Abstract
In 1971, Blobel proposed the first statement of the Signal Hypothesis which suggested that proteins have amino-terminal sequences that dictate their export and localization in the cell. A cytosolic binding factor was predicted, and later the protein conducting channel was discovered that was proposed in 1975 to align with the large ribosomal tunnel. The 1975 Signal Hypothesis also predicted that proteins targeted to different intracellular membranes would possess distinct signals and integral membrane proteins contained uncleaved signal sequences which initiate translocation of the polypeptide chain. This review summarizes the central role that the signal peptides play as address codes for proteins, their decisive role as targeting factors for delivery to the membrane and their function to activate the translocation machinery for export and membrane protein insertion. After shedding light on the navigation of proteins, the importance of removal of signal peptide and their degradation are addressed. Furthermore, the emerging work on signal peptidases as novel targets for antibiotic development is described.
Keywords: SecYEG translocase, SecA, signal peptide, protein transport, YidC, Tat pathway, signal peptidase, antibiotic targets
1 Introduction
The transport of proteins across cell membranes is fundamentally significant to many biological processes. Protein export also finds a special interest in biotechnology for production of hormones/enzymes and recombinant proteins, in laboratory techniques and disease diagnosis. Considerable progress has been made during the last several decades in understanding the characteristics of the folded state of substrates during translocation in the cytosol, membrane targeting, the structure and function of translocation devices, the insertion of membrane proteins into the lipid bilayer, and the role of energy in protein export. Insight into these fundamental concepts is highly appreciated and anticipated by scientists in the protein export field.
Protein integration and transport across the membranes are ubiquitous in every organism. Typically, these proteins are synthesized with a stretch of amino acids called the “signal peptide” that can be recognized by the cytosolic proteins for sorting and then targeting to the membrane. After being transferred to the translocation machinery, the proteins are membrane inserted or translocated across the membrane. In the final step, the signal peptide is proteolytically removed from the exported protein by signal peptidase.
The signal peptide plays center stage in this export process with a myriad of functions (Hegde and Bernstein, 2006). The signal peptides can bind to chaperones to prevent premature folding of the protein in the cytosol. In addition to slowing down the folding of a mature domain of a preprotein, signal peptides act as a zip code for sorting the proteins from the cytosol to the membrane. Finally, the signal peptide activates the translocation machinery, initiating the translocation process.
This review highlights the function of signal peptides in Gram-negative bacteria in protein sorting and targeting to the inner membrane, and translocation across the membrane and insertion. After navigating the journey of proteins, their removal and degradation are discussed. Furthermore, the potential of the signal peptidases (endopeptidases which remove signal peptides) as antibacterial targets will be covered.
2 Signal Peptides
Most exported proteins in bacteria are transported across the inner membrane by the general secretion (Sec) pathway or the Twin arginine translocation (Tat) system or the simple membrane protein insertase YidC [reviewed in (Crane and Randall, 2017; Frain et al., 2019; Shanmugam and Dalbey, 2019; Oswald et al., 2021)]. The targeting of the preproteins to these pathways are dependent on the pathway selective for the respective signal peptide. These are the Sec signal peptide, the lipoprotein signal peptide, the Tat signal peptide and the prepilin signal peptide. Below we describe the properties of each of these signal peptides.
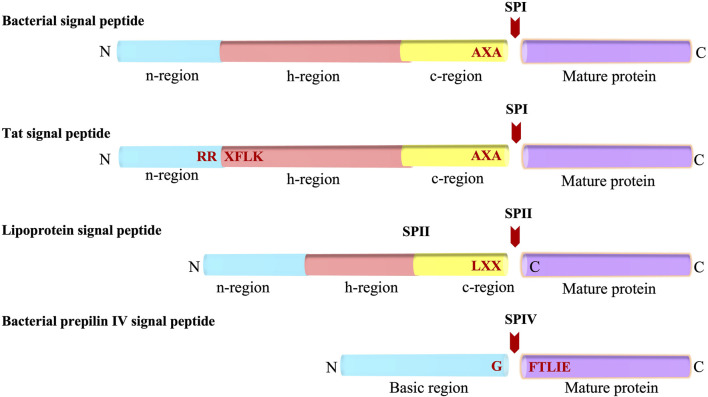
The Sec signal peptide targets the protein to the Sec machinery and is composed of three regions (Figure 1) (von Heijne and Abrahmsèn, 1989; Perlman and Halvorson, 1983; von Heijne, 1986a): 1) a positively charged N-terminal region (n), 2) a central hydrophobic region (h) and, 3) a rather polar C-terminal region which contains small amino acid residues at positions -1 and -3 (with respect to the cleavage site). Additionally, a helix breaking residue is often found at the -4 to -6 positions of the C-terminal region (von Heijne, 1986a). Genetic and mutagenesis studies have shown that the apolar region of the signal peptide is essential for the function of a cleavable signal peptide (Emr et al., 1978; Bassford and Beckwith, 1979; Michaelis and Beckwith, 1982). Moreover, the basic amino terminus can be important for making translocation more efficient (Vlasuk et al., 1983; Iino et al., 1987).
FIGURE 1.
Bacterial Signal peptides. Schematic representations of the Sec-type signal peptide, the twin-arginine (Tat) signal peptide, the lipoprotein signal peptide, and the prepilin signal peptide. The various regions of the signal peptides (n, h, c and basic regions) are indicated. The SP cleavage site is represented with a red arrow. N and C indicates amino and carboxyl-terminus, respectively.
Similar to the Sec signal peptide, the lipoprotein signal peptide which is processed by signal peptidase 2 (SPase II, lipoprotein signal peptidase) has a positively charged n region and a hydrophobic central region (h region) (Figure 1). The main difference between Sec and lipoprotein signal sequences is that the c region of the lipoprotein contains the lipobox motif comprised of Leu-Ala/Ser-Gly/Ala-Cysteine at the −3 to +1 position (Sankaran and Wu, 1994). The lipobox motif is a structural determinant for lipid modification of the strictly conserved Cys at the +1 position of the mature domain that gets modified by diacylglyceride. The glyceride fatty acid lipid is attached by a preprolipoprotein diacylglycerol transferase (Lgt), prior to cleavage by SPase II (Sankaran and Wu, 1994). In Gram-negative and some Gram-positive bacteria, most lipoproteins are further modified by an acyl chain by N-acyl transferase (Lnt) after SPase II cleavage (Gupta and Wu, 1991). Analysis of the signal sequences have shown that the h regions are shorter for lipoprotein signal peptides as compared to that present in the Sec signal peptides (Klein et al., 1988; von Heijne, 1989; Tjalsma et al., 2000).
The Tat signal peptide targets proteins to the Tat machinery and has a tripartite arrangement similar to the Sec signal peptide (Figure 1). It was initially discovered in chloroplast in exported proteins transported into the thylakoid lumen independently of ATP hydrolysis. Later, Berks and others observed it in cofactor containing periplasmic proteins of bacteria (Chaddock et al., 1995; Berks, 1996; Bogsch et al., 1998; Sargent et al., 1998; Weiner et al., 1998). The “Tat” signal peptide takes its name from the invariant and essential twin arginines in the n-region of the signal peptide. The motif for Tat signal peptides is RRXFLK where X can be any residue and F, L and K are quite commonly found. Mutagenesis of the twin arginines even to a lysine pair can abolish or significantly reduce transport although single mutations of the arginines are largely tolerated (Stanley et al., 2000; Buchanan et al., 2001; DeLisa et al., 2002). Typically, the Tat signal peptides are longer than the Sec signal peptides, and the h-region is less hydrophobic than that present in the Sec signal peptides (Cristobal et al., 1999). Moreover, there is often a basic residue in the c-region that functions as a Sec avoidance sequence (Bogsch et al., 1997). While most of the Tat preproteins are processed by signal peptidase 1 (SPase I) (Lüke et al., 2009), some contain a lipobox and are therefore processed by SPase II.
A specialized signal peptide called the prepilin signal peptide is found on the type 4 pilus proteins. Similar to the Sec and most lipoprotein signal peptides, it targets the protein to the Sec machinery. Type 4 substrates are found on the surface of many Gram-negative bacteria such as Pseudomonas aeruginosa and Neisseria gonorrhoeae. Pilin subunits allow the bacteria to stick to the surface of the host epithelial cells during infection. The prepilin signal peptide is unique as it is cleaved at the border of the n-h region (Strom and Lory, 1993; Mattick, 2002). The processing is carried out by prepilin signal peptidase, which recognizes the GFTLIE motif and cleaves after the glycine (Nunn and Lory, 1991). After cleavage, the prepilin signal peptidase methylates the amino terminus of the mature pilin (Strom et al., 1993). This generates N-methylphenylalanine as the first amino acid of the mature pilin.
In addition to these cleavable signal peptides, uncleaved signal peptides containing a longer hydrophobic stretch target proteins to the translocation machinery but remain as a membrane anchor sequence. These uncleavable signal peptides are found in membrane proteins which span the bacterial inner membrane as an α-helix. These domains are enriched in hydrophobic residues such as Ala, Ile, Leu, and Val but mostly void of charged residues (von Heijne, 2006). The uncleaved signals can span the membrane in different orientations, dictated by the positive inside rule (von Heijne, 1986b; von Heijne, 1999). If there are positive charges preceding the hydrophobic stretch, then the transmembrane (TM) segment is oriented with the C-terminus facing the periplasm whereas if the hydrophobic stretches are followed by positively charged residues, then the amino-terminus of the TM segment is localized to the cytoplasm. The positive inside rule is based on the finding that the membrane proteins have cytoplasmic loops that are enriched in positively charged residues (Lys, Arg) as compared to the periplasmic/translocated loops (von Heijne, 1986b).
2.1 Signal Peptide Targeting to the Membrane
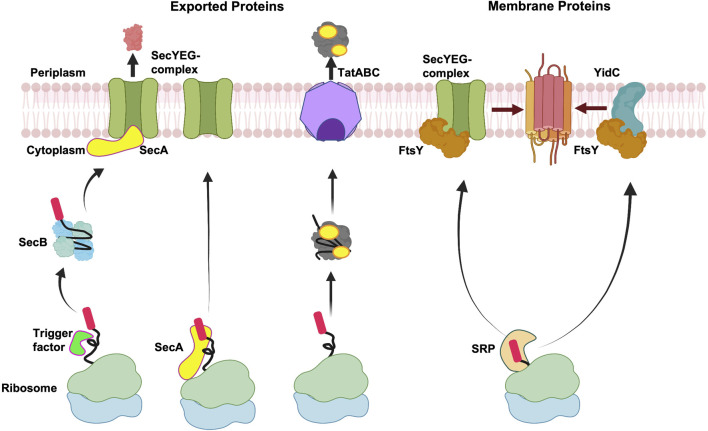
Targeting of exported and membrane proteins is initiated early on after the amino terminus of the nascent protein emerges from the ribosomal exit channel (Figure 2). The targeting pathway is decided by the interaction of the nascent protein with the ribosome-bound chaperones and targeting factors such as the Trigger Factor (TF) (Hoffmann et al., 2010; Castanié-Cornet et al., 2014), the signal recognition particle (SRP) (Grudnik et al., 2009; Akopian et al., 2013; Saraogi and Shan, 2014) and SecA (the ATPase motor of the Sec translocation machinery) in some cases (Kusters and Driessen, 2011; Chatzi et al., 2014). These chaperones and targeting factors facilitate the localization to the inner membrane of bacteria.
FIGURE 2.
Membrane targeting pathways. Overview of targeting of exported proteins and membrane proteins. After exported proteins are released from the ribosome, Sec-dependent proteins can be stabilized by the molecular chaperone SecB in an unfolded state and then targeted to SecA at the membrane, followed by translocation by the SecYEG complex. Alternatively, SecA can interact with the ribosome bound nascent chain and target the exported protein to the SecYEG complex. In case of Tat complex, the proteins fold in the cytoplasm before being exported by the Tat complex. In the event of co-translational targeting, the nascent membrane proteins form a complex with SRP, which target proteins to FtsY (SRP receptor) for membrane insertion either by the SecYEG complex or the YidC insertase. Created with BioRender.com.
In Gram-negative bacteria, the exported proteins are typically targeted to the Sec complex or the Tat translocase by the post-translational mechanism (Figure 2). Exported proteins which employ the Sec pathway contain moderately hydrophobic signal sequences and are transported through the Sec channel in a largely unfolded state. In contrast, the Tat substrates are translocated in the folded state after release of the protein from the ribosome and hence post-translational. Typically, the integral membrane proteins are targeted co-translationally either to the Sec machinery or the YidC insertase as soon as the hydrophobic TM segment emerges from the ribosomal tunnel.
2.1.1 Targeting of Exported Proteins
2.1.1.1 Sec Proteins
In the post-translational pathway, the TF is bound to the ribosome over the exit channel shielding the nascent chains from proteases (Figure 2) (Ferbitz et al., 2004). The ribosome-bound TF provides a protective environment preventing the premature folding and aggregation of the growing protein chain. Ribosome profiling studies have shown that the TF binds to the nascent chain only after approximately 100 amino acids are synthesized and play a role for the biogenesis of many β-barrel outer membrane proteins (Oh et al., 2011). After the protein is released from the ribosome, some proteins can form a complex with SecB (Kumamoto and Francetić, 1993), a dedicated molecular chaperone for export in bacteria. SecB is a tetramer (Xu et al., 2000) and how it keeps proteins in a non-native loosely-folded form (Randall and Hardy, 1986) is an intriguing mechanistic question. Recently, a state-of-the-art NMR study revealed that an unfolded preprotein wraps around the SecB. This is achieved by binding to the long hydrophobic grooves of SecB that run around the tetramer (Huang et al., 2016). The SecB delivers the preprotein to SecA bound to SecYEG at the membrane (Hartl et al., 1990). The targeting of the preprotein to SecA is achieved by SecA acting as a receptor that binds the signal peptide (Gelis et al., 2007) and the chaperone SecB (Zhou and Xu, 2003).
In an alternative scenario, cytosolic SecA can interact with the nascent chains emerging from the ribosome (Figure 2). Indeed, previous studies had suggested that SecA interacts with the nascent chains (Chun and Randall, 1994; Karamyshev and Johnson, 2005). In more recent studies, SecA was shown to interact with the ribosome near the ribosome exit channel (Huber et al., 2011). The binding of SecA to the ribosome is mediated by the ribosomal protein, L23. This interaction is important since mutations in L23 perturb SecA ribosome binding, significantly affecting the post-translational export of proteins in vivo (Huber et al., 2011). The isolation of mRNAs that copurify with SecA revealed that they encode both Sec exported and membrane proteins (Huber et al., 2017). The interaction of SecA with the nascent protein chains occurs only when the chains are longer than 110 residues. The interaction of SecA with the nascent chains is not dependent on TF or SecB. Notably, the authors found that SecB interaction with the nascent chains depended on SecA being bound to the nascent chains, suggesting that SecA interacts with a subset of SecB dependent substrates co-translationally (Huber et al., 2017). The emerging data suggests that SecA bound nascent chains can target proteins directly to the SecYEG complex or with the help of SecB (Figure 2). However, it is uncertain if the interaction of SecA with all SecA-dependent substrates occurs co-translationally.
In some cases, the preprotein is released from the ribosome with the TF still bound (not shown in Figure 2). In vitro, TF has been shown to form a stable 1 to 1 complex with proOmpA (Crooke et al., 1988), making proOmpA translocation competent. The TF functions as a holdase and foldase to bind its substrate in an unfolded state (Hoffmann et al., 2010). Saio et al. (2014) characterized the binding of the TF to the unfolded precursor of alkaline phosphatase (pre-PhoA) by NMR. With the help of multiple binding pockets, the TF engages with the nascent polypeptide and shields the emerging hydrophobic regions of pre-PhoA in solvent to prevent it from premature folding and aggregation. De Geyter et al. (2020) showed that the TF is a genuine export chaperone. Notably, they revealed that the TF bound preprotein can associate with the SecB, which then recruits SecA through its C-tail and promotes the transfer of the preprotein to SecA.
2.1.1.2 Tat Proteins
A different post-translational mechanism is used for targeting of Tat proteins to the inner membrane (Figure 2). These Tat proteins need to be folded in the cytoplasm prior to their translocation across the membrane (Palmer and Stansfeld, 2020). Many of the known substrates of the Tat pathway in bacteria bind a variety of redox cofactors, including molybdopterin centers and FeS cluster. There are specialized chaperones termed REMP (redox enzyme maturation proteins) to mediate cofactor insertion and proof reading (Turner et al., 2004; Robinson et al., 2011). For example, TorD is a REMP for TorA that encodes Trimethylamine-N-Oxide Reductase. TorD facilitates cofactor insertion and protects the TorA signal peptide from proteases (Ilbert et al., 2003) enabling the TorA to be delivered correctly to the Tat translocase (Jack et al., 2004). Another REMP is the DmsD that is involved in the biogenesis of dimethyl sulphoxide (DMSO) reductase (DmsA) (Ray et al., 2003). DmsD associates with the DmsA signal peptide (Oresnik et al., 2001) and also interacts with the molecular chaperones DnaK, DnaJ, GroE, GroEL, and TF (Li et al., 20101804; Castanié-Cornet et al., 2014). Finally, NapD is a REMP for the nitrate reductase complex localized in the periplasmic space. NapD binds to the Tat signal peptide of NapA (Maillard et al., 2007) and is involved in the insertion of the molybdenum cofactor.
2.1.2 Targeting of Membrane Proteins
For integral membrane proteins the hydrophobic segments in the nascent proteins interact with SRP at the ribosome exit channel and are sorted away from exported proteins that contain less hydrophobic sequences (Figure 2) (Lee and Bernstein, 2001). The inference for this comes from ribosome profiling studies examining the mRNAs that are bound to SRP engaged ribosome nascent chains (Schibich et al., 2016). The study revealed 87% of the SRP interactors are membrane proteins and only 6% are periplasmic/outer membrane proteins (Schibich et al., 2016). SRP can scan the ribosome with low affinity even before the nascent chain reaches the exit tunnel and interacts with the ribosomal binding proteins L23 and L29. This is called the stand-by mode (Holtkamp et al., 2012). When the nascent chain of 30–35 amino acids length reaches the exit site, SRP forms a high affinity complex with the translating ribosome and signal peptide (Bornemann et al., 2008; Holtkamp et al., 2012). Soon after forming this high affinity complex, the nascent chain is delivered to its receptor at the membrane (Figure 2, right side) to form a quaternary complex. The receptor FtsY (in prokaryotes) then transfers the ribosome nascent chain to the SecYEG complex by a mechanism involving the catalysis of GTPases.
The SRP has also been shown to target membrane proteins to the YidC insertase (Welte et al., 2012) (Figure 2). For example, MscL (Facey et al., 2007) and the tail anchored proteins SciP, DjlC, and Flk require both YidC and SRP for membrane protein insertion (Pross et al., 2016; Peschke et al., 2018). Ffh and FtsY can be crosslinked to the cytoplasmic loop of YidC, suggesting that the SRP-YidC nascent chains are targeted to FtsY that is in proximity to the YidC cytoplasmic loop (Petriman et al., 2018). The YidC cytoplasmic loop C2 and the C-tail of YidC binds to the ribosome supporting YidC activity (Geng et al., 2015).
Although the classical model predicts that SRP binds to the TM segment when it is exposed out of the ribosome exit channel, SRP can also interact with the hydrophobic regions that are not a part of the TM segment in some cases. Pross and Kuhn (2020) proved that there are two hydrophobic segments in the amino-terminal part of the C-tailed anchored protein SciP, which are recognized by SRP allowing it to target SciP to YidC . Additionally, in contrast to the classical view, ribosome profiling studies showed that 29% of the SRP interactors skipped interaction with the first TM segment of the membrane protein but were bound to C-terminal TM segments (Schibich et al., 2016). The SRP prefers to bind to ribosomes exposing ~ 12-17 amino acids enriched in hydrophobic and/or aromatic residues (Ile, Leu, Val, Met, Phe, Tyr, Typ) (Schibich et al., 2016).
