Abstract
The concepts of developing RNAs as new molecular entities for therapies have arisen again and again since the discoveries of antisense RNAs, direct RNA-protein interactions, functional noncoding RNAs, and RNA-directed gene editing. The feasibility was demonstrated with the development and utilization of synthetic RNA agents to selectively control target gene expression, modulate protein functions or alter the genome to manage diseases. Rather, RNAs are labile to degradation and cannot cross cell membrane barriers, making it hard to develop RNA medications. With the development of viable RNA technologies, such as chemistry and pharmaceutics, eight antisense oligonucleotides (ASOs) (fomivirsen, mipomersen, eteplirsen, nusinersen, inotersen, golodirsen, viltolarsen and casimersen), one aptamer (pegaptanib), and three small interfering RNAs (siRNAs) (patisiran, givosiran and lumasiran) have been approved by the United States Food and Drug Administration (FDA) for therapies, and two mRNA vaccines (BNT162b2 and mRNA-1273) under Emergency Use Authorization for the prevention of COVID-19. Therefore, RNAs have become a great addition to small molecules, proteins/antibodies, and cell-based modalities to improve the public health. In this article, we first summarize the general characteristics of therapeutic RNA agents, including chemistry, common delivery strategies, mechanisms of actions, and safety. By overviewing individual RNA medications and vaccines approved by the FDA and some agents under development, we illustrate the unique compositions and pharmacological actions of RNA products. A new era of RNA research and development will likely lead to commercialization of more RNA agents for medical use, expanding the range of therapeutic targets and increasing the diversity of molecular modalities.
Keywords: ASO, siRNA, miRNA, Aptamer, mRNA, gRNA
1. Introduction
The concept of developing RNAs as new molecular entities for therapies had arisen since the development and utilization of single-stranded oligonucleotides to act on target transcripts for the control of gene expression (Stephenson and Zamecnik, 1978; Zamecnik and Stephenson, 1978) and use of RNA molecules to directly modulate the functions of proteins or RNA-protein complexes (Stark et al., 1978; Kitajewski et al., 1986; Dingwall et al., 1989). The expectation became even higher following the discovery of genome-derived functional noncoding microRNAs (miRNAs or miRs) that govern posttranscriptional gene regulation (Lee et al., 1993; Wightman et al., 1993) and the development of double-stranded RNAs (dsRNAs) to achieve RNA interference (RNAi) (Elbashir et al., 2001; Fire et al., 1998; Zamore et al., 2000). Meanwhile, although RNA molecules derived from the human genome, including messenger RNAs (mRNAs) and functional noncoding RNAs (ncRNAs), outnumber the proteins enormously (Djebali et al., 2012), RNA macromolecules basically remain as undrugged targets. Indeed, the single-stranded antisense oligonucleotides (ASOs), double-stranded small interfering RNAs (siRNAs), and endogenous miRNAs can be unified in that the antisense strand directs particular cellular machineries to modulate posttranscriptional gene expression through the interference with target transcripts. Furthermore, exogenous mRNAs may be directly introduced into cells and translated by ribosomes into target proteins for replacement therapy or vaccination (Jirikowski et al., 1992; Mandl et al., 1998; Wolff et al., 1990). In addition, the discovery of clustered regularly interspaced short palindromic repeats-associated proteins (CRISPR-Cas) system for gene editing (Cong et al., 2013; Jinek et al., 2012; Mali et al., 2013; Mashiko et al., 2013) opens a new avenue to use guide RNAs (gRNAs) along with the Cas nuclease for the treatment of diseases. Therefore, RNA therapeutics and vaccines not only hold great promise to expand the range of druggable targets (e.g., from proteins to transcripts and the genome) but also increase the molecular diversity of medications (e.g., from small molecules and proteins to RNAs).
The development of RNA medications and vaccines has encountered many unprecedented challenges. Naked and unmodified RNAs are intrinsically susceptible to the cleavage and degradation by a variety of circulating ribonucleases (RNases) and hydrolases, and thus rapidly eliminated through renal clearance. As negatively-charged large molecules, RNAs are subjected to poor intracellular uptake and thus have insufficient access to the molecular targets within cells. With the development of viable technologies to improve the druggability of RNA molecules, including the improvement of metabolic stability through chemical modifications and utilization of efficient delivery systems (Crooke et al., 2018; Khvorova and Watts, 2017; Roberts et al., 2020; Yu et al., 2019; Yu et al., 2020b), a number of RNA medications and vaccines with specific pharmacological actions have been approved by the United States Food and Drug Administration (FDA) for the treatment and prevention of various human diseases (Fig. 1). Among them, eight are therapeutic ASOs (fomivirsen, mipomersen, eteplirsen, nusinersen, inotersen, golodirsen, viltolarsen and casimersen), one is RNA aptamer (pegaptanib), three are siRNAs (patisiran, givosiran and lumasiran), and two are mRNA vaccines (BNT162b2 and mRNA-1273).
Fig. 1.
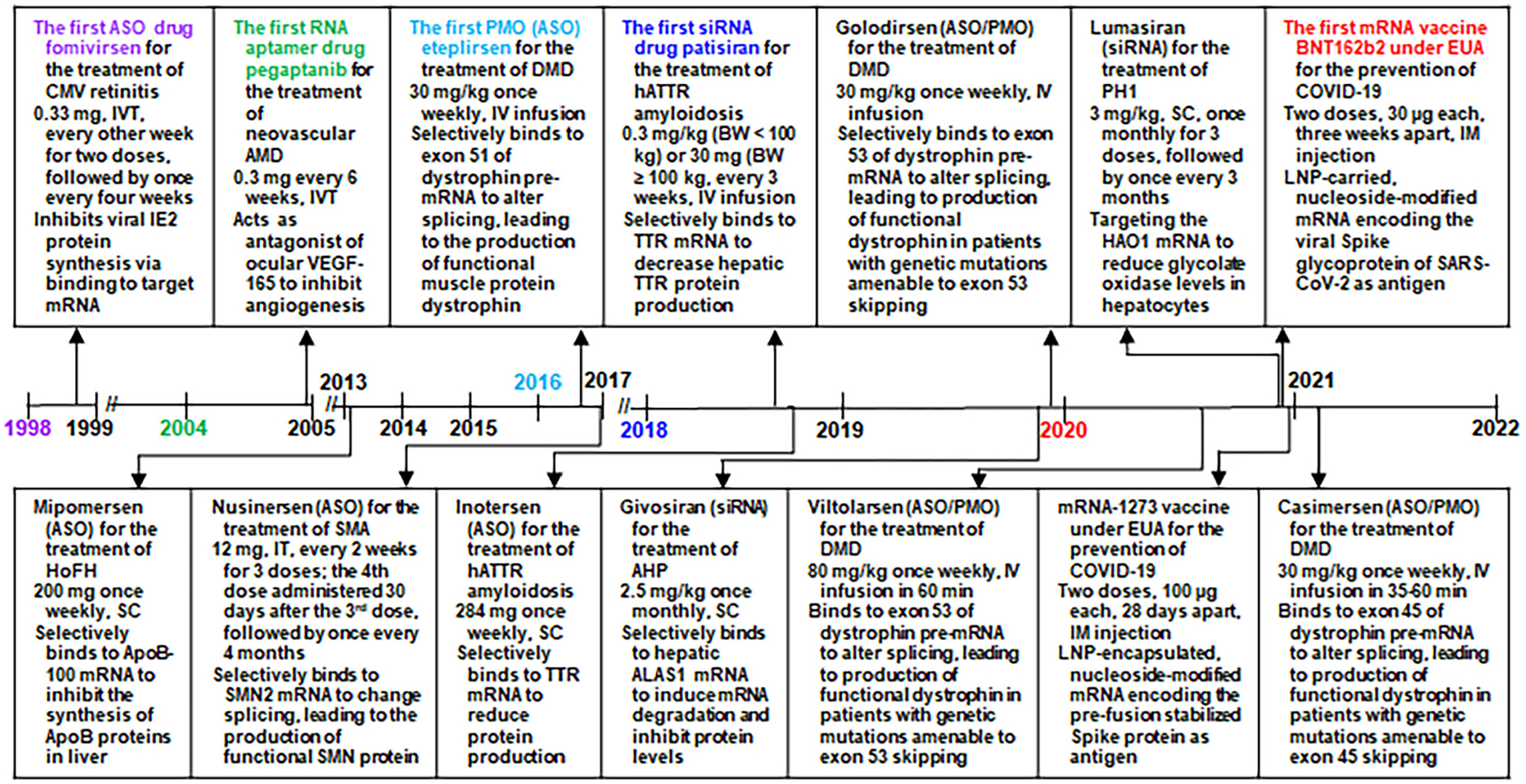
Timeline of the approval of RNA medications and vaccines by the US FDA and their indications, dosage regimens, and pharmacological actions. Note that (1) fomivirsen is an oligodeoxynucleotide acting as an ASO and it was withdrawn from the US market in 2006, (2) mipomersen was withdrawn from the US market in 2019, (3) the four PMO drugs consist of nucleobase thymine or 5-methyluracil, and (4) the two mRNA vaccines with FDA approval under emergency use authorization (EUA) are not for therapy but prevention from disease. AHP, acute hepatic porphyria; ALAS1, aminolevulinic acid synthase 1; AMD, age-related macular degeneration; ApoB, apolipoprotein B; ASO, antisense oligonucleotide; BW, body weight; COVID-19, coronavirus disease 2019; DMD, Duchenne muscular dystrophy; EUA, emergency use authorization; HAO1, hydroxyacid oxidase 1; hATTR amyloidosis, hereditary transthyretin-mediated amyloidosis; HoFH, homozygous familial hypercholesterolemia; IM, intramuscular; IV, intravenous; IVT, intravitreal injection; IT, intrathecally; LNP, lipid nanoparticle; PH1, primary hyperoxaluria type 1; PMO, phosphorodiamidate morpholino oligomer; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SC, subcutaneous; siRNA, small interfering RNA; SMA, spinal muscular atrophy; SMN, survival motor neuron; TTR, transthyretin; VEGF, vascular endothelial growth factor.
Regulatory approval of fomivirsen on August 26, 1998 for the treatment of cytomegalovirus (CMV) retinitis in AIDS patients, who are intolerant of or have a contraindication to other treatments for CMV retinitis or who were insufficiently responsive to previous treatments for CMV retinitis (Roehr, 1998), is a key milestone in drug development in that fomivirsen becomes the first nucleic acid entity acting on mRNA target for medical use among humans (Fig. 1). Pegaptanib was approved by FDA on December 17, 2004 for the treatment of neovascular age-related macular degeneration (AMD) (Ng and Adamis, 2005; Ng et al., 2006), which represents another important milestone as pegaptanib is the first true RNA analog and the first RNA aptamer drug that directly acts on a protein target. After nearly a decade of silence, seven ASO drugs including four phosphorodiamidate morpholine oligonucleotides (PMOs) were successfully marketed between 2013 and 2021 (Aartsma-Rus, 2017; Heo, 2020; Iftikhar et al., 2021; Keam, 2018; Kesselheim and Avorn, 2016; Morrow, 2013; Shirley, 2021).
Another exciting milestone in RNA drug development is the approval of the first siRNA medication, patisiran, by the FDA on August 10, 2018 for the treatment of peripheral nerve disease (polyneuropathy) caused by hereditary transthyretin-mediated (hATTR) amyloidosis in adult patients (Fig. 1) (Wood, 2018). Subsequently, two other siRNA drugs, givosiran and lumasiran, were approved by the FDA in November 2019 and November 2020 for the treatment of acute hepatic porphyria (AHP) and primary hyperoxaluria type 1 (PH1), respectively (Scott, 2020; Scott and Keam, 2021). Moreover, two mRNA vaccines have been approved most recently by the US FDA under Emergency Use Authorization (EUA) for the prevention of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection or coronavirus disease 2019 (COVID-19) (Fig. 1) (Oliver et al., 2020; Oliver et al., 2021). While recent approval of RNA medications and vaccines benefits from the improved understanding of RNA biology and advances in RNA technologies, it provides incentives for the discovery and development of new RNA agents for medical use.
In this review, we summarize the general characteristics of therapeutic RNAs, including chemistry, common delivery strategies, mechanisms of actions, and safety. By focusing on the discussion of FDA-approved RNA medications and vaccines, we provide an overview of individual RNA products and respective pharmacological actions, and outline some agents and strategies under development and evaluation.
2. Characteristics of RNA medications and vaccines
Therapeutic RNAs have their intrinsic characteristics, different from conventional small-molecule and protein drugs in many regards (Table 1). The molecular weights of polymeric RNA medications (7–20 kDa) are much greater than small-molecule medications (< 1 kDa) but generally less than protein therapeutics (>100 kDa), except that full-length mRNA drugs/vaccines are much larger in size (e.g., >100 kDa). Both RNA and protein medications are not orally bioavailable, whereas many small-molecules are. Therefore, RNA therapeutics are generally administered via intravenous or local injections. Overall pharmacokinetic properties of RNA agents including tissue distribution and intracellular uptake are highly dependent on chemical modifications and formulations or delivery vehicles. Mainly acting on target transcripts within cells towards the control of disease, the efficacy of an RNA drug is generally more relevant to tissue or intracellular exposure (Chong et al., 2021; Nair et al., 2017), but it could be linked to systemic exposure (Yu et al., 2009). Furthermore, the risk of induction of allergic reactions, such as cytokine release syndrome or cytokine storm (Table 1), is a major concern during the development and clinical use of RNA drugs, while other types of adverse events, including systemic and injection site reactions, may occur among individuals (Balwani et al., 2020; van Meer et al., 2016).
Table 1.
General characteristics of RNA therapeutics as compared with small-molecule and protein drugs. ADME, absorption, metabolism, distribution, and excretion; pharmacokinetics; PD, pharmacodynamics.
| Properties | Small-molecule drugs | Protein therapeutics | RNA therapeutics |
|---|---|---|---|
| Chemistry | Molecular weight < 1 kDa; Hydrophobic | Molecular weight >100 kDa; Positive/negative/neutral | Molecularweight 7–20 kDa; Negative charge |
| Dosing | Oral primarily, intravenous, etc.; Often daily | Mainly intravenous and subcutaneous; Weekly to monthly | Intravenous, subcutaneous, or specific local injections; Weekly to once every 3–6 months |
| ADME or PK | Orally bioavailable; Distributed to all organs and tissues; Cell permeable; Metabolized by Phase I and II drug-metabolizing enzymes; Excreted mainly in bile and urine | Not orally bioavailable; Distributed mainly in plasma or extracellular fluids; Cell impermeable; Catabolized extensively to peptides or amino acids by peptidases; Mainly urine | Not orally bioavailable; Distributed extensively to liver, kidney and lung; Intracellular delivery using various strategies; Catabolized extensively to shorter oligos or mononucleotides by nucleases or hydrolases; Mainly urine |
| PD | Target extra—/intra-cellular or cell-surface proteins mainly, and RNAs & DNAs; Direct or indirect link to blood PK | Target predominantly the cell-surface or extracellular proteins; Direct or indirect models linked to blood PK | Target intracellular RNAs primarily, as well as extra—/intra-cellular proteins, and nuclear DNAs; More relevant to tissue PK, but can linked to blood PK |
| Safety | Risk of various off-target effects | Risk of immunogenicity or immune reactions | Risk of immunogenicity or immune responses |
2.1. Chemistry of RNA molecules
RNA molecules in variable length consist of four nucleobase building blocks, adenine (A), uracil (U), cytosine (C), and guanine (G), which are connected in particular orders through glycosidic and phosphodiester bonds (Fig. 2A). Pseudouridine (ψ), an isomer of uridine or modified uridine in which the uracil is linked to ribose via carbon-carbon bond rather than nitrogen-carbon glycosidic bond, is found frequently in genome-derived RNAs and is also called the fifth nucleotide (Li et al., 2016). The RNA ribonucleosides differ from DNA nucleoside subunits in that each ribose is comprised of a hydroxy group whereas deoxyribose is not (Fig. 2A). In addition, thymine (T) or 5-methyluracil (m5U) is the fourth nucleobase in DNA, in contrast to uracil in RNA.
Fig. 2.
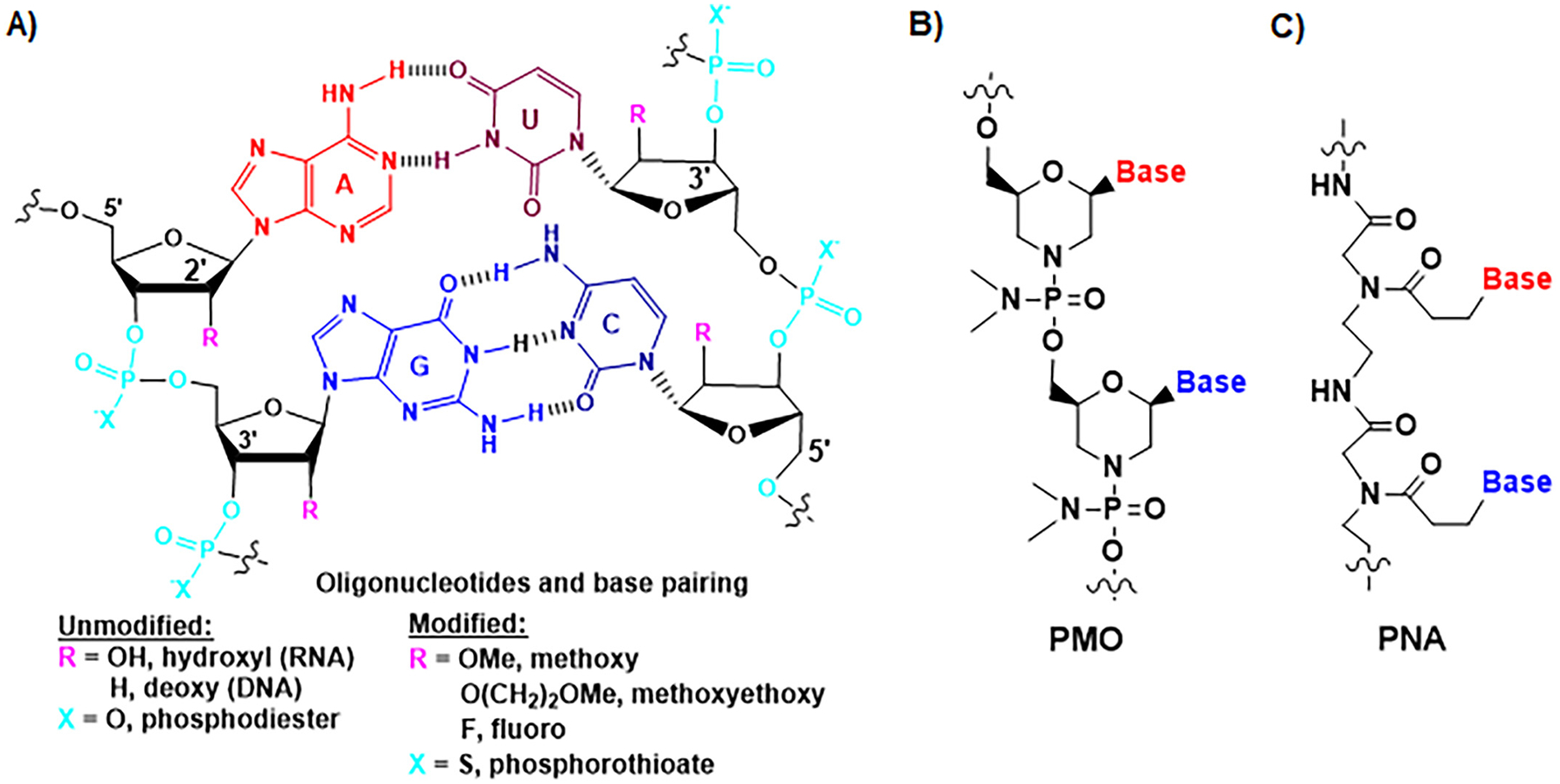
Chemical structures of natural and synthetic oligonucleotides. (A) The nucleobases are paired through hydrogen bonds (H-bonds). Adenine (A) and uracil (U) (or thymine (T) or 5-methyluracil (m5U)) pairs involve two H-bonds, and guanine (G) and cytosine (C) pairs consist of three H-bonds. The phosphodiester bond linkage and 2′-hydroxyl group on ribose ring within natural RNA molecules are two “soft spots” that are commonly modified as other forms towards a greater metabolic stability, such as phosphorothioate, 2′-methoxy, 2′-methoxyethoxy, and 2′-fluoro. (B) Synthetic phosphorodiamidate morpholine oligonucleotide (PMO) or morpholino oligomer is comprised of advanced modifications, in which a six-membered morpholine replaces the five-membered ribose ring and phosphoramidate substitutes the phosphodiester linkage while nucleobases are retained for pairings. (C) Peptide nucleic acid (PNA) is another type of artificial oligonucleotide in which nucleobases are connected with N-(2-aminoethyl)-glycine units linked by peptide bonds.
