Abstract
Transcription initiation with ς54-RNA polymerase holoenzyme (ς54-holoenzyme) has absolute requirements for an activator protein and ATP hydrolysis. ς54’s binding to core RNA polymerase and promoter DNA has been well studied, but little is known about its role in the subsequent steps of transcription initiation. Following random mutagenesis, we isolated eight mutant forms of Salmonella typhimurium ς54 that were deficient in transcription initiation but still directed ς54-holoenzyme to the promoter to form a closed complex. Four of these mutant proteins had amino acid substitutions in region I, which had been shown previously to be required for ς54-holoenzyme to respond to the activator. From the remaining mutants, we identified four residues in region III which when altered affect the function of ς54 at some point after closed-complex formation. These results suggest that in addition to its role in core and DNA binding, region III participates in one or more steps of transcription initiation that follow closed-complex formation.
Association of the ς subunit with core RNA polymerase results in a holoenzyme that recognizes specific promoter sequences. Multiple ς factors within a bacterial cell allow the holoenzyme to recognize different classes of promoters (17, 20). Some ς factors are primary ς factors that are responsible for transcription of most of the genes in the cell (e.g., Escherichia coli ς70), while others are alternative ς factors that are required for the expression of specific genes (20). In addition to binding core RNA polymerase and the promoter, ς factors have also been implicated in DNA melting, transcription pausing, and in some cases interactions with activator proteins (17–19, 22, 30). The majority of ς factors exhibit homology to ς70, the only exception being an alternative ς factor, ς54 (23).
ς54-RNA polymerase holoenzyme (ς54-holoenzyme) is responsible for the expression of genes whose products are involved in diverse metabolic processes, such as nitrogen assimilation and fixation, dicarboxylic acid transport, pilin and flagellin synthesis, toluene and xylene catabolism, and hydrogen metabolism (23). ς54-Holoenzyme binds to promoter elements in the −12 and −24 regions to form a closed complex but is unable to form a transcriptionally competent open complex in the absence of an activator protein (25, 28, 32). The activator binds to specific sites upstream of the promoter and makes transient contact with ς54-holoenzyme through DNA looping (31, 34). Protein cross-linking studies suggest that the activator contacts ς54 and the β subunit of ς54-holoenzyme during open-complex formation (19, 40). In addition to making productive contact with ς54-holoenzyme, the activator must also hydrolyze ATP to activate transcription (28, 41).
The role of ς54 in transcriptional initiation following formation of the closed promoter complex is poorly understood. Previous mutational studies of ς54 that were performed to help resolve this issue focused on specific regions of the protein (11, 15, 16, 26, 35–37). In this study, we mutagenized the entire ntrA gene (which encodes ς54) and isolated mutant forms of Salmonella typhimurium ς54 that were defective in transcription initiation but still directed holoenzyme to the promoter. We used a unique genetic screen to assess the ability of ς54 mutants to direct holoenzyme to a promoter that overlapped the phage P22 ant promoter and thereby repress transcription of an ant′-′lacZ reporter gene. Mutant forms of ς54 that retained promoter binding activity were very rare. After screening nearly 1,200 ς54 mutants that were defective in transcription initiation, we found only 8 mutants that repressed transcription of the ant′-′lacZ reporter gene.
MATERIALS AND METHODS
Media and chemicals.
Luria-Bertani broth was used for routine culture growth unless otherwise noted. For a minimal medium, we used either E minimal medium (38) supplemented with 1 mg of acid-hydrolyzed Casamino Acids/liter or M9 minimal medium (24) that contained 10 mM l-arginine as the primary nitrogen source and 50 μM leucine (M9-arginine medium). MacConkey agar was obtained from Difco Laboratories. When l-glutamine was added, it was filter sterilized and then added to autoclaved medium to a final concentration of 5 mM. Ampicillin, chloramphenicol, kanamycin, and tetracycline were added to final concentrations of 200, 20, 50, and 6.5 μg/ml, respectively. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 100 μM.
Bacterial strains.
BL21 (λDE3) [F− ompT (lon) hsdSB gal λDE3:lacI lacUV5-gene 1 (T7 polymerase)] carrying plasmid pLysE, which bears the gene encoding T7 lysozyme, was used for overexpression of histidine-tagged ς54 proteins. TRH107 {Δ(prt proAB)ataP::[(P22 int3 c2-ts29) sieA44 mnt::Kn9 PnifH arc(Am)H1605 ant′-′lacZYA Δ9-a1] ntrA209::Tn10} is a deletion prophage that carries the Sinorhizobium meliloti (formerly Rhizobium meliloti) nifH promoter overlapping the promoter of an ant′-′lacZ fusion (1). TRH107 also carries a Tn10 insertion in ntrA. TRH134 [leu414(Am) hsdL(r− m+) Fels− ΔntrA8455] is a ntrA deletion strain that lacks codons 8 through 455 of ntrA and was constructed as follows.
Plasmid pMK10 is a derivative of pMAK705 (13) which carries the chloramphenicol acetyltransferase (cat) gene and is temperature sensitive for replication. It carries 250 and 750 bp of DNA that flank the 5′ and 3′ ends, respectively, of a truncated version of S. typhimurium ntrA lacking codons 8 through 455. Plasmid pMK10 was introduced into S. typhimurium MS1868 [leuA414(Am) hsdSB(r− m+) Fels−] (10) by electroporation and maintained by growing the resulting transformant at 30°C. Allelic exchange between the partially deleted ntrA gene on pMK10 and the chromosomal copy of ntrA was carried out as described previously (13). A strain with a deletion in the chromosomal ntrA gene (TRH134) was identified by chloramphenicol sensitivity, indicating the loss of the plasmid, and glutamine auxotrophy, indicating the loss of a functional ntrA. To verify that the deletion had been introduced into the chromosomal copy of ntrA, oligonucleotide primers that flanked ntrA were used in a PCR to amplify this region from the chromosomal DNA of strain TRH134. The resulting PCR product was the predicted size for the deletion.
Plasmids.