In another variation, certain membrane proteins with internal TM segment can be co-translationally targeted to the membrane by SecA (Wang et al., 2017). SecA binds to the ribosome near the exit channel where it can recognize certain membrane proteins. SecA interacts with high specificity with nascent RodZ chains containing a TM segment far from the amino-terminus and targets the protein to the inner membrane (Wang et al., 2017). Previously, Rawat et al. (2015) had shown that SecA drives TM insertion and that Ffh and FtsY were not involved. SecA is sufficient for membrane targeting of RodZ both in vivo and in vitro (Wang et al., 2017). Interestingly, Knüpffer et al. (2019) found that SecA, just like SRP, deeply inserts into the exit tunnel of the ribosome to make contact with the intra-tunnel loop of L23 (Knüpffer et al., 2019). When the nascent chain is synthesized, SecA withdraws from the tunnel and the SecA bound to the L23 ribosome protein recruits the nascent TM segment. It is intriguing that the SecA amino-terminal amphipathic helix and the ribosomal L23 protein bind the nascent chain TM segment with the TM segment clustered in between, as revealed by Cro-EM studies (Wang et al., 2019). The SecA ribosome nascent chain complex is then targeted to the SecYEG complex, which repositions SecA on the ribosome, allowing the TM segment containing the nascent chain to be handed over to the SecYEG.
2.2 Crossing the Membrane
Once the signal peptide has navigated the transported protein to the membrane, it promotes interaction with the translocation machineries (see below). A vast majority of proteins are translocated by the SecYEG/SecDF system (Figure 3C) and SecA (Oliver and Beckwith, 1981; Crane and Randall, 2017; Tsirigotaki et al., 2017; Cranford-Smith and Huber, 2018), whereas the Tat machinery is involved in the export of around 30 proteins in E. coli (Berks, 2015; Palmer and Stansfeld, 2020) (Figure 3A). As mentioned before, the Tat machinery is radically distinct from the SecA/SecYEG/SecDF system as it can export fully folded proteins.
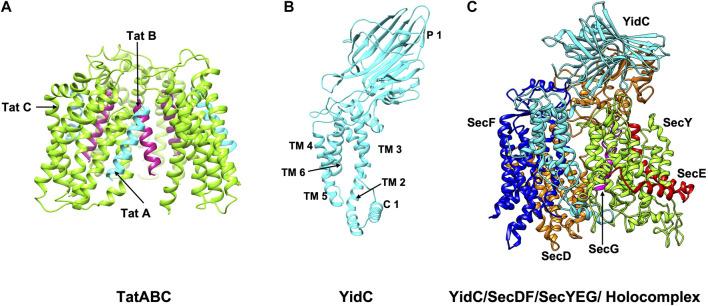
FIGURE 3.
The structures/models of the bacterial export and insertion machineries. Export of proteins across the membrane are catalyzed by (A) the Tat complex (resting complex shown) in a folded state (left side) (Habersetzer et al., 2017) or (C) by SecYEG/SecDF/YidC [adapted from Botte et al. (2016) PDB: 5MG3] energized by the SecA motor ATPase (not shown) in an unfolded state. TatA, TatB and TatC is shown in cyan, magenta and green, respectively. SecY, SecE, and SecG is shown in green, red, and magenta; SecD, SecF and YidC are shown in orange, blue and cyan. Membrane protein integration is catalyzed by the SecYEG/SecDF/YidC (C) complex or by the YidC insertase (B) [adapted from Kumazaki et al. (2014b) PDB: 3WVF]. The view is in the plane of the membrane with the periplasmic face at the top and the cytoplasmic face at the bottom.
The SecYEG/SecDF/YidC translocase (Figure 3C) plays the principal role for placing membrane proteins in the lipid bilayer with the correct topology (Cymer et al., 2015). Additionally, it functions to translocate hydrophilic domains of membrane proteins across the membrane and allows the hydrophobic regions to integrate into the lipid bilayer. The YidC insertase on its own or in cooperation with the Sec translocase can insert membrane proteins (Kiefer and Kuhn, 2018) (Figures 3B,C). The Tat machinery can act as insertase for the membrane proteins with C-terminal TM segments (Palmer and Stansfeld, 2020).
2.2.1 Protein Translocation Across the Membrane
2.2.1.1 SecYEG/SecA Translocase
SecA plays a crucial role for the export process both as a receptor and molecular motor (Cranford-Smith and Huber, 2018). The preprotein binds with high affinity to SecA/SecYEG but not to SecYEG (Hartl et al., 1990). SecA is also necessary for the translocation of proteins across the inner membrane (Oliver and Beckwith, 1981).
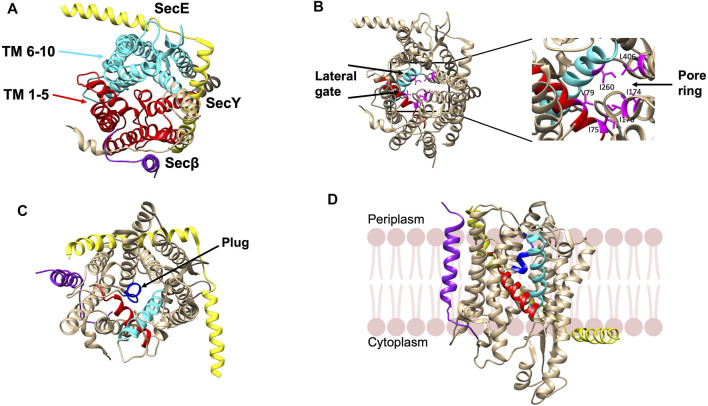
The structure of the SecYEβ protein-conducting channel, comprised of three subunits, was solved from Methanococcus jannashii in 2004 (van den Berg et al., 2004). SecY is the main channel forming unit that has a classic hourglass structure where TM 1–5 and TM 6–10 form two symmetric bundles held together by a linker (Figure 4A). The second subunit, SecE forms a clamp around SecY by wrapping around the two sides via its TM segment and cytoplasmic tail to stabilize the complex (Figure 4A). The SecE in E. coli is a 14 kDa essential 3TM protein. Sec61β (SecG of E. coli) is located on the third side of SecY (Figure 4A). Both SecY and SecE are evolutionarily conserved in bacteria, archaea and eukaryotes while SecG is not conserved in the three domains of life (Hartmann et al., 1994; Pohlschröder et al., 1997).
FIGURE 4.
Crystal structure of the SecYEβ complex in the resting state from Methanocaldococcus jannaschii [adapted from van den Berg et al. (2004) PDB: 1RHZ] (A). TM1-5 (red) and TM6-10 (cyan) are the halves of SecY. SecE and Sec61β are in yellow and purple, respectively. (B) The pore ring comprised of six residues (pink) and lateral gate (TM2b in red and TM7 in cyan) are highlighted. (C) The plug helix located above the pore ring is indicated in dark blue. (D) The SecYEβ complex from Thermus thermophiles (PDB: 5AWW). The lateral gate region comprised of TM2b (red) and TM7 (blue) is the site where the signal peptide or TM segments of membrane proteins exit the channel upon opening of the gate. The SecYEβ structures in (A–C) are shown perpendicular to the membrane.
There is a pore ring with a diameter of 4—6 Å (Figure 4B) at the center of the SecY channel (van den Berg et al., 2004). The pore ring is formed by 6 hydrophobic aliphatic residues and expands to accommodate the polypeptide chain during translocation (Bonardi et al., 2011). A short helix TM 2a termed as the “plug” keeps the pore closed (Figure 4C). The plug functions to maintain the integrity and preserve the permeability barrier of the membrane (Li et al., 2007). It has been shown that deleting the plug domain does not result in a major defect in protein export. However, channel experiments have shown that deletions of the plug compromise the membrane permeability of the channel as there are fluctuations between the open and closed state of the translocon (Li et al., 2007). This suggests that when the plug is present, the channel is stabilized in the closed state. Finally, on the front side of the channel, is the lateral gate (comprised of the TM 2a and TM 7) (Figure 4D) that can open sideways to allow signal peptides or TM segments to exit the channel (van den Berg et al., 2004).
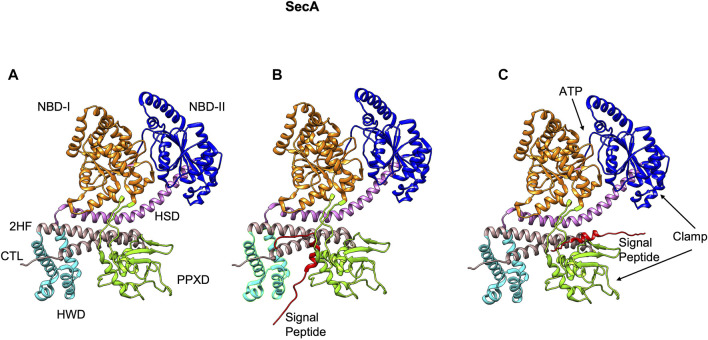
The peripheral subunit of the Sec complex is SecA which docks onto the SecYEG channel. It utilizes the energy from both ATP binding as well as ATP hydrolysis to drive the transport of unfolded proteins across the Sec channel. Structurally, SecA contains multiple domains with two ATP binding domains (NBD-1 and NBD-2) (Hunt et al., 2002), the HSD (helical scaffold domain), a preprotein crosslinking domain (PPXD) (Hunt et al., 2002), a helical wing domain (HWD), and a carboxyl-terminal linker domain (CTL) (Figure 5A). The HSD domain also has the central helix and the 2 helix finger (2HF) (Zimmer et al., 2006) or the regulator of ATPase (IRA1) (Karamanou et al., 1999) subdomains. PPXD and the 2HF have been implicated in binding the signal peptide and the mature region of the preprotein (Kourtz and Oliver, 2000; Papanikou et al., 2005; Musial-Siwek et al., 2007).
FIGURE 5.
The NMR structure of SecA from E. coli [adapted from Gelis et al. (2007) PDB: 2VDA]. (A) The various domains of SecA are highlighted (without the signal peptide). The nucleotide binding domains I (orange) and II (blue), the central helix subdomain of helical scaffold domain (HSD in purple), the preprotein crosslinking domain (PPXD green), the helical wing domain (HWD cyan), and the observed carboxyl-terminal linker domain (CTL). Also highlighted is the 2-helix finger (2HF tan) within the HSD domain. (B) The signal peptide (red) binds roughly perpendicular to 2HF based on NMR studies (Gelis et al., 2007). (C) The signal peptide is modeled parallel to the 2HF of the E. coli SecA NMR structure based on FRET, mutagenesis and genetic studies (Grady et al., 2012).
The crystal structures of the SecA-SecYEG complexes with and without substrate have shed light on how SecA moves the substrate polypeptide through the channel (Zimmer et al., 2008; Li et al., 2016; Ma et al., 2019). In the crystal structure of the SecA–SecYEG complex, a single SecA protein is bound to a single SecYEG protomer creating a groove for the passage of the preprotein (Zimmer et al., 2008). A clamp region can be observed at the interface of PPXD and NBD-2 domains (Figure 5A). Based on the crosslinking studies, the clamp has been proposed to bind the preprotein (Bauer and Rapoport, 2009). The 2HF region of SecA (Figure 6A) may push the preprotein into the channel (Bauer and Rapoport, 2009). Interestingly, while an NMR study (Gelis et al., 2007) showed the signal peptide was bound to a SecA groove formed at the interface of the 2HF and the PPXD (Figure 5B), it is possible that it would move from this region to align more parallel to the 2HF, such that it could be pushed into the channel. Indeed, based on the FRET, mutagenesis and genetic studies, Oliver and coworkers proposed a model where the signal peptide binds parallel to the 2HF (Figure 5C) (Grady et al., 2012).
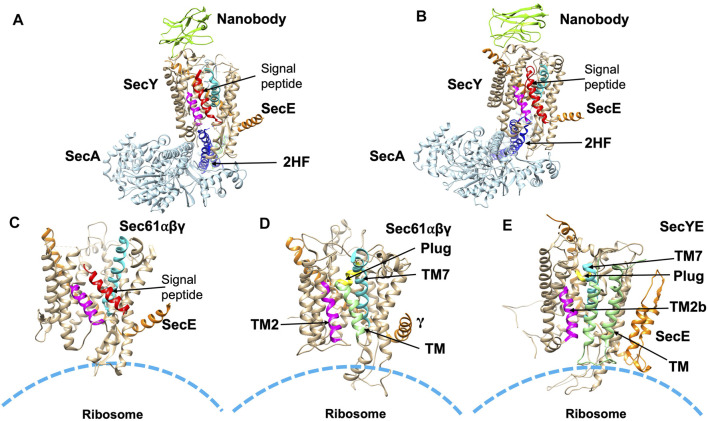
FIGURE 6.
Structures of substrate engaged SecYE or Sec61 complexes. (A) Crystal structure of SecYE-SecA [adapted from Li et al. (2016) PDB: 5EUL] with a portion of the preprotein (comprised of the OmpA signal sequence and a few residues in the mature region) fused into the 2HF (navy blue) by insertion between 741 and 744 of SecA. SecA (in light blue) was from B. subtilis and SecYE was from Geobacillus thermodenitrificans. Nanobody (chartreuse) bound to the periplasmic side of SecY (tan). (B) CryoEM structure of SecYEG-SecA complexed with a proOmpA sfGFP [adapted from Ma et al. (2019) PDB: 6ITC] fusion protein. The structure was performed with SecYE in a lipid nanodisc. An anti-GFP nanobody was inserted at the C-terminus of SecA to recognize and stabilize the fused sfGFP of the substrate. In addition, a disulfide was created between a cysteine at position 8 in the early mature region of the proOmpA GFP fusion protein and a cysteine placed in the plug domain of SecY. Finally, a SecY nanobody that recognizes the periplasmic SecY region was added to stabilize the complex. SecA was from B. subtilis and SecYE was from Geobacillus thermodenitrificans. The nanobody is shown in green in (A,B). (C) CryoEM structure [adapted from Voorhees and Hegde, 2016 PDB: 3JC2] in detergent of the canine ribosome Sec61 channel engaged with the N-terminal 86-amino acid preprolactin region. (D) CryoEM structure [adapted from Gogala et al. (2014) PDB: 4CG6] of the canine Sec61 channel engaged with a hydrophobic TM segment (light green) of a leader peptidase (lep) arrested intermediate. The TM segment was modeled within the opened TM2/TM7 lateral gate. (E) CryoEM structure (adapted from Bischoff et al. (2014) PDB: 5ABB) of a stalled E. coli ribosome SecYE complex engaged with proteorhodopsin (TM indicated in light green). TM2 and TM7 of the lateral gate are shown in magneta and cyan, respectively in (A–E). The signal peptide (red) is indicated in (A–C). The plug helix is indicated in yellow in (D,E).
To examine the structure of the SecY channel during active SecA-dependent translocation, X-ray crystallography was used to solve a substrate engaged SecA-SecYE complex (Figure 6A). The substrate sequence, which included the OmpA signal sequence and a short mature region, was inserted at the end of the 2HF of SecA (Li et al., 2016). The structure suggested that the signal peptide moved into the lateral gate facing the lipid bilayer and the mature region inserted into the channel as a loop, displacing the plug. Thus, the interaction of the signal sequence with the lateral gate induces conformational changes and movements of the plug domain. This leads the way for the substrate to move up the pore ring towards the periplasm by repeated ATP binding and hydrolysis events moving roughly 20–25 amino acids into the translocon in each step.
More recently, in order to gain insight into the path of a translocating polypeptide through SecA, another substrate engaged SecA-SecYE was solved in an active transition state of ATP hydrolysis with ADPBeFx bound (Figure 6B) (Ma et al., 2019). The SecA/SecYE translocation intermediate with SecA locked in an ATP bound state was generated using a substrate fusion protein consisting of the proOmpA signal sequence, a linker region, and a folded GFP. In order to stabilize the complex, a cysteine was added after the signal sequence to form a disufide bond to the cysteine introduced at the SecY plug. The protein was then reconstituted into nanodiscs and solved by cryo-EM with a resolution of about 3.5 Å. Tracing the substrate within the SecA-SecYE complex confirmed that in addition to the polypeptide being in proximity to the SecA 2HF, it also interacts with the SecA clamp region via a short β-strand. It also showed that the signal sequence forms a helix that is positioned in a groove outside the lateral gate of the SecY channel.
There are several models that have been proposed to account for the role of ATP energy in energizing SecA/SecYEG in protein transport, including (Hegde and Bernstein, 2006) power stroke, (Oswald et al., 2021), Brownian ratchet, (Frain et al., 2019), push and slide, and other mechanisms. According to the power stroke model, the SecA ATP hydrolysis causes conformational changes that result in mechanical pushing of the polypeptide chain through the SecYEG channel. Indeed, a large segment of SecA was proposed to move through the SecYEG channel to the periplasmic region in order to translocate the polypeptide to the trans side of the membrane (Ulbrandt et al., 1992; Economou and Wickner, 1994). Later versions of the power stroke model proposed that the 2HF, which is positioned at the entrance of the SecYEG channel, functions as a piston to push the polypeptide through the membrane. This rationale comes from the fact that the SecA 2HF interacts with the preprotein during protein translocation (Erlandson et al., 2008). Upon ATP binding, the 2HF undergoes a large conformational change that pushes the protein substrate chain into the SecY channel (Catipovic et al., 2019). After the 2HF releases the polypeptide substrate of the preprotein, the finger resets to its original position (Catipovic et al., 2019). This cycle of conformational changes occur multiple times until the polypeptide is translocated through the channel.
In a Brownian ratchet mechanism, the movement of a protein chain occurs via diffusion through the channel. SecA would mediate SecYEG channel opening thereby enabling the preprotein to diffuse through the SecYEG pore. The evidence for this action was presented in a model by Allen et al. (2016). The authors demonstrated that the SecYEG gate is wide open when ATP is bound to SecA and slightly open with ADP bound to SecA. The slightly open channel allows protein substrate regions with small side chains to slide through the pore, but larger side chains would require the pore to open. Interestingly, the SecYEG channel and the SecA 2HF are able to detect the presence of a protein chain which results in nucleotide exchange, allowing ATP to replace ADP. The binding of ATP to SecA leads to opening of the SecYEG channel through which the chain crosses by diffusion. Backsliding of the polypeptide chain is prevented by closure of the channel. More recently, ATP-driven translocation through the SecYEG channel was shown to be indirectly coupled to ATP hydrolysis providing further support to the Brownian ratchet model (Allen et al., 2020).
A push and slide mechanism combines the power strokes and the passive diffusion models. Bauer et al. (2014) found that certain protein chains can slide passively through the SecYEG channel without ATP hydrolysis. Passive sliding of the polypeptide chain takes place after the preprotein is released by the 2HF and SecA has bound to ADP. Under these conditions, the clamp region between PPXD domain and NBD-2 domain is open and cannot bind the mature domain. The polypeptide chain can passively slide in either direction. Power stroke would occur again after the binding of ATP to SecA. During the power stroke, segments of the polypeptide chain move deep into the SecY channel (Catipovic and Rapoport, 2020) as the SecA 2HF moves into the channel. Prior to the retraction of the 2HF to the original position, the SecA captures and tightens its clamp region around the mature domain of the preprotein substrate, thus preventing back sliding of the polypeptide chain (Catipovic et al., 2019). This tightening enables the forward translocation of the chain to be maintained. The clamp closure occurs before or during SecA ATP hydrolysis and that the 2HF resets all the way when the clamp is closed. Otherwise, as the 2HF resets, and moves away from the channel it would drag the polypeptide with it (Catipovic et al., 2019; Catipovic and Rapoport, 2020). One baffling fact is that the immobilization of the 2HF to SecYEG do not inhibit translocation (Whitehouse et al., 2012).
In addition to the energy of ATP binding and hydrolysis, the proton motive force (pmf) can also contribute to the translocation of preproteins across the SecYEG channel membrane (Date et al., 1980; Zimmermann and Wickner, 1983). SecD and SecF, which have 6 TM segments and a large periplasmic domain, are required for pmf stimulation of protein translocation (Arkowitz and Wickner, 1994). One model proposes that SecDF assists in translocation by binding the preprotein once it emerges from SecY and prevents back sliding (Tsukazaki et al., 2011). Then it swivels its head domain to translocate about 25 amino acids across the membrane with the help of the pmf. Therefore, SecDF functions in the pulling of preproteins across the membrane, and the release of preproteins from the SecYEG complex after complete translocation.
