Abstract
STUDY QUESTION
Is the use of ART, a proxy for infertility, associated with epigenetic age acceleration?
SUMMARY ANSWER
The epigenetic age acceleration measured by Dunedin Pace of Aging methylation (DunedinPoAm) differed significantly between non-ART and ART mothers.
WHAT IS KNOWN ALREADY
Among mothers who used ART, epigenetic age acceleration may be associated with low oocyte yield and poor ovarian response. However, the difference in epigenetic age acceleration between non-ART and ART mothers (or even fathers) has not been examined.
STUDY DESIGN, SIZE, DURATION
The Norwegian Mother, Father and Child Cohort Study (MoBa) recruited pregnant women and their partners across Norway at around 18 gestational weeks between 1999 and 2008. Approximately 95 000 mothers, 75 000 fathers and 114 000 children were included. Peripheral blood samples were taken from mothers and fathers at ultrasound appointments or from mothers at childbirth, and umbilical cord blood samples were collected from the newborns at birth.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Among the MoBa participants, we selected 1000 couples who conceived by coitus and 894 couples who conceived by IVF (n = 525) or ICSI (n = 369). We measured their DNA methylation (DNAm) levels using the Illumina MethylationEPIC array and calculated epigenetic age acceleration. A linear mixed model was used to examine the differences in five different epigenetic age accelerations between non-ART and ART parents.
MAIN RESULTS AND THE ROLE OF CHANCE
We found a significant difference in the epigenetic age acceleration calculated by DunedinPoAm between IVF and non-ART mothers (0.021 years, P-value = 2.89E−06) after adjustment for potential confounders. Further, we detected elevated DunedinPoAm in mothers with tubal factor infertility (0.030 years, P-value = 1.34E−05), ovulation factor (0.023 years, P-value = 0.0018) and unexplained infertility (0.023 years, P-value = 1.39E−04) compared with non-ART mothers. No differences in epigenetic age accelerations between non-ART and ICSI fathers were found. DunedinPoAm also showed stronger associations with smoking, education and parity than the other four epigenetic age accelerations.
LIMITATIONS, REASONS FOR CAUTION
We were not able to determine the directionality of the causal pathway between the epigenetic age accelerations and infertility. Since parents’ peripheral blood samples were collected after conception, we cannot rule out the possibility that the epigenetic profile of ART mothers was influenced by the ART treatment. Hence, the results should be interpreted with caution, and our results might not be generalizable to non-pregnant women.
WIDER IMPLICATIONS OF THE FINDINGS
A plausible biological mechanism behind the reported association is that IVF mothers could be closer to menopause than non-ART mothers. The pace of decline of the ovarian reserve that eventually leads to menopause varies between females yet, in general, accelerates after the age of 30, and some studies show an increased risk of infertility in females with low ovarian reserve.
STUDY FUNDING/COMPETING INTEREST(S)
This study was partly funded by the Research Council of Norway (Women’s fertility, project no. 320656) and through its Centres of Excellence Funding Scheme (project no. 262700). M.C.M. has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement number 947684). The authors declare no conflict of interest.
TRIAL REGISTRATION NUMBER
N/A.
Keywords: infertility, assisted reproductive technology, in vitro fertilization, intracytoplasmic sperm injection, epigenetic aging, Norwegian Mother, ather and Child Cohort Study
Introduction
Infertility is defined as the failure to achieve pregnancy after 12 months of regular and unprotected sexual intercourse due to an impairment of a person’s capacity to reproduce either as an individual or with his/her partner (Zegers-Hochschild et al., 2017). The prevalence of infertility in developed countries is ∼12% among women and 10% among men (Louis et al., 2013; Datta et al., 2016). Accordingly, as a treatment for infertility, the use of ART has vastly grown (Wyns et al., 2021), and the proportion of infants born after ART has reached 5% in some countries (Wyns et al., 2021). Although not all infertile couples use ART as a treatment (Te Velde et al., 2017), those who have used ART can be regarded as infertile. Previous literature has reported various risk factors for infertility in women (Gaskins and Chavarro, 2018; Boedt et al., 2021; Hernaez et al., 2021) and men (Jungwirth et al., 2013), such as obesity, diet, smoking, alcohol intake and aging. Among these, the effect of aging on infertility is of particular interest because a growing number of couples postpone their first childbirth to their 30s (Baird et al., 2005; Nabukera et al., 2006). Reduced fecundability (Zegers-Hochschild et al., 2017) is a well-known repercussion of aging (Baird et al., 2005) that partly explains why the prevalence of infertility increases with advanced chronological age. The intersection between these two trends, delaying childbearing and declining fecundability, highlights the public health urgency of developing a biomarker that accurately reflects individual age-related risk of infertility due to delayed parenthood.
Epigenetic biomarkers of aging, widely referred to as epigenetic clocks (Hannum et al., 2013; Horvath, 2013; Levine et al., 2018; Lu et al., 2019; Belsky et al., 2020), are compelling predictors of age-related conditions in the elderly. Despite having the same chronological age, individuals with epigenetic age acceleration are at higher risk of cancer, cardiovascular disease, neurodegenerative disease and all-cause mortality (Marioni et al., 2015; Horvath and Raj, 2018). The blood-based clock from Hannum et al. (2013) was devised based on the age-related changes in blood cell composition, and the pan-tissue clock was developed by Horvath (2013) to estimate the shared aging process across multiple human tissues. The PhenoAge clock by Levine et al. (2018), incorporating several age-related clinical measures, predicts cardiovascular disease and all-cause mortality, while the DNAmTL clock developed by Lu et al. (2019) estimates telomere length to predict mortality and coronary heart disease by reflecting cellular proliferation. Further, epigenetic biomarkers of aging are also associated with other age-dependent conditions among young individuals. For example, Suarez et al. (2018) reported that, in adolescence, accelerated epigenetic age was associated with advanced pubertal, neuroendocrine, psychiatric and cognitive maturity. Belsky et al. (2020) developed the Dunedin Pace of Aging methylation (DunedinPoAm), a new epigenetic biomarker reflecting the pace of aging (as a faster pace of aging results in epigenetic age acceleration), using longitudinal data that closely captures declines in physical and cognitive function among young adults who were 38 and 45 years old.
The relationships between epigenetic biomarkers of aging and male or female infertility are understudied. Monseur et al. (2020) reported an association between low oocyte yield and epigenetic age acceleration, and Hanson et al. (2020) revealed an association between poor ovarian response to ovarian stimulation in ART and epigenetic age acceleration in white blood cells. However, these studies were underpowered due to their small sample size (Monseur et al., n = 39 and Hanson et al., n = 175), lacked an important control group (e.g. women who conceived by coitus), and did not include fathers. In this study, based on data from the Norwegian Mother, Father and Child Cohort Study (MoBa), we compared epigenetic age accelerations stemming from five different epigenetic biomarkers of aging between mother–father pairs who conceived by coitus to those who conceived by ART. The ART group included those who used either IVF or ICSI. Further, we explored associations between the epigenetic age accelerations and various causes of infertility requiring the use of ART.