The nucleobases form base pairs through hydrogen bonds (H-bands), among which A and U (or T) base pairs consist of two H-bonds, and G and C pairs involve three H-bands (Fig. 2A). Intramolecular complementary base pairings, along with other types of physicochemical interactions, make RNA fold into higher orders of structures, including secondary (e.g., helices or stems, loops, and bulges), tertiary (e.g., motifs, pseudo-knot, and junctions), and quaternary (e.g., complexes) elements (Jones and Ferre-DAmare, 2015; Schlick, 2018), which are critical for RNA stability and function. By contrast, intermolecular complementary base pairing is the foundation for RNA therapeutics (e.g., ASOs, miRNAs, siRNAs, and gRNAs) to selectively act on target transcripts to achieve the control of gene expression and disease progression (Doudna, 2020; Roberts et al., 2020; Yu et al., 2020b).
Chemical modification is a proven means to improve RNA metabolic stability and pharmacokinetic property while maintaining or enhancing efficacy (Crooke et al., 2018; Khvorova and Watts, 2017; Roberts et al., 2020; Yu et al., 2019; Yu et al., 2020b). The phosphodiester bond is revealed as a major “soft spot” in RNA molecules, in which one non-bridging oxygen can be replaced by a sulfur atom to offer phosphorothioate oligonucleotides (Fig. 2A) towards a reduced nucleolytic degradation (Eckstein, 1985). The 2′-hydroxyl group on ribose ring is the other major “soft spot”, which is commonly changed to 2′-methoxy, 2′-methoxyethoxy, or 2′-fluoro (Fig. 2A) for greater serum stability (Bramsen and Kjems, 2012; Crooke et al., 2020; Ho and Yu, 2016; Khvorova and Watts, 2017). There are also some advanced modifications that aim at retaining nucleobases for complementary pairings while changing the ribose phosphodiester linkages. PMO or morpholino oligomer, in which a six-membered morpholine ring is used to substitute the five-membered ribose, and phosphoramidate linkage to replace the phosphodiester (Fig. 2B), has been proved as a successful strategy for the development of RNA therapeutics (Summerton, 2017; Summerton and Weller, 1997). In addition, peptide nucleic acids (PNAs) are under active development (Nielsen, 2000; Saarbach et al., 2019; Zhilina et al., 2005), among which the nucleobases are connected by using the N-(2-aminoethyl)-glycine units linked through peptide bonds (Fig. 2C). While chemical modifications are expected to retain pharmacological actions, structural and functional studies have revealed that modified RNAs may interact differently with binding proteins and targets, and exhibit greater or lower potency (Schirle et al., 2016).
Chemical synthesis and in vitro transcription are two common methods for the production of RNA agents. Phosphoramidite method is a well-established approach for chemical synthesis of oligonucleotides (Marshall and Kaiser, 2004), and specifically-modified building blocks can be incorporated precisely into the final products at desired locations during organic synthesis. By contrast, in vitro transcription is an enzymatic method for the synthesis of RNAs using the RNA polymerases (e.g., T7 and SP6) and proper templates (e.g., plasmid DNA, cDNA, or synthetic oligonucleotides) (Beckert and Masquida, 2011; Milligan et al., 1987). Some modified nucleosides, such as pseudouridine (Anderson et al., 2010; Kariko et al., 2008) or N1-methyl-pseudouridine (N1mψ) (Svitkin et al., 2017), can be incorporated systematically into the final RNA products during in vitro transcription, whereas it is not feasible to specifically assemble particularly-modified nucleosides into the products at defined locations. As chemical synthesis is a practical approach to the production of relatively shorter oligonucleotides (e.g., < 50 nt), in vitro transcription is a reliable and efficient method for the manufacture of longer RNA agents (e.g., mRNAs; > 100 nt). Rather, single-stranded ASOs and double-stranded siRNAs in shorter lengths can be made by in vitro transcription (Guiley et al., 2012; Kim et al., 2004; Wianny and Zernicka-Goetz, 2000; Yang et al., 2002). Interestingly, the 5′ initiating triphosphate was found to induce interferons, which need be removed to alleviate immune response (Kim et al., 2004).
Novel technologies are also emerging for in vivo production of functional RNA agents (Ho and Yu, 2016; Yu et al., 2019; Yu et al., 2020a). Bioengineered or recombinant RNA molecules are expected to better recapitulate the structures, functions, and safety of natural RNAs because both are produced, folded, and tolerated in living cells, and carrying only natural and necessary posttranscriptionally-modified nucleosides (Li et al., 2015; Wang et al., 2015). The use of stable scaffolds or carriers is the most reliable and versatile approach for heterogenous overexpression, among which the transfer RNA (tRNA) fused precursor miRNA (pre-miRNA) (tRNA/pre-miRNA) carriers are able to accommodate a wide variety of payload small RNAs (sRNAs), including miRNAs, siRNAs, and aptamers, to achieve a consistent, high-yield, and large-scale production of target ncRNAs for basic and translational research (Chen et al., 2015; Ho et al., 2018; Li et al., 2018; Li et al., 2019; Li et al., 2021; Petrek et al., 2019; Tu et al., 2019; Yi et al., 2020). In addition, direct heterogenous expression of specific RNAs in host bacteria lacking RNase III have been demonstrated recently (Hashiro et al., 2019a; Hashiro et al., 2019b). Such in vivo production approaches shall open new avenues for RNA manufacture, research, and development (Ho and Yu, 2016; Yu et al., 2019; Yu et al., 2020a).
2.2. Delivery of RNA agents
RNA drugs administered through systemic injections (e.g., intravenous), slightly different from local or other routes (e.g., subcutaneous, intravitreal, intrathecal, and intramuscular) (Table 1), are subjected to both systemic and cellular barriers that hinder their access to intracellular targets. As mentioned above, RNAs are highly susceptible to the degradation by RNases or hydrolases that are commonly found in blood and body fluids, and they are easily filtered through glomerulus and rapidly eliminated from the kidney. Therefore, unprotected RNAs have an extremely short metabolic or systemic half-life (e.g., less than 7 min) (de Smidt et al., 1991; Soutschek et al., 2004). In addition, negatively-charged RNA molecules over 7 kDa cannot freely passthrough the cell membrane to interact with target transcripts inside the cells. Therefore, it is essential to employ appropriate strategies to overcome these hurdles so that sufficient levels of RNAs can get into the cells and interact with intracellular targets to exert pharmacological effects. Among them, chemical modifications aforementioned have proven as viable means to enhance metabolic stability while the potency of RNA could be influenced to different degrees.
Conjugation with proper polymers or specific ligands (Fig. 3A) has found its success for the delivery of therapeutic RNAs (Juliano, 2016; Kowalski et al., 2019; Roberts et al., 2020; Yu et al., 2020b). The polyethylene glycol (PEG) is an FDA-approved polymer commonly used for drug delivery, including small molecules, polypeptides, proteins, and RNAs, which could lead to optimal pharmacokinetic property, efficacy, and safety profiles (Knop et al., 2010; Suk et al., 2016). Indeed, pegaptanib (Macugen), the first RNA aptamer drug approved by the FDA (Fig. 1), is an anti-vascular endothelial growth factor (VEGF) aptamer conjugated to 40-kDa PEG (Ng et al., 2006). Efforts have been made to introduce cleavable PEG to form siRNA conjugate (Jung et al., 2010) that could be released under lower pH or reductive environment. Nevertheless, the sense or guide strand is preferably subjected to conjugation because the 5′- and 3′-end of the antisense or passenger strand are critical for target recognition and anchoring the siRNA or miRNA in the Argonaute 2 (AGO2) protein (Elkayam et al., 2012; Ma et al., 2004; Schirle and MacRae, 2012). In addition, there are concerns about the formation of or pre-existing anti-PEG antibodies in subjects that may enhance drug clearance and lead to the reduction or loss of efficacy, as well as induction of hypersensitivity reactions (Kozma et al., 2020).
Fig. 3.
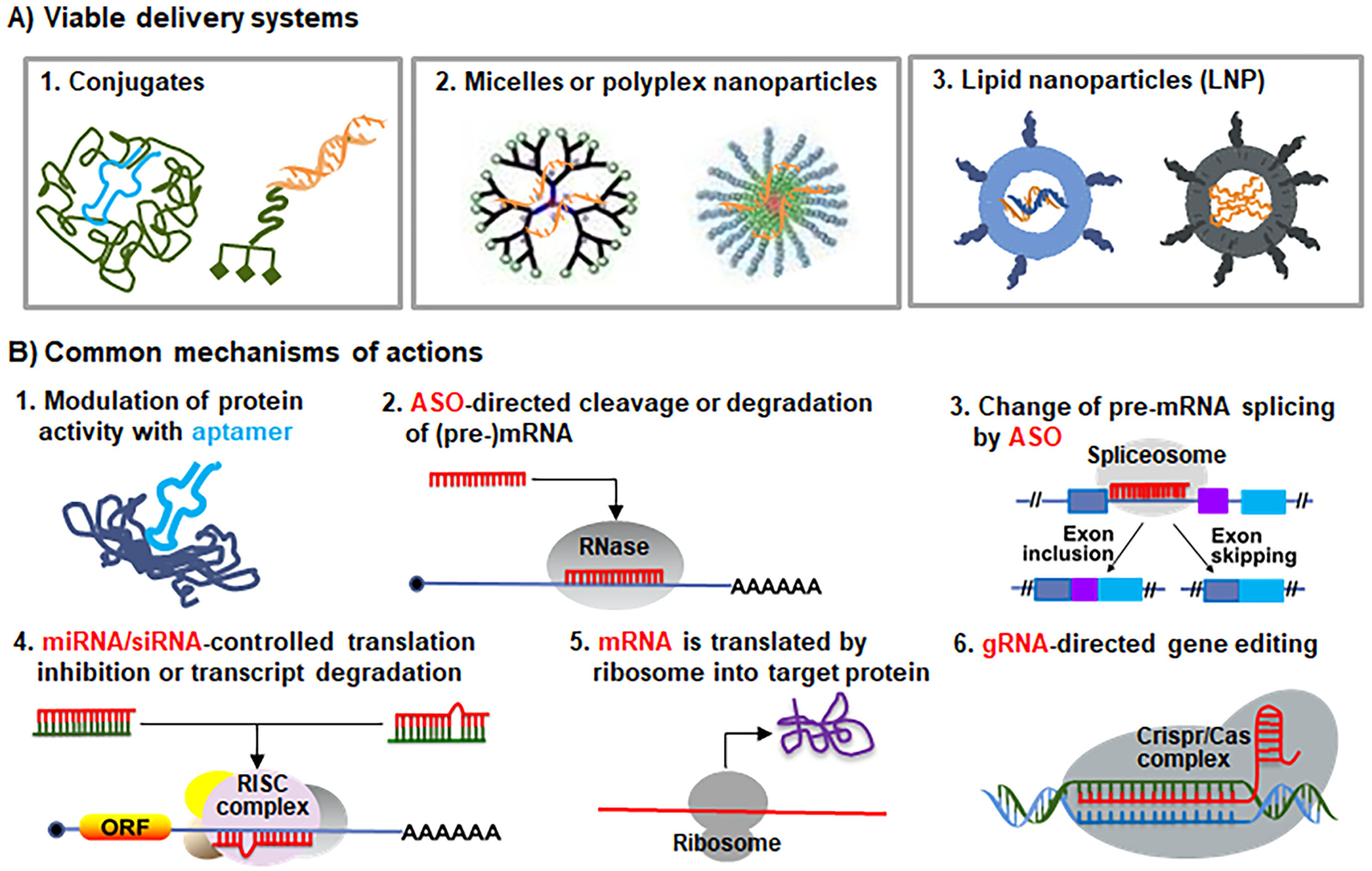
Common delivery systems and mechanisms of actions of therapeutic RNAs. (A) RNA modalities may be conjugated with specific polymers (e.g., PEG) or ligands (e.g., GalNAc), packaged with polymers (e.g., PEI), or formulation into lipid nanoparticles (e.g., using ionizable lipids) to improve the pharmacokinetics properties and enhance cellular uptake. See Figs. 4–5 for specific modifications used in RNA medications approved by the FDA. (B) Common mechanisms of actions of RNA therapeutics and vaccines marketed or under development. First, RNA aptamer drugs (e.g., pegaptanib) directly bind to extracellular, cell-surface or intracellular protein targets to block or activate their activities. Second, ASOs (e.g., DNA oligomer fomivirsen and “Gapmer” mipomersen and inotersen) direct RNases such as RNase H to cleave and degrade target transcripts within nucleus or cytoplasm. Third, ASOs (e.g., nusinersen and the PMOs (eteplirsen, golodirsen, viltolarsen and casimersen)) target the splice sites of pre-mRNAs and interact with splicing factors to alter RNA maturation in nucleus, causing exon inclusion or skipping. Fourth, the antisense or guide strands of siRNAs or miRNA mimics (e.g., patisiran, givosiran and lumasiran) are loaded into the cytoplasmic RISC complex to recognize target transcripts, leading to translation inhibition, mRNA cleavage or deadenylation. Indeed, both patisiran and lumasiran were designed to act on the 3′-untranslated regions of target transcripts, same as endogenous miRNAs. Fifth, exogenous mRNAs use cellular ribosomes to synthesize target proteins for replacement therapy or vaccination. Sixth, gRNAs interact with target gene sequences within nuclear CRISPR/Cas complex to achieve genome editing.
Besides PEG, RNA agents may be covalently conjugated to an array of entities, such as cholesterols (Soutschek et al., 2004), fatty acids and bile acids (Wolfrum et al., 2007), antibodies (Cuellar et al., 2015; Song et al., 2005), polypeptides (Klein et al., 2019; Ren et al., 2012), aptamers (Neff et al., 2011), and sugars (Nair et al., 2014; Nair et al., 2017). Polyconjugates are also developed in which an amphipathic polymer consisting of PEG and targeting ligands is attached to target siRNA (Rozema et al., 2007). The resulting conjugates are anticipated to have greater serum stability and facilitate intracellular uptake of payload RNAs, including tissue- or cell-specific delivery. Interestingly, the cell penetrating peptides (CPPs) (Abedi-Gaballu et al., 2018; Guidotti et al., 2017; Taylor and Zahid, 2020) are commonly linked to PNA by using disulfide or other covalent linkages to achieve antisense effects (Dragulescu-Andrasi et al., 2006; Pooga et al., 1998), while CPPs can directly conjugated to therapeutic RNAs (Srimanee et al., 2018; Ye et al., 2017) to improve intracellular delivery. The amino sugar, N-acetylgalactosamine (GalNAc), is a hepatocyte asialoglycoprotein receptor-binding ligand (Matsuda et al., 2015; Nair et al., 2014; Springer and Dowdy, 2018) that has been successfully applied to RNA drug delivery and found in two siRNA medications, givosiran and lumasiran. Specifically, the 3′-terminus of sense strand is attached to a trivalent GalNAc moiety to improve the delivery to hepatocytes (Nair et al., 2014; Nair et al., 2017).
RNA therapeutics may be encompassed in polymer-based carriers to form polymeric nanoparticles, micelles, dendrimers or polyplexes to achieve efficient delivery (Fig. 3A). RNA-polymer complexes are assembled through physicochemical or electrostatic interactions. Both natural and synthetic polymers, such as PEG (Jhaveri and Torchilin, 2014), polysaccharides including chitosan, hyaluronic acids, and cyclodextrins (Mousazadeh et al., 2021; Ragelle et al., 2013; Serrano-Sevilla et al., 2019), polyethylenimine (PEI) (Neuberg and Kichler, 2014), and polypeptides or their derivatives (Abedi-Gaballu et al., 2018; Guidotti et al., 2017; Koren and Torchilin, 2012; Taylor and Zahid, 2020), have been widely and continuously investigated. CALAA-01, a siRNA designed to reduce the expression of the M2 subunit of ribonucleotide reductase (RRM2) and complexed with a PEGylated and human transferrin protein (hTF)-modified linear cationic cyclodextrin-based polymer, represents a polymer-based siRNA nanotherapeutics evaluated among cancer patients in the clinic, and the human pharmacokinetics and safety data generally correlate to those from animals (Zuckerman et al., 2014). PEI has been shown as an effective material for the delivery of a siRNA targeting hypusinated eukaryotic translation initiation factor 5A (eIF5A) to inhibit tumor growth in multiple animal models of B-cell cancers without damaging normal tissues (Francis et al., 2014). As another example, CPPs are able to form complexes with RNA agents non-covalently and facilitate intracellular delivery, either alone or connected to other polymers (Malhotra et al., 2013; Schiroli et al., 2019; Srimanee et al., 2018).
RNA agents are formulated into lipid nanoparticles (LNP) (Fig. 3A) the most frequently through self-assembly, driven by a combination of electrostatic and hydrophobic interactions, for in vitro and in vivo delivery (Cullis and Hope, 2017; Kowalski et al., 2019; Samaridou et al., 2020). Indeed, a number of liposomal, small-molecule products have been approved by the FDA for medical use, such as hydrogenated soybean phosphatidylcholine (HSPC) and PEGylated 1,2-distearoyl-snglycero-3-phosphoethanolamine (PEG-DSPE)-packaged doxorubicin (Doxil®) (Baker, 1995; James, 1995), DSPC-formulated daunorubicin (DaunoXome®) (FDA, 1996), HSPC and PEG-DSPE-complexed amphotericin B (Ambisome®) (Baker, 1995), and egg sphingomyelin (ESM)-carried vincristine (Marqibo®) (Raj et al., 2013). Rather, cationic or ionizable lipids are preferably used for the delivery of anionic RNAs (Cullis and Hope, 2017; Kowalski et al., 2019; Samaridou et al., 2020). Cationic lipid-formed lipoplexes may retain their electronic charge during circulation, and ionizable lipids can become protonated in a low pH environment to facilitate endosomal escape. Other hydrophobic moieties, such as cholesterol and PEGylated lipids, are usually included at optimal ratios to enhance the stability of LNPs as well as the delivery efficacy and therapeutic efficacy of RNA agents (Chen et al., 2012; Mui et al., 2013). Lipids are also used to coat polyplexes to offer lipopolyplexes to improve the stability RNAs and reduce the use and toxicity of polymers (Rezaee et al., 2016). Patisiran, the first FDA-approved siRNA drug (Zhang, Goel, & Robbie, 2019; Zhang et al., 2020) (Fig. 1), is complexed with a novel ionizable lipid, dilinoleylmethyl-4-dimethylaminobutyrate (DLin-MC3-DMA or MC3), as well as α-(3′-((1C-di(myristyloxy)proponoxy)-carbonylamino)propyl)-ω-methoxy-polyoxyethylene (PEG2000-C-DMG) as a lipid PEG anchor, and DSPC and cholesterol as helper lipids at a molar ratio of 50/1.5/10/38.5 to enable delivery into hepatocytes (Jayaraman et al., 2012). In addition, ionizable cationic lipids, PEGylated lipids, phosphatidylcholine, and cholesterol carriers (Hassett et al., 2019; Jayaraman et al., 2012; Maier et al., 2013; Pardi et al., 2015; Richner et al., 2017) have found their success for the delivery of mRNA vaccines that have been approved under EUA for the prevention of unprecedented COVID-19.
As delivery remains a big challenge for the development of RNA therapeutics and vaccines, enormous efforts are still underway to explore and evaluate new effective and safe delivery materials and platforms. Natural biological nanoparticles, namely extracellular vesicles or exosomes, are also of particular interest (Barile and Vassalli, 2017; O’Brien et al., 2020). Cell-derived exosomes, especially those produced autologously, are expected to be more biocompatible than synthetic materials, and exosomes are able to pass through biological membranes to achieve intracellular delivery of RNA agents. Nevertheless, the heterogeneity of exosomes and batch-to-batch variations, such as size and composition, indicate the need for stringent quality control. It is also essential to obtain sufficient quantity of exosomes to ensure efficient loading of target RNAs for the administration of projected doses and examine the safety of the final products.
Utilization of therapeutic RNAs for the treatment of the central nervous system (CNS) disorders encounters additional challenge due to the presence of blood-brain barrier (BBB), the highly selective and permeable membrane that prevents xenobiotics and biologic molecules from entering the brain (Bennett et al., 2019; Hammond et al., 2021). While particular RNA agents such as PMOs exhibited a unidirectional influx across the BBB, fewer than 1% of the PMOs administered intravenously were actually found in the brain of mouse models and appropriate doses were warranted to effectively lessen CNS deficits (Banks et al., 2001). Based on ligand-receptor interactions, specific ligands or CPPs may be used to achieve more effective delivery to the CNS (Dastpeyman et al., 2021; Du et al., 2011; Kanazawa et al., 2013; Min et al., 2020). Further, given the important roles of miRNA-containing exosomes in cell-cell and organ-organ communications, neurophysiology and neuropathology, exosomes were shown to facilitate the delivery of siRNAs across BBB for effective knockdown of target gene expression in the brain (Alvarez-Erviti et al., 2011; Yang et al., 2017). While these approaches are promising, more extensive evaluation and validation studies are necessary before the application to medical use.
