Summary
Human broadly neutralizing antibodies (bnAbs) targeting the hemagglutinin stalk of group 1 influenza A viruses (IAVs) are biased for IGHV1-69 alleles that use phenylalanine (F54) but not leucine (L54) within their CDRH2 loops. Despite this, we demonstrated that both alleles encode for human IAV bnAbs that employ structurally convergent modes of contact to the same epitope. To resolve differences in lineage-expandability, we compared F54 vs L54 as substrate within humanized mice where antibodies develop with human-like CDRH3 diversity but are restricted to single VH-genes. While both alleles encoded for bnAb precursors, only F54 IGHV1-69 supported elicitation of heterosubtypic serum bnAbs following immunization with a stalk-only nanoparticle vaccine. L54 IGHV1-69 was unproductive, co-encoding for anergic B cells and autoreactive stalk antibodies that were cleared from B cell memory. Moreover, human stalk antibodies also demonstrated L54-dependent autoreactivity. Therefore, IGHV1-69 polymorphism, which is skewed ethnically, gates tolerance and vaccine-expandability of influenza bnAbs.
Graphical Abstract
eTOC:
Human broadly neutralizing antibodies (bnAbs) against influenza virus preferentially use F54 IGHV1-69 as opposed to the L54 allele of this antibody VH-gene. Sangesland et al. show that while both alleles encode for affinity to the same target, L54 IGHV1-69 also imparts autoreactivity, dampening vaccine-expandability in a humanized mouse model.
Introduction
Antibodies possesses exceptional diversity and can recognize essentially any antigen. In humans, this is enabled by a repertoire of ~1012 B cell receptors (BCRs) in which each BCR presents a unique configuration of hypervariable and antibody variable (V) gene-encoded antigen complementarity determining regions (CDRs) (Briney et al., 2019; Mora, 2019). Diversity is concentrated in the centrally positioned and hypervariable heavy chain CDRH3, which typically serves as the principal determinant of antigen recognition (Glanville et al., 2009; Xu and Davis, 2000). However, CDRH3-dominant antigen recognition often fails to explore the antigenic space evenly as hypervariable ‘vaccine-resistant’ viruses, such as HIV or influenza virus, establish complex immunodominance hierarchies that prevent the reproducible expansion of broadly neutralizing antibodies (bnAbs) targeting conserved sites of vulnerability on these pathogens (Altman et al., 2018; Kwong and Mascola, 2018). While the mechanisms governing B cell immunodominance are not fully understood, subdominance or immuno-recessive responses can occur when low frequencies of site-specific (on-target) BCRs are unable to compete for expansion within B cell germinal centers (GCs), allowing ‘immunodistracted’ BCRs targeting other sites (off-target) to dominate affinity maturation (Abbott and Crotty, 2020; Sangesland and Lingwood, 2021a).
In addition to CDRH3-dominant antibodies, some human bnAbs rely on V gene-encoded CDRs for principal contact to antigen, resulting in genetically reproducible or ‘public’ development pathways (Henry Dunand and Wilson, 2015; Lerner, 2011; Lingwood et al., 2012; Zhou et al., 2015). This has catalyzed the development of germline-stimulating vaccines, tasked with amplifying these genetically hardwired bnAb lineages (Abbott and Crotty, 2020; Burton, 2019; Kwong and Mascola, 2018; Peterhoff and Wagner, 2017; Sangesland and Lingwood, 2021b; Stamatatos et al., 2017). In the context of influenza A viruses (IAVs), we have recently demonstrated that public targeting provides a genetically reproducible foundation for vaccine-amplifying an otherwise immunologically subdominant human influenza bnAb response, which can be selectively expanded using a single rationally designed immunogen (Amitai et al., 2020; Sangesland et al., 2019).
IAV remains a major public health burden that continues to defy traditional approaches to vaccination due to ongoing viral evolution and immune-distractive properties (Nachbagauer and Palese, 2020; Paules et al., 2018). IAVs are divided into two phylogenetic and antigenically distinct groups (group 1 and group 2) encompassing subtype diversity of the viral surface protein hemagglutinin (HA), the current seasonal vaccine antigen and primary target of the antibody response (Erbelding et al., 2018; Paules and Subbarao, 2017). Structurally, HA is composed of a variable head domain and relatively invariant membrane proximal stalk region that displays a functionally conserved hydrophobic groove upon which human heterosubtypic group 1 and group 2 IAV bnAbs can center (Lee and Wilson, 2015; Sangesland and Lingwood, 2021a). However, HA stalk bnAbs are immunologically recessive after viral infection or immunization with conventional HA vaccines, requiring rational immunogen design concepts for elicitation at higher titer (Angeletti et al., 2019; Boyoglu-Barnum et al., 2021; Nachbagauer et al., 2021; Wei et al., 2020).
Despite being immunologically subdominant, humans do harbor a public or multidonor bnAb response with specificity to the central HA stalk bnAb epitope of group 1 IAVs, which emanates from a CDRH2 contact encoded by the antibody VH-gene IGHV1-69 (Avnir et al., 2014; Corti et al., 2010; Lingwood et al., 2012; Pappas et al., 2014; Throsby et al., 2008; Wheatley et al., 2015; Whittle et al., 2014; Wrammert et al., 2011). Analogous VH-gene endowed pathways to this vaccine target have also been described in non-human primates (Darricarrere et al., 2021). Using HC2 transgenic mice, which contain human-like CDRH3 diversity but are constrained to single user defined human VH-genes, we demonstrated that unlike other VH sequences, IGHV1-69 endows the germline repertoire with a reproducible source of B cells bearing affinity for the group 1 IAV bnAb epitope, which enables vaccine-amplification of immunodominant stalk bnAbs following immunization with the stalk-only nanoparticle immunogen SS-np (Amitai et al., 2020; Sangesland et al., 2019). The bnAb response could not be amplified in IGHV1-69 animals when immuno-distractive features of HA, namely the globular head domain, were incorporated into the nanoparticle immunogen (Amitai et al., 2020; Sangesland et al., 2019). We therefore introduced the principle of bnAb elicitation through pathway-amplification, wherein genetically deterministic affinity for cognate antigen can be selectively harnessed through rational vaccine design to expand normally immuno-recessive human bnAbs.
IGHV1-69 is also polymorphic, encoding for allelic variants that are categorized as 51p1-like or hv1263-like based on the presence of either a phenylalanine (F) or leucine (L) at position 54 within CDRH2, the critical apex residue which forms the initial contact to the bnAb epitope on the HA stalk (Avnir et al., 2016; Lingwood et al., 2012; Pappas et al., 2014; Wheatley et al., 2015). Given that the vast majority of human IGHV1-69 stalk bnAbs derive from the F54 allele, our previous studies in HC2 mice focused on the F54 variant. Similarly broad L54 stalk bnAbs do exist in humans, however L54 usage in group 1 IAV bnAbs is markedly depressed in B cell memory and L54 carriers exhibit lower serum titers of such bnAbs (Avnir et al., 2016; Pappas et al., 2014; Wheatley et al., 2015), which could indicate a hampered capacity to pathway-amplify the public bnAb response in these individuals. Notably, ~13% of the global population is homozygous for L54 IGHV1-69 (Lingwood et al., 2012), but L54 homozygosity appears elevated within certain ethnicities (Avnir et al., 2016). Hence, it is important to experimentally define whether L54 carriers will be able to support vaccine-elicitation of influenza bnAbs.
Results
F54 and L54 Allelic Usage Varies Across Global Populations
Homozygosity of L54 allelic usage is roughly 13% globally (Genomes Project et al., 2010; Lingwood et al., 2012), a frequency that varies across ethnic groups (Avnir et al., 2016; Genomes Project et al., 2010). Using the same public dataset, we assessed if there were statistically significant differences in the frequencies of the F54/F54, L54/L54, and F54/L54 genotypes across the 26 ethnic sub-populations recorded in the 1000 genomes project (Figure S1). Amongst homozygotes, we note statistically significant skewing for F54/F54 usage across the subpopulations, with the exception of South Asians, which maintain a distinct preference for the L54/L54 genotype, consistent with the previous report (Avnir et al., 2016; Genomes Project et al., 2010). We also note that the majority of humans, including amongst South Asians, utilize at least one copy of F54 IGHV1-69 as heterozygotes (Figure S1).
Human L54 Stalk bnAbs Reveal Structurally Convergent Contact Solutions to the Same Epitope
The paratopes of F54 IGHV1-69 stalk bnAbs are marked by contact through hydrophobic I53 and F54 CDRH2 residues as well as CDRH3 tyrosine residues which donate hydrogen bonds at Kabat positions 98–100 (Avnir et al., 2014; Ekiert et al., 2009; Lingwood et al., 2012; Pappas et al., 2014; Sui et al., 2009). We applied cryo-electron microscopy to solve the liganded structures of two L54 stalk bnAbs, 310-39G10 and 310-63E6, which were isolated from different donors (Wheatley et al., 2015). The antibodies were complexed with influenza hemagglutinin (H1N1 NC99) and were resolved to 3.37Å (HA+ 310-39G10) and 3.09 Å (HA+ 310-63E6) (Figures 1A–C, S2, S3A–B, Table S1). We find that both antibodies engaged the HA stalk with minimal light chain contact (Figures 1A–C, S3A–B), akin to the VH-constrained contact exhibited by F54 bnAbs (Ekiert et al., 2009; Sui et al., 2009). Moreover, 310-39G10 exhibited analogous germline contact through I53 in CDRH2 and Y98 in CDRH3 (Figures 1A, S3A). The 310-63E6 bnAb bound to an expanded epitope, where antigen engagement by the heavy chain was supported by contact through CDRH1, FR3 and CDRH3 (F100, Y100a, Y100b) (Figure 1B and S3B). In all cases, the contact angle was similarly constrained by IGHV1-69 with an overlapping epitope footprint between L54 and F54 bnAbs (Figures 1D–E, S3C, Table S2).
Figure 1. Structurally convergent VH-constrained targeting by human L54 and F54 IGHV1-69 bnAbs.
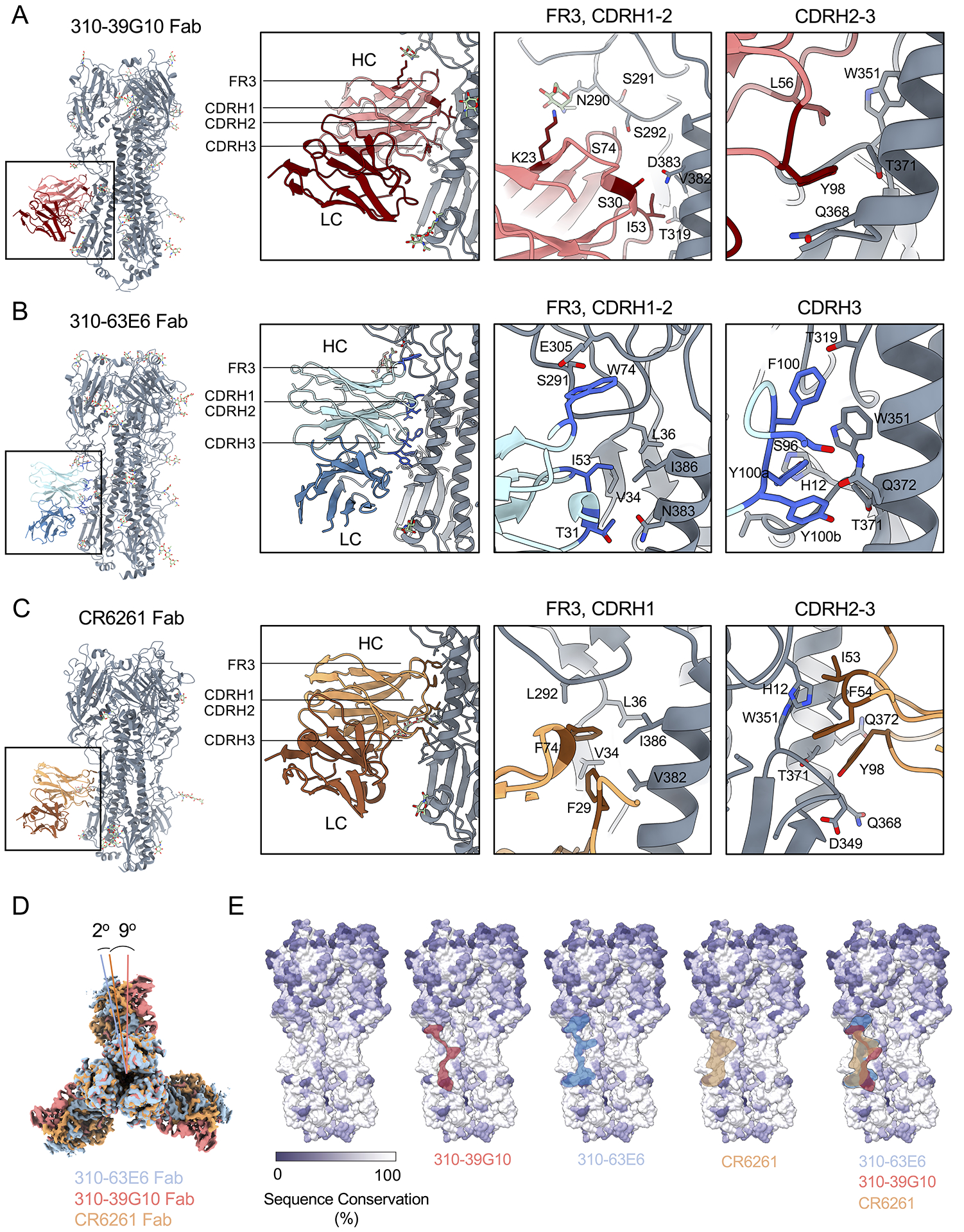
Structures of HA trimers in complex with (A) 310-39G10, (B) 310-63E6, (C) and CR6261 (PDB ID: 3gbm). HA trimers are shown in grey with heavy (HC) and light chains (LC) for each antibody represented as different colors and labeled. Relevant epitope and paratope regions are indicated in the panels. Antibody residues in direct contact with HA are represented as sticks and labeled. (D) Differences in angles of approach of the antibodies 310-39G10 and 310-63E6 relative to CR6261 were calculated using UCSF Chimera by superimposition of the two cryo-EM models of HA bound to 310-39G10 Fab, 310-63E6 Fab and the x-ray crystal structure of HA bound to CR6261 Fab (PDB 3GBM). (E) Surface representation of HA NC99 colored based on sequence conservation. The location of 310-39G10, 310-63E6 and CR6261 epitopes are outlined in red, blue and tan, respectively. HA sequences used are represented in Table S2. See also Figures S2, S3, and Table S1.
The HC2 Locus Enables Experimental Evaluation of F54 vs L54 IGHV1-69 Contribution to Vaccine-Amplification of Human IAV bnAbs
We deployed humanized mice homozygous for the integrated HC2 locus, as in previous studies (Amitai et al., 2020; Ronsard et al., 2021; Sangesland et al., 2019; Sangesland et al., 2020). The HC2 locus restricts antibodies to single user defined VH-genes but simultaneously ensures human-like diversity in the antibody CDRH3 loops (Sangesland et al., 2019; Sangesland et al., 2020). In this study, the VH-genes were fully restricted to the germline sequences of either IGHV1-69*01 (F54) or IGHV1-69*09 (L54), commonly used forms of the F54 and L54 alleles (Sasso et al., 1993; Wheatley et al., 2015) (Figures 2A, S4A). In both cases, germline CDRH3 diversity was confirmed by deep sequencing the naïve IgM BCR repertoires, which we found to mirror the human antibody repertoire in terms of CDRH3 length distribution and amino acid composition (Figure 2B–C).
Figure 2. L54 IGHV1-69 paucity is not due to bnAb precursor frequency or affinity in HC2 mice.
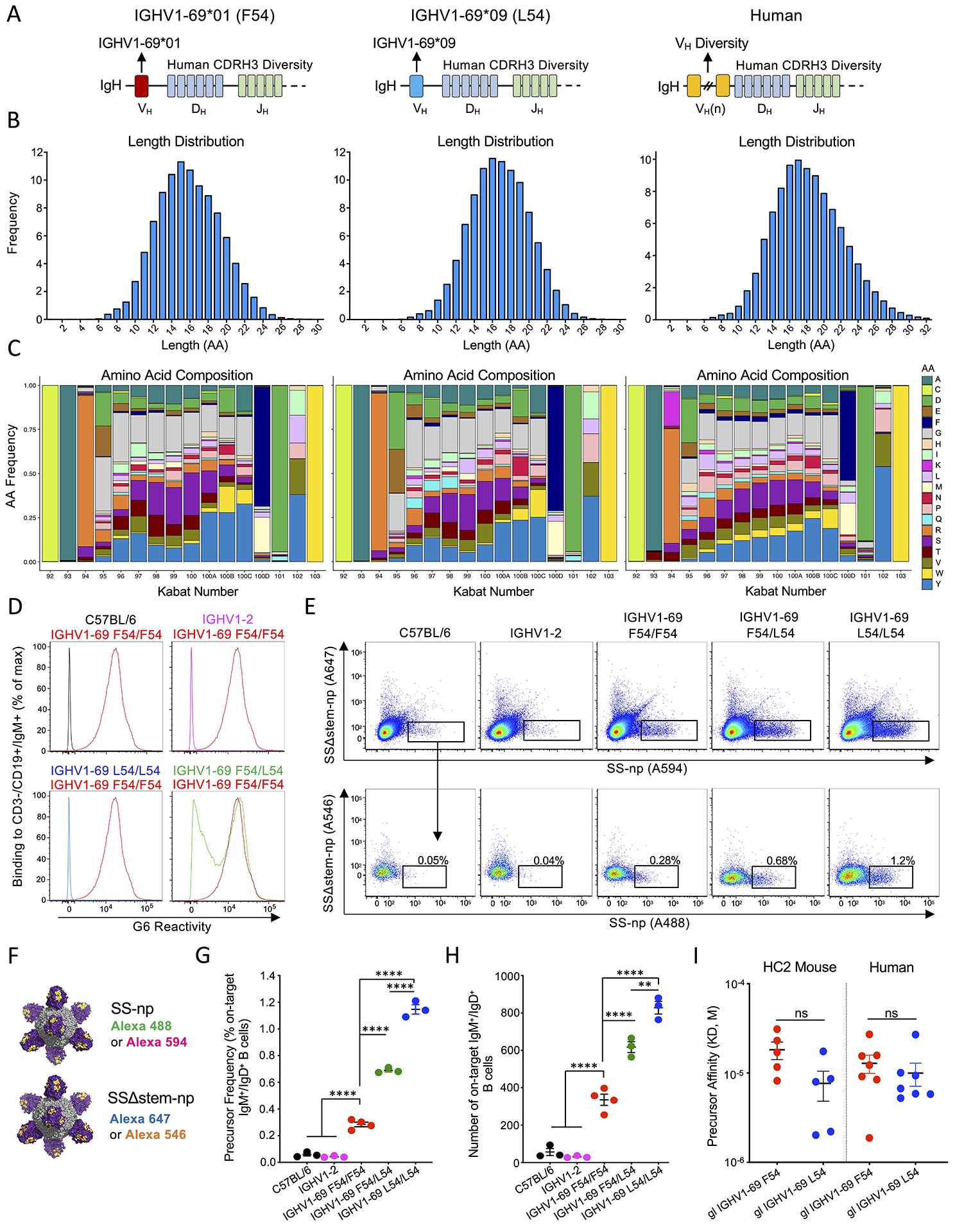
(A) IgH loci of F54 and L54 IGHV1-69 HC2 mice as compared to humans. (B) CDRH3 length distribution and (C) amino acid composition in the HC2 mice as compared to humans (>5 million BCR reads). HC2 sequences were obtained from naïve IgM B cells and human sequences were mined from a publicly available dataset (DeWitt et al., 2016). (D) F54 usage in mouse genotypes: C57BL/6 (black); IGHV1-2 (pink); F54/F54 IGHV1-69 (red); L54/L54 IGHV1-69 (blue); and F54/L54 IGHV1-69 (green), as measured by reactivity to G6, an anti-idiotypic antibody specific to F54 IGHV1-69 (Avnir et al., 2017; Wheatley et al., 2015). (E) SS-np probe reactivity to CD3−/CD4−/CD19+/IgG−/IgD+/IgM+ splenocytes (germline B cells) in C57BL/6, IGHV1-2, IGHV1-69 F54/F54, IGHV1-69 F54/L54, and IGHV1-69 L54/L54 animals. On-target B cells (black box, denotes the group 1 IAV bnAb site) are identified as germline BCRs that bind both positive selector probes but not the negative selectors (see also Figure S4 B–H for gating and additional analyses of functional B cell subsets). (F) Positive selector (SS-np Alexa-488 and SS-np Alexa-594) and negative selector probes containing the I45R/T49R mutations at the stalk bnAb epitope (SSΔstem-np Alexa-647 and SSΔstem-np Alexa 546). (G-H) Frequency and number of on-target responses across the mouse genotypes. Data presented as mean and SEM for n=3 or n=4 mice per genotype (**P<002, ****P<0.0001, ANOVA with Tukey’s test). (I) Stalk-affinity of germline on-target F54 and L54 Fabs isolated from HC2 mice and human (see also Tables S3 and S4). See also Figure S1 for human frequencies of F54/F54, L54/L54, and F54/L54 genotypes and Figure S5A–B for on-target SS-np probe gating in humans.
