Abstract
Massively parallel reporter assays (MPRAs) test the capacity of putative gene regulatory elements to drive transcription on a genome-wide scale. Most gene regulatory activity occurs within accessible chromatin, and recently described methods have combined assays that capture these regions—such as assay for transposase-accessible chromatin using sequencing (ATAC-seq)—with self-transcribing active regulatory region sequencing (STARR-seq) to selectively assay the regulatory potential of accessible DNA (ATAC-STARR-seq). Here, we report an integrated approach that quantifies activating and silencing regulatory activity, chromatin accessibility, and transcription factor (TF) occupancy with one assay using ATAC-STARR-seq. Our strategy, including important updates to the ATAC-STARR-seq assay and workflow, enabled high-resolution testing of ∼50 million unique DNA fragments tiling ∼101,000 accessible chromatin regions in human lymphoblastoid cells. We discovered that 30% of all accessible regions contain an activator, a silencer, or both. Although few MPRA studies have explored silencing activity, we demonstrate that silencers occur at similar frequencies to activators, and they represent a distinct functional group enriched for unique TF motifs and repressive histone modifications. We further show that Tn5 cut-site frequencies are retained in the ATAC-STARR plasmid library compared to standard ATAC-seq, enabling TF occupancy to be ascertained from ATAC-STARR data. With this approach, we found that activators and silencers cluster by distinct TF footprint combinations, and these groups of activity represent different gene regulatory networks of immune cell function. Altogether, these data highlight the multilayered capabilities of ATAC-STARR-seq to comprehensively investigate the regulatory landscape of the human genome all from a single DNA fragment source.
Transcription is regulated by transcription factors (TFs) and the DNA sequences they bind, called cis-regulatory elements. Enhancers, which are a class of cis-regulatory elements, are distally located from the genes they target and serve as key drivers of cell type–specific gene expression (Heinz et al. 2015). Because enhancers require TF binding, they are largely dependent on chromatin accessibility to elicit transcriptional activity. Therefore, chromatin accessibility is a vital regulator of enhancer function, and this is evidenced by the observation that ∼94% of all ENCODE TF ChIP-seq peaks fall within accessible chromatin (Klemm et al. 2019). In any given cell type, only a small fraction (∼2%) of the genome is accessible to TF binding (Thurman et al. 2012; Klemm et al. 2019). In this way, most enhancers are inaccessible and are less likely to drive transcription endogenously.
Enhancers are difficult to identify and validate because they lack uniform features and are less constrained by gene proximity than promoters (Gasperini et al. 2020). Massively parallel reporter assays (MPRAs) were developed to test the regulatory potential of thousands to millions of DNA sequences in parallel, providing high-throughput identification of putative enhancers. Overall, MPRAs test the regulatory potential of genomic regions by cloning them en masse into a reporter plasmid and leveraging high-throughput sequencing to quantify regulatory activity (Santiago-Algarra et al. 2017). Among the variety of different vector backbones and assay designs applied to MPRAs, self-transcribing active regulatory region sequencing (STARR-seq) is uniquely designed to assay an entire genome for regulatory activity (Melnikov et al. 2012; Patwardhan et al. 2012; Arnold et al. 2013; Inoue et al. 2017; Maricque et al. 2017; Muerdter et al. 2018; Kircher et al. 2019). STARR-seq quantifies regulatory activity genome-wide by cloning randomly fragmented genomic DNA into the 3′ UTR of the reporter plasmid. Thus, active enhancers drive transcription of themselves, and activity is quantified by the abundance of their own sequence in the transcript pool, removing the need for barcodes that some MPRAs employ. One major limitation of STARR-seq is that it is technically challenging to accommodate the massive size of the human genome; it requires large-scale cloning procedures and produces shallow sequencing coverage of human regulatory elements (Johnson et al. 2018). In addition, STARR-seq assays both accessible and inaccessible chromatin. Thus, many assayed regions are derived from heterochromatin and are less likely to be transcriptionally active in the cell type in question.
To narrow the scope of the assay, recent methods have combined STARR-seq with techniques that capture accessible chromatin to specifically test the regulatory potential of accessible DNA (Wang et al. 2018; Chaudhri et al. 2020; Glaser et al. 2021). As a result, these methods only sample a fraction of the human genome (∼2%) while assaying nearly all regulatory elements capable of driving transcription endogenously, because they are derived from open chromatin. This approach remains comprehensive while enabling deeper sequencing coverage of biologically relevant genomic regions. Furthermore, integrated approaches have recently been described that combine measurements of chromatin accessibility with analysis of transcription and other epigenomic features from a single population of cells (Kelly et al. 2012; Clark et al. 2018; Barnett et al. 2020; Chen et al. 2022). Similarly, ATAC-STARR-seq has the potential to reveal multiple levels of gene regulatory information simultaneously, but this potential has not been explored. In addition, a complete understanding of gene regulatory activity is lacking with most MPRA approaches because silencing activity is largely overlooked, with a few recent exceptions (Doni Jayavelu et al. 2020; Pang and Snyder 2020; Kim et al. 2021); this is potentially due to technical caveats of distinguishing silencers from either that of missing data or interference from head-on transcriptional conflicts or post-transcriptional silencing mechanisms.
Here, we demonstrate a new workflow that substantially expands the capabilities of ATAC-STARR-seq to extract and measure gene regulatory information. Using this approach, we aimed to identify both activators and silencers, as well as to simultaneously profile chromatin accessibility, and perform TF footprinting. From a single ATAC-STARR-seq data set, a multilayered, integrated view of the human genome can be captured—a feature that has not been explored previously. We provide a protocol and code repository so that this new ATAC-STARR-seq workflow may be easily used and adopted by the field.
Results
ATAC-STARR-seq experimental design
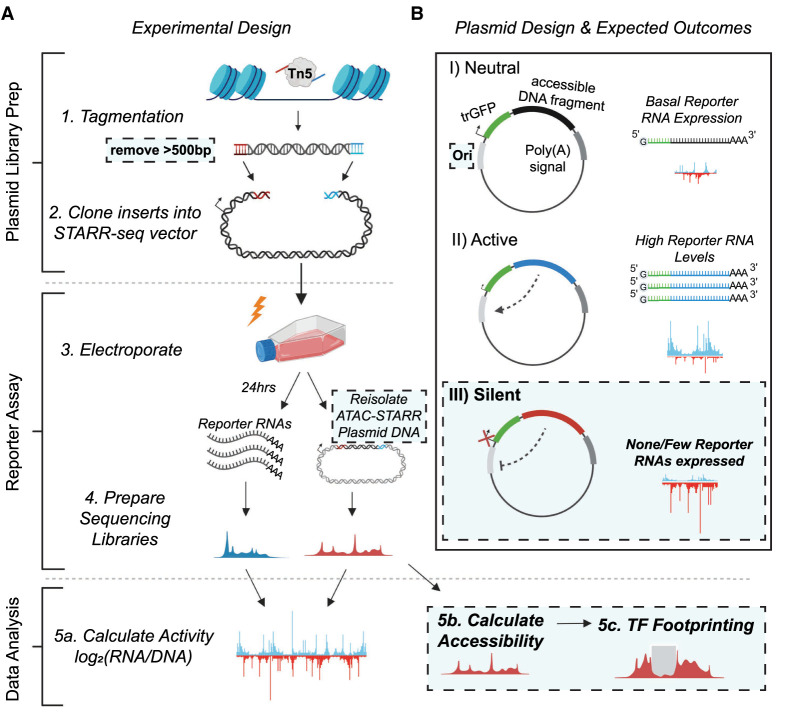
The ATAC-STARR-seq approach is divided into the three main parts: (1) ATAC-STARR-seq plasmid library generation; (2) reporter assay; and (3) data analysis (Fig. 1A). To generate ATAC-STARR-seq plasmid libraries, nuclei are isolated from a cell type of interest and exposed to Tn5, the cut-and-paste transposase used in the ATAC-seq method (Buenrostro et al. 2013). Tn5 simultaneously cleaves DNA fragments within accessible chromatin and attaches customizable sequence adapters to their 5′ ends. ATAC-STARR-seq adapters are designed to serve as homology arms for direct Gibson cloning into the STARR-seq reporter plasmid, which enables cloning of accessible DNA fragments en masse. The resulting ATAC-STARR-seq plasmid library consists of millions of unique plasmids each harboring their own unique open chromatin-derived DNA fragment.
Figure 1.
