Abstract
A key aspect of neuroscience research is the development of powerful, general-purpose data analyses that process large datasets. Unfortunately, modern data analyses have a hidden dependence upon complex computing infrastructure (e.g. software and hardware), which acts as an unaddressed deterrent to analysis users. While existing analyses are increasingly shared as open source software, the infrastructure and knowledge needed to deploy these analyses efficiently still pose significant barriers to use. In this work we develop Neuroscience Cloud Analysis As a Service (NeuroCAAS ): a fully automated open-source analysis platform offering automatic infrastructure reproducibility for any data analysis. We show how NeuroCAAS supports the design of simpler, more powerful data analyses, and that many popular data analysis tools offered through NeuroCAAS outperform counterparts on typical infrastructure. Pairing rigorous infrastructure management with cloud resources, NeuroCAAS dramatically accelerates the dissemination and use of new data analyses for neuroscientific discovery.
eTOC Blurb:
Computing infrastructure is a fundamental part of neural data analysis. Abe et al. present an open source, cloud based platform called NeuroCAAS to automatically build reproducible computing infrastructure for neural data analysis. They show that NeuroCAAS supports novel analysis design and can improve the efficiency of popular existing methods.
1. Introduction
Driven by the constant evolution of new recording technologies and the vast quantities of data that they generate, neural data analysis — which aims to build the path from these datasets to scientific understanding — has grown into a centrally important part of modern neuroscience, enabling significant new insights into the relationships between neural activity, behavior, and the external environment (Paninski and Cunningham, 2018). Accompanying this growth however, neural data analyses have become much more complex. Historically, the software implementation of a data analysis (what we call the core analysis- Figure 1A) was typically a small, isolated code script with few dependencies. In stark contrast, modern core analyses routinely incorporate video processing algorithms (Pnevmatikakis et al., 2016, Pachitariu et al., 2017, Mathis et al., 2018, Zhou et al., 2018, Giovannucci et al., 2019), deep neural networks (Batty et al., 2016, Gao et al., 2016, Lee et al., 2017, Parthasarathy et al., 2017, Mathis et al., 2018, Pandarinath et al., 2018, Giovannucci et al., 2019), sophisticated graphical models (Yu et al., 2009, Wiltschko et al., 2015, Gao et al., 2016; Wu et al., 2020), and other cutting-edge machine learning methods (Pachitariu et al., 2016, Lee et al., 2017) to create general purpose tools applicable to many datasets.
Figure 1: Data Analysis Infrastucture.
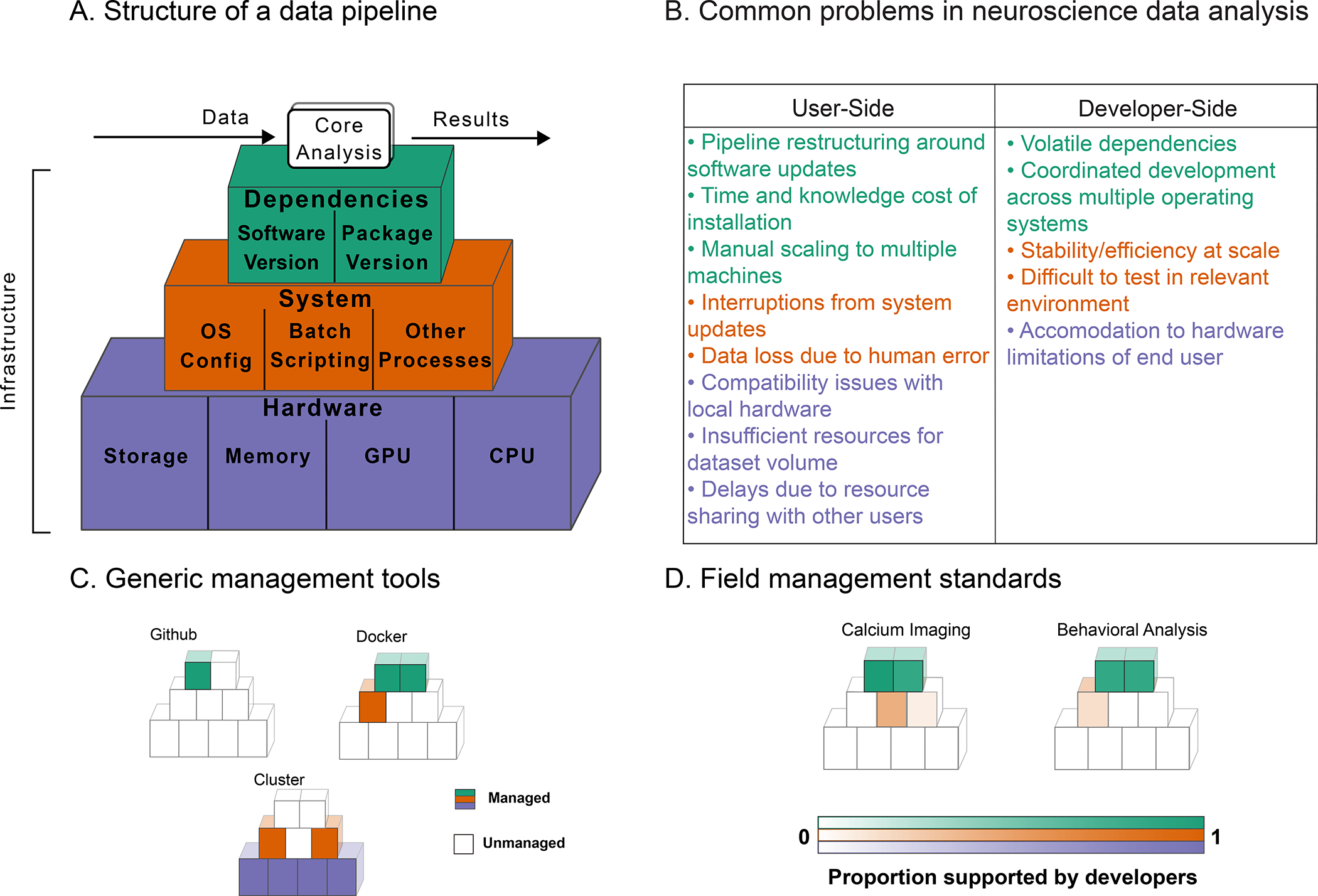
A. Core analysis code depends upon an infrastructure stack. B. Common problems arise at each layer of this infrastructure stack for analysis users and developers. C. Many common management tools deal only with one or two layers in the infrastructure stack, leaving gaps that users and developers must fill manually. D. In common neural data analysis tools for calcium imaging and behavioral analysis many infrastructure components are not managed by analysis developers and implicitly delegated to the user (see §9 for full details and supporting data in Tables S2,S1).
To support this increasing complexity, core analysis software is increasingly coupled to underlying analysis infrastructure (Figure 1A): software dependencies like the deep learning libraries PyTorch and TensorFlow (Abadi et al., 2016, Paszke et al., 2019), system level dependencies to manage jobs and computing resources (Merkel, 2014), and hardware dependencies such as a precisely configured CPU (central processing unit), access to a GPU (graphics processing unit), or a required amount of device memory. Figure 1A shows how these individual components form a full infrastructure stack: the necessary, but largely ignored foundation of resources enabling all data analyses (Demchenko et al., 2013, Jararweh et al., 2016, Zhou et al., 2016).
Neglected infrastructure has immediate implications already familiar to the neuroscience community: for every novel analysis, analysis users must spend labor and financial resources on hardware setup, software troubleshooting, unexpected interruptions during long analysis runs, processing constraints due to limited “on-premises” computational resources, and more (Figure 1B). However, far from simply being a nuisance, neglected infrastructure has wide reaching and urgent scientific consequences. Most prominently, infrastructure impacts analysis reproducibility. As data analyses become more dependent on complex infrastructure stacks, it becomes extremely difficult for analysis developers to work reproducibly (Monajemi et al., 2019, Nowogrodzki, 2019). The current treatment of analysis infrastructure is a major contributor to the endemic lack of reproducibility suffered by modern data analysis (Crook et al., 2013, Hinsen, 2015, Stodden et al., 2018, Krafczyk et al., 2019, Raff, 2019), and infrastructure-based barriers have been noted to impede the proliferation of new neuroscientific tools (Magland et al., 2020). Specific cases where seemingly small infrastructure issues directly affect the representation of data-derived quantities have been documented across the biological sciences (Ghosh et al., 2017, Miller, 2006, Glatard et al., 2015). Analogously, in machine learning, infrastructure components can dictate model performance (Sculley et al., 2015, Radiuk, 2017) and a recent survey of this literature observed that although local compute clusters claim to address the issue of hardware availability, none of the studies that required use of a compute cluster were reproducible (Raff, 2019).
Major efforts have been made by journals (Donoho, 2010, Hanson et al., 2011, ) and funding agencies (Carver et al., 2018) to encourage the sharing of core analysis software. Additionally, new tools have been developed to address related neuroscientific challenges like the formatting (Teeters et al., 2015, Rübel et al., 2019, Rübel et al., 2021) and storage of data (Dandi Team, 2019), or workflow management on existing infrastructure (Yatsenko et al., 2015, Gorgolewski et al., 2011) (see §3 for a detailed overview). However, these important efforts still neglect key issues in the configuration of infrastructure stacks. Despite calls to improve standards of practice in the field (Vogelstein et al., 2016), and work in fields such as astronomy, genomics, and high energy physics (Hoffa et al., 2008, Riley, 2010, Goecks et al., 2010, Zhou et al., 2016, Chen et al., 2019, Monajemi et al., 2019), there has been little concrete progress in our field towards a scientifically acceptable infrastructure solution for many popular core analyses. Some tools – compute clusters, versioning tools like Github (https://github.com), and containerization services like Docker (Merkel, 2014) – provide various infrastructure components (Figure 1C), but it is nontrivial to combine these components into a complete infrastructure stack. The ultimate effect of these partially used toolsets is a hodgepodge of often slipshod infrastructure practices (Figure 1D; supporting data in Tables S1, S2).
Critically, management of these issues most often falls upon trainees who are neither scientifically rewarded (Landhuis, 2017, Chan Zuckerberg Initiative, 2014), nor specifically instructed (Merali, 2010) on how to set up increasingly complex core analyses with infrastructure stacks, reliant on whatever resources are available on hand. We term this conventional model Infrastructure-as-Graduate-Student, or IaGS – infrastructure stacks treated as a scientific afterthought, delegated entirely to underresourced trainees and operating as a silent source of errors and inefficiency. The IaGS status quo fails any reasonable standard of scientific rigor, reduces the accessibility of valuable analytical tools, and impedes scientific training and progress.
Of course, infrastructure challenges are not specific to neuroscience, or even science generally. Entities that deploy software at industrial scale have recently adopted the Infrastructure-as-Code (IaC) paradigm, automating the creation and management of infrastructure stacks (Morris, 2016, Aguiar et al., 2018). In contrast to IaGS approaches, the IaC paradigm begins with a code document that completely specifies the infrastructure stack supporting any given core software. From this code document, the corresponding infrastructure stack can be assembled automatically (most often on a cloud platform), in a process called deployment. After deployment, anyone with access to the platform can use the core software in question without knowledge of its underlying infrastructure stack, while still having the assurance that core software is functioning exactly as indicated in the corresponding code document. Altogether, IaC enables reproducible usage at scale, skirting all of the issues shown in Figure 1B. Despite these benefits, there has been no previous effort to extend IaC to general-purpose neuroscience data analyses and associated infrastructure stacks.
In response, we developed Neuroscience Cloud Analysis as a Service (NeuroCAAS ), an IaC platform that pairs core analyses for neuroscience data with bespoke infrastructure stacks through deployable code documents. NeuroCAAS assigns each core analysis a corresponding infrastructure stack, using a set of modular components concisely specified in code (see §2.1 for details). NeuroCAAS stores the specification of this core analysis and infrastructure stack in a code document called a blueprint, which any analysis user can then deploy to analyze their data. To maximize the scale and accessibility benefits of our platform, we provide an open source web interface to NeuroCAAS (§2.2), available to the neuroscience community at large. The result is scalable, reproducible, drag-and-drop usage of neural data analysis: neuroscientists can log on to the NeuroCAAS website, set some parameters for an analysis, and simply submit their data. A new infrastructure stack is then deployed on the cloud according to a specified blueprint and autonomously produces analysis results, which are returned to the user. This aspect of NeuroCAAS warrants emphasis, as it diverges starkly from traditional scientific practice: NeuroCAAS is not only a platform design, or suggestion that the reader can attempt to recreate on their own; instead, NeuroCAAS is offered as an open source infrastructure platform available for immediate use, via a website (www.neurocaas.org).
We first describe IaC analysis infrastructure on NeuroCAAS (§2.1), and how it addresses common engineering challenges related to analysis reproducibility, accessibility and scale (§2.2). In (§2.3) we compare NeuroCAAS ‘s solution to these engineering challenges with features of existing data analysis platforms. Next, in §2.4,2.5, we show how NeuroCAAS can enable novel analyses designed to take advantage of the platform’s infrastructure benefits. Finally, in §2.6, we quantify the performance of popular data analyses on NeuroCAAS, and find that analyses encoded in blueprints are cheaper and faster than analogues run on local infrastructure (e.g. a compute cluster).
2. Results
NeuroCAAS ‘s primary technical contribution is a method to precisely specify the entire infrastructure stack underlying any core analysis, and reproduce it on demand. Treating core analysis and infrastructure as a unified whole within NeuroCAAS makes analyses more reproducible and accessible at scale than existing alternatives.
In the simplest use case, users simply log in to the platform and drag and drop their dataset(s) into a web browser (Figure 2, top left), sending it to cloud based user storage. After specifying a set of developer-defined parameters to apply to the selected dataset(s), they can submit a NeuroCAAS “job.” No further user input is needed: given the relevant datasets(s) and parameters, NeuroCAAS sets up core analysis for each dataset on an entirely new infrastructure stack from the corresponding blueprint (Figure 2, black arrows). NeuroCAAS then pulls data and parameters independently into each infrastructure stack (Figure 2, blue arrows), providing scalable and reproducible computational processing as needed (Figure 2, bottom right). Analysis outputs (including live status logs and a complete record of the job’s inputs and infrastructure) are then delivered back to timestamped folders in user storage for inspection by analysis users (Figure 2, bottom left), and finally infrastructure stacks are dissolved when data processing is complete (Figure 2, bottom right). As an example, Supplementary Video 1 shows how users can train three separate DeepGraphPose models (Wu et al., 2020) on three separate datasets simultaneously using the NeuroCAAS web interface.
Figure 2: Overview of NeuroCAAS User Workflow.
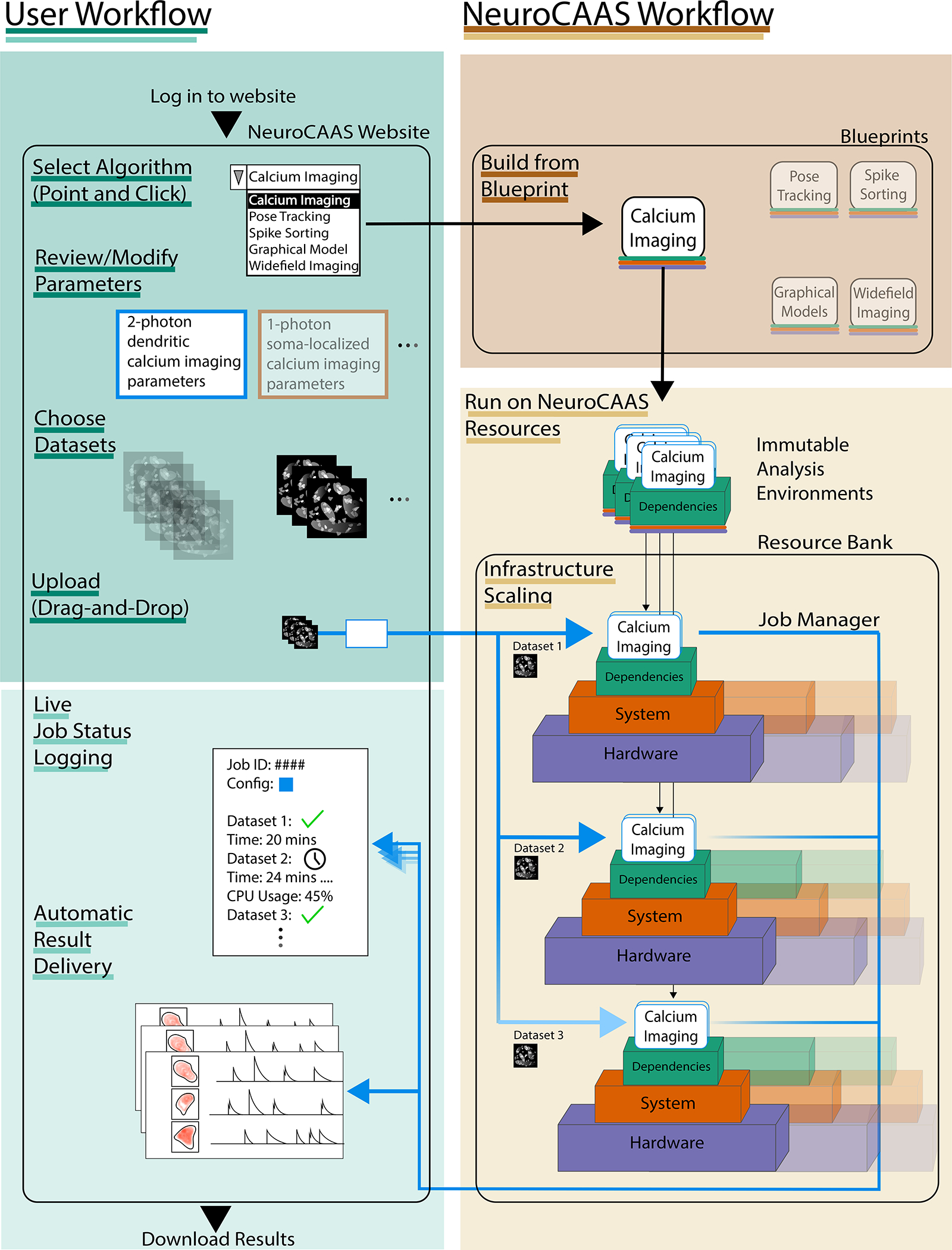
Left indicates the user’s experience; right indicates the work that NeuroCAAS performs. The user chooses from the analyses encoded in NeuroCAAS. They then modify corresponding configuration parameters as needed. Finally, the user uploads dataset(s) and a configuration file for analysis. NeuroCAAS detects upload event and deploys the requested analysis using an infrastructure blueprint (§2.1.4). NeuroCAAS builds the appropriate number of IAEs (§2.1.1) and corresponding hardware instances (§2.1.3). Multiple infrastructure stacks may be deployed in parallel for multiple datasets and the job manager (§2.1.2) automatically handles input and output scaling. The deployed resources persist only as necessary, and results, as well as diagnostic information, are automatically routed back to the user. See Figure S1 for comparison with IaGS, and Figure S3 for IAE list.