Plasmid pSA4 carries the S. typhimurium ntrA gene under the control of the E. coli lac promoter and operator along with lacIq (1). Plasmid pMK10, which was used to construct TRH134, was made as follows. Plasmid pJES82 (27) carries the S. typhimurium ntrA gene along with 250 and 750 bp of flanking DNA at the 5′ and 3′ ends of ntrA, respectively. Plasmid pJES82 was digested with BlpI and AscI, filled in with T4 DNA polymerase, and then religated to form pMK9. This resulted in the deletion of codons 8 through 455 of ntrA. A 1.2-kb EcoRI-HindIII fragment from pMK9, which had been filled in at the EcoRI site with T4 DNA polymerase, was cloned into the HincII and HindIII sites of pMAK705, resulting in plasmid pMK10. Plasmid pMK11, a derivative of pALTER-1 (Promega), was used for site-directed mutagenesis experiments. Plasmid pMK11 was constructed by cloning a 2.5-kb EcoRI-HindIII fragment from pSA4 that carried lacIq and ntrA under the control of the lac promoter and operator into the EcoRI and HindIII sites of pALTER-1. Plasmid pJES937 (supplied by S. Kustu) is a derivative of pET-28a(+) (Novagen) and carries the S. typhimurium ntrA gene fused at the 5′ end to a sequence encoding six histidine residues. Expression of this fusion protein was under the control of a T7 promoter. Selected ntrA alleles were subcloned as 1.3-kb NdeI-AscI fragments into the same sites of the ntrA allele on pJES937.
Generation and isolation of ς54 mutants.
Random mutagenesis of ntrA was carried out by either PCR or spontaneous mutagenesis in Epicurian Coli XL1-Red competent cells (Stratagene). For PCR mutagenesis, we used pJES82, which carries ntrA, as the template and the same primers and cloning strategy as described for the construction of pSA4 (1). We relied on the inherent error frequency of Taq DNA polymerase (Promega) for the introduction of mutations into ntrA. Thirty cycles were carried out, using a regime that included a denaturation temperature of 94°C, an annealing temperature of 45°C, and an elongation temperature of 72°C. Reaction mixtures contained the buffer provided with the enzyme, supplemented with 2 mM MgCl2. For generating spontaneous mutations, the Epicurian Coli XL1-Red strain was used as described by the supplier. Plasmid pSA4 was transformed into the mutator strain, and the resulting transformants were grown in Luria-Bertani broth or E minimal medium supplemented with ampicillin and l-glutamine. The cultures were subcultured twice in the same medium, and plasmid DNA was isolated.
For both methods, mutagenized plasmids were transformed into TRH107 by electroporation. The resulting transformants were plated on E minimal medium supplemented with l-glutamine and the appropriate antibiotics. Two colony sizes were observed on this medium. Approximately 10% of the smaller colonies were glutamine auxotrophs, while <0.2% of the larger colonies were glutamine auxotrophs. Glutamine auxotrophy, indicative of strains that lacked a functional ς54, was identified by patching colonies onto E minimal medium. These glutamine auxotrophs were screened on MacConkey agar supplemented with appropriate antibiotics and IPTG to induce overexpression of the plasmid-borne ntrA alleles. Strains which produced mutant ς54 proteins that repressed transcription of the ant′-′lacZ reporter gene yielded white or pale colonies (Lac−) on MacConkey agar.
Site-directed mutagenesis was done by using the Altered Sites II in vitro mutagenesis system as described by the supplier (Promega). Plasmid pMK11 was used as the template for these experiments. The ntrA alleles were subcloned into pSA4 so that the plasmid copy numbers were comparable to those of the other mutagenized plasmids.
Sequencing of ntrA alleles.
The ntrA alleles present on derivatives of pSA4 and pMK11 were sequenced with the primers 5′-GTGTGGAATTGTGAG-3′, 5′-CATTCAGCGTTTTGAT-3′, and 5′-GCCGTAACGACACGCT-3′. The first primer is complementary to a sequence within the lac promoter region, while the last two primers are complementary to sequences within S. typhimurium ntrA. DNA sequencing was done at the Molecular Genetics Instrumentation Facility at the University of Georgia.
β-Galactosidase assays.
To assess the degree to which the mutant ς54 proteins repressed transcription of the ant′-′lacZ reporter gene, β-galactosidase activities were determined in TRH107 as described previously (1). Mutant forms of ς54 were overexpressed by induction with IPTG. For each mutant, at least three separate assays were carried out, and activities were expressed as Miller units (24).
Glutamine synthetase assays.
Glutamine synthetase activities were determined by the γ-glutamyltransferase assay as described previously (2). Cultures of strain TRH134 bearing plasmids with the various ntrA alleles were grown to mid-log phase in a modified E minimal medium that lacked sodium ammonium phosphate and was supplemented with acid-hydrolyzed Casamino Acids, 1 mM l-glutamine, and 100 μM IPTG to induce the expression of ntrA. Cells were permeabilized by including hexadecyltrimethylammonium bromide in the assay buffer as described elsewhere (2). Protein concentrations were determined from whole cells by Lowry protein assays with bovine serum albumin as a standard (21). Glutamine synthetase activities were expressed as micromoles of γ-glutamyl hydroxymate produced per minute per milligram of protein, and all assays were done at least twice.
Purification of N-terminally histidine-tagged ς54 proteins.
Selected histidine-tagged ς54 proteins were overexpressed in BL21 (DE3) by induction with 1 mM IPTG. Cells were harvested after a 3-h induction period, resuspended in 50 mM Tris-acetate (pH 8.2)–200 mM KCl–1 mM EDTA–1 mM dithiothreitol (breakage buffer), and lysed in a French press cell at 12,000 lb/in2. Following centrifugation at 12,400 × g for 40 min, a majority of each histidine-tagged protein was in the insoluble fraction. Pellets were washed with a solution containing 1 M NaCl and 1% Triton X-100, after which the histidine-tagged ς54 proteins were solubilized in 50 mM Tris-HCl (pH 8.0)–50 mM NaCl–0.1 mM EDTA–1 mM dithiothreitol–5% glycerol–1% sarkosyl as described previously (7). Solubilized proteins were loaded onto an Ni-nitrilotriacetic acid resin column (Qiagen) and eluted with 250 mM imidazole. For storage, purified proteins were dialyzed against 50 mM Tris-HCl (pH 8.0)–0.5 M NaCl–0.1 mM EDTA–1 mM dithiothreitol–50% glycerol.
Gel mobility shift assays.
