Abstract
The yeast-to-hypha morphological transition (dimorphism) is typical of many pathogenic fungi. Dimorphism has been attributed to changes in temperature and nutritional status and is believed to constitute a mechanism of response to adverse conditions. We have isolated and characterized a gene, MHY1, whose transcription is dramatically increased during the yeast-to-hypha transition in Yarrowia lipolytica. Deletion of MHY1 is viable and has no effect on mating, but it does result in a complete inability of cells to undergo mycelial growth. MHY1 encodes a C2H2-type zinc finger protein, Mhy1p, which can bind putative cis-acting DNA stress response elements, suggesting that Mhy1p may act as a transcription factor. Interestingly, Mhy1p tagged with a hemagglutinin epitope was concentrated in the nuclei of actively growing cells found at the hyphal tip.
Several fungal species can undergo a yeast-to-hypha transition (dimorphism). Fungal dimorphism has received increasing attention because of its implication in pathogenesis and its potential as a simple experimental model of eukaryotic cell differentiation (2, 25, 32, 35). To date, only a few species have been systematically investigated with the aim of understanding the molecular aspects of dimorphism. Studies have concentrated on Candida albicans, the most common human fungal pathogen, and Saccharomyces cerevisiae, the most extensively studied fungus at the genetic, biochemical, and physiological levels.
Despite the development of specific molecular and genetic tools in the last few years (5, 6, 10, 16, 18, 33, 36), a better understanding of the yeast-to-hypha transition in C. albicans has largely been hampered by the diploid nature of this microorganism and its lack of a known sexual cycle (27, 41). S. cerevisiae has been used as a model of dimorphic transition, and results with this yeast have been extrapolated to other microorganisms, particularly C. albicans. Although significant advances in our understanding of dimorphic transition have been made by using S. cerevisiae, the inability of this yeast to form true hyphae, and the suggestion that true hyphal formation and pseudohyphal growth may occur by means of separate pathways (20), limit this approach. In order to overcome some of these difficulties and shortcomings, other fungal species have been proposed as alternative systems for the study of dimorphism. We have chosen to study dimorphic transition in Yarrowia lipolytica because it can reproduce sexually (42), it is amenable to genetic (26) and molecular biological (8, 24) analyses, and its response to the induction of mycelial growth is highly reproducible (14, 30).
Here we report the isolation of the gene MHY1 encoding a protein, Mhy1p, involved in the yeast-to-hypha transition in Y. lipolytica. Mhy1p shows strong homology in its zinc finger domain to the S. cerevisiae stress response factors Msn2p and Msn4p. Like these factors, Mhy1p is able to bind to putative stress response elements (STREs), cis-acting DNA sequences identified upstream of a number of genes conferring tolerance to a variety of stresses (cross-protection) (15, 21, 39), suggesting that Mhy1p may act as a transcription factor.
MATERIALS AND METHODS
Yeast strains and microbial techniques.
The Y. lipolytica strains used in this study are listed in Table 1. The mhy1-1 mutant strain was isolated after chemical mutagenesis of Y. lipolytica E122 cells with 1-methyl-3-nitro-1-nitrosoguanidine, as previously described (24). The medium components were as follows: YEPD, 1% yeast extract, 2% peptone, and 2% glucose; YNA, 0.67% yeast nitrogen base without amino acids and 2% sodium acetate; YNBGlc, 1.34% yeast nitrogen base without amino acids and 1% glucose; and YNBGlcNAc, 1.34% yeast nitrogen base without amino acids, 1% N-acetylglucosamine, and 50 mM citric acid (pH 6.0). YNA was supplemented with uracil, leucine, lysine, and histidine, each at 50 μg/ml, as required. YNBGlc and YNBGlcNAc were supplemented with 2× complete supplement mixture (Bio101, Vista, Calif.) or 2× complete supplement mixture minus leucine, as required. Media, growth conditions, and procedures for mating, sporulation, and transformation of Y. lipolytica have been described elsewhere (4, 24).
TABLE 1.
Y. lipolytica strains used in this study
| Straina | Genotype |
|---|---|
| E122 | MATA ura3-302 leu2-270 lys8-11 |
| 22301-3 | MATB ura3-302 leu2-270 his1 |
| mhy1-1 | MATA ura3-302 leu2-270 lys8-11 mhy1-1 |
| mhy1K09 | MATA ura3-302 leu2-270 lys8-11 mhy1∷URA3 |
| mhy1K09-B4 | MATB ura3-302 leu2-270 his1 mhy1∷URA3 |
| E122//22301-3 | MATA/MATB ura3-302/ura3-302 leu2-270/leu2-270 lys8-11/+ his1/+ |
| E122//mhy1K09-B4 | MATA/MATB ura3-302/ura3-302 leu2-270/leu2-270 lys8-11/+ his1/+ mhy1∷URA3/+ |
| 22301-3//mhy1K09 | MATA/MATB ura3-302/ura3-302 leu2-270/leu2-270 lys8-11/+ his1/+ mhy1∷URA3/+ |
| mhy1K09//mhy1K09-B4 | MATA/MATB ura3-302/ura3-302 leu2-270/leu2-270 lys8-11/+ his1/+ mhy1∷URA3/mhy1∷URA3 |
Strains E122 and 22301-3 were from C. Gaillardin, Thiverval-Grignon, France. All other strains were from this study.
DNA manipulation and growth of Escherichia coli were performed as previously described (1).
Mycelial induction.