Germline Precursor Frequency and Affinity Does Not Account for Subdominance of bnAbs in L54 Carriers
We first assessed the extent to which L54 vs F54 IGHV1-69 usage genetically endows for on-target bnAb precursors within the polyclonal germline repertoire. Accordingly, we generated HC2 mice encompassing all possible IGHV1-69 genotypes: homozygous F54/F54 and L54/L54, and heterozygous F54/L54, which were compared to C57BL/6 and IGHV1–2 HC2 mice, where antibody restriction to IGHV1–2 serves as an additional human VH gene control. F54 IGHV1-69 usage in these genotypes was measured by germline B cell reactivity to the anti-idiotypic antibody G6, which recognizes the F54 motif within CDRH2 (Avnir et al., 2016; Sangesland et al., 2019; Wheatley et al., 2015). Applying G6 staining across our animal genotypes confirmed that 100% of the BCRs were G6+ in F54/F54, 50% were G6+ in F54/L54, and 0% were G6+ in L54/L54, IGHV1–2, and WT C57BL/6 mice (Figure 2D, S4B).
The frequency of group 1 IAV bnAb epitope-targeting precursors was then measured as the proportion of germline B cells that reacted against a series of positive and negative selector SS-np probes (Sangesland et al., 2019) (Figure 2E–F). SS-np displays the trimeric stalk domain of NC99 HA as an 8mer on a ferritin nanoparticle scaffold (Yassine et al., 2015). On-target B cells were defined as those that bound both positive selector probes (SS-np-Alexa 488 and SS-np-Alexa 594) but not the negative selector versions (SSΔstem-np Alexa 647 and SSΔstem-np Alexa 546), where Δstem denotes the presence of the I45R and T49R mutations that occlude access to the bnAb site (Figure 2E–F, S4B). As with our previous findings (Sangesland et al., 2019), F54 IGHV1-69 usage endowed for a small but significantly elevated frequency and number of on-target germline B cells as compared to IGHV1–2 HC2 and C57BL/6 controls (Figure 2G–H). Notably, on-target BCRs in L54/L54 mice were significantly elevated relative to F54/F54 mice. In F54/L54 heterozygotes, the frequency and number of these B cells was diluted as would be expected by Mendelian genetics (Figure 2G–H). Differences in on-target frequencies were not correlated with abnormal numbers of B and T cells in our mice and were maintained throughout the functional B cells subsets (Figure S4B–H).
As antibody precursor affinity can also regulate the participation of B cell lineages within the humoral response (Abbott and Crotty, 2020; Sangesland and Lingwood, 2021a), we isolated, sequenced and expressed individual on-target mAbs from the germline repertories of F54/F54 and L54/L54 mice (Figure 2E–F), and from human subjects bearing F54 (IGHV1-69*01) and/or L54 (IGHV1-69*09) alleles (Figure S5). We found that the corresponding L54 and F54 Fabs bound antigen with similar affinities (Figures 2I, Table S3). As with all in solution measures of precursor affinity, it will not account for apparent affinity resulting from avidity effects in vivo (i.e. multimeric membrane BCR engaging multivalent SS-np) (Boonyaratanakornkit and Taylor, 2019; Lingwood et al., 2012).
Only F54 Genotypes Enable Pathway-Amplification of Protective Serum bnAbs Against Heterosubtypic IAVs
To evaluate F54 and L54 as substrate for amplification of group 1 IAV bnAbs, we sequentially vaccinated HC2 mice with SS-np, a regimen we have previously shown amplifies the broadly protective response in F54/F54 animals, but not in IGHV1–2 HC2 mice (Amitai et al., 2020; Sangesland et al., 2019) (Figure 3A). One hallmark of bnAb amplification in the HC2 mice is the IGHV1-69-dependent elicitation of serum antibodies that react with strain matched HA but not with HAΔstem probes (Figure 3B). We found that while homozygous F54 mice productively amplified serum antibodies targeting the bnAb epitope (~half of IgG elicited at post-boost 2), the L54 homozygotes were unable to do so (Figure 3C). F54/L54 genotypes also expanded the bnAb response, indicating that one copy of the F54 allele can partially rescue the response. C57BL/6 littermates and IGHV1-2 HC2 mice generated equivalent titers of anti-stalk responses, but did not expand serum antibodies to the group 1 bnAb epitope (Figure 3C).
Figure 3. SS-np elicits serum bnAbs against group 1 IAVs in F54 but not in L54 IGHV1-69 mice.
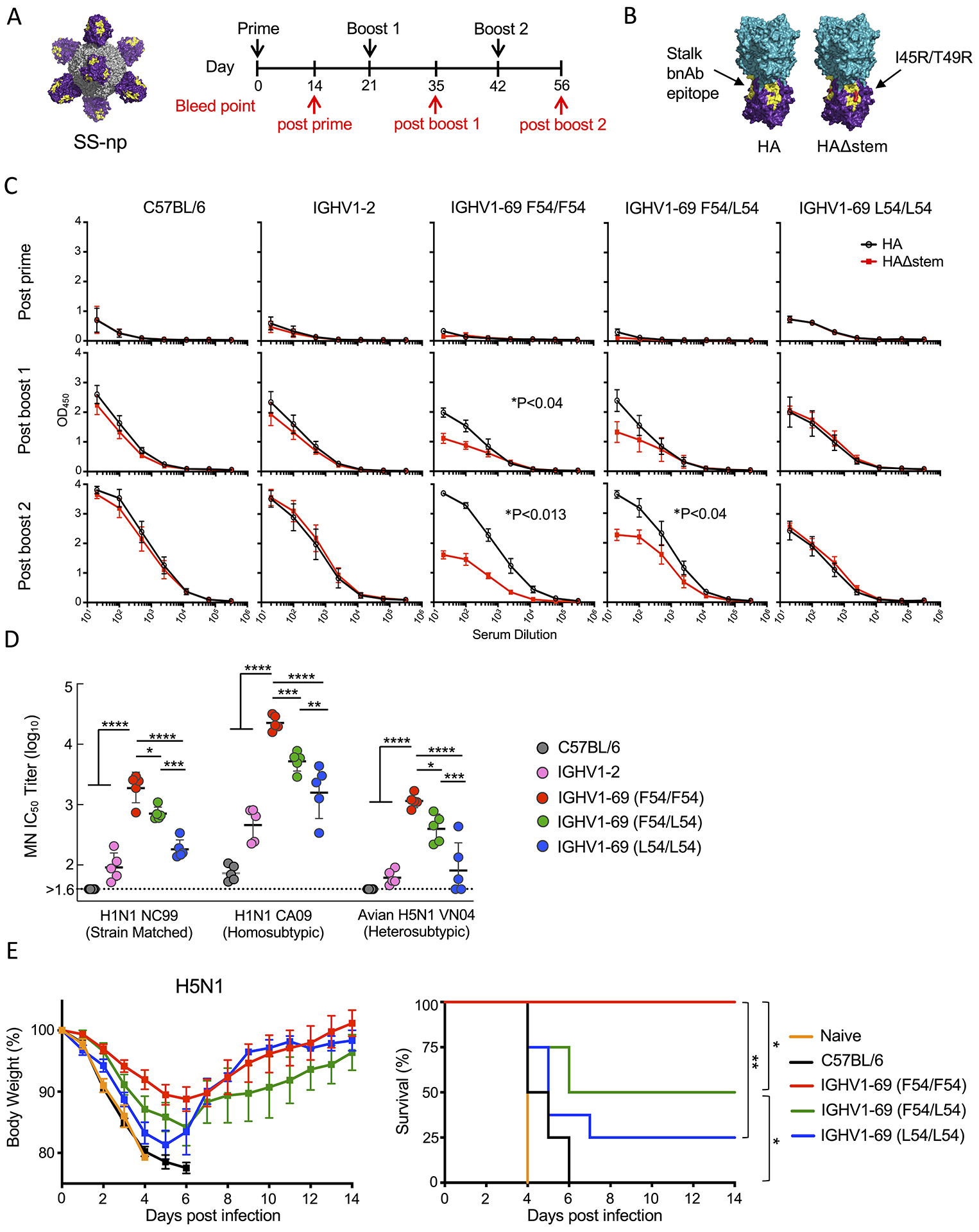
(A) Sequential immunization scheme with SS-np. (B) Full length HA trimer and HAΔstem (I45R+T49R) ELISA probes presenting the stalk domain (purple), head domain (teal) and group 1 IAV bnAb epitope (yellow). (C) Serum IgG responses in C57BL/6, IGHV1-2, F54/F54 IGHV169, F54/L54 IGHV1-69, and L54/L54 IGHV1-69 at post-prime (D14), post-boost 1 (D35), and post-boost 2 (D56) to HA (black line) and HAΔstem (red line). Shown as mean and SEM, n=5 mice per genotype. Stalk targeting bnAb responses are denoted as the difference in binding to HA vs HAΔstem (area under the curve, *P<0.05, Paired T-test). (D) Serum microneutralization against strain matched H1N1 (NC99), homosubtypic strain unmatched H1N1 (CA09), and heterosubtypic H5N1 (avian influenza VN04). Data presented as mean and SD, n=5 mice per genotype (****P<0.0001. ***P<0.001, **P<0.005, *P<0.03, ANOVA with Tukey’s test). (E) Heterosubtypic protection following lethal challenge to H5N1 avian influenza virus (n=8 mice per group, **P=0.0024 between IGHV1-69 F54/F54 and L54/L54, *P<0.03 between IGHV1-69 F54/F54 vs F54/L54, and *P<0.02 between IGHV1-69 F54/L54 and C57BL/6 (Mantel-Cox test of survivorship).
F54 IGHV1-69 dependent refocusing of the serum antibody response was further resolved as a capacity to neutralize diverse group 1 IAV strains (Figure 3D). Using microneutralization, we found that immune sera from F54/F54 animals neutralized vaccine matched virus (NC99, H1N1), vaccine unmatched virus within subtype (pandemic CA09, H1N1), and subtype unmatched virus (avian VN04, H5N1) (Figure 3D). However, increased L54 copy number resulted in a stepwise reduction in bnAb activity across group 1 IAVs, where F54/L54 heterozygotes exhibited intermediate neutralizing activities and L54/L54 homozygotes exhibited lower neutralization activities (Figure 3D). As expected, the serum antibody responses from C57BL/6 and IGHV1-2 mice did not confer broad neutralization across group 1 IAVs, despite equivalent titers of stalk antibodies. F54-dependence to vaccine protection was also observed following lethal challenge with heterosubtypic H5N1 avian influenza virus, where the L54 containing genotypes showed significantly reduced protective efficacy (Figure 3E).
L54 IGHV1-69 Encodes for Polyreactive and Autoreactive B cells in HC2 mice
We next evaluated if L54 and F54 germline stalk mAbs isolated from the HC2 mice (Figure 2E–F, Table S4) were reactive to autoantigens. We found that L54 mAbs were significantly more polyreactive than their F54 counterparts (Figure 4A, S6A), which was characterized as elevated reactivity to dsDNA, human recombinant insulin and bacterial lipopolysaccharide (LPS) (Andrews et al., 2015; Guthmiller et al., 2020). As anti-phospholipid antibodies often serve as markers of autoimmunity (Delgado Alves et al., 2002; Radway-Bright et al., 2000; Zhu et al., 1999), we further examined reactivity to sphingomyelin, POPC (1-palmitoyl-2-oleoyl-glycero-3-phosphocholine) and cardiolipin, a well-established autoimmune antigen (Haynes et al., 2005). Binding to these lipids was again significantly elevated in the L54 mAbs (Figure 4A, S6A). Likewise, L54 mAb binding to Hep-2 cells was enhanced (Figure 4B).
Figure 4. The L54 IGHV1-69 sequence encodes for autoreactivity in HC2 mice (A-C) and humans (D-F).
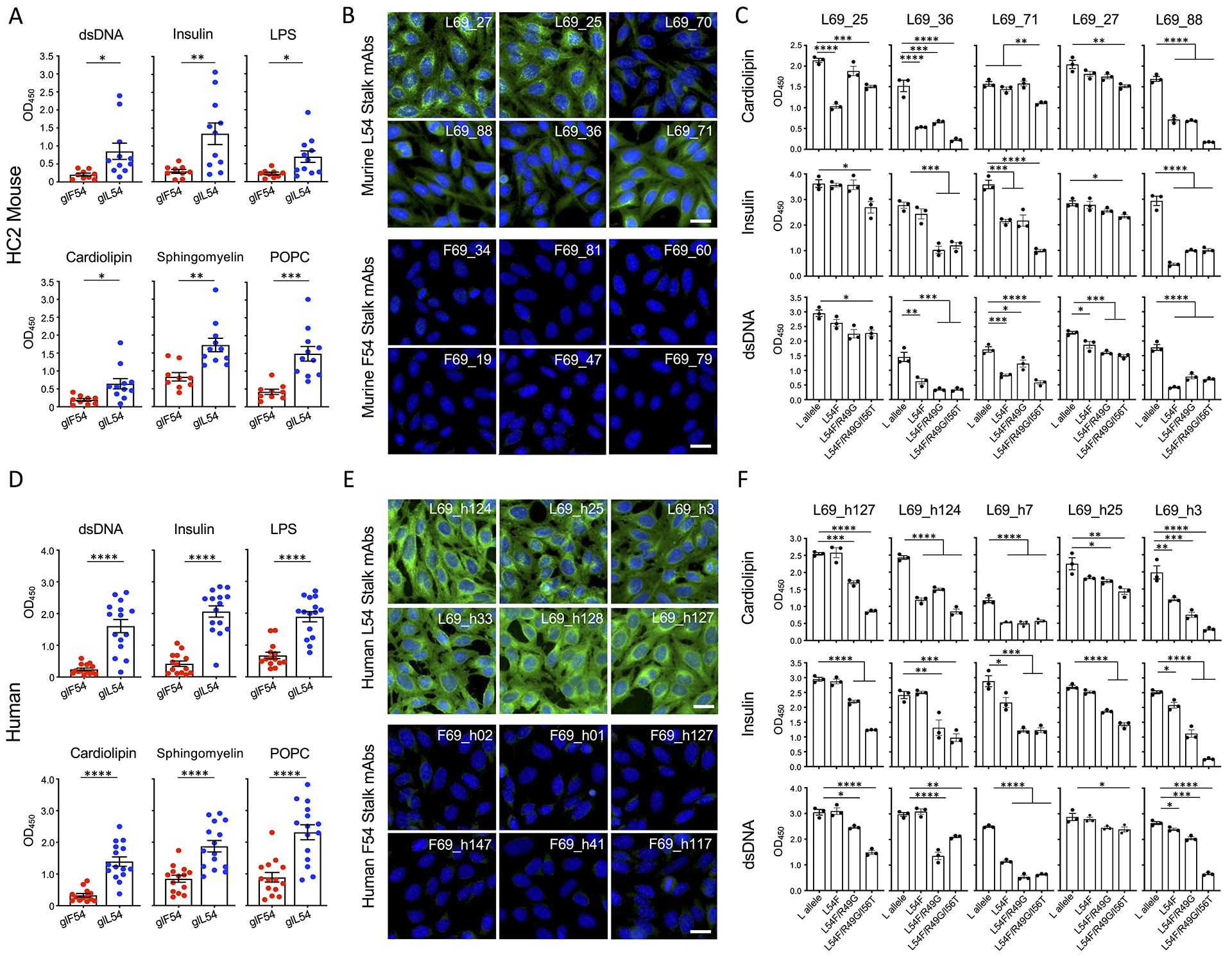
(A) Reactivity of HC2 L54 and F54 on-target germline stalk mAbs (see Figure 2E–F and Table S4) to dsDNA, human recombinant insulin, LPS, cardiolipin, sphingomyelin, and POPC (mean and SEM, n=3 replicates, *P<0.02, **P<0.004, ***P<0.004, T-test). (B) HC2 F54 and L54 germline mAb reactivity to Hep-2 cells. Samples were stained with anti-human IgG FITC secondary (green) and co-stained with DAPI (blue). Scale bar denotes 25μm. (C) Polyreactivity and autoreactivity by HC2 germline L54 stalk mAbs with the inclusion of F54 substitutions L54F, L54F/R49G, L54F/R49G/I56T (mean and SEM, n=3 replicates, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, ANOVA with Tukey’s test, See Table S5). (D) Reactivity of human F54 and L54 on-target stalk germline mAbs (see Figure S5, Table S4) to dsDNA, human recombinant insulin, LPS, cardiolipin, sphingomyelin, and POPC (Mean and SEM, n=3 replicates, ****P<0.0001, T-test). (E) Hep-2 cell staining with human L54 and F54 germline stalk mAbs. (F) Polyreactivity and autoreactivity by human germline L54 stalk mAbs with F54 substitutions (L54F, L54F/R49G, L54F/R49G/I56T, see Table S5) (mean and SEM, n=3 replicates, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, ANOVA with Tukey’s test). See also Figure S6.
