Abstract
Somatic hotspot mutations and structural amplifications and fusions that affect fibroblast growth factor receptor 2 (encoded by FGFR2) occur in multiple types of cancer1. However, clinical responses to FGFR inhibitors have remained variable1–9, emphasizing the need to better understand which FGFR2 alterations are oncogenic and therapeutically targetable. Here we apply transposon-based screening10,11 and tumour modelling in mice12,13, and find that the truncation of exon 18 (E18) of Fgfr2 is a potent driver mutation. Human oncogenomic datasets revealed a diverse set of FGFR2 alterations, including rearrangements, E1–E17 partial amplifications, and E18 nonsense and frameshift mutations, each causing the transcription of E18-truncated FGFR2 (FGFR2ΔE18). Functional in vitro and in vivo examination of a compendium of FGFR2ΔE18 and full-length variants pinpointed FGFR2-E18 truncation as single-driver alteration in cancer. By contrast, the oncogenic competence of FGFR2 full-length amplifications depended on a distinct landscape of cooperating driver genes. This suggests that genomic alterations that generate stable FGFR2ΔE18 variants are actionable therapeutic targets, which we confirmed in preclinical mouse and human tumour models, and in a clinical trial. We propose that cancers containing any FGFR2 variant with a truncated E18 should be considered for FGFR-targeted therapies.
Subject terms: Breast cancer, Cancer models, Growth factor signalling, Oncogenes
Truncation of exon 18 of FGFR2 (FGFR2ΔE18) is a potent driver mutation in mice and humans, and FGFR-targeted therapy should be considered for patients with cancer expressing stable FGFR2ΔE18 variants.
Main
FGFR2 is a receptor tyrosine kinase (RTK) that consists of an extracellular ligand-binding domain, intracellular tyrosine kinase domains and a carboxy (C)-terminal tail relevant for receptor activity fine-tuning14. In human cancers, FGFR2 can be affected by hotspot mutations and structural variants, namely fusions and amplifications1, some of which produce truncated FGFR2 isoforms15–19. FGFR2 structural variants have been considered to be oncogenic and actionable due to the resulting overexpression and increased stabilization of the receptor2,20–22. However, in patients with cancer with such structural variants, ATP-competitive small-molecule inhibitors targeting FGFRs have produced inconsistent clinical benefits1–9. A better understanding of the determinants defining the oncogenicity and clinical actionability of FGFR2 structural variants is therefore critical for precise matching of cancer patients to FGFR-targeted therapies.
A SB screen identified Fgfr2ΔE18
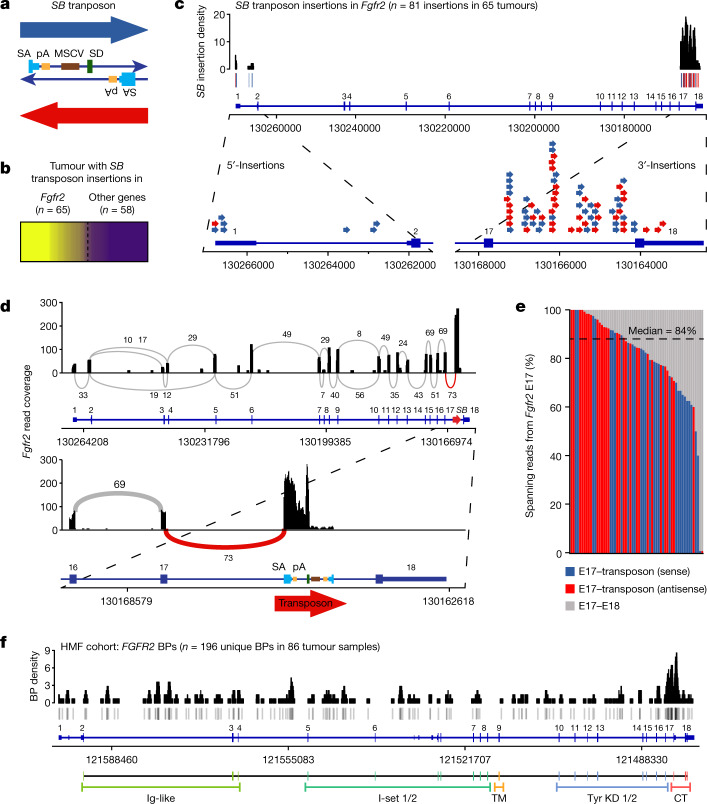
Sleeping Beauty (SB) transposon-based insertional mutagenesis screening has revealed potential tumour drivers in mice through transcriptional activation and/or truncation of target genes, and identified Fgfr2 as a top candidate driver in mammary tumorigenesis10,11 (Fig. 1a,b). Mapping of the SB insertions in Fgfr2 showed strong enrichment for insertions in intron 17 (I17; Fig. 1c and Extended Data Fig. 1a). Analysis of RNA sequencing (RNA-seq) data of SB tumours revealed that Fgfr2-I17 insertions enforce splicing of Fgfr2-E17 into the transposon. This led to Fgfr2 transcripts that lacked E17–E18 spanning reads, and expression of Fgfr2ΔE18 was confirmed by reverse transcription with quantitative PCR (RT–qPCR; Fig. 1d,e and Extended Data Fig. 1b–d). Tumours with SB insertions in the 5′ region of Fgfr2 either contained a second SB insertion in I17 or contained rearrangements (REs) in Fgfr2-I17, producing gene fusions11 and therefore also expressing Fgfr2ΔE18 (Extended Data Fig. 1d). E18 of both mouse and human FGFR2 encodes the C terminus of this RTK14. We observed an overall upregulation of Fgfr2 transcripts in tumours with SB insertions (Extended Data Fig. 1e), suggesting a loss of regulatory elements that are presumably encoded by the Fgfr2 3′-untranslated region (3′-UTR)21 and/or positive oncogenic selection of C-terminally truncated FGFR2.
Fig. 1. SB transposon screen and WGS analysis identifies recurrent FGFR2 E18 truncation.
a, Schematic of the SB transposon, which encodes splice acceptors (SA) followed by polyadenylation (pA) signals on both strands and a murine stem cell virus (MSCV) promoter followed by a splice donor (SD) on the plus strand. b, Mammary tumours with SB transposon insertions in Fgfr2 as identified in an insertional mutagenesis screen10. The relative clonality of SB insertions in Fgfr2 is shown by a colour gradient (yellow to purple, clonality of 1 to 0). SB insertions in Fgfr2 were called for tumours with a Fgfr2 relative insertion clonality of ≥0.25. c, The SB transposon insertions found in Fgfr2 (chromosome 7). The SB insertion density was calculated using a 500 bp sliding window. The blue bars/arrows show sense SB insertions; the red bars/arrows show antisense SB insertions. d, Sashimi plot showing Fgfr2 read coverage and junction reads plotted as arcs with the indicated junction read counts of a tumour with an I17 antisense SB insertion. e, The ratio of spanning reads from Fgfr2-E17 to E18 versus SB transposon in tumours with I17 SB insertions. n = 31 (sense) and n = 30 (antisense). f, BPs generating FGFR2 (chromosome 10) genomic REs identified in 86 out of 2,112 analysed WGS profiles from the HMF cohort on metastatic solid tumours23. n = 266 (total) and n = 196 (unique) BPs. BP density was calculated using a 500 bp sliding window. The grey bars show BPs. Corresponding protein domains are indicated. CT, C terminus; TM, transmembrane; Tyr KD, tyrosine kinase domain.
Extended Data Fig. 1. SB-insertions in Fgfr2 and FGFR2 REs found in the FMI cohort.
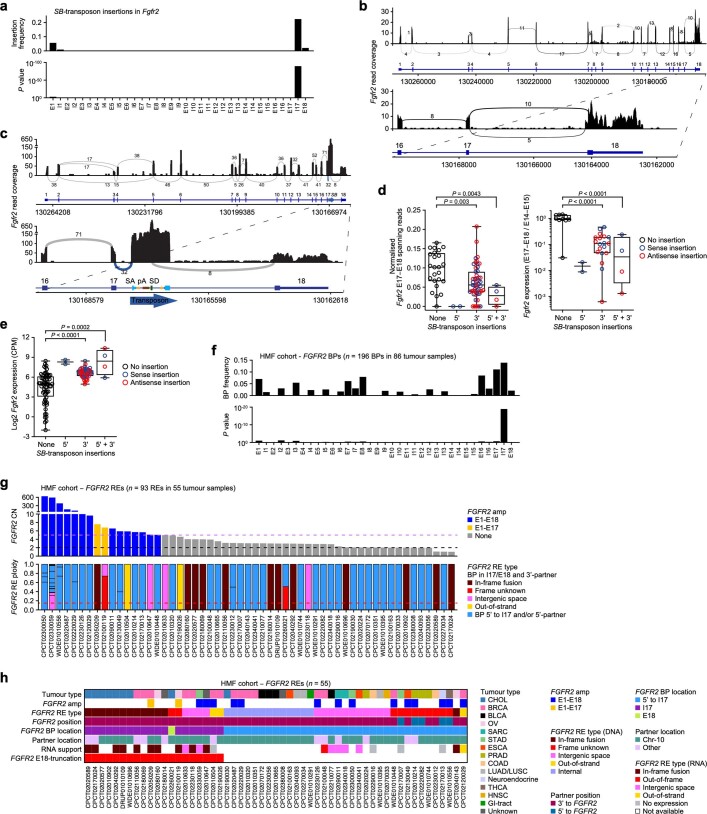
a, Normalized frequency (top panel) and enrichment significance (P values, bottom panel) of Sleeping Beauty (SB) transposon insertions (n = 81 insertions in 65 tumours) in each Fgfr2 exon (E) and intron (I) as identified in mammary tumours from a SB-transposon in vivo screen10. SB insertion frequency was normalized by the kilobase of feature (exon/intron) length and total number of SB-insertions. b, c, Sashimi plots showing Fgfr2 read coverage and junction reads plotted as arcs with indicated junction read counts of tumours with no SB-insertion (b) and an I17 sense SB insertion (c) in Fgfr2. SA, splice acceptor; SD, splice donor; pA, polyadenylation signal. d, Left panel, counts of Fgfr2-E17–E18 spanning reads (counts per million, CPM) normalized to Fgfr2 expression (CPM) in SB tumour RNA sequencing (RNA-seq) profiles (none, n = 24; 5′-sense, n = 2; 3′-sense, n = 22; 3′-antisense, n = 27; 5′ + 3′-sense, n = 2; 5′ + 3′-antisense, n = 2); right panel, RT-qPCR to quantify Fgfr2-E17–E18 over Fgfr2-E14–E15 expression in SB-tumours (none, n = 10; 5′-sense, n = 2; 3′-sense, n = 8; 3′-antisense, n = 11; 5′ + 3′-sense, n = 2; 5’ + 3′-antisense, n = 2; individual dots represent mean of 3 independent measurements). e, Expression of Fgfr2 (CPM) in SB tumour RNA-seq profiles (none, n = 64; 5′-sense, n = 2; 3′-sense, n = 30; 3′-antisense, n = 28; 5′ + 3′-sense, n = 2; 5′ + 3′-antisense, n = 2). f, Normalized frequency (top panel) and enrichment significance (P values, bottom panel) of FGFR2 genomic rearrangement (RE) breakpoints (BPs) identified in 2,112 whole-genome sequencing (WGS) profiles from the Hartwig Medical Foundation (HMF) cohort in each exon/intron. BP frequency was normalized by the kilobase of feature (exon/intron) length and total number of BPs. g, FGFR2 copy numbers (CN, top panel) and RE ploidy frequencies (bottom panel) in samples with FGFR2 REs. FGFR2 BPs resulting in unresolved REs were excluded generating a refined list of REs (n = 93 REs in 55 tumour samples). Dotted lines, black, normal CN; purple, amplified CN (> 5); red, RE ploidy frequency threshold (> 0.15) to call samples with E18-truncating FGFR2 REs (E18-truncating, n = 20; others, n = 35). Amp, amplification. h, FGFR2 RE types found in WGS profiles from HMF. RNA support indicates evidence for FGFR2 REs in matching RNA-seq profiles. Empty fields, no RNA-seq data available. BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CHOL, cholangiocarcinoma; chr, chromosome; COAD, colon adenocarcinoma; ESCA, oesophageal carcinoma; GI, gastro-intestinal; HNSC, head and neck squamous cell carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; PRAD, prostate adenocarcinoma; SARC, sarcoma; STAD, stomach adenocarcinoma; THCA, thyroid carcinoma90. Data in d, e are represented as median (centre line) ± interquartile range (IQR, 25th to 75th percentile, box) and ± full range (minimum to maximum, whiskers). P values were calculated with one-tailed binomial tests (a, f) or one-tailed one-way analysis of variance (ANOVA) and Tukey’s multiple-testing corrections (d, e).
FGFR2ΔE18 variants in human cancer
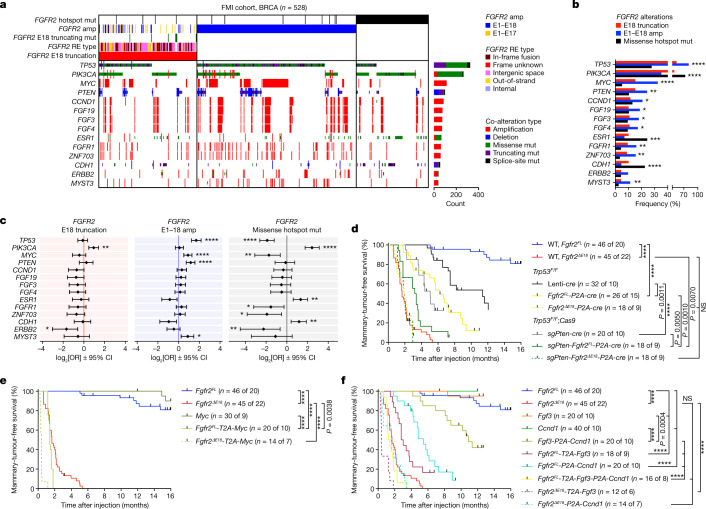
To assess whether genomic alterations producing FGFR2ΔE18 occur in human cancers, we first analysed whole-genome sequencing (WGS) data of metastatic solid tumours from the Hartwig Medical Foundation (HMF)23. Examination of structural variants affecting FGFR2 in 2,112 HMF WGS profiles revealed a significant enrichment of RE breakpoints (BPs) in I17 (Fig. 1f and Extended Data Fig. 1f), coinciding with reported FGFR2 fusion BPs20–22. Recurring chromosomal REs, such as breakage–fusion–bridge cycles, can produce focal FGFR2 amplifications (FGFR2amp)16,17,24, which we observed in a fraction of tumours with FGFR2 REs (Extended Data Fig. 1g). Some FGFR2-I17 REs implicated canonical in-frame fusions with 3′-partner genes, but we also found non-canonical REs in which the reading frame of the partner gene was undeterminable (frame unknown), the partner gene was out of strand, the partner sequence was derived from intergenic space or the REs occurred internally in FGFR2 (Extended Data Fig. 1h and Supplementary Table 1).
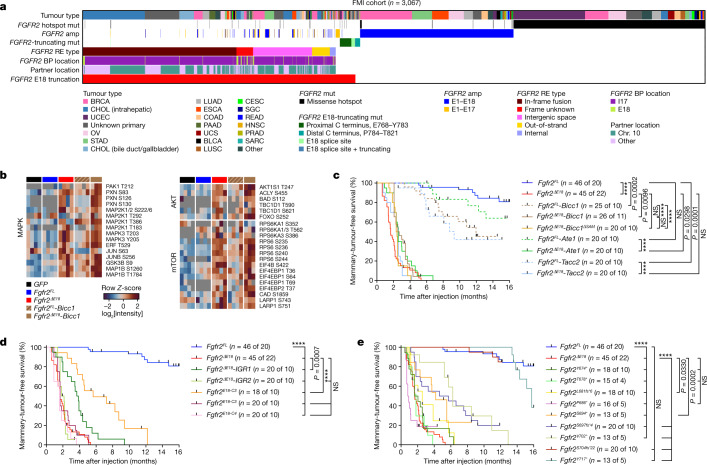
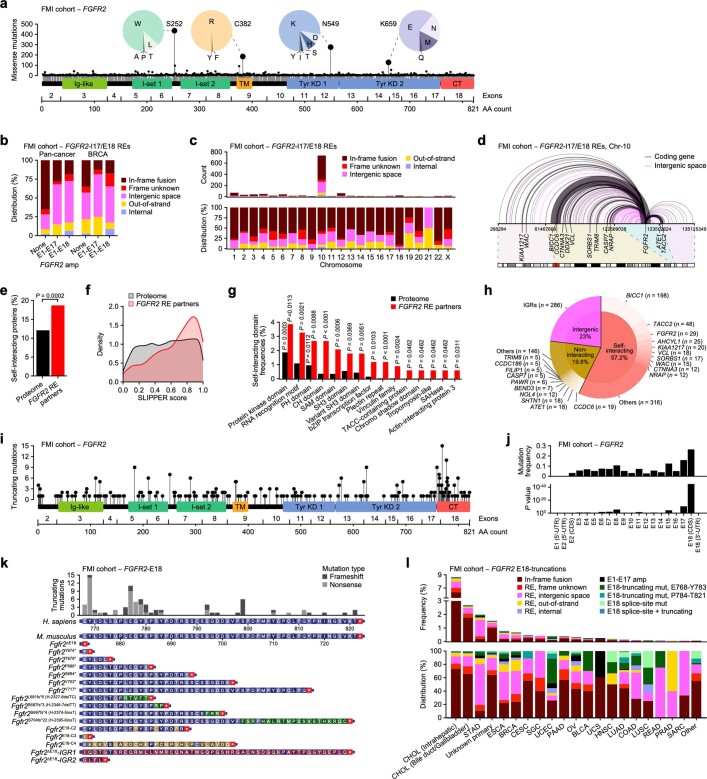
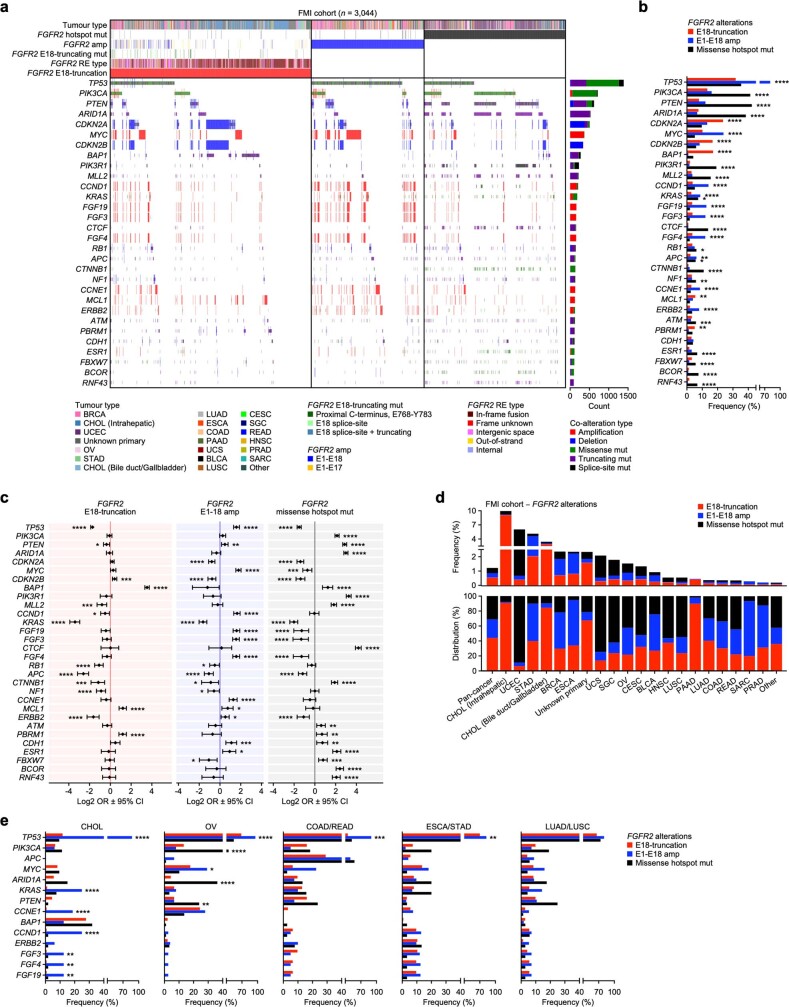
We next analysed oncogenomic data from Foundation Medicine (FMI) derived from 249,570 targeted tumour sequencing assays for the occurrence of FGFR2 alterations. Across cancers, we identified 1,367 samples with alterations potentially producing FGFR2ΔE18 (0.55% incidence). These were mutually exclusive to samples with FGFR2amp (n = 838, 0.34% incidence) or FGFR2 missense hotspot mutations (FGFR2hotspot; n = 978, 0.39% incidence; Fig. 2a and Extended Data Fig. 2a). Alterations potentially perturbing E18 were made up of 55.4% FGFR2-I17/E18 in-frame fusions. The remaining 44.6% were classified as variants of unknown significance and comprised FGFR2-I17/E18 frame-unknown, out-of-strand, intergenic space and internal REs, some of which coincided with focal amplifications (Fig. 2a, Extended Data Fig. 2b and Supplementary Table 2). Across the FGFR2 REs, intrachromosomal REs and known fusion partner genes (for example, BICC1, TACC2 and ATE1)20–22 were enriched (Extended Data Fig. 2c,d). FGFR2 RE partner genes encoded 337 unique proteins. Among these, the ability to self-interact was enriched as compared to the human proteome (Extended Data Fig. 2e–g). Nevertheless, 42.8% of all REs made use of a partner without evident self-interaction ability (Extended Data Fig. 2h). Other variants of unknown significance were FGFR2-E1–E17 partial amplifications, E18 splice-acceptor-site mutations and protein-truncating mutations significantly enriched in the coding sequence of FGFR2-E18 (Fig. 2a and Extended Data Fig. 2i–k). The identified FGFR2ΔE18 variants were most frequent in cholangiocarcinoma, but we also found considerable frequencies of in-frame fusions and especially structural variants of unknown significance in gastroesophageal and breast cancer (Extended Data Fig. 2l).
Fig. 2. Human FGFR2 E18-truncating alterations are oncogenic drivers in mice.
a, Analysis of 3,067 samples (1.23% incidence) containing FGFR2-I17/E18 in-frame fusions (n = 757, 0.30% incidence), frame unknown REs (n = 82, 0.03% incidence), intergenic space REs (n = 291, 0.12% incidence), out-of-strand REs (n = 88, 0.04% incidence), internal REs (n = 29, 0.01% incidence), FGFR2-E18 splice-site mutations (mut; n = 21, 0.01% incidence), E18-truncating nonsense and frameshift mutations (proximal, n = 59, 0.02% incidence; distal, n = 23, 0.01% incidence), FGFR2-E1–E17 partial amplifications (amp; n = 73, 0.03% incidence), E1–E18 full-length amplifications (n = 838, 0.34% incidence), and/or FGFR2 missense hotspot mutations affecting Ser252, Cys382, Asn549 or Lys659 (n = 978, 0.39% incidence) found in 249,570 pan-cancer diagnostic panel-seq profiles from FMI. BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; chr, chromosome; COAD, colon adenocarcinoma; ESCA, oesophageal carcinoma; HNSC, head and neck squamous cell carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SGC, salivary gland carcinoma; STAD, stomach adenocarcinoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterus carcinosarcoma. b, Global phosphoproteomic analysis of NMuMG cells expressing GFP or the indicated Fgfr2 variants. Groups were compared in a pairwise manner using the robust kinase activity inference (RoKAI) tool, including two-tailed hypothesis testing on Z-scores and false-discovery rate (FDR) multiple-testing correction using the Benjamini–Hochberg method. Group-comparison fold change (FC) values of −1.5 ≥ FC ≥ 1.5 and P < 0.05 were considered. The heatmaps show phosphosites subselected from the RoKAI output and grouped into the indicated signalling pathways guided by RoKAI as colour-coded row Z-scores calculated from log2-transformed intensity values. c–e, Kaplan–Meier curves showing mammary-tumour-free survival of female mice intraductally injected with lentiviruses encoding the indicated Fgfr2 variants. Cohort counts (n) are injected mammary glands (MGs) per number of mice. The Fgfr2FL and Fgfr2ΔE18 curves in c are duplicated in d and e. P values were calculated using log-rank (Mantel–Cox) tests; ****P < 0.0001; NS, not significant (P ≥ 0.05).
Extended Data Fig. 2. FGFR2 alterations found in the HMF cohort.
a, Lollipop plot of FGFR2 missense mutations identified in the Foundation Medicine (FMI) pan-cancer cohort (249,570 diagnostic hybrid-capture panel-seq profiles). The top four recurrent mutations (Ser 252, Cys 382, Asn 549, Lys 659) are referred to as hotspots in this study. b, Distribution of FGFR2-I17/E18 RE types in FMI samples with FGFR2 normal CN, E1-E17 amp, and E1-E18 amp. c, Total numbers and distributions of FGFR2-I17/E18 RE types across chromosomes according to RE partner location. d, Linear chr-10 map depicting intrachromosomal FGFR2-I17/E18 REs. Thickness of arcs is proportional to the recurrence of the corresponding RE partners. Light / dark grey and red bars denote ideogram and centromere of chr-10. e, Percentage of unique proteins with self-interacting capacity among FGFR2 RE partners (n = 337) versus the human proteome (n = 20,385). Based on the SLIPPER Golden Standard Dataset of self-interactors108. f, Distribution of self-interaction scores among FGFR2 RE partners using the SLIPPER algorithm108. g, Enrichment of self-interacting protein domains among FGFR2 RE partners using DAVID111. h, Recurrence of FGFR2 RE partners grouped by presence of self-interacting domains. Full list of RE partners is disclosed in Supplementary Table 2. IGRs, intergenic regions. i, j, Lollipop plot (i) and normalized frequency (top panel) and enrichment significance (P values, bottom panel) (j) of FGFR2-truncating mutations identified in the FMI cohort. Mutation frequency was normalized by the kilobase of feature (exon/intron) length and total number of mutations. AA, amino acid; CDS, coding sequence; CT, C terminus; TM, trans-membrane; UTR, untranslated region. k, Distribution of FGFR2-E18-truncating mutations identified in the FMI cohort and corresponding cloned mouse Fgfr2 variants representing most frequent human (H) FGFR2-E18 nonsense and frameshift (fs) mutations. C terminus sequences of cloned noncanonical E18-truncated Fgfr2 (Fgfr2ΔE18) variants are also displayed. IGR1 and IGR2 are based on TCGA-A8-A08A and TCGA-BH-A203 in Extended Data Fig. 5f, g. l, Frequencies (top panel) and distributions (bottom panel) per tumour type of E18-truncating FGFR2 alterations found in the FMI cohort. CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; mut, mutation; PAAD, pancreatic adenocarcinoma; READ, rectum adenocarcinoma; SGC, salivary gland carcinoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterus carcinosarcoma. P values were calculated with a one-tailed proportion z-test (e), one-tailed Fisher’s exact tests (g), or one-tailed binomial tests (j).