Mycelial growth was induced as described previously (14). Cells were grown in YNBGlc for 12 h, harvested by centrifugation at room temperature, washed with sterile distilled water, kept at 4°C for 15 min in YNB medium without a carbon source, and inoculated at a final density of 107/ml in YNBGlcNAc (for induction of the yeast-to-hypha transition) or YNBGlc medium (for growth as the yeast form).
Cloning, characterization, and disruption of the MHY1 gene.
The MHY1 gene was isolated from a Y. lipolytica genomic DNA library contained in the replicative E. coli shuttle vector pINA445 (24) by functional complementation of the mhy1-1 mutation. Plasmids were introduced into yeast cells by electroporation, and Leu+ transformants were screened on YNA agar plates for their ability to give rise to rough colonies.
Complementing plasmids were recovered by transformation of E. coli, and the smallest fragment capable of restoring hyphal growth was determined. Restriction fragments prepared from the genomic insert of one of these constructs (pMHY1) were subcloned into the vectors pGEM-7Zf(+) (Promega, Madison, Wis.) or pBluescript II SK(+) (Stratagene, La Jolla, Calif.) for dideoxynucleotide sequencing of both strands. The deduced polypeptide sequence, Mhy1p, was compared to other known protein sequences by using the BLAST Network Service of the National Center for Biotechnology Information (Bethesda, Md.).
To disrupt the MHY1 gene, a 0.8-kbp NcoI-NdeI fragment of pMHY1 containing the entire MHY1 gene was replaced by a 1.6-kbp NcoI-NdeI fragment containing the Y. lipolytica URA3 gene. This construct was digested with HpaI to liberate a 3.3-kbp fragment containing the entire URA3 gene flanked by 1.3 and 0.4 kbp of the 5′ and 3′ regions of the MHY1 gene, respectively. This linear fragment was used to transform the wild-type Y. lipolytica strain E122 to uracil prototrophy. Of 246 Ura+ transformants obtained, 2 showed a fully smooth phenotype after 3 days on YEPD agar plates. One of these transformants, mhy1K09 (Table 1), was confirmed by Southern blot analysis to have had its MHY1 gene correctly replaced by the URA3 gene.
Nucleic acid manipulation.
Genomic DNA, plasmid DNA, and total RNA were prepared from Y. lipolytica, as described previously (1). Southern and Northern blot analyses were carried out with DNA probes prepared with the ECL direct nucleic acid labeling and detection system (Amersham Life Sciences, Oakville, Ontario). Electrophoresis conditions and transfer to nitrocellulose membranes were as described previously (1). Hybridization, stringency of washes, and signal generation and detection were performed as recommended by the manufacturer.
Transcription-translation in vitro.
Coupled transcription-translation in vitro was carried out with the TnT T7-coupled reticulocyte lysate system (Promega). A 1.8-kbp EaeI-XbaI fragment containing the full-length MHY1 coding region was cloned under control of the T7 RNA polymerase promoter in pGEM-7Zf(+) to generate plasmid pG7-MHY1. Synthesis in vitro of Mhy1p was assessed by incorporation of L-[35S]methionine, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and fluorography.
Electrophoretic mobility shift analysis (EMSA).
A synthetic double-stranded oligonucleotide containing two tandem copies of the sequence located at nucleotides −419 to −399 from the A nucleotide of the initiator methionine codon of the Y. lipolytica CDC42 gene (forward strand, 5′-gatcTGACCCCTTGTTGTGCTGACCCCTTGTTGTG; reverse strand, 5′-gatcCACAACAAGGGGTCAGCACAACAAGGGGTCAG [putative STREs are underlined; the nucleotides designated in lowercase were added to provide cohesive BglII-BamHI ends]) and the same oligonucleotide containing single-base mutations (bold italics) in the putative STREs (underlined) (forward strand, 5′-gatcTGACCCCATGTTGTGCTGACCCCATGTTGTG; reverse strand, 5′-gatcCACAACATGGGGTCAGCACAACATGGGGTCAG) were end labeled with [α-32P]dATP and the Klenow fragment of DNA polymerase I. The same unlabeled oligonucleotides were used as specific competitors. Binding was performed in 15-μl (2 μl for Mhy1p translated in vitro in reticulocyte lysate) reaction mixtures containing 6 mM HEPES (pH 7.9), 120 mM NaCl, 0.4 mM MgCl2, 0.1 mM EDTA, 7% (vol/vol) glycerol, 4 μg of bovine serum albumin, 4 μg of salmon sperm DNA, and 4 μg of poly(dI-dC) · poly(dI-dC). Protein was incubated with unlabeled competitor at room temperature for 5 min. Radiolabeled probe was then added, and the reaction was continued for an additional 15 min at 25°C. Electrophoresis was carried out at 4°C on prerun 3.5% polyacrylamide gels (30:1 acrylamide/N,N′-methylene bisacrylamide weight ratio) in 22 mM Tris, 22 mM boric acid, and 1 mM EDTA as a running buffer. The gels were dried and subjected to autoradiography.
Stress treatment of cells.
Fresh cultures of Y. lipolytica E122 were transferred to YEPD medium, incubated at 28°C until exponential growth was reached (optical density at 600 nm, 1.0), and exposed to several stress conditions, as indicated. For carbon source starvation, the cells were washed with sterile distilled water, resuspended in YNB medium without glucose, and incubated at 28°C for 5 h; for heat shock, the cells were transferred to YEPD medium prewarmed to 35°C, followed by incubation at 35°C for 2 h; for osmotic shock, NaCl was added to the YEPD medium to a final concentration of 0.4 M, and the cells were maintained at 28°C for a further 2 h; for oxidative shock, hydrogen peroxide was added to the YEPD medium to a final concentration of 0.8 mM, and the cells were maintained at 28°C for a further 2 h.
