Summary
Background
Clinical application of artificial intelligence is limited due to the lack of interpretability and expandability in complex clinical settings. We aimed to develop an eye diseases screening system with improved interpretability and expandability based on a lesion-level dissection and tested the clinical expandability and auxiliary ability of the system.
Methods
The four-hierarchical interpretable eye diseases screening system (IEDSS) based on a novel structural pattern named lesion atlas was developed to identify 30 eye diseases and conditions using a total of 32,026 ultra-wide field images collected from the Second Affiliated Hospital of Zhejiang University, School of Medicine (SAHZU), the First Affiliated Hospital of University of Science and Technology of China (FAHUSTC), and the Affiliated People's Hospital of Ningbo University (APHNU) in China between November 1, 2016 to February 28, 2022. The performance of IEDSS was compared with ophthalmologists and classic models trained with image-level labels. We further evaluated IEDSS in two external datasets, and tested it in a real-world scenario and an extended dataset with new phenotypes beyond the training categories. The accuracy (ACC), F1 score and confusion matrix were calculated to assess the performance of IEDSS.
Findings
IEDSS reached average ACCs (aACC) of 0·9781 (95%CI 0·9739-0·9824), 0·9660 (95%CI 0·9591-0·9730) and 0·9709 (95%CI 0·9655-0·9763), frequency-weighted average F1 scores of 0·9042 (95%CI 0·8957-0·9127), 0·8837 (95%CI 0·8714-0·8960) and 0·8874 (95%CI 0·8772-0·8972) in datasets of SAHZU, APHNU and FAHUSTC, respectively. IEDSS reached a higher aACC (0·9781, 95%CI 0·9739-0·9824) compared with a multi-class image-level model (0·9398, 95%CI 0·9329-0·9467), a classic multi-label image-level model (0·9278, 95%CI 0·9189-0·9366), a novel multi-label image-level model (0·9241, 95%CI 0·9151-0·9331) and a lesion-level model without Adaboost (0·9381, 95%CI 0·9299-0·9463). In the real-world scenario, the aACC of IEDSS (0·9872, 95%CI 0·9828-0·9915) was higher than that of the senior ophthalmologist (SO) (0·9413, 95%CI 0·9321-0·9504, p = 0·000) and the junior ophthalmologist (JO) (0·8846, 95%CI 0·8722-0·8971, p = 0·000). IEDSS remained strong performance (ACC = 0·8560, 95%CI 0·8252-0·8868) compared with JO (ACC = 0·784, 95%CI 0·7479-0·8201, p= 0·003) and SO (ACC = 0·8500, 95%CI 0·8187-0·8813, p = 0·789) in the extended dataset.
Interpretation
IEDSS showed excellent and stable performance in identifying common eye conditions and conditions beyond the training categories. The transparency and expandability of IEDSS could tremendously increase the clinical application range and the practical clinical value of it. It would enhance the efficiency and reliability of clinical practice, especially in remote areas with a lack of experienced specialists.
Funding
National Natural Science Foundation Regional Innovation and Development Joint Fund (U20A20386), Key research and development program of Zhejiang Province (2019C03020), Clinical Medical Research Centre for Eye Diseases of Zhejiang Province (2021E50007).
Keywords: Artificial intelligence, Eye diseases screening system, Ultra-wide field fundus image, Interpretability, Enpandability
Research in context.
Evidence before this study
Pubmed was searched on May 30, 2022, with the terms of “artificial intelligence” OR “deep learning” OR “convolutional neural network” AND “eye diseases” OR “retina” AND “screen” for the papers describing deep-learning based eye diseases screening system, without date and language restrictions. Most of previous studies focused on developing screening systems using image-level labels. They recognised specific diseases restricted to limited training categories and visualised the algorithms using heatmaps that could be hard to explain clinically. The search results revealed that no eye disease screening system could directly output pathological and anatomical information and apply to new categories beyond the training samples.
Added value of this study
The system was embedded with lesion atlas, a structural pattern that could extract and visualize pathological features and their anatomical locations to improve the efficiency, interpretability and applicability of the system. In the multicentre datasets, it showed excellent and stable performance in diagnosing 30 eye conditions and new phenotypes beyond the 30 conditions. The system showed better performance than traditional models trained with image-level labels. A website tool was constructed for real-world clinical interaction. In the real-world scenario, the performance of the system was better than that of a senior ophthalmologist and a junior ophthalmologist. And it could significantly improve the performance of the junior ophthalmologist.
Implications of all the available evidence
The transparency and expandability of the system could tremendously increase the practical clinical value and the application range of it in complex clinical settings where only limited diseases classes could be available in the training data. It would enhance the efficiency and reliability of deep–learning–based screening system in clinical practice, especially in remote areas with a lack of experienced specialists.
Alt-text: Unlabelled box
Introduction
Vision impairment is a major health issue worldwide with over 2·2 billion of people suffering from vision impairments and ophthalmic conditions resulting in loss of sight,1,2 including cataracts,3 diabetic retinopathy (DR)4, age-related macular degeneration (AMD).5 glaucoma,6 among others. It not only affects life quality but also increases the socioeconomic burden and even the risk of death without accurate diagnosis and timely interventions.7,8 Therefore, it is essential to implement timely detection to prevent vision damage.
The new ultra-wide field (UWF) fundus images could provide a panoramic image of retina with 200° views using a red laser (532 nm) and a green laser (633 nm). Compared with traditional colorful fundus images with 30° to 60° views of the posterior pole using white light, more detailed retinal substructures could be observed in their individual laser separations from UWF images. UWF images were evaluated to be more likely to become the standard-of-care for screening, diagnosis, telemedicine and even treatment.9,10 It provides a new perspective on screening eye diseases. However, clinical application of UWF images could be limited owing to the insufficiency of well-trained retinal specialists, particularly in developing countries where there is a shortage of ophthalmic service.
In recent years, artificial intelligence (AI), especially deep learning (DL) has been effectively applied to detect vision-threatening diseases, including DR,11,12 AMD,13 glaucoma,14 etc. The improvement of diagnosis efficiency, and the expanded coverage of screening programs furthered the integration between AI and medical. However, there were still several challenges that needed to be considered since image-level DL-based methods generated fixed outputs directly from an input image in the manner of end-to-end. On the one hand, models would collapse when facing classes that were not available during training. A lot of studies have attempted to add more target classes in the training dataset to prevent this,15, 16, 17 but it was still difficult to train one model that could be applied to almost all clinical diseases. On the other hand, the limited interpretation of DL-based models discouraged clinical applications. Several studies tried to visualize the algorithms using heatmaps.18, 19, 20 But it was difficult to explain whether the highlight region was a new finding or a model error.21,22 And highlighted regions were not precise enough to locate small abnormalities in retina.
To address these limitations, an eye diseases screening system with improved interpretability and expandability was proposed in this study to facilitate AI-based diagnosis in complex clinical settings more accurately and reasonably. The system aimed to mimic clinical diagnosis process by analyzing the fine-grained pathological changes and their distribution, and precisely provide the logic evidence of algorithms’ outputs.
Methods
Datasets and annotation
This study was approved by the human research ethics committee of (HREC) the Second Affiliated Hospital of Zhejiang University, School of Medicine (SAHZU), the First Affiliated Hospital of University of Science and Technology of China (FAHUSTC), and the Affiliated People's Hospital of Ningbo University (APHNU). All procedures adhered to the principles of the Declaration of Helsinki. Informed consent was exempted by the HREC of SAHZU, FAHUSTC and APHNU in the retrospective sets. Written informed consent was obtained in the prospective sets.
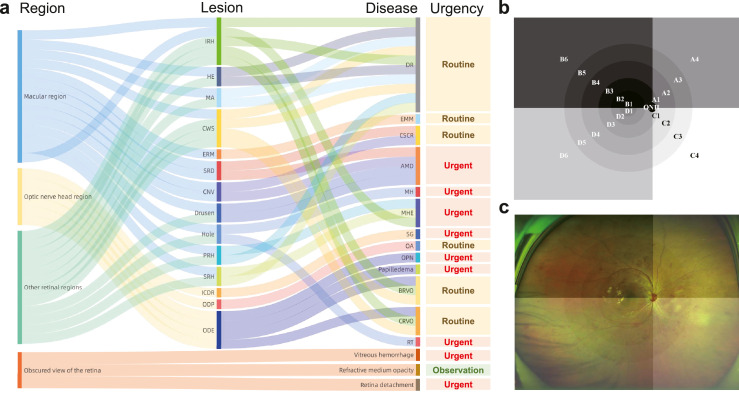
We tried to identify 15 common retinal lesions according to The Wills Eye manual,23 EyeWiki24 and the Digital Reference of Ophthalmology.25 The retinal lesions including choroidal neovascularization (CNV), hard exudates (HE), cotton wool spots (CWS), microaneurysm (MA), intraretinal hemorrhage (IRH), subretinal hemorrhage (SRH), preretinal hemorrhage (PRH), serous retinal detachment (SRD), epiretinal membrane (ERM), macular holes (MH), retinal tears (RT), optic disc edema (ODE), optic disc pallor (ODP), and increased cup-to-disc ratio (ICDR). On the basis of retinal signs and their position, 15 common eye diseases including DR, wet-AMD, ODE, MH, RT, refractive media opacity (RMO), retinal detachment (RD), vitreous hemorrhage (VH), central serous chorioretinopathy (CSCR), epimacular membrane (EMM), central retinal vein occlusion (CRVO), branch retinal vein occlusion (BRVO), optic atrophy (OA), suspected glaucoma (SG), and macula hemorrhage (MHE), and numerous multimorbidity were further recognized. A behavioral health urgency determination status of observation, routine and urgent was given after diagnosis (Figure 1a). Some diseases presented with same abnormalities were clustered into a broad category: diseases with optic disc swelling were classified into ODE, including papilledema, optic perineuritis, pseudopapilloedema, etc; diseases with ODP were classified as OA; diseases with ICDR were classified as SG; diseases with IRH, SRH, PRH in macular region were classified as MHE. The definitions and basis of judgement of retinal signs and eye diseases were provided in Supplementary Text S1.
Figure 1.
The clinical logical relation of lesion atlas. The lesion atlas integrated the pathological and anatomical information (a). The anatomical location was determined by dividing the retina into 21 regions as the prototype (b) and the overlaid image (c) displayed. DR, diabetic retinopathy; AMD, age-related macular degeneration; CRVO, central retinal vein occlusion; BRVO, branch retinal vein occlusion; CSCR, central serous chorioretinopathy; RT, retinal tears; EMM, epimacular membrane; MH, macular Hole; MHE, macular hemorrhage; ODE, optic disc edema; OA, optic atrophy; OPN, optic perineuritis; SG, suspected glaucoma; CNV, choroidal neovascularization; HE, hard exudates; CWS, cotton wool spots; MA, microaneurysm; IRH, intraretinal hemorrhage; SRH, subretinal hemorrhage; PRH, preretinal hemorrhage; SRD, serous retinal detachment; ERM, epiretinal membrane; ICDR increased cup-to-disc ratio; ODP, optic disc pallor.