F54 IGHV1-69*01 and L54 IGHV1-69*09 alleles differ by a minimal number of amino acids within the CDRH2 region (Sasso et al., 1993; Wheatley et al., 2015). To determine if autoreactivity is directly encoded by the VH-gene, we converted the germline L54 stalk antibodies into the F54 IGHV1-69*01 background by sequentially applying changes at these amino acid positions: L54F, L54F/R49G, and L54F/R49G/I56T (Figure 4C, Table S5). We find that incorporation of these changes sequentially attenuated the polyreactivity and autoreactivity of L54 germline stalk antibodies (Figure 4C).
Humans Show Similar L54-Encoded Autoreactivity
To determine if L54-encoded polyreactivity and autoreactivity is also present in the human repertoire, we evaluated F54 and L54 on-target germline antibodies isolated from human peripheral blood mononuclear cells (PBMCs) (Figure S5, Table S4). Akin to our results in HC2 mice, we found that the L54 antibodies displayed broad reactivity to all polyreactive and autoreactive antigen baits including insulin, dsDNA, cardiolipin, LPS, sphingomyelin, POPC, and showed elevated reactivity to Hep-2 cells, in contrast to their F54 counterparts (Figure 4D–E, S6B). Converting these germline antibodies into the F54 IGHV1-69*01 background (L54F, L54F/R49G, and L54F/R49G/I56T) also resulted in the attenuation of this polyreactive and autoreactive phenotype (Figure 4F, Table S5).
L54-encoded autoreactivity is not associated with glycans on the HA stalk.
Influenza HA displays N-linked glycans on both the head and stalk regions of this viral protein (Bajic et al., 2019a; Hutter et al., 2013). While these glycans do not impede access to the group 1 IAV bnAb epitope as engaged by 310-39G10, 310-63E6 or CR6261 (Figure S7A–D), their activity as autoantigens is not clear. To evaluate glycans as substrate for polyreactive and autoreactive L54 mAbs, we deglycosylated the N-glycans on denatured SS-np using the amidase PNGase F (Keating et al., 2020). We found that amidase treatment enhanced L54 mAb reactivity (Figure S7E), indicating that stalk glycans alone do not support polyreactivity and autoreactivity, although glycan binding in the context of conformationally presented carbohydrates cannot be ruled out.
L54-encoded polyreactivity and autoreactivity invokes tolerance and B cell anergy
We next evaluated B cell development in L54/L54 and F54/F54 mice at various stages: immature B cells in the bone marrow, transitional and mature B cells in the periphery (Figure 5A). We found that at each stage, the number of B cells were comparable between L54/L54 and F54/F54 genotypes (Figure 5B). However, L54 B cells were marked by a ~50% reduction in surface IgM as well as reduced CD79b expression, consistent with the downregulation of functional IgM BCR (Figure 5C). IgD surface expression was however comparable between both genotypes (Figure 5C). Notably, this L54-associated reduction in surface IgM BCR expression could be traced back to the bone marrow (Figure 5D) and continued through the transitional T1, T2, T3 (and T3/IgDhigh) subsets in the periphery (Figure 5E), indicative of central editing (Cambier et al., 2007; Cooper, 2015). To test the functional consequence of reduced surface IgM, we purified resting splenic B cells from L54/L54 and F54/F54 mice and then evaluated their capacity to respond to BCR stimulation after receptor crosslinking. We observed reduced receptor triggering in L54 B cells (Figure 5F–G).
Figure 5. L54-IGHV1-69 usage results in central editing and reduced responsiveness to antigen in the periphery.
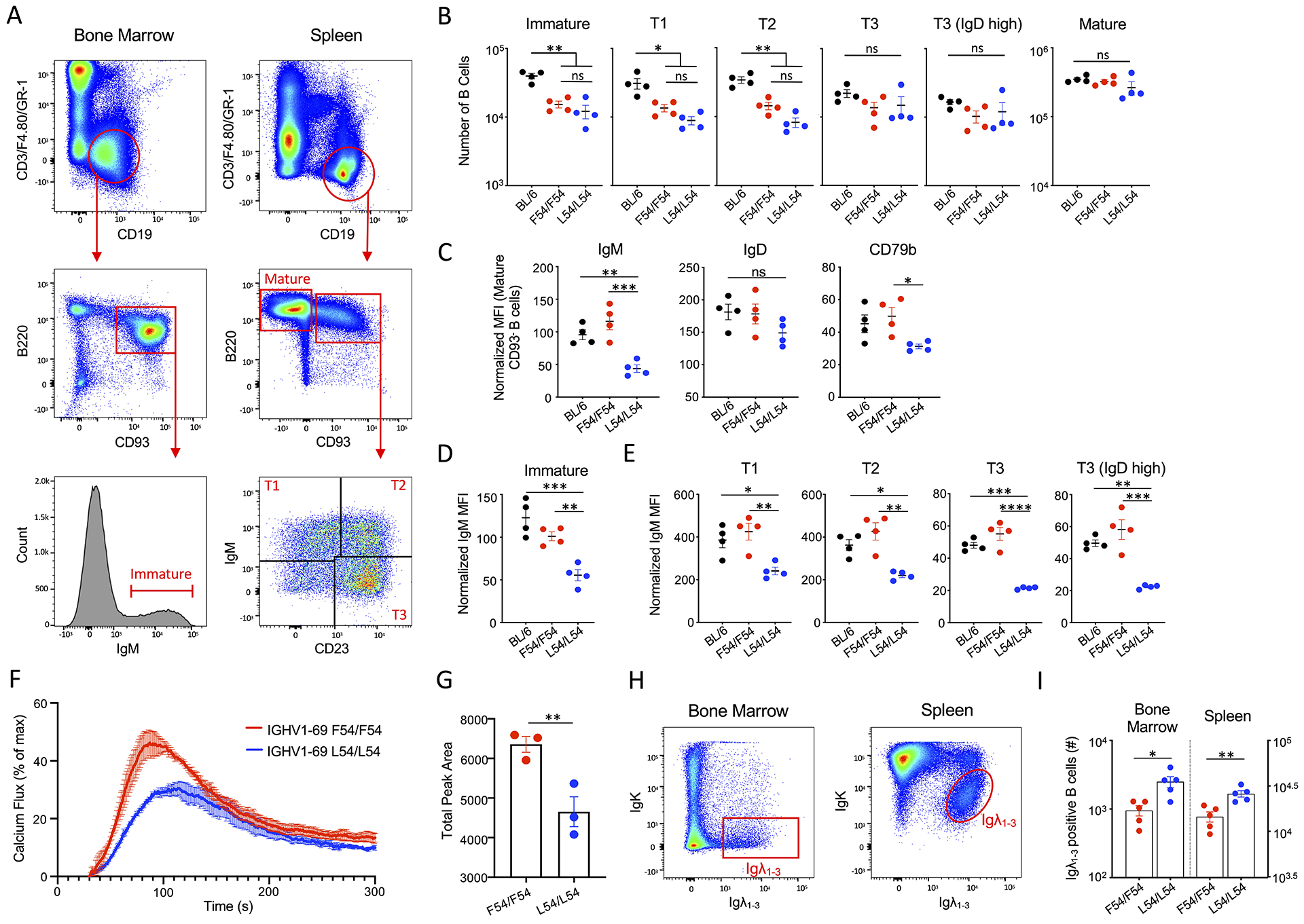
(A) Gating to evaluate L54 vs F54 B cell development: immature B cells in the bone marrow and mature, transitional (T1, T2 and T3) stages in the spleen. (B) B cell numbers in each developmental stage (mean and SEM, n=4 mice per genotype, *P<0.02, **P<0.004, ANOVA with Tukey’s test). (C) BCR surface expression in mature B cells as evaluated by mean fluorescent intensity (MFI) for IgM, IgD and CD79b normalized to CD19−/B220− cells (Bancroft et al., 2019) (mean and SEM, n=4 mice per genotype, *P<0.05, **P<0.009, ***P<0.001, ANOVA with Tukey’s test). (D) Normalized IgM MFI for immature B cells in the bone marrow and (E) T1, T2, T3 (T3/IgDhigh) stages in the spleen (mean and SEM, n=4 mice per genotype, *P<0.03, **P<0.008, ***P<0.0003, ****P<0.0001, ANOVA with Tukey’s Test). (F) Calcium fluxing after receptor crosslinking with anti-IgM in F54/F54 or L54/L54 resting B cells (mean and SEM, n=3 mice per genotype). (G) AUC from (F). Mean and SEM, n=3 mice per genotype, **P<0.02, T-test. (H) Gating to distinguish between IgK and IgL usage in L54 and F54 B cells present in the bone marrow and spleen. (I) Quantification of IgL usage from (H) (mean and SEM, n=5 mice per genotype, *P<0.02, **P<0.004, T-test).
Mice normally have Ig kappa (IgK) to Ig lambda (IgL) ratios of ~95:5, however when autoreactive features are present in the antibody heavy chain, increased IgL usage can promote immune tolerance through altered receptor diversification (Luning Prak et al., 2011; Tiegs et al., 1993; Vela et al., 2008). We find that IgL usage is significantly elevated in L54 B cells within the bone marrow and the spleen (Figure 5H–I).
Vaccine-expanded L54 B cells against the HA stalk are cleared from the B cell memory compartment
L54 B cell responsiveness to receptor crosslinking, while reduced, was not abolished, suggesting that vaccine expansion of L54 lineages would likely still be possible (Figure 5F–G). To investigate this, we examined the antigen specific B cells expanded during the SS-np vaccine regimen by applying H1-HA and H5-HA flow cytometry probes after each exposure to the immunogen. H1 reactivity denotes specificity to the HA stalk and H1+/H5+ cross-reactivity marks specificity to the stalk-bnAb epitope (Sangesland et al., 2019; Wheatley et al., 2015; Whittle et al., 2014). We found that while H1 reactive and H1+/H5+ cross-reactive IgG B cells were expanded after each exposure to SS-np in the F54-containing genotypes (Figure 6A–B), a tolerizing profile was observed in the L54/L54 mice, where H1 and H1+/H5+ IgG B cells were expanded and then cleared over the course of the immunization regimen (Figure 6A–B). The accumulation of stalk-specific HA B cells could be rescued by inclusion of the F54 allele, as seen in the F54/L54 heterozygote (Figure 6A–B).
Figure 6. Vaccine expanded L54 IGHV1-69 memory B cells are cleared limiting boosting to the HA stalk.
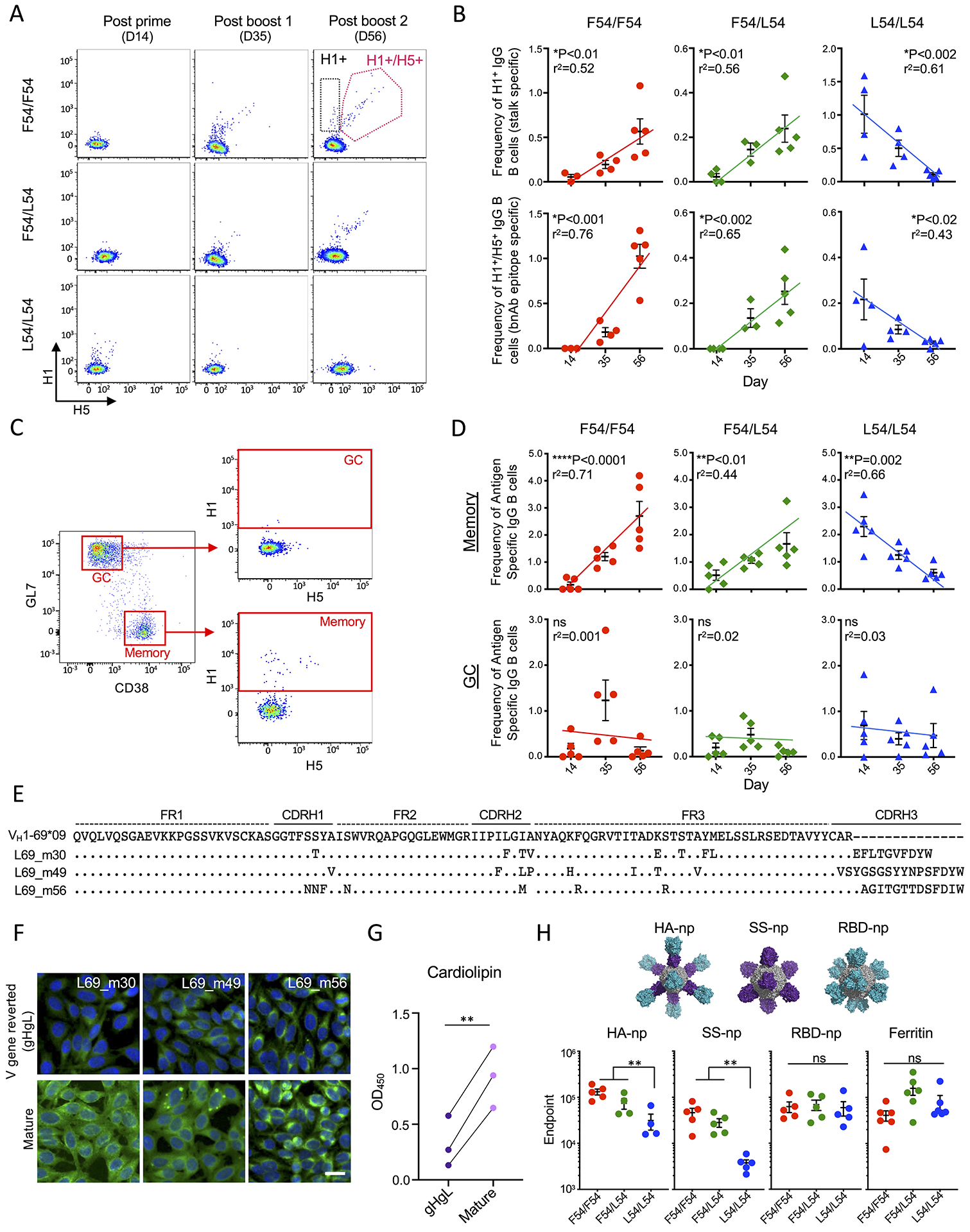
(A) Antigen specific IgG B cells were measured by flow cytometry during SS-np immunization. Reactivity to H1 trimer (black box) and reactivity to H1 + H5 trimers (pink box) (B) Quantification of H1+ and H1+/H5+ reactive IgG B cells following SS-np immunization at D14, D35, D56 (mean and SEM, n=3–5 mice per genotype, linear regression). (C) Gating strategy for distinguishing antigen specific B cells (H1+ and H1+/H5+ reactive) in the GC and memory compartments. Gated on CD4−/F4.80−/Gr-1−/CD19+/IgD− B cells (Tan et al., 2019). (D) Frequency of stalk reactive (H1+ and H1+/H5+ reactive) IgG B cells in GC and memory compartments at D14, D35, and D56 (mean and SEM, n=5 mice per genotype, linear regression). (E) Individual L54 stalk IgG (H1+/H5+) B cells expanded during the SS-np immunization regimen. (F) Hep2 reactivity of antibodies from (E) and their V gene-reverted (VHVL) sequences. (G) Cardiolipin reactivity of antibodies from (E) and their V gene-reverted (VHVL) sequences (**P<0.006, Paired T-test). (H) Endpoint titer of antigen specific IgG following sequential immunization with full length HA nanoparticle (HA-np); stalk only HA nanoparticle (SS-np), receptor binding domain nanoparticle (RBD-np). All HA sequences are derived from H1N1 NC99 and display the stalk domain (purple) and/or head domain (teal), and ferritin scaffold (grey). Endpoint titers following sequential immunization are shown at post boost 2 (D56) (mean and SEM, n=5 mice per genotype, **P<0.01, ANOVA with Tukey’s test). NC99 HA trimer baits are used for endpoints to HA-np, SS-np, and RBD-np and ferritin bait was used for SS-np immune sera.
To further define these effects, we tracked antigen specific B cells in the GC and memory subsets (Tan et al., 2019) (Figure 6C). In all genotypes, B cells entered GCs after each exposure to SS-np (Figure 6D), however, they did not accumulate in this compartment, as expected following sequential antigen exposure (Mesin et al., 2020). By contrast, antigen-specific memory B cells accumulated, but only within the F54 containing genotypes, while boosting of stalk-specific B cell memory was prevented in the L54/L54 mice (Figure 6D). While considerably less frequent, we also isolated H1+/H5+ BCRs expanded in L54/L54 mice. We found that L54 clones accumulated amino acid changes through somatic hypermutation (SHM) which enhanced their polyreactivity and autoreactivity, suggesting a basis for immune tolerance and clearance in the periphery (Figure 6E–6G, Table S6).
The inability to accumulate B cell memory blocks boosting of serum antibody responses
The clearance of B cell memory against the HA stalk could provide an explanation for the reduced secondary antibody responses observed in L54/L54 mice following sequential immunization with SS-np (Figure 3C). To further assess this, we sequentially immunized our IGHV1-69 HC2 genotypes with different presentations of influenza HA: HA-np, a full-length HA nanoparticle containing the trimeric head and stalk domain of NC99 HA (Kanekiyo et al., 2013); SS-np; or RBD-np, a ferritin nanoparticle displaying the trimeric receptor binding domain (RBD) from NC99 HA (Kanekiyo et al., 2019). Similar IgG titers against ferritin and RBD were elicited in L54/L54 mice, however IgG titers against HA were significantly reduced when this genotype was vaccinated with antigens containing the HA stalk domain (HA-np or SS-np) (Figure 6H).
Clonal Redemption Pathways for L54 B cell Lineages: VH-gene encoded polyreactivty and autoreactivity can be uncoupled from HA stalk engagement
To better understand if polyreactivity and autoreactivity is a feature of antibodies that recognize the group 1 bnAb epitope, we examined the L54/R49/I56 positions conferring autoreactivity to germline L54 mAbs (Figure 4C, F). We note that these residues are either mutated and/or not present in the binding sites of 310-39G10 and 310-63E6 (Figure 7A–B). In 310-39G10, residue L56 forms part of the epitope, interacting directly with G350 and W351, while A54 contributes both hydrophobic and van der waals interactions (Figure 7A). In 310-63E6, these residue positions do not participate in HA binding, as the closest amino acid to Q54 (Val34) is >4.5Å and there is no density in the EM map (Figure 7B). Moreover, incorporation of L54F/R49G/I56T into germline L54 stalk mAbs does not reduce germline affinity for HA (Figure 7C, Tables S3, S7)
Figure 7. Clonal redemption in L54/L54 individuals.
