Abstract
Pathogenic fungi use diverse infection strategies to obtain nutrients from plants. Biotrophic fungi feed only on living plant tissue, whereas necrotrophic fungi kill host cells to extract nutrients. To prevent disease, plants need to distinguish between pathogens with different life cycles, as a successful defense against a biotroph, which often involves programmed cell-death around the site of infection, is not an appropriate response to some necrotrophs. Plants utilize a vast collection of extracellular and intracellular receptors to detect the signatures of pathogen attack. In turn, pathogens are under strong selection to mask or avoid certain receptor responses while enhancing or manipulating other receptor responses to promote virulence. In this review, we focus on the plant receptors involved in resistance responses to fungal pathogens and highlight, with examples, how the infection strategy of fungal pathogens can determine if recognition responses are effective at preventing disease.
Keywords: biotroph, necrotroph, plant immunity
Introduction
Crop-infecting fungi are estimated to be responsible for an annual pre-harvest yield-loss of ∼20% worldwide [1]. The infection strategies used by fungi to obtain plant nutrients can be grouped into three broad categories, biotrophic, hemibiotrophic, and necrotrophic. Biotrophic pathogens extract nutrients from living cells, necrotrophic pathogens kill plant cells to access nutrients, and hemibiotrophs initially extract nutrients from living tissue before switching to a necrotrophic phase.
Plants rely on an innate immune system that is activated by receptor proteins at the cell surface and within the cell [2,3]. Extracellular pathogen recognition is controlled by plasma-membrane embedded receptors, while intracellular recognition often utilizes cytosolic nucleotide binding (NB) leucine-rich repeat (LRR) receptors (NLRs). Receptors are responsible for sensing molecules characteristic of pathogen infection, including microbe-associated molecular patterns (MAMPs), damage-associated molecular patterns (DAMPs), phytocytokines and pathogen-derived virulence molecules, known as effectors (Box 1). Effectors are secreted by adapted pathogens to promote virulence and are utilized by biotrophic, hemibiotrophic, and necrotrophic fungal pathogens to aid in colonization of host plants [4]. The responses activated by plant receptors can include the production of reactive oxygen species (ROS) and anti-microbial compounds, the influx of Ca2+ into the cytosol, immune signaling via mitogen-activated protein kinase (MAPK) cascades, transcriptional reprogramming, and a hypersensitive response (HR) leading to localized plant cell death [5,6].
Box 1. Definitions for the types of molecules that are characteristic of infection and can be sensed by plant receptor proteins to activate defense responses.
| Term | Definition |
|---|---|
| MAMP | Microbe-associated molecular patterns are compounds derived from molecules required for microbial survival. MAMPs do not actively promote infection, examples include chitin oligomers from fungi and flagellin peptides from bacteria. |
| DAMP | Damage-associated molecular patterns are plant molecules that are released by cellular damage resulting from pathogen infection. |
| Phytocytokine | Plant peptides produced in the cytosol and secreted into the apoplast to stimulate plant immunity. |
| Effector | A secreted molecule, typically a small protein, that functions to promote infection. |
In this review, we summarize recent findings concerning pathogen recognition via plant receptors, with a focus on their capacity to activate appropriate defense responses against biotrophic, hemibiotrophic, and necrotrophic pathogens.
Cell-surface recognition
Most cell-surface pathogen-recognition receptors are categorized as either receptor-like proteins (RLPs) or receptor-like kinases (RLKs), depending on the absence or presence of an intracellular kinase domain. Both receptor classes have a membrane-embedded domain and an extracellular ligand-binding domain. RLPs and RLKs can be further classified into subfamilies based on their extracellular domain (ECD). Identified ECDs on plant pathogen recognition receptors include lysin motif (LysM), leucine-rich repeat (LRR), lectin (lec), epidermal growth factor (EGF)-like, galacturonan-binding wall-associated-kinase (GubWAK) and WAK-associated C-terminal domain [7,8] (Figure 1). Cell-surface receptors are essential components to many cellular and developmental functions, and provide the first opportunity for detection of pathogen attack. They are therefore subject to manipulation by plant pathogens to facilitate infection and prevent appropriate resistance responses.
