Abstract
Purpose:
Despite extensive genomic and transcriptomic profiling, it remains unknown how signaling pathways are differentially activated and how tumors are differentially sensitized to certain perturbations. Here, we aim to characterize AKT signaling activity and its association with other genomic or IHC-based PI3K/AKT pathway biomarkers as well as the clinical activity of ipatasertib (AKT inhibitor) in the FAIRLANE trial.
Experimental Design:
In FAIRLANE, 151 patients with early triple-negative breast cancer (TNBC) were randomized 1:1 to receive paclitaxel with ipatasertib or placebo for 12 weeks prior to surgery. Adding ipatasertib did not increase pathologic complete response rate and numerically improved overall response rate by MRI. We used reverse-phase protein microarrays (RPPA) to examine the total level and/or phosphorylation states of over 100 proteins in various signaling or cell processes including PI3K/AKT and mTOR signaling. One hundred and twenty-five baseline and 127 on-treatment samples were evaluable by RPPA, with 110 paired samples at both time points.
Results:
Tumors with genomic/protein alterations in PIK3CA/AKT1/PTEN were associated with higher levels of AKT phosphorylation. In addition, phosphorylated AKT (pAKT) levels exhibited a significant association with enriched clinical benefit of ipatasertib, and identified patients who received benefit in the absence of PIK3CA/AKT1/PTEN alterations. Ipatasertib treatment led to a downregulation of AKT/mTORC1 signaling, which was more pronounced among the tumors with PIK3CA/AKT1/PTEN alterations or among the responders to the treatment.
Conclusions:
We showed that the high baseline pAKT levels are associated with the alterations of PI3K/AKT pathway components and enriched benefit of ipatasertib in TNBC.
Translational Relevance.
Triple-negative breast cancer (TNBC) is an aggressive form of breast cancer with poor prognosis and high recurrence and metastasis rate, highlighting the need for more effective therapeutic approaches with appropriate diagnostic biomarkers. Due to the molecular heterogeneity of TNBC, a key aspect for targeted therapy is identifying tumors that are most likely to be sensitive to the specific oncogenic signaling perturbation to maximize the clinical benefit. Here, we showed that PIK3CA/AKT1/PTEN alterations, together with multiple cell signaling activities, modulate the level of phosphorylated AKT (pAKT) on Serine473 and Threonine308. Importantly, tumors with high pAKT levels exhibited the strongest association with enriched ipatasertib activity, suggesting that the pAKT-high tumors are most addicted to AKT signaling. This study provides proof-of-concept that the baseline phosphorylation levels of AKT, the direct target of ipatasertib, could have predictive value and may possess an improved means of biomarker-based patient selection for AKT inhibitors and diagnostic utility for precision medicine.
Introduction
The PI3K/AKT signaling pathway, which regulates multiple cellular processes including mRNA translation, metabolism, survival, proliferation, and differentiation, is one of the most frequently dysregulated pathways in many types of human cancers (1). Aberrant activation of the PI3K/AKT signaling axis is commonly observed in breast cancer and emerges as a potential target in triple-negative breast cancer (TNBC; refs. 2, 3). Combined activating mutations in PIK3CA and AKT1, with loss-of-function or low expression in PTEN, occur in approximately 30% to 50% of TNBC (4–6).
Ipatasertib is a potent and highly selective oral small-molecule inhibitor of all three isoforms of AKT (7, 8) and is being evaluated in cancers with a high prevalence of genomic alterations in PI3K/AKT pathway including breast cancer (4, 5) and prostate cancer (9, 10). FAIRLANE is a double-blinded, placebo-controlled randomized phase II trial of neoadjuvant ipatasertib plus paclitaxel (IPAT+PAC) for early TNBC (eTNBC; ref. 5). In this trial, pathologic complete response (pCR) rate with IPAT+PAC versus placebo plus paclitaxel (PBO+PAC) were 17% and 13%, respectively (5). Adding ipatasertib showed a numeric increase in overall response rate (ORR) assessed by MRI, from 56% with PBO+PAC to 67% with IPAT+PAC (5). The antitumor effect of ipatasertib was enriched in a subset of patients with PIK3CA/AKT1/PTEN-altered tumors by next-generation sequencing (NGS) or tumors with low PTEN expression assessed by IHC (5), similarly observed in other studies (4, 9, 10). On the other hand, although genomic/protein alteration(s) of PIK3CA/AKT1/PTEN represents a key mechanism of PI3K/AKT pathway activation, there is a myriad of other mechanisms with a potential to lead to signaling activation (11). Given that the signal transduction pathway is exquisitely modulated at multiple levels, it remains unclear how the PI3K pathway genomic/protein alterations actually translate into the functional PI3K/AKT signaling activity outputs in patients’ tumors. Importantly, while the known mechanism of action of ipatasertib is to physically bind with AKT and modulate its kinase activity, it also remains to be understood whether the pretreatment baseline state of phosphorylation-modulated PI3K/AKT signaling activities in the tumor might predict response to ipatasertib treatment in the clinic.
TNBC is a clinically heterogeneous subtype of breast cancer with distinct molecular subtypes that vary in prognosis and show differential response to targeted, immune, or chemotherapeutic agents (12, 13). Although gene expression together with genomic profiling has generated many insights in the biology of TNBC (14–16), there remains a strong need for protein-based assays that can assess the state of signaling networks which cannot be accurately described at the genomic or transcriptomic level.
Here, we deployed reverse-phase protein microarray (RPPA), a quantitative proteomics approach that allows simultaneous measurement of numerous protein phosphorylation states in small quantities of patients’ tumor samples (17–20). Using this approach, coupled with Laser Capture Microdissection (LCM) to enrich for tumor epithelial cells, we analyzed over 100 proteins/phosphoproteins representing a variety of key “hubs” in important signaling pathways and cellular processes that are commonly dysregulated in human cancers. LCM-based cellular enrichment is key to ensure accurate measurement of protein and protein phosphorylation in tumor cells since signaling pathway components like AKT are ubiquitously expressed and differentially activated in every human cell type (21, 22). Based on the known mechanism of action of ipatasertib as an AKT kinase inhibitor, we first sought to assess the association of PI3K/AKT pathway alterations and AKT or mTORC1 activities measured by protein phosphorylation via RPPA, as well as the predictive value of AKT and mTORC1 activities with ipatasertib treatment effect. We also assessed the pharmacodynamic effect of ipatasertib on PI3K/AKT and other cell signaling activities from the paired pretreatment and on-treatment tumor biopsies.