Materials and methods
Study population
MoBa recruited pregnant women and their partners across Norway at 18 gestational weeks between 1999 and 2008 (Magnus et al., 2006, 2016). Approximately 95 000 mothers, 75 000 fathers and 114 000 children were included. The MoBa participants completed a series of questionnaires during pregnancy and at multiple time points after delivery. Peripheral blood samples were taken from mothers and fathers at ultrasound appointments or from mothers at childbirth (Ronningen et al., 2006), and umbilical cord blood samples were collected from the newborns at birth (Ronningen et al., 2006).
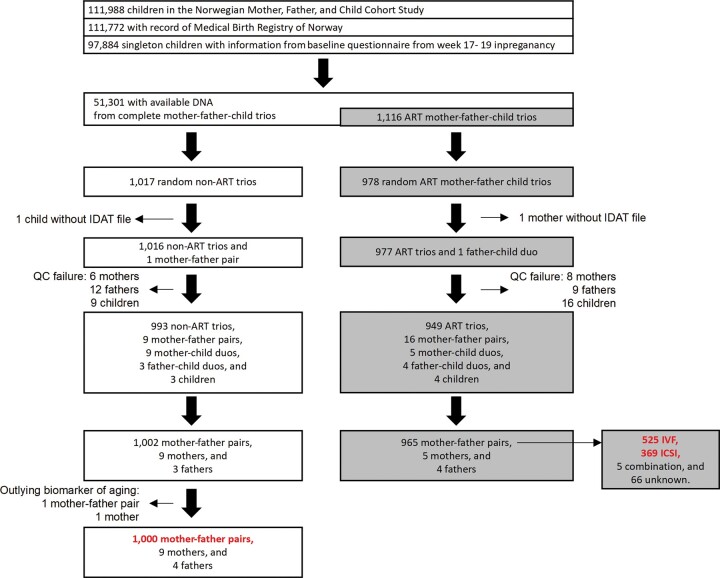
This study focused on a subset of mother–father–newborn trios in MoBa who met the following criteria: (i) the children must be singletons with full records from the Medical Birth Registry of Norway, (ii) the mothers must have completed the first MoBa questionnaire at the 17th week of gestation and (iii) DNA samples must be available for the complete trios. Among the 52 417 trios who met these criteria, we randomly selected 1017 non-ART and 978 ART trios, i.e. a total of 1995 trios (Fig. 1). For the analyses, we arrived at 1000 non-ART mother-father pairs, 525 mother–father pairs who used IVF, and 369 mother–father pairs who used ICSI after excluding outlying subjects with poor data quality (Fig. 1).
Figure 1.
Selection of study participants. The numbers in red refer to the mother–father pairs that were the focus of this study. The samples used in subsequent analyses could be slightly fewer because of missing data in the adjusting variables. IDAT, Intensity Data.
Blood-based DNA methylation
Blood-based DNA methylation of the 1995 mother–father–child trios was measured using the Illumina Infinium MethylationEPIC array (Pidsley et al., 2016). DNA samples were shipped to the Institute of Life & Brain Sciences at the University of Bonn in Germany for processing. The EZ-96DNA methylation-Lightning™MagPrep kit (Zymo Research, Irvine, USA) was used for bisulfite conversion.
Quality control was performed using the RnBeads R package in four batches separately (Muller et al., 2019). We excluded 44 210 cross-hybridizing probes (McCartney et al., 2016), 16 117 probes near single-nucleotide polymorphisms (within three base pairs), and probes with a high detection P-value (>0.01) using the greedy-cut algorithm. This resulted in 770 586 probes on the autosomes and 19 627 probes on the sex chromosomes. Our study focused on the 770 586 autosomal probes. We also excluded a total of 14 mothers, 21 fathers and 25 children due to poor data quality (Fig. 1). The fluorescence intensities were corrected for background noise using enmix.oob and normalized using the Beta-mixture quantile normalization (Ziller et al., 2013) from the wateRmelon R package (Pidsley et al., 2013).
Calculation of epigenetic biomarkers of aging and epigenetic age acceleration
The epigenetic biomarkers of aging were derived by taking weighted averages over the DNAm levels at selected CpGs of the MoBa-START parents. The weights, i.e. the coefficient estimates from penalized regressions, were obtained from the previous publications (Hannum et al., 2013; Horvath, 2013; Levine et al., 2018; Lu et al., 2019; Belsky et al., 2020). In this process, some CpGs of the epigenetic biomarkers of aging were excluded because they did not pass the quality control procedure described above. The number of the excluded CpGs are as follows: 5 out of 46 for DunedinPoAm by Belsky et al. (2020), 6 out of 513 for PhenoAge by Levine et al. (2018), 21 out of 140 for DNAmTL by Lu et al. (2019), 24 out of 353 for DNAmAge by Horvath (2013) and 9 out of 71 for DNAmAge by Hannum et al. (2013). Here, the epigenetic biomarker of aging by DunedinPoAm reflects the pace of aging (in years) from the time of blood sampling to 10–15 years back, whereas the other biomarkers estimate epigenetic age (in years) or telomere length (in kilobases) at the time of blood sampling.
We calculated the epigenetic biomarkers of aging ourselves without using the Horvath online calculator (http://dnamage.genetics.ucla.edu/). This was because uploading the individual level data of MoBa-START to an external website is not part of informed consent from the MoBa participants. Hence, Lu et al.’s GrimAge, which was only available through the Horvath online calculator, could not be included in this study.
We regressed each of the epigenetic biomarkers of aging on chronological age and derived the resultant residual term. For DunedinPoAm and DNAmTL, their residual terms were named as age-adjusted DunedinPoAm and age-adjusted DNAmTL, respectively. For the rest, a suffix of Accel was added, e.g. PhenoAgeAccel and DNAmAgeAccel. This is because these residual terms have been widely referred to as ‘epigenetic age acceleration’ in the literature. Despite the different dynamics of the biomarkers, we use the expression ‘epigenetic age acceleration’ as an umbrella term for the residual terms hereafter.
Assisted reproductive technologies
Information on the use of ART was obtained from the Medical Birth Registry of Norway. The reporting included the specific types of ART procedures used, e.g. IVF or ICSI, and the causes of infertility reported by ART physicians. The ART physicians could report several contributing or subsidiary causes in addition to a ‘main’ cause of infertility for each couple who underwent ART (Supplementary Table SI).
Covariates
Information on pre-pregnancy smoking, alcohol intake and BMI was derived from the MoBa questionnaire collected at the 15th week of gestation, while chronological age at blood sampling and parity were obtained from the Medical Birth Registry of Norway. Detailed information on previous gestations, for example medical complications during pregnancy and birth, was not obtained. Maternal alcohol intake was calculated by multiplying the quantity by the frequency of alcohol consumption (never: alcohol intake = 0, moderate: 0 < alcohol intake ≤ and heavy: alcohol intake > 3), while paternal alcohol intake was defined as the frequency of alcohol consumption (barely: alcohol intake ≤ 0.25, moderate: 0.25 < alcohol intake ≤ 2.5 and heavy: alcohol intake > 2.5) (Tollanes et al., 2016).