Expression of FGFR2ΔE18 in human cancer
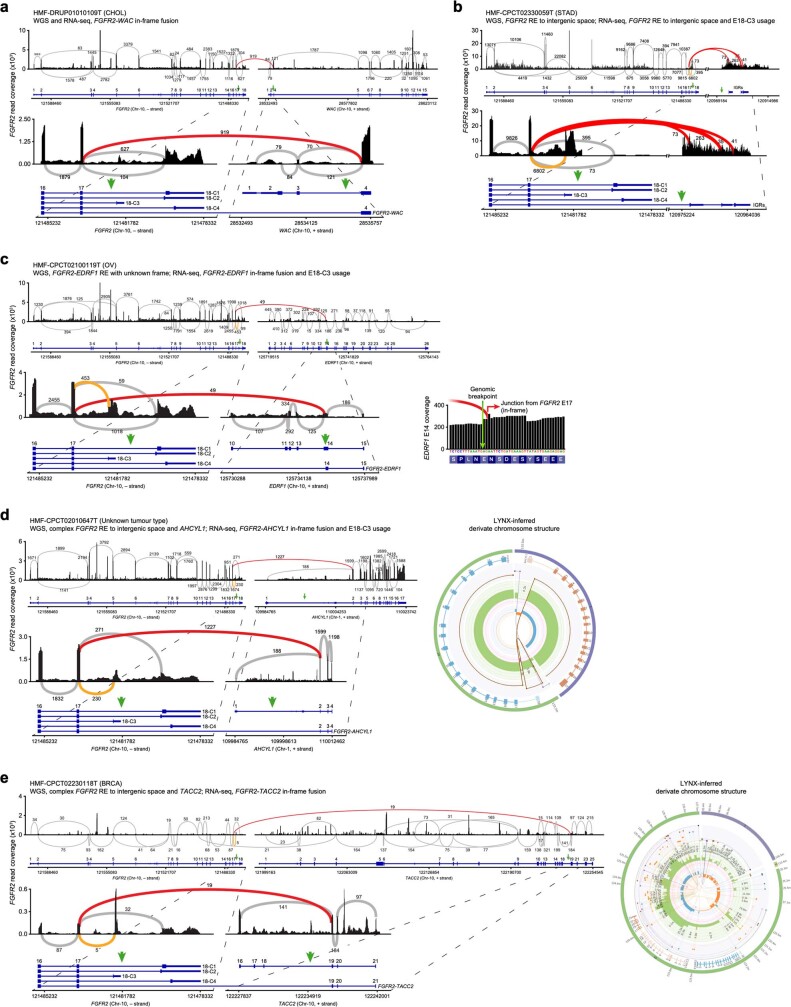
To validate expression of FGFR2ΔE18 variants, we analysed RNA-seq profiles matched to the HMF WGS samples. In the majority of the cases in which RNA-seq data was available, the predicted FGFR2 RE types were robustly expressed; REs with intergenic space produced FGFR2 transcripts terminating in intergenic region (IGR) pseudoexons encoding splice acceptors, a coding sequence and stop codons (Extended Data Figs. 1h and 3a–c). We also observed splicing to an alternative FGFR2-E18, termed C3 (Extended Data Fig. 3b–e), which is located in I17 and encodes a single isoleucine followed by a 3′-UTR. Two more FGFR2 isoforms make use of an alternative E18. The encoded C termini either overlap with the proximal part of the canonical FGFR2 C terminus (C2) or are different to it (C4)16–18. Thus, splicing to E18-C3 or E18-C4 generates FGFR2 isoforms that encode dysfunctional C termini resembling E18 truncation (Extended Data Fig. 2k). In a few cases with IGR REs, we found FGFR2 in-frame fusions at the RNA level. Reconstruction of derivate chromosomes revealed complex FGFR2 REs with several BPs that ultimately yielded in-frame fusions with protein-coding genes (Extended Data Fig. 3d,e).
Extended Data Fig. 3. Expression of E18-truncating FGFR2 variants in HMF samples.
a, Sashimi plot showing FGFR2 read coverage and junction reads of the HMF sample DRUP01010109T (CHOL). FGFR2-WAC in-frame fusion identified with WGS and FGFR2-E17 to WAC-E4 junction confirmed with RNA-seq. b, Sashimi plot showing FGFR2 read coverage and junction reads of the HMF sample CPCT02330059T (STAD). FGFR2-I17 RE to intergenic space identified with WGS and FGFR2-E17 to intergenic region (IGR) junctions and FGFR2-E18-C3 usage found with RNA-seq. c, Sashimi plot showing FGFR2 read coverage and junction reads of the HMF sample CPCT02100119T (OV). FGFR2-EDRF1 frame unknown RE identified with WGS and FGFR2-E17 to EDRF1-E14 in-frame junction and FGFR2–E18-C3 usage found with RNA-seq. d, Sashimi plot showing FGFR2 read coverage and junction reads of the HMF sample CPCT02010647T (unknown tumour type). FGFR2-I17 RE to intergenic space identified with WGS and discordant FGFR2-AHCYL1 in-frame fusion with FGFR2-E17 to AHCYL1-E2 junction and FGFR2-E18-C3 usage found with RNA-seq. e, Sashimi plot showing FGFR2 read coverage and junction reads of the HMF sample CPCT02230118T (BRCA). FGFR2-I17 RE to intergenic space identified with WGS and discordant FGFR2-TACC2 in-frame fusion with FGFR2-E17 to TACC2-E19 junction found with RNA-seq. Reconstructed derivate chromosomes using LINX88 are displayed for CPCT02010647T (d) and CPCT02230118T (e) and depict complex FGFR2 REs involving intergenic space and ultimately resolving to AHCYL1-E2 (d) and TACC2-E19 (e). Green arrows indicate BPs identified with WGS. E18-C1, canonical E18 of FGFR2FL; E18-C2/C3/C4, alternative FGFR2-E18.
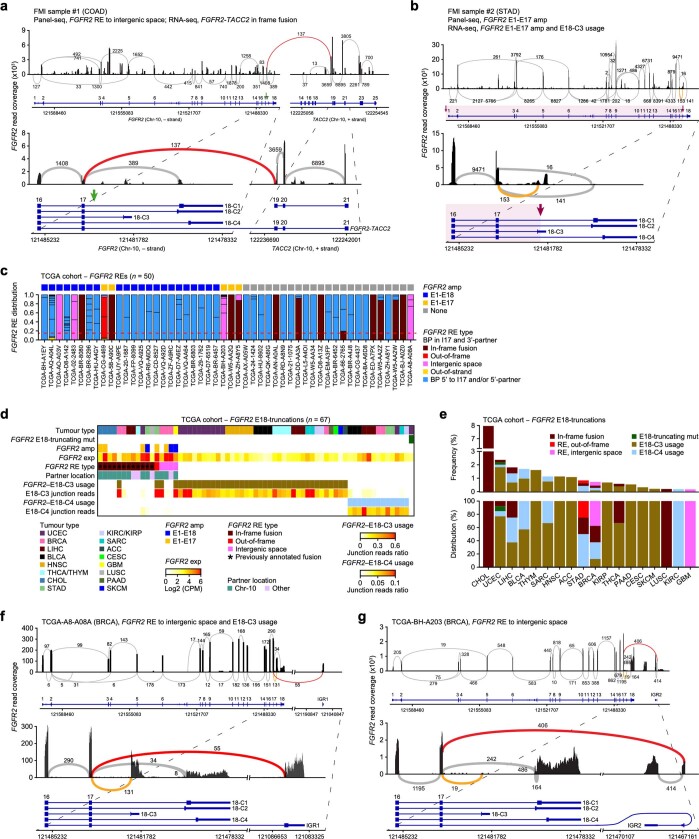
Next, we performed hybrid-capture RNA-seq analysis of two tumour samples, which were diagnosed by FMI to contain structural variants of unknown significance in FGFR2. One sample contained an FGFR2-I17 RE with intergenic space and the other contained an FGFR2amp involving E1–E17 only. RNA-seq profiling revealed a FGFR2 in-frame fusion in the first tumour, whereas the second tumour showed high FGFR2-E1–E17 expression with splicing to E18-C3 (Extended Data Fig. 4a,b). Comprehensive analysis of The Cancer Genome Atlas (TCGA) RNA-seq data identified tumours expressing FGFR2 in-frame fusions as well as non-canonical REs (Extended Data Fig. 4c–e). We found a few tumours containing FGFR2-I17 REs and concomitantly using E18-C3. However, a larger fraction of tumours used FGFR2-E18-C3 or FGFR2-E18-C4 in a mutually exclusive manner (Extended Data Fig. 4d–g and Supplementary Table 3). Taken together, we demonstrated that human tumours express diverse FGFR2ΔE18 transcripts derived from a variety of genomic alterations and alternative splicing events.
Extended Data Fig. 4. Expression of E18-truncating FGFR2 variants in FMI and TCGA samples.
a, Sashimi plot showing FGFR2 read coverage and junction reads of the FMI sample #1 (COAD). FGFR2-I17 RE to intergenic space was diagnosed by FMI, and discordant FGFR2-TACC2 in-frame fusion with FGFR2-E17 to TACC2-E19 junction was found with hybrid-capture RNA-seq. Green arrows indicate FGFR2 BP identified with panel-seq. b, Sashimi plot showing FGFR2 read coverage and junction reads of the FMI sample #2 (STAD). FGFR2-E1–E17 partial amp was diagnosed by FMI, and high FGFR2 expression with few E17–E18 junction reads but E18-C3 usage was found with hybrid-capture RNA-seq. Purple arrows indicate partially amplified FGFR2 region identified with panel-seq. c, FGFR2 amp status and RE type distribution in samples with FGFR2 REs (n = 50) found in the pan-cancer cohort from The Cancer Genome Atlas (TCGA, n = 10,344 samples). Dotted red line, RE read frequency threshold (> 0.15) to call samples expressing FGFR2ΔE18 REs (n = 17). d, 67 samples (0.65% incidence) containing FGFR2ΔE18 in-frame fusions (n = 12, 0.12% incidence), FGFR2ΔE18 non-canonical REs (n = 5, 0.05% incidence), proximal FGFR2-E18-truncating mutations (n = 1, 0.01% incidence), and cases with significant FGFR2-E18-C3 (n = 40; E18-C3 usage only, n = 36, 90% of total, 0.35% incidence; E18-C3 usage + RE, n = 4, 10% of total, 0.05% incidence) and/or E18-C4 (n = 13, 0.13% incidence) usage found in TCGA cohort. Asterisks mark previously annotated FGFR2 in-frame fusions91. exp, expression; GBM, glioblastoma multiforme; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; SKCM, skin cutaneous melanoma; THYM, thymoma. e, Frequencies (top panel) and distributions (bottom panel) per tumour type of expressed FGFR2ΔE18 alterations found in TCGA cohort. f, g, Sashimi plots showing FGFR2 read coverage and junction reads of TCGA-BRCA samples A8-A08A (f) and BH-A203 (g) with identified FGFR2-I17 REs to intergenic space and FGFR2-E18-C3 usage.
E18 loss is key to FGFR2 oncogenicity
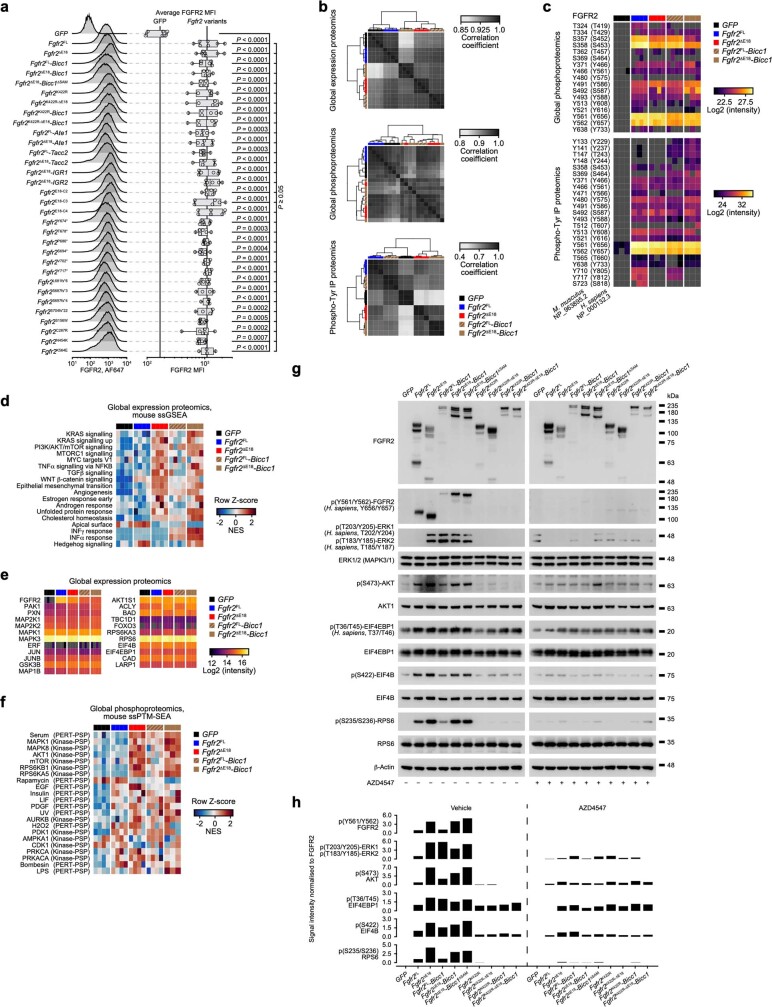
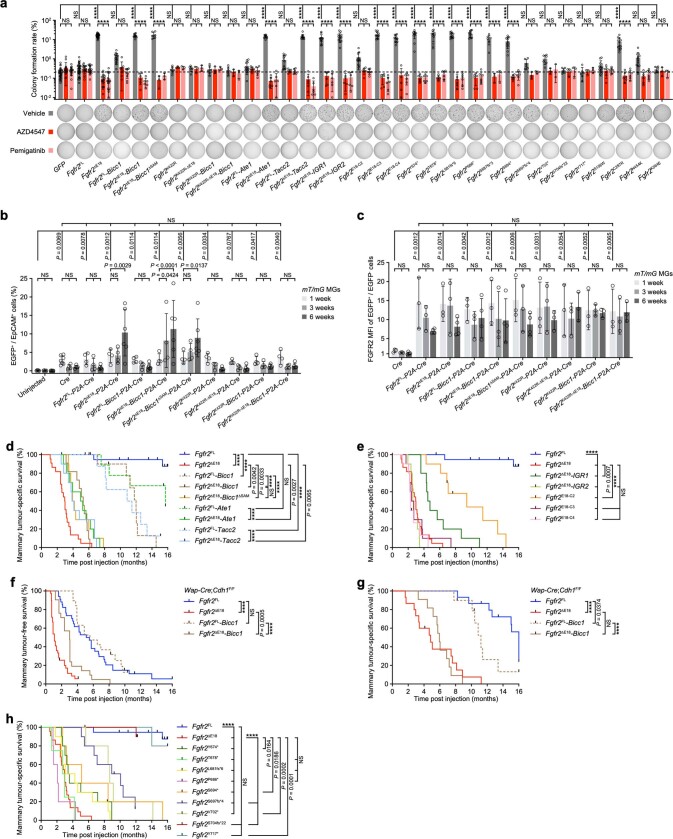
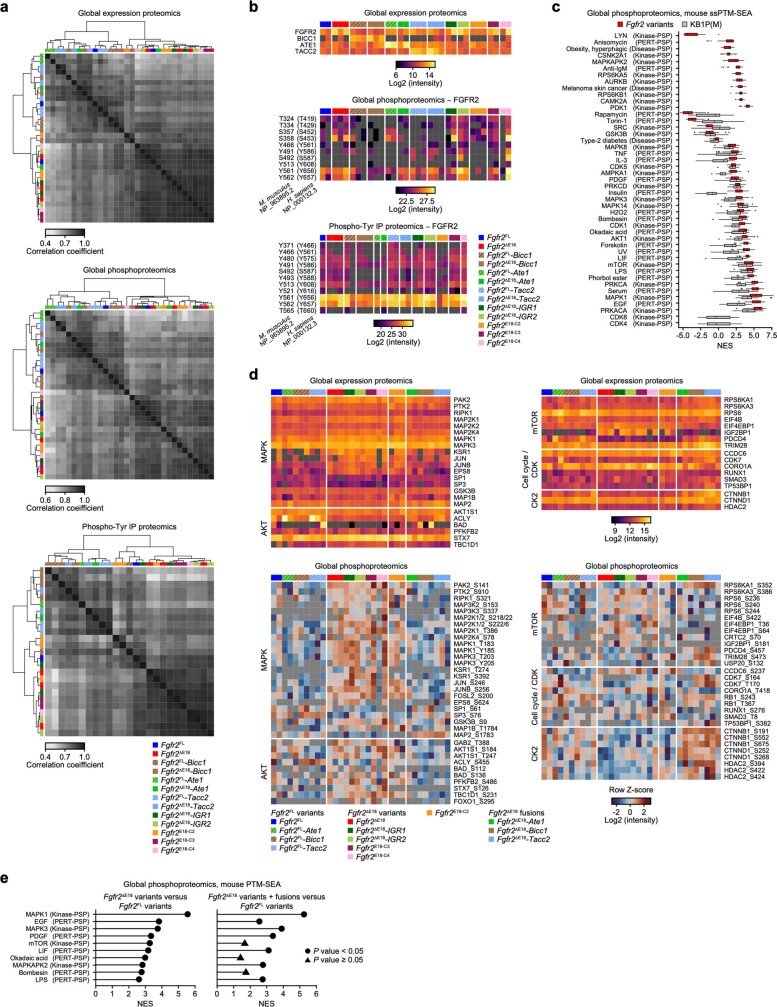
Previous research showed in vitro transforming abilities of C-terminally truncated FGFR2 isoforms17–19,25–27. Our in vivo screening data and analyses of human oncogenomic datasets similarly suggested that exclusion of E18 is a critical determinant to render FGFR2 REs oncogenic. To test this, we introduced mouse Fgfr2ΔE18 variants into mouse mammary epithelial cells. These were Fgfr2ΔE18 alone or fused to Ate1, Bicc1, Tacc2, two of the IGRs found in TCGA (Extended Data Fig. 4f,g), or the human E18-C2, E18-C3 or E18-C4 sequences, as well as Fgfr2 bearing E18 nonsense and frameshift mutations. The corresponding controls were full-length (FL) Fgfr2 (representing FGFR2amp), Fgfr2FL fusions, Fgfr2hotspot variants and kinase-domain-dead Fgfr2K422R variants (Extended Data Figs. 2k and 5a and Supplementary Table 4). Mass-spectrometry-based expression proteomics and phosphoproteomics revealed that overexpressed Fgfr2ΔE18 and Fgfr2ΔE18-Bicc1 both induced FGFR2 signalling resulting in the activation of the MAPK and PI3K–AKT–mTOR pathways (Fig. 2b and Extended Data Fig. 5b–f). This depended on a functional FGFR2 kinase domain, whereas the BICC1–SAM oligomerization domain28 was dispensable for Fgfr2ΔE18-Bicc1 activity (Extended Data Fig. 5g,h). Comparably, all of the tested Fgfr2ΔE18 variants, including proximal E18-truncating mutations and hotspot Fgfr2C287R, promoted colony formation in a 3D soft agar assay (Extended Data Fig. 6a). By contrast, overexpression of Fgfr2FL, its fusion variants that retain E18, and distal E18-truncating mutations and the remaining Fgfr2hotspot variants had limited potential to promote FGFR2 signalling or soft agar colonies (Fig. 2b and Extended Data Figs. 5 and 6a).
Extended Data Fig. 5. (Phospho)-proteomic analyses of NMuMG cells expressing Fgfr2 variants.
a, Fluorescence-activated cell sorting (FACS) to analyse FGFR2 mean fluorescence intensity (MFI) in NMuMG cells expressing GFP or indicated Fgfr2 variants. Fgfr2FL, full-length (FL) Fgfr2. Ate1, Bicc1, and Tacc2 correspond to the top-recurrent ATE1, BICC1, and TACC2 fusion partner genes in Extended Data Fig. 2h. Bicc1ΔSAM encodes BICC1 lacking its SAM oligomerisation domain. Fgfr2K422R variants encode tyrosine kinase domain (KD)-dead FGFR2 variants. Truncated or alternative C-termini encoded by IGR1/IGR2, E18-C2/C3/C4, Fgfr2Y674*, Fgfr2T678*, Fgfr2P686*, Fgfr2S694*, Fgfr2V702*, Fgfr2Y717*, Fgfr2L681fs*6, Fgfr2S687fs*3, Fgfr2S697fs*4, and Fgfr2S704fs*22 are displayed in Extended Data Fig. 2k. Fgfr2S156W, Fgfr2C287R, Fgfr2N454K, and Fgfr2K564E correspond to the human FGFR2S252W, FGFR2C382R, FGFR2N549K, and FGFR2K659E missense hotspot mutations in Extended Data Fig. 2a. Validation of overexpression of Fgfr2 variants using RT-qPCR is in Supplementary Table 4. Data are represented as median (centre line) ± IQR (25th to 75th percentile, box) and ± full range (minimum to maximum, whiskers) of GFP, n = 6; Fgfr2FL, Fgfr2ΔE18, n = 7; Fgfr2V702*, Fgfr2Y717*, Fgfr2S687fs*3, Fgfr2S697fs*4, Fgfr2S704fs*22, Fgfr2K564E, n = 4; other Fgfr2 variants, n = 6 independent replica. P values were calculated with one-tailed one-way ANOVA and false discovery rate (FDR) multiple-testing correction using the two-stage step-up method from Benjamini, Krieger, and Yekutieli. For FACS gating strategy, see Supplementary Fig. 2a. b, Mass spectrometry-based proteomic data showing correlation of NMuMG cells expressing GFP or indicated Fgfr2 variants for global protein expression, global phosphoproteomic analysis after enrichment with IMAC, and phospho-Tyr immunoprecipitation (IP)-enriched samples. Pearson’s R correlation coefficients are depicted and heatmaps were clustered unsupervised. c, Heatmaps visualizing FGFR2 phosphosites identified in (b). d, Single-sample gene set enrichment analysis (ssGSEA) based on hallmark gene sets from MSigDB79 and the global protein expression dataset. Significant single-sample normalized enrichment scores (NES) were calculated using GSEA standard settings77,78. NES are visualised as colour-coded row Z-scores and depicted terms are based on Fgfr2ΔE18 versus Fgfr2FL two-group comparisons using two-tailed unpaired Student’s t-tests. Significant terms are shown (P < 0.05). e, Relative candidate protein expression levels corresponding to MAPK, AKT, and mTOR substrates displayed in Fig. 2b and based on the global protein expression. f, Single-sample phosphosite signature enrichment analysis (ssPTM-SEA) based on murine kinase/pathway definitions of PTMsigDB82 and the global phosphoproteomic dataset. Significant single-sample NES were calculated using gene permutation (n = 1,000) and one-tailed permutation testing with FDR multiple-testing correction using the Benjamini-Hochberg method by applying PTM-SEA standard settings82. NES are visualised as colour-coded row Z-scores and depicted terms are based on Fgfr2ΔE18 versus GFP, Fgfr2ΔE18 versus Fgfr2FL, and/or Fgfr2FL versus GFP two-group comparisons using two-tailed unpaired Student’s t-tests. Terms significant for either of the three two-group comparisons are shown (P < 0.05). g, Western blots showing expression and phosphorylation of indicated proteins in NMuMG cells expressing GFP or indicated Fgfr2 variants and treated for 3 h with vehicle or 100 nM AZD4547. β-Actin was run on separate gels as sample processing control, and each blot was stained with Ponceau S to ensure equal loading of total protein. Blots stained with the same antibody were developed and recorded in parallel and subjected to equal post-imaging processing. For gel source data, see Supplementary Fig. 1a–g. h, Quantifications of relative phosphoprotein band intensities in (g) normalized to β-actin, corresponding total protein, and FGFR2 band intensities. Data in g, h represent 1 replica of 2 independent experiments.
