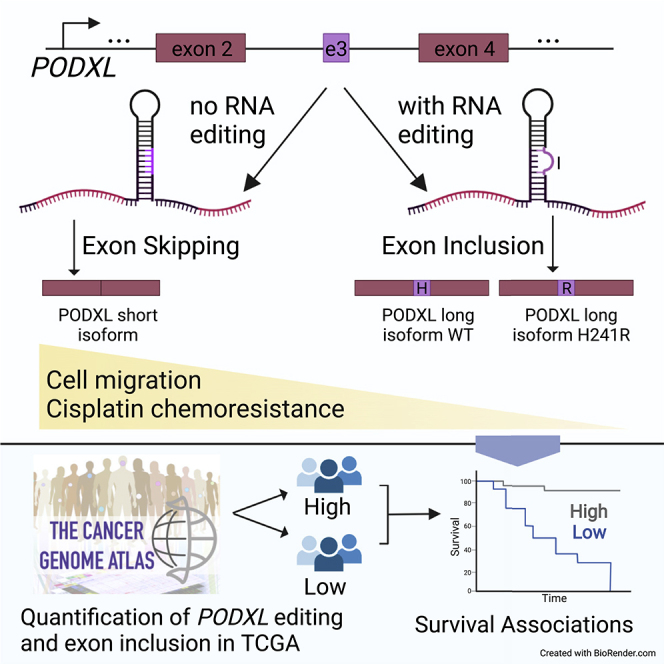
Summary
PODXL, a protein that is dysregulated in multiple cancers, plays an important role in promoting cancer metastasis. In this study, we report that RNA editing promotes the inclusion of a PODXL alternative exon. The resulting edited PODXL long isoform is more prone to protease digestion and has the strongest effects on reducing cell migration and cisplatin chemoresistance among the three PODXL isoforms (short, unedited long, and edited long isoforms). Importantly, the editing level of the PODXL recoding site and the inclusion level of the PODXL alternative exon are strongly associated with overall patient survival in Kidney Renal Clear Cell Carcinoma (KIRC). Supported by significant enrichment of exonic RNA editing sites in alternatively spliced exons, we hypothesize that exonic RNA editing sites may enhance proteomic diversity through alternative splicing, in addition to amino acid changes, a previously under-appreciated aspect of RNA editing function.
Subject areas: Biological sciences, bioinformatics, cancer, omics
Graphical abstract
Highlights
-
•
RNA editing promotes PODXL alternative exon inclusion
-
•
RNA recoding site promotes PODXL loss-of-function via splicing and codon change
-
•
High PODXL editing and exon inclusion are associated with better survival in KIRC
-
•
Exonic editing sites are enriched in alternatively spliced exons
Biological sciences; Bioinformatics; Cancer; Omics.
Introduction
RNA editing is a fundamental process in gene expression that introduces deletions, insertions, and base substitutions in the RNA transcripts. This process can happen as soon as the nascent RNA arises, thus potentially impacting transcriptome diversity by altering splicing (Rueter et al., 1999; Schoft et al., 2007; Hsiao et al., 2018), modification (Xiang et al., 2018; Rengaraj et al., 2021), localization (Zhang and Carmichael, 2001; Prasanth et al., 2005), abundance (Kawahara et al., 2008; Brümmer et al., 2017; Chan et al., 2020), and translation (Higuchi et al., 2000; Licht et al., 2019b) of RNA molecules. The most common type of RNA editing is adenosine-to-inosine (A-to-I) editing, which is also called A-to-G editing because inosine is interpreted as guanosine by the cellular machinery. A-to-I editing is carried out by the adenosine deaminase acting on RNA (ADAR) enzymes (Bass and Weintraub, 1988; Nishikura, 2010). In humans, all ADAR proteins (ADAR1, ADAR2, and ADAR3) share the dsRNA-binding domains as well as the deaminase domain that exert the catalytic function (Nishikura, 2016).
A-to-I RNA editing is dysregulated in cancer (Han and Liang, 2016; Jain et al., 2019). Analysis of RNA-seq data of human tumor samples revealed substantial changes in RNA editing in multiple cancer types (Han et al., 2015; Paz-Yaacov et al., 2015). Much attention has been given to editing sites located in the protein-coding regions, considering their potential in altering amino acid sequences (i.e., recoding). Indeed, a few functional recoding editing sites have been reported in regulating tumorigenesis (Chen et al., 2013, 2017; Han et al., 2014; Chan et al., 2016; Gumireddy et al., 2016; Fu et al., 2017; Peng et al., 2018; Takeda et al., 2019). One such example is the recoding editing event (H241R) in the gene PODXL (podocalyxin-like), first studied in gastric cancer (Chan et al., 2016). This editing site confers a loss of function of PODXL, which results in decreased cell invasion and tumor growth compared to the wild-type protein when overexpressed in the MKN28 cells (Chan et al., 2016). Yet it remains unclear how RNA editing alters the function of PODXL.
PODXL is a type I transmembrane protein expressed in various tissues including kidney podocytes, hematopoietic progenitor cells, vascular endothelia, and a subset of neurons (Nielsen and McNagny, 2009). The extracellular domain of PODXL is highly glycosylated and sialylated, contributing to the negatively charged cell surface coat of the glomerular epithelium that maintains the proper structure of the glomeruli in the kidney (Kerjaschki et al., 1984; Doyonnas et al., 2001; Le Tran, Wang and Nie, 2021). PODXL is also found to be abnormally upregulated in more than 10 types of cancer (Wu et al., 2013; Lin et al., 2014; Snyder et al., 2015; Lee et al., 2017; Itai et al., 2018; Tamayo-Orbegozo et al., 2020; Le Tran, Wang and Nie, 2021). Association studies suggested that PODXL is a potential biomarker for cancer diagnosis and prognosis in multiple cancers (Borg et al., 2016; Kusumoto et al., 2017; Lee et al., 2017; Taniuchi et al., 2018; Zhang et al., 2019; He et al., 2020). In addition, PODXL plays important role in cancer metastasis by promoting cell migration (Lin et al., 2014; Kusumoto et al., 2017; Lee et al., 2017, 2021), cell invasion (Wu et al., 2013; Lin et al., 2014; Chan et al., 2016; Kusumoto et al., 2017), cell extravasation (Fröse et al., 2018), immune evasion (Amo et al., 2015), and chemoresistance (Huang et al., 2015; Zhou et al., 2015; Lee et al., 2017; Tamayo-Orbegozo et al., 2020). Therefore, PODXL is also a valuable therapeutic target for cancer metastasis (Snyder et al., 2015; Chijiiwa et al., 2016).
Most previous studies on PODXL editing pursued the assumption that the RNA recoding site functions mainly through amino acid changes in the protein products. Nonetheless, it is known that some exonic editing sites rely on intronic editing complementary sequences to form double-stranded RNA substrates for ADARs (Higuchi et al., 1993; Schoft et al., 2007; Hsiao et al., 2018). These structures (and their interactions with ADAR) may impact downstream RNA processing steps. As the recoding site of PODXL is in the alternatively spliced exon, we hypothesized that it may also be involved in RNA splicing regulation. We observed that the two exonic RNA editing sites of PODXL synergistically enhance alternative exon inclusion. Through alternative splicing and editing-mediated amino acid changes, the PODXL gene can give rise to three isoforms that are functionally distinct in protease digestion patterns, cell migration, and cisplatin chemoresistance.
Results
RNA editing can potentially affect PODXL alternative splicing
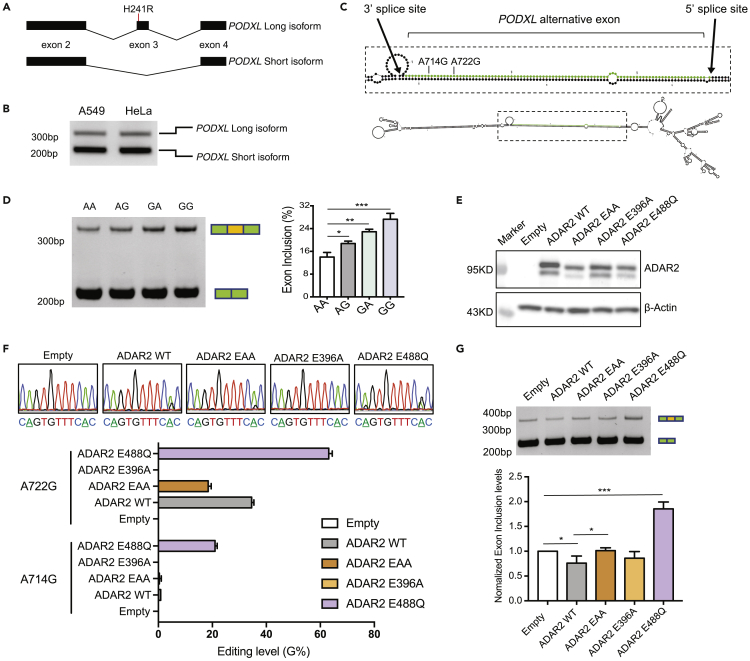
PODXL encodes for two transcript isoforms. The third exon of PODXL can be alternatively skipped, leading to an in-frame short isoform (Figure 1A). Both isoforms are endogenously expressed in A549 and HeLa cell lines with the shorter isoform being dominant (Figure 1B). Interestingly, the alternative exon of PODXL forms dsRNA structures with its upstream intronic sequences (Figure 1C). A previous study reported two A-to-I RNA editing sites (A722G and A714G) in the alternative exon of PODXL (Chan et al., 2016), one of which induces an amino acid change (namely, recoding site, H241R resulting from A722G).
Figure 1.
RNA editing and ADAR2 regulate PODXL alternative splicing
(A) The long and short isoforms of PODXL. Partial gene structures (exon 2 ∼ 4) are shown. The H241R recoding event is labeled in the alternative exon (exon 3) of the long isoform.
(B) Agarose gel image of the endogenous PODXL PCR products amplified from the cDNA of A549 and HeLa cells, respectively.
(C) Predicted RNA structure of the PODXL alternative exon (green) with its flanking introns (black). Locations of the two RNA editing sites are labeled.
(D) Left: PAGE gel resolving the PCR amplicons of transcripts derived from the PODXL splicing reporters in HeLa cells with four combinations of the A714G and A722G editing events (AA, AG, GA, GG, G represents edited and A, unedited). In the AG reporter, G was introduced at the A722G site. In the GA reporter, G was introduced at the A714G site. Right: Quantification of the PODXL alternative exon inclusion rate for each reporter based on the PAGE gel result (measured by ImageJ). Three biological replicates were included. Data are plotted as mean ± SEM. The p-values were calculated using Student’s t-test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). See also Figure S1A.
(E) Western blot showing overexpression of ADAR2 and its mutants in HeLa cells. The upper bands represent the FLAG-ADAR2 fusion proteins (see also Figure S1B). The lower bands represent ADAR2 or mutant proteins without FLAG tagging, which may result from alternative translation start sites in the overexpression constructs.
(F) Top: Sanger sequencing to detect RNA editing of the A714G and A722G editing sites (underlined As) in the minigene reporters after co-transfection with ADAR2 overexpression vectors or the empty control in HeLa cells. Bottom: Quantification of the RNA editing levels based on the Sanger sequencing peaks (measured using 4Peaks). Three biological replicates were included. Data are plotted as mean ± SEM. See also Figure S1C.
