SUMMARY
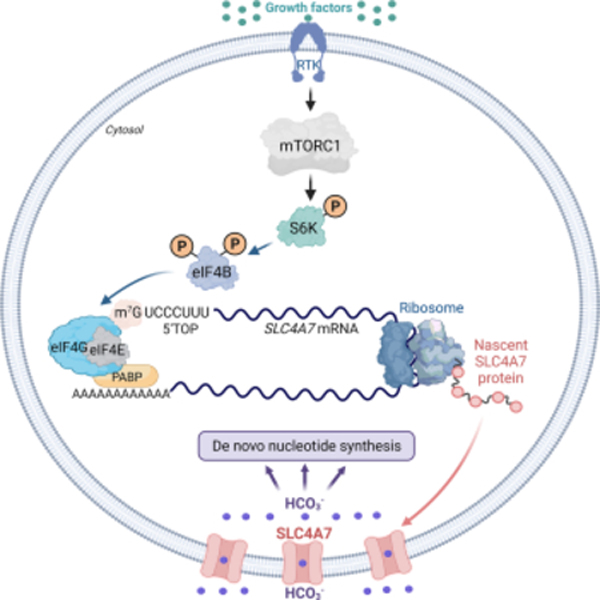
Bicarbonate (HCO3−) ions maintain pH homeostasis in eukaryotic cells and serve as a carbonyl donor to support cellular metabolism. However, whether the abundance of HCO3− is regulated or harnessed to promote cell growth is unknown. The mechanistic target of rapamycin complex 1 (mTORC1) adjusts cellular metabolism to support biomass production and cell growth. We find that mTORC1 stimulates the intracellular transport of HCO3− to promote nucleotide synthesis through the selective translational regulation of the sodium bicarbonate cotransporter SLC4A7. Downstream of mTORC1, SLC4A7 mRNA translation required the S6K-dependent phosphorylation of the translation factor eIF4B. In mTORC1-driven cells, loss of SLC4A7 resulted in reduced cell and tumor growth, and decreased flux through de novo purine and pyrimidine synthesis in human cells and tumors without altering the intracellular pH. Thus, mTORC1 signaling, through the control of SLC4A7 expression, harnesses environmental bicarbonate to promote anabolic metabolism, cell biomass, and growth.
eTOC blurb:
Ali et al. showed that bicarbonate (HCO3−) is a limiting substrate for nucleotide synthesis in proliferating cells. In response to growth factors stimuli, the mTORC1-S6K-eIF4B axis promotes an increase in the sodium bicarbonate cotransporter SLC4A7 mRNA translation to harness environmental HCO3− for cellular nucleotide synthesis and growth.
Graphical Abstract
INTRODUCTION
Cell metabolism is critical for numerous physiological functions of all life forms. As such, understanding the mechanisms that regulate cellular metabolism both in physiology and in human pathologies is key for the elucidation of biological function and eventual treatments for diseases such as cancer (DeBerardinis and Chandel, 2016; Liberti and Locasale, 2016; Vander Heiden and DeBerardinis, 2017). Anabolic metabolism enables the assembly of macromolecules such as protein, lipids, and nucleotides, required for cancer cell growth and proliferation (DeBerardinis et al., 2008; Faubert et al., 2020).
The mechanistic target of rapamycin complex 1 (mTORC1) is an exemplar in coordinating environmental signals with the control of anabolic pathways, including protein synthesis, de novo lipid, and nucleotide synthesis (Ben-Sahra and Manning, 2017; Saxton and Sabatini, 2017; Valvezan and Manning, 2019). In particular, mTORC1 has emerged as a major regulator for nucleotide synthesis, either via an acute manner through posttranslational control of the pyrimidine synthesis enzyme carbamoyl-phosphate synthetase 2, aspartate transcarbamoylase, and dihydroorotase (CAD) (Ben-Sahra et al., 2013) or in a long-term fashion through transcriptional control of de novo purine synthesis enzymes (Ben-Sahra et al., 2016). The de novo nucleotide synthesis is a major anabolic process that requires not only ample energy but also many small metabolite precursors, including glucose-derived ribose (5’-phosphoribosylpyrophosphate (PRPP)), amino acids (glutamine, glycine, aspartate), one-carbon units (5,10-methylene-THF, formyl-THF), and bicarbonate ions (HCO3−) to build purine and pyrimidine nucleotides (Lane and Fan, 2015; Villa et al., 2019). Thus, cells must ensure a sufficient supply of all the metabolites and nutrients for the successful production of macromolecules. While the molecular mechanisms regulating the abundance of glucose, amino acids, and one-carbon units are emerging (Ben-Sahra et al., 2016; DeNicola et al., 2015; Hay, 2011; Nogueira et al., 2018; Orozco et al., 2020; Torrence et al., 2021), whether cells regulate the availability of bicarbonate ions for nucleotide synthesis is not known.
Sodium bicarbonate (NaHCO3), commonly known as baking soda, is a weak base that regulates blood pH and renders it more alkaline (Calvo et al., 2021). Modulation of intracellular proton concentration (H+) is essential for ion homeostasis and for optimal metabolic cell function. To reduce the toxic effects of free H+, NaHCO3 combines with H+ to neutralize it (Aoi and Marunaka, 2014; Chiche et al., 2010). Although the effects of NaHCO3 on buffering intracellular pH are well known (Levraut et al., 2001), its role as a critical substrate for nucleotide metabolism has been overlooked.
NaHCO3 molecules enter cell membranes primarily by active transport through solute carriers such as SLC4A1-A11 (Cordat and Casey, 2009; Toft et al., 2021). Here, we report that one specific bicarbonate transporter, SLC4A7, exhibits gene coessentiality with the de novo nucleotide metabolic enzymes across a large panel of diverse cancer cell lines, suggesting that these genes are required for optimal cancer cell growth and survival in similar biological contexts. Among ten bicarbonate transporters of SLC4A family expressed in human cells, SLC4A7 protein abundance is selectively stimulated by mTORC1 in response to growth signals to specifically increase bicarbonate inflow and support flux through de novo purine and pyrimidine synthesis in human cells and tumors. Downstream of mTORC1 signaling, the S6 kinase-eIF4B axis promotes SLC4A7 mRNA translation and flux into nucleotide synthesis. Moreover, we demonstrate that SLC4A7 is required for mTORC1-driven cell proliferation and de novo nucleotide synthesis in proliferating cells and tumors. Thus, we reveal for the first time that bicarbonate levels are limiting for cell growth and proliferation and show that the mTORC1 pathway engages its downstream effector S6 kinase to regulate the entry of this important molecule in cells.
RESULTS
Decreased bicarbonate levels limit de novo nucleotide synthesis and cell growth.
To identify the effects of bicarbonate levels on cellular metabolism, we performed a steady-state metabolic profiling experiment in HeLa cells cultured in pH-buffered media and subjected to sodium bicarbonate (NaHCO3) depletion over time. Remarkably, intermediates from purine and pyrimidine metabolism were decreased in response to acute NaHCO3 depletion (Figure 1A, Table S1), despite an increase in some of the amino acid, glucose, and NAD metabolic intermediates (Figure 1B, Table S1). Because the de novo nucleotide synthesis pathways require HCO3− to effectively assemble the purine and pyrimidine rings (Figure 1C), we hypothesized that the intracellular bicarbonate levels can be limiting for nucleotide synthesis. To determine whether the effects of NaHCO3 levels on the abundance of purines and pyrimidines reflect the regulation of flux through the de novo nucleotide synthesis pathway, we measured relative flux in cells starved for NaHCO3 for 1 hour or 8 hours followed by a 1-hour pulse of stable isotope-labeled 13C6-glucose or 15N-glutamine (labeled on the amide nitrogen). These tracers label purine and pyrimidine nucleotide intermediates through specific pathways (Figure S1A), and their uptake is not affected in response to NaHCO3 depletion (Figure S1B). Short-term (1 hour) and long-term (8 hours) bicarbonate starvation significantly reduced the abundance and fractional enrichment of labeled purine and pyrimidine intermediates from 13C6-glucose or 15N-glutamine (Figures 1D, E, and Figures S1C–E). To determine whether NaHCO3-dependent regulation of cellular nucleotide levels affects the newly assembled nucleic acids, we used various radiotracers to evaluate whether bicarbonate levels induce specific alterations in the de novo purine synthesis (U-14C-glycine and 14C-formate) and the de novo pyrimidine pathways (U-14C-aspartate) (Figure 1F). Reducing concentrations of extracellular NaHCO3 led to a dose-dependent decrease in de novo purine and pyrimidine synthesis flux in HeLa cells, as evidenced by the reduction in the incorporation of 14C from glycine, formate, and aspartate into RNA (Figure 1F) without any visible changes in the substrate uptake (Figure S1F). A similar reduction in de novo nucleotide synthesis was also observed with a 3-14C-serine tracer, which specifically measures the contribution of one-carbon metabolism into de novo purine and deoxythymidine synthesis (Figures 1C, G, and Figure S1G) (Ducker and Rabinowitz, 2017). Collectively, these data indicate a global decrease in de novo nucleotide synthesis in response to bicarbonate depletion. Interestingly, bicarbonate starvation resulted in reduced cell proliferation, which could be rescued by supplementing the media with purine and pyrimidine nucleosides such as inosine and uridine (Figure 1H). Therefore, bicarbonate ions support cell proliferation through the preservation of nucleotide synthesis. Moreover, long-term bicarbonate starvation (8–12 h) induced a slight accumulation of cells in S and G2/M phases (Figure S1H) without any significant effects on DNA damage (Figure S1I). Collectively, these results establish that bicarbonate levels are limiting for nucleotide synthesis and cell proliferation.
Figure 1. Influence of sodium bicarbonate levels on nucleotide synthesis activity.
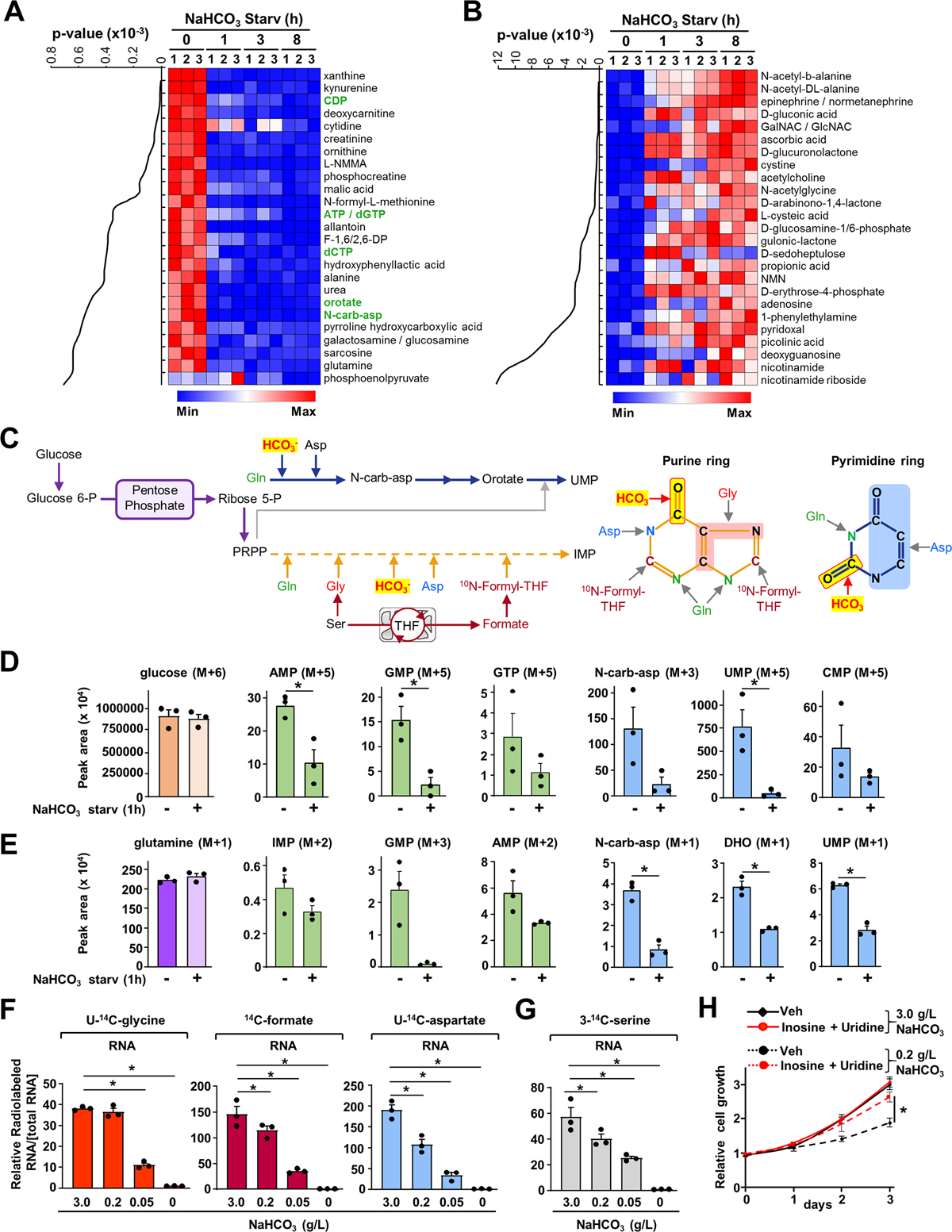
(A, B) Steady-state metabolite profiles from HeLa cells cultured in buffered DMEM medium (20 mM HEPES) in the presence or absence of sodium bicarbonate (NaHCO3) over the indicated times. Intracellular metabolites from three independent samples per condition were profiled by means of liquid chromatography‒mass spectrometry LC-MS/MS and ranked by those significantly decreased upon NaHCO3 depletion (8 h) relative to non-depleted condition (A) or increased upon NaHCO3 depletion (8 h) (B). The top 25 most significant metabolites are shown as row-normalized heat maps (A, B) ranked according to p-value. The complete metabolite profiles from these samples are provided in Table S1.
(C) Schematic illustrating the fate of the carbon and oxygen from bicarbonate and other metabolites into the pyrimidine and the purine rings.
(D) Normalized peak areas of 13C-labeled purine and pyrimidine intermediates, as measured by targeted LC-MS/MS, in HeLa cells grown in dialyzed serum and depleted of NaHCO3 for 1 h and labeled with 13C6-glucose for 1 h.
(E) Normalized peak areas of 15N-labeled purine intermediates from HeLa cells treated as in (D) and labeled with 15N-(amide)-glutamine for 1 h.
(F, G) Radiolabeling was performed for 3 h in the presence of the indicated concentrations of NaHCO3 (g/L) reflecting de novo purine synthesis (U-14C-glycine, 14C-formate), pyrimidine synthesis (U-14C-aspartate), or one-carbon metabolism (3-14C-serine).
(H) Proliferation of HeLa cells grown in dialyzed serum with indicated NaHCO3 concentrations in buffered medium supplemented or not with purine nucleoside (inosine 60 μM) and pyrimidine nucleoside (uridine 100 μM).
(D)-(H) The data are graphed as the means ± SDs of biological triplicates and are representative of at least two independent experiments. *p < 0.05, by two-tailed Student’s t-test for pairwise comparisons in (D, E) or one-way ANOVA with Tukey’s post hoc test for multiple pairwise comparisons (F, G, H).
The bicarbonate transporter SLC4A7 provides intracellular bicarbonate to support de novo nucleotide synthesis.
