Abstract
Background
This study was designed to unravel the genomic landscape and evolution of early-stage subsolid lung adenocarcinomas (SSN-LUADs) manifesting as pure ground-glass nodules (pGGNs), heterogeneous ground-glass nodules (HGGNs) and part-solid nodules (PSNs).
Methods
Samples subjected to either broad-panel next-generation sequencing (NGS) or whole-exome sequencing (WES) were included. Clinicopathologic and genomic features were compared among pGGN, HGGN and PSN, while tumour evolutionary trajectories and mutational signatures were evaluated in the entire cohort.
Results
In total, 247 SSN-LUAD samples subjected to broad-panel NGS and 125 to WES were identified. Compared with PSNs, HGGNs had significantly lower tumour mutation count (P < 0.001), genomic alteration count (P < 0.001), and intra-tumour heterogeneity (P = 0.005). Statistically significant upward trends were observed in alterations involving driver mutations and oncogenic pathways from pGGNs to HGGNs to PSNs. EGFR mutation was proved to be a key early event in the progression of SSN-LUADs, with subsequently two evolutionary trajectories involving either RBM10 or TP53 mutation in the cancer-evolution models.
Conclusions
This study provided evidence for unravelling the previously unknown genomic underpinnings associated with SSN-LUAD evolution from pGGN to HGGN to PSN, proving that HGGN was an intermediate SSN form between pGGN and PSN genetically.
Subject terms: Non-small-cell lung cancer, Cancer epigenetics
Background
The prevalence of early-stage lung adenocarcinoma (LUAD) manifesting as a subsolid nodule (SSN)/ ground-glass nodule (GGN) radiologically keeps increasing with the widespread application of chest CT for lung cancer screening [1, 2]. Different guidelines for the management of SSNs emphasise the importance of solid components accordantly. The solid component substantially affects the prognosis, clinical T stage, follow-up interval and resection extension of LUAD manifesting as SSN (SSN-LUAD) [3–6].
However, the criteria to define a solid component remains controversial [7]. One major disagreement is whether solid components should be measured in the lung or mediastinal window. In 2016, Kakinuma et al. classified SSN into pure ground-glass nodule (pGGN: without solid component), heterogeneous ground-glass nodule (HGGN: solid component detected only in lung window) and part-solid nodule (PSN: solid component detected both in lung and mediastinal windows) in a prospective study [8]. The results of long-term follow-up showed significantly higher proportion and shorter duration of HGGNs progressing to PSNs than those of pGGNs, and that invasive LUADs could only been detected in PSNs [8]. Thus, they proposed HGGN as an intermediate existing form between pGGN and PSN from radiological perspective. Recently, Yin et al. reported that LUADs manifesting as PSNs possessed a much higher proportion of micropapillary or solid (MIP/SOL) predominant pathologic subtypes and a significantly worse recurrence-free survival than HGGNs [9]. Thus, it could be speculated that the radiological solid components detected in the lung or mediastinal window differed significantly in tumour behaviour, pathological features and prognosis.
To date, there is no study on the genomic differences between pGGN, HGGN and PSN, while further studies of the molecular heterogeneity could help in understanding the evolution of SSN-LUADs. In 2020, our group completed the whole-exome sequencing (WES) of 154 SSNs and provided the first comprehensive description of the mutational landscape of SSNs. The results revealed that mixed GGNs (mGGNs) harboured higher tumour mutation burden (TMB) and higher frequency of EGFR and TP53 mutation than pGGNs [10]. However, mGGNs were not further subdivided into HGGNs and PSNs in that study.
To address this knowledge gap, we compared the results of broad-panel next-generation sequencing (NGS) in 247 early-stage SSN-LUADs (including 77 pGGNs, 48 HGGNs and 122 PSNs), and further validated the results in a whole-exome sequencing (WES) cohort. By interpreting their general genomic features, driver genes and oncogenic pathways, we unravelled the distinction between pGGNs, HGGNs and PSNs at the genomic level and further shed light on the evolution of SSN-LUADs.
Methods
SSN cohorts
We identified patients who underwent complete surgical resection for stage 0-I SSN-LUADs and broad-panel NGS performed on the primary tumours between January 2018 and June 2020 in the Department of Thoracic Surgery with accessible preoperative chest CT scan images (Supplementary Fig. 1). Identical inclusion criteria were as previous [10]. Notably, samples pathologically diagnosed as atypical adenomatous hyperplasia (AAH) were excluded from this WES cohort. Clinicopathologic features of the broad-panel NGS cohort (n = 247) and the WES cohort (n = 125) were summarised.
Radiological evaluation
Preoperative chest CT scan within 4 weeks prior to the surgery were reviewed by two radiologists (QQ and PY) with more than 5 years of experience in thoracic imaging. SSNs were classified into pGGNs, HGGNs and PSNs based on their radiological textures (Supplementary Fig. 2) [8]. The maximum diameters and solid components of SSNs were measured in both lung window (window width: 1500 Hounsfield units [HU]; window level: −500 HU) and mediastinal windows (window width: 350 HU; window level: 40 HU) under default display settings.
Tumour genomic analyses
Target capture of broad-panel NGS was performed using any one of the three commercial panels consisting of 363 (HR363, Berry Oncology, Beijing, China), 457 (HR457, Berry Oncology, Beijing, China), or 520 (RS520, Burning Rock Biotech, Guangzhou, China) cancer-related genes, spanning 1.18, 1.21 or 1.64 megabases (Mb) of the human genome, respectively (Supplementary Table 1). Protocols for genomic DNA extraction, targeted/whole-exome sequencing library preparation, and variant calling were described in previous publications [10–13].
Tumour mutation count was defined as the amount of nonsynonymous somatic mutations (single-nucleotide variants [SNVs] and insertions/deletions [InDels]) per sample, while TMB was calculated only in the WES cohort by the number of nonsynonymous somatic mutations per Mb of the coding regions. Genomic alteration count, defined as the sum of nonsynonymous somatic mutations, copy number variations (CNVs), and gene fusions, was further assessed in the broad-panel NGS cohort. Mutant-allele tumour heterogeneity (MATH) score was calculated based on the dispersion of variant allele frequency (VAF) distribution [14]. Higher MATH score is associated with higher intratumor heterogeneity (ITH).
