Abstract
Widespread polybrominated diphenyl ethers (PBDEs) contamination poses risks to human health and ecosystems. Bioremediation is widely considered to be a less ecologically disruptive strategy for remediation of organohalide contamination, but bioremediation of PBDE-contaminated sites is limited by a lack of knowledge about PBDE-dehalogenating microbial populations. Here we report anaerobic PBDE debromination in microcosms established from geographically distinct e-waste recycling sites. Complete debromination of a penta-BDE mixture to diphenyl ether was detected in 16 of 24 investigated microcosms; further enrichment of these 16 microcosms implicated microbial populations belonging to the bacterial genera Dehalococcoides, Dehalogenimonas, and Dehalobacter in PBDE debromination. Debrominating microcosms tended to contain either both Dehalogenimonas and Dehalobacter or Dehalococcoides alone. Separately, complete debromination of a penta-BDE mixture was also observed by axenic cultures of Dehalococcoides mccartyi strains CG1, CG4, and 11a5, suggesting that this phenotype may be fairly common amongst Dehalococcoides. PBDE debromination in these isolates was mediated by four reductive dehalogenases not previously known to debrominate PBDEs. Debromination of an octa-BDE mixture was less prevalent and less complete in microcosms. The PBDE reductive dehalogenase homologous genes in Dehalococcoides genomes represent plausible molecular markers to predict PBDE debromination in microbial communities via their prevalence and transcriptions analysis.
Subject terms: Environmental microbiology, Environmental sciences
Introduction
Recent decades have seen an exponential increase in the global market for consumer electronics [1]. The social and ecological impacts of decreases in the lifespans of many electronic devices have attracted a great deal of public attention as the annual production volume of electronic waste (e-waste) continues to rise [2]. Despite increased regulation and monitoring of the collection, disposal, and recycling of e-waste, an estimated 82.6% of global e-waste flows remain poorly documented [3]. Much of this undocumented e-waste is dumped, traded, or recycled by small-scale operations, leading to environmental contamination and posing health risks to human populations and ecosystems. E-waste often contains high levels of polybrominated diphenyl ethers (PBDEs), which were used extensively as additive flame retardants in a variety of manufactured products until the implementation of multiple bans in the mid-to-late-2000s [4]. Leaching of PBDEs from these sites, which have some of the highest reported environmental PBDE concentrations, can introduce severe contamination into nearby soils and sediments and affect land-use patterns (e.g., restricting agricultural activities over large areas) [5, 6]. Because e-waste disposal and recycling mostly occur in developing countries that lack incentives and funding for dedicated site remediation, environmental degradation or dehalogenation of PBDEs typically occurs via natural attenuation.
PBDEs are inherently resistant to natural attenuation, and efforts to identify microbial processes suitable for PBDEs bioremediation have been hindered by the low biodegradability of PBDEs in sediments and soils [7]. Nevertheless, two independent studies have recently reported the isolation of distinct Dehalococcoides mccartyi strains that completely debrominate penta-BDE mixtures. Both isolates were derived from soils contaminated by e-waste [8, 9], suggesting that long-term exposure to PBDEs may foster adaptions that enable metabolic conversion of PBDEs in some microbial populations. Therefore, it is of ecological relevance and scientific interest to comprehensively evaluate biological debromination of different PBDEs by the microbial communities present at e-waste contaminated sites as a better understanding of PBDE-debrominating bacterial populations and debromination pathways would facilitate the development of in situ biological remediation strategies. Another challenge to the implementation of in situ bioremediation of PBDEs is the lack of suitable genetic markers to evaluate and monitor microbial activity associated with PBDEs debromination. Currently, several anaerobic organohalide respiring bacteria (OHRB) capable of partial or complete debromination of PBDEs have been identified [7], but few PBDE-dehalogenating reductive dehalogenases (RDase) have been described. Genes encoding RDases with known dehalogenation capacity can be used as genetic markers to indicate in situ dehalogenation potential or to monitor metabolic activity of OHRB [10, 11], so the paucity of known PBDE RDases limits site characterization and bioaugmentation of PBDE contaminated sites.
This study investigated dehalogenation of commercially used penta- and octa-BDE mixtures by microcosms established from sediments and soils from geographically distinct e-waste recycling sites that are heavily contaminated with PBDEs. PBDE-debrominating OHRB populations (i.e., Dehalococcoides, Dehalogenimonas, and Dehalobacter) were identified and interactions among the different dehalogenating populations were inferred. The prevalence of Dehalococcoides in PBDE-debrominating microcosms prompted an investigation of PBDE debromination by previously isolated D. mccartyi strains CG1, CG4, and 11a5. These investigations identified additional reductive dehalogenase genes that could be used to predict and monitor PBDE debromination by Dehalococcoides populations in the microcosms and enrichments derived from e-waste contaminated sediments and soils.
Materials and methods
Chemical analyses and identification of debromination pathways
An octa-BDE mixture comprising nona- through hexa-BDEs (BDE-207, −203, −197, and 196, 183, and 153) and the individual hepta-BDE congener (BDE-183) were purchased from Sigma-Aldrich (St. Louis, MO, USA). A customized penta-BDE mixture comprising the predominant tetra- and penta-BDEs congeners in the commercial penta-BDE mixture (e.g., DE-71) (i.e., tetra-BDE 47, penta-BDE 99, and penta-BDE 100 in a mass ratio of 1:1:1) was purchased from Agilent Technologies, Inc. (Santa Clara, CA, USA). Deca-bromobiphenyl (DBB) was purchased from AccuStandard (New Haven, CT, USA) and used as an internal quantification standard.
To monitor the abundance of different PBDE congeners during cultivation, 1 ml samples were collected from culture bottles and extracted with isooctane (1:1 volume ratio) by vigorous agitation for 1 h. Extracted PBDEs were quantified on an Agilent gas chromatograph-mass spectrometer (GC6890-MSD5975) equipped with a BD-5MS column (15 m × 0.25 mm × 0.25 μm; Restek, Bellefonte, PA, USA) as previously described [12, 13]. Each PBDE congener present in the penta- and octa-BDE mixtures was quantified against five-point calibration curves generated for each congener. Intermediate metabolites and debromination pathways were identified as previously described [13]. Briefly, 39 hepta- to mono-BDE congeners (minimum purity 98%; Cambridge Isotope Laboratories, Inc., Andover, MA) were combined in a technical mixture and used as standards to identify PBDE congeners produced by debromination. The debromination products were assigned to PBDE congener homolog groups (e.g., tetra-BDEs) and quantified using the average area of peaks corresponding to each PBDE congener homolog (e.g., the average of all peaks corresponding to tetra-BDEs) (Supplementary Material Table S1). This approach was adopted to approximate the quantity of PBDE congeners for which standards are not available. Instrument detection limits for PBDE congeners ranged from 0.070–0.224 nM. Congener names, positions and number of bromine substituents, IUPAC nomenclature, CAS number, and GC retention time of PBDE congeners mentioned in this study are available in the Supplementary Dataset (Table S9).
