Abstract
Recent advances in interrogating RNA folding dynamics have shown the classical model of RNA folding to be incomplete. Here, we pose three prominent questions for the field that are at the forefront of our understanding of the importance of RNA folding dynamics for RNA function. The first centers on the most appropriate biophysical framework to describe changes to the RNA folding energy landscape that a growing RNA chain encounters during transcriptional elongation. The second focuses on the potential ubiquity of strand displacement – a process by which RNA can rapidly change conformations – and how this process may be generally present in broad classes of seemingly different RNAs. The third raises questions about the potential importance and roles of cellular protein factors in RNA conformational switching. Answers to these questions will greatly improve our fundamental knowledge of RNA folding and function, drive biotechnological advances that utilize engineered RNAs, and potentially point to new areas of biology yet to be discovered.
Keywords: energy landscapes, conformational switching, strand displacement, cotranscriptional RNA folding, protein-mediated RNA folding
Introduction
RNA function has long been known to be deeply intertwined with RNA structure [1]. Recently, progress in high resolution static structural determination from x-ray crystallography [2], NMR [3], and cryo-EM [4,5] have given us deep insights into the intricacies of RNA structure beyond simple base pairing. These studies have revealed complex and diverse RNA folding interactions that give rise to the atomic architectures of ligand-binding pockets [6,7], protein docking sites [8,9], and catalytic active sites [10,11] that are at the center of RNA functions ranging from gene expression control [12,13], RNA processing [14–16] and protein translation [17,18].
However, a new appreciation for the dynamics of RNA folding suggests that a static viewpoint of RNA structures is only the tip of the iceberg [19]. Whether from RNA folding transitions during transcription [20], or RNA switching conformations once synthesized [21], new studies of RNA folding dynamics are proving critical for understanding known RNA function [22,23] and for drug discovery [24]. Well-known examples of these important features are riboswitch RNAs that change structural conformations in response to ligand binding to regulate various aspects of gene expression [25], attenuator RNAs such as the TRAP system that similarly conformationally switch in response to protein binding to regulate transcription [26,27], and many other RNA regulatory mechanisms. Beyond explicit regulation there are also new appreciations for the role of RNA conformational switching in the activation of the spliceosome [28], the assembly of the ribosome [29,30], and as we later speculate for heterogeneous nuclear RNA proteins to guide dynamic RNA folding pathways in the cell. Even relatively simple RNA hairpin structures, such as the one found in the signal recognition particle (SRP) RNA, have been shown to undergo conformational switching during transcription to establish their functional folds [31,32] (Figure 1). These are but a few of a growing list of examples revealing that dynamic RNA folding is important for broad arrays of RNA functions.
Figure 1. Transcription elongation leads to shifting free energy landscapes traversed by dynamically changing RNA structures.
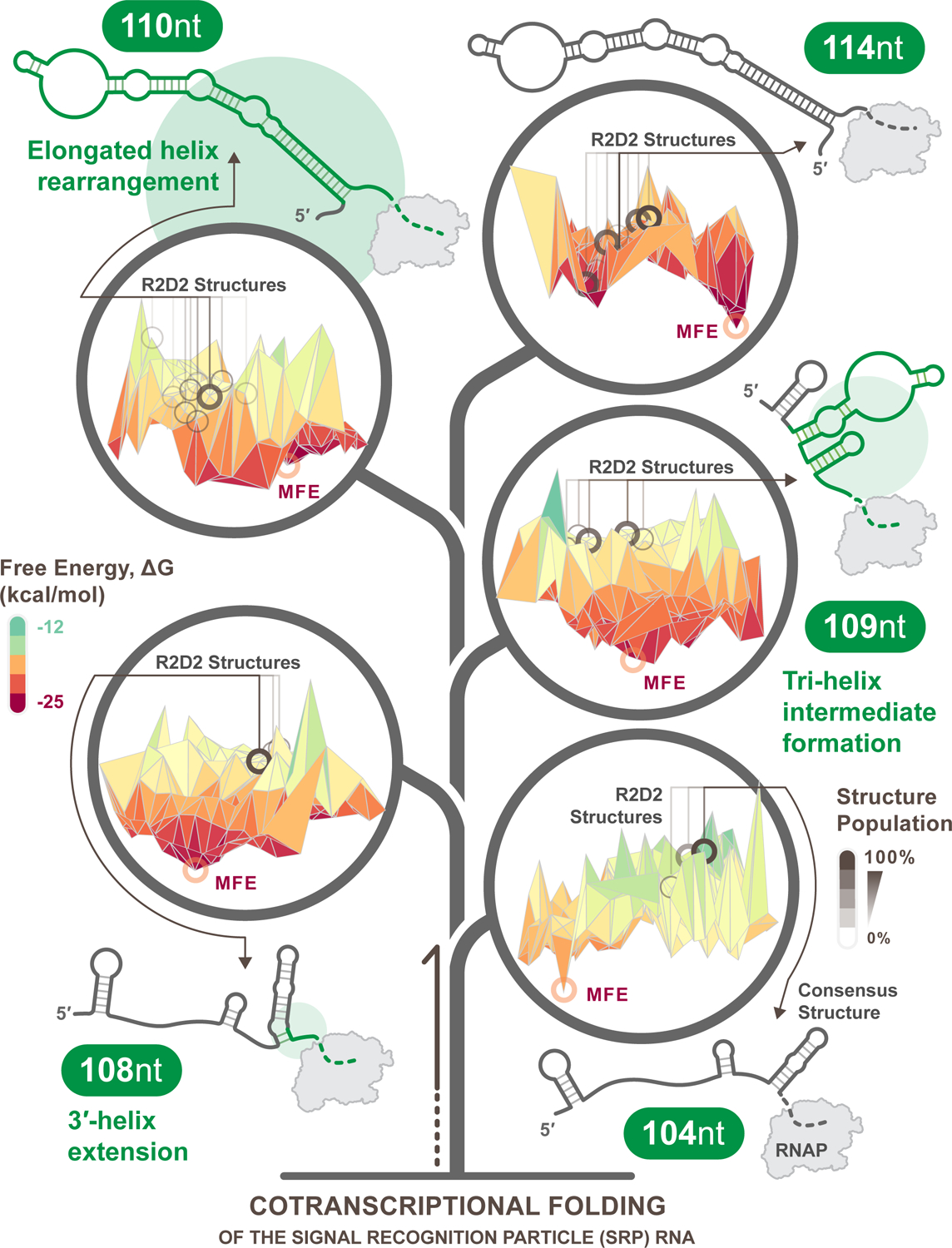
Free energy landscapes were estimated from experimentally informed structural reconstructions of the E. coli SRP cotranscriptional folding pathway using Reconstructing RNA Dynamics from Data (R2D2) [32]. R2D2 samples possible nascent RNA structures at each intermediate length and selects structures that are most consistent with experimental cotranscriptional RNA structure chemical probing data from each length. To model the possible free energy landscape of folding at each intermediate length, multidimensional scaling (MDS) was performed on the sampled structures at each length and plotted against predicted free energies. For visual clarity, the resulting plots were smoothed by calculating the mean free energy of all structures within a 10×10 coordinate area of the MDS plot. Each unique R2D2-selected structure is marked with a gray circle on the landscapes, where the shading corresponds to how frequently a structure is selected. These structures are notably far from the minimum free-energy structure (red circles) suggesting that the cotranscriptional folding process traverses out-of-equilibrium folding pathways on shifting free energy landscapes as nucleotides are added to the growing RNA chain. The illustrated secondary structures are consensus structures over 100 folding pathway predictions at each length by R2D2, with green highlighting and circles demonstrating the degree of change between previous structures. The addition of every nucleotide in the transcription elongation presents an opportunity for significant conformational switching as the entire energy landscape reshapes.
