In this study, Abuhashem et al. investigated the phenotypic and mechanistic details of the requirement for Pol II pausing during mammalian development. The authors found that loss of Pol II pausing stalls pluripotent state transitions within the epiblast of the early mouse embryo, and their work defines the acute and long-term molecular consequences of NELF loss and reveals a role for Pol II pausing in the pluripotency continuum as a modulator of cell state transitions.
Keywords: pluripotency, mouse embryo, epiblast, embryonic stem cells, NELF, pausing, transcription, degron, dTAG
Abstract
Promoter-proximal RNA Pol II pausing is a critical step in transcriptional control. Pol II pausing has been predominantly studied in tissue culture systems. While Pol II pausing has been shown to be required for mammalian development, the phenotypic and mechanistic details of this requirement are unknown. Here, we found that loss of Pol II pausing stalls pluripotent state transitions within the epiblast of the early mouse embryo. Using Nelfb−/− mice and a NELFB degron mouse pluripotent stem cell model, we show that embryonic stem cells (ESCs) representing the naïve state of pluripotency successfully initiate a transition program but fail to balance levels of induced and repressed genes and enhancers in the absence of NELF. We found an increase in chromatin-associated NELF during transition from the naïve to later pluripotent states. Overall, our work defines the acute and long-term molecular consequences of NELF loss and reveals a role for Pol II pausing in the pluripotency continuum as a modulator of cell state transitions.
Transcriptional regulation is a hallmark of cell fate specification (Johnston and Desplan 2010; Cramer 2019). Upstream cell-extrinsic inputs, such as growth factor signaling, mediate cell-intrinsic responses that converge on the transcriptional machinery to regulate recruitment of RNA polymerase II (Pol II) at specific gene targets, and thereby gene expression (Adelman and Lis 2012; Pope and Medzhitov 2018; Core and Adelman 2019). Pol II promoter-proximal pausing (Pol II pausing) has been identified as a key rate-limiting step of transcription in metazoans (Shao and Zeitlinger 2017; Core and Adelman 2019). Pol II pausing represents a brief halt of transcription 30–60 nt downstream from the transcription start site (TSS). This pause is regulated by two protein complexes, the DRB sensitivity-inducing factor (DSIF) and the negative elongation factor (NELF), which is composed of four essential proteins: NELFA, NELFB, NELFC/D, and NELFE (Yamaguchi et al. 1999; Chen et al. 2018). Release of paused Pol II is achieved by phosphorylation of NELF, DSIF, and Pol II by CDK9 (Adelman and Lis 2012). These phosphorylation events result in the dissociation of NELF and progression of DSIF and Pol II into productive elongation.
The functional role of Pol II pausing has been studied in a variety of contexts, predominantly in vitro. Genomic and structural studies have revealed that paused Pol II sterically hinders new initiation events and demonstrated that NELF occupies a large interaction surface with Pol II that is substituted for elongation factors, such as the PAF complex, upon pause release (Shao and Zeitlinger 2017; Vos et al. 2018a,b; Gressel et al. 2019). Kinetically, the stability of the paused polymerase, estimated at a time scale of minutes, highlights the importance of regulating this step (Krebs et al. 2017; Shao and Zeitlinger 2017; Steurer et al. 2018; Gressel et al. 2019). Several transcription factors and signaling components can act specifically on the pause release step to regulate gene expression (Gilchrist et al. 2012; Danko et al. 2013; Henriques et al. 2013; Liu et al. 2015; Williams et al. 2015; Yu et al. 2020). These include the heat shock response, glucocorticoid, TGF-β, and ERK signaling pathways. Attempts to perturb pausing have been achieved primarily via loss-of-function studies of NELF proteins, which play an exclusive role in Pol II pausing but not elongation (Chen et al. 2018). These studies have revealed that NELF is required for early development in Drosophila, zebrafish, and mice (Amleh et al. 2009; Wang et al. 2010; Yang et al. 2016; Abuhashem et al. 2022a). Despite several studies revealing broad requirements of NELF during embryonic development and a variety of tissue-specific contexts in mice, the underlying molecular mechanisms remain largely unknown (Pan et al. 2014; Williams et al. 2015; Hewitt et al. 2019; Robinson et al. 2021).
Development represents a dynamic period of gene regulation in which cells must constantly change their gene expression patterns as they adopt new states (Johnston and Desplan 2010). Consistent with this notion, NELF knockout mice exhibit embryonic lethality at peri-implantation (Amleh et al. 2009; Williams et al. 2015). Given the advantage of mouse embryonic stem cells (mESCs), the in vitro counterpart of the pluripotent epiblast of the embryo, to model key aspects of mouse early embryo development, NELF knockout and knockdown studies in mESCs revealed that Pol II pausing is essential for cellular differentiation (Amleh et al. 2009; Williams et al. 2015). However, interpretation of results from these studies has been complicated due to potential secondary effects resulting from long-term NELF knockout and compounding proliferation defects (Williams et al. 2015). Additionally, the cellular and molecular details of the developmental arrest of embryos remain unclear.
In this study, we performed a comprehensive characterization of the role of NELF in early mouse development, with a focus on pluripotent cell state transitions. We used a Nelfb knockout mouse model to show that Nelfb−/− embryos exhibit normal preimplantation development; they were recovered at Mendelian ratios with cell lineage specification comparable with wild-type stage-matched embryos. We show that postimplantation lineages are properly assigned and positioned, except for the posterior epiblast, and that mutant embryos fail pregastrulation at around embryonic day (E) 5.75. The epiblast lineage is specified during the blastocyst stage, at approximately E3.5, and transitions from a naïve state in the blastocyst to a primed state prior to gastrulation at approximately E6.5 in a step-wise manner (Morgani et al. 2017). To further investigate the molecular basis of the defect observed in embryos lacking NELFB, we took advantage of mESCs as a paradigm for modeling pluripotency transitions from the naïve to the subsequent formative and primed states (Hayashi et al. 2011). To allow efficient, rapid, and reversible depletion of NELFB protein, we designed mESCs harboring homozygous knock-in Nelfb degron alleles using the dTAG system (Nabet et al. 2018). This model recapitulated in vitro the defects of pluripotency transitions and priming observed in vivo in Nelfb−/− embryos and highlighted a requirement for NELFB during pluripotency transitions.
To gain mechanistic insights into the defects observed within the epiblast layer of the embryo, we used the mESC model and coupled chromatin immunoprecipitation followed by sequencing (ChIP-seq) and nascent transcriptomic analyses (PRO-seq) to show widespread binding of NELF at both promoters and enhancers, in support of previous studies of NELF binding in other contexts (Core et al. 2012; Henriques et al. 2018). Our NELFB degron cells enabled acute degradation of the NELF complex in mESCs, which resulted in global loss of Pol II pausing at both gene promoters and enhancers within 30 min. Degrading NELF transiently in the context of pluripotency transitions from naïve to the formative state caused a hyperinduction of genes associated with the formative state accompanied by hypersilencing of genes down-regulated as cells exited the naïve state. This observation is in agreement with recent studies suggesting that the absence of NELF predominantly perturbs state transitions, rather than steady-state cellular functions, in a variety of biological contexts (Hewitt et al. 2019; Yu et al. 2020; Robinson et al. 2021). Accordingly, we observed increased recruitment of NELF to chromatin during pluripotent state transitions. These data lead us to propose a model in which Pol II pausing facilitates transitions between pluripotent states by attenuating and buffering the expression of genes associated with cell identity, thereby enabling coordinated transitioning between successive cell states.
Results
Nelfb−/− embryos display defects in pluripotent epiblast state transitions
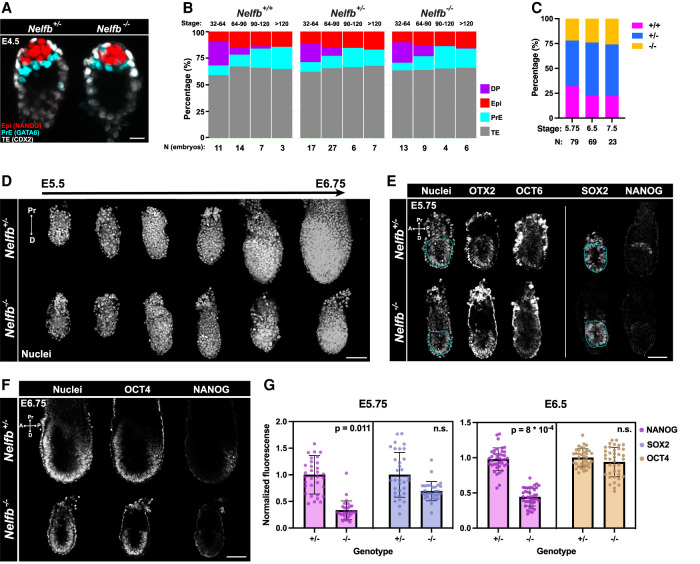
Nelfb−/− mouse embryos exhibit embryonic lethality at postimplantation stages (Amleh et al. 2009). To further characterize the defects observed in Nelfb−/− embryos, we used a mouse model that harbors a deletion of the first four exons of Nelfb, resulting in a protein-null allele (Supplemental Fig. S1A; Williams et al. 2015). Since previous reports suggested that blastocyst stage Nelfb−/− embryos might exhibit defects in cell fate specification, we initiated our analysis at preimplantation stages of embryonic development (Amleh et al. 2009; Williams et al. 2015). We collected early (E3.25) to late (E4.5) stage blastocysts and immunostained them for lineage-specific markers NANOG, GATA6, and CDX2 to identify the pluripotent epiblast (Epi), primitive endoderm (PrE), and trophectoderm (TE) cell lineages, respectively. Embryos were genotyped retrospectively after imaging (Supplemental Fig. S1B). Nelfb−/− blastocysts were morphologically indistinguishable from heterozygous and wild-type littermates and displayed the correct spatial distribution of their three cell lineages (Fig. 1A). To assess the developmental progression of blastocysts, we staged embryos based on their total cell number as an accurate metric of stage and assigned a lineage identity to each cell based on its relative expression of lineage-specific markers (Lou et al. 2014; Saiz et al. 2016a,b). Nelfb−/− embryos did not exhibit a defect in total cell number; ratio of TE, Epi, or PrE; or the gradual assignment of inner cell mass cells (NANOG/GATA6 double positive [DP]) to epiblast and primitive endoderm fates (Fig. 1B; Supplemental Fig. S1C,D). Moreover, we found that Nelfb−/− embryos could be recovered at Mendelian ratios up until postimplantation, but they did show significant defects by E7.5 (Fig. 1C; Supplemental Fig. S1E; Amleh et al. 2009). Thus, our analysis of preimplantation stage mutant mouse embryos suggests that Nelfb is dispensable for cell lineage specification, survival, and embryo implantation.
Figure 1.
Nelfb−/− embryos display defects in pluripotent epiblast state transitions. (A) Immunofluorescence of E4.5 blastocysts labeling epiblast (NANOG), primitive endoderm (GATA6), and trophectoderm (CDX2). Several Z slices are shown in maximum intensity projection (MIP) to show the ICM. Scale bar, 15 μm. (B) Stacked bar plot representing the percentage of each lineage in blastocysts sorted by stage, total cell number per blastocyst, and genotype. (C) Stacked bar plot representing percentage of each Nelfb genotype at different postimplantation stages. (D) Maximum intensity projection (MIP) of embryos dissected at stages between E5.5 and E6.75 at 0.25-d increments. Nuclei are shown to reflect whole embryo. Nuclei were labeled with Hoechst. Scale bar, 100 μm. (E) Immunofluorescence of E5.75 embryos of select pluripotency markers. The bordered region highlights the epiblast cup. The vertical line means separate embryos. Single Z slices are shown. Scale bar, 50 μm. (F) Immunofluorescence of E6.75 embryos of select pluripotency markers. Nuclei were labeled with Hoechst. Single Z slices are shown. Scale bar, 100 μm. (G) Normalized immunofluorescence intensity per epiblast nucleus for pluripotency markers. Single dots are single nuclei. Quantifications show four embryos per group. Statistical testing using t-test was performed on embryo averages. Error bars show standard deviation. P < 0.05 was used to determine significance.
