Abstract
Purpose:
PNOC003 is a multicenter precision medicine trial for children and young adults with newly diagnosed diffuse intrinsic pontine glioma (DIPG).
Patients and Methods:
Patients (3–25 years) were enrolled on the basis of imaging consistent with DIPG. Biopsy tissue was collected for whole-exome and mRNA sequencing. After radiotherapy (RT), patients were assigned up to four FDA-approved drugs based on molecular tumor board recommendations. H3K27M-mutant circulating tumor DNA (ctDNA) was longitudinally measured. Tumor tissue and matched primary cell lines were characterized using whole-genome sequencing and DNA methylation profiling. When applicable, results were verified in an independent cohort from the Children's Brain Tumor Network (CBTN).
Results:
Of 38 patients enrolled, 28 patients (median 6 years, 10 females) were reviewed by the molecular tumor board. Of those, 19 followed treatment recommendations. Median overall survival (OS) was 13.1 months [95% confidence interval (CI), 11.2–18.4] with no difference between patients who followed recommendations and those who did not. H3K27M-mutant ctDNA was detected at baseline in 60% of cases tested and associated with response to RT and survival. Eleven cell lines were established, showing 100% fidelity of key somatic driver gene alterations in the primary tumor. In H3K27-altered DIPGs, TP53 mutations were associated with worse OS (TP53mut 11.1 mo; 95% CI, 8.7–14; TP53wt 13.3 mo; 95% CI, 11.8–NA; P = 3.4e−2), genome instability (P = 3.1e−3), and RT resistance (P = 6.4e−4). The CBTN cohort confirmed an association between TP53 mutation status, genome instability, and clinical outcome.
Conclusions:
Upfront treatment-naïve biopsy provides insight into clinically relevant molecular alterations and prognostic biomarkers for H3K27-altered DIPGs.
Translational Relevance.
PNOC003 is one of the first to report on a biopsy-driven, biology-based combination therapy for children and young adults with DIPG. The cohort offers insight into molecular biomarkers for DIPG and provides support of TP53 mutations as markers of radiotherapy (RT) resistance in H3K27-altered DIPG/DMG. Molecular characterization further reveals that TP53 mutations associate with newly described molecular findings such as loss of 10q/PTEN and that this combined molecular signature correlates with the worst survival outcomes. The work provides a potential new molecular stratification for H3K27-altered DIPG/DMG and offers support for therapeutic considerations, such as RT sensitizers in patients with pertinent TP53 alterations. Finally, PNOC003 contributes to the growing application of circulating tumor DNA in pediatric central nervous system tumors and the development of cell lines with associated molecular comparison with tumor tissue.
Introduction
Despite many approaches being used to treat diffuse intrinsic pontine glioma (DIPG) over many decades, no therapy has successfully improved average survival beyond one year (1–4). The current standard-of-care treatment is up-front radiotherapy (RT), commonly coupled with or followed by novel therapies within a clinical trial (2, 3, 5). Trial options include targeted therapies, convection enhanced delivery with direct intratumoral drug infusion into the tumor, and immunotherapy (3, 6). Large-scale molecular profiling studies have revealed critical oncogenic somatic driver alterations and highlighted intertumoral heterogeneity in DIPG. Somatic mutations in H3F3A and HIST1H3B, resulting in a lysine-to-methionine substitution at position 27 on the H3.3/H3.1 histone tail (H3K27M), are present in 80%–90% of DIPG tumors (7–9) and among H3 subtypes there is non-random, co-segregation with partner mutations and distinct epigenetic signatures (10–16). H3.3K27M mutations frequently co-occur with alterations in the p53 pathway (e.g., TP53, PPM1D), along with receptor tyrosine kinase amplification/mutation (e.g., PDGFRA). In contrast, H3.1K27M mutations carry alterations in the TGFß/BMP receptor (ACVR1) and downstream components of the PI3-kinase pathway (e.g., PIK3CA, PIK3R1; refs. 7, 10, 17, 18). On the basis of the pathognomonic molecular characteristics, DIPG is now classified as H3K27-altered diffuse midline glioma (DMG) and defined by somatic mutations in H3F3A, HIST1H3B/C, EGFR, or EZHIP overexpression (19). The diverse range of molecular pathways contributing to the oncogenesis of DIPG suggests that single-agent therapy is unlikely to provide durable disease control.
Driven by advances in genome sequencing, the safety of surgical biopsy in the current era, and anticipation that multiagent approaches will be necessary to improve survival, we developed PNOC003 (NCT02274987), a precision medicine trial for DIPG. The trial used Clinical Laboratory Improvement Amendments (CLIA)–generated tumor-normal whole-exome sequencing (WES) and tumor mRNA sequencing (mRNA-seq) data to generate individualized therapy plans based on tumor-specific alterations, which were then applied in children and young adults with newly diagnosed DIPG after standard-of-care, upfront RT. Within the trial, collection of plasma H3K27M-mutant circulating tumor DNA (ctDNA), patient-derived cell line generation, whole genome sequencing (WGS), and DNA methylation profiling were performed. When feasible, molecular and clinical outcomes were retrospectively corroborated against an external dataset of pediatric patients with DMG from the Children's Brain Tumor Network (CBTN; Pediatric Brain Tumor Atlas, PBTA, https://doi.org/10.24370/SD_BHJXBDQK). Here, we report the results and exploratory biologic correlates from the multicenter clinical trial PNOC003.
Patients and Methods
Clinical trial design
PNOC003 was open to enrollment between September 2014 and January 2018. Study design and methods have been previously described (20). Patients participated across five Pacific Pediatric Neuro-Oncology Consortium (PNOC) institutions listed in the Supplementary Methods. Eligible patients were ≥3 and ≤25 years of age with newly diagnosed DIPG based on radiographic imaging and without disseminated disease. Patients underwent biopsy with local pathology review to confirm ≥50% tumor content. Fresh-frozen tissue samples were sent to Ashion Analytics (now part of Genomic Health, an Exact Sciences Laboratory) for CLIA WES, and mRNA-seq.
Patients were monitored by standard-of-care clinical examinations and laboratory and clinical assessments that aligned with anticipated toxicities based on specific drug combinations from specialized tumor board treatment recommendations. Treatment-related adverse events (TRAE) were collected from the time of study enrollment (n = 38) and throughout the completion of protocol-defined toxicity follow-up. RT-related toxicities were not included as these were considered part of standard of care. Patients underwent MRI assessments every odd cycle of study therapy (i.e., cycle 3, 5, etc.). Post hoc central radiology review was performed by a board-certified neuro-radiologist (J. Villanueva-Meyer). Protocol defined progressive disease on MRI was defined as a greater than 25% increase in the sum of perpendicular diameters and/or development of new enhancing or non-enhancing lesions, as previously described (21).
Before trial activation, necessary approvals were obtained by the FDA and institutional Investigational Review Boards (IRB) at enrolling sites. All patients and/or parents/guardians provided informed consent/assent before study enrollment and by IRB guidelines. The UCSF Data Safety and Monitoring Committee served as clinical trial oversight to monitor for safety and protocol conduct.
Biopsy collection and processing
Each enrolled patient underwent stereotactic biopsy with the collection as per local institutional standards. Details of the stereotactic approach and selection of at least 50% tumor content have been previously described (20, 22).
Clinical WES and transcriptome sequencing
Ashion Analytics performed DNA and RNA extractions on tumor biopsies and performed library preparations as previously described (20). Clinical WES (∼256X) was performed on biopsy tissue and matched normal blood (diagnosis, n = 29; progression, n = 2). For P-05, a 562-gene targeted exome panel [Ashion's Genomic-Enabled Medicine (GEM) Cancer Panel] was performed. Poly-A–selected RNA sequencing (RNA-Seq ∼200M reads) was performed (diagnosis, n = 30; progression, n = 2). WES and RNA-Seq libraries were sequenced at 2 × 100 bp on an Illumina HiSeq 2500.
Specialized tumor boards and treatment recommendations
Each tumor molecular profile was reviewed at a specialized tumor board, and a precision medicine approach of up to four FDA-approved drugs was determined. Treatment was initiated after completion of standard-of-care RT. Details of tumor boards and drug selection guidelines and administration have been previously described (20, 23, 24).
ctDNA analyses
Plasma ctDNA was collected at standard-of-care biopsy, the start of any molecular treatment recommendations, each MRI timepoint, progression, and end of treatment (25, 26). Plasma ctDNA at baseline versus post-RT was compared using Wilcoxon matched-pairs signed-rank test (GraphPad Prism 9 software). ctDNA survival analyses were performed using log-rank (Mantel–Cox) tests in R.
Cell line generation, propagation, and maintenance
Cell line generation was attempted, when feasible (27), from patient biopsy samples (P-06, P-07, P-09, P-16, P-26, P-31, P-33, P-37, and P-38), biopsy needle wash (P-05) or patient tumor–derived mouse xenograft tissue (P-04).
WGS data generation and processing for PNOC003 and CBTN
WGS was performed at NantHealth Sequencing Center (Culver City, CA) on a post hoc basis for biopsy tissue (∼60X) with matched controls (∼30X; diagnosis, n = 33; progression, n = 2; post-mortem, n = 4; external CBTN cohort, n = 22) and on DNA derived from PNOC003 patient-derived cell lines. The libraries for WGS were 2×150 bp and sequencing was done on an Illumina HiSeq platform (X/400). Details of CBTN sequencing were previously described in the OpenPBTA project (28).