Statistical analyses
Each of the standardized epigenetic age accelerations was regressed on a binary variable for (i) non-ART versus IVF, (ii) non-ART versus ICSI and (iii) non-ART versus subsidiary/main cause of infertility, e.g. endometriosis, fallopian tubal factor, ovulation factor, sperm factor and unexplained factor. We used linear mixed models (the lme function from the nlme R package (Pinheiro et al., 2007)) to adjust the associations for pre-pregnancy smoking, alcohol intake, BMI, education, parity and plate (random effect). We reported raw P-values from two-sided Wald tests (in all figures) but capitalized on the Bonferroni threshold (0.05/5 = 0.01) to report the significances of the P-values in the result section.
Ethics
This study was approved by the institutional review board of the Norwegian Institute of Public Health and by the Regional Committee for Medical and Health Research Ethics, southeast Norway (#2017/1362). The establishment of MoBa and initial data collection were based on a license from the Norwegian Data Protection Agency and approval from the Regional Committees for Medical and Health Research Ethics. The MoBa cohort is now regulated by the Norwegian Health Registry Act. All participants provided written informed consent.
Results
Epigenetic biomarkers of aging in MoBa
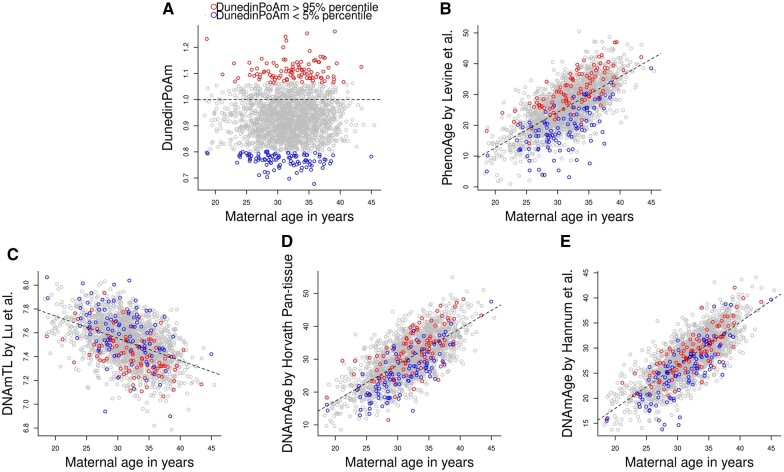
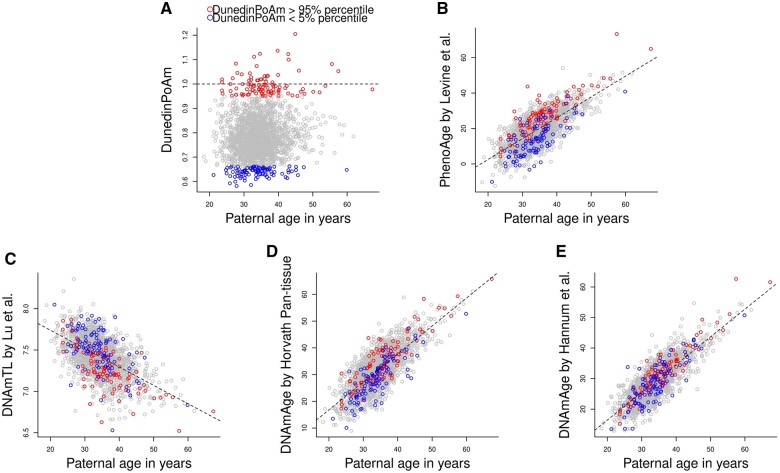
We measured DNAm of 1995 mother–father–child trios in MoBa, using the Illumina MethylationEPIC array. After excluding outliers and samples who did not pass quality control, we retained 1965 mother–father pairs (1000 non-ART and 965 ART pairs, Fig. 1). We used five epigenetic biomarkers of aging, including DNAm-estimated ‘pace of aging’ (referred to as DunedinPoAm hereafter) by Belsky et al. (2020), PhenoAge by Levine et al. (2018), DNAm-estimated telomere length (DNAmTL) by Lu et al. (2019), DNAmAge by Horvath (2013) and DNAmAge by Hannum et al. (2013) (Figs 2 and 3). Age-adjusted DunedinPoAm was moderately correlated with PhenoAgeAccel by Levine et al. (2018) (the Pearson correlation coefficient ranged from 0.46 to 0.48) and weakly correlated with the other epigenetic age accelerations (Absolute Pearson correlation coefficient ranged from 0.16 to 0.26) (Supplementary Fig. S1).
Figure 2.
Scatter plots of maternal age at blood sampling against maternal biomarkers of aging. (A) DNAm-estimated ‘Pace of Aging’ (referred to as DunedinPoAm) by Belsky et al. (2020), (B) PhenoAge by Levine et al. (2018), (C) DNAm-estimated telomere length (DNAmTL) by Lu et al. (2019), (D) DNAmAge by Horvath (2013) and (E) DNAmAge by Hannum et al. (2013). The red dots refer to the mothers with 95% percentile of DunedinPoAm, while the blue dots refer to the mothers with 5% percentile of DunedimPoAm. The dotted line indicates the linear regression of each biomarker of aging on maternal age at blood sampling. DNAm, DNA methylation; PhenoAge, DNAm-estimated phenotypic age; DNAmTL, DNAm-estimated telomere length; DNAmAge, DNA methylation age.
Figure 3.
Scatter plots of paternal age at blood sampling against paternal biomarkers of aging. (A) DNAm-estimated ‘Pace of Aging’ (referred to as DunedinPoAm) by Belsky et al. (2020), (B) PhenoAge by Levine et al. (2018), (C) DNAm-estimated telomere length (DNAmTL) by Lu et al. (2019), (D) DNAmAge by Horvath (2013) and (E) DNAmAge by Hannum et al. (2013). The red dots refer to the fathers with 95% percentile of DunedinPoAm, while the blue dots refer to the fathers with 5% percentile of DunedimPoAm. The dotted line indicates the linear regression of each biomarker of aging on paternal age at blood sampling. DNAm, DNA methylation; PhenoAge, DNAm-estimated phenotypic age; DNAmTL, DNAm-estimated telomere length; DNAmAge, DNA methylation age.
Epigenetic age accelerations between non-ART mothers and IVF mothers and between non-ART fathers and ICSI fathers
Assuming that the use of IVF or ICSI is a proxy for female or male infertility, respectively, we compared the epigenetic age accelerations between non-ART mothers and IVF mothers and between non-ART fathers and ICSI fathers. The characteristics of the study participants in each group can be found in Table I.
Table I.