2.3. Mechanisms of actions
RNA medications and vaccines are remarkably different from small-molecule and protein drugs in mechanisms of actions (Fig. 3B), in addition to chemistry and pharmacokinetics (Table 1). Therapeutic small molecules predominantly interact with intra- or extra-cellular or cell-surface proteins, e.g., acting as inhibitors to enzymes, kinases, transporters, and ion channels, or antagonists or agonists of receptors, to exert pharmacological effects, while some small-molecule drugs have direct interactions with DNA and RNA targets (Yu et al., 2020b). On the other hand, protein and antibody drugs mainly target cell-surface proteins or extracellular factors to reach therapeutic outcomes (Kintzing et al., 2016; Leader et al., 2008). By contrast, while particular forms of RNAs are capable of binding to proteins to alter their functions (e.g., aptamers), targeting the genome to achieve gene editing (gRNAs), or using ribosomes to produce target proteins or antigens (mRNAs), therapeutic RNAs (e.g., ASOs, siRNAs, and miRNAs) primarily interfere with intracellular transcripts to alter target gene expression for the control of diseases (Fig. 3B).
One group of therapeutic RNAs, namely aptamers (Bouchard et al., 2010; Kaur et al., 2018; Nimjee et al., 2017), mimic the actions of some cellular RNA molecules that have been revealed to bind to proteins to modulate their functions (Dingwall et al., 1989; Kitajewski et al., 1986). Through intermolecular base pairings and other physicochemical interactions, single-stranded RNA aptamers (e.g., pegaptanib) can fold into higher order structures which allow them directly and specifically to interact with protein targets (e.g., VEGF) to exert pharmacological effects (Fig. 3B). RNA aptamers are usually screened through a process called systematic evolution of ligands by exponential enrichment (SELEX) to achieve greater or optimal affinity and specificity (Ellington and Szostak, 1990; Robertson and Joyce, 1990; Tuerk and Gold, 1990). Besides therapy by inhibiting or activating a variety of potential protein targets, such as cytokines, proteases, kinases, cell-surface receptors, and cell-adhesion molecules (Nimjee et al., 2017), RNA aptamers have been identified for other applications such as the detection of specific small-molecule metabolites (Jaffrey, 2018; Rothlisberger et al., 2017).
Most therapeutic RNAs mechanistically act on mRNAs to modulate target gene expression for the management of diseases. Among them, single-stranded ASOs are a major group of RNAs under development and some have been approved for clinical practice (e.g., mipomersen and inotersen) (Bennett, 2019; Crooke et al., 2020; Kole et al., 2012), besides being used as research tools. This is following the discovery of functional antisense RNAs in the control of posttranscriptional gene regulation in cells across species (Ecker and Davis, 1986; Izant and Weintraub, 1984; Light and Molin, 1983; Simons and Kleckner, 1983; Spiegelman et al., 1972) and the development of synthetic ASOs to selectively control target gene expression via complementary base pairings with targeted transcripts (Stephenson and Zamecnik, 1978; Zamecnik and Stephenson, 1978). ASO-directed target gene suppression may not only include RNase H-mediated cleavage or degradation of mRNAs and/or precursor mRNAs (pre-mRNAs) (e.g., fomivirsen and “Gapmer” ASOs (mipomersen and inotersen)) within nucleus and cytoplasm (Fig. 3B) but also involve RNase-independent translation inhibition (Crooke et al., 2020) or spliceosome-controlled pre-mRNA splicing as discussed below.
Some therapeutic ASOs (e.g., nusinersen, eteplirsen, golodirsen, viltolarsen and casimersen) alter mRNA maturation by targeting pre-mRNAs and the nuclear spliceosome (Fig. 3B) (Crooke et al., 2020; Havens and Hastings, 2016). As such, these ASOs are also called splice-switching oligonucleotides (SSOs). The pre-mRNAs transcribed within the nucleus must undergo further processing to become mature mRNAs, in which the pre-mRNA splicing involves selective removal of interspersed noncoding sequences or introns, and precise ligation or assembly of individual exons or coding sequences through sequential trans-esterification reactions (Fica et al., 2013; Galej et al., 2018; Kramer, 1996). Distinct mRNAs may be generated from the same pre-mRNA through alternative splicing (Berget et al., 1977; Early et al., 1980), and some diseases are attributed to dysregulated splicing (Faustino and Cooper, 2003; Lee and Rio, 2015). Through base pairing with target pre-mRNA sequence and blocking RNA-spliceosome interactions (Fig. 3B), a SSO can be designed and used to effectively disrupt the normal splicing repertoire of the pre-mRNA, leading to exon inclusion or skipping and thus the production of functional proteins or preferred genetic products towards the control of diseases.
Therapeutic siRNAs or miRNA mimics (e.g., patisiran, givosiran and lumasiran) are double-stranded RNAs that rely on the RNA-induced silencing complex (RISC or miRISC) to modulate target gene expression (Fig. 3B). In particular, the antisense or guide strands of siRNAs or miRNAs are preferably loaded into cytoplasmic RISC to recognize target transcripts through complementary base pairings with targeted sequences, leading to translation inhibition, mRNA cleavage, or deadenylation. The concept of developing therapeutic siRNAs and miRNA mimics risen after the discovery of RNAi phenomena using double-stranded exogenous RNA agents (Elbashir et al., 2001; Fire et al., 1998; Zamore et al., 2000), as well as the genome-derived functional miRNAs that govern posttranscriptional gene regulation across species including humans (Lee et al., 1993; Pasquinelli et al., 2000; Reinhart et al., 2000; Wightman et al., 1993). While siRNAs are usually designed to interact with the coding sequences of target transcripts through perfect base pairings, endogenous miRNAs generally act on the 3′-untranslated regions (3’UTR) via imperfect base pairings. Interestingly, two of the three marketed siRNA drugs, patisiran and lumasiran, interfere with the 3’UTRs of target transcripts to achieve knockdown of gene expression. Furthermore, while both single-stranded ASOs and double-stranded siRNAs can be converged or unified into the posttranscriptional gene regulation mechanisms (Fig. 3B), siRNAs are more effective to knock down cytoplasmic targets and ASOs generally show greater efficacy in suppressing nuclear targets (Lennox and Behlke, 2016). Indeed, the actions of ASOs depend on RNases H and P-mediated cleavage or degradation of target transcripts which are highly abundant in the nucleus, and SSOs are to manipulate nuclear spliceosome and pre-mRNA processing. By contrast, the RISCs on which siRNAs and miRNAs depend for translation inhibition or transcript degradation are generally enriched within the cytoplasm while functional RNAi factors are present within human cell nuclei for selective and effective knockdown of nuclear targets by siRNAs (Berezhna et al., 2006; Gagnon et al., 2014). It is also noteworthy that, similar as some miRNAs inducing the expression of particular genes with complementary promoter sequences (Place et al., 2008), some double-stranded RNAs are designed to activate target gene expression and thus named as small activating RNAs (saRNAs), which generally target the promoter regions or transcriptional machinery to turn on transcription or improve transcription efficiency (Kwok et al., 2019; Voutila et al., 2017).
Exogenous mRNAs, generally made by in vitro transcription, may be introduced into cells and read by cellular ribosomes (Fig. 3B) towards the synthesis of targeted proteins/antibodies for replacement therapy or antibody therapy, or antigens for vaccination (Pardi et al., 2018; Van Hoecke and Roose, 2019; Weissman and Kariko, 2015). Because the single-stranded mRNA must have all essential components to achieve efficient translation, including the intact coding sequence (e.g., > 1000 nt), 5′ cap, and 3′ poly(A) tail, mRNA is unsurprisingly the largest therapeutic RNA molecule in size or length under development. Following the proof of concepts in animals that direct injection of mRNAs could lead to the expression of encoded proteins or vaccination (Jirikowski et al., 1992; Martinon et al., 1993; Wolff et al., 1990), many therapeutic mRNAs and vaccines have entered into clinical investigations to combat against various diseases (clinicaltrials.gov). Two mRNA vaccines (BNT162b2 and mRNA-1273) have been granted EUA for the prevention of COVID-19, and both encode the membrane Spike protein critical for the SARS-CoV-2 viral infection (Li et al., 2020; Ou et al., 2020; Shang et al., 2020). Following the production by ribosomes, the Spike proteins are expected to serve as antigens to stimulate the immune system to generate specific antibodies for the prevention of SARS-CoV-2 viral infection or COVID-19 and the emerging variants (Collier et al., 2021; Corbett et al., 2020; Kalnin et al., 2021; Walsh et al., 2020b; Wang et al., 2021).
Treating diseases through direct alteration of target gene within the genome is under resurgence, especially for genetic disorders (Long et al., 2016; Nelson et al., 2016; Tabebordbar et al., 2016; Xu et al., 2019; Xue et al., 2014; Zhang et al., 2017), with the discovery and development of CRISPR-Cas-based gene-editing mechanism and technologies (Cong et al., 2013; Jinek et al., 2012; Mali et al., 2013; Mashiko et al., 2013). Besides the use of 160-kDa Cas nuclease, a single-stranded gRNA is essential for selective and targeted gene editing. The small hairpin-structured gRNA binds to Cas protein to form Cas-gRNA ribonucleoprotein (RNP) complex to recognize a 20-nt sequence of target gene through complementary base pairings (Fig. 3B). The Cas nuclease is thus directed to create double-stranded DNA break (DSB) or single-stranded break (nick) towards genome engineering. Of particular note, while ASOs, siRNAs, miRNAs, and mRNAs all modulate the expression of targeted genes transiently, therapeutic gRNA is anticipated to irreversibly change target gene expression to achieve complete cure of a monogenic disorder. Rather, efficient delivery of both gRNA and Cas protein into the nucleus for targeted gene editing is undoubtedly more challenging than other forms of therapeutic RNAs discussed above.
2.4. Safety of RNA medications
Safety is another critical element of a medication, besides efficacy. Being designed to selectively modulate target gene expression, alter protein function, or edit the genome (Fig. 3B), single- or double-stranded, exogenous RNAs could be recognized by toll-like receptors (TLRs), such as TLR3, TLR7, and TLR8 (Diebold et al., 2004; Heil et al., 2004; Kleinman et al., 2008; Tanji et al., 2015), to provoke innate immune responses or mild to severe and even fatal cytokine release syndrome (Table 1). Such adverse effects caused by “unspecific” interactions with the organism’s self-protective machineries against pathogen infections such as viruses and bacteria are undoubtedly related to the dose, sequence, size, and folding of RNA agent. Some cytosolic sensors, such as pattern recognition receptor (PRR) and RNA-dependent protein kinase (PKR) (Garcia et al., 2006; Hornung et al., 2006), are also able to detect viral or exogenous RNA molecules to activate immune responses. In addition, high-mobility group box (HMGB) proteins 1, 2 and 3 have been revealed as universal sentinels for nucleic acids, and disruption of HMGBs severely impairs TLR activation by their cognate nucleic acids (Yanai et al., 2009). Therefore, risk of allergic reactions especially the induction of severe immune responses or cytokine storm often hinders the development of RNA medications. As some chemical modifications evoke immune responses (Bramsen and Kjems, 2012; Robbins et al., 2007), posttranscriptional modifications generally reduce or compromise on immunogenicity (Gehrig et al., 2012; Kariko et al., 2008; Kariko and Weissman, 2007; Nallagatla et al., 2008; Richner et al., 2017; Robbins et al., 2007). As such, natural modifications such as 2’-MeO and pseudouridine are preferably used for the development of therapeutic RNAs and commonly found in marketed RNA medications (Crooke et al., 2020; Yu et al., 2019). There is also growing interest in developing recombinant RNAs made and folded in living cells in vivo (Ho and Yu, 2016; Yu et al., 2019; Yu et al., 2020a).
The excipients or materials used for RNA formulation should be tolerable to ensure the safety of therapeutic RNA products. While lipids and liposomes within the RNA-LNPs (Fig. 3B) are usually biocompatible and safe, some TLRs such as TLR2 and TLR4 are known to recognize multiple glycolipids, lipopeptides, and lipopolysaccharides, and thus become activated to induce immune responses (Jin et al., 2007; Opitz et al., 2001; Park et al., 2009; Schroder et al., 2000). Further, although PEG is generally safe to humans and PEGylation enhances RNA delivery efficiency, cases of mild to life-threatening adverse effects associated with PEG conjugates or formulations, such as hypersensitivity, have been documented (Bruusgaard-Mouritsen et al., 2021; Stone Jr. et al., 2019; Wenande and Garvey, 2016). Indeed, concerns have been raised that the PEG ingredients used for delivery of mRNA vaccines might be the cause of some cases of allergic reactions following vaccination (Cabanillas, Akdis, & Novak, 2021; Klimek et al., 2021a; Ortega Rodriguez et al., 2021; Pitlick, Sitek, Kinate, Joshi, & Park, 2021). In addition, myocarditis (inflammation of the heart muscle) and pericarditis (inflammation of the tissue surrounding the heart) have been identified among some subjects following the receipt of an mRNA COVID-19 vaccine (Kim et al., 2021; Montgomery et al., 2021), and the FDA also issued warning label for the two mRNA vaccines on June 25, 2021. Caution is advised that all medications involve risks of adverse effects and should be used rationally and wisely.
3. RNA medications and vaccines in human use and under development
A total of ten therapeutic RNAs have been successfully marketed for the treatment of various diseases, and two mRNA vaccines have been approved very recently by the FDA under EUA to combat against the ongoing COVID-19 pandemic (Fig. 1). There are also many other therapeutic RNA agents and vaccines under clinical and preclinical development (Pardi et al., 2018; Quemener et al., 2020; Roberts et al., 2020; Van Hoecke and Roose, 2019; Yu et al., 2020b; Zhang et al., 2021). Based on their mechanisms of actions (Fig. 3B), therapeutic RNAs are classified as aptamers, ASOs, siRNAs or miRNA mimics, mRNAs, gRNAs, and miscellanea that are discussed individually below.
3.1. Aptamers
Pegaptanib is the first and currently the only RNA aptamer approved for medical use in humans, acting as a VEGF antagonist for the treatment of neovascular AMD (Ng et al., 2006) (Fig. 1). Pegaptanib is also the first angiogenic inhibitor approved for the treatment of human disease, followed by regulatory approval of many others antiangiogenic agents (e.g., ranibizumab, bevacizumab, sorafenib, regorafenib, and aflibercept) for different indications (e.g., AMD and cancer) (Al-Abd et al., 2017). Anti-VEGF became a potential therapeutic strategy (Bergers and Hanahan, 2008; Ng et al., 2006) since the discovery of VEGF and its critical roles in angiogenesis and vascular permeability during pathogenesis of related diseases (Ferrara and Henzel, 1989; Folkman et al., 1971; Keck et al., 1989; Leung et al., 1989; Senger et al., 1983; Tischer et al., 1991). Alternative splicing of human VEGF pre-mRNA leads to the production of several isoforms, among them an abundant 165-amino acid form, namely VEGF165, was targeted by anti-VEGF aptamers for anti-angiogenesis therapy (Green et al., 1995; Jellinek et al., 1994; Ruckman et al., 1998). High-affinity anti-VEGF aptamers were identified through SELEX, and the incorporation of particularly modified nucleosides, including 2′-fluoro and 2′-methoxy, not only improved the resistance to nucleases (e.g., by several orders of magnitude) but also enhanced binding affinity for VEGF165 (e.g., several fold) (Green et al., 1995; Jellinek et al., 1994; Ruckman et al., 1998). The PEGylated, 28-nt, single-stranded RNA aptamer (Fig. 4A) was subsequently moved into clinical investigations after extensive preclinical evaluations (Bell et al., 1999; Drolet et al., 2000; Eyetech Study G, 2002).
Fig. 4.
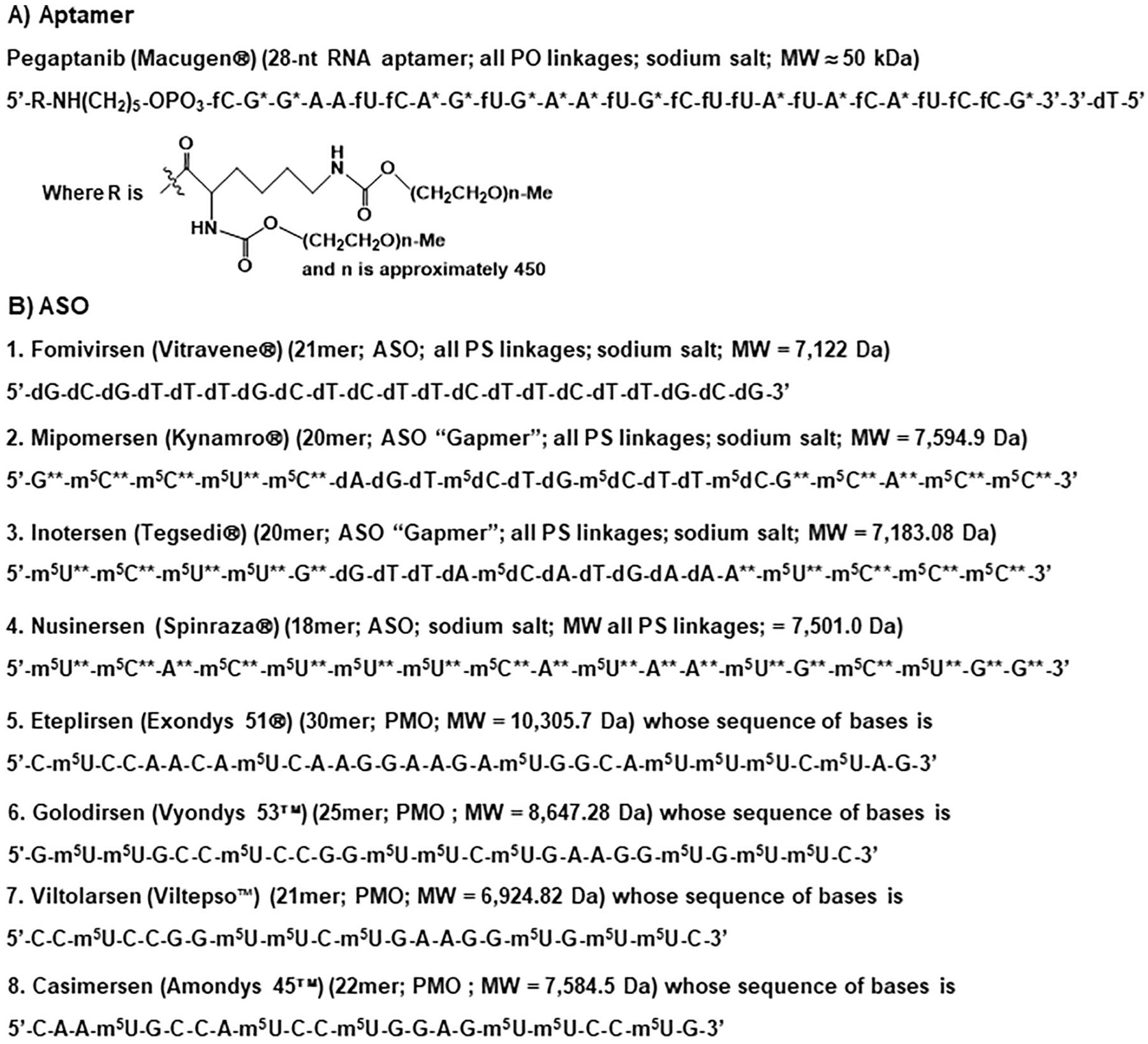
Chemical structural formulas of aptamer and ASO drugs approved by the FDA. (A) Pegaptanib is a single-stranded RNA aptamer drug consisting of two 20-kDa monomethoxy PEG units that are covalently connected to the two amino groups on a lysine residue attached to the 5′-end via a pentylamino linker. (B) Eight ASO medications, among which fomivirsen is a DNA oligomer, mipomersen and inotersen are “Gapmers” containing a segment of oligodeoxynucleotide in the middle of RNA oligomers, and eteplirsen, golodirsen, viltolarsen and casimersen are PMOs. A = adenine; U = uracil; G = guanine; C = cytosine; T = thymine that is also named as 5-methyluracil or m5U; f = 2′-fluoro; * = 2′-methoxy; ** = 2′-methoxyethoxy; m5 = 5-methyl; d = deoxyribose; PO = phosphodiester; PS = phosphorothioate; MW, molecular weight.
The first clinical study on pegaptanib therapy involved intravitreal, single ascending dose treatment in 15 patients with subfoveal choroidal neovascularization secondary to exudative AMD, and the results showed that 80% of patients exhibited stable or improved vision 3 months after treatment, while no significant safety issues related to the drug were noted (Eyetech Study G, 2002). The benefits of pegaptanib treatment for AMD patients were further demonstrated by a double-blind, multicenter, clinical study among approximately 1200 patients using broad entry criteria, including being less likely to lose visual acuity and more patients gaining vision (Gragoudas et al., 2004; Group VISiONCT et al., 2006). Some adverse events were observed in the clinic, among them endophthalmitis, traumatic injury to the lens, and retinal detachment were the most serious and required vigilance, ranging from 0.6% to 1.3% among the patients (Gragoudas et al., 2004). Furthermore, with the reports of possible hypersensitivity reactions to pegaptanib, studies identified a low incidence of individuals (e.g., 4 out of 131) who were negative at baseline showed positive IgG response to nonpegylated pegaptanib (https://www.ema.europa.eu/en/documents/withdrawal-report/withdrawal-assessment-report-macugen_en.pdf).