To determine when embryonic development became dysregulated in Nelfb−/− mutants, we collected postimplantation stage embryos just prior to and soon after the onset of gastrulation (E5.5–E6.75). By E6.75, when gastrulation had just initiated, Nelfb−/− embryos were smaller than their wild-type or heterozygous littermates (Fig. 1D). Prior to this stage at E5.75, Nelfb−/− embryos did not display defects in size or cell proliferation, as assayed by measuring the epiblast section area and staining for phosphorylated H3, respectively (Supplemental Fig. S1F,G). To determine whether specification of cell fates was affected, we analyzed the expression and distribution of lineage-specific transcription factors for epiblast (SOX2), visceral endoderm (GATA6), and extraembryonic ectoderm (CDX2). All three lineages were present in mutant embryos, with cells organized in the expected spatial arrangement (Supplemental Fig. S1H,I). We next crossed Nelfb+/− mice to the Afp-GFPTg pan-visceral endoderm and Hex-tdTomatoTg anterior visceral endoderm reporters (Kwon et al. 2006; Wu et al. 2017). Visualization of these lineage-specific reporters revealed that Nelfb−/− embryos possessed a visceral endoderm layer and had successfully specified the distal/anterior visceral endoderm population and that this was able to migrate anteriorly (Supplemental Fig. S1I,J). These results suggest that at E5.75, when the anterior visceral endoderm has completed its migration and prior to the onset of gastrulation, Nelfb−/− embryos are indistinguishable from their wild-type and heterozygous littermates by morphology, lineage-specific marker expression, and localization.
Given previous reports suggesting that Nelfb−/− mESCs, the in vitro counterpart of the epiblast of the embryo, exhibit defects in differentiation, we sought to examine the epiblast population further. Pluripotent epiblast cells are specified in the mid to late blastocyst and subsequently progress through a series of distinct pluripotent states before they exit pluripotency and differentiate at gastrulation (Morgani et al. 2017). Distinct stages in the pluripotency continuum include the early naïve state (E4.5, NANOG+), the subsequent formative state (E5.5, NANOG− and OTX2+), and the later posterior primed state (posterior epiblast at E5.75–E6.5, NANOG+OTX2+). At E5.75, we found that cells of the epiblast of Nelfb−/− embryos successfully induced expression of the formative state markers OTX2 and OCT6 (Fig. 1E). However, mutant embryos lacked a weak NANOG+ population representing the posterior primed state. By E6.75, the posterior primed population expressed NANOG robustly and surrounded the primitive streak in heterozygous and wild-type embryos but remained largely absent in Nelfb−/− embryos despite expression of comparable levels of the pan-pluripotency marker OCT4 (Fig. 1F,G). Subsequently, mutant embryos failed to induce a T+ primitive streak marking the site of gastrulation at E6.75 (Supplemental Fig. S1K). These results show that Nelfb−/− embryos exhibit defects at early postimplantation, after the AVE has migrated but before the onset of gastrulation (at approximately E5.75), where cells of the posterior epiblast are unable to attain a posterior primed state and progress to gastrulation.
NELFB-depleted mESCs recapitulate defects in pluripotent state transitions observed in the embryo
To further characterize the pluripotency transition defects observed in vivo in mutant embryos, we sought to develop an in vitro model of NELFB loss in mESCs. Naïve mESCs can be cultured under defined conditions in the presence of FGF and ACTIVIN to model pluripotency transitions to the subsequent formative and primed states (Hayashi et al. 2011; Morgani et al. 2017). We failed to derive mESCs from Nelfb−/− embryos, consistent with previous reports (Williams et al. 2015). Although previous studies of NELFB in cell culture models used either knockdown or conditional knockout systems, these methods require days to achieve successful depletion or deletion, resulting in an inability to discern primary versus secondary effects of NELF loss (Wu et al. 2020). We therefore took advantage of the dTAG protein degron system (Nabet et al. 2018). By fusing a protein of interest to a FKBP12F36V tag, the target protein can be acutely degraded using a heterobifunctional small molecule, such as dTAG-13, that targets FKBP12F36V for proteasomal degradation (Fig. 2A).
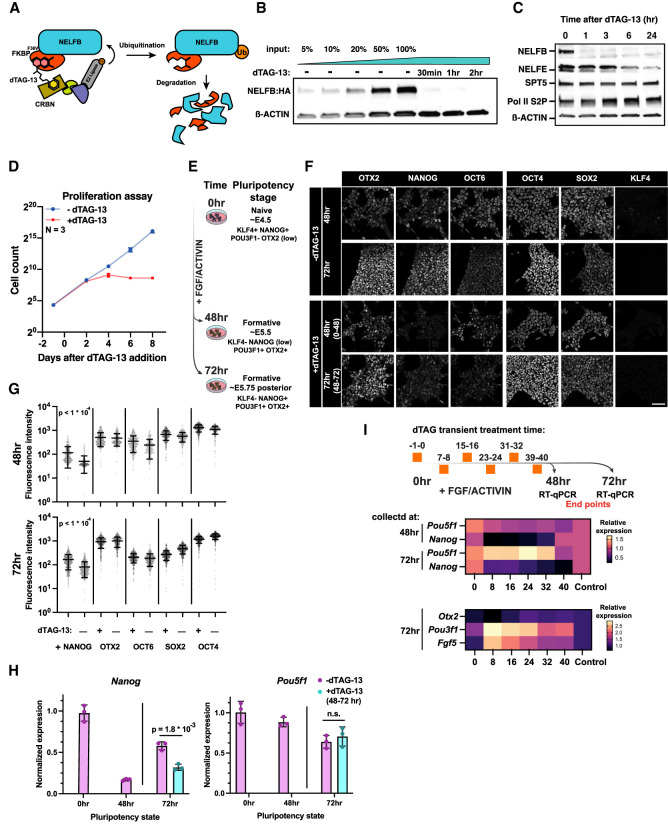
Figure 2.
NELFB-depleted mESCs recapitulate defects in pluripotent state transitions observed in the embryo. (A) Schematic of the dTAG targeted protein degradation system. (B) Western blot of NELFB degradation efficiency and dynamics following 500 nM dTAG-13 treatment. Input refers to relative amount of protein loaded to the gel. (C) Western blot of transcription-associated proteins following NELFB degradation for varying time periods. (D) Proliferation assay of Nelfbdeg mESCs in the presence and absence of 500 nM dTAG-13. Cells were counted and passaged every 2 d. (E) Schematic of the pluripotency transition protocol in vitro. The schematic shows corresponding in vivo stages and marker expression. (F) Immunofluorescence of Nelfbdeg mESCs following pluripotency transitions with and without dTAG-13 at 48 and 72 h. The time interval in parentheses in the treatment panels refers to the time of adding dTAG-13. Scale bar, 50 μm. (G) Quantification of immunofluorescence data in F. The quantification was performed automatically using MINS (see the Materials and Methods). Mean and standard deviation are shown. Statistical testing was performed using a t-test. (H) Normalized RT-qPCR of select factors from the experiment in F. The +dTAG-13 marks the addition of dTAG-13 between hours 48 and 72 of pluripotency transitions. Data were normalized to Actb levels. Statistical testing using t-test was performed on embryo averages. Error bars show standard deviation. P < 0.05 was used to determine significance. (I, top) Schematic of the experiment showing different times of adding dTAG-13 for 1 h followed by washing. Each time point represents one condition. Cells were collected for RT-qPCR at hours 48 and 72 of transitions. (Middle) Heat map of normalized RT-qPCR expression relative to control. Naïve factors are shown. (Bottom) Heat map of normalized RT-qPCR expression relative to control. Formative factors are shown.
We generated a Nelfb-FKBP12F36V-2xHA homozygous knock-in mESC line (referred to here as Nelfbdeg) using CRISPR editing with homology-directed repair (HDR) (Supplemental Fig. S2A,B,E; Ran et al. 2013). We noted that our system is capable of degrading NELFB to undetectable levels within 30 min following the addition of the degradation-inducing small molecule dTAG-13 (Fig. 2B; Supplemental Fig. S2C). Upon dTAG-13 washing, NELFB levels recovered significantly within 3–5 h, demonstrating the reversibility of the system (Supplemental Fig. S2D). Notably, NELFB degradation did not affect the levels of other proteins associated with transcription machinery, such as SPT5 and Pol II S2P (Fig. 2C). However, NELFE levels were markedly reduced 24 h after inducing degradation, as expected given the interdependence between the NELF complex proteins NELFA, NELFB, NELFC/D, and NELFE (Fig. 2C; Narita et al. 2007). Thus, degrading one protein within the complex would lead to the dissolution of the entire NELF complex. The mESCs did not display any toxicity to the edited allele or to dTAG-13 treatment in the absence of the edited allele, as assayed by their morphology and proliferation capacity (Supplemental Fig. S2F). Continuous degradation of NELFB resulted in reduced proliferation after 3–4 d and did not affect the expression of pluripotency markers, as previously reported (Fig. 2D; Supplemental Fig. S2G; Amleh et al. 2009; Williams et al. 2015). These data demonstrate that when applied in mESCs, the NELFB degron system achieves specific, rapidly inducible, and reversible protein depletion.
To assess whether NELFB depletion can affect transitions between pluripotent states in vitro, we used a protocol for directing mESCs, representing the naïve state of pluripotency, into epiblast-like cells (EpiLC), representing a subsequent formative/primed pluripotent state (Hayashi et al. 2011). Naïve mESCs were maintained in naïve conditions—basal medium with 2i (MEK and GSK-3β inhibitors) + LIF—and transferred to basal medium with FGF2 + ACTIVIN for 48–72 h to induce pluripotency transitions (Fig. 2E). By 48 h, cells had down-regulated KLF4 and NANOG, two markers associated with the naïve state of pluripotency, and activated expression of the formative pluripotency markers OTX2 and OCT6 (Fig. 2F). At 72 h, cells maintained expression of formative markers while up-regulating NANOG, consistent with their transition to a posterior-like primed pluripotent state (Fig. 2F). Degron-induced NELFB depletion from 0 to 72 h did not affect the onset of expression of formative markers and resulted in a marked loss of NANOG at 48 h, but without subsequent up-regulation at 72 h (Fig. 2F; Supplemental Fig. S2H). Given that continuous degradation of NELFB from 0 to 72 h resulted in reduced cell proliferation, we degraded NELFB for only a 24-h window, at 48–72 h after initiation of FGF2 + ACTIVIN exposure (representing the posterior priming phase). Under these conditions, we found that at 72 h cells recapitulated the failure in NANOG reactivation, but without affecting cell proliferation (Fig. 2F–H). These data demonstrate that we generated a system that faithfully recapitulates our in vivo findings in embryos in vitro in a pluripotent stem cell model, with a fine temporal control that can uncouple acute from secondary effects resulting from NELFB loss. Furthermore, these data define a 24-h window of time when NELFB is required within the epiblast and reveal that acute loss of NELFB specifically affects epiblast cells as they transition between OTX2+OCT6+NANOG− and subsequent OTX2+OCT6+NANOG+ states.
To further define the temporal requirement of NELFB during this process, we took advantage of the reversibility of our protein degradation system. mESCs were cultured in the presence of FGF2 + ACTIVIN to transition them from naïve to formative pluripotent states over a 72 h period of time. During this transition, we performed 1 h of NELFB degradation at 0, 8, 16, 24, 32, 40 h (for example, treating with dTAG-13 between −1 and 0 h, 7 and 8 h, and so forth). Samples were collected for RT-qPCR analysis at 48 and 72 h (the end point of the experiment). We found that treatments at 16, 24, and 32 h reproduced the effect on Nanog expression at both the 48- and 72-h time points, whereas no change was observed when degradation was performed at 0 h, immediately before initiating the transition (Fig. 2I; Supplemental Fig. S2I). Concomitantly, certain formative markers, including Fgf5 and Pou3f1 (encoding OCT6 protein), were further up-regulated at the same time points, with little to no change observed in the level of expression of the pan-pluripotency marker Pou5f1 (encoding OCT4 protein) (Fig. 2I; Supplemental Fig. S2I). Notably, Nanog expression was not reduced when cells maintained in the naïve state were treated with dTAG-13 for 72 h, in agreement with previous studies (Supplemental Fig. S2J; Amleh et al. 2009; Williams et al. 2015). These results suggest that NELFB and Pol II pausing are required for fine-tuning of gene regulatory networks when cells transition between successive pluripotent states, rather than during steady states of pluripotency, but are dispensable for the induction of formative state transition upon FGF2 + ACTIVIN treatment. Indeed, pretreating naïve cells with dTAG-13 for 30 min followed by addition of FGF + ACTIVIN for 30 min did not negatively affect the induction of immediate FGF targets, such as Fos and Dusp1 (Supplemental Fig. S2K).