Establishment and enrichment of microcosms
To investigate PBDE-debromination and identify potential debrominating bacterial populations in environmental samples, microcosms were established from 24 soil samples (30–50 cm subsurface) collected from e-waste contaminated sites around Asia (dubbed EW-1 to EW-24). The PBDEs in these soil samples were extracted based on a modified ultrasonically assisted method [5] and detected by GC-MS as described above. Briefly, 4 g freeze-dried and homogenized soils were weighed and PBDEs were extracted using isooctane (1:2, w/v) after vigorous vortexing for 16 h and ultrasonication for 15 min. The extracts were flushed with N2 until dried and then dissolved in 100 µL isooctane. Microcosms were established by adding 6 g soil to 60 ml serum bottles containing 30 ml bicarbonate-buffered defined mineral salts medium (DCB1) prepared as previously described [14]. Lactate (10 mM) was amended to the microcosms as a carbon source and electron donor. A total of either ~1.4 μM customized penta-BDE mixture or ~0.3 μM octa-BDE mixture was added as the electron acceptor source. The octa-BDE mixture comprised nona- through hexa-BDEs, in a mass ratio of 2:4:30:1. Debromination of the penta- and octa-BDE mixtures was monitored at regular intervals over periods of 2 and 9 months, respectively. Microcosms that debrominated the penta-BDE mixture after 2 months’ cultivation were sub-cultured in DCB1 medium amended with lactate and the penta-BDE mixture until sediment-free cultures were obtained. Lactate was replaced by acetate (10 mM) and hydrogen (0.3 ppv) as a carbon source and an electron donor, respectively, in enrichments to foster growth of PBDE-debrominating obligate OHRB populations.
Total bacterial abundance and abundance of OHRB were inferred by real-time quantitative PCR (qPCR) targeting the 16 S rRNA gene using universal bacterial (338F-518R) and genus-specific primers, respectively, as previously described [9–12, 14]. The presence of known PBDE-debrominating facultative OHRB populations in enrichment cultures that debrominated the penta-BDE mixture was assayed using genus-specific primers (described in Table S2) to elicit the potential involvement of these genera in the observed debromination. Briefly, 1 ml samples were collected from cultures and centrifuged at 12,000 × g (10 min, 4 °C); DNA was extracted from samples using the QIAGEN DNeasy Blood and Tissue Kit (GmbH, Hilden, Germany) according to the manufacturer’s instructions. Target amplicons were quantified based on 6-point standard calibration curves in the range of 102–107 /reaction; plasmids containing target gene fragments were constructed using the pGEM-T Easy Vector system (Promega, Madison, WI, USA) to generate the standard curves, all of which had an r > 0.999. The amplification efficiencies of all qPCR experiments were in the range of 95–105%. The limit of blank (LOB) and standard deviation of blank (σB) were calculated from 10 reactions using non-target DNA as template. The limit of detection (LOD) and limit of quantification (LOQ) were calculated as LOD = LOB + 3 × σB and LOQ = LOB + 10 × σB, respectively [12] (Table S3).
Growth and cultivation of Dehalococcoides strains
Dehalogenation of the penta- and octa- BDE mixtures by previously isolated D. mccartyi strains 11a, 11a5, 11 G, ANAS2, GEO, CG1, CG4, and CG5 [7, 13, 15, 16] were evaluated in cultures grown in 160 ml serum bottles containing 100 ml DCB1 medium [17]. Details of these D. mccartyi strains are available in Table S4. Cultures were amended with acetate (10 mM) as the sole carbon source, hydrogen (0.33 ppv) as an electron donor, and either chloroethenes (~0.5 mM) or PBDEs as indicated. All experiments involving debromination of the penta-BDE mixture were performed in cultures that had been cultivated with the penta-BDE mixture for at least six consecutive transfers (5% v/v) to mitigate residual effects of cultivation with alternate electron acceptors, i.e., tetrachloroethene (PCE) or trichloroethene (TCE), from the inoculum in the initial transfer. Debromination rate was calculated as the total number of bromine substituents removed per unit time, i.e., the difference between the total number of bromine substituents on all PBDE congeners (calculated as the product of the number of bromine substituents on a congener and the concentration of that congener) present in the initial amendment and in cultures after incubation for some period. Abiotic controls, comprising medium without inoculum or amended with autoclaved inoculum, were established alongside all cultures and microcosms to detect abiotic transformation of amended organohalides. All cultures and microcosms were incubated at 30 °C in the dark.
Enzymatic assays and characterization of PBDE RDases in Dehalococcoides
Proteins involved in penta-BDE mixture debromination mediated by D. mccartyi strains CG1, CG4, and 11a5 were elucidated by proteomics analyses. For proteomics, cells of strains CG1, CG4, and 11a5 were harvested by centrifugation (12,000 × g, 20 min, 4 °C) from 500 ml cultures amended with the penta-BDE mixture and resuspended in de-gassed tris-HCl (100 mM, pH 7.0). Proteins were extracted from resuspended pellets by three freeze-thaw cycles (−80 °C followed by 40 °C for 1 min) and digested with trypsin. Peptide samples were desalted using ZipTip-μC18 material (Merck Millipore) and later assayed on an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific, Waltham, MA) equipped with a nanoLC system (Dionex Ultimate 3000RSLC; Thermo Fisher Scientific), as previously described [18]. Proteins were identified against the UniProt protein database with Proteome Discover (v2.2, Thermo Fisher Scientific) using the SequestHT search engine. Protein and peptide abundances were calculated by label-free quantification based on area counts using the Minora node implemented in Proteome Discoverer.
In vitro enzymatic assays were performed to provide physical evidence to augment findings from proteomics analyses and to investigate potential co-metabolic debromination of the octa-BDE mixture. Dehalogenation of the penta-BDE mixture by crude cell extracts of D. mccartyi strains CG1, CG4, and 11a5 cultivated with PCE or TCE was conducted as previously described [13, 19]; briefly, cells were disrupted using a VCX 130 sonicator (130 W; 20% duty cycle; 3 min) to obtain crude cell extracts. Each in vitro activity assay was carried out in 4 ml vials containing 2 ml assay solution (2 mM methyl viologen; 1.5 mM titanium (III) citrate; 100 mM Tris-HCl buffer (pH 7.0)) inside an anaerobic chamber. Enzymatic activity was tested at indicated intervals over 6 h.
RDase-encoding genes involved in PBDE debromination
The abundances of genes encoding previously described PBDE RDases (i.e., pbrA1, pbrA2, pbrA3, pteATZ50, tceATZ50, and bdeA) and PBDE RDases identified in this study (i.e., pcbA1, pcbA4, tceA11a5, and 11a5_e001) were assayed in the 24 microcosms (EW-1 to EW-24). Gene abundances were enumerated by qPCR using gene-specific primers (Table S2). Transcription of each PBDE RDase homolog genes (rdh) present in enrichments was inferred by reverse-transcription qPCR (RT-qPCR), as previously described [16]. Briefly, 1 ml and 1.5 ml samples were collected for DNA and RNA extraction, respectively, at defined time points during cultivation. Samples were concentrated by centrifugation (12, 000 × g, 10 min) and stored at −80 °C for later processing. Samples for RNA extraction were resuspended in TRIzol before freezing. DNA was extracted using the QIAGEN Blood & Tissue kit. RNA was extracted and reverse transcribed to cDNA using the QIAGEN RNeasy and QIAGEN QuantiTect Reverse Transcription kits according to the manufacturer’s instructions. Defined concentrations of luciferase control RNA (Promega) were added to samples before RNA extraction as an internal standard to normalize RNA loss during extraction.
