Highlights
-
•
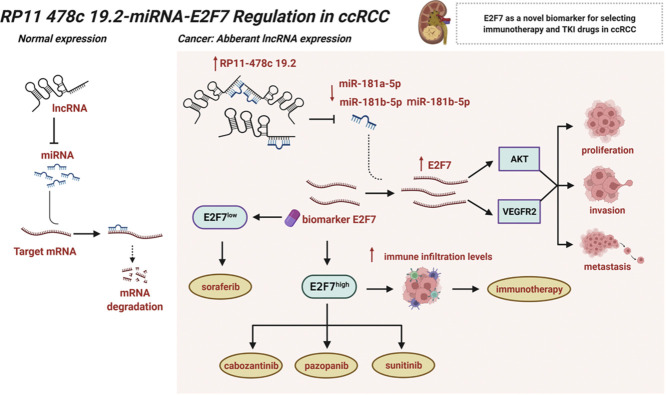
LncRNA RP11–478C19.2 promotes the progression of ccRCC via the RP11–478C19.2/hsa-miR-181b-5p, hsa-miR-181a-5p, hsa-miR-181c-5p/E2F7 axis.
-
•
E2F7 could be detected in patients with ccRCC to aid treatment regimen selection, particularly in advanced and metastatic disease.
-
•
Individuals who overexpress E2F7 may have a greater response to some first-line therapy for ccRCC, including cabozantinib, pazopanib, and sunitinib.
-
•
Patients with higher expression of E2F7 may have a higher percentage of ICB responses.
Keywords: Competing endogenous RNAs, E2F7, Immune infiltration analysis, ccRCC, Treatment decision, Pan-cancer analysis
Abstract
Clear cell renal cell carcinoma (ccRCC), accounting for 70–80% of all renal cell carcinomas, is a common malignancy. Survival rates decrease significantly in patients with advanced and metastatic ccRCC. Furthermore, ccRCC is less responsive to radiation and chemotherapy than other cancers. Therefore, targeted therapy and immunotherapy are particularly important for ccRCC management. A growing body of literature recognizes that competitive endogenous RNA (ceRNA) regulatory networks play a crucial role in various cancers. However, the biological functions of the ceRNA network in ccRCC require further investigation. In this study, we built the ceRNA network for ccRCC using the “GDCRNATools” package. After survival analysis, the RP11–478C19.2/hsa-miR-181b-5p, hsa-miR-181a-5p, and hsa-miR-181c-5p/E2F7 axes were obtained for further analysis. Unsupervised clustering was conducted basing on this ceRNA network. The results indicated that the prognosis and immune infiltration levels differed between the two clusters. Furthermore, we conducted correlation analysis, immune infiltration analysis, tumor mutation burden analysis, GSEA analysis, drug sensitivity analysis and pan-cancer analysis of E2F7 to explore its potential role in oncogenesis. Experiments in vitro were performed to confirm the pro-oncogenic impact of E2F7. The results suggest that the RP11–478C19.2/E2F7 axis might be a biomarker for the inclusion of cabozantinib, pazopanib, sunitinib, and immunotherapy in the therapeutic regimen. In summary, we found that the ceRNA-based RP11–478C19.2/E2F7 axis is involved in ccRCC and that it could be a novel biomarker for treatment decisions and a possible therapeutic target to increase the success of targeted therapy and immunotherapy in ccRCC.
Graphical abstract
Introduction
Kidney cancer is a common type of cancer, accounting for 431,288 new cases and 179,368 deaths globally in 2020 [1]. Among kidney malignancies, 90% are renal cell carcinoma (RCC). Among these, clear cell renal cell carcinoma (ccRCC) is the most prevalent subtype, accounting for approximately 70% to 80% of RCC cases, and its incidence has grown over the past years [2,3]. Despite breakthrough achievements in surgical operation, chemotherapy, radiotherapy, targeted therapy, and immunological therapy in the last few years, ccRCC patients still suffer from unsatisfactory survival outcomes, especially those with advanced pathology and clinical stage, or distant metastasis [4], [5], [6]. Compared to other cancers, ccRCC is less responsive to radiation and chemotherapy. Therefore, targeted therapy and immunotherapy are particularly important in the treatment of ccRCC [4,7]. Therefore, it is vital to formulate personalized strategies. Both the characteristics of carcinoma and the condition of patients should be taken into consideration when formulating a comprehensive scheme.
Immunotherapies, particularly immune checkpoint blockade (ICB), have revolutionized cancer therapy in recent years [8]. Removing immune suppression by ICB results in a remarkable clinical response in certain patients with ccRCC, and the combination of ICB with additional anticancer agents is currently the first-line treatment for advanced ccRCC patients [9]. However, a significant number of patients are ineligible for checkpoint blockers. Therefore, reliable biomarkers are critical for the rapid adjustment of therapy regimens. Additionally, identifying target sites that may decrease immunotherapeutic resistance and improve patient response to immunotherapy has the potential to revolutionize ccRCC treatment [10,11].
Long non-coding RNAs (lncRNAs) are transcripts with a length of more than 200 bases and a limited capacity to code for proteins. Numerous studies have shown that lncRNAs may play crucial roles in cancer development, often resulting in dysregulation of gene products that promote tumor growth [3,[12], [13], [14]]. Additionally, lncRNAs are used as diagnostic or prognostic indicators of poor prognosis, tumor recurrence, and metastasis. MicroRNAs (miRNAs) are encoded by endogenous genes. They regulate gene expression by targeting the 3′ untranslated region (UTR) of the target gene mRNA. Recent evidence suggests that most miRNAs participate in tumor development. However, the biological function and expression pattern of miRNAs in ccRCC are still unclear, and the effects of miRNAs on the progression and metastasis of tumors and their mechanisms are unknown [15,16]. Given the critical involvement of miRNAs and lncRNAs in tumor growth, the theory that they function a competitive endogenous RNAs (ceRNAs) was introduced in 2011 [17]. According to this theory, lncRNAs, miRNAs, and other RNAs can regulate each other functionally. Normally, all types of RNAs in the ceRNA network are in a dynamic balance [18]. However, an imbalance in this network could lead to disease occurrence. A ceRNA network has been constructed in prostate cancer [19], lung adenocarcinoma [20] and breast cancer [21]. However, ceRNA networks that play vital roles in ccRCC progression and treatment require further research.
In this study, we constructed a ceRNA network for ccRCC using the “GDCRNATools” package. This network included RP11–478C19.2/hsa-miR-181b-5p, hsa-miR-181a-5p, and hsa-miR-181c-5p/E2F7. Correlation analysis, immune infiltration analysis, tumor mutation burden analysis, GSEA analysis, drug sensitivity analysis and pan-cancer analysis were performed to explore the possible involvement of E2F7 in oncogenesis and cancer treatment.
Materials and methods
Data collection and preprocessing
The Cancer Genome Atlas (TCGA) database was used to retrieve transcriptome data (read counts), including mRNA, lncRNA, and miRNA expression data for KIRC patients, as well as associated clinical information and somatic mutation data (Table S1). Additionally, the Gene Expression Omnibus (GEO) database was queried for GSE36895 [[22], [23]], GSE40435 [24], GSE53757 [25]. R 4.0.4 was used to standardize the raw data.
Screening of differentially expressed genes
We identified differentially expressed mRNAs (DEmRNAs), DEmiRNAs, and DElncRNAs in data from TCGA database using the “edgeR” package [26]. DEmRNAs and DElncRNAs were chosen based on |logFC| >1 and FDR 〈 0.05, and the DEmiRNAs were obtained based on |logFC| 〉 0.3 and FDR < 0.05. DEmRNAs were identified in the GEO database by using the “limma” package [27] with |logFC| >1 and FDR < 0.05. Intersection of DEmRNAs from TCGA and three GEO series was taken for further analysis.
Construction of the ceRNA network
ceRNA networks were constructed based on the identified DEmRNAs, DEmiRNAs, and DElncRNAs. The “GDCRNATools” package [28] was applied to predict miRNA–mRNA and miRNA–lncRNA pairs using the hypergeometric test, Pearson correlation analysis and regulation pattern analysis. Three criteria were used to identify competing endogenous interactions: (1) the lncRNA and mRNA must share a significant number of miRNAs, (2) the expression of lncRNA and mRNA should be positively correlated, and (3) the common miRNAs play similar roles in regulating the expression of lncRNAs and mRNAs. Furthermore, we visualized the ceRNA network using Cytoscape software [29].
Survival analysis
Univariate Cox regression analysis was used to identify the mRNAs, miRNAs, and lncRNAs associated with overall survival. Thereafter, we selected networks in which the mRNAs, miRNAs, and lncRNAs were all related to prognosis for further analysis. ceRNA networks with high correlations between lncRNAs and mRNAs were investigated. Considering that mRNAs serve important roles in ceRNA networks, lncRNAs and miRNAs may regulate mRNA expression to exert biological effects. We evaluated the association between these mRNAs in the networks with patient clinical characteristics and immune infiltration levels. Additionally, the target sites in miRbase and LncBase Predicted v.2 were used to predict the potential target sites of miRNA-mRNA and miRNA-lncRNA.
Unsupervised consensus clustering based on the ceRNA network
The K-means algorithm in the “ConsensusClusterplus” R package [30] was employed to perform unsupervised consensus clustering based on ceRNA networks. ssGSEA was conducted to assess the degrees of infiltration of immune cell types between the two subclasses using the R package “GSVA” [31]. Immune and stromal scores were calculated using the R package “ESTIMATE” [32]. Furthermore, the expression analysis of immune checkpoint genes and survival analysis were carried out to compare the characteristics of the two clusters. Differential expression analysis was performed using |logFC| > 1 and p < 0.05. Additionally, GO and KEGG enrichment analyses were carried out by the R package “clusterProfiler” [33] with FDR < 0.05 and q value < 0.05.