While dynamic RNA structure and folding has long been appreciated, advances in biophysical techniques are fueling new discoveries through probing RNA structural dynamics [33–35] and detecting conformational changes tied to function. These studies raise new questions about how to conceptualize RNA folding and how to identify new mechanisms for which RNA dynamics are a core component.
Here we pose three prominent questions for the field that we believe are at the forefront of our knowledge of the importance of RNA folding dynamics and the role of conformational switching in RNA function. The first centers on the most appropriate biophysical framework to describe the complexities that are introduced when considering non-equilibrium conformational switching, especially in the context of a growing RNA chain during transcriptional elongation. The second is about the potential ubiquity of strand displacement – a process by which RNA can rapidly change conformations – and how this process may be present in broad classes of seemingly different RNAs. The third raises questions about the potential importance and roles of cellular protein factors in RNA conformational switching. Answers to these questions will greatly improve our fundamental knowledge of RNA folding and function, drive biotechnological advances that utilize engineered RNAs, and potentially point to new areas of biology yet to be discovered.
What is the appropriate biophysical framework to describe non-equilibrium conformational switching?
A biophysical framework for understanding “How RNA Folds” according to the thermodynamics of base pairing has long been established [1]. In thermodynamic renaturation conditions, RNA is understood to fold hierarchically, with secondary structures stabilizing first, creating an architecture to then establish tertiary interactions. Such a folding regime is well described by a free energy landscape, a multi-dimensional surface that represents the free energies of all possible folds of a given RNA sequence [36] (Figure 1). While powerful, this framework does not capture essential details of cellular RNA folding such as transient structures that can form during cellular RNA synthesis [35].
RNA folding begins during transcription, where due to the kinetics of RNA base pairing, nascent chains fold before the molecule is completely synthesized [37,38]. While these folds may undergo processing and other cellular interactions, recent studies suggest that for a large fraction of RNAs, these initial folds persist throughout the RNA molecule’s lifetime in the cell [39]. Moreover, nascent RNA structures are important for a wide array of cellular processes, such as the regulation of transcription, translation, and mRNA degradation in prokaryotes [40–42], and splicing, polyadenylation, and 3’ end processing in eukaryotes [43–46]. Thus, understanding how RNAs fold during transcription is essential for understanding fundamental RNA biology.
Cellular RNA folding is different than the prevailing biophysical paradigm in two important aspects: i) RNAs do not fold all at once from a denatured full-length state, making the renaturation folding model incomplete, and ii) the sequence of the RNA changes during transcription, making the concept of a fixed free energy landscape of an RNA sequence incomplete. Overall, this raises a deep conceptual question: What is the appropriate biophysical framework to describe non-equilibrium nascent RNA folding?
Progress on this question has come primarily from computational studies of kinetic RNA folding processes. Computational modeling has chiefly considered kinetic RNA folding to be a stochastic process, where the RNA transitions to different folding states with different probabilities. These probabilities are governed by transition rates that arise from local energy barriers between nearby possible RNA structures [47,48]. For example, Kinefold simulates the folding of a growing RNA chain by computationally searching for nearby possible structures, and stochastically making or breaking base pairs based on estimated rates to these nearby structures [49]. The Kinfold algorithm uses a similar framework, but chooses between the formation, breaking, or shifting of individual base pairs [50]. However, while these algorithms have great practical utility to simulate an RNA folding trajectory quickly and easily, they do not reveal a deeper understanding of general cotranscriptional RNA folding landscapes. Specifically, the stochastic approach samples one possible folding trajectory at a time out of a myriad of possibilities, making it intractable to gain a global view of possible folding trajectories of a given RNA molecule (Figure 1).
Recently, experimental techniques are also beginning to provide insights into non-equilibrium nascent RNA folding. Single molecule techniques, such as single molecule optical force spectroscopy, are able to measure structural transitions of a growing RNA chain at millisecond time resolution, revealing the switching mechanisms of riboswitches [51] and structural rearrangements of the signal recognition particle (SRP) RNA during transcription [31]. New methods utilizing Förster resonance energy transfer are being used to label nascent RNAs to understand folding properties during key moments in the folding pathway [20,52]. And whole new approaches are using engineered superhelicases to simulate transcription elongation and characterize kinetically controlled decision landscapes [34,53]. Atomic structure determination techniques have also been applied to study transient structures and structural transitions [5,54]. For example, the coupling of NMR with photo-cage labeling of RNA strands has allowed researchers to trigger the conformational switching of an adenine riboswitch by uncaging part of the expression platform and using NMR to watch it invade the aptamer [54]. Recently, single particle cryo-EM has been used to reveal local conformational dynamics in an exterior helix of the Tetrahymena ribozyme, and rotational flexibility of the R-loop in the target-bound CRISPR Cas9 complex [5,55]. And high throughput chemical probing approaches have been developed that can probe nascent RNA structures in transcription elongation complexes [35], that when combined with new modeling algorithms can reconstruct possible cotranscriptional folding pathways from this experimental data [32]. These approaches are revealing details of riboswitch mechanisms, such as strand invasion pathways that allow expression platform folding in the absence of ligand but are blocked by aptamer-ligand interactions [35,56,57], and rearrangement pathways of the SRP RNA molecule that were corroborated by single molecule techniques [32]. Collectively, these methods have pushed the field forward in experimentally understanding the physical parameters of cotranscriptional folding pathways and are permitting researchers to better hypothesize how RNA undergoes dynamic conformational switches during transcription.
As an example of how these techniques can reveal new insights into co-transcriptional RNA folding, we combined experimental cotranscriptional structure probing of the SRP RNA folding pathway [35] with computational structure prediction algorithms [32] to reconstruct possible free energy landscapes that the SRP RNA may traverse during transcription (Figure 1). In this reconstruction, we observe that the elongating transcript traverses vastly changing landscapes, where every transcribed nucleotide transforms the conformational ensemble space. At shorter RNA lengths the SRP landscape is shallow, with few minima creating many possible folds with similar free energies. However, as the RNA chain grows longer, deeper minima emerge in the free energy landscape, reflecting the resolution of folding possibilities into fewer states. In between these two regimes the SRP RNA dramatically rearranges its fold from a tri-helix intermediate at 109 nt to an elongated helix at 110 nt, though it is not clear from the energy landscape reconstructions alone how this may happen. Here a deeper understanding of the mechanisms by which RNAs may interconvert between states, and the measurements of the kinetics of these processes, may be important for understanding how RNAs can traverse rugged and changing free energy landscapes.
As a field, we appear poised to significantly elaborate our biophysical framework of RNA folding by incorporating the physics of cotranscriptional RNA folding as it traverses free energy landscapes. In this view, the RNA fold does not move along a fixed free energy landscape, but instead navigates a shifting landscape that transforms with each transcribed nucleotide [19] (Figure 1). Each new nucleotide could reveal new peaks and troughs in the landscape, allowing dynamical switching between conformations that on a fixed landscape may appear to be separated by insurmountable free energy barriers. Deeper research from a range of approaches is needed to confirm and refine this model. Applying this framework to a diverse set of RNA sequences may help us understand general principles of rapid dynamical conformational switching at the heart of many RNA functions.