Regulatory approval of intravitreal pegaptanib therapy supports the development of RNA medications that interact directly with protein targets (Fig. 3B), complimentary to existing approaches by using small molecules and proteins/antibodies. Rather, there are just a limited number of completed and active clinical trials that investigate the interventions with RNA or DNA aptamers (Maier and Levy, 2016). Several synthetic L-aptamers, also known as Spiegelmers that are resistant to nucleases as compared to corresponding natural D-formed aptamers, have been under development and showed some promising results (Menne et al., 2017; Steurer et al., 2019). An ongoing Phase IIa trial (NCT 04677803) is to investigate the safety, tolerability, and pharmacologic activity of BT200 among patients with hereditary bleeding disorders, which is a PEGylated RNA aptamer designed to bind to the A1 domain of human von Willebrand factor, inhibit its interactions with platelet glycoprotein Ib, and subsequently prevent arterial thrombosis (Kovacevic et al., 2021; Zhu et al., 2020). In addition to therapy, aptamers may serve as “targeting ligands” for the delivery of other therapeutic agents, such as siRNAs and miRNAs (Neff et al., 2011; Zhou and Rossi, 2017).
3.2. Antisense oligonucleotides
Six ASO medications have been approved by the FDA for medical use (Fig. 1) (Quemener et al., 2020; Roberts et al., 2020; Yu et al., 2020b). Fomivirsen is the first marketed ASO medication for the treatment of CMV retinitis among individuals with weakened immune systems (Roehr, 1998). Fomivirsen is an oligodeoxynucleotide in which the 21 deoxynucleotides are connected through phosphorothioate linkages (Fig. 4B). Fomivirsen was designed to complementarily bind to a specific segment of the immediate early region 2 (IE2) mRNA of CMV, leading to the suppression of viral protein synthesis and subsequently inhibition of viral replication (Anderson et al., 1996; Azad et al., 1993). Through intravitreal administration, fomivirsen was shown to be effective for the treatment of CMV retinitis in patients with AIDS (Vitravene Study G, 2002a; Vitravene Study G, 2002b).
Mipomersen and inotersen (Fig. 4B) are the second-generation of ASO drugs, namely “Gapmers” or chimeric ASOs (Bennett, 2019; Quemener et al., 2020; Roberts et al., 2020). Comprised of two oligoribonucleotides at the 5′ and 3′ ends with a “gap” of 10-mer oligodeoxynucleotide in the middle, a Gapmer binds to the target transcript through complementary base pairing to form a hybrid DNA/RNA duplex, resulting in the cleavage of target transcript by RNase H and thus knockdown of gene expression. Besides the use of phosphorothioate linkages to increase nuclease resistance, the 2′-hydroxyl group of each ribose ring is modified as 2′-O-methoxyethyl or 2′-methoxyethoxyl (2′-MOE), along with other modifications on particular nucleobases (Fig. 4B), towards an enhanced metabolic stability, lower cytotoxicity, and greater efficacy (Freier and Altmann, 1997). Mipomersen was shown to effectively inhibit the synthesis of apolipoprotein B-100 (ApoB-100) through the degradation of ApoB mRNA (Haddley, 2011; Yu et al., 2009), the principal Apo of low-density lipoprotein (LDL) and necessary for the assembly and secretion of very low-density lipoprotein (VLDL). Clinical studies further demonstrated the benefits of mipomersen treatment for patients with homozygous familial hypercholesterolemia (HoFH) (Crooke and Geary, 2013; Stein et al., 2012; Thomas et al., 2013). Likewise, inotersen was developed to reduce hepatic transthyretin (TTR) protein synthesis and serum TTR levels through RNase H-mediated degradation of mutant and wild-type TTR mRNAs following complementary base pairings (Ackermann et al., 2012; Ackermann et al., 2016). As autosomal dominant mutations in TTR gene cause aggregation of monomers into insoluble, extracellular amyloid deposits which accumulate in multiple organs and thus lead to hATTR amyloidosis (Lobato et al., 2003; Sekijima, 2015), inotersen showed therapeutic benefits for patients with hATTR amyloidosis (Benson et al., 2018; Coelho et al., 2020).
Nusinersen is another therapeutic ASO approved for the treatment of a rare autosomal recessive neuromuscular disorder called spinal muscular atrophy (SMA) (Aartsma-Rus, 2017). In particular, the genetic mutations in chromosome 5q alter survival motor neuron (SMN) pre-mRNA processing and result in SMN protein deficiency (Brzustowicz et al., 1990; Lefebvre et al., 1995). Nusinersen (or ASO-10–27), comprised of only phosphorothioate linkage, 2′-MOE on each ribose, and some methylated nucleobases (Fig. 4B), was designed to act as a SSO (Fig. 3B) to modulate SMN mRNA maturation (Hua et al., 2011). By increasing exon 7 inclusion and promoting the production of full-length functional SMN protein, intrathecal nusinersen treatment showed acceptable safety and tolerability, with significant and clinically meaningful improvement in motor function for children with SMA (Finkel et al., 2016; Finkel et al., 2017; Mercuri et al., 2018).
Eteplirsen (Stein, 2016; Syed, 2016), golodirsen (Heo, 2020), viltolarsen (Iftikhar et al., 2021) and casimersen (Shirley, 2021) represent the third-generation of ASO medications with advanced chemical modifications (Bennett, 2019; Quemener et al., 2020; Roberts et al., 2020). In particular, the four drugs all belong to PMOs whose nucleobases (Fig. 4B) are connected by using phosphoramidate morpholino linkages (Fig. 2). Interestingly, all of the PMO drugs were approved for the treatment of a lethal genetic neuromuscular disorder, Duchenne muscular dystrophy (DMD), which is caused by particular mutations altering normal processing of X-linked dystrophin pre-mRNA (Cirak et al., 2011). Eteplirsen, golodirsen, viltolarsen and casimersen were all designed to act as SSOs to modulate mRNA maturation (Fig. 3B), leading to exon 51, 53 or 45 skipping, for the production of internally-truncated, yet functional dystrophin proteins (Iftikhar et al., 2021; Popplewell et al., 2010; van Deutekom et al., 2007). As a result, DMD patients with particular splicing defects receiving eteplirsen (Cirak et al., 2011; Khan et al., 2019; Mendell et al., 2013; Mendell et al., 2016), golodirsen (Frank et al., 2020; Heo, 2020), viltolarsen (Clemens et al., 2020) or casimersen (Shirley, 2021) treatments showed an increased dystrophin production from baseline, slower rate of decline in ambulation or timed function tests.
Many other therapeutic ASOs are under development for the treatment of various diseases (Quemener et al., 2020; Roberts et al., 2020; Yu et al., 2020b). Intrathecal therapy with IONIS-HTTRx/RG6042, an ASO designed to direct RNase H1-mediated degradation of wild-type and mutant huntingtin pre-mRNA, was found to be well-tolerated and produce dose-dependent reduction in cerebrospinal fluid mutant huntingtin among early Huntington’s disease (HD) patients in the Phase I/II clinical trials (Tabrizi et al., 2019). An ongoing Phase III clinical study (NCT03761849) involves a randomized, double-blind, placebo-controlled design to evaluate the efficacy, safety, and biomarker effects of RG6042 among patients with manifest HD (Rodrigues et al., 2019). After the efficacy of ASO-directed knockdown of dynamin2 (DNM2) for the prevention and reversal of muscle pathology in mouse models (Tasfaout et al., 2017), a Phase I/II clinical trial (NCT04033159) has been initiated to evaluate the safety, tolerability, pharmacokinetics, and preliminary efficacy of intravenous Gapmer ASO, namely DYN101, in patients ≥16 years of age with centronuclear myopathy caused by mutations in DNM2 or myotubularin 1. As another example, an ASO-based therapeutic strategy for the treatment of Angelman syndrome by targeting an underlying long noncoding RNA was demonstrated in preclinical models (Meng et al., 2015), and a Phase I clinical trial (NCT04428281) is underway to investigate the safety, pharmacokinetics, and pharmacodynamics of a locked nucleic acid ASO, namely RO7248824, among patients with Angelman syndrome. Therefore, one can speculate more ASO medications would be commercialized in the next decades.
3.3. Small interfering RNAs and microRNAs
Patisiran (ALN-TTR02) (Fig. 5) is the first double-stranded siRNA drug approved by the FDA for the treatment of the polyneuropathy of hATTR amyloidosis in adults (Wood, 2018). As forementioned, the TTR mRNA is a verified therapeutic target for the treatment of genetic disorder hATTR amyloidosis (e.g., by the ASO drug inotersen) (Ackermann et al., 2012; Ackermann et al., 2016), on which patisiran acts through RNA interference mechanism (Fig. 3B), leading to the reduction of serum TTR protein and TTR protein deposits in tissues (Butler et al., 2016). It is also noteworthy that patisiran was designed to follow miRNA mechanistic actions by targeting the 3’UTR of mutant and wild-type TTR mRNA, instead of the coding region. Consisting of some specific chemical modifications, such as 2′-methoxy or 2′-O-methyl on some ribose rings, and two overhang deoxynucleotides at the 3′ termini of both strands (Fig. 5), patisiran represents the only siRNA medication on the market that is formulated into LNP to achieve favorable pharmacokinetics and pharmacodynamics (Zhang, Goel, & Robbie, 2019; Zhang et al., 2020). The effectiveness and safety of intravenous patisiran nanomedicine for the treatment of patients with hATTR amyloidosis was further demonstrated by clinical studies (Adams et al., 2018; Zhang et al., 2020).
Fig. 5.
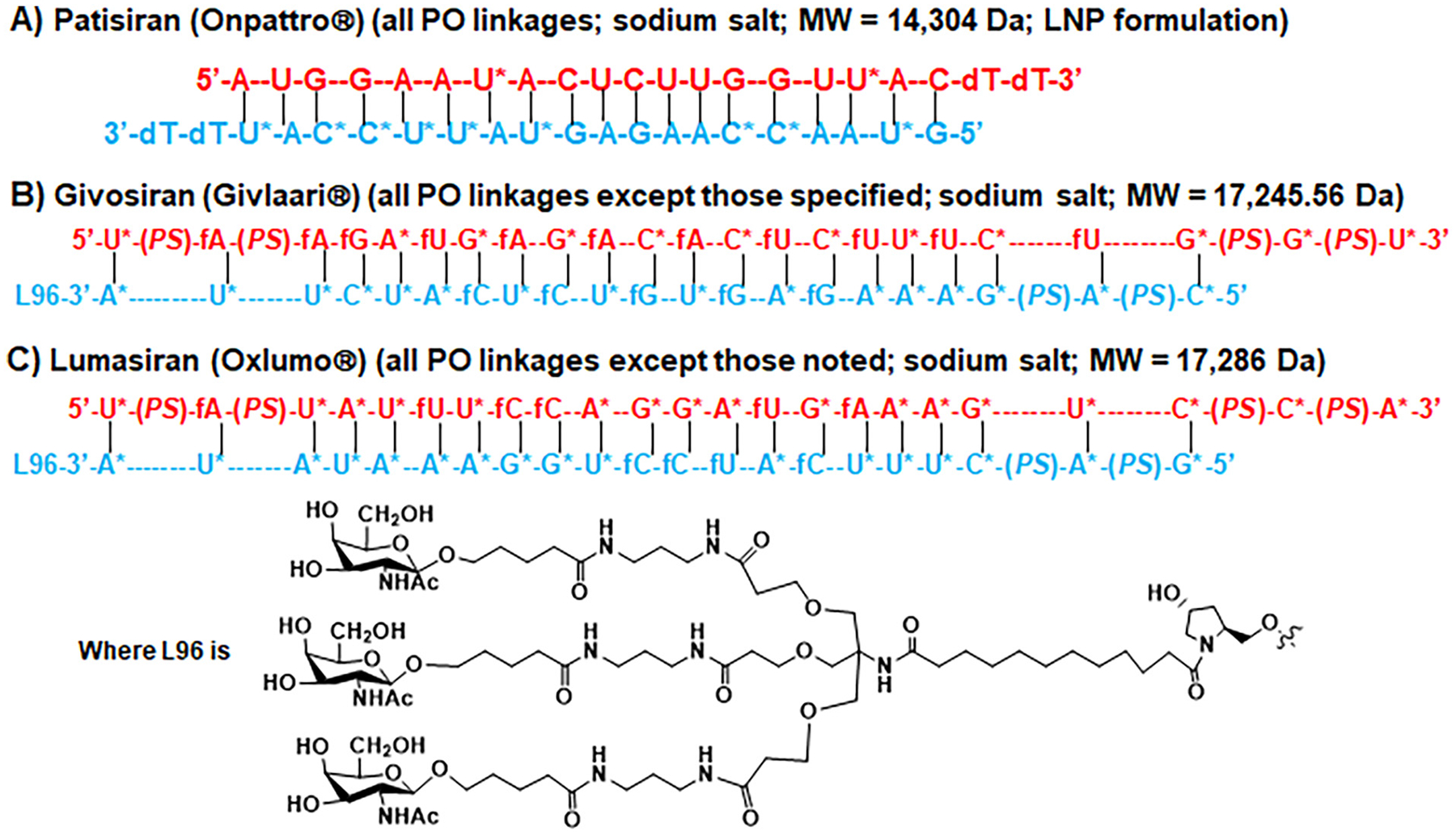
Chemical structural formulas of siRNA therapeutics approved by the FDA. (A) Patisiran is a double-stranded siRNA medication formulated into LNP for the delivery to hepatocytes. (B) Givosiran and (C) lumasiran are both double stranded siRNAs, in which the sense (or passenger) strand is covalently linked to a ligand consisting of three GalNAc moieties facilitating the delivery to hepatocytes, and the antisense (or guide) strand contains two 3′-overhang ribonucleotides connected through phosphoramidate linkages. T = thymine that is also known as 5-methyluracil or m5U; d = deoxy; f = 2′-fluoro; * = 2′-methoxy; PO = phosphodiester; PS = phosphorothioate. Note that both patisiran and lumasiran follow miRNA mechanism of actions to interfere with the 3’UTR of targeted transcripts (TTR and ALAS1, respectively).
Givosiran (ALN-AS1) (Fig. 5) is the second therapeutic siRNA agent approved by the FDA for the treatment of adult patients with acute hepatic porphyria (AHP) (Scott, 2020). Acute attacks and chronic symptoms among AHP patients are attributable to the up-regulation of hepatic δ-aminolevulinic acid synthase 1 (ALAS1), a rate-limiting enzyme in heme biosynthesis that converts succinyl-CoA and glycine into neurotoxic heme intermediates δ-aminolevulinic acid (ALA). The 3′ end of givosiran sense strand is covalently conjugated to a triantennary GalNAc ligand for an enhanced delivery of siRNAs into hepatocytes, while every other end is comprised of two phosphoramidate linkages, along with 2′-methoxy and 2′-fluoro on particular ribose rings (Fig. 5), to reduce nuclease-mediated cleavage and degradation. The antisense strand of givosiran, with two overhang ribonucleotides, was designed to target the coding sequence of hepatic ALAS1 mRNA, leading to the knockdown of ALAS1 gene expression, and thus lower circulating levels of ALA as well as porphobilinogen (de Paula Brandao, Titze-de-Almeida, & Titze-de-Almeida, 2020; Sardh et al., 2019). Clinical studies demonstrated the effectiveness of givosiran for the reduction of circulating ALA levels (Sardh et al., 2019) and porphyria attacks (Balwani et al., 2020), as compared with placebo treatment. Nevertheless, the efficacy was accompanied by a higher frequency of some hepatic and renal adverse events, such as increases in serum aminotransferase and creatinine levels as well as decrease of glomerular filtration rate (Balwani et al., 2020).
Regulatory approval of the third siRNA medication, lumasiran (ALN-GO1), for the treatment of PH1, a rare genetic disease caused by hepatic overproduction of oxalate (Garrelfs et al., 2021; Zhang et al., 2021), further illustrates the utility of GalNAc conjugate-based delivery (Fig. 3A) and unique chemical modifications (Fig. 2) in the development of RNA therapeutics. Similar as givosiran, the 3′ end of lumasiran sense strand is covalently attached to a triantennary GalNAc ligand, while every other terminal consists of two phosphoramidate linkages, as well as 2′-methoxy and 2′-fluoro modifications on particular ribose rings (Fig. 5). Similar as patisiran, lumasiran was designed to follow miRNA mechanism of action to target the 3’UTR of hydroxyacid oxidase or glycolate oxidase 1 (HAO1) transcript, and lumasiran was showed to effectively inhibit oxalate production in preclinical models of PH1 (Liebow et al., 2017). Clinical study further demonstrated that subcutaneous lumasiran treatment significantly reduced urinary oxalate excretion among patients with PH1, with mild and transient injection-site reactions noted in 38% of drug-treated patients (Garrelfs et al., 2021).
With the commercialization of three siRNA drugs, many therapeutic siRNAs and miRNAs are under development and their indications are also extended from genetic disorders to a broader range of infectious, metabolic, and cancerous diseases (Yu et al., 2020b). The records can be found on the ClinicalTrials.gov web site. A number of siRNAs, including vutrisiran (ALN-TTRsc02), nedosiran (DCR PHXC), inclisiran (KJX839), fitusiran (ALN-AT3SC-004), teprasiran (QPI-1002), cosdosiran (QPI-1007), and tivanisiran (SYL1001), have successfully entered into Phase III clinical trials, which is summarized most recently (Zhang et al., 2021). Many others are under early clinical development. One siRNA designed to target the ephrin type-A receptor 2 (EphA2) and formulated with neutral liposome into LNP for the treatment of solid tumors was verified in preclinical animal models (Wagner et al., 2017), whose safety and tolerability is presently under investigation in a Phase I clinical trial (NCT01591356) in patients with advanced solid tumors. As another example, mesenchymal cell-derived exosomes were used to carry a siRNA against the Kirsten rat sarcoma viral oncogene homolog (KRAS) G12D mutant, and the resultant iExosomes were effective to lessen tumor burden in pancreatic cancer xenograft mouse models and improve overall survival (Kamerkar et al., 2017). This iExosome is now under Phase I trial (NCT03608631) in pancreatic cancer patients with KRAS G12D mutation.
Restoration of endogenous miRNA expression and function lost or downregulated in diseased cells represents another promising therapeutic strategy. Many oncolytic miRNAs have been identified, and corresponding chemo-engineered miRNA mimics or biologic reagents were effective for the control of tumor progression and metastasis in animal models (see reviews (Bader et al., 2010; Petrek and Yu, 2019; Rupaimoole and Slack, 2017)). Among them, miR-34a mimics encapsulated with liposome, namely MRX34, was evaluated among patients with advanced solid tumors including unresectable liver cancer (Beg et al., 2017; Hong et al., 2020). While intravenous MRX34 showed antitumor activity in some patients, immune response-related adverse events including fever, chills, fatigue, back pain, nausea, and dyspnea, as well as a few cases of fatality were documented (Beg et al., 2017; Hong et al., 2020), leading to the termination of MRX34. Another miRNA entered into clinical trial is the miR-16-based mimics loaded into minicells (EnGeneIC Dream Vectors), called TargomiRs, to counteract the loss of miR-15/16 family miRNAs in patients with malignant pleural mesothelioma (van Zandwijk et al., 2017). TargomiRs infusion treatment showed some signs of antitumor activity and was tolerated, with cases of dose-limiting toxicities such as anaphylaxis and cardiomyopathy that required dexamethasone prophylaxis (van Zandwijk et al., 2017). These studies provide insights into the development of therapeutic miRNAs concerning about both efficacy and safety. As the knowledge of miRNA mechanism of action has benefited the design of siRNA drugs, the development of miRNA therapeutics shall benefit from the chemistry and pharmaceutics approaches found success in the development of siRNA medications (Fig. 5).