Genes with key roles in the same pathway are often most important for cellular fitness in the same contexts, and thus can be identified as “coessential” when comparing gene dependencies across cancer cell lines (Wang et al., 2017). We thus performed a bottom-up coessentiality network analysis for each de novo purine and pyrimidine metabolism gene using the FIREWORKS web tool (Amici et al., 2021) to probe for potential links between nucleotide and bicarbonate metabolism. This analysis revealed that several of the de novo pyrimidine and purine synthesis genes, including ATIC, PAICS, DHODH, CAD, UMPS, PAICS, and GART, are strikingly coessential with the bicarbonate transporter SLC4A7 (Figure 2A, Figures S2A, B, Table S2). A similar approach was also employed for nine other bicarbonate transporters (SLC4A1, SLC4A2, SLC4A3, SLC4A4, SLC4A5, SLC4A8, SLC4A9, SLC4A10, and SLC4A11), however with the exception of SLC4A7, none of the other bicarbonate transporters showed coessentiality with nucleotide synthesis genes (Figure S2C). We also compared the absolute gene dependency estimates (CERES scores) for bicarbonate transporters and nucleotide synthesis enzymes across a panel of 342 cancer cell lines (Meyers et al., 2017). As predicted, most of the purine and pyrimidine synthesis enzymes are critical for optimal cell fitness. Interestingly, from all the bicarbonate transporters, SLC4A7 displayed the strongest effect on cell fitness (Figure 2B, Table S3). To determine whether SLC4A7 directly impacts bicarbonate and nucleotide metabolism, we employed small interfering RNA (siRNA)–mediated depletion of SLC4A7 in HeLa cells. Depletion of SLC4A7 resulted in a ~20–25 % decrease in NaHCO3 uptake, without any significant changes in intracellular pH (Figure 2C). Conversely, SLC4A7 overexpression led to a ~20 % increase in bicarbonate uptake (Figure S2D). Importantly, SLC4A7 knockdown decreased the overall purine and pyrimidine abundance measured by a steady-state metabolite profiling (Figures 2D, E, Figure S2E and Table S4), and induced a minor cell cycle arrest in the G0/G1 phase (Figure S2F). Using radiolabeled Na+,14C-HCO3−, we confirmed that direct utilization of bicarbonate ions for nucleotide synthesis is reduced in SLC4A7-depleted cells (Figure S2G).
Figure 2. The sodium bicarbonate carrier SLC4A7 provides cellular bicarbonate for de novo purine and pyrimidine synthesis.
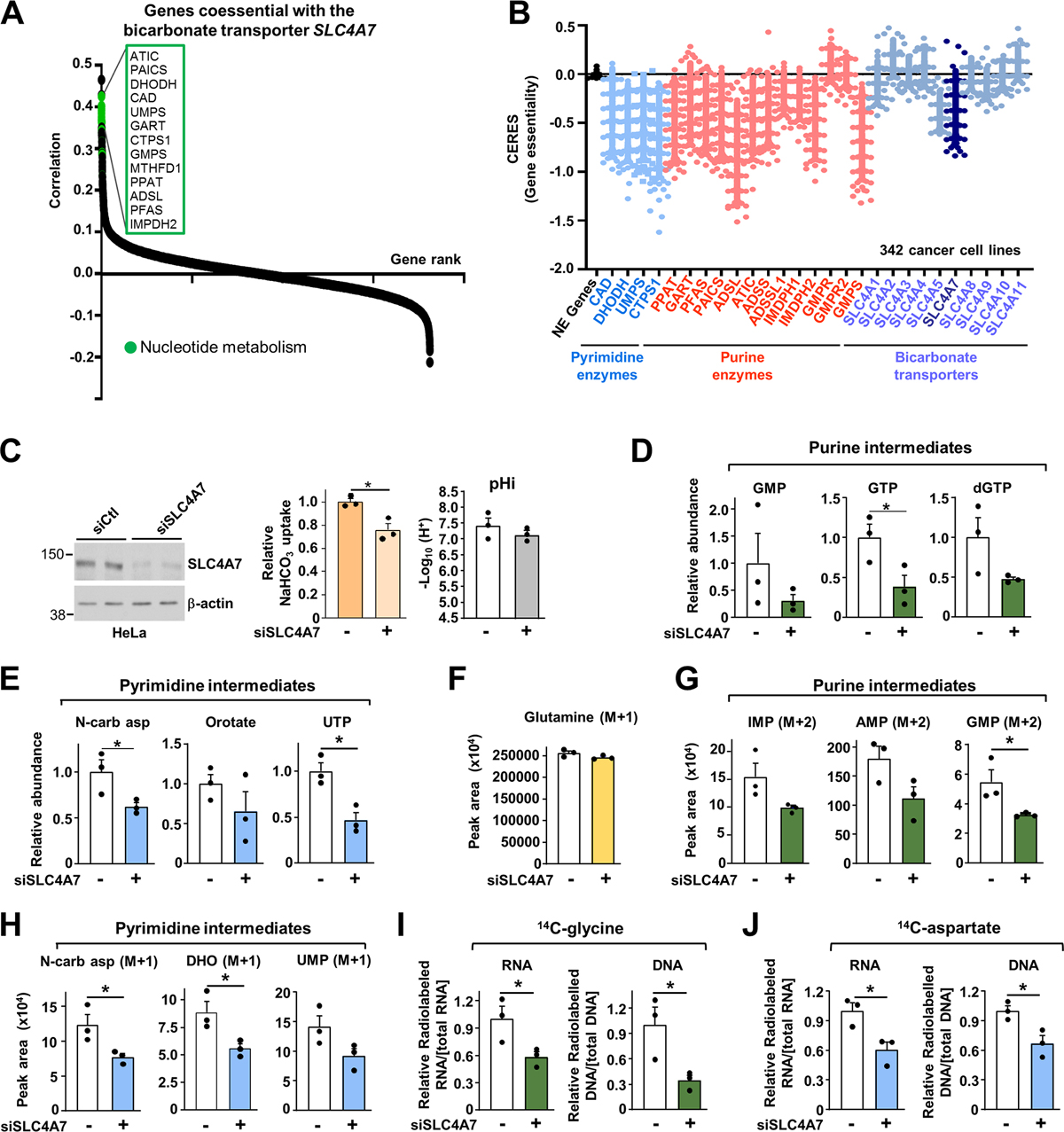
(A) Genes coessential with the sodium bicarbonate carrier SLC4A7.
(B) CERES dependency score for the de novo purine, pyrimidine synthesis and bicarbonate transporter enzymes. A CERES score less than 0 indicates a higher likelihood that the gene of interest is essential in a given cell line. A score of 0 indicates a gene is not essential (NE Genes: Non-essential genes).
(C) Immunoblots of HeLa cells transfected with siRNA targeting SLC4A7 or nontargeting controls, and measurement of intracellular pH (pHi) and NaHCO3 uptake.
(D, E) Purine and pyrimidine levels measured via LC-MS/MS in HeLa cells transfected as in (C)
(F) Normalized peak areas of 15N-glutamine levels measured as in (D) in HeLa cells treated as in (C) and labeled with 15N-amide-glutamine for 1 h.
(G, H) Normalized peak areas of 15N-labeled nucleotide intermediates from HeLa cells measured and treated as in (F).
(I, J) Radiolabeling was performed for 6 h in HeLa cells transfected as in (C) to measure de novo purine synthesis (14C-glycine) (I) or pyrimidine synthesis activity (14C-aspartate) (J).
(C)-(J) The data are graphed as the means ± SDs of biological triplicates and are representative of at least two independent experiments. *p < 0.05, by two-tailed Student’s t-test for pairwise comparisons.
To determine whether the effects of SLC4A7 knockdown on the steady-state levels of nucleotide intermediates reflected changes in the activity of de novo purine and pyrimidine synthesis, we employed a 1-hour pulse label with 15N-(amide)-glutamine. SLC4A7 depletion decreased the newly produced purine and pyrimidine intermediates without inducing changes in glutamine uptake (Figures 2F–H, Figure S2H). A similar reduction in de novo nucleotide synthesis was observed with radiolabeled tracers such as 14C-glycine and 14C-aspartate, where the 14C is incorporated into purines and pyrimidines, respectively (Figures 2I, J, Figure S2H). We did not observe any effects of SLC4A7 loss on the purine and pyrimidine salvage pathways measured with 3H-hypoxanthine and 3H-uridine into RNA, respectively, in agreement with the specific role of bicarbonate for the de novo nucleotide synthesis (Figures S2I, S2J). Collectively, these findings demonstrate that SLC4A7 is required to promote de novo nucleotide synthesis by enabling increased bicarbonate entry in cells.
SLC4A7 protein abundance is enhanced by growth factors stimuli to support the stimulation of de novo nucleotide synthesis.
Nucleotide synthesis is a highly regulated process. Physiological growth factors such as insulin-like growth factor 1 (IGF1), epidermal growth factor (EGF), or hormonal signals (e.g., insulin) stimulate the de novo nucleotide metabolism to support cell growth and proliferation (Ali et al., 2020; Ben-Sahra et al., 2013; Tsuchiya et al., 1990). Bicarbonate abundance correlated with the ability of insulin to stimulate nucleotide synthesis, as diminished extracellular NaHCO3 levels decreased the ability of insulin to promote flux through de novo purine (14C-glycine) and pyrimidine (14C-aspartate) synthesis (Figure 3A). To determine whether cellular bicarbonate transport, which contributes to nucleotide synthesis, is regulated by similar signals, we first measured bicarbonate uptake in HeLa cells treated with a time course of insulin. Notably, insulin increased bicarbonate uptake within 30 min treatment and plateaued after 2 hours (Figure 3B).
Figure 3. Growth factor signaling stimulates SLC4A7 expression and promotes bicarbonate uptake to promote nucleotide synthesis.
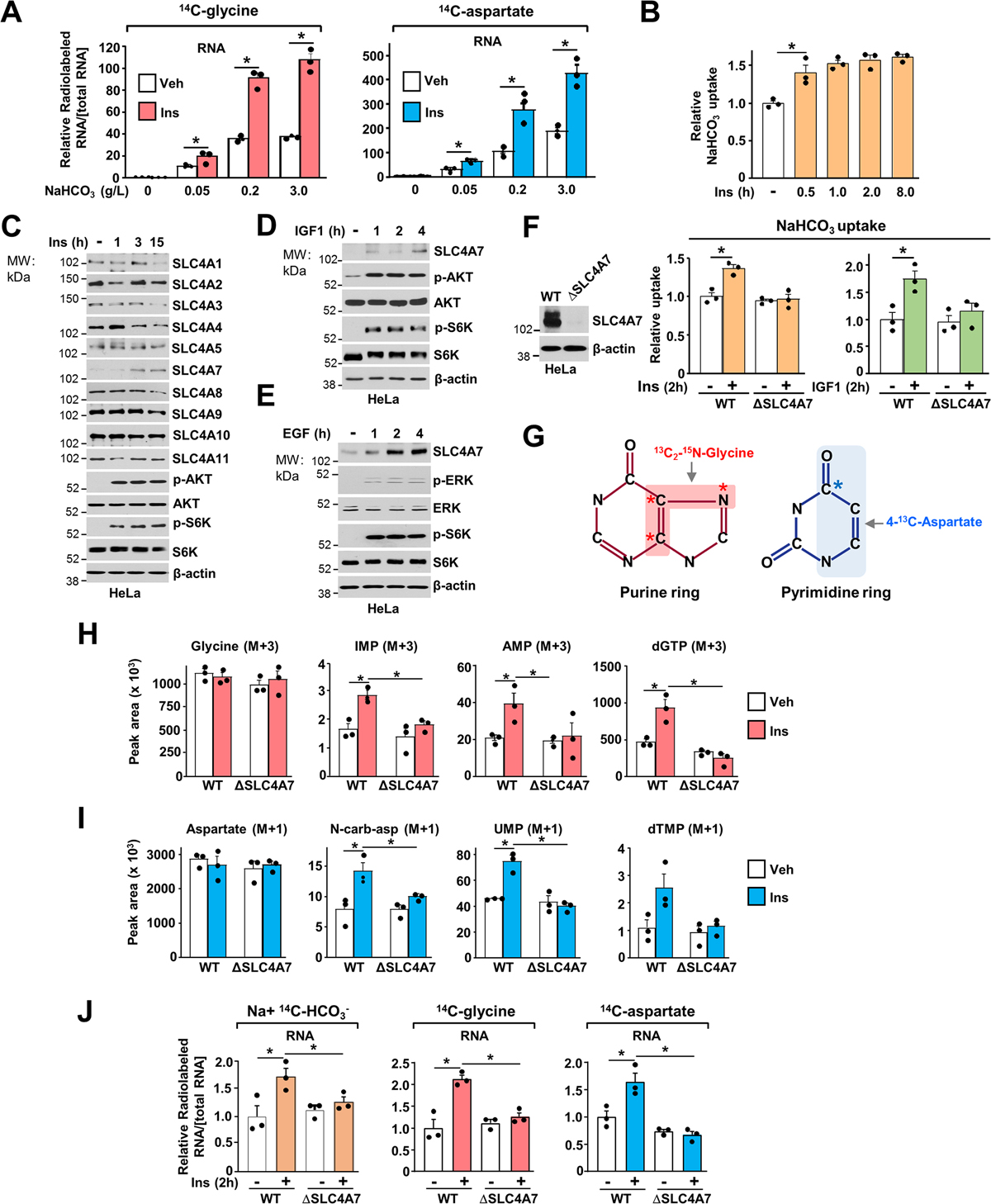
(A) Relative incorporation of radiolabels from 14C-glycine or 14C-aspartate (3 h labeling) into RNA from HeLa cells serum starved for 15 h and stimulated or not with insulin (100 nM) for 3 h in the presence of the indicated concentration of NaHCO3 (g/L).
(B) NaHCO3 uptake in HeLa cells serum starved for 15 h and stimulated or not with insulin (100 nM) over a time course.
(C) Immunoblots of HeLa cells serum starved for 15 h and stimulated or not with Insulin (100 nM) over a time-course.
(D, E) Immunoblots of HeLa cells serum starved for 15 h and stimulated or not with IGF1 (50 nM) (D) or EGF (50 ng/mL) (E) over a time-course
(F) Immunoblots of HeLa cells validating SLC4A7 loss (ΔSLC4A7) after gene editing through CRISPR-Cas9. NaHCO3 uptake in wild-type or ΔSLC4A7 HeLa cells serum starved for 15 h and stimulated or not with Insulin (100 nM) or IGF1 (50 nM) for 2 h and labeled for 5 min with Na+, 14C-HCO3− to measure. Uptake measurements are shown as relative to the vehicle-treated wildtype cell line.
(G) Illustration of the carbon and nitrogen from glycine and aspartate incorporated into the purine and pyrimidine ring, respectively.
(H) Normalized peak areas of 13C, 15N-labeled purine intermediates measured via LC-MS/MS in wild-type and ΔSLC4A7 HeLa cells, serum starved for 15 h and stimulated with insulin for 6 h and labeled with 13C2-15N-glycine (M+3) for the last hour.
(I) Normalized peak areas of 13C-labeled pyrimidine intermediates measured and treated as in (H) but stimulated with insulin for 2 h and labeled with 4-13C-aspartate (M+1) for the last hour.
(J) Wild-type and ΔSLC4A7 HeLa cells were serum starved for 15 h and stimulated or not with insulin (100 nM) for 2 h in the presence of the indicated radiotracers.
(A), (B), (F), (H)-(J) The data are plotted as the means ± SDs of biological triplicates and are representative of at least two independent experiments. *p < 0.05, by one-way ANOVA with Tukey’s post hoc test for multiple pairwise comparisons.
Given the rapid stimulation of bicarbonate transport, we tested whether the protein expression of bicarbonate transporters was also altered in response to insulin. Remarkably, among the ten bicarbonate transporters of SLC4A family expressed in mammalian cells, only SLC4A7 protein levels were increased in response to insulin stimulation (Figure 3C, Figure S3A), without any significant alterations in mRNA abundance (Figure S3B). An increase in SLC4A7 protein levels was also observed when cells were stimulated with other growth factors, including IGF1 (Figure 3D, Figure S3C) and epidermal growth factor (EGF) stimulation (Figure 3E, Figure S3C). Consistently, insulin and IGF1 also stimulated bicarbonate uptake, and this stimulation was blocked in SLC4A7 knockout (ΔSLC4A7) cells generated by CRISPR-Cas9 methodology (Figure 3F). Of note, when cells are starved of growth factors, we observed that loss of SLC4A7 does not affect basal NaHCO3 uptake, suggesting that SLC4A7 expression is mainly required to boost HCO3− entry in cells in response to growth signals (Figure 3F).