(G) Top: PAGE gel image of the amplicons of PODXL transcripts derived from the splicing reporters co-transfected with the ADAR2 overexpression vectors or the empty control in HeLa cells. Bottom: Normalized exon inclusion levels based on the PAGE gel band intensity (measured by ImageJ). Three biological replicates were included. For each replicate, the exon inclusion levels were normalized against the empty control. Data are represented as mean ± SEM. The p-values were calculated using Student’s t-test (∗p < 0.05, ∗∗∗p < 0.001).
As the two RNA editing sites are relatively close to the 3′ splice site of the alternative exon (+8, +16, Figure 1C), we hypothesized that RNA editing may affect the splicing of PODXL. To test this hypothesis, we sub-cloned the alternative exon and its flanking intronic regions (∼500bp on each side, encompassing the predicted dsRNA structure) into a splicing reporter that was developed previously (Wang et al., 2004; Xiao et al., 2009; Hsiao et al., 2018), with modifications (Figure S1A, see STAR Methods). This reporter contains two exons (together encoding the GFP) that are upstream and downstream of the tested alternative exon, respectively. We generated four constructs representing the four possible combinations of the editing status of the two RNA editing sites (AA, AG, GA, and GG, A and G representing the unedited and edited versions, respectively). These splicing reporters were then transfected into HeLa cells individually to test for exon inclusion rate. HeLa cells were used here because they are easy to transfect and show alternative splicing patterns for the endogenous PODXL (indicating the presence of trans-factors for its alternative splicing). Note that the minigenes were not edited in the presence of endogenous ADAR proteins (Figures S1B and S1C). We observed a general increase in exon inclusion rate when the G allele was introduced (Figure 1D). The GA version had a larger effect on splicing compared to the AG version, and the combination of two editing sites (GG) gave the largest increase in PODXL exon inclusion (Figure 1D). These results support the hypothesis that RNA editing events on the PODXL alternative exon affect its splicing.
ADAR2-dependent PODXL alternative splicing
Next, we asked whether splicing of the PODXL minigene depends on ADAR expression. According to previous literature, both A722G and A714G are regulated by ADAR2, whereas ADAR1 only affects A714G (Chan et al., 2016). Upon co-transfection of an ADAR1 overexpression vector and the PODXL minigene (AA version), we did not observe changes in editing levels at either editing site (Figures S1B and S1C). However, the A722G editing responded to ADAR2 overexpression. Thus, we focused on the impact of ADAR2 on PODXL splicing.
We generated three ADAR2 mutants including the binding mutant EAA (Valente and Nishikura, 2007), the editing-enhanced mutant E488Q (Kuttan and Bass, 2012; Phelps et al., 2015), and the editing-deficient mutant E396A (Valente and Nishikura, 2007). We co-transfected each ADAR2 mutant or the wild-type (WT) ADAR2 and the PODXL splicing reporter (AA version) into HeLa cells (Figure 1E). First, we confirmed that the PODXL reporter is not edited when co-transfected with an empty backbone vector or the editing-deficient mutant E396A. In contrast, co-expression of the WT ADAR2 enhanced the editing of the A722G site, and co-expression of the editing-enhanced mutant E488Q greatly increased the editing levels of both A714G and A722G sites in the reporter (Figure 1F). Interestingly, co-expression of the RNA binding mutant EAA also enhanced the editing level of the A722G site, indicating that this ADAR2 mutant can induce editing of the PODXL transcripts without the RNA binding domains. This editing activity may be enabled by RNA binding through endogenous ADAR2 in complex with the ADAR2 mutant. Alternatively, the deaminase domain of the ADAR2 mutant may facilitate RNA binding, as previously reported (Matthews et al., 2016; Wang et al., 2018).
Next, we quantified the splicing level of the PODXL minigene in the co-transfection assays. The editing-enhanced mutant E488Q of ADAR2 showed an increase in exon inclusion compared to empty control or the editing-deficient mutant E396A (Figure 1G). This result is consistent with the above finding where the GG version of the minigene had the highest exon inclusion level (Figure 1D). Curiously, overexpression of ADAR2 WT led to a slightly reduced exon inclusion rate compared to the empty control or its binding mutant EAA. We hypothesize that RNA binding by ADAR2 may inhibit the exon inclusion of PODXL, which counteracts the effect of increased RNA editing levels.
Edited PODXL long isoform is more prone to protease digestion
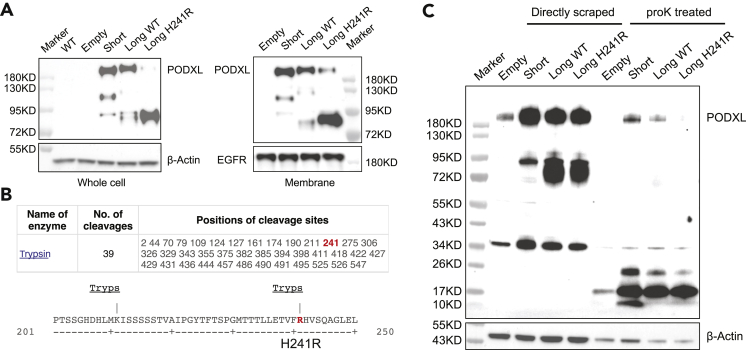
In addition to the regulation of PODXL splicing, one of the RNA editing sites (A722G) also introduces a recoding event (H241R) to the PODXL protein (Figure 1A). PODXL is a transmembrane protein that plays important role in maintaining the filtration slit in the glomerulus (Kerjaschki et al., 1984; Nielsen and McNagny, 2009). Previous studies showed PODXL enrichment in the leading edges of A549 cells during cell migration (Meng et al., 2011) and PODXL overexpression increased cell migration in A549 cells (Kusumoto et al., 2017). Interestingly, the PODXL alternative exon containing the recoding site is located in the extracellular domain. Thus, we first examined whether the alternative PODXL isoforms are located on the cell membrane. We generated stable A549 cell lines overexpressing different PODXL isoforms (short isoform where the alternative exon is skipped, wild-type long isoform, long isoform with the H241R recoding event) as well as the control vectors (empty). We performed cell fractionation on the PODXL-overexpressing (PODXL-OE) A549 cells and examined the protein expression levels of different PODXL isoforms in the cytoplasmic and membrane fractions. As expected, PODXL showed robust expression in the membrane fraction and no substantial difference was observed in the cellular localization of different PODXL isoforms (Figure S2).
Next, we asked if the presence of the alternative exon with or without the recoding event could change the protein conformation on the cell membrane. To test this, we treated the live PODXL-overexpressing A549 cells with the protease Trypsin and proteinase K. We reasoned that if different PODXL isoforms differ in their extracellular regions, they may possess different digestion patterns after the treatment. Strikingly, we observed a distinct trypsin digestion pattern for the PODXL long isoform with the H241R recoding event. Almost all of the upper band of the PODXL long isoform with H241R was digested while the upper band of the other two isoforms remained relatively intact (Figure 2A).
Figure 2.
PODXL long isoform with the H241R recoding event is more prone to protease digestion than other isoforms
(A) Western blot of trypsinized A549 cells overexpressing PODXL isoforms. Left: whole cell lysates. Right: cell lysates from the cell membrane fraction. The upper bands (∼180KD) represent intact PODXL proteins with posttranslational modifications. The middle and lower bands represent truncated PODXL proteins owing to trypsin digestion. See also Figure S2.
(B) Recoding RNA editing event H241R creates a novel trypsin digestion site on the PODXL long isoform. Top: Predicted trypsin cleavage sites on the edited PODXL long isoform. Bottom: Amino acid sequences around the H241R site with trypsin digestion sites labeled.
(C) Western blot of protease K treated A549 cells overexpressing PODXL isoforms. Whole cell lysates were used for Western blot. The upper bands (∼180KD) represent intact PODXL proteins with posttranslational modifications. The bands at 72KD ∼ 95KD have likely truncated PODXL proteins owing to protein degradation. The bands at ∼34KD are likely PODXL proteins lacking posttranslational modifications. The 10KD-to-26KD bands are digested PODXL proteins owing to protease K treatment.
Upon the examination of the amino acid sequences of each isoform, we observed that the recoding event H241R on the PODXL long isoform, indeed, creates a novel trypsin digestion site (Figure 2B). However, it should be noted that there are 38 trypsin digestion sites already present in the PODXL short and long isoforms without the H241R event. Thus, it is unlikely that the additional trypsin site led to a dramatic digestion change in the H241R isoform. Rather, we hypothesize that the PODXL long isoform with H241R editing has a different protein conformation that renders it more accessible to protease digestion. To test this hypothesis, we treated the live PODXL-overexpressing A549 cells with proteinase K, which does not have differential digestion sites between different PODXL isoforms. Interestingly, cells expressing the PODXL long isoform with the H241R recoding event showed higher sensitivity to protease K digestion (Figure 2C). These findings suggest that the H241R recoding event on the PODXL long isoform may alter protein conformation and thereby render them more prone to protease digestions.
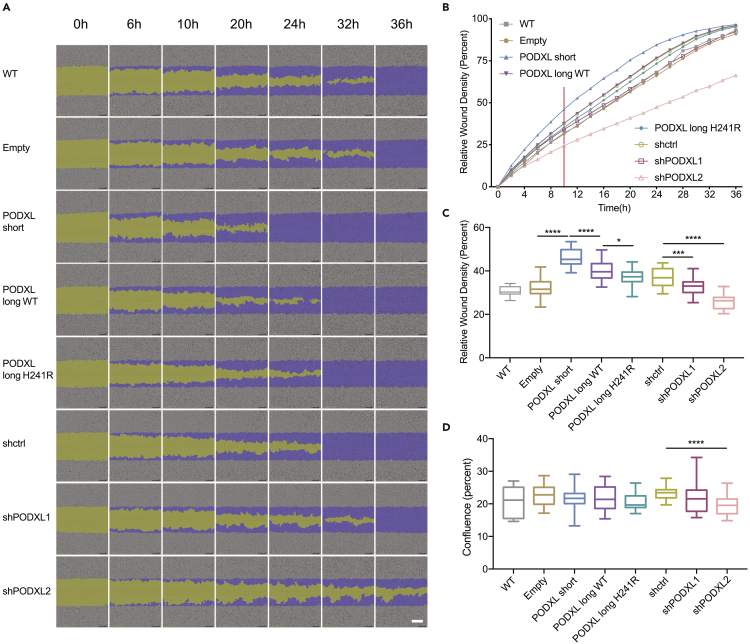
PODXL isoforms regulate cell migration
PODXL was reported to regulate cell migration and invasion in various cell line models, including A549 cells (Kusumoto et al., 2017; Tamayo-Orbegozo et al., 2020). Thus, we next asked whether PODXL isoforms impact cell migration and cell invasion abilities differently. We performed scratch wound cell migration assays using A549 cells with either stable PODXL overexpression or knockdown (KD) (Figure S3). Wounds were introduced to each well of confluent cells in 96-well plates, which were then imaged using the Incucyte® S3 live-cell analysis system at 2-h intervals (Figure 3A, showing a subset of time points to save space). We used the relative wound density as a measurement for the cell migration ability. Starting from 10 h post-wound generation, we observed increased cell migration in cells overexpressing PODXL and a decrease in cell migration in cells with PODXL KD (Figure 3B), which is consistent with previous literature (Kusumoto et al., 2017). Interestingly, the PODXL short isoform was associated with the highest cell migration ability, whereas cells overexpressing the long isoform with H241R had the lowest migration ability among the three isoforms (Figure 3C). To exclude the possible confounding effect of cell proliferation differences associated with the three isoforms, we seeded these cells at equal cell density and monitored their proliferation (Figure S3D). We observed no significant differences in cell proliferation associated with the three isoforms as measured by confluence at 10 h post-wound generation (Figure 3D). Overall, these findings suggest that the PODXL isoforms have functional differences in regulating cell migration.