2.2.1.2 Tat Translocase
The Tat machinery exports fully folded proteins of different sizes. It is comprised of TatA, TatB, and TatC (Tat complex) in E. coli and TatAC in B. subtilis (Jongbloed et al., 2004; Berks, 2015; Palmer and Stansfeld, 2020). The Tat components in E. coli are assembled on the cytoplasmic membrane as a TatBC complex and a cytoplasmic TatA pool. Interestingly, TatC membrane insertion is a SecYEG and YidC dependent event (Welte et al., 2012; Zhu et al., 2012). After forming the TatBC complex, the TatA oligomers are recruited to the TatBC complex in a pmf dependent event before substrate translocation. The recognition of the twin arginine motif by a conserved patch on TatC (Palmer and Stansfeld, 2020) initiates the architectural reorganization of the active complex assembly. The low stability, size and transient nature of the active complex presents a daunting challenge to identify the precise assembled active complex needed for Tat translocation.
The Tat components TatA and TatB have similar features each possessing a small periplasmic N-terminal region, a single short TM helix (TMH) followed by an amphipathic helix (APH), and a cytoplasmic domain. TatA is assumed to form the translocation complex with the substrate as it is capable of forming oligomeric rings of different sizes (Gohlke et al., 2005). TatB functions with TatC as a receptor to bind Tat substrates (Cline and Mori, 2001; Behrendt and Brüser, 2014). TatB and TatC form a 1:1 complex and have an oligomeric structure with a size range of 360–700 kDa (Bolhuis et al., 2001).
In the resting state, the TatBC receptor complex most likely has some TatA associated and recent studies suggest there are three to four copies each of TatA, TatB and TatC in a ratio of 1:1:1 (Bolhuis et al., 2001; Alcock et al., 2016; Habersetzer et al., 2017). Based primarily on crosslinking studies it has been proposed recently that in the resting state, this TatABC complex is organized such that TatB binds to the TatC TM5 and TatA binds to the TatC TM6. Upon activation of the complex by substrate addition, TatA and TatB switch positions which may be triggered by the substrate with the signal peptide moving further into the membrane interior (Habersetzer et al., 2017).
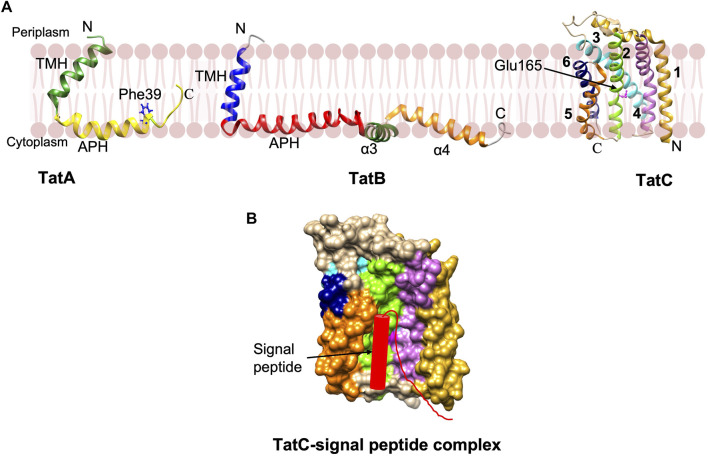
NMR studies reveal that TatA and TatB proteins possess a short α-helical TM segment followed by the amphipathic helix (APH) (Figure 7A) (Rodriguez et al., 2013; Zhang et al., 2014). TatA forms oligomeric rings with variable number of TatA molecules (Rodriguez et al., 2013). Interestingly, structural studies and molecular dynamic (MD) simulations predict that the TatA structure causes significant thinning of the membrane due to its short TM segments (Rodriguez et al., 2013). This may be true with TatB as well since it has a short TM segment. The structure of the 6-membrane spanning TatC from Aquifex aeolicus revealed that the protein has a “cup hand” or “glove-like structure” (Figure 7B), where TatC assembles into a concave structure that can accommodate a TM segment of TatB or the neighboring TatC (Rollauer et al., 2012; Ramasamy et al., 2013). Remarkably, the TatC surface has an ionizable Glu165 side chain that is expected to be buried in the hydrophobic interior of the bilayer. MD simulations show that the TatC has a hydrated cavity facing the cytosol with the conserved Glu/Gln at this position inside the membrane. This hydrophilic cavity and the short TM segments 5 and 6, cause thinning of the membrane.
FIGURE 7.
The Tat complex components and a model of TatC-signal peptide complex. (A) The single span TatA (PDB: 2MN7) and TatB (PDB: 2MI2) proteins were determined by NMR (Rodriguez et al., 2013; Zhang et al., 2014). The structure of 6 membrane spanning TatC [adapted from Rollauer et al. (2012) PDB: 4B4A] was solved by X-ray crystallography (Rollauer et al., 2012; Ramasamy et al., 2013). (B) The model of TatC binding with the Tat signal peptide in the groove adapted from Ramasamy et al. (2013). Only signal peptide and early mature region of the preprotein are indicated.
Export by the Tat pathway begins by the recognition of the Tat signal peptide of the preprotein substrate by TatC within the TatBC complex. TatC recognizes the RR motif via its N-terminal domain and a cytoplasmic loop 1 (Figure 7B) (Alami et al., 2003; Gérard and Cline, 2006). Subsequently, insertion of the signal sequence into the interior of the membrane takes place by contacting the periplasmic side of TatA. Following the substrate insertion into the TatBC complex, the oligomeric TatA complex is formed in a step that requires a TM pmf. An oligomeric complex of TatA facilitates the translocation of the folded substrate. TatA protomers are predicted to form oligomeric ring-like pores of varying diameters in the cytoplasmic membrane, permitting the movement of fully-folded proteins into the periplasm (Lausberg et al., 2012).
The precise mechanism of translocation is still debatable, but we will discuss the two main hypotheses documented in literature. The trap door model postulates that the amphipathic helix (APH) domain of TatA flips into the lipid bilayer with the help of membrane potential on contact with the substrate carrying the TatA interactive motif (Patel et al., 2014). Initially, TatA oligomers self-arrange to form a pore of ∼8.5–13 nm in diameter to accommodate the folded protein (Frain et al., 2019). The APH of the TatA oligomers at the cytoplasmic face mirrors a “trap door” that regulates the transient channel for translocation of the substrate. This would essentially depend on the flexible hinge (the conserved Gly residue) between the APH and TMH (Frain et al., 2019). When the APH swings up to align with the TMH, the polar residues may interact with the hydrophilic protein to be translocated and thus the folded protein is promoted across the formfitting passive conduit.
The second model proposed a weakening of the lipid bilayer when TatA oligomerizes with its polar N-tail destabilizing the membrane, allowing translocation of the Tat substrate (Brüser and Sanders, 2003). This model where transient bilayer disruptions occur, is gaining more support with the NMR structure of TatA and suggests that the TatA topology may not be as flexible as predicted by the trap door model. MD simulations reveal the phospholipids are highly distorted and the membrane thickness is dramatically shortened (Rodriguez et al., 2013). It is believed that the thinning of the membrane is due to the short TatA TM segment and the presence of the conserved glutamine in the oligomer. However, this model does not clarify what drives the translocation of the substrate across the membrane.
2.3 The Insertion of Proteins Into the Membrane
The insertion of proteins in their proper conformation and orientation into the lipid bilayer is crucial for the functional integrity of the membrane proteins [for recent reviews see (Cymer et al., 2015; Tsirigotaki et al., 2017; Hegde and Keenan, 2021)].
2.3.1 SecYEG/YidC
For membrane protein topogenesis, the nascent membrane protein chain is presumed to be driven across the membrane utilizing the GTP hydrolysis energy from protein synthesis. This is possible because the ribosome binds to the SecYEG complex and may form a single aqueous conduit that stretches across most of the inner membrane.
As the membrane protein enters the SecYEG channel, the hydrophobic TM sequence or signal peptide may first enter the hydrophilic channel and then exit the lateral gate (van den Berg et al., 2004) or it can slide into the membrane via the lateral gate by thermodynamically partitioning between the lipid and the aqueous pore (Cymer et al., 2015). Rather than the sequence of amino acids of the TM segment, the overall hydrophobic character of the segment is important for insertion into the membrane (Hessa et al., 2005; Hessa et al., 2007). The hydrophobic stretch can stabilize the open lateral gate (Zhang and Miller, 2010).
As seen with the substrate engaged SecYEG/SecA complex, the ribosome bound-Sec translocon showed the signal peptide in the lateral gate region. A cryoEM structure of the canine ribosome Sec61 translocon engaged with a preprolactin substrate revealed the signal peptide in the lateral gate (Figure 6C) (Voorhees and Hegde, 2016). The pore region of the channel would allow the polar polypeptide chains to be translocated to the trans side of the membrane (Erlandson et al., 2008). After translocation, a simple membrane protein with 1 TM segment would completely exit the channel and partition into the lipid bilayer.
Similarly, the lateral gate accommodates the TM segment of the ribosome bound membrane protein inserting into the Sec61 complex, as revealed by cryo-EM study (Figure 6D) (Gogala et al., 2014). Notably, another cryo-EM study of a nascent membrane protein-SecYE complex demonstrated that the first 2 TM α-helices of proteorhodopsin had exited the lateral gate to face the lipid with the N-terminus at the periphery of SecY (Figure 6E) (Bischoff et al., 2014). Kater et al. (2019) further elucidated that a partially inserted hydrophobic region can cause opening of the lateral gate.
SecA is always required for translocation of large loops and occasionally for small loops of membrane proteins (Kuhn, 1988; Andersson and von Heijne, 1993; Deitermann et al., 2005; Soman et al., 2014). However, the mechanism by which this task is carried out has not been elucidated, as the ribosome is expected to be already bound to the SecYEG complex when the membrane protein inserts co-translationally. In order to perform the translocation function by SecA, the ribosome should be dissociated partly or completely from the SecYEG. This illustrates the dynamic nature of the insertion process and the interplay of the various SecYEG binding partners.
An interesting method to study in vivo insertion and co-translational folding of membrane proteins is the application of translational arrest peptides to measure forces acting on a nascent protein during membrane insertion (Ismail et al., 2012; Sandhu et al., 2021). In this approach, the arrest peptide binds to the ribosomal tunnel and induces ribosomal stalling at a specific amino acid. SecYEG mediated membrane insertion and folding of a nascent chain is followed by examining the release of stalling and resumption of protein synthesis. This technique has been used to study the co-translational insertion of simple to complex proteins spanning the membrane ten times, showing that the surface helices and re-entrant loops that flank a TM segment can either advance or delay membrane protein insertion (Nicolaus et al., 2021). Moreover, the results supported a sliding mechanism where the inserting TM segment moved into the membrane along the outer part of the lateral gate.
The mechanism of insertion of multispanning membrane proteins is complicated with most of the findings coming from studies of the endoplasmic reticulum (ER) membrane system (Cymer et al., 2015; Hegde and Keenan, 2021). In some cases, a TM segment is inserted into the translocase and then reoriented within the channel (Goder and Spiess, 2003). Remarkably, some TM segments of the membrane proteins such as the aquaporin 4 channel, exit the channel but apparently interact again at a later stage in membrane biogenesis, validating the dynamic nature of membrane protein biogenesis. The Sec machinery can handle the internal TM segments by various mechanisms. Some TM segments integrate into the lipid bilayer spontaneously (Heinrich et al., 2000), others integrate into the lipid bilayer only after the protein synthesis is terminated (Do et al., 1996; McCormick et al., 2003), some pairs of TM segments co-integrate into the membrane (Skach and Lingappa, 1993; Heinrich and Rapoport, 2003), while the rest can be stabilized by chaperones such as TRAM and YidC (Do et al., 1996; Heinrich et al., 2000; Beck et al., 2001; Urbanus et al., 2001; Nagamori et al., 2004).
SecYEG plays the primary role in membrane insertion in the plasma membrane in bacteria. The accessory component, YidC actively participates in membrane protein biogenesis for several different Sec dependent proteins (Figures 2, 3). Substrates that require the synergistic action of both YidC and SecYEG for insertion include, subunit a and b of the F1Fo ATP synthase (Yi et al., 2004) and TatC of the twin arginine translocase (Welte et al., 2012; Zhu et al., 2012). Moreover, YidC can bind to the TM segment of membrane proteins after the TM segment exits the SecYEG channel (Urbanus et al., 2001). This case is exemplified by the TM segments of FtsQ and leader peptidase (Houben et al., 2004) which were shown to initially contact SecYEG followed by contact with YidC at a later stage during its translocation process. This latter finding implied that YidC may facilitate Sec substrates to partition into the bilayer and assist in the clearing the Sec channel of its substrates. Remarkably, in the case of CyoA (subunit of cytochrome bo3 oxidase), the amino-terminal domain is inserted by the sole action of the YidC insertase whereas the large C-terminal domain requires SecYEG operating with the SecA motor ATPase for insertion (Celebi et al., 2006; van Bloois et al., 2006; Kol et al., 2009).
Furthermore, YidC acts as a chaperone (Nagamori et al., 2004) and assembler of multi-TM complexes (Saller et al., 2012). Studies with LacY biogenesis showed that YidC acts in the late stages of membrane protein biogenesis and is crucial for the correct folding of the protein but nonessential for its insertion (Nagamori et al., 2004; Zhu et al., 2013). Wagner et al. (2008) discovered a similar trend with MalF, a subunit of the maltose binding complex. Upon YidC depletion, the stability of the complex was affected without compromising the insertion of the TM segments of MalF mediated by the SecYEG complex.
In order to perform these multi-functions, YidC must be located close to the SecYEG complex. Indeed, YidC, SecDF/YajC may associate with SecYEG to form a holo-translocon (Schulze et al., 2014). This has been validated by copurification of YidC with the SecYEG and SecDF/YajC. The purified complex is capable of inserting in vitro synthesized membrane proteins (Komar et al., 2016). A low-resolution structure of the holocomplex SecYEG/SecDFYidC revealed that the SecYEG-YidC interface is a lipid filled cavity (Martin et al., 2019). Although a YidC holocomplex can be isolated, YidC is capable of dynamic interaction with SecYEG. When YidC contacts SecYEG, it is in proximity to the lateral gate of SecYEG and can contact helices on either side of the lateral gate (TMs 2b, 3, 7 and 8) (Sachelaru et al., 2017). This contact is maintained even in the absence of SecDF. Furthermore, SecYEG contacts TM1 and cytosolic loop 1 of YidC (Petriman et al., 2018). The Sec lateral gate can contact the YidC TM3-TM5 region which forms the greasy slide (Steudle et al., 2021). Taken together, these studies suggest that the insertion of Sec-dependent protein substrates occurs at the interface of YidC/SecYEG.
2.3.2 YidC-Only Pathway
In addition to assisting SecYEG and acting as a chaperone for membrane insertion, YidC can also operate independently. Examples of the Sec-independent proteins include M13 phage coat protein (PC) and the Pf3 coat protein, which were earlier presumed to be inserted by an unassisted mechanism. Depletion of YidC resulted in the accumulation of these proteins in the cytoplasm (Samuelson et al., 2000; Chen et al., 2002; Serek et al., 2004). Moreover, crosslinking studies revealed that YidC interacts with the inserting Pf3 coat (Chen et al., 2002). Subunit c of F1F0 ATPase was shown to be dependent on YidC for membrane insertion both in vivo (Yi et al., 2003; van Bloois et al., 2004) and in vitro (Van Der Laan et al., 2004). Interestingly, YidC proteoliposomes were capable of forming the subunit c oligomer whereas the liposomes were not (Kol et al., 2006). The indispensable nature of YidC in cells is still speculative. One of the reasons may be attributed to the fact that YidC is required for the biogenesis of the respiratory complexes (van der Laan et al., 2003).
Other substrates for the YidC-only pathway are MscL (Mechanosensitive channel of large conductance), which inserts co-translationally (Facey et al., 2007) and the tail anchored membrane proteins TssL (SciP), DjlK, and Flk (Aschtgen et al., 2012; Pross et al., 2016; Peschke et al., 2018). In eukaryotes, the ER tail anchored membrane proteins with a high hydrophobic TM segment are inserted by the Get pathway while those proteins with low hydrophobic TM segment are inserted by the ER membrane protein complex (EMC) (Hegde and Keenan, 2021). Interestingly, Get1 and EMC3 of the Get complex and EMC, respectively, are YidC homologs found in the ER (Anghel et al., 2017; Chen and Dalbey, 2018; McDowell et al., 2021).
The common feature of substrates of the YidC only pathway is that they have a short translocated region (Hennon et al., 2015), suggesting that the YidC insertase has limited translocase activity. Indeed, if the polarity of the translocated domain exceeds a certain threshold by introduction of charged/polar residues, then YidC requires the assistance of the SecYEG complex, implying that YidC is incapable to translocate these substrates unaided (Soman et al., 2014; Hariharan et al., 2021). The switching from YidC-only pathway to the YidC/Sec pathway indicates that both the YidC and the SecYEG are surveying the polypeptide chain during the membrane insertion process. This is feasible by dynamic interaction of YidC with SecYEG (Sachelaru et al., 2017; Steudle et al., 2021) or, a certain portion of YidC is a part of the SecYEG/SecDFYajC/YidC holocomplex (Schulze et al., 2014).
Structural studies have shown that YidC is a monomer and contains a hydrophilic cavity within the 5 TM core region (Figure 8) (Kumazaki et al., 2014a; Kumazaki et al., 2014b). This aqueous groove with a conserved positively charged residue is open both to the cytoplasm and the lipid bilayer but closed from the periplasmic side. The existence of a hydrophilic groove located within the inner leaflet of the membrane was supported by in vivo solvent accessibility and MD simulation studies. The study also revealed that YidC shapes the membrane with significant membrane thinning around the protein (Chen et al., 2017). The presence of the hydrophilic groove in the membrane decreases the membrane crossing distance which would in turn reduce the energy cost of translocating a polypeptide chain. Wu and Rapoport (2021) have recently proposed that protein translocation through a locally thinned membrane is a new paradigm for lowering the energy cost for translocation.
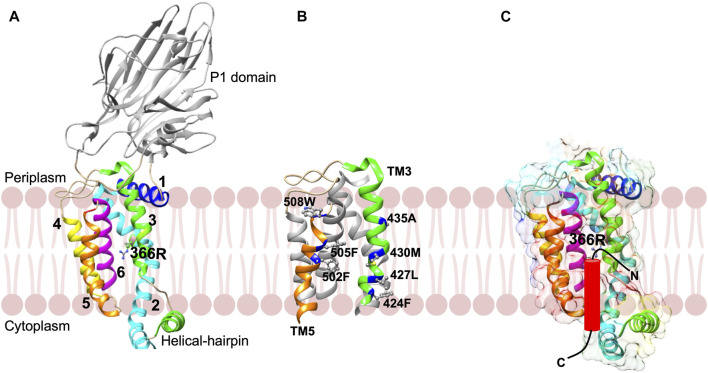
FIGURE 8.
The YidC insertase [adapted from Kumazaki et al. (2014b) PDB: 3WVF]. (A) The E. coli YidC has a large periplasmic domain, a coiled cytoplasmic domain, and a conserved core region comprising of 5 TM helices (TMs 2–6) that form a hydrophilic groove open to the cytoplasm and lipid bilayer. The hydrophilic groove has a strictly conserved arginine that helps to keep the region hydrated. (B) A close-up view of the greasy slide (TM3 and TM5) that contacts the TM region of YidC substrates during insertion. The residues that contact the substrate TM segment (s) are indicated in dark blue. (C) During membrane insertion of the Pf3 coat, the TM segment moves up the greasy slide with the N-tail region captured transiently in the hydrophilic groove. The periplasmic domain of YidC is omitted in (B,C).
