Abstract
Introduction:
Survivors of sepsis exhibit persistent immunosuppression. Epigenetic events may be responsible for some of these immunosuppressive changes. During sepsis circulating exosomes contain large quantities of DNA methyltransferase (DNMT) mRNAs. We hypothesized that exosomes directly transfer DNMT mRNAs to recipient monocytes with resultant methylation events and immunosuppression.
Methods:
Exosomes containing DNMT mRNA were generated by stimulating monocytes with LPS. Confocal microscopy was used to determine uptake kinetics in the presence of pharmacologic inhibition. Expression and packaging of specific DNMT mRNA was controlled using DNMT siRNAs. Whole genome and gene specific methylation was assessed using bisulfite sequencing. Ingenuity pathway analysis was performed to determine the biological function of significance of differentially methylated regions.
Results:
Exosomes effectively transferred DNMT mRNA to recipient monocytes. Pharmacologic inhibition of exosome uptake prevented this increase in DNMT mRNA expression. Recipient monocytes exhibited hypermethylation changes and gene suppression. siRNAs decreased the packaging of DNMT mRNAs and prevented TNFα gene suppression, restoring immunocompetence.
Conclusion:
These data support a role for exosome-mediated transfer of DNMT mRNA with resultant methylation and gene silencing. Pharmacologic uptake inhibition or targeted siRNA mediated DNMT gene silencing prevented DNMT mRNA transfer and maintained the cell’s ability to express TNFα in response to LPS. This highlights the potential therapeutic value of targeting these exosome-mediated epigenetic events to maintain the host immune response during sepsis.
Keywords: DNA methylation, exosomes, immunosuppression, severe sepsis
INTRODUCTION
Sepsis syndrome is mediated by an elegant network of cellular and circulating regulators (1, 2). In the acute phase, inflammatory cytokines, including TNFα, IL-1β, and IL-6, are released into the circulation where they mediate fever, leukocytosis, organ failure, and distributive shock (3, 4). Concomitant with the pro-inflammatory phase is a robust counter-regulatory, anti-inflammatory response that inhibits inflammatory cytokine production and suppresses innate immune function (5). Principal mediators of this immunosuppressive phase include IL-1ra, IL-4, and IL-10 whereas gene expression of TNFα and other pro-inflammatory mediators is suppressed (6, 7). Some patient’s exhibit a characteristic paradoxical constellation of symptoms referred to as persistent inflammatory, immunosuppressed catabolic syndrome (PICS) (6, 8). Other patients develop a prolonged immunosuppressive period that is characterized by continued expression of these counter-inflammatory cytokines, suppression of pro-inflammatory cytokines and profound innate immune dysfunction (9, 10).
This biphasic response is adaptive and necessary for a return to normal physiology, but can be maladaptive if the counter-inflammatory phase is excessive or prolonged. Prolonged immunosuppression in sepsis complicates recovery and results in multiple organ failure, prolonged disability, and increased morbidity (11, 12). Patients with post-sepsis immunosuppression syndrome show dramatically reduced survival for at least 5-years following their septic insult compared to age and comorbidity matched control patients, suggesting a persistent and potentially permanent alteration in the function of these patients (11). The mechanisms that regulate the gene expression that controls the inflammatory and counter-inflammatory phase is not well understood but are thought to include epigenetic and posttranslational gene modifications (13-16).
Circulating monocytes from patients with sepsis exhibit changes in DNA methylation, a cellular response leading to gene-silencing, and the signaling components of immunosuppression associated with sepsis exposure (15). Recent clinical evidence and animal model studies support the notion that epigenetic-mediated monocyte dysfunction plays a causative role in the delayed recovery of the body’s immune response post-sepsis (17-19). Our previous work has shown that circulating exosomes from patients with sepsis contain regulatory elements including mRNAs and miRNAs (20, 21). We have also shown that during sepsis, circulating exosomes contain high levels of DNA methyltransferase (DNMT) mRNA which is necessary for gene silencing (22).
The aim of the present study was to examine the functional role and mechanisms of cell-cell transfer of exosomal contained DMNT mRNAs. To address this, we generated exosomes containing high levels of DNMT mRNA and determined the mechanisms of DNMT mRNA transfer. Our data show that exosomes from human monocytes stimulated with LPS contain high levels of DNMT mRNA and that exosomes transfer these mRNAs to recipient monocytes resulting in DNA methylation and gene silencing. These data support the concept that DNA methylation events are initiated by circulating exosomes that harbor DNMT mRNAs that result in a counter regulatory anti-inflammatory response that potentially mediates the delayed morbidity and mortality of severe sepsis.
MATERIALS AND METHODS
Cell culture and exosome generation
Exosomes were generated using our previous described methods (20). A schematic of this process is included in the supplement (Supplemental Fig. 1, http://links.lww.com/SHK/B437). Briefly, human peripheral blood mononuclear cells (PBMCs) were isolated using fresh blood leukocyte source packs (American Red Cross, Columbus, OH) and isolation was performed using Ficoll-Hypaque density gradient centrifugation (GE Healthcare Cleveland Ohio). Magnetic beads conjugated with CD14 antibody were used for positive selection (MiltenyiBiotec, Auburn, CA). Monocytes were cultured in RPMI 1640 in a humidified atmosphere (5% CO2, 37°C). Cells were stimulated with 6 ng/mL LPS or PBS for 12 h. For qualitative assessment exosomes were isolated from cell culture supernatant using Exoquick Exosome Precipitation Solution ( System Biosciences Palo Alto, CA, cat# EXOTC10A). Sixty-three microliters 63 μL of the solution was added to 250 μL of media, incubated at 4°C for 2 h and centrifuged at 1500g for 30 min. The supernatant was aspirated and the pellet was centrifuged at 1500g for 5 min. For functional experiments, exosomes were isolated using high speed ultracentrifugation as previously described (23). Briefly, the supernatant was subjected to further centrifugation at 4,500g for 15 min at 4°C to remove cell debris followed by 21,000g for 30 min then at 100,000g at 4°C for EV isolation.
Quantitative PCR analysis
Cellular and exosomal RNA was isolated using previously described methods (21, 23). RNA was isolated from cell pellets and from exosome pellet using Trizol: chloroform extraction (Invitrogen, Carlsbad, CA, #15596-018). These were then precipitated in isopropanol for 24 to 48 h. Nanodrop spectrophotometry (ND-1000, Nanodrop Walthan MA) was used to identify concentrations. cDNA was synthesized using qScriptSuperMix (cat. no. 101414; Quanta Biosciences Gaithersburg, MD) and qPCR was performed using SYBR green-based chemistry and validated primers: DNMT1 (Cat. #PPH01055), DNMT3A (Cat. #PPH02339B), DNMT3B (Cat. #PPH01054F), 18s (Cat. #4333760T), TNF-α (Cat. #A15629). Gene expression was quantified using the ΔΔCt relative quantization method using 18S as a normalization control. The human Hippo signaling pathway was quantified using RT2 Profiler PCR Array (cat. no. PAHS-172ZC, Qiagen Germantown MD) using manufacturer recommendations.