The patients with target diseases were recruited in this study. We excluded images of poor quality due to non-pathological factors including: (1) Poor-location images, referring to images of which the optic nerve head and macula were off centre owing to the patient was not straight ahead. (2) Poor-field images, referring to images of which over one-third of the field was obscured by the eyelids. Besides, images with laser scars were excluded. A total of 26, 286 UWF images from 17, 389 subjects were obtained at the Eye Centre of SAHZU in China between November 1, 2016 to October 31, 2021 for training, validation and internal testing. We collected 3,414 images from FAHUSTC and 4,626 images from APHNU in China between October 1, 2021 to January 31, 2022 for external validation. Besides, 2,535 images of patients with target simple disease from SAHZU between November 1, 2021 to February 28, 2022 were prospectively collected for a real-world test. And 500 images of patients with multimorbidity consisting of target diseases were collected from SAHZU between June 1, 2021 to February 28, 2022 for an extended multimorbidity test. The patients with diseases beyond the type of target diseases or satisfied the exclusion criteria were excluded in the prospective collection process. The number of images in the internal dataset and two external datasets used for the training, validation and test was described in Table 1 and Supplementary Table S1. The images were acquired using a Daytona ultra-wide-field retinal camera (OPTOS Daytona, Dunefermline, UK) without mydriasis in SAHZU, FAHUSTC and APHNU.
Table 1.
Demographic statistics of the Second Affiliated Hospital of Zhejiang University (SAHZU) datasets and two external datasets.
| Total | SAHZU | External datasets |
Real-world test | ||||
|---|---|---|---|---|---|---|---|
| APHNU | FAHUSTC | ||||||
| Total no. of UWF images | 36,861 | 26,286 | 3,414 | 4,626 | 2,535 | ||
| Total no. of gradable images (%) | 31,526 (85·53) |
22,700 (86·36) |
2,597 (76·06) |
3,694 (79·85) | 2,535 (100) |
||
| Total no. of right eyes | 16,236 | 11,831 | 1,288 | 1,932 | 1,185 | ||
| Total no. of left eyes | 15,290 | 10,869 | 1,309 | 1,762 | 1,350 | ||
| No. of patients | 24,249 | 17,389 | 1,853 | 3,021 | 1,986 | ||
| Age, mean ± SD | 63 ± 11·71 | 66 ± 13·95 | 61 ± 12·18 | 65 ± 10·92 | 60 ± 12·45 | ||
| No. of women (%) | 13,615 (56·15) |
10,095 (58·05) |
872 (47·06) | 1652 (54·68) | 996 (50·15) | ||
| Training set | Validation set | Test set | Total | ||||
| DR | 3449 | 1150 | 1150 | 5749 | 840 | 1027 | 336 |
| wet-AMD | 864 | 288 | 288 | 1440 | 73 | 215 | 138 |
| CRVO | 484 | 161 | 161 | 806 | 136 | 162 | 104 |
| BRVO | 689 | 229 | 229 | 1147 | 120 | 237 | 186 |
| CSCR | 585 | 195 | 195 | 975 | 49 | 93 | 86 |
| RT | 1286 | 429 | 429 | 2144 | 215 | 241 | 235 |
| EMM | 914 | 305 | 305 | 1524 | 106 | 146 | 132 |
| MH | 434 | 145 | 144 | 723 | 48 | 58 | 58 |
| ODE | 315 | 105 | 105 | 525 | 39 | 61 | 38 |
| OA | 198 | 99 | 99 | 396 | 68 | 76 | 85 |
| Glaucoma | 607 | 202 | 202 | 1011 | 229 | 419 | 234 |
| RD | 2531 | 844 | 843 | 4218 | 364 | 532 | 422 |
| VH | 606 | 202 | 202 | 1010 | 208 | 313 | 251 |
| RMO | 620 | 206 | 206 | 1032 | 102 | 124 | 230 |
| Total | 13582 | 4560 | 4558 | 22700 | 2597 | 3694 | 2535 |
APHNU, the Affiliated People's Hospital of Ningbo University; FAHUSTC, the First Affiliated Hospital of University of Science and Technology of China; DR, diabetic retinopathy; wet-AMD, wet age-related macular degeneration; CRVO, central retinal vein occlusion; BRVO, branch retinal vein occlusion; CSCR, central serous chorioretinopathy; RT, retinal tears; EMM, epimacular membrane; MH, macular Hole; ODE, optic disc edema; OA, optic atrophy; RD, retinal detachment; VH, vitreous hemorrhage; RMO, refractive media opacity.
We recruited a professional labelling team (team 1) to generate the ground truth of annotations. The team consisted of two junior ophthalmologists (JO) with more than three years of clinical experience, two senior ophthalmologists (SO) with more than six years of clinical experience and one specialized ophthalmologist with more than 20 years of clinical experience. The team was divided into two groups which consisted of one JO and one SO, and the datasets were divided into two labelling quarters randomly and equally. Each quarter was firstly labelled by the JO and then checked by the SO. The divergences were finally confirmed by the specialized ophthalmologist.
The UWF images were annotated with image-level and lesion-level labels. The ground truth of images was identified based on clinical diagnosis and records including fundus fluorescein angiography, optical coherence tomography, optical coherence tomography angiography, visual field reports, etc. The detailed annotation process was provided in Supplementary Text S2.
Development of Interpretable eye diseases screening system (IEDSS)
Clinically, diagnosis and treatment fundamentally depended on the lesions’ symptoms and distribution. Different diseases could share similar lesions but their distribution pattern could be different. Therefore, the lesion atlas was designed to construct a structured description of lesion distribution in UWF images. It divided images into 21 anatomy regions based on anatomical location of optic nerve head and macula fovea as illustrated in 2.2 below, and counted the number of 15 types of lesions mentioned in Datasets and annotation section in each region. It was a process including lesion-level annotation, anatomical localization, distribution calculation and generalization, etc. The lesion atlas transformed qualitative definitions into computerized parametric information. Thus, the variability between different diseases were magnified and disease-to-disease interference in multimorbidity was weaken. It led classifiers to stay efficient in comorbidity diagnosis even if such samples were absent during training.
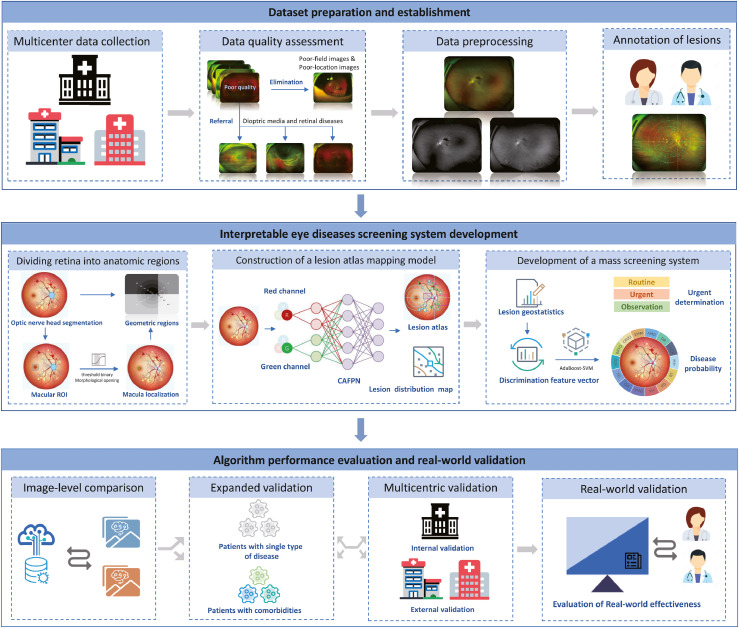
IEDSS was built based on the lesion atlas and consisted of four modules. Figure 2 depicted the workflow of the study. Firstly, the preliminary screening module was constructed to identify the diseases disturbing the image quality. A ResNetXt-50 was constructed for classifying diseases reducing image quality before lesions detection, including VH, RMO and RD. The aggregated transformation named bottleneck in ResNetXt-50 made it outperform ResNet-50 with the same model capacity.26 The ResNetXt-50 takes UWF images as input and outputs the probabilities of four classes (VH, RMO, RD and others).
Figure 2.
The workflow of overall study. Images were collected from three clinical centres and filtered by quality assessment. The preliminary screening module was constructed to provide automatic referral for patients with retinal detachment and vitreous hemorrhage, with the rest followed by preprocessing and lesion-level annotation. The dominant screening module consisted of three parts: (1) the location algorithm locating the optic disc region and macula region, and dividing the retina into 21 regions; (2) the lesion atlas mapping algorithm extracting the pathological and anatomical information of lesions automatically; (3) the mass screening algorithm integrating extracted features and making a final decision. The performance of the system was compared with image-level models and evaluated in a multicentric scenario, a real-world scenario and an expanded scenario in multimorbidity dataset.
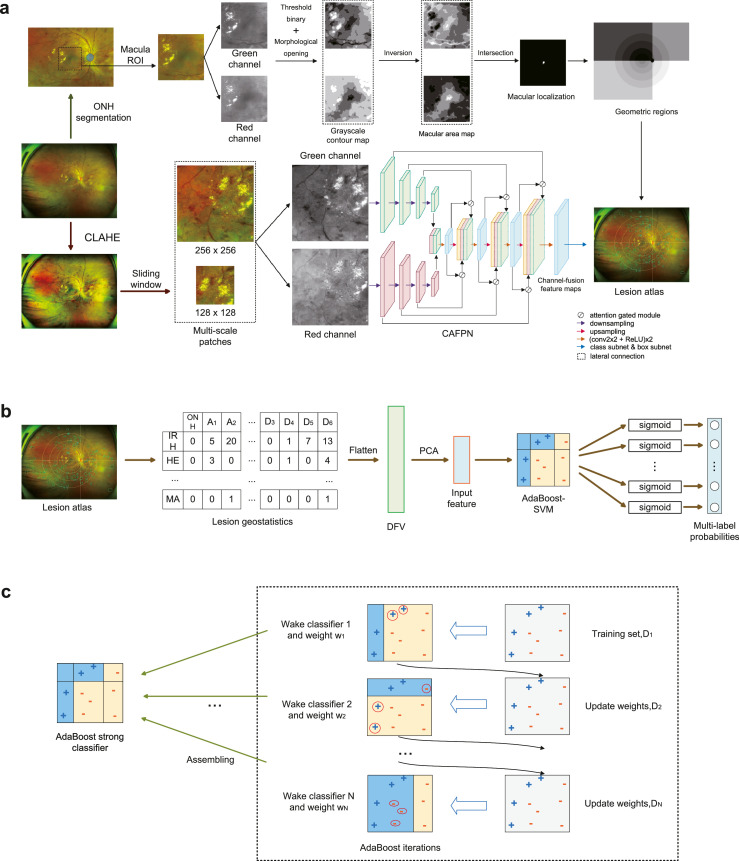
Secondly, a localization module was built to locate optic nerve head (ONH) and macular fovea. In order to divide the retina into 21 anatomical zones, the ONH was segmented by an UNet27 and the region of interest (ROI) of macular was found at 2·5 optic disc diameter (ρ) from the ONH centre.28 Grayscale contour maps (GCMs) of the macular ROI's green and red channels were generated by multi-thresholds and morphological opening. The centre point of the intersection of the darkest regions in these two GCMs was regarded as the macular fovea (Figure 3a). Taking the ONH and the macular fovea as references, and ρ as the unit of distance, the retina was divided into 21 geometric regions. The retina was firstly divided into six areas by five circles of radius with ρ, 2ρ, 3.5ρ, 5.5ρ and 7.5ρ surrounding the fovea. The areas were then separated into four sections (A, B, C, D) according to the line connecting ONH centre and macular fovea and the perpendicular passing through the ONH centre. The retina was finally divided into macular quadrant (B1 and D1), superotemporal quadrant (B2 to B6), inferotemporal quadrant (D2 to D6), superonasal quadrants (A1 to A4), and inferonasal quadrants (C1 to C4) (Figure 1b and 1c).