NELF marks active promoters and enhancers in mESCs
To investigate the function of pausing during pluripotency transitions, we first determined the chromatin occupancy of the NELF complex. We performed ChIP-seq of NELFB, NELFE, and SPT5 in Nelfbdeg mESCs maintained in serum/LIF conditions. NELFB and NELFE are essential components of the NELF complex, along with NELFA and NELFCD, and are expected to be present solely at Pol II pausing sites, while SPT5 plays an important role in Pol II pausing as well as productive elongation upon phosphorylation by CDK9, making it detectable at both pausing and productive elongation regions (Chen et al. 2018). NELFB, NELFE, and SPT5 showed correlated signals at transcription start sites (TSSs) of protein-coding genes (Fig. 3A; Supplemental Fig. S3A,G). NELFB and NELFE peaks (Padj < 0.05) highly overlapped, suggesting that our NELFB degron protein fusion maintained its normal chromatin binding capacity (Supplemental Fig. S3B). Annotation of NELFE and NELFB peaks revealed that a subset of called peaks (∼25%) did not correspond to gene TSSs, but instead mapped to intronic and intergenic regions (Supplemental Fig. S3E). We hypothesized that active, transcribed enhancers may show NELF binding in mESCs and that these likely represented the ∼25% of peaks not associated with gene promoters. Indeed, a large proportion of these peaks mapped to known mESC enhancers and also correlated with SPT5 occupancy (Fig. 3D–F; Supplemental Fig. S3F; Whyte et al. 2013). Notably, nearly all annotated superenhancers contained NELF peaks (Fig. 3E). Since superenhancers generally have higher levels of transcription than typical enhancers, we suspect that NELF peaks correlate with levels of transcription at enhancers (Henriques et al. 2018). These data are in agreement with reports suggesting that Pol II pausing is widespread at enhancers and suggest that, as with gene TSSs, NELF is a component of the pausing complex at enhancers in mammalian cells (Core et al. 2012; Henriques et al. 2018). Notably, the identification of NELF at enhancers, as well as promoters, in our system reveals a potential role for Pol II pausing/transcription in enhancer regulation during pluripotent state transitions.
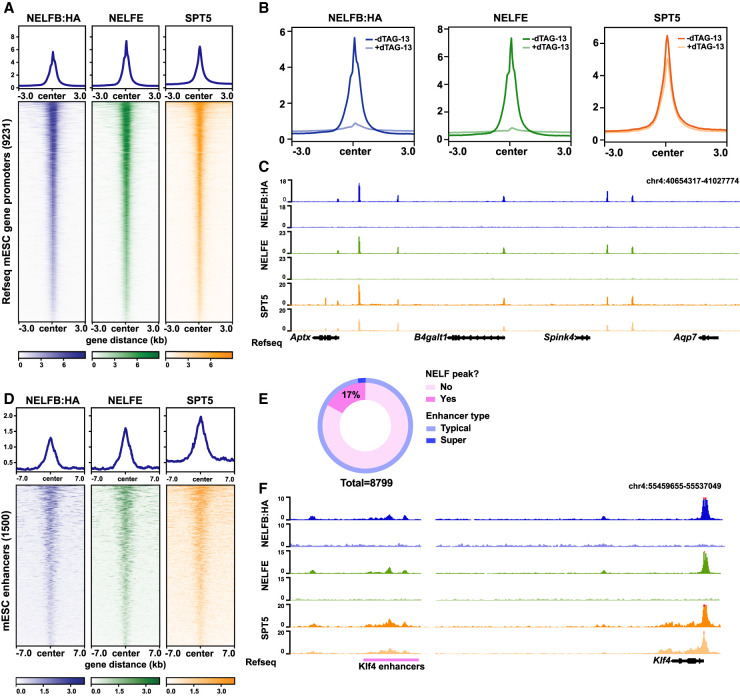
Figure 3.
NELF displays widespread binding at promoters and enhancers, and Nelfbdeg enables acute clearance of the NELF complex from chromatin. (A) Heat map of NELFB, NELFE, and SPT5 ChIP-seq signal at active protein-coding genes’ promoters in mESCs. Active promoters were designated as TSSs that contain an SPT5 peak (Q-value < 0.05). (B) Metaplot of ChIP-seq signals at promoters defined in A with and without 30 min of dTAG-13. (C) Genome browser shot of a representative region for metaplots in B. (D) Heat map of NELFB, NELFE, and SPT5 ChIP-seq signal at mESC-specific enhancers (Whyte et al. 2013). Enhancers with NELF peaks (Q-value < 0.05) are shown. (E) The ratio of enhancers and superenhancers that contain NELF peaks. (F) Genome browser shot of a representative enhancer region showing NELF peaks.
NELFB degradation results in acute clearance of the complex from chromatin
To test the immediate effect of degrading NELFB on the NELF complex and SPT5, we performed ChIP-seq in matched samples after 30 min of mESC culture in the presence of dTAG-13. As expected, NELFB peaks were abolished within 30 min (Fig. 3B,C; Supplemental Fig. S3A,D). Consistent with the interdependence of individual NELF complex subunits, NELFE peaks were similarly abolished (Fig. 3B,C; Supplemental Fig. S3A,D). Spike-in-normalized SPT5 peaks around TSSs showed a global reduction of ∼25% (Fig. 3B,C; Supplemental Fig. S3A). The reduced SPT5 signal suggests that acute disruption of the NELF complex perturbs Pol II pausing but does not abolish transcription entirely. Overall, these results show that Nelfbdeg mESCs can rapidly and specifically remove the NELF complex from chromatin with dTAG treatment and degradation of NELFB and thus can be used to study the consequences of an acute loss of Pol II pausing. Our results are consistent with recent experiments degrading NELFC/D in a human DLD-1 cell line (Aoi et al. 2020).
NELFB stabilizes Pol II pausing and transcription at promoters and enhancers
Our observations prompted us to study changes in nascent transcription globally upon NELFB depletion in Nelfbdeg mESCs. To assess nascent transcription, we used precision run-on sequencing (PRO-seq) (Kwak et al. 2013; Mahat et al. 2016). PRO-seq identifies the position of transcriptionally engaged RNA polymerases at approximately single-base resolution and allows an assessment of transcription at TSSs, gene bodies, and regulatory elements, including enhancers (Wissink et al. 2019). We were particularly interested in identifying the immediate, direct effects of NELFB loss on transcription. To do so, we treated mESCs in serum/LIF with dTAG-13 for 30 and 60 min and then collected nuclei for analysis (Fig. 4A). We analyzed two to three replicates per condition and used a spike-in to normalize for general transcriptional changes. Replicates showed good correlation (Supplemental Table S1). Metagene plots revealed a loss of signal at TSSs and gene bodies at both the 30- and 60-min time points (Fig. 4B).
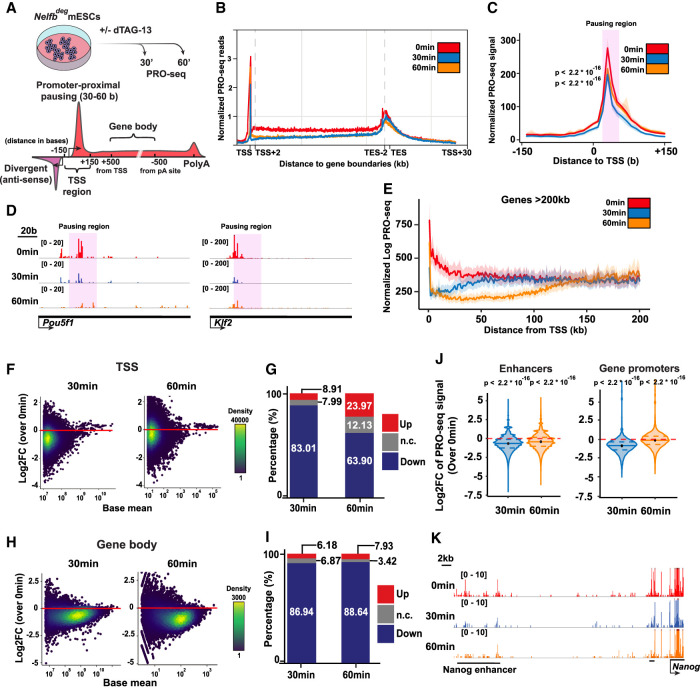
Figure 4.
NELF stabilizes Pol II pausing and transcription at promoters and enhancers. (A) Schematic of treatments of 30 and 60 min before PRO-seq analysis (top), and regions of each defined DNA element in the following analysis (bottom). (B) Metaplot of scaled protein-coding genes’ PRO-seq signal relative to TSSs and TESs. (C) Metaplot of PRO-seq signal at TSSs. The highlighted region marks the proximal-pausing region. Statistical testing was performed using Wilcoxon and paired t-tests with similar results. (D) Genome browser shot of TSS regions of example pluripotency genes. The highlighted region marks the proximal-pausing region. (E) Metaplot of PRO-seq signal at genes >200 kb. (F) Log2 fold change of PRO-seq signal at TSSs calculated using DEseq2. (G) Bar plot showing the percentage of up, down, and unchanged loci in F. Padj of 0.05 was used as a cutoff. (H) Log2 fold change of PRO-seq signal at gene bodies calculated using DEseq2. (I) Bar plot showing the percentage of up, down, and unchanged loci in H. Padj of 0.05 was used as a cutoff. (J) Violin plot of TSS log2 fold change data in F separated by enhancer versus protein-coding gene TSSs. Plots show mean, 25th percentile, and 75th percentile inside each violin plot. Statistical testing was performed using Wilcoxon and paired t-tests with similar results. (K) Genome browser shot of an example enhancer signal across treatments.
To investigate these changes further, we focused on TSSs. We used published mESC START-seq data to define the exact positions of TSSs at both gene promoters and regulatory elements (Henriques et al. 2018). TSS metaprofiles revealed the expected Pol II pause peak 30–50 bases downstream from the TSS (Fig. 4C). This peak was significantly and globally reduced when NELFB was depleted (Fig. 4C,D). Importantly, we identified a drop in PRO-seq signal on gene bodies that extended from the TSSs and corresponded with each treatment time and an elongating Pol II speed of ∼1–2 kb/min—a drop across the first ∼40 kb of gene bodies in the 30-min treatment group (Fig. 4E; Supplemental Fig. S4A,C). This phenomenon, previously termed clearing waves, is a result of having elongating polymerases that escape the TSS region prior to dTAG-13 treatment. The presence of clearing waves points to the negative effect of NELFB degradation on transcription, which primarily terminates Pol II around the TSS such that a drop could be seen along gene bodies corresponding to length of treatment. These results suggest that NELF acts on polymerases close to the TSS to enable their efficient transition from pausing to productive elongation. Our findings place NELF as a positive effector required for transcription to proceed effectively and highlight the power and specificity of our degron system.
To determine signal changes at each locus in a pair-wise manner, we assessed differential expression at all active TSSs in mESCs and gene bodies using DESeq2 (Love et al. 2014). In agreement with our previous observations, we noted a global reduction in transcription at TSSs, on average, within 30 min (Fig. 4F–J; Supplemental Fig. S4B,D). Notably, the reduction at 60 min was conserved at enhancer TSSs and gene bodies, but not at gene TSSs (Fig. 4J). This recovery of transcription at gene TSSs from 30 min to 60 min was not found in the canonical Pol II pausing region (∼30 bases from TSSs) but further downstream in an apparent widespread redistribution of the pause peak in the absence of NELF, which presumably stabilizes the pause position to 30–50 bases from TSSs (Fig. 4C; Aoi et al. 2020).
