Abstract
A growing need for organs and novel cell-based therapies has provided a niche for approaches like interspecies chimeras. To generate organs from one donor species in another host species requires techniques such as blastocyst complementation and gene editing to successfully create an embryo that has cells from both the donor and the host. However, the task of developing highly efficacious and competent interspecies chimeras is met by many challenges. These interspecies chimeric barriers impede the formation of chimeras, often leading to lower levels of chimeric competency. The barriers that need to be addressed include the evolutionary distance between species, stage-matching, temporal and spatial synchronization of developmental timing, interspecies cell competition and the survival of pluripotent stem cells and embryos, compatibility of ligand–receptor signaling between species, and the ethical concerns of forming such models. By overcoming the interspecies chimera barriers and creating highly competent chimeras, the technology of organ and cellular generation can be honed and refined to develop fully functioning exogenic organs, tissues, and cells for transplantation.
Keywords: interspecies chimeras, xenogeneic barriers, chimera competency, interspecies organogenesis
Introduction
As an established and practical treatment option for patients with end-stage organ dysfunction, organ transplantation is standard of care to replace liver, kidney, pancreas, heart, and lung. However, several barriers exist and can still impede the success of organ transplantation. Organ supply from both deceased and living donors is unable to address the increasing demand both within the United States and around the world. Although the success of immunosuppression for transplantation has advanced significantly, the available therapies are not always sufficient to prevent graft rejection 1 . The field of regenerative medicine has begun exploring the possibility of new technologies such as stem cells and tissue engineering to repair damaged organs and restore cellular function. Regenerative medicine now has the potential to provide transplantation of tissues and organs that is free of the requirements for immunosuppression and to create an unlimited source of organs 2 . The clinical relevance and application of generating autologous, on-demand organs to combat these major barriers of supply-demand and immunoreactivity would be enormous.
Intraspecies and interspecies chimeras are useful tools with the potential for clinical implementation due to their broad and basic translational applications. Chimeras contain cell types with two or more distinct genetic lineages used to study cell potency, signaling pathways, cell-to-cell communication, production of exogenic organs, and the generalized early development of animals and humans. Chimeric models make the engineering of exogenic organs possible by taking advantage of stem cell technologies and the process of blastocyst complementation as a creative tool. The technology of blastocyst complementation allows us to generate organs via the injection of normal pluripotent stem cells (PSCs) into a dysorganogenetic embryo 3 . The absent organ is produced by the exogenous cells and maintained and grown by the host embryo. This approach allows for exogenic organs of one species to be created within the body of another species as a biological incubator.
In the last decade, PSC-derived organs have been at the forefront of chimeric studies. By exploiting genetically engineered developmental niches, these studies demonstrated the ability to generate both mouse and rat pancreases in Pdx1−/− mouse blastocysts 4 . The goal in exogenic organ development is to have adequate contribution of the injected PSCs into the preimplantation blastocyst or postimplantation embryos4–6 (Fig. 1). The ability to form exogenic organs in intraspecies PSC-derived tissues is far more efficient compared with interspecies PSC-derived organs, which incorporate barriers to impede chimera formation. These barriers include (1) evolutionary distance between species; (2) stage-matching for temporal and spatial synchronization of developmental timing; (3) cell competition and the survival of PSCs and embryos; (4) compatibility of ligand–receptor signaling between species; and (5) the ethical concerns of creating such models. Successfully overcoming interspecies barriers allows the field to better utilize the chimeric animal model and technology to produce organs and tissues for transplantation.
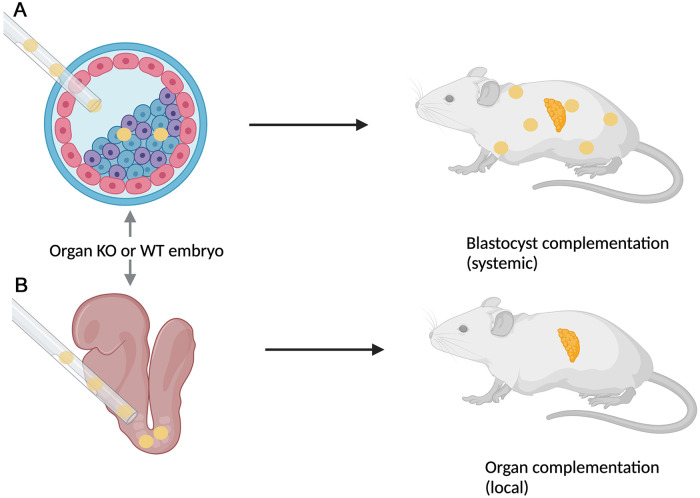
Figure 1.
Generation of interspecies chimeras and organs. (A) Using blastocyst complementation, donor stem cells (yellow) from one species are injected into a host blastocyst. The donor stem cells contribute more robustly to host animals, often leading to systemic contribution, represented by the donor cells found throughout the host animal. (B) Injection of donor stem cells (yellow) into postimplantation embryos results in more limited, localized contribution to the host animal, represented by the donor stem cells in a single organ. WT: wild type; KO: knockout.