To identify driver mutations of SSN-LUADs, dNdScv algorithm [15, 16] was applied to infer significantly mutated genes (q < 0.100 in any caller and nonsilent mutations n ≥ 5) in the broad-panel NGS cohort. For oncogenic pathway analyses, only functional alterations labelled as oncogenic, likely oncogenic or predicted oncogenic in OncoKB database were retained, discarding variants of unknown significance [17–19]. Therapeutic actionability information was also annotated using OncoKB database. Each genomic alteration was stratified into one of four levels according to its clinical implication [20].
Cancer-evolution models were tested in the broad-panel NGS cohort and the WES cohort separately using Cancer Progression Inference (CAPRI) algorithm with 100 non-parametric bootstrap (NPB) iterations [21–23]. Non-negative matrix factorisation (NMF) algorithm was used for de novo discovery of mutational signatures associated with SSN-LUADs in the WES cohort [24, 25]. Cosine similarity analyses were conducted to measure the similarity between de novo mutational profiles and previously reported signatures [26, 27]. At last, we performed unsupervised consensus clustering using k-means algorithm with 1000 bootstraps and 80% item resampling of the genomic features [28].
Statistical analyses
The distribution of clinicopathologic features were summarised as median (interquartile range [IQR]) or frequency (percentage). Then the features were compared between three radiological subtypes using Kruskal–Wallis test for continuous variables and Fisher’s exact test or chi-square test for categorical variables. Genomic variables were similarly compared across radiological subtypes using Cochran–Armitage test for trend or Fisher’s exact test with the application of false discovery rate (FDR) to account for multiple testing.
Univariate and multivariate negative binomial regression (NBR) models were constructed to determine independent predictive factors for high tumour mutation count or TMB. Least absolute shrinkage and selection operator (LASSO) penalised NBR model with cross-validation was further built to identify clinicopathologic and radiological features that contributed most to tumour genomic characteristics.
Statistical analyses were conducted using R 4.0.2 (R Core Team, Vienna, Austria). All statistical tests were two-sided, and P < 0.050 (or FDR q < 0.100) was considered statistically significant in this study.
Results
Clinicopathologic Characteristics
A total of 247 SSNs (77 pGGNs, 48 HGGNs and 122 PSNs) in 223 patients were included in this study (Table 1). The median tumour size at resection (9.3 vs. 13.6 vs. 23.8 mm for pGGN vs. HGGN vs. PSN, respectively, P < 0.001) and the proportion of lobectomy (25% vs. 27% vs. 65% for pGGN vs. HGGN vs. PSN, respectively, P < 0.001) were much higher in PSNs. Pathologically, the percentage of IAC (40% vs. 65% vs. 94% for pGGN vs. HGGN vs. PSN, respectively, P < 0.001) and MIP/SOL/Mucinous predominant histologic subtype (0% vs. 4% vs. 4% for pGGN vs. HGGN vs. PSN, respectively, P < 0.001) were significantly lower in pGGNs. STAS, lymphovascular invasion and pathologic stage IB disease only presented in PSNs. The high IAC proportion (40%) of the pGGN subgroup was probably due to the large tumour size (9.3 [7.0–15.6] mm) and that NGS could only be applied to large pGGNs with enough tissue for sampling.
Table 1.
Clinicopathologic characteristics by radiological subtype.
| Characteristic | Total (N = 247) | pGGN (n = 77 (31%)) | HGGN (n = 48 (20%)) | PSN (n = 122 (49%)) | P value |
|---|---|---|---|---|---|
| Age at resection, y | 59.0 (50.5–66.0) | 56.0 (45.0–64.0) | 54.5 (48.5–64.0) | 62.0 (56.0–67.0) | <0.001* |
| Sex | 0.429 | ||||
| Male | 91 (37) | 24 (31) | 20 (42) | 47 (39) | |
| Female | 156 (63) | 53 (69) | 28 (58) | 75 (61) | |
| Smoking status | 0.552 | ||||
| Nonsmoker | 199 (81) | 65 (84) | 37 (77) | 97 (80) | |
| Smoker | 48 (19) | 12 (16) | 11 (23) | 25 (20) | |
| Total size on CT, mm | 16.1 (10.2–25.0) | 9.3 (7.0–15.6) | 13.6 (10.3–16.3) | 23.8 (17.1–29.5) | <0.001* |
| Procedure type | <0.001* | ||||
| Wedge resection | 106 (43) | 47 (61) | 30 (63) | 29 (24) | |
| Segmentectomy | 29 (12) | 11 (14) | 5 (10) | 13 (11) | |
| Lobectomy | 112 (45) | 19 (25) | 13 (27) | 80 (65) | |
| Pathological subtype | <0.001* | ||||
| AIS | 7 (3) | 7 (9) | 0 (0) | 0 (0) | |
| MIA | 63 (25) | 39 (51) | 17 (35) | 7 (6) | |
| IAC | 177 (72) | 31 (40) | 31 (65) | 115 (94) | |
| Predominant histology | <0.001* | ||||
| LEP | 85 (34) | 47 (61) | 23 (48) | 15 (12) | |
| ACI/PAP | 155 (63) | 30 (39) | 23 (48) | 102 (84) | |
| MIP/SOL/Mucinous | 7 (3) | 0 (0) | 2 (4) | 5 (4) | |
| MIP/SOL component | 0.042* | ||||
| No | 237 (96) | 76 (99) | 48 (100) | 113 (93) | |
| Yes | 10 (4) | 1 (1) | 0 (0) | 9 (7) | |
| STAS | 0.012* | ||||
| No | 239 (97) | 77 (100) | 48 (100) | 114 (93) | |
| Yes | 8 (3) | 0 (0) | 0 (0) | 8 (7) | |
| VPI | 0.191 | ||||
| No | 243 (98) | 77 (100) | 48 (100) | 118 (97) | |
| Yes | 4 (2) | 0 (0) | 0 (0) | 4 (3) | |
| LVI | <0.001* | ||||
| No | 233 (94) | 77 (100) | 48 (100) | 108 (89) | |
| Yes | 14 (6) | 0 (0) | 0 (0) | 14 (11) | |
| Pathological stage | <0.001* | ||||
| 0 | 7 (3) | 7 (9) | 0 (0) | 0 (0) | |
| IA | 230 (93) | 70 (91) | 48 (100) | 112 (92) | |
| IB | 10 (4) | 0 (0) | 0 (0) | 10 (8) | |
| NGS panel | 0.559 | ||||
| HR363 | 45 (18) | 17 (22) | 10 (21) | 18 (15) | |
| HR457 | 78 (32) | 25 (32) | 12 (25) | 41 (34) | |
| RS520 | 124 (50) | 35 (46) | 26 (54) | 63 (51) |
Note: Data are number (%) or median (interquartile range).