Figure 3.
PODXL isoforms regulate cell migration to different degrees
(A) Cell migration assay of A549 cells overexpressing PODXL isoforms or with PODXL knockdown (PODXL-OE/KD A549 cells). WT: wild-type A549 cells. Empty: A549 cells overexpressing the empty backbone. shctrl: A549 cells with scrambled control shRNA. Two alternative shRNAs for PODXL were used (shPODXL1, shPODXL2). Phase-contrast images at different time points were shown (purple: initial scratch wound mask, yellow: wound region). Scale bar: white line at the bottom right, 300 μm. See also Figure S3.
(B) Cell migration curve of the PODXL-OE/KD A549 cells described in (A). The plot shows the average values of one set of experiments performed with three biological replicates. Red vertical line highlights data at 10 h post wound creation.
(C) Quantification of cell migration with relative wound density for the PODXL-OE/KD A549 cells. Data at 10 h post wound creation are shown, which was the earliest time point when we observed significant differences in cell migration among PODXL isoforms. Two independent sets of experiments were performed with three biological replicates included in each experiment. Data are plotted as mean ± SEM. The p-values were calculated using Student’s t-test (∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
(D) Quantification of cell proliferation using cell confluence levels for the PODXL OE/KD A549 cells. Data at 10 h post wound creation are shown, to exclude the possible effect of cell proliferation on cell migration differences shown in C. Two independent sets of experiments were performed with three biological replicates included in each experiment. Data are plotted as mean ± SEM. The p-values were calculated using Student’s t-test (∗∗∗∗p < 0.0001). See also Figure S3.
To test the effects of PODXL isoforms on cell invasion, we performed the three-dimensional (3D) 96-well scratch wound invasion assay. Similar to the cell migration assay, we introduced wounds to confluent cells in 96-wells and then added Matrigel on top of the cells to create a 3D matrix mimicking the extracellular matrix. However, we did not find significant differences in cell invasion associated with different PODXL isoforms (Figure S3E). Although we observed a decrease in cell invasion for cells with PODXL KD (Figure S3E), this may reflect the reduced cell proliferation upon PODXL KD (Figure S3D).
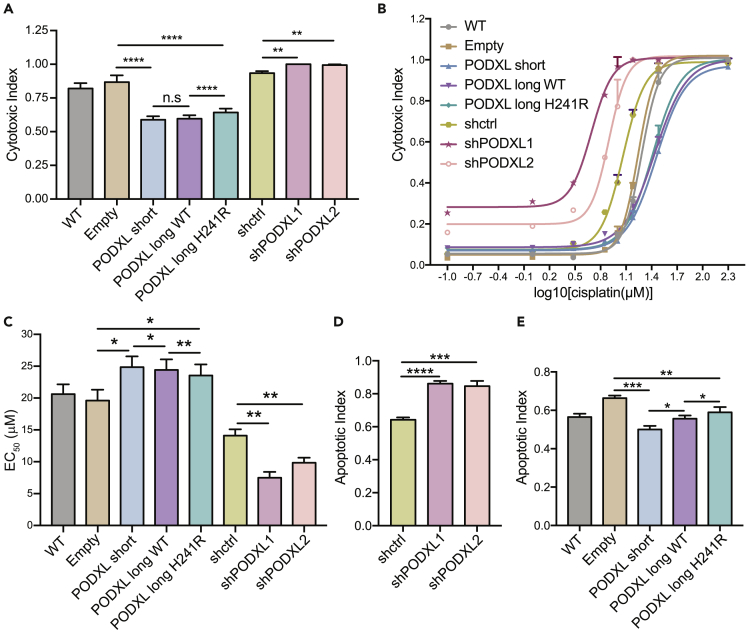
PODXL isoforms regulate cisplatin chemoresistance
Cisplatin is a wide-spectrum anti-tumor drug that has been applied to treat various human solid tumors. Previous studies reported that PODXL promotes cell resistance to cisplatin in oral tongue squamous cell carcinoma and osteosarcoma (OS) (Huang et al., 2015; Zhou et al., 2015). To further understand the functional differences of PODXL isoforms, we examined cisplatin chemoresistance of human OS cell line U2OS overexpressing different PODXL isoforms as well as U2OS cells with PODXL KD (Figure S4). We conducted the cell cytotoxicity assay with cisplatin at a concentration of 30 μM, which confers around a 50% death rate for the U2OS cells overexpressing PODXL isoforms after 48 h treatment (Figure 4A). To remove bias in cell proliferation, we calculated cytotoxic indexes by normalizing the dead cell object counts against the total number of DNA-containing object counts. We observed significantly higher cytotoxic index values (∼1) in U2OS cells with PODXL KD compared to those treated with scrambled controls (shctrl) (Figure 4A), confirming a lower level of cisplatin chemoresistance in KD cells. We also found that cells overexpressing all three PODXL isoforms showed significantly lower cytotoxic index values compared to cells expressing the empty backbone (Figure 4A). These results are consistent with the previously reported the function of PODXL in promoting cisplatin chemoresistance (Huang et al., 2015). Interestingly, cells overexpressing the PODXL long isoform with the H241R recoding event showed a slightly higher cytotoxic index compared to cells overexpressing the unedited version (Figure 4A). Thus, the H241R recoding event dampened the function of PODXL in promoting cisplatin chemoresistance.
Figure 4.
PODXL isoforms regulate cell sensitivity to cisplatin to different degrees
(A) Cytotoxic index values of U2OS WT cells, U2OS cells overexpressing empty (control) backbone, the PODXL isoforms, scrambled control shRNA (shctrl), or PODXL shRNAs (shPODXL1, shPODXL2) were generated (See also Figure S4). Cells were treated with 30 μM cisplatin for 48 h. Cytotoxic index was calculated by normalizing the dead cell object counts against the total number of DNA-containing object counts. Three independent sets of experiments were performed with three biological replicates included in each experiment. Data are plotted as mean ± SEM. The p-values were calculated using Student’s t-test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
(B) Dose-response curves for the U2OS cells with PODXL overexpression or KD, and controls (WT, Empty, shctrl). The plot shows one set of experiment performed with two biological replicates. Data are plotted as mean ± SEM.
(C) EC50 of U2OS cells with PODXL overexpression or KD, and control cell lines. Three independent sets of experiments were performed with two biological replicates included in each experiment. Data are plotted as mean ± SEM. The p-values were calculated using Student’s t-test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
(D) Apoptotic Index values of U2OS cells with PODXL KD. The assay was ended at 26 h after cisplatin treatment (30 μM). Two independent sets of experiments were performed with three biological replicates included in each experiment. Data are plotted as mean ± SEM. The p-values were calculated using Student’s t-test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
(E) Similar to (D), for PODXL OE cells. The assay was ended at 40 h after cisplatin treatment (30 μM). Data are plotted as mean ± SEM. The p-values were calculated using Student’s t-test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
To further compare the cisplatin chemoresistance associated with different PODXL isoforms, we performed the cytotoxicity assay under various concentrations of cisplatin. Figures 4B and 4C show the dose-response curves and the half maximal effective concentration (EC50) of cisplatin. A higher EC50 value reflects higher cisplatin chemoresistance. The wild-type U2OS cells and U2OS cells overexpressing empty controls had mean EC50 values of 20.6 and 19.6 μM, respectively (Figure 4C). In contrast, U2OS cells overexpressing PODXL wild-type isoforms showed increased mean EC50 values (24.9 μM for the short isoform and 24.4 μM for the long isoform) (Figure 4C). Cells overexpressing the PODXL long isoform with the H241R recoding event had a mean EC50 value of 23.5 μM, which was slightly lower than its wild-type counterpart (Figure 4C). In addition, the mean EC50 values for U2OS cells with PODXL KDs were 7.5 μM (shPODXL1) and 9.9 μM (shPODXL2), both of which were lower than the U2OS shctrl cells (14.1 μM) (Figure 4C). The above results again confirm the role of PODXL in promoting cisplatin chemoresistance. In addition, the edited PODXL long isoform showed reduced capacity in this role compared to the short or unedited isoform.
Upon the addition of cisplatin to the U2OS cells, we observed the loss of cell-to-cell contact and cell shrinkage, indicating the induction of cell apoptosis. To directly measure the effect of PODXL isoforms on cisplatin-induced cell apoptosis, we performed the cell apoptosis assay in U2OS cells at a fixed cisplatin concentration of 30 μM. Following cisplatin treatment, we calculated the normalized apoptotic index, defined as the number of caspase-3/7 positive objects divided by the total number of DNA-containing objects. Consistent with the cytotoxic index results, KD of PODXL increased the cell sensitivity to cisplatin as reflected by the increased apoptotic indexes in U2OS shPODXL1/2 cells (Figure 4D). We also observed decreased apoptotic index in U2OS cells overexpressing PODXL isoforms than in the U2OS empty control cells (Figure 4E). Among the three PODXL isoforms, the PODXL long isoform with H241R showed the highest apoptotic index, whereas the short PODXL isoform had the lowest apoptotic index values (Figure 4E). These results suggest that the PODXL short isoform has the strongest resistance against cisplatin, followed by the PODXL long isoform, then the PODXL long isoform with the H241R recoding site, respectively.
PODXL editing and splicing are clinically informative
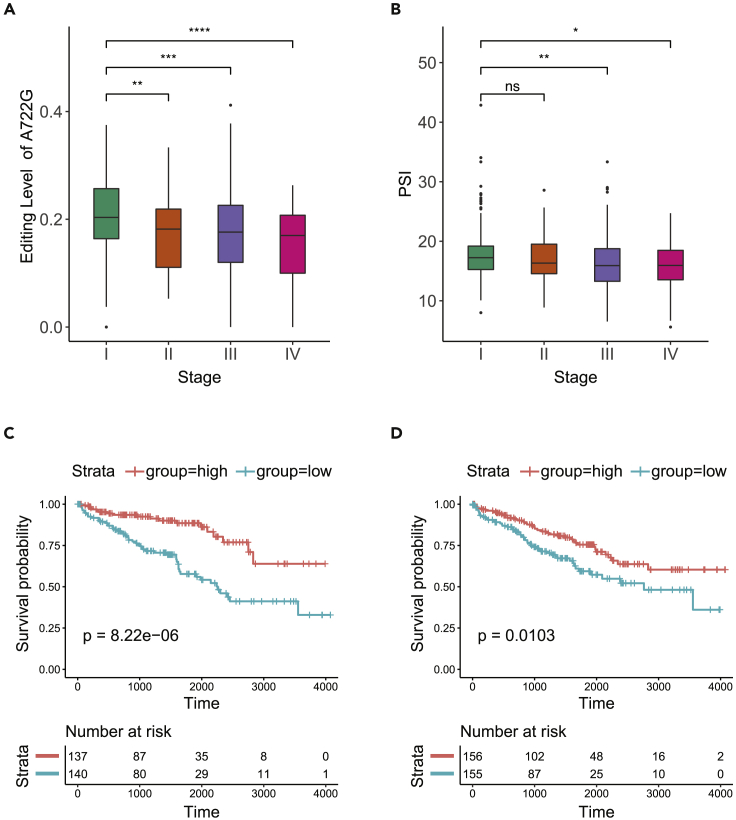
Given the distinct effects of PODXL isoforms on cancer cell migration and sensitivity to cisplatin, we investigated the clinical relevance of the PODXL alternative exon and its editing levels. For this analysis, we quantified editing (A722G and A714G) and inclusion of the alternative exon in Kidney Renal Clear Cell Carcinoma (KIRC) tumors from The Cancer Genome Atlas (TCGA), as PODXL is closely involved in kidney function (Kerjaschki et al., 1984; Doyonnas et al., 2001; Nielsen and McNagny, 2009; Le Tran, Wang and Nie, 2021). Comparisons across clinical stages revealed significantly reduced editing levels of A722G in patients with advanced disease (Figure 5A). Parallel with editing levels of this recoding site, the inclusion of the alternative exon significantly decreased over the progression of tumor stages (Figure 5B). These coordinated changes across tumor stages are consistent with the effect of editing on alternative splicing (Figure 1D) and regulation of cell migration by PODXL isoforms (Figure 3). Considering next the prognostic value of these PODXL alternative exon features, we observed that low editing (A722G) and low alternative exon inclusion were each significantly associated with worse overall survival (Figures 5C and 5D). Low inclusion of the alternative exon, regulated by diminished editing levels, corresponds to a relatively higher abundance of the short isoform. Consequently, the presence of this short isoform may contribute to enhanced migratory capacity, which may lead to poorer patient prognosis.