Descriptive statistics of the study samples.
| N (%) or mean (SD) |
|||||
|---|---|---|---|---|---|
| Mother |
Father |
||||
| Non-ART (n = 1000) | IVF (n = 525) | Non-ART (n = 1000) | ICSI (n = 369) | ||
| Chronological age1 | 30.1 ± 4.63 | 33.57 ± 3.6 | 32.74 ± 5.5 | 36.19 ± 5.72 | |
| Smoking2,3 | No | 643 (64.3%) | 376 (71.6%) | 684 (68.4%) | 265 (71.8%) |
| Sometimes | 100 (10%) | 56 (10.7%) | 98 (9.8%) | 38 (10.3%) | |
| Daily | 177 (17.7%) | 56 (10.7%) | 192 (19.2%) | 58 (15.7%) | |
| NA | 80 (8%) | 37 (7%) | 26 (2.6%) | 8 (2.2%) | |
| Alcohol2,4 | No or barely | 116 (11.6%) | 80 (15.2%) | 211 (21.1%) | 80 (21.7%) |
| Moderate | 571 (57.1%) | 333 (63.4%) | 545 (54.5%) | 176 (47.7%) | |
| Severe | 298 (29.8%) | 99 (18.9%) | 158 (15.8%) | 76 (20.6%) | |
| NA | 15 (1.5%) | 13 (2.5%) | 86 (8.6%) | 37 (10%) | |
| BMI2,5 | Underweight (<18.5 kg/m2) | 27 (2.7%) | 11 (2.1%) | – | – |
| Normal (18.5–25 kg/m2) | 636 (63.6%) | 334 (63.6%) | 412 (41.2%) | 136 (36.9%) | |
| Overweight (25–30 kg/m2) | 218 (21.8%) | 122 (23.2%) | 446 (44.6%) | 175 (47.4%) | |
| Obese (30–35 kg/m2) | 74 (7.4%) | 39 (7.4%) | 92 (9.2%) | 38 (10.3%) | |
| Excessive obese (>35 kg/m2) | 31 (3.1%) | 8 (1.5%) | 16 (1.6%) | 17 (4.6%) | |
| NA | 14 (1.4%) | 11 (2.1%) | 34 (3.4%) | 3 (0.8%) | |
| Education2 | <Vocational school | 161 (16.1%) | 63 (12%) | 340 (34%) | 95 (25.7%) |
| High school | 213 (21.3%) | 82 (15.6%) | 191 (19.1%) | 67 (18.2%) | |
| University ≤4 years | 382 (38.2%) | 223 (42.5%) | 259 (25.9%) | 106 (28.7%) | |
| University >4 years | 230 (23%) | 152 (29%) | 205 (20.5%) | 99 (26.8%) | |
| NA | 14 (1.4%) | 5 (1%) | 5 (0.5%) | 2 (0.5%) | |
| Parity2,6 | 0 | 477 (47.7%) | 364 (69.3%) | 477 (47.7%) | 256 (69.4%) |
| 1 | 346 (34.6%) | 130 (24.8%) | 346 (34.6%) | 94 (25.5%) | |
| 2 | 147 (14.7%) | 27 (5.1%) | 147 (14.7%) | 15 (4.1%) | |
| 3+ | 30 (3%) | 4 (0.8%) | 30 (3%) | 4 (1.1%) | |
Mean (SD).
N (%).
For mothers, self-reported smoking status 3 months before pregnancy was considered. For fathers, self-reported smoking status before and during their partner’s pregnancy was considered.
For mothers, self-reported alcohol consumption 3 months before pregnancy was considered. For fathers, self-reported alcohol consumption 6 months before their partner’s pregnancy was considered.
For mothers, self-reported pre-pregnancy BMI was considered.
The parity of mothers.
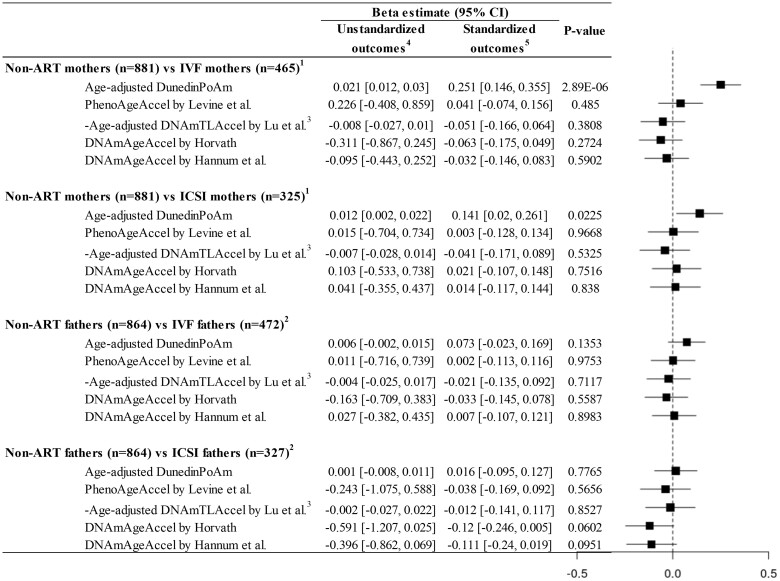
We found a minute but significant difference in the age-adjusted DunedinPoAm between non-ART and IVF mothers (0.021 years, 95% CI 0.012, 0.03, P-value = 2.89E−06, Fig. 4) with adjustment for pre-pregnancy smoking, alcohol intake, BMI, education, parity and batch number used in measuring DNAm. This indicates that IVF mothers showed a faster pace of aging of 0.021 years compared to non-ART mothers. However, there were no associations between the other epigenetic age accelerations and the use of IVF in mothers. Furthermore, none of the epigenetic age accelerations were associated with the use of ICSI in the fathers. For a clearer comparison of the different epigenetic age accelerations (as they are in different scales, Figs 2 and 3), the differences in standardized epigenetic age accelerations between non-ART and IVF group were added to Fig. 4.
Figure 4.
Differences in epigenetic age accelerations between non-ART and IVF/ICSI parents. The plot on the right side was generated based on the beta coefficient estimates with the standardized outcomes. DNAm, DNA methylation; DunedinPoAm, DNAm-estimated ‘Pace of Aging’ by Belsky et al. (2020); PhenoAgeAccel, DNAm-estimated phenotypic age acceleration; DNAmTL, DNAm-estimated telomere length; DNAmAgeAccel, DNA methylation age acceleration. 1In mothers, the associations were adjusted for smoking (pre-pregnancy), alcohol intake (pre-pregnancy), BMI (pre-pregnancy), education, parity and plate. 2In fathers, the associations were adjusted for smoking, alcohol intake, BMI, education, partner’s parity and plate. 3Age-adjusted DNAm-estimated telomere length (DNAmTL) by Lu et al. was multiplied by −1. DNAmTL declines with advanced chronological age. 4The epigenetic age accelerations as outcome variables were not standardized. 5The epigenetic age accelerations as outcome variables were standardized.
Epigenetic age accelerations between non-ART group and ART groups with various causes of infertility
For each couple who underwent ART, ART physicians could report several contributing ‘subsidiary causes’ and one ‘main’ cause of infertility including endometriosis, tubal factor, ovulation factor, sperm factor and unexplained infertility. The distributions of the various subsidiary and main causes of infertility can be found in Supplementary Table SI.
For the subsidiary causes of infertility, we found significant differences in the age-adjusted DunedinPoAm between non-ART mothers and ART (IVF and ICSI) mothers with tubal factor (0.030 years, P-value = 1.34E−05, Supplementary Fig. S2), and between non-ART mothers and ART mothers with ovulation factor (0.023 years, P-value = 0.0018, Supplementary Fig. S2). Additionally, the age-adjusted DunedinPoAm differed between non-ART mothers and ART mothers with unexplained infertility (0.023 years, P-value = 1.39E−04, Supplementary Fig. S2). Again, in fathers, none of the epigenetic age accelerations were associated with sperm factor or unexplained infertility.