ACI acinar, AIS adenocarcinoma in situ, CT computed tomography, IAC invasive adenocarcinoma, LEP lepidic, LVI lymphovascular invasion, MIA minimally invasive adenocarcinoma, MIP micropapillary, NGS next-generation sequencing, PAP papillary, SOL solid, STAS spread through air spaces, VPI visceral pleural invasion.
Significant differences are labelled with asterisks (P < 0.050).
Genomic features
No significant difference was found regarding the distribution of NGS panels between three SSN subgroups (P = 0.559). Seven hundred and fifty-eight nonsynonymous somatic mutations were identified in all SSNs, including 496 SNVs, 31 splice-site, 51 stop-gain/start lost, 73 frame-shift indels and 107 in-frame indels, insertions and deletions (Fig. 1a). The mutational landscapes of SSNs in WES cohort are summarised in Supplementary Fig. 4A.
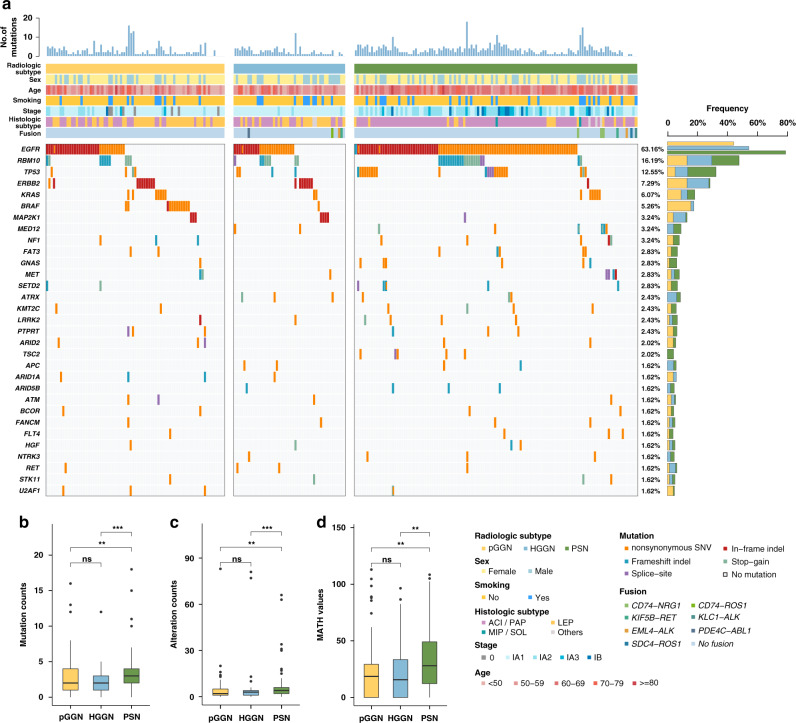
Fig. 1. Association between radiological subtype and genomic features in the broad-panel NGS cohort.
a Oncoprint of the most frequently mutated genes (top 30) by radiological subtype. b Box plot of tumour mutation count versus radiological subtype. c Box plot of genomic alteration count versus radiological subtype. d Box plot of the MATH score versus radiological subtype. MATH mutant-allele tumour heterogeneity.
The tumour mutation count (Fig. 1b), genomic alteration count (Fig. 1c) and MATH score (Fig. 1d) were compared to further reveal the genomic difference of the radiologically different SSNs. In NGS panel cohort, the median tumour mutation count of all SSNs was 3 (IQR, 1–4). The median tumour mutation count of PSNs was significantly higher than that of pGGNs and HGGNs, while no difference was detected between pGGNs and HGGNs (2 [IQR, 1–4] vs. 2 [IQR, 1–3] vs. 3 [IQR, 2–4] for pGGN vs. HGGN vs. PSN, respectively; P value: pGGN vs. HGGN: 0.723, pGGN vs. PSN: 0.001, HGGN vs. PSN < 0.001). The results of negative binomial regression models revealed that age, smoking status and solid components in the mediastinal window were still independent risk factors for tumour mutation count in multivariate analyses (Table 2 and Supplementary Fig. 3). Similar results were found in WES cohort. The median TMB of HGGNs and PSNs were significantly higher than that of pGGNs (pGGN vs. HGGN, P = 0.037; pGGN vs. PSN, P < 0.001). The median TMB of PSNs was higher than that of HGGNs, though without a significant difference (HGGN vs. PSN, P = 0.127, Supplementary Fig. 4B).
Table 2.
Negative binomial regression analysis on association with mutation counts.
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| IRR | 95% CI | P value | IRR | 95% CI | P value | |
| Age at resection, per 1 y increased | 1.026 | 1.017–1.035 | <0.001* | 1.018 | 1.009–1.028 | <0.001* |
| Sex | ||||||
| Male | Reference | Reference | ||||
| Female | 0.707 | 0.583–0.858 | <0.001* | 1.041 | 0.827–1.314 | 0.734 |
| Smoking | ||||||
| No | Reference | Reference | ||||
| Yes | 1.747 | 1.408–2.168 | <0.001* | 1.560 | 1.203–2.026 | <0.001* |
| Pathological subtype | ||||||
| AIS | Reference | Reference | ||||
| MIA | 1.176 | 0.598–2.455 | 0.650 | 1.467 | 0.774–2.990 | 0.266 |
| IAC | 2.043 | 1.066–4.176 | 0.038* | 1.606 | 0.769–3.530 | 0.223 |
| Predominant histology | ||||||
| LEP | Reference | Reference | ||||
| ACI/PAP | 1.721 | 1.397–2.126 | <0.001* | 1.221 | 0.823–1.851 | 0.333 |
| MIP/SOL/mucinous | 2.722 | 1.657–4.499 | <0.001* | 1.492 | 0.820–2.723 | 0.191 |
| Total size on CT (L), per 1 mm increased | 1.026 | 1.017–1.035 | <0.001* | 1.007 | 0.993–1.022 | 0.313 |
| Solid size on CT (L), per 1 mm increased | 1.023 | 1.012–1.033 | <0.001* | 0.978 | 0.950–1.005 | 0.117 |
| Solid size on CT (M), per 1 mm increased | 1.029 | 1.017–1.042 | <0.001* | 1.032 | 1.001–1.064 | 0.042* |
ACI acinar, AIS adenocarcinoma in situ, CT computed tomography, IAC invasive adenocarcinoma, IRR incidence rate ratio, L Lung window, LEP lepidic, M mediastinal (soft-tissue) window, MIA minimally invasive adenocarcinoma, MIP micropapillary, PAP papillary, SOL solid.