Epitope tagging of Mhy1p.
An ApaI site was introduced before the stop codon of the MHY1 gene by replacement of the 0.8-kbp NdeI-XbaI fragment of pMHY1 with a 0.8-kbp NdeI-XbaI fragment obtained by PCR with the oligonucleotides 5′-CGCCCAGCATATGCGTACGCATCCTCGGGCCCAGAGGTAGAGCGCC and 5′-CCAATGCATCTAGACTGGACATACGTGAATCTACACTGCCAAACCAG (the ApaI site is italicized, the NdeI site is underlined, and the XbaI site is doubly underlined), generating the plasmid pMHY1-ApaI. A fragment with ApaI termini, encoding the peptide PLAMYPYDVPDYAAMYPYDVPDYAAMGKGE, which contains two repeats of the influenza virus hemagglutinin (HA) epitope (underlined residues) (17), was generated by PCR with the oligonucleotides 5′-TTAGGGCCCCGCTAGCCATGTACCCATACGACGTCCCAGACTAC and 5′-TTAGGGCCCTCTTCTATTCACCCTTACCCATGGCAGCGTAGTCT (the ApaI sites are italicized) and ligated into the unique ApaI site of pMHY1-ApaI to obtain the plasmid pMHY1-HA encoding a Mhy1p tagged at its carboxyl terminus with two repeats of the HA epitope (Mhy1p-HA). The integrity of the final construct was confirmed by sequencing, and pMHY1-HA was found to fully reproduce the phenotype produced by pMHY1 upon introduction into strain mhy1-1.
Indirect immunofluorescence microscopy.
Indirect immunofluorescence microscopy was performed as previously described (28). Mhy1p-HA was detected with monoclonal antibody 12CA5 (Berkeley Antibody Co., Richmond, Calif.), which recognizes a 9-amino-acid epitope of the influenza virus HA protein, and rhodamine (tetramethyl rhodamine isothiocyanate)-conjugated goat anti-mouse immunoglobulin antibodies (Sigma Chemical Co., St. Louis, Mo.). Nuclei were stained by the addition of 4,6-diamidino-2-phenylindole (DAPI) to the mounting medium at a final concentration of 1 μg/ml. Images were scanned with SPOT software 1.2.1 (Diagnostic Instruments, Inc., Sterling Heights, Mich.), processed in Photoshop 4.0.1 (Adobe Systems Inc., San Jose, Calif.), and printed on a DS8650 PS color printer (Eastman Kodak Co., Rochester, N.Y.).
Nucleotide sequence accession number.
The sequence data reported here are available from EMBL, GenBank, and DDBJ under accession no. AF124404.
RESULTS
Isolation and characterization of the MHY1 gene.
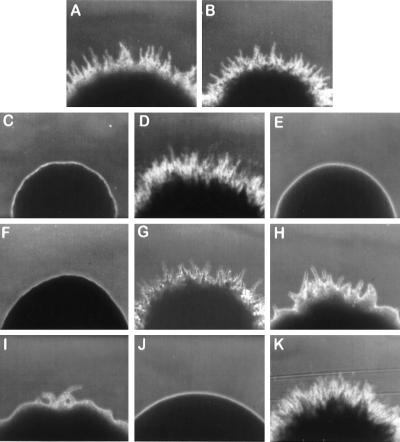
The Y. lipolytica mutant strain mhy1-1 (Fig. 1C) was initially isolated by its inability to form wild-type rough-surfaced colonies (Fig. 1A and B) after 3 days of incubation at 28°C. Further analysis revealed that the mhy1-1 strain was able to grow only in the budding form on both rich and minimal media and that this attribute was stably maintained through multiple generations.
FIG. 1.
Colony morphology of various Y. lipolytica strains. (A and B) Wild-type strains E122 and 22301-3; (C) original mutant strain mhy1-1; (D) strain mhy1K09 transformed with plasmid pMHY1; (E and F) MHY1 disruptant strains mhy1K09 and mhy1K09-B4; (G) MHY1/MHY1 diploid strain E122//22301-3; (H and I) MHY1/mhy1 diploid strains 22301-3/mhy1K09 and E122//mhy1K09-B4; (J) mhy1/mhy1 diploid strain mhy1K09//mhy1K09-B4; (K) mhy1/mhy1 diploid strain mhy1K09//mhy1K09-B4 carrying plasmid pMHY1. The colonies were photographed at ×100 magnification after 3 days of incubation at 28°C on YNA agar plates.