Extended Data Fig. 6. In vitro and in vivo oncogenic capacities of Fgfr2 variants.
a, Representative images of 12-well plate wells and quantification of 3D soft agar colony formation assay using NMuMG cells expressing GFP or indicated Fgfr2 variants and treated with vehicle, 100 nM AZD4547, or 100 nM pemigatinib for 15 days. Data are represented as mean ± standard deviation (s.d.) of GFP, Fgfr2FL, Fgfr2ΔE18, vehicle, n = 33; AZD4547, pemigatinib, n = 18 independent replica from 4 individual experiments. Fgfr2FL-Bicc1, vehicle, n = 18; AZD4547, pemigatinib, n = 9 independent replica from 3 individual experiments. Fgfr2ΔE18-Bicc1, Fgfr2FL-Ate1, Fgfr2ΔE18-Ate1, Fgfr2FL-Tacc2, Fgfr2ΔE18-Tacc2, Fgfr2ΔE18-IGR1, Fgfr2ΔE18-IGR2, Fgfr2E18-C2, Fgfr2E18-C3, Fgfr2E18-C4, Fgfr2Y674*, Fgfr2T678*, Fgfr2L681fs*6, Fgfr2P686*, Fgfr2S694*, Fgfr2S156W, Fgfr2C287R, Fgfr2N454K, vehicle, n = 12; AZD4547, pemigatinib, n = 6 independent replica from 2 individual experiments. Fgfr2ΔE18-Bicc1ΔSAM, Fgfr2K422R, Fgfr2K422R-ΔE18, Fgfr2K422R-Bicc1, Fgfr2K422R-ΔE18-Bicc1, Fgfr2S687fs*3, Fgfr2S697fs*4, Fgfr2V702*, Fgfr2S704fs*22, Fgfr2Y717*, Fgfr2K564E, n = 6; AZD4547, pemigatinib, n = 3 independent replica from 1 experiment. b, c, FACS to quantify traced EGFP+ EpCAM+ epithelial cells (b) and their FGFR2 MFI (c). Rosa26-mT/mG female reporter mice were intraductally injected with lentiviruses encoding Cre or indicated Fgfr2-P2A-Cre variants resulting in Cre-mediated mT/mG allele switching, thus cell membrane-localized tdTomato (mT) expression was replaced by membrane-localized EGFP (mG) expression. Mammary glands (MGs) were subjected to FACS analysis at indicated timepoints post injection. Data are represented as mean ± s.d. and each data point represents a MG pool of an individual mouse. Analyses were done in batches of 1–2 mice of each Fgfr2 variant and one timepoint. In (b), 1 week, uninjected MGs, n = 5; Cre, n = 7; other Fgfr2 variants, n = 4; 3 weeks, all groups, n = 4; 6 weeks, uninjected MGs, Cre, Fgfr2K422R-P2A-Cre, Fgfr2K422R-ΔE18-P2A-Cre, Fgfr2K422R-Bicc1-P2A-Cre, Fgfr2K422R-ΔE18-Bicc1-P2A-Cre, n = 5; other Fgfr2 variants, n = 6 mice. In (c), 1 week and 3 weeks, all groups, n = 3; 6 weeks, Cre, Fgfr2FL-P2A-Cre, Fgfr2ΔE18-P2A-Cre, Fgfr2FL-Bicc1-P2A-Cre, Fgfr2ΔE18-Bicc1-P2A-Cre, Fgfr2ΔE18-Bicc1ΔSAM-P2A-Cre, n = 4; other Fgfr2 variants, n = 3 mice. For FACS gating strategy, see Supplementary Fig. 2b. d, e, Kaplan-Meier curves showing mammary tumour-specific survival of female wild-type (WT) mice intraductally injected with lentiviruses encoding indicated Fgfr2 variants. Fgfr2FL, n = 20; Fgfr2ΔE18, n = 22; Fgfr2FL-Bicc1, Fgfr2ΔE18-Bicc1ΔSAM, Fgfr2FL-Ate1, Fgfr2ΔE18-Ate1, Fgfr2FL-Tacc2, Fgfr2ΔE18-Tacc2, Fgfr2ΔE18-IGR1, Fgfr2ΔE18-IGR2, Fgfr2E18-C2, Fgfr2E18-C3, Fgfr2E18-C4, n = 10; Fgfr2ΔE18-Bicc1, n = 11 mice. Fgfr2FL and Fgfr2ΔE18 curves in a are duplicated in d, h. f, g, Kaplan-Meier curves showing mammary tumour-free (c) and -specific (d) survival of female Wap-Cre;Cdh1F/F mice intraductally injected with lentiviruses encoding indicated Fgfr2 variants. Fgfr2FL, n = 34 of 15; Fgfr2ΔE18, n = 39 of 15; Fgfr2FL-Bicc1, n = 19 of 10; Fgfr2ΔE18-Bicc1, n = 21 injected MGs of 11 mice. h, Kaplan-Meier curves showing mammary tumour-specific survival of female wild-type (WT) mice intraductally injected with lentiviruses encoding indicated Fgfr2 variants. Fgfr2Y674*, Fgfr2L681fs*6, Fgfr2S697fs*4, Fgfr2S704fs*22, n = 10; Fgfr2T678*, n = 4; Fgfr2P686*, Fgfr2S694*, Fgfr2V702*, Fgfr2Y717*, n = 5 mice. P values were calculated with one-tailed two-way ANOVA and FDR multiple-testing corrections using the two-stage step-up method from Benjamini, Krieger, and Yekutieli (a, c), one-tailed Kruskal-Wallis tests and Dunn’s multiple-testing corrections (b), or log rank (Mantel-Cox) tests (b–h). ****P < 0.0001; NS, not significant (P ≥ 0.05).
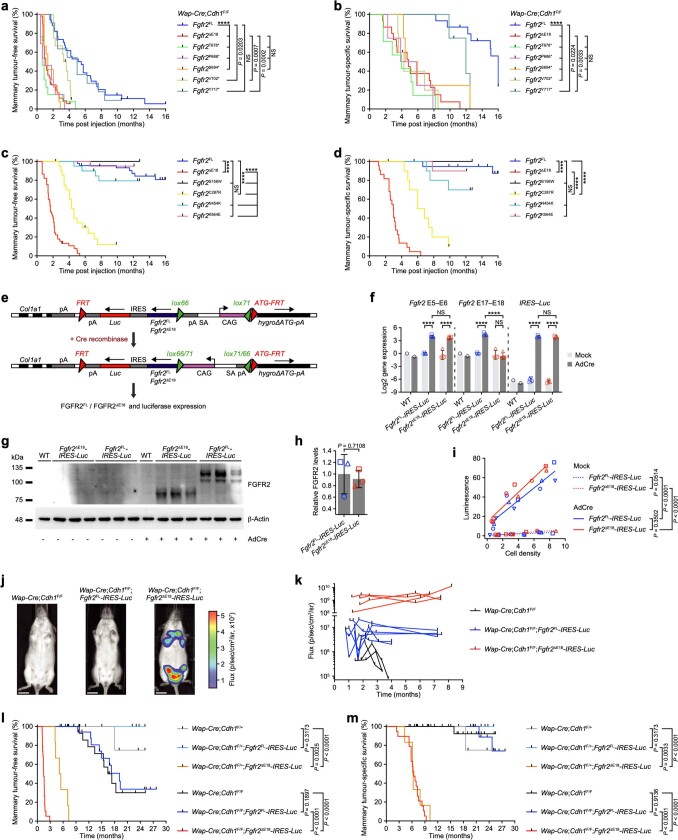
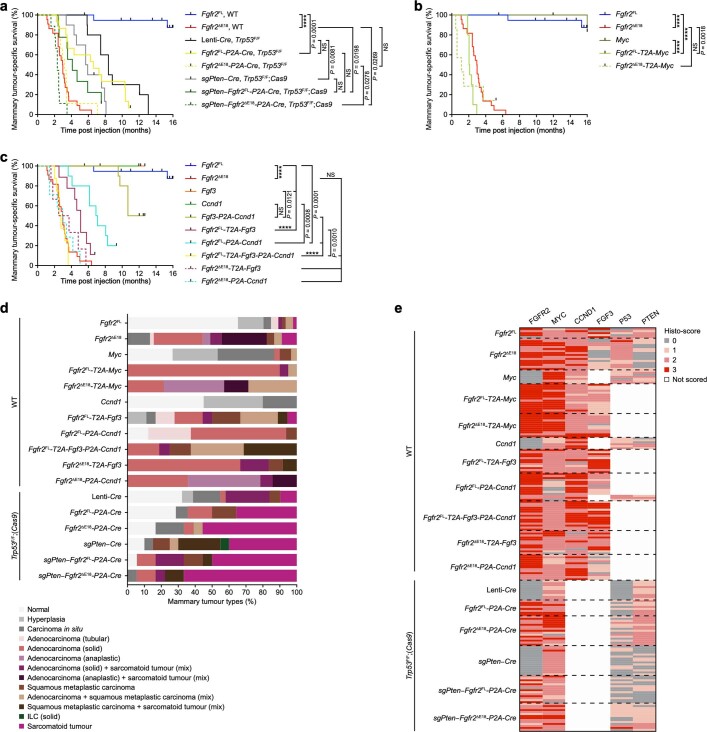
Next, we evaluated the in vivo oncogenicity of Fgfr2 variants using somatic delivery to mouse mammary glands through intraductal injection of lentiviruses12,13. Lineage tracing using lentiviral Fgfr2-P2A-cre constructs and mT/mG female mice showed comparable mammary epithelial transduction rates and FGFR2 expression levels across the Fgfr2 variants tested. However, only Fgfr2ΔE18 variants drove clonal expansion of the mammary epithelium, which depended on the FGFR2 kinase domain but not on the BICC1–SAM oligomerization domain (Extended Data Fig. 6b,c). To assess Fgfr2ΔE18 oncogenicity in mammary tumour models representative of different breast cancer subtypes—including invasive lobular carcinoma, a hallmark of which is E-cadherin loss29—we intraductally delivered Fgfr2 variants to wild-type (WT) or Wap-cre;Cdh1F/F mice. Fgfr2ΔE18 variants rapidly induced mammary tumours regardless of Cdh1 mutation status (Fig. 2c,d and Extended Data Fig. 6d–g), and progressive truncation of Fgfr2-E18 gradually decreased tumour onset (Fig. 2e and Extended Data Figs. 6h and 7a,b). By contrast, mammary glands injected with Fgfr2FL variants displayed no or slow tumorigenesis in WT and Wap-cre;Cdh1F/F mice (Fig. 2c–e and Extended Data Figs. 6d–h and 7a,b). Fgfr2hotspot variants were also non-tumorigenic, except for Fgfr2C287R, which drove marked mammary tumour formation (Extended Data Fig. 7c,d). Furthermore, we generated genetically engineered mouse models (GEMMs) bearing Cre-inducible Fgfr2-IRES-Luc alleles (Extended Data Fig. 7e–i), in which Wap-cre-mediated induction of Fgfr2FL-IRES-Luc had comparably little effect on mammary tumorigenesis. However, induction of Fgfr2ΔE18-IRES-Luc led to increased mammary gland bioluminescence, which coincided with rapid and multifocal tumour formation in Wap-cre;Cdh1F/+;Fgfr2ΔE18-IRES-Luc and Wap-cre;Cdh1F/F;Fgfr2ΔE18-IRES-Luc females (Extended Data Fig. 7j–m). Histopathological evaluation of the mammary glands of Fgfr2FL somatic models and GEMMs revealed mostly healthy tissue or low-grade lesions. By contrast, the majority of Fgfr2ΔE18 glands contained FGFR2-positive high-grade adenocarcinomas or E-cadherin-negative invasive lobular carcinomas or sarcomatoid tumours (Extended Data Fig. 8). Proteomic analyses of tumours induced by Fgfr2 variants demonstrated consistent expression and phosphorylation of FGFR2 variants along with downstream signalling activities, which were distinct from the phosphoproteome of FGFR2-independent K14-cre;Brca1F/F;Trp53F/F;(Mdr1a/b−/−) tumours (Extended Data Fig. 9a–c). Notably, MAPK and AKT–mTOR signalling pathways were particularly active in tumours driven by Fgfr2ΔE18 variants (Extended Data Fig. 9d,e). Together, these data establish that E18 truncation of Fgfr2 is a bona fide tumour-driver alteration and the loss of the C terminus is a key determinant of FGFR2 oncogenicity.
Extended Data Fig. 7. In vivo oncogenic capacities of Fgfr2 variants in somatic models and GEMMs.
a, b, Kaplan-Meier curves showing mammary tumour-free (a) and -specific (b) survival of female Wap-Cre;Cdh1F/F mice intraductally injected with lentiviruses encoding indicated Fgfr2 variants. Fgfr2T678*, n = 13 of 7; Fgfr2P686*, n = 13 of 4; Fgfr2S694*, n = 12 of 4; Fgfr2V702*, n = 12 of 5; Fgfr2Y717*, n = 11 injected MGs of 4 mice. Fgfr2FL and Fgfr2ΔE18 curves in a, b are duplicates from Extended Data Fig. 6f, g. c, d, Kaplan-Meier curves showing mammary tumour-free (c) and -specific (d) survival of female WT mice intraductally injected with lentiviruses encoding indicated Fgfr2 variants. Fgfr2S156W, Fgfr2C287R, Fgfr2N454K, Fgfr2K564E, n = 20 injected MGs of 10 mice. Fgfr2FL and Fgfr2ΔE18 curves in c, d are duplicates from Fig. 2c and Extended Data Fig. 6d. e, Schematic representation of the engineered Fgfr2FL and Fgfr2ΔE18 alleles. Frt-invCAG-Fgfr2FL-IRES-Luc and Frt-invCAG-Fgfr2ΔE18-IRES-Luc were inserted into the Col1a1 locus using the genetically engineered mouse model – embryonic stem cell (GEMM-ESC) methodology54. Cre activity inverts the CAG promoter resulting in coherent FGFR2 and luciferase (Luc) expression. IRES, internal ribosome entry site. f, RT-qPCR quantification of Fgfr2 and Luc expression in mouse mammary epithelial cells (MMECs) isolated from pooled MGs of 10-week-old WT control, Fgfr2FL-IRES-Luc, and Fgfr2ΔE18-IRES-Luc female mice and mock-treated or treated with adenoviral Ad5CMVCre (AdCre) to switch Fgfr2 alleles in vitro. Data are represented as mean ± s.d. of WT, n = 1; Fgfr2FL-IRES-Luc, Fgfr2ΔE18-IRES-Luc, n = 4 MMEC cultures each from MG pools of individual mice. g, h, Western blot showing FGFR2 expression of mock- or AdCre-treated MMEC cultures (g) and quantification of relative FGFR2 intensities normalized to β-actin (h). β-Actin was run on a separate gel as sample processing control, and membranes were stained with Ponceau S to ensure equal loading of total protein. For gel source data, see Supplementary Fig. 1h, i. In h, data are represented as mean ± s.d. of WT, n = 1; Fgfr2FL-IRES-Luc, Fgfr2ΔE18-IRES-Luc, n = 3 MMEC cultures each from MG pools of individial mice. i, Luciferase activity measured using luciferin and bioluminescence imaging on mock- or AdCre-treated MMEC cultures. Data are represented as simple linear regressions across Fgfr2FL-IRES-Luc, n = 4; Fgfr2ΔE18-IRES-Luc, n = 3 MMEC cultures (each from MG pools of individual mice) at indicated cell densities. j, Representative in vivo bioluminescence images showing luciferase activity following luciferin administration measured as photon flux in 10-week-old Wap-Cre;Cdh1F/F, Wap-Cre;Cdh1F/F;Fgfr2FL-IRES-Luc, and Wap-Cre;Cdh1F/F;Fgfr2ΔE18-IRES-Luc female mice. Scale bars, 1 cm. k, Quantification of luciferase activity using recurrent bioluminescence imaging in indicated GEMMs. Wap-Cre;Cdh1F/F female mice show background luminescence. Wap-Cre;Cdh1F/F, n = 3; Wap-Cre;Cdh1F/F;Fgfr2FL-IRES-Luc, n = 6; Wap-Cre;Cdh1F/F;Fgfr2ΔE18-IRES-Luc, n = 4 mice. l, m, Kaplan-Meier curves showing mammary tumour-free (l) and -specific (m) survival of indicated GEMMs. Wap-Cre;Cdh1F/+, n = 12; Wap-Cre;Cdh1F/+;Fgfr2FL-IRES-Luc, n = 5; Wap-Cre;Cdh1F/+;Fgfr2ΔE18-IRES-Luc, n = 6; Wap-Cre;Cdh1F/F, n = 16; Wap-Cre;Cdh1F/F;Fgfr2FL-IRES-Luc, Wap-Cre;Cdh1F/F;Fgfr2ΔE18-IRES-Luc, n = 19 mice. P values were calculated with log rank (Mantel-Cox) tests (a–d, l, m), one-tailed two-way ANOVA and FDR multiple-testing corrections using the two-stage step-up method from Benjamini, Krieger, and Yekutieli (f), a two-tailed unpaired Student’s t-test (h), or one-way analysis of covariance (ANCOVA) to compare linear regression slopes (i). ****P < 0.0001.
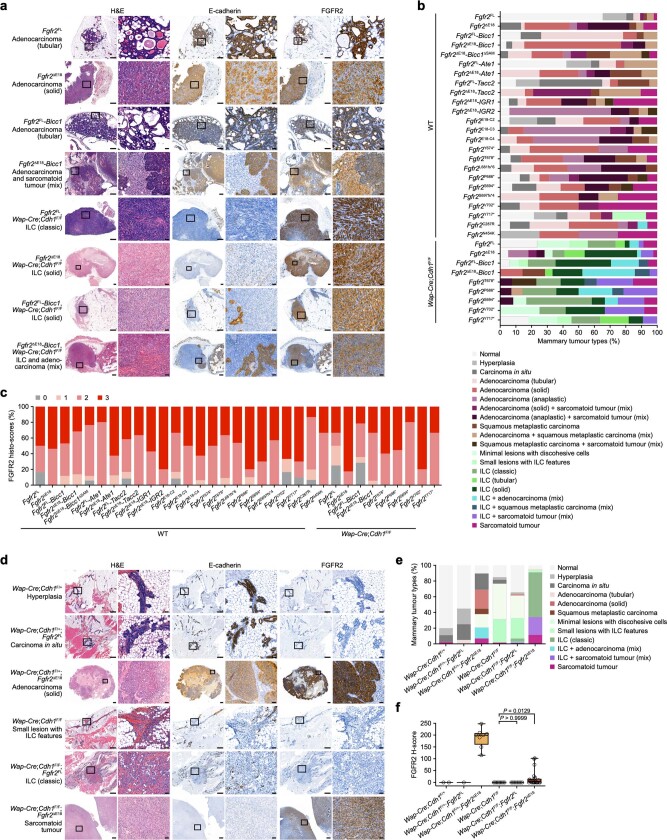
Extended Data Fig. 8. Mammary tumour types observed in Fgfr2 mouse models.
a, Representative hematoxylin and eosin (H&E) histochemistry and FGFR2 and E-cadherin immunohistochemistry (IHC) stains on mammary tissue sections from indicated Fgfr2 somatic mouse models. Per MG one tissue section was stained and quantified for each of the indicated stains acquired in multiple independent randomized batches across all Fgfr2 variants. Numbers of stained and quantified MGs are in (b, c). ILC, invasive lobular carcinoma. b, Mammary tumour type classifications of Fgfr2 somatic mouse models based on H&Es and E-cadherin IHC stains. WT, Fgfr2FL, n = 46 of 20; Fgfr2ΔE18, n = 45 of 22; Fgfr2FL-Bicc1, n = 23 of 10; Fgfr2ΔE18-Bicc1, n = 26 of 11; Fgfr2ΔE18-Bicc1ΔSAM, Fgfr2ΔE18-Ate1, Fgfr2FL-Tacc2, Fgfr2E18-C3, n = 20 of 10; Fgfr2FL-Ate1, Fgfr2ΔE18-Tacc2, Fgfr2E18-C4, n = 19 of 10; Fgfr2ΔE18-IGR1, n = 18 of 9; Fgfr2ΔE18-IGR2, Fgfr2E18-C2, n = 17 of 10; Fgfr2Y674*, Fgfr2C287R, n = 16 of 8; Fgfr2T678*, n = 15 of 4; Fgfr2L681fs*6, n = 14 of 7; Fgfr2P686*, n = 16 of 5; Fgfr2S694*, n = 13 of 5; Fgfr2S697fs*4, n = 8 of 4; Fgfr2V702*, n = 8 of 3; Fgfr2Y717*, n = 14 of 5; Fgfr2N454K, n = 4 injected MGs of 2 mice. Wap-Cre;Cdh1F/F, Fgfr2FL, n = 34 of 15; Fgfr2ΔE18, n = 39 of 15; Fgfr2FL-Bicc1, n = 17 of 9; Fgfr2ΔE18-Bicc1, n = 21 of 11; Fgfr2T678*, n = 13 of 7; Fgfr2P686*, n = 14 of 4; Fgfr2S694*, n = 12 of 4; Fgfr2V702*, n = 12 of 5; Fgfr2Y717*, n = 11 injected MGs of 4 mice. c, Histo-scoring of FGFR2 IHC stains on mammary tumours from Fgfr2 somatic mouse models. WT, Fgfr2FL, n = 6 of 5; Fgfr2ΔE18, n = 39 of 22; Fgfr2FL-Bicc1, n = 17 of 10; Fgfr2ΔE18-Bicc1, n = 22 of 11; Fgfr2ΔE18-Bicc1ΔSAM, n = 17 of 9; Fgfr2FL-Ate1, n = 5 of 5; Fgfr2ΔE18-Ate1, Fgfr2E18-C4, n = 16 of 9; Fgfr2FL-Tacc2, n = 12 of 8; Fgfr2ΔE18-Tacc2, n = 11 of 8; Fgfr2ΔE18-IGR1, Fgfr2E18-C3, n = 14 of 9; Fgfr2ΔE18-IGR2, n = 15 of 10; Fgfr2E18-C2, n = 12 of 9; Fgfr2Y674*, n = 14 of 8; Fgfr2T678*, n = 11 of 4; Fgfr2L681fs*6, n = 13 of 8; Fgfr2P686*, Fgfr2S694*, n = 10 of 5; Fgfr2S697fs*4, n = 7 of 4; Fgfr2V702*, n = 6 of 3; Fgfr2Y717*, n = 10 of 4; Fgfr2C287R, n = 15 of 8; Fgfr2N454K, n = 3 tumours of 2 mice. Wap-Cre;Cdh1F/F, Fgfr2FL, n = 12 of 8; Fgfr2ΔE18, n = 23 of 9; Fgfr2FL-Bicc1, n = 14 of 7; Fgfr2ΔE18-Bicc1, n = 18 of 10; Fgfr2T678*, Fgfr2V702*, n = 10 of 5; Fgfr2P686*, n = 9 of 4; Fgfr2S694*, n = 10 of 4; Fgfr2Y717*, n = 6 tumours of 3 mice. d, Representative H&E histochemistry and E-cadherin and FGFR2 IHC stains on mammary tissue sections from indicated GEMMs. Per MG one tissue section was stained and quantified for each of the indicated stains acquired in two independent randomized batches across all genotypes. Numbers of stained and quantified MGs are in (e, f). e, Mammary tumour type classifications of GEMMs based on H&Es and E-cadherin IHC stains. Wap-Cre;Cdh1F/+, n = 45 of 12; Wap-Cre;Cdh1F/+;Fgfr2FL, n = 20 of 5; Wap-Cre;Cdh1F/+;Fgfr2ΔE18, n = 29 of 6; Wap-Cre;Cdh1F/F, n = 60 of 16; Wap-Cre;Cdh1F/F;Fgfr2FL, n = 73 of 19; Wap-Cre;Cdh1F/F;Fgfr2ΔE18, n = 79 MGs of 19 mice. f, Histo (H)-score quantifications of FGFR2 IHC stains on mammary tumours from GEMMs. Wap-Cre;Cdh1F/+, n = 2 of 2; Wap-Cre;Cdh1F/+;Fgfr2FL, n = 1 of 1; Wap-Cre;Cdh1F/+;Fgfr2ΔE18, n = 8 of 5; Wap-Cre;Cdh1F/F, n = 9 of 8; Wap-Cre;Cdh1F/F;Fgfr2FL, n = 8 of 6; Wap-Cre;Cdh1F/F;Fgfr2ΔE18, n = 24 tumours of 19 mice. Data are represented as median (centre line) ± IQR (25th to 75th percentile, box) and ± full range (minimum to maximum, whiskers) and P values were calculated with one-tailed Kruskal-Wallis tests and Dunn’s multiple-testing corrections. Scale bars, overview, 500 μm; inset, 50 μm.
Extended Data Fig. 9. (Phospho)-proteomic analyses of tumours from Fgfr2 somatic mouse models.
a, Mass spectrometry-based proteomic data showing correlation of indicated Fgfr2 somatic mouse models for global protein expression, global phosphoproteomic analysis after enrichment with IMAC, and phospho-Tyr IP-enriched mammary tumours. Pearson’s R correlation coefficients are depicted and heatmaps were clustered unsupervised. b, Relative protein expression of FGFR2 and its fusion partners BICC1, ATE1, and TACC2 next to FGFR2 phosphosites identified in datasets from a. Heatmaps colour-code relative intensities of protein expression and phosphorylation. c, ssPTM-SEA based on murine kinase/pathway definitions of PTMsigDB and the global phosphoproteomic dataset in a as well as phosphoproteomic data generated from mammary tumours from K14-Cre;BrcaF/F;Trp53F/F (KB1P) and KB1P;Mdr1a/b−/− (KB1PM) GEMMs. Each boxplot represents one ssPTM-SEA term and shows NES of individual Fgfr2 variant tumours or KB1P(M) tumours. ssPTM-SEA terms enriched in the Fgfr2 variant and/or the KB1P(M) tumour cohorts are shown. Significant single-sample NES were calculated using gene permutation (n = 1,000) and permutation-derived P values by applying PTM-SEA standard settings82. No further statistical selections were applied. Boxplots are represented as median (centre line) ± IQR (25th to 75th percentile, box) and IQR ± 1.5 x IQR (whiskers). Fgfr2 variants, n = 32; KB1P, n = 14; KB1PM, n = 10 tumours. d, Relative candidate protein expression (top panels) and phosphorylation (bottom panels) levels of MAPK, AKT, mTOR, cell cycle / CDK, and CK2 substrates identified in a. For the phosphoproteomic analysis, samples were grouped into Fgfr2FL variants, Fgfr2ΔE18 variants, and Fgfr2ΔE18 fusion variants and compared pairwise using the robust kinase activity inference (RoKAI) tool at default settings83 including two-tailed hypothesis testing on Z-scores and FDR multiple-testing correction using the Benjamini-Hochberg method. Group comparison fold change (FC) values of −1.5 ≥ FC ≥ 1.5 and P < 0.05 were considered. The RoKAI output was used to manually curate phosphosites of interest, and phosphosites were manually grouped into indicated signalling pathways guided by RoKAI. The heatmaps depict relative expression intensities (top panels) and Z-scores of phosphosite intensities calculated per row from log2-transformed intensity values (bottom panels). e, PTM-SEA based on murine kinase/pathway definitions of PTMsigDB and performed with global phosphoproteomic data and limma-based two-group comparisons of Fgfr2ΔE18 variants versus Fgfr2FL variants groups (left panel) and Fgfr2ΔE18 variants including fusions versus Fgfr2FL variants groups (right panel). Significant NES were calculated by using gene permutation (n = 1,000) and one-tailed permutation testing without multiple-testing correction by applying PTM-SEA standard settings82. Lollipops show NES of terms significantly enriched in either of the two comparisons (P < 0.05).
FGFR2 oncogenicity depends on co-drivers
Compared with Fgfr2ΔE18, our in vivo modelling efforts showed limited oncogenic competences of Fgfr2FL and Fgfr2hotspot variants in WT and Cdh1-deficient mammary glands. Yet, besides FGFR2ΔE18, FGFR2amp and FGFR2hotspot made up considerable fractions of human FGFR2 alterations. The oncogenic ability of specific FGFR2 alterations might be affected by the tissue of origin as well as the mutational context. To examine possible cooperation between FGFR2 variants and other genes, we analysed driver gene alterations diagnosed by FMI oncogenomic profiling and their incidence in FGFR2-altered cancers (Extended Data Fig. 10a and Supplementary Table 2). FGFR2ΔE18, FGFR2amp and FGFR2hotspot showed co-occurrences and mutual exclusivities with distinct sets of driver alterations (Extended Data Fig. 10b,c). The proportions of FGFR2ΔE18, FGFR2amp and FGFR2hotspot varied across cancer entities, suggesting differential selections of FGFR2 aberrations and concurrent driver alterations among tissues of origin (Extended Data Fig. 10d). We evaluated driver-gene enrichments among the three FGFR2 alteration categories in a tumour-type-specific manner. In breast cancers with FGFR2amp, TP53 driver mutations, MYC amplifications, PTEN loss-of-function alterations, and CCND1 and FGF3/4/19 co-amplifications were significantly more enriched compared with the other classes of FGFR2 aberration (Fig. 3a,b). Accordingly, FGFR2amp showed co-occurrence with TP53, PTEN and MYC alterations in breast cancer (Fig. 3c). In several other cancer types, we also observed enrichments of TP53 and MYC driver alterations in FGFR2-amplified cases (Extended Data Fig. 10e). By contrast, FGFR2ΔE18 and FGFR2hotspot samples did not co-occur with these drivers (Fig. 3b,c and Extended Data Fig. 10e). This suggested that the oncogenic competence of full-length FGFR2amp depends on specific cooperating driver genes.