3.4. Messenger RNAs
The BNT162b2, developed by Pfizer, BioNTech, and Fosun Pharma, is the first mRNA vaccine granted for EUA by the FDA for the prevention of unprecedented COVID-19 among individuals 16 years of age and older (Fig. 1) (Oliver et al., 2020). BNT162b2 is an LNP-encapsulated mRNA encoding the full-length SARS-CoV-2 membrane Spike glycoprotein that is modified by 2 proline residues to lock it in the prefusion conformation to increase the potential to provoke virus-neutralizing antibodies (Walsh et al., 2020a). The mRNA sequence consists of pseudouridines incorporated systematically during in vitro transcription towards an enhanced stability and translational capacity as well as lower immunogenicity (Kariko et al., 2008), and LNP components include cationic ((4-hydroxybutyl)azanediyl)di(hexane-6,1-diyl)-bis(2-hexyldecanoate), 2-[(polyethylene glycol)-2000]-N, N-ditetradecylacetamide, 1,2-distearoyl-sn-glycero-3-phosphocholine, and cholesterol (Pardi et al., 2015). Phase I clinical study among healthy adults showed that BNT162b2 immunization provoked dose-dependent SARS-CoV-2-neutralizing antibody titers, while associated with a lower incidence and severity of systemic reactions than the other candidate mRNA vaccine named BNT162b1 (Walsh et al., 2020b). A collection of data from the Phase II/III trials revealed that there were 8 confirmed cases of COVID-19 onset at least 7 days after the second injection among 18,198 participants vaccinated with BNT162b2, as compared to 162 confirmed COVID-19 illnesses among 18,325 participants administered with placebo, indicating BNT162b2 vaccine was 95% more effective than placebo treatment in preventing from COVID-19, or 95% vaccine efficacy by definition (Polack et al., 2020). A 2-month follow-up after the second dose of BNT162b2 vaccine identified some mild-to-moderate adverse events in participants, including pain at the injection site, fatigue, headache, chills, and joint pain, as well as a low incidence of serious adverse events that were showed to be similar between the vaccine and placebo groups (Polack et al., 2020). Although the long-term benefits and risks are unknown (Polack et al., 2020) and concerns remain about possible causes of severe and even life-threatening allergic reactions (Klimek et al., 2021b; Turner et al., 2021), the mRNA vaccines are hoped to mitigate against the ongoing COVID-19 pandemic.
The Moderna mRNA-1273 is the second mRNA vaccine, approved under EUA by the FDA one week after BNT162b2, for the prevention of SARS-CoV-2 infection in individuals 18 years of age and older (Fig. 1) (Oliver et al., 2021). Likewise, mRNA-1273 vaccine is an LNP-encapsulated and 1-methylpseudouridine-substituted (Richner et al., 2017) mRNA that encodes the pre-fusion stabilized Spike protein of SARS-CoV-2 virus (Corbett et al., 2020; Jackson et al., 2020). The open-label, dose-escalation (25–100 μg, twice, 28 days apart), Phase I clinical trial carried out among 45 healthy adults showed that mRNA-1273 vaccine induced anti-SARS-CoV-2 antibody titers and immune responses, without trial-limiting safety concerns amid mild-to-moderate adverse events identified in more than half the participants (Jackson et al., 2020). The randomized, observer-blinded, placebo-controlled Phase III clinical trial (Baden et al., 2021) showed that, starting 14 days after the second dose, a total of 185 symptomatic COVID-19 cases were confirmed out of 14,073 participants received placebo treatment, while 11 COVID-19 cases were identified from 14,134 participants administered mRNA-1273 vaccine, indicating a 94.1% reduction in COVID-19 among the vaccinated group or 94.1% vaccine efficacy (Baden et al., 2021). Similar to previous report (Jackson et al., 2020), moderate and transient local and systemic adverse events, including fever, headache, fatigue, myalgia, arthralgia, chills, and pain at the injection site, were more frequently observed in the mRNA-1273 vaccination group. While serious adverse events were rare and the incidence was similar between the mRNA-1273 vaccine and placebo groups (Baden et al., 2021), the long-term safety and efficacy in protecting against COVID-19 including the emerging new variants remains to be determined.
Many clinical trials are still underway to evaluate BNT162b2 and mRNA-1273 vaccines, their variants or different formulations among healthy subjects or specific populations, as well as new COVID-19 mRNA vaccines, such as CVnCoV by CureVac AG (NCT04838847) and VAW00001 by Sanofi Pasteur (NCT04798027) (clinicaltrials.gov). Actually, before the COVID-19 pandemic, many mRNA vaccines against other infectious and malignant diseases had been investigated in the clinic (Bahl et al., 2017; Cafri et al., 2020; Leal et al., 2018; Papachristofilou et al., 2019; Rittig et al., 2011; Van Gulck et al., 2012; Weide et al., 2009), amid many challenges including mRNA production, quality (e.g., purity), delivery and translation efficiency. Therefore, studies on those experimental vaccines were limited to Phase I/II trials and their benefits (efficacy) versus risks (adverse effects) warrant further investigations among much larger populations. Nevertheless, related concepts and platform technologies ultimately facilitated the quick development and EUA approval of COVID-19 mRNA vaccines. Furthermore, mRNAs encoding specific proteins or antibodies have been under development for protein replacement therapy or antibody therapy (Fig. 3B) for rare genetic disorders as well as more common infection and cancer diseases (Martini and Guey, 2019; Schlake et al., 2019), complementary to conventional protein/antibody therapies with recombinant or bio-engineered proteins/antibodies and gene therapies using DNA materials or gene editing technologies. As one example, defect of propionyl-CoA carboxylase (PCC), an enzyme catalyzing the bio-transformation of propionyl-CoA to methylmalonyl-CoA, leads to the rare genetic and metabolic disorder, namely propionic acidemia (Wongkittichote et al., 2017); an open-label, dose-escalation, Phase I/II study is underway to evaluate the safety, pharmacodynamics, and pharmacokinetics of intravenous mRNA-3927 in participants 1 year of age and older with propionic acidemia amenable to PCC mutations, which encodes functional mitochondrial PCC proteins and is encapsulated within LNP (NCT04159103). Another active Phase Ib clinical trial is to assess the safety, tolerability and pharmacokinetics of intravenous ARCT-810, an LNP product comprising the ornithine transcarbamylase (OTC) mRNA, among adults with documented diagnosis of late onset OTC deficiency (NCT04442347), an X-linked genetic disorder impaired in metabolizing and disposing ammonia (Redant et al., 2021). With limited treatment options, such diseases could ultimately benefit from the treatments with new mRNA therapeutics or vaccines. Moreover, the approval and use of COVID-19 mRNA vaccines has brought great enthusiasm about developing mRNA therapeutics for the treatment cancer and genetic diseases, as well as more vaccines for the prevention of cancer and other infectious diseases.
3.5. Guide RNAs
CRISPR-Cas-based gene editing holds great promise for the treatment of disorders caused by genetic mutations or defects; however, co-delivery of gRNA with the large-size Cas protein or simply their complex (ribonucleoprotein) into the nucleus to achieve efficacious and safe therapy is challenging. Chemo-engineered gRNAs bearing specific chemical modifications towards greater stabilities were showed to improve the efficiency and specificity in genome editing (Hendel et al., 2015; Rahdar et al., 2015). Rather, therapeutic Cas-gRNAs are commonly introduced by using conventional gene delivery systems (e.g., plasmid DNAs and virus vector-based expression) or integrated into cell-based therapies (Doudna, 2020; Manriquez-Roman et al., 2021; Sahel et al., 2019; Zeballos & Gaj, 2021). One completed Phase I trial evaluated the safety and efficacy of CRISPR-edited T cells among 22 participants with refractory non-small-cell lung cancer, in which the programmed cell death protein 1 (PD-1) was altered by using CRISPR-Cas gene editing technology (Lu et al., 2020). In particular, the Cas9 and small gRNA expressing plasmids were co-transfected into ex vivo T cells by electroporation. After infusion, the edited T cells were found to be detectable among 11 of the 12 treated patients, as manifested by the presence of the edited PD-1 gene in peripheral blood mononuclear cells determined by the next-generation sequencing. While the next generation sequencing analyses identified the presence of a fraction of off-target mutants and effects, this study demonstrated the feasibility of therapy with CRISPR-Cas9-gRNA-edited T cells (Lu et al., 2020). Although it is not as advanced as other RNA-based therapies, the CRISPR-Cas-gRNA system has gained momentum in recent years and is progressing from bench to bedside direction.
3.6. Miscellaneous RNAs
Other types of therapeutic RNAs under development include short hairpin RNAs (shRNAs) (Acharya, 2019), saRNAs (Kwok et al., 2019), catalytic RNAs or ribozymes (Burnett and Rossi, 2012; Lewin and Hauswirth, 2001), and miscellaneous ncRNAs (Baptista et al., 2021). Among them, ribozymes are RNA enzymes catalyzing a variety of reactions; both natural and artificial ribozymes may be used to modulate target gene expression towards disease management (James and Gibson, 1998; Lewin and Hauswirth, 2001). Angiozyme is the first synthetic ribozyme entered into clinical trials, which was designed to target the mRNA of VEGF receptor 1 (VEGFR1) to inhibit the angiogenesis towards the control of tumor growth (Kobayashi et al., 2005; Sandberg et al., 2000). However, a Phase II study among patients with metastatic breast cancer revealed that Angiozyme, while showing a tolerable safety profile, did not exhibit any significant efficacy which, might be related to multiple factors such as prior chemotherapies among the enrolled patients, importance of VEGFR1 in overall survival of breast cancer patients, and potency of Angiozyme, precluded it from further development (Morrow et al., 2012).
In contrast to siRNAs that suppress target gene expression, double-stranded saRNAs elevate target gene expression by acting on specific sequences within the promoter regions or factors in transcriptional regulation (Kwok et al., 2019). One saRNA was designed to induce the expression of the transcription factor CCAAT/enhancer-binding protein alpha (CEBPα), a tumor suppressor and critical regulator of hepatocyte function (Reebye et al., 2018). This saRNA is formulated into LNP and named as MTL-CEBPA, and it has entered into clinical investigations for the treatment of hepatocellular carcinoma (HCC) (Sarker et al., 2020; Voutila et al., 2017), as well as advanced solid tumors when combined with PD-1 inhibitor (NCT04105335). Although such forms of RNAs are still at early stages of development, these studies support the concept that RNAs with diverse functions can be employed to control target gene expression for the treatment of human diseases.
4. Conclusions and perspectives
RNAs were once considered as “undruggable” molecules due to their vulnerability to abundant nucleases in body fluids, making them less attractive than other molecular entities. Limited cellular uptake and potential of off-target effects are two other major barriers for the development of RNA therapeutics and vaccines. The approval of the first RNA aptamer drug, pegaptanib, proved the feasibility of developing RNAs as a new molecular entity for therapy. Having experienced ups and downs in the past two decades, RNA therapy is delivering the promise of increasing the diversity of molecular entities of medications and expanding the range of therapeutic targets. Since 2013, seven ASO drugs (mipomersen, eteplirsen nusinersen, inotersen, golodirsen, viltolarsen and casimersen) and three siRNA medications (patisiran, givosiran, and lumasiran) have been successfully marketed, with the development of new RNA technologies, including chemistry, biomaterials, pharmaceutics, and biotechnology, as well as an improved understanding of RNA biology and pathogenesis. In addition, two mRNA vaccines (BNT162b2 and mRNA-1273) have been approved most recently under EUA to combat against the unprecedented COVID-19 pandemic. With the booming of RNA-based therapies, many candidates have been being under clinical and preclinical development, and thus more RNA therapeutics and vaccines are expected to be approved in the future for the treatment or prevention of diverse diseases ranging from rare genetic disorders to more common ones, such as infection and cancer. Meanwhile, one should be mindful of the fact that long-term safety and efficacy of RNA therapeutics and vaccines warrants critical examination and verification, including post-market studies.
New strategies and concepts are also emerging in the development of RNA-based therapies. The success of GalNAc-based siRNA conjugates offers an incentive to pursue ligand-directed delivery of therapeutic RNAs. Efforts are also underway to employ in vivo fermentation approach for the production of true biologic RNA agents and subsequent evaluation of their functions and therapeutic potentials (Yu et al., 2019; Yu et al., 2020a). While mRNA encodes a target protein as antigen for immunization, we speculate that functional ncRNAs may directly serve as antigens to stimulate the immune system for the production of effective antibodies against RNA viruses or pathogens. On the other hand, there is growing interest in exploring and developing small molecules to directly target RNAs (Costales, Childs-Disney, Haniff, & Disney, 2020), which are not only supported by the presence of many RNA-targeted small-molecule antibiotics (Yu et al., 2020b) but also the most recent regulatory approval of risdiplam for the treatment of SMA through its interaction with SMN2 pre-mRNA to alter RNA splicing and increase functional SMN protein levels (Dhillon, 2020). Moreover, new molecular entities may be developed to modulate RNA-binding proteins or RNA-protein complexes critical for disease progression (; Gebauer et al., 2021; Mohibi et al., 2019). The world of RNAs has been opened up; a new era of RNA research and development shall be witnessed, and therapeutic RNAs and vaccines will become a great addition to small molecules, proteins/antibodies, and cell-based modalities to improve the public health.
Acknowledgments
Dr. Yu is supported by grants from the National Cancer Institute [R01CA225958 and R01CA253250], National Institutes of Health, United States.
Abbreviations:
- A
adenine
- AGO2
Argonaute 2
- AHP
acute hepatic porphyria
- ALA
δ-aminolevulinic acid
- ALAS1
δ-aminolevulinic acid synthase 1
- AMD
age-related macular degeneration
- ApoB-100
apolipoprotein B-100
- ASO
antisense oligonucleotide
- asRNA
antisense RNA
- BBB
blood-brain barrier
- CEBPα
CCAAT/enhancer-binding protein alpha
- C
cytosine
- Cas
CRISPR-associated protein
- CMV
cytomegalovirus
- COVID-19
coronavirus disease 2019
- CNS
central nervous system
- CPP
cell penetrating peptide
- CRISPR
clustered regularly interspaced short palindromic repeats
- DLin-MC3-DMA or MC3
dilinoleylmethyl-4-dimethylaminobutyrate
- DMD
Duchenne muscular dystrophy
- DNM2
dynamin2
- DSB
double-stranded DNA break
- DSPE
1,2-distearoyl-sn-glycero-3-phosphoethanolamine
- eIF5A
eukaryotic translation initiation factor 5A
- EphA2
ephrin type-A receptor 2
- ESM
egg sphingomyelin
- EUA
Emergency Use Authorization
- FDA
Food and Drug Administration
- G
guanine
- GalNAc
N-acetylgalactosamine
- gRNA
guide RNA
- HAO1
hydroxyacid oxidase or glycolate oxidase 1
- hATTR amyloidosis
hereditary transthyretin-mediated amyloidosis
- HCC
hepatocellular carcinoma
- HD
Huntington’s disease
- HMGB
high-mobility group box
- HoFH
homozygous familial hypercholesterolemia
- HSPC
hydrogenated soybean phosphatidylcholine
- hTF
human transferrin protein
- IE2
immediate early region 2
- KRAS
Kirsten rat sarcoma viral oncogene homolog
- LDL
low-density lipoprotein
- LNP
lipid nanoparticles
- m5U
5-methyluridine
- miRNA
microRNA
- mRNA
messenger RNA
- ncRNA
noncoding RNA
- N1mΨ
N1-methyl-pseudouridine
- OTC
ornithine transcarbamylase
- PCC
propionyl-CoA carboxylase
- PD-1
programmed cell death protein 1
- PEG
polyethylene glycol
- PEG2000-C-DMG
α-(3′-((1C-di(myristyloxy)proponoxy)-carbonylamino)propyl)-ω-methoxy-polyoxyethylene
- PEG-DSPE
PEGylated DSPE
- PEI
polyethylenimine
- PH1
primary hyperoxaluria type 1
- PKR
RNA-dependent protein kinase
- ψ
pseudouridine
- PMO
phosphorodiamidate morpholine oligonucleotide
- PNA
peptide nucleic acid
- pre-miRNA
precursor miRNA
- PRR
pattern recognition receptor
- PS
phosphorothioate
- RISC
RNA-induced silencing complex
- RNAi
RNA interference
- RNase
ribonuclease
- RNP
ribonucleoprotein
- RRM2
M2 subunit of ribonucleotide reductase
- saRNA
small activating RNA
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- SELEX
systematic evolution of ligands by exponential enrichment
- shRNA
short-hairpin RNA
- siRNA
small interfering RNA
- SMA
spinal muscular atrophy
- SMN
survival motor neuron
- sRNA
small RNA
- SSO
splice-switching oligonucleotides.
- T
thymine
- TLRs
toll-like receptors
- tRNA
transfer RNA
- TTR
transthyretin
- U
uracil
- VEGF
vascular endothelial growth factor
- VEGFR1
VEGF receptor 1
- VLDL
very low -density lipoprotein
- 2’-MOE
2′-methoxyethoxyl
- 3’UTR
3′-untranslated regions
Footnotes
Declaration of Competing Interest
The authors are named inventors of issued and pending patents related to RNA bioengineering technology and use that are owned by the University of California, Davis; and Dr. Yu is a founder of AimRNA, Inc. that intends to license the intellectual property.