We then examined the associations between the epigenetic age accelerations and the main causes of infertility. Unlike the subsidiary causes of infertility presented above, only one main cause of infertility could be assigned to each couple. Strikingly, despite its underpowered comparison due to the fewer cases, the difference in the age-adjusted DunedinPoAm between non-ART mothers and ART mothers with tubal factor appeared to be larger and more significant (0.033 years, P-value = 7.88E−06, Supplementary Fig. S3) than the corresponding effect size (0.030 years, P-value = 1.34E−05) reported in Supplementary Fig. S2. The age-adjusted DunedinPoAm also differed between the non-ART mothers and the ART mothers with unexplained infertility (0.021 years, P-value = 7.95E−04, Supplementary Fig. S3). Again, in fathers, none of the epigenetic age accelerations were associated with sperm factor or unexplained infertility.
As additional analyses, we focused on the comparison (i) between non-ART mothers and IVF mothers whose partner was not reported to have any sperm factor infertility and (ii) between non-ART fathers and ICSI fathers where sperm factor infertility was reported as a main cause of infertility. A small fraction (12%) of the IVF couples were reported to have sperm factor as a subsidiary cause of infertility, and most (77%) ICSI couples had sperm factor as a main cause of infertility. The difference in DunedinPoAm between non-ART mothers and IVF mothers who were not influenced by any sperm factor was 0.023 years (P-value = 1.44E−06, Supplementary Fig. S4). This difference is larger and more significant than that shown in Fig. 4 (0.021 years, P-value = 2.89E−06), even if the sample size was reduced. In fathers, we found no association between epigenetic age accelerations and the use of ICSI even after restricting the analysis to the ICSI fathers with sperm factor as the main cause of infertility (Supplementary Fig. S5).
Replication of associations between covariates and epigenetic age accelerations
As previously reported by others, we also found associations between the epigenetic age accelerations and environmental exposures such as smoking, alcohol intake, BMI, education and parity (Supplementary Figs S6 and S7). The distributions of the environmental factors can be found in Table I. Expectedly, heavy smokers, obese and poorly educated individuals showed accelerated epigenetic aging profiles (DunedinPoAm, PhenoAge by Levine et al. (2018) and DNAmTL by Lu et al. (2019)). Interestingly, the individuals with higher parity showed accelerated aging compared to the primiparous ones.
Evaluation of the CpGs included in DunedinPoAm
We explored whether the 46 CpGs included in DunedinPoAm have been associated with any fertility-related health outcomes in previous studies. We queried each of the 46 CpGs in the epigenome-wide association study catalog (http://www.ewascatalog.org/). There were 38 CpGs associated with age or gestational age in whole blood, fetal cord blood or fetal brain tissue, and 12 CpGs (near AHRR, ETV6 and others) were associated with smoking or maternal smoking in whole blood or fetal cord blood (Supplementary Table SII). This is in line with our findings; DunedinPoAm was highly sensitive to smoking in both mothers and fathers (Supplementary Figs S6 and S7). However, DunedinPoAm is not merely a proxy for smoking as maternal DunedinPoAm remained strongly associated with IVF, tubal factor and unexplained infertility, even with adjustment for smoking (Fig. 4, Supplementary Figs S2 and S3). Moreover, when restricting our analysis to non-smoking mothers, we still found strong associations between maternal DunedinPoAm and use of IVF (0.016 years, P-value = 0.0012) and tubal factor as the main cause of infertility (0.027 years, P-value = 0.0007).
Discussion
This study examined the differences in five epigenetic age accelerations between non-ART mothers (n = 1000) and IVF mothers (n = 525) and between non-ART fathers (n = 1000) and ICSI fathers (n = 369) using microarray DNAm data. Among the epigenetic age accelerations tested, DunedinPoAm, an epigenetic estimator of the ‘Pace of Aging’, differed significantly between non-ART mothers and IVF mothers (Fig. 4). This effect persisted in more granular comparisons of the non-ART mothers versus ART mothers with female infertility, e.g. tubal factor and ovulation factor (Supplementary Figs S2 and S3). However, the associations of the other epigenetic age accelerations with the use of IVF in mothers were weak to moderate. In fathers, no epigenetic age accelerations were associated with the use of ICSI or sperm factor infertility.
According to our findings, DunedinPoAm might surpass the other epigenetic biomarkers of aging in terms of estimating the biological aging profile of individuals who are in their reproductive years. Here, the novelty of DunedinPoAm lies in its quantification of the ‘Pace of Aging’. Using the Dunedin study (Belsky et al., 2015), Belsky et al. (2020), the inventors of DunedinPoAm, quantified the ‘Pace of Aging’ based on 18 clinical measures collected longitudinally (at age 26, 32 and 38 years, which broadly covers the reproductive age in developed countries (Khandwala et al., 2017; Beaujouan, 2020)). Then, they regressed the Pace of Aging on DNAm at CpG sites collected at age 38. In their validation process, DunedinPoAm was highly predictive of the longitudinal decline of functional capabilities, e.g. balance, grip strength, perceptual reasoning and so forth (see Fig. 2 of Belsky et al. (2020)), among individuals at age 38 and 45. In addition, as shown in our analyses (Supplementary Figs S6 and S7), DunedinPoAm was more sensitive to environmental exposures such as smoking, education attainment and parity than the other biomarkers of aging.
The other epigenetic age accelerations showed no significant associations with the use of IVF or causes of infertility in mothers. In other studies, these biomarkers were significantly associated with cancer, cardiovascular disease, neurodegenerative disease and all-cause mortality (Horvath and Raj, 2018). As both male and female infertility are age-dependent (Baird et al., 2005; Sharma et al., 2015) and associated with cardiovascular disease later in life (Hanson et al., 2017), it is sensible to infer some extent of associations between these epigenetic biomarkers of aging and infertility. However, except for DunedinPoAm, the other epigenetic age accelerations were only weakly associated with the causes of infertility in mothers (Supplementary Figs S2 and S3). This observation may be explained by the fact that the other epigenetic biomarkers of aging, by their construction, estimate an individual’s aging status compared to his/her peers using the age-adjusted epigenetic age, i.e. the residual term from a regression of epigenetic age on chronological age (Horvath, 2013; Horvath et al., 2018; Levine et al., 2018). This way of inference should always include a true biological signal and a random noise term by their construction. Because of this random noise term, the other epigenetic biomarkers of aging may require a greater sample size to capture the decline of fecundity in young individuals (Zhang et al., 2019) than DunedinPoAm. Indeed, the other epigenetic biomarkers of aging mostly showed the same directions of associations with tubal factor, smoking status and education attainment as DunedinPoAm, but presented weaker associations than DunedinPoAm.