Extended Data Fig. 10. Top driver genes co-occurring in samples with FGFR2 alterations.
a, 3,044 samples classified as either FGFR2-E18-truncated (n = 1,344, 44.2% of total, 0.54% incidence), amplified (n = 757, 24.8% of total, 0.30% incidence), or missense hotspot mutant (n = 943, 31.0% of total, 0.38% incidence) and top-30 co-enriched tumour driver alterations found in the FMI pan-cancer cohort (n = 249,570). b, Enrichments of top-30 tumour driver co-alterations in the indicated FGFR2 alteration categories in the FMI pan-cancer cohort. c, Odds ratios (OR) of top-30 tumour driver co-alterations in the indicated FGFR2 alteration categories (E18-truncation, n = 1,344; E1-E18 amp, n = 757; missense hotspot mut, n = 943) versus FGFR2 WT samples (n = 224,711) of the FMI pan-cancer cohort. Data are represented as log2-transformed OR ± 95% confidence interval (CI). Co-occurrence, OR > 1; mutual exclusivity, OR < 1. d, Frequencies (top panel) and distributions (bottom panel) per tumour type of the indicated FGFR2 alteration categories in the FMI pan-cancer cohort. e, Enrichment of top tumour driver co-alterations in the indicated FGFR2 alteration categories in the FMI-CHOL, OV, COAD/READ, ESCA/STAD, and LUAD/LUSC cohorts. P values were calculated with one-tailed proportion z-tests (b, e) or two-tailed Fisher’s exact tests (c) and FDR multiple-testing corrections using the Benjamini-Hochberg method (b, c, e). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Sample sizes and statistical details for b, c, e are in Supplementary Table 2.
Fig. 3. The oncogenic competence of FGFR2 alterations depends on co-occurring drivers.
a, Analysis of 528 breast cancer samples classified as either FGFR2 E18-truncated (n = 157, 29.7% of total, 0.70% incidence), E1–E18 amplified (n = 256, 48.5% of total, 1.14% incidence) or missense hotspot mutant (n = 115, 21.8% of total, 0.51% incidence), and the top co-enriched tumour driver alterations found in 22,380 breast cancer profiles from FMI. b, Enrichment of the top tumour driver co-alterations in the indicated FGFR2 alteration categories in the FMI breast cancer cohort. c, The odds ratios (ORs) of the top tumour driver co-alterations in the indicated FGFR2 alteration categories (E18 truncation, n = 157; E1–E18 amplification, n = 256; missense hotspot mutation, n = 115) versus FGFR2 WT samples (n = 22,307) of the FMI breast cancer cohort. Data are represented as log2-transformed OR ± 95% confidence interval (CI). Co-occurrence, OR > 1; mutual exclusivity, OR < 1. P values were calculated using one-tailed proportion Z-tests (b) or two-tailed Fisher’s exact tests (c) with FDR multiple-testing corrections using the Benjamini–Hochberg method (b and c). Sample sizes and statistical details for b and c are shown in Supplementary Table 2. d–f, Kaplan–Meier analysis of the mammary-tumour-free survival of Trp53F/F and Trp53F/F;Rosa26-Cas9 (d) or WT (e,f) female mice that were intraductally injected with lentiviruses encoding the indicated variants. Cohort counts (n) represent injected mammary glands (MGs) per number of mice. The Fgfr2FL and Fgfr2ΔE18 curves in d–f are duplicates from Fig. 2c. P values were calculated using log-rank (Mantel–Cox) tests. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We therefore combined lentiviral Fgfr2FL with cre to delete floxed Trp53 (Trp53F) alleles and a single-guide RNA against Pten (sgPten) to disrupt the endogenous Pten locus. Intraductal delivery of Fgfr2FL-P2A-cre or sgPten-Fgfr2FL-P2A-cre lentiviruses into mammary glands of Trp53F/F or Trp53F/F;Cas9 mice, respectively, significantly increased Fgfr2FL tumorigenicity. Fgfr2FL became nearly as oncogenic as Fgfr2ΔE18 when Trp53 and Pten were concomitantly lost, whereas Fgfr2ΔE18 oncogenicity was unaffected by the loss of Trp53 and/or Pten (Fig. 3d and Extended Data Fig. 11a). Similarly, combinations of Fgfr2FL with Myc, Fgf3 and/or Ccnd1 cDNAs into single lentiviral constructs cooperatively shortened tumour onset after intraductal delivery, with the latencies of the Fgfr2FL-T2A-Myc and Fgfr2FL-T2A-Fgf3-P2A-Ccnd1 combinations matching Fgfr2ΔE18 single-driver latency. Notably, Myc, Fgf3 and Ccnd1 alone were effectively non-tumorigenic (Fig. 3e,f and Extended Data Fig. 11b,c). Evaluation of mammary glands containing Fgfr2FL and co-driver alterations confirmed targeting or expression of the driver combinations and revealed high-grade tumours comparable to Fgfr2ΔE18-driven lesions (Extended Data Fig. 11d,e). Thus, Fgfr2FL oncogenicity relied on a cooperative oncogenomic network, whereas Fgfr2ΔE18 acted as a context-independent oncogene.
Extended Data Fig. 11. Somatic modelling of Fgfr2 variants and co-occurring driver alterations.
a-c, Kaplan-Meier curves showing mammary tumour-specific survival of Trp53F/F and Trp53F/F;Rosa26-Cas9 (a) and WT (b, c) female mice intraductally injected with lentiviruses encoding indicated variants. Trp53F/F;(Rosa26-Cas9), Lenti-Cre, sgPten–Cre, n = 10; Fgfr2FL-P2A-Cre, n = 15; Fgfr2ΔE18-P2A-Cre, sgPten–Fgfr2FL-P2A-Cre, sgPten–Fgfr2ΔE18-P2A-Cre, n = 9 mice. WT, Myc, Fgfr2FL-T2A-Fgf3, n = 9; Fgfr2FL-T2A-Myc, Fgf3, Ccnd1, Fgf3-P2A-Ccnd1, Fgfr2FL-P2A-Ccnd1, n = 10; Fgfr2ΔE18-T2A-Myc, Fgfr2ΔE18-P2A-Ccnd1, n = 7; Fgfr2FL-T2A-Fgf3-P2A-Ccnd1, n = 8; Fgfr2ΔE18-T2A-Fgf3, n = 6 mice. Fgfr2FL and Fgfr2ΔE18 curves are duplicates from Extended Data Fig. 6d. P values were calculated with log rank (Mantel-Cox) tests. ****P < 0.0001. d, Mammary tumour type classifications of somatic mouse models based on H&Es. WT, Myc, n = 30 of 9; Fgfr2FL-T2A-Myc, n = 20 of 10; Fgfr2ΔE18-T2A-Myc, Fgfr2ΔE18-P2A-Ccnd1, n = 14 of 7; Ccnd1, n = 40 of 10; Fgfr2FL-T2A-Fgf3, n = 18 of 9; Fgfr2FL-P2A-Ccnd1, Fgfr2FL-T2A-Fgf3-P2A-Ccnd1, n = 16 of 8; Fgfr2ΔE18-T2A-Fgf3, n = 12 injected MGs of 6 mice. Trp53F/F;(Rosa26-Cas9), Lenti-Cre, n = 31 of 10; Fgfr2FL-P2A-Cre, n = 14 of 7; Fgfr2ΔE18-P2A-Cre, sgPten–Fgfr2FL-P2A-Cre, sgPten–Fgfr2ΔE18-P2A-Cre, n = 18 of 9; sgPten–Cre, n = 20 injected MGs of 10 mice. WT Fgfr2FL and Fgfr2ΔE18 classifications are duplicates from Extended Data Fig. 8b. e, Histo-scoring of indicated IHC stains on mammary tumours from somatic mouse models. WT, Fgfr2FL, n = 5 of 5; Fgfr2ΔE18, n = 16 of 11; Myc, n = 7 of 3; Fgfr2FL-T2A-Myc, n = 15 of 10; Fgfr2ΔE18-T2A-Myc, n = 12 of 6; Ccnd1, n = 6 of 2; Fgfr2FL-T2A-Fgf3, n = 12 of 8; Fgfr2FL-P2A-Ccnd1, n = 14 of 9 ; Fgfr2FL-T2A-Fgf3-P2A-Ccnd1, n = 15 of 8; Fgfr2ΔE18-T2A-Fgf3, n = 12 of 7; Fgfr2ΔE18-P2A-Ccnd1, n = 13 tumours of 7 mice. Trp53F/F;(Rosa26-Cas9), Lenti-Cre, n = 10 of 8; Fgfr2FL-P2A-Cre, n = 8 of 5; Fgfr2ΔE18-P2A-Cre, n = 15 of 10; sgPten–Cre, n = 15 of 9; sgPten–Fgfr2FL-P2A-Cre, n = 14 of 7; sgPten–Fgfr2ΔE18-P2A-Cre, n = 14 tumours of 9 mice. In d, e, one tissue section per MG was stained and quantified for each of the indicated stains acquired in 4 independent randomized batches across all Fgfr2 variants and genotypes.
FGFR2ΔE18 tumours are sensitive to FGFRi
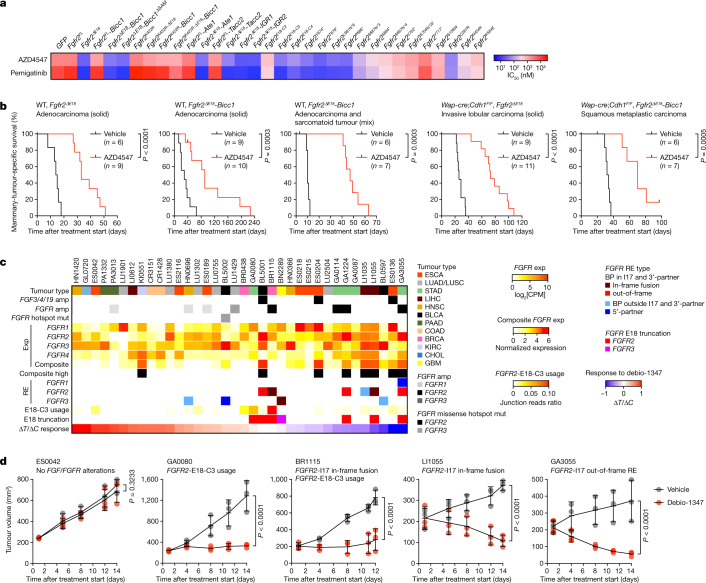
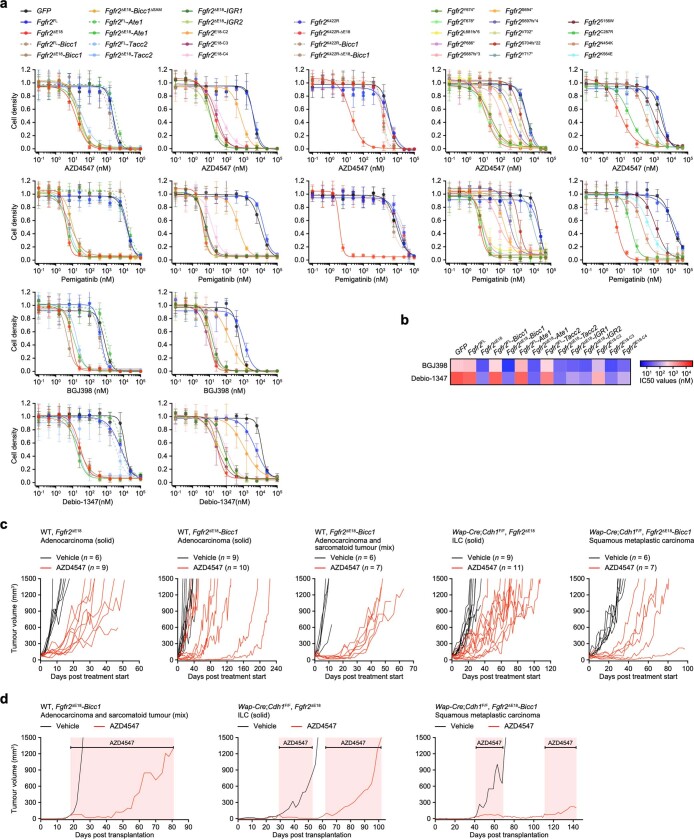
We next tested whether different Fgfr2 variants were sensitive to the clinical FGFR inhibitors (FGFRi) AZD4547, pemigatinib, BGJ398 and debio-1347. Expression of Fgfr2ΔE18 variants and Fgfr2C287R rendered mouse mammary epithelial cells highly sensitive to FGFR2 inhibition (Fig. 4a and Extended Data Fig. 12a,b). As a consequence, FGFRi suppressed both Fgfr2ΔE18-variant-induced signalling and soft agar clonogenicity (Extended Data Figs. 5g,h and 6a). By contrast, cells expressing Fgfr2FL and the remaining Fgfr2hotspot variants were less sensitive to FGFRi (Fig. 4a and Extended Data Fig. 12a,b). We also orthotopically transplanted tumours driven by Fgfr2ΔE18 variants and treated the recipient mice with AZD4547, which significantly suppressed tumour growth (Fig. 4b and Extended Data Fig. 12c,d).
Fig. 4. Human and mouse FGFR2 alteration cancer models are sensitive to FGFRi.
a, Half-maximum inhibitory concentration (IC50) value quantifications of 2D-grown NMuMG cells expressing GFP or the indicated Fgfr2 variants and treated with AZD4547 or pemigatinib for 4 days. Data are the mean of 5 independent experiments (GFP, Fgfr2FL, Fgfr2ΔE18) or 1 experiment (other Fgfr2 variants). b, Kaplan–Meier analysis of mammary-tumour-specific survival of female syngeneic WT mice bearing mammary fat pad transplants derived from the indicated tumour donors and treated daily orally with vehicle or 12.5 mg per kg AZD4547 using an intermittent dosing regimen. P values were calculated using log-rank (Mantel–Cox) tests. c, Collection of PDX models (n = 36) rank-ordered according to debio-1347 ΔT/ΔC response ratios. FGF/FGFR copy number alteration and mutation data and RNA-seq profiles to analyse FGF/FGFR expression (exp) were obtained from CrownBio-HuPrime, and had been generated from non-treated PDXs. Composite FGFR expression was defined as high if normalized expression > 3. FGFR2-E18-C3 use and FGFR RE types were identified in RNA-seq profiles. GBM, glioblastoma multiforme; KIRC, kidney renal clear cell carcinoma; LIHC, liver hepatocellular carcinoma. d, Growth curves of the indicated PDXs engrafted in female BALB/c nude mice and treated daily orally with vehicle or debio-1347 (BR1115 and LI1050, 60 mg per kg; ES0042, GA0080 and GA3055, 80 mg per kg). n = 3 mice per PDX model and treatment group. Data are mean ± s.d. P values were calculated using one-tailed two-way analysis of variance with FDR multiple-testing corrections using the two-stage step-up method from Benjamini, Krieger and Yekutieli.
Extended Data Fig. 12. In vitro and in vivo sensitivity of Fgfr2 variants to FGFRi.
a, Dose-response curves of 2D-grown NMuMG cells expressing GFP or indicated Fgfr2 variants and treated with AZD4547, pemigatinib, BGJ398, or debio-1347 for 4 days. Data are represented as mean ± s.d. of n = 5 replica per group collected across 5 independent experiments. b, Half-maximum inhibitory concentration (IC50) value quantifications of BGJ398 and debio-1347 dose-response curves in a. Data are represented as mean of 3 independent experiments (GFP, Fgfr2FL, Fgfr2ΔE18) or 1 experiment (other Fgfr2 variants). IC50 values for AZD4547 and pemigatinib are displayed in Fig. 4a. c, Individual growth curves of indicated tumour donors transplanted into the mammary fat pad of female syngeneic WT mice and treated daily orally with vehicle or 12.5 mg/kg AZD4547 using a previously established55 intermittent dosing regimen. d, Selected tumour transplant growth curves of mice in c. Durations of AZD4547 treatments according to intermittent dosing regimen are indicated.
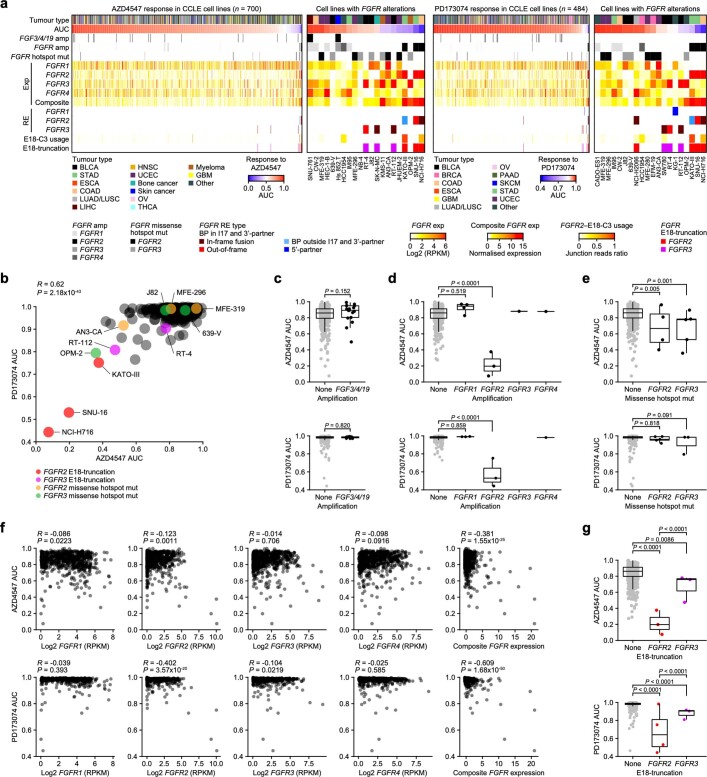
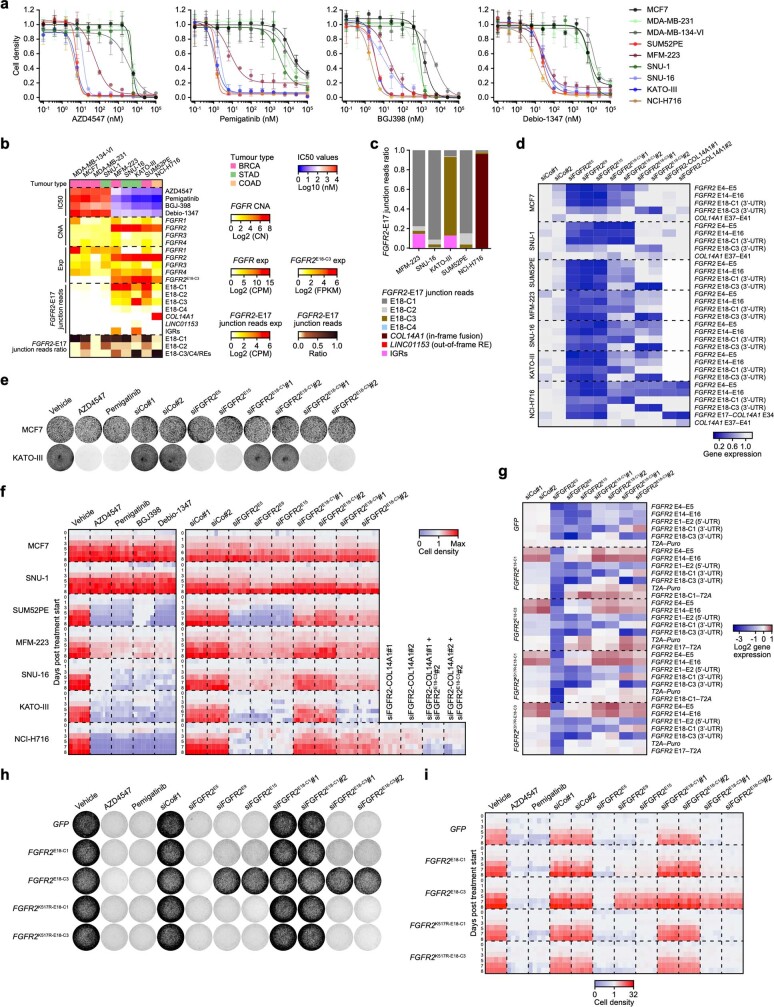
To further investigate the connection between distinct FGFR2 alterations and the FGFRi response in human tumour models, we analysed human cancer cell line pharmacogenomic datasets30–32. We evaluated the association between dose–responses to the FGFRi AZD4547 and PD173074 and genomic and transcriptomic features that potentially affect FGFR signalling, that is, FGF3/4/19 amplification, FGFR1–4 mutation, amplification, RE and expression, and use of FGFR2-E18-C3 (Extended Data Fig. 13a,b and Supplementary Table 5). Among these, FGFR2/3 missense hotspot mutations, FGFR2 expression and composite FGFR expression—a biomarker of FGFRi response33—modestly correlated with FGFRi sensitivity (Extended Data Fig. 13c–f), whereas cell lines with concurrent FGFR2amp and E18 truncations through I17 REs and/or E18-C3 use exhibited high sensitivity to FGFRi (Extended Data Fig. 13a,g). Cell lines expressing E18-truncating FGFR3 REs were less sensitive to FGFRi compared with cells with FGFR2ΔE18 (Extended Data Fig. 13b,g). On the basis of these correlates, we obtained the human SUM52PE, MFM-223, SNU-16, KATO-III and NCI-H716 cancer cell lines, each expressing amplified FGFR2 variants15–18,34,35. These cells highly expressed full-length FGFR2E18-C1 but also E18-truncated transcripts, namely FGFR2E18-C3 and FGFR2-I17 REs, and were sensitive to FGFRi (Extended Data Fig. 14a–c). To functionally dissect the dependence on FGFR2E18-C1 versus E18-truncated transcripts, we used small interfering RNAs (siRNAs) targeting either shared or unique FGFR2 exons (Extended Data Fig. 14d). Silencing of all FGFR2 transcripts suppressed the growth of cell lines with FGFR2amp (Extended Data Fig. 14e,f). Regardless of expression prevalence (Extended Data Fig. 14c), the growth of these cell lines could also be inhibited by specific silencing of E18-truncated FGFR2 RE or E18-C3 transcripts. By contrast, siRNAs specifically targeting FGFR2E18-C1 only marginally suppressed cell line growth (Extended Data Fig. 14e,f). Importantly, in KATO-III cells mainly expressing FGFR2E18-C3 (Extended Data Fig. 14c), overexpression of FGFR2E18-C3 fully rescued silencing of any FGFR2 transcripts, which depended on a functional FGFR2 kinase domain. However, full-length FGFR2E18-C1 was hardly able to rescue silencing of E18-truncated FGFR2E18-C3 (Extended Data Fig. 14g–i and Supplementary Table 4).
Extended Data Fig. 13. Correlation of FGFR2 alterations to FGFRi sensitivity across CCLE.
a, The Broad Institute Cancer Cell Line Encyclopedia (CCLE) cell lines rank-ordered according to their AZD4547 or PD173074 FGFRi area under the sigmoid-fit concentration-response curve (AUC) values derived from the Cancer Therapeutics Response Portal (CTRP) v2 deposited in the PharmacoDB database (n = 700) and the Genomics of Drug Sensitivity in Cancer (GDSC) database (n = 484), respectively. Data on FGF/FGFR mutations, CN status, REs, and expression and FGFR2–E18-C3 usage was obtained from CCLE. RPKM, reads per kilobase of transcript per million mapped reads. b, Correlation of AZD4547 versus PD173074 AUC values across shared CCLE cell lines (n = 384). c, AZD4547 and PD173074 AUC values in CCLE cell lines with FGF3/4/19 amp (AZD4547, n = 17; PD173074, n = 16) versus unaltered cell lines (AZD4547, n = 658; PD173074, n = 455). d, AZD4547 and PD173074 AUC values in CCLE cell lines with FGFR1 amp (AZD4547, n = 4; PD173074, n = 3), FGFR2 amp (AZD4547, n = 3; PD173074, n = 3), FGFR3 amp (AZD4547, n = 1; PD173074, n = 0), or FGFR4 amp (AZD4547, n = 1; PD173074, n = 1) versus unaltered cell lines (AZD4547, n = 666; PD173074, n = 464). e, AZD4547 and PD173074 AUC values in CCLE cell lines with FGFR2 missense hotspot mut (AZD4547, n = 4; PD173074, n = 5) or FGFR3 missense hotspot mut (AZD4547, n = 5; PD173074, n = 3) versus unaltered cell lines (AZD4547, n = 691; PD173074, n = 476). f, Correlations of FGFR1, FGFR2, FGFR3, FGFR4, or composite FGFR expression versus AZD4547 or PD173074 AUC values across CCLE cell lines. g, AZD4547 or PD173074 AUC values in CCLE cell lines expressing E18-truncated FGFR2 (AZD4547, n = 3; PD173074, n = 4) or FGFR3 (AZD4547, n = 3; PD173074, n = 3) versus no truncation (AZD4547, n = 694; PD173074, n = 477). Data in c–e, g are represented as median (centre line) ± IQR (25th to 75th percentile, box) and IQR ± 1.5 x IQR (whiskers) . P values were calculated with two-tailed t-transformations of Pearson’s R correlation coefficients (b, f), two-tailed Wilcoxon rank-sum tests (c), or one-tailed one-way ANOVA and Tukey’s multiple-testing corrections (d, e, g).
