Abstract
In eubacteria, green algae, and plant chloroplasts, isopentenyl diphosphate, a key intermediate in the biosynthesis of isoprenoids, is synthesized by the methylerythritol phosphate pathway. The five carbons of the basic isoprenoid unit are assembled by joining pyruvate and d-glyceraldehyde 3-phosphate. The reaction is catalyzed by the thiamine diphosphate-dependent enzyme 1-deoxy-d-xylulose 5-phosphate synthase. In Rhodobacter capsulatus, two open reading frames (ORFs) carry the genes that encode 1-deoxy-d-xylulose 5-phosphate synthase. ORF 2816 is located in the photosynthesis-related gene cluster, along with most of the genes required for synthesis of the photosynthetic machinery of the bacterium, whereas ORF 2895 is located elsewhere in the genome. The proteins encoded by ORF 2816 and ORF 2895, 1-deoxy-d-xylulose 5-phosphate synthase A and B, containing a His6 tag, were synthesized in Escherichia coli and purified to greater than 95% homogeneity in two steps. 1-Deoxy-d-xylulose 5-phosphate synthase A appears to be a homodimer with 68 kDa subunits. A new assay was developed, and the following steady-state kinetic constants were determined for 1-deoxy-d-xylulose 5-phosphate synthase A and B: Kmpyruvate = 0.61 and 3.0 mM, Kmd-glyceraldehyde 3-phosphate = 150 and 120 μM, and Vmax = 1.9 and 1.4 μmol/min/mg in 200 mM sodium citrate (pH 7.4). The ORF encoding 1-deoxy-d-xylulose 5-phosphate synthase B complemented the disrupted essential dxs gene in E. coli strain FH11.
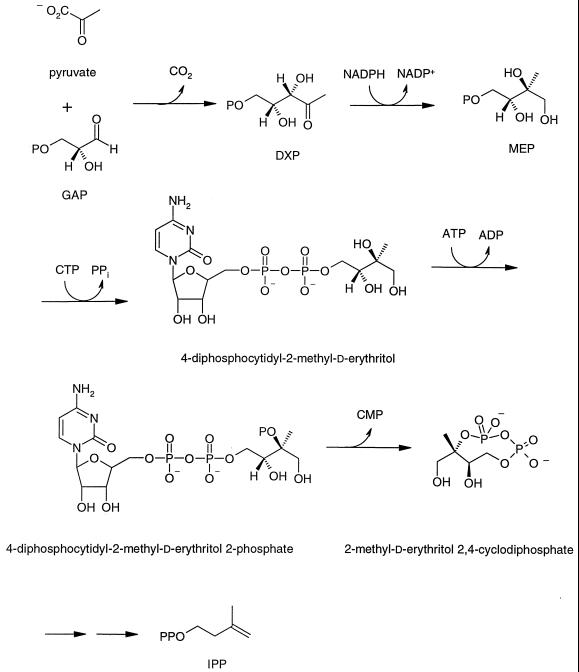
Isoprenoid compounds form a large, ubiquitous class of natural products consisting of over 30,000 individual members. They have a wide variety of cellular functions—such as components of cell membranes (sterols), electron transport (ubiquinones), signal transduction (prenylated proteins), photosynthetic pigments (chlorophylls), and cell wall biosynthesis (dolichols)—essential for viability (39). Until recently, all isoprenoid compounds were thought to be synthesized from acetyl coenzyme A by the widely accepted mevalonate (MVA) pathway found in eukaryotes and archaebacteria (33). Work stimulated by finding labeling patterns in bacterial hopanoids and ubiquinones inconsistent with their biosynthesis by the MVA pathway (32, 37) led to the discovery of a new, independent route to these molecules in many bacteria, green algae, and plants (for reviews, see references 10 and 36). In the newly discovered pathway, the five carbon atoms in the basic isoprenoid unit are derived from pyruvate and d-glyceraldehyde 3-phosphate (GAP). As shown in Fig. 1, pyruvate and GAP are condensed to give 1-deoxy-d-xylulose 5-phosphate (DXP). In addition to serving as a precursor for the biosynthesis of thiamine and pyridoxol, DXP undergoes rearrangement and reduction to form 2-methylerythritol 4-phosphate (MEP), the first committed intermediate in the MEP pathway for biosynthesis of isoprenoids (19, 47). MEP is then appended to CMP to form a cytidine derivative (35), and this is followed by phosphorylation of the C-2 hydroxyl group (28) and elimination of CMP to form a 2,4-cyclic diphosphate (16). The remaining steps to the fundamental five-carbon isoprenoid building blocks, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), have not been established. For most organisms reported to date, the enzymes in the MEP pathway are encoded by essential single-copy genes. The exception is a strain of Streptomyces, which contains both the MEP and MVA pathways (44).
FIG. 1.
MEP pathway for isoprenoid biosynthesis.
DXP synthase (DXPase) lies just before the branch point to the B vitamins and isoprenoids. Genes encoding the enzyme have been cloned from Escherichia coli (27, 46), Streptomyces sp. strain CL190 (18), Mentha × piperita (21), and Capsicum annuum (5). Disruption of the E. coli dxs gene is lethal. DXPase catalyzes the decarboxylation of pyruvate and the subsequent condensation of the thiamine-bound two-carbon intermediate with GAP in a reaction similar to those catalyzed by transketolases. Interestingly, DXPases contain regions with strong homology to the E1 subunit of pyruvate dehydrogenases and to transketolases (46). Although recombinant forms of the enzyme can use either GAP or d-glyceraldehyde as cosubstrates with pyruvate, the phosphorylated form of the deoxy-sugar appears to be the normal intermediate in the pathway (47).
Rhodobacter capsulatus is a purple nonsulfur bacterium that can grow by respiration in an aerobic environment or utilize its photosynthesis machinery to grow anaerobically. The genes for photosynthesis are located in a 46-kb cluster that encodes all of the enzymes needed to synthesize the carotenoids and the C20 isoprenoid chain in bacteriochlorophyll from IPP. Although most of the genes are present as single copies in the bacterial genome, the probable existence of two copies of idi, the gene for IPP isomerase, one located in the photosynthesis cluster and one located elsewhere on the chromosome, has been previously reported (13). We now report that R. capsulatus has two DXPase genes as well. One, dxsA (open reading frame [ORF] 2816) is located in the photosynthesis cluster, and the other, dxsB (ORF 2895), is located elsewhere in the chromosome. Plasmid-located R. capsulatus dxsB can complement E. coli FH11, in which the chromosomal copy of dxs is disrupted. FH11 can also be complemented by growth on 2-methylerythritol (ME) or MVA when transformed with a plasmid containing an “operon” consisting of the three yeast genes required for biosynthesis of IPP from MVA. The recombinant R. capsulatus DXPases encoded by dxsA and dxsB were purified, and the steady-state kinetic studies were conducted using a new assay.
MATERIALS AND METHODS
Materials.
Sodium [2-14C]pyruvate was purchased from Dupont/NEN. Triosephosphate isomerase (TIM), d-glyceraldehyde, dihydroxyacetone phosphate (DHAP), pyruvic acid, isopropyl-β-d-thiogalactoside (IPTG), leupeptin, GAP, and pepstatin were purchased from Sigma. Thiamine diphosphate (TPP) was purchased from ICN Biomedicals, Inc. Imidazole was purchased from Acros Organics. Ni-nitrilotriacetate silica resin was purchased from Qiagen. All restriction endonucleases, Klenow, and T7 DNA polymerase were purchased from New England Biolabs. Deoxynucleoside triphosphates and T4 DNA ligase were purchased from Pharmacia. Shrimp alkaline phosphatase (SAP) and dithiothreitol were purchased from U.S. Biochemical Corp. Plasmid pFL226 was provided by J. E. Hearst (Lawrence Berkeley Laboratories, Berkeley, Calif.). Phage DD92 was provided by F. R. Blattner (University of Wisconsin, Madison, Wis.). Plasmid pMAK705 was provided by S. R. Kushner (University of Georgia, Athens, Ga.). Plasmid pNGH1-amp was provided by C. R. H. Raetz (Duke University, Durham, N.C.). Oligonucleotide primers and linkers were synthesized by the Protein/DNA Core Facility of the Utah Regional Cancer Center. ME was synthesized by the procedure of Duvold et al. (8). DXP and 1-deoxy-d-xylulose (DXS) were synthesized by the procedure of Blagg and Poulter (3).
Bacterial strains and growth conditions.
