Abstract
The germline cells are essential for the propagation of human beings, thus essential for the survival of mankind. The germline stem cells, as a unique cell type, generate various states of germ stem cells and then differentiate into specialized cells, spermatozoa and ova, for producing offspring, while self-renew to generate more stem cells. Abnormal development of germline stem cells often causes severe diseases in humans, including infertility and cancer. Primordial germ cells (PGCs) first emerge during early embryonic development, migrate into the gentile ridge, and then join in the formation of gonads. In males, they differentiate into spermatogonial stem cells, which give rise to spermatozoa via meiosis from the onset of puberty, while in females, the female germline stem cells (FGSCs) retain stemness in the ovary and initiate meiosis to generate oocytes. Primordial germ cell-like cells (PGCLCs) can be induced in vitro from embryonic stem cells or induced pluripotent stem cells. In this review, we focus on current advances in these embryonic and adult germline stem cells, and the induced PGCLCs in humans, provide an overview of molecular mechanisms underlying the development and differentiation of the germline stem cells and outline their physiological functions, pathological implications, and clinical applications.
Subject terms: Cell biology, Molecular medicine, Pluripotent stem cells
Introduction
In mammals, an organism consists mainly of two cell types, somatic cells, and germ cells. Based on the concept of the germline proposed by early biologist August Weismann,1 the somatic cells die along with the individual, in contrast, the germ cells can pass both genetic and epigenetic information from one generation to the next. The germline is a lineage of cells in an organism from which both oocytes and sperm cells arise, which is thus essential for the propagation of species. As such, abnormal development of the germline cells will lead to severe diseases in humans, including infertility and cancer. For example, the incidence rate of ovarian cancer is 11.5 per 100,000 women during 2010–2014,2 there are 22,530 new cases in 2019,3 and an estimated 19,880 people will be diagnosed with ovarian cancer in the United State.4 Ovarian cancers are also among incidences of top 10 cancers for females, with 50,000 cases in China.5 In females, an increasing prevalence of disease type is ovarian dysfunction, which includes altered frequency, and duration of the menstrual cycle,6 with or without premature ovarian failure or polycystic ovary syndrome. Testicular germ cell cancers account for ~1% of all solid cancers in Caucasian males, in particular, 60% are diagnosed in adolescents and young adults.7–9 In addition, infertility is common and affects around 8–17% of reproductive-aged couples worldwide.6,10,11 Thus, the development and regulation of germline cells play a fundamental role in survival, health, and disease in humans.
During early embryonic development, first emerged germline cells are called primordial germ cells (PGCs).12 The PGCs are the founder cells of the germline, to some extent, also the source of germline totipotency, ensuring the creation of new organisms.13 In humans, when, where, and how first PGCs are specified within an early embryo remain a central challenge. There are probably two sets of constraints for this: One is ethical and technical limitations in obtaining and manipulating human (h) PGCs (hPGCs) from early embryos at the peri-implantation stage, and the other is differences in PGC development among species for comparative studies. While animal models have provided instructive knowledge for hPGCs, cell and molecular mechanisms underlying PGCs specification observed in model animals are generally incapable of recapitulating essential points of those in humans.
In humans, early investigations by light microscopy found that hPGCs have a large size with a large nucleus and prominent nucleolus in the yolk-sac endoderm of embryonic day 24 (E24), which migrated into the developing genital ridges at E28.14 Witschi suggested that the hPGCs migrated within the embryo by active movements,14 which was confirmed by a time-lapse analysis of living mouse (m) PGCs (mPGCs) migration 50 years later.15 However, passive movement associated with morphogenesis has also been observed.16 Fine morphology, migration, and origin of hPGCs were then observed by transmission electron microscopy, which was characterized by the presence of abundant glycogen particles and lipid droplets in the cytoplasm.17 This observation has metabolic implications for hPGCs, as the term glycolysis is often used to describe stemness.18 Alkaline phosphatase activity was also observed on the plasma membrane of the hPGCs, indicating a characteristic marker of hPGCs.17,19 To overcome technical limitations in manipulating early hPGCs in vivo, approaches of in vitro induction of hPGCs have been established from embryonic stem cells and induced pluripotent stem cells.20–22 These induced hPGCs are called hPGC-Like Cells (hPGCLCs), which are a bona fide in vitro counterpart of hPGCs.23 Lineage trajectory and mechanistic insights of hPGCs specification using the hPGCLCs induction system have been further clarified by means of single-cell transcriptomics and cell lineage tracing.24 Recently, the hPGCLCs have also been used for in vitro gametogenesis,23 which has important implications in reproductive medicine.
As precursors of the gametes, hPGCs continue to divide mitotically when arriving at genital ridges. In the following processes of gonad development, hPGCs will go through a distinct process of development depending on their sex chromosome composition (XX/XY) in embryos. In female embryos (XX), some hPGCs enter into the meiotic division phase and subsequently differentiate into oocytes in the ovary, thus ending their stem cell potential, while the others keep stemness and become FGSCs.25 In contrast to those in the female, male hPGCs (XY) enter the seminiferous cords, become gonocytes, and are arrested in G0/G1 phase of cell cycles until birth.26–28 In neonatal testis, the gonocytes resume to divide and differentiate into spermatogonial stem cells (SSCs), which then give rise to spermatozoa via meiosis from the onset of puberty. Manipulation and transplantation of the SSCs provide a powerful system to study stem cell biology, preserve individual genomes, modify germ lines and treat male infertility.27,29,30 For example, SSCs transplantation can recover male fertility when the SSCs of patients are damaged upon irradiation or chemotherapy in cancer treatment.
In mammals, it has long been believed that the total number of ovarian follicles is determined during the perinatal period, and production of ovarian oocytes is thought to stop in adult female.31–33 However, accumulating evidence shows that there are female germline stem cells (FGSCs) in ovaries in mice and humans, which are able to undergo postnatal neo-oogenesis.25,34–36 The newly found FGSCs provide an alternative way to investigate the development of germline stem cells by oogenesis, not just by spermatogenesis. More importantly, FGSCs have important clinical implications, for example, in the expansion of the follicle reserve for fertility preservation and treatment of infertility and premature ovarian failure.
Given the above historical and developmental overview (Fig. 1), the germline stem cell (GSC) is a unique cell type that produces more stem cells via self-renewal or different states/subtypes of stem cells during germline development, and finally differentiate into specialized cells, spermatozoa and ova, for producing offspring. In mammals, the GSCs mainly include (1) primordial germ cells (PGCs) from embryos, being embryonic pluripotent stem cells, (2) induced PGC-like cells (PGCLCs) from embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs), (3) spermatogonial stem cells (SSCs), and (4) female germline stem cells (FGSCs). Both SSCs and FGSCs belong to adult pluripotent stem cells (ASCs). Here, we summarize these germline stem cells in humans, provide an overview of molecular mechanisms underlying GSC development and disease, and outline their physiological functions, pathological implications, and potential clinical applications.
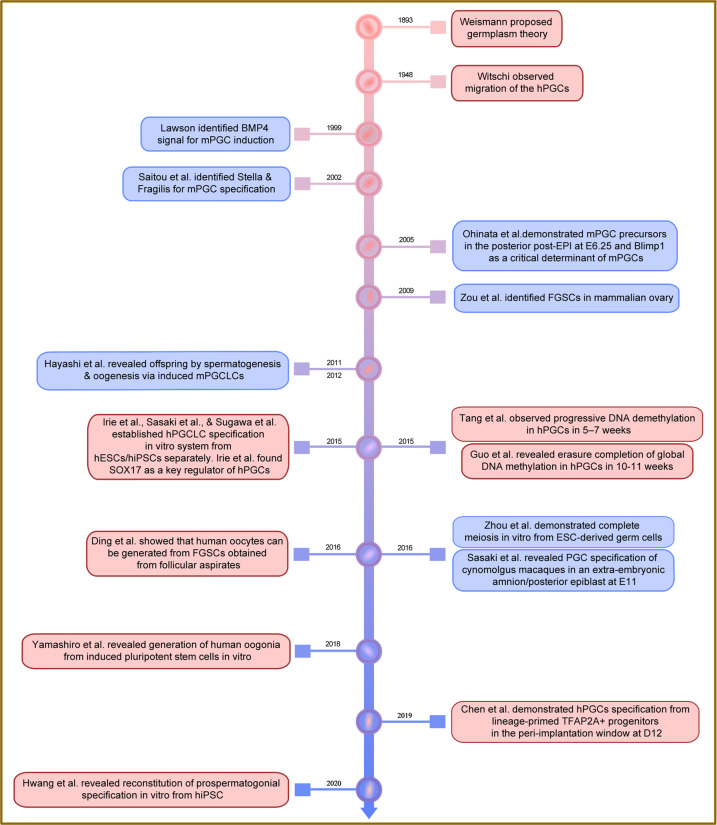
Fig. 1.
History and main events of the studies in human germline stem cells. A glance of the discovery and advances starts from 1893 and the most of advances in human germline stem cells have been made since 2015
Precursors for the gametes: human primordial germ cells
Origin and specification of hPGCs
Development of human early embryos
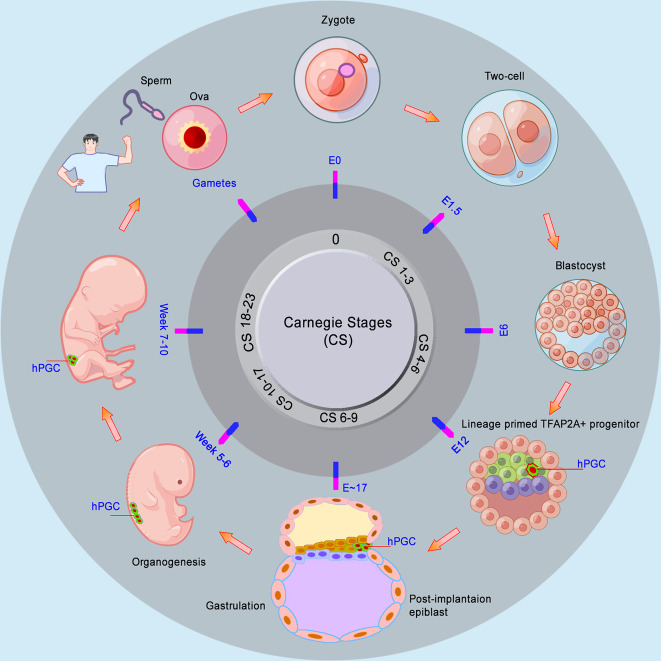
Primordial germ cells are early embryonic pluripotent stem cells. Embryonic development begins after fertilization. Eggs support the following embryonic development until the new organism can feed on most animals, including zebrafish, frogs, and chickens. In contrast, in mammals, early embryos must obtain essential nourishment support from the mother by implantation into the uterus. In humans, implantation occurs at the end of the blastocyst stage during embryonic day E7–E8.37,38 The inner cell mass splits into the hypoblast, which forms the yolk sac, and the epiblast, which generates the embryo properly. The post-implanted embryo in humans is flat with a bilaminar germ disc, consisting of the epiblast and hypoblast, in addition to the trophoblast, whereas it is cylindrical in mice.39,40 The following formation of the primitive streak begins in the posterior part of the embryonic epiblast at E14, which means the start of gastrulation, and the cells gradually lose their pluripotency. Then the epiblast cells move through the primitive streak to give rise to mesoderm and endoderm. At the end of gastrulation, the other epiblast cells become ectoderm.38,39 The formation of the three germ layers starts the subsequent organogenesis of embryos. The timing of human embryo development is often referred to as Carnegie stages (CS), based on the appearance of morphological structures rather than time.41 Nevertheless, the CS can loosely be corresponding to days after fertilization or embryonic day.37,38,42–44 Based on this, the timing of hPGC development is determined (Fig. 2).
Fig. 2.
Development of human embryos and the timing of hPGC specification. Human early embryos must obtain essential nourishment from mother by implantation into uterus, which occurs at the end of blastocyst stage. The Carnegie stages (CS) can be corresponding to days after fertilization or embryonic day (E). hPGCs are indicated in green (cytoplasm) and red (nucleus). The main parts of the figure were drawn by Figdraw
Origin of hPGCs
Primordial germ cells are the first population of germ cells established during early embryo development in animals,12 yet the origin of PGCs differs among animals. Single-cell transcriptome shows that the germline of Xenopus is evolutionarily closer to that of zebrafish than to humans and mice.45 In some model organisms, including Xenopus,46–58 zebrafish,59–69 and chicken,70–82 their PGCs are specified via the maternally inherited germ plasm, including vasa and nanos, during the first several cleavages. In contrast, in mammals, the PGCs are specified by induction during early embryo development. In mice, the PGCs were observed at the base of the incipient allantois in the extraembryonic mesoderm (ExM) at E7.25.12,83,84 Lineage tracing shows that the mPGC precursors with Blimp1+ were detected in the posterior post-epiblast (EPI) at E6.25, indicating the origin of the mPGCs from the posterior post-EPI in mice.85,86 In cynomolgus monkeys, cyPGCs are specified in the early amnion at E11 prior to gastrulation.87 Nevertheless, it has been a long journey to determine the origin of hPGCs in human embryos. Early observations by histology and microscopy of human embryos showed the hPGCs identifiable as early as E24.14,17,19 Recently, the origin of hPGCs was accurately determined by single-cell RNA sequencing and lineage trajectory mapping of the hPGCLC. The study shows that hPGCs were specified beginning at E12 from lineage-primed TFAP2A+ progenitors, which share characteristics with pre- and postimplantation epiblasts24 (Fig. 2). In addition, rabbit PGCs show a similar developmental pattern and mechanism with hPGCs, including bilaminar-disc embryos, PGC origin of epiblasts, and SOX17 as a key regulator of PGCs,88 suggesting a valuable model for studies of human PGC development.
Regulation of hPGC specification
Specification genes of hPGCs
Knowledge about how PGCs are induced comes largely from studies in model mammals, including nonhuman primates,86,87,89–95 mice,12,83,85,96–108 and pigs.109–118 Studies in the other group of model organisms, including chicken,119–126 zebrafish,63,64,66,67,127 medaka,128–136 frogs,55–57,137 Drosophila,138–146 and Caenorhabditis elegans,147–156 provide abundant information about PGC specification by preformation or maternally inherited determinants. The PGCs in these two groups of organisms show a similarity that is the presence of some kind of aggregate of electron-dense, basophilic bodies containing the proteins and RNAs, (e.g., VASA, NANOS, and MAGO) in their cytoplasm.70 Nevertheless, even between mice and humans, accumulated evidence shows that there are differences in cell and molecular mechanisms underlying PGC specification, especially, in expressed genes and signaling pathways.24,86,157 For example, SOX17 is critical for hPGC specification, but not for mPGC induction.20 SOX2 is expressed in mPGCs, but not in hPGCs.158 In humans, SOX2 exerts its roles mainly in adult tissues and cancers through regulating self-renewal and stemness of cancer stem cells.159 In recent years, studies in nonhuman primates provide some information for hPGC specification.87,89,91,95,160
In PGCs of humans and nonhuman primates, a core group of primate-specific PGC markers is expressed, including SOX17, BLIMP1, TFAP2C, NANOG, and POU5F1, but the absence of SOX2,87,90,95,160,161 suggesting that these factors are conserved and are important for PGC specification in primates. Nevertheless, it is perhaps different from the origin of hPGCs, that PGCs in cynomolgus monkey might emerge in amnion.87,95 In general, hPGCs/hPGCLCs express a range of types of key genes, including (1) pluripotency markers: NANOG, POU5F1, ALPL, KLF4, LIN28, KIT, NANOS3, SSEA-1, SSEA-4, DPPA3 (also called as STELLA), and ZFP42 (also known as REX1),17,20,21,161–167 (2) cell-surface makers: CD38, EPCAM, ITGA6 (INTEGRIN alpha 6), ITGB3, FGFR3, KIT, and ALPL,20,21,167,168 (3) germline markers: SOX17, BLIMP1 (also known as PRDM1), TFAP2C, PRDM14, DDX4 (also known as VASA), DAZL, and TCL1A,20,21,167,169–172 (4) amnion-related genes: CDX2 and GATA3,24 (5) mesoderm markers: EOMES, NODAL, SP5, and T,20,21 (6) transcription factors: SOX17, BLIMP1, SOX15, GATA4, PRDM14, SALL4, and UTF1,20,21,172–175 and (7) epigenetic regulation factors: DNA demethylation dioxygenases (TET1, TET2, and TET3), protein arginine methyltransferase 5(PRMT5), and DND microRNA-mediated repression inhibitor 1(DND1).20,167,176,177 The types and expression patterns of these marker genes reflect corresponding states of hPGC development and cell identity. For example, expression levels of pluripotency genes gradually decrease in hPGCs from embryos of 4–19 weeks,167 indicating a slow loss of pluripotency. Thus, not all these types of genes are expressed in a certain state of hPGC at a certain developmental time. Of course, there are other factors important for hPGC development, that remain to be identified. For example, TRIM71, an E3 ubiquitin ligase, is associated with the proliferation of hPGC-like TCam-2 cells.178 NOD-like receptor Nlrp14 knockout inhibits SSC differentiation in mice.179
Signaling pathways and regulations for hPGC specification
BMP-SMAD signaling
The bone morphogenetic proteins (BMP) are members of the transforming growth factor-beta superfamily and play important roles in embryo development.180 Further studies in knockout mice reveal that BMP proteins are required for PGC induction.96,107 Some upstream factors that regulate BMP expression play important roles in PGC formation, for example, LncBMP4, a long noncoding RNA that targets BMP4, has similar functions as BMP4.119 BMP proteins originate from the extraembryonic tissues but exert their roles through their receptors (BMPRs) on the membrane of epiblast cells. Upon binding to BMPRs that phosphorylate intracellular signaling molecules SMAD1/SMAD5 in the cytoplasm, the activated SMAD1/SMAD5 dimerize with SMAD4, translocate into the nucleus, and regulate the key transcriptional regulators of PGCs.98,181 In mice, BMP4 induces PGCs with an expression of both BLIMP1 and PRDM14 in the epiblast, which can be induced to generate functional sperm cells in vivo by gonad reconstruction and seminiferous tubule injection.13 Bmp4 homozygous KO embryos lack PGC development, demonstrating a key role of BMP4 in PGC induction in vivo.96 The authors also showed that the response of epiblast cells to BMP is dose-dependent during PGC induction. Further analysis of the roles of intracellular signaling molecules in PGC induction indicates that SMAD1 signaling is critical for the initial commitment of PGCs, as evidenced by the fact that the knockout of Smad1 led to the complete absence of PGCs in mice.98 In cultured epiblast cells, BMP4 is also sufficient to induce PGCs in a dose-responsive manner.13 In a culture of hPGCs from fetal gonads at 8–11 weeks, the addition of BMP4 increases the number of hPGCs in a dose-responsive manner, whereas the addition of an antagonist of the BMP4 pathway decreases PGC proliferation.182 WNT3 is expressed in the epiblast at around E5.5 and this ensures its responsiveness to BMP4 signalling.13,183 In addition, activin A induces the expression of OCT4, NANOG, NODAL, WNT3, bFGF, and FGF8, and suppresses the BMP signaling,184 but it shows high competence to differentiate to hPGCLCs when transiently converted to the 4i-state prior to differentiation in culture,185 thus induces an increased differentiation potential of germ cells.186 These studies clearly suggest that BMP-SMAD signaling is key and indispensable for PGC specification in mammals. Thus, in the following studies of hPGCLCs induction in vitro, BMP4 is widely used as an essential factor.20,21 Upstream regulation of BMP4 will be another layer for hPGC specification. In nonhuman primates, transcription factor ISL1 acts upstream of BMP4 and plays an indispensable for amnion formation.187 Given amnion as a signaling center during mesoderm formation, it is possible that ISL1 might function in hPGC specification, which needs to be explored further.
SOX17- BLIMP1
SOX17, a member of the SOX (SRY-related HMG-box) family of transcription factors, is originally identified as a transcription factor for spermatogenesis.188,189 Later studies reveal an important role of SOX17 in endoderm development of the post-implanted embryos in mice, as knockout embryos are deficient in gut endoderm.190 Induction in vitro of hPGCLCs reveals that SOX17 is a key regulator of hPGC fate, loss of SOX17 impairs hPGC specification, and BLIMP1 works downstream of SOX17, which then represses endodermal and somatic genes.20 This pathway works only in hPGCs, but it is not necessary for mPGC fate. The transcription factor EOMES (T-box gene Eomesodermin) functions upstream of SOX17 for hPGCLC specification, which is upregulated in incipient mesoderm-like cells (iMeLCs) and activates SOX17 in response to WNT signaling.191 EOMES knockout impairs hPGCLC differentiation from human embryonic stem cells (hESCs).192 These data suggest an essential role of EOMES for hPGCLC specification. In mice, loss-of-function reveals that Eomes mutants arrest at implantation, suggesting a critical role of EOMES in the specification of the definitive endoderm lineage.193 The mesodermal protein T (TBXT) is a downstream effector of WNT3 signaling and essential for mPGC specification in mice,194 but it is dispensable for hPGCs.191 Further studies revealed that SOX17, TFAP2C, and BLIMP1 are not sufficient to generate hPGCLCs, in contrast, transcription factors GATA3/GATA2 as key BMP effectors, combined with SOX17 and TFAP2C, drive the hPGCLC program.195 Nonetheless, the precise molecular mechanisms involved in the transcription factors during hPGC specification in vivo remain to be explored further.
BLIMP1, also known as PRDM1, encodes a zinc finger transcriptional repressor required for anterior endomesodermal cell fate and head induction196 and can bind directly to repress somatic cell proliferation genes.174 In mice, BLIMP1 is expressed in the most proximal layer of the epiblast at E6.25, BLIMP1-positive cells are lineage-restricted to mPGCs, and in Blimp1 mutants, formation and immigration of mPGC are impaired.85 But, BLIMP1 is dispensable for the derivation and maintenance of ESCs and postimplantation epiblast stem cells.197 In humans, BLIMP1 expression is detected in human fetal gonocytes in 12th week.176 Accumulated evidence shows that BLIMP1 is essential for hPGCLC specification,20–22,171,191 and this function is also conserved in mPGCs.85,198 Mechanistically, BLIMP1 acts downstream of SOX17 to suppress neuron differentiation and both endodermal and mesodermal genes and initiate the transcriptional network of human germ cells, including NANOS3.20,21,195 In the knockout cells of BLIMP1 or SOX17, the gene expression network of hPGC specification is also abrogated, including NANOS3.20 Thus, the SOX17-BLIMP1 axis initiates hPGC program from competent cells upon induction by BMP signaling.20 As a complex and programmed developmental process, hPGC development needs many other genes and a coordinated network. For example, another two transcription factors TFAP2C and PRDM14 play indispensable roles in hPGC specification.
TFAP2C
TFAP2C (also known as AP2-GAMMA) is a sequence-specific DNA-binding transcription factor involved in the activation of several developmental genes. In hESCs, TFAP2C binds to a naive-specific POU5F1 (OCT4) enhancer to maintain pluripotency and repress neuroectodermal differentiation during the transition from primed to naive in preimplantation embryos.199 In humans, TFAP2C functions upstream of SOX17 for germline specification, through binding to SOX17 promoter.24 On both sides of the binding site, there is also the coordinately enriched H3K27ac in hPGCLCs, and this kind of epigenetic regulation of TFAP2C might enable SOX17 expression at the point of hPGCLC specification.24 Through lineage tracking and mapping of the human germline trajectory, Chen and collaborators demonstrated that the TFAP2A-expressing progenitors exhibited the potential for both hPGC specification and amnion/gastrulation development at around day 11, and loss of TFAP2C led to exiting of the germline pathway, but toward differentiation of primitive streak or amnion-like somatic cells.24 One of the mechanisms of the TFAP2C-regulated hPGC formation is through the opening of enhancers proximal to pluripotency factor OCT4.199,200 Thus, these data suggest that TFAP2C plays an essential role in hPGC specification by directly regulating SOX17 expression at the critical point of hPGC specification. Of particular note is that BMP signaling also activates TFAP2C in a SOX17-independent manner, and both SOX17 and TFAP2C act upstream of BLIMP1 in human germ cell specification.191 In mice, the Tfap2c knockout impaired mPGCLC generation from ESCs,201 suggesting a similar role of TFAP2C in the maintenance of the PGC specification. However, there is a difference in regulatory mechanisms of TFAP2C in PGC specification between hPGCs and mPGCs. For example, in mice, TFAP2C is a direct target of BLIMP1, which cooperates with PRDM14 to induce PGC gene expression.174,191 Yet, TFAP2C regulates other cellular processes, including cell cycle (CDKN1A/P21 and CDK6), in addition to germline development (NANOS3 and c-KIT) in mice.201 Together, TFAP2C plays a critical role in PGC specification, but through distinct regulation modes between mice and humans.
