Abstract
Aneuploidy is a hallmark of human cancers, but the effects of aneuploidy on protein expression remain poorly understood. To uncover how chromosome copy number changes influence the cancer proteome, we conducted an analysis of hundreds of human cancer cell lines and tumors with matched copy number, RNA expression, and protein expression data. We found that a majority of proteins show dosage compensation and fail to change by the degree expected based on chromosome copy number alone. We uncovered a variety of gene groups that were recurrently buffered upon both chromosome gain and loss, including protein complex subunits and cell cycle genes. Several genetic and biophysical factors were predictive of protein buffering, highlighting complex post-translational regulatory mechanisms that maintain appropriate gene product dosage. Finally, we established that chromosomal aneuploidy has a moderate effect on the expression of oncogenes and tumor suppressors, showing that these key cancer drivers can be subject to dosage compensation as well. In total, our comprehensive analysis of aneuploidy and dosage compensation across cancers will help identify the key driver genes encoded on altered chromosomes and will shed light on the overall consequences of aneuploidy during tumor development.
A majority of cancers have chromosome arm gains or losses (Weaver and Cleveland 2006), a state called aneuploidy. Aneuploidy has been shown to contribute to tumor evolution (Sansregret and Swanton 2017; Salgueiro et al. 2020; Shukla et al. 2020), drug resistance (Selmecki et al. 2009; Davoli et al. 2017; Replogle et al. 2020; Shukla et al. 2020; Ippolito et al. 2021; Lukow et al. 2021), and metastatic dissemination (Gao et al. 2016; Bakhoum et al. 2018; Vasudevan et al. 2020). High levels of aneuploidy are also associated with poor patient survival (Kheir et al. 1988; Xu et al. 2016; Hieronymus et al. 2018; Smith and Sheltzer 2018, 2022; Stopsack et al. 2019; Shukla et al. 2020; van Dijk et al. 2021). It is hypothesized that aneuploidy drives tumor development by increasing the dosage of oncogenes (OGs) and decreasing the dosage of tumor suppressor genes (TSGs) (Davoli et al. 2013; Giam and Rancati 2015; Smith and Sheltzer 2018), which may be reflected in the recurrent aneuploid karyotypes found in certain cancer lineages (Bailey et al. 2018). Despite these findings, aneuploidy itself has also been observed to induce substantial tumor-suppressive stresses (Sheltzer et al. 2017; Vasudevan et al. 2020, 2021). Aneuploid cells have increased metabolic requirements (Williams et al. 2008; Tang et al. 2011; Stingele et al. 2012; Sheltzer 2013; Dephoure et al. 2014; Schukken et al. 2020), display high levels of senescence (Estrada et al. 2013; Sheltzer et al. 2017; He et al. 2018; Macedo et al. 2018; Giam et al. 2020), show significant genomic instability (Sheltzer et al. 2011; Passerini et al. 2016), and are sensitive to compounds that interfere with protein folding and turnover (Torres et al. 2010; Tang et al. 2011; Donnelly et al. 2014; Donnelly and Storchová 2015). Aneuploidy-associated stresses may be caused by the deregulation of gene expression, which leads to the imbalanced production of key cellular proteins. How cells adapt to and compensate for these aneuploidy-induced proteome imbalances is a key area of ongoing research.
Genome-wide studies in yeast strains engineered to harbor single extra chromosomes have revealed that copy number gains result in the increased expression of most genes encoded on that chromosome (Torres et al. 2007; Dephoure et al. 2014). However, proteomic analysis of these aneuploid yeast strains showed that ∼20% of proteins encoded on extra chromosomes are subject to dosage compensation (Dephoure et al. 2014). Protein dosage compensation is particularly strong for ribosomal subunits and genes that encode subunits of protein complexes (Dephoure et al. 2014; Brennan et al. 2019; Taggart et al. 2020; Senger and Schaefer 2021).
We have significantly less insight into how aneuploidy shapes the proteome of human cancers. Experiments performed on single cancer cell lines harboring a few chromosome gains have suggested that human cells overexpress most proteins on gained chromosomes, with ribosomes and some protein complex subunits showing dosage compensation (Stingele et al. 2012; McShane et al. 2016; Viganó et al. 2018; Brennan et al. 2019; Hwang et al. 2021). Similarly, studies of subchromosomal amplifications in cancers suggest that up to ∼30% of DNA copy number changes do not result in corresponding changes in protein abundance (Zhang et al. 2014, 2016; Mertins et al. 2016; Gonçalves et al. 2017; Sousa et al. 2019). Although proportional synthesis has been reported to control complex stoichiometry abundance in lower eukaryotes, mammalian cells seem to rely on post-translational degradation for expression attenuation (Buccitelli and Selbach 2020; Taggart et al. 2020). Additionally, although chromosome loss events outnumber chromosome gain events in most tumors (Duijf et al. 2013; Ben-David and Amon 2020), the cellular effects of chromosome losses in human cancers have not been comprehensively explored (Chunduri et al. 2021).
Currently, we lack a genome-wide understanding of the effects of arm-length aneuploidy on protein expression in cancer. To surmount this limitation, we studied aneuploidy-associated changes in gene expression across a cohort of 367 human cancer cell lines and 168 ovarian tumors with known chromosome copy numbers, RNA expression levels, and protein expression levels (Nusinow et al. 2020).
Results
Mean protein expression increases upon chromosome gain and decreases upon chromosome loss
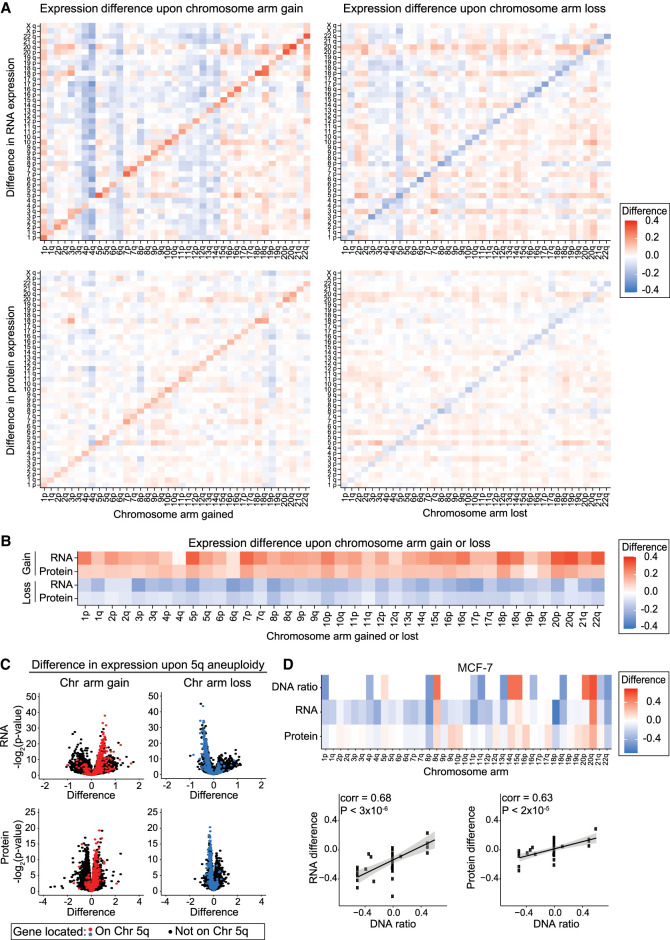
To develop a genome-wide understanding of the effects of arm-length aneuploidy on protein expression in cancer, we examined data from 367 human cancer cell lines from the Cancer Cell Line Encyclopedia (CCLE) with matched DNA copy number, RNA expression, and protein expression measurements (Supplemental Table S1; Barretina et al. 2012; Ghandi et al. 2019; Broad DepMap [https://figshare.com/articles/dataset/DepMap_20Q4_Public/13237076/2] [accessed June 3, 2021], [https://figshare.com/articles/dataset/DepMap_21Q2_Public/14541774/2] [accessed June 6, 2021]; Nusinow and Gygi 2020; Nusinow et al. 2020). We used a recently described data set in which each chromosome arm in a cell line was classified as “neutral,” “lost,” or “gained” relative to that cancer cell line's basal ploidy (Cohen-Sharir et al. 2021). We calculated mean RNA and protein expression differences along each chromosome arm between cell lines with neutral ploidy for that arm and cell lines in which that arm was either gained or lost (Supplemental Table S2). We found that average RNA and protein expression increases upon chromosome arm gain and decreases upon chromosome arm loss (Fig. 1A,B). For instance, in cancer cell lines with Chromosome 5q gains, the genes located on Chromosome 5q tend to show increased expression, whereas in cell lines with 5q losses, the genes located upon 5q tend to show decreased expression relative to cell lines in which 5q shows a neutral ploidy (Fig. 1C). Although mean gene expression differences correlated with chromosome copy number, we observed a large range of expression differences within the genes located on single aneuploid chromosomes, with some genes changing significantly upon chromosome gain or loss, whereas other genes showed little to no change in expression (Fig. 1C). The effects of aneuploidy on gene and protein expression were also apparent when analyzing gene expression in individual cancer cell lines (Fig. 1D; Supplemental Fig. S1A–E).
Figure 1.
Change in RNA and protein expression upon chromosome arm gain or loss. (A) Heatmaps displaying the difference in RNA expression (top) or protein expression (bottom) between cell lines with a chromosome arm gain (left) or chromosome arm loss (right) compared with cell lines with neutral ploidy for that arm. The mean RNA or protein expression differences per chromosome arm are displayed. (B) Heatmaps displaying the same analysis as in A, but only for each aneuploid chromosome. (C) Volcano plots displaying the difference in RNA expression (top) and protein expression (bottom) versus the P-value, per gene, between cells with Chromosome 5q gain (left) and cells with Chromosome 5q loss (right) compared with cell lines with neutral ploidy for Chromosome 5q. Genes encoded on Chromosome 5q are indicated in red (upon gain) or blue (upon loss), and all other genes are indicated in black. (D) Heatmap of the DNA ratio, RNA expression difference, and protein expression difference, per chromosome arm, in MCF-7 cells relative to cells neutral for each chromosome arm (top). Scatterplots showing the relationship between the DNA ratio and the RNA or protein expression differences (bottom). Linear regression with 95% confidence intervals is plotted against the data.
We found that aneuploidy-driven effects on gene expression were more pronounced at the RNA level than at the protein level. For chromosome gains, we found that the mean transcript encoded on that chromosome increased by an average of 22%, whereas protein expression increased by only 12%. A similar pattern was apparent upon chromosome loss, which we found to decrease RNA expression by 15% and protein expression by 8.4%. These differences suggest that both transcriptional and post-transcriptional dosage compensation can buffer the effects of aneuploidy on gene expression in cancer.
Ploidy gains buffer the effects of aneuploidy
We next investigated whether the difference in gene expression associated with aneuploidy was affected by a cell line's basal ploidy. Aneuploidy had a significant effect on RNA and protein expression in both near-diploid and near-triploid cell lines (Supplemental Fig. S1F). However, the increase in protein expression upon chromosome gain was significantly larger in diploid relative to triploid cell lines (14% vs. 10% increase, respectively, P < 3 × 10−16), and the decrease in protein expression upon chromosome loss was significantly larger in diploid relative to triploid cell lines (−11% vs. −10%, P = 0.00315). These results show that ploidy gain has a small but significant buffering effect on aneuploidy-driven gene expression changes.
