Abstract
A 5.9-kb region of the Bacillus subtilis chromosome is transcribed as a single transcript that is predicted to encode seven membrane-spanning proteins. Homologues of the first gene of this operon, for which the designation mrp (multiple resistance and pH adaptation) is proposed here, have been suggested to encode an Na+/H+ antiporter or a K+/H+ antiporter. In the present studies of the B. subtilis mrp operon, both polar and nonpolar mutations in mrpA were generated. Growth of these mutants was completely inhibited by concentrations of added Na+ as low as 0.3 M at pH 7.0 and 0.03 M at pH 8.3; there was no comparable inhibition by added K+. A null mutant that was constructed by full replacement of the mrp operon was even more Na+ sensitive. A double mutant with mutations in both mrpA and the multifunctional antiporter-encoding tetA(L) gene was no more sensitive than the mrpA mutants to Na+, consistent with a major role for mrpA in Na+ resistance. Expression of mrpA from an inducible promoter, upon insertion into the amyE locus, restored significant Na+ resistance in both the polar and nonpolar mrpA mutants but did not restore resistance in the null mutant. The mrpA disruption also resulted in an impairment of cytoplasmic pH regulation upon a sudden shift in external pH from 7.5 to 8.5 in the presence of Na+ and, to some extent, K+ in the range from 10 to 25 mM. By contrast, the mrpA tetA(L) double mutant, like the tetA(L) single mutant, completely lost its capacity for both Na+- and K+-dependent cytoplasmic pH regulation upon this kind of shift at cation concentrations ranging from 10 to 100 mM; thus, tetA(L) has a more pronounced involvement than mrpA in pH regulation. Measurements of Na+ efflux from the wild-type strain, the nonpolar mrpA mutant, and the complemented mutant indicated that inducible expression of mrpA increased the rate of protonophore- and cyanide-sensitive Na+ efflux over that in the wild-type in cells preloaded with 5 mM Na+. The mrpA and null mutants showed no such efflux in that concentration range. This is consistent with MrpA encoding a secondary, proton motive force-energized Na+/H+ antiporter. Studies of a polar mutant that leads to loss of mrpFG and its complementation in trans by mrpF or mrpFG support a role for MrpF as an efflux system for Na+ and cholate. Part of the Na+ efflux capacity of the whole mrp operon products is attributable to mrpF. Neither mrpF nor mrpFG expression in trans enhanced the cholate or Na+ resistance of the null mutant. Thus, one or more other mrp gene products must be present, but not at stoichiometric levels, for stability, assembly, or function of both MrpF and MrpA expressed in trans. Also, phenotypic differences among the mrp mutants suggest that functions in addition to Na+ and cholate resistance and pH homeostasis will be found among the remaining mrp genes.
The sequence of a locus that was reported as part of the Bacillus subtilis genome project (21; GenBank accession no. Z93937 and Z93932 [some corrections to the original sequence were noted during the present study and entered into the databank]) is of interest in connection with monovalent cation resistance and cytoplasmic pH regulation in adaption to high pH. This unusual cluster of genes is 5.9 kb long and is predicted to encode seven hydrophobic proteins that are likely to be coordinately expressed as an operon. The special interest in connection with alkali and monovalent cation resistance is based on studies reported by others on diverse homologues of this locus, i.e., a homologue from alkaliphilic Bacillus strain C-125 encompassing only the first three genes (10) and full operons designated pha (pH adaptation) from Rhizobium meliloti (23) and designated mnh from Staphylococcus aureus that complemented a Na+/H+ antiporter-deficient Escherichia coli strain (12). In the alkaliphilic Bacillus strain C-125 in which this gene family was first identified (10, 16), a crossover event involving the first gene of the partially cloned operon corrected a point mutation in the chromosomal copy of a mutant that had a nonalkaliphilic, pH homeostasis-negative, and Na+/H+ antiporter-negative phenotype. By contrast, the recently reported studies of the full homologue from R. meliloti (23) arose from characterization of a transposition mutant whose disruption of the first gene in the operon, phaA, rendered it unable to invade nodule tissue, exquisitely sensitive to inhibition by K+, and deficient in diethanolamine-induced K+ efflux but normal with respect to Na+-related properties. The data presented by both sets of studies support the respective suggestions that the first gene of the alkaliphile operon encodes an Na+/H+ antiporter and that of the pha operon encodes a K+/H+ antiporter. However, neither study included complementation of mutant phenotypes in trans without a recombination event. This is particularly important with this operon because of its unusual complexity compared to other monovalent cation/H+ antiporter-encoding loci. Indeed, Hiramatsu et al. (12) demonstrated that Na+-related functions of the S. aureus mnh operon expressed in E. coli were dependent on the presence of more than just the first gene (12). Also, as noted by the other investigators (10, 23) and observed in our sequence analysis of the B. subtilis operon, there is a striking similarity between several of the genes of these operons and those encoding subunits of proton-translocating NADH dehydrogenases (26, 28) and a recently analyzed, putative proton-translocating formate hydrogenlyase system from E. coli (2). Hiramatsu et al. (12) proposed that the Na+/H+ antiporter is a novel multisubunit secondary transporter that is energized conventionally by the proton motive force. It should not be ruled out, though, that products of these new antiporter-encoding loci may form complexes that under some conditions function as a primary ion extrusion or exchange system that is energized directly, e.g., by electron transport through Mrp components. Alternatively, the complexity of the operon may reflect the presence of genes that encode diverse transporters whose functions all relate to a particular stress. Such an operon might also include specific regulators, sensors, assembly factors, or chaperones that are stable under that stress condition and allow the function of the transporters. This would result in an interdependence among the individual gene products of the type suggested for mnh (12).
In the present studies on the B. subtilis operon, we have examined the properties of a mutant with a null mutation in the operon. We have also focused on the first gene and the final two genes of the operon, generating mutants that could be complemented in trans. Since roles in pH homeostasis and Na+ and cholate resistance were found, the name mrp is proposed for the operon, for “multiple resistance and pH adaptation locus.”