2.1. NeuroCAAS Builds Complete Infrastructure Stacks
The structure of NeuroCAAS naturally solves the issues of reproducibility, accessibility, and scale that burden existing infrastructure tools and platforms. NeuroCAAS partitions a complete infrastructure stack into three decoupled parts that together are sufficient to support virtually any given core analysis. First, to address all software level infrastructure, NeuroCAAS offers all analyses in immutable analysis environments (§2.1.1). Second, to address system configuration, each NeuroCAAS analysis has a built-in job manager (§2.1.2) that automates all of the logistical tasks associated with analyzing data: configuring hardware, logging outputs, parallelizing jobs and more. Third, to provide reproducible computing hardware on demand, NeuroCAAS manages a resource bank (§2.1.3) built on the public cloud, making the service globally accessible at unmatched scale. For a given core analysis, the configuration of these three infrastructure components is concisely summarized in a NeuroCAAS blueprint, from which it can be automatically rebuilt (§2.1.4). We describe component implementation in further depth in §9.2.
2.1.1. Immutable Analysis Environments for Software Infrastructure
On NeuroCAAS, all core analyses run inside immutable analysis environments (IAEs). An IAE is an isolated computing environment containing the installed core analysis code and all necessary software dependencies, similar to a Docker container (Merkel, 2014). Importantly, an IAE also contains a single script that parses input and parameters in a prescribed way, and runs the steps of core analysis workflow (Figure 2, right; Figure S6, top left). The fact that analysis workflow is entirely governed by this script (i.e. non-interactive) makes our analysis environments immutable. IAEs eliminate the possibility of bugs resulting from incompatible dependencies, mid-analysis misconfiguration (Figure S1A, installation and troubleshooting), or other so-called “user degrees of freedom” and ensure that analyses are run within developer-defined workflows. Immutability has a long history as a principle of effective programming and resource management in computer science (Bloch, 2008, Morris, 2016), and in this context is closely related to the view that data analysis should be automated as much as possible (Tukey, 1962, Waltz and Buchanan, 2009). These views are justified by observed benefits to analysis at scale, which we leverage in (§2.4.,2.5).
Each IAE has a unique ID, and analysis updates can be recorded in IAEs linked through blueprint versions (see §2.1.4 for details). We have currently implemented 22 analyses in a series of immutable analysis environments (Table S3) and are actively developing more (see www.neurocaas.org for current options).
2.1.2. Job Managers for System Infrastructure
Given a dataset and analysis parameters, how does NeuroCAAS set up the right IAE and computing hardware to process these inputs? This configuration is the responsibility of the NeuroCAAS job manager, which monitors analysis progress and returns timestamped job outputs to user storage from the IAE (including live job logs). Although similar in these regards to a cluster workload manager like slurm (Yoo et al., 2003) (Figure 2, blue arrows) the NeuroCAAS job manager does not assign jobs to running infrastructure, but rather set up all other infrastructure components “on the fly,” removing the need for manual infrastructure maintenance (Figure 2; black arrows). The job manager for each analysis functions according to a code “protocol” that describes what steps should be taken when a new NeuroCAAS job is requested. Importantly, protocols can be customized for each analysis, allowing developers to implement simple features like input parsing, or complex multi-stack workflows as shown in §2.4.,2.5.
2.1.3. Resource Banks for Hardware Infrastructure
To automatically reproduce infrastructure on demand, we crucially need a way to create identical hardware configurations across multiple users of the same analysis, who may be analyzing data simultaneously at many different locations around the world. This key requirement is handled by the NeuroCAAS resource bank. The NeuroCAAS resource bank can make hardware available through pre-specified instances: bundled collections of virtual CPUs, memory, and GPUs that can emulate any number of familiar hardware configurations (e.g. personal laptop, workstation, on premise cluster). However unlike these persistent computing resources, the NeuroCAAS resource bank is built upon globally available, virtualized compute hardware offered through the public cloud (currently Amazon Web Services). At any time, the resource bank can provide a large number of effectively identical hardware instances to execute a particular task (Figure 2, bottom right). The reproducible nature of hardware instances in the resource bank complements the immutability of workflows imposed by IAEs. By default, we fix a single instance type per analysis in order to facilitate reproducibility (see 9.2 for details).
2.1.4. Blueprints for Instant Reproducibility
For any given analysis, each of NeuroCAAS ‘s infrastructure components (§2.1.1–2.1.3) has a specification in code (IAEs and resource bank instances have IDs, job managers have protocol scripts). The collection of all infrastructure identifying code associated with a given NeuroCAAS analysis is stored in the blueprint of that analysis (Figure 2, top right- for an example see Figure S2), from which new instances of the infrastructure stack can be deployed at will, providing reproducibility by design. Despite sustained efforts to promote reproducible research, (Buckheit and Donoho, 1995), in many typical cases data analysis remains frustratingly non-reproducible (Crook et al., 2013, Gorgolewski et al., 2017, Stodden et al., 2018, Raff, 2019). NeuroCAAS sidesteps all of the typical barriers to reproducible research by tightly coupling the creation and function of infrastructure stacks to their documentation. For transparency, NeuroCAAS stores all currently available and developing analysis blueprints in a public code repository (see §9.2). Updates made to any component of an infrastructure stack on NeuroCAAS (IAEs, job managers, or hardware instances) can only be implemented through subsequent deployments of updated blueprints. We record these changes systematically using simple version control on the blueprint itself, ensuring a publicly visible record of analysis development.
2.2. NeuroCAAS Supports Simple Use and Development
Users analyze data on NeuroCAAS solely through interactions with cloud storage. Therefore, NeuroCAAS supports any interface that allows users to transfer data files to and from cloud storage. The standard interface to NeuroCAAS is a website, www.neurocaas.org, where users can sign up for an account, browse analyses, deposit data and monitor analysis progress until results are returned to them as described in Figure 2. We will describe other interfaces to NeuroCAAS in §2.4 and §3. Regardless of interface, there is no need to manage persistent compute resources during or after analysis, and costs directly reflect usage time.
For comparison, IaGS begins with a number of time-consuming manual steps, including hardware acquisition, hardware setup, and software installation. With a functional infrastructure stack in hand, the user must prepare datasets for analysis, manually recording analysis parameters and monitoring the system for errors as they work. While parallel processing is possible, it must be scripted by the user, and in many cases datasets are run serially. What results from IaGS is massive inefficiency of time and resources. Users must also support the cost of new hardware “up front,” before ever seeing the scientific value of the infrastructure that they are purchasing. Likewise, labs or institutions must pay support costs to maintain infrastructure when it is not being used, and replace components when they fail or become obsolete (see Figure S1 for a side-by-side comparison with NeuroCAAS ). Two editorial remarks bear mentioning at this point: first, the stark difference laid out in these workflows is the essence of IaGS vs IaC, and explains the dominance of IaC in modern industrial settings. Second, NeuroCAAS is and will remain an open-source tool for the scientific community, in keeping with its sole purpose of improving the reproducibility and dissemination of neuroscience analysis tools.
For analysis developers interested in NeuroCAAS, we designed a developer workflow and companion python package that streamlines the process of migrating an existing analysis to the NeuroCAAS platform (see §9.2 and Figure S6 for an overview). Our developer workflow abstracts away the cloud infrastructure that NeuroCAAS is built on, allowing for analysis development entirely from the developer’s command line. We highlight several key features of this workflow here. Importantly, we do not expect any previous experience in containerization technology, cloud tools, or IaC from developers.
Curated deployments.
After setting up an IAE and initializing a corresponding blueprint, developers submit blueprints and test data to our code repository in publicly visible pull requests. NeuroCAAS team members then review the submitted blueprint and deploy the corresponding infrastructure stack in “test mode,” reviewing outputs, requesting updates, and ensuring stated function in a public forum before releasing the analysis to users.
Improving analysis robustness.
A central challenge of building general-purpose data analysis is the difficulty of anticipating all analysis use cases as a developer and ensure robustness to all possible datasets. This is true even with the testing, review, and development practices that NeuroCAAS puts in place. However, NeuroCAAS ‘s design has several features intended to accelerate the process of improving analysis robustness both during and after initial deployment. When errors occur, users can refer the analysis developer to version controlled analysis outputs using standard interfaces like Github issues, greatly simplifying error replication. Developers can then set up fixes on the same infrastructure, and update the analysis blueprint in subsequent deployments. Since infrastructure is rebuilt from the blueprint for each NeuroCAAS job, an updated blueprint fixes the bug, for all future analysis runs of all analysis users. Importantly, updates can be made to a public analysis without influencing the reproducibility of past results (see 9.2 and Figure S6 for details, and §9.4 for links to a full developer guide).
2.2.1. Testing the NeuroCAAS Usage Model
Next, we study how the design of NeuroCAAS translates into quantifiable analysis benefits. We confirmed the accessibility of data analyses on NeuroCAAS by opening the platform to a group of alpha testers (users and developers) over a period of 22 months. In Figure 3A, we see that while some users analyzed a handful of datasets, others analyzed hundreds and spent days of compute with the platform. Figure 3B further studies the co-occurrence of different usage patterns: a large number of single dataset jobs are suggestive of one-off exploratory use, while there is also a considerable proportion of jobs that leverage NeuroCAAS ‘s capacity for parallelism, analyzing anywhere from 2 to 70 datasets in a single job. We also grouped usage by data analysis (Figure 3C). We classified different analyses as follows: dark blue bars indicate existing analysis adapted for NeuroCAAS by manuscript authors. These analyses were developed collaboratively, and in many cases, we iterated on an initial “dev” version of an analysis adapted for NeuroCAAS with feedback from users, before releasing a “public” version that was robust to various differences in workflow and dataset type. Light green bars indicate analyses developed by independent researchers following our developer workflow. We highlight the fact that analyses built following the developer workflow are well used, indicating the viability of the workflow that we have built. Dark green bars indicate analyses that we introduce in this paper specifically for NeuroCAAS infrastructure, described further in §2.4, 2.5. Finally, light blue bars indicate “custom” analyses that we built for particular user groups. NeuroCAAS authors built custom analyses through simple copying and editing of existing, general purpose blueprints. While per-user custom analyses are not a focus of our platform, these results demonstrate the ease with which different variants of an analysis can be provided within NeuroCAAS ‘s design, and we discuss how users can leverage NeuroCAAS for custom use cases in §3.
Figure 3: Usage statistics NeuroCAAS Platform.
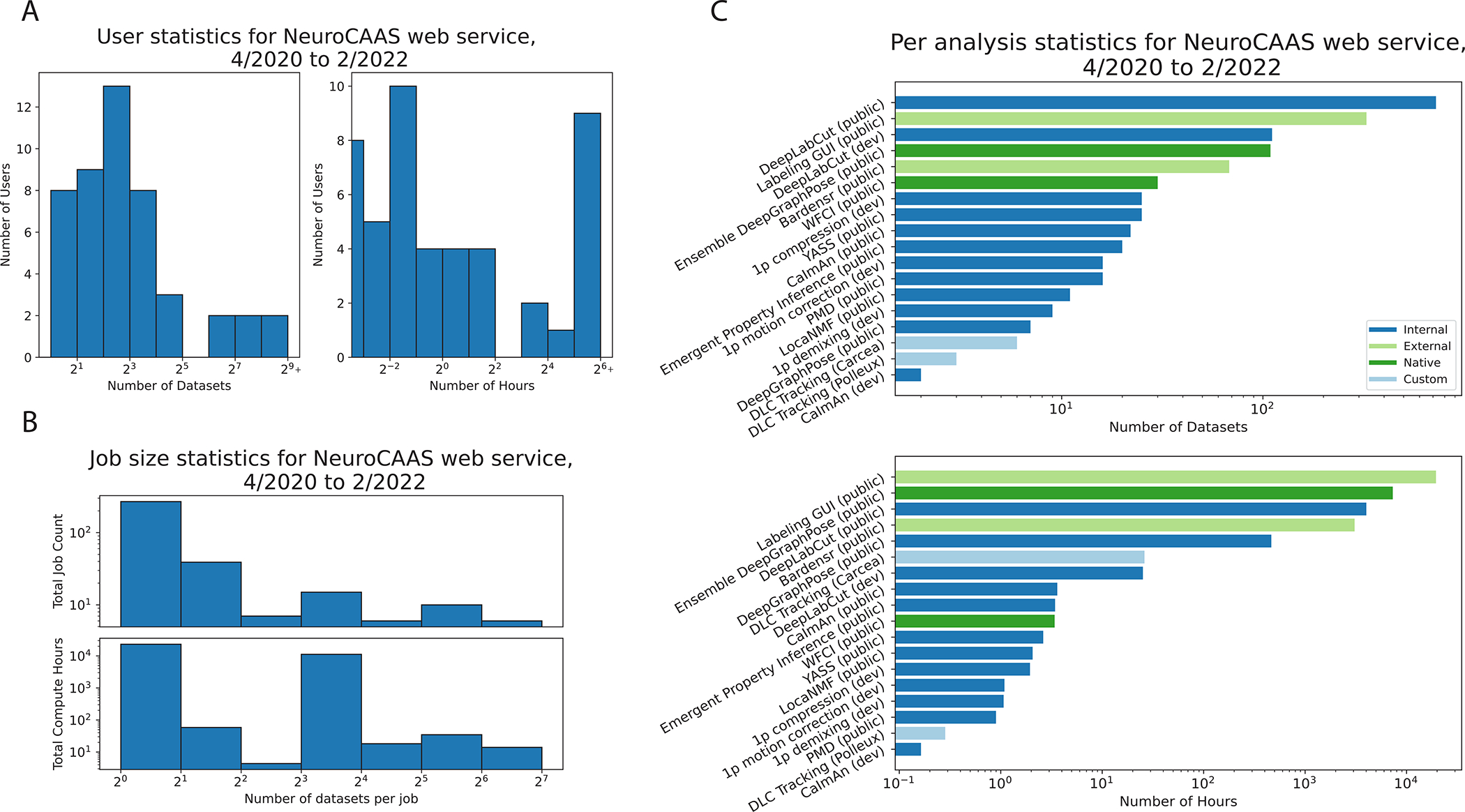
Usage data over a 22-month alpha test period. A. Histogram for number of datasets (left) and corresponding compute hours (right) spent by each active user of NeuroCAAS. B. Histograms for job size indicates the number of datasets (top) and corresponding compute hours (bottom) concurrently analyzed in jobs. C. Usage grouped by platform developer. Dark blue: analyses adapted for NeuroCAAS by paper authors. Light green: analyses that were not developed by NeuroCAAS authors. Dark green: NeuroCAAS native analyses (§2.4, 2.5). Light blue: custom versions of generic analyses built for individual alpha users. We exclude usage attributed to NeuroCAAS team members.
Next we confirmed that the design of data analysis using NeuroCAAS ‘s IaC approach does indeed provide robust reproducibility. We selected two analyses available on our platform and we analyzed the similarity of analysis outputs across multiple runs in Table 2 (see Figure S3 for in depth analysis, and Figure S4 for corresponding analysis of timings). Fixing a single dataset, configuration file, and blueprint for each analysis, we evaluated reproducibility of results in terms of differences between analysis outputs (see Table 2 caption for metrics.) First, we observe that over multiple runs conducted by the same researcher in the United States, (Table 2, vs Run 2), results showed no scientifically relevant differences. Second, we varied the identity and physical location of the person requesting NeuroCAAS jobs: compared to jobs started by independent researchers in India and Switzerland (Table 2, vs Run 5, Run 10 respectively) we once again note no meaningful differences in the outputs of these analyses. Whereas physical location might bias or restrict researchers to use specific analysis infrastructure on other platforms, they have access to the exact same analysis infrastructure through NeuroCAAS. Finally, we conducted a test to measure if our platform was truly IaC: given dataset, configuration file, and analysis blueprint, there should be no reliance on the compute resources that we used to develop these analysis and perform reference runs. For a final run, we automatically deployed a complete clone of the NeuroCAAS platform on a new set of cloud resources, as any user of our platform can do in a few simple steps (details in §3, Figure S5). We then ran jobs with the blueprints, datasets and configuration files for the corresponding analyses (Table 2, vs Run 14), showing that results from this cloned platform are indistinguishable from those generated by the original platform.