Figure 5.
Clinical relevance of PODXL editing and splicing in KIRC
(A) Significant decrease in editing level of the A722G site over stage progression of KIRC. The p-values were calculated using Wilcoxon rank-sum test (∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001).
(B) Significant decrease in PODXL alternative exon inclusion (measured by PSI) over stage progression of KIRC. The p-values were calculated using Wilcoxon rank-sum test (∗p ≤ 0.05, ∗∗p ≤ 0.01, ns p > 0.05).
(C) Lower editing levels of the A722G site are associated with worse overall survival in KIRC. Patients were grouped into high (red) and low (blue) groups by editing level tertiles. The p-value was calculated by the log-rank test.
(D) Lower PODXL alternative exon inclusion is associated with overall survival in KIRC. Patients were grouped into high (red) and low (blue) groups by PSI tertiles. The p-value was calculated by the log-rank test. See also Figure S6.
Known exonic editing sites are enriched in alternative exons
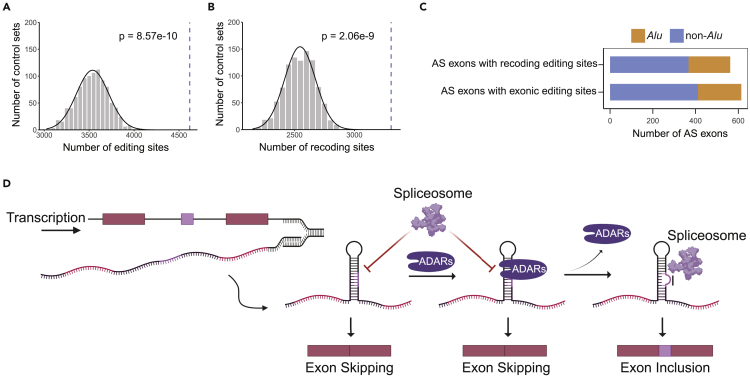
Our data suggest a coupling between RNA editing and splicing in PODXL, both of which are functionally relevant. Similar to PODXL, the RNA recoding sites in the glutamate receptor subunit B (GluR-B) pre-mRNA modulate its splicing (Schoft et al., 2007). RNA editing also plays important role in the exonization of Alu exons (Lev-Maor et al., 2007). Thus, we hypothesize that the coupling between RNA editing and alternative splicing is a widespread phenomenon. To support this hypothesis, we examined all known exonic editing sites cataloged by the REDIportal database (Mansi et al., 2021). Strikingly, alternative exons encompassed a significantly higher number of known exonic editing sites compared to random control exons (p = 8.6e-10, Figure 6A). Similar enrichment was also found if only recoding editing sites were included (from REDIportal) (p = 2.1e-9, Figure 6B).
Figure 6.
Exonic editing sites are enriched in alternatively spliced exons
(A) Number of exonic editing sites from REDIportal overlapping alternative exons (blue dashed line), compared to the numbers of exonic editing sites in 1000 sets of random control exons (gray histogram). Black curve represents the normal distribution fit to the histogram. The p-value was calculated using the normal fit.
(B) Similar to A, but for recoding exonic editing sites only.
(C) Number of editing-containing alternative exons overlapping with Alu elements. See also Figure S5.
(D) The multifaceted model of the impact of ADARs/RNA editing on alternative splicing. In the pre-mRNA transcript, the alternative exon forms dsRNA structures with the flanking introns. ADARs bind to the dsRNA structure, which may compete with the spliceosome and prevent splicing. On the other hand, ADARs introduce RNA editing sites to the transcript that destabilize the dsRNA structure. The edited pre-mRNA thus allows access of the spliceosome to the splice sites of the alternative exon, promoting exon inclusion. The illustration was created using BioRender.com.
This enrichment of editing sites in alternatively spliced exons may reflect an apparent relationship resulting from the prevalence of RNA editing in Alus and enrichment of alternative splicing among Alu exons. To assess this possibility, we checked the overlap between the editing-harboring alternative exons and Alu annotations. Only ∼30% of these alternative exons showed overlap with the Alu elements (Figure 6C), indicating that the observation of enriched exonic editing sites in alternative exons is not completely associated with Alu exonizations. Rather, it may also reflect the functional roles of RNA editing in regulating alternative splicing (Figure 6D). These findings support the coupling between RNA editing and alternative splicing. Although the exact mechanisms remain unknown, it is possible that a large number of exonic editing sites, in addition to inducing potential codon changes, may also enhance proteomic diversity by regulating RNA splicing. The alternative exons overlapping with exonic editing sites are enriched in gene ontology terms such as platelet activation, metal ion binding, and response to DNA damage (Figure S5). Dysregulated editing and alternative splicing may thus further contribute to molecular phenotypes characteristic of human diseases.
Discussion
Among the vast number of RNA editing sites in human transcriptomes, recoding sites, i.e., those that alter amino acid sequences, have been the focus of many studies owing to their readily appreciated impact on protein sequences (Chen et al., 2013, 2017; Han et al., 2014; Chan et al., 2016; Gumireddy et al., 2016; Fu et al., 2017; Peng et al., 2018; Takeda et al., 2019). Here we report a novel mechanism in which the recoding site in PODXL, a gene abnormally expressed in cancer, promotes PODXL loss-of-function via both alternative splicing and protein-recoding. PODXL is a transmembrane protein expressed in various tissues including the kidney podocytes (Nielsen and McNagny, 2009). Abnormally expressed in multiple types of cancer (Wu et al., 2013; Lin et al., 2014; Snyder et al., 2015; Lee et al., 2017; Itai et al., 2018; Tamayo-Orbegozo et al., 2020; Le Tran, Wang and Nie, 2021), PODXL is a potential biomarker for cancer diagnosis and prognosis (Borg et al., 2016; Kusumoto et al., 2017; Lee et al., 2017; Taniuchi et al., 2018; Zhang et al., 2019; He et al., 2020), and a therapeutic target for cancer metastasis in multiple cancers (Snyder et al., 2015; Chijiiwa et al., 2016).
We showed that the recoding editing site, residing in an alternatively skipped exon of PODXL, promotes the inclusion of this exon. The long isoform of PODXL results from exon inclusion, and furthermore, the edited version of the long isoform, reduces the protein’s function in promoting cell migration and cisplatin chemoresistance. Consistently, higher editing and higher exon inclusion were associated with better patient survival in KIRC. Although the function of PODXL and its recoding site has been reported previously (Chan et al., 2016), our study affords an in-depth functional comparison of the alternative isoforms and edited versions of PODXL, an aspect that had not been fully appreciated and may have clinical relevance.
RNA editing may affect alternative splicing through multiple mechanisms such as by creating splice site sequences, altering exonic splicing enhancers or silencers, or via a kinetic competition between the splicing machinery and ADARs (Lev-Maor et al., 2007; Schoft et al., 2007; Licht et al., 2016; Hsiao et al., 2018; Licht et al., 2019a). For the two RNA editing sites of PODXL, using the Mutation Analysis tool provided by Human Spicing Finder (Desmet et al., 2009), we did not detect any significant alterations in splicing signals that would explain our observations. Alternatively, the impact of RNA editing on PODXL splicing may be explained by a two-facet model. First, ADARs and the spliceosome compete for the dsRNA substrate formed between the alternative exon and the flanking intron, which explains the inhibitory effect of ADAR2 binding on the PODXL alternative exon inclusion (Figures 1G and 6D). Second, once edited by the ADARs, the dsRNA structure is more accessible to the spliceosome, which enhances exon inclusion (Figure 6D). In this model, the two aspects of ADAR function, RNA binding, and RNA editing, have opposing impacts on alternative splicing. Future work is needed to examine the kinetic and binding properties of ADAR and its relationship to the spliceosome.
Our study highlighted a previously under-appreciated aspect that RNA editing sites promote proteomic diversity not only through amino acid changes but also through alternative splicing. Interestingly, we found a strong enrichment of known exonic editing sites in alternatively spliced exons (Figure 6A). This observation could be the consequence of Alu exonization, which gives rise to alternative exons that are prone to RNA editing owing to the dsRNA structure of Alu. However, only ∼30% of the editing-containing alternative exons overlap with Alu elements (Figure 6C), indicating that the enrichment cannot be fully explained by Alu exonization. Rather, it may reflect the general roles of exonic editing sites and ADARs in regulating alternative splicing.
Interestingly, we found that the PODXL long isoform with the H241R recoding event is more prone to protease digestion (Figure 2). The H241R recoding site directly creates a new trypsin digestion site (Figure 2B). It is possible that the long isoform with the H241R recoding site undergoes conformational changes that further expose the digestion sites to the proteases, explaining its higher protease sensitivity compared to the other isoforms (Figures 2A–2C). This observation may have close relevance to cancer metastasis. The tumor microenvironment exhibits abnormal protease activities that modulate tumor invasion and metastasis (Park et al., 2020). Although the digestion of the extracellular matrix facilitates cell invasion, cleavage of the extracellular domain of transmembrane receptors also modulates the associated intracellular signaling pathways that are important in tumor cell survival and drug resistance (Miller et al., 2017; Park et al., 2020). It is possible that the PODXL long isoform with the H241R recoding event is subject to higher proteolytic pressure, thus altering the intracellular binding complex (NHERF1/2-Ezrin-Actin) (Sizemore et al., 2007; Nielsen and McNagny, 2008) or downstream signaling pathways (PI3K/Akt (Sizemore et al., 2007; Huang et al., 2015) and Bmi1/FAK (Zhou et al., 2015), and so forth) that regulate its function in cell migration and drug resistance.
In summary, our study highlights the functional importance of PODXL editing in multiple cancer-related processes. We showed that RNA editing in PODXL affects the protein function by inducing changes in both alternative splicing and protein sequences. Such multifaceted roles of RNA editing were previously under-appreciated and may exist for many editing sites in the coding regions.