PRDM14
As a member of the PRDI-BF1 and RIZ homology domain containing (PRDM) family of transcriptional regulators, PRDM14 is expressed in preimplantation embryos and PGCs in mice and humans.20,22,202,203 In hPGC-competent pluripotent cells, PRDM14 is highly expressed.172 Accumulating evidence shows that PRDM14 plays important roles in the maintenance and induction of pluripotency of stem cells and PGC development in a range of species, including humans,22,172,204,205 mice,174,202,203,206–212 rats,213 and chicken.214 In mice, PRDM14 has critical roles for mPGC specification by upregulation of germline-specific genes, suppression of somatic genes, regulation of global epigenetic reprogramming, for example, maintenance of global DNA hypomethylation, histone modifications, and X-chromosomal reprogramming.174,202,206,212,215–218 In human ES cells, knockdown of PRDM14 induced expression of early differentiation marker genes and suppressed expression of stem cell markers, whereas overexpression of PRDM14 showed a remarkable suppression of the expression of differentiation marker genes, suggesting a role of PRDM14 in the maintenance of pluripotency in human ES cells by suppressing of expression of differentiation genes.204 A genome-wide RNAi screen shows that PRDM14 binds to the proximal enhancer of pluripotency gene POU5F1/OCT4 to regulate its expression in human ES cells, and functional analysis reveals that PRDM14 is also required for reprogramming of fibroblasts to iPSCs.219 Nevertheless, the hPGCLCs induced from hPSCs show a minimal PRDM14 expression, which is different from that observed in mPGCs, suggesting that human PGCs could not require PRDM14, or, alternatively, that low levels of PRDM14 expression is enough for hPGC development.22 Indeed, inducible loss of PRDM14 affects the efficiency of specification and leads to downregulation of hPGC marker genes, including UTF1 and NANOG, and re-expression of PRDM14 rescues hPGCLC differentiation,172 suggesting a critical role of PRDM14 in hPGC fate. Notably, PRDM14 regulates hPGC development probably through coordination with both TFAP2C and BLIMP1, as it shares a subset of transcription targets with TFAP2C and BLIMP1,172 although the exact position of PRDM14 in the regulatory network of hPGC specification remains unknown.
Regulation network of hPGC specification
In response to BMP4, hPGCLCs can be induced from hESCs.20 Transcription factors GATA3 and GATA2 are BMP effectors and promote the hPGCLC specification, together with SOX17 and TFAP2C. BMP signaling could also activate SOX17 and TFAP2C expression, probably independent from GATA3/2.195 SOX17 is a critical regulator of hPGCLCs.20 BLIMP1 is activated by SOX17, suggesting that SOX17 acts upstream of BLIMP1.20,191,195 In addition, BLIMP1 promotes germline transcription and represses the neuronal differentiation program.21
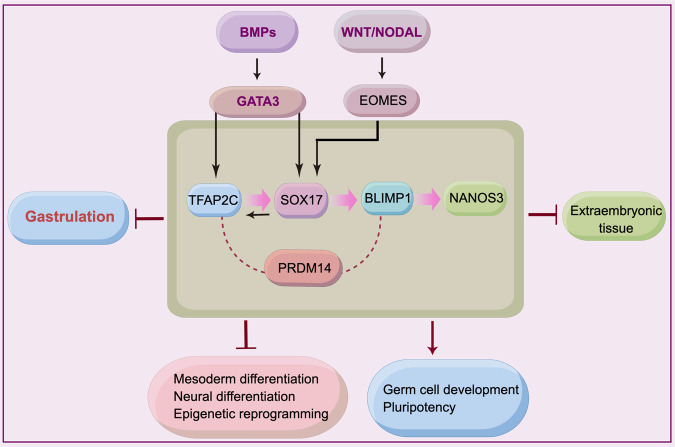
SOX17 and BLIMP1 together are necessary and sufficient for inducing PGCs and initiating the germline-specific epigenetic program.109 TFAP2C activates SOX17 expression through binding to SOX17 promoter, indicating that TFAP2C functions upstream of SOX17 for germline specification.24 A recent report shows that SOX17 and TFAP2C activate the expression of PRDM1, POU5F1, and NANOG.220 PRDM14 cooperates with TFAP2C and BLIMP1 to induce hPGCLC formation, yet repress WNT signaling and somatic markers.172 Nodal signaling is also required for PGCLC specification.221 In addition, SOX17, TFAP2C, and PRDM14 upregulate expression of themselves, respectively.172,195 The synergetic effects of these factors, on the one hand, induce PGC fate and ultimately, on the other hand, suppress the somatic program (Fig. 3).
Fig. 3.
Regulation network of hPGC specification. BMP- and WNT signaling promote the hPGCLC specification via regulating TFAP2C and SOX17. SOX17 is a critical regulator for hPGC specification and works upstream of BLIMP1. TFAP2C activates SOX17 expression. Final effects of hPGC specification promote germline development and pluripotency, while suppress somatic programs. The figure was drawn by Figdraw. Arrows and blunt-ended arrows depict positive and negative regulation, respectively. Dashed lines indicate synergetic role
Epigenetic reprogramming of hPGCs
Epigenetic reprogramming is another layer of regulation in hPGC development. During hPGCs development, shortly after specification, throughout the migration, and towards gonad colonization, epigenetic reprogramming takes place in hPGCs. At genomic DNA levels, globe genomic DNA demethylation is one of the major epigenetic events during hPGCs development, which occurs at week 7.167 The inactivated X chromosome is reactivated in female hPGCs of 5.5–9 weeks,167,171 which is similar to that in mPGCs.222 The lowest level of hypomethylation occurs at week 10 (female) and week 11 (male), respectively.167 The low levels of methylation are maintained till week 19, but global re-methylation already starts in female PGCs at week 11 and male PGCs at week 19, respectively.167 Consistent with this in hPGCs, DNA demethylation dioxygenase TET1 is highly expressed in hPGCs from week 4 to 11, TET2 and TET3 are also mildly expressed, while the 5hmC level is very low in 7–11 weeks.167 A similar pattern of globe DNA demethylation is also detected in hPGCLCs.20 As loss of BLIMP1 affects the initiation of DNA demethylation, SOX17 and BLIMP1 pathway is proposed to drive extensive DNA demethylation and chromatin reorganization in hPGC specification.171 Nevertheless, global changes in gene expression might not correlate with global changes in DNA methylation in developing prenatal germline cells.223 In addition, chromatin modifications are involved in hPGC specification. The hPGCs at week 4 show a remarkable enrichment of H3K27me3, then a declined trend, and hPGCs of 7–11 weeks retain a certain level of H3K9me3, a constitutive heterochromatin marker, indicating its role in hPGC development.171 Histone lysine demethylase KDM2B would regulate the demethylation of histone marks, for example, H3K4me3 and H3K36me2 for hPGCLC specification.224 A recent report indicates that the hominidae-specific transposable elements (LTR5Hs) are expressed in both hPGCs and hPGCLCs, which are involved in chromatin accessibility and localized DNA demethylation. LTR5Hs retain an open chromatin state for binding by key PGC factors, including NANOG, TFAP2C, SOX17, and SOX15 after hPGCLC induction, and serve as TE embedded enhancers necessary for germ cell specification.225 In addition, LTR5Hs play an important role in the gene regulatory network shared between hPGCLCs and naïve ESCs.226 Thus, hPGC development is a complexly coordinated process involved in both genetic and epigenetic regulations, along with the spatiotemporal dynamic change of these regulators (Fig. 3).
Migration and colonization of hPGCs
Migration route
PGCs are migratory cells during embryogenesis, which originate from the epiblast, move toward, and finally colonize the developing genital ridges. The PGCs eventually participate in gonad construction, together with somatic cells from intermediate mesoderm and visceral mesoderm.39 Our knowledge about mammalian PGC migration is mostly drawn from mPGCs in mice. After their specification in the epiblast, at around E8, the mPGCs begin to move actively into the visceral endoderm, go through the hindgut at E9.5, and during the E10.0–E10.5 period, migrate directionally from the dorsal body wall into the genital ridges.15,227–231 Wylie’s group clearly recorded the migration process by time-lapse analysis of living mPGCs from OCT4:GFP transgenic mice.15,227 In humans, the hPGC migration is mainly observed by morphology, histochemistry, and immunohistochemistry using the PGC markers, including alkaline phosphatase,19 glucosaminoglycans,231 and OCT4.232 The migration route from the hindgut epithelium towards the genital ridges is generally as follows, starting at around four weeks, out of the wall of the hindgut, through the dorsal mesentery to the midline of the dorsal wall, finally migrating into the developing gonads at 6 weeks14,17,231–233 (Fig. 2).
Signaling pathways and regulations for hPGC migration
Accumulating evidence shows that hPGC migration is both active and passive. Active movement of PGCs is considerable, although the migration along with passive translocation,16,229 as evidenced by (1) possessing pseudopodia,17 and (2) guiding of signaling molecules.234 For example, culture in vitro showed that the genital ridge tissue from 8.5 dpc mouse embryos could attract mPGCs and exert long-range effects on the migrating population of mPGCs.235 Screening for the factors involved in PGC migration has identified a number of signaling molecules essential for the migration from a variety of animals, including Drosophila,138,236 Xenopus laevis,237 zebrafish,119,238–240 chicken,119,241 and mice.15,227,230,242–251 Several signaling pathways of PGC migration are conserved in humans. Main signaling molecules and their pathways involved in PGC migration include SDF1–CXCR4, KIT-KITLG, HMGCR, and cholesterol. In addition, the extracellular matrix and sympathetic nerve fibers of the autonomous system play important roles in PGC migration.231,232,251,252
SDF1–CXCR4 signaling
SDF1 (also known as CXCL12) is a member of the alpha chemokine protein family. It is expressed in the body wall mesenchyme and genital ridges and acts as the ligand for the G-protein coupled receptor, and chemokine receptor 4 with the C-X-C motif (CXCR4) is expressed in migrating germ cells.249 Zebrafish have another SDF1/CXCL12 receptor, CXCR7, which is also crucial for PGC migration toward their targets.253 In mice, mPGCs have the cell-surface expression of the receptor CXCR4, and loss of the ligand SDF1 results in a delayed migration of mPGC.244 Embryos carrying targeted mutations in the receptor CXCR4 show defects in PGC migration and a reduced number in the genital ridges.249 Thus, the interaction through direct binding of SDF1 with CXCR4 plays a critical role in the directed migration of PGC towards the genital ridges. In fact, SDF1/CXCR4 pathway has also involved the migration of various cell types in humans, including cancer cells.254 TCam-2, a human germline seminoma, has a global similarity in gene expression pattern with hPGCs and hPGCLCs, including expression of chemokine members, CXCR4 and CXCR7, in addition to hPGC markers SOX17, BLIMP1, and CD38.20,255 CXCL12 supplement on matrigel-simulated basement membrane in culture shows a greater cell invasion in TCam-2.256 Nevertheless, direct evidence of SDF1–CXCR4 signaling in hPGC migration in vivo remains to be explored.
KIT-KITLG signaling
KIT is a receptor tyrosine kinase expressed on PGC surface for migration, and upon activation by its cytokine ligand KITLG in somatic cells, KIT phosphorylates intracellular proteins that could play a role in PGC migration. PGC motility and survival require both KIT and its ligand KITLG.257 Mice homozygous for KIT mutation are usually sterile, and their mPGCs are markedly reduced in number and showed a delayed and ectopic migration.258 The ligand steel (KITLG) is continuously expressed by somatic cells surrounding PGCs throughout migration,234 and the lacking KITLG results in cessation of motility and, finally, death of the ectopic germ cells, suggesting that KITLG protein promotes PGC migration.242,257,259 In addition, the transmembrane protein steel favors PGC adhesion to somatic cells via KITLG-KIT interaction, which may be independent of KITLG-induced tyrosine autophosphorylation of KIT receptor.248 Nevertheless, the culture of 11.5 dpc mPGCs showed that the ligand KITLG and 740Y-P peptide (an activator of PI3 kinase) rapidly increased autophosphorylation of its receptor KIT and caused phosphorylation of the serine–threonine kinase AKT through the action of PI3K and stimulated PGC migration, while the inhibitor of PI3K (LY294002) and inhibitor of the MEK/ERK signaling (U0126) impaired the PGC migration.260 In humans, genome-wide association studies have revealed that KITLG on chromosome 12 is a key susceptibility locus for testicular germ cell tumors in the populations of UK261 and US.262 An abnormally high expression of KIT has been observed in both testicular germ cell tumors and malignant ovarian.263,264 High expression of KIT is also detected in extragonadal testicular germ cell tumors, indicating an association of ectopic PGCs with extragonadal tumors, for example, in the central nervous system.265 It is worth mentioning that extragonadal tumors have been linked to the KIT-KITLG signaling, because of aberrantly migrated ectopic PGCs.266 These data indicate the importance of KIT-KITLG signaling in hPGC migration and neoplastic transformation of the germ cells derived.
HMGCR and cholesterol
Hydroxymethylglutaryl coenzyme A reductase, HMGCR (also known as HMGCoAR or LDLCQ3), is a rate-limiting enzyme for the cholesterol synthesis pathway. HMGCR inhibition by atorvastatin, an HMG-CoA reductase inhibitor that also has the ability to effectively decrease blood lipids, exhibits germ cell migration defects in zebrafish embryos, which can be rescued by mevalonate, the product of HMGCR activity.239 In mice, the genital ridges could accumulate high levels of cholesterol by localized uptake, and inhibition of the HMGCR activity in the culture of the genital ridges resulted in defects of germ cell survival and migration, suggesting that cholesterol is required for PGC survival and motility.243 Although direct evidence of the roles of cholesterol in hPGC migration is lacking in humans, recent studies show that cholesterol has essential roles in the specific ligand binding mode in the CX3CR1 chemokine.267 Given that the receptor CXCR4 for the ligand SDF1 shares a similar structure of the key region ECL2 with CX3CR1,267 thus, cholesterol molecules probably play essential roles in the receptor activation of SDF1–CXCR4 signaling for hPGC migration. As a novel link between cholesterol metabolism and hPGC development, this is a particularly interesting topic, which remains to be elucidated further.
hPGC-related diseases: infertility and cancer
The hPGC development is an essential event during embryogenesis. Dysregulation of hPGCs in origin, migration, colonization, and differentiation will lead to major diseases in humans. The main types of diseases related hPGCs include infertility and cancer. Infertility is one of the main disorders in humans, which is mentioned throughout this topic. Thus, we will mainly discuss human germ cell tumors (GCTs) as follows. Ovarian cancer is one of the five deadliest cancers in women.4 Human GCTs are generally derived from germline cells, stem cells in particular, in the early embryos. GCTs occur not only in gonads (ovary and testis) but also in various organs in humans, although the most common types of GCTs are testicular germ cell cancer and ovarian cancer. Human germ cell tumors have been classed into seven GCT types, from type 0 to type VI, based on their developmental potential.268 Extragonadal GCTs are involved in a wide range of organs, for example, brain, head/neck, heart/mediastinum, lung, thymus, sacrococcygeal region, abdomen, retroperitoneum, vagina, and placenta, which are also sites of germ cell tumors266,268,269 (Fig. 4). It is widely accepted that extragonadal GCTs mainly originate from mismigration of hPGCs that failed to undergo apoptosis.265,270–272 Approximately 3% of malignant pediatric tumors are GCTs, and most of them are brain cancers.265,273,274 Interestingly, central nervous system GCTs express pluripotency marker genes PLAP, TFAP2C, NANOG, and KIT, in addition to markers for Sertoli/granulosa cells, MIC2 and AMH, and cancer-related markers MAGE-A4 and TSPY.265 Mutations are detected in intracranial germ cell tumors, including KIT, its downstream mediators KRAS and NRAS, copy number gains of the AKT1, and tumor suppressor BCORL1.275 In addition, mismigration of hPGCs in progenitor cells in the pancreas during early embryogenesis has been suggested as the main pathogeny of mucinous cystic neoplasms of the pancreas in humans.276 In general, as being pluripotential tumors, it is a very important and common understanding that the GCTs express germline markers, including OCT4, SOX17, NANOG, VASA, KIT, CXCR4, and TSPY, which may be used for diagnosis and treatment of extragonadal GCTs.263,265,268,277–281 For example, knockdown of CXCR4 expression suppresses proliferation, adhesion, and migration,282 and CXCR4 antagonists will be a promising therapy in antitumor activity in patients with various malignancies.278
Fig. 4.
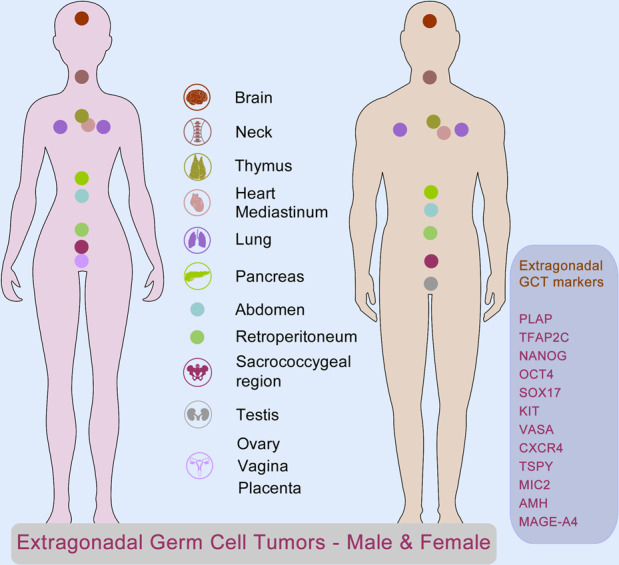
Extragonadal germ cell tumors (GCTs) in humans, related to PGC mismigration. As being pluripotential tumors, extragonadal germ cell tumors occur in a wide range of organs from central nervous system to ovary and testis indicated in the central panel, with expression of PGC markers, including PLAP, TFAP2C, NANOG, OCT4, SOX17, KIT, VASA, CXCR4, TSPY, MIC2, AMH, and MAGE-A4 listed in the right panel. Some elements of the figure were derived from Soehui
In addition, hPGC-related disorders include other types of diseases. For example, Fanconi anemia, a recessive congenital disease, has characteristics of progressive bone marrow failure, mismigration of hPGCs, and predisposition to cancer, including acute myeloid leukemia and squamous cell carcinoma. FANC family of genes and related DNA interstrand crosslink pathway are identified for Fanconi anemia pathology,283 and FANCG is responsible for the PGC migration.245 Overexpression of hPGC marker gene PRDM14 is detected in lymphoblastic lymphomas,284 suggesting PRDM14 as a proto-oncogene involved in lymphoblastic lymphoma formation. It has been suggested that depletion of PRDM14 expression may be an effective and radical therapy for solid cancers.285,286
Most germ cell tumors are not caused by gene mutations, but instead by reprogramming their germ cells of the origin in the target niches. For example, testicular germ cell tumors prefer retention of PGC-lineage erasure of both maternal and paternal DNA imprints272,287,288 and histone modifications like H3K27ac.289 Somatic gene mutations and chromosomal mutations may also result in some germ cell tumors. For example, isochromosome 12p is common in seminomas and non-seminomas.280,290–294 KIT-KITLG mutations or their signaling activation,262,264,275,281,295–299 deletions of genes (e.g. SOX17, the gr/gr deletion on chromosome Y),279,300 structural variation, duplication, and loss of chromosomes,301–304 and aneuploidy302 are frequently detected in germ cell tumors. Recent reports show that super-enhancers are preferentially amplified in ovarian cancer.305 Indeed gene amplification occurs frequently in ovarian cancer.306 In addition, loss of the PGC gene TFAP2C leads to a high rate of germ cell tumors in mice, resembling pediatric Type I germ cell tumors in humans.201 DMRT1, a spermatogonia marker, is highly expressed in germ cell neoplasia in situ, and drives in vivo reprogramming and propagation of GCT-like tumor cells,307 indicating a shared feature of DMRT1-mediated reprogramming in germ cell tumors. Furthermore, germ cell tumors might be composed of somatic tumor cells and PGC-like tumor cells,308 which may bring difficulties to medical treatment. In clinics, cisplatin-based chemotherapy is a major means for the treatment of GCTs, which has a high cure rate.268 However, resistance to cisplatin often occurs in a proportion of 10–20%.280 The deubiquitinase USP11 is an important determinant of ovarian cancer chemoresistance.306 JMJD6 inhibitor SKLB325 has a significant effect in suppressing proliferation and promoting apoptosis of ovarian cancer cells.309 New targeted treatment approaches are needed for germ cell tumors. In particular, diagnosis and treatment based on pluripotency markers and targets of GCTs will be promising strategies. For example, the hPGC gene LIN28B plays a very important role in the inhibition of apoptosis through regulation of the AKT2/FOXO3A/BIM axis in ovarian cancer cells,310 indicating a novel target based on hPGC pluripotency in the diagnosis and therapeutics of ovarian cancer.
Human PGCLCs, induced PGC-like cells
It is inaccessible to early hPGCs in vivo for the study of the human PGC development because of ethical issues. Fortunately, in vitro induction systems for differentiating hESCs/iPSCs into hPGC-Like Cells (hPGCLCs) have been established,20–22 which are not only a way of circumventing the issues, but also provide an approach to producing functional human gametes from hPGCLCs in vitro in the future. The in vitro reconstitution of human germ cell development will be instrumental in developing innovative medical applications in infertility and cancer.311,312 Thus, hPGCLC advances facilitate our understanding of human germ cell development and provide a new therapeutic means for treating infertility and cancer.
Features of hPGCLCs
Early studies in mice showed that ES cells possess the ability to contribute to the germ cell lineage when cultured in vitro.313–315 Mouse ES cells derived from the ICM (inner cell mass) of the blastocyst are coaxed to differentiate into oogonia and sperm cells in vitro.314,315 Following the studies of the induced PGCs in mice, human ICM cells were also induced to differentiate into embryoid bodies, and some of the induced cells expressed markers of germ cells, including VASA,316 indicating that human ES cells could also be induced into the PGC-like cells in vitro. Based on the principle of hPGC development, robust approaches for hPGCLC specification in vitro from germ cell competent hESCs/hiPSCs under defined conditions have been developed.20–22 In summary, hPGCLCs possess several features of in vivo hPGCs. (1). These hPGCLCs show a similar pattern of gene expression to that of early hPGCs, including core PGC genes, SOX17, BLIMP1, and TFAP2C, but do not express late PGC markers including DAZL and DDX4.20,21 (2). Both hPGCLCs and hPGCs also share expression of pluripotency genes (NANOG and OCT4) and cell-surface markers (CD38, EPCAM, and ALPL).20 (3). The hPGCLCs exhibit upregulation of 5hmC (5-hydroxymethylacytosine) and TET1(a demethylase that belongs to the TETs), and a decline in the expression of de novo DNA methyltransferase 3 A and 3B (DNMT3A and DNMT3B),20,171 indicating an early pattern of DNA demethylation. Global loss of DNA methylation in the hPGCLC genome reveals the progress of epigenetic reprogramming similar to hPGCs in vivo.22 (4). Thusly, the hPGCLCs would correspond to early-stage hPGCs in vivo and probably represent pre-migratory hPGCs. (5). Finally, the hPGCLCs and hPGCs share a functional similarity in differentiating into oogonia and prospermatogonia.317–319 In induction culture systems, hPGCLCs are cultured with mouse fetal testicular somatic cells in long-term cultured xenogeneic reconstituted testes317 or with mouse ovarian somatic cells in xenogeneic reconstituted ovaries.318,319 hPGCLCs are also co-cultured with somatic cells from postnatal rat testes.320 These somatic cells provide an appropriate niche for hPGCLCs to mature into oogonia or prospermatogonia similar to those of hPGC differentiation in vivo, respectively. Nevertheless, differentiation in vitro from hPGCLCs to mature gametes (ova and spermatozoa) and function testing remain to be explored further.
Methodologies for inducing hPGCLCs
hPGCLCs can be induced from both embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs). Two strategies for inducing hPGCLCs from hESCs and hiPSCs have been developed, the iMeLCs strategy and the 4i strategy.20,21 The basic principle of both strategies is based on the development rules of hPGCs, maintaining hPGC pluripotency, inducing hPGC fate, and inhibiting endodermal and other somatic genes.
The iMeLCs strategy
The strategy is a two-phase induction process from hiPSCs to iMeLCs and then to hPGCLCs (hiPSCs- iMeLCs- hPGCLCs)21 (Fig. 5a). During the first phase, hiPSCs are cultured under a conventional condition, and induced into incipient mesoderm-like cells (iMeLCs), a similar state to EpiLCs induced from ESCs/iPSCs in mice.321 EpiLCs bear a cellular state similar to pregastrulating epiblasts with high competence for the PGC fate.321 In the phase, Activin A and CHIR (a WNT signaling agonist322) are added to KSR medium to stimulate hiPSCs for 48 h, which is critical for hiPSCs to acquire a capacitated iMeLC state.21 iMeLCs are incipient mesoderm/primitive streak-like cells, express genes for pluripotency and mesoderm, but do not express PGC markers.21
Fig. 5.