Figure 3.
Overview pipeline of Interpretable eye diseases screening system (IEDSS). There were two paths in the lesion atlas mapping module. In the upper path, geometric regions were generated after ONH segmentation and macula localization. In the lower path, multi-scale patches were cropped from a CLAHE-ed UWF image, and sent into the CAFPN. Results of the lesion detection were output from the CAPFN, and summarized into the lesion atlas with geometric regions (a). The lesion geostatisitcs showed the lesion distribution in the lesion atlas and was flatten to get the DFV. The input feature was generated by DFV dimensionality reduction through PCA and input into the AdaBoost-SVM to get multi-label probabilities in the mass screening system (b). In the training process of the AdaBoost-SVM, Weak classifiers were trained iteratively, and assembled to the strong classifier (c). ONH, optic nerve head; HE, hard exudates; MA, microaneurysm; IRH, intraretinal hemorrhage; CAFPN, channel-attention feature pyramid network; CLAHE, contrast limited adaptive histogram equalization; AdaBoost-SVM, AdaBoost-support vector machine; DFV, discrimination feature vectors; PCA, principal components analysis; ROI, region of interest.
Thirdly, a lesion atlas mapping module was constructed to differentiate the abnormal findings and locate their anatomical positions. In this paper, a channel-attention feature pyramid network (CAFPN) was designed to detect ONH-excluded lesions in UWF images (Figure 3a). It adopted feature pyramid network backbone to emphasize small lesions in UWF.29 The UWF images were preprocessed using contrast limited adaptive histogram equalization (CLAHE) before being fed into CAFPN (Supplementary Text S3). Since the information was in green and red channels, features in these channels were firstly extracted by two bottom-up pathways. Each pathway contained four downsampling modules. Then, final feature maps from bottom-up pathways were merged and sent into the top-down pathway. The top-down pathway combined information from green and red channels, and generated the channel-fusion feature map by attention gated modules30 and lateral connections. The following class and box subnets mapped the channel-fusion feature map to lesion detection results. Lastly, the lesion atlas was produced by uniting detection results and geometric regions. And the lesion atlas was transformed to a table named lesion geostatistics.
Finally, a mass screening system was developed to classify diseases and further recognize multimorbidity. Since different diseases could coexist in the same UWF image (except CRVO and BRVO), the diagnosis problem was regarded as a multilabel classification problem in this paper. AdaBoost-support vector machine (SVM)31 was constructed for multi-label classification of DR, AMD, CRVO, BRVO, CSCR, MH, EMM and RT in the mass screening system. It integrated the advantages of weak classifiers and achieve a better accuracy for multiple categories (Figure 3b and 3c). Since CRVO and BRVO could not coexist, a penalty term was added to the loss function (Supplementary Text S4). We took discrimination feature vectors (DFVs) from lesion geostatistics in lesion atlas as features and the diagnosis annotations as labels to optimize the AdaBoost-SVM. The detailed training process was shown in Supplementary Text S4. We applied principal components analysis (PCA) to reduce the dimensionality of DFVs and chose variables with a cumulative variance contribution rate of 90% before sending them into the AdaBoost-SVM. Besides, a ResNetXt-50 was built to classify patterns of normal, ODE, OA and SG. The bounding box area based on the ONH contour was cropped and sent into the model. The ONH classifier output probabilities of four class, and chose the one with the highest probabilities as the prediction.
Evaluation in two external datasets
An external test in datasets from APHNU and FAHUSTC was conducted to validate the generalization performance of IEDSS. A total of 2,597 gradable images from 1,853 patients of APHNU and 3,694 gradable images from 3,021 patients in FAHUSTC were included for the test (Table 1). The annotations and the ground truth were generated by the procedures mentioned above.
Comparison with image-level identification models
To evaluate the effect of lesion atlas, the performance of IEDSS was compared with traditional models that took images as input, including a multi-class ResNetXt-50, a multi-label ResNetXt-50 and a novel multi-label vision transformer (ViT).32 Besides, to validate the function of AdaBoost-SVM, we compared IEDSS with a multi-label SVM without AdaBoost (CAFPN+SVM). All the models were trained and tested in the SAHZU dataset.
Development of a cloud platform and evaluation in a prospective real-world scenario
The system was integrated into a cloud platform to improve the auxiliary diagnostic practicability. The platform was constructed using Python 3·7 and vue·js 2·9·6. The UWF image could be uploaded and the pathological features could be automated analysed by the back-end server. The detected lesions, their anatomy locations, the lesion distribution diagram, the predicted probabilities for candidate diseases, the dominant predicted diagnosis of IEDSS and the referral advance were presented to the clinicians to assist them make decisions.
To investigate the practicability of IEDSS in assisting clinical decision making, a JO and a SO outside the labelling team (team 2) were invited to perform the comparison task using the web interface. We prospectively collected 2,535 images from patients with only one kind of disease from SAHZU for the real-world test (Table 1). They were required to read the images without the assistance of IEDSS and read the images with the assistance of IEDSS in a reverse order after a washout period of one month.33
Evaluation of expandability of IEDSS in a multimorbidity scenario
Theoretically, IEDSS was applicable to identify not only a single disease but also the multimorbidity based on lesion atlas. Thus, the expandability of IEDSS was evaluated in a dataset containing challenging images of multimorbidity that the system had not encountered during training (Supplementary Table S1). Team 2 was invited to give a diagnosis without (with) the assistance of IEDSS to test the assistance capability of IEDSS. There was a washout time of one month between the two judgements.
Statistical analysis
The precision-recall curve and the average precision were generated to quantize the performance of the lesion atlas mapping module. The diagnostic accuracy (ACC), confusion matrix, sensitivity, specificity, receiver operating characteristics curve (ROC) and the area under the ROC curve (AUROC) were calculated for assessing the rest of IEDSS modules. Considering of imbalanced datasets, F1 scores and frequency-weighted average F1 was also introduced to evaluate IEDSS. The comparison test with human was evaluated by sensitivity, specificity and ACC in the multimorbidity scenario and the real-world scenario. We applied Chi-Squared Test to assess the performance between the system, clinicians with and without IEDSS in the diagnosis task. A p-value of 0·05 was considered significant. All statistical analyses for the study were conducted using STATISTICAL PACKAGE FOR THE SOCIAL SCIENCES Version 22·0 (SPSS, Inc., Chicago, IL, USA) and Python 3·7.
Role of the funding source
The funder played no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. All authors had full access to all the data in the study and accept responsibility to submit for publication.
Results
Data characteristics
In total, 26,286 images were obtained from SAHZU and 22,700 images were gradable and annotated for preliminary algorithm development and validation. The dataset was divided into the training, validation and internal test set in a 3:1:1 ratio. A total of 8,040 UWF images from two other clinical centres, APHNU and FAHUSTC, were collected and annotated for external validation and 2,535 prospective collected images from SAHZU were used for evaluation in a real-world scenario. An extra dataset with 500 images was built for extended algorithm validation in competence of recognizing multimorbidity. The characteristics of patients and the distribution of collected images were summarized in Table 1.
Evaluation of the performance and interpretability of IEDSS in the internal dataset
It's showed that based on a four-hierarchical framework, IEDSS achieved an average ACC of 0·9781 (95%CI 0·9739–0·9824), sensitivity of 0·9635 (95%CI 0·9580-0·9689), specificity of 0·9792 (95%CI 0·9751-0·9833) and AUROC of 0·9904 (95%CI 0·9876-0·9932) (Table 2, Supplementary Figure S1). It showed better performance than JO and SO, with a higher average ACC (0·9781, 95%CI 0·9739-0·9824 for IEDSS vs 0·8891, 95%CI 0·8800-0·8983 for JO and 0·9540, 95%CI 0·9479-0·9601 for SO) and sensitivity (0·9634, 95%CI 0·9580-0·9689 for IEDSS vs 0·8666, 95%CI 0·8568-0·8765 for JO and 0·9434, 95%CI 0·9366-0·9501 for SO) (Supplementary Table S2). The highest ACC were reached in classes with obvious features such as MH (0·9909, 95%CI 0·9877-0·9942) and pathological changes in the ONH region were well differentiated with ACCs over 0·99. The detailed evaluation of preliminary screening module and lesion atlas mapping module of IEDSS was elaborated in Supplementary Text S5, Figure S2 and Figure S3. F1 scores and precision were also calculated in consideration of the imbalance datasets (Table 2 and Supplementary Table S3). The frequency-weighted average F1 score achieved 0·9042 (95%CI 0·8957-0·9127). F1 scores were above 0·84 for all diseases except CSCR (0·7088, 95%CI 0·7028-0·7147), AMD (0·7813, 95%CI 0·7789-0·7866) and CRVO (0·6245, 95%CI 0·6182-0·6308). The precisions were over 0·74 except CSCR (0·5878, 95%CI 0·5177-0·6546), AMD (0·6611, 95%CI 0·6046-0·7133) and CRVO (0·4580, 95%CI 0·3829-0·5350). It seemed that IEDSS obtained lower F1 scores and precision in seriously imbalanced subclasses. For example, F1 score and precision reached 0·9443 (95%CI 0·9414-0·9473) and 0·9300 (95%CI 0·9138-0·9434) for DR, whose positive to negative ratio was 1: 1·88, but was 0·6245 (95%CI 0·6182-0·6308) and 0·4580 (95%CI 0·3829-0·5350) for CRVO, whose positive to negative ratio was 1:19·54. In our multi-label dataset, the negative samples were vastly outnumbered by positive samples, which would magnify the shortcoming of F1 score and precision that the false-positives would dominate the results.34,35
Table 2.
