Vesus Radiation Alone in Stage III non-Small-Cell Lung Cancer. N Engl J Med (1990) 323:940–5. doi: 10.1056/NEJM199010043231403 [DOI] [PubMed] [Google Scholar]
- 4. Arroyo-Hernandez M, Maldonado F, Lozano-Ruiz F, Munoz-Montano W, Nunez-Baez M, Arrieta O. Radiation-Induced Lung Injury: Current Evidence. BMC Pulm Med (2021) 21(1):9. doi: 10.1186/s12890-020-01376-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Daly ME, Singh N, Ismaila N, Antonoff MB, Arenberg DA, Bradley J, et al. Management of Stage III Non-Small-Cell Lung Cancer: ASCO Guideline. J Clin Oncol (2022) 40(12):1356–84. doi: 10.1200/JCO.21.02528 [DOI] [PubMed] [Google Scholar]
- 6. Barnett GC, West CML, Dunning AM, Elliott RM, Coles CE, Pharoah PDP, et al. Normal Tissue Reactions to Radiotherapy: Towards Tailoring Treatment Dose by Genotype. Nat Rev Cancer (2009) 9(2):134–42. doi: 10.1038/nrc2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Taylor AM, Harnden DG, Arlett CF, Harcourt SA, Lehmann AR, Stevens S, et al. Ataxia Telangiectasia: A Human Mutation With Abnormal Radiation Sensitivity. Nature (1975) 258:427–9. doi: 10.1038/258427a0 [DOI] [PubMed] [Google Scholar]
- 8. Kleinerman RA. Radiation-Sensitive Genetically Susceptible Pediatric Sub-Populations. Pediatr Radiol (2009) 39(Suppl 1):S27–31. doi: 10.1007/s00247-008-1015-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dayal R, Singh A, Pandey A, Mishra KP. Reactive Oxygen Species as Mediator of Tumor Radiosensitivity. J Cancer Res Ther (2014) 10(4):811–8. doi: 10.4103/0973-1482.146073 [DOI] [PubMed] [Google Scholar]
- 10. Multhoff G, Radons J. Radiation, Inflammation, and Immune Responses in Cancer. Front Oncol (2012) 2:58. doi: 10.3389/fonc.2012.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tang Y, Liu B, Li J, Wu H, Yang J, Zhou X, et al. Genetic Variants in PI3K/AKT Pathway are Associated With Severe Radiation Pneumonitis in Lung Cancer Patients Treated With Radiation Therapy. Cancer Med (2016) 5(1):24–32. doi: 10.1002/cam4.564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cordes N, Hildebrandt MAT, Komaki R, Liao Z, Gu J, Chang JY, et al. Genetic Variants in Inflammation-Related Genes Are Associated With Radiation-Induced Toxicity Following Treatment for Non-Small Cell Lung Cancer. PloS One (2010) 5(8):e12402. doi: 10.1371/journal.pone.0012402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He K, Zhang S, Shao LL, Yin JC, Wu X, Shao YW, et al. Developing More Sensitive Genomic Approaches to Detect Radioresponse in Precision Radiation Oncology: From Tissue DNA Analysis to Circulating Tumor DNA. Cancer Lett (2020) 472:108–18. doi: 10.1016/j.canlet.2019.12.004 [DOI] [PubMed] [Google Scholar]
- 14. Cox JD, Stetz J, Pajak TF. Toxicity Criteria of the Radiation Therapy Oncology Group (RTOG) and the European Organization for Research and Treatment of Cancer (EORTC). Int J Radiat Oncol Biol Phys (1995) 31(5):1341–6. doi: 10.1016/0360-3016(95)00060-C [DOI] [PubMed] [Google Scholar]
- 15. Bolger AM, Lohse M, Usadel B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics (2014) 30(15):2114–20. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li H, Durbin R. Fast and Accurate Short Read Alignment With Burrows-Wheeler Transform. Bioinformatics (2009) 25(14):1754–60. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: Somatic Mutation and Copy Number Alteration Discovery in Cancer by Exome Sequencing. Genome Res (2012) 22(3):568–76. doi: 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-Wide Copy Number Detection and Visualization From Targeted DNA Sequencing. PloS Comput Biol (2016) 12(4):e1004873. doi: 10.1371/journal.pcbi.1004873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fang W, Ma Y, Yin JC, Hong S, Zhou H, Wang A, et al. Comprehensive Genomic Profiling Identifies Novel Genetic Predictors of Response to Anti-PD-(L)1 Therapies in Non-Small Cell Lung Cancer. Clin Cancer Res (2019) 25(16):5015–26. doi: 10.1158/1078-0432.CCR-19-0585 [DOI] [PubMed] [Google Scholar]