Schematic of the ATAC-STARR-seq methodology. (A) The experimental design of ATAC-STARR-seq consists of three parts: plasmid library generation; reporter assay; and data analysis. Open chromatin is isolated from cells with the cut-and-paste transposase Tn5 and only large DNA fragments (>500 bp) are removed. The open chromatin fragments are cloned into a reporter plasmid and the resulting clones—called an ATAC-STARR-seq plasmid library—are electroporated into cells. Twenty-four hours later, both reporter RNAs (blue)—which are transcribed directly off the ATAC-STARR-seq plasmid—and ATAC-STARR-seq plasmid DNA (red) are harvested, and Illumina sequencing libraries are prepared and sequenced. The resulting ATAC-STARR-seq data are analyzed to extract regulatory activity, chromatin accessibility, and transcription factor footprints. (B) Reporter plasmid design and the expected outcomes for neutral, active, and silent regulatory elements. Each ATAC-STARR-seq plasmid within a library contains a truncated GFP (trGFP) coding sequence, a polyadenylation signal sequence, an origin of replication (Ori) (which moonlights as a minimal core promoter), and the unique open chromatin fragment being assayed. Because the accessible region is contained in the 3′ UTR, the abundance of itself in the transcript pool reflects its activity. In this way, neutral elements do not affect the system and reporter RNAs are expressed at a basal expression level dictated by the minimal core promoter, the Ori. Accessible chromatin fragments that are active express reporter RNAs at a higher level than the basal expression level, whereas silent elements repress the Ori and reporter RNAs are expressed at a lower level than basal expression. Dashed boxes represent new components of the ATAC-STARR-seq assay design and workflow.
In our updated ATAC-STARR-seq workflow, we employ the STARR-seq Ori backbone, where the origin of replication (Ori) functions as the minimal promoter (Supplemental Table S1; Muerdter et al. 2018). Each plasmid in the ATAC-STARR-seq plasmid library contains a truncated GFP (trGFP) coding sequence, a polyadenylation signal sequence, the Ori, and the unique accessible DNA fragment being assayed (Fig. 1B). Critically, the accessible region is cloned into the 3′ UTR, so if the accessible region is active, it interacts with the Ori to drive self-transcription. Thus, an accessible region's level of activity is reflected by its own level of expression. Transcripts from ATAC-STARR-seq plasmids, termed “reporter RNAs,” are expressed at basal levels from the activity of the Ori itself. This allows detection of silencing activity—the inhibition of the basal expression—in this assay.
Following its creation, the ATAC-STARR-seq plasmid library is transfected via electroporation into a given cell line. From the same flask of cells, both reporter RNAs and plasmid DNA are harvested 24 h later, then prepared as Illumina sequencing libraries and sequenced. Activity is calculated as the log2 ratio between normalized read counts from the reporter RNA and plasmid DNA data sets. The re-isolation of plasmid DNA recovers only the ATAC-STARR-seq plasmids that were successfully transfected, thus providing a more accurate representation of the “input” sample than sequencing without transfection. Supplemental Table S1 provides a comparison of experimental and analytical features as well as reported data metrics for the current ATAC-STARR design and previously reported approaches (Wang et al. 2018; Chaudhri et al. 2020).
ATAC-STARR-seq maintains library complexity and nucleosome profiles of Tn5-selected DNA fragments
Following the experimental design outlined above, we tagmented GM12878 cells and generated an ATAC-STARR-seq plasmid library that yielded about 50 million unique accessible DNA fragments (Supplemental Text; Supplemental Fig. S1A). For a total of three replicates, we then transfected the library into GM12878 cells and harvested both reporter RNAs and plasmid DNA from the same flask of cells 24 h later. We chose 24 h post-transfection to avoid significant effects from the plasmid-induced interferon gene response and to ensure that the data reflects steady-state regulatory properties of GM12878 accessible regions (Supplemental Text; Supplemental Fig. S1B; Muerdter et al. 2018). Using the captured reporter RNAs and plasmid DNA, we prepared Illumina sequencing libraries for each replicate and submitted for sequencing.
The size distribution of the accessible DNA fragments remained consistent throughout the ATAC-STARR-seq procedure and displayed the characteristic nucleosome banding and DNA pitch typified by ATAC-seq fragment libraries (Supplemental Fig. S2A,B). Analysis of library complexity between replicates revealed an average maximum complexity of 90 million unique fragments for input DNA and 10 million unique fragments for reporter RNAs (Supplemental Fig. S2C). The difference between RNA and DNA complexities is likely due to higher duplication rates in the RNA samples (Supplemental Table S2) driven by both the expression of multiple transcripts per plasmid and more PCR cycles required for the RNA samples. In addition, for both RNA and DNA samples, replicates displayed high Pearson (r2: 0.96–0.99) and Spearman's (ρ: 0.77–0.93) correlation coefficients indicating strong agreement among the three replicates assayed (Supplemental Fig. S3). Altogether, the ATAC-STARR-seq sequence libraries demonstrated the necessary quality and complexity for downstream analysis.
ATAC-STARR-seq faithfully captures chromatin accessibility with high signal-to-noise
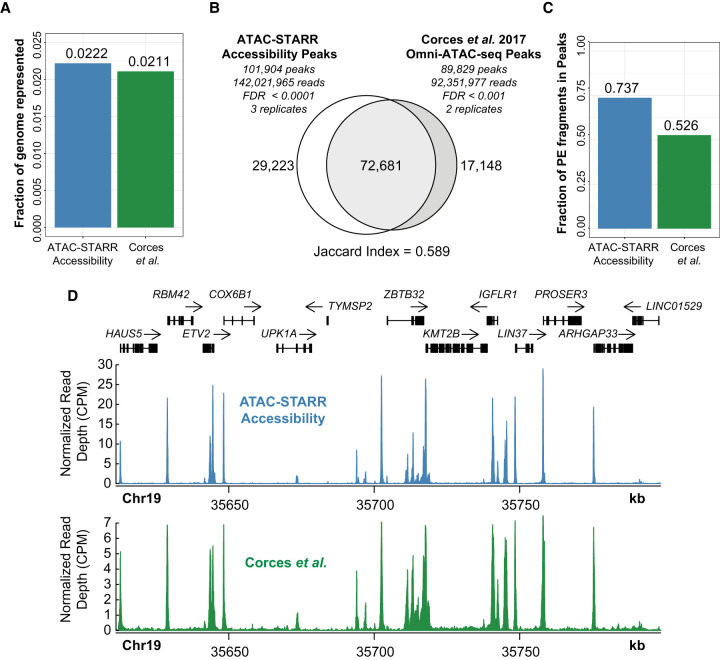
The use of Tn5 on native chromatin to selectively clone chromatin accessible DNA fragments provides the opportunity to quantify not only reporter activity but also chromatin accessibility simultaneously from the same plasmid library. This is because the same DNA fragments sequenced in a typical ATAC-seq workflow are contained in the ATAC-STARR-seq plasmids. Given the insert fragments from re-isolated plasmids are sequenced, we asked if the resulting peak profiles recapitulate native ATAC-seq to measure chromatin accessibility. This is important because, in contrast to a typical ATAC-seq procedure, ATAC-STARR-seq involves several additional steps, including cloning, transfection, and re-isolation, which could distort the content of the library such that it no longer represents its native profile in the genome. Specifically, mapped sequence reads derived from inserts of re-isolated plasmids are counted at a given locus and this estimate infers the accessibility of the region at the time of tagmentation. This also reflects the number of plasmids that represent a given region within the re-isolated ATAC-STARR-seq plasmid library. To test this, we processed the re-isolated plasmid DNA as an Omni-ATAC-seq data set and benchmarked against the GM12878 Omni-ATAC-seq data set from Corces et al. (2017). Raw sequences obtained for both data sets were processed through identical workflows (see Methods). After collapsing read duplicates, we called peaks for each data set using a variety of false-discovery rates (FDRs) (Supplemental Table S3). To closely match the number of peaks previously reported by Corces et al. (2017) (108,433), we chose two separate FDR thresholds—0.0001 for ATAC-STARR-seq and 0.001 for the Corces data—yielding 101,904 and 89,829 accessible chromatin peaks, respectively (Corces et al. 2017). The ATAC-STARR-seq and Corces et al. peak sets represent 2.22% and 2.11% of the genome, respectively, which agrees with previous reports (Fig. 2A; Thurman et al. 2012; Klemm et al. 2019). Overall, 71% of ATAC-STARR-seq peaks are reproduced in the Corces et al. (2017) data set, whereas 81% of Corces et al. peaks overlap the ATAC-STARR-seq data set (Jaccard index = 0.589) (Fig. 2B), indicating strong agreement between these data despite substantial differences in ATAC-STARR DNA sample preparation. Furthermore, the fraction of reads in peaks (FRiP) score, an ENCODE ATAC-seq standard measure of noise, is considerably higher for both ATAC-STARR-seq (0.74) and Corces et al. (2017) (0.526) than the ENCODE accepted standard (>0.2) (Fig. 2C), indicating minimal background in our data set. The high signal-to-noise is also evident when looking at normalized read pileups at a representative locus (Fig. 2D), where the signal mirrors the Corces et al. accessibility signal patterns. Based on these results, we conclude that ATAC-STARR-seq can accurately retain chromatin accessible peaks in the human genome with high signal-to-noise.
