Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a highly transmissible and virulent human-infecting coronavirus that emerged in late December 2019 in Wuhan, China, causing a respiratory disease called coronavirus disease 2019 (COVID-19), which has massively impacted global public health and caused widespread disruption to daily life. The crisis caused by COVID-19 has mobilized scientists and public health authorities across the world to rapidly improve our knowledge about this devastating disease, shedding light on its management and control, and spawned the development of new countermeasures. Here we provide an overview of the state of the art of knowledge gained in the last 2 years about the virus and COVID-19, including its origin and natural reservoir hosts, viral etiology, epidemiology, modes of transmission, clinical manifestations, pathophysiology, diagnosis, treatment, prevention, emerging variants, and vaccines, highlighting important differences from previously known highly pathogenic coronaviruses. We also discuss selected key discoveries from each topic and underline the gaps of knowledge for future investigations.
Keywords: clinical features, diagnosis, SARS-CoV-2, variants, transmission, treatment, vaccines, reservoir hosts, pathophysiology, prevention
Coronaviruses (CoVs) are enveloped RNA viruses that belong to the Coronaviridae family within the order Nidovirales. They are a diverse group of enveloped positive-sense single-stranded RNA (+ssRNA) viruses that widely infect humans and animals and cause respiratory, hepatic, neurological, and enteric diseases.1 In humans, four coronaviruses (CoV-229E, CoV-OC43, CoV-NL63, and CoV-HKU1) are endemic and typically are associated with mild respiratory disease in healthy individuals.2 However, in the last two decades, three zoonotic coronaviruses originating from bats have emerged and caused severe respiratory disease in humans: severe acute respiratory syndrome coronavirus (SARS-CoV),3,4 Middle East respiratory syndrome coronavirus (MERS-CoV),5 and, most recently, the pandemic coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).6−8
SARS-CoV-2 was first reported in early December 2019 in Wuhan, Hubei Province, China, causing an outbreak of respiratory illness later named coronavirus disease 2019 (COVID-19).8,9 The rapid spread of the disease outside China led the World Health Organization (WHO) to declare a Public Health Emergency of International Concern (PHEIC) on January 30, 2020, and subsequently, a pandemic on March 11, 2020.10 As of July 14, 2022, more than 559.5 million cases of COVID-19 infection and 6.3 million deaths have been reported worldwide, most of which involved people living in the USA, followed by those living in India, Brazil, and France.11
SARS-CoV-2 shows sustained person-to-person transmission through direct contact and the air (as respiratory droplets and/or aerosols).12,13 The infection has a median incubation period of approximately 4–5 days, but it can be as long as 14 days.14,15 The most common symptoms reported in patients with COVID-19 are fever, fatigue, and dry cough.14,16,17 Other less common symptoms include headache, sore throat, myalgia, diarrhea, vomiting, chills, loss of smell, and loss of taste.14,16−18 Clinically, many COVID-19 patients present mild to moderate symptoms (81%). However, approximately 14% of infected patients progress to pneumonia and may require ventilation in an intensive care unit (ICU), and 5% eventually develop more critical manifestations such as acute respiratory distress syndrome (ARDS), septic shock, and multiple organ dysfunction or failure.19−21
Since the emergence of the virus and its subsequent spread across the world, rapid progress has been made toward understanding many features of COVID-19. Based on the current scientific knowledge, this comprehensive review outlines the latest information on many topics related COVID-19 generated over the last 2 years, including origin and natural reservoir hosts, viral etiology, epidemiology, routes of transmission, clinical manifestations, pathophysiology, diagnosis, treatment, prevention, emerging variants, and vaccines.
Brief History
In the early 1930s, members of the Coronaviridae family were identified as responsible for infectious illness in several animal species, including mice, chickens, and pigs.22 In the 1960s, Tyrrell and other virologists visualized morphological features of mouse hepatitis virus, bronchitis virus, and swine gastroenteritis virus using electron microscopy.23,24 This novel group of viral agents were called coronaviruses (referring to the crownlike appearance similar to a solar corona), and later this designation was officially recognized.24,25 During the same decade, human coronaviruses, including OC43 and 229E, were first isolated from human patients displaying upper respiratory disease.26,27
For approximately 40 years, no additional coronaviruses capable of infecting humans had been described. Later, the coronaviruses NL63 and HKU1 were first identified in 2004 and 2005, respectively, and have been added to the spectrum of viruses that cause the common cold.28,29 These CoVs are considered to be of low pathogenicity, with infection typically resulting in mild or moderate symptoms in immunocompetent individuals.30 In addition to these four endemic CoVs, three highly pathogenic respiratory betacoronaviruses have emerged from bats and caused severe outbreaks in humans during the 21st century. In 2002, severe acute respiratory syndrome coronavirus (SARS-CoV) emerged in Guangdong Province, China, and caused a disease with the same name.3,31 The virus rapidly spread to more than 27 countries, resulting in more than 8000 human infections and 774 deaths between 2002 and 2004, with a lethality rate of approximately 10%.4,32 In 2012, a novel betacoronavirus called Middle East respiratory syndrome coronavirus (MERS-CoV) was first identified in a Saudi Arabian patient suffering from a severe respiratory disease that was later named Middle East respiratory syndrome (MERS).5,33 MERS-CoV has been reported in 27 countries to date, mainly in the Middle East region, resulting in more than 2500 laboratory-confirmed cases and 927 deaths, with a mortality rate of 35%.34 Whereas there have been no reported SARS cases anywhere in the world since late 2004,35 MERS-CoV is still actively circulating in the Middle East.34
Despite the concerns raised by SARS-CoV and MES-CoV, both viruses failed to establish efficient transmission in the human population. However, the third of the highly pathogenic emergent CoVs proved to be different. On December 31, 2019, health officials reported to the WHO China Country Office cases of pneumonia of unknown etiology detected in Wuhan City, Hubei Province, China. The cluster of cases was epidemiologically linked to the Wuhan Huanan Seafood Wholesale Market, a large public market that commercializes seafood and several species of domestic and wild animals. An epidemiological and etiological investigation was initiated, and highly pathogenic CoVs were suspected on the basis of the clinical presentation, the time of the year (winter), and the link of the patients with a wet market, which was similar to SARS infections. A series of patients were sampled and submitted to pan-CoV PCR testing. Five samples turned out be PCR-positive for CoVs, and metagenomics analysis of one sample using next-generation sequencing (NGS) identified a novel CoV, which was first called 2019-nCoV. Other researchers independently discovered SARS-CoV-2 at the same time from different patients, and most cases had been at the market in Wuhan, suggesting that the earliest documented COVID-19 cases were indeed linked to the market.8,9 The International Committee on Taxonomy of Viruses (ICTV) and the WHO officially named the virus as SARS-CoV-2 and the disease as COVID-19.36
Origin and Natural Reservoir Hosts
Despite the passage of 2 years since the beginning of the pandemic, there still exists a great mystery about the origins of SARS-CoV-2. While there has been speculation that SARS-CoV-2 had been created in a laboratory, a comparative genomic study suggested that this was not the case and supported two scenarios for the origin of SARS-CoV-2: (i) natural selection in an animal reservoir before zoonotic spillover and (ii) natural selection in humans following zoonotic spillover.37 These results suggested that SARS-CoV-2 is not a pathogen purposely manipulated or constructed in the laboratory. Phylogenetic analysis showed that SARS-CoV-2 is clustered with SARS-related coronaviruses (SARSr-CoVs) and the SARS-CoV previously reported in bats, placing it in the subgenus Sarbecovirus and genus Betacoronavirus.8,9 SARS-CoV-2 shares 79% genome identity with SARS-CoV and 50% with MERS-CoV.7,8 Although the origin and direct ancestral virus of SARS-CoV-2 are yet to be discovered, RaTG13, a CoV detected in the horseshoe bat Rhinolophus affinis in Yunnan Province, China, has 96.2% genome similarity with SARS-CoV-2 and is the closest relative of SARS-CoV-2 identified to date.7 Notably, the high genetic similarity between SARS-CoV-2 and related bat CoVs likely represents more than two decades of evolution, suggesting that these bat CoVs are the most probable evolutionary progenitor of SARS-CoV-2, while other intermediate hosts might have played a crucial role in the process of transmission to humans.38,39 Recently, scientists have identified SARSr-CoVs in Rhinolophus shameli bats sampled in Cambodia in the year 2010. They showed that these viruses share 92.6% nucleotide identity with SARS-CoV-2 and are closely related to SARS-CoV-2 in most genomic regions, except for the spike (S) protein that binds to the ACE2 receptor in human cells.40
Three recent new studies have added evidence of the role of the Huanan Seafood Wholesale Market in Wuhan in the emergence of SARS-CoV-2. Gao and co-workers tested 1380 environmental and animal samples collected within the market in early 2020 and found 73 environmental samples positive for SARS-CoV-2, whereas none of the animal samples were positive. They were able to isolate live SARS-CoV-2 from three environmental samples. Worobey and co-workers used spatial analysis and genomic data to show that the earliest known COVID-19 cases were geographically and epidemiologically linked to the Huanan seafood market. Pekar and co-workers used Bayesian phylogenetic analysis in early SARS-CoV-2 sequences to show that the emergence of SARS-CoV-2 occurred via multiple zoonotic events in a similar way as SARS-CoV in 2002 and 2003. Together, these studies suggest that the market played a major role as the epicenter of SARS-CoV-2 emergence and further weakened the lab-leak hypothesis.41−43
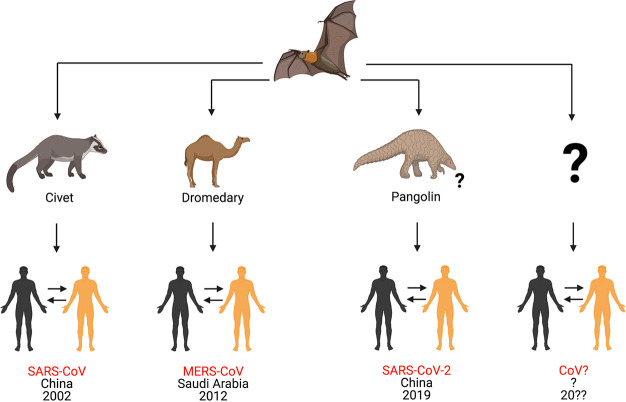
With regard to intermediate hosts, both SARS-CoV and MERS-CoV emerged from bats and were transmitted directly to humans from civets and dromedary camels, respectively.2,6 However, knowledge about the intermediate host(s) for SARS-CoV-2 remains incomplete and requires further studies (Figure 1).7,44 The identification of intermediate hosts is crucial for public health measures to prevent future outbreaks of SARS-CoV-2 or related viruses.37,45,46 Some studies have suggested that pangolins can host SARS-CoV-2.47−50 SARS-CoV-2-related viruses have been detected in and isolated from tissues of Malayan pangolins from China with clinical signs of disease and histological alterations.50 In that study, Xiao and colleagues revealed that a CoV isolated from the Malayan pangolin showed 100%, 98.6%, 97.8%, and 90.7% amino acid identity with SARS-CoV-2 envelope [E], membrane [M], nucleocapsid [N], and spike [S] proteins, respectively.50 Lam and co-workers used metagenomics and phylogenetic analysis to show that the viruses from pangolins were associated with two distinct sublineages of SARS-CoV-2-related coronaviruses, including one that exhibited strong similarity (97.4% amino acid similarity) in the receptor-binding domain (RBD) to SARS-CoV-2.47 Similarly, Liu and colleagues assembled the complete genome of a coronavirus identified in three sick Malayan pangolins and demonstrated that it was genetically related to SARS-CoV-2.49 Taken together, these results suggest that pangolins have the potential to act as an intermediate host of SARS-CoV-2, although more studies are needed to confirm this hypothesis.
Figure 1.
Origins of different coronaviruses. In the 21st century, three highly pathogenic betacoronaviruses have emerged from bats to cause respiratory disease in humans. In 2002, a betacoronavirus called severe acute respiratory syndrome coronavirus (SARS-CoV) emerged in Guangdong Province, China, and caused respiratory disease in humans. One decade later, another betacoronavirus called Middle East respiratory syndrome coronavirus (MERS-CoV) was reported in Saudi Arabia. Both SARS-CoV and MERS-CoV emerged from bats and were transmitted to humans via civets and dromedary camels, respectively. Later, in December 2019, a novel betacoronavirus called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged from bats and caused a pandemic disease called coronavirus disease 2019 (COVID-19). It was likely transmitted to humans by pangolins, although its origin is still being investigated. In summary, coronaviruses represent an example of emerging zoonotic viruses that have crossed the species barrier to cause disease in the human population. The possibility of a new, highly pathogenic coronavirus emerging from wild animals in the next few years cannot be ruled out. This figure was created with Biorender.com.
Since the emergence of SARS-CoV-2, research groups from across the world have investigated the susceptibility of domestic animals to SARS-CoV-2 infection.51−53 In this context, Shi and co-workers provided and discussed important insights into the animal reservoirs of SARS-CoV-2.53 They investigated the susceptibility of ferrets and other animals to SARS-CoV-2 infection, most of them traditionally having close contact with humans, including cats, dogs, pigs, ducks, and chickens, and demonstrated that SARS-CoV-2 replicates effectively in cats and ferrets but poorly in dogs, pigs, ducks, and chickens.53 Additionally, it was demonstrated that SARS-CoV-2 can be transmitted easily among cats through respiratory droplets.53 Similarly, Halfmann and colleagues evaluated the transmission of SARS-CoV-2 in domestic cats and provided evidence of the potential human–cat–human transmission chain.52 In that study, none of the infected cats showed any clinical signs of disease, such as fever, substantial weight loss, or conjunctivitis, suggesting that cats might be a silent intermediate host for SARS-CoV-2. However, there is no clear evidence supporting the hypothesis that SARS-CoV-2 can be transmitted from infected animals to humans,54 and further studies are required to understand the role of cats and other domestic animals in the transmission of SARS-CoV-2 to humans.
In addition to domestic animals, many studies have been conducted to establish experimental animal models for SARS-CoV-2.51,55 The development and identification of animal models for studying SARS-CoV-2 are crucial for the study of virus biology, transmission, and COVID-19 pathogenesis and to evaluate potential therapeutic agents and vaccines.55 The susceptibility of many animal species, including hamsters, mice, ferrets, rabbits, bats, ducks, pigs, chickens, minks, and non-human primates, to SARS-CoV-2 infection has been investigated.53,55−59 In general, the results demonstrate that susceptibility varies according to animal species and that hamsters, human ACE2-transgenic mice, ferrets, and non-human primates seem to be more promising in vivo models. To date, our knowledge of the intermediate hosts of SARS-CoV-2 remains incomplete, and all reservoir hosts of the virus have not been clearly established. Therefore, experimental studies using animal models aiming to determine potential reservoir hosts should be addressed to elucidate other routes for the spread of SARS-CoV-2 within and among humans and animals.51,60,61
Etiology and Replication Cycle
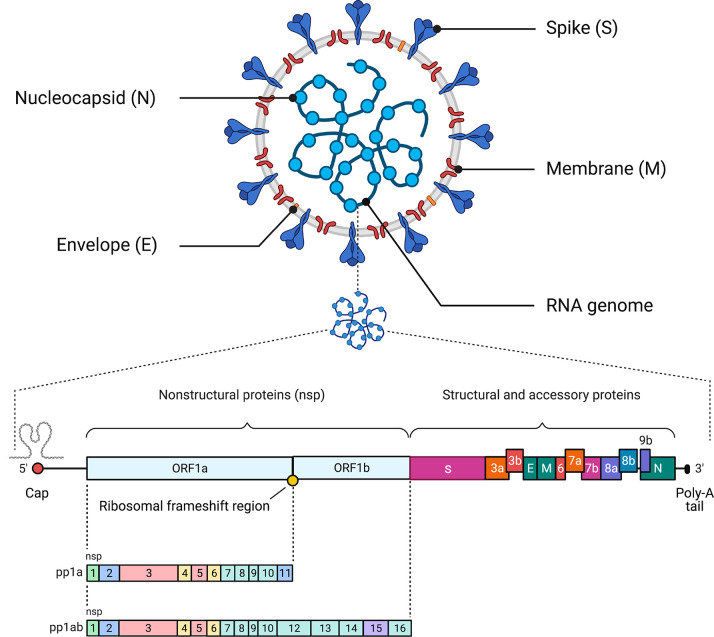
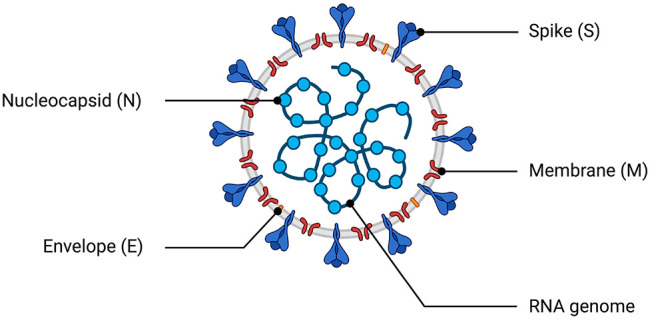
SARS-CoV-2 is a CoV member of order Nidovirales, family Coronaviridae, subfamily Orthocoronavirinae. This subfamily is subdivided into four genera on the basis of genetic characteristics: Alphacoronavirus (α-CoV), Betacoronavirus (β-CoV), Gammacoronavirus (γ-CoV), and Deltacoronavirus (δ-CoV).62 Similar to SARS-CoV and MERS-CoV, SARS-CoV-2 belongs to the β-CoV cluster and has a diameter of 80–160 nM and an RNA genome that is approximately 30 kilobases (kb) in length (Figure 2).7,63−65 All viruses in order Nidovirales are enveloped with a genome consisting of a single +ssRNA with a 5′-cap structure and a 3′-poly-A tail, allowing it to function as an mRNA for the replicase proteins.62 The ORF1a and ORF1b genes occupy two-thirds of the 5′ genome and are translated into polyprotein 1a (pp1a) and polyprotein 1ab (pp1ab), respectively.66 The resulting polyproteins 1a and 1ab are cleaved by the 3C-like protease (3CLpro) and the papain-like protease (PLpro). As a result of this process, pp1a is cleaved into 11 individual nonstructural proteins (nsps), and pp1ab is translated after a ribosomal frameshift takes place in the −1 position of the ORF1a stop codon and is then cleaved into 16 nsps (Table 1).60,66 The SARS-CoV-2 structural proteins (the spike [S], membrane [M], envelope [E], and nucleocapsid [N] proteins) are encoded by one-third of the viral genome, and these proteins are required for the assembly of new viral particles.1,62,67 The S protein encodes the signal peptide (SP), RBD, subdomain 1 (SD1), and subdomain 2 (SD2) in the S1 subunit and the fusion peptide (FP), heptad repeat 1 (HR1), heptad repeat 2 (HR2), and transmembrane (TM) in membrane-fusion subunit (S2).68 SARS-CoV-2 also encodes accessory proteins, including ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8a, ORF8b, and ORF9b, all of which are distributed among the structural genes.38,69 In a rapidly moving field of study, studies have suggested the presence of other accessory proteins (ORF3c, ORF3d, ORF9c, and ORF10).70,71 Additionally, the 5′ end contains an untranslated region (UTR), which forms multiple stem–loop structures needed for RNA transcription and replication.62
Figure 2.
Schematic of the SARS-CoV-2 virus particle and genome architecture. The top panel illustrates the general structure of the SARS-CoV-2 viral particle, indicating its structural proteins and genome. The bottom panel illustrates the genome organization of SARS-CoV-2, including the 5′ cap, the region that encodes the nonstructural proteins required for viral replication (nsp1–nsp16), the region that encodes accessory and structural proteins (spike [S] protein, membrane [M] protein, envelope [E] protein, and nucleocapsid [N] proteins), and the poly-A tail. This figure was created with Biorender.com.
Table 1. Roles of Nonstructural Proteins (nsps) of SARS-CoV-2.
| nsp | functionsa | refs |
|---|---|---|
| nsp1 | disrupting the mRNA export machinery to inhibit host gene expression; inhibiting host protein translation; inhibiting IFN pathway | (633−636) |
| nsp2 | linking viral transcription within the viral replication–transcription complex (RTC) to the initiation of translation | (637) |
| nsp3 | essential component of the replication/transcription complex; also responsible for inhibiting host innate immune response, promoting cytokine expression, and cleaving viral polypeptides | (638−640) |
| nsp4 | critical role in the organization and stability of DMVs | (641−643) |
| nsp5 | 3CLpro and Mpro protease activity; blocking the IFN pathway | (644−646) |
| nsp6 | organizing DMVs; restoring autophagosome expansion; involved in autophagy | (643), (647), (648) |
| nsp7 | cofactor with nsp12; forming the hexadecameric complex with nsp8; exhibiting primer-independent RNA polymerase activity | (649−651) |
| nsp8 | cofactor with nsp12; forming the hexadecameric complex with nsp7; primase | (649), (650), (652) |
| nsp9 | RNA binding; enhances dimerization with diverse modes; interacts with nsp8; probably involved in viral RNA replication | (653−655) |
| nsp10 | cofactor for the N7-guanine-methyltransferase/exoribonuclease activities (nsp14) and for the 2′-O-methyltransferase activity (nsp16) | (656−659) |
| nsp11 | dispensable for viral replication in cultured cells; intrinsically disordered protein | (660), (661) |
| nsp12 | replication enzyme (RNA-dependent RNA polymerase) | (649,662−664) |
| nsp13 | RNA helicase activity; RNA 5′ triphosphatase; blocking the IFN pathway | (636), (665−667) |
| nsp14 | exoribonuclease activity; N7-methyltransferase activity | (559), (668−671) |
| nsp15 | viral endoribonuclease activity; evasion of dsRNA sensors | (672−674) |
| nsp16 | 2′-O-methyltransferase activity; regulating host immunity response; RNA cap formation | (657), (658), (675−678) |
Abbreviations: DMV, double-membrane vesicle; IFN, interferon; 3CLpro, 3C-like protease; Mpro, main protease.
As previously mentioned, coronavirus particles are composed of four main structural proteins, among which the S protein has an essential role during the initial attachment, fusion, and entry of the viral particle into the host cell.1,72−74 Moreover, the S protein plays a critical role in determining transmission ability and host tropism, and it is also the major target for vaccines and therapeutic antibodies.72,74−80 Structural studies of the S protein have identified residues in the protein’s RBD that are essential for binding to the host cell receptor, the majority of which are highly conserved or share similar side-chain characteristics compared to the SARS-CoV RBD.81,82 Other motifs and domains of the S protein are key mediators of viral entry into host cells: the S1 subunit is used for binding to a host receptor and the S2 subunit for fusing the viral envelope and host cell membrane.73 Similar to other highly pathogenic viruses such as the avian influenza virus (AIV), the S protein of SARS-CoV-2 harbors a polybasic cleavage site (RRAR).83 This motif enables effective cleavage of S protein by furin and other proteases and is required for transmission of SARS-CoV-2.84,85 A growing body of data has demonstrated that furin cleavage of the SARS-CoV-2 spike protein promotes viral entry and allows cleavage during virus packaging.84 SARS-COV-2 interacts with the host protein angiotensin-converting enzyme II (ACE2), which is also a receptor for SARS-CoV, suggesting that these two CoVs share many steps in their replication cycles.7,8,87−90 SARS-CoV-2 and SARS-CoV have been shown to use the cell-surface transmembrane protease serine protease (TMPRSS2) for priming and entry, although other proteases such as cathepsin B (CatB) and CatL can also assist in this mechanism.90 The expression and tissue distribution of ACE2 consequently influence the tropism and pathogenicity of SARS-CoV-2.91,92 More recently, it was demonstrated that the SARS-CoV-2 S protein also binds to the surface receptor CD147 on the host cell, suggesting an alternative route to mediate the cell invasion of SARS-CoV-2.93 In contrast, MERS-CoV uses CD26 (also known as dipeptidyl peptidase 4, DPP4) as the cell receptor.94
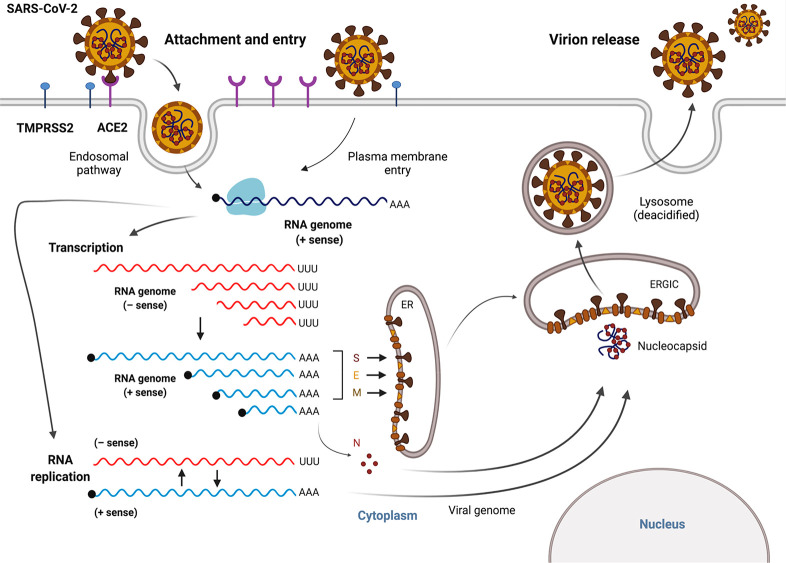
SARS-CoV-2 is highly transmissible and displays broad tissue tropism, which is determined by the susceptibility and permissiveness of specific host cells to the virus.95 During the infection process, the SARS-CoV-2 replication cycle starts with binding of the S protein to the host receptor ACE2, which together with host factors (e.g., furin, TMPRSS2, and Cat B/L) results in conformational changes in the S protein followed by viral uptake.85,91,96 SARS-CoV-2 enters the host cell by direct fusion of the viral envelope protein with the host cell membrane or membrane fusion within the endosome after endocytosis.97 Next, viral replication takes place in the cytoplasm,61 where viral RNA utilizes the host and its own enzymatic machinery to replicate its genome, express viral proteins, and assemble new SARS-CoV-2 particles.90,98 More specifically, viral RNA is released into the host cytoplasm, and ORF1a and ORF1b are translated, after which the resulting products then go on to form the viral replication and transcription complex (RTC).91 In all coronaviruses, the translation of ORF1b requires a programmed −1 ribosomal frameshift, an alternative mechanism of translation to merge proteins encoded by two overlapping ORFs.99 After ribosomes reach the end of the ORF1a coding sequence, they encounter a frameshifting element that causes the ribosomes to backtrack by one nucleotide and reposition in the −1 reading frame before continuing the translation and producing a full-length ORF1ab polyprotein.100 As a result of the translation of nonstructural proteins, viral genomic RNA replication and transcription of subgenomic mRNAs (sg mRNAs) are initiated.91 These subgenomic RNAs are then transcribed and translated to produce accessory and structurally relevant proteins for the replication cycle and the production of new viral particles.101 Assembly of virions occurs via the interaction between viral genomic RNA and structural proteins located in the endoplasmic reticulum (ER) and the ER–Golgi intermediate compartment (ERGIC). Finally, these virions are released to the plasma membrane via deacidified lysosomes102 and secreted from the infected cell via exocytosis (Figure 3).90,101
Figure 3.
The SARS-CoV-2 replication cycle. SARS-CoV-2 enters the host cell via an endosomal pathway or through fusion of the viral envelope with the host cell membrane. Briefly, viral entry is initiated by binding of the RBD of the spike (S) protein to the human host cell receptor (ACE2). After the RBD–receptor interaction, the S protein undergoes proteolytic cleavage, which can be catalyzed by several host proteases, such as TMPRSS2, furin, and cathepsin B/L. Following viral entry, SARS-CoV-2 releases its genomic RNA into the cytoplasm and utilizes both the host’s and its own enzymatic machinery to replicate its genetic material and assemble new viral particles. The viral RNA genome is first translated into viral replicase polyproteins (pp1a and pp1ab), which are then cleaved into 16 nsps. In the process of genome replication and transcription mediated by the replication–transcription complex (RTC), the negative-sense (− sense) genomic RNA is synthesized and used as a template to generate a positive-sense (+ sense) genomic RNA and subgenomic RNAs. Viral assembly is aided by the interaction between viral genomic RNA and structural proteins located in the endoplasmic reticulum (ER) and ER–Golgi intermediate compartment (ERGIC). Finally, these virions are released to the plasma membrane via deacidified lysosomes and secreted from the infected cell via exocytosis. This figure was created with Biorender.com.
Epidemiology
Since the emergence of SARS-CoV-2 in China, the virus has rapidly spread worldwide and has shaken our health care and economic systems.10 Two years after the beginning of the COVID-19 pandemic, the virus remains a public health threat, although the number of cases and deaths has declined globally thanks mainly to the large scale deployment of effective vaccines.11 To date, the USA leads the number of laboratory-confirmed cases, followed by India, Brazil, and France. The global case fatality rate of COVID-19 is approximately 1.2%.11 A modeling study suggested that approximately 24% of fatal COVID-19 cases are underreported in the USA, representing more than 180 000 deaths, suggesting that almost a quarter of deaths attributable to COVID-19 are currently not reported as such on the death certificate.103 The Americas are currently responsible for almost half of all COVID-19 deaths throughout the world, followed by Europe, despite the fact that the latter reported higher total numbers of COVID-19 cases. A possible reason for this apparent disparity is test shortage in most Latin American countries, which may have underestimated the true incidence of COVID-19 (https://www.worldometers.info/coronavirus/). The observed differences may also be attributed to patient access to high-quality care. According to a study done in Brazil with the first 250 000 patients admitted to hospitals with COVID-19, 80% of the patients who needed invasive ventilation died, which is higher than the mortality reported for intubated patients in Europe (51.7% to 69%).104
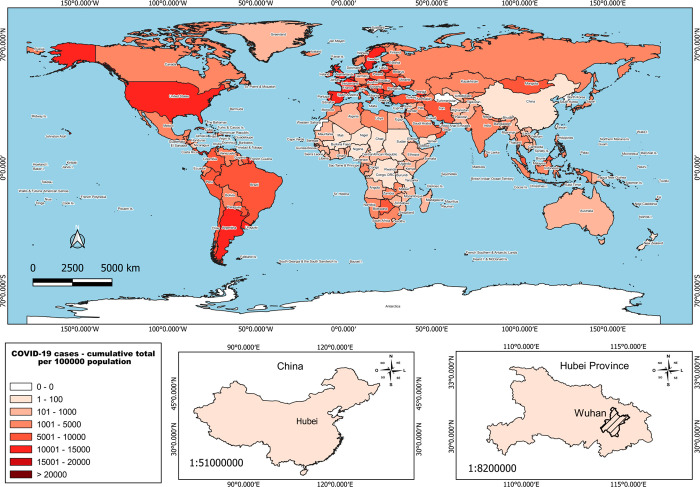
Compared with the global trend, the currently least affected continent is Africa. The first case of COVID-19 in Africa was reported on February 14, 2020, and to date, the epidemic curve on the continent has remained stable compared with the Americas and Europe. The African continent reported 8 488 173 cases and 170 610 deaths from COVID-19 as of March 15, 2022 (https://covid19.who.int/table). Attempts have been made to explain the low rate of COVID-19 morbidity and mortality in Africa, and possible associated factors have emerged, such as population demography, climate, urbanization, and economic level.105 Compounding the challenge of such studies, the impact of the pandemic appears to be poorly characterized in low- and middle-income countries. Insufficient diagnostic capabilities and inadequate infrastructure have limited the availability of robust data, resulting in uncertainty about the status of the pandemic.106Figure 4 shows the cumulative cases of COVID-19 in all countries across the world according to data from the WHO.
Figure 4.
Epidemiology map of COVID-19. Cumulative cases of COVID-19 in all countries throughout the world. The bottom panels indicate the geographic location of Wuhan and Hubei Province in China, where the first COVID-19 cases were identified. The data were obtained from the World Health Organization (WHO).
The transmissibility of a virus is indicated by the reproduction number (R0), which represents the average number of new infections generated by an infected person throughout their infectious period in a totally naive population. Therefore, for R0 < 1 the number of infections declines or remains constant, whereas for R0 > 1 the number of infections is likely to increase. SARS-CoV-2 studies have suggested that an exponential increase of SARS-CoV-2 infection occurs when R0 ranges from 1.4 to 6.49 with an average of 3.28,107,108 which corresponds to each infected person transmitting the virus to over three individuals.107 However, further studies are required to better understand the epidemiology and ability of SARS-CoV-2 to spread in the human population, especially after the deployment of effective vaccines and the emergence of more transmissible variants of SARS-CoV-2.
Modes of Transmission
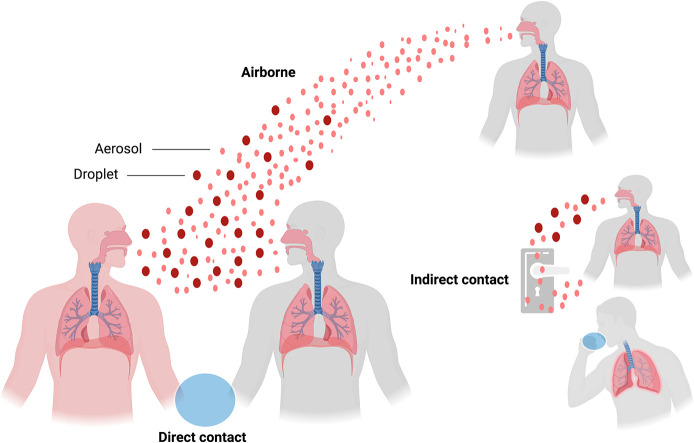
According to current evidence, SARS-CoV-2 is transmitted from person to person when the infectious particles are released from the respiratory tract of an infected individual and reach the respiratory tract of a susceptible individual.12,109 Briefly, SARS-CoV-2 can be transmitted through three main routes that are not mutually exclusive: (i) airborne transmission (respiratory droplets and aerosols), (ii) direct contact (infectious virus deposited on persons), and (iii) indirect contact (infectious virus deposited on fomites) (Figure 5).110 Notably, SARS-CoV-2 has a high human-to-human transmission rate through close contact with infected persons,111 especially when the infectious virus is expelled during talking, breathing, coughing, or sneezing by an infected individual.112−114 SARS-CoV-2 enters the body through the mucous membranes of the eyes, mouth, or nose and spreads to the sinus cavity, throat, and nose lining until deposition along the human respiratory tract.115 After infection, the viral load in the upper respiratory tract appears to peak together with symptom onset, and viral shedding starts nearly 2 to 3 days before symptoms begin.116 Epidemiological and modeling studies have shown that transmission of SARS-CoV-2 may occur from symptomatic, asymptomatic, and presymptomatic persons,117−121 which suggests that the identification and isolation of individuals with symptomatic COVID-19 alone will not control the ongoing spread of SARS-CoV-2.117
Figure 5.
Major modes of SARS-CoV-2 human-to-human transmission. Transmission can be through direct contact of airborne infectious particles deposited in respiratory droplets and aerosols. Indirect contact by infectious particles deposited on fomites represents another potential route for viral transmission. This figure was created with Biorender.com.
Airborne and direct contact are considered the dominant routes for spreading of SARS-CoV-2 among persons.12,13,122−124 Prolonged exposure to any infected individual (6 foot proximity for at least 15 min) and short exposures to symptomatic (e.g., coughing) infected persons are associated with successful virus transmission.125
SARS-CoV-2 transmission may also occur via contact with contaminated surfaces.109,126 Notably, the risk of infection through this route is probably multifactorial and is influenced by the distance from the viral source, the amount of virus to which an individual is exposed, and the length of time since the virus has been deposited on the surface. The viability of SARS-CoV-2 on surfaces over time is affected by several environmental stressors, including humidity, temperature, and level of ultraviolet radiation.12,127,128 Previous studies conducted under laboratory conditions have shown the ability of SARS-CoV-2 to remain infectious on various surfaces (e.g., paper, glass, and stainless steel) for up to 28 days at 20° C depending on the type of material129 and in aerosols for up to 3 h.128 To address this question in real-life settings, many recent studies have investigated the presence of SARS-CoV-2 contamination in the air and on environmental surfaces, including health care units123,124,130−132 and urban settings.126,133−135 The results demonstrate different levels of viral contamination varying from high130,136 to low132 or even no contamination by SARS-CoV-2 RNA.134 Additionally, most of the reported positive environmental samples were found to have high reverse transcription quantitative polymerase chain reaction (RT-qPCR) cycle threshold (Ct) values (>30) for most of the positive samples,130,136 indicating low viral load and the labile nature of SARS-CoV-2 in the environment. Importantly, the majority of these studies did not investigate the capacity of SARS-CoV-2 to be cultured from an environmental surface sample, which is crucial for understanding the role of SARS-CoV-2 RNA-positive samples in terms of infectious potential toward the human population.130,132,136 Taken together, recent aggregated studies reinforce the potential of environmental samples for SARS-CoV-2 transmission (i.e., indirect contact), although virus spread via close contact remains the primary route for SARS-CoV-2 transmission.
Other possible routes of transmission are being evaluated by research groups around the world, including the fecal–oral and blood-borne routes and vertical transmission from mothers to neonates. SARS-CoV-2 RNA has been detected in stool samples of COVID-19 patients, suggesting that viral shedding in the stool could be a potential route of fecal–oral transmission.137,138 In addition, SARS-CoV-2 has also been reported in blood samples, but the risk of blood-borne transmission was shown to be negligible.138,139 With regard to vertical transmission, several meta-analysis studies based on the current scientific evidence have suggested a low risk of such transmission for the spread of SARS-CoV-2.140−142 There is no evidence that the infection may lead to vertical transmission of SARS-CoV-2 or serious adverse outcomes in newborns.143−145
Clinical Manifestations
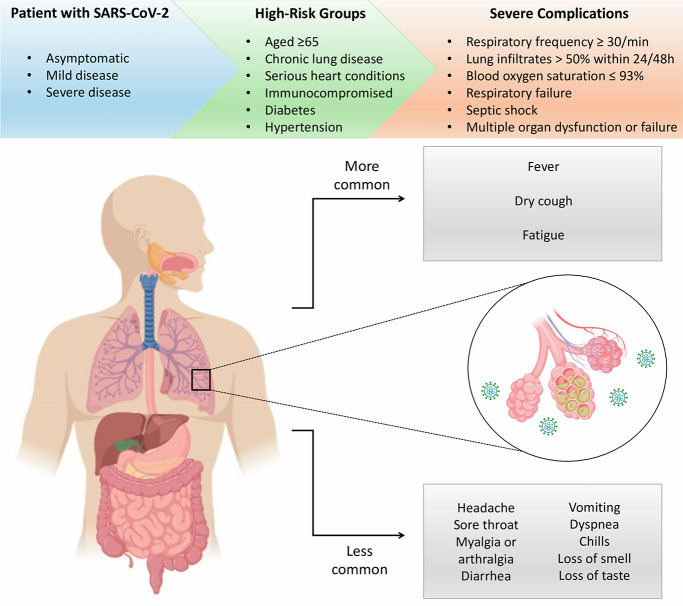
The mean incubation period (the time of exposure to symptom onset) of COVID-19 is approximately 5 days (95% confidence interval [CI], 4.1–7.0 days) and, when it occurs, pneumonia within a median time of 8 days from disease onset.15,20 Approximately 97% of infected persons who develop clinical manifestations will do so within 11 days of infection.146 The median interval from symptom onset to hospital admission is 7 days (3–9 days).147 Recent studies have demonstrated that people of all ages are susceptible to SARS-CoV-2 infection, although the median age of infection is around 50 years.14,16−18,20 Overall, men ≥65 years old with comorbidities are more likely to be susceptible to develop a severe respiratory illness that requires hospital admission, while most young people and children experience asymptomatic infection or mild disease (Figure 6).14,18
Figure 6.
Clinical manifestations of COVID-19. Patients infected with SARS-CoV-2 can be asymptomatic, develop mild disease with diverse symptoms, or progress to severe illness. COVID-19 cases with severe complications are more frequently presented by patients from the high-risk group. This figure was created with Biorender.com.
The initial clinical presentations of SARS-CoV-2 infection are varied and often similar to symptoms caused by other respiratory viruses such as influenza and parainfluenza viruses, therefore representing a challenge for clinical diagnosis.148 The most common symptoms of SARS-CoV-2 infection are fever, dry cough, and fatigue.14,16−18,149 Less common symptoms include headache, sore throat, myalgia or arthralgia, shortness of breath, diarrhea, vomiting, dyspnea, chills, and alterations in smell (anosmia, hyposmia) and taste (ageusia, dysgeusia).14,16−18,150 In a recent study of 417 mild-to-moderate European COVID-19 patients, 85.6% and 88.0% reported olfactory and gustatory disorders, respectively.151 It was demonstrated that these dysfunctions persisted after the resolution of other symptoms and that women were significantly more affected by olfactory and gustatory dysfunctions than men,151 although the prevalence of these disorders may occur with varying intensities and should be considered as part of the clinical presentations of COVID-19.152 A meta-analysis showed that anosmia or hyposmia is significantly associated with positive COVID-19 infections.153 Other atypical presentations of COVID-19 include cutaneous manifestations, where individuals can present different types of lesions such as urticarial, livedoid, purpuric, maculopapular, thromboticischemic, and papulovesicular.154−157
According to current evidence, COVID-19 is recognized as a multiorgan disease with a broad spectrum of clinical presentations.158,159 In a large study including 72 314 individuals with COVID-19 in China, 81% of the cases presented mild or moderate symptoms, 14% of infected patients eventually developed severe pneumonia that required ventilation in an ICU, and approximately 5% of the cases had critical manifestations, which included patients with respiratory failure, septic shock, and/or multiple organ dysfunction or failure.20,21 Like post-acute viral syndromes reported in survivors of other pathogenic coronaviruses, there are increasing reports of persistent and prolonged effects after acute COVID-19, which are characterized by persistent symptoms and/or delayed or long-term complications beyond 3–4 weeks from the onset of symptoms.159−161 Moreover, studies have suggested that COVID-19 patients can develop a chronic disease or post-COVID-19 syndrome, which includes symptoms and abnormalities persisting beyond 12 weeks of the disease onset.161−163 COVID-19 complications in patients with severe disease may include impaired function of the lung, liver, heart, brain, coagulation system, and kidney.158,159 Risk factors for the development of severe COVID-19 include age ≥65 years and comorbidities such as hypertension, diabetes, chronic pulmonary disease, chronic kidney disease, immunodeficiencies, chronic liver disease, cancer, cardiovascular disease, and obesity.164−169 Typical characteristics of patients with severe COVID-19 include respiratory frequency ≥ 30/min, lung infiltrates > 50% within 20/48 h, blood oxygen saturation < 93%, and an altered PaO2/FiO2 ratio, which is associated with high mortality and morbidity.19,20
COVID-19 is usually a mild disease in children, including infants. When infected, most children remain asymptomatic.170 However, a small proportion (4%) of children with COVID-19 develop severe disease requiring ICU admission and prolonged ventilation, although fatal outcomes are overall rare.171 Approximately 2–5% of infected patients with COVID-19 are younger than 18 years, with a median age of 11 years.171 The underlying mechanisms of the severity of COVID-19 in children are being rapidly unraveled.172 The presence of comorbidities, immunological response, and genetic factors have been investigated to understand the spectrum of disease in children and adults.172 As the pandemic progressed, a cumulative body of data reported a multisystem inflammatory syndrome in children (MIS-C) due to SARS-CoV-2 infection.173−176 The pathogenesis of this rare MIS-C is still unclear but shares some characteristics with Kawasaki disease, suggesting a vascular and likely autoimmune etiology.174 A recent meta-analysis study evaluated 783 cases of MIS-C between March and June 2020.177 The results revealed that patients with MIS-C have a high frequency of gastrointestinal symptoms (71%), including abdominal pain (34%) and diarrhea (27%). Other common symptoms include cough and respiratory distress, which were found in 4.5% and 9.6% of the cases, respectively.177 While exhibiting a low lethality rate (1.5%), MIS-C appears to be a condition of higher severity for infected patients.177
Pathophysiology
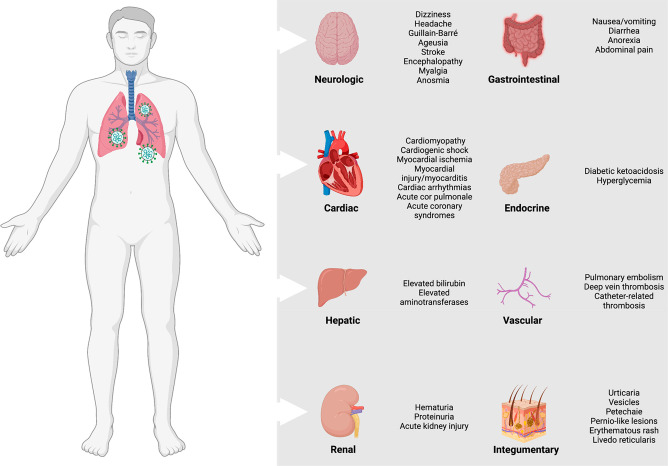
While SARS-CoV-2 infection is known to cause substantial pulmonary disease, including pneumonia and ARDS in most patients, extrapulmonary manifestations of the disease are also a quite common feature, especially in severe cases.178 These include associated complications in several systems, including the neurological, cardiac, hepatic, renal, gastrointestinal, endocrine, vascular, and integumentary systems (Figure 7).178,179 In short, there are key factors that may have critical roles in the pathophysiology of multiorgan injury secondary to infection with SARS-CoV-2. These mechanisms include endothelial cell damage, thromboinflammation, dysregulation of the renin–angiotensin–aldosterone system (RAAS), dysregulation of the immune response, and direct viral toxicity.178
Figure 7.
Extrapulmonary complications from COVID-19. The extrapulmonary complications include a wide spectrum of disorders in several systems, including the neurological, cardiac, hepatic, renal, gastrointestinal, endocrine, vascular, and integumentary systems, which may occur in severe and critically ill COVID-19 patients and are linked to prolonged hospitalization and increased mortality risk.178,679 This figure was created with Biorender.com.
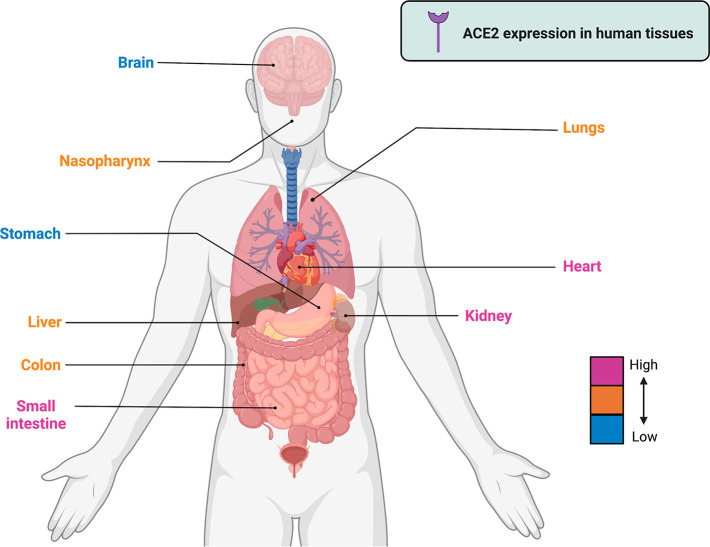
SARS-CoV-2 attachment and entry into alveolar epithelial cells are dependent on the presence of ACE2, a strategy that is shared with SARS-CoV.7,180 However, the affinity of SARS-CoV-2 S protein to human ACE2 is around 10- to 20-fold higher than that of the SARS-CoV S protein,181 which explains the higher transmissibility of the novel coronavirus.182 ACE2 is highly expressed in several human tissues, such as the small intestine, heart, kidneys, testis, thyroid, and adipose tissue, and is expressed at low levels in the brain, blood, bone marrow, spleen, muscle, and blood vessels. ACE2 is expressed at moderate levels in the lungs, liver, bladder, colon, and adrenal glands (https://www.proteinatlas.org/ENSG00000130234-ACE2/tissue) (Figure 8),183 which could explain the tropism of SARS-CoV-2 and the wide spectrum of clinical pulmonary and extrapulmonary manifestations associated with COVID-19 disease.
Figure 8.
Gene expression of the ACE2 receptor in human tissues. The level of expression in each organ is categorized from high to low using different colors. Sources: https://www.proteinatlas.org/ENSG00000130234-ACE2/tissue and Li et al. (2020).183 This figure was created with Biorender.com.
SARS-CoV-2 requires proteolytic processing of the S protein to promote viral entry. Recent studies have demonstrated that proteases, including TMPRSS2, furin, and CatB/L, participate in cleavage of the S protein, resulting in entry of SARS-CoV-2 and fusion of the viral envelope and endosome.86,90,184 More recently, it has been suggested that SARS-CoV-2 can bind to another surface receptor, CD147, which would provide an additional route for host cell invasion (CD147–S protein).93 It is presumed that primary viral replication takes place in the mucosal epithelium of the upper respiratory tract, followed by multiplication in the lower respiratory tract.185 During viral entry into cells, pattern recognition receptors (PRRs), such as the endosomal toll-like receptors (TLR), can detect viral genomic RNA, which triggers an inflammatory response.186 Activation of TLR-3 or TLR-7 triggers a signaling pathway that releases the main transcriptional regulator of inflammation, NF-κB, from its inhibitor.187 Recognition of the S protein by TLR-2 placed on the cell surface can also contribute to signaling that activates the host defense response.187 Despite being a part of the host defense system, overactivation of TLR and its adaptor protein MyD88 has been hypothesized as a predisposition factor for exacerbated inflammation observed in COVID-19 patients with obesity.188 Once released, NF-κB migrates to the nucleus and activates genes that codify proteins with immunological properties, such as cytokines, chemokines, and other immunological mediators.189
Thereafter, fusion between the viral envelope and endosomal membrane induces release of the viral genome into the cell, which can be identified by other PRRs in the cytosol like MDA-5 or RIG-1.190,191 These PRRs will also trigger the activation of NF-κB, although through a different signaling pathway,191 and activate transcriptional regulators including IRF-3 and IRF-7 that induce the expression of the class I interferon (IFN) genes.191 An efficient antiviral response relies on the production of the class I IFNs IFN-α and IFN-β.192,193 The IFN family of proteins play key roles in host immunological response against viruses and other pathogens.194 Briefly, canonical type I IFN signaling activates the Janus kinase (JAK)–signal transducer and activator of transcription (STAT) pathway, which leads to the transcription of IFN-stimulated genes (ISGs) that confer antiviral activities to host cells.194−198 In the context of SARS-CoV-2 infections, a cumulative body of data has demonstrated that severe COVID-19 cases are associated with the presence of auto-antibodies that block the action of specific IFNs,199,200 a reduction of IFN signaling,201,202 and genetic variants that impair IFN signaling.203,204In vivo studies suggested type I IFN signaling as a driver of pathology in mouse models for SARS-CoV and SARS-CoV-2 infections.205 In a rapidly moving field of study, recent reports suggested that type I and III IFNs are linked to a disruption of lung barrier function and increased susceptibility to secondary bacterial infections in mice.206,207 Additionally, type II IFN, also known as IFN-γ, is transmitted through a different receptor, has effects that are independent of type I IFNs, and also plays a relevant role in combating viral infection and modulating the antiviral immune response.208 In COVID-19 patients with moderate and severe infection, IFN-γ was documented as an independent risk factor associated with mortality.209 Taken together, these findings and results reported by others have revealed multiple roles of IFN signaling during the clinical course of COVID-19, meaning that it is possible to observe context-dependent variations and that IFN signaling may attenuate or exacerbate COVID-19 pathology.210
In the absence of a proper antiviral response mediated by IFNs, the host will rely on other innate immune mechanisms for defense, like the immune cells.211 Activation of NF-κB leads to the production of cytokines and chemokines that will recruit and activate immune cells from the bloodstream.198,212 The first immune cells to reach the infection site from the circulation are neutrophils and monocytes.213 Several chemokines, such as CCL2, CCL3, and CXCL10, together with CCL7, are potent chemokines for monocytes and have been found at high concentrations in COVID-19 patients with severe disease.211 Abnormal levels of monocyte population subsets have been demonstrated in COVID-19 patients, suggesting the migration of intermediate (CD14++ CD16+) and nonclassical monocytes (CD14+ CD16+++) to inflamed tissue.214−216 Nonclassical monocytes have previously been associated with an immune response against viral infection.216 Normal or nearly normal levels of those monocyte subsets have been associated with moderate illness, and this has been suggested as a favorable prognostic indicator.214,216
More recently, Chevrier and colleagues described important insights about the immune signatures involved during the progression from mild to severe COVID-19 disease.217 The authors used mass cytometry and targeted proteomics to profile the innate immune responses of patients with mild or severe COVID-19 and healthy individuals. The results showed that the production of CD169+ monocytes, combined with IFN-γ+ MCP-2+ monocytes, rapidly follows symptom onset. At later stages, they found a persistent inflammatory phenotype in patients with severe COVID-19, dominated by high CCL3 and CCL4 cytokine abundance correlated with the reappearance of CD16+ monocytes, while the response of mild COVID-19 patients was normalized.217
Macrophages and monocytes are also cell types that are susceptible to SARS-CoV-2 infection, which triggers the production and secretion of inflammatory cytokines.218−220 Increased levels of several cytokines have been reported in patients with severe COVID-19, including interleukin (IL)-1β, IL-2, IL-6, IL-7, IL-8, IL-10, IL-17, IFN-γ, IFN-γ-inducible protein 10, MCP-1, G-CSF, MIP-1α, and TNF-α.213 The term cytokine storm is used to define this pathological overproduction of cytokines that leads to systemic inflammatory response affecting several organs, such as the heart, liver, and kidney, and is the leading cause of death in COVID-19 patients.17,221,222 High levels of proinflammatory cytokines in the circulation trigger several symptoms, including fever, headache, rash, diarrhea, arthralgia, and myalgia.221 A high level of cytokines in the circulation can lead to hypotension, vascular leakage, disseminated intravascular coagulation, and multiorgan failure.223 Many of these symptoms were reported in critical patients with COVID-19, and cytokine storm has been associated as part of the pathology in severe cases,224 although the mechanism that triggers this exacerbated immunological response has not been completely characterized. Although antibody-dependent enhancement (ADE) of infection plays a critical role in the pathogenesis of many viral infections, antibodies induced by SARS-CoV-2 infection do not contribute to inducing aberrant cytokine production by macrophages.225 Recent findings have shown that SARS-CoV-2 infection in lung-resident human macrophages is a critical driver of the immunological response during COVID-19 disease.226 In response to SARS-CoV-2 infection, human macrophages activate inflammasomes, release IL-1 and IL-18, and undergo pyroptosis, thereby contributing to the hyperinflammatory state of the lungs.226 Together, these results suggest that the inflammasomes oppose host infection by production of inflammatory cytokines and suicide by pyroptosis to prevent a productive viral cycle.226
Neutrophils are also recruited to the sites of viral infection.227,228 They are effector cells that produce reactive oxygen species that damage infected tissues.229 The damage caused by the neutrophils contributes to the virus clearance and the elimination of invasive pathogens.230 The virus infection and excessive oxidative stress can induce the release of damage-associated molecular pattern (DAMP), which can act as a proinflammatory stimulus.230 Neutrophilia in the lungs has been associated with enhanced tissue injury and pneumonia in COVID-19 patients, and the increase in the neutrophil/lymphocyte ratio (NLR) has been suggested as a risk factor for disease severity.231 Activation of neutrophils can also promote the formation of neutrophil extracellular traps (NETs).213,230 NET release was also induced by infective SARS-CoV-2 in neutrophil culture cells in vitro but not by an inactive virus, suggesting that the process of viral infection may be a trigger for NET induction.232 As part of NETs, neutrophils die, releasing their DNA and other bioactive molecules, which further reinforce the inflammatory response and enhance the prothrombotic disbalance.233 The formation of NETs can constrain the spread of a pathogen through circulation as well as lead to the release of antimicrobial compounds.230,233 Post-mortem histological data from patients with severe COVID-19 described increased numbers of degenerated neutrophils, indicating NET formation.234 Additionally, the procoagulation stimulus of the NET has been associated with the systemic manifestation of vascular disbalance, thrombi formation in the microvasculature of several organs, and ARDS.233
The respiratory system is the primary site of virus-induced immunopathology.235 In some cases the virus reaches the lower respiratory tract and infects the alveolar cell lining and pneumocytes I and II,236 leading to the secretion of immune mediators that activate the endothelial cells.237 Activation of the endothelium weakens epithelial barrier function, which increases the influx of fluid from circulation to the surrounding interstitial area, leading to edema.237 The fluid is accompanied by the recruitment of immune cells such as monocytes and neutrophils, which cross the endothelial barrier driven by the secretion of chemokines such as CCL2, CCL3, CCL7, and CXCL10.211 In the surrounding tissue, these cells enhance the damage associated with exaggerating inflammation and contribute further to thrombi formation.215 The hyperinflammatory response leads to disorder of the pneumocyte’s alveolar lining and ARDS, disrupting the gas exchange function.238
Once the virus reaches the bloodstream, it can spread and infect other cells.235 It has been shown that vascular manifestations are caused not by SARS-CoV-2 infection of blood vessel cells but rather by viral-inflammation-induced endothelial activation and barrier disruption.239,240 This has been associated with the formation of thrombi in the microvasculature and ischemia in the limbs and extremities of COVID-19 patients.241 The resulting intravascular coagulation has stimulated the use of anticoagulant therapy in these patients.242
Abnormal blood parameters, such as D-dimer and fibrinogen levels, are also reported in critical COVID-19 patients.243 These abnormalities could reflect the excessive inflammation caused by elevated levels of the proinflammatory cytokine IL-6.178 Other abnormal parameters found in patients with severe illness are elevated C-reactive protein (CRP) and lactate.244−246 Once the endothelium becomes infected, the virus induces an immune response, mainly mediated by neutrophils and lymphocytes, against the related endothelial tissues, triggering endotheliitis.239 However, other reports have shown the opposite results, suggesting that endothelial cells are not efficiently infected by SARS-CoV-2.240 A cytokine storm can also drive vessel leakage syndrome, which is characterized by uncontrolled efflux of fluid from circulation to surrounding tissues.223,247
SARS-CoV-2 can spread to other organs such as the heart, kidney, and liver.178 Cardiac injury is a common condition among hospitalized COVID-19 patients, and it is associated with a higher risk of mortality.17,248 A cumulative body of data has demonstrated that SARS-CoV-2 infection can cause both direct and indirect cardiovascular sequels, including arrhythmias, cardiogenic shock, acute coronary syndrome (ACS), myocardial injury, cardiomyopathy, acute cor pulmonale, and thrombotic complications.249,250 Similarly, cardiac injuries have also been associated with infection by other highly pathogenic CoVs, including SARS-CoV and MERS-CoV.251 Autopsy results have shown interstitial myocardium infiltration of mononuclear cells,19 and myocarditis has been reported in more than 20% of the patients hospitalized in the ICU.252 Despite not being a specific marker, the increased level of troponin can suggest myocardial damage, and abnormal high levels are observed in patients with COVID-19.253 Other heart abnormalities in COVID-19 patients have also been reported, including cell necrosis, dysfunctionality of myocytes, arrhythmia, and even cardiac arrest.254
Acute kidney injury is a complication that is frequently reported in critical-condition patients and is associated with mortality, resulting in proteinuria, hematuria, and leukocyturia.255−257 The formation of thrombi in the microvasculature of the kidney in association with NET formation has also been reported.258 Unlike the lungs, SARS-CoV-2 infection of the kidneys occurs later in the course of the disease when other more severe symptoms have already been exhibited. As a result of this process, kidney damage can be a consequence of a hyperinflammation response, cytokine storm, and hypoxia.221,223,259 Hyponatremia, hypochloremia, hypocalcemia, and acidosis are common electrolyte abnormalities associated with the high cell turnover seen in COVID-19 patients with acute kidney injury.260,261 Direct SARS-CoV-2 infection of kidney cells has been reported using in vitro and post-mortem studies and may also contribute to local inflammation and kidney damage.262−264
The gastrointestinal (GI) tract is also a system affected by coronaviruses in animals and has been associated with life-threatening infections.265 GI symptoms in COVID-19 are usually self-limited and include diarrhea, vomiting, nausea, abdominal pain, and discomfort.266,267 Similarly, other coronaviruses, such as SARS-CoV and MERS-CoV, have been associated with GI symptoms in some patients.266 Enterocytes can be productively infected by SARS-CoV-2,268,269 and virus particles have been observed in stool samples.270 This suggests that direct viral damage could be the cause of enteric manifestations. In some patients, the development of enteric symptoms can precede respiratory symptoms.265,271 Although fecal–oral transmission is a possible route for SARS-CoV-2 infection of the GI tract,272 the virus spreads from person to person mainly through direct contact or airborne transmission.12,13
Liver damage can also result from systemic hyperinflammatory responses such as cytokine storms, NET-mediated coagulation, and hypoxia,178 and altered liver function has been identified in more than 50% of hospitalized patients.273 Elevated levels of aminotransferases, such as alanine aminotransferase and aspartate aminotransferase, together with a slight increase in bilirubin levels have been associated with SARS-CoV-2-related liver injuries.274 Müller and colleagues demonstrated that SARS-CoV-2 can infect cells of the human exocrine and endocrine pancreas in both in vivo and ex vivo models.275,276 However, recent evidence suggests that despite the susceptibility of all pancreatic cell types to SARS-CoV-2, viral infection leads to only modest cellular alterations and inflammatory responses. Interestingly, infection by SARS-CoV-2 could lead to new-onset diabetes,277 and further studies to explore this possibility are certainly warranted.