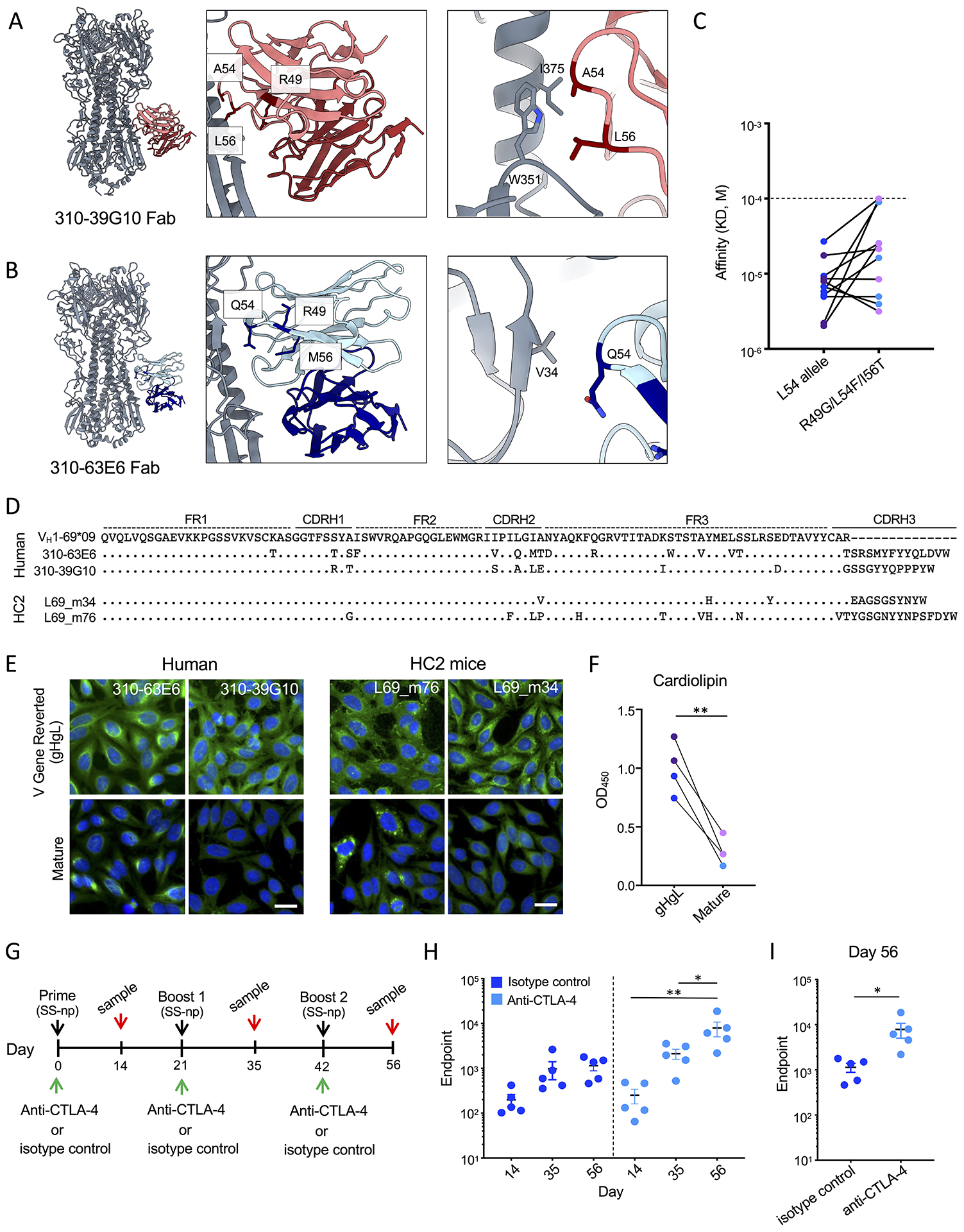
Residues implicated in L54-IGHV1-69 encoded poly- and autoreactivity, as shown for 310-39G10 Fab (A) and 310-36E6 Fab (B). HA trimers are shown in gray and antibodies are colored as red and blue. (C) HA stalk affinity for human and HC2 mouse stalk specific L54 germline Fabs with or without the L54F/R49G/I56T substitutions (See also Figures 2E, F, S5, and Tables S3, S4, S5, and S7). Human mAbs are denoted in blue while HC2 mAbs are denoted in purple (P>0.05, T-test). (D) Pathways of clonal redemption for affinity matured human (310-39G10, 310-36E6) and HC2 L54 stalk-targeting mAbs (L69_m76, and L69_m34). (E-F) Autoreactivity of mature and V gene-reverted (gHgL) antibodies as evaluated by binding to Hep-2 cells (E) and cardiolipin (F, **P<0.008, Paired T-test). Human mAbs are denoted in blue and HC2 mAbs are denoted in purple. Scale bar in (E) denotes 25μm. (G) Immunization scheme for CTLA-4 blockade in L54/L54 HC2 mice vaccinated with SS-np. (H) Endpoint titers for isotype control treated (dark blue) and anti-CTLA-4 treated (light blue) at day 14, 35 and 56 (mean and SEM, n=5 mice per group, *P<0.03, **P<0.002, ANOVA with Tukey’s test). (I) Day 56 endpoint dilution for stalk specific IgG (n=5 mice per group, mean and SEM, *P<0.05, T-test).
This flexibility suggests that L54 stalk mAbs should be able to accommodate a variety of mutations at these positions, potentially allowing for changes that would support clonal redemption. Consistent with this hypothesis, we found that the human L54 bnAbs 310-39G10 and 310-63E6 did indeed lose the autoreactivity that was present in their V gene-reverted (gHgL) BCRs (Figure 7D–F). We also observed analogous SHM that eliminated L54 encoded autoreactivity in some of the H1+/H5+ B cell lineages that were expanded after SS-np vaccination in HC2 mice (Figure 7D–F). Thus while L54 IGHV1-69 encodes for polyreactive and autoreactive BCRs in the repertoire, this reactivity can be biochemically separated from germline BCR specificity for the vaccine target, and in some cases SHM can be applied to ‘rescue’ the lineage.
To further evaluate whether such redemption may be enhanced by therapeutic intervention, we performed antibody blockade of the immune regulatory receptor CTLA-4 to dampen immune tolerance, a procedure that can elevate the expansion of some HIV bnAb lineages that predispose for autoreactivity (Bradley et al., 2020). We found that this treatment restores boosting of secondary L54 antibody responses against the HA stalk (Figure 7G–I).
Discussion
F54 alleles are favored within human HA stalk bnAbs against group 1 IAVs (Avnir et al., 2016; Pappas et al., 2014; Wheatley et al., 2015). However, L54 stalk bnAbs have also been isolated and do show similar neutralization breadth (Wheatley et al., 2015). One hypothesis for bias in allelic preference is that L54 versions of IGHV1-69 do not impart BCR affinity to the bnAb site. However, we found that human L54 bnAbs isolated from different donors engaged the same target via structurally convergent VH-gene encoded contact. Furthermore, the germline antibody repertories of F54/F54 and L54/L54 HC2 mice both encoded for polyclonal affinities to the bnAb site, and this precursor frequency was strongly elevated in the L54/L54 genotype. As we did not observe differences between F54 and L54 antibody precursor affinity, an advantage could be predicted for L54/L54 mice. However, in contrast to F54 IGHV1-69, the L54 allele was non-productive for protective serum bnAbs. Rather, our results indicate that the expansion of L54 B cell lineages is intrinsically limited by co-endowed polyreactivity and autoreactivity.
Self-reactive B cells can be eliminated by central tolerance in the bone marrow as well as peripheral tolerance in the spleen (Getahun, 2022; Nemazee, 2017; Platt et al., 2019). Indeed, polyreactivity and autoreactivity have been implicated in limiting the expansion of human bnAbs against HIV and influenza virus (Andrews et al., 2015; Bajic et al., 2019b; Haynes et al., 2005; Schroeder et al., 2017; Yang et al., 2013). However, as controlled vaccine elicitation of serum bnAbs against these viruses has itself been a largely unmet goal, modulation of bnAb immunodominance through autoreactivity has been difficult to experimentally demonstrate in vivo. We now observe the attenuation of protective serum bnAbs from a specific human VH-gene encoded pathway for antibody development.
Transgenic KI mouse models expressing polyreactive HIV bnAbs and some of their germline precursors have shown that a large fraction of the B cells are cleared by central tolerance, and those remaining in the periphery tend to be anergic or less responsive to antigen stimulation due to lowered expression of surface IgM and IgD (Bancroft et al., 2019; Chen et al., 2013; Doyle-Cooper et al., 2013; Finton et al., 2013; McGuire et al., 2016). However, we did not observe large-scale disruption in B cell development, perhaps due to the repertoire diversity in HC2 mouse system, which allows for CDRH3 diversification, the major engine of BCR plurality. Indeed, ~20% of the human peripheral B cell repertoire is normally self-reactive (Getahun, 2022; Wardemann et al., 2003). However, we do report central editing to favor anergy as L54 IGHV1-69 B cells emerge from the bone marrow with reduced IgM BCR expression and lowered responsiveness to receptor triggering. This phenotype is also seen in human BND cells, a germline B cell subset encompassing ~2.5% of human peripheral blood B cells, which are anergic, autoreactive and may represent precursors of autoantibody secreting plasma cells (Duty et al., 2009).
While anergy likely limits the overall capacity of L54 stalk specific B cells to respond following SS-np vaccination, L54 B cell responsiveness to antigen was not completely eliminated by central tolerance. Combined with a higher precursor frequency in the periphery, this likely allows for some L54 B cell expansion following immunization. Indeed, the increase in IgL expression and usage in L54 B cells points to furthered repertoire diversification and receptor editing to offset autoreactivity (Luning Prak et al., 2011; Tiegs et al., 1993; Vela et al., 2008). However, during sequential immunization with SS-np, L54-stalk B cells were cleared in the memory compartment. The prevention of accumulating L54 B cell memory provides a basis for sub-optimal boosting of stalk-specific serum antibody responses in L54/L54 mice. These results also suggest a reason for the marked reduction of IGHV1-69 in stalk-specific human memory B cells of individuals who are homozygous for the L54 allele (Wheatley et al., 2015).
Notably however, our results also indicate a capacity to biochemically uncouple recognition of the bnAb target from the L54 VH-gene encoded residues that predispose for reactivity to self-antigens. This flexibility likely enables clonal redemption and salvage pathways through SHM, which we have observed in some L54 B cell lineages. Administration of antibodies targeting immune checkpoint receptors likely enhances the opportunity for clonal redemption and suggests a clinical intervention for restoring autoreactive germline encoded stalk responses in L54/L54 individuals.
Collectively, our findings indicate that autoreactivity and anergy tunes the expandability of human influenza bnAbs in vivo and that subtle changes in the antibody gene sequence can gate these factors, restructuring antibody immunodominance hierarchies within a vaccine response. Notably, productivity of the response was rescued by genetically restoring the F54 allele, which resulted in the ‘recovery’ of bnAb elicitation and vaccine protection following heterosubtypic influenza viral challenge. These results underscore the genetic basis for immune tolerance and likely explains the paucity of L54 usage within human group 1 IAV bnAbs (Avnir et al., 2016; Pappas et al., 2014; Wheatley et al., 2015).
On a global scale, the inability to pathway-amplify bnAbs through L54 IGHV1-69 may hold significance for certain ethnicities with elevated L54 homozygosity. While it is unclear what drives these differences, most individuals carry at least one copy of F54 IGHV1-69 as F54/L54 heterozygotes (Genomes Project et al., 2010; Lingwood et al., 2012), which we show rescues amplification of protective serum bnAbs. We therefore predict that the ability to pathway-amplify the F54 IGHV1-69 encoded group 1 IAV bnAb response will be globally widespread but will also be tuned by allelic usage.
Limitations of the Study
It is not yet clear that antibody tolerance mechanisms in mice will fully represent those present in humans. Our study also does not account for the possibility of antibody VH gene duplication, which could lead to more than two allelic forms of IGHV1-69 in the genome. Additionally, the unrestricted antibody VH-gene repertoire of humans will ensure responses by non-IGHV1-69 B cell lineages, and our experiments did not account for this.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Lead Contact
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, Daniel Lingwood (dlingwood@mgh.harvard.edu).
Materials Availability
There are restrictions to the availability of the transgenic mice due to a MTA with Bristol-Myers Squibb.
Data and Code Availability
3D maps and models from the EM analysis have been deposited to the Electron Microscopy Databank (http://www.emdatabank.org/) and the Protein Data Bank (https://www.rcsb.org/), respectively. The accession numbers are listed Table S1 and in the Key Resources Table.
Antibody sequences used in this publication can be found in Tables S4–S6 or within NCBI’s Gene Expression Omnibus and are accessible through GEO accession number listed in the Key Resource Table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| G6 Anti-Idiotypic mAb | Produced in house | N/A |
| CH65 | Produced in house | N/A |
| CR6261 | Produced in house | N/A |
| Sheep Anti-Mouse IgG-HRP | GE Healthcare | Cat#NA931 |
| Sheep Anti-Human IgG-HRP | GE Healthcare | Cat#NA933 |
| Hamster Anti-Mouse CD3e-PE/Cy7 (clone 145-2C11) | BioLegend | Cat#100320 |
| Rat Anti-Mouse CD4-Alexa Fluor 700 (clone RM4-5) | BD Biosciences | Cat#557956; RRID: AB_396956 |
| Rat Anti-Mouse CD19-BV421 (clone 6D5) | BioLegend | Cat#115549; RRID: AB_2563066 |
| Rat Anti-Mouse IgM-BV605 (clone RMM-1) | BioLegend | Cat#406523; RRID: AB_2563358 |
| Goat Anti-Mouse IgG-PerCP/Cy5.5 (clone Poly4053) | BioLegend | Cat#405314; RRID: AB_10662053 |
| Rat Anti-Mouse IgM-BV650 (clone II/41) | BD Biosciences | Cat#743326; RRID: AB_2741427 |
| Rat Anti-Mouse CD23-BV786 (clone B3B4) | BD Biosciences | Cat#563988; RRID: AB_2738526 |
| Rat Anti-Mouse CD93-BV421 (clone AA4.1) | BD Biosciences | Cat#747716; RRID: AB_2872195 |
| Rat Anti-Mouse IgD-PerCP-Cy5.5 (clone 11–26C.1) | BD Biosciences | Cat#564273; RRID: 2738722 |
| Rat Anti-Mouse CD21/CD35-PE-Cyanine7 (clone 8D9) | eBioscience | Cat#25-0211-80; RRID: AB_1518767 |
| Rat Ani-Mouse CD45R/B220-BUV395 (clone RA3-6B2) | BD Biosciences | Cat#563793; RRID: AB_2738427 |
| Hamster Anti-Mouse CD3e-BV510 (clone 145-2C11) | BD Biosciences | Cat#563024; RRID: AB_2737959 |
| Rat Anti-Mouse F4/80-BV510 (clone BM8) | BioLegend | Cat#123135; RRID: AB_2562622 |
| Rat Anti-Mouse Ly-6G and Ly-6C-BV510 (clone RB6-8C5) | BD Biosciences | Cat#563040; RRID: AB_2722496 |
| Rat Anti-Mouse CD19-APC-eFlour780 (clone 1D3) | eBioscience | Cat#47-0193-82; RRID: AB_10853189 |
| Hamster Anti-Mouse CD79b-BV605 (clone HM79b) | BD Biosciences | Cat#740383; RRID: AB_2740113 |
| Rat Anti-Mouse Ig k Light chain-PE (clone 187.1) | BD Biosciences | Cat#559940; RRID: AB_397384 |
| Rat Anti-Mouse Ig, λ1, λ2, & λ3 Light chain-FITC (clone R26-46) | BD Biosciences | Cat#553434; RRID: AB_394854 |
| Rat Anti-Mouse CD38-Alexa Flour 594 (clone 90) | BioLegend | Cat#102725; RRID: AB_2566435 |
| Rat Anti-Mouse/human GL7-Alexa Flour 488 (clone GL7) | BioLegend | Cat#144612; RRID: AB_2563285 |
| Rat Anti-Mouse IgD-BUV395 (clone 11-26C.1) | BD Biosciences | Cat#564274; RRID: AB_2738723 |
| Hamster Anti-Mouse CD95-PE-Cyanine7 (clone Jo2) | BD Biosciences | Cat#557653; RRID: AB_396768 |
| Rat Anti-Mouse Ly-6G/Ly-6C-APC-eFlour780 (clone RB6–8C5) | eBiosciecne | Cat#47-5931-82; RRID: AB_1518804 |
| Rat Anti-Mouse CD4-APC-eFlour780 (clone RM4-5) | eBioscience | Cat#47-0042-82; RRID: AB_1272183 |
| Rat Anti-Mouse CD8a-APC-eFlour780 (clone 53–6.7) | eBioscience | Cat#47-0081-82; RRID: AB_1272185 |
| Rat Anti-Mouse/Human CD45R/B220-BV605 (clone RA3–6B2) | BioLegend | Cat#103243; RRID: AB_11203907 |
| Rat Anti-Mouse CD19-PerCP/Cyanine5.5 (clone 1D3) | BioLegend | Cat#152405; RRID: AB_2629814 |
| Rat Anti-Mouse F4/80-APC-eFlour780 (clone BM8) | eBioscience | Cat#47-4801-82; RRID: AB_2735036 |
| Goat Anti-Mouse IgG-BV421 (clone Poly4053) | BioLegend | Cat#405317; RRID: AB_10900419 |
| Rat Anti-Mouse IgD-Alexa Fluor 700 (clone 11–26c.2a) | BioLegend | Cat#405730; RRID: AB_2563341 |
| AffiniPure F(ab’)2 Fragment Goat Anti-Mouse IgM | Jackson ImmunoResearch | Cat#115-006-020; RRID: AB_2338469 |
| inVivoMAb Anti-mouse CTLA-4 (clone 9H10) | BioXCell | Cat#BE0131 |
| inVivoMAb Polyclonal Syrian Hamster | BioXCell | Cat#BE0087 |
| Mouse Anti-Human CD19-Alexa Fluor 700 (clone HIB19) | BioLegend | Cat#302226; RRID: AB_493751 |
| Mouse Anti-Human IgG-Brilliant Violet 421 (clone G18–145) | BD Biosciences | Cat# 562581; RRID: AB_2737665 |
| Mouse Anti-Human IgD-PE/Cy7 (clone IA6–2) | BD Biosciences | Cat#561314; RRID: AB_10642457 |
| Mouse Anti-Human CD3-APC-Cy7 (clone SK7) | BD Biosciences | Cat#557832; RRID: AB_396890 |
| Mouse Anti-Human IgM-PerCP/Cyanine5.5 (clone MHM-88) | BioLegend | Cat#314512; RRID: AB_2076098 |
| HC2 and Human Germline on-Target F54 and L54 Antibodies, see Table S4 | This Paper | N/A |
| HC2 and Human L54F/R49G/I56T Conversion Antibodies, see Table S5 | This Paper | N/A |
| HC2 and Human Vaccine Expanded Stalk Antibodies, see Table S6 | This Paper | N/A |
| Bacterial and virus strains | ||
| A/chicken/Vietnam/NCVD-016/2008(H5N1)-PR8-IDCDC-RG12 | International Reagent Resource | FR-725 |
| Influenza reporter viruses | Masaru Kanekiyo, NIH (Creanga et al., 2021) | N/A |
| Biological samples | ||
| PBMC Samples from Human Donors | Massachusetts General Hospital | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| H1 HA Trimer (NC99) | Produced in house (Whittle et al., 2014) | N/A |
| H1 Y98F HA Trimer (NC99) | Produced in house (Whittle et al., 2014) | N/A |
| H5 Y98F HA Trimer (Indo05) | Produced in house (Whittle et al., 2014) | N/A |
| H1 HA-np (NC99) | Produced in house (Kanekiyo et al., 2013) | N/A |
| SS-np (NC99) | Produced in house (Yassine et al., 2015) | N/A |
| SS-npΔstem (NC99) | Produced in house (Sangesland et al., 2019) | N/A |
| H. Pylori Ferritin | Aaron Schmidt, Ragon Institute of MGH, MIT, and Harvard | N/A |
| H1 RBD-np (NC99) | Masaru Kanekiyo, NIH (Kanekiyo et al., 2019) | N/A |
| Streptavidin-allophycocyanin (APC) Conjugate | Life Technologies | Cat#S32362 |
| Streptavidin-phycoerythrin (PE) Conjugate | Life Technologies | Cat#S21388 |
| Fura Red, AM, Cell Permeant | Thermo Fisher | Cat#F3021 |
| Ionomycin | Thermo Fisher | Cat#I24222 |
| Sigma Adjuvant System | Sigma-Aldrich | Cat#S6322 |
| Recombinant Human Insulin Protein | Fitzgerald | Cat# 30-Al51 |
| UltraPure Calf Thymus DNA solution | Invitrogen | Cat#15633019 |
| Sphingomyelin | Avanti Polar Lipids | Cat#860062 |
| Cardiolipin | Sigma-Aldrich | Cat#C1649 |