The performance of Interpretable Eye Diseases Screening System (IEDSS) for identifying common eye diseases in internal dataset and multicenter datasets.
| SAHZU |
APHNU |
FAHUSTC |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUROC (%)(95% CI) | ACC (%)(95%CI) | F1(95%CI) | Sensitivity (%) (95% CI) | Specificity (%)(95% CI) | AUROC (%)(95% CI) | ACC (%)(95%CI) | F1(95%CI) | Sensitivity (%)(95% CI) | Specificity (%) (95% CI) | AUROC (%)(95% CI) | ACC (%)(95%CI) | F1(95%CI) | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | |
| wet-AMD | 98·84(98·48-99·20) | 95·34(94·63-96·06) | 0·7813(0·7759-0·7866) | 95·49(92·22-97·47) | 95·33(94·50-96·04) | 98·62(98·36-99·32) | 94·70(93·69-95·70) | 0·5750(0·5560-0·5940) | 94·52(85·84-98·23) | 94·70(93·56-95·66) | 98·84(98·44-99·24) | 95·96(95·22-96·70) | 0·7860(0·7728-0·7992) | 93·95(89·65-96·61) | 96·14(95·29-96·84) |
| BRVO | 99·61(99·40-99·82) | 97·55(97·02-98·08) | 0·8469(0·8422-0·8516) | 97·82(94·70-99·19) | 97·53(96·90-98·04) | 99·24(99·33-99·89) | 96·93(96·16-97·70) | 0·7986(0·7832-0·8141) | 97·50(92·32-99·35) | 96·89(95·96-97·62) | 99·51(99·25-99·77) | 98·20(97·70-98·70) | 0·9026(0·8930-0·9121) | 95·78(92·14-97·84) | 98·43(97·84-98·87) |
| CRVO | 98·61(98·21-99·01) | 94·25(93·46-95·05) | 0·6245(0·6182-0·6308) | 98·14(94·22-99·52) | 94·06(93·16-94·84) | 97·74(98·09-99·13) | 87·26(85·77-88·75) | 0·5224(0·5032-0·5416) | 98·53(94·25-99·74) | 86·40(84·71-87·94) | 97·38(96·78-97·98) | 92·18(91·18-93·19) | 0·5958(0·5800-0·6117) | 96·91(92·57-98·86) | 91·88(90·74-92·90) |
| CSCR | 98·16(97·70-98·62) | 95·68(94·98-96·37) | 0·7088(0·7028-0·7147) | 89·23(83·80-93·06) | 96·08(95·32-96·72) | 98·54(97·56-98·76) | 98·13(97·52-98·73) | 0·7049(0·6874-0·7225) | 87·76(74·45-94·92) | 98·40(97·69-98·90) | 98·72(98·30-99·14) | 94·94(94·11-95·76) | 0·5175(0·5014-0·5336) | 89·16(79·94-94·62) | 95·12(94·21-95·89) |
| DR | 98·75(98·37-99·13) | 96·07(95·41-96·73) | 0·9443(0·9414-0·9473) | 95·91(94·56-96·95) | 96·15(95·23-96·91) | 98·56(98·25-99·25) | 93·45(92·34-94·55) | 0·9259(0·9158-0·936) | 93·69(91·77-95·20) | 93·26(91·56-94·65) | 98·25(97·76-98·74) | 95·82(95·06-96·57) | 0·9442(0·9368-0·9516) | 93·97(92·28-95·31) | 96·94(95·97-97·68) |
| EMM | 99·35(99·08-99·62) | 97·67(97·16-98·19) | 0·8824(0·8783-0·8866) | 94·75(91·45-96·87) | 97·97(97·38-98·43) | 97·29(98·99-99·71) | 93·03(91·89-94·17) | 0·5939(0·5751-0·6128) | 92·45(85·23-96·45) | 93·07(91·77-94·17) | 98·40(97·93-98·87) | 92·29(98·34-99·17) | 0·5607(0·5447-0·5767) | 91·78(85·77-95·49) | 92·32(91·21-93·31) |
| MH | 99·74(99·57-100·00) | 99·09(98·77-99·42) | 0·9045(0·9006-0·9083) | 98·61(94·56-99·76) | 99·11(98·70-99·40) | 99·50(99·51-99·97) | 98·18(97·58-98·78) | 0·7244(0·7072-0·7416) | 95·83(84·57-99·28) | 98·24(97·51-98·77) | 99·60(99·36-99·84) | 98·75(98·34-99·17) | 0·7671(0·7535-0·7808) | 96·55(87·05-99·40) | 98·80(98·29-99·16) |
| RT | 99·26(98·97-99·70) | 96·73(96·13-97·34) | 0·8846(0·8805-0·8888) | 96·50(94·17-97·96) | 96·77(96·04-97·37) | 99·08(98·88-99·64) | 94·70(93·69-95·70) | 0·8000(0·7846-0·8154) | 94·88(90·79-97·29) | 94·67(93·47-95·67) | 98·82(98·41-99·23) | 95·56(94·79-96·33) | 0·7858(0·7726-0·7991) | 92·12(87·77-95·06) | 95·89(95·02-96·62) |
| VH | NA | 99·39(99·16-99·61) | 0·9333(0·9301-0·9366) | 97·03(93·35-98·79) | 99·49(99·14-99·69) | NA | 99·19(98·85-99·53) | 0·9501(0·9541-0·9585) | 96·15(92·29-98·20) | 99·46(99·04-99·70) | NA | 99·38(99·13-99·63) | 0·9635(0·9575-0·9696) | 97·12(94·42-98·59) | 99·59(99·29-99·76) |
| RD | NA | 99·65(99·48-99·82) | 0·9905(0·9893-0·9918) | 99·17(98·22-99·64) | 99·76(99·44-99·90) | NA | 99·77(99·59-99·95) | 0·9863(0·9819-0·9908) | 99·18(97·41-99·79) | 99·69(99·32-99·86) | NA | 99·73(99·56-99·90) | 0·9906(0·9875-0·9937) | 99·44(98·22-99·85) | 99·78(99·52-99·90) |
| RMO | NA | 99·46(99·24-99·67) | 0·9395(0·9364-0·9426) | 94·17(89·80-96·82) | 99·71(99·43-99·86) | NA | 99·38(99·08-99·68) | 0·9252(0·9151-0·9353) | 97·06(91·02-99·24) | 99·48(99·09-99·71) | NA | 99·21(98·92-99·50) | 0·8889(0·8788-0·899) | 93·55(87·28-96·97) | 99·41(99·09-99·63) |
| ODE | NA | 99·52(99·28-99·75) | 0·9292(0·9259-0·9325) | 100·00(95·60-100·00) | 99·50(99·17-99·70) | NA | 99·69(99·48-99·91) | 0·9048(0·8935-0·9161) | 97·44(84·92-99·87) | 99·73(99·41-99·88) | NA | 99·70(99·53-99·88) | 0·9160(0·9071-0·9250) | 96·77(87·83-99·44) | 99·75(99·51-99·88) |
| OA | NA | 99·73(99·55-99·91) | 0·9557(0·953-0·9583) | 97·98(92·19-99·65) | 99·78(99·53-99·90) | NA | 99·58(99·33-99·83) | 0·9241(0·914-0·9343) | 98·53(90·99-99·92) | 99·60(99·25-99·80) | NA | 99·68(99·49-99·86) | 0·9250(0·9165-0·9335) | 97·37(89·95-99·54) | 99·72(99·47-99·86) |
| SG | NA | 99·24(98·95-99·54) | 0·9383(0·9351-0·9414) | 94·06(89·60-96·75) | 99·58(99·26-99·77) | NA | 98·46(97·99-98·93) | 0·9180(0·9075-0·9286) | 97·82(94·70-99·19) | 98·52(97·93-98·95) | NA | 97·83(97·36-98·30) | 0·9111(0·9019-0·9203) | 97·62(95·52-98·79) | 97·86(97·29-98·32) |
AUROC is not applicable for multi-class models since the multi-class models make decisions by selecting the highest probability in each sample instead of a threshold.
SAHZU, the Second Affiliated Hospital of Zhejiang University; APHNU, the Affiliated People's Hospital of Ningbo University; FAHUSTC, the First Affiliated Hospital of University of Science and Technology of China; wet-AMD, wet age-related macular degeneration; BRVO, branch retinal vein occlusion; CRVO, central retinal vein occlusion; CSCR, central serous chorioretinopathy; DR, diabetic retinopathy; EMM, epimacular membrane; MH, macular Hole; RT, retinal tears; VH, vitreous hemorrhage; RD, retinal detachment; RMO, refractive media opacity; ODE, optic disc edema; OA, optic atrophy; SG, suspected glaucoma; NA, not applicable.
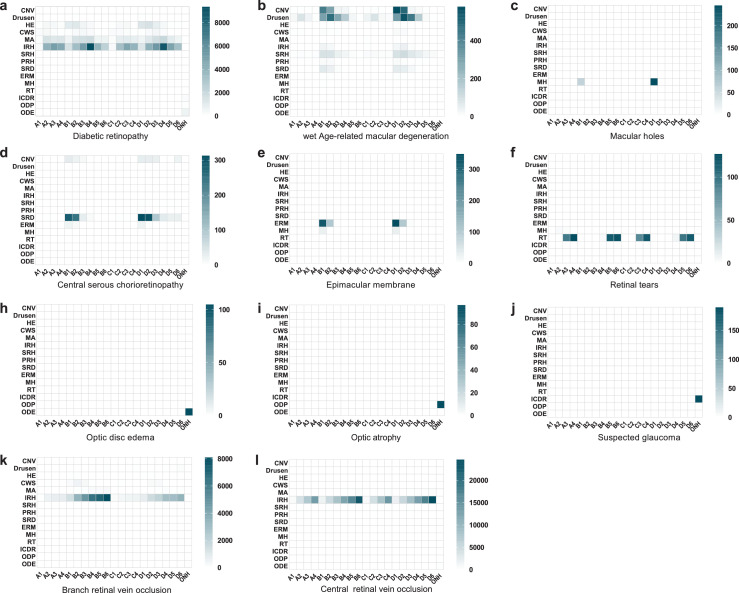
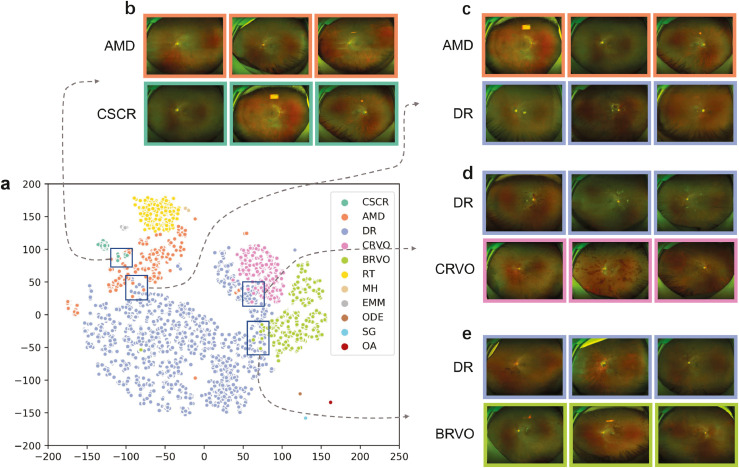
The screening system successfully localised and discerned the pathological features in different diseases as shown in Figure 4. It was revealed that the distribution and the amount of pathological features were significantly different between diseases, which visualised the principle of lesion atlas. Since the lesion type and the total number of lesions were similar in BRVO and CRVO subsets, we presented the distribution and number of IRH in each individual (Supplementary Figure S4). It was showed that identified IRH distributed in all retinal regions of individuals with CRVO while tended to be more prevalent in one or two regions of individuals with BRVO. To further visualize how the IEDSS discerned encoded features from different images, we mapped the 243-demension features into two-dimensional coordinates by the t-distributed stochastic neighbor embedding (t-SNE) (Figure 5). It showed that UWF images formed distinct clusters (Figure 5a). Examining query images selected from neighboring reference images helped to explain class confusions. It was demonstrated that the confusions were mainly in diseases shared with similar lesions (Figure 5b-5e). To be more specific, AMD and chronic CSCR would be confused due to same lesions like SRD and CNV. Meanwhile, retinal vascular diseases, including DR, CRVO and BRVO, could be misclassified because the distribution of shared lesions (IRH, HE, CWS) was similar.