Expression analysis and survival analysis of E2F7 in different stages
To explore whether E2F7 expression was affected by clinical features, we investigated E2F7 expression in tumors and paired normal tissues. Other clinical indicators, such as pathological stage, histological grade, and metastatic status, were also evaluated for their association with E2F7 expression. Additionally, GSEA analysis was performed to discover the differences in signaling pathways between the E2F7high and E2F7low groups to reveal the possible mechanism.
Correlation between immune infiltration, tumor mutation burden and E2F7 expression in KIRC
TIMER [34] was applied to investigate the association between E2F7 expression and immune -cell infiltration. Moreover, the association between immune checkpoint gene expression and the ceRNA network was analyzed to determine the potential effect of the ceRNA network on immunotherapy. Tumor mutation burden was estimated to determine the response to immune checkpoint blockade therapy. Finally, we evaluated the relationship between E2F7 expression and patient responses to ICB in an immunotherapy cohort (GSE67501 [35]).
Drug sensitivity analysis
RNA-seq and DTP NCI-60 data were downloaded from CellMiner, and drugs authorized by the FDA or currently being examined in clinical trials were selected. We calculated the correlation between E2F7 expression and drug sensitivity, and a p value < 0.05 was considered statistically significant. Additionally, transcriptional data of tumor cell lines and IC50 values for anticancer medicines from the GDSC database were utilized to accomplish drug sensitivity analysis using the "pRRophetic" package [36].
Pan-cancer analysis of E2F7
We conducted a pan-cancer investigation of E2F7 to elucidate its potential implications in various malignancies. TIMER was used to visualize the differential expression of E2F7 in 33 cancer types, and RNA-seq data from 33 cancer types in TCGA database were downloaded to determine the correlation between immune cell infiltration, immune checkpoint genes, and E2F7 expression. Additionally, univariate Cox regression analysis was used to determine whether there was a possible correlation between E2F7 expression and OS and PFS in the 33 cancer types. Finally, a correlation analysis was performed between tumor mutation burden (TMB), microsatellite instability (MSI), and E2F7 expression in the 33 cancer types.
Cell lines and reagents
Human renal cell carcinoma cell lines (786-O and OSRC-2) were acquired from the Shanghai Cell Bank Type Culture Collection Committee (Shanghai, China) and incubated in RPMI1640 medium containing 10% fetal bovine serum (FBS). Sunitinib, pazopanib, and cabozantinib were purchased from Selleck Chemicals Corporation (Shanghai, China) and were dissolved in DMSO.
SiRNA transfection
siRNA and the equivalent negative control were manufactured by RiboBio Company (Guangzhou, China). OSRC-2 and 786-O cells were transfected with 80 nM E2F7 or control siRNA using Lipofectamine 3000 (Thermo Fisher Scientific, Shanghai, China). The knockdown effectiveness was confirmed at the mRNA and protein levels.
RNA extraction and qRT-PCR
The cells were harvested 48 h after siRNA transfection. RNA was extracted and reverse transcribed into cDNA. qRT-PCR analysis was conducted using the SYBR Green Master Mix procedure in a Step One PCR system (Applied Biosystems, Foster, CA, USA), with GAPDH serving as the endogenous control for mRNA expression normalization. E2F7 upstream primer: GCAGTGGTTGTTTCTGTCAGG, downstream primer: TCTCTTAGTAGGACCACCAACG; CD274 upstream primer: TGCAGGGCATTCCAGAAAGA, downstream primer: TAGGTCCTTGGGAACCGTGA; GAPDH upstream primer: CGCTCTCTGCTCCTCCTGTTC, downstream primer: ATCCGTTGACTCCGACCTTCAC.
Western blot
Total protein was isolated on ice using RIPA buffer containing protease and phosphatase inhibitors. Equal quantities of the extracted proteins (20 ug) were separated using SDS-PAGE electrophoresis, transferred to PVDF membranes, and blocked for 90 min with 5% BSA at room temperature. The membranes were then incubated with primary rabbit antibodies overnight at 4 °C (anti-E2F7, 1:800, Proteintech, Wuhan, China; anti-AKT, 1:1000, Servicebio, Wuhan, China; anti-phospho AKTser473, 1:800, Boster, Wuhan, China; anti-VEGFR2, 1:1000, ABclonal, Wuhan, China; anti-phospho VEGFR2s473, 1:500, ABclonal; GAPDH, 1:1000, Boster). After incubating the membrane with the appropriate secondary antibodies for 90 min, the protein bands were visualized using a Pierce ECL substrate WB detection system (Bio-Rad Laboratories, Shanghai, China).
CCK-8 viability assay
Cell viability and cytotoxicity assays were carried out using a Cell Counting Kit-8 (Boster). Transfected cells were seeded at a density of 3 × 103 cells per well in 96-well plates. After 0, 24, 48, and 72 h of incubation, each well received a 10 µl CCK-8 solution and was incubated for 2 h at 37 °C. OD values at 450 nm were recorded using a microplate reader (Thermo Fisher Scientific, Rochester, NY, USA).
Wound healing assay
Transfected cells were seeded at a density of 2.0 × 105 cells/well in 6-well plates. Three parallel wound lines were scratched on the cell monolayer after reaching 90% confluency. The relative width of the wound at 0 and 24 h was measured using the ImageJ software.
Colony formation assay
Transfected cells were seeded at a density of 1000 cells per well in 6-well plates and incubated for two weeks. The cells were fixed with 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet for 30 min. The number of cell colonies was calculated and analyzed using ImageJ software.
Transwell assay for migration and invasion
Transfected cells were collected and resuspended in serum-free media before being placed in the upper chamber of 24-well Transwell plates (8 µm; Costar, Corning Inc. Corning, NY, USA) with or without Matrigel (BD Biosciences, San Jose, CA, USA). The lower chamber was filled with medium containing 10% FBS. After 24 h at 37 °C, the cells on the upper membrane surface were fixed for 30 min with 4% paraformaldehyde and stained with 0.1% crystal violet for 60 min. Photographs were captured at × 200 magnification, and the cells were counted in five randomly selected areas.
Flow cytometry
Apoptotic cell death was assessed using annexin V/PI staining (Yeasen, Shanghai, China). Transfected cells were fixed in 75% ethanol at 4 °C for 12 h and stained with propidium iodide (PI) (Yeasen) solution for cell cycle analysis. The proportion of cells in each cell cycle phase was analyzed using a CytoFlex cytometer (Beckman Coulter, USA). All data were analyzed using the FlowJo V10 software.
Statistical analysis
RStudio 4.0.4 and GraphPad 8.0 were used for all analyses. Spearman correlation analysis was used to determine correlations. For two-group comparisons, Student's t-test and Wilcoxon test were used. For multiple groups, the Kruskal–Wallis test and one-way ANOVA were employed. Statistical significance was set at p < 0.05.
Results
Identification of significant DEmRNAs, DEmiRNAs, and DElncRNAs in KIRC
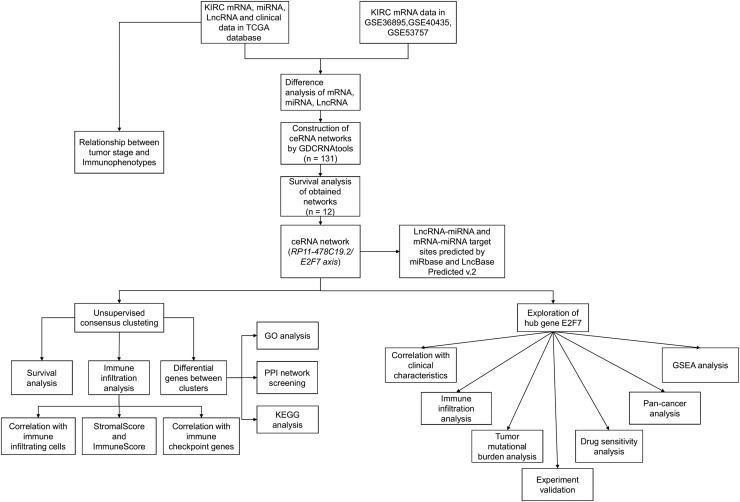
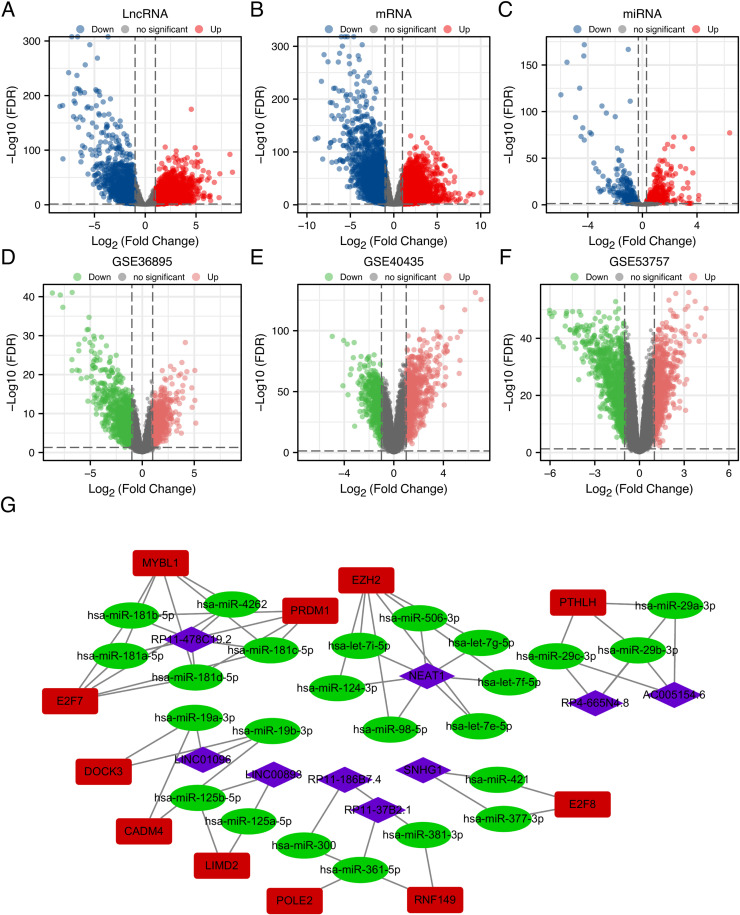
Fig. 1 shows the roadmap for this study. A total of 5811 DEmRNAs, 683 DEmiRNAs, and 5735 DElncRNAs were identified (Fig. 2A-C). Moreover, we identified 1625, 1076 and 1690 DEmRNAs in GSE36895, GSE40435 and GSE53757, respectively. (Fig. 2D-F). We intersected DEmRNAs identified from TCGA database and these three GEO series. 1714 DEmRNAs were obtained for further analysis.