To define the properties of the promoters that displayed Pol II redistribution, we selected a list of significantly recovering gene TSSs (404 genes; at 30 min: down, Padj < 0.05; at 60 min: up, Padj < 0.05) and measured levels of NELF and H3K4me3, which marks active promoters. Additionally, we inferred the rates of initiation and pause release at these promoters using a recently described statistical model (see the Materials and Methods; Siepel 2021). Of note, the initiation and release rate model has been developed to function under steady-state conditions without perturbation. The rates calculated are relative and do not reflect absolute numbers of initiation or release events, which allows for intrasample comparison only. We found that genes exhibiting a redistribution of Pol II consistently harbored high signals for NELFB, NELFE, and H3K4me3, suggesting that these are highly active promoters with significant occurrence of pausing (Supplemental Fig. S4E,F). Measuring the initiation and pause release rates showed that these promoters have a higher initiation rate and a lower release rate, indicating that these genes may have high initiation rates, whereas the rate of pause release is rate limiting to transcriptional activation (Supplemental Fig. S4G). Transient transcriptome sequencing (TT-seq) can detect nascent transcription as well as terminated transcripts, allowing for measurement of initiation rates experimentally (Schwalb et al. 2016). Measuring the signal of recovering genes in publicly available mESCs, TT-seq data confirmed that these promoters are more active in untreated mESCs, indicating higher initiation rates (Supplemental Fig. S4H; Shao et al. 2021). Additionally, the positive correlation between transcriptional activity, NELF ChIP-seq signal, and pause index holds true globally (Supplemental Fig. S4I,J). Overall, we found that Pol II pausing correlates with transcriptional activity globally and that NELF plays a specific role in stabilizing paused polymerases at a defined position 30–50 bases downstream from TSSs, thereby enabling the efficient transition from initiation to productive elongation.
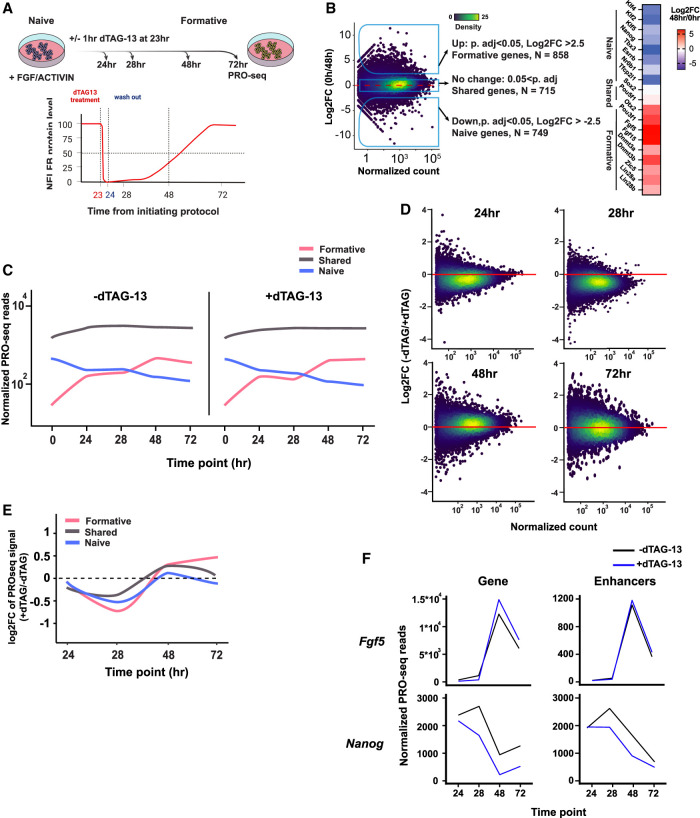
Pol II pausing balances induced and repressed gene regulatory networks during pluripotency transitions
Having established the validity of the Nelfbdeg mESC model and acute molecular consequences of NELFB depletion on Pol II pausing and transcription, we sought to analyze the effect of depleting NELF during pluripotent state transitions. We opted to use a transient pulsed NELF degradation approach in Nelfbdeg mESCs. As described earlier, this treatment regimen was able to recapitulate the state transition defects observed in embryos while minimizing secondary effects. This experimental design enabled us to assess how a minimal perturbation of Pol II pausing during transitions would affect transcription in transitioning cells. Transitioning cells were treated with dTAG-13 for 1 h between hours 23 and 24 of the transitioning protocol in the presence of FGF2 + ACTIVIN, followed by washing and continued culture in the presence of FGF2 + ACTIVIN but in the absence of dTAG-13 for a total of 72 h (Fig. 5A). This treatment resulted in acute depletion of NELFB at the 24-h time point and a recovery over the following 24 h, as we have shown previously (Supplemental Fig. S2D). Samples were collected for PRO-seq at 24, 28, 48, and 72 h. The first two time points represent intermediate points during the transitions, while the latter two time points represent fully transitioned EpiLCs/formative states. Our analysis focused on pluripotency-associated genes that are differentially expressed and genes that maintain a comparative level of expression during state transitions. We identified genes that were up-regulated, down-regulated, and shared between the naïve (0 h) and formative (48 h) stages using DEseq2 in untreated samples (−2.5 > log2FC > 2.5, Padj < 0.05). These groups included many expected genes that are specific to or shared between states, thereby validating the transition of these cells (up: Otx2, Pou3f1, Fgf5, and Fgf15; down: Klf4, Klf2, Nr0b1, and Nanog; shared: Sox2 and Pou5f1) (Fig. 5B).
Figure 5.
NELF balances gene induction and repression during pluripotency transitions. (A, top) Schematic of experiment and analysis time points. (Bottom) Schematic of NELFB protein levels during the experiment following transient depletion. (B, left) Log2 fold change of PRO-seq data for gene expression between 0 and 48 h, which was used to define naïve genes, formative genes, and shared genes. (Right) Heat map of log2 fold change of known naïve and formative markers. (C) Mean normalized PRO-seq reads per gene in each gene class during the transition. Full data range is shown in Supplemental Figure S5A, and heat maps are in Supplemental Figure S5B. (D) Log2 fold change of PRO-seq data for gene expression at each time point of the analysis using DEseq2. (E) Mean log2 fold change of PRO-seq data for gene expression at each time point of the analysis per gene group. Full data range is shown in Supplemental Figure S5C. (F) Normalized gene expression/reads from PRO-seq data at candidate genes and their associated enhancers during the transition protocol. Other genes are shown in Supplemental Figure S5F.
To assess whether NELFB depletion during transitions influenced the cells’ ability to initiate transitions, we generated metaplots and heat maps of naïve and formative genes at each time point with and without dTAG-13 treatment (Fig. 5C; Supplemental Fig. S5A,B). The general trend suggested that treated cells maintained expression of the same genes as untreated cells, in agreement with the ability of Nelfbdeg mESCs to induce formative markers when cultured in dTAG-13 and Nelfb−/− embryos up-regulating formative epiblast markers.
To quantify these observations, we performed a pair-wise differential expression analysis using DEseq2 and tracked the trend of each gene group during the transition. In agreement with our previous results, we found that the 24- and 28-h time points showed a global decrease in transcription when compared with nontreated time-point-matched controls (Fig. 5D). This global decrease was largely recovered by the terminal time points at 48 and 72 h, most likely due to recovery of NELFB protein (Fig. 5D). To directly determine the effect of NELFB depletion on induced (formative-specific), repressed (naïve-specific), and shared genes between both states during the transition, we compared the change in expression of these groups of genes at each time point with and without dTAG-13. While all groups showed initial down-regulation, state-specific groups (naïve and formative) were more severely affected (Fig. 5E; Supplemental Fig. S5C). By 72 h, genes shared between naïve and formative states showed minimal change, while genes induced as cells entered the formative state showed a stronger induction, and genes repressed in the naïve state showed stronger silencing, with several candidate genes showing this trend (Fig. 5E,F; Supplemental Fig. S5F). These data offer evidence for an involvement of Pol II pausing in mediating the levels of expression of genes that are either up-regulated or down-regulated during pluripotency transitions (Fig. 5E; Supplemental Fig. S5C). In the absence of pausing, gene activation and repression are misregulated during pluripotent state transitions.
Previous studies have linked enhancer transcription to target gene promoter activity (Kim et al. 2010; Hah et al. 2013). Given that NELF and Pol II pausing can occur at enhancers, we wanted to assess the enhancer landscape during pluripotent state transitions. To do so, we used two approaches. First, we used the dREG algorithm to identify transcriptional regulatory elements (TREs)—genomic regions that have putative roles in gene regulation at the formative stage (Wang et al. 2019). Overall, TREs showed down-regulated expression in samples treated with dTAG-13 at most time points, re-emphasizing the role of NELF and pausing in maintaining enhancer activity (Supplemental Fig. S5D). To identify specific changes at putative enhancers for genes of interest, we selected TREs that fall within a topologically associated domain (TAD) of a gene of interest and were marked by H3K27ac histone modifications for active enhancers. This strategy enabled us to identify several high-confidence putative enhancers for genes (Supplemental Fig. S5E; see the Materials and Methods). We applied this approach to the Nanog and Fgf5 loci as representative genes that are repressed and induced, respectively, during the transition from naïve to a formative pluripotent state. We found that Nanog and Fgf5 enhancer activities mirrored the trend observed in their respective gene expression (Fig. 5F). The observed changes are consistent with the presence of Pol II pausing at enhancers, as well as the coupling between transcription at enhancers and associated target genes. Overall, these results detail the effects of perturbing pausing during pluripotency transitions at the transcriptional level, where Pol II pausing plays an essential role in balancing gene and enhancer induction and repression during state transitions.
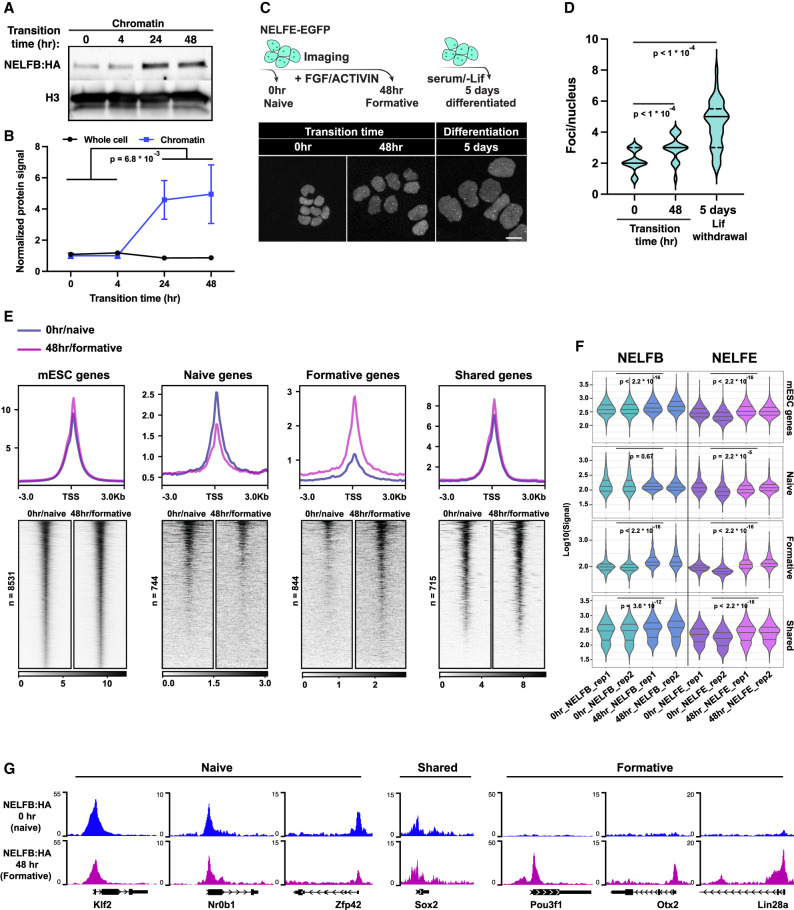
NELF recruitment to chromatin is enhanced during pluripotency transitions
Previous studies on the function of Pol II pausing and NELF in mESCs have suggested that Pol II pausing is not required to maintain pluripotency (Amleh et al. 2009; Williams et al. 2015). Our results in embryos and mESCs using controlled NELFB depletion are in agreement with these observations and extend them by suggesting that Pol II pausing plays a key role as cells change states. We hypothesized that if this is the case, de novo recruitment of NELF to chromatin might be observed during pluripotency transitions. To test this hypothesis, we measured NELFB levels in the chromatin fraction during the transitioning period. In support of our hypothesis, we found a significant increase in chromatin-bound NELFB but not in whole-cell lysates observed at 24 and 48 h of transitioning in FGF2 + ACTIVIN (Fig. 6A,B; Supplemental Fig. S6A). The stable expression of whole-cell NELFB matches the stable expression of all NELF complex components during pluripotency transitions in publicly available data sets (Supplemental Fig. S6B; Yang et al. 2019). Notably, this increase was not observed at 4 h of transitioning, suggesting that NELFB recruitment is not initiated during the acute phase of FGF2 + ACTIVIN stimulation, but rather during the rewiring of transcriptional networks that follows.