Somatic driver gene discovery
Using the MAF file from our consensus SNV/indel callset, we performed a de novo driver gene discovery using the R package dndscv (29) with default parameters (q < 0.2) and combined with prior knowledge about high-grade glioma driver genes from IntOGen (30).
DNA methylation array
DNA was extracted from tumor tissue specimens and cell pellets (500,000 cells) and was quantified using Qubit dsDNA Broad Range Assay. DNA was bisulfite converted using the EZ DNA Methylation-Gold kit (Zymo Research) and hybridized onto Infinium MethylationEPIC BeadChip using Infinium MethylationEPIC BeadChip Kit per manufacturer instructions (Illumina). BeadChip arrays were scanned using the iScan Reader (Illumina). IDAT files were uploaded and analyzed using the DKFZ brain tumor methylation classifier (v11b4; https://www.molecularneuropathology.org/mnp; ref. 31).
Chromosomal instability
The consensus PBTA CNV callset was queried for large gains and losses with full/partial chromosomal alteration defined as events >5Mb. A chromosome instability (CIN) score was computed for each patient based on the number of chromosomes affected by large-scale events and previously described as: Chromosomal gain/loss events combined, only chromosomal gain events, and only chromosomal loss events.
Statistical analysis
At the completion of enrollment of the feasibility cohort for PNOC003 (20), the protocol was amended to evaluate clinical response in a total accrual of 19 patients, as defined by OS at 12 months (OS12, primary objective). Secondary objectives were to describe the toxicity and safety of the biopsy. Exploratory objectives were to compare the fidelity of WGS with WES and mRNA-seq analyses and between molecular profiles of longitudinal tumor samples over disease course and to evaluate ctDNA as a biomarker of treatment response or resistance. To compare characteristics of patients that did or did not follow treatment recommendations, χ2 tests were used for binary variables (gender, race, and ethnicity) and Kruskal–Wallis for non-normally distributed variables (age). Survival outcomes were compared using Kaplan–Meier survival analysis and significance calculated by the log-rank test. Cox proportional hazards regression models were used to assess the combination of genomic markers on survival outcomes. Violations of the non-proportional hazards assumptions of log-rank tests were tested and ruled out using Schoenfeld Residuals Test. Mann–Whitney tests were used to compare RT response based on individual gene alterations as well as chromosome gains and losses in the setting of CIN. The Fisher's exact test was used to compare chromosome gains and losses based on individual gene alterations.
Data availability
Access to raw data can be requested from CBTN (https://cbtn.org). Code for the somatic workflows can be found at https://github.com/d3b-center/OpenPBTA-workflows. Code for downstream analyses can be found at https://github.com/AlexsLemonade/OpenPBTA-analysis/. Processed files are publicly available on CAVATICA (https://cavatica.sbgenomics.com/u/cavatica/pbta-pnoc003; https://cavatica.sbgenomics.com/u/cavatica/openpbta). Processed data can be visualized in PedcBioPortal (https://pedcbioportal.kidsfirstdrc.org).
Additional details on all Patients and Methods can be found in Supplementary Methods.
Results
Multi-omics tumor profiling is safe and feasible and informs personalized treatment recommendations in patients with newly diagnosed DIPG
Thirty-eight patients were enrolled in PNOC003 between 2014 and 2018 (White/Non-Hispanic, n = 11; White/Unknown, n = 2; Black/African American/Non-Hispanic, n = 4; Asian/Non-Hispanic, n = 3; Unknown/Non-Hispanic, n = 4; Unknown/Hispanic/Latino, n = 10; Unknown/Unknown, n = 4). A total of 28 out of 38 patients were included in analyses for clinical trial outcomes after removal of 10 patients due to: Family changing decision about undergoing biopsy (n = 1), failure to collect sufficient tissue for CLIA molecular analysis (n = 3), ineligible pathology diagnosis [pilocytic astrocytoma (n = 1); embryonal tumor with multi-layered rosettes (n = 1); embryonal tumor (n = 1)], withdrawal of participation after the biopsy but before study required treatment (n = 1), and death before completion of CLIA molecular profiling or RT (n = 2; Fig. 1A; Supplementary Table S1). WES was completed in all 28 patients, except P-05, for whom gene panel sequencing was substituted. CLIA mRNA-seq was completed for all, except P-17, due to failed required quality control. Nineteen of 28 patients [10 (36%) females; median age of 6 years at diagnosis (range, 4–25 years; Supplementary Table S2)] followed biology-based, multiagent combination therapy. Two patients underwent tissue collection at progression (P-06 and P-07). Four patients underwent postmortem tissue collection (P-04, P-07, P-13, and P-18).
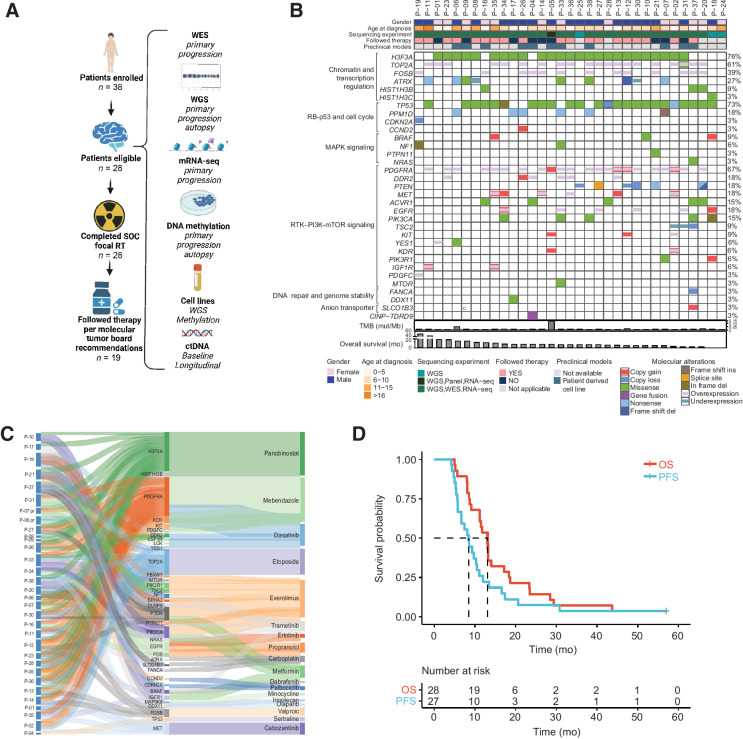
Figure 1.
Overview of the PNOC003 clinical trial, molecular alterations identified, assigned therapy recommendations based on molecular data, and clinical outcomes. A, Left shows the clinical trial outline with the total number of patients in each treatment phase of the trial (includes 38 enrolled patients and 10 patients removed from outcome analyses due to the family changing decision about undergoing biopsy; n = 1), failure to collect sufficient tissue for CLIA molecular analysis (n = 3), ineligible pathology diagnosis, withdrawal of participation after the biopsy but before study required treatment (n = 1), and death before completion of CLIA molecular profiling or radiotherapy (n = 2). Right provides an overview of the completed multi-omic profiling of tumor tissue, germline, CSF, and cell lines. B, Oncoprint representation of alterations identified for all patients that successfully underwent WGS, WES, or RNA-seq in primary DIPG tumors (n = 33; WES and mRNA-seq, CLIA; WGS, non-CLIA). Patients P-18, P-24, and P-25 were removed from trial due to insufficient tissue availability for WES and mRNA-seq; however, these patients completed WGS (not used for treatment decision-making). “Tier 1” targetable alteration listed. “Not applicable” for “Followed Therapy” row indicates patients that came off therapy before initiation of therapy recommendations due to family preference (n = 1), the family changed mind about continuing therapy recommendations (n = 1), patient death before rendering therapy recommendations (n = 1), or did not have sufficient tissue to perform CLIA molecular analyses required to render therapy recommendations (n = 2). Patients are represented in columns, and genes are labeled in rows. Percentages on the right column represent the proportion of patients in the cohort with molecular alterations. Tumor mutation burden (TMB) and overall survival (OS) are represented below the oncoprint. C, Sankey diagram illustrates the individualized, targeted therapy recommendations for each PNOC003 patient who underwent molecular tumor board (n = 30) and based on gene alterations identified via molecular profiling. The first node shows the patient identifier connected to the therapeutically informative genes in the second node. The third node depicts targeted therapy agents recommended by the molecular tumor board. Abbreviations used for drugs are shown in parentheses, “pr” indicates targeted recommendations from repeat biopsy at progression (n = 2). Two patients underwent molecular tumor board but were removed from therapy due to patient/family preference (n = 1; P-28) or patient death during RT (n = 1; P-20). D, Kaplan–Meier OS and PFS of all patients followed for survival outcomes (n = 28 for OS, n = 27 for PFS; based on missing PFS for P-31). Median OS of 13.1 months and median PFS of 8.5 months. WES, whole-exome sequencing; WGS, whole-genome sequencing; mRNA-seq, mRNA sequencing; ctDNA, circulating tumor DNA; TMB, tumor mutation burden; OS, overall survival; PFS, progression-free survival.