Limitations of study
We showed that higher editing and higher exon inclusion in PODXL were associated with better patient survival in KIRC. The cell-based assays in this study yielded converging results using multiple cell lines. In addition, previous literature supports the ubiquitous involvement of PODXL in multiple cancer types (Tamayo-Orbegozo et al., 2020). Thus, we hypothesize that PODXL editing and splicing levels may have clinical relevance in multiple cancer types. However, a significant association of PODXL editing and splicing with patient survival was not observed in other cancer types (e.g., lung adenocarcinoma, Figure S6). This may be owing to the lack of accurate measures of editing and splicing levels that demand high coverage of PODXL transcripts in the RNA-seq data, as the other tissues (e.g., lung) had much lower PODXL expression than kidney (Figure S6E) (Uhlén et al., 2015). In the future, the relevance of PODXL editing and splicing to a wide range of cancer types needs to be further examined.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-ADAR1 Antibody (15.8.6) | Santa Cruz Biotechnology | Cat# sc-73408; RRID: AB_2222767 |
| Anti-ADAR2 Antibody (1.3.1) | Santa Cruz Biotechnology | Cat# sc-73409; RRID: AB_2289194 |
| Monoclonal ANTI-FLAG® M2 antibody | Sigma-Aldrich | Cat# F1804; RRID: AB_262044 |
| Anti-Podocalyxin-like 1 Antibody (3D3) | Santa Cruz Biotechnology | Cat# sc-23904; RRID: AB_2166006 |
| Anti-HSP 90α/β Antibody (F-8) | Santa Cruz Biotechnology | Cat# sc-13119; RRID: AB_675659 |
| EGF Receptor (D38B1) XP® Rabbit mAb | Cell Signaling | Cat# 4267; RRID: AB_2246311 |
| Anti-β-Actin Antibody (C4) | Santa Cruz Biotechnology | Cat# sc-47778; RRID: AB_626632 |
| goat anti-rabbit IgG-HRP | Santa Cruz Biotechnology | Cat# sc-2004; RRID: AB_631746 |
| goat anti-mouse IgG-HRP | Santa Cruz Biotechnology | Cat# sc-2005; RRID: AB_631736 |
| Bacterial and virus strains | ||
| NEB Stable Competent E.coli (High Efficiency) | NEB | Cat# C3040H |
| NEB 10-beta Competent E.coli (High Efficiency) | NEB | Cat# C3019H |
| NEB 5-alpha Competent E.coli (High Efficiency) | NEB | Cat# C2987H |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM, high glucose, pyruvate | Gibco | Cat# 11995065 |
| Fetal Bovine Serum, qualified, United States | Gibco | Cat# 26140079 |
| Antibiotic-Antimycotic (100X) | Gibco | Cat# 15240062 |
| Trypsin-EDTA (0.05%), phenol red | Gibco | Cat# 25300120 |
| DPBS, no calcium, no magnesium | Gibco | Cat# 14190144 |
| Puromycin Dihydrochloride | Fisher BioReagents | Cat# BP2956100 |
| Lipofectamine™ 3000 Transfection Reagent | Thermo Fisher Scientific | Cat# L3000015 |
| Polybrene | Santa Cruz Biotechnology | Cat# sc-134220 |
| TRIzol Reagent | Thermo Fisher Scientific | Cat# 15596026 |
| Chloroform | Fisher Chemical | Cat# C298-500 |
| Ethanol, Absolute (200 Proof), Molecular Biology Grade | Fisher BioReagents | Cat# BP2818500 |
| SuperScript™ IV Reverse Transcriptase | Thermo Fisher Scientific | Cat# 18090010 |
| DreamTaq™ Green PCR Master Mix | Thermo Fisher Scientific | Cat# K1082 |
| PowerUp™ SYBR® Green Master Mix | Thermo Fisher Scientific | Cat# A25742 |
| Q5® Hot Start High-Fidelity 2X Master Mix | NEB | Cat# M0492L |
| Pierce™ Protease Inhibitor Tablets, EDTA-free | Thermo Fisher Scientific | Cat# A32965 |
| PageRuler™ Prestained Protein Ladder, 10 to 180 kDa | Thermo Fisher Scientific | Cat# 26617 |
| SuperSignal West Pico PLUS Chemiluminescent Substrate | Thermo Fisher Scientific | Cat# 34580 |
| Matrigel | Corning | Cat# 356255 |
| Cisplatin | Selleck Chemical LLC | Cat# S1166 |
| Incucyte® Cytotox Dye for Counting Dead Cells, Red | Sartorius | Cat# 4632 |
| Vybrant™ DyeCycle™ Green Stain | Thermo Fisher Scientific | Cat# V35004 |
| Incucyte® Caspase 3/7 Green Dye for apoptosis, Green | Sartorius | Cat# 4440 |
| AgeI-HF | NEB | Cat# R3552S |
| EcoRI-HF | NEB | Cat# R3101S |
| NotI-HF | NEB | Cat# R3189S |
| BstBI | NEB | Cat# R0519S |
| XbaI | NEB | Cat# R0145S |
| Critical commercial assays | ||
| Direct-zol RNA Miniprep Plus kit | Zymo Research | Mfr.No. R2072 |
| Pierce™ Detergent Compatible Bradford Assay Kit | Thermo Fisher Scientific | Cat# 23246 |
| Zymoclean™ Gel DNA Recovery Kit | Zymo Research | Cat# D4002 |
| Subcellular Protein Fractionation Kit for Cultured Cells | Thermo Fisher Scientific | Cat# 78840 |
| Deposited data | ||
| TCGA | GDC Data portal | https://portal.gdc.cancer.gov/ |
| Experimental models: Cell lines | ||
| A549 | ATCC | Cat# CCL-185 |
| HeLa | Grace Xiao Lab, UCLA | N/A |
| U2OS | Oliver I Fregoso Lab, UCLA | N/A |
| HEK293T | Grace Xiao Lab, UCLA | N/A |
| Oligonucleotides | ||
| See Table S1 for oligonucleotides used in this study. | ||
| Recombinant DNA | ||
| pLJM1-EGFP | Addgene | Plasmid #19319 |
| pLJM1-Empty | this manuscript | N/A |
| pLJM1-PODXL-short-isoform | this manuscript | N/A |
| pLJM1-PODXL-long-isoform-A | this manuscript | N/A |
| pLJM1-PODXL-long-isoform-G | this manuscript | N/A |
| dR8.91 | Grace Xiao Lab, UCLA | N/A |
| VSVG | Grace Xiao Lab, UCLA | N/A |
| pLKO.1-TRC cloning vector | Addgene | Plasmid #10878 |
| PLKO.1-Scrambled | Addgene | Plasmid #136035 |
| pLKO.1-shPODXL1 | this manuscript | N/A |
| pLKO.1-shPODXL2 | this manuscript | N/A |
| pZW1-GFP | Wang et al. (2004) | N/A |
| pZW1-PODXL-AA | this manuscript | N/A |
| pZW1-PODXL-AG | this manuscript | N/A |
| pZW1-PODXL-GA | this manuscript | N/A |
| pZW1-PODXL-GG | this manuscript | N/A |
| pcDNA4-TO-ADAR1 | Bahn et al. (2015) | N/A |
| pcDNA4-TO-FLAG-myc-His | Grace Xiao Lab, UCLA | N/A |
| pcDNA4-ADAR2-WT | Tran et al. (2019) | N/A |
| pcDNA4-ADAR1-p110 | this manuscript | N/A |
| pcDNA4-ADAR1-p150 | this manuscript | N/A |
| pcDNA4-ADAR2-EAA | this manuscript | N/A |
| pcDNA4-ADAR2-E396A | this manuscript | N/A |
| pcDNA4-ADAR2-E488Q | this manuscript | N/A |
| Software and algorithms | ||
| GeneSys v1.8.5 | Syngene | https://www.syngene.com/support/software-downloads/ |
| CFX Maestro v4.1.2434.0124 | Bio-Rad Laboratories | N/A |
| mFold | Zuker (2003) | http://www.unafold.org/mfold/applications/rna-folding-form.php |
| 4Peaks v1.8 | Nucleobytes | https://nucleobytes.com/4peaks/ |
| PeptideCutter | Expasy | https://web.expasy.org/peptide_cutter/ |
| Incucyte® software | Sartorius | N/A |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| GraphPad Prism 7 | GraphPad | https://www.graphpad.com/support/prism-7-updates/ |
| ANNOVAR | Wang et al. (2010) | https://annovar.openbioinformatics.org/en/latest/ |
| Python 2.7.18 | Python Software Foundation | https://www.python.org/ |
| R 4.1.0 | The R Foundation | https://www.r-project.org/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Xinshu Xiao (gxxiao@ucla.edu).
Materials availability
Plasmids generated in this study are available from the lead contact upon request.
Experimental model and subject details
A549 (male), HeLa (female), U2OS (female), and HEK293T (female) cells were maintained in DMEM (Gibco) with 10% FBS (Gibco) and antibiotic-antimycotic reagent (Gibco) at 37 °C with 5% CO2 supply. Cell lines have not been authenticated.
Method details
PODXL overexpression and knockdown
To generate the PODXL overexpression stable cell lines, the coding region of each PODXL isoforms was cloned into the pLJM1-EGFP vector (Addgene) using the restriction enzymes AgeI-HF (NEB) and EcoRI-HF (NEB). The primers used for cloning are listed in Table S1. For empty control (pLJM1-Empty), the EGFP coding sequences in the pLJM1-EGFP construct were replaced with a short fragment of multi-cloning sites that do not express any proteins. The resulting pLJM1 vectors (pLJM1-Empty, pLJM1-PODXL-short-isoform, pLJM1-PODXL-long-isoform-A, pLJM1-PODXL-long-isoform-G) were separately co-transfected with the dR8.91 and VSVG plasmids into the HEK293T cells using lipofectamine3000 (Invitrogen) according to the manufacturer’s protocol. The lentivirus-containing media were collected every 24 h for a total of 72 h. The lentivirus-containing media were then filtered through 0.2 μm PES filters (VWR). Prior to lentiviral transduction, the A549 cells or the U2OS cells were seeded at 0.1 M per well in 6-well plates. 500 μL of virus-containing media were added to each well with the addition of polybrene (Santa Cruz Biotechnology) at a final concentration of 8 μg/mL 24 h post cell transduction, fresh media were added to the cells with puromycin (Fisher BioReagents) at a final concentration of 1 μg/mL. The transduced cells were maintained in the puromycin-containing media for at least 7 days prior to any experiments. To generate PODXL knockdown stable cells, two PODXL-targeting shRNA constructs (TRCN0000296029, targets 3'UTR; TRCN0000310117, targets CDS; primers provided in Table S1) were cloned into the pLKO.1-TRC cloning vector. The two PODXL shRNA constructs (pLKO.1-shPODXL1 and pLKO.1-shPODXL2), together with the pLKO.1-scramble shRNA control, were co-transfected with the dR8.91 and VSVG plasmids into HKE293T cells for lentiviral packaging, individually. The viruses were then used to transduce A549 or U20S cells to create PODXL knockdown stable cells using similar procedures as described above. For all plasmid constructions described above, NEB Stable Competent E.coli were used.
RNA isolation and cDNA generation
Cells were washed with PBS (Gibco) and lysed with TRIzol (Thermo Fisher Scientific). Each 500 μL TRIzol-lysed solution was mixed with 100 μL chloroform (Fisher Chemical) to allow phase separation. The upper aqueous phase was transferred and mixed with equal volume ethanol (200 proof, Fisher BioReagents). The mixture was loaded to the column supplied by the Direct-zol RNA Miniprep Plus kit (Zymo Research) to isolate total RNA following the manufacturer’s protocol. 1∼2 μg of total RNA was used for cDNA synthesis with Super-Script IV (Thermo Fisher Scientific) using random hexamers.