The binding of mutant forms of ς54-holoenzyme to the Sinorhizobium meliloti nifH promoter was assessed by a modification of the method described by Guo and Gralla (12). Oligonucleotides that covered the −9 through −29 region of the Sinorhizobium meliloti nifH promoter (5′-GGCTGGCACGACTTTTGCACG-3′, 5′-GGCTGGCACGACTTTTGC-3′, and 5′-CGTGCAAAAGTCGTGCCAGCC-3′) were used. Two different DNA probes were generated from these oligonucleotides for the binding assays. One probe consisted of 21 bp of double-stranded DNA. The second probe had 18 bp of double-stranded DNA plus a 3-base 5′ overhang of the template strand which corresponded to residues −9 through −11. For each probe, the template strand was labeled with [γ-32P]ATP and polynucleotide kinase before being annealed to the respective nontemplate strand. Binding reaction mixtures contained 300 nM core RNA polymerase (Epicentre Technologies), 600 nM histidine-tagged ς54, and 5 nM DNA probe in a solution consisting of 50 mM HEPES-HCl (pH 7.9), 100 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 0.05 μg of bovine serum albumin/ml, 2.8% polyethylene glycol 8000, and 6 μg of sonicated calf thymus DNA/ml. Reaction mixtures were incubated on ice for 30 min, loaded onto a chilled 5% native polyacrylamide gel, and then subjected to electrophoresis at 10 V/cm for 2 h. DNA bands were visualized by exposing the gels to X-ray film.
RESULTS
Isolation of mutant forms of S. typhimurium ς54.
Expression of glutamine synthetase (encoded by glnA) in enteric bacteria is regulated from two promoters. The major promoter, glnAp2, is ς54 dependent and requires the nitrogen-regulatory protein C (NtrC) as an activator; the minor promoter, glnAp1, is ς70 dependent and subject to catabolite repression (14, 29). Strains that lack ς54 are glutamine auxotrophs, since they are unable to initiate transcription from glnAp2. Utilization of several forms of nitrogen, including arginine, is also dependent on ς54 and NtrC (29).
S. typhimurium TRH107 failed to grow on E minimal medium supplemented with acid-hydrolyzed Casamino Acids, since this medium is deficient in glutamine. Introduction of plasmid pSA4, which carries a copy of S. typhimurium ntrA under the control of the lac promoter and operator, allowed this strain to grow on E minimal and M9-arginine media. IPTG induction of ntrA from pSA4 was not necessary to allow growth on either of these media. The ntrA gene carried on pSA4 was subjected to random mutagenesis, and the resulting plasmids were introduced into TRH107. Transformants were screened for glutamine auxotrophy on E minimal medium to identify ς54 mutants that had reduced activity at glnAp2. Nearly 1,200 glutamine auxotrophs were isolated in this screen.
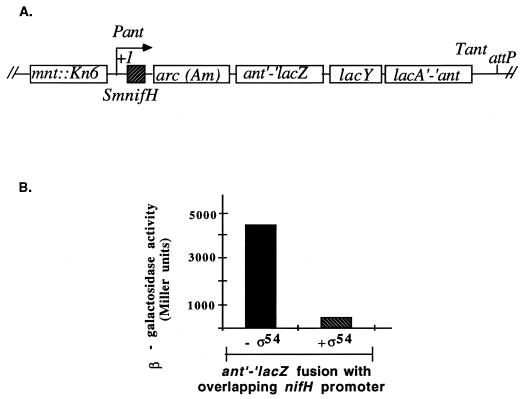
TRH107 carries a partially deleted P22 prophage bearing an ant′-′lacZ reporter gene with the ς54-dependent Sinorhizobium meliloti nifH promoter overlapping the ant promoter (Fig. 1A). Overexpression of ς54 from pSA4 in this strain allowed ς54-holoenzyme to repress transcription of the ant′-′lacZ reporter gene (Fig. 1B). Therefore, the binding of ς54-holoenzyme to the nifH promoter in vivo could be easily assessed by examining the Lac phenotype of the strain.
FIG. 1.
Repression of the ant′-′lacZ fusion in S. typhimurium by ς54. (A) The P22 prophage deletion within TRH107 carries the overlapping Pant and Sinorhizobium meliloti nifH (SmnifH) promoters and an ant′-′lacZ fusion. The GC doublet of the nifH promoter is at position +4 relative to the transcriptional start site of Pant. (B) IPTG induction of wild-type ntrA from pSA4 represses expression from the ant′-′lacZ reporter gene, indicative of ς54-holoenzyme binding to the Sinorhizobium meliloti nifH promoter. In TRH107, overexpression of ς54 caused a ∼10-fold repression relative to TRH107 that lacked ς54.
The glutamine auxotrophs isolated as described above were streaked on MacConkey agar supplemented with IPTG to induce expression of the ntrA alleles. This allowed us to assess the abilities of the mutant forms of ς54-holoenzyme to repress transcription from the ant′-′lacZ reporter gene. Only eight of the ς54 mutants repressed transcription when overexpressed, as indicated by white or pale-colored colonies. Plasmids from these strains were isolated and designated pNTRA1 to pNTRA8.
Given that we overexpressed the ς54 mutants to examine their abilities to repress transcription from the ant′-′lacZ reporter gene, we reexamined the phenotypes of the strains overexpressing the mutant ς54 proteins. Strains that carried pNTRA2, pNTRA3, or pNTRA5 grew as well as the strain that overexpressed wild-type ς54 on E minimal medium supplemented with IPTG (Table 1). These three strains, however, did exhibit growth defects on M9-arginine medium supplemented with IPTG, indicating that the mutant ς54 proteins did indeed have reduced activities. Strains that carried pNTRA1 or pNTRA4 grew on E minimal medium supplemented with IPTG, but not as well as the strain with wild-type ς54. Neither of these strains grew on M9-arginine medium supplemented with IPTG. Strains that carried pNTRA6, pNTRA7, or pNTRA8 failed to grow on either E minimal or M9-arginine medium supplemented with IPTG.
TABLE 1.