Fig. 1.
Flow diagram of the construction and analysis of the ceRNA network.
Fig. 2.
Volcano plots of DElncRNAs, DEmiRNAs, DEmRNAs, and construction of ceRNA network: (A–C) The volcano plots of TCGA data show (A) 5735 DElncRNAs (|logFC| > 1, FDR < 0.05), (B) 683 DEmiRNAs (|logFC| > 0.3, FDR < 0.05), and (C) 5811 DEmRNAs (|logFC| > 1, FDR< 0.05). (D-F) Volcano plots of data from three GEO series (|logFC| > 1, FDR< 0.05) showing (D) 1625 DEmRNAs in GSE36895, (E) 1076 DEmRNAs in GSE40435, and (F) 1690 DEmRNAs in GSE53757. (G) Construction of ceRNA network. Purple diamonds represent lncRNAs. Green ellipses represent miRNAs. Red rectangles represent mRNAs.
Construction of the ceRNA network and survival analysis
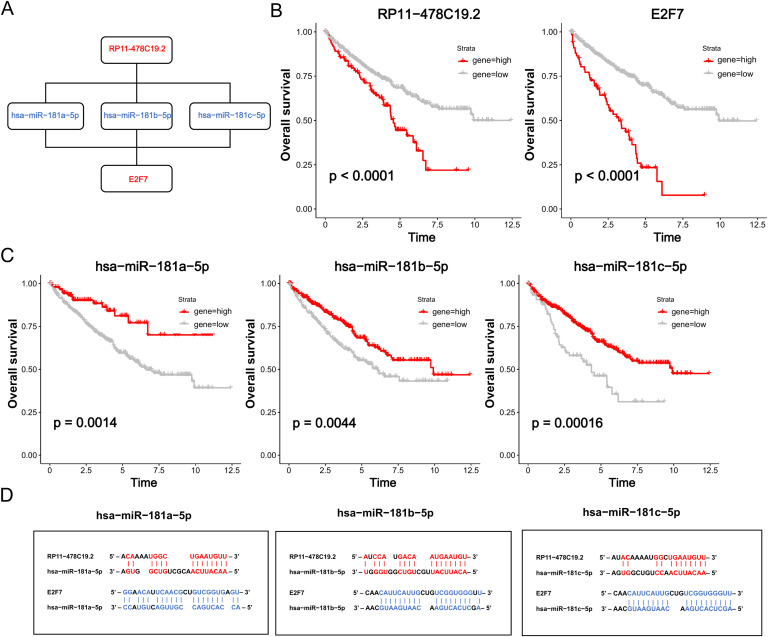
We constructed the ceRNA networks based on identified DEmRNAs, DEmiRNAs, and DElncRNAs. In total, 131 ceRNA networks, 118 miRNA nodes, 103 mRNA nodes, and 31 lncRNA nodes were identified as differentially expressed profiles (Table S2). Furthermore, survival analysis revealed associations between 16 miRNAs (Fig. S1), 51 mRNAs (Fig. S2), and 19 lncRNAs (Fig. S3), and ccRCC prognosis. As shown in Fig. 2G, 12 ceRNA networks were obtained after survival analysis (Table S3). Among them, RP11–478C19.2/hsa-miR-181b-5p, hsa-miR-181a-5p, and hsa-miR-181c-5p/E2F7 (Fig. 3A) showed a high correlation between mRNA and lncRNA and have not been previously reported in ccRCC. Therefore, it was selected for further analysis.
Fig. 3.
Overall survival analysis of the ceRNA network. (A) Schematic model of ceRNA. (B) Expression of AC005154.6 and E2F7 was compared by a Kaplan–Meier survival curve. (C) Expression of hsa-miR-181b-5p, hsa-miR-181a-5p, and hsa-miR-181c-5p was compared by a Kaplan–Meier curve. (D) Potential target sites of miRNA-mRNA (blue) and miRNA-lncRNA (red) predicted by miRbase and LncBase Predicted v.2.
Survival analysis revealed that high expression of RP11–478C19.2 and E2F7 were risk factors for ccRCC patients, while hsa-miR-181b-5p, hsa-miR-181a-5p, and hsa-miR-181c-5p were protective factors (Fig. 3B-C), indicating that RP11–478C19.2 may act as a ceRNA to upregulate the expression of E2F7 by sponging hsa-miR-181b-5p, hsa-miR-181a-5p, and hsa-miR-181c-5p. Finally, the target sites in the RP11–478C19.2 and E2F7 3′UTRs were predicted to pair with hsa-miR-181b-5p, hsa-miR-181a-5p, and hsa-miR-181c-5p using miRbase and LncBase Predicted v.2 (Fig. 3D).
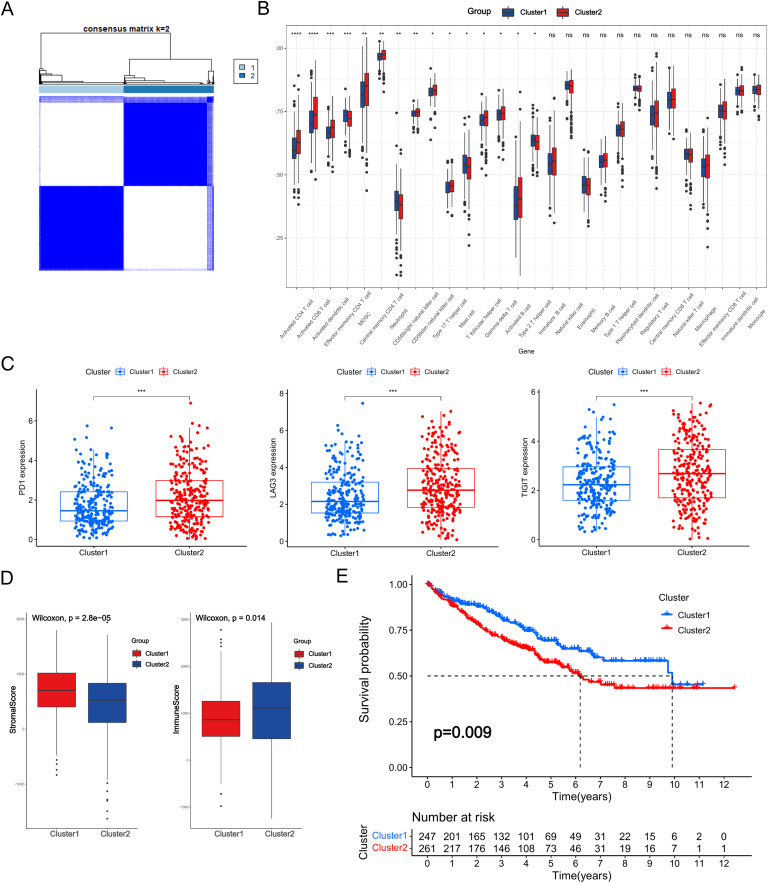
Unsupervised consensus clustering based on the ceRNA network
Unsupervised consensus clustering was performed on the basis of the ceRNA network. Since the boundary between the heatmaps of the consistency matrix remained clear when k = 2, it was chosen as the optimal cluster number (Fig. 4A, Table S4). Immune cell infiltration was evaluated between the two subclasses and a substantial difference in the degree of infiltration of the majority of immune cells was observed (Fig. 4B). Moreover, the expression of immune checkpoint genes such as PD-1, LAG3, and TIGIT was distinct between the two subclasses (Fig. 4C). Additionally, the results of the ESTIMATE algorithm indicated that differences existed in the tumor microenvironment of the two clusters (Fig. 4D). Furthermore, Kaplan–Meier analysis indicated that cluster2 had a statistically significant survival advantage over cluster1 (Fig. 4E).
Fig. 4.
Unsupervised consensus clustering based on the ceRNA network. (A) Heatmap of k-means clustering based on the ceRNA network. (B) Proportions of TME cells in the two clusters. (C) Expression of immune checkpoint genes in the two subgroups. (D) Immune and stromal scores of the two subgroups. (E) Kaplan–Meier survival curve of OS for KIRC patients in the two subgroups. (ns: not significant, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001).
Although the two clusters, based on the ceRNA network, performed differently in terms of the characteristics of the tumor microenvironment, the roles of each gene in the ceRNA network were not clear. Therefore, we estimated the correlation between each gene in the ceRNA network and TME-infiltrating cells, and the results revealed that most immune cells were upregulated in patients with high E2F7 expression (Fig. S4).
To explore potential genetic changes in the two clusters, 1639 differentially expressed genes were identified. GO and KEGG enrichment analyses revealed the biological roles of differentially expressed genes (Fig. S5). Biological processes were mainly enriched in functions of the immune system, such as complement activation, humoral immune response, and regulation of humoral immune response. KEGG pathway analysis revealed that the differentially expressed genes were mainly involved in pathways such as retinol metabolism and steroid hormone biosynthesis.