Infection by SARS-CoV-2 has been associated with a range of neurological complications, and viral RNA and virus particles have been found in post-mortem analysis of brain tissue, suggesting that SARS-CoV-2 is neuroinvasive and neurovirulent.278,279 During SARS-CoV-2 infection, the most common neurological symptoms reported range widely in severity; these include mild symptoms, such as sensorial disturbance, headache, hyposmia, hypogeusia, confusion, and dizziness,278,280 and severe symptoms, such as consciousness disorders, seizures, and paralysis.278,280−282 Neurological disorders such as acute inflammatory demyelinating polyneuropathy (Guillain-Barré syndrome) have also been documented in some COVID-19 patients.283,284 The virus’s main route to the nervous system is through the bloodstream, although alternative pathways through the cribriform plate or ethmoid bone have been suggested.285 Direct viral damage associated with the hyperinflammatory response, hypoxia, and metabolic disorders are the mechanisms thought to be involved in neurological COVID-19.285,286 Additionally, other severe neurological complications documented in COVID-19 patients include hemorrhagic posterior reversible encephalopathy syndrome, meningoencephalitis, and acute necrotizing encephalopathy.281,287−289
The mechanisms underlying the taste and olfactory dysfunctions have been the focus of many scientific studies. It has been shown that SARS-CoV-2 can result in downregulation of olfactory receptors and their signaling pathways,290 although the virus does not seem to directly infect the sensory neurons of the olfactory epithelium in COVID-19 patients.291 SARS-CoV-2 can infect a myriad of cells in the oral cavity, including human type II taste cells,292,293 which may explain the taste dysfunction during and after acute COVID-19.
Despite ethical concerns of studies that challenge humans with SARS-CoV-2, scientists recently infected 36 healthy naïve volunteers (male and female) in the U.K. aged 18–29 years with a low dose of SARS-CoV-2 (10 TCID50 of a wild-type virus) intranasally under controlled conditions.294 That study provided detailed insights into SARS-CoV-2 infection. Symptoms started to develop very quickly, on average about 2 days after contact with the virus.294 Interestingly, the infection first appeared in the throat, and the infectious virus peaked at about 5 days during the clinical course of the infection, and at that stage it was significantly higher in the nose (peaking at ∼8.87 log10 copies/mL) than in the throat.294 In addition, the results demonstrated that mild to moderate symptoms were reported in 89% of the infected participants (n = 16), beginning 2–4 days after inoculation, whereas 11% of the participants (n = 2) remained asymptomatic.294 Together, these results provide relevant insights into viral kinetics throughout primary infection with SARS-CoV-2 and represent the first SARS-CoV-2 human challenge study in young adults.
Diagnosis
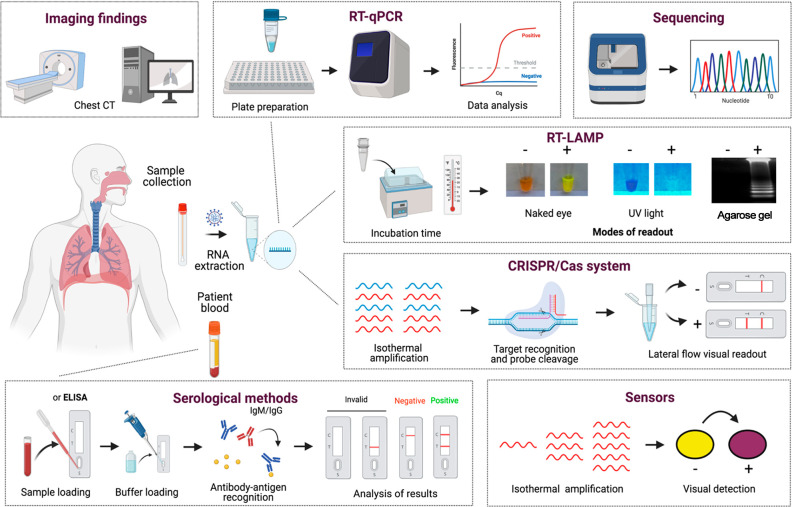
Early diagnosis is essential for contact tracing, identification of hot-spot areas with active community transmission, and control of the spread of SARS-CoV-2.150,295,296 Current confirmation of COVID-19 disease can be achieved through clinical symptoms, imaging findings, biomarker evaluation, nucleic acid tests, and serological methods. Briefly, direct tests are used to detect the presence of viral particles, virus antigens, or viral RNA, while indirect tests are used to detect the immunological response against SARS-CoV-2 in infected patients, particularly to detect immunoglobulin M (IgM) and IgG antibodies.150 In this section, we provide an overview of the clinical course of COVID-19 reported in infected patients. Moreover, we explore the different detection methods being developed or used for SARS-CoV-2 diagnosis, discussing their advances, principles, advantages, and limitations (Figure 9).
Figure 9.
Overview of different methods for COVID-19 diagnosis. SARS-CoV-2 can be directly detected in humans using molecular approaches, such as RT-qPCR, DNA sequencing, RT-LAMP, CRISPR/Cas systems, and sensors. Imaging tests, including chest computed tomography (CT), have been widely used as a complementary approach to diagnose COVID-19 patients. Additionally, human antibodies produced against SARS-CoV-2 antigens can be detected in blood samples via serological methods, including enzyme-linked immunosorbent assay (ELISA), chemiluminescence immunoassay (CLIA), immunofluorescence assay (IFA), and lateral flow assay (LFA). This figure was created with Biorender.com.
Clinical Course
Understanding the temporal dynamics of viral shedding and immune response in patients with COVID-19 is critical to correctly diagnose the SARS-CoV-2 infection. Since the beginning of the pandemic, viral shedding profiles of COVID-19 patients have been investigated.116,297−299 A recent meta-analysis study analyzed the viral load dynamics, duration of viral RNA shedding, and viable virus shedding of SARS-CoV-2 in several body fluids.300 Using 79 studies (5340 individuals), the report demonstrated that the mean durations of SARS-CoV-2 RNA shedding were 14.6 days (95% CI 9.3–20.0 days; seven studies, 260 individuals) in the lower respiratory tract, 17.0 days (95% CI 15.5–18.6 days; 43 studies, 3229 individuals) in the upper respiratory tract, 16.6 days (95% CI 3.6–29.7 days; two studies, 108 individuals) in serum samples, and 17.2 days (95% CI 14.4–20.1 days; 13 studies, 586 individuals) in stool.300 The longest durations of SARS-CoV-2 RNA shedding were 59 days in the lower respiratory tract, 83 days in the upper respiratory tract, 60 days in serum, and 126 days in stool.300 It was found that the peak occurs in the first week of the disease, while for SARS-CoV and MERS-CoV RNA the peaks occur in the ranges of 10–14 and 7–10 days, respectively.300 In patients with severe disease, the viral load appears to reach its highest level in the third and fourth weeks, while in patients with comorbidities, viral persistence is continuous.301,302 However, recent findings showed that infectious particles could not be detected beyond day 9 of disease.300 Therefore, SARS-CoV-2 isolation from respiratory samples should use specimens collected during the initial stages of COVID-19 that present a low cycle threshold (Ct < 24) on RT-qPCR.303 Typically, nucleic acid tests are commonly used to detect and amplify the SARS-CoV-2 genome from several types of specimens, including nasopharyngeal swabs, oropharyngeal swabs, or other upper respiratory tract samples.304 By the use of RT-qPCR, SARS-CoV-2 RNA is detected as early as day 1 of symptoms and peaks within the first week of symptoms onset.304 This positivity starts to decline by week 3, and subsequently SARS-CoV-2 RNA becomes undetectable.304 However, the viral load of severe COVID-19 cases was estimated to be 60 times higher than that of mild cases,299 and subsequently SARS-CoV-2-positive RT-qPCR may persist beyond 3 weeks after disease onset, while most mild cases will present a negative result.304,305
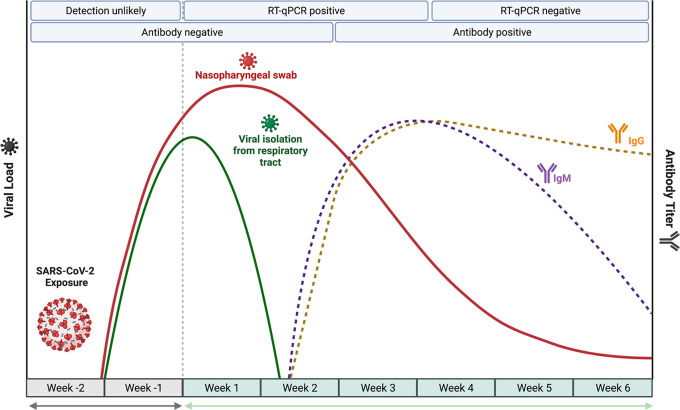
The host immune response to SARS-CoV-2 infection has also been investigated.306−310 In most COVID-19 patients, IgM levels increase during the first week after SARS-CoV-2 infection, reach their peak after 2 weeks, and subsequently fall back to near-background levels.311 Similarly, IgG is detectable 1 week after disease onset and is maintained at a high level for a long period, even more than 48 days.312 Recent studies found that seroconversion for IgG and IgM occurred simultaneously or sequentially, and both IgG and IgM titers plateaued within 6 days after seroconversion.307 In a large study with 285 patients with COVID-19, 100% of patients tested positive for IgG within 19 days after symptom onset.307 During the host immune response against SARS-CoV-2 infection, COVID-19 can be detected indirectly using serological methods, particularly detection of IgM and IgG. Briefly, Figure 10 describes how to interpret two types of diagnostic approaches commonly used for SARS-CoV-2 diagnosis (RT-qPCR and serological methods) and how the results may vary over time during COVID-19 clinical progression.
Figure 10.
Kinetics of viral load and immune response during SARS-CoV-2 infection. During the first week after SARS-CoV-2 exposure, a period when patients are typically presymptomatic, the viral load increases and reaches its peak during the initial days after symptom onset. Seroconversion in infected patients begins in the second week after symptom onset. Three to four weeks after symptom onset, the IgM and IgG levels both reach their peaks and then begin to drop—more rapidly for IgM than for IgG. To avoid false-negative results when COVID-19 diagnostic tests are performed, the kinetics of viral load and immune response should be taken into consideration. The figure was adapted from the template in Biorender.com.
Imaging Findings in COVID-19 Patients
In the early stage of SARS-CoV-2 infection, symptoms are usually nonspecific. This makes clinical diagnosis difficult, especially in areas with the circulation of other respiratory viruses, such as influenza virus and human rhinovirus (HRV), and even nonrespiratory pathogens such as dengue virus (DENV).148,295,313 Because of the nonspecific clinical manifestation of the disease, chest computed tomography (CT) has been widely used as a complementary tool in the investigation of COVID-19 patients. It has been used to evaluate the disease progression and assess the impairment of the lower respiratory tract and other anatomical areas by the disease.314,315 Chest CT scan images have been used to check for possible abnormalities suggestive of lower respiratory tract disease, such as viral pneumonia, eventually caused by SARS-CoV-2.316 Typically, the most important imaging changes observed in COVID-19 patients are multilobe lesions in both lungs and bilateral and peripheral ground-glass opacity (GGO) with or without consolidated changes.314 Other findings include rounded opacities, a crazy-paving pattern, an air bronchogram, and septal thickening mainly distributed in peripheral and posterior areas.317,318 It has been suggested that during patient clinical management, CT scanning combined with RT-qPCR should be used in the routine for the diagnosis of patients with a high clinical suspicion of COVID-19 who had a negative result on the RT-qPCR assay.319
Clinical Biomarkers in COVID-19 Patients
Besides the laboratory methods for detecting SARS-CoV-2 discussed throughout this review, many studies have demonstrated that hematological, biochemical, and blood chemical alterations in COVID-19 patients are possible markers of disease progression and patient health.244,320−323 The levels of these markers fluctuate depending on the clinical course of the disease, and since they can be assessed by routine blood tests, additional testing can be ordered by physicians on the basis of patients’ clinical evolution. Patients with increasing SARS-CoV-2 severity often have leukocytosis, leukopenia, decreased albumin levels, increased levels of lactate dehydrogenase (LDH), CRP, bilirubin, and creatinine kinase, and a high erythrocyte sedimentation rate (ESR).324 In general, no individual biomarker can be used to confirm or discard COVID-19 diagnosis, and diagnostic testing should be conducted for all suspected cases.
It should be noted that some clinical biomarkers have an important value for patient management since they can be used to assess the progression of the disease and its severity and even act as risk factors for death. Compared with healthy individuals, clinical biomarkers associated with increased disease severity in patients with COVID-19 include lymphopenia, thrombocytopenia, and high levels of liver alanine aminotransferase (ALT), aspartate aminotransferase (AST), LDH, CRP, and ferritin.321,325 A meta-analysis study demonstrated that patients with fatal COVID-19 disease progression had significantly increased white blood cell (WBC) count and decreased lymphocyte and platelet counts compared with nonsevere illnesses and survivors.326 Additionally, it was found that biomarkers of inflammation, cardiac and muscle injury, liver and kidney dysfunction, and coagulation measures were also significantly elevated in patients with both severe and fatal COVID-19.326 Elevated levels of serum biomarkers IL-6, IL-10, ferritin, CRP, and cardiac troponin acted as strong discriminators for disease severity and were associated with an increased risk of death.246,326,327
RT-qPCR
RT-qPCR is currently considered the gold-standard lab method for the diagnosis of SARS-CoV-2.150 Because of its high sensitivity and specificity, this technique allows detection of viral RNA in the first days after symptom onset during the initial stages of the disease or even during presymptomatic or postsymptomatic phases.311,328 Choosing the correct specimen for testing is a critical step to produce a reliable diagnosis.150 SARS-CoV-2 RT-qPCR is most often performed on upper respiratory specimens, which include nasopharyngeal or oropharyngeal swabs, aspirates or washes, sputum, and bronchoalveolar fluids.138,328−330 In addition to respiratory tract samples, SARS-CoV-2 detection by RT-qPCR has been documented in other specimens such as blood, urine, anal swabs, ocular secretions, breast milk, semen, and feces.138,331−335 Because of the discomfort associated with respiratory tract sampling, the need for trained healthcare personnel, and the risk of aerosol or droplet production, there is a great interest in alternative methods to collect samples from COVID-19 patients. Less invasive samples such as saliva and gargle lavages (“mouthwashes”) are promising alternatives for use in the routine, especially in patients with a high viral load.336−339
Since the emergence of SARS-CoV-2, many molecular assays based on RT-qPCR have been developed or are being used by clinical, research, and public health laboratories for the diagnosis of COVID-19.340−343 In addition to assays recommended by the WHO, many molecular RT-qPCR kits have been approved by the U.S. Food and Drug Administration (FDA) and are widely available for the detection and amplification of SARS-CoV-2 RNA (https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations). A variety of molecular targets within the SARS-CoV-2 genome have been used, with most assays targeting one or more genes, such as the spike (S), envelope (Env), nucleocapsid (N), RNA-dependent RNA polymerase (RdRp), and open reading frame (ORF) genes.304 In the initial stages of the COVID-19 pandemic, dual- or multigene detection strategies were adopted for RT-qPCR assays to ensure assay specificity.344 As the pandemic evolved and the disease prevalence increased, many laboratories around the world implemented a workflow using single-target detection of SARS-COV-2.315 To minimize false negatives associated with technical errors, internal control (IC) targeting human “housekeeping” transcripts like RNase P mRNA should be included during the testing of patient’s samples by RT-qPCR.345
Four of the most commonly used SARS-CoV-2 RT-qPCR assays have been developed by the China National Institute for Viral Disease Control and Prevention,342 the U.S. Centers for Disease Control and Prevention (CDC),341 Charité Institute of Virology, Universitätsmedizin Berlin (Charité),340 and Hong Kong University (HKU).343 In this context, Vogels and colleagues evaluated the analytical efficiencies and sensitivities of these four primer–probe sets to detect SARS-CoV-2.346 The results demonstrated that all of the primer–probe sets can be used to detect SARS-CoV-2 at 500 viral RNA copies per reaction, except for the RdRp-SARSr (Charité), which presented low sensitivity.346 In another related study, Nalla and co-workers evaluated the performance of seven RT-qPCR protocols recommended by the WHO for detecting SARS-CoV-2 RNA from patient samples.347 It was found that the most sensitive assays were those that used the E-gene primer–probe set described by Corman et al.(340) and the N2 set developed by the CDC.341 In addition, the results demonstrated that all of the RT-qPCR assays evaluated were highly specific for SARS-CoV-2 detection, and no cross-reactivity against other respiratory viruses was reported.347
Throughout the COVID-19 pandemic, reference laboratories have faced global shortages of diagnostic supplies, especially for the RNA extraction step.348−351 To meet this need, simplification of nucleic acid tests by eliminating the RNA extraction step is being explored.150,349 Studies showed that skipping RNA extraction by simple direct heating of samples for 5 min at 95–98 °C resulted in sensitivity and specificity comparable to the standard RT-qPCR method,349,351 suggesting that direct RT-qPCR without an RNA extraction step is a viable option to perform the diagnosis of COVID-19 patients. Although this strategy is promising and has great potential for the application of diagnostic workflows in low-resource settings, particular attention should be given to the increase of false-negative results.352
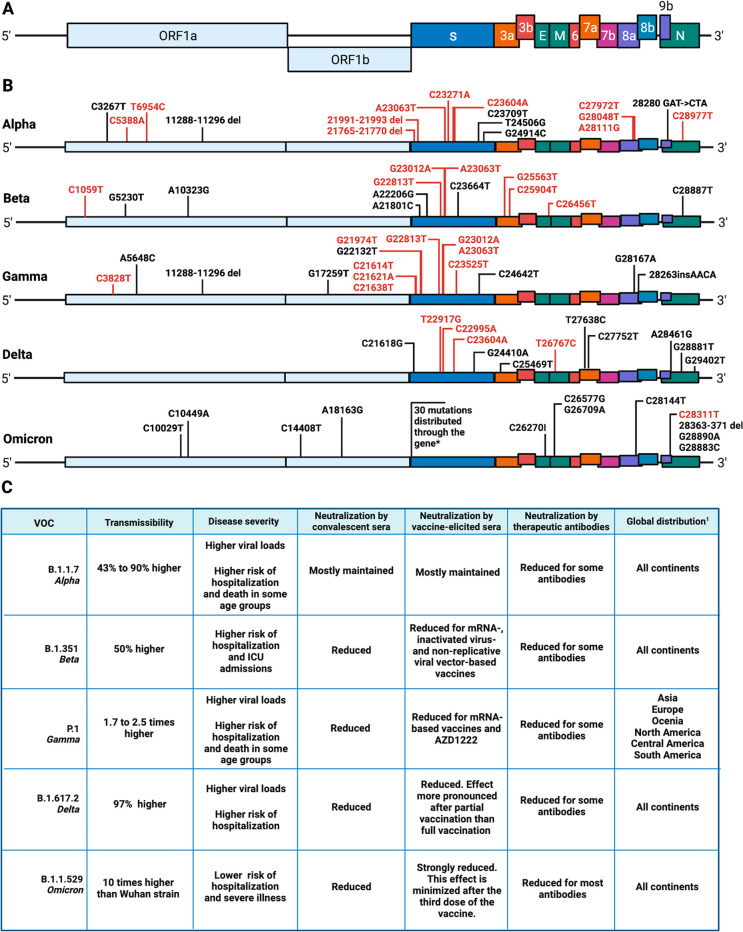
More recently, with the emergence of SARS-CoV-2 variants that may increase transmissibility and/or cause escape from immune responses, there is an urgent need for the targeted surveillance of these circulating variants in laboratories around the world.353 Vogels and colleagues designed and validated a multiplex RT-qPCR assay to detect SARS-CoV-2 variants of concern (VOCs).353 Using detection of the deletion Δ3675–3677 in the ORF1a gene, they were able to indicate the presence of the emerging variants B.1.1.7 [alpha], B.1.351 [beta], and P.1 [gamma] since this mutation had not already been detected in other SARS-CoV-2 variants. Detection of the deletion Δ69–70 in the S gene was applied to differentiate these three lineages.353 It will be crucial that optimization and validation of diagnostic tests continue as the COVID-19 pandemic evolves, since new variants of SARS-CoV-2 may emerge and existing assays must be constantly evaluated to ensure that the diagnosis is performed with high efficiency and accuracy.
Genome Sequencing
Although genome sequencing does not play a critical role in routine laboratory diagnostics for SARS-CoV-2, it has gained relevance with the emergence of new viral variants because it is essential for tracking changes in the viral genome over time and tracing transmission patterns.150,354,355 To date, only a limited number of reports have explored the use of NGS for SARS-CoV-2 detection with diagnostic purposes.356−358 For instance, Bloom and colleagues developed a protocol using NGS, called SwabSeq, to detect SARS-CoV-2 RNA in patient samples, including nasal or saliva samples, in a single run without the need for RNA extraction.357 The authors incorporated a synthetic RNA standard that facilitates end-point quantification and the calling of true negatives and reduces the requirement for automation, purification, and sample normalization. After 80 000 tests performed within 2 months, the results revealed an analytical sensitivity and specificity comparable to or better than the RT-qPCR reference method. Recent findings support the potential of NGS as a diagnostic tool for SARS-CoV-2 detection, although practical difficulties like high cost, shortage of globally available supplies, the need for a specialized laboratory infrastructure, sophisticated instrumentation, bioinformatics expertise, and well-trained staff may pose significant bottlenecks to its use for confirming COVID-19 infection, especially in low-resource countries.359 Another limitation of NGS technologies is their low analytical sensitivity, which may negatively affect the analysis of results.315 For NGS sequencing, the use of specimens with high viral loads (low Ct values) is recommended to generate high-quality results. Samples with low viral loads (high Ct values) can generate insufficient data or poor-quality results for subsequent analyses.356,360
Notably, sequencing protocols based on NGS (e.g., Illumina, BGI MGISEQ2000, and Nanopore [MinION]) and Sanger methods are being used to rapidly generate SARS-CoV-2 genome sequences from patient samples.7−9 As of June 15, 2022, 11 416 875 genome sequences have been deposited on the Global Initiative on Sharing All Influenza Data (GISAID) (https://www.gisaid.org/), including whole-genome sequences from COVID-19 patients from different countries around the world. To shed light on the ongoing evolution of the SARS-CoV-2 genome during the pandemic, sequencing is essential in order to identify mutations that may be associated with escape from vaccine-induced immunity or natural-induced immunities, diminishment of the performance of current diagnostics, and increased transmissibility and/or lethality.361,362,353,363−365 Thus, the development of rapid and low-cost sequencing protocols is crucial for discriminating all emerging SARS-CoV-2 variants as the pandemic evolves. To address this question, Bezerra and co-workers proposed a rapid and accessible protocol based on Sanger sequencing of a single PCR fragment that can identify and discriminate SARS-CoV-2 VOCs.359 They evaluated 12 samples from Brazilian patients using both NGS and Sanger sequencing approaches. Taken together, the findings from the Sanger sequencing matched the NGS results 100%.359 Moreover, this protocol allows a much broader network of laboratories to perform molecular surveillance of SARS-CoV-2 VOCs and report results within a shorter timeline, especially to increase sequencing capacity in low- and middle-income countries. In view of the evolving nature of the SARS-CoV-2 genome, genomic surveillance should be conducted and implemented on a large scale to allow early identification of new variants and to help establish policies for controlling the viral spread.
Even though it is not routinely used for SARS-CoV-2 detection in reference laboratories, genome sequencing of SARS-CoV-2-positive samples combined with computational tools has paved the way for many applications, including investigations of disease pathogenesis, diagnostics, vaccines, antiviral drugs, molecular epidemiology, viral evolution, cell receptor binding, possible viral hosts, and host antiviral immune response.8,60,366−372
RT-LAMP
Although RT-qPCR is currently the reference method for the diagnosis of SARS-CoV-2, it has several limitations, including long processing time, the requirement of highly specialized manpower, and the involvement of costly and specialized equipment for amplification and detection of the viral genome.150,373 These barriers make the technique unsuitable for large-scale applications and negatively impact the establishment of effective disease control programs in low- and middle-income countries. A recent modeling study concluded that effective screening is more impacted by the frequency of testing and sample-to-answer time than the analytical sensitivity of the test, highlighting the role of rapid tests for COVID-19 control.374 With this in mind, a variety of techniques have been developed to detect SARS-CoV-2 at the point of need.315 Isothermal techniques like reverse transcription loop-mediated isothermal amplification (RT-LAMP) are perhaps among the most promising methods for rapid detection of SARS-CoV-2. RT-LAMP has several advantages compared with RT-qPCR, since reactions are conducted at a constant temperature, eliminating the need for expensive equipment to perform the assay and allowing the test to be carried out in the field.373,375−377 Following the isothermal incubation in as little as 20 to 60 min, usually the results can be easily interpreted by naked-eye analysis through a color change of the reaction tube,373,377 although other different approaches can be used to visualize the results of the RT-LAMP reaction.376
Considering its advantages of high specificity and sensitivity, rapid amplification, simple operation, and low cost, RT-LAMP has potential applications for the diagnosis of many infectious diseases.376,378 Since the emergence of SARS-CoV-2, many RT-LAMP assays have been developed for the diagnosis of this virus in several types of clinical samples, including saliva, serum, nasopharyngeal swabs, oropharyngeal swabs, and urine.370,379−386 In general, RT-LAMP assays have been designed for different targets in the SARS-COV-2 genome (ORF1ab, N protein, E protein, RdRP, and M protein), and the clinical performances of many RT-LAMP assays have been compared with that of RT-qPCR.315,385,387 Efforts to decrease the cost and simplify the RT-LAMP workflow for testing patient samples are in progress using protocols without RNA extraction from patient specimens and in-house-produced enzymes,388−390 with the possibility to scale up COVID-19 diagnostics. With regard to the limit of detection (LoD), the majority of RT-LAMP assays should have LoDs ranging from 200 to 100 copies per reaction,382,383 while a few have demonstrated LoDs as low as 10 copies per reaction or even 1 copy per reaction.391−393 Currently, there are 14 diagnostic devices based on RT-LAMP assays that have been granted Emergency Use Authorization (EUA) by the FDA (https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-molecular-diagnostic-tests-sars-cov-2). Taken together, these developments highlight the potential of RT-LAMP-based methods to improve SARS-CoV-2 diagnosis on site in nearly real time, mainly for hot spots (e.g., care homes, small cities) and remote areas (e.g., rural communities) with limited laboratory infrastructure.
CRISPR/Cas-Based Systems
Another category of nucleic acid tests that could be used to detect SARS-CoV-2 RNA is the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas machinery.394 CRISPR systems are a fundamental part of a microbial adaptive immune system against foreign nucleic acids, and when activated, the machinery guides Cas proteins to recognize and cleave specific nucleic acid sequences.395−398 Thus, understanding of the mechanism and features of the CRISPR/Cas system over the past few years has led to many technological advances in genome editing and highlighted the use of the CRISPR/Cas system for diagnostic applications such as the detection of RNA viruses.394,399−404 Briefly, the CRISPR/Cas system is programmed to cleave specific nucleic acid sequences in the RNA/DNA target, and their cleavage can be detected by fluorescence or lateral-flow readouts.405 One of the first CRISPR/Cas-based detection methods, specific high-sensitivity enzymatic reporter unlocking (SHERLOCK), was described in 2017. In combination with isothermal preamplification, SHERLOCK was applied to the detection of specific strains of Zika and dengue viruses, distinguishing pathogenic bacteria, genotyping human DNA, and identifying mutations in cell-free tumor DNA from patients’ liquid biopsy samples.402,403 The same system was rapidly adapted to detect SARS-CoV-2 RNA.405−407 The suitability of SHERLOCK technology for the detection of SARS-CoV-2 was evaluated using 154 nasopharyngeal and throat swab samples. With a LoD of 42 RNA copies per reaction, the SHERLOCK platform was 100% specific and 96% sensitive in agreement with RT-qPCR.405 More recently, De Puig and colleagues developed a low-cost test based on the SHERLOCK system to perform diagnosis of SARS-CoV-2 and emerging variants.401 The authors achieved high sensitivity within 1 h and demonstrated multiplex detection of SARS-CoV-2 and mutations associated with emerging variants, including B.1.1.7 (alpha), B.1.351 (beta), and P.1 (gamma).401
Another technology based on the CRISPR/Cas system called DETECTR (for endonuclease-targeted CRISPR trans reporter) was developed by Mammoth Biosciences Company to detect any DNA or RNA target. More recently, this technology was combined with an RT-LAMP assay for the detection of SARS-CoV-2 from patient samples including nasopharyngeal or oropharyngeal swabs within 40 min.408 A clinical performance study using 78 samples from COVID-19 patients demonstrated that the DETECTR technology had 95% positive predictive agreement and 100% negative predictive agreement with RT-qPCR.408 In a larger patient cohort, the authors compared DETECTR with RT-qPCR to diagnose SARS-CoV-2 using samples from 378 patients.409 The results demonstrated a value of 95% reproducibility among methods and showed that when combined with RT-LAMP to detect SARS-CoV-2 RNA, DETECTR reached equal sensitivity in comparison to RT-qPCR.409 These findings highlight the promising potential of CRISPR/Cas-based diagnostic systems for the diagnosis of COVID-19 patients. However, most CRISPR-based workflows still require multiple liquid-handling steps including RNA extraction, reliable access to electricity, technical skills, and laboratory equipment like centrifuges, pipettes, and heating blocks,401 limiting its applicability in remote areas.
Sensors
Sensors are devices that detect chemical or biological components by generating signals,410 and they represent cost-effective alternatives for use in clinical practice. The use of sensors eliminates the limitations faced by RT-qPCR and provides a decentralized, high-capacity, and low-cost diagnostic tool for use in low-resource settings.411,412 Given their advantages, many studies have focused on alternative sensor-based modalities for the diagnosis of COVID-19 patients. In summary, most sensors developed for SARS-CoV-2 are based on platforms previously used for the diagnosis of other viral pathogens and are currently being adapted for SARS-CoV-2 diagnostics.150 Detection of SARS-CoV-2 has been made with various types of sensors, including genosensors, immunosensors, electrochemical sensors, and electrical immunosensors.411
Paper-based sensors offer another promising alternative for COVID-19 diagnosis since they provide high sensitivity and specificity, simple operation, easy adaptability, low cost, and absence of cold-chain distribution requirements.400,413 Our previous studies on synthetic biology-based diagnostics have paved the road and opened doors for the use of cell-free (CF) reactions for the detection of emerging and re-emerging pathogens such as Zika virus, chikungunya virus, norovirus, and Ebola virus (EV).400,414−417 Importantly, our toehold switch sensors are programmable synthetic riboregulators that control the translation of a gene via the binding of a trigger RNA. Briefly, the switch sensors contain a hairpin structure that blocks gene translation by sequestration of the ribosome binding site (RBS) and start codon. If the target sequence is present, it activates the translation of a reporter (e.g., LacZ) to create an optical signal through an enzymatic reaction that mediates a color change by converting a yellow substrate (chlorophenol red-b-d-galactopyranoside) to a purple product (chlorophenol red).400 In a effort to provide accessible diagnostics for the COVID-19 pandemic, we adapted this system for SARS-CoV-2 diagnostics and achieved high sensitivity (as few as 100 RNA copies) and high specificity against other respiratory pathogens, including H1N1, H7N9, and MERS-CoV.418 These results illustrate the potential of sensor-based tools to respond to global crises.
Serological Methods
Since the emergence of SARS-CoV-2, a variety of serological methods have been developed and commercialized, and a list of authorized COVID-19 serological assays in the U.S. is updated daily.419 They play an adjunct role to molecular assays to allow the identification of suspected patients that have tested negative by nucleic acid tests.315 Moreover, serological measurements could also be used in seroprevalence studies to determine past exposures to SARS-CoV-2 in the human population, assess attack rates in defined populations or geographical areas, and evaluate vaccine efficacy.150,315
Different serology-based assay platforms have been developed to date, including lateral flow assay (LFA), enzyme-linked immunosorbent assay (ELISA), chemiluminescence enzyme assay (CLIA), and immunofluorescence assay (IFA).420 Of these assays, LFA, ELISA, and CLIA are used as first-line methods in the routine to confirm COVID-19 infection.150 Serological methods often use recombinant antigens to detect one or more immunoglobulin isotypes (i.e., IgA, IgM, or IgG), with the S protein, S RBD, and N protein being the most commonly used antigens because of their high antigenicity.80,421,422 IgM and IgG are widely used in serological methods for SARS-CoV-2, while IgA detection is less common.423 Recent reports have shown that serological methods using the S antigen are more sensitive than the N-antigen-based methods.420 Moreover, it was suggested that an N-based assay is more cross-reactive with other anti-human coronavirus antibodies than an S-based assay.307,424 Within the S protein, the S1 subunit is a major immunodominant epitope produced in the response against SARS-CoV-2 infection, suggesting that this subunit is a promising candidate for use during the development of serological assays.425,426 The typical time required to detect immune responses to SARS-CoV-2 infection is around 1 to 2 weeks. Therefore, serological methods have limited utility for SARS-CoV-2 diagnostics in the acute stages of illness but have the best performance when used during the late phase of SARS-CoV-2 infection.315 High sensitivity, specificity, and overall accuracy are desired for serological assays, but concerns about the presence of high rates of false-positive and false-negative results have been raised, and the overall accuracy of many tests has not been well-defined.427,428 As a result, the understanding of the key parameters in terms of development and validation of diagnostic assays is critical for the correct interpretation of results.352,427 To overcome these drawbacks, serological methods must be developed following a standard guide as a reference, and after its development, the assay should be validated with adequate patient specimens representative of a real-world scenario.427,428 Assays that measure neutralizing antibody titers such as microneutralization, plaque reduction neutralization test (PRNT), and their variations are largely used to assess immunity against SARS-CoV-2 since neutralizing antibodies induced by natural infection or vaccination are highly preditive of protection.429,430
Antigen-Based Tests
Antigen detection tests are based on the identification of SARS-CoV-2 proteins by immunological reactions. Despite their lower sensitivity compared with molecular methods, they provide short turnaround times and are very useful for virus detection in samples with moderate to high viral loads.431,432 Since the emergence of SARS-CoV-2, extraordinary efforts by research groups and companies around the world have resulted in the development of many antigen tests to detect SARS-CoV-2.433−437 The suitability of antigen-based rapid tests (Ag-RDTs) for SARS-CoV-2 detection has been evaluated in a recent meta-analysis study using 214 clinical datasets including 112 323 patient specimens.432 For comparison, patient samples were also tested using RT-qPCR as a standard method. The results demonstrated a clinical sensitivity of 71.2% (95% CI 68.2% to 74.0%) and clinical specificity of 98.9% (95% CI 98.6% to 99.1%).432 The clinical sensitivity was considerably better in samples with lower RT-qPCR Ct values, i.e., <20 (96.5%, 95% CI 92.6% to 98.4%) and <25 (95.8%, 95% CI 92.3% to 97.8%) in relative to those with Ct ≥ 25 (50.7%, 95% CI 35.6% to 65.8%) and ≥30 (20.9%, 95% CI 12.5% to 32.8%).432 In addition, it was found that when the test was performed in the first week of symptoms onset, the sensitivity was substantially higher (83.8%, 95% CI 76.3% to 89.2%) compared with testing after 1 week (61.5%, 95% CI 52.2% to 70.0%).432 These findings suggest that Ag-RDTs can detect SARS-CoV-2-infected persons within the first week of symptoms onset but have low utility for diagnostic purposes in the late phase of COVID-19 and are therefore of limited value.
Treatment
Despite active research, few antivirals have shown potential benefit for COVID-19 patients. As of July 14, 2022, there were more than 700 drugs under development for COVID-19, and approximately 460 were in human clinical trials being reviewed by the FDA.438 Repurposed antiviral agents originally developed against influenza virus, human immunodeficiency virus (HIV), EV, and SARS-CoV/MERS-CoV viruses as well as antibiotics, antiprotozoals, and anthelmintic drugs are being investigated as potential therapeutic options for treating SARS-CoV-2 infection. These antivirals are proposed to inhibit the SARS-CoV-2 infectious cycle by targeting human cell receptors or viral proteins. The leading viral proteins targeted by anti-SARS-CoV-2 drugs include the RdRp, the helicase complex, and the 3-chymotrypsin-like (3CLpro) and papain-like (PLpro) viral proteases. Inhibitors for the human cellular receptor ACE2 and human proteases such as TMPRSS4, TMPRSS2, furin, and CatL have also been developed to block SARS-CoV-2 infection. Drugs and monoclonal/polyclonal antibodies designed for modulating the host response to the infection are an important additional part of the therapeutic arsenal that has been tested for treating COVID-19 patients. Despite the multitude of treatments that have been investigated throughout the past 2 years of the SARS-CoV-2 pandemic, which have been reviewed in detail elsewhere,439 a limited number of drugs have been investigated clinically, and an even smaller number have been approved for treating COVID-19.438
The U.S. National Institutes of Health (NIH) gathered a panel of experts to provide clinicians with evidence-based recommendations on the management of COVID-19. Immunomodulatory drugs have been tested for the treatment of COVID-19 with the goal of targeting the effects associated with the cytokine storm syndrome. According to the NIH COVID-19 treatment guidelines panel, the following immunomodulators are recommended for hospitalized patients according to their disease severity: corticosteroids (dexamethasone), IL-6 inhibitors (tocilizumab or sarilumab), and JAK inhibitors (baricitinib or tofacitinib). The panel does not recommend the use of colchicine for hospitalized patients and states that there is insufficient evidence for it to recommend either for or against the use of the following immunomodulatory drugs, except if done in a clinical trial: baricitinib plus tocilizumab, canakinumab, colchicine, intravenous immunoglobulin (except for MIS-C or when it is otherwise indicated), Bruton’s tyrosine kinase inhibitors (acalabrutinib, ibrutinib, zanubrutinib), JAK inhibitors other than baricitinib and tofacitinib (e.g., ruxolitinib), and siltuximab (https://www.covid19treatmentguidelines.nih.gov/therapies/immunomodulators/summary-recommendations/). The early use of IFN has been shown to have a highly protective effect, but its use during the inflammatory phase or in the severe stages of the disease results in immunopathology and long-lasting harm for patients. Thus, the NIH panel recommends against the clinical use of systemic IFN-α, -β, or -λ for the treatment of COVID-19 (https://www.covid19treatmentguidelines.nih.gov/therapies/antiviral-therapy/interferons/).
On the basis of the most updated scientific evidence, the panel recommends five available treatment options as preferred or alternative therapies (listed in order of preference): nirmatrelvir combined with ritonavir (Paxlovid); sotrovimab; remdesivir; bebtelovimab; or molnupiravir (https://www.covid19treatmentguidelines.nih.gov/about-the-guidelines/whats-new/). In the section below, we describe the major approved and promising drugs for COVID-19 treatment. Importantly, as the pandemic evolves, we can still anticipate the discovery of new therapeutic options for use in clinical practice.
Remdesivir
Remdesivir (GS-5734, Veklury) (Gilead Sciences) was the first drug to be approved by the main regulatory agencies around the world to treat COVID-19 patients (Table 2). Remdesivir is a bis(S-acyl-2-thioethyl) monophosphate prodrug that after chemical modification showed antiviral activity against hepatitis C virus (HCV), yellow fever virus (YFV), dengue virus 2 (DENV-2), influenza A virus (IAV), parainfluenza 3 virus (HPIV-3), and SARS-CoV in vitro, probably due to inhibition of viral RdRp by its triphosphate derivative.440 Remdesivir is a broad spectrum in vitro inhibitor for viruses from different families: EV and Marburg virus (MARV) (Filoviridae family); Nipah virus (NV), Hendra virus (HeV), measles virus (MV), and mumps virus (MuV) (Paramyxoviridae family); respiratory syncytial virus (RSV) and human metapneumovirus (hMPV) (Pneumoviridae family); Junín virus (JUNV) and Lassa virus (LASV) (Arenaviridae family); and SARS-CoV, MERS-CoV, and other human and bat CoVs (Coronaviridae family).441−446 Its triphosphate derivative inhibited RSV and HCV RdRp without inhibiting human RNA and DNA polymerases, and this activity is predicted to occur for RdRp from other viruses that present sequence similarity in motifs A and B of the nucleotide-binding regions of this enzyme.441−443 RdRp inhibition by a remdesivir triphosphate derivative is achieved by its incorporation into the nascent viral RNA, leading to delayed chain termination.442,447 For CoVs, the presence of exoribonuclease (ExoN)-mediated proofreading could diminish the sensitivity of the virus to remdesivir triphosphate derivative in vitro.445In vivo infection models showed that remdesivir is effective for treating EV infection in non-human primates,441,442,448 NV in non-human primates,449 and SARS-CoV and MERS-CoV infections in mice and non-human primates.444,450,451 In February 2020, Wang and colleagues published the first in vitro evidence of the efficacy of remdesivir in reducing SARS-CoV-2 infection in Vero E6 cells.452 Later, this activity was confirmed by additional studies using the same cell line453,454 and in human lung cells.455 Preclinical tests confirmed the anti-SARS-CoV-2 activity of remdesivir in mice and non-human primate models.455,456 Since the first in vitro/in vivo evidence, remdesivir has been clinically tested in patients with COVID-19. Four major randomized controlled clinical trials have investigated the benefits of remdesivir versus placebo/standard care for treating hospitalized COVID-19 patients.457−460 These studies showed that treatment of hospitalized COVID-19 patients with an intravenous infusion of 200 mg of remdesivir on the first day followed by 100 mg once daily for 5–10 days diminished the time to clinical improvement, enhanced the rates of recovered and discharged patients, and reduced the development of serious adverse events with no impact on mortality.461 These data supported the conditional or definitive approval of remdesivir as a therapeutic option for hospitalized COVID-19 patients in numerous countries (Table 2). Recently, efforts have been made to develop remdesivir derivatives for oral administration and with improved efficacy, but current data are preliminary.462−464
Table 2. Therapies Approved by the Main Regulatory Agencies Worldwide for Treatment of COVID-19 Patientsa.
| approval
by regulatory agencies |
||||||||
|---|---|---|---|---|---|---|---|---|
| drug | category | FDA | EMA | PMDA | ANVISA | indication | route of administration/therapeutic scheme | benefits |
| Antiviral Therapies | ||||||||
| remdesivir (Veklury) | viral RdRp inhibitor | approved (2020/10/22) | conditional marketing authorization (2020/07/03) | special approval for emergency (2020/05/07) | approved (2021/03/12) | 12-year-old or older COVID-19 patients that are hospitalized | intravenous infusion/attack dose of 200 mg on day 1 followed by 100 mg once daily | improves rates of recovery and discharge and reduces the development of serious adverse events461 |
| casirivimab and imdevimab (REGEN-COV or Ronapreve) | recombinant human IgG1 monoclonal antibodies against SARS-CoV-2 spike protein | EUA (2020/11/21) | marketing authorization (2021/11/12) | special approval for emergency (2021/07/19) | approved for emergency use (2021/04/20) | 12-year-old or older COVID-19 patients with mild to moderate disease who are at high risk for progression to severe COVID-19 or as postexposure prophylaxis | intravenous infusion or subcutaneous injection/single dose of 600 mg of casirivimab and 600 mg of imdevimab | reduces disease-related hospitalizations or deaths when administrated postinfection474 |
| reduces the risk of developing symptomatic or asymptomatic COVID-19 when administred as postexposure prophylaxis477 | ||||||||
| bamlanivimab and etesevimab | human IgG1 monoclonal antibodies against SARS-CoV-2 spike protein | EUA (2021/02/09) | none (recommended) | none | approved for emergency use (2021/05/13) | 12-year-old or older COVID-19 patients with mild to moderate disease who are at high risk for progression to severe COVID-19 or as postexposure prophylaxis | intravenous infusion/single dose of 700 mg of bamlanivimab and 1400 mg of etesevimab | reduces the number of related hospitalizations or deaths and improves viral load reduction from baseline484 |
| sotrovimab | recombinant human IgG1κ monoclonal antibody against SARS-CoV-2 spike protein | EUA (2021/05/26) | none (recommended) | special approval for emergency (2021/09/27) | approved for emergency use (2021/09/08) | 12-year-old or older COVID-19 patients with mild to moderate disease who are at high risk for progression to severe COVID-19 | intravenous infusion/single dose of 500 mg | reduces the risk of hospitalizations or deaths of any cause494 |
| regdanvimab (Regkirona) | human IgG monoclonal antibody anti- SARS-CoV-2 spike protein | none | marketing authorization (2021/11/12) | none | approved for emergency use (2021/08/11) | adult COVID-19 patients with mild to moderate disease who are at high risk for progression to severe COVID-19 | intravenous infusion/single dose of 40 mg/kg | accelerates viral clearance and clinical recovery498 |
| baricitinib (Olumiant) | inhibitor of JAK1/JAK2 (anti-inflammatory effect) and AAK1 and GAK (endocytosis inhibitor) | EUA (2020/02/04) | none | special approval for emergency (2021/04/23) | approved for emergency use (2021/09/17) | hospitalized COVID-19 patients in need of supplemental oxygen | oral/4 mg per day for 14 days or until discharge | reduces mortality rates, intensive care unit admissions, requirement for invasive mechanical ventilation, and risk of serious adverse events508 |
| Immunomodulatory Therapies | ||||||||
| tocilizumab (Actemra) | recombinant humanized monoclonal antibody against IL-6 receptor | EUA (2021/06/24) | none | none | none | hospitalized 2-year-old or older COVID-19 patients who are receiving systemic corticosteroids and require supplemental oxygen, noninvasive or invasive mechanical ventilation, or extracorporeal membrane oxygenation | intravenous infusion/single dose of 8 mg/kg (patients with weight ≥ 30 kg) or 12 mg/kg (patients with weight < 30 kg) | reduces risk of needing mechanical ventilation and poor outcome520 |
Legend: FDA, U.S. Food and Drug Administration; EMA, European Medicines Agency (European Union); PMDA, Pharmaceuticals and Medical Devices Agency (Japan); ANVISA, Agência Nacional de Vigilância Sanitária (Brazil); EUA, Emergency Use Authorization; RdRp, RNA-dependent RNA polymerase; IgG, immunoglobulin G; JAK, Janus kinase; AAK1, AP2-associated kinase 1; GAK, cyclin G-associated kinase; IL-6, interleukin 6.
Casirivimab and Imdevimab
The casirivimab and imdevimab cocktail (REGEN-COV or Ronapreve) (Regeneron Pharmaceuticals/Roche) was the first antibody-based therapy approved for COVID-19 by the FDA. Both are recombinant human IgG1 monoclonal antibodies that bind to the RBD of the SARS-CoV-2 S protein with high affinity, blocking S–ACE2 binding and thus preventing viral entry. They were discovered during a large antibody screen of genetically humanized mice that were challenged with SARS-CoV-2 S protein and by identifying antibodies from human COVID-19 survivors.465 Casirivimab and imdevimab bind to distinct and nonoverlapping regions of the RBD, and their combination (REGEN-COV) minimizes the escape of viral mutants that adapt to single-antibody-based treatments.466−468 REGEN-COV blocks the entry of important SARS-CoV-2 variants, such as B.1.1.7 (alpha), B.1.351 (beta), P.1 (gamma), and B.1.617.2 (delta), but the individual antibodies lose effectiveness over time, which supports the advantage of the dual-antibody treatment.469−472 COVID-19 animal models of mild (rhesus macaque) and severe (golden hamster) disease showed that REGEN-COV is effective in reducing viral loads in the upper and lower respiratory tracts, diminishing virus-induced pathology in rhesus macaque, and preventing weight loss in hamsters.473 A phase 3 randomized, double-blind, placebo-controlled trial was conducted with nonhospitalized patients above 18 years of age presenting at least one risk factor for developing severe COVID-19. After a single intravenous infusion of 2400 mg (1200 mg of each antibody) or 1200 mg (600 mg of each antibody) of the cocktail, REGEN-COV was shown to be safe and effective in reducing disease-related hospitalizations or deaths, time to symptoms resolution, time of hospital stay, and incidence of admission to an intensive care unit.474 Retrospective studies also showed that REGEN-COV reduces hospitalization rates in patients with mild to moderate disease in clinical practice.475,476 Another randomized, double-blind, placebo-controlled trial was conducted with asymptomatic subjects 12 years of age or older who were household contacts of a person infected with SARS-CoV-2. The study aimed to evaluate the capacity of a single subcutaneous injection of 1200 mg of REGEN-COV (600 mg of each antibody) to prevent SARS-CoV-2 infection. In exposed individuals, treatment reduced the risk of developing symptomatic or asymptomatic COVID-19, the average duration of symptoms, the average duration of RT-qPCR-detectable SARS-CoV-2 infection, and the viral load in exposed individuals.477 The clinical efficacy of REGEN-COV was maintained even in the presence of the SARS-CoV-2 delta variant.478 Different regulatory agencies approved REGEN-COV for treatment of people 12 years of age or older with mild to moderate COVID-19 who are at high risk for progression to severe disease and as prophylactic therapy for people exposed to SARS-CoV-2-infected individuals (Table 2).
Bamlanivimab and Etesevimab
The bamlanivimab and etesevimab cocktail (Eli Lilly and Company) is composed of two human monoclonal antibodies identified from convalescent sera of different COVID-19 patients that bind to different but partially overlapping RBD regions of SARS-CoV-2 S protein and consequently block S-ACE2 binding and virus entry.479,480 Bamlanivimab (LY-CoV555) was identified from more than 400 antibodies and had the best neutralization capacity of different SARS-CoV-2 isolates. Preinfection treatment of rhesus macaques with bamlanivimab was able to reduce viral replication and viral loads in the upper and lower respiratory tracts.479 Etesevimab (CB6 or LY-CoV016) was also the best neutralizer of SARS-CoV-2 pseudoviruses and infectious viruses among the antibodies discovered concomitantly. Pre- and postinfection treatments of rhesus macaques with etesevimab reduced viral titers in the upper respiratory tract and diminished infection-related lung damage.480 The two antibodies have been tested alone or in association in clinical trials. Healthy adults tolerated a single intravenous infusion of etesevimab at 2.5 to 50 mg/kg.481 The blocking viral attachment and cell entry with SARS-CoV-2 neutralizing antibodies study (BLAZE) is a randomized, double-blind, placebo-controlled trial currently in progress to evaluate the efficacy of bamlanivimab monotherapy and bamlanivimab + etesevimab combined therapy for treating COVID-19 patients. Preliminary data evaluating bamlanivimab treatment alone in nonhospitalized patients with mild to moderate COVID-19 showed that after 11 days, patients who received a single intravenous infusion of 2800 mg presented a larger decrease in viral loads compared with patients who received a placebo. After 29 days, treatment with bamlanivimab was considered safe and was associated with reduced hospitalization and symptom severity.482 Results from phase 2/3 of the same trial that included the analysis of the combined therapy showed that a single intravenous infusion of 2800 mg of bamlanivimab plus 2800 mg of etesevimab generated a larger decrease in viral load after 11 days and lowered hospitalization rates after 29 days compared with a placebo.483 Among patients with mild to moderate COVID-19 and a high risk to develop severe disease, the combined therapy was able to reduce the number of COVID-19-related hospitalizations or deaths and reduced the viral load compared with the baseline at two different dosages: 2800 mg each484 and 700 mg of bamlanivimab plus 1400 mg of etesevimab.485 A retrospective study reported a reduction in the rate of hospital admissions or the need for supplementary oxygen in patients who received treatment with 700 mg of bamlanivimab and 1400 mg of etesevimab within 5 days of symptoms onset compared with patients who received the treatment after 5 days.486 Currently, the FDA authorizes bamlanivimab + etesevimab for emergency use to treat patients that 12 years of age or older with mild to moderate COVID-19 who are at high risk of progression to severe disease and as a prophylactic therapy (Table 2). However, different mutants of SARS-CoV-2 S protein have been reported to confer viral resistance to bamlanivimab, etesevimab and bamlanivimab + etesevimab therapies. Some of these mutations are found in important circulating SARS-CoV-2 variants, including B.1.351 (beta), P.1 (gamma), and B.1.617.2 (delta plus).467,487−491 For this reason, the combined therapy is no longer authorized for use in the U.S. in jurisdictions where the combined frequency of variants resistant to bamlanivimab and etesevimab exceeds 5% (https://www.phe.gov/emergency/events/COVID19/investigation-MCM/Bamlanivimab-etesevimab/Pages/resumption-in-distribution-bamlanivimabetesevimab.aspx).
Sotrovimab
Sotrovimab (Vir Biotechnology/GlaxoSmithKline) is an engineered human monoclonal antibody derived from the S309 antibody identified from sera of a SARS-CoV-infected individual that is able to neutralize SARS-CoV, SARS-CoV-2 pseudoviruses and infectious viruses. S309 recognizes the SB domain of the S protein, leading to noncompetitive inhibition of S–ACE2 binding and consequently virus entry. Fc-dependent effector mechanisms also play a role in S309 neutralization activity.492 Sotrovimab (VIR-7831) was obtained after modification of the S309 variable region for enhanced developability and the addition of an “LS” mutation in its Fc portion to confer extended half-life and enhanced distribution to the respiratory mucosa. Sotrovimab maintained S309 binding characteristics, effector mechanisms, and efficacy in neutralizing SARS-CoV-2. It was also capable of neutralizing important SARS-CoV-2 variants, including alpha, beta, gamma, delta, and kappa, with a significant shift in IC50 and IC90 values for only the alpha variant. A hamster model was used to test sotrovimab preinfection treatment in vivo. Sotrovimab reduced weight loss and viral loads when the dosage was ≥5 mg/kg and reduced viral titers in the lungs when the dosage was ≥0.5 mg/kg.493 A randomized, double-blind, controlled phase 3 trial has been evaluating the efficacy of a single intravenous infusion of 500 mg of sotrovimab in reducing hospitalizations and deaths in nonhospitalized and symptomatic adult COVID-19 patients with at least one risk factor for disease progression. Sotrovimab reduced 85% of the risk of hospitalization or death in the evaluated population.494 Sotrovimab is approved for emergency use by the FDA for treating patients 12 years of age or older with mild to moderate COVID-19 who are at high risk for progression to severe disease (Table 2).