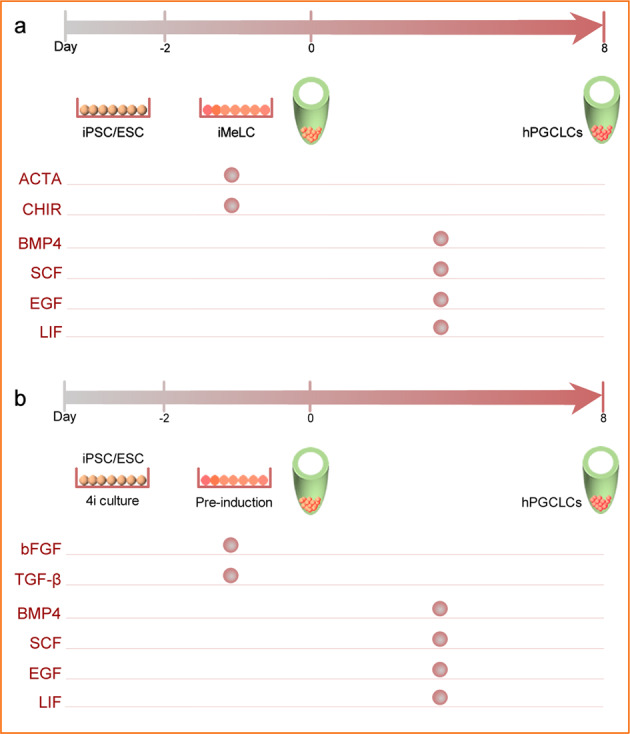
Methodology of hPGCLCs induction. hPGCLCs are induced from both embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs). a The iMeLCs strategy. b The 4i strategy. Induction culture timelines and added factors are indicated on the upper and the left in each panel, respectively
During the second phase from iMeLCs to hPGCLCs, the iMeLCs are cultured in GMEM + 15% KSR with BMP4, SCF, LIF, EGF, and Y-27632. The Y-27632 is a selective inhibitor of p160-Rho-associated coiled-coil kinase (ROCK) and plays roles in antiapoptosis and increases cloning efficiency.323 For hPGCLC induction, BMP4 is essential, which acts through activin receptor-like kinase 2/3 and upregulates GATA3.21,195 SCF, LIF, and EGF play additive roles in the proliferation and survival of hPGCLCs.21,321 The hPGCLCs are induced by plating iMeLCs into a well of a low-cell binding U-bottom 96-well plate. The induced hPGCLCs exhibit upregulation of the regulators for hPGCLC specification, including TFAP2C, PRDM1, SOX17, SOX15, KLF4, KIT, TCL1A, and DND1, whereas downregulated genes are those involved in pattern specification processes and neuron development.21 Epigenetic change shows low levels of H3K9me2 and DNA methylation, including the imprint erasure of H19, but imprints of MEG3, KCNQ1, and PEG10 are not affected,21 which are similar to those of mPGCLCs.198,321 These hPGCLCs also express cell-surface markers EpCAM and INTEGRINα6,21 which can be used to identify and purify the hPGCLCs using immunofluorescence and fluorescence-activated cell sorting analyses.
The 4i strategy
To be a competent state for hPGC fate, hESCs and iPSCs are first cultured on MEFs with 4i (inhibitors for MAPK, GSK3, p38, and JNK) and preinduced by TGFβ and bFGF for 2 days.20 The inhibition culture has been used in hPSC culture, which represents a naive state of human pluripotency.324,325 These preinduced cells are further induced into hPGCLCs by adding BMP2/BMP4, LIF, SCF, EGF, and Y-27632.20 The cells are generally induced in ultra-low cell attachment U-bottom 96-well plates. In the induction system, 4i culture is a key step, which makes the cells to be a competent state for hPGC fate (Fig. 5b). The induced hPGCLCs express key hPGC genes, including SOX17, BLIMP1, TFAP2C, PRDM14, STELLA, TNAP, and KIT, pluripotency genes, OCT4 and NANOG, and cell-surface markers, TNAP and CD38.20 The proportion of hPGCLCs (both TNAP and CD38 positive cells) in the culture of day 4 embryoids induced from 4i hESCs is close to 46%,20 indicating a high competency for hPGCLC fate in the induction system. The combination of cell-surface markers TNAP and CD38 can also be used to identify and purify the hPGCLCs using immunofluorescence and fluorescence-activated cell sorting analyses.
hPGCLCs and infertility treatment
Infertility affects over one-fifth of human couples worldwide.10,11 An increasing tendency to postpone child-bearing age often leads to difficulty to get children. There is a subsistent need for infertile patients who have an alternative choice, in vitro fertilization (IVF) with gametes derived from stem cells, or even somatic cells. There are great attempts to induce meiosis and haploid cells using hESCs/hiPSCs at different induction conditions.326–330 However, the induction is inefficient. hPGCLCs will be among the most promising cell types for producing gametes. hPGCLCs can be induced to differentiate into both oogonia and prospermatogonia by co-culturing with mouse embryonic ovarian or testicular somatic cells, respectively.317–319,331,332 Both oocytes and spermatozoa will be obtained via hPGCLCs induction for the treatment of infertile females and males. As mentioned above, hPGCLCs could also be induced from iPSC derived from somatic cells. Thus, in principle, oocytes and spermatozoa can be produced from somatic cells, not only from germline cells, in the future (Fig. 6). Of course, somatic cells as a new source of gametes via hPGCLCs will change our understanding of the continuity of life through germ cells,23 which needs wide discussions before application. Technology for the mouse in vitro gametogenesis has been closer to establishment. The oocytes induced from mPGCLCs are subjected to IVF, and viable pups have been obtained.333–335 The in vitro oogenesis system might also be used to explore molecular mechanisms for genetic diseases, for example, chromosomal aneuploidy.336 In addition, mPGCLCs start proper spermatogenesis after being transplanted into the testis, and relevant offspring are produced via IVF.321 Moreover, complete in vitro meiosis to generate male gametes from mouse ESC-derived mPGCLCs has also been reported.337 Spermatid-like cells derived from the mPGCLCs are subjected to intracytoplasmic injection into oocytes, and viable and fertile offspring have been obtained.337 The basic framework of human in vitro gametogenesis is roughly the same as that in mice, and further advances will benefit diagnosing, modeling, and treating infertility in humans.
Fig. 6.
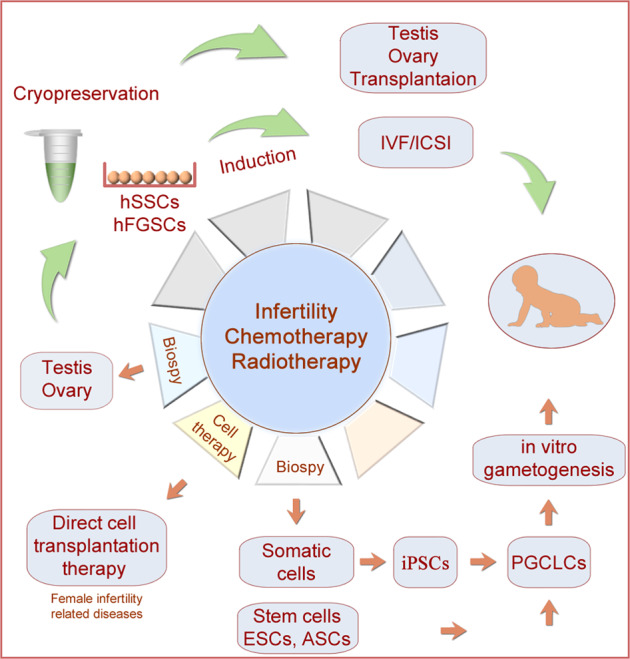
Fertility preservation and treatment of infertility and cancer in humans. Germ cell/gonad tissue transplantation is an alternative therapeutic means for treating infertility, and it is also a promising treatment strategy for both pubertal and prepubertal boys/girls diagnosed with cancers who will suffer from irradiation and chemotherapy. Cell sources for transplantation could include germ stem cells and hPGCLCs. As new progress in cell induction, transplanted cells might come from somatic cells of patients, which can be induced into gametes via hPGCLCs in the future. Some parts of the figure were drawn by Figdraw
Adult germline stem cells
Spermatogonial stem cells for spermatogenesis
Features of human SSCs
Both spermatogonial stem cells (SSCs) and female germline stem cells (FGSCs) are adult pluripotent stem cells (ASCs) in the germline. SSCs are pluripotent stem cells for generating spermatozoa in the testis, while FGSCs are newly identified pluripotent stem cells for producing oocytes in the ovary. Once gonadal colonization, hPGCs cease to proliferate, and the cells are called gonocytes or prespermatogonia in males.338 hPGCs will differentiate directly into fetal state 0-like spermatogonia starting at week 14 after fertilization.339 In humans, spermatogenesis begins 10–13 years after birth at puberty.340 During the first wave of spermatogenesis, some gonocytes resume proliferation, begin to move toward the periphery (basement membrane) of seminiferous tubules in the testis, and differentiate into spermatogonial stem cells.341 The SSCs are essential for generating spermatozoa throughout life. As germline stem cells, SSCs have features of stem cells, in addition to maintaining spermatogenesis. The main features of human SSCs include, (1) self-renewing to maintain SSCs population, (2) production of spermatogonia to support daily production of sperm cells,342 (3) SSC heterogeneity,342–345 as discussed below, (4) SSCs can transdifferentiate into other cell types, for example, oocytes, which is functional because the induced ovarian organoids derived from SSCs produced offspring.346 SSCs could be generated from induction from hPSCs,347 prepubertal SSCs are also induced to initiate meiosis and produce haploid germ cells in vitro.348 Somatic Sertoli cells are converted to become into SSCs by overexpression of DAZL, DAZ2, and BOULE,349 (5) SSCs are rare and account for about 0.02–0.03% of all cells in the testis,350 and (6) de novo mutations in SSCs increase as men age, which often are associated with congenital disorders.351
SSC heterogeneity and regulation
The earlier work has defined two types of A spermatogonia in humans, the Adark and Apale spermatogonia, which are considered undifferentiated stem cells, the reserve stem cells, and renewing stem cells, respectively.352–354 Single-cell RNA sequencing analysis of human testis has further exhibited spermatogonia heterogeneity by determining subtypes or states, and developmental trajectory.343–345,355 During the developmental process, SSCs first form progenitors that undergo proliferative expansion, then generate differentiated spermatogonia. Single-cell RNA clustering also shows three stages of spermatogonia, from SSCs to differentiating spermatogonia and then to differentiated spermatogonia.345 These spermatogonia at different developmental stages show three-dimensional chromatin architectural differences356 and express distinct marker genes.343–345,355 For example, SSCs cluster expresses markers, including GFRA1, RET, NANOS2, NANOS3, ZBTB16, SALL4, POU3F1, FGFR3, UTF1, PAX7, UCHL1, PLZF, and ID4, differentiating spermatogonia show expression of KIT, MKI67, DMRT1, and SOHLH1, indicating the proliferation of active spermatogonia, and STRA8, KIT, and MAGE-A4 are expressed in differentiated spermatogonia.344,345,357–360 The activation of EGR4, KLF6, KLF7, and/or SOX4 might be involved in the differentiation process from hPGCs to prespermatogonia.175 Nevertheless, functions in vivo of these SSC genes in humans remain to be determined further.
Using animal models, functions of important regulatory genes and pathways in human SSCs could be clarified. For example, in a mouse model, mTORC1 and FOXO1 signaling have been shown to be key regulators for regenerative undifferentiated spermatogonia.361 The hepatic stellate cell activation pathway is upregulated in SSCs, suggesting its role in the specification and maintenance of SSC fate.343 The SSC niche is another regulatory layer for SSC development. The seminiferous tubule and the interstitial tissue provide a local niche for SSCs.362,363 Somatic Sertoli cells are the supporting cells in the testis, and their differentiation is regulated by waves of transcription factors SRY, SOX9, AMH, and DHH.364 Sertoli cells finally generate and secrete specific factors for SSC development.365 The secreted glial cell-line-derived neurotrophic factor (GDNF) promotes the self-renewal of SSCs, while inhibiting their differentiation.366 GDNF activates RET tyrosine kinase in undifferentiated type A-spermatogonia collaborated with GFRA1, a ligand-specific co-receptor.367,368 Thus, GDNF signaling is essential for SSC self-renewal.369 CXCL12–CXCR4 signaling not only play an important role in PGC migration, as mentioned earlier but also in the establishment of the SSC niche. Sertoli cells express CXCR12, while SSCs express CXCR4 receptors on their membrane. Inhibition of CXCR4 signaling in mouse testes impaired SSC maintenance, leading to germline loss.370 Somatic Leydig cells are another type of supporting cells in the testis. Spatial transcriptome analysis shows distinct microenvironment compositions surrounding the undifferentiated versus differentiating spermatogonia between Leydig cells and Sertoli cells.371 Retinoic acid signaling regulates the differentiation process of the undifferentiated spermatogonia to differentiated spermatogonia.372 IGF1 and FGF9 signaling might also be associated with SSCs development.373 A recent report shows that H3K79 methyltransferase DOT1L is essential for SSC self-renewal in mice, which is associated with HOXC expression,374 indicating the importance of histone modifications in SSC regulation. At genomic DNA levels, ZBTB43 safeguards genomic integrity by regulating de novo DNA methylation at CG-containing purine–pyrimidine repeats, removing Z-DNA, and preventing DNA double-strand breaks in mouse prospermatogonia.375 It is interesting that ZBTB43 expression is also involved in cancer stemness in humans.376 In addition, male reproductive aging is associated with the capacity decline of SSC niche,363,377 thus, older males are often accompanied by a decrease in reproductive function.
SSC transplantation for fertility preservation in cancers and gene therapies
Spermatogonial stem cell transplantation is a promising approach to restore fertility for patients who need chemotherapy and radiation treatment, such as in cancer therapies, because these treatments often lead to damage to gonadal cells, thus infertility,361,378,379 although some antioxidants can reduce the damage to germ cells.380 Prepubertal boys who suffer from gonadotoxic treatment under pediatric cancer circumstances, e.g., acute lymphoblastic leukemia and testicular cancer, might cause sterile for the rest of their life.381 Autologous transplantation of SSCs or testicular tissue has been proposed as a strategy for fertility preservation and therapy382–385 (Fig. 6). In Europe, Canada, and USA, over 1033 young patients between 3 months and 18 years of age have already joined in fertility preservation by testicular tissue storage for late use.382,386 SSC transplantation has been considered in the fertility preservation of other genetic diseases, such as Klinefelter syndrome.387 Transgender women might also cryopreserve their germ cells before hormonal treatment, as a small percentage of transgender women have immature male germ cells.388
Testis-derived germ cell microinjection into seminiferous tubules of infertile recipients was first reported in mice, and transplanted cells colonized seminiferous tubules and initiated spermatogenesis.389,390 Germ cell transplantation in interspecies and intraspecies has been applied to zebrafish, rats, dogs, farm animals (goats, sheep, pigs, and cattle), nonhuman primates, and humans, in addition to mice.391–408 In primates, postpubertal SSC transplantation in infertile rhesus monkeys restored functional sperm production after puberty.409 In humans, the first clinical trial of testis-cell transplantation using cryopreserved single-cell suspension from patients’ testis with lymphoma before chemotherapy was reported in 1999.410 Nevertheless, it is difficult to evaluate the outcome, as endogenous spermatogenesis can occasionally escape from radiation or chemotherapy. Transplanted tissue/cells for fertility preservation may be testicular cell suspension, testicular prepubertal tissue fragments, or SSCs.382 SSCs can be isolated from the testis and proliferate in culture or induced from stem cells or Sertoli cells.349,411–413 Actual clinical implementation and safe should be carefully considered in the near future, for example, xenofree, clinical grade media, culture condition, and protocols.414,415 One of the concerned main issues about transplantation in cancer patients is the risk of reintroducing malignant cells present within tissue fragments to the patients. In fact, hPGCLCs will be a potential cell source for transplantation (Fig. 6). As mentioned earlier, hPGCLCs can be derived from iPSC, thus adult tissues from patients are an actual source of the cells, yet avoiding testis biopsy.
In addition, SSC transplantation is a promising treatment strategy for patients with genetic diseases, for example, Klinefelter syndrome, thalassemia, and drepanocytosis.29 For patients who carry gene mutations, their SSCs or hPGCLCs may be corrected before transplantation using CRISPR/CAS9 technology, which has potential applications for the treatment of genetic diseases in humans. Germline gene therapy via SSCs has achieved success in correcting an X-linked testis-expressed 11 (TEX11) mutation in mice with azoospermia phenotype.416 The mutant SSCs were isolated, and the TEX11 mutation was corrected by CRISPR-CAS9 technology. The final repaired SSCs were implanted back into the testis, which restored spermatogenesis in infertile males and gave rise to fertile offspring.416,417 The treatment technology might be used to cure azoospermia patients with TEX11 or other gene mutations in the future. However, the strategy needs to wait for a long-term discussion, public acceptance, and ethical argument, because of considerable ethical concerns for gene therapy through germline in humans.
Female germline stem cells for oogenesis
In contrast to well-known SSCs, FGSCs are newly identified germline stem cells in mammals, including humans. It is reported that juvenile and adult mouse ovaries have mitotically active germ cells, but the same group subsequently indicates that both bone marrow and peripheral blood serve as a source of these germ cells in adulthood.34,418 FGSCs are identified in neonatal and adult ovaries, which are isolated from ovaries and differentiate into functional oocytes after transplantation into mouse ovaries.25 Thus, FGSCs are able to undergo postnatal neo-oogenesis,25,35 possibly providing oocytes for reproductive life. The finding of FGSCs has updated the traditional idea that the ovary possesses a finite oocyte reserve before birth in female mammals.31–33 Thus, proliferation and differentiation of FGSCs may replenish the gradual exhaustion of reserved primordial follicles throughout the female’s fertile life. It has been shown that infertility and death of women are partly attributed to ovarian function-related diseases, for example, polycystic ovarian syndrome, premature ovarian failure, and ovarian cancer.419 Applications of FGSCs in reproductive medicine have significant clinical implications in the treatment of female reproductive aging and ovarian function-related disorders. For example, FGSCs transplantation might be applied to patients with ovarian cancers or premature ovarian failure as a strategy for fertility preservation and therapy in females (Fig. 6). Actually, ovary tissue possesses several types of somatic stem cells yet, for example, human OSE (ovarian surface epithelium) stem cells, which express SOX-2 and SSEA-4, but FGSCs do not, although they can form oocyte-like cells in culture,420 and human VSELs, a very small embryonic-like stem cells with nuclear OCT4 expression and LGR5+,421,422 in addition to Thecal stem cells and granulosa stem cells.423 However, FGSCs are germline stem cells, which will be discussed as follows.
Features of FGSCs
As germline stem cells, FGSCs have characteristics of adult pluripotent stem cells, in addition to maintaining oogenesis. The main features of FGSCs in mice and humans are summarized as follows. (1) FGSCs show similar morphology to those of SSCs with large nuclei and little cytoplasm.424 (2) FGSCs isolated from neonatal mice display the string-forming cell configuration, and E-cadherin mediates the cell–cell contact at membrane connection sites.425 (3) FGSCs express membrane marker Fragilis/IFITM3, while MVH and alkaline phosphatase are mainly localized to the cytoplasm and also expressed on the membrane.424,426 (4) IFITM3, DAZL, MVH, OCT4, PRDM1/BLIMP1, and DPPA3 are markers for FGSCs, in addition to TERT and alkaline phosphatase, and cell cycle-related transcription factors c-MYC and EGR-1 are also expressed in FGSCs.25,424,427 However, NANOG, SSEA-1, and SOX2 are not expressed in both neonatal and adult FGSCs.424 In addition, OCT4, BLIMP1, and DAZL are common markers in both PGC and FGSCs. (5) FGSCs have high telomerase activity.25 (6) FGSCs could be converted to female ES-like cells under ESC culture conditions with the addition of vitamin C and valproic acid, which show similar characteristics to ESCs in genomic imprinting, formation of the three germ layers and chimeras, and germline transmission capacity.428 (7) FGSCs self-renew to maintain proliferation with vigorous mitosis capacity.25,425 (8) FGSCs have the capacity to produce normal oocytes to support the generation of fertile offspring after transplantation into mouse ovaries.25 Primordial follicle-like structures form in vitro co-culture with granulosa cells from neonatal mouse ovaries.429 Oocytes are also generated in xenografted human ovary tissue after being injected into adult human ovarian cortical tissue biopsies and then xenografted into NOD-SCID female mice.427 (9) Given the germline transmission ability of FGSCs, genome editing of FGSCs might be used to treat genetic diseases in humans and to alter specific traits in animals. For example, transgenic animals, through introducing genes with functional importance and commercial value into FGSCs have been obtained.430–433 Transgenic rats with fat-1 gene, a Caenorhabditis elegans gene for the synthesis of N-3 polyunsaturated fatty acids from N-6 fatty acids, have been generated using FGSCs.430 N-3 polyunsaturated fatty acids are essential for human development, and their deficiency is associated with human diseases, including cardiovascular disease, hyperinsulinemia, and type 2 diabetes.434–436 Most mammals do not have fat-1 gene in their genomes, thus cannot convert n-6 into n-3 polyunsaturated fatty acids, which should be acquired by food intake. The fat-1 transgenic farm animals will provide an alternative food source of N-3 polyunsaturated fatty acids for humans.
Isolation and characterization of FGSCs
FGSCs exist in neonatal and adult mouse ovaries, especially enriched in neonatal ovaries of 1–3-day postpartum. To isolate the FGSCs, the two-step enzymatic digestion method (collagenase and trypsin) is often used for the efficient digestion of ovary tissue. MVH- or Fragilis-positive cells are then separated by MACS or FACS technology using antibodies against MVH25,427 or Fragilis,426 respectively. The germline-specific Fragilis is a membrane marker in FGSCs, thus, the efficiency of FGSC purification using anti-Fragilis and FACS technology is higher than that using anti-MVH.426 Differential adherence selection with passaging enrichment from postnatal ovaries without any antibody has also been used to isolate FGSCs.425 FGSCs are often cultured in MEM‑α medium supplied with FBS, non-essential amino acids, transferrin, insulin, EGF, GDNF, and bFGF on an inactive STO feeder layer.25 Isolated FGSCs are characterized by testing FGSCs markers, OCT4, MVH, IFITM3, DAZL, and BLIMP1, and differentiating into oocytes in vivo or in vitro. As stem cells, the proliferation capacity of FGSCs should be tested using mitotic markers, including cell cycle-related transcription factors c-MYC and EGR-1. Due to a string-forming characteristic, the mitotic ability may be tested using the mitotic antagonistic agent mitomycin C to treat the cells, which results in a decrease in string-formation.425 Germline transmission and oogenesis capacity are essential functional tests, including the ability to produce fully functional oocytes and fertile offspring after transplantation into chemotherapy-damaged mouse ovaries.25
Regulation of FGSCs development
FGSCs not only self-renew but also differentiate to initiate meiosis. During FGSC development, they first differentiate into germinal vesicle oocytes, then into the prophase of meiosis II via the prophase of meiosis I processes. In these developmental processes, the FGSC genome undergoes dramatically reorganization.437 In addition, the X chromosome shows a smaller proportion of the active compartment in comparison with that of autosomes, because the X inactivation might take place.437
The stem cell niche in the ovary is an important aspect of the regulation of FGSCs development, which provides essential microenvironments and signaling pathways for FGSCs. Disruption of the niche or related signaling leads to stem cell loss.438,439 The niche aging in the ovary is associated with the decline in ovarian reproductive function.440–442 CADHERIN-22, a member of the cadherin superfamily, promotes FGSC self-renewal through interaction with the JAK and β-CATENIN.443 Meanwhile, CADHERIN-22 enhances PI3K-AKT3 signaling, thus, upregulating the expression of N-MYC and CYCLIN, and GDNF-GFRA1 activates AKT3 via PI3K or SFK, subsequently promoting self-renewal of FGSCs.444 GSK3 inhibitor BIO promotes proliferation of FGSCs through activation of β-CATENIN and E-CADHERIN,445 indicating an important role of GSK3 signaling in FGSCs proliferation. The Hippo effector YAP1 also regulates the proliferation of FGSCs in mice.446 In addition, Hedgehog signaling pathway plays an essential role in FGSCs development. Inhibition of the hedgehog signaling pathway with GANT61 leads to follicular atresia and reduction in FGSC proliferation capacity in the mouse ovary and in vitro culture of FGSCs.447 Accumulated evidence shows the importance of the hedgehog signaling in ovary development,448–455 supporting the role of the hedgehog signaling in FGSCs development.
Anti-cancer agent C89, one kind of benzoborazoles, induces FGSC autophagy by inhibiting the activity of Akt and PI3K in vitro, thus inhibiting the proliferation of FGSCs.456 ZCL-082, another kind of benzoborazoles, has a similar effect in promoting autophagy and inhibiting the proliferation of FGSCs, but via regulating GAS5(long noncoding RNA)/miR-21a expression.457 Whereas, spermidine induces cytoprotective autophagy via inhibition of AKT/mTOR phosphorylation in FGSCs.458 The AKT signaling pathway is activated by daidzein through upregulating the stem cell growth factor CLEC11A, thus promoting FGSC proliferation.459 It has been shown that autophagy regulation is closely associated with testis development, spermatogenesis,460 ovary development,461–464 oogenesis, and gonad diseases.465–469 Thus, regulation of the autophagy pathway in FGSCs through small molecules will be promising and potential therapeutic targets for ovary diseases, for example, premature ovarian failure and ovarian cancers.