Pro-oncogenic role of E2F7 in KIRC
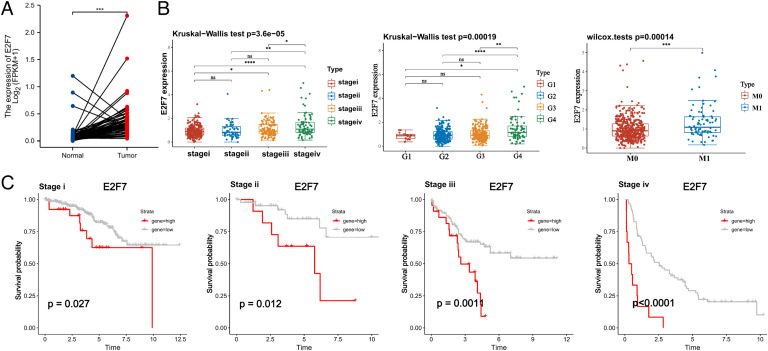
To gain a better understanding of the role of the RP11–478C19.2/E2F7 axis in KIRC, we analyzed E2F7 in detail. First, we discovered that E2F7 expression was higher in tumor samples than in normal tissues (Fig. 5A). E2F7 expression was abnormally elevated as malignancies progressed. Patients with advanced pathological stage, pathological grade, and lymph node metastasis overexpressed E2F7 (Fig. 5B). Correlations between clinical features and E2F7 expression were analyzed, and the results indicated that increased E2F7 expression was associated with T stage (p = 0.008), N stage (p = 0.013), M stage (p = 0.007), pathological stage (p = 0.013), and histological grade (p = 0.015) (Table 1). Additionally, as the pathological stage progressed, E2F7 seemed to play an increasingly crucial role in patient survival (Fig. 5C). Moreover, GSEA was carried out using samples with high and low expression levels of E2F7 to uncover the enriched pathways (Fig. S6). The upregulation of E2F7 was significantly involved in pathways such as the PI3K−Akt, JAK−STAT, and TGF−beta signaling pathways, which are considered to contribute to tumorigenesis. Additionally, the upregulation of E2F7 was significantly associated with pathways such as ECM-receptor interaction, natural killer cell-mediated cytotoxicity, and the T cell receptor signaling pathway, indicating its potential key roles in the immune response of ccRCC patients.
Fig. 5.
Differential expression and survival analysis of E2F7 (A) Expression of E2F7 in tumor samples and paired normal tissues. (B) Expression of E2F7 in different pathologic stages, histologic grades, and metastasis statuses. (C) Kaplan–Meier survival curve of OS for KIRC patients in different pathologic stages (ns: not significant, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001).
Table 1.
Correlations between E2F7 expression and clinicopathological parameters.
| Characteristic | Low expression of E2F7 | High expression of E2F7 | p |
|---|---|---|---|
| n | 269 | 270 | |
| Age, n (%) | 0.322 | ||
| <=60 | 128 (23.7%) | 141 (26.2%) | |
| >60 | 141 (26.2%) | 129 (23.9%) | |
| Race, n (%) | 0.235 | ||
| Asian | 3 (0.6%) | 5 (0.9%) | |
| Black or African American | 34 (6.4%) | 23 (4.3%) | |
| White | 229 (43%) | 238 (44.7%) | |
| Gender, n (%) | 0.164 | ||
| Female | 101 (18.7%) | 85 (15.8%) | |
| Male | 168 (31.2%) | 185 (34.3%) | |
| T stage, n (%) | 0.008 | ||
| T1 | 151 (28%) | 127 (23.6%) | |
| T2 | 40 (7.4%) | 31 (5.8%) | |
| T3 | 76 (14.1%) | 103 (19.1%) | |
| T4 | 2 (0.4%) | 9 (1.7%) | |
| N stage, n (%) | 0.013 | ||
| N0 | 115 (44.7%) | 126 (49%) | |
| N1 | 2 (0.8%) | 14 (5.4%) | |
| M stage, n (%) | 0.007 | ||
| M0 | 222 (43.9%) | 206 (40.7%) | |
| M1 | 27 (5.3%) | 51 (10.1%) | |
| Pathologic stage, n (%) | 0.013 | ||
| Stage I | 149 (27.8%) | 123 (22.9%) | |
| Stage II | 33 (6.2%) | 26 (4.9%) | |
| Stage III | 58 (10.8%) | 65 (12.1%) | |
| Stage IV | 29 (5.4%) | 53 (9.9%) | |
| Histologic grade, n (%) | 0.015 | ||
| G1 | 9 (1.7%) | 5 (0.9%) | |
| G2 | 125 (23.5%) | 110 (20.7%) | |
| G3 | 105 (19.8%) | 102 (19.2%) | |
| G4 | 25 (4.7%) | 50 (9.4%) | |
| OS event, n (%) | 0.003 | ||
| Alive | 199 (36.9%) | 167 (31%) | |
| Dead | 70 (13%) | 103 (19.1%) | |
| DSS event, n (%) | < 0.001 | ||
| Alive | 226 (42.8%) | 194 (36.7%) | |
| Dead | 35 (6.6%) | 73 (13.8%) | |
| PFI event, n (%) | < 0.001 | ||
| Alive | 207 (38.4%) | 171 (31.7%) | |
| Dead | 62 (11.5%) | 99 (18.4%) |
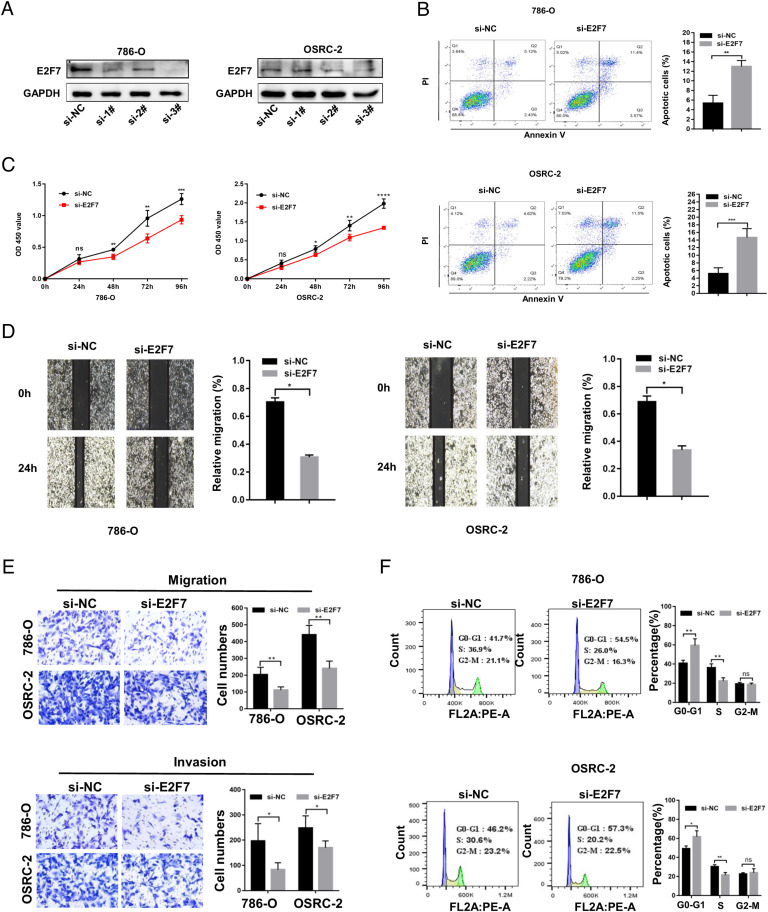
To validate the cancer-promoting role of E2F7 in ccRCC, we downregulated E2F7 in OSRC-2 and 786-O cells by siRNA transfection and verified the downregulation of E2F7 using qRT-PCR (Fig. S7) and western blotting (Fig. 6A). These results indicated that siRNA-3 had a higher efficiency, and thus, it was used in subsequent experiments. The CCK-8 assay demonstrated that silencing E2F7 greatly reduced OSRC-2 and 786-O cell growth (Fig. 6C), which was also validated by colony formation assay (Fig. S8). Additionally, cell apoptosis was evaluated and knockdown of E2F7 noticeably enhanced apoptosis in cancer cells (Fig. 6B).
Fig. 6.
The cancer-promoting role of E2F7 in ccRCC (A) Validation of the knockdown effect of E2F7 via western blotting. (B) Effect of E2F7 knockdown on apoptosis in OSRC-2 and 786-O cells. (C) Proliferation of OSRC-2 and 786-O cells detected by CCK8 assay. (D) Scratch assay in OSRC-2 and 786-O cells. (E) Migration and invasion ability of OSRC-2 and 786-O cells measured via transwell assay. (F) Cell cycle estimated by flow cytometry in OSRC-2 and 786-O cells.
To investigate the effect of E2F7 silencing on cancer cell migration and invasion, wound healing and transwell assays were performed. Consistent with prior findings, the knockdown of E2F7 decreased the migration and invasion abilities of OSRC-2 and 786-O cells (Fig. 6D-E). Considering that E2F7 belongs to the E2F family, which correlates strongly with the cell cycle [37], [38], [39], we performed cell cycle analysis using flow cytometry. In OSRC-2 and 786-O cells, downregulation of E2F7 expression led to G1/S phase arrest, the percentage of cells in G1 phase significantly increased, and the percentage of cells in S phase decreased (Fig. 6F).