To determine the contribution of SLC4A7-dependent bicarbonate transport in the insulin-mediated stimulation of nucleotide synthesis, we performed isotope tracing experiments using 13C2-15N-glycine or 4-13C-aspartate to measure purine and pyrimidine synthesis, respectively (Figure 3G). While insulin stimulates newly synthesized purines and pyrimidines in the wild-type conditions, it failed to promote flux through de novo nucleotide synthesis in ΔSLC4A7 cells, suggesting that SLC4A7 is required to promote growth factor-dependent stimulation of de novo nucleotide synthesis (Figures 3H, 3I). Similar results were also observed with radiolabeled tracers such as Na+, 14C-HCO3, which measures the bicarbonate contribution to nucleotide synthesis, and 14C-glycine and 14C-aspartate incorporation to assess de novo purine and pyrimidine synthesis, respectively (Figure 3J, and Figure S3D). These findings suggest that SLC4A7 expression is regulated by growth signals and participates in growth signal-dependent induction of de novo nucleotide synthesis.
SLC4A7 is a target of mTORC1 signaling and sustains the mTORC1-dependent control of nucleotide synthesis.
Since insulin and EGF signaling converge paths on TSC-mTORC1 signaling to promote anabolic metabolism (Figure 4A) (Foster and Fingar, 2010), we asked whether the TSC-mTORC1 axis controls SLC4A7 protein levels downstream of insulin and EGF signaling. To render mTORC1 hyperactive and insensitive to growth and stress signal inputs, we previously generated TSC2-deficient (ΔTSC2) cells using the CRISPR Cas 9 technique (Villa et al., 2021). TSC2 is a crucial component of the TSC complex and a negative regulator of mTORC1 signaling (Carsillo et al., 2000; Huang and Manning, 2008; Ran et al., 2013; Villa et al., 2021). Loss of TSC2 rendered mTORC1 constitutively active in the absence of growth factors, as shown by the high level of S6K phosphorylation and increased in 4E-BP1 electrophoretic mobility shift (Figures 4B, C). The protein levels of SLC4A7, but not those of the other bicarbonate transporters, were significantly increased in ΔTSC2 cells in an mTORC1 dependent manner, as treatment with specific mTORC1 inhibitor rapamycin ablated this increase (Figure 4B and Figure S4A). Short-term (1–2 hours) of rapamycin treatment were sufficient to decrease SLC4A7 protein levels in TSC2-deficient cell settings, including ΔTSC2 HeLa cells (Figure 4C) and TSC2-null human kidney-derived angiomyolipoma (AML) cells (Figure 4D). Consistent with a decrease in SLC4A7 levels, mTORC1 inhibition also decreased NaHCO3 uptake in wildtype and TSC2-deficient cells (Figure 4E). Next, we measured the effects of SLC4A7 knockdown on de novo purine and pyrimidine synthesis through 15N-(amide)-glutamine tracing in wild-type and ΔTSC2 cells (Figure 4F). While glutamine import was not altered upon SLC4A7 knockdown (Figure 4G), 15N-glutamine flux through de novo purine (Figure 4H) and pyrimidine synthesis (Figure 4I) was increased in ΔTSC2 cells compared to their wild-type counterparts and reduced when SLC4A7 was depleted (Figures 4H, I and Figure S4B). SLC4A7 knockdown also prevented the mTORC1-dependent stimulation of bicarbonate incorporation into RNA as well as de novo purine (14C-glycine) and pyrimidine (14C-aspartate) synthesis flux into RNA in ΔTSC2 cells (Figures 4J, K) without any significant effects on the purine and pyrimidine salvage pathway (Figure S4C). These findings demonstrate that SLC4A7 protein abundance is regulated by mTORC1 and is required for the mTORC1-dependent induction of de novo nucleotide synthesis.
Figure 4. mTORC1 stimulates SLC4A7 protein abundance and bicarbonate uptake to promote de novo nucleotide synthesis.
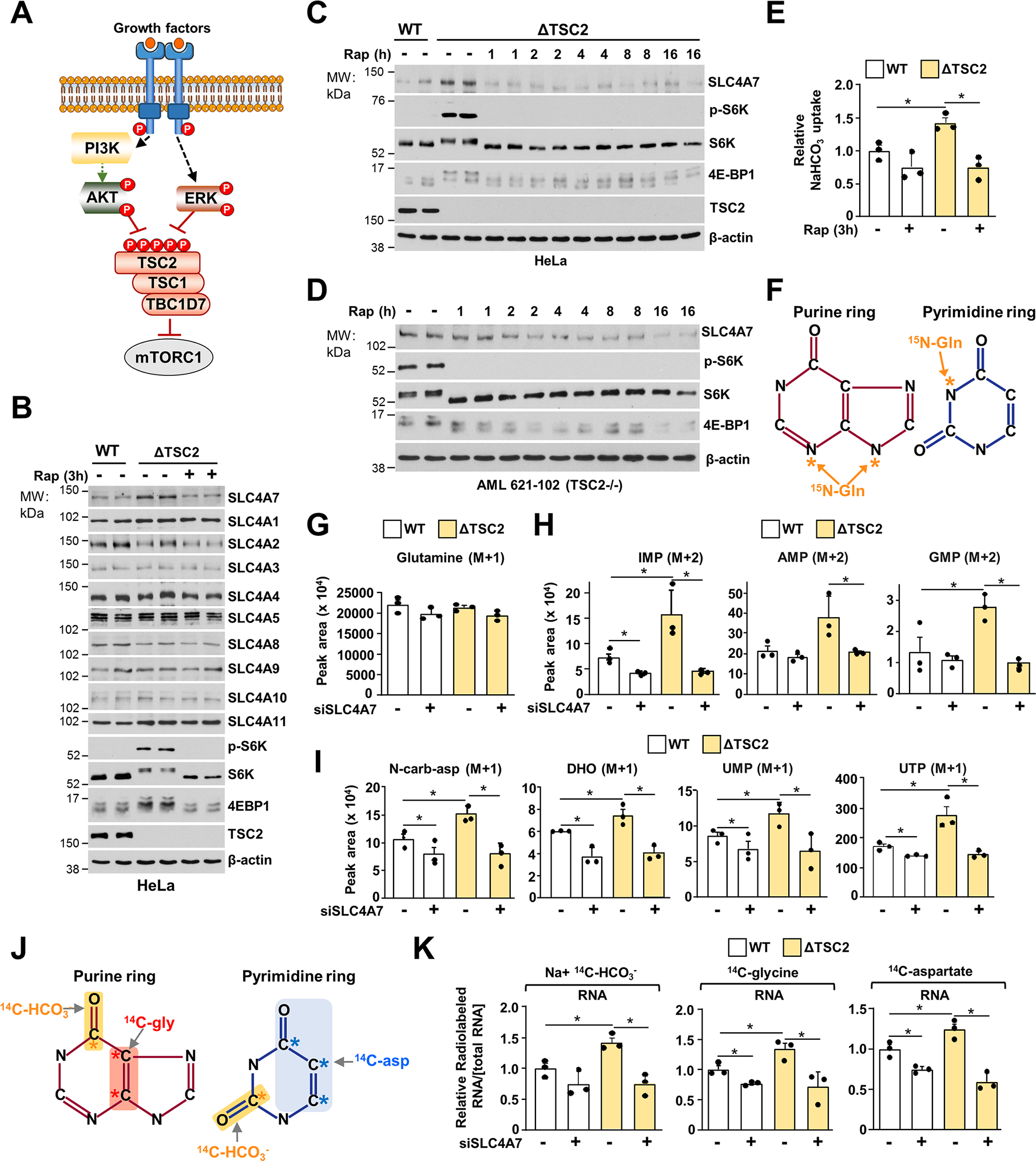
(A) Schematic illustrating the convergence of Akt and ERK signaling on the TSC-mTORC1 axis.
(B) Immunoblots of serum-deprived wild-type or ΔTSC2 HeLa cells for 15 h and treated with vehicle (DMSO) or rapamycin (20 nM) for 3 h.
(C, D) Immunoblots of HeLa (C) or human angiomyolipoma (AML) 621–102 TSC2−/− (D) cells prepared as in (B) but treated with rapamycin (20 nM) over a time course.
(E) NaHCO3 uptake from serum-deprived wild-type or ΔTSC2 HeLa cells treated with vehicle (DMSO) or rapamycin (20 nM) for 3 h.
(F) Illustration of the nitrogen from glutamine incorporated into the purine and pyrimidine rings.
(G) Normalized peak areas of 15N-glutamine measured via LC-MS/MS in HeLa cells transfected with siRNA targeting SLC4A7 or nontargeting controls after labeling cells with 15N-(amide)-glutamine for 1 h.
(H) Normalized peak areas of 15N-labeled purine intermediates measured via LC-MS/MS in HeLa cells transfected with siRNA targeting SLC4A7 or nontargeting treated as in (F).
(I) Normalized peak areas of 15N-labeled pyrimidine intermediates measured as in (F) and (G).
(J) Schematic of the carbons from glycine, aspartate and bicarbonate incorporated into the purine and pyrimidine rings.
(K) Relative incorporation of 14C from HCO3−, glycine, aspartate into RNA. 48 h after transfection with SLC4A7 siRNAs or nontargeting controls, wild-type or ΔTSC2 HeLa cells were serum starved for 15 h and labeled with the indicated radiotracers for 6 h.
(E),(G),(H)-(K) The data are plotted as the means ± SDs of biological triplicates and are representative of at least two independent experiments. *p < 0.05, by one-way ANOVA with Tukey’s post hoc test for multiple pairwise comparisons (E)-(I).
The mTORC1-S6K1-eIF4B axis controls SLC4A7 through translational-dependent mechanisms.
To identify the molecular mechanisms by which mTORC1 acutely controls SLC4A7 protein levels, we assessed whether S6 ribosomal kinase, a canonical mTORC1 target, could regulate SLC4A7. Knockdown of S6K1, S6K2, or both decreased SLC4A7 protein abundance (Figure 5A). Moreover, an S6K-specific inhibitor (PF-4708671) rapidly reduced SLC4A7 protein levels in ΔTSC2 HeLa and AML TSC2−/− cells, suggesting that S6K is responsible for the regulation of SLC4A7 (Figure 5B and Figure S5A). Expression of S6K1-constituvely active (S6K1-CA) in HEK293E cells cultured in the absence of growth factors also led to an increase in SLC4A7 (Figure 5C). These results indicate that the mTORC1-S6K signaling axis controls SLC4A7 expression. In contrast to protein levels, the mRNA expression levels of SLC4A7 were not affected by inhibition of mTORC1 (Figure S5B, Table S5), suggesting that mTORC1-S6K signaling increases SLC4A7 protein abundance via post-transcriptional mechanisms. To test whether translation of SLC4A7 is regulated downstream of mTORC1, we used cycloheximide (CHX) to block translation elongation in wild-type or ΔTSC2 cells in the absence of serum. Indeed, CHX treatment prevented the increase in SLC4A7 protein levels in ΔTSC2 cells (Figure 5D and Figure S5C), suggesting SLC4A7’s protein stability is reduced in ΔTSC2 cells. mTORC1-S6K has been shown to promote translation of mRNAs with highly structured 5′ UTR via recruitment of eukaryotic initiation factor complex 4A/B (eIF4A/4B) into the translation preinitiation complex (Holz et al., 2005). In response to mTORC1 activation, S6K1 phosphorylates the translation factor eIF4B on S406 and S422 (Figure S5D), enabling its recruitment into the eIF3 complex to promote mRNA translation (Holz et al., 2021) (Figure 5E). eIF4A unwinds highly structured 5′ UTR with its RNA helicase activity, thereby enhancing the efficiency of target mRNA translation (Jin et al., 2013; Waldron et al., 2019). Computational analysis based on the primary RNA sequence (Lorenz et al., 2011) showed that the 5′ UTR of SLC4A7 is highly structured, with very low Gibbs free energy (ΔG = −112.00 kcal/mol) (Figure 5F), suggesting that SLC4A7 mRNA is translated at a high rate (Leppek et al., 2018). This complexity of 5′ UTR structure is similar to that of well-established eIF4A/4B target genes such as cMyc, ODC1, and CDC25C (Figure 5F) (Csibi et al., 2014; Shahbazian et al., 2010). Knockdowns of eIF4A, eIF4B, or both reduced SLC4A7 protein levels (Figure 5G). Furthermore, expression of nonphosphorylatable eIF4B S406A or eIF4B S422A reduced SLC4A7 protein levels in ΔTSC2 cells (Figure 5H), which was accompanied by a decrease in the bicarbonate-dependent flux into nucleotide synthesis (Figure 5I), suggesting that the S6K-dependent phosphorylation of eIF4B is required for SLC4A7 translation and function. To better resolve the translation efficiency of SLC4A7 downstream of mTORC1, we undertook a polysome profiling assay (Chassé et al., 2017) in ΔTSC2 HeLa cells (Figures S5E, F). mTORC1 inhibition by rapamycin or knockdown of eIF4A/B shifted SLC4A7 mRNA toward lighter polysome fractions (Figures 5J, K), indicating that SLC4A7 mRNA translation is suppressed by mTORC1 and eIF4A/B inhibition. In contrast, the mRNA distribution of a control gene actin, did not display a similar shift (Figures S5G, H). Collectively, these results demonstrate that mTORC1-S6K signaling induces SLC4A7 protein expression via eIF4A/B-mediated mRNA translation to support the cellular entry of HCO3− molecules for the de novo nucleotide synthesis (Figure 5L).
Figure 5. mTORC1-S6 kinase promotes SLC4A7 mRNA translation through the control of eIF4B phosphorylation.