How widespread is strand displacement as a general feature of RNA conformational switching?
Large RNA conformational changes can present significant free energy barriers to structural rearrangement, requiring milliseconds to hours to occur – a time window that may limit cellular functions [19]. An intriguing solution to this challenge is ‘strand displacement’, a process by which RNA structures can efficiently rearrange through small nucleotide fluctuations that facilitate base pair exchange [58]. ‘Strand displacement’ or ‘strand exchange’ refers to a process whereby a double-stranded region comprising a substrate strand and incumbent strand is disrupted by a third invader strand, resulting in the formation of a new invader-substrate duplex (Figure 2a) [58]. While strand displacement can occur with three separate nucleic acid strands, in the context of RNA conformational switching, the incumbent, substrate, and invader strands may all be regions of the same RNA molecule.
Figure 2. Strand displacement is a widely observed mechanism of RNA conformational switching.
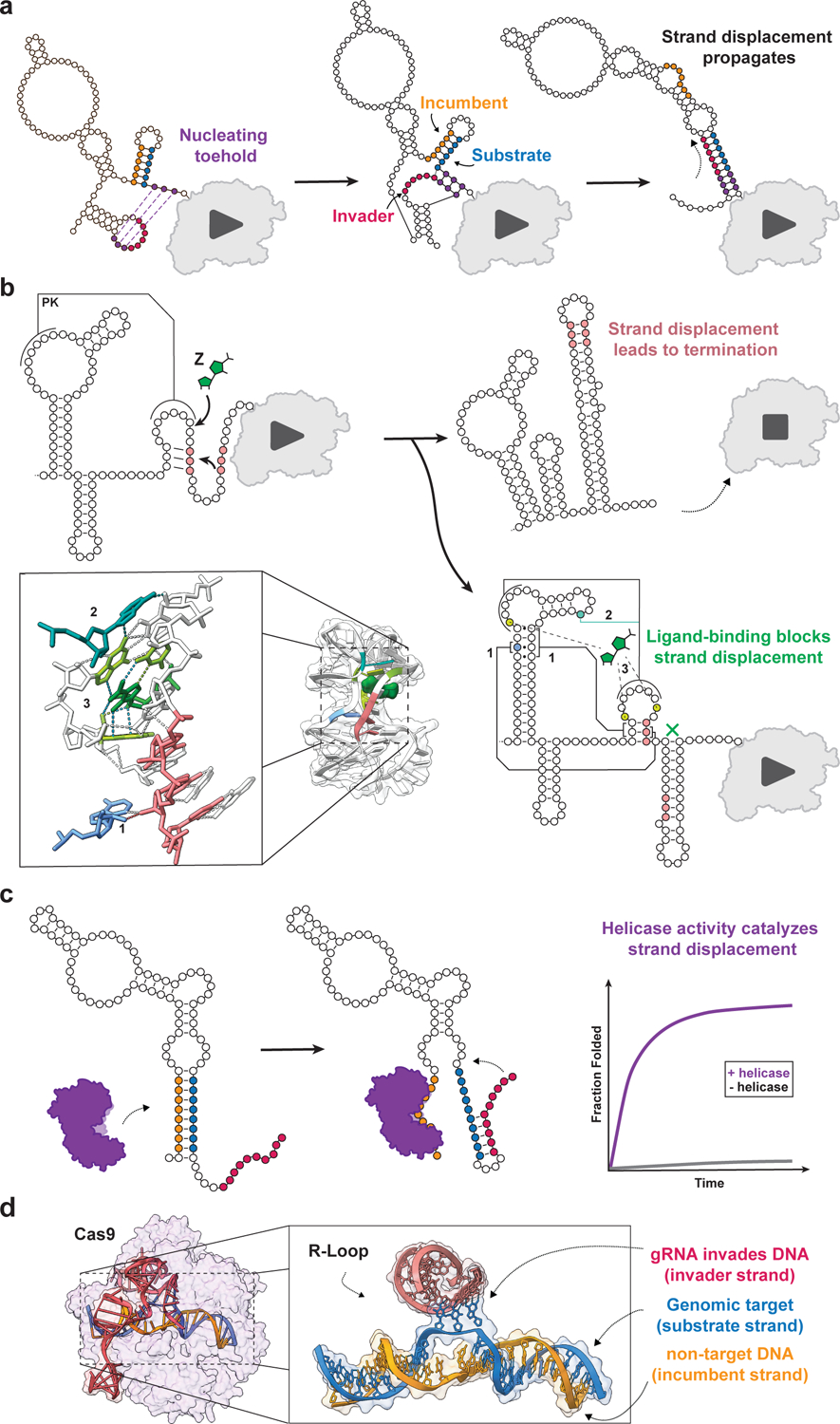
Illustrations of several well-studied strand displacement processes. (A) Cotranscriptional strand displacement for structural rearrangement during transcription of the E. coli SRP RNA. After transcription of 110 nucleotides, three nucleating base pairs can form between the 3’ end and the loop of a 5’ hairpin structure (purple). This interaction seeds a strand displacement reaction in which an invading strand (red) outcompetes an incumbent strand (orange) for binding to a substrate strand (blue), causing a global conformational change that remodels the nascent RNA from a triple helix structure to a single elongated helix [32]. (B) Strand displacement in RNA genetic switches. Secondary structure diagram of the cotranscriptional folding pathway of the Clostridium beijerinckii pfl ZTP/ZMP riboswitch. In the absence of ligand, a strand displacement event seeds the formation of a strong intrinsic terminator. In the presence of ligand, numerous tertiary structural interactions, including: (1) ribose zippers, (2) an A-minor triple, and (3) hydrogen bonds between ligand and the binding pocket stabilize the holo-aptamer structure such that strand displacement is unable to efficiently proceed. This results in the formation of an alternative anti-terminating structure [57,101] (PDB ID: 4ZNP). (C) Protein-catalyzed strand displacement. The Mss116p non-processive RNA helicase has been shown to catalyze strand displacement by capturing the incumbent strand (orange), freeing the substrate strand (blue) to anneal to the invading strand (red) [68,102] (PDB ID: 3I5X). (D) CRISPR gene editing. Cryo-EM structure model of target-bound Cas9 illustrates the molecular basis of strand displacement in Cas9-gRNA-DNA recognition [55] (PDB ID: 7S38). The gRNA (red) forms a heteroduplex with the DNA target sequence substrate (blue) by invading the dsDNA, forcing the incumbent non-target strand (orange) to dissociate.
Strand displacement was first identified by LeCuyer and Crothers, who proposed that this mechanism would allow mutually exclusive helices with similar free energy to quickly interconvert in a ‘break-one-form-one’ fashion comparable to branch migration [59,60]. Since then, strand displacement has been observed in two major areas of RNA function: i) dynamically during cotranscriptional folding [32,35,56,57], and ii) within some post-transcriptional rearrangements and cellular factor binding events [61,62] (Figure 2).
Since cotranscriptional folding favors local structures that can form immediately after synthesis, there is a potential for kinetic traps that consist of metastable RNA structures that must be rearranged into more thermodynamically stable or functional folds. In cases of conformational switching during the timescale of transcription, RNAs appear to utilize strand displacement mechanisms. This is especially true for transcriptional riboswitches, cis-acting non-coding RNAs that regulate transcription elongation through ligand-mediated conformational switching [25]. Several studies of transcriptional riboswitches reveal that, in these cases, the genetic decision is the result of a competition between two mutually exclusive structures that exchange via a strand displacement mechanism (Figure 2b). Ligand binding can then bias the folding pathway by stabilizing certain structures that prevent strand displacement by a downstream intrinsic terminator structure, allowing ligand binding to directly lead to the enactment of a genetic regulatory decision (Figure 2b) [35,57]. Furthermore, for some transcriptional riboswitches, the presence of an additional competing strand can flip this logic, with strand displacement leading to the retention of a strand that competes with the terminator structure [56].
