ABSTRACT
Rivers have a significant role in global carbon and nitrogen cycles, serving as a nexus for nutrient transport between terrestrial and marine ecosystems. Although rivers have a small global surface area, they contribute substantially to worldwide greenhouse gas emissions through microbially mediated processes within the river hyporheic zone. Despite this importance, research linking microbial and viral communities to specific biogeochemical reactions is still nascent in these sediment environments. To survey the metabolic potential and gene expression underpinning carbon and nitrogen biogeochemical cycling in river sediments, we collected an integrated data set of 33 metagenomes, metaproteomes, and paired metabolomes. We reconstructed over 500 microbial metagenome-assembled genomes (MAGs), which we dereplicated into 55 unique, nearly complete medium- and high-quality MAGs spanning 12 bacterial and archaeal phyla. We also reconstructed 2,482 viral genomic contigs, which were dereplicated into 111 viral MAGs (vMAGs) of >10 kb in size. As a result of integrating gene expression data with geochemical and metabolite data, we created a conceptual model that uncovered new roles for microorganisms in organic matter decomposition, carbon sequestration, nitrogen mineralization, nitrification, and denitrification. We show how these metabolic pathways, integrated through shared resource pools of ammonium, carbon dioxide, and inorganic nitrogen, could ultimately contribute to carbon dioxide and nitrous oxide fluxes from hyporheic sediments. Further, by linking viral MAGs to these active microbial hosts, we provide some of the first insights into viral modulation of river sediment carbon and nitrogen cycling.
IMPORTANCE Here we created HUM-V (hyporheic uncultured microbial and viral), an annotated microbial and viral MAG catalog that captures strain and functional diversity encoded in these Columbia River sediment samples. Demonstrating its utility, this genomic inventory encompasses multiple representatives of dominant microbial and archaeal phyla reported in other river sediments and provides novel viral MAGs that can putatively infect these. Furthermore, we used HUM-V to recruit gene expression data to decipher the functional activities of these MAGs and reconstruct their active roles in Columbia River sediment biogeochemical cycling. Ultimately, we show the power of MAG-resolved multi-omics to uncover interactions and chemical handoffs in river sediments that shape an intertwined carbon and nitrogen metabolic network. The accessible microbial and viral MAGs in HUM-V will serve as a community resource to further advance more untargeted, activity-based measurements in these, and related, freshwater terrestrial-aquatic ecosystems.
KEYWORDS: hyporheic zone, Binatia, microbiome, metagenomics, mineralization, greenhouse gas, viruses, auxiliary metabolic genes, Thermoproteota
INTRODUCTION
The hyporheic zone (HZ) is a transitional space between river compartments where the mixing of nutrients and organic carbon from river and groundwater stimulate microbial activity (1–3). Characterized as the permanently saturated interface between the river surface channel and underlying sediments, the HZ is considered a biogeochemical hot spot for microbial biogeochemistry (1–3), ultimately contributing to the majority of river greenhouse gas (GHG) fluxes. For instance, it is estimated that rivers contribute up to 85% of inland water carbon dioxide and 30% of nitrous oxide emissions (4–6). Microorganisms in the HZ also catalyze the transformation of pollutants and natural solutes, all while microbial biomass itself supports benthic food webs (7). Together, these findings highlight that microbial metabolism in HZ sediments has a substantial influence on overall river biogeochemistry and health.
Despite the importance of HZ microorganisms, research linking microbial identity to specific biogeochemical reactions in the carbon and nitrogen cycles is still nascent in sediments. In conjunction with geochemistry, microbial functional genes or gene products (e.g., nirS and nrfA) have been quantified to denote microbial contributions to specific biogeochemical pathways (e.g., nitrate reduction) (8). However, these studies often do not identify the microorganisms catalyzing the process and focus on only a few enzymatic reactions. Thus, a comprehensive assessment of the interconnected microbial metabolisms that fuel carbon and nitrogen cycling in river sediments is underexplored.
More recently, 16S rRNA amplicon sequencing has shed new light on the identity of bacterial and archaeal members in river sediments. These studies revealed that cosmopolitan and dominant members in river sediments often belong to six main phyla: Acidobacteria, Actinobacteriota, Firmicutes, Nitrospirota, Proteobacteria, and Thaumarchaeota (9, 10). In some instances, cultivation paired with amplicon sequencing has assigned some of these microorganisms (e.g., Proteobacteria) to specific biogeochemical process (e.g., denitrification) (11). Yet, most functional inferences from taxonomic data alone are unreliable due to the dissociation between microbial taxonomy and metabolic function (12, 13). Thus, many key biogeochemical pathways in rivers (e.g., plant biomass deconstruction, denitrification, nitrogen mineralization) are not holistically interrogated alongside microbial communities (14). Furthermore, amplicon sequencing fails to sample viral communities. While it is likely that viruses are key drivers of HZ microbial mortality and biogeochemical cycling by dynamics of predation and auxiliary metabolic genes, the evidence is even more sparse than for their bacterial and archaeal counterparts (15–18).
Cultivation-independent, community-wide, and genome-resolved approaches are key to addressing the knowledge gap of how microbial and viral communities influence river biogeochemical cycling. However, metagenomic studies in river sediments are limited and have focused primarily on gene content rather than the reconstruction of genomes (19, 20). To our knowledge, only two river sediment studies have generated microbial genomes to link taxonomy to functional processes, and these have focused on the impacts of nitrate-oxidizing and comammox microorganisms on nitrification (21, 22). As such, despite these recent advances, the chemical exchange points that interconnect the carbon and nitrogen cycles cannot be discerned from existing HZ microbiome studies.
With the overarching goal of providing enhanced resolution to microbial and viral contributions to carbon and nitrogen cycling in the HZ, we created the first of its kind Hyporheic Uncultured Microbial and Viral (HUM-V) genomic catalog. We then used HUM-V to recruit metaproteomic data collected from 33 laterally and depth-distributed HZ sediment samples. We further supported this gene expression data using chemical data from paired metabolomics and geochemical measurements. Our results (i) profiled expressed microbial metabolisms that support organic and inorganic carbon and nitrogen cycling in the HZ, (ii) uncovered roles for viruses that could modulate microbial activity in the HZ, and (iii) created a roadmap of a microbial metabolic circuitry that potentially contributes to greenhouse gas fluxes from rivers. We anticipate that this publicly available community resource will advance future microbial activity-based studies in HZ sediments and is a step toward the development of biologically aware, hydro-biogeochemical predictive models.
RESULTS AND DISCUSSION
HUM-V greatly expands the genomic sampling of HZ microbial members.
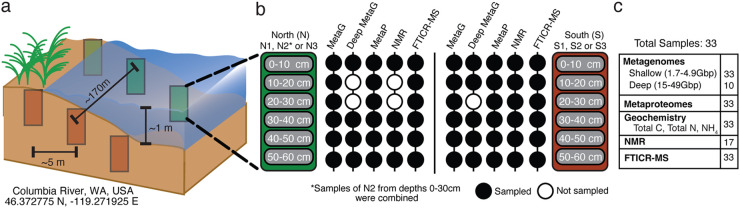
We used previously collected samples from HZ sediment cores from the Hanford Reach of the Columbia River in eastern Washington State, USA (23), an 80-km stretch of cobble-bed river that often experiences rapid discharge fluctuations (24). From this system, six samples per transect were collected, and each core was subsampled into six 10-cm-depth increments (0 to 60 cm) (Fig. 1a and b). Of these 36 samples, 33 were subsequently processed for metagenomic sequencing, geochemistry, metaproteomics, and Fourier transform ion cyclotron resonance mass spectrometry (FTICR-MS) (Fig. 1c; see Table S1 in the supplemental material) (see also Materials and Methods). A subset of these (n = 17) were also analyzed for nuclear magnetic resonance (NMR) metabolites. For our metagenomics data, we obtained 379 Gbp of sequencing across all 33 samples, which included (i) the original shallow sequencing of all samples (1.7 to 4.9 Gbp/sample) (23) and (ii) an additional deeper sequencing of 10 samples (15.3 to 49.2 Gbp/sample), which are reported here for the first time. We reconstructed 655 metagenome-assembled genomes (MAGs), of which 102 were denoted as medium or high quality based on current standards (25). MAGs from both transects were then dereplicated (99% identity; see Materials and Methods) into the 55 unique genomic representatives that constitute the microbial component of HUM-V (Table S2) (see “Data availability”). Of the MAGs retained in HUM-V, 36% were obtained from deeply sequenced, assembled, and binned samples, 27% were from coassemblies performed across samples, and the remaining 37% came from single assemblies of shallow sequences. The ability to recover additional MAGs relative to our group’s prior effort, which used only shallow sequencing (23), demonstrates how sequencing depth and integration of coassembly methods enhanced our ability to sample microbial HZ communities, corroborating findings from other studies with similar methods (26).
FIG 1.
Overview of hyporheic zone sampling and the microbial MAGs included in the HUM-V database. (a) Samples were collected from two transects, each with three sediment cores. Each core was subsectioned into 10-cm depth segments spanning 0 to 60 cm. This experiment included 33 samples total because a subsample (50 to 60 cm) was not obtained from core N1 and the top 3 subsamples from core N2 were combined (0 to 30 cm). (b) Schematic view of the data types available for each of the depth samples within a core. Black-filled circles indicate that there was a sample for that data type; open circles denote missing analyses. (c) Deeper metagenomic sequencing and NMR were performed on the remaining sediment samples, explaining why there are only 10 and 17 samples, respectively. The total number of samples per method is reported, with additional information provided in Table S1 in the supplemental material.
Metadata. Metagenome read counts, accession numbers, biosamples, geochemistry data, NMR metabolite data, and FTICR-MS data. Download Table S1, XLSX file, 4.0 MB (4.1MB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
MAGs. Completion information, accession numbers, annotations, read mapping information, metabolism summary from DRAM, correlations to geochemistry. Download Table S2, XLSX file, 1.4 MB (1.4MB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
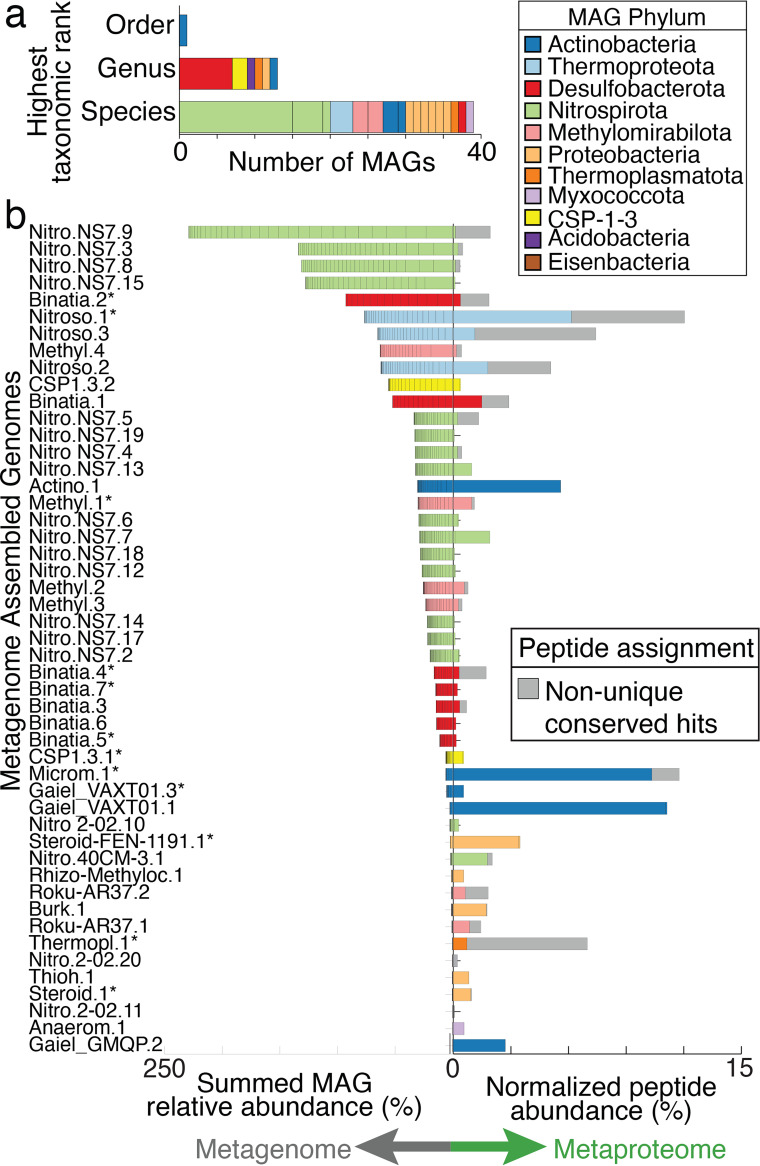
Given the few metagenomic studies in HZ sediments, it was not surprising that HUM-V contained the first MAG representatives of highly prevalent microorganisms (Fig. 2a and b; Table S2). Taxonomic assignment of the 55 unique HUM-V MAGs revealed that they spanned two archaeal and nine bacterial phyla and that most MAGs (n = 35) belonged to a subset of three bacterial phyla (Desulfobacterota, Nitrospirota, and Proteobacteria). To our knowledge, the eight Desulfobacterota (class Binatia) and seven Proteobacteria (orders Rhizobiales, Burkholderiales, Steroidobacterales, Thiohalobacterales, and Woeseiales) MAGs identified here represent the first HZ MAGs sampled from these commonly reported lineages. For the Nitrospirota, a prior study reported 21 MAGs that we dereplicated into 12 unique MAGs (99% average nucleotide identity [ANI]) (21), a sampling we further expanded by an additional 20 MAGs. The Nitrospirota MAGs sampled here spanned three genera that to our knowledge have not been previously sampled from rivers (Nitrospiraceae 2-02-FULL-62-14, 40CM-3-62-11, and NS7). Moreover, HUM-V contains one MAG of the Actinobacteriota that may represent a new order, as well as six new genera from Acidobacteriota, Actinobacteriota, CSP1-3, Desulfobacterota, Proteobacteria, and Thermoplasmatota (Fig. 2a and b). Further highlighting the genomic novelty of this ecosystem, HUM-V contains MAGs from entirely uncultivated members of different phyla (9 MAGs from CSP1-3 and Eisenbacteria) and classes (10 MAGs from Binatia and MOR-1). Ultimately, HUM-V is a public MAG resource that can be leveraged to enable taxonomic analyses and metabolic reconstruction of microbial metabolisms in HZ sediments.