Correlation between immune infiltration, tumor mutation burden, and E2F7 expression
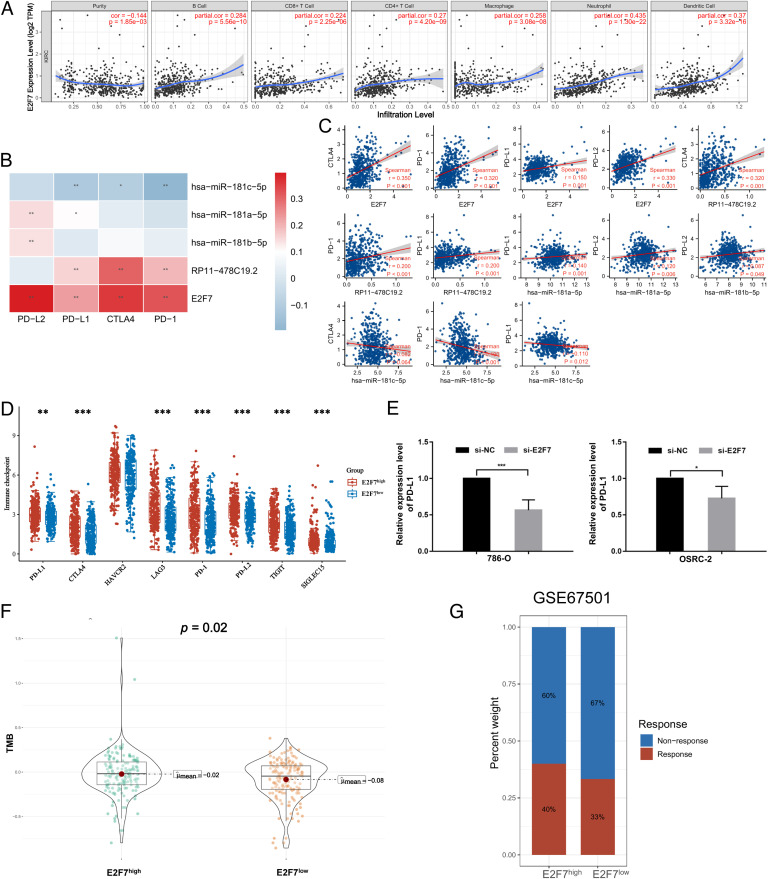
According to the TIMER results, the expression of E2F7 was significantly associated with tumor purity and positively related to B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells (Fig. 7A). Furthermore, the correlation between immune checkpoint genes and the ceRNA network was evaluated (Fig. 7B-C), and PD-1, CTLA4, LAG3, PD-L1, PD-L2, TIGIT, and SIGLEC15 were closely associated with E2F7 (Fig. 7D). The differential expression of PD-L1 between the si-E2F7 and si-NC groups was also validated in OSRC-2 and 786-O cells. As shown in Fig. 7E, PD-L1 expression was significantly decreased after E2F7 knockdown.
Fig. 7.
Association between immune cell infiltration, tumor mutation burden, and E2F7 expression. (A) Association between E2F7 expression and immune infiltration levels in KIRC. (B-D) Association between the expression of RP11–478C19.2, hsa-miR-181b-5p, hsa-miR-181a-5p, hsa-miR-181c-5p, and E2F7 and immune checkpoint gene expression. (E) Relative expression of PD-L1 between the si-E2F7 and si-NC groups in OSRC-2 and 786-O cells. (F) Comparison of tumor mutation burden between the E2F7high and E2F7low groups. (G) Preliminary validation in GSE67501 (* p < 0.05, ** p < 0.01, *** p < 0.001).
Additionally, we observed that TMB was significantly higher in the E2F7high group than in the E2F7low group (Fig. 7F), suggesting the potential role of E2F7 as a biomarker for predicting the clinical response to immunotherapy. Finally, we performed preliminary validation in an immunotherapy cohort (GSE67501), which provided the response to nivolumab in patients with renal cell carcinoma. The results indicated that patients in the E2F7high group had a higher percentage of ICB response (Fig. 7G).
As E2F7 was upregulated in patients with a high pathological stage, we evaluated whether immune infiltration levels were also distinct at different stages. We found differences in the expression of most immune cells, and CTLA4, LAG3, PD-1, and TIGIT were significantly associated with the tumor stage (Fig. S9).
Drug sensitivity analysis
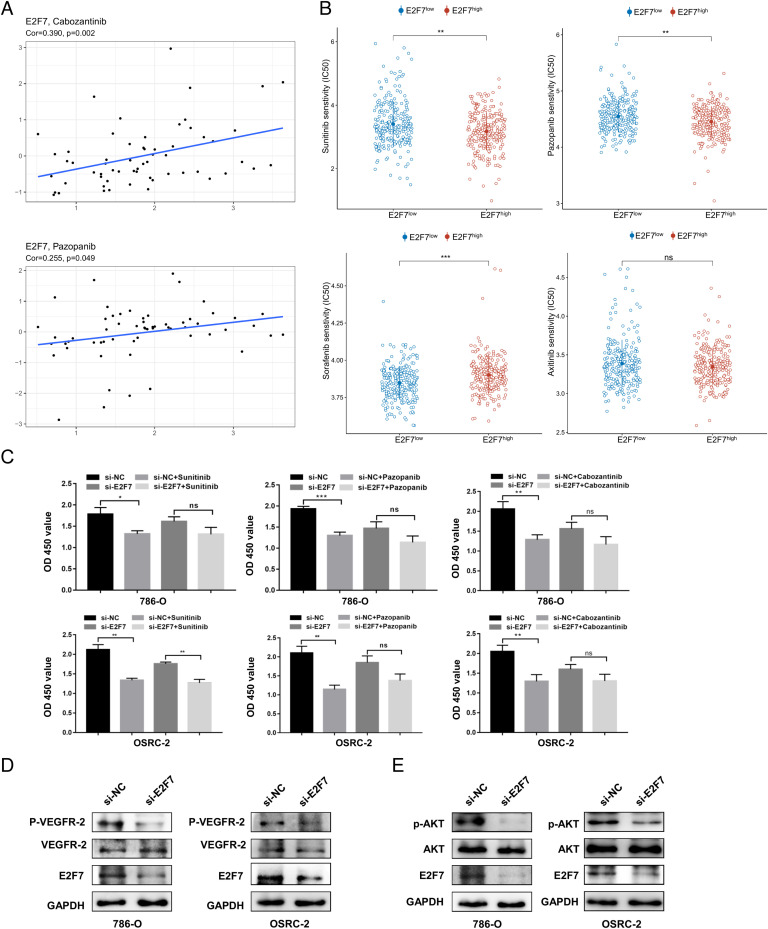
To evaluate the potential effects of E2F7 on drug response, we first estimated the correlation between drug sensitivity in CellMiner and the expression of E2F7. All results are presented in Table S5, and the top nine drugs used in the treatment of cancer are shown in Fig. S10. According to the results, patients with high E2F7 expression might have a better response to drugs such as cabozantinib and pazopanib (Fig. 8A), which are widely chosen as a treatment regimen for ccRCC patients in clinical practice. We observed that most of these drugs targeted the PI3K/AKT1/mTOR pathway, suggesting that E2F7 might regulate tumor progression through this mechanism. Therefore, we evaluated the correlation between E2F7 expression and this pathway. We discovered that the expression of E2F7 was positively correlated with PI3KCA, AKT1, and MTOR (Fig. S11), and the western blot results also suggested that E2F7 knockdown decreased the expression of phosphorylated AKT, which might result in the inhibition of PI3K/AKT1/mTOR signaling pathway activation (Fig. 8E).
Fig. 8.
Drug sensitivity analysis (A) Correlation between sensitivity to cabozantinib and pazopanib and the expression of E2F7. (B) Predicted IC50 values of sorafenib, sunitinib, pazopanib, and axitinib between the E2F7high and E2F7low groups. (C) Effectiveness of sunitinib, pazopanib, and cabozantinib in the si-E2F7 and si-NC groups of OSRC-2 and 786-O cells. Detection of p-VEGFR2 and VEGFR2 expression (D) and p-AKT and AKT expression (E) in OSRC-2 and 786-O cells after E2F7 knockdown.
Given that the data in the CellMiner database did not include all the first-line targeted therapies, we used the GDSC database to evaluate the association between the drug sensitivity of sorafenib, sunitinib, pazopanib, and axitinib and E2F7 expression. These results indicated that the estimated IC50 of axitinib was not significantly different between the E2F7high and E2F7low groups. However, substantial differences were detected regarding sorafenib, sunitinib, and pazopanib (Fig. 8B). Patients in the E2F7high group showed a higher sensitivity to sunitinib and pazopanib but seemed to be insensitive to sorafenib.
We also performed CCK8 assays to validate the effect of E2F7 on patient responsiveness to these targeted therapies. The viability of cancer cells was tested at multiple drug concentrations (Fig. S12). In both, the OSRC-2 and 786-O cell lines, the efficacy of cabozantinib and pazopanib was reduced after the knockdown of E2F7 (Fig. 8C). However, this effect was only observed in the 786-O cells treated with sunitinib. Downregulation of E2F7 did not influence the sensitivity of OSRC-2 cells to sunitinib (Fig. 8C). Considering that all these drugs target the VEGF signaling pathway, we also examined whether E2F7 might affect VEGF signaling pathway activation in ccRCC cells. The western blot results suggested that the knockdown of E2F7 reduced the expression of phosphorylated VEGFR2, indicating that E2F7 may influence drug sensitivity through the VEGF signaling pathway (Fig. 8D).