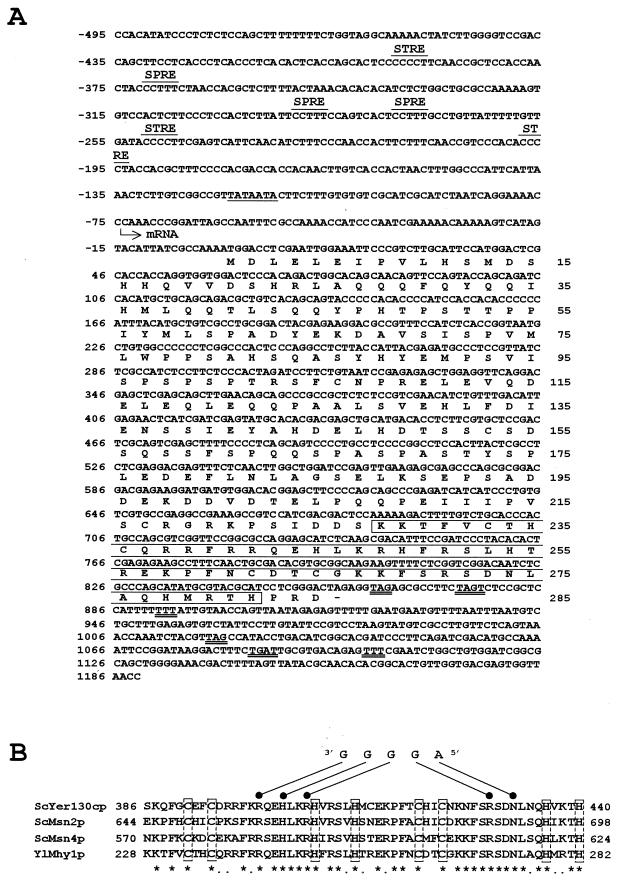
The MHY1 gene was isolated from a Y. lipolytica genomic DNA library contained in the replicative E. coli shuttle vector pINA445 (24) by its ability to restore filamentous growth to mhy1-1 cells. Of approximately 40,000 transformants screened, 4 showed an enhanced filamentous phenotype (Fig. 1D). Restriction enzyme analysis demonstrated that all complementing plasmids shared a 3.5-kbp BglII-HindIII fragment capable of complementing the mhy1-1 mutation. Sequencing of this fragment revealed that the largest open reading frame, the MHY1 gene, contained 855 bp coding for a 285-amino-acid protein, Mhy1p, with a predicted molecular mass of 32,636 Da (Fig. 2A).
FIG. 2.
Characteristics of the MHY1 gene and its encoded protein, Mhy1p. (A) Nucleotide sequence of the MHY1 gene and deduced amino acid sequence of Mhy1p. A putative TATA box is underlined. Putative transcription termination signals are doubly underlined. The transcriptional start site of the MHY1 gene is indicated by the arrow. SPRE, putative stationary-phase response elements. The segment of Mhy1p containing the two C2H2-type zinc finger motifs is boxed. (B) Amino acid sequence alignment of the two zinc finger domains of Mhy1p and of S. cerevisiae Msn2p, Msn4p, and Yer130cp. Identical (asterisks) and conserved (dots) amino acids are indicated. Cys and His residues capable of binding zinc ions are boxed. The STRE sequence 5′-AGGGG and the amino acids that bind it are indicated.
A putative TATA box, TATAATA, is found between nucleotides −119 and −113 from the A nucleotide of the potential initiating codon of the MHY1 gene. Initial analysis has shown that transcription of the MHY1 gene preferentially starts at position −74 from the A nucleotide of the first ATG codon (data not shown) within a CCAAA sequence, a common structural feature of Y. lipolytica genes (3). A nucleotides are also observed at positions −1 and −3 from the A nucleotide of the first ATG, a feature often observed in genes that are highly expressed in Y. lipolytica (3). The upstream regulatory region of the MHY1 gene contains consensus sequences for the binding of several transcription factors implicated in the regulation of fungal development and in the response of cells of S. cerevisiae to specific environmental conditions, including multiple copies of the putative STRE, AGGGG (15), and multiple copies of a putative stationary-phase response element, AAAGG, commonly found upstream of stationary-phase-responsive genes (40).
Mhy1p contains a glutamine-rich tract (52% of all residues between amino acids 27 and 45) at its amino terminus and two putative C2H2-type zinc finger motifs at its carboxyl terminus (Fig. 2A and B). Similar structural elements are found in transcription factors like the S. cerevisiae calcineurin-responsive zinc finger protein, Crz1p (23, 37), and Drosophila melanogaster Sp1, which is required for the development of the antennal, intercalary, and mandibular segments of the head (43). Moreover, the region in Mhy1p connecting these motifs (amino acids 46 to 175) is very rich in serine and proline (33%) and acidic residues (28%), a structural composition that has been implicated in protein-protein interactions (12). Three putative PEST regions, commonly found in rapidly degraded proteins (7, 29), are predicted at residues 49 to 65, 89 to 102, and 149 to 188 of Mhy1p.
Mhy1p binds in vitro to putative STRE elements.
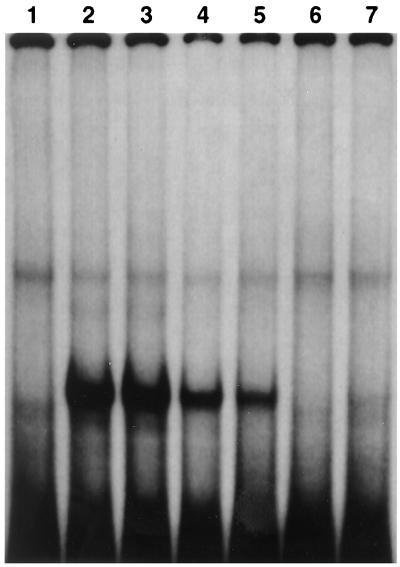
A search with the BLAST Network Service of the National Center for Biotechnology Information identified a number of S. cerevisiae transcription factors with homology to Mhy1p within the zinc finger motifs. Further analysis revealed that the two C2H2-type zinc fingers of Mhy1p displayed the same arrangement and main structural features as those found in the stress-responsive transcription factors Msn2p and Msn4p and in the putative protein encoded by the open reading frame Yer130cp (Fig. 2B). Predictions based on computer-assisted molecular modeling of Msn2p and Msn4p (22), combined with the presence of residues in Mhy1p (Arg242, His245, Arg248, Arg271, and Asn273) identical to those shown to be responsible for specific DNA binding by Msn2p and Msn4p (Fig. 2B), suggested that Mhy1p could also recognize and bind the AGGGG pentanucleotide of putative STREs. Specific binding of Mhy1p to the AGGGG pentanucleotide was demonstrated by EMSA. In vitro-synthesized Mhy1p bound to a radiolabeled double-stranded probe containing two tandemly repeated copies of one of the three AGGGG pentanucleotide sequences found in the upstream region of the CDC42 gene (Fig. 3, lane 2), whose transcription is increased during the dimorphic transition in Y. lipolytica (14a). This binding was efficiently competed by increasing amounts of unlabeled probe (Fig. 3, lanes 3 to 6). Mhy1p failed to bind to a probe containing mutations (italic) in the two AGGGG motifs (TGGGG) (Fig. 3, lane 7), thereby demonstrating the specificity of the interaction of Mhy1p and the putative STREs.