FIG 2.
A coupled metagenome and metaproteome approach revealed active members in the hyporheic zone microbiome. (a) The taxonomic novelty of the dereplicated MAGs is colored by phylum and stacked according to the first empty position within the taxonomic string provided by the Genome Taxonomy Database GTDB-Tk (Table S2). Each color represents a MAG phylum according to the MAG phylum legend, a coloring scheme that is maintained throughout this article. (b) The summed genomic relative abundance across all samples (left side) and the normalized mean proteomic relative abundance (right side) for dereplicated MAGs (55 total, 49 shown), with bars colored by phylum. MAGs that contain a partial or complete 16S rRNA sequence are denoted with an asterisk. Nonunique specialized peptide assignment is defined in Materials and Methods and is indicated by gray bars.
HUM-V recruits metaproteomes offering new insights into HZ microbiomes.
Leveraging paired metaproteomes collected with the metagenomes allowed us to assign gene expression to each MAG in HUM-V (Table S3). These MAGs recruited 13,102 total peptides to 1,313 proteins. Because our genome analyses revealed that there were closely related strains (Table S2), we analyzed the proteomic data using two approaches. First, we considered the “unique” peptides that were assigned only to proteins from a single MAG. These represented 67% of the genes expressed in our proteome. Next, we considered proteins that recruited “nonunique but conserved” peptides, which we defined as those assigned to proteins that (i) have identical functional annotation and (ii) are from more than one MAG within the same genus. These proteins are shown in gray in Fig. 2b, and although they accounted for a smaller fraction of the genes expressed (14%), this method prevented us from excluding data due to strain overlap in our database.
Metaproteomes. MAG unique peptides, MAG unique peptide NSAF relative abundance, MAG NSAF relative abundance, vMAG unique peptides, MAG expressed and encoded metabolism summary, Binatia MAG metabolism information, assembly-mapped metaproteomics information. Download Table S3, XLSX file, 1.3 MB (1.3MB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
In microbiome studies, dominance is often used as a proxy for microbial activity. Here, we evaluated this assumption by using our paired metagenome and metaproteome data. When comparing the MAG relative abundances to protein expression patterns, we observed that the most abundant MAGs were not necessarily those that were most actively expressing proteins at the time of sampling. The most abundant MAGs included members of the Binatia, Nitrospiraceae NS7, and Nitrososphaeraceae TA-21 (formerly Thaumarchaeota) (Fig. 2b). However, only the dominant Nitrososphaeraceae MAGs had high recruitment of the uniquely assigned proteome. On the other hand, some low-abundance members (e.g., Actinobacteriota) accounted for a sizeable fraction (30%) of the uniquely assigned proteome.
Leveraging these metagenomic and metaproteomic data sets, we first examined metabolic traits that were conserved across nearly all HUM-V MAGs. Notably, all but one (CSP1_3_1) of the MAGs recovered from this site encoded the genomic capacity for aerobic respiration. We defined this capability by the recovery of genes indicating a complete electron transport chain and some form of terminal oxidase within each MAG. Consistent with these genomic data, resazurin reduction assays indicated that the bulk of sediments were oxygenated and could likely support aerobic microbial respiration (Fig. S1a) (27). However, while proteomic evidence for aerobic respiration (cytochrome c oxidase aa3) was detected in nearly 40% of samples, it could be confidently assigned only to the Nitrososphaeraceae. This is likely due to the highly conserved nature of this gene, as well as the limitations of detecting membrane, heme-containing cytochromes with metaproteomic data (28). As such, we consider it likely that this metabolism was more active than was captured in the metaproteomic data.
Subset of geochemical measurements used to contextualize the multi-omics data, including the resazurin reduction assay (a), percent carbon (b), percent nitrogen (c), C:N ratio (d), and ammonium (e). Box and whisker plots (n = 33 samples) with points colored by sample transect and each box representing the sample depths spanned (0 to 60 cm). No significant differences were observed by site and depth, except for ammonium (P = 0.026), which was significantly different by bulk depths (i.e., surface = 0 to 30 cm, and deep = 30 to 60 cm). All geochemical measurements are included in Table S1. Download FIG S1, PDF file, 0.1 MB (145.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
We performed ordination analyses of our MAG-resolved metaproteomic recruitment and revealed that the recruited gene expression in each sample did not cluster significantly by sediment depth or transect position (Fig. S2a and b). These MAG-resolved results agree with those previously published (23) using an unbinned metaproteome approach. That is, neither study observed any structuring at the transect level based on any microbial data type (i.e., metagenomics or metaproteomics). Consistent with this, over 90% of the measured gene expression was shared across both transects (Fig. S2c). In contrast, significant differences at the transect level were observed for metabolite concentrations and nonbiotic data like molecular weight and carbon types, as previously reported (23). As such, when considering explanations for this lack of microbiological spatial structuring, it is possible that (i) the microbial gene expression heterogeneity in these samples occurred over a finer spatial resolution (pore or biofilm scale, <10 cm) or a larger one (>60 cm) than those sampled here and thus were not captured in our analyses and/or (ii) that while the chemical data show changes across transects, these changes do not differentially shape the metabolic processes of the microorganisms, as the same substrate types are still mainly present in both regions.
NMDS and Venn diagram of differences in transect (N/S) or depth (0 to 60 cm) for MAG proteome. (a) NMDS (n = 29) of north (green) and south (orange) transects of recruited proteome peptides for MAGs. Analysis of similarity (ANOSIM) is reported. (b) NMDS of sediment core depth (0 to 60 cm) of recruited proteome peptides for MAGs. ANOSIM is reported. (c) Euler diagram showing the number of total proteins recruiting peptides in each transect, where the overlap represents the proportion of these proteins recruiting peptides in both transects. Only proteins recruiting two or more total peptides were included (n = 898) to allow for recruitment of at least one peptide for a given protein to both transects or both depths and to reduce false positives. Proteome abundances for all MAGs can be found in Table S3. Download FIG S2, PDF file, 0.3 MB (317.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
An inventory of processes contributing to microbial carbon dioxide production and consumption.
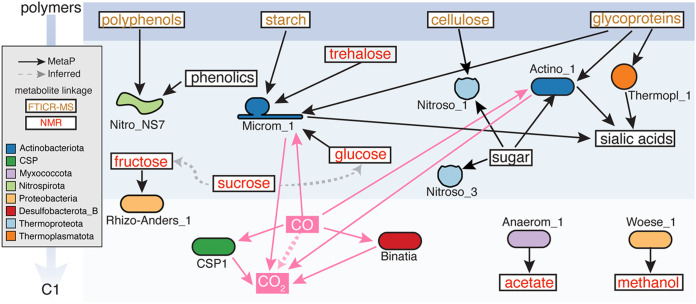
To uncover the microbial food web contributing to organic carbon decomposition in these HZ sediments, we reconstructed a carbon degradation network using coordinated genome potential, expression, and carbon metabolite data. Based on linkages to specific substrate classes, MAGs were assigned to the following trophic levels in carbon decomposition: (i) plant polymers, (ii) smaller organic compounds (e.g., sugars, alcohols, and fatty acids), and (iii) single-carbon compounds (carbon monoxide, carbon dioxide, methane) (Fig. 3).
FIG 3.
Integrated metaproteomics and metabolomics suggest carbon transformations expressed by bacterial and archaeal lineages in river sediments. Background blue shading spans polymers (top) to single-carbon compounds (C1; bottom) (arrow on left). Detected metabolites are shown in boxes, with NMR-detected compounds indicated in red type, polymers from FTICR-MS in orange type, and undetected metabolites in black type. MAG-resolved metaproteome information is indicated by solid arrows, with MAG icons colored by phylum. Pink arrows indicate processes that are expressed and could contribute to CO2 production, while black arrows indicate other carbon-transforming genes expressed in the proteome. Dotted arrows represent transformations supported by only one method: (i) gray dotted arrow indicates metabolites and downstream products (but no proteome support) and (ii) pink dotted arrow indicates metaproteomic evidence (but no geochemistry or metabolite support).
It is well recognized that heterotrophic oxidation of organic carbon derived in HZ sediments largely contributes to river respiration (2). Despite generally low organic concentrations in our sediments (<10 mg/g), FTICR-MS analysis showed that lignin-like compounds were the most abundant biochemical class detected in all samples regardless of transect or depth, suggesting that plant litter was a likely source of organic carbon (Fig. S3a). In support of this, 38% of the HUM-V MAGs encoded proteins for degradation of phenolic/aromatic monomers, while 11% could degrade the larger, more recalcitrant polyphenolic polymers. In fact, our analyses revealed that seven unique MAGs constituting a new genus within the uncultivated Binatia encoded novel pathways for the decomposition of aromatic compounds from plant biomass (phenylpropionic acid, phenylacetic acid, salicylic acid) and xenobiotics (phthalic acid) (Tables S2 and S3).
Percentage of identified formula classes by FTICR-MS and prevalence of NMR metabolites across samples. (a) The percentage of identified formula classes identified in FTICR-MS across samples is not statistically structured by depth or site. Bar plots show the average percentage of identified formulas for each class calculated as the number of formulas identified per class divided by the total number of formulas identified per sample, with error bars representing 1 standard deviation (n = 33). Classes were determined using criteria reported previously (E. B. Graham, A. R. Crump, D. W. Kennedy, E. Arntzen, et al., Sci Total Environ 642:742–753, 2018, https://doi.org/10.1016/j.scitotenv.2018.05.256). Individual data points are plotted for each sample, with point size increasing with depth. Peaks were classified as described in Materials and Methods. Raw, full data are provided in Table S1. (b) Bar graphs showing specific NMR metabolites and ammonium with the percentage of total samples in which they were found. Shading denotes the different categories of detected compounds. For detailed information on how these were collected, see Materials and Methods. Only metabolites detected in more than four samples are shown, with the complete NMR data provided in Table S1. Download FIG S3, PDF file, 0.5 MB (530.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Gene expression of carbohydrate-active enzymes (CAZymes) also supported the degradation of plant polymers like starch and cellulose. We detected the expression of putative extracellular glucoamylase (GH15) and endoglucanase (GH5) from an Actinobacteriota (Microm_1) and Nitrososphaeraceae (Nitroso_2) MAG, respectively. Additionally, using our unbinned assembled fractions, we detected expression of three GH33 CAZymes which could further contribute to the oxidation of organic carbon and were likely assigned to unbinned Rokubacteria and an unknown Actinobacteria. The integration of our chemical and biological data revealed that heterotrophic metabolism in these sediments could in part be maintained by inputs of plant biomass. In support of carbon depolymerization, sugars like glucose and sucrose were detected by nuclear magnetic resonance (NMR) (Fig. S3b).
We next sought to identify microorganisms that could utilize these sugars and found that members expressed transporters for fructose (Rhizo-Anders_1), glucose (Microm_1), and general sugar uptake (Actino_1, Nitroso_2, Nitroso_3). In support of further decomposition, we detected organic acids (acetate, butyrate, lactate, pyruvate, propionate) and alcohols (ethanol, methanol, isopropanol) by NMR (Fig. S3b). Similarly, proteomic data supported interconversions of these smaller carbon molecules, with the Myxococcota (Anaerom_1) expressing genes for aerobic acetate respiration and the archaeal Woeseia (Woese_1) respiring methanol. In summary, the chemical scaffolding and overlaid gene expression patterns support an active heterotrophic metabolic network in these HZ microorganisms, likely driven by plant biomass decomposition.
In addition to heterotrophy, our proteomic data revealed that autotrophy was also active in these sediments. Dehydrogenase genes for the aerobic oxidation of carbon monoxide (CO) were among the most prevalent across these sediments. This metabolism was expressed by phylogenetically distinct lineages, including members of uncultivated lineages Binatia (Binatia_2) and CSP1-3 (CSP1_3_1), as well as members of Actinobacteriota (Actino_1, Microm_1), Methylomirabilota (Roku_AR37_2), and Proteobacteria (Burk_1, Thioh_1). The wide range of bacteria and archaea that contained CO dehydrogenase genes, combined with gene expression data, suggests that carbon monoxide oxidation may be an important metabolism for persistence in HZ sediments.