Evolutionary Distance
Understanding evolutionary distance between species provides insights into molecular and cellular mechanisms that are critical to the formation of interspecies chimeras for exogenic organ development. Orthologous genes conserve molecular and cellular mechanisms associated with tissues, diseases, and aging between species, suggesting that comparable genes in different species encode for similar functions that drive development 7 . Orthologous genes evolved from a common ancestry but developed separately in individual species. While they preserve the overall outcome of a given protein, the mechanisms involved in their synthesis and/or function can vary significantly. Thus, there are differences in the gene and protein operating mechanisms between species that may contribute to inefficient chimera formation. Given the increased conservation among certain genes, one would predict a successful outcome in interspecies chimera formation within these developmental niches. However, despite the high levels of conservation of these cellular and molecular mechanisms, interspecies chimera formation remains a challenging outcome. Additional issues that must be considered in the deconstruction of the interspecies barriers to evolutionary distance include time divergence estimates and genomic and epigenomic factors.
Due to an ever-increasing amount of molecular data and fossil records, databases have been developed to make information readily accessible to the scientific community. TimeTree is an example of a database that acts as a resource for collecting information related to time of divergence, rates of change, and consensus among studies 8 . According to the TimeTree database, molecular time estimates from 75 different studies comparing rodents with humans produced an average estimated divergence time of 90 million years ago9,10. Rat and mouse estimated time of divergence from 84 studies is 20.9 million years 9 . Estimating divergence between species can be difficult using molecular methods and fossil records due to varying rates of species evolution. Some protein sequence studies utilize modeling that takes advantage of large data sets to determine the estimated divergence between species. The estimated time of divergence for the various species provides insight into the evolutionary distinctions that make each species unique. Most notably, it also addresses the complexity of creating interspecies chimeras. The genomic variations between species observed from previous studies further complicate the formation of interspecies chimeras due to the operational variances of molecular and cellular mechanisms for a specific animal. Rats and mice are far more similar than humans and rodents, observed by the roughly 70-million-year time gap between rat-mouse time divergence compared with that of human-rodent. These genomic differences are even further entangled and complicated by the myriad of variations in the epigenome.
Epigenomic modifications create an additional layer of information that is available for understanding the evolutionary genomic distance between species. Comparative epigenomic exploration is in its infancy, and many of the effects of those modifications are unknown11,12. These epi-modifications comprise a number of chemical changes affecting an array of functions and regulatory mechanisms within the cell11,13. Additional studies indicate the importance of epigenomes in regulating early vertebrate development. Because epigenetic modifications affect most DNA-dependent processes, they have the potential to mark genes for transcriptional activity or repression in a cell-specific fashion, leading to distinct cell fate(s) 13 . The gene regulatory networks epigenetically constrain or overcome those that are creating species change. The epigenome modifications early in development remain potentially critical factors that must be considered for ensuring successful chimera formation. Epigenetic modifications contribute to the complexity of evolutionary distance and the development of interspecies chimeras. Fewer than 37% of the tissue-specific epigenetic marks observed are conserved between human and mouse 12 . However, in recent years, the possibility to reprogram somatic cells to a pluripotent state has become a reality. Most notably, that seminal discovery demonstrates the ability to defy the epigenetic barriers by ectopically expressing pluripotency regulators that are normally expressed in both DNA and histone methylation14–16. Breaking through the epigenetic barrier by reprogramming somatic cells now makes it possible to alter the epigenome, thereby improving the formation and efficiency of interspecies chimeras. Based on the differences observed between species’ genomes and epigenomes, the next step becomes management and matching of these developmental changes between species.
Matched Developmental Timing Through Temporal and Spatial Synchronization
Early embryonic developmental timelines are different among species. For example, a human’s developmental time will be much longer than that of a rodent. The ability to match or manipulate these timelines could potentially increase levels of chimeric competency. The concept of stage-matching developmental timelines for multiple species demonstrates the ability to align both temporally and spatially for synchronization of species development. Temporal and spatial alignment of cellular and molecular mechanisms between species via stage-matching potentially allows for the correct developmental cues in the host embryo to trigger the appropriate response in the donor cells. The genome, transcriptome, and proteome data available for species-specific cell lines and embryos provide invaluable genetic information that is involved in early embryonic development.
Transcriptome data of human and mouse embryogenesis demonstrate the preservation of transcriptional changes in stage-specific time points. Many of the gene networks are highly conserved in maintaining and expressing the transcriptional architecture that is needed for development. Further interrogation with gene ontology analyses indicates that stage-specific time points between mouse and human have many functional similarities. The core transcriptional components are shared among early embryogenesis between humans and mice. However, a divergence in the species’ stage-specificity and timing occurs, which is likely influenced by species-specific differences in human and mouse gestational periods. Transcription factor expression in these species establishes initial cell fates in early embryogenesis via mechanisms such as autoregulation, reciprocal feedback loops, and lineage-specific transcription factors17,18. In addition, transcriptome data obtained from single-cell RNA sequencing (scRNA-seq) provide further information into the sequence, that is, temporal and spatial expression of genes activated and genetically programmed in early embryogenesis19–21.