Pan-cancer analysis of E2F7
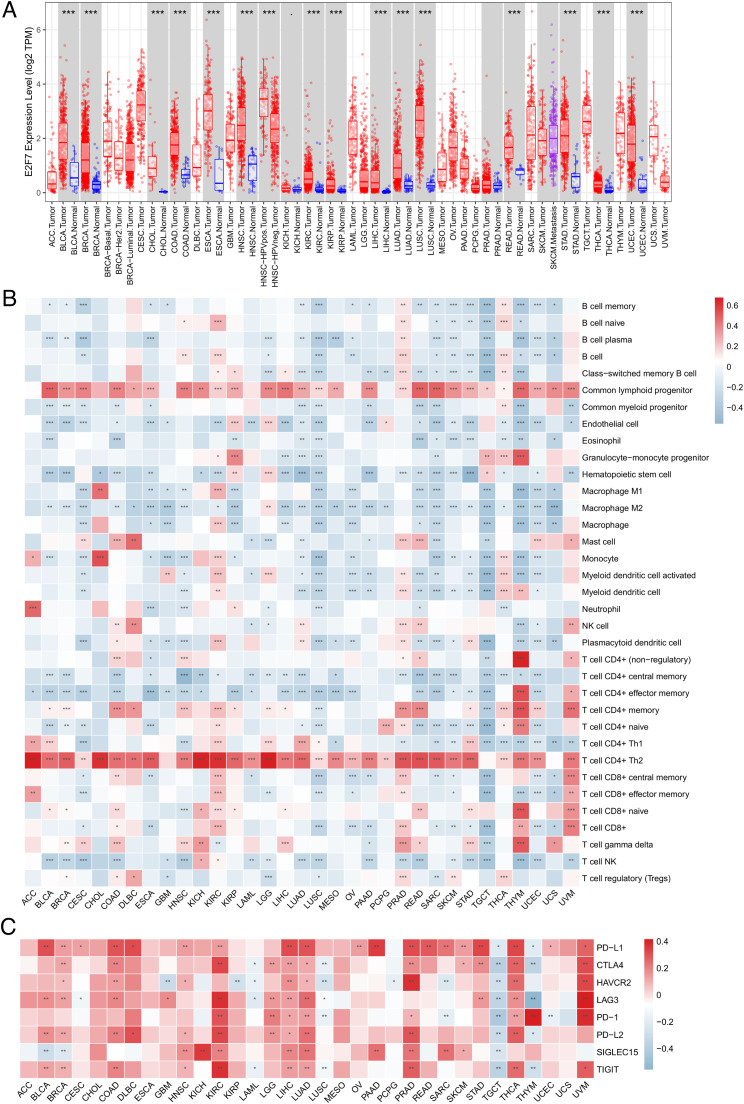
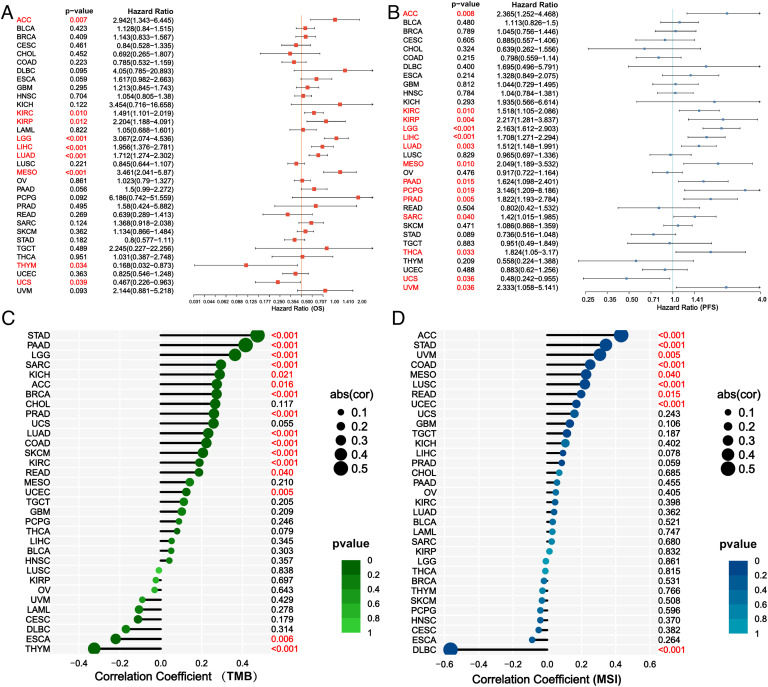
Differential expression was observed in BLCA, BRCA, CHOL, COAD, ESCA, HNSC, KIRC, KIRP, LIHC, LUAD, LUSC, READ, STAD, THCA, and UCEC (Fig. 9A). Furthermore, correlations between E2F7 expression and immune cell infiltration levels and immune checkpoint genes in 33 cancer types were observed (Fig. 9B-C). E2F7 showed a strong association with different immune cells and immune checkpoint genes, indicating that E2F7 may play vital roles in the tumor microenvironment and act as a target site for the enhancement of immunotherapy. Furthermore, the results of the survival analysis revealed that E2F7 expression was related to the OS of ACC, KIRC, KIRP, LGG, LIHC, LUAD, MESO, THYM, and UCS patients, as well as the PFS of ACC, KIRC, KIRP, LGG, LIHC, LUAD, MESO, PAAD, PCPG, PRAD, SARC, THCA, UCS, and UVM patients (Fig. 10A-B). Moreover, the expression of E2F7 was associated with TMB in STAD, PAAD, LGG, SARC, KICH, ACC, BRCA, PRAD, LUAD, COAD, SKCM, KIRC, READ, UCEC, ESCA, and THYM, as well as with MSI in ACC, STAD, UVM, COAD, MESO, LUSC, READ, UCEC, and DLBC (Fig. 10C-D).
Fig. 9.
Pan-cancer analysis of the association of E2F7 with immune infiltration levels (A) Expression of E2F7 in 33 cancer types. (B) Association between E2F7 expression and the immune infiltration levels in 33 cancer types. (C) Association between E2F7 expression and immune checkpoint gene expression in 33 cancer types. (* p < 0.05, ** p < 0.01, *** p < 0.001).
Fig. 10.
Association between E2F7 expression and OS (A), PFS (B), TMB (C), and MSI (D) in 33 cancer types.
Discussion
ccRCC is the most common type of kidney cancer. Owing to the lack of sensitive diagnostic biomarkers and limitations in effective screening detection, nearly 25–30% of patients exhibit unresectable tumor or distant metastasis upon diagnosis [40]. Although localized ccRCC can be successfully cured by surgery, poor survival is manifested in metastatic ccRCC, which is insensitive to conventional chemotherapy [41]. In the last few years, targeted medicines, such as sunitinib, bevacizumab, and pazopanib, which block VEGF and VEGFR, have revolutionized the treatment of metastatic ccRCC. Additionally, drugs such as everolimus, which inhibits mTOR complex 1, have been adopted in clinical practice. Since 2015, agents other than VEGFR inhibitors have been approved, including cabozantinib and lenvatinib [41].
Recently, extensive research has shown that the immune microenvironment formed by tumor immune cells can regulate cancer progression. Researchers have elucidated that infiltrating CD4+ T cells can affect ccRCC cell proliferation by regulating the TGFβ1/YBX1/HIF2α pathway [42]. In recent years, immune checkpoint blockade has been the most promising strategy for cancer immunotherapy [10,11,43]. Numerous studies have confirmed that PD-1 and PD-L1 are expressed on activated T cells, and inhibiting PD-1 and its ligand expression can enhance T cell function, and thus, inhibit tumor growth [44,45]. Immunotherapies, such as nivolumab, have already been added to the treatment regimens for metastatic ccRCC [41]. However, in the context of the extreme biological and genetic heterogeneity of the tumor, metastatic or advanced RCC patients still suffer from poor survival, owing to the lack of effective treatment options that can generate durable responses [46,47]. Therefore, well-validated predictive biomarkers with high specificity and sensitivity for advanced or distant metastatic ccRCC have become increasingly urgent [11].
Recently, ceRNAs have been discovered as critical regulators of tumor progression in gastric cancer [48], colorectal cancer [49], breast cancer [21] and pancreatic cancer [50]. To the best of our knowledge, similar studies, especially on the roles of ceRNAs in treatment decisions, are currently lacking in ccRCC. In the present study, we established a ceRNA network (RP11–478C19.2/hsa-miR-181b-5p, hsa-miR-181a-5p, and hsa-miR-181c-5p/E2F7) in ccRCC. Furthermore, correlation analysis, immune infiltration analysis, tumor mutation burden analysis, GSEA analysis, drug sensitivity analysis and pan-cancer analysis were performed to elucidate the potential regulatory mechanisms and explore the possibility of E2F7 as a novel biomarker for treatment decisions in ccRCC.
Among the miRNAs identified, hsa-miR-181a-5p has been reported to act as a tumor suppressor in various malignancies. In acute myeloid leukemia, increased expression of hsa-miR-181a-5p was associated with a higher incidence of complete remission, longer overall survival, and disease-free survival [51,52]. E2F7 is a member of the E2F transcription factor family. According to current research, E2F7 is implicated in tumor angiogenesis and enhances tumor growth and metastasis in hepatocellular carcinoma [53]. Moreover, E2F7 has been identified as a target of miR-424–5p, and E2F7 overexpression increased the activity of the VEGFR-2 pathway [54]. E2F7 functions as a tumor promoter in various malignancies, including gallbladder cancer [55], prostate cancer [56] and colon cancer [57]. This was also validated by our pan-cancer analysis results.
In the current study, E2F7 expression was remarkably higher in ccRCC tissues than that in normal tissues. The oncogenic role of E2F7 was validated using multiple in vitro assays. We observed that E2F7 was abnormally overexpressed during tumor progression. The expression of E2F7 seems to be upregulated in patients with a higher pathological stage, pathological grade, and lymph node metastasis. Moreover, elevated E2F7 expression was associated with poor prognosis, particularly in individuals diagnosed with stage III or stage IV cancer. All these findings showed that E2F7 may be critical in determining the prognosis of patients with advanced and metastatic ccRCC who may have already missed the opportunity for surgical resection.