Remarkably, the positively charged residue in the hydrophilic groove is essential for SpoIIIJ (YidC1) function in B. subtilis. It was proposed that the hydrophilic groove participates in an electrostatic step necessary to translocate the negatively charged N-terminal tail region of MifM across the membrane (Kumazaki et al., 2014a). However, the positive charge is not essential for the E. coli YidC (Chen et al., 2014). Rather, the positively charged residues plays a role in maintaining the hydrophilic microenvironment in the groove, which is necessary for the activity of YidC (Chen et al., 2022).
In addition to the hydrophilic groove, YidC has a cytoplasmic helical hairpin-like domain (Figure 8A) (Kumazaki et al., 2014a) which was predicted to be involved in the initial recruitment of the substrate. The arrangement of two antiparallel helices in the C1 region of EcYidC is rotated by 35° with respect to the core region, as compared to that in the BhYidC structure. Moreover, in both the structures the B factor for this region is high, demonstrating the flexibility of the C1 cytoplasmic loop region. Crosslinking studies of the essential C1 loop show contacts not only with the targeting proteins SRP and FtsY but also the Sec translocon (Geng et al., 2015; Petriman et al., 2018).
The mechanism of the substrate TM recognition by YidC is fascinating. Crosslinking studies have indicated that the TM3 of YidC contacts the TM domain of FtsQ, leader peptidase, subunit c of the F1FoATPase (Yu et al., 2008). Contacts are also observed with TM3 and TM5 of YidC to Pf3 coat and MscL (Klenner et al., 2008; Klenner and Kuhn, 2012). It has been proposed that the substrate enters the YidC groove between these TM3 and TM5 segments, which constitutes a greasy slide where the TM segment moves across the membrane (Figure 8B). Kedrov et al. (2016), performed cryo-EM studies on a YidC-ribosome Foc nascent chain complex where YidC was reconstituted in nanodiscs. The study revealed that the Foc nascent chain was in proximity to TM3 facing the lipid region.
He et al. (2020) elucidated the pathway employed by the single spanning Pf3 coat to provide insight into the YidC insertion mechanism of simple membrane proteins (Figure 8C). The tracking of the Pf3 coat protein through YidC was obtained by “freezing” each step of the insertion process by creating translational arrested intermediates of different sizes and investigating them by thiol crosslinking (He et al., 2020). The results divulged that the TM segment of Pf3 moves up the YidC greasy slide during membrane insertion. After the TM reached the top of the slide, the N-tail transiently enters the YidC hydrophilic groove. In the next step, the N-tail is released from the groove and translocated across the periplasmic leaflet of the membrane.
2.3.3 TatC and TatC/SecYEG
The Tat substrates of E. coli include the five integral membrane proteins including HybO, FdnH, FdoH, HyaA and HybA (Hatzixanthis et al., 2003). The genes encode subunits of NiFe hydrogenase or formate dehydrogenase. They are encoded with a Tat signal peptide and possess a C-terminal TM segment that functions as a stop transfer domain. The recognition of these membrane proteins by the Tat complex is achieved by the interaction of the Tat signal peptide with the TatBC complex.
Although mechanistically different, surprisingly, in some bacteria, cooperation is observed between the Tat and the Sec pathway for the insertion and assembly of polytopic membrane proteins (Keller et al., 2012). The first evidence for this was from the analysis of the iron-sulfur membrane bound Rieske proteins from Gram-positive actinobacteria that has 3 TM segments. While the first 2 TM segments are inserted by the SecYEG translocase, the third TM segment, which is preceded by a Tat motif was inserted by the Tat system. To understand the mechanism of insertion further and to decipher the handover process from Sec to Tat, the S. coelicolor Rieske protein, Sco2149 was examined (Tooke et al., 2017). The authors observed that a moderate hydrophobicity of the TM3 segment and the presence of several C-terminal positively charged residues promote the release of the TM3 from the SecYEG apparatus. This further allows the Tat TM segment to engage with the Tat translocase and stimulate translocation across the membrane. Other examples of the dual participation by Sec/Tat machineries targeting membrane proteins include the five spanning membrane proteins S. coelicolor Molybdenum cofactor protein Sco3746, and the delta proteobacterium MLMS-1 FeS containing polyferredoxin. In these cases, the first 4 TM segments are inserted by the SecYEG complex, and the Tat system inserts the last TM segment and translocates the folded C-terminal domain. In each case, the fifth TM segment has moderate hydrophobicity and an amino terminal Tat RR-motif (Tooke et al., 2017).
2.4 Removal of the Signal Peptide and Degradation
The last step in the translocation pathway is the removal of the signal peptide. This allows the exported proteins to be released from the membrane so that they can continue their journey to the periplasm, outer membrane or to the extracellular medium. The removed signal peptides are degraded by enzymes having signal peptide hydrolase activity.
Signal peptidases cleave off the signal peptides and play crucial roles as endopeptidases with clear cut substrate specificities (Paetzel, 2019). Type I signal peptidase (SPase I; also known as leader peptidase) processes the majority of preproteins while Type II signal peptidases (SPase II; also known as lipoprotein signal peptidase) process lipoprotein precursors (Paetzel, 2014).
2.4.1 Signal Peptidase I
The first signal peptidase to be characterized was E. coli signal peptidase I (SPase I). It was overproduced and purified to homogeneity (Wolfe et al., 1982). The purified protease in detergent was shown to cleave a wide variety of preproteins, including eukaryotic secretory preproteins (Watts et al., 1983). Similarly, the eukaryotic signal peptidase can cleave bacterial preproteins, demonstrating that the cleavage specificity is evolutionarily conserved (Watts et al., 1983). Subsequently, the E. coli protease enzyme was shown to be localized to the inner membrane with its catalytic domain facing the periplasmic space (Wolfe et al., 1983). Moreover, it was shown to be an essential enzyme for the bacteria (Date, 1983).
Signal peptidases are indispensable for the secretion process. Disruption of the signal peptide processing prevents the preproteins from arriving to their correct destination in the cell (Dalbey and Wickner, 1985). Under decreased SPase I activity in a depletion strain, the accumulated preproteins were translocated but were anchored to the inner membrane by the uncleaved signal peptide. Therefore, the function of signal peptidase is to release the exported protein from the membrane by removing the signal peptide so that they proceed to their destination. It is now established that SPase I processes the majority of non-lipoproteins that are exported by the Sec pathway or by the Tat pathway (Lüke et al., 2009).
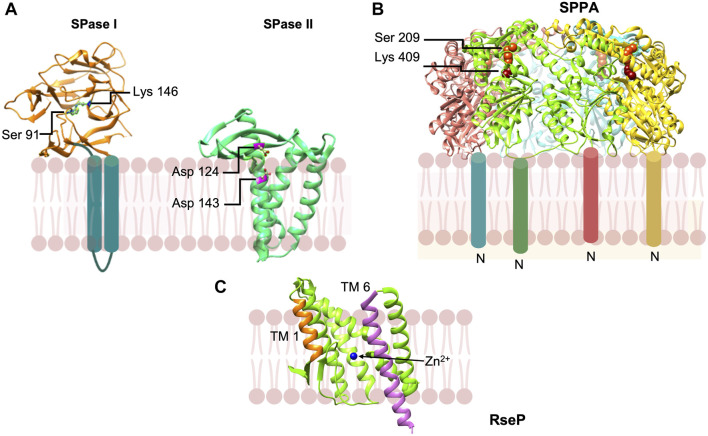
To understand how SPase cleaves and binds substrates at the active site, the structure of the E. coli signal peptidase periplasmic domain (Δ2-75) (Tschantz et al., 1995) was solved to high resolution of 1.9 Å (Paetzel et al., 1998) (Figure 9A). The catalytic serine (Ser 91) was covalently attached to the cleaved β-lactam inhibitor and the lysine 146 amino group was within hydrogen bond distance. This corroborates with the mutagenesis studies displaying the indispensable mechanism of active Ser and Lys dyad for catalysis (Sung and Dalbey, 1992; Tschantz et al., 1993; Paetzel et al., 1997) (Figure 9A) in contrast to the canonical Ser-His-Asp mechanism (Paetzel and Dalbey, 1997).
FIGURE 9.
Peptidases involved in the removal of signal peptides and their degradation. (A) Signal peptidase 1 [adapted from Paetzel et al. (2004) PDB: 1T7D] is a novel Ser-Lys protease that cleaves the preprotein at the membrane surface on the periplasmic side. Signal peptidase 2 [adapted from Vogeley et al. (2016) PDB: 5DIR] is an aspartic acid protease that cleaves a diacyl glyceride modified preproteins within the plane of the membrane. (B) SppA [adapted from Kim et al. (2008) PDB: 3BF0] is a tetrameric protein that degrades signal peptides which are released from the membrane into the periplasmic space. SppA employs a Ser-Lys dyad and is anchored to the membrane by an amino terminal TM segment. (C) RseP signal peptide peptidase [from Methanocaldococcus jannaschii adapted from Feng et al. (2007) PDB: 3B4R] in open state. Both water molecules and peptide substrates reach the active site containing Zn2+ ion (blue) during its open state.
Intensive analysis of the structure of the active site region with the inhibitor (Paetzel et al., 1998) and a signal peptide modeled into the binding site of the apo enzyme (Paetzel et al., 2002) revealed the binding pockets at the S1 and S3 positions that account for the “-3 and -1” or “Ala-X-Ala” rule for processing based on conserved residues in preproteins (Heijne, 1983). The S1 pocket is quite small and the S3 pocket is slightly bigger, fitting well with the known substrate specificity. The -2, -4 and -5 residues are solvent exposed, consistent with almost any residue found at these positions. The substrate binding pocket was further supported by the structure of the S. aureus SPase I (SpsB) with a portion of the signal peptide and the early mature region sequence binding to the active site (Ting et al., 2016).
2.4.2 Signal Peptidase II
As mentioned earlier, the substrate for signal peptidase II (SPase II) is a diacylglycerol modified prolipoprotein (Tokunaga et al., 1982). Following cleavage and further maturation, the bacterial lipoproteins possess a N-acyl diacylglycerylcysteine at the N-terminal end of the protein, which serves to anchor lipoproteins to the inner membrane or the outer membrane.
The gene for SPase II (lsp) was cloned by the Mizushima and the Wu labs independently (Tokunaga et al., 1983; Yamagata et al., 1983). The SPase protein spans the membrane four times with the protein ends facing the cytoplasm. The initial evidence of this peptidase as an aspartic protease was the fact that it was inhibited by pepstatin (Dev and Ray, 1984). Also, in B. subtilis, several aspartic acid residues located at the ends of the TM segments were shown to be important for activity (Tjalsma et al., 1999).
The structure of SPase II from Pseudomonas aeruginosa in complex with the inhibitor globomycin solved to 2.8 Å provided evidence that SPase II was an aspartic acid protease (Figure 9) (Vogeley et al., 2016). Along with mutagenesis studies, the work revealed that Asp 124 and Asp 143 comprise a catalytic dyad (Figure 9A). Interestingly, the aspartic residues are located within the predicted membrane region confirming that SPase catalyzes intramembrane proteolysis. These findings validate the fact that lipoprotein signal peptides typically have short hydrophobic regions. Caffrey and coworkers proposed a model for how SPase II binds the preprotein and carries out catalysis (Vogeley et al., 2016). The signal peptide helix of the preprotein enters the SPase II active region via TM segments 2 and 4, and then binds to the protein to position the preprotein lipobox residues analogous to the Leu-Ile-Ser tripeptide of the globomycin inhibitor. The signal peptide region immediately following the Leu residue in the lipobox is in an extended conformation with the Gly-Cys scissile bond positioned in proximity to the Asp catalytic dyad Asp124 and Asp143. The mature domain of the preprotein is located in the periplasmic region.
2.4.3 Signal Peptide Degradation
The final step “in the life and death of signal peptides (see ref von Heijne, 1998)” is their degradation. Degradation of the signal peptides is important because in many cases they can be toxic to the cell or interfere with protein export (Wickner et al., 1987; Bolhuis et al., 1999). The first protease discovered to possess signal peptide hydrolase activity was SppA (or protease IV), which was shown to degrade the lipoprotein signal peptide (Hussain et al., 1982). SppA is an inner protein that forms a tetrameric structure (Hussain et al., 1982; Ichihara et al., 1984). The catalytic domain of SppA containing the active site Ser-Lys dyad (Figure 9B) (Kim et al., 2008; Wang et al., 2008) is found within the periplasmic region at a large distance from the membrane indicating that the signal peptide is released from the membrane prior to its degradation (Kim et al., 2008). Apparently, SppA would cleave a wide variety of signal peptides that are released into the periplasmic space. Other proteases such as oligopeptidase A would hydrolyze signal peptides that are released into the cytoplasm (Novak et al., 1986).
The bacterial RseP, like signal peptide peptidase in eukaryotes (Lyko et al., 1995; Brown and Goldstein, 1997; Weihofen et al., 2002), can catalyze cleavage of membrane spanning signal peptides (Saito et al., 2011). RseP is a site-2 protease that can cleave within TM segments of membrane proteins as well (Akiyama et al., 2004). It is a zinc metalloprotease (Figure 9C). The active site is in an aqueous environment close to the cytoplasmic surface of the membrane. It binds zinc and has an essential catalytic glutamic acid. Saito et al. (2011) showed that RseP is capable of degrading a number of signal peptides from a wide variety of preproteins such as OmpA, M13 procoat, LivJ, LivK, PhoA, TolC, SecM, suggesting that it significantly contributes to signal peptide degradation in E. coli. This group of proteases is fascinating since they catalyze proteolysis within the membrane.
3 Targeting the Signal Peptide Proteases as Antibiotic Target
Signal peptidase I and II are attractive antibacterial drug targets (Paetzel et al., 2000; Rao et al., 2014; Craney and Romesberg, 2015; El Arnaout and Soulimane, 2019; Upert et al., 2021). SPase I is conserved in bacterial pathogens and has a novel active site architecture (Ser-Lys dyad) that can be targeted. Its active site location on the periplasmic side of the membrane makes it readily accessible (Smith and Romesberg, 2012). SPase II presents as another target candidate as lipoprotein signal peptidases are absent in the eukaryotic organism. The reduced efficacy of the existing antibiotics and the emergence of drug-resistant bacterial pathogens has led to the urgent demand for new treatments. This warrants the study of novel antibacterial targets such as SPase I and SPase II.
3.1 SPase I as Antibiotic Target
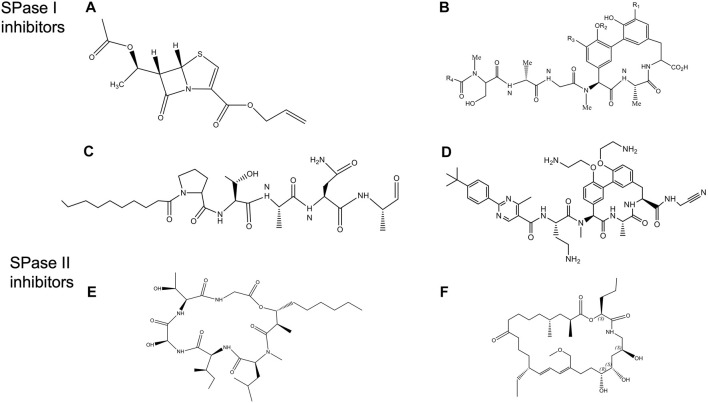
Various companies have centered on SPase I as an antibacterial target including Smithkline Beecham pharmaceuticals (now GlaxoSmithKline), Merck, Eli Lilly and Genentech. Some of the first inhibitors were the β-lactams, including clavams, thioclavams, penem carboxylate C6 Substituted esters, and allyl (5S,6S)-6-[R)-acetoxyethyl]penem-3-carboxylate (Figure 10A) and (5S)-tricyclic penem (Black and Bruton, 1998). Some of the peptide inhibitors such as, α-ketoamide peptides and decanoyl-PTANA-aldehyde (Figure 10C) were effective (Bruton et al., 2003; Buzder-Lantos et al., 2009).
FIGURE 10.
Structures of Signal peptidase inhibitors (Rao et al., 2014). (A) allyl (5S,6S)-6 [(R)-acetoxyethyl]-penem-3-carboxylate. (B) Arylomycin A. (C) Decanoyl PTANA aldehyde. (D) GO775, an optimized arylomycin. (E) Globomycin. (F) Myxovirescin.
Another promising class of inhibitors are the natural products Krisynomycin and Arylomycins. They are produced in Streptomyces by the non-ribosomal peptide synthesis. The Arylomycin family includes Arylomycin A and B (Höltzel et al., 2002; Schimana et al., 2002), and Arylomycin C (a lipoglycopeptide) (Kulanthaivel et al., 2004) discovered in the beginning of the 21st century. More recently, Arylomycin D was discovered (see below). Arylomycin D and its derivative M131, as well as Krisynomycin displayed significant antibacterial activity against the methicillin resistant S. aureus (MRSA) (Therien et al., 2012; Tan et al., 2020). Romesberg and others have developed a total synthesis of Arylomycin A2 (Roberts et al., 2007), Arylomycins B2 (Dufour et al., 2010; Roberts et al., 2011), Arylomycin C (Liu et al., 2011) and members of the Arylomycin D class (Therien et al., 2012; Tan et al., 2020).
Noteworthy, Smith and Romesberg (2012) proposed that Arylomycins may represent a class of latent antibiotics whose activity can be masked by mutations in the SPase I protease which otherwise would have rendered them susceptible. While Arylomycins normally have a narrow spectrum of antibiotic activity (Figure 10B), these antibiotics have the potential to have a much broader spectrum antibiotic activity against both Gram-positive and Gram-negative bacteria. Romesberg and coworkers initially investigated Staphylococcus epidermidis that was sensitive to Arylomycin A and isolated resistant strains which had mutations in SPase I (Ser to Pro changes at position 29). Intriguingly, the analogous Pro mutations in SPases that occurred during evolution, accounted for the natural resistance that is observed in E. coli, P. aeruginosa, and S. aureus. Strikingly, they found that a wide variety of bacteria that lacked this Pro substitution were sensitive to Arylomycin.
To gain insight into how to improve the potency of Ayrlomycin the structure of the E. coli SPase-Arylomycin complex was solved (Paetzel et al., 2004). Paetzel and coworkers revealed that the carboxylate of Arylomycin was bound to the catalytic serine, and the penultimate alanine of the inhibitor was localized within the S3 pocket. The structure also shows that residue 84 of E. coli SPase that confers resistance to the antibiotic is positioned near the amino-terminal part of the Arylomycin lipopeptide. A proline at the 84 position prevents the donation of one potential hydrogen bond from the backbone amide group on the β-strand 1 of SPase I to the carbonyl oxygen of the fatty acid of arylomycin. The presence of this extra H bond interaction would presumably increase the affinity of arylomycin to the SPase I proteins, making the bacteria more susceptible to the antibiotic.
A potential breakthrough in the antibiotic field by scientists at Genentech was the production of G0775, which represents a new class of Gram-negative antibiotics, that targets SPase I (Figure 10D) (Smith et al., 2018). This optimized arylomycin had several modifications including a replacement for the natural aliphatic tail, modification of the phenol groups of the tripeptide ring, and the introduction of an electrophilic warhead at the C-terminal carboxylate. GO775 was 500 times more potent than the arylomycin A-C16 against E. coli and K. pneumoniae which was normally not inhibited by arylomycin. It also had a potent activity against other pathogens. Additionally, it was effective in treating mice that were infected with Gram-negative bacterial pathogens, without any toxic impact on the mammalian cells. Intriguingly, the electrophilic warhead that was expected to covalently modify the active site serine instead modified the catalytic base lysine, by a novel mechanism. High affinity interaction between the target protein and the inhibitor made G0775 extremely active against multidrug resistant bacteria.
3.2 SPase II as Antibiotic Target
SPase II has been an attractive antibiotic target since the natural products globomycin (Figure 10E) and myxovirescin (Figure 10F) have been shown to have antibacterial activity (Inukai et al., 1978a; Inukai et al., 1978b; Nakajima et al., 1978; Xiao et al., 2012). Globomycin is a cyclic depsipeptide produced in Streptomyces. Myxovirescin is a secondary metabolite with a 28-membered macrocyclic lactone that is made in myxobacteria. Inhibition of SPase II is lethal in all Gram-negative bacteria.