FGSCs implantation and fertility preservation
As germline stem cells for females, FGSCs provide a new strategy for preserving fertility and delaying menopause, which will, of course, benefit female patients with reproduction dysfunctions (Fig. 6). FGSCs can be implanted into the ovary and initiate oogenesis in vivo for production of fertile offspring.25 FGSCs can also develop in vitro and differentiate into oocytes after injection into human ovarian cortical tissues xenografted into adult immunodeficient female mice.470 Alternatively, FGSCs are cultured into 3D ovarian organoids to produce oocytes for transplantation.471 in addition, functional oocytes are obtained through transdifferentiated from SSCs in vitro and also produce offspring in mice.346
In the clinic, cryopreserved ovarian tissues for females are being carried out.472 Autografting of frozen-thawed ovarian tissue fragments is already used to restore fertility from both adult473 and prepubertal, a 9-year-old girl suffering from thalassemia and a girl with sickle-cell anemia at age 14 years.474,475 A clinic study shows a high live birth rate (33%) after transplantation of ovarian tissue fragments.473 In humans, FGSCs have been obtained from scarce ovarian cortical tissues from follicular aspirates. These FGSCs differentiated into germinal vesicle stage oocytes in vitro for transplantation.470 Thus, the technology from scarce ovarian tissues to oocytes has clinical implications for fertility preservation for women of reproductive age before cancer treatment. In addition, FGSCs are transplanted into the ovary of infertile chemotherapy-treated mice, a premature ovarian failure model, finally restoring ovary function and generating offspring.476 This study provides a technology blueprint for clinic application in humans in the future, for example, in the treatment of premature ovarian failure, early menopause, and infertility, in addition to cancers.
Conclusions and perspectives
We have displayed the fantastic features of germline stem cells in humans, self-renewing, generating ova or sperm cells via halving the genome, and passing genetic information from one generation to the next, in contrast to those of somatic cells. Germline stem cells are pluripotent from embryonic PGCs to adult germ stem cells, SSCs, and FGSCs, with different developmental states. Later, they are the only cells to undergo meiosis. hPGCs are specified in the early stage of embryos, then migrate into the genital ridge, where they meet somatic gonadal cells (e.g., Sertoli cells and Leydig cells in XY embryos, granulosa cells and theca cells in XX embryos) together to assemble testis or ovary, respectively. The XY embryos of age between 41 and 44 days start to express the sex-determining gene SRY on Y chromosome,477 which triggers differentiation of bipotent gonad into the testis, otherwise, in the XX embryos without SRY, the gonad will differentiate into ovary.478 In XY embryos, SSCs gradually develop and start the first time of meiosis for spermatogenesis in adolescent boys at the age of 10–13 years,340 while XX embryos initiate meiosis for oocyte production. FGSCs are newly identified germline stem cells in neonatal and adult ovaries, which support self-renewing and differentiating into oocytes for the production of offspring.25 The finding of FGSCs demonstrates a new concept that adult women can continuously generate oocytes in their reproductive life. Notably, another important advance is that hPGCLCs can be induced in vitro from germ cell competent hESCs/hiPSCs under defined conditions.20–22 These advances have opened up new avenues to understand human germ cell development and provide new approaches to in vitro gametogenesis23 and therapeutic means for treating infertility and cancer (Fig. 6).
However, there are also multiple issues that need to be solved to understand human germline stem cell fate and develop new diagnosis and therapy approaches for medical applications. First, developmental mechanisms of hPGCs in vivo at the peri-implantation stage are not well known, because of the difficulty of access to human early embryos. This issue might partially be resolved through hPGCLCs developmental processes, adopting 3D organoid culture together with machine learning and artificial intelligence in particular, which will provide tractable in vitro models of human physiology and pathology.471,479–481 There is a general consensus that differences are remarkable in mechanisms underlying PGC development between mice and humans,24,86,157 while germ cell development in nonhuman primates better mimics the relevant processes in humans.89–91,95,160,187,482 Further studies using nonhuman primate models will provide new insight into germ cell development and differentiation. Second, cell therapy based on germline stem cells for infertility needs to be further explored, including selection and induction of donor cell type/state, cell transplantation, and quality control. Human in vitro gametogenesis has not been reached yet, although complete in vitro meiosis from mESC-derived mPGCLCs has been reported in mice.337 Nevertheless, hPGCLCs are a promising source for gamete production in vitro in the future, and more importantly, they can be induced from somatic cells.23 Nevertheless, social and ethical issues concerning in vitro gametogenesis and following IVF using these germ cells should be seriously discussed before applications in the clinic. Third, fertility preservation has also become a pressing issue.483–485 Several factors, including gonadotoxic therapies, environmental exposures, aging, genetic diseases, and cancers, might cause subfertility or infertility.6,486,487 Germ cell transplantation is a promising strategy for both pubertal and prepubertal boys/girls diagnosed with cancers who will suffer from irradiation and chemotherapy,488–499 because these therapies often lead to damage to SSCs and FGSCs of the patients.483,500–510 It is also important to consider the purity of transplanted germ cells, and cancer cell contamination must be completely eliminated.511,512 Of course, fertility preservation raises several ethical issues,513–516 which should be carefully considered with discussion and debate. Fourth, understanding extragonadal germ cell tumors has guided us to consider the diagnosis and treatments of cancers, extragonadal cancers in particular. Most extragonadal germ cell tumors occur in many organs other than the testis and ovary, for example, brain cancers.517–523 These cancers have some features of hPGCs,524,525 thus hPGC markers should be used to diagnose and treat these cancers in the future. Actually, targeting the WNT signaling pathway for cancer therapy has been in preclinical testing and clinical trials.526–528 Lastly, genome editing in germline stem cells in humans is a very cautious approach because of its germline transmission to the next generation. Genome editing has been used to correct gene mutations in mice and mimic human genetic diseases,529–533 and correct pathogenic gene mutations in human embryos.534,535 CRISPR-edited T cells in patients with cancers have been tested in clinical trials.536–538 Genome editing technology in the targeted therapy has shown a promising prospect for human diseases,539,540 especially, in fertility restoration in cancer survivors and prevention of paternal transmission of diseases. Yet, there are several major obstacles to be overcome, including off-targets, social, and ethical issues. Technically, precise gene editing, for example, spatiotemporal control of CRISPR/Cas9 editing541 and non-viral strategy,542 will provide new hope in medical applications in the future.
Acknowledgements
We apologize to the many researchers whose work is not referenced due to space limitations. This work was supported by the National Key R&D Program of China (2019YFA0802500) and the National Natural Science Foundation of China (31970539, 31771487, and 31771370).
Author contributions
R.Z. and H.C.: the conceptualization and design of this study. D.S.: the data analysis, methodology, and writing of the manuscript. R.Z. and H.C.: the funding acquisition and project administration of this study. R.Z. and H.C.: writing—review and editing of the manuscript. All authors have read and approved the article.
Competing interests
The authors declare no competing interests.
Contributor Information
Hanhua Cheng, Email: hhcheng@whu.edu.cn.
Rongjia Zhou, Email: rjzhou@whu.edu.cn.
References
- 1.Weismann, A. The Germ-plasma: Theory of Heredity. (Scribner’s Sons, 1893).
- 2.Torre LA, et al. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018;68:284–296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J. Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 5.Zheng R, Zeng H, Zhang S, Chen W. Estimates of cancer incidence and mortality in China, 2013. Chin. J. Cancer. 2017;36:66. doi: 10.1186/s40880-017-0234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farquhar CM, et al. Female subfertility. Nat. Rev. Dis. Prim. 2019;5:7. doi: 10.1038/s41572-018-0058-8. [DOI] [PubMed] [Google Scholar]
- 7.Ulbright TM. Germ cell neoplasms of the testis. Am. J. Surg. Pathol. 1993;17:1075–1091. doi: 10.1097/00000478-199311000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Looijenga LH. Human testicular (non)seminomatous germ cell tumours: the clinical implications of recent pathobiological insights. J. Pathol. 2009;218:146–162. doi: 10.1002/path.2522. [DOI] [PubMed] [Google Scholar]
- 9.Rijlaarsdam MA, Looijenga LH. An oncofetal and developmental perspective on testicular germ cell cancer. Semin. Cancer Biol. 2014;29:59–74. doi: 10.1016/j.semcancer.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Ombelet W, Cooke I, Dyer S, Serour G, Devroey P. Infertility and the provision of infertility medical services in developing countries. Hum. Reprod. Update. 2008;14:605–621. doi: 10.1093/humupd/dmn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inhorn MC, Patrizio P. Infertility around the globe: new thinking on gender, reproductive technologies and global movements in the 21st century. Hum. Reprod. Update. 2015;21:411–426. doi: 10.1093/humupd/dmv016. [DOI] [PubMed] [Google Scholar]
- 12.Saitou M, Yamaji M. Primordial germ cells in mice. Cold Spring Harb. Perspect. Biol. 2012;4:a008375. doi: 10.1101/cshperspect.a008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohinata Y, et al. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137:571–584. doi: 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 14.Witschi E. Migration of the germ cells of human embryos from the yolk sac to the primitive gonadal folds. Contr. Embryol. Carne. Inst. 1948;32:67–80. [Google Scholar]
- 15.Molyneaux KA, Stallock J, Schaible K, Wylie C. Time-lapse analysis of living mouse germ cell migration. Dev. Biol. 2001;240:488–498. doi: 10.1006/dbio.2001.0436. [DOI] [PubMed] [Google Scholar]
- 16.Kanamori M, Oikawa K, Tanemura K, Hara K. Mammalian germ cell migration during development, growth, and homeostasis. Reprod. Med. Biol. 2019;18:247–255. doi: 10.1002/rmb2.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujimoto T, Miyayama Y, Fuyuta M. The origin, migration and fine morphology of human primordial germ cells. Anat. Rec. 1977;188:315–330. doi: 10.1002/ar.1091880305. [DOI] [PubMed] [Google Scholar]
- 18.Navas LE, Carnero A. NAD(+) metabolism, stemness, the immune response, and cancer. Signal Transduct. Target. Ther. 2021;6:2. doi: 10.1038/s41392-020-00354-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKay DG, Hertig AT, Adams EC, Danziger S. Histochemical observations on the germ cells of human embryos. Anat. Rec. 1953;117:201–219. doi: 10.1002/ar.1091170206. [DOI] [PubMed] [Google Scholar]
- 20.Irie N, et al. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–268. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sasaki K, et al. Robust In vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell. 2015;17:178–194. doi: 10.1016/j.stem.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 22.Sugawa F, et al. Human primordial germ cell commitment in vitro associates with a unique PRDM14 expression profile. EMBO J. 2015;34:1009–1024. doi: 10.15252/embj.201488049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitou M, Hayashi K. Mammalian in vitro gametogenesis. Science. 2021;374:eaaz6830. doi: 10.1126/science.aaz6830. [DOI] [PubMed] [Google Scholar]
- 24.Chen D, et al. Human primordial germ cells are specified from lineage-primed progenitors. Cell Rep. 2019;29:4568–4582. doi: 10.1016/j.celrep.2019.11.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou K, et al. Production of offspring from a germline stem cell line derived from neonatal ovaries. Nat. Cell Biol. 2009;11:631–636. doi: 10.1038/ncb1869. [DOI] [PubMed] [Google Scholar]
- 26.de Rooij DG. Stem cells in the testis. Int. J. Exp. Pathol. 1998;79:67–80. doi: 10.1046/j.1365-2613.1998.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brinster RL. Germline stem cell transplantation and transgenesis. Science. 2002;296:2174–2176. doi: 10.1126/science.1071607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikolic A, Volarevic V, Armstrong L, Lako M, Stojkovic M. Primordial germ cells: current knowledge and perspectives. Stem Cells Int. 2016;2016:1741072. doi: 10.1155/2016/1741072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goossens E, Van Saen D, Tournaye H. Spermatogonial stem cell preservation and transplantation: from research to clinic. Hum. Reprod. 2013;28:897–907. doi: 10.1093/humrep/det039. [DOI] [PubMed] [Google Scholar]
- 30.Shetty G, et al. Restoration of functional sperm production in irradiated pubertal rhesus monkeys by spermatogonial stem cell transplantation. Andrology. 2020;8:1428–1441. doi: 10.1111/andr.12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson LD, Hirshfield AN. An overview of follicular development in the ovary: from embryo to the fertilized ovum in vitro. Md. Med. J. 1992;41:614–620. [PubMed] [Google Scholar]
- 32.Borum K. Oogenesis in the mouse. A study of the meiotic prophase. Exp. Cell Res. 1961;24:495–507. doi: 10.1016/0014-4827(61)90449-9. [DOI] [PubMed] [Google Scholar]
- 33.Green SH, Zuckerman S. Further observations on oocyte numbers in mature rhesus monkeys (Macaca mulatta) J. Endocrinol. 1954;10:284–290. doi: 10.1677/joe.0.0100284. [DOI] [PubMed] [Google Scholar]
- 34.Johnson J, Canning J, Kaneko T, Pru JK, Tilly JL. Germline stem cells and follicular renewal in the postnatal mammalian ovary. Nature. 2004;428:145–150. doi: 10.1038/nature02316. [DOI] [PubMed] [Google Scholar]
- 35.Zhang C, Wu J. Production of offspring from a germline stem cell line derived from prepubertal ovaries of germline reporter mice. Mol. Hum. Reprod. 2016;22:457–464. doi: 10.1093/molehr/gaw030. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, et al. Retinoic acid induced meiosis initiation in female germline stem cells by remodelling three-dimensional chromatin structure. Cell Prolif. 2022;55:e13242. doi: 10.1111/cpr.13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhai J, Xiao Z, Wang Y, Wang H. Human embryonic development: from peri-implantation to gastrulation. Trends Cell Biol. 2022;32:18–29. doi: 10.1016/j.tcb.2021.07.008. [DOI] [PubMed] [Google Scholar]
- 38.Shahbazi MN. Mechanisms of human embryo development: from cell fate to tissue shape and back. Development. 2020;147:dev190629. doi: 10.1242/dev.190629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalthoff, K. Analysis of Biological Development. (McGRAW-Hill, INC., 1996).
- 40.Wolpert, L. Principles of Development. (Oxford University Press, 2002).
- 41.O’Rahilly R, Müller F. Developmental stages in human embryos: revised and new measurements. Cells Tissues Organs. 2010;192:73–84. doi: 10.1159/000289817. [DOI] [PubMed] [Google Scholar]
- 42.Tyser, R. C. V. & Srinivas, S. Recent advances in understanding cell types during human gastrulation. Semin. Cell Dev. Biol. 10.1016/j.semcdb.2022.05.004 (2022). [DOI] [PMC free article] [PubMed]
- 43.Rossant J, Tam PPL. Early human embryonic development: blastocyst formation to gastrulation. Dev. Cell. 2022;57:152–165. doi: 10.1016/j.devcel.2021.12.022. [DOI] [PubMed] [Google Scholar]
- 44.Haniffa M, et al. A roadmap for the human developmental cell atlas. Nature. 2021;597:196–205. doi: 10.1038/s41586-021-03620-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao Y, et al. Cell landscape of larval and adult Xenopus laevis at single-cell resolution. Nat. Commun. 2022;13:4306. doi: 10.1038/s41467-022-31949-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kataoka K, et al. Visualization of the Xenopus primordial germ cells using a green fluorescent protein controlled by cis elements of the 3’ untranslated region of the DEADSouth gene. Mech. Dev. 2006;123:746–760. doi: 10.1016/j.mod.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 47.Butler AM, Aguero T, Newman KM, King ML. Primordial germ cell isolation from Xenopus laevis embryos. Methods Mol. Biol. 2017;1463:115–124. doi: 10.1007/978-1-4939-4017-2_9. [DOI] [PubMed] [Google Scholar]
- 48.Blitz IL. Primordial germ cell transplantation for CRISPR/Cas9-based Leapfrogging in Xenopus. J. Vis. Exp. 2018;132:56035. doi: 10.3791/56035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oh D, Houston DW. Role of maternal Xenopus syntabulin in germ plasm aggregation and primordial germ cell specification. Dev. Biol. 2017;432:237–247. doi: 10.1016/j.ydbio.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J, Aguero T, King ML. The Xenopus maternal-to-zygotic transition from the perspective of the germline. Curr. Top. Dev. Biol. 2015;113:271–303. doi: 10.1016/bs.ctdb.2015.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tada H, Mochii M, Orii H, Watanabe K. Ectopic formation of primordial germ cells by transplantation of the germ plasm: direct evidence for germ cell determinant in Xenopus. Dev. Biol. 2012;371:86–93. doi: 10.1016/j.ydbio.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 52.Butler AM, Owens DA, Wang L, King ML. A novel role for sox7 in Xenopus early primordial germ cell development: mining the PGC transcriptome. Development. 2018;145:dev155978. doi: 10.1242/dev.155978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tada H, Taira Y, Morichika K, Kinoshita T. Mitochondrial trafficking through Rhot1 is involved in the aggregation of germinal granule components during primordial germ cell formation in Xenopus embryos. Dev. Growth Differ. 2016;58:641–650. doi: 10.1111/dgd.12310. [DOI] [PubMed] [Google Scholar]
- 54.Kodama M, et al. Nanos3 of the frog Rana rugosa: molecular cloning and characterization. Dev. Growth Differ. 2018;60:112–120. doi: 10.1111/dgd.12421. [DOI] [PubMed] [Google Scholar]
- 55.Sekizaki H, et al. Tracing of Xenopus tropicalis germ plasm and presumptive primordial germ cells with the Xenopus tropicalis DAZ-like gene. Dev. Dyn. 2004;229:367–372. doi: 10.1002/dvdy.10448. [DOI] [PubMed] [Google Scholar]
- 56.Horvay K, Claussen M, Katzer M, Landgrebe J, Pieler T. Xenopus Dead end mRNA is a localized maternal determinant that serves a conserved function in germ cell development. Dev. Biol. 2006;291:1–11. doi: 10.1016/j.ydbio.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 57.Houston DW, King ML. A critical role for Xdazl, a germ plasm-localized RNA, in the differentiation of primordial germ cells in Xenopus. Development. 2000;127:447–456. doi: 10.1242/dev.127.3.447. [DOI] [PubMed] [Google Scholar]
- 58.Yamaguchi T, Taguchi A, Watanabe K, Orii H. Germes is involved in translocation of germ plasm during development of Xenopus primordial germ cells. Int. J. Dev. Biol. 2013;57:439–443. doi: 10.1387/ijdb.120215ty. [DOI] [PubMed] [Google Scholar]
- 59.Aalto A, Olguin-Olguin A, Raz E. Zebrafish primordial germ cell migration. Front. Cell Dev. Biol. 2021;9:684460. doi: 10.3389/fcell.2021.684460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Braat AK, Speksnijder JE, Zivkovic D. Germ line development in fishes. Int. J. Dev. Biol. 1999;43:745–760. [PubMed] [Google Scholar]
- 61.Olsen LC, Aasland R, Fjose A. A vasa-like gene in zebrafish identifies putative primordial germ cells. Mech. Dev. 1997;66:95–105. doi: 10.1016/S0925-4773(97)00099-3. [DOI] [PubMed] [Google Scholar]
- 62.Yoon C, Kawakami K, Hopkins N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 1997;124:3157–3165. doi: 10.1242/dev.124.16.3157. [DOI] [PubMed] [Google Scholar]
- 63.Zhang X, et al. Transcriptomic profile of early zebrafish PGCs by single cell sequencing. PLoS ONE. 2019;14:e0220364. doi: 10.1371/journal.pone.0220364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Riesco MF, Valcarce DG, Alfonso J, Herráez MP, Robles V. In vitro generation of zebrafish PGC-like cells. Biol. Reprod. 2014;91:114. doi: 10.1095/biolreprod.114.121491. [DOI] [PubMed] [Google Scholar]
- 65.Jin Y, et al. Maternal miR-202-5p is required for zebrafish primordial germ cell migration by protecting small GTPase Cdc42. J. Mol. Cell Biol. 2020;12:530–542. doi: 10.1093/jmcb/mjz103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li M, et al. IGF-2 mRNA binding protein 2 regulates primordial germ cell development in zebrafish. Gen. Comp. Endocrinol. 2021;313:113875. doi: 10.1016/j.ygcen.2021.113875. [DOI] [PubMed] [Google Scholar]
- 67.Raz E. Primordial germ-cell development: the zebrafish perspective. Nat. Rev. Genet. 2003;4:690–700. doi: 10.1038/nrg1154. [DOI] [PubMed] [Google Scholar]
- 68.Fan L, Moon J, Wong TT, Crodian J, Collodi P. Zebrafish primordial germ cell cultures derived from vasa::RFP transgenic embryos. Stem Cells Dev. 2008;17:585–597. doi: 10.1089/scd.2007.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paksa A, Raz E. Zebrafish germ cells: motility and guided migration. Curr. Opin. Cell Biol. 2015;36:80–85. doi: 10.1016/j.ceb.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 70.Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- 71.Tsunekawa N, Naito M, Sakai Y, Nishida T, Noce T. Isolation of chicken vasa homolog gene and tracing the origin of primordial germ cells. Development. 2000;127:2741–2750. doi: 10.1242/dev.127.12.2741. [DOI] [PubMed] [Google Scholar]
- 72.Tagami T, Miyahara D, Nakamura Y. Avian primordial germ cells. Adv. Exp. Med. Biol. 2017;1001:1–18. doi: 10.1007/978-981-10-3975-1_1. [DOI] [PubMed] [Google Scholar]
- 73.Kim YM, Han JY. The early development of germ cells in chicken. Int. J. Dev. Biol. 2018;62:145–152. doi: 10.1387/ijdb.170283jh. [DOI] [PubMed] [Google Scholar]
- 74.Ginsburg M. Primordial germ cell formation in birds. Ciba Found. Symp. 1994;182:52–61. doi: 10.1002/9780470514573.ch4. [DOI] [PubMed] [Google Scholar]
- 75.Nakamura Y, Kagami H, Tagami T. Development, differentiation and manipulation of chicken germ cells. Dev. Growth Differ. 2013;55:20–40. doi: 10.1111/dgd.12026. [DOI] [PubMed] [Google Scholar]
- 76.Zhao R, et al. Production of viable chicken by allogeneic transplantation of primordial germ cells induced from somatic cells. Nat. Commun. 2021;12:2989. doi: 10.1038/s41467-021-23242-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 77.Chen D, et al. GSK-3 signaling is involved in proliferation of chicken primordial germ cells. Theriogenology. 2020;141:62–67. doi: 10.1016/j.theriogenology.2019.09.002. [DOI] [PubMed] [Google Scholar]
- 78.Chen D, et al. Cholesterol induces proliferation of chicken primordial germ cells. Anim. Reprod. Sci. 2016;171:36–40. doi: 10.1016/j.anireprosci.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 79.Jin SD, et al. Regulatory elements and transcriptional control of chicken vasa homologue (CVH) promoter in chicken primordial germ cells. J. Anim. Sci. Biotechnol. 2017;8:6. doi: 10.1186/s40104-016-0133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zuo Q, et al. BMP4 activates the Wnt-Lin28A-Blimp1-Wnt pathway to promote primordial germ cell formation via altering H3K4me2. J. Cell Sci. 2021;134:jcs.249375. doi: 10.1242/jcs.249375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Z, et al. Crucial genes and pathways in chicken germ stem cell differentiation. J. Biol. Chem. 2015;290:13605–13621. doi: 10.1074/jbc.M114.601401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jung HG, et al. Role of epigenetic regulation by the REST/CoREST/HDAC corepressor complex of moderate NANOG expression in chicken primordial germ cells. Stem Cells Dev. 2018;27:1215–1225. doi: 10.1089/scd.2018.0059. [DOI] [PubMed] [Google Scholar]
- 83.Ginsburg M, Snow MH, McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development. 1990;110:521–528. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- 84.Chiquoine AD. The identification, origin, and migration of the primordial germ cells in the mouse embryo. Anat. Rec. 1954;118:135–146. doi: 10.1002/ar.1091180202. [DOI] [PubMed] [Google Scholar]