Given that these sediments have relatively low total carbon concentrations (Fig. S1b), we consider it possible that carbon monoxide may act as a supplemental microbial energy and/or carbon source. Based on genomic content, we cautiously infer that members of Actinobacteriota (Microm_1), Binatia, and CSP1-3 may be capable of carboxydotrophy (i.e., using carbon monoxide as sole energy and carbon source), while the Actinobacteriota (Actino_1) is a likely carboxydovore (i.e., oxidizes carbon monoxide, while requiring organic carbon). While this metabolism is poorly resolved environmentally, recent efforts have shown that it is induced by organic carbon starvation to mediate aerobic respiration, thereby enhancing survival in oligotrophic conditions (29). Here, we add river sediments to the list of oxygenated environments (e.g., ocean and soils) where this metabolism may act as a sink or regulate the emission of this indirect greenhouse gas (GHG) (30, 31).
Since proteomics indicated that heterotrophy and carbon monoxide oxidation could generate carbon dioxide, we next tracked microorganisms in HUM-V that could fix this compound, sequestering its release. Analyses revealed that four pathways for carbon fixation were encoded by 75% of HUM-V MAGs, including the (i) Calvin-Benson-Bassham cycle, (ii) reductive tricarboxylic acid (TCA) cycle, (iii) 3-hydroxypropionate/4-hydroxybutyrate (3HP/4HB) cycle, and (iv) 3-hydroxypropionate bicycle. The two nitrifying lineages were inferred chemolithoautotrophs, with Nitrososphaeraceae encoding 3HP/4HB and the Nitrospiraceae encoding the reductive TCA cycle. Additionally, phylogenetically diverse lineages, Acidobacteriota, Binatia, CSP1_3, Proteobacteria, and Woeseiaceae, encoded redundant fixation pathways. Although expression was not detected for these metabolisms in our MAGs or unbinned data, we hypothesize that these are likely relevant given the distribution of this metabolism across the microbial community.
Our genomic and proteomic data revealed the prevalence and activity of single-carbon metabolism in these sediments. Carbon monoxide and dioxide are likely the primary substrates, as HUM-V had only minimal evidence for methanol oxidation (Woeseia), no methanotrophs, and no methanogens. Along these lines, the unbinned metaproteomics approach also did not detect evidence for methanotrophy or methanogenesis in these samples. Together, our findings hint at the importance of carbon monoxide and carbon dioxide in sustaining microbial metabolism in these oxygenated but low, or fluctuating, carbon environments. Further work is needed to understand physiochemical factors controlling carbon monoxide oxidation and carbon dioxide fixation activity and the balance between production (via heterotrophy and carbon monoxide oxidation) and consumption (fixation) on overall river sediment carbon dioxide emissions.
Ammonium exchange can support coordinated nitrogen mineralization and nitrification pathways.
The ratio of total carbon (C) (Fig. S1b) to total nitrogen (N) (Fig. S1c) (e.g., C/N) is a geochemical proxy used to denote the possible microbial metabolisms that can be supported in a habitat (32, 33). Our HZ sediments had C/N ratios with a mean of 6.5 ± 1.1 (maximum, 8.4) (Fig. S1d). Geochemical theory posits that sediments with low C/N ratios (<15) support organic mineralization that yields sufficient ammonium such that heterotrophic bacteria are not N limited and nitrifying bacteria are able to compete successfully for ammonium, enabling nitrification (33–35). Based on our sediment C/N ratios, we hypothesized that organic nitrogen mineralization and nitrification co-occured in these sediments. Here, we profiled the microbial substrates (organic nitrogen metabolites, ammonium) and expressed pathways (mineralization and nitrification) to provide biological validation of this established geochemical theory.
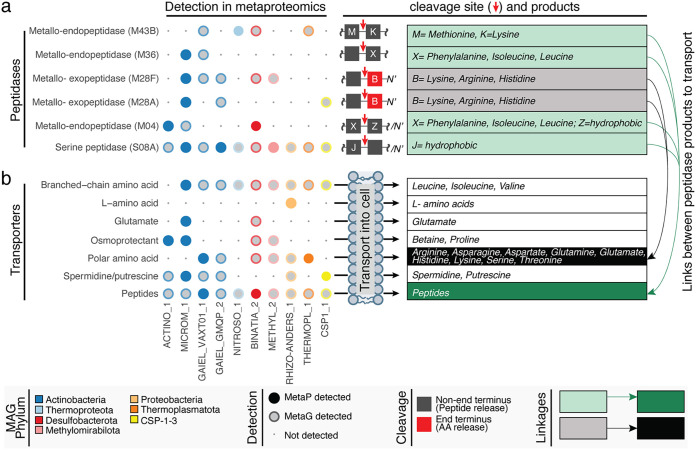
To examine the microbial contributions to organic nitrogen mineralization, we examined metaproteomic data for peptidases, i.e. genes that mineralize organic nitrogen into amino acids, and free ammonium. In support of active microbial N mineralization, FTICR-MS revealed that protein-like and amino sugar-like organic nitrogen compounds were correlated with high microbial activity (23), while here we show that hydrophobic, polar, and hydrophilic amino acids were prevalent in the 1H-NMR characterized metabolites (Fig. S3b). The expression of peptidases in situ, combined with our genomic resolution of their hosts, provided a new opportunity to interrogate the mechanisms underpinning nitrogen mineralization. We first noted which microorganisms expressed extracellular peptidases (inferred from references 36 to 38), as these enzymes could shape the external organic nitrogen pools in the sediment. We categorized these expressed peptidases as either releasing free amino acids (end terminus-cleaving families, e.g., M28) or releasing peptides (endocleaving families, e.g., S08A, M43B, M36, and MO4) (Fig. 4a). Members of the Actinobacteriota, Binatia, Methylomirabilota, and Thermoproteota were found to express extracellular peptidases and, as such, are likely candidates that contribute to sediment N mineralization.
FIG 4.
Organic nitrogen mineralization and cellular transport are expressed processes in river sediments. Bubble plots indicate the genes that were uniquely assigned to specific MAGs, including those coding for (i) extracellular peptidases and (ii) cellular transporters for organic nitrogen, with expression in metaproteomics (colored circles) and potential (gray circle with colored outline) noted. Unique peptides detected in at least three samples are represented by bubbles and colored by phylum. The table on the right shows putative amino acids cleaved or transported by the respective peptidases (green or gray) or transporters (dark green/black). The shading denotes peptides that are cleaved into amino acids that could be transported, providing linkages between extracellular organic nitrogen transformation and the transport of nitrogen into the cell. No shading (white boxes) indicates an organic nitrogen transporter that recruited peptides but could not be linked to outputs of specific peptidases.
We then profiled the expressed amino acid transporters in our MAGs, i.e., genes for the cellular uptake of these smaller organic nitrogen compounds (e.g., branched-chain amino acids, glutamate, amines, and peptides were examined) (Fig. 4b). At the time of sampling, some members expressed peptidases for cleavage of organic N to liberate smaller peptides, yet we could not detect expressed genes for the transport of these produced compounds. Other taxa like Actinobacteriota and Binatia expressed genes for external peptidases and for transporting the organic N products into the cell. Alternatively, members of the CSP1-3, Proteobacteria, and Thermoplasmatota expressed genes for assimilating peptidase products, but did not express genes that would contribute to their production.
While it is tempting to speculate that at the time of sampling some members of the community may have been operating as producers, and thus extending this concept to organic nitrogen processing as others have noted for carbon decomposition (39–41), we note that the metabolic potential of these microorganisms is more diverse than their expressed patterns, as shown in Fig. 4. Thus, we could have missed the expression of these genes, as they may be below detection in the proteome data or they may be missed entirely because our MAGs are draft genomes (inferred completion mean is 82%, with a maximum of 99%). As such, the absence of data here should be interpreted cautiously, and future research using model cultivated strains analogous to the work of Pollak et al. for polysaccharides as a public good (39) is necessary to classify producers and cheaters, and the conditions under which they operate, in organic nitrogen processing.
Finally, we examined the proteomes for evidence that nitrification co-occured with organic nitrogen mineralization. Supporting this possibility, the substrate ammonium (NH4+) was detected in all 33 sediment samples (Fig. S1e and S3b). We did not detect genomic evidence or expression for comammox or anammox metabolisms in HUM-V MAGs and did not identify these metabolisms in our metaproteomes mapped to the unbinned, assembled data. This suggests that aerobic nitrification by different organisms drives nitrification in our metabolic network. Proteomics confirmed that aerobic ammonium oxidation to nitrite was performed by archaeal Nitrososphaeraceae. In fact, ammonia monooxygenase (amo) subunits were within the top 5% most highly expressed functional proteins in this data set. The next step in nitrification, nitrite oxidation to nitrate, was inferred from nitrite oxidoreductase (nxr), which was expressed by members of Nitrospiraceae. Demonstrating that new lineages first discovered in HUM-V could shape in situ biogeochemistry, we confirmed that five MAGs from two new species of Nitrospiraceae expressed nitrification genes (Nitro_40CM-3_1, Nitro_NS7_3, Nitro_NS7_4, Nitro_NS7_5, and Nitro_NS7_14).
The proteome-supported archaeal-bacterial nitrifying mutualism outlined here appears well adapted to the low-nutrient conditions present in many HZ sediments, warranting future research on the universal variables that constrain nitrification rates (i.e., ammonium availability, dissolved oxygen, pH) and their role in driving nitrogen fluxes from these systems (42). In conclusion, our microbial data support the idea that nitrification is concomitant with mineralization in these samples, providing biological evidence to substantiate inferences made from the C/N ratio of these sediments.
Metabolic contributions from less characterized taxa actively shape nitrogen cycling.
Our proteomics suggests that aerobic nitrification could complement allochthonous nitrate from groundwater discharges, contributing to measured nitrate concentrations in excess of 20 mg/L (2, 43). Based on this, we next sought to summarize the metabolic potential and expressed gene content related to the use of nitrate or oxidized nitrogen species in our metagenome data set.
HUM-V MAGs with the genomic capacity for denitrification were phylogenetically diverse (Table S3). Nitrate reductases (narG or napX) were encoded in an additional 11 MAGs from the Actinobacteriota, Binatia, Myxococcota, and Proteobacteria, of which 2 had the subsequent capacity to reduce nitrite to nitric oxide gas. Notably, the conversion of nitric oxide to nitrous oxide, a potent greenhouse gas, was encoded by two Gammaproteobacteria (Steroid-FEN-1191_1 and Steroid_1) and a member of the Myxococcota (Anaerom_1). We also detected that the dissimilatory nitrite reduction to ammonium (DNRA) was encoded by multiple members of the Binatia and Nitrospiraceae (Table S3).
On the basis of the gene expression data, three members were assumed to be actively contributing to nitrogen reduction at the time these samples were collected. The only narG protein detected in our proteome for our MAGs was uniquely assigned to the Binatia, while proteins for nitrite reduction were assigned to the denitrifying Burkholderia and nitrifying Nitrososphaeraceae (Table S3). We did detect expression of narG and nirk within our unbinned data, but they were taxonomically inferred to be from Nitrospiraceae and Nitrososphaeraceae represented in HUM-V. We did not detect the expression of DNRA in our HUM-V MAGs or the unbinned fractions, but due to the high number of genomes encoding this functionality, we hypothesize that it could play important roles in ammonia generation by the microbes in these sediments. Additionally, while nitrous oxide production genes were not expressed in either the binned or unbinned fractions, we did detect nos gene expression for reducing nitrous oxide to nitrogen gas by the nondenitrifying Desulfobacterota_D (Desulf_UBA2774_1), formerly Dadabacteria (44). Our phylogenetic analysis (see “Data availability”) revealed that this sequence was a “clade II” nosZ sequence, an atypical variant adapted for lower or atmospheric nitrous oxide concentrations that is often encoded in nondenitrifying lineages (45).
Our metaproteome data add to the emerging interest on the use of untargeted approaches to identify the nitrogen-transforming genes that are expressed in hyporheic sediments. To our knowledge, we provide the first gene expression evidence for two lineages lacking cultured representatives, the Binatia and a member of the Desulfobacterota, in hyporheic zone denitrification. We also provide expression data supporting the notion that clade II nosZ gene expression could act as a nitrous oxide sink (without contributing to its production) (46, 47). Importantly, the activity of this enzyme would have been missed using traditional nosZ primers, indicating the value of our untargeted, expression-based approach (48). In summary, our gene expression data pinpoint new metabolic contributions from less characterized taxa that actively shape nitrogen cycling in the hyporheic zone.
Viral influence on sediment carbon and nitrogen cycling.