Table 2: Quantifying reproducibility via output comparisons for two analyses on NeuroCAAS.
For CaImAn, (an algorithm to analyze calcium imaging data) we independently characterized differences in the spatial and temporal components recovered by the model. Differences in spatial components are measured by the average Jaccard Distance over pairs of spatial components. A Jaccard distance of 0 corresponds to two spatial components that perfectly overlap. Differences in temporal components were calculated as the average root mean squared error (RMSE) taken over paired time series of component activity. For Ensemble DeepGraphPose (an algorithm to track body parts of animals during behavior from video), we considered multiple sets of outputs from a single, pretrained model. RMSE takes units of pixels, so differences of order 1e-8 are not relevant for behavioral quantification. For both analyses, we fixed a single dataset, configuration file and blueprint across runs. See Figures S3,S4 for more.
| Reference Run Output | vs. Run 2 | vs. Run 5 | vs. Run 10 | vs. Run 14 | |
|---|---|---|---|---|---|
| Analysis | (Comparison Metric) | (US) | (India) | (Switzerland) | (Platform Clone) |
| Spatial Components | 0.0 | 0.0 | 0.0 | 0.0 | |
| CaImAn | (Jaccard Distance) | ||||
| (Giovannucci et al., 2019) | Temporal Components | 0.0 | 0.0 | 0.0 | 0.0 |
| (RMSE) | |||||
| Ensemble | Body Part Traces | 1.2e-8 | 1.2e-8 | 2.3e-8 | 1.4e-8 |
| DeepGraphPose (§2.5) | (RMSE) |
2.3. Existing Platforms Leave Infrastructure Gaps
Although we do not attempt an exhaustive review of existing analysis platforms in neuroscience here, we characterize some exemplars in order to contrast NeuroCAAS from typical alternatives. In Figure 4, we plot a variety of popular neuroscience analyses onto a space defined by 1) their place in the adoption lifecycle and 2) corresponding infrastructure needs. We overlay several exemplar platforms on this graph, with shading representing the kinds of analyses they are able to support. The degree to which a platform’s support extends to the right defines its accessibility, or the ease with which developers and users can configure analyses on the platform, and begin to process data. Accessibility is a key feature for analyses that are still early in the adoption lifecycle with active development and a growing user base. Likewise, the degree to which a platform’s support extends upwards defines its scale, a one dimensional approximation of the infrastructure needs for which it can provide. While the exact positioning of these analyses and platforms is subjective and dynamic, there are general features of the analysis platform landscape that we discuss in what follows.
Figure 4: Landscape of Cellular/Circuit-Level Neuroscience Analysis Platforms.
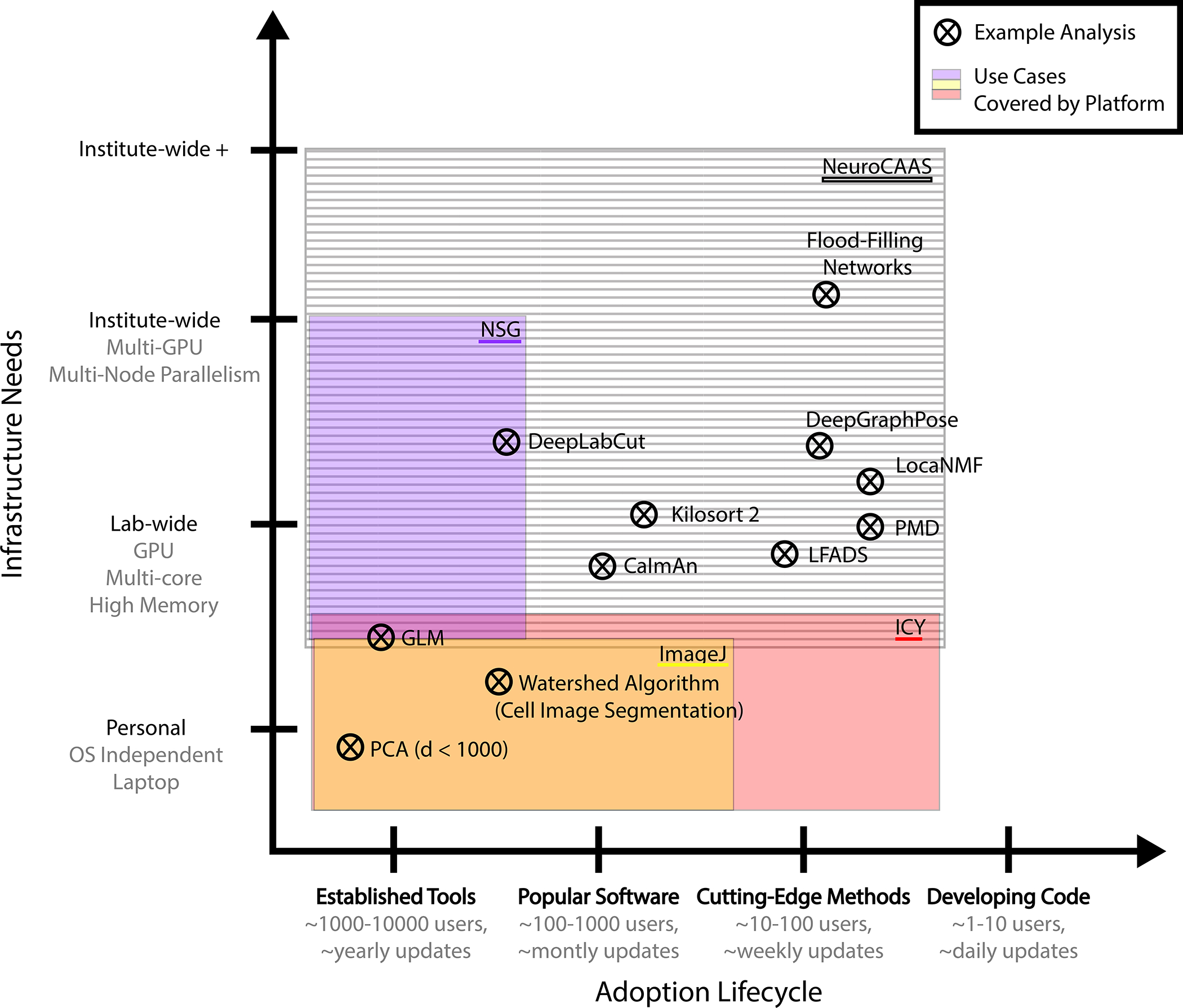
Crosses: popular analyses in terms of their place in the adoption lifecycle (number of users, rate of software updates), and their infrastructure needs. Coloring: representative platforms, indicating the parts of analysis space that are covered by a given platform. (Example analyses: (Goodman and Brette, 2009; Pnevmatikakis et al., 2016; Mathis et al., 2018; Pachitariu et al., 2016; Pandarinath et al., 2018; Januszewski et al., 2018; Saxena et al., 2020; Buchanan et al., 2018; Graving et al., 2019); Representative platforms: (Sanielevici et al., 2018; Chaumont et al., 2012; Schneider et al., 2012).
Local platforms like CellProfiler (Carpenter et al., 2006), Ilastik (Sommer et al., 2011) (cell-based image processing), Icy (Chaumont et al., 2012), ImageJ (Schneider et al., 2012) (generic bioimage analysis), BIDS Apps (Gorgolewski et al., 2017) (MRI analyses for Brain Imaging Data Structure format), and Bioconductor (Amezquita et al., 2020) (genomics) have all achieved success in the field by packaging together popular analyses with necessary software dependencies and intuitive, streamlined user interfaces. Most of these local platforms also have an open contribution system for interested developers. Local platforms are thus highly accessible to both developers and users, but are in the large majority of cases designed only for use on a user’s local hardware, limiting their scale (Figure 4, bottom).
In contrast, remote platforms like the Neuroscience Gateway (NSG) (Sanielevici et al., 2018) (specializing in neural simulators), Flywheel (flywheel.io) (emphasizing fMRI and medical imaging), and neuroscience-focused research computing clusters offer powerful hardware through the XSEDE (Extreme Science and Engineering Discovery Environment) portal (Towns et al., 2014), the public cloud and on-premises hardware, respectively. These remote platforms offer powerful compute, but at the cost of accessibility to users, who must adapt their software and workflow to new conventions (i.e. wait times for jobs to run on shared resources, hardware specific installation, custom scripting environments, limitations on concurrency) in order to make use of offered hardware. As a particular example, NSG requires users to submit a script that they would like to have run on existing compute nodes in the XSEDE cluster, making it more similar to a traditional on-premises cluster in usage than NeuroCAAS. NSG also restricts jobs to run serially, and does not have an open system for contributing new analyses, making it incompatible with the usage model and analyses that we present here. Likewise, while Flywheel () (with a focus on human brain imaging tools) offers the option of cloud compute, the platform is not structured in terms of infrastructure stacks for given analyses, in effect leaving many infrastructure design choices to individual users. Altogether, remote platforms are best for committed, experienced users who are already familiar with the analyses available on the remote platform and understand how to optimize them for available hardware. It is also more difficult to contribute new analyses to these platforms than their locally hosted counterparts (see Figure 4, left side). This difficulty makes them less suitable for actively developing or novel analyses, as updates may be slow to be incorporated, or introduce breaking changes to user-written scripts, making remote platforms altogether less accessible than local ones.
Some platforms provide both local or remote style usage: Galaxy (Goecks et al., 2010) and Brainlife (Avesani et al., 2019) offer a set of default compute resources, but can also be used to run analyses on personal computing resources, or the cloud. These mixed-compute models provide a useful way to increase the accessibility of many analyses, and can provide levels of reproducibility similar to that provided by the IAE and job manager of NeuroCAAS. However, without having an IaC framework that makes a reproducible configuration of compute resources available to all analysis users, we lose the guarantee that all platform users will be able to use analyses that depend on specific infrastructure configurations. As noted in (Goecks et al., 2010), it is a challenge even for these mixed-compute platforms to ensure accessibility for an analysis developed on a given set of local compute resources-significant work must be done to make this analysis functional on other computing platforms, or to maintain these local compute resources in order to ensure that others can use them whenever needed. These challenges are only exacerbated by the increasing reliance of analysis tools upon more powerful and specific infrastructure configurations, such as high performance GPUs.
Although undeniably useful, all available platforms operate on a tradeoff that forces researchers to choose between accessibility and scale. While these platforms often concentrate on applications that mitigate the effects of this tradeoff, there are many popular analyses that would not be suitable for existing analysis platforms (see Figure 4, center). Furthermore, critically for reproducibility, across all existing platforms analysis users and developers are still required to manually configure analysis infrastructure, whether by installing new tools onto one’s personal infrastructure, or porting code and dependencies to run in a remote (and sometimes variably allocated) infrastructure stack.
Some platforms in cellular and molecular biology (Riley, 2010), as well as bioinformatics (Simonyan and Mazumder, 2014,,Terra, 2022) have shown that IaC approaches are feasible to handle these infrastructure issues. A notable difference in the design of our platform is that these other platforms assume that individual users will themselves manually configure analyses- that they will choose relevant resources to support the analyses that they want to conduct, or compose an analysis of interest out of a set of small modular parts, in each case performing the work of a NeuroCAAS developer. While resulting analyses can be shared with other individuals on a case-by-case basis, this is very different from NeuroCAAS. Our platform is geared towards a heterogeneous community of researchers, where some researchers are developing general purpose analyses that only need to be configured or updated once before being used by a large group of potential users.
Next, we concretely demonstrate how the infrastructure benefits of the NeuroCAAS platform address ongoing challenges in neuroscience data analysis. We show two examples of NeuroCAAS native analyses that would not be feasible without the simultaneous benefits to accessibility, scale, and reproducibility that we provide.
2.4. NeuroCAAS Simplifies Large Data Pipelines: Widefield Imaging Protocol
Often, big data pipelines demand many individual preprocessing steps, creating the need for unwieldy multi-analysis infrastructure stacks— infrastructure stacks that support the needs of multiple core analyses at the same time. A notable example is widefield calcium imaging (WFCI)— a high-throughput imaging technique that can collect activity dependent fluorescence signals across the entire dorsal cortex of an awake, behaving mouse (Couto et al., 2021), potentially generating terabytes of data across chronic experiments. The protocol paper Couto et al. (2021) describes a complete WFCI analysis that links together cutting-edge data compression/denoising with demixing techniques designed explicitly for WFCI (via Penalized Matrix Decomposition, or PMD (Buchanan et al., 2018) and LocaNMF (Saxena et al., 2020), respectively). Each of these analyses depends upon its own specialized hardware and installation, creating many competing requirements on a multi-analysis infrastructure stack that are difficult to satisfy in practice. While we offer a NeuroCAAS implementation of the described WFCI analysis in Couto et al. (2021), we do not discuss how NeuroCAAS addresses the issue of multi-analysis infrastructure stacks, which can pose IaGS challenges even to our blueprint based infrastructure.
Instead of working with multi-analysis infrastructure stacks, for this analysis NeuroCAAS extends the function of a standard job manager (see Figure 5A) to trigger multiple jobs, built from separate blueprints, in sequence (Figure 5B). We employ this design to dramatically simplify the infrastructure requirements for a complete WFCI pipeline. First the initial steps of motion correction, denoising, and hemodynamic correction of the data are run from a blueprint that emphasizes multicore parallelism (64 CPU cores) to suit the matrix decomposition algorithms employed by PMD. Upon termination of this first step, analysis results are not only returned to user storage, but also used as inputs to a second job, performing demixing with LocaNMF on infrastructure supporting a high performance GPU. This modular organization improves the performance and efficiency of each analysis component (see Figure 7), and also allows users to run steps individually if desired, giving them the freedom to interleave existing analysis pipelines with the components offered here. As an alternative to the standard NeuroCAAS interface, this WFCI analysis can be controlled from a custom-built graphical user interface (GUI). This GUI further extends NeuroCAAS ‘s accessibility with features such as interactive alignment of a brain atlas to user data as part of parameter configuration (in the process validating input data as well). Following parameter configuration, this GUI interacts with NeuroCAAS programmatically, using locally run code scripts to perform data upload and job submission, and to detect and retrieve results once analysis is complete. Results can be visualized directly in this GUI as well. Altogether, this GUI can be used as a model for researchers who would like to take advantage of our computational infrastructure within a more sophisticated user interface, or integrate NeuroCAAS programmatically with other software tools. Importantly, despite its interactivity, the performance of our WFCI analysis does not depend on the infrastructure available to the user. For example, users could simultaneously launch many analyses and have them run in parallel through this GUI, easily conducting a hyperparameter search over their entire multi-step analysis. Researchers can find the GUI for this WFCI analysis with NeuroCAAS integration at https://github.com/jcouto/wfield.
Figure 5:
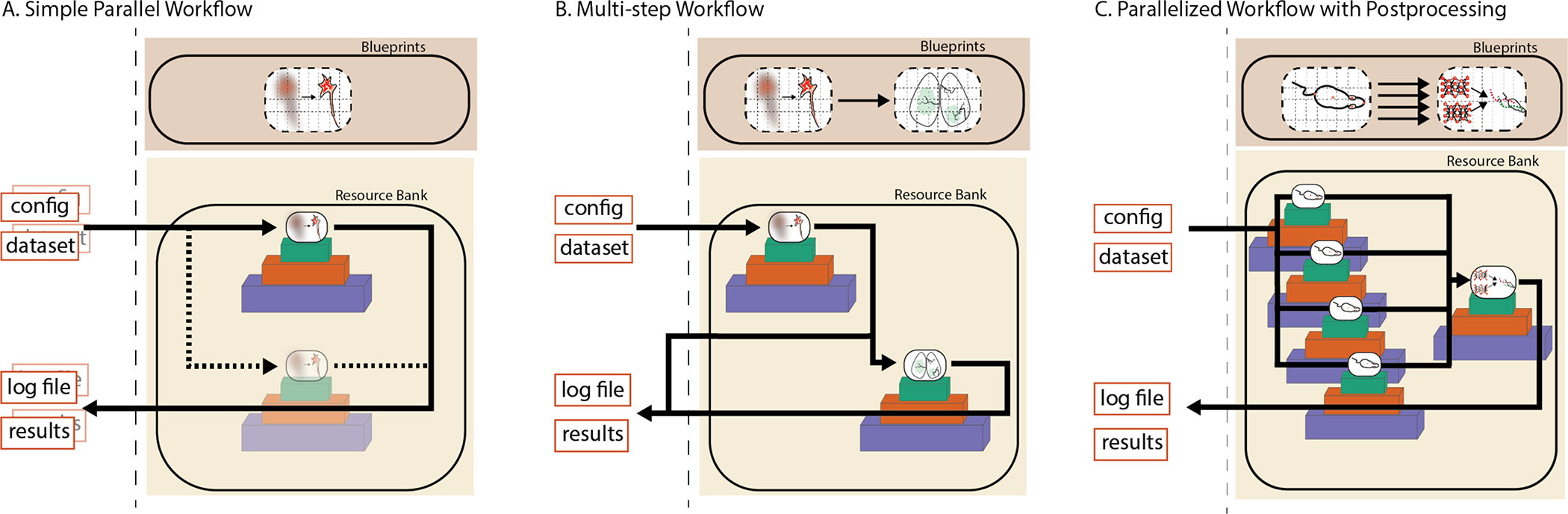
NeuroCAAS Supports Multi-Stack Design Patterns. A. Default workflow: If more than one dataset is submitted, NeuroCAAS automatically creates separate infrastructure for each. B. Chained workflow: Multiple analysis components with different infrastructure needs are seamlessly combined on demand. Intermediate results are returned to the user so that they can be examined and visualized as well (§2.4). C. Parallelism + chained workflow: Workflows A and B can also be combined to support batch processing pipelines with a separate postprocessing step (§2.5).