Figure 2.
ATAC-STARR-seq accurately quantifies chromatin accessibility. ATAC-seq data from Corces et al. (2017) is compared with ATAC-STARR-seq plasmid DNA data. (A) Fraction of the human genome represented by each peak set. (B) Venn diagram of peak overlap between the two data sets and the associated Jaccard index. (C) Fraction of paired-end (PE) fragments in peaks—FRiP scores—for both samples. (D) Signal tracks comparing counts per million (CPM) normalized read count at a representative locus.
A sliding windows approach increases activity region calling sensitivity
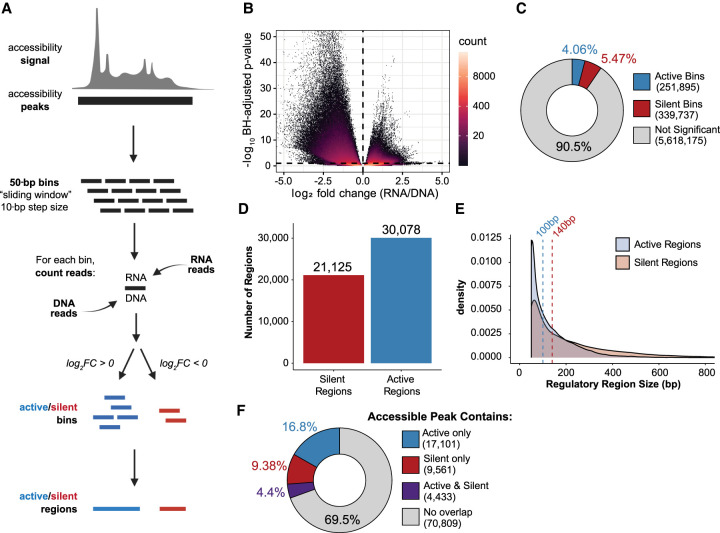
ATAC-STARR-seq tests regulatory activity in DNA enriched for accessible chromatin. Unlike whole genome STARR-seq or other MPRAs, where the genomic DNA fragment distribution is relatively constant, read coverage varies substantially from peak-to-peak in ATAC-STARR-seq. In this way, ATAC-STARR-seq requires an analysis strategy that calls active and silent regulatory regions within accessibility peaks. To address this “peaks-within-peaks” problem, we developed an analytical approach using DESeq2 to normalize reporter RNA read counts to re-isolated plasmid DNA read counts. DESeq2 additionally performs an independent filtering step which removes low count data confounders that can influence ratios and result in false positive peak calls (Love et al. 2014).
We tested two different approaches for regulatory activity analysis. The two approaches differ in how genomic regions are defined prior to differential analysis with DESeq2. Our “sliding window” method defines regions by slicing accessible peaks into 50-bp sliding bins with a 10-bp step size (Fig. 3A). Alternatively, the “fragment group” method, which is the approach used in Wang et al. (2018), synthesizes regions by grouping paired-end sequencing fragments by 75% or greater overlap (Supplemental Fig. S4A). Using a different set of genomic regions, both methods assign and count overlapping RNA and DNA reads to each genomic region and, using DESeq2, identify regions where the RNA count is statistically different from the DNA count at a Benjamini–Hochberg (BH) adjusted P-value < 0.1. The “sliding window” method yielded ∼30,000 distinct active regions, whereas the “fragment groups” method yielded ∼20,000 distinct active regions (Supplemental Fig. S4B). In addition, nearly all active regions defined using the fragment group method (95%) are also captured in the sliding window method regions (Supplemental Fig. S4C). Given this overlap and a 50% greater recovery with the sliding windows approach, we used the sliding windows method to call active ATAC-STARR-seq regulatory regions.
Figure 3.
ATAC-STARR-seq quantifies regulatory activity within accessible chromatin. (A) Schematic of the sliding window peak calling method. Accessibility peaks are chopped into 50-bp bins at a 10-bp step size with the BEDTools makewindows function (options -w 50, -s 10). For each window, RNA and DNA reads are counted using Subread's featureCounts function. Differential analysis comparing RNA and DNA read count is performed with DESeq2. Significant bins are called at a Benjamini–Hochberg (BH) adjusted P-value < 0.1 and parsed into active or silent depending on log2 fold-change (FC) value (± zero). Finally, bins are collapsed into regions using the BEDTools merge function. Log2FC scores are averaged across merged bins. (B) Volcano plot of log2FC scores against –log10-transformed BH adjusted P-value from DESeq2 for all bins analyzed. (C) The proportion of bins called as active or silent. (D) The number of regions defined as either active or silent. (E) Overlapping density plots of active and silent regulatory region size; dashed lines represent the medians in each case. (F) The proportion of accessible peaks that overlap an active or silent region, or both.
Because significance is the primary threshold in our region calling strategy, we examined the influence of replicate count on the number of active regions called (Supplemental Text; Supplemental Fig. S5). We found that, as expected, more replicates result in more active regions. However, we caution that these additional regions may represent a disproportionate number of false positives and may affect the outcomes of certain accuracy-sensitive applications like computational modeling. In this way, we believe that three replicates are sufficient for most purposes. We also investigated the impact of read duplicates on replicate correlations and activity region calling, finding that duplicate removal substantially hindered region calling sensitivity despite yielding higher correlation coefficients between replicates (described in more detail in Supplemental Text; Supplemental Fig. S6).
Both short and long DNA fragments are required for comprehensive region calling
Because DNA fragment synthesis for MPRAs is limited to 200 bp including the adapters and barcode, a significant advantage of ATAC-STARR-seq and other capture-based MPRAs is the ability to measure activity of longer DNA sequences (Santiago-Algarra et al. 2017). To investigate the effect of fragment length on regulatory region calls, we divided mapped reads into short (>125 bp) and long (<125 bp) fragments and independently called active and silent regulatory regions; 125 bp was chosen as it bisects the bimodal peak distribution displayed by RNA and DNA libraries (Supplemental Fig. S2B). Overall, read counts were similar for each sample after splitting into short and long groups (Supplemental Fig. S7A). Two to three times as many active and silent regions were called in the long fragment group compared to the short group (20,833 vs. 10,789 for active and 16,872 vs. 6213 for silent). Nonetheless, a substantial number of regions are called within the short fragment group, although both fell short of the number of active and silent regions called when both long and short were used (Supplemental Fig. S7B). The regulatory regions called using long DNA fragments are larger than those called with short fragments, as expected (Supplemental Fig. S7C); however, they display little difference in TSS distance, indicating that these groups are not comprised of different genomic annotations (Supplemental Fig. S7D). A critical observation is that only 23% of active regions called using short reads overlap active regions called using longer reads, revealing that the two groups identify different regulatory regions in the genome (Supplemental Fig. S7E); this is also true for the silent regulatory regions, although to a lesser extent. Altogether, this analysis reveals that short and long DNA fragments identify different regulatory region sets both in number and similarity. Therefore, to be as comprehensive as possible, STARR-seq assays should be designed to include both short and long DNA fragments rather than impose a size selection to remove smaller fragments.
ATAC-STARR-seq quantifies regulatory activity of open chromatin
In the sliding window approach, bins are classified as active or silent depending on whether RNA is enriched or depleted, respectively, and then like-bins are merged to collapse overlaps (Fig. 3A). Using this approach, we identified ∼590,000 bins where RNA and DNA counts were significantly different (Fig. 3B). More specifically, this analysis identified 251,895 (4.1%) active bins and 339,737 (5.5%) silent bins from the ∼5.6 million total bins measured (Fig. 3C). Overlapping bins were merged into 30,078 active and 21,125 silent regulatory regions (Fig. 3D). It is important to note that more silent than active bins are called; however, because silent regions are generally larger (Fig. 3E), merging overlapping bins results in fewer silent regions than active. Collectively, the active and silent bins represent ∼9.5% of all bins measured, indicating that the majority of accessible DNA is transcriptionally neutral. Moreover, most accessible peaks do not have an active or silent region contained within them (69.5%), suggesting that most accessible regions are neutral regulatory regions according to our assay (Fig. 3F). This suggests that the majority of accessible DNA has no regulatory potential in this cellular context or, alternatively, that ATAC-STARR-seq is not sensitive enough to measure weakly active or weakly silent regions. A recent study in mouse embryonic stem cells made the same observation using an orthogonal approach, suggesting this phenomenon is present in other mammalian species (Glaser et al. 2021). We note that a small percentage of accessible peaks (4.4%) contain both active and silent regions, demonstrating that there can be competing regulatory regions within the same accessible peak.