The strains used in this study are listed in Table 1 and were grown at 30, 37, or 44°C in Luria-Bertani (LB) medium supplemented with the following antibiotics as necessary: ampicillin (50 μg/ml), chloramphenicol (30 μg/ml), or kanamycin (25 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source |
|---|---|---|
| E. coli | ||
| DH5λ | Host for cloning vectors | Life Technologies |
| CJ236 | Host for mutagenesis experiments | Bio-Rad |
| BL21-DE3 | Host for protein synthesis | Novagen |
| LE392 | Host for phage DD92 propagation | Stratagene |
| JM101 | Host for disruption experiments | Stratagene |
| FH11 | JM101 dxs::Kanr/pDX4 | This work |
| Plasmids and phages | ||
| pFL226 | pBR325 with the 9.7-kb BamHI fragment corresponding to bp 36280–45954 of the 46-kb photosynthesis gene cluster (EMBL accession no. Z11165) | J. E. Hearst |
| DD92 | Double-stranded λ-phagemid with a 19.8-kb EcoRI fragment of E. coli genomic DNA containing the dxs gene | F. R. Blattner |
| pMAK705 | Camr, temperature-sensitive origin of replication | S. R. Kushner |
| pNGH1-Amp | pACYC177 derivative for complementation studies | C. R. H. Raetz |
| pUC4K | Contains Kanr cassette | Pharmacia |
| pBS(SK+) | pBluescript cloning vector; Ampr | Stratagene |
| pET11a | E. coli expression vector; Ampr; T7 promoter | Novagen |
| pGEM-T Easy | Cloning vector for PCR products; Ampr | |
| pDX1 | pBS(SK+) with the 2.5-kb BamHI-SalI fragment of pFL226 | This work |
| pDX2 | pDX1 with an NdeI site introduced by Kunkel mutagenesis | This work |
| pFMH28 | pET11a with the 2.0-kb NdeI-BamHI fragment of pDX2 containing ORF 2816 | This work |
| pDX3 | pDX3 with ORF 2816 modified by Kunkel mutagenesis to add 6 His residues to the C terminus of R. capsulatus DXPase-A | This work |
| pFMH30 | pET11a with the 2.0-kb NdeI-BamHI fragment of pDX3 | This work |
| pDX4 | pMAK705 with the 2.0-kb SphI-SmaI fragment of DD92 containing E. coli dxs | This work |
| pDX5 | pMAKDXS with the 1.3-kb EcoRI Kanr cassette from pUC4K inserted into E. coli dxs | This work |
| pFMH39 | pNGH1-Amp with the 1.9-kb EcoRI-BamHI fragment containing ORF 2895 | This work |
| pBRG12 | pBS(SK+) with the PCR product containing ERG12 adjacent to 54 bp of the 5′ end of ERG19 | This work |
| pERG8 | pGEM-T Easy with the PCR product containing ERG8 | This work |
| pERG19 | pGEM-T Easy with the PCR product containing ERG19 | This work |
| pBRG812 | pBRG12 with the 1.4-kb NotI-PstI fragment of pERG8 containing ERG8 | This work |
| pFCO1 | pBRG812 with the 1.1-kb AflII-XhoI fragment of pERG12 containing the rest of ERG19 | This work |
| pFCO2 | pNGH1-Amp with the 4.1-kb SacI-XhoI fragment of pFCO1 containing ERG8, ERG12, and ERG19 | This work |
| pLME10 | pET11a with the 2.0-kb NdeI-BamHI fragment containing ORF 2895-6 His | This work |
Camr, chloramphenicol resistance; Kanr, kanamycin resistance; Ampr, ampicillin resistance.
General methods.
Minipreparations of plasmid DNA for restriction analysis were obtained by the boiling method as described by Sambrook et al. (40). Large-scale plasmid preparations (>100 μg) were made using the Qiagen plasmid Midi kit. DNA fragments were purified on agarose gels (Bio-Rad) using a Geneclean II kit from Bio 101. Restriction digestions, ligations, and transformations of competent E. coli cells were conducted as described by Sambrook et al. (40). Site-directed mutagenesis was performed by the procedure of Kunkel (17) using the Muta-Gene Phagemid in vitro mutagenesis kit (version 2; Bio-Rad). PCR was performed using a Perkin-Elmer GeneAmp PCR System 2400 DNA thermal cycler and the polymerase mix provided in the Advantage PCR Kit (Clontech). DXPase was assayed by the procedure reported below. Radioactivity was measured in Ultima Gold scintillation fluid (Packard) using a Packard Tri-Carb 2300TR liquid scintillation analyzer. Size exclusion and affinity chromatography was conducted at 4°C using a Pharmacia fast protein liquid chromatography system. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using the discontinuous buffer system of Laemmli (20), and the gels were stained with Coomassie brilliant blue R (Sigma). Protein concentrations were determined by the method of Bradford (6) using bovine serum albumin (BSA) as a standard.
Sequence analysis.
Sequence searches were performed at the National Center for Biotechnology Information (NCBI). Pairwise sequence alignments were performed at the ExPASy Proteomics Tools website using the SIM program or with Vector NTI. Multiple-sequence alignments were performed using Vector NTI and the ClustalW 1.7 and PIMA 1.4 alignment methods at the Baylor College of Medicine Search Launcher website. DNA was sequenced at the Health Sciences Center Sequencing Facility, Eccles Institute of Human Genetics, University of Utah.
Plasmid constructions.
The plasmids used in this study are listed in Table 1. Plasmid pFL226 is a derivative of pBR325 and contains a 9.8-kb BamHI fragment of the R. capsulatus photosynthesis gene cluster. pFL226 was restricted with SalI and BamHI, and the purified 2.5-kb fragment was ligated into pBluescript SK(+) to yield pDX1. Site-directed mutagenesis was employed to introduce a unique NdeI site at the start of ORF 2816 using primer 5′-GCTCATCGGAGGACGCACATATGAGCGCCACGCCATCCC-3′ to yield pDX2. The new NdeI site is underlined, and the start codon of ORF 2816 is in bold. The purified 2.0-kb fragment was subcloned into the NdeI and BamHI sites of E. coli expression vector pET11a to give pFMH28. The newly introduced ORF was sequenced and was identical to the deposited sequence for ORF 2816 (GenBank accession no. Z11165).
Plasmid pDX2 was modified to introduce a glycine and six histidine residues at the C terminus of the protein using site-directed mutagenesis and primer 5′-CGGTGCGGATCGTGCACAGCGCGGGG(CAT)6TAACTCGAGGCGACACTGCAAAGATGCACGG-3′ to yield pDX3. A unique XhoI site introduced for screening is underlined, the codons for the new Gly and His residues are in bold, and the stop codon for the ORF is in italics. pDX3 was restricted with NdeI and BamHI, and the 2.0-kb fragment was ligated into pET11a to yield pFMH30.
ORF 2895 was isolated from R. capsulatus genomic DNA by PCR using the primers (sense) 5′-CGGAATTCATGAGCCAGCCCCCAGTGACCCCGATCCTCGACCGCGTGCGTCTG-3′ and (antisense) 5′-CGGGATCCTCAGGC GCGTTTCGCCACCGCGACGCCGAGCAGATCGAGCACTTTCG-3′. The ∼2-kb PCR product was restricted with EcoRI-BamHI and subcloned into the pACYC177 derivative pNGH1-amp to give pFMH39. The ORF contained within pFMH39 was sequenced and was identical to the sequence deposited at the Rhodobacter Capsulapedia website (see above).
The ORF encoding DXPase-B was then modified to introduce six histidine residues at the C terminus of the protein by PCR using the primers (sense) 5′-GAGATATACATATGAGCCAGCCCCCAGTG-3′ and (antisense) 5′-TCTAGGATCCTCA(ATG)6GGCGCGTTTCGCCTC-3′. Unique NdeI (sense) and BamHI (antisense) sites are underlined, the codons for the new His residues are in bold, and the stop codon is in italics. The ∼2-kb PCR product was restricted with NdeI-BamHI, purified, and subcloned into E. coli expression vector pET11a to give pLME10.
Within phagemid DD92 is a 19.8-kb EcoRI fragment of E. coli genomic DNA containing dxs, the gene for DXPase (Table 1). Following the isolation of the phage from E. coli strain LE392, DD92 was restricted with SphI and SmaI to give a 2.0-kb fragment containing E. coli dxs. The purified DNA fragment was ligated into the SphI and HindII sites of pMAK705, a plasmid containing a temperature-sensitive origin of replication (14). The resulting plasmid, pDX4, was restricted with SapI at a unique site located in the middle of the dxs gene, and the 5′ overhangs were filled in with the Klenow fragment and treated with SAP as specified by the manufacturer. A 1.3-kb blunt-ended DNA fragment containing the gene for Kanr from pUC4K (51) (Amersham Pharmacia Biotech catalog, 1999) was then ligated into the linearized pDX4 DNA to create pDX5 with a disruption in E. coli dxs.
Plasmids pFCO1 and pFCO2 containing a synthetic operon for the in vivo synthesis of IPP from mevalonate were constructed as follows: ERG12, ERG8, and ERG19 were isolated from S. cerevisiae genomic DNA by PCR using the respective primer sets FH0129-2 (sense) 5′-GGACTAGTCTGCAGGAGGAGTTTTAATGTCATTACCGTTCTTAACTTCTGCACCGGG-3′ and FH0129-1 (antisense) 5′-TTCTCGAGCTTAAGAGTAGCAATATTTACCGGAGCAGTTACA CTAGCAGTATATACAGTCATTAAAACTCCTCCTGTGAAGTCCATGGTAA ATTCG-3′, FH0211-2 (sense) 5′-TAGCGGCCGCAGGAGGAGTTCATATGTCAGAGTTGAGAGCCTTCAGTGCCCCAGGG-3′ and FH0211-1 (antisense) 5′-TTTCTGCAGTTTATCAAGATAAGTTTCCGGATCTTT-3′, and CT0419 (sense) 5′-GGAATTCATGACCGTTTACACAGCATCCGTTACCGCACCCG-3′ and CT0419-2 (antisense) 5′-GGCTCGAGTTAAAACTCCTCTTCCTTTGGTAGACCAGTCTTTGCG-3′. Primer FH0129-2 included a SpeI site (underlined). Primer FH0129-1 contained a XhoI site (underlined), an AflII site (double underlined), and 54 nucleotides corresponding to the 5′ end of ERG19 (bold italics). Following restriction with SpeI-XhoI, the PCR product containing ERG12 was subcloned into pBluescript SK(+) to create pBRG12. Primers FH0211-1 and FH0211-2 contained a NotI site (underlined) and a PstI site (underlined), respectively. The purified ERG8 PCR product was restricted with NotI-PstI and subcloned into pGEM-T Easy to create pERG8. The PCR product containing ERG19 was ligated directly into pGEM-T Easy to create pERG19. Restriction of pERG8 with NotI-PstI yielded a 1.4-kb DNA fragment containing ERG8. Restriction of pBRG12 with NotI-PstI was followed by insertion of the NotI-PstI DNA fragment to create pBRG812 containing both ERG8 and ERG12. Restriction of pERG19 with AflII-XhoI yielded a 1.2-kb DNA fragment containing the 3′ end of ERG19 missing in pBRG812. Ligation of the 1.2-kb DNA fragment into the AflII-XhoI sites of pBRG812 gave pFCO1 containing ERG8, ERG12, and ERG19. Restriction of pFCO1 with XhoI was followed by treatment with the Klenow fragment and deoxynucleoside triphosphates to create blunt ends. Subsequent restriction of pFCO1/XhoI/Klenow with SacI yielded a 3.9-kb DNA fragment containing ERG8, ERG12, and ERG19. The 3.9-kb fragment was inserted into the SmaI-SacI sites of pNGH1-amp to create pFCO2.