- 20. Kansanen E, Kuosmanen SM, Leinonen H, Levonen A-L. The Keap1-Nrf2 Pathway: Mechanisms of Activation and Dysregulation in Cancer. Redox Biol (2013) 1(1):45–9. doi: 10.1016/j.redox.2012.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jeong Y, Hoang NT, Lovejoy A, Stehr H, Newman AM, Gentles AJ, et al. Role of KEAP1/NRF2 and TP53 Mutations in Lung Squamous Cell Carcinoma Development and Radiation Resistance. Cancer Discov (2017) 7(1):86–101. doi: 10.1158/2159-8290.CD-16-0127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yahyanejad S, Theys J, Vooijs M. Targeting Notch to Overcome Radiation Resistance. Oncotarget (2015) 7(7):7610–28. doi: 10.18632/oncotarget.6714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. SenthilKumar G, Fisher MM, Skiba JH, Miller MC, Brennan SR, Kaushik S, et al. FGFR Inhibition Enhances Sensitivity to Radiation in Non–Small Cell Lung Cancer. Mol Cancer Ther (2020) 19(6):1255–65. doi: 10.1158/1535-7163.MCT-19-0931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Im JH, Buzzelli JN, Jones K, Franchini F, Gordon-Weeks A, Markelc B, et al. FGF2 Alters Macrophage Polarization, Tumour Immunity and Growth and can be Targeted During Radiotherapy. Nat Commun (2020) 11(1):4064. doi: 10.1038/s41467-020-17914-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cohen PA, Donini CF, Nguyen NT, Lincet H, Vendrell JA. The Dark Side of ZNF217, a Key Regulator of Tumorigenesis With Powerful Biomarker Value. Oncotarget (2015) 6(39):41566–81. doi: 10.18632/oncotarget.5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee D-F, Walsh MJ, Aguiló F. ZNF217/ZFP217 Meets Chromatin and RNA. Trends Biochem Sci (2016) 41(12):986–8. doi: 10.1016/j.tibs.2016.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cameron BD, Sekhar KR, Ofori M, Freeman ML. The Role of Nrf2 in the Response to Normal Tissue Radiation Injury. Radiat Res (2018) 190(2):99. doi: 10.1667/RR15059.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goeman F, De Nicola F, Scalera S, Sperati F, Gallo E, Ciuffreda L, et al. Mutations in the KEAP1-NFE2L2 Pathway Define a Molecular Subset of Rapidly Progressing Lung Adenocarcinoma. J Thorac Oncol (2019) 14(11):1924–34. doi: 10.1016/j.jtho.2019.07.003 [DOI] [PubMed] [Google Scholar]
- 29. Hassan KA, Wang L, Korkaya H, Chen G, Maillard I, Beer DG, et al. Notch Pathway Activity Identifies Cells With Cancer Stem Cell–like Properties and Correlates With Worse Survival in Lung Adenocarcinoma. Clin Cancer Res (2013) 19(8):1972–80. doi: 10.1158/1078-0432.CCR-12-0370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hasslacher S, Schneele L, Stroh S, Langhans J, Zeiler K, Kattner P, et al. Inhibition of PI3K Signalling Increases the Efficiency of Radiotherapy in Glioblastoma Cells. Int J Oncol (2018) 53(5):1881–96. doi: 10.3892/ijo.2018.4528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sklar MD. The Ras Oncogenes Increase the Intrinsic Resistance of NIH 3T3 Cells to Ionizing Radiation. Science (1988) 239(4840):645–7. doi: 10.1126/science.3277276 [DOI] [PubMed] [Google Scholar]