Significant differences in the multivariate analysis are labelled with asterisks (P < 0.050).
Next, the genomic alteration counts were compared between pGGNs, HGGNs and PSNs (Fig. 1c). The median genomic alteration count of PSNs was significantly higher (2 [IQR, 1–5] vs. 3 [IQR, 1–4] vs. 4 [IQR, 2–6] for pGGN vs. HGGN vs. PSN, respectively; P value: pGGN vs. HGGN: 0.839, pGGN vs. PSN: 0.001, HGGN vs. PSN < 0.001). Finally, we compared the tumour heterogeneity between pGGNs, HGGNs and PSNs (Fig. 1d). The MATH score of PSNs was also significantly higher, indicating relatively high ITH in PSNs (18.54 [IQR, 0–29.24] vs. 15.67 [IQR, 0–33.37] vs. 28.04 [IQR, 12.22–49.07] for pGGN vs. HGGN vs. PSN, respectively; P value: pGGN vs. HGGN: 0.624, pGGN vs. PSN: 0.003, HGGN vs. PSN: 0.005).
To further diminish the confounding effects caused by pathological subtype, we conducted subgroup analysis on IAC samples. A total of 177 IAC samples were incorporated in the subgroup analysis, including 31 pGGNs, 31 HGGNs, and 115 PSNs, respectively (Table 1). Tumour mutation count, genomic alteration count, and MATH score were further compared among the three radiological subtypes (see Supplementary Fig. 5A–C). The median tumour mutation count (3 [IQR, 1–5] vs. 2 [IQR, 1–3] vs. 3 [IQR, 2–4.5] for pGGN vs. HGGN vs. PSN; Wilcoxon P value: pGGN vs. HGGN: 0.173, pGGN vs. PSN: 0.614, HGGN vs. PSN: 0.009; Kruskal–Wallis P value: 0.039) and genomic alteration count (3 [IQR, 2–7.5] vs. 3 [IQR, 2–4] vs. 4 [IQR, 2–6.5] for pGGN vs. HGGN vs. PSN; Wilcoxon P value: pGGN vs. HGGN: 0.256, pGGN vs. PSN: 0.663, HGGN vs. PSN: 0.021; Kruskal–Wallis P value: 0.084) of PSNs was significantly higher than that of HGGNs. In terms of intratumor heterogeneity, there was an increasing trend regarding MATH score among the three radiological subtypes; however, no significant difference was found probably due to limited sample size and thus insufficient statistical power (20.3 [IQR, 13.3–25] vs. 22.4 [IQR, 0–33.9] vs. 28.6 [IQR, 12.6–47.8] for pGGN vs. HGGN vs. PSN; Wilcoxon P value: pGGN vs. HGGN: 0.977, pGGN vs. PSN: 0.097, HGGN vs. PSN: 0.078; Kruskal–Wallis P value: 0.088).
Driver genes
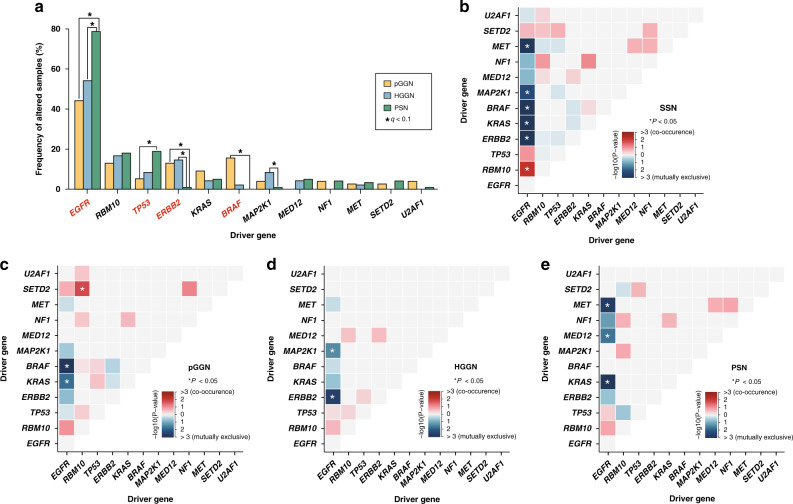
In the entire broad-panel NGS cohort, twelve driver genes at FDR of 0.100 were identified (Fig. 2a). The most frequent drivers were EGFR (71%), TP53 (26%), RBM10 (15%) and KRAS (11%). PSNs harboured higher EGFR mutation frequency than pGGNs and HGGNs (44% vs. 54% vs. 79% for pGGN vs. HGGN vs. PSN, respectively. q value for Fisher test: pGGN vs. HGGN: 0.697; pGGN vs. PSN: < 0.001; HGGN vs. PSN: 0.014). Similarly, TP53 mutation in PSNs was significantly higher than that in pGGNs (5% vs. 8% vs. 19% for pGGN vs. HGGN vs. PSN, respectively. q value for Fisher test: pGGN vs. HGGN: 0.697; pGGN vs. PSN: 0.017; HGGN vs. PSN: 0.319). The results of Cochran–Armitage test revealed a significantly increasing trend in EGFR (P < 0.001) and TP53 (P = 0.004) mutations from pGGNs to HGGNs to PSNs, while a significantly decreasing trend in ERBB2 (P < 0.001) and BRAF (P < 0.001) mutations was identified. The driver genes of the WES cohort further confirmed that PSNs owned the highest EGFR mutation frequency, and that an increasing trend existed in mutation frequencies of EGFR, TP53 and RBM10 from pGGNs to HGGNs to PSNs (Supplementary Fig. 4C).
Fig. 2. Analyses of driver genes detected by dNdScv algorithm in the broad-panel NGS cohort.
a Comparison of mutation frequency of driver genes among radiological subtypes. Drivers with significant differences using Fisher’s exact test were labelled with asterisks (FDR q < 0.100), while those with significant differences in Cochran–Armitage test for trend were highlighted in red (P < 0.050). b–e shows co-occurrence (red) and mutual exclusivity (blue) of driver genes in SSN, pGGN, HGGN, and PSN cohorts. FDR false discovery rate.