ς54 mutants and their affects on growth
| Plasmid | Growth phenotype on mediuma:
|
Amino acid substitution(s)b | |
|---|---|---|---|
| E + IPTG | M9 + Arg + IPTG | ||
| None | − | − | |
| pSA4 (wild type) | +++ | +++ | |
| Initial isolates | |||
| pNTRA1 | ++ | − | L37P |
| pNTRA2 | +++ | + | L46P |
| pNTRA3 | +++ | + | L46P |
| pNTRA4 | + | − | L333P |
| pNTRA5 | +++ | + | E32K G189V |
| pNTRA6 | − | − | L124P V148A |
| pNTRA7 | − | − | W126R L199P D225E D231G I428V |
| pNTRA8 | − | − | E42G I56T G189SL235W F318S |
| Derivatives of pNTRA5 | |||
| pNTRA9 | +++ | +++ | E32K |
| pNTRA10 | +++ | +++ | G189V |
| Derivatives of pNTRA6 | |||
| pNTRA11 | +++ | − | L124P |
| pNTRA12 | +++ | +++ | V148A |
| Derivatives of pNTRA7 | |||
| pNTRA13 | +++ | +++ | W126R |
| pNTRA14 | +++ | +++ | D225E D231G I428V |
| pNTRA15 | − | − | L199P D225E D231G I428V |
| pNTRA16 | +++ | +++ | W126R D225E D231G I428V |
| pNTRA17 | +++ | +++ | L199P |
| pNTRA18 | − | − | L199P D231G |
| pNTRA19 | +++ | +++ | D231G |
| Derivatives of pNTRA8 | |||
| pNTRA20 | +++ | +++ | E42G I56T G189S |
| pNTRA21 | +++ | +++ | L235W F318S |
| pNTRA22 | +++ | +++ | E42G I56T L235W F318S |
| pNTRA23 | +++ | +++ | G189S |
| pNTRA24 | +++ | +++ | F318S |
| pNTRA25 | +++ | +++ | G189S F318S |
Growth phenotypes are as follows: +++, wild-type growth; ++, slower growth, but colonies eventually attain wild-type size; +, poor growth with small colonies; and −, no growth.
Amino acids which are identical or similar in at least 26 or 29 ς54 proteins from various bacteria are underlined.
Analysis of mutant ntrA alleles.
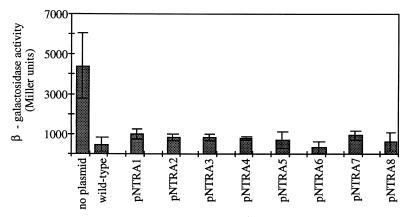
The degrees to which the eight ς54 mutants repressed transcription from the ant′-′lacZ reporter gene were assessed by assaying β-galactosidase (LacZ) activities. For these assays, the ς54 mutants were overexpressed by including IPTG in the culture medium. When wild-type ς54 was overexpressed from pSA4 in TRH107, there was a ∼10-fold repression of the ant′-′lacZ reporter gene relative to a strain that lacked ς54 (Fig. 1 and 2). When the mutant forms of ς54 were overexpressed in TRH107, equivalent levels of repression of the ant′-′lacZ reporter gene were observed (Fig. 2).
FIG. 2.
Repression of the ant′-′lacZ fusion in S. typhimurium by mutant ς54 proteins. β-Galactosidase activities were determined in TRH107 carrying the plasmids indicated. Values are averages of at least three assays. Error bars show the standard deviations for the data sets. no plasmid, control (activity for TRH107 in the absence of a plasmid-borne ntrA allele); wild-type, activity for TRH107 with the wild-type ntrA allele on pSA4. The plasmids and the ntrA alleles that they carry are indicated in Table 1.
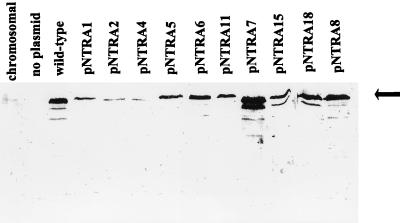
We verified that the eight ς54 mutants were stably expressed in TRH107 by immunoblotting with antiserum directed against S. typhimurium ς54. All eight ς54 mutant proteins were overexpressed, although the levels of some of the proteins were not as high as that of wild-type ς54 (Fig. 3). All of the ς54 mutant proteins accumulated to levels that were higher than the level of protein expressed from the chromosomal copy of ntrA (Fig. 3). The mutant proteins expressed from pNTRA2 and pNTRA4, however, accumulated to levels only a fewfold higher than that of ς54 expressed from the chromosomal copy of ntrA.
FIG. 3.
Overexpression of the mutant ς54 proteins. The mutant and wild-type ς54 proteins, indicated by the arrow, were overexpressed in TRH134. Cultures were grown to mid-log phase, and then expression from ntrA was induced for 3 h with 100 μM IPTG. Whole-cell extracts were analyzed by immunoblotting with antiserum directed against S. typhimurium ς54. no plasmid, TRH134 without any plasmid; wild-type, TRH134 bearing pSA4; chromosomal, S. typhimurium MS1868, the parental strain of TRH134, which contains a chromosomal copy of ntrA but no plasmid-borne copy of the gene. The plasmids and the ntrA alleles that they carry are indicated in Table 1.
All eight ntrA alleles were sequenced to identify the mutations. Four of the ntrA alleles had single missense mutations, two alleles had two missense mutations, and two alleles had five missense mutations (Table 1). The deduced amino acid sequences of the ntrA alleles revealed that the alleles carried on plasmids pNTRA2 and pNTRA3 had the same amino acid substitution, a proline for the leucine at position 46 (L46P). These two mutant plasmids were isolated independently of each other. Of the 16 different positions with amino acid substitutions in the mutant proteins, 10 positions were either identical or similar in at least 26 of 29 ς54 proteins from various bacteria.
To quantitate the activities of the ς54 mutants at glnAp2, we measured glutamine synthetase activities in vivo. We used the γ-glutamyltransferase assay for glutamine synthetase, which measures the conversion of glutamine and hydroxylamine to γ-glutamyl hydroxymate (2). Glutamine synthetase is regulated by adenylylation, but the γ-glutamyltransferase activities of both the adenylylated and unadenylylated forms of the enzyme are supported in the presence of Mn2+. Glutamine synthetase activities were measured at pH 7.15, a pH at which all forms of the enzyme have equivalent activities in the presence of 0.3 mM Mn2+ (33). This allowed us to compare the total amounts of glutamine synthetase produced in the various strains. Cultures were grown under nitrogen-limited conditions in a modified E minimal medium that lacked sodium ammonium phosphate and contained 1 mM l-glutamine, the lowest concentration of glutamine which permitted growth of the glutamine auxotrophs. IPTG was included in the medium for overexpression of ς54.