A specialized molecular tumor board reviewed WES and mRNA-seq data for each patient (n = 28) and issued biology-informed treatment recommendations in a median of 18 business days (range, 15–20 days; ref. 20). WES and mRNA-seq data identified alterations affecting H3F3A (82%, n = 23) and TP53 (68%, n = 19) as the most frequent gene alterations. High-level DNA amplifications were recurrently seen in PDGFRA (n = 4) and MET (n = 6). The most frequent gene expression outliers were seen for TOP2A (68%, n = 19) and PDGFRA (68%, n = 19; Fig. 1B). Eighteen different FDA-approved molecular-targeted drugs were recommended across all patients (Fig. 1C; ref. 20). The top recommended drugs were the HDAC inhibitor, panobinostat, to target histone H3K27M induced epigenetic alterations (68%, n = 19; ref. 32), mebendazole to target PDGFRA amplification and/or overexpression (50%, n = 14; refs. 33, 34), and everolimus to target PI3K/PTEN/mTOR pathway activation (43%, n = 12; refs. 35–37). Nineteen patients (68%) followed treatment recommendations (Supplementary Table S2). There were no differences in gender, age at diagnosis, race, ethnicity, or institution of enrollment between patients that followed versus those that did not follow treatment recommendations.
Surgical AEs were collected on all patients that underwent biopsy (n = 37), and medication-related AEs were collected on all patients that initiated therapy as per specialized tumor board treatment recommendations (n = 19). Most TRAEs were grade 1 and 2, including surgery-related (Supplementary Tables S3 and S4). The most frequently reported medication-related TRAEs were hematologic, including grade 3 and 4 leukopenia (37%, n = 7), lymphopenia (26%, n = 5), neutropenia (53%, n = 10), and thrombocytopenia (37%, n = 7). Surgical TRAEs included grade 3 abducens nerve disorder, dysarthria, and nystagmus, all of which were existing grade 2 AEs at each patient's baseline (n = 1 each) and resolved back to baseline. Related serious AEs occurred in one patient with grade 3 hypokalemia and hypertension (both resolved with medical management). Two patients underwent repeat biopsy without associated toxicity related to the second biopsy. Overall, there were no treatment- or surgical toxicity-related deaths.
Driver gene alterations in TP53, PTEN, and PDGFRA are molecular biomarkers predictive of overall survival in DIPG
Median overall survival (OS) for the cohort (inclusive of patients that met eligibility criteria and were not replaced, n = 28, Supplementary Table S2) was 13.1 months [95% confidence interval (CI), 11.2–18.4]. Median OS did not differ between patients that followed specialized tumor board treatment recommendations (n = 19; 11.8 mo; 95% CI, 11.0–21.8) and those who did not (n = 9, 13.1 mo; 95% CI, 8.2–20.0; P = 5.9e−1). OS at 12 months (OS12) for the entire cohort was 54% (95% CI, 38–76; Fig. 1D). To better understand the impact of tumor heterogeneity on outcome in our cohort, we investigated genetic and molecular biomarkers of treatment response and survival outcomes in patients with H3K27-altered DIPG (inclusive of patients with available WGS and survival data, n = 30, Supplementary Tables S1 and S2). We used for our biomarker analyses WGS data, both for samples with available CLIA WES and mRNA-seq and samples in which CLIA analyses were not done.
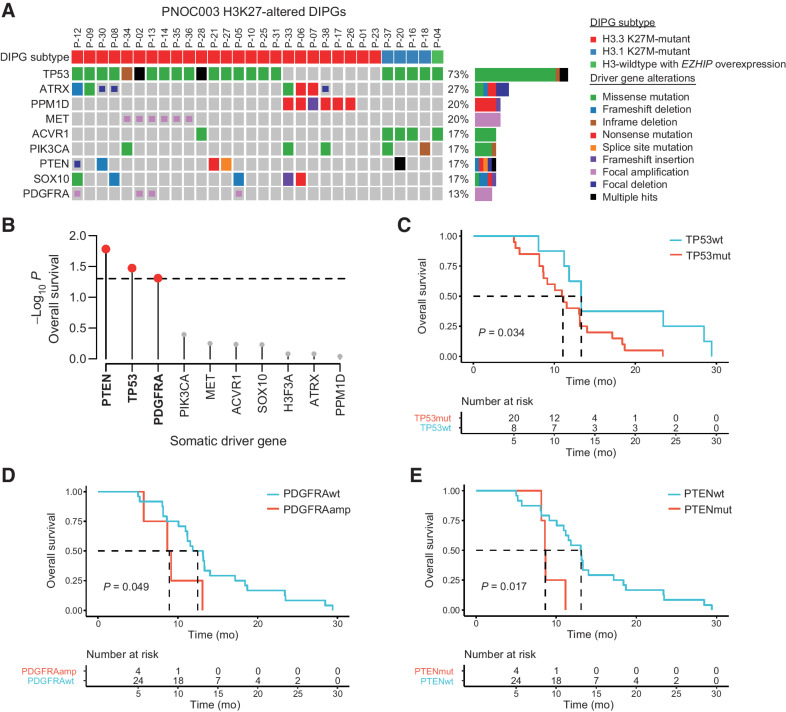
We identified nine recurrently mutated driver genes in H3K27-altered DIPG tumors, including TP53 (73%), ATRX (27%), PPM1D (20%), MET (20%), ACVR1, PIK3CA, PTEN, SOX10, and PDGFRA (17% each; Fig. 2A). Association between driver gene mutation status and OS revealed PTEN (P = 1.7e−2), TP53 (P = 3.4e−2), and PDGFRA (P = 4.9e−2) to be significantly associated with worse clinical outcomes (Fig. 2B). Patients with somatic TP53 driver mutations (TP53mut, n = 20; OS, 11.1 mo; 95% CI, 8.7–14) demonstrated worse OS compared with TP53 wild-type tumors (TP53wt, n = 8, OS, 13.3 mo; 95% CI, 11.8–NA; P = 3.4e−2; n = 28 with survival data; Fig. 2C). This finding was corroborated in 22 patients with H3K27M-mutant DMG from the CBTN (TP53mut, n = 15, OS, 9.0 mo; 95% CI, 7.4–15.8; TP53wt, n = 7, OS, 17.6 mo; 95% CI, 8.9–NA; P = 2.4e−2; Supplementary Fig. S1A and S1B). The survival outcomes of the CBTN cohort were not statistically different when compared with the PNOC003 cohort (Supplementary Fig. S1A); however, the prolonged OS of patients with TP53wt tumors in the CBTN cohort may be due inclusion of tumors in midline structures outside of the pons (e.g., thalamus).
Figure 2.
Somatic TP53, PTEN, and PDGFRA alterations are associated with clinical outcomes in H3K27-altered DIPG. A, Oncoprint representation of recurrent somatic driver gene alterations in H3K27-altered DIPGs with available WES, WGS, and mRNA-seq, regardless of the availability of survival outcomes (n = 30). H3K27-altered DIPG subtyping based on the 2021 WHO Classification of Central Nervous System Tumors system: H3F3A (p.K27M), HIST1H3B (p.K27M), and EZHIP overexpression. B, Association between somatic driver gene status and OS in H3K27-altered DIPG patients (n = 28). Lollipop plot shows the −log10 log-rank test P value for all tested driver genes (n = 8). Red colored dots mark genes significantly (P < 0.05) associated with OS. C–E, Kaplan–Meier survival curves and log-rank P values for H3K27-altered DIPG patients stratified by TP53 (C), PDGFRA (D), and PTEN (E) alteration status. WES, whole-exome sequencing; WGS, whole-genome sequencing; mRNA-seq, mRNA sequencing; OS, overall survival; PFS, progression-free survival; wt, wild-type; mut, mutant; amp, amplification.
In addition to TP53 mutation status as a negative predictor of survival outcome, patients with PDGFRA amplification (n = 4, OS, 8.9 mo; 95% CI, 5.7–NA) showed worse OS (n = 24, PDGFRAwt OS, 12.5 mo; 95% CI, 11.2–17.2; P = 4.9e−2; Fig. 2D). Furthermore, patients with PTEN-altered tumors, including somatic mutations (n = 3) and focal deletions (n = 1), demonstrated worse survival (n = 4, OS, 8.6 mo; 95% CI, 8.3−NA) compared with patients with PTEN wild-type tumors (n = 24, OS, 13.1 mo; 95% CI, 11.2–17.2; P = 1.7e−2; Fig. 2E). These trends persisted even when tested independent of TP53 mutations (Supplementary Fig. S2).