FIG. 3.
Mhy1p binds specifically in vitro to the putative STRE pentanucleotide AGGGG. Mhy1p was translated in vitro, incubated with a radiolabeled probe which is derived from the upstream region of the CDC42 gene and which contains two copies of the AGGGG pentanucleotide, and analyzed by EMSA. The specificity of the DNA-protein interaction was determined by competition analysis with unlabeled probe and by EMSA with a mutant probe containing single-base mutations (AGGGG to TGGGG) in the two putative STREs of the wild-type probe. Lane 1, wild-type probe incubated with unprogrammed lysate. Lane 2, wild-type probe incubated with in vitro-translated Mhy1p. Lanes 3 to 6, wild-type probe incubated with in vitro-translated Mhy1p and 10-, 50-, 100-, and 500-fold molar excesses of unlabeled wild-type oligonucleotide, respectively. Lane 7, mutant probe incubated with in vitro-translated Mhy1p.
Disruption of the MHY1 gene does not affect mating of Y. lipolytica.
Mating is a phenomenon that involves dramatic changes in cell morphology in response to environmental conditions, and it has been found to be intimately connected to dimorphism (19). We therefore investigated whether disruption of the MHY1 gene had any effect on the mating ability of Y. lipolytica. The B mating type strain, mhy1K09-B4 (Table 1), with its MHY1 gene deleted, was obtained by crossing strain mhy1K09 with the isogenic wild-type strain 22301-3, followed by sporulation of the resultant diploid and selection for the inability to undergo dimorphic transition. The mhy1∷URA3 genotype of the mhy1K09-B4 strain was confirmed by cosegregation of the Fil− and Ura+ phenotypes in random spore analysis (data not shown). MHY1/mhy1 (Fig. 1H and I) and mhy1/mhy1 (Fig. 1J) diploid strains were readily obtained by mating any combination of the mutant haploid strains mhy1K09 and mhy1K0-B4 and the wild-type strains E122 and 22301-3, indicating that no defect in mating ability was associated with the loss of MHY1.
Filamentation is affected by gene dosage of MHY1.
Since transformation of mhy1 mutant strains with the pINA445-based autonomously replicating plasmid pMHY1, which is believed to be present in two to five copies per cell (11), resulted in an enhanced filamentous phenotype (Fig. 1D), crossings of the mutant strains mhy1K09 and mhy1K09-B4 were performed with the wild-type strains E122 and 22301-3 to determine the effects of gene dosage on the filamentous growth of diploid strains. Diploid strains containing a single copy of MHY1 (Fig. 1H and I) gave rise to colonies with significantly reduced filamentation compared to that of wild-type haploid strains (Fig. 1A and B), while transformation of an mhy1/mhy1 diploid strain (Fig. 1J) with the plasmid pMHY1 resulted in an enhanced filamentous phenotype (Fig. 1K). Colonies formed by MHY1/mhy1 diploid strains (Fig. 1H and I) showed a substantial increase in the proportion of yeast-like cells compared to colonies of the MHY1/MHY1 strain (Fig. 1G).
Transcription of the MHY1 gene is increased during dimorphic transition.
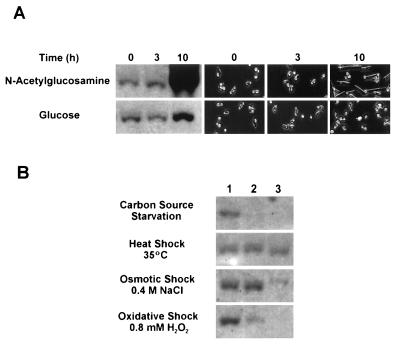
The yeast-to-hypha transition in exponentially growing E122 cells was induced by a 15-min carbon source starvation period at 4°C, followed by transfer to YNBGlcNAc medium. Under these conditions, more than 90% of cells gave rise to germ tubes after 10 h of incubation at 28°C (Fig. 4A). In contrast, cells transferred to fresh glucose-containing (YNBGlc) medium grew almost exclusively as the budding form (Fig. 4A), as previously described (14). Northern blot analysis carried out with total RNA extracted from cells harvested following 3 and 10 h of incubation showed that MHY1 mRNA levels dramatically increased during the dimorphic transition but remained essentially constant during the first hours of incubation in YNBGlcNAc or YNBGlc (Fig. 4A). A smaller increase in the levels of MHY1 mRNA (approximately twofold) was observed after 10 h of incubation in YNBGlc (Fig. 4A), probably due to the small number of germ tubes (less than 1%) present.
FIG. 4.