Methylation analysis
Genome-wide methylation analysis was performed on blood monocytes with following treatments: PBS or LPS and PBS EV or LPS EV treatment using reduced representation bisulfite sequencing (RRBS). RRBS and sequencing was done using the Ohio State University Genomics Shared Resource facility (22). This approach utilizes the methylation-insensitive restriction enzyme MspI to cut DNA at CCGG sites in the genome. This effectively enriches for fragments derived from CpG-rich regions. RRBS libraries were made and sequenced on Illumina sequencer as described previously (22). RRBS data in fastq format was preprocessed with Trim Galore (22, 24). Trimmed reads were aligned to the bisulfite converted human genome (GRCh38) using the Bismark aligner (25). Post-alignment quality control statistics was done on FastQC and Bismark output files. Differential methylation analysis was performed using methylKit for extraction of CpG methylation values using q-value cutoff of 0.05 for all of the differentially methylated CpGs (DMCs), and a difference in methylation of at least 10% (26). Gene specific methylation was assessed using the Epitect Methyl II PCR Primer Assay (Qiagen), specific to each gene as previously described (23, 27).
Ingenuity pathway analysis
The biological functions of significant DMCs were analyzed using Ingenuity pathway analysis (IPA) ( Ingenuity Systems Redwood City, CA) as described previously (20, 26, 28). Briefly, the excel file containing the list of DMCs along with their respective methylation difference value was loaded as a dataset in IPA software. Core analysis was then performed to obtain enriched pathways represented by DMCs using Ingenuity Knowledge Base with default settings in IPA. Both direct and indirect relationships that could affect networks and upstream regulators were considered. A total of 2,892 analysis-ready DMCs (1,790 hypermethylated and 1,102 hypomethylated) were obtained. HIPPO signaling was found to be a significantly associated pathway (−log(P-value)=1.9; Z-score=−2.11; 10 genes hypermethylated) using canonical pathway feature of IPA. The IPA pathway tool was then used to generate a graphical representation of the biological relationships between differentially methylated genes in a HIPPO pathway, where nodes represented genes and edges represented the biological relationships.
DNMT siRNA transfection
Cultured human monocytes were seeded (0.1 × 106 cells) per well in a 12-well plate in antibiotic-free medium. Transfection was performed at 70% confluence using DharmaFECT reagent (GE Dharmacon Lafayette, CO) and OptiMEM (Thermo Fisher Scientific, Waltham, MA) as reported previously (29, 30) using siRNA (Thermo Fisher Scientific, Waltham, MA) specific for human DNMT1 (siRNA ID 110914), DNMT3A (siRNA ID HSS176224), and DNMT3B (siRNA ID 111744).
Flow cytometry and confocal microscopy
EVs isolated from cultured monocytes were tagged with lipophilic dye PKH26 (cat. no. PKH26GL, Sigma Aldrich, St. Louis, MO) and fed to the naïve cells in absence and presence of wortmannin (cat. no. W1628, Sigma Aldrich) and latrunculin B (cat. no. L5288, Sigma Aldrich). After 24 h, treated cells were then subjected to PKH67 (cat. no. PKH26GL, Sigma Aldrich) for assessment of EV uptake through flow cytometry and confocal microscopy analysis. The fluorescence and light-scattering properties (forward scatter and side scatter) of the cells were determined by using BD Accuri C6 as reported (29). Confocal imaging was done using an inverted fluorescence/confocal microscope (Olympus FV1000). Image analysis was performed as described previously (29, 30).
Statistical analysis
Statistical analyses were performed using SPSS 28.0 (Armonk, NY). The ΔΔCt value was used for statistical analysis of all RT-qPCR data. An unpaired Student’s one and two-sided t tests were used to analyze data between two groups as indicated in the figures. P < 0.05 was considered statistically significant. P values and significance levels are included in the relevant figures. Data were assumed to be normally distributed for all analyses conducted. Data are listed as mean±standard deviation.
Study approval
This study was approved under the institutional IRB #2016H0410.
RESULTS
LPS stimulated monocytes (monocyteLPS) produce exosomes containing high levels of DNMT mRNAs
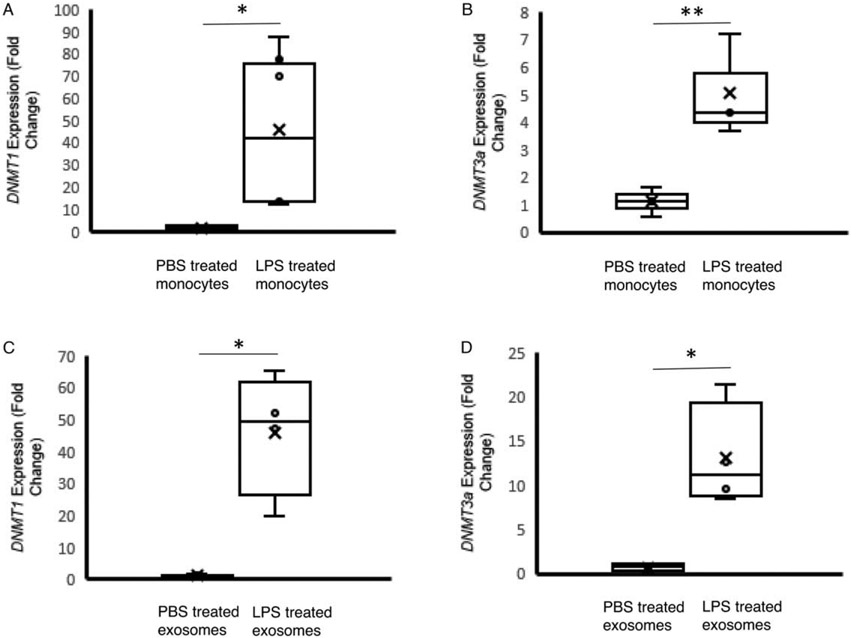
We have previously shown that patients with sepsis and septic shock generate exosomes containing increasing amounts of DNMT1, 3a, and 3b mRNA as disease severity increases (23). In order to develop a model for these DNMT containing exosomes, we utilized primary human monocytes treated with LPS. RT-qPCR analyses show a 45-fold increase in DNMT1 levels (P=0.01) and a 5-fold increase in DNMT3a levels (P=0.03) in monocytes treated with LPS (Fig. 1A). The exosomes generated from these LPS treated monocytes also demonstrated 46-fold increase in DNMT1 mRNA (P=0.009) and a 13-fold increase in DNMT3a mRNA (P=0.01) (Fig. 1B). Exosome size and concentration did not differ significantly between monocytes treated with LPS and monocytes treated with PBS (Supplemental Fig. 2, http://links.lww.com/SHK/B438). These data demonstrate that monocytes stimulated with LPS produced high levels on intracellular DNMT mRNA and shedding exosomes contain high levels of DNMT mRNA
Fig. 1. DNMT mRNA abundance in monocytes and monocyte-derived exosomes after exposure of the cells to LPS.