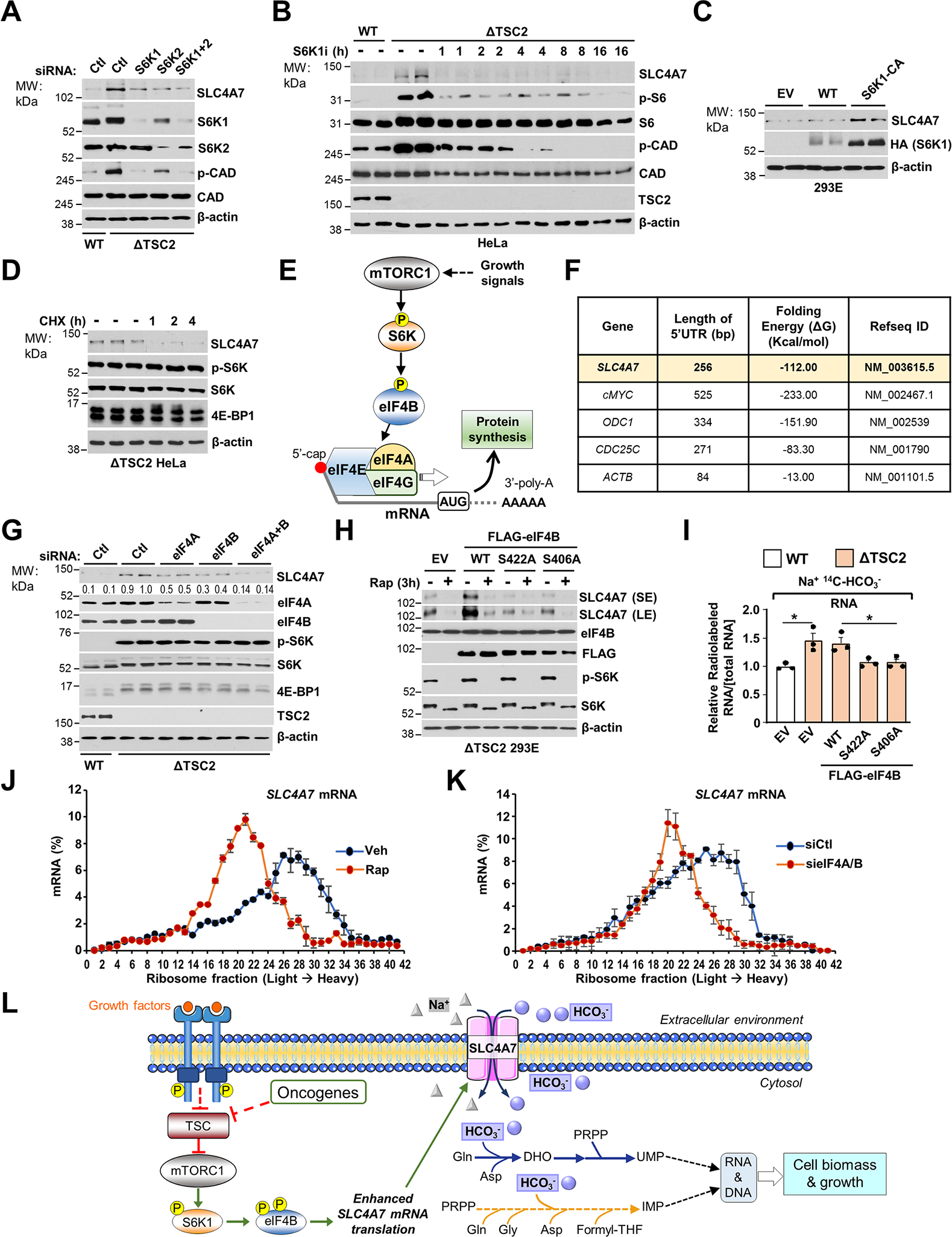
(A) Immunoblots of wild-type or ΔTSC2 HeLa cells transfected with siRNA targeting S6K1, S6K2, both or nontargeting controls for 48 h followed by a 15 h serum starvation.
(B) Immunoblots of wild-type or ΔTSC2 HeLa cells grown in serum depleted condition for 15 h and treated with either vehicle (DMSO) or S6K1 inhibitor (S6K1i, PF470871 10 μM).
(C) Immunoblots of wild-type HEK293E cells transfected with empty vector (EV), wild-type S6K1 (S6K1-WT) or constitutively active S6K1 (S6K1-CA) for 36 h and grown in serum depleted condition for 15 h.
(D) Immunoblots of ΔTSC2 HeLa cells grown in serum-depleted condition for 15 h and treated with either vehicle (DMSO) or cycloheximide (CHX, 10 μM) over a time-course.
(E) Schematic illustrating the role of the S6K-eIF4 complex in the regulation of mRNA translation downstream of mTORC1 signaling.
(F) Analysis of the Gibbs free energy (ΔG) of 5′ UTR of mRNA.
(G) Immunoblots of wild-type or ΔTSC2 HeLa cells transfected with siRNA targeting eIF4A, eIF4B, both or nontargeting controls for 48 h.
(H) Immunoblots of ΔTSC2 HEK293E cells transfected with empty vector, wild-type eIF4B, eIF4B S422A, or eIF4B S406A for 36 h and serum starved for 15 h prior to protein extraction.
(I) Relative incorporation of 14C from HCO3− into RNA from wild-type or ΔTSC2 HEK293E cells treated as in (H) and serum starved for 15 h and incubated for 6 h in the presence of the Na+, 14C-HCO3−.
(J) qPCR analysis of sucrose-gradient polysome fractions of ΔTSC2 HeLa cells treated with vehicle (DMSO) or rapamycin (20 nM) for 3 hours.
(K) qPCR analysis of sucrose-gradient polysome fractions of ΔTSC2 HeLa cells transfected with siRNA targeting eIF4A/B or nontargeting controls for 48 h.
(L) Model of regulation and function of SLC4A7 downstream of mTORC1 signaling.
(I)-(K) The data are plotted as the means ± SDs of biological triplicates. (A)-(D), (G), (H) are representative of at least two independent experiments *p < 0.05, by one-way ANOVA with Tukey’s post hoc test for multiple pairwise comparisons (I).
Tumor formation requires SLC4A7 for growth and maintenance of nucleotide metabolism.
Given the dependence of de novo nucleotide synthesis for SLC4A7 expression and bicarbonate levels, we set out to determine whether SLC4A7 is required for cell proliferation, tumor growth, and de novo purine and pyrimidine synthesis in vivo. To define whether SLC4A7 could represent a metabolic dependency in mTORC1-driven tumor cells, we depleted SLC4A7 levels in several proliferating cells. SLC4A7 loss or knockdown reduced proliferation in various cell settings, such as HEK293E, HeLa, CAL-51, A549, and AML TSC2−/− cells (Figures 6A, B, and Figure S6A). In addition, supplementation with nucleosides partially rescues the proliferation of ΔSLC4A7 cells, suggesting that loss of SLC4A7 partly limits nucleotide availability for cell proliferation (Figure S6B). Cells with hyperactive mTORC1 due to TSC2 loss were more sensitive to SLC4A7 depletion than wild-type cells (Figure 6C). Interestingly, mTORC1 inhibition together with SLC4A7 depletion in CAL-51 cells, resulted in a more pronounced decrease in cell proliferation than each of these perturbations alone (Figure 6D). Moreover, SLC4A7 depletion decreased the anchorage-independent growth of A549 and CAL-51 cancer cells (Figure 6E), and a low-dose of rapamycin further sensitized ΔSLC4A7 cells under these conditions. Given the antiproliferative effects of SLC4A7 depletion in a variety of cancer cell lines (Figure 6A), we sought to evaluate the antitumor effect of SLC4A7 in tumor models in mice. Since SLC4A7 expression is associated with unfavorable overall survival in patients with various cancer types (Figure S6C) and that upregulation of SLC4A7 has been observed in breast cancer (Gorbatenko et al., 2014), we sought to directly evaluate the role of SLC4A7 in breast cancer progression and nucleotide synthesis in vivo using a xenograft mouse model. We implanted CAL-51 breast cancer cell line deleted or not for SLC4A7 into the flanks of nude mice and treated them with low-dose rapamycin (1 mg/kg) when tumors reached an average volume of 100 mm3. Notably, SLC4A7 deletion reduced tumor growth to a similar level to the low dose rapamycin treatment, while rapamycin treatment further sensitized the ΔSLC4A7 CAL-51-derived tumors (Figures 6F, G), suggesting a crucial role for SLC4A7 in tumor growth. To determine the contribution of SLC4A7 for the de novo nucleotide synthesis in vivo, we applied 15N-(amide)-glutamine intraperitoneally (Lane et al., 2015) in mice bearing breast cancer tumor indicated in Figure 6F, and quantified purine and pyrimidine intermediates via LC-MS/MS in these tumors. Mice showed 60% enrichment of 15N-glutamine in the plasma, and similar levels 15N-glutamine in tumors across the different conditions (Figures S6D, E). However, purine and pyrimidine intermediates derived from 15N-glutamine were reduced in ΔSLC4A7 tumors compared to wild-type tumors (Figures 6H, I, and Figures S6F, G), suggesting that SLC4A7 is required to sustain de novo nucleotide synthesis in tumors. Moreover, rapamycin treatment also resulted in reduced nucleotide synthesis in tumors. The combination of rapamycin with SLC4A7 depletion, further reduced the rate of nucleotide synthesis in vivo, likely due to other mechanisms of nucleotide synthesis inhibition in response to rapamycin (Ben-Sahra and Manning, 2017; Villa et al., 2019). Collectively, this study supports a critical role for SLC4A7 in the control of cell proliferation and tumor growth through the regulation of de novo nucleotide synthesis.
Figure 6. SLC4A7 is required for tumor growth and de novo nucleotide synthesis in tumors.
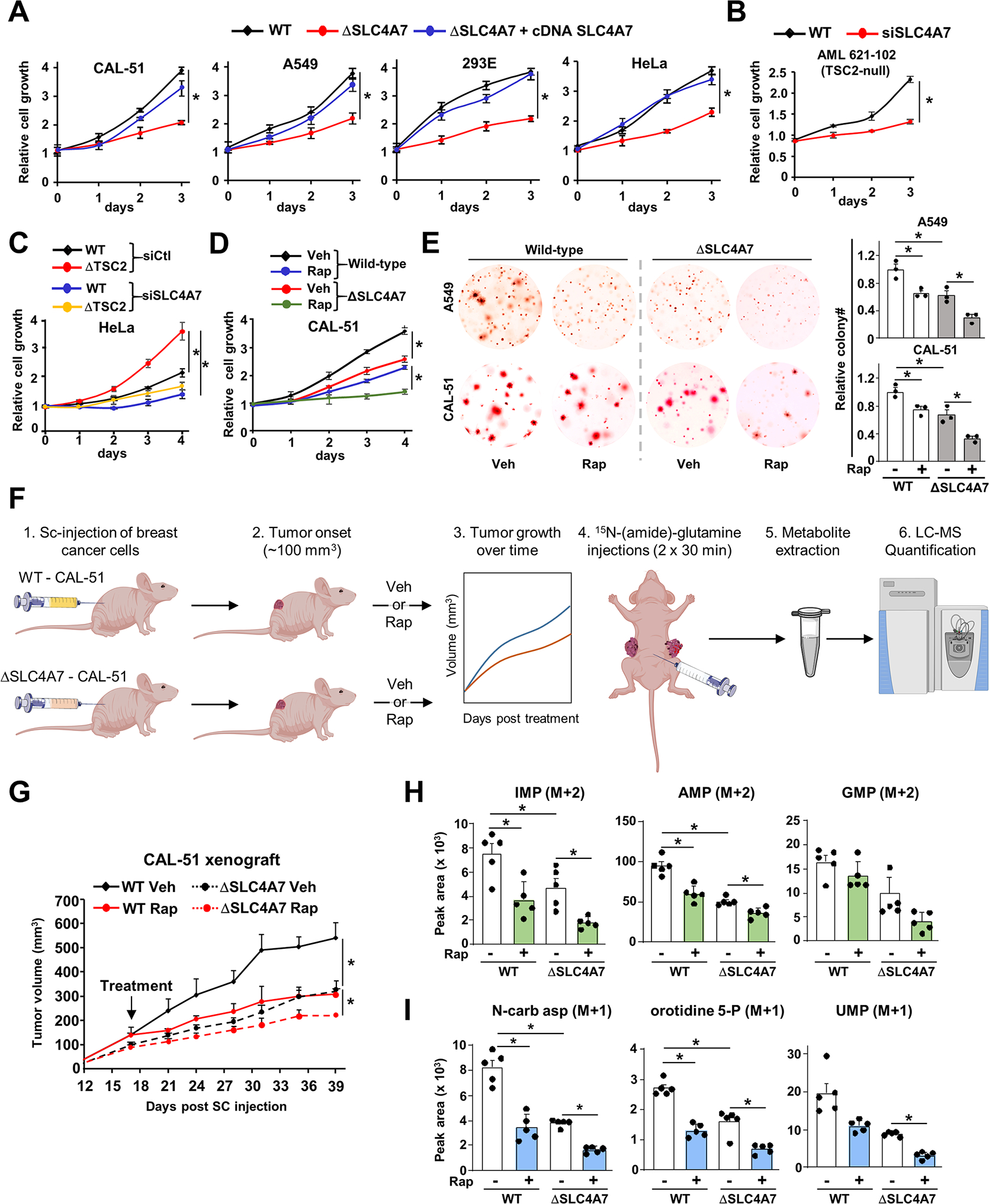
(A) Indicated wild-type or ΔSLC4A7 cell lines were transfected with either empty vector or SLC4A7 cDNA for 48 h and grown in 1 % serum (CAL-51 and A549 cells) or 10 % dialyzed serum (HEK293E and HeLa cells) for 72 h, and cell number was measured via crystal violet staining every 24 h.
(B, C) AML 621–102 (TSC2−/− cells) (B), wild-type and ΔTSC2 HeLa cells (C) grown in 1 % serum for 72 h (B) or 96 h (C), and cell number was measured via crystal violet staining every 24 h.
(D) Wild-type and ΔSLC4A7 CAL-51 cells treated with vehicle or rapamycin (20 nM) for 96 h, and cell number was measured via crystal violet staining every 24 h.
(E) Soft agar colony formation assay with indicated cells treated with vehicle (DMSO) or rapamycin (10 nM) for three weeks. Cell images were acquired at 3× magnification. Colony quantification from three independent biological replicates is shown.
(F) Workflow for the xenograft and in vivo 15N-(amide)-glutamine stable isotope tracing experiments.
(G) Wild-type and ΔSLC4A7 CAL-51 cells were subcutaneously injected into athymic nude mice (n = 5 per group). Mice were treated with vehicle or rapamycin (1 mg/kg), and tumor volume was monitored over time.
(H, I) Mice bearing wild-type or ΔSLC4A7 CAL-51-derived tumors (n = 5) injected with 15N-glutamine (0.8 g/kg) through the intraperitoneal route for 60 min prior to tumor collection and metabolite extraction and analysis via LC-MS/MS.
(A)-(E) The data are plotted as the means ± SDs of biological triplicates and are representative of at least two independent experiments. *p < 0.05, by two-tailed Student’s t-test for pairwise comparisons (B) or one-way ANOVA with Tukey’s post hoc test for multiple pairwise comparisons (A), (C)-(E), (G)-(I).
DISCUSSION
This study demonstrates that mTORC1 signaling controls bicarbonate availability to regulate nucleotide synthesis and cell growth in proliferating cells and tumors. Until now, cellular bicarbonate inflow was primarily known to influence intracellular pH to maintain ion homeostasis. However, the role of the bicarbonate molecule as a critical metabolic precursor for nucleotide synthesis or cell growth has not been recognized. Our finding that reduced bicarbonate levels result in a stark decrease in both purine and pyrimidine nucleotides and cell growth, reveals an anabolic role for bicarbonate ions in the maintenance of cell growth and proliferation.
While mammalian cells express more than ten bicarbonate transporters, we found that only SLC4A7 was significantly coessential with the nucleotide biosynthesis genes. Depleting SLC4A7 resulted in a 20% reduction in bicarbonate uptake, and while this was sufficient to attenuate nucleotide synthesis, it did not significantly alter intracellular pH. Our results are in agreement with a previous study showing a role for SLC4A7 in mediating RAS-dependent induction of macropinocytosis in cancer cells without impacting intracellular pH (Ramirez et al., 2019).
While we found that mTORC1 stimulates the import of bicarbonate through SLC4A7, cells can produce bicarbonate through carbonic anhydrase (CA) reactions (Effros et al., 1978). Indeed, CAs reversibly use carbon dioxide (CO2) and water to produce carbonic acid (H2CO3), which can spontaneously produce bicarbonate ions. CAs are primarily expressed in red blood cells, gastric mucosa, and renal tubules and they serve to maintain basal cellular pH (Kivelä et al., 2005; Mboge et al., 2018). CAs have also been implicated in pH regulation in primary tumor cells (Chiche et al., 2009; Mboge et al., 2018). However, it remains to be determined whether CAs are regulated in response to pro-growth signals akin to SLC4A7, and whether bicarbonate derived from CAs is channeled towards nucleotide biosynthesis and cell biomass (White et al., 2017).
Our findings that SLC4A7 is specifically controlled by mTORC1 signaling, a major regulator of de novo nucleotide synthesis (Ben-Sahra and Manning, 2017), support the concept that SLC4A7 is selectively regulated to provide sufficient bicarbonate as a substrate for de novo nucleotide synthesis.