Regdanvimab
Celltrion Healthcare announced acceptance and priority review by Health Canada for its monoclonal antibody treatment for COVID-19, regdanvimab (originally CT-P59), a human monoclonal antibody obtained from sera of a convalescent COVID-19 patient. It binds to the RBD of SARS-CoV-2 S protein and competitively blocks S–ACE2 binding and consequently virus entry.495 Regdanvimab was able to inhibit SARS-CoV-2 infection in vitro.496 The antibody was tested in three different animal models: ferrets, golden Syrian hamsters, and rhesus monkeys.495 Postinfection treatment of ferrets with regdanvimab reduced viral titers in nasal washes and lungs, especially after administration of a high dosage (30 mg/kg). This reduction was followed by an improvement in clinical symptoms and lung pathology. In golden Syrian hamsters, postinfection treatment reduced viral titers in the lungs at all dosages tested, but with 30 mg/kg complete inhibition of SARS-CoV-2 replication was achieved. In rhesus monkeys, treatment with two doses of regdanvimab (45 and 90 mg/kg) reduced viral titers in nasal and throat swabs.495 Regdanvimab is effective against SARS-CoV-2 B.1.351 (beta), P.1 (gamma), and B.1.617. 2 (delta) variants in vivo, although in vitro assays have suggested different results.496,497 Two randomized, double-blind, placebo-controlled phase 1 studies have investigated the safety and efficacy of regdanvimab in a small cohort of healthy (study 1.1) or mildly symptomatic (study 1.2) adult COVID-19 patients that received a 10, 20, 40, or 80 mg/kg dose of the antibody in a single intravenous infusion. After a 90 day follow-up, no drug-related serious adverse event was detected in any patients who received therapy. Patients with high viral loads at baseline (>105 copies/mL) who received 20, 40, or 80 mg/mL regdanvimab exhibited faster viral clearance than patients who received a placebo. Patients receiving regdanvimab also exhibited faster clinical recovery in a dose-dependent fashion. To date, phase 2 and 3 studies of regdanvimab efficacy are ongoing in nonhospitalized patients with SARS-CoV-2 infection.498 The European Medicines Agency (EMA) currently authorizes regdanvimab for COVID-19 treatment in adults with mild to moderate disease who are at high risk of developing severe outcomes (Table 2).
Baricitinib
Baricitinib (Eli Lilly) is an anti-inflammatory drug inhibitor of Janus kinases 1 and 2 (JAK1/2) and is approved for treatment of rheumatoid arthritis.499 Baricitinib has been suggested as a possible antiviral drug for treatment of COVID-19 because of its capacity to inhibit the important clathrin-mediated endocytosis regulators AP2-associated protein kinase 1 (AAK1) and cyclin G-associated kinase (GAK), which would block viral entry through this route.500 In addition, the anti-inflammatory activity of this drug would be beneficial to COVID-19 patients because inflammation causes lung damage and subsequent mortality.501 In fact, exogenous treatment of blood cells from COVID-19 patients with baricitinib reduced SARS-CoV-2-specific immune response in vitro.502 Baricitinib was also able to inhibit IFN-α2-mediated transcriptome switch and ACE2 induction as well as SARS-CoV-2 replication in liver organoids but not in Vero and A549 cells, indicating a tissue-specific response.503 Although baricitinib was not effective in reducing SARS-CoV-2 loads in nasal/throat swabs and bronchoalveolar lavages of rhesus monkeys, daily oral administration of 4 mg for 8–9 days was able to reduce lung pathology and inflammation in infected animals.504 In a pilot study with adults presenting moderate COVID-19, baricitinib treatment was considered safe and did not lead to any serious adverse events in patients receiving 4 mg/day together with lopinavir–ritonavir therapy for 2 weeks. Clinical parameters and respiratory functions were improved, and discharges were higher in baricitinib-treated patients compared with patients receiving standard care (lopinavir–ritonavir plus hydroxychloroquine).505 Two randomized, double-blind, placebo-controlled phase 3 trials have evaluated the efficacy of oral baricitinib treatment at 4 mg/day for 14 days (or until discharge) in COVID-19 patients. The COV-BARRIER trial was conducted in hospitalized adult COVID-19 patients with at least one elevated inflammatory marker. In this clinical trial, baricitinib treatment resulted in lower mortality rates after 28 or 60 days and a similar frequency of serious adverse events compared with placebo.506 The ACTT-2 trial evaluated baricitinib treatment in combination with remdesivir in hospitalized moderate to severe COVID-19 patients. Baricitinib + remdesivir treatment resulted in a shorter recovery time, especially for patients receiving noninvasive ventilation or high-flow oxygen, and fewer serious adverse events than remdesivir alone.507 A recent systematic review and meta-analysis evaluating the clinical efficacy of baricitinib included the previously mentioned randomized clinical trials in addition to nonrandomized trials and observational studies. The authors concluded that baricitinib reduced mortality rates, intensive care unit admissions, and the requirement for invasive mechanical ventilation, improved the oxygenation index, and reduced the risk of serious adverse events.508 Baricitinib received an EUA from the FDA for treatment of COVID-19-hospitalized patients in need of supplemental oxygen (Table 2).
Tocilizumab
Tocilizumab (Genentech/Roche) is a recombinant humanized monoclonal antibody directed against the IL-6 receptor (IL-6R) that was initially reported to inhibit IL-6-dependent multiple myeloma cell growth.509 It can bind both soluble and membrane-bound IL-6R, leading to the blockade of IL-6 signaling.510 The FDA approved the use of tocilizumab to treat adult patients with moderately to severely active rheumatoid arthritis or giant cell arteritis and to treat people 2 years of age or older with active polyarticular or systemic juvenile idiopathic arthritis or chimeric antigen receptor (CAR) T cell-induced cytokine release syndrome (https://www.accessdata.fda.gov/drugsatfda_docs/label/2017/125276s114lbl.pdf).511−514 Tocilizumab was proposed as a treatment option for severe COVID-19 cases at the beginning of the pandemic on the basis of evidence of its involvement during cytokine storms in disease progression.515 Gu and co-workers observed that in a mouse model the IL-6 pathway is important to initiate the cytokine storm syndrome in SARS-CoV-2-infected animals. They also showed that treatment with an anti-IL-6R antibody resulted in a reduction of lung neutrophil infiltration, supporting a benefit from anti-IL-6R therapy to COVID-19 patients.516 To date, three randomized, double-blind, placebo-controlled trials have been published evaluating the effect of tocilizumab treatment (8 mg/kg, single intravenous infusion) on hospitalized COVID-19 patients. The BACC Bay trial evaluating moderately ill patients did not find any benefit of tocilizumab treatment in preventing intubation, death, or other secondary/tertiary outcomes.517 The EMPACTA trial was conducted in hospitalized patients with COVID-19 pneumonia who were not receiving mechanical ventilation. In that trial, treatment with tocilizumab reduced the risk of receiving mechanical ventilation or death on day 28.518 In the COVACTA trial, treatment with tocilizumab did not improve the clinical status or mortality rates in patients with severe COVID-19 pneumonia.519 By analyzing data from open-label and double-blind randomized trials and cohort studies, Tleyjeh and co-workers concluded that tocilizumab treatment reduced the risk of mechanical ventilation and poor outcomes (randomized trials) and decreased the short-term mortality in COVID-19 patients.520 Tocilizumab received an EUA from the FDA for treatmento of hospitalized COVID-19 patients 2 years of age or older who are receiving systemic corticosteroids and require supplemental oxygen, noninvasive or invasive mechanical ventilation, or extracorporeal membrane oxygenation (Table 2).
Promising COVID-19 Therapies
Systemic Corticosteroids
Systemic corticosteroids have been widely used in COVID-19 patients and are recommended by WHO for treating patients with severe and critical illness.521 Corticosteroids are nonexpensive and broadly available molecules that diminish inflammation because of their capacity to stimulate the synthesis/release of anti-inflammatory mediators and to inhibit the synthesis/release of proinflammatory proteins through genomic and nongenomic pathways.522 Systemic inflammation is one of the main players in the development of severe outcomes in COVID-19, which suggested that anti-inflammatory drugs, such as corticosteroids, could be an effective treatment option for patients.523 Several observational studies and randomized controlled trials have evaluated the efficacy of systemic corticosteroids in COVID-19 patients.521 The CoDEX trial (randomized, open-label) evaluated dexamethasone treatment (10 or 20 mg daily for 5 days) of hospitalized moderate-to-severe COVID-19 patients receiving mechanical ventilation. In that trial, patients receiving dexamethasone showed longer ventilator-free periods than patients receiving standard care.524 The RECOVERY trial (randomized, open-label) evaluated dexamethasone treatment (6 mg daily for 10 days) of hospitalized moderate-to-severe COVID-19 patients. Patients receiving invasive mechanical ventilation or oxygen without invasive mechanical ventilation that were treated with dexamethasone showed lower mortality rates than those treated with standard care.525 Methylprednisolone is another corticosteroid used in COVID-19 treatment. The GLUCOCOVID trial (randomized, open-label) analyzed the efficacy of methylprednisolone in treating COVID-19 patients receiving oxygen without mechanical ventilation and showing evidence of systemic inflammatory response. Treatment with methylprednisolone (40 mg for 3 days followed by 20 mg for 3 days) reduced the risk of death, admission to the intensive care unit, or requirement for noninvasive ventilation.526 The Metcovid trial (randomized, double-blind, placebo-controlled phase 2b) evaluated the treatment of hospitalized COVID-19 patients with 0.5 mg/kg methylprednisolone versus placebo. Patients 60 years of age or older receiving methylprednisolone had a lower mortality rate compared with patients receiving placebo.527 A recent meta-analysis study including observational studies and randomized trials involving different drugs suggested that treatment with corticosteroids reduces mortality and risk of progression to invasive mechanical ventilation in severe COVID-19 patients.528 Treatment with methylprednisolone has resulted in better clinical outcomes than treatment with dexamethasone, but larger randomized controlled studies are needed to confirm this finding.529,530
AZD7442
AZD7442 (AstraZeneca) is a cocktail of two human monoclonal antibodies previously designated COV2-2196 and COV2-2130 that after engineering were called AZD8895 and AZD1061, respectively. Both antibodies were obtained from sera of convalescent COVID-19 patients and are directed against SARS-CoV-2 S protein.531 They recognize nonoverlapping regions of the S RBD and can inhibit S–ACE2 binding and consequently neutralize SARS-CoV-2 in a synergistic manner. Prophylactic treatment of ACE2-mice with both antibodies (isolated or in combination) prevented SARS-CoV-2-induced weight loss, reduced viral loads in the lungs, heart, and spleen, diminished the expression of inflammation mediators in the lung, and reduced lung pathology.532 The combination of the two antibodies prevents mutational escape of SARS-CoV-2 variants, and the AZD7442 cocktail maintains its neutralization capacity even against different VOCs.533 Phase 3 clinical trials are currently being conducted to test AZD7442 for COVID-19 prevention (ClinicalTrials.gov ID: NCT04625725), postexposure prophylaxis (NCT04625972), outpatient treatment (NCT04723394 and NCT04518410), and inpatient treatment (NCT04315948 and NCT04501978). The company claims that pre-exposure prophylactic treatment with intramuscular AZD7442 (300 mg) reduced the risk of developing symptomatic COVID-19 by 77% (https://www.astrazeneca.com/media-centre/press-releases/2021/azd7442-prophylaxis-trial-met-primary-endpoint.html). Treatment of nonhospitalized patients who had been symptomatic for ≤7 days and ≤5 days with intramuscular AZD7442 (600 mg) reduced the risk of severe illness or death by 50% and 67%, respectively (https://www.astrazeneca.com/media-centre/press-releases/2021/azd7442-phiii-trial-positive-in-covid-outpatients.html). However, these clinical data have not been published to date.
Molnupiravir
Molnupiravir (MK-4482, EIDD-2801) (Ridgeback Biotherapeutics/MSD) is a 5′-isopropyl ester of the nucleoside analogue N4-hydroxycytidine (NHC), a prodrug originally developed to treat influenza A and B viruses by the oral route. This drug has better oral bioavailability in non-human primates and ferrets than its precursor NHC and is efficiently hydrolyzed in vivo after absorption, releasing the active molecule in the plasma.534 NHC is capable of inhibiting SARS-CoV-2 and other related coronaviruses using in vitro models.535,536 This nucleoside analogue is incorporated by the viral RdRp into the nascent viral RNA, leading to error catastrophe and RNA synthesis inhibition.537 When administered to Syrian hamsters and human lung-only mice before or after SARS-CoV-2 infection, molnupiravir was able to decrease viral loads in the lungs and lung pathology.535,538 In ferrets, molnupiravir treatment after SARS-CoV-2 infection reduced viral loads in nasal lavages and inhibited the spread to untreated contact animals.539 Molnupiravir was also able to inhibit SARS-CoV-2 infection in hamsters infected with the B.1.1.7 (alpha) and B.1.351 (beta) variants of the Wuhan virus.540 A randomized, double-blind, placebo-controlled phase 1 trial with healthy volunteers attested that oral administration of 50–1600 mg of molnupiravir was well-tolerated and that only a few mild adverse events were observed in the treated population.541 Phase 3 clinical trials are currently being conducted to test molnupiravir for COVID-19 postexposure prophylaxis (NCT04939428), outpatient treatment (NCT04575584), and inpatient treatment (NCT04575597). MSD and Ridgeback Biotherapeutics announced that molnupiravir reduced the risk of hospitalization or death by approximately 50% in nonhospitalized adult patients with mild-to-moderate COVID-19 disease (https://www.merck.com/news/merck-and-ridgebacks-investigational-oral-antiviral-molnupiravir-reduced-the-risk-of-hospitalization-or-death-by-approximately-50-percent-compared-to-placebo-for-patients-with-mild-or-moderat/). Recently, the U.K.’s Medicines and Healthcare Products Regulatory Agency was the first agency to temporary authorize molnupiravir for the treatment of mild to moderate COVID-19 in adults with at least one risk factor for severe illness (https://www.gov.uk/government/publications/regulatory-approval-of-lagevrio-molnupiravir).
PF-07321332
PF-07321332 (Paxlovid) (Pfizer Inc.) is a reversible covalent inhibitor of the 3CLpro protease of SARS-CoV-2.542 It is a second-generation orally available 3CLpro inhibitor developed by Pfizer during the pandemic. To date, there are three clinical trials evaluating the association of PF-07321332 and ritonavir for COVID-19 treatment of patients with low risk (NCT05011513) or high risk (NCT04960202) to develop severe illness and as postexposure prophylaxis (NCT05047601). Pfizer recently announced that PF-07321332 combined with ritonavir significantly reduced by 89% hospitalization and death of nonhospitalized adult patients with COVID-19 who are at high risk of progressing to severe illness (https://www.pfizer.com/news/press-release/press-release-detail/pfizers-novel-covid-19-oral-antiviral-treatment-candidate). Ritonavir is coadministered in low doses to reduce metabolism of PF-07321332.
Prevention
At present, vaccination represents the most effective long-term strategy for controlling and preventing COVID-19.111 Besides effective vaccines, the prophylaxis of SARS-CoV-2 infection is based on a series of countermeasures. Reducing the spread of COVID-19 requires two factors: reducing the transmission probability per contact and limiting contact between persons via physical distancing.543,544 Thus, prevention practices together with the implementation of effective measures represent a crucial strategy to contain the rapid spread of SARS-CoV-2 among humans.
Recommendations to prevent SARS-CoV-2 infection established by the CDC include the following: (i) Masks should be worn, as they reduce transmissibility through exhaled air from infected respiratory particles in both clinical and laboratory scenarios and subsequently could result in a large reduction in the risk of SARS-CoV-2 infection at the population level.125,543,545 (ii) The 2 meter social distancing rule should be applied. Inside the home, close contact with people who are sick should be avoided. When outside, a distance of 2 meters from other people should be maintained. (iii) Crowds and poorly ventilated areas should be avoided. (iv) Hands should be washed often with soap and water for at least 20 seconds, especially after staying in a public place or after blowing the nose, coughing, or sneezing. If soap and water are not readily available, a hand sanitizer that contains at least 60% alcohol should be used. Eyes, nose, and mouth should not be touched with unwashed hands. (v) Coughs and sneezes should be covered. Those wearing a mask can cough or sneeze into the mask. If a mask is not being worn, the mouth and nose should be covered with a tissue during coughing or sneezing, or the inside of the elbow should be used without spitting. (vi) Highly touched surfaces should be cleaned and disinfected daily, including handles, desks, phones, keyboards, toilets, faucets, tables, doorknobs, light switches, countertops, and sinks. (vii) Health should be monitored daily.546 Notably, these measures are most effective at reducing the spread of the virus when adherence is high among the human population.545 For healthcare and frontline professionals, additional precautions are required, including airborne precautions (use of N95 respirators), contact precautions (use of gown and gloves), and eye protection (use of goggles or a face shield).547 Additionally, early diagnosis, quarantine, and supportive treatments are essential for the clinical management of infected patients.
SARS-CoV-2 can remain for a long time on various types of surfaces, including aerosols, plastic, stainless steel, copper, and cardboard,128,129 so several agents can be used to inactivate the virus.547 In this context, Chin and colleagues reported the stability of SARS-CoV-2 when exposed to several disinfectants agents such as household bleach (1:49–1:99), hand soap solution (1:49), ethanol (70%), povidone iodine (7.5%), chloroxylenol (0.05%), chlorhexidine (0.05%), and benzalkonium chloride (0.1%).127 These agents were evaluated at different times of exposition (5, 15, and 30 min) followed by a 50% tissue culture infective dose (TCID50) assay for viral titration to confirm the presence of infectious viruses. The results revealed that viral inactivation was observed after only 5 min of exposure using all of the disinfectant agents except for hand soap.127 In addition, it was shown that SARS-CoV-2 is stable at a wide range of pH values (3–10) at room temperature.127
Vaccines
As for other infectious diseases, vaccination is the main approach to preventing COVID-19. Since the emergency of SARS-CoV-2, several vaccine platforms have been developed, and as of July 14, 2022, 40 vaccines have been approved by at least one country in the world (Figure 11). There are 153 vaccines in clinical development and 196 in preclinical development. Currently approved vaccines are based on protein subunits (n = 16), inactivated virus (n = 11), nonreplicated viral vectors (n = 7), RNA (n = 4), DNA (n = 1), or viruslike particles (VLPs) (n = 1) (https://covid19.trackvaccines.org/vaccines/approved/). Among these, 10 vaccines were granted Emergency Use Listing (EUL) by the WHO, and they are discussed below.548
Figure 11.
Vaccine platforms currently being developed against SARS-CoV-2 infection. Since the emergence of SARS-CoV-2, great efforts have been made by research groups and companies around the world toward the development of effective vaccines. Briefly, the graph in (A) shows current vaccine candidates in the clinical phase. Vaccine platforms currently being developed against SARS-CoV-2 infection include those based on (B) RNA, (C) DNA, (D) replicating viral vectors, (E) inactivated viruses, (F) attenuated viruses, (G) viruslike particles (VLPs), (H) nonreplicating viral vectors, (I) protein subunits, and (J) modified antigen-presenting cells (APCs). Abbreviations: S, spike protein; LNP, lipid nanoparticle; LV, lentiviral vector; DC, dendritric cell. This figure was created with Biorender.com.
Vaccines based on protein subunits consist of antigenic pathogen fragments and have the potential to exhibit efficacy in protecting humans from viral infection.549 However, because only a few viral fragments are included, they do not display the full antigenic complexity of the virus and are therefore of limited value because the protective efficacy can be reduced.550 Protein subunit vaccines, such as Nuvaxovid (Novavax) and COVOVAX (manufactured by Serum Institute of India, Novavax formulation), are based on the recombinant nanoparticle S protein associated with Matrix-M adjuvant. Stabilizing mutations have been introduced into the S protein that are intended to prevent the intrinsic problem of its conformational instability.551
Inactivated vaccines such as Covaxin (Bharat Biotech), Covilo (Sinopharm), and CoronaVac (Sinovac) are based on whole-virus preparations made in cells followed by chemical inactivation, purification, and mixing with specific compounds that act as stimulants of immune cells and amplifiers of immune responses, such as aluminum hydroxide adjuvant.552 Notably, chemically inactivated, irradiated, or heat-inactivated pathogens sometimes lose their immunogenicity, rendering this platform less efficient than live attenuated pathogen platforms.552
Nonreplicated viral vector vaccines approved for human use are based on either animal or human replication-defective adenovirus vectors. Vaxzevria (Oxford/AstraZeneca) and Covishield (manufactured with Oxford and AstraZeneca formulation by Serum Institute of India and Fiocruz-Brazil) are based on chimpanzee adenovirus encoding the SARS-CoV-2 S glycoprotein. Ad26.COV2.S (Janssen/Johnson & Johnson) is based on a replication-incompetent recombinant human adenovirus type 26 vector expressing the S protein in a stabilized conformation.553
RNA-based vaccines have been approved for the first time for human use, and they have displayed excellent safety and efficacy profiles, placing this platform at the forefront of the fast development of vaccines against emerging diseases.554−556 Although there are some differences in how they were engineered, Comirnaty (Pfizer/BioNTech) and Spikevax (Moderna) are both lipid-nanoparticle-formulated, nucleoside-modified RNA vaccines that encode full-length SARS-CoV-2 S protein modified by two proline mutations to ensure that it remains in the prefusion conformation.
A recent systematic review and meta-regression on COVID-19 vaccine efficacy and/or effectiveness concluded that most vaccines (81%) had efficacy or effectiveness against severe disease that remained greater than 70% after full vaccination with a minimal decrease (∼10%) 6 months after immunization.557 Most of these vaccines have been manufactured on the basis of the prototype Wuhan-Hu-1 strain, and their efficacy and effectiveness are lower toward the VOCs that have emerged since the beginning of the pandemic. Thus, updates of vaccine composition to reflect the current most prevalent variant(s) of SARS-CoV-2 must be considered to provide optimum protection against these circulating SARS-CoV-2 variants. With the ephemeral nature of COVID-19 vaccine-induced immunity, novel prophylactic approaches that elicit long-term protection are warranted.
Emerging Variants of SARS-CoV-2
Since the discovery of SARS-CoV-2 in December 2019, the viral genomes from clinical samples have been sequenced daily and worldwide, and thousands of complete genomes have been deposited in databanks to date. RNA viruses, such as SARS-CoV-2, are known to present high mutation rates due to the low capacity of the RdRp to correct errors during genome replication. However, the Coronaviridae family is an exception since the replication machinery of these viruses contains a 3′-5′ exoribonuclease domain that is able to proofread.558 This domain has also been detected in SARS-CoV-2 as a component of the nonstructural protein nsp14.559 In fact, following analysis of different genomes published through November 2020, SARS-CoV-2 showed low global nucleotide diversity. However, nucleotide diversity tends to increase as virus incidence enhances.560 Virus mutation over time has culminated in the emergence of SARS-CoV-2 variants, i.e., virus specimens that are genetically different from the main or initial SARS-CoV-2 lineage. These mutations could be phenotypically neutral, with no major impacts on viral biology, or confer advantages to the variants that possess them, improving viral adaptation and fitness.561 The first important variation of the Wuhan reference strain observed possessed the D614G mutation on the S protein. This mutation was first detected in March 2020 and rapidly became globally dominant, being present in most of the current circulating viral lineages. D614G confers replicative advantages to the virus that could explain its rapid dissemination worldwide.562,563
The CDC has classified SARS-CoV-2 variants into three groups: variants of interest (VOIs); VOCs, and variants of high consequence (VOHCs). VOIs are variants with limited prevalence or dissemination that possess mutations predicted to affect the transmission, diagnostics, therapeutics, or sensibility to antibodies produced after previous exposition or vaccination. VOCs are defined as those who have evidence of increased incidence/transmission, diagnostic or therapeutic failure, or reduced neutralization by antibodies produced after previous exposition or vaccination. VOHCs are those for which prevention measures or medical countermeasures have reduced effectiveness. As the COVID-19 pandemic evolved, SARS-CoV-2 has been characterized by the repeated identification of different variants over time.564 Since its emergence, five SARS-CoV-2 variants have been classified as VOCs (alpha, beta, gamma, delta, and omicron) at different times during the course of the pandemic.354,361,564 To date, CDC has not classified any variant as a VOHC. All of these variants share the D614G mutation, which is involved in increased virus replication in the upper respiratory tract and higher transmissibility.361,563 There are different schemes for variant naming,565,566 but here we will use the nomenclature based on the phylogenetic framework proposed by Rambaut and colleagues, known as the Pango lineage,36 and the label defined by the World Health Organization (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/). The VOCs will be discussed more deeply in the sections below and are summarized in Figure 12.
Figure 12.
SARS-CoV-2 variants of concern (VOCs). (A) Schematic of the SARS-CoV-2 genome architecture. (B) Definition of nonsynonymous mutations and deletions of each VOC. Nucleotide alterations without predicted or confirmed impact on protein structure and/or function are shown in black. Nucleotide alterations with predicted or confirmed impact on protein structure and/or function are shown in red. (C) Phenotypic characteristics of VOCs. Global distributions are according to the PANGO lineages website (https://cov-lineages.org/index.html). This figure was created with Biorender.com.
Alpha
In December 2020, after epidemiological and genomic surveillance, the U.K. reported a new SARS-CoV-2 variant, initially called VUI-202012/01 (first variant under investigation in December 2020).567 This variant, renamed B.1.1.7 according to Pango lineages and subsequently as alpha according to WHO, genetically differs from others by 23 nucleotide alterations: 14 nonsynonymous substitutions, six synonymous substitutions, and three deletions (Table S1).568 The alpha variant was estimated to be more transmissible than previous variants, becoming the most prevalent in the U.K. over time and associated with higher mortality.569,570 The reproduction number of the alpha variant is 43% to 90% higher than those of previous variants in the U.K. and other countries, depending on the model used to calculate the data.569 Recent studies have proposed that the enhanced transmissibility could be related to higher viral loads or a longer infectious period.569 In fact, patients infected with the alpha variant have presented higher viral RNA levels and a more lasting positivity for the virus.571,572 Some commercial RT-qPCR kits targeting the S gene could not detect infection with the alpha variant because of the presence of the 21765–21770 deletion in the viral genome, a phenomenon usually called S gene target failure (SGTF).573 Using the SGTF as an approach to differentiate the alpha variant from others, Funk and colleagues observed a higher risk of hospitalization of European patients in the age groups of 20–39 and 40–59 years and ICU admission in the age group of 40–59 years.574 Recent studies have shown that the alpha variant was associated with a high risk (55–64%) of death in infected patients with SARS-CoV-2.570,575 Mutations on the S protein of the alpha variant have been linked to reduced neutralization by monoclonal antibodies.576,577 However, their impact on viral neutralization by convalescent sera or sera obtained from vaccinated subjects seems to be small or absent.576−579 The efficiencies of the mRNA-based vaccines BNT162b2 (BioNTech, Pfizer) and mRNA-1273 (Moderna) against the alpha VOC were shown to be similar to those against the previous variant.580,581 The inactivated-virus-based vaccines BBIBP-CorV (Sinopharm) and BBV152/COVAXIN (Bharat Biotech) were shown to be effective against alpha,582,583 while CoronaVac (Sinovac) reduced the neutralization capacity against this variant by a factor of 0.5.583 The nonreplicative-viral-vector-based vaccines ChAdOx1 nCoV-19/AZD1222 (Oxford, AstraZeneca) and Ad26.COV2.S (Janssen) also showed a reduced neutralization capacity against the alpha variant584 (Assessment Report EMA/158424/2021), although the overall efficacy of AZD1222 against symptomatic and asymptomatic cases was preserved (61.7% against the alpha variant and 77.3% against other variants).584 The efficacy of Ad26.COV2.S in preventing COVID-19 caused by the alpha variant still needs to be evaluated. Sputnik V Ad26/Ad5 (Gamaleya Institute) is still effective in neutralizing the alpha variant.585
Beta
In the same month that the alpha variant was first reported in the U.K., South African researchers described a new variant of SARS-CoV-2 that emerged in the country after the first epidemic wave.586 Initially called S501.V2, this new variant was named B.1.351 by Pango lineages and beta according to the WHO.36 The beta VOC was first characterized as containing 31 mutations, of which four are shared with the parent B.1 variant. Among the 27 specific variations found in this lineage, 21 are nonsynonymous mutations while 12 have been fixed in the variant population over time (Table S1).586 This emerging variant shares with the alpha VOC the N501Y substitution on the S protein, an important mutation for virus phenotype. It was estimated that beta VOC was 50% more transmissible than previously circulating variants.587 Compared with non-VOCs, the beta VOC showed a higher risk of hospitalization in European patients in the age groups of 40–59 and 60–79 years and ICU admission in the age group of 40–59 years, but this did not lead to an increase in the mortality rate.574 Until now, what seems to be the most important characteristic of beta VOC is its reduced sensitivity to neutralization by convalescent and vaccine-elicited sera.577,578,583,588−591 The BNT162b2, mRNA-1273, BBIBP-CorV, CoronaVac, ChAdOx1 nCoV-19/AZD1222, and Sputnik V Ad26/Ad5 vaccines showed reduced neutralization capacity against this variant.577,578,583,585,588−592 In a population-based study, an important reduction in the vaccine efficacy was also observed for ChAdOx1 nCoV-19/AZD1222.592 BNT162b2, for instance, seems to maintain its efficacy to prevent severe forms of the disease.580 BBV152/COVAXIN and Ad26.COV2.S were estimated to be effective against the beta VOC593 (Assessment Report EMA/158424/2021). Reduced neutralization by therapeutic monoclonal antibodies was also observed for this variant.578,591 Thus, the beta VOC could be implicated in the augmentation of reinfection frequency and vaccine or therapy failure and must be closely tracked by genomic surveillance.
Gamma
In December 2020, a new SARS-CoV-2 variant called P.1 (gamma) was detected in Manaus, Brazil, and was potentially linked to an important increase in COVID-19 frequency in that city.594,595 The same variant was also detected in infected travelers who originated from that state and arrived in Tokyo, Japan, in January 2021.596 This new variant was originally characterized by 35 mutations distributed across the entire genome. Ten nonsynonymous variations are located in the S gene, of which three (K417T, E484 K and N501Y) are shared with the B.1.351 variant and one (N501Y) is shared with both the B.1.1.7 and B.1.351 variants (Table S1).594 The transmissibility of the gamma VOC was estimated to be 1.7 to 2.5 times higher than those of non-gamma variants circulating in Manaus, which made it the dominant variant in the city in January 2021.594,595 Higher viral loads in people infected with the gamma variant were also reported, which could be a contributing factor to its more infectious behavior.595 Infection with the gamma variant was associated with a high risk of hospitalization and ICU admission.574 Reinfection cases597,598 and resurgence of the disease in places where herd immunity was probably achieved by previous variants354,599 could also be explained by the rise of this new variant. Gamma is little to completely resistant to neutralization by therapeutic monoclonal antibodies and convalescent plasma.600,601 The BNT162b2 and mRNA-1273 vaccines were the best assessed, showing modest to moderate reductions in their neutralization capacities against this variant.589,600−602 In fact, a case of a fully BNT162b2-vaccinated man that developed mild symptoms after gamma infection was reported.603 CoronaVac was estimated to be effective against gamma,604 and the capacity of AZD1222 to neutralize this virus is reduced.602
Delta
In December 2020 and the first months of 2021, India reported an increase in COVID-19 cases associated with the emergence of the new SARS-CoV-2 variants B.1.617.1, B.1.617.2, and B.1.617.3 which were exported to other countries by Indian travelers.605 B.1.617.2 (delta) rapidly became the dominant lineage in India and established a community transmission chain in other countries worldwide, rapidly increasing its proportion.605,606 With a reproduction number 97% higher than the observed number for non-VOCs and at least 30% higher than those for other VOCs, the delta VOC was estimated to become the dominant circulating lineage worldwide until the emergence of the omicron variant.606 Twelve nonsynonymous mutations characterize this variant, five of them being in the S gene (Table S1). The increased transmissibility presented by delta could be related to higher viral loads,607−609 possibly due to a higher replication rate compared with other variants.610 Also, two spike mutations presented by this variant, L452R and T478K, are predicted to improve its interaction with ACE2 and possibly increase the ability of the virus to enter human cells,611,612 although this hypothesis should be further evaluated. The delta VOC was linked to a higher risk of hospitalization and disease severity.607,613 As observed for other VOCs, the delta VOC is resistant to neutralization by some therapeutic monoclonal antibodies578,614 and to convalescent sera.578,614 BNT162b2-vaccine-elicited sera also showed a reduced neutralization capacity against delta, especially after partial vaccination.578,614−617 Full vaccination with BNT162b2 seems to generate immunity against delta comparable to that against other SARS-CoV-2 variants.578,614,617 In fact, full vaccination had a similar efficacy against the delta VOC compared to the B.1.1.7 variant in population-based studies.618,619 mRNA-1273 vaccination against the development of symptomatic cases derived from delta infection was less effective than that from B.1.1.7 infection.619 However, mRNA-based vaccines (BNT162b2 and mRNA-1273 analyzed together) were able to protect vaccinated subjects from the development of moderate to severe illness.620 Inactivated-virus-based vaccines, including HB02 and WIV04 from Sinopharm, CoronaVac, and Biokangtai’s inactivated COVID19 vaccine, were evaluated together regarding their efficacy against the delta VOC in a Chinese population. They achieved efficacies of 69.5% against COVID-19-associated pneumonia and 100% against severe illness.621 Neutralization of the delta variant by BBV152/COVAXIN-elicited sera was slightly reduced, which suggests that these sera maintained their efficacy against this strain.622 Sera elicited by nonreplicative-viral-vector-based vaccines had their neutralization capacity reduced against the delta VOC.578,614,623,624 The efficacy of AZD122 obtained from population-based studies diverges between authors and must be further investigated.618,619
Omicron
The SARS-CoV-2 omicron variant (B.1.1.529) first emerged in Botswana and South Africa and has been associated with a steep increase in the incidence of COVID-19; it was classified as a VOC by the WHO on November 26, 2021. Its emergence has contributed to the fourth wave of the COVID-19 pandemic in many countries worldwide.625 The omicron VOC displays distinct biological characteristics, including strong binding to human ACE2 receptor and high transmissibility,626−628 despite being less virulent than the original strain or the previous VOCs. In addition, the omicron VOC displays high environmental stability and high resistance against clinically approved monoclonal antibodies and immunity elicited by natural infection or vaccination.629−631 More recently, we have revised in detail the SARS-CoV-2 omicron variant and several characteristics related to this novel variant.632
Final Considerations and Public Health Perspectives
The emergence of this novel coronavirus in the human population precipitated a global threat and was designated a pandemic by the WHO on March 11, 2020. Two years later, it can certainly be considered one of the greatest public health crises. Notably, global society was unprepared for the emergence of SARS-CoV-2 and the serious and widespread consequences of COVID-19 infections. However, the rapid response to the COVID-19 crisis, including efforts made by the WHO, health authorities, industry, governments, and global researchers, have improved public health resilience and helped to mitigate societal impacts. Clinically these actions included the open sharing of information on infection rates and deaths. Open research, like the early publication of the viral genome, and industry engagement with the development and patient trial validation of vaccine candidates as well as governments’ rapid approval of novel diagnostic tests and vaccines—all within a short time—helped to blunt the severity of SARS-CoV-2. The pandemic has highlighted both our ability to respond and our shortcomings in pandemic preparedness for events of this scale.
The pandemic has taught several lessons, including the need for rapid large-scale production and distribution of vaccines, the production and availability of point-of-care diagnostic tests, and last but not least, the need to address the trade of wildlife and ecosystem loss as critical factors in the emergence of infectious diseases. The lessons learned from the COVID-19 pandemic will be critical for dealing with future public health threats, especially for the emergence of new pathogens.
In addition to seeing so much scientific knowledge generated during the last 2 years, society also many experienced lessons. During the course of the COVID-19 pandemic, the human population showed incredible resilience in the face of losses and economic uncertainty. We are still learning to live with SARS-CoV-2, but we are certainly more prepared for future biothreats, and there is reason to hope that human ingenuity will ensure that this virus will not be a threat over time.
Acknowledgments
This work was supported by the CIHR Canada Research Chair Program (950-231075) to K.P. and Canada’s International Development Research Centre (Grant 109434-001) through the Canadian 2019 Novel Coronavirus (COVID-19) Rapid Research Funding Opportunity to K.P. and L.P. The work in Dr. Pena’s lab was funded by the Fiocruz Inova Program, IDRC-Canada, and the Foundation for Science and Technology of Pernambuco (FACEPE) (Grant APQ-0560-2.12/19). S.J.R.d.S. was the recipient of a Ph.D. fellowship sponsored by FACEPE (Grant IBPG-1321-2.12/18) and a postdoctoral fellowship sponsored by the University of Toronto. A.K. was funded by the U.K. Medical Research Council (MC_UU_12014/8). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank Dr. Allyson Andrade Mendonça for helping to design the review and Dr. Reza Mohamadi for his critical comments on the manuscript. Figures (except Figure 4) were created with Biorender.com under academic license to L.P. and K.P. We apologize to those researchers whose related work we were not able to cite in this review.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.2c00204.
Definitions of nonsynonymous mutations and deletions of each variant of concern of SARS-CoV-2 classified by the U.S. CDC and their individual impacts on the virus phenotype (Table S1) (PDF)
Author Contributions
S.J.R.d.S., L.P., K.P., A.K., and A.S.-J. conceived the work. S.J.R.d.S., J.C.F.d.N., and J.J.F.d.M. created the figures. S.J.R.d.S., J.C.F.d.N., R.P.G.M., K.M.G., C.T.A.d.S., P.G.d.S., J.J.F.d.M. and L.P. wrote the original draft. S.J.R.d.S., J.C.F.d.N., J.R.J.V., A.S.-J., A.K., K.P., and L.P. reviewed the final manuscript. S.J.R.d.S. and L.P. supervised the work. All of the authors critically revised the manuscript and approved the final version of the submitted manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Weiss S. R.; Leibowitz J. L. Coronavirus Pathogenesis. Adv. Virus Res. 2011, 81, 85–164. 10.1016/B978-0-12-385885-6.00009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S.; Wong G.; Shi W.; Liu J.; Lai A. C. K.; Zhou J.; Liu W.; Bi Y.; Gao G. F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24 (6), 490–502. 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J. S. M.; Guan Y.; Yuen K. Y. Severe Acute Respiratory Syndrome. Nat. Med. 2004, 10 (S12), S88–S97. 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-Yeung M.; Xu R.-H. SARS: Epidemiology. Respirology 2003, 8 (S1), S9–S14. 10.1046/j.1440-1843.2003.00518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A. M.; Van Boheemen S.; Bestebroer T. M.; Osterhaus A. D. M. E.; Fouchier R. A. M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367 (19), 1814–1820. 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Cui J.; Li F.; Shi Z. L. Origin and Evolution of Pathogenic Coronaviruses. Nat. Rev. Microbiol. 2019, 17 (3), 181–192. 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P.; Yang X.-L.; Wang X.-G.; Hu B.; Zhang L.; Zhang W.; Si H.-R.; Zhu Y.; Li B.; Huang C.-L.; Chen H.-D.; Chen J.; Luo Y.; Guo H.; Jiang R.-D.; Liu M.-Q.; Chen Y.; Shen X.-R.; Wang X.; Zheng X.-S.; Zhao K.; Chen Q.-J.; Deng F.; Liu L.-L.; Yan B.; Zhan F.-X.; Wang Y.-Y.; Xiao G.-F.; Shi Z.-L. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579 (7798), 270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R.; Zhao X.; Li J.; Niu P.; Yang B.; Wu H.; Wang W.; Song H.; Huang B.; Zhu N.; Bi Y.; Ma X.; Zhan F.; Wang L.; Hu T.; Zhou H.; Hu Z.; Zhou W.; Zhao L.; Chen J.; Meng Y.; Wang J.; Lin Y.; Yuan J.; Xie Z.; Ma J.; Liu W. J.; Wang D.; Xu W.; Holmes E. C.; Gao G. F.; Wu G.; Chen W.; Shi W.; Tan W. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Lancet 2020, 395 (10224), 565–574. 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N.; Zhang D.; Wang W.; Li X.; Yang B.; Song J.; Zhao X.; Huang B.; Shi W.; Lu R.; Niu P.; Zhan F.; Ma X.; Wang D.; Xu W.; Wu G.; Gao G. F.; Tan W. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronavirus Disease 2019 (COVID-19): Situation Report—51. World Health Organization, 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200311-sitrep-51-covid-19.pdf?sfvrsn=1ba62e57_10 (accessed 2022-04-18). [PubMed]
- Dong E.; Du H.; Gardner L. An Interactive Web-Based Dashboard to Track COVID-19 in Real Time. Lancet Infect. Dis. 2020, 20 (5), 533–534. 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R.; Li Y.; Zhang A. L.; Wang Y.; Molina M. J. Identifying Airborne Transmission as the Dominant Route for the Spread of COVID-19. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (26), 14857–14863. 10.1073/pnas.2009637117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falahi S.; Kenarkoohi A. Transmission Routes for SARS-CoV-2 Infection: Review of Evidence. New Microbes New Infect. 2020, 38, 100778. 10.1016/j.nmni.2020.100778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W. J.; Ni Z. Y.; Hu Y.; Liang W. H.; Ou C. Q.; He J. X.; Liu L.; Shan H.; Lei C. L.; Hui D. S. C.; Du B.; Li L. J.; Zeng G.; Yuen K. Y.; Chen R. C.; Tang C. L.; Wang T.; Chen P. Y.; Xiang J.; Li S. Y.; Wang J. L.; Liang Z. J.; Peng Y. X.; Wei L.; Liu Y.; Hu Y. H.; Peng P.; Wang J. M.; Liu J. Y.; Chen Z.; Li G.; Zheng Z. J.; Qiu S. Q.; Luo J.; Ye C. J.; Zhu S. Y.; Zhong N. S. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Guan X.; Wu P.; Wang X.; Zhou L.; Tong Y.; Ren R.; Leung K. S. M.; Lau E. H. Y.; Wong J. Y.; Xing X.; Xiang N.; Wu Y.; Li C.; Chen Q.; Li D.; Liu T.; Zhao J.; Liu M.; Tu W.; Chen C.; Jin L.; Yang R.; Wang Q.; Zhou S.; Wang R.; Liu H.; Luo Y.; Liu Y.; Shao G.; Li H.; Tao Z.; Yang Y.; Deng Z.; Liu B.; Ma Z.; Zhang Y.; Shi G.; Lam T. T. Y.; Wu J. T.; Gao G. F.; Cowling B. J.; Yang B.; Leung G. M.; Feng Z. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N.; Zhou M.; Dong X.; Qu J.; Gong F.; Han Y.; Qiu Y.; Wang J.; Liu Y.; Wei Y.; Xia J.; Yu T.; Zhang X.; Zhang L. Epidemiological and Clinical Characteristics of 99 Cases of 2019 Novel Coronavirus Pneumonia in Wuhan, China: A Descriptive Study. Lancet 2020, 395 (10223), 507–513. 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.; Wang Y.; Li X.; Ren L.; Zhao J.; Hu Y.; Zhang L.; Fan G.; Xu J.; Gu X.; Cheng Z.; Yu T.; Xia J.; Wei Y.; Wu W.; Xie X.; Yin W.; Li H.; Liu M.; Xiao Y.; Gao H.; Guo L.; Xie J.; Wang G.; Jiang R.; Gao Z.; Jin Q.; Wang J.; Cao B. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395 (10223), 497–506. 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.; Hu B.; Hu C.; Zhu F.; Liu X.; Zhang J.; Wang B.; Xiang H.; Cheng Z.; Xiong Y.; Zhao Y.; Li Y.; Wang X.; Peng Z. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA - J. Am. Med. Assoc. 2020, 323 (11), 1061–1069. 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z.; Shi L.; Wang Y.; Zhang J.; Huang L.; Zhang C.; Liu S.; Zhao P.; Liu H.; Zhu L.; Tai Y.; Bai C.; Gao T.; Song J.; Xia P.; Dong J.; Zhao J.; Wang F.-S. Pathological Findings of COVID-19 Associated with Acute Respiratory Distress Syndrome. Lancet Respir. Med. 2020, 8 (4), 420–422. 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.; McGoogan J. M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China. JAMA 2020, 323 (13), 1239. 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Chen T.; Wu D.; Chen H.; Yan W.; Yang D.; Chen G.; Ma K.; Xu D.; Yu H.; Wang H.; Wang T.; Guo W.; Chen J.; Ding C.; Zhang X.; Huang J.; Han M.; Li S.; Luo X.; Zhao J.; Ning Q. Clinical Characteristics of 113 Deceased Patients with Coronavirus Disease 2019: Retrospective Study. BMJ. 2020, m1091. 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K.; Chao R. K.; Krause H. E.; Wasil R.; Mocega H. E.; Mufson M. A. Coronavirus Infection in Acute Lower Respiratory Tract Disease of Infants. J. Infect. Dis. 1974, 130 (5), 502–507. 10.1093/infdis/130.5.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte K. H.; Tajima M.; Easterday B. C. Morphologic Characteristics and Nucleic Acid Type of Transmissible Gastroenteritis Virus of Pigs. Arch. Gesamte Virusforsch. 1968, 23 (1-2), 53–70. 10.1007/BF01242114. [DOI] [PubMed] [Google Scholar]
- Almeida J. Virology: Coronaviruses. Nature 1968, 220, 650. 10.1038/220650b0. [DOI] [Google Scholar]
- Tyrrell D. A. J.; Almeida J. D.; Cunningham C. H.; Dowdle W. R.; Hofstad M. S.; McIntosh K.; Tajima M.; Zakstelskaya L. Y.; Easterday B. C.; Kapikian A.; Bingham R. W. Coronaviridae. Intervirology 2004, 5 (1-2), 76–82. 10.1159/000149883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamre D.; Procknow J. J. A New Virus Isolated from the Human Respiratory Tract. Exp. Biol. Med. 1966, 121 (1), 190–193. 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- McIntosh K. Coronaviruses in the Limelight. J. Infect. Dis. 2005, 191 (4), 489–491. 10.1086/428510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L.; Pyrc K.; Jebbink M. F.; Vermeulen-Oost W.; Berkhout R. J. M.; Wolthers K. C.; Wertheim-van Dillen P. M. E.; Kaandorp J.; Spaargaren J.; Berkhout B. Identification of a New Human Coronavirus. Nat. Med. 2004, 10 (4), 368–373. 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C. Y.; Lau S. K. P.; Chu C.; Chan K.; Tsoi H.; Huang Y.; Wong B. H. L.; Poon R. W. S.; Cai J. J.; Luk W.; Poon L. L. M.; Wong S. S. Y.; Guan Y.; Peiris J. S. M.; Yuen K. Characterization and Complete Genome Sequence of a Novel Coronavirus, Coronavirus HKU1, from Patients with Pneumonia. J. Virol. 2005, 79 (2), 884–895. 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K.; Berkhout B.; van der Hoek L. The Novel Human Coronaviruses NL63 and HKU1. J. Virol. 2007, 81 (7), 3051–3057. 10.1128/JVI.01466-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C.; Günther S.; Preiser W.; van der Werf S.; Brodt H.-R.; Becker S.; Rabenau H.; Panning M.; Kolesnikova L.; Fouchier R. A. M.; Berger A.; Burguière A.-M.; Cinatl J.; Eickmann M.; Escriou N.; Grywna K.; Kramme S.; Manuguerra J.-C.; Müller S.; Rickerts V.; Stürmer M.; Vieth S.; Klenk H.-D.; Osterhaus A. D. M. E.; Schmitz H.; Doerr H. W. Identification of a Novel Coronavirus in Patients with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003, 348 (20), 1967–1976. 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Cheng V. C. C.; Lau S. K. P.; Woo P. C. Y.; Yuen K. Y. Severe Acute Respiratory Syndrome Coronavirus as an Agent of Emerging and Reemerging Infection. Clin. Microbiol. Rev. 2007, 20 (4), 660–694. 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V. S.; Osterhaus A. D.; Fouchier R. A.; Haagmans B. L. MERS: Emergence of a Novel Human Coronavirus. Curr. Opin. Virol. 2014, 5, 58–62. 10.1016/j.coviro.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. MERS situation update, June 2022. http://www.emro.who.int/health-topics/mers-cov/mers-outbreaks.html.