Moreover, cotranscriptional strand displacement can be guided by higher order transcriptional dynamics. For example, while the E. coli SRP RNA must form a long hairpin structure for proper function, it was shown that efficient folding of this structure utilizes RNA polymerase (RNAP) pausing to promote the formation of a labile 5′ hairpin structure that then rearranges to form the extended functional fold [63]. Recent studies strongly suggest that this rearrangement proceeds via a strand displacement pathway [31,32] (Figure 2a).
In addition to mediating structural rearrangement during transcription, strand displacement could play a role in many post-transcriptional cellular contexts where RNA conformational switching is functionally important. For example, structure-probing studies showed that the NEAT1 lncRNA has two functionally distinct folds – one with many short helices that has unknown biological function, and another with a long-range helix that facilitates paraspeckle formation to regulate gene expression [64]. Here, strand displacement could be an advantageous mechanism of conformational switching, Such a mechanism may offer a much faster route to rearranging RNA structures that occur on rugged free energy landscapes (Figure 1) – rather than having to melt local structures within two complementary regions prior to re-folding, strand displacement could allow local structural interconversions that would each have relatively smaller energy barriers to traverse [61]. In fact, DNA nanotechnology studies have found that strand displacement kinetics can occur on the microseconds timescale [65], similar to the timescales of local nucleotide fluctuations [66]. Moreover, because strand displacement can rely on only a few nucleating base pairs for initiation (Figure 2a), conformational switching could be tightly regulated by controlling the formation of these base pairs [58]. For example, large-scale conformational changes that proceed via strand displacement can be catalyzed by nonprocessive RNA helicases like DEAD-box RNA helicases [67,68] (Figure 2c). Thus, strand displacement may underlie the regulation of some RNAs with post-transcriptional function.
Strand displacement also appears to play a role in biological mechanisms that utilize RNA for sequence recognition such as CRISPR-Cas9. CRISPR-Cas9 is a bipartite system composed of the Cas9 endonuclease and a guide RNA (gRNA) which targets the complex to complementary genomic loci. After the Cas9-gRNA complex binds tightly to a protospacer-adjacent motif, the single-stranded seed region of the gRNA is positioned to invade the target genomic site, opening up the dsDNA duplex and forming a RNA:DNA heteroduplex via strand displacement [55,62,69] (Figure 2d). In this way, Cas9 facilitates strand displacement, which can typically only disrupt double-stranded structures if there are transient fluctuations in base pairing at the ends. Structural studies of other Cas enzymes have shown similar dynamics [70]. While different from Cas9, argonaute proteins also pre-organize the 5′ end of their miRNA (seed region), allowing increased discrimination between sites with and without a single mismatch in this region at rates much higher than would be expected from simple nearest-neighbor thermodynamic rules [71]. Thus, while strand displacement was first identified for RNA helical switching, it appears to have also been inherited in RNA-protein binding and recognition mechanisms.
Strand displacement allows RNAs to efficiently traverse large energetic barriers to enact conformational switching [61]. The process can be tightly regulated by controlling the spatial proximity of the three participating strands, and by controlling the formation of nucleating base pairs. This raises the intriguing question of how broadly-distributed is strand displacement as a mechanism for RNA conformational switching? An evolutionary study of bacterial P RNA, SRP RNA, the trp operator leader, hepatitis delta virus ribozyme, Levivirus maturation gene, and a class of S-adenosylmethionine riboswitches, found evidence that the cotranscriptional folding pathways of these RNAs likely feature evolutionarily conserved transient metastable structures that guide the RNA to adopt its final fold, potentially by helping the RNA avoid kinetic traps [72]. Further bioinformatic analysis could be used to investigate other RNA systems that have sequences that could facilitate strand displacement, with experimental techniques used to corroborate these possibilities. As techniques develop to understand how RNA dynamically switches conformations in different contexts, we predict strand displacement will be shown to be a general-purpose strategy for guiding RNA structural transitions throughout biology.
How do proteins mediate cotranscriptional RNA conformational switching?
The biophysics of cotranscriptional folding creates a key challenge for cellular RNAs – the propensity of nascent RNAs to fold into non-functional kinetically trapped states at local minima in a rugged and changing free energy landscape (Figure 1). However, cellular RNA folding does not occur within a vacuum, but in a complex milieu filled with other cellular components such as proteins that can bind to RNA, influencing its folding and function [73]. For example, RNA folding chaperones can prevent RNAs from folding into non-functional states by mitigating kinetic folding traps [74] (Figure 3a). This raises the fundamental biological question: How do proteins mediate dynamic RNA folding through RNA conformational switching?
Figure 3. Protein-RNA interactions guide cotranscriptional folding, influence transcription dynamics, and determine gene expression outcomes.
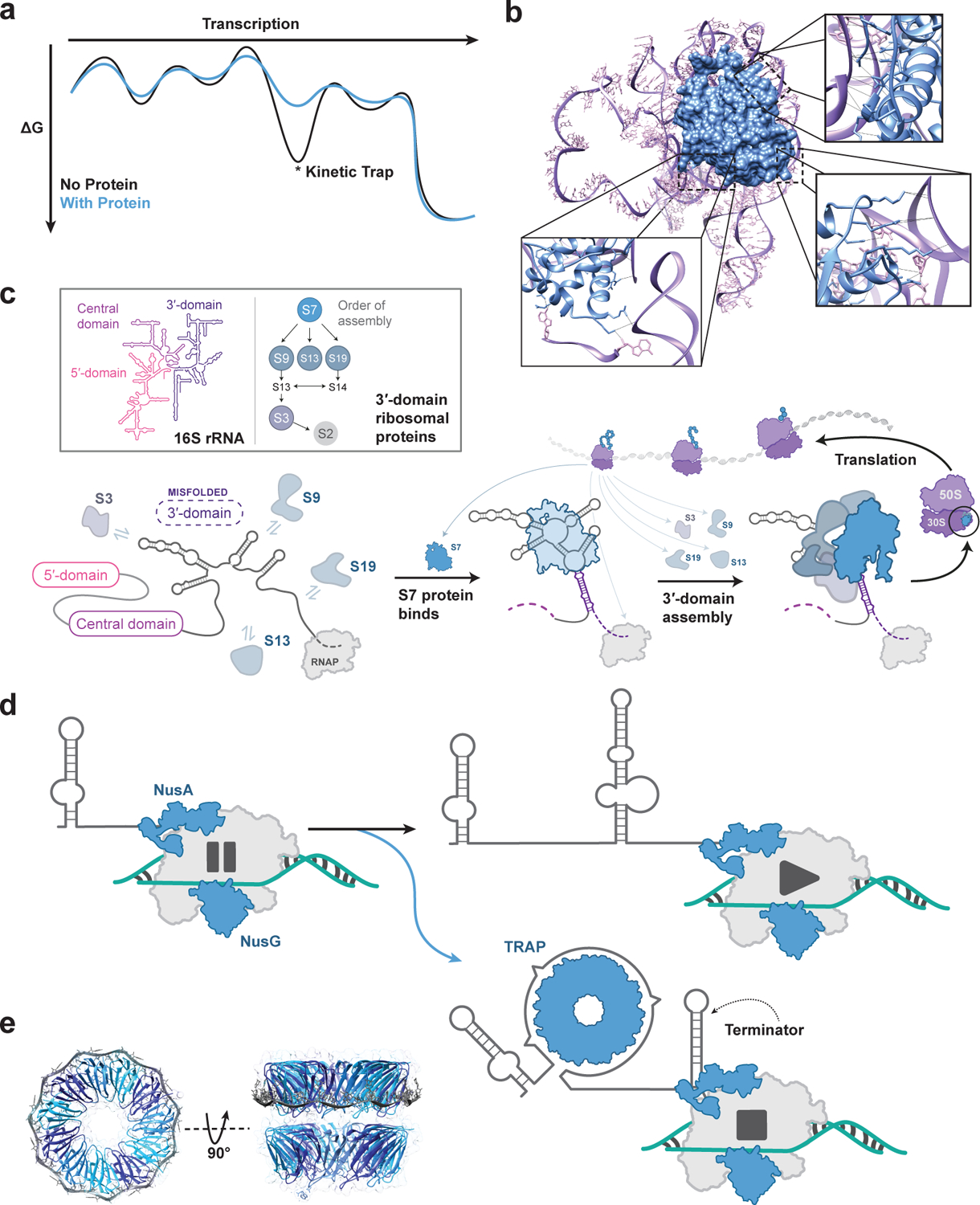