Extensive protein buffering occurs within single cancer types and is detectable on other measurement platforms
As aneuploidy has been widely reported to significantly alter the expression of genes encoded on altered chromosomes (Pollack et al. 2002; Tsafrir et al. 2006; Grade et al. 2007; Torres et al. 2007; Gu et al. 2008; Fontanillo et al. 2012; Stingele et al. 2012; Davoli et al. 2013), we were surprised by our discovery that chromosome copy number changes affected average protein expression by only ∼10%. In light of this finding, we conducted several control analyses to investigate alternate explanations for these results.
We considered it possible that cancer-associated mutations might cause nonsense-mediated decay or changes in protein stability, which in turn could affect protein abundance. Therefore, we eliminated from consideration all genes harboring point mutations in a given cell line and recalculated the effects of aneuploidy on protein abundance.
Cancer genomes can harbor both chromosome-length and subchromosomal copy number changes. We reasoned that certain subchromosomal changes, like focal amplifications on lost chromosomes and deletions on gained chromosomes, could drive apparent protein dosage compensation. We eliminated from consideration these “flipped” genes that were located on a gained chromosome but whose individual gene copy number was decreased (2.4% of gained genes) and genes located on lost chromosomes but whose individual gene copy number was increased (10.4% of lost genes).
Low-expression genes may be difficult to detect at the protein level. We therefore calculated average RNA expression for each gene and removed the 20% of genes with the lowest average transcript levels.
A recent report identified proteins that show low reproducibility between technical mass spectrometry replicates and suggested that these proteins could contribute to artifactual results in proteomics experiments (Upadhya and Ryan 2021). We acquired their list of protein-reproducibility scores and eliminated the 20% of proteins with the lowest technical reproducibility.
We combined the previous four filters to generate a “merged-control” data set in which all mutated genes, “flipped” genes, low RNA expression genes, and low reproducibility proteins were removed from consideration.
Finally, to control for lineage-specific differences in protein expression, we conducted a subanalysis on non-small cell lung cancer (NSCLC) cell lines, which was the lineage with the largest number of cell lines in our data set.
We calculated RNA and protein expression differences upon chromosome gain and loss for each of these control data sets (Supplemental Fig. S2A; Supplemental Table S3). We found that the aneuploidy-associated expression differences for each of these control groups were similar to the bulk analysis (Supplemental Fig. S2B,C). For instance, in the “merged-control” data set, we observed that average protein expression decreased by 9.0% upon chromosome loss, comparable to the bulk analysis that showed an 8.4% decrease.
We considered the possibility that this protein buffering phenotype was caused by the technical limits or normalization methods of mass spectrometry. We note that the CCLE proteomics data set was generated using triple-stage mass spectrometry (MS3), which has been shown to almost completely eliminate ratio compression compared to standard tandem mass spectrometry (Ting et al. 2011). Nevertheless, we also assessed protein buffering in reverse-phase protein array (RPPA) expression data (Ghandi et al. 2019) that used antibodies to measure the abundance of 145 proteins in the CCLE. We found that the effects of aneuploidy on protein expression as determined via RPPA were not significantly different than the effects as determined by mass spectrometry (Supplemental Fig. S2D; Supplemental Table S3). We also found a strong and significant correlation between individual protein expression changes as measured by RPPA and mass spectrometry upon chromosome gain or loss (Supplemental Fig. S2E). Together, these data show that extensive protein buffering upon aneuploidy is independent of several potentially confounding factors and is consistent across measurement platforms.
mRNA changes strongly influence, but do not fully explain, the effects of aneuploidy on protein expression
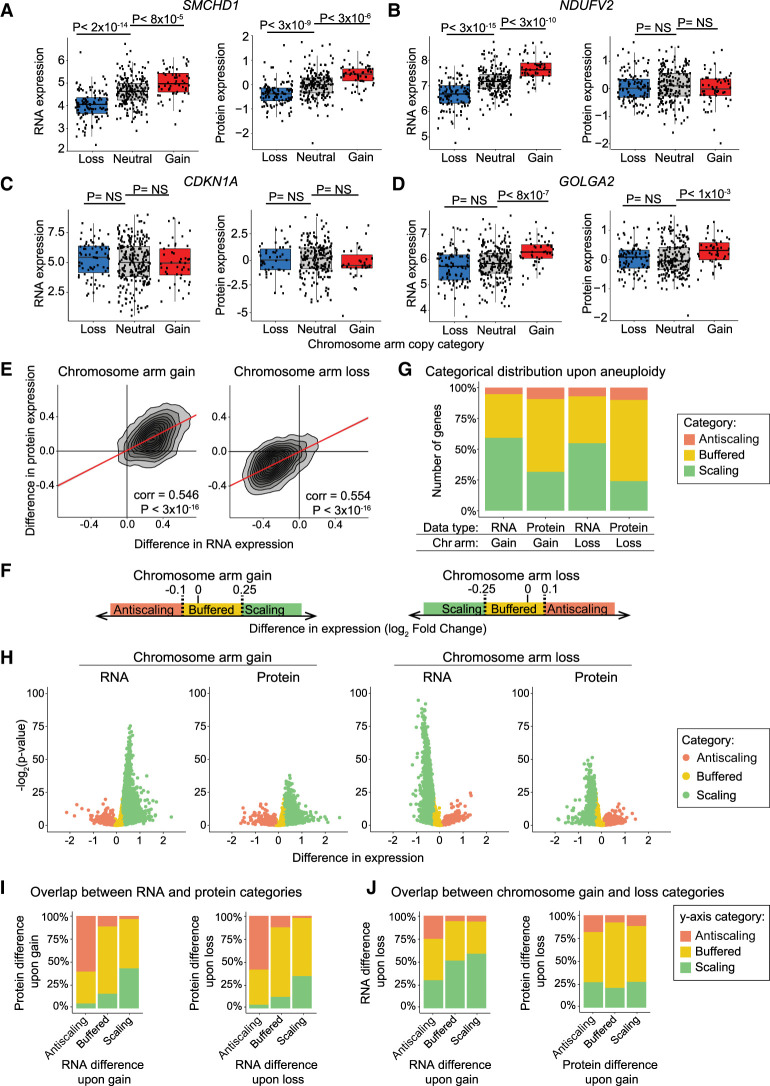
Next, we sought to understand the effects of aneuploidy on the expression of individual genes. We compared expression levels between cell lines in which a gene of interest was present on a lost, neutral, or gained chromosome, and we observed several different aneuploidy-driven expression patterns. The expression levels of some genes were significantly increased upon gain of the corresponding chromosome arm and significantly decreased upon loss of that chromosome arm, at both RNA and protein levels (e.g., SMCHD1, 9% of genes) (Fig. 2A). Other genes significantly changed at the RNA level but not at the protein level (e.g., NDUFV2, 13% of genes) (Fig. 2B). Many genes had no significant change in expression upon chromosome arm gain or loss (e.g., CDKN1A, 19% of genes) (Fig. 2C). Finally, the remaining 59% of genes showed more complex and/or variable expression patterns. For instance, GOLGA2 protein expression significantly increased upon chromosome gain, but expression levels of this protein were not significantly affected by chromosome loss (Fig. 2D).
Figure 2.
Protein expression differences are frequently buffered upon chromosome gain or loss. (A–D) Normalized RNA and protein expression levels of the indicated genes (SMCHD1, NDUFV2, CDKN1A, and GOLGA2) are displayed for cell lines in which the chromosome that that gene is encoded on is either lost, neutral, or gained. Boxplots display the 25th, 50th, and 75th percentiles of the data, and the whiskers indicate a 1.5 interquartile range. P-values were calculated using two-sided t-tests. (E) A density plot comparing the difference in RNA expression versus the difference in protein expression levels upon the gain (left) or loss (right) of the chromosome arm the gene is located on. The Pearson correlation coefficient and P-value are displayed. Linear regressions (red) with 95% confidence intervals are displayed. (F) Diagram displaying the categorical cutoff points for gene difference in expression. The cutoffs are −0.1 and 0.25 for chromosome gain and −0.25 and 0.1 for chromosome loss. The categorical cutoffs are labeled with dashed lines. (G) The percentage of RNAs and proteins that fall into each difference category upon chromosome gain and loss are displayed. (H) Volcano plots displaying the difference in RNA expression or protein expression upon chromosome arm gain (left) or chromosome arm loss (right) versus the P-value for each gene. Genes are color-coded based on a categorical distribution as either scaling, buffered, or antiscaling. (I) Bar graphs displaying the percentage of genes in each RNA difference category (x-axis) whose corresponding proteins fall into each of the indicated expression categories upon chromosome gain (left) and loss (right). (J) Bar graphs displaying the percentage of RNAs in each RNA difference category upon chromosome gain (x-axis) that fall into each of the indicated expression categories upon chromosome loss (y-axis; left). Percentage of proteins, per protein difference category upon chromosome gain (x-axis), that fall into each expression category upon chromosome loss (y-axis; right).
We investigated the degree to which these protein expression changes were determined by the effects of aneuploidy on mRNA expression. We found that RNA expression differences show a significant but moderate genome-wide correlation with protein expression differences upon chromosome arm gain (Pearson correlation coefficient = 0.546, P < 3 × 10−16) and upon chromosome arm loss (Pearson correlation coefficient = 0.554, P < 3 × 10−16) (Fig. 2E). These correlations indicate that, although protein expression differences are strongly driven by RNA expression differences, additional factors influence the expression of proteins encoded on aneuploid chromosomes.
Certain genes show consistent dosage compensation upon chromosome gain or chromosome loss
We categorized genes as either “scaling,” “buffered,” or “antiscaling” based on their expression changes upon chromosome gain or loss (Fig. 2F–H). At the RNA level, a majority of genes scaled with aneuploidy, whereas at the protein level, a majority of proteins were classified as buffered (Fig. 2G). For instance, upon chromosome gains, 59% of genes scaled at the RNA level, whereas 59% of genes showed buffering at the protein level. Additionally, we found that ∼5% of RNA and ∼10% of proteins showed an antiscaling expression pattern (e.g., decreased in expression upon chromosome gain or increased in expression upon chromosome loss). Similar results were observed in the merged-control data set described above (Supplemental Fig. S3A) and in the RPPA protein expression data set (Supplemental Fig. S3B).
We observed a significant correlation between RNA and protein difference categories upon chromosome gain (chi-squared = 2447, P < 3 × 10−16) and loss (chi-squared = 2580, P < 3 × 10−16) (Fig. 2I). For example, upon chromosome gain, 73% of genes that were buffered at RNA level were also buffered at the protein level. Next, we investigated whether genes that were buffered upon loss were more likely to be buffered upon gain. We found that there was a small but significant correlation between gene expression categories upon chromosome gain and loss at the RNA level (chi-squared = 359, P < 3 × 10−16) and at the protein level (chi-squared = 182, P < 3 × 10−16) (Fig. 2J). These results suggest that certain genes show consistent expression patterns, independent of whether the chromosome that they are encoded on is gained or lost.
Finally, we examined how aneuploidy affected the expression of noncoding RNAs on aneuploid chromosomes. We found that pseudogenes and microRNAs showed greater compensation than RNAs encoding protein-coding genes (Supplemental Fig. S4A–D; Wright and Bruford 2011; Barretina et al. 2012). Although protein-coding gene expression can be regulated at two levels (RNA and protein), noncoding genes can only be regulated at the RNA level, and we speculate that cells have evolved mechanisms to more carefully constrain RNA expression for noncoding loci.