Data deposition
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [20] partner repository with the dataset identifier PXD026932.
Results
PBDE debromination in microcosms with soil collected from e-waste recycling sites
Diverse PBDE congeners were present at detectable concentrations in the soil samples collected for this study. The average in situ ∑PBDEs was 1641.74 ± 1345.34 ng/g dry soil; minimum and maximum in situ ∑PBDEs were 103.90 (EW-10) and 4812.90 ng/ g dry soil (EW-18), respectively (Fig. S1). Bacterial populations associated with obligate OHRB genera (i.e., Dehalococcoides, Dehalogenimonas, and Dehalobacter) were found in all investigated sediments and soils collected from e-waste recycling sites. The average abundances of Dehalococcoides and Dehalobacter were similar in all sediment and soil samples (1.1 ± 2.9 × 105 and 3.9 ± 3.5 × 104 16 S rRNA gene copies/g dry soil, respectively) and both approximately one order of magnitude lower than that of Dehalogenimonas (1.3 ± 1.0 × 106 16 S rRNA gene copies /g dry soil). No obligate OHRB genera comprised greater than 1% of the total bacterial community in any one soil sample (Figure S2, Table S5).
After incubation for 2 months, 16 of 24 microcosms debrominated a BDE mixture comprising tetra-BDE 47, penta-BDE 99, and penta-BDE 100 (hereafter referred to as “penta-BDE mixture”) to lower congeners (Fig. 1a). Among the 16 debrominating microcosms, 14 completely debrominated the penta-BDE mixture to diphenyl ether, one (EW-4) debrominated tetra-BDE 47 to diphenyl ether but did not debrominate penta-BDE 99 or 100, and one (EW-15) debrominated tetra-BDE 47 and penta-BDE 99 to diphenyl ether but did not debrominate penta-BDE 100; the other eight microcosms exhibited no debromination. Partial debromination (removal of 14.1–36.1% of parent congeners) of a BDE mixture comprising nona-BDE 207, octa-BDE 203, 197, and 196, hepta-BDE 183, and hexa-BDE 153 (hereafter referred to as “octa-BDE mixture”) was observed in only four microcosms (EW-4, −6, −17, and −19) after cultivation for 9 months (Fig. 1b). The predominant end-products of octa-BDE mixture debromination were hexa- through tetra-BDEs, among which hexa-BDE 154, penta-BDE 103, and tetra-BDE 49 were detected as representative congeners of each congener homolog group (Table 1). However, neither the ∑PBDEs nor the concentration of any one PBDE congener in the e-waste soils collected for this study was a reliable predictor of whether microcosms could debrominate either the penta- or octa-BDE mixtures (t-test, p > 0.05; Fig. S3).
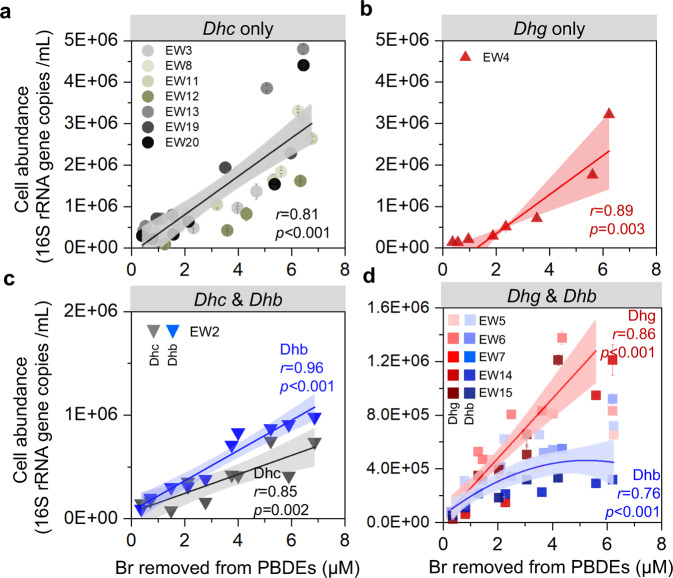
Fig. 1. Debromination of PBDEs in microcosms established from soil and sediments collected from 24 geographically distinct e-waste contaminated sites.
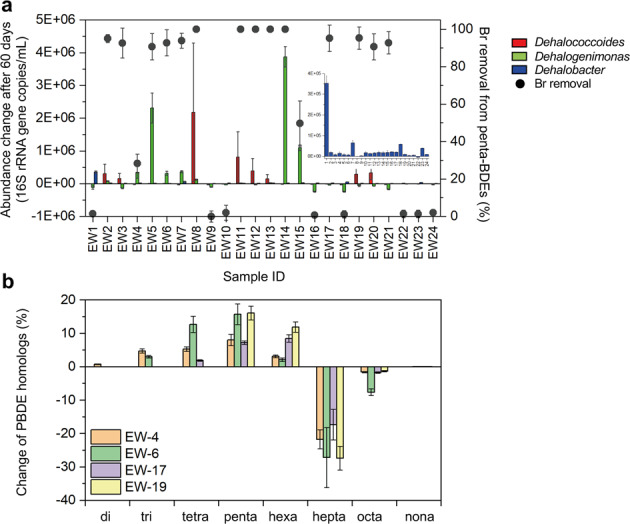
a Debromination of a penta-BDE mixture (BDE 47, 99, 100) and abundance change of Dehalococcoides, Dehalogenimonas, and Dehalobacter in microcosms after 2 months of incubation. b Change in molar fraction of PBDE homologs in an octa-BDE mixture (nona- through hexa-BDEs) in debrominating microcosms after 9 months of incubation.
Table 1.
Hepta-BDE 183 debromination metabolites in microcosms established with soils collected from e-waste contaminated sites after cultivation for 9 months at 30 °C.
| Name | IUPAC | EW-4, % | EW-6, % | EW-17, % | EW-19, % |
|---|---|---|---|---|---|
| di-4 | 2-2 | 0.9 | |||
| tri-32 | 26-4 | 4.0 | |||
| tri-17 | 24-2 | 0.9 | |||
| tri-25 | 24-3 | 0.7 | 1.4 | ||
| tri-28 | 24-4 | 0.9 | 1.3 | ||
| tetra-49 | 24-25 | 3.2 | 5.5 | 2.3 | |
| tetra-48 | 245-2 | 1.5 | |||
| tetra-47 | 24-24 | 1.6 | 3.9 | ||
| tetra-66 | 24-34 | 1.6 | 4.4 | ||
| penta-103 | 246-25 | 7.4 | 2.5 | 11.9 | |
| penta-95 | 236-25 | 1.9 | 2.9 | ||
| penta-101 | 245-25 | 1.1 | 3.8 | ||
| penta-118 | 245-34 | 1.3 | 1.1 | ||
| penta-85 | 234-24 | 5.2 | 1.4 | ||
| hexa-154 | 245-246 | 3.9 | 4.3 | 10.5 | 8.8 |
| hexa-144 | 2346-25 | 2.6 | 4.3 | ||
| hexa-149 | 236-245 | 1.7 | |||
| hepta-183 | 2346-245 | 74.7 | 66.5 | 80.1 | 67.9 |
Percentage indicated is the molar fraction of each congener relative to the total abundance of PBDEs.