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The wild-type and mutant strains of B. subtilis used in this study are listed in Table 1. The mrp mutants included a null strain (VKN1), mutants with polar and nonpolar mutations in mrpA (VK6 and VK1), a mutant with a polar mutation in mrpF (VK15), and a double mutant with mutations in mrpA and tetA(L) (VK123); these mutants were all made as described below. B. subtilis JC112 is a wild-type strain in which the chromosomal tetA(L) locus was replaced with a chloramphenicol resistance cassette (5); this strain was included in some of the growth studies and in the pH homeostasis studies. Routine growth of all these strains was carried out at 30°C, with shaking, in TKM medium (Tris-potassium malate, no added Na+). For growth experiments, pH shift experiments, and solute efflux experiments, either TTM medium (Tris-Tris malate, 1 mM potassium phosphate, no added Na+) or TKM medium, described previously (5), served as the base to which the indicated additions were made for specific protocols; yeast extract was added to these media at 0.1% (wt/vol). In completely synthetic media (including Spizizen salts-based media [24]) that supported rapid growth of the wild type and substantial growth of JC112, VK1, and VK15, VK6 did not exhibit significant growth. The inability of VK6 to grow in such media was not overcome by the use of glucose instead of malate as the carbon source or by elevation of the phosphate concentration (data not shown). For experiments including constructs into which mrp genes were introduced into the amyE locus under control of the pspac promoter, 200 μM isopropyl-β-d-thiogalactoside (IPTG) was included in the growth media as well as in the dilution buffers for efflux experiments. If the IPTG was omitted, there was only marginally significant complementation by the constructs that complemented substantially or completely when IPTG was added.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli | ||
| DH5αMCR | F−mcrAΔ1 (mrr-hsd RMS-mcrBC) φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 supE44 λ thi-1 gyr-496 relA1 | GIBCO-BRL |
| B. subtilis | ||
| BD99 (wild type) | hisA1 thrS trpC2 | A. Garro |
| JC112 | BD99 tetA(L)::Cmr | 5 |
| VK123 | BD99 tetA(L)::CmrmrpA::Spcr | This study |
| VK1 | BD99 ΔmrpA | This study |
| VK6 | BD99 mrpA::Spcr | This study |
| VK15 | BD99 mrpF::Spcr | This study |
| VK1/mrpA | VK1 amyE::pspac-mrpA | This study |
| VK6/mrpA | VK6 amyE::pspac-mrpA | This study |
| VK6/mrpF | VK6 amyE::pspac-mrpF | This study |
| VK6/mrpFG | VK6 amyE::pspac-mrpFG | This study |
| VK15/mrpF | VK15 amyE::pspac-mrpF | This study |
| VK15/mrpFG | VK15 amyE::pspac-mrpFG | This study |
| VKN1 | BD99ΔmrpA–G::Spcr | |
| VKN1/mrpA | VKNI amyE::pspac-mrpA | |
| VKN1/mrpF | VKNI amyE::pspac-mrpF | |
| VKN1/mrpFG | VKN1 amyE::pspac-mrpFG | |
| pGEM11Zf(+) | Cloning vector (Apr) | Promega |
| pGEM9Zf(−) | Cloning vector (Apr) | Promega |
| pDH88 | Cmr vector for cloning B. subtilis chromosomal DNA and integration into the corresponding locus | 11 |
| pDR67 | amyE integration vector with Cmr gene and pspac promoter upstream of multiple cloning site | 14 |
| pDHA1 | pDH88 + ΔmrpA | This study |
| pDRA1 | pDR67 + mrpA | This study |
| pDRF1 | pDR67 + mrpF | This study |
| pDRFG1 | pDR67 + mrpFG | This study |
Complementation and resistance studies.
For the determinations of NaCl sensitivity, TKM medium at pH 7.0 or pH 8.3 was supplemented with different concentrations of NaCl. Cultures (2 ml) were grown in 15-ml conical tubes with shaking at 30°C. They were inoculated with 10 μl of an 8-h culture grown in TKM medium (pH 7.0), and the absorbance at 600 nm was read after 15 h. The MIC was defined as the minimal NaCl concentration that completely prevented growth after 15 h of incubation. For the determination of growth sensitivity to cholate, 2 ml of TKM (pH 7.0), with or without the addition of 0.08% (wt/vol) cholate, was inoculated with 50 μl of an 8-h culture grown on TKM (pH 7.0). The incubation, in 15-ml conical tubes, was conducted with shaking at 30°C. The absorbance at 600 nm was recorded after 6 h of incubation.
Construction of mutant strains.
For each type of mutant, the phenotype of the strain used in the studies was the same as several others from the construction protocol. Each mutant that was used in the subsequent studies was shown to contain the expected sequence. The sequencing was conducted at the Utah State Biotechnology Center (Logan, Utah) with an ABI-100 model 377 Sequencer.
(i) The mrp operon null strain, VKN1.
Strain VKN1 was constructed by gene splicing via overlap extension (gene SOEing), as described previously (13). Two independent PCRs were performed on wild-type DNA with the sets of primers shown in Fig. 1, BSMRPNE1 and BSMRPNR, and BSMRPNF and BSMRPNB2. BSMRPNE1 (5′-GGAATCCAGCTGCGGCTGTCAAGTAT-3′) corresponded to the complementary sequence of bp 9563 to 9581 of the database entry GenBank accession no. Z93937 and additional nucleotides containing an EcoRI site. The restriction enzyme sites are underlined. BSMRPNR (5′-TTCTCATCAAGCTTGACCCGGGCGCTTCGAACTGCTGTAATGGA-3′) corresponded to the complementary sequence of bp 7154 to 7171 of the database entry GenBank accession no. Z93932, bp 10358 to 10337 of the database entry GenBank accession no. Z93937, and additional nucleotides containing a SmaI site in the middle of the sequence. BSMRPNF (5′-TCCATTAACAGCAGTTCGAAGCGTCCCGGGTCAAGCTTGATGAGAGAA-3′) corresponded to the complementary sequence of bp 10337 to 10359 of the database entry GenBank accession no. Z93937, bp 7171 to 7154 of the database entry GenBank accession no. Z93932, and additional nucleotides containing a SmaI site in the middle of the sequence. BSMRPNB2 (5′-CGCGGATCCATCAGCAAAACGGAATCT-3′) corresponded to the complementary sequence of bp 6313 to 6330 of the database entry GenBank accession no. Z93932 and additional nucleotides containing a BglII site. The two purified PCR products were used as a template for a second PCR with primers BSMRPNE1 and BSMRPNB2. The purified PCR product of this reaction was digested with EcoRI and BglII, and then cloned into EcoRI-BamHI-digested pGEM11Zf(+) (Apr; Promega). The recombinant plasmid was digested with SmaI. A gene encoding Spcr (20) was ligated to this linear plasmid, resulting in a recombinant plasmid containing fragments upstream and downstream of the mrp operon with a Spcr gene between them instead of mrp. After isolation, the recombinant plasmid was digested with EcoRI and the linear plasmid was introduced into wild-type B. subtilis. Mutants with deletions in the mrp operon were identified by spectinomycin resistance (150 μg/ml) and confirmed by PCR and then sequencing for the strain used in the studies.
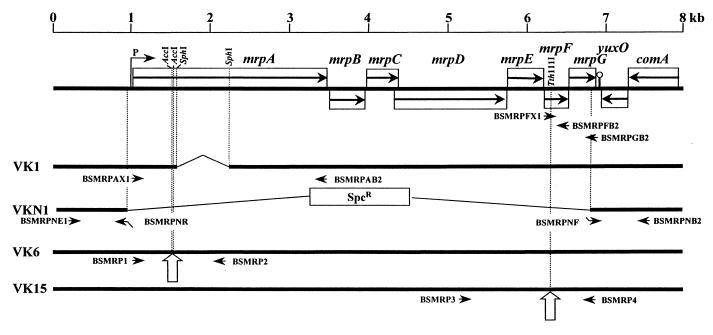
FIG. 1.
Schematic diagram of the mrp locus showing the sizes of the predicted open reading frames, the site of the nonpolar mutation made in mutant strain VK1, and the sites of the disruptions made in mutant strains VKN1, VK6, and VK15. Seven open reading frames (mrpABCDEFG) are indicated, using homologues from other bacteria, especially the R. meliloti pha region, as part of the frame of reference. The open reading frames are shown as open boxes, with the direction of transcription indicated by the horizontal arrows within the boxes. An arrow emerging upward from the fragment upstream of mrpA and pointing in the direction of transcription indicates the putative promoter (P). The SphI fragment deleted in VK1 to create a nonpolar in-frame deletion is indicated by the thin bent line flanked by dotted vertical lines. The sites disrupted by a spectinomycin resistance cassette in strains VK6 and VK15 are shown by the open arrows pointing upward. The replacement of the entire mrp locus with a spectinomycin resistance cassette is indicated for VKN1. The primers used to construct mutant strains and integration vectors in this work are shown by horizontal arrows (see Materials and Methods).
(ii) Mutants of the wild type (VK6) and JC112 (VK123) with polar disruptions in mrpA.