Construction of JM101 dxs::Kanr/pDX4(FH11).
An E. coli mutant with a disruption of the chromosomal dxs gene was constructed as described by Hamilton et al. (14). Competent E. coli JM101 cells were transformed with pDX5 and grown to an optical density at 600 nm of 0.6 at 30°C. Approximately 10,000 cells were plated out on LB-chloramphenicol medium prewarmed to 44°C. The plates were incubated at 44°C, and several of the resulting colonies were picked and grown at 44°C in 4 ml of LB-chloramphenicol medium. Four 50-ml LB-chloramphenicol cultures were then started with 0.5 ml from four of the 4-ml cultures and grown overnight at 30°C. Four fresh 50-ml LB-chloramphenicol cultures were then started with 100 μl of the previous cultures and grown overnight at 30°C. An aliquot of one of the 50-ml cultures was serially diluted 5 × 105-fold, and 5 μl was plated on LB-chloramphenicol medium. Following incubation at 30°C, the resulting colonies were used to individually inoculate 3 ml of LB-chloramphenicol/kanamycin medium. Twelve LB-chloramphenicol/kanamycin cultures were grown overnight at 30°C and used for plasmid DNA isolation. E. coli cells where the disrupted copy of dxs was incorporated into the genome were identified by restriction analysis of the isolated plasmid DNA and verified by sequence analysis of the DNA contained in the plasmids. The strain was designated FH11.
Construction of FH11/pFMH39, FH11/pFCO2, and FH11/pNGH1-Amp.
Colonies of E. coli strain FH11 transformed with pFMH39, pFCO2, or pNGH1-Amp were isolated by incubation at 30°C on LB-kanamycin/ampicillin medium. Several of the transformants were then grown in LB-kanamycin/ampicillin medium to stationary phase. Restriction analysis of plasmid DNA verified the presence of pDX4 and pFMH39, pDX4 and pFCO2, or pDX4 and pNGH1-Amp.
Growth of FH11 at 30 and 44°C.
Two 50-ml LB-kanamycin samples were inoculated with 0.5 ml of FH11-(4) and FH11-(7) cultures of cells with the chromosomal dxs::Kanr knockout. Following growth at 30°C overnight, the cultures were diluted 10,000-fold, and 20-μl portions were spread on LB-kanamycin plates at room temperature (RT) or prewarmed to 44°C. A sample of FH11 was also spread on a prewarmed LB-kanamycin plate containing ∼7 mg of ME. The prewarmed plates were incubated overnight at 44°C, and the RT plates were incubated at 30°C overnight.
Growth of FH11/pFMH3, pFH11/pFCO2, and FH11/pNGH1-Amp at 30 and 44°C.
Using the procedure described in the preceding section, FH11/pFMH39, pFH11/pFCO2, and FH11/pNGH1-Amp were spread on LB-kanamycin/ampicillin plates that were prewarmed to 44°C or kept at RT. Approximately 1.3 mg of MVA was also spread on the plates used for FH11/pFCO2. The samples were incubated as described above.
Expression of R. capsulatus dxs and purification of DXPase.
Plasmid pFMH30 or pLME10 was used to transform E. coli BL21-DE3-competent cells. LB-ampicillin cultures (3 ml) were inoculated with single colonies, grown overnight at 37°C, and used to inoculate 500 ml of M9-CAGM-ampicillin (40). The cultures were grown at 37°C to an optical density at 600 nm of ∼0.6, the temperature was lowered to 30°C, IPTG was added to a final concentration of 1 mM, and incubation was continued for 5 h. The cells were harvested by centrifugation.
All steps in the purification were done at 4°C. Cell paste (∼8 g) from BL21-DE3/pFMH30 or BL21-DE3/pLME10 was suspended in 40 ml of extraction buffer consisting of 50 mM sodium phosphate (pH 8), 5 mM β-mercaptoethanol (BME), 1 mM EDTA, leupeptin (2 μg/ml), pepstatin (1 μg/ml), and 2 mM phenylmethylsulfonyl fluoride (PMSF) and disrupted by sonication. The resulting homogenate was centrifuged at 23,700 × g to remove cellular debris. (NH4)2SO4 was added to 46% saturation (26.7 g/100 ml), and the sample was centrifuged. The resulting pellet was resuspended in 30 ml of extraction buffer and centrifuged, and the supernatant was dialyzed against 50 mM sodium phosphate (pH 8)–10 mM imidazole–150 μM PMSF–5 mM BME to remove the (NH4)2SO4. A solution of 2 M NaCl was added to the dialyzed supernatant to a final concentration of 0.5 M. The sample was loaded onto a 1.0- by 10.0-cm Ni-nitrilotriacetic acid silica column previously equilibrated with 50 mM sodium phosphate (pH 8)–0.5 M NaCl–10 mM imidazole–5 mM BME (buffer A). The column was first washed at 1.0 ml/min with 90 ml of 50 mM sodium phosphate (pH 8)–0.5 M NaCl–30 mM imidazole–5 mM BME (buffer B) and eluted at 1.0 ml/min with a linear gradient of 100% buffer B to 100% buffer C (50 mM sodium phosphate [pH 8], 0.5 M NaCl, 0.5 M imidazole, 5 mM BME) over 60 ml. Active fractions were combined and dialyzed against 50 mM Tris-HCl (pH 7.5)–150 μM PMSF–5 mM BME. The protein was concentrated to 3 to 4 mg/ml by centrifugation in a Centriprep-30 (Amicon) concentrator and stored in 30% glycerol at −80°C.
Gel filtration of DXPase-A.
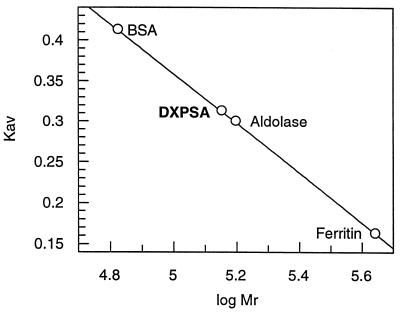
Size exclusion chromatography was performed on a 1.6- by 60-cm HiLoad Superdex 200 prep grade gel filtration column (Pharmacia). All protein samples were kept in 50 mM Tris (pH 7.5) buffer containing 150 mM NaCl, 10 mM MgCl2, and 10 mM BME. The void volume of the column (40 ml) was measured with Blue Dextran 2000, and the column was calibrated with BSA (67 kDa), aldolase (158 kDa), and ferritin (440 kDa). The molecular mass of recombinant R. capsulatus DXPase-A was estimated from a plot of the logarithm of the molecular mass (log Mr) versus the retention volumes (Kav) of the eluted proteins by using Grafit (22).
Product analysis.
Affinity-purified R. capsulatus DXPase was incubated with 50 μl of 200 mM citrate buffer (pH 6.0) containing 1 mM TPP, 10 mM MgCl2, 50 mM d-glyceraldehyde, and 50 mM pyruvate. After 45 min, a 10-μl portion of the mixture was loaded onto a silica thin-layer chromatography (TLC) plate and a second lane was spotted with an authentic sample of 1-deoxy-d-xylulose (DXS) (3). The plate was developed by published procedures using n-propyl alcohol–ethyl acetate–H2O (6:1:3) (27). DXP was synthesized from fructose-1,6-diphosphate, pyruvate, and TPP with a coupled enzyme system consisting of rabbit muscle aldolase, TIM, and DXPase-A by the method of Taylor et al. (48). Alternatively, DXP was synthesized from DHAP, pyruvate, and TPP with a coupled enzyme assay system consisting of TIM and DXPase-B as described below. Enzymatic and chemically synthesized DXP samples were spotted in adjacent lanes and analyzed by TLC as described above.
Assays for DXPase. (i) Pyruvate and d-Glyceraldehyde.
DXPase activity with pyruvate and d-glyceraldehyde as substrates was measured by incorporation of radioactivity into DXS from [2-14C]pyruvate. Pyruvate concentrations were determined using an end-point assay with lactate dehydrogenase and NADH. Assay mixtures contained 200 mM sodium citrate (pH 7.4), 5 mM dithiothreitol, 2 mM MgCl2, 2 mM TPP, and various concentrations of [2-14C]pyruvate and d-glyceraldehyde in a final volume of 50 μl. The mixtures were preincubated for 5 min at 37°C, and the reactions were initiated by addition of enzyme. After incubation at 37°C for 10 min, the reactions were quenched by the addition of either 50 μl of 0.5 M EDTA or 325 μl of 1:1 (vol/vol) methanol-CHCl3. The reaction mixtures were clarified by centrifugation, and 50 or 125 μl was loaded onto a 0.5- by 7.0-cm silica gel grade 60, 200- to 400-mesh column containing 1.0 cm of Na2SO4. [2-14C]DXS was eluted with 11 to 12 ml of 1:3 (vol/vol) methanol-CHCl3. The solvent was removed with a gentle stream of nitrogen, the residue was resuspended in 1 ml of methanol, and the mixture was vortexed for 20 s. ULTIMA GOLD scintillation fluid (10 ml) was added, the solutions were vortexed for 30 s, and radioactivity was measured by liquid scintillation spectrometry.
(ii) Pyruvate and GAP.