Materials and Methods
Patients and molecular assays
The design and outcomes of the placebo-controlled double-blind randomized phase II FAIRLANE trial (ClinicalTrials.gov: NCT02301988) was described previously (5). The study conformed with the International Conference on Harmonization (ICH) E6 guideline for Good Clinical Practice and principles of the Declaration of Helsinki (or local laws and regulations, whichever afforded greater protection to individuals). All study-related materials were approved by each participating center's Institutional Review Board or Ethics Committee prior to study initiation. All patients provided written informed consent. Tumor sample collections, NGS testing, IHC assay, and RNA sequencing (RNA-seq) of tumor samples were described (5).
Microdissection and cellular lysate arraying
Enriched epithelial cell populations were isolated from 8-μm cryosections (>95% purity) of tissue using an Arcturus Pixcell IIe LCM system (Arcturus) as previously described (23). Approximately 10,000 epithelial cells were captured for each sample at the pretreatment and C1D8 time points. Microdissected material was stored at −80°C and samples were lysed as described (19). Cell lysates were printed in triplicate spots (approximately 10-nL per spot) onto nitrocellulose coated slides (Grace Biolabs) using a Quanterix 2470 Arrayer (Quanterix). Standard curves of control cell lysates were also included for quality assurance purposes. The proteins and phosphoproteins measured in this study (endpoints, 110 in total) are listed in Supplementary Table S1. Immunostaining and scanning of arrays was performed as described (19).
Analysis of RPPA data
RPPA data was generated from scanned arrays as previously described (18). For each data point, the signal intensity was normalized first by subtraction of the negative control spot intensity and then divided by the total protein amount. The normalized expression values from three technical replicates were averaged using geometric mean. Data was then log2 transformed after adding an offset of 1. Endpoints with high variabilities [>15 out of 260 samples with coefficient of variation (CV) greater than 5%] were filtered out. Samples with missing values in more than 20 out of 110 endpoints were also removed. Endpoints with zero expression were imputed with the minimal expression of a given endpoint across all the samples and missing values were imputed with the median expression.
Bioinformatic analysis
Differential analysis of expression of individual endpoints between PIK3CA/AKT1/PTEN genomic/protein altered and nonaltered was conducted using the limma package (24) with vooma observational-level weights (25). Analysis of signaling scores [phosphorylated AKT (pAKT) activity, AKT score, and mTORC1 score] was performed with a standard linear model using the lm function, where P values were computed based on the two-sided t test. Differential expression analysis between baseline and C1D8 was performed with limma mixed model (24) for the individual endpoints, where subject ID was used as a blocking variable to account for the paired nature of this data. P values were adjusted using the Benjamini–Hochberg procedure for multi-testing and results with adjusted P values were shown.
Data availability statement
Data has been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001005892.
Results
Analysis of cell signaling pathways with integrated activity scores
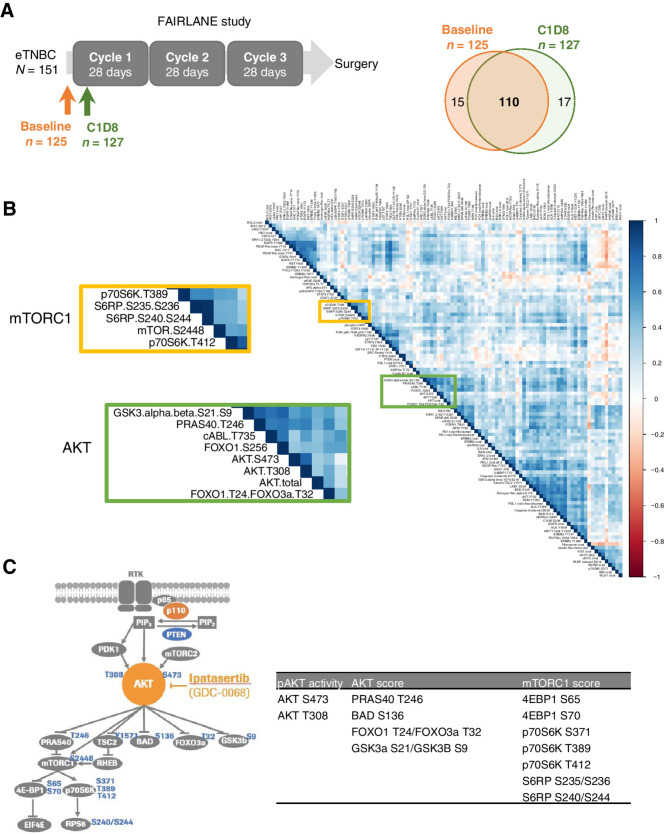
Among the 151 patients enrolled in the FAIRLANE study, baseline tissue samples from 125 patients and on-treatment (C1D8) samples from 127 patients were evaluable by RPPA, with 110 paired samples at both time points (Fig. 1A). Baseline characteristic and clinical outcomes in the RPPA-evaluable populations were representative of the overall population (Supplementary Fig. S1; Supplementary Table S2).
Figure 1.
RPPA analysis of cell signaling proteins from frozen tumor samples in FAIRLANE study. A, Schematic showing the collection of baseline and cycle 1 day 8 (C1D8) tumor samples for RPPA analysis. Venn diagram shows the number of the baseline and C1D8 RPPA samples. B, Correlation plot showing the pair-wise correlation between all endpoints measured by RPPA at baseline. The AKT and mTORC1 downstream components cluster tightly together shown in the zoomed plot. C, Diagram of the AKT/mTORC1 signaling pathway highlighting the phosphorylation sites measured by RPPA in this study. pAKT activity, AKT score, and mTORC1 score were calculated by the phosphorylation levels of AKT itself, AKT, and mTORC1 direct substrates, respectively.
First, we analyzed the baseline samples and found that phosphoproteins involved in the same signaling pathway tend to exhibit the highest levels of correlations. For instance, all the major AKT signaling components measured in this study, including AKT and its known kinase substrates (PRAS40, FOXO, GSK3), cluster together; likewise, all the components of the mTORC1 kinase and its known substrates that we measured (p70S6K, S6RP) were found in another cluster (Fig. 1B), consistent with them being directly regulated by AKT and mTORC1, respectively (Fig. 1C). Although AKT and mTORC1 are often depicted in the linear PI3K-AKT-mTORC1 pathway, the activity of mTORC1 is actually modulated by many other signaling inputs in addition to AKT (26–28), explaining the AKT and mTORC1 substrates being in two different clusters.