A further explanation for the different results from DunedinPoAm and the others is that epigenetic biomarkers of aging reflect different aspects of biological aging (Horvath and Raj, 2018; Bell et al., 2019). Epigenetic biomarkers of aging are weakly correlated (Supplementary Fig. S1) and do not share a bulk of CpGs with one another (Horvath and Raj, 2018; Liu et al., 2020). Single-cell analysis, as an attempt to link epigenetic aging with existing hallmarks of aging, has gained attention. Lowe et al. (2016) reported that, in epithelial cells, epigenetic aging captured by Horvath (2013) was accompanied with replicative and oncogene-induced cellular senescence but was distinct from senescence induced by telomere attrition or DNA damage. Liu et al. (2020) reported that epigenetic aging by Levine et al. (2018) did not capture oncogene-induced senescence but replicative senescence in fibroblast cells. Further analysis of single cells is expected to unveil what mechanism underlies DunedinPoAm.
A plausible biological mechanism behind the faster pace of epigenetic aging in IVF mothers is that they could be closer to menopause than non-ART mothers. The pace of decline of the ovarian reserve that eventually leads to menopause varies between females, yet in general, accelerates after the age of 30 (Baird et al., 2005), and some studies show an increased risk for infertility in females with low ovarian reserve (Korsholm et al., 2018; Lin et al., 2021). Regrettably, no markers for ovarian reserve such as pre-pregnancy serum anti-Mullerian hormone measurements or antral follicle counts were available in our dataset. Neither could low ovarian reserve be registered as a main or subsidiary cause of infertility by ART-clinicians. Amongst the causes of infertility that could be registered, unexplained infertility is known to be closely intertwined with age-related infertility and low ovarian reserve (Somigliana et al., 2016) and was indeed associated with DunedinPoAm in our study. Although the use of IVF is not causal for ovarian aging and early menopause (Elder et al., 2008), it is noteworthy that one study found epigenetic age acceleration later in life to be associated with having experienced an early menopause (Levine et al., 2016). Another study found that mural granulosa cells and leukocytes in women with diminished ovarian response show a specific epigenetic profile (Olsen et al., 2021).
DunedinPoAm also showed epigenetic age acceleration in ART mothers with ovulation factor infertility and those with tubal factor infertility. Ovulation factor infertility is often caused by polycystic ovary syndrome, a syndrome characterized by androgen excess (Deswal et al., 2018), and some studies have linked androgen excess to epigenetic aging. For example, women who experienced early menopause (i.e. relative androgen excess) showed epigenetic age acceleration (Levine et al., 2016). Further, an animal study revealed that castration of male sheep might delay epigenetic aging (Sugrue et al., 2021). The association between tubal factor infertility and epigenetic age acceleration could also be medicated by pelvic infection or previous surgery in the pelvis. Women with pelvic infection and surgery are more likely to experience tubal factor infertility (Wiesenfeld et al., 2012; Hoenderboom et al., 2019) and showed significant epigenetic age acceleration (Kananen et al., 2015; Sadahiro et al., 2020). However, given that the above-mentioned studies preceded DunedinPoAm, further investigation is necessary to verify this suggested mechanism.
Our results showed that mothers with parity of three or more had higher epigenetic age acceleration compared to those with parity of zero (Supplementary Figs S6 and S7). Yet, this finding should be interpreted with caution due to the low number of the mothers with parity of three or more (Table I). One previous study was in line with our finding (Kresovich et al., 2019), whereas another study found no association between epigenetic age acceleration and parity (Harville et al., 2021). This discrepancy between studies could be due to inclusion or not of medically complicated pregnancies. On the other hand, the association between previously having fathered three children or more and epigenetic age acceleration is novel. Previous studies in demography suggested that the observed U-shaped fertility–mortality relationship in males is partly an effect of reproductive behavior on lifestyle (Grundy and Kravdal, 2010).
As limitations, we were not able to determine the directionality of the causal pathway between the epigenetic age accelerations and infertility. Importantly, parents’ peripheral blood samples were collected after conception (in the majority of cases, blood samples were collected at ultrasound appointments scheduled around the 17th to 18th week of gestation (Paltiel et al., 2014)). Therefore, we cannot rule out the possibility that the epigenetic profile of ART mothers was influenced by the ART treatment or pregnancy. Hence, the results should be interpreted with caution as they might not be generalizable to non-pregnant women. According to Gruzieva et al. (2019), the methylation levels at 196 CpGs change before, during and after pregnancy. As another limitation, this study lacked couples who were unsuccessful at conceiving even after undergoing ART. Therefore, we were not able to investigate the extent to which epigenetic aging was altered among severely infertile couples who never conceived, compared to those who conceived by coitus or ART.
Conclusions
The Pace of Aging measured by DunedinPoAm differed significantly between non-ART mothers and IVF mothers. There were minute but significant differences in DunedinPoAm between non-ART mothers and ART mothers with tubal factor, ovulation factor and unexplained infertility. We found no differences in epigenetic age accelerations between non-ART and ICSI fathers.
Supplementary Material
Acknowledgements
The Norwegian Mother, Father and Child Cohort Study is supported by the Norwegian Ministry of Health and Care Services and the Ministry of Education and Research. The authors are grateful to all the families in Norway who take part in this ongoing cohort study.
Authors’ roles
Y.L., J.B., J.R.H., P.M., A.J., M.C.M., S.E.H. and H.I.H. designed the study. Y.L. conducted the statistical analyses. C.M.P. and H.E.N. performed the quality control for DNA methylation data. Y.L., J.R.H. and H.I.H. wrote the manuscript. S.E.H. and P.M. acquired funding and administrated the study. All the authors interpreted the results and contributed to manuscript preparation.
Funding
This study was partly funded by the Research Council of Norway (Women’s fertility, project no. 320656) and through its Centres of Excellence Funding Scheme (project no. 262700). M.C.M. has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement number 947684).
Conflict of interests
The authors declare no conflict of interest.
Contributor Information
Yunsung Lee, Centre for Fertility and Health, Norwegian Institute of Public Health, Oslo, Norway.
Jon Bohlin, Centre for Fertility and Health, Norwegian Institute of Public Health, Oslo, Norway.
Christian M Page, Centre for Fertility and Health, Norwegian Institute of Public Health, Oslo, Norway; Department of Mathematics, Faculty of Mathematics and Natural Sciences, University of Oslo, Oslo, Norway.
Haakon E Nustad, Centre for Fertility and Health, Norwegian Institute of Public Health, Oslo, Norway; Deepinsight, Oslo, Norway.
Jennifer R Harris, Centre for Fertility and Health, Norwegian Institute of Public Health, Oslo, Norway.
Per Magnus, Centre for Fertility and Health, Norwegian Institute of Public Health, Oslo, Norway.
Astanand Jugessur, Centre for Fertility and Health, Norwegian Institute of Public Health, Oslo, Norway; Department of Global Public Health and Primary Care, University of Bergen, Bergen, Norway.
Maria C Magnus, Centre for Fertility and Health, Norwegian Institute of Public Health, Oslo, Norway.
Siri E Håberg, Centre for Fertility and Health, Norwegian Institute of Public Health, Oslo, Norway.
Hans I Hanevik, Centre for Fertility and Health, Norwegian Institute of Public Health, Oslo, Norway; Fertility Department Sør, Telemark Hospital Trust, Porsgrunn, Norway.