Assay mixtures contained 200 mM sodium citrate (pH 7.4), 10 mM MgCl2, 2 mM TPP, 0.025 to 25 mM [2-14C]pyruvate (0.10 to 7.9 μCi/μmol), and 1.0 to 40 mM DHAP. TIM (15 U) was added, and the tubes were equilibrated at RT for 45 min. The assay was initiated by the addition of affinity-purified DXPase to a final volume of 50 μl. Incubations were carried out at 37°C for 2 to 13 min. The reaction was quenched by heating at 80°C for 2 min. The samples were cooled at 0°C for 5 min, the MgCl2 concentration was adjusted to 50 mM, and 10 U of SAP was added. The reaction mixture was incubated at 37°C for 90 min and clarified by centrifugation. A 25-μl portion was loaded onto a silica column as described above for DXS. The radioactivity in [2-14C]DXS was measured as described above. Initial velocities were determined from the slope of a plot of product versus time.
Steady-state kinetic constants for DXPase.
Steady-state kinetic constants determined for pyruvate and d-glyceraldehyde were measured at saturating concentrations of the second substrate, 2.5 to 100 mM d-glyceraldehyde, and 0.25 to 25 mM [2-14C]pyruvate (0.10 μCi/μmol). Reactions were run at 37°C in buffer containing 200 mM sodium citrate (pH 7.4), 2 mM MgCl2, 2 mM TPP, and affinity-purified DXPase (6.0 to 7.5 μg) and were quenched by the addition of EDTA to a final concentration of 250 mM. Conversion of the limiting substrate was 10% or less. The Km and Vmax of d-glyceraldehyde and pyruvate were obtained by fitting the data to the Michaelis-Menten equation using Grafit (22).
Steady-state kinetic constants determined for pyruvate and GAP were measured at saturating concentrations of the second substrate. Reactions were run at 37°C in mixtures containing 200 mM sodium citrate (pH 7.4), 10 mM MgCl2, 2 mM TPP, 0.025 to 15 mM [2-14C]pyruvate (0.10 to 7.9 μCi/μmol), 0.25 to 35 mM DHAP (0.04 to 1.4 mM GAP), 15 U of TIM, and affinity-purified DXPase (1 to 3 μg). The reactions were quenched by heating at 80°C for 2 min, and the products were treated with SAP as described above. Initial velocities were determined from the slope of a plot of product versus time. Km and Vmax for GAP and pyruvate were obtained by fitting the data to the Michaelis-Menten equation using Grafit (22).
Metal ion dependence.
DXPase-A activity was measured in 200 mM sodium citrate (pH 7.4)–2 mM TPP–50 mM [2-14C]pyruvate (0.084 μCi/μmol)–50 mM d-glyceraldehyde–0.01 to 20 mM Mg2+, Mn2+, or Ca2+ in a total volume of 50 μl. The assay mixtures were preincubated at 37°C for 5 min, the reactions were initiated by the addition of 7.5 μg of DXPase-A, and the mixtures were incubated at 37°C for 10 min. The reactions were quenched by the addition of 325 μl of 1:1 (vol/vol) methanol-CHCl3, and the radioactivity in [2-14C]DXS was measured as described above.
Inhibition by DHAP.
Assay mixtures containing 200 mM sodium citrate (pH 7.4), 2 mM MgCl2, 2 mM TPP, 20 mM [2-14C]pyruvate (0.034 μCi/μmol), 25 mM d-glyceraldehyde, and 0 to 80 mM DHAP were preincubated at 37°C for 5 min. The reaction was initiated by the addition of 6 μg of DXPase-A. The assay mixtures were incubated at 37°C for 10 min before the addition of EDTA to a final concentration of 250 mM. The radioactivity in [2-14C]DXS was measured as described above.
RESULTS
R. capsulatus ORF 2816 carries dxsA, the gene for DXPase-A.
Database searches were conducted using the entire translated sequence of ORF 2816, corresponding to nucleotides 43929 to 45854 (previously designated ORF 641 [GenBank accession no. Z11165]) in the photosynthesis gene cluster of R. capsulatus. The initial results revealed a strong similarity to both transketolases and the E1 component of pyruvate dehydrogenases involved in pyruvate decarboxylation. These motifs were consistent for an enzyme that catalyzes the synthesis of DXP by condensation of GAP the two-carbon acetyl acyl anion produced by a thiamine-mediated decarboxylation of pyruvate. This mechanism, initially proposed by Rohmer et al. (38), is similar to those for reactions catalyzed by transketolases. Several hypothetical proteins were also found with substantial similarity to the protein encoded by ORF 2816, including gene products from Arabidopsis thaliana CLA1 (accession no. U2709), Synechocystis sp. hypothetical protein PID:d1017822 (accession no. D90903), Haemophilus influenzae hypothetical protein HI1439 (Swiss-Prot accession no. P45205), Escherichia coli hypothetical protein PID:g1773104 (accession no. U82664), Bacillus subtilis hypothetical protein (Swiss-Prot accession no. P54523), and Mycobacterium tuberculosis hypothetical protein (Swiss-Prot accession no. O07184). Shortly after our search was conducted, two other groups reported that the hypothetical E. coli protein was a DXPase (27, 46). Their results provided strong evidence that R. capsulatus ORF 2816 encoded DXPase-A. We discovered several proteins labeled as transketolases, for example, Mycobacterium leprae (accession no. U15181) and Streptomyces coelicolor (accession no. AL049485), that appear to be DXPases based on the presence of a highly conserved “DRAG” sequence (see below). Other putative DXPases identified in subsequent searches included a second M. tuberculosis protein (accession no. AL009198), the 626-amino-acid translation product of Neisseria gonorrhoeae (Contig320, an unfinished fragment of the genome deposited at NCBI), and a protein from the unicellular eukaryote Plasmodium falciparum (accession no. AF111814).
R. capsulatus ORF 2895 carries dxsB, a second gene for DXPase.
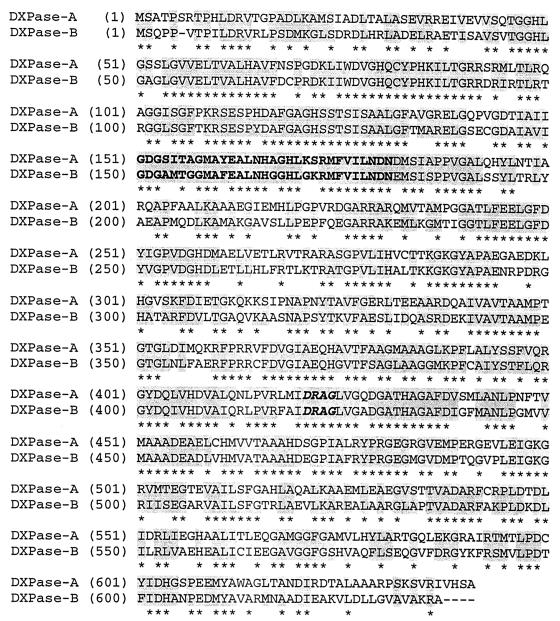
A BLAST search at the University of Chicago R. capsulatus website (see above) using E. coli dxs as the query sequence uncovered a second ORF in R. capsulatus, which we subsequently identified as DXPase-B biochemically. DXPase-B is a 636-amino-acid protein, compared to DXPase-A with 641 amino acids. DXPase-A and DXPase-B have 64.3% identity over a sequence of 622 residues (Fig. 2).
FIG. 2.
Pairwise alignment of R. capsulatus DXPase-A and DXPase-B. Regions of homology are shown by a shaded background. The 402 identical residues are indicated by asterisks, the thiamine diphosphate binding motif is in bold, and the DRAG signature sequence is in bold italics.
Sequence comparisons.
DXPases vary in length from 536 amino acids in M. tuberculosis to 739 residues in A. thaliana. However, most of the proteins are similar in length to the two Rhodobacter proteins. A multiple sequence alignment was conducted using DXPase-A from R. capsulatus and 10 of the proteins described above to identify conserved amino acids and regions of significance. A total of 93 residues in the set were invariant. A highly conserved region with a high level of similarity to the consensus TPP binding motif (15, 34) corresponds to residues 151 to 181 in the Rhodobacter DXPase-A protein and residues 150 to 180 in the DXPase-B protein (Fig. 2).
DXPase-A also has a substantial degree of similarity to transketolases. Identical residues are distributed throughout the entire sequences of transketolases and the Rhodobacter DXPases. Many residues known to be important for binding and/or catalysis in transketolases are also present in the Rhodobacter proteins. Table 2 correlates key amino acids in yeast transketolase, along with their roles assigned in catalysis, with their counterparts in Rhodobacter DXPase-A and DXPase-B. Schenk et al. (41) have designated a highly conserved region consisting of 36 amino acids important for structure and function in transketolases as a “transketolase motif.” An analogous region was found for residues 419 to 454 in Rhodobacter DXPase-A. A cluster of four residues, THDS, located at the beginning of the transketolase motif is replaced by a DRAG cluster in all of the sequences unambiguously shown to be DXPases. The product of a putative dxs gene in Plasmodium falciparum has a four-residue GRSG sequence instead of the DRAG motif; however, the protein also lacks the His residue of THDS conserved in all transketolases. We identified additional putative dxs genes in NCBI databases using the Rhodobacter DXPase-A sequence as a probe. These include ORFs from Neisseria meningitidis, Pseudomonas aeruginosa, and Deinococcus radiodurans. A key element to our identification of putative DXPases is the DRAG signature sequence (Fig. 2), which has been identified in a wide variety of organisms, including Synechocystis sp., Bacillus subtilis, Rhodobacter capsulatus, Arabidopsis thaliana, Oryza sativa, Mentha × piperita, Escherichia coli, Haemophilus influenzae, Pseudomonas aeruginosa, Neisseria meningitidis, Neisseria gonorrhoeae, Porphyromonas gingivalis, Helicobacter pylori, Campylobacter jejuni, Aquifex aeolicus, Clostridium acetobutylicum, Deinococcus radiodurans, Treponema pallidum, Mycobacterium leprae, Mycobacterium tuberculosis, Streptomyces coelicolor, Mycobacterium tuberculosis, Chlamydia trachomatis, and Plasmodium falciparum.