CD14+ monocytes were isolated from human PBMC. A and B, DNMT1 and DNMT3a mRNA levels measured in monocytes treated with PBS (monocytePBS, n=6) or LPS (monocyteLPS, 6ng/mL, n=4). C and D, DNMT1 and DNMT3a mRNA levels measured in exosomes produced from PBS treated monocytes (n=4) or LPS (n=4) treated monocytes. The data shown are mean±SD with quartiles. One-tailed t test analysis was performed for comparison between group. *P<0.01, **P=0.03.
Monocytes take up monocyte-derived exosomes containing DNMT mRNA via phagocytotic and endocytotic pathways
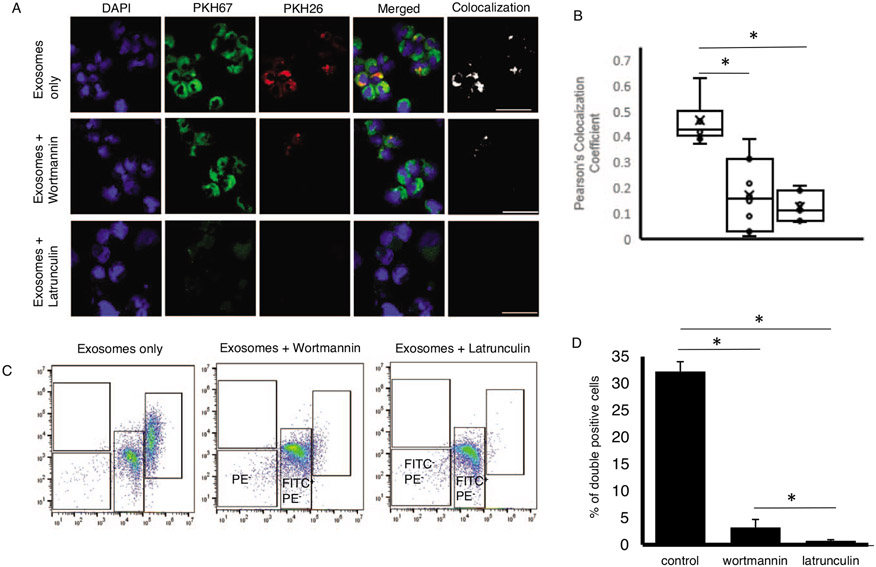
It has been shown in existing literature and in our previous work that exosomes contain regulatory elements including mRNAs that are transferred to recipient cells to alter their function (20, 21, 23). Utilizing confocal microscopy and flow cytometry, we were able to quantify exosome uptake via human monocytes (Fig. 2). Monocytes stained with PKH26 were exposed to LPS to generate exosomes. Hence, red (PKH26) labeled exosomes were isolated from LPS-stimulated monocytes. Recipient monocytes were labeled green using PKH67. Significant co-localization of PKH67 and PKH26 fluorescence signals were observed in the primary human monocytes at 12 h after incubation with the LPS-stimulated exosomes (Fig. 2A). Analysis of co-localization demonstrated a Pearson correlation coefficient of 0.45 for recipient monocytes treated with exosomes from LPS-stimulated monocytes, 0.17 for monocytes also treated with wortmannin (P<0.001), and 0.15 for monocytes treated with lantrunculin (P<0.001) (Fig. 2B). Flow cytometry analysis was used to quantify uptake efficiency (Fig. 2C). On average, 32.2+/1.8% of cells were positive for PKH67 and PKH26 indicating high efficiency of exosome fusion with the monocytes (Fig. 2D). Utilizing lantrunculin to inhibit phagocytosis, reduced the positivity for PKH67 and PKH26 to <5% (P<0.001). Endocytosis inhibition with wortmannin also reduced positivity for PKH67 and PKH26 to <5% (P<0.001) (Fig. 2D).
Fig. 2. Uptake of exosomes by monocytes.
CD14+ monocytes were isolated from human PBMC. Exosomes were isolated from LPS treated monocytes. A, Monocytes stained with PKH26 (red) were stimulated with LPS (monocyteLPS) to generate stained exosomes containing DNMT mRNA. Primary monocytes stained with PKH67 (green) were used as recipient cells and exposed to PKH26 stained exosomes (monocyteLPS-exosome). After incubation for 12 h, confocal microscopy was used to assess and quantify exosome uptake. B, Pearson’s coefficient for colocalization was determined for each image. Control n=11, wortmannin n=11, latrunculin n=8. C, Flow cytometry was used to quantify assess uptake. D, The percentage of PKH26 (red) and PKH67 (green) double positive cells was assessed to confirm the uptake efficiency (n=3). Data shown are mean±standard deviation with quartiles. One-tailed t test analysis was performed for comparisons. *P<0.001.
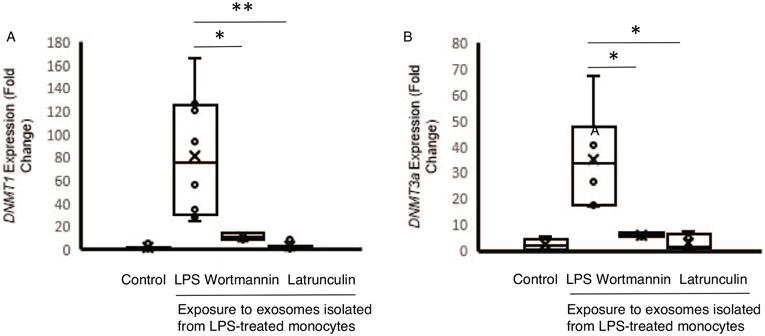
We next assessed the impact of pharmacologic uptake inhibition on DNMT mRNA expression within recipient cells. Primary human monocytes were incubated for 12 h with exosomes derived from LPS-stimulated monocytes (monocytesLPS-exosome) and mRNA expression was measured using RT-qPCR (Fig. 2). Compared to monocytes treated with exosomes derived from PBS treated (monocytesPBS-exosomes) monocytes, cells treated with monocytesLPS-exosomes demonstrated a 84-fold increase in DNMT1 mRNA levels (P<0.001) (Fig. 3A) and a 35-fold increase in DNMT3a mRNA levels (P<0.001) (Fig. 3B). Pharmacologic treatment with wortmannin resulted in significantly reduced DNMT1 mRNA (84.6±56.1 vs. 10.8±3.1, fold change, P=0.03) and DNMT3a mRNA (34.9±15.2 vs. 5.9±2.6, fold change, P=0.01) compared to the increases seen in cells without uptake inhibition (Fig. 3, A and B). Latrunculin treatment also reduced DNMT1 mRNA levels (2.3±3.1, fold change, P<0.01) and DNMT3a mRNA levels (3.1±1.3, fold change, P<0.01). These data suggest that upon LPS stimulation, monocyte-derived exosomes can deliver DNMT mRNA through endocytic or phagocytic pathways.