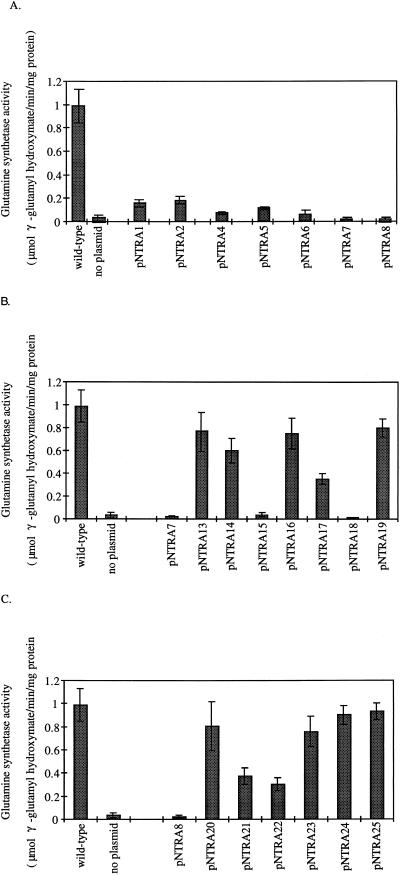
The glutamine synthetase activity of the strain that overexpressed wild-type ς54 was 0.988 μmol of γ-glutamyl hydroxymate produced/min/mg of protein (Fig. 4). The glutamine synthetase activity in TRH107 in the absence of ς54 was 0.034 μmol of γ-glutamyl hydroxymate produced/min/mg of protein. This background level of glutamine synthetase activity represented the level of expression from the ς70-dependent glnAp1 promoter. Strains carrying mutant plasmids that allowed growth on E minimal medium supplemented with IPTG (pNTRA1, pNTRA2, pNTRA4, and pNTRA5) had glutamine synthetase activities that ranged from 8 to 19% of that observed for the strain carrying pSA4. Strains carrying mutant plasmids that did not allow growth on E minimal medium even in the presence of IPTG (pNTRA6, pNTRA7, and pNTRA8) had glutamine synthetase activities that ranged from 2 to 7% of that observed for the strain carrying pSA4.
FIG. 4.
Glutamine synthetase activities of selected strains that overexpress ς54 mutants. The plasmids and the ntrA alleles that they carry are indicated in Table 1. Assays were done at least twice, and error bars show the standard deviations for the data sets. (A) The ntrA alleles originally isolated. (B) The pNTRA7-borne allele and the derivatives of this mutant allele. (C) The pNTRA8-borne allele and the derivatives of this mutant allele. no plasmid, strain TRH134 without a plasmid-borne ntrA allele; wild-type, TRH134 with the wild-type ntrA allele on pSA4.
Analysis of ntrA alleles with multiple mutations.
For each of the ς54 mutants with multiple amino acid substitutions, we wanted to determine if a single amino acid substitution or a combination of substitutions was responsible for the loss of function. For the mutants with two amino acid substitutions, E32K G189V (encoded on pNTRA5) and L124P V148A (encoded on pNTRA6), we separated the mutations by subcloning.
Separation of the two mutations on pNTRA5 by subcloning yielded plasmids pNTRA9 and pNTRA10 (Table 1). When the last two plasmids were introduced into TRH107, the growth phenotypes were identical to those of the strain with pSA4. Since the double mutant was overexpressed at a level comparable to that of the wild-type ς54 but allowed only poor growth on M9-arginine medium, the E32K and G189V substitutions appear to act synergistically to disrupt the function of ς54.
The two mutations on pNTRA6 were similarly separated by subcloning, yielding plasmids pNTRA11 and pNTRA12 (Table 1). When pNTRA12 was introduced into TRH107, the strain exhibited wild-type growth on both E minimal and M9-arginine media. In contrast, the growth phenotype of TRH107 carrying pNTRA11 was similar to that of the strain carrying pNTRA6. Consistent with the growth phenotypes, the glutamine synthetase activities of the strains carrying pNTRA6 or pNTRA11 were comparable (0.066 and 0.056 μmol of γ-glutamyl hydroxymate produced/min/mg of protein, respectively). Overexpression of the ntrA allele on pNTRA11 resulted in a level of repression of the ant′-′lacZ reporter gene which was comparable to that observed with pNTRA6 (103 and 305 Miller units, respectively). These data indicated that the L124P substitution was responsible for the loss of function in the original mutant.
The mutant allele carried on pNTRA7 had the amino acid substitutions W126R, L199P, D225E, D231G, and I428V. It was not possible to separate all of the mutations by subcloning. Nevertheless, by subcloning it was shown that the ς54 mutant with the W126R substitution allowed growth on both E minimal and M9-arginine media, as did the mutant with the substitutions D225E, D231G, and I428V (Table 1). The ς54 mutant with the substitutions L199P, D225E, D231G, and I428V (pNTRA15), however, did not allow growth on E minimal or M9-arginine media. The glutamine synthetase activity of the strain carrying pNTRA15 was comparable to that of the strain carrying pNTRA7 (Fig. 4B). These results suggested that L199P was responsible for the loss of function. Consistent with this, the proline at position 199 had reverted to the original leucine residue in four independent derivatives of pNTRA7 that complemented the growth defect of TRH107 on E minimal medium (represented by pNTRA16).
Introduction of proline at position 199 by site-directed mutagenesis, however, allowed wild-type growth on E minimal and M9-arginine media (Table 1). These data suggested that L199P in combination with D225E, D231G, or I428V was required for the loss of function of ς54. The D231G substitution was the most drastic, and we guessed that it might be required for loss of function. We generated an ntrA allele with both the L199P and D231G substitutions by site-directed mutagenesis (pNTRA18). The strain that overexpressed this ς54 mutant failed to grow on E minimal or M9-arginine medium and had a level of glutamine synthetase activity comparable to that of the strain carrying pNTRA7 (Fig. 4B). The L199P D231G double mutant also repressed transcription of the ant′-′lacZ reporter gene as effectively as the original mutant (214 and 642 Miller units, respectively). The mutant with the single substitution D231G allowed growth on both E minimal and M9-arginine media (Table 1). These data indicated that both the L199P and D231G substitutions were required for a loss of function. We cannot exclude the possibility that other combinations of amino acid substitutions could also have resulted in a loss of function.