- 32. Dent P, Yacoub A, Fisher PB, Hagan MP, Grant S. MAPK Pathways in Radiation Responses. Oncogene (2003) 22(37):5885–96. doi: 10.1038/sj.onc.1206701 [DOI] [PubMed] [Google Scholar]
- 33. Turner N, Grose R. Fibroblast Growth Factor Signalling: From Development to Cancer. Nat Rev Cancer (2010) 10(2):116–29. doi: 10.1038/nrc2780 [DOI] [PubMed] [Google Scholar]
- 34. Barnett GC, Coles CE, Elliott RM, Baynes C, Luccarini C, Conroy D, et al. Independent Validation of Genes and Polymorphisms Reported to be Associated With Radiation Toxicity: A Prospective Analysis Study. Lancet Oncol (2012) 13(1):65–77. doi: 10.1016/S1470-2045(11)70302-3 [DOI] [PubMed] [Google Scholar]
- 35. Alsner J, Andreassen CN, Overgaard J. Genetic Markers for Prediction of Normal Tissue Toxicity After Radiotherapy. Semin Radiat Oncol (2008) 18(2):126–35. doi: 10.1016/j.semradonc.2007.10.004 [DOI] [PubMed] [Google Scholar]
- 36. Georgakilas AG, Pavlopoulou A, Louka M, Nikitaki Z, Vorgias CE, Bagos PG, et al. Emerging Molecular Networks Common in Ionizing Radiation, Immune and Inflammatory Responses by Employing Bioinformatics Approaches. Cancer Lett (2015) 368(2):164–72. doi: 10.1016/j.canlet.2015.03.021 [DOI] [PubMed] [Google Scholar]
- 37. Rosenstein B. Identification of SNPs Associated With Susceptibility for Development of Adverse Reactions to Radiotherapy. Pharmacogenomics (2011) 12(2):267–75. doi: 10.2217/pgs.10.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang L, Yang M, Bi N, Fang M, Sun T, Ji W, et al. ATM Polymorphisms Are Associated With Risk of Radiation-Induced Pneumonitis. Int J Radiat Oncol Biol Physics (2010) 77(5):1360–8. doi: 10.1016/j.ijrobp.2009.07.1675 [DOI] [PubMed] [Google Scholar]
- 39. Xiong H, Liao Z, Liu Z, Xu T, Wang Q, Liu H, et al. ATM Polymorphisms Predict Severe Radiation Pneumonitis in Patients With Non-Small Cell Lung Cancer Treated With Definitive Radiation Therapy. Int J Radiat Oncol Biol Physics (2013) 85(4):1066–73. doi: 10.1016/j.ijrobp.2012.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seibold P, Behrens S, Schmezer P, Helmbold I, Barnett G, Coles C, et al. XRCC1 Polymorphism Associated With Late Toxicity After Radiation Therapy in Breast Cancer Patients. Int J Radiat Oncol Biol Phys (2015) 92(5):1084–92. doi: 10.1016/j.ijrobp.2015.04.011 [DOI] [PubMed] [Google Scholar]
- 41. Moullan N, Cox DG, Angele S, Romestaing P, Gerard JP, Hall J. Polymorphisms in the DNA Repair Gene XRCC1, Breast Cancer Risk, and Response to Radiotherapy. Cancer Epidemiol Biomark Prev (2003) 12:1168–74. [PubMed] [Google Scholar]
- 42. Yin M, Liao Z, Liu Z, Wang L-E, Gomez D, Komaki R, et al. Functional Polymorphisms of Base Excision Repair Genes XRCC1 and APEX1 Predict Risk of Radiation Pneumonitis in Patients With Non–Small Cell Lung Cancer Treated With Definitive Radiation Therapy. Int J Radiat Oncol Biol Physics (2011) 81(3):e67–73. doi: 10.1016/j.ijrobp.2010.11.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mangoni M, Bisanzi S, Carozzi F, Sani C, Biti G, Livi L, et al. Association Between Genetic Polymorphisms in the XRCC1, XRCC3, XPD, GSTM1, GSTT1, MSH2, MLH1, MSH3, and MGMT Genes and Radiosensitivity in Breast Cancer Patients. Int J Radiat Oncol Biol Physics (2011) 81(1):52–8. doi: 10.1016/j.ijrobp.2010.04.023 [DOI] [PubMed] [Google Scholar]