Diverse obligate organohalide-respiring bacteria are involved in PBDE debromination
Involvement of each OHRB genera in PBDE debromination was inferred by correlating changes in cell abundance with penta-BDE mixture debromination activity (Fig. 1a). In debrominating microcosms, the abundance of either Dehalococcoides or Dehalogenimonas was strongly correlated with penta-BDE mixture debromination (r = 0.73 and 0.52, respectively, Fig. S4) in microcosms. That is, the abundance of at least one of these genera increased when debromination was observed in microcosms. EW-17 and EW-21 were the two exceptions, where significant debromination occurred but there was no change in Dehalococcoides or Dehalogenimonas abundance. Other microcosms in which Dehalococcoides and Dehalogenimonas abundance decreased exhibited only limited penta-BDE mixture debromination (<2%; microcosms EW-1, −9, −10, −16, −18, −22, −23, and −24). Increased Dehalobacter abundance was observed in all microcosms regardless of debromination, making its role in debromination unclear. Correlations between Dehalococcoides and Dehalogenimonas abundance and octa-BDE mixture debromination were comparatively poor, but Dehalobacter abundance increased in two of four octa-BDE mixture debrominating microcosms (EW-4 and −6). The octa-BDE mixture debrominating microcosms had comparatively low abundance of both Dehalococcoides and Dehalobacter (3.74 ± 1.5 × 103 – 7.21 ± 0.42 × 103 and 3.10 ± 0.84 × 103 – 2.21 ± 0.22 × 104 16 S rRNA gene copies /ml, respectively; Fig. S5) and no increase in Dehalogenimonas abundance was observed in any of the octa-BDE mixture debrominating microcosms. These data do not exclude the involvement of other microbial lineages in the observed octa-BDEs debromination.
Interactions of organohalide-respiring bacteria in penta-BDE mixture debrominating enrichments
The explicit roles of obligate OHRB in penta-BDE mixture debromination were investigated in enrichment cultures obtained via consecutive sub-culturing of the 16 debrominating microcosms. Debromination rates accelerated markedly in enrichment cultures, with final enrichments completely debrominating the penta-BDE mixture in 12–28 d (Fig. S6). Debromination was maintained during enrichment in most cultures, except for microcosms EW-17 and −21, which ceased debromination when acetate was substituted for lactate as the available carbon source. Meanwhile, more extensive and complete penta-BDE mixture debromination was observed in two enrichments (EW-4 and −15) than in their respective source microcosms.
Positive correlations between bromine removal and abundance of Dehalococcoides, Dehalogenimonas, and Dehalobacter in debrominating enrichment cultures (Fig. 2, Table S6) suggest that debromination occurred metabolically in these cultures. Dehalococcoides, Dehalogenimonas, and Dehalobacter were more prevalent in debrominating enrichment cultures, which tended to be dominated by either Dehalogenimonas and Dehalobacter together (enrichments EW-5, −6, −7, −14, and −15) or Dehalococcoides alone (enrichments EW-3, −8, −11, −12, −13, −19, and −20) (Fig. 2). Dehalobacter was enriched alongside Dehalococcoides or Dehalogenimonas populations in some debrominating cultures despite the poor correlation between Dehalobacter abundance and debromination in microcosms. While some enrichment cultures comprised only OHRB belonging to the genera Dehalogenimonas or Dehalococcoides, none comprised Dehalobacter alone. Furthermore, Dehalobacter growth occurred only during the initial stages of debromination in Dehalogenimonas-containing enrichment cultures, suggesting involvement in debromination of more highly substituted congeners (Fig. 2d).
Fig. 2. Correlations between the abundance of Dehalococcoides, Dehalogenimonas and Dehalobacter and removal of bromine from a penta-BDE mixture in enrichment cultures.
The cultures were dominated by a Dehalococcoides only, b Dehalogenimonas only, c Dehalococcoides and Dehalobacter, and d Dehalogenimonas and Dehalobacter. Dhc Dehalococcoides, Dhg Dehalogenimonas, Dhb Dehalobacter.
Debromination of penta- and octa- BDE mixtures by Dehalococcoides
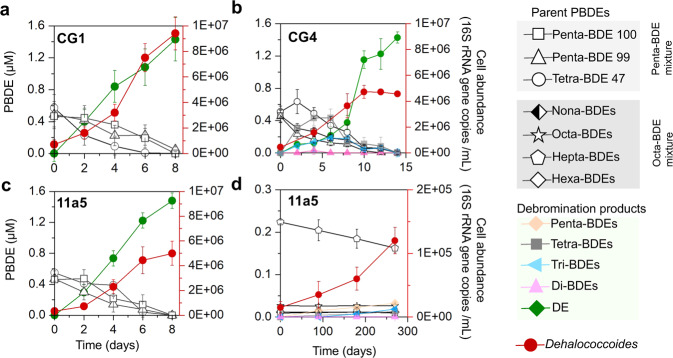
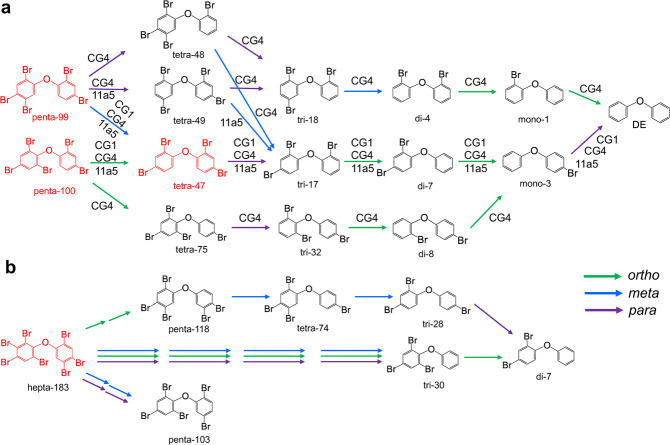
Complete debromination of the penta-BDE mixture by enrichment cultures containing Dehalococcoides as the only OHRB genera prompted investigation of penta-BDE debromination by other previously isolated Dehalococcoides strains. Among eight D. mccartyi strains tested, no debromination was detected in strains 11a, 11 G, CG5, ANAS2, and GEO and complete debromination of the penta-BDE mixture to diphenyl ether was detected in strains CG1, CG4, and 11a5. The rate of debromination varied in each strain, with strain CG4 (0.50 μM Br- removal /d [average]) being notably slower than CG1 and 11a5 (0.88 and 0.83 μM Br- removal /d [average], respectively; Fig. 3), though the cell yield from debromination was similar among all three strains (6.48 ± 0.06 × 107–1.31 ± 0.19 × 108 cells /µmole Br− released). Of these, only strain 11a5 debrominated the octa-BDE mixture after prolonged incubation (9 months). However, octa-BDE mixture debromination by strain 11a5 was incomplete, producing penta- through di-BDEs with an average decrease in bromine substituents /BDE of 0.83 ± 0.15 (Fig. 3d). Hepta-BDE 183 was the most readily debrominated congener from the octa-BDE mixture (Table S7) and was used to evaluate debromination pathways by D. mccartyi strain 11a5. D. mccartyi strains CG1, CG4, and 11a5 each produced different intermediate debromination products during penta-BDE debromination, with no apparent preference for ortho-, meta-, or para-substitution, whereas the incomplete debromination of hepta-BDE 183 by strain 11a5 proceeded primarily via meta- and para-substitution (Fig. 4).