Active and silent ATAC-STARR-seq regions represent both proximal and distal cis-regulatory elements, and lie within functional chromatin states
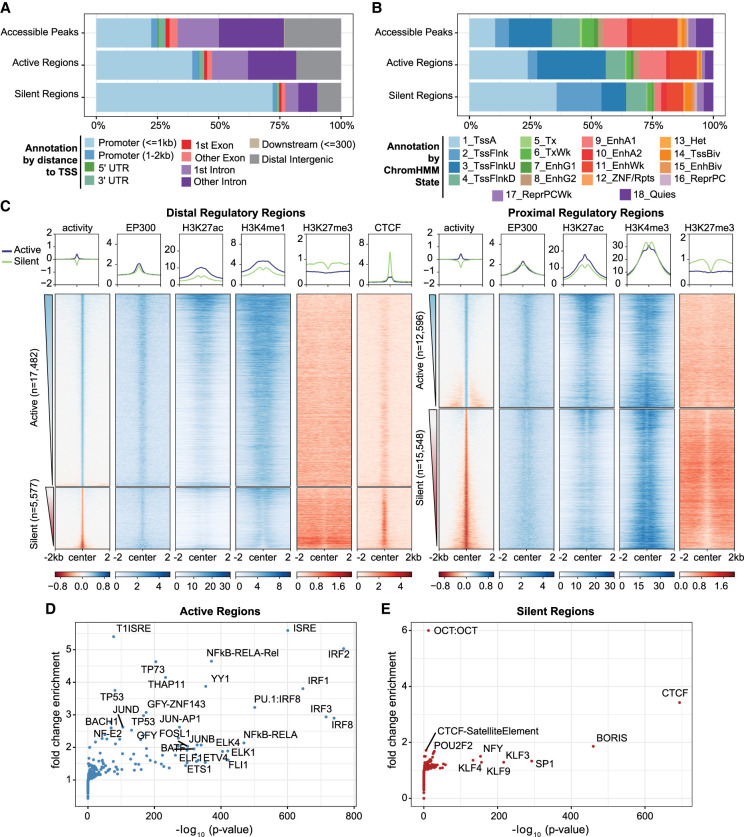
To gain insight into the regulatory features of active regions, we annotated both active and silent regions according to genomic location. Active regions are found in both promoter proximal and distal areas of the genome, with a majority occurring in intronic and intergenic sites (∼55%), whereas silent regions coincide primarily with promoters (∼75%) (Fig. 4A). Functional classification of active and silent regions by the 18-state ChromHMM model (Roadmap Epigenomics Consortium et al. 2015) revealed that active regions consist of TSS active, TSS flanking upstream, and Enhancer Active 1 chromatin states and are devoid of repressive states like Repressed Polycomb Weak and Quiescent (Fig. 4B). In contrast, silent regions are slightly enriched for bivalent chromatin states (TSSBiv, EnhBiv), consistent with the observation that they are accessible but not active. Most silent regions also coincide with TSS Active and TSSFlnk ChromHMM states, which corroborates their promoter proximal locations; however, their designation as “active” by ChromHMM is somewhat puzzling considering these DNA fragments do not drive transcription in our assay. One explanation is that silent regulatory activity, as measured by episomal-based reporter assays, does not fully copy regulatory activity as predicted by ChromHMM. Alternatively, active promoters may confound the reporter assay by initiating transcription from the 3′ UTR of the plasmid, causing conflicts with active transcription from the Ori.
Figure 4.
Regulatory regions defined by ATAC-STARR exhibit annotations, histone modifications, and TFs characteristic of their function. (A) Annotation of regulatory regions relative to the transcriptional start site (TSS). The promoter is defined as 2 kb upstream and 1 kb downstream of the TSS. (B) Annotation of regulatory regions by the ChromHMM 18-state model for GM12878 cells. (C) Heat maps of GM12878 ENCODE ChIP-seq signal and regulatory activity for proximal and distal ATAC-STARR-defined regulatory regions. Proximal regions were classified as within 2 kb upstream and 1 kb downstream of a TSS; all other regions were annotated as distal. Active and silent regions were ranked by mean activity signal for both proximal and distal regions. (D,E) Transcription factor motif enrichment analysis as quantified by HOMER. Fold-change values are relative to the default background calculated by HOMER.
To further investigate if silent regions are a result of 3′ UTR transcription initiation, we considered if an orientation bias existed in reporter RNA levels. If 3′ UTR transcription conflicts exist, we would expect many fewer reporter RNAs when transcription results in head-on conflicts rather than occurring in the same direction as the Ori. We therefore subset reads based on whether they arose from an insert cloned in a 3′–5′ direction or in a 5′–3′ direction (Supplemental Fig. S8A). We then assigned read counts to all bins analyzed (Supplemental Fig. S8B,C), the bins called active (Supplemental Fig. S8D,E), or the bins called silent (Supplemental Fig. S8F,G). Because this is expected to be a promoter-specific effect, we also split bins into proximal and distal based on location to the nearest transcription start site. In all cases, more than 95% of the bins do not display an orientation bias, which we defined as a normalized read count difference greater than five between orientations (Supplemental Methods; Supplemental Fig. S8H). Moreover, we observe high Pearson and Spearman's correlation coefficients between orientations for all conditions (r2: 0.80–0.91 and ρ: 0.73–0.90), and the minimal contribution of orientation bias to silent regions is in agreement with a previous report (Klein et al. 2020). For the <5% of regions that do display orientation bias, proximal bins are more affected than distal bins, as expected. Altogether, ATAC-STARR-seq does not display a significant orientation bias and most of the 21,000 silent regions we observe result from legitimate silencing activity or another source.
Active and silent ATAC-STARR-seq regions are distinct functional classes, and are enriched for specific histone modifications and TF motifs
To further investigate the chromatin landscape of the active and silent regions, we plotted ENCODE GM12878 ChIP-seq signal (The ENCODE Project Consortium et al. 2020) for EP300, CTCF, and histone modifications associated with active and repressed chromatin states (Fig. 4C). As expected, active regions contain EP300 at their center with histone 3 lysine 27 acetylation (H3K27ac) more broadly distributed across the center; histone 3 lysine 4 mono-methylation (H3K4me1) is also present at distal regions, whereas histone 3 lysine 4 tri-methylation (H3K4me3) is at proximal regions. In addition, histone 3 lysine 27 tri-methylation (H3K27me3)—a bivalent repressive mark—is largely absent from active regions. Proximal silent regions, on the other hand, are enriched for H3K27me3 and H3K4me3. This suggests many of the proximal silent regions are accessible bivalent regulatory elements in lymphoblastoid cells. To support their designation as silent calls, we compared histone modification signal at accessible peaks that contain either a silent region, an active region, both a silent and active region, or neither, which we define as neutral accessible peaks (Supplemental Fig. S9A). Consistent with the observations above, silent accessible peaks contain more H3K27me3 signal and are devoid of H3K27ac signal relative to the other accessible peak types.
It is important to note that silent regions are distinct from neutral regions, which are defined as regions failing to reach significance in the RNA-DNA differential analysis. Overall, neutral regions exhibit baseline levels of histone modifications and distribution in genomic annotations like that of all accessible peaks (Supplemental Figs. S4A,B, S9B,C). Although neutral regions represent the majority of accessible peaks, it is possible that a subset are weak enhancers, as indicated by overlap with ChromHMM states, or regulatory elements that display activity in a different cellular context.
Our analysis of TF motifs within active and silent regions revealed prominent differences in motif enrichment. Distal silent regions are strongly enriched for CTCF and its counterpart BORIS, which is associated with diverse functions including gene repression and insulator activity (Fig. 4D,E; Kim et al. 2015). In addition, we found enrichment for the SP/KLF family, several of which are known to be transcriptional repressors (Cao et al. 2010). In contrast, the most enriched TFs in active regions were the IRF family, the ETS family, subunits of the NF-kB complex, and general promoter TFs such as THAP11 and YY1. These data are consistent with our current understanding of immune gene regulation and regulatory element function, which together corroborates the quantification of regulatory activity with ATAC-STARR-seq.