Extended Data Fig. 14. Human cancer cell lines depend on FGFR2ΔE18 variants.
a, Dose-response curves of indicated human cancer cell lines treated with AZD4547, pemigatinib, BGJ398, or debio-1347 for 4 days. Data are represented as mean ± s.d. of n = 5 replica per group collected across 2 independent experiments. b, Human cancer cell lines rank-ordered according to IC50 values for indicated FGFRi in (a). FGFR1-4 CNA and expression is based on low-coverage WGS and RNA-seq profiles. FGFR2E18-C3 isoform expression, FGFR2-E17 junction reads expression, and FGFR2 RE types were identified in RNA-seq profiles. FPKM, fragments per kilobase of transcript per million mapped reads. c, Distribution of FGFR2-E17 junction reads to canonical full-length E18-C1 versus noncanonical E18-C2/C3/C4, RE partners, and IGRs in indicated human cancer cell lines. d, Heatmap showing silencing of FGFR2 variants in indicated human cancer cell lines using small interfering (si) RNAs. Cells were transfected with the following siRNAs: non-targeting siRNAs (siCo), siRNAs targeting shared exons among FGFR2 isoforms (siFGFR2E5,E9,E15), siRNAs specifically targeting canonical E18-C1 of FGFR2FL (siFGFR2E18-C1) or E18-C3 of truncated FGFR2E18-C3 (siFGFR2E18-C3), or siRNAs specifically targeting the FGFR2-COL14A1 fusion (siFGFR2-COL14A1). Silencing of specific FGFR2 variants was detected with RT-qPCR using primers spanning indicated cDNA segments. Expression of each cDNA segment is normalized to USF1 expression and cDNA segment expression in siCo condition of each cell line (average of siCo#1 and siCo#2). Data are represented as mean of n = 3 technical replica per group. Data represent 1 replica of 2 independent experiments. e, f, Representative images of 6-well plate wells at 8 days post treatment start (e) and cell density quantifications over 8 days (f) of 2D-grown indicated cell lines treated with vehicle or 100 nM AZD4547, pemigatinib, BGJ398, or debio-1347 or (co)-transfected with siCo, siFGFR2E5, siFGFR2E9, siFGFR2E15, siFGFR2E18-C1, siFGFR2E18-C3, and/or siFGFR2-COL14A1. Data in f represent n = 6 independent replica collected across 1 experiment (MCF7, siRNA treatments), n = 10 independent replica collected across 2 independent experiments (KATO-III vehicle, AZD4547, and siRNA treatments), or n = 5 independent replica collected across 2 independent experiments (other cell lines and/or treatment conditions). g, Heatmap showing silencing of FGFR2 isoforms using indicated siRNAs in KATO-III cells expressing GFP or indicated FGFR2 variants. Validation of overexpression of FGFR2 variants using RT-qPCR is in Supplementary Table 4. FGFR2K517R variants encode KD-dead FGFR2 variants. siFGFR2E5 targets endogenous FGFR2 transcripts and FGFR2 transcripts derived from lentiviral constructs. Other siRNAs specifically target endogenous FGFR2 transcripts. Silencing of specific FGFR2 variants was detected with RT-qPCR using primers spanning indicated cDNA segments. E4–E5 and E14–E16 primers detect endogenous FGFR2 transcripts and FGFR2 transcripts derived from lentiviral constructs. E1–E2 (5′-UTR), E18-C1 (3′-UTR), and E18-C3 (3′-UTR) primers specifically detect endogenous FGFR2 transcripts. E18-C1–T2A, E17–T2A, and T2A–Puro primers specifically detect FGFR2 transcripts derived from lentiviral constructs. Log2-transformed expression of each cDNA segment is normalized to USF1 expression and cDNA segment expression in siCo-treated GFP-expressing cells (average of siCo#1 and siCo#2). Data are are represented as mean of n = 3 technical replica per group of 1 experiment. h, i, Representative images of 6-well plate wells at 8 days post treatment start (h) and cell density quantifications over 8 days (i) of 2D-grown KATO-III cells expressing GFP or indicated FGFR2 variants and treated with vehicle, 100 nM AZD4547, or 100 nM pemigatinib or transfected with siCo, siFGFR2E5, siFGFR2E9, siFGFR2E15, siFGFR2E18-C1, or siFGFR2E18-C3. Data in i represent n = 6 independent replica per group collected across 1 experiment.
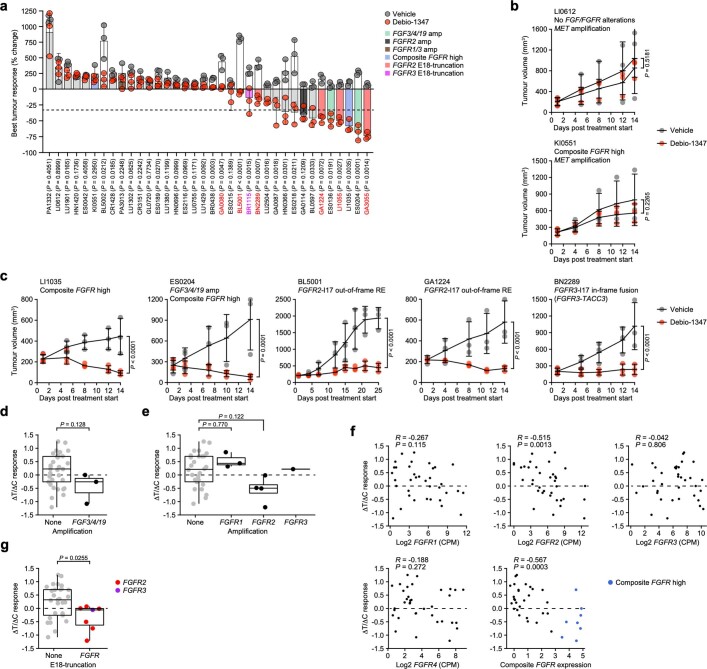
We next screened the CrownBio-HuPrime patient-derived xenograft (PDX) collection for the occurrence of genomic and transcriptomic alterations in the FGFR signalling pathway and enrolled the PDXs into a drug-intervention study using debio-1347 (Fig. 4c and Supplementary Table 6). All PDXs with FGFR2/3-I17 in-frame fusions as well as those with noncanonical FGFR2 REs or E18-C3 use strongly responded to FGFR blockade (Fig. 4d and Extended Data Fig. 15a–c). Among the correlates with debio-1347 treatment response were FGFR2 and composite FGFR expression (Extended Data Fig. 15d–f), but especially truncation of FGFR2/3-E18 exhibited substantial correlation with debio-1347-mediated growth inhibition (Extended Data Fig. 15g). Thus, human tumour models express and are dependent on E18-truncated FGFR2 and FGFR3 variants and are actionable by FGFR-targeted therapies.
Extended Data Fig. 15. PDXs expressing FGFR2ΔE18 variants are sensitive to debio-1347.
a, Best percentage change from baseline tumour volume in patient-derived xenograft (PDX) models (n = 36) engrafted in female NOD-SCID (BL5001, BL5002) or BALB/c Nude (all other PDX models) mice and treated daily orally with vehicle or debio-1347 (n = 3 mice per PDX model and treatment group). Coloured bars indicate identified FGF/FGFR2 alterations detailed in Fig. 4c. b, c, Growth curves of indicated PDXs models engrafted in NOD-SCID or BALB/c Nude mice and treated daily orally with vehicle or debio-1347 (ES0204, Li0612, LI1035, BN2289, 60 mg/kg; GA1224, KI0551, 80 mg/kg; BL5001, day 1-14, 40 mg/kg; day 15-25, 60 mg/kg; n = 3 mice per PDX model and treatment group). d, Debio-1347 ΔT/ΔC response ratios in PDXs with FGF3/4/19 amp (n = 3) versus normal CN (n = 33). e, Debio-1347 ΔT/ΔC response ratios in PDXs with FGFR1 amp (n = 3), FGFR2 amp (n = 4), or FGFR3 amp (n = 1) versus normal CN (n = 28). f, Correlations of FGFR1, FGFR2, FGFR3, FGFR4, or composite FGFR expression versus debio-1347 ΔT/ΔC response ratios across PDXs. Composite FGFR expression was defined as high, if normalized expression > 3. g, Debio-1347 ΔT/ΔC response ratios in PDXs expressing E18-truncated FGFR2 (n = 6) or FGFR3 (n = 1) versus no truncation (n = 29). Data are represented as mean ± s.d. (a, b) or as median (centre line) ± IQR (25th to 75th percentile, box) and IQR ± 1.5 x IQR (whiskers) (d, f). P values were calculated with two-tailed unpaired Student’s t-tests (a), one-tailed two-way ANOVA and FDR multiple-testing corrections using the two-stage step-up method from Benjamini, Krieger, and Yekutieli (b, c), two-tailed Wilcoxon rank-sum tests (d, g), one-tailed one-way ANOVA and Tukey’s multiple-testing corrections (e), or two-tailed t-transformations of Pearson’s R correlation coefficients (f).
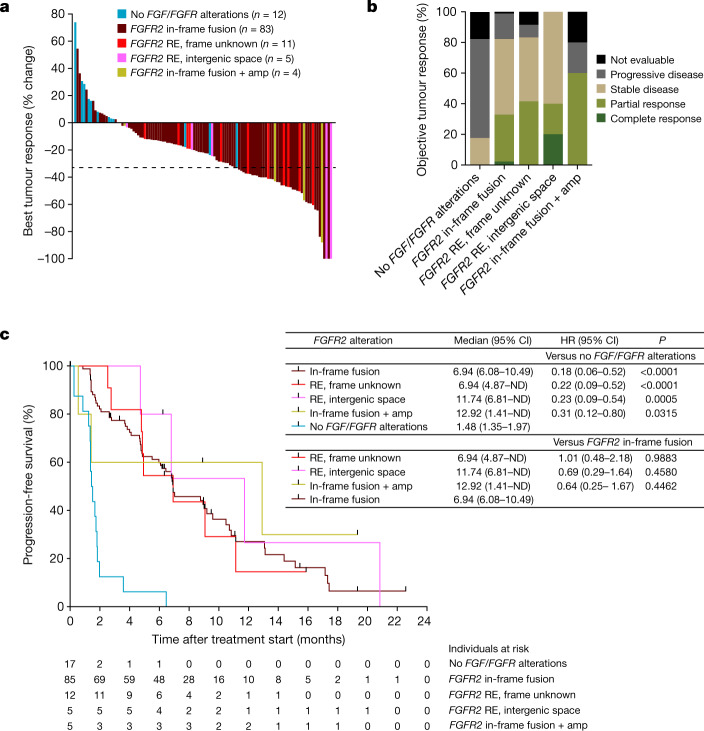
These findings suggest that patients with cancer with any type of FGFR2ΔE18 variant might respond to FGFR2 targeting. We therefore re-examined FIGHT-202 (NCT02924376), a phase II trial of pemigatinib in patients with advanced cholangiocarcinoma. Patients with fusions or REs in the FGFR2-I17/E18 hotspot had an objective response rate of 35.5%, whereas those with other or no FGF/FGFR alterations had no response9. To determine which classes of FGFR2 REs benefit from pemigatinib, we stratified individual patients according to their FGFR2amp status and E18-damaging RE types, namely in-frame fusions, frame unknown REs and REs with IGRs. Patients with FGFR2 in-frame fusions, frame unknown REs and IGR REs—independently of FGFR2amp status—showed strong tumour responses to pemigatinib therapy (Fig. 5a), resulting in objective responses or stable disease in 80–100% of patients, irrespective of the diagnosed FGFR2 RE type (Fig. 5b). As a consequence, although the patient cohort with no FGF/FGFR alterations quickly progressed during pemigatinib treatment (Fig. 5a–c), the four patient cohorts with non-amplified and amplified FGFR2 in-frame fusions and non-canonical REs showed equally prolonged progression-free survival times (Fig. 5c). Taken together, FGFR2ΔE18, generated by either in-frame fusions or other REs, is a clinically actionable oncogene in patients with cholangiocarcinoma and probably in patients with other types of cancer.
Fig. 5. Patients with cholangiocarcinoma respond to FGFR-targeted therapy irrespective of FGFR2 RE type.
a, Centrally assessed best percentage change from the baseline in target lesion size of 115 (92%) of 125 individual patients with cholangiocarcinoma treated with pemigatinib, who had post-baseline scans. Data are from the FIGHT-202 study9 and the coloured bars indicate FGFR2-I17/E18 RE types and FGFR2 amplification status as diagnosed by FoundationOne. b, Objective tumour responses observed in the FIGHT-202 study assessed according to the Response Evaluation Criteria in Solid Tumours v.1.1 (RECIST 1.1) and grouped according to FGFR2 RE types/amplification status. No FGF/FGFR alterations (n = 17), FGFR2 in-frame fusion (n = 85), frame unknown RE (n = 12), intergenic space RE (n = 5), in-frame fusion + amplification (n = 5). ‘Not evaluable’ indicates that the patient was not evaluable for response using RECIST. c, Kaplan–Meier analysis of the progression-free survival of patients with cholangiocarcinoma treated with pemigatinib from the FIGHT-202 study and grouped according to FGFR2 RE types/amplification status. Data are median ± 95% CI for each cohort, and log-rank hazard ratios (HR) ± 95% CI for the indicated comparisons are shown. P values were calculated using log-rank (Mantel–Cox) tests. ND, not defined.
Discussion
The C-terminal tail of FGFR2 encoded by E18 proposedly moderates RTK signalling14. The proximal part of the C terminus includes the RTK internalization motif27, a tyrosine residue that is relevant for PI3K-pathway attenuation25,36, and serine residues that bind to ERK1/2 and RSK2 to stimulate receptor endocytosis and suppress MAPK signalling37,38. The distal C terminus contains proline-rich motifs that bind to GRB2 and mitigate kinase domain activity39,40. We found that especially proximal C-terminal truncation is critical for oncogenic signal transduction, as evidenced by the gradually accelerated tumorigenesis observed for the Fgfr2E18-C2 variant and proximal E18-truncating mutations. Notably, C-terminally truncated isoforms of EGFR, HER2 and other RTKs have also shown elevated transforming activities41–44. Thus, the paradigm of C-terminal FGFR2 truncation identified here might be key to the pathogenicity of multiple RTKs.
FGFR2 amplification and fusion structural variants have been considered to be relevant tumour drivers owing to the consequential receptor overexpression and constitutive dimerization mediated by oligomerization domains in the fusion partners2,20–22. We identified that the tumour-driver potential of C-terminally truncated FGFR2 is independent of specific fusion partners, whereas full-length FGFR2 overexpression was marginally tumorigenic in the absence of other driver alterations. The oncogenicity of FGFR2amp might therefore depend on the ability to generate FGFR2ΔE18 transcripts, for example, through complex REs16,17,24. As shown in our study, FGFR2ΔE18 acts as a potent single-driver alteration. By contrast, the oncogenic competence of full-length FGFR2 relied on co-drivers that may augment canonical FGFR2 signalling45–48 and thereby phenocopy the strong signalling induction observed for C-terminally truncated FGFR2. In clinical trials, objective responses to FGFRi were scarce in patients with FGFR2amp tumours2–4. Interestingly, tumours with overexpression of E18-truncated FGFR2E18-C3 responded particularly well to FGFR2 targeting2. In cohorts of mixed FGFR alterations, patients with FGFR2-E18-truncating fusions displayed favourable responses over patients with other FGF/FGFR alterations5–9. Thus, FGFRi efficacy might be dictated by the expression of single-driver FGFR2ΔE18 versus FGFR2 alterations that depend on oncogenic co-drivers. In FGFR-aberrant cancers, MYC or CCND1 amplifications can indeed confer resistance to FGFRi48–50. Combination therapies might therefore elevate the response rates in FGFR2amp tumours, as proposed for FGFRi–CDK4/6i combination therapy in patients with breast cancer with FGFR2 and CCND1 amplifications50.
Our findings have fundamental implications for the selection of patients for FGFR2 targeting therapies. Instead of considering patients on the basis of FGFR2 mutation, fusion or amplification status alone, our data suggest that expression of oncogenic FGFR2 transcripts and co-mutational landscapes should also be considered. Importantly, identifying cancers with structural variants or mutations that result in expression of FGFR2ΔE18 variants will be a highly relevant biomarker for FGFR-targeted therapeutics, and may substantially expand the number of cancer patients who may benefit from such therapy.
Methods
Mouse models
GEMMs
The FVB/NRj Wap-cre;Cdh1F/F, Trp53F/F, Trp53F/F;Rosa26-Cas9, and Rosa26-mT/mG mouse strains were maintained at the NKI animal facility and PCR-genotyped as previously described12,13,51–53. To generate GEMMs bearing Fgfr2-IRES-Luc alleles, mouse Fgfr2 (NM_201601.2) was isolated from a cDNA clone (MC221076, OriGene) using the primer sequences listed in Supplementary Table 7 amplifying Fgfr2-E1–E18 (FL) or Fgfr2-E1–E17 (ΔE18) and the sequences were verified and inserted with FseI-PmeI fragments into the Frt-invCag-IRES-Luc vector (shuttle vector). This resulted in the Frt-invCAG-Fgfr2FL-IRES-Luc and Frt-invCAG-Fgfr2ΔE18-IRES-Luc alleles. Flp-mediated integration of the shuttle vectors in Wap-cre;Cdh1F/F;Col1a1frt/+ GEMM-derived embryonic stem cell (ESC) clones (FVB/NRj background) and subsequent blastocyst injections of the modified ESCs were performed using the GEMM–ESC methodology54. Chimeric animals were mated with Cdh1F/+ and Cdh1F/F mice on the FVB/NRj background to generate the experimental cohorts. The Col1a1frt-invCAG-Fgfr2-IRES-Luc/+ and WT alleles were detected using standard PCR with an annealing temperature of 58 °C using the primer sequences listed in Supplementary Table 8 generating the following PCR products: Col1a1frt-invCAG-Fgfr2-FL-IRES-Luc, 585 bp; Col1a1frt-invCAG-Fgfr2-ΔE18-IRES-Luc, 420 bp; and WT, 234 bp. Here, Col1a1frt-invCAG-Fgfr2-FL-IRES-Luc and Col1a1frt-invCAG-Fgfr2-ΔE18-IRES-Luc are referred to as Fgfr2FL-IRES-Luc and Fgfr2ΔE18-IRES-Luc, respectively. The GEMM cohorts were monitored weekly and mammary-tumour-free survival was scored (event) when the first palpable tumour was detected, whereas mice that did not develop any mammary tumours were censored. Tumour volume was measured in two dimensions using callipers as follows: volume = length × width2 × 0.5.
Somatic mouse models
To somatically model Fgfr2 variants in the mouse mammary gland, 6-week-old FVB/NRj WT, Wap-cre;Cdh1F/F, Trp53F/F, Trp53F/F;Rosa26-Cas9 or Rosa26-mT/mG female mice were intraductally injected as previously described12 with lentiviruses encoding Fgfr2 variants in combination with cre, Myc, Ccnd1, Fgf3 and/or a previously validated sgRNA targeting E7 of Pten (sgPten)12,13. In brief, 20 μl of high-titre lentiviruses were injected into the fourth and/or the third mammary glands using a 34G needle. Lentiviral titres ranging from 2 × 108 to 2 × 109 transfection units (TU) per ml were used. The somatic model cohorts were monitored twice weekly and mammary-tumour-free survival was scored (event) for each injected mammary gland individually when palpable tumours were detected, whereas mammary glands that did not develop any tumours were censored. Tumour volume was measured in two dimensions using callipers as follows: volume = length × width2 × 0.5.
AZD4547 intervention study
To allograft tumours, DMSO-preserved 1 mm3 tumour fragments derived from somatic Fgfr2 models were orthotopically transplanted into the right mammary fat pad of 8-week-old syngeneic FVB/NRj female mice (Janvier Labs) as previously described55. The mice were twice weekly weighed and monitored for mammary tumour development and, as soon as tumours reached a volume of 62.5 mm3 (5 × 5 mm, measured in two dimensions using callipers; volume = length × width2 × 0.5), the mice were randomly allocated to vehicle versus AZD4547 FGFRi treatment arms. The treatments were performed daily through oral gavage using vehicle (1% Tween-80 in demineralized water) or 12.5 mg per kg AZD4547 (AstraZeneca) according to a previously optimized intermittent dosing regimen55. Mice were euthanized 1 h after the last dosing.
General guidelines
For all mouse models, mammary-tumour-specific survival was scored when a single mammary tumour burden reached a volume of 1,500 mm3, the total mammary tumour burden reached a volume of 2,000 mm3 or the mice suffered from clinical signs of distress, such as respiratory distress, ascites, distended abdomen, rapid weight loss and severe anaemia, caused by primary tumour burden or metastatic disease. Mice that were euthanized due to other circumstances were censored. The maximal permitted disease end points were not exceeded in any of the experiments. Mammary glands were collected and analysed for histological abnormalities. Sample sizes were determined using G*Power software (v.3.1)56 and were large enough to measure the effect sizes. Tumour measurements and post mortem analyses were performed in a blinded manner. The mouse colony was housed in a certified animal facility under a 12 h–12 h light–dark cycle in a temperature- and humidity-controlled room set to 21 °C and 55% relative humidity. The mice were kept in individually ventilated cages, and food and water were provided ad libitum. All of the animal experiments were approved by the Animal Ethics Committee of the Netherlands Cancer Institute and performed in accordance with institutional, national and European guidelines for Animal Care and Use.
In vivo bioluminescence imaging
In vivo bioluminescence imaging of luciferase expression was performed as previously described57 by intraperitoneally injecting 150 mg per kg beetle luciferin (E1601, Promega). Signal intensity was measured on the whole body of the mouse (excluding the head and tail) using an IVIS Spectrum In Vivo Imaging System (124262, PerkinElmer) operated by Living Image Software (v.4.5.2, PerkinElmer) and a size-fixed square. Signal intensity was quantified as flux (photons per second per cm2 per steradian).
Histology and IHC
Tissues were formalin-fixed and paraffin-embedded (FFPE), sectioned and processed for haematoxylin and eosin (H&E) histochemical and immunohistochemistry (IHC) staining using routine procedures. For IHC staining, antigen retrieval was performed with citrate buffer (CBB999, ScyTek) at pH 6 (FGF3, FGFR2, PTEN) or Tris-EDTA at pH 9 (MYC, Cyclin D1, E-cadherin, P53). Sections were incubated with primary antibodies (Supplementary Table 9) overnight at 4 °C. Primary antibodies were labelled with the EnVision+ HRP Labelled Polymer Anti-Rabbit System (K4003, Dako), visualized with the Liquid DAB+ Substrate Chromogen System (K3468, Dako) and counterstained with haematoxylin. The antibodies used were independently validated by a certified pathologist by evaluation of IHC results in positive and negative biological control FFPE tissues to ensure specificity and sensitivity. Moreover, negative technical controls were performed by omission of the primary antibody in extra sections for a randomly selected subset of the samples. H&E and E-cadherin slides were used to classify mammary tumour lesion types according to the international consensus of mammary pathology58. IHC stains were quantitatively analysed by evaluating tumour cell-specific positivity using a histo-scoring system (0, negative; 1, weakly positive; 2, moderately positive; 3, strongly positive) or by calculating a histo (H)-score for each tumour defined as follows: H-score = 1 × (the percentage of tumour cells with weak staining intensity) + 2 × (the percentage of tumour cells with moderate staining intensity) + 3 × (the percentage of tumour cells with strong staining intensity), resulting in a score between 0 and 300. All slides were reviewed and quantified by a comparative pathologist (S.K.) in a blinded manner. Slides were digitally processed using a Pannoramic 1000 whole-slide scanner (3DHISTECH) and captured using CaseViewer software (v.2.2.1, 3DHISTECH).
Isolation of MMECs
Primary mouse mammary epithelial cells (MMECs) were isolated from 10-week-old WT, Fgfr2FL-IRES-Luc and Fgfr2ΔE18-IRES-Luc female mice as previously described53. In brief, mammary glands were minced and digested with 4 mg ml−1 collagenase A (11088793001, Roche) and 25 μg ml−1 DNase I (DN25, Sigma-Aldrich) in Dulbecco’s modified Eagle medium/nutrient mixture F-12 (DMEM/F-12, 31331, Thermo Fisher Scientific) containing 100 IU ml−1 penicillin–streptomycin (15070, Thermo Fisher Scientific) for 1 h at 37 °C. Digests were passed through a 70 μm cell strainer that was prewetted with PBS containing 10% fetal bovine serum (FBS, S-FBS-EU-015, Serana) and 2 mM EDTA, and cells were cultured in DMEM/F-12 supplemented with 10% FBS, 100 IU ml−1 penicillin–streptomycin, 5 ng ml−1 epidermal growth factor (EGF, E4127, Sigma-Aldrich), 5 ng ml−1 insulin (I0516, Sigma-Aldrich) and 10 μM Y-27632 (M1817, AbMole). Contaminating fibroblasts were removed from MMEC cultures by differential trypsinization. Confluent wells were transduced with adenoviral Ad5CMVCre particles (AdCre, 1 × 109 TU ml−1; University of Iowa Viral Vector Core) in the presence of 8 μg ml−1 Polybrene (H9268, Sigma-Aldrich) for 48 h before subjection to subsequent assays. Switching of Fgfr2-IRES-Luc alleles was confirmed using 1 mg ml−1 beetle luciferin and bioluminescence imaging with an Infinite M Plex plate reader (Tecan) operated with Tecan i-control software (v.3.9.1, Tecan).
FACS analysis of mammary glands
Mammary glands of Rosa26-mT/mG female mice injected with Fgfr2-P2A-cre lentiviruses were processed as described for MMEC isolation. Single cells were stained with the fluorescence-activated cell sorting (FACS)-validated BV650-conjugated anti-EPCAM antibody (1:100, 740559, BD Biosciences) in FACS buffer (PBS with 10% FBS and 2 mM EDTA), labelled with the LIVE/DEAD Fixable Violet Dead Cell Stain Kit (405 nm excitation, L34964, Thermo Fisher Scientific), fixed with BD Phosflow Fix Buffer I (557870, BD Biosciences) and permeabilized with BD Phosflow Perm Buffer III (558050, BD Biosciences), each for 30 min at 4 °C. Cells were incubated with primary antibodies (anti-FGFR2, 1:200, 11835, Cell Signaling Technology; anti-GFP, 1:200, ab6673, Abcam) overnight and subsequently with secondary antibodies for 1 h both in FACS buffer at 4 °C. Anti-FGFR2 and anti-GFP antibodies were validated for FACS using NMuMG cells overexpressing GFP or FGFR2 versus control cells negative for these proteins. Details of the antibodies used are provided in Supplementary Table 9. FACS was performed using the BD LSRFortessa Cell Analyzer (BD Biosciences) equipped with the BD FACSDiva Software (v.8.0.2, BD Biosciences) and with 405 nm (450/50, 670/30 pass filters), 488 nm (530/30 pass filters) and 638 nm (670/30 pass filters) lasers to measure 405-Live/Dead, BV650–EPCAM, EGFP–AF488 and FGFR2–AF647, respectively. Data were analysed using FlowJo (v.10.7.1, BD Biosciences).