Detection of PODXL isoforms via PCR
Primers used for PODXL isoform detection are listed in Table S1. For endogenous PODXL isoform detection, 1 μL of the cDNA was used for PCR using DreamTaq™ Green PCR Master Mix (2X) (Thermo Fisher Scientific). The PCR reaction was carried out with an annealing temperature of 55 °C for 28 cycles. The PCR products were resolved in a 1% agarose gel with Ethidium bromide (Sigma) staining and visualized under the imager (Syngene PXi). For PODXL isoform detection in splicing reporters, 1 μL of the cDNA was used for PCR using DreamTaq™ Green PCR Master Mix (2X) (Thermo Fisher Scientific), which underwent 28 cycles with an annealing temperature of 60 °C. The PCR products were resolved in a 6% PAGE gel, stained with SYBR Safe DNA Gel Stain (Thermo Fisher Scientific), and visualized under the imager (Syngene PXi). Images were analyzed using ImageJ to quantify band intensity for both short and long isoforms. The exon inclusion rate was calculated as (short/(short + long)). To measure the expression of PODXL (all isoforms) or just the long isoforms via qPCR, 1 μL of cDNA was used, together with the PowerUp™ SYBR® Green Master Mix (Thermo Fisher Scientific). The reaction was performed in the CFX96 Touch Real-Time PCR detection system (Bio-Rad) with the following settings: 50 °C for 10 min, 95 °C for 2 min, 95 °C for 15 s, 60 °C for 30 s, and with the last two steps repeated for 45 cycles. For the PODXL overexpression samples, the expression of PODXL was normalized against the expression of 18S or TBP. For the PODXL knockdown samples, the expression of PODXL was normalized against the expression of TBP.
RNA structure predictions
The sequences of the PODXL alternative exon and its flanking introns (∼500bp each) were folded using mFold (Zuker, 2003) with default settings.
Construction of splicing minigene reporters
The PODXL alternative exon and its flanking intronic sequences (∼500bp upstream and downstream of the exon) were cloned into the pZW1-GFP splicing reporter, which contains two exons (together encoding the GFP) that are upstream and downstream of the tested alternative exon, respectively (Wang et al., 2004; Xiao et al., 2009; Hsiao et al., 2018). To generate an in-frame transcript when PODXL alternative exon is included, we introduced two insertions (+1c, +ag) at the splice sites of the two GFP sub-exons. A T-to-G mutation (named as 3ssTg) was also introduced to the PODXL 3’splice site to increase the basal exon inclusion rate so that it approximately matches the endogenous exon inclusion level. Primers used for making modifications to the splicing reporter and introducing the PODXL editing sites are listed in Table S1. NEB 5-alpha Competent E.coli were used for plasmid construction.
ADAR overexpressing constructs
The coding sequences for the ADAR1 p110 isoform and ADAR1 p150 isoform were amplified from the constructs previously generated in our lab (Bahn et al., 2015) and cloned into the pcDNA4-TO-FLAG-myc-His vector (Invitrogen) using restriction enzymes NotI-HF (NEB) and BstBI (NEB). ADAR2 mutant constructs (EAA, E396A, and E488Q) were generated by introducing the recoding mutations to the pcDNA4-ADAR2-WT construct previously generated in our lab (Tran et al., 2019). In general, the ADAR2 coding sequences were reamplified to introduce mutations using overlap extension PCR, followed by digestion and ligation into the pcDNA4-TO-FLAG-myc-His vector via the restriction enzymes NotI-HF (NEB) and XbaI (NEB). All PCR reactions were performed using the Q5® Hot Start High-Fidelity 2X Master Mix (NEB). The primers used for PCR reactions are listed in Table S1. NEB 10-beta Competent E.coli were used for plasmid construction.
Western blot
The cells were washed with cold 1x PBS and then lysed with cold RIPA buffer with Pierce™ Protease Inhibitor Tablets (EDTA-free, Thermo Fisher Scientific) freshly added. After incubation at 4 °C for 30 min, the whole cell lysate was collected and centrifuged for 30 min at 12,000 g, 4 °C. The supernatants were transferred to a new tube for protein concentration measurement using the Pierce™ Detergent Compatible Bradford Assay Kit (Thermo Fisher Scientific). After mixing with the 4× SDS protein loading dye, the protein samples were boiled at 95 °C for 5 min and loaded onto SDS-PAGE gels with the PageRuler™ Prestained Protein Ladder, 10 to 180 kDa (Thermo Fisher Scientific), followed by protein transfer to nitrocellulose membranes (GE Healthcare) and antibody incubations. Antibodies used were as follows: ADAR1 antibody (Santa Cruz Biotechnology, sc-73408, 1:200), ADAR2 antibody (Santa Cruz Biotechnology, sc-73409, 1:200), FLAG antibody (Sigma, F1804, 1:1000), PODXL antibody (Santa Cruz Biotechnology, sc-23904, 1:500), HSP 90α/β antibody(Santa Cruz Biotechnology, sc-13119, 1: 500), EGFR antibody (Cell Signaling, #4267, 1:1000), β-actin-HRP antibody (Santa Cruz Biotechnology, sc-47778, 1: 2000), goat anti-rabbit IgG-HRP (Santa Cruz Biotechnology, sc-2004, 1:2000), and goat anti-mouse IgG-HRP (Santa Cruz Biotechnology, sc-2005, 1:2000). To visualize, the membrane blots were incubated with SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific) and then imaged using the Syngene Pxi imager.
Splicing reporter assay
HeLa cells were seeded in 12-well plates to reach 90% confluency by the time of cell transfection. 375 ng of reporter plasmids were transfected into each well with lipofectamine 3000 (Invitrogen). For ADAR co-transfection experiments, 1,250 ng of ADAR-overexpressing plasmids and 375 ng of reporter plasmids were transfected into each well with lipofectamine 3000 (Invitrogen). The total RNA was harvested 24 h post cell transfection and processed to detect PODXL isoforms generated from the splicing reporters, as described above.
Quantification of RNA editing levels
The PODXL alternative exon was amplified from the cDNA of Hela cells transfected with the splicing reporters. The primers used are listed in Table S1. PCR reactions were performed with DreamTaq™ Green PCR Master Mix (2X) (Thermo Fisher Scientific) for 28 cycles. PCR products were resolved in 1% agarose gel and then purified using Zymoclean™ Gel DNA Recovery Kit (Zymo Research). The reverse primer was mixed with the amplicons and sent for Sanger sequencing. To quantify the RNA editing levels, the peak signals of both A alleles and G alleles were measured using 4Peaks, followed by editing level calculation (G/(A + G)).
Cell fractionation and protease digestion
Cell fractionation was performed using the Subcellular Protein Fractionation Kit for Cultured Cells (Thermo Fisher Scientific). The PODXL-OE A549 cells were washed with cold 1x PBS and directly lysed with the Cytoplasmic Extraction Buffer provided by the kit. For protease digestion assay, the PODXL-OE A549 cells were washed with warm 1x PBS, and then treated with either TrypLE™ Express (Gibco) or protease K (100 μg/mL, Zymo Research) at 37 °C for 5 min. The digested cells were then collected with growth media and centrifuged at 500 g for 5 min. The cell pellet was further processed using the Subcellular Protein Fractionation Kit for Cultured Cells (Thermo Fisher Scientific) following the manufacturer’s protocol.
Prediction of protease cleavage sites
The protease cleavage sites of the PODXL long isoforms (WT and H241R) were predicted using PeptideCutter (Gasteiger et al., 2005).
Cell proliferation assay
The PODXL-OE A549 cells were seeded at 3,000 cells per well in the 96-well plates. After 24 h incubation at 37 °C, the plate was transferred to the Incucyte® S3 live-cell analysis system (Sartorius) to monitor cell proliferation. Images were taken every 2 h and analyzed for confluency.
Cell migration assay
The PODXL-OE A549 cells were seeded at 30,000 cells per well in the Incucyte® Imagelock 96-well plate (Sartorius) to reach 100% confluency after 24 h incubation. A scratch wound was created on each well using the WoundMaker™ (Sartorius) followed by fresh media change. The 96-well plate was monitored for wound closure by imaging every 2 h. The images were analyzed to calculate the relative wound density, a measure of the density of the wound region relative to the density of the cell region, as recommended by the Incucyte® manual for 96-well scratch wound cell migration assay.
Cell invasion assay
Prior to cell seeding, the Incucyte® Imagelock 96-well plate (Sartorius) was pre-coated with 50 μL of 100 μg/mL Matrigel (Corning) in each well at 37 °C for at least 2 h. The PODXL-OE A549 cells were seeded at 30,000 cells per well in the coated plate and incubated at 37 °C for 24 h to reach 100% confluency. After scratch wound introduction using WoundMaker™ (Sartorius), 50 μL of 8 mg/mL Matrigel (Corning) were added to each well and incubated at 37 °C for 20 min until the Matrigel is solidified. Additional 250 μL of cell growth media were added to each well before the plate was transferred to the Incucyte® S3 live-cell analysis system (Sartorius). Images were taken every 2 h to monitor cell invasion activities. The images were analyzed to calculate the relative wound density as described above.
Cell cytotoxicity assay
The U2OS cells with PODXL-OE or PODXL-KD were seeded at 3,000 cells per well in the 96-well plate and incubated at 37 °C for 24 h. The cells were then treated with 100 μL cell growth media containing 30 μM cisplatin (Selleck Chemical LLC) and the Incucyte® Cytotox Red Dye for counting dead cells. Both phase-contrast and red-fluorescence images were taken for each well every 2 h under the Incucyte® S3 live-cell analysis system (Sartorius). At 48 h after treatment, the assay was terminated by adding 20 μL of 12 μM (diluted in 1x PBS) Vybrant™ DyeCycle™ Green Stain (Invitrogen™) directly to each well (final dye conc. at 2 μM) and imaged using the Incucyte® S3 live-cell analysis system (Sartorius) for phase-contrast and green-fluorescence images. The cytotoxic index was calculated by dividing the dead cell numbers (red-fluorescence object counts) by the total number of DNA-containing cells (green-fluorescence object counts).
Determination of the EC50 value of cisplatin
The cell cytotoxicity assay was performed under a range of concentrations (0.1 μM, 1 μM, 3 μM, 7 μM, 10 μM, 15 μM, 30 μM, and 200 μM) of cisplatin (Selleck Chemical LLC). The cytotoxic index at 48 h of cisplatin treatment was calculated and plotted against the cisplatin concentrations in GraphPad Prism. The EC50 value was calculated for each cell line based on the dose-response curve (nonlinear regression) using the “find ECanything” function in GraphPad Prism.
Cell apoptosis assay
The U2OS cells with PODXL-OE or PODXL-KD were seeded at 3,000 cells per well in the 96-well plate. After 24 h incubation in the cell incubator, each well was replaced with 100 μL cell growth media containing 30 μM cisplatin (Selleck Chemical LLC) and the Incucyte® Caspase 3/7 Green Dye for apoptosis (Sartorius). Images in phase-contrast and green-fluorescence were taken every 2 h using the Incucyte® S3 live-cell analysis system (Sartorius). At either 26 h or 40 h post cisplatin addition, the assay was ended by adding 20 μL of 12 μM (diluted in 1x PBS) Vybrant™ DyeCycle™ Green Stain (Invitrogen™) directly to each well and imaged again. The apoptotic index was calculated by dividing the number of apoptotic objects (green-fluorescence object counts before the addition of Vybrant™ DyeCycle™ Green Stain) by the total number of DNA-containing objects (green-fluorescence object counts after the addition of Vybrant™ DyeCycle™ Green Stain).
Quantification and statistical analysis
Statistics for cell-based assays
Data were analyzed and plotted using Graphpad Prism 7. Data are plotted as mean ± SEM. The p-values were calculated using Student’s t-test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). P-value < 0.05 was used to call significance.