| 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) | Avanti Polar Lipids | Cat#850457P |
| Lipopolysaccharide from Escherichia coli O55:B5 | Sigma-Aldrich | Cat#L2880 |
| 293fectin Reagent | Invitrogen | Cat#12347019 |
| Ni-Sepharose excel Affinity Medium | GE Healthcare | Cat#GE17-3712-02 |
| Erythrina Cristagalli (ECA) Immobilized Lectin | EY Laboratories | Cat#A-5901-2 |
| Protein G Sepharose | GE Healthcare | Cat#17061802 |
| IgG Elution Buffer | Pierce | Cat#21009 |
| LIVE/DEAD Fixable Aqua Dead Cell Stain | Thermo Fisher | Cat#L34957 |
| LIVE/DEAD Fixable Blue Dead Cell Stain | Thermo Fisher | Cat#L34961 |
| Critical commercial assays | ||
| NOVA Lite Hep-2 IgG kit | Werfen | Cat#708100 |
| BirA Biotin-Protein Ligase Bulk Reaction Kit | Avidity | Cat#Bulk BirA |
| Alexa Fluor 647 Protein Labeling Kit | Thermo Fisher | Cat#A20173 |
| Alexa Fluor 488 Protein Labeling Kit | Thermo Fisher | Cat#A10235 |
| Alexa Fluor 594 Protein Labeling Kit | Thermo Fisher | Cat#A10239 |
| Alexa Fluor 546 Protein Labeling Kit | Thermo Fisher | Cat#A20183 |
| MiSeq Reagent Kit, V2 500 cycles | Illumina | Cat#MS-102-2003 |
| Deposited data | ||
| IGHV1-69 HC2 CDRH3 sequences | This Paper | GEO: GSE207054 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE207054 |
| CryoEM structures | This Paper | PBD IDs: 7SCO, 7SCN |
| Experimental models: Cell lines | ||
| Human: FreeStyle 293F | Thermo Fisher | Cat#R79007; RRID: CVCL_D603 |
| Human: Expi293F | Thermo Fisher | Cat#A14527; RRID: CVCL_D615 |
| Canine: MDCK | ATCC | Cat#CCL-34; RRID: CVCL_0422 |
| Experimental models: Organisms/strains | ||
| Mouse: IGHV1-69*01+/+ IgH Restricted Mouse Model with Diverse Human CDRH3 (C57Bl/6 Strain) | Sangesland et al., 2019 | N/A |
| Mouse: IGHV1-2*02+/+ IgH Restricted Mouse Model with Diverse Human CDRH3 (C57Bl/6 Strain) | Sangesland et al., 2019 | N/A |
| Mouse: IGHV1-69*09+/+ IgH Restricted Mouse Model with Diverse Human CDRH3 (C57Bl/6 Strain) | This Paper | N/A |
| Mouse: IGHV1-69*01+/−/IGHV1-69*09+/− IgH (C57Bl/6 Strain) | This Paper | N/A |
| Oligonucleotides | ||
| Primers for Single Cell and Bulk BCR Amplification | Sangesland et al., 2019 | N/A |
| Recombinant DNA | ||
| Software and algorithms | ||
| Flowjo v10.6.2 | TreeStar | https://www.flowjo.com; RRID: SCR_008520 |
| Prism v.8.4.3 | GraphPad | https://www.graphpad.com; RRID: SCR_002798 |
| Other | ||
| Superdex 200 10/300 Column | GE Healthcare | Cat#17517501 |
| Superose 6 10/300 Column | GE Healthcare | Cat#17517201 |
| SPF Embryonated Chicken Eggs | Charles River Laboratories | Cat#10100335 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Transgenic mice
The transgenic mice in this study were generated as described previously (Amitai et al., 2020; Ronsard et al., 2021; Sangesland et al., 2019; Sangesland et al., 2020). Using HC2 recombineering technology, mice were constrained to single user defined VH-genes while also containing diverse human D and J gene segments that were capable of recombination (Fishwild et al., 1996; Lonberg et al., 1994). We have previously used this method to generate mice with germline CDRH3 repertories that are similar to humans, while at the same time, being completely restricted to IGHV1-69*01 (F54) and IGHV1-2*02 (Amitai et al., 2020; Ronsard et al., 2021; Sangesland et al., 2019; Sangesland et al., 2020). The light chain repertoire in these mice is provided by the endogenous murine kappa and lambda loci. In this study, we constrained mice to IGHV1-69*09 (L54) with otherwise normal human-like CDRH3 diversity. As with IGHV1-69*01 and IGHV1-2*02, a synthetic genomic DNA construct of ~6.6kb derived from human IGHV1-69*09 was synthesized at Geneart (Regensburg, Germany) using codon sequences obtained from IMGT “IG” and “TR” Repertoire (http://www.imgt.org/). Coordinates of the IGHV1-69*09 allele included in the HC2 construct correspond to NCBI GRCh38.p12 Primary Assembly of huCHr14 (nts 106714682 – 106715120). The animals were generated as described (Fishwild et al., 1996; Lonberg et al., 1994) as per standard techniques (Pease, Shirley, and Thomas L. Saunders. Advanced Protocols for Animal Transgenesis: an ISTT Manual. Springer, 2011) and were a gift to D.L. from Bristol-Myers Squibb (Redwood City, CA). The transgenic mice used in this study consisted of the following genotypes: IGHV1-69*01+/+; IGHV1-69*09+/+; IGHV1-69*01+/IGHV1-69*09+; IGHV1-2*02+/+; and all were wild-type C57Bl/6 for light chain expression.
All animals were maintained within the HPPF barrier facility at the Ragon Institute of MGH, MIT and Harvard and all experiments were conducted with institutional IACUC approval (MGH protocol 2014N000252). Both male and female animals were used in this study. Ages of the animals ranged from 6–10 weeks.
Human PBMC
Human PBMC were isolated from Leukopacs obtained from the MGH blood donor center (5 donors). Before blood donation, the donors signed an attestation/consent statement, as per hospital requirements. It states: “I give permission for my blood to be used for transfusion to patients or for research”. The samples were de-identified and the gender and age of the patients is not recorded by the MGH blood donor center. Eligible donors must be a minimum of 16 years of age and weigh a minimum of 110lbs. All experiments with human PBMC were conducted with institutional approval (MGH protocols 2014B000035, 2005P001218).
METHODS DETAILS
Recombinant immunogens and B cell probes
Recombinant soluble HA trimers, stalk-only trimers, stalk-only ferritin nanoparticles (SS-np), full-length HA ferritin nanoparticles (HA-np), and receptor binding domain ferritin nanoparticles (RBD-np), all derivations of HA from A/New Caledonia/20/1999 (NC99), were generated as previously described (Kanekiyo et al., 2019; Kanekiyo et al., 2013; Weaver et al., 2016; Yassine et al., 2015). Expression vectors encoding the HA constructs were transfected into 293F cells at 500μg/L using 293fectin reagent. The cells were grown in Freestyle 293 expression medium for five days after which culture supernatants were harvested and filtered. Soluble HA trimers and stalk trimers were purified using Ni Sepharose resin (GE Healthcare) with 20mM and 500mM imidazole serving as the washing and elution steps. The trimers were then were then exchanged into PBS and by size exclusion chromatography (SEC) using an AKTA pure protein purification system and Superdex 200 10/300 column (GE Healthcare). SS-np and HA-np were bound to Erythrina cristagalli Gel-ECA-Immobilized Lectin (EY Laboratories), washed in PBS, eluted in 0.2M lactose, and then resolved by SEC using a Superose 6 10/300 column in PBS. RBD-np was purified using ion-exchange chromatography (HiTrap Q HP columns at pH 8.0; GE Healthcare) followed by with SEC (Superose 6 XK 16/70 PG column in PBS; GE Healthcare). Naked H. Pylori ferritin was a gift from Aaron Schmidt (The Ragon Institute of MGH, MIT, and Harvard).
SS-np B cell probes were generated by labeling with Alexa 488 or Alexa 594, while SSΔstem-np (I45R+ T49R) probes were generated by labeling with Alexa 647 or Alexa 546, using commercial kits as we have performed previously (Sangesland et al., 2019). After labeling, the fluorescent probes were then re-purified by SEC. For the full-length HA trimer probes, avi-tagged H1 (NC99) and H5 (A/Indonesia/05/2005) were biotinylated and labeled with streptavidin-phycoerythrin (H1-PE) or streptavidin-allophycocyanin (H5-APC) (Weaver et al., 2016; Whittle et al., 2014). These HA trimers contained the Y98F mutation which prevents surface sialic acid binding (Whittle et al., 2014).
Recombinant Antibodies
The heavy and light chain variable regions from the monoclonal antibodies generated in this study were synthesized as gene blocks (GenScript) and cloned into expression vectors expressing human IgG1 heavy and light chains (Sangesland et al., 2019; Whittle et al., 2014). All monoclonal antibodies were transfected into 293F cells as described above, cultured for 6 days, and the harvested supernatant was incubated with Protein G Sepharose resin. IgG was eluted with IgG elution buffer into 1M Tris, pH 8 and purified and buffer exchanged to PBS using SEC with a Superdex 200 10/300 column (GE Healthcare). The monoclonal antibodies: CR6261 (Ekiert et al., 2009; Throsby et al., 2008); G6 (Avnir et al., 2017); CH65 (Whittle et al., 2011); 310-39G10, 310-63E6 (Wheatley et al., 2015) were purified in the same way. The mAb G6 was also labeled with Alexa 647.
Cryo-EM
The 310-63E6 and 310-39G10 IgGs were digested to Fabs using papain. Papain was activated for 15 min at 37°C in digestion buffer (100mM Tris, 2mM EDTA, 10mM L-Cysteine, 1 mg/ml papain). Next, 5 mg of polyclonal IgG were incubated in digestion buffer with papain (20 mM sodium phosphate, 10 mM EDTA, 20 mM cysteine, 0.1 mg/ml papain, pH 7.4). The reaction was incubated for 4–5 h at 37°C. Iodacetamide was added to the sample at a final concentration of 0.03 M to quench the reaction. The digested IgG was concentrated and buffer exchanged to TBS pH 7.4 using 10 kDa cutoff Amicon ultrafiltration units. The undigested IgGs were removed by SEC using a Superose 200 increase column (GE Healthcare Biosciences) in TBS buffer. The fractions containing purified Fabs were concentrated using 10 kDa Amicon ultrafiltration units.
HA was concentrated to 2 mg/ml and incubated with 310-63E6 and 310-39G10 Fabs at a 1:3 molar ratio (HA:Fab) for 30 min at room temperature. The HA-Fab complexes were mixed with octyl-beta-glucoside (OBG) such that the final concentration of the HA-Fab complexes was 0.2 mg/ml and the OBG was 0.3%. UltrAufoil R 1.2/1.3 300 mesh gold grids were plasma cleaned for 7 seconds using solarus advanced plasma cleaning system (Gatan) before loading the sample. Subsequently, 3 μl of the sample were loaded onto the grid and plunge-frozen into nitrogen-cooled liquid ethane using the Vitrobot mark IV (Thermo Fischer Scientific). The settings for the Vitrobot were as follows: Temperature: 4°C, humidity: 100%, blotting time: 5.5 seconds, blotting force: 0 and wait time: 7 seconds.
For data collection, the grids were loaded into a Talos Arctica electron microscope (Thermo Fischer Scientific) operating at 200kV and K2 Summit direct electron detector camera (Gatan). The data were collected at a total cumulative dose of 49.88e−/Å2. Magnification was set at 36,000x with a resulting pixel size of 1.15Å/pix at the specimen plane. Automated data collection was performed using Leginon software (Suloway et al., 2005). The data collection details are presented in Table S1.
The acquired movie micrographs were aligned and dose-weighted using MotionCor2 (Zheng et al., 2017) and uploaded to cryoSPARC v3.1.0 (Punjani et al., 2017). GCTF was applied and particles were picked from micrographs using template picker. Particle extraction and 2D classification was performed. Subsequently, particles from selected 2D subsets were transferred to Relion/3.0. Multiple rounds of 3D refinement and 3D classification were performed to sort out the particles used for the final reconstruction. A soft solvent mask around the HA-Fab was used in the last 3D refinement and post-processing step. All the steps after the second 3D refinement were performed using C3 symmetry. A summary of the data processing approach for each complex are displayed in Figure S2. Final post-processed maps, half-maps and masks used for refinement and post-processing were submitted to the Electron Microscopy Data Bank (EMDB).
The post-processed maps were then used to build the final atomic models. The crystal structure of the HA trimer A/California/07/09 (PDB entry 4m4y) served as the HA initial model. ABodyBuilder was used to produce the models of both antibody Fv regions (Leem et al., 2016). Multiple rounds of Rosetta relaxed refinement (Wang et al., 2016) and manual Coot refinement (Emsley and Crispin, 2018) were performed to build the final models (Figure S2A–G). Model validation was done by using EMRinger (Barad et al., 2015) and MolProbity (Williams et al., 2018) analyses. The final refined models were submitted to the Protein Data Bank (PDB).
The epitope footprints of the antibodies were also depicted on a composite HA model assembled from 59 sequences drawn from H1N1 viruses spanning 103 years and downloaded from the Influenza Research Database (Table S2). These representative sequences are an updated version of an H1N1 dataset assembled previously (Guthmiller et al., 2021).
Immunization regimens
For vaccination studies, mice were sequentially immunized with either 15 μg of SS-np, equimolar HA-np, or RBD-np. Immunizations were administered in 100ul of inoculum containing 50% w/v Sigma adjuvant through the intraperitoneal route (Amitai et al., 2020; Sangesland et al., 2019). The mice were pre-bled prior to each experiment and were immunized at day 0, 21, and 42, and bled two weeks (14 days) after each exposure. For antibody blockade studies 250 μg of CTLA-4 antibody (BioXCell #BE0131) or 250 μg isotype control (BioXCell #BE0087) was administered by intraperitoneal injection concurrently with SS-np immunization (Bradley et al., 2020).
ELISA
Serum antibody responses from vaccinated mice were measured as performed previously (Sangesland et al., 2019). Nunc MaxiSorp plates (96 well) were coated at 4°C overnight at 200ng per well with trimeric NC99 HA and NC99 HAΔstem (containing I45R and T49R point mutations) in PBS. Plates were blocked with 2% BSA in PBS for 1 hour and washed with PBST (PBS containing 0.05% Tween 20). Serum samples were initially diluted at 1:20 and serially diluted 1:5 in PBS and incubated for an hour. The monoclonal antibodies, 310-16B5, 310-29C9, 310-39G10, 310-63E6, CR6261, CH65 were tested at 10 μg/ml and serially diluted at 1:5 in PBS. Plates were then washed with PBST and incubated with either sheep anti-mouse IgG-HRP (GE Healthcare) or sheep anti-human IgG-HRP (GE Healthcare) at 1:5000 dilution in PBS. Plates were again washed in PBST and then developed using TMB solution and quenched with 1N sulfuric acid. Samples were read at 450nm using the Teacan Infinite m1000 Pro microplate absorbance reader (Mannedorf, Switzerland). HA and HAΔstem loading was normalized by reactivity to the RBS specific mAb CH65. To quantify reactivity to HA vs HAΔstem, area under the curve was calculated using GraphPad PRISM software. The difference in binding to HA vs HAΔstem denotes the fraction of the serum antibody response that is specific to the group 1 IAV bnAb epitope on the HA stalk (Sangesland et al., 2019).
Microneutralization Assay
All reporter viruses were prepared as described previously (Creanga et al., 2021). Briefly, all H1N1 viruses were made with a modified PB1 segments expressing the TdKatushka reporter gene (R3ΔPB1) and propagated in MDCK-SIAT-PB1 cells, while H5N1 reporter virus was made with a modified HA segment expressing the reporter (R3ΔHA) and produced in cells stably expressing H5 HA. Virus stocks were stored at −80°C. Mouse sera were treated with receptor destroying enzyme (RDE II; Denka Seiken) and heat-inactivated before use in neutralization assays. Immune sera were serially diluted and incubated for 1 h at 37°C with pre-titrated viruses (H1N1 NC99, H1N1 Ca09, H5N1 VN04). Serum-virus mixtures were then transferred to 96-well plates (PerkinElmer), and 1.0×104 MDCK-SIAT1-PB1 cells (Bloom et al., 2010; Creanga et al., 2021) were added to each well. After overnight incubation at 37°C, the number of fluorescent cells in each well was counted automatically using a Celigo image cytometer (Nexcelom Biosciences).