Figure 7: Quantitative Comparison of NeuroCAAS vs. Local Processing for Three Different Analyses.
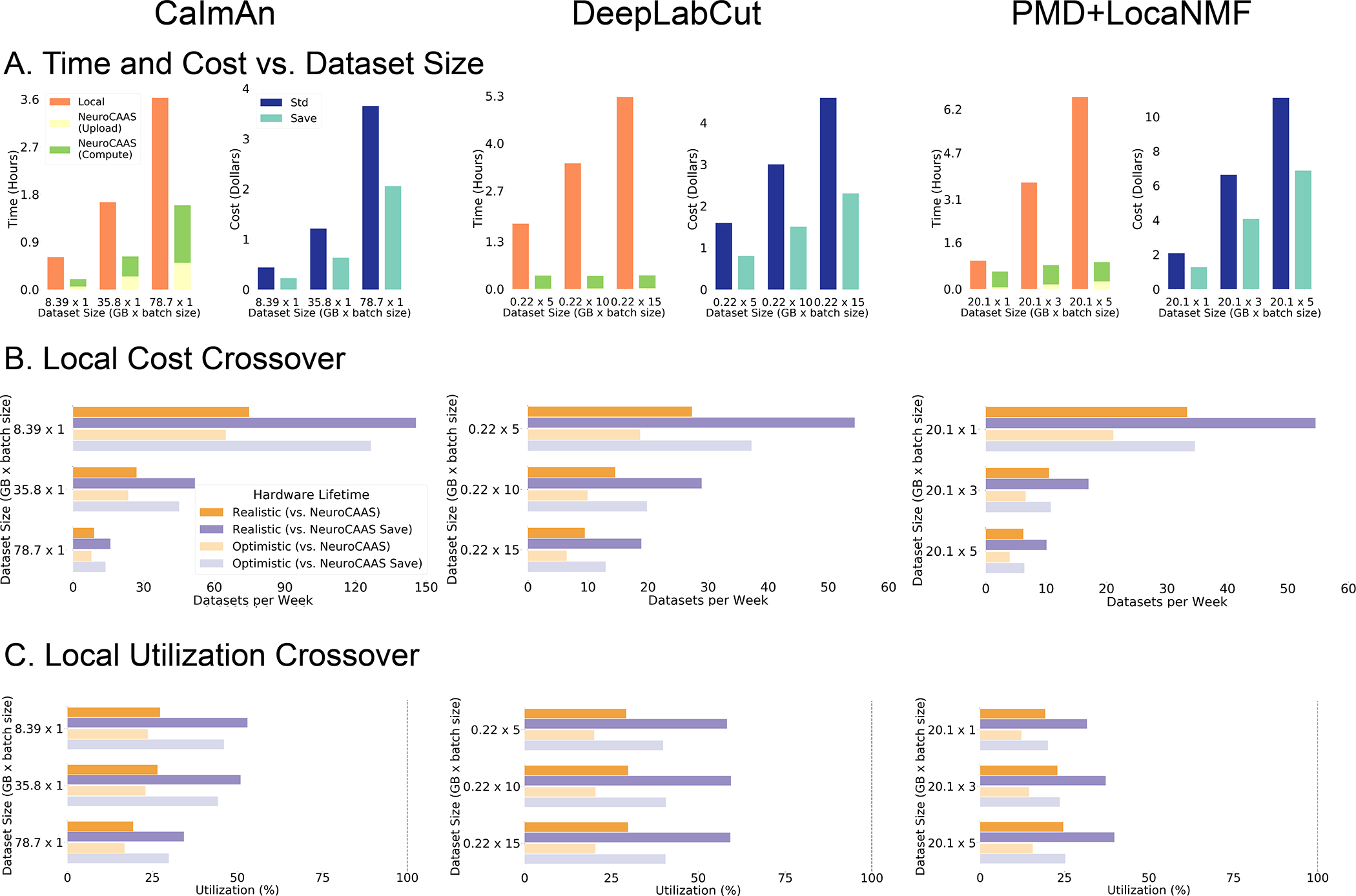
A. Simple quantifications of NeuroCAAS performance. Left graphs compare total processing time on NeuroCAAS vs. local infrastructure (orange). NeuroCAAS processing time is broken into two parts: Upload (yellow) and Compute (green). Right graphs quantify cost of analyzing data on NeuroCAAS with two different pricing schemes: Standard (dark blue) or Save (light blue). B. Cost comparison with local infrastructure (LCC). Figure compares local pricing against both Standard and Save prices, with Realistic (2 year) and Optimistic (4 year) lifecycle times for local hardware. C. Achieving Crossover Analysis Rates. Local Utilization Crossover gives the minimum utilization required to achieve crossover rates shown in B. Dashed vertical line indicates maximum feasible utilization rate at 100% (utilizing local infrastructure 24 hours, every day). See Figure S7 for cluster analysis, and Tables S4–S8 for supporting data.
For developers, this analysis presents a counterpart to existing domain specific projects such as CaImAn (Giovannucci et al., 2019) for cellular resolution calcium imaging or SpikeInterface (Buccino et al., 2020) for electrophysiology, which explicitly make multi-step data analyses compatible with optimized hardware. In contrast, our WFCI analysis is made directly available to users on powerful remote hardware without the need for anyone to revise existing analysis or infrastructure. To our knowledge, there is no other neuroscience platform with the accessibility, scale, and support for reproducibility to link together cutting-edge analyses across separate infrastructures, and make this exact configuration available directly to the research community.
2.5. NeuroCAAS Stabilizes Deep Learning Models: Ensemble Markerless Tracking
The black box nature of deep learning can generate sparse, difficult to detect errors that reduce the benefits of deep learning based tools in sensitive applications. For modern markerless tracking analyses built on deep neural networks (Mathis et al., 2018, Graving et al., 2019, Nilsson et al., 2020, Wu et al., 2020), these errors can manifest as “glitches” (Wu et al., 2020), where a marker point will jump to an incorrect location, often without registering as an error in the network’s generated likelihood metrics (see Figure 6).
Figure 6: Ensemble Markerless Tracking.
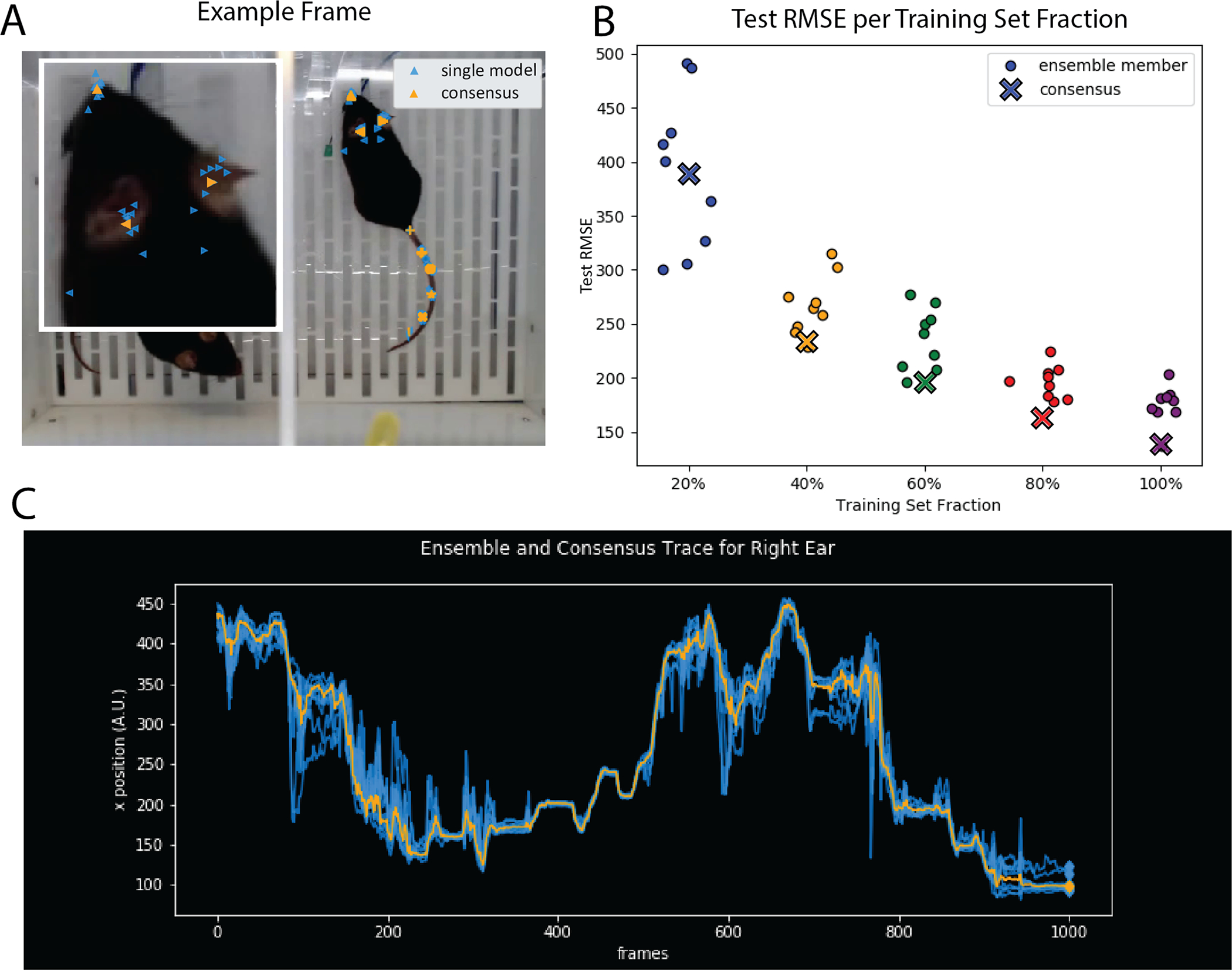
A. Example frame from mouse behavior dataset (courtesy of Erica Rodriguez and C. Daniel Salzman) tracking keypoints on the top down view of a mouse, as analyzed in Wu et al. (2020). Marker shapes track different body parts: blue markers representing the output of individual tracking models, and orange markers representing the consensus. Inset image shows tracking performance on the nose and ears of the mouse. B. consensus test performance vs. test performance of individual networks on a dataset with ground truth labels as measured via root mean squared error (RMSE). C. traces from 9 networks (blue) + consensus (orange). Across the entire figure, ensemble size = 9. A and C correspond to traces taken from the 100% split in B corresponding to 20 training frames.
One general purpose approach to combat the unreliable nature of individual machine learning models is ensembling (Dietterich, 2000): instead of working with a single model, a researcher simultaneously prepares multiple models on the same task, subsequently aggregating their outputs into a single consensus output. Ensemble methods have been shown to be effective for deep networks in a variety of contexts, (Lakshminarayanan et al., 2017, Fort et al., 2019, Ovadia et al., 2019), but they confer a massive infrastructure burden if run on limited local compute resources: researchers must simultaneously train, manage, and aggregate outputs across many different deep learning models, incurring either prohibitively large commitments to deep learning specific infrastructure and/or infeasibly long wait times.
In contrast, NeuroCAAS enables easy and routine implementation of ensemble methods. By modifying the NeuroCAAS job manager, we designed an analysis which takes input training data, and distributes it to N identical sets of IAEs and resource bank instances (Figure 5C). For the application shown here, we used an IAE with DeepGraphPose (Wu et al., 2020) as our core analysis; the N infrastructures differ only in the minibatch order of data used to train models. The results from each trained model are then used to produce a consensus tracking output, taking each individual model’s estimate of part location across the entire image (i.e. the confidence map output) and averaging these estimates. Even with this relatively simple approach, we find the consensus tracking output is robust to the errors made by individual models (Figure 6A,C). This consensus performance is maintained even when we significantly reduce the size of the training set (Figure 6B). Finally, in Figure 6C, we can see that there are portions of the dataset where the individual model detections fluctuate around the consensus detection. This fluctuation offers an empirical readout of tracking difficulty within any given dataset; frames with large diversity in the ensemble outputs are good candidates for further labeling, and could be easily incorporated in an active learning loop. After training, models can be kept in user storage, and used to analyze further behavioral data, without moving these models out of NeuroCAAS. Overall, Figure 6 shows that with the scale of infrastructure available on NeuroCAAS, ensembling can easily improve the robustness of markerless tracking, naturally complementing the infrastructure reproducibility provided by the platform.
NeuroCAAS is uniquely capable of providing the flexible infrastructure necessary to support a generally available, on-demand ensemble markerless tracking application. To our knowledge, none of the platforms with the scale to support markerless tracking on publicly available resources (e.g. on premise clusters, Google Colab, Galaxy Goecks et al. (2010), NSG (Sanielevici et al., 2018), Brainlife Avesani et al. (2019)) can satisfactorily alleviate the burden of a deep ensembling approach, still forcing the user to accept either long wait times or manual management of infrastructure. These limitations also prohibit use cases involving the quantification of ensemble behavior across different parameter settings (c.f. Figure 6B, where we trained 45 networks simultaneously).
2.6. NeuroCAAS is Faster and Cheaper than IaGS Analogues
NeuroCAAS offers a number of major advantages over IaGS : reproducibility, accessibility, and scale, whether we compare against a personal workstation or resources allocated from a locally available cluster. However, since NeuroCAAS is based on a cloud computing architecture, one might worry that data transfer times (i.e., uploading and downloading data to and from the cloud) could potentially lead to slower overall processing or that the cost of cloud compute could outweigh that of local infrastructure.
Figure 7 considers this question quantitatively, comparing NeuroCAAS to a simulated personal workstation (see §9.3 for details). For the analogous comparisons (with similar conclusions) against a simulated local cluster, see Figure S7. Figure 7 presents time and cost benchmark results on four popular analyses that cover a variety of data modalities: CaImAn (Giovannucci et al., 2019) for cellular resolution calcium imaging; DeepLabCut (DLC) (Mathis et al., 2018) for markerless tracking in behavioral videos; and a two-step analysis consisting of PMD (Buchanan et al., 2018) and LocaNMF (Saxena et al., 2020) for analysis of widefield imaging data. To be (extremely) conservative, we assume local infrastructure is set up, neglecting all of the time associated with installing and maintaining software and hardware.
Across all analyses and datasets considered in Figure 7, analyses run on NeuroCAAS were significantly faster than those run on the selected local infrastructure, even accounting for the time taken to stage data to the cloud (Figure 7A, left panes). We batched data to take advantage of both compute optimization offered by individual core analyses, and NeuroCAAS ‘s scale (see 9.3 for details). These examples show that many analyses can be used efficiently on NeuroCAAS regardless of the degree to which they have been intrinsically optimized for parallelism. Additionally NeuroCAAS upload time can be ignored if analyzing data that is already in a user storage — for example if there is a need to reprocess data with an updated algorithm or parameter setting — leading to further speedups. Finally, although we found download speeds negligible (see 9.3 for full details of timing quantifications) this could vary significantly based on user internet speeds and analyses. Across our platform, we have attempted to design analyses with much smaller outputs than input data- a point we will return to in the discussion (§3).
Next we turn to cost analyses. Over the range of algorithms and datasets considered here, we found that the total baseline NeuroCAAS analysis cost was on the order of a few US dollars per dataset (Figure 7A, right panels)- see Table S7 for pricing details. We observe that for the most part, costs are approximately linear in compute time, ensuring that even compute intensive operations like training a deep network for DLC (~12 hours on the same machines used here) can be accomplished for ~ $10-trained networks can be maintained in cloud storage to reduce data transfer cost. In addition to our baseline implementation, we also offer an option to run analyses at a significantly lower price (indicated as “Std” and “Save” respectively in the cost barplots in Figure 7), if the user can upper bound the expected runtime of their analysis to anything lower than 6 hours (i.e. from previous runs of similar data).
Finally, we compare the cost of NeuroCAAS directly to the cost of purchasing local infrastructure. We use a total cost of ownership (TCO) metric (Morey and Nambiar, 2009) that includes the purchase cost of local hardware, plus reasonable maintenance costs over estimates of hardware lifetime; see §9.3 for full details. We first ask how frequently one would have to run the analyses presented in Figure 7 before it becomes worthwhile to purchase dedicated local infrastructure. This question is answered by the Local Cost Crossover (LCC): the threshold weekly rate at which a user would have to analyze data for NeuroCAAS costs to exceed the TCO of local hardware. As an example, the top two bars of Figure 7B, left, show that in order for a local machine to be cost effective for CaImAn, one must analyze ~ 100 datasets of 8.39 GB per week, every week for several years (see Table S4 for a conversion to data dimensions). In all use cases, the LCC rates in Figure 7B show that a researcher would have to consistently analyze ~ 10–100 datasets per week for several years before it becomes cost effective to use local infrastructure. While such use cases are certainly feasible, managing these use cases on local infrastructure via IaGS would involve an significant amount of human labor.