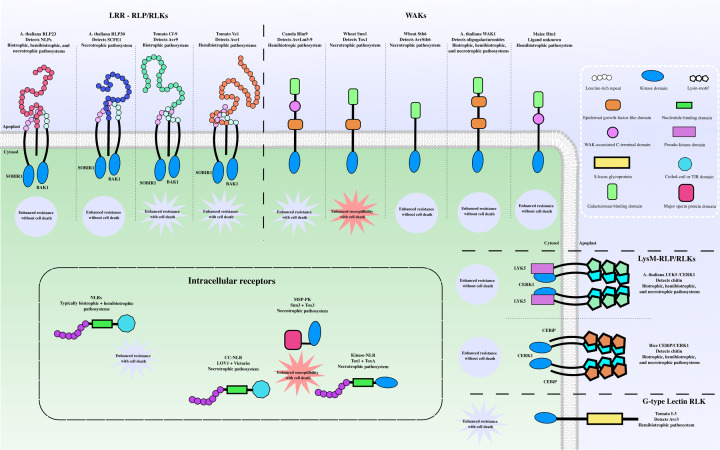
Figure 1. Extracellular and intracellular receptor proteins can promote disease resistance or susceptibility to fungal pathogens.
Cell-surface receptors are embedded in the plasma-membrane while intracellular receptors are localized to the cytosol. The cell-surface receptors are grouped into subfamilies based on their extracellular domain/s. Lysin-motif receptor-like proteins and receptor-like kinases (LysM-RLP/RLKs) can detect chitin oligomers released from the fungal cell wall to activate defense responses that promote disease resistance against biotrophic, hemibiotrophic, and necrotrophic fungi without leading to plant cell death. Leucine-rich repeat (LRR) receptors can detect secreted proteins (effectors) from pathogenic fungi and mediate downstream immune signaling by forming receptor complexes with other LRR-RLKs (SOBIR1 and BAK1). Notably, the LRR-RLP/RLKs that detect effectors from necrotrophic fungi and promote disease resistance do not lead to plant cell death. Likewise, wall-associated kinase (WAK) receptor proteins activate responses that typically promote disease resistance. However, Snn1 from wheat detects a necrotrophic effector (Tox1) and triggers a cell-death response that supports the growth of the invading necrotroph. Inside the cell, intracellular receptors detect effectors and typically promote disease resistance by initiating a localized cell-death response that restricts the growth of biotrophic and hemibiotrophic fungi. However, multiple necrotrophic fungi use effectors to manipulate intracellular receptors into activating cell death, ultimately promoting disease susceptibility. Only the receptors discussed in this review are depicted in the figure.
LysM receptors
LysM-RLP/RLKs are essential for microorganism perception and the activation of various plant responses facilitating defense and symbiosis. The ECDs of LysM receptors typically bind to polysaccharides, including chitin oligomers cleaved from the cell wall of invading fungi. Chitin elicitor receptor kinase1 (CERK1) is a LysM-RLK crucial for chitin-induced defense responses and is present in both monocot and dicot plants [9,10]. In rice, CERK1 is recruited by the chitin elicitor binding protein (CEBiP), a LysM-RLP, following homodimerization of CEBiP promoted by binding to long-chain chitin oligomers [11]. The CERK1 and CEBiP complex activates immune signaling pathways and ultimately promotes fungal disease resistance [11] (Figure 1 and Table 1). Rice CERK1 is also implicated in symbiosis with arbuscular mycorrhizal fungi [12,13] and can detect short-chain chitin oligomers produced by mycorrhizal fungi when in complex MYR1 (another LysM-RLK) [14,15]. A recent discovery demonstrated that MYR1/CERK1 complex formation reduces the chitin-induced immune response by inhibiting the recruitment of CERK1 by CEBiP [16], emphasising the complex nature of cell surface perception and signaling.