Multiple studies have analyzed scRNA-seq data of embryos at different developmental stages and across multiple species for in silico stage-matching in interspecies chimerism and blastocyst complementation. In addition to embryos, scRNA-seq data of different embryonic stem cell (ESC) and induced pluripotent stem cell (iPSC) lines have also been used for interspecies stage-matching. The overarching hypothesis behind these in silico studies is that the higher the similarity in transcriptomes between any two developmental stages, the greater the probability of generating efficient chimera. Using the in silico approach, numerous studies have stage-matched different combinations of human, nonhuman primate, mouse, and pig early embryos. For example, the human inner cell mass (ICM) is best matched with the marmoset ICM and the pig late blastocyst stage embryos22,23. These in silico stage-matching results agree with the successful in vivo and ex vivo interspecies chimerism and blastocyst complementation studies published to date6,24,25. Further analysis of the scRNA-seq datasets using Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) provides insight into the genes and developmental pathways that are similar as well as distinct between different species. These insights can be used to modulate the genes in the grafted cells or the host embryos to increase the efficiency of chimerism.
In addition to in silico stage-matching methods, techniques to relate the temporal development between the species of the grafted cells and host embryos are also being explored. Early embryos develop at different rates across different species, providing an opportunity for aligning important timepoints of embryo development between species. For example, the early embryo undergoes implantation around embryonic day 5 in the mouse and around embryonic day 8 in human. Previous studies have explored the causes for interspecies differences in the oscillation speed of the segmentation clock. These studies have shown that although the genes are conserved across different species, specific biochemical factors, translational rates, and protein degradation rates account for the observed differences 26 . Decreasing the temporal development of the mouse to match that of the human or other grafted cells’ species could ensure that those cells develop at the same rate as in the host embryo and increase the efficiency of chimerism. Altering the temporal and spatial development in the donor cells or host embryo may play a critical role in interspecies chimera formation. Gene activation and gene repression may provide tools to achieve the temporal development alignment, and location of key gene networks may supply information on spatial alignment required during interspecies embryogenesis.
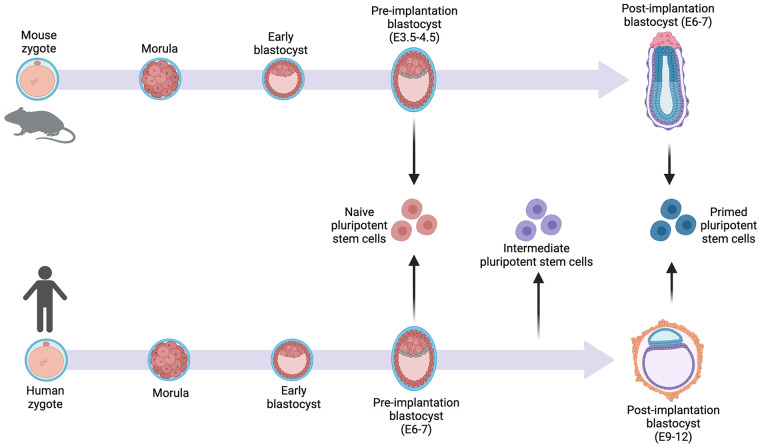
Further comparisons between host embryos and donor iPSCs or ESCs have revealed that PSC lines more closely match with certain embryonic stages. This is observed in the naïve or primed state of the donor stem cells. Naïve human stem cells resemble preimplantation human day 6 blastocysts and primed human iPSCs resemble postimplantation human day 10 epiblasts. An additional stem cell line was determined to be closer to day 8 epiblasts, making it an intermediate stage stem cell 27 . Comparisons among mice and human cells demonstrated that human pluripotent stem cells more closely resembled mouse epiblast-derived stem cells from the early postimplantation stage of development28,29. The naïve, primed, or intermediate states of human stem cells matched with the host embryo cells at the appropriate time may form more successful chimeras (Fig. 2). However, when trying to create interspecies chimeras, the state of both species must be considered. Mouse ESCs derived from murine embryo cultures were found to be in the naïve ground state 30 . The naïve ground state of the mouse should be matched with the naïve state in human stem cells to match the developmental cues between the species. The differences observed between various species stem cell states represent the incongruencies of activated and repressed gene networks at a given time in each species. In addition, cell cultures are maintained at the same status of pluripotency, while a species embryogenesis is dynamic. The ability to stage-match and continue to meet the needs of the host’s embryonic environment and donor stem cells is crucial for synchronization.
Figure 2.
The naïve state of pluripotent stem cells aligns with the best match embryonic stage of development. Mice and human morphologic development are similar through the blastocyst stage, with human development being slower than that of mouse. The naïve stem cells and primed stem cells most resemble the preimplantation blastocyst and postimplantation blastocyst, respectively, for the different species. Human intermediate stem cells fall between the preimplantation and postimplantation blastocyst stage of development.