Lentiviral vectors and virus production
The SIN.LV.SF, SIN.LV.SF-T2A-Puro, SIN.LV.SF-GFP-T2A-Puro lentiviral vectors and the SIN.LV.SF-Cre (Lenti-Cre), SIN.LV.SF-P2A-Cre and pGIN sgPten–P2A-Cre lentivectors all encoding improved cre with mammalian codon usage (derived from pBOB-CAG-iCRE-SD, 12336, Addgene) and the last-mentioned encoding a validated sgRNA targeting E7 of Pten (sgPten) were all previously described10,12,13,59. Mouse Fgfr2 (NM_201601.2) and Myc (NM_010849.4) were isolated from cDNA clones (Fgfr2, MC221076, OriGene; Myc, 8861953, Source BioScience) using Q5 High-Fidelity DNA Polymerase (M0491S, New England Biolabs) and primers with AgeI–SalI overhangs amplifying Fgfr2FL or Fgfr2ΔE18, or BamHI–AgeI overhangs amplifying Myc. Amplicons were inserted into the SIN.LV.SF, SIN.LV.SF-T2A-Puro and SIN.LV.SF-P2A-Cre vectors, resulting in SIN.LV.SF-Fgfr2FL, SIN.LV.SF-Fgfr2ΔE18, SIN.LV.SF-Fgfr2FL-T2A-Puro, SIN.LV.SF-Fgfr2ΔE18-T2A-Puro, SIN.LV.SF-Fgfr2FL-P2A-Cre, SIN.LV.SF-Fgfr2ΔE18-P2A-Cre, and SIN.LV.SF-Myc. Custom-synthesized gBlocks gene fragments of mouse Ccnd1 (NM_001379248.1), Fgf3 (NM_008007.2), Ate1-E11–E12 (NM_001271343.1), Bicc1-E3–E21 (NM_001347189.1) and Tacc2-E15–E21 (NM_206856.4), which were the homologues of human FGFR2 fusion partner genes (Extended Data Fig. 2h), as well as human full-length FGFR2E18-C1 (NM_000141.4), FGFR2E18-C2 (XM_017015921.2), FGFR2E18-C3 (NM_001144913.1), FGFR2E18-C4 (NM_001144915.2), the IGR1 sequence (identified in TCGA-A8-A08A; Extended Data Fig. 4f) and E18 sequences resulting from frameshift mutations (Extended Data Fig. 2k) were purchased (Integrated DNA Technologies). gBlocks gene fragments or parts thereof were assembled in the respective lentivectors using PCR amplification of backbone and insert(s) and the In-Fusion HD Cloning Plus kit (638911, Takara Bio) according to the manufacturer’s recommendations. Point mutations in FGFR2, short deletions/insertions to generate gradual Fgfr2 E18 truncations and introduction of the IGR2 sequence (identified in TCGA-BH-A203; Extended Data Fig. 4g) to Fgfr2 were performed using the QuikChange Lightning Site-Directed Mutagenesis Kit (210519, Agilent). In-Fusion and site-directed mutagenesis primers were designed using SnapGene (v.5.2) and QuikChange Primer Design60, respectively. All lentivectors were verified using Sanger sequencing. Concentrated lentiviral stocks were produced by transient co-transfection of four plasmids in HEK293T cells as previously described61. Viral titres were determined using the qPCR Lentivirus Titre Kit (LV900, Applied Biological Materials).
Cell culture
HEK293T cells (CRL-3216, ATCC) were cultured in Iscove’s modified Dulbecco’s medium (31980, Thermo Fisher Scientific) containing 10% FBS and 100 IU ml−1 penicillin–streptomycin. MCF7 (HTB-22), MDA-MB-134-VI (HTB-23), MDA-MB-231 (HTB-26), NCI-H716 (CCL-251), NMuMG (CRL-1636), KATO-III (HTB-103), SNU-1 (CRL-5971) and SNU-16 (CRL-5974, all ATCC) as well as MFM-223 (98050130, ECACC) and SUM52PE (HUMANSUM-0003018, BioIVT) cells were cultured in DMEM/F-12 containing 10% FBS and 100 IU ml−1 penicillin–streptomycin. All cell lines were previously authenticated by providers. No re-authentication was carried out for this study. To stably express the lentiviral GFP-T2A-Puro or FGFR2-T2A-Puro constructs, NMuMG and KATO-III cells were transduced with lentiviral supernatants at equal TU per ml in the presence of 8 μg ml−1 Polybrene for 24 h. Transduced cells were selected with 2 μg ml−1 puromycin (A11138, Thermo Fisher Scientific) for 5 days and subsequently grown in DMEM/F-12 containing 10% FBS, 100 IU ml−1 penicillin–streptomycin, 1 μg ml−1 puromycin and 10 μM Y-27632. Overexpression of lentiviral constructs was verified using RT–qPCR (Supplementary Table 4). All cell lines were cultured in standard incubators at 37 °C with 5% CO2 and routinely tested for mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (LT07-218, Lonza).
Gene silencing using siRNA
Human cells were transfected with Silencer Select Negative Control 1 or 2 siRNAs (siCo#1 and #2, 4390844, 4390847, Thermo Fisher Scientific) or Silencer Select siRNAs designed with the GeneAssist Custom siRNA Builder (Thermo Fisher Scientific) to target shared exons (E5, E9, E15) among FGFR2 isoforms (siFGFR2E5/E9/E15), the 3′-UTR of E18-C1 of full-length FGFR2 (siFGFR2E18-C1), the 3′-UTR of E18-C3 of truncated FGFR2E18-C3 (siFGFR2E18-C3) or the FGFR2-COL14A1 fusion (siFGFR2-COL14A1). siFGFR2E5 targeted endogenous FGFR2 transcripts as well as FGFR2 transcripts derived from lentiviral constructs. All other siRNAs specifically targeted endogenous FGFR2 transcripts, because the FGFR2 cDNA sequences used in the lentivectors lacked 3′-UTRs and contained silent mutations in E9 and E15 to prevent the binding of siFGFR2E9/E15. A list of the custom-designed siRNA sequences is provided in Supplementary Table 10. siRNA (50 nM) was used in combination with the jetPRIME transfection reagent (114-15, Polyplus Transfection) as previously described62.
Drug-response curves
A total of 800 NMuMG or SNU-1 cells; 2,000 MCF7, MDA-MB-231, KATO-III, SNU-16, or SUM52PE cells; 3,000 MFM-223 or NCI-H716 cells; or 4,000 MDA-MB-134-VI cells per well were seeded in 96-well plates using DMEM/F-12 supplemented with penicillin–streptomycin and 10% FBS for human cell lines or 3% FBS for NMuMG. After 24 h, cells were treated with FGFRi for 4 days using vehicle (DMSO), AZD4547 (AstraZeneca), or pemigatinib (HY-109099), BGJ398 (HY-13311) or debio-1347 (HY-19957, all MedChemExpress) with a range of 0.1 nM to 100 μM. Usage of AZD4547, pemigatinib, BGJ398 and debio-1347 was previously described63–66. Cell viability was assayed using CellTiter-Blue Reagent (G808A, Promega) for 4 h and subsequently measuring fluorescence on the Infinite M Plex plate reader operated using the Tecan i-control software. Drug-response curves were modelled using [inhibitor] versus response with variable slope (four parameters) and least-squares regression in Prism (v.9.3.1, GraphPad Software).
2D colony-formation assay
A total of 5,000 KATO-III or MCF7 cells per well were seeded in six-well plates and, after 24 h, were treated with vehicle, 100 nM AZD4547 or 100 nM pemigatinib, or transfected with 50 nM siRNAs and cultured for 7 days. For KATO-III cells, six-well plates were precoated with laminin using RAC-11P cells67 as previously described53. Cells were stained with crystal violet as previously described53 and the plates were imaged using the GelCount colony counter (Oxford Optronix).
96-well cell growth assay
A total of 800 SNU-1 cells; 1,500 MCF7, MFM-223, KATO-III, SNU-16 or SUM52PE cells; or 3,000 NCI-H716 cells per well were seeded in 96-well plates and, after 24 h, were treated with vehicle or 100 nM AZD4547, pemigatinib, BGJ398 or debio-1347, or transfected with 50 nM siRNAs. Cell density was assayed over 8 days on sister plates using the CellTiter-Blue Reagent for 4 h and the Infinite M Plex plate reader operated using the Tecan i-control software.
3D soft agar colony-formation assay
Six- or twelve-well plates were precoated with 2 ml or 1 ml of 0.6% low-gelling-temperature agarose (A9414, Sigma-Aldrich) by diluting 3% agarose solution (in PBS) in DMEM/F-12 supplemented with 3% FBS and penicillin–streptomycin. The bottom layer was solidified at 4 °C for 30 min. NMuMG cells were passed through a 70 μm cell strainer and 20,000 or 10,000 single cells per well of the six- or twelve-well plates, respectively, were suspended in 2 ml or 1 ml of 0.35% low-gelling-temperature agarose in DMEM/F-12 supplemented with 3% FBS, penicillin–streptomycin and vehicle, 100 nM AZD4547 or 100 nM pemigatinib, and plated on top. The top layer was solidified at 4 °C for 30 min before transferring the plates to the incubator. The plates were imaged after 15 days using the GelCount colony counter and anchorage-independent growth was quantified using the integrated GelCount colony counting platform (v.1.1.2, Oxford Optronix).
FACS analysis of cells
Cultured NMuMG cells were collected with 2 mM EDTA and passed through a 70 μm cell strainer prewetted with FACS buffer. Single cells were labelled with the LIVE/DEAD Fixable Violet Dead Cell Stain Kit and fixed, permeabilized, incubated with primary and secondary antibodies, and analysed as described for FACS analysis of mammary glands.
RNA isolation and RT–qPCR
RNA from frozen mammary tumour pieces was isolated as previously described10,68. Cultured cells were lysed (72 h after siRNA transfection in case of human cell lines) in buffer RLY (BIO-52079, Bioline) containing 1% 2-mercaptoethanol. Total RNA extraction and DNase treatment of samples was performed using the ISOLATE II RNA Mini Kit (BIO-52072, Bioline) according to manufacturer’s guidelines. Purified RNA was quantified using the DS-11 Series Spectrophotometer/Fluorometer (DeNovix) and subjected to reverse transcriptase reaction using the Tetro cDNA Synthesis Kit (BIO-65042, Bioline) with oligo (dT)18 primers (tumour pieces) or random hexamer primers (cells). qPCR was performed using the SensiFAST SYBR Hi-ROX Kit (BIO-92005, Bioline) and the QuantStudio 6 Flex Real-Time PCR System (4485691, Thermo Fisher Scientific) operated with the QuantStudio Real-Time PCR Software (v.1.7.2, Thermo Fisher Scientific). Primers used were designed using Primer-BLAST69 and a list of which is provided in Supplementary Table 11. Relative quantified cDNA was normalized using either mouse Hprt (tumour pieces) or Usf1 (cells) or human USF1 as the housekeeping transcript.
Protein isolation and western blotting
NMuMG cells were cultured in DMEM/F-12 starvation medium (0% FBS) for 48 h and treated with vehicle or 100 nM AZD4547 for 3 h. Cells were lysed in previously described RIPA buffer53 containing Halt protease and phosphatase inhibitor cocktail (78440, Thermo Fisher Scientific). Protein concentrations were determined using the BCA Protein Assay Kit (23227, Thermo Fisher Scientific) and by measuring absorbance using the Infinite M Plex plate reader operated with the Tecan i-control software. Equal amounts of protein and the BlueEye Prestained Protein Marker (PS-104, Jena Bioscience) were separated on NuPAGE 4–12% Bis-Tris Mini Protein Gels (NP0323, NP0329, Thermo Fisher Scientific) and transferred overnight at 4 °C onto nitrocellulose membranes (88018, Thermo Fisher Scientific) in previously described transfer buffer53. The membranes were stained with Ponceau S solution (ab270042, Abcam) and imaged using Fusion FX (Vilber), blocked in 5% bovine serum albumin (BSA, A8022, Sigma-Aldrich) in PBS-T (0.05% Tween-20) and incubated with primary antibodies in 5% BSA in PBS-T overnight at 4 °C. The membranes were washed with PBS-T and incubated with secondary antibodies in 5% BSA in PBS-T for 1 h at room temperature. A list of the antibodies used (all validated for western blotting by the manufacturers) is provided in Supplementary Table 9. The membranes were washed in PBS-T and developed using SuperSignal West Pico PLUS Chemiluminescent Substrate or Femto Maximum Sensitivity Substrate (34580, 34095, Thermo Fisher Scientific). The membranes were imaged using Fusion FX operated with the Fusion FX7 Edge imaging system (v.18.05, Vilber), post-imaging processed with Photoshop 2022 (v.23.2.2, Adobe) using input levels and output levels, and band-intensities were measured with mean grey value in Fiji (v.1.0)70. Protein band intensities were normalized to β-actin and phosphoprotein bands were further normalized to corresponding total protein bands and to FGFR2 intensity.
Proteomics of mouse cells and tumours
Sample preparation
Two (global phosphoproteomics) or three (phosphorylated-Tyr immunoprecipitation (p-Tyr IP) proteomics) 15 cm dishes of NMuMG cells expressing GFP or Fgfr2 variants were collected in 3 ml urea lysis buffer71. Fresh-frozen samples of Fgfr2 tumours collected in this study and K14-cre;BrcaF/F;Trp53F/F (KB1P) and KB1P;Mdr1a/b−/− (KB1PM) tumours collected elsewhere72 were mounted with Milli-Q H2O and processed using a cryotome. Sections were collected to a final wet weight of up to 250 mg in urea lysis buffer (40× wet weight). Lysates were sonicated and cleared by centrifugation as previously described73. Protein concentrations were determined using the BCA Protein Assay Kit, and protein phosphorylation integrity was verified using western blotting and the p-Tyr-1000 antibody (8954, Cell Signaling Technology). To create a spectral library for protein expression analysis, for each setting, a ten-band in-gel-digestion experiment was performed and SDS gels were processed as described previously74. Per cell lysate sample, 45 µg total protein was loaded. Furthermore, 45 µg total protein of a mouse liver lysate71 was added. Tumour lysates were prepared in 6 pools consisting of 4–7 individual samples each, and 60 µg total protein was loaded per pool. For global phosphoproteomics and p-Tyr IP experiments, in-solution protein digestion of an equivalent of 500 µg total protein (p-Tyr IP cells, 5 mg; p-Tyr IP tumours, 4 mg) using trypsin and desalting with Oasis HLB 1 cm3 Vac Cartridge (186000383, Waters) was performed as previously described71,73. For global phosphoproteomic experiments, phosphopeptide enrichment was performed on the AssayMAP Bravo Platform (Agilent Technologies) using 5 µl Fe(III)-NTA immobilized metal affinity chromatography (IMAC) cartridges (G5496-60085, Agilent Technologies) starting from 200 µg desalted peptides in 0.1% trifluoroacetic acid and 80% acetonitrile. Phosphopeptides were eluted in 25 µl 5% NH4OH/30% acetonitrile. Phosphopeptide enrichment for KB1P(M) tumours was performed using titanium dioxide beads as previously described71. IP of p-Tyr-containing peptides was performed using the PTMScan p-Tyr-1000 Kit (8803, Cell Signaling Technology) as previously described73.
MS measurements
For Fgfr2 samples, phosphopeptides were separated using the Ultimate 3000 nanoLC–mass spectrometry (MS)/MS system (Thermo Fisher Scientific) equipped with a 50 cm × 75 μm ID Acclaim Pepmap (C18, 1.9 μm) column. After injection, peptides were trapped at 3 μl min−1 on a 10 mm × 75 μm ID Acclaim Pepmap trap at 2% buffer B (80% acetonitrile, 0.1% formic acid) and separated at 300 nl min−1 in a 10–40% gradient of buffer B over 110 min at 35 °C. Eluting peptides were ionized at a potential of +2 kVa into a Q Exactive HF mass spectrometer (Thermo Fisher Scientific) operated by Tune (v.2.11) and Xcalibur Software (v.4.3.73.11, OPTON-30965, both Thermo Fisher Scientific). Intact masses were measured at m/z 350–1,400 at a resolution of 120,000 (at m/z 200) in the Orbitrap system using an AGC target value of 3 × 106 charges and a maxIT of 100 ms. The top 15 peptide signals (charge-states 2+ and higher) were submitted to MS/MS in the higher-energy collision cell (1.4 amu isolation width, 26% normalized collision energy). MS/MS spectra were acquired at a resolution of 15,000 (at m/z 200) in the Orbitrap system using an AGC target value of 1 × 106 charges, a maxIT of 64 ms and an underfill ratio of 0.1%, resulting in an intensity threshold of 1.3 × 105. Peptide separation for KB1P(M) samples was performed using a 40 cm × 75 μm (inner diameter) fused silica column custom packed with 1.9 μm 120 Å ReproSil Pur C18 aqua (Dr. Maisch). After injection, peptides were trapped at 6 μl min−1 on a 10 mm × 100 μm (inner diameter) trap column packed with 5 μm 120 Å ReproSil Pur C18 aqua at 2% buffer B and separated at 300 nl min−1 in a gradient of 10–40% buffer B over 90 min. The LC column was maintained at 50 °C using a pencil column heater (Phoenix S&T). Eluting peptides were ionized at a potential of +2 kVa into a Q Exactive HF mass spectrometer operated by Tune and Xcalibur Software. Intact masses were measured at a resolution of 70,000 (at m/z 200) in the Orbitrap system using an AGC target value of 3 × 106 charges. The top 10 peptide signals (charge-states 2+ and higher) were submitted to MS/MS in the higher-energy collision cell (1.6 amu isolation width, 25% normalized collision energy). MS/MS spectra were acquired at a resolution of 17,500 (at m/z 200) in the Orbitrap system using an AGC target value of 1 × 106 charges, a maxIT of 80 ms and an underfill ratio of 0.1%, resulting in an intensity threshold of 1.3 × 104. For Fgfr2 and KB1P(M) samples, a dynamic exclusion was applied with a repeat count of 1 and an exclusion time of 30 s. For protein expression experiments, peptides (1 µg total peptides, desalted) were separated and eluted as described for Fgfr2 phosphopeptides. The data independent acquisition (DIA)-MS method consisted of an MS1 scan from 350 to 1,400 m/z at a resolution of 120,000 (AGC target of 3 × 106 and 60 ms injection time). For MS2, 24 variable-size DIA segments were acquired at 30,000 resolution (AGC target 3 × 106 and auto for injection time). The DIA-MS method starting at 350 m/z included one window of 35 m/z, 20 windows of 25 m/z, 2 windows of 60 m/z and one window of 418 m/z, which ended at 1,400 m/z. Normalized collision energy was set at 28. The spectra were recorded in centroid mode with a default charge state for MS2 set to 3+ and a first mass of 200 m/z. Spectral library data files were acquired with the same acquisition settings as for the phosphoproteomic experiments.
(Phospho)-peptide quantification and data analysis
For protein expression experiments, MS/MS spectra derived from data-dependent acquisition (DDA) mode of the in-gel digestion experiment were searched against the Swiss-Prot Mus musculus reference proteome (25,374 entries, canonical and isoforms, release 2021_10) using MaxQuant (v.2.0.3.0)75,76 software with the default settings. Peptide identifications were propagated across samples with the match between runs (MBR) option enabled. The MaxQuant msms.txt file was used to generate a spectral library using Spectronaut software (v.15.4.210913, Biognosys). Spectra derived from single sample measurements in DIA mode were first analysed library-free in Spectronaut (directDIA) using the Biognosys factory settings to create a second spectral library. For the final search of DIA data in Spectronaut, both libraries were assigned using the default settings using the protein LFQ method set to MaxLFQ, imputation option switched off and an automatic normalization strategy. The Spectronaut report was further processed with R. Single-sample gene set enrichment analysis (ssGSEA) was performed by the GenePattern platform77 using the ssGSEA module (v.10.0.11)78 and Hallmark gene sets from MSigDB (v.7.0)79. Missing values were imputed with a zero. For phosphoproteome experiments, phosphopeptide identification and quantification was performed as previously described71 using the Swiss-Prot (Fgfr2 samples) or the UniProt (KB1P(M) samples) Mus musculus reference proteomes (UniProt, 34,331 entries, canonical and isoforms, release 2015_06) and MaxQuant software. Phosphosites with a localization probability of <0.75 (class 1)80 were discarded. The R package limma (v.3.52.1)81 was used to perform differential expression analysis on class 1 phosphosite intensity data. For two-group comparisons, phosphosite intensity data were filtered for high data presence in at least one of the groups under comparison (cells, ≥75%; tumours, ≥50%). In the case of data presence in one group and absence in the other (phosphosite on/off behaviour), only observations with a very high data presence in the ‘phosphosite on’ group were allowed (cells, 100%; tumours, ≥90%). In these cases, missing values were imputed in the ‘phosphosite off’ group with a zero. Fold change values were determined using the mean of each treatment group and the antilog value was calculated. If downstream analysis did not allow the presence of duplicated phosphosite amino acid windows, the entry with the lowest P value was used. Phosphosite signature enrichment analysis (PTM-SEA)82 was performed with the GenePattern platform77 using a seven-amino-acid sequence flanking the phosphosite as an identifier and the mouse kinase/pathway definitions of PTMsigDB (v.1.9.0)82 with the default settings. When PTM-SEA was performed following a two-group comparison, the rank metric was derived by multiplying the sign of FCs with the −log10-transformed P values calculated by limma. When PTM-SEA was performed on single samples, duplicated phosphosite amino acid windows were filtered for entries with the highest row-sum of intensities over all of the samples. The samples were ranked using the phosphosite intensities and missing values were imputed with a zero. To assign probable upstream kinases to differentially regulated phosphosites, the robust kinase activity inference (RoKAI) tool83 was used with the default settings and the UniProt Mus musculus reference proteome. RoKAI kinase and kinase target tables were shortlisted (cells, FDR < 0.05, number of substrates ≥ 3; tumours, number of substrates ≥ 2), assigned to significantly changed phosphosites (−1.5 ≥ FC ≥ 1.5, P < 0.05) and selected subsets of these phosphosites were visualized.
Analysis of SB transposon insertions in the mutagenesis screen
For the SB transposon insertional mutagenesis screen in ref. 10, mapping of SB insertions and calculation of insertion clonalities using next-generation sequencing of genomic DNA from SB-containing tumours was described in detail10. In brief, the relative clonality scores of SB insertions were calculated by normalizing each unique ligation score between genomic DNA and a SB cassette insertion to the highest ligation score within a given tumour sample. Then, each SB insertion was assigned a score between 0 (no insertion) and 1 (fully clonal insertion). Tumours with at least one relative insertion clonality score for Fgfr2 of ≥0.25 were defined as tumours containing SB insertion(s) in Fgfr2 (n = 65 tumours; total, n = 123 tumours).
Analysis of RNA-seq data from SB tumours
Published RNA-seq data generated from tumours of the SB transposon insertional mutagenesis screen10 were used to derive Fgfr2 gene- and exon-level expression as well as splice junction information. Gene fusions affecting Fgfr2 in tumours with SB insertions were previously identified11. To quantify the expression of SB transposons in Fgfr2, customized fasta and gtf files were constructed for individual tumours by inserting the SB transposon sequence at the genomic position and according to its orientation as previously mapped10. Sequencing reads were then mapped on the basis of the customized fasta and gtf files using STAR (v.2.7.2)84. Splice junctions between Fgfr2-E17 and the SB transposon were quantified using SJ.out.tab obtained from STAR alignment. To determine the usage of Fgfr2-I17-inserted SB transposons as splice acceptors, the ratio of junction reads spanning from E17 to the SB transposon versus E18 was computed. Integrated Genomics Viewer (IGV, v.1.11.0)85 was used to generate sashimi plots.
Analysis of WGS data from the HMF
WGS data on metastatic solid tumours were obtained from the HMF (data access request DR-138) through their Google cloud computing platform and analysed based on their bioinformatics pipeline (https://github.com/hartwigmedical/pipeline5) designed to detect all types of somatic alterations including structural variants and CNAs as previously described23. In brief, sequencing reads were mapped against the human reference genome GRCh37 using Burrows–Wheeler Alignment (BWA-MEM, v.0.7.5a)86. Somatic structural variants were called with GRIDSS (v.1.8.0)87 and CNAs and tumour purity were estimated using PURPLE (v.2.43)88. Finally, LINX (v.1.9)88 was performed to annotate events and to construct derivate chromosome structures. On the basis of the PURPLE output, samples containing structural variant BPs within the FGFR2 genomic region were considered for further structural variant analyses (n = 266 total BPs and 196 unique BPs in 86 tumour samples; Fig. 1f). To annotate structural variants, the location and orientation of both BP sides (FGFR2 and its partner) were used to determine RE types. Among the RE partners, the gene encoding the longer protein sequence was used as RE classification backbone. The following RE types were defined: (1) in-frame fusion, both BP sides were located in the intronic regions of coding genes and the upstream and downstream exons adjacent to the BP were both in-frame (complete reading frame) or both BP sides were located in the exonic regions of coding genes and the fused sequence was in-frame; (2) frame unknown RE, both BP sides were located in the intronic regions of coding genes and either the upstream or the downstream exon adjacent to the BP was out of frame (incomplete reading frame), or one or both BP sides were located in exonic regions of coding genes and the fused sequence was out of frame. Any of these cases made the reading frame unpredictable (unknown). (3) RE with intergenic space, one BP side located to FGFR2 and the other BP side located to a non-coding IGR; (4) out-of-strand RE, both BP sides were located in the coding regions of genes. The gene upstream to the BP (FGFR2) was supported by a sense-oriented read sequence, whereas the gene downstream to the BP was supported by an antisense-oriented read sequence; (5) internal RE, both BP sides were located within the genomic region of FGFR2; (6) unresolved REs, the gene upstream to the BP was supported by antisense-oriented read sequences or the REs contained single breakends. Unresolved REs were excluded, resulting in a refined list of samples containing FGFR2 REs (n = 93 REs in 55 tumour samples; Extended Data Fig. 1g,h). For the samples with multiple FGFR2 REs, the relative allele frequency of each RE was computed using the ploidy level inferred by LINX. An I17/E18 RE allele frequency of >15% was used as a threshold to define samples with FGFR2 REs causing E18 truncations (E18-truncating, n = 20; others, n = 35; Extended Data Fig. 1g,h and Supplementary Table 1). FGFR2 copy number (CN) gains of >5 were defined as amplifications. Among the samples with FGFR2 CN segment BPs at I17, samples with E1–E17 CN (CNE1–E17) > 5 and CNE1–E17 − CNE18 > 2 were defined as FGFR2-E1–E17 partially amplified. A few samples were expressing an FGFR2 in-frame fusion gene based on RNA-seq, but showed discordant RE types in WGS. In these cases, in-depth annotation of the WGS data was performed using LINX to infer the plausible structures of derivate chromosomes constructed by complex RE events.