PCR was performed on purified wild-type B. subtilis chromosomal DNA by using the PCR primers BSMRP1 and BSMRP2. The forward primer, BSMRP1 (5′-AGGAGGTCTTATCTTTGCAGCTC-3′), corresponded to the complementary sequence of bp 10449 to 10473 of the database entry GenBank accession no. Z93937 and is at the 5′ end of the region of interest (Fig. 1). The reverse primer, BSMRP2 (5′-GGCATAATCGCCATCAGGCCGCC-3′), corresponded to the complementary sequence of bp 11603 to 11581 of the same database entry. After 25 cycles of amplification, the purified PCR product (1,155 bp) was first ligated into HincII-digested pGEM11Zf(+) (Apr; Promega). The recombinant plasmid was identified by blue-white screening in E. coli DH5α. After isolation, the plasmid was digested with AccI and a 46-bp AccI-AccI fragment was removed from the middle of mrpA. The plasmid was then treated with mung bean nuclease to generate blunt ends. A Spcr gene (20) was ligated to this linear plasmid, resulting in a recombinant plasmid containing a fragment of mrpA with a small deletion in a region into which a Spcr gene had been introduced. After isolation, the recombinant plasmid was digested with ScaI, whose site was located in the Apr gene, and the linear plasmid was introduced into wild-type B. subtilis and into the tetA(L) mutant strain JC112 by competent cell transformation (24). Mutants of each of these strains that were disrupted in the mrpA locus were identified by spectinomycin resistance (150 μg/ml) and confirmed by PCR analysis.
(iii) Mutant strain with a polar disruption of mrpF (VK15).
The strategy was the same as in the construction of the polar mrpA mutants above, except that PCR primers BSMRP3 and BSMRP4 (Fig. 1) were used. The forward primer, BSMRP3 (5′-GTACTGTACTCTGTGCTGAGGATC-3′), corresponded to the complementary sequence of bp 8437 to 8414 of the database entry GenBank accession no. Z93932. The reverse primer, BSMRP4 (5′-AGCAAGAGAGGCTGATCTGTATATCCAGA-3′), corresponded to the complementary sequence of bp 6992 to 7020. The purified PCR product was ligated into pGEM11Zf(+), and a recombinant plasmid was isolated as described above. This plasmid was digested with Tth111I, whose site was located in the middle of mrpF. The digested plasmid was then treated with mung bean nuclease to generate blunt ends, and the same Spcr gene was ligated into the disruption site. The introduction into wild-type B. subtilis and isolation and characterization of the mutants followed the procedures used for isolation of mrpA mutants.
(iv) Nonpolar mutant VK1 with a deletion in mrpA.
An in-frame deletion in mrpA of wild-type B. subtilis BD99 was made as follows. PCR was performed on wild-type DNA using the PCR primers BSMRPAX1 and BSMRPAB2 (Fig. 1). The forward primer, BSMRPAX1 (5′-CTAGTCTAGAAAGGAGGTCTTATCTTTGCAGCTC-3′), corresponded to the complementary sequence of bp 10449 to 10473 of the database entry GenBank accession no. Z93937 and additional nucleotides containing an XbaI site. The reverse primer, BSMRPAB2 (5′-GAAGATCTCATTCATTCACCGCTTTCCCCTCCT-3′), corresponded to the complementary sequence of bp 12848 to 12871 of the same database entry and additional nucleotides creating a BglII site. The purified PCR product was cloned into HincII-digested pGEM9Zf(−) (Apr; Promega). This recombinant plasmid was digested with SphI, and a 705-bp SphI-SphI fragment was removed from the middle of mrpA. The plasmid was ligated to itself, resulting in a recombinant plasmid containing a mrpA fragment with an in-frame deletion. For isolation of that fragment, the recombinant plasmid was digested with BglII and XbaI. The fragment was then cloned into BglII-XbaI-digested pDH88. The resulting plasmid, pDHA1, was integrated into the mrpA locus in the chromosome by a single crossover with chloramphenicol resistance for selection (11). To prepare strains that had lost the plasmid sequences, leaving a single mutant mrpA allele, several independent recombinants from the transformation with pDHA1 were grown under nonselective conditions (i.e., in the absence of chloramphenicol), and plated on LBK (Luria broth with KCl) plates. Colonies were screened for sensitivity to 100 mM Na+, and such strains were further tested for chloramphenicol sensitivity, which would indicate loss of the plasmid. PCR analyses were used for initial confirmation of the deletion and were followed by sequencing.
Integration of selected mrp genes into the amyE loci of particular mutant strains under a pspac promoter.
Particular mutant strains that had been prepared as described above were constructed with one or more mrp genes integrated into the chromosomal amyE locus behind an IPTG-inducible pspac promoter. Plasmid pDR67 was used for these constructions (14). This plasmid contains fragments of the front and back ends of the amyE gene flanking a chloramphenicol resistance (Cmr) gene and also contains the pspac promoter upstream of a multiple-cloning site. For construction of a plasmid that was carrying an intact mrpA gene, PCR was performed on wild-type DNA with the PCR primers BSMRPAX1 and BSMRPAB2 (Fig. 1). For construction of a plasmid that was carrying an intact mrpF gene, PCR was performed on wild-type DNA with the PCR primers BSMRPFX1 and BSMRPFB2 (Fig. 1). The forward primer BSMRPFX1 (5′-CTAGTCTAGAAAAAGCCATACAGGAGGTGAGCC-3′) corresponded to the complementary sequence of bp 7733 to 7757 of the database entry GenBank accession no. Z93932 and additional nucleotides containing an XbaI site. The reverse primer BSMRPFB2 (5′-GAAGATCTTAGCGGTTTCGATCATTTTCG-3′) corresponded to the complementary sequence of bp 7443 to 7465 of the same database entry plus additional nucleotides containing a BglII site. For construction of a plasmid that was carrying intact mrpF and mrpG genes together, PCR was performed on wild-type DNA with PCR primers BSMRPFX1 and BSMRPGB2 (Fig. 1). The reverse primer BSMRPGB2 (5′-GAAGATCTAGCAAGAGAGGCTGATCTGTATATCCAGATG-3′) corresponded to the complementary sequence of bp 6991 to 7022 of the database entry GenBank accession no. Z93932 plus additional nucleotides containing a BglII site. Each amplified fragment was cloned into XbaI-BglII-digested pDR67, yielding pDRA1 (mrpA), pDRF1 (mrpF), and pDRFG1 (mrpFG). Each plasmid DNA was linearized with NruI and used to transform particular mutants to a Cmr Amy− phenotype. The plasmids used in this study are listed together with the bacterial strains in Table 1; the ones used were all confirmed to have the correct sequence.
Northern analyses.
Northern analyses were as described previously (7). An oligonucleotide probe was used. Oligonucleotide BSMRPA (5′-TCATTCACCGCTTTTCCCCTCCT-3′) corresponded to the complementary sequence of bp 12848 to 12871 of the database entry GenBank accession no. Z93937, a region downstream of the point of disruption of the polar mrpA mutants. The oligonucleotide was radiolabeled by incubation of [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs) at 37°C for 30 min. Polynucleotide kinase was inactivated by heating to 65°C for 5 min. The radiolabeled oligonucleotide was separated from [γ-32P]ATP by Microcon YM-3 centrifugal filter devices (Amicon). Additional analyses were conducted with a probe of 157 bp corresponding to a 5′ region of mrpA (sequence of bp 10450 to 10606 of GenBank accession no. Z93937).
Determination of the cytoplasmic pH after a pH shift from 7.5 to 8.5.