We also conducted a subgroup analysis on IAC cohort to further diminish the confounding effects caused by the pathological subtype. In IAC cohort (Supplementary Fig. 5D and Supplementary Table 7), there were increasing trends regarding EGFR (68% vs. 68% vs. 82% for pGGN vs. HGGN vs. PSN; Cochran–Armitage P value: 0.052) and TP53 (6% vs. 13% vs. 20% for pGGN vs. HGGN vs. PSN; Cochran–Armitage P value: 0.058) mutations among the three radiological subtypes. Moreover, significantly decreasing trends in KRAS (19% vs. 6% vs. 5% for pGGN vs. HGGN vs. PSN; Cochran–Armitage P value: 0.017) and U2AF1 (10% vs. 0% vs. 1% for pGGN vs. HGGN vs. PSN; Cochran–Armitage P value: 0.011) mutations were observed.
In terms of co-occurrence and mutual exclusivity (COME), EGFR-RBM10 co-occurred with statistical significance, while EGFR-KRAS and EGFR-STK11 showed mutual exclusiveness in the entire broad-panel NGS cohort (Fig. 2b). In pGGNs, the only pair of co-occurrence drivers was RBM10-SETD2, while another two pairs (EGFR- KRAS, EGFR-BRAF) showed mutual exclusiveness (Fig. 2c). In HGGNs and PSNs, no statistically significant mutual co-occurrence was identified. Two pairs of drivers (EGFR-ERBB2, EGFR-MAP2K1) in HGGNs (Fig. 2d) and three pairs of drivers (EGFR-KRAS, EGFR-MED12, EGFR-MET) in PSNs (Fig. 2e) showed mutual exclusiveness. The difference in COME provided further evidence for genomic discordance between radiologically different SSN subgroups.
Oncogenic pathways
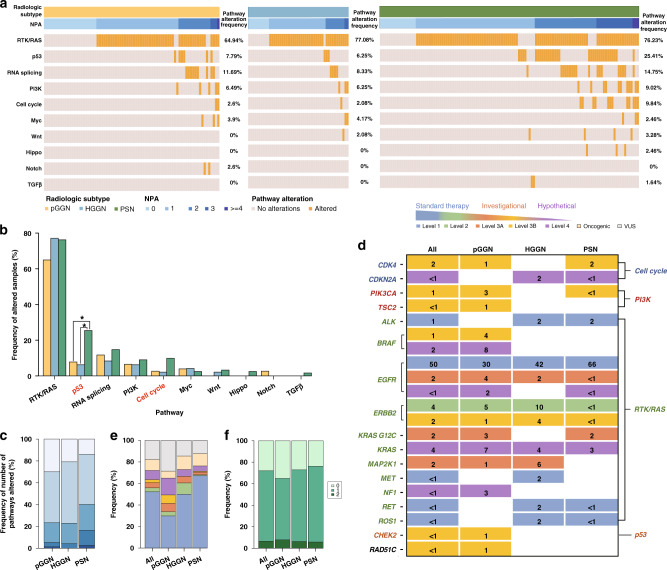
At the sample level, we compared the alteration frequencies of oncogenic signalling pathways by radiological subtype in both the broad-panel NGS cohort (Fig. 3a) and WES cohort (Supplementary Fig. 6A). In broad-panel NGS cohort, the most frequently altered pathway was RTK/RAS (73%), followed by p53 (16%) and RNA splicing/processing (13%). The results of Cochran–Armitage test presented an increasing trend in the alteration frequencies of p53 (P < 0.001) and Cell cycle (P = 0.028) pathways from pGGNs to HGGNs to PSNs (Fig. 3b). In terms of the number of pathways altered (NPA), the mean NPA of the entire broad-panel NGS cohort was 1.23. PSNs possessed a significantly higher NPA than pGGNs and HGGNs (1.00 vs. 1.06 vs. 1.45 for pGGN vs. HGGN vs. PSN, respectively. P value: pGGN vs. HGGN: 0.521, pGGN vs. PSN: 0.001, HGGN vs. PSN: 0.021, Fig. 3c). Furthermore, the COME of oncogenic pathways in the entire broad-panel NGS cohort was displayed in Supplementary Fig. 7. In WES cohort, the results demonstrated an increasing trend in the alteration frequencies of RTK/RAS (P = 0.011), p53 (P = 0.006) and RNA splicing (P = 0.008) pathways from pGGNs to HGGNs to PSNs (Supplementary Fig. 6B). Consistently, the PSNs in WES cohort also obtained higher NPA than pGGNs and HGGNs (0.75 vs. 1.00 vs. 1.38 for pGGN vs. HGGN vs. PSN, respectively. P value: pGGN vs. HGGN: 0.305, pGGN vs. PSN < 0.001, HGGN vs. PSN: 0.075, Supplementary Fig. 6C).
Fig. 3. Oncogenic pathway and therapeutic actionability analyses in the broad-panel NGS cohort.
a Oncoprint of altered oncogenic pathways by radiological subtype. Pathway alterations were presented by increasing NPA. b Comparison of alteration frequency of oncogenic pathways among radiological subtypes. Pathways with significant differences using Fisher’s exact test were labelled with asterisks (FDR q < 0.100), while those with significant differences in Cochran–Armitage test for trend are highlighted in red (P < 0.050). c Frequency of NPA versus radiological subtype. d Frequency of samples with actionable alteration by radiological subtype. e Level of evidence versus radiological subtype. Samples were classified by the alteration that carried the highest level of evidence. f Frequency of the number of actionable alteration versus radiological subtype. FDR false discovery rate, NPA number of pathway alteration.
We also conducted a subgroup analysis on IAC cohort to further diminish the confounding effects caused by the pathological subtype. In IAC cohort (Supplementary Fig. 5E and Supplementary Table 14), RTK/RAS (77%) was still the most frequently altered pathway, followed by p53 (21%) and RNA splicing/processing pathway (14%). A significantly increasing trend in the alteration frequency of p53 pathway (13% vs. 10% vs. 27% for pGGN vs. HGGN vs. PSN; Cochran–Armitage P value: 0.034) was identified in IAC cohort.
Therapeutic actionabilities
In total, one hundred and ninety-four actionable alterations across 17 genes were identified using OncoKB database, including 132 (68%) level 1, 10 (5%) level 2, 13 (7%) level 3A, 17 (9%) level 3B and 22 (11%) level 4 alterations (Fig. 3d). The RTK/RAS pathway harboured the most actionable alterations (94% [182/194]), of which (73% [132/182]) had level 1 evidence. At sample level, actionable alterations were identified in 72% (178/247) in 247 SSN samples, of which (74% [132/178]) had level 1 evidence. PSNs possessed a significantly higher frequency of level 1 actionable target than pGGNs and HGGNs (30% vs. 50% vs. 67% for pGGN vs. HGGN vs. PSN, respectively, P < 0.001). The difference in numbers of actionable alterations per sample among three subgroups was not statistically significant (0.73 vs. 0.79 vs. 0.82, for pGGN vs. HGGN vs. PSN, respectively, all P value > 0.050).