Protein complex subunits and cell cycle genes tend to be buffered upon both chromosome gain and chromosome loss
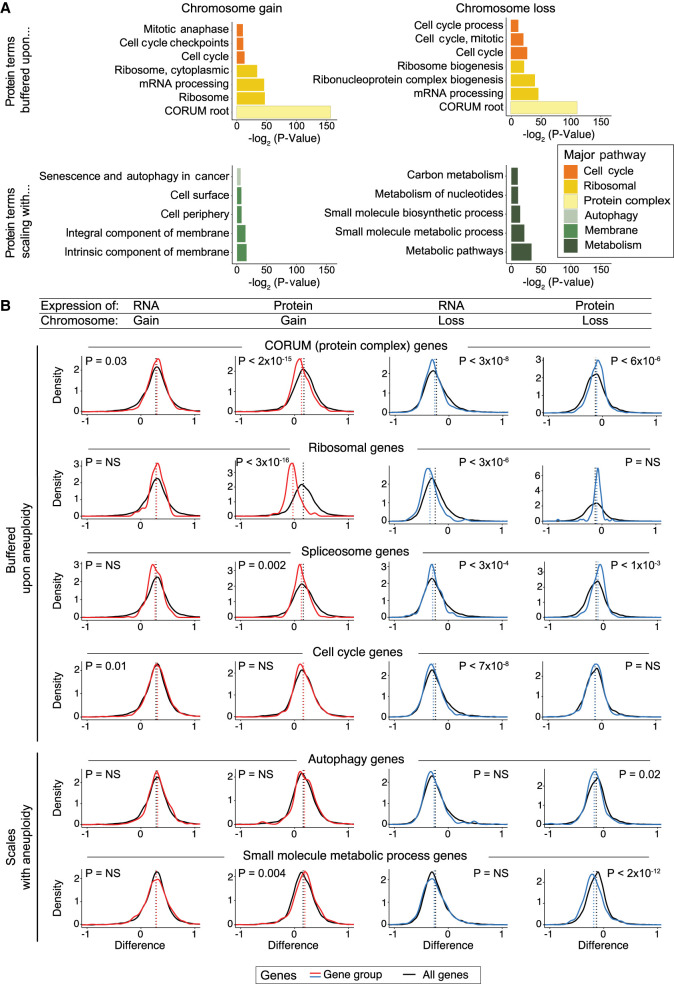
We investigated whether genes with shared functions displayed similar expression patterns when encoded on aneuploid chromosomes. Gene Ontology enrichment analysis (GOEA) was used to identify biological terms enriched among scaling, buffered, and antiscaling genes (Fig. 3A; Supplemental Fig. S5A,B; Supplemental Tables S4, S5; Raudvere et al. 2019). To visualize these expression patterns, we generated density plots of the expression differences of subsets of genes that were encoded on aneuploid chromosomes (Fig. 3B; Supplemental Table S6).
Figure 3.
Specific gene groups tend to show buffering or scaling upon chromosome gain and loss. (A) Bar graphs displaying the biological terms enriched in proteins buffered or scaling upon chromosome gain or loss, categorized by the major overarching pathway. The complete GO term lists are included in Supplemental Tables S4 and S5. (B) Density graphs displaying the difference in RNA or protein expression per gene group upon chromosome gain (red) or loss (blue). The difference in expression for all other genes is shown in black. The mean difference per gene group is indicated by the dotted lines. P-values represent two-sided t-tests between the indicated gene groups and the background set of all other genes. The list of genes in gene groups is in Supplemental Table S6.
Consistent with previous results (Stingele et al. 2012; Dephoure et al. 2014; McShane et al. 2016; Brennan et al. 2019), we observed that ribosomal genes, RNA processing genes, spliceosome components, and other genes that encode protein complex subunits were enriched among genes that were buffered upon chromosome gain. We also found that these proteins tended to be buffered upon chromosome loss and decreased in expression less than other proteins encoded on lost chromosomes. Similar enrichments were observed upon GOEA analysis of the merged-control data set (Supplemental Fig. S5C; Supplemental Table S7). We observed that, on average, protein complex subunits (CORUM genes) increase 21% less than other genes upon chromosome gain and decrease 13% less than other genes upon chromosome loss. Additionally, we found that a majority of genes encoding cell cycle components showed protein buffering upon chromosome gain and loss (64% and 71% of proteins, respectively). These findings suggest that dosage compensation is not simply a response to protein overproduction from extra chromosomes, and cancers have the ability to rebalance proteins and protein complex subunits when those genes are encoded on lost chromosomes as well.
Although many biological pathways were enriched among buffered proteins, we found that few Gene Ontology (GO) terms were enriched among buffered RNAs (Supplemental Fig. S5B; Supplemental Tables S4, S5). Indeed, mRNAs coding for protein complex subunits and cell cycle genes were found to be moderately enriched among RNAs that scaled with chromosome copy number, rather than among the buffered subset. This lack of dosage compensation at the RNA level indicates that translational or post-translational regulation, rather than transcriptional regulation, drives the pervasive dosage compensation of buffered proteins and protein complex subunits that we have observed in aneuploid cancer cells.
Although a significant percentage of proteins scale with chromosome gain (31%) and loss (24%), we found that few GO terms were enriched among the “scaling” protein category (Fig. 3A). One exception was metabolic pathways, especially small-molecule metabolic pathways, which were found to scale in expression with chromosome loss (Fig. 3A,B).
Only 5%–10% of genes showed an expression difference in the opposite direction of the DNA copy number change, which we classified as “antiscaling.” We found that genes associated with the extracellular environment and cell adhesion were enriched among antiscaling genes at both the RNA level and the protein level and upon chromosome gain and chromosome loss (Supplemental Fig. S5B–D). This may reflect the wide variability in the expression within these gene sets (discussed in more detail below).
In total, we observed that specific gene groups and pathways show similar expression patterns when present on aneuploid chromosomes, including both gained and lost chromosomes. These shared regulatory patterns may arise as a result of genetic and biophysical features that are shared among these genes (discussed in more detail below).
Patterns of dosage compensation are conserved between aneuploid cancers, trisomic primary cell lines, and disomic yeast
We hypothesized that the expression differences we observed between cell lines that have or lack certain aneuploidies represent a consequence of that chromosome copy number change. Alternatively, these expression patterns could be dictated by a cell line's lineage or genetic background. If these patterns of scaling and buffering represent a fundamental consequence of aneuploidy, then we would expect to observe similar expression patterns in other aneuploid cell lines. To investigate this possibility, we examined published transcriptome and proteome data from stably aneuploid human cell lines (Stingele et al. 2012), Down syndrome fibroblasts (Letourneau et al. 2014; Liu et al. 2017), and aneuploid yeast (Supplemental Fig. S6; Dephoure et al. 2014).
We first assessed the difference in protein expression between four published cell lines engineered to have a stable gain of Chromosome 5, normalized to their corresponding euploid controls (Stingele et al. 2012). We found that there was a highly significant correlation between the expression difference of proteins encoded on Chromosome 5 among CCLE lines with Chromosome 5 gains and the expression of those same proteins in the engineered cell lines (corr = 0.431, P < 5 × 10−10) (Supplemental Fig. S6A). Next, we examined protein differences in fibroblasts from Down syndrome patients (with Trisomy 21) normalized to matched euploid fibroblasts. We observed a strong and significant correlation between protein expression differences of proteins located on Chromosome 21 in Down syndrome patients and in CCLE cell lines with gains of Chromosome 21 (corr = 0.539, P < 4 × 10−4) (Supplemental Fig. S6B).
To investigate whether these patterns of protein dosage compensation are evolutionarily conserved outside of humans, we examined protein expression differences in haploid yeast that had been engineered to harbor single disomic chromosomes (Torres et al. 2007; Dephoure et al. 2014). We identified one-to-one orthologs between budding yeast and human proteins, and we found a significant correlation between the expression of proteins encoded on aneuploid chromosomes in human cancers and in budding yeast (corr = 0.214, P < 5 × 10−9) (Supplemental Fig. S6C; Supplemental Table S3). For instance, the ribosomal subunit gene RPL38, the splicing factor gene SF3A1, and the ER membrane complex gene EMC3 showed consistent dosage compensation at the protein level when they were encoded on a chromosome that was gained in cancer cells and when their orthologs were encoded on a chromosome that was gained in yeast (Supplemental Fig. S6D–F). Proteins displaying dosage compensation in both aneuploid cancers and yeast disomies were enriched for protein complex, ribosomal, and RNA processing terms (Supplemental Fig. S6L; Supplemental Table S8). In contrast, mRNA expression differences showed 1.5- to threefold lower correlations between aneuploid cell types relative to the same genes at the protein level (Supplemental Fig. S6G–K). In total, these data show that the gene expression changes observed on aneuploid chromosomes are conserved across aneuploid cell types and even across species, particularly at the protein level.
Effects of cellular aneuploidy on global patterns of protein expression
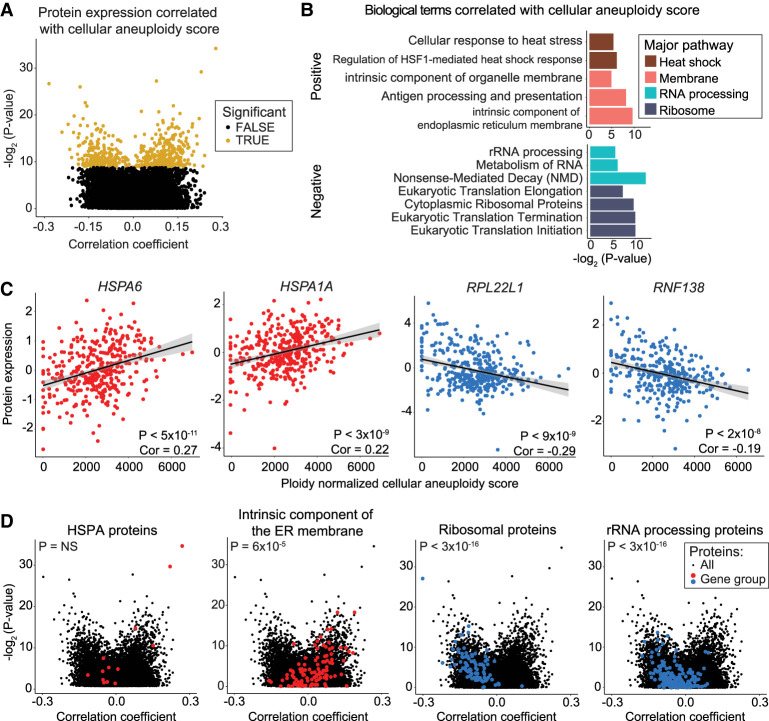
It has previously been reported that aneuploid cells show a set of shared transcriptional changes, independent of the identity of the extra chromosome (e.g., in trans) (Sheltzer et al. 2012; Dürrbaum et al. 2014). To investigate whether aneuploidy is associated with gene expression changes in trans across a large panel of cancer cell lines and to determine where these patterns are maintained at the protein level, we calculated a cellular aneuploidy score based on the total number of genes encoded on aneuploid chromosomes in each cell line, normalized to that line's basal ploidy. We then performed Pearson correlation analysis to identify genes whose expression was positively or negatively correlated with this cellular aneuploidy score. Positive correlations represent genes that were up-regulated in highly aneuploid cell lines relative to cell lines with lower total aneuploidy, whereas negative correlations represent genes that were down-regulated in highly aneuploid cell lines (Fig. 4A; Supplemental Fig. S7A; Supplemental Table S9).
Figure 4.