The pH of the cytoplasm was determined 10 min after a sudden shift in external pH as described previously (5). Briefly, cells were grown in, and then washed and equilibrated with, TTM medium at pH 7.5. Additions of various concentrations of choline chloride, KCl, or NaCl were made to the equilibration buffer as indicated. Then the external pH was rapidly adjusted to 8.5. After 10 min, the distribution of radiolabeled methylamine was used to assay the presence or absence of a transmembrane pH gradient, acid in; measurement of the precise external pH during the assay then allowed calculation of the cytoplasmic pH after corrections for binding that were conducted as described previously (5).
Solute efflux assays.
For assays of Na+ or cholate efflux, cells were grown to the mid-logarithmic phase in TKM medium (pH 7.0), harvested, washed twice with 50 mM potassium morpholinepropanesulfonate (MOPS) (pH 7.5), and resuspended to 20 mg of protein/ml in the same buffer. The cells were starved and loaded with 5 mM 22NaCl (10 μCi/ml) or 20 μM sodium [14C]cholate (1 μCi/ml) for 2 h. After starvation and loading, the cells in Na+ efflux experiments were diluted 1:100 into 50 mM potassium–MOPS (pH 7.5) plus 5 mM NaCl. For cholate efflux experiments, 5 μl of cells was diluted into 500 μl of 50 mM potassium–MOPS (pH 7.5). The samples were vacuum filtered at various times onto Millipore HAWP 0.45-μm-pore-size filters, washed with 5 ml of buffer, dried, and counted by liquid scintillation. Where indicated, either 10 μM carbonyl cyanide p-chlorophenylhydrazone (CCCP) or 10 mM KCN was added for 15 min of preincubation to the cell suspension and was also included in the dilution buffer. Also, where indicated, 10 mM glucose was added to the dilution buffer. Protein concentrations in this and other assays were determined by the method of Lowry et al. (17) with egg white lysozyme as the standard.
RESULTS
Northern analyses of the wild type and potentially polar mutants with mutations in the mrp locus.
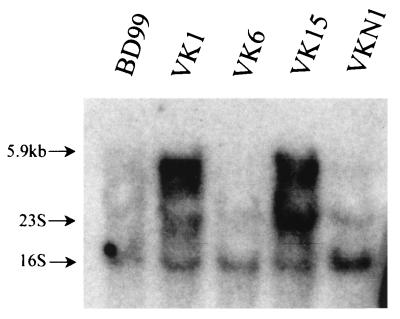
Northern analyses were conducted to confirm the expectation that the mrp genes were expressed as an operon, VKN1 was a null strain for mrp, and VK6 was a polar mrpA mutant in contrast to VK1. As shown in Fig. 2, it proved difficult to visualize Northern data from the wild type, VK6, and VKN1, as well as VK1 and VK15. Under conditions of long exposures with both the oligonucleotide probe and DNA probes to 3′ and 5′ regions of mrpA, the wild type exhibited a band of 5.9 kb, which would correspond to the expected length of a transcript of all seven mrp genes. Under such conditions, VK6 exhibited a faint band at the predicted position for that construct. VK1 and VK15 evidently showed great overexpression of a species of the same size as each other and a little smaller than that of the wild type; the degree of overexpression made its measurement difficult. In other experiments, the size of the VK1 transcript was more clearly seen as predicted and that of VK15 was consistent with a polar mrpF mutation. In no instance was a band attributable to mrp observed in the null strain VKN1 (data not shown). In the particular experiment shown in Fig. 2, the oligonucleotide probe to a region at the 3′ end of mrpA was used and conditions were chosen such that the wild type was underexposed, with the 5.9-kb band just discernible in the wild type but not in VKN1 or even the polar VK6 mutant. This condition made it possible to see the distinction between the large top band and the ribosomal bands in VK1 and VK15; these highly expressed top bands were a little less than 5.9 kb and of similar size. Ultimately it will be of interest to investigate the mechanism of mrp overexpression in nonfunctional mutants and whether it is mediated by cytoplasmic Na+ levels.
FIG. 2.
Northern blot analysis of mRNA in the wild type (BD99), mrpA mutants VK1 and VK6, mrpF mutant VK15, and null mutant VKN1. The formaldehyde-agarose isolated RNA was probed with the end-labeled 5′ oligonucleotide probe described in Materials and Methods. The 23S and 16S rRNA bands are indicated. The top arrow indicates the position corresponding to 5.9 kb.
Na+ sensitivity.
As shown in Table 2, the null mutant VKN1 was exquisitely sensitive to growth inhibition by Na+. All the other VK mutant strains, while not as sensitive as VKN1, exhibited pronounced Na+ sensitivity relative to both the wild type and the moderately Na+-sensitive tetA(L)-disrupted strain, JC112. TetA(L) is a multifunctional antiporter that catalyzes the efflux of a cobalt-tetracycline complex, Na+, or K+ in exchange for protons (3, 5, 6). The double mutant, VK123, in which a polar mrpA mutation identical to that in VK6 was introduced into JC112, exhibited the same Na+ sensitivity as VK6. The Na+ sensitivity in mrp mutants was particularly increased at pH 8.3, indicating that the Na+ extrusion function of the operon is particularly important at elevated pH. Interestingly, VK1 was more sensitive than VK6 at pH 7.0. This suggests that one of the mrp genes downstream of mrpA, whose expression is reduced in polar VK6 relative to VK1, might catalyze the uptake of Na+ at neutral pH, e.g., in symport with another solute. An inducible mrpA construct was introduced into the amyE loci of the null mutant (VKN1) as well as the polar (VK6) and nonpolar (VK1) mutants that had disruptions in their chromosomal mrpA. VKN1 exhibited no complementation, suggesting that one or more additional mrp gene products are required for mrpA function. VK1 was complemented almost to wild-type levels at pH 7.0 and also exhibited substantial complementation at pH 8.3. VK6 was also significantly complemented at both pH 7.0 and 8.3, although this complementation was lower than that observed with VK1. The significant complementation of VK6, with its reduced levels of mrpB to mrpG, by induced expression of mrpA in trans is important. It indicates that MrpA produced in stoichiometric excess with respect to the other mrp gene products is functional in Na+ resistance. As shown below, the MrpA-dependent transport Na+ activity in VK1/mrpA is elevated over that observed in the wild type, consistent with the same conclusion.
TABLE 2.
Na+ resistance of B. subtilis strains with mutations in the tetA(L) and/or selected mrp genes
| Strain | MIC (M)a of Na+ at:
|
|
|---|---|---|
| pH 7.0 | pH 8.3 | |
| Wild type | 1.30 ± 0.05 | 0.71 ± 0.03 |
| JC112 | 0.80 ± 0.03 | 0.40 ± 0.02 |
| VK123 | 0.27 ± 0.02 | 0.025 ± 0.01 |
| VK1 | 0.15 ± 0.02 | 0.026 ± 0.01 |
| VK1/mrpA | 1.15 ± 0.04 | 0.45 ± 0.02 |
| VK15 | 0.27 ± 0.03 | 0.026 ± 0.01 |
| VK15/mrpF | 0.45 ± 0.03 | 0.15 ± 0.02 |
| VK15/mrpFG | 0.65 ± 0.03 | 0.25 ± 0.02 |
| VK6 | 0.29 ± 0.03 | 0.025 ± 0.01 |
| VK6/mrpA | 0.62 ± 0.04 | 0.15 ± 0.02 |
| VK6/mrpF | 0.46 ± 0.02 | 0.16 ± 0.01 |
| VK6/mrpFG | 0.45 ± 0.03 | 0.15 ± 0.02 |
| VKN1 | 0.09 ± 0.03 | 0.024 ± 0.01 |
| VKN1/mrpA | 0.12 ± 0.02 | 0.035 ± 0.02 |
| VKN1/mrpFG | 0.14 ± 0.03 | 0.027 ± 0.02 |
| VKN1/mrpF | 0.092 ± 0.02 | 0.026 ± 0.02 |
Minimal Na+ concentration at which no growth was observed after 15 h. The values are means ± standard deviations.