Levels of MHY1 mRNA during the dimorphic transition (A) and under stress conditions (B). Total RNA was isolated from E122 cells that were incubated at 28°C in YNBGlcNAc (for induction of filamentous growth) or YNBGlc (for control culture, yeast-like cells) for the times indicated (A) or that were exposed to several stress conditions (B). For carbon source starvation, cultures were sampled at 0, 1, and 5 h (lanes 1, 2, and 3, respectively). For heat shock and osmotic and oxidative stresses, cultures were sampled at 0, 30, and 120 min (lanes 1, 2, and 3, respectively). RNA (10 μg) from each time point was separated on a formaldehyde agarose gel, transferred to nitrocellulose, and subjected to Northern blot analysis. The blots were hybridized with a probe specific for the MHY1 gene. Equal loading of RNA was ensured by ethidium bromide staining (data not shown). In panel A, cell morphologies at t = 0, 3, and 10 h are shown.
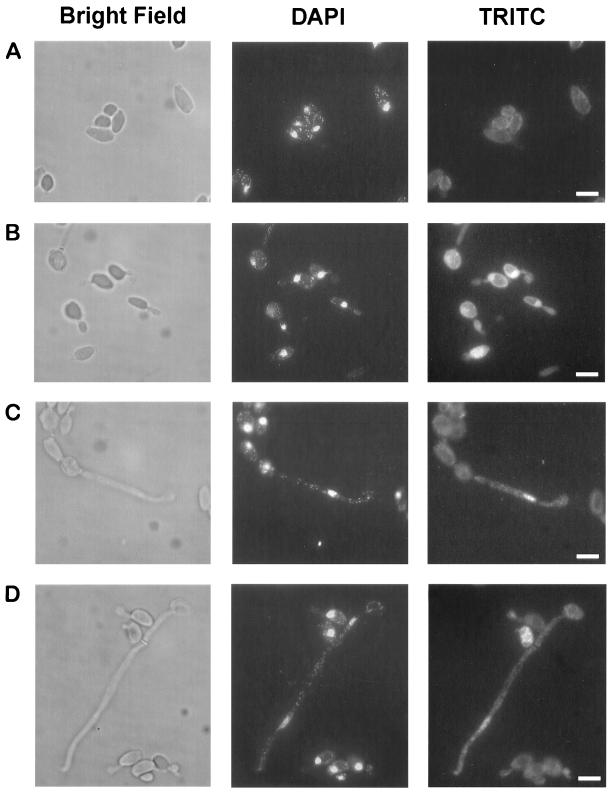
Mhy1p is localized to the nucleus during the dimorphic transition and is concentrated in actively growing hyphal cells.
To determine whether the subcellular localization of Mhy1p is consistent with its potential function as a transcription factor, indirect in situ immunofluorescence of a carboxyl-terminal HA-tagged version of Mhy1p was carried out in cells cultivated in YNBGlcNAc. Mhy1p-HA was undetectable in the nuclei of cells growing as the yeast form (Fig. 5A) but was readily detected in the nuclei of cells undergoing dimorphic transition (Fig. 5B to D). Strikingly, Mhy1p-HA was found to be concentrated in actively growing hyphal cells. Thus, while Mhy1p-HA was detected in the nuclei of cells emitting germ tubes (Fig. 5B), once the transition step was completed, Mhy1p-HA was concentrated in the filamentous cells located at the growing hyphal tip (98% of nuclei labeled; n = 150 cells), with no, or a barely detectable, signal being detected in the other cells of the filamentous chain (14% of nuclei labeled; n = 250 cells) (Fig. 5C and D).
FIG. 5.
Nuclear localization of Mhy1p during filamentous growth of Y. lipolytica. Indirect immunofluorescence of HA-tagged Mhy1p was carried out on yeast-like cells (A) and at different stages of filamentation (early germ tube formation [B], late germ tube formation [C], and hyphal growth [D]) as described in Materials and Methods. Left panels, cells visualized by bright-field microscopy; middle panels, DAPI staining of nuclei; right panels, rhodamine (tetramethyl rhodamine isothiocyanate) staining observed with an anti-HA monoclonal antibody. Bars = 5 μm.
Transcription of MHY1 during the stress response.
To investigate possible links between the dimorphic transition program and the stress response in Y. lipolytica via MHY1, Northern blotting of total RNA prepared from E122 cells exposed to several stress conditions was carried out with a MHY1 fragment as a probe (Fig. 4B). Surprisingly, MHY1 mRNA levels were found to decrease to barely detectable levels after 2 h of exposure to osmotic stress (0.4 M NaCl) and were undetectable after 1 h of carbon source starvation or 2 h of exposure to oxidative stress (0.8 mM H2O2). In contrast, transcription of MHY1 remained essentially constant even after 2 h of exposure to thermal stress at 35°C. Interestingly, the viability of E122 cells and mhy1K09 cells was essentially the same for both strains following exposure to a particular type of stress (data not shown).