- Liang W.-N.; Zhao T.; Liu Z.-J.; Guan B.-Y.; He X.; Liu M.; Chen Q.; Liu G.-F.; Wu J.; Huang R.-G.; Xie X.-Q.; Wu Z.-L. Severe acute respiratory syndrome-retrospect and lessons of 2004 outbreak in China. Biomed Environ. Sci. 2006, 19 (6), 445–451. [PubMed] [Google Scholar]
- Rambaut A.; Holmes E. C.; O’Toole Á.; Hill V.; McCrone J. T.; Ruis C.; du Plessis L.; Pybus O. G. A Dynamic Nomenclature Proposal for SARS-CoV-2 Lineages to Assist Genomic Epidemiology. Nat. Microbiol. 2020, 5 (11), 1403–1407. 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K. G.; Rambaut A.; Lipkin W. I.; Holmes E. C.; Garry R. F. The Proximal Origin of SARS-CoV-2. Nat. Med. 2020, 26 (4), 450–452. 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. F.-W.; Kok K.-H.; Zhu Z.; Chu H.; To K. K.-W.; Yuan S.; Yuen K.-Y. Genomic Characterization of the 2019 Novel Human-Pathogenic Coronavirus Isolated from a Patient with Atypical Pneumonia after Visiting Wuhan. Emerg. Microbes Infect. 2020, 9 (1), 221–236. 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-Z.; Holmes E. C. A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. Cell 2020, 181 (2), 223–227. 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaune D.; Hul V.; Karlsson E. A.; Hassanin A.; Ou T. P.; Baidaliuk A.; Gámbaro F.; Prot M.; Tu V. T.; Chea S.; Keatts L.; Mazet J.; Johnson C. K.; Buchy P.; Dussart P.; Goldstein T.; Simon-Lorière E.; Duong V. A Novel SARS-CoV-2 Related Coronavirus in Bats from Cambodia. Nat. Commun. 2021, 12 (1), 6563. 10.1038/s41467-021-26809-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G.; Liu W.; Liu P.; Lei W.; Jia Z.; He X.; Liu L.L.; Shi W.; Tan Y.; Zou S.; Zhao X.; Wong X.; Wong G.; Wang J.; Wang F.; Wang G.; Qin K.; Gao R.; Zhang J.; Li M.; Xiao W.; Guo Y.; Xu Z.; Zhao; Song J.; Zhang J.; Zhen W.; Zhou W.; Ye B.; Song J.; Yang M.; Zhou W.; Bi Y.; Cai K.; Wang D.; Tan W.; Han J.; Xu W.; Wu G. Surveillance of SARS-CoV-2 in the environment and animal samples of the Huanan Seafood Market. Research Square 2022, 10.21203/rs.3.rs-1370392/v1. [DOI] [Google Scholar]
- Worobey M. The Huanan market was the epicenter of SARS-CoV-2 emergence. Zenodo 2022, 10.5281/zenodo.6299600. [DOI] [Google Scholar]
- Pekar J. E.; Magee A.; Parker E.; Moshiri N.; Izhikevich K.; Havens J. L.; Gangavarapu K.; Malpica Serrano L. M.; Crits-Christoph A.; Matteson N. L.; Zeller M.; Levy J. I.; Wang J. C.; Hughes S.; Lee J.; Park H.; Park M.-S.; Yan K. C. Z.; Lin R. T. P.; Mat Isa M. N.; Noor Y. M.; Vasylyeva T. I.; Garry R. F.; Holmes E. C.; Rambaut A.; Suchard M. A.; Andersen K. G.; Worobey M.; Wertheim J. O. SARS-CoV-2 emergence very likely resulted from at least two zoonotic events. Zenodo 2022, 10.5281/zenodo.6291628. [DOI] [Google Scholar]
- Xu Y. Unveiling the Origin and Transmission of 2019-NCoV. Trends Microbiol. 2020, 28 (4), 239–240. 10.1016/j.tim.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman D. A. To Stop the next Pandemic, We Need to Unravel the Origins of COVID-19. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (47), 29246–29248. 10.1073/pnas.2021133117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S. Another Decade, Another Coronavirus. N. Engl. J. Med. 2020, 382 (8), 760–762. 10.1056/NEJMe2001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T. T.-Y.; Jia N.; Zhang Y.-W.; Shum M. H.-H.; Jiang J.-F.; Zhu H.-C.; Tong Y.-G.; Shi Y.-X.; Ni X.-B.; Liao Y.-S.; Li W.-J.; Jiang B.-G.; Wei W.; Yuan T.-T.; Zheng K.; Cui X.-M.; Li J.; Pei G.-Q.; Qiang X.; Cheung W. Y.-M.; Li L.-F.; Sun F.-F.; Qin S.; Huang J.-C.; Leung G. M.; Holmes E. C.; Hu Y.-L.; Guan Y.; Cao W.-C. Identifying SARS-CoV-2-Related Coronaviruses in Malayan Pangolins. Nature 2020, 583 (7815), 282–285. 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Wacharapluesadee S.; Tan C. W.; Maneeorn P.; Duengkae P.; Zhu F.; Joyjinda Y.; Kaewpom T.; Chia W. N.; Ampoot W.; Lim B. L.; Worachotsueptrakun K.; Chen V. C.-W.; Sirichan N.; Ruchisrisarod C.; Rodpan A.; Noradechanon K.; Phaichana T.; Jantarat N.; Thongnumchaima B.; Tu C.; Crameri G.; Stokes M. M.; Hemachudha T.; Wang L.-F. Evidence for SARS-CoV-2 Related Coronaviruses Circulating in Bats and Pangolins in Southeast Asia. Nat. Commun. 2021, 12 (1), 972. 10.1038/s41467-021-21240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P.; Jiang J.-Z.; Wan X.-F.; Hua Y.; Li L.; Zhou J.; Wang X.; Hou F.; Chen J.; Zou J.; Chen J. Are Pangolins the Intermediate Host of the 2019 Novel Coronavirus (SARS-CoV-2)?. PLoS Pathog. 2020, 16 (5), e1008421. 10.1371/journal.ppat.1008421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K.; Zhai J.; Feng Y.; Zhou N.; Zhang X.; Zou J.-J.; Li N.; Guo Y.; Li X.; Shen X.; Zhang Z.; Shu F.; Huang W.; Li Y.; Zhang Z.; Chen R.-A.; Wu Y.-J.; Peng S.-M.; Huang M.; Xie W.-J.; Cai Q.-H.; Hou F.-H.; Chen W.; Xiao L.; Shen Y. Isolation of SARS-CoV-2-Related Coronavirus from Malayan Pangolins. Nature 2020, 583 (7815), 286–289. 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- Rosa R. B.; Dantas W. M.; do Nascimento J. C. F.; da Silva M. V.; de Oliveira R. N.; Pena L. J. In Vitro and In Vivo Models for Studying SARS-CoV-2, the Etiological Agent Responsible for COVID-19 Pandemic. Viruses 2021, 13 (3), 379. 10.3390/v13030379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann P. J.; Hatta M.; Chiba S.; Maemura T.; Fan S.; Takeda M.; Kinoshita N.; Hattori S.; Sakai-Tagawa Y.; Iwatsuki-Horimoto K.; Imai M.; Kawaoka Y. Transmission of SARS-CoV-2 in Domestic Cats. N. Engl. J. Med. 2020, 383 (6), 592–594. 10.1056/NEJMc2013400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J.; Wen Z.; Zhong G.; Yang H.; Wang C.; Huang B.; Liu R.; He X.; Shuai L.; Sun Z.; Zhao Y.; Liu P.; Liang L.; Cui P.; Wang J.; Zhang X.; Guan Y.; Tan W.; Wu G.; Chen H.; Bu Z. Susceptibility of Ferrets, Cats, Dogs, and Other Domesticated Animals to SARS-Coronavirus 2. Science 2020, 368 (6494), 1016–1020. 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.; Zhang H.; Gao J.; Huang K.; Yang Y.; Hui X.; He X.; Li C.; Gong W.; Zhang Y.; Zhao Y.; Peng C.; Gao X.; Chen H.; Zou Z.; Shi Z.-L.; Jin M. A Serological Survey of SARS-CoV-2 in Cat in Wuhan. Emerg. Microbes Infect. 2020, 9 (1), 2013–2019. 10.1080/22221751.2020.1817796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Fontela C.; Dowling W. E.; Funnell S. G. P.; Gsell P.-S.; Riveros-Balta A. X.; Albrecht R. A.; Andersen H.; Baric R. S.; Carroll M. W.; Cavaleri M.; Qin C.; Crozier I.; Dallmeier K.; de Waal L.; de Wit E.; Delang L.; Dohm E.; Duprex W. P.; Falzarano D.; Finch C. L.; Frieman M. B.; Graham B. S.; Gralinski L. E.; Guilfoyle K.; Haagmans B. L.; Hamilton G. A.; Hartman A. L.; Herfst S.; Kaptein S. J. F.; Klimstra W. B.; Knezevic I.; Krause P. R.; Kuhn J. H.; Le Grand R.; Lewis M. G.; Liu W.-C.; Maisonnasse P.; McElroy A. K.; Munster V.; Oreshkova N.; Rasmussen A. L.; Rocha-Pereira J.; Rockx B.; Rodríguez E.; Rogers T. F.; Salguero F. J.; Schotsaert M.; Stittelaar K. J.; Thibaut H. J.; Tseng C.-T.; Vergara-Alert J.; Beer M.; Brasel T.; Chan J. F. W.; García-Sastre A.; Neyts J.; Perlman S.; Reed D. S.; Richt J. A.; Roy C. J.; Segalés J.; Vasan S. S.; Henao-Restrepo A. M.; Barouch D. H. Animal Models for COVID-19. Nature 2020, 586 (7830), 509–515. 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-I.; Kim S.-G.; Kim S.-M.; Kim E.-H.; Park S.-J.; Yu K.-M.; Chang J.-H.; Kim E. J.; Lee S.; Casel M. A. B.; Um J.; Song M.-S.; Jeong H. W.; Lai V. D.; Kim Y.; Chin B. S.; Park J.-S.; Chung K.-H.; Foo S.-S.; Poo H.; Mo I.-P.; Lee O.-J.; Webby R. J.; Jung J. U.; Choi Y. K. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe 2020, 27 (5), 704–709. 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A. O.; Case J. B.; Winkler E. S.; Thackray L. B.; Kafai N. M.; Bailey A. L.; McCune B. T.; Fox J. M.; Chen R. E.; Alsoussi W. B.; Turner J. S.; Schmitz A. J.; Lei T.; Shrihari S.; Keeler S. P.; Fremont D. H.; Greco S.; McCray P. B.; Perlman S.; Holtzman M. J.; Ellebedy A. H.; Diamond M. S. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell 2020, 182 (3), 744–753. 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreshkova N.; Molenaar R. J.; Vreman S.; Harders F.; Oude Munnink B. B.; Hakze-van der Honing R. W.; Gerhards N.; Tolsma P.; Bouwstra R.; Sikkema R. S.; Tacken M. G.; de Rooij M. M.; Weesendorp E.; Engelsma M. Y.; Bruschke C. J.; Smit L. A.; Koopmans M.; van der Poel W. H.; Stegeman A. SARS-CoV-2 Infection in Farmed Minks, The Netherlands, April and May 2020. Eurosurveillance 2020, 25 (23), 2001005 10.2807/1560-7917.ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.; Zhao Y.; Yu W.; Yang Y.; Gao J.; Wang J.; Kuang D.; Yang M.; Yang J.; Ma C.; Xu J.; Qian X.; Li H.; Zhao S.; Li J.; Wang H.; Long H.; Zhou J.; Luo F.; Ding K.; Wu D.; Zhang Y.; Dong Y.; Liu Y.; Zheng Y.; Lin X.; Jiao L.; Zheng H.; Dai Q.; Sun Q.; Hu Y.; Ke C.; Liu H.; Peng X. Comparison of Nonhuman Primates Identified the Suitable Model for COVID-19. Signal Transduct. Target. Ther. 2020, 5 (1), 157. 10.1038/s41392-020-00269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S. J. R.; Alves da Silva C. T.; Mendes R. P. G.; Pena L. Role of Nonstructural Proteins in the Pathogenesis of SARS-CoV-2. J. Med. Virol. 2020, 92 (9), 1427–1429. 10.1002/jmv.25858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S. J. R. da; Germano Mendes R. P.; Alves da Silva C. T.; Lorusso A.; Kohl A.; Pena L. Insights into SARS-CoV-2, the Coronavirus Underlying COVID-19: Recent Genomic Data and the Development of Reverse Genetics Systems. J. Gen. Virol. 2020, 101 (10), 1021–1024. 10.1099/jgv.0.001458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A. R.; Perlman S. Coronaviruses: An Overview of Their Replication and Pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W. B.; Kwon N.-J.; Choi S.-J.; Kang C. K.; Choe P. G.; Kim J. Y.; Yun J.; Lee G.-W.; Seong M.-W.; Kim N. J.; Seo J.-S.; Oh M. Virus Isolation from the First Patient with SARS-CoV-2 in Korea. J. Korean Med. Sci. 2020, 35 (7), e84 10.3346/jkms.2020.35.e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B. W.; Adair B. D.; Yoshioka C.; Quispe J. D.; Orca G.; Kuhn P.; Milligan R. A.; Yeager M.; Buchmeier M. J. Supramolecular Architecture of Severe Acute Respiratory Syndrome Coronavirus Revealed by Electron Cryomicroscopy. J. Virol. 2006, 80 (16), 7918–7928. 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.; Mendonça L.; Yang Y.; Gao Y.; Shen C.; Liu J.; Ni T.; Ju B.; Liu C.; Tang X.; Wei J.; Ma X.; Zhu Y.; Liu W.; Xu S.; Liu Y.; Yuan J.; Wu J.; Liu Z.; Zhang Z.; Liu L.; Wang P.; Zhang P. The Architecture of Inactivated SARS-CoV-2 with Postfusion Spikes Revealed by Cryo-EM and Cryo-ET. Structure 2020, 28 (11), 1218–1224. 10.1016/j.str.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Z.; Chan S. Y.; Liu W. J.; Li P.; Huang W. Recent Insights into Emerging Coronavirus: SARS-CoV-2. ACS Infect. Dis. 2021, 7 (6), 1369–1388. 10.1021/acsinfecdis.0c00646. [DOI] [PubMed] [Google Scholar]
- Kim J.-M.; Chung Y.-S.; Jo H. J.; Lee N.-J.; Kim M. S.; Woo S. H.; Park S.; Kim J. W.; Kim H. M.; Han M.-G. Identification of Coronavirus Isolated from a Patient in Korea with COVID-19. Osong Public Heal. Res. Perspect. 2020, 11 (1), 3–7. 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu Y. L.; Teoh K. T.; Lo J.; Chan C. M.; Kien F.; Escriou N.; Tsao S. W.; Nicholls J. M.; Altmeyer R.; Peiris J. S. M.; Bruzzone R.; Nal B. The M, E, and N Structural Proteins of the Severe Acute Respiratory Syndrome Coronavirus Are Required for Efficient Assembly, Trafficking, and Release of Virus-Like Particles. J. Virol. 2008, 82 (22), 11318–11330. 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenian E.; Nandakumar D.; Lari A.; Ly M.; Tucker J. M.; Glaunsinger B. A. The Molecular Virology of Coronaviruses. J. Biol. Chem. 2020, 295 (37), 12910–12934. 10.1074/jbc.REV120.013930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J.; Han N.; Chen Z.; Peng Y.; Li L.; Zhou H.; Ji C.; Meng J.; Jiang T.; Wu A. Compositional Diversity and Evolutionary Pattern of Coronavirus Accessory Proteins. Brief. Bioinf. 2021, 22 (2), 1267–1278. 10.1093/bib/bbaa262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo N.; Zaldívar-López S.; Garrido J. J.; Montoya M. SARS-CoV-2 Accessory Proteins in Viral Pathogenesis: Knowns and Unknowns. Front. Immunol. 2021, 12, 708264 10.3389/fimmu.2021.708264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L.; He Y.; Zhou Y.; Liu S.; Zheng B.-J.; Jiang S. The Spike Protein of SARS-CoV—A Target for Vaccine and Therapeutic Development. Nat. Rev. Microbiol. 2009, 7 (3), 226–236. 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.; Li W.; Farzan M.; Harrison S. C. Structure of SARS Coronavirus Spike Receptor-Binding Domain Complexed with Receptor. Science 2005, 309 (5742), 1864–1868. 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3 (1), 237–261. 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L.; Yang Y.; Zhou Y.; Lu L.; Li F.; Jiang S. MERS-CoV Spike Protein: A Key Target for Antivirals. Expert Opin. Ther. Targets 2017, 21 (2), 131–143. 10.1080/14728222.2017.1271415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Wong G.; Lu G.; Yan J.; Gao G. F. MERS-CoV Spike Protein: Targets for Vaccines and Therapeutics. Antiviral Res. 2016, 133, 165–177. 10.1016/j.antiviral.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.; Liu Q.; Zhu Y.; Chan K.-H.; Qin L.; Li Y.; Wang Q.; Chan J. F.-W.; Du L.; Yu F.; Ma C.; Ye S.; Yuen K.-Y.; Zhang R.; Jiang S. Structure-Based Discovery of Middle East Respiratory Syndrome Coronavirus Fusion Inhibitor. Nat. Commun. 2014, 5 (1), 3067. 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L.; Tai W.; Yang Y.; Zhao G.; Zhu Q.; Sun S.; Liu C.; Tao X.; Tseng C.-T. K.; Perlman S.; Jiang S.; Zhou Y.; Li F. Introduction of Neutralizing Immunogenicity Index to the Rational Design of MERS Coronavirus Subunit Vaccines. Nat. Commun. 2016, 7 (1), 13473. 10.1038/ncomms13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.; Xiao G.; Chen Y.; He Y.; Niu J.; Escalante C. R.; Xiong H.; Farmar J.; Debnath A. K.; Tien P.; Jiang S. Interaction between Heptad Repeat 1 and 2 Regions in Spike Protein of SARS-Associated Coronavirus: Implications for Virus Fusogenic Mechanism and Identification of Fusion Inhibitors. Lancet 2004, 363 (9413), 938–947. 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A. C.; Park Y.-J.; Tortorici M. A.; Wall A.; McGuire A. T.; Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181 (2), 281–292. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J.; Ge J.; Yu J.; Shan S.; Zhou H.; Fan S.; Zhang Q.; Shi X.; Wang Q.; Zhang L.; Wang X. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 2020, 581 (7807), 215–220. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Tai W.; He L.; Zhang X.; Pu J.; Voronin D.; Jiang S.; Zhou Y.; Du L. Characterization of the Receptor-Binding Domain (RBD) of 2019 Novel Coronavirus: Implication for Development of RBD Protein as a Viral Attachment Inhibitor and Vaccine. Cell. Mol. Immunol. 2020, 17 (6), 613–620. 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker G. R. SARS-CoV-2 Spike and Its Adaptable Furin Cleavage Site. Lancet Microbe 2021, 2 (10), e488–e489. 10.1016/S2666-5247(21)00174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock T. P.; Goldhill D. H.; Zhou J.; Baillon L.; Frise R.; Swann O. C.; Kugathasan R.; Penn R.; Brown J. C.; Sanchez-David R. Y.; Braga L.; Williamson M. K.; Hassard J. A.; Staller E.; Hanley B.; Osborn M.; Giacca M.; Davidson A. D.; Matthews D. A.; Barclay W. S. The Furin Cleavage Site in the SARS-CoV-2 Spike Protein Is Required for Transmission in Ferrets. Nat. Microbiol. 2021, 6 (7), 899–909. 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- Coutard B.; Valle C.; de Lamballerie X.; Canard B.; Seidah N. G.; Decroly E. The Spike Glycoprotein of the New Coronavirus 2019-NCoV Contains a Furin-like Cleavage Site Absent in CoV of the Same Clade. Antiviral Res. 2020, 176, 104742. 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J.; Wan Y.; Luo C.; Ye G.; Geng Q.; Auerbach A.; Li F. Cell Entry Mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (21), 11727–11734. 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.; Moore M. J.; Vasilieva N.; Sui J.; Wong S. K.; Berne M. A.; Somasundaran M.; Sullivan J. L.; Luzuriaga K.; Greenough T. C.; Choe H.; Farzan M. Angiotensin-Converting Enzyme 2 Is a Functional Receptor for the SARS Coronavirus. Nature 2003, 426 (6965), 450–454. 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W.; He L.; Zhang X.; Pu J.; Voronin D.; Jiang S.; Zhou Y.; Du L. Characterization of the Receptor-Binding Domain (RBD) of 2019 Novel Coronavirus: Implication for Development of RBD Protein as a Viral Attachment Inhibitor and Vaccine. Cell. Mol. Immunol. 2020, 17 (6), 613–620. 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R.; Zhang Y.; Li Y.; Xia L.; Guo Y.; Zhou Q. Structural Basis for the Recognition of SARS-CoV-2 by Full-Length Human ACE2. Science 2020, 367 (6485), 1444–1448. 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Kleine-Weber H.; Schroeder S.; Krüger N.; Herrler T.; Erichsen S.; Schiergens T. S.; Herrler G.; Wu N.-H.; Nitsche A.; Müller M. A.; Drosten C.; Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181 (2), 271–280. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski P.; Kratzel A.; Steiner S.; Stalder H.; Thiel V. Coronavirus Biology and Replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19 (3), 155–170. 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Pöhlmann S. How SARS-CoV-2 Makes the Cut. Nat. Microbiol. 2021, 6 (7), 828–829. 10.1038/s41564-021-00931-x. [DOI] [PubMed] [Google Scholar]
- Wang K.; Chen W.; Zhang Z.; Deng Y.; Lian J.-Q.; Du P.; Wei D.; Zhang Y.; Sun X.-X.; Gong L.; Yang X.; He L.; Zhang L.; Yang Z.; Geng J.-J.; Chen R.; Zhang H.; Wang B.; Zhu Y.-M.; Nan G.; Jiang J.-L.; Li L.; Wu J.; Lin P.; Huang W.; Xie L.; Zheng Z.-H.; Zhang K.; Miao J.-L.; Cui H.-Y.; Huang M.; Zhang J.; Fu L.; Yang X.-M.; Zhao Z.; Sun S.; Gu H.; Wang Z.; Wang C.-F.; Lu Y.; Liu Y.-Y.; Wang Q.-Y.; Bian H.; Zhu P.; Chen Z.-N. CD147-Spike Protein Is a Novel Route for SARS-CoV-2 Infection to Host Cells. Signal Transduct. Target. Ther. 2020, 5 (1), 283. 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G.; Hu Y.; Wang Q.; Qi J.; Gao F.; Li Y.; Zhang Y.; Zhang W.; Yuan Y.; Bao J.; Zhang B.; Shi Y.; Yan J.; Gao G. F. Molecular Basis of Binding between Novel Human Coronavirus MERS-CoV and Its Receptor CD26. Nature 2013, 500 (7461), 227–231. 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison A. G.; Lin T.; Wang P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020, 41 (12), 1100–1115. 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S.; Taguchi F. Two-Step Conformational Changes in a Coronavirus Envelope Glycoprotein Mediated by Receptor Binding and Proteolysis. J. Virol. 2009, 83 (21), 11133–11141. 10.1128/JVI.00959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.; Xiang R.; Huo S.; Zhou Y.; Jiang S.; Wang Q.; Yu F. Molecular Mechanism of Interaction between SARS-CoV-2 and Host Cells and Interventional Therapy. Signal Transduct. Target. Ther. 2021, 6 (1), 233. 10.1038/s41392-021-00653-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Kleine-Weber H.; Pöhlmann S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78 (4), 779–784. 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant E. P.; Pérez-Alvarado G. C.; Jacobs J. L.; Mukhopadhyay B.; Hennig M.; Dinman J. D. A Three-Stemmed MRNA Pseudoknot in the SARS Coronavirus Frameshift Signal. PLoS Biol. 2005, 3 (6), e172. 10.1371/journal.pbio.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.; Abriola L.; Niederer R. O.; Pedersen S. F.; Alfajaro M. M.; Silva Monteiro V.; Wilen C. B.; Ho Y.-C.; Gilbert W. V.; Surovtseva Y. V.; Lindenbach B. D.; Guo J. U. Restriction of SARS-CoV-2 Replication by Targeting Programmed -1 Ribosomal Frameshifting. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (26), e2023051118 10.1073/pnas.2023051118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y.; Ng Y.; Tam J.; Liu D. Human Coronaviruses: A Review of Virus-Host Interactions. Diseases 2016, 4 (4), 26. 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S.; Dellibovi-Ragheb T. A.; Kerviel A.; Pak E.; Qiu Q.; Fisher M.; Takvorian P. M.; Bleck C.; Hsu V. W.; Fehr A. R.; Perlman S.; Achar S. R.; Straus M. R.; Whittaker G. R.; de Haan C. A. M.; Kehrl J.; Altan-Bonnet G.; Altan-Bonnet N. β-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway. Cell 2020, 183 (6), 1520–1535. 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuliano A. D.; Chang H. H.; Patel N. N.; Threlkel R.; Kniss K.; Reich J.; Steele M.; Hall A. J.; Fry A. M.; Reed C. Estimating Under-Recognized COVID-19 Deaths, United States, March 2020-May 2021 Using an Excess Mortality Modelling Approach. Lancet Reg. Health - Am. 2021, 1, 100019. 10.1016/j.lana.2021.100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranzani O. T.; Bastos L. S. L.; Gelli J. G. M.; Marchesi J. F.; Baião F.; Hamacher S.; Bozza F. A. Characterisation of the First 250 000 Hospital Admissions for COVID-19 in Brazil: A Retrospective Analysis of Nationwide Data. Lancet Respir. Med. 2021, 9 (4), 407–418. 10.1016/S2213-2600(20)30560-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsinda E. K.; Mmbando G. S. Recent Updates on the Possible Reasons for the Low Incidence and Morbidity of COVID-19 Cases in Africa. Bull. Natl. Res. Cent. 2021, 45 (1), 133. 10.1186/s42269-021-00589-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi S. A.; Nel J. Epidemiology of Severe COVID-19 from South Africa. Lancet HIV 2021, 8 (9), e524–e526. 10.1016/S2352-3018(21)00183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Gayle A. A.; Wilder-Smith A.; Rocklöv J. The Reproductive Number of COVID-19 Is Higher Compared to SARS Coronavirus. J. Travel Med. 2020, 27 (2), taaa021 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billah M. A.; Miah M. M.; Khan M. N. Reproductive Number of Coronavirus: A Systematic Review and Meta-Analysis Based on Global Level Evidence. PLoS One 2020, 15 (11), e0242128. 10.1371/journal.pone.0242128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung N. H. L. Transmissibility and Transmission of Respiratory Viruses. Nat. Rev. Microbiol. 2021, 19 (8), 528–545. 10.1038/s41579-021-00535-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquès M.; Domingo J. L. Contamination of Inert Surfaces by SARS-CoV-2: Persistence, Stability and Infectivity. A Review. Environ. Res. 2021, 193, 110559. 10.1016/j.envres.2020.110559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B.; Guo H.; Zhou P.; Shi Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19 (3), 141–154. 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung N. H. L.; Chu D. K. W.; Shiu E. Y. C.; Chan K.-H.; McDevitt J. J.; Hau B. J. P.; Yen H.-L.; Li Y.; Ip D. K. M.; Peiris J. S. M.; Seto W.-H.; Leung G. M.; Milton D. K.; Cowling B. J. Respiratory Virus Shedding in Exhaled Breath and Efficacy of Face Masks. Nat. Med. 2020, 26 (5), 676–680. 10.1038/s41591-020-0843-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadnytskyi V.; Bax C. E.; Bax A.; Anfinrud P. The Airborne Lifetime of Small Speech Droplets and Their Potential Importance in SARS-CoV-2 Transmission. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (22), 11875–11877. 10.1073/pnas.2006874117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lednicky J. A.; Lauzardo M.; Fan Z. H.; Jutla A.; Tilly T. B.; Gangwar M.; Usmani M.; Shankar S. N.; Mohamed K.; Eiguren-Fernandez A.; Stephenson C. J.; Alam M. M.; Elbadry M. A.; Loeb J. C.; Subramaniam K.; Waltzek T. B.; Cherabuddi K.; Morris J. G.; Wu C.-Y. Viable SARS-CoV-2 in the Air of a Hospital Room with COVID-19 Patients. Int. J. Infect. Dis. 2020, 100, 476–482. 10.1016/j.ijid.2020.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S.; Bherwani H.; Gulia S.; Vijay R.; Kumar R. Understanding COVID-19 Transmission, Health Impacts and Mitigation: Timely Social Distancing Is the Key. Environ. Dev. Sustainability 2021, 23 (5), 6681–6697. 10.1007/s10668-020-00884-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X.; Lau E. H. Y.; Wu P.; Deng X.; Wang J.; Hao X.; Lau Y. C.; Wong J. Y.; Guan Y.; Tan X.; Mo X.; Chen Y.; Liao B.; Chen W.; Hu F.; Zhang Q.; Zhong M.; Wu Y.; Zhao L.; Zhang F.; Cowling B. J.; Li F.; Leung G. M. Temporal Dynamics in Viral Shedding and Transmissibility of COVID-19. Nat. Med. 2020, 26 (5), 672–675. 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Johansson M. A.; Quandelacy T. M.; Kada S.; Prasad P. V.; Steele M.; Brooks J. T.; Slayton R. B.; Biggerstaff M.; Butler J. C. SARS-CoV-2 Transmission From People Without COVID-19 Symptoms. JAMA Netw. Open 2021, 4 (1), e2035057. 10.1001/jamanetworkopen.2020.35057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W. E.; Li Z.; Chiew C. J.; Yong S. E.; Toh M. P.; Lee V. J. Presymptomatic Transmission of SARS-CoV-2 — Singapore, January 23-March 16, 2020. Morb. Mortal. Wkly. Rep. 2020, 69 (14), 411–415. 10.15585/mmwr.mm6914e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y.; Yao L.; Wei T.; Tian F.; Jin D.-Y.; Chen L.; Wang M. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA 2020, 323 (14), 1406. 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe C.; Schunk M.; Sothmann P.; Bretzel G.; Froeschl G.; Wallrauch C.; Zimmer T.; Thiel V.; Janke C.; Guggemos W.; Seilmaier M.; Drosten C.; Vollmar P.; Zwirglmaier K.; Zange S.; Wölfel R.; Hoelscher M. Transmission of 2019-NCoV Infection from an Asymptomatic Contact in Germany. N. Engl. J. Med. 2020, 382 (10), 970–971. 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H.; Lu X.; Deng Y.; Tang Y.; Lu J. COVID-19: Asymptomatic Carrier Transmission Is an Underestimated Problem. Epidemiol. Infect. 2020, 148, e116. 10.1017/S0950268820001235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh T.; Jimenez J. L.; Prather K. A.; Tufekci Z.; Fisman D.; Schooley R. Ten Scientific Reasons in Support of Airborne Transmission of SARS-CoV-2. Lancet 2021, 397 (10285), 1603–1605. 10.1016/S0140-6736(21)00869-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarpia J. L.; Rivera D. N.; Herrera V. L.; Morwitzer M. J.; Creager H. M.; Santarpia G. W.; Crown K. K.; Brett-Major D. M.; Schnaubelt E. R.; Broadhurst M. J.; Lawler J. V.; Reid S. P.; Lowe J. J. Aerosol and Surface Contamination of SARS-CoV-2 Observed in Quarantine and Isolation Care. Sci. Rep. 2020, 10 (1), 12732. 10.1038/s41598-020-69286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Ning Z.; Chen Y.; Guo M.; Liu Y.; Gali N. K.; Sun L.; Duan Y.; Cai J.; Westerdahl D.; Liu X.; Xu K.; Ho K.; Kan H.; Fu Q.; Lan K. Aerodynamic Analysis of SARS-CoV-2 in Two Wuhan Hospitals. Nature 2020, 582 (7813), 557–560. 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- Chu D. K.; Akl E. A.; Duda S.; Solo K.; Yaacoub S.; Schünemann H. J.; Chu D. K.; Akl E. A.; El-harakeh A.; Bognanni A.; Lotfi T.; Loeb M.; Hajizadeh A.; Bak A.; Izcovich A.; Cuello-Garcia C. A.; Chen C.; Harris D. J.; Borowiack E.; Chamseddine F.; Schünemann F.; Morgano G. P.; Muti Schünemann G. E. U.; Chen G.; Zhao H.; Neumann I.; Chan J.; Khabsa J.; Hneiny L.; Harrison L.; Smith M.; Rizk N.; Giorgi Rossi P.; AbiHanna P.; El-khoury R.; Stalteri R.; Baldeh T.; Piggott T.; Zhang Y.; Saad Z.; Khamis A.; Reinap M.; Duda S.; Solo K.; Yaacoub S.; Schünemann H. J. Physical Distancing, Face Masks, and Eye Protection to Prevent Person-to-Person Transmission of SARS-CoV-2 and COVID-19: A Systematic Review and Meta-Analysis. Lancet 2020, 395 (10242), 1973–1987. 10.1016/S0140-6736(20)31142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L.; Liu D.; Zhang H.; Li Z.; Zhen R.; Zhang X.; Xie H.; Song W.; Liu J.; Huang Q.; Liu J.; Yang X.; Chen Z.; Mao C. Air and Surface Contamination in Non-Health Care Settings among 641 Environmental Specimens of 39 COVID-19 Cases. PLoS Negl. Trop. Dis. 2020, 14 (10), e0008570. 10.1371/journal.pntd.0008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A. W. H.; Chu J. T. S.; Perera M. R. A.; Hui K. P. Y.; Yen H.-L.; Chan M. C. W.; Peiris M.; Poon L. L. M. Stability of SARS-CoV-2 in Different Environmental Conditions. Lancet Microbe 2020, 1 (1), e10. 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N.; Bushmaker T.; Morris D. H.; Holbrook M. G.; Gamble A.; Williamson B. N.; Tamin A.; Harcourt J. L.; Thornburg N. J.; Gerber S. I.; Lloyd-Smith J. O.; de Wit E.; Munster V. J. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382 (16), 1564–1567. 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddell S.; Goldie S.; Hill A.; Eagles D.; Drew T. W. The Effect of Temperature on Persistence of SARS-CoV-2 on Common Surfaces. Virol. J. 2020, 17 (1), 145. 10.1186/s12985-020-01418-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.; Otter J. A.; Price J. R.; Cimpeanu C.; Meno Garcia D.; Kinross J.; Boshier P. R.; Mason S.; Bolt F.; Holmes A. H.; Barclay W. S. Investigating Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Surface and Air Contamination in an Acute Healthcare Setting During the Peak of the Coronavirus Disease 2019 (COVID-19) Pandemic in London. Clin. Infect. Dis. 2021, 73 (7), e1870–e1877. 10.1093/cid/ciaa905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia P. Y.; Coleman K. K.; Tan Y. K.; Ong S. W. X.; Gum M.; Lau S. K.; Lim X. F.; Lim A. S.; Sutjipto S.; Lee P. H.; Son T. T.; Young B. E.; Milton D. K.; Gray G. C.; Schuster S.; Barkham T.; De P. P.; Vasoo S.; Chan M.; Ang B. S. P.; Tan B. H.; Leo Y.-S.; Ng O.-T.; Wong M. S. Y.; Marimuthu K. Detection of Air and Surface Contamination by SARS-CoV-2 in Hospital Rooms of Infected Patients. Nat. Commun. 2020, 11 (1), 2800. 10.1038/s41467-020-16670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaneri M.; Seminari E.; Novati S.; Asperges E.; Biscarini S.; Piralla A.; Percivalle E.; Cassaniti I.; Baldanti F.; Bruno R.; Mondelli M. U.; Bruno R.; Mondelli M. U.; Brunetti E.; Di Matteo A.; Seminari E.; Maiocchi L.; Zuccaro V.; Pagnucco L.; Ludovisi S.; Lissandrin R.; Parisi A.; Sacchi P.; Patruno S. F. A.; Michelone G.; Gulminetti R.; Zanaboni D.; Novati S.; Maserati R.; Orsolini P.; Vecchia M. Severe Acute Respiratory Syndrome Coronavirus 2 RNA Contamination of Inanimate Surfaces and Virus Viability in a Health Care Emergency Unit. Clin. Microbiol. Infect. 2020, 26 (8), 1094.e1–1094.e5. 10.1016/j.cmi.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahão J. S.; Sacchetto L.; Rezende I. M.; Rodrigues R. A. L.; Crispim A. P. C.; Moura C.; Mendonça D. C.; Reis E.; Souza F.; Oliveira G. F. G.; Domingos I.; de Miranda Boratto P. V.; Silva P. H. B.; Queiroz V. F.; Machado T. B.; Andrade L. A. F.; Lourenço K. L.; Silva T.; Oliveira G. P.; de Souza Alves V.; Alves P. A.; Kroon E. G.; de Souza Trindade G.; Drumond B. P. Detection of SARS-CoV-2 RNA on Public Surfaces in a Densely Populated Urban Area of Brazil: A Potential Tool for Monitoring the Circulation of Infected Patients. Sci. Total Environ. 2021, 766, 142645. 10.1016/j.scitotenv.2020.142645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Carlo P.; Chiacchiaretta P.; Sinjari B.; Aruffo E.; Stuppia L.; De Laurenzi V.; Di Tomo P.; Pelusi L.; Potenza F.; Veronese A.; Vecchiet J.; Falasca K.; Ucciferri C. Air and Surface Measurements of SARS-CoV-2 inside a Bus during Normal Operation. PLoS One 2020, 15 (11), e0235943. 10.1371/journal.pone.0235943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S. J. R.; Nascimento J. C. F.; Santos Reis W. P. M.; Silva C. T. A.; Silva P. G.; Mendes R. P. G.; Mendonça A. A.; Santos B. N. R.; Magalhães J. J. F.; Kohl A.; Pena L. Widespread Contamination of SARS-CoV-2 on Highly Touched Surfaces in Brazil during the Second Wave of the COVID-19 Pandemic. Environ. Microbiol. 2021, 23 (12), 7382–7395. 10.1111/1462-2920.15855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva S. J. R. Widespread Contamination of SARS-CoV-2 on Highly Touched Surfaces in Brazil During the Second Wave of the COVID-19 Pandemic. medRxiv 2021, 10.1111/1462-2920.15855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S. W. X.; Tan Y. K.; Chia P. Y.; Lee T. H.; Ng O. T.; Wong M. S. Y.; Marimuthu K. Air, Surface Environmental, and Personal Protective Equipment Contamination by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) From a Symptomatic Patient. JAMA 2020, 323 (16), 1610. 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Xu Y.; Gao R.; Lu R.; Han K.; Wu G.; Tan W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323 (18), 1843–1844. 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu M.; Sylverken A. A.; El-Duah P.; Ayisi-Boateng N. K.; Yeboah R.; Adu E.; Asamoah J.; Frimpong M.; Senyo J.; Acheampong G.; Mutocheluh M.; Amuasi J.; Owusu-Dabo E.; Adu-Sarkodie Y.; Phillips R. O. Low Risk of SARS-CoV-2 in Blood Transfusion. PLoS One 2021, 16 (4), e0249069. 10.1371/journal.pone.0249069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capobianco G.; Saderi L.; Aliberti S.; Mondoni M.; Piana A.; Dessole F.; Dessole M.; Cherchi P. L.; Dessole S.; Sotgiu G. COVID-19 in Pregnant Women: A Systematic Review and Meta-Analysis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020, 252, 543–558. 10.1016/j.ejogrb.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellos I.; Pandita A.; Panza R. Maternal and Perinatal Outcomes in Pregnant Women Infected by SARS-CoV-2: A Meta-Analysis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2021, 256, 194–204. 10.1016/j.ejogrb.2020.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero-Castillero A.; Beam K. S.; Bernardini L. B.; Ramos E. G. C.; Davenport P. E.; Duncan A. R.; Fraiman Y. S.; Frazer L. C.; Healy H.; Herzberg E. M.; Keyes M. L.; Leeman K. T.; Leone K.; Levin J. C.; Lin M.; Raju R. M.; Sullivan A. COVID-19: Neonatal-Perinatal Perspectives. J. Perinatol. 2021, 41 (5), 940–951. 10.1038/s41372-020-00874-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashraath P.; Wong J. L. J.; Lim M. X. K.; Lim L. M.; Li S.; Biswas A.; Choolani M.; Mattar C.; Su L. L. Coronavirus Disease 2019 (COVID-19) Pandemic and Pregnancy. Am. J. Obstet. Gynecol. 2020, 222 (6), 521–531. 10.1016/j.ajog.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.; Guo J.; Wang C.; Luo F.; Yu X.; Zhang W.; Li J.; Zhao D.; Xu D.; Gong Q.; Liao J.; Yang H.; Hou W.; Zhang Y. Clinical Characteristics and Intrauterine Vertical Transmission Potential of COVID-19 Infection in Nine Pregnant Women: A Retrospective Review of Medical Records. Lancet 2020, 395 (10226), 809–815. 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L.; Xia S.; Yuan W.; Yan K.; Xiao F.; Shao J.; Zhou W. Neonatal Early-Onset Infection With SARS-CoV-2 in 33 Neonates Born to Mothers With COVID-19 in Wuhan, China. JAMA Pediatr. 2020, 174 (7), 722. 10.1001/jamapediatrics.2020.0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer S. A.; Grantz K. H.; Bi Q.; Jones F. K.; Zheng Q.; Meredith H. R.; Azman A. S.; Reich N. G.; Lessler J. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Ann. Int. Med. 2020, 172 (9), 577–582. 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L.; Whitaker M.; O’Halloran A.; Kambhampati A.; Chai S. J.; Reingold A.; Armistead I.; Kawasaki B.; Meek J.; Yousey-Hindes K.; Anderson E. J.; Openo K. P.; Weigel A.; Ryan P.; Monroe M. L.; Fox K.; Kim S.; Lynfield R.; Bye E.; Shrum Davis S.; Smelser C.; Barney G.; Spina N. L.; Bennett N. M.; Felsen C. B.; Billing L. M.; Shiltz J.; Sutton M.; West N.; Talbot H. K.; Schaffner W.; Risk I.; Price A.; Brammer L.; Fry A. M.; Hall A. J.; Langley G. E.; Garg S.; Coates A.; Daily Kirley P.; Libby T.; Roland J.; Alden N.; Herlihy R.; McLafferty S.; Clogher P.; Kayalioglu H.; Maslar A.; Misiorski A.; Niccolai L.; Olson D.; Parisi C.; Fawcett E.; Gretzinger S.; Lengacher K.; Williams J.; Blythe D.; Brooks A.; Park R.; Wilson M.; Como-Sabetti K.; Danila R.; Cline C.; Angeles K.; Eisenberg N.; Flores K.; Habrun C.; Hancock E.; Khanlian S.; Novi M.; Phipps E.; Salazar-Sanchez Y.; Dufort E.; Muse A.; Bushey S.; Gaitan M.; Kurtz R.; Owusu-Dommey A.; Snyder L.; Michaelis K.; Seeley K.; Markus T.; Chatelain R.; George A.; Hill M.; McCullough L.; Spencer M.; Swain A.; McCaffrey K.; Holstein R.; Meador S.; Wortham J. Hospitalization Rates and Characteristics of Children Aged <18 Years Hospitalized with Laboratory-Confirmed COVID-19 — COVID-NET, 14 States, March 1-July 25, 2020. Morb. Mortal. Wkly. Rep. 2020, 69 (32), 1081–1088. 10.15585/mmwr.mm6932e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordi L.; Nicastri E.; Scorzolini L.; Di Caro A.; Capobianchi M. R.; Castilletti C.; Lalle E. Differential Diagnosis of Illness in Patients under Investigation for the Novel Coronavirus (SARS-CoV-2), Italy, February 2020. Eurosurveillance 2020, 25 (8), 2000170 10.2807/1560-7917.ES.2020.25.8.2000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhães J. J. F. de; Mendes R. P. G.; Silva C. T. A. da; Silva S. J. R. da; Guarines K. M.; Pena L. Epidemiological and Clinical Characteristics of the First 557 Successive Patients with COVID-19 in Pernambuco State, Northeast Brazil. Travel Med. Infect. Dis. 2020, 38, 101884. 10.1016/j.tmaid.2020.101884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva S. J. R.; Silva C. T. A. da; Guarines K. M.; Mendes R. P. G.; Pardee K.; Kohl A.; Pena L. Clinical and Laboratory Diagnosis of SARS-CoV-2, the Virus Causing COVID-19. ACS Infect. Dis. 2020, 6 (9), 2319–2336. 10.1021/acsinfecdis.0c00274. [DOI] [PubMed] [Google Scholar]
- Lechien J. R.; Chiesa-Estomba C. M.; De Siati D. R.; Horoi M.; Le Bon S. D.; Rodriguez A.; Dequanter D.; Blecic S.; El Afia F.; Distinguin L.; Chekkoury-Idrissi Y.; Hans S.; Delgado I. L.; Calvo-Henriquez C.; Lavigne P.; Falanga C.; Barillari M. R.; Cammaroto G.; Khalife M.; Leich P.; Souchay C.; Rossi C.; Journe F.; Hsieh J.; Edjlali M.; Carlier R.; Ris L.; Lovato A.; De Filippis C.; Coppee F.; Fakhry N.; Ayad T.; Saussez S. Olfactory and Gustatory Dysfunctions as a Clinical Presentation of Mild-to-Moderate Forms of the Coronavirus Disease (COVID-19): A Multicenter European Study. Eur. Arch. Oto-Rhino-Laryngol. 2020, 277 (8), 2251–2261. 10.1007/s00405-020-05965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa K. V. T. da; Carnaúba A. T. L.; Rocha K. W.; Andrade K. C. L. de; Ferreira S. M. S.; Menezes P. de L. Olfactory and Taste Disorders in COVID-19: A Systematic Review. Braz. J. Otorhinolaryngol. 2020, 86 (6), 781–792. 10.1016/j.bjorl.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariyanto T. I.; Rizki N. A.; Kurniawan A. Anosmia/Hyposmia Is a Good Predictor of Coronavirus Disease 2019 (COVID-19) Infection: A Meta-Analysis. Int. Arch. Otorhinolaryngol. 2021, 25 (01), e170–e174. 10.1055/s-0040-1719120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammaro A.; Adebanjo G. A. R.; Parisella F. R.; Pezzuto A.; Rello J. Cutaneous Manifestations in COVID-19: The Experiences of Barcelona and Rome. J. Eur. Acad. Dermatol. Venereol. 2020, 34 (7), e306–e307. 10.1111/jdv.16530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Santas M.; Diaz-Guimaraens B.; Garcia Abellas P.; Moreno-Garcia del Real C.; Burgos-Blasco P.; Suarez-Valle A. Cutaneous Small-vessel Vasculitis Associated with Novel 2019 Coronavirus SARS-CoV-2 Infection (COVID-19). J. Eur. Acad. Dermatol. Venereol. 2020, 34 (10), e536–e537. 10.1111/jdv.16663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recalcati S. Cutaneous Manifestations in COVID-19: A First Perspective. J. Eur. Acad. Dermatol. Venereol. 2020, 34 (5), e212–e213. 10.1111/jdv.16387. [DOI] [PubMed] [Google Scholar]
- Elmas Ö. F.; Demirbaş A.; Özyurt K.; Atasoy M.; Türsen Ü. Cutaneous Manifestations of COVID-19: A Review of the Published Literature. Dermatol. Ther. 2020, 33 (4), e13696 10.1111/dth.13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga W. J.; Rhodes A.; Cheng A. C.; Peacock S. J.; Prescott H. C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19). JAMA 2020, 324 (8), 782. 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- Nalbandian A.; Sehgal K.; Gupta A.; Madhavan M. V.; McGroder C.; Stevens J. S.; Cook J. R.; Nordvig A. S.; Shalev D.; Sehrawat T. S.; Ahluwalia N.; Bikdeli B.; Dietz D.; Der-Nigoghossian C.; Liyanage-Don N.; Rosner G. F.; Bernstein E. J.; Mohan S.; Beckley A. A.; Seres D. S.; Choueiri T. K.; Uriel N.; Ausiello J. C.; Accili D.; Freedberg D. E.; Baldwin M.; Schwartz A.; Brodie D.; Garcia C. K.; Elkind M. S. V.; Connors J. M.; Bilezikian J. P.; Landry D. W.; Wan E. Y. Post-Acute COVID-19 Syndrome. Nat. Med. 2021, 27 (4), 601–615. 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S. D.; Talwar A.; Lee J. T. A Proposed Framework and Timeline of the Spectrum of Disease Due to SARS-CoV-2 Infection. JAMA 2020, 324 (22), 2251. 10.1001/jama.2020.22717. [DOI] [PubMed] [Google Scholar]
- Greenhalgh T.; Knight M.; A’Court C.; Buxton M.; Husain L. Management of Post-Acute Covid-19 in Primary Care. BMJ. 2020, m3026. 10.1136/bmj.m3026. [DOI] [PubMed] [Google Scholar]
- Shah W.; Hillman T.; Playford E. D.; Hishmeh L. Managing the Long Term Effects of Covid-19: Summary of NICE, SIGN, and RCGP Rapid Guideline. BMJ. 2021, n136. 10.1136/bmj.n136. [DOI] [PubMed] [Google Scholar]
- Moreno-Pérez O.; Merino E.; Leon-Ramirez J.-M.; Andres M.; Ramos J. M.; Arenas-Jiménez J.; Asensio S.; Sanchez R.; Ruiz-Torregrosa P.; Galan I.; Scholz A.; Amo A.; González-delaAleja P.; Boix V.; Gil J. Post-Acute COVID-19 Syndrome. Incidence and Risk Factors: A Mediterranean Cohort Study. J. Infect. 2021, 82 (3), 378–383. 10.1016/j.jinf.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du R.-H.; Liang L.-R.; Yang C.-Q.; Wang W.; Cao T.-Z.; Li M.; Guo G.-Y.; Du J.; Zheng C.-L.; Zhu Q.; Hu M.; Li X.-Y.; Peng P.; Shi H.-Z. Predictors of Mortality for Patients with COVID-19 Pneumonia Caused by SARS-CoV-2: A Prospective Cohort Study. Eur. Respir. J. 2020, 55 (5), 2000524. 10.1183/13993003.00524-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W.; Guan W.; Chen R.; Wang W.; Li J.; Xu K.; Li C.; Ai Q.; Lu W.; Liang H.; Li S.; He J. Cancer Patients in SARS-CoV-2 Infection: A Nationwide Analysis in China. Lancet Oncol. 2020, 21 (3), 335–337. 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg S.; Kim L.; Whitaker M.; O’Halloran A.; Cummings C.; Holstein R.; Prill M.; Chai S. J.; Kirley P. D.; Alden N. B.; Kawasaki B.; Yousey-Hindes K.; Niccolai L.; Anderson E. J.; Openo K. P.; Weigel A.; Monroe M. L.; Ryan P.; Henderson J.; Kim S.; Como-Sabetti K.; Lynfield R.; Sosin D.; Torres S.; Muse A.; Bennett N. M.; Billing L.; Sutton M.; West N.; Schaffner W.; Talbot H. K.; Aquino C.; George A.; Budd A.; Brammer L.; Langley G.; Hall A. J.; Fry A. Hospitalization Rates and Characteristics of Patients Hospitalized with Laboratory-Confirmed Coronavirus Disease 2019 — COVID-NET, 14 States, March 1-30, 2020. Morb. Mortal. Wkly. Rep. 2020, 69 (15), 458–464. 10.15585/mmwr.mm6915e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S.; Hirsch J. S.; Narasimhan M.; Crawford J. M.; McGinn T.; Davidson K. W.; Barnaby D. P.; Becker L. B.; Chelico J. D.; Cohen S. L.; Cookingham J.; Coppa K.; Diefenbach M. A.; Dominello A. J.; Duer-Hefele J.; Falzon L.; Gitlin J.; Hajizadeh N.; Harvin T. G.; Hirschwerk D. A.; Kim E. J.; Kozel Z. M.; Marrast L. M.; Mogavero J. N.; Osorio G. A.; Qiu M.; Zanos T. P. Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA 2020, 323 (20), 2052. 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasselli G.; Zangrillo A.; Zanella A.; Antonelli M.; Cabrini L.; Castelli A.; Cereda D.; Coluccello A.; Foti G.; Fumagalli R.; Iotti G.; Latronico N.; Lorini L.; Merler S.; Natalini G.; Piatti A.; Ranieri M. V.; Scandroglio A. M.; Storti E.; Cecconi M.; Pesenti A.; Agosteo E.; Alaimo V.; Albano G.; Albertin A.; Alborghetti A.; Aldegheri G.; Antonini B.; Barbara E.; Belgiorno N.; Belliato M.; Benini A.; Beretta E.; Bianciardi L.; Bonazzi S.; Borelli M.; Boselli E.; Bronzini N.; Capra C.; Carnevale L.; Casella G.; Castelli G.; Catena E.; Cattaneo S.; Chiumello D.; Cirri S.; Citerio G.; Colombo S.; Coppini D.; Corona A.; Cortellazzi P.; Costantini E.; Covello R. D.; De Filippi G.; Dei Poli M.; Della Mura F.; Evasi G.; Fernandez-Olmos R.; Forastieri Molinari A.; Galletti M.; Gallioli G.; Gemma M.; Gnesin P.; Grazioli L.; Greco S.; Gritti P.; Grosso P.; Guatteri L.; Guzzon D.; Harizay F.; Keim R.; Landoni G.; Langer T.; Lombardo A.; Malara A.; Malpetti E.; Marino F.; Marino G.; Mazzoni M. G.; Merli G.; Micucci A.; Mojoli F.; Muttini S.; Nailescu A.; Panigada M.; Perazzo P.; Perego G. B.; Petrucci N.; Pezzi A.; Protti A.; Radrizzani D.; Raimondi M.; Ranucci M.; Rasulo F.; Riccio M.; Rona R.; Roscitano C.; Ruggeri P.; Sala A.; Sala G.; Salvi L.; Sebastiano P.; Severgnini P.; Sforzini I.; Sigurtà F. D.; Subert M.; Tagliabue P.; Troiano C.; Valsecchi R.; Viola U.; Vitale G.; Zambon M.; Zoia E. Baseline Characteristics and Outcomes of 1591 Patients Infected With SARS-CoV-2 Admitted to ICUs of the Lombardy Region, Italy. JAMA 2020, 323 (16), 1574. 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrilli C. M.; Jones S. A.; Yang J.; Rajagopalan H.; O’Donnell L.; Chernyak Y.; Tobin K. A.; Cerfolio R. J.; Francois F.; Horwitz L. I. Factors Associated with Hospital Admission and Critical Illness among 5279 People with Coronavirus Disease 2019 in New York City: Prospective Cohort Study. BMJ. 2020, m1966. 10.1136/bmj.m1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaythorpe K. A. M.; Bhatia S.; Mangal T.; Unwin H. J. T.; Imai N.; Cuomo-Dannenburg G.; Walters C. E.; Jauneikaite E.; Bayley H.; Kont M. D.; Mousa A.; Whittles L. K.; Riley S.; Ferguson N. M. Children’s Role in the COVID-19 Pandemic: A Systematic Review of Early Surveillance Data on Susceptibility, Severity, and Transmissibility. Sci. Rep. 2021, 11 (1), 13903. 10.1038/s41598-021-92500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götzinger F.; Santiago-García B.; Noguera-Julián A.; Lanaspa M.; Lancella L.; Calò Carducci F. I.; Gabrovska N.; Velizarova S.; Prunk P.; Osterman V.; Krivec U.; Lo Vecchio A.; Shingadia D.; Soriano-Arandes A.; Melendo S.; Lanari M.; Pierantoni L.; Wagner N.; L’Huillier A. G.; Heininger U.; Ritz N.; Bandi S.; Krajcar N.; Roglić S.; Santos M.; Christiaens C.; Creuven M.; Buonsenso D.; Welch S. B.; Bogyi M.; Brinkmann F.; Tebruegge M.; Pfefferle J.; Zacharasiewicz A.; Berger A.; Berger R.; Strenger V.; Kohlfürst D. S.; Zschocke A.; Bernar B.; Simma B.; Haberlandt E.; Thir C.; Biebl A.; Vanden Driessche K.; Boiy T.; Van Brusselen D.; Bael A.; Debulpaep S.; Schelstraete P.; Pavic I.; Nygaard U.; Glenthoej J. P.; Heilmann Jensen L.; Lind I.; Tistsenko M.; Uustalu Ü.; Buchtala L.; Thee S.; Kobbe R.; Rau C.; Schwerk N.; Barker M.; Tsolia M.; Eleftheriou I.; Gavin P.; Kozdoba O.; Zsigmond B.; Valentini P.; Ivaškeviciene I.; Ivaškevicius R.; Vilc V.; Schölvinck E.; Rojahn A.; Smyrnaios A.; Klingenberg C.; Carvalho I.; Ribeiro A.; Starshinova A.; Solovic I.; Falcón L.; Neth O.; Minguell L.; Bustillo M.; Gutiérrez-Sánchez A. M.; Guarch Ibáñez B.; Ripoll F.; Soto B.; Kötz K.; Zimmermann P.; Schmid H.; Zucol F.; Niederer A.; Buettcher M.; Cetin B. S.; Bilogortseva O.; Chechenyeva V.; Demirjian A.; Shackley F.; McFetridge L.; Speirs L.; Doherty C.; Jones L.; McMaster P.; Murray C.; Child F.; Beuvink Y.; Makwana N.; Whittaker E.; Williams A.; Fidler K.; Bernatoniene J.; Song R.; Oliver Z.; Riordan A. COVID-19 in Children and Adolescents in Europe: A Multinational, Multicentre Cohort Study. Lancet Child Adolesc. Health 2020, 4 (9), 653–661. 10.1016/S2352-4642(20)30177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J.; Thomas P. G.; Randolph A. G. Immunology of SARS-CoV-2 Infection in Children. Nat. Immunol. 2022, 23 (2), 177–185. 10.1038/s41590-021-01123-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldstein L. R.; Tenforde M. W.; Friedman K. G.; Newhams M.; Rose E. B.; Dapul H.; Soma V. L.; Maddux A. B.; Mourani P. M.; Bowens C.; Maamari M.; Hall M. W.; Riggs B. J.; Giuliano J. S.; Singh A. R.; Li S.; Kong M.; Schuster J. E.; McLaughlin G. E.; Schwartz S. P.; Walker T. C.; Loftis L. L.; Hobbs C. V.; Halasa N. B.; Doymaz S.; Babbitt C. J.; Hume J. R.; Gertz S. J.; Irby K.; Clouser K. N.; Cvijanovich N. Z.; Bradford T. T.; Smith L. S.; Heidemann S. M.; Zackai S. P.; Wellnitz K.; Nofziger R. A.; Horwitz S. M.; Carroll R. W.; Rowan C. M.; Tarquinio K. M.; Mack E. H.; Fitzgerald J. C.; Coates B. M.; Jackson A. M.; Young C. C.; Son M. B. F.; Patel M. M.; Newburger J. W.; Randolph A. G. Characteristics and Outcomes of US Children and Adolescents With Multisystem Inflammatory Syndrome in Children (MIS-C) Compared With Severe Acute COVID-19. JAMA 2021, 325 (11), 1074. 10.1001/jama.2021.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consiglio C. R.; Cotugno N.; Sardh F.; Pou C.; Amodio D.; Rodriguez L.; Tan Z.; Zicari S.; Ruggiero A.; Pascucci G. R.; Santilli V.; Campbell T.; Bryceson Y.; Eriksson D.; Wang J.; Marchesi A.; Lakshmikanth T.; Campana A.; Villani A.; Rossi P.; Landegren N.; Palma P.; Brodin P. The Immunology of Multisystem Inflammatory Syndrome in Children with COVID-19. Cell 2020, 183 (4), 968–981. 10.1016/j.cell.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoni L.; Mazza A.; Gervasoni A.; Martelli L.; Ruggeri M.; Ciuffreda M.; Bonanomi E.; D’Antiga L. An Outbreak of Severe Kawasaki-like Disease at the Italian Epicentre of the SARS-CoV-2 Epidemic: An Observational Cohort Study. Lancet 2020, 395 (10239), 1771–1778. 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker E.; Bamford A.; Kenny J.; Kaforou M.; Jones C. E.; Shah P.; Ramnarayan P.; Fraisse A.; Miller O.; Davies P.; Kucera F.; Brierley J.; McDougall M.; Carter M.; Tremoulet A.; Shimizu C.; Herberg J.; Burns J. C.; Lyall H.; Levin M. Clinical Characteristics of 58 Children With a Pediatric Inflammatory Multisystem Syndrome Temporally Associated With SARS-CoV-2. JAMA 2020, 324 (3), 259. 10.1001/jama.2020.10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radia T.; Williams N.; Agrawal P.; Harman K.; Weale J.; Cook J.; Gupta A. Multi-System Inflammatory Syndrome in Children & Adolescents (MIS-C): A Systematic Review of Clinical Features and Presentation. Paediatr. Respir. Rev. 2021, 38, 51–57. 10.1016/j.prrv.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A.; Madhavan M. V.; Sehgal K.; Nair N.; Mahajan S.; Sehrawat T. S.; Bikdeli B.; Ahluwalia N.; Ausiello J. C.; Wan E. Y.; Freedberg D. E.; Kirtane A. J.; Parikh S. A.; Maurer M. S.; Nordvig A. S.; Accili D.; Bathon J. M.; Mohan S.; Bauer K. A.; Leon M. B.; Krumholz H. M.; Uriel N.; Mehra M. R.; Elkind M. S. V.; Stone G. W.; Schwartz A.; Ho D. D.; Bilezikian J. P.; Landry D. W. Extrapulmonary Manifestations of COVID-19. Nat. Med. 2020, 26 (7), 1017–1032. 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M. M.; Haagmans B. L. SARS-CoV-2 Pathogenesis. Nat. Rev. Microbiol. 2022, 20 (5), 270–284. 10.1038/s41579-022-00713-0. [DOI] [PubMed] [Google Scholar]
- Hui D. S.; I Azhar E.; Madani T. A.; Ntoumi F.; Kock R.; Dar O.; Ippolito G.; Mchugh T. D.; Memish Z. A.; Drosten C.; Zumla A.; Petersen E. The Continuing 2019-NCoV Epidemic Threat of Novel Coronaviruses to Global Health — The Latest 2019 Novel Coronavirus Outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D.; Wang N.; Corbett K. S.; Goldsmith J. A.; Hsieh C.-L.; Abiona O.; Graham B. S.; McLellan J. S. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science 2020, 367 (6483), 1260–1263. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Liu S.-M.; Yu X.-H.; Tang S.-L.; Tang C.-K. Coronavirus Disease 2019 (COVID-19): Current Status and Future Perspectives. Int. J. Antimicrob. Agents 2020, 55 (5), 105951. 10.1016/j.ijantimicag.2020.105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.-Y.; Li L.; Zhang Y.; Wang X.-S. Expression of the SARS-CoV-2 Cell Receptor Gene ACE2 in a Wide Variety of Human Tissues. Infect. Dis. Poverty 2020, 9 (1), 45. 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X.; Liu Y.; Lei X.; Li P.; Mi D.; Ren L.; Guo L.; Guo R.; Chen T.; Hu J.; Xiang Z.; Mu Z.; Chen X.; Chen J.; Hu K.; Jin Q.; Wang J.; Qian Z. Characterization of Spike Glycoprotein of SARS-CoV-2 on Virus Entry and Its Immune Cross-Reactivity with SARS-CoV. Nat. Commun. 2020, 11 (1), 1620. 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F.; Tang M.; Zheng X.; Liu Y.; Li X.; Shan H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology 2020, 158 (6), 1831–1833. 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totura A. L.; Baric R. S. SARS Coronavirus Pathogenesis: Host Innate Immune Responses and Viral Antagonism of Interferon. Curr. Opin. Virol. 2012, 2 (3), 264–275. 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouayad A. Innate Immune Evasion by SARS-CoV-2: Comparison with SARS-CoV. Rev. Med. Virol. 2020, 30 (6), 1–9. 10.1002/rmv.2135. [DOI] [PubMed] [Google Scholar]