Somatic TP53 alterations predict response to RT
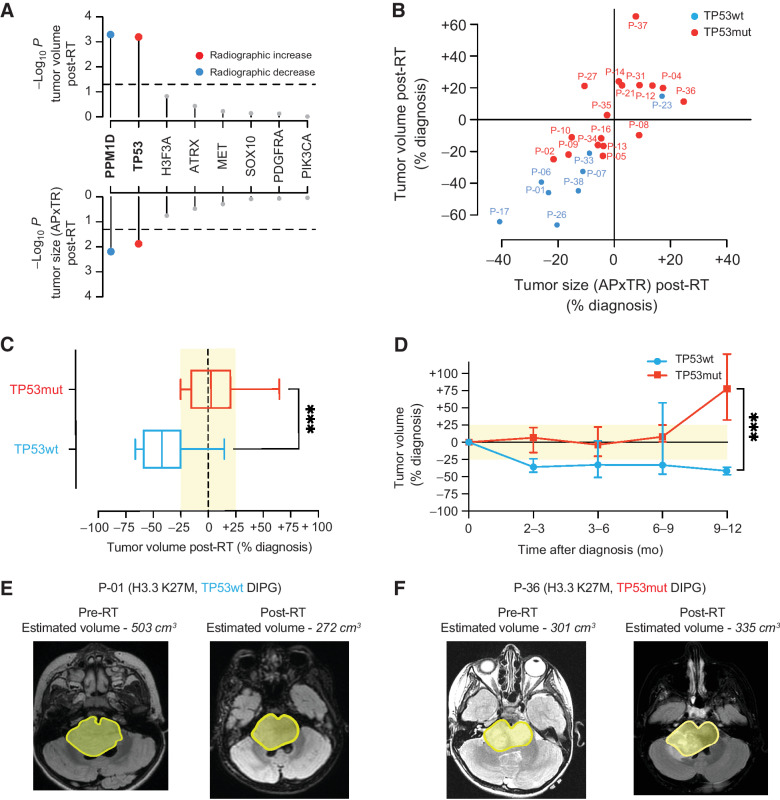
TP53 mutations have been previously associated with poor RT response in DIPG (38). Therefore, we investigated the association between mutations in TP53 and other driver genes with RT response across H3K27-altered DIPG patients, using pre- and post-RT MRI. The median time between pre- and post-RT MRI was 2.6 months (range, 2.2–4.2). Across all driver genes, only TP53mut H3K27-altered DIPGs showed stable tumor volumes after RT when comparing pre- to post-RT images (median +3%, n = 17). In contrast, TP53wt H3K27-altered tumors demonstrated a marked reduction in tumor volumes (median −42%, n = 8, Fig. 3A). Notably, PPM1D mutations were mutually exclusive with TP53 mutations and associated with reduced tumor burden post-RT, consistent with published in vitro observations (Fig. 3A; Supplementary Fig. S3; ref. 38). Most TP53mut tumors showed tumor volume measurement differences in the range of −25% to +25% relative to pre-RT measurement, whereas TP53wt tumors demonstrated a 25% or greater decrease in tumor volume (Fig. 3B and C). Moreover, tumor volume estimates remained stable among TP53wt patients up to 12 months, whereas TP53mut tumor volumes increased at six to nine months post-RT (Fig. 3D). Representative imaging pre- and post-RT are shown for P-01 (H3.3 K27M, TP53wt; Fig. 3E) and P-36 (H3.3 K27M, TP53mut; Fig. 3F).
Figure 3.
Somatic TP53 mutations predict poor radiographic response after radiotherapy in patients with H3K27-altered DIPG. A, Association between somatic driver gene status and change in tumor volume (top) and tumor size measured by anterior-posterior (AP) and transverse (TR) dimensions (bottom) post-RT (n = 25 H3K27-altered DIPG patients with available pre- and post-RT MRI data). Colored dots show mutant driver genes that are significantly (P < 0.05) associated with an increase (red) or decrease (blue) in tumor volume/size post-RT. B, Scatter plot comparing the percentage of change pre- and post-RT tumor volume versus tumor size across TP53mut (n = 17; red dots) and TP53wt (n = 8, blue dots) H3K27-altered DIPG. C, Box plot comparing tumor volumes stratified by TP53 mutation status based on post-RT MR images in patients enrolled in PNOC003 (TP53mut, n = 17, red box; TP53wt, n = 8, blue box). D, Line graph showing longitudinal changes in tumor volume from time of initial diagnosis up to 12 months from subjects enrolled in PNOC003 based on volumetric tumor assessment on MRI (n = 99 MRI scans; TP53mut, n = 17, red line; TP53wt, n = 8, blue line). E and F show a representative example of pre- and post-RT MRI tumor volume for patients with a TP53wt (C) and TP53mut (F) H3.3 K27M-mutant DIPG. Yellow area marks tumor outline. RT, radiotherapy; wt, wild-type; mut, mutant; MRI, magnetic resonance imaging; AP, anterior–posterior; TR, transverse. ***, P < 0.001.
TP53-associated genome instability is associated with recurrent loss of 10q/PTEN
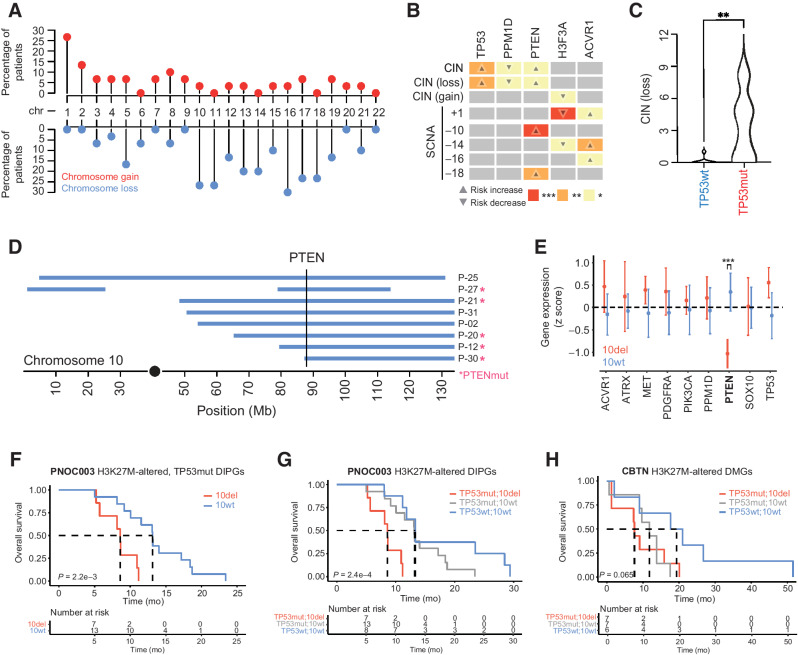
To better understand the mechanism behind TP53 as a biomarker for worse clinical outcomes and given the role of TP53 in genome instability across cancers (39), we next analyzed chromosomal gain and loss events and CIN in H3K27-altered DIPGs. Recurrent chromosomal alterations affected several chromosomes with the highest frequency of loss events on chromosomes 10, 11, 13, 14, 16, 17, and 18, and recurrent gain events on chromosome 1 (Fig. 4A). TP53 mutations showed the strongest association with CIN and, more specifically with chromosomal losses, consistent with similar observations in TP53mut SHH-medulloblastoma (40) and other pediatric brain tumor entities (ref. 41; Fig. 4B and C).
Figure 4.
Somatic driver gene alterations are associated with distinct patterns of chromosome instability in H3K27-altered DIPG. A, Lollipop plot showing the frequency of somatic chromosomal gain and loss events in H3K27-altered DIPGs (n = 30). Percentages on the left show the proportion of primary tumors with chromosomal gains or losses (middle row). Red dots represent full/partial chromosome gains; blue dots represent full/partial chromosome losses. B, Association between somatic driver gene alterations (top side), CIN (left side), and SCNAs (left side) in H3K27-altered DIPGs. Box color and associated number of asterisks indicate the degree of statistical significance (colored boxes). Direction of the arrow indicates an increased risk of association (up-arrow) or decreased risk of association (down-arrow). C, Plot shows the total number of chromosomal losses in TP53mut (n = 20) and TP53wt (n = 8) H3K27-altered DIPGs. D, Somatic PTEN alterations are associated with SCNAs on 10q. Plot shows the genomic position of somatic deletions (blue bars) on chromosome 10 and somatic PTEN alterations (pink asterisk). The vertical line marks the genomic location of the PTEN gene. E, Association between driver gene expression and 10q deletion status in H3K27M-altered DIPG. PTEN expression is significantly reduced in DIPGs that harbor a 10q deletion (Mann–Whitney U test). F, Kaplan–Meier survival curves show poor clinical outcomes in H3K27-altered, TP53-mutant DIPG patients in PNOC003. G and H, Kaplan–Meier survival curves for PNOC003 (C) and CBTN (D) H3K27-altered DIPG/DMG patients after stratification into three genetically defined risk groups: TP53mut/10del (red, highest risk), TP53mut/10wt (gray, intermediate risk), and TP53wt/10wt (blue, lowest risk). SCNA, somatic copy-number alterations; CIN, chromosomal instability; wt, wild-type; mut, mutant; del, deletion; CBTN, Children's Brain Tumor Network; ***, P < 0.001; **, P < 0.01; *, P < 0.05.