Figure 6.
NELF is recruited to chromatin during pluripotency transitions. (A) Chromatin fraction Western blot of cells during pluripotency transitions. (B) Quantification of NELFB in chromatin and whole-cell lysates during pluripotency transitions. Statistical testing was performed using a t-test. Each time point includes two biological replicates. (C) Imaging of NELFE-EGFP in naïve, formative, and randomly differentiated mESCs. (Top) Schematic of the experiment. (Bottom) Images of select time points. (D) Violin plot of the number of NELF bodies per nucleus in conditions presented in C. Statistical testing was performed using a t-test. Mean, 25th percentile, and 75th percentile are shown inside each violin plot. (E) Heat map and metaplot of NELFB ChIP-seq signal at the promoters of select gene groups, as defined previously. (F) Quantification of cumulative NELFB and NELFE signal across replicates at each gene group shown in E. Statistical testing was performed using a Wilcoxon test. (G) Genome browser shots of candidate genes representing each group in E and F showing NELFB ChIP-seq signal.
To extend these observations, we took advantage of work that identified a putatively liquid–liquid phase-separated compartment (referred to as NELF bodies) as sites of NELF-mediated transcriptional regulation (Narita et al. 2007; Rawat et al. 2021). To visualize NELF bodies, we generated a clonal transgenic NELFE-EGFP fusion on our Nelfbdeg mESC line background (Supplemental Fig. S6C). We speculated that tagging NELFE in Nelfbdeg would allow us to monitor NELF bodies with and without degradation, given that NELFE should be present but not functional in the absence of NELFB. In dTAG-13-untreated conditions, distinct foci (approximately two to four per nucleus) could be visualized, consistent with previous observations. However, NELFB depletion resulted in complete dissolution of NELF bodies without affecting overall fluorescence levels of NELFE-EGFP, further demonstrating an interdependence between the subunits of the NELF complex and suggesting that these bodies represent hubs of transcriptional regulation (Supplemental Fig. S6D,E). We hypothesized that cells would display a greater number of NELF bodies during periods of transition (for example, when transitioning pluripotent states or changing their fate), as opposed to steady-state conditions. Indeed, we found a significant increase in the number of NELF bodies per nucleus upon pluripotency transition, as well as during the differentiation of mESCs maintained in serum-containing medium in the absence of LIF. This is matched by a step-wise increase in chromatin fraction-localized NELFB and is stable up to 5 d after transitions (Fig. 6C,D; Supplemental Fig. S6F–H).
To expand on these findings, we performed ChIP-seq of NELFB and NELFE at both naïve and formative stages. We found an overall higher signal at formative stages, in agreement with previous results suggesting higher NELF chromatin binding during transitions (Fig. 6E,F). To probe this further, we assessed binding at stage-specific and shared genes defined previously. As expected, we found that NELF binding was dynamic at stage-specific naïve and formative genes and higher at the respective state of these gene groups (i.e., higher signal at formative genes in the formative state). However, we found that after naïve pluripotency exit, NELF maintained significant binding at naïve genes such as Klf2 and Nr0b1, but the opposite was not true for formative genes such as Pou3f1 and Otx2 at the naïve state (Fig. 6E–G; Supplemental Fig. S7A). Our results suggest that increased NELF recruitment to chromatin occurs during pluripotency transitions at least in part due to maintenance of NELF binding at naïve genes simultaneously with de novo binding at formative genes, presumably to attenuate and buffer gene induction and repression to ensure a smooth transition between sequential cell states.
Discussion
The discovery of Pol II pausing at heat shock genes represented an additional layer of gene regulation (Rougvie and Lis 1988). Subsequent work defined the protein complexes involved in this step, including NELF and DSIF, as major regulators of Pol II pausing (Wu et al. 2003; Gilchrist et al. 2012). Further work demonstrated that Pol II pausing occurs globally in metazoans and that it can regulate the transcriptional output of a variety of signaling pathways (Nechaev et al. 2010; Danko et al. 2013; Liu et al. 2015; Abuhashem et al. 2022a). Recent structural studies have provided high-resolution maps of the paused Pol II complex, revealing how NELF and unphosphorylated SPT5 can block elongation of Pol II and sterically inhibit the formation of new preinitiation complexes (PICs), confirming that Pol II pausing acts as a bottleneck step in transcription (Vos et al. 2018a,b).
Previous studies identified roles of Pol II pausing in cultured cells, as well as model organisms such as Drosophila, zebrafish, and mice. These roles revolved around modulating responses to several signaling pathways. In mice, NELF was found to have an essential role in embryonic development (whereby loss of function results in embryonic lethality) and for enabling the differentiation of mESCs in culture via regulating FGF signaling (Amleh et al. 2009; Williams et al. 2015). These studies relied on long-term genetic knockout or siRNA approaches, which result in secondary defects that may mask primary and acute functions of NELF. Here, we sought to understand the direct function of Pol II pausing in early mammalian development by applying acute protein depletion to interrogate the molecular and temporal requirements of Pol II pausing in vitro in mouse ESCs, in parallel with studies in vivo in mutant embryos. We identified cell state transitions within the pluripotent epiblast tissue of the embryo—as modeled by pluripotent stem cells in culture preceding pluripotency exit and the onset of germ layer differentiation—as a key process that requires Pol II pausing to achieve smooth transitions between consecutive states and, ultimately, differentiation of pluripotent cells.
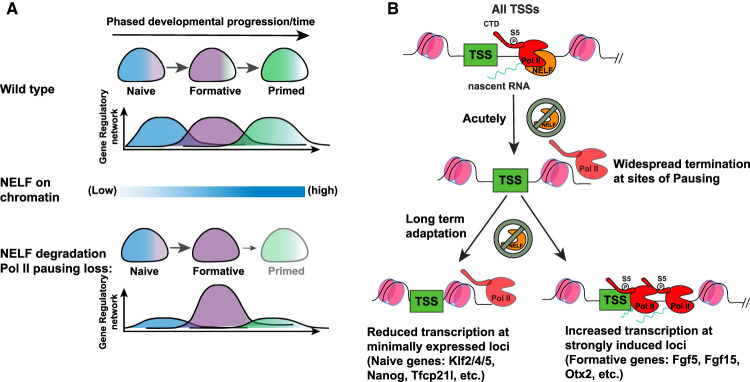
The timing of the defect characterized in mouse postimplantation embryos is consistent with previous studies identifying a role for Nelfb in mESC differentiation in vitro (Amleh et al. 2009; Williams et al. 2015). However, it is notable that initial cell fate specification events in the blastocyst and peri-implantation stages were unaffected in the absence of NELFB. As pluripotent cells progress from their initial naïve to a later primed state, they prepare to exit pluripotency in favor of germ layer specification and differentiation. Pluripotent cells therefore need to calibrate gene expression for precise spatiotemporal control of cellular differentiation. Our data suggest that Pol II pausing mediates cell state transitions by balancing gene regulatory networks during transitions (Fig. 7A). This model is supported by previous studies at the molecular and cellular levels. Molecularly, profiling of Pol II pausing across preimplantation mouse development has identified a reduction in Pol II pausing following zygotic genome activation (ZGA) at the two- to four-cell stage and continuing until the late blastocyst stage, at which point it is re-established (Liu et al. 2020). We recently reported that NELF is required prior to this reduction at ZGA to regulate the major ZGA wave in mouse embryos (Abuhashem et al. 2022b). At the cellular level, several studies investigating tissue-specific loss of Nelfb have revealed that functional defects are observed when Nelfb−/− tissues are challenged by an external stimulus, such as an injury or an infection, or the need to regenerate in the context of muscle stem cells, the uterine and intestinal walls, and macrophages (Hewitt et al. 2019; Yu et al. 2020; Ou et al. 2021; Robinson et al. 2021). Our model—suggesting that Pol II pausing acts to fine-tune transcription during state transitions—explains the defects observed in both the present and previous studies.
Figure 7.
Model of Pol II pausing function during fate transitions. (A) Schematic of Pol II pausing function at the cellular level. (B) Schematic of Pol II pausing function at the molecular level.
Leveraging the dTAG system to acutely deplete NELFB at specific time points allowed us to address why Pol II pausing may be particularly important during pluripotent state transitions and, potentially, in other contexts where cells transition between different states. By combining the fine temporal control of protein expression with the resolution of PRO-seq data, we were able to perform an unbiased assessment of the direct effects of NELFB depletion on global transcriptional activity while bypassing the secondary effects of disrupting Pol II pausing on cell proliferation. Our data suggest that disrupting Pol II pausing as cells transition between successive states results in dysregulation of induced and repressed genes and their enhancers. Specifically, Pol II pausing appears to limit the induction of gene networks and delay the loss of repressed gene networks as cells transition to new states. Superinduction of state-specific genes in the absence of NELF, as observed in our data, effectively functions as an overexpression of state-specific genes, limiting the ability of cells to exit their starting state and attain their subsequent state (Fig. 7A). This conclusion is supported by the observation of increased chromatin recruitment of NELF during pluripotency transitions. Notably, a similar loss of Pol II pausing at the earliest stage of state transitioning, corresponding to hour 0, did not result in a defect, and we concomitantly did not observe increased NELF chromatin recruitment at this stage, corresponding to 0–4 h of transitioning. These data suggest that Pol II pausing is not necessarily required for acute responses to the FGF2 and NODAL cytokines used here to drive pluripotent state transitions and, potentially, other signals. This is in line with normal induction of early-release genes, such as Fos, after NELFB degradation. One inherent limitation to this analysis is that the acute degradation of NELFB during pluripotency transitions results initially in global down-regulation, in agreement with our acute stage experiments, which could partially confound the effects we are reporting on stage-specific genes during pluripotency transition.
Superinduction of highly active loci in the absence of Pol II pausing has been observed previously, and molecularly may be due to Pol II pausing acting as a rate-limiting step at highly active loci (Henriques et al. 2018; Yu et al. 2020). Conversely, loss of minimally expressed/repressed genes could result from increased nucleosome occupancy in the absence of a paused Pol II (Fig. 7B; Gilchrist et al. 2010; Henriques et al. 2018). Indeed, our ChIP-seq at formative states revealed an unexpected residual binding of NELF at naïve state genes that can help sustain a nucleosome-free zone longer. Furthermore, we could observe both effects, up-regulation and down-regulation in the absence of NELFB, at the same locus (Nanog) depending on its expression status in the naïve or formative state, further supporting a link between Pol II pausing role and the level of gene expression rather than the locus itself. Importantly, our analysis does not refute previous results suggesting that FGF signaling is attenuated in Nelfb−/− mESCs, but rather suggests that these defects are most likely secondary and not limited to FGF signaling (Williams et al. 2015).
Our data suggest that NELF-enforced Pol II pausing is widespread at promoters and enhancers. Depleting NELF destabilizes and terminates paused transcripts. These observations highlight a general positive effect of NELF-enforced Pol II pausing on transcription. The presence of paused Pol II can regulate and limit transcription from a certain locus; however, its loss results in destabilizing this important regulatory step and not in release of productive elongating polymerases. Furthermore, at gene promoters that have high initiation rates, NELF centers the paused polymerase 30–50 bases downstream from the TSS, and upon its depletion, polymerases extend further downstream but do not produce productive elongation. These observations are consistent with a study performing acute depletion of NELFC/D, which resulted in the formation of a “second pause” position of promoter-proximal Pol II (Aoi et al. 2020).
In summary, by performing a comprehensive investigation of Pol II pausing function in a relevant developmental context using comparative in vivo (embryo) and in vitro (mESCs) models, we propose a model in which pausing functions as a rheostat for changing transcriptomes during cell state transitions (Fig. 7A).
Material and methods
Availability of materials
Requests for reagents should be directed to and will be fulfilled by A.-K.H. (hadj@mskcc.org).