- Cuevas A. M.; Clark J. M.; Potter J. J. Increased TLR/MyD88 Signaling in Patients with Obesity: Is There a Link to COVID-19 Disease Severity?. Int. J. Obes. 2021, 45 (5), 1152–1154. 10.1038/s41366-021-00768-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan A.; Hakeem A. R.; Radhakrishnan S.; Reddy M. S.; Rela M. The Role and Therapeutic Potential of NF-Kappa-B Pathway in Severe COVID-19 Patients. Inflammopharmacology 2021, 29 (1), 91–100. 10.1007/s10787-020-00773-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalinger Z. B.; Elliott R.; Rose K. M.; Weiss S. R. MDA5 Is Critical to Host Defense during Infection with Murine Coronavirus. J. Virol. 2015, 89 (24), 12330–12340. 10.1128/JVI.01470-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.; McGoogan J. M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China. JAMA 2020, 323 (13), 1239. 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Shokri S.; Mahmoudvand S.; Taherkhani R.; Farshadpour F. Modulation of the Immune Response by Middle East Respiratory Syndrome Coronavirus. J. Cell. Physiol. 2019, 234 (3), 2143–2151. 10.1002/jcp.27155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R.; Fehr A. R.; Zheng J.; Wohlford-Lenane C.; Abrahante J. E.; Mack M.; Sompallae R.; McCray P. B.; Meyerholz D. K.; Perlman S. IFN-I Response Timing Relative to Virus Replication Determines MERS Coronavirus Infection Outcomes. J. Clin. Invest. 2019, 129 (9), 3625–3639. 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv L. B.; Donlin L. T. Regulation of Type I Interferon Responses. Nat. Rev. Immunol. 2014, 14 (1), 36–49. 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien T. R.; Thomas D. L.; Jackson S. S.; Prokunina-Olsson L.; Donnelly R. P.; Hartmann R. Weak Induction of Interferon Expression by Severe Acute Respiratory Syndrome Coronavirus 2 Supports Clinical Trials of Interferon-λ to Treat Early Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71 (6), 1410–1412. 10.1093/cid/ciaa453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H.; Shi P.-Y. Antagonism of Type I Interferon by Severe Acute Respiratory Syndrome Coronavirus 2. J. Interface Cytokine Res. 2020, 40 (12), 543–548. 10.1089/jir.2020.0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya D.; Liu G.; Gack M. U. Dysregulation of Type I Interferon Responses in COVID-19. Nat. Rev. Immunol. 2020, 20 (7), 397–398. 10.1038/s41577-020-0346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze J. L.; Aschenbrenner A. C. COVID-19 and the Human Innate Immune System. Cell 2021, 184 (7), 1671–1692. 10.1016/j.cell.2021.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P.; Rosen L. B.; Zhang Q.; Michailidis E.; Hoffmann H.-H.; Zhang Y.; Dorgham K.; Philippot Q.; Rosain J.; Béziat V.; et al. Autoantibodies against Type I IFNs in Patients with Life-Threatening COVID-19. Science 2020, 370 (6515), eabd4585 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J.; Mommert M.; Mouton W.; Pizzorno A.; Brengel-Pesce K.; Mezidi M.; Villard M.; Lina B.; Richard J.-C.; Fassier J.-B.; Cheynet V.; Padey B.; Duliere V.; Julien T.; Paul S.; Bastard P.; Belot A.; Bal A.; Casanova J.-L.; Rosa-Calatrava M.; Morfin F.; Walzer T.; Trouillet-Assant S. Early Nasal Type I IFN Immunity against SARS-CoV-2 Is Compromised in Patients with Autoantibodies against Type I IFNs. J. Exp. Med. 2021, 218 (10), e20211211 10.1084/jem.20211211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J.; Yatim N.; Barnabei L.; Corneau A.; Boussier J.; Smith N.; Péré H.; Charbit B.; Bondet V.; Chenevier-Gobeaux C.; Breillat P.; Carlier N.; Gauzit R.; Morbieu C.; Pène F.; Marin N.; Roche N.; Szwebel T.-A.; Merkling S. H.; Treluyer J.-M.; Veyer D.; Mouthon L.; Blanc C.; Tharaux P.-L.; Rozenberg F.; Fischer A.; Duffy D.; Rieux-Laucat F.; Kernéis S.; Terrier B. Impaired Type I Interferon Activity and Inflammatory Responses in Severe COVID-19 Patients. Science 2020, 369 (6504), 718–724. 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C. G. K.; Miao V. N.; Owings A. H.; Navia A. W.; Tang Y.; Bromley J. D.; Lotfy P.; Sloan M.; Laird H.; Williams H. B.; George M.; Drake R. S.; Christian T.; Parker A.; Sindel C. B.; Burger M. W.; Pride Y.; Hasan M.; Abraham G. E.; Senitko M.; Robinson T. O.; Shalek A. K.; Glover S. C.; Horwitz B. H.; Ordovas-Montanes J. Impaired Local Intrinsic Immunity to SARS-CoV-2 Infection in Severe COVID-19. Cell 2021, 184 (18), 4713–4733. 10.1016/j.cell.2021.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E.; Clohisey S.; Klaric L.; Bretherick A. D.; Rawlik K.; Pasko D.; Walker S.; Parkinson N.; Fourman M. H.; Russell C. D.; Furniss J.; Richmond A.; Gountouna E.; Wrobel N.; Harrison D.; Wang B.; Wu Y.; Meynert A.; Griffiths F.; Oosthuyzen W.; Kousathanas A.; Moutsianas L.; Yang Z.; Zhai R.; Zheng C.; Grimes G.; Beale R.; Millar J.; Shih B.; Keating S.; Zechner M.; Haley C.; Porteous D. J.; Hayward C.; Yang J.; Knight J.; Summers C.; Shankar-Hari M.; Klenerman P.; Turtle L.; Ho A.; Moore S. C.; Hinds C.; Horby P.; Nichol A.; Maslove D.; Ling L.; McAuley D.; Montgomery H.; Walsh T.; Pereira A. C.; Renieri A.; Shen X.; Ponting C. P.; Fawkes A.; Tenesa A.; Caulfield M.; Scott R.; Rowan K.; Murphy L.; Openshaw P. J. M.; Semple M. G.; Law A.; Vitart V.; Wilson J. F.; Baillie J. K. Genetic Mechanisms of Critical Illness in COVID-19. Nature 2021, 591 (7848), 92–98. 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Bastard P.; Liu Z.; Le Pen J.; Moncada-Velez M.; Chen J.; Ogishi M.; Sabli I. K. D.; Hodeib S.; Korol C.; et al. Inborn Errors of Type I IFN Immunity in Patients with Life-Threatening COVID-19. Science 2020, 370 (6515), eabd4570 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelow B.; Song E.; Mao T.; Lu P.; Meir A.; Liu F.; Alfajaro M. M.; Wei J.; Dong H.; Homer R. J.; Ring A.; Wilen C. B.; Iwasaki A. Mouse Model of SARS-CoV-2 Reveals Inflammatory Role of Type I Interferon Signaling. J. Exp. Med. 2020, 217 (12), e20201241 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major J.; Crotta S.; Llorian M.; McCabe T. M.; Gad H. H.; Priestnall S. L.; Hartmann R.; Wack A. Type I and III Interferons Disrupt Lung Epithelial Repair during Recovery from Viral Infection. Science 2020, 369 (6504), 712–717. 10.1126/science.abc2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broggi A.; Ghosh S.; Sposito B.; Spreafico R.; Balzarini F.; Lo Cascio A.; Clementi N.; De Santis M.; Mancini N.; Granucci F.; Zanoni I. Type III Interferons Disrupt the Lung Epithelial Barrier upon Viral Recognition. Science 2020, 369 (6504), 706–712. 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. J.; Ashkar A. A. The Dual Nature of Type I and Type II Interferons. Front. Immunol. 2018, 9, 2061 10.3389/fimmu.2018.02061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadotti A. C.; de Castro Deus M.; Telles J. P.; Wind R.; Goes M.; Garcia Charello Ossoski R.; de Padua A. M.; de Noronha L.; Moreno-Amaral A.; Baena C. P.; Tuon F. F. IFN-γ Is an Independent Risk Factor Associated with Mortality in Patients with Moderate and Severe COVID-19 Infection. Virus Res. 2020, 289, 198171. 10.1016/j.virusres.2020.198171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith M. D.; Kinning K. T.; Sullivan K. D.; Araya P.; Smith K. P.; Granrath R. E.; Shaw J. R.; Baxter R.; Jordan K. R.; Russell S.; Dzieciatkowska M.; Reisz J. A.; Gamboni F.; Cendali F.; Ghosh T.; Guo K.; Wilson C. C.; Santiago M. L.; Monte A. A.; Bennett T. D.; Hansen K. C.; Hsieh E. W. Y.; D’Alessandro A.; Espinosa J. M. Specialized Interferon Action in COVID-19. Proc. Natl. Acad. Sci. U. S. A. 2022, 119 (11), e2116730119 10.1073/pnas.2116730119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M.; Martin J. C. Pathological Inflammation in Patients with COVID-19: A Key Role for Monocytes and Macrophages. Nat. Rev. Immunol. 2020, 20 (6), 355–362. 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.; Choi W. J. Overview of COVID-19 Inflammatory Pathogenesis from the Therapeutic Perspective. Arch. Pharm. Res. 2021, 44 (1), 99–116. 10.1007/s12272-020-01301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes B. J.; Adrover J. M.; Baxter-Stoltzfus A.; Borczuk A.; Cools-Lartigue J.; Crawford J. M.; Daßler-Plenker J.; Guerci P.; Huynh C.; Knight J. S.; Loda M.; Looney M. R.; McAllister F.; Rayes R.; Renaud S.; Rousseau S.; Salvatore S.; Schwartz R. E.; Spicer J. D.; Yost C. C.; Weber A.; Zuo Y.; Egeblad M. Targeting Potential Drivers of COVID-19: Neutrophil Extracellular Traps. J. Exp. Med. 2020, 217 (6), e20200652 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.; Guo R.; Lei L.; Liu H.; Wang Y.; Wang Y.; Qian H.; Dai T.; Zhang T.; Lai Y.; Wang J.; Liu Z.; Chen T.; He A.; O’Dwyer M.; Hu J. Frontline Science: COVID-19 Infection Induces Readily Detectable Morphologic and Inflammation-related Phenotypic Changes in Peripheral Blood Monocytes. J. Leukoc. Biol. 2021, 109 (1), 13–22. 10.1002/JLB.4HI0720-470R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarzadeh A.; Chauhan P.; Saha B.; Jafarzadeh S.; Nemati M. Contribution of Monocytes and Macrophages to the Local Tissue Inflammation and Cytokine Storm in COVID-19: Lessons from SARS and MERS, and Potential Therapeutic Interventions. Life Sci. 2020, 257, 118102. 10.1016/j.lfs.2020.118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti A.; Radrizzani D.; Viganò P.; Mazzone A.; Brando B. Decrease of Non-Classical and Intermediate Monocyte Subsets in Severe Acute SARS-CoV -2 Infection. Cytometry, Part A 2020, 97 (9), 887–890. 10.1002/cyto.a.24188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier S.; Zurbuchen Y.; Cervia C.; Adamo S.; Raeber M. E.; de Souza N.; Sivapatham S.; Jacobs A.; Bachli E.; Rudiger A.; Stüssi-Helbling M.; Huber L. C.; Schaer D. J.; Nilsson J.; Boyman O.; Bodenmiller B. A Distinct Innate Immune Signature Marks Progression from Mild to Severe COVID-19. Cell Rep. Med. 2021, 2 (1), 100166. 10.1016/j.xcrm.2020.100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. B.; June C. H. Cytokine Release Syndrome in Severe COVID-19. Science 2020, 368 (6490), 473–474. 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- Zhang C.; Wu Z.; Li J.-W.; Zhao H.; Wang G.-Q. Cytokine Release Syndrome in Severe COVID-19: Interleukin-6 Receptor Antagonist Tocilizumab May Be the Key to Reduce Mortality. Int. J. Antimicrob. Agents 2020, 55 (5), 105954. 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machhi J.; Herskovitz J.; Senan A. M.; Dutta D.; Nath B.; Oleynikov M. D.; Blomberg W. R.; Meigs D. D.; Hasan M.; Patel M.; Kline P.; Chang R. C.-C.; Chang L.; Gendelman H. E.; Kevadiya B. D. The Natural History, Pathobiology, and Clinical Manifestations of SARS-CoV-2 Infections. J. Neuroimmune Pharmacol. 2020, 15 (3), 359–386. 10.1007/s11481-020-09944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajgenbaum D. C.; June C. H. Cytokine Storm. N. Engl. J. Med. 2020, 383 (23), 2255–2273. 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P.; McAuley D. F.; Brown M.; Sanchez E.; Tattersall R. S.; Manson J. J. COVID-19: Consider Cytokine Storm Syndromes and Immunosuppression. Lancet 2020, 395 (10229), 1033–1034. 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimabukuro-Vornhagen A.; Gödel P.; Subklewe M.; Stemmler H. J.; Schlößer H. A.; Schlaak M.; Kochanek M.; Böll B.; von Bergwelt-Baildon M. S. Cytokine Release Syndrome. J. Immunother. Cancer 2018, 6 (1), 56. 10.1186/s40425-018-0343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B.; Huang S.; Yin L. The Cytokine Storm and COVID-19. J. Med. Virol. 2021, 93 (1), 250–256. 10.1002/jmv.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maemura T.; Kuroda M.; Armbrust T.; Yamayoshi S.; Halfmann P. J.; Kawaoka Y. Antibody-Dependent Enhancement of SARS-CoV-2 Infection Is Mediated by the IgG Receptors FcγRIIA and FcγRIIIA but Does Not Contribute to Aberrant Cytokine Production by Macrophages. mBio 2021, 12 (5), e01987-21 10.1128/mBio.01987-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefik E.; Qu R.; Junqueira C.; Kaffe E.; Mirza H.; Zhao J.; Brewer J. R.; Han A.; Steach H. R.; Israelow B.; Blackburn H. N.; Velazquez S. E.; Chen Y. G.; Halene S.; Iwasaki A.; Meffre E.; Nussenzweig M.; Lieberman J.; Wilen C. B.; Kluger Y.; Flavell R. A. Inflammasome Activation in Infected Macrophages Drives COVID-19 Pathology. Nature 2022, 606, 585–593. 10.1038/s41586-022-04802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges L.; Pithon-Curi T. C.; Curi R.; Hatanaka E. COVID-19 and Neutrophils: The Relationship between Hyperinflammation and Neutrophil Extracellular Traps. Mediators Inflamm. 2020, 2020, 1–7. 10.1155/2020/8829674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcante-Silva L. H. A.; Carvalho D. C. M.; Lima É. d. A.; Galvão J. G. F. M.; da Silva J. S. d. F.; de Sales-Neto J. M.; Rodrigues-Mascarenhas S. Neutrophils and COVID-19: The Road so Far. Int. Immunopharmacol. 2021, 90, 107233. 10.1016/j.intimp.2020.107233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforge M.; Elbim C.; Frère C.; Hémadi M.; Massaad C.; Nuss P.; Benoliel J.-J.; Becker C. Tissue Damage from Neutrophil-Induced Oxidative Stress in COVID-19. Nat. Rev. Immunol. 2020, 20 (9), 515–516. 10.1038/s41577-020-0407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönrich G.; Raftery M. J.; Samstag Y. Devilishly Radical NETwork in COVID-19: Oxidative Stress, Neutrophil Extracellular Traps (NETs), and T Cell Suppression. Adv. Biol. Regul. 2020, 77, 100741. 10.1016/j.jbior.2020.100741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Li Q.; Yin Y.; Zhang Y.; Cao Y.; Lin X.; Huang L.; Hoffmann D.; Lu M.; Qiu Y. Excessive Neutrophils and Neutrophil Extracellular Traps in COVID-19. Front. Immunol. 2020, 11, 2063 10.3389/fimmu.2020.02063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veras F. P.; Pontelli M. C.; Silva C. M.; Toller-Kawahisa J. E.; de Lima M.; Nascimento D. C.; Schneider A. H.; Caetité D.; Tavares L. A.; Paiva I. M.; Rosales R.; Colón D.; Martins R.; Castro I. A.; Almeida G. M.; Lopes M. I. F.; Benatti M. N.; Bonjorno L. P.; Giannini M. C.; Luppino-Assad R.; Almeida S. L.; Vilar F.; Santana R.; Bollela V. R.; Auxiliadora-Martins M.; Borges M.; Miranda C. H.; Pazin-Filho A.; da Silva L. L.; Cunha L. D.; Zamboni D. S.; Dal-Pizzol F.; Leiria L. O.; Siyuan L.; Batah S.; Fabro A.; Mauad T.; Dolhnikoff M.; Duarte-Neto A.; Saldiva P.; Cunha T. M.; Alves-Filho J. C.; Arruda E.; Louzada-Junior P.; Oliveira R. D.; Cunha F. Q. SARS-CoV-2-Triggered Neutrophil Extracellular Traps Mediate COVID-19 Pathology. J. Exp. Med. 2020, 217 (12), e20201129 10.1084/jem.20201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton E. A.; He X.-Y.; Denorme F.; Campbell R. A.; Ng D.; Salvatore S. P.; Mostyka M.; Baxter-Stoltzfus A.; Borczuk A. C.; Loda M.; Cody M. J.; Manne B. K.; Portier I.; Harris E. S.; Petrey A. C.; Beswick E. J.; Caulin A. F.; Iovino A.; Abegglen L. M.; Weyrich A. S.; Rondina M. T.; Egeblad M.; Schiffman J. D.; Yost C. C. Neutrophil Extracellular Traps Contribute to Immunothrombosis in COVID-19 Acute Respiratory Distress Syndrome. Blood 2020, 136 (10), 1169–1179. 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox S. E.; Akmatbekov A.; Harbert J. L.; Li G.; Quincy Brown J.; Vander Heide R. S. Pulmonary and Cardiac Pathology in African American Patients with COVID-19: An Autopsy Series from New Orleans. Lancet Respir. Med. 2020, 8 (7), 681–686. 10.1016/S2213-2600(20)30243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik M.; Kuppalli K.; Kindrachuk J.; Peiris M. Virology, Transmission, and Pathogenesis of SARS-CoV-2. BMJ. 2020, m3862. 10.1136/bmj.m3862. [DOI] [PubMed] [Google Scholar]
- Chu H.; Chan J. F.-W.; Wang Y.; Yuen T. T.-T.; Chai Y.; Hou Y.; Shuai H.; Yang D.; Hu B.; Huang X.; Zhang X.; Cai J.-P.; Zhou J.; Yuan S.; Kok K.-H.; To K. K.-W.; Chan I. H.-Y.; Zhang A. J.; Sit K.-Y.; Au W.-K.; Yuen K.-Y. Comparative Replication and Immune Activation Profiles of SARS-CoV-2 and SARS-CoV in Human Lungs: An Ex Vivo Study With Implications for the Pathogenesis of COVID-19. Clin. Infect. Dis. 2020, 71 (6), 1400–1409. 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colling M. E.; Kanthi Y. COVID-19-Associated Coagulopathy: An Exploration of Mechanisms. Vasc. Med. 2020, 25 (5), 471–478. 10.1177/1358863X20932640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangalmurti N. S.; Reilly J. P.; Cines D. B.; Meyer N. J.; Hunter C. A.; Vaughan A. E. COVID-19-Associated Acute Respiratory Distress Syndrome Clarified: A Vascular Endotype?. Am. J. Respir. Crit. Care Med. 2020, 202 (5), 750–753. 10.1164/rccm.202006-2598LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z.; Flammer A. J.; Steiger P.; Haberecker M.; Andermatt R.; Zinkernagel A. S.; Mehra M. R.; Schuepbach R. A.; Ruschitzka F.; Moch H. Endothelial Cell Infection and Endotheliitis in COVID-19. Lancet 2020, 395 (10234), 1417–1418. 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel L.; Chew K. Y.; Stocks C. J.; Yordanov T. E.; Essebier P.; Kulasinghe A.; Monkman J.; dos Santos Miggiolaro A. F. R.; Cooper C.; de Noronha L.; Schroder K.; Lagendijk A. K.; Labzin L. I.; Short K. R.; Gordon E. J. Endothelial Cells Are Not Productively Infected by SARS-CoV-2. Clin. Transl. Immunol. 2021, 10 (10), e1350 10.1002/cti2.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikdeli B.; Madhavan M. V.; Jimenez D.; Chuich T.; Dreyfus I.; Driggin E.; Nigoghossian C. D.; Ageno W.; Madjid M.; Guo Y.; Tang L. V.; Hu Y.; Giri J.; Cushman M.; Quéré I.; Dimakakos E. P.; Gibson C. M.; Lippi G.; Favaloro E. J.; Fareed J.; Caprini J. A.; Tafur A. J.; Burton J. R.; Francese D. P.; Wang E. Y.; Falanga A.; McLintock C.; Hunt B. J.; Spyropoulos A. C.; Barnes G. D.; Eikelboom J. W.; Weinberg I.; Schulman S.; Carrier M.; Piazza G.; Beckman J. A.; Steg P. G.; Stone G. W.; Rosenkranz S.; Goldhaber S. Z.; Parikh S. A.; Monreal M.; Krumholz H. M.; Konstantinides S. V.; Weitz J. I.; Lip G. Y. H. COVID-19 and Thrombotic or Thromboembolic Disease: Implications for Prevention, Antithrombotic Therapy, and Follow-Up. J. Am. Coll. Cardiol. 2020, 75 (23), 2950–2973. 10.1016/j.jacc.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N.; Bai H.; Chen X.; Gong J.; Li D.; Sun Z. Anticoagulant Treatment Is Associated with Decreased Mortality in Severe Coronavirus Disease 2019 Patients with Coagulopathy. J. Thromb. Haemost. 2020, 18 (5), 1094–1099. 10.1111/jth.14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arachchillage D. R. J.; Laffan M. Abnormal Coagulation Parameters Are Associated with Poor Prognosis in Patients with Novel Coronavirus Pneumonia. J. Thromb. Haemost. 2020, 18 (5), 1233–1234. 10.1111/jth.14820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F.; Yu T.; Du R.; Fan G.; Liu Y.; Liu Z.; Xiang J.; Wang Y.; Song B.; Gu X.; Guan L.; Wei Y.; Li H.; Wu X.; Xu J.; Tu S.; Zhang Y.; Chen H.; Cao B. Clinical Course and Risk Factors for Mortality of Adult Inpatients with COVID-19 in Wuhan, China: A Retrospective Cohort Study. Lancet 2020, 395 (10229), 1054–1062. 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.; Chen X.; Cai Y.; Xia J.; Zhou X.; Xu S.; Huang H.; Zhang L.; Zhou X.; Du C.; Zhang Y.; Song J.; Wang S.; Chao Y.; Yang Z.; Xu J.; Zhou X.; Chen D.; Xiong W.; Xu L.; Zhou F.; Jiang J.; Bai C.; Zheng J.; Song Y. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Int. Med. 2020, 180 (7), 934. 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q.; Yang K.; Wang W.; Jiang L.; Song J. Clinical Predictors of Mortality Due to COVID-19 Based on an Analysis of Data of 150 Patients from Wuhan, China. Intensive Care Med. 2020, 46 (5), 846–848. 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl L.; Pink I.; Kühne J. F.; Beushausen K.; Keil J.; Christoph S.; Sauer A.; Boblitz L.; Schmidt J.; David S.; Jäck H.-M.; Roth E.; Cornberg M.; Schulz T. F.; Welte T.; Höper M. M.; Falk C. S. Endothelial Dysfunction Contributes to Severe COVID-19 in Combination with Dysregulated Lymphocyte Responses and Cytokine Networks. Signal Transduct. Target. Ther. 2021, 6 (1), 418. 10.1038/s41392-021-00819-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S.; Qin M.; Shen B.; Cai Y.; Liu T.; Yang F.; Gong W.; Liu X.; Liang J.; Zhao Q.; Huang H.; Yang B.; Huang C. Association of Cardiac Injury With Mortality in Hospitalized Patients With COVID-19 in Wuhan, China. JAMA Cardiol. 2020, 5 (7), 802. 10.1001/jamacardio.2020.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjpe I.; Fuster V.; Lala A.; Russak A. J.; Glicksberg B. S.; Levin M. A.; Charney A. W.; Narula J.; Fayad Z. A.; Bagiella E.; Zhao S.; Nadkarni G. N. Association of Treatment Dose Anticoagulation With In-Hospital Survival Among Hospitalized Patients With COVID-19. J. Am. Coll. Cardiol. 2020, 76 (1), 122–124. 10.1016/j.jacc.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driggin E.; Madhavan M. V.; Bikdeli B.; Chuich T.; Laracy J.; Biondi-Zoccai G.; Brown T. S.; Der Nigoghossian C.; Zidar D. A.; Haythe J.; Brodie D.; Beckman J. A.; Kirtane A. J.; Stone G. W.; Krumholz H. M.; Parikh S. A. Cardiovascular Considerations for Patients, Health Care Workers, and Health Systems During the COVID-19 Pandemic. J. Am. Coll. Cardiol. 2020, 75 (18), 2352–2371. 10.1016/j.jacc.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capotosto L.; Nguyen B. L.; Ciardi M. R.; Mastroianni C.; Vitarelli A. Heart, COVID-19, and Echocardiography. Echocardiography 2020, 37 (9), 1454–1464. 10.1111/echo.14834. [DOI] [PubMed] [Google Scholar]
- Oleszak F.; Maryniak A.; Botti E.; Abrahim C.; Salifu M. O.; Youssef M.; Henglein V. L.; McFarlane S. I. Myocarditis Associated With COVID-19. Am. J. Med. Case Rep. 2020, 8 (12), 498–502. 10.12691/ajmcr-8-12-19.33088905 [DOI] [Google Scholar]
- Zhao B.-C.; Liu W.-F.; Lei S.-H.; Zhou B.-W.; Yang X.; Huang T.-Y.; Deng Q.-W.; Xu M.; Li C.; Liu K.-X. Prevalence and Prognostic Value of Elevated Troponins in Patients Hospitalised for Coronavirus Disease 2019: A Systematic Review and Meta-Analysis. J. Intensive Care 2020, 8 (1), 88. 10.1186/s40560-020-00508-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol E. J. COVID-19 Can Affect the Heart. Science 2020, 370 (6515), 408–409. 10.1126/science.abe2813. [DOI] [PubMed] [Google Scholar]
- Chan L.; Chaudhary K.; Saha A.; Chauhan K.; Vaid A.; Zhao S.; Paranjpe I.; Somani S.; Richter F.; Miotto R.; Lala A.; Kia A.; Timsina P.; Li L.; Freeman R.; Chen R.; Narula J.; Just A. C.; Horowitz C.; Fayad Z.; Cordon-Cardo C.; Schadt E.; Levin M. A.; Reich D. L.; Fuster V.; Murphy B.; He J. C.; Charney A. W.; Böttinger E. P.; Glicksberg B. S.; Coca S. G.; Nadkarni G. N. AKI in Hospitalized Patients with COVID-19. J. Am. Soc. Nephrol. 2021, 32 (1), 151–160. 10.1681/ASN.2020050615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naicker S.; Yang C.-W.; Hwang S.-J.; Liu B.-C.; Chen J.-H.; Jha V. The Novel Coronavirus 2019 Epidemic and Kidneys. Kidney Int. 2020, 97 (5), 824–828. 10.1016/j.kint.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.; Luo R.; Wang K.; Zhang M.; Wang Z.; Dong L.; Li J.; Yao Y.; Ge S.; Xu G. Kidney Disease Is Associated with In-Hospital Death of Patients with COVID-19. Kidney Int. 2020, 97 (5), 829–838. 10.1016/j.kint.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolai L.; Leunig A.; Brambs S.; Kaiser R.; Weinberger T.; Weigand M.; Muenchhoff M.; Hellmuth J. C.; Ledderose S.; Schulz H.; Scherer C.; Rudelius M.; Zoller M.; Höchter D.; Keppler O.; Teupser D.; Zwißler B.; von Bergwelt-Baildon M.; Kääb S.; Massberg S.; Pekayvaz K.; Stark K. Immunothrombotic Dysregulation in COVID-19 Pneumonia Is Associated With Respiratory Failure and Coagulopathy. Circulation 2020, 142 (12), 1176–1189. 10.1161/CIRCULATIONAHA.120.048488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum J. A.; van Till J. W. O.; Mulligan G. Targeting Acute Kidney Injury in COVID-19. Nephrol. Dial. Transplant. 2020, 35 (10), 1652–1662. 10.1093/ndt/gfaa231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malieckal D. A.; Uppal N. N.; Ng J. H.; Jhaveri K. D.; Hirsch J. S.; Abate M.; Andrade H. P.; Barnett R. L.; Bellucci A.; Bhaskaran M. C.; Corona A. G.; Flores Chang B. S.; Finger M.; Fishbane S.; Gitman M.; Halinski C.; Hasan S.; Hazzan A. D.; Hirsch J. S.; Hong S.; Jhaveri K. D.; Khanin Y.; Kuan A.; Madireddy V.; Malieckal D.; Muzib A.; Nair G.; Nair V. V.; Ng J. H.; Parikh R.; Ross D. W.; Sakhiya V.; Sachdeva M.; Schwarz R.; Shah H. H.; Sharma P.; Singhal P. C.; Uppal N. N.; Wanchoo R. Electrolyte Abnormalities in Patients Hospitalized with COVID-19. Clin. Kidney J. 2021, 14 (6), 1704–1707. 10.1093/ckj/sfab060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano G.; Fontana F.; Mori G.; Giaroni F.; Ferrari A.; Giovanella S.; Ligabue G.; Ascione E.; Cazzato S.; Ballestri M.; Di Gaetano M.; Meschiari M.; Menozzi M.; Milic J.; Andrea B.; Franceschini E.; Cuomo G.; Magistroni R.; Mussini C.; Cappelli G.; Guaraldi G.; Mussini C.; Guaraldi G.; Bacca E.; Bedini A.; Borghi V.; Burastero G.; Carli F.; Ciusa G.; Corradi L.; Cuomo G.; Digaetano M.; Dolci G.; Faltoni M.; Fantini R.; Franceschi G.; Franceschini E.; Iadisernia V.; Larné D.; Menozzi M.; Meschiari M.; Milic J.; Orlando G.; Pellegrino F.; Raimondi A.; Rogati C.; Santoro A.; Tonelli R.; Tutone M.; Volpi S.; Yaacoub D.; Cappelli G.; Magistroni R.; Alfano G.; Ferrari A.; Fontana F.; Marco B.; Mori G.; Pulizzi R.; Ascione E.; Leonelli M.; Facchini F.; Damiano F.; Girardis M.; Andreotti A.; Biagioni E.; Bondi F.; Busani S.; Chierego G.; Scotti M.; Serio L.; Cossarizza A.; Bellinazzi C.; Borella R.; De Biasi S.; De Gaetano A.; Fidanza L.; Gibellini L.; Iannone A.; Tartaro D. Lo; Mattioli M.; Nasi M.; Paolini A.; Pinti M. Acid Base Disorders in Patients with COVID-19. Int. Urol. Nephrol. 2022, 54 (2), 405–410. 10.1007/s11255-021-02855-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B.; Wang C.; Wang R.; Feng Z.; Zhang J.; Yang H.; Tan Y.; Wang H.; Wang C.; Liu L.; Liu Y.; Liu Y.; Wang G.; Yuan Z.; Hou X.; Ren L.; Wu Y.; Chen Y. Human Kidney Is a Target for Novel Severe Acute Respiratory Syndrome Coronavirus 2 Infection. Nat. Commun. 2021, 12 (1), 2506. 10.1038/s41467-021-22781-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalejaiye T. D.; Bhattacharya R.; Burt M. A.; Travieso T.; Okafor A. E.; Mou X.; Blasi M.; Musah S. BSG/CD147 and ACE2 receptors facilitate SARS-CoV-2 infection of human iPS cell-derived kidney podocytes. bioRxiv 2021, 10.1101/2021.11.16.468893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand M.; Bell S.; Forni L.; Joannidis M.; Koyner J. L.; Liu K.; Cantaluppi V. Pathophysiology of COVID-19-Associated Acute Kidney Injury. Nat. Rev. Nephrol. 2021, 17 (11), 751–764. 10.1038/s41581-021-00452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S.; Liang T. J. Is SARS-CoV-2 Also an Enteric Pathogen With Potential Fecal-Oral Transmission? A COVID-19 Virological and Clinical Review. Gastroenterology 2020, 159 (1), 53–61. 10.1053/j.gastro.2020.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariyawasam J. C.; Jayarajah U.; Riza R.; Abeysuriya V.; Seneviratne S. L. Gastrointestinal Manifestations in COVID-19. Trans. R. Soc. Trop. Med. Hyg. 2021, 115 (12), 1362–1388. 10.1093/trstmh/trab042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S. H.; Lui R. N.; Sung J. J. Covid-19 and the Digestive System. J. Gastroenterol. Hepatol. 2020, 35 (5), 744–748. 10.1111/jgh.15047. [DOI] [PubMed] [Google Scholar]
- Lamers M. M.; Beumer J.; van der Vaart J.; Knoops K.; Puschhof J.; Breugem T. I.; Ravelli R. B. G.; Paul van Schayck J.; Mykytyn A. Z.; Duimel H. Q.; van Donselaar E.; Riesebosch S.; Kuijpers H. J. H.; Schipper D.; van de Wetering W. J.; de Graaf M.; Koopmans M.; Cuppen E.; Peters P. J.; Haagmans B. L.; Clevers H. SARS-CoV-2 Productively Infects Human Gut Enterocytes. Science 2020, 369 (6499), 50–54. 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R.; Castro M. F. G.; McCune B. T.; Zeng Q.; Rothlauf P. W.; Sonnek N. M.; Liu Z.; Brulois K. F.; Wang X.; Greenberg H. B.; Diamond M. S.; Ciorba M. A.; Whelan S. P. J.; Ding S. TMPRSS2 and TMPRSS4 Promote SARS-CoV-2 Infection of Human Small Intestinal Enterocytes. Sci. Immunol. 2020, 5 (47), eabc3582 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva F. A. F.; de Brito B. B.; Santos M. L. C.; Marques H. S.; da Silva Júnior R. T.; de Carvalho L. S.; Vieira E. S.; Oliveira M. V.; de Melo F. F. COVID-19 Gastrointestinal Manifestations: A Systematic Review. Rev. Soc. Bras. Med. Trop. 2020, 53, e20200714 10.1590/0037-8682-0714-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amico F.; Baumgart D. C.; Danese S.; Peyrin-Biroulet L. Diarrhea During COVID-19 Infection: Pathogenesis, Epidemiology, Prevention, and Management. Clin. Gastroenterol. Hepatol. 2020, 18 (8), 1663–1672. 10.1016/j.cgh.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo C.; Kaushal S.; Yeo D. Enteric Involvement of Coronaviruses: Is Faecal-Oral Transmission of SARS-CoV-2 Possible?. Lancet Gastroenterol. Hepatol. 2020, 5 (4), 335–337. 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saviano A.; Wrensch F.; Ghany M. G.; Baumert T. F. Liver Disease and Coronavirus Disease 2019: From Pathogenesis to Clinical Care. Hepatology 2021, 74 (2), 1088–1100. 10.1002/hep.31684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin M. COVID-19 and the Liver: Overview. Eur. J. Gastroenterol. Hepatol. 2021, 33 (3), 309–311. 10.1097/MEG.0000000000001808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-T.; Lidsky P. V.; Xiao Y.; Lee I. T.; Cheng R.; Nakayama T.; Jiang S.; Demeter J.; Bevacqua R. J.; Chang C. A.; Whitener R. L.; Stalder A. K.; Zhu B.; Chen H.; Goltsev Y.; Tzankov A.; Nayak J. V.; Nolan G. P.; Matter M. S.; Andino R.; Jackson P. K. SARS-CoV-2 Infects Human Pancreatic β Cells and Elicits β Cell Impairment. Cell Metab. 2021, 33 (8), 1565–1576. 10.1016/j.cmet.2021.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J. A.; Groß R.; Conzelmann C.; Krüger J.; Merle U.; Steinhart J.; Weil T.; Koepke L.; Bozzo C. P.; Read C.; Fois G.; Eiseler T.; Gehrmann J.; van Vuuren J.; Wessbecher I. M.; Frick M.; Costa I. G.; Breunig M.; Grüner B.; Peters L.; Schuster M.; Liebau S.; Seufferlein T.; Stenger S.; Stenzinger A.; MacDonald P. E.; Kirchhoff F.; Sparrer K. M. J.; Walther P.; Lickert H.; Barth T. F. E.; Wagner M.; Münch J.; Heller S.; Kleger A. SARS-CoV-2 Infects and Replicates in Cells of the Human Endocrine and Exocrine Pancreas. Nat. Metab. 2021, 3 (2), 149–165. 10.1038/s42255-021-00347-1. [DOI] [PubMed] [Google Scholar]
- van der Heide V.; Jangra S.; Cohen P.; Rathnasinghe R.; Aslam S.; Aydillo T.; Geanon D.; Handler D.; Kelley G.; Lee B.; Rahman A.; Dawson T.; Qi J.; D’Souza D.; Kim-Schulze S.; Panzer J. K.; Caicedo A.; Kusmartseva I.; Posgai A. L.; Atkinson M. A.; Albrecht R. A.; García-Sastre A.; Rosenberg B. R.; Schotsaert M.; Homann D. Limited Extent and Consequences of Pancreatic SARS-CoV-2 Infection. Cell Rep. 2022, 38 (11), 110508. 10.1016/j.celrep.2022.110508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriguchi T.; Harii N.; Goto J.; Harada D.; Sugawara H.; Takamino J.; Ueno M.; Sakata H.; Kondo K.; Myose N.; Nakao A.; Takeda M.; Haro H.; Inoue O.; Suzuki-Inoue K.; Kubokawa K.; Ogihara S.; Sasaki T.; Kinouchi H.; Kojin H.; Ito M.; Onishi H.; Shimizu T.; Sasaki Y.; Enomoto N.; Ishihara H.; Furuya S.; Yamamoto T.; Shimada S. A First Case of Meningitis/Encephalitis Associated with SARS-Coronavirus-2. Int. J. Infect. Dis. 2020, 94, 55–58. 10.1016/j.ijid.2020.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan D.; Du T.; Hong W.; Chen L.; Que H.; Lu S.; Peng X. Neurological Complications and Infection Mechanism of SARS-CoV-2. Signal Transduct. Target. Ther. 2021, 6 (1), 406. 10.1038/s41392-021-00818-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli A.; Pezzati L.; Conti F.; Bernacchia D.; Siano M.; Oreni L.; Rusconi S.; Gervasoni C.; Ridolfo A. L.; Rizzardini G.; Antinori S.; Galli M. Self-Reported Olfactory and Taste Disorders in Patients With Severe Acute Respiratory Coronavirus 2 Infection: A Cross-Sectional Study. Clin. Infect. Dis. 2020, 71 (15), 889–890. 10.1093/cid/ciaa330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyiadji N.; Shahin G.; Noujaim D.; Stone M.; Patel S.; Griffith B. COVID-19-Associated Acute Hemorrhagic Necrotizing Encephalopathy: Imaging Features. Radiology 2020, 296 (2), E119–E120. 10.1148/radiol.2020201187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniz-Mondolfi A.; Bryce C.; Grimes Z.; Gordon R. E.; Reidy J.; Lednicky J.; Sordillo E. M.; Fowkes M. Central Nervous System Involvement by Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2). J. Med. Virol. 2020, 92 (7), 699–702. 10.1002/jmv.25915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toscano G.; Palmerini F.; Ravaglia S.; Ruiz L.; Invernizzi P.; Cuzzoni M. G.; Franciotta D.; Baldanti F.; Daturi R.; Postorino P.; Cavallini A.; Micieli G. Guillain-Barré Syndrome Associated with SARS-CoV-2. N. Engl. J. Med. 2020, 382 (26), 2574–2576. 10.1056/NEJMc2009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H.; Shen D.; Zhou H.; Liu J.; Chen S. Guillain-Barré Syndrome Associated with SARS-CoV-2 Infection: Causality or Coincidence?. Lancet Neurol. 2020, 19 (5), 383–384. 10.1016/S1474-4422(20)30109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig A. M. Neurological Manifestations in COVID-19 Caused by SARS-CoV-2. CNS Neurosci. Ther. 2020, 26 (5), 499–501. 10.1111/cns.13372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E.; Zhang C.; Israelow B.; Lu-Culligan A.; Prado A. V.; Skriabine S.; Lu P.; Weizman O.-E.; Liu F.; Dai Y.; Szigeti-Buck K.; Yasumoto Y.; Wang G.; Castaldi C.; Heltke J.; Ng E.; Wheeler J.; Alfajaro M. M.; Levavasseur E.; Fontes B.; Ravindra N. G.; Van Dijk D.; Mane S.; Gunel M.; Ring A.; Kazmi S. A. J.; Zhang K.; Wilen C. B.; Horvath T. L.; Plu I.; Haik S.; Thomas J.-L.; Louvi A.; Farhadian S. F.; Huttner A.; Seilhean D.; Renier N.; Bilguvar K.; Iwasaki A. Neuroinvasion of SARS-CoV-2 in Human and Mouse Brain. J. Exp. Med. 2021, 218 (3), e20202135 10.1084/jem.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms J.; Kremer S.; Merdji H.; Clere-Jehl R.; Schenck M.; Kummerlen C.; Collange O.; Boulay C.; Fafi-Kremer S.; Ohana M.; Anheim M.; Meziani F. Neurologic Features in Severe SARS-CoV-2 Infection. N. Engl. J. Med. 2020, 382 (23), 2268–2270. 10.1056/NEJMc2008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi A. M.; Ahmed O.; Giliberto L.; Castillo M. Hemorrhagic Posterior Reversible Encephalopathy Syndrome as a Manifestation of COVID-19 Infection. Am. J. Neuroradiol. 2020, 41 (7), 1173–1176. 10.3174/ajnr.A6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal R.; Ganguly U.; Deb S.; Shome G.; Pramanik S.; Bandyopadhyay D.; Lahiri D. Meningoencephalitis Associated with COVID-19: A Systematic Review. J. Neurovirol. 2021, 27 (1), 12–25. 10.1007/s13365-020-00923-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zazhytska M.; Kodra A.; Hoagland D. A.; Frere J.; Fullard J. F.; Shayya H.; McArthur N. G.; Moeller R.; Uhl S.; Omer A. D.; Gottesman M. E.; Firestein S.; Gong Q.; Canoll P. D.; Goldman J. E.; Roussos P.; TenOever B. R.; Overdevest J. B.; Lomvardas S. Non-Cell-Autonomous Disruption of Nuclear Architecture as a Potential Cause of COVID-19-Induced Anosmia. Cell 2022, 185 (6), 1052–1064. 10.1016/j.cell.2022.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.; Yoo S.-J.; Clijsters M.; Backaert W.; Vanstapel A.; Speleman K.; Lietaer C.; Choi S.; Hether T. D.; Marcelis L.; Nam A.; Pan L.; Reeves J. W.; Van Bulck P.; Zhou H.; Bourgeois M.; Debaveye Y.; De Munter P.; Gunst J.; Jorissen M.; Lagrou K.; Lorent N.; Neyrinck A.; Peetermans M.; Thal D. R.; Vandenbriele C.; Wauters J.; Mombaerts P.; Van Gerven L. Visualizing in Deceased COVID-19 Patients How SARS-CoV-2 Attacks the Respiratory and Olfactory Mucosae but Spares the Olfactory Bulb. Cell 2021, 184 (24), 5932–5949. 10.1016/j.cell.2021.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M. E.; Appleton A.; Liu Q.-R.; Yao Q.; Mazucanti C. H.; Egan J. M. Human Type II Taste Cells Express Angiotensin-Converting Enzyme 2 and Are Infected by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Am. J. Pathol. 2021, 191 (9), 1511–1519. 10.1016/j.ajpath.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N.; Pérez P.; Kato T.; Mikami Y.; Okuda K.; Gilmore R. C.; Conde C. D.; Gasmi B.; Stein S.; Beach M.; Pelayo E.; Maldonado J. O.; Lafont B. A.; Jang S.-I.; Nasir N.; Padilla R. J.; Murrah V. A.; Maile R.; Lovell W.; Wallet S. M.; Bowman N. M.; Meinig S. L.; Wolfgang M. C.; Choudhury S. N.; Novotny M.; Aevermann B. D.; Scheuermann R. H.; Cannon G.; Anderson C. W.; Lee R. E.; Marchesan J. T.; Bush M.; Freire M.; Kimple A. J.; Herr D. L.; Rabin J.; Grazioli A.; Das S.; French B. N.; Pranzatelli T.; Chiorini J. A.; Kleiner D. E.; Pittaluga S.; Hewitt S. M.; Burbelo P. D.; Chertow D.; Frank K.; Lee J.; Boucher R. C.; Teichmann S. A.; Warner B. M.; Byrd K. M. SARS-CoV-2 Infection of the Oral Cavity and Saliva. Nat. Med. 2021, 27 (5), 892–903. 10.1038/s41591-021-01296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killingley B.; Mann A. J.; Kalinova M.; Boyers A.; Goonawardane N.; Zhou J.; Lindsell K.; Hare S. S.; Brown J.; Frise R.; Smith E.; Hopkins C.; Noulin N.; Löndt B.; Wilkinson T.; Harden S.; McShane H.; Baillet M.; Gilbert A.; Jacobs M.; Charman C.; Mande P.; Nguyen-Van-Tam J. S.; Semple M. G.; Read R. C.; Ferguson N. M.; Openshaw P. J.; Rapeport G.; Barclay W. S.; Catchpole A. P.; Chiu C. Safety, Tolerability and Viral Kinetics during SARS-CoV-2 Human Challenge in Young Adults. Nat. Med. 2022, 28 (5), 1031–1041. 10.1038/s41591-022-01780-9. [DOI] [PubMed] [Google Scholar]
- da Silva S. J. R.; de Magalhães J. J. F.; Pena L. Simultaneous Circulation of DENV, CHIKV, ZIKV and SARS-CoV-2 in Brazil: An Inconvenient Truth. One Health 2021, 12, 100205. 10.1016/j.onehlt.2020.100205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews Q.; da Silva S. J. R.; Norouzi M.; Pena L. J.; Pardee K. Adaptive, Diverse and de-Centralized Diagnostics Are Key to the Future of Outbreak Response. BMC Biol. 2020, 18 (1), 153. 10.1186/s12915-020-00891-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A. T.; Tong Y. X.; Zhang S. Profile of RT-PCR for SARS-CoV-2: A Preliminary Study From 56 COVID-19 Patients. Clin. Infect. Dis. 2020, 71 (16), 2249–2251. 10.1093/cid/ciaa460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y.; Zhang D.; Yang P.; Poon L. L. M.; Wang Q. Viral Load of SARS-CoV-2 in Clinical Samples. Lancet Infect. Dis. 2020, 20 (4), 411–412. 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Yan L.-M.; Wan L.; Xiang T.-X.; Le A.; Liu J.-M.; Peiris M.; Poon L. L. M.; Zhang W. Viral Dynamics in Mild and Severe Cases of COVID-19. Lancet Infect. Dis. 2020, 20 (6), 656–657. 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik M.; Tate M.; Lloyd O.; Maraolo A. E.; Schafers J.; Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV Viral Load Dynamics, Duration of Viral Shedding, and Infectiousness: A Systematic Review and Meta-Analysis. Lancet Microbe 2021, 2 (1), e13–e22. 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R.; Yi S.; Zhang J.; Lv Z.; Zhu C.; Zhang Y. Viral Load Dynamics in Sputum and Nasopharyngeal Swab in Patients with COVID-19. J. Dent. Res. 2020, 99 (11), 1239–1244. 10.1177/0022034520946251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yongchen Z.; Shen H.; Wang X.; Shi X.; Li Y.; Yan J.; Chen Y.; Gu B. Different Longitudinal Patterns of Nucleic Acid and Serology Testing Results Based on Disease Severity of COVID-19 Patients. Emerg. Microbes Infect. 2020, 9 (1), 833–836. 10.1080/22221751.2020.1756699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard J.; Dust K.; Funk D.; Strong J. E.; Alexander D.; Garnett L.; Boodman C.; Bello A.; Hedley A.; Schiffman Z.; Doan K.; Bastien N.; Li Y.; Van Caeseele P. G.; Poliquin G. Predicting Infectious Severe Acute Respiratory Syndrome Coronavirus 2 From Diagnostic Samples. Clin. Infect. Dis. 2020, 71 (10), 2663–2666. 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethuraman N.; Jeremiah S. S.; Ryo A. Interpreting Diagnostic Tests for SARS-CoV-2. JAMA 2020, 323 (22), 2249. 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- Zheng S.; Fan J.; Yu F.; Feng B.; Lou B.; Zou Q.; Xie G.; Lin S.; Wang R.; Yang X.; Chen W.; Wang Q.; Zhang D.; Liu Y.; Gong R.; Ma Z.; Lu S.; Xiao Y.; Gu Y.; Zhang J.; Yao H.; Xu K.; Lu X.; Wei G.; Zhou J.; Fang Q.; Cai H.; Qiu Y.; Sheng J.; Chen Y.; Liang T. Viral Load Dynamics and Disease Severity in Patients Infected with SARS-CoV-2 in Zhejiang Province, China, January-March 2020: Retrospective Cohort Study. BMJ. 2020, m1443. 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.; Yuan Q.; Wang H.; Liu W.; Liao X.; Su Y.; Wang X.; Yuan J.; Li T.; Li J.; Qian S.; Hong C.; Wang F.; Liu Y.; Wang Z.; He Q.; Li Z.; He B.; Zhang T.; Fu Y.; Ge S.; Liu L.; Zhang J.; Xia N.; Zhang Z. Antibody Responses to SARS-CoV-2 in Patients With Novel Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71 (16), 2027–2034. 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X.; Liu B.-Z.; Deng H.-J.; Wu G.-C.; Deng K.; Chen Y.-K.; Liao P.; Qiu J.-F.; Lin Y.; Cai X.-F.; Wang D.-Q.; Hu Y.; Ren J.-H.; Tang N.; Xu Y.-Y.; Yu L.-H.; Mo Z.; Gong F.; Zhang X.-L.; Tian W.-G.; Hu L.; Zhang X.-X.; Xiang J.-L.; Du H.-X.; Liu H.-W.; Lang C.-H.; Luo X.-H.; Wu S.-B.; Cui X.-P.; Zhou Z.; Zhu M.-M.; Wang J.; Xue C.-J.; Li X.-F.; Wang L.; Li Z.-J.; Wang K.; Niu C.-C.; Yang Q.-J.; Tang X.-J.; Zhang Y.; Liu X.-M.; Li J.-J.; Zhang D.-C.; Zhang F.; Liu P.; Yuan J.; Li Q.; Hu J.-L.; Chen J.; Huang A.-L. Antibody Responses to SARS-CoV-2 in Patients with COVID-19. Nat. Med. 2020, 26 (6), 845–848. 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Jiang S.; Hillyer C.; Du L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 2020, 41 (5), 355–359. 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L.; Ye F.; Cheng M.-L.; Feng Y.; Deng Y.-Q.; Zhao H.; Wei P.; Ge J.; Gou M.; Li X.; Sun L.; Cao T.; Wang P.; Zhou C.; Zhang R.; Liang P.; Guo H.; Wang X.; Qin C.-F.; Chen F.; Dong C. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity 2020, 52 (6), 971–977. 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran R.; McReynolds C.; Yang J.; Hammock B. D.; Ikram A.; Ali A.; Bashir A.; Zohra T.; Chang W. L. W.; Hartigan-O’Connor D. J.; Rashidi H. H.; Khan I. H. Immune Response Dynamics in COVID-19 Patients to SARS-CoV-2 and Other Human Coronaviruses. PLoS One 2021, 16 (7), e0254367. 10.1371/journal.pone.0254367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevadiya B. D.; Machhi J.; Herskovitz J.; Oleynikov M. D.; Blomberg W. R.; Bajwa N.; Soni D.; Das S.; Hasan M.; Patel M.; Senan A. M.; Gorantla S.; McMillan J.; Edagwa B.; Eisenberg R.; Gurumurthy C. B.; Reid S. P. M.; Punyadeera C.; Chang L.; Gendelman H. E. Diagnostics for SARS-CoV-2 Infections. Nat. Mater. 2021, 20 (5), 593–605. 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H.; Wang T.; Zhang B.; Luo Y.; Mao L.; Wang F.; Wu S.; Sun Z. Detection of IgM and IgG Antibodies in Patients with Coronavirus Disease 2019. Clin. Transl. Immunol. 2020, 9 (5), e1136 10.1002/cti2.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G.; Lee C. K.; Lam L. T. M.; Yan B.; Chua Y. X.; Lim A. Y. N.; Phang K. F.; Kew G. S.; Teng H.; Ngai C. H.; Lin L.; Foo R. M.; Pada S.; Ng L. C.; Tambyah P. A. Covert COVID-19 and False-Positive Dengue Serology in Singapore. Lancet Infect. Dis. 2020, 20 (5), 536. 10.1016/S1473-3099(20)30158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.; Kang S.; Tian R.; Zhang X.; Zhang X.; Wang Y. Imaging Manifestations and Diagnostic Value of Chest CT of Coronavirus Disease 2019 (COVID-19) in the Xiaogan Area. Clin. Radiol. 2020, 75 (5), 341–347. 10.1016/j.crad.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safiabadi Tali S. H.; LeBlanc J. J.; Sadiq Z.; Oyewunmi O. D.; Camargo C.; Nikpour B.; Armanfard N.; Sagan S. M.; Jahanshahi-Anbuhi S. Tools and Techniques for Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)/COVID-19 Detection. Clin. Microbiol. Rev. 2021, 34 (3), e00228-20 10.1128/CMR.00228-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udugama B.; Kadhiresan P.; Kozlowski H. N.; Malekjahani A.; Osborne M.; Li V. Y. C.; Chen H.; Mubareka S.; Gubbay J. B.; Chan W. C. W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14 (4), 3822–3835. 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- Liu R.; Yi S.; Zhang J.; Lv Z.; Zhu C.; Zhang Y. Viral Load Dynamics in Sputum and Nasopharyngeal Swab in Patients with COVID-19. J. Dent. Res. 2020, 99 (11), 1239–1244. 10.1177/0022034520946251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.; Guo D.; Li C.; Fang Z.; Chen L.; Yang R.; Li X.; Zeng W. Coronavirus Disease 2019: Initial Chest CT Findings. Eur. Radiol. 2020, 30 (8), 4398–4406. 10.1007/s00330-020-06816-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X.; Zhong Z.; Zhao W.; Zheng C.; Wang F.; Liu J. Chest CT for Typical Coronavirus Disease 2019 (COVID-19) Pneumonia: Relationship to Negative RT-PCR Testing. Radiology 2020, 296 (2), E41–E45. 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W.; Xie K.; Lu H.; Xu L.; Zhou S.; Fang S. Initial Clinical Features of Suspected Coronavirus Disease 2019 in Two Emergency Departments Outside of Hubei, China. J. Med. Virol. 2020, 92 (9), 1525–1532. 10.1002/jmv.25763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhimraj A.; Morgan R. L.; Shumaker A. H.; Lavergne V.; Baden L.; Cheng V. C.-C.; Edwards K. M.; Gandhi R.; Muller W. J.; O’Horo J. C.; Shoham S.; Murad M. H.; Mustafa R. A.; Sultan S.; Falck-Ytter Y. Infectious Diseases Society of America Guidelines on the Treatment and Management of Patients With Coronavirus Disease 2019 (COVID-19). Clin. Infect. Dis. 2020, ciaa478 10.1093/cid/ciaa478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea P. M.; Lee G. R.; Griffin T. P.; Tormey V.; Hayat A.; Costelloe S. J.; Griffin D. G.; Srinivasan S.; O’Kane M.; Burke C. M.; Faul J.; Thompson C. J.; Curley G.; Tormey W. P. COVID-19 in Adults: Test Menu for Hospital Blood Science Laboratories. Irish J. Med. Sci. (1971 -) 2020, 189 (4), 1147–1152. 10.1007/s11845-020-02252-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegeman I.; Ochodo E. A.; Guleid F.; Holtman G. A.; Yang B.; Davenport C.; Deeks J. J.; Dinnes J.; Dittrich S.; Emperador D.; Hooft L.; Spijker R.; Takwoingi Y.; Van den Bruel A.; Wang J.; Langendam M.; Verbakel J. Y.; Leeflang M. M. G. Routine Laboratory Testing to Determine If a Patient Has COVID-19. Cochrane Database Syst. Rev. 2020, 2020 (11), CD013787 10.1002/14651858.CD013787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Morales A. J.; Cardona-Ospina J. A.; Gutiérrez-Ocampo E.; Villamizar-Peña R.; Holguin-Rivera Y.; Escalera-Antezana J. P.; Alvarado-Arnez L. E.; Bonilla-Aldana D. K.; Franco-Paredes C.; Henao-Martinez A. F.; Paniz-Mondolfi A.; Lagos-Grisales G. J.; Ramírez-Vallejo E.; Suárez J. A.; Zambrano L. I.; Villamil-Gómez W. E.; Balbin-Ramon G. J.; Rabaan A. A.; Harapan H.; Dhama K.; Nishiura H.; Kataoka H.; Ahmad T.; Sah R. Clinical, Laboratory and Imaging Features of COVID-19: A Systematic Review and Meta-Analysis. Travel Med. Infect. Dis. 2020, 34, 101623. 10.1016/j.tmaid.2020.101623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik P.; Patel U.; Mehta D.; Patel N.; Kelkar R.; Akrmah M.; Gabrilove J. L.; Sacks H. Biomarkers and Outcomes of COVID-19 Hospitalisations: Systematic Review and Meta-Analysis. BMJ. Evidence-Based Med. 2021, 26 (3), 107–108. 10.1136/bmjebm-2020-111536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry B. M.; de Oliveira M. H. S.; Benoit S.; Plebani M.; Lippi G. Hematologic, Biochemical and Immune Biomarker Abnormalities Associated with Severe Illness and Mortality in Coronavirus Disease 2019 (COVID-19): A Meta-Analysis. Clin. Chem. Lab. Med. 2020, 58 (7), 1021–1028. 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- Young B. E.; Ong S. W. X.; Ng L. F. P.; Anderson D. E.; Chia W. N.; Chia P. Y.; Ang L. W.; Mak T.-M.; Kalimuddin S.; Chai L. Y. A.; Pada S.; Tan S. Y.; Sun L.; Parthasarathy P.; Fong S.-W.; Chan Y.-H.; Tan C. W.; Lee B.; Rötzschke O.; Ding Y.; Tambyah P.; Low J. G. H.; Cui L.; Barkham T.; Lin R. T. P.; Leo Y.-S.; Renia L.; Wang L.-F.; Lye D. C.; Lim P. L.; Peng Ang B. S.; Lee C. C.; U Lee L. S.; Ling L. M.; Ng O. T.; Chan M.; Marimuthu K.; Vasoo S.; Wong C. S.; Lee T. H.; Sadarangani S.; Lin R. J.; Sadasiv M. S.; Ling Ng D. H.; Choy C. Y.; En Tan G. S.; Tan Y. K.; Sutjipto S.; Lee P. H.; Tay J. Y.; Yeo T. W.; Khoo B. Y.; Tay W. C.; Ng G.; Mah Y. Y.; Tan W.; De P. P.; Pooja R.; Chia J. W. Z.; Constance Chen Y. Y.; Mendis S.; Toh B. K.; Choon Fong R. K.; Lin Oh H. M.; Fong Chien J. M.; Shafi H.; Cheong H. Y.; Tan T. Y.; Tan T. T.; Tan B. H.; Wijaya L.; Venkatachalam I.; Chua Y. Y.; Zhi Cherng B. P.; Zi Chan Y. F.; Wong H. M.; Thien S. Y.; Meng Goh K. C.; Ling Tan S. Y.; Ean Oon L. L.; Chan K. S.; Lin L.; Gin Chan D. S.; Ooi S. T.; Narayana D. R.; Somani J.; Ling Oon J. E.; Yan G. Z.; Allen D. M.; Jureen R.; Yan B.; Foo R.; Kang A.; Sivalingam V.; How W.; Fernandez N. L.; Yeo N. K.-W.; Chee R. S.-L.; Amrun S. N. Viral Dynamics and Immune Correlates of Coronavirus Disease 2019 (COVID-19) Severity. Clin. Infect. Dis. 2021, 73 (9), e2932–e2942. 10.1093/cid/ciaa1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R.; Corman V. M.; Guggemos W.; Seilmaier M.; Zange S.; Müller M. A.; Niemeyer D.; Jones T. C.; Vollmar P.; Rothe C.; Hoelscher M.; Bleicker T.; Brünink S.; Schneider J.; Ehmann R.; Zwirglmaier K.; Drosten C.; Wendtner C. Virological Assessment of Hospitalized Patients with COVID-2019. Nature 2020, 581 (7809), 465–469. 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Zou L.; Ruan F.; Huang M.; Liang L.; Huang H.; Hong Z.; Yu J.; Kang M.; Song Y.; Xia J.; Guo Q.; Song T.; He J.; Yen H.-L.; Peiris M.; Wu J. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020, 382 (12), 1177–1179. 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Persons for Coronavirus Disease 2019 (COVID-19); U.S. Centers for Disease Control and Prevention, 2020. [Google Scholar]
- Kipkorir V.; Cheruiyot I.; Ngure B.; Misiani M.; Munguti J. Prolonged SARS-CoV-2 RNA Detection in Anal/Rectal Swabs and Stool Specimens in COVID-19 Patients after Negative Conversion in Nasopharyngeal RT-PCR Test. J. Med. Virol. 2020, 92 (11), 2328–2331. 10.1002/jmv.26007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L.; Liu J.; Xu W.; Luo Q.; Chen D.; Lei Z.; Huang Z.; Li X.; Deng K.; Lin B.; Gao Z. SARS-CoV-2 Can Be Detected in Urine, Blood, Anal Swabs, and Oropharyngeal Swabs Specimens. J. Med. Virol. 2020, 92 (9), 1676–1680. 10.1002/jmv.25936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colavita F.; Lapa D.; Carletti F.; Lalle E.; Bordi L.; Marsella P.; Nicastri E.; Bevilacqua N.; Giancola M. L.; Corpolongo A.; Ippolito G.; Capobianchi M. R.; Castilletti C. SARS-CoV-2 Isolation From Ocular Secretions of a Patient With COVID-19 in Italy With Prolonged Viral RNA Detection. Ann. Int. Med. 2020, 173 (3), 242–243. 10.7326/M20-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groß R.; Conzelmann C.; Müller J. A.; Stenger S.; Steinhart K.; Kirchhoff F.; Münch J. Detection of SARS-CoV-2 in Human Breastmilk. Lancet 2020, 395 (10239), 1757–1758. 10.1016/S0140-6736(20)31181-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.; Jin M.; Bao P.; Zhao W.; Zhang S. Clinical Characteristics and Results of Semen Tests Among Men With Coronavirus Disease 2019. JAMA Netw. Open 2020, 3 (5), e208292. 10.1001/jamanetworkopen.2020.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricic T.; Nickel O.; Aximu-Petri A.; Essel E.; Gansauge M.; Kanis P.; Macak D.; Richter J.; Riesenberg S.; Bokelmann L.; Zeberg H.; Meyer M.; Borte S.; Pääbo S. A Direct RT-QPCR Approach to Test Large Numbers of Individuals for SARS-CoV-2. PLoS One 2020, 15 (12), e0244824. 10.1371/journal.pone.0244824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb D. M.; Tilley P.; Al-Rawahi G. N.; Srigley J. A.; Ford G.; Pedersen H.; Pabbi A.; Hannam-Clark S.; Charles M.; Dittrick M.; Gadkar V. J.; Pernica J. M.; Hoang L. M. N. Self-Collected Saline Gargle Samples as an Alternative to Health Care Worker-Collected Nasopharyngeal Swabs for COVID-19 Diagnosis in Outpatients. J. Clin. Microbiol. 2021, 59 (4), e02427-20 10.1128/JCM.02427-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzi L.; Carcano G.; Gianfagna F.; Grossi P.; Gasperina D. D.; Genoni A.; Fasano M.; Sessa F.; Tettamanti L.; Carinci F.; Maurino V.; Rossi A.; Tagliabue A.; Baj A. Saliva Is a Reliable Tool to Detect SARS-CoV-2. J. Infect. 2020, 81 (1), e45–e50. 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie A. L.; Fournier J.; Casanovas-Massana A.; Campbell M.; Tokuyama M.; Vijayakumar P.; Warren J. L.; Geng B.; Muenker M. C.; Moore A. J.; Vogels C. B. F.; Petrone M. E.; Ott I. M.; Lu P.; Venkataraman A.; Lu-Culligan A.; Klein J.; Earnest R.; Simonov M.; Datta R.; Handoko R.; Naushad N.; Sewanan L. R.; Valdez J.; White E. B.; Lapidus S.; Kalinich C. C.; Jiang X.; Kim D. J.; Kudo E.; Linehan M.; Mao T.; Moriyama M.; Oh J. E.; Park A.; Silva J.; Song E.; Takahashi T.; Taura M.; Weizman O.-E.; Wong P.; Yang Y.; Bermejo S.; Odio C. D.; Omer S. B.; Dela Cruz C. S.; Farhadian S.; Martinello R. A.; Iwasaki A.; Grubaugh N. D.; Ko A. I. Saliva or Nasopharyngeal Swab Specimens for Detection of SARS-CoV-2. N. Engl. J. Med. 2020, 383 (13), 1283–1286. 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V. M.; Landt O.; Kaiser M.; Molenkamp R.; Meijer A.; Chu D. K.; Bleicker T.; Brünink S.; Schneider J.; Schmidt M. L.; Mulders D. G.; Haagmans B. L.; van der Veer B.; van den Brink S.; Wijsman L.; Goderski G.; Romette J.-L.; Ellis J.; Zambon M.; Peiris M.; Goossens H.; Reusken C.; Koopmans M. P.; Drosten C. Detection of 2019 Novel Coronavirus (2019-NCoV) by Real-Time RT-PCR. Eurosurveillance 2020, 25 (3), 2000045 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Research use only 2019-Novel Coronavirus (2019-nCoV) Real-time RT-PCR primers and probes. U.S. Centers for Disease Control and Protection, June 6, 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html (accessed 2022-04-18).