(A) Cartoon representation of how a protein can alter an RNA folding free energy landscape, allowing alternate folding pathways to, for example, avoid kinetic traps. (B) Three-dimensional structure of the E. coli ribosomal protein S7 binding to 16S rRNA [79] (PDB ID: 4V9P). Hydrogen bonding interactions with the rRNA are shown in the insets. (C) Cartoon of ribosomal protein S7 binding to nascent 16S rRNA during transcription as a portion of ribosome biogenesis leading to a fully functional ribosome. The ribosome can then perform its function of protein translation. In some cases, such as for S7 depicted, the translated protein can bind to nascent RNA during transcription, participating in hierarchical ribosome assembly pathways. (D) Cartoon example of transcription attenuation through the 11-mer trp RNA-binding attenuation protein (TRAP) mechanism. As the 5′ UTR of the trp attenuator is transcribed, RNA polymerase reaches a NusA-NusG mediated pause site. In excess tryptophan conditions, tryptophan cooperatively binds between adjacent subunits of the 11-mer TRAP complex, which can bind the 5′ UTR allowing an intrinsic terminator to form. In limiting tryptophan conditions, TRAP does not form nor bind the 5′ UTR. Instead, an anti-terminating stem forms, allowing the RNA polymerase to continue transcribing the downstream operon [87,88]. (E) Three-dimensional structure of the Geobacillus stearothermophilus Trp RNA-Binding Attenuation Protein bound to single-stranded RNA (PDB ID: 1C9S).
A confluence of biophysical and biochemical methods are now revealing the details of how cellular factors associate with transient intermediate structures of nascent RNAs to help the RNA navigate its complex conformational landscape towards its functional form [75]. Several aspects of biological processes appear to be particularly influenced by RNA-protein interactions: the assembly of ribonucleoprotein complexes (RNPs) (Figure 3b,c), the feedback between nascent RNAs and the elongation complex (Figure 3d), eukaryotic RNA processing and gene expression, and RNA-driven formation of nuclear compartments.
RNPs can assemble cotranscriptionally from their molecular components and involve many RNA-protein interactions, including those that influence the fold of the RNA molecule as part of the assembly process [76] (Figure 3c). Bacterial ribosomal biogenesis is among the most well-studied systems exemplifying the importance of timely incorporation of cellular factors during RNP assembly. Ribosomal assembly begins during transcription, and is intimately coupled with processing and maturation of the pre-rRNA [77,78]. The hierarchical addition of ribosomal proteins (RPs) to ribosomal subunits can lead to structural changes in the pre-rRNA, with each newly incorporated RP stabilizing the folded structure of its immediate RNA binding site, in turn stimulating structural changes in adjacent pre-rRNA residues that help recruit other proteins to the complex [29,30]. For example, the nascent 16S pre-rRNA 3’ domain initially misfolds, but the binding of the S7 RP structurally remodels 16S to enable subsequent hierarchical assembly of the 30S ribosomal subunit [79] (Figure 3b, c). Overall, ribosome biogenesis is illustrative of how proteins can smooth the RNA folding landscape by preferentially stabilizing productive folding intermediates [80].
Another key area where RNAs and proteins intimately interact is within the transcription elongation complex itself. Transcriptional dynamics arising from the two-way interplay between nascent RNA folding and the transcription elongation complex have been shown to play important roles in the temporal coupling of RNA synthesis and conformational switching [54,81]. Furthermore, RNAP pausing has been shown to influence RNA folding pathways, either by giving time for the RNA to rearrange into more thermodynamically stable structures or by selecting for certain metastable structures that prevent downstream sequence from interfering with labile folds [63]. Elongation factors that affect the rate of transcription or bind to nascent RNA structures themselves can also play roles in guiding nascent RNA through its free energy landscape. For example, the elongation factor NusA forms transient contacts with the elongating RNA chain, allowing this protein to modulate transcriptional dynamics in response to the formation of certain RNA structures and vice versa [63,82–85]. This points to broader roles for elongation factors such as NusA, NusG and others in facilitating cotranscriptional RNA folding (Figure 3d).
Principles from RNP assembly and protein-mediated modulation of transcriptional dynamics can both be seen in the intricate trp operator leader system, which employs RNAP pauses and binding of the 11-mer trp RNA-binding attenuation protein (TRAP, Figure 3e) to nascent RNA for the regulation of gene expression [86,87]. During transcription of the trp leader, NusA- and NusG-stimulated RNAP pauses allow enough time for holo-TRAP to bind the elongating transcript at repeating tryptophan codons, conformationally switching the RNA to form a terminator hairpin that attenuates transcription of the downstream tryptophan biosynthetic operons [86–89] (Figure 3d,e). These examples reveal the importance of a tight interplay between protein binding and dynamic RNA folding, as the timely addition of proteins can aid other segments of RNA to fold into place, carving a direct pathway towards functionality through an otherwise incredibly rugged energy landscape.
Eukaryotes have evolved additional mechanisms that leverage RNA-protein interactions to influence RNA folding. In eukaryotes, nascent transcripts bind a diverse set of proteins throughout their lifetimes in the nucleus, forming heterogeneous ribonucleoprotein complexes (hnRNPs) [90,91]. Due to their ability to both recognize DNA and RNA structures and recruit necessary protein components for particular processes, hnRNPs play crucial roles in almost every aspect of RNA biogenesis, from transcription and splicing to nuclear export and translation [92–95]. In addition, there is now evidence that nascent RNA structures themselves may play an equally valuable role by driving the formation of macromolecular assemblies within the nucleus, for example acting as seeds to drive spatial localization of otherwise diffusive non-coding RNA and protein molecules [96]. Ultimately, much work remains to elucidate the precise mechanisms by which hnRNPs promote, suppress, and guide nascent RNA folding dynamics, as well as the role of RNA structure and conformational switching in these processes.