In Figure 7C, we characterize this labor cost via the Local Utilization Crossover (LUC): the actual time cost of analyzing data on a local machine at the corresponding LCC rate. Across the analyses that we considered, local infrastructure would have to be dedicated to the indicated analysis for 25–50% of the infrastructure’s total lifetime (i.e. ~ 6–12 hours per day, every day) to achieve its corresponding LCC threshold, requiring an inordinate amount of work on the part of the researcher to manually run datasets, monitor analysis progress for errors, or build the computing infrastructure required to automate this process– in essence forcing researchers to perform by hand the large scale infrastructure management that NeuroCAAS achieves automatically. These calculations demonstrate that even without considering all of the IaGS issues that our solution avoids, or explicitly assigning a cost to researcher time, it is difficult to use local infrastructure more efficiently than NeuroCAAS for a variety of popular analyses. Given the diversity of IaGS solutions, we also provide a tool for users to benchmark their available infrastructure options against NeuroCAAS (see the instructions at https://github.com/cunningham-lab/neurocaas).
2.7. NeuroCAAS is Offered as a Free Service for Many Users
In many cases, researchers may use infrastructure available on hand to test out analyses before purchasing components for a dedicated infrastructure stack. Given the low per-dataset cost and the major advantages summarized above of NeuroCAAS compared to the current IaGS status quo, we have decided to mirror this model on the NeuroCAAS platform, and subsidize a large part of NeuroCAAS usage by the community. Users do not need to set up any billing information or worry about incurring any costs when starting work on NeuroCAAS; we cover all costs up to a per-user cap (initially set at $300). This subsidization removes one final friction point that might slow adoption of NeuroCAAS, and protects NeuroCAAS as a non-commercial open-source effort. Since NeuroCAAS is relatively inexpensive, many users will not hit the cap; thus, for these users, NeuroCAAS is offered as a free service. We note that we are also open to consider budget increases for researchers as they become necessary.
3. Discussion
NeuroCAAS integrates rigorous infrastructure practices into neural data analysis while also respecting current development and use practices. The fundamental choice made by NeuroCAAS is to provide analysis infrastructure with as much automation as possible. This choice naturally makes NeuroCAAS into a service, and in the simplest case neither analysis users nor analysis developers have to manage infrastructure directly; rather, NeuroCAAS removes the infrastructure burden entirely. However, as an open source project, NeuroCAAS also acknowledges the possibility that some users may want to accept some degree of responsibility for computing infrastructure, in return for a greater degree of flexibility in how they use the platform. We highlight two notable alternative use cases here:
Working at scale: large datasets/many jobs.
Although our drag-and-drop console removes the need for users to have previous experience with coding, some users may find the console restrictive when working with large datasets, or managing many jobs at once- both important facets of analysis use that NeuroCAAS is poised to improve. These restrictions can be reduced by working with NeuroCAAS from the command line, or by integrating calls to NeuroCAAS within locally run applications, as is done in §2.4. Since NeuroCAAS can be used solely by interacting with cloud storage, these interfaces to NeuroCAAS are easily supported by general purpose data transfer tools. We provide instructions for this use case in our developer documentation (see §9.4).
In order to streamline data transfer in cases where input or output data are unavoidably large, we have also implemented a “storage bypass” option for our CLI interface. Using this option, public data stored elsewhere in the AWS cloud can be analyzed, and results can be written back directly to this location without incurring additional data transfer time and costs, laying the groundwork for the integration of NeuroCAAS analyses with external data sources. This option is intended for analyses which handle especially large input or output data, where CLI use is preferable, but we plan to extend this functionality to all analyses and our standard interface soon. We believe these additional features will better equip NeuroCAAS to handle the ever increasing scale of neuroscience data (e.g. Steinmetz et al., 2021; Couto et al., 2021), as well as methods that consider multiple data modalities simultaneously (e.g. Batty et al., 2019), and faciliate sharing of analysis outputs across many users.
Working independently: private management of costs/compute resources.
A major benefit of NeuroCAAS ‘s IaC construction is that the entire platform (except private user data) can be reconstructed automatically given the code in the NeuroCAAS source repository (§9.2, Figure S5): there is no dependence of the platform upon specifics of infrastructure configuration that are not recorded in a blueprint. This benefit means that if users anticipate very high costs, or would like to use IaC management for their own custom analyses, it is easy for them to switch from using our public implementation of NeuroCAAS, to one that they pay for themselves, maintaining all the benefits of NeuroCAAS ‘s infrastructure management. We provide detailed instructions on this process in our developer documentation (§9.4), describing platform setup and cloning of individual analyses.
Finally, we revisit NeuroCAAS ‘s stated objectives of supporting reproducible, accessible, and scalable data analyses. These are fundamentally multifaceted issues, and will manifest in different ways across a variety of use cases. To this end, we identify strengths and limitations of NeuroCAAS ‘s approach to these issues (and related costs) so that researchers can evaluate the suitability of NeuroCAAS to their particular use case.
Reproducibility.
What are the benefits and limits of analysis reproducibility in NeuroCAAS? In section §2.2.1, we show that a dataset, configuration file, and analysis blueprint form a set of sufficient resources to reproduce an analysis output against a set of practically relevant interventions. We note some qualifications to this performance: First, it can be non-trivial to maintain a dataset across multiple analysis runs. Importantly, when data is uploaded it will not be versioned by default, creating the potential for it to be overwritten. For dataset provenance, we recommend data infrastructure projects like DANDI Dandi Team (2019). Integration with a data archive is an important future direction to extend reproducibility for NeuroCAAS. Second, we are limited by the inherent computational reproducibility of the core analysis we offer- for example, random computations can introduce significant differences from run to run (although ensemble methods can mitigate these issues). Finally, we can consider the lifecycle of different resources on the AWS cloud. For example, reproducibility could be affected if support for certain hardware instances become deprecated, and can no longer be used to run analyses. Given the large scale reliance of industrial applications on the AWS cloud, such events are very rare and announced well in advance, but we can take steps to address such a contingency. In particular, an important future direction is to consider how we can expand our approach outside of a particular cloud provider (see 9.2 for details).
Accessibility.
NeuroCAAS aims to improve the accessibility of data analysis by removing the need for users to independently configure infrastructure stacks, as is the de facto standard with IaGS approaches. By default, NeuroCAAS does not aim to improve other aspects of usability, such as the scientific use of core analysis algorithms. For example, if a user has data that is incorrectly formatted for a particular algorithm, the same error will happen with NeuroCAAS as it would with conventional usage, although curated deployments and blueprint based updates can significantly mitigate these issues.
Another approach towards achieving robust and general purpose analyses focuses on the explicit standardization of data formats and workflow. As mentioned, we plan to integrate with data archiving projects like DANDI (distributed archives for neurophysiology data integration) (Dandi Team, 2019) which enforces the NWB (Teeters et al., 2015, Rübel et al., 2019, Rübel et al., 2021) data standard, providing both a stable set of expectations for analysis developers, while also improving reproducibility of analysis results. Likewise, workflow management systems for neuroscience such as Datajoint (Yatsenko et al., 2015) or more general tools like snakemake (Koster and Rahmann, 2012) and the Common Workflow Language (Amstutz et al., 2016) codify the sequential steps that make up a data analysis on given infrastructure, ensuring data integrity and provenance. Other platforms both within (NeuroScout, 2022, Avesani et al., 2019) and outside of neuroscience (Seven Bridges, 2019) provide well-designed examples of how standardized data formats paired with workflow management systems can be used to make analyses more modular and easy to use.
Scale.
Although NeuroCAAS offers analyses at scale, it does not offer unstructured access to cloud computational resources. The concept of IAEs should clarify this fact: NeuroCAAS serves a set of analyses that are configured to a particular specification, as established by the analysis developer. This constraint is often ideal, since the specification is in many cases established by the analysis method’s original authors. Without specific structure to manage the near infinite scale of resources available on the cloud, the management of resources on the cloud easily becomes susceptible to the issues of IaGS that motivated the development of NeuroCAAS to begin with (Monajemi et al., 2019). The constraint of immutability distinguishes NeuroCAAS from interactive data analysis offerings that offer cloud computing like Pan-neuro (Rokem et al., 2021) or Google Colab, in keeping with their differing intended use cases. While interactive computing plays a key role in data analysis applications, we believe there is fundamental value in immutable data analyses as well.
Importantly, immmutability does not suggest that analyses on NeuroCAAS are a black box. All NeuroCAAS analyses are built from open source projects, the workflow scripts used to parse datasets and config files inside an IAE are made available to all analysis users, and jobs constantly print live status logs back to users. Furthermore, our novel analyses show that there are means of comprehensively characterizing analysis performance that only become available at scale (i.e. full parameter searches over a multi-step analysis, or ensembling to evaluate reliability of analysis outputs).
Cost.
The cost quantifications that we present in this manuscript are intended to demonstrate that the cost of using NeuroCAAS ‘s computing infrastructure is practically feasible when compared against the cost of computing on typical IaGS infrastructure. One point to note is that for individual research groups, the cost of using local infrastructure may vary significantly across institutions. Our quantifications are best fit to the case where a research group is supporting its own computing costs and resources. While the relative cost of using NeuroCAAS may thus differ from group to group, it is our hope that offering analyses at a uniform (and highly subsidized) cost will increase analysis accessibility to a significant portion of the neuroscience community, and potentially provide a more concrete understanding of the costs associated with the development and adoption of new analysis tools.
Beyond compute, we do not discuss the costs of storing and retrieving data from the cloud in depth. Without restricting data sizes on user storage, we found that data storage costs were small enough that we could support them without counting them towards user budgets. A common theme of the analyses that we discuss is that we can minimize data retrieval costs by designing workflows such that analysis results that the user actually needed to retrieve were far smaller than input data, specifically by modifying IAEs, and by maintaining large intermediate results on the cloud for use in future analyses. NeuroCAAS ‘s cost benefits may be reduced if these conditions are harder to achieve for a given analysis, although we believe that alternative use cases, such as our CLI interface with “storage bypass” are well poised to handle these contingencies, especially when paired with future directions such as integration with a data archive. Importantly, on private resources users will have to pay for cloud storage. This cost can be minimized by deleting input data and storing all results locally when not in use.
Beyond our proposed improvements above, NeuroCAAS will naturally continue to evolve by virtue of its open source code and public cloud construction. First, we hope to build a community of developers who will add more analysis algorithms to NeuroCAAS, with an emphasis on subfields of computational analysis that we do not yet support. Throughout this manuscript, we focus largely on analyses in systems neuroscience and neurophysiology, in accordance with the previous experience of the authors, and the opinion that analyses in this area are in great need of the platform design implemented by NeuroCAAS. We also plan to add support for real-time processing (e.g., Giovannucci et al. (2017) for calcium imaging, or Schweihoff et al. (2021), Kane et al. (2020) for closed-loop experiments, or Lopes et al. (2015) for the coordination of multiple data streams), using blueprint based methods to design fast, reliable infrastructure for closed loop analyses, in the same spirit as these batch mode analyses. Second, other tools have brought large-scale distributed computing to neural data analyses (Freeman, 2015, Rocklin, 2015) in ways that conform to more traditional high performance computing ideas of scalability for applications that are less easily parallelized than those presented here. Integrating more elaborate scaling into NeuroCAAS while maintaining development accessibility will be an important goal going forwards. Third, we aim to take inspiration from other computing platforms both within and beyond neuroscience to improve the usability of our platform, such as reporting the expected runtime and success rate of analyses Avesani et al. (2019), indicating the compatibility of different analysis steps in a sequence (Seven Bridges, 2019), or improving user and developer resources to include forums and full time support Goecks et al. (2010). We also aim to identify platforms and tools that could potentially be integrated with NeuroCAAS resources, in order to provide the infrastructure reliability that we prioritize. Finally, a major opportunity for future work is the integration of NeuroCAAS with modern visualization tools. We have emphasized above that immutable analysis environments on NeuroCAAS are designed with the ideal of fully automated data analyses in mind, because of the virtues that automation brings to data analyses. However, we recognize that for some of the core analyses on NeuroCAAS, and indeed most of those popular in the field, some user interaction is required to speed up analysis and optimize results. We have already demonstrated the compatibility of interactive interfaces with NeuroCAAS in our widefield calcium imaging analysis, and we will aim to establish a general purpose interface toolbox for developers in the same spirit, without sacrificing the benefits of cost efficiency, scalability, and reproducibility that distinguish NeuroCAAS in its current form.
Longer term, we hope to build a sustainable and open source user and developer community around NeuroCAAS. We welcome suggestions for improvements from users, and new analyses as well as extensions from interested developers, with the goal of creating a sustainable community-driven resource that will enable new large-scale neural data science in the decade to come.
9. STAR Methods
9.1. Resource Availability
Lead Contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, John P. Cunningham (jpc2181@columbia.edu)
Materials Availability
This study did not generate new unique reagents.
Data/Code Availability
Quantifications of performance and reproducibility of NeuroCAAS have been deposited at Zenodo and are publicly available as of the date of publication. DOIs are listed in the key resources table.
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
All other data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| Raw data used for the benchmarking of CaImAn (Giovannucci et al. 2019) | Zenodo | DOI: 10.5281/zenodo.1659149 |
| Performance quantification data used to report timing and cost of analyses on NeuroCAAS (related to Table 1, Figure 8) | Zenodo | DOI: 10.5281/zenodo.6512118 |
| Raw data used for to test WFCI analysis. | Cold Spring Harbor Repository | DOI: 10.14224/1.38599 |
| Software and Algorithms | ||
| Source repository used to build analyses from blueprints. | Zenodo | DOI: 10.5281/zenodo.6512118 |
| Contributor repository used to help developers add analyses to NeuroCAAS. | Zenodo | DOI: 10.5281/zenodo.6512121 |
| Interface repository used to build the website www.neurocaas.org | Zenodo | DOI: 10.5281/zenodo.6512125 |
| Repository used to generate ensemble outputs from individually trained models | Zenodo | DOI: 10.5281/zenodo.6513057 |
9.2. Method Details
NeuroCAAS architecture specifics
The software supporting the NeuroCAAS platform has been divided into three separate Github repositories. The first, https://github.com/cunningham-lab/neurocaas is the main repository that hosts the Infrastructure-as-Code implementation of NeuroCAAS. We will refer to this repository as the source repo throughout this section. The source repo is supported by two additional repositories: https://github.com/cunningham-lab/neurocaas_contrib hosts contribution tools to assist in the development and creation of new analyses on NeuroCAAS, and https://github.com/jjhbriggs/neurocaas_frontend hosts the website interface to NeuroCAAS. We will refer to these as the contrib repo and the interface repo respectively throughout this section. We discuss the relationship between these repositories in the following section, and in Figure S5.
Source Repo
Section 2.1 gives an overview of how NeuroCAAS encodes individual analyses into blueprints, and deploys them into full infrastructure stacks, following the principle of Infrastructure-as-Code (IaC). This section presents blueprints in more depth and show how the whole NeuroCAAS platform can be managed through IaC, encoding features such as user data storage, credentials, and logging infrastructure in code documents analogous to analysis blueprints as well. All of these code documents, together with code to deploy them, make up NeuroCAAS ‘s source repo. There is a one-to-one correspondence between NeuroCAAS ‘s source repo and infrastructure components: deploying the source repo provides total coverage of all the infrastructure needed to analyze data on NeuroCAAS (Figure S5, bottom). Although much of the code to translate blueprints and other infrastructure code necessarily references AWS resources, NeuroCAAS blueprints and other IaC artefacts are not tied to AWS, except in their reliance on particular hardware instance configurations. We can potentially recreate these hardware instances in other public clouds, using existing tools to support cloud-agnostic IaC approaches, as suggested by Brikman (2019). Doing so will further improve the scale and robustness of our platform.
Within the source repo, each NeuroCAAS blueprint (see Figure S2 for an example) is formatted as a JSON document with predefined fields. The expected values for most of these fields identify a particular cloud resource, such as the ID for an immutable analysis environment, or a hardware identifier to specify an instance within the resource bank (Lambda.LambdaConfig.AMI and Lambda.LambdaConfig.INSTANCE_TYPE in Figure S2, respectively). Upon deployment, these fields determine the creation of certain cloud resources: AWS EC2 Amazon Machine Images in the case of IAE IDs, and AWS EC2 Instances in the case of hardware identifiers. One notable exception is the protocol specifying behavior of a corresponding NeuroCAAS job manager (Lambda.CodeUri and Lambda.Handler in Figure S2). Instead of identifying a particular cloud resource, each blueprint’s protocol is a python module within the source repo that contains functions to execute tasks on the cloud in response to user input. The ability to specify protocols in python allows NeuroCAAS to support the complex workflows shown in Figure 5. Job managers are deployed from these protocols as AWS Lambda functions that execute the protocol code for a particular analysis whenever users submit data and parameters. Since all parts of NeuroCAAS workflow can be managed with python code (i.e. through a programmatic interface, job manager protocol, or within the IAE itself), external workflow management tools can easily be integrated to analyses on a case-by-case basis in order to deploy the scale of NeuroCAAS in parallel or sequentially, as needed.