Furthermore, we validated the association between TP53 mutations and CIN in 21 H3K27M-mutant DMGs from CBTN (Supplementary Fig. S4A and S4B). PTEN alterations were also associated with CIN (P = 2.0e−2) and specifically chromosome losses (P = 2.3e−2; Fig. 4B). All PTEN-altered tumors were, however, also positive for TP53 mutations and specifically associated with loss of chromosome 10 (P = 3.9e−4; Fig. 4B). In contrast, PPM1D mutations were associated with genome stability (P = 3.1e−2; Fig. 4B). Given the observed co-occurrence of TP53 and PTEN mutations with chromosome 10 loss events, we evaluated patterns of somatic copy-number alterations along chromosome 10. Chromosomal breakpoints consistently converged on the full or terminal loss of 10q (Fig. 4D), associated with PTEN loss of heterozygosity (n = 5 out of 8 are PTEN-altered tumors; Fig. 4D), and associated with reduced PTEN expression (Fig. 4E). We assessed the clinical relevance of CIN and observed that loss of chromosome 10q was significantly associated with poor clinical outcome in H3K27-altered DIPGs (n = 28, OS 8.6 vs. 13.2 mo, P = 1.1e−4, Supplementary Fig. S1C and S1D).
Loss of 10q associates with poor clinical outcome in H3K27-altered, TP53-mutant DIPGs
We next assessed the joint clinical relevance of genomic biomarkers found in our exploratory analyses. We observed that loss of 10q occurred almost exclusively in TP53-mutant tumors in the PNOC003 H3K27-altered DIPG cohort (Supplementary Fig. S4C), which we validated in the CBTN H3K27-altered DMG cohort (Supplementary Fig. S4C). We, therefore, assessed the clinical impact of chromosome 10q loss among TP53-mutant H3K27-altered DMGs and found that this event correlates with worse OS among this subgroup (Fig. 4F). These results suggest that even within H3K27-altered, TP53-mutant DMGs, distinct molecular subgroups with unique clinical outcomes exist. Survival analyses were then expanded to include patients with both genomic biomarkers. Patients with TP53 mutations and loss of chromosome 10q demonstrated the shortest OS, whereas patients with wild-type TP53 and retention of chromosome 10q demonstrated the most prolonged OS (Fig. 4G and H).
Joint analysis of PNOC003 and CBTN cohorts (n = 49) further demonstrated that TP53mut [hazards ratio (HR), 2.33; P = 3.2e−2] and loss of chromosome 10q (HR, 2.34; P = 2.2e−2) are independent prognostic biomarkers of clinical outcome (TP53mut/10qdel, n = 14, OS, 8.4; 95% CI, 7.4–15.8); TP53mut/10qwt, n = 20, OS, 13.1 mo; 95% CI, 10.1–17.2; TP53wt/10qwt, n = 14, OS, 15.5 mo; 95% CI, 11.8–29.4; P = 2.2e−3; Supplementary Fig. S1E and S1F). These results suggest that loss of chromosome 10q and/or PTEN is an added adverse genomic event in H3K27-altered, TP53-mutant DIPG/DMGs and warrant validation in future, larger patient cohorts.
Exploratory molecular profiling and contemporary diagnostic criteria result in updated diagnoses in three patients
In addition to revealing impacts on clinical outcome, WGS, mRNA-seq, and DNA methylation profiling led to updated diagnoses in three patients. Given that our trial was initiated before the 2016 and 2021 WHO classification of central nervous system tumors (19, 42), we re-analyzed patient clinical data with newly published diagnostic criteria and, according to the study determined histopathology, combined with molecular profiling. Patient P-04 was initially diagnosed with diffuse astrocytoma, IDH- and H3-wildtype, WHO grade 2 based on immunohistochemistry. Gene expression profiling identified overexpression of EZHIP, and DNA methylation profiling classified this tumor as “DMG, H3K27M-mutant” with a calibrated score of 0.96 (Supplementary Fig. S5; ref. 19). The clinical course and survival were consistent with this diagnosis with an OS of 13.2 months.
In contrast, patients P-11 and P-19 were diagnosed with anaplastic astrocytoma, H3-wild-type, WHO grade 3, and diffuse astrocytoma, NOS, respectively, and demonstrated exceptional OS. P-11 survived 43.7 months, and P-19 remained alive past data cutoff. Molecular analysis of a biopsied specimen from patient P-11 (25 years old at diagnosis) did not reveal any molecular alterations consistent with a contemporary diagnosis of DMG but harbored a somatic IDH1 R132H mutation consistent with adult-type IDH-mutant astrocytoma. Similarly, the tumor from patient P-19 lacked identifiers consistent with H3K27-altered DMG and instead demonstrated biallelic NF1 alterations and a focal homozygous deletion of CDKN2A/B. DNA methylation profiling of tumor tissue clustered this patient with “anaplastic pilocytic astrocytoma” with a calibrated score of 0.85.
Impact of H3K27M-mutant plasma ctDNA level on RT response and survival outcome
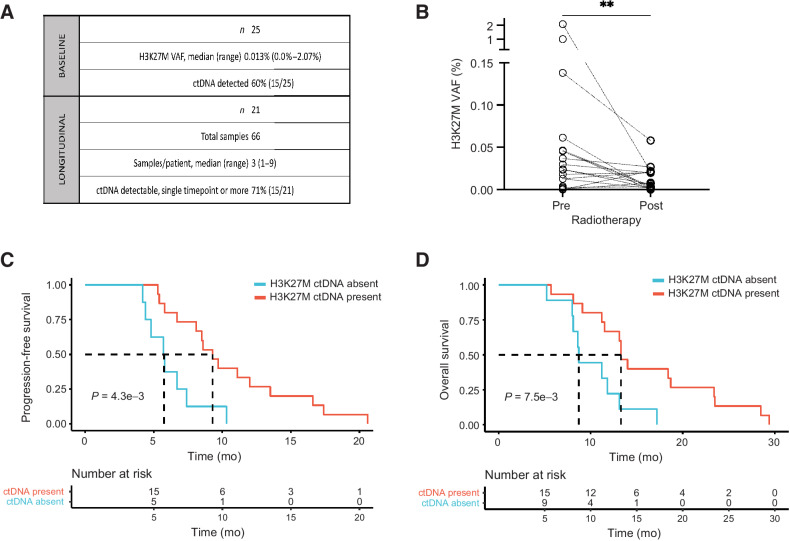
For additional biomarker analysis, plasma was collected from patients at initial diagnosis (n = 25) and longitudinally during therapy (n = 21; Fig. 5A). H3K27M-mutant ctDNA was detected in 60% (n = 15) of patients at baseline. We observed a significant decrease in plasma H3K27M ctDNA between upfront diagnosis and the first timepoint post-RT (Fig. 5B). Although three patients showed markedly high upfront ctDNA levels (Fig. 5B), our associations between ctDNA level and treatment response were maintained in the absence of these exemplary cases (Supplementary Fig. S6A–S6C). Moreover, the absence of detectable H3K27M ctDNA at baseline was significantly associated with shorter PFS (P = 4.3e−3, Fig. 5C) and OS (P = 7.5e−3, Fig. 5D). Among patients with detectable H3K27M ctDNA at baseline, those who exhibited a decrease in ctDNA (>0.01%) post-RT showed slightly longer PFS and OS (not significant, Supplementary Fig. S6D–S6G). The value of Δ 0.01% was selected on the basis of median variant allele frequency (VAF) change from pre- to post-RT ctDNA among patients with detectable H3K27M ctDNA at baseline; this value is above the previously published cutoff value of true positive plasma samples (0.001%; ref. 25).
Figure 5.
H3K27M-mutant plasma ctDNA associates with clinical outcomes in DIPG. A, Summary table with baseline and longitudinal plasma ctDNA collection in PNOC003. B, Change in plasma H3K27M-mutant ctDNA VAF pre- and post-RT in PNOC003 cohort. C and D, Kaplan–Meier PFS (C) and OS (D) curves after stratification of patients with (present) and without (absent) detectable plasma H3K27M-mutant ctDNA at baseline. VAF, variant allele frequency; ctDNA, circulating tumor DNA; RT, radiotherapy; PFS, progression-free survival; OS, overall survival. **, P < 0.01.