- Specific primers and probes for detection 2019 novel coronavirus. China National Institute for Viral Disease Control and Prevention, 2020. http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.html (accessed 2022-04-18).
- Chu D. K. W.; Pan Y.; Cheng S. M. S.; Hui K. P. Y.; Krishnan P.; Liu Y.; Ng D. Y. M.; Wan C. K. C.; Yang P.; Wang Q.; Peiris M.; Poon L. L. M. Molecular Diagnosis of a Novel Coronavirus (2019-NCoV) Causing an Outbreak of Pneumonia. Clin. Chem. 2020, 66 (4), 549–555. 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc J. J.; Gubbay J. B.; Li Y.; Needle R.; Arneson S. R.; Marcino D.; Charest H.; Desnoyers G.; Dust K.; Fattouh R.; Garceau R.; German G.; Hatchette T. F.; Kozak R. A.; Krajden M.; Kuschak T.; Lang A. L. S.; Levett P.; Mazzulli T.; McDonald R.; Mubareka S.; Prystajecky N.; Rutherford C.; Smieja M.; Yu Y.; Zahariadis G.; Zelyas N.; Bastien N. Real-Time PCR-Based SARS-CoV-2 Detection in Canadian Laboratories. J. Clin. Virol. 2020, 128, 104433. 10.1016/j.jcv.2020.104433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y.; Shimizu T.; Noguchi T.; Nakano S.; Yamamoto M.; Nagao M. Comparison of 12 Molecular Detection Assays for Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Mol. Diagnostics 2021, 23 (2), 164–170. 10.1016/j.jmoldx.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels C. B. F.; Brito A. F.; Wyllie A. L.; Fauver J. R.; Ott I. M.; Kalinich C. C.; Petrone M. E.; Casanovas-Massana A.; Catherine Muenker M.; Moore A. J.; Klein J.; Lu P.; Lu-Culligan A.; Jiang X.; Kim D. J.; Kudo E.; Mao T.; Moriyama M.; Oh J. E.; Park A.; Silva J.; Song E.; Takahashi T.; Taura M.; Tokuyama M.; Venkataraman A.; Weizman O.-E.; Wong P.; Yang Y.; Cheemarla N. R.; White E. B.; Lapidus S.; Earnest R.; Geng B.; Vijayakumar P.; Odio C.; Fournier J.; Bermejo S.; Farhadian S.; Dela Cruz C. S.; Iwasaki A.; Ko A. I.; Landry M. L.; Foxman E. F.; Grubaugh N. D. Analytical Sensitivity and Efficiency Comparisons of SARS-CoV-2 RT-QPCR Primer-Probe Sets. Nat. Microbiol. 2020, 5 (10), 1299–1305. 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalla A. K.; Casto A. M.; Huang M.-L. W.; Perchetti G. A.; Sampoleo R.; Shrestha L.; Wei Y.; Zhu H.; Jerome K. R.; Greninger A. L. Comparative Performance of SARS-CoV-2 Detection Assays Using Seven Different Primer-Probe Sets and One Assay Kit. J. Clin. Microbiol. 2020, 58 (6), e00557-20 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcoba-Florez J.; González-Montelongo R.; Íñigo-Campos A.; de Artola D. G.-M.; Gil-Campesino H.; Ciuffreda L.; Valenzuela-Fernández A.; Flores C. Fast SARS-CoV-2 Detection by RT-QPCR in Preheated Nasopharyngeal Swab Samples. Int. J. Infect. Dis. 2020, 97, 66–68. 10.1016/j.ijid.2020.05.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyrlaki I.; Ekman M.; Lentini A.; Rufino de Sousa N.; Papanicolaou N.; Vondracek M.; Aarum J.; Safari H.; Muradrasoli S.; Rothfuchs A. G.; Albert J.; Högberg B.; Reinius B. Massive and Rapid COVID-19 Testing Is Feasible by Extraction-Free SARS-CoV-2 RT-PCR. Nat. Commun. 2020, 11 (1), 4812. 10.1038/s41467-020-18611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merindol N.; Pépin G.; Marchand C.; Rheault M.; Peterson C.; Poirier A.; Houle C.; Germain H.; Danylo A. SARS-CoV-2 Detection by Direct RRT-PCR without RNA Extraction. J. Clin. Virol. 2020, 128, 104423. 10.1016/j.jcv.2020.104423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomsgaard A. S.; Rosenstierne M. W. An Alternative Workflow for Molecular Detection of SARS-CoV-2 - Escape from the NA Extraction Kit-Shortage, Copenhagen, Denmark, March 2020. Eurosurveillance 2020, 25 (14), 2000398 10.2807/1560-7917.ES.2020.25.14.2000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woloshin S.; Patel N.; Kesselheim A. S. False Negative Tests for SARS-CoV-2 Infection — Challenges and Implications. N. Engl. J. Med. 2020, 383 (6), e38. 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- Vogels C. B. F.; Breban M. I.; Ott I. M.; Alpert T.; Petrone M. E.; Watkins A. E.; Kalinich C. C.; Earnest R.; Rothman J. E.; Goes de Jesus J.; Morales Claro I.; Magalhães Ferreira G.; Crispim M. A. E.; Singh L.; Tegally H.; Anyaneji U. J.; Hodcroft E. B.; Mason C. E.; Khullar G.; Metti J.; Dudley J. T.; MacKay M. J.; Nash M.; Wang J.; Liu C.; Hui P.; Murphy S.; Neal C.; Laszlo E.; Landry M. L.; Muyombwe A.; Downing R.; Razeq J.; de Oliveira T.; Faria N. R.; Sabino E. C.; Neher R. A.; Fauver J. R.; Grubaugh N. D. Multiplex QPCR Discriminates Variants of Concern to Enhance Global Surveillance of SARS-CoV-2. PLoS Biol. 2021, 19 (5), e3001236. 10.1371/journal.pbio.3001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva S. J. R.; Pena L. Collapse of the Public Health System and the Emergence of New Variants during the Second Wave of the COVID-19 Pandemic in Brazil. One Health 2021, 13, 100287. 10.1016/j.onehlt.2021.100287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva M. H. S.; Guedes D. R. D.; Docena C.; Bezerra M. F.; Dezordi F. Z.; Machado L. C.; Krokovsky L.; Helvecio E.; da Silva A. F.; Vasconcelos L. R. S.; Rezende A. M.; da Silva S. J. R.; Sales K. G. da S.; de Sá B. S. L. F.; da Cruz D. L.; Cavalcanti C. E.; Neto A. de M.; da Silva C. T. A.; Mendes R. P. G.; da Silva M. A. L.; Gräf T.; Resende P. C.; Bello G.; Barros M. da S.; do Nascimento W. R. C.; Arcoverde R. M. L.; Bezerra L. C. A.; Brandão-Filho S. P.; Ayres C. F. J.; Wallau G. L. Multiple Introductions Followed by Ongoing Community Spread of SARS-CoV-2 at One of the Largest Metropolitan Areas of Northeast Brazil. Viruses 2020, 12 (12), 1414. 10.3390/v12121414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir J. A.; Kozak R. A.; Aftanas P.; Raphenya A. R.; Smith K. M.; Maguire F.; Maan H.; Alruwaili M.; Banerjee A.; Mbareche H.; Alcock B. P.; Knox N. C.; Mossman K.; Wang B.; Hiscox J. A.; McArthur A. G.; Mubareka S. A Comparison of Whole Genome Sequencing of SARS-CoV-2 Using Amplicon-Based Sequencing, Random Hexamers, and Bait Capture. Viruses 2020, 12 (8), 895. 10.3390/v12080895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J. S.; Sathe L.; Munugala C.; Jones E. M.; Gasperini M.; Lubock N. B.; Yarza F.; Thompson E. M.; Kovary K. M.; Park J.; Marquette D.; Kay S.; Lucas M.; Love T.; Sina Booeshaghi A.; Brandenberg O. F.; Guo L.; Boocock J.; Hochman M.; Simpkins S. W.; Lin I.; LaPierre N.; Hong D.; Zhang Y.; Oland G.; Choe B. J.; Chandrasekaran S.; Hilt E. E.; Butte M. J.; Damoiseaux R.; Kravit C.; Cooper A. R.; Yin Y.; Pachter L.; Garner O. B.; Flint J.; Eskin E.; Luo C.; Kosuri S.; Kruglyak L.; Arboleda V. A. Massively Scaled-up Testing for SARS-CoV-2 RNA via next-Generation Sequencing of Pooled and Barcoded Nasal and Saliva Samples. Nat. Biomed. Eng. 2021, 5 (7), 657–665. 10.1038/s41551-021-00754-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoyar R. C.; Jain A.; Sehgal P.; Divakar M. K.; Sharma D.; Imran M.; Jolly B.; Ranjan G.; Rophina M.; Sharma S.; Siwach S.; Pandhare K.; Sahoo S.; Sahoo M.; Nayak A.; Mohanty J. N.; Das J.; Bhandari S.; Mathur S. K.; Kumar A.; Sahlot R.; Rojarani P.; Lakshmi J. V.; Surekha A.; Sekhar P. C.; Mahajan S.; Masih S.; Singh P.; Kumar V.; Jose B.; Mahajan V.; Gupta V.; Gupta R.; Arumugam P.; Singh A.; Nandy A.; P. V. R.; Jha R. M.; Kumari A.; Gandotra S.; Rao V.; Faruq M.; Kumar S.; Reshma G. B.; Varma G. N.; Roy S. S.; Sengupta A.; Chattopadhyay S.; Singhal K.; Pradhan S.; Jha D.; Naushin S.; Wadhwa S.; Tyagi N.; Poojary M.; Scaria V.; Sivasubbu S. High Throughput Detection and Genetic Epidemiology of SARS-CoV-2 Using COVIDSeq next-Generation Sequencing. PLoS One 2021, 16 (2), e0247115. 10.1371/journal.pone.0247115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra M. F.; Machado L. C.; De Carvalho V. do C. V.; Docena C.; Brandão-Filho S. P.; Ayres C. F. J.; Paiva M. H. S.; Wallau G. L. A Sanger-Based Approach for Scaling up Screening of SARS-CoV-2 Variants of Interest and Concern. Infect. Genet. Evol. 2021, 92, 104910. 10.1016/j.meegid.2021.104910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith L. W.; Hamilton W. L.; Warne B.; Houldcroft C. J.; Hosmillo M.; Jahun A. S.; Curran M. D.; Parmar S.; Caller L. G.; Caddy S. L.; Khokhar F. A.; Yakovleva A.; Hall G.; Feltwell T.; Forrest S.; Sridhar S.; Weekes M. P.; Baker S.; Brown N.; Moore E.; Popay A.; Roddick I.; Reacher M.; Gouliouris T.; Peacock S. J.; Dougan G.; Török M. E.; Goodfellow I. Rapid Implementation of SARS-CoV-2 Sequencing to Investigate Cases of Health-Care Associated COVID-19: A Prospective Genomic Surveillance Study. Lancet Infect. Dis. 2020, 20 (11), 1263–1271. 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SARS-CoV-2 Variant Classifications and Definitions; U.S. Centers for Disease Control and Prevention, 2021. [Google Scholar]
- Rana D. R.; Pokhrel N. Sequence Mismatch in PCR Probes May Mask the COVID-19 Detection in Nepal. Mol. Cell. Probes 2020, 53, 101599. 10.1016/j.mcp.2020.101599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V. M.; Rasche A.; Baronti C.; Aldabbagh S.; Cadar D.; Reusken C. B.; Pas S. D.; Goorhuis A.; Schinkel J.; Molenkamp R.; Kümmerer B. M.; Bleicker T.; Brünink S.; Eschbach-Bludau M.; Eis-Hübinger A. M.; Koopmans M. P.; Schmidt-Chanasit J.; Grobusch M. P.; de Lamballerie X.; Drosten C.; Drexler J. F. Assay Optimization for Molecular Detection of Zika Virus. Bull. World Health Organ. 2016, 94 (12), 880–892. 10.2471/BLT.16.175950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toms D.; Li J.; Cai H. Y. Evaluation of WHO listed COVID-19 qPCR primers and probe in silico with 375 SERS-CoV-2 full genome sequences. medRxiv 2020, 10.1101/2020.04.22.20075697. [DOI] [Google Scholar]
- Peñarrubia L.; Ruiz M.; Porco R.; Rao S. N.; Juanola-Falgarona M.; Manissero D.; López-Fontanals M.; Pareja J. Multiple Assays in a Real-Time RT-PCR SARS-CoV-2 Panel Can Mitigate the Risk of Loss of Sensitivity by New Genomic Variants during the COVID-19 Outbreak. Int. J. Infect. Dis. 2020, 97, 225–229. 10.1016/j.ijid.2020.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.-R.; Cao Q.-D.; Hong Z.-S.; Tan Y.-Y.; Chen S.-D.; Jin H.-J.; Tan K.-S.; Wang D.-Y.; Yan Y. The Origin, Transmission and Clinical Therapies on Coronavirus Disease 2019 (COVID-19) Outbreak—An Update on the Status. Mil. Med. Res. 2020, 7 (1), 11. 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevis D.; Kostaki E. G.; Magiorkinis G.; Panayiotakopoulos G.; Sourvinos G.; Tsiodras S. Full-Genome Evolutionary Analysis of the Novel Corona Virus (2019-NCoV) Rejects the Hypothesis of Emergence as a Result of a Recent Recombination Event. Infect. Genet. Evol. 2020, 79, 104212. 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S. F.; Quadeer A. A.; McKay M. R. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses 2020, 12 (3), 254. 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A.; Sidney J.; Zhang Y.; Scheuermann R. H.; Peters B.; Sette A. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe 2020, 27 (4), 671–680. 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi V. L. D.; Herbst K.; Boerner K.; Meurer M.; Kremer L. P.; Kirrmaier D.; Freistaedter A.; Papagiannidis D.; Galmozzi C.; Stanifer M. L.; Boulant S.; Klein S.; Chlanda P.; Khalid D.; Barreto Miranda I.; Schnitzler P.; Kräusslich H.-G.; Knop M.; Anders S. A Colorimetric RT-LAMP Assay and LAMP-Sequencing for Detecting SARS-CoV-2 RNA in Clinical Samples. Sci. Transl. Med. 2020, 12 (556), eabc7075 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik Y. S.; Sircar S.; Bhat S.; Sharun K.; Dhama K.; Dadar M.; Tiwari R.; Chaicumpa W. Emerging Novel Coronavirus (2019-NCoV)—Current Scenario, Evolutionary Perspective Based on Genome Analysis and Recent Developments. Vet. Q. 2020, 40 (1), 68–76. 10.1080/01652176.2020.1727993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahmi M.; Kubota Y.; Ito M. Nonstructural Proteins NS7b and NS8 Are Likely to Be Phylogenetically Associated with Evolution of 2019-NCoV. Infect. Genet. Evol. 2020, 81, 104272. 10.1016/j.meegid.2020.104272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S. J. R. da; Paiva M. H. S.; Guedes D. R. D.; Krokovsky L.; Melo F. L. de; Silva M. A. L. da; Silva A. da; Ayres C. F. J.; Pena L. J. Development and Validation of Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) for Rapid Detection of ZIKV in Mosquito Samples from Brazil. Sci. Rep. 2019, 9 (1), 4494. 10.1038/s41598-019-40960-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larremore D. B.; Wilder B.; Lester E.; Shehata S.; Burke J. M.; Hay J. A.; Tambe M.; Mina M. J.; Parker R. Test Sensitivity Is Secondary to Frequency and Turnaround Time for COVID-19 Screening. Sci. Adv. 2021, 7 (1), eabd5393 10.1126/sciadv.abd5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T.; Okayama H.; Masubuchi H.; Yonekawa T.; Watanabe K.; Amino N.; Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28 (12), e63 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S. J. R. da; Pardee K.; Pena L. Loop-Mediated Isothermal Amplification (LAMP) for the Diagnosis of Zika Virus: A Review. Viruses 2020, 12 (1), 19. 10.3390/v12010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva S. J. R.; Pardee K.; Balasuriya U. B. R.; Pena L. Development and Validation of a One-Step Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) for Rapid Detection of ZIKV in Patient Samples from Brazil. Sci. Rep. 2021, 11 (1), 4111. 10.1038/s41598-021-83371-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y.; Notomi T. Loop-Mediated Isothermal Amplification (LAMP): A Rapid, Accurate, and Cost-Effective Diagnostic Method for Infectious Diseases. J. Infect. Chemother. 2009, 15 (2), 62–69. 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C.; Cui J.; Huang L.; Du B.; Chen L.; Xue G.; Li S.; Zhang W.; Zhao L.; Sun Y.; Yao H.; Li N.; Zhao H.; Feng Y.; Liu S.; Zhang Q.; Liu D.; Yuan J. Rapid and Visual Detection of 2019 Novel Coronavirus (SARS-CoV-2) by a Reverse Transcription Loop-Mediated Isothermal Amplification Assay. Clin. Microbiol. Infect. 2020, 26 (6), 773–779. 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W. E.; Lim B.; Hsu C.; Xiong D.; Wu W.; Yu Y.; Jia H.; Wang Y.; Zeng Y.; Ji M.; Chang H.; Zhang X.; Wang H.; Cui Z. RT-LAMP for Rapid Diagnosis of Coronavirus SARS-CoV-2. Microb. Biotechnol. 2020, 13 (4), 950–961. 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek Y. H.; Um J.; Antigua K. J. C.; Park J.-H.; Kim Y.; Oh S.; Kim Y.; Choi W.-S.; Kim S. G.; Jeong J. H.; Chin B. S.; Nicolas H. D. G.; Ahn J.-Y.; Shin K. S.; Choi Y. K.; Park J.-S.; Song M.-S. Development of a Reverse Transcription-Loop-Mediated Isothermal Amplification as a Rapid Early-Detection Method for Novel SARS-CoV-2. Emerg. Microbes Infect. 2020, 9 (1), 998–1007. 10.1080/22221751.2020.1756698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R.; Wu X.; Wan Z.; Li Y.; Jin X.; Zhang C. A Novel Reverse Transcription Loop-Mediated Isothermal Amplification Method for Rapid Detection of SARS-CoV-2. Int. J. Mol. Sci. 2020, 21 (8), 2826. 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G.-S.; Ku K.; Baek S.-H.; Kim S.-J.; Kim S. Il; Kim B.-T.; Maeng J.-S. Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays Targeting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Mol. Diagnostics 2020, 22 (6), 729–735. 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb L. E.; Bartolone S. N.; Ward E.; Chancellor M. B. Rapid Detection of Novel Coronavirus/Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by Reverse Transcription-Loop-Mediated Isothermal Amplification. PLoS One 2020, 15 (6), e0234682. 10.1371/journal.pone.0234682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig K. U.; Schmithausen R. M.; Li D.; Jacobs M. L.; Hollstein R.; Blumenstock K.; Liebing J.; Słabicki M.; Ben-Shmuel A.; Israeli O.; Weiss S.; Ebert T. S.; Paran N.; Rüdiger W.; Wilbring G.; Feldman D.; Lippke B.; Ishorst N.; Hochfeld L. M.; Beins E. C.; Kaltheuner I. H.; Schmitz M.; Wöhler A.; Döhla M.; Sib E.; Jentzsch M.; Moench E.-M. C.; Borrajo J. D.; Strecker J.; Reinhardt J.; Cleary B.; Geyer M.; Hölzel M.; Macrae R.; Nöthen M. M.; Hoffmann P.; Exner M.; Regev A.; Zhang F.; Schmid-Burgk J. L. LAMP-Seq Enables Sensitive, Multiplexed COVID-19 Diagnostics Using Molecular Barcoding. Nat. Biotechnol. 2021, 39 (12), 1556–1562. 10.1038/s41587-021-00966-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.; Duplat D.; Benitez-Bolivar P.; León C.; Villota S. D.; Veloz-Villavicencio E.; Arévalo V.; Jaenes K.; Guo Y.; Cicek S.; Robinson L.; Peidis P.; Pearson J. D.; Woodgett J.; Mazzulli T.; Ponce P.; Restrepo S.; González J. M.; Bernal A.; Guevara-Suarez M.; Pardee K.; Cevallos V. E.; González C.; Bremner R. Multicenter International Assessment of a SARS-CoV-2 RT-LAMP Test for Point of Care Clinical Application. PLoS One 2022, 17 (5), e0268340. 10.1371/journal.pone.0268340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soroka M.; Wasowicz B.; Rymaszewska A. Loop-Mediated Isothermal Amplification (LAMP): The Better Sibling of PCR?. Cells 2021, 10 (8), 1931. 10.3390/cells10081931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseenko A.; Barrett D.; Pareja-Sanchez Y.; Howard R. J.; Strandback E.; Ampah-Korsah H.; Rovšnik U.; Zuniga-Veliz S.; Klenov A.; Malloo J.; Ye S.; Liu X.; Reinius B.; Elsässer S. J.; Nyman T.; Sandh G.; Yin X.; Pelechano V. Direct Detection of SARS-CoV-2 Using Non-Commercial RT-LAMP Reagents on Heat-Inactivated Samples. Sci. Rep. 2021, 11 (1), 1820. 10.1038/s41598-020-80352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S.; Kohl E.; Djandji A.; Morgan S.; Whittier S.; Mansukhani M.; Hod E.; D’Alton M.; Suh Y.; Williams Z. Direct Diagnostic Testing of SARS-CoV-2 without the Need for Prior RNA Extraction. Sci. Rep. 2021, 11 (1), 2402. 10.1038/s41598-021-81487-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli M. A.; Langmade J. S.; Chen X.; Fronick C. C.; Sawyer C. S.; Burcea L. C.; Wilkinson M. N.; Fulton R. S.; Heinz M.; Buchser W. J.; Head R. D.; Mitra R. D.; Milbrandt J. Rapid and Extraction-Free Detection of SARS-CoV-2 from Saliva by Colorimetric Reverse-Transcription Loop-Mediated Isothermal Amplification. Clin. Chem. 2021, 67 (2), 415–424. 10.1093/clinchem/hvaa267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral C.; Antunes W.; Moe E.; Duarte A. G.; Lima L. M. P.; Santos C.; Gomes I. L.; Afonso G. S.; Vieira R.; Teles H. S. S.; Reis M. S.; da Silva M. A. R.; Henriques A. M.; Fevereiro M.; Ventura M. R.; Serrano M.; Pimentel C. A Molecular Test Based on RT-LAMP for Rapid, Sensitive and Inexpensive Colorimetric Detection of SARS-CoV-2 in Clinical Samples. Sci. Rep. 2021, 11 (1), 16430. 10.1038/s41598-021-95799-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L.; Wu S.; Hao X.; Dong X.; Mao L.; Pelechano V.; Chen W.-H.; Yin X. Rapid Detection of COVID-19 Coronavirus Using a Reverse Transcriptional Loop-Mediated Isothermal Amplification (RT-LAMP) Diagnostic Platform. Clin. Chem. 2020, 66 (7), 975–977. 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa Y.; Orihara Y.; Kawamura R.; Imai K.; Sakai J.; Tarumoto N.; Matsuoka M.; Takeuchi S.; Maesaki S.; Maeda T. Evaluation of Rapid Diagnosis of Novel Coronavirus Disease (COVID-19) Using Loop-Mediated Isothermal Amplification. J. Clin. Virol. 2020, 129, 104446. 10.1016/j.jcv.2020.104446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski M. M.; Abudayyeh O. O.; Gootenberg J. S.; Zhang F.; Collins J. J. CRISPR-Based Diagnostics. Nat. Biomed. Eng. 2021, 5 (7), 643–656. 10.1038/s41551-021-00760-7. [DOI] [PubMed] [Google Scholar]
- Mojica F. J. M.; DΔez-VillaseΔor C.; GarcΔa-MartΔnez J.; Soria E. Intervening Sequences of Regularly Spaced Prokaryotic Repeats Derive from Foreign Genetic Elements. J. Mol. Evol. 2005, 60 (2), 174–182. 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Hille F.; Richter H.; Wong S. P.; Bratovič M.; Ressel S.; Charpentier E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172 (6), 1239–1259. 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- Wright A. V.; Nuñez J. K.; Doudna J. A. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell 2016, 164 (1-2), 29–44. 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- Sander J. D.; Joung J. K. CRISPR-Cas Systems for Editing, Regulating and Targeting Genomes. Nat. Biotechnol. 2014, 32 (4), 347–355. 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manghwar H.; Li B.; Ding X.; Hussain A.; Lindsey K.; Zhang X.; Jin S. CRISPR/Cas Systems in Genome Editing: Methodologies and Tools for sgRNA Design, Off-Target Evaluation, and Strategies to Mitigate Off-Target Effects. Adv. Sci. 2020, 7 (6), 1902312. 10.1002/advs.201902312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee K.; Green A. A.; Takahashi M. K.; Braff D.; Lambert G.; Lee J. W.; Ferrante T.; Ma D.; Donghia N.; Fan M.; Daringer N. M.; Bosch I.; Dudley D. M.; O’Connor D. H.; Gehrke L.; Collins J. J. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165 (5), 1255–1266. 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- de Puig H.; Lee R. A.; Najjar D.; Tan X.; Soenksen L. R.; Angenent-Mari N. M.; Donghia N. M.; Weckman N. E.; Ory A.; Ng C. F.; Nguyen P. Q.; Mao A. S.; Ferrante T. C.; Lansberry G.; Sallum H.; Niemi J.; Collins J. J. Minimally Instrumented SHERLOCK (miSHERLOCK) for CRISPR-Based Point-of-Care Diagnosis of SARS-CoV-2 and Emerging Variants. Sci. Adv. 2021, 7 (32), eabh2944 10.1126/sciadv.abh2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J. S.; Abudayyeh O. O.; Lee J. W.; Essletzbichler P.; Dy A. J.; Joung J.; Verdine V.; Donghia N.; Daringer N. M.; Freije C. A.; Myhrvold C.; Bhattacharyya R. P.; Livny J.; Regev A.; Koonin E. V.; Hung D. T.; Sabeti P. C.; Collins J. J.; Zhang F. Nucleic Acid Detection with CRISPR-Cas13a/C2c2. Science 2017, 356 (6336), 438–442. 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J. S.; Abudayyeh O. O.; Kellner M. J.; Joung J.; Collins J. J.; Zhang F. Multiplexed and Portable Nucleic Acid Detection Platform with Cas13, Cas12a, and Csm6. Science 2018, 360 (6387), 439–444. 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch N. L.; Zhu M.; Hua C.; Weller J.; Mirhashemi M. E.; Nguyen T. G.; Mantena S.; Bauer M. R.; Shaw B. M.; Ackerman C. M.; Thakku S. G.; Tse M. W.; Kehe J.; Uwera M.-M.; Eversley J. S.; Bielwaski D. A.; McGrath G.; Braidt J.; Johnson J.; Cerrato F.; Moreno G. K.; Krasilnikova L. A.; Petros B. A.; Gionet G. L.; King E.; Huard R. C.; Jalbert S. K.; Cleary M. L.; Fitzgerald N. A.; Gabriel S. B.; Gallagher G. R.; Smole S. C.; Madoff L. C.; Brown C. M.; Keller M. W.; Wilson M. M.; Kirby M. K.; Barnes J. R.; Park D. J.; Siddle K. J.; Happi C. T.; Hung D. T.; Springer M.; MacInnis B. L.; Lemieux J. E.; Rosenberg E.; Branda J. A.; Blainey P. C.; Sabeti P. C.; Myhrvold C. Multiplexed CRISPR-Based Microfluidic Platform for Clinical Testing of Respiratory Viruses and Identification of SARS-CoV-2 Variants. Nat. Med. 2022, 28 (5), 1083–1094. 10.1038/s41591-022-01734-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patchsung M.; Jantarug K.; Pattama A.; Aphicho K.; Suraritdechachai S.; Meesawat P.; Sappakhaw K.; Leelahakorn N.; Ruenkam T.; Wongsatit T.; Athipanyasilp N.; Eiamthong B.; Lakkanasirorat B.; Phoodokmai T.; Niljianskul N.; Pakotiprapha D.; Chanarat S.; Homchan A.; Tinikul R.; Kamutira P.; Phiwkaow K.; Soithongcharoen S.; Kantiwiriyawanitch C.; Pongsupasa V.; Trisrivirat D.; Jaroensuk J.; Wongnate T.; Maenpuen S.; Chaiyen P.; Kamnerdnakta S.; Swangsri J.; Chuthapisith S.; Sirivatanauksorn Y.; Chaimayo C.; Sutthent R.; Kantakamalakul W.; Joung J.; Ladha A.; Jin X.; Gootenberg J. S.; Abudayyeh O. O.; Zhang F.; Horthongkham N.; Uttamapinant C. Clinical Validation of a Cas13-Based Assay for the Detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4 (12), 1140–1149. 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- Schermer B.; Fabretti F.; Damagnez M.; Di Cristanziano V.; Heger E.; Arjune S.; Tanner N. A.; Imhof T.; Koch M.; Ladha A.; Joung J.; Gootenberg J. S.; Abudayyeh O. O.; Burst V.; Zhang F.; Klein F.; Benzing T.; Müller R.-U. Rapid SARS-CoV-2 Testing in Primary Material Based on a Novel Multiplex RT-LAMP Assay. PLoS One 2020, 15 (11), e0238612 10.1371/journal.pone.0238612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J.; Ladha A.; Saito M.; Kim N.-G.; Woolley A. E.; Segel M.; Barretto R. P. J.; Ranu A.; Macrae R. K.; Faure G.; Ioannidi E. I.; Krajeski R. N.; Bruneau R.; Huang M.-L. W.; Yu X. G.; Li J. Z.; Walker B. D.; Hung D. T.; Greninger A. L.; Jerome K. R.; Gootenberg J. S.; Abudayyeh O. O.; Zhang F. Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N. Engl. J. Med. 2020, 383 (15), 1492–1494. 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J. P.; Deng X.; Yu G.; Fasching C. L.; Servellita V.; Singh J.; Miao X.; Streithorst J. A.; Granados A.; Sotomayor-Gonzalez A.; Zorn K.; Gopez A.; Hsu E.; Gu W.; Miller S.; Pan C.-Y.; Guevara H.; Wadford D. A.; Chen J. S.; Chiu C. Y. CRISPR-Cas12-Based Detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38 (7), 870–874. 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandsma E.; Verhagen H. J. M. P.; van de Laar T. J. W.; Claas E. C. J.; Cornelissen M.; van den Akker E. Rapid, Sensitive, and Specific Severe Acute Respiratory Syndrome Coronavirus 2 Detection: A Multicenter Comparison Between Standard Quantitative Reverse-Transcriptase Polymerase Chain Reaction and CRISPR-Based DETECTR. J. Infect. Dis. 2021, 223 (2), 206–213. 10.1093/infdis/jiaa641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N.; Jolly P.; Formisano N.; Estrela P. Introduction to Biosensors. Essays Biochem. 2016, 60 (1), 1–8. 10.1042/EBC20150001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli I. A.; Hassan A.; Oliveira O. N.; Crespilho F. N. On the Challenges for the Diagnosis of SARS-CoV-2 Based on a Review of Current Methodologies. ACS Sensors 2020, 5 (12), 3655–3677. 10.1021/acssensors.0c01382. [DOI] [PubMed] [Google Scholar]
- Qin Z.; Peng R.; Baravik I. K.; Liu X. Fighting COVID-19: Integrated Micro- and Nanosystems for Viral Infection Diagnostics. Matter 2020, 3 (3), 628–651. 10.1016/j.matt.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinafar A.; Jaenes K.; Pardee K. Synthetic Biology Goes Cell-Free. BMC Biol. 2019, 17 (1), 64. 10.1186/s12915-019-0685-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee K.; Green A. A.; Ferrante T.; Cameron D. E.; DaleyKeyser A.; Yin P.; Collins J. J. Paper-Based Synthetic Gene Networks. Cell 2014, 159 (4), 940–954. 10.1016/j.cell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green A. A.; Silver P. A.; Collins J. J.; Yin P. Toehold Switches: De-Novo-Designed Regulators of Gene Expression. Cell 2014, 159 (4), 925–939. 10.1016/j.cell.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlikow M.; da Silva S. J. R.; Guo Y.; Cicek S.; Krokovsky L.; Homme P.; Xiong Y.; Xu T.; Calderón-Peláez M.-A.; Camacho-Ortega S.; Ma D.; de Magalhães J. J. F.; Souza B. N. R. F.; de Albuquerque Cabral D. G.; Jaenes K.; Sutyrina P.; Ferrante T.; Benitez A. D.; Nipaz V.; Ponce P.; Rackus D. G.; Collins J. J.; Paiva M.; Castellanos J. E.; Cevallos V.; Green A. A.; Ayres C.; Pena L.; Pardee K. Field Validation of the Performance of Paper-Based Tests for the Detection of the Zika and Chikungunya Viruses in Serum Samples. Nat. Biomed. Eng. 2022, 6 (3), 246–256. 10.1038/s41551-022-00850-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D.; Shen L.; Wu K.; Diehnelt C. W.; Green A. A. Low-Cost Detection of Norovirus Using Paper-Based Cell-Free Systems and Synbody-Based Viral Enrichment. Synth. Biol. 2018, 3 (1), ysy018 10.1093/synbio/ysy018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amalfitano E.; Karlikow M.; Norouzi M.; Jaenes K.; Cicek S.; Masum F.; Sadat Mousavi P.; Guo Y.; Tang L.; Sydor A.; Ma D.; Pearson J. D.; Trcka D.; Pinette M.; Ambagala A.; Babiuk S.; Pickering B.; Wrana J.; Bremner R.; Mazzulli T.; Sinton D.; Brumell J. H.; Green A. A.; Pardee K. A Glucose Meter Interface for Point-of-Care Gene Circuit-Based Diagnostics. Nat. Commun. 2021, 12 (1), 724. 10.1038/s41467-020-20639-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroneman A.; Vega E.; Vennema H.; Vinjé J.; White P. A.; Hansman G.; Green K.; Martella V.; Katayama K.; Koopmans M. Proposal for a Unified Norovirus Nomenclature and Genotyping. Arch. Virol. 2013, 158, 2059–2068. 10.1007/s00705-013-1708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontou P. I.; Braliou G. G.; Dimou N. L.; Nikolopoulos G.; Bagos P. G. Antibody Tests in Detecting SARS-CoV-2 Infection: A Meta-Analysis. Diagnostics 2020, 10 (5), 319. 10.3390/diagnostics10050319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.-W.; Schmitz J. E.; Persing D. H.; Stratton C. W. Laboratory Diagnosis of COVID-19: Current Issues and Challenges. J. Clin. Microbiol. 2020, 58 (6), e00512-20 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M.; Song L. Novel Antibody Epitopes Dominate the Antigenicity of Spike Glycoprotein in SARS-CoV-2 Compared to SARS-CoV. Cell. Mol. Immunol. 2020, 17 (5), 536–538. 10.1038/s41423-020-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejo A. P.; Akgun Y.; Al Mana A. F.; Tjendra Y.; Millan N. C.; Gomez-Fernandez C.; Cray C. Review of Current Advances in Serologic Testing for COVID-19. Am. J. Clin. Pathol. 2020, 154 (3), 293–304. 10.1093/ajcp/aqaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. E.; Tan C. W.; Chia W. N.; Young B. E.; Linster M.; Low J. H.; Tan Y.-J.; Chen M. I.-C.; Smith G. J. D.; Leo Y. S.; Lye D. C.; Wang L.-F. Lack of Cross-Neutralization by SARS Patient Sera towards SARS-CoV-2. Emerg. Microbes Infect. 2020, 9 (1), 900–902. 10.1080/22221751.2020.1761267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar L.; Segovia-Chumbez B.; Jadi R.; Martinez D. R.; Raut R.; Markmann A. J.; Cornaby C.; Bartelt L.; Weiss S.; Park Y.; Edwards C. E.; Weimer E.; Scherer E. M.; Rouphael N.; Edupuganti S.; Weiskopf D.; Tse L. V.; Hou Y. J.; Margolis D.; Sette A.; Collins M. H.; Schmitz J.; Baric R. S.; de Silva A. M. The Receptor-Binding Domain of the Viral Spike Protein Is an Immunodominant and Highly Specific Target of Antibodies in SARS-CoV-2 Patients. Sci. Immunol. 2020, 5 (48), eabc8413 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okba N. M. A.; Müller M. A.; Li W.; Wang C.; GeurtsvanKessel C. H.; Corman V. M.; Lamers M. M.; Sikkema R. S.; de Bruin E.; Chandler F. D.; Yazdanpanah Y.; Le Hingrat Q.; Descamps D.; Houhou-Fidouh N.; Reusken C. B. E. M.; Bosch B.-J.; Drosten C.; Koopmans M. P. G.; Haagmans B. L. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease Patients. Emerg. Infect. Dis. 2020, 26 (7), 1478–1488. 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S. J. R. da; Pena L. J. A Word of Caution in Interpreting COVID-19 Diagnostics Tests. J. Med. Virol. 2021, 93 (2), 717–718. 10.1002/jmv.26531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson K. E.; Caliendo A. M.; Arias C. A.; Englund J. A.; Hayden M. K.; Lee M. J.; Loeb M.; Patel R.; Altayar O.; El Alayli A.; Sultan S.; Falck-Ytter Y.; Lavergne V.; Morgan R. L.; Murad M. H.; Bhimraj A.; Mustafa R. A. Infectious Diseases Society of America Guidelines on the Diagnosis of Coronavirus Disease 2019 (COVID-19): Serologic Testing. Clin. Infect. Dis. 2020, 10.1093/cid/ciaa1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D. S.; Cromer D.; Reynaldi A.; Schlub T. E.; Wheatley A. K.; Juno J. A.; Subbarao K.; Kent S. J.; Triccas J. A.; Davenport M. P. Neutralizing Antibody Levels Are Highly Predictive of Immune Protection from Symptomatic SARS-CoV-2 Infection. Nat. Med. 2021, 27 (7), 1205–1211. 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Cromer D.; Steain M.; Reynaldi A.; Schlub T. E.; Wheatley A. K.; Juno J. A.; Kent S. J.; Triccas J. A.; Khoury D. S.; Davenport M. P. Neutralising Antibody Titres as Predictors of Protection against SARS-CoV-2 Variants and the Impact of Boosting: A Meta-Analysis. Lancet Microbe 2022, 3 (1), e52–e61. 10.1016/S2666-5247(21)00267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habli Z.; Saleh S.; Zaraket H.; Khraiche M. L. COVID-19 in-Vitro Diagnostics: State-of-the-Art and Challenges for Rapid, Scalable, and High-Accuracy Screening. Front. Bioeng. Biotechnol. 2021, 8, 605702 10.3389/fbioe.2020.605702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brümmer L. E.; Katzenschlager S.; Gaeddert M.; Erdmann C.; Schmitz S.; Bota M.; Grilli M.; Larmann J.; Weigand M. A.; Pollock N. R.; Macé A.; Carmona S.; Ongarello S.; Sacks J. A.; Denkinger C. M. Accuracy of Novel Antigen Rapid Diagnostics for SARS-CoV-2: A Living Systematic Review and Meta-Analysis. PLoS Med. 2021, 18 (8), e1003735 10.1371/journal.pmed.1003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak G. C.; Cheng P. K.; Lau S. S.; Wong K. K.; Lau C.; Lam E. T.; Chan R. C.; Tsang D. N. Evaluation of Rapid Antigen Test for Detection of SARS-CoV-2 Virus. J. Clin. Virol. 2020, 129, 104500. 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlbauer D.; Amanat F.; Chromikova V.; Jiang K.; Strohmeier S.; Arunkumar G. A.; Tan J.; Bhavsar D.; Capuano C.; Kirkpatrick E.; Meade P.; Brito R. N.; Teo C.; McMahon M.; Simon V.; Krammer F. SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr. Protoc. Microbiol. 2020, 57 (1), e100 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porte L.; Legarraga P.; Vollrath V.; Aguilera X.; Munita J. M.; Araos R.; Pizarro G.; Vial P.; Iruretagoyena M.; Dittrich S.; Weitzel T. Evaluation of a Novel Antigen-Based Rapid Detection Test for the Diagnosis of SARS-CoV-2 in Respiratory Samples. Int. J. Infect. Dis. 2020, 99, 328–333. 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrikou S.; Moschopoulou G.; Tsekouras V.; Kintzios S. Development of a Portable, Ultra-Rapid and Ultra-Sensitive Cell-Based Biosensor for the Direct Detection of the SARS-CoV-2 S1 Spike Protein Antigen. Sensors 2020, 20 (11), 3121. 10.3390/s20113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q.; Wen D.; Wu J.; Liu L.; Wu W.; Fang X.; Kong J. Microfluidic Immunoassays for Sensitive and Simultaneous Detection of IgG/IgM/Antigen of SARS-CoV-2 within 15 Min. Anal. Chem. 2020, 92 (14), 9454–9458. 10.1021/acs.analchem.0c01635. [DOI] [PubMed] [Google Scholar]
- Coronavirus Treatment Acceleration Program (CTAP); U.S. Food and Drug Administration, 2022. [Google Scholar]
- Tao K.; Tzou P. L.; Nouhin J.; Bonilla H.; Jagannathan P.; Shafer R. W. SARS-CoV-2 Antiviral Therapy. Clin Microbiol Rev. 2021, 34 (4), e0010921 10.1128/CMR.00109-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho A.; Saunders O. L.; Butler T.; Zhang L.; Xu J.; Vela J. E.; Feng J. Y.; Ray A. S.; Kim C. U. Synthesis and Antiviral Activity of a Series of 1′-Substituted 4-Aza-7,9-Dideazaadenosine C-Nucleosides. Bioorg. Med. Chem. Lett. 2012, 22 (8), 2705–2707. 10.1016/j.bmcl.2012.02.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel D.; Hui H. C.; Doerffler E.; Clarke M. O.; Chun K.; Zhang L.; Neville S.; Carra E.; Lew W.; Ross B.; Wang Q.; Wolfe L.; Jordan R.; Soloveva V.; Knox J.; Perry J.; Perron M.; Stray K. M.; Barauskas O.; Feng J. Y.; Xu Y.; Lee G.; Rheingold A. L.; Ray A. S.; Bannister R.; Strickley R.; Swaminathan S.; Lee W. A.; Bavari S.; Cihlar T.; Lo M. K.; Warren T. K.; Mackman R. L. Discovery and Synthesis of a Phosphoramidate Prodrug of a Pyrrolo[2,1- f][Triazin-4-Amino] Adenine C -Nucleoside (GS-5734) for the Treatment of Ebola and Emerging Viruses. J. Med. Chem. 2017, 60 (5), 1648–1661. 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- Warren T. K.; Jordan R.; Lo M. K.; Ray A. S.; Mackman R. L.; Soloveva V.; Siegel D.; Perron M.; Bannister R.; Hui H. C.; Larson N.; Strickley R.; Wells J.; Stuthman K. S.; Van Tongeren S. A.; Garza N. L.; Donnelly G.; Shurtleff A. C.; Retterer C. J.; Gharaibeh D.; Zamani R.; Kenny T.; Eaton B. P.; Grimes E.; Welch L. S.; Gomba L.; Wilhelmsen C. L.; Nichols D. K.; Nuss J. E.; Nagle E. R.; Kugelman J. R.; Palacios G.; Doerffler E.; Neville S.; Carra E.; Clarke M. O.; Zhang L.; Lew W.; Ross B.; Wang Q.; Chun K.; Wolfe L.; Babusis D.; Park Y.; Stray K. M.; Trancheva I.; Feng J. Y.; Barauskas O.; Xu Y.; Wong P.; Braun M. R.; Flint M.; McMullan L. K.; Chen S.-S.; Fearns R.; Swaminathan S.; Mayers D. L.; Spiropoulou C. F.; Lee W. A.; Nichol S. T.; Cihlar T.; Bavari S. Therapeutic Efficacy of the Small Molecule GS-5734 against Ebola Virus in Rhesus Monkeys. Nature 2016, 531 (7594), 381–385. 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M. K.; Jordan R.; Arvey A.; Sudhamsu J.; Shrivastava-Ranjan P.; Hotard A. L.; Flint M.; McMullan L. K.; Siegel D.; Clarke M. O.; Mackman R. L.; Hui H. C.; Perron M.; Ray A. S.; Cihlar T.; Nichol S. T.; Spiropoulou C. F. GS-5734 and Its Parent Nucleoside Analog Inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017, 7 (1), 43395. 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T. P.; Sims A. C.; Graham R. L.; Menachery V. D.; Gralinski L. E.; Case J. B.; Leist S. R.; Pyrc K.; Feng J. Y.; Trantcheva I.; Bannister R.; Park Y.; Babusis D.; Clarke M. O.; Mackman R. L.; Spahn J. E.; Palmiotti C. A.; Siegel D.; Ray A. S.; Cihlar T.; Jordan R.; Denison M. R.; Baric R. S. Broad-Spectrum Antiviral GS-5734 Inhibits Both Epidemic and Zoonotic Coronaviruses. Sci. Transl. Med. 2017, 9 (396), eaal3653 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M. L.; Andres E. L.; Sims A. C.; Graham R. L.; Sheahan T. P.; Lu X.; Smith E. C.; Case J. B.; Feng J. Y.; Jordan R.; Ray A. S.; Cihlar T.; Siegel D.; Mackman R. L.; Clarke M. O.; Baric R. S.; Denison M. R. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio 2018, 9 (2), e00221-18 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. J.; Won J. J.; Graham R. L.; Dinnon K. H.; Sims A. C.; Feng J. Y.; Cihlar T.; Denison M. R.; Baric R. S.; Sheahan T. P. Broad Spectrum Antiviral Remdesivir Inhibits Human Endemic and Zoonotic Deltacoronaviruses with a Highly Divergent RNA Dependent RNA Polymerase. Antiviral Res. 2019, 169, 104541. 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesnokov E.; Feng J.; Porter D.; Götte M. Mechanism of Inhibition of Ebola Virus RNA-Dependent RNA Polymerase by Remdesivir. Viruses 2019, 11 (4), 326. 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T. K.; Kane C. D.; Wells J.; Stuthman K. S.; Van Tongeren S. A.; Garza N. L.; Donnelly G.; Steffens J.; Gomba L.; Weidner J. M.; Norris S.; Zeng X.; Bannister R.; Cihlar T.; Bavari S.; Porter D. P.; Iversen P. L. Remdesivir Is Efficacious in Rhesus Monkeys Exposed to Aerosolized Ebola Virus. Sci. Rep. 2021, 11 (1), 19458. 10.1038/s41598-021-98971-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M. K.; Feldmann F.; Gary J. M.; Jordan R.; Bannister R.; Cronin J.; Patel N. R.; Klena J. D.; Nichol S. T.; Cihlar T.; Zaki S. R.; Feldmann H.; Spiropoulou C. F.; de Wit E. Remdesivir (GS-5734) Protects African Green Monkeys from Nipah Virus Challenge. Sci. Transl. Med. 2019, 11 (494), eaau9242 10.1126/scitranslmed.aau9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E.; Feldmann F.; Cronin J.; Jordan R.; Okumura A.; Thomas T.; Scott D.; Cihlar T.; Feldmann H. Prophylactic and Therapeutic Remdesivir (GS-5734) Treatment in the Rhesus Macaque Model of MERS-CoV Infection. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (12), 6771–6776. 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T. P.; Sims A. C.; Leist S. R.; Schäfer A.; Won J.; Brown A. J.; Montgomery S. A.; Hogg A.; Babusis D.; Clarke M. O.; Spahn J. E.; Bauer L.; Sellers S.; Porter D.; Feng J. Y.; Cihlar T.; Jordan R.; Denison M. R.; Baric R. S. Comparative Therapeutic Efficacy of Remdesivir and Combination Lopinavir, Ritonavir, and Interferon Beta against MERS-CoV. Nat. Commun. 2020, 11 (1), 222. 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Cao R.; Zhang L.; Yang X.; Liu J.; Xu M.; Shi Z.; Hu Z.; Zhong W.; Xiao G. Remdesivir and Chloroquine Effectively Inhibit the Recently Emerged Novel Coronavirus (2019-NCoV) in Vitro. Cell Res. 2020, 30 (3), 269–271. 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy K.-T.; Wong A. Y.-L.; Kaewpreedee P.; Sia S. F.; Chen D.; Hui K. P. Y.; Chu D. K. W.; Chan M. C. W.; Cheung P. P.-H.; Huang X.; Peiris M.; Yen H.-L. Remdesivir, Lopinavir, Emetine, and Homoharringtonine Inhibit SARS-CoV-2 Replication in Vitro. Antiviral Res. 2020, 178, 104786. 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runfeng L.; Yunlong H.; Jicheng H.; Weiqi P.; Qinhai M.; Yongxia S.; Chufang L.; Jin Z.; Zhenhua J.; Haiming J.; Kui Z.; Shuxiang H.; Jun D.; Xiaobo L.; Xiaotao H.; Lin W.; Nanshan Z.; Zifeng Y. Lianhuaqingwen Exerts Anti-Viral and Anti-Inflammatory Activity against Novel Coronavirus (SARS-CoV-2). Pharmacol. Res. 2020, 156, 104761. 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruijssers A. J.; George A. S.; Schäfer A.; Leist S. R.; Gralinksi L. E.; Dinnon K. H.; Yount B. L.; Agostini M. L.; Stevens L. J.; Chappell J. D.; Lu X.; Hughes T. M.; Gully K.; Martinez D. R.; Brown A. J.; Graham R. L.; Perry J. K.; Du Pont V.; Pitts J.; Ma B.; Babusis D.; Murakami E.; Feng J. Y.; Bilello J. P.; Porter D. P.; Cihlar T.; Baric R. S.; Denison M. R.; Sheahan T. P. Remdesivir Inhibits SARS-CoV-2 in Human Lung Cells and Chimeric SARS-CoV Expressing the SARS-CoV-2 RNA Polymerase in Mice. Cell Rep. 2020, 32 (3), 107940. 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson B. N.; Feldmann F.; Schwarz B.; Meade-White K.; Porter D. P.; Schulz J.; van Doremalen N.; Leighton I.; Yinda C. K.; Pérez-Pérez L.; Okumura A.; Lovaglio J.; Hanley P. W.; Saturday G.; Bosio C. M.; Anzick S.; Barbian K.; Cihlar T.; Martens C.; Scott D. P.; Munster V. J.; de Wit E. Clinical Benefit of Remdesivir in Rhesus Macaques Infected with SARS-CoV-2. Nature 2020, 585 (7824), 273–276. 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J. H.; Tomashek K. M.; Dodd L. E.; Mehta A. K.; Zingman B. S.; Kalil A. C.; Hohmann E.; Chu H. Y.; Luetkemeyer A.; Kline S.; Lopez de Castilla D.; Finberg R. W.; Dierberg K.; Tapson V.; Hsieh L.; Patterson T. F.; Paredes R.; Sweeney D. A.; Short W. R.; Touloumi G.; Lye D. C.; Ohmagari N.; Oh M.; Ruiz-Palacios G. M.; Benfield T.; Fätkenheuer G.; Kortepeter M. G.; Atmar R. L.; Creech C. B.; Lundgren J.; Babiker A. G.; Pett S.; Neaton J. D.; Burgess T. H.; Bonnett T.; Green M.; Makowski M.; Osinusi A.; Nayak S.; Lane H. C. Remdesivir for the Treatment of Covid-19 — Final Report. N. Engl. J. Med. 2020, 383 (19), 1813–1826. 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H.; Peto R.; Henao-Restrepo A. M.; Preziosi M. P.; Sathiyamoorthy V.; Abdool Karim Q.; Alejandria M. M.; Hernández García C.; Kieny M. P.; Malekzadeh R.; et al. Repurposed Antiviral Drugs for Covid-19 - Interim WHO Solidarity Trial Results. N. Engl. J. Med. 2021, 384 (6), 497–511. 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinner C. D.; Gottlieb R. L.; Criner G. J.; Arribas López J. R.; Cattelan A. M.; Soriano Viladomiu A.; Ogbuagu O.; Malhotra P.; Mullane K. M.; Castagna A.; Chai L. Y. A.; Roestenberg M.; Tsang O. T. Y.; Bernasconi E.; Le Turnier P.; Chang S.-C.; SenGupta D.; Hyland R. H.; Osinusi A. O.; Cao H.; Blair C.; Wang H.; Gaggar A.; Brainard D. M.; McPhail M. J.; Bhagani S.; Ahn M. Y.; Sanyal A. J.; Huhn G.; Marty F. M. Effect of Remdesivir vs Standard Care on Clinical Status at 11 Days in Patients With Moderate COVID-19. JAMA 2020, 324 (11), 1048. 10.1001/jama.2020.16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Zhang D.; Du G.; Du R.; Zhao J.; Jin Y.; Fu S.; Gao L.; Cheng Z.; Lu Q.; Hu Y.; Luo G.; Wang K.; Lu Y.; Li H.; Wang S.; Ruan S.; Yang C.; Mei C.; Wang Y.; Ding D.; Wu F.; Tang X.; Ye X.; Ye Y.; Liu B.; Yang J.; Yin W.; Wang A.; Fan G.; Zhou F.; Liu Z.; Gu X.; Xu J.; Shang L.; Zhang Y.; Cao L.; Guo T.; Wan Y.; Qin H.; Jiang Y.; Jaki T.; Hayden F. G.; Horby P. W.; Cao B.; Wang C. Remdesivir in Adults with Severe COVID-19: A Randomised, Double-Blind, Placebo-Controlled, Multicentre Trial. Lancet 2020, 395 (10236), 1569–1578. 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Abdouh A.; Bizanti A.; Barbarawi M.; Jabri A.; Kumar A.; Fashanu O. E.; Khan S. U.; Zhao D.; Antar A. A. R.; Michos E. D. Remdesivir for the Treatment of COVID-19: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Contemp. Clin. Trials 2021, 101, 106272. 10.1016/j.cct.2021.106272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y.; Yin W.; Zhang Y.; Shang W.; Wang Z.; Luan X.; Tian G.; Aisa H. A.; Xu Y.; Xiao G.; Li J.; Jiang H.; Zhang S.; Zhang L.; Xu H. E.; Shen J. Design and Development of an Oral Remdesivir Derivative VV116 against SARS-CoV-2. Cell Res. 2021, 31 (11), 1212–1214. 10.1038/s41422-021-00570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. M.; Wolf J. D.; Lieber C. M.; Sourimant J.; Lin M. J.; Babusis D.; DuPont V.; Chan J.; Barrett K. T.; Lye D.; Kalla R.; Chun K.; Mackman R. L.; Ye C.; Cihlar T.; Martinez-Sobrido L.; Greninger A. L.; Bilello J. P.; Plemper R. K. Oral Prodrug of Remdesivir Parent GS-441524 Is Efficacious against SARS-CoV-2 in Ferrets. Nat. Commun. 2021, 12 (1), 6415. 10.1038/s41467-021-26760-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D.; Hu T.; Zhang Y.; Zheng W.; Xue H.; Shen J.; Xie Y.; Aisa H. A. Potency and Pharmacokinetics of GS-441524 Derivatives against SARS-CoV-2. Bioorg. Med. Chem. 2021, 46, 116364. 10.1016/j.bmc.2021.116364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J.; Baum A.; Pascal K. E.; Russo V.; Giordano S.; Wloga E.; Fulton B. O.; Yan Y.; Koon K.; Patel K.; Chung K. M.; Hermann A.; Ullman E.; Cruz J.; Rafique A.; Huang T.; Fairhurst J.; Libertiny C.; Malbec M.; Lee W.; Welsh R.; Farr G.; Pennington S.; Deshpande D.; Cheng J.; Watty A.; Bouffard P.; Babb R.; Levenkova N.; Chen C.; Zhang B.; Romero Hernandez A.; Saotome K.; Zhou Y.; Franklin M.; Sivapalasingam S.; Lye D. C.; Weston S.; Logue J.; Haupt R.; Frieman M.; Chen G.; Olson W.; Murphy A. J.; Stahl N.; Yancopoulos G. D.; Kyratsous C. A. Studies in Humanized Mice and Convalescent Humans Yield a SARS-CoV-2 Antibody Cocktail. Science 2020, 369 (6506), 1010–1014. 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A.; Fulton B. O.; Wloga E.; Copin R.; Pascal K. E.; Russo V.; Giordano S.; Lanza K.; Negron N.; Ni M.; Wei Y.; Atwal G. S.; Murphy A. J.; Stahl N.; Yancopoulos G. D.; Kyratsous C. A. Antibody Cocktail to SARS-CoV-2 Spike Protein Prevents Rapid Mutational Escape Seen with Individual Antibodies. Science 2020, 369 (6506), 1014–1018. 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T. N.; Greaney A. J.; Dingens A. S.; Bloom J. D. Complete Map of SARS-CoV-2 RBD Mutations That Escape the Monoclonal Antibody LY-CoV555 and Its Cocktail with LY-CoV016. Cell Reports Med. 2021, 2 (4), 100255. 10.1016/j.xcrm.2021.100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellas C.; Del Bello A.; Debard A.; Steinmeyer Z.; Tribaudeau L.; Ranger N.; Jeanne N.; Martin-Blondel G.; Delobel P.; Kamar N.; Izopet J. Influence of Treatment with Neutralizing Monoclonal Antibodies on the SARS-CoV-2 Nasopharyngeal Load and Quasispecies. Clin. Microbiol. Infect. 2022, 28 (1), 139.e5–139.e8. 10.1016/j.cmi.2021.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Arora P.; Groß R.; Seidel A.; Hörnich B. F.; Hahn A. S.; Krüger N.; Graichen L.; Hofmann-Winkler H.; Kempf A.; Winkler M. S.; Schulz S.; Jäck H.-M.; Jahrsdörfer B.; Schrezenmeier H.; Müller M.; Kleger A.; Münch J.; Pöhlmann S. SARS-CoV-2 Variants B.1.351 and P.1 Escape from Neutralizing Antibodies. Cell 2021, 184 (9), 2384–2393. 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widera M.; Wilhelm A.; Hoehl S.; Pallas C.; Kohmer N.; Wolf T.; Rabenau H. F.; Corman V. M.; Drosten C.; Vehreschild M. J. G. T.; Goetsch U.; Gottschalk R.; Ciesek S. Limited Neutralization of Authentic Severe Acute Respiratory Syndrome Coronavirus 2 Variants Carrying E484K In Vitro. J. Infect. Dis. 2021, 224 (7), 1109–1114. 10.1093/infdis/jiab355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J.; Jang U. S.; Soh S. M.; Lee J.-Y.; Lee H.-R. The Impact on Infectivity and Neutralization Efficiency of SARS-CoV-2 Lineage B.1.351 Pseudovirus. Viruses 2021, 13 (4), 633. 10.3390/v13040633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm A.; Toptan T.; Pallas C.; Wolf T.; Goetsch U.; Gottschalk R.; Vehreschild M. J. G. T.; Ciesek S.; Widera M. Antibody-Mediated Neutralization of Authentic SARS-CoV-2 B.1.617 Variants Harboring L452R and T478K/E484Q. Viruses 2021, 13 (9), 1693. 10.3390/v13091693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A.; Ajithdoss D.; Copin R.; Zhou A.; Lanza K.; Negron N.; Ni M.; Wei Y.; Mohammadi K.; Musser B.; Atwal G. S.; Oyejide A.; Goez-Gazi Y.; Dutton J.; Clemmons E.; Staples H. M.; Bartley C.; Klaffke B.; Alfson K.; Gazi M.; Gonzalez O.; Dick E.; Carrion R.; Pessaint L.; Porto M.; Cook A.; Brown R.; Ali V.; Greenhouse J.; Taylor T.; Andersen H.; Lewis M. G.; Stahl N.; Murphy A. J.; Yancopoulos G. D.; Kyratsous C. A. REGN-COV2 Antibodies Prevent and Treat SARS-CoV-2 Infection in Rhesus Macaques and Hamsters. Science 2020, 370 (6520), 1110–1115. 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich D. M.; Sivapalasingam S.; Norton T.; Ali S.; Gao H.; Bhore R.; Xiao J.; Hooper A. T.; Hamilton J. D.; Musser B. J.; Rofail D.; Hussein M.; Im J.; Atmodjo D. Y.; Perry C.; Pan C.; Mahmood A.; Hosain R.; Davis J. D.; Turner K. C.; Baum A.; Kyratsous C. A.; Kim Y.; Cook A.; Kampman W.; Roque-Guerrero L.; Acloque G.; Aazami H.; Cannon K.; Simón-Campos J. A.; Bocchini J. A.; Kowal B.; DiCioccio A. T.; Soo Y.; Geba G. P.; Stahl N.; Lipsich L.; Braunstein N.; Herman G.; Yancopoulos G. D. REGEN-COV Antibody Combination and Outcomes in Outpatients with Covid-19. N. Engl. J. Med. 2021, 385 (23), e81 10.1056/NEJMoa2108163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razonable R. R.; Pawlowski C.; O’Horo J. C.; Arndt L. L.; Arndt R.; Bierle D. M.; Borgen M. D.; Hanson S. N.; Hedin M. C.; Lenehan P.; Puranik A.; Seville M. T.; Speicher L. L.; Tulledge-Scheitel S. M.; Venkatakrishnan A.; Wilker C. G.; Badley A. D.; Ganesh R. Casirivimab-Imdevimab Treatment Is Associated with Reduced Rates of Hospitalization among High-Risk Patients with Mild to Moderate Coronavirus Disease-19. EClinicalMedicine 2021, 40, 101102. 10.1016/j.eclinm.2021.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierle D. M.; Ganesh R.; Razonable R. R. Breakthrough COVID-19 and Casirivimab-Imdevimab Treatment during a SARS-CoV-2 B1.617.2 (Delta) Surge. J. Clin. Virol. 2021, 145, 105026. 10.1016/j.jcv.2021.105026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien M. P.; Forleo-Neto E.; Musser B. J.; Isa F.; Chan K.-C.; Sarkar N.; Bar K. J.; Barnabas R. V.; Barouch D. H.; Cohen M. S.; Hurt C. B.; Burwen D. R.; Marovich M. A.; Hou P.; Heirman I.; Davis J. D.; Turner K. C.; Ramesh D.; Mahmood A.; Hooper A. T.; Hamilton J. D.; Kim Y.; Purcell L. A.; Baum A.; Kyratsous C. A.; Krainson J.; Perez-Perez R.; Mohseni R.; Kowal B.; DiCioccio A. T.; Stahl N.; Lipsich L.; Braunstein N.; Herman G.; Yancopoulos G. D.; Weinreich D. M. Subcutaneous REGEN-COV Antibody Combination to Prevent Covid-19. N. Engl. J. Med. 2021, 385 (13), 1184–1195. 10.1056/NEJMoa2109682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V J.; Banu S.; Sasikala M.; Parsa K. V. L.; Sowpati D. T.; Yadav R.; Tallapaka K. B.; Siva A. B.; Vishnubhotla R.; Rao G. V.; Reddy D. N. Effectiveness of REGEN-COV Antibody Cocktail against the B.1.617.2 (Delta) Variant of SARS-CoV-2: A Cohort Study. J. Int. Med. 2022, 291 (3), 380–383. 10.1111/joim.13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B. E.; Brown-Augsburger P. L.; Corbett K. S.; Westendorf K.; Davies J.; Cujec T. P.; Wiethoff C. M.; Blackbourne J. L.; Heinz B. A.; Foster D.; Higgs R. E.; Balasubramaniam D.; Wang L.; Zhang Y.; Yang E. S.; Bidshahri R.; Kraft L.; Hwang Y.; Žentelis S.; Jepson K. R.; Goya R.; Smith M. A.; Collins D. W.; Hinshaw S. J.; Tycho S. A.; Pellacani D.; Xiang P.; Muthuraman K.; Sobhanifar S.; Piper M. H.; Triana F. J.; Hendle J.; Pustilnik A.; Adams A. C.; Berens S. J.; Baric R. S.; Martinez D. R.; Cross R. W.; Geisbert T. W.; Borisevich V.; Abiona O.; Belli H. M.; de Vries M.; Mohamed A.; Dittmann M.; Samanovic M. I.; Mulligan M. J.; Goldsmith J. A.; Hsieh C.-L.; Johnson N. V.; Wrapp D.; McLellan J. S.; Barnhart B. C.; Graham B. S.; Mascola J. R.; Hansen C. L.; Falconer E. The Neutralizing Antibody, LY-CoV555, Protects against SARS-CoV-2 Infection in Nonhuman Primates. Sci. Transl. Med. 2021, 13 (593), eabf1906 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R.; Shan C.; Duan X.; Chen Z.; Liu P.; Song J.; Song T.; Bi X.; Han C.; Wu L.; Gao G.; Hu X.; Zhang Y.; Tong Z.; Huang W.; Liu W. J.; Wu G.; Zhang B.; Wang L.; Qi J.; Feng H.; Wang F.-S.; Wang Q.; Gao G. F.; Yuan Z.; Yan J. A Human Neutralizing Antibody Targets the Receptor-Binding Site of SARS-CoV-2. Nature 2020, 584 (7819), 120–124. 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Wu X.; Li N.; Wang G.; Liu W.; Yu J.; Cao G.; Wang J.; Chen Y.; Ma J.; Wu J.; Yang H.; Mao X.; He J.; Yu Y.; Qiu C.; Li N.; Yao S.; Feng H.; Yan J.; Zhang W.; Zhang J. Tolerability, Safety, Pharmacokinetics, and Immunogenicity of a Novel SARS-CoV-2 Neutralizing Antibody, Etesevimab, in Chinese Healthy Adults: A Randomized, Double-Blind, Placebo-Controlled, First-in-Human Phase 1 Study. Antimicrob. Agents Chemother. 2021, 65 (8), e00350-21 10.1128/AAC.00350-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.; Nirula A.; Heller B.; Gottlieb R. L.; Boscia J.; Morris J.; Huhn G.; Cardona J.; Mocherla B.; Stosor V.; Shawa I.; Adams A. C.; Van Naarden J.; Custer K. L.; Shen L.; Durante M.; Oakley G.; Schade A. E.; Sabo J.; Patel D. R.; Klekotka P.; Skovronsky D. M. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N. Engl. J. Med. 2021, 384 (3), 229–237. 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb R. L.; Nirula A.; Chen P.; Boscia J.; Heller B.; Morris J.; Huhn G.; Cardona J.; Mocherla B.; Stosor V.; Shawa I.; Kumar P.; Adams A. C.; Van Naarden J.; Custer K. L.; Durante M.; Oakley G.; Schade A. E.; Holzer T. R.; Ebert P. J.; Higgs R. E.; Kallewaard N. L.; Sabo J.; Patel D. R.; Klekotka P.; Shen L.; Skovronsky D. M. Effect of Bamlanivimab as Monotherapy or in Combination With Etesevimab on Viral Load in Patients With Mild to Moderate COVID-19. JAMA 2021, 325 (7), 632. 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan M.; Nirula A.; Azizad M.; Mocherla B.; Gottlieb R. L.; Chen P.; Hebert C.; Perry R.; Boscia J.; Heller B.; Morris J.; Crystal C.; Igbinadolor A.; Huhn G.; Cardona J.; Shawa I.; Kumar P.; Adams A. C.; Van Naarden J.; Custer K. L.; Durante M.; Oakley G.; Schade A. E.; Holzer T. R.; Ebert P. J.; Higgs R. E.; Kallewaard N. L.; Sabo J.; Patel D. R.; Dabora M. C.; Klekotka P.; Shen L.; Skovronsky D. M. Bamlanivimab plus Etesevimab in Mild or Moderate Covid-19. N. Engl. J. Med. 2021, 385 (15), 1382–1392. 10.1056/NEJMoa2102685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan M.; Azizad M.; Mocherla B.; Gottlieb R. L.; Chen P.; Hebert C.; Perry R.; Boscia J.; Heller B.; Morris J.; Crystal C.; Igbinadolor A.; Huhn G.; Cardona J.; Shawa I.; Kumar P.; Blomkalns A.; Adams A. C.; Van Naarden J.; Custer K. L.; Knorr J.; Oakley G.; Schade A. E.; Holzer T. R.; Ebert P. J.; Higgs R. E.; Sabo J.; Patel D. R.; Dabora M. C.; Williams M.; Klekotka P.; Shen L.; Skovronsky D. M.; Nirula A. A Randomized, Placebo-Controlled Clinical Trial of Bamlanivimab and Etesevimab Together in High-Risk Ambulatory Patients with COVID-19 and Validation of the Prognostic Value of Persistently High Viral Load. Clin. Infect. Dis. 2021, 10.1093/cid/ciab912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vena A.; Cenderello G.; Balletto E.; Mezzogori L.; Santagostino Barbone A.; Berruti M.; Ball L.; Battaglini D.; Bonsignore A.; Dentone C.; Giacobbe D. R.; Eldin T. K.; Mikulska M.; Rebesco B.; Robba C.; Scintu A.; Stimamiglio A.; Taramasso L.; Pelosi P.; Artioli S.; Bassetti M. Early Administration of Bamlanivimab in Combination with Etesevimab Increases the Benefits of COVID-19 Treatment: Real-World Experience from the Liguria Region. J. Clin. Med. 2021, 10 (20), 4682. 10.3390/jcm10204682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focosi D.; Novazzi F.; Genoni A.; Dentali F.; Gasperina D. D.; Baj A.; Maggi F. Emergence of SARS-COV-2 Spike Protein Escape Mutation Q493R after Treatment for COVID-19. Emerg. Infect. Dis. 2021, 27 (10), 2728–2731. 10.3201/eid2710.211538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora P.; Kempf A.; Nehlmeier I.; Graichen L.; Sidarovich A.; Winkler M. S.; Schulz S.; Jäck H.-M.; Stankov M. V.; Behrens G. M. N.; Pöhlmann S.; Hoffmann M. Delta Variant (B.1.617.2) Sublineages Do Not Show Increased Neutralization Resistance. Cell. Mol. Immunol. 2021, 18 (11), 2557–2559. 10.1038/s41423-021-00772-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurini E.; Marson D.; Aulic S.; Fermeglia A.; Pricl S. Molecular Rationale for SARS-CoV-2 Spike Circulating Mutations Able to Escape Bamlanivimab and Etesevimab Monoclonal Antibodies. Sci. Rep. 2021, 11 (1), 20274. 10.1038/s41598-021-99827-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.; Huynh T.; Stauft C. B.; Wang T. T.; Luan B. Structure-Function Analysis of Resistance to Bamlanivimab by SARS-CoV-2 Variants Kappa, Delta, and Lambda. J. Chem. Inf. Model. 2021, 61 (10), 5133–5140. 10.1021/acs.jcim.1c01058. [DOI] [PubMed] [Google Scholar]