Quite surprisingly, the structures of the S. aureus SPase II in complex with either globomycin or myxovirescin (Olatunji et al., 2020) revealed that the mode of inhibition is similar despite the two antibiotics interacting mostly with the protein on opposite sides of the substrate binding pocket. While both inhibitors bind to the catalytic Asp dyad with the hydroxyl group wedging in between (the β-hydroxy group of serine residue of globomycin and a 6 OH group from myxovirescin), most of the remaining parts of the molecule were on opposite sides of the substrate binding region. The interaction of the OH with the aspartic acid behaved like a non-cleavable tetrahedral analog (Olatunji et al., 2020). The hydroxyl groups of the antibiotics inhibited the enzyme by targeting the catalytic dyad aspartic acid residues.
To identify new inhibitors of SPase II, a high throughput screen was performed (Kitamura et al., 2018) where 646,275 molecules were analyzed using a SPase II FRET substrate assay. To validate their assay, they showed globomycin inhibited SPase II with an IC50 of 1.2 nM. Myxovirescin had a comparable or even better IC50. After identifying the best molecules from this initial screening, further optimization of the compound by medicinal chemistry resulted in an inhibitor of IC50 of 99 nM. Although this was a potent SPase II inhibitor, it did not accrue antibacterial activity in E. coli unless it was used in combination with polymyxin B nonapeptide, which made the outer membrane more permeable to compounds.
More recently, the structure of Globomycin was optimized to improve its antibacterial activity against E. coli (Garland et al., 2020) and its permeability across the outer membrane. Taking advantage of the SPase-globomycin structure, modifications were made to alter the lipophilic side chains, the n-hexyl group, and the backbone to introduce a salt-bridge that interacted with the SPase II catalytic aspartic residues. Several compounds were obtained that had increased potency against several Gram-negative pathogens.
4 Summary and Outlook
In conclusion, the signal peptide plays a universal central role in protein export in the three kingdoms of life. It orchestrates the sorting of proteins from the cytosol to the membrane. After targeting the protein to the membrane by binding to the receptor, the signal peptide activates the translocase such that the preprotein can make its way to the other side of the membrane.
Activation of the SecYEG channel occurs by binding of the signal peptide to the lateral gate, leading to a conformational change in the channel. Thus, signal peptide binding unlocks the Sec channel for translocation. In case of the Tat translocation machinery, the signal peptide provides the signal for the assembly of an oligomeric Tat translocase capable of transporting fully folded protein substrates. The key to triggering this process is the twin arginines within the signal peptide that binds to the TatBC receptor which switches on the Tat assembly process.
The final step in the translocation process is the removal of the signal peptide and its degradation. The hydrophobic region of the signal peptide positions the cleavage site for proteolysis by signal peptidases. After having served its purpose of protein navigation, the signal peptide is degraded by signal peptide hydrolases.
Owing to the decisive role the signal peptidases play in protein transport process, they have been appreciated recently as novel targets for antibiotics. The proteins so inhibited are involved in an array of bacterial fundamental processes essential for growth and viability of the bacteria/pathogen. The recent studies present Arylomycin and its derivatives such as G0775 as promising candidates for translation into new medicine to treat multidrug resistant pathogens. These compounds and the next generation of synthetic analogs will hopefully prove to be successful antibiotics to combat bacterial infections.
Yet, much remains to be discovered in the protein targeting and export field even 50 years after the Signal Hypothesis was proposed by Günter Blobel. Snapshots of the machineries engaged in substrate translocation are expected to provide new mechanistic insight into the processes of translocation dynamics and orientations of polytopic membrane proteins. Protein export has entered an exciting chapter, and more is anticipated in the days to come.
Acknowledgments
I would like to thank Andreas Kuhn for valuable comments on the paper. In addition, I would like to thank Tracy Palmer for providing the coordinates of the TatABC model in the resting state.
Author Contributions
SK, HH, and RD all contributed to writing the paper.
Funding
This research is supported by a grant from National Science Foundation Grant MCB-1052033.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Akiyama Y., Kanehara K., Ito K. (2004). RseP (YaeL), an Escherichia coli RIP Protease, Cleaves Transmembrane Sequences. Embo J. 23, 4434–4442. 10.1038/sj.emboj.7600449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopian D., Shen K., Zhang X., Shan S.-o. (2013). Signal Recognition Particle: An Essential Protein-Targeting Machine. Annu. Rev. Biochem. 82, 693–721. 10.1146/annurev-biochem-072711-164732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alami M., Lüke I., Deitermann S., Eisner G., Koch H.-G., Brunner J., et al. (2003). Differential Interactions between a Twin-Arginine Signal Peptide and its Translocase in Escherichia coli . Mol. Cell 12, 937–946. 10.1016/s1097-2765(03)00398-8 [DOI] [PubMed] [Google Scholar]
- Alcock F., Stansfeld P. J., Basit H., Habersetzer J., Baker M. A., Palmer T., et al. (2016). Assembling the Tat Protein Translocase. Elife 5, e20718. 10.7554/eLife.20718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen W. J., Corey R. A., Oatley P., Sessions R. B., Baldwin S. A., Radford S. E., et al. (2016). Two-Way Communication between SecY and SecA Suggests a Brownian Ratchet Mechanism for Protein Translocation. Elife 5, e15598. 10.7554/eLife.15598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen W. J., Watkins D. W., Dillingham M. S., Collinson I. (2020). Refined Measurement of SecA-Driven Protein Secretion Reveals that Translocation Is Indirectly Coupled to ATP Turnover. Proc. Natl. Acad. Sci. U.S.A. 117, 31808–31816. 10.1073/pnas.2010906117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson H., von Heijne G. (1993). Sec Dependent and Sec Independent Assembly of E. coli Inner Membrane Proteins: The Topological Rules Depend on Chain Length. EMBO J. 12, 683–691. 10.1002/j.1460-2075.1993.tb05702.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anghel S. A., McGilvray P. T., Hegde R. S., Keenan R. J. (2017). Identification of Oxa1 Homologs Operating in the Eukaryotic Endoplasmic Reticulum. Cell Rep. 21, 3708–3716. 10.1016/j.celrep.2017.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkowitz R. A., Wickner W. (1994). SecD and SecF Are Required for the Proton Electrochemical Gradient Stimulation of Preprotein Translocation. EMBO J. 13, 954–963. 10.1002/j.1460-2075.1994.tb06340.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschtgen M. S., Zoued A., Lloubès R., Journet L., Cascales E. (2012). The C-Tail Anchored TssL Subunit, an Essential Protein of the Enteroaggregative Escherichia coli Sci-1 Type VI Secretion System, Is Inserted by YidC. Microbiologyopen 1, 71–82. 10.1002/mbo3.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassford P., Beckwith J. (1979). Escherichia coli Mutants Accumulating the Precursor of a Secreted Protein in the Cytoplasm. Nature 277, 538–541. 10.1038/277538a0 [DOI] [PubMed] [Google Scholar]
- Bauer B. W., Rapoport T. A. (2009). Mapping Polypeptide Interactions of the SecA ATPase during Translocation. Proc. Natl. Acad. Sci. U. S. A. 106, 20800–20805. 10.1073/pnas.0910550106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer B. W., Shemesh T., Chen Y., Rapoport T. A. (2014). A “Push and Slide” Mechanism Allows Sequence-Insensitive Translocation of Secretory Proteins by the SecA ATPase. Cell 157, 1416–1429. 10.1016/j.cell.2014.03.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck K., Eisner G., Trescher D., Dalbey R. E., Brunner J., Müller M. (2001). YidC, an Assembly Site for Polytopic Escherichia coli Membrane Proteins Located in Immediate Proximity to the SecYE Translocon and Lipids. EMBO Rep. 2, 709–714. 10.1093/embo-reports/kve154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrendt J., Brüser T. (2014). The TatBC Complex of the Tat Protein Translocase in Escherichia coli and its Transition to the Substrate-Bound TatABC Complex. Biochemistry 53, 2344–2354. 10.1021/bi500169s [DOI] [PubMed] [Google Scholar]
- Berks B. C. (1996). A Common Export Pathway for Proteins Binding Complex Redox Cofactors? Mol. Microbiol. 22, 393–404. 10.1046/j.1365-2958.1996.00114.x [DOI] [PubMed] [Google Scholar]
- Berks B. C. (2015). The Twin-Arginine Protein Translocation Pathway. Annu. Rev. Biochem. 84, 843–864. 10.1146/annurev-biochem-060614-034251 [DOI] [PubMed] [Google Scholar]
- Bischoff L., Wickles S., Berninghausen O., van der Sluis E. O., Beckmann R. (2014). Visualization of a Polytopic Membrane Protein during SecY-Mediated Membrane Insertion. Nat. Commun. 5, 4103. 10.1038/ncomms5103 [DOI] [PubMed] [Google Scholar]
- Black M. T., Bruton G. (1998). Inhibitors of Bacterial Signal Peptidases. Curr. Pharm. Des. 4, 133–154. [PubMed] [Google Scholar]
- Bogsch E., Brink S., Robinson C. (1997). Pathway Specificity for a ΔpH-dependent Precursor Thylakoid Lumen Protein Is Governed by a ‘Sec-Avoidance' Motif in the Transfer Peptide and a ‘Sec-Incompatible' Mature Protein. Embo J. 16, 3851–3859. 10.1093/emboj/16.13.3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogsch E. G., Sargent F., Stanley N. R., Berks B. C., Robinson C., Palmer T. (1998). An Essential Component of a Novel Bacterial Protein Export System with Homologues in Plastids and Mitochondria. J. Biol. Chem. 273, 18003–18006. 10.1074/jbc.273.29.18003 [DOI] [PubMed] [Google Scholar]
- Bolhuis A., Mathers J. E., Thomas J. D., Barrett C. M. L., Robinson C. (2001). TatB and TatC Form a Functional and Structural Unit of the Twin-Arginine Translocase from Escherichia coli . J. Biol. Chem. 276, 20213–20219. 10.1074/jbc.m100682200 [DOI] [PubMed] [Google Scholar]
- Bolhuis A., Matzen A., Hyyryläinen H.-L., Kontinen V. P., Meima R., Chapuis J., et al. (1999). Signal Peptide Peptidase- and ClpP-like Proteins of Bacillus Subtilis Required for Efficient Translocation and Processing of Secretory Proteins. J. Biol. Chem. 274, 24585–24592. 10.1074/jbc.274.35.24585 [DOI] [PubMed] [Google Scholar]
- Bonardi F., Halza E., Walko M., Du Plessis F., Nouwen N., Feringa B. L., et al. (2011). Probing the SecYEG Translocation Pore Size with Preproteins Conjugated with Sizable Rigid Spherical Molecules. Proc. Natl. Acad. Sci. U. S. A. 108, 7775–7780. 10.1073/pnas.1101705108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornemann T., Jöckel J., Rodnina M. V., Wintermeyer W. (2008). Signal Sequence-independent Membrane Targeting of Ribosomes Containing Short Nascent Peptides within the Exit Tunnel. Nat. Struct. Mol. Biol. 15, 494–499. 10.1038/nsmb.1402 [DOI] [PubMed] [Google Scholar]
- Botte M., Zaccai N. R., Nijeholt J. L., Martin R., Knoops K., Papai G., et al. (2016). A Central Cavity Within the Holo-Translocon Suggests a Mechanism for Membrane Protein Insertion. Sci. Rep. 6, 38399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. S., Goldstein J. L. (1997). The SREBP Pathway: Regulation of Cholesterol Metabolism by Proteolysis of a Membrane-Bound Transcription Factor. Cell 89, 331–340. 10.1016/s0092-8674(00)80213-5 [DOI] [PubMed] [Google Scholar]
- Brüser T., Sanders C. (2003). An Alternative Model of the Twin Arginine Translocation System. Microbiol. Res. 158, 7–17. 10.1078/0944-5013-00176 [DOI] [PubMed] [Google Scholar]
- Bruton G., Huxley A., O'Hanlon P., Orlek B., Eggleston D., Humphries J., et al. (2003). Lipopeptide Substrates for SpsB, the Staphylococcus aureus Type I Signal Peptidase: Design, Conformation and Conversion to α-ketoamide Inhibitors. Eur. J. Med. Chem. 38, 351–356. 10.1016/s0223-5234(03)00040-0 [DOI] [PubMed] [Google Scholar]
- Buchanan G., Sargent F., Berks B., Palmer T. (2001). A Genetic Screen for Suppressors of Escherichia coli Tat Signal Peptide Mutations Establishes a Critical Role for the Second Arginine within the Twin-Arginine Motif. Archives Microbiol. 177, 107–112. 10.1007/s00203-001-0366-2 [DOI] [PubMed] [Google Scholar]
- Buzder-Lantos P., Bockstael K., Anné J., Herdewijn P. (2009). Substrate Based Peptide Aldehyde Inhibits Bacterial Type I Signal Peptidase. Bioorg. Med. Chem. Lett. 19, 2880–2883. 10.1016/j.bmcl.2009.03.064 [DOI] [PubMed] [Google Scholar]
- Castanié-Cornet M.-P., Bruel N., Genevaux P. (2014). Chaperone Networking Facilitates Protein Targeting to the Bacterial Cytoplasmic Membrane. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1843, 1442–1456. 10.1016/j.bbamcr.2013.11.007 [DOI] [PubMed] [Google Scholar]
- Catipovic M. A., Bauer B. W., Loparo J. J., Rapoport T. A. (2019). Protein Translocation by the SecA ATPase Occurs by a Power-Stroke Mechanism. EMBO J. 38, e101140. 10.15252/embj.2018101140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catipovic M. A., Rapoport T. A. (2020). Protease Protection Assays Show Polypeptide Movement into the SecY Channel by Power Strokes of the SecA ATPase. EMBO Rep. 21, e50905. 10.15252/embr.202050905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celebi N., Yi L., J. Facey S., Kuhn A., Dalbey R. E. (2006). Membrane Biogenesis of Subunit II of Cytochrome Bo Oxidase: Contrasting Requirements for Insertion of N-Terminal and C-Terminal Domains. J. Mol. Biol. 357, 1428–1436. 10.1016/j.jmb.2006.01.030 [DOI] [PubMed] [Google Scholar]
- Chaddock A. M., Mant A., Karnauchov I., Brink S., Herrmann R. G., Klösgen R. B., et al. (1995). A New Type of Signal Peptide: Central Role of a Twin-Arginine Motif in Transfer Signals for the Delta pH-dependent Thylakoidal Protein Translocase. EMBO J. 14, 2715–2722. 10.1002/j.1460-2075.1995.tb07272.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzi K. E., Sardis M. F., Economou A., Karamanou S. (2014). SecA-mediated Targeting and Translocation of Secretory Proteins. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1843, 1466–1474. 10.1016/j.bbamcr.2014.02.014 [DOI] [PubMed] [Google Scholar]
- Chen M., Samuelson J. C., Jiang F., Muller M., Kuhn A., Dalbey R. E. (2002). Direct Interaction of YidC with the Sec-independent Pf3 Coat Protein during its Membrane Protein Insertion. J. Biol. Chem. 277, 7670–7675. 10.1074/jbc.m110644200 [DOI] [PubMed] [Google Scholar]
- Chen Y., Capponi S., Zhu L., Gellenbeck P., Freites J. A., White S. H., et al. (2017). YidC Insertase of Escherichia coli: Water Accessibility and Membrane Shaping. Structure 25, 1403–1414. e1403. 10.1016/j.str.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Dalbey R. E. (2018). Oxa1 Superfamily: New Members Found in the ER. Trends Biochem. Sci. 43, 151–153. 10.1016/j.tibs.2017.12.005 [DOI] [PubMed] [Google Scholar]
- Chen Y., Soman R., Shanmugam S. K., Kuhn A., Dalbey R. E. (2014). The Role of the Strictly Conserved Positively Charged Residue Differs Among the Gram-Positive, Gram-Negative, and Chloroplast YidC Homologs. J. Biol. Chem. 289, 35656–35667. 10.1074/jbc.m114.595082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Sotomayor M., Capponi S., Hariharan B., Sahu I. D., Haase M., et al. (2022). A Hydrophilic Microenvironment in the Substrate-Translocating Groove of the YidC Membrane Insertase Is Essential for Enzyme Function. J. Biol. Chem. 298, 101690. 10.1016/j.jbc.2022.101690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S. Y., Randall L. L. (1994). In Vivo studies of the Role of SecA during Protein Export in Escherichia coli . J. Bacteriol. 176, 4197–4203. 10.1128/jb.176.14.4197-4203.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K., Mori H. (2001). Thylakoid ΔpH-dependent Precursor Proteins Bind to a cpTatC-Hcf106 Complex before Tha4-dependent Transport. J. Cell Biol. 154, 719–730. 10.1083/jcb.200105149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane J. M., Randall L. L. (2017). The Sec System: Protein Export in Escherichia coli . EcoSal Plus 7, 1. 10.1128/ecosalplus.ESP-0002-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craney A., Romesberg F. E. (2015). The Inhibition of Type I Bacterial Signal Peptidase: Biological Consequences and Therapeutic Potential. Bioorg. Med. Chem. Lett. 25, 4761–4766. 10.1016/j.bmcl.2015.07.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranford-Smith T., Huber D. (2018). The Way Is the Goal: How SecA Transports Proteins across the Cytoplasmic Membrane in Bacteria. FEMS Microbiol. Lett. 365, fny093. 10.1093/femsle/fny093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristobal S., de Gier J. W., Nielsen H., von Heijne G. (1999). Competition between Sec- and TAT-dependent Protein Translocation in Escherichia coli . Embo J. 18, 2982–2990. 10.1093/emboj/18.11.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke E., Guthrie B., Lecker S., Lill R., Wickner W. (1988). ProOmpA Is Stabilized for Membrane Translocation by Either Purified E. coli Trigger Factor or Canine Signal Recognition Particle. Cell 54, 1003–1011. 10.1016/0092-8674(88)90115-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cymer F., von Heijne G., White S. H. (2015). Mechanisms of Integral Membrane Protein Insertion and Folding. J. Mol. Biol. 427, 999–1022. 10.1016/j.jmb.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbey R. E., Wickner W. (1985). Leader Peptidase Catalyzes the Release of Exported Proteins from the Outer Surface of the Escherichia coli Plasma Membrane. J. Biol. Chem. 260, 15925–15931. 10.1016/s0021-9258(17)36347-0 [DOI] [PubMed] [Google Scholar]