PNOC003 patient-derived cell lines exhibit varied molecular fidelity to corresponding human tumor
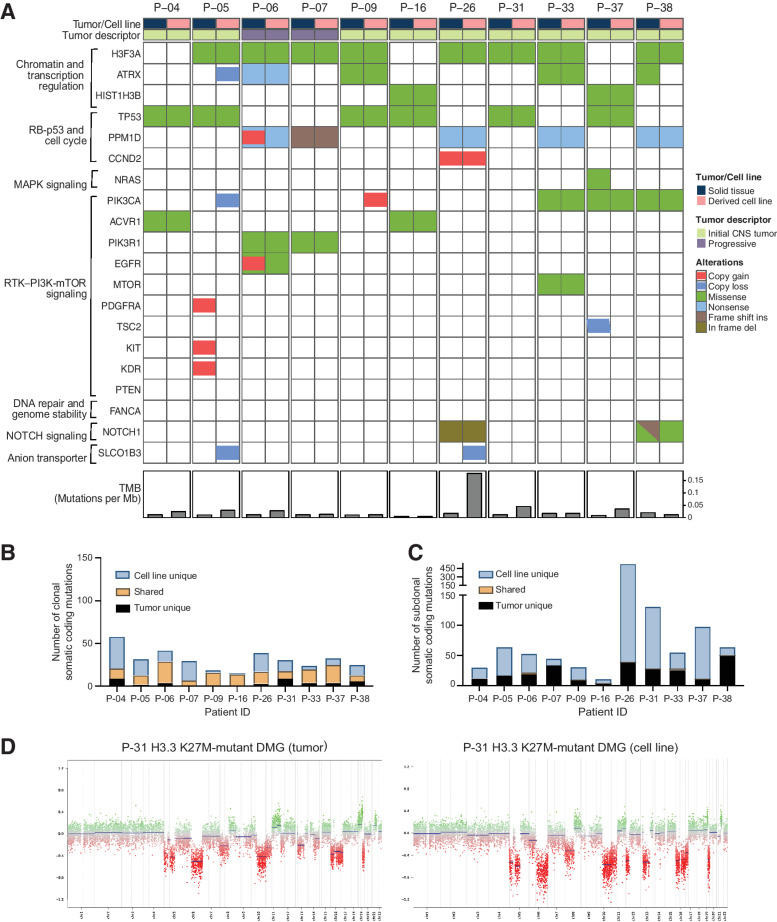
As a part of tissue collection for all patients, primary cell line generation was attempted when sufficient tissue was available. We established 11 primary DIPG cell lines (diagnosis, n = 9; progression, n = 2), subsequently evaluated by WGS and DNA methylation profiling. Key somatic driver gene alterations representative of DIPG were retained in all cell lines, including H3K27M (n = 10 out of 10), p53 pathway mutations (TP53, n = 6 out of 6; PPM1D, n = 5 out of 5), and oncogenic alterations in several components of the RTK/PI3K/mTOR pathway (Fig. 6A). Overall, tumor mutation burden was higher in 54% of cell lines when compared with paired biopsy tissue, consistent with other reports (refs. 43, 44; n = 6; Fig. 6A, bottom). We compared somatic coding mutations between paired tumors and derived cell lines, both at the clonal (Fig. 6B) and subclonal level (Fig. 6C). We found that although the majority of clonal DIPG driver gene alterations were retained, several unique subclonal alterations were identified in cell lines (Fig. 6B and C; Supplementary Tables S5 and S6), suggesting divergence at the genetic level in patient-derived models. In contrast, analyses of DNA methylation array-derived genomic profiles revealed concordant global chromosome-level events (Supplementary Fig. S7). A representative copy-number profile for a primary DIPG tumor and derived cell line is Fig. 6D.
Figure 6.
Genomic fidelity of DIPG cell lines derived from primary and progressive tumor biopsies. A, Oncoprint of WGS-derived somatic driver gene alterations for 11 DIPG cell lines and matched tumor tissue samples (biopsy at diagnosis, n = 9; biopsy at progression, n = 2). B and C, Total number of nonsynonymous gene mutations in 11 DIPG cell lines and matched tumors with clonal (B) and subclonal mutations (C). Somatic mutations in DIPG cell lines and matched tumors are shown in orange, and mutations present only in cell lines and matched tumors are shown in blue and black, respectively. D, DNA methylation-based somatic copy-number profile of a representative H3.3K27M-mutant DIPG cell line and matched primary tumor biopsy sample. WGS, whole-genome sequencing; TMB, tumor mutation burden; VAF, variant allele frequency; clonal, VAF >0.20; sub-clonal, VAF 0.05–0.20.
Genomic analysis of progressive and post-mortem tissue revealed conservation of major somatic driver mutations targeted in the clinical trial
We performed WES and mRNA-seq on biopsies at the progression from two patients (P-06 and P-07; ref. 20) and WGS on post-mortem tissue from different anatomical locations from four patients (P-04, P-07, P-13, and P-18; Supplementary Fig. S8). We observed retention of key oncogenic mutations between biopsy and post-mortem tissues, including TP53 (P-04, P-13, and P-18) and H3F3A (P-07, P-13). In the case of P-07 (PFS 4.4 mo, OS 8.0 mo), a subclonal TP53 mutation (5% VAF) was identified at diagnosis, yet lost at progression and in post-mortem tissue. In contrast, this patient harbored a clonal PPM1D mutation (33% VAF) at diagnosis, which was retained at progression and post-mortem. Comparison of mRNA-seq between diagnostic tissue and at time of progression revealed new overexpression of FOSB and TOP2A in P-06 and P-07 and DDR2 in P-07.
Discussion
Despite diverse approaches in early-phase clinical trials, there has been no progress in improving survival for children with DIPG over decades. The advent of molecular profiling in clinically meaningful timeframes, including multi-omics analyses incorporating WGS/WES, mRNA-seq, and DNA methylation profiling, offers great promise in how we diagnose, understand, and treat DIPG and, more contemporarily, H3K27-altered DMG. The current study harnessed multi-omics profiling to develop a precision medicine approach for children and young adults with newly diagnosed DIPG.
The primary goal of PNOC003 was to assess the impact of biology-based, multi-agent therapy in newly diagnosed pediatric DIPG. The study indicates the feasibility of such an approach (20), supports the safety of surgical biopsy for DIPG tumors, and highlights biopsy in ensuring accurate diagnosis and treatment decision making. These findings align with recent publications on the feasibility of biopsy in patients with DIPG and agree with earlier reports (22, 45–47). In addition, the tolerability of multi-drug regimens supports future combinatorial strategies and consideration of combinations with RT, which may further survival impact.
The lack of clinical benefit of PNOC003 raises the question of how the approach is failing, including the limited availability of drugs that effectively address critical driver genes such as TP53. The drug decision-making in our trial was based on a structured prioritization of drug selection and preclinical and/or clinical work that supported targeting specific molecular aberrations (20). Recent efforts like CNS-TAP now incorporate algorithms, which include blood–brain-barrier penetration and compare clinical promise across multiple drugs from the same class (48). Hopefully, these efforts combined with more effective exploration of tumor penetration and target inhibition and drug development aimed at key driver pathways highlighted in our work will further the clinical benefit of precision medicine-based efforts for DIPG/DMG.
Perhaps, our most significant molecular finding was the identification of a novel genomic subtype with H3K27 alteration, TP53 mutations, and associated loss of 10q/PTEN. This molecular combination demonstrated the worst survival outcome among DIPG/DMGs in PNOC003. Although we recognize that our small sample size requires confirmation in larger cohorts, TP53 mutations are associated with both worse OS and RT resistance in our cohort. This was consistent with prior findings (38, 49) and corroborated in an independent CBTN H3K27-altered DMG cohort. Our findings indicate that patients with H3K27-altered, TP53-mutant DMG may most directly benefit from radio-sensitizing agents and warrant investigation into the impact of reirradiation at progression across subtypes.
Of additional clinical interest is that TP53 mutations are associated with significant genomic instability and primarily chromosomal losses, which may lead to loss of 10q/PTEN. Together, these alterations predicted the worst clinical outcome in our cohort. Although we attempted to target PTEN alterations with everolimus in PNOC003 patients, this agent is not specific for PTEN. We are hopeful that newer, more specific agents (e.g., alpelisib) will improve clinical practice responses. We advocate that ongoing work confirms this new high-risk molecular stratification system in larger sample size, verifies that it should be aggressively targeted in our patient population, and hones focus on future drug development.
Two subjects enrolled on PNOC003 underwent repeat biopsy at the time of disease progression, without experiencing toxicity related to the second biopsy. We recognize this is a small sample size but provides early support for consideration of serial biopsies if the information obtained can affect subsequent therapy options. The timing of such biopsies remains to be determined but should coincide with the collection of circulating biomarkers to help validate this approach. We demonstrated that collection of plasma ctDNA enabled detection of clinically relevant driver mutations (H3F3A and HIST1H3B K27M) at diagnosis in our patient population. Interestingly, the absence of detectable ctDNA at baseline correlated with worse PFS. One possible explanation is that lack of ctDNA in the periphery is a marker of a denser tumor with a lower likelihood for drug penetration or could be due to an intact BBB restricting passing of ctDNA into the periphery. We attempted to correlate imaging characteristics with ctDNA to answer this question; however, our small sample size was limiting. Regardless, given the association between upfront ctDNA level and survival outcome and observed decreases in post-RT H3K27M-mutant ctDNA, the clinical impact of ctDNA warrants ongoing investigation and should be expanded to additional circulating tumor biomarkers in both blood and CSF (50).
Our study is the first to complete multi-omic profiling of patient samples to direct therapy and integrate results across multiple platforms, including CLIA WES, mRNA-seq, WGS, and DNA methylation, allowing comparison of molecular findings. This work provides insight into the variability in diagnoses and treatments that may occur based on the molecular platform used and indicates that larger-scale studies are needed to elucidate which platforms are most clinically informative. Furthermore, cases where diagnoses were updated highlight the benefit of more extensive molecular profiling when standard diagnostic criteria for DIPG are not met, particularly for clinical trial eligibility, treatment decision-making, and prognostication. Our work also highlights the successful development of patient-derived preclinical models and the molecular variability that can occur between patient samples and patient-derived models, particularly with divergent partner alterations. Such variability informs on possible pitfalls of preclinical models in translational efforts and preclinical drug discovery. In depth RNA sequencing, methylation profiling, and large-scale drug screen studies are actively underway using the PNOC003 cell lines in addition to an expanded cohort of DMG primary derived cell lines. We anticipate that these findings will grow our understanding of potential mechanisms of drug resistance and tumor escape from targeted therapies in our patient cohort and inform the next iteration of precision-based therapies for DMG.