Fig. 3. Debromination of a penta- and an octa- BDE mixture by Dehalococcoides isolates.
Debromination of a penta-BDE mixture by Dehalococcoides mccartyi strains a CG1, b CG4 and c 11a5; d debromination of an octa-BDE mixture by strain 11a5. Kinetics data are available in Supplementary Dataset Table S12.
Fig. 4. Debromination pathways of a penta-BDE mixture and hepta-BDE 183 by Dehalococcoides isolates.
a Pathway of a penta-BDE mixture mediated by D. mccartyi strains CG1, CG4 and 11a5; b Pathway of hepta-BDE 183 carried by D. mccartyi 11a5. Removal of ortho-, meta-, and para- bromines are indicated with green, blue, and purple lines, respectively. Red and black color font indicate PBDE congeners used as substrates and those formed as metabolites, respectively.
Prevalence and activity of PBDE rdh in environmental samples
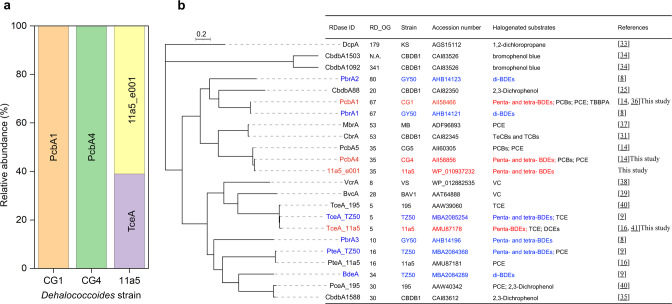
Proteomic profiles of D. mccartyi strains CG1, CG4, and 11a5 during cultivation with the penta-BDE mixture were constructed to implicate more highly expressed RDases in specific debromination activity. Each strain was cultivated with the penta-BDE mixture for at least six consecutive transfers (5% v/v) before cells were harvested for proteomics analyses. During penta-BDE debromination, PcbA1 and PcbA4, previously shown to catalyze dechlorination of polychlorinated biphenyls and PCE, were the only RDases detected in strains CG1 and CG4, respectively, while TceA11a5 (11a5_1352) and 11a5_e001, were expressed by strain 11a5 (Fig. 5, Table S8). Crude cell extracts from cultures of strains CG1, CG4, and 11a5 cultivated with PCE (strains CG1 and CG4) or TCE (strain 11a5) to induce expression of PcbA1, PcbA4, and TceA11a5, respectively, confirmed the involvement of these RDases in all or part of the debromination observed in each strain (Fig. S7). 11a5_e001 was not expressed by strain 11a5 during cultivation with TCE and may be involved in ortho-substitution during PBDE debromination, which was observed in live cells but not detected in in vitro assays (i.e., penta-BDE 100 to tetra-BDE 47 and tri-BDE 17 to diphenyl ether via di-BDE 7 and mono-BDE 3; Fig. 4 and S7c).
Fig. 5. Identification of PBDEs reductive dehalogenases (RDases).
a Relative abundances of the RDases in crude cell lysates of Dehalococcoides mccartyi strains CG1, CG4 and 11a5 cultivated with the penta-BDE mixture. b Dendrogram of PBDE RDases described in this study (red) and in previous studies (blue), as well as selected functionally characterized Dehalococcoides RDases.
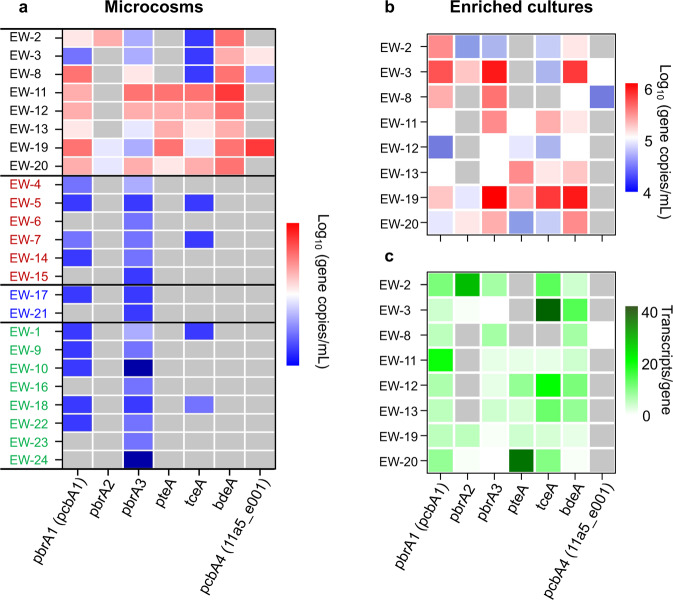
To investigate whether the genes encoding PBDE RDases could serve as genetic markers of debromination activity in mixed microbial communities, correlations between debromination activity and presence and expression of the four rdh found to debrominate PBDEs in the current study (11a5_e001, tceA11a5, pcbA1, and pcbA4) as well as six previously characterized PBDE rdh (pbrA1, pbrA2, and pbrA3 from D. mccartyi strain GY50 and pteATZ50, tceATZ50, and bdeA from D. mccartyi strain TZ50) in the 24 microcosms were investigated (Fig. 6). The presence and abundance of these rdh in the 24 microcosms after 60 d incubation were determined by qPCR. Due to the high degree of sequence identity between several of these rdh (≥98.6% over the full sequence), primers targeting pbrA1/pcbA1, pcbA4/11a5_e001, and tceA11a5/tceATZ50 were designed (Table S2). The other rdh share much lower identity (≤40.23%) and were targeted individually. Eight of the penta-BDE mixture debrominating microcosms (EW-2, −3, −8, −11, −12, −13, −19, and −20) that also contained Dehalococcoides as the dominant OHRB genus had greater abundance and diversity of these genes, harboring at least five of seven PBDE rdh (Fig. 6a). The most common and abundant PBDE rdh was bdeA, which was present in all eight Dehalococcoides-containing microcosms, while the others were less abundant and present in only some. pteATZ50 was detected in five of these eight microcosms at comparatively high levels. Other than pbrA3 and pbrA1/pcbA1, which were present at low levels (1 × 102 – 3 × 103 copies /ml), the PBDE rdh were infrequently detected above quantification limits in the 16 microcosms that did not contain Dehalococcoides.
Fig. 6. Prevelance and activity of identified PBDE reductive dehalogenase homologous genes (rdh).
a Abundance of PBDE rdh in microcosms established with soils and sediments from 24 e-waste contaminated sites after incubation for 2 months. b Abundance of PBDE rdh following complete debromination of a penta-BDE mixture by Dehalococcoides-containing enrichment cultures derived from PBDE debrominating microcosms. c Maximum levels of PBDE rdh transcription in Dehalococcoides-containing enrichments. Grey boxes indicate values below detection limits. In panel a, microcosms are grouped based on debromination activity and taxonomy of organohalide bacteria present in the microcosm. Microcosms are grouped as: penta-BDE debrominating microcosms dominated by Dehalococcoides (black font), Dehalogenimonas/Dehalobacter (red font) and unknown dechlorinating bacteria (blue font); microcosms with no penta-BDE debrominating activity (green font).