Regardless of the environment, the propensity of RNA to fold into local kinetically trapped states that prevent proper folding and function creates a need for mechanisms to resolve misfolding. While conformational switching mechanisms like strand displacement are nucleic acid-centric, the mechanisms described above demonstrate how proteins have evolved to also address nucleic acid folding challenges. Utilizing proteins as dynamic RNA folding chaperones could allow advantageous modularity, allowing the RNA sequence to develop into highly specialized final structural states that ultimately influence cellular events. And while this question is limited in scope to proteins, as techniques develop, the same questions could be taken further to understand how chemical modifications [97], temperature [98], small molecules [22], and even other RNAs [99] influence RNA conformational dynamics. Ultimately, more work investigating a connection between proteins and the dynamic folding free energy landscape of RNAs could reveal an exciting new area of RNA biology to explore.
Discussion
Innovative computational and experimental techniques are shedding new light on the complexities of the cellular RNA folding problem. An increased appreciation for the predominance of kinetically-driven processes necessitated by RNA biogenesis is causing a re-thinking of classic RNA folding paradigms.
Previous conceptualizations of the RNA folding problem posited that the minimum free energy structure was likely the final folded state of most RNAs [1]. However, later developments have established that RNAs are more accurately described as adopting an ensemble of structures occupying multiple local minima of a rugged energy landscape [19]. Detailed studies of specific RNA systems are revealing how the process of transcription guides structural ensembles through ever-shifting energy landscapes, prompting us to revisit our conceptual framework for understanding the principles of RNA folding, and how these principles manifest in RNA function. Interestingly, cotranslational protein folding may serve as a guide for a deeper understanding of cotranscriptional RNA folding. For example, cotranslational folding is commonly described as a nested energy landscape, where the volume of conformational space is proportional to the length of the nascent polypeptide [100]. How a similar conceptualization of cotranscriptional folding can be woven in with the inherent ruggedness of RNA folding landscapes will be interesting to develop further.
Ultimately, the dynamic aspects of cellular RNA folding provide nature with an expanded complexity of RNA folds beyond those attainable in equilibrium. It is our hope that the three central questions posed above will inspire the next wave of research that probes a deeper understanding of this RNA folding regime and how it facilitates myriad cellular functions. We anticipate answers to these questions will drive fundamental understanding and accelerate developments in RNA-based biotechnologies.
Acknowledgements
D.Z.B. was supported in part by a NIH Training Grant (T32GM008382) through the Northwestern University Molecular Biophysics Training Program. L.M.H. was supported in part by a National Institutes of Health (NIH) Training Grant (T32GM008449) through the Northwestern University Biotechnology Training Program. This work was also supported by NIH Research Project grants R01GM130901 and R03HG011113.
Footnotes
Declaration of Competing Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Tinoco I & Bustamante C (1999) How RNA folds. J. Mol. Biol 293, 271–281. [DOI] [PubMed] [Google Scholar]
- 2.Yusupova G & Yusupov M (2021) A Path to the Atomic-Resolution Structures of Prokaryotic and Eukaryotic Ribosomes. Biochemistry (Mosc) 86, 926–941. [DOI] [PubMed] [Google Scholar]
- 3.Liu B, Shi H & Al-Hashimi HM (2021) Developments in solution-state NMR yield broader and deeper views of the dynamic ensembles of nucleic acids. Curr. Opin. Struct. Biol 70, 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kappel K, Zhang K, Su Z, Watkins AM, Kladwang W, Li S, Pintilie G, Topkar VV, Rangan R, Zheludev IN, et al. (2020) Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures. Nat. Methods 17, 699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su Z, Zhang K, Kappel K, Li S, Palo MZ, Pintilie GD, Rangan R, Luo B, Wei Y, Das R, et al. (2021) Cryo-EM structures of full-length Tetrahymena ribozyme at 3.1 Å resolution. Nature 596, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones CP & Ferré-D’Amaré AR (2015) Recognition of the bacterial alarmone ZMP through long-distance association of two RNA subdomains. Nature Structural and Molecular Biology 22, 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren A, Rajashankar KR & Patel DJ (2012) Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature 486, 85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, et al. (2014) Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343, 1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen THD, Tam J, Wu RA, Greber BJ, Toso D, Nogales E & Collins K (2018) Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature 557, 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DasGupta S, Suslov NB & Piccirilli JA (2017) Structural Basis for Substrate Helix Remodeling and Cleavage Loop Activation in the Varkud Satellite Ribozyme. J. Am. Chem. Soc 139, 9591–9597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krochmal D, Shao Y, Li N-S, DasGupta S, Shelke SA, Koirala D & Piccirilli JA (2022) Structural basis for substrate binding and catalysis by a self-alkylating ribozyme. Nat. Chem. Biol [DOI] [PubMed] [Google Scholar]
- 12.Chełkowska-Pauszek A, Kosiński JG, Marciniak K, Wysocka M, Bąkowska-Żywicka K & Żywicki M (2021) The Role of RNA Secondary Structure in Regulation of Gene Expression in Bacteria. Int. J. Mol. Sci 22, 7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serganov A & Nudler E (2013) A Decade of Riboswitches. Cell 152, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perederina A, Li D, Lee H, Bator C, Berezin I, Hafenstein SL & Krasilnikov AS (2020) Cryo-EM structure of catalytic ribonucleoprotein complex RNase MRP. Nat. Commun 11, 3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T & Mondragón A (2010) Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 468, 784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S, Aibara S, Vos SM, Agafonov DE, Lührmann R & Cramer P (2021) Structure of a transcribing RNA polymerase II-U1 snRNP complex. Science 371, 305–309. [DOI] [PubMed] [Google Scholar]
- 17.Wang C, Molodtsov V, Firlar E, Kaelber JT, Blaha G, Su M & Ebright RH (2020) Structural basis of transcription-translation coupling. Science 369, 1359–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson ZL, Ward FR, Méheust R, Ad O, Schepartz A, Banfield JF & Cate JH (2020) Structure of the bacterial ribosome at 2 Å resolution. eLife 9, e60482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganser LR, Kelly ML, Herschlag D & Al-Hashimi HM (2019) The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell. Biol 20, 474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chauvier A, St-Pierre P, Nadon J-F, Hien EDM, Pérez-González C, Eschbach SH, Lamontagne A-M, Penedo JC & Lafontaine DA (2021) Monitoring RNA dynamics in native transcriptional complexes. Proc. Natl. Acad. Sci. USA 118, e2106564118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganser LR, Chu C-C, Bogerd HP, Kelly ML, Cullen BR & Al-Hashimi HM (2020) Probing RNA Conformational Equilibria within the Functional Cellular Context. Cell Rep. 30, 2472–2480.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Widom JR, Nedialkov YA, Rai V, Hayes RL, Brooks CL, Artsimovitch I & Walter NG (2018) Ligand Modulates Cross-Coupling between Riboswitch Folding and Transcriptional Pausing. Molecular Cell 72, 541–552.