The remaining mutant allele with five mutations (encoded on pNTRA8) was analyzed in a similar manner. These mutations were divided into groups of three (E42G, I56T, and G189S) and two (L235W and F318S) mutations by subcloning. Both of these alleles allowed growth on E minimal and M9-arginine media (Table 1). We selected for revertants of pNTRA8 that complemented the growth defect of TRH107 on E minimal medium. Two independent isolates were sequenced, and in both cases the serine at position 189 had reverted to the original glycine (represented by pNTRA22). These data suggested that the G189S substitution was responsible for the loss of function of the original mutant. When a serine was introduced at position 189 by site-directed mutagenesis, however, the resulting mutant ς54 allowed growth on both E minimal and M9-arginine media. This implied that a combination of G189S and either L235W or F318S was required for loss of function. F318 had been previously identified as a functionally important residue in E. coli ς54 (11). However, the G189S F318S double mutant as well as the F318S single mutant allowed wild-type growth on E minimal and M9-arginine media (Table 1). We did not construct the G189S L235W double mutant.
In vitro analysis of core binding activities of the ς54 mutants.
We assumed that the ς54 mutants were able to bind core RNA polymerase and direct the holoenzyme to the nifH promoter in vivo given that the affinity of free ς54 for promoter DNA is very low (3). To test this assumption, selected mutant ς54 proteins were purified and tested for their abilities to bind to the nifH promoter in a gel mobility shift assay in conjunction with core RNA polymerase. Histidine tags were placed at the amino termini of these proteins to facilitate their purification.
For these gel shift assays, we used labeled oligonucleotides that corresponded to the −9 to −29 region of the Sinorhizobium meliloti nifH promoter. One of these oligonucleotides was double stranded over its entire length (double-stranded probe), while the other oligonucleotide had a 5′ overhang of 3 bp that corresponded to residues −11 to −9 of the template strand (fork junction probe). E. coli ς54 had been shown previously to bind to this fork junction probe to form a heparin-resistant complex (12). We found that ς54-holoenzyme shifted the fork junction probe much more effectively than it did the double-stranded probe (Fig. 5, lanes 1 and 2). Free ς54 also shifted the fork junction probe, but this species had a faster mobility than the species shifted with holoenzyme (data not shown). Unlike the complex formed with ς54, the complex formed by ς54-holoenzyme and the fork junction probe was heparin sensitive (data not shown). Core RNA polymerase did not bind to the fork junction probe under the assay conditions used in the gel shift assay (Fig. 5, lane 20). These data confirmed that the supershifted species was a complex of ς54-holoenzyme and the fork junction probe and indicated that this gel shift assay could be used to assess the binding of the ς54 mutants to core RNA polymerase.
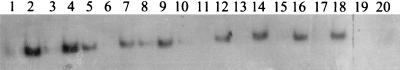
FIG. 5.
Gel mobility shift assays with mutant forms of ς54-holoenzyme. The double-stranded probe was used in the odd-numbered lanes, while the fork junction probe was used in the even-numbered lanes. The free probe is not shown. Binding reaction mixtures contained 300 nM core RNA polymerase plus either wild-type ς54 (lanes 1 and 2), histidine-tagged ς54 (lanes 3 and 4), the L37P mutant (lanes 5 and 6), the L46P mutant (lanes 7 and 8), the L333P mutant (lanes 9 and 10), the L199P D231G mutant (lanes 11 and 12), the E32K G189V mutant (lanes 13 and 14), the E42G I56T G189S L235W F318S mutant (lanes 15 and 16), the L124P V148A mutant (lanes 17 and 18), or no ς54 protein (lanes 19 and 20). For assays with wild-type ς54, >50% of the fork junction probe was shifted by holoenzyme.
The histidine tag at the amino terminus of ς54 did not interfere with the ability of ς54-holoenzyme to bind to either probe (Fig. 5, lanes 3 and 4). Interestingly, the holoenzymes formed with three of the ς54 mutants, L37P, L46P, and L333P, bound the double-stranded probe better than wild-type holoenzyme (Fig. 5, lanes 5, 7, and 9). These mutant forms of ς54-holoenzyme, however, bound very poorly to the fork junction probe (Fig. 5, lanes 6, 8, and 10). The remaining mutant forms of ς54-holoenzyme behaved like the wild-type holoenzyme in that they had higher affinities for the fork junction probe than for the double-stranded probe (Fig. 5, lanes 11 to 18). These data demonstrated that the mutant ς54 proteins retained core binding activity. The data also showed that the ς54 mutants could be divided into at least two classes on the basis of their affinities for the fork junction probe.
DISCUSSION
Transcription initiation involves several discrete steps, including formation of a closed complex between polymerase and the promoter, isomerization of the closed complex to an open complex, and promoter clearance. ς54 could play important roles at any of these steps.
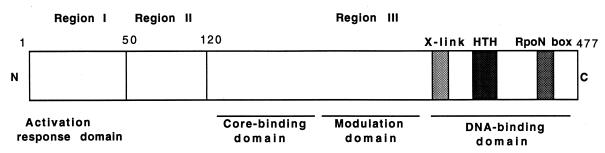
The ς54 protein consists of three functional regions (Fig. 6). The highly conserved region I of ς54 consists of approximately 50 amino-terminal residues and is rich in glutamine and leucine (23). Mutational analysis of region I has indicated that it is required for ς54-holoenzyme to respond to the activator protein (15, 16, 35, 39). Region I is not required for binding to either core or DNA (6, 43). Given these properties of region I mutants, we expected our screen to yield some ς54 mutants with substitutions in region I. Indeed, four of our original eight ς54 mutants had substitutions within this region. We identified E32, L37, and L46 as potentially important residues in S. typhimurium ς54. Previous studies had suggested that L33, E36, and L37 were functionally important in E. coli ς54 (35). Region I-deleted holoenzyme protects a larger region of the promoter from S1 nuclease cleavage than the wild-type holoenzyme, indicating that region I influences the conformation of the holoenzyme (5). The L37P and L46P mutant proteins may have affected the conformation of the holoenzyme similarly, which could explain the reduced affinities of the mutant holoenzymes formed with these ς54 mutants for the fork junction probe.
FIG. 6.
Structure of ς54. Regions I, II, and III are the three functional regions of ς54. X-link, a region that cross-links to promoter DNA on UV irradiation (4); HTH, a putative DNA-binding helix-turn-helix motif; RpoN, the signature motif of ς54 proteins that has the consensus sequence ARRTVAKYRE.