An interesting set of observations on Na+ sensitivity was made for VK15 and for complementation of both VK15 and VK6 by introduction of an inducible mrpF or mrpFG construct into the amyE locus. First, VK15 was just as sensitive to Na+ as VK6 was. Induced expression of mrpF increased the Na+ resistance of VK6 at pH 7.0 and 8.3, and the increase was not augmented in the strain expressing mrpFG in trans. In VK15 itself, which has a functional chromosomal mrpA, the induced expression of mrpF in trans increased the Na+ resistance, but in this strain there was an augmentation when mrpFG was induced instead of mrpF. These findings suggest that MrpF enhances Na+ resistance both in the presence and in the absence of the putative antiporter-encoding MrpA whereas MrpG enhances only in association with a functional MrpA.
Work on an earlier Na+ extrusion system of B. subtilis, the ATP-binding cassette-type natAB transport system for Na+ extrusion, had indicated that this system was inducible by ethanol and other membrane perturbants and contributed to solvent resistance in part by excluding Na+ that might leak in adversely. Several of the mrp mutants were examined for their sensitivity to ethanol. Although not shown, there was a qualitatively but not quantitatively consistent sensitivity of the mutants relative to the wild type.
pH homeostasis phenotypes.
Earlier studies had shown that pH 7.5-equilibrated cells of JC112 were completely unable to regulate their cytoplasmic pH upon a sudden shift in the external pH to 8.5 in the presence of 100 mM K+ or Na+. These results indicated a major role for TetA(L) in pH homeostasis at that level of monovalent cation (5). Given the much greater Na+ sensitivity of all the mrp mutants than of JC112, it seemed likely that any mrp-encoded antiporter activity might establish lower cytoplasmic Na+ concentrations than could be accomplished by TetA(L) alone. Therefore, pH shift experiments were conducted at 100, 25, and 10 mM Na+ or K+ as well as in the absence of added Na+ or K+; in the latter experiments, choline chloride was substituted for the sodium and potassium salts. Although the results are not shown, none of the strains exhibited any capacity for pH homeostasis after the pH shift when the choline salts were used; i.e., after the shift, the cytoplasmic pH was the same as the new external pH. As shown in Table 3, the wild-type strain was capable of excellent pH homeostasis in the presence of only 10 mM Na+, maintaining a cytoplasmic pH close to the preshift level. Significant but more modest acidification of the cytoplasm relative to the new external pH was observed in the presence of 10 mM K+. The double mutant VK123, like the tetA(L) mutant JC112 (data not shown), exhibited neither Na+- nor K+-dependent pH homeostasis at any of the monovalent cation concentrations. By contrast, VKN1, VK1, VK6, and VK15 all showed a small deficit in K+-dependent pH homeostasis. These strains also all exhibited significant and comparable deficits in cytoplasmic pH regulation relative to the wild type in the presence of 25 mM and, especially, 10 mM Na+. Expression of mrpA in trans in VK1 but not in VKN1 restored a capacity for Na+-dependent pH homeostasis that was comparable to that of the wild type.
TABLE 3.
Cytoplasmic pH measured in cells of wild type and various mrp constructs after a shift in the external pH from 7.5 to 8.5a
| Concn of indicated salt added (mM) | Cytoplasmic pH of strain:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type
|
VK1
|
VK1/mrpA
|
VK6
|
VK15
|
VK123
|
VKNI
|
VKNI/mrpA
|
|||||||||
| NaCl | KCl | NaCl | KCl | NaCl | KCl | NaCl | KCl | NaCl | KCl | NaCl | KCl | NaCl | KCl | NaCl | KCl | |
| 100 | 7.5 ± 0.08 | 7.70 ± 0.07 | 7.6 ± 0.15 | 7.8 ± 0.17 | 7.5 ± 0.11 | 7.7 ± 0.04 | 7.60 ± 0.07 | 8.00 ± 0.09 | 7.7 ± 0.05 | 7.90 ± 0.10 | 8.5 ± 0.12 | 8.5 ± 0.13 | 7.6 ± 0.14 | 7.8 ± 0.16 | 7.5 ± 0.15 | 7.7 ± 0.16 |
| 25 | 7.50 ± 0.12 | 7.90 ± 0.11 | 8.2 ± 0.12 | 8.0 ± 0.15 | 7.5 ± 0.14 | 7.9 ± 0.11 | 7.90 ± 0.14 | 8.20 ± 0.11 | 8.00 ± 0.15 | 8.10 ± 0.15 | 8.5 ± 0.11 | 8.5 ± 0.12 | 8.3 ± 0.13 | 8.1 ± 0.14 | 8.2 ± 0.13 | 8.1 ± 0.15 |
| 10 | 7.50 ± 0.09 | 8.10 ± 0.13 | 8.4 ± 0.12 | 8.3 ± 0.15 | 7.8 ± 0.15 | 8.3 ± 0.12 | 8.50 ± 0.08 | 8.50 ± 0.10 | 8.50 ± 0.05 | 8.40 ± 0.15 | 8.5 ± 0.12 | 8.4 ± 0.15 | 8.5 ± 0.11 | 8.4 ± 0.13 | 8.5 ± 0.15 | 8.4 ± 0.14 |
All strains were grown at pH 7.0 in TTM medium before being subjected to a sudden shift in the external pH to 8.5 as described under Materials and Methods. The values shown, means ± standard deviations, were measurements taken in duplicate from at least six independent experiments.
Na+ efflux.
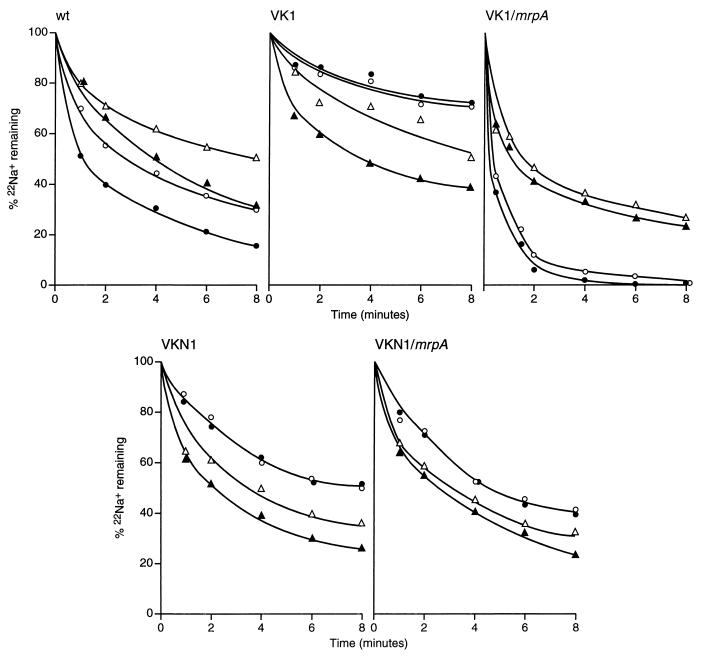
The apparent roles of MrpA in both Na+ resistance and Na+-dependent pH homeostasis were consistent with its being an Na+/H+ antiporter, as has been proposed for the homologues in alkaliphilic Bacillus strain C-125 (10) and S. aureus (12). To more directly assay such an activity, cells of the wild type, VK1, VK1/mrpA, VKN1, and VKN1/mrpA were partially energy depleted and loaded with 5 mM 22Na+. The cells were then diluted into buffers containing 5 mM nonradioactive Na+ in the presence or absence of other additions. The concentration of 5 mM Na+ was chosen because it is well below the concentration at which the TetA(L) Na+/H+ antiporter activity is optimally active (8). Indeed, as shown in Fig. 3, wild-type B. subtilis exhibited fast efflux of Na+ that was significantly stimulated by glucose addition and significantly inhibited by the addition of either cyanide or the protonophore CCCP. By contrast, Na+ efflux from VK1 cells exhibited a much slower Na+ efflux which was not stimulated by glucose but was enhanced by both cyanide and CCCP; most probably, in the only partially starved cells, the cytoplasmic Na+ concentration slightly exceeded the external Na+ concentration after loading, and the stimulation by cyanide and CCCP represents a stimulation of a leak of cytoplasmic Na+ down its concentration gradient once the electrical potential component of the proton motive force, positive out, is dissipated. In the mrpA-complemented VK1, the Na+ efflux was faster than in the wild-type cells, in both the absence and presence of added glucose. Cyanide and CCCP both inhibited efflux significantly but not completely. The Na+ efflux pattern of VKN1 was similar to that of VK1 except for a slightly faster Na+ efflux in VKN1; this, again, could be explained if the complete deletion of VKN1 removes a Na+-coupled uptake protein as well as Na+ efflux systems. Restoration of mrpA to VKN1 increased the efflux rate slightly, but that efflux rate was still stimulated rather than inhibited by both cyanide and CCCP.