Quantification of editing and PSI in TCGA
Using the BAM slicing functionality of the Genomic Data Commons (GDC) Application Programming Interface (API), we downloaded PODXL-overlapping bam files for tumors of Kidney Renal Clear Cell Carcinoma (KIRC) and Lung Adenocarcinoma (LUAD) patients in The Cancer Genome Atlas (TCGA). After retaining only uniquely mapped reads, we counted reads with or without PODXL editing events (Bahn et al., 2012; Lee et al., 2013; Tran et al., 2019; Chan et al., 2020) to calculate editing ratios at two A-to-I RNA editing sites in the alternative exon of PODXL (A722G and A714G). To quantify inclusion levels of the PODXL alternative exon, we calculated its percent spliced in (PSI) using only exon-exon junction reads (Barbosa-Morais et al., 2012). Comparison between tumor stages were done using Wilcoxon rank-sum test and p-value ≤ 0.05 was used to determine significance.
Survival associations
We downloaded the TCGA Pan-Cancer Clinical Data Resource (Liu et al., 2018) (https://api.gdc.cancer.gov/data/1b5f413e-a8d1-4d10-92eb-7c4ae739ed81) to obtain survival times for KIRC and LUAD patients. High and low editing groups of patients were defined based on tertiles of PODXL A722G editing levels in each cancer type. Similarly, patients were categorized into high and low alternative exon inclusion groups in each cancer type, based on PSI tertiles. With the R package survival, we used the log-rank test to compare overall survival between high and low PODXL A722G editing groups, as well as between high and low alternative exon inclusion groups in KIRC and LUAD. P-value < 0.05 was used to call significance. We visualized the Kaplan Meier survival curves using the R package survminer.
Enrichment of editing in alternative exons
To test whether recoding sites were enriched in alternatively spliced exons, we first annotated recoding sites within the REDIportal V2 database (Mansi et al., 2021) by running ANNOVAR (Wang et al., 2010). After defining alternative and constitutive exons based on gene annotations from the Consensus Coding Sequence (CCDS) project (Pruitt et al., 2009), we counted the number of recoding sites within alternative exons. We also counted overlapping recoding sites within 1000 sets of control exons, which were randomly selected from the same genes containing alternative exons. An enrichment p-value for recoding events within alternative exons was calculated from a normal distribution fit to the recoding site counts within control exons. We similarly tested the enrichment of editing sites (not limited to recoding sites) within alternative exons by using counts of editing sites overlapping alternative exons and 1000 sets of control exons. P-value < 0.05 was used to call significance. The alternative exons were overlapped with the Alu annotations downloaded from the UCSC Genome Browser (Fernandes et al., 2020).
Gene ontology (GO) enrichment analysis
For each query gene that contains AS exons overlapping with RNA editing sites, a control gene was randomly chosen among the background genes (excluding the query genes) in the CCDS database (Pruitt et al., 2009), 10,000 times. The p-value of the enrichment of each GO term in the query genes was calculated using the normal distribution fit to the occurrence of the GO term in the 10,000 sets of control genes. To call significance, FDR < 0.05 and occurrence (number of genes associated with a term) ≥ 10 were used.
Additional resources
This paper does not report additional resources.
Acknowledgments
We thank members of the Xiao laboratory for helpful discussions and comments on this work. The results published here are in part based upon data generated by The Cancer Genome Atlas managed by the NCI and NHGRI. This work was supported in part by grants from the National Institutes of Health (U01HG009417, R01CA262686 to X.X.) and the Jonsson Comprehensive Cancer Center at UCLA. T.F. was supported by the UCLA Hyde Fellowship and Dissertation Year Fellowship. T.W.C. was supported by the NIH T32LM012424.
Author contributions
T.F. and X.X. designed the study with inputs from all other authors. T.F., J.H.B., and T.-H.K. conducted molecular, cellular, and biochemical experiments. T.W.C. and T.F. conducted the bioinformatics analyses. X.X. and A.C.R. provided supervisory inputs. All authors contributed to the writing of the article. All authors approved the final article.
Declaration of interests
The authors declare no competing interests.
Published: August 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104836.
Supplemental information
Data and code availability
This paper analyzes existing, publicly available data. The accession link for the datasets is provided in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Amo L., Tamayo-Orbegozo E., Maruri N., Buque A., Solaun M., Rinon M., Arrieta A., Larrucea S. Podocalyxin-like protein 1 functions as an immunomodulatory molecule in breast cancer cells. Cancer Lett. 2015;368:26–35. doi: 10.1016/j.canlet.2015.06.029. [DOI] [PubMed] [Google Scholar]
- Bahn J.H., Ahn J., Lin X., Zhang Q., Lee J.H., Civelek M., Xiao X. Genomic analysis of ADAR1 binding and its involvement in multiple RNA processing pathways. Nat. Commun. 2015;6:1–13. doi: 10.1038/ncomms7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn J.H.J.H., Lee J.H., Li G., Greer C., Peng G., Xiao X. Accurate Identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res. 2012;22:142–150. doi: 10.1101/gr.124107.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais N.L., Irimia M., Pan Q., Xiong H.Y., Gueroussov S., Lee L.J., Slobodeniuc V., Kutter C., Watt S., Colak R., et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/SCIENCE.1230612. [DOI] [PubMed] [Google Scholar]
- Bass B.L., Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-X. [DOI] [PubMed] [Google Scholar]
- Borg D., Hedner C., Nodin B., Larsson A., Johnsson A., Eberhard J., Jirstrom K. Expression of podocalyxin-like protein is an independent prognostic biomarker in resected esophageal and gastric adenocarcinoma. BMC Clin. Pathol. 2016;16:13. doi: 10.1186/s12907-016-0034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brümmer A., Yang Y., Chan T.W., Xiao X. Structure-mediated modulation of mRNA abundance by A-to-I editing. Nat. Commun. 2017;8:1255. doi: 10.1038/s41467-017-01459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T.H.M., Qamra A., Tan K.T., Guo J., Yang H., Qi L., Lin J.S., Ng V.H.E., Song Y., Hong H., et al. ADAR-mediated RNA editing predicts progression and prognosis of gastric cancer. Gastroenterology. 2016;151:637–650.e10. doi: 10.1053/j.gastro.2016.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T.W., Fu T., Bahn J.H., Jun H.I., Lee J.H., Quinones-Valdez G., Cheng C., Xiao X. RNA editing in cancer impacts mRNA abundance in immune response pathways. Genome Biol. 2020;21:268. doi: 10.1186/s13059-020-02171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Li Y., Lin C.H., Chan T.H.M., Chow R.K.K., Song Y., Liu M., Yuan Y.-F., Fu L., Kong K.L., et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 2013;19:209–216. doi: 10.1038/nm.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.B., Liao X.L., Zhang J.B., Wang F., Qin H., Zhang L., Shugart Y.Y., Zeng Y.X., Jia W.H. ADAR2 functions as a tumor suppressor via editing IGFBP7 in esophageal squamous cell carcinoma. Int. J. Oncol. 2017;50:622–630. doi: 10.3892/ijo.2016.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chijiiwa Y., Moriyama T., Ohuchida K., Nabae T., Ohtsuka T., Miyasaka Y., Fujita, H., Maeyama R., Manabe T., Abe A., et al. Overexpression of microRNA-5100 decreases the aggressive phenotype of pancreatic cancer cells by targeting PODXL. Int. J. Oncol. 2016;48:1688–1700. doi: 10.3892/ijo.2016.3389. [DOI] [PubMed] [Google Scholar]
- Desmet F.O., Hamroun D., Lalande M., Collod-Béroud G., Clausters M., Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/NAR/GKP215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyonnas R., Kershaw D.B., Duhme C., Merkens H., Chelliah S., Graf T., McNagny K.M. Anuria, omphalocele, and perinatal lethality in mice lacking the Cd34-related protein podocalyxin. J. Exp. Med. 2001;194:13. doi: 10.1084/JEM.194.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J.D., Zamudio-Hurtado A., Clawson H., Kent W.J., Haussler D., Salama S.R., Haeussler M. The UCSC repeat browser allows discovery and visualization of evolutionary conflict across repeat families. Mob. DNA. 2020;11:13. doi: 10.1186/S13100-020-00208-W/FIGURES/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröse J., Chen M.B., Hebron K.E., Reinhardt F., Hajal C., Zijlstra A., Kamm R.D., Weinberg R.A. Epithelial-Mesenchymal transition induces podocalyxin to promote extravasation via ezrin signaling. Cell Rep. 2018;24:962–972. doi: 10.1016/j.celrep.2018.06.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Qin Y.R., Ming X.Y., Zuo X.B., Diao Y.W., Zhang L.Y., Ai J., Liu B.L., Huang T.X., Cao T.T., et al. RNA editing of SLC22A3 drives early tumor invasion and metastasis in familial esophageal cancer. Proc. Natl. Acad. Sci. USA. 2017;114:E4631–E4640. doi: 10.1073/pnas.1703178114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. In: The Proteomics Protocols Handbook. Walker J.M., editor. Humana Press; 2005. Protein identification and analysis tools on the ExPASy server; pp. 571–607. [Google Scholar]
- Gumireddy K., Li A., Kossenkov A.V., Sakurai M., Yan J., Li Y., Xu H., Wang J., Zhang P.J., Zhang L., et al. The mRNA-edited form of GABRA3 suppresses GABRA3-mediated Akt activation and breast cancer metastasis. Nat. Commun. 2016;7:10715. doi: 10.1038/ncomms10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Diao L., Yu S., Xu X., Li J., Zhang R., Yang Y., Werner H.M., Eterovic A.K., Yuan Y., Li J. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015;28:515–528. doi: 10.1016/j.ccell.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Liang H. RNA editing in cancer: mechanistic, prognostic, and therapeutic implications. Mol. Cell Oncol. 2016;3:e1117702. doi: 10.1080/23723556.2015.1117702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.W., Kim H.P., Shin J.Y., Jeong E.G., Lee W.C., Kim K.Y., Park S.Y., Lee D.W., Won J.K., Jeong S.Y., et al. RNA editing in RHOQ promotes invasion potential in colorectal cancer. J. Exp. Med. 2014;211:613–621. doi: 10.1084/jem.20132209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Du W., Li M., Yan M., Zheng F. PODXL might be a new prognostic biomarker in various cancers: a meta-analysis and sequential verification with TCGA datasets. BMC Cancer. 2020;20:620. doi: 10.1186/s12885-020-07108-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M., Single F.N., Kohler M., Sommer B., Sprengel R., Seeburg P.H. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-W. [DOI] [PubMed] [Google Scholar]
- Higuchi M., Maas S., Single F.N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- Hsiao Y.-H.E., Bahn J.H., Yang, Y., Lin X., Tran S., Yang E.W., Quinones-Valdez G., Xiao X. RNA editing in nascent RNA affects pre-mRNA splicing. Genome Res. 2018;28:812–823. doi: 10.1101/gr.231209.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Huang Y., He H., Ni J. Podocalyxin promotes cisplatin chemoresistance in osteosarcoma cells through phosphatidylinositide 3-kinase signaling. Mol. Med. Rep. 2015;12:3916–3922. doi: 10.3892/mmr.2015.3859. [DOI] [PubMed] [Google Scholar]