- Date T. (1983). Demonstration by a Novel Genetic Technique that Leader Peptidase Is an Essential Enzyme of Escherichia coli . J. Bacteriol. 154, 76–83. 10.1128/jb.154.1.76-83.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date T., Goodman J. M., Wickner W. T. (1980). Procoat, the Precursor of M13 Coat Protein, Requires an Electrochemical Potential for Membrane Insertion. Proc. Natl. Acad. Sci. U.S.A. 77, 4669–4673. 10.1073/pnas.77.8.4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Geyter J., Portaliou A. G., Srinivasu B., Krishnamurthy S., Economou A., Karamanou S. (2020). Trigger Factor Is a Bona Fide Secretory Pathway Chaperone that Interacts with SecB and the Translocase. EMBO Rep. 21, e49054. 10.15252/embr.201949054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitermann S., Sprie G. S., Koch H.-G. (2005). A Dual Function for SecA in the Assembly of Single Spanning Membrane Proteins in Escherichia coli . J. Biol. Chem. 280, 39077–39085. 10.1074/jbc.m509647200 [DOI] [PubMed] [Google Scholar]
- DeLisa M. P., Samuelson P., Palmer T., Georgiou G. (2002). Genetic Analysis of the Twin Arginine Translocator Secretion Pathway in Bacteria. J. Biol. Chem. 277, 29825–29831. 10.1074/jbc.m201956200 [DOI] [PubMed] [Google Scholar]
- Dev I. K., Ray P. H. (1984). Rapid Assay and Purification of a Unique Signal Peptidase that Processes the Prolipoprotein from Escherichia coli B. J. Biol. Chem. 259, 11114–11120. 10.1016/s0021-9258(18)90629-0 [DOI] [PubMed] [Google Scholar]
- Do H., Falcone D., Lin J., Andrews D. W., Johnson A. E. (1996). The Cotranslational Integration of Membrane Proteins into the Phospholipid Bilayer Is a Multistep Process. Cell 85, 369–378. 10.1016/s0092-8674(00)81115-0 [DOI] [PubMed] [Google Scholar]
- Dufour J., Neuville L., Zhu J. (2010). Intramolecular Suzuki-Miyaura Reaction for the Total Synthesis of Signal Peptidase Inhibitors, Arylomycins A2 and B2. Chem. Eur. J. 16, 10523–10534. 10.1002/chem.201000924 [DOI] [PubMed] [Google Scholar]
- Economou A., Wickner W. (1994). SecA Promotes Preprotein Translocation by Undergoing ATP-Driven Cycles of Membrane Insertion and Deinsertion. Cell 78, 835–843. 10.1016/s0092-8674(94)90582-7 [DOI] [PubMed] [Google Scholar]
- El Arnaout T., Soulimane T. (2019). Targeting Lipoprotein Biogenesis: Considerations towards Antimicrobials. Trends Biochem. Sci. 44, 701–715. 10.1016/j.tibs.2019.03.007 [DOI] [PubMed] [Google Scholar]
- Emr S. D., Schwartz M., Silhavy T. J. (1978). Mutations Altering the Cellular Localization of the Phage Lambda Receptor, an Escherichia coli Outer Membrane Protein. Proc. Natl. Acad. Sci. U.S.A. 75, 5802–5806. 10.1073/pnas.75.12.5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlandson K. J., Miller S. B. M., Nam Y., Osborne A. R., Zimmer J., Rapoport T. A. (2008). A Role for the Two-Helix Finger of the SecA ATPase in Protein Translocation. Nature 455, 984–987. 10.1038/nature07439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facey S. J., Neugebauer S. A., Krauss S., Kuhn A. (2007). The Mechanosensitive Channel Protein MscL Is Targeted by the SRP to the Novel YidC Membrane Insertion Pathway of Escherichia coli . J. Mol. Biol. 365, 995–1004. 10.1016/j.jmb.2006.10.083 [DOI] [PubMed] [Google Scholar]
- Feng L., Yan H., Wu Z., Yan N., Wang Z., Jeffrey P. D., et al. (2007). Structure of a Site-2 Protease Family Intramembrane Metalloprotease. Science 318, 1608–1612. [DOI] [PubMed] [Google Scholar]
- Ferbitz L., Maier T., Patzelt H., Bukau B., Deuerling E., Ban N. (2004). Trigger Factor in Complex with the Ribosome Forms a Molecular Cradle for Nascent Proteins. Nature 431, 590–596. 10.1038/nature02899 [DOI] [PubMed] [Google Scholar]
- Frain K. M., Robinson C., van Dijl J. M. (2019). Transport of Folded Proteins by the Tat System. Protein J. 38, 377–388. 10.1007/s10930-019-09859-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland K., Pantua H., Braun M.-G., Burdick D. J., Castanedo G. M., Chen Y.-C., et al. (2020). Optimization of Globomycin Analogs as Novel Gram-Negative Antibiotics. Bioorg. Med. Chem. Lett. 30, 127419. 10.1016/j.bmcl.2020.127419 [DOI] [PubMed] [Google Scholar]
- Gelis I., Bonvin A. M. J. J., Keramisanou D., Koukaki M., Gouridis G., Karamanou S., et al. (2007). Structural Basis for Signal-Sequence Recognition by the Translocase Motor SecA as Determined by NMR. Cell 131, 756–769. 10.1016/j.cell.2007.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y., Kedrov A., Caumanns J. J., Crevenna A. H., Lamb D. C., Beckmann R., et al. (2015). Role of the Cytosolic Loop C2 and the C Terminus of YidC in Ribosome Binding and Insertion Activity. J. Biol. Chem. 290, 17250–17261. 10.1074/jbc.m115.650309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérard F., Cline K. (2006). Efficient Twin Arginine Translocation (Tat) Pathway Transport of a Precursor Protein Covalently Anchored to its Initial cpTatC Binding Site. J. Biol. Chem. 281, 6130–6135. 10.1074/jbc.m512733200 [DOI] [PubMed] [Google Scholar]
- Goder V., Spiess M. (2003). Molecular Mechanism of Signal Sequence Orientation in the Endoplasmic Reticulum. EMBO J. 22, 3645–3653. 10.1093/emboj/cdg361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogala M., Becker T., Beatrix B., Armache J.-P., Barrio-Garcia C., Berninghausen O., et al. (2014). Structures of the Sec61 Complex Engaged in Nascent Peptide Translocation or Membrane Insertion. Nature 506, 107–110. 10.1038/nature12950 [DOI] [PubMed] [Google Scholar]
- Gohlke U., Pullan L., McDevitt C. A., Porcelli I., de Leeuw E., Palmer T., et al. (2005). The TatA Component of the Twin-Arginine Protein Transport System Forms Channel Complexes of Variable Diameter. Proc. Natl. Acad. Sci. U.S.A. 102, 10482–10486. 10.1073/pnas.0503558102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady L. M., Michtavy J., Oliver D. B. (2012). Characterization of the Escherichia coli SecA Signal Peptide-Binding Site. J. Bacteriol. 194, 307–316. 10.1128/jb.06150-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grudnik P., Bange G., Sinning I. (2009). Protein Targeting by the Signal Recognition Particle. Biol. Chem. 390, 775–782. 10.1515/bc.2009.102 [DOI] [PubMed] [Google Scholar]
- Gupta S. D., Wu H. C. (1991). Identification and Subcellular Localization of apolipoproteinN-Acyltransferase inEscherichia Coli. FEMS Microbiol. Lett. 78, 37–42. 10.1111/j.1574-6968.1991.tb04413.x [DOI] [PubMed] [Google Scholar]
- Habersetzer J., Moore K., Cherry J., Buchanan G., Stansfeld P. J., Palmer T. (2017). Substrate-triggered Position Switching of TatA and TatB during Tat Transport in Escherichia coli . Open Biol. 7, 170091. 10.1098/rsob.170091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan B., Pross E., Soman R., Kaushik S., Kuhn A., Dalbey R. E. (2021). Polarity/charge as a Determinant of Translocase Requirements for Membrane Protein Insertion. Biochimica Biophysica Acta (BBA) - Biomembr. 1863, 183502. 10.1016/j.bbamem.2020.183502 [DOI] [PubMed] [Google Scholar]
- Hartl F.-U., Lecker S., Schiebel E., Hendrick J. P., Wickner W. (1990). The Binding Cascade of SecB to SecA to SecYE Mediates Preprotein Targeting to the E. coli Plasma Membrane. Cell 63, 269–279. 10.1016/0092-8674(90)90160-g [DOI] [PubMed] [Google Scholar]
- Hartmann E., Sommer T., Prehn S., Görlich D., Jentsch S., Rapoport T. A. (1994). Evolutionary Conservation of Components of the Protein Translocation Complex. Nature 367, 654–657. 10.1038/367654a0 [DOI] [PubMed] [Google Scholar]
- Hatzixanthis K., Palmer T., Sargent F. (2003). A Subset of Bacterial Inner Membrane Proteins Integrated by the Twin-Arginine Translocase. Mol. Microbiol. 49, 1377–1390. 10.1046/j.1365-2958.2003.03642.x [DOI] [PubMed] [Google Scholar]
- He H., Kuhn A., Dalbey R. E. (2020). Tracking the Stepwise Movement of a Membrane-Inserting Protein In Vivo . J. Mol. Biol. 432, 484–496. 10.1016/j.jmb.2019.10.010 [DOI] [PubMed] [Google Scholar]
- Hegde R. S., Bernstein H. D. (2006). The Surprising Complexity of Signal Sequences. Trends Biochem. Sci. 31, 563–571. 10.1016/j.tibs.2006.08.004 [DOI] [PubMed] [Google Scholar]
- Hegde R. S., Keenan R. J. (2021). The Mechanisms of Integral Membrane Protein Biogenesis. Nat. Rev. Mol. Cell Biol. 23 (2), 107–124. 10.1038/s41580-021-00413-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijne G. (1983). Patterns of Amino Acids Near Signal-Sequence Cleavage Sites. Eur. J. Biochem. 133, 17–21. 10.1111/j.1432-1033.1983.tb07424.x [DOI] [PubMed] [Google Scholar]
- Heinrich S. U., Mothes W., Brunner J., Rapoport T. A. (2000). The Sec61p Complex Mediates the Integration of a Membrane Protein by Allowing Lipid Partitioning of the Transmembrane Domain. Cell 102, 233–244. 10.1016/s0092-8674(00)00028-3 [DOI] [PubMed] [Google Scholar]
- Heinrich S. U., Rapoport T. A. (2003). Cooperation of Transmembrane Segments during the Integration of a Double-Spanning Protein into the ER Membrane. EMBO J. 22, 3654–3663. 10.1093/emboj/cdg346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennon S. W., Soman R., Zhu L., Dalbey R. E. (2015). YidC/Alb3/Oxa1 Family of Insertases. J. Biol. Chem. 290, 14866–14874. 10.1074/jbc.r115.638171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessa T., Kim H., Bihlmaier K., Lundin C., Boekel J., Andersson H., et al. (2005). Recognition of Transmembrane Helices by the Endoplasmic Reticulum Translocon. Nature 433, 377–381. 10.1038/nature03216 [DOI] [PubMed] [Google Scholar]
- Hessa T., Meindl-Beinker N. M., Bernsel A., Kim H., Sato Y., Lerch-Bader M., et al. (2007). Molecular Code for Transmembrane-Helix Recognition by the Sec61 Translocon. Nature 450, 1026–1030. 10.1038/nature06387 [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Bukau B., Kramer G. (2010). Structure and Function of the Molecular Chaperone Trigger Factor. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1803, 650–661. 10.1016/j.bbamcr.2010.01.017 [DOI] [PubMed] [Google Scholar]
- Holtkamp W., Lee S., Bornemann T., Senyushkina T., Rodnina M. V., Wintermeyer W. (2012). Dynamic Switch of the Signal Recognition Particle from Scanning to Targeting. Nat. Struct. Mol. Biol. 19, 1332–1337. 10.1038/nsmb.2421 [DOI] [PubMed] [Google Scholar]
- Höltzel A., Schmid D. G., Nicholson G. J., Stevanovic S., Schimana J., Gebhardt K., et al. (2002). Arylomycins A and B, New Biaryl-Bridged Lipopeptide Antibiotics Produced by Streptomyces Sp. Tue 6075. II. Structure Elucidation. J. Antibiot. 55, 571–577. 10.7164/antibiotics.55.571 [DOI] [PubMed] [Google Scholar]
- Houben E. N. G., ten Hagen‐Jongman C. M., Brunner J., Oudega B., Luirink J. (2004). The Two Membrane Segments of Leader Peptidase Partition One by One into the Lipid Bilayer via a Sec/YidC Interface. EMBO Rep. 5, 970–975. 10.1038/sj.embor.7400261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Rossi P., Saio T., Kalodimos C. G. (2016). Structural Basis for the Antifolding Activity of a Molecular Chaperone. Nature 537, 202–206. 10.1038/nature18965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber D., Jamshad M., Hanmer R., Schibich D., Döring K., Marcomini I., et al. (2017). SecA Cotranslationally Interacts with Nascent Substrate Proteins In Vivo . J. Bacteriol. 199, e00622–16. 10.1128/JB.00622-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber D., Rajagopalan N., Preissler S., Rocco M. A., Merz F., Kramer G., et al. (2011). SecA Interacts with Ribosomes in Order to Facilitate Posttranslational Translocation in Bacteria. Mol. Cell 41, 343–353. 10.1016/j.molcel.2010.12.028 [DOI] [PubMed] [Google Scholar]
- Hunt J. F., Weinkauf S., Henry L., Fak J. J., McNicholas P., Oliver D. B., et al. (2002). Nucleotide Control of Interdomain Interactions in the Conformational Reaction Cycle of SecA. Science 297, 2018–2026. 10.1126/science.1074424 [DOI] [PubMed] [Google Scholar]
- Hussain M., Ichihara S., Mizushima S. (1982). Mechanism of Signal Peptide Cleavage in the Biosynthesis of the Major Lipoprotein of the Escherichia coli Outer Membrane. J. Biol. Chem. 257, 5177–5182. 10.1016/s0021-9258(18)34652-0 [DOI] [PubMed] [Google Scholar]
- Ichihara S., Beppu N., Mizushima S. (1984). Protease IV, a Cytoplasmic Membrane Protein of Escherichia coli, Has Signal Peptide Peptidase Activity. J. Biol. Chem. 259, 9853–9857. 10.1016/s0021-9258(17)42778-5 [DOI] [PubMed] [Google Scholar]
- Iino T., Takahashi M., Sako T. (1987). Role of Amino-Terminal Positive Charge on Signal Peptide in Staphylokinase Export across the Cytoplasmic Membrane of Escherichia coli . J. Biol. Chem. 262, 7412–7417. 10.1016/s0021-9258(18)48252-x [DOI] [PubMed] [Google Scholar]
- Ilbert M., Méjean V., Giudici-Orticoni M.-T., Samama J.-P., Iobbi-Nivol C. (2003). Involvement of a Mate Chaperone (TorD) in the Maturation Pathway of Molybdoenzyme TorA. J. Biol. Chem. 278, 28787–28792. 10.1074/jbc.m302730200 [DOI] [PubMed] [Google Scholar]
- Inukai M., Nakajima M., Osawa M., Haneishi T., Arai M. (1978). Globomycin, a New Peptide Antibiotic with Spheroplast-Forming Activity. II. Isolation and Physico-Chemical and Biological Characterization. J. Antibiot. 31, 421–425. 10.7164/antibiotics.31.421 [DOI] [PubMed] [Google Scholar]
- Inukai M., Takeuchi M., Shimizu K., Arai M. (1978). Mechanism of Action of Globomycin. J. Antibiot. 31, 1203–1205. 10.7164/antibiotics.31.1203 [DOI] [PubMed] [Google Scholar]
- Ismail N., Hedman R., Schiller N., von Heijne G. (2012). A Biphasic Pulling Force Acts on Transmembrane Helices during Translocon-Mediated Membrane Integration. Nat. Struct. Mol. Biol. 19, 1018–1022. 10.1038/nsmb.2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack R. L., Buchanan G., Dubini A., Hatzixanthis K., Palmer T., Sargent F. (2004). Coordinating Assembly and Export of Complex Bacterial Proteins. Embo J. 23, 3962–3972. 10.1038/sj.emboj.7600409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongbloed J. D. H., Grieger U., Antelmann H., Hecker M., Nijland R., Bron S., et al. (2004). Two Minimal Tat Translocases in Bacillus. Mol. Microbiol. 54, 1319–1325. 10.1111/j.1365-2958.2004.04341.x [DOI] [PubMed] [Google Scholar]
- Karamanou S., Vrontou E., Sianidis G., Baud C., Roos T., Kuhn A., et al. (1999). A Molecular Switch in SecA Protein Couples ATP Hydrolysis to Protein Translocation. Mol. Microbiol. 34, 1133–1145. 10.1046/j.1365-2958.1999.01686.x [DOI] [PubMed] [Google Scholar]
- Karamyshev A. L., Johnson A. E. (2005). Selective SecA Association with Signal Sequences in Ribosome-Bound Nascent Chains. J. Biol. Chem. 280, 37930–37940. 10.1074/jbc.m509100200 [DOI] [PubMed] [Google Scholar]
- Kater L., Frieg B., Berninghausen O., Gohlke H., Beckmann R., Kedrov A. (2019). Partially Inserted Nascent Chain Unzips the Lateral Gate of the Sec Translocon. EMBO Rep. 20, e48191. 10.15252/embr.201948191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedrov A., Wickles S., Crevenna A. H., van der Sluis E. O., Buschauer R., Berninghausen O., et al. (2016). Structural Dynamics of the YidC:Ribosome Complex during Membrane Protein Biogenesis. Cell Rep. 17, 2943–2954. 10.1016/j.celrep.2016.11.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R., de Keyzer J., Driessen A. J. M., Palmer T. (2012). Co-operation between Different Targeting Pathways during Integration of a Membrane Protein. J. Cell Biol. 199, 303–315. 10.1083/jcb.201204149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer D., Kuhn A. (2018). YidC-mediated Membrane Insertion. FEMS Microbiol. Lett. 365, 1. 10.1093/femsle/fny106 [DOI] [PubMed] [Google Scholar]
- Kim A. C., Oliver D. C., Paetzel M. (2008). Crystal Structure of a Bacterial Signal Peptide Peptidase. J. Mol. Biol. 376, 352–366. 10.1016/j.jmb.2007.11.080 [DOI] [PubMed] [Google Scholar]
- Kitamura S., Owensby A., Wall D., Wolan D. W. (2018). Lipoprotein Signal Peptidase Inhibitors with Antibiotic Properties Identified through Design of a Robust In Vitro HT Platform. Cell Chem. Biol. 25, 301–308. e312. 10.1016/j.chembiol.2017.12.011 [DOI] [PubMed] [Google Scholar]
- Klein P., Somorjai R. L., Lau P. C. K. (1988). Distinctive Properties of Signal Sequences from Bacterial Lipoproteins. Protein Eng. Des. Sel. 2, 15–20. 10.1093/protein/2.1.15 [DOI] [PubMed] [Google Scholar]
- Klenner C., Kuhn A. (2012). Dynamic Disulfide Scanning of the Membrane-Inserting Pf3 Coat Protein Reveals Multiple YidC Substrate Contacts. J. Biol. Chem. 287, 3769–3776. 10.1074/jbc.m111.307223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenner C., Yuan J., Dalbey R. E., Kuhn A. (2008). The Pf3 Coat Protein Contacts TM1 and TM3 of YidC during Membrane Biogenesis. FEBS Lett. 582, 3967–3972. 10.1016/j.febslet.2008.10.044 [DOI] [PubMed] [Google Scholar]
- Knüpffer L., Fehrenbach C., Denks K., Erichsen V., Petriman N. A., Koch H. G. (2019). Molecular Mimicry of SecA and Signal Recognition Particle Binding to the Bacterial Ribosome. mBio 10, 1. 10.1128/mBio.01317-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kol S., Majczak W., Heerlien R., van der Berg J. P., Nouwen N., Driessen A. J. M. (2009). Subunit a of the F1F0 ATP Synthase Requires YidC and SecYEG for Membrane Insertion. J. Mol. Biol. 390, 893–901. 10.1016/j.jmb.2009.05.074 [DOI] [PubMed] [Google Scholar]
- Kol S., Turrell B. R., de Keyzer J., van der Laan M., Nouwen N., Driessen A. J. M. (2006). YidC-mediated Membrane Insertion of Assembly Mutants of Subunit C of the F1F0 ATPase. J. Biol. Chem. 281, 29762–29768. 10.1074/jbc.m605317200 [DOI] [PubMed] [Google Scholar]