The PBDE rdh were also present in Dehalococcoides-containing enrichment cultures derived from debrominating microcosms (Fig. 6b). However, there was a notable lack of correspondence between the identities of predominant rdh in microcosms and their associated enrichment cultures, suggesting a degree of variability in OHRB populations during enrichment. For example, the pbrA3 gene was present at comparatively lower abundance in microcosm EW-3 but was the most abundant rdh in enrichment culture EW-3. Moreover, multiple PBDE rdh were expressed above detection limits in the debrominating enrichment cultures at some point during cultivation, which may indicate involvement of more than one rdh during PBDE debromination (Fig. 6c).
Discussion
This study found extensive debromination of a penta-BDE mixture to diphenyl ether or lesser brominated BDEs by 16 out of 24 microcosms established from soils and sediments of geographically distinct e-waste recycling sites. At least one of three obligate OHRB genera, Dehalococcoides, Dehalogenimonas, and Dehalobacter, present in all investigated environmental samples. These genera were differentially abundant in microcosms that debrominated penta- and tetra-BDEs; positive correlations between growth and abundance of these genera and debromination in enrichment cultures provided further evidence of their involvement in the observed debromination. Debromination of PBDEs has been reported in both obligate OHRB (i.e., Dehalococcoides, Dehalogenimonas, and Dehalobacter), facultative OHRB (i.e., members of the genera Desulfitobacterium and Sulfurospirillum), and Acetobacterium. Unlike obligate OHRB, other PBDE-debrominating bacterial populations require auxiliary substrates for PBDE debromination. For example, Desulfitobacterium PCP-1 and Sulfurospirillum multivorans DSM12446 require pentachlorophenol and trichloroethylene, respectively, as auxiliary substrates [21, 22]. The absence of such auxiliary substrates in the microcosms established in this study suggests that known PBDE-dehalogenating bacterial populations other than obligate OHRB are unlikely to be involved in the observed debromination but does not preclude the involvement of other unknown PBDE-dehalogenating facultative OHRB. Additionally, because facultative OHRB populations can generate energy from electron acceptors other than organohalides (e.g., NO3−, SO42−), it is difficult to definitively link proliferation of bacterial populations other than obligate OHRB to dehalogenation. Contrarily, obligate OHRB derive energy solely from organohalide respiration and any observed increase in the abundance of obligate OHRB populations can be directly attributed to dehalogenation.
Enrichment of the debrominating microcosms yielded several cultures in which Dehalococcoides was the only OHRB genus (7 of 14 enrichments) and a single culture in which Dehalogenimonas was the only OHRB genus; the remainder had Dehalobacter as well as either Dehalococcoides (1 of 14 enrichments) or Dehalogenimonas (5 of 14 enrichments), indicating that Dehalobacter is likely performing a cooperative function with the other OHRB to complete debromination of PBDEs. The profile of OHRB genera in the debrominating enrichment cultures could suggest that the Dehalococcoides in the enrichments were more apt to completely debrominate the penta-BDE mixture alone whereas some type of syntrophic partnership facilitated debromination by Dehalogenimonas and Dehalobacter populations. The prevalence of Dehalogenimonas in the debrominating enrichment cultures is notable, as no members of this genus have been previously found to dehalogenate brominated organohalides [23–26], making the unidentified Dehalogenimonas in enrichment culture EW-4 the first non-Dehalococcoides obligate OHRB found to do so. Our results suggest that Dehalogenimonas, together with syntrophic partners (e.g., Dehalobacter), may play a wider role in environmental organohalide removal than is reflected in current literature. Indeed, a recent survey of sites contaminated with chlorinated solvents reported a higher abundance of Dehalogenimonas than Dehalococcoides at many of the sites [23], corroborating results from the microcosms in our current study and lending credence to the hypothesis that Dehalogenimonas is involved in dehalogenation of a wide array of halogenated contaminants.
While at least some debromination of the penta-BDE mixture was observed in 16 of the 24 microcosms established in this study, debromination of the octa-BDE mixture, which was observed in just 4 microcosms, was comparatively rare and was incomplete in all cases (producing hexa- through tetra-BDEs, with hexa-BDE 154, penta-BDE 103, and tetra-BDE 49 the most prevalent end-products in octa-BDE mixture debrominating cultures). Nona- through hexa-BDEs comprise a major fraction of many commercially used octa-BDE mixtures (e.g., DE-79) and are known to be more resistant to microbial dehalogenation in anaerobic environmental compartments than lesser-brominated BDEs [7, 22]. Yet, toxic, lesser-brominated congeners frequently accumulate in PBDE-contaminated environments [4, 27, 28]. Together with earlier studies showing microbial debromination of higher PBDE congeners by members of the genera Acetobacterium, Desulfitobacterium, and Sulfurospirillum [17, 21], our results suggest that at least some natural attenuation of highly-brominated PBDEs is due to in situ microbial dehalogenation. Furthermore, our results demonstrate that microbial PBDE bioremediation strategies must consider the potential for producing and eliminating intermediate debromination products. For example, while cultures EW-4, −6, −17, and −19 completely debrominated the penta-BDE mixture to non-toxic diphenyl ether, the incomplete debromination of the octa-BDE mixture by these cultures produced penta- and tetra-BDE compounds that could not be further dehalogenated and accumulated in the system. Hence, the construction of microbial consortia for detoxification of higher PBDE congeners must consider the nature of intermediate dehalogenation products and include microbial populations capable of catalyzing dehalogenation of the intermediates produced.
Bioremediation is often discounted as a viable strategy for in situ remediation of PBDEs due to a lack of suitable candidate bacteria for bioaugmentation or biostimulation. Though no strong correlations were apparent between debromination and in situ ∑PBDEs or the concentration of any one PBDE congener, the comparable abundance of obligate OHRB detected in all 24 soil samples could suggest the presence of other organohalides in situ. Contaminated soils, particularly those at e-waste contaminated sites, often contain multiple dissimilar organohalide pollutants that could conceivably support OHRB growth (e.g., polychlorinated biphenyls or Tetrabromobisphenol A). The microcosms and enrichments described in the current study demonstrate that PBDE-contaminated soils and sediments themselves are a repository of potential PBDE-dehalogenating microbes that can be exploited as a resource for the development of PBDE bioremediation technologies. Further, our results indicate that previously described D. mccartyi strains may have dehalogenation potential beyond what is currently reported. Considering that over 500 functionally uncharacterized RDases have been described in the more than 40 isolated or identified D. mccartyi strains, it seems likely that further investigation could identify additional D. mccartyi strains capable of dehalogenating a variety of PBDE congeners [29].
In addition to identifying PBDE-dehalogenating Dehalogenimonas and PBDE debromination by previously isolated D. mccartyi strains, the current study also described previously unknown PBDE dehalogenation activity by four Dehalococcoides RDases. With the four identified in this study, a total of ten distinct RDases in Dehalococcoides are now known to dehalogenate PBDEs. Yet, dissimilarities in the debromination pathways mediated by each RDase and the lack of identity in the amino acid sequences of the different RDases preclude broad predictive statements about putative PBDE dehalogenation by other RDases. For example, our data show that dissimilar RDases can catalyze similar debromination reactions, as in the cases of PbrA1 and PbrA2 (in strain GY50), and BdeA (in strain TZ50), all of which debrominate di-BDE 7 to diphenyl ether via para- substitution. Each of these PBDE RDases can dehalogenate both aromatic and aliphatic halogenated compounds, which may indicate a certain degree of latitude in the stereospecificity of these of RDases [9, 30, 31]. Although the mechanism underlying this phenomenon remains unclear.