A limitation of our study is the small sample size. PNOC003 was intended to confirm the feasibility of a new treatment paradigm. Yet, even with a small sample size, our comprehensive molecular findings can inform future translational efforts and treatment decision-making for DIPG. We aimed to address the shortcomings of a small cohort through validation of key molecular findings in an external, independent H3K27-altered DMG cohort. We also recognize that we limited ourselves to FDA-approved drugs and drugs that were delivered orally or intravenously. By broadening treatment recommendations to therapies that may still be undergoing clinical investigation and with a focus on the aggressive genotype-phenotype relationships in our cohort, we may better extend the clinical benefit to patients with DIPG. Our experience in PNOC003 supports the exploration of novel drug delivery systems and drug combinations in future clinical trials and addressing drug penetration and pharmacodynamics (PD) a priori. We are currently exploring the impact of drug penetration, PD biomarkers, and more comprehensive preclinical work before clinical translation in follow-up trials to PNOC003, specifically DMG-ACT (PNOC023, NCT04732056; PNOC022, NCT05009992) and PNOC008 (NCT03739372). We also recognize that we did not pre-emptively explore the impact of our therapy on targeted pathways in longitudinal DNA and RNA sequencing and were limited by the number of patients that underwent follow up biopsy at progression. However, we found ongoing PDGFRA overexpression in P-07, despite aiming to target this with mebendazole. We also identified MAP3K8 overexpression in the patient's tumor after upfront targeting of a PIK3R1 alteration. This could be indicative of alternative pathway activation driven by use of everolimus. We intend to overcome future limitations in pathway analysis by implementing treat-biopsy-treat approaches in larger sample sizes, such as PNOC016, a target validation study inclusive of DIPG and DMG (NCT03893487), and DMG-ACT trials (NCT05009992; NCT04732065). These larger cohorts will further investigate RNA expression patterns and to correlate with driver pathways. In addition, ongoing trials, such as DMG-ACT, are assessing active molecular pathways in DMG a priori via antibody staining of tissue both at diagnosis and post-therapy.
In summary, PNOC003 is the first clinical trial to bring together a complement of clinical, multi-omic profiling to determine a combination therapy approach while exploring biologic endpoints that inform the next generation of therapy for children and young adults with newly diagnosed DIPG. Together, our work (i) confirms proof of concept for multi-targeted, multi-agent combinations in DIPG, (ii) highlights TP53 and 10q/PTEN alterations as potential mechanisms of therapeutic resistance with uniquely poor prognoses in H3K27-altered DMG, and (iii) supports future investigation of next-generation approaches and drug delivery systems to target the most aggressive subtypes of DIPG/DMG.
Supplementary Material
Acknowledgments
The authors want to acknowledge all patients and families for participating in this study, the clinical staff at the PNOC sites who cared for these patients; the Brain Tumor Center Tissue Core at UCSF for managing samples collected as part of the clinical trial; members of the Children's Brain Tumor Network (CBTN) and D3b team, including Komal S. Rathi, Yuankun Zhu, Nicholas Van Kuren, Meen Chul Kim, Bailey Farrow, Allison P. Heath, Alexander Sickler, Miguel A. Brown, Tejaswi Koganti; all members of PNOC; members of the TGEN team, including Daniel Enriquez, Tyler Izatt, and Emily Cannon; and the UCSF Department of Neurological Surgery for continual support. This work was funded by the V foundation (Atlanta, GA); the Pacific Pediatric Neuro-Oncology Foundation (Larkspur, CA), the Pediatric Brain Tumor Foundation (Asheville, NC); Charlie Kerr and Isabella Kerr Molina Foundation; TGEN Foundation; Dragon Master Foundation; Kortney Rose Foundation; Musella Foundation; Project Open DIPG; The Gabriella Miller Kids First Data Resource Center; Goldwin Foundation; Smashing Walnuts Foundation; Piedmont Community Foundation; Matthew Larson Foundation; Kaminsky Foundation; Jenny's Quest Foundation; Research Council of Norway; University of Oslo; South-Eastern Norway Regional Health Authority; and generous donations from families and patients.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
C. Kline reports other support from Regeneron Pharmaceuticals, Bristol Myers Squibb, Day One Biopharmaceuticals, Midatech Pharma, Ipsen, Curis Inc., Chimerix, Kazia, and Secura Bio outside the submitted work. L. Kilburn reports grants and other support from UCSF/V Foundation during the conduct of the study. J. Villanueva-Meyer reports grants from GE Healthcare outside the submitted work. J. Nazarian reports grants from ChadTough Defeat DIPG Foundation and The Lilabean Foundation during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
C. Kline: Data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. P. Jain: Data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. L. Kilburn: Conceptualization, data curation, supervision, investigation, methodology, project administration, writing–review and editing. E.R. Bonner: Data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. N. Gupta: Conceptualization, data curation, supervision, investigation, methodology, writing–review and editing. J.R. Crawford: Conceptualization, data curation, supervision, investigation, methodology, writing–review and editing. A. Banerjee: Data curation, investigation, writing–review and editing. R.J. Packer: Data curation, formal analysis, investigation, writing–original draft, writing–review and editing. J. Villanueva-Meyer: Formal analysis, writing–original draft, writing–review and editing. T. Luks: Formal analysis, validation, writing–original draft, writing–review and editing. Y. Zhang: Formal analysis, validation, investigation, methodology, writing–original draft, writing–review and editing. M. Kambhampati: Formal analysis, investigation, methodology, writing–original draft, writing–review and editing. J. Zhang: Investigation, methodology, writing–original draft, writing–review and editing. S. Yadavilli: Software, formal analysis, supervision, validation, investigation, methodology, writing–original draft, writing–review and editing. B. Zhang: Software, formal analysis, supervision, validation, investigation, visualization, methodology, writing–review and editing. K.S. Gaonkar: Conceptualization, data curation, software, formal analysis, validation, investigation, methodology, writing–original draft, writing–review and editing. J.L. Rokita: Conceptualization, data curation, formal analysis, supervision, validation, investigation, methodology, writing–original draft, writing–review and editing. A. Kraya: Conceptualization, data curation, software, formal analysis, supervision, validation, investigation, methodology, writing–original draft, writing–review and editing. J. Kuhn: Conceptualization, data curation, formal analysis, supervision, funding acquisition, validation, investigation, methodology, writing–original draft, writing–review and editing. W. Liang: Conceptualization, resources, data curation, formal analysis, supervision, validation, investigation, methodology, writing–original draft, writing–review and editing. S. Byron: Conceptualization, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. M. Berens: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. A. Molinaro: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. M. Prados: Conceptualization, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. A. Resnick: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. S.M. Waszak: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. J. Nazarian: Data curation, software, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. S. Mueller: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing.