Interspecies Ligand–Receptor Interactions
The embryonic environment of an interspecies chimera is further complicated by associated ligand–receptor interactions. Ligands and receptors co-evolved with one another in different species, which improved the binding affinity and specificity of these interactions 31 . The co-evolution of ligands and receptors in different species is called parallel evolution, in which binding affinity and specificity are maintained in divergent species 32 . Due to the co-evolution of species-specific ligands and receptors, the ligand–receptor incompatibilities between species may, in part, be responsible for the observed low-level chimeric competency. To better understand interspecies ligand and receptor interactions, recent studies compared host embryos and chimeric embryos to determine the potential for cellular interactions between species in a chimera33,34. A greater number of ligand–receptor interactions were found in chimeric embryos than in the control embryos 35 . The interactions between different cell types among varying species in embryological development have shown cellular interactions to be more robust between some species compared with others. A previous study reported the amount of ligand–receptor interactions for humans, monkeys, and pigs; these interactions were 89, 98, and 7, respectively. The epiblasts in monkeys and humans shared more human–ligand pairings (eg, fibroblast growth factor-2 and fibroblast growth receptor-3) than those shared by pigs and humans (eg, nerve growth factor receptor and brain-derived neurotrophic factor) 27 . By analyzing these cell–cell interactions with KEGG and GO, specific signaling pathways highlight the differences observed in chimeras and control embryos27,35. The ligand–receptor interactions and pathways derived from these studies can now be considered as future pathways to target for improving chimeric competency (Fig. 3). Humanizing pathways in the host embryo had the potential for narrowing the divergence previously observed between species and their ligand–receptor pairs.
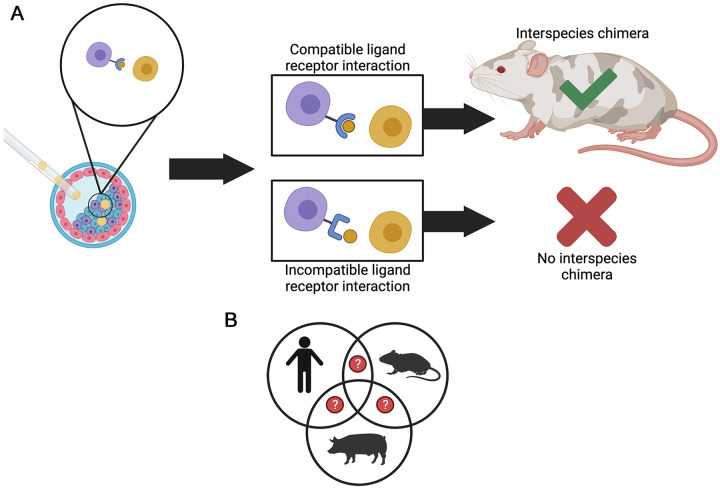
Figure 3.
Compatibility of ligands and receptors may influence the formation of interspecies chimeras. Ligand–receptor pairs found within a developing embryo participate in signaling pathways involved in embryogenesis. These ligand–receptor pairs cue subsequent steps of development to occur in an embryo. The same logic can apply to successful chimeric embryo development. (A) Injected donor stem cells (yellow cells) and host cells (blue, purple, and pink cells) may have similar or different ligand–receptor pairs that can influence the success of chimeras formed through blastocyst complementation. Chimera competency will likely increase when the ligand (brown circle) and receptor (blue shape attached to the purple cell) from two different species are compatible with one another. In contrast, incompatibility will likely prevent the formation of interspecies chimeras. (B) In addition, the cell–cell interactions between different species and tissues are currently being explored to better understand the compatibility of ligands and receptors, which are involved in the formation of interspecies chimera. The Venn diagram depicts human, rodent, and pig cell–cell relationships. The question marks allude to the unknown ligand–receptor pairs that may be crucial for development in chimeric animals. The species may have similarities or differences in these ligand–receptor pairs that may provide insight into interspecies chimera formation.
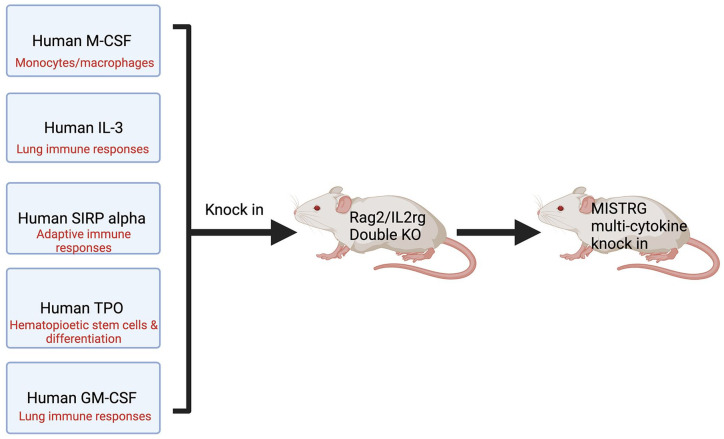
Figure 4.