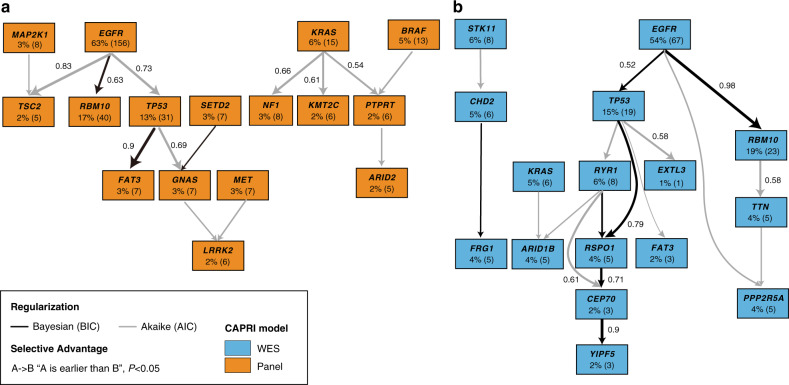
Cancer-evolution models
To gain further insights into the temporal heterogeneity of somatic mutations in SSN-LUAD evolution, we applied CAPRI algorithm in both broad-panel and WES cohorts (Fig. 4 and Supplementary Fig. 8). In the broad-panel cohort, our model captured early somatic mutational events including EGFR (63%), KRAS (6%) and BRAF (5%). The selection of RBM10 (non-parametric bootstrap score [NPB] 63%), TP53 (NPB 73%) and TSC2 (NPB 83%) mutations by EGFR were observed. Moreover, FAT3 mutation occurred in a specific order following EGFR and TP53 mutations with an NPB score of 90%. In the WES cohort, EGFR (54%), STK11 (6%), and KRAS (5%) were identified as early mutations. Similar postulated selective advantage relations, including the selection of RBM10 (NPB 98%) and TP53 (NPB 52%) mutations by EGFR, were confirmed in this cohort. In summary, our models revealed EGFR mutation as a key early event in the progression of SSN-LUADs, with subsequently two evolutionary trajectories involving either RBM10 or TP53 mutation.
Fig. 4. Cancer-evolution models of early-stage SSN-LUADs.
a, b show simplified version of evolution models identified in the broad-panel NGS cohort (orange) and the WES cohort (blue) using CAPRI algorithm. Edges with NPB score >50% were shown. Both AIC and BIC were applied for regularisation to prevent overfitting. AIC Akaike information criteria, BIC Bayesian information criteria, CAPRI cancer progression inference, NPB non-parametric bootstrap.
Mutational signatures
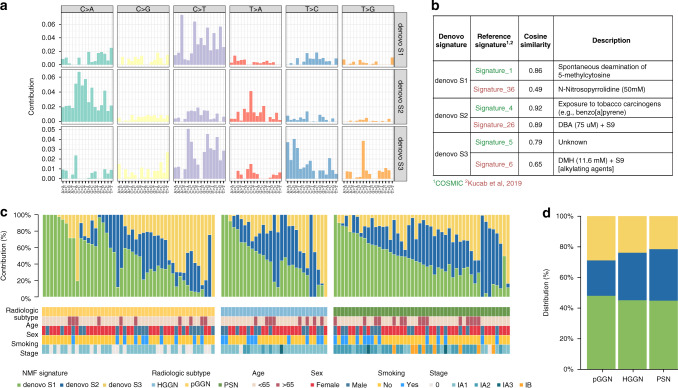
Using the NMF algorithm, we discovered three mutational signatures in the WES cohort, and defined them as de novo S1, S2, and S3 respectively (Fig. 5a and Supplementary Table 19). To reveal the potential contribution of endogenous and exogenous mutagens to these de novo signatures, cosine similarity analyses against 30 validated human cancer signatures from COSMIC database (https://cancer.sanger.ac.uk/cosmic/signatures_v2) and 53 environmental carcinogen signatures investigated by Kucab et al. were performed [29]. Mutational signatures best matching those de novo ones were (1) exposure to tobacco carcinogens, (2) exposure to dibenz[a,h]anthracene (DBA), and (3) deamination of 5-methylcytosine (Fig. 5b). DBA are polycyclic aromatic hydrocarbons (PAHs) produced by incomplete combustion of organic matter, primarily found in gasoline exhaust, tobacco smoke, coal tar and soot (https://pubchem.ncbi.nlm.nih.gov). More importantly, these airborne particulate PAHs adsorbed by fine particulate matter (PM2.5-bound PAHs) are highly carcinogenic, and have relatively high regional concentration in Eastern Asia [30]. Thus, our mutational signature analyses suggested that cumulative exposure to ambient air pollutants potentially played a role during tumorigenesis of SSN-LUADs in the Chinese population [31, 32].
Fig. 5. Mutational signature analyses in the WES cohort.
a Trinucleotide motif frequency plot of the three de novo mutational profiles associated with SSN-LUADs. b Cosine similarity comparison between de novo mutational profiles and mutation patterns of 30 validated cancer signatures reported from the COSMIC database (green) and 53 carcinogen signatures reported from Kucab et al. (red). c Bar plots of de novo mutational signatures by individual tumours. d Distribution of de novo mutational signatures by radiological subtype. COSMIC catalogue of somatic mutations in cancer.
Finally, relative percentage of each de novo signature and corresponding clinical features of individual patients were illustrated in Fig. 5c. The proportion of samples enriched in each de novo signature was not significantly different among the three radiological subtypes (Fig. 5d).