Fig. 3. DNMT mRNA expression in monocytes exposed to exosomes isolated from LPS stimulated monocytes (monocyteLPS-exosome).
CD14+ monocytes were isolated from human PBMC. A, DNMT1 levels were measured using RT-PCR in naïve recipient human monocytes after 12 h incubation with exosomes derived from human monocytes treated with PBS (control, n=5) or with LPS (6 ng/mL, n=7). Recipient cells were also treated with pharmacologic uptake inhibitors wortmannin (n=4) or latrunculin (n=10). B, DNMT3a levels were measured under the same conditions. The data shown are mean±SD with quartiles. One-tailed t test analysis was performed for comparisons. *P<0.01, **P=0.03.
Exosome-mediated delivery of DNMT mRNA facilitates DNA methylation to induce immunosuppression in host monocytes
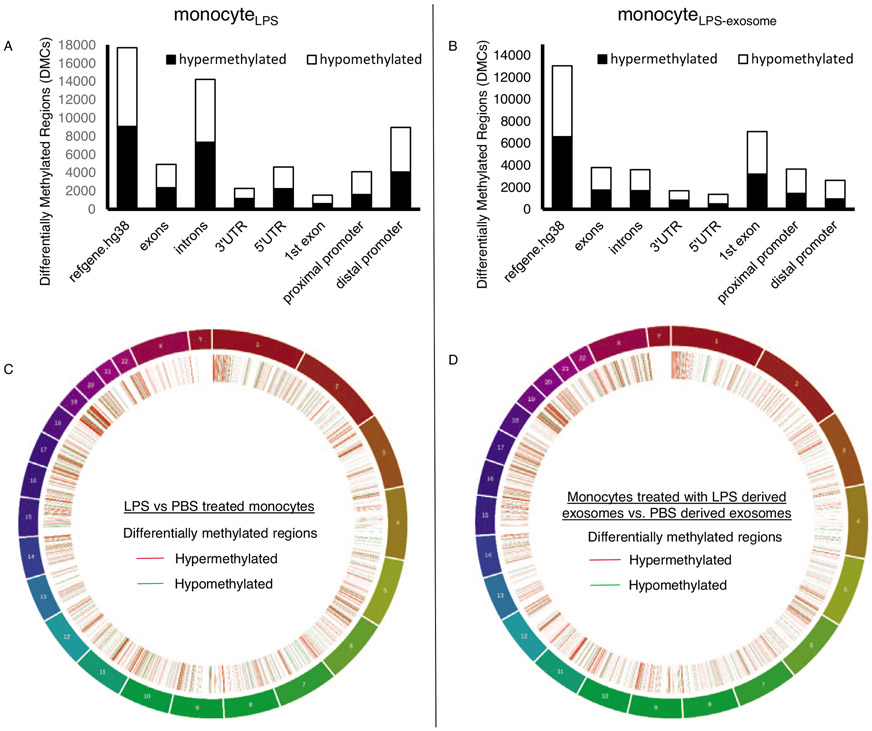
Exosomes from patients with sepsis and exosomes derived from primary monocytes stimulated with LPS (monocyteLPS) are known to deliver protein cargo with immuno-modulatory effects in recipient monocytes (20). The effect of this protein transfer results in decreased TNFα production in response to LPS stimulation and proteomic pathway analysis also suggested an immunosuppressed phenotype (20). In order to examine the effects of the DNMT mRNA transfer on recipient cell function, we analyzed the methylation effects of recipient monocytes treated with exosomes derived from LPS-stimulated monocytes (monocytesLPS-exosome). Bioinformatics analysis identified 9,070 hyper-methylated CpG sites in human monocytes exposed to LPS (monocyteLPS) (Fig. 4A). These hypermethylation events were recapitulated in naïve human monocytes upon exposure to monocytesLPS-exosome, where 6,602 hypermethylated CpG sites were identified (Fig. 4B). In these cells, roughly 20% were confined to the promoter regions (Fig. 4, A and B). These methylation events appear genome wide (Fig. 4, C and D). The top 10 genes with methylation events from monocyteLPS and monocytes treated with monocytesLPS-exosome are shown in Figure 5A and B respectively. Examining gene specific promoter methylation events showed significant methylation for genes involved in the inflammatory response (TNFα, TGFβ, IL-12, and IL-13) in monocytes treated with monocytesLPS-exosome (Fig. 6). In order to assess exosome-mediated promoter methylation on gene silencing related to immunosuppression, human monocytes exposed to exosomes were challenged with LPS and their capability to release TNFα was assayed using RT-qPCR (Fig. 7). Compared to cells that were treated with control exosomes, monocytes that were treated with monocytesLPS-exosome exhibited reduced TNFα production 1.11±0.53 vs. 0.69±0.23-fold change, P=0.007 (Fig. 7A). Inhibition of exosome uptake with wortmannin (1.22±0.78 vs. 0.92±0.61-fold change, P=0.44) or latrunculin (1.0±0.55 (n = 7) vs. 0.96±0.59-fold change, P=0.81) effectively maintained the ability of the monocyte to generate TNFα in response to LPS stimulation (Fig. 7, B and C). This provides direct support for our hypothesis that exosome-derived DMNT mRNA delivery facilitate DNA methylation of recipient cells as a contributing mechanism to gene silencing and post-sepsis immune-suppression.
Fig. 4. Exosomes isolated from monocytes exposed to LPS induced epigenetic changes in naïve recipient monocytes.
CD14+ monocytes were isolated from human PBMC. A, Differentially methylated CpGs (DMCs) observed in direct treatment of LPS to cultured primary monocytes (monocyteLPS) as compared to PBS. B, DMCs of cells treated with exosomes derived from LPS stimulated monocytes (monocyteLPS-exosome) compared to exosomes derived from PBS treated monocytes. C, Genomic distribution of DMCs observed in direct exposure of LPS/PBS to cultured primary monocytes. Red represents the number of genes which were hypermethylated while green represents the number of genes which were hypomethylated in LPS treated monocytes. D, Genomic distribution of DMCs observed in monocytes exposed to exosomes LPS/PBS derived exosomes.