Implementation of bioremediation strategies requires suitable molecular tools to predict in situ capacity for dehalogenation and monitor the activity of augmented microbial populations. Many studies have shown that quantitation of rdh encoding functionally characterized RDases can provide suitably reliable information for site characterization related to a narrow range of specific organohalide compounds [11, 32–41], however, the structural variability of some halogenated aromatic compounds, like PBDEs, and the comparatively few characterized PBDE RDases limit the value of rdh as genetic markers to predict PBDE dehalogenation potential. The microcosms and enrichments cultivated in this study were probed using a suite of seven primers targeting the ten different PBDE rdh to determine whether the presence and abundance of these rdh were correlated with observed PBDE debromination activity.The presence and abundance of at least one of the PBDE rdh correlate well with PBDE debromination in microcosms and enrichments dominated by Dehalococcoides but the same is not true for debrominating microcosms that lack any members of this genus. All rdh detected in the Dehalococcoides-dominant debrominating enrichments were expressed during debromination, which may suggest the involvement of multiple RDases in PBDE debromination in the enrichments. However, further study is needed to determine whether this is true and to what extent each RDase is participating in different debromination steps. Further, none of the identified PBDE RDases were detected at a 100% similarity in the non-PBDEs debrominating D. mccartyi strains investigated in this study (Fig. S8). Therefore, while specific PBDE debromination pathways cannot be reliably inferred from nucleotide or amino acid sequences of RDases, the identified PBDE rdh can be used as indicators of potential PBDE debrominating activity in environmental samples.
Supplementary information
Acknowledgements
This study was supported by the Ng Teng Fong Charitable Foundation (NTFCF) fund (No.: R-302-000-198-720) and the Ministry of Education, Singapore under Academic Research Fund Tier 2 project MOE-000033-01 and Tier 1 project R302000239114.We would also like to acknowledge the help kindly provided by Prof. Lorenz Adrian from Helmholtz-Centre for Environmental Research for proteomics analysis.
Author contributions
SZ and JH designed the study. SZ, DC, GX, and RR performed the experiments. SZ analyzed the data and wrote the manuscript. SZ, MR and JH contributed to the revision and finalization of the manuscript.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01257-0.
References
- 1.Statista (2020). ‘Consumer Electronics’: Statista, Hamburg, Germany.
- 2.Bacher J, Dams Y, Duhoux T, Deng Y, Teittinen T, Mortensen LF. Electronic products and obsolescence in a circular economy. European Topic Centre Waste and Materials in a Green Economy: Belgiu; 2020.
- 3.Forti V, Bald CP, Kuehr R, Bel G. The Global E-waste Monitor 2020: Quantities, flows and the circular economy potential. Bonn, Geneva and Rotterdam: United Nations University/United Nations Institute for Training and Research, International Telecommunication Union, and International Solid Waste Association; 2020. [Google Scholar]
- 4.McGrath TJ, Ball AS, Clarke BO. Critical review of soil contamination by polybrominated diphenyl ethers (PBDEs) and novel brominated flame retardants (NBFRs); concentrations, sources and congener profiles. Environ Pollut. 2017;230:741–57. doi: 10.1016/j.envpol.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Lu Q, Liang Y, Fang W, Guan KL, Huang C, Qi X, et al. Spatial distribution, bioconversion and ecological risk of PCBs and PBDEs in the surface sediment of contaminated urban rivers: a nationwide study in China. Environ Sci Technol. 2021;55:9579–90. doi: 10.1021/acs.est.1c01095. [DOI] [PubMed] [Google Scholar]
- 6.Li WL, Ma WL, Jia HL, Hong WJ, Moon HB, Nakata H, et al. Polybrominated diphenyl ethers (PBDEs) in surface soils across five Asian countries: levels, spatial distribution, and source contribution. Environ Sci Technol. 2016;50:12779–88. doi: 10.1021/acs.est.6b04046. [DOI] [PubMed] [Google Scholar]
- 7.Zhao S, Rogers MJ, Ding C, He J. Reductive debromination of polybrominated diphenyl ethers - microbes, processes and dehalogenases. Front Microbiol. 2018;9:1292. doi: 10.3389/fmicb.2018.01292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding C, Rogers MJ, Yang KL, He J. Loss of the ssrA genome island led to partial debromination in the PBDE respiring Dehalococcoides mccartyi strain GY50. Environ Microbiol. 2017;19:2906–15. doi: 10.1111/1462-2920.13817. [DOI] [PubMed] [Google Scholar]
- 9.Zhao S, Rogers MJ, Cao L, Ding C, He J. Identification of reductive dehalogenases that mediate complete debromination of penta- and tetrabrominated diphenyl ethers in Dehalococcoides spp. Appl Environ Microbiol. 2021;87:e0060221. doi: 10.1128/AEM.00602-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding C, He J. Molecular techniques in the biotechnological fight against halogenated compounds in anoxic environments. Micro Biotechnol. 2012;5:347–67. doi: 10.1111/j.1751-7915.2011.00313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee PK, Johnson DR, Holmes VF, He J, Alvarez-Cohen L. Reductive dehalogenase gene expression as a biomarker for physiological activity of Dehalococcoides spp. Appl Environ Microbiol. 2006;72:6161–8. doi: 10.1128/AEM.01070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shrivastava A, Gupta V. Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Chron Young- Scientists. 2011;2:21. doi: 10.4103/2229-5186.79345. [DOI] [Google Scholar]
- 13.Wang S, Chng KR, Wilm A, Zhao S, Yang K-L, Nagarajan N, et al. Genomic characterization of three unique Dehalococcoides that respire on persistent polychlorinated biphenyls. Proc Natl Acad Sci USA. 2014;111:12103–8. doi: 10.1073/pnas.1404845111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He J, Holmes VF, Lee PK, Alvarez-Cohen L. Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl Environ Microbiol. 2007;73:2847–53. doi: 10.1128/AEM.02574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding C, Rogers MJ, He J. Dehalococcoides mccartyi strain GEO12 has a natural tolerance to chloroform inhibition. Environ Sci Technol. 2020;54:8750–9. doi: 10.1021/acs.est.0c00993. [DOI] [PubMed] [Google Scholar]
- 16.Lee PK, Cheng D, West KA, Alvarez-Cohen L, He J. Isolation of two new Dehalococcoides mccartyi strains with dissimilar dechlorination functions and their characterization by comparative genomics via microarray analysis. Environ Microbiol. 2013;15:2293–305. doi: 10.1111/1462-2920.12099. [DOI] [PubMed] [Google Scholar]