Region II is poorly conserved and is in fact missing in Rhodobacter capsulatus ς54 (9). In ς54 proteins from enteric bacteria, region II has a high proportion of acidic amino acid residues. Deletions within region II of E. coli ς54 appear to decrease the rate of open-complex formation (42). None of the residues that we identified as functionally important in S. typhimurium ς54 in this study was in region II.
Determinants for core and DNA binding are located within region III of ς54 (6, 11, 36, 37, 43). Our results clearly show that in addition to these roles, region III is involved in one or more subsequent steps in transcription initiation. We identified four residues within region III—L124, L199, D231, and L333—which when altered disrupted transcription initiation at some point following closed-complex formation.
Substitution of proline for leucine at position 124 resulted in the loss of function. L124 lies within the minimal core binding domain (residues 120 to 215) as defined by deletion analysis (6, 43). Core protects the region between residues 36 and 140 of ς54 from hydroxyl-radical cleavage (8), suggesting that L124 is in a region of the protein that contacts core. Given that the L124P mutant retains core binding activity, interactions between the region around L124 and core may be required for open-complex formation rather than binding of ς54 to core. The L199P and D231G mutations acted synergistically to disrupt the function of ς54. L199 is within the minimal core binding domain. The region around L199, however, does not appear to closely contact core, since it is not protected from hydroxyl-radical cleavage by core (8). D231 is within a domain that modulates the DNA-binding activity of the protein (7). Like the region around L124, the regions around L199 and D231 had not been shown previously to participate in steps following closed-complex formation.
L333 is located within the DNA-binding domain of ς54 (residues 332 to 462) (6), and a proline substitution at this position caused a loss of function. The region around L333 had been suggested previously to play a role in later steps in transcription initiation. Deletion of residues 293 through 332 disrupts the function of E. coli ς54, and like the L333P mutant isolated in our study, this deletion mutant retained its core and DNA-binding activities (43). L333P mutations in E. coli ς54 were described previously (11), but in this earlier publication it was not specified whether these proteins had other substitutions. In addition, the core and DNA-binding activities of the E. coli mutants were not reported. Like the holoenzymes formed with L37P and L46P, the L333P holoenzyme had a low affinity for the fork junction probe, suggesting that the region around Leu-333 could influence the conformation of holoenzyme.
The frequency with which we isolated ς54 mutants that retained core and DNA-binding activities was very low. Many of these ς54 mutants had substitutions of proline for leucine. This may reflect the fact that such mutations require a single base change. Alternatively, this may have been due to the stringency of the genetic screen used to isolate the ς54 mutants. Proline substitutions could have disrupted the secondary structure and severely impaired the function of ς54. Consistent with this idea, strains with as little as 8% of the wild-type glutamine synthetase activity were glutamine prototrophs, indicating that the ς54 mutants isolated were severely impaired in their function at the glnAp2 promoter. Determining the biochemical basis for the failure of these mutant proteins to function will help clarify the roles of ς54 in transcription initiation.
ACKNOWLEDGMENTS
We thank Ellen Neidle and Jonathan Olson for comments on the manuscript.
This work was supported by award MCB-9630454 to T.R.H. from the National Science Foundation.
REFERENCES
- 1.Ashraf S I, Kelly M T, Wang Y-K, Hoover T R. Genetic analysis of the Rhizobium meliloti nifH promoter, using the P22 challenge phage system. J Bacteriol. 1997;179:2356–2362. doi: 10.1128/jb.179.7.2356-2362.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender R A, Janssen K A, Resnick A D, Blumenberg M, Foor F, Magasanik B. Biochemical parameters of glutamine synthetase from Klebsiella aerogenes. J Bacteriol. 1977;129:1001–1009. doi: 10.1128/jb.129.2.1001-1009.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cannon W, Claverie-Martin F, Austin S, Buck M. Core RNA polymerase assists binding of the transcription factor ς54 to promoter DNA. Mol Microbiol. 1993;8:287–298. doi: 10.1111/j.1365-2958.1993.tb01573.x. [DOI] [PubMed] [Google Scholar]
- 4.Cannon W, Claverie-Martin F, Austin S, Buck M. Identification of a DNA-contacting surface in the transcription factor sigma-54. Mol Microbiol. 1994;11:227–236. doi: 10.1111/j.1365-2958.1994.tb00303.x. [DOI] [PubMed] [Google Scholar]
- 5.Cannon W, Gallegos M-T, Casaz P, Buck M. Amino-terminal sequences of ςN (ς54) inhibit RNA polymerase isomerization. Genes Dev. 1999;13:357–370. doi: 10.1101/gad.13.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon W, Missailidis S, Smith C, Cottier A, Austin S, Moore M, Buck M. Core RNA polymerase and promoter DNA interactions of purified domains of ςN: bipartite functions. J Mol Biol. 1995;248:781–803. doi: 10.1006/jmbi.1995.0260. [DOI] [PubMed] [Google Scholar]
- 7.Cannon W V, Chaney M K, Wang X-Y, Buck M. Two domains within ςN (ς54) cooperate for DNA binding. Proc Natl Acad Sci USA. 1997;94:5006–5011. doi: 10.1073/pnas.94.10.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casaz P, Buck M. Region I modifies DNA-binding domain conformation of sigma 54 within the holoenzyme. J Mol Biol. 1999;285:507–514. doi: 10.1006/jmbi.1998.2328. [DOI] [PubMed] [Google Scholar]
- 9.Cullen P J, Forster-Hartnett D, Gabbert K K, Kranz R G. Structure and expression of the alternative sigma factor, RpoN, in Rhodobacter capsulatus; physiological relevance of an autoactivated nifU2-rpoN superoperon. Mol Microbiol. 1994;11:51–65. doi: 10.1111/j.1365-2958.1994.tb00289.x. [DOI] [PubMed] [Google Scholar]