ATAC-STARR-seq retains the ability to map in vivo TF binding
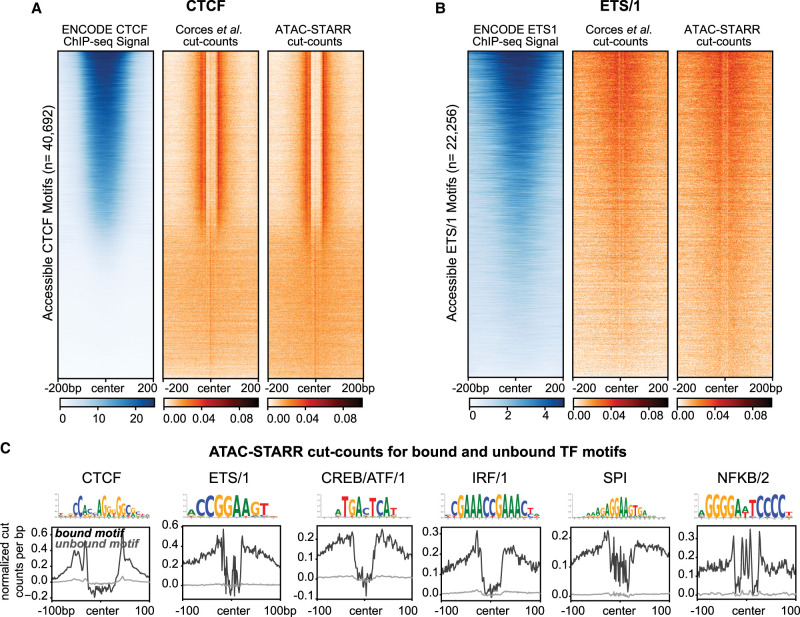
An inherent advantage of an ATAC-seq-based approach is the ability to perform TF footprinting. Computational footprinting methods identify Tn5 cleavage events or “cut sites” from ATAC-seq data and, when combined with motif analysis, can identify TF binding sites with high accuracy (Bentsen et al. 2020; Yan et al. 2020). Because ATAC-STARR-seq produces similar high-quality chromatin accessibility peak profiles as standard ATAC-seq, we explored whether TF footprints were also preserved. We generated Tn5-bias corrected cut site signal files for both Corces et al. (2017) and ATAC-STARR-seq accessibility data sets and plotted cut site signal at all accessible CTCF motifs (Fig. 5A; Bentsen et al. 2020). As a control, we also plotted GM12878 CTCF ChIP-seq signal from ENCODE and ranked region order by the highest mean ChIP-seq signal. We observed consistent depletion of Tn5 cut sites for both Corces et al. (2017) and ATAC-STARR-seq accessibility at CTCF sites. Moreover, we only observe footprints at motifs with CTCF ChIP-seq signal, demonstrating the utility of TF footprinting to determine motifs that are bound or unbound by TFs. Given the importance of TFs in driving enhancer function, this distinction is vital when dissecting transcriptional regulation in human cells.
Figure 5.
ATAC-STARR-seq identifies transcription factor footprints. (A) Comparison of ENCODE CTCF ChIP-seq signal to Corces et al. (2017) and ATAC-STARR-seq cut count signal for all accessible CTCF motifs. (B) Comparison of ENCODE ETS1 ChIP-seq signal to Corces et al. (2017) and ATAC-STARR-seq cut count signal for all accessible motifs with the ETS/1 motif archetype. For both, regions were ranked by largest mean ChIP-seq signal. (C) Aggregate plots representing mean signal for the TOBIAS-defined bound and unbound motif archetypes: CTCF, ETS/1, CREB/ATF/1, IRF/1, SPI, NFKB/2.
TF motif enrichment analysis pointed to multiple ETS family members, including ETS1, which is an important immune cell regulator (Fig. 4D; Garrett-Sinha 2013). So, we asked whether ETS1 footprints are also present in our data. Unlike CTCF, ETS1 shares its motif with many other transcription factors, such as ETV4; therefore, footprinting cannot distinguish ETS1 and ETV4 binding sites. For this reason, we refer to TFs using their ENCODE-defined “archetypes,” which reflects the group of TFs that share the same motif (Vierstra et al. 2020). For each archetype, we performed footprinting against one of the TFs within an archetype to infer motifs bound by members of the group, such as ETS1 for the ETS/1 archetype. To assess the extent to which ETS1 footprints can be explained by ETS1 binding, we plotted GM12878 ETS1 ChIP-seq signal from ENCODE within both Corces et al. (2017) and ATAC-STARR-seq cut sites (Fig. 5B). Indeed, ETS1 ChIP-seq signal explains the majority but not all the ETS/1 footprints present. We observe similar cut site signal to Corces et al. (2017), further indicating that ATAC-STARR-seq can detect in vivo binding of transcription factors despite the additional cloning and transfection steps involved in producing ATAC-STARR-seq DNA libraries.
We performed footprinting for several more immune-related TF archetypes to identify bound or unbound TF motifs (Fig. 5C). For all TFs, bound motifs display substantially larger footprint depth than unbound motifs. Together, this indicates that ATAC-STARR-seq, when combined with footprinting, can identify regions of the genome where TFs are bound. This additional level of information can be leveraged in conjunction with accessibility and activity to understand the context of TF binding while circumventing the need to perform individual chromatin immunoprecipitations.
Collective profiling of accessibility, in vivo TF binding, and activity with ATAC-STARR-seq reveals distinct networks of gene regulation
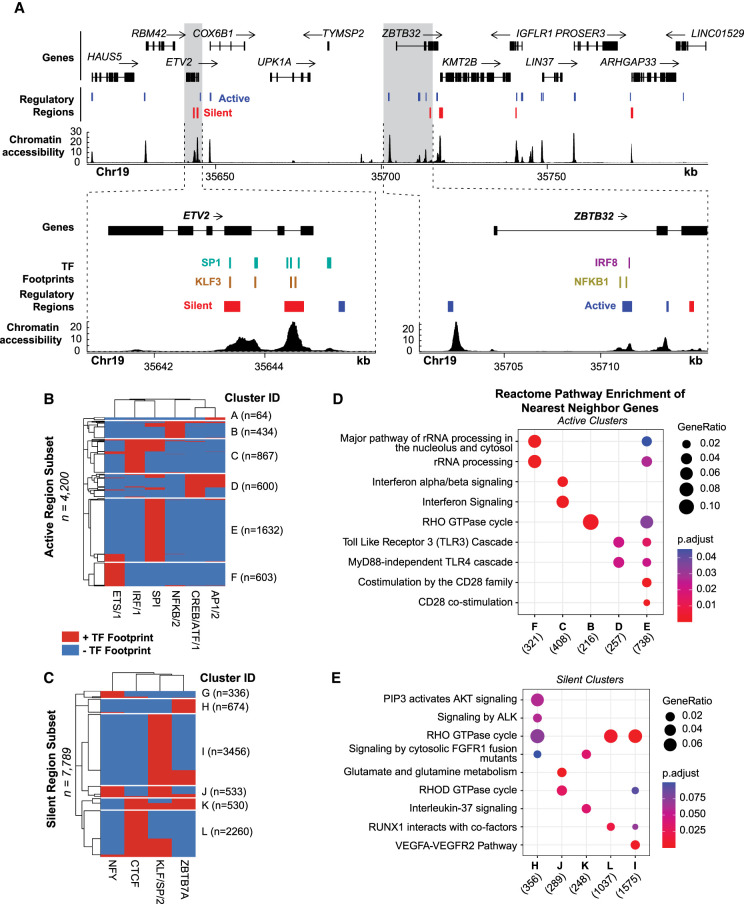
Interrogating chromatin accessibility, TF binding, and regulatory activity together can be used to interpret locus-specific gene regulatory mechanisms. For example, active regulatory elements surrounding the B cell–specific expressed gene ZBTB32 overlap IRF8 and NFKB1 footprints, suggesting these regions are regulated by IRF8 and NFKB1 binding (Fig. 6A). We also observe SP1 and KLF3 footprints overlapping a silent region at the ETV2 locus, a gene lowly expressed in B cells, according to the Human Protein Atlas (Uhlen et al. 2015, 2019). Together, this indicates that active and silent regions can, in part, be explained by the occupancy of these TFs.
Figure 6.