Specific protein expression levels correlate with total cellular aneuploidy. (A) A volcano plot displaying the Pearson correlation coefficient between protein expression levels and cellular aneuploidy score versus the P-value for that comparison. Genes that are significant at a P < 0.05 threshold after Benjamini–Hochberg correction with a 5% FDR are labeled in gold. (B) A bar graph displaying the GO terms that are enriched among genes positively (top) or negatively (bottom) correlated with total cellular aneuploidy. The complete list of GO terms is included in Supplemental Table S10. (C) Scatterplots displaying the expression of the two proteins that are most significantly positively correlated with aneuploidy score (left) and the two proteins that are most significantly negatively correlated with aneuploidy score (right). Protein expression is plotted by cellular aneuploidy score. Pearson correlation coefficients and P-values are displayed. Linear regressions and 95% confidence intervals are plotted against the data. (D) Volcano plots displaying the correlation coefficient between protein expression and cellular aneuploidy score versus the corresponding P-value. The background set of all genes are plotted in black, and genes belonging to indicated groups are labeled in red or blue. P-values correspond to two-sided t-tests between correlation coefficients of gene groups and all other genes. The list of genes in gene groups is in Supplemental Table S6.
Consistent with prior publications (Stingele et al. 2012; Brennan et al. 2019), we observed that ribosomal and rRNA processing terms were strongly negatively correlated with cellular aneuploidy scores at both the RNA and protein level (Fig. 4B; Supplemental Fig. S7B; Supplemental Table S10). Indeed, the two genes most significantly negatively correlated at the protein level are the ribosome-associated genes RPL22L1 and RNF138 (Fig. 4C). Another ribosomal gene, RPL3, is the most strongly negatively correlated gene at the RNA level (Supplemental Fig. S7C). The two genes most positively correlated with aneuploidy scores, HSPA6 and HSPA1A, both encode heat shock proteins (Fig. 4C). HSPA1A has previously been reported to be up-regulated in aneuploid murine fibroblasts (Tang et al. 2011). Additionally, genes annotated to “heat assimilation” and “cellular response to heat stress” were enriched among the genes whose expression positively correlated with aneuploidy, which may reflect an aneuploid cell's increased reliance on protein folding chaperones (Fig. 4B; Supplemental Fig. S7B; Supplemental Table S10; Oromendia et al. 2012; Donnelly et al. 2014). Finally, membrane-related genes were enriched among genes positively correlated with cellular aneuploidy (Fig. 4B). For example, intrinsic components of the endoplasmic reticulum (ER) membrane genes were significantly up-regulated at the RNA (Supplemental Fig. S7D) and protein level (Fig. 4D). Together, these data show that the expression of certain genes and pathways is associated with the levels of total cellular aneuploidy, independent of whether the genes themselves are encoded on an aneuploid chromosome.
Both cis and trans effects contribute to dosage compensation in aneuploid cancer cell lines
In the above analysis, we noted that certain gene groups, particularly the ribosome and translation-associated processes, tended to be down-regulated in both highly aneuploid cells and when those genes were encoded on a gained chromosome. We therefore considered the possibility that the “dosage compensation” phenotype that we described above was a consequence of this global response to cellular aneuploidy, rather than a consequence of the altered copy number of these genes in cis.
Three analyses indicate that trans effects are insufficient to fully account for the protein buffering that we have observed. First, we found that protein complexes and ribosome subunits were up-regulated, rather than down-regulated, relative to the mean protein when encoded on a lost chromosome (Fig. 3B). This change is in the opposite direction of the trans effects of aneuploidy, suggesting that this effect must occur in cis. Second, we found that the patterns of protein buffering and scaling were maintained in fibroblasts with natural single-chromosome trisomies and congenic cancer cell lines engineered to harbor single extra chromosomes, indicating that cell lines with very low total aneuploidy display dosage compensation of protein complexes (Supplemental Fig. S6). Third, we conducted an additional analysis, in which we isolated and analyzed the quartile of cell lines with the lowest cellular aneuploidy scores and the quartile of cell lines with the highest cellular aneuploidy scores. We then calculated the difference in gene expression upon chromosome arm gain and loss specifically within these quartiles (Supplemental Table S11). We found that ribosome proteins and protein complex subunits were similarly enriched among buffered proteins within both the lowest-aneuploidy quartile and the highest-aneuploidy quartile (Supplemental Fig. S8A,B; Supplemental Table S12). Thus, although the genome-wide effects of aneuploidy on protein expression can influence the expression landscapes of cancer cell lines, these analyses indicate that dosage effects in cis significantly contribute to the buffering phenotype that we have described, independent of total cellular aneuploidy levels.
Post-translational modifications, protein complex formation, and RNA expression variance contribute to protein buffering upon aneuploidy
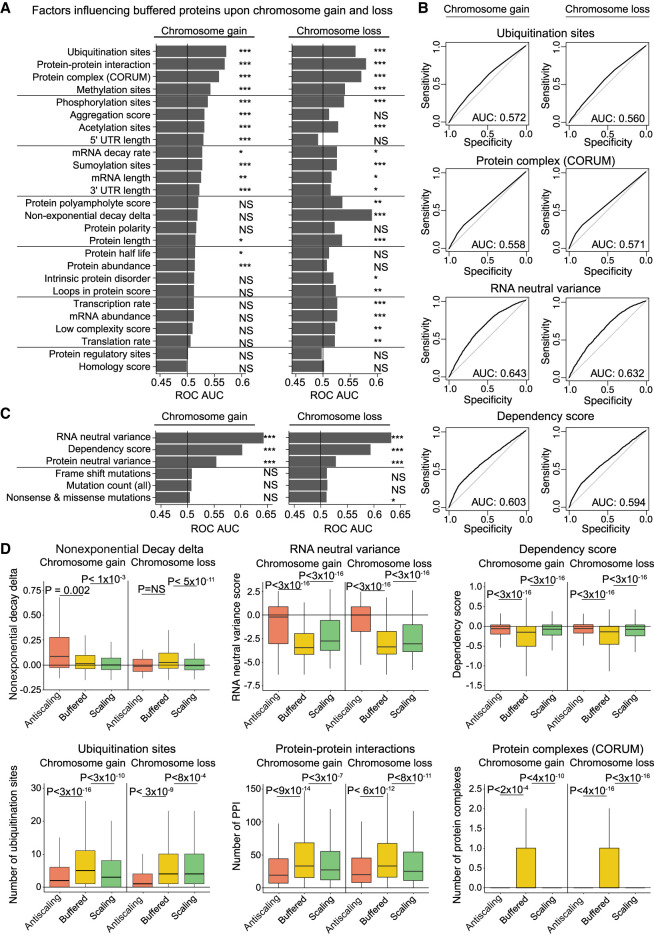
We next sought to identify the factors that drive protein dosage compensation. To do this, we calculated the receiver operator characteristic area under the curve (ROC AUC) to estimate a factor's ability to predict protein buffering. We examined 30 different factors that capture various genetic, biochemical, and biophysical features of each gene or protein, and we assessed their correlation with buffering upon chromosome gain and chromosome loss (see Supplemental Methods).
The strongest predictors of buffering upon chromosome gain were the number of ubiquitination sites within a protein, the number of protein–protein interactions that a protein showed, and the number of protein complexes a protein is incorporated into (AUC = 0.56–0.57) (Fig. 5A,B). The same features were also strongly correlated with buffering upon chromosome loss. Several other post-translational modifications, including protein methylation, phosphorylation, and acetylation, were also correlated with protein buffering upon either chromosome gain or loss. This suggests that protein complex subunits and proteins that are regulated by post-translational modifications tend to show dosage compensation in aneuploid cells.
Figure 5.
Multiple genetic and biochemical factors predict protein buffering upon aneuploidy. (A) Bar graphs displaying ROC area under the curve values for each independent factor upon chromosome gain or loss. Genes classified as “buffered” upon chromosome gain or loss were set as the true positive fraction. Significance was calculated by performing 10,000 random permutations and bootstrapping P-values, where (*) indicates < 0.05, (**) < 0.005, and (***) < 0.0005. (B) ROC curves are displayed for certain key factors. Genes classified as “buffered” upon chromosome gain or loss were set as the true positive fraction. (C) ROC AUC values for data set–specific factors upon chromosome gain or loss. Genes classified as “buffered” upon chromosome gain or loss were set as the true positive fraction. (D) Boxplots displaying buffering factor scores, per difference category upon chromosome gain or loss. Six buffering factors are displayed: nonexponential decay delta, RNA neutral ploidy variance, dependency score, number of ubiquitination sites, number of protein–protein interactions, and number of protein complexes. P-values are from two-sided t-tests. Boxplots display the 25th, 50th, and 75th percentiles of the data, and the whiskers indicate 1.5 interquartile ranges.
Several aspects of protein regulation, including aggregation scores and 5′ UTR length, were significantly associated with buffering upon chromosome gain but not loss. Inversely, of the factors tested, the nonexponential decay (NED) delta (McShane et al. 2016) was the strongest predictor of buffering upon chromosome loss but did not significantly predict buffering upon chromosome gain.
We also examined several factors that were specific to this CCLE data set. We found that buffered proteins showed lower expression variation at both the RNA and protein levels within cell lines in which that gene was encoded on a nonaneuploid chromosome (Fig. 5B–D). This indicates that proteins buffered in aneuploid cells tend to be more tightly regulated even in euploid conditions. A gene's dependency score was also observed to predict protein buffering (AUC = 0.59–0.60) (Fig. 5B,C). Finally, we found that the frequency of mutations per gene did not affect their likelihood to be buffered upon aneuploidy, suggesting that nonsense-mediated decay was not a significant factor in dosage compensation.
Next, we examined several of the predictive factors identified above individually. We verified that buffered proteins tended to show lower dependency scores (indicative of essential genes), more ubiquitination sites, and more protein–protein interactions than nonbuffered proteins (Fig. 5D). In fact, buffered proteins tended to be significantly different from both the scaling and antiscaling gene groups. For instance, 31% of buffered proteins were members of one or more protein complexes, which was significantly more than we observed among either scaling or antiscaling proteins. These differences suggest that the antiscaling gene group is not simply a more extreme instance of protein buffering and is instead driven by separate factors. Indeed, we observed that antiscaling genes show significantly higher expression variation when encoded on nonaneuploid chromosomes compared to either buffered or scaling genes. Thus, the expression changes within this gene group may reflect inherent noise or variability in their regulation.