Genomic clustering analyses
The above-mentioned results showed that different subtypes of SSN-LUADs harboured distinctive genomic landscape. However, whether genomic features could distinguish radiological subtype remained unknown. We thus performed unsupervised clustering using genomic features regardless of radiological subtype. As shown in Supplementary Fig. 9, two distinct genomic “clusters” emerged in the SSN-LUADs using consensus clustering on high-frequency somatic mutations. Compared with cluster 2, samples from cluster 1 not only possessed higher tumour mutation count (3 [IQR, 2–4] vs. 2 [IQR, 1–3], P < 0.001), but also had significantly higher genomic alteration count (4 [IQR, 2–6] vs. 2 [IQR, 1–5], P < 0.001) and higher intratumor heterogeneity (the MATH score: 29.2 [IQR, 15.4–51.1] vs. 12.2 [IQR, 0–23.7], P < 0.001). Thus, SSN-LUADs allocated in cluster 1 harboured significantly higher complexity in genomic architecture compared with those allocated in cluster 2. The distribution of radiological subtypes between the two genomic clusters was then assessed. We found that unlike pGGNs/HGGNs, PSNs were significantly enriched in cluster 1 (OR: 0.470, [95% CI, 2.524–8.828], P < 0.001). Moreover, the results of Cochran–Armitage test revealed a significantly increasing trend in cluster 1 proportion from pGGNs (44%) to HGGNs (54%) to PSNs (79%), confirming a progressively complex genomic architecture among the three radiological subtypes during the acquisition of solid component on chest CT.
Discussion
To deepen the understanding of the heterogeneity among radiologically different SSNs, for the first time, we compared the genomic features of resected LUADs manifesting as pGGNs, HGGNs and PSNs using broad-panel NGS, and further validated the results in the WES cohort. Results of both the broad-panel NGS and WES cohorts accordantly reflected HGGNs harboured much lower complexity in genomic architecture compared with PSNs. A slight uptrend in genomic architecture complexity was identified from pGGNs to HGGNs in WES cohort, although similarly lower level of malignancy was revealed for pGGNs and HGGNs in the broad-panel NGS cohort, penetrating that HGGN was an intermediate form in the evolution of SSN between pGGN and PSN. The stepwise evolutionary genomic characteristic of pGGN, HGGN and PSN offered valuable biological insights into the management of radiologically different SSNs, and alert clinicians to the significance of solid component in both the mediastinal window and lung window which may prompt better management of SSNs.
Currently, most guidelines take only solid component in the lung window into consideration in SSNs management. Now that previous studies have reported differences in the natural history and long-term survival between HGGN and PSN [8, 9], it is worthwhile to explore in depth the molecular mechanism underlying the clinicopathological heterogeneity between pGGN, HGGN and PSN. On the one hand, HGGNs possessed lower complexity in genomic architecture compared with PSNs. In the broad-panel NGS cohort, we identified significantly lower tumour mutation count, genomic alteration count and MATH score in HGGNs compared with PSNs. Moreover, lower driver gene mutation frequencies (including EGFR and TP53), oncogenic pathway mutation frequencies (including p53) and NPA were detected in HGGNs than in PSNs, which were validated in the WES cohort. These results are in favour of previous studies reporting worse clinicopathological outcome in PSNs [9].
On the other hand, although similarly stable genomic characteristics in HGGNs and pGGNs were identified in broad-panel NGS cohort, a slight upgoing trend from pGGNs to HGGNs was revealed in WES cohort. In broad-panel NGS cohort, HGGNs harboured similar tumour mutation count, genomic alteration count, MATH score and NPA compared with pGGNs. Meanwhile, in WES cohort, HGGNs possessed higher TMB and NPA than pGGNs. In both cohorts, HGGNs harboured higher frequencies of EGFR and TP53 mutations than pGGNs, though without statistically significant difference. The slightly inconsistent results were likely due to the incomplete overlap of two sequencing methods. The broad panel covered only the hot-spot mutations but not rare mutations included in WES. To sum up, the evidence above is detailed enough to sustain that HGGN represents an intermediate form between pGGN and PSN deep from genomic level. SSN with solid component only in lung window may already reflect the onset of progression in genomic complexity.
Remarkably, stepwise evolution from pGGN to HGGN to PSN was uncovered by the uptrend of pivotal driver mutations and pathway alterations. As for driver mutation frequencies, an increasing trend in EGFR and TP53 from pGGNs to HGGNs to PSNs was revealed using Cochran–Armitage test in both cohorts. As for pathways alteration frequencies, an increasing trend in p53 and Cell cycle was identified from pGGNs to HGGNs to PSNs in broad-panel NGS cohort. In WES cohort, the alteration frequencies of RTK/RAS, p53 and RNA splicing raised from pGGNs to HGGNs to PSNs. The similar differences in frequencies of driver gene mutation and oncogenic pathway alteration by radiological subtype were revealed in IAC subgroup. Those results implied EGFR and TP53 mutations, and pathway alterations including p53, RTK/RAS, Cell cycle and RNA splicing, played important roles in driving the invasiveness of SSNs, both from pGGN to HGGN and from HGGN to PSN. The stepwise evolutionary genomic characteristics from pGGN to HGGN to PSN echoed the radiological natural history of SSNs [8].
Supplementary information
Author contributions
HL: conceptualisation, project administration, formal analysis and writing—original draft; ZS: methodology, software, formal analysis and writing—original draft; RX: formal analysis and writing—original draft; QQ: resources, investigation and data curation; XL: project administration and supervision; HH: methodology, software, formal analysis and visualisation; XW: investigation; JZ: investigation; ZW: investigation; PY: investigation; FY: project administration and supervision; JW: project administration and supervision.
Funding
This work was financially supported by the National Natural Science Foundation of China (grant 82002410, HL), the Major Research Plan of National Natural Science of China (grant 92059203, JW) and Peking University People’s Hospital Scientific Research Development Funds (grant RDY2020-02, HL).
Data availability
All data are available from the authors upon reasonable request.
Competing interests
HH and KL were employed by the company Berry Oncology, Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board at Peking University People’s Hospital (2020PHB363-01). The study was performed in accordance with the Declaration of Helsinki.
Consent to publish
No individual person’s data is presented in this manuscript.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Hao Li, Zewen Sun, Rongxin Xiao, Qingyi Qi.
Contributor Information
Xiao Li, Email: dr.lixiao@163.com.
Jun Wang, Email: wangjun@pkuph.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-01821-7.