Cell lines
The ATCC E14 ES cell line was cultured on 0.1% gelatin-coated (Millipore) tissue culture-grade plates in a humidified 37°C incubator with 5% CO2. For routine culture, cells were grown in serum/LIF conditions: DMEM (Gibco) supplemented with 2 mM L-glutamine (Gibco), 1× MEM nonessential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), 100 U/mL penicillin/100 U/mL streptomycin (Gibco), 0.1 mM 2-mercaptoethanol (Gibco), 15% fetal bovine serum (Gibco), and 1000 U/mL recombinant leukemia inhibitory factor (LIF).
To model different stages of pluripotency, cells were initially cultured in N2B27 + 2i/LIF for 4 d to induce naïve pluripotency, equivalent to 0 h in this study. N2B27 comprised 50% neurobasal medium (Gibco) with 100× N2 supplement (Gibco), 50% DMEM/F12 (Gibco) with 50× B27 supplement (Gibco), 2 mM L-glutamine (Gibco), 100 U/mL penicillin/100 U/mL streptomycin (Gibco), 0.1 mM 2-mercaptoethanol (Gibco), and 1% knockout serum replacement (Gibco). To initiate transitions, we followed the EpiLC conversion protocol (Hayashi et al. 2011). Plates were coated with 16 μg/mL fibronectin (Millipore) in PBS for 30 min at 37°C, followed by two washes of PBS. Naïve cells were plated at 25 × 103 cells/cm2 in N2B27 supplemented with 12 ng/mL FGF2 and 20 ng/mL ACTIVIN A (Peprotech). Medium changes were done daily for all conditions.
Plasmid generation
Three plasmids were generated for this study: (1) Cas9 vector to target the C terminus of Nelfb gene: PX459 vector (Addgene 62988) was digested using BbsI-HF (NEB) and single guide RNA targeting Nelfb was annealed (Ran et al. 2013). (2) Homology-directed repair (HDR) vector containing the insert FKBPF36V tag, 2× HA tag, self-cleaving P2A sequence, and puromycin resistance, flanked by 1-kb Nelfb HDR sequences: The insert was obtained from pCRIS-PITCHv2-dTAG-BSD (Addgene 91795) (Nabet et al. 2018). The plasmid backbone (pBluescript), Nelfb HDR sequences, and the insert were amplified using Q5 polymerase (NEB), and the plasmid was constructed using NEBuilder HiFi DNA assembly (NEB). (3) Nelfe-EGFP vector as a fluorescent reporter of NELF bodies: Nelfe cDNA was amplified using Q5 polymerase (NEB). Linker-EGFP and PGK backbone were amplified from pHaloTag-EGFP (Addgene 86629) and PGKneobpa (Addgene 13442), respectively. The plasmid was constructed using NEBuilder HiFi DNA assembly (NEB).
Genome editing
To generate Nelfbdeg mESCs, 3 million cells were transfected with 10 µg of PX459-Nelfb_sgRNA and 10 µg of Nelfb_left-FKBPF36V-2xHA-P2A-BSD-Nelfb_right. Cells were transfected using Lonza P3 primary cell 4D-Nucleofector X 100-µL cuvettes (Lonza). Following transfection, cells were plated on a 10-cm dish (Falcon) coated with MEFs. Forty-eight hours after transfection, correctly targeted cells were selected for in 6 µg/mL Blasticidin (InvivoGen) for 5 d. Surviving cells were split into 1000 cells/10-cm dish and maintained for 9 d under puromycin selection. Surviving clones were picked under a stereomicroscope, expanded, and genotyped for the insert.
Mouse strains and husbandry
All animal work was approved by the Memorial Sloan Kettering Cancer Center (MSKCC) Institutional Animal Care and Use Committee (IACUC). Animals were housed in a pathogen-free facility under a 12-h light cycle. Mouse strains used in this study were Nelfb+/− and wild-type CD-1/ICR (Charles River). Nelfb+/− mice were imported from the Karen Adelman laboratory (JAX 033115). The imported mice had a floxed allele. Following crossing with Zp3-cre (Jax 003651), heterozygous knockout progeny were identified and expanded.
Cell dTAG treatment
dTAG-13 (Bio-Techne) was reconstituted in DMSO (Sigma) at 5 mM. dTAG-13 was diluted in maintenance medium to 500 nM and added to cells with medium changes for the specified amounts of time.
Embryo collection
For all experiments, embryos were obtained via natural mating of females 6–12 wk of age with males 7–16 wk of age. For preimplantation stages, embryos were recovered by flushing the uterine horns (E3.25–E4.5). These dissections were carried out in flushing and holding medium (FHM; Millipore) as described (Behringer et al. 2014).
For postimplantation embryos (E5.5–E7.5), uterine horns were retrieved and cut into single decidual swellings in 5% newborn calf serum in DMEM/F12 (Gibco). Embryos were dissected out by removing the uterine wall and decidual tissue. The parietal endoderm was removed carefully with the ectoplacental cone.
Immunofluorescence
For cultured mESCs, cells were plated on u-Slide eight-well plates (Ibidi), washed with PBS+/+, and fixed in 4% PFA (electron microscopy sciences) in PBS+/+ for 10 min at room temperature. Fixed cells were washed twice with PBS+/+, once with wash buffer and 0.1% Triton X-100 (Sigma) in PBS+/+, and then permeabilized in 0.5% Triton X-100 (Sigma) in PBS+/+ for 10 min. Cells were then blocked with 3% donkey serum (Sigma) and 1% BSA (Sigma) for 1 h at room temperature. Cells were then incubated with primary antibodies in blocking buffer overnight at 4°C (antibodies and concentrations are listed in Supplemental Table S1). Cells were then washed three times in wash buffer and incubated with suitable donkey Alexa Fluor (1:500; Invitrogen) for 1 h at room temperature. Cells were then washed three times with wash buffer, the last containing 5 μg/mL Hoechst 33342 (Invitrogen), and then imaged.
For E3.25–E4.5 preimplantation embryos, the zona pellucida was removed by incubation in acid Tyrode's solution (Sigma) for 2 min at 37°C. Embryos were subsequently washed briefly in PBS+/+ before fixation in 4% PFA for 10 min at room temperature. Fixed embryos were washed in 0.1% Triton X-100 in PBS+/+ (PBX) for 5 min, permeabilized in 0.5% Triton X-100 (Sigma) in PBS+/+ for 5 min, washed again for 5 min in PBX, and blocked in 2% horse serum (Sigma) in PBS+/+ for 1 h at room temperature. Embryos were incubated in primary antibodies diluted in blocking solution overnight at 4°C. Embryos were then washed three times for 5 min each in PBX and blocked again for 1 h at room temperature prior to incubation with secondary antibodies. Secondary antibodies diluted in blocking solution were applied for 1 h at 4°C. Embryos were then washed twice for 5 min each in PBX and incubated with 5 ug/mL Hoechst 33342 (Invitrogen) in PBS for 5 min or until mounting for imaging. The following primary antibodies were used: goat anti-GATA6 (1:100; R&D Systems), mouse anti-CDX2 (1:200; BioGenex), and rabbit anti-NANOG (1:500; CosmoBio). Secondary Alexa Fluor-conjugated antibodies (Invitrogen) were used at a dilution of 1:500. DNA was visualized using Hoechst 33342.
E6.5 and E7.5 embryos were washed briefly in PBS+/+ before fixation in 4% PFA for 20 min at room temperature. Fixed embryos were washed in 0.1% Triton X-100 in PBS+/+ (PBX) for 5 min, permeabilized in 0.5% Triton X-100 (Sigma) in PBS+/+ for 20 min, washed again for 5 min in PBX, and blocked in 3% horse serum (Sigma) in PBX for 1 h at room temperature. Embryos were incubated in primary antibodies diluted in blocking solution overnight at 4°C. Embryos were then washed three times for 10 min each in PBX and blocked again for 1 h at room temperature prior to incubation with secondary antibodies. Secondary antibodies diluted in blocking solution were applied overnight at 4°C. Embryos were then washed three times for 5 min each in PBX and incubated with 5 µg/mL Hoechst 33342 (Invitrogen) in PBX for 1 h or until mounting for imaging.
Image data acquisition
Fixed immunostained samples were imaged on a Zeiss LSM880 laser scanning confocal microscope. Preimplantation embryos were mounted in microdrops of 5 μg/mL Hoechst 33342 in PBS+/+ on glass-bottomed dishes (MatTek) coated with mineral oil (Sigma). Embryos were imaged along the entire Z-axis with a 1-μm step using an oil-immersion Zeiss EC Plan-Neofluar 40×/NA 1.3 with a 0.17-mm working distance. For postimplantation embryos, a similar setup was used but with an air plan-apochromat 20×/NA 0.75 objective.
Superresolution imaging of Nelfe-EGFP was performed on a Zeiss Elyra 7 with lattice SIM using an oil-immersion Zeiss plan-apochromat 63×/NA 1.4 objective.
Western blotting
For cells, 350 µL of lysis buffer (1× cell lysis buffer [Cell Signaling] with 1 mM PMSF [Cell Signaling] and cOmplete Ultra protease inhibitor [Sigma]) was added to a 90% confluent six-well dish (Falcone) after washing with PBS−/−. Cells were incubated with lysis buffer for 5 min on ice, scraped, and collected. Samples were sonicated for 15 sec to complete lysis and then spun down at 12,000g for 10 min at 4°C. The supernatant was collected, and protein concentration was measured using Pierce BCA protein assay kit (Thermo). Ten micrograms to 20 µg of protein was mixed with Blue loading buffer (Cell Signaling) and 40 mM DTT (Cell Signaling). Samples were boiled for 5 min at 95°C for denaturation. To prepare cellular compartment fractions, subcellular protein fractionation kit was used (Thermo) according to the manufacturer's instructions.
Samples were run on a Bio-Rad Protean system and transferred using transblot semidry transfer cells (Bio-Rad) to a nitrocellulose membrane (Cell Signaling) following the manufacturer's instructions and reagents. Membranes were then washed briefly with ddH2O and stained with Ponceau S (Sigma) for 1 min to check for transfer quality and as a loading control. Membranes were then washed three times with TBST (0.1% Tween 20 [Fisher] in TBS). Membranes were blocked with 4% BSA in TBST for 1 h at room temperature and subsequently incubated with primary antibodies diluted in blocking buffer overnight at 4°C. They were then washed three times with TBST, incubated with secondary antibodies in blocking buffer for 1 h, washed three times with TBST, incubated with ECL reagent SignalFire for 1–2 min, and imaged using a ChemiDoc (Bio-Rad). Primary and secondary antibodies are listed in Supplemental Table S1.
RT-qPCR
RNA was extracted from samples using TRIzol (Thermo) following the manufacturer's instructions. One microgram of RNA was used to generate cDNA using the QuantiTect reverse transcription kit (Qiagen). qPCR reaction was performed using PowerUp SYBR Green master mix (Thermo) and a Bio-Rad CFX96. The primers used are listed in Supplemental Table S1.
ChIP-seq
Twenty-five million cells were collected for each sample/replicate. Cells were cross-linked in 1% PFA (Electron Microscopy Sciences) in PBS for 10 min at room temperature. Following quenching with 125 mM glycine (Sigma) for 5 min at room temperature, cells were washed twice with PBS and then suspended in lysis buffer (10 mM Tris at pH 8, 1 mM EDTA, 0.5% SDS [Sigma]) at 20 × 106 cells per 400 μL. To shear chromatin, samples were sonicated using a Bioruptor Pico sonication device (Diagenode) for 12 cycles, 30 sec on/30 sec off, and then pelleted at the maximum speed for 10 min at 4°C. The supernatant was diluted five times with dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris at pH 8, 167 mM NaCl [Sigma]) and then incubated with primary antibodies overnight at 4°C. Protein G Dynabeads (Thermo) were blocked overnight at 4°C using 100 ng per 10 μL of beads. The next day, beads were added to samples at 20 μL per sample for 3 h at 4°C. Using a magnet to stabilize the beads, they were washed twice in low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris at pH 8), twice in high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris at pH 8), twice in LiCl buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholic acid, 1 mM EDTA, 10 mM Tris at pH 8), and once in TE buffer (10 mM Tris at pH 8, 0.1 mM EDTA). Subsequently, the DNA was eluted from the beads by incubation with 150 μL of elution buffer (100 mM NaHCO3, 1% SDS) for 20 min at 65°C with vortexing using Eppendorf ThermoMixer C (Eppendorf). The supernatant was collected, reverse cross-linked by incubation overnight at 65°C in the presence of proteinase K (Roche), and cleaned by RNase A (Thermo) treatment for 1 h at 37°C. The DNA was purified using a DNA clean and concentrate kit (Zymo Research). Spt5 ChIP samples were spiked-in with 10% human HEK293T cells to perform normalized quantification of signal.