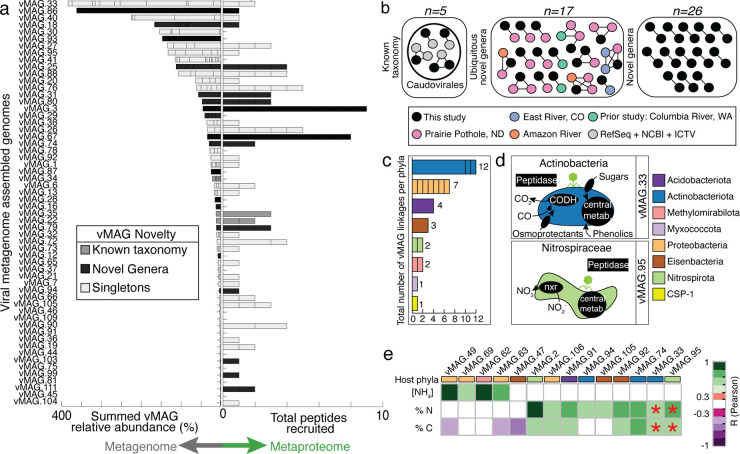
We reconstructed 2,482 viral MAGs (vMAGs) that dereplicated into 111 viral populations (>10 kb) in HUM-V (Fig. 5a; Table S4), making this one of only a handful of genome-resolved studies that include viruses derived from rivers (16, 49, 50). To our knowledge, this is the first study to provide a coordinated analysis of microbial and viral community MAGs, and given their sparse sampling, only 5 of the 111 HUM-V vMAGs had taxonomic assignments established from standard reference databases. To better understand whether the remaining vMAGs had been previously detected in other virally sampled freshwater systems, we compared the protein content of the vMAGs in our system to that of an additional 1,861 vMAGs we reconstructed de novo or obtained from public metagenomes from North and South America (Fig. 5b; Table S4). Of the 106 nontaxonomically assigned viruses, 15% (n = 17) clustered with these freshwater-derived viral genomic sequences, indicating possible cosmopolitan viruses, and another 23% (n = 26) clustered only with vMAGs recovered in this data set, indicating multiple samplings of the same virus across different sites and depths. The remaining 57% (n = 63) of the vMAGs were singletons, meaning that they were sampled only from these sediments once. Combined, these results hint at the possible biogeographically diverse as well as endemic viral lineages, warranting further exploration in river sediments.
FIG 5.
Viruses in HUM-V are taxonomically novel, express genes, and may play roles in microbial host metabolism and river geochemistry. (a) The genomic relative abundance (left side) and total peptides recruited (right side) for each vMAG population. Bars are colored by clustering of vMAGs from this study with (i) viruses of known taxonomy (dark gray), (ii) novel genera (black), and (iii) no clustering with members from any database (light gray, singletons), as defined in Materials and Methods. (b) Similarity network of the few vMAGs from our study (denoted in black) that clustered to viruses with known taxonomy (gray) or to viruses in other freshwater, publicly available data sets (denoted in pink, purple, orange, and turquoise) and the remaining clusters of viruses that were novel (e.g., did not cluster with prior vMAGs). The full network table including singletons is shown in Table S4. (c) The total number of vMAGs (n = 32) with putative host linkages. Each bar represents a phylum, and lines within bars indicate the number of linkages for specific MAGs within that phylum. (d) Subset of cartoons of microbial metabolisms for two representative MAGs with putative viral linkages, with the genes shown in black text boxes denoting processes detected in proteomics. (e) Correlations between a subset of vMAGs, with rectangle colors denoting the putative host taxonomy. Correlations between these vMAGs and ecosystem geochemistry (concentration of NH4 [μg/g], %N, and %C) are reported, with significant correlation coefficients denoted by purple-green shading according to the legend. Red asterisks indicate that the vMAG relative abundance predicted a key environmental variable by sparse partial least-squares (sPLS) regression; note that these two vMAGs are shown in panel d.
vMAGs. MIUViG information, accession numbers, annotations, read mapping information, vConTACT2 results, downloaded MetaG information for vConTACT2 analyses, VirHostMatcher2 results, correlations to geochemistry, and sPLS results. Download Table S4, XLSX file, 1.2 MB (1.2MB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
We then assessed peptide recruitment to the viral portion of HUM-V (Fig. 5a; Table S3). For viruses and microbes alike, the most abundant vMAGs did not have the highest gene expression. While viral gene expression was not structured by edaphic or spatial factors (Fig. S4a and b), it was strongly coordinated to the microbial abundance patterns (Fig. S4c). Like our microbial MAG peptide recruitment, 66% of the vMAGs uniquely recruited peptides. This exceeded prior viral metaproteome recruitment from other environmental systems (e.g., wastewater, saliva, and rumen, with ranges from 0.4 to 15%) (51–53). From this, we infer that a relatively large portion of the viral community was active at the time of sampling.
vMAG recruited peptides combined with virus-host linkages indicate possible impacts on river sediment carbon and nitrogen cycling. (a and b) NMDS (n = 29) of north (green) and south (orange) transects (a) or sediment core depth (0 to 60 cm) (b) of vMAG recruited peptides show no significant clustering, with stress and ANOSIM reported. (c) Procrustes ordination of MAG and vMAG NMDS ordinations with relative abundance data across the 28 shallow sequenced samples that recruited reads to our MAG and vMAG communities. (d) Gray dotted circles represent the number of significant vMAGs that putatively infect each MAG, and MAG cartoons are colored based on phylum. (e) Assignment of vMAGs to their putatively infected host MAGs. Colors match host MAG phylum assignment. (f) Different AMGs identified in vMAGs with putative hosts, with the predicted AMG substrate type denoted by colored boxes on the y axis. (g) Inferred cleavage activities for glycoside hydrolase (GH74) and a pectin lyase (PL1) AMG are shown. (h) A conceptual model for how viral AMGs could impact the metabolism of their hosts is shown. One putative bacterial host, Steroidobacteraceae, has a linkage to a vMAG (vMAG.63) that encodes a PL1. Integration of this vMAG into the host MAG would provide the capacity to cleave the pectin backbone (PL1), a functionality not encoded in the host genome. This would create a new resource for the host which does encode genes for the oligo cleavage but not for backbone cleavage of pectin (via GH4 and GH2). This cleavage would result in galactose monomers that can further support host cell-encoded metabolism. Download FIG S4, PDF file, 0.5 MB (547.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The proteomic recruitment of viruses sampled in HUM-V hinted at the possibility that viruses could structure the microbial biogeochemistry through predation. In silico analysis assigned a putative host to 29% of the 111 vMAGs. Viruses were linked to 18 microbial MAGs that belong to bacterial members in Acidobacteriota, Actinobacteriota, CSP1-3, Eisenbacteria, Methylomirabilota, Myxococcota, Nitrospirota, and Proteobacteria (Fig. 5c; Fig. S4d and e). Analysis of the metaproteomes for these putative phage-impacted MAGs revealed that these hosts expressed genes for carbon monoxide oxidation (Actinobacteriota), carbon fixation (CSP1-3), nitrogen mineralization (Acidobacteriota and Methylomirabilota), methanol respiration (Myxococcota), nitrification (Nitrospirota), and ammonia oxidation (Proteobacteria) (Fig. 5d). Thus, viral predation in HZs could impact carbon and nitrogen biogeochemistry and may explain some of the strain and functional redundancy we observe in the microbial communities of these sediments.
We next inventoried HUM-V vMAGs for auxiliary metabolic genes (AMGs) with the potential to augment biogeochemistry. We detected 14 AMGs which we confirmed were not bacterial in origin and had virus-like genes on both flanks (54). These putative AMGs had the potential to augment carbon (CAZymes), sulfur (sulfate adenylyltransferase), and nitrogen (amidase to cleave ammonium) metabolism (Fig. S4f). One of our vMAGs that was putatively linked to a Steroidobacteraceae (Steroid_1) contained a pectin lyase gene (PL1). This viral PL1 could enable its host to cleave the backbone of pectin, generating pectin oligosaccharides that could be used via two host-encoded glycoside hydrolases (GH4 and GH2), ultimately freeing galactose for energy metabolism (Fig. S4g and h). While theoretical, we include this as an example to illustrate how virally encoded genes could expand the substrate ranges for their hosts and alter biogeochemical cycling in river sediments.
In support of their importance for modulating microbial activity and sediment biogeochemistry, we noted that the use of vMAG abundance patterns in addition to MAG abundance patterns improved our predictions of river sediment carbon and nitrogen concentrations (Fig. S5). In summary, these results indicate that viral predation and AMGs may contribute to river sediment biogeochemistry, either through top-down or bottom-up controls on the microbial community.
sPLS regressions show vMAG relative abundance-enhanced carbon and nitrogen predictions relative to those of MAG relative abundance alone. The predicted versus measured values for percent carbon (c_per) and percent nitrogen (n_per) with vMAGs only (a), MAGs only (b), and vMAGs and MAGs combined (c) are shown. Values corresponding to t, degrees of freedom, P values, confidence intervals, and correlation are also shown for each plot. Download FIG S5, PDF file, 0.4 MB (391KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
A multi-omics-informed roadmap of carbon and nitrogen cycling in HZ sediments.
Despite the importance of the HZ and its relative accessibility in terms of sampling locations, HZ microbial and viral communities are surprisingly undersampled in a genomic context. Previous studies pertaining to this ecosystem are not genome resolved and used 16S rRNA amplicons or unbinned metagenomes (8–11, 19, 20, 22), thus limiting the predictive and explanative power of the study and oftentimes not discriminating between metabolically active and inactive organisms. Furthermore, the few studies which are genome resolved (21, 22) often focus on specific lineages or single processes and not the entire microbial community, missing the complex interplays between the carbon and nitrogen cycles.
Here we created HUM-V, a MAG-resolved database, to expand on prior non-genome-resolved analyses done in a previous publication by our team (23). Our MAG-resolved proteomic data further provided some of the first activity indicators for members of hyporheic microbiomes. As an example, we focus on new insights gleaned from the seven recovered Binatia MAGs (one genome included a complete 16S rRNA gene), which are uncultured and have been described only in terms of metabolic potential by one previous publication (55). Proteomics demonstrated that these bacteria (i) aerobically oxidized carbon monoxide, (ii) mineralized organic nitrogen, and (iii) denitrified, contributing to carbon and nitrogen cycling in these sediments (Fig. S6a). Using 16S rRNA recovered from these MAGs, we show that closely related strains are biogeographically widespread (Fig. S6b), and thus it is possible that the gene expression findings we illuminate here are more widespread across other habitats. These results illustrate the power of HUM-V in uncovering new roles for members of uncultivated, previously enigmatic microbial lineages in hyporheic zone biogeochemistry.
Uncultured Binatia are widely dispersed across ecosystems and express nitrogen and carbon cycling genes in situ. (a) Metabolic MAG cartoon for the major functions encoded by the Binatia MAGs. Dotted arrows indicate functions encoded in the metagenome, and solid arrows correspond to metaproteome-detected enzymes. BCAA, branched-chain amino acids. (b) Using the 16S rRNA gene (from Binatia_7), we inventoried the distribution of closely related species to our HUM-V MAGs (>97% similarity) in the Sequence Read Archive (SRA) samples, uncovering the ecological distribution of these microorganisms from soils, as well as a wide variety of terrestrial, terrestrial-aquatic, and marine samples, indicating that the expressed metabolisms in panel a are likely applicable to a wide range of ecosystems. Download FIG S6, PDF file, 0.1 MB (137KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
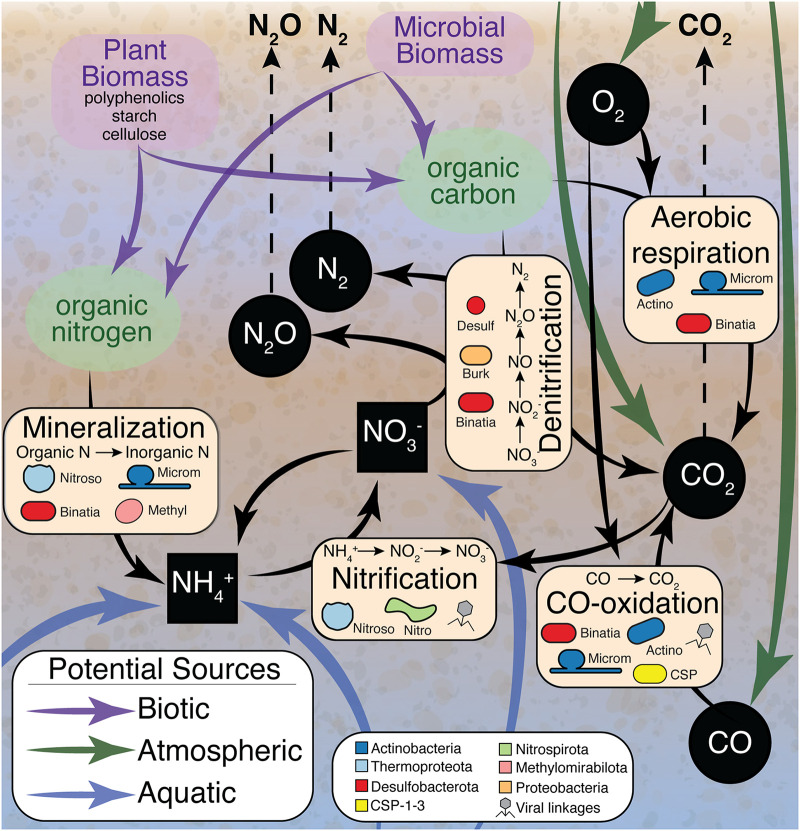
Empowered by our process-based metaproteomic analyses (Fig. 2 to 5), we present a conceptual model outlining microbial and viral contributions to carbon and nitrogen biogeochemistry in these sediments (Fig. 6). Together, our results demonstrated how these pathways could result in the formation and depletion of nutrients in shared resource pools. From gene expression data, we suggest a network of metabolisms that affected organic and inorganic carbon cycling and that is intertwined with nitrogen mineralization, nitrification, and denitrification pathways.
FIG 6.