Table 1. A summary of the receptors used by plants to detect invading fungi and the outcome of receptor activation as discussed in this review and depicted in Figure 1.
| Plant receptor and species | Identified co-receptor/s | Molecule detected | Infection strategy of associated pathogen/s | Plant cell death | Outcome (enhanced disease resistance or susceptibility) |
|---|---|---|---|---|---|
| LysM-RLP/RLKs | |||||
| LYK5 – A. thaliana | CERK1 | Chitin oligomers | All | No | Resistance |
| CEBiP – Rice | CERK1 | Chitin oligomers | All | No | Resistance |
| LRR-RLP/RLKs | |||||
| RLP30 – A. thaliana | SOBIR1/BAK1 | A small secreted protein | Necrotroph | No | Resistance |
| RLP23 – A. thaliana | SOBIR1/BAK1 | NLPs | All | No | Resistance |
| Ve1 - Tomato | SOBIR1/BAK1 | Ave1 | Hemibiotroph | Yes | Resistance |
| Cf-9 - Tomato | SOBIR1/BAK1 | Avr9 | Biotroph | Yes | Resistance |
| WAKs | |||||
| Stb6 – Wheat | None | AvrStb6 | Necrotroph | No | Resistance |
| Snn1 – Wheat | None | Tox1 | Necrotroph | Yes | Susceptibility |
| Rlm-9 – Canola | None | AvrLm5-9 | Hemibiotroph | Yes | Resistance |
| WAK1 - A. thaliana | None | Oligogalacturonides | All | No | Resistance |
| Htn1 - Maize | None | Unknown | Hemibiotroph | No | Resistance |
| G-type lectin RLKs | |||||
| I-3 – Tomato | None | Avr3 | Hemibiotroph | Yes | Resistance |
| NLRs | |||||
| LOV1 – A. thaliana | None | Victorin | Necrotroph | Yes | Susceptibility |
| Tsn1 – Wheat | None | ToxA | Necrotroph | Yes | Susceptibility |
| MSP-kinase | |||||
| Snn3 – Wheat | None | Tox3 | Necrotroph | Yes | Susceptibility |
This table is not an exhaustive list of all plant receptors that detect pathogenic fungi.
Arabidopsis thaliana has three LysM-RLP homologs of CEBiP, but none of these are required for general chitin-induced immune responses [17]. In A. thaliana, two LysM-RLKs with pseudo-kinase domains (LYK4 and LYK5), in addition to CERK1, are indispensable for the typical immune responses following chitin perception [9,18] (Figure 1 and Table 1). Like CEBiP, LYK4/LYK5 interact with CERK1 to form hetero-oligomers and mediate downstream immune signaling following chitin binding [10,18]. It is believed that the oligomerization of AtLYK4/LYK5 and CEBiP with CERK1 facilitates CERK1 homodimerization and the subsequent phosphorylation of CERK1 that is required for effective downstream signaling [19–21]. The kinase domain of CERK1 potentiates immune signaling by phosphorylating target proteins, including various receptor-like cytoplasmic kinases (RLCKs) [22–27]. In addition to typical intracellular immune responses (ROS production, Ca2+ influx, MAPK signaling, transcriptional changes), recent work demonstrated that chitin perception dependent on CERK1 induces stomatal closure, likely inhibiting the entry of certain foliar pathogens [28,29]. For further details concerning CERK1 mediated defense and the methods fungi use to suppress chitin-triggered immunity, we refer readers to a recent review [19].
By recognizing highly conserved fungal cell wall components, LysM-RLPs/RLKs can inhibit the growth of a diverse range of pathogenic fungi, including biotrophs, hemibiotrophs, and necrotrophs. The fact that a single LysM-RLK can both promote the establishment of symbiotic fungi and restrict the growth of pathogenic fungi implies a complex signaling system which probably requires additional receptors and/or co-receptors to differentiate friend from foe.