- Guigon A.; Faure E.; Lemaire C.; Chopin M.-C.; Tinez C.; Assaf A.; Lazrek M.; Hober D.; Bocket L.; Engelmann I.; Alidjinou E. K. Emergence of Q493R Mutation in SARS-CoV-2 Spike Protein during Bamlanivimab/Etesevimab Treatment and Resistance to Viral Clearance. J. Infect. 2022, 84 (2), 248–288. 10.1016/j.jinf.2021.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D.; Park Y.-J.; Beltramello M.; Walls A. C.; Tortorici M. A.; Bianchi S.; Jaconi S.; Culap K.; Zatta F.; De Marco A.; Peter A.; Guarino B.; Spreafico R.; Cameroni E.; Case J. B.; Chen R. E.; Havenar-Daughton C.; Snell G.; Telenti A.; Virgin H. W.; Lanzavecchia A.; Diamond M. S.; Fink K.; Veesler D.; Corti D. Cross-Neutralization of SARS-CoV-2 by a Human Monoclonal SARS-CoV Antibody. Nature 2020, 583 (7815), 290–295. 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Cathcart A. L. The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. bioRxiv 2021, 10.1101/2021.03.09.434607. [DOI] [Google Scholar]
- Gupta A.; Gonzalez-Rojas Y.; Juarez E.; Crespo Casal M.; Moya J.; Falci D. R.; Sarkis E.; Solis J.; Zheng H.; Scott N.; Cathcart A. L.; Hebner C. M.; Sager J.; Mogalian E.; Tipple C.; Peppercorn A.; Alexander E.; Pang P. S.; Free A.; Brinson C.; Aldinger M.; Shapiro A. E. Early Treatment for Covid-19 with SARS-CoV-2 Neutralizing Antibody Sotrovimab. N. Engl. J. Med. 2021, 385 (21), 1941–1950. 10.1056/NEJMoa2107934. [DOI] [PubMed] [Google Scholar]
- Kim C.; Ryu D.-K.; Lee J.; Kim Y.-I.; Seo J.-M.; Kim Y.-G.; Jeong J.-H.; Kim M.; Kim J.-I.; Kim P.; Bae J. S.; Shim E. Y.; Lee M. S.; Kim M. S.; Noh H.; Park G.-S.; Park J. S.; Son D.; An Y.; Lee J. N.; Kwon K.-S.; Lee J.-Y.; Lee H.; Yang J.-S.; Kim K.-C.; Kim S. S.; Woo H.-M.; Kim J.-W.; Park M.-S.; Yu K.-M.; Kim S.-M.; Kim E.-H.; Park S.-J.; Jeong S. T.; Yu C. H.; Song Y.; Gu S. H.; Oh H.; Koo B.-S.; Hong J. J.; Ryu C.-M.; Park W. B.; Oh M.; Choi Y. K.; Lee S.-Y. A Therapeutic Neutralizing Antibody Targeting Receptor Binding Domain of SARS-CoV-2 Spike Protein. Nat. Commun. 2021, 12 (1), 288. 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu D.-K.; Kang B.; Noh H.; Woo S.-J.; Lee M.-H.; Nuijten P. M.; Kim J.-I.; Seo J.-M.; Kim C.; Kim M.; Yang E.; Lim G.; Kim S.-G.; Eo S.-K.; Choi J.; Song M.; Oh S.-S.; Chung H.-Y.; Tijsma A. S.; van Baalen C. A.; Kwon K.-S.; Lee S.-Y. The in Vitro and in Vivo Efficacy of CT-P59 against Gamma, Delta and Its Associated Variants of SARS-CoV-2. Biochem. Biophys. Res. Commun. 2021, 578, 91–96. 10.1016/j.bbrc.2021.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu D.-K.; Song R.; Kim M.; Kim Y.-I.; Kim C.; Kim J.-I.; Kwon K.-S.; Tijsma A. S.; Nuijten P. M.; van Baalen C. A.; Hermanus T.; Kgagudi P.; Moyo-Gwete T.; Moore P. L.; Choi Y. K.; Lee S.-Y. Therapeutic Effect of CT-P59 against SARS-CoV-2 South African Variant. Biochem. Biophys. Res. Commun. 2021, 566, 135–140. 10.1016/j.bbrc.2021.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Y.; Jang Y. R.; Hong J. H.; Jung J. G.; Park J.-H.; Streinu-Cercel A.; Streinu-Cercel A.; Săndulescu O.; Lee S. J.; Kim S. H.; Jung N. H.; Lee S. G.; Park J. E.; Kim M. K.; Jeon D. B.; Lee Y. J.; Kim B. S.; Lee Y. M.; Kim Y.-S. Safety, Virologic Efficacy, and Pharmacokinetics of CT-P59, a Neutralizing Monoclonal Antibody Against SARS-CoV-2 Spike Receptor-Binding Protein: Two Randomized, Placebo-Controlled, Phase I Studies in Healthy Individuals and Patients With Mild SARS-CoV-2. Clin. Ther. 2021, 43 (10), 1706–1727. 10.1016/j.clinthera.2021.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechman K.; Subesinghe S.; Norton S.; Atzeni F.; Galli M.; Cope A. P.; Winthrop K. L.; Galloway J. B. A Systematic Review and Meta-Analysis of Infection Risk with Small Molecule JAK Inhibitors in Rheumatoid Arthritis. Rheumatology 2019, 58 (10), 1755–1766. 10.1093/rheumatology/kez087. [DOI] [PubMed] [Google Scholar]
- Richardson P.; Griffin I.; Tucker C.; Smith D.; Oechsle O.; Phelan A.; Rawling M.; Savory E.; Stebbing J. Baricitinib as Potential Treatment for 2019-NCoV Acute Respiratory Disease. Lancet 2020, 395 (10223), e30–e31. 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing J.; Phelan A.; Griffin I.; Tucker C.; Oechsle O.; Smith D.; Richardson P. COVID-19: Combining Antiviral and Anti-Inflammatory Treatments. Lancet Infect. Dis. 2020, 20 (4), 400–402. 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrone L.; Petruccioli E.; Alonzi T.; Vanini V.; Cuzzi G.; Najafi Fard S.; Castilletti C.; Palmieri F.; Gualano G.; Vittozzi P.; Nicastri E.; Lepore L.; Grifoni A.; Antinori A.; Vergori A.; Ippolito G.; Cantini F.; Goletti D. In-Vitro Evaluation of the Immunomodulatory Effects of Baricitinib: Implication for COVID-19 Therapy. J. Infect. 2021, 82 (4), 58–66. 10.1016/j.jinf.2021.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing J.; Sánchez Nievas G.; Falcone M.; Youhanna S.; Richardson P.; Ottaviani S.; Shen J. X.; Sommerauer C.; Tiseo G.; Ghiadoni L.; Virdis A.; Monzani F.; Rizos L. R.; Forfori F.; Avendaño Céspedes A.; De Marco S.; Carrozzi L.; Lena F.; Sánchez-Jurado P. M.; Lacerenza L. G.; Cesira N.; Caldevilla Bernardo D.; Perrella A.; Niccoli L.; Méndez L. S.; Matarrese D.; Goletti D.; Tan Y.-J.; Monteil V.; Dranitsaris G.; Cantini F.; Farcomeni A.; Dutta S.; Burley S. K.; Zhang H.; Pistello M.; Li W.; Romero M. M.; Andrés Pretel F.; Simón-Talero R. S.; García-Molina R.; Kutter C.; Felce J. H.; Nizami Z. F.; Miklosi A. G.; Penninger J. M.; Menichetti F.; Mirazimi A.; Abizanda P.; Lauschke V. M. JAK Inhibition Reduces SARS-CoV-2 Liver Infectivity and Modulates Inflammatory Responses to Reduce Morbidity and Mortality. Sci. Adv. 2021, 7 (1), eabe4724 10.1126/sciadv.abe4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T. N.; Pino M.; Boddapati A. K.; Viox E. G.; Starke C. E.; Upadhyay A. A.; Gumber S.; Nekorchuk M.; Busman-Sahay K.; Strongin Z.; Harper J. L.; Tharp G. K.; Pellegrini K. L.; Kirejczyk S.; Zandi K.; Tao S.; Horton T. R.; Beagle E. N.; Mahar E. A.; Lee M. Y. H.; Cohen J.; Jean S. M.; Wood J. S.; Connor-Stroud F.; Stammen R. L.; Delmas O. M.; Wang S.; Cooney K. A.; Sayegh M. N.; Wang L.; Filev P. D.; Weiskopf D.; Silvestri G.; Waggoner J.; Piantadosi A.; Kasturi S. P.; Al-Shakhshir H.; Ribeiro S. P.; Sekaly R. P.; Levit R. D.; Estes J. D.; Vanderford T. H.; Schinazi R. F.; Bosinger S. E.; Paiardini M. Baricitinib Treatment Resolves Lower-Airway Macrophage Inflammation and Neutrophil Recruitment in SARS-CoV-2-Infected Rhesus Macaques. Cell 2021, 184 (2), 460–475. 10.1016/j.cell.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantini F.; Niccoli L.; Matarrese D.; Nicastri E.; Stobbione P.; Goletti D. Baricitinib Therapy in COVID-19: A Pilot Study on Safety and Clinical Impact. J. Infect. 2020, 81 (2), 318–356. 10.1016/j.jinf.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconi V. C.; Ramanan A. V.; de Bono S.; Kartman C. E.; Krishnan V.; Liao R.; Piruzeli M. L. B.; Goldman J. D.; Alatorre-Alexander J.; de Cassia Pellegrini R.; Estrada V.; Som M.; Cardoso A.; Chakladar S.; Crowe B.; Reis P.; Zhang X.; Adams D. H.; Ely E. W.; Ahn M.-Y.; Akasbi M.; Alatorre-Alexander J.; Altclas J. D.; Ariel F.; Ariza H. A.; Atkar C.; Bertetti A.; Bhattacharya M.; Briones M. L.; Budhraja A.; Burza A.; Camacho Ortiz A.; Caricchio R.; Casas M.; Cevoli Recio V.; Choi W. S.; Cohen E.; Comulada-Rivera A.; Cook P.; Cornejo Juarez D. P.; Daniel C.; Degrecci Relvas L. F.; Dominguez Cherit J. G.; Ellerin T.; Enikeev D.; Erico Tanni Minamoto S.; Estrada V.; Fiss E.; Furuichi M.; Giovanni Luz K.; Goldman J. D.; Gonzalez O.; Gordeev I.; Gruenewald T.; Hamamoto Sato V. A.; Heo E. Y.; Heo J. Y.; Hermida M.; Hirai Y.; Hutchinson D.; Iastrebner C.; Ioachimescu O.; Jain M.; Juliani Souza Lima M. P.; Khan A.; Kremer A. E.; Lawrie T.; MacElwee M.; Madhani-Lovely F.; Malhotra V.; Martínez Resendez M. F.; McKinnell J.; Milligan P.; Minelli C.; Moran Rodriguez M. A.; Parody M. L.; Paulin P.; Pellegrini R. de C.; Pemu P.; Procopio Carvalho A. C.; Puoti M.; Purow J.; Ramesh M.; Rea Neto A.; Rea Neto A.; Robinson P.; Rodrigues C.; Rojas Velasco G.; Saraiva J. F. K.; Scheinberg M.; Schreiber S.; Scublinsky D.; Sevciovic Grumach A.; Shawa I.; Simon Campos J.; Sofat N.; Som M.; Spinner C. D.; Sprinz E.; Stienecker R.; Suarez J.; Tachikawa N.; Tahir H.; Tiffany B.; Vishnevsky A.; Westheimer Cavalcante A.; Zirpe K. Efficacy and Safety of Baricitinib for the Treatment of Hospitalised Adults with COVID-19 (COV-BARRIER): A Randomised, Double-Blind, Parallel-Group, Placebo-Controlled Phase 3 Trial. Lancet Respir. Med. 2021, 9 (12), 1407–1418. 10.1016/S2213-2600(21)00331-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalil A. C.; Patterson T. F.; Mehta A. K.; Tomashek K. M.; Wolfe C. R.; Ghazaryan V.; Marconi V. C.; Ruiz-Palacios G. M.; Hsieh L.; Kline S.; Tapson V.; Iovine N. M.; Jain M. K.; Sweeney D. A.; El Sahly H. M.; Branche A. R.; Regalado Pineda J.; Lye D. C.; Sandkovsky U.; Luetkemeyer A. F.; Cohen S. H.; Finberg R. W.; Jackson P. E. H.; Taiwo B.; Paules C. I.; Arguinchona H.; Erdmann N.; Ahuja N.; Frank M.; Oh M.; Kim E.-S.; Tan S. Y.; Mularski R. A.; Nielsen H.; Ponce P. O.; Taylor B. S.; Larson L.; Rouphael N. G.; Saklawi Y.; Cantos V. D.; Ko E. R.; Engemann J. J.; Amin A. N.; Watanabe M.; Billings J.; Elie M.-C.; Davey R. T.; Burgess T. H.; Ferreira J.; Green M.; Makowski M.; Cardoso A.; de Bono S.; Bonnett T.; Proschan M.; Deye G. A.; Dempsey W.; Nayak S. U.; Dodd L. E.; Beigel J. H. Baricitinib plus Remdesivir for Hospitalized Adults with Covid-19. N. Engl. J. Med. 2021, 384 (9), 795–807. 10.1056/NEJMoa2031994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z.; Niu J.; Xu Y.; Qin L.; Ding J.; Zhou L. Clinical Efficacy and Adverse Events of Baricitinib Treatment for Coronavirus Disease-2019 (COVID-19): A Systematic Review and Meta-analysis. J. Med. Virol. 2022, 94 (4), 1523–1534. 10.1002/jmv.27482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K.; Tsuchiya M.; Saldanha J.; Koishihara Y.; Ohsugi Y.; Kishimoto T.; Bendig M. M. Reshaping a human antibody to inhibit the interleukin 6-dependent tumor cell growth. Cancer Res. 1993, 53 (4), 851–856. [PubMed] [Google Scholar]
- Mihara M.; Kasutani K.; Okazaki M.; Nakamura A.; Kawai S.; Sugimoto M.; Matsumoto Y.; Ohsugi Y. Tocilizumab Inhibits Signal Transduction Mediated by Both MIL-6R and SIL-6R, but Not by the Receptors of Other Members of IL-6 Cytokine Family. Int. Immunopharmacol. 2005, 5 (12), 1731–1740. 10.1016/j.intimp.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Amarilyo G.; Tarp S.; Foeldvari I.; Cohen N.; Pope T. D.; Woo J. M. P.; Christensen R.; Furst D. E. Biological Agents in Polyarticular Juvenile Idiopathic Arthritis: A Meta-Analysis of Randomized Withdrawal Trials. Semin. Arthritis Rheum. 2016, 46 (3), 312–318. 10.1016/j.semarthrit.2016.07.001. [DOI] [PubMed] [Google Scholar]
- Gérard A.-L.; Simon-Tillaux N.; Yordanov Y.; Cacoub P.; Tubach F.; Saadoun D.; Dechartres A. Efficacy and Safety of Steroid-Sparing Treatments in Giant Cell Arteritis According to the Glucocorticoids Tapering Regimen: A Systematic Review and Meta-Analysis. Eur. J. Int. Med. 2021, 88, 96–103. 10.1016/j.ejim.2021.03.040. [DOI] [PubMed] [Google Scholar]
- Navarro G.; Taroumian S.; Barroso N.; Duan L.; Furst D. Tocilizumab in Rheumatoid Arthritis: A Meta-Analysis of Efficacy and Selected Clinical Conundrums. Semin. Arthritis Rheum. 2014, 43 (4), 458–469. 10.1016/j.semarthrit.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Si S.; Teachey D. T. Spotlight on Tocilizumab in the Treatment of CAR-T-Cell-Induced Cytokine Release Syndrome: Clinical Evidence to Date. Ther. Clin. Risk Manage. 2020, 16, 705–714. 10.2147/TCRM.S223468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Zhong Y.; Pan L.; Dong J. Treat 2019 Novel Coronavirus (COVID-19) with IL-6 Inhibitor: Are We Already That Far?. Drug Discovery Ther. 2020, 14 (2), 100–102. 10.5582/ddt.2020.03006. [DOI] [PubMed] [Google Scholar]
- Gu T.; Zhao S.; Jin G.; Song M.; Zhi Y.; Zhao R.; Ma F.; Zheng Y.; Wang K.; Liu H.; Xin M.; Han W.; Li X.; Dong C. D.; Liu K.; Dong Z. Cytokine Signature Induced by SARS-CoV-2 Spike Protein in a Mouse Model. Front. Immunol. 2021, 11, 621441 10.3389/fimmu.2020.621441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J. H.; Frigault M. J.; Serling-Boyd N. J.; Fernandes A. D.; Harvey L.; Foulkes A. S.; Horick N. K.; Healy B. C.; Shah R.; Bensaci A. M.; Woolley A. E.; Nikiforow S.; Lin N.; Sagar M.; Schrager H.; Huckins D. S.; Axelrod M.; Pincus M. D.; Fleisher J.; Sacks C. A.; Dougan M.; North C. M.; Halvorsen Y.-D.; Thurber T. K.; Dagher Z.; Scherer A.; Wallwork R. S.; Kim A. Y.; Schoenfeld S.; Sen P.; Neilan T. G.; Perugino C. A.; Unizony S. H.; Collier D. S.; Matza M. A.; Yinh J. M.; Bowman K. A.; Meyerowitz E.; Zafar A.; Drobni Z. D.; Bolster M. B.; Kohler M.; D’Silva K. M.; Dau J.; Lockwood M. M.; Cubbison C.; Weber B. N.; Mansour M. K. Efficacy of Tocilizumab in Patients Hospitalized with Covid-19. N. Engl. J. Med. 2020, 383 (24), 2333–2344. 10.1056/NEJMoa2028836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama C.; Han J.; Yau L.; Reiss W. G.; Kramer B.; Neidhart J. D.; Criner G. J.; Kaplan-Lewis E.; Baden R.; Pandit L.; Cameron M. L.; Garcia-Diaz J.; Chávez V.; Mekebeb-Reuter M.; Lima de Menezes F.; Shah R.; González-Lara M. F.; Assman B.; Freedman J.; Mohan S. V. Tocilizumab in Patients Hospitalized with Covid-19 Pneumonia. N. Engl. J. Med. 2021, 384 (1), 20–30. 10.1056/NEJMoa2030340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas I. O.; Bräu N.; Waters M.; Go R. C.; Hunter B. D.; Bhagani S.; Skiest D.; Aziz M. S.; Cooper N.; Douglas I. S.; Savic S.; Youngstein T.; Del Sorbo L.; Cubillo Gracian A.; De La Zerda D. J.; Ustianowski A.; Bao M.; Dimonaco S.; Graham E.; Matharu B.; Spotswood H.; Tsai L.; Malhotra A. Tocilizumab in Hospitalized Patients with Severe Covid-19 Pneumonia. N. Engl. J. Med. 2021, 384 (16), 1503–1516. 10.1056/NEJMoa2028700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tleyjeh I. M.; Kashour Z.; Damlaj M.; Riaz M.; Tlayjeh H.; Altannir M.; Altannir Y.; Al-Tannir M.; Tleyjeh R.; Hassett L.; Kashour T. Efficacy and Safety of Tocilizumab in COVID-19 Patients: A Living Systematic Review and Meta-Analysis. Clin. Microbiol. Infect. 2021, 27 (2), 215–227. 10.1016/j.cmi.2020.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annane D. Corticosteroids for COVID-19. J. Intensive Med. 2021, 1 (1), 14–25. 10.1016/j.jointm.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain D. W.; Cidlowski J. A. Immune Regulation by Glucocorticoids. Nat. Rev. Immunol. 2017, 17 (4), 233–247. 10.1038/nri.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polidoro R. B.; Hagan R. S.; de Santis Santiago R.; Schmidt N. W. Overview: Systemic Inflammatory Response Derived From Lung Injury Caused by SARS-CoV-2 Infection Explains Severe Outcomes in COVID-19. Front. Immunol. 2020, 11, 1626 10.3389/fimmu.2020.01626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomazini B. M.; Maia I. S.; Cavalcanti A. B.; Berwanger O.; Rosa R. G.; Veiga V. C.; Avezum A.; Lopes R. D.; Bueno F. R.; Silva M. V. A. O.; Baldassare F. P.; Costa E. L. V.; Moura R. A. B.; Honorato M. O.; Costa A. N.; Damiani L. P.; Lisboa T.; Kawano-Dourado L.; Zampieri F. G.; Olivato G. B.; Righy C.; Amendola C. P.; Roepke R. M. L.; Freitas D. H. M.; Forte D. N.; Freitas F. G. R.; Fernandes C. C. F.; Melro L. M. G.; Junior G. F. S.; Morais D. C.; Zung S.; Machado F. R.; Azevedo L. C. P. Effect of Dexamethasone on Days Alive and Ventilator-Free in Patients With Moderate or Severe Acute Respiratory Distress Syndrome and COVID-19. JAMA 2020, 324 (13), 1307. 10.1001/jama.2020.17021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P.; Lim W. S.; Emberson J. R.; Mafham M.; Bell J. L.; Linsell L.; Staplin N.; Brightling C.; Ustianowski A.; Elmahi E.; et al. Dexamethasone in Hospitalized Patients with Covid-19. N. Engl. J. Med. 2021, 384 (8), 693–704. 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral-Gudino L.; Bahamonde A.; Arnaiz-Revillas F.; Gómez-Barquero J.; Abadía-Otero J.; García-Ibarbia C.; Mora V.; Cerezo-Hernández A.; Hernández J. L.; López-Muñíz G.; Hernández-Blanco F.; Cifrián J. M.; Olmos J. M.; Carrascosa M.; Nieto L.; Fariñas M. C.; Riancho J. A. Methylprednisolone in Adults Hospitalized with COVID-19 Pneumonia. Wien. Klin. Wochenschr. 2021, 133 (7-8), 303–311. 10.1007/s00508-020-01805-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C. M. P.; Farias M. E. L.; Val F. F. A.; Sampaio V. S.; Alexandre M. A. A.; Melo G. C.; Safe I. P.; Borba M. G. S.; Netto R. L. A.; Maciel A. B. S.; Neto J. R. S.; Oliveira L. B.; Figueiredo E. F. G.; Oliveira Dinelly K. M.; de Almeida Rodrigues M. G.; Brito M.; Mourão M. P. G.; Pivoto João G. A.; Hajjar L. A.; Bassat Q.; Romero G. A. S.; Naveca F. G.; Vasconcelos H. L.; de Araújo Tavares M.; Brito-Sousa J. D.; Costa F. T. M.; Nogueira M. L.; Baía-da-Silva D. C.; Xavier M. S.; Monteiro W. M.; Lacerda M. V. G.; de Lemos Vasconcelos A.; Praia Marins A. F.; de Oliveira Trindade A.; Mendes Záu A. S.; de Oliveira A. C.; Azevedo Furtado A. C.; Coelho Rocha A. P.; da Silva Souza A.; de Souza Dias A.; Belém A.; dos Santos A. G. R.; da Silva Sousa A. M.; da Silva B. F.; Franco B. L.; da Silva B. M.; da Costa B. L. G.; Sato Barros do Amaral C. M. S.; Judice C. C.; de Morais C. E. P.; Camilo C. C.; Sena da Silva D. S.; Gomes Duarte D. C.; da Silva E. G. N.; da Silva Lemos E.; de Fátima Ponte Frota E.; do Nascimento E. F.; de Almeida E. S.; Marques E. A.; de Almeida E. M. M.; da Silva E. L.; dos Santos E. G.; da Silva Oliveira E.; Martins Shimizu F. M.; de Souza F. R. F.; da Silva do Vale F.; dos Santos de Almeida Lima F.; da Fonseca F. H. J.; Fontenelle F. A.; de Azevedo Furtado F.; Da Silva Pereira G.; Bezerra G. A.; Maciel Salazar G. K.; da Silva Pereira H.; de Melo H. F.; Oliveira I. N.; Pereira Filho I. V.; Gomes J. V.; e Silva Rosa J.; Lemos J. M.; Brutus J. N.; Pessoa K. P.; Costa Rodrigues L. D.; Barros Cirino L. E.; Mourão Filho L. F.; Moura L.; Barbosa L. R. P.; de Souza L. P.; Oliveira L. B.; de Lima Ferreira L. C.; dos Santos M. M.; da Silva M. V. R.; Rodrigues M. P.; de Menezes M. T.; dos Santos Mota M. M.; Freire M.; Corrêa N. F.; Rocha N. M.; Bittencourt N.; de Melo Silva N. G.; de Oliveira Saraiva P.; de Sousa Monteiro Q.; dos Santos R. T.; Freire R. S.; de Araújo Pinto R. A.; Ferreira R. B.; de Lima R. S.; de Melo R. F. T.; Saenz S. T.; Alvarez Fernandes S. S.; Vítor-Silva S.; de Oliveira T. M. R.; Tavella T. A.; Câmara T. T.; Santos T. C.; Pinto T. S.; dos Santos T. W. R.; do Nascimento V. A.; Sousa Barbosa W. P.; de Melo W. F.; Salgado Sobrinho W. B. Methylprednisolone as Adjunctive Therapy for Patients Hospitalized With Coronavirus Disease 2019 (COVID-19; Metcovid): A Randomized, Double-Blind, Phase IIb, Placebo-Controlled Trial. Clin. Infect. Dis. 2021, 72 (9), e373–e381. 10.1093/cid/ciaa1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G.-Q.; Jiang Z.-H.; Yang Z.-B.; Jiang S.-Q.; Quan X.-Q. The Effect of Glucocorticoids on Mortality in Severe COVID-19 Patients. Medicine (Baltimore). 2021, 100 (40), e27373 10.1097/MD.0000000000027373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzón M. A.; Ortiz S.; Holguín H.; Betancur J. F.; Cardona Arango D.; Laniado H.; Arias Arias C.; Muñoz B.; Quiceno J.; Jaramillo D.; Ramirez Z. Dexamethasone vs Methylprednisolone High Dose for Covid-19 Pneumonia. PLoS One 2021, 16 (5), e0252057 10.1371/journal.pone.0252057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjbar K.; Moghadami M.; Mirahmadizadeh A.; Fallahi M. J.; Khaloo V.; Shahriarirad R.; Erfani A.; Khodamoradi Z.; Saadi M. H. G. Correction to: Methylprednisolone or Dexamethasone, Which One Is Superior Corticosteroid in the Treatment of Hospitalized COVID-19 Patients: A Triple-Blinded Randomized Controlled Trial. BMC Infect. Dis. 2021, 21 (1), 436. 10.1186/s12879-021-06130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S. J.; Gilchuk P.; Chen R. E.; Case J. B.; Reidy J. X.; Trivette A.; Nargi R. S.; Sutton R. E.; Suryadevara N.; Chen E. C.; Binshtein E.; Shrihari S.; Ostrowski M.; Chu H. Y.; Didier J. E.; MacRenaris K. W.; Jones T.; Day S.; Myers L.; Eun-Hyung Lee F.; Nguyen D. C.; Sanz I.; Martinez D. R.; Rothlauf P. W.; Bloyet L.-M.; Whelan S. P. J.; Baric R. S.; Thackray L. B.; Diamond M. S.; Carnahan R. H.; Crowe J. E. Rapid Isolation and Profiling of a Diverse Panel of Human Monoclonal Antibodies Targeting the SARS-CoV-2 Spike Protein. Nat. Med. 2020, 26 (9), 1422–1427. 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S. J.; Gilchuk P.; Case J. B.; Binshtein E.; Chen R. E.; Nkolola J. P.; Schäfer A.; Reidy J. X.; Trivette A.; Nargi R. S.; Sutton R. E.; Suryadevara N.; Martinez D. R.; Williamson L. E.; Chen E. C.; Jones T.; Day S.; Myers L.; Hassan A. O.; Kafai N. M.; Winkler E. S.; Fox J. M.; Shrihari S.; Mueller B. K.; Meiler J.; Chandrashekar A.; Mercado N. B.; Steinhardt J. J.; Ren K.; Loo Y.-M.; Kallewaard N. L.; McCune B. T.; Keeler S. P.; Holtzman M. J.; Barouch D. H.; Gralinski L. E.; Baric R. S.; Thackray L. B.; Diamond M. S.; Carnahan R. H.; Crowe J. E. Potently Neutralizing and Protective Human Antibodies against SARS-CoV-2. Nature 2020, 584 (7821), 443–449. 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J.; Zost S. J.; Greaney A. J.; Starr T. N.; Dingens A. S.; Chen E. C.; Chen R. E.; Case J. B.; Sutton R. E.; Gilchuk P.; Rodriguez J.; Armstrong E.; Gainza C.; Nargi R. S.; Binshtein E.; Xie X.; Zhang X.; Shi P.-Y.; Logue J.; Weston S.; McGrath M. E.; Frieman M. B.; Brady T.; Tuffy K. M.; Bright H.; Loo Y.-M.; McTamney P. M.; Esser M. T.; Carnahan R. H.; Diamond M. S.; Bloom J. D.; Crowe J. E. Genetic and Structural Basis for SARS-CoV-2 Variant Neutralization by a Two-Antibody Cocktail. Nat. Microbiol. 2021, 6 (10), 1233–1244. 10.1038/s41564-021-00972-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toots M.; Yoon J.-J.; Cox R. M.; Hart M.; Sticher Z. M.; Makhsous N.; Plesker R.; Barrena A. H.; Reddy P. G.; Mitchell D. G.; Shean R. C.; Bluemling G. R.; Kolykhalov A. A.; Greninger A. L.; Natchus M. G.; Painter G. R.; Plemper R. K. Characterization of Orally Efficacious Influenza Drug with High Resistance Barrier in Ferrets and Human Airway Epithelia. Sci. Transl. Med. 2019, 11 (515), eaax5866 10.1126/scitranslmed.aax5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenke K.; Hansen F.; Schwarz B.; Feldmann F.; Haddock E.; Rosenke R.; Barbian K.; Meade-White K.; Okumura A.; Leventhal S.; Hawman D. W.; Ricotta E.; Bosio C. M.; Martens C.; Saturday G.; Feldmann H.; Jarvis M. A. Orally Delivered MK-4482 Inhibits SARS-CoV-2 Replication in the Syrian Hamster Model. Nat. Commun. 2021, 12 (1), 2295. 10.1038/s41467-021-22580-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T. P.; Sims A. C.; Zhou S.; Graham R. L.; Pruijssers A. J.; Agostini M. L.; Leist S. R.; Schäfer A.; Dinnon K. H.; Stevens L. J.; Chappell J. D.; Lu X.; Hughes T. M.; George A. S.; Hill C. S.; Montgomery S. A.; Brown A. J.; Bluemling G. R.; Natchus M. G.; Saindane M.; Kolykhalov A. A.; Painter G.; Harcourt J.; Tamin A.; Thornburg N. J.; Swanstrom R.; Denison M. R.; Baric R. S. An Orally Bioavailable Broad-Spectrum Antiviral Inhibits SARS-CoV-2 in Human Airway Epithelial Cell Cultures and Multiple Coronaviruses in Mice. Sci. Transl. Med. 2020, 12 (541), eabb5883 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C. J.; Tchesnokov E. P.; Schinazi R. F.; Götte M. Molnupiravir Promotes SARS-CoV-2 Mutagenesis via the RNA Template. J. Biol. Chem. 2021, 297 (1), 100770. 10.1016/j.jbc.2021.100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl A.; Gralinski L. E.; Johnson C. E.; Yao W.; Kovarova M.; Dinnon K. H.; Liu H.; Madden V. J.; Krzystek H. M.; De C.; White K. K.; Gully K.; Schäfer A.; Zaman T.; Leist S. R.; Grant P. O.; Bluemling G. R.; Kolykhalov A. A.; Natchus M. G.; Askin F. B.; Painter G.; Browne E. P.; Jones C. D.; Pickles R. J.; Baric R. S.; Garcia J. V. SARS-CoV-2 Infection Is Effectively Treated and Prevented by EIDD-2801. Nature 2021, 591 (7850), 451–457. 10.1038/s41586-021-03312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. M.; Wolf J. D.; Plemper R. K. Therapeutically Administered Ribonucleoside Analogue MK-4482/EIDD-2801 Blocks SARS-CoV-2 Transmission in Ferrets. Nat. Microbiol. 2021, 6 (1), 11–18. 10.1038/s41564-020-00835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelnabi R.; Foo C. S.; De Jonghe S.; Maes P.; Weynand B.; Neyts J. Molnupiravir Inhibits Replication of the Emerging SARS-CoV-2 Variants of Concern in a Hamster Infection Model. J. Infect. Dis. 2021, 224 (5), 749–753. 10.1093/infdis/jiab361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter W. P.; Holman W.; Bush J. A.; Almazedi F.; Malik H.; Eraut N. C. J. E.; Morin M. J.; Szewczyk L. J.; Painter G. R. Human Safety, Tolerability, and Pharmacokinetics of Molnupiravir, a Novel Broad-Spectrum Oral Antiviral Agent with Activity against SARS-CoV-2. Antimicrob. Agents Chemother. 2021, 65 (5), e02428-20 10.1128/AAC.02428-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Fang C.; Zhang Q.; Zhang R.; Zhao X.; Duan Y.; Wang H.; Zhu Y.; Feng L.; Zhao J.; Shao M.; Yang X.; Zhang L.; Peng C.; Yang K.; Ma D.; Rao Z.; Yang H. Crystal Structure of SARS-CoV-2 Main Protease in Complex with Protease Inhibitor PF-07321332. Protein Cell 2022, 13 (9), 689–693. 10.1007/s13238-021-00883-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J.; Huang A.; Li Z.; Tufekci Z.; Zdimal V.; van der Westhuizen H.-M.; von Delft A.; Price A.; Fridman L.; Tang L.-H.; Tang V.; Watson G. L.; Bax C. E.; Shaikh R.; Questier F.; Hernandez D.; Chu L. F.; Ramirez C. M.; Rimoin A. W. An Evidence Review of Face Masks against COVID-19. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (4), e2014564118 10.1073/pnas.2014564118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal T. A Review of Coronavirus Disease-2019 (COVID-19). Indian J. Pediatr. 2020, 87 (April), 281–286. 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.; Ma N.; Witt C.; Rapp S.; Wild P. S.; Andreae M. O.; Pöschl U.; Su H. Face Masks Effectively Limit the Probability of SARS-CoV-2 Transmission. Science 2021, 372 (6549), 1439–1443. 10.1126/science.abg6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M. P.; Hall A. J. Norovirus Illnesses in Children and Adolescents. Infect. Dis. Clin. N. Am. 2018, 32, 103–118. 10.1016/j.idc.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungaro R. C.; Sullivan T.; Colombel J.-F.; Patel G. What Should Gastroenterologists and Patients Know About COVID-19?. Clin. Gastroenterol. Hepatol. 2020, 18 (7), 1409–1411. 10.1016/j.cgh.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiolet T.; Kherabi Y.; MacDonald C.-J.; Ghosn J.; Peiffer-Smadja N. Comparing COVID-19 Vaccines for Their Characteristics, Efficacy and Effectiveness against SARS-CoV-2 and Variants of Concern: A Narrative Review. Clin. Microbiol. Infect. 2022, 28 (2), 202–221. 10.1016/j.cmi.2021.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.; Li Y.; Dai L.; Wang J.; He P.; Li C.; Fang X.; Wang C.; Zhao X.; Huang E.; Wu C.; Zhong Z.; Wang F.; Duan X.; Tian S.; Wu L.; Liu Y.; Luo Y.; Chen Z.; Li F.; Li J.; Yu X.; Ren H.; Liu L.; Meng S.; Yan J.; Hu Z.; Gao L.; Gao G. F. Safety and Immunogenicity of a Recombinant Tandem-Repeat Dimeric RBD-Based Protein Subunit Vaccine (ZF2001) against COVID-19 in Adults: Two Randomised, Double-Blind, Placebo-Controlled, Phase 1 and 2 Trials. Lancet Infect. Dis. 2021, 21 (8), 1107–1119. 10.1016/S1473-3099(21)00127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes L.; Zuñiga S.; Castaño-Rodriguez C.; Gutierrez-Alvarez J.; Canton J.; Sola I. Molecular Basis of Coronavirus Virulence and Vaccine Development. Adv. Virus Res. 2016, 96, 245–286. 10.1016/bs.aivir.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz F. X.; Stiasny K. Distinguishing Features of Current COVID-19 Vaccines: Knowns and Unknowns of Antigen Presentation and Modes of Action. npj Vaccines 2021, 6 (1), 104. 10.1038/s41541-021-00369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakidis N. C.; López-Cortés A.; González E. V.; Grimaldos A. B.; Prado E. O. SARS-CoV-2 Vaccines Strategies: A Comprehensive Review of Phase 3 Candidates. npj Vaccines 2021, 6 (1), 28. 10.1038/s41541-021-00292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados-Riveron J. T.; Aquino-Jarquin G. Engineering of the Current Nucleoside-Modified MRNA-LNP Vaccines against SARS-CoV-2. Biomed. Pharmacother. 2021, 142, 111953. 10.1016/j.biopha.2021.111953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F. P.; Thomas S. J.; Kitchin N.; Absalon J.; Gurtman A.; Lockhart S.; Perez J. L.; Pérez Marc G.; Moreira E. D.; Zerbini C.; Bailey R.; Swanson K. A.; Roychoudhury S.; Koury K.; Li P.; Kalina W. V.; Cooper D.; Frenck R. W.; Hammitt L. L.; Türeci Ö.; Nell H.; Schaefer A.; Ünal S.; Tresnan D. B.; Mather S.; Dormitzer P. R.; Şahin U.; Jansen K. U.; Gruber W. C. Safety and Efficacy of the BNT162b2MRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383 (27), 2603–2615. 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan N.; Barda N.; Kepten E.; Miron O.; Perchik S.; Katz M. A.; Hernán M. A.; Lipsitch M.; Reis B.; Balicer R. D. BNT162b2MRNA Covid-19 Vaccine in a Nationwide Mass Vaccination Setting. N. Engl. J. Med. 2021, 384 (15), 1412–1423. 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L. R.; El Sahly H. M.; Essink B.; Kotloff K.; Frey S.; Novak R.; Diemert D.; Spector S. A.; Rouphael N.; Creech C. B.; McGettigan J.; Khetan S.; Segall N.; Solis J.; Brosz A.; Fierro C.; Schwartz H.; Neuzil K.; Corey L.; Gilbert P.; Janes H.; Follmann D.; Marovich M.; Mascola J.; Polakowski L.; Ledgerwood J.; Graham B. S.; Bennett H.; Pajon R.; Knightly C.; Leav B.; Deng W.; Zhou H.; Han S.; Ivarsson M.; Miller J.; Zaks T. Efficacy and Safety of the MRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384 (5), 403–416. 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feikin D. R.; Higdon M. M.; Abu-Raddad L. J.; Andrews N.; Araos R.; Goldberg Y.; Groome M. J.; Huppert A.; O’Brien K. L.; Smith P. G.; Wilder-Smith A.; Zeger S.; Deloria Knoll M.; Patel M. K. Duration of Effectiveness of Vaccines against SARS-CoV-2 Infection and COVID-19 Disease: Results of a Systematic Review and Meta-Regression. Lancet 2022, 399 (10328), 924–944. 10.1016/S0140-6736(22)00152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando N. S.; Ferron F.; Decroly E.; Canard B.; Posthuma C. C.; Snijder E. J. The Curious Case of the Nidovirus Exoribonuclease: Its Role in RNA Synthesis and Replication Fidelity. Front. Microbiol. 2019, 10, 1813 10.3389/fmicb.2019.01813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando N. S.; Zevenhoven-Dobbe J. C.; van der Meer Y.; Bredenbeek P. J.; Posthuma C. C.; Snijder E. J.; Gallagher T. The Enzymatic Activity of the Nsp14 Exoribonuclease Is Critical for Replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020, 94 (23), e01246-20 10.1128/JVI.01246-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Alanis A.; Cruz-Rangel A.; Rodríguez-Gómez F.; González J.; Torres-Guerrero C. A.; Delgado G.; Cravioto A.; Morales-Espinosa R. Molecular Epidemiology Surveillance of SARS-CoV-2: Mutations and Genetic Diversity One Year after Emerging. Pathogens 2021, 10 (2), 184. 10.3390/pathogens10020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring A. S.; Hodcroft E. B. Genetic Variants of SARS-CoV-2—What Do They Mean?. JAMA 2021, 325 (6), 529. 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- Korber B.; Fischer W. M.; Gnanakaran S.; Yoon H.; Theiler J.; Abfalterer W.; Hengartner N.; Giorgi E. E.; Bhattacharya T.; Foley B.; Hastie K. M.; Parker M. D.; Partridge D. G.; Evans C. M.; Freeman T. M.; de Silva T. I.; McDanal C.; Perez L. G.; Tang H.; Moon-Walker A.; Whelan S. P.; LaBranche C. C.; Saphire E. O.; Montefiori D. C.; Angyal A.; Brown R. L.; Carrilero L.; Green L. R.; Groves D. C.; Johnson K. J.; Keeley A. J.; Lindsey B. B.; Parsons P. J.; Raza M.; Rowland-Jones S.; Smith N.; Tucker R. M.; Wang D.; Wyles M. D. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182 (4), 812–827. 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J. A.; Liu Y.; Liu J.; Xia H.; Johnson B. A.; Lokugamage K. G.; Zhang X.; Muruato A. E.; Zou J.; Fontes-Garfias C. R.; Mirchandani D.; Scharton D.; Bilello J. P.; Ku Z.; An Z.; Kalveram B.; Freiberg A. N.; Menachery V. D.; Xie X.; Plante K. S.; Weaver S. C.; Shi P.-Y. Spike Mutation D614G Alters SARS-CoV-2 Fitness. Nature 2021, 592 (7852), 116–121. 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva S. J. R.; de Lima S. C.; da Silva R. C.; Kohl A.; Pena L. Viral Load in COVID-19 Patients: Implications for Prognosis and Vaccine Efficacy in the Context of Emerging SARS-CoV-2 Variants. Front. Med. 2022, 8, 836826 10.3389/fmed.2021.836826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. ‘A Bloody Mess’: Confusion Reigns over Naming of New COVID Variants. Nature 2021, 589 (7842), 339–339. 10.1038/d41586-021-00097-w. [DOI] [PubMed] [Google Scholar]
- Abdool Karim S. S.; de Oliveira T.; Loots G. Appropriate Names for COVID-19 Variants. Science 2021, 371 (6535), 1215–1215. 10.1126/science.abh0836. [DOI] [PubMed] [Google Scholar]
- Rahimi F.; Talebi Bezmin Abadi A. Implications of the Emergence of a New Variant of SARS-CoV-2, VUI-202012/01. Arch. Med. Res. 2021, 52 (5), 569–571. 10.1016/j.arcmed.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsumi T.; Lusida M. I.; Dinana Z.; Wahyuni R. M.; Yamani L. N.; Juniastuti; Soetjipto; Matsui C.; Deng L.; Abe T.; Doan Y. H.; Fujii Y.; Kimura H.; Katayama K.; Shoji I. Occurrence of Norovirus Infection in an Asymptomatic Population in Indonesia. Infect. Genet. Evol. 2017, 55, 1–7. 10.1016/j.meegid.2017.08.020. [DOI] [PubMed] [Google Scholar]
- Davies N. G.; Abbott S.; Barnard R. C.; Jarvis C. I.; Kucharski A. J.; Munday J. D.; Pearson C. A. B.; Russell T. W.; Tully D. C.; Washburne A. D.; Wenseleers T.; Gimma A.; Waites W.; Wong K. L. M.; van Zandvoort K.; Silverman J. D.; Diaz-Ordaz K.; Keogh R.; Eggo R. M.; Funk S.; Jit M.; Atkins K. E.; Edmunds W. J. Estimated Transmissibility and Impact of SARS-CoV-2 Lineage B.1.1.7 in England. Science 2021, 372 (6538), eabg3055 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N. G.; Jarvis C. I.; Edmunds W. J.; Jewell N. P.; Diaz-Ordaz K.; Keogh R. H. Increased Mortality in Community-Tested Cases of SARS-CoV-2 Lineage B.1.1.7. Nature 2021, 593 (7858), 270–274. 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calistri P.; Amato L.; Puglia I.; Cito F.; Di Giuseppe A.; Danzetta M. L.; Morelli D.; Di Domenico M.; Caporale M.; Scialabba S.; Portanti O.; Curini V.; Perletta F.; Cammà C.; Ancora M.; Savini G.; Migliorati G.; D’Alterio N.; Lorusso A. Infection Sustained by Lineage B.1.1.7 of SARS-CoV-2 Is Characterised by Longer Persistence and Higher Viral RNA Loads in Nasopharyngeal Swabs. Int. J. Infect. Dis. 2021, 105, 753–755. 10.1016/j.ijid.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton D.; Rampling T.; Cross A.; Bailey H.; Heaney J.; Byott M.; Scott R.; Sconza R.; Price J.; Margaritis M.; Bergstrom M.; Spyer M. J.; Miralhes P. B.; Grant P.; Kirk S.; Valerio C.; Mangera Z.; Prabhahar T.; Moreno-Cuesta J.; Arulkumaran N.; Singer M.; Shin G. Y.; Sanchez E.; Paraskevopoulou S. M.; Pillay D.; McKendry R. A.; Mirfenderesky M.; Houlihan C. F.; Nastouli E. Genomic Characteristics and Clinical Effect of the Emergent SARS-CoV-2 B.1.1.7 Lineage in London, UK: A Whole-Genome Sequencing and Hospital-Based Cohort Study. Lancet Infect. Dis. 2021, 21 (9), 1246–1256. 10.1016/S1473-3099(21)00170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal A.; Destras G.; Gaymard A.; Stefic K.; Marlet J.; Eymieux S.; Regue H.; Semanas Q.; D’Aubarede C.; Billaud G.; Laurent F.; Gonzalez C.; Mekki Y.; Valette M.; Bouscambert M.; Gaudy-Graffin C.; Lina B.; Morfin F.; Josset L. Two-Step Strategy for the Identification of SARS-CoV-2 Variant of Concern 202012/01 and Other Variants with Spike Deletion H69-V70, France, August to December 2020. Eurosurveillance 2021, 26 (3), 2100008 10.2807/1560-7917.ES.2021.26.3.2100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk T.; Pharris A.; Spiteri G.; Bundle N.; Melidou A.; Carr M.; Gonzalez G.; Garcia-Leon A.; Crispie F.; O’Connor L.; Murphy N.; Mossong J.; Vergison A.; Wienecke-Baldacchino A. K.; Abdelrahman T.; Riccardo F.; Stefanelli P.; Di Martino A.; Bella A.; Lo Presti A.; Casaca P.; Moreno J.; Borges V.; Isidro J.; Ferreira R.; Gomes J. P.; Dotsenko L.; Suija H.; Epstein J.; Sadikova O.; Sepp H.; Ikonen N.; Savolainen-Kopra C.; Blomqvist S.; Möttönen T.; Helve O.; Gomes-Dias J.; Adlhoch C. Characteristics of SARS-CoV-2 Variants of Concern B.1.1.7, B.1.351 or P.1: Data from Seven EU/EEA Countries, Weeks 38/2020 to 10/2021. Eurosurveillance 2021, 26 (16), 2100348 10.2807/1560-7917.ES.2021.26.16.2100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen R.; Brooks-Pollock E.; Read J. M.; Dyson L.; Tsaneva-Atanasova K.; Danon L. Risk of Mortality in Patients Infected with SARS-CoV-2 Variant of Concern 202012/1: Matched Cohort Study. BMJ. 2021, n579. 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees-Spear C.; Muir L.; Griffith S. A.; Heaney J.; Aldon Y.; Snitselaar J. L.; Thomas P.; Graham C.; Seow J.; Lee N.; Rosa A.; Roustan C.; Houlihan C. F.; Sanders R. W.; Gupta R. K.; Cherepanov P.; Stauss H. J.; Nastouli E.; Doores K. J.; van Gils M. J.; McCoy L. E. The Effect of Spike Mutations on SARS-CoV-2 Neutralization. Cell Rep. 2021, 34 (12), 108890. 10.1016/j.celrep.2021.108890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.; Nair M. S.; Liu L.; Iketani S.; Luo Y.; Guo Y.; Wang M.; Yu J.; Zhang B.; Kwong P. D.; Graham B. S.; Mascola J. R.; Chang J. Y.; Yin M. T.; Sobieszczyk M.; Kyratsous C. A.; Shapiro L.; Sheng Z.; Huang Y.; Ho D. D. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature 2021, 593 (7857), 130–135. 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Planas D.; Bruel T.; Grzelak L.; Guivel-Benhassine F.; Staropoli I.; Porrot F.; Planchais C.; Buchrieser J.; Rajah M. M.; Bishop E.; Albert M.; Donati F.; Prot M.; Behillil S.; Enouf V.; Maquart M.; Smati-Lafarge M.; Varon E.; Schortgen F.; Yahyaoui L.; Gonzalez M.; De Sèze J.; Péré H.; Veyer D.; Sève A.; Simon-Lorière E.; Fafi-Kremer S.; Stefic K.; Mouquet H.; Hocqueloux L.; van der Werf S.; Prazuck T.; Schwartz O. Sensitivity of Infectious SARS-CoV-2 B.1.1.7 and B.1.351 Variants to Neutralizing Antibodies. Nat. Med. 2021, 27 (5), 917–924. 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- Rondinone V.; Pace L.; Fasanella A.; Manzulli V.; Parisi A.; Capobianchi M. R.; Ostuni A.; Chironna M.; Caprioli E.; Labonia M.; Cipolletta D.; Della Rovere I.; Serrecchia L.; Petruzzi F.; Pennuzzi G.; Galante D. VOC 202012/01 Variant Is Effectively Neutralized by Antibodies Produced by Patients Infected before Its Diffusion in Italy. Viruses 2021, 13 (2), 276. 10.3390/v13020276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Raddad L. J.; Chemaitelly H.; Butt A. A. Effectiveness of the BNT162b2 Covid-19 Vaccine against the B.1.1.7 and B.1.351 Variants. N. Engl. J. Med. 2021, 385 (2), 187–189. 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik A.; Wallisch A.-K.; Sänger B.; Swanson K. A.; Mühl J.; Chen W.; Cai H.; Maurus D.; Sarkar R.; Türeci Ö.; Dormitzer P. R.; Şahin U. Neutralization of SARS-CoV-2 Lineage B.1.1.7 Pseudovirus by BNT162b2 Vaccine-Elicited Human Sera. Science 2021, 371 (6534), 1152–1153. 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkal G. N.; Yadav P. D.; Ella R.; Deshpande G. R.; Sahay R. R.; Gupta N.; Vadrevu K. M.; Abraham P.; Panda S.; Bhargava B. Inactivated COVID-19 Vaccine BBV152/COVAXIN Effectively Neutralizes Recently Emerged B.1.1.7 Variant of SARS-CoV-2. J. Travel Med. 2021, 28 (4), taab051 10.1093/jtm/taab051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.-L.; Wang Z.-Y.; Duan L.-J.; Meng Q.-C.; Jiang M.-D.; Cao J.; Yao L.; Zhu K.-L.; Cao W.-C.; Ma M.-J. Susceptibility of Circulating SARS-CoV-2 Variants to Neutralization. N. Engl. J. Med. 2021, 384 (24), 2354–2356. 10.1056/NEJMc2103022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emary K. R. W.; Golubchik T.; Aley P. K.; Ariani C. V.; Angus B.; Bibi S.; Blane B.; Bonsall D.; Cicconi P.; Charlton S.; Clutterbuck E. A.; Collins A. M.; Cox T.; Darton T. C.; Dold C.; Douglas A. D.; Duncan C. J. A.; Ewer K. J.; Flaxman A. L.; Faust S. N.; Ferreira D. M.; Feng S.; Finn A.; Folegatti P. M.; Fuskova M.; Galiza E.; Goodman A. L.; Green C. M.; Green C. A.; Greenland M.; Hallis B.; Heath P. T.; Hay J.; Hill H. C.; Jenkin D.; Kerridge S.; Lazarus R.; Libri V.; Lillie P. J.; Ludden C.; Marchevsky N. G.; Minassian A. M.; McGregor A. C.; Mujadidi Y. F.; Phillips D. J.; Plested E.; Pollock K. M.; Robinson H.; Smith A.; Song R.; Snape M. D.; Sutherland R. K.; Thomson E. C.; Toshner M.; Turner D. P. J.; Vekemans J.; Villafana T. L.; Williams C. J.; Hill A. V. S.; Lambe T.; Gilbert S. C.; Voysey M.; Ramasamy M. N.; Pollard A. J. Efficacy of ChAdOx1 NCoV-19 (AZD1222) Vaccine against SARS-CoV-2 Variant of Concern 202012/01 (B.1.1.7): An Exploratory Analysis of a Randomised Controlled Trial. Lancet 2021, 397 (10282), 1351–1362. 10.1016/S0140-6736(21)00628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegame S.; Siddiquey M. N. A.; Hung C.-T.; Haas G.; Brambilla L.; Oguntuyo K. Y.; Kowdle S.; Chiu H.-P.; Stevens C. S.; Vilardo A. E.; Edelstein A.; Perandones C.; Kamil J. P.; Lee B. Neutralizing Activity of Sputnik V Vaccine Sera against SARS-CoV-2 Variants. Nat. Commun. 2021, 12 (1), 4598. 10.1038/s41467-021-24909-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H.; Wilkinson E.; Giovanetti M.; Iranzadeh A.; Fonseca V.; Giandhari J.; Doolabh D.; Pillay S.; San E. J.; Msomi N.; Mlisana K.; von Gottberg A.; Walaza S.; Allam M.; Ismail A.; Mohale T.; Glass A. J.; Engelbrecht S.; Van Zyl G.; Preiser W.; Petruccione F.; Sigal A.; Hardie D.; Marais G.; Hsiao N.; Korsman S.; Davies M.-A.; Tyers L.; Mudau I.; York D.; Maslo C.; Goedhals D.; Abrahams S.; Laguda-Akingba O.; Alisoltani-Dehkordi A.; Godzik A.; Wibmer C. K.; Sewell B. T.; Lourenço J.; Alcantara L. C. J.; Kosakovsky Pond S. L.; Weaver S.; Martin D.; Lessells R. J.; Bhiman J. N.; Williamson C.; de Oliveira T. Detection of a SARS-CoV-2 Variant of Concern in South Africa. Nature 2021, 592 (7854), 438–443. 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- Pearson C. A. B.; Russell T. W.; Davies N.; et al. Estimates of severity and transmissibility of novel SARS-CoV-2 variant 501Y.V2 in South Africa. CMMID Repository, November 1, 2021. https://cmmid.github.io/topics/covid19/sa-novel-variant.html#:~:text=Assuming%20complete%20cross%2Dprotection%2C%20we,%25)%20of%20previously%20acquired%20immunity. (accessed 2022-04-18).