Systemic therapy, specifically targeted therapy and immunotherapy, is critical in patients with advanced or metastatic ccRCC. According to the European Association of Urology guidelines, VEGF-targeted agents such as sunitinib [58], pazopanib [59] or bevacizumab (combined with IFN) [60] constitute the standard of care for patients with good-risk ccRCC. For intermediate-risk patients, the new first-line therapy is a combination of nivolumab and ipilimumab [61]. Additionally, cabozantinib is recommended for these patients [62]. The standard treatment for poor-risk patients is similar to that for intermediate-risk patients. When cabozantinib became available, it was recognized as a desirable alternative. Indeed, the combination of anti-angiogenic therapy and immune checkpoint blockade has been validated as an effective treatment strategy to improve cancer immunity [63]. As a result, we performed drug sensitivity and immune infiltration analyses to determine whether E2F7 could be used as a biomarker for treatment choices. These findings suggest that individuals who overexpress E2F7 may have a greater response to certain medicines, including cabozantinib, pazopanib, and sunitinib, which are also highly recommended as first-line treatment regimen for patients with ccRCC. Western blot analysis revealed that E2F7 might influence patient responsiveness to these drugs via the VEGF and PI3K/AKT1/mTOR signaling pathways. However, further research is required to fully understand this mechanism. According to immune infiltration analysis, most immune cells were upregulated in patients overexpressing E2F7. Moreover, the expression of most immune checkpoint genes appeared to be discrepant between the E2F7high and E2F7low groups. TMB was also higher in patients with high E2F7 expression, indicating that immunotherapy may benefit patients with elevated E2F7 expression. Based on these findings, we suggest determining the expression levels of E2F7 in patients with ccRCC prior to treatment regimen selection, particularly in advanced and metastatic disease. High E2F7 expression may serve as a biomarker for the selection of cabozantinib, pazopanib, sunitinib, and immunotherapy as components of an effective therapeutic strategy.
Because E2F7 has been investigated in a variety of tumor types, we performed pan-cancer analysis and discovered that E2F7 is differentially expressed and correlates with immune infiltration levels in multiple cancers. Additionally, E2F7 expression is linked to TMB and MSI in many cancers, indicating that it may also serve as a biomarker for treatment choices in different types of cancer.
Although we constructed a ceRNA network in ccRCC that might provide potential biomarkers to select a suitable treatment regimen, some limitations exist. First, the miRNA-mRNA and miRNA-lncRNA binding sites obtained from the database should be validated further. Moreover, animal experiments are vital to further validate the biological functions of the RP11–478C19.2/E2F7 axis.
Conclusion
In summary, we identified a ceRNA network (RP11–478C19.2/hsa-miR-181b-5p, hsa-miR-181a-5p, and hsa-miR-181c-5p/E2F7) associated with the prognosis and treatment of ccRCC. The RP11–478C19.2/E2F7 axis may serve as a biomarker for taking cabozantinib, pazopanib, sunitinib, and immunotherapy into the therapeutic regimen.
CRediT authorship contribution statement
Kai Zeng: Writing – review & editing. Guoda Song: Writing – review & editing. Bingliang Chen: Writing – review & editing. Xintao Gao: Writing – review & editing. Chaofan Liu: Writing – review & editing. Jianping Miao: Writing – review & editing. Yajun Ruan: Writing – review & editing. Yang Luan: Writing – review & editing. Xin Chen: Writing – review & editing. Jihong Liu: Writing – review & editing. Qinyu Li: Visualization, Writing – review & editing. Bo Liu: Visualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Availability of data and materials
The datasets used in this study can be found on the online website or obtained by contacting the corresponding author.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Grant Number 81902619) and the National Natural Science Foundation of Hubei Province (2020CFB591).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101525.
Contributor Information
Qinyu Li, Email: qinyuli2022tjh@163.com.
Bo Liu, Email: boliu888@hotmail.com.
Appendix. Supplementary materials
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Hsieh J.J., Le V.H., Oyama T., Ricketts C.J., Ho T.H., Cheng E.H. Chromosome 3p loss-orchestrated VHL, HIF, and epigenetic deregulation in clear cell renal cell carcinoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2018 doi: 10.1200/JCO.2018.79.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X., Meng X., Wei C., Zhou Y., Chen H., Huang H., Chen M. Dissecting LncRNA roles in renal cell carcinoma metastasis and characterizing genomic heterogeneity by single-cell RNA-seq. Mol. Cancer Res. MCR. 2018;16:1879–1888. doi: 10.1158/1541-7786.MCR-17-0776. [DOI] [PubMed] [Google Scholar]
- 4.Tacconi E.M.C., Tuthill M., Protheroe A. Review of adjuvant therapies in renal cell carcinoma: evidence to date. OncoTargets Ther. 2020;13:12301–12316. doi: 10.2147/OTT.S174149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y., Ellinger J., Ritter M., Schmidt-Wolf I.G.H. Clinical studies applying cytokine-induced killer cells for the treatment of renal cell carcinoma. Cancers. 2020;12:E2471. doi: 10.3390/cancers12092471. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aeppli S., Schmaus M., Eisen T., Escudier B., Grünwald V., Larkin J., McDermott D., Oldenburg J., Porta C., Rini B.I., et al. First-line treatment of metastatic clear cell renal cell carcinoma: a decision-making analysis among experts. ESMO Open. 2021;6 doi: 10.1016/j.esmoop.2020.100030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makhov P., Joshi S., Ghatalia P., Kutikov A., Uzzo R.G., Kolenko V.M. Resistance to systemic therapies in clear cell renal cell carcinoma: mechanisms and management strategies. Mol. Cancer Ther. 2018;17:1355–1364. doi: 10.1158/1535-7163.MCT-17-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi T., Song X., Wang Y., Liu F., Wei J. Combining oncolytic viruses with cancer immunotherapy: establishing a new generation of cancer treatment. Front. Immunol. 2020;11:683. doi: 10.3389/fimmu.2020.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Díaz-Montero C.M., Rini B.I., Finke J.H. The immunology of renal cell carcinoma. Nat. Rev. Nephrol. 2020;16:721–735. doi: 10.1038/s41581-020-0316-3. [DOI] [PubMed] [Google Scholar]
- 10.Deleuze A., Saout J., Dugay F., Peyronnet B., Mathieu R., Verhoest G., Bensalah K., Crouzet L., Laguerre B., Belaud-Rotureau M.A., et al. Immunotherapy in renal cell carcinoma: the future is now. Int. J. Mol. Sci. 2020;21:E2532. doi: 10.3390/ijms21072532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith C.C., Beckermann K.E., Bortone D.S., De Cubas A.A., Bixby L.M., Lee S.J., Panda A., Ganesan S., Bhanot G., Wallen E.M., et al. Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J. Clin. Invest. 2018;128:4804–4820. doi: 10.1172/JCI121476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L., Meng X., Zhu X.W., Yang D.C., Chen R., Jiang Y., Xu T. Long non-coding RNAs in Oral squamous cell carcinoma: biologic function, mechanisms and clinical implications. Mol. Cancer. 2019;18:102. doi: 10.1186/s12943-019-1021-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braga E.A., Fridman M.V., Filippova E.A., Loginov V.I., Pronina I.V., Burdennyy A.M., Karpukhin A.V., Dmitriev A.A., Morozov S.G. LncRNAs in the regulation of genes and signaling pathways through miRNA-mediated and other mechanisms in clear cell renal cell carcinoma. Int. J. Mol. Sci. 2021;22:11193. doi: 10.3390/ijms222011193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang Y., Zhang H., Li W., Yan Y., Yao X., Gu W. LINC01426 contributes to clear cell renal cell carcinoma progression by modulating CTBP1/miR-423-5p/FOXM1 axis via interacting with IGF2BP1. J. Cell. Physiol. 2021;236:427–439. doi: 10.1002/jcp.29871. [DOI] [PubMed] [Google Scholar]
- 15.Yamada Y., Nohata N., Uchida A., Kato M., Arai T., Moriya S., Mizuno K., Kojima S., Yamazaki K., Naya Y., et al. Replisome genes regulation by antitumor miR-101-5p in clear cell renal cell carcinoma. Cancer Sci. 2020;111:1392–1406. doi: 10.1111/cas.14327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cochetti G., Cari L., Nocentini G., Maulà V., Suvieri C., Cagnani R., Rossi De Vermandois J.A., Mearini E. Detection of urinary miRNAs for diagnosis of clear cell renal cell carcinoma. Sci. Rep. 2020;10:21290. doi: 10.1038/s41598-020-77774-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA Hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong K., Xie T., Luo Y., Guo H., Chen J., Tan Z., Yang Y., Xie L. Comprehensive analysis of lncRNA biomarkers in kidney renal clear cell carcinoma by lncRNA-mediated ceRNA network. PLoS ONE. 2021;16 doi: 10.1371/journal.pone.0252452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Y., Gao F., He Q., Li G., Ding G. lncRNA UCA1 functions as a ceRNA to promote prostate cancer progression via sponging miR143. Mol. Ther. Nucleic Acids. 2020;19:751–758. doi: 10.1016/j.omtn.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X., Sui Z., Zhang H., Wang Y., Yu Z. Integrated analysis of lncRNA-mediated ceRNA network in lung adenocarcinoma. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.554759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin X., Wang P., Yang T., Li G., Teng X., Huang W., Yu H. Identification of key modules and genes associated with breast cancer prognosis using WGCNA and ceRNA network analysis. Aging. 2020;13:2519–2538. doi: 10.18632/aging.202285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peña-Llopis S., Vega-Rubín-de-Celis S., Liao A., Leng N., Pavía-Jiménez A., Wang S., Yamasaki T., Zhrebker L., Sivanand S., Spence P., et al. BAP1 loss defines a new class of renal cell carcinoma. Nat. Genet. 2012;44:751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peña-Llopis S., Brugarolas J. Simultaneous isolation of high-quality DNA, RNA, miRNA and proteins from tissues for genomic applications. Nat. Protoc. 2013;8:2240–2255. doi: 10.1038/nprot.2013.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wozniak M.B., Le Calvez-Kelm F., Abedi-Ardekani B., Byrnes G., Durand G., Carreira C., Michelon J., Janout V., Holcatova I., Foretova L., et al. Integrative genome-wide gene expression profiling of clear cell renal cell carcinoma in Czech Republic and in the United States. PLoS ONE. 2013;8:e57886. doi: 10.1371/journal.pone.0057886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Roemeling C.A., Radisky D.C., Marlow L.A., Cooper S.J., Grebe S.K., Anastasiadis P.Z., Tun H.W., Copland J.A. Neuronal pentraxin 2 supports clear cell renal cell carcinoma by activating the AMPA-selective glutamate receptor-4. Cancer Res. 2014;74:4796–4810. doi: 10.1158/0008-5472.CAN-14-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinforma. Oxf. Engl. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li R., Qu H., Wang S., Wei J., Zhang L., Ma R., Lu J., Zhu J., Zhong W.D., Jia Z. GDCRNATools: an R/bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC. Bioinforma. Oxf. Engl. 2018;34:2515–2517. doi: 10.1093/bioinformatics/bty124. [DOI] [PubMed] [Google Scholar]