LRR receptors
LRR ECDs commonly bind to peptides or proteins (although a well-studied exception is the detection of brassinosteroids (BRs) (reviewed in [30])). Like the LysM-RLP/RLK heteromeric receptor complexes, LRR-RLP/RLKs interact with each other to perceive ligands and potentiate intracellular signaling. To identify LRR-RLK receptor complexes, Smakowska-Luzan et al. used high-throughput assays investigating pair-wise interactions between 200 A. thaliana LRR-RLKs [31]. A key finding from the interaction network was that LRR-RLKs with small ECDs (<12 leucine rich repeats) typically interact with LRR-RLKs with large ECDs (>12 repeats) [31]. The concept that small LRR-RLKs are more likely to act as co-receptors for multiple receptor complexes is demonstrated by the well-characterized BAK1, a small LRR-RLK (4 repeats) that forms receptor complexes with multiple large LRR-RLKs, including those required for the perception of BRs (BRI1, 25 repeats) and peptides from bacterial flagellin (FLS2, 28 repeats) [32–36]. Immune signaling via BAK1 and FLS2 also requires the malectin-like receptor kinase FERONIA, which acts as a scaffold that can be regulated by plant RAPID ALKALINIZATION FACTOR propeptides to inhibit plant immune responses [37].
Most LRR-RLP/RLKs that perceive fungal-specific molecules detect effector proteins. Fulvia fulva (syn. Cladosporium fulvum) is a biotrophic pathogen responsible for leaf mold in tomato. The first effector-detecting LRR-RLP gene identified was Cf-9 (28 repeats) from tomato, that detects the F. fulva effector Avr9 [38]. Subsequent work identified more LRR-RLPs responsible for detecting F. fulva effectors, in all cases, the receptors lack intracellular kinase domains typically used for signal transduction [39–42]. It was therefore assumed that the effector-detecting receptors form complexes with RLKs to initiate immune signaling. SOBIR1, a small LRR-RLK (5 repeats), was the first Cf-9 interacting receptor identified [43]. Additional research determined that BAK1 and SERK1 (both small LRR-RLKs) are recruited to the SOBIR1/Cf receptor complex following ligand binding and are required for immune activation [44]. The Cf receptors trigger a local HR that restricts biotrophic pathogen growth [45] (Figure 1 and Table 1). HR activating LRR-RLPs are also effective at preventing the growth of certain hemibiotrophic pathogens. The Ave1 effector from Verticillium, Fusarium and Colletotrichum spp. is recognized by the LRR-RLP Ve1 (38 repeats) in tomato [46,47]. Like the Cf receptors, Ve1 requires SOBIR1 [43] and BAK1 [48] for complete immune activation (Figure 1 and Table 1). LRR-RLPs are also important for disease resistance against necrotrophic pathogens; however in this case the activation of plant immunity does not result in a HR (Figure 1). The LRR-RLP RLP30 (21 repeats) from A. thaliana detects a small (16–22 kDa) secreted protein from the necrotrophic pathogen Sclerotinia sclerotiorum, activating immune responses and decreasing fungal growth without inducing plant cell-death [49]. Likewise, RLP23 (27 repeats) detects a small peptide from Nep1-like proteins (NLPs) encoded by diverse microbes to activate plant immunity without a HR [50,51] (Figure 1 and Table 1). NLPs can also act as cytotoxic effectors that interact with plasma-membrane embedded sphingolipids and an extracellular plant LRR protein to cause plant cell death, this is believed to promote the virulence of certain necrotrophic and hemibiotrophic pathogens [52–55]. However, NLPs are also encoded by biotrophic pathogens and many are not cytotoxic, suggesting that they likely have another unknown function [55]. By detecting NLPs without inducing a HR, RLP23 increases disease resistance against various biotrophic, hemibiotrophic, and necrotrophic pathogens [50,51,56].
As illustrated in Figure 1, RLP23, RLP30, Ve1, and the Cf-9 receptors all rely on similar RLK co-receptors (SOBIR1 and BAK1) for immune signaling despite differences in the outcome of the immune response [49,51]. The signaling output from LRR-RLP/RLKs is often controlled by the intracellular domain (ICD) and can be uncoupled from the ECD [57]. For example, Wu et al. recently reported that swapping the ECD of Cf-9 with the ECD of EFR (an LRR-RLK that does not activate a HR), leads to a HR when the chimeric protein is expressed in tobacco and exposed to the ligand detected by EFR [58]. The exact mechanism by which the ICDs of RLP/RLKs fine-tune immune signaling is currently unknown. Potentially the different responses may be achieved by the different receptor complexes influencing the phosphorylation of the ICD of BAK1 and/or SOBIR1 which in turn affects downstream signaling. Perraki et al. demonstrated that BAK1 has phosphosites that are required for immune responses activated by FLS2 but are not necessary for BRI1 signaling [59]. Future studies characterizing the phosphorylation of BAK1 and/or SOBIR1 in different receptor complexes may uncover how the receptors effective against biotrophs and hemibiotrophs signal for a HR, while the receptors detecting effectors from necrotrophs signal for immune activation without cell death.