FIG. 3.
Efflux of Na+ from cells of wild type (wt), VK1, VK1/mrpA, VKN1, and VKN1/mrpA. The cells were washed, energy depleted, and loaded with 5 mM 22NaCl as described in Materials and Methods. Efflux was initiated by diluting the suspension 100-fold into buffer (containing 5 mM NaCl) and no further additions (○), buffer containing 10 mM glucose (●), buffer containing glucose plus 10 mM cyanide (▵), or buffer containing glucose plus 10 μM CCCP (▴). Samples were taken at various times, filtered, and washed, and the radioactivity was determined by liquid scintillation counting.
MrpF-dependent resistance to cholate and efflux of Na+ and cholate.
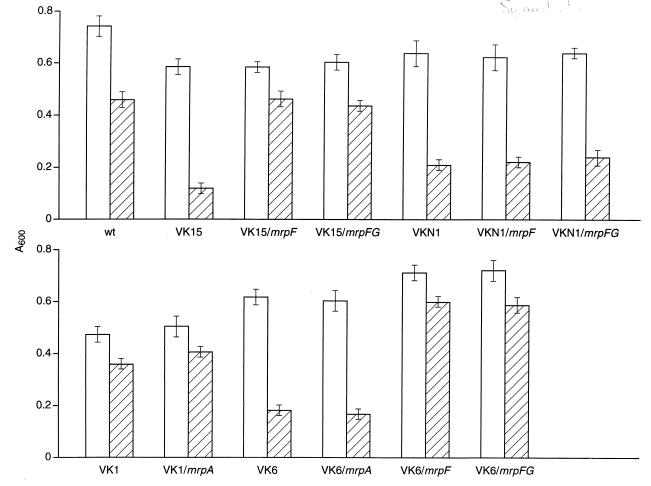
The determinations of Na+ resistance in mrpF-complemented VK6 indicated that there was an enhancement of Na+ resistance when MrpF expression was strongly induced in this MrpA− mutant at pH 7.0. Concomitant expression of MrpG did not increase the enhancement. These results suggested that MrpF might be a transporter that catalyzed Na+ efflux independently of MrpA. The greater Na+ sensitivity of VKN1 relative to VK1 or VK6 (Table 2) was similarly consistent with there being an mrp-encoded Na+ efflux system in addition to MrpA. Since sequence similarity was noted between MrpF and several eukaryotic Na+-coupled bile acid transporters (9, 15, 26, 28) in BLAST analyses (1), the possibility was raised that the Na+ and a bile salt type of compound might be cosubstrates for an efflux system which could be energized by the proton motive force. Cholate was used as the probe to assess this hypothesis. The cholate resistance of various mrp mutant strains was determined in comparison to the wild type and to each other, with and without induced expression of mrpA or mrpF in trans. The differences among the strains examined with respect to growth inhibition by cholate were not sufficient to affect the MIC in the same pronounced manner as was observed among the strains for Na+ resistance. However, at particular concentrations of cholate, e.g., 0.08% (wt/vol), significant and highly reproducible differences were observed. As shown in Fig. 4, VKN1, VK1, VK6, and VK15 all exhibited somewhat less growth than the wild-type strain at pH 7.0 in the absence of added cholate; this is likely to have resulted in part from inevitable contamination of the media with Na+. In addition, and especially in the strains strongly expressing mrpA or mrpF, overexpression of a hydrophobic protein may account for some of this small growth deficit. As expected, the nonpolar mutant VK1 showed no increase in cholate inhibition of growth relative to the wild type and the mrpA status of the strain was similarly not a significant factor in cholate sensitivity. In contrast, the cholate sensitivity of VKN1 and of the polar VK6 mutant was more similar to that of VK15, i.e., significantly greater than the sensitivity of the wild type. The same difference in sensitivity was not observed when taurocholate was used instead of cholate (results not shown). The mrpA status was again irrelevant with respect to significant effects on cholate sensitivity in VK6. Expression of mrpF or of mrpFG restored a comparable level of resistance to both VK6 and VK15 that was even greater than that of the wild type. Neither mrpF nor mrpFG, however, restored cholate resistance to the mrp-null strain VKN1. These findings were consistent with the function of MrpF as an Na+-coupled cholate efflux system whose functional expression requires at least low levels of one or more additional mrp gene products but not of mrpA.
FIG. 4.
Effect of cholate on the growth of wild-type (wt) B. subtilis and several uncomplemented and complemented strains with mutations in the mrp gene. Cells were grown in TKM medium (pH 7.0) in the presence (hatched bars) or absence (open bars) of 0.08% (wt/vol) cholate. The absorbance at 600 nm (A600) was determined after 6 h of shaking at 30°C. The results represent the mean of at least six determinations; standard deviations are shown as error bars.
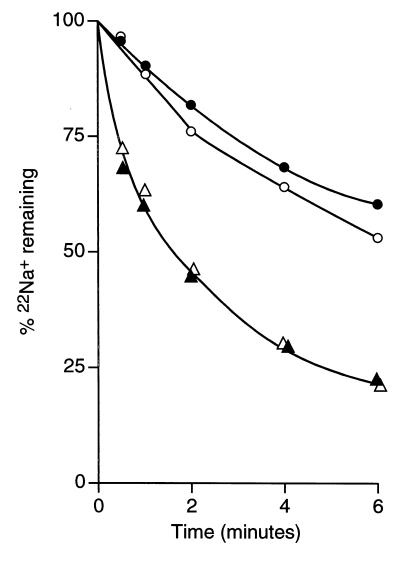
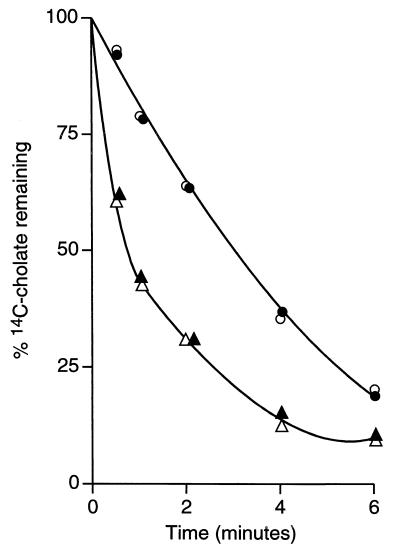
Efflux assays were undertaken to further document such an MrpF-dependent activity. MrpF-dependent efflux of 22Na+ was examined in VK6 cells with and without mrpF expression from the amyE locus. VK6 was used to eliminate the contribution of MrpA to the Na+ efflux, even though a modest level of residual MrpF function may exist in VK6. The efflux was measured in cells that were partially energy depleted and loaded with 22Na+ in either the presence or absence of 0.08% (wt/vol) cholate. As shown in Fig. 5, VK6/mrpF exhibited significantly faster Na+ efflux than VK6 did. No stimulation of Na+ efflux was observed in the presence of cholate. Efflux of cholate was monitored in cells of VK15, with and without mrpF expression from the amyE locus. The cells were preloaded with 20 μM [14C]cholate in the presence or absence of 5 mM Na+. As shown in Fig. 6, cholate efflux was significantly faster in VK15/mrpF than in VK15. No effect of Na+ addition was observed.