- 29.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinforma. Oxf. Engl. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., Treviño V., Shen H., Laird P.W., Levine D.A., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li T., Fan J., Wang B., Traugh N., Chen Q., Liu J.S., Li B., Liu X.S. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ascierto M.L., McMiller T.L., Berger A.E., Danilova L., Anders R.A., Netto G.J., Xu H., Pritchard T.S., Fan J., Cheadle C., et al. The intratumoral balance between metabolic and immunologic gene expression is associated with anti-PD-1 response in patients with renal cell carcinoma. Cancer Immunol. Res. 2016;4:726–733. doi: 10.1158/2326-6066.CIR-16-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geeleher P., Cox N., Huang R.S. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kent L.N., Leone G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer. 2019;19:326–338. doi: 10.1038/s41568-019-0143-7. [DOI] [PubMed] [Google Scholar]
- 38.Fischer M., Müller G.A. Cell cycle transcription control: dREAM/MuvB and RB-E2F complexes. Crit. Rev. Biochem. Mol. Biol. 2017;52:638–662. doi: 10.1080/10409238.2017.1360836. [DOI] [PubMed] [Google Scholar]
- 39.Wang L., Chen H., Wang C., Hu Z., Yan S. Negative regulator of E2F transcription factors links cell cycle checkpoint and DNA damage repair. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E3837–E3845. doi: 10.1073/pnas.1720094115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kotecha R.R., Motzer R.J., Voss M.H. Towards individualized therapy for metastatic renal cell carcinoma. Nat. Rev. Clin. Oncol. 2019;16:621–633. doi: 10.1038/s41571-019-0209-1. [DOI] [PubMed] [Google Scholar]
- 41.Hsieh J.J., Purdue M.P., Signoretti S., Swanton C., Albiges L., Schmidinger M., Heng D.Y., Larkin J., Ficarra V. Renal cell carcinoma. Nat. Rev. Dis. Primer. 2017;3:17009. doi: 10.1038/nrdp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., Wang Y., Xu L., Lu X., Fu D., Su J., Geng H., Qin G., Chen R., Quan C., et al. CD4 + T cells promote renal cell carcinoma proliferation via modulating YBX1. Exp. Cell Res. 2018;363:95–101. doi: 10.1016/j.yexcr.2017.12.026. [DOI] [PubMed] [Google Scholar]
- 43.Ribas A., Wolchok J.D. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freeman G.J., Long A.J., Iwai Y., Bourque K., Chernova T., Nishimura H., Fitz L.J., Malenkovich N., Okazaki T., Byrne M.C., et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J. Exp. Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baraibar I., Melero I., Ponz-Sarvise M., Castanon E. Safety and tolerability of immune checkpoint inhibitors (PD-1 and PD-L1) in cancer. Drug Saf. 2019;42:281–294. doi: 10.1007/s40264-018-0774-8. [DOI] [PubMed] [Google Scholar]
- 46.Mitchell A.P., Hirsch B.R., Harrison M.R., Abernethy A.P., George D.J. Deferred systemic therapy in patients with metastatic renal cell carcinoma. Clin. Genitourin. Cancer. 2015;13:e159–e166. doi: 10.1016/j.clgc.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 47.Park I., Lee J.L., Ahn J.H., Lee D.H., Lee K.H., Jeong I.G., Song C., Hong B., Hong J.H., Ahn H. Active surveillance for metastatic or recurrent renal cell carcinoma. J. Cancer Res. Clin. Oncol. 2014;140:1421–1428. doi: 10.1007/s00432-014-1680-9. [DOI] [PubMed] [Google Scholar]
- 48.Zhang B., Wu Q., Li B., Wang D., Wang L., Zhou Y.L. m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol. Cancer. 2020;19:53. doi: 10.1186/s12943-020-01170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pancione M., Giordano G., Remo A., Febbraro A., Sabatino L., Manfrin E., Ceccarelli M., Colantuoni V. Immune escape mechanisms in colorectal cancer pathogenesis and liver metastasis. J. Immunol. Res. 2014 doi: 10.1155/2014/686879. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morrison A.H., Byrne K.T., Vonderheide R.H. Immunotherapy and prevention of pancreatic cancer. Trends Cancer. 2018;4:418–428. doi: 10.1016/j.trecan.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seipel K., Messerli C., Wiedemann G., Bacher U., Pabst T. MN1, FOXP1 and hsa-miR-181a-5p as prognostic markers in acute myeloid leukemia patients treated with intensive induction chemotherapy and autologous stem cell transplantation. Leuk. Res. 2020;89 doi: 10.1016/j.leukres.2020.106296. [DOI] [PubMed] [Google Scholar]
- 52.Schwind S., Maharry K., Radmacher M.D., Mrózek K., Holland K.B., Margeson D., Whitman S.P., Hickey C., Becker H., Metzeler K.H., et al. Prognostic significance of expression of a single microRNA, miR-181a, in cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B STUDY. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2010;28:5257–5264. doi: 10.1200/JCO.2010.29.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morse M.A., Sun W., Kim R., He A.R., Abada P.B., Mynderse M., Finn R.S. The role of angiogenesis in hepatocellular Carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019;25:912–920. doi: 10.1158/1078-0432.CCR-18-1254. [DOI] [PubMed] [Google Scholar]
- 54.Teng F., Zhang J.X., Chang Q.M., Wu X.B., Tang W.G., Wang J.F., Feng J.F., Zhang Z.P., Hu Z.Q. LncRNA MYLK-AS1 facilitates tumor progression and angiogenesis by targeting miR-424-5p/E2F7 axis and activating VEGFR-2 signaling pathway in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. CR. 2020;39:235. doi: 10.1186/s13046-020-01739-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiang S., Wang Z., Ye Y., Zhang F., Li H., Yang Y., Miao H., Liang H., Zhang Y., Jiang L., et al. E2F1 and E2F7 differentially regulate KPNA2 to promote the development of gallbladder cancer. Oncogene. 2019;38:1269–1281. doi: 10.1038/s41388-018-0494-7. [DOI] [PubMed] [Google Scholar]
- 56.He Y., Lu J., Ye Z., Hao S., Wang L., Kohli M., Tindall D.J., Li B., Zhu R., Wang L., et al. Androgen receptor splice variants bind to constitutively open chromatin and promote abiraterone-resistant growth of prostate cancer. Nucleic Acids Res. 2018;46:1895–1911. doi: 10.1093/nar/gkx1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo X., Liu L., Zhang Q., Yang W., Zhang Y. E2F7 transcriptionally inhibits MicroRNA-199b expression to promote USP47, thereby enhancing colon cancer tumor stem cell activity and promoting the occurrence of colon cancer. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.565449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Motzer R.J., Hutson T.E., Tomczak P., Michaelson M.D., Bukowski R.M., Rixe O., Oudard S., Negrier S., Szczylik C., Kim S.T., et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N. Engl. J. Med. 2007;356:115–124. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 59.Sternberg C.N., Davis I.D., Mardiak J., Szczylik C., Lee E., Wagstaff J., Barrios C.H., Salman P., Gladkov O.A., Kavina A., et al. Pazopanib in locally advanced or metastatic renal cell carcinoma: results of a randomized phase III trial. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2010;28:1061–1068. doi: 10.1200/JCO.2009.23.9764. [DOI] [PubMed] [Google Scholar]
- 60.Escudier B., Pluzanska A., Koralewski P., Ravaud A., Bracarda S., Szczylik C., Chevreau C., Filipek M., Melichar B., Bajetta E., et al. Bevacizumab plus interferon alfa-2a for treatment of metastatic renal cell carcinoma: a randomised, double-blind phase III trial. Lancet Lond. Engl. 2007;370:2103–2111. doi: 10.1016/S0140-6736(07)61904-7. [DOI] [PubMed] [Google Scholar]
- 61.Motzer R.J., Tannir N.M., McDermott D.F., Arén Frontera O., Melichar B., Choueiri T.K., Plimack E.R., Barthélémy P., Porta C., George S., et al. Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N. Engl. J. Med. 2018;378:1277–1290. doi: 10.1056/NEJMoa1712126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choueiri T.K., Hessel C., Halabi S., Sanford B., Michaelson M.D., Hahn O., Walsh M., Olencki T., Picus J., Small E.J., et al. Cabozantinib versus sunitinib as initial therapy for metastatic renal cell carcinoma of intermediate or poor risk (Alliance A031203 CABOSUN randomised trial): progression-free survival by independent review and overall survival update. Eur. J. Cancer Oxf. Engl. 2018;94:115–125. doi: 10.1016/j.ejca.2018.02.012. 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee W.S., Yang H., Chon H.J., Kim C. Combination of anti-angiogenic therapy and immune checkpoint blockade normalizes vascular-immune crosstalk to potentiate cancer immunity. Exp. Mol. Med. 2020;52:1475–1485. doi: 10.1038/s12276-020-00500-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used in this study can be found on the online website or obtained by contacting the corresponding author.