The presented examples demonstrate that LRR receptor complexes have the capacity to increase disease resistance against a broad range of fungal pathogens by detecting effectors. Interestingly, the detection of biotrophic/hemibiotrophic effectors typically activates a localized HR preventing biotrophic growth whereas the detection of necrotrophic effectors stimulates the immune system without causing plant cell death that could promote necrotrophic infection [45,46,49,51].
Lectin domain receptors
Receptors with lectin (carbohydrate binding) domains (Lec-RLKs) are more commonly associated with developmental process rather than defense. Lec-RLKs can be divided into three classes: L-type, C-type and G-type. Only G-type Lec-RLKs are known to detect molecules of fungal origin, for a detailed overview of all types of Lec-RLKs and their potential ligands we refer readers to recent reviews [60,61]. The ECD of G-type Lec-RLKs includes a G-lectin domain, plasminogen-apple-nematode (PAN) motif, and a S-locus glycoprotein (SLG) domain. G-type Lec-RLKs are best known for their role in self-incompatibility (SI) systems [62–64]. To prevent self-fertilization, pollen and pistil cells recognize ‘self’ using a secreted small cysteine-rich protein (SCR) and extracellular SLG-domain containing receptor capable of binding to the SCR [65,66]. SCR-binding can trigger homodimerization of the receptor, likely causing transphosphorylation of the ICDs that initiates downstream signaling [66,67]. Studies in field poppy, reviewed in [68], have demonstrated that SI activation can result in ROS production, increased cytosolic [Ca2+], irreversible oxidation of plant proteins, cytosolic acidification, and cell death [69–73].
The I-3 protein from tomato is currently the only G-type Lec-RLK known to recognize a fungal molecule [74]. I-3 detects the Avr3 effector from the hemibiotroph Fusarium oxysporum f. sp. lycopersici and inhibits pathogen growth (Table 1). Transient expression of Avr3 in I-3 containing tomato or co-expression of Avr3 and I-3 in Nicotiana benthamiana does not lead to a HR [74]. However, it was recently demonstrated that infiltration of purified recombinant Avr3 protein into I-3 tomato results in an I-3-dependent cell-death response [75]. Whether I-3 mediated immunity and plant cell death depends on the same signaling components as SI activation remains to be determined. Avr3 is a small cysteine-rich protein, analogous to the SCRs involved in SI, suggesting that Avr3 may bind directly to the I-3 SLG domain [76]. While not sufficient to rule out a direct interaction, previous yeast-two-hybrid experiments were unable to detect binding of Avr3 to the I-3 ectodomain [74]. Avr1, which is recognized by the LRR-RLP I [77], bears structural similarity to Avr3 [75] and is able to inhibit Avr3 resistance responses [78]. The mechanism of Avr3 mediated resistance suppression by Avr1 is unknown, but in-light of the structural similarity it is tempting to speculate that Avr1 interferes with Avr3 mediated recognition by binding to I-3 or a common ‘guarded’ host target. Given the ability of S-locus glycoproteins to detect small cysteine-rich proteins and activate plant cell death, it is likely that future research will identify more effector detecting G-type Lec-RLKs involved in promoting plant immunity against biotrophic and hemibiotrophic pathogens.