- 10.Grana D, Youderian P, Susskind M M. Mutations that improve the ant promoter of Salmonella phage P22. Genetics. 1985;110:1–16. doi: 10.1093/genetics/110.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo Y, Gralla J D. DNA-binding determinants of sigma 54 as deduced from libraries of mutations. J Bacteriol. 1997;179:1239–1245. doi: 10.1128/jb.179.4.1239-1245.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Y, Gralla J D. Promoter opening via a DNA fork junction binding activity. Proc Natl Acad Sci USA. 1998;95:11655–11660. doi: 10.1073/pnas.95.20.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirschmam J, Wang P-K, Sei K, Keener J, Kustu S. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a ς factor. Proc Natl Acad Sci USA. 1985;82:7525–7529. doi: 10.1073/pnas.82.22.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh M, Gralla J D. Analysis of the N-terminal leucine heptad and hexad repeats of sigma 54. J Mol Biol. 1994;239:15–24. doi: 10.1006/jmbi.1994.1347. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh M, Tintut Y, Gralla J D. Functional roles for the glutamines within the glutamine-rich region of the transcription factor ς54. J Biol Chem. 1994;269:373–378. [PubMed] [Google Scholar]
- 17.Ishihama A. Protein-protein communication within the transcription apparatus. J Bacteriol. 1993;175:2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juang Y L, Helmann J D. A promoter melting region in the primary sigma factor of Bacillus subtilis. Identification of functionally important aromatic amino acids. J Mol Biol. 1994;235:1470–1488. doi: 10.1006/jmbi.1994.1102. [DOI] [PubMed] [Google Scholar]
- 19.Lee J H, Hoover T R. Protein crosslinking studies suggest that Rhizobium meliloti C4-dicarboxylic acid transport protein D, a ς54-dependent transcriptional activator, interacts with ς54 and the β subunit of RNA polymerase. Proc Natl Acad Sci USA. 1995;92:9702–9706. doi: 10.1073/pnas.92.21.9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lonetto M, Gribskov M, Gross C A. The ς70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 22.Mei L, Moyle H, Susskind M M. Target of the transcriptional activator function of phage λ cI protein. Science. 1994;263:75–77. doi: 10.1126/science.8272867. [DOI] [PubMed] [Google Scholar]
- 23.Merrick M J. In a class of its own—the RNA polymerase sigma factor ς54 (ςN) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 25.Morett E, Buck M. In vivo studies on the interaction of RNA polymerase-ς54 with the Klebsiella pneumoniae and Rhizobium meliloti nifH promoters: the role of NIFA in the formation of an open promoter complex. J Mol Biol. 1989;210:65–77. doi: 10.1016/0022-2836(89)90291-x. [DOI] [PubMed] [Google Scholar]
- 26.Oguiza J A, Buck M. DNA-binding domain mutant of sigma-N (ςN, ς54) defective between closed and stable open promoter complex formation. Mol Microbiol. 1997;26:655–664. doi: 10.1046/j.1365-2958.1997.5861954.x. [DOI] [PubMed] [Google Scholar]
- 27.Popham D, Keener J, Kustu S. Purification of the alternative ς factor, ς54, from Salmonella typhimurium and characterization of ς54-holoenzyme. J Biol Chem. 1991;266:19510–19518. [PubMed] [Google Scholar]
- 28.Popham D, Szeto D, Keener J, Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989;243:629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- 29.Reitzer L J. Sources of nitrogen and their utilization. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 380–390. [Google Scholar]
- 30.Ring B Z, Yarnell W S, Roberts J W. Function of the E. coli RNA polymerase sigma factor sigma 70 in promoter-proximal pausing. Cell. 1996;86:485–493. doi: 10.1016/s0092-8674(00)80121-x. [DOI] [PubMed] [Google Scholar]
- 31.Rippe K, Guthold M, von Hippel P H, Bustamante C. Transcriptional activation via DNA-looping: visualization of intermediates in the activation pathway of E. coli RNA polymerase ς54 holoenzyme by scanning force microscopy. J Mol Biol. 1997;270:125–138. doi: 10.1006/jmbi.1997.1079. [DOI] [PubMed] [Google Scholar]
- 32.Sasse-Dwight S, Gralla J D. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc Natl Acad Sci USA. 1988;85:8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stadtman E R, Ginsburg A, Ciardi J E, Yeh J, Hennig S B, Shapiro B M. Multiple molecular forms of glutamine synthetase produced by enzyme catalyzed adenylylation and deadenylylation reactions. Adv Enzyme Regul. 1970;8:99–118. doi: 10.1016/0065-2571(70)90011-7. [DOI] [PubMed] [Google Scholar]
- 34.Su W, Porter S, Kustu S, Echols H. DNA-looping and enhancer activity: association between DNA-bound NTRC activator and RNA polymerase at the bacterial glnA promoter. Proc Natl Acad Sci USA. 1990;87:5504–5508. doi: 10.1073/pnas.87.14.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Syed A, Gralla J D. Identification of an N-terminal region of sigma 54 required for enhancer responsiveness. J Bacteriol. 1998;180:5619–5625. doi: 10.1128/jb.180.21.5619-5625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor M, Butler R, Chambers S, Casimiro M, Badii F, Merrick M. The RpoN-box motif of the RNA polymerase sigma factor ςN plays a role in promoter recognition. Mol Microbiol. 1996;22:1045–1054. doi: 10.1046/j.1365-2958.1996.01547.x. [DOI] [PubMed] [Google Scholar]
- 37.Tintut Y, Gralla J D. PCR mutagenesis identifies a polymerase-binding sequence of sigma 54 that includes a sigma 70 homology region. J Bacteriol. 1995;177:5818–5825. doi: 10.1128/jb.177.20.5818-5825.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogel H J, Bonner D M. Acetyl ornithase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 39.Wang J T, Syed A, Gralla J D. Multiple pathways to bypass the enhancer requirement of sigma 54 RNA polymerase: roles for DNA and protein determinants. Proc Natl Acad Sci USA. 1997;94:9538–9543. doi: 10.1073/pnas.94.18.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y-K, Lee J H, Brewer J M, Hoover T R. A conserved region in the ς54-dependent activator DctD is involved in both binding to RNA polymerase and coupling ATP hydrolysis to activation. Mol Microbiol. 1997;26:373–386. doi: 10.1046/j.1365-2958.1997.5851955.x. [DOI] [PubMed] [Google Scholar]
- 41.Weiss D S, Batut J, Klose K E, Keener J, Kustu S. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell. 1991;67:155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- 42.Wong C, Gralla J D. A role for the acidic trimer repeat region of transcription factor ς54 in setting the rate and temperature dependence of promoter melting in vivo. J Biol Chem. 1992;267:24762–24768. [PubMed] [Google Scholar]
- 43.Wong C, Tintut Y, Gralla J D. The domain structure of sigma 54 as determined by analysis of a set of deletion mutants. J Mol Biol. 1994;236:81–90. doi: 10.1006/jmbi.1994.1120. [DOI] [PubMed] [Google Scholar]