Generation of MISTRG mice requires the knocking in of human cytokine genes. When knocked in and expressed, human M-CSF, IL-3, SIRP alpha, TPO, and human GM-CSF provide the appropriate microenvironment for human hematopoietic cells to successfully engraft into the mouse. By so doing, this prevents the engulfment of the human cells by phagocytes in the mouse. MISTRG: M-CSF, IL-3, SIRP alpha, thrombopoietin, and human GM-CSF; M-CSF: macrophage colony-stimulating factor; IL-3: interleukin 3; SIRP: signal regulatory protein α; TPO: thrombopoietin; KO: knockout.
Another example of humanization of ligand–receptor interactions to enhance chimera competency is the generation of the human immune system in mice. A mouse model that demonstrates the compatibility to humanize ligand–receptor interactions is the MISTRG mouse, with knock-in human cytokine genes in place of their mouse gene counterparts 36 . MISTRG mice are genetically altered to express human genes for M -CSF, I L-3, S IRP alpha, t h r ombopoietin, and human G M-CSF (MISTRG), which function to increase the tolerance of the mouse phagocytes and deter them from engulfing grafted human cells (Fig. 4)37,38. By targeting homologous cytokine genes in the mouse, this ensures location and functional specificity of the human cytokines. The MISTRG mouse model allows for the induction and maintenance of human hematopoiesis, and further supports the development of human innate immune cells. The human cells exist within the mouse without provoking an immune response. The concept of humanized ligand–receptor interactions provides a nuanced approach to improving chimera competency through the humanization of key embryo developmental time points. The humanized ligand–receptor interactions have the potential to influence the early cell lineages, which may allow for better temporal and spatial synchronization of donor stem cells in a host embryo. In addition, the immune system of the host embryo would theoretically be humanized at an early stage of development and therefore more accepting of the donor stem cells.
Anti-apoptotic Cells/Enhancing Survival of Cells and Embryo
Recent successes in interspecies chimera formation have been hindered by the low chimeric competency that may be driven by death of donor stem cells or the host embryo39–41. The studies by Zheng et al. 40 examined interspecies cell competition between a variety of paired combinations in tissue culture aggregates. They observed that cells of distantly related species exhibited robust competition leading to the survival of cells from the “winning” species and the apoptotic death of cells from the “losing” species. Anti-apoptotic compounds can combat this perceived barrier by interfering with cell death. Anti-apoptotic activity necessary for chimera formation increases the levels of integration and survival of chimeras by improving the survival rate of donor stem cells and host embryos, especially in interspecies chimeras. Anti-apoptotic expression of genes and proteins or other nonprotein compounds increases cell survival or decreases apoptosis in the cells of interest.
Anti-apoptotic compounds target mechanisms and functions of cells to prevent or delay cell death. Under normal conditions, anti-apoptotic proteins are modulated throughout development, and downregulation allows for apoptosis and directs cells toward specific lineages. Some anti-apoptotic proteins identified in interspecies chimera embryos include BCL-2, BCL-XL, MCL1, and BCL-W42,43. In fact, the BCL family demonstrates anti-apoptotic activities through a variety of different mechanisms41,43. The overexpression of BCL-2 has been studied by several groups, and only varying levels of success were achieved in generating chimeras from different donor cell types. BCL-2 overexpression induced Sox17+ endoderm precursor cells to integrate into embryos after blastocyst complementation and formed interspecies chimeras that survived to adulthood 41 . The overexpression of BCL-2 in neural crest cells also formed chimeras with functionally integrated blastocysts. However, these findings were complicated by BCL-2 overexpression delaying differentiation of the ESCs into neural crest cells. Overexpression of BCL-2 in primary neural crest cells injected into blastocysts failed to produce donor cell contributions in embryos 44 . These studies suggested that the anti-apoptotic activity relies on the different cell types, the anti-apoptotic protein expression levels within the cells, and the timing of injection. Ultimately, the anti-apoptotic compounds influence chimera formation through several different mechanisms. It is not quite as simple as only extending the survival of donor stem cells or the embryo through anti-apoptotic compounds. Even in in vitro cultures of pancreatic progenitor cells derived from human PSCs, the levels of expression of BCL-XL become upregulated when apoptosis requires prevention 43 . Those studies demonstrated that the anti-apoptotic activity occurs in PSCs that are used in creating chimeras. Anti-apoptotic activity contributed to the role of cell competition observed when generating chimeras, suggesting a type of survival of the fittest 40 . The role of gene expression in regulating cell death in chimeras demonstrated one important anti-apoptotic activity to consider in developing chimera competency and efficiency. In addition to the importance of genes, the embryonic environment during development remains a crucial factor to consider, and this includes the stress of embryonic development.