To describe the state of signaling networks, we calculated an integrated pathway activation score for each of the key oncogenic signaling pathways, including AKT, mTORC1, insulin-like growth factor-1 receptor (IGF-1R), EGFR, HER2, HER3, MAPK, apoptosis, cell cycle, immune, and JAK-STAT signaling (Supplementary Table S3; ref. 29). In addition, given that AKT is a central node in the PI3K/AKT pathway and the target of ipatasertib, we developed two scores with one to describe the phosphorylation levels on AKT itself and the other score for AKT activity reflected by the phosphorylation levels of its direct substrates (Fig. 1C; Supplementary Table S3). Signaling scores were computed as an average of z-score normalized expression of individual endpoints included in the module definition (Supplementary Table S3). Within each pathway, individual components tend to exhibit positive correlations to each other (Supplementary Fig. S2).
pAKT levels are modulated by PI3K/AKT pathway alterations and additional cell signaling activities
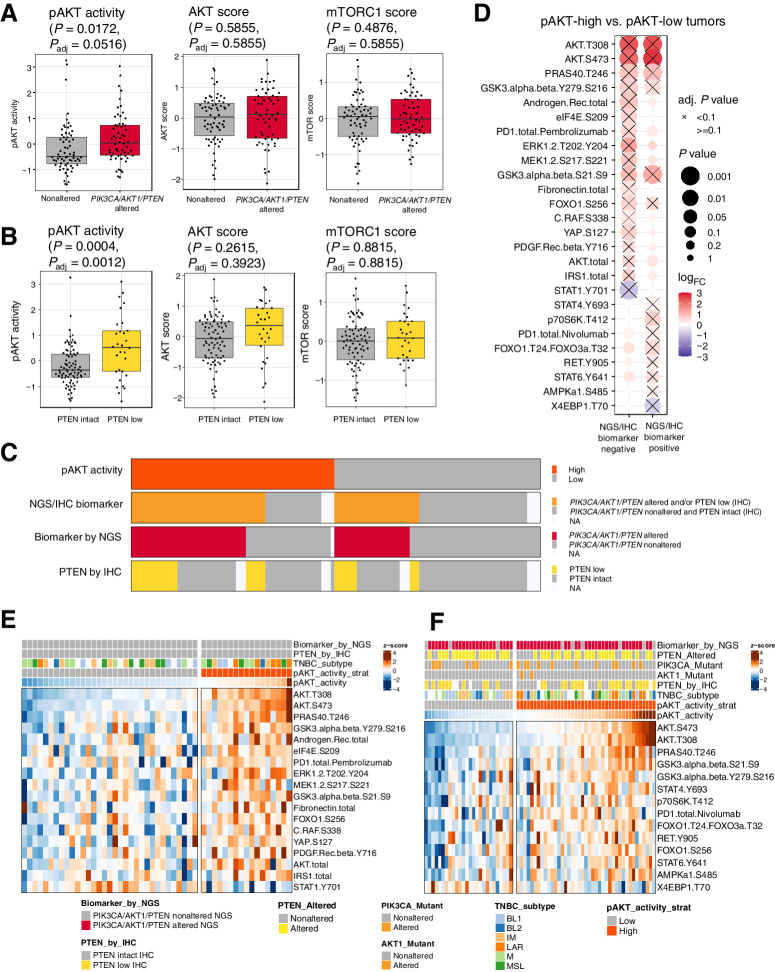
To identify the signaling pathways that were differentially activated in different biomarker subgroups, we performed a differential expression analysis first comparing PIK3CA/AKT1/PTEN genomically altered tumors versus PIK3CA/AKT1/PTEN nonaltered tumors. Of all endpoints measured, AKT (T308 and S473) and PTEN (S380 and total) were significantly higher or lower [P value < 0.05, log2 fold change (FC) > 0.5 or log2FC < −0.5] in PIK3CA/AKT1/PTEN-altered tumors (Fig. 2A; Supplementary Fig. S3A), whereas low PTEN expression levels were observed specifically among the PTEN-altered tumors (Supplementary Fig. S3B). While AKT phosphorylation (T308 and S473) levels were elevated at a population level, the total AKT levels were not significantly altered in PIK3CA/AKT1/PTEN-altered tumors (Supplementary Fig. S3B), consistent with posttranslational modification of AKT by activating oncogenic PIK3CA/AKT1/PTEN alterations. Consistently, AKT (T308 and S473) and PTEN (S380 and total) also showed a similar pattern comparing PTEN low versus PTEN intact tumors assessed by IHC (Fig. 2B; Supplementary Fig. S3C). On the other hand, although some AKT substrates showed a trend of elevation, the integrated AKT pathway score was not significantly higher in PIK3CA/AKT1/PTEN-altered (NGS) or PTEN-low (IHC) tumors (Fig. 2A and B).
Figure 2.
pAKT levels are modulated by PI3K/AKT pathway alterations and additional cell signaling activities. Boxplot showing the levels of pAKT activity, AKT score, and mTORC1 scores in PIK3CA/AKT1/PTEN-nonaltered compared with -altered tumors assessed by NGS in A, and in PTEN-intact compared with PTEN-loss tumors assessed by IHC in B. C, Tile plot showing the overlap of pAKT activity, the status of PIK3CA/AKT1/PTEN alteration by NGS, and PTEN expression by IHC. D, Differential expression analysis between pAKT-high and pAKT-low tumors, stratified by the status of PI3K/AKT pathway biomarkers assessed by NGS and IHC. NGS/IHC biomarker negative: PIK3CA/AKT1/PTEN nonaltered and PTEN intact; NGS/IHC biomarker positive: PIK3CA/AKT1/PTEN altered and/or PTEN low. Heat maps showing the differentially expressed endpoints comparing pAKT-high and pAKT-low tumors (adjusted P value < 0.1), in NGS/IHC biomarker–negative (E) and NGS/IHC biomarker–positive (F) samples, respectively. pAKT_activity_strat, stratifying pAKT activity using a median cut point. TNBC subtypes include: basal-like 1 (BL1); basal-like 2 (BL2); immunomodulatory (IM); LAR; mesenchymal (M); mesenchymal stem–like (MSL). Adj., adjusted.
Despite the elevated pAKT levels, these tumors did not display significantly elevated levels of downstream mTORC1 activities (Fig. 2A and B). This is likely due to the multiple additional signaling inputs (e.g., cellular nutrient and energy levels) that also modulate the activity of mTORC1 (26–28), resulting in the differential activities of AKT and mTORC1.
We then focused on the association of genomic PIK3CA/AKT1/PTEN alterations directly with activation/phosphorylation of the ipatasertib drug target AKT. We dichotomized pAKT levels into pAKT-high or pAKT-low using a median cut point. Among the pAKT-high and pAKT-low samples, 71% and 44% have alterations in the PIK3CA/AKT1/PTEN NGS/IHC biomarkers, respectively (Fig. 2C).