Data Availability
The individual-level DNAm and phenotypic data from the Medical Registry of Norway and the MoBa questionnaires are accessible upon request and after approval by the Norwegian Institute of Public Health (https://www.fhi.no/en/studies/moba/).
References
- Baird DT, Collins J, Egozcue J, Evers LH, Gianaroli L, Leridon H, Sunde A, Templeton A, Van Steirteghem A, Cohen J. et al. ; ESHRE Capri Workshop Group. Fertility and ageing. Hum Reprod Update 2005;11:261–276. [DOI] [PubMed] [Google Scholar]
- Beaujouan E. Latest-late fertility? Decline and resurgence of late parenthood across the low-fertility countries. Popul Dev Rev 2020;46:219–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CG, Lowe R, Adams PD, Baccarelli AA, Beck S, Bell JT, Christensen BC, Gladyshev VN, Heijmans BT, Horvath S. et al. DNA methylation aging clocks: challenges and recommendations. Genome Biol 2019;20:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, Caspi A, Arseneault L, Baccarelli A, Corcoran DL, Gao X, Hannon E, Harrington HL, Rasmussen LJ, Houts R. et al. Quantification of the pace of biological aging in humans through a blood test, the DunedinPoAm DNA methylation algorithm. Elife 2020;9:e54870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, Caspi A, Houts R, Cohen HJ, Corcoran DL, Danese A, Harrington H, Israel S, Levine ME, Schaefer JD. et al. Quantification of biological aging in young adults. Proc Natl Acad Sci USA 2015;112:E4104–E4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedt T, Vanhove A-C, Vercoe MA, Matthys C, Dancet E, Lie Fong S.. Preconception lifestyle advice for people with infertility. Cochrane Database Syst Rev 2021;4:CD008189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta J, Palmer MJ, Tanton C, Gibson LJ, Jones KG, Macdowall W, Glasier A, Sonnenberg P, Field N, Mercer CH. et al. Prevalence of infertility and help seeking among 15 000 women and men. Hum Reprod 2016;31:2108–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deswal R, Yadav A, Dang AS.. Sex hormone binding globulin—an important biomarker for predicting PCOS risk: a systematic review and meta-analysis. Syst Biol Reprod Med 2018;64:12–24. [DOI] [PubMed] [Google Scholar]
- Elder K, Mathews T, Kutner E, Kim E, Espenberg D, Faddy M, Gosden R.. Impact of gonadotrophin stimulation for assisted reproductive technology on ovarian ageing and menopause. Reprod Biomed Online 2008;16:611–616. [DOI] [PubMed] [Google Scholar]
- Gaskins AJ, Chavarro JE.. Diet and fertility: a review. Am J Obstet Gynecol 2018;218:379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy E, Kravdal O.. Fertility history and cause-specific mortality: a register-based analysis of complete cohorts of Norwegian women and men. Soc Sci Med 2010;70:1847–1857. [DOI] [PubMed] [Google Scholar]
- Gruzieva O, Merid SK, Chen S, Mukherjee N, Hedman AM, Almqvist C, Andolf E, Jiang Y, Kere J, Scheynius A. et al. DNA methylation trajectories during pregnancy. Epigenet Insights 2019;12:2516865719867090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y. et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 2013;49:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson B, Johnstone E, Dorais J, Silver B, Peterson CM, Hotaling J.. Female infertility, infertility-associated diagnoses, and comorbidities: a review. J Assist Reprod Genet 2017;34:167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson BM, Tao X, Zhan Y, Jenkins TG, Morin SJ, Scott RT, Seli EU.. Young women with poor ovarian response exhibit epigenetic age acceleration based on evaluation of white blood cells using a DNA methylation-derived age prediction model. Hum Reprod 2020;35:2579–2588. [DOI] [PubMed] [Google Scholar]
- Harville EW, Mishra PP, Kahonen M, Raitoharju E, Marttila S, Raitakari O, Lehtimaki T.. Reproductive history and blood cell DNA methylation later in life: the Young Finns Study. Clin Epigenetics 2021;13:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernaez A, Rogne T, Skara KH, Haberg SE, Page CM, Fraser A, Burgess S, Lawlor DA, Magnus MC.. Body mass index and subfertility: multivariable regression and Mendelian randomization analyses in the Norwegian Mother, Father and Child Cohort Study. Hum Reprod 2021;36:3141–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenderboom BM, van Benthem BHB, van Bergen J, Dukers-Muijrers N, Gotz HM, Hoebe C, Hogewoning AA, Land JA, van der Sande MAB, Morre SA. et al. Relation between Chlamydia trachomatis infection and pelvic inflammatory disease, ectopic pregnancy and tubal factor infertility in a Dutch cohort of women previously tested for chlamydia in a chlamydia screening trial. Sex Transm Infect 2019;95:sextrans-2018-053778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol 2013;14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Oshima J, Martin GM, Lu AT, Quach A, Cohen H, Felton S, Matsuyama M, Lowe D, Kabacik S. et al. Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies. Aging (Albany NY) 2018;10:1758–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Raj K.. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet 2018;19:371–384. [DOI] [PubMed] [Google Scholar]
- Jungwirth A, Diemer T, Dohle G, Giwercman A, Kopa Z, Tournaye H, Krausz C.. EAU guidelines on male infertility. Eur Urol 2013;7:226–241. [DOI] [PubMed] [Google Scholar]
- Kananen L, Nevalainen T, Jylhava J, Marttila S, Hervonen A, Jylha M, Hurme M.. Cytomegalovirus infection accelerates epigenetic aging. Exp Gerontol 2015;72:227–229. [DOI] [PubMed] [Google Scholar]
- Khandwala YS, Zhang CA, Lu Y, Eisenberg ML.. The age of fathers in the USA is rising: an analysis of 168 867 480 births from 1972 to 2015. Hum Reprod 2017;32:2110–2116. [DOI] [PubMed] [Google Scholar]
- Korsholm AS, Petersen KB, Bentzen JG, Hilsted LM, Andersen AN, Hvidman HW.. Investigation of anti-Mullerian hormone concentrations in relation to natural conception rate and time to pregnancy. Reprod Biomed Online 2018;36:568–575. [DOI] [PubMed] [Google Scholar]
- Kresovich JK, Harmon QE, Xu Z, Nichols HB, Sandler DP, Taylor JA.. Reproduction, DNA methylation and biological age. Hum Reprod 2019;34:1965–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Lu AT, Chen BH, Hernandez DG, Singleton AB, Ferrucci L, Bandinelli S, Salfati E, Manson JE, Quach A. et al. Menopause accelerates biological aging. Proc Natl Acad Sci USA 2016;113:9327–9332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, Hou L, Baccarelli AA, Stewart JD, Li Y. et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 2018;10:573–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Jing M, Zhu W, Tu X, Chen Q, Wang X, Zheng Y, Zhang R.. The value of anti-mullerian hormone in the prediction of spontaneous pregnancy: a systematic review and meta-analysis. Front Endocrinol (Lausanne) 2021;12:695157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Leung D, Thrush K, Zhao W, Ratliff S, Tanaka T, Schmitz LL, Smith JA, Ferrucci L, Levine ME.. Underlying features of epigenetic aging clocks in vivo and in vitro. Aging Cell 2020;19:e13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis JF, Thoma ME, Sorensen DN, McLain AC, King RB, Sundaram R, Keiding N, Buck Louis GM.. The prevalence of couple infertility in the United States from a male perspective: evidence from a nationally representative sample. Andrology 2013;1:741–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe D, Horvath S, Raj K.. Epigenetic clock analyses of cellular senescence and ageing. Oncotarget 2016;7:8524–8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu AT, Seeboth A, Tsai PC, Sun D, Quach A, Reiner AP, Kooperberg C, Ferrucci L, Hou L, Baccarelli AA. et al. DNA methylation-based estimator of telomere length. Aging (Albany NY) 2019;11:5895–5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnus P, Birke C, Vejrup K, Haugan A, Alsaker E, Daltveit AK, Handal M, Haugen M, Hoiseth G, Knudsen GP. et al. Cohort profile update: the Norwegian Mother and Child Cohort Study (MoBa). Int J Epidemiol 2016;45:382–388. [DOI] [PubMed] [Google Scholar]