DISCUSSION
Here we report the identification of a novel gene, MHY1, and initial characterization of its product, Mhy1p, involved in the yeast-to-hypha transition of the dimorphic yeast Y. lipolytica. Although the exact role of Mhy1p in the dimorphic transition remains undetermined, several features of Mhy1p suggest that it may function as a transcription factor. The most striking of these is the presence near its carboxyl terminus of two C2H2-type zinc fingers, which are strongly homologous to zinc fingers found in the proteins encoded by the S. cerevisiae genes MSN2 and MSN4 and the open reading frame of unknown function, YER130C. Msn2p and Msn4p are transcriptional activators of the multistress response in S. cerevisiae. They act via upstream STREs, which have the consensus core sequence AGGGG (or CCCCT) and which are able to mediate transcription induced by a broad range of environmental and physiological conditions, thereby enabling cells to develop tolerance to different forms of stress (cross-protection) (15, 21, 22, 31, 34, 39). Msn2p and Msn4p have been found to be constitutively synthesized during growth on glucose (9), and they are activated by their translocation from the cytosol into the nucleus in response to stress conditions, such as heat shock, carbon source starvation, osmotic stress, and the presence of ethanol or sorbate (13). Interestingly, conditions promoting the yeast-to-hypha transition led to a redistribution of Mhy1p from the cytosol, followed by its concentration in the nuclei of cells undergoing dimorphic transition.
Like Msn2p and Msn4p, Mhy1p specifically recognizes, and binds to, sequences containing the AGGGG pentanucleotide, strongly suggestive of a role for Mhy1p in the transcriptional regulation of genes containing this sequence in their promoter regions. Database analysis has revealed that most promoters of Y. lipolytica genes contain AGGGG sequences, but they are particularly abundant in the genes HOY1 (six copies) and ICL1 (five copies). HOY1 has been shown to be directly involved in the yeast-to-hypha transition (38), while ICL1 encodes one of the two main enzymes of the glyoxylate pathway, a strongly regulated anaplerotic cycle that in Y. lipolytica is under the control of GPR1, a gene also implicated in the dimorphic transition (3). We are currently conducting experiments aimed at determining whether Mhy1p is a transcription factor and, if it is, whether it modulates gene expression through its interaction with STREs.
MHY1 expression is dramatically increased during the yeast-to-hypha transition. Surprisingly, MHY1 mRNA levels were significantly decreased under conditions that otherwise would activate Msn2p- and Msn4p-mediated expression of stress-responsive genes in S. cerevisiae, i.e., carbon source starvation and osmotic and oxidative shock. However, transcription of MHY1 was unaffected by thermal stress. It has been suggested that heat shock, and not starvation, may act synergistically with N-acetylglucosamine to achieve full induction of mycelial growth (14). Our results are in agreement with this hypothesis. Overall, analysis of the upstream region of MHY1 suggests a rather complex regulation of expression, involving feedback regulatory loops with possible connections to nitrogen starvation and stationary-phase maintenance. We are currently investigating whether these putative regulatory elements are functional and searching for proteins that can recognize these sequences.
In closing, it is important to point out that the study of stress response in Y. lipolytica is still incipient. Isolation of stress-responsive genes, and knowledge of the conditions they regulate, will be important for determining possible links between stress response and filamentous growth in dimorphic yeasts, two phenomena with large implications for the development of virulence by fungal pathogens.
ACKNOWLEDGMENTS
This work was supported by an International Research Scholarship from the Howard Hughes Medical Institute to R.A.R. R.A.R. is a Medical Research Council of Canada Senior Scientist.
REFERENCES
- 1.Ausubel F J, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates; 1989. [Google Scholar]
- 2.Banuett F. Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu Rev Genet. 1995;29:179–208. doi: 10.1146/annurev.ge.29.120195.001143. [DOI] [PubMed] [Google Scholar]
- 3.Barth G, Gaillardin C. Yarrowia lipolytica. In: Wolf K, editor. Non-conventional yeasts in biotechnology. A handbook. New York, N.Y: Springer-Verlag; 1996. pp. 313–388. [Google Scholar]
- 4.Barth G, Weber H. Improvement of sporulation in the yeast Yarrowia lipolytica. Antonie Leeuwenhoek J Microbiol Serol. 1986;51:167–177. doi: 10.1007/BF02310010. [DOI] [PubMed] [Google Scholar]
- 5.Birse C E, Irwin M Y, Fonzi W A, Sypherd P S. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect Immun. 1993;61:3648–3655. doi: 10.1128/iai.61.9.3648-3655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun B R, Johnson A D. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- 7.Chevaillier P. PEST sequences in nuclear proteins. Int J Biochem. 1993;25:479–482. doi: 10.1016/0020-711x(93)90653-v. [DOI] [PubMed] [Google Scholar]
- 8.Davidow L S, Apostolakos D, O’Donnell M M, Proctor A R, Ogrydziak D M, Wing R A, Stasko I, DeZeeuw J R. Integrative transformation of the yeast Yarrowia lipolytica. Curr Genet. 1985;10:39–48. [Google Scholar]
- 9.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 10.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fournier P, Abbas A, Chasles M, Kudla B, Ogrydziak D M, Yaver D, Xuan J-W, Peito A, Ribet A-M, Feynerol C, He F, Gaillardin C. Colocalization of centromeric and replicative functions on autonomously replicating sequences from the yeast Yarrowia lipolytica. Proc Natl Acad Sci USA. 1993;90:4912–4916. doi: 10.1073/pnas.90.11.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frankel A D, Kim P S. Modular structure of transcription factors: implications for gene regulation. Cell. 1991;65:717–719. doi: 10.1016/0092-8674(91)90378-c. [DOI] [PubMed] [Google Scholar]
- 13.Görner W, Durchschlag E, Martinez-Pastor M T, Estruch F, Ammerer G, Hamilton B, Ruis H, Schüller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guevara-Olvera L, Calvo-Mendez C, Ruiz-Herrera J. The role of polyamine metabolism in dimorphism of Yarrowia lipolytica. J Gen Microbiol. 1993;193:485–493. doi: 10.1099/00221287-139-3-485. [DOI] [PubMed] [Google Scholar]
- 14a.Hurtado, C. A. R., and R. A. Rachubinski. Unpublished data.