Fig. 5. Top 10 genes in monocytes that were differentially methylated in response to LPS stimulation (monocyteLPS) or exposure to exosomes isolated from LPS treated cells (monocyteLPS-exosome).
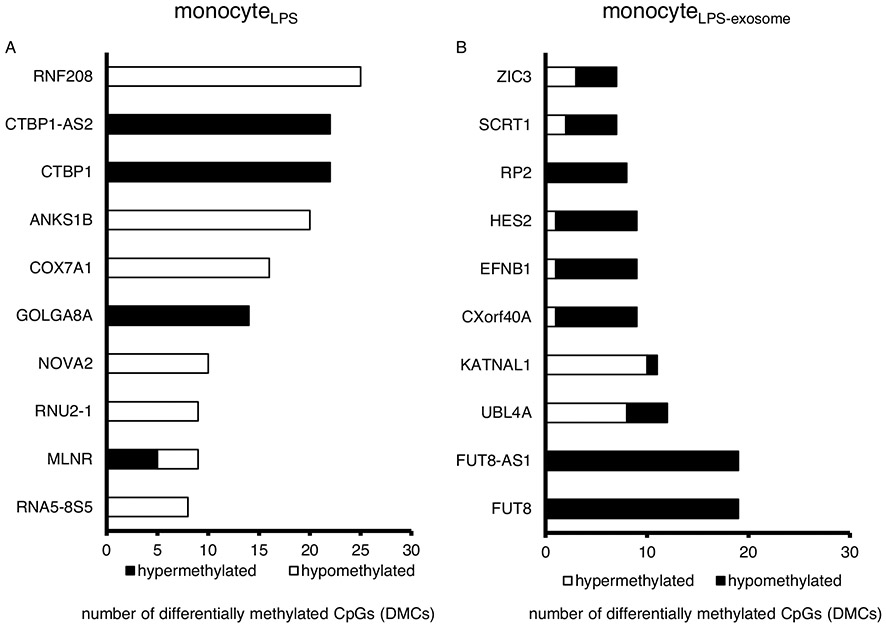
A, Top 10 genes having highest number of differentially methylated CpGs (DMCs) in monocytes treated with LPS (monocyteLPS) compared to PBS. B, Top 10 genes having highest number of DMCs in monocyteLPS-exosome.
Fig. 6. Gene specific promoter methylation in response to treatment with exosomes derived from LPS stimulated monocytes (monocyteLPS-exosome).
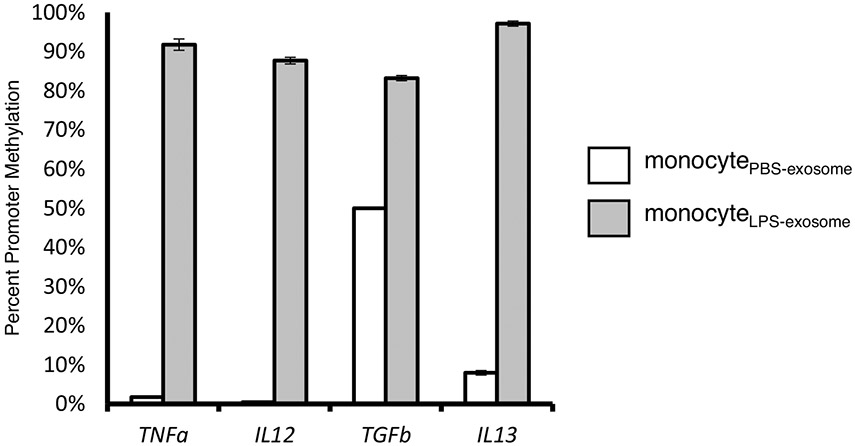
Gene specific analysis of promoter methylation was performed using Epitect Methyl II PCR assay at TNF-α, IL-12, TGF-β, and IL-13 (n=3). Data shown are mean±SD. One-tailed t test analysis was performed for comparisons. P<0.001 for all.
Fig. 7. TNFα expression in monocytes exposed to exosomes isolated from LPS stimulated cells in the presence of pharmacologic exosome uptake inhibition.
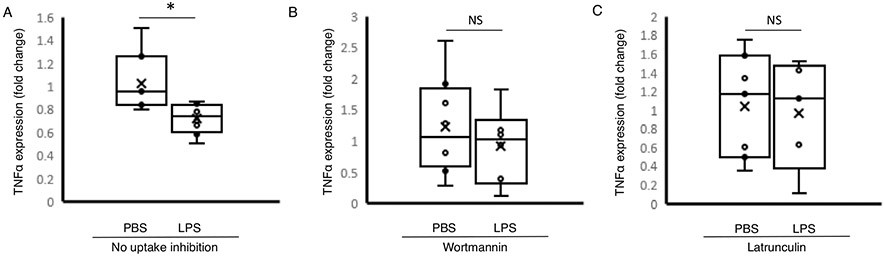
A, Human monocytes were treated with exosomes derived from PBS stimulated monocytes (n=18) or LPS stimulated monocytes (monocyteLPS-exosome, n=16). TNFα expression was assessed using RT-qPCR. B, Human monocytes were treated with exosomes isolated from PBS or LPS stimulated monocytes in the presence of pharmacologic uptake inhibitor wortmannin and (C) latrunculin. The data shown are mean±SD with quartiles. One-tailed t test analysis was performed for comparisons. *P<0.01, NS=non-significant.
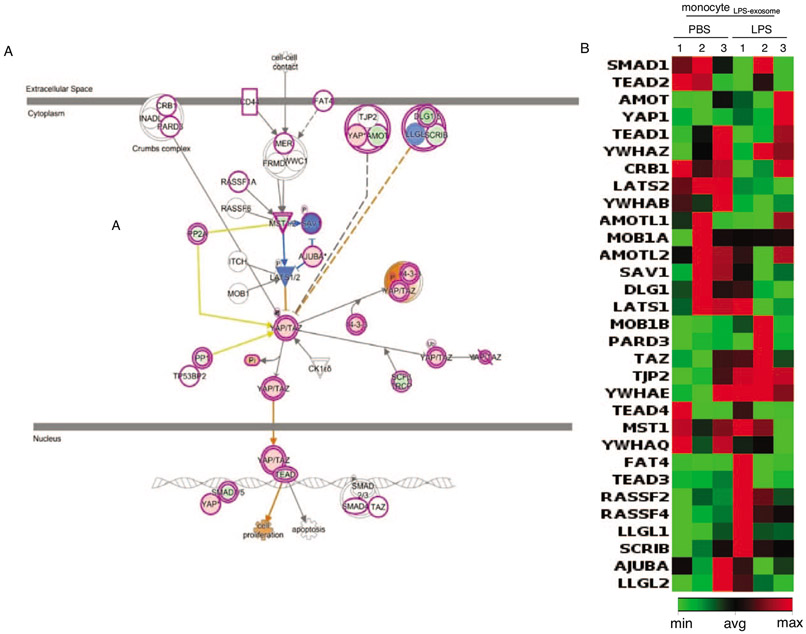
Based on the methylation patterns identified, we performed Ingenuity pathway analysis for classification (20, 29). Primary monocytes treated with monocytesLPS-exosome demonstrated significant hypermethylation in the HIPPO-YAP signaling pathway (Fig. 7A). We utilized RT-qPCR for 31 genes involved in the HIPPO-YAP pathway to confirm gene expression changes that were predicted from IPA (Fig. 8, Supplemental Fig. 3, http://links.lww.com/SHK/B439). These data confirm the effects of DNMT mRNA transfer and gene regulation in recipient monocytes.