TABLE 2.
Comparison of conserved transketolase residues with those in DXPase-A and DXPase-Ba
| DXPase-A residue | DXPase-B residue | Transketolase residue | Function | Reference(s) |
|---|---|---|---|---|
| H80 | H79 | H69 | Stabilization of reaction intermediate; binds C-1 hydroxyl of donor substrate | 53 |
| H49 | H48 | H30 | Acid-base catalyst (proton acceptor or donor) | 53 |
| H258 | H257 | H263 | Acid-base catalyst (proton acceptor or donor) | 53 |
| D152 | D151 | D157 | Metal binding (side chain) | 42 |
| N181 | N180 | N187 | Metal binding (side chain) | 42 |
| M183 | M182 | I189 | Metal binding (main-chain oxygen) | 42 |
| D429 | D428 | D477 | Binds C-2 hydroxyl of acceptor substrate; enantioselectivity of enzyme | 30, 42 |
| H433 | H432 | H481 | Transition-state stabilization | 53 |
| E372 | E372 | E418 | Binds N-1′ nitrogen of TPP cofactor (essential for catalytic activity) | 42, 52 |
| R480 | R479 | R528 | Phosphate binding of substrate | 31, 42 |
| Y394 | Y393 | F442 | Interaction with pyrimidine ring of TPP | 42 |
| F397 | F396 | F445 | Interaction with pyrimidine ring of TPP | 42 |
| Y402 | Y401 | Y448 | Interaction with pyrimidine ring of TPP | 42 |
Important conserved residues from S. cerevisiae transketolase and the similarly conserved residues in R. capsulatus DXPase-A and DXPase-B are compared. The proposed function of these residues is based on numerous studies of transketolases reported in the literature.
A multiple sequence alignment of the Rhodobacter DXPase-A sequence with several pyruvate dehydrogenase E1 component sequences was also performed. The most striking feature of the alignment was the homology of the N-terminal part of DXPase-A to the λ subunit of the E1 component and the homology of the C-terminal end of the Rhodobacter sequence to the β subunit of the E1 component.
R. capsulatus ORF 2895 complements E. coli dxs::Kanr.
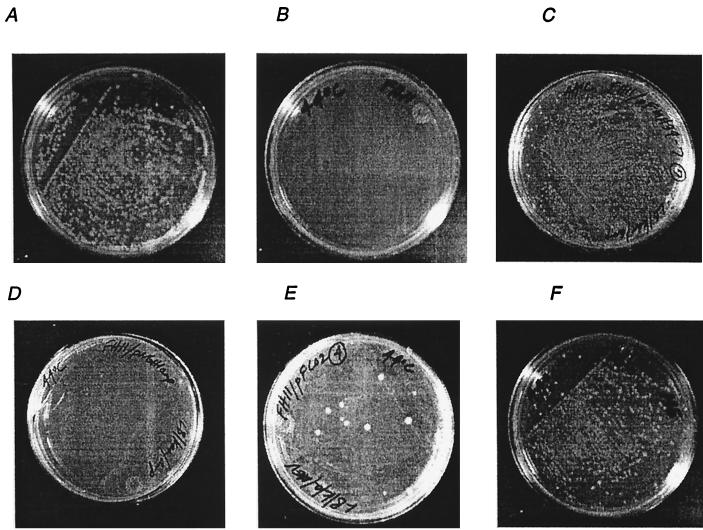
Following the construction of strain FH11, the episomal copy of dxs contained in pDX4 was turned off at 44°C. The inability of FH11 to grow at the restrictive temperature demonstrates that dxs is an essential single-copy gene in E. coli. The strain was rescued at 44°C by addition of ME, an advanced intermediate in the pathway, to the medium (8). To establish that ORF 2895 encodes a functional DXPase, FH11 cells were transformed with pFMH39. FH11/pFMH39 cells were able to grow at the restrictive temperature of 44°C, whereas FH11/pNGH1-Amp cells, containing the expression vector without the dxs insert, did not grow. The results are illustrated in Fig. 3 for ORF 2895. The ability of R. capsulatus ORF 2895 to complement E. coli dxs::Kanr provides genetic evidence that it encodes DXPase-B.
FIG. 3.
Growth of E. coli FH11 at 30 and 44°C. FH11 cells with chromosomal dxs::Kanr and episomal dxs under the control of the temperature-sensitive origin of replication contained in pMAK705 (14) were grown under the following conditions. (A) FH11 cells were able to grow at 30°C on LB-kanamycin medium. (B) FH11 cells were unable to grow at the nonpermissive temperature of 44°C on LB-kanamycin medium, establishing that E. coli dxs is an essential gene. (C) FH11 cells with R. capsulatus ORF 2895 contained in pFMH39 were able to grow at the nonpermissive temperature of 44°C on LB-kanamycin/ampicillin medium, establishing that R. capsulatus ORF 2895 carries dxsB. (D) FH11/pNGH1-Amp cells containing the parent vector of pFMH39 and pFCO2 were unable to grow at 44°C on LB-kanamycin/ampicillin medium. (E) FH11/pFCO2 cells were able to grow at 44°C on LB-kanamycin/ampicillin medium supplemented with 50 mg of mevalonic acid per ml, establishing the ability of the synthetic operon contained within pFCO2 to synthesize IPP. (F) FH11 cells were able to grow at the nonpermissive temperature of 44°C on LB-kanamycin medium containing ∼0.3 mg of ME per ml.
Synthesis of IPP from MVA in E. coli.
The three yeast genes ERG8, ERG12, and ERG19 were isolated by PCR and were translationally coupled with overlapping start and stop codons (29) in the synthetic operon contained within pFCO1. The PCR primers for all three yeast genes contained additional nucleotides for the ribosome binding site AGGAGGAG and the insertion of Gln-Glu-Glu-Phe at the C terminus of the encoded proteins to facilitate their purification using a monoclonal antibody column (29). The gene for MVA kinase, ERG12, served as the foundation for the construction of the operon. ERG8, the gene for phospho-MVA kinase, was added upstream of ERG12, and ERG19, the gene for MVA diphosphate decarboxylase, was appended to the 3′ end of ERG12. To ensure specific annealing to ERG12, the additional nucleotides corresponding to codons for the N terminus of MVA diphosphate decarboxylase were altered during the design of primer FH0129 to direct the synthesis of the correct amino acids using codons as different as possible from wild-type ERG19. A cassette containing the coupled yeast genes was removed from pFCO1 and inserted into pNGH1-Amp to give pFCO2 in order to test the ability of the operon to complement the dxs::Kanr disruption in FH11 grown on MVA. FH11/pFCO2 transformants did not grow at the restrictive temperature of 44°C unless MVA (50 mg/liter) was added to the growth medium (Fig. 3), thus establishing the ability of the synthetic operon to direct the synthesis of IPP from MVA in E. coli.
Expression and purification of DXPase.
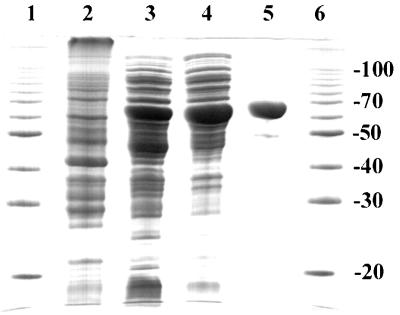
E. coli strains BL21-DE3/pFMH30 and BL21-DE3/pLME10 produce recombinant R. capsulatus DXPase-A and DXPase-B, respectively, to approximately 5 and 3% of total cellular protein. Both enzymes were purified to >95% homogeneity in two steps by (NH4)2SO4 precipitation followed by Ni2+-silica affinity chromatography (results for DXPase-A are given in Table 3). The purified enzymes were isolated in 45% (DXPase-A) and 30% (DXPase-B) yields and stored at −20°C in dialysis buffer (50 mM Tris-HCl [pH 7.5], 5 mM BME, 150 μm PMSF) containing 30% glycerol until needed, with no significant decrease in enzyme activity for 4 to 6 weeks. An SDS-PAGE gel of samples from each purification step of DXPase-A is shown (Fig. 4). After Ni2+-silica affinity chromatography, SDS-PAGE of the sample gave a single band at 68 kDa, as expected for the protein encoded by dxsA (Fig. 4). Similar results were obtained for dxsB (data not shown).
TABLE 3.
Summary of purification of R. capsulatus DXPase-A
| Step | Amt of protein (mg) | Sp acta (μmol/min/ mg) | Total activity (μmol/min) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|---|
| Supernatant | 493 | 0.18 | 89 | 1 | 100 |
| 40% (NH4)2SO4 precipitate | 213 | 0.35 | 75 | 2 | 84 |
| Ni2+-silica affinity column | 25 | 1.6 | 40 | 10 | 45 |
DXPase-A activity was determined with 25 mM [2-14C]pyruvate (0.06 μCi/μmol), 100 mM d-glyceraldehyde, and 2 mM MgCl2. Reactions were quenched by the addition of EDTA to a final concentration of 250 mM as described in Materials and Methods.
FIG. 4.
Purification of R. capsulatus DXPase-A. Samples from each purification step were electrophoresed on a 0.1% SDS–12% polyacrylamide gel and stained with Coomassie blue. Lanes: 1 and 6, molecular mass standards (at 20, 30, 40, 50, 70, and 100 kDa); 2, cell extract of BL21/pFMH30 without IPTG induction (∼40 μg); 3, cell extract of BL21/pFMH30 with IPTG induction (∼70 μg); 4, ammonium sulfate precipitation (∼50 μg); 5, Ni2+-silica affinity chromatography (∼9 μg).
Product analysis.