To further understand mechanisms beyond PIK3CA/AKT1/PTEN genomic/protein alteration(s) that might impact the pAKT levels, we examined other protein or phosphoprotein markers associated with the pAKT levels, stratified by the status of PIK3CA/AKT1/PTEN NGS/IHC biomarkers (Fig. 2D). Among tumors without genomic/protein alterations in PIK3CA/AKT1/PTEN (NGS/IHC biomarker negative), we found an enrichment of multiple markers, including IRS1 total, PDGFR β Y716, CRAF S338, MEK1/2 S217/S221, ERK1/2 T202/Y204, AKT total, and androgen receptor (AR) among pAKT-high samples (Fig. 2D and E). The samples with high levels of these markers were not entirely overlapping (Fig. 2E), indicating possible heterogeneous mechanisms underlying the high levels of pAKT. For example, the high levels of phosphorylated MEK (pMEK)/phosphorylated ERK (pERK) may indicate a common upstream signal, possibly an RTK, that activates both the PI3K/AKT and MAPK downstream signaling activities. On the other hand, a subset of samples might have high pAKT levels due to an upregulation of total AKT rather than posttranslational modification. Lastly, the high AR expression observed in a subset of pAKT-high samples and its enrichment in the luminal AR (LAR) subtype prompted us to assess the potential association of TNBC subtypes (defined by RNA-seq) and pAKT levels. There was an enrichment of LAR subtypes among the pAKT-high samples regardless of the status NGS/IHC biomarkers (Supplementary Fig. S4), indicating a potential association between LAR subtype and PI3K/AKT pathway activities beyond known enrichment of PI3KCA/AKT1 alterations among LAR subtype (30). On the other hand, among the NGS/IHC biomarker positive samples, we found a different set of markers, including RET Y905 (Fig. 2D and F), that were enriched among the pAKT-high samples. This suggests that even in the tumors with genomic/protein alterations in the PIK3CA/PTEN/AKT1 biomarkers assessed by NGS/IHC, the pAKT levels are still impacted by many other factors, such as activation by RTK signaling activities as an example. Additionally, intratumor heterogeneity can also play a role: among samples with alterations in PIK3CA/AKT1, the normalized allele frequency of PIK3CA/AKT1 mutations appear to be lower among the pAKT-low compared with pAKT-high tumors (Supplementary Fig. S5).
pAKT levels are associated with enriched clinical benefit of ipatasertib
Next, to understand which protein signaling biomarker(s) were associated with the tumor response to ipatasertib treatment, we first focused on the several PI3K/AKT-related biomarkers measured by RPPA.
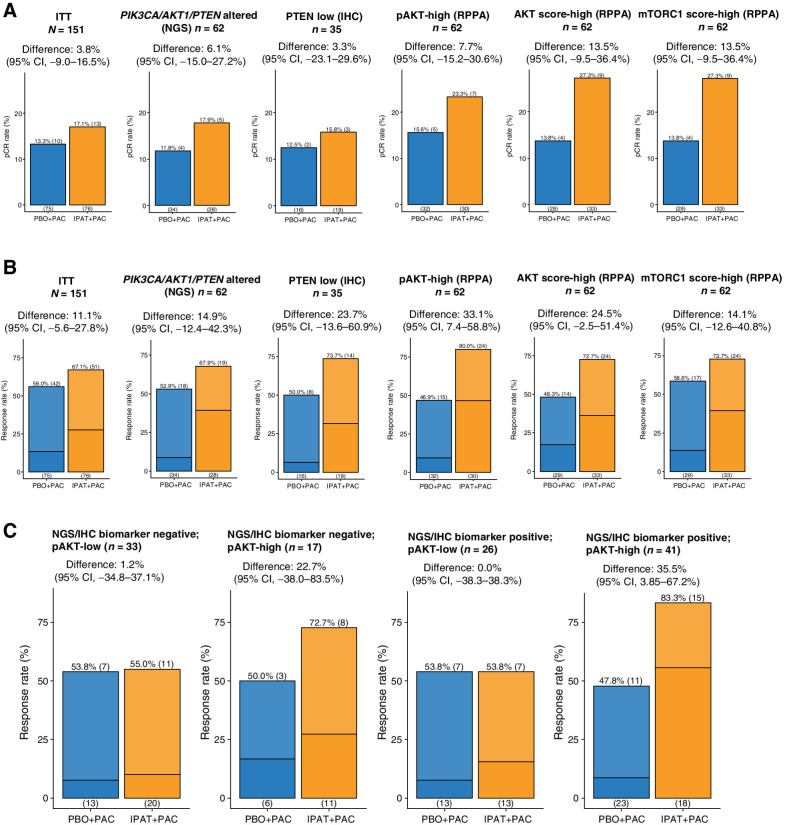
The pCR rate with paclitaxel alone in FAIRLANE was lower than typically observed after neoadjuvant chemotherapy in TNBC (31–33), likely due to the absence of anthracycline and cyclophosphamide. Despite the numeric increase in pCR among the pAKT-high, AKT score–high, and mTORC1 score–high subgroups, the difference between the two arms was not statistically significant (Fig. 3A), which could be at least partially due to the very low pCR rate in the control arm.
Figure 3.
High pAKT levels are associated with enriched benefit of ipatasertib treatment. A, Bar plot showing the pCR in the IPAT+PAC versus PBO+PAC arms, among different biomarker-selected patient subpopulations. B, Bar plot showing the ORR (MRI) in the IPAT+PAC versus PBO+PAC arms, among different biomarker-selected patient subpopulations. Darker color indicates CR and lighter color indicates PR. C, Analysis showing the ORR (MRI) in the pAKT-low and pAKT-high, among the NGS/IHC biomarker subgroups. NGS/IHC biomarker negative: PIK3CA/AKT1/PTEN nonaltered and PTEN intact; NGS/IHC biomarker positive: PIK3CA/AKT1/PTEN altered and/or PTEN low. 95% CI, 95% confidence interval; ITT, intention-to-treat.
We then examined tumor ORR by MRI and found that the pAKT-high but not mTORC1 score–high subgroup showed a significantly higher ORR with ipatasertib (Fig. 3B). The AKT score–high group also exhibited a trend of higher ORR with ipatasertib, however it was not statistically significant (Fig. 3B). In contrast, the level of total AKT was not associated with enriched benefit of ipatasertib (Supplementary Fig. S6). We also examined the change in sum of the longest diameters (SLD) of the target lesions, stratified by the levels of pAKT. Consistently, only the pAKT-high but not the pAKT-low subgroup showed a significantly deeper decrease in SLD with ipatasertib (Supplementary Fig. S7).
We continued to ask whether the enriched benefit of ipatasertib among pAKT-high subgroup was actually attributed to the genomic/protein alterations in PI3K/AKT components. To this end, we analyzed the ORR among the four subgroups with the two biomarkers (PIK3CA/AKT1/PTEN NGS/IHC biomarker status and pAKT level): NGS/IHC biomarker negative, pAKT-low (n = 33); NGS/IHC biomarker negative, pAKT-high (n = 17); NGS/IHC biomarker positive, pAKT-low (n = 26); and NGS/IHC biomarker positive, pAKT-high (n = 41). Remarkably, increased ORR with ipatasertib was observed in the two pAKT-high subgroups regardless of the NGS/IHC biomarker status (Fig. 3C). The NGS/IHC biomarker negative, pAKT-high subgroup had a small sample size (n = 17) and did not reach statistical significance despite the clear trend. In contrast, pAKT-low subgroups showed no difference in ORR between the two arms, even with the NGS/IHC biomarker–positive subgroup (Fig. 3C). Taken together, these findings suggest that high pAKT levels are ultimately associated with the ipatasertib benefit, independently of the presence of PI3KCA/AKT1/PTEN genomic alterations or PTEN IHC status.