Ethical Concerns
The production of interspecies chimeras has been an instrumental animal model in the pursuit of one day achieving exogenic organs for transplantation. However, the use of these models to treat a myriad of human disease presents challenges, not the least of which are ethical in nature. Chimeras derived from blastocyst complementation have organs that develop from both the host embryo and donor stem cells, and the long-term goal of developing autologous human organs within a different species is to then transplant them into the human host. The concern of crossing the species boundaries between humans and animals may encounter public resistance, as the thought of mixing human and animal genes or cells is potentially worrisome45,46. In addition, laws and regulations within various countries regulate the production of interspecies chimeras in research. In fact, some forms of interspecies chimeras are actually illegal in some countries, such as human interspecies chimeras45,47.
The introduction of human cells and genome into animals drives two major ethical concerns that focus primarily on brain chimerism and gamete production. No doubt, there may be a risk of potentially “humanizing” animals created from the integration of human PSCs into an animal’s blastocyst stage. There is a palpable concern for producing human-like “consciousness” or “sentience” in the brain of the host animal or contributing to its reproductive system leading to hybrid embryos 48 . Methods to mitigate the risk of humanizing the host brain include using pluripotent stem cells that are either unable to differentiate into neural cells or undergo a type of programmed cell death referred to as “targeted organ generation”45,48. However, a recent report by Crane et al. 49 , on the transplantation of neurological chimeras with human progenitor cells, suggests that the resulting differentiated human neuronal cells were able to modulate the brain connectome of the recipient species. However, and most importantly, no humanized behaviors emerged from the integration of these human neuronal cells with the neural network of the host brain.
It is important to note that interspecies offspring development via gamete transmission is low due to the strong interspecies reproductive barriers that are in place. This problem can be ceded by the sterilization of these organ donating animals or by using a similar notion proposed for the integration into the brain in which the human PSCs are modified not to differentiate into gametes or undergo apoptosis if differentiation occurs 45 . Despite these concerns regarding the potential contribution of human cells to the brain and reproductive organs of host chimeras, the attitudes of both the Japanese 50 and American 51 publics were positive toward interspecies chimera research to generate organs for transplantation. The technology is literally at the forefront of personalized medicine.
Conclusion
Developing interspecies chimeras is a crucial step toward the progress and application of autologous organ transplantations, and a more accessible interspecies chimeric model to begin with is that of human and mouse. With many interspecies chimeric models, there are current challenges and questions that complicate the ability to produce interspecies chimeras with high efficiency and efficacy. By increasing our understanding of these barriers, the potential to mitigate and strategize ways to overcome the human–animal barriers and obtain successful interspecies chimeras is attainable. As interspecies chimera research continues to advance, the scientific questions, ethical concerns, and public perceptions must be routinely and continually considered in the development of this new and cutting-edge technology for developing organs.
Footnotes
Ethical Approval: This study was approved by our institutional review board.
Statement of Human and Animal Rights: This article does not contain any studies with human or animal subjects.
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported in part from NIH grants R01 DK117286 (CJS) and R01 DK117286-03S1 (CJS and WCL) and from the Suzanne M. Schwarz Fund.
ORCID iDs: Phoebe Strell  https://orcid.org/0000-0002-1700-015X
https://orcid.org/0000-0002-1700-015X
Anala Shetty  https://orcid.org/0000-0003-1185-3183
https://orcid.org/0000-0003-1185-3183
Walter C. Low  https://orcid.org/0000-0001-8593-0175
https://orcid.org/0000-0001-8593-0175
References
- 1. Black CK, Termanini KM, Aguirre O, Hawksworth JS, Sosin M. Solid organ transplantation in the 21st century. Ann Transl Med. 2018;6(20):409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jain A, Bansal R. Applications of regenerative medicine in organ transplantation. J Pharm Bioallied Sci. 2015;7(3):188–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eckardt S, McLaughlin KJ, Willenbring H. Mouse chimeras as a system to investigate development, cell and tissue function, disease mechanisms and organ regeneration. Cell Cycle. 2011;10(13):2091–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kobayashi T, Yamaguchi T, Hamanaka S, Kato-Itoh M, Yamazaki Y, Ibata M, Sato H, Lee Y, Usui J, Knisley AS, Hirabayashi M, et al. Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell. 2010;142(5):787–99. [DOI] [PubMed] [Google Scholar]