Another major aspect of NeuroCAAS ‘s source repo that is not discussed in §2 is the management of individual users. NeuroCAAS applies the same IaC principles to user creation and management as it does to individual analyses. To add a new user to the platform, NeuroCAAS first creates a corresponding user profile in the source repo (Figure S5, right), that specifies user budgets, creates private data storage space, generates their (encrypted) security credentials, and identifies other users who they collaborate with. Users resources are created using the AWS Identity and Accesss Management (IAM) service.
Contrib and Interface Repos.
Given only the NeuroCAAS source repo, analyses can be hosted on the NeuroCAAS platform and new users can be added to the platform simply by deploying the relevant code documents. However, interacting directly with resources provided by the NeuroCAAS source repo can be challenging for both analysis users and developers. For developers, the steps required to fill in a new analysis blueprint may not be clear, and the scripting steps necessary within an IAE to retrieve user data and parameters requires knowledge of specific resources on the Amazon Web Services cloud. For users, the NeuroCAAS source repo on its own does not support an intuitive interface or analysis documentation, requiring users to interact with NeuroCAAS through generic cloud storage browsers, forcing them to engage in tedious tasks like navigating file storage and downloading logs before examining them. Collectively, these tasks lower the accessibility that is a key part of NeuroCAAS ‘s intended design. To handle these challenges, we created two additional code repositories, the NeuroCAAS contrib repo and interface repo, for developers and users, respectively.
The NeuroCAAS contrib repo supports a command line tool and python code to streamline the process of developing and creating new NeuroCAAS analyses. During the development process, the NeuroCAAS contrib repo can create infrastructure stacks independently of input-triggered job managers for a limited time, allowing developers to build and test IAEs interactively on powerful hardware instances in “debug mode” (Figure S5, bottom right), and populate the analysis blueprint as they go. Then, when a new analysis is ready to be used on NeuroCAAS, the NeuroCAAS contrib repo automatically versions the entire source repository after integrating and deploying the new blueprint, generating a unique analysis version ID. All NeuroCAAS analyses can be updated only by directly editing blueprints, and blueprints are assigned a new analysis version ID every time that they are updated. By enforcing a tight correspondence between blueprints and analyses, we ensured the reproducibility of all analyses conducted via NeuroCAAS, regardless of ongoing updates to the underlying infrastructure or algorithm (Figure S5, top right). With an analysis version ID, it is possible to replicate results that were generated with older versions of some analysis algorithm, making this a particularly useful feature for users processing data with an analysis that is still actively being developed. The NeuroCAAS developer documentation §9.4 contains a detailed guide for developers to get started with NeuroCAAS.
The NeuroCAAS interface repo supports the website interface to NeuroCAAS, hosted at www.neurocaas.org. In addition to providing documentation and a simpler user interface, (Figure S5, bottom left) the interface repo interacts with the source repo to automatically create and deploy user profiles when users sign up, significantly increasing the potential scale of the platform (Figure S5, top left). This website based user credentialing system can be referenced by other user interfaces as well, as is done in https://github.com/jcouto/wfield. If users wish to share analysis access and data with other users, they can also use the website to create and request unique “group codes” at sign up, that they can use to invite other users into the same group. Doing so allows them to easily share analysis access with others.
Developer Workflow
In this section we give more specific steps of how developers and authors built analyses for NeuroCAAS. See also Figure S6 for a corresponding schematic.
Flexible installation and scripting. Developers first install their core analysis into an IAE and hardware instance, just as they would with a local computer. Within an IAE any programming language can be supported, although certain precautions must be taken when working with licensed software such as Matlab (see developer documentation for more details https://neurocaas.readthedocs.io/en/latest/index.html). We provide tools to help developers write a workflow script to make their core analysis immutable (Figure S6). Post installation, the configured IAE is automatically saved in a blueprint (Figure S6, right).
Simple input/output handling. All NeuroCAAS analyses take a dataset file and a YAML formatted configuration file as input. Datasets can be parallelized over, and configuration files can specify any kind of parameter, including paths to supplemental data files (we provide an example of such a workflow in our custom analyses). NeuroCAAS handles transfer of data files from users into an IAE, and likewise writes any results back to users in timestamped folders that ensure version control for analysis outputs. All messages that would be printed to the IAE console during analysis are delivered back to the user in real time as a set of automatically formatted log files, along with job success/failure status messages, CPU usage, and memory usage. Developers can specify any files to be returned to the user, including custom logs, or intermediate results that would be useful to examine as analysis proceeds.
Testing with private data storage. Each user of a given analysis has a password protected account that authorizes them to interface with their own private cloud storage. Users have separated input and output areas in cloud storage, where they can maintain datasets for re-analysis, or keep intermediate analysis results as convenient. Although we have not capped the size of data we allow each user to store overall, we restrict both as follows: users cannot download data from input areas (although they can delete), and they cannot upload to output areas. These restrictions have distinct benefits for cost and reproducibility. In the later stages of testing, developers can upload test data and configuration files to private data storage exactly as a user would, and ensure that results and logging information appear as intended before releasing their analysis to the public. Developers can also set up selective access for designated test users before releasing the analysis to the general public. At this stage, we also work with developers to determine the optimal hardware instance type from the resource bank for their analysis and to determine if additional configuration of a custom job manager is necessary. We note, importantly that our current storage solution does not meet HIPAA standards and should not be used for sensitive health records.
Reproducible use and development By default, each analysis on NeuroCAAS provisions a single type of hardware for all data. However, if needed instances can be provisioned in a dataset-dependent manner, adjusting the size of storage volumes, memory, or other computing resources. These per-job changes are still recorded in versioned logs to ensure reproducibility of all jobs (see §2.2 for details). Such dataset dependent changes can be triggered by users, with the CLI interface, or programmed by developers through the job manager. In cases where users report bugs, analysis developers can then access the exact same IAE, resource bank instance, and inputs in interactive “debug mode” once again, making changes, and redeploying the blueprint exactly as they did in the initial deployment. Furthermore, if developers would like to continue updating their analysis, they can do so without impacting the reproducibility of existing results, because each NeuroCAAS job produces an analysis ID identifying a particular blueprint version.
Novel Analyses
For each novel analysis §2.4 §2.5, we provide details on its component infrastructure stacks, as well as details on relevant development outside the NeuroCAAS framework we have already presented.
Widefield Imaging.
The Widefield Calcium Imaging analysis that we present involves two independent infrastructure stacks, with the second taking as input the results of the first. The first infrastructure stack performs motion correction, denoising, compression, and hemodynamic correction, and is performed on an instance with 64 virtual cores (further infrastructure details are identical to the “PMD” row of Table S5). The second infrastructure stack performs demixing of denoised, corrected widefield imaging data, and is performed on an instance with a Tesla V100 GPU (further infrastructure details are identical to the “LocaNMF” row of Table S5). In addition to these two infrastructure stacks, we support a custom graphical user interface (available at https://github.com/jcouto/wfield). This user interface integrates with the credentials generated for users on the NeuroCAAS website, allowing users who have signed up via the website to use the GUI with an existing account. The GUI hosts a number of initialization steps on the user’s local machine, involving selection of parameters and alignment of data to landmarks on a given brain atlas. The GUI is also able to upload data directly to NeuroCAAS cloud storage, submit jobs, and monitor their progress. Next, the GUI is able to detect when the first step of processing is completed, and submits the relevant results files as input to the second step, mimicking the steps a user would take manually to manage this process. Finally, when all processing is complete the GUI retrieves analysis results back to the user’s local machine. For more details on implementation of each analysis step, please see Couto et al. (2021).
Ensemble Markerless Tracking.
The deep ensembling analysis that we present is also performed in two separate infrastructure stacks, but both the initial training and the consensus output generation steps are performed on the same IAE and resource bank instance. In both cases, we use an instance equipped with a Tesla V100 GPU, otherwise identical to the infrastructure shown in the DeepLabCut row of Table S5). We trained DeepGraphPose with the default training settings provided in the file run_dgp_demo.py within the core DeepGraphPose analysis code, on the “twomice-top-down” data from the DeepGraphPose paper (Wu et al., 2020). That paper provides full videos of analysis of this dataset using a single DeepGraphPose model. To enable ensembling, we built a separate set of ensembling tools that work with DeepGraphPose (Wu et al., 2020) - they can be found at https://github.com/cunningham-lab/neurocaas_ensembles. In order to create a consensus output, we averaged the confidence maps from each model in an ensemble in the following way: Assume a set of N trained DGP networks, ϕi, i∈1…N, and a video frame, . Assume that the network has been trained to track a single body part (the general case follows immediately), and take the scoremap outputs (unnormalized likelihoods) on this image from the output convolutional layer, denoted , where each scoremap . These scoremap outputs are unnormalized likelihoods representing the probability that the body part of interest is located in any individual pixel of the image. Then, we can compute the mean scoremap for a given image as:
| (1) |
Where S is the elementwise sigmoid function. The consensus output is then calculated from the softargmax function of this mean scoremap.
Furthermore, to calculate the rmse error, we use the following metric: Assume we have detections for all of the test frames in a video as a tensor, , with entries xtdc, where t represents the frame index, d the part index, and c the coordinate ∈[x, y]. Likewise, we have groundtruth data g with entries gtdc of the same dimension. Then the error is calculated as follows:
| (2) |
Details and implementation can be found in the repository https://github.com/cunningham-lab/neurocaas_ensembles, and the full analysis is available for use at http://neurocaas.org/analysis/14.
9.3. Quantification and Statistical Analysis
Quantifying reproducibility on NeuroCAAS
In order to quantify the reproducibility of analyses on NeuroCAAS, we selected two analyses already available on NeuroCAAS; CaImAn Giovannucci et al. (2019), and Ensemble DeepGraphPose (§2.5). We fixed in place the blueprint version for these analyses, as well as a dataset and configuration file, and compared the results of 15 independent runs for each of these analyses. These runs capture a variety of different real world interventions that can affect analysis reproducibility in practice. In particular, Runs 1–5 were performed by a paper author in the United States, Runs 6–10 were performed by a non-author researcher in India, and Runs 11–14 were performed by a non-author researcher in Switzerland. Within each of these sets of runs, we can test for the variation in analysis outputs over analysis runs conducted by the same researcher. Across these sets of runs, we can test for variation in analysis outputs over the physical location of the researcher performing experiments. Finally, for both analyses Run 15 was performed using an entirely separate instatiation of the NeuroCAAS platform, as described in the Discussion §3, and for which detailed instructions are provided in our developer documentation. This run ensures that there is no dependence of our analysis results on the particular set of infrastructure reosurces used to build and implement NeuroCAAS analyses blueprints.
In the absence of a generic metric to compare analysis outputs, we chose specific metrics for each analysis.
CaImAn.
We benchmarked CaImAn on the YST dataset introduced in the CaImAn paper Giovannucci et al. (2019). To compare the outputs of CaImAn, we independently looked at the differences between the spatial components (referred to as the “A” matrix in the CaImAn software package) and the temporal components (referred to as the “C” matrix in the CaImAn software package) found by the system independently. For both temporal and spatial components, we assumed a deterministic ordering of components across runs. We compared components pairwise, and reported average quantities across paired components. To determine the variation of spatial components across runs, we quantified the Jaccard similarity coefficient pairwise between detected components, as has been done previously in Giovannucci et al. (2017). In order to compare the variation of temporal components across runs, we quantified the root mean square error between paired temporal components. In both cases, we found no detectable variation across runs.
Ensemble DeepGraphPose.
We benchmarked Ensemble DeepGraphPose on the “twomice-top-down” data described in the methods above with the introduction of Ensemble Markerless Tracking. We compared the outputs of each run as the time series describing the temporal evolution of each individual body part. We quantified differences between two runs, x1, x2 as the root mean squared error between time series describing the evolution of each individual body part, averaged overall body parts. RMSE has previously been used in the literature to quantify the similarity between the tracked positions of behavioral markers, as in Mathis et al. (2018); Wu et al. (2020). We expect this variation to come from non-deterministic computations used to speed up the computation of convolutions employed in the relevant behavioral tracking model.
Timings.
As a separate but related question, we also quantified the variation in the time taken to analyze data on NeuroCAAS across these same 15 runs. We quantified timings as follows:
The Setup Time of an analysis was calculated as the time between the moment when a job was submitted to user storage (measured as the upload timestamp associated with a job’s “submit.json” file) and when the job was marked as started in the corresponding “certificate.txt” log in user storage (reported as a delta following the statement “JOB MONITOR LOG COMPLETE”.)
The Analysis Time was calculated as the time between the end of a job’s Setup Time and the time when the last analysis output of a particular run is uploaded to user storage (measured using upload timestamps of those analysis results).
The Shutdown Time of an analysis is the time between the end of a job’s Analysis Time and the time when all resources associated with the job have been stopped. It is measured based upon the time when a job end marker file is written to user storage (called “end.txt”).
The total timing of an analysis job is the sum of these three contributions. In practice, we care most about variation in the first two components, as these are most relevant to a user’s experience of NeuroCAAS.
Quantifying usage
By default, NeuroCAAS records metadata for each job that is requested, such as the requester of the job, the time at which it was requested, and the datasets and config file that were analyzed. These quantities are necessary to enable per-user budgeting.
We aggregated this per-user metadata in order to generate the usage statistics shown in Figure 3. Aggregation and analysis of user data is now offered to developers through the contrib repo described above, and the data for Figure 3 panels were generated by running the command:
$ neurocaas-contrib monitor visualize-parallelism
This command is included with the developer CLI, which lists the usage of a single analysis bucket. We ran this command for the following analyses:
epi-ncap-web
dlc-ncap-web
pmd-ncap-web
caiman-ncap-web
locanmf-ncap-web
bardensr
dlc-ncap-stable
caiman-ncap-stable
polleuxmonitored
carceamonitored
wfield-preprocess
yass-ncap-stable
one-photon-compress
one-photon-demix
one-photon-mcorr
dgp-refactor
ensemble-dgp
label-job-create-web
We then iterated through all of these logs, and aggregated information across all platform usage for these analyses, using metadata for individual jobs to group results by user, by parallelism, and by developer. For each aggregated collection of jobs, we extracted the total corresponding number of compute hours as well, using the script figures/parallelized.py included in the contrib repo. Critically, here we exclude usage by NeuroCAAS team members testing analyses for deployment, which is run from the user accounts ”reviewers”,“debuggers,“examplegroup2”. In rare cases where metadata logging failed, we excluded the corresponding analysis from this quantification.
Benchmarking algorithms on NeuroCAAS
For each analysis currently on NeuroCAAS, the specific infrastructure choices in the corresponding blueprint (Figure S5, right) are given in Table S5. To meaningfully benchmark NeuroCAAS against current standards, we simulated corresponding local infrastructure. Local infrastructure was also built on AWS, and spans resources comparable to personal hardware and cluster compute, depending on the use case (see Table S6). As a general guideline, we chose local infrastructure representatives that would reasonably be available to a typical researcher, unless the datasets we considered required more powerful resources. To account for the diversity of resources available to neuroscience users, we offer alternative quantifications to those presented in Figure 7 in the supplementary methods (see Figure S7), and make performance quantification data and calculations available to users who would like to compare to their own infrastructure through a custom tool on our project repository (see README: https://github.com/cunningham-lab/neurocaas).
For each analysis that we benchmarked on NeuroCAAS, we chose three datasets of increasing size as representative use cases of the algorithms in question. The size differences of these datasets reflect the diversity of potential use cases among different users of the same algorithm. The CaImAn benchmarking data consists of datasets N.02.00, J_123, J_115 from the data shared with the CaImAn paper (Giovannucci et al., 2019). Benchmark analysis is based on a script provided to regenerate Figure 7 of the CaImAn paper. Note that although this data could be batched, we choose to maintain all three datasets as contiguous wholes. We took advantage of the fact that the algorithm was built to parallelize across multiple cores of the same machine, and chose hardware to make effective use of this implementation across data sizes (for details see Giovannucci et al. (2019), Figure 8). DeepLabCut benchmarking data consists of behavioral video capturing social interactions between two mice in their home cage. Data is provided courtesy of Robert C. Froemke and Ioana Carcea, as analyzed and presented in Carcea et al. (2019). Data processing consisted of analyzing these videos with a model that had previously been trained on other images from the same dataset. The same dataset was used to benchmark PMD and LocaNMF as a single analysis pipeline with two discrete parts. Input data consist of the dataset (“mSM30”), comprising widefield calcium imaging data videos, provided courtesy of Simon Musall and Anne Churchland, as used in Musall et al. (2019) and Saxena et al. (2020). The full dataset is available in a denoised format at http://repository.cshl.edu/id/eprint/38599/. Data processing on NeuroCAAS consisted of first processing the raw videos with PMD, then passing the resulting output to LocaNMF. For DLC and PMD+LocaNMF, the NeuroCAAS compute time was effectively constant across increasing total dataset size, as we assumed data was evenly batched into subsets of approximately equal size and each batch was analyzed in its own independent infrastructure stack (as in Figure 5A).
Further details on the datasets used can be found in Table S4.