- Edara V. V.; Norwood C.; Floyd K.; Lai L.; Davis-Gardner M. E.; Hudson W. H.; Mantus G.; Nyhoff L. E.; Adelman M. W.; Fineman R.; Patel S.; Byram R.; Gomes D. N.; Michael G.; Abdullahi H.; Beydoun N.; Panganiban B.; McNair N.; Hellmeister K.; Pitts J.; Winters J.; Kleinhenz J.; Usher J.; O’Keefe J. B.; Piantadosi A.; Waggoner J. J.; Babiker A.; Stephens D. S.; Anderson E. J.; Edupuganti S.; Rouphael N.; Ahmed R.; Wrammert J.; Suthar M. S. Infection- and Vaccine-Induced Antibody Binding and Neutralization of the B.1.351 SARS-CoV-2 Variant. Cell Host Microbe 2021, 29 (4), 516–521. 10.1016/j.chom.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W. F.; Lam E. C.; St. Denis K.; Nitido A. D.; Garcia Z. H.; Hauser B. M.; Feldman J.; Pavlovic M. N.; Gregory D. J.; Poznansky M. C.; Sigal A.; Schmidt A. G.; Iafrate A. J.; Naranbhai V.; Balazs A. B. Multiple SARS-CoV-2 Variants Escape Neutralization by Vaccine-Induced Humoral Immunity. Cell 2021, 184 (9), 2372–2383. 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmina A.; Khalaila Y.; Voloshin O.; Keren-Naus A.; Boehm-Cohen L.; Raviv Y.; Shemer-Avni Y.; Rosenberg E.; Taube R. SARS-CoV-2 Spike Variants Exhibit Differential Infectivity and Neutralization Resistance to Convalescent or Post-Vaccination Sera. Cell Host Microbe 2021, 29 (4), 522–528. 10.1016/j.chom.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D.; Dejnirattisai W.; Supasa P.; Liu C.; Mentzer A. J.; Ginn H. M.; Zhao Y.; Duyvesteyn H. M. E.; Tuekprakhon A.; Nutalai R.; Wang B.; Paesen G. C.; Lopez-Camacho C.; Slon-Campos J.; Hallis B.; Coombes N.; Bewley K.; Charlton S.; Walter T. S.; Skelly D.; Lumley S. F.; Dold C.; Levin R.; Dong T.; Pollard A. J.; Knight J. C.; Crook D.; Lambe T.; Clutterbuck E.; Bibi S.; Flaxman A.; Bittaye M.; Belij-Rammerstorfer S.; Gilbert S.; James W.; Carroll M. W.; Klenerman P.; Barnes E.; Dunachie S. J.; Fry E. E.; Mongkolsapaya J.; Ren J.; Stuart D. I.; Screaton G. R. Evidence of Escape of SARS-CoV-2 Variant B.1.351 from Natural and Vaccine-Induced Sera. Cell 2021, 184 (9), 2348–2361. 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi S. A.; Baillie V.; Cutland C. L.; Voysey M.; Koen A. L.; Fairlie L.; Padayachee S. D.; Dheda K.; Barnabas S. L.; Bhorat Q. E.; Briner C.; Kwatra G.; Ahmed K.; Aley P.; Bhikha S.; Bhiman J. N.; Bhorat A. E.; du Plessis J.; Esmail A.; Groenewald M.; Horne E.; Hwa S.-H.; Jose A.; Lambe T.; Laubscher M.; Malahleha M.; Masenya M.; Masilela M.; McKenzie S.; Molapo K.; Moultrie A.; Oelofse S.; Patel F.; Pillay S.; Rhead S.; Rodel H.; Rossouw L.; Taoushanis C.; Tegally H.; Thombrayil A.; van Eck S.; Wibmer C. K.; Durham N. M.; Kelly E. J.; Villafana T. L.; Gilbert S.; Pollard A. J.; de Oliveira T.; Moore P. L.; Sigal A.; Izu A. Efficacy of the ChAdOx1 NCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021, 384 (20), 1885–1898. 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturriza-Gomara M.; O’Brien S. J. Foodborne Viral Infections. Curr. Opin. Infect. Dis. 2016, 29 (5), 495–501. 10.1097/QCO.0000000000000299. [DOI] [PubMed] [Google Scholar]
- Faria N. R.; Mellan T. A.; Whittaker C.; Claro I. M.; Candido D. da S.; Mishra S.; Crispim M. A. E.; Sales F. C. S.; Hawryluk I.; McCrone J. T.; Hulswit R. J. G.; Franco L. A. M.; Ramundo M. S.; de Jesus J. G.; Andrade P. S.; Coletti T. M.; Ferreira G. M.; Silva C. A. M.; Manuli E. R.; Pereira R. H. M.; Peixoto P. S.; Kraemer M. U. G.; Gaburo N.; Camilo C. da C.; Hoeltgebaum H.; Souza W. M.; Rocha E. C.; de Souza L. M.; de Pinho M. C.; Araujo L. J. T.; Malta F. S. V.; de Lima A. B.; Silva J. do P.; Zauli D. A. G.; Ferreira A. C. de S.; Schnekenberg R. P.; Laydon D. J.; Walker P. G. T.; Schlüter H. M.; dos Santos A. L. P.; Vidal M. S.; Del Caro V. S.; Filho R. M. F.; dos Santos H. M.; Aguiar R. S.; Proença-Modena J. L.; Nelson B.; Hay J. A.; Monod M.; Miscouridou X.; Coupland H.; Sonabend R.; Vollmer M.; Gandy A.; Prete C. A.; Nascimento V. H.; Suchard M. A.; Bowden T. A.; Pond S. L. K.; Wu C.-H.; Ratmann O.; Ferguson N. M.; Dye C.; Loman N. J.; Lemey P.; Rambaut A.; Fraiji N. A.; Carvalho M. do P. S. S.; Pybus O. G.; Flaxman S.; Bhatt S.; Sabino E. C. Genomics and Epidemiology of the P.1 SARS-CoV-2 Lineage in Manaus, Brazil. Science 2021, 372 (6544), 815–821. 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveca F. G.; Nascimento V.; de Souza V. C.; Corado A. de L.; Nascimento F.; Silva G.; Costa Á.; Duarte D.; Pessoa K.; Mejía M.; Brandão M. J.; Jesus M.; Gonçalves L.; da Costa C. F.; Sampaio V.; Barros D.; Silva M.; Mattos T.; Pontes G.; Abdalla L.; Santos J. H.; Arantes I.; Dezordi F. Z.; Siqueira M. M.; Wallau G. L.; Resende P. C.; Delatorre E.; Gräf T.; Bello G. COVID-19 in Amazonas, Brazil, Was Driven by the Persistence of Endemic Lineages and P.1 Emergence. Nat. Med. 2021, 27 (7), 1230–1238. 10.1038/s41591-021-01378-7. [DOI] [PubMed] [Google Scholar]
- Barclay L.; Park G. W.; Vega E.; Hall A.; Parashar U.; Vinjé J.; Lopman B. Clin. Microbiol. Infect. 2014, 20 (8), 731–740. 10.1111/1469-0691.12674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levidiotou S.; Gartzonika C.; Papaventsis D.; Christaki C.; Priavali E.; Zotos N.; Kapsali E.; Vrioni G. CMI Viral Agents of Acute Gastroenteritis in Hospitalized Children in Greece. Clin. Microbiol. Infect. 2009, 15 (6), 596–598. 10.1111/j.1469-0691.2009.02855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano C. M.; Felix A. C.; de Paula A. V.; de Jesus J. G.; Andrade P. S.; Cândido D.; de Oliveira F. M.; Ribeiro A. C.; da Silva F. C.; Inemami M.; Costa A. A.; Leal C. O. D.; Figueiredo W. M.; Pannuti C. S.; de Souza W. M.; Faria N. R.; Sabino E. C. SARS-CoV-2 Reinfection Caused by the P.1 Lineage in Araraquara City, Sao Paulo State, Brazil. Rev. Inst. Med. Trop. Sao Paulo 2021, 63, e36 10.1590/s1678-9946202163036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino E. C.; Buss L. F.; Carvalho M. P. S.; Prete C. A.; Crispim M. A. E.; Fraiji N. A.; Pereira R. H. M.; Parag K. V.; da Silva Peixoto P.; Kraemer M. U. G.; Oikawa M. K.; Salomon T.; Cucunuba Z. M.; Castro M. C.; de Souza Santos A. A.; Nascimento V. H.; Pereira H. S.; Ferguson N. M.; Pybus O. G.; Kucharski A.; Busch M. P.; Dye C.; Faria N. R. Resurgence of COVID-19 in Manaus, Brazil, despite High Seroprevalence. Lancet 2021, 397 (10273), 452–455. 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. E.; Zhang X.; Case J. B.; Winkler E. S.; Liu Y.; VanBlargan L. A.; Liu J.; Errico J. M.; Xie X.; Suryadevara N.; Gilchuk P.; Zost S. J.; Tahan S.; Droit L.; Turner J. S.; Kim W.; Schmitz A. J.; Thapa M.; Wang D.; Boon A. C. M.; Presti R. M.; O’Halloran J. A.; Kim A. H. J.; Deepak P.; Pinto D.; Fremont D. H.; Crowe J. E.; Corti D.; Virgin H. W.; Ellebedy A. H.; Shi P.-Y.; Diamond M. S. Resistance of SARS-CoV-2 Variants to Neutralization by Monoclonal and Serum-Derived Polyclonal Antibodies. Nat. Med. 2021, 27 (4), 717–726. 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.; Casner R. G.; Nair M. S.; Wang M.; Yu J.; Cerutti G.; Liu L.; Kwong P. D.; Huang Y.; Shapiro L.; Ho D. D. Increased Resistance of SARS-CoV-2 Variant P.1 to Antibody Neutralization. Cell Host Microbe 2021, 29 (5), 747–751. 10.1016/j.chom.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W.; Zhou D.; Supasa P.; Liu C.; Mentzer A. J.; Ginn H. M.; Zhao Y.; Duyvesteyn H. M. E.; Tuekprakhon A.; Nutalai R.; Wang B.; López-Camacho C.; Slon-Campos J.; Walter T. S.; Skelly D.; Costa Clemens S. A.; Naveca F. G.; Nascimento V.; Nascimento F.; Fernandes da Costa C.; Resende P. C.; Pauvolid-Correa A.; Siqueira M. M.; Dold C.; Levin R.; Dong T.; Pollard A. J.; Knight J. C.; Crook D.; Lambe T.; Clutterbuck E.; Bibi S.; Flaxman A.; Bittaye M.; Belij-Rammerstorfer S.; Gilbert S. C.; Carroll M. W.; Klenerman P.; Barnes E.; Dunachie S. J.; Paterson N. G.; Williams M. A.; Hall D. R.; Hulswit R. J. G.; Bowden T. A.; Fry E. E.; Mongkolsapaya J.; Ren J.; Stuart D. I.; Screaton G. R. Antibody Evasion by the P.1 Strain of SARS-CoV-2. Cell 2021, 184 (11), 2939–2954. 10.1016/j.cell.2021.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabiani M.; Margiotti K.; Viola A.; Mesoraca A.; Giorlandino C. Mild Symptomatic SARS-CoV-2 P.1 (B.1.1.28) Infection in a Fully Vaccinated 83-Year-Old Man. Pathogens 2021, 10 (5), 614. 10.3390/pathogens10050614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchings M. D. T.; Ranzani O. T.; Torres M. S. S.; de Oliveira S. B.; Almiron M.; Said R.; Borg R.; Schulz W. L.; de Oliveira R. D.; da Silva P. V.; de Castro D. B.; Sampaio V. d. S.; de Albuquerque B. C.; Ramos T. C. A.; Fraxe S. H. H.; da Costa C. F.; Naveca F. G.; Siqueira A. M.; de Araújo W. N.; Andrews J. R.; Cummings D. A. T.; Ko A. I.; Croda J. Effectiveness of CoronaVac among Healthcare Workers in the Setting of High SARS-CoV-2 Gamma Variant Transmission in Manaus, Brazil: A Test-Negative Case-Control Study. Lancet Reg. Health - Am. 2021, 1, 100025. 10.1016/j.lana.2021.100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threat Assessment Brief: Emergence of SARS-CoV-2 B.1.617 variants in India and situation in the EU/EEA. European Centre for Disease Prevention and Control, May 11, 2021. https://www.ecdc.europa.eu/en/publications-data/threat-assessment-emergence-sars-cov-2-b1617-variants (accessed 2022-04-18).
- Campbell F.; Archer B.; Laurenson-Schafer H.; Jinnai Y.; Konings F.; Batra N.; Pavlin B.; Vandemaele K.; Van Kerkhove M. D.; Jombart T.; Morgan O.; le Polain de Waroux O. Increased Transmissibility and Global Spread of SARS-CoV-2 Variants of Concern as at June 2021. Eurosurveillance 2021, 26 (24), 2100509 10.2807/1560-7917.ES.2021.26.24.2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S. W. X.; Chiew C. J.; Ang L. W.; Mak T.-M.; Cui L.; Toh M. P. H. S.; Lim Y. D.; Lee P. H.; Lee T. H.; Chia P. Y.; Maurer-Stroh S.; Lin R. T. P.; Leo Y.-S.; Lee V. J.; Lye D. C.; Young B. E. Clinical and Virological Features of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Variants of Concern: A Retrospective Cohort Study Comparing B.1.1.7 (Alpha), B.1.351 (Beta), and B.1.617.2 (Delta). Clin. Infect. Dis. 2021, 10.1093/cid/ciab721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G. H.; Llewelyn A.; Brandao R.; Chowdhary K.; Hardisty K.-M.; Loddo M. SARS-CoV-2 Testing and Sequencing for International Arrivals Reveals Significant Cross Border Transmission of High Risk Variants into the United Kingdom. EClinicalMedicine 2021, 38, 101021. 10.1016/j.eclinm.2021.101021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Wintersdorff C.; Dingemans J.; Van Alphen L.; Wolffs P.; Van der Veer B.; Hoebe C.; Savelkoul P.. Infections caused by the Delta variant (B.1.617.2) of SARS-CoV-2 are associated with increased viral loads compared to infections with the Alpha variant (B.1.1.7) or non-Variants of Concern. Research square 2021, 10.21203/rs.3.rs-777577/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlcochova P.; Kemp S. A.; Dhar M. S.; Papa G.; Meng B.; Ferreira I. A. T. M.; Datir R.; Collier D. A.; Albecka A.; Singh S.; Pandey R.; Brown J.; Zhou J.; Goonawardane N.; Mishra S.; Whittaker C.; Mellan T.; Marwal R.; Datta M.; Sengupta S.; Ponnusamy K.; Radhakrishnan V. S.; Abdullahi A.; Charles O.; Chattopadhyay P.; Devi P.; Caputo D.; Peacock T.; Wattal C.; Goel N.; Satwik A.; Vaishya R.; Agarwal M.; Chauhan H.; Dikid T.; Gogia H.; Lall H.; Verma K.; Dhar M. S.; Singh M. K.; Soni N.; Meena N.; Madan P.; Singh P.; Sharma R.; Sharma R.; Kabra S.; Kumar S.; Kumari S.; Sharma U.; Chaudhary U.; Sivasubbu S.; Scaria V.; Oberoi J. K.; Raveendran R.; Datta S.; Das S.; Maitra A.; Chinnaswamy S.; Biswas N. K.; Parida A.; Raghav S. K.; Prasad P.; Sarin A.; Mayor S.; Ramakrishnan U.; Palakodeti D.; Seshasayee A. S. N.; Thangaraj K.; Bashyam M. D.; Dalal A.; Bhat M.; Shouche Y.; Pillai A.; Abraham P.; Potdar V. A.; Cherian S. S.; Desai A. S.; Pattabiraman C.; Manjunatha M. V.; Mani R. S.; Udupi G. A.; Nandicoori V.; Tallapaka K. B.; Sowpati D. T.; Kawabata R.; Morizako N.; Sadamasu K.; Asakura H.; Nagashima M.; Yoshimura K.; Ito J.; Kimura I.; Uriu K.; Kosugi Y.; Suganami M.; Oide A.; Yokoyama M.; Chiba M.; Saito A.; Butlertanaka E. P.; Tanaka Y. L.; Ikeda T.; Motozono C.; Nasser H.; Shimizu R.; Yuan Y.; Kitazato K.; Hasebe H.; Nakagawa S.; Wu J.; Takahashi M.; Fukuhara T.; Shimizu K.; Tsushima K.; Kubo H.; Shirakawa K.; Kazuma Y.; Nomura R.; Horisawa Y.; Takaori-Kondo A.; Tokunaga K.; Ozono S.; Baker S.; Dougan G.; Hess C.; Kingston N.; Lehner P. J.; Lyons P. A.; Matheson N. J.; Owehand W. H.; Saunders C.; Summers C.; Thaventhiran J. E. D.; Toshner M.; Weekes M. P.; Maxwell P.; Shaw A.; Bucke A.; Calder J.; Canna L.; Domingo J.; Elmer A.; Fuller S.; Harris J.; Hewitt S.; Kennet J.; Jose S.; Kourampa J.; Meadows A.; O’Brien C.; Price J.; Publico C.; Rastall R.; Ribeiro C.; Rowlands J.; Ruffolo V.; Tordesillas H.; Bullman B.; Dunmore B. J.; Fawke S.; Gräf S.; Hodgson J.; Huang C.; Hunter K.; Jones E.; Legchenko E.; Matara C.; Martin J.; Mescia F.; O’Donnell C.; Pointon L.; Pond N.; Shih J.; Sutcliffe R.; Tilly T.; Treacy C.; Tong Z.; Wood J.; Wylot M.; Bergamaschi L.; Betancourt A.; Bower G.; Cossetti C.; De Sa A.; Epping M.; Fawke S.; Gleadall N.; Grenfell R.; Hinch A.; Huhn O.; Jackson S.; Jarvis I.; Krishna B.; Lewis D.; Marsden J.; Nice F.; Okecha G.; Omarjee O.; Perera M.; Potts M.; Richoz N.; Romashova V.; Yarkoni N. S.; Sharma R.; Stefanucci L.; Stephens J.; Strezlecki M.; Turner L.; De Bie E. M. D. D.; Bunclark K.; Josipovic M.; Mackay M.; Rossi S.; Selvan M.; Spencer S.; Yong C.; Allison J.; Butcher H.; Caputo D.; Clapham-Riley D.; Dewhurst E.; Furlong A.; Graves B.; Gray J.; Ivers T.; Kasanicki M.; Le Gresley E.; Linger R.; Meloy S.; Muldoon F.; Ovington N.; Papadia S.; Phelan I.; Stark H.; Stirrups K. E.; Townsend P.; Walker N.; Webster J.; Scholtes I.; Hein S.; King R.; Mavousian A.; Lee J. H.; Bassi J.; Silacci-Fegni C.; Saliba C.; Pinto D.; Irie T.; Yoshida I.; Hamilton W. L.; Sato K.; Bhatt S.; Flaxman S.; James L. C.; Corti D.; Piccoli L.; Barclay W. S.; Rakshit P.; Agrawal A.; Gupta R. K. SARS-CoV-2 B.1.617.2 Delta Variant Replication and Immune Evasion. Nature 2021, 599 (7883), 114–119. 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian S.; Potdar V.; Jadhav S.; Yadav P.; Gupta N.; Das M.; Rakshit P.; Singh S.; Abraham P.; Panda S.; Team N. SARS-CoV-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. Microorganisms 2021, 9 (7), 1542. 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascarella S.; Ciccozzi M.; Zella D.; Bianchi M.; Benedetti F.; Benvenuto D.; Broccolo F.; Cauda R.; Caruso A.; Angeletti S.; Giovanetti M.; Cassone A. SARS-CoV-2 B.1.617 Indian Variants: Are Electrostatic Potential Changes Responsible for a Higher Transmission Rate?. J. Med. Virol. 2021, 93 (12), 6551–6556. 10.1002/jmv.27210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh A.; McMenamin J.; Taylor B.; Robertson C. SARS-CoV-2 Delta VOC in Scotland: Demographics, Risk of Hospital Admission, and Vaccine Effectiveness. Lancet 2021, 397 (10293), 2461–2462. 10.1016/S0140-6736(21)01358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.; Ginn H. M.; Dejnirattisai W.; Supasa P.; Wang B.; Tuekprakhon A.; Nutalai R.; Zhou D.; Mentzer A. J.; Zhao Y.; Duyvesteyn H. M. E.; López-Camacho C.; Slon-Campos J.; Walter T. S.; Skelly D.; Johnson S. A.; Ritter T. G.; Mason C.; Costa Clemens S. A.; Gomes Naveca F.; Nascimento V.; Nascimento F.; Fernandes da Costa C.; Resende P. C.; Pauvolid-Correa A.; Siqueira M. M.; Dold C.; Temperton N.; Dong T.; Pollard A. J.; Knight J. C.; Crook D.; Lambe T.; Clutterbuck E.; Bibi S.; Flaxman A.; Bittaye M.; Belij-Rammerstorfer S.; Gilbert S. C.; Malik T.; Carroll M. W.; Klenerman P.; Barnes E.; Dunachie S. J.; Baillie V.; Serafin N.; Ditse Z.; Da Silva K.; Paterson N. G.; Williams M. A.; Hall D. R.; Madhi S.; Nunes M. C.; Goulder P.; Fry E. E.; Mongkolsapaya J.; Ren J.; Stuart D. I.; Screaton G. R. Reduced Neutralization of SARS-CoV-2 B.1.617 by Vaccine and Convalescent Serum. Cell 2021, 184 (16), 4220–4236. 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Liu Y.; Xia H.; Zou J.; Weaver S. C.; Swanson K. A.; Cai H.; Cutler M.; Cooper D.; Muik A.; Jansen K. U.; Sahin U.; Xie X.; Dormitzer P. R.; Shi P.-Y. BNT162b2-Elicited Neutralization of B.1.617 and Other SARS-CoV-2 Variants. Nature 2021, 596 (7871), 273–275. 10.1038/s41586-021-03693-y. [DOI] [PubMed] [Google Scholar]
- Lustig Y.; Zuckerman N.; Nemet I.; Atari N.; Kliker L.; Regev-Yochay G.; Sapir E.; Mor O.; Alroy-Preis S.; Mendelson E.; Mandelboim M. Neutralising Capacity against Delta (B.1.617.2) and Other Variants of Concern Following Comirnaty (BNT162b2, BioNTech/Pfizer) Vaccination in Health Care Workers, Israel. Eurosurveillance 2021, 26 (26), 2100557 10.2807/1560-7917.ES.2021.26.26.2100557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall E. C.; Wu M.; Harvey R.; Kelly G.; Warchal S.; Sawyer C.; Daniels R.; Hobson P.; Hatipoglu E.; Ngai Y.; Hussain S.; Nicod J.; Goldstone R.; Ambrose K.; Hindmarsh S.; Beale R.; Riddell A.; Gamblin S.; Howell M.; Kassiotis G.; Libri V.; Williams B.; Swanton C.; Gandhi S.; Bauer D. L. Neutralising Antibody Activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 Vaccination. Lancet 2021, 397 (10292), 2331–2333. 10.1016/S0140-6736(21)01290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Bernal J.; Andrews N.; Gower C.; Gallagher E.; Simmons R.; Thelwall S.; Stowe J.; Tessier E.; Groves N.; Dabrera G.; Myers R.; Campbell C. N. J.; Amirthalingam G.; Edmunds M.; Zambon M.; Brown K. E.; Hopkins S.; Chand M.; Ramsay M. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021, 385 (7), 585–594. 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasreen S.; Chung H.; He S.; Brown K. A.; Gubbay J. B.; Buchan S. A.; Fell D. B.; Austin P. C.; Schwartz K. L.; Sundaram M. E.; Calzavara A.; Chen B.; Tadrous M.; Wilson K.; Wilson S. E.; Kwong J. C. Effectiveness of COVID-19 Vaccines against Symptomatic SARS-CoV-2 Infection and Severe Outcomes with Variants of Concern in Ontario. Nat. Microbiol. 2022, 7 (3), 379–385. 10.1038/s41564-021-01053-0. [DOI] [PubMed] [Google Scholar]
- Chia P. Y.; Ong S. W. X.; Chiew C. J.; Ang L. W.; Chavatte J.-M.; Mak T.-M.; Cui L.; Kalimuddin S.; Chia W. N.; Tan C. W.; Chai L. Y. A.; Tan S. Y.; Zheng S.; Lin R. T. P.; Wang L.; Leo Y.-S.; Lee V. J.; Lye D. C.; Young B. E. Virological and Serological Kinetics of SARS-CoV-2 Delta Variant Vaccine Breakthrough Infections: A Multicentre Cohort Study. Clin. Microbiol. Infect. 2022, 28 (4), 612.e1–612.e7. 10.1016/j.cmi.2021.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M.; Yi Y.; Li Y.; Sun L.; Deng A.; Hu T.; Zhang J.; Liu J.; Cheng M.; Xie S.; Luo M.; Jiang J.; Jiang Y.; Tang S.; He J. Effectiveness of Inactivated COVID-19 Vaccines Against Illness Caused by the B.1.617.2 (Delta) Variant During an Outbreak in Guangdong, China. Ann. Int. Med. 2022, 175 (4), 533–540. 10.7326/M21-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav P. D.; Sahay R. R.; Sapkal G.; Nyayanit D.; Shete A. M.; Deshpande G.; Patil D. Y.; Gupta N.; Kumar S.; Abraham P.; Panda S.; Bhargava B. Comparable Neutralization of SARS-CoV-2 Delta AY.1 and Delta with Individuals Sera Vaccinated with BBV152. J. Travel Med. 2021, 28 (8), taab154 10.1093/jtm/taab154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gushchin V. A.; Dolzhikova I. V.; Shchetinin A. M.; Odintsova A. S.; Siniavin A. E.; Nikiforova M. A.; Pochtovyi A. A.; Shidlovskaya E. V.; Kuznetsova N. A.; Burgasova O. A.; Kolobukhina L. V.; Iliukhina A. A.; Kovyrshina A. V.; Botikov A. G.; Kuzina A. V.; Grousova D. M.; Tukhvatulin A. I.; Shcheblyakov D. V.; Zubkova O. V.; Karpova O. V.; Voronina O. L.; Ryzhova N. N.; Aksenova E. I.; Kunda M. S.; Lioznov D. A.; Danilenko D. M.; Komissarov A. B.; Tkachuck A. P.; Logunov D. Y.; Gintsburg A. L. Neutralizing Activity of Sera from Sputnik V-Vaccinated People against Variants of Concern (VOC: B.1.1.7, B.1.351, P.1, B.1.617.2, B.1.617.3) and Moscow Endemic SARS-CoV-2 Variants. Vaccines 2021, 9 (7), 779. 10.3390/vaccines9070779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongeneelen M.; et al. Ad26.COV2.S elicited neutralizing activity against Delta and other SARS-CoV-2 variants of concern. bioRxiv 2021, 10.1101/2021.07.01.450707. [DOI] [Google Scholar]
- Karim S. S. A.; Karim Q. A. Omicron SARS-CoV-2 Variant: A New Chapter in the COVID-19 Pandemic. Lancet 2021, 398 (10317), 2126–2128. 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Krüger N.; Schulz S.; Cossmann A.; Rocha C.; Kempf A.; Nehlmeier I.; Graichen L.; Moldenhauer A.-S.; Winkler M. S.; Lier M.; Dopfer-Jablonka A.; Jäck H.-M.; Behrens G. M. N.; Pöhlmann S. The Omicron Variant Is Highly Resistant against Antibody-Mediated Neutralization: Implications for Control of the COVID-19 Pandemic. Cell 2022, 185 (3), 447–456. 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W.; Huo J.; Zhou D.; Zahradník J.; Supasa P.; Liu C.; Duyvesteyn H. M. E.; Ginn H. M.; Mentzer A. J.; Tuekprakhon A. Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. bioRxiv 2021, 10.1101/2021.12.03.471045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P.; Li L.; Liu S.; Wang Q.; Zhang D.; Xu Z.; Han P.; Li X.; Peng Q.; Su C.; Huang B.; Li D.; Zhang R.; Tian M.; Fu L.; Gao Y.; Zhao X.; Liu K.; Qi J.; Gao G. F.; Wang P. Receptor Binding and Complex Structures of Human ACE2 to Spike RBD from Omicron and Delta SARS-CoV-2. Cell 2022, 185 (4), 630–640. 10.1016/j.cell.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W. F.; St. Denis K. J.; Hoelzemer A.; Lam E. C.; Nitido A. D.; Sheehan M. L.; Berrios C.; Ofoman O.; Chang C. C.; Hauser B. M.; Feldman J.; Roederer A. L.; Gregory D. J.; Poznansky M. C.; Schmidt A. G.; Iafrate A. J.; Naranbhai V.; Balazs A. B. MRNA-Based COVID-19 Vaccine Boosters Induce Neutralizing Immunity against SARS-CoV-2 Omicron Variant. Cell 2022, 185 (3), 457–466. 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W.; Shaw R. H.; Supasa P.; Liu C.; Stuart A. S.; Pollard A. J.; Liu X.; Lambe T.; Crook D.; Stuart D. I.; Mongkolsapaya J.; Nguyen-Van-Tam J. S.; Snape M. D.; Screaton G. R. Reduced Neutralisation of SARS-CoV-2 Omicron B.1.1.529 Variant by Post-Immunisation Serum. Lancet 2022, 399 (10321), 234–236. 10.1016/S0140-6736(21)02844-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose R.; Itoh Y.; Ikegaya H.; Miyazaki H.; Watanabe N.; Yoshida T.; Bandou R.; Daidoji T.; Nakaya T. Differences in Environmental Stability among SARS-CoV-2 Variants of Concern: Both Omicron BA.1 and BA.2 Have Higher Stability. Clin. Microbiol. Infect. 2022, 10.1016/j.cmi.2022.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva S. J. R.; Kohl A.; Pena L.; Pardee K. Recent Insights into SARS-CoV-2 Omicron Variant. Rev. Med. Virol. 2022, 10.1002/rmv.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K.; Miorin L.; Makio T.; Dehghan I.; Gao S.; Xie Y.; Zhong H.; Esparza M.; Kehrer T.; Kumar A.; Hobman T. C.; Ptak C.; Gao B.; Minna J. D.; Chen Z.; García-Sastre A.; Ren Y.; Wozniak R. W.; Fontoura B. M. A. Nsp1 Protein of SARS-CoV-2 Disrupts the MRNA Export Machinery to Inhibit Host Gene Expression. Sci. Adv. 2021, 7 (6), eabe7386 10.1126/sciadv.abe7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert K.; Karousis E. D.; Jomaa A.; Scaiola A.; Echeverria B.; Gurzeler L.-A.; Leibundgut M.; Thiel V.; Mühlemann O.; Ban N. SARS-CoV-2 Nsp1 Binds the Ribosomal MRNA Channel to Inhibit Translation. Nat. Struct. Mol. Biol. 2020, 27 (10), 959–966. 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- Narayanan K.; Huang C.; Lokugamage K.; Kamitani W.; Ikegami T.; Tseng C.-T. K.; Makino S. Severe Acute Respiratory Syndrome Coronavirus Nsp1 Suppresses Host Gene Expression, Including That of Type I Interferon, in Infected Cells. J. Virol. 2008, 82 (9), 4471–4479. 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez C.; Swanson S. E.; Negatu S. G.; Dittmar M.; Miller J.; Ramage H. R.; Cherry S.; Jurado K. A. SARS-CoV-2 Viral Proteins NSP1 and NSP13 Inhibit Interferon Activation through Distinct Mechanisms. PLoS One 2021, 16 (6), e0253089 10.1371/journal.pone.0253089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M.; Azumaya C. M.; Moritz M.; Pourmal S.; Diallo A.; Merz G. E.; Jang G.; Bouhaddou M.; Fossati A.; Brilot A. F. CryoEM and AI reveal a structure of SARS-CoV-2 Nsp2, a multifunctional protein involved in key host processes. bioRxiv 2021, 10.1101/2021.05.10.443524. [DOI] [Google Scholar]
- Lei J.; Kusov Y.; Hilgenfeld R. Nsp3 of Coronaviruses: Structures and Functions of a Large Multi-Domain Protein. Antiviral Res. 2018, 149, 58–74. 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano P.; Johnson M. A.; Chatterjee A.; Neuman B. W.; Joseph J. S.; Buchmeier M. J.; Kuhn P.; Wüthrich K. Nuclear Magnetic Resonance Structure of the Nucleic Acid-Binding Domain of Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein 3. J. Virol. 2009, 83 (24), 12998–13008. 10.1128/JVI.01253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Liu Q.; Guo D. Emerging Coronaviruses: Genome Structure, Replication, and Pathogenesis. J. Med. Virol. 2020, 92 (4), 418–423. 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachboard D. C.; Anderson-Daniels J. M.; Denison M. R. Mutations across Murine Hepatitis Virus Nsp4 Alter Virus Fitness and Membrane Modifications. J. Virol. 2015, 89 (4), 2080–2089. 10.1128/JVI.02776-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadlage M. J.; Sparks J. S.; Beachboard D. C.; Cox R. G.; Doyle J. D.; Stobart C. C.; Denison M. R. Murine Hepatitis Virus Nonstructural Protein 4 Regulates Virus-Induced Membrane Modifications and Replication Complex Function. J. Virol. 2010, 84 (1), 280–290. 10.1128/JVI.01772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini M. M.; Akhlaghpour M.; Neuman B. W.; Buchmeier M. J. Severe Acute Respiratory Syndrome Coronavirus Nonstructural Proteins 3, 4, and 6 Induce Double-Membrane Vesicles. mBio 2013, 4 (4), e00524-13 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobart C. C.; Sexton N. R.; Munjal H.; Lu X.; Molland K. L.; Tomar S.; Mesecar A. D.; Denison M. R. Chimeric Exchange of Coronavirus Nsp5 Proteases (3CLpro) Identifies Common and Divergent Regulatory Determinants of Protease Activity. J. Virol. 2013, 87 (23), 12611–12618. 10.1128/JVI.02050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.; Fang L.; Wang D.; Yang Y.; Chen J.; Ye X.; Foda M. F.; Xiao S. Porcine Deltacoronavirus Nsp5 Inhibits Interferon-β Production through the Cleavage of NEMO. Virology 2017, 502, 33–38. 10.1016/j.virol.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.; Wang D.; Zhou J.; Pan T.; Chen J.; Yang Y.; Lv M.; Ye X.; Peng G.; Fang L.; Xiao S. Porcine Deltacoronavirus Nsp5 Antagonizes Type I Interferon Signaling by Cleaving STAT2. J. Virol. 2017, 91 (10), e00003-17 10.1128/JVI.00003-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E. M.; Whelband M. C.; Wileman T. Coronavirus NSP6 Restricts Autophagosome Expansion. Autophagy 2014, 10 (8), 1426–1441. 10.4161/auto.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuto D.; Angeletti S.; Giovanetti M.; Bianchi M.; Pascarella S.; Cauda R.; Ciccozzi M.; Cassone A. Evolutionary Analysis of SARS-CoV-2: How Mutation of Non-Structural Protein 6 (NSP6) Could Affect Viral Autophagy. J. Infect. 2020, 81 (1), e24–e27. 10.1016/j.jinf.2020.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R. N.; Ward A. B. Structure of the SARS-CoV Nsp12 Polymerase Bound to Nsp7 and Nsp8 Co-Factors. Nat. Commun. 2019, 10 (1), 2342. 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y.; Sun F.; Li X.; Pang H.; Xu X.; Bartlam M.; Rao Z. Insights into SARS-CoV Transcription and Replication from the Structure of the Nsp7-Nsp8 Hexadecamer. Nat. Struct. Mol. Biol. 2005, 12 (11), 980–986. 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y.; Ma Q.; Restle T.; Shang W.; Svergun D. I.; Ponnusamy R.; Sczakiel G.; Hilgenfeld R. Nonstructural Proteins 7 and 8 of Feline Coronavirus Form a 2:1 Heterotrimer That Exhibits Primer-Independent RNA Polymerase Activity. J. Virol. 2012, 86 (8), 4444–4454. 10.1128/JVI.06635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velthuis A. J. W.; van den Worm S. H. E.; Snijder E. J. The SARS-Coronavirus Nsp7+nsp8 Complex Is a Unique Multimeric RNA Polymerase Capable of Both de Novo Initiation and Primer Extension. Nucleic Acids Res. 2012, 40 (4), 1737–1747. 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.-P.; Ferron F.; Campanacci V.; Longhi S.; Rancurel C.; Dutartre H.; Snijder E. J.; Gorbalenya A. E.; Cambillau C.; Canard B. The Severe Acute Respiratory Syndrome-Coronavirus Replicative Protein Nsp9 Is a Single-Stranded RNA-Binding Subunit Unique in the RNA Virus World. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (11), 3792–3796. 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z.; Deng F.; Shi K.; Ye G.; Wang G.; Fang L.; Xiao S.; Fu Z.; Peng G. Dimerization of Coronavirus Nsp9 with Diverse Modes Enhances Its Nucleic Acid Binding Affinity. J. Virol. 2018, 92 (17), e00692-18 10.1128/JVI.00692-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton G.; Fry E.; Carter L.; Sainsbury S.; Walter T.; Nettleship J.; Berrow N.; Owens R.; Gilbert R.; Davidson A.; Siddell S.; Poon L. L. M.; Diprose J.; Alderton D.; Walsh M.; Grimes J. M.; Stuart D. I. The Nsp9 Replicase Protein of SARS-Coronavirus, Structure and Functional Insights. Structure 2004, 12 (2), 341–353. 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M.; Lugari A.; Posthuma C. C.; Zevenhoven J. C.; Bernard S.; Betzi S.; Imbert I.; Canard B.; Guillemot J.-C.; Lécine P.; Pfefferle S.; Drosten C.; Snijder E. J.; Decroly E.; Morelli X. Coronavirus Nsp10, a Critical Co-Factor for Activation of Multiple Replicative Enzymes. J. Biol. Chem. 2014, 289 (37), 25783–25796. 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Su C.; Ke M.; Jin X.; Xu L.; Zhang Z.; Wu A.; Sun Y.; Yang Z.; Tien P.; Ahola T.; Liang Y.; Liu X.; Guo D. Biochemical and Structural Insights into the Mechanisms of SARS Coronavirus RNA Ribose 2′-O-Methylation by Nsp16/Nsp10 Protein Complex. PLoS Pathog. 2011, 7 (10), e1002294 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E.; Debarnot C.; Ferron F.; Bouvet M.; Coutard B.; Imbert I.; Gluais L.; Papageorgiou N.; Sharff A.; Bricogne G.; Ortiz-Lombardia M.; Lescar J.; Canard B. Crystal Structure and Functional Analysis of the SARS-Coronavirus RNA Cap 2′-O-Methyltransferase Nsp10/Nsp16 Complex. PLoS Pathog. 2011, 7 (5), e1002059 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.; Wu L.; Shaw N.; Gao Y.; Wang J.; Sun Y.; Lou Z.; Yan L.; Zhang R.; Rao Z. Structural Basis and Functional Analysis of the SARS Coronavirus Nsp14-Nsp10 Complex. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (30), 9436–9441. 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S. G.; Shen H.; Wang J.; Tay F. P. L.; Liu D. X. Proteolytic Processing of Polyproteins 1a and 1ab between Non-Structural Proteins 10 and 11/12 of Coronavirus Infectious Bronchitis Virus Is Dispensable for Viral Replication in Cultured Cells. Virology 2008, 379 (2), 175–180. 10.1016/j.virol.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadhave K.; Kumar P.; Kumar A.; Bhardwaj T.; Garg N.; Giri R. Conformational Dynamics of 13 Amino Acids Long NSP11 of SARS-CoV-2 under Membrane Mimetics and Different Solvent Conditions. Microb. Pathog. 2021, 158, 105041. 10.1016/j.micpath.2021.105041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn D.-G.; Choi J.-K.; Taylor D. R.; Oh J.-W. Biochemical Characterization of a Recombinant SARS Coronavirus Nsp12 RNA-Dependent RNA Polymerase Capable of Copying Viral RNA Templates. Arch. Virol. 2012, 157 (11), 2095–2104. 10.1007/s00705-012-1404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velthuis A. J. W.; Arnold J. J.; Cameron C. E.; van den Worm S. H. E.; Snijder E. J. The RNA Polymerase Activity of SARS-Coronavirus Nsp12 Is Primer Dependent. Nucleic Acids Res. 2010, 38 (1), 203–214. 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen H. S.; Kokic G.; Farnung L.; Dienemann C.; Tegunov D.; Cramer P. Structure of Replicating SARS-CoV-2 Polymerase. Nature 2020, 584 (7819), 154–156. 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- Shu T.; Huang M.; Wu D.; Ren Y.; Zhang X.; Han Y.; Mu J.; Wang R.; Qiu Y.; Zhang D.-Y.; Zhou X. SARS-Coronavirus-2 Nsp13 Possesses NTPase and RNA Helicase Activities That Can Be Inhibited by Bismuth Salts. Virol. Sin. 2020, 35 (3), 321–329. 10.1007/s12250-020-00242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W.; Wojdyla J. A.; Zhao R.; Han R.; Das R.; Zlatev I.; Manoharan M.; Wang M.; Cui S. Crystal Structure of Middle East Respiratory Syndrome Coronavirus Helicase. PLoS Pathog. 2017, 13 (6), e1006474 10.1371/journal.ppat.1006474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z.; Yan L.; Ren Z.; Wu L.; Wang J.; Guo J.; Zheng L.; Ming Z.; Zhang L.; Lou Z.; Rao Z. Delicate Structural Coordination of the Severe Acute Respiratory Syndrome Coronavirus Nsp13 upon ATP Hydrolysis. Nucleic Acids Res. 2019, 47 (12), 6538–6550. 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerle L. D.; Becker M. M.; Halpin R. A.; Li K.; Venter E.; Lu X.; Scherbakova S.; Graham R. L.; Baric R. S.; Stockwell T. B.; Spiro D. J.; Denison M. R. Infidelity of SARS-CoV Nsp14-Exonuclease Mutant Virus Replication Is Revealed by Complete Genome Sequencing. PLoS Pathog. 2010, 6 (5), e1000896 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M.; Imbert I.; Subissi L.; Gluais L.; Canard B.; Decroly E. RNA 3’-End Mismatch Excision by the Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein Nsp10/Nsp14 Exoribonuclease Complex. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (24), 9372–9377. 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Cai H.; Pan J.; Xiang N.; Tien P.; Ahola T.; Guo D. Functional Screen Reveals SARS Coronavirus Nonstructural Protein Nsp14 as a Novel Cap N7Methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (9), 3484–3489. 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minskaia E.; Hertzig T.; Gorbalenya A. E.; Campanacci V.; Cambillau C.; Canard B.; Ziebuhr J. Discovery of an RNA Virus 3′→5′ Exoribonuclease That Is Critically Involved in Coronavirus RNA Synthesis. Proc. Natl. Acad. Sci. U. S. A. 2006, 103 (13), 5108–5113. 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K.; Sun J.; Holzenburg A.; Guarino L. A.; Kao C. C. RNA Recognition and Cleavage by the SARS Coronavirus Endoribonuclease. J. Mol. Biol. 2006, 361 (2), 243–256. 10.1016/j.jmb.2006.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X.; Hackbart M.; Mettelman R. C.; O’Brien A.; Mielech A. M.; Yi G.; Kao C. C.; Baker S. C. Coronavirus Nonstructural Protein 15 Mediates Evasion of DsRNA Sensors and Limits Apoptosis in Macrophages. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (21), E4251–E4260. 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Li L.; Yan L.; Ming Z.; Jia Z.; Lou Z.; Rao Z. Structural and Biochemical Characterization of Endoribonuclease Nsp15 Encoded by Middle East Respiratory Syndrome Coronavirus. J. Virol. 2018, 92 (22), e00893-18 10.1128/JVI.00893-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vithani N.; Ward M. D.; Zimmerman M. I.; Novak B.; Borowsky J. H.; Singh S.; Bowman G. R. SARS-CoV-2 Nsp16 Activation Mechanism and a Cryptic Pocket with Pan-Coronavirus Antiviral Potential. Biophys. J. 2021, 120 (14), 2880–2889. 10.1016/j.bpj.2021.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafcikova P.; Silhan J.; Nencka R.; Boura E. Structural Analysis of the SARS-CoV-2 Methyltransferase Complex Involved in RNA Cap Creation Bound to Sinefungin. Nat. Commun. 2020, 11 (1), 3717. 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P.; Su Y.; Li R.; Liang Z.; Dong S.; Huang J. PEDV Nsp16 Negatively Regulates Innate Immunity to Promote Viral Proliferation. Virus Res. 2019, 265, 57–66. 10.1016/j.virusres.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L.-J.; Chen T.-H. NSP16 2′-O-MTase in Coronavirus Pathogenesis: Possible Prevention and Treatments Strategies. Viruses 2021, 13 (4), 538. 10.3390/v13040538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Q.; Wu D.; Wang X.; Xi D.; Chen T.; Chen G.; Wang H.; Lu H.; Wang M.; Zhu L.; Hu J.; Liu T.; Ma K.; Han M.; Luo X. The Mechanism Underlying Extrapulmonary Complications of the Coronavirus Disease 2019 and Its Therapeutic Implication. Signal Transduct. Target. Ther. 2022, 7 (1), 57. 10.1038/s41392-022-00907-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.