- 15.Kobayashi N, McEntee K. Identification of cis and trans components of a novel heat shock stress regulatory pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:248–256. doi: 10.1128/mcb.13.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohler J R, Fink G R. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci USA. 1996;93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolodziej P A, Young R A. Epitope tagging and protein surveillance. Methods Enzymol. 1991;194:508–519. doi: 10.1016/0076-6879(91)94038-e. [DOI] [PubMed] [Google Scholar]
- 18.Liu H, Kohler J, Fink G R. Suppression of hyphal formation in Candida albicans by a mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 19.Madhani H D, Fink G R. The control of filamentous differentiation and virulence in fungi. Trends Cell Biol. 1998;8:348–353. doi: 10.1016/s0962-8924(98)01298-7. [DOI] [PubMed] [Google Scholar]
- 20.Magee P T. Which came first, the hypha or the yeast? Science. 1997;277:52–53. doi: 10.1126/science.277.5322.52. [DOI] [PubMed] [Google Scholar]
- 21.Marchler G, Schüller C, Adam G, Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez-Pastor M T, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress-response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 23.Matheos D P, Kingsbury T J, Ahsan U S, Cunningham K W. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 1997;11:3445–3458. doi: 10.1101/gad.11.24.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nuttley W M, Brade A M, Gaillardin C, Eitzen G A, Glover J R, Aitchison J D, Rachubinski R A. Rapid identification and characterization of peroxisomal assembly mutants in Yarrowia lipolytica. Yeast. 1993;9:507–517. [Google Scholar]
- 25.Odds F C. Candida and candidosis. A review and bibliography. London, United Kingdom: Bailliere Tindal; 1988. pp. 42–59. [Google Scholar]
- 26.Ogrydziak D, Bassel J, Contopoulou R, Mortimer R. Development of genetic techniques and the genetic map of the yeast Saccharomyces lipolytica. Mol Gen Genet. 1978;163:229–239. [Google Scholar]
- 27.Olaiya A F, Sogin S J. Ploidy determination of Candida albicans. J Bacteriol. 1979;140:1043–1049. doi: 10.1128/jb.140.3.1043-1049.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pringle J R, Adams A E M, Drubin D G, Haarer B K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 29.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 30.Rodriguez C, Dominguez A. The growth characteristics of Saccharomycopsis lipolytica: morphology and induction of mycelium formation. Can J Microbiol. 1984;30:605–612. [Google Scholar]
- 31.Ruis H, Schüller C. Stress signaling in yeast. Bioessays. 1995;17:959–965. doi: 10.1002/bies.950171109. [DOI] [PubMed] [Google Scholar]
- 32.San-Blas G, San-Blas F. Molecular aspects of fungal dimorphism. Crit Rev Microbiol. 1984;11:101–127. doi: 10.3109/10408418409105474. [DOI] [PubMed] [Google Scholar]
- 33.Saporito-Irwin S M, Birse C E, Sypherd P S, Fonzi W A. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol Cell Biol. 1995;15:601–613. doi: 10.1128/mcb.15.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schüller C, Brewster J L, Alexander M R, Gustin M C, Ruis H. The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 1994;13:4382–4389. doi: 10.1002/j.1460-2075.1994.tb06758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shepherd M G. Morphogenetic transformation in fungi. Curr Top Med Mycol. 1988;2:278–304. doi: 10.1007/978-1-4612-3730-3_8. [DOI] [PubMed] [Google Scholar]
- 36.Staab J F, Ferrer C A, Sundstrom P. Developmental expression of a tandemly repeated, proline- and glutamine-rich amino acid motif on hyphal surfaces of Candida albicans. J Biol Chem. 1996;271:6298–6305. doi: 10.1074/jbc.271.11.6298. [DOI] [PubMed] [Google Scholar]
- 37.Stathopoulos A M, Cyert M S. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997;11:3432–3444. doi: 10.1101/gad.11.24.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torres-Guzman J C, Dominguez A. HOY1, a homeo gene required for hyphal formation in Yarrowia lipolytica. Mol Cell Biol. 1997;17:6283–6293. doi: 10.1128/mcb.17.11.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Treger J M, Magee T R, McEntee K. Functional analysis of the stress response element and its role in the multistress response of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1998;243:13–19. doi: 10.1006/bbrc.1997.8061. [DOI] [PubMed] [Google Scholar]
- 40.Wang W, Nishikawa T, Isono K. Isolation and characterization of Saccharomyces cerevisiae genes differentially expressed under different growth conditions. J Gen Appl Microbiol. 1997;43:217–224. doi: 10.2323/jgam.43.217. [DOI] [PubMed] [Google Scholar]
- 41.Whelan W L, Partridge R M, Magee P T. Heterozygosity and segregation in Candida albicans. Mol Gen Genet. 1980;180:107–113. doi: 10.1007/BF00267358. [DOI] [PubMed] [Google Scholar]
- 42.Wickerham L J. Sexual reproduction in Candida lipolytica. Science. 1970;167:1141. doi: 10.1126/science.167.3921.1141. [DOI] [PubMed] [Google Scholar]
- 43.Wimmer E A, Jackle H, Pfeifle C, Cohen S M. A Drosophila homologue of human Sp1 is a head-specific segmentation gene. Nature. 1993;366:690–694. doi: 10.1038/366690a0. [DOI] [PubMed] [Google Scholar]