Conceptual model summarizing the microbial and viral contributions to HZ carbon and nitrogen cycling identified in this study. Black arrows signify microbial transformations uncovered in our MAG-resolved metaproteomic data. Specific processes (e.g., mineralization, nitrification, CO oxidation, denitrification, and aerobic respiration) are highlighted in beige boxes, with microorganisms inferred to carry out these processes denoted by overlaid cell shapes colored by phylum. Possible biotic, atmospheric, and aquatic carbon and nitrogen sources are indicated by purple, green, and blue arrows, respectively. Inorganic carbon and nitrogen sources are shown by black squares (aqueous) and black circles (gaseous), with white text and dashed arrows indicating possible gasses that could be released to the atmosphere. Processes that could be impacted by viruses are marked with gray virus symbols.
In conclusion, while river carbon and nitrogen budgets are often quantified by direct measurements of inputs and the concentrations of inorganic and organic compounds exported from rivers, our findings put forth an integrated framework that advances the resolution of microbial roles in hyporheic carbon and nitrogen transformations. It yields insights that could inform research strategies to reduce existing predictive uncertainties in river corridor models and resolves some of the microbial contributions that were thought to occur but were poorly defined in river sediments (e.g., nitrogen mineralization). We also highlight previously enigmatic processes that could directly impact river GHG fluxes in unappreciated ways (e.g., carbon dioxide fixation, carbon monoxide oxidation, type II nitrous oxide reduction). Ultimately, we show that a MAG-resolved database allows us to track the consumption and production of carbon dioxide, ammonium, and inorganic nitrogen and helps us explain how these transformations might contribute to overall GHG fluxes.
MATERIALS AND METHODS
Sample collection, DNA isolation, and chemical characterization.
Samples were collected from the hyporheic zone of the Columbia River (46°22′15.80″N, 119°16′31.52″W) in March 2015 as previously described (23). Briefly, sediment profiles (0 to 60 cm) were collected along two transects separated by approximately 170 m (Fig. 1). At each transect, three sediment cores up to 60 cm in depth were collected at 5-m intervals perpendicular to the river flow. Liquid-N2-frozen sediment profiles were collected as detailed previously (56), with a pointed stainless-steel tube (152-cm length, 3.3-cm outside diameter, 2.4-cm inside diameter) driven into the riverbed and liquid N2 poured down the tube for ~15 min. Once the sediments were frozen, the tube and attached material were removed from the riverbed with a chain hoist suspended beneath a tripod. Profiles were placed over an aluminum foil-lined cooler containing dry ice. The material was then wrapped in the foil and transported on dry ice to storage at −80°C. In the lab, each core was then sectioned into 10-cm segments from 0- to 60-cm depths for downstream analyses, except for core “N2,” which had the first 3 depths (0 to 30 cm) subsampled together, and for core “N1” (50 to 60 cm), which was damaged and did not pass quality control. For processing, samples were transferred to an anaerobic glove bag with 95% N2 and 5% H2 (Coy Laboratory Products, Grass Lake, MI) and thawed on clean 2-mm stainless steel sieves prior to processing. Approximately 5 g was transferred to a 40-mL borosilicate glass vial and stored at −80°C for chemical analyses. An additional sample was taken for elemental analysis (organic carbon [OC], and nitrogen [N]), and the remaining material was divided into 20-g samples collected into 40-mL borosilicate glass vials and stored at −80°C.
DNA isolation was carried out as previously described (23). To release biomass from sediment particles, thawed samples were suspended in 20 mL of chilled phosphate-buffered saline (PBS)–0.1% Na pyrophosphate solution and vortexed for 1 min. The suspended fraction was decanted to a fresh tube and centrifuged for 15 min at 7,000 × g and 10°C. DNA was extracted from the resulting pellets using the MoBio PowerSoil kit in plate format (MoBio Laboratories, Inc., Carlsbad, CA) in accordance with the manufacturer's instructions, with the addition of a 2-h proteinase K incubation at 55°C prior to bead beating to facilitate cell lysis. DNA was then sent to the Joint Genome Institute (JGI; n = 33) for sequencing. The additional deep sequencing described here was performed at the Genomics Shared Resource facility at The Ohio State University (OSU; n = 10) using a Nextera XT library system. Libraries at both facilities were sequenced using an Illumina HiSeq 2500 platform. Table S1 in the supplemental material details all sequencing information, including accession numbers for metagenomes.
Chemical analyses included geochemical and metabolite data, for which geochemistry and Fourier transform ion cyclotron resonance mass spectrometry (FTICR-MS) methods were performed as previously described (23). Regarding FTICR-MS, sediments were extracted with three solvents with different polarities—water (H2O), methanol (CH3OH; abbreviated MeOH), and chloroform (CHCl3)—to sequentially extract a large diversity of organic compounds from samples, according to previous publications (57, 58). Water extractions were performed first, followed by extractions with MeOH and then CHCl3. Ultra-high-resolution mass spectrometry of the three different extracts from each sample was carried out using a 12-T Bruker SolariX FTICR-MS located at the Environmental Molecular Sciences Laboratory (EMSL) in Richland, WA.
The total nitrogen, sulfur, and carbon content was determined using an Elementar vario El cube (Elementar Co., Germany). NH4+ was extracted with KCl and measured with a Hach kit (Hach, Loveland, Co). Aerobic metabolism was inferred by the resazurin reduction assay, based on a method previously described (27). FTICR-MS compounds are reported as relative abundance values based on counts of C, H, and O. The relative abundance of a biochemical class is defined as follows: (number of formulas in a class per sample)/(total number of formulas per sample). This was done for the following H:C and O:C ranges: lipids (0 < O:C ≤ 0.3, 1.5 ≤ H:C ≤ 2.5), unsaturated hydrocarbons (0 ≤ O:C ≤ 0.125, 0.8 ≤ H:C < 2.5), proteins (0.3 < O:C ≤ 0.55, 1.5 ≤ H:C ≤ 2.3), amino sugars (0.55 < O:C ≤ 0.7, 1.5 ≤ H:C ≤ 2.2), lignin (0.125 < O:C ≤ 0.65, 0.8 ≤ H:C < 1.5), tannins (0.65 < O:C ≤ 1.1, 0.8 ≤ H:C < 1.5), and condensed hydrocarbons (0 ≤ 200 O:C ≤ 0.95, 0.2 ≤ H:C < 0.8) (57).
Additional metabolite data were obtained through 1H nuclear magnetic resonance (NMR) spectroscopy on water-extracted sediments. Thawed sediment samples were mixed with 200, 300, or 600 μL of Milli-Q water depending on the sediment mass (Table S1) and centrifuged to remove the sediment (58–60). Supernatant (180 μL) was then diluted by 10% (vol/vol) with 5 mM 2,2-dimethyl-2-silapentane-5-sulfonate-d6 (DSS-d6) as an internal standard. All NMR spectra were collected using a Varian Direct Drive 600-MHz NMR spectrometer equipped with a 5-mm triple-resonance salt-tolerant cold probe. Chemical shifts were referenced to the 1H or 13C methyl signal in DSS-d6 at 0 ppm. The one-dimensional (1D) 1H NMR spectra of all samples were processed, assigned, and analyzed using Chenomx NMR Suite 8.3 with quantification based on spectral intensities relative to the internal standard as described previously (61, 62). For the NMR, while obtaining concentrations is possible, many compounds were below the limit of quantitation (2 μM) but above the limit of detection (1 μM). To still derive meaning from these data, we reported NMR-identified compounds as present (detected) or absent (not >1 μM), with “relative abundance” reported as the number of formulas in a class per sample divided by the total number of formulas per sample. All geochemical and metabolite data can be found in Table S1.
Metagenome assembly and binning.
Raw reads were trimmed for length and quality using Sickle v1.33 (https://github.com/najoshi/sickle) and then subsequently assembled using IDBA-UD 1.1.0 (63) with an initial kmer of 40. Two of our samples did not assemble with IDBA-UD 1.1.0 at this kmer and were assembled with metaSPAdes 3.13.0 (64) using default parameters (Table S1). To further increase genomic recovery, for the 10 samples that had shallow and deep sequencing, metagenomic reads were coassembled using IDBA-UD 1.1.0 with an initial kmer of 40 (Table S1). All assemblies, including coassemblies, were then individually binned using MetaBAT2 v2.12.1 (65) with default parameters to obtain microbial MAGs.
For each bin, MAG completion was estimated based on the presence of core gene sets using Amphora2 (E value = 1e−3, which used RaxML [v8.2.9], HMMER [v3.3], and EMBOSS [v6.6.0.0]) and CheckM v1.1.2 (66, 67). To ensure that only quality MAGs were utilized for metabolic analyses, we discarded all MAGs that had completion of <70% and contamination of >10%. This equates to “high-quality” (HQ) bins and a more stringent “medium-quality” (MQ) bin cutoff than those used by the genome consortium standard (25). These 102 MAGs (6 HQ, 96 MQ) were then dereplicated using dRep (68) with default parameters to result in a final set of 55 MAGs (>99% ANI, which represents strain-level MAG distinctions) (Table S2). To further assess bin quality, we used the Distilled and Refined Annotation of MAGs (DRAM) v1.0 tool (54) to identify rRNAs and tRNAs to ensure they were taxonomically consistent with overall taxonomic assignments. To further curate our bins, we used bowtie2 (69) to calculate the coverage of each scaffold within a MAG and manually checked scaffolds that had 10% higher coverage than the mean, confirming consistency in taxonomic assignment and annotations of the scaffold in question with the overall bin. MAG quality and taxonomic information are reported in Table S2.
Metabolic and taxonomic analyses of MAGs.
Medium- and high-quality MAGs were taxonomically classified using the Genome Taxonomy Database (GTDB) Toolkit v1.3.0 using reference data r95 on September 2020 (70). Novel taxonomy was identified as the first taxonomic level with no designation using GTDB taxonomy. MAG scaffolds were annotated using the DRAM v1.0 tool, which uses the PFAM (v33.1), KEGG (v89.1), dbCAN (v9), MEROPS (v120), and VOGDB database for annotations (54) (https://github.com/WrightonLabCSU/DRAM). Phylogenetic analyses were performed on genes annotated as respiratory nitrate reductase (nar) and nitrite oxidoreductase (nxr) to resolve the novel Binatia role in nitrogen cycling. Specifically, sequences from reference 71 were downloaded and combined with nar and nxr amino acid sequences from dereplicated bins, aligned using MUSCLE (v3.8.31), and run through an in-house script for generating phylogenetic trees (https://github.com/WrightonLabCSU/columbia_river). Phylogenetic trees are provided in Zenodo at https://doi.org/10.5281/zenodo.6339808. For polyphenol and carbon polymer degradation, we used predicted secretion and functional annotations to characterize these metabolisms. To determine if the predicted genes encoded a secreted protein, pSortb (36) and SignalP (38) were used to predict location; if those methods did not detect a signal peptide, the amino acid sequence was queried using SecretomeP (with a SecP score of >0.5 [37] used as a threshold to report noncanonical secretion signals). Metabolic characterization for each MAG discussed in this paper is available in Table S3.
Viral analyses.
Metagenomic assemblies (n = 43) were screened for DNA viral sequences using VirSorter v1.0.3 with the ViromeDB database option (72), retaining viral contigs ranked 1, 2, 4, or 5, where categories 1 and 2 indicate high-confidence predicted lytic viruses and categories 4 and 5 indicate high-confidence prophage sequences from VirSorter output (72). Viral sequences were filtered based on size to retain those greater than or equal to 10 kb on the basis of current standards (73). Viral scaffolds were then clustered into vMAGs at 95% ANI across 85% of the shortest contig using ClusterGenomes 5.1 (https://github.com/simroux/ClusterGenomes) (73). After clustering, vMAGs were manually confirmed to be viral by looking at DRAM-v annotations and assessing the total virus-like genes relative to nonviral genes per scaffold. Using DRAM-v, vMAGs that were assigned the J-flag (which indicates vMAGs containing more than 18% of nonviral genes) were deemed suspicious, manually confirmed to contain no viral hallmark genes, and subsequently discarded (54). All vMAG information can be found in Table S4.
To determine taxonomic affiliation, vMAGs were clustered with viruses belonging to the viral reference taxonomy databases in NCBI Bacterial and Archaeal Viral RefSeq V85, and viruses from the International Committee on Taxonomy of Viruses (ICTV) by using the network-based protein classification software vConTACT2 v0.9.8 with default settings (74, 75). To determine the geographic distribution of viruses in freshwater ecosystems, we included viruses mined from publicly available freshwater metagenomes in the following vConTACT2 analyses: (i) East River, CO (PRJNA579838); (ii) a previous Columbia River, WA, study (PRJNA375338); (iii) Prairie Potholes, ND (PRJNA365086); and (iv) the Amazon River (PRJNA237344). The viral sequences that were identified from these systems and the genes used for vConTACT2 are deposited in Zenodo under https://doi.org/10.5281/zenodo.6310084 with more information about downloaded data sets provided in Table S4.