- Magnus P, Irgens LM, Haug K, Nystad W, Skjaerven R, Stoltenberg C; MoBa Study Group. Cohort profile: the Norwegian Mother and Child Cohort Study (MoBa). Int J Epidemiol 2006;35:1146–1150. [DOI] [PubMed] [Google Scholar]
- Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, Gibson J, Henders AK, Redmond P, Cox SR. et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol 2015;16:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney DL, Walker RM, Morris SW, McIntosh AM, Porteous DJ, Evans KL.. Identification of polymorphic and off-target probe binding sites on the Illumina Infinium MethylationEPIC BeadChip. Genom Data 2016;9:22–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monseur B, Murugappan G, Bentley J, Teng N, Westphal L.. Epigenetic clock measuring age acceleration via DNA methylation levels in blood is associated with decreased oocyte yield. J Assist Reprod Genet 2020;37:1097–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F, Scherer M, Assenov Y, Lutsik P, Walter J, Lengauer T, Bock C.. RnBeads 2.0: comprehensive analysis of DNA methylation data. Genome Biol 2019;20:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabukera S, Wingate MS, Alexander GR, Salihu HM.. First-time births among women 30 years and older in the United States: patterns and risk of adverse outcomes. J Reprod Med 2006;51:676–682. [PubMed] [Google Scholar]
- Olsen KW, Castillo-Fernandez J, Chan AC, la Cour Freiesleben N, Zedeler A, Bungum M, Cardona A, Perry JRB, Skouby SO, Hoffmann ER. et al. Identification of a unique epigenetic profile in women with diminished ovarian reserve. Fertil Steril 2021;115:732–741. [DOI] [PubMed] [Google Scholar]
- Paltiel L, Anita H, Skjerden T, Harbak K, Bækken S, Kristin SN, Knudsen GP, Magnus P.. The biobank of the Norwegian Mother and Child Cohort Study—present status. Nor J Epidemiol 2014;24:29–35. [Google Scholar]
- Pidsley R, Y Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC.. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics 2013;14:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, Van Djik S, Muhlhausler B, Stirzaker C, Clark SJ.. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol 2016;17:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D; R Core Team. Linear and nonlinear mixed effects models. R Package Version 2007;3:1–89. [Google Scholar]
- Ronningen KS, Paltiel L, Meltzer HM, Nordhagen R, Lie KK, Hovengen R, Haugen M, Nystad W, Magnus P, Hoppin JA.. The biobank of the Norwegian Mother and Child Cohort Study: a resource for the next 100 years. Eur J Epidemiol 2006;21:619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadahiro R, Knight B, James F, Hannon E, Charity J, Daniels IR, Burrage J, Knox O, Crawford B, Smart NJ. et al. Major surgery induces acute changes in measured DNA methylation associated with immune response pathways. Sci Rep 2020;10:5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Agarwal A, Rohra VK, Assidi M, Abu-Elmagd M, Turki RF.. Effects of increased paternal age on sperm quality, reproductive outcome and associated epigenetic risks to offspring. Reprod Biol Endocrinol 2015;13:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somigliana E, Paffoni A, Busnelli A, Filippi F, Pagliardini L, Vigano P, Vercellini P.. Age-related infertility and unexplained infertility: an intricate clinical dilemma. Hum Reprod 2016;31:1390–1396. [DOI] [PubMed] [Google Scholar]
- Suarez A, Lahti J, Czamara D, Lahti-Pulkkinen M, Girchenko P, Andersson S, Strandberg TE, Reynolds RM, Kajantie E, Binder EB. et al. The epigenetic clock and pubertal, neuroendocrine, psychiatric, and cognitive outcomes in adolescents. Clin Epigenetics 2018;10:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugrue VJ, Zoller JA, Narayan P, Lu AT, Ortega-Recalde OJ, Grant MJ, Bawden CS, Rudiger SR, Haghani A, Bond DM. et al. Castration delays epigenetic aging and feminizes DNA methylation at androgen-regulated loci. Elife 2021;10:e64932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Velde E, Habbema D, Nieschlag E, Sobotka T, Burdorf A.. Ever growing demand for in vitro fertilization despite stable biological fertility—a European paradox. Eur J Obstet Gynecol Reprod Biol 2017;214:204–208. [DOI] [PubMed] [Google Scholar]
- Tollanes MC, Strandberg-Larsen K, Forthun I, Petersen TG, Moster D, Andersen AM, Stoltenberg C, Olsen J, Wilcox AJ.. Cohort profile: cerebral palsy in the Norwegian and Danish birth cohorts (MOBAND-CP). BMJ Open 2016;6:e012777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenfeld HC, Hillier SL, Meyn LA, Amortegui AJ, Sweet RL.. Subclinical pelvic inflammatory disease and infertility. Obstet Gynecol 2012;120:37–43. [DOI] [PubMed] [Google Scholar]
- Wyns C, De Geyter C, Calhaz-Jorge C, Kupka MS, Motrenko T, Smeenk J, Bergh C, Tandler-Schneider A, Rugescu IA, Vidakovic S. et al. ; European IVF-Monitoring Consortium (EIM) for the European Society of Human Reproduction and Embryology (ESHRE). ART in Europe, 2017: results generated from European registries by ESHRE. Hum Reprod Open 2021;2021:hoab026.34377841 [Google Scholar]
- Zegers-Hochschild F, Adamson GD, Dyer S, Racowsky C, de Mouzon J, Sokol R, Rienzi L, Sunde A, Schmidt L, Cooke ID. et al. The International Glossary on Infertility and Fertility Care, 2017. Fertil Steril 2017;108:393–406. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Vallerga CL, Walker RM, Lin T, Henders AK, Montgomery GW, He J, Fan D, Fowdar J, Kennedy M. et al. Improved precision of epigenetic clock estimates across tissues and its implication for biological ageing. Genome Med 2019;11:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Müller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE. et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 2013;500:477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The individual-level DNAm and phenotypic data from the Medical Registry of Norway and the MoBa questionnaires are accessible upon request and after approval by the Norwegian Institute of Public Health (https://www.fhi.no/en/studies/moba/).