Analysis of RNA-seq data from the HMF
Raw RNA-seq data on metastatic solid tumours were obtained from the HMF (data access request DR-138). Sequencing reads were mapped to the human reference genome GRCh38 (Gencode v32 CTAT) using STAR (v.2.7.2)84 with the recommended parameters to subsequently run STAR-Fusion (v.1.8.1)89. STAR-Fusion was executed with chimeric alignment information (Chimeric.out.junction) obtained from STAR and GRCh38 Gencode v32 CTAT. Chimeric alignments from STAR and gene fusions from STAR-Fusion were inspected for RNA-seq alignments supporting the REs identified in WGS. For the samples with in-frame fusions identified in WGS, upstream and downstream exon numbers of the fusion gene inferred from STAR-Fusion were matched to the fusion found in WGS. For the samples with other types of REs identified in WGS, chimeric reads spanning the upstream exon and the downstream exon (out-of-frame REs), the downstream intergenic sequence (REs with intergenic space) or the downstream antisense gene sequence (out-of-strand REs) were mined from the ‘Chimeric.out.junction’ file and matched to the REs found in WGS. Genome coordinates were converted from GRCh37 to GRCh38 using UCSC Lift Genome Annotations (https://genome.ucsc.edu/cgi-bin/hgLiftOver). IGV (v.1.11.0)85 was used to generate sashimi plots.
Analysis of RNA-seq data from TCGA
Among the 10,344 TCGA samples90, we preselected samples potentially expressing FGFR2ΔE18 on the basis of several criteria: the presence of (1) FGFR2 amplifications or (2) truncating mutations in FGFR2-E18, (3) shifts in CN segment values in FGFR2-I17, (4) a lack of FGFR2-E18 expression, (6) usage of FGFR2-E18-C3 or -E18-C4, and/or (6) previously annotated FGFR2 fusions91. FGFR2 amplification and mutation information was obtained from the cBioPortal92. CN segment files for CN break information and exon-level expression data were obtained from the NCI-GDC data portal (https://portal.gdc.cancer.gov/). Among the samples with FGFR2 CN BPs at I17, samples with CNE1–E17 segment values (log2[CN/2]) > 0.3 (typical GISTIC threshold for amplifications) and CNE1–E17 − CNE18 > 0.3 were defined as FGFR2 E1–E17 partially amplified. To select samples with loss of FGFR2-E18 expression, E18 expression was normalized to the median expression of E1–E17. Tumour samples showing lower FGFR2-E18 expression compared with the minimum expression observed in TCGA normal tissue samples were selected. To evaluate E18-C3 and E18-C4 use, we obtained splice junction read counts from the NCI-GDC data portal. FGFR2-E17 to E18-C3 and E18-C4 spanning read counts were divided by total junction reads from FGFR2-E17 to calculate E18-C3 and E18-C4 use. Tumour samples showing higher FGFR2-E18-C3 or -E18-C4 use compared with the maximum usage observed in TCGA normal tissue samples were selected. In total, the selection process yielded 165 samples for which raw RNA-seq data were downloaded from the NCI-GDC data portal using TCGAbiolinks (v. 2.14.1)93. Sequencing reads were mapped to the human reference genome GRCh38 (Gencode v32 CTAT) using STAR (v.2.7.2)84 with the recommended parameters to subsequently run STAR-Fusion (v.1.8.1)89. STAR-Fusion was executed with chimeric alignment information (Chimeric.out.junction) derived from STAR to obtain high-confidence in-frame and out-of-frame (frameshift or fusion with non-coding RNA) gene fusions. STAR-Fusion uses only exon–exon spanning reads to detect gene fusions; we therefore used exon–intron/intron–intron spanning reads from the Chimeric.out.junction file to find non-canonical types of out-of-frame fusions applying several filtering steps. Chimeric spanning reads with FGFR2 BPs were discarded, if we found (1) multiple chimeric alignments, (2) PCR duplicates and/or (3) mitochondrial/Immunoglobulin/HLA mapping. Out-of-frame REs were defined by either exon–exon spanning reads resulting in frameshift or fusion with non-coding RNA (STAR-Fusion) or exon–intron/intron–intron spanning reads (STAR chimeric alignments). Intergenic REs were defined by spanning reads between FGFR2 and an IGR. Out-of-strand REs were defined by spanning reads between FGFR2 and an antisense partner gene. REs with recurrent BP support were considered (spanning read count > 2). For the samples with multiple FGFR2 REs, the relative expression of each RE was computed on the basis of the supporting junction read counts. An E17 junction read frequency of >15% was used as the threshold to define samples with FGFR2ΔE18 REs. IGV (v.1.11.0)85 was used to generate sashimi plots.
Analyses of CCLE, CTRPv2 and GDSC pharmacogenomic datasets
Mutation, CN, gene expression, exon usage ratio and fusion data for cell lines of the Broad Institute Cancer Cell Line Encyclopedia (CCLE) were obtained from the CCLE data portal30. FGFR2/3 missense hotspot mutations were selected in agreement with previous annotations92,94 and, in the case of FGFR2, based on the FMI cohort (Extended Data Fig. 2a). Missense mutations affecting the following amino acids were considered to be hotspots: FGFR2, Ser252, Cys382, Asn549, Lys659; FGFR3, Arg248, Ser249, Tyr373, Lys650. CN data were obtained as log2[CN/2] values, and log2[CN/2] ≥ 2 was considered to be an amplification. FGFR fusion data (CCLE_Fusions_unfiltered_20181130.txt) were further cleaned by applying the following filters: (1) FFPM > 0.1, (2) spanning fragment count ≥ 5 and (3) expression value RPKM ≥ 1. FGFR2/3 was considered to be E18-truncated if cell lines contained FGFR2/3 fusions with I17 BPs or exhibited high FGFR2-E18-C3 use (P < 0.01 derived from Z-score normalization of exon usage ratio) among the samples with robust expression of FGFR2. To compute composite expression of FGF receptors, FGFR1–4 expression was normalized by the geometric mean of each receptor among all of the samples and summed as previously described33. Drug-response data for AZD4547 and PD173074 were obtained from the Cancer Therapeutics Response Portal (CTRP) v2 deposited in the PharmacoDB database31,95 and from the Genomics of Drug Sensitivity in Cancer (GDSC) database32, respectively. Integrated area under the sigmoid-fit concentration-response curve values were used to evaluate the association between FGF/FGFR status and drug sensitivity.
Low-coverage WGS of human cancer cell lines
Genomic DNA from cultured cells was isolated using the ISOLATE II Genomic DNA Kit (BIO-52066, Bioline) according to the manufacturer’s guidelines. Low-coverage WGS was performed as previously described57. Libraries were sequenced with 65 bp single reads using the HiSeq 2500 System with V4 chemistry (Illumina) operated by the HiSeq Control Software (v.2.2.68, Illumina). Sequencing reads were mapped to the human reference genome GRCh38 using BWA-MEM (v.0.7.5a)86. Reads with mapping quality lower than 37 were excluded. The resulting alignments were analysed with QDNAseq (v.1.14.0) using sequence mappability and GC content correction and a bin size of 20,000 bp to generate segmented CN values96.
RNA-seq analysis of human cancer cell lines
RNA-seq analysis of cultured cells was performed as previously described68. In brief, cells were lysed in Buffer RLT (79216, Qiagen) containing 1% 2-mercaptoethanol. Total RNA extraction was performed using the RNeasy Mini Kit (74104, Qiagen) according to the manufacturer’s guidelines. The quality and quantity of RNA was assessed using the 2100 Bioanalyzer system and a Nano chip (Agilent). RNA samples with RIN > 8 were processed for polyA-stranded library preparation using the TruSeq RNA Library Prep Kit v2 (RS-122-2001/2, Illumina) according to the manufacturer’s instructions, quality-checked with the 2100 Bioanalyzer system using a 7500 chip, and pooled equimolar into a 10 nM sequencing stock solution. Libraries were sequenced with 100 bp paired-end reads using the HiSeq 2500 System with V4 chemistry and operated by HiSeq Control Software. Sequencing reads were mapped to the human reference genome GRCh38 (Gencode v32 CTAT) using STAR (v.2.7.2)84 with the recommended parameters to subsequently run STAR-Fusion (v.1.8.1)89. Gene- and exon-level expression read counts were quantified by featureCounts (v.1.6.2)97 on the basis of gene structures defined in GRCh38. Genes with CPM values greater than 1 in at least 10% of the total number of samples were considered expressed and used for downstream analysis. Read counts for expressed genes were normalized by trimmed mean of M-value (TMM) method using edgeR (v.3.26.6)98,99. To detect FGFR2 gene fusions and REs from RNA-seq, we followed the approach as described for TCGA RNA-seq analysis.
Hybrid-capture RNA-seq analysis of FFPE samples
Total RNA from FFPE samples was isolated using the RNeasy DSP FFPE Kit (73604, Qiagen) according to the manufacturer’s guidelines. The quality and quantity of RNA was assessed using the Agilent TapeStation system and High Sensitivity D1000 Reagents (Agilent). A total of 20 ng of fragmented total RNA was used for Illumina-compatible cDNA library preparation. First, total RNA was used for reverse transcription and first-strand cDNA synthesis. After end-repair and adapter ligation, cDNA sequences were selected for enrichment of exonic sequences using biotinylated target specific probes as provided in the TruSeq RNA Exome kit (Illumina). Standard RNA-seq libraries were generated using captured/exome-enriched cDNA. Purified cDNA sequences were amplified using barcoded primers for different samples. Purified libraries were quantified using Qubit Flex Fluorometer (Thermo Fisher Scientific) and sequenced with 2 × 150 bp configurations using the NextSeq 500 or the NovaSeq 6000 Systems (Illumina) operated by NextSeq (v.2.0.2) and NovaSeq (v.1.7.5) control software, respectively. STAR-Fusion (v.1.8.1)89 and the human reference genome GRCh37 were used for RNA fusion detection with the default parameters. STAR (v.2.7.3a)84 and RSEM (v.1.3.0)100 were used for gene and transcript quantification using the default parameters. LeafCutter (v.0.2.9)101 and STAR-produced bam files were used to examine intron excision counts for splicing variants. IGV (v.1.11.0)85 was used to generate sashimi plots.
PDX models
Model selection and analysis
PDX models were previously characterized by Crown Bioscience102 and are described in the HuPrime PDX collection (https://www.crownbio.com/oncology/in-vivo-services/patient-derived-xenograft-pdx-tumor-models). PDX models were selected on the basis of (1) FGF3/4/19 amplification, (2) FGFR2/3 missense hotspot mutations, (3) FGFR1/2/3 amplification, (4) high expression of FGFR1/2/3/4 and/or (5) expression of an FGFR fusion gene. The PDX models KI0551, LI0612 and LU1901 were included as controls, because each contained a MET oncogenic amplification potentially rendering tumours resistant to FGFRi55,103. CN and mutation data generated by whole-exome sequencing and raw RNA-seq data of the selected PDX models (Fig. 4c) were obtained from the CrownBio-HuPrime data portal. Sequencing data were derived from non-treated PDXs. Sequencing reads were mapped to the human (GRCh38 Gencode v32 CTAT) and mouse (mm10 Gencode M23) reference genomes to filter out mouse-derived reads using Disambiguate (v.2018.05.03-6)104. The remaining human reads were analysed as described for the analysis of human cell line RNA-seq data, composite FGFR1-4 expression was computed as described for the CCLE RNA-seq analysis, and FGFR2 gene fusions and REs were detected as described for TCGA RNA-seq analysis. We also implemented fusions/REs previously annotated by Crown Biosciences and deposited in CrownBio-HuPrime into our analysis.
Debio-1347 intervention study
PDX fragments of 2–3 mm in diameter were injected subcutaneously into the right flank of 8-week-old female BALB/cAnNRj-Foxn1nu/nu mice (HFK Bioscience and Shanghai Laboratory Animal Center), except for BL5001 and BL5002, for which 8-week-old female NOD.CB17-Prkdcscid/NCrHsd mice (Envigo) were used. Mice were twice weekly weighed and monitored for tumour development and, as soon as tumours reached a volume of 200–250 mm3 (measured in two dimensions using callipers; volume = length × width2 × 0.5), the mice were randomly allocated to vehicle versus debio-1347 FGFRi treatment arms. The treatments were performed daily through oral gavage for 12–25 consecutive days using vehicle (1% Kollidon VA64 in demineralized water), 40 mg per kg debio-1347 (Debiopharm) and increased to 60 mg per kg during treatment (BL5001, BL5002), 60 mg per kg debio-1347 (BN2289, BL0597, BR0438, BR1115, CR3151, ES0136, ES0189, ES0204, ES0215, ES0218, ES2116, GA0114, LI0612, LI1035, LI1055, LU0755, PA1332) or 80 mg per kg debio-1347 (CR1428, ES0042, GA0080, GA0087, GA1224, GA3055, GL0720, HN0366, HN0696, HN1420, KI0551, LU1302, LU1380, LU1429, LU1901, LU2504, PA3013). The treatment response was determined by relative treatment-to-control ratios (ΔT/ΔC). ΔT and ΔC are the mean volume difference between last treatment day and initial treatment day of the treated and control groups, respectively. All of the animal procedures were conducted at a Crown Bioscience SPF facility. All of the procedures related to animal handling, care and the treatment in this intervention study were performed according to guidelines approved by the Institutional Animal Care and Use Committee (IACUC) of Crown Bioscience following the guidance of the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC).
Analysis of CGP data from FMI
Hybrid-capture-based CGP
Comprehensive genomic profiling (CGP) was performed on FFPE tumour tissue or blood samples prospectively collected during routine clinical care. Testing was performed in a Clinical Laboratory Improvement Amendments-certified, College of American Pathologists-accredited, New York State-regulated reference laboratory (Foundation Medicine). Approval for this study, including a waiver of informed consent and a Health Insurance Portability and Accountability Act waiver of authorization, was obtained from the Western Institutional Review Board (protocol 20152817). For 217,017 tumour tissue specimens, DNA (>50 ng) was extracted from FFPE specimens and next-generation sequencing was performed by FoundationOne companion diagnostic testing using hybridization-captured, adapter-ligation-based libraries to high, uniform coverage (>500×) for all coding exons of 315 or 324 cancer-related genes plus selected introns of 28 or 36 genes that are frequently rearranged in cancer, as previously described105. A total of 26,289 samples were similarly assayed; DNA was sequenced for 406 genes and selected introns of 31 genes involved in REs, and RNA was sequenced for 265 genes106. For 6,264 liquid samples, plasma was isolated from 20 ml of peripheral whole blood and ≥20 ng of circulating tumour DNA was extracted to create adapted sequencing libraries for coding exons of 70 genes before hybrid-capture and sample-multiplexed sequencing107. Results were analysed for base substitutions, short insertions and deletions, CN gains or losses, and REs. The companion diagnostic tests included probes against all FGFR2 exons and FGFR2-I17.
Data analysis
FMI classifies FGFR2 REs as fusions if the genomic BP is in the I17/E18 hotspot, if the predicted chimeric protein includes both an N terminus and a C terminus (in strand), and if the gene partner is either a previously described fusion partner (in-frame or frame unknown) or a novel gene partner predicted to be in-frame with FGFR2. I17/E18 hotspot out-of-strand REs, any REs with a BP in intergenic space and any REs with a BP in E1–E17 of FGFR2 were classified as REs. Here we reclassified FGFR2 REs as described for WGS data analysis. In brief, REs were defined as in-frame fusions if the genomic BP was in I17/E18 and if the frame of the fusion partner was predictable and in-strand. Frame-unknown REs, out-of-strand REs and REs with a BP in intergenic space were classified as non-canonical REs. FGFR2 amplifications were called if ≥80% of the FGFR2 targets were at an amplified CN (defined as ≥4 + median ploidy of the sample). Differential CN gains of E18 targets < E1–E17 targets were defined as FGFR2-E1–E17 partial amplifications. In samples with FGFR2 REs and co-amplification, low-level REs were discarded at a read threshold dependent on the amplification CN. FGFR2-E18-truncating nonsense and frameshift mutations were subgrouped into mutations affecting the proximal (E768–Y783) versus the distal (P784–T821) C terminus (encoded by E18) on the basis of the functional classifications of truncating mutations in this study (Fig. 2e and Extended Data Figs. 6a,h, 7a,b and 8a–c). I17/E18 in-frame fusions or non-canonical REs, E1–E17 partial amplifications, E18 splice-site mutations and/or proximal E18-truncating mutations were grouped as FGFR2-E18-truncating alterations. The four most common FGFR2 missense mutations affecting Ser252, Cys382, Asn549 and Lys659 are referred to as hotspots throughout this study (Extended Data Fig. 2a), in agreement with previous annotations92,94. To establish the co-driver landscape of FGFR2-altered tumours, the top 30 driver genes concurrently altered (amplifications, deletions, and missense, truncating and splice-site mutations) in samples with FGFR2 alterations (E18 truncations, E1–E18 full-length amplifications and/or missense hotspot mutations) were identified. The samples were grouped according to FGFR2-E18-truncating alterations, FGFR2-E1–E18 full-length amplifications or FGFR2 missense hotspot mutations, and proportion Z-tests were used to identify co-driver genes significantly enriched in either of the 3 FGFR2 alteration categories, both in the pan-cancer cohort as well as in the BRCA, CHOL, OV, COAD/READ, ESCA/STAD and LUAD/LUSC cohorts specifically. Fisher’s exact tests were used to evaluate co-occurrence (odds ratio > 1) or mutual exclusivity (odds ratio < 1) of the co-driver genes in each of the 3 FGFR2 alteration categories versus FGFR2 WT samples in the pan-cancer cohort and in the BRCA cohort specifically.
Analysis of self-interacting capacity among FGFR2 RE partners
The SLIPPER algorithm predicts the interaction capacities of proteins108. It has been trained with seven different proteome databases (DIP, IntAct, MINT, BioGRID, PDB, MatrixDB and 12D) to establish the SLIPPER Golden Standard Dataset of potentially self-interacting proteins108. On the basis of this dataset, the proportion of self-interactors among unique proteins encoded by FGFR2 RE partner genes identified in the FMI dataset was calculated. The self-interacting ability of FGFR2 RE partners was also evaluated using the SLIPPER algorithm108 itself. To identify specific self-interacting domains in FGFR2 RE partners, domain–domain interaction information was obtained from the 3did109 and PPIDM110 databases, and domain enrichment analysis was performed with DAVID bioinformatic resources111. The Swiss-Prot Homo sapiens proteome (release 2021_04) was used as reference dataset for these analyses.
Re-examination of FIGHT-202 study
Details on the study design, eligibility criteria, and efficacy and safety findings of FIGHT-202 (NCT02924376), a phase II, open-label, multicentre, global study of pemigatinib in patients with previously treated advanced or metastatic cholangiocarcinoma, with or without FGF/FGFR alterations, were previously published9. Before entering screening for trial eligibility, the patients were either prescreened for FGF/FGFR status using FoundationOne or patients provided a commercial FoundationOne report or an FGF/FGFR status report based on local testing, the latter of which required retrospective central confirmation through FoundationOne. In FIGHT-202, FGFR2 REs were classified (fusions versus REs) on the basis of the FoundationOne report and biomarker definition as described above. In this reanalysis of the FIGHT-202 oncogenomic data, we used the alteration data provided by FMI to classify FGFR2 amplification status and FGFR2 REs by frame only. Five patients who were classified as having fusions on their FoundationOne report had FGFR2 amplifications in conjunction with fusions in the alteration data; four patients classified as having fusions on their FoundationOne report had an unknown frame in the structured data; and two patients who were classified as having REs on their FoundationOne report were classified as in-frame in the alteration data. These discrepancies are due to FoundationOne reporting rules and ongoing updates to the analysis and annotation pipeline used by FMI from the time of the original report to the time of the generation of the alteration data. Importantly, these changes do not affect the results from the primary efficacy cohort from FIGHT-202 but, rather, provide an alternative classification for this subset analysis.
Statistics and reproducibility
Data of in vitro and in vivo experiments were analysed using Prism (v.9.3.1, GraphPad Software). Genomic and proteomic data were analysed using R (v.3.6.3–4.1.2). In vitro experiments were independently repeated at least twice, and all attempts at replication were successful. Across these, data on n ≥ 3 independent replica were collected. No sample size calculations were performed. Sample sizes of mouse cohorts and for ex vivo analyses thereof (FACS analyses, H&E and IHC analyses, proteomics, RNA-seq and RT–qPCR) were based on previous calculations10 or determined using G*Power software (v.3.1)56, and were large enough to measure the effect sizes. Data were reproducible across mice or batches analysed, and all attempts at replication were successful. Sample sizes in the FIGHT-202 trial were based on previous calculations9 and were large enough to measure the effect sizes. The statistical tests and multiple-testing correction models used are described in the corresponding figure legends. P < 0.05 was considered to be statistically significant. Except for P < 0.0001 and P ≥ 0.05, exact P values are always shown in the corresponding figure panels or, where indicated, in Supplementary Table 2.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-022-05066-5.
Supplementary information
Supplementary Figs. 1–2, Supplementary Tables 7–11 and the legends for Supplementary Tables 1–6. Supplementary Fig. 1: uncropped western blots related to Extended Data Figs. 5g and 7g. Supplementary Fig. 2: FACS gating strategies related to Extended Data Figs. 5a and 6b,c. Supplementary Table 7: the Fgfr2 GEMM cloning primer sequences. Supplementary Table 8: the mouse genotyping primer sequences. Supplementary Table 9: a list of the antibodies used. Supplementary Table 10: siRNA sequences. Supplementary Table 11: qPCR primer sequences.
A list of FGFR2 REs identified in the HMF cohort.
A list of FGFR2 alterations identified in the FMI cohort, and the sample size and statistical details related to Fig. 3b,c and Extended Data Fig. 10b,c,e.
List of samples expressing FGFR2ΔE18 variants identified in the TCGA cohort.
Expression of lentiviral FGFR2 constructs.
A list of FGF/FGFR alterations identified in CCLE cell lines with FGFRi responses.
A list of FGF/FGFR alterations identified in PDX models with debio-1347 response.
Acknowledgements
We thank D. Zimmerli and I. van der Heijden for experimental support and J. Houthuijzen and A. Moises Da Silva for providing mice; the staff at the NKI animal facility, the animal pathology facility, the flow cytometry facility and the genomics core facility and Rutgers Cancer Institute of New Jersey Biospecimen Repository and Histopathology Service Shared Resource for their technical support; the people from the transgenic and the preclinical intervention units of the Mouse Clinic for Cancer and Ageing (MCCA) at the NKI for their technical support performing the animal experiments; and the staff at Crown Bioscience for their technical support with performing the PDX experiments. This publication and the underlying study have been made possible in part on the basis of the data that the Hartwig Medical Foundation and the Center of Personalised Cancer Treatment (CPCT) have made available to the study as well as the data from the Broad Institute (CCLE and CTRPv2), the Wellcome Sanger Institute (GDSC) and TCGA Research Network. This work was funded by the Dutch Cancer Society (KWF 2017-61169 to J.B., L.F.A.W. and J.J.; KWF 2020-12894 to D.Z., E.C., L.F.A.W. and J.J.), the European Research Council (ERC Synergy project CombatCancer to J.J.; ERC AdG-883877 to S.R.), the Netherlands Organisation for Scientific Research (NWO-Middelgroot project 91116017 to C.R.J.), the VUmc-Cancer Center Amsterdam (infrastructure grant to C.R.J.), the Swiss National Science Foundation (SNF P2ZHP3_175027 to D.Z.; SNF 310030_179360 to S.R.), the German Research Foundation (DFG 319175447 to F.R.), and the US National Cancer Institute (NCI P30CA072720 to C.S.C. and S.G.). This work is part of the Oncode Institute, which is partly financed by the Dutch Cancer Society.
Extended data figures and tables
Source data
Author contributions
D.Z., J.B., S.M.K., L.F.A.W. and J.J. conceived the ideas and designed the experiments. D.Z., J.Y., S.M.K., F.R., C.L., S.A. and E.W. performed the laboratory experiments. J.B. and F.R. conceived and performed the bioinformatic analyses. F.R., S.R.P, E.G. and R.R.d.G.-d.H. performed the MS-based proteomics experiments. C.L., T.E., M.B., B.S., J.S., B.d.K., A.P.D., E.v.d.B., E.S., N.P., U.B., R.d.K.-G. and B.v.G. performed the animal experiments. J.K.L., R.M., J.S.R. and S.M.A. provided FMI data. S.K. assessed the histology of mouse tumours and quantified IHC slides. I.M.S., H.Z., G.K.A.-A., L.F. and T.C.B. provided re-evaluation of FIGHT-202. C.S.C., G.M.R. and H.K. performed and analysed hybrid-capture RNA-seq. L.H. and M.H.v.M. generated Fgfr2 GEMMs. R.d.B. supported RNA-seq analyses. J.R.d.R. mapped the SB insertion sites. M.v.d.V. supervised the animal experiments. S.R. and C.R.J. supervised the MS-based proteomics experiments. E.C. supported bioinformatic analyses of HMF data. A.V.C. designed and supervised the PDX experiments. D.Z., S.G., L.F.A.W. and J.J. supervised the bioinformatic analyses and the experiments. D.Z., J.B., S.G., L.F.A.W. and J.J. wrote the manuscript.
Peer review
Peer review information
Nature thanks Masaru Katoh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
The low-coverage WGS and RNA-seq data of human cell lines generated in this study are available at the European Nucleotide Archive (ENA) under accession number PRJEB42514. The MS proteomic data and MaxQuant-generated text files generated in this study are available at the ProteomeXchange Consortium through the PRIDE database112 under accession numbers PXD031711 for Fgfr2 samples and PXD032007 for KB1P(M) samples. Sequencing data of SB tumours were previously published10 and are available at ENA under accession number PRJEB14134. WGS and RNA-seq data from the HMF were downloaded from their Google cloud computing platform under data-sharing agreement DR-138, and can be obtained through standardized procedures and request forms online (https://www.hartwigmedicalfoundation.nl/en/). CGP data can be obtained from FMI on reasonable request (https://www.foundationmedicine.com/service/genomic-data-solutions). TCGA data90 can be obtained from the NCI-GDC data portal (https://portal.gdc.cancer.gov/). Data from CCLE, CTRPv2 and GDSC are available through the respective data portals30–32,95. Details on PDXs can be obtained from the CrownBio-HuPrime data portal (https://www.crownbio.com/oncology/in-vivo-services/patient-derived-xenograft-pdx-tumor-models). The FIGHT-202 study was previously published9. Information on Incyte’s clinical trial data sharing policy and instructions for submitting clinical trial data requests are available online (https://www.incyte.com/Portals/0/Assets/Compliance%20and%20Transparency/clinical-trial-data-sharing.pdf?ver=2020-05-21-132838-960). The human reference genome (GRCh38 Gencode v32 CTAT) used for RNA-seq data analysis is available in CTAT Genome Lib data resources (https://data.broadinstitute.org/Trinity/CTAT_RESOURCE_LIB). The SLIPPER list of self-interacting proteins was previously published108. Domain–domain interaction information from 3did109 and PPIDM110 are available online (https://3did.irbbarcelona.org and http://ppidm.loria.fr, respectively). Reference human and mouse Swiss-Prot proteome information is available in the UniProt database (https://www.uniprot.org). All data generated or analysed during this study are included in this Article and its Supplementary Information. Source data are provided with this paper.