Viral Challenge
WT C57BL/6, IGHV1-69 F54/F54, IGHV1-69 F54/L54, and IGHV1-69 L/54/L54 mice were vaccinated 3x with SS-np. At week 8 (two weeks after the final immunization), all mice were then infected intranasally with the low path H5N1 avian influenza virus A/chicken/Vietnam/NCVD-016/2008(H5N1)-PR8-IDCDC-RG12 at 105.25 TCID50 units/ml (Sangesland et al., 2019). Naïve unvaccinated mice were also infected as above. All mice were monitored daily for survival and body weight loss. The humane endpoint was set at 20% of the starting body weight. This virus was cultured in 10 day-old embryonated chicken eggs. Seed stocks were obtained from the Centers for Disease Control and Prevention International Reagent Resource (IRR).
B Cell Subsets and Flow Cytometry
To measure immature B cells in our transgenic animals, bone marrow cells were flushed from femur and tibia using PBS. Red blood cells were then depleted using ACK lysis buffer (Lonza) and the suspension was passed through a 70 μm strainer. These cells were then stained with blue viability dye and the following panel of antibodies: IgM BV650 (BD Biosciences); CD93 BV421 (BD Biosciences); IgD PerCP/cy5.5 (BD Biosciences); B220 BUV395 (BD Biosciences); CD3 BV510 (BD Bioscience), F4.80 BV510 (Biolegend); GR-1 BV510 (BD Biosciences); CD19 APC-eFluor780 (eBioscience); Igλ1–3 – FITC (BD Biosciences); and IgK – PE (BD Biosciences).
B cells in the periphery were evaluated in the spleen. Single cell suspensions from mouse splenocytes were obtained using a 70μm cell strainer and red blood cells were removed by ACK lysis buffer. To measure G6 reactivity, splenocytes were stained with blue viability dye and a panel of B and T cell specific antibodies: B220 BV605 (Biolegend), CD4 APC-eFluor780 (Invitrogen), CD8 APC-eFluor780 (Invitrogen), IgM BV650 (BD Biosciences), CD19 PerCP/cy5.5 (Biolegend), and G6 Alexa647 (0.25μg). B cell subsets in the periphery (follicular, marginal zone, T1, T2, T3, T3/BND) were assessed by staining splenocytes with blue viability day and a modified antibody panel: IgM BV650 (BD Biosciences); CD23 BV786 (BD Biosciences); CD93 BV421 (BD Biosciences); IgD PerCP/cy5.5 (BD Biosciences); CD21 PE/cy7 (eBioscience); B220 BUV395 (BD Biosciences); CD3 BV510 (BD Bioscience); F4.80 BV510 (Biolegend); GR-1 BV510 (BD Biosciences); CD19 APC-eFlour 780 (eBioscience); and CD79b BV605 (BD Biosciences); Igλ1–3 – FITC (BD Biosciences); and IgK – PE (BD Biosciences). This panel also included SS-np Alexa 488, SS-np Alexa 594, SSΔstem-np Alexa 647, and SSΔstem-np Alexa 546 probes (each at 0.25 μg).
In all cases, flow cytometry was performed on a 5 Laser LSR Fortessa (BD Biosciences) and the data analyzed using FlowJo software version 9.3.2 (TreeStar).
Antigen specific B cell FACS
To measure the frequency of H1+/H5+ reactive IgG B cells expanded after each immunization step, single cell suspensions of mouse splenocytes were stained with Aqua Live/Dead amine-reactive dye along with the following antibodies: CD3 PE/cy7 (Biolegend), CD4 Alexa Fluor 700 (BD Biosciences), CD19 BV421 (Biolegend), IgM BV605 (Biolegend), IgG PerCP/Cy5.5 (Biolegend) with the addition of 0.25 μg each of H1-PE and H5-APC, as we have performed previously (Sangesland et al., 2019; Weaver et al., 2016; Whittle et al., 2014). The H1+/H5+ reactive B cells were also evaluated within a panel that distinguishes between the germinal center and B cell memory compartment in mice (Tan et al., 2019): CD19 BV421 (Biolegend); F4/80 APCeFlour 780 (eBioscience); CD4 APCeFlour 780 (eBioscience); CD8a APCeFlour 780 (eBioscience); Ly-6G/Ly-6C APCeFlour 780 (eBioscience); GL7 Alexa 488 (Biolegend); CD95 - PE/cy7 (BD Biosciences); IgM BV605 (BioLegend); IgD - BuV395 (BD Biosciences); CD38 - Alexa 594 (Biolegend). Single H1+/H5+ reactive B cells were sorted using a FACS Aria Fusion Sorter (BD Biosciences) into 96 well plates containing RLT lysis buffer with 1% BME and then immediately frozen and stored at −80°C.
The repertoire frequencies of germline B cells specific for the group 1 IAV bnAb site (on target) in our transgenic mice were measured as previously described (Sangesland et al., 2019). Mouse splenocytes were processed as above and stained with blue live/dead amine-reactive dye. Cells were further stained with a panel containing 0.25 μg each of SS-np Alexa 488, SS-np Alexa 594, SS-npΔstem Alexa 647, and SS-npΔstem Alexa 546 probes as previously described (Sangesland et al., 2019), along with the following antibodies: IgG BV421 (Biolegend), IgD Alexa Fluor 700 (Biolegend), CD3 PE/cy7 (Biolegend), IgM BV605 (Biolegend), CD19 PerCP/cy5.5 (Biolegend). Germline on-target B cells were defined as CD3−/CD19+/IgG−/IgM+/IgD+/SS-np Alexa 488+/SS-np Alexa 594+/SSΔstem-np Alexa 647−/SSΔstem-np Alexa 546−. Single B cells were sorted using the FACS Aria Fusion Sorter (BD Biosciences) into 96 well plates containing RLT lysis buffer with 1% BME and immediately frozen and stored at −80°C.
For human germline B cells PBMC from five subjects, cells were stained with Aqua Live/Dead amine-reactive dye, washed, and then incubated with the following antibodies: CD19 Alexa 700 (Biolegend), IgG BV421 (BD Biosciences), IgD PE-Cy7 (BD Biosciences), IgM PerCP-Cy5.5 (BioLegend), CD3 APC-Cy7 (BD Biosciences); and 0.25μg each of SS-np Alexa 488, SS-np Alexa 594, SSΔstem-np Alexa 647 and SSΔstem-np Alexa 546. Individual on-target cells were isolated by FACS and processed for BCR sequencing. Analyses of the FACS data were performed using FlowJo software version 9.3.2 (TreeStar).
BCR sequencing
Bulk CDRH3 diversity within our transgenic mice was measured by deep sequencing antigen naïve IgM B cells (Sangesland et al., 2019; Sangesland et al., 2020; Trombetta et al., 2014). BCR libraries were generated from whole transcriptome amplification (WTA) products using the Smart-Seq2 protocol (Trombetta et al., 2014). BCR heavy chains (FR3 to CDR3) were PCR amplified from WTA using forward and reverse primers specific to the FR3 region (0.5μM final concentration) of IGHV1-69 and the heavy chain constant region (1 uM final concentration), respectively. These primers were also attached to the Illumina P7 (FR3 region) and P5 (constant region) sequences. Heavy chain sequences were further amplified by step-out PCR, with the addition of Illumina sequencing adaptors. The resulting PCR products (FR3 to CDRH3) were sequenced on the Illumina Miseq and the paired end FASTQs were aligned using MIXCR (Bolotin et al., 2015). The following parameters were used for alignment: OvParameters.geneFeatureToAlign={FR3Begin:Vend}, and restricted alignment to heavy chain matches. Low quality sequence reads were trimmed by MIXCR using the default settings. Human CDRH3 repertoire diversity was mined from donor D2 using naïve BCR sequences (DeWitt et al., 2016). Using custom scripts within immunarch (v0.5.5) tcR (Nazarov et al., 2015), we analyzed the BCR sequences from IGHV1-69*01 and IGHV1-69*09 mice and naïve human B cells to generate frequency plots of CDRH3 lengths and assigned Kabat numbering for amino acid composition at 16aa CDRH3 length. The number of aligned reads were 7.54×106 for IGHV1-69*01 and 8.17×106 for IGHV1-69*09 mice.
For single cell BCR sequencing, we again amplified WTA from germline SS-np on-target and H1+/H5+ reactive B cells as previously described (Sangesland et al., 2019; Trombetta et al., 2014). BCR sequences from heavy and light chains were enriched from single B cells using a pool of partially degenerate V region specific primers against all possible human IGHV or mouse IGLV and IGKV segments (FR1 region) and reverse primers against the heavy or light chain constant regions, again attached to Illumina P7 and P5 sequences (Sangesland et al., 2019). The step out PCR was also used to add cellular barcodes and Illumina sequencing adapters (based on Nextera XT Index Adapters) to each heavy and light chain. Single cell heavy and light chain samples were then pooled and sequenced using paired end V2–500 cycle (250×250 read and 8×8 index reads) Illumina Miseq System. Following demultiplexing, BCR heavy and light chains were paired, overlapping sequencing reads were reconstructed using PandaSeq (Masella et al., 2012), and aligned against the human IMGT database (Shi et al., 2014) with sequencing error correction using MigMAP, a wrapper for IgBlast (https://github.com/mikessh/migmap). Consensus VH and VL/VK chain for each single cell was determined by collapsing all reads with the same CDR3 sequence and calling the top heavy and light chain sequences by frequency. Heavy and light chains sequences have at least 25 reads or a frequency two-times greater than the next sequence of the same chain in order to be given a consensus sequence.
Biolayer Interferometry (BLI)
For BLI experiments the Fabs were generated by digesting the recombinant IgGs with the endoproteinase Lys-C (New England Biolabs Cat. # P8109S), as performed previously (Borst et al., 2018; Gadgil et al., 2006). This reaction consisted of 5μg Lys-C per milligram of IgG within PBS supplemented with 1mM EDTA. After 12h at room temperature, a 1× complete protease inhibitor cocktail (Roche, Cat # 11697498001) was added to the mixture and the uncleaved IgG was removed by the addition of Protein A/G-agarose. The beads were washed in PBS and the supernatant was concentrated using Amicon Ultra concentrators (10 kDa cut off) and the Fabs were then further isolated by SEC using a Superdex 200 10/300 column (GE Healthcare).
The binding affinities of antibody Fabs were measured by biolayer interferometry using the Personal Assay BLItz System (Fortebio). His-tagged stalk only-trimer loaded onto Ni-NTA biosensors (Fortebio). After acquiring a baseline in kinetic buffer (PBS + 0.02% Tween20 + 0.1% BSA), Fabs were added at 1.25 μM, 2.5 μM, 5 μM, and 10 μM. The binding measurements used 120 sec association and 120 sec dissociation periods. The equilibrium dissociation constant (KD) values were then calculated by applying a 1:1 binding isotherm using vendor-supplied software (Bajic et al., 2020; Schmidt et al., 2015).
Polyreactivity and Autoreactivity assays
For autoantigen ELISAs, germline L54 and F54 on-target IGHV1-69 mAbs, isolated from HC2 mice and human PBMC (Patients 1–5), were tested both at 10 μg/ml and 200 μg/ml (Andrews et al., 2015; Haynes et al., 2005). For insulin and dsDNA reactivity, plates were coated with 200ng per well with recombinant human insulin (Fitzgerald) or 50 μg/ml double stranded calf thymus DNA (Invitrogen) in 100ul of PBS. ELISAs were conducted as described in the previous section. For cardiolipin, plates were coated with 5ug per well of cardiolipin (Sigma) in 30ul of ethanol and evaporated overnight in a vacuum sealed desiccator (Pierangeli and Harris, 2008). Plates were washed with PBS and blocked in PBS with 10% FBS for 1 hour. The primary and secondary antibodies were incubated in PBS with 1% FBS. Plates were again washed in PBS and then developed using TMB solution and quenched with 1N sulfuric acid. Samples were read at 450nm using the Teacan Infinite m1000 Pro microplate absorbance reader (Mannedorf, Switzerland). For other lipid antigens, ELISAs were conducted according to a previously described method (Sangesland et al., 2020; Tobias et al., 1989). Plates were coated with 30 μg/ml of LPS (Sigma), POPC (Avanti Lipids) or Sphingomyelin (Avanti Lipids) in carbonate buffer (100mM Na2CO3, 20mM EDTA, pH 9.6) for 3 hours at 37°C. The plates were then washed with water and dried overnight in a vacuum sealed desiccator. The next day the plates were blocked with HS buffer (50mM Hepes, 0.15mM NaCl, pH 7.4) with the addition of 10 mg/ml BSA. All washing steps were performed with HS buffer with 1mg/ml BSA in PBS. All primary and secondary antibodies were incubated in HS buffer with 1mg/ml BSA and then developed as above.
To evaluate the contribution of HA stalk N-linked glycans to the binding of poly/autoreactive mAbs, SS-np was heat denatured and deglycosylated using PNGase F (New England BioLabs, P0704S), as per the manufacturer’s instructions (Keating et al., 2020). The PNGase F was then separated from the denatured SS-np using size exclusion chromatography (Keating et al., 2020). We then evaluated binding of germline L54 on-target IGHV1-69 mAbs (human and HC2 mouse) to denatured SS-np (pre- and post- deglycosylation) using the same conditions described for human insulin above.
For Hep2 cell immunofluorescence, germline on-target IGHV1-69 F54 and L54 monoclonal antibodies were tested at a concentration of 200 μg/ml in PBS using the NOVA Lite Hep-2 ANA kit (Inova Diagnostics) according to the manufacturer’s instructions. Cells were also co-stained with DAPI for 5 min. Images were acquired using the TissueFAXs whole slide scanning system (TissueGnostics) at 20X magnification with extended focus.
Ca2+ fluxing
IgM BCR signaling was evaluated in resting F54 and L54 IGHV1-69 B cells using calcium flux, as per our established protocol (Weaver et al., 2016). Resting B cells were isolated from the spleens of F54/F54 and L54/L54 IGHV1-69 mice using MACS B cell isolation kit according to the manufacturer’s instructions (Miltenyo Biotec). The cells were then reconstituted in RPMI (lacking FBS) and stained with membrane permeable calcium sensing dye Fura Red at 1.5 μg/ml. After a 20 minute incubation at 37°C, the cells were washed in RPMI and then subjected to flow cytometry (LSR II, BD) where BCR stimulation was measured kinetically as the ratio of the Ca2+ bound/unbound states of Fura Red (Weaver et al., 2016). Ca2+ fluxing was measured three times (n=3 per mouse genotype) using 2×106 B cells per run. During each run, ratiometric flux measurements were made before and after exposure to 10 μg/ml anti-IgM [(Fab’)2, Jackson Immunoresearch]. These values were then standardized to the total/maximum Ca2+ flux capacity in each run, as defined by exposure of the cells to 10 μg/ml ionomycin (Weaver et al., 2016).
Datasets and statistical analyses
Human IGHV1-69 F54/L54 allelic usage was mined from the 1000 genomes project (rs 55891010) (Avnir et al., 2016; Genomes Project et al., 2010). All statistical analysis were conducted using Prism Graphpad, R 3.6.0 and SAS 9.4. Sample sizes and statistical tests are indicated in the figure legends. Data were considered statistically significant at P<0.05.
Supplementary Material
Table S1 – Cryo-EM data collection, processing, model refinement and validation – related to Figures 1, S2 and S3.
Table S3 – HC2 mouse and human germline on-target antibody affinities for the HA stalk – related to Figures 2 and 7.
Table S4 – HC2 mouse and human germline on-target stalk mAbs – related to Figures 2, 4, 7, S4, S5, S6 and S7.
Table S5 – Conversion of HC2 mouse and human L54 germline stalk mAbs into the F54 IGHV1-69 background – related to Figures 4 and 7.
Table S7 – HC2 mouse and human germline antibody affinities for the HA stalk after L54 to F54 conversion - related to Figure 7.
Table S6 – HC2 mouse and human stalk mAbs expanded after vaccination– related to Figures 6 and 7.
Highlights.
Both F54 and L54 IGHV1-69 alleles encode for human bnAbs against influenza virus
Only F54 IGHV1-69 can provide substrate for vaccine-amplifying bnAbs to high titer
L54 IGHV1-69 co-endows for poly/autoreactive B cells which become tolerized
Immune cell checkpoint inhibitors can restore L54 IGHV1-69 antibody responses
Acknowledgments
D.L. was supported by NIH (DP2DA042422, R01AI124378, R01AI153098, R01AI155447, U19AI057229, P30AI060354), the Harvard University Milton Award, The Gilead Research Scholars Program. M.S. was supported by the NSF Graduate Research Fellowship Program and an NIH fellowship (F31Al138368). A.T.P is a recipient of a Rubicon fellowship from the Netherlands Organization for Scientific Research (NWO). We are indebted to BMS for gifting transgenic mice to D.L. We thank Julia Bals, Vintus Okonkwo and Nathania Hartojo for recombinant protein production, the MGH Center for Comparative Medicine and Daniel Tapia for help with animal experiments. We also thank Adam Wheatley (Peter Doherty Institute) for providing the sequences of 310-63E6 and 310-39G10 and Julie Zikherman (University of California, San Francisco) for helpful suggestions on this study. We gratefully acknowledge Aaron Schmidt at the Ragon Institute for providing the ferritin-only nanoparticle and Patrick McTamney for assistance with some of the figure graphics. Lastly, we thank the Lingwood lab for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
Authors declare no competing interests. M.K is listed as inventor of patents and patent applications on vaccine immunogens used in this study filed by the U.S. Department of Health and Human Services.
Inclusion and Diversity
One or more of the authors of this paper identifies as an underrepresented ethnic minority in science.