We split the time taken to run analyses on NeuroCAAS into two separate quantities. First, we quantified the time taken to upload data from local machines to NeuroCAAS, denoted as NeuroCAAS (Upload) in Figure 7. This time depends upon the specifics of the internet connection that is being used. It is also a one time cost: once data is uploaded to NeuroCAAS, it can be reanalyzed many times without incurring this cost again. Upload times were measured from the same NeuroCAAS interface made available to the user. (This upload time was skipped in the quantification of local processing time.) Second, we automatically quantified the total time elapsed between job submission and job termination, when results have been delivered back to the end user in the NeuroCAAS interface (denoted as NeuroCAAS (Compute) in Figure 7) via AWS native tools (see Supplemental Information for details, and use of this data for Figure 3). Finally, we do not include time taken to download data back to a local machine in these quantifications because we found that this time was negligible across all analyses that we considered- at most ~ 2 minutes, and in most cases on the order of a few seconds. Local timings were measured on automated portions of workflow in the same manner as NeuroCAAS (Compute).
We quantified the cost of running analysis on NeuroCAAS by enumerating costs of each of the AWS resources used in the course of a single analysis. Costs can be found in Table S7. We provide the raw quantification data and corresponding prices in Table S7. To further reduce costs, we also offer the option to utilize AWS Spot Instances (dedicated duration); these are functionally identical to standard compute instances, but are provisioned for a pre-determined amount of time with the benefit of significantly reduced prices. We provide the estimated cost of running analyses with both of these options in Figure 7, with quantifications labeled “NeuroCAAS Save” corresponding to analyses run with dedicated duration spot instances, and those labeled “NeuroCAAS Std” corresponding to those run with standard instances. For more on Spot Instance price quantification, see Supplemental Information.
With simulated local infrastructures on AWS in hand, costs were calculated by pricing analogous computing resources as if the user had purchased them for a personal workstation, or as if they had been allocated to the user on an on-premises cluster (Table S8, https://calculator.aws/). In Figure 7, we assume that the local infrastructures considered are hosted on typical local laptop or desktop computing resources, supplemented with the resources necessary to run analyses as they were done on NeuroCAAS (additional storage, memory, GPU, etc), while maintaining approximate parity in processor power. We referred to (Morey and Nambiar, 2009) to convert pricetag costs of local machines to Equivalent Annual Costs, i.e. the effective cost per year if we assume our local machines will remain in service for a given number of years, as our implementation of a TCO calculation (as is often done in industry). Given a price tag cost xlocal, an assumed lifetime n, an annuity rate r, and cs (n) defined as the estimated annual cost of local machine support given a lifetime n, we follow Mahvi and Zarfaty (2009), Morey and Nambiar (2009) in calculating the Equivalent Annual Cost as:
Here cs (n) is provided in the cited paper (Morey and Nambiar, 2009), estimated from representative data across many different industries. The denominator of the first term is an annuity factor. We consider two different values for n, which we label as “realistic” (2 years) and “optimistic” (4 years) in the text. In industry, 3–4 years is the generally accepted optimal lifespan for computers, after which support costs outweigh the value of maintaining an old machine (, Mahvi and Zarfaty, 2009, Morey and Nambiar, 2009). Some have argued that with more modern hardware, the optimal refresh cycle has shortened to 2 years (J.Gold Associates LLC, 2014). By providing quantifications assuming two and four year refresh cycle, we consider the short and long end of this generally discussed optimal range.
Given a per-dataset NeuroCAAS cost xNeuroCAAS, we further derive the Local Cost Crossover (LCC), the threshold weekly data analysis rate at which it becomes cost-effective to buy a local machine. The LCC is given by:
Furthermore, given the per-dataset local analysis time, we can estimate the corresponding Local Utilization Crossover (LUC). The LUC considers the LCC in the context of the maximal achievable data analysis rate on local infrastructure as calculated in the previous section. If the time taken to analyze a dataset on a local machine is given by tlocal (in seconds), The LUC is given by:
Survey of Analyses and Platforms
We characterized data analysis infrastructure stacks as consisting of three hierarchical parts (Dependencies, System, Hardware), segmented consistently with infrastructure descriptions referenced elsewhere (Demchenko et al., 2013, Zhou et al., 2016). In several different subfields of neuroscience, we then selected 10 recent or prominent analysis techniques, and asked how they fulfilled each component of data analysis infrastructure to generate Figure 1D. We denoted a particular infrastructure component as supported if it is referenced in the relevant installation and usage guides as being provided in a reliable, automated manner (e.g., automatic package installation via pip), offering a conservative estimate of lack of infrastructure support. Survey details are provided in Tables S1, S2. We addressed the question of how data analyses are installed and used with these surveys in the tradition of the open source usability literature. Surveys such as these are standard methodology in this field, which relies upon empirical data from studies of user’s usage habits (Nichols et al., 2001, Zhao and Deek, 2005), developer sentiment (Terry et al., 2010), and observation of user-developer interactions via platforms like Github (Cheng and Guo, 2018).
To generate Figure 4, we first quantified the traffic and infrastructure experienced by individual analyses by examining their Github pages, and taking the maximum of the number of forks, stars, and watchers, as well as the listed hardware requirements of each analysis (numbers as of September 2020). We then overlaid several exemplar platforms based on the analyses that they supported, as well as restrictions based on the accessibility and scale requirements imposed by each (local hardware, limitation to one analysis at a time), taking care to include analyses that the platforms supported in practice.
9.4. Additional Resources
Standard interface for users to work with analyses on NeuroCAAS : www.neurocaas.org
-
Documentation for developer workflow (and CLI usage):
https://neurocaas.readthedocs.io/en/latest/develop/installation.html
Supplementary Material
Table 1: Quantifying reproducibility via output comparisons for two analyses on NeuroCAAS.
For CaImAn, (an algorithm to analyze calcium imaging data) we independently characterized differences in the spatial and temporal components recovered by the model. Differences in spatial components are measured by the average Jaccard Distance over pairs of spatial components. A Jaccard distance of 0 corresponds to two spatial components that perfectly overlap. Differences in temporal components were calculated as the average root mean squared error (RMSE) taken over paired time series of component activity. For Ensemble DeepGraphPose (an algorithm to track body parts of animals during behavior from video), we considered multiple sets of outputs from a single, pretrained model. RMSE takes units of pixels, so differences of order 1e-8 are not relevant for behavioral quantification. For both analyses, we fixed a single dataset, configuration file and blueprint across runs. See Figures S3,S4 for more.
| Reference Run Output | vs. Run 2 | vs. Run 5 | vs. Run 10 | vs. Run 14 | |
|---|---|---|---|---|---|
| Analysis | (Comparison Metric) | (US) | (India) | (Switzerland) | (Platform Clone) |
| Spatial Components | 0.0 | 0.0 | 0.0 | 0.0 | |
| CaImAn | (Jaccard Distance) | ||||
| (Giovannucci et al., 2019) | Temporal Components | 0.0 | 0.0 | 0.0 | 0.0 |
| (RMSE) | |||||
| Ensemble | Body Part Traces | 1.2e-8 | 1.2e-8 | 2.3e-8 | 1.4e-8 |
| DeepGraphPose (§2.5) | (RMSE) |
Highlights:
The NeuroCAAS platform provides reprodicible data analysis infrastructure at scale
Reproducible, cloud based infrastructure enables novel analysis design
Popular data analyses adapted to NeuroCAAS are faster and cheaper than alternatives
4. Acknowledgements
We thank Ioana Carcea and Robert C. Froemke for the use of benchmarking data for DeepLabCut, Simon Musall and Anne Churchland for the use of benchmarking data for Penalized Matrix Decomposition and LocaNMF, and Erica Rodriguez and C. Daniel Salzman for the use of data to demonstrate ensemble markerless tracking. We thank Larry Abbott for feedback and support regarding the goals and usage of our platform, Danil Tyulmankov for useful references and discussions of benchmarking, Dan Biderman and Michelle Stackmann for paper comments, and Jian Wang and Yakov Stern for helpful discussions on NeuroCAAS ‘s architecture and extensions. We thank Dmitri Yatsenko, Edgar Walker, Raphael Guzman, Thinh Nguyen, and other members of the Datajoint team, as well as Ben Dichter, Alessio Buccino and other members of the CatalystNeuro and DANDI teams for discussions of how our platform might integrate with others, as well as general advice. We thank Anjali Agarwal and Catalin Mitelut for submitting neurocaas jobs as independent researchers to study reproducibility. We thank Peter Lee, Jackson Loper, Shuonan Chen, and Nick Greenspan for development of additional algorithms early in development and for help prototyping relevant extensions for job managers, Selmaan Chettih for suggesting ensembling of pose tracking, and Zahra Adahman, Ioana Carcea, Claire Everett, Andres Bendesky, Andres Villegas, Franck Polleux, Vivek Athalye, Darcy Peterka, and Avner Wallach for discussion and feedback on the use of NeuroCAAS during development, as well as all users and developers during alpha testing. T.A. is supported by NIH training grant 2T32NS064929-11, NSF DBI-1707398, and the Gatsby Charitable Foundation (Gatsby Charitable Foundation GAT3708). S.S. is supported by the Swiss National Science Foundation P2SKP2_178197 and 5U19NS104649. E.K.B. is supported by NIH training grant T32NS064929, NIH U19NS107613, NSF DBI-1707398, and the Gatsby Charitable Foundation (Gatsby Charitable Foundation GAT3708). J.P.C. is supported by Simons 542963. L.P. is funded by IARPA MICRONS D16PC00003, NIH 5U01NS103489, 5U19NS107613, 1UF1NS107696, 1UF1NS108213, 1RF1MH120680, DARPA NESD N66001-17-C-4002, 1U19NS123716-01 and Simons Foundation 543023. L.P. and J.P.C. are supported by NSF Neuronex Award DBI-1707398 and NIH 5U19NS104649.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M et al. (2016), Tensorflow: A system for large-scale machine learning, in ‘12th Symposium on Operating Systems Design and Implementation (OSDI 16)’, pp. 265–283. [Google Scholar]
- Aguiar A, Díaz J, Almaraz R, Pérez J and Garbajosa J (2018), DevOps in Practice – An Exploratory Case Study, in ‘Proceedings of the 19th International Conference on Agile Software Development: Companion, XP ‘18’, pp. 1–3. [Google Scholar]
- Amezquita RA, Lun AT, Becht E, Carey VJ, Carpp LN, Geistlinger L, Marini F, Rue-Albrecht K, Risso D, Soneson C et al. (2020), ‘Orchestrating single-cell analysis with bioconductor’, Nature methods 17(2), 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amstutz P, Crusoe MR, Tijanic N. s., Chapman B, Chilton J, Heuer M, Kartashov A, Kern J, Leehr D, Menager H, Nedeljkovich M, Scales M, Soiland-Reyes S and Stojanovic L (2016), ‘Common Workflow Language, v1.0’. 10.6084/m9.figshare.3115156.v2 [DOI]
- Avesani P, McPherson B, Hayashi S, Caiafa CF, Henschel R, Garyfallidis E, Kitchell L, Bullock D, Patterson A, Olivetti E et al. (2019), ‘The open diffusion data derivatives, brain data upcycling via integrated publishing of derivatives and reproducible open cloud services’, Scientific data 6(1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batty E, Merel J, Brackbill N, Heitman A, Sher A, Litke A, Chichilnisky E and Paninski L (2016), ‘Multilayer recurrent network models of primate retinal ganglion cell responses’, International Conference on Learning Representations. [Google Scholar]
- Batty E, Whiteway M, Saxena S, Biderman D, Abe T, Musall S, Gillis W, Markowitz J, Churchland A, Cunningham JP et al. (2019), BehaveNet: nonlinear embedding and Bayesian neural decoding of behavioral videos, in ‘Advances in Neural Information Processing Systems’, pp. 15680–15691. [Google Scholar]
- Bittner SR, Palmigiano A, Piet AT, Duan CA, Brody CD, Miller KD and Cunningham JP (2019), ‘Interrogating theoretical models of neural computation with deep inference’, bioRxiv p. 837567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch J (2008), Effective java (the java series), Prentice Hall PTR. [Google Scholar]
- Brikman Y (2019), Terraform: up and running: writing infrastructure as code, O’Reilly Media. [Google Scholar]
- Buccino AP, Hurwitz CL, Garcia S, Magland J, Siegle JH, Hurwitz R and Hennig MH (2020), ‘SpikeInterface, a unified framework for spike sorting’, eLife 9, e61834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan EK, Kinsella I, Zhou D, Zhu R, Zhou P, Gerhard F, Ferrante J, Ma Y, Kim S, Shaik M et al. (2018), ‘Penalized matrix decomposition for denoising, compression, and improved demixing of functional imaging data’, arXiv preprint arXiv:1807.06203. [Google Scholar]
- Buckheit JB and Donoho DL (1995), Wavelab and reproducible research, in ‘Wavelets and statistics’, Springer, pp. 55–81. [Google Scholar]
- Business Intelligence (2004), Pilot Study: Optimum Refresh Cycle and Method for Desktop Outsourcing, Technical report, Intel Business Center. [Google Scholar]
- Carcea I, Caraballo NL, Marlin BJ, Ooyama R, Navarro JMM, Opendak M, Diaz VE, Schuster L, Torres MIA, Lethin H et al. (2019), ‘Oxytocin Neurons Enable Social Transmission of Maternal Behavior’, bioRxiv p. 845495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J et al. (2006), ‘CellProfiler: image analysis software for identifying and quantifying cell phenotypes’, Genome biology 7(10), R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver JC, Gesing S, Katz DS, Ram K and Weber N (2018), ‘Conceptualization of a us research software sustainability institute (URSSI)’, Computing in Science \and Engineering 20(3), 4–9. [Google Scholar]
- Chan Zuckerberg Initiative (2019), ‘Essential Open Source Software for Science (EOSS) - Chan Zuckerberg Initiative’. https://chanzuckerberg.com/eoss/
- Chaumont FD, Dallongeville S, Chenouard N, Herve N, Pop S, Provoost T, Meas-Yedid V, Pankajakshan P, Lecomte T, Montagner YL et al. (2012), ‘Icy: an open bioimage informatics platform for extended reproducible research’, Nature methods 9(7), 690. [DOI] [PubMed] [Google Scholar]
- Chen S, Loper J, Chen X, Zador T and Paninski L (2020), ‘BARcode DEmixing through Non-negative Spatial Regression (BarDensr)’, bioRxiv p. 2020.08.17.253666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Dallmeier-Tiessen S, Dasler R, Feger S, Fokianos P, Gonzalez JB, Hirvonsalo H, Kousidis D, Lavasa A, Mele S et al. (2019), ‘Open is not enough’, Nature Physics 15(2), 113–119. [Google Scholar]
- Cheng J and Guo JL (2018), How do the open source communities address usability and ux issues?: An exploratory study, in ‘Extended Abstracts of the 2018 CHI Conference on Human Factors in Computing Systems’, p. LBW523. [Google Scholar]
- Couto J, Musall S, Sun XR, Khanal A, Gluf S, Saxena S, Kinsella I, Abe T, Cunningham JP, Paninski L and Churchland AK (2021), ‘Chronic, cortex-wide imaging of specific cell populations during behavior’, Nature Protocols 16(7), 3241–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook SM, Davison AP and Plesser HE (2013), Learning from the past: approaches for reproducibility in computational neuroscience, in ‘20 Years of Computational Neuroscience’, Springer, pp. 73–102. [Google Scholar]
- Dandi Team (2019), ‘Dandi Archive’. https://www.dandiarchive.org/
- Demchenko Y, Grosso P, Laat CD and Membrey P (2013), Addressing big data issues in scientific data infrastructure, in ‘2013 International Conference on Collaboration Technologies and Systems (CTS)’, pp. 48–55. [Google Scholar]
- Dietterich TG (2000), ‘Multiple Classifier Systems, First International Workshop, MCS 2000 Cagliari, Italy, June 21–23, 2000 Proceedings’, Lecture Notes in Computer Science pp. 1–15. [Google Scholar]
- Donoho DL (2010), ‘An invitation to reproducible computational research’, Biostatistics 11(3), 385–388. [DOI] [PubMed] [Google Scholar]
- Editorial (2014), ‘Code share : Nature News and Comment’. https://www.nature.com/news/code-share-1.16232
- Flywheel Exchange (2019), ‘Flywheel: Informatics Platform for Biomedical Research and Collaboration’. https://flywheel.io/