Viral contigs were annotated with DRAM-v (54). Genes that were identified by DRAM-v as being high-confidence possible auxiliary metabolic genes (auxiliary scores of 1 to 3) (54) were subjected to protein modeling using the Protein Homology/analogY Recognition Engine (PHYRE2) (76). Auxiliary scores were assigned by DRAM (54), based on the following ranking system. A gene is given an auxiliary score of 1 if there is at least one hallmark gene on both the left and right flanks, indicating that the gene is likely viral. An auxiliary score of 2 is assigned when the gene has a viral hallmark gene on one flank and a virus-like gene on the other flank. An auxiliary score of 3 is assigned to genes that have a virus-like gene on both flanks. To identify likely vMAG hosts, oligonucleotide frequencies between virus (n = 111) and nondereplicated hosts (n = 102) were analyzed using VirHostMatcher using a threshold of d2* measurements of <0.25 (77). The lowest d2* value for each viral contig of <0.25 was used. All vMAG information is reported in Table S4.
MAG relative abundance calculations and their use in predictions.
To estimate the relative abundance of each MAG and vMAG, the metagenomic reads for each sample were rarefied to 3 Gbp and mapped to 55 unique MAGs via Bowtie2 (60, 61, 69). For MAGs, a minimum scaffold coverage of 75% and a depth of 3× coverage were required for read recruitment at 7 mismatches. For vMAGs, reads were mapped using Bowtie2 (69) at a maximum mismatch of 15, a minimum contig coverage of 75%, and a minimum depth coverage of 2×. Relative abundances for each MAG and vMAG were calculated as their coverage proportion from the sum of the whole coverage of all bins for each set of metagenomic reads. MAG relative abundances per sample for MAGs and vMAGs are reported in Tables S2 and S4. Correlations and sparse partial least squares regression (sPLS) predictions (PLS R package [78]) used mapping data pertaining to only the 10 deeply sequenced metagenomes rarefied to 4.8 Gbp (Tables S2 and S4).
Metaproteome generation and peptide mapping.
Metaproteomic mapping results for MAGs and vMAGs are shown in Table S3. Sediment samples were prepared for metaproteome analysis as previously reported by Graham et al. (23) and using the protocol outlined by Nicora et al. (79). As previously described (61, 80), tandem mass spectrometry (MS/MS) spectra from all liquid chromatography-tandem mass spectrometry (LC-MS/MS) data sets were converted to ASCII text (.dta format) using MSConvert (http://proteowizard.sourceforge.net/tools/msconvert.html), and the data files were then interrogated via a target-decoy approach (81) using MSGF+ (82). For protein identification, spectra were searched against two files that included (i) 55 dereplicated MAG and (ii) 111 clustered vMAG amino acid sequences. Peptide recruitment for each MAG amino acid sequence per sample is reported in Table S3. Hits were divided into three categories: (i) unique peptide hits to a single protein, (ii) nonunique specialized peptide hits to multiple amino acid sequences that all had same annotation and MAG taxonomy, and (iii) nonunique peptide hits to multiple amino acid sequences with different annotations or from MAGs with different taxonomies. This last designation was necessary, as several hits could not be resolved to the MAG level due to functional conservation across closely related MAGs (strains) in the HUM-V database. Data in Fig. 2 showcase categories (i) and (ii). Microbial metaproteomes were converted to normalized spectral abundance frequency (NSAF) values and subsequently divided into unique, nonunique specialized, and nonunique categories and only reported as expressed when detected in more than three samples (unless otherwise noted and subsequently manually inspected to be good hits). Viral metaproteomes were analyzed using peptide counts only from unique hits due to low recruitment.
To understand changes in the metaproteome recruited to MAGs over spatial gradients, nonmetric multidimensional scaling (NMDS) ordinations were performed on the subset of 29 samples that recruited sufficient peptides. Analyses were run with multiple transformations and with presence/absence and relative abundance data. Relevant code for Fig. S2a and b, Fig. S4a and b, and input data used can be found on GitHub (https://github.com/WrightonLabCSU/columbia_river).
In addition to our binned approach, we also analyzed all proteins that recruited peptides across all assembled contigs (i.e., both binned and unbinned). Proteins that recruited peptides uniquely and in more than three samples were then annotated using DRAM. To determine if the predicted carbon cycling genes encoded a secreted protein, pSortb (36) and SignalP (38) were used to predict location; if those methods did not detect a signal peptide, the amino acid sequence was queried using SecretomeP (with a SecP score of >0.5 [37] used as a threshold to report noncanonical secretion signals). The genes that were relevant to the article and their annotations are included in Table S3, along with the mapping results for all unique genes. The annotations for all unique hits, without QC, are available on Zenodo at https://doi.org/10.5281/zenodo.6607647.
Data availability.
The data sets supporting the conclusions of this article are publicly available. All sequencing data information can be found in Table S1 and are available in NCBI under BioProject no. PRJNA576070. The reads sequenced at JGI are also available on JGI/M ER under Gold ID Gs0114663 alongside their respective JGI Assembly Pipeline data (https://img.jgi.doe.gov/mer/). MAG accession numbers and quality information can all be found in Table S2 and are deposited under BioSample no. SAMN18867633 to SAMN18867734. The accession numbers and quality for 111 vMAGs can be found in Table S4, and the sequences are deposited in NCBI under BioProject no. PRJNA576070.
Raw annotations for each MAG are deposited in Zenodo at https://doi.org/10.5281/zenodo.5128772, with the corresponding DRAM interactive heat map at https://zenodo.org/record/5124964. Additionally, the data set of freshwater viruses used to cluster to the HUM-V vMAGs is provided on Zenodo at https://doi.org/10.5281/zenodo.6310084. Metaproteomic data are deposited in the MassIVE database under accession no. MSV000087330. Metabolomics data are publicly available and deposited in Zenodo at https://doi.org/10.5281/zenodo.5076253. Phylogenetic trees are provided on Zenodo at https://doi.org/10.5281/zenodo.6339808. GTDB-Tk phylogenetic analysis output is provided on Zenodo at https://doi.org/10.5281/zenodo.6502149. The unbinned metaproteomic mapping annotation data is hosted on Zenodo at https://doi.org/10.5281/zenodo.6607647. All scripts along with the input files used in this paper are available at https://github.com/WrightonLabCSU/columbia_river.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Subsurface Biogeochemical Research (SBR) program (DE-SC0018170) and the Environmental System Science (ESS) program in the Office of Biological and Environmental Research, Office of Science, U.S. Department of Energy (DOE), through a subcontract from the River Corridor Scientific Focus Area project at the Pacific Northwest National Laboratory. These analyses were also supported by technology developed in the Wrighton laboratory supported by the National Sciences Foundation Division of Biological Infrastructure (grant no. 1759874) and the Department of Energy Office of Biological & Environmental Research (grant no. DE-SC0021350).
J.A.R.-R. was partially supported by the National Science Foundation (NRT-DESE) (grant no. 1450032) via a Trans-Disciplinary Graduate Training Program in Biosensing and Computational Biology at Colorado State University. B.B.M. and K.C.W. were supported by an NSF early career award to K.C.W. (grant no. 1912915). The NMR data, FTICR-MS data, and MS-proteomic data in this work were collected using instrumentation in the Environmental Molecular Science Laboratory (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory. Pacific Northwest National Laboratory is operated by Battelle for the DOE under contract no. DE-AC05-76RL01830. This work (proposal no. 10.46936/jejc.proj.2014.48473/60005497) conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science in the U.S. Department of Energy operated under contract no. DE-AC02-05CH11231. A portion of the metagenomic sequencing for this research was performed at the Genomics Shared Resource Core at The Ohio State University Comprehensive Cancer Center, supported by grant no. P30 CA016058.
We thank Tyson Claffey and Richard Wolfe for Colorado State University server management, Sandy Shew for management of computing resources retained from The Ohio State University Unity cluster, Pearlly Yan at the Genomics Shared Resource Core at The Ohio State University Comprehensive Cancer Center for management of metagenomic sequencing, and J. John for continuous support.
We declare we have no competing interests.
Contributor Information
Kelly C. Wrighton, Email: Kelly.Wrighton@colostate.edu.
Laura A. Hug, University of Waterloo
REFERENCES
- 1.Boulton AJ, Findlay S, Marmonier P, Stanley EH, Valett HM. 1998. The functional significance of the hyporheic zone in streams and rivers. Annu Rev Ecol Syst 29:59–81. doi: 10.1146/annurev.ecolsys.29.1.59. [DOI] [Google Scholar]
- 2.Stegen JC, Johnson T, Fredrickson JK, Wilkins MJ, Konopka AE, Nelson WC, Arntzen EV, Chrisler WB, Chu RK, Fansler SJ, Graham EB, Kennedy DW, Resch CT, Tfaily M, Zachara J. 2018. Influences of organic carbon speciation on hyporheic corridor biogeochemistry and microbial ecology. Nat Commun 9:585. doi: 10.1038/s41467-018-02922-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newcomer ME, Hubbard SS, Fleckenstein JH, Maier U, Schmidt C, Thullner M, Ulrich C, Flipo N, Rubin Y. 2018. Influence of hydrological perturbations and riverbed sediment characteristics on hyporheic zone respiration of CO2 and N2. J Geophys Res Biogeosci 123:902–922. doi: 10.1002/2017JG004090. [DOI] [Google Scholar]
- 4.Hu M, Chen D, Dahlgren RA. 2016. Modeling nitrous oxide emission from rivers: a global assessment. Glob Chang Biol 22:3566–3582. doi: 10.1111/gcb.13351. [DOI] [PubMed] [Google Scholar]
- 5.Raymond PA, Hartmann J, Lauerwald R, Sobek S, McDonald C, Hoover M, Butman D, Striegl R, Mayorga E, Humborg C, Kortelainen P, Dürr H, Meybeck M, Ciais P, Guth P. 2013. Global carbon dioxide emissions from inland waters. Nature 503:355–359. doi: 10.1038/nature12760. [DOI] [PubMed] [Google Scholar]
- 6.Gómez-Gener L, Rocher-Ros G, Battin T, Cohen MJ, Dalmagro HJ, Dinsmore KJ, Drake TW, Duvert C, Enrich-Prast A, Horgby Å, Johnson MS, Kirk L, Machado-Silva F, Marzolf NS, McDowell MJ, McDowell WH, Miettinen H, Ojala AK, Peter H, Pumpanen J, Ran L, Riveros-Iregui DA, Santos IR, Six J, Stanley EH, Wallin MB, White SA, Sponseller RA. 2021. Global carbon dioxide efflux from rivers enhanced by high nocturnal emissions. Nat Geosci 14:289–294. doi: 10.1038/s41561-021-00722-3. [DOI] [Google Scholar]
- 7.Lewandowski J, Arnon S, Banks E, Batelaan O, Betterle A, Broecker T, Coll C, Drummond JD, Gaona Garcia J, Galloway J, Gomez-Velez J, Grabowski RC, Herzog SP, Hinkelmann R, Höhne A, Hollender J, Horn MA, Jaeger A, Krause S, Löchner Prats A, Magliozzi C, Meinikmann K, Mojarrad BB, Mueller BM, Peralta-Maraver I, Popp AL, Posselt M, Putschew A, Radke M, Raza M, Riml J, Robertson A, Rutere C, Schaper JL, Schirmer M, Schulz H, Shanafield M, Singh T, Ward AS, Wolke P, Wörman A, Wu L. 2019. Is the hyporheic zone relevant beyond the scientific community? Water 11:2230. doi: 10.3390/w11112230. [DOI] [Google Scholar]
- 8.Raes EJ, Karsh K, Kessler AJ, Cook PLM, Holmes BH, van de Kamp J, Bodrossy L, Bissett A. 2020. Can we use functional genetics to predict the fate of nitrogen in estuaries? Front Microbiol 11:1261. doi: 10.3389/fmicb.2020.01261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou Z, Nelson WC, Stegen JC, Murray CJ, Arntzen E, Crump AR, Kennedy DW, Perkins MC, Scheibe TD, Fredrickson JK, Zachara JM. 2017. Geochemical and microbial community attributes in relation to hyporheic zone geological facies. Sci Rep 7:12006. doi: 10.1038/s41598-017-12275-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson AR, Sawyer AH, Gabor RS, Saup CM, Bryant SR, Harris KD, Briggs MA, Williams KH, Wilkins MJ. 2019. Heterogeneity in hyporheic flow, pore water chemistry, and microbial community composition in an alpine streambed. J Geophys Res Biogeosci 124:3465–3478. doi: 10.1029/2019JG005226. [DOI] [Google Scholar]
- 11.Sackett JD, Shope CL, Bruckner JC, Wallace J, Cooper CA, Moser DP. 2019. Microbial community structure and metabolic potential of the hyporheic zone of a large mid-stream channel bar. Geomicrobiol J 36:765–776. doi: 10.1080/01490451.2019.1621964. [DOI] [Google Scholar]
- 12.Sun S, Jones RB, Fodor AA. 2020. Inference-based accuracy of metagenome prediction tools varies across sample types and functional categories. Microbiome 8:46. doi: 10.1186/s40168-020-00815-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Djemiel C, Maron P-A, Terrat S, Dequiedt S, Cottin A, Ranjard L. 2022. Inferring microbiota functions from taxonomic genes: a review. Gigascience 11:giab090. doi: 10.1093/gigascience/giab090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thornton PE, Lamarque J-F, Rosenbloom NA, Mahowald NM. 2007. Influence of carbon-nitrogen cycle coupling on land model response to CO2 fertilization and climate variability. Global Biogeochem Cycles 21:GB4018. [Google Scholar]