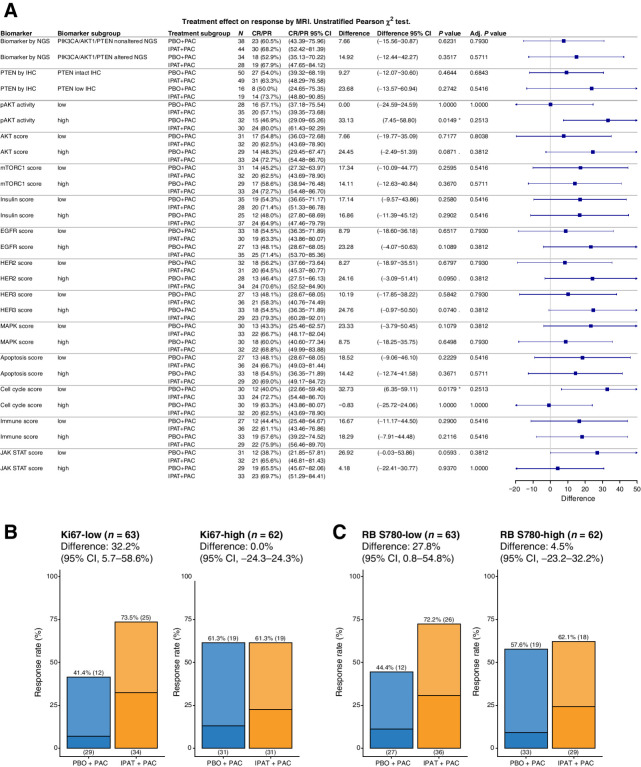
Ipatasertib increases the response rate of slow-dividing tumors
To explore what additional biomarkers are associated with the clinical benefit of ipatasertib, we asked what biomarker subgroup(s) could have a significantly higher ORR in the IPAT+PAC arm compared with the PBO+PAC arm. Among all the signaling modules evaluated, pAKT-high and cell cycle score–low subgroups showed significantly higher ORR when ipatasertib was added to the treatment (Fig. 4A). Of note, there was no obvious association between the cell cycle score and pAKT levels or PIK3CA/AKT1/PTEN NGS/IHC biomarker status, indicating that their associations with ipatasertib treatment benefit were independent (Supplementary Fig. S8). Low levels of Ki-67 and RB S780, two markers composed of the cell cycle score, were both associated with lower ORR in the PBO+PAC arm (Fig. 4B and C). Since paclitaxel can kill tumor cells as a consequence of mitotic arrest through stabilization of microtubules, slow-dividing tumors might be less sensitive to this mode of action. On the other hand, the low ORR in the cell cycle–low subgroup was rescued by the addition of ipatasertib to the treatment (Fig. 4B and C), suggesting that at least a subset of these tumors is still vulnerable to the inhibition of AKT signaling.
Figure 4.
Ipatasertib increases the response rate of slow-dividing tumors. A, Forest plot showing the improvement in ORR with ipatasertib treatment among different biomarker subgroup(s). Signaling scores calculated from RPPA was dichotomized into the low and high subgroups using a median cut point. Pearson χ2 test was used to assess the difference in ORR with 95% confidence intervals (CI), comparing IPAT+PAC with PBO+PAC. (*) P value < 0.05, (.) 0.05 < P value <0.1. Bar plot showing the ORR (MRI) in the IPAT+PAC versus PBO+PAC arms, among Ki67-low and Ki-67 subgroups (B) or RB S780-low and RB S780-high subgroups (C). Darker color indicates CR and lighter color indicates PR. Adj., adjusted.
Although not statistically significant, tumors with HER2-high or HER3-high scores also showed a trend of improved ORR in the IPAT+PAC arm compared with the PBO+PAC arm (Fig. 4A). Activation of HER family phosphoproteins among a subset of TNBC tumors has been reported to be associated with response to neratinib treatment (19). These tumors might also be sensitized to the inhibition of AKT which acts downstream of HER2 family RTKs. Furthermore, JAK/STAT-low but not JAK/STAT-high tumors tend to be associated with higher ORR when ipatasertib was added to the treatment (Fig. 4A). Consistent with this observation, it has been reported that a JAK2/STAT5-evoked feedback loop can dampen the efficacy of PI3K/mTOR inhibition (34). Therefore, cotargeting both pathways might be a therapeutic strategy to maximize the treatment efficacy.
Ipatasertib treatment leads to robust downregulation of AKT and mTORC1 signaling activities
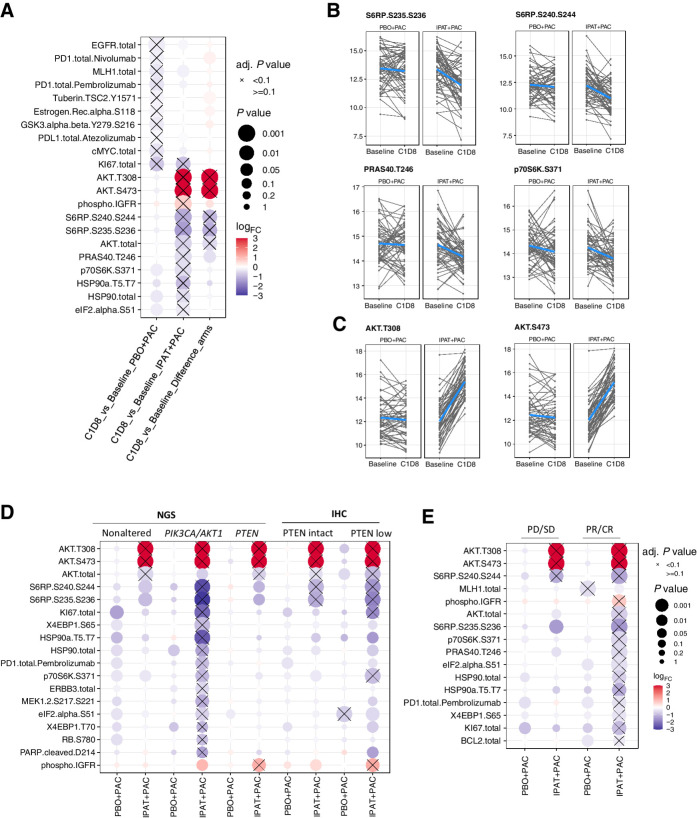
On-treatment changes in signaling activities were evaluated in the 110 samples with paired baseline and C1D8 samples evaluable by RPPA (Fig. 1A). Indeed, phosphorylation levels of multiple AKT or mTORC1 signaling components were significantly downregulated with the ipatasertib treatment, including S6RP S235/236 and S6RP S240/244, PRAS40 T246, and p70S6K S371 (Fig. 5A and B). Meanwhile, phosphorylation levels of AKT T308 and AKT S473 were significantly upregulated with ipatasertib treatment (Fig. 5C), consistent with the previous study showing that ipatasertib restricts phosphatase accessibility of AKT (35).