Another interesting aspect lies in the mechanism by which mTORC1 signaling controls SLC4A7 expression. mTORC1 activation, through the S6K-dependent eIF4B phosphorylation, stimulates SLC4A7 mRNA translation to increase SLC4A7 protein abundance and promote bicarbonate inflow in proliferating cells. mTORC1 orchestrates the selective translation of a family of mRNA transcripts defined by a 5′ terminal oligopyrimidine (TOP) motif (Jefferies et al., 1997; Thoreen et al., 2012). High throughput 5′TOP mRNA analysis and ribosome profiling experiments showed that the ribosome protein RPL13A has the highest 5′TOP score (Philippe et al., 2020). In the same dataset, SLC4A7 mRNA exhibits a similar mTORC1-dependent ribosome footprint on mRNA to that of RPL13A mRNA, further supporting the translational regulation of SLC4A7 downstream of mTORC1 (Philippe et al., 2020).
Interestingly, high throughput phosphoproteomic studies determining downstream substrates of mTORC1 have shown that mTORC1 signaling could induce SLC4A7 phosphorylation at the S89 site (Hsu et al., 2011). Therefore, we cannot exclude that mTORC1 signaling post-translationally regulates SLC4A7 activity in addition to the translational regulation reported in our study. Interestingly, among the SLC4A family, only SLC4A7 was seen phosphorylated in response to mTORC1 activation, supporting that there is a specific link between mTORC1 signaling and SLC4A7.
Our findings suggest that mobilizing key substrates such as bicarbonate ions is critical for the mTORC1-dependent stimulation of macromolecular synthesis and cell biomass accumulation. However, it remains to be determined whether SLC4A7 regulates other bicarbonate-dependent metabolic pathways that are important for cell growth, such as pyruvate carboxylase (PC) or carbamoyl phosphate synthase 1 (CPS1) from the urea cycle. The pyrimidine enzyme CAD, the purine enzyme PAICS, CPS1, and PC, which all use HCO3− as a substrate, exhibit similar binding affinity constants for HCO3− in the millimolar range (Diez-Fernandez et al., 2013; Jitrapakdee et al., 1999; Shaw and Carrey, 1992). Therefore, the cellular decrease in HCO3− uptake in response to SLC4A7 depletion is likely to alter metabolic processes beyond de novo nucleotide synthesis. In fact, our metabolite profile experiment (Table S4) revealed that urea cycle intermediates such as citrulline, ornithine, and urea, were reduced upon SLC4A7 knockdown, suggesting that the SLC4A7-dependent control of bicarbonate levels is critical for the function of the urea cycle. Our data showed that bicarbonate depletion did induce a reduction in the intermediates of the urea cycle (Table S1), however, the decrease in cell fitness in response to HCO3− starvation occurs mostly through a reduction in de novo nucleotide synthesis.
While our results in proliferating cells indicate a role for SLC4A7 in regulating nucleotide synthesis and cell growth in a pH-independent manner, loss of SLC4A7 has been implicated to increase cytoplasmic acidification that is necessary for phagosome acidification (Sedlyarov et al., 2018). Further studies remain to be performed to define the importance of this regulation in differentiated human cells and pathological conditions.
Finally, our study also reveals a key role for SLC4A7 in supporting nucleotide synthesis in tumors and tumor growth. Indeed, targeting SLC4A7 showed significant antiproliferative and antitumoral effects, indicating that bicarbonate metabolism can be targeted in cancers. These in vivo results suggest that developing therapeutic strategies targeting SLC4A7 could expose metabolic vulnerabilities in cancers that have high levels of mTORC1 signaling and SLC4A7. Collectively, our observations implicate SLC4A7 as a key mediator of de novo nucleotide synthesis downstream of mTORC1 signaling in proliferating cells and tumors. While bicarbonate is a byproduct of metabolism and a regulator of pH homeostasis, our study reveals that bicarbonate levels, akin to other nutrients such as glucose and amino acids, are regulated and limiting for anabolic pathways to support cell biomass production and growth.
LIMITATION OF STUDY
We have found that mTORC1 activation stimulates SLC4A7 mRNA translation through the S6K-dependent phosphorylation of eIF4B. While this mechanism occurs in proliferating and cancer cells, the significance of the mTORC1-SLC4A7 axis in physiological contexts remains to be determined. Given the similarities in metabolic requirements between the immune and cancer cells (Andrejeva and Rathmell, 2017), it is tempting to speculate that such mechanisms could also occur in immune cells. Moreover, since SLC4A7 levels are sensitive to the immunosuppressant drug rapamycin (Brizuela et al., 1991), it is important to evaluate whether the stimulation of SLC4A7 expression by mTORC1 is required to activate the immune cells in response to infection. While a previous study showed that SLC4A7 promotes the phagosome acidification of macrophages (Sedlyarov et al., 2018), it remains to be demonstrated if SLC4A7 regulates de novo nucleotide synthesis in macrophages and other immune cell settings to enable proper physiological response.
SLC4A7 exhibits more than 10 different isoforms, which vary in surface abundance and transport activity. These isoforms originate from different promoter regions, three structural elements (cassettes I-III), and the respective alternative splicing (Liu et al., 2013; Liu et al., 2015; Romero et al., 2013). SLC4A7 isoforms are also expressed in a tissue-specific manner. Additional studies will be needed to address whether the expression of a particular SLC4A7 isoform is regulated by mTORC1 and promotes the regulation of nucleotide synthesis in specific tissues.
With respect to the physiological relevance of these findings, it will be important to take into consideration the concentration of bicarbonate found in plasma (~24–30 mM) and tissues for follow-up studies (Hirai et al., 2019). In fact, measurement of bicarbonate levels in vivo will be the next step to determine whether bicarbonate levels correlate with alterations in nucleotide metabolism and cell function in physiological and pathological conditions. For example, it will be critical to address whether increasing bicarbonate abundance in the tumor microenvironment participates in tumor growth and proliferation.
Another outstanding question is why SLC4A7, but not the other SLC4A carriers, is linked to the synthesis of nucleotides? Our data suggest that SLC4A7 is uniquely downstream of mTORC1 signaling because of its highly structured 5′ UTR and 5′ TOP mRNA sequence. Therefore, the acute increase of SLC4A7 protein abundance downstream of mTORC1 signaling can represent one of the arms by which anabolic metabolism is sustained in growing cells. However, we cannot rule out other mechanisms specifically linking SLC4A7 to nucleotide metabolism and compensation mechanisms from other SLC4A bicarbonate carrier family members.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Requests of information and reagents should be directed to and will be fulfilled by the Lead Contact, Issam Ben-Sahra (issam.ben-sahra@northwestern.edu).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact without restriction.
Data and Code Availability
Original western blots have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table. Mass spectrometry data are presented in the table S1.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-S6K1 | Cell Signaling Technology | Cat#2708; RRID: AB_390722 |
| Rabbit monoclonal anti-pS6 (S240/S244) | Cell Signaling Technology | Cat#5364; RRID: AB_10694233 |
| Rabbit Polyclonal anti-SLC4A7 | Invitrogen (Thermo Fisher) | Cat#PA5-75807; RRID:AB_2719535 |
| Band 3/ AE1 Polyclonal antibody (SLC4A1) | ProteinTech Group, Inc | Cat#28131-1-AP |
| AE2 Polyclonal antibody (SLC4A2) | ProteinTech Group, Inc | Cat#26332-1-AP |
| SLC4A3 Polyclonal antibody | ProteinTech Group, Inc | Cat#26883-1-AP |
| SLC4A4 Polyclonal antibody | ProteinTech Group, Inc | Cat#11885-1-AP |
| Rabbit Polyclonal anti-SLC4A5 Antibody | Novus Biologicals | Cat#NBP1-59528 |
| Rabbit Polyclonal anti-SLC4A8 | ProteinTech Group, Inc | Cat#12531-1-AP |
| Rabbit AE4 Polyclonal Antibody (SLC4A9) | Mybiosci | Cat#MBS821050 |
| SLC4A10 Rabbit pAb | Abclonal Technology | Cat#A13810 |
| Rabbit Polyclonal anti-SLC4A11 | BiorByt | Cat#orb215316 |
| Phospho-p70 S6 Kinase (Thr389) (108D2) Rabbit mAb | Cell Signaling Technology | Cat# 9234S; RRID:AB_2269803 |
| Rabbit Anti-p70 S6 Kinase Monoclonal Antibody | Cell Signaling Technology | Cat# 2708S; RRID:AB_390722 |
| Phospho-S6 Ribosomal Protein (Ser240/244) Antibody | Cell Signaling Technology | Cat# 2215S; RRID:AB_331682 |
| S6 Ribosomal Protein (5G10) Rabbit mAb | Cell Signaling Technology | Cat# 2217S; RRID:AB_331355 |
| Phospho-Akt (Ser473) (D9E) Rabbit mAb | Cell Signaling Technology | Cat# 11962; RRID:AB_2797780 |
| Akt (pan) (C67E7) Rabbit mAb | Cell Signaling Technology | Cat# 4691, RRID:AB_915783 |
| 4E-BP1 Antibody | Cell Signaling Technology | Cat# 9452S; RRID:AB_331692 |
| Tuberin/TSC2 (D93F12) XP_ Rabbit mAb | Cell Signaling Technology | Cat# 4308S; RRID:AB_10547134 |
| Anti-mouse IgG, HRP-linked Antibody | Cell Signaling Technology | Cat#7076; RRID:AB_330924 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling Technology | Cat#7074; RRID:AB_2099233 |
| β-actin Antibody | Sigma-Aldrich | Cat#A5316; RRID: AB_476743 |
| Phospho-CAD (Ser1859) Antibody | Cell Signaling Technology | Cat#12662; RRID:AB_2750934 |
| CAD Antibody | Cell Signaling Technology | Cat#11933; RRID:AB_2797772 |
| S6K2 Polyclonal antibody | ProteinTech Group, Inc | Cat#15268-1-AP; RRID:AB_2182250 |
| Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (E10) Mouse mAb antibody | Cell Signaling Technology | Cat# 9106S; RRID: AB_331768 |
| p44/42 MAPK (Erk1/2) Antibody | Cell Signaling Technology | Cat# 9102S; RRID: AB_330744 |
| eIF4B Antibody | Cell Signaling Technology | Cat# 3592; AB_2293388 |
| eIF4A (C32B4) Rabbit mAb | Cell Signaling Technology | Cat#2013; RRID:AB_2097363 |
| DYKDDDDK Tag (D6W5B) Rabbit mAb | Cell Signaling Technology | Cat#14793; RRID:AB_2572291 |
| HA-Tag (C29F4) Rabbit mAb | Cell Signaling Technology | Cat#3724; RRID:AB_10693385 |
| Phospho-eIF4B (S422) Antibody | Cell Signaling Technology | Cat#3591; RRID:AB_2097522 |
| Phospho-eIF4B (Ser406) Antibody | Cell Signaling Technology | Cat#5399; RRID:AB_10695246 |
| eIF4A Antibody | Cell Signaling Technology | Cat#2490; AB_2097363 |
| H2AX Antibody | Thermo Fisher (product of: Bethyl) | Cat#A300-083A-M |
| Bacterial and virus strains | ||
| NEB Stable Competent E. coli | NEB | Cat#C3040 |
| DH5 alpha | Fisher Scientific | Cat#18258012 |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM without Bicarbonate | US Biological | Cat#D9812-07 |
| DMEM without glucose, bicarbonate | US Biological | Cat#D9800-02B |
| DMEM without glycine (and serine) | US Biological | Cat#D9800-16 |
| Rapamycin | Calbiochem/Sigma-Aldrich | Cat#553210 |
| Cycloheximide | EMD Millipore/Merck | Cat#239764 |
| Glycine | Sigma-Aldrich | Cat#G5417 |
| PF-9366 | MedChemExpress | Cat#HY-107778 |
| Insulin human | Alpha diagnostic | Cat#INSL16-N-5 |
| EGF (human recombinant) | Life Technologies/GIBCO | Cat#PHG0311 |
| Insulin-like Growth Factor-I human | Sigma-Aldrich | Cat#I3769 |
| Hypoxanthine | Sigma-Aldrich | Cat#H9636 |
| Inosine | Sigma-Aldrich | Cat#I4125 |
| 13C2-15N-glycine | Sigma-Aldrich | Cat#489522 |
| L-glutamine-(amide-15N) | Sigma-Aldrich | Cat#490024 |
| 1,2-3H-2-deoxy-D-Glucose | Perkin-Elmer | Cat#NET328A250UC |
| U-14C-glycine | Perkin-Elmer | Cat#NEC276E050UC |
| U-14C-glucose | Perkin-Elmer | Cat#NEC042V250UC |
| 3H-hypoxanthine | Perkin-Elmer | Cat#NET177001MC |
| 14C-glutamine | Perkin Elmer | Cat#NEC451050UC |
| 14C-formate | American Radiolabeled Chemicals | Cat#ARC 0163A |
| 3-14C-serine | Perkin-Elmer | Cat#NEC853050UC |
| 3H-uridine | Perkin Elmer | Cat#NET367250UC |
| 4-13C aspartate | Sigma-Aldrich | Cat#489999 |
| 14C-aspartate | Perkin Elmer | Cat#NEC268E050UC |
| Sodium, 14C- bicarbonate | American Radiolabeled Chemicals | Cat#ARC 0138C-250 μCi |
| DMSO | Sigma-Aldrich | Cat#D2650-100ML |
| L-glutamine | Corning | Cat#MT25005Cl |
| 2-mercaptoethanol | Sigma-Aldrich | Cat#M6250 |
| Thiazolyl Blue Tetrazolium Bromide | Sigma-Aldrich | Cat#M5655 |
| Polyethylenimine (PEI) | Polysciences | Cat#23966-2 |
| Opti-MEM media | Thermo Fisher | Cat#31985062 |
| Lipofectamine 3000 Transfection reagent | Invitrogen | Cat#L3000008 |
| Lipofectamine RNAi max | Invitrogen | Cat#13778150 |
| SuperSignal West Femto Chemiluminescent | Thermo Fisher scientific | Cat#PI34096 |
| SuperSignal West Pico PLUS | Thermo Fisher scientific | Cat#PI34580 |
| Fetal bovine serum (FBS) | Sigma-Aldrich | Cat#F2442 |
| Fetal Bovine Serum dialyzed | Sigma-Aldrich | Cat#F0392 |
| Microcystin-LR | Fisher Scientific | Cat#ALX350012C500 |
| Protease inhibitor cocktail | Sigma-Aldrich | Cat#P8340 |
| Bio-Rad Braford | Bio-Rad | Cat#5000006 |
| Acrylamide Protogel 30% | Fisher Scientific | Cat#EC-890 |
| UltraPure LMP Agarose | Invitrogen | Cat#16520-050 |
| Uridine | Sigma Aldrich | Cat#U3003 |
| Glutamine-free DMEM | GIBCO | Cat#11960-044 |
| Glucose-free DMEM | Invitrogen | Cat#11966025 |
| Propidium iodide | Sigma | Cat#P4170 |
| Critical commercial assays | ||
| Fluorometric Intracellular pH Assay Kit | Sigma-Aldrich | Cat# MAK150 |
| Intracellular pH Calibration Buffer Kit | Invitrogen | Cat# P35379 |
| SYBR Green PCR Master Mix | Life Technologies | Cat# 4312704 |
| RNeasy Mini Kit | QIAGEN | Cat#74104 |
| HiSpeed Plasmid Maxi Kit (25) | QIAGEN | Cat#12663 |
| AllPrep DNA/RNA Mini Kit | QIAGEN | Cat#80204 |
| KOD Extreme Hot Start DNA polymerase kit | Merck Millipore | Cat#71975-3 |
| GeneJET Plasmid Miniprep Kit | Thermo Fisher | Cat# K0503 |
| Deposited data | ||
| Ali et al. unprocessed files | Mendeley Data | http://dx.doi.org/10.17632/9zd4vncrf4.1 |
| Experimental models: Cell lines | ||
| HEK293E | John Blenis lab (Weill Cornell Medicine) | N/A |
| ΔTSC2 HEK293E | Villa et al. 2021 | N/A |
| HeLa | ATCC | Cat#CCL-2 |
| ΔTSC2 HeLa | Villa et al. 2021 | N/A |
| A549 | ATCC | Cat#CCL-185 |
| CAL51 | DSMZ | Cat#ACC 302 |
| AML 621-102 (TSC2-null-Empty Vector) | Dr. David Kwiatkowski (Brigham and Women’s Hospital) | N/A |
| Experimental models: Organisms/strains | ||
| Athymic nude Foxn1nu | The Jackson laboratory | Cat#002019 |
| Oligonucleotides | ||
| See Table S4 for list of primers used in this study | Integrated DNA Technologies | N/A |
| siRNA targeting human SLC4A7 | Dharmacon | Cat#L-007586-01-0005 |
| siRNA targeting human S6K1 | Dharmacon | Cat#L-003616-00-0005 |
| siRNA targeting human S6K2 | Dharmacon | Cat#L-004671 -00-0005 |
| siRNA targeting human eIF4A | Dharmacon | Cat#L-020178-00-0005 |
| siRNA targeting human eIF4B | Dharmacon | Cat#L-020179-00-0005 |
| Non-targeting siRNA pool | Dharmacon | Cat#D-001810-10-20 |
| Recombinant DNA | ||
| SLC4A7 (NM_003615) Human Tagged ORF Clone | OriGene | Cat#RC220734 |
| EIF4B (NM_001417) Human Tagged ORF Clone | OriGene | Cat#RC209955 |
| pcDNA3 Flag HA | Addgene | Cat#10792 |
| pCMV6-Entry Tagged Cloning Vector | OriGene | Cat#PS100001 |
| PX458 CRISPR | Addgene | Cat#48138) |
| pRK7-HA-S6K1-WT | Addgene | Cat#8984 |
| pRK7/HA-S6K1-F5A-E389-R3A | Addgene | Cat#8991 |
| pCMV6/Flag-EIF4B-S422A | This study | N/A |
| pCMV6/Flag-EIF4B-S406A | This study | N/A |
| Software and algorithms | ||
| Image J | National Institutes of Health | RRID: SCR_003070 |
| Prism 9 | GraphPad Software | https://www.graphpad.com/ |
| FlowJo Software | BD Biosciences | https://www.flowjo.com/ |
| Other | ||
| Microplate Reader | Tecan | Infinite M1000 Pro |
| Multipurpose Scintillation Counter | Beckman Coulter | Model: LS 6500 |
| Nitrocellulose membrane 0.2 μm | GE Healthcare | Cat# 10600001 |
| Polypropylene centrifuge tubes | Beckman Coulter | Cat# 331372 |
| PCR plates | Bio-Rad | Cat#HSP9601 |
| UV transparent flat bottom 96 well plate | Corning | Cat#3635 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines and Tissue Culture
HeLa, and A549 cell lines were obtained from American Type Culture Collection (ATCC) and CAL-51 cells were obtained from DSMZ. The Human ΔTSC2 HeLa and ΔTSC2 HEK-293E cells was generated in our previous study (Villa et al., 2021). HEK293E cells were kindly provided by Dr. John Blenis (Weill Cornell Medicine). HeLa, HEK293E, and A549 cell lines were cultured in DMEM with 25 mM glucose (CellGro), 10 % Fetal bovine serum (FBS), 37°C, and 5 % CO2. CAL-51 cells were cultured in analogous medium but with 20 % FBS. Media were changed to serum free, 15 hours prior to metabolite extraction, pH measurement, and polysome profiling assay. Viable cells were counted using a TC-20 Automated Cell Counter (Bio-Rad).