References
- Aartsma-Rus A (2017). FDA approval of nusinersen for spinal muscular atrophy makes 2016 the year of splice modulating oligonucleotides. Nucleic Acid Therapeutics 27, 67–69. [DOI] [PubMed] [Google Scholar]
- Abedi-Gaballu F, Dehghan G, Ghaffari M, Yekta R, Abbaspour-Ravasjani S, Baradaran B, … Hamblin MR (2018). PAMAM dendrimers as efficient drug and gene delivery nanosystems for cancer therapy. Applied Materials Today 12, 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya R (2019). The recent progresses in shRNA-nanoparticle conjugate as a therapeutic approach. Materials Science & Engineering. C, Materials for Biological Applications 104, 109928. [DOI] [PubMed] [Google Scholar]
- Ackermann EJ, Guo S, Booten S, Alvarado L, Benson M, Hughes S, & Monia BP (2012). Clinical development of an antisense therapy for the treatment of transthyretin-associated polyneuropathy. Amyloid 19(Suppl. 1), 43–44. [DOI] [PubMed] [Google Scholar]
- Ackermann EJ, Guo S, Benson MD, Booten S, Freier S, Hughes SG, … Monia BP (2016). Suppressing transthyretin production in mice, monkeys and humans using 2nd-generation antisense oligonucleotides. Amyloid 23, 148–157. [DOI] [PubMed] [Google Scholar]
- Adams D, Gonzalez-Duarte A, O’Riordan WD, Yang CC, Ueda M, Kristen AV, … Suhr OB (2018). Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N Engl J Med 379, 11–21. [DOI] [PubMed] [Google Scholar]
- Al-Abd AM, Alamoudi AJ, Abdel-Naim AB, Neamatallah TA, & Ashour OM (2017). Anti-angiogenic agents for the treatment of solid tumors: Potential pathways, therapy and current strategies - a review. Journal of Advanced Research 8, 591–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, & Wood MJ (2011). Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nature Biotechnology 29, 341–345. [DOI] [PubMed] [Google Scholar]
- Anderson KP, Fox MC, Brown-Driver V, Martin MJ, & Azad RF (1996). Inhibition of human cytomegalovirus immediate-early gene expression by an antisense oligonucleotide complementary to immediate-early RNA. Antimicrobial Agents and Chemotherapy 40, 2004–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson BR, Muramatsu H, Nallagatla SR, Bevilacqua PC, Sansing LH, Weissman D, & Kariko K (2010). Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Research 38, 5884–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad RF, Driver VB, Tanaka K, Crooke RM, & Anderson KP (1993). Antiviral activity of a phosphorothioate oligonucleotide complementary to RNA of the human cytomegalovirus major immediate-early region. Antimicrobial Agents and Chemotherapy 37, 1945–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, … Group CS (2021). Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. The New England Journal of Medicine 384, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG, Brown D, & Winkler M (2010). The promise of microRNA replacement therapy. Cancer Research 70, 7027–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl K, Senn JJ, Yuzhakov O, Bulychev A, Brito LA, Hassett KJ, … Ciaramella G (2017). Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Molecular Therapy 25, 1316–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R (1995). Early approval for two lipid-based drugs. BETA 4. [PubMed] [Google Scholar]
- Balwani M, Sardh E, Ventura P, Peiro PA, Rees DC, Stolzel U, … Investigators E (2020). Phase 3 trial of RNAi therapeutic Givosiran for acute intermittent porphyria. The New England Journal of Medicine 382, 2289–2301. [DOI] [PubMed] [Google Scholar]
- Banks WA, Farr SA, Butt W, Kumar VB, Franko MW, & Morley JE (2001). Delivery across the blood-brain barrier of antisense directed against amyloid beta: Reversal of learning and memory deficits in mice overexpressing amyloid precursor protein. The Journal of Pharmacology and Experimental Therapeutics 297, 1113–1121. [PubMed] [Google Scholar]
- Baptista B, Riscado M, Queiroz JA, Pichon C, & Sousa F (2021). Non-coding RNAs: Emerging from the discovery to therapeutic applications. Biochem Pharmacol, 114469. [DOI] [PubMed] [Google Scholar]
- Barile L, & Vassalli G (2017). Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacology & Therapeutics 174, 63–78. [DOI] [PubMed] [Google Scholar]
- Beckert B, & Masquida B (2011). Synthesis of RNA by in vitro transcription. Methods in Molecular Biology 703, 29–41. [DOI] [PubMed] [Google Scholar]
- Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, … Hong DS (2017). Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investigational New Drugs 35, 180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell C, Lynam E, Landfair DJ, Janjic N, & Wiles ME (1999). Oligonucleotide NX1838 inhibits VEGF165-mediated cellular responses in vitro. In Vitro Cellular & Developmental Biology. Animal 35, 533–542. [DOI] [PubMed] [Google Scholar]
- Bennett CF (2019). Therapeutic antisense oligonucleotides are coming of age. Annual Review of Medicine 70, 307–321. [DOI] [PubMed] [Google Scholar]
- Bennett CF, Krainer AR, & Cleveland DW (2019). Antisense oligonucleotide therapies for neurodegenerative diseases. Annual Review of Neuroscience 42, 385–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson MD, Waddington-Cruz M, Berk JL, Polydefkis M, Dyck PJ, Wang AK, … Coelho T (2018). Inotersen Treatment for Patients with Hereditary Transthyretin Amyloidosis. N Engl J Med 379, 22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezhna SY, Supekova L, Supek F, Schultz PG, & Deniz AA (2006). siRNA in human cells selectively localizes to target RNA sites. Proceedings of the National Academy of Sciences of the United States of America 103, 7682–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergers G, & Hanahan D (2008). Modes of resistance to anti-angiogenic therapy. Nature Reviews. Cancer 8, 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget SM, Moore C, & Sharp PA (1977). Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proceedings of the National Academy of Sciences of the United States of America 74, 3171–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard PR, Hutabarat RM, & Thompson KM (2010). Discovery and development of therapeutic aptamers. Annual Review of Pharmacology and Toxicology 50, 237–257. [DOI] [PubMed] [Google Scholar]
- Bramsen JB, & Kjems J (2012). Development of therapeutic-grade small interfering RNAs by chemical engineering. Frontiers in Genetics 3, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruusgaard-Mouritsen MA, Johansen JD, & Garvey LH (2021). Clinical manifestations and impact on daily life of allergy to polyethylene glycol (PEG) in ten patients. Clinical and Experimental Allergy 51, 463–470. [DOI] [PubMed] [Google Scholar]
- Brzustowicz LM, Lehner T, Castilla LH, Penchaszadeh GK, Wilhelmsen KC, Daniels R, … Wood D, et al. (1990). Genetic mapping of chronic childhood-onset spinal muscular atrophy to chromosome 5q11.2–13.3. Nature 344, 540–541. [DOI] [PubMed] [Google Scholar]
- Burnett JC, & Rossi JJ (2012). RNA-based therapeutics: Current progress and future prospects. Chemistry & Biology 19, 60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JS, Chan A, Costelha S, Fishman S, Willoughby JL, Borland TD, … Zimmermann TS (2016). Preclinical evaluation of RNAi as a treatment for transthyretin-mediated amyloidosis. Amyloid 23, 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabanillas B, Akdis C, & Novak N (2021). Allergic reactions to the first COVID-19 vaccine: A potential role of polyethylene glycol? Allergy. 76, 1617–1618. [DOI] [PubMed] [Google Scholar]
- Cafri G, Gartner JJ, Zaks T, Hopson K, Levin N, Paria BC, … Rosenberg SA (2020). mRNA vaccine-induced neoantigen-specific T cell immunity in patients with gastrointestinal cancer. The Journal of Clinical Investigation 130, 5976–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Love KT, Chen Y, Eltoukhy AA, Kastrup C, Sahay G, … Anderson DG (2012). Rapid discovery of potent siRNA-containing lipid nanoparticles enabled by controlled microfluidic formulation. Journal of the American Chemical Society 134, 6948–6951. [DOI] [PubMed] [Google Scholar]
- Chen QX, Wang WP, Zeng S, Urayama S, & Yu AM (2015). A general approach to high-yield biosynthesis of chimeric RNAs bearing various types of functional small RNAs for broad applications. Nucleic Acids Research 43, 3857–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S, Agarwal S, Agarwal S, Aluri KC, Arciprete M, Brown C, & Castellanos-Rizaldos E (2021). The nonclinical disposition and PK/PD properties of GalNA-cconjugated siRNA are highly predictable and build confidence in translation to man. Drug Metabolism and Disposition. 10.1124/dmd.121.000428 Online ahead of print. [DOI] [PubMed] [Google Scholar]
- Cirak S, Arechavala-Gomeza V, Guglieri M, Feng L, Torelli S, Anthony K, … Muntoni F (2011). Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: An open-label, phase 2, dose-escalation study. Lancet 378, 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens PR, Rao VK, Connolly AM, Harper AD, Mah JK, Smith EC, … Investigators CD (2020). Safety, tolerability, and efficacy of Viltolarsen in boys with Duchenne muscular dystrophy amenable to exon 53 skipping: A phase 2 randomized clinical trial. JAMA Neurology 77, 982–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho T, Yarlas A, Waddington-Cruz M, White MK, Sikora Kessler A, Lovley A, & Berk JL (2020). Inotersen preserves or improves quality of life in hereditary transthyretin amyloidosis. Journal of Neurology 267, 1070–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier DA, De Marco A, Ferreira I, Meng B, Datir RP, Walls AC, … Gupta RK (2021). Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature 593, 136–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, … Zhang F (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KS, Edwards DK, Leist SR, Abiona OM, Boyoglu-Barnum S, Gillespie RA, … Graham BS (2020). SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586, 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costales MG, Childs-Disney JL, Haniff HS, & Disney MD (2020). How we think about targeting RNA with small molecules. Journal of Medicinal Chemistry 63, 8880–8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke ST, & Geary RS (2013). Clinical pharmacological properties of mipomersen (Kynamro), a second generation antisense inhibitor of apolipoprotein B. British Journal of Clinical Pharmacology 76, 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke ST, Witztum JL, Bennett CF, & Baker BF (2018). RNA-targeted therapeutics. Cell Metabolism 27, 714–739. [DOI] [PubMed] [Google Scholar]
- Crooke ST, Liang XH, Crooke RM, Baker BF, & Geary RS (2020). Antisense drug discovery and development technology considered in a pharmacological context. Biochem Pharmacol, 114196. [DOI] [PubMed] [Google Scholar]
- Cuellar TL, Barnes D, Nelson C, Tanguay J, Yu SF, Wen X, … Siebel CW (2015). Systematic evaluation of antibody-mediated siRNA delivery using an industrial platform of THIOMAB-siRNA conjugates. Nucleic Acids Research 43, 1189–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullis PR, & Hope MJ (2017). Lipid nanoparticle Systems for Enabling Gene Therapies. Molecular Therapy 25, 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastpeyman M, Sharifi R, Amin A, Karas JA, Cuic B, Pan Y, … Shabanpoor F (2021). Endosomal escape cell-penetrating peptides significantly enhance pharmacological effectiveness and CNS activity of systemically administered antisense oligonucleotides. International Journal of Pharmaceutics 599, 120398. [DOI] [PubMed] [Google Scholar]
- de Paula Brandao PR, Titze-de-Almeida SS, & Titze-de-Almeida R (2020). Leading RNA interference therapeutics part 2: Silencing Delta-Aminolevulinic acid synthase 1, with a focus on Givosiran. Molecular Diagnosis & Therapy 24, 61–68. [DOI] [PubMed] [Google Scholar]
- de Smidt PC, Le Doan T, de Falco S, & van Berkel TJ (1991). Association of antisense oligonucleotides with lipoproteins prolongs the plasma half-life and modifies the tissue distribution. Nucleic Acids Research 19, 4695–4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon S (2020). Risdiplam: First approval. Drugs 80, 1853–1858. [DOI] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, & Reis e Sousa C (2004). Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303, 1529–1531. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Ernberg I, Gait MJ, Green SM, Heaphy S, Karn J, … Valerio R (1989). Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proceedings of the National Academy of Sciences of the United States of America 86, 6925–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, … Gingeras TR (2012). Landscape of transcription in human cells. Nature 489, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA (2020). The promise and challenge of therapeutic genome editing. Nature 578, 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragulescu-Andrasi A, Rapireddy S, He G, Bhattacharya B, Hyldig-Nielsen JJ, Zon G, & Ly DH (2006). Cell-permeable peptide nucleic acid designed to bind to the 5′-untranslated region of E-cadherin transcript induces potent and sequence-specific anti-sense effects. Journal of the American Chemical Society 128, 16104–16112. [DOI] [PubMed] [Google Scholar]
- Drolet DW, Nelson J, Tucker CE, Zack PM, Nixon K, Bolin R, … Bendele RA (2000). Pharmacokinetics and safety of an anti-vascular endothelial growth factor aptamer (NX1838) following injection into the vitreous humor of rhesus monkeys. Pharmaceutical Research 17, 1503–1510. [DOI] [PubMed] [Google Scholar]
- Du L, Kayali R, Bertoni C, Fike F, Hu H, Iversen PL, & Gatti RA (2011). Arginine-rich cell-penetrating peptide dramatically enhances AMO-mediated ATM aberrant splicing correction and enables delivery to brain and cerebellum. Human Molecular Genetics 20, 3151–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early P, Rogers J, Davis M, Calame K, Bond M, Wall R, & Hood L (1980). Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell 20, 313–319. [DOI] [PubMed] [Google Scholar]
- Ecker JR, & Davis RW (1986). Inhibition of gene expression in plant cells by expression of antisense RNA. Proceedings of the National Academy of Sciences of the United States of America 83, 5372–5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein F (1985). Nucleoside phosphorothioates. Annual Review of Biochemistry 54, 367–402. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, & Tuschl T (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, & Joshua-Tor L (2012). The structure of human argonaute-2 in complex with miR-20a. Cell 150, 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington AD, & Szostak JW (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822. [DOI] [PubMed] [Google Scholar]
- Eyetech Study G (2002). Preclinical and phase 1A clinical evaluation of an anti-VEGF pegylated aptamer (EYE001) for the treatment of exudative age-related macular degeneration. Retina 22, 143–152. [DOI] [PubMed] [Google Scholar]
- Faustino NA, & Cooper TA (2003). Pre-mRNA splicing and human disease. Genes & Development 17, 419–437. [DOI] [PubMed] [Google Scholar]
- FDA (1996). FDA approves DaunoXome as first-line therapy for Kaposi’s sarcoma. Food and Drug Administration. J Int Assoc Physicians AIDS Care 2, 50–51. [PubMed] [Google Scholar]
- Ferrara N, & Henzel WJ (1989). Pituitary follicular cells secrete a novel heparin-binding growth factor specific for vascular endothelial cells. Biochemical and Biophysical Research Communications 161, 851–858. [DOI] [PubMed] [Google Scholar]
- Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, … Piccirilli JA (2013). RNA catalyses nuclear pre-mRNA splicing. Nature 503, 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel RS, Chiriboga CA, Vajsar J, Day JW, Montes J, De Vivo DC, … Bishop KM (2016). Treatment of infantile-onset spinal muscular atrophy with nusinersen: A phase 2, open-label, dose-escalation study. Lancet 388, 3017–3026. [DOI] [PubMed] [Google Scholar]
- Finkel RS, Mercuri E, Darras BT, Connolly AM, Kuntz NL, Kirschner J, … Group ES (2017). Nusinersen versus sham control in infantile-onset spinal muscular atrophy. The New England Journal of Medicine 377, 1723–1732. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, & Mello CC (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Folkman J, Merler E, Abernathy C, & Williams G (1971). Isolation of a tumor factor responsible for angiogenesis. The Journal of Experimental Medicine 133, 275–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis SM, Taylor CA, Tang T, Liu Z, Zheng Q, Dondero R, & Thompson JE (2014). SNS01-T modulation of eIF5A inhibits B-cell cancer progression and synergizes with bortezomib and lenalidomide. Molecular Therapy 22, 1643–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DE, Schnell FJ, Akana C, El-Husayni SH, Desjardins CA, Morgan J, … Group S-NS (2020). Increased dystrophin production with golodirsen in patients with Duchenne muscular dystrophy. Neurology 94, e2270–e2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier SM, & Altmann KH (1997). The ups and downs of nucleic acid duplex stability: Structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Research 25, 4429–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon KT, Li L, Chu Y, Janowski BA, & Corey DR (2014). RNAi factors are present and active in human cell nuclei. Cell Reports 6, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galej WP, Toor N, Newman AJ, & Nagai K (2018). Molecular mechanism and evolution of nuclear pre-mRNA and group II intron splicing: Insights from Cryo-electron microscopy structures. Chemical Reviews 118, 4156–4176. [DOI] [PubMed] [Google Scholar]
- Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, & Esteban M (2006). Impact of protein kinase PKR in cell biology: From antiviral to antiproliferative action. Microbiology and Molecular Biology Reviews 70, 1032–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrelfs SF, Frishberg Y, Hulton SA, Koren MJ, O’Riordan WD, Cochat P, … Collaborators, I. -A. (2021). Lumasiran, an RNAi therapeutic for primary Hyperoxaluria type 1. The New England Journal of Medicine 384, 1216–1226. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Schwarzl T, Valcarcel J, & Hentze MW (2021). RNA-binding proteins in human genetic disease. Nature Reviews. Genetics 22, 185–198. [DOI] [PubMed] [Google Scholar]
- Gehrig S, Eberle ME, Botschen F, Rimbach K, Eberle F, Eigenbrod T, … Helm M (2012). Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. The Journal of Experimental Medicine 209, 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gragoudas ES, Adamis AP, Cunningham ET Jr., Feinsod M, Guyer DR, & Group VISiONCT (2004). Pegaptanib for neovascular age-related macular degeneration. The New England Journal of Medicine 351, 2805–2816. [DOI] [PubMed] [Google Scholar]
- Green LS, Jellinek D, Bell C, Beebe LA, Feistner BD, Gill SC, … Janjic N (1995). Nuclease-resistant nucleic acid ligands to vascular permeability factor/vascular endothelial growth factor. Chemistry & Biology 2, 683–695. [DOI] [PubMed] [Google Scholar]
- Group VISiONCT, Chakravarthy U, Adamis AP, Cunningham ET Jr., Goldbaum M, Guyer DR, … Patel M (2006). Year 2 efficacy results of 2 randomized controlled clinical trials of pegaptanib for neovascular age-related macular degeneration. Ophthalmology 113(1508), e1501–e1525. [DOI] [PubMed] [Google Scholar]
- Guidotti G, Brambilla L, & Rossi D (2017). Cell-penetrating peptides: From basic research to clinics. Trends in Pharmacological Sciences 38, 406–424. [DOI] [PubMed] [Google Scholar]
- Guiley KZ, Pratt AJ, & MacRae IJ (2012). Single-pot enzymatic synthesis of dicer-substrate siRNAs. Nucleic Acids Research 40, Article e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddley K (2011). Mipomersen sodium: A new option for the treatment of familial hypercholesterolemia. Drugs Today (Barc) 47, 891–901. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Aartsma-Rus A, Alves S, Borgos SE, Buijsen RAM, Collin RWJ, … Arechavala-Gomeza V (2021). Delivery of oligonucleotide-based therapeutics: Challenges and opportunities. EMBO Molecular Medicine 13, Article e13243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiro S, Mitsuhashi M, Chikami Y, Kawaguchi H, Niimi T, & Yasueda H (2019a). Construction of Corynebacterium glutamicum cells as containers encapsulating dsRNA overexpressed for agricultural pest control. Applied Microbiology and Biotechnology 103, 8485–8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiro S, Mitsuhashi M, & Yasueda H (2019b). Overexpression system for recombinant RNA in Corynebacterium glutamicum using a strong promoter derived from corynephage BFK20. Journal of Bioscience and Bioengineering 128, 255–263. [DOI] [PubMed] [Google Scholar]
- Hassett KJ, Benenato KE, Jacquinet E, Lee A, Woods A, Yuzhakov O, … Brito LA (2019). Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol Ther Nucleic Acids 15, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens MA, & Hastings ML (2016). Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Research 44, 6549–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, … Bauer S (2004). Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303, 1526–1529. [DOI] [PubMed] [Google Scholar]
- Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, … Porteus MH (2015). Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nature Biotechnology 33, 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo YA (2020). Golodirsen: First approval. Drugs 80, 329–333. [DOI] [PubMed] [Google Scholar]
- Ho PY, & Yu AM (2016). Bioengineering of noncoding RNAs for research agents and therapeutics. Wiley Interdiscip Rev RNA 7, 186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PY, Duan Z, Batra N, Jilek JL, Tu MJ, Qiu JX, … Yu AM (2018). Bioengineered noncoding RNAs selectively change cellular miRNome profiles for cancer therapy. The Journal of Pharmacology and Experimental Therapeutics 365, 494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong DS, Kang YK, Borad M, Sachdev J, Ejadi S, Lim HY, … Beg MS (2020). Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. British Journal of Cancer 122, 1630–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, … Hartmann G (2006). 5′-triphosphate RNA is the ligand for RIG-I. Science 314, 994–997. [DOI] [PubMed] [Google Scholar]
- Hua Y, Sahashi K, Rigo F, Hung G, Horev G, Bennett CF, & Krainer AR (2011). Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature 478, 123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iftikhar M, Frey J, Shohan MJ, Malek S, & Mousa SA (2021). Current and emerging therapies for Duchenne muscular dystrophy and spinal muscular atrophy. Pharmacology & Therapeutics 220, 107719. [DOI] [PubMed] [Google Scholar]
- Izant JG, & Weintraub H (1984). Inhibition of thymidine kinase gene expression by anti-sense RNA: A molecular approach to genetic analysis. Cell 36, 1007–1015. [DOI] [PubMed] [Google Scholar]
- Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, … m RNASG (2020). An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N Engl J Med 383, 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey SR (2018). RNA-based fluorescent biosensors for detecting metabolites in vitro and in living cells. Advances in Pharmacology 82, 187–203. [DOI] [PubMed] [Google Scholar]
- James JS (1995). DOXIL approved for KS. AIDS Treat News 6. [PubMed] [Google Scholar]
- James HA, & Gibson I (1998). The therapeutic potential of ribozymes. Blood 91, 371–382. [PubMed] [Google Scholar]