- 44. Venkatesh GH, Manjunath VB, Mumbrekar KD, Negi H, Fernandes DJ, Sharan K, et al. Polymorphisms in Radio-Responsive Genes and Its Association With Acute Toxicity Among Head and Neck Cancer Patients. PloS One (2014) 9(3):e89079. doi: 10.1371/journal.pone.0089079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gupta A, Mathew D, Bhat SA, Ghoshal S, Pal A. Genetic Variants of DNA Repair Genes as Predictors of Radiation-Induced Subcutaneous Fibrosis in Oropharyngeal Carcinoma. Front Oncol (2021) 11:652049. doi: 10.3389/fonc.2021.652049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Burnet NG, Barnett GC, Elliott RM, Dearnaley DP, Pharoah PD, Dunning AM, et al. RAPPER: The Radiogenomics of Radiation Toxicity. Clin Oncol (R Coll Radiol) (2013) 25(7):431–4. doi: 10.1016/j.clon.2013.04.001 [DOI] [PubMed] [Google Scholar]
- 47. Eschrich S, Zhang H, Zhao H, Boulware D, Lee JH, Bloom G, et al. Systems Biology Modeling of the Radiation Sensitivity Network: A Biomarker Discovery Platform. Int J Radiat Oncol Biol Phys (2009) 75(2):497–505. doi: 10.1016/j.ijrobp.2009.05.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Eschrich SA, Pramana J, Zhang H, Zhao H, Boulware D, Lee JH, et al. A Gene Expression Model of Intrinsic Tumor Radiosensitivity: Prediction of Response and Prognosis After Chemoradiation. Int J Radiat Oncol Biol Phys (2009) 75(2):489–96. doi: 10.1016/j.ijrobp.2009.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Torres-Roca JF, Eschrich S, Zhao H, Bloom G, Sung J, McCarthy S, et al. Prediction of Radiation Sensitivity Using a Gene Expression Classifier. Cancer Res (2005) 65(16):7169–76. doi: 10.1158/0008-5472.CAN-05-0656 [DOI] [PubMed] [Google Scholar]
- 50. Scott JG, Berglund A, Schell MJ, Mihaylov I, Fulp WJ, Yue B, et al. A Genome-Based Model for Adjusting Radiotherapy Dose (GARD): A Retrospective, Cohort-Based Study. Lancet Oncol (2017) 18(2):202–11. doi: 10.1016/S1470-2045(16)30648-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Eschrich SA, Fulp WJ, Pawitan Y, Foekens JA, Smid M, Martens JW, et al. Validation of a Radiosensitivity Molecular Signature in Breast Cancer. Clin Cancer Res (2012) 18(18):5134–43. doi: 10.1158/1078-0432.CCR-12-0891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ahmed KA, Chinnaiyan P, Fulp WJ, Eschrich S, Torres-Roca JF, Caudell JJ. The Radiosensitivity Index Predicts for Overall Survival in Glioblastoma. Oncotarget (2015) 6:34413–22. doi: 10.18632/oncotarget.5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Strom T, Hoffe SE, Fulp W, Frakes J, Coppola D, Springett GM, et al. Radiosensitivity Index Predicts for Survival With Adjuvant Radiation in Resectable Pancreatic Cancer. Radiother Oncol (2015) 117(1):159–64. doi: 10.1016/j.radonc.2015.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Torres-Roca JF, Fulp WJ, Caudell JJ, Servant N, Bollet MA, van de Vijver M, et al. Integration of a Radiosensitivity Molecular Signature Into the Assessment of Local Recurrence Risk in Breast Cancer. Int J Radiat Oncol Biol Phys (2015) 93(3):631–8. doi: 10.1016/j.ijrobp.2015.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ahmed KA, Fulp WJ, Berglund AE, Hoffe SE, Dilling TJ, Eschrich SA, et al. Differences Between Colon Cancer Primaries and Metastases Using a Molecular Assay for Tumor Radiation Sensitivity Suggest Implications for Potential Oligometastatic SBRT Patient Selection. Int J Radiat Oncol Biol Phys (2015) 92(4):837–42. doi: 10.1016/j.ijrobp.2015.