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeller MJ, Nuthanakanti A, Li K, Aubé J, Serganov A & Weeks KM (2022) Subsite Ligand Recognition and Cooperativity in the TPP Riboswitch: Implications for Fragment-Linking in RNA Ligand Discovery. ACS Chem. Biol 17, 438–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stelzer AC, Frank AT, Kratz JD, Swanson MD, Gonzalez-Hernandez MJ, Lee J, Andricioaei I, Markovitz DM & Al-Hashimi HM (2011) Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat. Chem. Biol 7, 553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roth A & Breaker RR (2009) The Structural and Functional Diversity of Metabolite-Binding Riboswitches. Annu. Rev. Biochem 78, 305–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ihms EC, Zhou M, Zhang Y, Kleckner IR, McElroy CA, Wysocki VH, Gollnick P & Foster MP (2014) Gene regulation by substoichiometric heterocomplex formation of undecameric TRAP and trimeric anti-TRAP. Proc. Natl. Acad. Sci. USA 111, 3442–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merino E, Babitzke P & Yanofsky C (1995) trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translational as well as transcriptional regulation of the Bacillus subtilis trp operon. J. Bacteriol 177, 6362–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodgers ML, Didychuk AL, Butcher SE, Brow DA & Hoskins AA (2016) A multi-step model for facilitated unwinding of the yeast U4/U6 RNA duplex. Nucleic Acids Res. 44, 10912–10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abeysirigunawardena SC, Kim H, Lai J, Ragunathan K, Rappé MC, Luthey-Schulten Z, Ha T & Woodson SA (2017) Evolution of protein-coupled RNA dynamics during hierarchical assembly of ribosomal complexes. Nat. Commun 8, 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim H, Abeysirigunawarden SC, Chen K, Mayerle M, Ragunathan K, Luthey-Schulten Z, Ha T & Woodson SA (2014) Protein-guided RNA dynamics during early ribosome assembly. Nature 506, 334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukuda S, Yan S, Komi Y, Sun M, Gabizon R & Bustamante C (2020) The Biogenesis of SRP RNA Is Modulated by an RNA Folding Intermediate Attained during Transcription. Mol. Cell 77, 241–250. [DOI] [PubMed] [Google Scholar]
- 32.Yu AM, Gasper PM, Cheng L, Lai LB, Kaur S, Gopalan V, Chen AA & Lucks JB (2021) Computationally reconstructing cotranscriptional RNA folding from experimental data reveals rearrangement of non-native folding intermediates. Mol. Cell 81, 870–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chauvier A, Cabello-Villegas J & Walter NG (2019) Probing RNA structure and interaction dynamics at the single molecule level. Methods 162–163, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hua B, Panja S, Wang Y, Woodson SA & Ha T (2018) Mimicking Co-Transcriptional RNA Folding Using a Superhelicase. J. Am. Chem. Soc 140, 10067–10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watters KE, Strobel EJ, Yu AM, Lis JT & Lucks JB (2016) Cotranscriptional folding of a riboswitch at nucleotide resolution. Nat. Struct. Mol. Biol 23, 1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen SJ & Dill KA (2000) RNA folding energy landscapes. Proc. Natl. Acad. Sci. USA 97, 646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kramer FR & Mills DR (1981) Secondary structure formation during RNA synthesis. Nucleic Acids Res. 9, 5109–5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan T & Sosnick T (2006) RNA folding during transcription. Annu. Rev. Biophys. Biomol. Struct 35, 161–175. [DOI] [PubMed] [Google Scholar]
- 39.Sun L, Fazal FM, Li P, Broughton JP, Lee B, Tang L, Huang W, Kool ET, Chang HY & Zhang QC (2019) RNA structure maps across mammalian cellular compartments. Nat. Struct. Mol. Biol 26, 322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ray-Soni A, Bellecourt MJ & Landick R (2016) Mechanisms of Bacterial Transcription Termination: All Good Things Must End. Annu. Rev. Biochem 85, 319–347. [DOI] [PubMed] [Google Scholar]
- 41.de Smit MH & van Duin J (1990) Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci U S A 87, 7668–7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carrier TA & Keasling JD (1999) Library of synthetic 5’ secondary structures to manipulate mRNA stability in Escherichia coli. Biotechnol Prog 15, 58–64. [DOI] [PubMed] [Google Scholar]
- 43.Buratti E & Baralle FE (2004) Influence of RNA Secondary Structure on the Pre-mRNA Splicing Process. MCB 24, 10505–10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saldi T, Riemondy K, Erickson B & Bentley DL (2021) Alternative RNA structures formed during transcription depend on elongation rate and modify RNA processing. Molecular Cell S1097276521000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saldi T, Fong N & Bentley DL (2018) Transcription elongation rate affects nascent histone pre-mRNA folding and 3’ end processing. Genes Dev 32, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu X & Bartel DP (2017) Widespread Influence of 3′-End Structures on Mammalian mRNA Processing and Stability. Cell 169, 905–917.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danilova LV, Pervouchine DD, Favorov AV & Mironov AA (2006) RNAKinetics: a web server that models secondary structure kinetics of an elongating RNA. J. Bioinform. Comput. Biol 4, 589–596. [DOI] [PubMed] [Google Scholar]
- 48.Xayaphoummine A, Bucher T, Thalmann F & Isambert H (2003) Prediction and statistics of pseudoknots in RNA structures using exactly clustered stochastic simulations. Proc. Natl. Acad. Sci. USA 100, 15310–15315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Isambert H & Siggia ED (2000) Modeling RNA folding paths with pseudoknots: Application to hepatitis delta virus ribozyme. Proc. Natl. Acad. Sci. USA 97, 6515–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flamm C, Fontana W, Hofacker IL & Schuster P (2000) RNA folding at elementary step resolution. RNA 6, 325–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Freida KL & Block SM (2012) Direct Observation of Cotranscriptional Folding in an Adenine Riboswitch. Science 338, 397–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holden SJ, Uphoff S, Hohlbein J, Yadin D, Le Reste L, Britton OJ & Kapanidis AN (2010) Defining the limits of single-molecule FRET resolution in TIRF microscopy. Biophys. J 99, 3102–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hua B, Jones CP, Mitra J, Murray PJ, Rosenthal R, Ferré-D’Amaré AR & Ha T (2020) Real-time monitoring of single ZTP riboswitches reveals a complex and kinetically controlled decision landscape. Nat. Commun 11, 4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Helmling C, Klötzner DP, Sochor F, Mooney RA, Wacker A, Landick R, Fürtig B, Heckel A & Schwalbe H (2018) Life times of metastable states guide regulatory signaling in transcriptional riboswitches. Nat. Commun 9, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cofsky JC, Soczek KM, Knott GJ, Nogales E & Doudna JA (2022) CRISPR–Cas9 bends and twists DNA to read its sequence. Nat Struct Mol Biol 29, 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng L, White EN, Brandt NL, Yu AM, Chen AA & Lucks JB (2022) Cotranscriptional RNA strand exchange underlies the gene regulation mechanism in a purine-sensing transcriptional riboswitch. Nucleic Acids Research gkac102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strobel EJ, Cheng L, Berman KE, Carlson PD & Lucks JB (2019) A ligand-gated strand displacement mechanism for ZTP riboswitch transcription control. Nat. Chem. Biol 15, 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hong F & Šulc P (2019) An emergent understanding of strand displacement in RNA biology. J Struct Biol 207, 241–249. [DOI] [PubMed] [Google Scholar]
- 59.LeCuyer KA & Crothers DM (1993) The Leptomonas collosoma spliced leader RNA can switch between two alternate structural forms. Biochemistry 32, 5301–5311. [DOI] [PubMed] [Google Scholar]