- Itai S., Yamada S., Kaneko M.K., Sano M., Nakamura T., Yanaka M., Handa S., Hisamatsu K., Nakamura Y., Furusawa Y., et al. Podocalyxin is crucial for the growth of oral squamous cell carcinoma cell line HSC-2. Biochem. Biophys. Rep. 2018;15:93–96. doi: 10.1016/j.bbrep.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Jantsch M.F., Licht K. The editor’s I on disease development. Trends Genet. 2019;35:903–913. doi: 10.1016/J.TIG.2019.09.004. [DOI] [PubMed] [Google Scholar]
- Kawahara Y., Megraw M., Kreider E., Iizasa H., Valente L., Hatzigeorgiou A.G., Nishikura K. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270. doi: 10.1093/NAR/GKN479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerjaschki D., Sharkey D.J., Farquhar M.G. Identification and characterization of podocalyxin--the major sialoprotein of the renal glomerular epithelial cell. J. Cell Biol. 1984;98:1591–1596. doi: 10.1083/JCB.98.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumoto H., Shintani Y., Kanzaki R., Kawamura T., Funaki S., Minami M., Nagatomo I., Morii E., Okumura M. Podocalyxin influences malignant potential by controlling epithelial–mesenchymal transition in lung adenocarcinoma. Cancer Sci. 2017;108:528–535. doi: 10.1111/cas.13142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuttan A., Bass B.L. Mechanistic insights into editing-site specificity of ADARs. Proc. Natl. Acad. Sci. USA. 2012;109 doi: 10.1073/PNAS.1212548109/SUPPL_FILE/PNAS.201212548SI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Kong J.S., Lee S.S., Kim A. Radiation-induced overexpression of TGFβ and PODXL contributes to colorectal cancer cell radioresistance through enhanced motility. Cells. 2021;10:2087. doi: 10.3390/cells10082087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Ang J.K., Xiao X. Analysis and design of RNA sequencing experiments for identifying RNA editing and other single-nucleotide variants. RNA. 2013;19:725–732. doi: 10.1261/rna.037903.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.Y., Kuo C.C., Lin B.X., Cheng C.H., Chen K.C., Lin C.W. Podocalyxin-like protein 1 regulates TAZ signaling and stemness properties in colon cancer. Int. J. Mol. Sci. 2017;18:2047. doi: 10.3390/ijms18102047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev-Maor G., Sorek R., Levanon E.Y., Paz N., Eisenberg E., Ast G. RNA-editing-mediated exon evolution. Genome Biol. 2007;8:R29. doi: 10.1186/gb-2007-8-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht K., Kapoor U., Mayrhofer E., Jantsch M.F. Adenosine to Inosine editing frequency controlled by splicing efficiency. Nucleic Acids Res. 2016;44:6398–6408. doi: 10.1093/nar/gkw325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht K., Kapoor U., Amman F., Picardi E., Martin D., Bajad P., Jantsch M.F. A high resolution A-to-I editing map in the mouse identifies editing events controlled by pre-mRNA splicing. Genome Res. 2019;29:1453–1463. doi: 10.1101/gr.242636.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht K., Hartl M., Hartl M., Amman F., Anrather D., Janisiw M.P., Jantsch M.F. Inosine induces context-dependent recoding and translational stalling. Nucleic Acids Res. 2019;47:3–14. doi: 10.1093/NAR/GKY1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.-W., Sun M.-S., Wu H.-C. Podocalyxin-like 1 is associated with tumor aggressiveness and metastatic gene expression in human oral squamous cell carcinoma. Int. J. Oncol. 2014;45:710–718. doi: 10.3892/ijo.2014.2427. [DOI] [PubMed] [Google Scholar]
- Liu J., Lichtenberg T., Hoadley K.A., Poisson L.M., Lazar A.J., Cherniack A.D., Kovatich A.J., Benz C.C., Levine D.A., Lee A.V., et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome article clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansi L., Tangaro M.A., Lo Giudice C., Flati T., Kopel E., Schaffer A.A., Castrignanò T., Chillemi G., Pesole G., Picardi E. REDIportal: millions of novel A-to-I RNA editing events from thousands of RNAseq experiments. Nucleic Acids Res. 2021;49:D1012–D1019. doi: 10.1093/NAR/GKAA916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews M.M., Thomas J.M., Zheng Y., Tran K., Phelps K.J., Scott A.I., Havel J., Fisher A.J., Beal P.A. Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity. Nat. Struct. Mol. Biol. 2016;23:426–433. doi: 10.1038/NSMB.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Ezzati P., Wilkins J.A. Requirement of podocalyxin in TGF-beta induced epithelial mesenchymal transition. PLoS One. 2011;6:e18715. doi: 10.1371/journal.pone.0018715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.A., Sullivan R.J., Lauffenburger D.A. Molecular pathways: receptor ectodomain shedding in treatment, resistance, and monitoring of cancer. Clin. Cancer Res. 2017;23:623–629. doi: 10.1158/1078-0432.CCR-16-0869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J.S., McNagny K.M. Novel functions of the CD34 family. J. Cell Sci. 2008;121(Pt 22):3683–3692. doi: 10.1242/jcs.037507. [DOI] [PubMed] [Google Scholar]
- Nielsen J.S., McNagny K.M. The role of podocalyxin in health and disease. J. Am. Soc. Nephrol. 2009;20:1669–1676. doi: 10.1681/ASN.2008070782. [DOI] [PubMed] [Google Scholar]
- Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Ann. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016;17:83–96. doi: 10.1038/nrm.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K.C., Dharmasivam M., Richardson D.R. The role of extracellular proteases in tumor progression and the development of innovative metal ion chelators that inhibit their activity. Int. J. Mol. Sci. 2020;21:1–22. doi: 10.3390/IJMS21186805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Yaacov N., Bazak L., Buchumenski I., Porath H.T., Danan-Gotthold M., Knisbacher B.A., Eisenberg E., Levanon E.Y. Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Rep. 2015;13:267–276. doi: 10.1016/J.CELREP.2015.08.080. [DOI] [PubMed] [Google Scholar]
- Peng X., Xu X., Wang Y., Hawke D.H., Yu S., Han L., Zhou Z., Mojumdar K., Jeong K.J., Labrie M., et al. A-to-I RNA editing contributes to proteomic diversity in cancer. Cancer Cell. 2018;33:817–828.e7. doi: 10.1016/j.ccell.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps K.J., Tran K., Eifler T., Erickson A.I., Fisher A.J., Beal P.A. Recognition of duplex RNA by the deaminase domain of the RNA editing enzyme ADAR2. Nucleic Acids Res. 2015;43:1123–1132. doi: 10.1093/NAR/GKU1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth K.V., Prasanth S.G., Xuan Z., Hearn S., Freier S.M., Bennett C.F., Zhang M.Q., Spector D.L. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/J.CELL.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Pruitt K.D., Harrow J., Harte R.A., Wallin C., Diekhans M., Maglott D.R., Searle S., Farrell C.M., Loveland J.E., Ruef B.J., et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/GR.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengaraj P., Obrdlik A., Vukic D., Varadarajan N.M., Keegan L.P., Vanacova S., O’Connell M.A. Interplays of different types of epitranscriptomic mRNA modifications. RNA Biol. 2021;18:19–30. doi: 10.1080/15476286.2021.1969113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueter S.M., Dawson T.R., Emeson R.B. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- Schoft V.K., Schopoff S., Jantsch M.F. Regulation of glutamate receptor B pre-mRNA splicing by RNA editing. Nucleic Acids Res. 2007;35:3723. doi: 10.1093/NAR/GKM314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizemore S., Cicek M., Sizemore N., Ng K.P., Casey G. Podocalyxin increases the aggressive phenotype of breast and prostate cancer cells in vitro through its interaction with ezrin. Cancer Res. 2007;67:6183–6191. doi: 10.1158/0008-5472.CAN-06-3575. [DOI] [PubMed] [Google Scholar]
- Snyder K.A., Hughes M.R., Hedberg B., Brandon J., Hernaez D.C., Bergqvist P., Cruz F., Po K., Graves M.L., Turvey M.E., et al. Podocalyxin enhances breast tumor growth and metastasis and is a target for monoclonal antibody therapy. Breast Cancer Res. 2015;17:46. doi: 10.1186/s13058-015-0562-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S., Shigeyasu K., Okugawa Y., Yoshida K., Mori Y., Yano S., Noma K., Umeda Y., Kondo Y., Kishimoto H., et al. Activation of AZIN1 RNA editing is a novel mechanism that promotes invasive potential of cancer-associated fibroblasts in colorectal cancer. Cancer Lett. 2019;444:127–135. doi: 10.1016/J.CANLET.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo-Orbegozo E., Amo L., Diez-Garcia J., Amutio E., Rinon M., Alonso M., Arana P., Maruri N., Larrucea S. Emerging role of podocalyxin in the progression of mature B-cell non-hodgkin lymphoma. Cancers. 2020;12:396. doi: 10.3390/cancers12020396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi K., Tsuboi M., Sakaguchi M., Saibara T. Measurement of serum PODXL concentration for detection of pancreatic cancer. OncoTargets Ther. 2018;11:1433–1445. doi: 10.2147/OTT.S155367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Tran N., Wang Y., Nie G. Podocalyxin in normal tissue and epithelial cancer. Cancers. 2021;13:2863. doi: 10.3390/CANCERS13122863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran S.S., Jun H.I., Bahn J.H., Azghadi A., Ramaswami G., Van Nostrand E.L., Nguyen T.B., Hsiao Y.H.E., Lee C., Pratt G.A., et al. Widespread RNA editing dysregulation in brains from autistic individuals. Nat. Neurosci. 2019;22:25–36. doi: 10.1038/s41593-018-0287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/SCIENCE.1260419/SUPPL_FILE/1260419. [DOI] [PubMed] [Google Scholar]
- Valente L., Nishikura K. RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J. Biol. Chem. 2007;282:16054–16061. doi: 10.1074/jbc.M611392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/NAR/GKQ603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Park S.H., Beal P.A. Selective recognition of RNA substrates by ADAR deaminase domains. Biochemistry. 2018;57:1640–1651. doi: 10.1021/acs.biochem.7b01100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Rolish M.E., Yeo G., Tung V., Mawson M., Burge C.B. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Wu H., Yang L., Liao D., Chen Y., Wang W., Fang J. Podocalyxin regulates astrocytoma cell invasion and survival against temozolomide. Exp. Ther. Med. 2013;5:1025–1029. doi: 10.3892/etm.2013.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J.F., Yang Q., Liu C.X., Wu M., Chen L.L., Yang L. N 6-methyladenosines modulate A-to-I RNA editing. Mol. Cell. 2018;69:126–135.e6. doi: 10.1016/J.MOLCEL.2017.12.006. [DOI] [PubMed] [Google Scholar]
- Xiao X., Wang Z., Jang M., Nutiu R., Wang E.T., Burge C.B. Splice site strength-dependent activity and genetic buffering by poly-G runs. Nat. Struct. Mol. Biol. 2009;16:1094–1100. doi: 10.1038/nsmb.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhu Z., Wu H., Yu Z., Rong Z., Luo Z., Xu Y., Huang K., Qiu Z., Huang C. PODXL, negatively regulated by KLF4, promotes the EMT and metastasis and serves as a novel prognostic indicator of gastric cancer. Gastric Cancer. 2019;22:48–59. doi: 10.1007/s10120-018-0833-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Carmichael G.G. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–476. doi: 10.1016/S0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhang L., Pan H., Wang B., Yan F., Fang X., Munnee K., Tang Z. Bmi1 essentially mediates podocalyxin-enhanced cisplatin chemoresistance in oral tongue squamous cell carcinoma. PLoS One. 2015;10:e0123208. doi: 10.1371/journal.pone.0123208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/NAR/GKG595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper analyzes existing, publicly available data. The accession link for the datasets is provided in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.