Figure 5.
Pharmacodynamic biomarker analysis showing that ipatasertib treatment leads to downregulation of AKT and mTORC1 activities. A, Pharmacodynamic biomarker analysis showing the endpoints with differential expressions at C1D8 compared with its baseline levels in the PBO+PAC arm, IPAT+PAC arm, and the difference between the two arms. B, Downregulation of S6RP 235/236, S6RP S240/244, PRAS40 T246, and p70S6K S371 levels from baseline to C1D8 in the IPAT+PAC arm. C, Upregulation of AKT T308 and AKT S473 levels from baseline to C1D8 in the IPAT+PAC arm. D, The changes in phosphoprotein markers are more pronounced among the PIK3CA/AKT1/PTEN-altered (NGS) and PTEN-low (IHC) subgroups. E, The changes in phosphoprotein markers are more pronounced among responders (CR or PR) compared with nonresponders (PD or SD). Adj., adjusted; PD, progressive disease; SD, stable disease.
Besides the downregulation of the downstream AKT or mTORC1 signaling activities, we observed a significant upregulation of phosphorylation on the insulin receptor (IR) or IGF-1R (Fig. 5A; Supplementary Fig. S9A), possibly due to the feedback reactivation of the upstream signaling (36–38). Interestingly, robust upregulation of IR/IGF-1R phosphorylation was observed among the responders [patients with complete response (CR) or partial response (PR); Supplementary Fig. S9B and S9C]. This suggests that IR/IGF-1R phosphorylation is not simply a biomarker of resistance, and its effect in conferring resistance could depend on the potency of the drug and levels of pathway reactivation.
We continued to ask whether the degree of signaling inhibition could be more profound among the patients with PIK3CA/ATK1/PTEN NGS or IHC biomarker–positive tumors. Indeed, these biomarker-selected subgroups overall exhibit a more significant downregulation of AKT and mTORC1 activity (Fig. 5D). Furthermore, baseline pAKT-high tumors also showed a more robust downregulation of AKT and mTORC1 activity (Supplementary Fig. S10). Consistently, we also observed that tumors in responders (CR or PR) overall exhibited a stronger level of AKT and mTORC1 signaling inhibition with ipatasertib treatment (Fig. 5E).
Discussion
Driver mutations in key signaling components have been extensively studied over the past few decades, however, the actual association of underpinning genetic alterations and signaling activities have yet to be fully characterized in the clinic. This is partially due to the operational and technical challenges of performing comprehensive multi-omic analysis on small biopsy samples. In the FAIRLANE IPAT+PAC trial, we collected fresh frozen tumor biopsies pre- and post-treatment. This allowed us to perform in-depth molecular profiling at the genomic, transcriptomic, and most importantly, proteomic and phosphoproteomic levels in parallel. We found that tumors with PIK3CA/AKT1/PTEN alterations either by NGS or IHC are indeed associated with higher levels of AKT protein phosphorylation indicative of AKT activation. However, PIK3CA/AKT1/PTEN alterations at the genomic level and PTEN expression status at the protein level only partially explain the functional protein activation/phosphorylation of pAKT. A significant subset (34%, 17/50) of patients had tumors without PIK3CA/AKT1/PTEN genomic/protein alterations yet exhibited pAKT-high measurements and had a significantly increased response rate associated with ipatasertib treatment (73% with IPAT+PAC vs. 50% with PBO+PAC; Fig. 3C). Conversely, another subset (39%, 26/67) of patients had tumors with PIK3CA/AKT1/PTEN genomic/protein alterations but had pAKT-low measurement, and in these patients no increase in response rate was observed when ipatasertib was added to the chemotherapy treatment (54% with IPAT+PAC vs. 54% with PBO+PAC).
We found that multiple additional factors, including the upstream RTK activities, total AKT amount, tumor heterogeneity in the driver mutations, as well as the molecular subtypes (e.g., LAR), all can impact the overall pAKT levels. As a result, the “genotype” (PIK3CA/AKT1/PTEN status) only potentiates, but imperfectly predicts the “phenotype” (pAKT-high), likely due to the additional layers of regulation in AKT activities. Importantly, high pAKT levels rather than the status of PIK3CA/AKT1/PTEN genomic/protein alterations are ultimately associated with the ipatasertib benefit. This corroborates previous findings with preclinical models (7, 39).
Although AKT and mTORC1 are often depicted in the same PI3K-AKT-mTORC1 pathway, many recent studies have shown that this is actually not a simple linear signal transduction cascade (40, 41). Here, we found that the proximal but not the distal biomarker is associated with the alterations in PIK3CA/AKT1/PTEN. Furthermore, the baseline levels of pAKT, as the proximal and most direct marker of AKT itself showed the best predictive value of ipatasertib treatment response.
Our study has several limitations. The relatively small sample size rendered the study underpowered to evaluate several endpoints. The study is retrospective in nature of biomarker analysis and lacks the long-term survival data after the surgery. To further validate the findings from this study, additional exploratory biomarker analysis of the randomized phase III trial IPATunity130 Cohort A (NCT03337724) is ongoing. Additional prospective studies with long-term survival follow-up are also needed to further validate the predictive value of pAKT for ipatasertib treatment.
In conclusion, using pAKT as a potential biomarker in this study, we can identify patients who do not have genomic or protein alterations in PIK3CA/AKT1/PTEN yet respond to ipatasertib, as well as patients who have the alterations in PIK3CA/AKT1/PTEN yet do not benefit from the ipatasertib treatment. This study provides the first proof-of-concept evidence that the baseline functional protein phosphorylation levels of AKT might have predictive value for ipatasertib and may possesses utility for patient selection for AKT inhibitors like ipatasertib.