Figure 4.
The lesion geostatistics heatmap generated by lesion atlas illustrating the type, number and distribution of lesions in different diseases. CNV, choroidal neovascularization; HE, hard exudates; CWS, cotton wool spots; MA, microaneurysm; IRH, intraretinal hemorrhage; SRH, subretinal hemorrhage; PRH, preretinal hemorrhage; SRD, serous retinal detachment; ERM, epiretinal membrane; MH, macular holes; RT, retinal tears; ODE, optic disc edema; ODP, optic disc pallor; ICDR, increased cup-to-disc ratio.
Figure 5.
The t-Distributed stochastic neighbor embedding visualizations of discrimination feature vectors in the screening model for the classification of 11 diseases (a). The map preserved the topologic features, and adjacent coordinates meant they shared similar features in the original feature space. A selection of adjacent cases showing similar clinical features (b-e). DR, diabetic retinopathy; CSCR, central serous chorioretinopathy; AMD, age-related macular degeneration; MH, macular Hole; EMM, epimacular membrane; CRVO, central retinal vein occlusion; BRVO, branch retinal vein occlusion; RT, retinal tears; ODE, optic disc edema; OA, optic atrophy; SG, suspected glaucoma.
Performance in identification of a single disease in external datasets
The evaluate the generalization ability of IEDSS, the task was performed in other two heterogeneous datasets. The system worked well in external datasets with the average ACCs of 0·9660 (95%CI 0·9591-0·9730) and 0·9709 (95%CI 0·9655-0·9763), sensitivities of 0·9581 (95%CI 0·9504-0·9658) and 0·9515 (95%CI 0·9432-0·9598), specificities of 0·9658 (95%CI 0·9588-0·9728) and 0·9726 (95%CI 0·9663-0·9789), AUROCs of 0·9857 (95%CI 0·9811-0·9903) and 0·9869 (95%CI 0·9825-0·9913), and frequency-weighted average F1 scores of 0·8837 (95%CI 0·8714-0·8960) and 0·8874 (95%CI 0·8752-0·8996) in the APHNU dataset and the FAHUSTC dataset, respectively (Table 2, Supplementary Table S3, Figure S1 and Figure S2).
Comparison with image-level identification
The comparisons of model performance among IEDSS, a classic image-level classification model, a classic image-level multi-label model, a novel image-level multi-label model and a lesion atlas-based multi-label model without AdaBoost were shown in Table 3, Supplementary Table S3 and Table S4. The average sensitivity of CAFPN+SVM (0·8892, 95%CI 0·8784-0·8999) was higher than that of image-level multi-class ResNetXt-50 (0·8196, 95%CI 0·8081-0·8305), multi-label ResNetXt-50 (0·8180, 95%CI 0·8048-0·8311) and multi-label ViT (0·8498, 95%CI 0·8376-0·8620), which approved the effectiveness of CAFPN (Supplementary Table S4). The advantage of the framework of IEDSS and the effect of AdaBoost were further investigated in comparison with classic and novel image-level multi-label models, multi-class model, and the model without AdaBoost module. IEDSS showed better performance than other models with the average ACCs of 0·9781 (95%CI 0·9739-0·9824), 0·9398 (95%CI 0·9329-0·9467), 0·9278 (95%CI 0·9189-0·9366), 0·9241 (95%CI 0·9151-0·9331) and 0·9381 (95%CI 0·9299-0·9463), sensitivities of 0·9635 (95%CI 0·9580-0·9689), 0·8196 (95%CI 0·8081-0·8305), 0·8180 (95%CI 0·8035-0·8316), 0·8498 (95%CI 0·8376-0·8620) and 0·8892 (95%CI 0·8785-0·8999), specificities of 0·9792 (95%CI 0·9743-0·9841), 0·9494 (95%CI 0·9476-0·9511), 0·9406 (95%CI 0·9325-0·9487), 0·9333 (95%CI 0·9248-0·9418) and 0·9431 (95%CI 0·9352-0·9510), and frequency-weighted average F1 scores of 0·9042 (95%CI 0·8957-0·9127), 0·6923 (95%CI 0·6766-0·7080), 0·7337 (95%CI 0·7186-0·7488), 0·6391 (95%CI 0·6227-0·6555) and 0·7761 (95%CI 0·7619-0·7903) for IEDSS, multi-class ResNetXt50, multi-label ResNetXt50, multi-label ViT and CAFPN+SVM, respectively (Table 3).
Table 3.
The performance comparison experiment of Interpretable Eye Diseases Screening System (IEDSS) and classic algorithms.
| Multi-class ResNetXt-50 |
Multi-label ResNetXt-50 |
Multi-label ViT |
CAFPN+SVM |
IEDSS |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACC (%)(95% CI) | AUROC(%)(95% CI) | F1 (95% CI) | ACC (%)(95% CI) | AUROC(%)(95% CI) | F1 (95% CI) | ACC (%)(95% CI) | AUROC(%)(95% CI) | F1 (95% CI) | ACC (%)(95% CI) | AUROC(%)(95% CI) | F1 (95% CI) | ACC (%)(95% CI) | AUROC(%)(95% CI) | F1 (95% CI) | |
| wet-AMD | 90·63(89·79-91·48) | 99·12(98·80-99·44) | 0·5052(0·4858-0·5247) | 92·08(91·16-93·00) | 93·39(92·54-94·23) | 0·6225(0·6036-0·6413) | 91·65(90·71-92·60) | 96·17(95·51-96·82) | 0·6416(0·6229-0·6602) | 92·20(91·28-93·11) | 96·62(96·00-97·24) | 0·6632(0·6448-0·6816) | 95·34(94·63-96·06) | 98·84(98·48-99·20) | 0·7813(0·7759-0·7866) |
| BRVO | 94·98(94·34-95·61) | 99·39(99·12-99·66) | 0·6514(0·6329-0·6700) | 95·71(95·02-96·40) | 98·23(97·78-98·68) | 0·7482(0·7313-0·7651) | 96·92(96·33-97·5) | 98·46(98·04-98·88) | 0·8097(0·7944-0·8250) | 95·62(94·92-96·31) | 98·62(98·22-99·02) | 0·7504(0·7336-0·7673) | 97·55(97·02-98·08) | 99·61(99·40-99·82) | 0·8469(0·8422-0·8516) |
| CRVO | 96·09(95·53-96·66) | 98·29(97·84-98·73) | 0·5762(0·5570-0·5954) | 90·23(89·22-91·24) | 94·14(93·34-94·94) | 0·4590(0·4396-0·4784) | 89·23(88·18-90·29) | 94·75(93·99-95·51) | 0·4385(0·4192-0·4578) | 91·53(90·58-92·48) | 96·97(96·39-97·56) | 0·5035(0·4841-0·523) | 94·25(93·46-95·05) | 98·61(98·21-99·01) | 0·6245(0·6182-0·6308) |
| CSCR | 91·97(91·18-92·76) | 98·23(97·78-98·68) | 0·3624(0·3437-0·3811) | 91·05(90·08-92·02) | 94·76(94·00-95·52) | 0·4825(0·4631-0·502) | 91·84(90·9-92·77) | 93·34(92·49-94·19) | 0·5018(0·4824-0·5213) | 93·62(92·79-94·45) | 96·56(95·94-97·18) | 0·5903(0·5711-0·6094) | 95·68(94·98-96·37) | 98·16(97·70-98·62) | 0·7088(0·7028-0·7147) |
| DR | 87·80(86·85-88·75) | 96·65(96·03-97·26) | 0·7470(0·7301-0·7640) | 89·23(88·18-90·29) | 94·11(93·30-94·91) | 0·8429(0·8287-0·8571) | 87·72(86·6-88·84) | 92·78(91·90-93·66) | 0·8227(0·8078-0·8376) | 91·53(90·58-92·48) | 96·35(95·71-96·99) | 0·8801(0·8675-0·8928) | 96·07(95·41-96·73) | 98·75(98·37-99·13) | 0·9443(0·9414-0·9473) |
| EMM | 92·26(91·48-93·03) | 93·55(92·71-94·38) | 0·5537(0·5344-0·5731) | 93·80(92·98-94·62) | 96·24(95·59-96·89) | 0·6861(0·6680-0·7041) | 91·84(90·9-92·77) | 95·98(95·31-96·65) | 0·6260(0·6072-0·6449) | 93·44(92·59-94·28) | 97·14(96·57-97·71) | 0·7031(0·6854-0·7209) | 97·67(97·16-98·19) | 99·35(99·08-99·62) | 0·8824(0·8783-0·8866) |
| MH | 97·30(96·83-97·77) | 93·97(93·16-94·78) | 0·6720(0·6537-0·6903) | 97·01(96·43-97·59) | 99·59(99·37-99·81) | 0·7227(0·7053-0·7401) | 97·64(97·12-98·16) | 99·21(98·91-99·51) | 0·7784(0·7622-0·7946) | 98·61(98·21-99·01) | 99·59(99·37-99·81) | 0·8571(0·8435-0·8708) | 99·09(98·77-99·42) | 99·74(99·57-100·00) | 0·9045(0·9006-0·9083) |
| RT | 95·46(94·85-96·06) | 97·10(96·53-97·68) | 0·7708(0·7544-0·7871) | 93·11(92·24-93·97) | 97·21(96·65-97·78) | 0·7625(0·7459-0·7791) | 92·44(91·54-93·34) | 97·43(96·90-97·97) | 0·7596(0·743-0·7763) | 93·95(93·14-94·76) | 97·48(96·95-98·02) | 0·7984(0·7828-0·814) | 96·73(96·13-97·34) | 99·26(98·97-99·70) | 0·8846(0·8805-0·8888) |
| VH | 97·19(96·71-97·67) | 94·43(93·65-95·21) | 0·7500(0·7331-0·7669) | NA | NA | NA | NA | NA | NA | NA | NA | NA | 99·39(99·16-99·61) | NA | 0·9333(0·9301-0·9366) |
| RD | 97·26(96·78-97·73) | 93·13(92·27-93·99) | 0·9297(0·9198-0·9397) | NA | NA | NA | NA | NA | NA | NA | NA | NA | 99·65(99·48-99·82) | NA | 0·9905(0·9893-0·9918) |
| RMO | 96·20(95·65-96·76) | 93·90(93·08-94·72) | 0·6826(0·6644-0·7007) | NA | NA | NA | NA | NA | NA | NA | NA | NA | 99·46(99·24-99·67) | NA | 0·9395(0·9364-0·9426) |
| ODE | 95·46(94·85-96·06) | 95·63(94·93-96·32) | 0·4838(0·4643-0·5032) | NA | NA | NA | NA | NA | NA | NA | NA | NA | 99·52(99·28-99·75) | NA | 0·9292(0·9259-0·9325) |
| OA | 93·73(93·02-94·43) | 98·80(98·43-99·17) | 0·3755(0·3567-0·3944) | NA | NA | NA | NA | NA | NA | NA | NA | NA | 99·73(99·55-99·91) | NA | 0·9557(0·953-0·9583) |
| SG | 89·38(88·49-90·28) | 97·79(97·29-98·29) | 0·3858(0·3668-0·4047) | NA | NA | NA | NA | NA | NA | NA | NA | NA | 99·24(98·95-99·54) | NA | 0·9383(0·9351-0·9414) |
The multi-label models including Multi-label ResNetXt-50, Multi-label ViT and CAFPN+SVM were not tested on the multi-class labels, including VH, RD, RMO, ODE, OA, SG.
wet-AMD, wet age-related macular degeneration; BRVO, branch retinal vein occlusion; CRVO, central retinal vein occlusion; CSCR, central serous chorioretinopathy; DR, diabetic retinopathy; EMM, epimacular membrane; MH, macular Hole; RT, retinal tears; VH, vitreous hemorrhage; RD, retinal detachment; RMO, refractive media opacity; ODE, optic disc edema; OA, optic atrophy; SG, suspected glaucoma; CAFPN, channel-attention feature pyramid network; SVM, support vector machine; NA, not applicable.