- Fort S, Hu H and Lakshminarayanan B (2019), ‘Deep Ensembles: A Loss Landscape Perspective’, arXiv preprint arXiv:1912.02757. [Google Scholar]
- Freeman J (2015), ‘Open source tools for large-scale neuroscience’, Current opinion in neurobiology 32, 156–163. [DOI] [PubMed] [Google Scholar]
- Gao Y, Archer EW, Paninski L and Cunningham JP (2016), Linear dynamical neural population models through nonlinear embeddings, in ‘Advances in neural information processing systems’, pp. 163–171. [Google Scholar]
- Genomics, S. B. (2019), ‘The seven bridges platform. (retrieved february 28,2022)’. https://www.sbgenomics.com/
- Ghosh SS, Poline J-B, Keator DB, Halchenko YO, Thomas AG, Kessler DA and Kennedy DN (2017), ‘A very simple, re-executable neuroimaging publication’, F1000Research 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci A, Friedrich J, Gunn P, Kalfon J, Brown BL, Koay SA, Taxidis J, Najafi F, Gauthier JL, Zhou P et al. (2019), ‘CaImAn an open source tool for scalable calcium imaging data analysis’, Elife 8, e38173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci A, Friedrich J, Kaufman M, Churchland A, Chklovskii D, Paninski L and Pnevmatikakis EA (2017), Onacid: Online analysis of calcium imaging data in real time, in ‘Advances in neural information processing systems’, pp. 2381–2391. [Google Scholar]
- Glatard T, Lewis LB, Silva R. F. d., Adalat R, Beck N, Lepage C, Rioux P, Rousseau M-E, Sherif T, Deelman E, Khalili-Mahani N and Evans AC (2015), ‘Reproducibility of neuroimaging analyses across operating systems’, Frontiers in Neuroinformatics 9, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A and Taylor J (2010), ‘Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences’, Genome biology 11(8), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman DFM and Brette R (2009), ‘The Brian Simulator’, Frontiers in Neuroscience 3(2), 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski K, Burns CD, Madison C, Clark D, Halchenko YO, Waskom ML and Ghosh SS (2011), ‘Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in python’, Frontiers in neuroinformatics 5, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski KJ, Alfaro-Almagro F, Auer T, Bellec P, Capota M, Chakravarty MM, Churchill NW, Cohen AL, Craddock RC, Devenyi GA et al. (2017), ‘BIDS apps: Improving ease of use, accessibility, and reproducibility of neuroimaging data analysis methods’, PLoS computational biology 13(3), e1005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graving JM, Chae D, Naik H, Li L, Koger B, Costelloe BR and Couzin ID (2019), ‘DeepPoseKit, a software toolkit for fast and robust animal pose estimation using deep learning’, eLife 8, e47994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson B, Sugden A and Alberts B (2011), ‘Making data maximally available’, Science 331(6018), 649. [DOI] [PubMed] [Google Scholar]
- Hinsen K (2015), ‘Technical Debt in Computational Science’, Computing in Science and Engineering 17(6), 103–107. [Google Scholar]
- Hoffa C, Mehta G, Freeman T, Deelman E, Keahey K, Berriman B and Good J (2008), On the use of cloud computing for scientific workflows, in ‘2008 IEEE fourth international conference on eScience’, pp. 640–645. [Google Scholar]
- Januszewski M, Kornfeld J, Li PH, Pope A, Blakely T, Lindsey L, Maitin-Shepard J, Tyka M, Denk W and Jain V (2018), ‘High-precision automated reconstruction of neurons with flood-filling networks’, Nature Methods 15(8), 605–610. [DOI] [PubMed] [Google Scholar]
- Jararweh Y, Al-Ayyoub M, Benkhelifa E, Vouk M, Rindos A et al. (2016), ‘Software defined cloud: Survey, system and evaluation’, Future Generation Computer Systems 58, 56–74. [Google Scholar]
- Gold Associates LLC J (2014), Replacing Enterprise PCs: The Fallacy of the 3–4 Year Upgrade Cycle [White Paper], Technical report, J.Gold Associates LLC. [Google Scholar]
- Kane GA, Lopes G, Sanders JL, Mathis A and Mathis M (2020), ‘Real-time, low-latency closed-loop feedback using markerless posture tracking’, eLife 9, e61909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster J and Rahmann S (2012), ‘Snakemake—a scalable bioinformatics workflow engine’, Bioinformatics 28(19), 2520–2522. [DOI] [PubMed] [Google Scholar]
- Krafczyk M, Shi A, Bhaskar A, Marinov D and Stodden V (2019), Scientific Tests and Continuous Integration Strategies to Enhance Reproducibility in the Scientific Software Context, in ‘Proceedings of the 2nd International Workshop on Practical Reproducible Evaluation of Computer Systems’, pp. 23–28. [Google Scholar]
- Lakshminarayanan B, Pritzel A and Blundell C (2017), ‘Simple and scalable predictive uncertainty estimation using deep ensembles’, Advances in neural information processing systems 30. [Google Scholar]
- Landhuis E (2017), ‘Neuroscience: Big brain, big data’, Nature 541(7638), 559–561. [DOI] [PubMed] [Google Scholar]
- Lee JH, Carlson DE, Razaghi HS, Yao W, Goetz GA, Hagen E, Batty E, Chichilnisky E, Einevoll GT and Paninski L (2017), Yass: Yet another spike sorter, in ‘Advances in neural information processing systems’, pp. 4002–4012. [Google Scholar]
- Lopes G, Bonacchi N, Frazao J, Neto JP, Atallah BV, Soares S, Moreira L, Matias S, Itskov PM, Correia PA et al. (2015), ‘Bonsai: an event-based framework for processing and controlling data streams’, Frontiers in neuroinformatics 9, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magland J, Jun JJ, Lovero E, Morley AJ, Hurwitz CL, Buccino AP, Garcia S and Barnett AH (2020), ‘SpikeForest, reproducible web-facing ground-truth validation of automated neural spike sorters’, eLife 9, e55167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahvi J and Zarfaty A (2009), ‘Using TCO to Determine PC Upgrade Cycles’, Intel Corporation. [Google Scholar]
- Mathis A, Mamidanna P, Cury KM, Abe T, Murthy VN, Mathis MW and Bethge M (2018), ‘DeepLabCut: markerless pose estimation of user-defined body parts with deep learning’, Nature Neuroscience 21(9), 1281–1289. [DOI] [PubMed] [Google Scholar]
- Merali Z (2010), ‘Computational science: …Error’, Nature 467(7317), 775–777. [DOI] [PubMed] [Google Scholar]
- Merkel D (2014), ‘Docker: lightweight linux containers for consistent development and deployment’, Linux journal 2014(239), 2. [Google Scholar]
- Miller G (2006), ‘A Scientist’s Nightmare: Software Problem Leads to Five Retractions’, Science 314(5807), 1856–1857. [DOI] [PubMed] [Google Scholar]
- Minka TP (1999), ‘From Hidden Markov Models to Linear Dynamical Systems’.
- Monajemi H, Murri R, Jonas E, Liang P, Stodden V and Donoho DL (2019), ‘Ambitious Data Science Can Be Painless’, arXiv preprint arXiv:1901.08705. [Google Scholar]
- Morey T and Nambiar R (2009), Using Total Cost of Owner-ship to Determine Optimal PC Refresh Lifecycles [White Paper], Technical report, Wipro Ltd. [Google Scholar]
- Morris K (2016), Infrastructure as code: managing servers in the cloud, ” O’Reilly Media, Inc.”. [Google Scholar]
- Musall S, Kaufman MT, Juavinett AL, Gluf S and Churchland AK (2019), ‘Single-trial neural dynamics are dominated by richly varied movements’, Nature neuroscience 22(10), 1677–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NeuroScout (2022), ‘Neuroscout. (retrieved february 28, 2022)’. https://neuroscout.github.io/neuroscout
- Nichols DM, Thomson K and Yeates SA (2001), Usability and open-source software development, in ‘CHINZ’01’, pp. 49–54. [Google Scholar]
- Nilsson SR, Goodwin NL, Choong JJ, Hwang S, Wright HR, Norville ZC, Tong X, Lin D, Bentzley BS, Eshel N, McLaughlin RJ and Golden SA (2020), ‘Simple Behavioral Analysis (SimBA) – an open source toolkit for computer classification of complex social behaviors in experimental animals’, bioRxiv p. 2020.04.19.049452. [Google Scholar]
- Nowogrodzki A (2019), ‘How to support open source software and stay sane’, Nature 571, 133–134. [DOI] [PubMed] [Google Scholar]
- Ovadia Y, Fertig E, Ren J, Nado Z, Sculley D, Nowozin S, Dillon J, Lakshminarayanan B and Snoek J (2019), ‘Can you trust your model’s uncertainty? evaluating predictive uncertainty under dataset shift’, Advances in neural information processing systems 32. [Google Scholar]
- Pachitariu M, Steinmetz NA, Kadir SN, Carandini M and Harris KD (2016), Fast and accurate spike sorting of high-channel count probes with KiloSort, in ‘Advances in Neural Information Processing Systems’, pp. 4448–4456. [Google Scholar]
- Pachitariu M, Stringer C, Dipoppa M, Schroder S, Rossi LF, Dalgleish H, Carandini M and Harris KD (2017), ‘Suite2p: beyond 10,000 neurons with standard two-photon microscopy’, Bioarxiv p. 061507. [Google Scholar]
- Pandarinath C, O’Shea DJ, Collins J, Jozefowicz R, Stavisky SD, Kao JC, Trautmann EM, Kaufman MT, Ryu SI, Hochberg LR et al. (2018), ‘Inferring single-trial neural population dynamics using sequential auto-encoders’, Nature methods 15(10), 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paninski L and Cunningham J (2018), ‘Neural data science: accelerating the experiment-analysis-theory cycle in large-scale neuroscience’, Current opinion in neurobiology 50, 232–241. [DOI] [PubMed] [Google Scholar]
- Parthasarathy N, Batty E, Falcon W, Rutten T, Rajpal M, Chichilnisky E and Paninski L (2017), Neural networks for efficient bayesian decoding of natural images from retinal neurons, in ‘Advances in Neural Information Processing Systems’, pp. 6434–6445. [Google Scholar]
- Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L et al. (2019), PyTorch: An imperative style, high-performance deep learning library, in ‘Advances in Neural Information Processing Systems’, pp. 8024–8035. [Google Scholar]
- Pnevmatikakis EA, Soudry D, Gao Y, Machado TA, Merel J, Pfau D, Reardon T, Mu Y, Lacefield C, Yang W et al. (2016), ‘Simultaneous denoising, deconvolution, and demixing of calcium imaging data’, Neuron 89(2), 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radiuk PM (2017), ‘Impact of training set batch size on the performance of convolutional neural networks for diverse datasets’, Information Technology and Management Science 20(1), 20–24. [Google Scholar]
- Raff E (2019), A step toward quantifying independently reproducible machine learning research, Vol. 32. [Google Scholar]
- Riley J (2010), Starcluster-numpy/scipy computing on amazon’s elastic compute cloud (ec2), in ‘SciPY2010: Python for Scientific Computing Conference,(Austin Texas)’. [Google Scholar]
- Rocklin M (2015), Dask: Parallel Computation with Blocked algorithms and Task Scheduling, in ‘Proceedings of the 14th Python in Science Conference’, pp. 130–136. [Google Scholar]
- Rokem A, Dichter B, Holdgraf C and Ghosh S (2021), ‘Pan-neuro: interactive computing at scale with brain datasets’.
- Rübel O, Tritt A, Dichter B, Braun T, Cain N, Clack N, Davidson T, Dougherty M, Fillion-Robin J-C, Graddis N et al. (2019), ‘Nwb: N 2.0: an accessible data standard for neurophysiology’.
- Rübel O, Tritt A, Ly R, Dichter BK, Ghosh S, Niu L, Soltesz I, Svoboda K, Frank L and Bouchard KE (2021), ‘The neurodata without borders ecosystem for neurophysiological data science’, bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanielevici S, Sivagnanam S, Yoshimoto K, Carnevale NT and Majumdar A (2018), The Neuroscience Gateway: Enabling Large Scale Modeling and Data Processing in Neuroscience, in ‘PEARC ‘18: Proceedings of the Practice and Experience on Advanced Research Computing’, p. 52. [Google Scholar]
- Saxena S, Kinsella I, Musall S, Kim SH, Meszaros J, Thibodeaux DN, Kim C, Cunningham J, Hillman EM, Churchland A et al. (2020), ‘Localized semi-nonnegative matrix factorization (LocaNMF) of widefield calcium imaging data’, PLOS Computational Biology 16(4), e1007791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS and Eliceiri KW (2012), ‘NIH Image to ImageJ: 25 years of image analysis’, Nature Methods 9(7), 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweihoff JF, Loshakov M, Pavlova I, Kück L, Ewell LA and Schwarz MK (2021), ‘Deeplabstream enables closed-loop behavioral experiments using deep learning-based markerless, real-time posture detection’, Communications biology 4(1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sculley D, Holt G, Golovin D, Davydov E, Phillips T, Ebner D, Chaudhary V, Young M, Crespo J-F and Dennison D (2015), ‘Hidden technical debt in machine learning systems’, Advances in neural information processing systems 28. [Google Scholar]
- Simonyan V and Mazumder R (2014), ‘High-performance integrated virtual environment (hive) tools and applications for big data analysis’, Genes 5(4), 957–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer C, Straehle C, Koethe U and Hamprecht FA (2011), Ilastik: Interactive learning and segmentation toolkit, in ‘2011 IEEE international symposium on biomedical imaging: From nano to macro’, pp. 230–233. [Google Scholar]
- Steinmetz NA, Aydin C, Lebedeva A, Okun M, Pachitariu M, Bauza M, Beau M, Bhagat J, Böhm C, Broux M et al. (2021), ‘Neuropixels 2.0: A miniaturized high-density probe for stable, long-term brain recordings’, Science 372(6539), eabf4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stodden V, Seiler J and Ma Z (2018), ‘An empirical analysis of journal policy effectiveness for computational reproducibility’, Proceedings of the National Academy of Sciences 115(11), 2584–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussillo D, Jozefowicz R, Abbott LF and Pandarinath C (2016), ‘LFADS - Latent Factor Analysis via Dynamical Systems’, arXiv preprint arXiv:1608.06315. [Google Scholar]
- Teeters J, Godfrey K, Young R, Dang C, Friedsam C, Wark B, Asari H, Peron S, Li N, Peyrache A, Denisov G, Siegle J, Olsen S, Martin C, Chun M, Tripathy S, Blanche T, Harris K, Buzsáki G, Koch C, Meister M, Svoboda K and Sommer F (2015), ‘Neurodata Without Borders: Creating a Common Data Format for Neurophysiology’, Neuron 88(4), 629–634. [DOI] [PubMed] [Google Scholar]
- Terra (2022), ‘Terra. (retrieved february 28, 2022)’. https://app.terra.bio
- Terry M, Kay M and Lafreniere B (2010), Perceptions and practices of usability in the free/open source software (FoSS) community, in ‘Proceedings of the SIGCHI Conference on Human Factors in Computing Systems’, pp. 999–1008. [Google Scholar]
- Towns J, Peterson GD, Roskies R, Scott JR, Wilkins-Diehr N, Cockerill T, Dahan M, Foster I, Gaither K, Grimshaw A, Hazlewood V, Lathrop S and Lifka D (2014), ‘XSEDE: Accelerating Scientific Discovery’, Computing in Science and Engineering 16(5), 62–74. [Google Scholar]
- Tukey JW (1962), ‘The future of data analysis’, The annals of mathematical statistics 33(1), 1–67. [Google Scholar]
- Vogelstein JT, Mensh B, Hausser M, Spruston N, Evans AC, Kording K, Amunts K, Ebell C, Muller J, Telefont M et al. (2016), ‘To the cloud! A grassroots proposal to accelerate brain science discovery’, Neuron 92(3), 622–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltz D and Buchanan BG (2009), ‘Automating science’, Science 324(5923), 43–44. [DOI] [PubMed] [Google Scholar]
- Whiteway MR, Biderman D, Friedman Y, Dipoppa M, Buchanan EK, Wu A, Zhou J, Noel J-P, Laboratory TIB, Cunningham J and Paninski L (2021), ‘Partitioning variability in animal behavioral videos using semi-supervised variational autoencoders’, bioRxiv p. 2021.02.22.432309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltschko AB, Johnson MJ, Iurilli G, Peterson RE, Katon JM, Pashkovski SL, Abraira VE, Adams RP and Datta SR (2015), ‘Mapping sub-second structure in mouse behavior’, Neuron 88(6), 1121–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A, Buchanan EK, Whiteway MR, Schartner M, Meijer G, Noel J-P, Rodriguez E, Everett C, Norovich A, Schaffer E, Mishra N, Salzman CD, Angelaki D, Bendesky A, Laboratory TIB, Cunningham J and Paninski L (2020), ‘Deep Graph Pose: a semi-supervised deep graphical model for improved animal pose tracking’, bioRxiv p. 2020.08.20.259705. [Google Scholar]
- Yatsenko D, Reimer J, Ecker AS, Walker EY, Sinz F, Berens P, Hoenselaar A, Cotton RJ, Siapas AS and Tolias AS (2015), ‘DataJoint: managing big scientific data using MATLAB or Python’, bioRxiv. [Google Scholar]
- Yoo AB, Jette MA and Grondona M (2003), ‘Job Scheduling Strategies for Parallel Processing, 9th International Workshop, JSSPP 2003, Seattle, WA, USA, June 24, 2003. Revised Paper’, Lecture Notes in Computer Science pp. 44–60. [Google Scholar]
- Yu BM, Cunningham JP, Santhanam G, Ryu SI, Shenoy KV and Sahani M (2009), ‘Gaussian-process factor analysis for low-dimensional single-trial analysis of neural population activity’, J Neurophysiol 102(1), 614–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L and Deek FP (2005), ‘Improving open source software usability’, AMCIS 2005 Proceedings p. 430. [Google Scholar]
- Zhou A, He B, Ibrahim S, Buyya R, Calheiros R and Dastjerdi A (2016), ‘eScience and Big Data Workflow in Clouds: A Taxonomy and Survey’, Big data: Principles and paradigms pp. 431–456. [Google Scholar]
- Zhou P, Resendez SL, Rodriguez-Romaguera J, Jimenez JC, Neufeld SQ, Giovannucci A, Friedrich J, Pnevmatikakis EA, Stuber GD, Hen R et al. (2018), ‘Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data’, Elife 7, e28728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