- Komar J., Alvira S., Schulze R. J., Martin R., Lycklama a Nijeholt J. A., Lee S. C., et al. (2016). Membrane Protein Insertion and Assembly by the Bacterial Holo-Translocon SecYEG-SecDF-YajC-YidC. Biochem. J. 473, 3341–3354. 10.1042/bcj20160545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtz L., Oliver D. (2000). Tyr-326 Plays a Critical Role in Controlling SecA-Preprotein Interaction. Mol. Microbiol. 37, 1342–1356. 10.1046/j.1365-2958.2000.02078.x [DOI] [PubMed] [Google Scholar]
- Kuhn A. (1988). Alterations in the Extracellular Domain of M13 Procoat Protein Make its Membrane Insertion Dependent on secA and secY. Eur. J. Biochem. 177, 267–271. 10.1111/j.1432-1033.1988.tb14371.x [DOI] [PubMed] [Google Scholar]
- Kulanthaivel P., Kreuzman A. J., Strege M. A., Belvo M. D., Smitka T. A., Clemens M., et al. (2004a). Novel Lipoglycopeptides as Inhibitors of Bacterial Signal Peptidase I. J. Biol. Chem. 279, 36250–36258. 10.1074/jbc.m405884200 [DOI] [PubMed] [Google Scholar]
- Kumamoto C. A., Francetić O. (1993). Highly Selective Binding of Nascent Polypeptides by an Escherichia coli Chaperone Protein In Vivo . J. Bacteriol. 175, 2184–2188. 10.1128/jb.175.8.2184-2188.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazaki K., Chiba S., Takemoto M., Furukawa A., Nishiyama K.-i., Sugano Y., et al. (2014a). Structural Basis of Sec-independent Membrane Protein Insertion by YidC. Nature 509, 516–520. 10.1038/nature13167 [DOI] [PubMed] [Google Scholar]
- Kumazaki K., Kishimoto T., Furukawa A., Mori H., Tanaka Y., Dohmae N., et al. (2014b). Crystal Structure of Escherichia coli YidC, a Membrane Protein Chaperone and Insertase. Sci. Rep. 4, 7299. 10.1038/srep07299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusters I., Driessen A. J. M. (2011). SecA, a Remarkable Nanomachine. Cell. Mol. Life Sci. 68, 2053–2066. 10.1007/s00018-011-0681-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lausberg F., Fleckenstein S., Kreutzenbeck P., Fröbel J., Rose P., Müller M., et al. (2012). Genetic Evidence for a Tight Cooperation of TatB and TatC during Productive Recognition of Twin-Arginine (Tat) Signal Peptides in Escherichia coli . PloS one 7, e39867. 10.1371/journal.pone.0039867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. C., Bernstein H. D. (2001). The Targeting Pathway of Escherichia coli Presecretory and Integral Membrane Proteins Is Specified by the Hydrophobicity of the Targeting Signal. Proc. Natl. Acad. Sci. U.S.A. 98, 3471–3476. 10.1073/pnas.051484198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Chang L., Howell J. M., Turner R. J. (2010). DmsD, a Tat System Specific Chaperone, Interacts with Other General Chaperones and Proteins Involved in the Molybdenum Cofactor Biosynthesis. Biochimica Biophysica Acta (BBA) - Proteins Proteomics 1804, 1301–1309. 10.1016/j.bbapap.2010.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Park E., Ling J., Ingram J., Ploegh H., Rapoport T. A. (2016). Crystal Structure of a Substrate-Engaged SecY Protein-Translocation Channel. Nature 531, 395–399. 10.1038/nature17163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Schulman S., Boyd D., Erlandson K., Beckwith J., Rapoport T. A. (2007). The Plug Domain of the SecY Protein Stabilizes the Closed State of the Translocation Channel and Maintains a Membrane Seal. Mol. Cell 26, 511–521. 10.1016/j.molcel.2007.05.002 [DOI] [PubMed] [Google Scholar]
- Liu J., Luo C., Smith P. A., Chin J. K., Page M. G. P., Paetzel M., et al. (2011). Synthesis and Characterization of the Arylomycin Lipoglycopeptide Antibiotics and the Crystallographic Analysis of Their Complex with Signal Peptidase. J. Am. Chem. Soc. 133, 17869–17877. 10.1021/ja207318n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüke I., Handford J. I., Palmer T., Sargent F. (2009). Proteolytic Processing of Escherichia coli Twin-Arginine Signal Peptides by LepB. Arch. Microbiol. 191, 919–925. 10.1007/s00203-009-0516-5 [DOI] [PubMed] [Google Scholar]
- Lyko F., Martoglio B., Jungnickel B., Rapoport T. A., Dobberstein B. (1995). Signal Sequence Processing in Rough Microsomes. J. Biol. Chem. 270, 19873–19878. 10.1074/jbc.270.34.19873 [DOI] [PubMed] [Google Scholar]
- Ma C., Wu X., Sun D., Park E., Catipovic M. A., Rapoport T. A., et al. (2019). Structure of the Substrate-Engaged SecA-SecY Protein Translocation Machine. Nat. Commun. 10, 2872. 10.1038/s41467-019-10918-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard J., Spronk C. A. E. M., Buchanan G., Lyall V., Richardson D. J., Palmer T., et al. (2007). Structural Diversity in Twin-Arginine Signal Peptide-Binding Proteins. Proc. Natl. Acad. Sci. U.S.A. 104, 15641–15646. 10.1073/pnas.0703967104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R., Larsen A. H., Corey R. A., Midtgaard S. R., Frielinghaus H., Schaffitzel C., et al. (2019). Structure and Dynamics of the Central Lipid Pool and Proteins of the Bacterial Holo-Translocon. Biophysical J. 116, 1931–1940. 10.1016/j.bpj.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. S. (2002). Type IV Pili and Twitching Motility. Annu. Rev. Microbiol. 56, 289–314. 10.1146/annurev.micro.56.012302.160938 [DOI] [PubMed] [Google Scholar]
- McCormick P. J., Miao Y., Shao Y., Lin J., Johnson A. E. (2003). Cotranslational Protein Integration into the ER Membrane Is Mediated by the Binding of Nascent Chains to Translocon Proteins. Mol. Cell 12, 329–341. 10.1016/s1097-2765(03)00304-6 [DOI] [PubMed] [Google Scholar]
- McDowell M. A., Heimes M., Sinning I. (2021). Structural and Molecular Mechanisms for Membrane Protein Biogenesis by the Oxa1 Superfamily. Nat. Struct. Mol. Biol. 28, 234–239. 10.1038/s41594-021-00567-9 [DOI] [PubMed] [Google Scholar]
- Michaelis S., Beckwith J. (1982). Mechanism of Incorporation of Cell Envelope Proteins in Escherichia coli . Annu. Rev. Microbiol. 36, 435–465. 10.1146/annurev.mi.36.100182.002251 [DOI] [PubMed] [Google Scholar]
- Musial-Siwek M., Rusch S. L., Kendall D. A. (2007). Selective Photoaffinity Labeling Identifies the Signal Peptide Binding Domain on SecA. J. Mol. Biol. 365, 637–648. 10.1016/j.jmb.2006.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamori S., Smirnova I. N., Kaback H. R. (2004). Role of YidC in Folding of Polytopic Membrane Proteins. J. Cell Biol. 165, 53–62. 10.1083/jcb.200402067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M., Inukai M., Haneishi T., Terahara A., Arai M., Kinoshita T., et al. (1978). Globomycin, a New Peptide Antibiotic with Spheroplast-Forming Activity. III. Structural Determination of Globomycin. J. Antibiot. 31, 426–432. 10.7164/antibiotics.31.426 [DOI] [PubMed] [Google Scholar]
- Nicolaus F., Metola A., Mermans D., Liljenström A., Krč A., Abdullahi S. M., et al. (2021). Residue-by-residue Analysis of Cotranslational Membrane Protein Integration In Vivo . Elife 10, e64302. 10.7554/eLife.64302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak P., Ray P. H., Dev I. K. (1986). Localization and Purification of Two Enzymes from Escherichia coli Capable of Hydrolyzing a Signal Peptide. J. Biol. Chem. 261, 420–427. 10.1016/s0021-9258(17)42489-6 [DOI] [PubMed] [Google Scholar]
- Nunn D. N., Lory S. (1991). Product of the Pseudomonas aeruginosa Gene pilD Is a Prepilin Leader Peptidase. Proc. Natl. Acad. Sci. U.S.A. 88, 3281–3285. 10.1073/pnas.88.8.3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Becker A. H., Sandikci A., Huber D., Chaba R., Gloge F., et al. (2011). Selective Ribosome Profiling Reveals the Cotranslational Chaperone Action of Trigger Factor In Vivo . Cell 147, 1295–1308. 10.1016/j.cell.2011.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olatunji S., Yu X., Bailey J., Huang C.-Y., Zapotoczna M., Bowen K., et al. (2020). Structures of Lipoprotein Signal Peptidase II from Staphylococcus aureus Complexed with Antibiotics Globomycin and Myxovirescin. Nat. Commun. 11, 140. 10.1038/s41467-019-13724-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D. B., Beckwith J. (1981). E. coli Mutant Pleiotropically Defective in the Export of Secreted Proteins. Cell 25, 765–772. 10.1016/0092-8674(81)90184-7 [DOI] [PubMed] [Google Scholar]
- Oresnik I. J., Ladner C. L., Turner R. J. (2001). Identification of a Twin-Arginine Leader-Binding Protein. Mol. Microbiol. 40, 323–331. 10.1046/j.1365-2958.2001.02391.x [DOI] [PubMed] [Google Scholar]
- Oswald J., Njenga R., Natriashvili A., Sarmah P., Koch H.-G. (2021). The Dynamic SecYEG Translocon. Front. Mol. Biosci. 8, 664241. 10.3389/fmolb.2021.664241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetzel M. (2019). Bacterial Signal Peptidases. Subcell. Biochem. 92, 187–219. 10.1007/978-3-030-18768-2_7 [DOI] [PubMed] [Google Scholar]
- Paetzel M., Dalbey R. E. (1997). Catalytic Hydroxyl/amine Dyads within Serine Proteases. Trends Biochem. Sci. 22, 28–31. 10.1016/s0968-0004(96)10065-7 [DOI] [PubMed] [Google Scholar]
- Paetzel M., Dalbey R. E., Strynadka N. C. J. (2002). Crystal Structure of a Bacterial Signal Peptidase Apoenzyme. J. Biol. Chem. 277, 9512–9519. 10.1074/jbc.m110983200 [DOI] [PubMed] [Google Scholar]
- Paetzel M., Dalbey R. E., Strynadka N. C. J. (1998). Crystal Structure of a Bacterial Signal Peptidase in Complex with a β-lactam Inhibitor. Nature 396, 186–190. 10.1038/24196 [DOI] [PubMed] [Google Scholar]
- Paetzel M., Dalbey R. E., Strynadka N. C. J. (2000). The Structure and Mechanism of Bacterial Type I Signal Peptidases. Pharmacol. Ther. 87, 27–49. 10.1016/s0163-7258(00)00064-4 [DOI] [PubMed] [Google Scholar]
- Paetzel M., Goodall J. J., Kania M., Dalbey R. E., Page M. G. P. (2004). Crystallographic and Biophysical Analysis of a Bacterial Signal Peptidase in Complex with a Lipopeptide-Based Inhibitor. J. Biol. Chem. 279, 30781–30790. 10.1074/jbc.m401686200 [DOI] [PubMed] [Google Scholar]
- Paetzel M. (2014). Structure and Mechanism of Escherichia coli Type I Signal Peptidase. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1843, 1497–1508. 10.1016/j.bbamcr.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Paetzel M., Strynadka N. C. J., Tschantz W. R., Casareno R., Bullinger P. R., Dalbey R. E. (1997). Use of Site-Directed Chemical Modification to Study an Essential Lysine in Escherichia coli Leader Peptidase. J. Biol. Chem. 272, 9994–10003. 10.1074/jbc.272.15.9994 [DOI] [PubMed] [Google Scholar]
- Palmer T., Stansfeld P. J. (2020). Targeting of Proteins to the Twin‐arginine Translocation Pathway. Mol. Microbiol. 113, 861–871. 10.1111/mmi.14461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanikou E., Karamanou S., Baud C., Frank M., Sianidis G., Keramisanou D., et al. (2005). Identification of the Preprotein Binding Domain of SecA. J. Biol. Chem. 280, 43209–43217. 10.1074/jbc.m509990200 [DOI] [PubMed] [Google Scholar]
- Patel R., Smith S. M., Robinson C. (2014). Protein Transport by the Bacterial Tat Pathway. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1843, 1620–1628. 10.1016/j.bbamcr.2014.02.013 [DOI] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. (1983). A Putative Signal Peptidase Recognition Site and Sequence in Eukaryotic and Prokaryotic Signal Peptides. J. Mol. Biol. 167, 391–409. 10.1016/s0022-2836(83)80341-6 [DOI] [PubMed] [Google Scholar]
- Peschke M., Le Goff M., Koningstein G. M., Karyolaimos A., de Gier J.-W., van Ulsen P., et al. (2018). SRP, FtsY, DnaK and YidC Are Required for the Biogenesis of the E. coli Tail-Anchored Membrane Proteins DjlC and Flk. J. Mol. Biol. 430, 389–403. 10.1016/j.jmb.2017.12.004 [DOI] [PubMed] [Google Scholar]
- Petriman N.-A., Jauss B., Hufnagel A., Franz L., Sachelaru I., Drepper F., et al. (2018). The Interaction Network of the YidC Insertase with the SecYEG Translocon, SRP and the SRP Receptor FtsY. Sci. Rep. 8, 578. 10.1038/s41598-017-19019-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlschröder M., Prinz W. A., Hartmann E., Beckwith J. (1997). Protein Translocation in the Three Domains of Life: Variations on a Theme. Cell 91, 563–566. 10.1016/s0092-8674(00)80443-2 [DOI] [PubMed] [Google Scholar]
- Pross E., Kuhn A. (2020). Two Signal Recognition Particle Sequences Are Present in the Amino-Terminal Domain of the C-Tailed Protein SciP. J. Bacteriol. 203, e00312–20. 10.1128/JB.00312-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pross E., Soussoula L., Seitl I., Lupo D., Kuhn A. (2016). Membrane Targeting and Insertion of the C-Tail Protein SciP. J. Mol. Biol. 428, 4218–4227. 10.1016/j.jmb.2016.09.001 [DOI] [PubMed] [Google Scholar]
- Ramasamy S., Abrol R., Suloway C. J. M., Clemons W. M., Jr. (2013). The Glove-like Structure of the Conserved Membrane Protein TatC Provides Insight into Signal Sequence Recognition in Twin-Arginine Translocation. Structure 21, 777–788. 10.1016/j.str.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall L. L., Hardy S. J. S. (1986). Correlation of Competence for Export with Lack of Tertiary Structure of the Mature Species: a Study In Vivo of Maltose-Binding Protein in E. coli . Cell 46, 921–928. 10.1016/0092-8674(86)90074-7 [DOI] [PubMed] [Google Scholar]
- Rao S., De Waelheyns E., Economou A., Anné J. (2014). Antibiotic Targeting of the Bacterial Secretory Pathway. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1843, 1762–1783. 10.1016/j.bbamcr.2014.02.004 [DOI] [PubMed] [Google Scholar]
- Rawat S., Zhu L., Lindner E., Dalbey R. E., White S. H. (2015). SecA Drives Transmembrane Insertion of RodZ, an Unusual Single-Span Membrane Protein. J. Mol. Biol. 427, 1023–1037. 10.1016/j.jmb.2014.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray N., Oates J., Turner R. J., Robinson C. (2003). DmsD Is Required for the Biogenesis of DMSO Reductase inEscherichia Colibut Not for the Interaction of the DmsA Signal Peptide with the Tat Apparatus. FEBS Lett. 534, 156–160. 10.1016/s0014-5793(02)03839-5 [DOI] [PubMed] [Google Scholar]
- Roberts T. C., Smith P. A., Cirz R. T., Romesberg F. E. (2007). Structural and Initial Biological Analysis of Synthetic Arylomycin A2. J. Am. Chem. Soc. 129, 15830–15838. 10.1021/ja073340u [DOI] [PubMed] [Google Scholar]
- Roberts T. C., Smith P. A., Romesberg F. E. (2011). Synthesis and Biological Characterization of Arylomycin B Antibiotics. J. Nat. Prod. 74, 956–961. 10.1021/np200163g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C., Matos C. F. R. O., Beck D., Ren C., Lawrence J., Vasisht N., et al. (2011). Transport and Proofreading of Proteins by the Twin-Arginine Translocation (Tat) System in Bacteria. Biochimica Biophysica Acta (BBA) - Biomembr. 1808, 876–884. 10.1016/j.bbamem.2010.11.023 [DOI] [PubMed] [Google Scholar]
- Rodriguez F., Rouse S. L., Tait C. E., Harmer J., De Riso A., Timmel C. R., et al. (2013). Structural Model for the Protein-Translocating Element of the Twin-Arginine Transport System. Proc. Natl. Acad. Sci. U. S. A. 110, E1092–E1101. 10.1073/pnas.1219486110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollauer S. E., Tarry M. J., Graham J. E., Jääskeläinen M., Jäger F., Johnson S., et al. (2012). Structure of the TatC Core of the Twin-Arginine Protein Transport System. Nature 492, 210–214. 10.1038/nature11683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachelaru I., Winter L., Knyazev D. G., Zimmermann M., Vogt A., Kuttner R., et al. (2017). YidC and SecYEG Form a Heterotetrameric Protein Translocation Channel. Sci. Rep. 7, 101. 10.1038/s41598-017-00109-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saio T., Guan X., Rossi P., Economou A., Kalodimos C. G. (2014). Structural Basis for Protein Antiaggregation Activity of the Trigger Factor Chaperone. Science 344, 1250494. 10.1126/science.1250494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A., Hizukuri Y., Matsuo E.-i., Chiba S., Mori H., Nishimura O., et al. (2011). Post-Liberation Cleavage of Signal Peptides Is Catalyzed by the Site-2 Protease (S2P) in Bacteria. Proc. Natl. Acad. Sci. U.S.A. 108, 13740–13745. 10.1073/pnas.1108376108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saller M. J., Wu Z. C., de Keyzer J., Driessen A. J. M. (2012). The YidC/Oxa1/Alb3 Protein Family: Common Principles and Distinct Features. Biol. Chem. 393, 1279–1290. 10.1515/hsz-2012-0199 [DOI] [PubMed] [Google Scholar]
- Samuelson J. C., Chen M., Jiang F., Möller I., Wiedmann M., Kuhn A., et al. (2000). YidC Mediates Membrane Protein Insertion in Bacteria. Nature 406, 637–641. 10.1038/35020586 [DOI] [PubMed] [Google Scholar]
- Sandhu H., Hedman R., Cymer F., Kudva R., Ismail N., von Heijne G. (2021). Cotranslational Translocation and Folding of a Periplasmic Protein Domain in Escherichia coli . J. Mol. Biol. 433, 167047. 10.1016/j.jmb.2021.167047 [DOI] [PubMed] [Google Scholar]
- Sankaran K., Wu H. C. (1994). Lipid Modification of Bacterial Prolipoprotein. Transfer of Diacylglyceryl Moiety from Phosphatidylglycerol. J. Biol. Chem. 269, 19701–19706. 10.1016/s0021-9258(17)32077-x [DOI] [PubMed] [Google Scholar]
- Saraogi I., Shan S.-o. (2014). Co-translational Protein Targeting to the Bacterial Membrane. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1843, 1433–1441. 10.1016/j.bbamcr.2013.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent F., Bogsch E. G., Stanley N. R., Wexler M., Robinson C., Berks B. C., et al. (1998). Overlapping Functions of Components of a Bacterial Sec-independent Protein Export Pathway. Embo J. 17, 3640–3650. 10.1093/emboj/17.13.3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibich D., Gloge F., Pöhner I., Björkholm P., Wade R. C., von Heijne G., et al. (2016). Global Profiling of SRP Interaction with Nascent Polypeptides. Nature 536, 219–223. 10.1038/nature19070 [DOI] [PubMed] [Google Scholar]