ChIP-seq analysis
Reads were aligned to mm10 and filtered using the following pipeline (https://github.com/soccin/ChIP-seq). Briefly, reads were aligned using Bowtie 2.3.5 and then filtered using a MAPQ of >30. Properly paired reads were kept. Resulting BAMs were used to generate BigWigs using DeepTools (https://deeptools.readthedocs.io/en/develop). BigWigs were normalized to 10 million. For SPT5, the samples were aligned to an index with both mm10 and hg38 to normalize to human cell spike-ins. The normalization was applied as a scale factor during BigWig generation, where the scale factor is the multiple required for each spike-in to be equal to the average of all spike-ins. NELFB and NELFE ChIP-seqs at the naïve and formative states were also normalized to a spike-in similar to SPT5 samples. Peak calling was performed using MACS2 with paired reads mode on and Q-value < 0.05. Shared peaks across replicates were analyzed. Downstream analysis was performed in Rstudio 4.1.2 using Bioconductor packages and DeepTools to generate heat maps.
PRO-seq (sample preparation and library preparation)
Cells (5 × 106 to 15 × 106) were detached using tryspin (Thermo) and then resuspended in 500 μL of wash buffer (10 mM Tris-Cl at pH 8.0, 300 mM sucrose, 10 mM NaCl, 2 mM MgAc2 [all from Sigma]). All following steps were performed at 4°C. Next, 500 μL of lysis buffer (10 mM Tris-Cl at pH 8.0, 300 mM sucrose, 10 mM NaCl, 2 mM MgAc2, 6 mM CaCl2, 0.2% NP-40/Igepal [all from Sigma]) was added to the resuspended cells, followed by pipetting the cells up and down 10 times. The total volume was then brought to 10 mL by adding 4.5 mL of wash buffer and 4.5 mL of lysis buffer. The tubes were mixed by inverting gently for 1 min, and then nuclei were pelleted at 800g for 5 min. The nuclei were then washed with 1 mL of storage buffer (50 mM Tris-HCL at pH 8.3, 40% glycerol, 5 mM MgCl2, 0.1 mM EDTA [all from Sigma]). Nuclei were counted, and 5 × 106 were pelleted per replicate in 1.5-mL Eppendorf tubes. Pellets were resuspended in 42 μL of storage buffer. A similar procedure was performed separately for D. melanogaster S2 cells. In the final step, 8 μL of storage buffer with 35 × 103 S2 spike-in cells was added to the 42 μL of storage buffer with mESC nuclei and frozen in LN2 until the run-on reaction.
PRO-seq libraries were prepared according to published protocols (Mahat et al. 2016). Adjustments made to the original protocol were as follows: (1) In the run-on master mix, biotinylated nucleotides were provided at the following concentrations: 10 mM Biotin-11-ATP, 10 mM Biotin-11-GTP, 100 mM Biotin-11-CTP, and 100 mM Biotin-11-UTP. (2) Trizol LS (Invitrogen 10296-010) was replaced by TRI reagent-LS (MRC TS 120). (3) Trizol (Invitrogen 15596-026) was replaced by TRI reagent (MRC TR 118). (4) Digestion of RNA by base hydrolysis in 0.2 N NaOH on ice was reduced from 8 min to 6 min. (5) Nascent RNA was purified by binding streptavidin beads (NEB S1421S) and washed as described. Hydrophilic streptavidin magnetic beads (NEB S1421S) were replaced by streptavidin magnetic beads (NEB S1420S). (6) SuperScript III reverse transcriptase (Invitrogen 18080-044) was replaced by SuperScript IV reverse transcriptase (Invitrogen 18090050). Libraries were prepared using adapters that contained a 6-bp unique molecular identifier sequence on read 1.
PRO-seq analysis
PRO-seq libraries were competitively aligned to a genome resulting from merging mm10 assembly with D. melanogaster dm3 genome assembly. Alignment was performed using the proseq2.0 pipeline (https://github.com/Danko-Lab/proseq2.0) using the parameters -PE –RNA5=R2_5prime –UMI1=6. Downstream analysis was performed in R using genomic ranges (Lawrence et al. 2013) and BRgenomics 1.1.3 (https://mdeber.github.io/index.html).
To account for global changes in nascent RNA production as well as technical variations between libraries, spike-in D. melanogaster S2 nuclei were used as internal controls. The ratio between fly and mouse nuclei was 1:150. For normalization, we divided the mouse reads in each sample by the total number of fly reads in the same sample.
We quantified changes in gene expression using the GENCODE v20 annotations in mice. To compute differential expression between treatments, we used DEseq2. First, we used unnormalized BigWigs to count the total number of reads around each TSS or within gene bodies of annotated GENCODE v20 genes. For TSSs, we took a 300-bp window centered on gene start sites, while gene bodies were defined as the entirety of the gene excluding the first and last 300 bp from the TSS and TES, respectively. Next, we provided the raw PRO-seq counts as input to DEseq2. We used the total number of Drosophila reads as scaling factors. For generating metaprofiles, Drosophila spike-in-normalized counts were used. TSS metaprofiles in Figure 4, B and C, were aligned to mESC START-seq data due to better accuracy than GENCODE v20 (Henriques et al. 2018).
Heat maps
Heat maps were generated using Drosophila spike-in-normalized reads. We sorted GENCODE v20 genes by length and depicted the number of spike-in-normalized reads per 1-kb bin from 1 kb upstream of the annotated TSS to 200 kb downstream.
dREG peaks
We called regulatory element peaks using dREG gateway (https://dreg.dnasequence.org; Danko et al. 2015; Wang et al. 2019).
Analysis of Micro-C data
Publicly available Micro-C data were downloaded from GSE130275 (Hsieh et al. 2020). HiC profiles were plotted using hicPlotTADs and used to define TAD boundaries for Nanog and Fgf5 (Wolff et al. 2020). Regulatory regions for Nanog and Fgf5 were called using the custom virtual4C script developed by the Danko laboratory (https://github.com/Danko-Lab/HS_transcription_regulation) using parameters -w 4000000 -b 5000 -q 30. The obtained regulatory regions were overlapped with dREG calls to define putative enhancers that are in contact with the four promoters of interest.
Initiation release rate estimation
An algorithm was used to estimate initiation rates for each gene (Siepel 2021). Specifically, initiation rate was estimated by
where α is initiation rate, SB is the number of read counts within gene body, l is gene length, and λ is a library-specific scaling factor determined by the number of spike-in reads mapped to the D. melanogaster genome. Pause release rate was estimated by
where β is pause release rate, Sp is the number of read counts within the pause peak, and k is the length of it. The first protein coding annotations from GENCODE (version vM20) were used for each gene, and regions 1 kb downstream from TSSs to the end of the gene (up to 90 kb) were used as gene body for read counting. Note that α is the maximum likelihood estimator of initiation rate when assuming read counts following Poisson distribution, and it is also widely used in many literatures to represent transcriptional activity with some heuristic justifications (Siepel 2021).
Image processing and quantification
For preimplantation embryos, semiautomated 3D nuclear segmentation for cell counting and quantification of fluorescence intensity was carried out using MINS, a Matlab-based algorithm (http://katlab-tools.org; Lou et al. 2014).The same imaging parameters were used for all experiments consisting of the same primary and secondary antibody combinations to minimize quantitative variance due to image acquisition. The MINS output was checked for oversegmentation or undersegmentation, and tables were corrected manually using ImageJ (National Institutes of Health [NIH], https://imagej.nih.gov/ij) (Schindelin et al. 2012). Undersegmented nuclei (two or more nuclei detected as one, or nuclei that were not detected) were assigned fluorescence intensity values that were directly measured using ImageJ (NIH). To correct fluorescence decay along the Z-axis, we used a linear regression method to calculate the global average of the regression coefficients in the HA channel (Saiz et al. 2016b). This slope was then used to adjust the logarithm values of HA fluorescence intensity for each nucleus. Trophectoderm (TE) versus inner cell mass (ICM) cell assignment was achieved by a threshold for CDX2, which is present exclusively in TE. To avoid batch variability, directly compared embryos were stained and imaged in the same session.
Statistical analysis
All statistical tests of immunofluorescence data were carried out in PRISM 9 (GraphPad). Statistical significance was established using a Student's t-test with P-value threshold of 0.05. The P-value range for each experiment is indicated in the figure legends.
For sequencing data, analysis of differentially expressed genes was done in R using the DEseq2 method with 0.05 P adjusted (Love et al. 2014). Other comparisons between gene groups were performed using two-way paired t-tests.
Data and code availability
Raw and processed sequencing data from this work have been deposited in the Gene Expression Omnibus under the accession numbers GSE196543 for ChIP-seq and GSE196653 for PRO-seq.
Supplementary Material
Acknowledgments
We thank Memorial Sloan Kettering's Integrated Genomics Operation and Bioinformatics Core Facility for assistance in sequencing and sequence data analysis, Dr. James Muller of the Sloan Kettering Institute light microscopy shared instrument cluster for assistance with use of the Zeiss Elyra 7, and Dr. Karen Adelman and Dr. Lucy Williams for sharing their Nelfb−/− mouse model. We are grateful to Dr. Effie Apostolou for providing feedback and guidance on this work, and members of the Hadjantonakis laboratory for stimulating discussion and critical feedback. A.A. is supported by a Medical Scientist Training Program training grant from the National Institutes of Health (NIH; T32GM007739) awarded to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program, and NIH F30HD103398. Work in A.S.’s laboratory is supported by the NIH (R01HG010346 and R35GM127070). Work in C.G.D.’s laboratory is supported by the NIH (R01HG009309) and NASA (17-EXO-17-2-0112). Work in A.-K.H.’s laboratory is supported by the NIH (R01HD094868, R01DK127821, R01HD086478, and P30CA008748).
Author contributions: A.A. and A.-K.H. conceptualized the study. A.A. designed, performed, and analyzed most experiments. A.G.C. and C.G.D. analyzed and interpreted PRO-seq data. E.J.R. constructed PRO-seq libraries. Y.Z. and A.S. estimated the initiation release rates. C.G.D. supervised all bioinformatics analyses. A.A. wrote the manuscript with input from all authors. A.A., A.-K.H., and C.G.D. acquired funding. A.-K.H. supervised the work.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.349565.122.
Competing interest statement
The authors declare no competing interest.