TF footprints stratify ATAC-STARR-defined regulatory regions into gene regulatory networks. (A) ATAC-STARR-defined chromatin accessibility, TF footprints, and regulatory regions at Chr 19: 35,611,232–35,798,446 (hg38). Signal tracks represent counts per million normalized read depth of chromatin accessibility. Zooms into ETV2 and ZBTB32 show that some regulatory regions are occupied by a SP1, KLF3, IRF8, or NFKB1 footprint. (B,C) Heat maps of clustered (B) active and (C) silent regions based on presence or absence of footprints for select TF motif archetypes. (D,E) Reactome pathway enrichment analysis for nearest-neighbor gene sets for each of the clusters. Genes counts for each cluster are displayed below their group identifier.
To demonstrate the power of integrating TF footprints and regulatory regions on a global scale, we clustered active and silent regions based on the presence or absence of several TF footprints (Fig. 6B,C). Footprints were selected based on top hits from the previous motif enrichment analysis (Fig. 4D,E). Regulatory activity may be driven by one or multiple TF binding events that define the cluster and are representative of a gene regulatory network in the genome. Indeed, we find that the putative target genes regulated by each unique group are enriched for distinct gene regulatory pathways and are often related to the TFs in the cluster (Fig. 6D,E). For example, cluster C is primarily defined by the presence of IRF/1 and is enriched for interferon alpha/beta signaling. It is interesting that active clusters tend to be more associated with B cell function than silent clusters, which are more associated with general, non–B cell–related pathways.
Altogether, these distinct gene regulatory networks provide an additional layer of insight into the mechanisms that control gene expression and showcase how integration of the multiple layers of gene regulatory information provided by ATAC-STARR-seq can narrow the focus of gene targets for active and silent regions. We envision such an analysis could be used to interpret the functional consequences of a dysregulated transcription factor or disease-associated genetic variants. We provide this level of detail from a single data set, which further demonstrates the strong potential of our workflow to reveal distinct functional layers of human gene regulation. The resolution we achieve here would not be possible without all three levels of regulatory information provided by ATAC-STARR-seq.
Discussion
Genome-wide approaches that integrate measurements of multiple layers of gene regulation are needed to better understand enhancer function. By combining ATAC-seq with STARR-seq, ATAC-STARR-seq assays regulatory activity only within the context of accessible chromatin. This allows deeper coverage of regulatory elements by narrowing the scope but remaining inclusive of nearly all active regulatory elements. In this report, we substantially expand the capabilities of ATAC-STARR-seq and present an improved workflow which uniquely permits simultaneous profiling of accessibility, TF occupancy, and regulatory activity from a single DNA fragment source. Specifically, we implement key experimental and analytical improvements and present data rationalizing the decisions we make. Experimentally, we adapt a modified tagmentation protocol (Omni-ATAC) to remove mitochondrial DNA from the DNA fragment pool. We also utilize the Ori as the minimal promoter on the STARR-seq backbone which improves reporter RNA expression, recovery, and dynamic range over the super core promoter (SCP1) backbone (Muerdter et al. 2018; Klein et al. 2020). Furthermore, we re-isolate the transfected plasmid DNA to capture only the DNA that is available to cells, which is a more accurate measure of the input than sequencing prior to transfection. Re-isolating plasmid DNA drives a greater degree of variance between samples and better reflects a true experimental replicate than sequencing the same DNA sample for each RNA replicate. Finally, we show that replicate number and inclusion of long and short fragment sizes are critical for comprehensive region calling.
Critically, we developed and tested a simple and sensitive region calling strategy that improves detection of regulatory regions including silencers. We also quantify chromatin accessibility and identify TF footprints, which is surprising given the added processing steps in ATAC-STARR-seq including cloning, transfection, and recapture of DNA libraries that can dull or degrade footprint signal. This enabled us to stratify the active and silent regulatory regions into distinct gene regulatory networks defined by the presence of one or multiple TF footprints. Such an analysis typically requires multiple genomic sequencing assays, but we do this using a single data set.
With this improved workflow, we identified 30,078 active regions and 21,125 silent regions in lymphoblastoid cells. Most active regions were distal to transcription start sites, enriched for functional active ChromHMM states, and were enriched for known B cell–regulating TF motifs such as IRF8 and NF-kB. In contrast, the silencers are proximal to transcription start sites and enriched for CTCF and the SP/KLF TF family. Silent regions are also enriched for the bivalent marks H3K27me3 and H3K4me3 and may represent regulatory regions that are poised, particularly at promoters. Because our plasmid design places regulatory regions within the 3′ UTR of the truncated reporter gene, it is possible that the lack of observed reporter RNAs at silent regions is a result of head-on transcriptional conflicts that arise from antisense transcription initiation from the 3′ UTR. However, we show this minimally occurs in our system and the silent regions reflect true silencing activity or another source that has yet to be identified. Although further studies may be needed to validate these silent regions, this work confirms that the silencers are a distinct class of regulatory element with distinct properties compared to active and neutral regions and warrant further investigation. Even with an increasing number of studies targeted at identifying silencers in the human genome, silencing regulatory regions remain an understudied aspect of gene regulation and our approach provides a new strategy to investigate these elements on a global scale (Doni Jayavelu et al. 2020; Pang and Snyder 2020; Kim et al. 2021).
ATAC-STARR-seq data has several distinct attributes that require a tailored analysis strategy. Current MPRA bioinformatic tools and pipelines are not tractable for these data, because in ATAC-STARR-seq, the input itself is enriched for accessible chromatin and the read pileup varies considerably within these loci. In this way, the analysis of our data required calling essentially “peaks within peaks.” For this reason, it was critical to (1) normalize RNA to DNA, and (2) avoid regions of low count data, which is why we adapted approaches using DESeq2. We also showed that including PCR duplicates was preferred over collapsing duplicates. In the future, it would be beneficial to introduce a unique molecular identifier to the system—such as the strategy employed by UMI-STARR-seq (Neumayr et al. 2019)—to collapse only the duplicates arising from PCR. Although we show comparisons of analysis strategies here, we believe that more information could be extracted from this and future ATAC-STARR-seq data sets with improved analysis strategies. In recent years, we have seen the development of tailor-made peak callers for whole genome STARR-seq, such as CRADLE (Kim et al. 2021) and STARRPeaker (Lee et al. 2020); a similarly tailored ATAC-STARR-seq peak caller could further improve the capabilities of the method.
Although this study was limited to one condition, there are many potential applications of ATAC-STARR-seq. With the ability to subset enhancers by TF occupancy, ATAC-STARR-seq could be leveraged to investigate enhancer grammar by pairing measurable regulatory activity with multiple TF footprints that drive enhancer function. This approach also has the potential to identify dysfunctional gene regulatory networks in diseases like cancer where neoplastic transformation can be driven by the dysfunction of a specific TF. Additionally, an ATAC-STARR-seq plasmid library may be generated from one cell type and tested in another. This flexibility could be used as a tool to determine context dependent activity or investigate enhancer and TF usage patterns during a differentiation time course.
In this study, we demonstrated that our improved ATAC-STARR-seq workflow is a powerful approach enabling joint quantification of chromatin accessibility, transcription factor occupancy, and regulatory activity. We further demonstrate how this single assay can characterize the human genome at many functional levels from chromatin accessibility to distinct gene regulatory networks. This method provides a state-of-the-art approach to deeply investigate transcriptional regulation of the human genome. We provide a detailed protocol and a well-documented code repository so that ATAC-STARR-seq may be easily used and adapted by the field.
Methods
Cell culture
GM12878 cells were obtained from Coriell and cultured with RPMI 1640 media containing 15% fetal bovine serum, 2 mM GlutaMAX, 100 units/mL penicillin, and 100 μg/mL streptomycin. Cells were cultured at 37°C, 80% relative humidity, and 5% CO2. Cell density was maintained between 0.2 × 106 and 1 × 106 cells/mL with a 50% media change every 2–4 d. All cell lines were regularly screened for mycoplasma contamination using the MycoAlert kit (Lonza).
Plasmids
The hSTARR-seq_ORI plasmid vector was a gift from Alexander Stark (Addgene, plasmid #99296), and the pcDNA3-EGFP was a gift from Doug Golenbock (Addgene, plasmid #13031). The bacterial stabs from Addgene were spread onto an LB agar plate containing 100 μg/mL ampicillin and incubated overnight at 37°C. For each, a single colony was picked and grown in 50 mL LB containing 100 μg/mL ampicillin overnight at 37°C while shaking at 225 rpm. Plasmid DNA was extracted using the ZymoPURE II Plasmid Midiprep kit (Zymo Research, #D4200).