Fig. 8. Ingenuity pathway analysis (IPA) of differential methylated CpGs (DMCs) in monocytes in response to exposure of exosomes isolated from LPS stimulated monocytes (monocyteLPS-exosome).
A, Ingenuity pathway analysis of genes which were differentially methylated in primary monocytes exposed to exosomes derived from LPS treated monocytes identified HIPPO signaling as a dominant pathway that was hypermethylated. B, Gene expression changes in HIPPO signaling were validated using RT-qPCR (n=3).
Inhibition of exosome-mediated DNMT delivery reduces LPS-tolerance
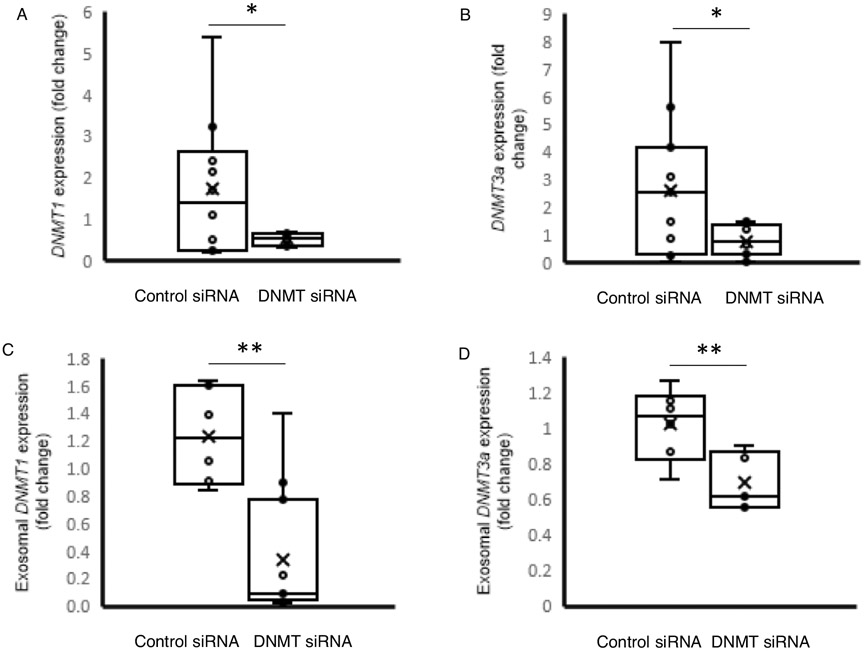
To further define the role of DNMT mRNA in exosome-mediated methylation events as a contributing factor for post-immune suppression, we obtained a siRNA cocktail for silencing of all 3 isoforms of DNMTs (1, 3A, and 3B, Thermo Fisher Scientific, Waltham, MA). Monocytes treated with the siRNA cocktail then stimulated with LPS had significant reductions in DNMT1 mRNA levels compared to monocytes treated with control siRNAs (1.71±1.65 vs. 0.30±0.15-fold change, P<0.01) (P<0.01) and DNMT3a (0.29-fold change, P<0.01) mRNA levels following treatment with LPS (Fig. 9, A and B). DNMT3B transcripts were not detected in all the samples. Exosomes derived from these monocytes also exhibited decrease DNMT1 (1.23±0.35 vs. 0.33±0.47-fold change, P<0.01) and DNMT3a (1.02±0.20 vs. 0.69±0.16-fold change, P<0.01) mRNA levels (Fig. 9, C and D). In order to test the effect on recipient cells were first measured TNFα expression. When the recipient cells were re-stimulated with LPS, TNFαexpression was higher in cells exposed to exosomes-derived from DNMT siRNA-treated cells compared with those derived from cells treated with the control siRNA (1.37±0.59 vs. 3.20±1.15-fold change, P<0.001), demonstrating a maintenance of the ability to generate TNFα in response to LPS (Fig. 10).
Fig. 9. DNMT and TNFα mRNA expression in monocytes exposed to exosomes isolated from cells that have been stimulated with LPS in presence of DNMT siRNAs.
A, DNMT1 and (B) DNMT3A mRNA expression was assessed in human monocytes stimulated with LPS after siRNA treatment for against DNMT1 and DNMT3a (n=10). C, DNMT1 mRNA and (D) DNMT3A mRNA expression was assessed in exosomes produced from LPS stimulated monocytes after siRNA treatment against DNMT1 and DNMT3a. E, TNFα mRNA expression was measured to assess for gene silencing. The data shown are mean±SD with quartiles. One-tailed t test analysis was performed for comparisons. *P=0.03, **P<0.01.
Fig. 10. TNFα mRNA expression in monocytes in response to LPS stimulation after exposure to exosomes isolated from cells that have been stimulated with LPS in presence of ONMT siRNAs.
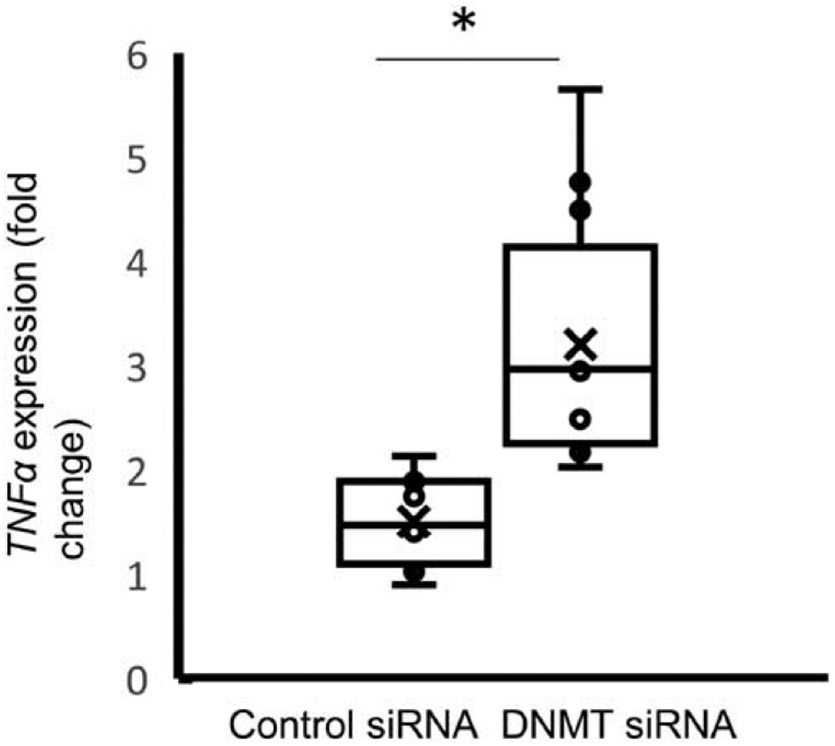
Monocytes were treated with exosomes produced from LPS stimulated monocytes treated with control (n=11 ) or DNMTs siRNAs (n=12) then stimulated with LPS. TNFa mRNA expression was measured to assess for gene silencing. The data shown are mean ± SO with quartiles. 1-tailed t test analysis was performed for comparisons. *p <0.01.