Code availability
WGS data from the HMF were analysed using published computer codes (https://github.com/hartwigmedical/pipeline5)23 including BWA-MEM (v.0.7.5a)86, GRIDSS (v.1.8.0)87, PURPLE (v.2.43)88 and LINX (v.1.9)88 to detect all types of somatic alterations, including structural variants and CNAs. BWA-MEM (v.0.7.5a)86, R package QDNAseq (v.1.14.0)96, R package TCGAbiolinks (v.2.14.1)93, STAR (v.2.7.2)84, STAR-Fusion (v.1.8.1)89, featureCounts (v.1.6.2)97, R package edgeR (v.3.26.6)98,99, RSEM (v.1.3.0)100, R package LeafCutter (v.0.2.9)101, Disambiguate (v.2018.05.03-6)104 and R package limma (v.3.52.1)81 were integrated into custom computer codes to analyse the genomic and proteomic data in this study, which are available at 10.5281/zenodo.6630874 and 10.5281/zenodo.6630632, respectively.
Competing interests
J.K.L. and R.M are employees of Foundation Medicine and have equity in Roche. I.M.S., H.Z., L.F. and T.C.B. are employees of and have equity in Incyte. G.K.A.-A. has performed research for ActaBiologica, Agios, Astra Zeneca, Bayer, Beigene, Berry Genomics, BMS, Casi, Celgene, Exelixis, Genentech/Roche, Halozyme, Incyte, Mabvax, Puma, QED, Sillajen and Yiviva, and has consulted for Agios, Astra Zeneca, Autem, Bayer, Beigene, Berry Genomics, Celgene, CytomX, Debio, Eisai, Eli Lilly, Exelixis, Flatiron, Genentech/Roche, Gilead, Helio, Incyte, Ipsen, Loxo, Merck, MINA, Polaris, QED, Redhill, Silenseed, Sillajen, Sobi, Therabionics, Twoxar, Vector and Yiviva. J.S.R. is an employee of Foundation Medicine, has equity in Roche and is a board member of Celsius Therapeutics. A.V.C. is an employee of Debiopharm International. S.M.A. is an employee of and has equity in EQRX, is a former employee of Foundation Medicine, is a scientific advisory board member of iN Therapeutics and has consulted for Takeda. S.G. has received research funding from M2Gen, has equity in Inspirata and Silagene, and has consulted for Foghorn Therapeutics, Foundation Medicine, Inspirata, Merck, Novartis, Roche and Silagene; his spouse is an employee of and has equity in Merck. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Daniel Zingg, Jinhyuk Bhin, Julia Yemelyanenko, Sjors M. Kas
Change history
9/1/2022
A Correction to this paper has been published: 10.1038/s41586-022-05287-8
Contributor Information
Shridar Ganesan, Email: ganesash@cinj.rutgers.edu.
Lodewyk F. A. Wessels, Email: l.wessels@nki.nl
Jos Jonkers, Email: j.jonkers@nki.nl.
Extended data
is available for this paper at 10.1038/s41586-022-05066-5.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-022-05066-5.
References
- 1.Katoh M. Fibroblast growth factor receptors as treatment targets in clinical oncology. Nat. Rev. Clin. Oncol. 2019;16:105–122. doi: 10.1038/s41571-018-0115-y. [DOI] [PubMed] [Google Scholar]
- 2.Pearson A, et al. High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. Cancer Discov. 2016;6:838–851. doi: 10.1158/2159-8290.CD-15-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Cutsem E, et al. A randomized, open-label study of the efficacy and safety of AZD4547 monotherapy versus paclitaxel for the treatment of advanced gastric adenocarcinoma with FGFR2 polysomy or gene amplification. Ann. Oncol. 2017;28:1316–1324. doi: 10.1093/annonc/mdx107. [DOI] [PubMed] [Google Scholar]
- 4.Chae YK, et al. Phase II study of AZD4547 in patients with tumors harboring aberrations in the FGFR pathway: results from the NCI-MATCH trial (EAY131) subprotocol W. J. Clin. Oncol. 2020;38:2407–2417. doi: 10.1200/JCO.19.02630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tabernero J, et al. Phase I dose-escalation study of JNJ-42756493, an oral pan–fibroblast growth factor receptor inhibitor, in patients with advanced solid tumors. J. Clin. Oncol. 2015;33:3401–3408. doi: 10.1200/JCO.2014.60.7341. [DOI] [PubMed] [Google Scholar]
- 6.Javle M, et al. Phase II study of BGJ398 in patients with FGFR-altered advanced cholangiocarcinoma. J. Clin. Oncol. 2018;36:276–282. doi: 10.1200/JCO.2017.75.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voss MH, et al. A phase I, open-label, multicenter, dose-escalation study of the oral selective FGFR inhibitor debio 1347 in patients with advanced solid tumors harboring FGFR gene alterations. Clin. Cancer Res. 2019;25:2699–2707. doi: 10.1158/1078-0432.CCR-18-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subbiah V, et al. FIGHT-101, a first-in-human study of potent and selective FGFR 1-3 inhibitor pemigatinib in pan-cancer patients with FGF/FGFR alterations and advanced malignancies. Ann. Oncol. 2022;33:522–533. doi: 10.1016/j.annonc.2022.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Abou-Alfa GK, et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol. 2020;21:671–684. doi: 10.1016/S1470-2045(20)30109-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kas SM, et al. Insertional mutagenesis identifies drivers of a novel oncogenic pathway in invasive lobular breast carcinoma. Nat. Genet. 2017;49:1219–1230. doi: 10.1038/ng.3905. [DOI] [PubMed] [Google Scholar]
- 11.de Ruiter JR, et al. Identifying transposon insertions and their effects from RNA-sequencing data. Nucleic Acids Res. 2017;45:7064–7077. doi: 10.1093/nar/gkx461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Annunziato S, et al. Modeling invasive lobular breast carcinoma by CRISPR/Cas9-mediated somatic genome editing of the mammary gland. Genes Dev. 2016;30:1470–1480. doi: 10.1101/gad.279190.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Annunziato S, et al. Comparative oncogenomics identifies combinations of driver genes and drug targets in BRCA1-mutated breast cancer. Nat. Commun. 2019;10:397. doi: 10.1038/s41467-019-08301-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szybowska P, Kostas M, Wesche J, Haugsten EM, Wiedlocha A. Negative regulation of FGFR (fibroblast growth factor receptor) signaling. Cells. 2021;10:1342. doi: 10.3390/cells10061342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tannheimer SL, Rehemtulla A, Ethier SP. Characterization of fibroblast growth factor receptor 2 overexpression in the human breast cancer cell line SUM-52PE. Breast Cancer Res. 2000;2:311. doi: 10.1186/bcr73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh H, et al. Preferential alternative splicing in cancer generates a K-sam messenger RNA with higher transforming activity. Cancer Res. 1994;54:3237–3241. [PubMed] [Google Scholar]
- 17.Ueda T, et al. Deletion of the carboxyl-terminal exons of K-sam/FGFR2 by short homology-mediated recombination, generating preferential expression of specific messenger RNAs. Cancer Res. 1999;59:6080–6086. [PubMed] [Google Scholar]
- 18.Sakaguchi K, Lorenzi MV, Matsushita H, Miki T. Identification of a novel activated form of the keratinocyte growth factor receptor by expression cloning from parathyroid adenoma tissue. Oncogene. 1999;18:5497–5505. doi: 10.1038/sj.onc.1202947. [DOI] [PubMed] [Google Scholar]
- 19.Lorenzi MV, Horii Y, Yamanaka R, Sakaguchi K, Miki T. FRAG1, a gene that potently activates fibroblast growth factor receptor by C-terminal fusion through chromosomal rearrangement. Proc. Natl Acad. Sci. USA. 1996;93:8956–8961. doi: 10.1073/pnas.93.17.8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y-M, et al. Identification of targetable fgfr gene fusions in diverse cancers. Cancer Discov. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jusakul A, et al. Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov. 2017;7:1116–1135. doi: 10.1158/2159-8290.CD-17-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin A, et al. Detection of known and novel FGFR fusions in non–small cell lung cancer by comprehensive genomic profiling. J. Thorac. Oncol. 2019;14:54–62. doi: 10.1016/j.jtho.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 23.Priestley P, et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature. 2019;575:210–216. doi: 10.1038/s41586-019-1689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, et al. Patterns of somatic structural variation in human cancer genomes. Nature. 2020;578:112–121. doi: 10.1038/s41586-019-1913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorenzi MV, Castagnino P, Chen Q, Chedid M, Miki T. Ligand-independent activation of fibroblast growth factor receptor-2 by carboxyl terminal alterations. Oncogene. 1997;15:817–826. doi: 10.1038/sj.onc.1201242. [DOI] [PubMed] [Google Scholar]
- 26.Moffa AB, Tannheimer SL, Ethier SP. Transforming potential of alternatively spliced variants of fibroblast growth factor receptor 2 in human mammary epithelial cells. Mol. Cancer Res. 2004;2:643–652. [PubMed] [Google Scholar]
- 27.Cha JY, Maddileti S, Mitin N, Harden TK, Der CJ. Aberrant receptor internalization and enhanced FRS2-dependent signaling contribute to the transforming activity of the fibroblast growth factor receptor 2 IIIb C3 isoform. J. Biol. Chem. 2009;284:6227–6240. doi: 10.1074/jbc.M803998200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rothé B, et al. Crystal structure of Bicc1 SAM polymer and mapping of interactions between the ciliopathy-associated proteins Bicc1, ANKS3, and ANKS6. Structure. 2018;26:209–224. doi: 10.1016/j.str.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciriello G, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghandi M, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seashore-Ludlow B, et al. Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov. 2015;5:1210–1223. doi: 10.1158/2159-8290.CD-15-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iorio F, et al. A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166:740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sánchez-Guixé M, et al. High FGFR1–4 mRNA expression levels correlate with response to selective fgfr inhibitors in breast cancer. Clin. Cancer Res. 2022;28:137–149. doi: 10.1158/1078-0432.CCR-21-1810. [DOI] [PubMed] [Google Scholar]
- 34.Turner N, et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29:2013–2023. doi: 10.1038/onc.2009.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathur A, et al. FGFR2 is amplified in the NCI-H716 colorectal cancer cell line and is required for growth and survival. PLoS ONE. 2014;9:e98515. doi: 10.1371/journal.pone.0098515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hart KC, Robertson SC, Donoghue DJ. Identification of tyrosine residues in constitutively activated fibroblast growth factor receptor 3 involved in mitogenesis, stat activation, and phosphatidylinositol 3-kinase activation. Mol. Biol. Cell. 2001;12:931–942. doi: 10.1091/mbc.12.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nadratowska-Wesolowska B, et al. RSK2 regulates endocytosis of FGF receptor 1 by phosphorylation on serine 789. Oncogene. 2014;33:4823–4836. doi: 10.1038/onc.2013.425. [DOI] [PubMed] [Google Scholar]
- 38.Szybowska P, Kostas M, Wesche J, Wiedlocha A, Haugsten EM. Cancer mutations in FGFR2 prevent a negative feedback loop mediated by the ERK1/2 pathway. Cells. 2019;8:518. doi: 10.3390/cells8060518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin C-C, et al. Inhibition of basal FGF receptor signaling by dimeric Grb2. Cell. 2012;149:1514–1524. doi: 10.1016/j.cell.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 40.Lin, C.-C. et al. Regulation of kinase activity by combined action of juxtamembrane and C-terminal regions of receptors. Preprint at bioRxiv10.1101/2020.10.01.322123 (2020).
- 41.Khazaie K, et al. Truncation of the human EGF receptor leads to differential transforming potentials in primary avian fibroblasts and erythroblasts. EMBO J. 1988;7:3061–3071. doi: 10.1002/j.1460-2075.1988.tb03171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akiyama T, et al. The transforming potential of the c-erbB-2 protein is regulated by its autophosphorylation at the carboxyl-terminal domain. Mol. Cell. Biol. 1991;11:833–842. doi: 10.1128/mcb.11.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woolford J, McAuliffe A, Rohrschneider LR. Activation of the feline c-fms proto-oncogene: multiple alterations are required to generate a fully transformed phenotype. Cell. 1988;55:965–977. doi: 10.1016/0092-8674(88)90242-5. [DOI] [PubMed] [Google Scholar]
- 44.Niu X-L, Peters KG, Kontos CD. Deletion of the carboxyl terminus of Tie2 enhances kinase activity, signaling, and function. J. Biol. Chem. 2002;277:31768–31773. doi: 10.1074/jbc.M203995200. [DOI] [PubMed] [Google Scholar]
- 45.Hung KL, et al. ecDNA hubs drive cooperative intermolecular oncogene expression. Nature. 2021;600:731–736. doi: 10.1038/s41586-021-04116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ota S, Zhou Z-Q, Link JM, Hurlin PJ. The role of senescence and prosurvival signaling in controlling the oncogenic activity of FGFR2 mutants associated with cancer and birth defects. Hum. Mol. Genet. 2009;18:2609–2621. doi: 10.1093/hmg/ddp195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hertzler-Schaefer K, et al. Pten loss induces autocrine FGF signaling to promote skin tumorigenesis. Cell Rep. 2014;6:818–826. doi: 10.1016/j.celrep.2014.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koziczak M, Holbro T, Hynes NE. Blocking of FGFR signaling inhibits breast cancer cell proliferation through downregulation of D-type cyclins. Oncogene. 2004;23:3501–3508. doi: 10.1038/sj.onc.1207331. [DOI] [PubMed] [Google Scholar]
- 49.Liu H, et al. c-Myc alteration determines the therapeutic response to FGFR inhibitors. Clin. Cancer Res. 2017;23:974–984. doi: 10.1158/1078-0432.CCR-15-2448. [DOI] [PubMed] [Google Scholar]
- 50.Formisano L, et al. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer. Nat. Commun. 2019;10:1373. doi: 10.1038/s41467-019-09068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Derksen PWB, et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell. 2006;10:437–449. doi: 10.1016/j.ccr.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 52.Derksen PWB, et al. Mammary-specific inactivation of E-cadherin and p53 impairs functional gland development and leads to pleomorphic invasive lobular carcinoma in mice. Dis. Model. Mech. 2011;4:347–358. doi: 10.1242/dmm.006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schipper K, et al. Rebalancing of actomyosin contractility enables mammary tumor formation upon loss of E-cadherin. Nat. Commun. 2019;10:3800. doi: 10.1038/s41467-019-11716-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huijbers IJ, et al. Using the GEMM-ESC strategy to study gene function in mouse models. Nat. Protoc. 2015;10:1755–1785. doi: 10.1038/nprot.2015.114. [DOI] [PubMed] [Google Scholar]
- 55.Kas SM, et al. Transcriptomics and transposon mutagenesis identify multiple mechanisms of resistance to the FGFR inhibitor AZD4547. Cancer Res. 2018;78:5668–5679. doi: 10.1158/0008-5472.CAN-18-0757. [DOI] [PubMed] [Google Scholar]
- 56.Faul F, Erdfelder E, Buchner A, Lang A-G. Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behav. Res. Methods. 2009;41:1149–1160. doi: 10.3758/BRM.41.4.1149. [DOI] [PubMed] [Google Scholar]
- 57.Henneman L, et al. Selective resistance to the PARP inhibitor olaparib in a mouse model for BRCA1-deficient metaplastic breast cancer. Proc. Natl Acad. Sci. USA. 2015;112:8409–8414. doi: 10.1073/pnas.1500223112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cardiff RD, et al. The mammary pathology of genetically engineered mice: the consensus report and recommendations from the Annapolis meeting. Oncogene. 2000;19:968–988. doi: 10.1038/sj.onc.1203277. [DOI] [PubMed] [Google Scholar]
- 59.Montini E, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J. Clin. Invest. 2009;119:964–975. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaasterland T, et al. Computational principles of primer design for site directed mutagenesis. TechConnect Briefs. 2005;1:532–535. [Google Scholar]
- 61.Follenzi A, Ailles LE, Bakovic S, Geuna M, Naldini L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 2000;25:217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- 62.Zingg D, et al. EZH2-mediated primary cilium deconstruction drives metastatic melanoma formation. Cancer Cell. 2018;34:69–84. doi: 10.1016/j.ccell.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 63.Zhang J, et al. Translating the therapeutic potential of AZD4547 in FGFR1-amplified non-small cell lung cancer through the use of patient-derived tumor xenograft models. Clin. Cancer Res. 2012;18:6658–6667. doi: 10.1158/1078-0432.CCR-12-2694. [DOI] [PubMed] [Google Scholar]
- 64.Guagnano V, et al. FGFR genetic alterations predict for sensitivity to NVP-BGJ398, a selective pan-FGFR inhibitor. Cancer Discov. 2012;2:1118–1133. doi: 10.1158/2159-8290.CD-12-0210. [DOI] [PubMed] [Google Scholar]
- 65.Nakanishi Y, et al. The fibroblast growth factor receptor genetic status as a potential predictor of the sensitivity to CH5183284/Debio 1347, a novel selective FGFR inhibitor. Mol. Cancer Ther. 2014;13:2547–2558. doi: 10.1158/1535-7163.MCT-14-0248. [DOI] [PubMed] [Google Scholar]
- 66.Liu PCC, et al. INCB054828 (pemigatinib), a potent and selective inhibitor of fibroblast growth factor receptors 1, 2, and 3, displays activity against genetically defined tumor models. PLoS ONE. 2020;15:e0231877. doi: 10.1371/journal.pone.0231877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delwel GO, et al. Expression and function of the cytoplasmic variants of the integrin alpha 6 subunit in transfected K562 cells. Activation-dependent adhesion and interaction with isoforms of laminin. J. Biol. Chem. 1993;268:25865–25875. [PubMed] [Google Scholar]
- 68.Boelens MC, et al. PTEN loss in E-cadherin-deficient mouse mammary epithelial cells rescues apoptosis and results in development of classical invasive lobular carcinoma. Cell Rep. 2016;16:2087–2101. doi: 10.1016/j.celrep.2016.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ye J, et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rolfs F, Piersma SR, Dias MP, Jonkers J, Jimenez CR. Feasibility of phosphoproteomics on leftover samples after RNA extraction with guanidinium thiocyanate. Mol. Cell. Proteomics. 2021;20:100078. doi: 10.1016/j.mcpro.2021.100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gogola E, et al. Selective loss of PARG restores PARylation and counteracts PARP inhibitor-mediated synthetic lethality. Cancer Cell. 2018;33:1078–1093. doi: 10.1016/j.ccell.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 73.Beekhof R, et al. INKA, an integrative data analysis pipeline for phosphoproteomic inference of active kinases. Mol. Syst. Biol. 2019;15:e8250. doi: 10.15252/msb.20188250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Böttger F, et al. Tumor heterogeneity underlies differential cisplatin sensitivity in mouse models of small-cell lung cancer. Cell Rep. 2019;27:3345–3358. doi: 10.1016/j.celrep.2019.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 76.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 77.Reich M, et al. GenePattern 2.0. Nat. Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 78.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liberzon A, et al. The Molecular Signatures Database Hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 81.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krug K, et al. A curated resource for phosphosite-specific signature analysis. Mol. Cell. Proteomics. 2019;18:576–593. doi: 10.1074/mcp.TIR118.000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yılmaz S, et al. Robust inference of kinase activity using functional networks. Nat. Commun. 2021;12:1177. doi: 10.1038/s41467-021-21211-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Robinson JT, et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cameron DL, et al. GRIDSS: sensitive and specific genomic rearrangement detection using positional de Bruijn graph assembly. Genome Res. 2017;27:2050–2060. doi: 10.1101/gr.222109.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cameron, D. et al. GRIDSS, PURPLE, LINX: unscrambling the tumor genome via integrated analysis of structural variation and copy number. Preprint at bioRxiv10.1101/781013 (2019).
- 89.Haas BJ, et al. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol. 2019;20:213. doi: 10.1186/s13059-019-1842-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weinstein JN, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gao Q, et al. Driver fusions and their implications in the development and treatment of human cancers. Cell Rep. 2018;23:227–238. doi: 10.1016/j.celrep.2018.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cerami E, et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Colaprico A, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nakamura IT, et al. Comprehensive functional evaluation of variants of fibroblast growth factor receptor genes in cancer. NPJ Precis. Oncol. 2021;5:66. doi: 10.1038/s41698-021-00204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smirnov P, et al. PharmacoDB: an integrative database for mining in vitro anticancer drug screening studies. Nucleic Acids Res. 2018;46:D994–D1002. doi: 10.1093/nar/gkx911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Scheinin I, et al. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014;24:2022–2032. doi: 10.1101/gr.175141.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 98.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Robinson MD, McCarthy DJ, Smyth G,K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li YI, et al. Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet. 2018;50:151–158. doi: 10.1038/s41588-017-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen, D. et al. In vivo pharmacology models for cancer target research. Methods Mol. Biol.1953, 183–211 (2019). [DOI] [PubMed]
- 103.Kim S-M, et al. Activation of the Met kinase confers acquired drug resistance in FGFR-targeted lung cancer therapy. Oncogenesis. 2016;5:e241. doi: 10.1038/oncsis.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ahdesmäki MJ, Gray SR, Johnson JH, Lai Z. Disambiguate: an open-source application for disambiguating two species in next generation sequencing data from grafted samples. F1000Res. 2017;5:2741. doi: 10.12688/f1000research.10082.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Frampton GM, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.He J, et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood. 2016;127:3004–3014. doi: 10.1182/blood-2015-08-664649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Clark TA, et al. Analytical validation of a hybrid capture–based next-generation sequencing clinical assay for genomic profiling of cell-free circulating tumor DNA. J. Mol. Diagn. 2018;20:686–702. doi: 10.1016/j.jmoldx.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu Z, et al. Proteome-wide prediction of self-interacting proteins based on multiple properties. Mol. Cell. Proteomics. 2013;12:1689–700. doi: 10.1074/mcp.M112.021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stein A, Russell RB, Aloy P. 3did: interacting protein domains of known three-dimensional structure. Nucleic Acids Res. 2005;33:D413–D417. doi: 10.1093/nar/gki037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alborzi SZ, Ahmed Nacer A, Najjar H, Ritchie DW, Devignes M-D. PPIDomainMiner: inferring domain-domain interactions from multiple sources of protein-protein interactions. PLoS Comput. Biol. 2021;17:e1008844. doi: 10.1371/journal.pcbi.1008844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 112.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figs. 1–2, Supplementary Tables 7–11 and the legends for Supplementary Tables 1–6. Supplementary Fig. 1: uncropped western blots related to Extended Data Figs. 5g and 7g. Supplementary Fig. 2: FACS gating strategies related to Extended Data Figs. 5a and 6b,c. Supplementary Table 7: the Fgfr2 GEMM cloning primer sequences. Supplementary Table 8: the mouse genotyping primer sequences. Supplementary Table 9: a list of the antibodies used. Supplementary Table 10: siRNA sequences. Supplementary Table 11: qPCR primer sequences.
A list of FGFR2 REs identified in the HMF cohort.
A list of FGFR2 alterations identified in the FMI cohort, and the sample size and statistical details related to Fig. 3b,c and Extended Data Fig. 10b,c,e.
List of samples expressing FGFR2ΔE18 variants identified in the TCGA cohort.
Expression of lentiviral FGFR2 constructs.
A list of FGF/FGFR alterations identified in CCLE cell lines with FGFRi responses.
A list of FGF/FGFR alterations identified in PDX models with debio-1347 response.
Data Availability Statement
The low-coverage WGS and RNA-seq data of human cell lines generated in this study are available at the European Nucleotide Archive (ENA) under accession number PRJEB42514. The MS proteomic data and MaxQuant-generated text files generated in this study are available at the ProteomeXchange Consortium through the PRIDE database112 under accession numbers PXD031711 for Fgfr2 samples and PXD032007 for KB1P(M) samples. Sequencing data of SB tumours were previously published10 and are available at ENA under accession number PRJEB14134. WGS and RNA-seq data from the HMF were downloaded from their Google cloud computing platform under data-sharing agreement DR-138, and can be obtained through standardized procedures and request forms online (https://www.hartwigmedicalfoundation.nl/en/). CGP data can be obtained from FMI on reasonable request (https://www.foundationmedicine.com/service/genomic-data-solutions). TCGA data90 can be obtained from the NCI-GDC data portal (https://portal.gdc.cancer.gov/). Data from CCLE, CTRPv2 and GDSC are available through the respective data portals30–32,95. Details on PDXs can be obtained from the CrownBio-HuPrime data portal (https://www.crownbio.com/oncology/in-vivo-services/patient-derived-xenograft-pdx-tumor-models). The FIGHT-202 study was previously published9. Information on Incyte’s clinical trial data sharing policy and instructions for submitting clinical trial data requests are available online (https://www.incyte.com/Portals/0/Assets/Compliance%20and%20Transparency/clinical-trial-data-sharing.pdf?ver=2020-05-21-132838-960). The human reference genome (GRCh38 Gencode v32 CTAT) used for RNA-seq data analysis is available in CTAT Genome Lib data resources (https://data.broadinstitute.org/Trinity/CTAT_RESOURCE_LIB). The SLIPPER list of self-interacting proteins was previously published108. Domain–domain interaction information from 3did109 and PPIDM110 are available online (https://3did.irbbarcelona.org and http://ppidm.loria.fr, respectively). Reference human and mouse Swiss-Prot proteome information is available in the UniProt database (https://www.uniprot.org). All data generated or analysed during this study are included in this Article and its Supplementary Information. Source data are provided with this paper.
WGS data from the HMF were analysed using published computer codes (https://github.com/hartwigmedical/pipeline5)23 including BWA-MEM (v.0.7.5a)86, GRIDSS (v.1.8.0)87, PURPLE (v.2.43)88 and LINX (v.1.9)88 to detect all types of somatic alterations, including structural variants and CNAs. BWA-MEM (v.0.7.5a)86, R package QDNAseq (v.1.14.0)96, R package TCGAbiolinks (v.2.14.1)93, STAR (v.2.7.2)84, STAR-Fusion (v.1.8.1)89, featureCounts (v.1.6.2)97, R package edgeR (v.3.26.6)98,99, RSEM (v.1.3.0)100, R package LeafCutter (v.0.2.9)101, Disambiguate (v.2018.05.03-6)104 and R package limma (v.3.52.1)81 were integrated into custom computer codes to analyse the genomic and proteomic data in this study, which are available at 10.5281/zenodo.6630874 and 10.5281/zenodo.6630632, respectively.