- Jayaraman M, Ansell SM, Mui BL, Tam YK, Chen J, Du X, … Hope MJ (2012). Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angewandte Chemie (International Ed. in English) 51, 8529–8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellinek D, Green LS, Bell C, & Janjic N (1994). Inhibition of receptor binding by high-affinity RNA ligands to vascular endothelial growth factor. Biochemistry 33, 10450–10456. [DOI] [PubMed] [Google Scholar]
- Jhaveri AM, & Torchilin VP (2014). Multifunctional polymeric micelles for delivery of drugs and siRNA. Frontiers in Pharmacology 5, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, … Lee JO (2007). Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 130, 1071–1082. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, & Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirikowski GF, Sanna PP, Maciejewski-Lenoir D, & Bloom FE (1992). Reversal of diabetes insipidus in Brattleboro rats: Intrahypothalamic injection of vasopressin mRNA. Science 255, 996–998. [DOI] [PubMed] [Google Scholar]
- Jones CP, & Ferre-D’Amare AR (2015). RNA quaternary structure and global symmetry. Trends in Biochemical Sciences 40, 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano RL (2016). The delivery of therapeutic oligonucleotides. Nucleic Acids Research 44, 6518–6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Lee SH, Mok H, Chung HJ, & Park TG (2010). Gene silencing efficiency of siRNA-PEG conjugates: Effect of PEGylation site and PEG molecular weight. Journal of Controlled Release 144, 306–313. [DOI] [PubMed] [Google Scholar]
- Kalnin KV, Plitnik T, Kishko M, Zhang J, Zhang D, Beauvais A, … Casimiro D (2021). Immunogenicity and efficacy of mRNA COVID-19 vaccine MRT5500 in preclinical animal models. NPJ Vaccines 6, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, … Kalluri R (2017). Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 546, 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa T, Akiyama F, Kakizaki S, Takashima Y, & Seta Y (2013). Delivery of siRNA to the brain using a combination of nose-to-brain delivery and cell-penetrating peptide-modified nano-micelles. Biomaterials 34, 9220–9226. [DOI] [PubMed] [Google Scholar]
- Kariko K, & Weissman D (2007). Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: Implication for therapeutic RNA development. Current Opinion in Drug Discovery & Development 10, 523–532. [PubMed] [Google Scholar]
- Kariko K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, & Weissman D (2008). Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Molecular Therapy 16, 1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, Bruno JG, Kumar A, & Sharma TK (2018). Aptamers in the therapeutics and diagnostics pipelines. Theranostics 8, 4016–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keam SJ (2018). Inotersen: First global approval. Drugs 78, 1371–1376. [DOI] [PubMed] [Google Scholar]
- Keck PJ, Hauser SD, Krivi G, Sanzo K, Warren T, Feder J, & Connolly DT (1989). Vascular permeability factor, an endothelial cell mitogen related to PDGF. Science 246, 1309–1312. [DOI] [PubMed] [Google Scholar]
- Kesselheim AS, & Avorn J (2016). Approving a problematic muscular dystrophy drug: Implications for FDA policy. JAMA 316, 2357–2358. [DOI] [PubMed] [Google Scholar]
- Khan N, Eliopoulos H, Han L, Kinane TB, Lowes LP, Mendell JR, … the CDI (2019). Eteplirsen treatment attenuates respiratory decline in ambulatory and non-ambulatory patients with Duchenne muscular dystrophy. J Neuromuscul Dis 6, 213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, & Watts JK (2017). The chemical evolution of oligonucleotide therapies of clinical utility. Nature Biotechnology 35, 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Longo M, Han Y, Lundberg P, Cantin E, & Rossi JJ (2004). Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nature Biotechnology 22, 321–325. [DOI] [PubMed] [Google Scholar]
- Kim HW, Jenista ER, Wendell DC, Azevedo CF, Campbell MJ, Darty SN, & Kim RJ (2021). Patients with acute myocarditis following mRNA COVID-19 vaccination. JAMA Cardiol. 10.1001/jamacardio.2021.2828 Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintzing JR, Filsinger Interrante MV, & Cochran JR (2016). Emerging strategies for developing next-generation protein therapeutics for cancer treatment. Trends in Pharmacological Sciences 37, 993–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajewski J, Schneider RJ, Safer B, Munemitsu SM, Samuel CE, Thimmappaya B, & Shenk T (1986). Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2 alpha kinase. Cell 45, 195–200. [DOI] [PubMed] [Google Scholar]
- Klein AF, Varela MA, Arandel L, Holland A, Naouar N, Arzumanov A, … Wood MJ (2019). Peptide-conjugated oligonucleotides evoke long-lasting myotonic dystrophy correction in patient-derived cells and mice. The Journal of Clinical Investigation 129, 4739–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, … Ambati J (2008). Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature 452, 591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimek L, Novak N, Cabanillas B, Jutel M, Bousquet J, & Akdis CA (2021a). Allergenic components of the mRNA-1273 vaccine for COVID-19: Possible involvement of polyethylene glycol and IgG-mediated complement activation. Allergy. 10.1111/all.14794; Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimek L, Novak N, Hamelmann E, Werfel T, Wagenmann M, Taube C, … Worm M (2021b). Severe allergic reactions after COVID-19 vaccination with the Pfizer/BioNTech vaccine in Great Britain and USA: Position statement of the German allergy societies: Medical Association of German Allergologists (AeDA), German Society for Allergology and Clinical Immunology (DGAKI) and Society for Pediatric Allergology and Environmental Medicine (GPA). Allergo J Int 30, 51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop K, Hoogenboom R, Fischer D, & Schubert US (2010). Poly(ethylene glycol) in drug delivery: Pros and cons as well as potential alternatives. Angewandte Chemie (International Ed. in English) 49, 6288–6308. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Eckhardt SG, Lockridge JA, Rothenberg ML, Sandler AB, O’Bryant CL, … Basche ML (2005). Safety and pharmacokinetic study of RPI.4610 (ANGIOZYME), an anti-VEGFR-1 ribozyme, in combination with carboplatin and paclitaxel in patients with advanced solid tumors. Cancer Chemotherapy and Pharmacology 56, 329–336. [DOI] [PubMed] [Google Scholar]
- Kole R, Krainer AR, & Altman S (2012). RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nature Reviews. Drug Discovery 11, 125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren E, & Torchilin VP (2012). Cell-penetrating peptides: Breaking through to the other side. Trends in Molecular Medicine 18, 385–393. [DOI] [PubMed] [Google Scholar]
- Kovacevic KD, Greisenegger S, Langer A, Gelbenegger G, Buchtele N, Pabinger I, … Jilma B (2021). The aptamer BT200 blocks von Willebrand factor and platelet function in blood of stroke patients. Scientific Reports 11, 3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski PS, Rudra A, Miao L, & Anderson DG (2019). Delivering the messenger: Advances in Technologies for Therapeutic mRNA delivery. Molecular Therapy 27, 710–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozma GT, Shimizu T, Ishida T, & Szebeni J (2020). Anti-PEG antibodies: Properties, formation, testing and role in adverse immune reactions to PEGylated nanobiopharmaceuticals. Advanced Drug Delivery Reviews 154–155, 163–175. [DOI] [PubMed] [Google Scholar]
- Kramer A (1996). The structure and function of proteins involved in mammalian pre-mRNA splicing. Annual Review of Biochemistry 65, 367–409. [DOI] [PubMed] [Google Scholar]
- Kwok A, Raulf N, & Habib N (2019). Developing small activating RNA as a therapeutic: Current challenges and promises. Therapeutic Delivery 10, 151–164. [DOI] [PubMed] [Google Scholar]
- Leader B, Baca QJ, & Golan DE (2008). Protein therapeutics: A summary and pharmacological classification. Nature Reviews. Drug Discovery 7, 21–39. [DOI] [PubMed] [Google Scholar]
- Leal L, Guardo AC, Moron-Lopez S, Salgado M, Mothe B, Heirman C, … i Hc (2018). Phase I clinical trial of an intranodally administered mRNA-based therapeutic vaccine against HIV-1 infection. AIDS 32, 2533–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, & Rio DC (2015). Mechanisms and regulation of alternative pre-mRNA splicing. Annual Review of Biochemistry 84, 291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, & Ambros V (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, … Zeviani M, et al. (1995). Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80, 155–165. [DOI] [PubMed] [Google Scholar]
- Lennox KA, & Behlke MA (2016). Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Research 44, 863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW, Cachianes G, Kuang WJ, Goeddel DV, & Ferrara N (1989). Vascular endothelial growth factor is a secreted angiogenic mitogen. Science 246, 1306–1309. [DOI] [PubMed] [Google Scholar]
- Lewin AS, & Hauswirth WW (2001). Ribozyme gene therapy: Applications for molecular medicine. Trends in Molecular Medicine 7, 221–228. [DOI] [PubMed] [Google Scholar]
- Li MM, Addepalli B, Tu MJ, Chen QX, Wang WP, Limbach PA, … Yu AM (2015). Chimeric MicroRNA-1291 biosynthesized efficiently in Escherichia coli is effective to reduce target gene expression in human carcinoma cells and improve Chemosensitivity. Drug Metabolism and Disposition 43, 1129–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ma S, & Yi C (2016). Pseudouridine: The fifth RNA nucleotide with renewed interests. Current Opinion in Chemical Biology 33, 108–116. [DOI] [PubMed] [Google Scholar]
- Li PC, Tu MJ, Ho PY, Jilek JL, Duan Z, Zhang QY, … Yu AM (2018). Bioengineered NRF2-siRNA is effective to interfere with NRF2 pathways and improve Chemosensitivity of human cancer cells. Drug Metabolism and Disposition 46, 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Tian Y, Tu MJ, Ho PY, Batra N, & Yu AM (2019). Bioengineered miR-27b-3p and miR-328–3p modulate drug metabolism and disposition via the regulation of target ADME gene expression. Acta Pharmaceutica Sinica B 9, 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, … Wang Y (2020). The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell 182(1284–1294), Article e1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PC, Tu MJ, Ho PY, Batra N, Tran MML, Qiu JX, … Yu AM (2021). In vivo fermentation production of humanized noncoding RNAs carrying payload miRNAs for targeted anticancer therapy. Theranostics 11, 4858–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebow A, Li X, Racie T, Hettinger J, Bettencourt BR, Najafian N, … Knight J (2017). An investigational RNAi therapeutic targeting Glycolate oxidase reduces oxalate production in models of primary Hyperoxaluria. J Am Soc Nephrol 28, 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light J, & Molin S (1983). Post-transcriptional control of expression of the repA gene of plasmid R1 mediated by a small RNA molecule. The EMBO Journal 2, 93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobato L, Beirao I, Silva M, Bravo F, Silvestre F, Guimaraes S, … Sequeiros J (2003). Familial ATTR amyloidosis: Microalbuminuria as a predictor of symptomatic disease and clinical nephropathy. Nephrology, Dialysis, Transplantation 18, 532–538. [DOI] [PubMed] [Google Scholar]
- Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, … Olson EN (2016). Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351, 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Xue J, Deng T, Zhou X, Yu K, Deng L, … Mok T (2020). Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nature Medicine 26, 732–740. [DOI] [PubMed] [Google Scholar]
- Ma JB, Ye K, & Patel DJ (2004). Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429, 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier KE, & Levy M (2016). From selection hits to clinical leads: Progress in aptamer discovery. Mol Ther Methods Clin Dev 5, 16014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier MA, Jayaraman M, Matsuda S, Liu J, Barros S, Querbes W, … Akinc A (2013). Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Molecular Therapy 21, 1570–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra M, Tomaro-Duchesneau C, & Prakash S (2013). Synthesis of TAT peptide-tagged PEGylated chitosan nanoparticles for siRNA delivery targeting neurodegenerative diseases. Biomaterials 34, 1270–1280. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, … Church GM (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl CW, Aberle JH, Aberle SW, Holzmann H, Allison SL, & Heinz FX (1998). In vitro-synthesized infectious RNA as an attenuated live vaccine in a flavivirus model. Nature Medicine 4, 1438–1440. [DOI] [PubMed] [Google Scholar]
- Manriquez-Roman C, Siegler EL, & Kenderian SS (2021). CRISPR takes the front seat in CART-cell development. BioDrugs 35, 113–124. [DOI] [PubMed] [Google Scholar]
- Marshall WS, & Kaiser RJ (2004). Recent advances in the high-speed solid phase synthesis of RNA. Current Opinion in Chemical Biology 8, 222–229. [DOI] [PubMed] [Google Scholar]
- Martini PGV, & Guey LT (2019). A new era for rare genetic diseases: Messenger RNA therapy. Human Gene Therapy 30, 1180–1189. [DOI] [PubMed] [Google Scholar]
- Martinon F, Krishnan S, Lenzen G, Magne R, Gomard E, Guillet JG, … Meulien P (1993). Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. European Journal of Immunology 23, 1719–1722. [DOI] [PubMed] [Google Scholar]
- Mashiko D, Fujihara Y, Satouh Y, Miyata H, Isotani A, & Ikawa M (2013). Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Scientific Reports 3, 3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Keiser K, Nair JK, Charisse K, Manoharan RM, Kretschmer P, … Manoharan M (2015). siRNA conjugates carrying sequentially assembled trivalent N-acetylgalactosamine linked through nucleosides elicit robust gene silencing in vivo in hepatocytes. ACS Chemical Biology 10, 1181–1187. [DOI] [PubMed] [Google Scholar]
- Mendell JR, Rodino-Klapac LR, Sahenk Z, Roush K, Bird L, Lowes LP, … Eteplirsen Study G (2013). Eteplirsen for the treatment of Duchenne muscular dystrophy. Annals of Neurology 74, 637–647. [DOI] [PubMed] [Google Scholar]
- Mendell JR, Goemans N, Lowes LP, Alfano LN, Berry K, Shao J, … Eteplirsen Study G, and Telethon Foundation DMDIN (2016). Longitudinal effect of eteplirsen versus historical control on ambulation in Duchenne muscular dystrophy. Annals of Neurology 79, 257–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, & Rigo F (2015). Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 518, 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menne J, Eulberg D, Beyer D, Baumann M, Saudek F, Valkusz Z, … Emapticap Study G (2017). C-C motif-ligand 2 inhibition with emapticap pegol (NOX-E36) in type 2 diabetic patients with albuminuria. Nephrology, Dialysis, Transplantation 32, 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercuri E, Darras BT, Chiriboga CA, Day JW, Campbell C, Connolly AM, … Group CS (2018). Nusinersen versus sham control in later-onset spinal muscular atrophy. The New England Journal of Medicine 378, 625–635. [DOI] [PubMed] [Google Scholar]
- Milligan JF, Groebe DR, Witherell GW, & Uhlenbeck OC (1987). Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Research 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min HS, Kim HJ, Naito M, Ogura S, Toh K, Hayashi K, … Kataoka K (2020). Systemic brain delivery of antisense oligonucleotides across the blood-brain barrier with a glucose-coated polymeric Nanocarrier. Angewandte Chemie (International Ed. in English) 59, 8173–8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohibi S, Chen X, & Zhang J (2019). Cancer the’RBP’eutics-RNA-binding proteins as therapeutic targets for cancer. Pharmacology & Therapeutics 203, 107390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery J, Ryan M, Engler R, Hoffman D, McClenathan B, Collins L, & Cooper LT Jr. (2021). Myocarditis following immunization with mRNA COVID-19 vaccines in members of the US military. JAMA Cardiol. 10.1001/jamacardio.2021.2833 Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow T (2013). For patients who inherit homozygous familial hypercholesterolemia, 2 new treatments available. Managed Care 22, 47–48. [PubMed] [Google Scholar]
- Morrow PK, Murthy RK, Ensor JD, Gordon GS, Margolin KA, Elias AD, … Hortobagyi GN (2012). An open-label, phase 2 trial of RPI.4610 (Angiozyme) in the treatment of metastatic breast cancer. Cancer 118, 4098–4104. [DOI] [PubMed] [Google Scholar]
- Mousazadeh H, Pilehvar-Soltanahmadi Y, Dadashpour M, & Zarghami N (2021). Cyclodextrin based natural nanostructured carbohydrate polymers as effective non-viral siRNA delivery systems for cancer gene therapy. Journal of Controlled Release 330, 1046–1070. [DOI] [PubMed] [Google Scholar]
- Mui BL, Tam YK, Jayaraman M, Ansell SM, Du X, Tam YY, … Hope MJ (2013). Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol Ther Nucleic Acids 2, Article e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair JK, Willoughby JL, Chan A, Charisse K, Alam MR, Wang Q, … Manoharan M (2014). Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. Journal of the American Chemical Society 136, 16958–16961. [DOI] [PubMed] [Google Scholar]
- Nair JK, Attarwala H, Sehgal A, Wang Q, Aluri K, Zhang X, … Maier MA (2017). Impact of enhanced metabolic stability on pharmacokinetics and pharmacodynamics of GalNAc-siRNA conjugates. Nucleic Acids Research 45, 10969–10977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallagatla SR, Toroney R, & Bevilacqua PC (2008). A brilliant disguise for self RNA: 5′-end and internal modifications of primary transcripts suppress elements of innate immunity. RNA Biology 5, 140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff CP, Zhou J, Remling L, Kuruvilla J, Zhang J, Li H, … Akkina R (2011). An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4 (+) T cell decline in humanized mice. Sci Transl Med 3, 66ra66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, … Gersbach CA (2016). In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351, 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberg P, & Kichler A (2014). Recent developments in nucleic acid delivery with polyethylenimines. Advances in Genetics 88, 263–288. [DOI] [PubMed] [Google Scholar]
- Ng EW, & Adamis AP (2005). Targeting angiogenesis, the underlying disorder in neovascular age-related macular degeneration. Canadian Journal of Ophthalmology 40, 352–368. [DOI] [PubMed] [Google Scholar]
- Ng EW, Shima DT, Calias P, Cunningham ET Jr., Guyer DR, & Adamis AP (2006). Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nature Reviews. Drug Discovery 5, 123–132. [DOI] [PubMed] [Google Scholar]
- Nielsen PE (2000). Peptide nucleic acids: On the road to new gene therapeutic drugs. Pharmacology & Toxicology 86, 3–7. [DOI] [PubMed] [Google Scholar]
- Nimjee SM, White RR, Becker RC, & Sullenger BA (2017). Aptamers as therapeutics. Annual Review of Pharmacology and Toxicology 57, 61–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien K, Breyne K, Ughetto S, Laurent LC, & Breakefield XO (2020). RNA delivery by extracellular vesicles in mammalian cells and its applications. Nature Reviews. Molecular Cell Biology 21, 585–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SE, Gargano JW, Marin M, Wallace M, Curran KG, Chamberland M, … Dooling K (2020). The advisory Committee on immunization Practices’ interim recommendation for use of Pfizer-BioNTech COVID-19 vaccine - United States, December 2020. MMWR. Morbidity and Mortality Weekly Report 69, 1922–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SE, Gargano JW, Marin M, Wallace M, Curran KG, Chamberland M, … Dooling K (2021). The advisory Committee on immunization Practices’ interim recommendation for use of Moderna COVID-19 vaccine - United States, December 2020. MMWR. Morbidity and Mortality Weekly Report 69, 1653–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz B, Schroder NW, Spreitzer I, Michelsen KS, Kirschning CJ, Hallatschek W, … Schumann RR (2001). Toll-like receptor-2 mediates Treponema glycolipid and lipoteichoic acid-induced NF-kappaB translocation. The Journal of Biological Chemistry 276, 22041–22047. [DOI] [PubMed] [Google Scholar]
- Ortega Rodriguez NR, Audicana Berasategui MT, de la Hoz CB, & Valero Santiago A (2021). The century of mRNA vaccines: COVID-19 vaccines and allergy. Journal of Investigational Allergology & Clinical Immunology 31, 89–91. [DOI] [PubMed] [Google Scholar]
- Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, … Qian Z (2020). Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nature Communications 11, 1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papachristofilou A, Hipp MM, Klinkhardt U, Fruh M, Sebastian M, Weiss C, … Zippelius A (2019). Phase Ib evaluation of a self-adjuvanted protamine formulated mRNA-based active cancer immunotherapy, BI1361849 (CV9202), combined with local radiation treatment in patients with stage IV non-small cell lung cancer. Journal for Immunotherapy of Cancer 7, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N, Tuyishime S, Muramatsu H, Kariko K, Mui BL, Tam YK, … Weissman D (2015). Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. Journal of Controlled Release 217, 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N, Hogan MJ, Porter FW, & Weissman D (2018). mRNA vaccines - a new era in vaccinology. Nature Reviews. Drug Discovery 17, 261–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Song DH, Kim HM, Choi BS, Lee H, & Lee JO (2009). The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458, 1191–1195. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, … Ruvkun G (2000). Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408, 86–89. [DOI] [PubMed] [Google Scholar]
- Petrek H, & Yu AM (2019). MicroRNAs in non-small cell lung cancer: Gene regulation, impact on cancer cellular processes, and therapeutic potential. Pharmacology Research & Perspectives 7, Article e00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrek H, Batra N, Ho PY, Tu MJ, & Yu AM (2019). Bioengineering of a single long noncoding RNA molecule that carries multiple small RNAs. Applied Microbiology and Biotechnology 103, 6107–6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitlick MM, Sitek AN, Kinate SA, Joshi AY, & Park MA (2021). Polyethylene glycol and Polysorbate skin testing in the evaluation of COVID-19 vaccine reactions: Early report. Annals of Allergy, Asthma & Immunology 126, 735–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, & Dahiya R (2008). MicroRNA-373 induces expression of genes with complementary promoter sequences. Proceedings of the National Academy of Sciences of the United States of America 105, 1608–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, … Group CCT (2020). Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. The New England Journal of Medicine 383, 2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooga M, Soomets U, Hallbrink M, Valkna A, Saar K, Rezaei K, … Langel U (1998). Cell penetrating PNA constructs regulate galanin receptor levels and modify pain transmission in vivo. Nature Biotechnology 16, 857–861. [DOI] [PubMed] [Google Scholar]
- Popplewell LJ, Adkin C, Arechavala-Gomeza V, Aartsma-Rus A, de Winter CL, Wilton SD, … Dickson G (2010). Comparative analysis of antisense oligonucleotide sequences targeting exon 53 of the human DMD gene: Implications for future clinical trials. Neuromuscular Disorders 20, 102–110. [DOI] [PubMed] [Google Scholar]
- Quemener AM, Bachelot L, Forestier A, Donnou-Fournet E, Gilot D, & Galibert MD (2020). The powerful world of antisense oligonucleotides: From bench to bedside. Wiley Interdiscip Rev RNA 11, Article e1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragelle H, Vandermeulen G, & Preat V (2013). Chitosan-based siRNA delivery systems. Journal of Controlled Release 172, 207–218. [DOI] [PubMed] [Google Scholar]