Xenograft Experiments
5-week-old athymic nude mice (Female) were injected subcutaneously in the flank with 5 × 106 wild-type CAL-51 cells or 5 × 106 ΔSLC4A7 CAL-51 cells in a 1:1 mixture of Matrigel and PBS. When tumors reached a size of approximately 100 mm3, the mice were treated with either vehicle (10% EtOH, 40% PEG300, 5% Tween-80 and 45% saline (0.9 % NaCl)) or rapamycin (1 mg/kg). Either rapamycin or the vehicle was administered three times per week for 27 days through the intraperitoneal route. Tumor size was measured in three dimensions using an electronic caliper twice per week for four weeks. In vivo stable isotope tracing experiments were performed by two intraperitoneal bolus injections (2 × 30 min) of 15N-(amide)-glutamine (0.8 g/kg) dissolved in Hank’s Balanced Salt Solution (Sigma). Mice were euthanized 30 minutes after the last injection. Tissue were harvested, frozen in ice cold-methanol 80 %, and stored at −80 C until samples were ready for processing. All experiments were accomplished following appropriate guidelines and regulations. The Institutional Animal Care and Use Committee (IACUC) approved all animal procedures and studies at Northwestern University.
METHOD DETAILS
CRISPR/Cas9, siRNA and plasmids
To generate SLC4A7 knockout in HEK293E, HeLa, A549 and CAL-51 cells small guide RNA (sgRNA) sequence targeting the exon 3 of SLC4A7 was cloned into the PX458 CRISPR (Addgene, Plasmid # 48138) vector using the following oligonucleotide indicated in Table S5.
48 hours after transfection, GFP positive cells were sorted into 96-well plates as single cell per well. Clonal cells were screened by immunoblotting with anti-SLC4A7 antibody. For siRNA experiments, nontargeting control pool (D-001810-10-20), SLC4A7 (L-007586-01-0005), S6K1 (L-003616-00-0005), S6K2 (L-004671-00-0005) siRNAs (On TARGETplus SMARTpool) were acquired from Dharmacon (Horizon) and used at 35 nM (final concentration) with 6 μL of Lipofectamine RNAimax (Thermo fisher scientific # 13778150) (6-well plate) for each condition. The following plasmid constructs were used in this study; pCMV6/Flag-SLC4A7, pRK7/HA-S6K1-WT, pRK7/HA-S6K1-F5A-E389-R3A, pCMV6/Flag-EIF4B, pCMV6/Flag-EIF4B-S422A, pCMV6/Flag-EIF4B-S406A. Cells were transfected with siRNA or plasmids for 48 hours before proceeding to any specific treatments.
Metabolite profiling and flux analyses
To determine the relative abundances of intracellular metabolites, extracts were prepared and analyzed via LC-MS/MS. Briefly, for targeted steady-state samples, metabolites were extracted on dry ice with 4 mL 80% methanol (−80°C), as described previously (Ben-Sahra et al., 2013; Yuan et al., 2019). Insoluble material was pelleted by centrifugation at 3000 g for 5 min, followed by two consecutive extractions of the insoluble pellet with 0.5 mL 80 % methanol, with centrifugation at 20,000 g for 5 min. The 5-mL metabolite extract from the pooled supernatants was dried down under nitrogen gas using the N-EVAP (Organomation, Inc, Associates). 50% acetonitrile was added to the samples for reconstitution following by vortexing for 30 sec. Samples solution was centrifuged 20,000 g for 30 min at 4 °C. Supernatant was collected for LC-MS analysis.
For isotope tracing experiments, cells were seeded in biological triplicate (~80–90% confluent), washed once with serum-free DMEM and then incubated with either 10 mM 13C6-glucose, 4 mM 15N-(amide)-glutamine, or 400 μM 13C2-15N-glycine for 1 hour in either glucose-free, glutamine-free, or glycine-free media, respectively. Metabolites were extracted as described in the steady-state studies (Yuan et al., 2019). For 13C-aspartate tracing, 4-13C-aspartate (400 μM) was added in regular DMEM (which does not contain aspartate) and metabolites were extracted as described previously. For both, steady-state and isotope tracing experiments, samples were analyzed by High-Performance Liquid Chromatography and High-Resolution Mass Spectrometry and Tandem Mass Spectrometry (HPLC-MS/MS). In detail, the LC-MS/MS system is comprised of a Thermo Q-Exactive in line with an electrospray ionization (ESI) source and an Ultimate3000 (Thermo) series HPLC consisting of a binary pump, degasser, and auto-sampler outfitted with a Xbridge Amide column (Waters; dimensions of 2.3 mm × 100 mm and a 3.5 μm particle size). The mobile phase A contained 95% (vol/vol) water, 5% (vol/vol) acetonitrile, 10 mM ammonium hydroxide, 10 mM ammonium acetate, pH = 9.0; B was 100% Acetonitrile. The gradient was as following: 0 min, 15 % A; 2.5 min, 30 % A; 7 min, 43 % A; 16 min, 62 % A; 16.1–18 min, 75 % A; 18–25 min, 15 % A with a flow rate of 150 μL/min. The capillary of the ESI source was arranged for 275 °C, with sheath gas at 35 arbitrary units, auxiliary gas at 5 arbitrary units and the spray voltage at 4.0 kV. In positive/negative polarity switching mode, an m/z scan range from 60 to 900 was chosen and MS1 data was collected at a resolution of 70,000. The automatic gain control (AGC) target was set at 1 × 106 and the maximum injection time was 200 ms. The top 5 precursor ions were then fragmented, in a data-dependent manner, using the higher energy collisional dissociation (HCD) cell set to 30% normalized collision energy in MS2 at a resolution power of 17,500. Besides matching m/z, metabolites are identified by matching either retention time with analytical standards and/or MS2 fragmentation pattern. Data acquisition and analysis were carried out by Xcalibur 4.1 software and Tracefinder 4.1 software, respectively (both from Thermo Fisher Scientific).
Immunoblotting
Cells were lysed in ice-cold Triton lysis buffer (120 mM NaCl, 40 mM HEPES, pH 7.4, 1% Triton X-100, 1 mM EDTA, 10 mM sodium pyrophosphate, 10 mM glycerol 2-phosphate, 0.5 mM sodium orthovanadate and 50 mM NaF; 1 μM Microcystin-LR and protease inhibitor cocktail were added extemporaneously) and incubated on ice for 30 min. Lysates were centrifuged at 20,000 g for 15 min at 4°C. Protein concentrations were quantified using Bradford assay and protein lysates were normalized. Equal amounts of protein lysates (~10 to 20 μg) were prepared with the Laemmli sample buffer, incubated at 95°C on heat block for 5 min. Denaturated protein lysates were then separated on SDS-PAGE and transferred onto nitrocellulose blotting membranes. Membranes were blocked in 5 % non-fat milk in TBS tween (50 mM Tris pH 7.4, 150 mM NaCl, 0.1 % Tween 20) and incubated for 15 hours at 4°C with the indicated primary antibodies. Membranes were washed with TBST and incubated with horseradish peroxidase (HRP)-tagged anti-rabbit or anti-mouse secondary antibodies for one hour. Protein bands were developed with chemiluminescent substrates (Thermo fisher scientific # 34577, #34095). In all western blot experiments, β-actin was used as a loading control.
Measurement of 14C-bicarbonate, U-14C-glycine, L-3-14C-serine, 14C-formate, 3H-hypoxanthine and 3H-uridine incorporation into nucleic acids
80% confluent cells were serum starved for 15 hours, then treated and labeled as indicated in the figures. Cells were labeled with 2 μCi of either 14C-bicarbonate, U-14C-glycine, 3-14C-serine, 14C-formate, 3H-hypoxanthine, and 3H-uridine. RNA or DNA was purified using Allprep DNA/RNA kits according to the manufacturer’s instructions and quantified using a spectrophotometer. 30 μl of eluted RNA or 70 μl of eluted DNA were added to scintillation vials containing 3-mL of liquid scintillation emulsifier safe (PerkinElmer) and radioactivity was measured by liquid scintillation counting and normalized to the total RNA or DNA concentrations, respectively. All conditions were analyzed with biological triplicates and representative of at least three independent experiments.
mRNA expression analysis
Total cellular RNA was isolated (RNeasy kit, Qiagen, 74104) from the wild-type and ΔTSC2 HeLa cells grown in the indicated conditions and reverse-transcription (SuperScript II kit) was performed. The resulting cDNA was diluted in DNase free water (1:20) before quantification by real-time PCR. The transcript levels were normalized with the RPLP0 transcript level and data were represented as fold change relative to the average of control samples (2−ΔΔCT). Data are representative of at least two independent experiments.
Cell proliferation
Wild-type or SLC4A7 knockout cell lines (HEK-293E, HeLa, A549, and CAL-51) were seeded in 96-well plates at density of 3000 cells per well in media containing 1 % FBS (fetal bovine serum). ΔTSC2 HeLa cells were also transfected with nontargeted siRNA control (siCtl) or siRNA against SLC4A7 in media containing 0.5 % FBS. The CAL-51 cancer cell lines (wild-type or ΔSLC4A7) seeded in 96-well plates at density of 3000 cells per well in media containing 0.5 % dialyzed FBS treated with vehicle (DMSO) or rapamycin (10 nM). Cell proliferation was measured daily using crystal violet. The data were normalized to day 0. All cell proliferation assays were performed in biological triplicates seeded in technical quadruplicates for each condition.
Soft agar colony formation assay
Soft agar assays using wild-type or ΔSLC4A7 cancer cells (A549 and CAL-51) were performed. 5000 cells in 1.5 ml of growth media (DMEM with 10 % dFBS) containing 0.3 % agarose were seeded in 12-well plates with a bottom layer of 0.6 % agarose (1ml was prepared in same media as top layer). An additional 0.5 ml growth media was added over top layer, once a week. After 6 weeks, colonies were stained with 0.2 ml of 1 mg/ml of thiazolyl Blue Tetrazolium Bromide solution for 5 hours. Number of colonies was quantified using ImageJ software (Schneider et al., 2012) and represented as average of technical triplicates. Images shown in the manuscript (Figure 6E) are representative of biological triplicates from three independent experiments.
Bicarbonate, glucose, glycine, serine, formate and glutamine uptakes
Uptake was measured using a modification of the amino acid uptake protocol as previously described (Edinger and Thompson, 2002). Briefly, HeLa cells were cultured in serum-free DMEM for 15 hours and 50 μL of uptake medium containing 2 μCi of Na+, 14C-HCO3− or 1 μCi of specific activity (14C-glycine, 14C-serine, 14C-formate, 3H-2-deoxy-D-glucose, and 14C-glutamine) was added for 5 min. Cells take up the radiolabeled molecule for 5 min at 37°C. The uptake was stopped by washing the cells in ice cold PBS and lysing them in Triton 1 % buffer (40 mM HEPES, pH 7.4, 120 mM NaCl, 1 mM EDTA, 1 % Triton X-100, 10 mM glycerol 2-phosphate, 10 mM sodium pyrophosphate, 0.5 mM sodium orthovanadate and 50 mM NaF; protease inhibitor cocktail were added prior to the lysis of cells). Samples were centrifuged at 20,000 g and 4°C for 15 min. 75% of the supernatant was added to scintillation vials and radioactivity was measured by liquid scintillation counting (c.p.m) and normalized to the protein concentrations. Data points are presented as the mean of triplicate samples ± S.Ds.
Measurement of intracellular pH
Intracellular pH was measured using the Fluorometric Intracellular pH Assay Kit (Sigma-Aldrich, Cat# MAK150) according to the manufacturer’s instructions. Briefly, after 48 h siRNA transfection in 6 well plates, cells were further seeded overnight into 96-well plates (40,000 cells/well/100 μL) as quadruplicates in growth medium. Next day, growth medium was replaced with HHBS buffer (100 μl/well), subsequently Dye Loading Solution (BCFL-AM reagent in assay buffer) was added, and incubated the cells in a 5 % CO2, 37°C incubator for 30 min followed by incubation at room temperature for an additional 30 min. Fluorescence was measured at λex = 490 nm, λem = 535 nm using a Tecan Infinite M1000 Pro microplate reader. A calibration standard curve was prepared using pH calibration buffers (pH 4.5 to pH 7.5; increments of 1 pH units, Cat# P35379, Invitrogen/Molecular Probes) containing 10 μM of valinomycin and 10 μM nigericin. The calibration standard curve was used to determine the intracellular pH associated with the experimental conditions.