The linear vector used in the ATAC-STARR-seq Gibson cloning step was generated by a single 50-μL PCR reaction using NEBNext Ultra II Q5 Master Mix (NEB, #M0544S). Although not necessary for this study, primers were designed to add the i5 barcode to the linearized vector; this allows for different ATAC-STARR-seq plasmid libraries to be pooled and tracked. Following this approach, a universal forward primer (Fwd_universal_STARR) and a reverse primer (Rev_N504_STARR) designed to add the N504 barcode were used (primer sequences are provided in Supplemental Table S4). PCR products were purified with the Zymo Research DNA Clean & Concentrator-5 kit. DNA yield was determined by NanoDrop, and purity was analyzed by gel electrophoresis; the linearized vector was the only product observed on the gel.
Tagmentation
A total of eight tagmentation reactions were performed on 50,000 GM12878 cells each. We followed a slightly modified version of the Omni-ATAC approach used in Corces et al. (2017). Specifically, twice as much Tn5 than described in the protocol was used. Standard Tn5 transposase was prepared in-house following the method described in Picelli et al. (2014). Standard Tn5 transposome was assembled as described in Barnett et al. (2020) with the following oligos: Tn5_1, Tn5_2_ME_comp, and TN5MEREV. Tagmented products were pooled together and purified with the Zymo Research DNA Clean & Concentrator-5 kit (#D4013). The entire elution was split and amplified via five 10-μL PCR reactions. We used NEBNext High-Fidelity 2× PCR Master Mix (#M0541S)—which is not a hot-start formulation—to first extend tagments before the initial denaturation step of PCR via the following cycling parameters: 72°C for 5 min, 98°C for 30 sec; four cycles of 98°C for 10 sec, 62°C for 30 sec, 72°C for 60 sec; final extension 72°C for 2 min; hold at 10°C. Forward and reverse primer sequences, Fwd_atac-starr_tag and Rev_atac-starr_tag, are provided in Supplemental Table S3. Amplified products were purified with the Zymo Research DNA Clean & Concentrator-5 kit and then analyzed for concentration and size distribution with a HSD5000 screentape (Agilent, #5067) on an Agilent 4150 TapeStation system. After amplification, we selected PCR products <500 bp using SPRISelect beads (Beckman-Coulter, #B23317) at a 0.6× volume ratio of beads:sample. Selection was verified using a HSD5000 screentape.
Massively parallel cloning
Four 10-μL Gibson cloning reactions were performed with NEBuilder HiFi DNA Assembly Master Mix at a vector:insert molar ratio of 1:2. As a negative control, we performed one cloning reaction substituting tagments with nuclease-free water. Gibson products were pooled and purified via ethanol precipitation as previously described in Sambrook and Russell (2006); we used glycoblue (150 μg/mL) as a coprecipitant. Purified Gibson products were electroporated into MegaX DH10B T1R Electrocomp cells (Invitrogen, #C640003) using a Bio-Rad Gene Pulser. Three electroporations for the ATAC-STARR-seq sample (and one for the control) were performed with the following parameters: exponential decay pulse type, 2 kV, 200 Ω, 25 μF, and 0.1-cm gap distance. Prewarmed SOC media (1 mL) was added immediately following electroporation; the three reactions were pooled and incubated for 1 h at 37°C. We confirmed cloning success by plating a dilution series—using a small aliquot from the ATAC-STARR-seq and negative control samples—onto prewarmed LB agar plates containing 100 μg/mL ampicillin and visualizing colonies 24 h later. The remaining ATAC-STARR-seq transformation was added directly to a 1-L LB liquid culture with 100 μg/mL ampicillin and grown overnight at 37°C while shaking at 225 rpm. The next day, plasmid DNA was harvested from the 1-L culture using the ZymoPURE II Plasmid Gigaprep (Zymo Research, #D4204). Before prepping, we recorded a 1.633 optical density.
Electroporation
GM12878 cells were cultured so that cell density was between 400,000 and 800,000 cells/mL on the day of transfection. Three replicates were performed on separate days. For each replicate, a total of 20 electroporation reactions was performed using the Neon Transfection System 100 µL kit (Invitrogen, #MPK10025) and the associated Neon Transfection System (Invitrogen, #MPK5000); 121 million GM12878 cells were collected, washed with 45 mL PBS, and resuspended in 2178 μL Buffer R. For each reaction, 5 million cells (in 90 μL Buffer R) were electroporated with 5 μg of ATAC-STARR-seq plasmid DNA (in 10 μL nuclease-free water) in a total volume of 100 μL with the following parameters: 1100 V, 30 ms, and 2 pulses. Electroporated cells were dispensed immediately into a prewarmed T-75 flask containing 50 mL of RPMI 1640 with 20% fetal bovine serum and 2 mM GlutaMAX.
Cell harvest
Twenty-four hours after transfection, the 50-mL ATAC-STARR-seq flask was divided into two equal volumes; plasmid DNA was extracted from one volume, and reporter RNAs were extracted from the other. Plasmid DNA was isolated with the ZymoPURE II Plasmid Midiprep kit (#D4200) and eluted in 50 μL 10 mM Tris-HCL, pH 8.0. Prior to lysis, cells were washed with 25 mL PBS to remove any extracellular plasmid DNA. Reporter RNAs were extracted in a stepwise process. First, total RNA was isolated from the second volume of transfected cells using the TRIzol Reagent and Phasemaker Tubes Complete System (Invitrogen, #A33251). Specifically, 5 mL TRIzol were added to homogenize the washed and pelleted cells. Next, polyadenylated RNA was isolated from total RNA using oligo(dT)25 Magnetic Beads (NEB, #S1419S) at a 1 μg total RNA to 10 μg beads ratio. We performed this step at 4°C and eluted into 50 μL 10 mM Tris-HCl, pH 7.5. The extracted poly(A)+ RNA was treated with DNase I (NEB, #M0303S). This reaction was cleaned up using the Zymo Research RNA Clean & Concentrator-25 kit (Zymo Research, #R1018).
First-strand cDNA synthesis
For each sample, 10 50-μL reverse transcription reactions were carried out using PrimeScript Reverse Transcriptase (Takara, #2680) and a gene specific primer (STARR_GSP) as described by Muerdter et al. (2018). Single-stranded cDNA was treated with RNase A at a concentration of 20 μg/mL in low-salt concentrations and cleaned up with a Zymo Research DNA Clean & Concentrator-5 kit.
Illumina sequencing library preparation
For re-isolated plasmid and reporter RNA samples, Illumina-compatible libraries were generated using NEBNext Ultra II Q5 Master Mix and a unique combination of the following Nextera indexes: N504-N505 (i5) and N701-N702 (i7); see Supplemental Table S1 for primer sequences. DNA samples were amplified for eight PCR cycles, and RNA was amplified for 12–13 cycles. In both cases, products were purified with the Zymo Research DNA Clean & Concentrator-5 kit and analyzed for concentration and size distribution using a HSD5000 screentape. Purified products were sequenced on an Illumina NovaSeq, PE150, at a requested read depth of 50 or 75 million reads, for DNA and RNA samples, respectively, on an Illumina NovaSeq 6000 machine through the Vanderbilt Technology for Advanced Genomics (VANTAGE) sequencing core. Reads were processed and analyzed as described in the Supplemental Methods. We provide guidelines for ATAC-STAR-seq quality control in the Supplemental Text.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE181317. Python scripts and additional code for ATAC-STARR-seq data analysis are available at GitHub (https:// github.com/HodgesGenomicsLab/ATAC-STARR-seq) and as Supplemental Code. An interactive version of the protocol is posted on protocols.io (https://www.protocols.io/view/atac-starr-seq-b2nuqdew.html) and a PDF version of the protocol at publication date is included as a Supplemental File.