Many OGs are buffered upon aneuploidy but scale with gene amplification
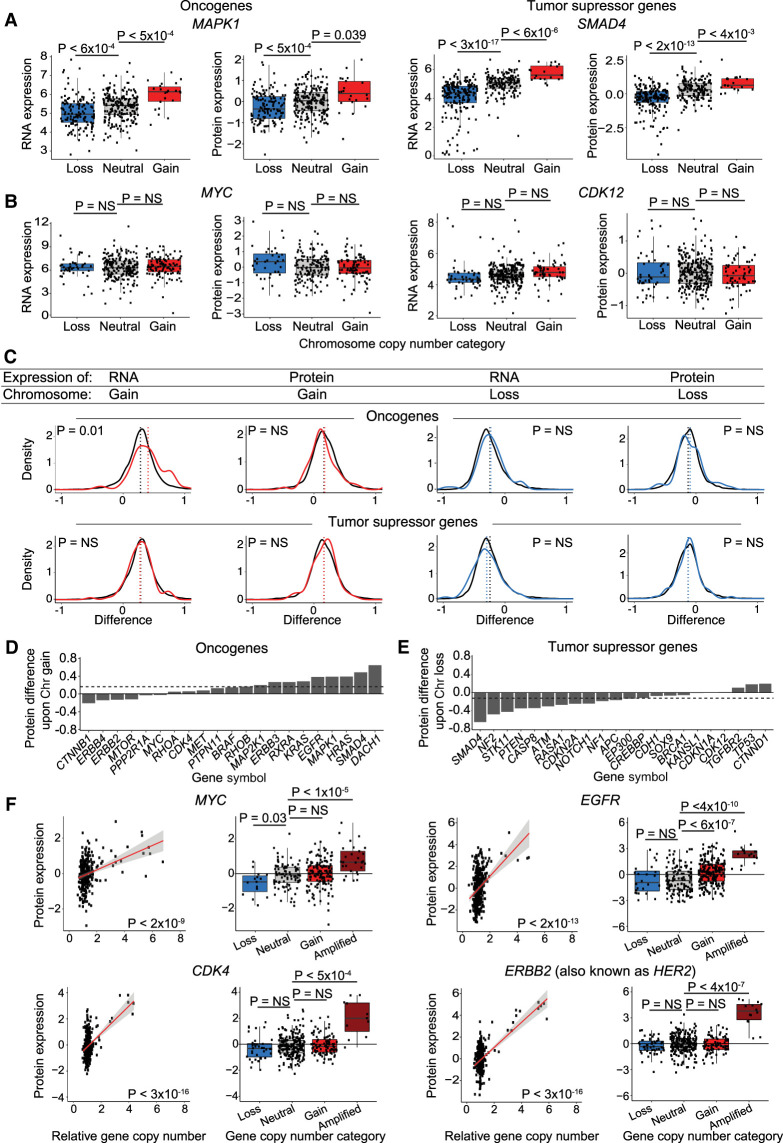
It is commonly hypothesized that aneuploidy drives tumorigenesis by increasing the expression of OGs and decreasing the expression of TSGs (Davoli et al. 2013; Giam and Rancati 2015; Smith and Sheltzer 2018). To investigate the relationship between aneuploidy and tumorigenesis, we examined the effects of aneuploidy on a set of verified TSGs and OGs (Bailey et al. 2018). At the individual gene level, some OGs and TSGs significantly scale with chromosome gain and loss (MAPK1 and SMAD4) (Fig. 6A), whereas others showed no significant difference in protein expression upon chromosome gain or loss (MYC and CDK12) (Fig. 6B). We discovered that the average protein expression of OGs and TSGs was not significantly different compared to the background expression of all proteins on aneuploid chromosomes (Fig. 6C). Thus, a chromosome gain event increased the expression of the average OG by only 12%, and a chromosome loss event decreased the expression of the average tumor suppressor by only 8%. No GO terms were found to be enriched among scaling, buffered, or antiscaling OGs or TSGs upon chromosome gain or loss.
Figure 6.
OGs and TSGs can be buffered upon chromosome gain or loss, though some OGs scale with gene copy number amplifications. (A,B) Boxplots displaying RNA and protein expression differences upon chromosome gain and loss for OGs (left) and TSGs (right). Boxplots display the 25th, 50th, and 75th percentiles of the data, and the whiskers indicate 1.5 interquartile ranges. P-values represent two-sided t-tests. (C) Density curve of OG and TSG expression at the RNA level and protein level upon chromosome gain (red) or chromosome loss (blue). Difference in expression for all other genes is displayed in black. Mean expression differences per condition and gene group are labeled by dotted lines. P-values are from two-sided t-tests between the indicated gene group and the background set of all other genes. The list of all OGs and TSGs used is in Supplemental Table S6. (D,E) Bar graphs displaying the mean protein expression difference for OGs upon chromosome gain (D) and TSGs upon chromosome loss (E). Not all OGs and TSGs are displayed. Dotted lines indicate the mean differences in expression for all genes. The complete list of OG and TSG expression differences upon chromosome gain and loss are available in Supplemental Table S2. (F) Scatterplots and boxplots displaying protein expression levels for four OGs, MYC, EGFR, CDK4, and ERBB2 (also known as HER2), relative to their gene copy number. Linear regressions (red) with 95% confidence intervals (gray) are plotted against the data. P-values in the scatterplots were calculated from the Pearson correlation coefficient, and P-values in the boxplots were calculated from two-sided t-tests.
Among individual OGs and TSGs, we noted that several potent OGs showed buffering or antiscaling upon chromosome gain (MYC, MTOR, ERBB4) (Fig. 6D). For instance, the MYC gene is located on Chromosome 8q, but we did not detect a significant increase in MYC expression in 110 cell lines with Chr8q gains compared to 211 cell lines that are neutral for Chr8q (Fig. 6B). Similarly, the expression of many tumor suppressors, including BRCA1, CDKN1A, and CDH1, did not significantly decrease when the chromosome that they were encoded on was lost (Fig. 6E).
If OGs are subjected to dosage compensation in aneuploid cells, then how could increases in OG expression levels arise to drive tumorigenesis? We observed that several OGs that were subject to dosage compensation upon single-chromosome gains were nonetheless sensitive to gene-level focal amplifications. For example, 11% of cancer cell lines showed focal amplifications of the MYC locus, and this was associated with a significant 86% increase in MYC protein expression (Fig. 6F). Similar results were obtained for other potent OGs, including EGFR, CDK4, and ERBB2 (also known as HER2) (Fig. 6F). Thus, OG dosage compensation is imperfect. Although genes like MYC can be resistant to expression changes resulting from arm-length aneuploidies, focal amplifications overcome these compensatory mechanisms and cause an increase in driver OG expression.
Ovarian tumors display extensive protein dosage compensation
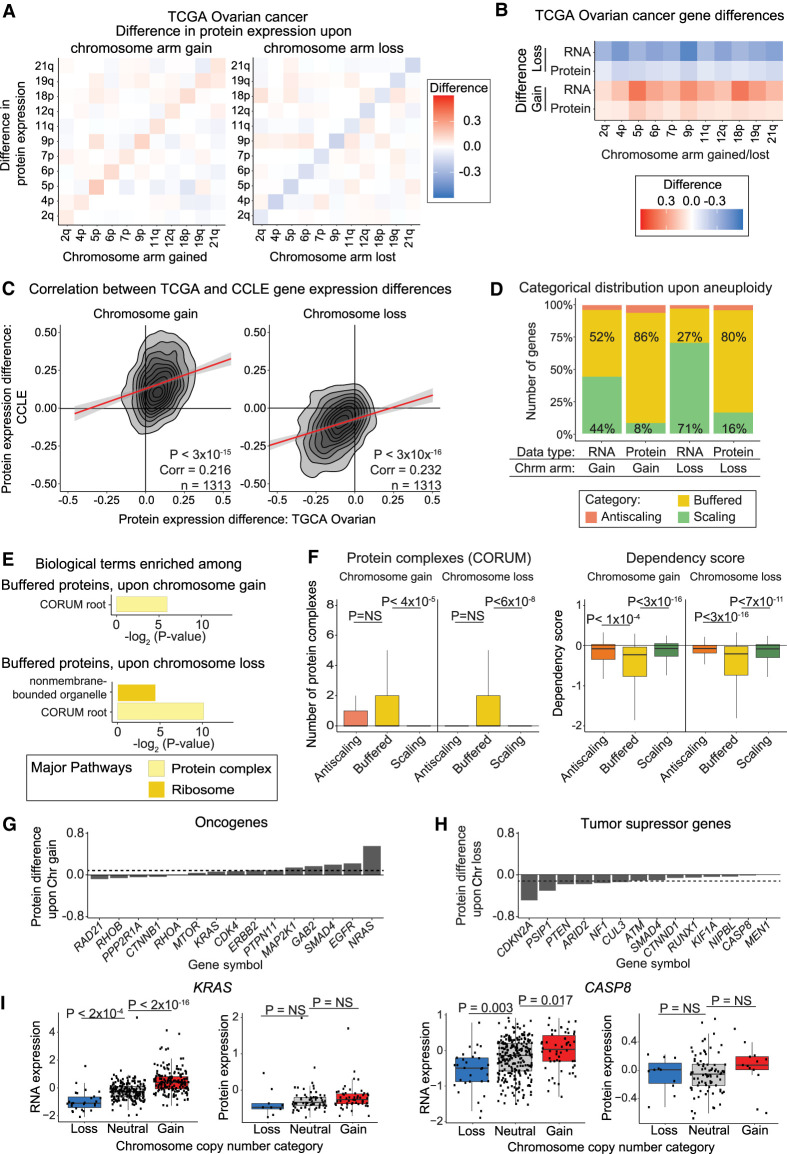
We sought to determine if the extensive dosage compensation we uncovered in cancer cell lines was maintained in vivo. To address this, we examined data from a cohort of 168 ovarian tumors with matched DNA copy number, RNA expression, and protein expression measurements (Edwards et al. 2015; Broad Institute TCGA Genome Data Analysis Center [http://gdac.broadinstitute.org/runs/stddata__2016_01_28] [accessed November 30, 2021]; Taylor et al. 2018). We calculated the average RNA and protein expression changes for all chromosome arms (Fig. 7A; Supplemental Fig. S9A; Supplemental Table S13). On average, RNA and protein expression changes increase 22% and 8% upon chromosome gain and decrease 33% and 12% upon chromosome loss, respectively (Fig. 7B).
Figure 7.
Dosage compensation in aneuploid ovarian tumors. (A) Heatmaps displaying the difference in protein expression between ovarian tumors with a chromosome arm gain (left) or chromosome arm loss (right) compared to tumors with neutral ploidy for that arm. The mean protein expression differences per chromosome arm are displayed. (B) Heatmaps displaying the same analysis as in A, but only for each aneuploid chromosome. (C) Density plots comparing the difference in CCLE and ovarian tumor protein expression differences upon the gain (left) or loss (right) of the chromosome arm on which the gene is located. Pearson correlation coefficient and P-value are displayed. Linear regressions (red) with 95% confidence intervals are displayed. (D) The percentage of ovarian tumor RNAs and proteins that fall into each difference category upon chromosome gain and loss are displayed. (E) Bar graphs displaying the biological terms enriched in ovarian tumor proteins buffered upon chromosome gain or loss, categorized by the major overarching pathway. (F) Boxplots displaying buffering factor scores, per difference category upon chromosome gain or loss using ovarian tumor data. Two buffering factors are displayed: number of protein complexes and dependency scores. P-values are from two-sided t-tests. Boxplots display the 25th, 50th, and 75th percentiles of the data, and the whiskers indicate 1.5 interquartile ranges. (G,H) Bar graphs displaying the mean protein expression difference for OGs upon chromosome gain (G) and TSGs upon chromosome loss (H), based on ovarian tumor data. Not all OGs and TSGs are displayed. Dotted lines indicate the mean differences in expression for all genes. Complete lists of OG and TSG expression differences upon chromosome gain and loss are available in Supplemental Table S2. (I) Boxplots displaying protein expression for KRAS and CASP8 by their chromosome copy number category. P-values in the boxplots were calculated from two-sided t-tests.