- Rahdar M, McMahon MA, Prakash TP, Swayze EE, Bennett CF, & Cleveland DW (2015). Synthetic CRISPR RNA-Cas9-guided genome editing in human cells. Proceedings of the National Academy of Sciences of the United States of America 112, E7110–E7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj TA, Smith AM, & Moore AS (2013). Vincristine sulfate liposomal injection for acute lymphoblastic leukemia. International Journal of Nanomedicine 8, 4361–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redant S, Empain A, Mugisha A, Kamgang P, Attou R, Honore PM, & De Bels D (2021). Management of late onset urea cycle disorders-a remaining challenge for the intensivist? Annals of Intensive Care 11, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reebye V, Huang KW, Lin V, Jarvis S, Cutilas P, Dorman S, … Habib NA (2018). Gene activation of CEBPA using saRNA: Preclinical studies of the first in human saRNA drug candidate for liver cancer. Oncogene 37, 3216–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, … Ruvkun G (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906. [DOI] [PubMed] [Google Scholar]
- Ren Y, Hauert S, Lo JH, & Bhatia SN (2012). Identification and characterization of receptor-specific peptides for siRNA delivery. ACS Nano 6, 8620–8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaee M, Oskuee RK, Nassirli H, & Malaekeh-Nikouei B (2016). Progress in the development of lipopolyplexes as efficient non-viral gene delivery systems. Journal of Controlled Release 236, 1–14. [DOI] [PubMed] [Google Scholar]
- Richner JM, Himansu S, Dowd KA, Butler SL, Salazar V, Fox JM, … Diamond MS (2017). Modified mRNA vaccines protect against Zika virus infection. Cell 169, 176. [DOI] [PubMed] [Google Scholar]
- Rittig SM, Haentschel M, Weimer KJ, Heine A, Muller MR, Brugger W, … Brossart P (2011). Intradermal vaccinations with RNA coding for TAA generate CD8+ and CD4+ immune responses and induce clinical benefit in vaccinated patients. Molecular Therapy 19, 990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins M, Judge A, Liang L, McClintock K, Yaworski E, & MacLachlan I (2007). 2’-O-methyl-modified RNAs act as TLR7 antagonists. Molecular Therapy 15, 1663–1669. [DOI] [PubMed] [Google Scholar]
- Roberts TC, Langer R, & Wood MJA (2020). Advances in oligonucleotide drug delivery. Nature Reviews. Drug Discovery 19, 673–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson DL, & Joyce GF (1990). Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 344, 467–468. [DOI] [PubMed] [Google Scholar]
- Rodrigues FB, Ferreira JJ, & Wild EJ (2019). Huntington’s disease clinical trials corner: June 2019. J Huntingtons Dis 8, 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehr B (1998). Fomivirsen approved for CMV retinitis. Journal of the International Association of Physicians in AIDS Care 4, 14–16. [PubMed] [Google Scholar]
- Rothlisberger P, Gasse C, & Hollenstein M (2017). Nucleic acid aptamers: Emerging applications in medical imaging, nanotechnology, neurosciences, and drug delivery. International Journal of Molecular Sciences 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozema DB, Lewis DL, Wakefield DH, Wong SC, Klein JJ, Roesch PL, … Wolff JA (2007). Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proceedings of the National Academy of Sciences of the United States of America 104, 12982–12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD, … Janjic N (1998). 2’-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. The Journal of Biological Chemistry 273, 20556–20567. [DOI] [PubMed] [Google Scholar]
- Rupaimoole R, & Slack FJ (2017). MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nature Reviews. Drug Discovery 16, 203–222. [DOI] [PubMed] [Google Scholar]
- Saarbach J, Sabale PM, & Winssinger N (2019). Peptide nucleic acid (PNA) and its applications in chemical biology, diagnostics, and therapeutics. Current Opinion in Chemical Biology 52, 112–124. [DOI] [PubMed] [Google Scholar]
- Sahel DK, Mittal A, & Chitkara D (2019). CRISPR/Cas system for genome editing: Progress and prospects as a therapeutic tool. The Journal of Pharmacology and Experimental Therapeutics 370, 725–735. [DOI] [PubMed] [Google Scholar]
- Samaridou E, Heyes J, & Lutwyche P (2020). Lipid nanoparticles for nucleic acid delivery: Current perspectives. Advanced Drug Delivery Reviews 154–155, 37–63. [DOI] [PubMed] [Google Scholar]
- Sandberg JA, Parker VP, Blanchard KS, Sweedler D, Powell JA, Kachensky A, … Blatt LM (2000). Pharmacokinetics and tolerability of an antiangiogenic ribozyme (ANGIOZYME) in healthy volunteers. Journal of Clinical Pharmacology 40, 1462–1469. [PubMed] [Google Scholar]
- Sardh E, Harper P, Balwani M, Stein P, Rees D, Bissell DM, … Anderson KE (2019). Phase 1 trial of an RNA interference therapy for acute intermittent porphyria. The New England Journal of Medicine 380, 549–558. [DOI] [PubMed] [Google Scholar]
- Sarker D, Plummer R, Meyer T, Sodergren MH, Basu B, Chee CE, … Habib N (2020). MTL-CEBPA, a small activating RNA therapeutic upregulating C/EBP-alpha, in patients with advanced liver cancer: A first-in-human, Multicenter, open-label, phase I trial. Clinical Cancer Research 26, 3936–3946. [DOI] [PubMed] [Google Scholar]
- Schirle NT, & MacRae IJ (2012). The crystal structure of human Argonaute2. Science 336, 1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, Kinberger GA, Murray HF, Lima WF, Prakash TP, & MacRae IJ (2016). Structural analysis of human Argonaute-2 bound to a modified siRNA guide. Journal of the American Chemical Society 138, 8694–8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiroli D, Gomara MJ, Maurizi E, Atkinson SD, Mairs L, Christie KA, … Moore T (2019). Effective in Vivo topical delivery of siRNA and gene silencing in intact corneal epithelium using a modified cell-penetrating peptide. Mol Ther Nucleic Acids 17, 891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlake T, Thran M, Fiedler K, Heidenreich R, Petsch B, & Fotin-Mleczek M (2019). mRNA: A novel avenue to antibody therapy? Molecular Therapy 27, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlick T (2018). Adventures with RNA graphs. Methods 143, 16–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder NW, Opitz B, Lamping N, Michelsen KS, Zahringer U, Gobel UB, & Schumann RR (2000). Involvement of lipopolysaccharide binding protein, CD14, and toll-like receptors in the initiation of innate immune responses by Treponema glycolipids. Journal of Immunology 165, 2683–2693. [DOI] [PubMed] [Google Scholar]
- Scott LJ (2020). Givosiran: First Approval. Drugs 80, 335–339. [DOI] [PubMed] [Google Scholar]
- Scott LJ, & Keam SJ (2021). Lumasiran: First approval. Drugs 81, 277–282. [DOI] [PubMed] [Google Scholar]
- Sekijima Y (2015). Transthyretin (ATTR) amyloidosis: Clinical spectrum, molecular pathogenesis and disease-modifying treatments. Journal of Neurology, Neurosurgery, and Psychiatry 86, 1036–1043. [DOI] [PubMed] [Google Scholar]
- Senger DR, Galli SJ, Dvorak AM, Perruzzi CA, Harvey VS, & Dvorak HF (1983). Tumor cells secrete a vascular permeability factor that promotes accumulation of as-cites fluid. Science 219, 983–985. [DOI] [PubMed] [Google Scholar]
- Serrano-Sevilla I, Artiga A, Mitchell SG, De Matteis L, & de la Fuente JM (2019). Natural polysaccharides for siRNA delivery: Nanocarriers based on chitosan, hyaluronic acid, and their derivatives. Molecules 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, & Li F (2020). Cell entry mechanisms of SARS-CoV-2. Proceedings of the National Academy of Sciences of the United States of America 117, 11727–11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley M (2021). Casimersen: First approval. Drugs 81, 875–879. [DOI] [PubMed] [Google Scholar]
- Simons RW, & Kleckner N (1983). Translational control of IS10 transposition. Cell 34, 683–691. [DOI] [PubMed] [Google Scholar]
- Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, … Lieberman J (2005). Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nature Biotechnology 23, 709–717. [DOI] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, … Vornlocher HP (2004). Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432, 173–178. [DOI] [PubMed] [Google Scholar]
- Spiegelman WG, Reichardt LF, Yaniv M, Heinemann SF, Kaiser AD, & Eisen H (1972). Bidirectional transcription and the regulation of phage lambda repressor synthesis. Proceedings of the National Academy of Sciences of the United States of America 69, 3156–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer AD, & Dowdy SF (2018). GalNAc-siRNA conjugates: Leading the way for delivery of RNAi therapeutics. Nucleic Acid Therapeutics 28, 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srimanee A, Arvanitidou M, Kim K, Hallbrink M, & Langel U (2018). Cell-penetrating peptides for siRNA delivery to glioblastomas. Peptides 104, 62–69. [DOI] [PubMed] [Google Scholar]
- Stark BC, Kole R, Bowman EJ, & Altman S (1978). Ribonuclease P: An enzyme with an essential RNA component. Proceedings of the National Academy of Sciences of the United States of America 75, 3717–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CA (2016). Eteplirsen approved for Duchenne muscular dystrophy: The FDA faces a difficult choice. Molecular Therapy 24, 1884–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein EA, Dufour R, Gagne C, Gaudet D, East C, Donovan JM, … McGowan M (2012). Apolipoprotein B synthesis inhibition with mipomersen in heterozygous familial hypercholesterolemia: Results of a randomized, double-blind, placebo-controlled trial to assess efficacy and safety as add-on therapy in patients with coronary artery disease. Circulation 126, 2283–2292. [DOI] [PubMed] [Google Scholar]
- Stephenson ML, & Zamecnik PC (1978). Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proceedings of the National Academy of Sciences of the United States of America 75, 285–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steurer M, Montillo M, Scarfo L, Mauro FR, Andel J, Wildner S, … Gobbi M (2019). Olaptesed pegol (NOX-A12) with bendamustine and rituximab: A phase IIa study in patients with relapsed/refractory chronic lymphocytic leukemia. Haematologica 104, 2053–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone CA Jr., Liu Y, Relling MV, Krantz MS, Pratt AL, Abreo A, … Phillips EJ (2019). Immediate hypersensitivity to polyethylene glycols and Polysorbates: More common than we have recognized. The Journal of Allergy and Clinical Immunology. In Practice 7(1533–1540), Article e1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk JS, Xu Q, Kim N, Hanes J, & Ensign LM (2016). PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Advanced Drug Delivery Reviews 99, 28–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerton JE (2017). Invention and Early history of Morpholinos: From pipe dream to practical products. Methods in Molecular Biology 1565, 1–15. [DOI] [PubMed] [Google Scholar]
- Summerton J, & Weller D (1997). Morpholino antisense oligomers: Design, preparation, and properties. Antisense & Nucleic Acid Drug Development 7, 187–195. [DOI] [PubMed] [Google Scholar]
- Svitkin YV, Cheng YM, Chakraborty T, Presnyak V, John M, & Sonenberg N (2017). N1-methyl-pseudouridine in mRNA enhances translation through eIF2alpha-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Research 45, 6023–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed YY (2016). Eteplirsen: First global approval. Drugs 76, 1699–1704. [DOI] [PubMed] [Google Scholar]
- Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX, … Wagers AJ (2016). In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabrizi SJ, Leavitt BR, Landwehrmeyer GB, Wild EJ, Saft C, Barker RA, … Phase 1–2a I-HSST (2019). Targeting huntingtin expression in patients with Huntington’s disease. The New England Journal of Medicine 380, 2307–2316. [DOI] [PubMed] [Google Scholar]
- Tanji H, Ohto U, Shibata T, Taoka M, Yamauchi Y, Isobe T, … Shimizu T (2015). Toll-like receptor 8 senses degradation products of single-stranded RNA. Nature Structural & Molecular Biology 22, 109–115. [DOI] [PubMed] [Google Scholar]
- Tasfaout H, Buono S, Guo S, Kretz C, Messaddeq N, Booten S, … Laporte J (2017). Antisense oligonucleotide-mediated Dnm2 knockdown prevents and reverts myotubular myopathy in mice. Nature Communications 8, 15661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RE, & Zahid M (2020). Cell penetrating peptides, novel vectors for gene therapy. Pharmaceutics 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GS, Cromwell WC, Ali S, Chin W, Flaim JD, & Davidson M (2013). Mipomersen, an apolipoprotein B synthesis inhibitor, reduces atherogenic lipoproteins in patients with severe hypercholesterolemia at high cardiovascular risk: A randomized, double-blind, placebo-controlled trial. Journal of the American College of Cardiology 62, 2178–2184. [DOI] [PubMed] [Google Scholar]
- Tischer E, Mitchell R, Hartman T, Silva M, Gospodarowicz D, Fiddes JC, & Abraham JA (1991). The human gene for vascular endothelial growth factor. Multiple protein forms are encoded through alternative exon splicing. J Biol Chem 266, 11947–11954. [PubMed] [Google Scholar]
- Tu MJ, Ho PY, Zhang QY, Jian C, Qiu JX, Kim EJ, … Yu AM (2019). Bioengineered miRNA-1291 prodrug therapy in pancreatic cancer cells and patient-derived xenograft mouse models. Cancer Letters 442, 82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C, & Gold L (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510. [DOI] [PubMed] [Google Scholar]
- Turner PJ, Ansotegui IJ, Campbell DE, Cardona V, Ebisawa M, El-Gamal Y, … Committee WAOA (2021). COVID-19 vaccine-associated anaphylaxis: A statement of the world allergy organization anaphylaxis Committee. World Allergy Organization Journal 14, 100517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, … van Ommen GJ (2007). Local dystrophin restoration with antisense oligonucleotide PRO051. The New England Journal of Medicine 357, 2677–2686. [DOI] [PubMed] [Google Scholar]
- Van Gulck E, Vlieghe E, Vekemans M, Van Tendeloo VF, Van De Velde A, Smits E, … Berneman ZN (2012). mRNA-based dendritic cell vaccination induces potent antiviral T-cell responses in HIV-1-infected patients. AIDS 26, F1–12. [DOI] [PubMed] [Google Scholar]
- Van Hoecke L, & Roose K (2019). How mRNA therapeutics are entering the monoclonal antibody field. Journal of Translational Medicine 17, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer L, Moerland M, Gallagher J, van Doorn MB, Prens EP, Cohen AF, … Burggraaf J (2016). Injection site reactions after subcutaneous oligonucleotide therapy. British Journal of Clinical Pharmacology 82, 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zandwijk N, Pavlakis N, Kao SC, Linton A, Boyer MJ, Clarke S, … Reid G (2017). Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. The Lancet Oncology 18, 1386–1396. [DOI] [PubMed] [Google Scholar]
- Vitravene Study G (2002a). A randomized controlled clinical trial of intravitreous fomivirsen for treatment of newly diagnosed peripheral cytomegalovirus retinitis in patients with AIDS. American Journal of Ophthalmology 133, 467–474. [DOI] [PubMed] [Google Scholar]
- Vitravene Study G (2002b). Randomized dose-comparison studies of intravitreous fomivirsen for treatment of cytomegalovirus retinitis that has reactivated or is persistently active despite other therapies in patients with AIDS. American Journal of Ophthalmology 133, 475–483. [DOI] [PubMed] [Google Scholar]
- Voutila J, Reebye V, Roberts TC, Protopapa P, Andrikakou P, Blakey DC, … Habib NA (2017). Development and mechanism of small activating RNA targeting CEBPA, a novel therapeutic in clinical trials for liver cancer. Molecular Therapy 25, 2705–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner MJ, Mitra R, McArthur MJ, Baze W, Barnhart K, Wu SY, … Sood AK (2017). Preclinical mammalian safety studies of EPHARNA (DOPC Nanoliposomal EphA2-targeted siRNA). Molecular Cancer Therapeutics 16, 1114–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh EE, Frenck R, Falsey AR, Kitchin N, Absalon J, Gurtman A, … Gruber WC (2020a). RNA-based COVID-19 vaccine BNT162b2 selected for a pivotal efficacy study. medRxiv. 10.1101/2020.08.17.20176651. [DOI] [Google Scholar]
- Walsh EE, Frenck RW Jr., Falsey AR, Kitchin N, Absalon J, Gurtman A, … Gruber WC (2020b). Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. The New England Journal of Medicine 383, 2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WP, Ho PY, Chen QX, Addepalli B, Limbach PA, Li MM, … Yu AM (2015). Bioengineering novel chimeric microRNA-34a for prodrug cancer therapy: High-yield expression and purification, and structural and functional characterization. The Journal of Pharmacology and Experimental Therapeutics 354, 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, … Nussenzweig MC (2021). mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 592, 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weide B, Pascolo S, Scheel B, Derhovanessian E, Pflugfelder A, Eigentler TK, … Garbe C (2009). Direct injection of protamine-protected mRNA: Results of a phase 1/2 vaccination trial in metastatic melanoma patients. Journal of Immunotherapy 32, 498–507. [DOI] [PubMed] [Google Scholar]
- Weissman D, & Kariko K (2015). mRNA: Fulfilling the promise of gene therapy. Molecular Therapy 23, 1416–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenande E, & Garvey LH (2016). Immediate-type hypersensitivity to polyethylene glycols: A review. Clinical and Experimental Allergy 46, 907–922. [DOI] [PubMed] [Google Scholar]
- Wianny F, & Zernicka-Goetz M (2000). Specific interference with gene function by double-stranded RNA in early mouse development. Nature Cell Biology 2, 70–75. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, & Ruvkun G (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862. [DOI] [PubMed] [Google Scholar]
- Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, & Felgner PL (1990). Direct gene transfer into mouse muscle in vivo. Science 247, 1465–1468. [DOI] [PubMed] [Google Scholar]
- Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, … Stoffel M (2007). Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nature Biotechnology 25, 1149–1157. [DOI] [PubMed] [Google Scholar]
- Wongkittichote P, Ah Mew N, & Chapman KA (2017). Propionyl-CoA carboxylase - a review. Molecular Genetics and Metabolism 122, 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood H (2018). FDA approves patisiran to treat hereditary transthyretin amyloidosis. Nature Reviews. Neurology 14, 570. [DOI] [PubMed] [Google Scholar]
- Xu S, Luk K, Yao Q, Shen AH, Zeng J, Wu Y, … Bauer DE (2019). Editing aberrant splice sites efficiently restores beta-globin expression in beta-thalassemia. Blood 133, 2255–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, … Jacks T (2014). CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai H, Ban T, Wang Z, Choi MK, Kawamura T, Negishi H, … Taniguchi T (2009). HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature 462, 99–103. [DOI] [PubMed] [Google Scholar]
- Yang D, Buchholz F, Huang Z, Goga A, Chen CY, Brodsky FM, & Bishop JM (2002). Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America 99, 9942–9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Fogarty B, LaForge B, Aziz S, Pham T, Lai L, & Bai S (2017). Delivery of small interfering RNA to inhibit vascular endothelial growth factor in zebrafish using natural brain endothelia cell-secreted exosome Nanovesicles for the treatment of brain cancer. The AAPS Journal 19, 475–486. [DOI] [PubMed] [Google Scholar]
- Ye J, Liu E, Gong J, Wang J, Huang Y, He H, & Yang VC (2017). High-yield synthesis of monomeric LMWP(CPP)-siRNA covalent conjugate for effective cytosolic delivery of siRNA. Theranostics 7, 2495–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi WR, Tu MJ, Liu Z, Zhang C, Batra N, Yu AX, & Yu AM (2020). Bioengineered miR-328–3p modulates GLUT1-mediated glucose uptake and metabolism to exert synergistic antiproliferative effects with chemotherapeutics. Acta Pharmaceutica Sinica B 10, 159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu RZ, Lemonidis KM, Graham MJ, Matson JE, Crooke RM, Tribble DL, … Geary RS (2009). Cross-species comparison of in vivo PK/PD relationships for second-generation antisense oligonucleotides targeting apolipoprotein B-100. Biochemical Pharmacology 77, 910–919. [DOI] [PubMed] [Google Scholar]
- Yu AM, Jian C, Yu AH, & Tu MJ (2019). RNA therapy: Are we using the right molecules? Pharmacology & Therapeutics 196, 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AM, Batra N, Tu MJ, & Sweeney C (2020a). Novel approaches for efficient in vivo fermentation production of noncoding RNAs. Applied Microbiology and Biotechnology 104, 1927–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AM, Choi YH, & Tu MJ (2020b). RNA drugs and RNA targets for small molecules: Principles, Progress, and challenges. Pharmacological Reviews 72, 862–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamecnik PC, & Stephenson ML (1978). Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proceedings of the National Academy of Sciences of the United States of America 75, 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, & Bartel DP (2000). RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101, 25–33. [DOI] [PubMed] [Google Scholar]
- Zhang X, Goel V, & Robbie GJ (2019). Pharmacokinetics of Patisiran, the first approved RNA interference therapy in patients with hereditary transthyretin-mediated amyloidosis. Journal of Clinical Pharmacology 60, 573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Long C, Li H, McAnally JR, Baskin KK, Shelton JM, … Olson EN (2017). CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Science Advances 3, Article e1602814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Goel V, Attarwala H, Sweetser MT, Clausen VA, & Robbie GJ (2020). Patisiran pharmacokinetics, pharmacodynamics, and exposure-response analyses in the phase 3 APOLLO trial in patients with hereditary transthyretin-mediated (hATTR) amyloidosis. Journal of Clinical Pharmacology 60, 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeballos CM, & Gaj T (2021). Next-generation CRISPR technologies and their applications in gene and Cell therapy. Trends in Biotechnology 39, 692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MM, Bahal R, Rasmussen TP, Manautou JE, & Zhong XB (2021). The growth of siRNA-based therapeutics: Updated clinical studies. Biochem. Pharmacol, 114432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhilina ZV, Ziemba AJ, & Ebbinghaus SW (2005). Peptide nucleic acid conjugates: Synthesis, properties and applications. Current Topics in Medicinal Chemistry 5, 1119–1131. [DOI] [PubMed] [Google Scholar]
- Zhou J, & Rossi J (2017). Aptamers as targeted therapeutics: Current potential and challenges. Nature Reviews. Drug Discovery 16, 440. [DOI] [PubMed] [Google Scholar]
- Zhu S, Gilbert JC, Liang Z, Kang D, Li M, Tarantino PM, & Jilma B (2020). Potent and rapid reversal of the von Willebrand factor inhibitor aptamer BT200. Journal of Thrombosis and Haemostasis 18, 1695–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerman JE, Gritli I, Tolcher A, Heidel JD, Lim D, Morgan R, … Yen Y (2014). Correlating animal and human phase Ia/Ib clinical data with CALAA-01, a targeted, polymer-based nanoparticle containing siRNA. Proceedings of the National Academy of Sciences of the United States of America 111, 11449–11454. [DOI] [PMC free article] [PubMed] [Google Scholar]