References
- Abbott RK, and Crotty S (2020). Factors in B cell competition and immunodominance. Immunological reviews 296, 120–131. 10.1111/imr.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman MO, Angeletti D, and Yewdell JW (2018). Antibody Immunodominance: The Key to Understanding Influenza Virus Antigenic Drift. Viral Immunology 31, 142–149. 10.1089/vim.2017.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitai A, Sangesland M, Barnes RM, Rohrer D, Lonberg N, Lingwood D, and Chakraborty AK (2020). Defining and Manipulating B Cell Immunodominance Hierarchies to Elicit Broadly Neutralizing Antibody Responses against Influenza Virus. Cell Systems 11, 573–588 10.1016/j.cels.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, Henry Dunand CJ, Taylor WM, Lim S, Huang M, et al. (2015). Immune history profoundly affects broadly protective B cell responses to influenza. Science translational medicine 7, 316ra192. 10.1126/scitranslmed.aad0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeletti D, Kosik I, Santos JJS, Yewdell WT, Boudreau CM, Mallajosyula VVA, Mankowski MC, Chambers M, Prabhakaran M, Hickman HD, et al. (2019). Outflanking immunodominance to target subdominant broadly neutralizing epitopes. Proceedings of the National Academy of Sciences of the United States of America 116, 13474–13479 10.1073/pnas.1816300116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avnir Y, Prachanronarong KL, Zhang Z, Hou S, Peterson EC, Sui J, Zayed H, Kurella VB, McGuire AT, Stamatatos L, et al. (2017). Structural Determination of the Broadly Reactive Anti-IGHV1-69 Anti-idiotypic Antibody G6 and Its Idiotope. Cell Reports 21, 3243–3255. 10.1016/j.celrep.2017.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avnir Y, Tallarico AS, Zhu Q, Bennett AS, Connelly G, Sheehan J, Sui J, Fahmy A, Huang CY, Cadwell G, et al. (2014). Molecular signatures of hemagglutinin stem-directed heterosubtypic human neutralizing antibodies against influenza A viruses. PLoS pathogens 10, e1004103. 10.1371/journal.ppat.1004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avnir Y, Watson CT, Glanville J, Peterson EC, Tallarico AS, Bennett AS, Qin K, Fu Y, Huang CY, Beigel JH, et al. (2016). IGHV1-69 polymorphism modulates anti-influenza antibody repertoires, correlates with IGHV utilization shifts and varies by ethnicity. Scientific reports 6, 20842. 10.1038/srep20842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic G, Maron MJ, Adachi Y, Onodera T, McCarthy KR, McGee CE, Sempowski GD, Takahashi Y, Kelsoe G, Kuraoka M, and Schmidt AG (2019a). Influenza Antigen Engineering Focuses Immune Responses to a Subdominant but Broadly Protective Viral Epitope. Cell host & microbe 25, 827–835 e826. 10.1016/j.chom.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic G, Maron MJ, Caradonna TM, Tian M, Mermelstein A, Fera D, Kelsoe G, Kuraoka M, and Schmidt AG (2020). Structure-Guided Molecular Grafting of a Complex Broadly Neutralizing Viral Epitope. ACS Infect Dis 6, 1182–1191. 10.1021/acsinfecdis.0c00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic G, van der Poel CE, Kuraoka M, Schmidt AG, Carroll MC, Kelsoe G, and Harrison SC (2019b). Autoreactivity profiles of influenza hemagglutinin broadly neutralizing antibodies. Scientific Reports 9, 3492. 10.1038/s41598-019-40175-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft T, DeBuysscher BL, Weidle C, Schwartz A, Wall A, Gray MD, Feng J, Steach HR, Fitzpatrick KS, Gewe MM, et al. (2019). Detection and activation of HIV broadly neutralizing antibody precursor B cells using anti-idiotypes. The Journal of Experimental Medicine 216, 2331–2347. 10.1084/jem.20190164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barad BA, Echols N, Wang RY, Cheng Y, DiMaio F, Adams PD, and Fraser JS (2015). EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nature Methods 12, 943–946. 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JD, Gong LI, and Baltimore D (2010). Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328, 1272–1275. 10.1126/science.1187816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, and Chudakov DM (2015). MiXCR: software for comprehensive adaptive immunity profiling. Nature methods 12, 380–381. 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- Borst AJ, Weidle CE, Gray MD, Frenz B, Snijder J, Joyce MG, Georgiev IS, Stewart-Jones GB, Kwong PD, McGuire AT, et al. (2018). Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core. Elife 7. 10.7554/eLife.37688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyoglu-Barnum S, Ellis D, Gillespie RA, Hutchinson GB, Park YJ, Moin SM, Acton OJ, Ravichandran R, Murphy M, Pettie D, et al. (2021). Quadrivalent influenza nanoparticle vaccines induce broad protection. Nature 592, 623–628. 10.1038/s41586-021-03365-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyaratanakornkit J, and Taylor JJ (2019). Techniques to Study Antigen-Specific B Cell Responses. Frontiers in immunology 10, 1694. 10.3389/fimmu.2019.01694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley T, Kuraoka M, Yeh CH, Tian M, Chen H, Cain DW, Chen X, Cheng C, Ellebedy AH, Parks R, et al. (2020). Immune checkpoint modulation enhances HIV-1 antibody induction. Nature Communications 11, 948. 10.1038/s41467-020-14670-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briney B, Inderbitzin A, Joyce C, and Burton DR (2019). Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature 566, 393–397. 10.1038/s41586-019-0879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR (2019). Advancing an HIV vaccine; advancing vaccinology. Nature reviews. Immunology 19, 77–78. 10.1038/s41577-018-0103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambier JC, Gauld SB, Merrell KT, and Vilen BJ (2007). B-cell anergy: from transgenic models to naturally occurring anergic B cells? Nature reviews. Immunology 7, 633–643. 10.1038/nri2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang J, Hwang KK, Bouton-Verville H, Xia SM, Newman A, Ouyang YB, Haynes BF, and Verkoczy L (2013). Common tolerance mechanisms, but distinct cross-reactivities associated with gp41 and lipids, limit production of HIV-1 broad neutralizing antibodies 2F5 and 4E10. Journal of immunology 191, 1260–1275. 10.4049/jimmunol.1300770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MD (2015). The early history of B cells. Nature reviews. Immunology 15, 191–197. 10.1038/nri3801. [DOI] [PubMed] [Google Scholar]
- Corti D, Suguitan AL Jr., Pinna D, Silacci C, Fernandez-Rodriguez BM, Vanzetta F, Santos C, Luke CJ, Torres-Velez FJ, Temperton NJ, et al. (2010). Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. The Journal of Clinical Investigation 120, 1663–1673. 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creanga A, Gillespie RA, Fisher BE, Andrews SF, Lederhofer J, Yap C, Hatch L, Stephens T, Tsybovsky Y, Crank MC, et al. (2021). A comprehensive influenza reporter virus panel for high-throughput deep profiling of neutralizing antibodies. Nature Communication 12, 1722. 10.1038/s41467-021-21954-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darricarrere N, Qiu Y, Kanekiyo M, Creanga A, Gillespie RA, Moin SM, Saleh J, Sancho J, Chou TH, Zhou Y, et al. (2021). Broad neutralization of H1 and H3 viruses by adjuvanted influenza HA stem vaccines in nonhuman primates. Science translational medicine 13. 10.1126/scitranslmed.abe5449. [DOI] [PubMed] [Google Scholar]
- Delgado Alves J, Ames PR, Donohue S, Stanyer L, Nourooz-Zadeh J, Ravirajan C, and Isenberg DA (2002). Antibodies to high-density lipoprotein and beta2-glycoprotein I are inversely correlated with paraoxonase activity in systemic lupus erythematosus and primary antiphospholipid syndrome. Arthritis Rheumatology 46, 2686–2694. 10.1002/art.10542. [DOI] [PubMed] [Google Scholar]
- DeWitt WS, Lindau P, Snyder TM, Sherwood AM, Vignali M, Carlson CS, Greenberg PD, Duerkopp N, Emerson RO, and Robins HS (2016). A Public Database of Memory and Naive B-Cell Receptor Sequences. PloS one 11, e0160853. 10.1371/journal.pone.0160853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle-Cooper C, Hudson KE, Cooper AB, Ota T, Skog P, Dawson PE, Zwick MB, Schief WR, Burton DR, and Nemazee D (2013). Immune tolerance negatively regulates B cells in knock-in mice expressing broadly neutralizing HIV antibody 4E10. Journal of immunology 191, 3186–3191. 10.4049/jimmunol.1301285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duty JA, Szodoray P, Zheng NY, Koelsch KA, Zhang Q, Swiatkowski M, Mathias M, Garman L, Helms C, Nakken B, et al. (2009). Functional anergy in a subpopulation of naive B cells from healthy humans that express autoreactive immunoglobulin receptors. The Journal of Experimental Medicine 206, 139–151. 10.1084/jem.20080611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, and Wilson IA (2009). Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–251. 1171491. 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Crispin M (2018). Structural analysis of glycoproteins: building N-linked glycans with Coot. Acta Crystallographica Section D 74, 256–263. 10.1107/S2059798318005119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbelding EJ, Post D, Stemmy E, Roberts PC, Augustine AD, Ferguson S, Paules CI, Graham BS, and Fauci AS (2018). A Universal Influenza Vaccine: The Strategic Plan for the National Institute of Allergy and Infectious Diseases. The Journal of infectious diseases 218, 347–354. 10.1093/infdis/jiy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finton KA, Larimore K, Larman HB, Friend D, Correnti C, Rupert PB, Elledge SJ, Greenberg PD, and Strong RK (2013). Autoreactivity and exceptional CDR plasticity (but not unusual polyspecificity) hinder elicitation of the anti-HIV antibody 4E10. PLoS pathogens 9, e1003639. 10.1371/journal.ppat.1003639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishwild DM, O’Donnell SL, Bengoechea T, Hudson DV, Harding F, Bernhard SL, Jones D, Kay RM, Higgins KM, Schramm SR, and Lonberg N (1996). High-avidity human IgG kappa monoclonal antibodies from a novel strain of minilocus transgenic mice. Nature Biotechnology 14, 845–851. 10.1038/nbt0796-845. [DOI] [PubMed] [Google Scholar]
- Gadgil HS, Bondarenko PV, Pipes GD, Dillon TM, Banks D, Abel J, Kleemann GR, and Treuheit MJ (2006). Identification of cysteinylation of a free cysteine in the Fab region of a recombinant monoclonal IgG1 antibody using Lys-C limited proteolysis coupled with LC/MS analysis. Analytical Biochemistry 355, 165–174. 10.1016/j.ab.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Genomes Project C, Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, and McVean GA (2010). A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073. 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getahun A (2022). Role of inhibitory signaling in peripheral B cell tolerance. Immunological reviews 307, 27–42. 10.1111/imr.13070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanville J, Zhai W, Berka J, Telman D, Huerta G, Mehta GR, Ni I, Mei L, Sundar PD, Day GM, et al. (2009). Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proceedings of the National Academy of Sciences of the United States of America 106, 20216–20221. 10.1073/pnas.0909775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthmiller JJ, Han J, Li L, Freyn AW, Liu STH, Stovicek O, Stamper CT, Dugan HL, Tepora ME, Utset HA, et al. (2021). First exposure to the pandemic H1N1 virus induced broadly neutralizing antibodies targeting hemagglutinin head epitopes. Science translational medicine 13. 10.1126/scitranslmed.abg4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthmiller JJ, Lan LY, Fernandez-Quintero ML, Han J, Utset HA, Bitar DJ, Hamel NJ, Stovicek O, Li L, Tepora M, et al. (2020). Polyreactive Broadly Neutralizing B cells Are Selected to Provide Defense against Pandemic Threat Influenza Viruses. Immunity 53, 1230–1244 e1235. 10.1016/j.immuni.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, et al. (2005). Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308, 1906–1908. 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- Henry Dunand CJ, and Wilson PC (2015). Restricted, canonical, stereotyped and convergent immunoglobulin responses. Philosophical transactions of the Royal Society of London. Series B, Biological sciences 370. 10.1098/rstb.2014.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter J, Rodig JV, Hoper D, Seeberger PH, Reichl U, Rapp E, and Lepenies B (2013). Toward animal cell culture-based influenza vaccine design: viral hemagglutinin N-glycosylation markedly impacts immunogenicity. Journal of Immunology 190, 220–230. 10.4049/jimmunol.1201060. [DOI] [PubMed] [Google Scholar]
- Kanekiyo M, Joyce MG, Gillespie RA, Gallagher JR, Andrews SF, Yassine HM, Wheatley AK, Fisher BE, Ambrozak DR, Creanga A, et al. (2019). Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nature immunology 20, 362–372. 10.1038/s41590-018-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo M, Wei CJ, Yassine HM, McTamney PM, Boyington JC, Whittle JR, Rao SS, Kong WP, Wang L, and Nabel GJ (2013). Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499, 102–106. nature12202 10.1038/nature12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating CL, Kuhn E, Bals J, Cocco AR, Yousif AS, Matysiak C, Sangesland M, Ronsard L, Smoot M, Barcamonte Moreno T, et al. (2020). Spontaneous glycan reattachment following N-glycanase treatment of influenza and HIV vaccine antigens. Journal of Proteome research. 10.1021/acs.jproteome.9b00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, and Mascola JR (2018). HIV-1 Vaccines Based on Antibody Identification, B Cell Ontogeny, and Epitope Structure. Immunity 48, 855–871. 10.1016/j.immuni.2018.04.029. [DOI] [PubMed] [Google Scholar]
- Lee PS, and Wilson IA (2015). Structural characterization of viral epitopes recognized by broadly cross-reactive antibodies. Current Topics in Microbiology and Immunology 386, 323–341. 10.1007/82_2014_413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leem J, Dunbar J, Georges G, Shi J, and Deane CM (2016). ABodyBuilder: Automated antibody structure prediction with data-driven accuracy estimation. MAbs 8, 1259–1268. 10.1080/19420862.2016.1205773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner RA (2011). Rare antibodies from combinatorial libraries suggests an S.O.S. component of the human immunological repertoire. Molecular bioSystems 7, 1004–1012. 10.1039/c0mb00310g. [DOI] [PubMed] [Google Scholar]
- Lingwood D, McTamney PM, Yassine HM, Whittle JR, Guo X, Boyington JC, Wei CJ, and Nabel GJ (2012). Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature 489, 566–570. 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonberg N, Taylor LD, Harding FA, Trounstine M, Higgins KM, Schramm SR, Kuo CC, Mashayekh R, Wymore K, McCabe JG, and et al. (1994). Antigen-specific human antibodies from mice comprising four distinct genetic modifications. Nature 368, 856–859. 10.1038/368856a0. [DOI] [PubMed] [Google Scholar]
- Luning Prak ET, Monestier M, and Eisenberg RA (2011). B cell receptor editing in tolerance and autoimmunity. Annals of the New York Academy of Sciences 1217, 96–121. 10.1111/j.1749-6632.2010.05877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masella AP, Bartram AK, Truszkowski JM, Brown DG, and Neufeld JD (2012). PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics 13, 31. 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Gray MD, Dosenovic P, Gitlin AD, Freund NT, Petersen J, Correnti C, Johnsen W, Kegel R, Stuart AB, et al. (2016). Specifically modified Env immunogens activate B-cell precursors of broadly neutralizing HIV-1 antibodies in transgenic mice. Nature communications 7, 10618. 10.1038/ncomms10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesin L, Schiepers A, Ersching J, Barbulescu A, Cavazzoni CB, Angelini A, Okada T, Kurosaki T, and Victora GD (2020). Restricted Clonality and Limited Germinal Center Reentry Characterize Memory B Cell Reactivation by Boosting. Cell 180, 92–106 e111. 10.1016/j.cell.2019.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora T, Walczak AM (2019). How many different clonotypes do immune repertoires contain? Current Opinion in Systems Biology 18, 104–110. [Google Scholar]
- Nachbagauer R, Feser J, Naficy A, Bernstein DI, Guptill J, Walter EB, Berlanda-Scorza F, Stadlbauer D, Wilson PC, Aydillo T, et al. (2021). A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nature medicine 27, 106–114. 10.1038/s41591-020-1118-7. [DOI] [PubMed] [Google Scholar]
- Nachbagauer R, and Palese P (2020). Is a Universal Influenza Virus Vaccine Possible? Annual Review of Medicine 71, 315–327. 10.1146/annurev-med-120617-041310. [DOI] [PubMed] [Google Scholar]
- Nazarov VI, Pogorelyy MV, Komech EA, Zvyagin IV, Bolotin DA, Shugay M, Chudakov DM, Lebedev YB, and Mamedov IZ (2015). tcR: an R package for T cell receptor repertoire advanced data analysis. BMC bioinformatics 16, 175. 10.1186/s12859-015-0613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemazee D (2017). Mechanisms of central tolerance for B cells. Nature reviews. Immunology 17, 281–294. 10.1038/nri.2017.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas L, Foglierini M, Piccoli L, Kallewaard NL, Turrini F, Silacci C, Fernandez-Rodriguez B, Agatic G, Giacchetto-Sasselli I, Pellicciotta G, et al. (2014). Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature 516, 418–422. 10.1038/nature13764. [DOI] [PubMed] [Google Scholar]
- Paules C, and Subbarao K (2017). Influenza. Lancet 390, 697–708. 10.1016/S0140-6736(17)30129-0. [DOI] [PubMed] [Google Scholar]
- Paules CI, Sullivan SG, Subbarao K, and Fauci AS (2018). Chasing Seasonal Influenza - The Need for a Universal Influenza Vaccine. The New England Journal of Medicine 378, 7–9. 10.1056/NEJMp1714916. [DOI] [PubMed] [Google Scholar]
- Pease S, and Saunders TL, eds. (2011). Advanced Protocols for Animal Transgenesis. An ISTT Manual (Springer; ). [Google Scholar]
- Peterhoff D, and Wagner R (2017). Guiding the long way to broad HIV neutralization. Current opinion in HIV and AIDS 12, 257–264. 10.1097/COH.0000000000000356. [DOI] [PubMed] [Google Scholar]