The identity of the product from pyruvate and d-glyceraldehyde was confirmed by a comparison of TLC mobility with an authentic sample of DXS. An attempt to use a commercial sample of GAP to establish the structure of DXP in a parallel experiment was unsuccessful. We then used the method of Taylor et al. (48) to synthesize GAP from fructose-1,6-diphosphate with rabbit muscle aldolase. The aldolase products, GAP and DHAP, were isomerized with TIM to generate a constant supply of GAP for DXPase. Alternatively, GAP was generated in situ from commercially available DHAP and TIM. The Rf of the DXPase product upon TLC was identical to that of authentic DXP.
R. capsulatus DXPase-A is a homodimer.
The molecular mass of affinity purified recombinant DXPase-A was estimated by gel filtration chromatography (Fig. 5). The molecular mass was determined to be 141 kDa, consistent with a homodimer of 68-kDa subunits calculated from the gene sequence.
FIG. 5.
Determination of the molecular mass of R. capsulatus DXPase-A by gel filtration chromatography. The retention volume of ∼2 mg of purified recombinant R. capsulatus DXPase-A was measured on a calibrated HiLoad Superdex 200 prep grade gel filtration column (Pharmacia) with BSA (Mr = 67,000), aldolase (Mr = 158,000), and ferritin (Mr = 440,000) as standards. Using the formula Kav = (column void volume − elution volume)/(column bed volume − column void volume), KavDXPase-A = 0.313 and Mr = 141,000. Since Mr = 67,940 for the protein encoded by dxsA, the enzyme is most probably a homodimer.
Biochemical properties of recombinant DXPase-A.
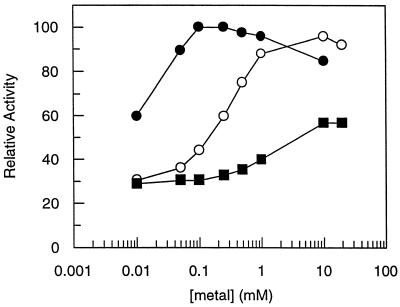
Maximal activity for R. capsulatus DXPase-A required the addition of a divalent metal, either Mn2+ or Mg2+ (Fig. 6). DXPase-A showed optimal activity at concentrations between 0.05 and 1 mM for Mn2+ and between 2 and 10 mM for Mg2+. Only 30 to 40% of the maximal activity was achieved with Ca2+ over a range of concentrations between 0.01 and 20 mM. A requirement for a divalent metal by DXPase-A is consistent with that by other TPP-dependent enzymes. Pyruvate decarboxylases, transketolases, benzoylformate decarboxylases, pyruvate oxidases, and acetolactate synthases all catalyze TPP-dependent reactions in the presence of Mg2+ (43).
FIG. 6.
Metal requirements for DXPase-A were determined under standard assay conditions with 50 mM [2-14C]pyruvate (0.08 μCi/μmol), 50 mM d-glyceraldehyde, 7 μg DXPase-A, and different concentrations of either Mn2+ (●), Mg2+ (○), or Ca2+ (■). Reactions were quenched by the addition of methanol-CHCl3 (1:1) as described in Materials and Methods.
Michaelis constants for pyruvate and GAP were determined by a coupled assay where GAP was generated in situ from DHAP by TIM. The DHAP/GAP equilibrium is displaced toward DHAP, and GAP concentrations were based on the 96:4 equilibrium ratio for DHAP to GAP reported for TIM (50). Thus, 50 mM DHAP gave 2 mM GAP in the coupled assay system when the activity of TIM was in substantial excess over DXPase. Due to the unfavorable ratio and the large amount of DHAP with respect to GAP, it was necessary to establish that DHAP does not inhibit DXPase. When DXPase was incubated with 25 mM pyruvate, 25 mM d-glyceraldehyde, and 0 to 80 mM DHAP, no decrease in DXPase activity due to inhibition by DHAP was seen (data not shown).
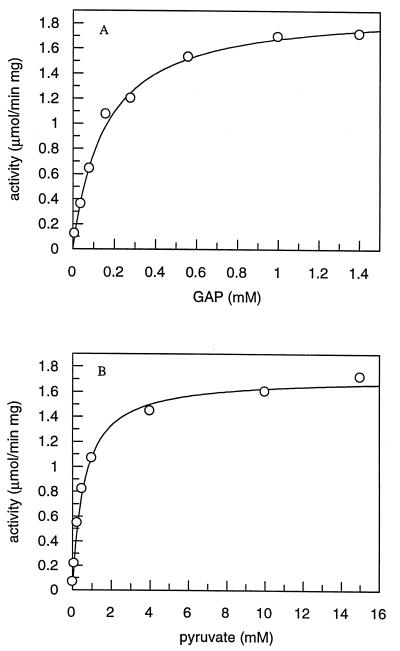
Michaelis constants for pyruvate and GAP were determined by fitting hyperbolic plots of initial velocities versus substrate concentration (Fig. 7) from initial velocities calculated from the slope of product (DXS) versus time plots at each concentration of varied substrate. DXPase-A gave Kmpyruvate = 610 ± 50 μM, KmGAP = 150 ± 10 μM, and Vmax = 1.9 ± 0.1 μmol/min/mg, while DXPase-B gave Kmpyruvate = 3.0 ± 0.2 mM, KmGAP = 120 ± 14 μM, and Vmax = 1.4 ± 0.1 μmol/min/mg. When GAP was replaced by d-glyceraldehyde, DXPase-A gave Kmpyruvate = 4.3 ± 0.2 mM, Kmd-glyceraldehyde = 14 ± 2 mM, and Vmax = 1.6 ± 0.1 μmol/min/mg and DXPase-B gave Kmpyruvate = 14 ± 1 mM, Kmd-glyceraldehyde = 10 ± 2 mM, and Vmax = 1.0 ± 0.1 μmol/min/mg. The aldehyde substrates d-glyceraldehyde and GAP exist primarily as aldehyde hydrates in solution, unlike pyruvate. For d-glyceraldehyde, the effective aldehyde concentration is only 4.4% of the total substrate concentration (2); thus, the Km values for the aldehyde substrates are considerably lower than Kmpyruvate. The Vmax values we measured for the R. capsulatus enzymes are similar to previously reported activities for DXPases from other organisms (21, 27, 46), except for E. coli, where Vmax = 300 to 370 μmol/min/mg and Kmpyruvate = 65 to 96 μM (18). The optimal pH for R. capsulatus DXPase-A in sodium citrate buffer was 7.0 to 7.5. The enzyme was noticeably less active at pH 7.5 in several other buffers (HEPES, morpholinepropanesulfonic acid [MOPS], and bicyclo[2.2.1]hept-5-en-2,3-dicarboxylic acid [BHDA]) that we examined. To ensure that the sodium citrate buffer was not inhibiting the enzyme, glycylglycine, a buffer commonly utilized in transketolase assays (45), was also investigated. For buffer concentrations of sodium citrate and glycylglycine between 50 and 200 mM, the activity of DXPase changed by less than 20%. Kmpyruvate was unaltered in sodium citrate and glycylglycine buffers.
FIG. 7.
Substrate saturation curves for DXPase-A determined in the presence of a fixed concentration of the second substrate utilizing a coupled enzyme assay. GAP is generated in situ by utilizing a coupled enzyme assay starting with dihydroxyacetone phosphate. (A) Different concentrations of GAP (0.03 to 1.0 mM) and 5 mM [2-14C]pyruvate (0.10 to 7.9 μCi/μmol). (B) Different concentrations of [2-14C]pyruvate (0.025 to 15 mM) and 1.2 mM GAP. The assay mixtures contained 2 mM MgCl2, 15 U of TIM, and 3 μg of DXPase-A, and the reactions were quenched by heating at 80°C and treated with SAP as described in Materials and Methods. The initial velocities were determined from the slope of a plot of time versus product for each concentration of GAP and pyruvate (data not shown).
DISCUSSION
Bacteria typically require isoprenoid compounds for synthesis of respiratory quinones, synthesis of cell walls, and modification of tRNAs. Photosynthetic bacteria also require chlorophyll for light harvesting and carotenoids for protection against UV radiation (1). Related photosynthesis compounds are produced in plant plastids, along with a wide variety of other monoterpenes (11) and diterpenes (9). Bacterial isoprenoids are synthesized by the MEP pathway in the species examined to date, except for a strain of Streptomyces (44), which has both the MEP and MVA pathways. Both pathways are also found in plants, where isoprenoid compounds synthesized in the cytosol are derived from MVA and those produced in plastids are derived from MEP (25, 26). The MEP and MVA pathways converge at IPP, and the reactions in the isoprenoid pathway are similar beyond that point in all organisms.
DXP is a key intermediate in bacterial metabolism. The deoxy sugar is the substrate for the first pathway-specific enzyme in the biosynthesis of thiamine, pyridoxol, and isoprenoids (38). Searches of the E. coli genome have provided no evidence for genes in the MVA pathway, and those in the MEP pathway that have been identified thus far appear to occur as single copies. We constructed a dxs knockout in E. coli that was complemented by wild-type dxs on a temperature-sensitive plasmid. The strain was not viable at the restrictive temperature but was rescued when grown on media supplemented with B vitamins and ME, thus confirming that dxs is an essential single-copy gene for the biosynthesis of isoprenoid compounds. Moreover, when cotransformed with a plasmid carrying the yeast genes necessary to convert MVA to IPP, the knockout strain was viable at the restrictive temperature when grown on MVA. While this experiment demonstrates that there is no special requirement for the MEP pathway in E. coli beyond synthesis of IPP, isoprenoid metabolism in E. coli appears to be more complex. In the MVA pathway, IPP is produced by fragmentation of phosphorylated MVA, and the subsequent isomerization of IPP to DMAPP is mandatory. However, when Hahn et al. disabled idi, the gene encoding IPP isomerase in E. coli, the resulting strain was viable (12). A careful search of E. coli databases using several different probe sequences did not retrieve any additional E. coli idi genes. These results are most easily explained if one assumes that the MEP pathway in E. coli produces both IPP and DMAPP and that IPP isomerase performs the nonessential role of balancing the pool of the two metabolites as they are converted to respiratory quinones and dolichols. Recent labeling studies for E. coli with intact cells (7) and with purified enzymes (23, 24) are consistent with this hypothesis.