RNA and protein expression differences in ovarian tumors were significantly positively correlated with RNA and protein expression differences in the CCLE upon both chromosome gain and loss (Fig. 7C; Supplemental Fig. S9B). Similar to the CCLE data set, the majority of proteins in ovarian tumor samples are buffered upon chromosome gain and loss (86% and 80%, respectively) (Fig. 7D). Next, we used GOEA to identify terms enriched in proteins buffered in ovarian tumors. The only terms enriched in buffered proteins were protein complexes and ribosome-associated features (Fig. 7E; Supplemental Table S13), whereas extracellular genes were enriched in buffered RNA upon chromosome loss (Supplemental Fig. S9C; Supplemental Table S13). Similar factors were associated with protein buffering in both the CCLE and ovarian tumor data sets, including the number of ubiquitination sites, protein complex formation, and gene dependency scores (Fig. 7F; Supplemental Fig. S9D; Supplemental Table S13).
Finally, we turned our attention to the question of OG and tumor suppressor dosage compensation in vivo. We found extensive dosage compensation among OGs and TSGs in ovarian tumors (Fig. 7G,H). For example, the expression of KRAS and CASP8 correlated with chromosome copy number at the RNA level but was dosage compensated at the protein level (Fig. 7I). In sum, these results show that the patterns of dosage compensation that we described in vitro are conserved in vivo.
Discussion
Here, we conducted a genome-wide analysis of protein expression changes in aneuploid cancer cell lines and ovarian tumors. We show that, although protein expression tends to increase upon chromosome gain, and protein expression tends to decrease upon chromosome loss, a majority of proteins are subjected to dosage compensation. These patterns of scaling and buffering upon aneuploidy are conserved between aneuploid yeast cells, primary human fibroblasts, immortalized cancer cell lines, and ovarian tumors. Dosage compensation in aneuploid cancers may counteract alterations in the expression of OGs and tumor suppressors, and therefore a comprehensive understanding of this phenomenon will shed light on how chromosomal alterations drive tumorigenesis.
Consistent with previous results (Geiger et al. 2010; McShane et al. 2016; Gonçalves et al. 2017; Brennan et al. 2019; Taggart et al. 2020; Senger and Schaefer 2021), we found that protein complex subunits showed significant buffering when they were encoded upon a gained chromosome arm. This trend is exemplified by ribosomal complex proteins (Stingele et al. 2012; Dephoure et al. 2014), which showed highly significant dosage compensation. A recent study showed that protein complex subunits located on nonaneuploid chromosomes tend to co-regulate with their co-complex partners on aneuploid chromosomes in human tumor samples (Senger et al. 2022). We discovered that protein complex subunits were also buffered upon chromosome arm loss. Protein complex RNA levels tend to scale with chromosome copy number, indicating that buffering occurs primarily at the post-transcriptional level. We hypothesize that the stabilization and reduced degradation that proteins experience when they are incorporated into complexes may contribute to expression buffering upon chromosome loss, whereas the rapid degradation of unincorporated subunits may contribute to protein buffering upon chromosome gain. In support of this hypothesis, NED kinetics (McShane et al. 2016) are more predictive for buffered proteins upon chromosome loss than gain. Additionally, the NED delta is an indirect measure of protein overproduction in euploid conditions, and protein overproduction can buffer gene down-regulation. Thus, cells may have unique pathways that are able to maintain appropriate complex stoichiometries in the event of both subunit overproduction and subunit underproduction.
Our study has several limitations. Our analysis was performed across genetically diverse human tumors and cancer cell lines. These cancers show varying degrees of aneuploidy, which are also capable of affecting gene expression in trans through the induction of aneuploidy-associated stresses (Sheltzer et al. 2012; Dürrbaum et al. 2014). Nonetheless, we observed striking expression similarities between our analysis of cancer cell lines, ovarian tumors, congenic cell lines engineered to harbor single extra chromosomes, primary fibroblasts naturally trisomic for Chromosome 21, and even yeast strains harboring additional yeast chromosomes. These findings indicate that, despite the genetic diversity and potential presence of tumor heterogeneity, the patterns of buffering and scaling that we describe reflect protein dosage changes that are shared in other aneuploid conditions. Moreover, the evolutionary conservation of these buffering patterns may reflect highly conserved methods for controlling the stoichiometry of protein complex subunits. The conservation of protein expression patterns between aneuploid cancers and Down syndrome fibroblasts suggests that the patterns we have described could also contribute to aneuploidy-associated developmental syndromes (Goel et al. 2019; Hwang et al. 2019; Bull 2020).
Several distinct genetic and biochemical factors were capable of predicting protein buffering. Although this analysis gives insights into commonalities among buffered proteins, these correlations do not necessarily imply causality. In addition to protein–protein interactions, these factors included a number of post-translational modifications, aggregation propensity, and gene essentiality. We also discovered that 10% of proteins encoded on an aneuploid chromosome showed an opposite directional change relative to the chromosome copy number alteration. Our analysis highlights that buffered proteins and these “antiscaling” proteins are differentially regulated by a variety of biological factors. For example, antiscaling genes are less likely to be part of a protein complex than buffered genes and show fewer total protein–protein interactions. More research will be needed to fully elucidate the regulation of these unusual genes and their effects on the physiology of aneuploid cancers.
It is commonly hypothesized that one of the main drivers of the evolution of aneuploid cancers is the copy number of OGs and tumor suppressors (Davoli et al. 2013). Although many OGs show increased expression upon chromosome gain, and many tumor suppressors show decreased expression upon chromosome loss, we discovered that this is not true for all OGs and tumor suppressors. Indeed, many important OGs, including ERBB2 (also known as HER2), MTOR, and MYC, do not increase in expression with the gain of the corresponding chromosome arm, and the overall effect of chromosome gains across all OGs was moderate in both cancer cell lines and in vivo. Although prior studies of subchromosomal amplifications have highlighted that these alterations are frequently buffered at the protein level (Zhang et al. 2014, 2016; Mertins et al. 2016; Gonçalves et al. 2017), we found that focal amplifications (but not chromosome-scale alterations) are associated with significant increases in the expression of key OGs like MYC and ERBB2. These results indicate that the buffering of OG expression is imperfect, and although single-copy gains may be subject to dosage compensation, high-copy amplifications result in OG overexpression, even though this expression may not be perfectly proportional to OG copy number. Finally, we suggest that the evolutionary pressure to overcome OG buffering during tumor development may promote selection for high-copy amplifications that escape compensatory down-regulation, leading to the highly complex karyotypes often found in advanced malignancies.
OG and TSG dosage compensation complicates, but does not contradict, the aneuploidy/driver gene dosage hypothesis. Developing a comprehensive picture of OG and tumor suppressor buffering will improve our understanding of cancer driver genes and their role in shaping aneuploid karyotypes in cancer. As aneuploidy has been recognized to be a highly significant prognostic factor across cancer types (Kheir et al. 1988; Xu et al. 2016; Hieronymus et al. 2018; Smith and Sheltzer 2018; Stopsack et al. 2019; Shukla et al. 2020; van Dijk et al. 2021), and as genomic and exomic sequencing costs have rapidly fallen (Mardis 2019), we expect that high-resolution cancer karyotyping will become increasingly routine in a clinical setting. Knowing which proteins are sensitive to chromosome gains and losses may help identify the specific drivers in a patient's tumor, leading to better personalized therapies and improved patient outcomes.
Methods
Data set filtering
We generated a data set measuring gene expression difference upon chromosome gain or loss. We only used cell lines with proteomics data, RNA expression data, and chromosome arm copy number data (367 cancer cell lines and 168 ovarian tumor samples). We only included a gene in our difference analysis if we had RNA and protein expression data for that gene from at least 10 cell lines in which the chromosome arm in which that gene was encoded on was gained, 10 cell lines in which the chromosome arm was lost, and 10 cell lines in which the chromosome arm had a neutral ploidy. Not all chromosome arms in the ovarian tumor data had more than 10 samples with a loss, 10 samples with a gain, and 10 samples neutral for that chromosome arm.
For the protein expression correlation with aneuploidy score data set used for Figure 4, all cell lines with protein expression and chromosome arm copy number data were used for protein aneuploidy score analysis. Similarly, for the RNA expression correlation with aneuploidy score data set used in Supplemental Figure S7, all cell lines with RNA expression measurements and chromosome arm copy number data were used.
Analyzing the effects of aneuploidy on gene expression
Gene expression difference upon aneuploidy was calculated for each gene, for both RNA and protein data. For each gene, the chromosome arm on which the gene was located was identified. Next, cell lines in the CCLE filtered data set (see above) were grouped as either having a chromosome arm gain, chromosome arm loss, or having a neutral ploidy for that chromosome arm. Then, the mean RNA and protein expression data for the gene of interest were taken per DNA aneuploidy category (gain, loss, or neutral). The mean gene expression in the neutral category was subtracted from the mean gene expression in the chromosome gain category to obtain the difference in expression upon chromosome gain. Similarly, mean gene expression in the neutral category was subtracted from the mean protein expression in the chromosome loss category to get the difference in expression upon chromosome loss. We found the difference in expression upon chromosome gain and loss for both protein expression data and RNA expression data. As protein and RNA expression data have already been log2 transformed, the difference [log2(A) − log2(B)] is equivalent to log2 fold change [log2(A/B)]. P-values per gene were calculated by using a two-sided t-test between gene expression in cell lines neutral for the corresponding chromosome arm and cell lines with either a chromosome gain or loss (Supplemental Tables S2, S11, S13).
Percent change in gene expression upon aneuploidy was calculated in various data sets. The mean difference in expression upon chromosome gain or loss was calculated and converted into percentages by using X = 2 (mean difference) − 1.
Cell line–specific difference was calculated for RNA and protein data in Figure 1 and Supplemental Figure S1. The mean RNA and protein expression data per chromosome arm were taken from cell lines neutral for chromosome arm gains or losses. This mean “neutral” gene expression was subtracted from the mean gene expression per chromosome arm in the cell line of interest.
Gene expression categories were based on the difference upon chromosome gain or loss in RNA or protein expression data. For chromosome gain events, genes were classified as “antiscaling” if their mean expression difference was less than −0.1 relative to cell lines that were neutral for that chromosome arm. Genes were classified as “buffered” if their mean expression ranged from −0.1 to 0.25 relative to cell lines that were neutral for that chromosome arm. Genes were classified as “scaling” if their mean expression was greater than 0.25 relative to cell lines that were neutral for that chromosome arm. For chromosome loss events, the values were reversed (greater than 0.1 for antiscaling, between 0.1 and −0.25 for buffered, and less than −0.25 for scaling).
For the analysis of protein expression via RPPA, antibodies that recognized specific post-translational modifications and antibodies that recognized proteins encoded by multiple genes were excluded from analysis. This left 149 proteins that were recognized by 151 antibodies. For each remaining antibody, we separately calculated the average expression when the gene encoding the protein recognized by the antibody was present on a neutral, gained, or lost chromosome. We then subtracted the mean expression of each protein when present on a neutral chromosome from the mean expression of the same protein when encoded on a gained or lost chromosome. For the two proteins recognized by two antibodies (BCL2L11 [also known as BIM] and RAF1), the values were collapsed by averaging.
Cellular aneuploidy scores were calculated by summing the number of genes on all aneuploid chromosome arms in a cell line and then dividing that total by the cell's basal ploidy. In this way, the gain or loss of a chromosome arm contributes equally to the aneuploidy score.
Aneuploidy score correlation was calculated per gene for both protein expression and RNA expression data. Pearson correlations between protein or RNA expression and a cell's aneuploidy score were measured for every gene. Associated P-values are Pearson correlation significance measurements (Supplemental Table S9).
GO term enrichment analysis
Biological term enrichments were identified using g:Profiler (Raudvere et al. 2019). Significance values were calculated against a custom background set of genes or proteins that were included in our processed and filtered data set.