Performance in identification of single disease on a prospective real-world scenario
A cloud platform that could present the abnormal findings, diagnostic reference and the referral recommendations was established for clinical interaction (Supplementary Figure S5, Video 1). In the test, IEDSS showed greater performance than JO, with average ACCs of 0·9872 (95%CI 0·9828-0·9915) and 0·8846 (95%CI 0·8722-0·8971) (p= 0·000), respectively. It also reached higher average ACC than SO (0·9872, 95%CI 0·9828-0·9915 vs 0·9413, 95%CI 0·9321-0·9504, p= 0·000) and especially worked better in identifying EMM (p = 0·025) and CRVO (p = 0·031). The diagnostic capability of JO was significantly improved with the assistance of IEDSS, with an average ACC increased from 0·8846 (95%CI 0·8722-0·8970) to 0·9906 (95%CI 0·9868-0·9944). The ACC of SO was also significantly increased with the help of IEDSS in MH (0·9972, 95%CI 0·9952-0·9993 vs 0·9996, 95%CI 0·9988-1·000, p = 0·028), EMM (0·9925, 95%CI 0·9891-0·9959 vs 0·9965, 95%CI 0·9941-0·9988, p = 0·046) and RT (0·9968, 95%CI 0·9947-0·9990 vs 0·9996, 95%CI 0·9988-1·000, p = 0·018) (Table 4).
Table 4.
The auxiliary performance of Interpretable Eye Diseases Screening System (IEDSS) in the real-world scenario.
| Diseases | IEDSS |
Junior ophthalmologist (JO) |
Senior ophthalmologist (SO) |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACC (%)(95%CI) | Sensitivity (%) (95%CI) | Specificity (%) (95%CI) |
P value |
Without IEDSS |
With IEDSS |
P value | Without IEDSS |
With IEDSS |
P value | ||||||||||
| IEDSS vs JO | IEDSS vs SO | ACC (%)(95%CI) | Sensitivity (%) (95%CI) | Specificity (%) (95%CI) | ACC (%)(95%CI) | Sensitivity (%) (95%CI) | Specificity (%) (95%CI) | ACC (%)(95%CI) | Sensitivity (%) (95%CI) | Specificity (%) (95%CI) | ACC (%)(95%CI) | Sensitivity (%) (95%CI) | Specificity (%) (95%CI) | ||||||
| DR | 97·36(96·73-97·98) | 97·62(95·18-98·89) | 97·32(96·53-97·93) | 0·001⁎⁎ | 0·268 | 97·83(97·26-98·40) | 91·67(88·05-94·30) | 98·77(98·19-99·17) | 98·62(98·17-99·07) | 93·75(90·46-96·00) | 99·36(98·91-99·64) | 0·299 | 98·93(98·54-99·33) | 96·13(93·31-97·84) | 99·36(98·91-99·64) | 99·45(99·16-99·74) | 97·92(95·57-99·08) | 99·68(99·31-99·86) | 0·173 |
| CSCR | 97·20(96·56-97·84) | 88·37(79·21-93·98) | 97·51(96·79-98·07) | 0·012* | 0·648 | 98·46(97·98-98·94) | 73·26(62·44-81·96) | 99·35(98·92-99·61) | 99·17(98·82-99·52) | 87·21(77·85-93·13) | 99·59(99·22-99·79) | 0·022 | 98·90(98·49-99·30) | 86·05(76·50-92·27) | 99·35(98·92-99·61) | 99·37(99·06-99·68) | 91·86(83·42-96·39) | 99·63(99·28-99·82) | 0·224 |
| MH | 99·41(99·11-99·71) | 96·55(87·05-99·40) | 99·48(99·08-99·71) | 0·015 * | 0·083 | 99·61(99·36-99·85) | 82·76(70·12-90·99) | 100·00(99·81-100·00) | 99·92(99·81-100·00) | 96·55(87·05-99·40) | 100·00(99·81-100·00) | 0·015* | 99·72(99·52-99·93) | 87·93(76·09-94·61) | 100·00(99·81-100·00) | 99·96(99·88-100·00) | 98·28(89·54-99·91) | 100·00(99·81-100·00) | 0·028* |
| EMM | 98·78(98·35-99·20) | 93·94(88·21-97·15) | 99·04(98·54-99·38) | 0·000⁎⁎ | 0·025* | 98·70(98·26-99·14) | 78·03(69·82-84·57) | 99·83(99·54-99·95) | 99·65(99·41-99·59) | 93·94(88·02-97·15) | 99·96(99·73-100·00) | 0·000** | 99·25(98·91-99·59) | 85·61(78·18-90·89) | 100·00(99·80-100·00) | 99·65(99·41-99·88) | 93·18(87·08-96·64) | 100·00(99·80-100·00) | 0·046* |
| Wet-AMD | 97·59(97·00-98·19) | 93·48(87·62-96·78) | 97·83(97·14-98·36) | 0·008⁎⁎ | 0·099 | 98·07(97·53-98·60) | 83·33(75·83-88·93) | 98·92(98·39-99·28) | 99·25(98·91-99·59) | 91·30(84·98-95·22) | 99·71(99·37-99·87) | 0·047* | 99·05(98·68-99·43) | 87·68(80·74-92·45) | 99·71(99·37-99·87) | 99·57(99·31-99·82) | 93·48(87·62-96·78) | 99·92(99·66-99·99) | 0·099 |
| CRVO | 95·94(95·17-96·71) | 99·04(93·99-99·95) | 95·80(94·91-96·55) | 0·010* | 0·031* | 98·86(98·44-99·27) | 91·35(83·78-95·72) | 99·18(98·71-99·48) | 99·17(98·82-99·52) | 94·23(87·36-97·63) | 99·38(98·96-99·64) | 0·421 | 99·33(99·01-99·65) | 93·27(86·15-97·02) | 99·59(99·22-99·79) | 99·68(99·47-99·90) | 97·12(91·19-99·25) | 99·79(99·49-99·92) | 0·195 |
| BRVO | 99·13(98·77-99·49) | 98·39(94·98-99·58) | 99·19(98·74-99·50) | 0·006 ⁎⁎ | 0·311 | 99·17(98·82-99·52) | 92·47(87·44-95·67) | 99·70(99·36-99·87) | 99·61(99·36-99·85) | 96·24(92·09-98·34) | 99·87(99·59-99·97) | 0·116 | 99·72(99·52-99·93) | 96·77(92·79-98·68) | 99·96(99·72-100·00) | 99·80(99·63-99·98) | 97·85(94·23-99·31) | 99·96(99·72-100·00) | 0·5210 |
| RT | 99·80(99·63-99·98) | 98·72(96·01-99·67) | 99·91(99·65-99·98) | 0·000 ⁎⁎ | 0·127 | 98·93(98·54-99·33) | 88·51(83·56-92·16) | 100·00(99·79-100·00) | 99·88(99·75-100·00) | 98·72(96·01-99·67) | 100·00(99·79-100·00) | 0·000⁎⁎ | 99·68(99·47-99·90) | 96·60(93·15-98·41) | 100·00(99·79-100·00) | 99·96(99·88-100·00) | 99·57(97·28-99·98) | 100·00(99·79-100·00) | 0·018* |
| ODE | 99·96(99·88-100·00) | 100·00(88·57-100·00) | 99·88(99·62-99·97) | 0·005⁎⁎ | 0·314 | 99·72(99·52-99·93) | 81·58(65·11-91·68) | 100·00(99·81-100·00) | 99·96(99·88-100·00) | 97·37(84·57-99·86) | 100·00(99·81-100·00) | 0·025* | 99·96(99·98-100·00) | 97·37(84·57-99·86) | 100·00(99·81-100·00) | 100·00(100·00-100·00) | 100·00(88·57-100·00) | 100·00(99·81-100·00) | 0·314 |
| OA | 99·61(99·36-99·85) | 98·82(92·71-99·94) | 99·63(99·28-99·82) | 0·096 | 0·560 | 99·80(99·63-99·98) | 94·12(86·20-97·81) | 100·00(99·80-100·00) | 99·96(99·88-100·00) | 98·82(92·71-99·94) | 100·00(99·80-100·00) | 0·096 | 99·92(99·81-100·00) | 97·65(90·96-99·59) | 100·00(99·80-100·00) | 99·96(99·88-100·00) | 98·82(92·71-99·94) | 100·00(99·80-100·00) | 0·560 |
| SG | 99·57(99·31-99·82) | 97·86(94·81-99·21) | 99·17(98·69-99·49) | 0·000⁎⁎ | 0·055 | 99·25(98·91-99·59) | 91·88(87·42-94·91) | 100·00(99·79-100·00) | 99·80(99·63-99·98) | 97·86(94·81-99·21) | 100·00(99·79-100·00) | 0·003⁎⁎ | 99·68(99·47-99·90) | 96·58(93·13-98·40) | 100·00(99·79-100·00) | 99·84(99·69-100·00) | 98·29(95·39-99·45) | 100·00(99·79-100·00) | 0·242 |
| RD | 99·84(99·69-100·00) | 99·29(97·77-99·82) | 99·95(99·69-99·99) | 0·033* | 0·563 | 99·72(99·52-99·93) | 98·34(96·46-99·27) | 100·00(99·77-100·00) | 99·96(99·88-100·00) | 99·76(98·48-99·99) | 100·00(99·77-100·00) | 0·033* | 99·92(99·81-100·00) | 99·53(98·11-99·92) | 100·00(99·77-100·00) | 99·96(99·88-100·00) | 99·76(98·48-99·99) | 100·00(99·77-100·00) | 0·563 |
| RMO | 98·07(97·53-98·60) | 95·65(91·91-97·78) | 98·31(97·67-98·77) | 0·662 | 0·189 | 99·17(98·82-99·52) | 94·78(90·84-97·15) | 99·61(99·23-99·81) | 99·53(99·26-99·79) | 95·65(91·91-98·77) | 99·91(99·65-99·99) | 0·662 | 99·68(99·47-99·90) | 97·83(94·72-99·20) | 99·87(99·59-99·97) | 99·76(99·88-100·00) | 97·83(94·72-99·20) | 99·87(99·59-99·97) | 1·000 |
| VH | 99·87(99·57-99·95) | 98·80(96·26-99·69) | 99·87(99·5-99·96) | 0·080 | 1·000 | 99·49(99·21-99·77) | 96·41(93·17-98·24) | 99·82(99·52-99·94) | 99·76(99·57-99·95) | 99·20(96·84-99·86) | 99·82(99·52-99·94) | 0·033* | 99·60(99·36-99·85) | 98·80(96·26-99·69) | 99·91(99·65-99·98) | 99·92(99·81-100·00) | 99·60(97·45-99·98) | 99·96(99·72-100·00) | 0·315 |
DR, diabetic retinopathy; CSCR, central serous chorioretinopathy; MH, macular hole; EMM, epimacular membrane; wet-AMD, wet age-related macular degeneration; CRVO, central retinal vein occlusion; BRVO, branch retinal vein occlusion; RT, retinal tears; ODE, optic disc edema; OA, optic atrophy; SG, suspected glaucoma; RD, retinal detachment; RMO, refractive media opacity; VH, vitreous hemorrhage.
p < 0·05,
p < 0·01.