Polysome profiling assay
After treatment of 100 μg/mL cycloheximide (Sigma-Aldrich) for 2 min, HeLa cells (around 80–90% confluent) were washed with ice-cold PBS. After thoroughly aspirating the PBS, ‘in-dish lysis with flash-freezing’ method was performed. Briefly, the dish (10 cm) was quickly immersed in liquid nitrogen, and immediately moved on to dry ice. 500 μL of polysome lysis buffer (20 mM Tris.Cl [pH 7.5], 10 mM MgCl2, 150 mM KCl, 1 % (vol/vol) Triton X-100, 1 mM DTT, 100 μg/mL cycloheximide, 30 U/mL DNase I (Qiagen, Cat# 79254), 20 U/mL SuperaseIn (Ambion, Cat# AM2694) was added onto the frozen dish, and transferred the dish to the wet ice to be thawed. Lysates were centrifuged for 10 min at 20,000 g at 4°C. Cell lysates containing the same amount (200 μg) of RNA were loaded onto 10 % to 50 % sucrose gradient prepared by a BioComp Gradient Master. The samples were centrifuged at 35,000 rpm at 4°C in a SW41 rotor in a Beckman Coulter Optima XPN-100 ultracentrifuge for 3 h. Fractionation was done using a Brandel fractionation system equipped with a UA-6 UV detector. 200 μL of per fraction was collected and RNA extraction was performed using Phenol/chloroform extraction process or RNeasy kit (Qiagen). RNA was subsequently reverse transcribed using SuperScript II kit (Qiagen). mRNA levels in the resulting cDNA were analyzed by qPCR. Using the cycle threshold (CT) values, the percent (%) distribution for the mRNAs across the gradients was calculated.
Cell cycle analysis
Flow cytometric analysis of cell cycle distribution based on DNA content was performed as previously described (Tran et al., 2021). Briefly, after reaching 85 % of confluency, HeLa cells were exposed to DMEM in the presence or absence of HCO3− over a time-course. Regarding the SLC4A7 knockdown experiments, HeLa cells were treated with either siRNA targeting SLC4A7 or nontargeting controls for 48 h, and subsequently synchronized in G0/G1 phase with 1.5 % DMSO treatment overnight as previously described (Fiore et al., 2002). In the following morning, cells were released from DMSO for 6 or 12 h in fresh DMEM supplemented with 10 % dialyzed serum. Cells were collected, immediately fixed with ice-cold ethanol (100%), and subjected to 500 μl staining solution containing 0.1 % Triton X-100, 0.2 mg/mL RNAase A and 50 μg/mL propidium iodide (Sigma, P4170) in PBS. Fluorescence was measured using a BD Symphony A5 cytometer (BD Biosciences), and collected data were analyzed, percentage of cells in the cell cycle phases was quantified using the FlowJo software (BD Biosciences, version 10.7.1 for Windows).
Overall survival analysis
Human protein atlas (https://www.proteinatlas.org/) or Kaplan-Meier Plotter (https://kmplot.com/) collects publicly available gene and protein expression data from the National Cancer Institute GDC Data Portal (https://portal.gdc.cancer.gov/). Based on the Fragments Per Kilobase Million (FPKM) counts, patients were ranked into two groups to inspect the prognostic value of SLC4A7 mRNA expression. The prognosis of each group of patients was examined by Kaplan-Meier survival estimators, and the survival outcomes of the two groups were compared by log-rank tests. The two patient cohorts showing differential gene expression were compared by a Kaplan-Meier survival plot, and the hazard ratio with 95 % confidence intervals and logrank P values were calculated.
Folding free energy of 5′ UTR of specific mRNA
5’ untranslated region (5’ UTR) of specific mRNA was retrieved from UTRdb database (Grillo et al., 2010). The accession Refseq IDs used: ACTB (Refseq: NM_001101.5), SLC4A7 (Refseq: NM_003615.5), cMYC (Refseq: NM_002467.1), ODC1 (Refseq: NM_002539.3) and CDC25C (Refseq: NM_001790). Then, the minimum free energy and secondary structure of 5’ UTR of mRNAs were calculated by the RNAfold web server (Lorenz et al., 2011).
QUANTIFICATION AND STATISTICAL ANALYSIS
One-way ANOVA followed by Tukey’s post hoc tests were performed in GraphPad Prism 9.0 to determine differences between each group when more than two conditions were present. A two-tailed Student’s t tests were performed in Microsoft Excel 2016 for two pairwise comparisons. All error bars represent standard deviation (SD). A value of P < 0.05 was considered significant. The number of independent experiments done is described in each figure legend.
Supplementary Material
Table S1. LC-MS/MS Peak areas of the steady-state metabolite profiling from HeLa cells exposed or not to NaHCO3 starvation. Related to Figure 1.
Table S2. Genes coessential with the sodium bicarbonate transporter SLC4A7. Related to Figure 2.
Table S3. CERES dependency score for nucleotide synthesis and NaHCO3− transporter genes from genome-wide CRISPR-Cas9 essentiality screens across 342 cancer cell lines. Related to Figure 2.
Table S4. LC-MS/MS Peak areas for the SLC4A7 knockdown experiment. Related to Figure 2.
Highlights:
Cellular bicarbonate levels are limiting for nucleotide synthesis.
Insulin elevates SLC4A7 levels to import bicarbonate and promote nucleotide synthesis.
The mTORC1-S6K-eIF4B signaling axis controls SLC4A7 mRNA translation.
Loss of SLC4A7 sensitizes tumors to mTORC1 inhibition.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health, R01GM135587 R01GM143334 (I.B.-S.), R01GM121359, R01AI150986 (M.-N.F.Y.), F30CA264513, T32GM008152 (D.R.A.), and the LAM Foundation Established Investigator Award LAM0151E01-22 (I.B.-S.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no conflict of interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ali ES, Sahu U, Villa E, O’Hara BP, Gao P, Beaudet C, Wood AW, Asara JM, and Ben-Sahra I (2020). ERK2 Phosphorylates PFAS to Mediate Posttranslational Control of De Novo Purine Synthesis. Mol Cell 78, 1178–1191.e1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amici DR, Jackson JM, Truica MI, Smith RS, Abdulkadir SA, and Mendillo ML (2021). FIREWORKS: a bottom-up approach to integrative coessentiality network analysis. Life Sci Alliance 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrejeva G, and Rathmell JC (2017). Similarities and Distinctions of Cancer and Immune Metabolism in Inflammation and Tumors. Cell Metab 26, 49–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoi W, and Marunaka Y (2014). Importance of pH homeostasis in metabolic health and diseases: crucial role of membrane proton transport. Biomed Res Int 2014, 598986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Howell JJ, Asara JM, and Manning BD (2013). Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science (New York, N.Y.) 339, 1323–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, and Manning BD (2016). mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science (New York, N.Y.) 351, 728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, and Manning BD (2017). mTORC1 signaling and the metabolic control of cell growth. Curr Opin Cell Biol 45, 72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizuela L, Chrebet G, Bostian KA, and Parent SA (1991). Antifungal properties of the immunosuppressant FK-506: identification of an FK-506-responsive yeast gene distinct from FKB1. Mol Cell Biol 11, 4616–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo JL, Xu H, Mon-López D, Pareja-Galeano H, and Jiménez SL (2021). Effect of sodium bicarbonate contribution on energy metabolism during exercise: a systematic review and meta-analysis. J Int Soc Sports Nutr 18, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carsillo T, Astrinidis A, and Henske EP (2000). Mutations in the tuberous sclerosis complex gene TSC2 are a cause of sporadic pulmonary lymphangioleiomyomatosis. Proc Natl Acad Sci U S A 97, 6085–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassé H, Boulben S, Costache V, Cormier P, and Morales J (2017). Analysis of translation using polysome profiling. Nucleic Acids Res 45, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiche J, Brahimi-Horn MC, and Pouysségur J (2010). Tumour hypoxia induces a metabolic shift causing acidosis: a common feature in cancer. J Cell Mol Med 14, 771–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiche J, Ilc K, Laferrière J, Trottier E, Dayan F, Mazure NM, Brahimi-Horn MC, and Pouysségur J (2009). Hypoxia-inducible carbonic anhydrase IX and XII promote tumor cell growth by counteracting acidosis through the regulation of the intracellular pH. Cancer Res 69, 358–368. [DOI] [PubMed] [Google Scholar]
- Cordat E, and Casey JR (2009). Bicarbonate transport in cell physiology and disease. Biochem J 417, 423–439. [DOI] [PubMed] [Google Scholar]
- Csibi A, Lee G, Yoon SO, Tong H, Ilter D, Elia I, Fendt SM, Roberts TM, and Blenis J (2014). The mTORC1/S6K1 pathway regulates glutamine metabolism through the eIF4B-dependent control of c-Myc translation. Curr Biol 24, 2274–2280. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- DeBerardinis RJ, and Chandel NS (2016). Fundamentals of cancer metabolism. Sci Adv 2, e1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, and Thompson CB (2008). The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7, 11–20. [DOI] [PubMed] [Google Scholar]
- DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, et al. (2015). NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet 47, 1475–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Fernandez C, Martínez AI, Pekkala S, Barcelona B, Pérez-Arellano I, Guadalajara AM, Summar M, Cervera J, and Rubio V (2013). Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as a key tool. Hum Mutat 34, 1149–1159. [DOI] [PubMed] [Google Scholar]
- Ducker GS, and Rabinowitz JD (2017). One-Carbon Metabolism in Health and Disease. Cell Metab 25, 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger AL, and Thompson CB (2002). Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol Biol Cell 13, 2276–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effros RM, Chang RS, and Silverman P (1978). Acceleration of plasma bicarbonate conversion to carbon dioxide by pulmonary carbonic anhydrase. Science (New York, N.Y.) 199, 427–429. [DOI] [PubMed] [Google Scholar]
- Faubert B, Solmonson A, and DeBerardinis RJ (2020). Metabolic reprogramming and cancer progression. Science (New York, N.Y.) 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore M, Zanier R, and Degrassi F (2002). Reversible G(1) arrest by dimethyl sulfoxide as a new method to synchronize Chinese hamster cells. Mutagenesis 17, 419–424. [DOI] [PubMed] [Google Scholar]
- Foster KG, and Fingar DC (2010). Mammalian target of rapamycin (mTOR): conducting the cellular signaling symphony. J Biol Chem 285, 14071–14077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbatenko A, Olesen CW, Mørup N, Thiel G, Kallunki T, Valen E, and Pedersen SF (2014). ErbB2 upregulates the Na+,HCO3(−)-cotransporter NBCn1/SLC4A7 in human breast cancer cells via Akt, ERK, Src, and Kruppel-like factor 4. Faseb j 28, 350–363. [DOI] [PubMed] [Google Scholar]
- Grillo G, Turi A, Licciulli F, Mignone F, Liuni S, Banfi S, Gennarino VA, Horner DS, Pavesi G, Picardi E, et al. (2010). UTRdb and UTRsite (RELEASE 2010): a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res 38, D75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N (2011). Akt isoforms and glucose homeostasis - the leptin connection. Trends Endocrinol Metab 22, 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai K, Minato S, Kaneko S, Yanai K, Ishii H, Kitano T, Shindo M, Miyazawa H, Ito K, Ueda Y, et al. (2019). Approximation of bicarbonate concentration using serum total carbon dioxide concentration in patients with non-dialysis chronic kidney disease. Kidney Res Clin Pract 38, 326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, and Blenis J (2005). mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 123, 569–580. [DOI] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, and Blenis J (2021). mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 184, 2255. [DOI] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. (2011). The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science (New York, N.Y.) 332, 1317–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, and Manning BD (2008). The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J 412, 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies HB, Fumagalli S, Dennis PB, Reinhard C, Pearson RB, and Thomas G (1997). Rapamycin suppresses 5’TOP mRNA translation through inhibition of p70s6k. Embo j 16, 3693–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Rajabi H, Rodrigo CM, Porco JA Jr., and Kufe D (2013). Targeting the eIF4A RNA helicase blocks translation of the MUC1-C oncoprotein. Oncogene 32, 2179–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitrapakdee S, Walker ME, and Wallace JC (1999). Functional expression, purification, and characterization of recombinant human pyruvate carboxylase. Biochem Biophys Res Commun 266, 512–517. [DOI] [PubMed] [Google Scholar]
- Kivelä AJ, Kivelä J, Saarnio J, and Parkkila S (2005). Carbonic anhydrases in normal gastrointestinal tract and gastrointestinal tumours. World J Gastroenterol 11, 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane AN, and Fan TW (2015). Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res 43, 2466–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane AN, Yan J, and Fan TW (2015). (13)C Tracer Studies of Metabolism in Mouse Tumor Xenografts. Bio Protoc 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K, Das R, and Barna M (2018). Functional 5’ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol 19, 158–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levraut J, Giunti C, Ciebiera JP, de Sousa G, Ramhani R, Payan P, and Grimaud D (2001). Initial effect of sodium bicarbonate on intracellular pH depends on the extracellular nonbicarbonate buffering capacity. Crit Care Med 29, 1033–1039. [DOI] [PubMed] [Google Scholar]
- Liberti MV, and Locasale JW (2016). The Warburg Effect: How Does it Benefit Cancer Cells? Trends in biochemical sciences 41, 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Qin X, Wang DK, Guo YM, Gill HS, Morris N, Parker MD, Chen LM, and Boron WF (2013). Effects of optional structural elements, including two alternative amino termini and a new splicing cassette IV, on the function of the sodium-bicarbonate cotransporter NBCn1 (SLC4A7). J Physiol 591, 4983–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yang J, and Chen LM (2015). Structure and Function of SLC4 Family [Formula: see text] Transporters. Front Physiol 6, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, and Hofacker IL (2011). ViennaRNA Package 2.0. Algorithms Mol Biol 6, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mboge MY, Mahon BP, McKenna R, and Frost SC (2018). Carbonic Anhydrases: Role in pH Control and Cancer. Metabolites 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, et al. (2017). Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet 49, 1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira V, Patra KC, and Hay N (2018). Selective eradication of cancer displaying hyperactive Akt by exploiting the metabolic consequences of Akt activation. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco JM, Krawczyk PA, Scaria SM, Cangelosi AL, Chan SH, Kunchok T, Lewis CA, and Sabatini DM (2020). Dihydroxyacetone phosphate signals glucose availability to mTORC1. Nat Metab 2, 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe L, van den Elzen AMG, Watson MJ, and Thoreen CC (2020). Global analysis of LARP1 translation targets reveals tunable and dynamic features of 5’ TOP motifs. Proc Natl Acad Sci U S A 117, 5319–5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez C, Hauser AD, Vucic EA, and Bar-Sagi D (2019). Plasma membrane V-ATPase controls oncogenic RAS-induced macropinocytosis. Nature 576, 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nature protocols 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero MF, Chen AP, Parker MD, and Boron WF (2013). The SLC4 family of bicarbonate (HCO₃−) transporters. Mol Aspects Med 34, 159–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, and Sabatini DM (2017). mTOR Signaling in Growth, Metabolism, and Disease. Cell 168, 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlyarov V, Eichner R, Girardi E, Essletzbichler P, Goldmann U, Nunes-Hasler P, Srndic I, Moskovskich A, Heinz LX, Kartnig F, et al. (2018). The Bicarbonate Transporter SLC4A7 Plays a Key Role in Macrophage Phagosome Acidification. Cell Host Microbe 23, 766–774.e765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian D, Parsyan A, Petroulakis E, Hershey J, and Sonenberg N (2010). eIF4B controls survival and proliferation and is regulated by proto-oncogenic signaling pathways. Cell Cycle 9, 4106–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SM, and Carrey EA (1992). Regulation of the mammalian carbamoyl-phosphate synthetase II by effectors and phosphorylation. Altered affinity for ATP and magnesium ions measured using the ammonia-dependent part reaction. Eur J Biochem 207, 957–965. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, and Sabatini DM (2012). A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toft NJ, Axelsen TV, Pedersen HL, Mele M, Burton M, Balling E, Johansen T, Thomassen M, Christiansen PM, and Boedtkjer E (2021). Acid-base transporters and pH dynamics in human breast carcinomas predict proliferative activity, metastasis, and survival. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrence ME, MacArthur MR, Hosios AM, Valvezan AJ, Asara JM, Mitchell JR, and Manning BD (2021). The mTORC1-mediated activation of ATF4 promotes protein and glutathione synthesis downstream of growth signals. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran DH, Kesavan R, Rion H, Soflaee MH, Solmonson A, Bezwada D, Vu HS, Cai F, Phillips JA 3rd, DeBerardinis RJ, et al. (2021). Mitochondrial NADP(+) is essential for proline biosynthesis during cell growth. Nat Metab 3, 571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya M, Yoshikawa H, Itakura M, and Yamashita K (1990). Increased de novo purine synthesis by insulin through selective enzyme induction in primary cultured rat hepatocytes. Am J Physiol 258, C841–848. [DOI] [PubMed] [Google Scholar]
- Valvezan AJ, and Manning BD (2019). Molecular logic of mTORC1 signalling as a metabolic rheostat. Nat Metab 1, 321–333. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, and DeBerardinis RJ (2017). Understanding the Intersections between Metabolism and Cancer Biology. Cell 168, 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa E, Ali ES, Sahu U, and Ben-Sahra I (2019). Cancer Cells Tune the Signaling Pathways to Empower de Novo Synthesis of Nucleotides. Cancers (Basel) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa E, Sahu U, O’Hara BP, Ali ES, Helmin KA, Asara JM, Gao P, Singer BD, and Ben-Sahra I (2021). mTORC1 stimulates cell growth through SAM synthesis and m(6)A mRNA-dependent control of protein synthesis. Mol Cell 81, 2076–2093.e2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron JA, Tack DC, Ritchey LE, Gillen SL, Wilczynska A, Turro E, Bevilacqua PC, Assmann SM, Bushell M, and Le Quesne J (2019). mRNA structural elements immediately upstream of the start codon dictate dependence upon eIF4A helicase activity. Genome Biol 20, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, Chen WW, Lander ES, and Sabatini DM (2017). Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell 168, 890–903.e815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KA, Grillo-Hill BK, and Barber DL (2017). Cancer cell behaviors mediated by dysregulated pH dynamics at a glance. J Cell Sci 130, 663–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Kremer DM, Huang H, Breitkopf SB, Ben-Sahra I, Manning BD, Lyssiotis CA, and Asara JM (2019). Ex vivo and in vivo stable isotope labelling of central carbon metabolism and related pathways with analysis by LC-MS/MS. Nature protocols 14, 313–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. LC-MS/MS Peak areas of the steady-state metabolite profiling from HeLa cells exposed or not to NaHCO3 starvation. Related to Figure 1.