OG and tumor suppressor buffering analysis
Relative gene copy number categories were classified as “loss” with a score less than 0.9, “neutral” at 0.9–1.1, “gain” at 1.1–1.75, and “amplified” at 1.75+. Relative gene copy numbers are log2-transformed data, with a pseudo-count of one.
Additional sources
See the Supplemental Methods for a complete list of data sources, additional data set filtering information, yeast ortholog identification methods, and methods used to identify genomic features that correlate with protein buffering. Several sources, in addition to those cited in the above text, were used for data analysis (Yang et al. 2003; Robin et al. 2011; Ciryam et al. 2013; Savitski et al. 2013; Hornbeck et al. 2015; Smedley et al. 2015; Alanis-Lobato et al. 2017; Mathieson et al. 2018; Giurgiu et al. 2019; Hausser et al. 2019; Piovesan et al. 2021).
Data access
The code and intermediate analysis data sets generated in this study are available at GitHub (https://github.com/kschukken/Genome_Research_Protein_Dosage) and as Supplemental Code. All code was written in R.
Supplementary Material
Acknowledgments
Research in the Sheltzer laboratory is supported by a National Institutes of Health (NIH) Early Independence award (1DP5OD021385), NIH grant R01CA237652, Department of Defense grant W81XWH-20-1-068, a Damon Runyon-Rachleff Innovation award, an American Cancer Society Research Scholar grant, and a grant from the New York Community Trust.
Author contributions: K.M.S. and J.M.S. conceived, designed, and performed the analysis described in this work. K.M.S. and J.M.S. wrote the manuscript and prepared the figures.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276378.121.
Competing interest statement
J.M.S. has received consulting fees from Ono Pharmaceuticals and Merck, is a member of the advisory board of Tyra Biosciences, and is a cofounder of Meliora Therapeutics.
References
- Alanis-Lobato G, Andrade-Navarro MA, Schaefer MH. 2017. HIPPIE v2.0: enhancing meaningfulness and reliability of protein–protein interaction networks. Nucleic Acids Res 45: D408–D414. 10.1093/nar/gkw985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, Colaprico A, Wendl MC, Kim J, Reardon B, et al. 2018. Comprehensive characterization of cancer driver genes and mutations. Cell 173: 371–385.e18. 10.1016/j.cell.2018.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Ngo B, Laughney AM, Cavallo J-A, Murphy CJ, Ly P, Shah P, Sriram RK, Watkins TBK, Taunk NK, et al. 2018. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553: 467–472. 10.1038/nature25432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, et al. 2012. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483: 603–607. 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, Amon A. 2020. Context is everything: aneuploidy in cancer. Nat Rev Genet 21: 44–62. 10.1038/s41576-019-0171-x [DOI] [PubMed] [Google Scholar]
- Brennan CM, Vaites LP, Wells JN, Santaguida S, Paulo JA, Storchova Z, Harper JW, Marsh JA, Amon A. 2019. Protein aggregation mediates stoichiometry of protein complexes in aneuploid cells. Genes Dev 33: 1031–1047. 10.1101/gad.327494.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buccitelli C, Selbach M. 2020. mRNAs, proteins and the emerging principles of gene expression control. Nat Rev Genet 21: 630–644. 10.1038/s41576-020-0258-4 [DOI] [PubMed] [Google Scholar]
- Bull MJ. 2020. Down syndrome. N Engl J Med 382: 2344–2352. 10.1056/NEJMra1706537 [DOI] [PubMed] [Google Scholar]
- Chunduri NK, Menges P, Zhang X, Wieland A, Gotsmann VL, Mardin BR, Buccitelli C, Korbel JO, Willmund F, Kschischo M, et al. 2021. Systems approaches identify the consequences of monosomy in somatic human cells. Nat Commun 12: 5576. 10.1038/s41467-021-25288-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciryam P, Tartaglia GG, Morimoto RI, Dobson CM, Vendruscolo M. 2013. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep 5: 781–790. 10.1016/j.celrep.2013.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Sharir Y, McFarland JM, Abdusamad M, Marquis C, Bernhard SV, Kazachkova M, Tang H, Ippolito MR, Laue K, Zerbib J, et al. 2021. Aneuploidy renders cancer cells vulnerable to mitotic checkpoint inhibition. Nature 590: 486–491. 10.1038/s41586-020-03114-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, Elledge SJ. 2013. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155: 948–962. 10.1016/j.cell.2013.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Uno H, Wooten EC, Elledge SJ. 2017. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355: eaaf8399. 10.1126/science.aaf8399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Hwang S, O'Sullivan C, Dodgson SE, Gygi SP, Amon A, Torres EM. 2014. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. eLife 3: e03023. 10.7554/eLife.03023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly N, Storchová Z. 2015. Causes and consequences of protein folding stress in aneuploid cells. Cell Cycle 14: 495–501. 10.1080/15384101.2015.1006043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly N, Passerini V, Dürrbaum M, Stingele S, Storchová Z. 2014. HSF1 deficiency and impaired HSP90-dependent protein folding are hallmarks of aneuploid human cells. EMBO J 33: 2374–2387. 10.15252/embj.201488648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duijf PHG, Schultz N, Benezra R. 2013. Cancer cells preferentially lose small chromosomes. Int J Cancer 132: 2316–2326. 10.1002/ijc.27924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürrbaum M, Kuznetsova AY, Passerini V, Stingele S, Stoehr G, Storchová Z. 2014. Unique features of the transcriptional response to model aneuploidy in human cells. BMC Genomics 15: 139. 10.1186/1471-2164-15-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards NJ, Oberti M, Thangudu RR, Cai S, McGarvey PB, Jacob S, Madhavan S, Ketchum KA. 2015. The CPTAC data portal: a resource for cancer proteomics research. J Proteome Res 14: 2707–2713. 10.1021/pr501254j [DOI] [PubMed] [Google Scholar]
- Estrada JC, Torres Y, Benguría A, Dopazo A, Roche E, Carrera-Quintanar L, Pérez RA, Enríquez JA, Torres R, Ramírez JC, et al. 2013. Human mesenchymal stem cell-replicative senescence and oxidative stress are closely linked to aneuploidy. Cell Death Dis 4: e691. 10.1038/cddis.2013.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanillo C, Aibar S, Sanchez-Santos JM, De Las Rivas J. 2012. Combined analysis of genome-wide expression and copy number profiles to identify key altered genomic regions in cancer. BMC Genomics 13: S5. 10.1186/1471-2164-13-S5-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Su Y, Koeman J, Haak E, Dykema K, Essenberg C, Hudson E, Petillo D, Khoo SK, Vande Woude GF. 2016. Chromosome instability drives phenotypic switching to metastasis. Proc Natl Acad Sci 113: 14793–14798. 10.1073/pnas.1618215113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T, Cox J, Mann M. 2010. Proteomic changes resulting from gene copy number variations in cancer cells. PLoS Genet 6: e1001090. 10.1371/journal.pgen.1001090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER, Barretina J, Gelfand ET, Bielski CM, Li H, et al. 2019. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569: 503–508. 10.1038/s41586-019-1186-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giam M, Rancati G. 2015. Aneuploidy and chromosomal instability in cancer: a jackpot to chaos. Cell Div 10: 3. 10.1186/s13008-015-0009-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giam M, Wong CK, Low JS, Sinelli M, Dreesen O, Rancati G. 2020. P53 induces senescence in the unstable progeny of aneuploid cells. Cell Cycle 19: 3508–3520. 10.1080/15384101.2020.1850968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giurgiu M, Reinhard J, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Ruepp A. 2019. CORUM: the comprehensive resource of mammalian protein complexes: 2019. Nucleic Acids Res 47: D559–D563. 10.1093/nar/gky973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel N, Morris JK, Tucker D, de Walle HEK, Bakker MK, Kancherla V, Marengo L, Canfield MA, Kallen K, Lelong N, et al. 2019. Trisomy 13 and 18—prevalence and mortality—a multi-registry population based analysis. Am J Med Genet A 179: 2382–2392. 10.1002/ajmg.a.61365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves E, Fragoulis A, Garcia-Alonso L, Cramer T, Saez-Rodriguez J, Beltrao P. 2017. Widespread post-transcriptional attenuation of genomic copy-number variation in cancer. Cell Syst 5: 386–398.e4. 10.1016/j.cels.2017.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grade M, Hörmann P, Becker S, Hummon AB, Wangsa D, Varma S, Simon R, Liersch T, Becker H, Difilippantonio MJ, et al. 2007. Gene expression profiling reveals a massive, aneuploidy-dependent transcriptional deregulation and distinct differences between lymph node–negative and lymph node–positive colon carcinomas. Cancer Res 67: 41–56. 10.1158/0008-5472.CAN-06-1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Choi H, Ghosh D. 2008. Global associations between copy number and transcript mRNA microarray data: an empirical study. Cancer Inform 6: 17–23. 10.4137/cin.s342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausser J, Mayo A, Keren L, Alon U. 2019. Central dogma rates and the trade-off between precision and economy in gene expression. Nat Commun 10: 68. 10.1038/s41467-018-07391-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Au B, Kulkarni M, Shen Y, Lim KJ, Maimaiti J, Wong CK, Luijten MNH, Chong HC, Lim EH, et al. 2018. Chromosomal instability-induced senescence potentiates cell non-autonomous tumourigenic effects. Oncogenesis 7: 62. 10.1038/s41389-018-0072-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H, Murali R, Tin A, Yadav K, Abida W, Moller H, Berney D, Scher H, Carver B, Scardino P, et al. 2018. Tumor copy number alteration burden is a pan-cancer prognostic factor associated with recurrence and death. eLife 7: e37294. 10.7554/eLife.37294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. 2015. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res 43: D512–D520. 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Williams JF, Kneissig M, Lioudyno M, Rivera I, Helguera P, Busciglio J, Storchova Z, King MC, Torres EM. 2019. Suppressing aneuploidy-associated phenotypes improves the fitness of trisomy 21 cells. Cell Rep 29: 2473–2488.e5. 10.1016/j.celrep.2019.10.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Cavaliere P, Li R, Zhu LJ, Dephoure N, Torres EM. 2021. Consequences of aneuploidy in human fibroblasts with trisomy 21. Proc Natl Acad Sci 118: e2014723118. 10.1073/pnas.2014723118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippolito MR, Martis V, Martin S, Tijhuis AE, Hong C, Wardenaar R, Dumont M, Zerbib J, Spierings DCJ, Fachinetti D, et al. 2021. Gene copy-number changes and chromosomal instability induced by aneuploidy confer resistance to chemotherapy. Dev Cell 56: 2440–2454.e6. 10.1016/j.devcel.2021.07.006 [DOI] [PubMed] [Google Scholar]
- Kheir SM, Bines SD, Vonroenn JH, Soong SJ, Urist MM, Coon JS. 1988. Prognostic significance of DNA aneuploidy in stage I cutaneous melanoma. Ann Surg 207: 455–461. 10.1097/00000658-198804000-00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneau A, Santoni FA, Bonilla X, Sailani MR, Gonzalez D, Kind J, Chevalier C, Thurman R, Sandstrom RS, Hibaoui Y, et al. 2014. Domains of genome-wide gene expression dysregulation in Down's syndrome. Nature 508: 345–350. 10.1038/nature13200 [DOI] [PubMed] [Google Scholar]