Supplementary Material
Acknowledgments
This study was sponsored by F. Hoffmann-La Roche Ltd.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
Z. Shi reports other support from Roche/Genentech outside the submitted work. J. Wulfkuhle reports other support from Theralink Technologies outside the submitted work. M. Nowicka reports personal fees from F. Hoffmann-La Roche Ltd during the conduct of the study, as well as personal fees from F. Hoffmann-La Roche Ltd outside the submitted work. C. Saura reports personal fees and other support from AstraZeneca, Daiichi Sankyo, Eisai, Exact Sciences, Novartis, Pfizer, Pierre Fabre, Puma, Roche Farma, and SeaGen, as well as personal fees from Byondis B.V., Exeter Pharma, F. Hoffmann-La Roche Ltd, MediTech, Merck Sharp & Dohme, Philips, PintPharma, Sanofi-Aventis, and Zymeworks outside the submitted work. P.G. Nuciforo reports grants from F. Hoffmann-La Roche Ltd during the conduct of the study, as well as personal fees from Bayer, MSD, and Novartis outside the submitted work. J. Andersen reports personal fees from Genentech, AstraZeneca/Daiichi Sankyo, Puma, Gilead/Immunomedics, SeaGen, Merck, Novartis, Athenex, Biotheranostics, Myriad, and Genomic Health outside the submitted work. J.L. Passos-Coelho reports personal fees from Roche Pharmaceuticals outside the submitted work. M.J. Gil-Gil reports personal fees from Novartis, Daiichi Sankyo, Pfizer, AstraZeneca, and F. Hoffmann-La Roche Ltd outside the submitted work. B. Bermejo reports personal fees from Novartis, F. Hoffmann-La Roche Ltd, Pierre Fabre, and MSD outside the submitted work. E.M. Ciruelos reports personal fees from Roche Farma, Eli Lilly and Company, Novartis, AstraZeneca DS, MSD, and Pfizer outside the submitted work. P. Villagrasa reports other support from GNE during the conduct of the study; P. Villagrasa also reports personal fees from Nanostring, as well as other support from REVEAL GENOMICS S.L. outside the submitted work. M.J. Wongchenko reports other support from Genentech and F. Hoffman-La Roche Ltd outside the submitted work. E.F. Petricoin reports personal fees from Theralink Technologies, Inc. and Perthera, Inc. during the conduct of the study, as well as personal fees from Theralink Technologies, Inc. and Perthera, Inc. outside the submitted work; in addition, E.F. Petricoin has a patent 6,969,614 issued and with royalties paid from The United States of America as represented by the Department of Health and Human Services. M. Oliveira reports grants from Genentech during the conduct of the study. M. Oliveira also reports grants, personal fees, and non-financial support from F. Hoffman-La Roche Ltd, AstraZeneca, and Novartis; grants and personal fees from SeaGen, GlaxoSmithKline, and PUMA Biotechnology; personal fees from Pfizer, Guardant Health, iTEOS, and MSD; grants from Genentech, Immunomedics, Boehringer Ingelheim, Cascadian Therapeutics, Sanofi-Aventis, Piqur, and Zenith Epigenetics; and personal fees and non-financial support from Eisai and Pierre Fabre. S.J. Isakoff reports personal fees from Mylan, Puma, Seattle Genetics, and Novartis, as well as other support from Oncopep, Genentech, Merck, and AstraZeneca outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
Z. Shi: Formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. J. Wulfkuhle: Data curation, investigation, methodology, writing–review and editing. M. Nowicka: Data curation, formal analysis, visualization, methodology, writing–review and editing. R.I. Gallagher: Data curation, writing–review and editing. C. Saura: Conceptualization, supervision, writing–review and editing. P.G. Nuciforo: Writing-review and editing. I. Calvo: Writing-review and editing. J. Andersen: Writing-review and editing. J.L. Passos-Coelho: Supervision, investigation, writing–review and editing. M.J. Gil-Gil: Resources, investigation, writing–review and editing. B. Bermejo: Resources, data curation, supervision, investigation, writing–review and editing. D.A. Pratt: Writing-review and editing. E.M. Ciruelos: Investigation, writing–review and editing. P. Villagrasa: Investigation, project administration, writing–review and editing. M.J. Wongchenko: Conceptualization, supervision, investigation, writing–review and editing. E.F. Petricoin: Conceptualization, supervision, investigation, methodology, writing–original draft, project administration, writing–review and editing. M. Oliveira: Conceptualization, resources, supervision, investigation, writing–review and editing. S.J. Isakoff: Conceptualization, data curation, formal analysis, writing–review and editing.
References
- 1. Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K pathway in human disease. Cell 2017;170:605–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Di Cosimo S, Baselga J. Management of breast cancer with targeted agents: Importance of heterogenicity. Nat Rev Clin Oncol 2010;7:139–47. [DOI] [PubMed] [Google Scholar]
- 3. Janku F, Yap TA, Meric-Bernstam F. Targeting the PI3K pathway in cancer: Are we making headway? Nat Rev Clin Oncol 2018;15:273–91. [DOI] [PubMed] [Google Scholar]
- 4. Kim S-B, Dent R, Im S-A, Espié M, Blau S, Tan AR, et al. . Ipatasertib plus paclitaxel versus placebo plus paclitaxel as first-line therapy for metastatic triple-negative breast cancer (LOTUS): a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol 2017;18:1360–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oliveira M, Saura C, Nuciforo P, Calvo I, Andersen J, Gil MG. FAIRLANE, a double-blind placebo-controlled randomized phase II trial of neoadjuvant ipatasertib plus paclitaxel for early triple-negative breast cancer. Ann Oncol 2019;30:1289–97. [DOI] [PubMed] [Google Scholar]
- 6. Pascual J, Turner NC. Targeting the PI3-kinase pathway in triple-negative breast cancer. Ann Oncol 2019;30:1051–60. [DOI] [PubMed] [Google Scholar]
- 7. Lin J, Sampath D, Nannini MA, Lee BB, Degtyarev M, Oeh J, et al. . Targeting activated Akt with GDC-0068, a novel selective Akt inhibitor that is efficacious in multiple tumor models. Clin Cancer Res 2013;19:1760–72. [DOI] [PubMed] [Google Scholar]
- 8. Saura C, Roda D, Roselló S, Oliveira M, Macarulla T, Pérez-Fidalgo JA, et al. . A first-in-human phase I study of the ATP-competitive AKT inhibitor ipatasertib demonstrates robust and safe targeting of AKT in patients with solid tumors. Cancer Discov 2017;7:102–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Bono JS, De Giorgi U, Nava Rodrigues D, Massard C, Bracarda S, Font A, et al. . Randomized phase II study of Akt blockade with or without ipatasertib in abiraterone-treated patients with metastatic prostate cancer with and without PTEN loss. Clin Cancer Res 2019;25:928–36. [DOI] [PubMed] [Google Scholar]
- 10. de Bono JS, Bracarda S, Sternberg CN, Chi KN, Olmos D, Sandhu S, et al. . LBA4 IPATential150: Phase III study of ipatasertib (ipat) plus abiraterone (abi) vs placebo (pbo) plus abi in metastatic castration-resistant prostate cancer (mCRPC) [abstract]. In: Proceedings of the ESMO Virtual Congress; 2020 Sept 19–Oct 18; Switzerland. Ann Oncol; 2020. [Google Scholar]