Table S2. Genes coessential with the sodium bicarbonate transporter SLC4A7. Related to Figure 2.
Table S3. CERES dependency score for nucleotide synthesis and NaHCO3− transporter genes from genome-wide CRISPR-Cas9 essentiality screens across 342 cancer cell lines. Related to Figure 2.
Table S4. LC-MS/MS Peak areas for the SLC4A7 knockdown experiment. Related to Figure 2.
Data Availability Statement
Original western blots have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table. Mass spectrometry data are presented in the table S1.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-S6K1 | Cell Signaling Technology | Cat#2708; RRID: AB_390722 |
| Rabbit monoclonal anti-pS6 (S240/S244) | Cell Signaling Technology | Cat#5364; RRID: AB_10694233 |
| Rabbit Polyclonal anti-SLC4A7 | Invitrogen (Thermo Fisher) | Cat#PA5-75807; RRID:AB_2719535 |
| Band 3/ AE1 Polyclonal antibody (SLC4A1) | ProteinTech Group, Inc | Cat#28131-1-AP |
| AE2 Polyclonal antibody (SLC4A2) | ProteinTech Group, Inc | Cat#26332-1-AP |
| SLC4A3 Polyclonal antibody | ProteinTech Group, Inc | Cat#26883-1-AP |
| SLC4A4 Polyclonal antibody | ProteinTech Group, Inc | Cat#11885-1-AP |
| Rabbit Polyclonal anti-SLC4A5 Antibody | Novus Biologicals | Cat#NBP1-59528 |
| Rabbit Polyclonal anti-SLC4A8 | ProteinTech Group, Inc | Cat#12531-1-AP |
| Rabbit AE4 Polyclonal Antibody (SLC4A9) | Mybiosci | Cat#MBS821050 |
| SLC4A10 Rabbit pAb | Abclonal Technology | Cat#A13810 |
| Rabbit Polyclonal anti-SLC4A11 | BiorByt | Cat#orb215316 |
| Phospho-p70 S6 Kinase (Thr389) (108D2) Rabbit mAb | Cell Signaling Technology | Cat# 9234S; RRID:AB_2269803 |
| Rabbit Anti-p70 S6 Kinase Monoclonal Antibody | Cell Signaling Technology | Cat# 2708S; RRID:AB_390722 |
| Phospho-S6 Ribosomal Protein (Ser240/244) Antibody | Cell Signaling Technology | Cat# 2215S; RRID:AB_331682 |
| S6 Ribosomal Protein (5G10) Rabbit mAb | Cell Signaling Technology | Cat# 2217S; RRID:AB_331355 |
| Phospho-Akt (Ser473) (D9E) Rabbit mAb | Cell Signaling Technology | Cat# 11962; RRID:AB_2797780 |
| Akt (pan) (C67E7) Rabbit mAb | Cell Signaling Technology | Cat# 4691, RRID:AB_915783 |
| 4E-BP1 Antibody | Cell Signaling Technology | Cat# 9452S; RRID:AB_331692 |
| Tuberin/TSC2 (D93F12) XP_ Rabbit mAb | Cell Signaling Technology | Cat# 4308S; RRID:AB_10547134 |
| Anti-mouse IgG, HRP-linked Antibody | Cell Signaling Technology | Cat#7076; RRID:AB_330924 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling Technology | Cat#7074; RRID:AB_2099233 |
| β-actin Antibody | Sigma-Aldrich | Cat#A5316; RRID: AB_476743 |
| Phospho-CAD (Ser1859) Antibody | Cell Signaling Technology | Cat#12662; RRID:AB_2750934 |
| CAD Antibody | Cell Signaling Technology | Cat#11933; RRID:AB_2797772 |
| S6K2 Polyclonal antibody | ProteinTech Group, Inc | Cat#15268-1-AP; RRID:AB_2182250 |
| Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (E10) Mouse mAb antibody | Cell Signaling Technology | Cat# 9106S; RRID: AB_331768 |
| p44/42 MAPK (Erk1/2) Antibody | Cell Signaling Technology | Cat# 9102S; RRID: AB_330744 |
| eIF4B Antibody | Cell Signaling Technology | Cat# 3592; AB_2293388 |
| eIF4A (C32B4) Rabbit mAb | Cell Signaling Technology | Cat#2013; RRID:AB_2097363 |
| DYKDDDDK Tag (D6W5B) Rabbit mAb | Cell Signaling Technology | Cat#14793; RRID:AB_2572291 |
| HA-Tag (C29F4) Rabbit mAb | Cell Signaling Technology | Cat#3724; RRID:AB_10693385 |
| Phospho-eIF4B (S422) Antibody | Cell Signaling Technology | Cat#3591; RRID:AB_2097522 |
| Phospho-eIF4B (Ser406) Antibody | Cell Signaling Technology | Cat#5399; RRID:AB_10695246 |
| eIF4A Antibody | Cell Signaling Technology | Cat#2490; AB_2097363 |
| H2AX Antibody | Thermo Fisher (product of: Bethyl) | Cat#A300-083A-M |
| Bacterial and virus strains | ||
| NEB Stable Competent E. coli | NEB | Cat#C3040 |
| DH5 alpha | Fisher Scientific | Cat#18258012 |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM without Bicarbonate | US Biological | Cat#D9812-07 |
| DMEM without glucose, bicarbonate | US Biological | Cat#D9800-02B |
| DMEM without glycine (and serine) | US Biological | Cat#D9800-16 |
| Rapamycin | Calbiochem/Sigma-Aldrich | Cat#553210 |
| Cycloheximide | EMD Millipore/Merck | Cat#239764 |
| Glycine | Sigma-Aldrich | Cat#G5417 |
| PF-9366 | MedChemExpress | Cat#HY-107778 |
| Insulin human | Alpha diagnostic | Cat#INSL16-N-5 |
| EGF (human recombinant) | Life Technologies/GIBCO | Cat#PHG0311 |
| Insulin-like Growth Factor-I human | Sigma-Aldrich | Cat#I3769 |
| Hypoxanthine | Sigma-Aldrich | Cat#H9636 |
| Inosine | Sigma-Aldrich | Cat#I4125 |
| 13C2-15N-glycine | Sigma-Aldrich | Cat#489522 |
| L-glutamine-(amide-15N) | Sigma-Aldrich | Cat#490024 |
| 1,2-3H-2-deoxy-D-Glucose | Perkin-Elmer | Cat#NET328A250UC |
| U-14C-glycine | Perkin-Elmer | Cat#NEC276E050UC |
| U-14C-glucose | Perkin-Elmer | Cat#NEC042V250UC |
| 3H-hypoxanthine | Perkin-Elmer | Cat#NET177001MC |
| 14C-glutamine | Perkin Elmer | Cat#NEC451050UC |
| 14C-formate | American Radiolabeled Chemicals | Cat#ARC 0163A |
| 3-14C-serine | Perkin-Elmer | Cat#NEC853050UC |
| 3H-uridine | Perkin Elmer | Cat#NET367250UC |
| 4-13C aspartate | Sigma-Aldrich | Cat#489999 |
| 14C-aspartate | Perkin Elmer | Cat#NEC268E050UC |
| Sodium, 14C- bicarbonate | American Radiolabeled Chemicals | Cat#ARC 0138C-250 μCi |
| DMSO | Sigma-Aldrich | Cat#D2650-100ML |
| L-glutamine | Corning | Cat#MT25005Cl |
| 2-mercaptoethanol | Sigma-Aldrich | Cat#M6250 |
| Thiazolyl Blue Tetrazolium Bromide | Sigma-Aldrich | Cat#M5655 |
| Polyethylenimine (PEI) | Polysciences | Cat#23966-2 |
| Opti-MEM media | Thermo Fisher | Cat#31985062 |
| Lipofectamine 3000 Transfection reagent | Invitrogen | Cat#L3000008 |
| Lipofectamine RNAi max | Invitrogen | Cat#13778150 |
| SuperSignal West Femto Chemiluminescent | Thermo Fisher scientific | Cat#PI34096 |
| SuperSignal West Pico PLUS | Thermo Fisher scientific | Cat#PI34580 |
| Fetal bovine serum (FBS) | Sigma-Aldrich | Cat#F2442 |
| Fetal Bovine Serum dialyzed | Sigma-Aldrich | Cat#F0392 |
| Microcystin-LR | Fisher Scientific | Cat#ALX350012C500 |
| Protease inhibitor cocktail | Sigma-Aldrich | Cat#P8340 |
| Bio-Rad Braford | Bio-Rad | Cat#5000006 |
| Acrylamide Protogel 30% | Fisher Scientific | Cat#EC-890 |
| UltraPure LMP Agarose | Invitrogen | Cat#16520-050 |
| Uridine | Sigma Aldrich | Cat#U3003 |
| Glutamine-free DMEM | GIBCO | Cat#11960-044 |
| Glucose-free DMEM | Invitrogen | Cat#11966025 |
| Propidium iodide | Sigma | Cat#P4170 |
| Critical commercial assays | ||
| Fluorometric Intracellular pH Assay Kit | Sigma-Aldrich | Cat# MAK150 |
| Intracellular pH Calibration Buffer Kit | Invitrogen | Cat# P35379 |
| SYBR Green PCR Master Mix | Life Technologies | Cat# 4312704 |
| RNeasy Mini Kit | QIAGEN | Cat#74104 |
| HiSpeed Plasmid Maxi Kit (25) | QIAGEN | Cat#12663 |
| AllPrep DNA/RNA Mini Kit | QIAGEN | Cat#80204 |
| KOD Extreme Hot Start DNA polymerase kit | Merck Millipore | Cat#71975-3 |
| GeneJET Plasmid Miniprep Kit | Thermo Fisher | Cat# K0503 |
| Deposited data | ||
| Ali et al. unprocessed files | Mendeley Data | http://dx.doi.org/10.17632/9zd4vncrf4.1 |
| Experimental models: Cell lines | ||
| HEK293E | John Blenis lab (Weill Cornell Medicine) | N/A |
| ΔTSC2 HEK293E | Villa et al. 2021 | N/A |
| HeLa | ATCC | Cat#CCL-2 |
| ΔTSC2 HeLa | Villa et al. 2021 | N/A |
| A549 | ATCC | Cat#CCL-185 |
| CAL51 | DSMZ | Cat#ACC 302 |
| AML 621-102 (TSC2-null-Empty Vector) | Dr. David Kwiatkowski (Brigham and Women’s Hospital) | N/A |
| Experimental models: Organisms/strains | ||
| Athymic nude Foxn1nu | The Jackson laboratory | Cat#002019 |
| Oligonucleotides | ||
| See Table S4 for list of primers used in this study | Integrated DNA Technologies | N/A |
| siRNA targeting human SLC4A7 | Dharmacon | Cat#L-007586-01-0005 |
| siRNA targeting human S6K1 | Dharmacon | Cat#L-003616-00-0005 |
| siRNA targeting human S6K2 | Dharmacon | Cat#L-004671 -00-0005 |
| siRNA targeting human eIF4A | Dharmacon | Cat#L-020178-00-0005 |
| siRNA targeting human eIF4B | Dharmacon | Cat#L-020179-00-0005 |
| Non-targeting siRNA pool | Dharmacon | Cat#D-001810-10-20 |
| Recombinant DNA | ||
| SLC4A7 (NM_003615) Human Tagged ORF Clone | OriGene | Cat#RC220734 |
| EIF4B (NM_001417) Human Tagged ORF Clone | OriGene | Cat#RC209955 |
| pcDNA3 Flag HA | Addgene | Cat#10792 |
| pCMV6-Entry Tagged Cloning Vector | OriGene | Cat#PS100001 |
| PX458 CRISPR | Addgene | Cat#48138) |
| pRK7-HA-S6K1-WT | Addgene | Cat#8984 |
| pRK7/HA-S6K1-F5A-E389-R3A | Addgene | Cat#8991 |
| pCMV6/Flag-EIF4B-S422A | This study | N/A |
| pCMV6/Flag-EIF4B-S406A | This study | N/A |
| Software and algorithms | ||
| Image J | National Institutes of Health | RRID: SCR_003070 |
| Prism 9 | GraphPad Software | https://www.graphpad.com/ |
| FlowJo Software | BD Biosciences | https://www.flowjo.com/ |
| Other | ||
| Microplate Reader | Tecan | Infinite M1000 Pro |
| Multipurpose Scintillation Counter | Beckman Coulter | Model: LS 6500 |
| Nitrocellulose membrane 0.2 μm | GE Healthcare | Cat# 10600001 |
| Polypropylene centrifuge tubes | Beckman Coulter | Cat# 331372 |
| PCR plates | Bio-Rad | Cat#HSP9601 |
| UV transparent flat bottom 96 well plate | Corning | Cat#3635 |