- Liu Y, Borel C, Li L, Müller T, Williams EG, Germain P-L, Buljan M, Sajic T, Boersema PJ, Shao W, et al. 2017. Systematic proteome and proteostasis profiling in human trisomy 21 fibroblast cells. Nat Commun 8: 1212. 10.1038/s41467-017-01422-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukow DA, Sausville EL, Suri P, Chunduri NK, Wieland A, Leu J, Smith JC, Girish V, Kumar AA, Kendall J, et al. 2021. Chromosomal instability accelerates the evolution of resistance to anti-cancer therapies. Dev Cell 56: 2427–2439.e4. 10.1016/j.devcel.2021.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo JC, Vaz S, Bakker B, Ribeiro R, Bakker PL, Escandell JM, Ferreira MG, Medema R, Foijer F, Logarinho E. 2018. FoxM1 repression during human aging leads to mitotic decline and aneuploidy-driven full senescence. Nat Commun 9: 2834. 10.1038/s41467-018-05258-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. 2019. The impact of next-generation sequencing on cancer genomics: from discovery to clinic. Cold Spring Harb Perspect Med 9: a036269. 10.1101/cshperspect.a036269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson T, Franken H, Kosinski J, Kurzawa N, Zinn N, Sweetman G, Poeckel D, Ratnu VS, Schramm M, Becher I, et al. 2018. Systematic analysis of protein turnover in primary cells. Nat Commun 9: 689. 10.1038/s41467-018-03106-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane E, Sin C, Zauber H, Wells JN, Donnelly N, Wang X, Hou J, Chen W, Storchova Z, Marsh JA, et al. 2016. Kinetic analysis of protein stability reveals age-dependent degradation. Cell 167: 803–815.e21. 10.1016/j.cell.2016.09.015 [DOI] [PubMed] [Google Scholar]
- Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, Wang X, Qiao JW, Cao S, Petrali F, et al. 2016. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534: 55–62. 10.1038/nature18003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow DP, Gygi SP. 2020. A guide to the quantitative proteomic profiles of the Cancer Cell Line Encyclopedia. bioRxiv 10.1101/2020.02.03.932384 [DOI]
- Nusinow DP, Szpyt J, Ghandi M, Rose CM, McDonald ER, Kalocsay M, Jané-Valbuena J, Gelfand E, Schweppe DK, Jedrychowski M, et al. 2020. Quantitative proteomics of the Cancer Cell Line Encyclopedia. Cell 180: 387–402.e16. 10.1016/j.cell.2019.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oromendia AB, Dodgson SE, Amon A. 2012. Aneuploidy causes proteotoxic stress in yeast. Genes Dev 26: 2696–2708. 10.1101/gad.207407.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passerini V, Ozeri-Galai E, de Pagter MS, Donnelly N, Schmalbrock S, Kloosterman WP, Kerem B, Storchová Z. 2016. The presence of extra chromosomes leads to genomic instability. Nat Commun 7: 10754. 10.1038/ncomms10754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovesan D, Necci M, Escobedo N, Monzon AM, Hatos A, Mičetić I, Quaglia F, Paladin L, Ramasamy P, Dosztányi Z, et al. 2021. MobiDB: intrinsically disordered proteins in 2021. Nucleic Acids Res 49: D361–D367. 10.1093/nar/gkaa1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack JR, Sørlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Børresen-Dale A-L, Brown PO. 2002. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci 99: 12963–12968. 10.1073/pnas.162471999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. 2019. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 47: W191–W198. 10.1093/nar/gkz369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Replogle JM, Zhou W, Amaro AE, McFarland JM, Villalobos-Ortiz M, Ryan J, Letai A, Yilmaz O, Sheltzer J, Lippard SJ, et al. 2020. Aneuploidy increases resistance to chemotherapeutics by antagonizing cell division. Proc Natl Acad Sci 117: 30566–30576. 10.1073/pnas.2009506117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, Müller M. 2011. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12: 77. 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgueiro L, Buccitelli C, Rowald K, Somogyi K, Kandala S, Korbel JO, Sotillo R. 2020. Acquisition of chromosome instability is a mechanism to evade oncogene addiction. EMBO Mol Med 12: e10941. 10.15252/emmm.201910941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansregret L, Swanton C. 2017. The role of aneuploidy in cancer evolution. Cold Spring Harb Perspect Med 7: a028373. 10.1101/cshperspect.a028373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitski MM, Mathieson T, Zinn N, Sweetman G, Doce C, Becher I, Pachl F, Kuster B, Bantscheff M. 2013. Measuring and managing ratio compression for accurate iTRAQ/TMT quantification. J Proteome Res 12: 3586–3598. 10.1021/pr400098r [DOI] [PubMed] [Google Scholar]
- Schukken KM, Lin Y-C, Bakker PL, Schubert M, Preuss SF, Simon JE, van den Bos H, Storchova Z, Colomé-Tatché M, Bastians H, et al. 2020. Altering microtubule dynamics is synergistically toxic with spindle assembly checkpoint inhibition. Life Sci Alliance 3: e201900499. 10.26508/lsa.201900499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki AM, Dulmage K, Cowen LE, Anderson JB, Berman J. 2009. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet 5: e1000705. 10.1371/journal.pgen.1000705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger G, Schaefer MH. 2021. Protein complex organization imposes constraints on proteome dysregulation in cancer. Front Bioinforma 1: 723482. 10.3389/fbinf.2021.723482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger G, Santaguida S, Schaefer MH. 2022. Regulation of protein complex partners as a compensatory mechanism in aneuploid tumors. eLife 11: e75526. 10.7554/eLife.75526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM. 2013. A transcriptional and metabolic signature of primary aneuploidy is present in chromosomally unstable cancer cells and informs clinical prognosis. Cancer Res 73: 6401–6412. 10.1158/0008-5472.CAN-13-0749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A. 2011. Aneuploidy drives genomic instability in yeast. Science 333: 1026–1030. 10.1126/science.1206412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Torres EM, Dunham MJ, Amon A. 2012. Transcriptional consequences of aneuploidy. Proc Natl Acad Sci 109: 12644–12649. 10.1073/pnas.1209227109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Ko JH, Replogle JM, Habibe Burgos NC, Chung ES, Meehl CM, Sayles NM, Passerini V, Storchova Z, Amon A. 2017. Single-chromosome gains commonly function as tumor suppressors. Cancer Cell 31: 240–255. 10.1016/j.ccell.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Nguyen THM, Moka SB, Ellis JJ, Grady JP, Oey H, Cristino AS, Khanna KK, Kroese DP, Krause L, et al. 2020. Chromosome arm aneuploidies shape tumour evolution and drug response. Nat Commun 11: 449. 10.1038/s41467-020-14286-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Haider S, Durinck S, Pandini L, Provero P, Allen J, Arnaiz O, Awedh MH, Baldock R, Barbiera G, et al. 2015. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res 43: W589–W598. 10.1093/nar/gkv350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JC, Sheltzer JM. 2018. Systematic identification of mutations and copy number alterations associated with cancer patient prognosis. eLife 7: e39217. 10.7554/eLife.39217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JC, Sheltzer JM. 2022. Genome-wide identification and analysis of prognostic features in human cancers. Cell Rep 38: 110569. 10.1016/j.celrep.2022.110569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A, Gonçalves E, Mirauta B, Ochoa D, Stegle O, Beltrao P. 2019. Multi-omics characterization of interaction-mediated control of human protein abundance levels. Mol Cell Proteomics MCP 18: S114–S125. 10.1074/mcp.RA118.001280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z. 2012. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol 8: 608. 10.1038/msb.2012.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopsack KH, Whittaker CA, Gerke TA, Loda M, Kantoff PW, Mucci LA, Amon A. 2019. Aneuploidy drives lethal progression in prostate cancer. Proc Natl Acad Sci 116: 11390–11395. 10.1073/pnas.1902645116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart JC, Zauber H, Selbach M, Li G-W, McShane E. 2020. Keeping the proportions of protein complex components in check. Cell Syst 10: 125–132. 10.1016/j.cels.2020.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y-C, Williams BR, Siegel JJ, Amon A. 2011. Identification of aneuploidy-selective antiproliferation compounds. Cell 144: 499–512. 10.1016/j.cell.2011.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, Schumacher SE, Wang C, Hu H, Liu J, et al. 2018. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33: 676–689.e3. 10.1016/j.ccell.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting L, Rad R, Gygi SP, Haas W. 2011. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods 8: 937–940. 10.1038/nmeth.1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916–924. 10.1126/science.1142210 [DOI] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. 2010. Identification of aneuploidy-tolerating mutations. Cell 143: 71–83. 10.1016/j.cell.2010.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsafrir D, Bacolod M, Selvanayagam Z, Tsafrir I, Shia J, Zeng Z, Liu H, Krier C, Stengel RF, Barany F, et al. 2006. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res 66: 2129–2137. 10.1158/0008-5472.CAN-05-2569 [DOI] [PubMed] [Google Scholar]
- Upadhya SR, Ryan CJ. 2021. Experimental reproducibility limits the correlation between mRNA and protein abundances in tumour proteomic profiles. bioRxiv 10.1101/2021.09.22.461108 [DOI] [PMC free article] [PubMed]
- van Dijk E, van den Bosch T, Lenos KJ, El Makrini K, Nijman LE, van Essen HFB, Lansu N, Boekhout M, Hageman JH, Fitzgerald RC, et al. 2021. Chromosomal copy number heterogeneity predicts survival rates across cancers. Nat Commun 12: 3188. 10.1038/s41467-021-23384-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan A, Baruah PS, Smith JC, Wang Z, Sayles NM, Andrews P, Kendall J, Leu J, Chunduri NK, Levy D, et al. 2020. Single-chromosomal gains can function as metastasis suppressors and promoters in colon cancer. Dev Cell 52: 413–428.e6. 10.1016/j.devcel.2020.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan A, Schukken KM, Sausville EL, Girish V, Adebambo OA, Sheltzer JM. 2021. Aneuploidy as a promoter and suppressor of malignant growth. Nat Rev Cancer 21: 89–103. 10.1038/s41568-020-00321-1 [DOI] [PubMed] [Google Scholar]
- Viganó C, von Schubert C, Ahrné E, Schmidt A, Lorber T, Bubendorf L, De Vetter JRF, Zaman GJR, Storchova Z, Nigg EA. 2018. Quantitative proteomic and phosphoproteomic comparison of human colon cancer DLD-1 cells differing in ploidy and chromosome stability. Mol Biol Cell 29: 1031–1047. 10.1091/mbc.E17-10-0577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BAA, Cleveland DW. 2006. Does aneuploidy cause cancer? Curr Opin Cell Biol 18: 658–667. 10.1016/j.ceb.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. 2008. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322: 703–709. 10.1126/science.1160058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MW, Bruford EA. 2011. Naming “junk”: human non-protein coding RNA (ncRNA) gene nomenclature. Hum Genomics 5: 90. 10.1186/1479-7364-5-2-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Huang L, Li J. 2016. DNA aneuploidy and breast cancer: a meta-analysis of 141,163 cases. Oncotarget 7: 60218–60229. 10.18632/oncotarget.11130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE. 2003. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res 13: 1863–1872. 10.1101/gr.1272403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, et al. 2014. Proteogenomic characterization of human colon and rectal cancer. Nature 513: 382–387. 10.1038/nature13438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu T, Zhang Z, Payne SH, Zhang B, McDermott JE, Zhou J-Y, Petyuk VA, Chen L, Ray D, et al. 2016. Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell 166: 755–765. 10.1016/j.cell.2016.05.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.