Expandability of IEDSS in multimorbidity datasets
In the expanded task, the performance of IEDSS was better than that of JO (ACC = 0·8560, 95%CI 0·8252-0·8868 vs 0·7840, 95%CI 0·7479-0·8201, p = 0·003) and comparable to that of SO (ACC = 0·8560, 95%CI 0·8252-0·8868 vs 0·850, 95%CI 0·9952-0·9993, p = 0·789). With the help of IEDSS, it achieved a higher ACC of 0·8480 (95%CI 0·8165-0·8795) by JO (p = 0·009) and 0·8740 (95%CI 0·8449-0·9031) by SO (p = 0·271) (Table 5). It was found that the misdiagnoses mainly existed in diseases sharing same pathological features such as AMD and chronic CSCR, CRVO and DR, etc. IEDSS was more likely to generate false positives when the lesion atlas of the sample was similar to other diseases. The diagnoses of 30 examples by IEDSS and two ophthalmologists with (without) the assistance of IEDSS were listed in Figure 6.
Table 5.
The auxiliary performance of Interpretable Eye Diseases Screening System (IEDSS) in identifying multimorbidity.
| Accuracy (95% CI) | P value(vs IEDSS) | P value(with IEDSS vs without IEDSS) | Correct diagnosis | Misidenti-fication | Undetected lesions | Misdetected lesions | |
|---|---|---|---|---|---|---|---|
| Junior ophthalmologist without IEDSS | 78·40 (74·79-82·01) |
0·003⁎⁎ | 0·009⁎⁎ | 392 | 61 | 29 | 22 |
| Junior ophthalmologist with IEDSS | 84·80 (81·65-87·95) |
0·722 | 424 | 51 | 15 | 14 | |
| Senior ophthalmologist without IEDSS | 85·00 (81·87-88·13) |
0·789 | 0·271 | 425 | 32 | 18 | 27 |
| Senior ophthalmologist with IEDSS | 87·40 (84·49-90·31) |
0·405 | 437 | 30 | 10 | 23 | |
| IEDSS | 85·60 (82·52-88·68) |
428 | 53 | 9 | 16 |
Correct diagnosis was defined as successfully identifying all types of diseases. The wrong samples were summarized into: (1) misidentification, which was defined as the lesion was correctly detected but the disease was misdiagnosed; (2) undetected lesions, which was defined as the missed detection of lesions; (3) misdetected lesions, including false positives and wrongly categorized lesions. *p < 0·05,
p < 0·01.
Figure 6.
The diagnosis of 30 examples of multimorbidity by the IEDSS and two ophthalmologists with (without) assistance of Interpretable eye diseases screening system (IEDSS). Correct diagnosis was defined as successfully identifying all types of diseases. The wrong samples were analysed and summarized the reasons into the following three types: 1. misidentification, which was defined as the lesion was correctly detected but the disease was misdiagnosed, 2. undetected lesions, which was defined as the missed detection of lesions, 3. misdetected lesions, including false positives and wrongly categorized lesions. DR, diabetic retinopathy; CSCR, central serous chorioretinopathy; AMD, age-related macular degeneration; MH, macular Hole; EMM, epimacular membrane; CRVO, central retinal vein occlusion; BRVO, branch retinal vein occlusion; ODE, optic disc edema; OA, optic atrophy; SG, suspected glaucoma.
Discussion
We developed a four-level hierarchical eye diseases screening system integrated with lesion atlas that could identify up to 30 abnormalities and eye diseases in UWF. The lesion atlas, an inductive pattern was introduced to the DL training process, to overcome the clinical challenge that the limited training samples could not cover diverse clinical settings. It also surmounted the drawback that the DL system often lacked fine-grained explainable information which limited the application of computer-aided diagnosis system. The system reached an average ACC of 0·987 in the real-world scenario and 0·856 in diagnosing multimorbidity. The performance of the system rivaled that of senior human ophthalmologists and could significantly increase the diagnostic accuracy of junior clinicians. It would greatly facilitate learning in medical knowledge and improve efficiency of clinical diagnosis especially in remote areas with few specialists.
Our system covered a wide range of diseases from UWF images and demonstrated favorable accuracy for all classes. It obtained higher AUCs of over 0·981 in the multi-label task of UWF images compared with previous study of identifying singles diseases, with 0·976 of BRVO.36, 0·915 of DR,37 0·953 of RT.38 The sensitivity was greatly improved in our study to reduce miss rates (0·980 vs 0·940 of BRVO,36 0·959 vs 0·834 of DR,37 0·965 vs 0·875 of RT).38 The sensitivity of IEDSS was higher than JO and comparable to that of SO (Supplementary Table S2). Several innovations were proposed in IEDSS to achieve a higher accuracy and sensitivity with less training data. Firstly, IEDSS was designed to extract lesions directly from original images to suppress irrelevant noise and amplify critical features. The t-SNE map exhibited prominent class separability of IEDSS (Figure 5). The extracted features and their distribution pattern could clearly distinguish different diseases (Figure 4 and Supplementary Figure S4). Secondly, the lesion-based diagnosis model could efficiently improve the fault-tolerant ability of the system. A few errors in detection would not notably take the edge off the system's performance because the judgement was based on the summarization of the lesions’ geostatistical information and the system did not need to detect every pathological change. In the comparative test with image-level models, it was proved that IEDSS achieved higher ACCs and F1 scores. The average sensitivity was dramatically increased by over 20% after extracting key features (Table 3 and Supplementary Table S4). Besides, compared with CAFPN+SVM, it could be found that the AdaBoost module could help to form a stronger SVM, with an average ACC increased from 0·9381 to 0·9781. The improvement of ACCs and sensitivities was particularly suitable for the diseases screening task especially when there was a significant shortage of ophthalmologists in developing countries.39
IEDSS demonstrated excellent expandability in complex clinical settings. Previously, several studies tried to identify multiple diseases from colorful fundus photographs (CFP). Son et al. developed 12 classification neural networks to screen 12 abnormal findings in CFP.40 Cen et al. built a multi-label model that could detect 39 classes of fundus diseases and conditions.41 Whereas, it was still hard for these models to apply to patients with more than one disease, which were common in the clinic. It was difficult for multi-class models to recognize multimorbidity since the outcomes were mutually exclusive. The task was also challenged for image-level multi-label classifiers due to noise and intertwined features. In view of these problems, lesion atlas was introduced to overcome the obstacles and make sound judgements by essentially discriminating the pathological changes of different diseases. It was demonstrated that IEDSS with lesion atlas could recognize more than one disease in one UWF image with high accuracy even if it was not trained for such samples before. The performance of IEDSS in recognizing multimorbidity was excellent and better than that of JO (0·856 vs 0·784, p = 0·003) and comparable to that of SO (ACC = 0·856 vs 0·850, p = 0·789) (Table 5).
The interpretability and acceptance of the system was also improved with lesion atlas proposed to visualize the analyzing progress of the DL system. With the dramatic advances of DL, the “black-box” nature has been the most challenging factor that limited large-scale adoption of AI in healthcare. Several technical advancements were provided to solve this problem, such as occlusion testing,42 class activation mappings (CAMs)43 and so on. However, the highlighted area was often hard for clinical interpretation44 and it was unclear that whether it was a new biomarker or just an erroneous correlation.45 There could be a considerable disagreement between highlighted regions and expert annotations.46 In addition, these techniques were not suitable for retinal diseases perfectly since they could not precisely highlight small targets. Herein, lesion atlas was raised to present the evidence of the DL-based diagnosis in a pathological and anatomical level. Compared with previous studies40,41 which highlighted a rough outline as potential pathological changes using heatmaps, the system focused on detailed pathological information, which mimicked human thought process. The visualized lesion feature bounding boxes, distribution diagrams and urgency determination were presented to doctors by a user-interface established in our study (Supplementary Figure S5). Ophthalmologists could combine the clinical experience and pathological changes provided by IEDSS to make the diagnosis and treatment decisions more reliably. In the prospective real-world test, the ACC of JO was significantly enhanced with the assistance of IEDSS (Table 4). The performance of SO in discerning MH, EMM and RT from UWF images could also be promoted by IEDSS. The system with high sensitivity for minor lesions could be suited to young doctors without abundant clinical experience and medical students in the early learning stage. The auxiliary pattern seemed to be particularly essential when there was an ophthalmological health service gap worldwide, with only one ophthalmologist per 110,000 people in developing countries and one ophthalmologist per 13,000 in developed countries.39
There were several limitations in our study. Firstly, it was hard to precisely detect subtle changes such as MA, tiny IRH and CWS. It was warranted to develop algorithms to detect subtle pathological changes in retinal images with higher accuracy. Secondly, it would often lead to confusion when diseases shared similar lesions and distribution, as showed in Figure 5. The confused diseases mostly focused on vascular diseases (DR, BRVO and CRVO) and CNV-related diseases (AMD and chronic CSC). To make a more confirmative diagnosis, a more delicate analysis of vascular morphology and a comprehensive clinical information were required. Thirdly, the concrete number and area of lesions were not calculated. Several neighbored lesions would be boxed into the same detection frame. In addition, only accuracy was calculated for evaluation of expandability of IEDSS in a multimorbidity scenario because it was hard to define false positive and false negative for the images with more than one diagnosis. Lastly, samples collected from three clinical centres this study included populations from different regions of China and could be representative of Asian populations, to a certain extent. The community-based study and multiethnic clinic-based study were needed to further investigated the generalization performance of IEDSS in other ethnics.
In conclusion, we designed an explainable and expansible DL-assisted eye diseases screening system and evaluated its performance in the multicentre scenario, the extended multimorbidity scenario and the real-world scenario. Lesion atlas was proposed to improve the accuracy, interpretability and potential of clinical application of IEDSS by visualizing fine-grained pathological and anatomical information. The system would be more valuable for clinical requirements. It could significantly enhance the efficiency and reliability of eye diseases diagnosis, and equilibrate medical resources, especially in remote areas with limited and uneven medical resources.