- Pierangeli SS, and Harris EN (2008). A protocol for determination of anticardiolipin antibodies by ELISA. Nat Protoc 3, 840–848. 10.1038/nprot.2008.48. [DOI] [PubMed] [Google Scholar]
- Platt JL, Garcia de Mattos Barbosa M, and Cascalho M (2019). The five dimensions of B cell tolerance. Immunological reviews 292, 180–193. 10.1111/imr.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nature methods 14, 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Radway-Bright EL, Ravirajan CT, and Isenberg DA (2000). The prevalence of antibodies to anionic phospholipids in patients with the primary antiphospholipid syndrome, systemic lupus erythematosus and their relatives and spouses. Rheumatology (Oxford) 39, 427–431. 10.1093/rheumatology/39.4.427. [DOI] [PubMed] [Google Scholar]
- Ronsard L, Yousif AS, Peabody J, Okonkwo V, Devant P, Mogus AT, Barnes RM, Rohrer D, Lonberg N, Peabody D, et al. (2021). Engineering an Antibody V Gene-Selective Vaccine. Frontiers in immunology 12, 730471. 10.3389/fimmu.2021.730471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangesland M, and Lingwood D (2021a). Antibody Focusing to Conserved Sites of Vulnerability: The Immunological Pathways for ‘Universal’ Influenza Vaccines. Vaccines (Basel) 9. 10.3390/vaccines9020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangesland M, and Lingwood D (2021b). Public Immunity: Evolutionary Spandrels for Pathway-Amplifying Protective Antibodies. Frontiers in immunology 12, 708882. 10.3389/fimmu.2021.708882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangesland M, Ronsard L, Kazer SW, Bals J, Boyoglu-Barnum S, Yousif AS, Barnes R, Feldman J, Quirindongo-Crespo M, McTamney PM, et al. (2019). Germline-Encoded Affinity for Cognate Antigen Enables Vaccine Amplification of a Human Broadly Neutralizing Response against Influenza Virus. Immunity 51, 735–749 e738. 10.1016/j.immuni.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangesland M, Yousif AS, Ronsard L, Kazer SW, Zhu AL, Gatter GJ, Hayward MR, Barnes RM, Quirindongo-Crespo M, Rohrer D, et al. (2020). A Single Human VH-gene Allows for a Broad-Spectrum Antibody Response Targeting Bacterial Lipopolysaccharides in the Blood. Cell Rep 32, 108065. 10.1016/j.celrep.2020.108065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasso EH, Willems van Dijk K, Bull AP, and Milner EC (1993). A fetally expressed immunoglobulin VH1 gene belongs to a complex set of alleles. The Journal of clinical investigation 91, 2358–2367. 10.1172/JCI116468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AG, Do KT, McCarthy KR, Kepler TB, Liao HX, Moody MA, Haynes BF, and Harrison SC (2015). Immunogenic Stimulus for Germline Precursors of Antibodies that Engage the Influenza Hemagglutinin Receptor-Binding Site. Cell Reports 13, 2842–2850. 10.1016/j.celrep.2015.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder KMS, Agazio A, Strauch PJ, Jones ST, Thompson SB, Harper MS, Pelanda R, Santiago ML, and Torres RM (2017). Breaching peripheral tolerance promotes the production of HIV-1-neutralizing antibodies. The Journal of Experimental Medicine 214, 2283–2302. 10.1084/jem.20161190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B, Ma L, He X, Wang X, Wang P, Zhou L, and Yao X (2014). Comparative analysis of human and mouse immunoglobulin variable heavy regions from IMGT/LIGM-DB with IMGT/HighV-QUEST. Theor Biol Med Model 11, 30. 10.1186/1742-4682-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L, Pancera M, and McGuire AT (2017). Germline-targeting immunogens. Immunological reviews 275, 203–216. 10.1111/imr.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, et al. (2009). Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nature Structural & Molecular Biology 16, 265–273. 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, and Carragher B (2005). Automated molecular microscopy: the new Leginon system. Journal of Structural Biology 151, 41–60. 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tan HX, Jegaskanda S, Juno JA, Esterbauer R, Wong J, Kelly HG, Liu Y, Tilmanis D, Hurt AC, Yewdell JW, et al. (2019). Subdominance and poor intrinsic immunogenicity limit humoral immunity targeting influenza HA stem. The Journal of Clinical Investigation 129, 850–862. 10.1172/JCI123366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, et al. (2008). Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One 3, e3942. 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiegs SL, Russell DM, and Nemazee D (1993). Receptor editing in self-reactive bone marrow B cells. The Journal of Experimental Medicine 177, 1009–1020. 10.1084/jem.177.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias PS, Soldau K, and Ulevitch RJ (1989). Identification of a lipid A binding site in the acute phase reactant lipopolysaccharide binding protein. The Journal of Biological Chemistry 264, 10867–10871. [PubMed] [Google Scholar]
- Trombetta JJ, Gennert D, Lu D, Satija R, Shalek AK, and Regev A (2014). Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Curr Protoc Mol Biol 107, 4 22 21–17. 10.1002/0471142727.mb0422s107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela JL, Ait-Azzouzene D, Duong BH, Ota T, and Nemazee D (2008). Rearrangement of mouse immunoglobulin kappa deleting element recombining sequence promotes immune tolerance and lambda B cell production. Immunity 28, 161–170. 10.1016/j.immuni.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RY, Song Y, Barad BA, Cheng Y, Fraser JS, and DiMaio F (2016). Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. Elife 5. 10.7554/eLife.17219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, and Nussenzweig MC (2003). Predominant autoantibody production by early human B cell precursors. Science 301, 1374–1377. 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- Weaver GC, Villar RF, Kanekiyo M, Nabel GJ, Mascola JR, and Lingwood D (2016). In vitro reconstitution of B cell receptor-antigen interactions to evaluate potential vaccine candidates. Nature protocols 11, 193–213. 10.1038/nprot.2016.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CJ, Crank MC, Shiver J, Graham BS, Mascola JR, and Nabel GJ (2020). Next-generation influenza vaccines: opportunities and challenges. Nature Reviews Drug Discovery 19, 239–252. 10.1038/s41573-019-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley AK, Whittle JR, Lingwood D, Kanekiyo M, Yassine HM, Ma SS, Narpala SR, Prabhakaran MS, Matus-Nicodemos RA, Bailer RT, et al. (2015). H5N1 Vaccine-Elicited Memory B Cells Are Genetically Constrained by the IGHV Locus in the Recognition of a Neutralizing Epitope in the Hemagglutinin Stem. Journal of immunology 195, 602–610. 10.4049/jimmunol.1402835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JR, Wheatley AK, Wu L, Lingwood D, Kanekiyo M, Ma SS, Narpala SR, Yassine HM, Frank GM, Yewdell JW, et al. (2014). Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. Journal of virology 88, 4047–4057. JVI.03422–1310.1128/JVI.03422–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, et al. (2011). Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proceedings of the National Academy of Sciences of the United States of America 108, 14216–14221. 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, et al. (2018). MolProbity: More and better reference data for improved all-atom structure validation. Protein Science : A Publication of the Protein Society 27, 293–315. 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, et al. (2011). Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. The Journal of Experimental Medicine 208, 181–193. 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JL, and Davis MM (2000). Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity 13, 37–45. S1074–7613(00)00006–6. [DOI] [PubMed] [Google Scholar]
- Yang G, Holl TM, Liu Y, Li Y, Lu X, Nicely NI, Kepler TB, Alam SM, Liao HX, Cain DW, et al. (2013). Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. Journal of Experimental Medicinee 210, 241–256. 10.1084/jem.20121977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, Gallagher JR, Wang L, Zhang Y, Joyce MG, et al. (2015). Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nature medicine 21, 1065–1070. 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature Methods 14, 331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, et al. (2015). Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell 161, 1280–1292. 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Olee T, Le DT, Roubey RA, Hahn BH, Woods VL Jr., and Chen PP (1999). Characterization of IgG monoclonal anti-cardiolipin/anti-beta2GP1 antibodies from two patients with antiphospholipid syndrome reveals three species of antibodies. British Journal of Haematology 105, 102–109. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 – Cryo-EM data collection, processing, model refinement and validation – related to Figures 1, S2 and S3.
Table S3 – HC2 mouse and human germline on-target antibody affinities for the HA stalk – related to Figures 2 and 7.
Table S4 – HC2 mouse and human germline on-target stalk mAbs – related to Figures 2, 4, 7, S4, S5, S6 and S7.
Table S5 – Conversion of HC2 mouse and human L54 germline stalk mAbs into the F54 IGHV1-69 background – related to Figures 4 and 7.
Table S7 – HC2 mouse and human germline antibody affinities for the HA stalk after L54 to F54 conversion - related to Figure 7.
Table S6 – HC2 mouse and human stalk mAbs expanded after vaccination– related to Figures 6 and 7.
Data Availability Statement
3D maps and models from the EM analysis have been deposited to the Electron Microscopy Databank (http://www.emdatabank.org/) and the Protein Data Bank (https://www.rcsb.org/), respectively. The accession numbers are listed Table S1 and in the Key Resources Table.
Antibody sequences used in this publication can be found in Tables S4–S6 or within NCBI’s Gene Expression Omnibus and are accessible through GEO accession number listed in the Key Resource Table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| G6 Anti-Idiotypic mAb | Produced in house | N/A |
| CH65 | Produced in house | N/A |
| CR6261 | Produced in house | N/A |
| Sheep Anti-Mouse IgG-HRP | GE Healthcare | Cat#NA931 |
| Sheep Anti-Human IgG-HRP | GE Healthcare | Cat#NA933 |
| Hamster Anti-Mouse CD3e-PE/Cy7 (clone 145-2C11) | BioLegend | Cat#100320 |
| Rat Anti-Mouse CD4-Alexa Fluor 700 (clone RM4-5) | BD Biosciences | Cat#557956; RRID: AB_396956 |
| Rat Anti-Mouse CD19-BV421 (clone 6D5) | BioLegend | Cat#115549; RRID: AB_2563066 |
| Rat Anti-Mouse IgM-BV605 (clone RMM-1) | BioLegend | Cat#406523; RRID: AB_2563358 |
| Goat Anti-Mouse IgG-PerCP/Cy5.5 (clone Poly4053) | BioLegend | Cat#405314; RRID: AB_10662053 |
| Rat Anti-Mouse IgM-BV650 (clone II/41) | BD Biosciences | Cat#743326; RRID: AB_2741427 |
| Rat Anti-Mouse CD23-BV786 (clone B3B4) | BD Biosciences | Cat#563988; RRID: AB_2738526 |
| Rat Anti-Mouse CD93-BV421 (clone AA4.1) | BD Biosciences | Cat#747716; RRID: AB_2872195 |
| Rat Anti-Mouse IgD-PerCP-Cy5.5 (clone 11–26C.1) | BD Biosciences | Cat#564273; RRID: 2738722 |
| Rat Anti-Mouse CD21/CD35-PE-Cyanine7 (clone 8D9) | eBioscience | Cat#25-0211-80; RRID: AB_1518767 |
| Rat Ani-Mouse CD45R/B220-BUV395 (clone RA3-6B2) | BD Biosciences | Cat#563793; RRID: AB_2738427 |
| Hamster Anti-Mouse CD3e-BV510 (clone 145-2C11) | BD Biosciences | Cat#563024; RRID: AB_2737959 |
| Rat Anti-Mouse F4/80-BV510 (clone BM8) | BioLegend | Cat#123135; RRID: AB_2562622 |
| Rat Anti-Mouse Ly-6G and Ly-6C-BV510 (clone RB6-8C5) | BD Biosciences | Cat#563040; RRID: AB_2722496 |
| Rat Anti-Mouse CD19-APC-eFlour780 (clone 1D3) | eBioscience | Cat#47-0193-82; RRID: AB_10853189 |
| Hamster Anti-Mouse CD79b-BV605 (clone HM79b) | BD Biosciences | Cat#740383; RRID: AB_2740113 |
| Rat Anti-Mouse Ig k Light chain-PE (clone 187.1) | BD Biosciences | Cat#559940; RRID: AB_397384 |
| Rat Anti-Mouse Ig, λ1, λ2, & λ3 Light chain-FITC (clone R26-46) | BD Biosciences | Cat#553434; RRID: AB_394854 |
| Rat Anti-Mouse CD38-Alexa Flour 594 (clone 90) | BioLegend | Cat#102725; RRID: AB_2566435 |
| Rat Anti-Mouse/human GL7-Alexa Flour 488 (clone GL7) | BioLegend | Cat#144612; RRID: AB_2563285 |
| Rat Anti-Mouse IgD-BUV395 (clone 11-26C.1) | BD Biosciences | Cat#564274; RRID: AB_2738723 |
| Hamster Anti-Mouse CD95-PE-Cyanine7 (clone Jo2) | BD Biosciences | Cat#557653; RRID: AB_396768 |
| Rat Anti-Mouse Ly-6G/Ly-6C-APC-eFlour780 (clone RB6–8C5) | eBiosciecne | Cat#47-5931-82; RRID: AB_1518804 |
| Rat Anti-Mouse CD4-APC-eFlour780 (clone RM4-5) | eBioscience | Cat#47-0042-82; RRID: AB_1272183 |
| Rat Anti-Mouse CD8a-APC-eFlour780 (clone 53–6.7) | eBioscience | Cat#47-0081-82; RRID: AB_1272185 |
| Rat Anti-Mouse/Human CD45R/B220-BV605 (clone RA3–6B2) | BioLegend | Cat#103243; RRID: AB_11203907 |
| Rat Anti-Mouse CD19-PerCP/Cyanine5.5 (clone 1D3) | BioLegend | Cat#152405; RRID: AB_2629814 |
| Rat Anti-Mouse F4/80-APC-eFlour780 (clone BM8) | eBioscience | Cat#47-4801-82; RRID: AB_2735036 |
| Goat Anti-Mouse IgG-BV421 (clone Poly4053) | BioLegend | Cat#405317; RRID: AB_10900419 |
| Rat Anti-Mouse IgD-Alexa Fluor 700 (clone 11–26c.2a) | BioLegend | Cat#405730; RRID: AB_2563341 |
| AffiniPure F(ab’)2 Fragment Goat Anti-Mouse IgM | Jackson ImmunoResearch | Cat#115-006-020; RRID: AB_2338469 |
| inVivoMAb Anti-mouse CTLA-4 (clone 9H10) | BioXCell | Cat#BE0131 |
| inVivoMAb Polyclonal Syrian Hamster | BioXCell | Cat#BE0087 |
| Mouse Anti-Human CD19-Alexa Fluor 700 (clone HIB19) | BioLegend | Cat#302226; RRID: AB_493751 |
| Mouse Anti-Human IgG-Brilliant Violet 421 (clone G18–145) | BD Biosciences | Cat# 562581; RRID: AB_2737665 |
| Mouse Anti-Human IgD-PE/Cy7 (clone IA6–2) | BD Biosciences | Cat#561314; RRID: AB_10642457 |
| Mouse Anti-Human CD3-APC-Cy7 (clone SK7) | BD Biosciences | Cat#557832; RRID: AB_396890 |
| Mouse Anti-Human IgM-PerCP/Cyanine5.5 (clone MHM-88) | BioLegend | Cat#314512; RRID: AB_2076098 |
| HC2 and Human Germline on-Target F54 and L54 Antibodies, see Table S4 | This Paper | N/A |
| HC2 and Human L54F/R49G/I56T Conversion Antibodies, see Table S5 | This Paper | N/A |
| HC2 and Human Vaccine Expanded Stalk Antibodies, see Table S6 | This Paper | N/A |
| Bacterial and virus strains | ||
| A/chicken/Vietnam/NCVD-016/2008(H5N1)-PR8-IDCDC-RG12 | International Reagent Resource | FR-725 |
| Influenza reporter viruses | Masaru Kanekiyo, NIH (Creanga et al., 2021) | N/A |
| Biological samples | ||
| PBMC Samples from Human Donors | Massachusetts General Hospital | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| H1 HA Trimer (NC99) | Produced in house (Whittle et al., 2014) | N/A |
| H1 Y98F HA Trimer (NC99) | Produced in house (Whittle et al., 2014) | N/A |
| H5 Y98F HA Trimer (Indo05) | Produced in house (Whittle et al., 2014) | N/A |
| H1 HA-np (NC99) | Produced in house (Kanekiyo et al., 2013) | N/A |
| SS-np (NC99) | Produced in house (Yassine et al., 2015) | N/A |
| SS-npΔstem (NC99) | Produced in house (Sangesland et al., 2019) | N/A |
| H. Pylori Ferritin | Aaron Schmidt, Ragon Institute of MGH, MIT, and Harvard | N/A |
| H1 RBD-np (NC99) | Masaru Kanekiyo, NIH (Kanekiyo et al., 2019) | N/A |
| Streptavidin-allophycocyanin (APC) Conjugate | Life Technologies | Cat#S32362 |
| Streptavidin-phycoerythrin (PE) Conjugate | Life Technologies | Cat#S21388 |
| Fura Red, AM, Cell Permeant | Thermo Fisher | Cat#F3021 |
| Ionomycin | Thermo Fisher | Cat#I24222 |
| Sigma Adjuvant System | Sigma-Aldrich | Cat#S6322 |
| Recombinant Human Insulin Protein | Fitzgerald | Cat# 30-Al51 |
| UltraPure Calf Thymus DNA solution | Invitrogen | Cat#15633019 |
| Sphingomyelin | Avanti Polar Lipids | Cat#860062 |
| Cardiolipin | Sigma-Aldrich | Cat#C1649 |
| 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) | Avanti Polar Lipids | Cat#850457P |
| Lipopolysaccharide from Escherichia coli O55:B5 | Sigma-Aldrich | Cat#L2880 |
| 293fectin Reagent | Invitrogen | Cat#12347019 |
| Ni-Sepharose excel Affinity Medium | GE Healthcare | Cat#GE17-3712-02 |
| Erythrina Cristagalli (ECA) Immobilized Lectin | EY Laboratories | Cat#A-5901-2 |
| Protein G Sepharose | GE Healthcare | Cat#17061802 |
| IgG Elution Buffer | Pierce | Cat#21009 |
| LIVE/DEAD Fixable Aqua Dead Cell Stain | Thermo Fisher | Cat#L34957 |
| LIVE/DEAD Fixable Blue Dead Cell Stain | Thermo Fisher | Cat#L34961 |
| Critical commercial assays | ||
| NOVA Lite Hep-2 IgG kit | Werfen | Cat#708100 |
| BirA Biotin-Protein Ligase Bulk Reaction Kit | Avidity | Cat#Bulk BirA |
| Alexa Fluor 647 Protein Labeling Kit | Thermo Fisher | Cat#A20173 |
| Alexa Fluor 488 Protein Labeling Kit | Thermo Fisher | Cat#A10235 |
| Alexa Fluor 594 Protein Labeling Kit | Thermo Fisher | Cat#A10239 |
| Alexa Fluor 546 Protein Labeling Kit | Thermo Fisher | Cat#A20183 |
| MiSeq Reagent Kit, V2 500 cycles | Illumina | Cat#MS-102-2003 |
| Deposited data | ||
| IGHV1-69 HC2 CDRH3 sequences | This Paper | GEO: GSE207054 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE207054 |
| CryoEM structures | This Paper | PBD IDs: 7SCO, 7SCN |
| Experimental models: Cell lines | ||
| Human: FreeStyle 293F | Thermo Fisher | Cat#R79007; RRID: CVCL_D603 |
| Human: Expi293F | Thermo Fisher | Cat#A14527; RRID: CVCL_D615 |
| Canine: MDCK | ATCC | Cat#CCL-34; RRID: CVCL_0422 |
| Experimental models: Organisms/strains | ||
| Mouse: IGHV1-69*01+/+ IgH Restricted Mouse Model with Diverse Human CDRH3 (C57Bl/6 Strain) | Sangesland et al., 2019 | N/A |
| Mouse: IGHV1-2*02+/+ IgH Restricted Mouse Model with Diverse Human CDRH3 (C57Bl/6 Strain) | Sangesland et al., 2019 | N/A |
| Mouse: IGHV1-69*09+/+ IgH Restricted Mouse Model with Diverse Human CDRH3 (C57Bl/6 Strain) | This Paper | N/A |
| Mouse: IGHV1-69*01+/−/IGHV1-69*09+/− IgH (C57Bl/6 Strain) | This Paper | N/A |
| Oligonucleotides | ||
| Primers for Single Cell and Bulk BCR Amplification | Sangesland et al., 2019 | N/A |
| Recombinant DNA | ||
| Software and algorithms | ||
| Flowjo v10.6.2 | TreeStar | https://www.flowjo.com; RRID: SCR_008520 |
| Prism v.8.4.3 | GraphPad | https://www.graphpad.com; RRID: SCR_002798 |
| Other | ||
| Superdex 200 10/300 Column | GE Healthcare | Cat#17517501 |
| Superose 6 10/300 Column | GE Healthcare | Cat#17517201 |
| SPF Embryonated Chicken Eggs | Charles River Laboratories | Cat#10100335 |