- 15.Weitz JS, Wilhelm SW. 2012. Ocean viruses and their effects on microbial communities and biogeochemical cycles. F1000 Biol Rep 4:17. doi: 10.3410/B4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peduzzi P. 2016. Virus ecology of fluvial systems: a blank spot on the map? Biol Rev Camb Philos Soc 91:937–949. doi: 10.1111/brv.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollard PC, Ducklow H. 2011. Ultrahigh bacterial production in a eutrophic subtropical Australian river: does viral lysis short-circuit the microbial loop? Limnol Oceanogr 56:1115–1129. doi: 10.4319/lo.2011.56.3.1115. [DOI] [Google Scholar]
- 18.Ma L, Sun R, Mao G, Yu H, Wang Y. 2013. Seasonal and spatial variability of virioplanktonic abundance in Haihe River, China. Biomed Res Int 2013:526362. doi: 10.1155/2013/526362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samson R, Shah M, Yadav R, Sarode P, Rajput V, Dastager SG, Dharne MS, Khairnar K. 2019. Metagenomic insights to understand transient influence of Yamuna River on taxonomic and functional aspects of bacterial and archaeal communities of River Ganges. Sci Total Environ 674:288–299. doi: 10.1016/j.scitotenv.2019.04.166. [DOI] [PubMed] [Google Scholar]
- 20.Huber DH, Ugwuanyi IR, Malkaram SA, Montenegro-Garcia NA, Lhilhi Noundou V, Chavarria-Palma JE. 2018. Metagenome sequences of sediment from a recovering industrialized Appalachian River in West Virginia. Genome Announc 6:e00350-18. doi: 10.1128/genomeA.00350-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S, Wang H, Chen L, Wang J, Zheng M, Liu S, Chen Q, Ni J. 2020. Comammox Nitrospira within the Yangtze River continuum: community, biogeography, and ecological drivers. ISME J 14:2488–2504. doi: 10.1038/s41396-020-0701-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Black EM, Just CL. 2018. The genomic potentials of NOB and comammox Nitrospira in river sediment are impacted by native freshwater mussels. Front Microbiol 9:2061. doi: 10.3389/fmicb.2018.02061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham EB, Crump AR, Kennedy DW, Arntzen E, Fansler S, Purvine SO, Nicora CD, Nelson W, Tfaily MM, Stegen JC. 2018. Multi’omics comparison reveals metabolome biochemistry, not microbiome composition or gene expression, corresponds to elevated biogeochemical function in the hyporheic zone. Sci Total Environ 642:742–753. doi: 10.1016/j.scitotenv.2018.05.256. [DOI] [PubMed] [Google Scholar]
- 24.Arntzen EV, Geist DR, Dresel PE. 2006. Effects of fluctuating river flow on groundwater/surface water mixing in the hyporheic zone of a regulated, large cobble bed river. River Res Applic 22:937–946. doi: 10.1002/rra.947. [DOI] [Google Scholar]
- 25.Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK, Schulz F, Jarett J, Rivers AR, Eloe-Fadrosh EA, Tringe SG, Ivanova NN, Copeland A, Clum A, Becraft ED, Malmstrom RR, Birren B, Podar M, Bork P, Weinstock GM, Garrity GM, Dodsworth JA, Yooseph S, Sutton G, Glöckner FO, Gilbert JA, Nelson WC, Hallam SJ, Jungbluth SP, Ettema TJG, Tighe S, Konstantinidis KT, Liu W-T, Baker BJ, Rattei T, Eisen JA, Hedlund B, McMahon KD, Fierer N, Knight R, Finn R, Cochrane G, Karsch-Mizrachi I, Tyson GW, Rinke C, Lapidus A, Meyer F, Yilmaz P, Parks DH, Eren AM, Genome Standards Consortium, et al. 2017. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 35:725–731. doi: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vosloo S, Huo L, Anderson CL, Dai Z, Sevillano M, Pinto A. 2021. Evaluating de novo assembly and binning strategies for time series drinking water metagenomes. Microbiol Spectr 9:e0143421. doi: 10.1128/Spectrum.01434-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haggerty R, Martí E, Argerich A, von Schiller D, Grimm NB. 2009. Resazurin as a “smart” tracer for quantifying metabolically active transient storage in stream ecosystems. J Geophys Res 114:G03014. [Google Scholar]
- 28.Yang F, Bogdanov B, Strittmatter EF, Vilkov AN, Gritsenko M, Shi L, Elias DA, Ni S, Romine M, Pasa-Tolić L, Lipton MS, Smith RD. 2005. Characterization of purified c-type heme-containing peptides and identification of c-type heme-attachment sites in Shewanella oneidenis cytochromes using mass spectrometry. J Proteome Res 4:846–854. doi: 10.1021/pr0497475. [DOI] [PubMed] [Google Scholar]
- 29.Cordero PRF, Bayly K, Man Leung P, Huang C, Islam ZF, Schittenhelm RB, King GM, Greening C. 2019. Atmospheric carbon monoxide oxidation is a widespread mechanism supporting microbial survival. ISME J 13:2868–2881. doi: 10.1038/s41396-019-0479-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zafiriou OC, Andrews SS, Wang W. 2003. Concordant estimates of oceanic carbon monoxide source and sink processes in the Pacific yield a balanced global “blue-water” CO budget. Global Biogeochem Cycles 17:1015. [Google Scholar]
- 31.Haszpra L, Ferenczi Z, Barcza Z. 2019. Estimation of greenhouse gas emission factors based on observed covariance of CO2, CH4, N2O and CO mole fractions. Environ Sci Eur 31:95. doi: 10.1186/s12302-019-0277-y. [DOI] [Google Scholar]
- 32.Xia X, Zhang S, Li S, Zhang L, Wang G, Zhang L, Wang J, Li Z. 2018. The cycle of nitrogen in river systems: sources, transformation, and flux. Environ Sci Process Impacts 20:863–891. doi: 10.1039/c8em00042e. [DOI] [PubMed] [Google Scholar]
- 33.Strauss EA, Lamberti GA. 2000. Regulation of nitrification in aquatic sediments by organic carbon. Limnol Oceanogr 45:1854–1859. doi: 10.4319/lo.2000.45.8.1854. [DOI] [Google Scholar]
- 34.Brust GE. 2019. Management strategies for organic vegetable fertility, p 193–212. In Biswas D, Micallef SA (ed), Safety and practice for organic food. Academic Press, Inc, New York, NY. [Google Scholar]
- 35.Verhagen FJ, Laanbroek HJ. 1991. Competition for ammonium between nitrifying and heterotrophic bacteria in dual energy-limited chemostats. Appl Environ Microbiol 57:3255–3263. doi: 10.1128/aem.57.11.3255-3263.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FSL. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bendtsen JD, Kiemer L, Fausbøll A, Brunak S. 2005. Non-classical protein secretion in bacteria. BMC Microbiol 5:58. doi: 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Armenteros JJA, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 39.Pollak S, Gralka M, Sato Y, Schwartzman J, Lu L, Cordero OX. 2021. Public good exploitation in natural bacterioplankton communities. Sci Adv 7:eabi4717. doi: 10.1126/sciadv.abi4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allison SD. 2005. Cheaters, diffusion and nutrients constrain decomposition by microbial enzymes in spatially structured environments. Ecol Lett 8:626–635. doi: 10.1111/j.1461-0248.2005.00756.x. [DOI] [Google Scholar]
- 41.Smith P, Schuster M. 2019. Public goods and cheating in microbes. Curr Biol 29:R442–R447. doi: 10.1016/j.cub.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 42.Strauss EA, Richardson WB, Bartsch LA, Cavanaugh JC, Bruesewitz DA, Imker H, Heinz JA, Soballe DM. 2004. Nitrification in the Upper Mississippi River: patterns, controls, and contribution to the NO3− budget. J North Am Benthol Soc 23:1–14. doi:. [DOI] [Google Scholar]
- 43.Stegen JC, Fredrickson JK, Wilkins MJ, Konopka AE, Nelson WC, Arntzen EV, Chrisler WB, Chu RK, Danczak RE, Fansler SJ, Kennedy DW, Resch CT, Tfaily M. 2016. Groundwater-surface water mixing shifts ecological assembly processes and stimulates organic carbon turnover. Nat Commun 7:11237. doi: 10.1038/ncomms11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, Thomas BC, Singh A, Wilkins MJ, Karaoz U, Brodie EL, Williams KH, Hubbard SS, Banfield JF. 2016. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun 7:13219. doi: 10.1038/ncomms13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones CM, Spor A, Brennan FP, Breuil M-C, Bru D, Lemanceau P, Griffiths B, Hallin S, Philippot L. 2014. Recently identified microbial guild mediates soil N2O sink capacity. Nat Clim Chang 4:801–805. doi: 10.1038/nclimate2301. [DOI] [Google Scholar]
- 46.Conthe M, Wittorf L, Kuenen JG, Kleerebezem R, van Loosdrecht MCM, Hallin S. 2018. Life on N2O: deciphering the ecophysiology of N2O respiring bacterial communities in a continuous culture. ISME J 12:1142–1153. doi: 10.1038/s41396-018-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hallin S, Philippot L, Löffler FE, Sanford RA, Jones CM. 2018. Genomics and ecology of novel N2O-reducing microorganisms. Trends Microbiol 26:43–55. doi: 10.1016/j.tim.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 48.Orellana LH, Rodriguez-R LM, Higgins S, Chee-Sanford JC, Sanford RA, Ritalahti KM, Löffler FE, Konstantinidis KT. 2014. Detecting nitrous oxide reductase (NosZ) genes in soil metagenomes: method development and implications for the nitrogen cycle. mBio 5:e01193-14. doi: 10.1128/mBio.01193-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moon K, Jeon JH, Kang I, Park KS, Lee K, Cha C-J, Lee SH, Cho J-C. 2020. Freshwater viral metagenome reveals novel and functional phage-borne antibiotic resistance genes. Microbiome 8:75. doi: 10.1186/s40168-020-00863-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolf YI, Silas S, Wang Y, Wu S, Bocek M, Kazlauskas D, Krupovic M, Fire A, Dolja VV, Koonin EV. 2020. Doubling of the known set of RNA viruses by metagenomic analysis of an aquatic virome. Nat Microbiol 5:1262–1270. doi: 10.1038/s41564-020-0755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Püttker S, Kohrs F, Benndorf D, Heyer R, Rapp E, Reichl U. 2015. Metaproteomics of activated sludge from a wastewater treatment plant—a pilot study. Proteomics 15:3596–3601. doi: 10.1002/pmic.201400559. [DOI] [PubMed] [Google Scholar]
- 52.Rudney JD, Xie H, Rhodus NL, Ondrey FG, Griffin TJ. 2010. A metaproteomic analysis of the human salivary microbiota by three-dimensional peptide fractionation and tandem mass spectrometry. Mol Oral Microbiol 25:38–49. doi: 10.1111/j.2041-1014.2009.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Solden LM, Naas AE, Roux S, Daly RA, Collins WB, Nicora CD, Purvine SO, Hoyt DW, Schückel J, Jørgensen B, Willats W, Spalinger DE, Firkins JL, Lipton MS, Sullivan MB, Pope PB, Wrighton KC. 2018. Interspecies cross-feeding orchestrates carbon degradation in the rumen ecosystem. Nat Microbiol 3:1274–1284. doi: 10.1038/s41564-018-0225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaffer M, Borton MA, McGivern BB, Zayed AA, La Rosa SL, Solden LM, Liu P, Narrowe AB, Rodríguez-Ramos J, Bolduc B, Gazitúa MC, Daly RA, Smith GJ, Vik DR, Pope PB, Sullivan MB, Roux S, Wrighton KC. 2020. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res 48:8883–8900. doi: 10.1093/nar/gkaa621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murphy CL, Sheremet A, Dunfield PF, Spear JR, Stepanauskas R, Woyke T, Elshahed MS, Youssef NH. 2021. Genomic analysis of the yet-uncultured Binatota reveals broad methylotrophic, alkane-degradation, and pigment production capacities. mBio 12:e00985-21. doi: 10.1128/mBio.00985-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moser DP, Fredrickson JK, Geist DR, Arntzen EV, Peacock AD, Li S-MW, Spadoni T, McKinley JP. 2003. Biogeochemical processes and microbial characteristics across groundwater-surface water boundaries of the Hanford Reach of the Columbia River. Environ Sci Technol 37:5127–5134. doi: 10.1021/es034457v. [DOI] [PubMed] [Google Scholar]
- 57.Tfaily MM, Chu RK, Tolić N, Roscioli KM, Anderton CR, Paša-Tolić L, Robinson EW, Hess NJ. 2015. Advanced solvent based methods for molecular characterization of soil organic matter by high-resolution mass spectrometry. Anal Chem 87:5206–5215. doi: 10.1021/acs.analchem.5b00116. [DOI] [PubMed] [Google Scholar]
- 58.Tfaily MM, Chu RK, Toyoda J, Tolić N, Robinson EW, Paša-Tolić L, Hess NJ. 2017. Sequential extraction protocol for organic matter from soils and sediments using high resolution mass spectrometry. Anal Chim Acta 972:54–61. doi: 10.1016/j.aca.2017.03.031. [DOI] [PubMed] [Google Scholar]
- 59.RoyChowdhury T, Bramer L, Hoyt DW, Kim Y-M, Metz TO, McCue LA, Diefenderfer HL, Jansson JK, Bailey V. 2018. Temporal dynamics of CO2 and CH4 loss potentials in response to rapid hydrological shifts in tidal freshwater wetland soils. Ecol Eng 114:104–114. doi: 10.1016/j.ecoleng.2017.06.041. [DOI] [Google Scholar]
- 60.Daly RA, Borton MA, Wilkins MJ, Hoyt DW, Kountz DJ, Wolfe RA, Welch SA, Marcus DN, Trexler RV, MacRae JD, Krzycki JA, Cole DR, Mouser PJ, Wrighton KC. 2016. Microbial metabolisms in a 2.5-km-deep ecosystem created by hydraulic fracturing in shales. Nat Microbiol 1:16146. doi: 10.1038/nmicrobiol.2016.146. [DOI] [PubMed] [Google Scholar]