Contributors
J. Cao, K. You and J. Ye conceptualized and designed the study. J. Cao, J.X. Zhou, M.Y. Xu, P.F. Xu, Y. Wang, L. Wen, S.Z. Wang and K. Jin collected and assessed the data. J. Cao, P.F. Xu and K. You contributed to the data analysis and data interpretation. J. Cao and K. You drafted the manuscript. J.X. Zhou, M.Y. Xu, P.F. Xu, Y. Wang, L. Wen, S.Z. Wang, L.X. Lou, K. Jin and J. Ye critically revised the manuscript. J. Ye, L. Wen, S.Z. Wang have accessed and verified the data. J. Ye provided research fundings, supervised the study and coordinated the research. All authors had full access to all the data in the study and accept responsibility to submit for publication. J. Cao and K. You contributed equally as first authors.
Data sharing statement
Due to the privacy of patients, the data related to patients cannot be available for public access but can be obtained from the corresponding author on reasonable request approved by the human research ethics committee of the Second Affiliated Hospital of Zhejiang University.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
We sincerely appreciated the support from the National Natural Science Foundation Regional Innovation and Development Joint Fund (U20A20386), Key research and development program of Zhejiang Province (2019C03020), Clinical Medical Research Centre for Eye Diseases of Zhejiang Province (2021E50007). The funding organisation played no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript for publication.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.eclinm.2022.101633.
Appendix. Supplementary materials
References
- 1.World Health Organization. “Universal eye health: A global action plan 2014-2019”, https://www.who.int/blindness/actionplan/en/. Accessed 20 March 2022.
- 2.World Health Organization. “World report on vision”, https://www.who.int/publications detail/world report on vision. Accessed 20 March 2022.
- 3.Liu YC, Wilkins M, Kim T, Malyugin B, Mehta JS. Cataracts. Lancet. 2017;390(10094):600–612. doi: 10.1016/S0140-6736(17)30544-5. [DOI] [PubMed] [Google Scholar]
- 4.Yau JW, Rogers SL, Kawasaki R, et al. Global prevalence and major risk factors of diabetic retinopathy. Diabetes Care. 2012;35(3):556–564. doi: 10.2337/dc11-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong WL, Su X, Li X, et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob health. 2014;2(2):e106–e116. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 6.Tham YC, Li X, Wong TY, Quigley HA, Aung T, Cheng CY. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014;121(11):2081–2090. doi: 10.1016/j.ophtha.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 7.Eckert KA, Carter MJ, Lansingh VC, et al. A Simple Method for Estimating the Economic Cost of Productivity Loss Due to Blindness and Moderate to Severe Visual Impairment. Ophthalmic Epidemiol. 2015;22(5):349–355. doi: 10.3109/09286586.2015.1066394. [DOI] [PubMed] [Google Scholar]
- 8.McCarty CA, Nanjan MB, Taylor HR. Vision impairment predicts 5 year mortality. Br J Ophthalmol. 2001;85(3):322–326. doi: 10.1136/bjo.85.3.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagiel A, Lalane RA, Sadda SR, Schwartz SD. Ultra-widefield fundus imaging: a review of clinical applications and future trends. Retina. 2016;36(4):660–678. doi: 10.1097/IAE.0000000000000937. [DOI] [PubMed] [Google Scholar]
- 10.Mrejen S, Sarraf D, Chexal S, Wald K, Freund KB. Choroidal involvement in acute posterior multifocal placoid pigment epitheliopathy. Ophthalmic Surg Lasers Imaging Retina. 2016;47(1):20–26. doi: 10.3928/23258160-20151214-03. [DOI] [PubMed] [Google Scholar]
- 11.Abràmoff MD, Lavin PT, Birch M, Shah N, Folk JC. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. NPJ Digit Med. 2018;1:39. doi: 10.1038/s41746-018-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ting DSW, Cheung CY, Lim G, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318(22):2211–2223. doi: 10.1001/jama.2017.18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng Y, Dharssi S, Chen Q, et al. DeepSeeNet: A deep learning model for automated classification of patient-based age-related macular degeneration severity from color fundus photographs. Ophthalmology. 2019;126(4):565–575. doi: 10.1016/j.ophtha.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh A, Dutta MK, ParthaSarathi M, Uher V, Burget R. Image processing based automatic diagnosis of glaucoma using wavelet features of segmented optic disc from fundus image. Comput Methods Programs Biomed. 2016;124:108–120. doi: 10.1016/j.cmpb.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Dong L, He W, Zhang R, et al. Artificial intelligence for screening of multiple retinal and optic nerve diseases. JAMA Netw Open. 2022;5(5) doi: 10.1001/jamanetworkopen.2022.9960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi JY, Yoo TK, Seo JG, Kwak J, Um TT, Rim TH. Multi-categorical deep learning neural network to classify retinal images: a pilot study employing small database. PLoS One. 2017;12(11) doi: 10.1371/journal.pone.0187336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Chen H, Zhang B, et al. Development and evaluation of a deep learning model for the detection of multiple fundus diseases based on colour fundus photography. Br J Ophthalmol. 2021;106(8):1079–1086. doi: 10.1136/bjophthalmol-2020-316290. [DOI] [PubMed] [Google Scholar]
- 18.Han J, Choi S, Park JI, et al. Classifying neovascular age-related macular degeneration with a deep convolutional neural network based on optical coherence tomography images. Sci Rep. 2022;12(1):2232. doi: 10.1038/s41598-022-05903-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko YC, Wey SY, Chen WT, et al. Deep learning assisted detection of glaucomatous optic neuropathy and potential designs for a generalizable model. PLoS One. 2020;15(5) doi: 10.1371/journal.pone.0233079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Qiang W, Chen H, et al. Artificial intelligence to detect malignant eyelid tumors from photographic images. NPJ Digit Med. 2022;5(1):23. doi: 10.1038/s41746-022-00571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gargeya R, Leng T. Automated identification of diabetic retinopathy using deep learning. Ophthalmology. 2017;124(7):962–969. doi: 10.1016/j.ophtha.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Zhang K, Liu X, Xu J, et al. Deep-learning models for the detection and incidence prediction of chronic kidney disease and type 2 diabetes from retinal fundus images. Nat Biomed Eng. 2021;5(6):533–545. doi: 10.1038/s41551-021-00745-6. [DOI] [PubMed] [Google Scholar]
- 23.Bagheri Nika, Wajda Brynn N. Lippincott Williams & Wilkins; Philadelphia: 2017. The Wills Eye Manual: Office and Emergency Room Diagnosis and Treatment of Eye Disease. [Google Scholar]
- 24.Cat Nguyen Burkat. EyeWiki: The Eye Encyclopedia written by Eye Physicians & Surgeons. https://eyewiki.org/Main_Page. Accessed 20 March 2022.
- 25.Columbia university department of ophthalmology. Digital-reference-of-ophthalmology. https://www.columbiaeye.org/education/digital-reference-of-ophthalmology. Accessed 20 March 2022.
- 26.Xie S, Girshick R, Dollár P, Tu Z, He K. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Aggregated residual transformations for deep neural networks. [Google Scholar]
- 27.Ronneberger O, Fischer P, Brox T, editors. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. U-net: Convolutional networks for biomedical image segmentation. [Google Scholar]
- 28.Larsen H.W. WB Saunders Company; Philadelphia: 1976. The Ocular Fundus: a Color Atlas. [Google Scholar]
- 29.Lin T-Y, Dollár P, Girshick R, He K, Hariharan B, Belongie S, editors. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Feature pyramid networks for object detection. [Google Scholar]
- 30.Schlemper J, Oktay O, Schaap M, et al. Attention gated networks: Learning to leverage salient regions in medical images. Med Image Anal. 2019;53:197–207. doi: 10.1016/j.media.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedman J, Hastie T, Tibshirani R. Additive logistic regression: a statistical view of boosting (with discussion and a rejoinder by the authors) Ann Stat. 2000;28(2):337–407. [Google Scholar]
- 32.Dosovitskiy A, Beyer L, Kolesnikov A, et al. ICLR; 2020. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. [Google Scholar]
- 33.Murre JM, Dros J. Replication and analysis of Ebbinghaus' forgetting curve. PLoS One. 2015;10(7) doi: 10.1371/journal.pone.0120644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chicco D, Jurman G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics. 2020;21(1):6. doi: 10.1186/s12864-019-6413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yedidia A. 2016. Against the F-score.https://adamyedidia.files.wordpress.com/2014/11/f_score.pdf Accessed 10 December 2019. [Google Scholar]
- 36.Nagasato D, Tabuchi H, Ohsugi H, et al. Deep-learning classifier with ultrawide-field fundus ophthalmoscopy for detecting branch retinal vein occlusion. Int J Ophthalmol. 2019;12(1):94–99. doi: 10.18240/ijo.2019.01.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh K, Kang HM, Leem D, Lee H, Seo KY, Yoon S. Early detection of diabetic retinopathy based on deep learning and ultra-wide-field fundus images. Sci Rep. 2021;11(1):1897. doi: 10.1038/s41598-021-81539-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang C, He F, Li B, et al. Development of a deep-learning system for detection of lattice degeneration, retinal breaks, and retinal detachment in tessellated eyes using ultra-wide-field fundus images: a pilot study. Graefe's Arch Clinic Exp Ophthalmol. 2021;259(8):2225–2234. doi: 10.1007/s00417-021-05105-3. [DOI] [PubMed] [Google Scholar]
- 39.Resnikoff S, Lansingh VC, Washburn L, et al. Estimated number of ophthalmologists worldwide (International Council of Ophthalmology update): will we meet the needs? Br J Ophthalmol. 2020;104(4):588–592. doi: 10.1136/bjophthalmol-2019-314336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Son J, Shin JY, Kim HD, Jung KH, Park KH, Park SJ. Development and validation of deep learning models for screening multiple abnormal findings in retinal fundus images. Ophthalmology. 2020;127(1):85–94. doi: 10.1016/j.ophtha.2019.05.029. [DOI] [PubMed] [Google Scholar]
- 41.Cen LP, Ji J, Lin JW, et al. Automatic detection of 39 fundus diseases and conditions in retinal photographs using deep neural networks. Nat Commun. 2021;12(1):4828. doi: 10.1038/s41467-021-25138-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li F, Yan L, Wang Y, et al. Deep learning-based automated detection of glaucomatous optic neuropathy on color fundus photographs. Graefe's Arch Clinic Exp Ophthalmol. 2020;258(4):851–867. doi: 10.1007/s00417-020-04609-8. [DOI] [PubMed] [Google Scholar]
- 43.Poplin R, Varadarajan AV, Blumer K, et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng. 2018;2(3):158–164. doi: 10.1038/s41551-018-0195-0. [DOI] [PubMed] [Google Scholar]
- 44.Ramanishka V, Das A, Zhang J. Top-down visual saliency guided by captions. https://arxiv.org/abs/1612.07360. Accessed 20 March 2022.
- 45.Keel S, Wu J, Lee PY, Scheetz J, He M. Visualizing deep learning models for the detection of referable diabetic retinopathy and glaucoma. JAMA Ophthalmol. 2019;137(3):288–292. doi: 10.1001/jamaophthalmol.2018.6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Craenendonck T, Elen B, Gerrits N, De Boever P. Systematic comparison of heatmapping techniques in deep learning in the context of diabetic retinopathy lesion detection. Transl Vis Sci Technol. 2020;9(2):64. doi: 10.1167/tvst.9.2.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.