References
- 1. Cohen KJ, Heideman RL, Zhou T, Holmes EJ, Lavey RS, Bouffet E, et al. Temozolomide in the treatment of children with newly diagnosed diffuse intrinsic pontine gliomas: a report from the Children's Oncology Group. Neuro Oncol 2011;13:410–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Macy ME, Kieran MW, Chi SN, Cohen KJ, MacDonald TJ, Smith AA, et al. A pediatric trial of radiation/cetuximab followed by irinotecan/cetuximab in newly diagnosed diffuse pontine gliomas and high-grade astrocytomas: A Pediatric Oncology Experimental Therapeutics Investigators' Consortium study. Pediatr Blood Cancer. 2017 Nov;64(11):10.1002/pbc.26621. doi: 10.1002/pbc.26621. Epub 2017 May 24. PMID: 28544128; PMCID: PMC5605460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vanan MI, Eisenstat DD. DIPG in Children—What Can We Learn from the Past? Front Oncol 2015;5:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Veldhuijzen van Zanten SEM, El-Khouly FE, Jansen MHA, Bakker DP, Sanchez Aliaga E, Haasbeek CJA, et al. A phase I/II study of gemcitabine during radiotherapy in children with newly diagnosed diffuse intrinsic pontine glioma. J Neurooncol 2017;135:307–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Veldhuijzen van Zanten SE, Jansen MH, Sanchez Aliaga E, van Vuurden DG, Vandertop WP, Kaspers GJ. A twenty-year review of diagnosing and treating children with diffuse intrinsic pontine glioma in the Netherlands. Expert Rev Anticancer Ther 2015;15:157–64. [DOI] [PubMed] [Google Scholar]
- 6. Souweidane MM, Kramer K, Pandit-Taskar N, Zhou Z, Haque S, Zanzonico P, et al. Convection-enhanced delivery for diffuse intrinsic pontine glioma: a single-centre, dose-escalation, phase 1 trial. Lancet Oncol 2018;19:1040–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fontebasso AM, Papillon-Cavanagh S, Schwartzentruber J, Nikbakht H, Gerges N, Fiset PO, et al. Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat Genet 2014;46:462–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol 2012;124:439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 2012;44:251–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Buczkowicz P, Hoeman C, Rakopoulos P, Pajovic S, Letourneau L, Dzamba M, et al. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet 2014;46:451–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 2013;27:985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jones C, Karajannis MA, Jones DTW, Kieran MW, Monje M, Baker SJ, et al. Pediatric high-grade glioma: biologically and clinically in need of new thinking. Neuro Oncol 2017;19:153–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mackay A, Burford A, Carvalho D, Izquierdo E, Fazal-Salom J, Taylor KR, et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell 2017;32:520–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saratsis AM, Yadavilli S, Magge S, Rood BR, Perez J, Hill DA, et al. Insights into pediatric diffuse intrinsic pontine glioma through proteomic analysis of cerebrospinal fluid. Neuro Oncol 2012;14:547–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yadavilli S, Scafidi J, Becher OJ, Saratsis AM, Hiner RL, Kambhampati M, et al. The emerging role of NG2 in pediatric diffuse intrinsic pontine glioma. Oncotarget 2015;6:12141–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nagaraja S, Quezada MA, Gillespie SM, Arzt M, Lennon JJ, Woo PJ, et al. Histone variant and cell context determine H3K27M Reprogramming of the enhancer landscape and oncogenic State. Mol Cell 2019;76:965–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Taylor KR, Mackay A, Truffaux N, Butterfield Y, Morozova O, Philippe C, et al. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat Genet 2014;46:457–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taylor KR, Vinci M, Bullock AN, Jones C. ACVR1 mutations in DIPG: lessons learned from FOP. Cancer Res 2014;74:4565–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO Classification of tumors of the central nervous system: a summary. Neuro Oncol 2021;23:1231–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mueller S, Jain P, Liang WS, Kilburn L, Kline C, Gupta N, et al. A pilot precision medicine trial for children with diffuse intrinsic pontine glioma-PNOC003: a report from the Pacific Pediatric Neuro-Oncology Consortium. Int J Cancer 2019;145:1889–901. [DOI] [PubMed] [Google Scholar]
- 21. Mueller S, Taitt JM, Villanueva-Meyer JE, Bonner ER, Nejo T, Lulla RR, et al. Mass cytometry detects H3.3K27M-specific vaccine responses in diffuse midline glioma. J Clin Invest 2020;130:6325–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gupta N, Goumnerova LC, Manley P, Chi SN, Neuberg D, Puligandla M, et al. Prospective feasibility and safety assessment of surgical biopsy for patients with newly diagnosed diffuse intrinsic pontine glioma. Neuro Oncol 2018;20:1547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nasser S, Kurdolgu AA, Izatt T, Aldrich J, Russell ML, Christoforides A, et al. An integrated framework for reporting clinically relevant biomarkers from paired tumor/normal genomic and transcriptomic sequencing data in support of clinical trials in personalized medicine. Pac Symp Biocomput 2015:56–67. [PubMed] [Google Scholar]
- 24. Byron SA, Tran NL, Halperin RF, Phillips JJ, Kuhn JG, de Groot JF, et al. Prospective feasibility trial for genomics-informed treatment in recurrent and progressive glioblastoma. Clin Cancer Res 2018;24:295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Panditharatna E, Kilburn LB, Aboian MS, Kambhampati M, Gordish-Dressman H, Magge SN, et al. Clinically relevant and minimally invasive tumor surveillance of pediatric diffuse midline gliomas using patient-derived liquid biopsy. Clin Cancer Res 2018;24:5850–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bonner ER, Saoud K, Lee S, Panditharatna E, Kambhampati M, Mueller S, et al. Detection and Monitoring of Tumor Associated Circulating DNA in Patient Biofluids. J Vis Exp. 2019Jun 8;(148). doi: 10.3791/59721. PMID: 31233019. [DOI] [PubMed] [Google Scholar]
- 27. Mueller S, Haas-Kogan DA. WEE1 Kinase as a target for cancer therapy. Journal Clin Oncol 2015;33:3485–7. [DOI] [PubMed] [Google Scholar]
- 28. Shapiro JA, Savonen CL, Bethell CJ, Gaonkar KS, Zhu Y, Brown MA, et al. An open pediatric brain tumor atlas. 2022. Available from: https://alexslemonade.github.io/OpenPBTA-manuscript/. [DOI] [PMC free article] [PubMed]
- 29. Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, et al. Universal patterns of selection in cancer and somatic tissues. Cell 2017;171:1029–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martinez-Jimenez F, Muinos F, Sentis I, Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, et al. A compendium of mutational cancer driver genes. Nat Rev Cancer 2020;20:555–72. [DOI] [PubMed] [Google Scholar]
- 31. Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation-based classification of central nervous system tumours. Nature 2018;555:469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grasso CS, Tang Y, Truffaux N, Berlow NE, Liu L, Debily MA, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med 2015;21:555–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nygren P, Fryknas M, Agerup B, Larsson R. Repositioning of the anthelmintic drug mebendazole for the treatment for colon cancer. J Cancer Res Clin Oncol 2013;139:2133–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nygren P, Larsson R. Drug repositioning from bench to bedside: tumour remission by the antihelmintic drug mebendazole in refractory metastatic colon cancer. Acta Oncol 2014;53:427–8. [DOI] [PubMed] [Google Scholar]
- 35. Du L, Li X, Zhen L, Chen W, Mu L, Zhang Y, et al. Everolimus inhibits breast cancer cell growth through PI3K/AKT/mTOR signaling pathway. Mol Med Rep 2018;17:7163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang XY, Hu QP, Shi HY, Zheng YY, Hu RR, Guo Q. Everolimus inhibits PI3K/Akt/mTOR and NF-kB/IL-6 signaling and protects seizure-induced brain injury in rats. J Chem Neuroanat 2021;114:101960. [DOI] [PubMed] [Google Scholar]
- 37. Ullrich NJ, Prabhu SP, Reddy AT, Fisher MJ, Packer R, Goldman S, et al. A phase II study of continuous oral mTOR inhibitor everolimus for recurrent, radiographic-progressive neurofibromatosis type 1-associated pediatric low-grade glioma: a Neurofibromatosis Clinical Trials Consortium study. Neuro Oncol 2020;22:1527–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Werbrouck C, Evangelista CCS, Lobon-Iglesias MJ, Barret E, Le Teuff G, Merlevede J, et al. TP53 Pathway alterations drive radioresistance in diffuse intrinsic pontine gliomas (DIPG). Clin Cancer Res 2019;25:6788–800. [DOI] [PubMed] [Google Scholar]
- 39. Eischen CM. Genome stability requires p53. Cold Spring Harb Perspect Med 2016;6:a026096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 2012;148:59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grobner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, et al. The landscape of genomic alterations across childhood cancers. Nature 2018;555:321–7. [DOI] [PubMed] [Google Scholar]
- 42. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016;131:803–20. [DOI] [PubMed] [Google Scholar]
- 43. Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun 2013;4:2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goodspeed A, Heiser LM, Gray JW, Costello JC. Tumor-derived cell lines as molecular models of cancer pharmacogenomics. Mol Cancer Res 2016;14:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Puget S, Blauwblomme T, Grill J. Is biopsy safe in children with newly diagnosed diffuse intrinsic pontine glioma? Am Soc Clin Oncol Educ Book 2012:629–33. doi: 10.14694/EdBook_AM.2012.32.59. PMID: 24451809. [DOI] [PubMed] [Google Scholar]
- 46. Puget S, Beccaria K, Blauwblomme T, Roujeau T, James S, Grill J, et al. Biopsy in a series of 130 pediatric diffuse intrinsic Pontine gliomas. Childs Nerv Syst 2015;31:1773–80. [DOI] [PubMed] [Google Scholar]
- 47. Hamisch C, Kickingereder P, Fischer M, Simon T, Ruge MI. Update on the diagnostic value and safety of stereotactic biopsy for pediatric brainstem tumors: a systematic review and meta-analysis of 735 cases. J Neurosurg Pediatr 2017;20:261–8. [DOI] [PubMed] [Google Scholar]
- 48. Linzey JR, Marini BL, Pasternak A, Smith C, Miklja Z, Zhao L, et al. Development of the CNS TAP tool for the selection of precision medicine therapies in neuro-oncology. J Neurooncol 2018;137:155–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Debily MA, Kergrohen T, Varlet P, Le Teuff G, Nysome K, Blomgren K, et al. PDTM-36. Whole-exome sequencing (WES) of DIPG patients from the biomede trial reveals new prognostic subgroups with specific oncogenic programmes Neuro Oncol 2019;21:vi195. [Google Scholar]
- 50. Huang TY, Piunti A, Lulla RR, Qi J, Horbinski CM, Tomita T, et al. Detection of Histone H3 mutations in cerebrospinal fluid-derived tumor DNA from children with diffuse midline glioma. Acta Neuropathol Commun 2017;5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Access to raw data can be requested from CBTN (https://cbtn.org). Code for the somatic workflows can be found at https://github.com/d3b-center/OpenPBTA-workflows. Code for downstream analyses can be found at https://github.com/AlexsLemonade/OpenPBTA-analysis/. Processed files are publicly available on CAVATICA (https://cavatica.sbgenomics.com/u/cavatica/pbta-pnoc003; https://cavatica.sbgenomics.com/u/cavatica/openpbta). Processed data can be visualized in PedcBioPortal (https://pedcbioportal.kidsfirstdrc.org).
Additional details on all Patients and Methods can be found in Supplementary Methods.