- 85.Ohinata Y, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 86.Hancock GV, Wamaitha SE, Peretz L, Clark AT. Mammalian primordial germ cell specification. Development. 2021;148:dev189217. doi: 10.1242/dev.189217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sasaki K, et al. The germ cell fate of Cynomolgus monkeys is specified in the nascent amnion. Dev. Cell. 2016;39:169–185. doi: 10.1016/j.devcel.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 88.Kobayashi T, et al. Tracing the emergence of primordial germ cells from bilaminar disc rabbit embryos and pluripotent stem cells. Cell Rep. 2021;37:109812. doi: 10.1016/j.celrep.2021.109812. [DOI] [PubMed] [Google Scholar]
- 89.Sharma S, et al. Male germline stem cells in non-human primates. Primate Biol. 2017;4:173–184. doi: 10.5194/pb-4-173-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sosa E, et al. Differentiation of primate primordial germ cell-like cells following transplantation into the adult gonadal niche. Nat. Commun. 2018;9:5339. doi: 10.1038/s41467-018-07740-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Clark AT, et al. Primate primordial germ cells acquire transplantation potential by carnegie stage 23. Stem Cell Rep. 2017;9:329–341. doi: 10.1016/j.stemcr.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sakai Y, et al. Induction of the germ cell fate from pluripotent stem cells in cynomolgus monkeys. Biol. Reprod. 2020;102:620–638. doi: 10.1093/biolre/ioz205. [DOI] [PubMed] [Google Scholar]
- 93.Zhang P, et al. Mapping developmental paths of monkey primordial germ-like cells differentiation from pluripotent stem cells by single cell ribonucleic acid sequencing analysis†. Biol. Reprod. 2022;107:237–249. doi: 10.1093/biolre/ioac133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Teramura T, et al. Primate embryonic stem cells proceed to early gametogenesis in vitro. Cloning Stem Cells. 2007;9:144–156. doi: 10.1089/clo.2006.0070. [DOI] [PubMed] [Google Scholar]
- 95.Niu Y, et al. Dissecting primate early post-implantation development using long-term in vitro embryo culture. Science. 2019;366:aaw5754. doi: 10.1126/science.aaw5754. [DOI] [PubMed] [Google Scholar]
- 96.Lawson KA, et al. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ishikura Y, et al. In vitro reconstitution of the whole male germ-cell development from mouse pluripotent stem cells. Cell Stem Cell. 2021;28:2167–2179. doi: 10.1016/j.stem.2021.08.005. [DOI] [PubMed] [Google Scholar]
- 98.Hayashi K, et al. SMAD1 signaling is critical for initial commitment of germ cell lineage from mouse epiblast. Mech. Dev. 2002;118:99–109. doi: 10.1016/S0925-4773(02)00237-X. [DOI] [PubMed] [Google Scholar]
- 99.Yu L, et al. Derivation of intermediate pluripotent stem cells amenable to primordial germ cell specification. Cell Stem Cell. 2021;28:550–567. doi: 10.1016/j.stem.2020.11.003. [DOI] [PubMed] [Google Scholar]
- 100.Tam PP, Zhou SX. The allocation of epiblast cells to ectodermal and germ-line lineages is influenced by the position of the cells in the gastrulating mouse embryo. Dev. Biol. 1996;178:124–132. doi: 10.1006/dbio.1996.0203. [DOI] [PubMed] [Google Scholar]
- 101.Saitou M, Barton SC, Surani MA. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- 102.Kurimoto K, et al. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 2008;22:1617–1635. doi: 10.1101/gad.1649908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Weber S, et al. Critical function of AP-2 gamma/TCFAP2C in mouse embryonic germ cell maintenance. Biol. Reprod. 2010;82:214–223. doi: 10.1095/biolreprod.109.078717. [DOI] [PubMed] [Google Scholar]
- 104.Zhao GQ. Consequences of knocking out BMP signaling in the mouse. Genesis. 2003;35:43–56. doi: 10.1002/gene.10167. [DOI] [PubMed] [Google Scholar]
- 105.Senft AD, Bikoff EK, Robertson EJ, Costello I. Genetic dissection of Nodal and Bmp signalling requirements during primordial germ cell development in mouse. Nat. Commun. 2019;10:1089. doi: 10.1038/s41467-019-09052-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.de Sousa Lopes SM, et al. BMP signaling mediated by ALK2 in the visceral endoderm is necessary for the generation of primordial germ cells in the mouse embryo. Genes Dev. 2004;18:1838–1849. doi: 10.1101/gad.294004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ying Y, Zhao GQ. Cooperation of endoderm-derived BMP2 and extraembryonic ectoderm-derived BMP4 in primordial germ cell generation in the mouse. Dev. Biol. 2001;232:484–492. doi: 10.1006/dbio.2001.0173. [DOI] [PubMed] [Google Scholar]
- 108.Ying Y, Liu XM, Marble A, Lawson KA, Zhao GQ. Requirement of Bmp8b for the generation of primordial germ cells in the mouse. Mol. Endocrinol. 2000;14:1053–1063. doi: 10.1210/mend.14.7.0479. [DOI] [PubMed] [Google Scholar]
- 109.Kobayashi T, et al. Principles of early human development and germ cell program from conserved model systems. Nature. 2017;546:416–420. doi: 10.1038/nature22812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Takagi Y, Talbot NC, Rexroad CE, Jr., Pursel VG. Identification of pig primordial germ cells by immunocytochemistry and lectin binding. Mol. Reprod. Dev. 1997;46:567–580. doi: 10.1002/(SICI)1098-2795(199704)46:4<567::AID-MRD14>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 111.Zhang MY, et al. The proliferation role of LH on porcine primordial germ cell-like cells (pPGCLCs) through ceRNA network construction. Clin. Transl. Med. 2021;11:e560. doi: 10.1002/ctm2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang H, et al. Induction of germ cell-like cells from porcine induced pluripotent stem cells. Sci. Rep. 2016;6:27256. doi: 10.1038/srep27256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yan HC, et al. RA promotes proliferation of primordial germ cell-like cells differentiated from porcine skin-derived stem cells. J. Cell Physiol. 2019;234:18214–18229. doi: 10.1002/jcp.28454. [DOI] [PubMed] [Google Scholar]
- 114.Pieri NCG, et al. Porcine primordial germ cell-like cells generated from induced pluripotent stem cells under different culture conditions. Stem Cell Rev. Rep. 2022;18:1639–1656. doi: 10.1007/s12015-021-10198-8. [DOI] [PubMed] [Google Scholar]
- 115.Klisch K, et al. The Sda/GM2-glycan is a carbohydrate marker of porcine primordial germ cells and of a subpopulation of spermatogonia in cattle, pigs, horses and llama. Reproduction. 2011;142:667–674. doi: 10.1530/REP-11-0007. [DOI] [PubMed] [Google Scholar]
- 116.Petkov SG, Reh WA, Anderson GB. Methylation changes in porcine primordial germ cells. Mol. Reprod. Dev. 2009;76:22–30. doi: 10.1002/mrd.20926. [DOI] [PubMed] [Google Scholar]
- 117.Hyldig SM, Ostrup O, Vejlsted M, Thomsen PD. Changes of DNA methylation level and spatial arrangement of primordial germ cells in embryonic day 15 to embryonic day 28 pig embryos. Biol. Reprod. 2011;84:1087–1093. doi: 10.1095/biolreprod.110.086082. [DOI] [PubMed] [Google Scholar]
- 118.Liu L, Moor RM, Laurie S, Notarianni E. Nuclear remodelling and early development in cryopreserved, porcine primordial germ cells following nuclear transfer into in vitro-matured oocytes. Int. J. Dev. Biol. 1995;39:639–644. [PubMed] [Google Scholar]
- 119.Zuo Q, et al. Dual regulatory actions of LncBMP4 on BMP4 promote chicken primordial germ cell formation. EMBO Rep. 2022;23:e52491. doi: 10.15252/embr.202152491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dunislawska A, Szczerba A, Siwek M, Bednarczyk M. Dynamics of the transcriptome during chicken embryo development based on primordial germ cells. BMC Res. Notes. 2020;13:441. doi: 10.1186/s13104-020-05286-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Choi HJ, et al. Differential transcriptional regulation of the NANOG gene in chicken primordial germ cells and embryonic stem cells. J. Anim. Sci. Biotechnol. 2021;12:40. doi: 10.1186/s40104-021-00563-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Aramaki S, Kubota K, Soh T, Yamauchi N, Hattori MA. Chicken dead end homologue protein is a nucleoprotein of germ cells including primordial germ cells. J. Reprod. Dev. 2009;55:214–218. doi: 10.1262/jrd.20154. [DOI] [PubMed] [Google Scholar]
- 123.Zhang C, et al. Narrow H3K4me2 is required for chicken PGC formation. J. Cell Physiol. 2021;236:1391–1400. doi: 10.1002/jcp.29945. [DOI] [PubMed] [Google Scholar]
- 124.Jung JG, et al. Development of novel markers for the characterization of chicken primordial germ cells. Stem Cells. 2005;23:689–698. doi: 10.1634/stemcells.2004-0208. [DOI] [PubMed] [Google Scholar]
- 125.Yu M, Ge C, Zeng W, Mi Y, Zhang C. Retinoic acid promotes proliferation of chicken primordial germ cells via activation of PI3K/Akt-mediated NF-κB signalling cascade. Cell Biol. Int. 2012;36:705–712. doi: 10.1042/CBI20110542. [DOI] [PubMed] [Google Scholar]
- 126.Li D, et al. Hedgehog-Gli1 signaling regelates differentiation of chicken (Gallus gallus) embryonic stem cells to male germ cells. Anim. Reprod. Sci. 2017;182:9–20. doi: 10.1016/j.anireprosci.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 127.Skvortsova K, et al. Retention of paternal DNA methylome in the developing zebrafish germline. Nat. Commun. 2019;10:3054. doi: 10.1038/s41467-019-10895-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li Z, Li M, Hong N, Yi M, Hong Y. Formation and cultivation of medaka primordial germ cells. Cell Tissue Res. 2014;357:71–81. doi: 10.1007/s00441-014-1867-z. [DOI] [PubMed] [Google Scholar]
- 129.Hong N, et al. Dnd Is a critical specifier of primordial germ cells in the medaka fish. Stem Cell Rep. 2016;6:411–421. doi: 10.1016/j.stemcr.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang X, Bhandari RK. The dynamics of DNA methylation during epigenetic reprogramming of primordial germ cells in medaka (Oryzias latipes) Epigenetics. 2020;15:483–498v. doi: 10.1080/15592294.2019.1695341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Song P, et al. Bucky ball induces primordial germ cell increase in medaka. Gene. 2021;768:145317. doi: 10.1016/j.gene.2020.145317. [DOI] [PubMed] [Google Scholar]
- 132.Herpin A, et al. Specification of primordial germ cells in medaka (Oryzias latipes) BMC Dev. Biol. 2007;7:3. doi: 10.1186/1471-213X-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li M, Zhu F, Li Z, Hong N, Hong Y. Dazl is a critical player for primordial germ cell formation in medaka. Sci. Rep. 2016;6:28317. doi: 10.1038/srep28317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li M, et al. Medaka vasa is required for migration but not survival of primordial germ cells. Mech. Dev. 2009;126:366–381. doi: 10.1016/j.mod.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 135.Herpin A, et al. Inhibition of primordial germ cell proliferation by the medaka male determining gene Dmrt I bY. BMC Dev. Biol. 2007;7:99. doi: 10.1186/1471-213X-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Herpin A, et al. A novel evolutionary conserved mechanism of RNA stability regulates synexpression of primordial germ cell-specific genes prior to the sex-determination stage in medaka. PLoS Biol. 2019;17:e3000185. doi: 10.1371/journal.pbio.3000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Porras-Gómez TJ, Villagrán-SantaCruz M, Moreno-Mendoza N. Biology of primordial germ cells in vertebrates with emphasis in urodeles amphibians. Mol. Reprod. Dev. 2021;88:773–792. doi: 10.1002/mrd.23533. [DOI] [PubMed] [Google Scholar]
- 138.Banisch TU, et al. A transitory signaling center controls timing of primordial germ cell differentiation. Dev. Cell. 2021;56:1742–1755. doi: 10.1016/j.devcel.2021.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Colonnetta MM, et al. Preformation and epigenesis converge to specify primordial germ cell fate in the early Drosophila embryo. PLoS Genet. 2022;18:e1010002. doi: 10.1371/journal.pgen.1010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fang J, Lerit DA. Drosophila pericentrin-like protein promotes the formation of primordial germ cells. Genesis. 2020;58:e23347. doi: 10.1002/dvg.23347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kistler KE, et al. Phase transitioned nuclear Oskar promotes cell division of Drosophila primordial germ cells. Elife. 2018;7:e37949. doi: 10.7554/eLife.37949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Syal S, et al. Reactive oxygen species signaling in primordial germ cell development in Drosophila embryos. Genesis. 2020;58:e23362. doi: 10.1002/dvg.23362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, Nakamura A. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature. 2008;451:730–733. doi: 10.1038/nature06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sato T, Ueda S, Niki Y. Wingless signaling initiates mitosis of primordial germ cells during development in Drosophila. Mech. Dev. 2008;125:498–507. doi: 10.1016/j.mod.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 145.Jones J, Macdonald PM. Neurl4 contributes to germ cell formation and integrity in Drosophila. Biol. Open. 2015;4:937–946. doi: 10.1242/bio.012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Deshpande G, et al. BMP signaling and the maintenance of primordial germ cell identity in Drosophila embryos. PLoS ONE. 2014;9:e88847. doi: 10.1371/journal.pone.0088847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fry AL, et al. DAF-18/PTEN inhibits germline zygotic gene activation during primordial germ cell quiescence. PLoS Genet. 2021;17:e1009650. doi: 10.1371/journal.pgen.1009650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mainpal R, Nance J, Yanowitz JL. A germ cell determinant reveals parallel pathways for germ line development in Caenorhabditis elegans. Development. 2015;142:3571–3582. doi: 10.1242/dev.125732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Andralojc KM, et al. ELLI-1, a novel germline protein, modulates RNAi activity and P-granule accumulation in Caenorhabditis elegans. PLoS Genet. 2017;13:e1006611. doi: 10.1371/journal.pgen.1006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Guven-Ozkan T, Robertson SM, Nishi Y, Lin R. zif-1 translational repression defines a second, mutually exclusive OMA function in germline transcriptional repression. Development. 2010;137:3373–3382. doi: 10.1242/dev.055327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Subramaniam K, Seydoux G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development. 1999;126:4861–4871. doi: 10.1242/dev.126.21.4861. [DOI] [PubMed] [Google Scholar]
- 152.Fujita M, et al. MRG-1, a mortality factor-related chromodomain protein, is required maternally for primordial germ cells to initiate mitotic proliferation in C. elegans. Mech. Dev. 2002;114:61–69. doi: 10.1016/S0925-4773(02)00058-8. [DOI] [PubMed] [Google Scholar]
- 153.Jadhav S, Rana M, Subramaniam K. Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells. Development. 2008;135:1803–1812. doi: 10.1242/dev.013656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Furuhashi H, et al. Trans-generational epigenetic regulation of C. elegans primordial germ cells. Epigenetics Chromatin. 2010;3:15. doi: 10.1186/1756-8935-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Miwa T, Inoue K, Sakamoto H. MRG-1 is required for both chromatin-based transcriptional silencing and genomic integrity of primordial germ cells in Caenorhabditis elegans. Genes Cells. 2019;24:377–389. doi: 10.1111/gtc.12683. [DOI] [PubMed] [Google Scholar]
- 156.Lebedeva LA, et al. Transcriptional quiescence in primordial germ cells. Crit. Rev. Biochem. Mol. Biol. 2018;53:579–595. doi: 10.1080/10409238.2018.1506733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tang WW, Kobayashi T, Irie N, Dietmann S, Surani MA. Specification and epigenetic programming of the human germ line. Nat. Rev. Genet. 2016;17:585–600. doi: 10.1038/nrg.2016.88. [DOI] [PubMed] [Google Scholar]
- 158.Perrett RM, et al. The early human germ cell lineage does not express SOX2 during in vivo development or upon in vitro culture. Biol. Reprod. 2008;78:852–858. doi: 10.1095/biolreprod.107.066175. [DOI] [PubMed] [Google Scholar]
- 159.Zhang S, Xiong X, Sun Y. Functional characterization of SOX2 as an anticancer target. Signal Transduct. Target. Ther. 2020;5:135. doi: 10.1038/s41392-020-00242-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bergmann S, et al. Spatial profiling of early primate gastrulation in utero. Nature. 2022;609:136–143. doi: 10.1038/s41586-022-04953-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tyser RCV, et al. Single-cell transcriptomic characterization of a gastrulating human embryo. Nature. 2021;600:285–289. doi: 10.1038/s41586-021-04158-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gaskell TL, Esnal A, Robinson LL, Anderson RA, Saunders PT. Immunohistochemical profiling of germ cells within the human fetal testis: identification of three subpopulations. Biol. Reprod. 2004;71:2012–2021. doi: 10.1095/biolreprod.104.028381. [DOI] [PubMed] [Google Scholar]
- 163.Kerr CL, Hill CM, Blumenthal PD, Gearhart JD. Expression of pluripotent stem cell markers in the human fetal ovary. Hum. Reprod. 2008;23:589–599. doi: 10.1093/humrep/dem411. [DOI] [PubMed] [Google Scholar]
- 164.Kerr CL, Hill CM, Blumenthal PD, Gearhart JD. Expression of pluripotent stem cell markers in the human fetal testis. Stem Cells. 2008;26:412–421. doi: 10.1634/stemcells.2007-0605. [DOI] [PubMed] [Google Scholar]
- 165.Childs AJ, Kinnell HL, He J, Anderson RA. LIN28 is selectively expressed by primordial and pre-meiotic germ cells in the human fetal ovary. Stem Cells Dev. 2012;21:2343–2349. doi: 10.1089/scd.2011.0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gillis AJ, et al. Expression and interdependencies of pluripotency factors LIN28, OCT3/4, NANOG and SOX2 in human testicular germ cells and tumours of the testis. Int. J. Androl. 2011;34:e160–e174. doi: 10.1111/j.1365-2605.2011.01148.x. [DOI] [PubMed] [Google Scholar]
- 167.Guo F, et al. The Transcriptome and DNA methylome landscapes of human primordial germ cells. Cell. 2015;161:1437–1452. doi: 10.1016/j.cell.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 168.Chitiashvili T, Hsu FM, Dror I, Plath K, Clark A. FGFR3 is expressed by human primordial germ cells and is repressed after meiotic initiation to form primordial oocytes. Stem Cell Rep. 2022;17:1268–1278. doi: 10.1016/j.stemcr.2022.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Castrillon DH, Quade BJ, Wang TY, Quigley C, Crum CP. The human VASA gene is specifically expressed in the germ cell lineage. Proc. Natl Acad. Sci. USA. 2000;97:9585–9590. doi: 10.1073/pnas.160274797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Anderson RA, Fulton N, Cowan G, Coutts S, Saunders PT. Conserved and divergent patterns of expression of DAZL, VASA and OCT4 in the germ cells of the human fetal ovary and testis. BMC Dev. Biol. 2007;7:136. doi: 10.1186/1471-213X-7-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tang WW, et al. A unique gene regulatory network resets the human germline epigenome for development. Cell. 2015;161:1453–1467. doi: 10.1016/j.cell.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sybirna A, et al. A critical role of PRDM14 in human primordial germ cell fate revealed by inducible degrons. Nat. Commun. 2020;11:1282. doi: 10.1038/s41467-020-15042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pierson Smela M, Sybirna A, Wong FCK, Surani MA. Testing the role of SOX15 in human primordial germ cell fate. Wellcome Open Res. 2019;4:122. doi: 10.12688/wellcomeopenres.15381.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Magnúsdóttir E, et al. A tripartite transcription factor network regulates primordial germ cell specification in mice. Nat. Cell Biol. 2013;15:905–915. doi: 10.1038/ncb2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Garcia-Alonso L, et al. Single-cell roadmap of human gonadal development. Nature. 2022;607:540–547. doi: 10.1038/s41586-022-04918-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Eckert D, et al. Expression of BLIMP1/PRMT5 and concurrent histone H2A/H4 arginine 3 dimethylation in fetal germ cells, CIS/IGCNU and germ cell tumors. BMC Dev. Biol. 2008;8:106. doi: 10.1186/1471-213X-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mall EM, et al. Heading towards a dead end: the role of DND1 in germ line differentiation of human iPSCs. PLoS ONE. 2021;16:e0258427. doi: 10.1371/journal.pone.0258427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Torres-Fernández LA, et al. TRIM71 deficiency causes germ cell loss during mouse embryogenesis and is associated with human male infertility. Front. Cell Dev. Biol. 2021;9:658966. doi: 10.3389/fcell.2021.658966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yin Y, et al. A noncanonical role of NOD-like receptor NLRP14 in PGCLC differentiation and spermatogenesis. Proc. Natl Acad. Sci. USA. 2020;117:22237–22248. doi: 10.1073/pnas.2005533117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Beppu H, et al. BMP type II receptor is required for gastrulation and early development of mouse embryos. Dev. Biol. 2000;221:249–258. doi: 10.1006/dbio.2000.9670. [DOI] [PubMed] [Google Scholar]
- 181.Magnúsdóttir E, Surani MA. How to make a primordial germ cell. Development. 2014;141:245–252. doi: 10.1242/dev.098269. [DOI] [PubMed] [Google Scholar]
- 182.Hiller M, Liu C, Blumenthal PD, Gearhart JD, Kerr CL. Bone morphogenetic protein 4 mediates human embryonic germ cell derivation. Stem Cells Dev. 2011;20:351–361. doi: 10.1089/scd.2010.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kemp C, Willems E, Abdo S, Lambiv L, Leyns L. Expression of all Wnt genes and their secreted antagonists during mouse blastocyst and postimplantation development. Dev. Dyn. 2005;233:1064–1075. doi: 10.1002/dvdy.20408. [DOI] [PubMed] [Google Scholar]
- 184.Xiao L, Yuan X, Sharkis SJ. Activin A maintains self-renewal and regulates fibroblast growth factor, Wnt, and bone morphogenic protein pathways in human embryonic stem cells. Stem Cells. 2006;24:1476–1486. doi: 10.1634/stemcells.2005-0299. [DOI] [PubMed] [Google Scholar]
- 185.Mishra S, et al. Activin A-derived human embryonic stem cells show increased competence to differentiate into primordial germ cell-like cells. Stem Cells. 2021;39:551–563. doi: 10.1002/stem.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Duggal G, et al. Influence of activin A supplementation during human embryonic stem cell derivation on germ cell differentiation potential. Stem Cells Dev. 2013;22:3141–3155. doi: 10.1089/scd.2013.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yang R, et al. Amnion signals are essential for mesoderm formation in primates. Nat. Commun. 2021;12:5126. doi: 10.1038/s41467-021-25186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kanai Y, et al. Identification of two Sox17 messenger RNA isoforms, with and without the high mobility group box region, and their differential expression in mouse spermatogenesis. J. Cell Biol. 1996;133:667–681. doi: 10.1083/jcb.133.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wang R, et al. Molecular cloning and expression of Sox17 in gonads during sex reversal in the rice field eel, a teleost fish with a characteristic of natural sex transformation. Biochem. Biophys. Res. Commun. 2003;303:452–457. doi: 10.1016/S0006-291X(03)00361-9. [DOI] [PubMed] [Google Scholar]
- 190.Kanai-Azuma M, et al. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development. 2002;129:2367–2379. doi: 10.1242/dev.129.10.2367. [DOI] [PubMed] [Google Scholar]
- 191.Kojima Y, et al. Evolutionarily distinctive transcriptional and signaling programs drive human germ cell lineage specification from pluripotent stem cells. Cell Stem Cell. 2017;21:517–532. doi: 10.1016/j.stem.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 192.Chen D, et al. Germline competency of human embryonic stem cells depends on eomesodermin. Biol. Reprod. 2017;97:850–861. doi: 10.1093/biolre/iox138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Arnold SJ, Hofmann UK, Bikoff EK, Robertson EJ. Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development. 2008;135:501–511. doi: 10.1242/dev.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Aramaki S, et al. A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev. Cell. 2013;27:516–529. doi: 10.1016/j.devcel.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 195.Kojima Y, et al. GATA transcription factors, SOX17 and TFAP2C, drive the human germ-cell specification program. Life Sci. Alliance. 2021;4:e202000974. doi: 10.26508/lsa.202000974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.de Souza FS, et al. The zinc finger gene Xblimp1 controls anterior endomesodermal cell fate in Spemann’s organizer. EMBO J. 1999;18:6062–6072. doi: 10.1093/emboj/18.21.6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bao S, et al. The germ cell determinant Blimp1 is not required for derivation of pluripotent stem cells. Cell Stem Cell. 2012;11:110–117. doi: 10.1016/j.stem.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kurimoto K, Saitou M. Mechanism and reconstitution in vitro of germ cell development in mammals. Cold Spring Harb. Symp. Quant. Biol. 2015;80:147–154. doi: 10.1101/sqb.2015.80.027425. [DOI] [PubMed] [Google Scholar]