- Schimana J., Gebhardt K., Höltzel A., Schmid D. G., Süssmuth R., Müller J., et al. (2002). Arylomycins A and B, New Biaryl-Bridged Lipopeptide Antibiotics Produced by Streptomyces Sp. Tue 6075. I. Taxonomy, Fermentation, Isolation and Biological Activities. J. Antibiot. 55, 565–570. 10.7164/antibiotics.55.565 [DOI] [PubMed] [Google Scholar]
- Schulze R. J., Komar J., Botte M., Allen W. J., Whitehouse S., Gold V. A. M., et al. (2014). Membrane Protein Insertion and Proton-motive-force-dependent Secretion through the Bacterial Holo-Translocon SecYEG-SecDF-YajC-YidC. Proc. Natl. Acad. Sci. U.S.A. 111, 4844–4849. 10.1073/pnas.1315901111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serek J., Bauer-Manz G., Struhalla G., van den Berg L., Kiefer D., Dalbey R., et al. (2004). Escherichia coli YidC Is a Membrane Insertase for Sec-independent Proteins. EMBO J. 23, 294–301. 10.1038/sj.emboj.7600063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam S. K., Dalbey R. E. (2019). The Conserved Role of YidC in Membrane Protein Biogenesis. Microbiol. Spectr. 7, 1. 10.1128/microbiolspec.PSIB-0014-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skach W. R., Lingappa V. R. (1993). Amino-terminal Assembly of Human P-Glycoprotein at the Endoplasmic Reticulum Is Directed by Cooperative Actions of Two Internal Sequences. J. Biol. Chem. 268, 23552–23561. 10.1016/s0021-9258(19)49498-2 [DOI] [PubMed] [Google Scholar]
- Smith P. A., Koehler M. F. T., Girgis H. S., Yan D., Chen Y., Chen Y., et al. (2018). Optimized Arylomycins Are a New Class of Gram-Negative Antibiotics. Nature 561, 189–194. 10.1038/s41586-018-0483-6 [DOI] [PubMed] [Google Scholar]
- Smith P. A., Romesberg F. E. (2012). Mechanism of Action of the Arylomycin Antibiotics and Effects of Signal Peptidase I Inhibition. Antimicrob. Agents Chemother. 56, 5054–5060. 10.1128/aac.00785-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soman R., Yuan J., Kuhn A., Dalbey R. E. (2014). Polarity and Charge of the Periplasmic Loop Determine the YidC and Sec Translocase Requirement for the M13 Procoat Lep Protein. J. Biol. Chem. 289, 1023–1032. 10.1074/jbc.m113.522250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley N. R., Palmer T., Berks B. C. (2000). The Twin Arginine Consensus Motif of Tat Signal Peptides Is Involved in Sec-independent Protein Targeting in Escherichia coli . J. Biol. Chem. 275, 11591–11596. 10.1074/jbc.275.16.11591 [DOI] [PubMed] [Google Scholar]
- Steudle A., Spann D., Pross E., Shanmugam S. K., Dalbey R. E., Kuhn A. (2021). Molecular Communication of the Membrane Insertase YidC with Translocase SecYEG Affects Client Proteins. Sci. Rep. 11, 3940. 10.1038/s41598-021-83224-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom M. S., Lory S. (1993). Structure-function and Biogenesis of the Type IV Pili. Annu. Rev. Microbiol. 47, 565–596. 10.1146/annurev.mi.47.100193.003025 [DOI] [PubMed] [Google Scholar]
- Strom M. S., Nunn D. N., Lory S. (1993). A Single Bifunctional Enzyme, PilD, Catalyzes Cleavage and N-Methylation of Proteins Belonging to the Type IV Pilin Family. Proc. Natl. Acad. Sci. U.S.A. 90, 2404–2408. 10.1073/pnas.90.6.2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M., Dalbey R. E. (1992). Identification of Potential Active-Site Residues in the Escherichia coli Leader Peptidase. J. Biol. Chem. 267, 13154–13159. 10.1016/s0021-9258(18)42186-2 [DOI] [PubMed] [Google Scholar]
- Tan Y. X., Peters D. S., Walsh S. I., Holcomb M., Santos-Martins D., Forli S., et al. (2020). Initial Analysis of the Arylomycin D Antibiotics. J. Nat. Prod. 83, 2112–2121. 10.1021/acs.jnatprod.9b01174 [DOI] [PubMed] [Google Scholar]
- Therien A. G., Huber J. L., Wilson K. E., Beaulieu P., Caron A., Claveau D., et al. (2012). Broadening the Spectrum of β-Lactam Antibiotics through Inhibition of Signal Peptidase Type I. Antimicrob. Agents Chemother. 56, 4662–4670. 10.1128/aac.00726-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting Y. T., Harris P. W. R., Batot G., Brimble M. A., Baker E. N., Young P. G. (2016). Peptide Binding to a Bacterial Signal Peptidase Visualized by Peptide Tethering and Carrier-Driven Crystallization. Int. Union Crystallogr. J. 3, 10–19. 10.1107/s2052252515019971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjalsma H., Zanen G., Bron S., van Dijl J. M. (2000). “The Eubacterial Lipoprotein-specific (Type II) Signal Peptidases,” in The Enzymes (Co- and Posttranslational Proteolysis Of Proteins). Editors Dalbey R. E., Sigman D. S.. third edition (London, UK): Academic Press; ), 3–26. XXII. [Google Scholar]
- Tjalsma H., Zanen G., Venema G., Bron S., van Dijl J. M. (1999). The Potential Active Site of the Lipoprotein-specific (Type II) Signal Peptidase of Bacillus Subtilis. J. Biol. Chem. 274, 28191–28197. 10.1074/jbc.274.40.28191 [DOI] [PubMed] [Google Scholar]
- Tokunaga M., Loranger J. M., Wu H. C. (1983). Isolation and Characterization of an Escherichia coli Clone Overproducing Prolipoprotein Signal Peptidase. J. Biol. Chem. 258, 12102–12105. 10.1016/s0021-9258(17)44136-6 [DOI] [PubMed] [Google Scholar]
- Tokunaga M., Tokunaga H., Wu H. C. (1982). Post-Translational Modification and Processing of Escherichia coli Prolipoprotein In Vitro . Proc. Natl. Acad. Sci. U.S.A. 79, 2255–2259. 10.1073/pnas.79.7.2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooke F. J., Babot M., Chandra G., Buchanan G., Palmer T. (2017). A Unifying Mechanism for the Biogenesis of Membrane Proteins Co-operatively Integrated by the Sec and Tat Pathways. Elife 6, e26577. 10.7554/eLife.26577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschantz W., Paetzel M., Cao G., Suciu D., Inouye M., Dalbey R. E. (1995). Characterization of a Soluble, Catalytically Active Form of Escherichia coli Leader Peptidase: Requirement of Detergent or Phospholipid for Optimal Activity. Biochemistry 34, 3935–3941. 10.1021/bi00012a010 [DOI] [PubMed] [Google Scholar]
- Tschantz W. R., Sung M., Delgado-Partin V. M., Dalbey R. E. (1993). A Serine and a Lysine Residue Implicated in the Catalytic Mechanism of the Escherichia coli Leader Peptidase. J. Biol. Chem. 268, 27349–27354. 10.1016/s0021-9258(19)74256-2 [DOI] [PubMed] [Google Scholar]
- Tsirigotaki A., De Geyter J., Šoštaric´ N., Economou A., Karamanou S. (2017). Protein Export through the Bacterial Sec Pathway. Nat. Rev. Microbiol. 15, 21–36. 10.1038/nrmicro.2016.161 [DOI] [PubMed] [Google Scholar]
- Tsukazaki T., Mori H., Echizen Y., Ishitani R., Fukai S., Tanaka T., et al. (2011). Structure and Function of a Membrane Component SecDF that Enhances Protein Export. Nature 474, 235–238. 10.1038/nature09980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner R. J., Papish A. L., Sargent F. (2004). Sequence Analysis of Bacterial Redox Enzyme Maturation Proteins (REMPs). Can. J. Microbiol. 50, 225–238. 10.1139/w03-117 [DOI] [PubMed] [Google Scholar]
- Ulbrandt N. D., London E., Oliver D. B. (1992). Deep Penetration of a Portion of Escherichia coli SecA Protein into Model Membranes Is Promoted by Anionic Phospholipids and by Partial Unfolding. J. Biol. Chem. 267, 15184–15192. 10.1016/s0021-9258(18)42163-1 [DOI] [PubMed] [Google Scholar]
- Upert G., Luther A., Obrecht D., Ermert P. (2021). Emerging Peptide Antibiotics with Therapeutic Potential. Med. Drug Discov. 9, 100078. 10.1016/j.medidd.2020.100078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanus M. L., Scotti P. A., Fröderberg L., Sääf A., de Gier J. W. L., Brunner J., et al. (2001). Sec‐dependent Membrane Protein Insertion: Sequential Interaction of Nascent FtsQ with SecY and YidC. EMBO Rep. 2, 524–529. 10.1093/embo-reports/kve108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bloois E., Haan G.-J., de Gier J.-W., Oudega B., Luirink J. (2006). Distinct Requirements for Translocation of the N-Tail and C-Tail of the Escherichia coli Inner Membrane Protein CyoA. J. Biol. Chem. 281, 10002–10009. 10.1074/jbc.m511357200 [DOI] [PubMed] [Google Scholar]
- van Bloois E., Jan Haan G., de Gier J.-W., Oudega B., Luirink J. (2004). F1F0ATP Synthase Subunit C Is Targeted by the SRP to YidC in the E. Coli Inner Membrane. FEBS Lett. 576, 97–100. 10.1016/j.febslet.2004.08.069 [DOI] [PubMed] [Google Scholar]
- van den Berg B., Clemons W. M., Collinson I., Modis Y., Hartmann E., Harrison S. C., et al. (2004). X-Ray Structure of a Protein-Conducting Channel. Nature 427, 36–44. 10.1038/nature02218 [DOI] [PubMed] [Google Scholar]
- Van Der Laan M., Bechtluft P., Kol S., Nouwen N., Driessen A. J. M. (2004). F1F0 ATP Synthase Subunit C Is a Substrate of the Novel YidC Pathway for Membrane Protein Biogenesis. J. Cell Biol. 165, 213–222. 10.1083/jcb.200402100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M., Urbanus M. L., ten Hagen-Jongman C. M., Nouwen N., Oudega B., Harms N., et al. (2003). A Conserved Function of YidC in the Biogenesis of Respiratory Chain Complexes. Proc. Natl. Acad. Sci. U.S.A. 100, 5801–5806. 10.1073/pnas.0636761100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasuk G. P., Inouye S., Ito H., Itakura K., Inouye M. (1983). Effects of the Complete Removal of Basic Amino Acid Residues from the Signal Peptide on Secretion of Lipoprotein in Escherichia coli . J. Biol. Chem. 258, 7141–7148. 10.1016/s0021-9258(18)32343-3 [DOI] [PubMed] [Google Scholar]
- Vogeley L., El Arnaout T., Bailey J., Stansfeld P. J., Boland C., Caffrey M. (2016). Structural Basis of Lipoprotein Signal Peptidase II Action and Inhibition by the Antibiotic Globomycin. Science 351, 876–880. 10.1126/science.aad3747 [DOI] [PubMed] [Google Scholar]
- von Heijne G. (1998). Life and Death of a Signal Peptide. Nature 396 (111), 111–113. 10.1038/24036 [DOI] [PubMed] [Google Scholar]
- von Heijne G. (1986). A New Method for Predicting Signal Sequence Cleavage Sites. Nucl. Acids Res. 14, 4683–4690. 10.1093/nar/14.11.4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G., Abrahmsèn L. (1989). Species-specific Variation in Signal Peptide Design Implications for Protein Secretion in Foreign Hosts. FEBS Lett. 244, 439–446. 10.1016/0014-5793(89)80579-4 [DOI] [PubMed] [Google Scholar]
- von Heijne G. (2006). Membrane-protein Topology. Nat. Rev. Mol. Cell Biol. 7, 909–918. 10.1038/nrm2063 [DOI] [PubMed] [Google Scholar]
- von Heijne G. (1999). Recent Advances in the Understanding of Membrane Protein Assembly and Structure. Quart. Rev. Biophys. 32, 285–307. 10.1017/s0033583500003541 [DOI] [PubMed] [Google Scholar]
- von Heijne G. (1986). The Distribution of Positively Charged Residues in Bacterial Inner Membrane Proteins Correlates with the Trans-membrane Topology. EMBO J. 5, 3021–3027. 10.1002/j.1460-2075.1986.tb04601.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. (1989). The Structure of Signal Peptides from Bacterial Lipoproteins. Protein Eng. Des. Sel. 2, 531–534. 10.1093/protein/2.7.531 [DOI] [PubMed] [Google Scholar]
- Voorhees R. M., Hegde R. S. (2016). Structure of the Sec61 Channel Opened by a Signal Sequence. Science 351, 88–91. 10.1126/science.aad4992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S., Pop O., Haan G.-J., Baars L., Koningstein G., Klepsch M. M., et al. (2008). Biogenesis of MalF and the MalFGK2 Maltose Transport Complex in Escherichia coli Requires YidC. J. Biol. Chem. 283, 17881–17890. 10.1074/jbc.m801481200 [DOI] [PubMed] [Google Scholar]
- Wang P., Shim E., Cravatt B., Jacobsen R., Schoeniger J., Kim A. C., et al. (2008). Escherichia coli Signal Peptide Peptidase A Is a Serine-Lysine Protease with a Lysine Recruited to the Nonconserved Amino-Terminal Domain in the S49 Protease Family. Biochemistry 47, 6361–6369. 10.1021/bi800657p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Jomaa A., Jaskolowski M., Yang C.-I., Ban N., Shan S.-o. (2019). The Molecular Mechanism of Cotranslational Membrane Protein Recognition and Targeting by SecA. Nat. Struct. Mol. Biol. 26, 919–929. 10.1038/s41594-019-0297-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Yang C.-I., Shan S.-o. (2017). SecA Mediates Cotranslational Targeting and Translocation of an Inner Membrane Protein. J. Cell Biol. 216, 3639–3653. 10.1083/jcb.201704036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts C., Wickner W., Zimmermann R. (1983). M13 Procoat and a Pre-immunoglobulin Share Processing Specificity but Use Different Membrane Receptor Mechanisms. Proc. Natl. Acad. Sci. U.S.A. 80, 2809–2813. 10.1073/pnas.80.10.2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihofen A., Binns K., Lemberg M. K., Ashman K., Martoglio B. (2002). Identification of Signal Peptide Peptidase, a Presenilin-type Aspartic Protease. Science 296, 2215–2218. 10.1126/science.1070925 [DOI] [PubMed] [Google Scholar]
- Weiner J. H., Bilous P. T., Shaw G. M., Lubitz S. P., Frost L., Thomas G. H., et al. (1998). A Novel and Ubiquitous System for Membrane Targeting and Secretion of Cofactor-Containing Proteins. Cell 93, 93–101. 10.1016/s0092-8674(00)81149-6 [DOI] [PubMed] [Google Scholar]
- Welte T., Kudva R., Kuhn P., Sturm L., Braig D., Müller M., et al. (2012). Promiscuous Targeting of Polytopic Membrane Proteins to SecYEG or YidC by the Escherichia Coli Signal Recognition Particle. MBoC 23, 464–479. 10.1091/mbc.e11-07-0590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse S., Gold V. A. M., Robson A., Allen W. J., Sessions R. B., Collinson I. (2012). Mobility of the SecA 2-Helix-Finger Is Not Essential for Polypeptide Translocation via the SecYEG Complex. J. Cell Biol. 199, 919–929. 10.1083/jcb.201205191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W., Moore K., Dibb N., Geissert D., Rice M. (1987). Inhibition of Purified Escherichia coli Leader Peptidase by the Leader (Signal) Peptide of Bacteriophage M13 Procoat. J. Bacteriol. 169, 3821–3822. 10.1128/jb.169.8.3821-3822.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe P. B., Silver P., Wickner W. (1982). The Isolation of Homogeneous Leader Peptidase from a Strain of Escherichia coli Which Overproduces the Enzyme. J. Biol. Chem. 257, 7898–7902. 10.1016/s0021-9258(18)34466-1 [DOI] [PubMed] [Google Scholar]
- Wolfe P. B., Zwizinski C., Wickner W. (1983). (3] Purification and Characterization of Leader Peptidase from Escherichia coli . Methods Enzymol. 97, 40–46. 10.1016/0076-6879(83)97116-1 [DOI] [PubMed] [Google Scholar]
- Wu X., Rapoport T. A. (2021). Translocation of Proteins through a Distorted Lipid Bilayer. Trends Cell Biol. 31, 473–484. 10.1016/j.tcb.2021.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Gerth K., Müller R., Wall D. (2012). Myxobacterium-produced Antibiotic TA (Myxovirescin) Inhibits Type II Signal Peptidase. Antimicrob. Agents Chemother. 56, 2014–2021. 10.1128/aac.06148-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Knafels J. D., Yoshino K. (2000). Crystal Structure of the Bacterial Protein Export Chaperone secB. Nat. Struct. Biol. 7, 1172–1177. 10.1038/82040 [DOI] [PubMed] [Google Scholar]
- Yamagata H., Daishima K., Mizushima S. (1983). Cloning and Expression of a Gene Coding for the Prolipoprotein Signal Peptidase of Escherichia coli . FEBS Lett. 158, 301–304. 10.1016/0014-5793(83)80600-0 [DOI] [PubMed] [Google Scholar]
- Yi L., Celebi N., Chen M., Dalbey R. E. (2004). Sec/SRP Requirements and Energetics of Membrane Insertion of Subunits a, B, and C of the Escherichia coli F1F0 ATP Synthase. J. Biol. Chem. 279, 39260–39267. 10.1074/jbc.m405490200 [DOI] [PubMed] [Google Scholar]
- Yi L., Jiang F., Chen M., Cain B., Bolhuis A., Dalbey R. E. (2003). YidC Is Strictly Required for Membrane Insertion of Subunits a and C of the F1F0ATP Synthase and SecE of the SecYEG Translocase. Biochemistry 42, 10537–10544. 10.1021/bi034309h [DOI] [PubMed] [Google Scholar]
- Yu Z., Koningstein G., Pop A., Luirink J. (2008). The Conserved Third Transmembrane Segment of YidC Contacts Nascent Escherichia coli Inner Membrane Proteins. J. Biol. Chem. 283, 34635–34642. 10.1074/jbc.m804344200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Miller T. F., 3rd. (2010). Hydrophobically Stabilized Open State for the Lateral Gate of the Sec Translocon. Proc. Natl. Acad. Sci. U.S.A. 107, 5399–5404. 10.1073/pnas.0914752107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Liu L., Maltsev S., Lorigan G. A., Dabney-Smith C. (2014). Investigating the Interaction between Peptides of the Amphipathic Helix of Hcf106 and the Phospholipid Bilayer by Solid-State NMR Spectroscopy. Biochimica Biophysica Acta (BBA) - Biomembr. 1838, 413–418. 10.1016/j.bbamem.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Xu Z. (2003). Structural Determinants of SecB Recognition by SecA in Bacterial Protein Translocation. Nat. Struct. Mol. Biol. 10, 942–947. 10.1038/nsb980 [DOI] [PubMed] [Google Scholar]
- Zhu L., Kaback H. R., Dalbey R. E. (2013). YidC Protein, a Molecular Chaperone for LacY Protein Folding via the SecYEG Protein Machinery. J. Biol. Chem. 288, 28180–28194. 10.1074/jbc.m113.491613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Klenner C., Kuhn A., Dalbey R. E. (2012). Both YidC and SecYEG Are Required for Translocation of the Periplasmic Loops 1 and 2 of the Multispanning Membrane Protein TatC. J. Mol. Biol. 424, 354–367. 10.1016/j.jmb.2012.09.026 [DOI] [PubMed] [Google Scholar]
- Zimmer J., Li W., Rapoport T. A. (2006). A Novel Dimer Interface and Conformational Changes Revealed by an X-Ray Structure of B. Subtilis SecA. J. Mol. Biol. 364, 259–265. 10.1016/j.jmb.2006.08.044 [DOI] [PubMed] [Google Scholar]
- Zimmer J., Nam Y., Rapoport T. A. (2008). Structure of a Complex of the ATPase SecA and the Protein-Translocation Channel. Nature 455, 936–943. 10.1038/nature07335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R., Wickner W. (1983). Energetics and Intermediates of the Assembly of Protein OmpA into the Outer Membrane of Escherichia coli . J. Biol. Chem. 258, 3920–3925. 10.1016/s0021-9258(18)32755-8 [DOI] [PubMed] [Google Scholar]