- 60.LeCuyer KA & Crothers DM (1994) Kinetics of an RNA conformational switch. Proc. Natl. Acad. Sci. USA 91, 3373–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Homann M, Nedbal W & Sczakiel G (1996) Dissociation of Long-Chain Duplex RNA Can Occur Via Strand Displacement in Vitro : Biological Implications. Nucleic Acids Res. 24, 4395–4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, Siksnys V & Seidel R (2014) Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc. Natl. Acad. Sci. USA 111, 9798–9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong TN, Sosnick TR & Pan T (2007) Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc. Natl. Acad. Sci. USA 104, 17995–18000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin Y, Schmidt BF, Bruchez MP & McManus CJ (2018) Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res. 46, 3742–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang DY & Winfree E (2010) Robustness and modularity properties of a non-covalent DNA catalytic reaction. Nucleic Acids Res 38, 4182–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Al-Hashimi HM & Walter NG (2008) RNA dynamics: it is about time. Current Opinion in Structural Biology 18, 321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rössler OG, Straka A & Stahl H (2001) Rearrangement of structured RNA via branch migration structures catalysed by the highly related DEAD-box proteins p68 and p72. Nucleic Acids Res. 29, 2088–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ruminski DJ, Watson PY, Mahen EM & Fedor MJ (2016) A DEAD-box RNA helicase promotes thermodynamic equilibration of kinetically trapped RNA structures in vivo. RNA 22, 416–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cofsky JC, Knott GJ, Gee CL & Doudna JA (2022) Crystal structure of an RNA/DNA strand exchange junction. PLoS ONE (Kursula P, ed.) 17, e0263547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pausch P, Soczek KM, Herbst DA, Tsuchida CA, Al-Shayeb B, Banfield JF, Nogales E & Doudna JA (2021) DNA interference states of the hypercompact CRISPR-CasΦ effector. Nat. Struct. Mol. Biol 28, 652–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salomon WE, Jolly SM, Moore MJ, Zamore PD & Serebrov V (2015) Single-Molecule Imaging Reveals that Argonaute Reshapes the Binding Properties of Its Nucleic Acid Guides. Cell 162, 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu JYA, Steif A, Proctor JR & Meyer IM (2013) Transient RNA structure features are evolutionarily conserved and can be computationally predicted. Nucleic Acids Res. 41, 6273–6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brion P & Westhof E (1997) Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct 26, 113–137. [DOI] [PubMed] [Google Scholar]
- 74.Herschlag D (1995) RNA chaperones and the RNA folding problem. J. Biol. Chem 270, 20871–20874. [DOI] [PubMed] [Google Scholar]
- 75.Woodson SA, Panja S & Santiago-Frangos A (2018) Proteins That Chaperone RNA Regulation. Microbiol. Spectr 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Woodson SA (2011) RNA folding pathways and the self-assembly of ribosomes. Acc. Chem. Res 44, 1312–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rodgers ML & Woodson SA (2021) A roadmap for rRNA folding and assembly during transcription. Trends Biochem. Sci 46, 889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rodgers ML & Woodson SA (2019) Transcription Increases the Cooperativity of Ribonucleoprotein Assembly. Cell 179, 1370–1381.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duss O, Stepanyuk GA, Puglisi JD & Williamson JR (2019) Transient Protein-RNA Interactions Guide Nascent Ribosomal RNA Folding. Cell 179, 1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Woodson SA (2010) Taming free energy landscapes with RNA chaperones. RNA Biol. 7, 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang J & Landick R (2016) A Two-Way Street: Regulatory Interplay between RNA Polymerase and Nascent RNA Structure. Trends Biochem. Sci 41, 293–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chauvier A, Ajmera P, Yadav R & Walter NG (2021) Dynamic competition between a ligand and transcription factor NusA governs riboswitch-mediated transcription regulation. Proc. Natl. Acad. Sci. USA 118, e2109026118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guo X, Myasnikov AG, Chen J, Crucifix C, Papai G, Takacs M, Schultz P & Weixlbaumer A (2018) Structural Basis for NusA Stabilized Transcriptional Pausing. Mol. Cell 69, 816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pan T, Artsimovitch I, Fang XW, Landick R & Sosnick TR (1999) Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proceedings of the National Academy of Sciences of the United States of America 96, 9545–9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wickiser JK, Winkler WC, Breaker RR & Crothers DM (2005) The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Molecular Cell 18, 49–60. [DOI] [PubMed] [Google Scholar]
- 86.Mondal S, Yakhnin AV & Babitzke P (2017) Modular Organization of the NusA- and NusG-Stimulated RNA Polymerase Pause Signal That Participates in the Bacillus subtilis trp Operon Attenuation Mechanism. J. Bacteriol (Gourse RL, ed.) 199, e00223–17, e00223–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shimotsu H, Kuroda MI, Yanofsky C & Henner DJ (1986) Novel form of transcription attenuation regulates expression the Bacillus subtilis tryptophan operon. J. Bacteriol 166, 461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Antson AA, Dodson EJ, Dodson G, Greaves RB, Chen X & Gollnick P (1999) Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature 401, 235–242. [DOI] [PubMed] [Google Scholar]
- 89.McElroy C, Manfredo A, Wendt A, Gollnick P & Foster M (2002) TROSY-NMR studies of the 91kDa TRAP protein reveal allosteric control of a gene regulatory protein by ligand-altered flexibility. J. Mol. Biol 323, 463–473. [DOI] [PubMed] [Google Scholar]
- 90.Singh G, Pratt G, Yeo GW & Moore MJ (2015) The Clothes Make the mRNA: Past and Present Trends in mRNP Fashion. Annu. Rev. Biochem 84, 325–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dreyfuss G, Matunis MJ, Piñol-Roma S & Burd CG (1993) hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem 62, 289–321. [DOI] [PubMed] [Google Scholar]
- 92.Krecic AM & Swanson MS (1999) hnRNP complexes: composition, structure, and function. Curr. Opin. Cell. Biol 11, 363–371. [DOI] [PubMed] [Google Scholar]
- 93.Busch A & Hertel KJ (2012) Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip. Rev. RNA 3, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jean-Philippe J, Paz S & Caputi M (2013) hnRNP A1: the Swiss army knife of gene expression. Int. J. Mol. Sci 14, 18999–19024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Geuens T, Bouhy D & Timmerman V (2016) The hnRNP family: insights into their role in health and disease. Hum. Genet 135, 851–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Quinodoz SA, Jachowicz JW, Bhat P, Ollikainen N, Banerjee AK, Goronzy IN, Blanco MR, Chovanec P, Chow A, Markaki Y, et al. (2021) RNA promotes the formation of spatial compartments in the nucleus. Cell 184, 5775–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harcourt EM, Kietrys AM & Kool ET (2017) Chemical and structural effects of base modifications in messenger RNA. Nature 541, 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Choi EK, Ulanowicz KA, Nguyen YAH, Frandsen JK & Mitton-Fry RM (2017) SHAPE analysis of the htrA RNA thermometer from Salmonella enterica. RNA 23, 1569–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kolb FA, Westhof E, Ehresmann B, Ehresmann C, Wagner EG & Romby P (2001) Four-way junctions in antisense RNA-mRNA complexes involved in plasmid replication control: a common theme? J. Mol. Biol 309, 605–614. [DOI] [PubMed] [Google Scholar]
- 100.Waudby CA, Dobson CM & Christodoulou J (2019) Nature and Regulation of Protein Folding on the Ribosome. Trends in Biochemical Sciences 44, 914–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ren A, Rajashankar KR & Patel DJ (2015) Global RNA Fold and Molecular Recognition for a pfl Riboswitch Bound to ZMP, a Master Regulator of One-Carbon Metabolism. Structure 23, 1375–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Del Campo M & Lambowitz AM (2009) Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol. Cell 35, 598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]