DISCUSSION
Our data show that LPS stimulation of human monocytes results in the release of exosomes that contain DMNT mRNA. Treatment of human monocytes with exosomes from LPS treated monocytes, results in transfer of the DMNT mRNA cargo to the recipient monocytes resulting in methylation events that alter gene expression. Pharmacologic inhibition of exosome uptake or ablation of DNMT packaging with siRNAs in the source monocytes ameliorates the DNMT gene expression and methylation events in the recipient monocytes. Notably, many of the silenced genes in the recipient monocytes are involved in the inflammatory response, suggesting a role for exosome-mediated methylation events that have been reported in patients with sepsis and septic shock (12, 15). These findings support the scientific premise that epigenetic methylation events occur as part of the inflammatory response during sepsis, and exosome mediated regulation of DNA methylation contributes to gene-silencing associated with post-sepsis immunosuppression.
These data indicate an important, previously unidentified mechanism of gene regulation that occurs during sepsis where circulating exosomes containing high levels of DNMT mRNA induce immunosuppression in patients with sepsis. The importance of DNA methylation events and gene silencing that occurs during sepsis is an emerging area of investigation. Within monocytes, epigenetic analysis has revealed a close link between changes in promoter methylation, a cellular response leading to gene-silencing, and the signaling components of immunosuppression associated with sepsis exposure. Clinical evidence and animal studies support the notion that epigenetic-mediated monocyte dysfunction plays a causative role in the delayed recovery of the body’s immune response post-sepsis. LPS stimulation of monocytes has been shown to effectively model these methylation events (31). The current work provides mechanistic evidence for the role of DNMTs and epigenetic regulation in the physiologic processes occurring in patients with sepsis.
A key piece of missing information is how methylation events are transduced systemically. The current work demonstrates the impact and importance of circulating exosomes and their ability to amplify the methylation signal systemically. Our data strongly support a role for exosome mediated transfer of DNMT mRNA cargo that results in methylation events and gene silencing important to the inflammatory response. We have identified that disruption of this mechanism restores immunologic function to human monocytes. Utilizing either pharmacologic inhibition of exosome uptake, or prevention of DNMT mRNA exosome packaging, we have identified multiple steps in the signal transduction process that may be ripe for therapeutic intervention. Existing in vivo animal studies support the concept of methylation inhibition as a means to treat sepsis with survival advantages for animals treated with DNA methylation inhibitors.
There are several limitations of the present study. The present data does not identify which DNMT mRNA is most important for the development of the methylation patterns seen. Both DNMT 1 and 3a are present in high quantities and each isoform may have differing effects. Another limitation is that our data were generated from in vitro experiments. While they provide considerable mechanistic support, they do not account for the myriad of covariates that are present in an in vivo system that may alter the findings. Therefore, it is necessary for future studies to conduct functional assessments an in vivo system. It is known that DNA methylation is not the only form of epigenetic regulation and exosomes are known to carry additional regulators including histone deacetylases and microRNAs. While our data support the role for DNMT-mediated DNA methylation as a contributing factor for the epigenetic modulation of post-septic immunosuppression, we need to test the role of other exosome-mediated epigenetic modulators. Additionally, we do not have a full understanding of the impact of siRNA inhibition in the exosome producing cells. It may be that inhibiting DNMT expression alters downstream gene expression and exosome packaging. We utilized pharmacologic methods of exosome uptake inhibition in addition to siRNA treatments to provide some control for these unobserved differences in exosome packaging, but this still may introduce an unknown variable.
In summary, in the present study we find that exosomes derived from LPS stimulated monocytes contain high levels of DNMT mRNAs that can be transferred to recipient monocytes with resultant methylation events and immunosuppression. Interruption of the exosome packaging or transfer events restored signaling and retained the ability to generate an immune response to LPS. Pharmacologic inhibition of phagocytosis and endocytosis effectively prevented this transferred and resulted in maintained immune function in recipient cells. Furthermore, prevention of DNMT mRNA packaging in exosomes prevented the methylation events and resultant alterations in inflammatory signaling. This provides strong mechanistic support to the hypothesis that circulating exosomes contribute to the methylation events and alterations in inflammatory signaling.
Supplementary Material
Sources of support:
GM137078 NIH KO8 (JRW), NIH R01HL137224 (JWC).
Footnotes
The authors have declared no conflict of interest exists.
Ethics statement: The material in this submission is not under consideration and has not been published elsewhere. The authors have all approved this submission. Sources of support and conflicts of interested are listed.