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yin M, Liao Z, Liu Z, Wang L-E, O'Reilly M, Gomez D, et al. Genetic Variants of the Nonhomologous End Joining Genelig4and Severe Radiation Pneumonitis in Nonsmall Cell Lung Cancer Patients Treated With Definitive Radiotherapy. Cancer (2012) 118(2):528–35. doi: 10.1002/cncr.26214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang Y, Li Z, Zhang J, Li H, Qiao Y, Huang C, et al. Genetic Variants in MTHFR Gene Predict ≥ 2 Radiation Pneumonitis in Esophageal Squamous Cell Carcinoma Patients Treated With Thoracic Radiotherapy. PloS One (2017) 12(1):e0169147. doi: 10.1371/journal.pone.0169147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mak RH, Alexander BM, Asomaning K, Heist RS, Liu CY, Su L, et al. A Single-Nucleotide Polymorphism in the Methylene Tetrahydrofolate Reductase (MTHFR) Gene is Associated With Risk of Radiation Pneumonitis in Lung Cancer Patients Treated With Thoracic Radiation Therapy. Cancer (2012) 118(14):3654–65. doi: 10.1002/cncr.26667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative Stress, Inflammation, and Cancer: How are They Linked? Free Radic Biol Med (2010) 49(11):1603–16. doi: 10.1016/j.freeradbiomed.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dinkova-Kostova AT, Talalay P. NAD(P)H:quinone Acceptor Oxidoreductase 1 (NQO1), a Multifunctional Antioxidant Enzyme and Exceptionally Versatile Cytoprotector. Arch Biochem Biophys (2010) 501(1):116–23. doi: 10.1016/j.abb.2010.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Egeblad MN, Nakasone ES, Werb Z. Tumors as Organs: Complex Tissues That Interface With the Entire Organism. Dev Cell Rev (2010) 18(6):884–901. doi: 10.1016/j.devcel.2010.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Thillainadesan G, Isovic M, Loney E, Andrews J, Tini M, Torchia J. Genome Analysis Identifies the P15ink4b Tumor Suppressor as a Direct Target of the ZNF217/CoREST Complex. Mol Cell Biol (2008) 28(19):6066–77. doi: 10.1128/MCB.00246-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Banck MS, Li S, Nishio H, Wang C, Beutler AS, Walsh MJ. The ZNF217 Oncogene is a Candidate Organizer of Repressive Histone Modifiers. Epigenetics (2009) 4(2):100–6. doi: 10.4161/epi.4.2.7953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang F, Zhao Q, Wang YN, Jin Y, He MM, Liu ZX, et al. Evaluation of POLE and POLD1 Mutations as Biomarkers for Immunotherapy Outcomes Across Multiple Cancer Types. JAMA Oncol (2019) 5(10):1504–6. doi: 10.1001/jamaoncol.2019.2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Church DN, Briggs SE, Palles C, Domingo E, Kearsey SJ, Grimes JM, et al. DNA Polymerase Epsilon and Delta Exonuclease Domain Mutations in Endometrial Cancer. Hum Mol Genet (2013) 22(14):2820–8. doi: 10.1093/hmg/ddt131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nolan E, Savas P, Policheni AN, Darcy PK, Vaillant F, Mintoff CP, et al. Combined Immune Checkpoint Blockade as a Therapeutic Strategy for BRCA1-Mutated Breast Cancer. Sci Trans Med (2017) 9(393):eaal4922. doi: 10.1126/scitranslmed.aal4922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li L, Tang S, Yin JC, Dong L, Yang Z, Liu Y, et al. Comprehensive Next-Generation Sequencing Reveals Novel Predictive Biomarkers of Recurrence and Thoracic Toxicity Risks After Chemoradiation Therapy in Limited Stage Small Cell Lung Cancer. Int J Radiat Oncol Biol Phys (2022) 112(5):1165–76. doi: 10.1016/j.ijrobp.2021.12.009 [DOI] [PubMed] [Google Scholar]
- 68. He J, Dong G, Deng Y, He J, Xiu Z, Feng F. Comparison of Application Value of Different Radiation Dose Evaluation Methods in Evaluating Radiation Dose of Adult Thoracic and Abdominal CT Scan. Front Surg (2022) 9. doi: 10.3389/fsurg.2022.860968 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: Genome Sequence Archive for Human (GSA- Human); HRA002626.