- 199.Pastor WA, et al. TFAP2C regulates transcription in human naive pluripotency by opening enhancers. Nat. Cell Biol. 2018;20:553–564. doi: 10.1038/s41556-018-0089-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chen D, et al. The TFAP2C-regulated OCT4 naive enhancer is involved in human germline formation. Cell Rep. 2018;25:3591–3602. doi: 10.1016/j.celrep.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Schemmer J, et al. Transcription factor TFAP2C regulates major programs required for murine fetal germ cell maintenance and haploinsufficiency predisposes to teratomas in male mice. PLoS ONE. 2013;8:e71113. doi: 10.1371/journal.pone.0071113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Burton A, et al. Single-cell profiling of epigenetic modifiers identifies PRDM14 as an inducer of cell fate in the mammalian embryo. Cell Rep. 2013;5:687–701. doi: 10.1016/j.celrep.2013.09.044. [DOI] [PubMed] [Google Scholar]
- 203.Yamaji M, et al. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 2008;40:1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- 204.Tsuneyoshi N, et al. PRDM14 suppresses expression of differentiation marker genes in human embryonic stem cells. Biochem. Biophys. Res. Commun. 2008;367:899–905. doi: 10.1016/j.bbrc.2007.12.189. [DOI] [PubMed] [Google Scholar]
- 205.Chan YS, et al. A PRC2-dependent repressive role of PRDM14 in human embryonic stem cells and induced pluripotent stem cell reprogramming. Stem Cells. 2013;31:682–692. doi: 10.1002/stem.1307. [DOI] [PubMed] [Google Scholar]
- 206.Yamaji M, et al. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell. 2013;12:368–382. doi: 10.1016/j.stem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 207.Yamamoto M, et al. The PRDM14-CtBP1/2-PRC2 complex regulates transcriptional repression during the transition from primed to naïve pluripotency. J. Cell Sci. 2020;133:jcs240176. doi: 10.1242/jcs.240176. [DOI] [PubMed] [Google Scholar]
- 208.Payer B, et al. Tsix RNA and the germline factor, PRDM14, link X reactivation and stem cell reprogramming. Mol. Cell. 2013;52:805–818. doi: 10.1016/j.molcel.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Grabole N, et al. Prdm14 promotes germline fate and naive pluripotency by repressing FGF signalling and DNA methylation. EMBO Rep. 2013;14:629–637. doi: 10.1038/embor.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kurimoto K, Yamaji M, Seki Y, Saitou M. Specification of the germ cell lineage in mice: a process orchestrated by the PR-domain proteins, Blimp1 and Prdm14. Cell Cycle. 2008;7:3514–351. doi: 10.4161/cc.7.22.6979. [DOI] [PubMed] [Google Scholar]
- 211.Okashita N, et al. PRDM14 drives OCT3/4 recruitment via active demethylation in the transition from primed to naive pluripotency. Stem Cell Rep. 2016;7:1072–1086. doi: 10.1016/j.stemcr.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mallol A, Guirola M, Payer B. PRDM14 controls X-chromosomal and global epigenetic reprogramming of H3K27me3 in migrating mouse primordial germ cells. Epigenetics Chromatin. 2019;12:38. doi: 10.1186/s13072-019-0284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kobayashi T, et al. Germline development in rat revealed by visualization and deletion of Prdm14. Development. 2020;147:dev183798. doi: 10.1242/dev.183798. [DOI] [PubMed] [Google Scholar]
- 214.Okuzaki Y, et al. PRDM14 and BLIMP1 control the development of chicken primordial germ cells. Dev. Biol. 2019;455:32–41. doi: 10.1016/j.ydbio.2019.06.018. [DOI] [PubMed] [Google Scholar]
- 215.Seki Y. PRDM14 is a unique epigenetic regulator stabilizing transcriptional networks for pluripotency. Front. Cell Dev. Biol. 2018;6:12. doi: 10.3389/fcell.2018.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Nakaki F, et al. Induction of mouse germ-cell fate by transcription factors in vitro. Nature. 2013;501:222–226. doi: 10.1038/nature12417. [DOI] [PubMed] [Google Scholar]
- 217.Okashita N, et al. PRDM14 promotes active DNA demethylation through the ten-eleven translocation (TET)-mediated base excision repair pathway in embryonic stem cells. Development. 2014;141:269–280. doi: 10.1242/dev.099622. [DOI] [PubMed] [Google Scholar]
- 218.Shirane K, et al. Global landscape and regulatory principles of DNA methylation reprogramming for germ cell specification by mouse pluripotent stem cells. Dev. Cell. 2016;39:87–103. doi: 10.1016/j.devcel.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 219.Chia NY, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- 220.Tang WWC, et al. Sequential enhancer state remodelling defines human germline competence and specification. Nat. Cell Biol. 2022;24:448–460. doi: 10.1038/s41556-022-00878-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Jo K, et al. Efficient differentiation of human primordial germ cells through geometric control reveals a key role for Nodal signaling. Elife. 2022;11:e72811. doi: 10.7554/eLife.72811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Chuva de Sousa Lopes SM, et al. X chromosome activity in mouse XX primordial germ cells. PLoS Genet. 2008;4:e30. doi: 10.1371/journal.pgen.0040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Gkountela S, et al. DNA demethylation dynamics in the human prenatal germline. Cell. 2015;161:1425–1436. doi: 10.1016/j.cell.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Yuan W, et al. The histone demethylase KDM2B regulates human primordial germ cell-like cells specification. Int. J. Biol. Sci. 2021;17:527–538. doi: 10.7150/ijbs.55873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Xiang X, et al. Human reproduction is regulated by retrotransposons derived from ancient Hominidae-specific viral infections. Nat. Commun. 2022;13:463. doi: 10.1038/s41467-022-28105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Ito J, et al. A hominoid-specific endogenous retrovirus may have rewired the gene regulatory network shared between primordial germ cells and naïve pluripotent cells. PLoS Genet. 2022;18:e1009846. doi: 10.1371/journal.pgen.1009846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Anderson R, Copeland TK, Schöler H, Heasman J, Wylie C. The onset of germ cell migration in the mouse embryo. Mech. Dev. 2000;91:61–68. doi: 10.1016/S0925-4773(99)00271-3. [DOI] [PubMed] [Google Scholar]
- 228.Leitch HG, Tang WW, Surani MA. Primordial germ-cell development and epigenetic reprogramming in mammals. Curr. Top. Dev. Biol. 2013;104:149–187. doi: 10.1016/B978-0-12-416027-9.00005-X. [DOI] [PubMed] [Google Scholar]
- 229.McLaren A. Primordial germ cells in the mouse. Dev. Biol. 2003;262:1–15. doi: 10.1016/S0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- 230.Fujimoto T, Yoshinaga K, Kono I. Distribution of fibronectin on the migratory pathway of primordial germ cells in mice. Anat. Rec. 1985;211:271–278. doi: 10.1002/ar.1092110307. [DOI] [PubMed] [Google Scholar]
- 231.Pereda J, Zorn T, Soto-Suazo M. Migration of human and mouse primordial germ cells and colonization of the developing ovary: an ultrastructural and cytochemical study. Microsc. Res. Tech. 2006;69:386–395. doi: 10.1002/jemt.20298. [DOI] [PubMed] [Google Scholar]
- 232.Mamsen LS, Brøchner CB, Byskov AG, Møllgard K. The migration and loss of human primordial germ stem cells from the hind gut epithelium towards the gonadal ridge. Int. J. Dev. Biol. 2012;56:771–778. doi: 10.1387/ijdb.120202lm. [DOI] [PubMed] [Google Scholar]
- 233.De Felici M. The formation and migration of primordial germ cells in mouse and man. Results Probl. Cell Differ. 2016;58:23–46. doi: 10.1007/978-3-319-31973-5_2. [DOI] [PubMed] [Google Scholar]
- 234.Richardson BE, Lehmann R. Mechanisms guiding primordial germ cell migration: strategies from different organisms. Nat. Rev. Mol. Cell Biol. 2010;11:37–49. doi: 10.1038/nrm2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Godin I, Wylie C, Heasman J. Genital ridges exert long-range effects on mouse primordial germ cell numbers and direction of migration in culture. Development. 1990;108:357–363. doi: 10.1242/dev.108.2.357. [DOI] [PubMed] [Google Scholar]
- 236.Deshpande G, et al. Role of the ABC transporter Mdr49 in Hedgehog signaling and germ cell migration. Development. 2016;143:2111–2120. doi: 10.1242/dev.133587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Owens DA, et al. High-throughput analysis reveals novel maternal germline RNAs crucial for primordial germ cell preservation and proper migration. Development. 2017;144:292–304. doi: 10.1242/dev.139220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Li H, Liang R, Lu Y, Wang M, Li Z. RTN3 regulates the expression level of chemokine receptor CXCR4 and is required for migration of primordial germ cells. Int. J. Mol. Sci. 2016;17:382. doi: 10.3390/ijms17040382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Thorpe JL, Doitsidou M, Ho SY, Raz E, Farber SA. Germ cell migration in zebrafish is dependent on HMGCoA reductase activity and prenylation. Dev. Cell. 2004;6:295–302. doi: 10.1016/S1534-5807(04)00032-2. [DOI] [PubMed] [Google Scholar]
- 240.Staton AA, Knaut H, Giraldez AJ. miRNA regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nat. Genet. 2011;43:204–211. doi: 10.1038/ng.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hen G, Friedman-Einat M, Sela-Donenfeld D. Primordial germ cells in the dorsal mesentery of the chicken embryo demonstrate left-right asymmetry and polarized distribution of the EMA1 epitope. J. Anat. 2014;224:556–563. doi: 10.1111/joa.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Gu Y, Runyan C, Shoemaker A, Surani MA, Wylie C. Membrane-bound steel factor maintains a high local concentration for mouse primordial germ cell motility, and defines the region of their migration. PLoS ONE. 2011;6:e25984. doi: 10.1371/journal.pone.0025984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ding J, et al. Inhibition of HMG CoA reductase reveals an unexpected role for cholesterol during PGC migration in the mouse. BMC Dev. Biol. 2008;8:120. doi: 10.1186/1471-213X-8-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ara T, et al. Impaired colonization of the gonads by primordial germ cells in mice lacking a chemokine, stromal cell-derived factor-1 (SDF-1) Proc. Natl Acad. Sci. USA. 2003;100:5319–5323. doi: 10.1073/pnas.0730719100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Jarysta A, et al. Abnormal migration behavior linked to Rac1 signaling contributes to primordial germ cell exhaustion in Fanconi anemia pathway-deficient Fancg-/- embryos. Hum. Mol. Genet. 2022;31:97–110. doi: 10.1093/hmg/ddab222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Anderson R, et al. Mouse primordial germ cells lacking beta1 integrins enter the germline but fail to migrate normally to the gonads. Development. 1999;126:1655–1664. doi: 10.1242/dev.126.8.1655. [DOI] [PubMed] [Google Scholar]
- 247.García-Castro MI, Anderson R, Heasman J, Wylie C. Interactions between germ cells and extracellular matrix glycoproteins during migration and gonad assembly in the mouse embryo. J. Cell Biol. 1997;138:471–480. doi: 10.1083/jcb.138.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Pesce M, Di Carlo A, De Felici M. The c-kit receptor is involved in the adhesion of mouse primordial germ cells to somatic cells in culture. Mech. Dev. 1997;68:37–44. doi: 10.1016/S0925-4773(97)00120-2. [DOI] [PubMed] [Google Scholar]
- 249.Molyneaux KA, et al. The chemokine SDF1/CXCL12 and its receptor CXCR4 regulate mouse germ cell migration and survival. Development. 2003;130:4279–4286. doi: 10.1242/dev.00640. [DOI] [PubMed] [Google Scholar]
- 250.De Felici M, Dolci S. In vitro adhesion of mouse fetal germ cells to extracellular matrix components. Cell Differ. Dev. 1989;26:87–96. doi: 10.1016/0922-3371(89)90011-7. [DOI] [PubMed] [Google Scholar]
- 251.Di Carlo A, De Felici M. A role for E-cadherin in mouse primordial germ cell development. Dev. Biol. 2000;226:209–219. doi: 10.1006/dbio.2000.9861. [DOI] [PubMed] [Google Scholar]
- 252.Bendel-Stenzel MR, Gomperts M, Anderson R, Heasman J, Wylie C. The role of cadherins during primordial germ cell migration and early gonad formation in the mouse. Mech. Dev. 2000;91:143–152. doi: 10.1016/S0925-4773(99)00287-7. [DOI] [PubMed] [Google Scholar]
- 253.Boldajipour B, et al. Control of chemokine-guided cell migration by ligand sequestration. Cell. 2008;132:463–473. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- 254.Sánchez-Sánchez AV, et al. The embryonic key pluripotent factor NANOG mediates glioblastoma cell migration via the SDF1/CXCR4 pathway. Int. J. Mol. Sci. 2021;22:10620. doi: 10.3390/ijms221910620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Gilbert DC, et al. Clinical and biological significance of CXCL12 and CXCR4 expression in adult testes and germ cell tumours of adults and adolescents. J. Pathol. 2009;217:94–102. doi: 10.1002/path.2436. [DOI] [PubMed] [Google Scholar]
- 256.McIver SC, et al. The chemokine CXCL12 and its receptor CXCR4 are implicated in human seminoma metastasis. Andrology. 2013;1:517–529. doi: 10.1111/j.2047-2927.2013.00081.x. [DOI] [PubMed] [Google Scholar]
- 257.Mahakali Zama A, Hudson FP, 3rd, Bedell MA. Analysis of hypomorphic KitlSl mutants suggests different requirements for KITL in proliferation and migration of mouse primordial germ cells. Biol. Reprod. 2005;73:639–647. doi: 10.1095/biolreprod.105.042846. [DOI] [PubMed] [Google Scholar]
- 258.Buehr M, McLaren A, Bartley A, Darling S. Proliferation and migration of primordial germ cells in We/We mouse embryos. Dev. Dyn. 1993;198:182–189. doi: 10.1002/aja.1001980304. [DOI] [PubMed] [Google Scholar]
- 259.Gu Y, Runyan C, Shoemaker A, Surani A, Wylie C. Steel factor controls primordial germ cell survival and motility from the time of their specification in the allantois, and provides a continuous niche throughout their migration. Development. 2009;136:1295–1303. doi: 10.1242/dev.030619. [DOI] [PubMed] [Google Scholar]
- 260.Farini D, La Sala G, Tedesco M, De Felici M. Chemoattractant action and molecular signaling pathways of Kit ligand on mouse primordial germ cells. Dev. Biol. 2007;306:572–583. doi: 10.1016/j.ydbio.2007.03.031. [DOI] [PubMed] [Google Scholar]
- 261.Rapley EA, et al. A genome-wide association study of testicular germ cell tumor. Nat. Genet. 2009;41:807–810. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Kanetsky PA, et al. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat. Genet. 2009;41:811–815. doi: 10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Hoei-Hansen CE, et al. Ovarian dysgerminomas are characterised by frequent KIT mutations and abundant expression of pluripotency markers. Mol. Cancer. 2007;6:12. doi: 10.1186/1476-4598-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Chou PM, Barquin N, Guinan P, Ridaura Sanz C, Gonzalez-Crussi F. Differential expression of p53, c-kit, and CD34 in prepubertal and postpubertal testicular germ cell tumors. Cancer. 1997;79:2430–2434. doi: 10.1002/(SICI)1097-0142(19970615)79:12<2430::AID-CNCR20>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 265.Hoei-Hansen CE, et al. New evidence for the origin of intracranial germ cell tumours from primordial germ cells: expression of pluripotency and cell differentiation markers. J. Pathol. 2006;209:25–33. doi: 10.1002/path.1948. [DOI] [PubMed] [Google Scholar]
- 266.Mosbech CH, Rechnitzer C, Brok JS, Rajpert-De Meyts E, Hoei-Hansen CE. Recent advances in understanding the etiology and pathogenesis of pediatric germ cell tumors. J. Pediatr. Hematol. Oncol. 2014;36:263–270. doi: 10.1097/MPH.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 267.Lu M, et al. Activation of the human chemokine receptor CX3CR1 regulated by cholesterol. Sci. Adv. 2022;8:eabn8048. doi: 10.1126/sciadv.abn8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Oosterhuis JW, Looijenga LHJ. Human germ cell tumours from a developmental perspective. Nat. Rev. Cancer. 2019;19:522–537. doi: 10.1038/s41568-019-0178-9. [DOI] [PubMed] [Google Scholar]
- 269.Kanneganti A, Bhadiraju P, Tong PSY. Extragonadal teratomas in women and adolescent girls: a systematic review. Eur. J. Obstet. Gynecol. Reprod. Biol. 2021;262:134–141. doi: 10.1016/j.ejogrb.2021.05.005. [DOI] [PubMed] [Google Scholar]
- 270.De Felici M, et al. To be or not to be a germ cell: the extragonadal germ cell tumor paradigm. Int. J. Mol. Sci. 2021;22:5982. doi: 10.3390/ijms22115982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Teilum G. Classification of endodermal sinus tumour (mesoblatoma vitellinum) and so-called “embryonal carcinoma” of the ovary. Acta Pathol. Microbiol. Scand. 1965;64:407–429. doi: 10.1111/apm.1965.64.4.407. [DOI] [PubMed] [Google Scholar]
- 272.Schneider DT, et al. Multipoint imprinting analysis indicates a common precursor cell for gonadal and nongonadal pediatric germ cell tumors. Cancer Res. 2001;61:7268–7276. [PubMed] [Google Scholar]
- 273.Göbel U, et al. Germ-cell tumors in childhood and adolescence. GPOH MAKEI and the MAHO study groups. Ann. Oncol. 2000;11:263–271. doi: 10.1023/A:1008360523160. [DOI] [PubMed] [Google Scholar]
- 274.Felix I, Becker LE. Intracranial germ cell tumors in children: an immunohistochemical and electron microscopic study. Pediatr. Neurosurg. 1990;16:156–162. doi: 10.1159/000120517. [DOI] [PubMed] [Google Scholar]
- 275.Wang L, et al. Novel somatic and germline mutations in intracranial germ cell tumours. Nature. 2014;511:241–245. doi: 10.1038/nature13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Elias KM, et al. Primordial germ cells as a potential shared cell of origin for mucinous cystic neoplasms of the pancreas and mucinous ovarian tumors. J. Pathol. 2018;246:459–469. doi: 10.1002/path.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Honecker F, et al. Germ cell lineage differentiation in non-seminomatous germ cell tumours. J. Pathol. 2006;208:395–400. doi: 10.1002/path.1872. [DOI] [PubMed] [Google Scholar]
- 278.Burger JA, Kipps TJ. CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood. 2006;107:1761–1767. doi: 10.1182/blood-2005-08-3182. [DOI] [PubMed] [Google Scholar]
- 279.Jostes SV, et al. Unique and redundant roles of SOX2 and SOX17 in regulating the germ cell tumor fate. Int. J. Cancer. 2020;146:1592–1605. doi: 10.1002/ijc.32714. [DOI] [PubMed] [Google Scholar]
- 280.de Vries G, Rosas-Plaza X, van Vugt M, Gietema JA, de Jong S. Testicular cancer: Determinants of cisplatin sensitivity and novel therapeutic opportunities. Cancer Treat. Rev. 2020;88:102054. doi: 10.1016/j.ctrv.2020.102054. [DOI] [PubMed] [Google Scholar]
- 281.Goddard NC, et al. KIT and RAS signalling pathways in testicular germ cell tumours: new data and a review of the literature. Int. J. Androl. 2007;30:337–348. doi: 10.1111/j.1365-2605.2007.00769.x. [DOI] [PubMed] [Google Scholar]
- 282.Guo S, et al. Interfering with CXCR4 expression inhibits proliferation, adhesion and migration of breast cancer MDA-MB-231 cells. Oncol. Lett. 2014;8:1557–1562. doi: 10.3892/ol.2014.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat. Rev. Mol. Cell Biol. 2016;17:337–349. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- 284.Dettman EJ, Justice MJ. The zinc finger SET domain gene Prdm14 is overexpressed in lymphoblastic lymphomas with retroviral insertions at Evi32. PLoS ONE. 2008;3:e3823. doi: 10.1371/journal.pone.0003823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Taniguchi H, Imai K. PRDM14, a zinc fnger protein, regulates cancer stemness. Methods Mol. Biol. 2018;1867:3–13. doi: 10.1007/978-1-4939-8799-3_1. [DOI] [PubMed] [Google Scholar]
- 286.Tracey LJ, Justice MJ. Off to a bad start: cancer initiation by pluripotency regulator PRDM14. Trends Genet. 2019;35:489–500. doi: 10.1016/j.tig.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Killian JK, et al. Imprints and DPPA3 are bypassed during pluripotency- and differentiation-coupled methylation reprogramming in testicular germ cell tumors. Genome Res. 2016;26:1490–1504. doi: 10.1101/gr.201293.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.van Gurp RJ, Oosterhuis JW, Kalscheuer V, Mariman EC, Looijenga LH. Biallelic expression of the H19 and IGF2 genes in human testicular germ cell tumors. J. Natl Cancer Inst. 1994;86:1070–1075. doi: 10.1093/jnci/86.14.1070. [DOI] [PubMed] [Google Scholar]
- 289.Kristensen DG, Skakkebæk NE, Rajpert-De Meyts E, Almstrup K. Epigenetic features of testicular germ cell tumours in relation to epigenetic characteristics of foetal germ cells. Int. J. Dev. Biol. 2013;57:309–317. doi: 10.1387/ijdb.130142ka. [DOI] [PubMed] [Google Scholar]
- 290.Buljubašić R, et al. Epigenetics and testicular germ cell tumors. Gene. 2018;661:22–33. doi: 10.1016/j.gene.2018.03.072. [DOI] [PubMed] [Google Scholar]
- 291.Fichtner A, et al. The detection of isochromosome i(12p) in malignant germ cell tumours and tumours with somatic malignant transformation by the use of quantitative real-time polymerase chain reaction. Histopathology. 2021;78:593–606. doi: 10.1111/his.14258. [DOI] [PubMed] [Google Scholar]
- 292.Freitag CE, et al. Assessment of isochromosome 12p and 12p abnormalities in germ cell tumors using fluorescence in situ hybridization, single-nucleotide polymorphism arrays, and next-generation sequencing/mate-pair sequencing. Hum. Pathol. 2021;112:20–34. doi: 10.1016/j.humpath.2021.03.008. [DOI] [PubMed] [Google Scholar]
- 293.Litchfield K, Levy M, Huddart RA, Shipley J, Turnbull C. The genomic landscape of testicular germ cell tumours: from susceptibility to treatment. Nat. Rev. Urol. 2016;13:409–419. doi: 10.1038/nrurol.2016.107. [DOI] [PubMed] [Google Scholar]
- 294.de Bruin TW, et al. Isochromosome 12p-positive pineal germ cell tumor. Cancer Res. 1994;54:1542–1544. [PubMed] [Google Scholar]
- 295.Looijenga LH, et al. Stem cell factor receptor (c-KIT) codon 816 mutations predict development of bilateral testicular germ-cell tumors. Cancer Res. 2003;63:7674–7678. [PubMed] [Google Scholar]
- 296.Cutcutache I, et al. Exome-wide sequencing shows low mutation rates and identifies novel mutated genes in seminomas. Eur. Urol. 2015;68:77–83. doi: 10.1016/j.eururo.2014.12.040. [DOI] [PubMed] [Google Scholar]
- 297.Litchfield K, et al. Whole-exome sequencing reveals the mutational spectrum of testicular germ cell tumours. Nat. Commun. 2015;6:5973. doi: 10.1038/ncomms6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Shen H, et al. Integrated molecular characterization of testicular germ cell tumors. Cell Rep. 2018;23:3392–3406. doi: 10.1016/j.celrep.2018.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Heaney JD, Lam MY, Michelson MV, Nadeau JH. Loss of the transmembrane but not the soluble kit ligand isoform increases testicular germ cell tumor susceptibility in mice. Cancer Res. 2008;68:5193–5197. doi: 10.1158/0008-5472.CAN-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Nathanson KL, et al. The Y deletion gr/gr and susceptibility to testicular germ cell tumor. Am. J. Hum. Genet. 2005;77:1034–1043. doi: 10.1086/498455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Giannoulatou E, et al. Whole-genome sequencing of spermatocytic tumors provides insights into the mutational processes operating in the male germline. PLoS ONE. 2017;12:e0178169. doi: 10.1371/journal.pone.0178169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Oosterhuis JW, et al. Ploidy of primary germ cell tumors of the testis. Pathogenetic and clinical relevance. Lab. Invest. 1989;60:14–21. [PubMed] [Google Scholar]
- 303.de Jong B, Oosterhuis JW, Castedo SM, Vos A, te Meerman GJ. Pathogenesis of adult testicular germ cell tumors. A cytogenetic model. Cancer Genet. Cytogenet. 1990;48:143–167. doi: 10.1016/0165-4608(90)90115-Q. [DOI] [PubMed] [Google Scholar]
- 304.Murty VV, Chaganti RS. A genetic perspective of male germ cell tumors. Semin. Oncol. 1998;25:133–144. [PubMed] [Google Scholar]
- 305.Kelly MR, et al. A multi-omic dissection of super-enhancer driven oncogenic gene expression programs in ovarian cancer. Nat. Commun. 2022;13:4247. doi: 10.1038/s41467-022-31919-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Zhu X, et al. The deubiquitinase USP11 promotes ovarian cancer chemoresistance by stabilizing BIP. Signal Transduct. Target. Ther. 2021;6:264. doi: 10.1038/s41392-021-00580-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Taguchi J, et al. DMRT1-mediated reprogramming drives development of cancer resembling human germ cell tumors with features of totipotency. Nat. Commun. 2021;12:5041. doi: 10.1038/s41467-021-25249-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Liu C, Moten A, Ma Z, Lin HK. The foundational framework of tumors: gametogenesis, p53, and cancer. Semin. Cancer Biol. 2022;81:193–205. doi: 10.1016/j.semcancer.2021.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Zheng H, et al. Jumonji domain-containing 6 (JMJD6) identified as a potential therapeutic target in ovarian cancer. Signal Transduct. Target. Ther. 2019;4:24. doi: 10.1038/s41392-019-0055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Lin X, et al. RNA-binding protein LIN28B inhibits apoptosis through regulation of the AKT2/FOXO3A/BIM axis in ovarian cancer cells. Signal Transduct. Target. Ther. 2018;3:23. doi: 10.1038/s41392-018-0026-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Saitou M. Mammalian germ cell development: from mechanism to in vitro reconstitution. Stem Cell Rep. 2021;16:669–680. doi: 10.1016/j.stemcr.2021.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Li L, Yuan Y, Sha J. Potential clinical value of in vitro spermatogenesis†. Biol. Reprod. 2022;107:95–100. doi: 10.1093/biolre/ioac076. [DOI] [PubMed] [Google Scholar]