REFERENCES
- 1.Torres LK, Pickkers P, van der Poll T: Sepsis-induced immunosuppression. Annu Rev Physiol 84:157–181, 2022. [DOI] [PubMed] [Google Scholar]
- 2.Xiao WZ, Mindrinos MN, Seok JH, Cuschieri J, Cuenca AG, Gao H, Hayden DL, Hennessy L, Moore EE, Minei JP, et al. : The inflammation and host response to injury large-scale collaborative research program: a genomic storm in critically injured humans. J Exp Med 208(13):2581–2590, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sriskandan S, Altmann DM: The immunology of sepsis. J Pathol 214(2):211–223, 2007. [DOI] [PubMed] [Google Scholar]
- 4.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernad GR, Chiche JD, Coopersmith CM, et al. : The third international consensus definitions for sepsis and septic shock. JAMA 315(8):801–810, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hotchkiss RS, Monnerat G, Payen D: Immunosuppression in sepsis: a novel understanding of the disorder and a new therapeutic approach. Lancet Inf Dis 13(3):260–268, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gentile LF, Cuenca AG, Efron PA, Ang D, Bihorac A, McKinley BA, Moldawer LL, Moore FA: Persistent inflammation and immunosuppression: a common syndrome and new horizon for surgical intensive care. J Trauma Acute Care Surg 72(6):1491–1501, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ertel W, Kremer JP, Kenney J, Steckholzer U, Jarrar D, Trentz O, Schildberg FW: Downregulation of proinflammatory cytokine release in whole blood from septic patients. Blood 85(5):1341–1347, 1995. [PubMed] [Google Scholar]
- 8.Mira JC, Brakenridge SC, Moldawer LL, Moore FA: Persistent inflammation, immunosuppression and catabolism syndrome. Crit Care Clin 33(2):245–258, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Vught LA, Klouwenberg CK, Spitoni C, Scicluna BP, Wiewel MA, Horn J, Schultz MJ, Nurnberg P, Boten MJ, Cremer OL, et al. : Incidence, risk factors, attributable mortality of secondary infections in the intensive care unit after admission for sepsis. JAMA 315(14):1469–1479, 2016. [DOI] [PubMed] [Google Scholar]
- 10.Wang HE, Szychowski JM, Griffin R, Safford MM, Shapiro NI, Howard G: Long-term mortality after community-acquired sepsis: a longitudinal population-based cohort study. BMJ 4(1):1–8, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasse KC, Nauenberg E, Long A, Anton B, Tucker HJ, Hu TW: Long-term survival after intensive care unit admission with sepsis. Crit Care Med 23(6):1040–1047, 1995. [DOI] [PubMed] [Google Scholar]
- 12.Mira J, Gentile L, Mathias BJ, Efron PA, Brakenridge SC, Mohr AM, Moore FA, Moldawer LL: Sepsis pathophysiology, chronic critical illness, and persistent inflammation-immunosuppresion and catabolism syndrome. Crit Care Med 45(2):253–262, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novakovic B, Habibi E, Wang S, Arts RJW, Davar R, Megchelenbrink W, Kim B, Kuznetsova T, Kox M, Zwaag J: B-glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell 167(5):1354–1386, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carson WF, Cavassani KA, Dou Y, Kunkel SL: Epigenetic regulation of immune cell functions during post-septic immunosuppression. Epigenetics 6(3):272–283, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiterer S, Uhle F, Lichtenstern C, Siegler BH, Bhuju S, Jarek M, Bartkuhn M, Weigand MA: Sepsis induces specific changes in histone modification patterns in human monocytes. PLoS ONE 10(3):e0121748, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCall CE, Yoza BK: Gene silencing in severe systemic inflammation. Am J Respir Crit Care Med 175(8):763–767, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weighardt H, Heidecke CD, Emmanuilidis K, Maier S, Bartels H, Siewart JR, Holzmann B: Sepsis after major visceral surgery is associated with sustained interferon-gamma-resistant defects of monocyte cytokine production. Surgery 127(3):309–315, 2000. [DOI] [PubMed] [Google Scholar]
- 18.Munoz C, Carlet J, Fitting C, Misset B, Bleriot JP, Cavaillon JM: Dysregulation of in vitro cytokine production by monocytes during sepsis. J Clin Invest 88(5):1747–1754, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shalova IN, Lim JY, Chittezhath M, Zinkernagel AS, Beasley F, Hernandez-jimenez E, Toledano V, Cubillos-zapata C, Rapisarda A, Chen J: Human monocytes undergo functional re-programming during sepsis mediated by hypoxia-inducible factor-1α. Immunity 42(3):484–498, 2015. [DOI] [PubMed] [Google Scholar]
- 20.Wisler JR, Singh K, McCarty A, Abouhashem ASE, Christman JW, Sen CK: Proteomic pathway analysis of monocyte-derived exosomes during surgical sepsis identifies immunoregulatory functions. Surgical Infections 21(2):101–111, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ismail N, Wang Y, Dakhlallah D, Moldovan L, Agarwal K, Batte K, Shah P, Wisler J, Eubank TD, Tridanadapani S: Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood 121(6):984–995, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caution K, Pan A, Krause K, Badr A, Hamilton K, Vaidya A, Gosu W, Daily K, Estfanous S, Gavrilin MA: Methylomic correlates of autophagy activity in cystic fibrosis. J Cyst Fibros 18(4):491–500, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dakhlallah DA, Wisler J, Gencheva M, Brown CM, Leatherman ER, Singh K, Brundage K, Karsies T, Daklallah A, Witwer KW: Circulating extracellular vesicle content reveals de novo DNA methyltransferase expression as a molecular method to predict septic shock. J Extracell Vesicles 8(1):1669881, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krueger T, Fisher PL, Becker S, Pontasch S, Dove S, Hoegh-Guldberg O, Leggat W, Davy S: Transcriptomic characterization of the enzymatic antioxidants FeSOD, MnSOD, APX and KatG in the dinoflagellate genus symbiodinium. BMC Evol Biol 15(48):s12862–s12915, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akalin A, Kormaksson M, Li S, eGarret-Bakelman FE, Figueroa ME, Melnick A, Mason CE: MethylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol 13(10):R87, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh K, Pal D, Sinha M, Ghatak S, Gnyawali SC, Khanna S, Roy S, Sen CK: Epigenetic modification of microRNA-200b contributes to diabetic vasculopathy. Mol Ther 25(12):2689–2704, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dakhlallah D, Batte K, Wang Y, Catemir-Stone CZ, Yan P, Nuovo G, Mikhail A, Hitchcock CL, Wright VP, Nan-Sinkam S: Epigenetic regulation of miR-17~92 contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med 187(4):397–405, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosh N, Das A, Biswas N, Gnyawali S, Singh K, Gorain M, Polcyn S, Roy S, Sen CK: Urolithin A augments angiogenic pathways in skeletal muscle by bolstering NAD and SIRT1. Sci Rep 10(1):20184, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh K, Sinha M, Pal D, Tabasum S, Gnyawali SC, Khona D, Sarkar S, Mohanty SK, Soto-Gonzalez F, Khanna S: Cutaneous epithelial to mesenchymal transition activator ZEB1 regulates wound angiogenesis and closure in a glycemic status-dependent manner. Diabetes 68(11):2175–2190, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinha M, Sen CK, Singh K, Das A, Ghatak S, Rhea B, Blackstone B, Powell HM, Khanna S, Roy S: Direct conversion of injury-site myeloid cells to fibroblast-like cells of granulation tissue. Nat Commun 9(1):936, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorente-Sorolla C, Garcia-Gomez A, Catala-Moll F, Toledano V, Ciudad L, Avendano-Ortiz J, Maroun-Eid C, Martin-Quiros A, Martinez-Gallo M, Ruiz-Sanmartin JC: Inflammatory cytokines and organ dysfunction associate with aberrant DNA methylome of monocytes in sepsis. Genome Med 11(1):66, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.