FIG. 5.
Efflux of Na+ from cells of VK6 and VK6/mrpF in the presence or absence of cholate. The cells were starved and loaded with 22NaCl, as described in Materials and Methods, in the absence (open symbols) or presence (solid symbols) of 0.08% (wt/vol) cholate. Efflux of Na+ from VK6 (○, ●) and VK6/mrpF (▵, ▴) was assayed in the presence of glucose, as in Fig. 3.
FIG. 6.
Efflux of cholate from cells of VK15 and VK15/mrpF. The cells were energy depleted and loaded with 20 μM [14C]cholate as described in Materials and Methods. Half of the cells were also loaded with 5 mM nonradioactive NaCl (solid symbols), and half had no further additions (open symbols). Cholate efflux was measured by sampling, as described in the legend to Fig. 3, from the assay mixtures with VK15 (○, ●) and VK15/mrpF (▵, ▴).
DISCUSSION
The name mrp is proposed for the group of genes whose analysis was initiated in this study because of the multiple resistance and pH homeostasis-related functions of this locus. MrpA is a strong candidate for a secondary Na+/H+ antiporter which probably also has some K+/H+ antiport capacity. A mutant with a nonpolar mutation in mrpA (VK1) is highly sensitive to Na+ and exhibits a defect in Na+-dependent pH homeostasis at moderate concentrations of cation. This mutant exhibits no proton motive force-dependent Na+ efflux from cells preloaded with 5 mM Na+ and diluted into energization buffer containing the same Na+ concentration. Induction of mrpA in the amyE locus from the pspac promoter restores close to wild-type levels of Na+ resistance, Na+-dependent pH homeostasis, and even faster protonophore-sensitive Na+ efflux than the wild type. Moreover, since mrpA expression in trans in the polar VK6 mutant also complemented significantly, MrpA is apparently catalytically competent in stoichiometric excess over the other mrp gene products. Together, these results are consistent with MrpA being an Na+/H+ antiporter that can be energized by the proton motive force and that can function independently of a fixed complex with other mrp gene products. This would make MrpA function similar to other prokaryotic Na+/H+ antiporters, three of which have rigorously been shown to catalyze antiport when purified and reconstituted alone in proteoliposomes (6, 22, 25). In studies of the S. aureus homologue that were conducted entirely in E. coli, Hiramatsu et al. (12) noted the requirement of multiple genes for antiport function and proposed that the antiporter functions as a multisubunit entity. The current finding that MrpA raises antiporter activity upon overexpression in the wild type and, most impressively, in the polar VK6 mutant with low levels of other mrp genes is difficult to reconcile with a strict dependency on a particular stoichiometric complex. The present studies do not rule out the possibility, however, that under particular conditions MrpA can also function within a specific stoichiometric relationship to other mrp gene products in a membrane complex. Moreover, it is conceivable that under these circumstances there is a primary mode of energization via electron transport through the complex. A more complete mutational and biochemical analysis of this complicated locus is needed to fully resolve this important issue.
It is likely that at least one other mrp gene product is needed as a chaperone or assembly factor for catalytic mrp gene products, e.g., MrpA and MrpF. This would account for the lack of complementation of VKN1. Expression of mrpA from an IPTG-inducible promoter would have obviated the need for any otherwise necessary transcriptional regulator. This level of overexpression might also have allowed the assembly of significant amounts of active MrpA even at lower levels of a chaperone or assembly factor (i.e., as in VK6) than are essential for significant activity when mrpA expression occurs at normal low levels. In VK15, which has an intact mrpA gene but no mrpFG function, the Na+ resistance was enhanced more by mrpFG expression than by mrpF expression. No difference was observed when the mrpA gene was absent. This suggests that MrpG enhances MrpA stability, assembly, or function when mrpA is expressed in its normal chromosomal setting. We hypothesize that MrpG may play chaperone or assembly roles for MrpA and perhaps for other mrp-encoded structural gene products.
MrpF also appears catalytically competent when expressed from an IPTG-inducible promoter in the VK15 mutant as well as in VK6, which has no MrpA and greatly reduced levels of all other mrp gene products. MrpF-dependent Na+ efflux and cholate efflux were evident, but coupling of the two fluxes was not demonstrated. It may be difficult to demonstrate cholate-dependent Na+ efflux, because additional Na+ efflux systems are present in the biological membranes. Because of the low concentration of cholate used, it is similarly unlikely that Na+-dependent cholate efflux could be demonstrated without developing a much more purified system. It is of interest that a soil bacterium such as B. subtilis has a cholate efflux system and, as assessed from the genome project annotations, may have more than this one (YocS; GenBank accession no. Z99114). Whether this relates to the need to extrude some endogenous substrate that has a related structure or to the presence of cholate-like animal or plant products in the natural environment is unknown. Other bacterial extrusion systems for cholate have been reported (18, 19), and one of them is induced by Na+, although the cation has not been shown to be a substrate (18).
Both MrpA and MrpF may contribute to the Na+ resistance role. If MrpF-dependent Na+-cholate efflux is coupled to H+ uptake, they may both contribute to pH homeostasis as well. Clarification of the relative contributions, under different conditions, will require nonpolar mutations in each gene. Elucidation of the roles of the other mrp gene products, including the hypothesized chaperone role for MrpG, will similarly require additional, single nonpolar mutations in those other genes.
Thus far, all the B. subtilis mrp gene functions are linked by a relationship to Na+, and by far the dominant phenotype associated with all the mutations in this locus is Na+ sensitivity. After clarification of the full panoply of functions of this locus, it will be of interest to examine the differences between the B. subtilis operon and those of other organisms in which the dominant physiological role is different or predominantly related to a different cation, e.g., Na+-specific pH homeostasis in Bacillus strain C-125 (10) and K+ sensitivity in R. meliloti (23). The mrp locus is the third locus of B. subtilis that has been shown to play a role in Na+ resistance. tetA(L) plays a dominant role in Na+ (K+)-dependent pH homeostasis in B. subtilis (5) and also plays a significant role in Na+ exclusion in a range of Na+ above 25 mM, but the present studies indicate that Mrp function is a crucial adjunct to TetA(L) for the purposes of Na+ resistance. Another Na+ extrusion system, the ATP-binding cassette-type natAB of B. subtilis, is probably more specialized, becoming important primarily under circumstances when the membrane integrity and/or the proton motive force are reduced (4). Preliminary experiments indicate, though, that mrp also plays a role under such circumstances.
ACKNOWLEDGMENTS
This work was supported by research grants GM28454 and GM52837 from the National Institute of General Medical Sciences (to T.A.K.) and a grant-in-aid for scientific research from the Ministry of Education, Science and Culture of Japan (to M.I.).