Supplementary Material
Acknowledgments
We thank Felix Muerdter, Sarah Fong, Tony Capra, and members of the Hodges Lab, especially Kelly Barnett, Tim Scott, Lindsey Guerin, Verda Agan, Elizabeth Dorans, and Ali Wilt for helpful feedback and discussions. We also thank Biorender.com for illustrations, Addgene for plasmids used in the study, the Dave Cortez Lab for use of their Bio-Rad Gene Pulser, and the Manny Ascano Lab for qPCR primers and helpful advice. We acknowledge support of the project and the time invested in producing this manuscript by the National Institutes of Health awards (K22 CA184308-03 to E.H.), Department of Defense Idea Award (W81XWH-20-1-0522 to E.H.), and American Cancer Society (ACS) Institutional Research Grant (#IRG-15-169-56).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276766.122.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Arnold CD, Gerlach D, Stelzer C, Boryń LM, Rath M, Stark A. 2013. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339: 1074–1077. 10.1126/science.1232542 [DOI] [PubMed] [Google Scholar]
- Barnett KR, Decato BE, Scott TJ, Hansen TJ, Chen B, Attalla J, Smith AD, Hodges E. 2020. ATAC-Me captures prolonged DNA methylation of dynamic chromatin accessibility loci during cell fate transitions. Mol Cell 77: 1350–1364.e6. 10.1016/j.molcel.2020.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsen M, Goymann P, Schultheis H, Klee K, Petrova A, Wiegandt R, Fust A, Preussner J, Kuenne C, Braun T, et al. 2020. ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat Commun 11: 4267. 10.1038/s41467-020-18035-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Sun X, Icli B, Wara AK, Feinberg MW. 2010. Role of Krüppel-like factors in leukocyte development, function, and disease. Blood 116: 4404–4414. 10.1182/blood-2010-05-285353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhri VK, Dienger-Stambaugh K, Wu Z, Shrestha M, Singh H. 2020. Charting the cis-regulome of activated B cells by coupling structural and functional genomics. Nat Immunol 21: 210–220. 10.1038/s41590-019-0565-0 [DOI] [PubMed] [Google Scholar]
- Chen AF, Parks B, Kathiria AS, Ober-Reynolds B, Goronzy JJ, Greenleaf WJ. 2022. NEAT-seq: simultaneous profiling of intra-nuclear proteins, chromatin accessibility and gene expression in single cells. Nat Methods 19: 547–553. 10.1038/s41592-022-01461-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Argelaguet R, Kapourani CA, Stubbs TM, Lee HJ, Alda-Catalinas C, Krueger F, Sanguinetti G, Kelsey G, Marioni JC, et al. 2018. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat Commun 9: 781. 10.1038/s41467-018-03149-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, et al. 2017. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14: 959–962. 10.1038/nmeth.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doni Jayavelu N, Jajodia A, Mishra A, Hawkins RD. 2020. Candidate silencer elements for the human and mouse genomes. Nat Commun 11: 1061. 10.1038/s41467-020-14853-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Moore JE, Purcaro MJ, Pratt HE, Epstein CB, Shoresh N, Adrian J, Kawli T, Davis CA, Dobin A, et al. 2020. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583: 699–710. 10.1038/s41586-020-2493-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Sinha LA. 2013. Review of Ets1 structure, function, and roles in immunity. Cell Mol Life Sci 70: 3375–3390. 10.1007/s00018-012-1243-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini M, Tome JM, Shendure J. 2020. Towards a comprehensive catalogue of validated and target-linked human enhancers. Nat Rev Genet 21: 292–310. 10.1038/s41576-019-0209-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser LV, Steiger M, Fuchs A, van Bommel A, Einfeldt E, Chung HR, Vingron M, Meijsing SH. 2021. Assessing genome-wide dynamic changes in enhancer activity during early mESC differentiation by FAIRE-STARR-seq. Nucleic Acids Res 49: 12178–12195. 10.1093/nar/gkab1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Glass CK. 2015. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 16: 144–154. 10.1038/nrm3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue F, Kircher M, Martin B, Cooper GM, Witten DM, McManus MT, Ahituv N, Shendure J. 2017. A systematic comparison reveals substantial differences in chromosomal versus episomal encoding of enhancer activity. Genome Res 27: 38–52. 10.1101/gr.212092.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GD, Barrera A, McDowell IC, D'Ippolito AM, Majoros WH, Vockley CM, Wang X, Allen AS, Reddy TE. 2018. Human genome-wide measurement of drug-responsive regulatory activity. Nat Commun 9: 5317. 10.1038/s41467-018-07607-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TK, Liu Y, Lay FD, Liang G, Berman BP, Jones PA. 2012. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res 22: 2497–2506. 10.1101/gr.143008.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Yu NK, Kaang BK. 2015. CTCF as a multifunctional protein in genome regulation and gene expression. Exp Mol Med 47: e166. 10.1038/emm.2015.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Johnson GD, Seo J, Barrera A, Cowart TN, Majoros WH, Ochoa A, Allen AS, Reddy TE. 2021. Correcting signal biases and detecting regulatory elements in STARR-seq data. Genome Res 31: 877–889. 10.1101/gr.269209.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Xiong C, Martin B, Schubach M, Inoue F, Bell RJA, Costello JF, Shendure J, Ahituv N. 2019. Saturation mutagenesis of twenty disease-associated regulatory elements at single base-pair resolution. Nat Commun 10: 3583. 10.1038/s41467-019-11526-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JC, Agarwal V, Inoue F, Keith A, Martin B, Kircher M, Ahituv N, Shendure J. 2020. A systematic evaluation of the design and context dependencies of massively parallel reporter assays. Nat Methods 17: 1083–1091. 10.1038/s41592-020-0965-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm SL, Shipony Z, Greenleaf WJ. 2019. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet 20: 207–220. 10.1038/s41576-018-0089-8 [DOI] [PubMed] [Google Scholar]
- Lee D, Shi M, Moran J, Wall M, Zhang J, Liu J, Fitzgerald D, Kyono Y, Ma L, White KP, et al. 2020. STARRPeaker: uniform processing and accurate identification of STARR-seq active regions. Genome Biol 21: 298. 10.1186/s13059-020-02194-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricque BB, Dougherty JD, Cohen BA. 2017. A genome-integrated massively parallel reporter assay reveals DNA sequence determinants of cis-regulatory activity in neural cells. Nucleic Acids Res 45: e16. 10.1093/nar/gkw942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, Feizi S, Gnirke A, Callan CG Jr, Kinney JB, et al. 2012. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol 30: 271–277. 10.1038/nbt.2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Boryń LM, Woodfin AR, Neumayr C, Rath M, Zabidi MA, Pagani M, Haberle V, Kazmar T, Catarino RR, et al. 2018. Resolving systematic errors in widely used enhancer activity assays in human cells. Nat Methods 15: 141–149. 10.1038/nmeth.4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumayr C, Pagani M, Stark A, Arnold CD. 2019. STARR-seq and UMI-STARR-seq: assessing enhancer activities for genome-wide-, high-, and low-complexity candidate libraries. Curr Protoc Mol Biol 128: e105. 10.1002/cpmb.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang B, Snyder MP. 2020. Systematic identification of silencers in human cells. Nat Genet 52: 254–263. 10.1038/s41588-020-0578-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan RP, Hiatt JB, Witten DM, Kim MJ, Smith RP, May D, Lee C, Andrie JM, Lee SI, Cooper GM, et al. 2012. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol 30: 265–270. 10.1038/nbt.2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Björklund AK, Reinius B, Sagasser S, Winberg G, Sandberg R. 2014. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res 24: 2033–2040. 10.1101/gr.177881.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. 2006. Standard ethanol precipitation of DNA in microcentrifuge tubes. CSH Protoc 2006: pdb.prot4456. 10.1101/pdb.prot4456 [DOI] [PubMed] [Google Scholar]
- Santiago-Algarra D, Dao LTM, Pradel L, Espana A, Spicuglia S. 2017. Recent advances in high-throughput approaches to dissect enhancer function. F1000Res 6: 939. 10.12688/f1000research.11581.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489: 75–82. 10.1038/nature11232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjöstedt E, Asplund A, et al. 2015. Tissue-based map of the human proteome. Science 347: 1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Uhlen M, Karlsson MJ, Zhong W, Tebani A, Pou C, Mikes J, Lakshmikanth T, Forsström B, Edfors F, Odeberg J, et al. 2019. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science 366: eaax9198. 10.1126/science.aax9198 [DOI] [PubMed] [Google Scholar]
- Vierstra J, Lazar J, Sandstrom R, Halow J, Lee K, Bates D, Diegel M, Dunn D, Neri F, Haugen E, et al. 2020. Global reference mapping of human transcription factor footprints. Nature 583: 729–736. 10.1038/s41586-020-2528-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, He L, Goggin SM, Saadat A, Wang L, Sinnott-Armstrong N, Claussnitzer M, Kellis M. 2018. High-resolution genome-wide functional dissection of transcriptional regulatory regions and nucleotides in human. Nat Commun 9: 5380. 10.1038/s41467-018-07746-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Powell DR, Curtis DJ, Wong NC. 2020. From reads to insight: a hitchhiker's guide to ATAC-seq data analysis. Genome Biol 21: 22. 10.1186/s13059-020-1929-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.