- 5. Masaki H, Kato-Itoh M, Umino A, Sato H, Hamanaka S, Kobayashi T, Yamaguchi T, Nishimura K, Ohtaka M, Nakanishi M, Nakauchi H. Interspecific in vitro assay for the chimera-forming ability of human pluripotent stem cells. Development. 2015;142(18):3222–30. [DOI] [PubMed] [Google Scholar]
- 6. Mascetti V, Pedersen R. Human-mouse chimerism validates human stem cell pluripotency. Cell Stem Cell. 2016;18(1):67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Monaco G, van Dam S, Casal Novo Ribeiro JL, Larbi A, de Magalhães JP. A comparison of human and mouse gene co-expression networks reveals conservation and divergence at the tissue, pathway and disease levels. BMC Evol Biol. 2015;15(1):259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22(23):2971–72. [DOI] [PubMed] [Google Scholar]
- 9. TimeTree: the timescale of life. Philadelphia (PA): Institute of Genomics and Evolutionary Medicine Temple University; 2021. http://www.timetree.org/ [accessed 2021 Aug 31]. [Google Scholar]
- 10. Nei M, Xu P, Glazko G. Estimation of divergence times from multiprotein sequences for a few mammalian species and several distantly related organisms. Proc Natl Acad Sci USA. 2001;98(5):2497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xiao S, Xie D, Cao X, Yu P, Xing X, Chen C-C, Musselman M, Xie M, West FD, Lewin HA, Wang T, et al. Comparative epigenomic annotation of regulatory DNA. Cell. 2012;149(6):1381–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou J, Sears RL, Xing X, Zhang B, Li D, Rockweiler NB, Jang HS, Choudhary MNK, Lee HJ, Lowdon RF, Arand J, et al. Tissue-specific DNA methylation is conserved across human, mouse, and rat, and driven by primary sequence conservation. BMC Genomics. 2017;18(1):724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bogdanović O, van Heeringen SJ, Veenstra GJ. The epigenome in early vertebrate development. Genesis. 2011;50(3):192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Athanasiadou R, de Sousa D, Myant K, Merusi C, Stancheva I, Bird A. Targeting of de novo DNA methylation throughout the Oct-4 gene regulatory region in differentiating embryonic stem cells. PLoS ONE. 2010;5(4):e9937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. [DOI] [PubMed] [Google Scholar]
- 17. Frum T, Ralston A. Cell signaling and transcription factors regulating cell fate during formation of the mouse blastocyst. Trends Genet. 2015;31(7):402–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Niwa H. How is pluripotency determined and maintained? Development. 2007;134(4):635–46. [DOI] [PubMed] [Google Scholar]
- 19. Boroviak T, Loos R, Lombard P, Okahara J, Behr R, Sasaki E, Nichols J, Smith A, Bertone P. Lineage-specific profiling delineates the emergence and progression of naive pluripotency in mammalian embryogenesis. Dev Cell. 2015;35(3): 366–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pijuan-Sala B, Griffiths JA, Guibentif C, Hiscock TW, Jawaid W, Calero-Nieto FJ, Mulas C, Ibarra-Soria X, Tyser RCV, Ho DLL, Reik W, et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature. 2019;566(7745):490–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xue Z, Huang K, Cai C, Cai L, Jiang C-y, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, Liu J-y, et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500(7464):593–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boroviak T, Stirparo GG, Dietmann S, Hernando-Herraez I, Mohammed H, Reik W, Smith A, Sasaki E, Nichols J, Bertone P. Single cell transcriptome analysis of human, marmoset and mouse embryos reveals common and divergent features of preimplantation development. Development. 2018;145(21): dev167833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramos-Ibeas P, Sang F, Zhu Q, Tang W, Withey S, Klisch D, Wood L, Loose M, Surani MA, Alberio R. Pluripotency and X chromosome dynamics revealed in pig pre-gastrulating embryos by single cell analysis. Nat Commun. 2019;10(1):500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cohen MA, Wert KJ, Goldmann J, Markoulaki S, Buganim Y, Fu D, Jaenisch R. Human neural crest cells contribute to coat pigmentation in interspecies chimeras after in utero injection into mouse embryos. Proc Natl Acad Sci USA. 2016;113(6): 1570–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cohen MA, Zhang S, Sengupta S, Ma H, Bell GW, Horton B, Sharma B, George RE, Spranger S, Jaenisch R. Formation of human neuroblastoma in mouse-human neural crest chimeras. Cell Stem Cell. 2020;26(4):579–92.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsuda M, Yamanaka Y, Uemura M, Osawa M, Saito MK, Nagahashi A, Nishio M, Guo L, Ikegawa S, Sakurai S, Kihara S, et al. Recapitulating the human segmentation clock with pluripotent stem cells. Nature. 2020;580(7801):124–29. [DOI] [PubMed] [Google Scholar]
- 27. Liu T, Li J, Yu L, Sun H-X, Li J, Dong G, Hu Y, Li Y, Shen Y, Wu J, Gu Y. Cross-species single-cell transcriptomic analysis reveals pre-gastrulation developmental differences among pigs, monkeys, and humans. Cell Discov. 2021;7(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448(7150):191–95. [DOI] [PubMed] [Google Scholar]