- 313.Geijsen N, et al. Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature. 2004;427:148–154. doi: 10.1038/nature02247. [DOI] [PubMed] [Google Scholar]
- 314.Hübner K, et al. Derivation of oocytes from mouse embryonic stem cells. Science. 2003;300:1251–1256. doi: 10.1126/science.1083452. [DOI] [PubMed] [Google Scholar]
- 315.Toyooka Y, Tsunekawa N, Akasu R, Noce T. Embryonic stem cells can form germ cells in vitro. Proc. Natl Acad. Sci. USA. 2003;100:11457–11462. doi: 10.1073/pnas.1932826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Clark AT, et al. Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum. Mol. Genet. 2004;13:727–739. doi: 10.1093/hmg/ddh088. [DOI] [PubMed] [Google Scholar]
- 317.Hwang YS, et al. Reconstitution of prospermatogonial specification in vitro from human induced pluripotent stem cells. Nat. Commun. 2020;11:5656. doi: 10.1038/s41467-020-19350-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Yamashiro C, et al. Generation of human oogonia from induced pluripotent stem cells in vitro. Science. 2018;362:356–360. doi: 10.1126/science.aat1674. [DOI] [PubMed] [Google Scholar]
- 319.Murase Y, et al. Long-term expansion with germline potential of human primordial germ cell-like cells in vitro. EMBO J. 2020;39:e104929. doi: 10.15252/embj.2020104929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Mall EM, et al. A novel xeno-organoid approach: exploring the crosstalk between human iPSC-derived PGC-like and rat testicular cells. Mol. Hum. Reprod. 2020;26:879–893. doi: 10.1093/molehr/gaaa067. [DOI] [PubMed] [Google Scholar]
- 321.Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 322.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Watanabe K, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 324.Gafni O, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 325.Nakamura T, et al. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature. 2016;537:57–62. doi: 10.1038/nature19096. [DOI] [PubMed] [Google Scholar]
- 326.Panula S, et al. Human germ cell differentiation from fetal- and adult-derived induced pluripotent stem cells. Hum. Mol. Genet. 2011;20:752–762. doi: 10.1093/hmg/ddq520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Kee K, Angeles VT, Flores M, Nguyen HN, Reijo Pera RA. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature. 2009;462:222–225. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Eguizabal C, et al. Complete meiosis from human induced pluripotent stem cells. Stem Cells. 2011;29:1186–1195. doi: 10.1002/stem.672. [DOI] [PubMed] [Google Scholar]
- 329.Medrano JV, Ramathal C, Nguyen HN, Simon C, Reijo Pera RA. Divergent RNA-binding proteins, DAZL and VASA, induce meiotic progression in human germ cells derived in vitro. Stem Cells. 2012;30:441–451. doi: 10.1002/stem.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Easley CAT, et al. Direct differentiation of human pluripotent stem cells into haploid spermatogenic cells. Cell Rep. 2012;2:440–446. doi: 10.1016/j.celrep.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Yang S, et al. Meiosis resumption in human primordial germ cells from induced pluripotent stem cells by in vitro activation and reconstruction of ovarian nests. Stem Cell Res. Ther. 2022;13:339. doi: 10.1186/s13287-022-03019-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Wang H, et al. Induction of meiosis by embryonic gonadal somatic cells differentiated from pluripotent stem cells. Stem Cell Res. Ther. 2021;12:607. doi: 10.1186/s13287-021-02672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Hikabe O, et al. Reconstitution in vitro of the entire cycle of the mouse female germ line. Nature. 2016;539:299–303. doi: 10.1038/nature20104. [DOI] [PubMed] [Google Scholar]
- 334.Hayashi K, et al. Offspring from oocytes derived from in vitro primordial germ cell-like cells in mice. Science. 2012;338:971–975. doi: 10.1126/science.1226889. [DOI] [PubMed] [Google Scholar]
- 335.Tian C, et al. Functional oocytes derived from granulosa cells. Cell Rep. 2019;29:4256–4267. doi: 10.1016/j.celrep.2019.11.080. [DOI] [PubMed] [Google Scholar]
- 336.Hamada N, et al. Germ cell-intrinsic effects of sex chromosomes on early oocyte differentiation in mice. PLoS Genet. 2020;16:e1008676. doi: 10.1371/journal.pgen.1008676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Zhou Q, et al. Complete meiosis from embryonic stem cell-derived germ cells in vitro. Cell Stem Cell. 2016;18:330–340. doi: 10.1016/j.stem.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 338.De Felici M. Primordial germ cell biology at the beginning of the XXI century. Int. J. Dev. Biol. 2009;53:891–894. doi: 10.1387/ijdb.082815mf. [DOI] [PubMed] [Google Scholar]
- 339.Guo J, et al. Single-cell analysis of the developing human testis reveals somatic niche cell specification and fetal germline stem cell establishment. Cell Stem Cell. 2021;28:764–778. doi: 10.1016/j.stem.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Dym M, Kokkinaki M, He Z. Spermatogonial stem cells: mouse and human comparisons. Birth Defects Res. C. Embryo Today. 2009;87:27–34. doi: 10.1002/bdrc.20141. [DOI] [PubMed] [Google Scholar]
- 341.Kolasa A, Misiakiewicz K, Marchlewicz M, Wiszniewska B. The generation of spermatogonial stem cells and spermatogonia in mammals. Reprod. Biol. 2012;12:5–23. doi: 10.1016/S1642-431X(12)60074-6. [DOI] [PubMed] [Google Scholar]
- 342.de Rooij DG. The nature and dynamics of spermatogonial stem cells. Development. 2017;144:3022–3030. doi: 10.1242/dev.146571. [DOI] [PubMed] [Google Scholar]
- 343.Hermann BP, et al. The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep. 2018;25:1650–1667. doi: 10.1016/j.celrep.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Guo J, et al. The adult human testis transcriptional cell atlas. Cell Res. 2018;28:1141–1157. doi: 10.1038/s41422-018-0099-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Wang M, et al. Single-cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell. 2018;23:599–614. doi: 10.1016/j.stem.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 346.Luo H, et al. Offspring production of ovarian organoids derived from spermatogonial stem cells by defined factors with chromatin reorganization. J. Adv. Res. 2021;33:81–98. doi: 10.1016/j.jare.2021.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Xu H, et al. Derivation and propagation of spermatogonial stem cells from human pluripotent cells. Stem Cell Res. Ther. 2020;11:408. doi: 10.1186/s13287-020-01896-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Ibtisham F, et al. In vitro production of haploid germ cells from murine spermatogonial stem cells using a two-dimensional cell culture system. Theriogenology. 2021;162:84–94. doi: 10.1016/j.theriogenology.2020.12.024. [DOI] [PubMed] [Google Scholar]
- 349.Zhang W, et al. Direct reprogramming of human Sertoli cells into male germline stem cells with the self-renewal and differentiation potentials via overexpressing DAZL/DAZ2/BOULE genes. Stem Cell Rep. 2021;16:2798–2812. doi: 10.1016/j.stemcr.2021.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Tegelenbosch RA, de Rooij DG. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat. Res. 1993;290:193–200. doi: 10.1016/0027-5107(93)90159-D. [DOI] [PubMed] [Google Scholar]
- 351.Breuss MW, Yang X, Gleeson JG. Sperm mosaicism: implications for genomic diversity and disease. Trends Genet. 2021;37:890–902. doi: 10.1016/j.tig.2021.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Clermont Y. The cycle of the seminiferous epithelium in man. Am. J. Anat. 1963;112:35–51. doi: 10.1002/aja.1001120103. [DOI] [PubMed] [Google Scholar]
- 353.Clermont Y. Renewal of spermatogonia in man. Am. J. Anat. 1966;118:509–524. doi: 10.1002/aja.1001180211. [DOI] [PubMed] [Google Scholar]
- 354.Clermont Y. Spermatogenesis in man. A study of the spermatogonial population. Fertil. Steril. 1966;17:705–721. doi: 10.1016/S0015-0282(16)36120-9. [DOI] [PubMed] [Google Scholar]
- 355.Tan K, Wilkinson MF. Human spermatogonial stem cells scrutinized under the single-cell magnifying glass. Cell Stem Cell. 2019;24:201–203. doi: 10.1016/j.stem.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Zheng Y, et al. Unraveling three-dimensional chromatin structural dynamics during spermatogonial differentiation. J. Biol. Chem. 2022;298:101559. doi: 10.1016/j.jbc.2021.101559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Fayomi AP, Orwig KE. Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Res. 2018;29:207–214. doi: 10.1016/j.scr.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Ibtisham F, Honaramooz A. Spermatogonial stem cells for in vitro spermatogenesis and in vivo restoration of fertility. Cells. 2020;9:745. doi: 10.3390/cells9030745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Xi HM, et al. Recent advances in isolation, identification, and culture of mammalian spermatogonial stem cells. Asian J. Androl. 2022;24:5–14. doi: 10.4103/aja.aja_41_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Wei, B. H., Hao, S. L. & Yang, W. X. Regulation of spermatogonial stem cell self-renewal and proliferation in mammals. Histol. Histopathol. 10.14670/hh-18-461 (2022). [DOI] [PubMed]
- 361.La HM, et al. Distinctive molecular features of regenerative stem cells in the damaged male germline. Nat. Commun. 2022;13:2500. doi: 10.1038/s41467-022-30130-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Diao L, Turek PJ, John CM, Fang F, Reijo Pera RA. Roles of spermatogonial stem cells in spermatogenesis and fertility restoration. Front. Endocrinol. (Lausanne) 2022;13:895528. doi: 10.3389/fendo.2022.895528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Oatley JM, Brinster RL. The germline stem cell niche unit in mammalian testes. Physiol. Rev. 2012;92:577–595. doi: 10.1152/physrev.00025.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Stévant I, et al. Deciphering cell lineage specification during male sex determination with single-cell rna sequencing. Cell Rep. 2018;22:1589–1599. doi: 10.1016/j.celrep.2018.01.043. [DOI] [PubMed] [Google Scholar]
- 365.Chen SR, Liu YX. Regulation of spermatogonial stem cell self-renewal and spermatocyte meiosis by Sertoli cell signaling. Reproduction. 2015;149:R159–R167. doi: 10.1530/REP-14-0481. [DOI] [PubMed] [Google Scholar]
- 366.Meng X, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- 367.Naughton CK, Jain S, Strickland AM, Gupta A, Milbrandt J. Glial cell-line derived neurotrophic factor-mediated RET signaling regulates spermatogonial stem cell fate. Biol. Reprod. 2006;74:314–321. doi: 10.1095/biolreprod.105.047365. [DOI] [PubMed] [Google Scholar]
- 368.Jing S, et al. GDNF-induced activation of the ret protein tyrosine kinase is mediated by GDNFR-alpha, a novel receptor for GDNF. Cell. 1996;85:1113–1124. doi: 10.1016/S0092-8674(00)81311-2. [DOI] [PubMed] [Google Scholar]
- 369.Parker N, et al. Spermatogonial stem cell numbers are reduced by transient inhibition of GDNF signaling but restored by self-Renewing replication when signaling resumes. Stem Cell Rep. 2021;16:597–609. doi: 10.1016/j.stemcr.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Yang QE, Kim D, Kaucher A, Oatley MJ, Oatley JM. CXCL12-CXCR4 signaling is required for the maintenance of mouse spermatogonial stem cells. J. Cell Sci. 2013;126:1009–1020. doi: 10.1242/jcs.119826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Chen H, et al. Dissecting mammalian spermatogenesis using spatial transcriptomics. Cell Rep. 2021;37:109915. doi: 10.1016/j.celrep.2021.109915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Nakamura Y, Jörg DJ, Kon Y, Simons BD, Yoshida S. Transient suppression of transplanted spermatogonial stem cell differentiation restores fertility in mice. Cell Stem Cell. 2021;28:1443–1456. doi: 10.1016/j.stem.2021.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Overeem AW, Chang YW, Spruit J, Roelse CM, Chuva De Sousa Lopes SM. Ligand-receptor interactions elucidate sex specific pathways in the trajectory from primordial germ cells to gonia during human development. Front. Cell. Dev. Biol. 2021;9:661243. doi: 10.3389/fcell.2021.661243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Lin H, et al. Histone methyltransferase DOT1L is essential for self-renewal of germline stem cells. Genes Dev. 2022;36:752–763. doi: 10.1101/gad.349550.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Meng Y, et al. Z-DNA is remodelled by ZBTB43 in prospermatogonia to safeguard the germline genome and epigenome. Nat. Cell Biol. 2022;24:1141–1153. doi: 10.1038/s41556-022-00941-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Zhang Z, et al. Integrated analysis of single-cell and bulk RNA sequencing data reveals a pan-cancer stemness signature predicting immunotherapy response. Genome Med. 2022;14:45. doi: 10.1186/s13073-022-01050-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Wang C, Leung A, Sinha-Hikim AP. Reproductive aging in the male brown-Norway rat: a model for the human. Endocrinology. 1993;133:2773–2781. doi: 10.1210/endo.133.6.8243304. [DOI] [PubMed] [Google Scholar]
- 378.Jahnukainen K, Ehmcke J, Hou M, Schlatt S. Testicular function and fertility preservation in male cancer patients. Best. Pract. Res. Clin. Endocrinol. Metab. 2011;25:287–302. doi: 10.1016/j.beem.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 379.Kubota H, Brinster RL. Technology insight: in vitro culture of spermatogonial stem cells and their potential therapeutic uses. Nat. Clin. Pract. Endocrinol. Metab. 2006;2:99–108. doi: 10.1038/ncpendmet0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Allen CM, Lopes F, Mitchell RT, Spears N. Can antioxidants protect against chemotherapy in a rat spermatogonial stem cell line? Reprod. Fertil. 2021;2:L7–l9. doi: 10.1530/RAF-21-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Wallace WH, Shalet SM, Lendon M, Morris-Jones PH. Male fertility in long-term survivors of childhood acute lymphoblastic leukaemia. Int. J. Androl. 1991;14:312–319. doi: 10.1111/j.1365-2605.1991.tb01098.x. [DOI] [PubMed] [Google Scholar]
- 382.Kanbar M, Delwiche G, Wyns C. Fertility preservation for prepubertal boys: are we ready for autologous grafting of cryopreserved immature testicular tissue? Ann. Endocrinol. (Paris) 2022;83:210–217. doi: 10.1016/j.ando.2022.04.006. [DOI] [PubMed] [Google Scholar]
- 383.Jensen CFS, et al. Fertility preservation in boys facing gonadotoxic cancer therapy. Nat. Rev. Urol. 2022;19:71–83. doi: 10.1038/s41585-021-00523-8. [DOI] [PubMed] [Google Scholar]
- 384.Delgouffe E, Braye A, Goossens E. Testicular tissue banking for fertility preservation in young boys: which patients should be included? Front. Endocrinol. (Lausanne) 2022;13:854186. doi: 10.3389/fendo.2022.854186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Abdelaal NE, et al. Cellular therapy via spermatogonial stem cells for treating impaired spermatogenesis, non-obstructive azoospermia. Cells. 2021;10:1779. doi: 10.3390/cells10071779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Goossens E, et al. Fertility preservation in boys: recent developments and new insights. Hum. Reprod. Open. 2020;2020:hoaa016. doi: 10.1093/hropen/hoaa016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Deebel NA, Bradshaw AW, Sadri-Ardekani H. Infertility considerations in klinefelter syndrome: from origin to management. Best. Pract. Res. Clin. Endocrinol. Metab. 2020;34:101480. doi: 10.1016/j.beem.2020.101480. [DOI] [PubMed] [Google Scholar]
- 388.de Nie I, et al. Histological study on the influence of puberty suppression and hormonal treatment on developing germ cells in transgender women. Hum. Reprod. 2022;37:297–308. doi: 10.1093/humrep/deab240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proc. Natl Acad. Sci. USA. 1994;91:11298–11302. doi: 10.1073/pnas.91.24.11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Brinster RL, Avarbock MR. Germline transmission of donor haplotype following spermatogonial transplantation. Proc. Natl Acad. Sci. USA. 1994;91:11303–11307. doi: 10.1073/pnas.91.24.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Kubota H, Brinster RL. Spermatogonial stem cells. Biol. Reprod. 2018;99:52–74. doi: 10.1093/biolre/ioy077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Liang D, et al. Xenotransplantation of human spermatogonia into various mouse recipient models. Front. Cell Dev. Biol. 2022;10:883314. doi: 10.3389/fcell.2022.883314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Azizi H, Niazi Tabar A, Skutella T. Successful transplantation of spermatogonial stem cells into the seminiferous tubules of busulfan-treated mice. Reprod. Health. 2021;18:189. doi: 10.1186/s12978-021-01242-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Jiang FX, Short RV. Male germ cell transplantation in rats: apparent synchronization of spermatogenesis between host and donor seminiferous epithelia. Int. J. Androl. 1995;18:326–330. doi: 10.1111/j.1365-2605.1995.tb00570.x. [DOI] [PubMed] [Google Scholar]
- 395.Ogawa T, Dobrinski I, Brinster RL. Recipient preparation is critical for spermatogonial transplantation in the rat. Tissue Cell. 1999;31:461–472. doi: 10.1054/tice.1999.0060. [DOI] [PubMed] [Google Scholar]
- 396.Honaramooz A, Megee SO, Dobrinski I. Germ cell transplantation in pigs. Biol. Reprod. 2002;66:21–28. doi: 10.1095/biolreprod66.1.21. [DOI] [PubMed] [Google Scholar]
- 397.Honaramooz A, Behboodi E, Blash S, Megee SO, Dobrinski I. Germ cell transplantation in goats. Mol. Reprod. Dev. 2003;64:422–428. doi: 10.1002/mrd.10205. [DOI] [PubMed] [Google Scholar]
- 398.Herrid M, Vignarajan S, Davey R, Dobrinski I, Hill JR. Successful transplantation of bovine testicular cells to heterologous recipients. Reproduction. 2006;132:617–624. doi: 10.1530/rep.1.01125. [DOI] [PubMed] [Google Scholar]
- 399.Kim Y, et al. Production of donor-derived sperm after spermatogonial stem cell transplantation in the dog. Reproduction. 2008;136:823–831. doi: 10.1530/REP-08-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Izadyar F, et al. Autologous and homologous transplantation of bovine spermatogonial stem cells. Reproduction. 2003;126:765–774. doi: 10.1530/rep.0.1260765. [DOI] [PubMed] [Google Scholar]
- 401.Schlatt S, et al. Germ cell transfer into rat, bovine, monkey and human testes. Hum. Reprod. 1999;14:144–150. doi: 10.1093/humrep/14.1.144. [DOI] [PubMed] [Google Scholar]
- 402.Mikkola M, et al. Transplantation of normal boar testicular cells resulted in complete focal spermatogenesis in a boar affected by the immotile short-tail sperm defect. Reprod. Domest. Anim. 2006;41:124–128. doi: 10.1111/j.1439-0531.2006.00651.x. [DOI] [PubMed] [Google Scholar]
- 403.Rodriguez-Sosa JR, Silvertown JD, Foster RA, Medin JA, Hahnel A. Transduction and transplantation of spermatogonia into the testis of ram lambs through the extra-testicular rete. Reprod. Domest. Anim. 2009;44:612–620. doi: 10.1111/j.1439-0531.2007.01030.x. [DOI] [PubMed] [Google Scholar]
- 404.Hermann BP, et al. Spermatogonial stem cell transplantation into rhesus testes regenerates spermatogenesis producing functional sperm. Cell Stem Cell. 2012;11:715–726. doi: 10.1016/j.stem.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Zhang F, et al. Surrogate production of genome-edited sperm from a different subfamily by spermatogonial stem cell transplantation. Sci. China Life Sci. 2022;65:969–987. doi: 10.1007/s11427-021-1989-9. [DOI] [PubMed] [Google Scholar]
- 406.Yuan H, et al. Primary culture of germ cells that portray stem cell characteristics and recipient preparation for autologous transplantation in the rhesus monkey. J. Cell Mol. Med. 2022;26:1567–1578. doi: 10.1111/jcmm.17197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Wang D, et al. Characterization and survival of human infant testicular cells after direct xenotransplantation. Front. Endocrinol. (Lausanne) 2022;13:853482. doi: 10.3389/fendo.2022.853482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Kanatsu-Shinohara M, et al. Regeneration of spermatogenesis by mouse germ cell transplantation into allogeneic and xenogeneic testis primordia or organoids. Stem Cell Rep. 2022;17:924–935. doi: 10.1016/j.stemcr.2022.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Shetty G, et al. Postpubertal spermatogonial stem cell transplantation restores functional sperm production in rhesus monkeys irradiated before and after puberty. Andrology. 2021;9:1603–1616. doi: 10.1111/andr.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Radford J, Shalet S, Lieberman B. Fertility after treatment for cancer. Questions remain over ways of preserving ovarian and testicular tissue. BMJ. 1999;319:935–936. doi: 10.1136/bmj.319.7215.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Cui, Y. H., Chen, W., Wu, S., Wan, C. L. & He, Z. Generation of male germ cells in vitro from the stem cells. Asian J. Androl. 10.4103/aja20226 (2022). [DOI] [PMC free article] [PubMed]
- 412.Bashiri Z, Zahiri M, Allahyari H, Esmaeilzade B. Proliferation of human spermatogonial stem cells on optimized PCL/Gelatin nanofibrous scaffolds. Andrologia. 2022;54:e14380. doi: 10.1111/and.14380. [DOI] [PubMed] [Google Scholar]
- 413.Mahboudi S, Parivar K, Mazaheri Z, Irani SH. Mir-106b cluster regulates primordial germ cells differentiation from human mesenchymal stem cells. Cell J. 2021;23:294–302. doi: 10.22074/cellj.2021.6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Sanou I, et al. Spermatogonial stem cell-based therapies: taking preclinical research to the next level. Front. Endocrinol. (Lausanne) 2022;13:850219. doi: 10.3389/fendo.2022.850219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Voigt AL, Thiageswaran S, de Lima EMLN, Dobrinski I. Metabolic requirements for spermatogonial stem cell establishment and maintenance in vivo and in vitro. Int. J. Mol. Sci. 2021;22:1998. doi: 10.3390/ijms22041998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Wang YH, et al. Rescue of male infertility through correcting a genetic mutation causing meiotic arrest in spermatogonial stem cells. Asian J. Androl. 2021;23:590–599. doi: 10.4103/aja.aja_97_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Lei Q, Hamer G. The use of spermatogonial stem cells to correct a mutation causing meiotic arrest. Asian J. Androl. 2021;23:600–601. doi: 10.4103/aja.aja_2_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Johnson J, et al. Oocyte generation in adult mammalian ovaries by putative germ cells in bone marrow and peripheral blood. Cell. 2005;122:303–315. doi: 10.1016/j.cell.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 419.Pouryousefi-Koodehi, T. et al. Can mesenchymal stem cells derived from adipose tissue and their conditioned medium improve ovarian functions? A mini-review. Zygote10.1017/S0967199422000235 (2022). [DOI] [PubMed]
- 420.Virant-Klun I, et al. Parthenogenetic embryo-like structures in the human ovarian surface epithelium cell culture in postmenopausal women with no naturally present follicles and oocytes. Stem Cells Dev. 2009;18:137–149. doi: 10.1089/scd.2007.0238. [DOI] [PubMed] [Google Scholar]
- 421.Bhartiya D. Ovarian stem cells are always accompanied by very small embryonic-like stem cells in adult mammalian ovary. J. Ovarian Res. 2015;8:70. doi: 10.1186/s13048-015-0200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Esmaeilian Y, Atalay A, Erdemli E. Putative germline and pluripotent stem cells in adult mouse ovary and their in vitro differentiation potential into oocyte-like and somatic cells. Zygote. 2017;25:358–375. doi: 10.1017/S0967199417000235. [DOI] [PubMed] [Google Scholar]
- 423.Wu J, Ding X, Wang J. Stem cells in mammalian gonads. Results Probl. Cell Differ. 2016;58:289–307. doi: 10.1007/978-3-319-31973-5_11. [DOI] [PubMed] [Google Scholar]
- 424.Xie W, Wang H, Wu J. Similar morphological and molecular signatures shared by female and male germline stem cells. Sci. Rep. 2014;4:558. doi: 10.1038/srep05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Liu J, et al. Isolation and characterization of string-forming female germline stem cells from ovaries of neonatal mice. J. Biol. Chem. 2017;292:16003–16013. doi: 10.1074/jbc.M117.799403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Zou K, Hou L, Sun K, Xie W, Wu J. Improved efficiency of female germline stem cell purification using fragilis-based magnetic bead sorting. Stem Cells Dev. 2011;20:2197–2204. doi: 10.1089/scd.2011.0091. [DOI] [PubMed] [Google Scholar]