ADDENDUM IN PROOF
While this article was under review, Kosono et al. (S. Kosono, S. Morotomi, M. Kitada, and T. Kudo, Biochim. Biophys. Acta 1409:171–175, 1999) reported the properties of a mrpA mutant that was probably equivalent to the polar mrpA mutant (VK6) described in this study; these investigators found sensitivity to Na+ similar to that observed in VK6.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E F, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Andrews S C, Berks B C, McClay J, Ambler A, Quail M A, Golby P, Guest J R. A 12-cistron Escherichia coli operon (hyf) encoding a putative proton-translocating formate hydrogenlyase system. Microbiology. 1997;143:3633–3647. doi: 10.1099/00221287-143-11-3633. [DOI] [PubMed] [Google Scholar]
- 3.Cheng J, Guffanti A A, Krulwich T A. The chromosomal tetracycline-resistance locus of Bacillus subtilis encodes a Na+/H+ antiporter that is physiologically important at elevated growth pH. J Biol Chem. 1994;269:27365–27371. [PubMed] [Google Scholar]
- 4.Cheng J, Guffanti A A, Krulwich T A. A two gene ABC-type transport system involved in Na+ extrusion by Bacillus subtilis is induced by ethanol and protonophore. Mol Microbiol. 1997;23:1107–1120. doi: 10.1046/j.1365-2958.1997.2951656.x. [DOI] [PubMed] [Google Scholar]
- 5.Cheng J, Guffanti A A, Wang W, Krulwich T A, Bechhofer D H. Chromosomal tetA(L) gene of Bacillus subtilis: regulation of expression and physiology of a tetA(L) deletion strain. J Bacteriol. 1996;178:2853–2860. doi: 10.1128/jb.178.10.2853-2860.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng J, Hicks D B, Krulwich T A. The purified Bacillus subtilis tetracycline efflux protein TetA(L) reconstitutes both tetracycline-cobalt/H+ and Na+(K+)/H+ exchange. Proc Natl Acad Sci USA. 1996;93:14446–14451. doi: 10.1073/pnas.93.25.14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimari J F, Bechhofer D. Initiation of mRNA decay in Bacillus subtilis. Mol Microbiol. 1993;7:705–717. doi: 10.1111/j.1365-2958.1993.tb01161.x. [DOI] [PubMed] [Google Scholar]
- 8.Guffanti A A, Krulwich T A. Tetracycline/H+ antiport and Na+/H+ antiport catalyzed by the Bacillus subtilis TetA(L) transporter expressed in Escherichia coli. J Bacteriol. 1995;177:4557–4561. doi: 10.1128/jb.177.15.4557-4561.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagenbuch B, Stieger B, Foguet M, Lubbert H, Meier P J. Functional expression cloning and characterization of the hepatocyte Na+/bile acid cotransport system. Proc Natl Acad Sci USA. 1991;88:10629–10633. doi: 10.1073/pnas.88.23.10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamamoto T, Hashimoto M, Hino M, Kitada M, Seto Y, Kudo T, Horikoshi K. Characterization of a gene responsible for the Na+/H+ antiporter system of alkalophilic Bacillus species strain C-125. Mol Microbiol. 1994;14:939–946. doi: 10.1111/j.1365-2958.1994.tb01329.x. [DOI] [PubMed] [Google Scholar]
- 11.Henner D J. Inducible expression of regulatory genes in Bacillus subtilis. Methods Enzymol. 1990;185:223–228. doi: 10.1016/0076-6879(90)85022-g. [DOI] [PubMed] [Google Scholar]
- 12.Hiramatsu T, Kodama K, Kuroda T, Mizushima T, Tsuchiya T. A putative multisubunit Na+/H+ antiporter from Staphylococcus aureus. J Bacteriol. 1998;180:6642–6648. doi: 10.1128/jb.180.24.6642-6648.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horton R M. In vitro recombination and mutagenesis of DNA. Methods Mol Biol. 1996;67:141–149. doi: 10.1385/0-89603-483-6:141. [DOI] [PubMed] [Google Scholar]
- 14.Ireton D D, Rudner Z, Siranosian K J, Grossman A D. Integration of multiple developmental signals in Bacillus subtilis through Spo0A transcription factor. Genes Dev. 1993;7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- 15.Jacquemin E, Hagenbuch B, Stieger B, Wolkoff A W, Meier P J. Expression cloning of a rat liver Na+-independent organic anion transporter. Proc Natl Acad Sci USA. 1994;91:133–137. doi: 10.1073/pnas.91.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kudo T, Hino M, Kitada M, Horikoshi K. DNA sequences required for the alkalophily of Bacillus sp. strain C-125 are located close together on its chromosomal DNA. J Bacteriol. 1990;172:7282–7283. doi: 10.1128/jb.172.12.7282-7283.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 18.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 19.Mallonee D H, Hylemon P B. Sequencing and expression of a gene encoding a bile acid transporter from Eubacterium sp. strain VPI 12708. J Bacteriol. 1996;178:7053–7058. doi: 10.1128/jb.178.24.7053-7058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy E, Huwyler L, Bastos M D D. Transposon Tn554: complete nucleotide sequence and isolation of transposition defective and antibiotic-sensitive mutants. EMBO J. 1985;4:3357–3365. doi: 10.1002/j.1460-2075.1985.tb04089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oudega B, Koningstein G, Rodriguez L, deSalas Ramon M, Hilbert H, Disterhoft A, Pohl T M, Weitzenegger T. Analysis of the Bacillus subtilis genome: cloning and nucleotide sequence of a 62 kb region between 275− (rrnB) and 284− (pai) Microbiology. 1997;143:2769–2774. doi: 10.1099/00221287-143-8-2769. [DOI] [PubMed] [Google Scholar]
- 22.Pinner E, Padan E, Schuldiner S. Kinetic properties of NhaB, a Na+/H+ antiporter from Escherichia coli. J Biol Chem. 1994;269:26274–26279. [PubMed] [Google Scholar]
- 23.Putnoky P, Kerezt A, Nakamura T, Endre G, Grosskopf E, Kiss P, Kondorosi A. The pha cluster of Rhizobium meliloti involved in pH adaptation and symbiosis encodes a novel type of K+ efflux system. Mol Microbiol. 1998;28:1091–1101. doi: 10.1046/j.1365-2958.1998.00868.x. [DOI] [PubMed] [Google Scholar]
- 24.Spizizen J. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc Natl Acad Sci USA. 1958;44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taglicht D, Padan E, Schuldiner S. Overproduction and purification of a functional Na+/H+ antiporter coded by nhaA(ant) from Escherichia coli. J Biol Chem. 1991;266:11289–11294. [PubMed] [Google Scholar]
- 26.Thanassi D G, Cheng L W, Nikaido H. Active efflux of bile salts by Escherichia coli. J Bacteriol. 1997;179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weidner U, Geier S, Ptock A, Friedrich T, Leif H, Weiss H. The gene locus of the proton-translocating NADH:ubiquinone oxidoreductase in Escherichia coli. J Mol Biol. 1993;233:109–122. doi: 10.1006/jmbi.1993.1488. [DOI] [PubMed] [Google Scholar]
- 28.Wong M H, Oelkers P, Craddock A L, Dawson P A. Expression cloning and characterization of the hamster ileal sodium-dependent bile acid transporter. J Biol Chem. 1994;269:1340–1347. [PubMed] [Google Scholar]
- 29.Yagi T. The bacterial energy-transducing NADH-quinone oxidoreductases. Biochim Biophys Acta. 1993;1141:1–17. doi: 10.1016/0005-2728(93)90182-f. [DOI] [PubMed] [Google Scholar]