R. capsulatus encodes all of the enzymes needed to synthesize photosynthetic isoprenoids from IPP in a 46-kb cluster. As observed for E. coli, disruption of the copy of idi located in the photosynthesis cluster of R. capsulatus is not lethal (4). A Southern analysis of R. capsulatus genomic DNA has provided evidence for a second gene for IPP isomerase (13). In addition, a more recent search of newly deposited DNA sequences using idi in the photosynthesis gene cluster as a probe revealed a second gene carried by ORF 735, which is located at a site remote from the cluster. Thus, the photosynthetic bacterium appears to have two genes that encode IPP isomerase, idiA in the photosynthesis cluster and idiB located elsewhere in the chromosome (13). We do not know if, like E. coli, R. capsulatus can survive without a functional copy of idi.
We initially identified the ORF for DXPase-A in the R. capsulatus photosynthesis cluster from the substantial similarity between regions in the encoded protein and those in pyruvate decarboxylases and transketolases, enzymes that catalyze similar reactions needed for synthesis of DXP from pyruvate and GAP. An ORF (ORF 2895) for a second synthase, DXPase-B, was subsequently identified from R. capsulatus after additional sequences had been placed in the database. Interestingly, the genes encoding the enzymes in the MEP pathway between DXPase and IPP isomerase that have been thus far characterized do not appear to be located in the photosynthesis cluster. For example, the putative gene for DXP reductoisomerase, the next enzyme in the pathway, is found on the 690-kb contig 1A01-1C09 located elsewhere in the chromosome. Like the second idi gene, the gene for DXPase-B is not in the photosynthesis cluster.
The DXPases represent a new class of TPP-dependent enzymes whose chemical properties combine traits of pyruvate decarboxylases and transketolases. They first catalyze a TPP-dependent decarboxylation of pyruvate, by analogy to pyruvate decarboxylases. The enzyme-bound thiamine-stabilized acetyl anion is then transferred to the aldehyde moiety in GAP in a reaction similar to the related transfer of the glycolyl anion by transketolases. A comparison of amino acid sequences reveals conserved elements of both pyruvate decarboxylases and transketolases in DXPases. Multiple sequence alignments show significant homology in the N-terminal region of R. capsulatus DXPases to a region in the γ subunit of the E1 component of pyruvate dehydrogenases and homology between sequences from the C-terminal region of DXPase with a region in the β subunit of the dehydrogenases (15). The homology to transketolases is even more substantial. Identical residues are distributed throughout the entire polypeptide chains of DXPase-A, DXPase-B, and transketolases, including many specifically identified as being important for binding and catalysis in transketolases (42). In particular, a 36-amino-acid sequence in the DXPases, beginning with I420 in DXPase-A, aligns with the highly conserved “transketolase motif” identified by Schenk et al. (41). However the conserved THDS four-residue block at the beginning of the 36-amino-acid motif in transketolases is replaced by a DRAG sequence in DXPases. The only exception to the DRAG signature found to date is a closely related GRSG block seen in the putative DXPase from Plasmodium falciparum.
The biochemical properties of the R. capsulatus DXPases are similar to those reported for the E. coli (27, 46) and Mentha × piperita (21) proteins. Although pyruvate and GAP are preferred substrates, the enzymes synthesize 1-deoxy-d-xylulose when incubated with pyruvate and d-glyceraldehyde. The major difference in the catalytic efficiencies of the two reactions results from a Km for glyceraldehyde that is approximately 100-fold higher than the Km for GAP. Similar activity has been reported for sugars involved in transketolization reactions (49). DXPase-A appears to be a homodimer. Given the substantial degree of similarity among DXPases, we anticipate that the other enzymes are homodimers as well.
In summary, isoprenoid compounds are synthesized from 1-deoxy-d-xylulose 5-phosphate in plant chloroplasts and most bacteria by the MEP pathway. In E. coli, DXPase is encoded by dxs, an essential single-copy gene. A disruption in dxs can be complemented chemically by ME, which is presumably phosphorylated and then processed normally by the MEP pathway. The disruption can also be complemented by mevalonate when the yeast genes required to convert MVA to IPP are present on a plasmid. In contrast to E. coli, R. capsulatus has two dxs genes. dxsA is located in the photosynthesis cluster, along with all of the other genes required to synthesize isoprenoid compounds necessary for photosynthesis, while dxsB is located elsewhere in the chromosome. A similar redundancy was found in R. capsulatus for idi, the gene encoding IPP isomerase. Interestingly the genes encoding other enzymes in the MEP pathway are apparently not located in the photosynthesis cluster. The occurrence of two genes for DXPase and for IPP isomerase, one in the photosynthesis cluster and one elsewhere, raise interesting questions about regulation of the biosynthesis of isoprenoids during aerobic and photosynthetic anaerobic growth.
ACKNOWLEDGMENTS
F.M.H. and L.M.E. contributed equally to this work.
This work was supported by NIH grant GM 255211. B.S.J.B. and L.M.E. are recipients of NIH Predoctoral Trainee grant GM08573.
REFERENCES
- 1.Armstrong G A, Alberti M, Leach F, Hearst J E. Nucleotide sequence, organization, and nature of the protein products of the carotenoid biosynthesis gene cluster of Rhodobacter capsulatus. Mol Gen Genet. 1989;216:254–268. doi: 10.1007/BF00334364. [DOI] [PubMed] [Google Scholar]
- 2.Barker R, Swenson C A. Proportion of keto and aldehydo forms in solutions of sugars and sugar phosphates. Biochemistry. 1971;10:3151–3154. doi: 10.1021/bi00792a026. [DOI] [PubMed] [Google Scholar]
- 3.Blagg B S J, Poulter C D. Synthesis of 1-deoxy-d-xylulose and 1- deoxy-d-xylulose-5-phosphate. J Org Chem. 1999;64:1508–1511. doi: 10.1021/jo981966k. [DOI] [PubMed] [Google Scholar]
- 4.Bollivar D W, Suzuki J Y, Beatty J T, Dobrowolski J M, Bauer C E. Directed mutational analysis of bacteriochlorophyll a biosynthesis in Rhodobacter capsulatus. J Mol Biol. 1994;237:622–640. doi: 10.1006/jmbi.1994.1260. [DOI] [PubMed] [Google Scholar]
- 5.Bouvier F, d'Harlingue A, Suire C, Backhaus R A, Camara B. Dedicated roles of plastid transketolases during the early onset of isoprenoid biogenesis in pepper fruits. Plant Physiol. 1998;117:14323–14331. doi: 10.1104/pp.117.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Charon L, Hoeffler J-F, Pale-Grosdemange C, Lois L-M, Campos N, Boronat A, Rhomer M. Deuterium-labelled isotopomers of 2-C-methyl-d-erythritol as tools for the elucidation of the 2-C-methyl-d-erythritol 4-phosphate pathway for isoprenoid biosynthesis. Biochem J. 2000;346:737–742. [PMC free article] [PubMed] [Google Scholar]
- 8.Duvold T, Cali P, Bravo J-M, Rohmer M. Incorporation of 2-C-methyl-d-erythritol, a putative isoprenoid precursor in the mevalonate-independent pathway, into ubiquinone and menaquinone of Escherichia coli. Tetrahedron Lett. 1997;38:6181–6184. [Google Scholar]
- 9.Eisenreich W, Menhard B, Hylands P J, Zenk M H, Bacher A. Studies on the biosynthesis of taxol: the taxane carbon skeleton is not of mevalonoid origin. Proc Natl Acad Sci USA. 1996;93:6431–6436. doi: 10.1073/pnas.93.13.6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisenreich W, Schwarz M, Cartayrade A, Arigoni D, Zenk M H, Bacher A. The deoxyxylulose phosphate pathway of terpenoid biosynthesis in plants and microorganisms. Chem Biol. 1998;5:R221–R233. doi: 10.1016/s1074-5521(98)90002-3. [DOI] [PubMed] [Google Scholar]
- 11.Eisenreich W, Sagner S, Zenk M H, Bacher A. Monoterpenoid essential oils are not mevalonoid origin. Tetrahedron Lett. 1997;38:3889–3892. [Google Scholar]
- 12.Hahn F M, Hulbert A P, Poulter C D. Escherichia coli open reading frame 696 is idi, a nonessential gene encoding isopentenyl diphosphate isomerase. J Bacteriol. 1999;181:4499–4504. doi: 10.1128/jb.181.15.4499-4504.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn F M, Baker J A, Poulter C D. Open reading frame 176 in the photosynthesis gene cluster of Rhodobacter capsulatus encodes idi, a gene for isopentenyl diphosphate isomerase. J Bacteriol. 1996;178:619–624. doi: 10.1128/jb.178.3.619-624.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hawkins C F, Borges A, Perham R N. A common structural motif in thiamin pyrophosphate-binding enzymes. FEBS Lett. 1989;255:77–82. doi: 10.1016/0014-5793(89)81064-6. [DOI] [PubMed] [Google Scholar]