- 427.White YA, et al. Oocyte formation by mitotically active germ cells purified from ovaries of reproductive-age women. Nat. Med. 2012;18:413–421. doi: 10.1038/nm.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Wang H, et al. Conversion of female germline stem cells from neonatal and prepubertal mice into pluripotent stem cells. J. Mol. Cell Biol. 2014;6:164–171. doi: 10.1093/jmcb/mju004. [DOI] [PubMed] [Google Scholar]
- 429.Pacchiarotti J, et al. Differentiation potential of germ line stem cells derived from the postnatal mouse ovary. Differentiation. 2010;79:159–170. doi: 10.1016/j.diff.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 430.Zhou L, et al. Production of fat-1 transgenic rats using a post-natal female germline stem cell line. Mol. Hum. Reprod. 2014;20:271–281. doi: 10.1093/molehr/gat081. [DOI] [PubMed] [Google Scholar]
- 431.Zhang Y, Wu J. Molecular cloning and characterization of a new gene, Oocyte-G1. J. Cell Physiol. 2009;218:75–83. doi: 10.1002/jcp.21569. [DOI] [PubMed] [Google Scholar]
- 432.Yang Z, Wu J. Mouse dynein axonemal intermediate chain 2: cloning and expression. DNA Cell Biol. 2008;27:479–488. doi: 10.1089/dna.2008.0752. [DOI] [PubMed] [Google Scholar]
- 433.Zhang Y, et al. Production of transgenic mice by random recombination of targeted genes in female germline stem cells. J. Mol. Cell Biol. 2011;3:132–141. doi: 10.1093/jmcb/mjq043. [DOI] [PubMed] [Google Scholar]
- 434.Simopoulos AP. Human requirement for N-3 polyunsaturated fatty acids. Poult. Sci. 2000;79:961–970. doi: 10.1093/ps/79.7.961. [DOI] [PubMed] [Google Scholar]
- 435.Simopoulos AP. Omega-3 fatty acids in health and disease and in growth and development. Am. J. Clin. Nutr. 1991;54:438–463. doi: 10.1093/ajcn/54.3.438. [DOI] [PubMed] [Google Scholar]
- 436.Leaf A, Weber PC. A new era for science in nutrition. Am. J. Clin. Nutr. 1987;45:1048–1053. doi: 10.1093/ajcn/45.5.1048. [DOI] [PubMed] [Google Scholar]
- 437.Tian GG, Hou C, Li J, Wu J. Three-dimensional genome structure shapes the recombination landscape of chromatin features during female germline stem cell development. Clin. Transl. Med. 2022;12:e927. doi: 10.1002/ctm2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Lin H. The stem-cell niche theory: lessons from flies. Nat. Rev. Genet. 2002;3:931–940. doi: 10.1038/nrg952. [DOI] [PubMed] [Google Scholar]
- 439.Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: stem cells and their niche. Cell. 2004;116:769–778. doi: 10.1016/S0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- 440.Ye H, et al. Ovarian stem cell nests in reproduction and ovarian aging. Cell Physiol. Biochem. 2017;43:1917–1925. doi: 10.1159/000484114. [DOI] [PubMed] [Google Scholar]
- 441.Massasa E, Costa XS, Taylor HS. Failure of the stem cell niche rather than loss of oocyte stem cells in the aging ovary. Aging (Albany NY) 2010;2:1–2. doi: 10.18632/aging.100119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Hong W, et al. Female germline stem cells: aging and anti-aging. J. Ovarian Res. 2022;15:79. doi: 10.1186/s13048-022-01011-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Zhang X, et al. Cadherin 22 participates in the self-renewal of mouse female germ line stem cells via interaction with JAK2 and β-catenin. Cell Mol. Life Sci. 2018;75:1241–1253. doi: 10.1007/s00018-017-2689-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Zhang X, et al. AKT3 Is a pivotal molecule of cadherin-22 and GDNF family receptor-α1 signal pathways regulating self-renewal in female germline stem cells. Stem Cells. 2019;37:1095–1107. doi: 10.1002/stem.3030. [DOI] [PubMed] [Google Scholar]
- 445.Hu Y, et al. GSK3 inhibitor-BIO regulates proliferation of female germline stem cells from the postnatal mouse ovary. Cell Prolif. 2012;45:287–298. doi: 10.1111/j.1365-2184.2012.00821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Ye H, et al. The Hippo signaling pathway regulates ovarian function via the proliferation of ovarian germline stem cells. Cell Physiol. Biochem. 2017;41:1051–1062. doi: 10.1159/000464113. [DOI] [PubMed] [Google Scholar]
- 447.Jiang Y, et al. Hedgehog pathway inhibition causes primary follicle atresia and decreases female germline stem cell proliferation capacity or stemness. Stem Cell Res. Ther. 2019;10:198. doi: 10.1186/s13287-019-1299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Li L, Shi X, Shi Y, Wang Z. The signaling pathways involved in ovarian follicle development. Front. Physiol. 2021;12:730196. doi: 10.3389/fphys.2021.730196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Richards JS, Ren YA, Candelaria N, Adams JE, Rajkovic A. Ovarian follicular theca cell recruitment, differentiation, and impact on fertility: 2017 update. Endocr. Rev. 2018;39:1–20. doi: 10.1210/er.2017-00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Pepling ME. Hedgehog signaling in follicle development. Biol. Reprod. 2012;86:173. doi: 10.1095/biolreprod.112.100859. [DOI] [PubMed] [Google Scholar]
- 451.Finco I, LaPensee CR, Krill KT, Hammer GD. Hedgehog signaling and steroidogenesis. Annu. Rev. Physiol. 2015;77:105–129. doi: 10.1146/annurev-physiol-061214-111754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Cowan RG, Quirk SM. Cells responding to hedgehog signaling contribute to the theca of ovarian follicles. Reproduction. 2021;161:437–448. doi: 10.1530/REP-20-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Russell MC, Cowan RG, Harman RM, Walker AL, Quirk SM. The hedgehog signaling pathway in the mouse ovary. Biol. Reprod. 2007;77:226–236. doi: 10.1095/biolreprod.106.053629. [DOI] [PubMed] [Google Scholar]
- 454.Asiabi P, et al. New insights into the GDF9-Hedgehog-GLI signaling pathway in human ovaries: from fetus to postmenopause. J. Assist. Reprod. Genet. 2021;38:1387–1403. doi: 10.1007/s10815-021-02161-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Ren Y, Cowan RG, Migone FF, Quirk SM. Overactivation of hedgehog signaling alters development of the ovarian vasculature in mice. Biol. Reprod. 2012;86:174. doi: 10.1095/biolreprod.112.099176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Li X, et al. C89 induces autophagy of female germline stem cells via inhibition of the PI3K-Akt pathway in vitro. Cells. 2019;8:606. doi: 10.3390/cells8060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Li, B. et al. GAS5/miR-21 axis as a potential target to rescue ZCL-082-induced autophagy of female germline stem cells in vitro. Mol. Ther. Nucleic Acids17, 436–447 (2019). [DOI] [PMC free article] [PubMed]
- 458.Yuan X, Tian GG, Pei X, Hu X, Wu J. Spermidine induces cytoprotective autophagy of female germline stem cells in vitro and ameliorates aging caused by oxidative stress through upregulated sequestosome-1/p62 expression. Cell Biosci. 2021;11:107. doi: 10.1186/s13578-021-00614-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Li, F., Hu, X. & Wu, J. Daidzein activates Akt pathway to promote the proliferation of female germline stem cells through upregulating Clec11a. Stem Cell Rev. Rep. 10.1007/s12015-022-10394-0 (2022). [DOI] [PubMed]
- 460.Zhang Y, et al. SPATA33 is an autophagy mediator for cargo selectivity in germline mitophagy. Cell Death Differ. 2021;28:1076–1090. doi: 10.1038/s41418-020-00638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Chen K, Huang C, Yuan J, Cheng H, Zhou R. Long-term artificial selection reveals a role of TCTP in autophagy in mammalian cells. Mol. Biol. Evol. 2014;31:2194–2211. doi: 10.1093/molbev/msu181. [DOI] [PubMed] [Google Scholar]
- 462.Luo M, Zhao X, Song Y, Cheng H, Zhou R. Nuclear autophagy: an evolutionarily conserved mechanism of nuclear degradation in the cytoplasm. Autophagy. 2016;12:1973–1983. doi: 10.1080/15548627.2016.1217381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Xu X, et al. RAB37 multiple alleles, transcription activation and evolution in mammals. Int. J. Biol. Sci. 2020;16:2964–2973. doi: 10.7150/ijbs.47959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Yuan J, et al. MYBL2 guides autophagy suppressor VDAC2 in the developing ovary to inhibit autophagy through a complex of VDAC2-BECN1-BCL2L1 in mammals. Autophagy. 2015;11:1081–1098. doi: 10.1080/15548627.2015.1040970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Kumariya S, Ubba V, Jha RK, Gayen JR. Autophagy in ovary and polycystic ovary syndrome: role, dispute and future perspective. Autophagy. 2021;17:2706–2733. doi: 10.1080/15548627.2021.1938914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Sutton MN, et al. RAS-related GTPases DIRAS1 and DIRAS2 induce autophagic cancer cell death and are required for autophagy in murine ovarian cancer cells. Autophagy. 2018;14:637–653. doi: 10.1080/15548627.2018.1427022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Witkin, S. S., Kanninen, T. T. & Sisti, G. in The Role of Heat Shock Proteins in Reproductive System Development and Function (ed. Daniel J. MacPhee) 117–127 (Springer International Publishing, 2017).
- 468.Xu X, et al. Inhibition of sestrin 1 alleviates polycystic ovary syndrome by decreasing autophagy. Aging (Albany NY) 2021;13:11774–1178. doi: 10.18632/aging.202872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Shang D, Wang L, Klionsky DJ, Cheng H, Zhou R. Sex differences in autophagy-mediated diseases: toward precision medicine. Autophagy. 2021;17:1065–1076. doi: 10.1080/15548627.2020.1752511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Ding X, et al. Human GV oocytes generated by mitotically active germ cells obtained from follicular aspirates. Sci. Rep. 2016;6:28218. doi: 10.1038/srep28218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Li X, et al. Generation of offspring-producing 3D ovarian organoids derived from female germline stem cells and their application in toxicological detection. Biomaterials. 2021;279:121213. doi: 10.1016/j.biomaterials.2021.121213. [DOI] [PubMed] [Google Scholar]
- 472.Anderson RA, Wallace WH. Fertility preservation in girls and young women. Clin. Endocrinol. 2011;75:409–419. doi: 10.1111/j.1365-2265.2011.04100.x. [DOI] [PubMed] [Google Scholar]
- 473.Jadoul P, et al. Efficacy of ovarian tissue cryopreservation for fertility preservation: lessons learned from 545 cases. Hum. Reprod. 2017;32:1046–1054. doi: 10.1093/humrep/dex040. [DOI] [PubMed] [Google Scholar]
- 474.Demeestere I, et al. Live birth after autograft of ovarian tissue cryopreserved during childhood. Hum. Reprod. 2015;30:2107–2109. doi: 10.1093/humrep/dev128. [DOI] [PubMed] [Google Scholar]
- 475.Matthews SJ, Picton H, Ernst E, Andersen CY. Successful pregnancy in a woman previously suffering from β-thalassemia following transplantation of ovarian tissue cryopreserved before puberty. Minerva Ginecol. 2018;70:432–435. doi: 10.23736/S0026-4784.18.04240-5. [DOI] [PubMed] [Google Scholar]
- 476.Wu C, et al. Tracing and characterizing the development of transplanted female germline stem cells in vivo. Mol. Ther. 2017;25:1408–1419. doi: 10.1016/j.ymthe.2017.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Hanley NA, et al. SRY, SOX9, and DAX1 expression patterns during human sex determination and gonadal development. Mech. Dev. 2000;91:403–407. doi: 10.1016/S0925-4773(99)00307-X. [DOI] [PubMed] [Google Scholar]
- 478.Stévant I, Nef S. Genetic control of gonadal sex determination and development. Trends Genet. 2019;35:346–358. doi: 10.1016/j.tig.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 479.Bock C, et al. The organoid cell atlas. Nat. Biotechnol. 2021;39:13–17. doi: 10.1038/s41587-020-00762-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Larsen BM, et al. A pan-cancer organoid platform for precision medicine. Cell Rep. 2021;36:109429. doi: 10.1016/j.celrep.2021.109429. [DOI] [PubMed] [Google Scholar]
- 481.Sun A, et al. 3D in vivo magnetic particle imaging of human stem cell-derived islet organoid transplantation using a machine learning algorithm. Front. Cell Dev. Biol. 2021;9:704483. doi: 10.3389/fcell.2021.704483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Wang D, et al. Gene delivery to nonhuman primate preimplantation embryos using recombinant adeno-associated virus. Adv. Sci. (Weinh.) 2019;6:1900440. doi: 10.1002/advs.201900440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Medrano JV, et al. Basic and clinical approaches for fertility preservation and restoration in cancer patients. Trends Biotechnol. 2018;36:199–215. doi: 10.1016/j.tibtech.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 484.Wyns C, Kanbar M, Giudice MG, Poels J. Fertility preservation for prepubertal boys: lessons learned from the past and update on remaining challenges towards clinical translation. Hum. Reprod. Update. 2021;27:433–459. doi: 10.1093/humupd/dmaa050. [DOI] [PubMed] [Google Scholar]
- 485.Kawwass JF, Shandley LM, Boulet SL, Hipp HS. Oncologic oocyte cryopreservation: national comparison of fertility preservation between women with and without cancer. J. Assist. Reprod. Genet. 2020;37:883–890. doi: 10.1007/s10815-020-01715-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Gauthier-Fisher A, Kauffman A, Librach CL. Potential use of stem cells for fertility preservation. Andrology. 2020;8:862–878. doi: 10.1111/andr.12713. [DOI] [PubMed] [Google Scholar]
- 487.De Sanctis V, et al. Testicular damage in children and adolescents treated for malignancy: a short review. Acta Biomed. 2018;89:7–17. doi: 10.23750/abm.v89i3-S.7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Fisch B, Abir R. Female fertility preservation: past, present and future. Reproduction. 2018;156:F11–F27. doi: 10.1530/REP-17-0483. [DOI] [PubMed] [Google Scholar]
- 489.Donnez J, Dolmans MM. Fertility preservation in men and women: where are we in 2021? Are we rising to the challenge? Fertil. Steril. 2021;115:1089–1090. doi: 10.1016/j.fertnstert.2021.03.028. [DOI] [PubMed] [Google Scholar]
- 490.Argyle CE, Harper JC, Davies MC. Oocyte cryopreservation: where are we now? Hum. Reprod. Update. 2016;22:440–449. doi: 10.1093/humupd/dmw007. [DOI] [PubMed] [Google Scholar]
- 491.Grin L, Girsh E, Harlev A. Male fertility preservation-methods, indications and challenges. Andrologia. 2021;53:e13635. doi: 10.1111/and.13635. [DOI] [PubMed] [Google Scholar]
- 492.Hussein RS, Khan Z, Zhao Y. Fertility preservation in women: indications and options for therapy. Mayo Clin. Proc. 2020;95:770–783. doi: 10.1016/j.mayocp.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 493.Gul M, et al. Review of injection techniques for spermatogonial stem cell transplantation. Hum. Reprod. Update. 2020;26:368–391. doi: 10.1093/humupd/dmaa003. [DOI] [PubMed] [Google Scholar]
- 494.Yasmin E, Mitchell R, Lane S. Preservation of fertility in teenagers and young adults treated for haematological malignancies. Lancet Haematol. 2021;8:e149–e160. doi: 10.1016/S2352-3026(20)30324-0. [DOI] [PubMed] [Google Scholar]
- 495.Jurewicz M, Hillelsohn J, Mehta S, Gilbert BR. Fertility preservation in pubertal and pre-pubertal boys with cancer. Pediatr. Endocrinol. Rev. 2018;15:234–243. doi: 10.17458/per.vol15.2018.jhmg.fertilitypubertalboys. [DOI] [PubMed] [Google Scholar]
- 496.Brannigan RE, Fantus RJ, Halpern JA. Fertility preservation in men: a contemporary overview and a look toward emerging technologies. Fertil. Steril. 2021;115:1126–1139. doi: 10.1016/j.fertnstert.2021.03.026. [DOI] [PubMed] [Google Scholar]
- 497.Iussig B, et al. A brief history of oocyte cryopreservation: arguments and facts. Acta Obstet. Gynecol. Scand. 2019;98:550–558. doi: 10.1111/aogs.13569. [DOI] [PubMed] [Google Scholar]
- 498.Kappy M, Lieman HJ, Pollack S, Buyuk E. Fertility preservation for cancer patients: treatment gaps and considerations in patients’ choices. Arch. Gynecol. Obstet. 2021;303:1617–1623. doi: 10.1007/s00404-021-05985-0. [DOI] [PubMed] [Google Scholar]
- 499.Moragón S, et al. Fertility and breast cancer: a literature review of counseling, preservation options and outcomes. Crit. Rev. Oncol. Hematol. 2021;166:103461. doi: 10.1016/j.critrevonc.2021.103461. [DOI] [PubMed] [Google Scholar]
- 500.Howell S, Shalet S. Gonadal damage from chemotherapy and radiotherapy. Endocrinol. Metab. Clin. North Am. 1998;27:927–943. doi: 10.1016/S0889-8529(05)70048-7. [DOI] [PubMed] [Google Scholar]
- 501.Chhabra S, Kutchi I. Fertility preservation in gynecological cancers. Clin. Med. Insights Reprod. Health. 2013;7:49–59. doi: 10.4137/CMRH.S10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Okada K, Fujisawa M. Recovery of spermatogenesis following cancer treatment with cytotoxic chemotherapy and radiotherapy. World J. Mens. Health. 2019;37:166–174. doi: 10.5534/wjmh.180043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Vo KCT, Kawamura K. Female oncofertility: current understandings, therapeutic approaches, controversies, and future perspectives. J. Clin. Med. 2021;10:5690. doi: 10.3390/jcm10235690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Dohle GR. Male infertility in cancer patients: review of the literature. Int. J. Urol. 2010;17:327–331. doi: 10.1111/j.1442-2042.2010.02484.x. [DOI] [PubMed] [Google Scholar]
- 505.Müller J. Impact of cancer therapy on the reproductive axis. Horm. Res. 2003;59(Suppl 1):12–20. doi: 10.1159/000067835. [DOI] [PubMed] [Google Scholar]
- 506.Voutsadakis IA. The chemosensitivity of testicular germ cell tumors. Cell Oncol. (Dordr.) 2014;37:79–94. doi: 10.1007/s13402-014-0168-6. [DOI] [PubMed] [Google Scholar]
- 507.DeWire M, et al. Pubertal development and primary ovarian insufficiency in female survivors of embryonal brain tumors following risk-adapted craniospinal irradiation and adjuvant chemotherapy. Pediatr. Blood Cancer. 2015;62:329–334. doi: 10.1002/pbc.25274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Kinsella TJ. Effects of radiation therapy and chemotherapy on testicular function. Prog. Clin. Biol. Res. 1989;302:157–171. [PubMed] [Google Scholar]
- 509.Mitchell RT, Saunders PTK, Sharpe RM, Kelnar CJH, Wallace WHB. Male fertility and strategies for fertility preservation following childhood cancer treatment. Endocr. Dev. 2009;15:101–134. doi: 10.1159/000207612. [DOI] [PubMed] [Google Scholar]
- 510.Arnon J, Meirow D, Lewis-Roness H, Ornoy A. Genetic and teratogenic effects of cancer treatments on gametes and embryos. Hum. Reprod. Update. 2001;7:394–403. doi: 10.1093/humupd/7.4.394. [DOI] [PubMed] [Google Scholar]
- 511.Jahnukainen K, Mitchell RT, Stukenborg JB. Testicular function and fertility preservation after treatment for haematological cancer. Curr. Opin. Endocrinol. Diabetes Obes. 2015;22:217–223. doi: 10.1097/MED.0000000000000156. [DOI] [PubMed] [Google Scholar]
- 512.Telfer EE. Fertility preservation: progress and prospects for developing human immature oocytes in vitro. Reproduction. 2019;158:F45–F54. doi: 10.1530/REP-19-0077. [DOI] [PubMed] [Google Scholar]
- 513.Klipstein S, Fallat ME, Savelli S. Fertility preservation for pediatric and adolescent patients with cancer: medical and ethical considerations. Pediatrics. 2020;145:e20193994. doi: 10.1542/peds.2019-3994. [DOI] [PubMed] [Google Scholar]
- 514.Rebar RW. Social and ethical implications of fertility preservation. Fertil. Steril. 2016;105:1449–1451. doi: 10.1016/j.fertnstert.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 515.McCracken K, Nahata L. Fertility preservation in children and adolescents: current options and considerations. Curr. Opin. Obstet. Gynecol. 2017;29:283–288. doi: 10.1097/GCO.0000000000000395. [DOI] [PubMed] [Google Scholar]
- 516.Petropanagos A, Cattapan A, Baylis F, Leader A. Social egg freezing: risk, benefits and other considerations. CMAJ. 2015;187:666–669. doi: 10.1503/cmaj.141605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Kubicek P, et al. Could aberrant migration explain metachronous germ cell tumors? Cancer Invest. 2021;39:195–201. doi: 10.1080/07357907.2020.1828447. [DOI] [PubMed] [Google Scholar]
- 518.Prall JA, McGavran L, Greffe BS, Partington MD. Intracranial malignant germ cell tumor and the Klinefelter syndrome. Case report and review of the literature. Pediatr. Neurosurg. 1995;23:219–224. doi: 10.1159/000120962. [DOI] [PubMed] [Google Scholar]
- 519.Nakatsuka S, et al. Primary extragonadal germinoma of the medulla oblongata. Int. J. Surg. Pathol. 2012;20:276–279. doi: 10.1177/1066896911424489. [DOI] [PubMed] [Google Scholar]
- 520.Ueno T, et al. Spectrum of germ cell tumors: from head to toe. Radiographics. 2004;24:387–404. doi: 10.1148/rg.242035082. [DOI] [PubMed] [Google Scholar]
- 521.Spunt SL, et al. Brain metastases of malignant germ cell tumors in children and adolescents. Cancer. 2004;101:620–626. doi: 10.1002/cncr.20411. [DOI] [PubMed] [Google Scholar]
- 522.Bassetto MA, Pasini F, Franceschi T, Mustacchi G, Cetto GL. Extragonadal germ cell tumor: a clinical study. Anticancer Res. 1995;15:2751–2754. [PubMed] [Google Scholar]
- 523.Utz DC, Buscemi MF. Extragonadal testicular tumors. J. Urol. 1971;105:271–274. doi: 10.1016/S0022-5347(17)61508-8. [DOI] [PubMed] [Google Scholar]
- 524.Ichimura K, et al. Recurrent neomorphic mutations of MTOR in central nervous system and testicular germ cell tumors may be targeted for therapy. Acta Neuropathol. 2016;131:889–901. doi: 10.1007/s00401-016-1557-x. [DOI] [PubMed] [Google Scholar]
- 525.Cheng L, et al. OCT4: biological functions and clinical applications as a marker of germ cell neoplasia. J. Pathol. 2007;211:1–9. doi: 10.1002/path.2105. [DOI] [PubMed] [Google Scholar]
- 526.Yang L, et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 2020;5:8. doi: 10.1038/s41392-020-0110-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Le PN, McDermott JD, Jimeno A. Targeting the Wnt pathway in human cancers: therapeutic targeting with a focus on OMP-54F28. Pharmacol. Ther. 2015;146:1–11. doi: 10.1016/j.pharmthera.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Jimeno A, et al. Phase I study of the Hedgehog pathway inhibitor IPI-926 in adult patients with solid tumors. Clin. Cancer Res. 2013;19:2766–2774. doi: 10.1158/1078-0432.CCR-12-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Wu Y, et al. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 2015;25:67–79. doi: 10.1038/cr.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Lin P, Jiang J, Wu M. CRISPR base editor treats premature-aging syndrome. Signal Transduct. Target. Ther. 2021;6:158. doi: 10.1038/s41392-021-00576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Koblan LW, et al. In vivo base editing rescues Hutchinson-Gilford progeria syndrome in mice. Nature. 2021;589:608–614. doi: 10.1038/s41586-020-03086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Chen M, et al. Mutations of MSH5 in nonobstructive azoospermia (NOA) and rescued via in vivo gene editing. Signal Transduct. Target. Ther. 2022;7:1. doi: 10.1038/s41392-021-00710-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Li X, Sun T, Wang X, Tang J, Liu Y. Restore natural fertility of Kit(w)/Kit(wv) mouse with nonobstructive azoospermia through gene editing on SSCs mediated by CRISPR-Cas9. Stem Cell Res. Ther. 2019;10:271. doi: 10.1186/s13287-019-1386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Ma H, et al. Correction of a pathogenic gene mutation in human embryos. Nature. 2017;548:413–419. doi: 10.1038/nature23305. [DOI] [PubMed] [Google Scholar]
- 535.de Melo-Martín I, Rosenwaks Z. Human embryo genetic editing: hope or pipe dream? Fertil. Steril. 2021;116:25–26. doi: 10.1016/j.fertnstert.2021.04.017. [DOI] [PubMed] [Google Scholar]
- 536.Stadtmauer EA, et al. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367:eaba7365. doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Lu Y, et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 2020;26:732–740. doi: 10.1038/s41591-020-0840-5. [DOI] [PubMed] [Google Scholar]
- 538.He S. The first human trial of CRISPR-based cell therapy clears safety concerns as new treatment for late-stage lung cancer. Signal Transduct. Target. Ther. 2020;5:168. doi: 10.1038/s41392-020-00283-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Li H, et al. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct. Target. Ther. 2020;5:1. doi: 10.1038/s41392-019-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Mulder CL, et al. Spermatogonial stem cell autotransplantation and germline genomic editing: a future cure for spermatogenic failure and prevention of transmission of genomic diseases. Hum. Reprod. Update. 2016;22:561–573. doi: 10.1093/humupd/dmw017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Zhuo C, et al. Spatiotemporal control of CRISPR/Cas9 gene editing. Signal Transduct. Target. Ther. 2021;6:238. doi: 10.1038/s41392-021-00645-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Fang R, et al. Highly efficient gene editing and single cell analysis of hematopoietic stem/progenitor cells from X-linked sideroblastic anemia patients. Signal Transduct. Target. Ther. 2021;6:248. doi: 10.1038/s41392-021-00622-3. [DOI] [PMC free article] [PubMed] [Google Scholar]