References
- Abuhashem A, Garg V, Hadjantonakis A-K. 2022a. RNA polymerase II pausing in development: orchestrating transcription. Open Biol 12: 210220. 10.1098/rsob.210220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abuhashem A, Lee AS, Joyner AL, Hadjantonakis A-K. 2022b. Rapid and efficient degradation of endogenous proteins in vivo identifies stage-specific roles of RNA Pol II pausing in mammalian development. Dev Cell 57: 1068–1080.e6. 10.1016/j.devcel.2022.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K, Lis JT. 2012. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 13: 720–731. 10.1038/nrg3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amleh A, Nair SJ, Sun J, Sutherland A, Hasty P, Li R. 2009. Mouse cofactor of BRCA1 (Cobra1) is required for early embryogenesis. PLoS One 4: e5034. 10.1371/journal.pone.0005034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoi Y, Smith ER, Shah AP, Rendleman EJ, Marshall SA, Woodfin AR, Chen FX, Shiekhattar R, Shilatifard A. 2020. NELF regulates a promoter-proximal step distinct from RNA Pol II pause-release. Mol Cell 78: 261–274.e5. 10.1016/j.molcel.2020.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer R, Gertsenstein M, Nagy KV, Nagy A. 2014. Manipulating the mouse embryo: a laboratory manual, 4th ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Chen FX, Smith ER, Shilatifard A. 2018. Born to run: control of transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 19: 464–478. 10.1038/s41580-018-0010-5 [DOI] [PubMed] [Google Scholar]
- Core L, Adelman K. 2019. Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev 33: 960–982. 10.1101/gad.325142.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Gilchrist DA, Fargo DC, Kwak H, Adelman K, Lis JT. 2012. Defining the status of RNA polymerase at promoters. Cell Rep 2: 1025–1035. 10.1016/j.celrep.2012.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P. 2019. Organization and regulation of gene transcription. Nature 573: 45–54. 10.1038/s41586-019-1517-4 [DOI] [PubMed] [Google Scholar]
- Danko CG, Hah N, Luo X, Martins AL, Core L, Lis JT, Siepel A, Kraus WL. 2013. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol Cell 50: 212–222. 10.1016/j.molcel.2013.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danko CG, Hyland SL, Core LJ, Martins AL, Waters CT, Lee HW, Cheung VG, Kraus WL, Lis JT, Siepel A. 2015. Identification of active transcriptional regulatory elements from GRO-seq data. Nat Methods 12: 433–438. 10.1038/nmeth.3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. 2010. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143: 540–551. 10.1016/j.cell.2010.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Fromm G, dos Santos G, Pham LN, McDaniel IE, Burkholder A, Fargo DC, Adelman K. 2012. Regulating the regulators: the pervasive effects of Pol II pausing on stimulus-responsive gene networks. Genes Dev 26: 933–944. 10.1101/gad.187781.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressel S, Schwalb B, Cramer P. 2019. The pause-initiation limit restricts transcription activation in human cells. Nat Commun 10: 3603. 10.1038/s41467-019-11536-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. 2013. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res 23: 1210–1223. 10.1101/gr.152306.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. 2011. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146: 519–532. 10.1016/j.cell.2011.06.052 [DOI] [PubMed] [Google Scholar]
- Henriques T, Gilchrist DA, Nechaev S, Bern M, Muse GW, Burkholder A, Fargo DC, Adelman K. 2013. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell 52: 517–528. 10.1016/j.molcel.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques T, Scruggs BS, Inouye MO, Muse GW, Williams LH, Burkholder AB, Lavender CA, Fargo DC, Adelman K. 2018. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev 32: 26–41. 10.1101/gad.309351.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt SC, Li R, Adams N, Winuthayanon W, Hamilton KJ, Donoghue LJ, Lierz SL, Garcia M, Lydon JP, DeMayo FJ, et al. 2019. Negative elongation factor is essential for endometrial function. FASEB J 33: 3010–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T-HS, Cattoglio C, Slobodyanyuk E, Hansen AS, Rando OJ, Tjian R, Darzacq X. 2020. Resolving the 3D landscape of transcription-linked mammalian chromatin folding. Mol Cell 78: 539–553.e8. 10.1016/j.molcel.2020.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJJ, Desplan C. 2010. Stochastic mechanisms of cell fate specification that yield random or robust outcomes. Annu Rev Cell Dev Biol 26: 689–719. 10.1146/annurev-cellbio-100109-104113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187. 10.1038/nature09033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs AR, Imanci D, Hoerner L, Gaidatzis D, Burger L, Schübeler D. 2017. Genome-wide single-molecule footprinting reveals high RNA polymerase II turnover at paused promoters. Mol Cell 67: 411–422.e4. 10.1016/j.molcel.2017.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT. 2013. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339: 950–953. 10.1126/science.1229386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon GS, Fraser ST, Eakin GS, Mangano M, Isern J, Sahr KE, Hadjantonakis A-K, Baron MH. 2006. Tg(Afp-GFP) expression marks primitive and definitive endoderm lineages during mouse development. Dev Dyn 235: 2549–2558. 10.1002/dvdy.20843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol 9: e1003118. 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Kraus WL, Bai X. 2015. Ready, pause, go: regulation of RNA polymerase II pausing and release by cellular signaling pathways. Trends Biochem Sci 40: 516–525. 10.1016/j.tibs.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Xu Q, Wang Q, Feng S, Lai F, Wang P, Zheng F, Xiang Y, Wu J, Nie J, et al. 2020. The landscape of RNA Pol II binding reveals a stepwise transition during ZGA. Nature 587: 139–144. 10.1038/s41586-020-2847-y [DOI] [PubMed] [Google Scholar]
- Lou X, Kang M, Xenopoulos P, Muñoz-Descalzo S, Hadjantonakis A-K. 2014. A rapid and efficient 2D/3D nuclear segmentation method for analysis of early mouse embryo and stem cell image data. Stem Cell Reports 2: 382–397. 10.1016/j.stemcr.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat DB, Kwak H, Booth GT, Jonkers IH, Danko CG, Patel RK, Waters CT, Munson K, Core LJ, Lis JT. 2016. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq). Nat Protoc 11: 1455–1476. 10.1038/nprot.2016.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgani S, Nichols J, Hadjantonakis A-K. 2017. The many faces of pluripotency: in vitro adaptations of a continuum of in vivo states. BMC Dev Biol 17: 7. 10.1186/s12861-017-0150-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabet B, Roberts JM, Buckley DL, Paulk J, Dastjerdi S, Yang A, Leggett AL, Erb MA, Lawlor MA, Souza A, et al. 2018. The dTAG system for immediate and target-specific protein degradation. Nat Chem Biol 14: 431–441. 10.1038/s41589-018-0021-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Yung TMC, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. 2007. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol Cell 26: 349–365. 10.1016/j.molcel.2007.04.011 [DOI] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. 2010. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327: 335–338. 10.1126/science.1181421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J, Guan X, Wang J, Wang T, Zhang B, Li R, Xu H, Hu X, Guo X-K. 2021. Epithelial NELF guards intestinal barrier function to ameliorate colitis by maintaining junctional integrity. Mucosal Immunol 15: 279–288. 10.1038/s41385-021-00465-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Qin K, Guo Z, Ma Y, April C, Gao X, Andrews TG, Bokov A, Zhang J, Chen Y, et al. 2014. Negative elongation factor controls energy homeostasis in cardiomyocytes. Cell Rep 7: 79–85. 10.1016/j.celrep.2014.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope SD, Medzhitov R. 2018. Emerging principles of gene expression programs and their regulation. Mol Cell 71: 389–397. 10.1016/j.molcel.2018.07.017 [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR–Cas9 system. Nat Protoc 8: 2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat P, Boehning M, Hummel B, Aprile-Garcia F, Pandit AS, Eisenhardt N, Khavaran A, Niskanen E, Vos SM, Palvimo JJ, et al. 2021. Stress-induced nuclear condensation of NELF drives transcriptional downregulation. Mol Cell 81: 1013–1026.e11. 10.1016/j.molcel.2021.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DCL, Ritso M, Nelson GM, Mokhtari Z, Nakka K, Bandukwala H, Goldman SR, Park PJ, Mounier R, Chazaud B, et al. 2021. Negative elongation factor regulates muscle progenitor expansion for efficient myofiber repair and stem cell pool repopulation. Dev Cell 56: 1014–1029.e7. 10.1016/j.devcel.2021.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54: 795–804. 10.1016/S0092-8674(88)91087-2 [DOI] [PubMed] [Google Scholar]
- Saiz N, Kang M, Schrode N, Lou X, Hadjantonakis A-K. 2016a. Quantitative analysis of protein expression to study lineage specification in mouse preimplantation embryos. J Vis Exp. 10.3791/53654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiz N, Williams KM, Seshan VE, Hadjantonakis A-K. 2016b. Asynchronous fate decisions by single cells collectively ensure consistent lineage composition in the mouse blastocyst. Nat Commun 7: 13463. 10.1038/ncomms13463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalb B, Michel M, Zacher B, Frühauf K, Demel C, Tresch A, Gagneur J, Cramer P. 2016. TT-seq maps the human transient transcriptome. Science 352: 1225–1228. 10.1126/science.aad9841 [DOI] [PubMed] [Google Scholar]
- Shao W, Zeitlinger J. 2017. Paused RNA polymerase II inhibits new transcriptional initiation. Nat Genet 49: 1045–1051. 10.1038/ng.3867 [DOI] [PubMed] [Google Scholar]
- Shao R, Kumar B, Lidschreiber K, Lidschreiber M, Cramer P, Elsässer SJ. 2021. Newly synthesized RNA sequencing characterizes transcription dynamics in three pluripotent states. bioRxiv. 10.1101/2021.06.11.448016 [DOI] [Google Scholar]
- Siepel A. 2021. A unified probabilistic modeling framework for eukaryotic transcription based on nascent RNA sequencing data. bioRxiv. 10.1101/2021.01.12.426408 [DOI] [Google Scholar]
- Steurer B, Janssens RC, Geverts B, Geijer ME, Wienholz F, Theil AF, Chang J, Dealy S, Pothof J, van Cappellen WA, et al. 2018. Live-cell analysis of endogenous GFP-RPB1 uncovers rapid turnover of initiating and promoter-paused RNA polymerase II. Proc Natl Acad Sci 115: E4368–E4376. 10.1073/pnas.1717920115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SM, Farnung L, Boehning M, Wigge C, Linden A, Urlaub H, Cramer P. 2018a. Structure of activated transcription complex Pol II–DSIF–PAF–SPT6. Nature 560: 607–612. 10.1038/s41586-018-0440-4 [DOI] [PubMed] [Google Scholar]
- Vos SM, Farnung L, Urlaub H, Cramer P. 2018b. Structure of paused transcription complex Pol II–DSIF–NELF. Nature 560: 601–606. 10.1038/s41586-018-0442-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hang S, Prazak L, Gergen JP. 2010. NELF potentiates gene transcription in the Drosophila embryo. PLoS One 5: e11498. 10.1371/journal.pone.0011498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Chu T, Choate LA, Danko CG. 2019. Identification of regulatory elements from nascent transcription using dREG. Genome Res 29: 293–303. 10.1101/gr.238279.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. 10.1016/j.cell.2013.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LH, Fromm G, Gokey NG, Henriques T, Muse GW, Burkholder A, Fargo DC, Hu G, Adelman K. 2015. Pausing of RNA polymerase II regulates mammalian developmental potential through control of signaling networks. Mol Cell 58: 311–322. 10.1016/j.molcel.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissink EM, Vihervaara A, Tippens ND, Lis JT. 2019. Nascent RNA analyses: tracking transcription and its regulation. Nat Rev Genet 20: 705–723. 10.1038/s41576-019-0159-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff J, Rabbani L, Gilsbach R, Richard G, Manke T, Backofen R, Grüning BA. 2020. Galaxy HiCExplorer 3: a web server for reproducible Hi-C, capture Hi-C and single-cell Hi-C data analysis, quality control and visualization. Nucleic Acids Res 48: W177–W184. 10.1093/nar/gkaa220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-H, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D. 2003. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev 17: 1402–1414. 10.1101/gad.1091403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Hadjantonakis A-K, Nowotschin S. 2017. Visualizing endoderm cell populations and their dynamics in the mouse embryo with a Hex-tdTomato reporter. Biol Open 6: 678–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Yoon H, Xiong Y, Dixon-Clarke SE, Nowak RP, Fischer ES. 2020. Targeted protein degradation as a powerful research tool in basic biology and drug target discovery. Nat Struct Mol Biol 27: 605–614. 10.1038/s41594-020-0438-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97: 41–51. 10.1016/S0092-8674(00)80713-8 [DOI] [PubMed] [Google Scholar]
- Yang Q, Liu X, Zhou T, Cook J, Nguyen K, Bai X. 2016. RNA polymerase II pausing modulates hematopoietic stem cell emergence in zebrafish. Blood 128: 1701–1710. 10.1182/blood-2016-02-697847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Humphrey SJ, Cinghu S, Pathania R, Oldfield AJ, Kumar D, Perera D, Yang JYH, James DE, Mann M, et al. 2019. Multi-omic profiling reveals dynamics of the phased progression of pluripotency. Cell Syst 8: 427–445.e10. 10.1016/j.cels.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Zhang B, Deochand D, Sacta MA, Coppo M, Shang Y, Guo Z, Zeng X, Rollins DA, Tharmalingam B, et al. 2020. Negative elongation factor complex enables macrophage inflammatory responses by controlling anti-inflammatory gene expression. Nat Commun 11: 2286. 10.1038/s41467-020-16209-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed sequencing data from this work have been deposited in the Gene Expression Omnibus under the accession numbers GSE196543 for ChIP-seq and GSE196653 for PRO-seq.