- 17.Ding C, Chow WL, He J. Isolation of Acetobacterium sp. strain AG, which reductively debrominates octa- and pentabrominated diphenyl ether technical mixtures. Appl Environ Microbiol. 2013;79:1110–7. doi: 10.1128/AEM.02919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schiffmann CL, Jehmlich N, Otto W, Hansen R, Nielsen PH, Adrian L, et al. Proteome profile and proteogenomics of the organohalide-respiring bacterium Dehalococcoides mccartyi strain CBDB1 grown on hexachlorobenzene as electron acceptor. J Proteom. 2014;98:59–64. doi: 10.1016/j.jprot.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Zhao S, Ding C, He J. Genomic characterization of Dehalococcoides mccartyi strain 11a5 reveals a circular extrachromosomal genetic element and a new tetrachloroethene reductive dehalogenase gene. FEMS Microbiol Ecol. 2017;93:fiw235. doi: 10.1093/femsec/fiw235. [DOI] [PubMed] [Google Scholar]
- 20.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He J, Robrock KR, Alvarez-Cohen L. Microbial reductive debromination of polybrominated diphenyl ethers (PBDEs) Environ Sci Technol. 2006;40:4429–34. doi: 10.1021/es052508d. [DOI] [PubMed] [Google Scholar]
- 22.Robrock KR, Korytar P, Alvarez-Cohen L. Pathways for the anaerobic microbial debromination of polybrominated diphenyl ethers. Environ Sci Technol. 2008;42:2845–52. doi: 10.1021/es0720917. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Higgins SA, Yan J, Simsir B, Chourey K, Iyer R, et al. Grape pomace compost harbors organohalide-respiring Dehalogenimonas species with novel reductive dehalogenase genes. ISME J. 2017;11:2767–80. doi: 10.1038/ismej.2017.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiao W, Luo F, Lomheim L, Mack EE, Ye S, Wu J, et al. A Dehalogenimonas population respires 1,2,4-trichlorobenzene and dichlorobenzenes. Environ Sci Technol. 2018;52:13391–8. doi: 10.1021/acs.est.8b04239. [DOI] [PubMed] [Google Scholar]
- 25.Moe WM, Yan J, Nobre MF, da Costa MS, Rainey FA. Dehalogenimonas lykanthroporepellens gen. nov., sp. nov., a reductively dehalogenating bacterium isolated from chlorinated solvent-contaminated groundwater. Int J Syst Evol Microbiol. 2009;59:2692–7. doi: 10.1099/ijs.0.011502-0. [DOI] [PubMed] [Google Scholar]
- 26.Key TA, Bowman KS, Lee I, Chun J, Albuquerque L, da Costa MS, et al. Dehalogenimonas formicexedens sp. nov., a chlorinated alkane-respiring bacterium isolated from contaminated groundwater. Int J Syst Evol Microbiol. 2017;67:1366–73. doi: 10.1099/ijsem.0.001819. [DOI] [PubMed] [Google Scholar]
- 27.Darnerud PO, Eriksen GS, Johannesson T, Larsen PB, Viluksela M. Polybrominated diphenyl ethers: occurrence, dietary exposure, and toxicology. Environ Health Perspect. 2001;109:49–68. doi: 10.1289/ehp.01109s149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu G, Zhao X, Zhao S, Chen C, Rogers MJ, Ramaswamy R, et al. Insights into the occurrence, fate, and impacts of halogenated flame retardants in municipal wastewater treatment plants. Environ Sci Technol. 2021;55:4205–26. doi: 10.1021/acs.est.0c05681. [DOI] [PubMed] [Google Scholar]
- 29.Molenda O, Puentes Jacome LA, Cao X, Nesbo CL, Tang S, Morson N, et al. Insights into origins and function of the unexplored majority of the reductive dehalogenase gene family as a result of genome assembly and ortholog group classification. Environ Sci Process Impacts. 2020;22:663–78. doi: 10.1039/C9EM00605B. [DOI] [PubMed] [Google Scholar]
- 30.Qiu L, Fang W, He H, Liang Z, Zhan Y, Lu Q, et al. Organohalide-respiring bacteria in polluted urban rivers employ novel bifunctional reductive dehalogenases to dechlorinate polychlorinated biphenyls and tetrachloroethene. Environ Sci Technol. 2020;54:8791–800. doi: 10.1021/acs.est.0c01569. [DOI] [PubMed] [Google Scholar]
- 31.Adrian L, Rahnenfuhrer J, Gobom J, Holscher T. Identification of a chlorobenzene reductive dehalogenase in Dehalococcoides sp. strain CBDB1. Appl Environ Microbiol. 2007;73:7717–24. doi: 10.1128/AEM.01649-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heavner GLW, Mansfeldt CB, Wilkins MJ, Nicora CD, Debs GE, Edwards EA, et al. Detection of organohalide-respiring enzyme biomarkers at a bioaugmented TCE-contaminated field site. Front Microbiol. 2019;10:1433. doi: 10.3389/fmicb.2019.01433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Padilla-Crespo E, Yan J, Swift C, Wagner DD, Chourey K, Hettich RL, et al. Identification and environmental distribution of dcpA, which encodes the reductive dehalogenase catalyzing the dichloroelimination of 1,2-dichloropropane to propene in organohalide-respiring chloroflexi. Appl Environ Microbiol. 2014;80:808–18. doi: 10.1128/AEM.02927-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang C, Kublik A, Weidauer C, Seiwert B, Adrian L. Reductive dehalogenation of oligocyclic phenolic bromoaromatics by Dehalococcoides mccartyi strain CBDB1. Environ Sci Technol. 2015;49:8497–505. doi: 10.1021/acs.est.5b01401. [DOI] [PubMed] [Google Scholar]
- 35.Morris RM, Fung JM, Rahm BG, Zhang S, Freedman DL, Zinder SH, et al. Comparative proteomics of Dehalococcoides spp. reveals strain-specific peptides associated with activity. Appl Environ Microbiol. 2007;73:320–6. doi: 10.1128/AEM.02129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramaswamy R, Zhao S, Bae S, He J. Debromination of TetraBromoBisphenol-A (TBBPA) depicting the metabolic versatility of Dehalococcoides. J Hazard Mater. 2021;419:126408. doi: 10.1016/j.jhazmat.2021.126408. [DOI] [PubMed] [Google Scholar]
- 37.Chow WL, Cheng D, Wang S, He J. Identification and transcriptional analysis of trans-DCE-producing reductive dehalogenases in Dehalococcoides species. ISME J. 2010;4:1020–30. doi: 10.1038/ismej.2010.27. [DOI] [PubMed] [Google Scholar]
- 38.Muller JA, Rosner BM, Von Abendroth G, Meshulam-Simon G, McCarty PL, Spormann AM. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl Environ Microbiol. 2004;70:4880–8. doi: 10.1128/AEM.70.8.4880-4888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krajmalnik-Brown R, Holscher T, Thomson IN, Saunders FM, Ritalahti KM, Löffler FE. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl Environ Microbiol. 2004;70:6347–51. doi: 10.1128/AEM.70.10.6347-6351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fung JM, Morris RM, Adrian L, Zinder SH. Expression of reductive dehalogenase genes in Dehalococcoides ethenogenes strain 195 growing on tetrachloroethene, trichloroethene, or 2,3-dichlorophenol. Appl Environ Microbiol. 2007;73:4439–45. doi: 10.1128/AEM.00215-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Magnuson JK, Stern RV, Gossett JM, Zinder SH, Burris DR. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl Environ Microbiol. 1998;64:1270–5. doi: 10.1128/AEM.64.4.1270-1275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.