- 11. Yuan TL, Cantley LC. PI3K pathway alterations in cancer: Variations on a theme. Oncogene 2008;27:5497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lehmann BD, Pietenpol JA. Clinical implications of molecular heterogeneity in triple negative breast cancer. Breast 2015;24:S36–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yin L, Duan JJ, Bian XW, Yu SC. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res 2020;22:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lehmann BDB, Bauer JaJ, Chen X, Sanders ME, Chakravarthy aB, Shyr Y, et al. . Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121:2750–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SAW, et al. . Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res 2015;21:1688–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu YR, Jiang YZ, Xu XE, Yu K, Da JX, Hu X, et al. . Comprehensive transcriptome analysis identifies novel molecular subtypes and subtype-specific RNAs of triple-negative breast cancer. Breast Cancer Res 2016;18:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pjaweletz CP, Charboneau L, Bichsel VE, Simone NL, Chen T, Gillespie JW, et al. . Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene 2001;20:1981–9. [DOI] [PubMed] [Google Scholar]
- 18. Liotta LA, Espina V, Mehta AI, Calvert V, Rosenblatt K, Geho D, et al. . Protein microarrays: Meeting analytical challenges for clinical applications. Cancer Cell 2003;3:317–25. [DOI] [PubMed] [Google Scholar]
- 19. Wulfkuhle JD, Yau C, Wolf DM, Vis DJ, Gallagher RI, Brown-Swigart L, et al. . Evaluation of the HER/PI3K/AKT family signaling network as a predictive biomarker of pathologic complete response for patients with breast cancer treated with neratinib in the I-SPY 2 TRIAL. JCO Precis Oncol 2018;2:PO.18.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pierobon M, Ramos C, Wong S, Hodge KA, Aldrich J, Byron S, et al. . Enrichment of PI3K-AKT–mTOR pathway activation in hepatic metastases from breast cancer. Clin Cancer Res 2017;23:4919–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baldelli E, Haura EB, Crinò L, Cress DW, Ludovini V, Schabath MB, et al. . Impact of upfront cellular enrichment by laser capture microdissection on protein and phosphoprotein drug target signaling activation measurements in human lung cancer: Implications for personalized medicine. Proteomics Clin Appl 2015;9:928–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mueller C, De Carvalho AC, Mikkelsen T, Lehman NL, Calvert V, Espina V, et al. . Glioblastoma cell enrichment is critical for analysis of phosphorylated drug targets and proteomic-genomic correlations. Cancer Res 2014;74:818–28. [DOI] [PubMed] [Google Scholar]
- 23. Akbani R, Becker KF, Carragher N, Goldstein T, De Koning L, Korf U, et al. . Realizing the promise of reverse phase protein arrays for clinical, translational, and basic research: a workshop report the RPPA (Reverse Phase Protein Array) Society. Mol Cell Proteomics 2014;13:1625–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. . Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Law CW, Chen Y, Shi W, Smyth GK. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014;15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell 2003;115:577–90. [DOI] [PubMed] [Google Scholar]
- 27. Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, et al. . The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 2004;6:91–9. [DOI] [PubMed] [Google Scholar]
- 28. Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell 2006;21:521–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Federici G, Gao X, Slawek J, Arodz T, Shitaye A, Wulfkuhle JD. Systems analysis of the NCI-60 cancer cell lines by alignment of protein pathway activation modules with “-OMIC” data fields and therapeutic response signatures. Mol Cancer Res 2013;11:676–85. [DOI] [PubMed] [Google Scholar]
- 30. Lehmann BD, Bauer JA, Schafer JM, Pendleton CS, Tang L, Johnson KC, et al. . PIK3CA mutations in androgen receptor-positive triple negative breast cancer confer sensitivity to the combination of PI3K and androgen receptor inhibitors. Breast Cancer Res 2014;16:406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sikov WM, Berry DA, Perou CM, Singh B, Cirrincione CT, Tolaney SM, et al. . Impact of the addition of carboplatin and/or bevacizumab to neoadjuvant once-per-week paclitaxel followed by dose-dense doxorubicin and cyclophosphamide on pathologic complete response rates in stage II to III triple-negative breast cancer: CALGB 40603 (Alliance). J Clin Oncol 2015;33:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Von Minckwitz G, Schneeweiss A, Loibl S, Salat C, Denkert C, Rezai M, et al. . Neoadjuvant carboplatin in patients with triple-negative and HER2-positive early breast cancer (GeparSixto; GBG 66): A randomised phase 2 trial. Lancet Oncol 2014;15:747–56. [DOI] [PubMed] [Google Scholar]
- 33. Mittendorf EA, Zhang H, Barrios CH, Saji S, Jung KH, Hegg R, et al. . Neoadjuvant atezolizumab in combination with sequential nab-paclitaxel and anthracycline-based chemotherapy versus placebo and chemotherapy in patients with early-stage triple-negative breast cancer (IMpassion031): a randomised, double-blind, phase 3 trial. Lancet 2020;396:1090–100. [DOI] [PubMed] [Google Scholar]
- 34. Britschgi A, Andraos R, Brinkhaus H, Klebba I, Romanet V, Müller U, et al. . JAK2/STAT5 inhibition circumvents resistance to PI3K/mTOR blockade: a rationale for cotargeting these pathways in metastatic breast cancer. Cancer Cell 2012;22:796–811. [DOI] [PubMed] [Google Scholar]
- 35. Lin K, Lin J, Wu WI, Ballard J, Lee BB, Gloor SL, et al. . An ATP-site on-off switch that restricts phosphatase accessibility of Akt. Sci Signal 2012;5:ra37. [DOI] [PubMed] [Google Scholar]
- 36. Wan X, Harkavy B, Shen N, Grohar P, Helman LJ. Rapamycin induces feedback activation of Akt signaling through an IGF-1R-dependent mechanism. Oncogene 2007;26:1932–40. [DOI] [PubMed] [Google Scholar]
- 37. O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. . mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res 2006;66:1500–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hopkins BD, Pauli C, Xing D, Wang DG, Li X, Wu D, et al. . Suppression of insulin feedback enhances the efficacy of PI3K inhibitors. Nature 2018;560:499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gris-Oliver A, Palafox M, Monserrat L, Braso-Maristany F, Odena A, Sanchez-Guixe M, et al. . Genetic alterations in the PI3K/AKT pathway and baseline AKT activity define AKT inhibitor sensitivity in breast cancer patient-derived xenografts. Clin Cancer Res 2020;26:3720–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, et al. . AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell 2009;16:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sonnenblick A, Venet D, Brohée S, Pondé N, Sotiriou C. pAKT pathway activation is associated with PIK3CA mutations and good prognosis in luminal breast cancer in contrast to p-mTOR pathway activation. NPJ Breast Cancer 2019;5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data has been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001005892.