- 61.Borton MA, Hoyt DW, Roux S, Daly RA, Welch SA, Nicora CD, Purvine S, Eder EK, Hanson AJ, Sheets JM, Morgan DM, Wolfe RA, Sharma S, Carr TR, Cole DR, Mouser PJ, Lipton MS, Wilkins MJ, Wrighton KC. 2018. Coupled laboratory and field investigations resolve microbial interactions that underpin persistence in hydraulically fractured shales. Proc Natl Acad Sci USA 115:E6585–E6594. doi: 10.1073/pnas.1800155115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. 2006. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal Chem 78:4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 63.Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 64.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kang D, Li F, Kirton ES, Thomas A, Egan RS, An H, Wang Z. 2019. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e27522v1. doi: 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu M, Scott AJ. 2012. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28:1033–1034. doi: 10.1093/bioinformatics/bts079. [DOI] [PubMed] [Google Scholar]
- 67.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Olm MR, Brown CT, Brooks B, Banfield JF. 2017. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J 11:2864–2868. doi: 10.1038/ismej.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. 2019. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36:1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Castelle CJ, Hug LA, Wrighton KC, Thomas BC, Williams KH, Wu D, Tringe SG, Singer SW, Eisen JA, Banfield JF. 2013. Extraordinary phylogenetic diversity and metabolic versatility in aquifer sediment. Nat Commun 4:2120. doi: 10.1038/ncomms3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roux S, Enault F, Hurwitz BL, Sullivan MB. 2015. VirSorter: mining viral signal from microbial genomic data. PeerJ 3:e985. doi: 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roux S, Adriaenssens EM, Dutilh BE, Koonin EV, Kropinski AM, Krupovic M, Kuhn JH, Lavigne R, Brister JR, Varsani A, Amid C, Aziz RK, Bordenstein SR, Bork P, Breitbart M, Cochrane GR, Daly RA, Desnues C, Duhaime MB, Emerson JB, Enault F, Fuhrman JA, Hingamp P, Hugenholtz P, Hurwitz BL, Ivanova NN, Labonté JM, Lee K-B, Malmstrom RR, Martinez-Garcia M, Mizrachi IK, Ogata H, Páez-Espino D, Petit M-A, Putonti C, Rattei T, Reyes A, Rodriguez-Valera F, Rosario K, Schriml L, Schulz F, Steward GF, Sullivan MB, Sunagawa S, Suttle CA, Temperton B, Tringe SG, Thurber RV, Webster NS, Whiteson KL, Wilhelm SW, Wommack KE, Woyke T, et al. 2019. Minimum Information about an Uncultivated Virus Genome (MIUViG). Nat Biotechnol 37:29–37. doi: 10.1038/nbt.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Merchant N, Lyons E, Goff S, Vaughn M, Ware D, Micklos D, Antin P. 2016. The iPlant Collaborative: cyberinfrastructure for enabling data to discovery for the life sciences. PLoS Biol 14:e1002342. doi: 10.1371/journal.pbio.1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bin Jang H, Bolduc B, Zablocki O, Kuhn JH, Roux S, Adriaenssens EM, Brister JR, Kropinski AM, Krupovic M, Lavigne R, Turner D, Sullivan MB. 2019. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat Biotechnol 37:632–639. doi: 10.1038/s41587-019-0100-8. [DOI] [PubMed] [Google Scholar]
- 76.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ahlgren NA, Ren J, Lu YY, Fuhrman JA, Sun F. 2017. Alignment-free oligonucleotide frequency dissimilarity measure improves prediction of hosts from metagenomically-derived viral sequences. Nucleic Acids Res 45:39–53. doi: 10.1093/nar/gkw1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lê Cao K-A, Rossouw D, Robert-Granié C, Besse P. 2008. A sparse PLS for variable selection when integrating omics data. Stat Appl Genet Mol Biol 7:35. doi: 10.2202/1544-6115.1390. [DOI] [PubMed] [Google Scholar]
- 79.Nicora CD, Burnum-Johnson KE, Nakayasu ES, Casey CP, White RA, Roy Chowdhury T, Kyle JE, Kim Y-M, Smith RD, Metz TO, Jansson JK, Baker ES. 2018. The MPLEx protocol for multi-omic analyses of soil samples. J Vis Exp 57343. doi: 10.3791/57343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McGivern BB, Tfaily MM, Borton MA, Kosina SM, Daly RA, Nicora CD, Purvine SO, Wong AR, Lipton MS, Hoyt DW, Northen TR, Hagerman AE, Wrighton KC. 2021. Decrypting bacterial polyphenol metabolism in an anoxic wetland soil. Nat Commun 12:2466. doi: 10.1038/s41467-021-22765-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Elias JE, Gygi SP. 2010. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol Biol 604:55–71. doi: 10.1007/978-1-60761-444-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim S, Pevzner PA. 2014. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun 5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metadata. Metagenome read counts, accession numbers, biosamples, geochemistry data, NMR metabolite data, and FTICR-MS data. Download Table S1, XLSX file, 4.0 MB (4.1MB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
MAGs. Completion information, accession numbers, annotations, read mapping information, metabolism summary from DRAM, correlations to geochemistry. Download Table S2, XLSX file, 1.4 MB (1.4MB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Metaproteomes. MAG unique peptides, MAG unique peptide NSAF relative abundance, MAG NSAF relative abundance, vMAG unique peptides, MAG expressed and encoded metabolism summary, Binatia MAG metabolism information, assembly-mapped metaproteomics information. Download Table S3, XLSX file, 1.3 MB (1.3MB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Subset of geochemical measurements used to contextualize the multi-omics data, including the resazurin reduction assay (a), percent carbon (b), percent nitrogen (c), C:N ratio (d), and ammonium (e). Box and whisker plots (n = 33 samples) with points colored by sample transect and each box representing the sample depths spanned (0 to 60 cm). No significant differences were observed by site and depth, except for ammonium (P = 0.026), which was significantly different by bulk depths (i.e., surface = 0 to 30 cm, and deep = 30 to 60 cm). All geochemical measurements are included in Table S1. Download FIG S1, PDF file, 0.1 MB (145.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
NMDS and Venn diagram of differences in transect (N/S) or depth (0 to 60 cm) for MAG proteome. (a) NMDS (n = 29) of north (green) and south (orange) transects of recruited proteome peptides for MAGs. Analysis of similarity (ANOSIM) is reported. (b) NMDS of sediment core depth (0 to 60 cm) of recruited proteome peptides for MAGs. ANOSIM is reported. (c) Euler diagram showing the number of total proteins recruiting peptides in each transect, where the overlap represents the proportion of these proteins recruiting peptides in both transects. Only proteins recruiting two or more total peptides were included (n = 898) to allow for recruitment of at least one peptide for a given protein to both transects or both depths and to reduce false positives. Proteome abundances for all MAGs can be found in Table S3. Download FIG S2, PDF file, 0.3 MB (317.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Percentage of identified formula classes by FTICR-MS and prevalence of NMR metabolites across samples. (a) The percentage of identified formula classes identified in FTICR-MS across samples is not statistically structured by depth or site. Bar plots show the average percentage of identified formulas for each class calculated as the number of formulas identified per class divided by the total number of formulas identified per sample, with error bars representing 1 standard deviation (n = 33). Classes were determined using criteria reported previously (E. B. Graham, A. R. Crump, D. W. Kennedy, E. Arntzen, et al., Sci Total Environ 642:742–753, 2018, https://doi.org/10.1016/j.scitotenv.2018.05.256). Individual data points are plotted for each sample, with point size increasing with depth. Peaks were classified as described in Materials and Methods. Raw, full data are provided in Table S1. (b) Bar graphs showing specific NMR metabolites and ammonium with the percentage of total samples in which they were found. Shading denotes the different categories of detected compounds. For detailed information on how these were collected, see Materials and Methods. Only metabolites detected in more than four samples are shown, with the complete NMR data provided in Table S1. Download FIG S3, PDF file, 0.5 MB (530.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
vMAGs. MIUViG information, accession numbers, annotations, read mapping information, vConTACT2 results, downloaded MetaG information for vConTACT2 analyses, VirHostMatcher2 results, correlations to geochemistry, and sPLS results. Download Table S4, XLSX file, 1.2 MB (1.2MB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
vMAG recruited peptides combined with virus-host linkages indicate possible impacts on river sediment carbon and nitrogen cycling. (a and b) NMDS (n = 29) of north (green) and south (orange) transects (a) or sediment core depth (0 to 60 cm) (b) of vMAG recruited peptides show no significant clustering, with stress and ANOSIM reported. (c) Procrustes ordination of MAG and vMAG NMDS ordinations with relative abundance data across the 28 shallow sequenced samples that recruited reads to our MAG and vMAG communities. (d) Gray dotted circles represent the number of significant vMAGs that putatively infect each MAG, and MAG cartoons are colored based on phylum. (e) Assignment of vMAGs to their putatively infected host MAGs. Colors match host MAG phylum assignment. (f) Different AMGs identified in vMAGs with putative hosts, with the predicted AMG substrate type denoted by colored boxes on the y axis. (g) Inferred cleavage activities for glycoside hydrolase (GH74) and a pectin lyase (PL1) AMG are shown. (h) A conceptual model for how viral AMGs could impact the metabolism of their hosts is shown. One putative bacterial host, Steroidobacteraceae, has a linkage to a vMAG (vMAG.63) that encodes a PL1. Integration of this vMAG into the host MAG would provide the capacity to cleave the pectin backbone (PL1), a functionality not encoded in the host genome. This would create a new resource for the host which does encode genes for the oligo cleavage but not for backbone cleavage of pectin (via GH4 and GH2). This cleavage would result in galactose monomers that can further support host cell-encoded metabolism. Download FIG S4, PDF file, 0.5 MB (547.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
sPLS regressions show vMAG relative abundance-enhanced carbon and nitrogen predictions relative to those of MAG relative abundance alone. The predicted versus measured values for percent carbon (c_per) and percent nitrogen (n_per) with vMAGs only (a), MAGs only (b), and vMAGs and MAGs combined (c) are shown. Values corresponding to t, degrees of freedom, P values, confidence intervals, and correlation are also shown for each plot. Download FIG S5, PDF file, 0.4 MB (391KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Uncultured Binatia are widely dispersed across ecosystems and express nitrogen and carbon cycling genes in situ. (a) Metabolic MAG cartoon for the major functions encoded by the Binatia MAGs. Dotted arrows indicate functions encoded in the metagenome, and solid arrows correspond to metaproteome-detected enzymes. BCAA, branched-chain amino acids. (b) Using the 16S rRNA gene (from Binatia_7), we inventoried the distribution of closely related species to our HUM-V MAGs (>97% similarity) in the Sequence Read Archive (SRA) samples, uncovering the ecological distribution of these microorganisms from soils, as well as a wide variety of terrestrial, terrestrial-aquatic, and marine samples, indicating that the expressed metabolisms in panel a are likely applicable to a wide range of ecosystems. Download FIG S6, PDF file, 0.1 MB (137KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
The data sets supporting the conclusions of this article are publicly available. All sequencing data information can be found in Table S1 and are available in NCBI under BioProject no. PRJNA576070. The reads sequenced at JGI are also available on JGI/M ER under Gold ID Gs0114663 alongside their respective JGI Assembly Pipeline data (https://img.jgi.doe.gov/mer/). MAG accession numbers and quality information can all be found in Table S2 and are deposited under BioSample no. SAMN18867633 to SAMN18867734. The accession numbers and quality for 111 vMAGs can be found in Table S4, and the sequences are deposited in NCBI under BioProject no. PRJNA576070.
Raw annotations for each MAG are deposited in Zenodo at https://doi.org/10.5281/zenodo.5128772, with the corresponding DRAM interactive heat map at https://zenodo.org/record/5124964. Additionally, the data set of freshwater viruses used to cluster to the HUM-V vMAGs is provided on Zenodo at https://doi.org/10.5281/zenodo.6310084. Metaproteomic data are deposited in the MassIVE database under accession no. MSV000087330. Metabolomics data are publicly available and deposited in Zenodo at https://doi.org/10.5281/zenodo.5076253. Phylogenetic trees are provided on Zenodo at https://doi.org/10.5281/zenodo.6339808. GTDB-Tk phylogenetic analysis output is provided on Zenodo at https://doi.org/10.5281/zenodo.6502149. The unbinned metaproteomic mapping annotation data is hosted on Zenodo at https://doi.org/10.5281/zenodo.6607647. All scripts along with the input files used in this paper are available at https://github.com/WrightonLabCSU/columbia_river.