References
- 1.Zhang Y, Jheon S, Li H, Zhang H, Xie Y, Qian B, et al. Results of low-dose computed tomography as a regular health examination among Chinese hospital employees. J Thorac Cardiovasc Surg. 2020;160:824–31. [DOI] [PubMed]
- 2.Kim YW, Kwon BS, Lim SY, Lee YJ, Park JS, Cho YJ, et al. Lung cancer probability and clinical outcomes of baseline and new subsolid nodules detected on low-dose CT screening. Thorax. 2021;76:980–8. [DOI] [PMC free article] [PubMed]
- 3.Sawada S, Yamashita N, Sugimoto R, Ueno T, Yamashita M. Long-term outcomes of patients with ground-glass opacities detected using CT scanning. Chest. 2017;151:308–15. [DOI] [PubMed]
- 4.Travis WD, Asamura H, Bankier AA, Beasley MB, Detterbeck F, Flieder DB, et al. The IASLC lung cancer staging project: proposals for coding T categories for Subsolid Nodules and assessment of tumor size in part-solid tumors in the forthcoming eighth edition of the TNM classification of lung cancer. J Thorac Oncol. 2016;11:1204–23. [DOI] [PubMed]
- 5.MacMahon H, Naidich DP, Goo JM, Lee KS, Leung A, Mayo JR, et al. Guidelines for management of incidental pulmonary nodules detected on CT images: from the Fleischner Society 2017. Radiology. 2017;284:228–43. [DOI] [PubMed]
- 6.Asamura H, Hishida T, Suzuki K, Koike T, Nakamura K, Kusumoto M, et al. Radiographically determined noninvasive adenocarcinoma of the lung: survival outcomes of Japan Clinical Oncology Group 0201. J Thorac Cardiovasc Surg. 2013;146:24–30. [DOI] [PubMed]
- 7.Bankier AA, MacMahon H, Goo JM, Rubin GD, Schaefer-Prokop CM, Naidich DP. Recommendations for measuring pulmonary nodules at CT: A statement from the Fleischner Society. Radiology. 2017;285:584–600. [DOI] [PubMed]
- 8.Kakinuma R, Noguchi M, Ashizawa K, Kuriyama K, Maeshima AM, Koizumi N, et al. Natural history of pulmonary subsolid nodules: a prospective multicenter study. J Thorac Oncol. 2016;11:1012–28. [DOI] [PubMed]
- 9.Yin J, Xi J, Liang J, Zhan C, Jiang W, Lin Z, et al. Solid components in the mediastinal window of computed tomography define a distinct subtype of subsolid nodules in clinical stage I lung cancers. Clin Lung Cancer. 2021;22:324–31. [DOI] [PubMed]
- 10.Li Y, Li X, Li H, Zhao Y, Liu Z, Sun K, et al. Genomic characterisation of pulmonary subsolid nodules: mutational landscape and radiological features. Eur Respir J. 2020;55. 10.1183/13993003.01409-2019 [DOI] [PubMed]
- 11.Wang Y, Yu M, Yang JX, Cao DY, Zhang Y, Zhou HM, et al. Genomic comparison of endometrioid endometrial carcinoma and its precancerous lesions in Chinese patients by high-depth next generation sequencing. Front Oncol. 2019;9:123. [DOI] [PMC free article] [PubMed]
- 12.Cheng Y, Zhang Y, Yuan Y, Wang J, Liu K, Yu B, et al. The comprehensive analyses of genomic variations and assessment of TMB and PD-L1 expression in Chinese lung adenosquamous carcinoma. Front Genet. 2020;11:1794. [DOI] [PMC free article] [PubMed]
- 13.Xiao W, Zhang G, Chen B, Chen X, Wen L, Lai J, et al. Characterization of frequently mutated cancer genes and tumor mutation burden in Chinese breast cancer. Front Oncol. 2021;11:1107. [DOI] [PMC free article] [PubMed]
- 14.Mroz EA, Rocco JW. A novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol. 2013;49:211–5. [DOI] [PMC free article] [PubMed]
- 15.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013; 10.1038/nature12213 [DOI] [PMC free article] [PubMed]
- 16.Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173:371–85. [DOI] [PMC free article] [PubMed]
- 17.Network CGAR. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. [DOI] [PMC free article] [PubMed]
- 18.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell. 2018;173:321–37. [DOI] [PMC free article] [PubMed]
- 19.Zhou J, Sanchez-Vega F, Caso R, Tan KS, Brandt WS, Jones GD, et al. Analysis of tumor genomic pathway alterations using broad-panel next-generation sequencing in surgically resected lung adenocarcinoma. Clin Cancer Res. 2019;14:2763–7. [DOI] [PMC free article] [PubMed]
- 20.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017; 10.1200/PO.17.00011 [DOI] [PMC free article] [PubMed]
- 21.Ramazzotti D, Caravagna G, Olde Loohuis L, Graudenzi A, Korsunsky I, Mauri G, et al. CAPRI: efficient inference of cancer progression models from cross-sectional data. Bioinformatics. 2015;31:3016–26. [DOI] [PubMed]
- 22.De Sano L, Caravagna G, Ramazzotti D, Graudenzi A, Mauri G, Mishra B, et al. TRONCO: an R package for the inference of cancer progression models from heterogeneous genomic data. Bioinformatics. 2016;32:1911–3. [DOI] [PMC free article] [PubMed]
- 23.Caravagna G, Graudenzi A, Ramazzotti D, Sanz-Pamplona R, De Sano L, Mauri G, et al. Algorithmic methods to infer the evolutionary trajectories in cancer progression. Proc Natl Acad Sci USA. 2016;113:E4025–34. [DOI] [PMC free article] [PubMed]
- 24.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci USA. 2004;101:4164–9. [DOI] [PMC free article] [PubMed]
- 25.Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010;11:1–9. [DOI] [PMC free article] [PubMed]
- 26.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;534:47–54.
- 27.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;35:951–9. [DOI] [PMC free article] [PubMed]
- 28.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–3. [DOI] [PMC free article] [PubMed]
- 29.Kucab JE, Zou X, Morganella S, Joel M, Nanda AS, Nagy E, et al. A compendium of mutational signatures of environmental agents. Cell. 2019;177:821–36. [DOI] [PMC free article] [PubMed]
- 30.Yan D, Wu S, Zhou S, Tong G, Li F, Wang Y, et al. Characteristics, sources and health risk assessment of airborne particulate PAHs in Chinese cities: a review. Environ Pollut. 2019;248:804–14. [DOI] [PubMed]
- 31.Chen YJ, Roumeliotis TI, Chang YH, Chen CT, Han CL, Lin MH, et al. Proteogenomics of non-smoking lung cancer in East Asia delineates molecular signatures of pathogenesis and progression. Cell. 2020;182:226–44. [DOI] [PubMed]
- 32.Myers R, Brauer M, Dummer T, Atkar-Khattra S, Yee J, Melosky B, et al. High-ambient air pollution exposure among never smokers versus ever smokers with lung cancer. J Thorac Oncol. 2021;16:1850–8. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the authors upon reasonable request.