- 16.Herz S, Wungsintaweekul J, Schuhr C A, Hecht S, Lüttgen H, Sagner S, Fellermeier M, Eisenreich W, Zenk M H, Bacher A, Rohdich F. Biosynthesis of terpenoids: YgbB protein converts 4-diphosphocytidyl-2C-methyl-d-erythritol 2-phosphate to 2C-methyl-d-erythritol 2,4-cyclodiphosphate. Proc Natl Acad Sci USA. 2000;97:2486–2490. doi: 10.1073/pnas.040554697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotype selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuzuyama T, Takagi M, Takahashi S, Seto H. Cloning and characterization of 1-deoxy-d-xylulose 5-phosphate synthase from Streptomyces sp. strain CL190, which uses both the mevalonate and nonmevalonate pathways for isopentyl diphosphate biosynthesis. J Bacteriol. 2000;182:891–897. doi: 10.1128/jb.182.4.891-897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuzuyama T, Takahashi S, Watanabe H, Seto H. Direct formation of 2-C-methyl-d-erythritol 4-phosphate from 1-deoxy-d-xylulose 5-phosphate by 1-deoxy-d-xylulose 5-phosphate reductoisomerase, a new enzyme in the non-mevalonate pathway to isopentenyl diphosphate. Tetrahedron Lett. 1998;39:4509–4512. [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lange B M, Wildung M R, McCaskill D, Croteau R. A family of transketolases that directs isoprenoid biosynthesis via a mevalonate-independent pathway. Proc Natl Acad Sci USA. 1998;95:2100–2104. doi: 10.1073/pnas.95.5.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leatherbarrow R J. Grafit version 3.01. Staines, England: Erithicus Software Ltd.; 1992. [Google Scholar]
- 23.Leyes A E, Baker J A, Poulter C D. Biosynthesis of isoprenoids in Escherichia coli: stereochemistry of the reaction catalyzed by farnesyl diphosphate synthase. Org Lett. 1999;1:1071–1073. doi: 10.1021/ol990876e. [DOI] [PubMed] [Google Scholar]
- 24.Leyes A E, Baker J A, Hahn F M, Poulter C D. Biosynthesis of isoprenoids in Escherichia coli: stereochemistry of the reaction catalyzed by isopentenyl diphosphate: dimethylallyl diphosphate isomerase. J Chem Soc Chem Commun. 1999;1999:717–718. [Google Scholar]
- 25.Lichtenthaler H K, Schwender J, Disch A, Rohmer M. Biosynthesis of isoprenoids in higher plant chloroplasts proceeds via a mevalonate-independent pathway. FEBS Lett. 1997;400:271–274. doi: 10.1016/s0014-5793(96)01404-4. [DOI] [PubMed] [Google Scholar]
- 26.Lichtenthaler H K, Rohmer M, Scwender J. Two independent biochemical pathways for isopentenyl diphosphate and isoprenoid biosynthesis in higher plants. Physiol Plant. 1997;101:643–652. [Google Scholar]
- 27.Lois L M, Campos N, Putra S R, Danielsen K, Rohmer M, Boronat A. Cloning and characterization of a gene from Escherichia coli encoding a transketolase-like enzyme that catalyzes the synthesis of d-1-deoxyxylulose 5-phosphate, a common precursor for isoprenoid, thiamin and pyridoxol biosynthesis. Proc Natl Acad Sci USA. 1998;95:2105–2110. doi: 10.1073/pnas.95.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lüttgen H, Rohdich F, Herz S, Wungsintaweekul J, Hecht S, Schuhr C A, Fellermeier M, Sagner S, Zenk M H, Bacher A, Eisenreich W. Biosynthesis of terpenoids: YchB protein of Escherichia coli phosphorylates the 2-hydroxy group of 4-diphosphocytidyl-2C-methyl-d-erythritol. Proc Natl Acad Sci USA. 2000;97:1062–1067. doi: 10.1073/pnas.97.3.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayer M P, Prestwich G P, Dolence J M, Bond P D, Wu H Y, Poulter C D. Protein farnesyltransferase: production in Escherichia coli and immunoaffinity purification of the heterodimer from Saccharomyces cerevisiae. Gene. 1998;132:41–47. doi: 10.1016/0378-1119(93)90512-2. [DOI] [PubMed] [Google Scholar]
- 30.Nilsson U, Hecquet L, Gefflaut T, Guerard C, Scheider G. Asp477 is a determinant of the enantioselectivity in yeast transketolase. FEBS Lett. 1998;424:49–52. doi: 10.1016/s0014-5793(98)00136-7. [DOI] [PubMed] [Google Scholar]
- 31.Nilsson U, Meshalkina L, Lindqvist Y, Schneider G. Examination of substrate binding in thiamin diphosphate-dependent transketolase by protein crystallography and site-directed mutagenesis. J Biol Chem. 1997;272:1864–1869. doi: 10.1074/jbc.272.3.1864. [DOI] [PubMed] [Google Scholar]
- 32.Putra S R, Disch A, Bravo J-M, Rohmer M. Distribution of mevalonate and glyceraldehyde 3-phosphate/pyruvate routes for isoprenoid biosynthesis in some Gram-negative bacteria and mycobacteria. FEMS Microbiol Lett. 1998;164:169–175. doi: 10.1111/j.1574-6968.1998.tb13082.x. [DOI] [PubMed] [Google Scholar]
- 33.Qureshi N, Porter J W. In: Biosynthesis of isoprenoid compounds. Porter J W, Spurgeon S L, editors. New York, N.Y: John Wiley & Sons, Inc.; 1981. [Google Scholar]
- 34.Reynen M, Sahm H. Comparison of the structural genes for pyruvate decarboxylase in different Zymomonas mobilis strains. J Bacteriol. 1988;170:3310–3313. doi: 10.1128/jb.170.7.3310-3313.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohdich F, Wungsintaweekul J, Fellermeier M, Sagner S, Herz S, Kis K, Eisinreich W, Bacher A, Zenk M H. Cytidine 5′-triphosphate-dependent biosynthesis of isoprenoids: YgbB protein of Escherichia coli catalyzes the formation of 4-diphosphocytidyl-2-C-methylerythritol. Proc Natl Acad Sci USA. 1999;96:11758–11763. doi: 10.1073/pnas.96.21.11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohmer M. Isoprenoid biosynthesis via the mevalonate-independent route, a novel target for antibacterial drugs? Prog Drug Res. 1998;50:135–154. doi: 10.1007/978-3-0348-8833-2_3. [DOI] [PubMed] [Google Scholar]
- 37.Rohmer M, Knani M, Simonin P, Sutter B, Sahm H. Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem J. 1993;295:517–524. doi: 10.1042/bj2950517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohmer M, Seeman M, Horbach S, Bringer-Meyer S, Sahm H. Glyceraldehyde 3-phosphate and pyruvate as precursors of isoprenic units in an alternative non-mevalonate pathway for terpenoid biosynthesis. J Am Chem Soc. 1996;118:2564–2566. [Google Scholar]
- 39.Saccettini J C, Poulter C D. Creating isoprenoid diversity. Science. 1997;277:1788–1789. doi: 10.1126/science.277.5333.1788. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Schenk G, Layfield R, Candy J M, Duggleby R G, Nixon P F. Molecular evolutionary analysis of the thiamine-diphosphate-dependent enzyme, transketolase. J Mol Evol. 1997;44:552–572. doi: 10.1007/pl00006179. [DOI] [PubMed] [Google Scholar]
- 42.Schneider G, Lindqvist Y. Crystallography and mutagenesis of transketolase: mechanistic implications for enzymatic thiamin catalysis. Biochem Biophys Acta. 1998;1385:387–398. doi: 10.1016/s0167-4838(98)00082-x. [DOI] [PubMed] [Google Scholar]
- 43.Schowen R L. Thiamin-dependent enzymes. In: Sinnot M, editor. Comprehensive biological catalysis. Vol. 2. New York, N.Y: Academic Press, Inc.; 1998. pp. 217–266. [Google Scholar]
- 44.Seto H, Watanabe H, Furihata K. Simultaneous operation of the mevalonate and non-mevalonate pathways in the biosynthesis of isopentenyl diphosphate in Streptomyces aeriouvifer. Tetrahedron Lett. 1996;37:7979–7982. [Google Scholar]
- 45.Sprenger G A, Schörken U, Sprenger G, Sahm H. Transketolase A of Escherichia coli K12: purification and properties of the enzyme from recombinant strains. Eur J Biochem. 1995;230:525–532. doi: 10.1111/j.1432-1033.1995.0525h.x. [DOI] [PubMed] [Google Scholar]
- 46.Sprenger G A, Schörken U, Wiegert T, Grolle S, de Graaf A A, Taylor S V, Begley T P, Bringer-Meyer S, Sahm H. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-d-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc Natl Acad Sci USA. 1997;94:12857–12862. doi: 10.1073/pnas.94.24.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi S, Kuzuyama T, Watanabe H, Seto H. A 1-deoxy-d-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-d-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc Natl Acad Sci USA. 1998;95:9879–9884. doi: 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor S V, Vu L D, Begley T P. Chemical and enzymatic synthesis of 1-deoxy-d-xylulose-5-phosphate. J Org Chem. 1998;63:2375–2377. [Google Scholar]
- 49.Usmanov R A, Kochetov G. Function of the arginine residue in the active center of baker's yeast transketolase. Biochemistry (Engl Transl Biokhimiia) 1983;48:772–781. [PubMed] [Google Scholar]
- 50.Veech R L, Rajman L, Dalziel K, Krebs H H. Disequilibrium in the triose phosphate isomerase system in rat liver. Biochem J. 1969;115:837–842. doi: 10.1042/bj1150837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 52.Wikner C, Meshalkina L, Nilsson U, Nikkola M, Lindqvist Y, Schneider G. Analysis of an invariant cofactor-protein interaction in thiamin diphosphate-dependent enzymes by site-directed mutagenesis. J Biol Chem. 1994;269:32144–32150. [PubMed] [Google Scholar]
- 53.Wikner C, Nilsson U, Meshalkina L, Udekwu C, Lindqvist Y, Schneider G. Identification of catalytically important residues in yeast transketolase. Biochemistry. 1997;36:15643–15649. doi: 10.1021/bi971606b. [DOI] [PubMed] [Google Scholar]