- 29. Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448(7150):196–99. [DOI] [PubMed] [Google Scholar]
- 30. Hu Z, Li H, Jiang H, Ren Y, Yu X, Qiu J, Stablewski AB, Zhang B, Buck MJ, Feng J. Transient inhibition of mTOR in human pluripotent stem cells enables robust formation of mouse-human chimeric embryos. Sci Adv. 2020;6(20):eaaz0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Markov GV, Paris M, Bertrand S, Laudet V. The evolution of the ligand/receptor couple: a long road from comparative endocrinology to comparative genomics. Mol Cell Endocrinol. 2008;293(1–2):5–16. [DOI] [PubMed] [Google Scholar]
- 32. Moyle WR, Campbell RK, Myers RV, Bernard MP, Han Y, Wang X. Co-evolution of ligand-receptor pairs. Nature. 1994;368(6468):251–55. [DOI] [PubMed] [Google Scholar]
- 33. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15(4):1484–506. [DOI] [PubMed] [Google Scholar]
- 34. Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, Nie Q. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12(1):1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tan T, Wu J, Si C, Dai S, Zhang Y, Sun N, Zhang E, Shao H, Si W, Yang P, Wang H, et al. Chimeric contribution of human extended pluripotent stem cells to monkey embryos ex vivo. Cell. 2021;184(8):2020–2032.e14. [DOI] [PubMed] [Google Scholar]
- 36. Rongvaux A, Willinger T, Martinek J, Strowig T, Gearty SV, Teichmann LL, Saito Y, Marches F, Halene S, Palucka AK, Manz MG, et al. Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol. 2014;32(4):364–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Radtke S, Chan YY, Sippel TR, Kiem HP, Rongvaux A. MISTRG mice support engraftment and assessment of nonhuman primate hematopoietic stem and progenitor cells. Exp Hematol. 2019;70:31–41.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van den Pol AN, Zhang X, Maher SE, Bothwell ALM. Immune cells enhance Zika virus-mediated neurologic dysfunction in brain of mice with humanized immune systems. Dev Neurobiol. 2021;81(4):389–99. [DOI] [PubMed] [Google Scholar]
- 39. Aksoy I, Rognard C, Moulin A, Marcy G, Masfaraud E, Wianny F, Cortay V, Bellemin-Ménard A, Doerflinger N, Dirheimer M, Mayère C, et al. Apoptosis, G1 phase stall, and premature differentiation account for low chimeric competence of human and Rhesus monkey naive pluripotent stem cells. Stem Cell Reports. 2021;16(1):56–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zheng C, Hu Y, Sakurai M, Pinzon-Arteaga CA, Li J, Wei Y, Okamura D, Ravaux B, Barlow HR, Yu L, Sun HX, et al. Cell competition constitutes a barrier for interspecies chimerism. Nature. 2021;592(7853):272–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Masaki H, Kato-Itoh M, Takahashi Y, Umino A, Sato H, Ito K, Yanagida A, Nishimura T, Yamaguchi T, Hirabayashi M, Era T, et al. Inhibition of apoptosis overcomes stage-related compatibility barriers to chimera formation in mouse embryos. Cell Stem Cell. 2016;19(5):587–92. [DOI] [PubMed] [Google Scholar]
- 42. Haouzi D, Hamamah S. Pertinence of apoptosis markers for the improvement of in vitro fertilization (IVF). Curr Med Chem. 2009;16(15):1905–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Loo LS, Soetedjo AA, Lau HH, Ng NH, Ghosh S, Nguyen L, Krishnan VG, Choi H, Roca X, Hoon S, Teo AKK. BCL-xL/BCL2L1 is a critical anti-apoptotic protein that promotes the survival of differentiating pancreatic cells from human pluripotent stem cells. Cell Death Dis. 2020;11(5):378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cohen MA, Markoulaki S, Jaenisch R. Matched developmental timing of donor cells with the host Is crucial for chimera formation. Stem Cell Reports. 2018;10(5):1445–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bourret R, Martinez E, Vialla F, Giquel C, Thonnat-Marin A, De Vos J. Human–animal chimeras: ethical issues about farming chimeric animals bearing human organs. Stem Cell Res Ther. 2016;7(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Porsdam Mann S, Sun R, Hermeren G. Ethical considerations in crossing the xenobarrier. In: Hyun I, De Los Angeles A, editors. Chimera research: methods and protocols. New York (NY): Humana Press; 2005, p. 175–94. [DOI] [PubMed] [Google Scholar]
- 47. Wu J, Greely HT, Jaenisch R, Nakauchi H, Rossant J, Belmonte JC. Stem cells and interspecies chimaeras. Nature. 2016;540(7631):51–59. [DOI] [PubMed] [Google Scholar]
- 48. Rashid T, Kobayashi T, Nakauchi H. Revisiting the flight of Icarus: making human organs from PSCs with large animal chimeras. Cell Stem Cell. 2014;15(4):406–409. [DOI] [PubMed] [Google Scholar]
- 49. Crane AT, Voth JP, Shen FX, Low WC. Concise review: human-animal neurological chimeras: humanized animals or human cells in an animal? Stem Cells. 2019;37(4):444–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sawai T, Hatta T, Fujita M. Public attitudes in Japan towards human-animal chimeric embryo research using human induced pluripotent stem cells. Regen Med. 2017;12(3):233–48. [DOI] [PubMed] [Google Scholar]
- 51. Crane AT, Shen FX, Brown JL, Cormack W, Ruiz-Estevez M, Voth JP, Sawai T, Hatta T, Fujita M, Low WC. The American public is ready to accept human-animal chimera research. Stem Cell Reports. 2020;15(4):804–10. [DOI] [PMC free article] [PubMed] [Google Scholar]