WAK receptors
Wall-associated kinases (WAKs) are characterized by a galacturonan-binding GubWAK ECD and often also have an extracellular EGF-like domain. There are 22 members of the WAK and WAK-like gene family in Arabidopsis [79], but these families have expanded in other plant species with 99 members in Gossypium hirsutum (cotton) [80], 91 members in barley [81], 125 in rice [82] and 555 in wheat [83]. The expansion of WAKs in both monocot and dicot species and the presence of unique structural families within species suggests that WAKs may have evolved following the divergence of monocots and dicots and during speciation. WAK1 from A. thaliana was first described to associate with the plant cell wall [84], specifically via pectin [85]. In addition, a recombinantly produced fragment of the WAK1 ectodomain was able to interact with cell wall extracted polygalacturonic acid, oligogalacturonides and pectin [86]. WAK1 was then shown to activate immunity by recognising plant-derived oligogalacturonides [87,88], and recent evidence demonstrates that WAKs can also detect fungal effectors [89–91]. Rlm9 from canola is a WAK immune receptor that perceives the AvrLm5-9 effector from the hemibiotrophic fungus Leptosphaeria maculans, the causal agent of blackleg disease [90]. Recognition of AvrLm5-9 by Rlm9 results in a HR and inhibits infection, however whether other immune responses are activated, how Rlm-9 potentiates immune signals, or if Rlm-9 requires co-receptors is unknown [90]. Rlm4 and Rlm7 are allelic variants of Rlm9 and detect the L. maculans effectors AvrLm4 and AvrLm4-7 to activate a HR and prevent further infection [91]. AvrLm4-7 and AvrLm5-9 are structurally similar [92], and AvrLm4-7 can inhibit AvrLm5-9 mediated resistance [93]. Like the Avr1 and Avr3 effectors mentioned previously, the mode of inhibition is unknown, but a common means of recognition evasion by pathogens could come from the selection of structurally similar effector proteins that are able to negate established forms of pathogen detection.
A WAK from wheat, Stb6, recognizes the AvrStb6 effector from the necrotrophic pathogen Zymoseptoria tritici [89]. Stb6 lacks an EGF-like domain, suggesting that this domain is not critical for the activation of immune responses by WAKs [89]. As opposed to Rlm9, Stb6 does not induce a HR following effector recognition [89]. The Tox1 effector from the necrotroph Parastagonospora nodorum is also recognized by a wheat WAK, Snn1 [94]. In contrast to all effector detecting receptors discussed above, the recognition of Tox1 by Snn1 promotes disease through a model known as effector-triggered susceptibility [94–96]. Snn1 activation results in typical immune responses, including the production of ROS, the expression of defense-related genes, and plant-cell death [95]. Both Rlm9 and Snn1 activation leads to plant cell death. However, due to different infection strategies, Snn1 enhances disease-susceptibility in the wheat-P. nodorum pathosystem, whereas Rlm9 promotes disease-resistance in the canola-L. maculans pathosystem (Figure 1 and Table 1). An exciting avenue for future research could be aiming to engineer Stb6, a WAK that enhances immunity against a necrotrophic pathogen without inducing a HR, to detect Tox1 by swapping ECDs with Snn1. Proof-of-concept domain-swapping experiments using WAK1 and the LRR-RLK EFR have demonstrated that WAK1 immune signaling by the ICD can be uncoupled from ligand-binding by the ECD [87].
Several other WAKs involved in fungal-disease resistance have been described, but their means of pathogen perception and ability to confer resistance require further investigation (Figure 1). Agriculturally important alleles of the maize WAK ZmWAK-RLK1 are able to delay the progression of northern corn leaf blight (NCLB), but not completely halt the disease [97,98]. A resistance allele of ZmWAK-RLK1 has been reported to modify the expression of genes involved in jasmonic acid (JA) and ethylene defense pathways, lignin synthesis, and cell wall maintenance, with most of these genes differentially expressed before infection occurs [99]. Unlike the previous WAK examples, alleles of ZmWAK-RLK1 may therefore function independently of the recognition of pathogen derived components to confer resistance.
WAKs are fast emerging as important receptor proteins capable of promoting disease resistance against biotrophic, hemibiotrophic, and necrotrophic pathogens (Figure 1 and Table 1). Similar to LRR receptors, the WAKs that detect effector proteins can activate pathways that lead to cell death or to enhanced immunity without cell death. Whether the response reduces or promotes infection depends on the lifestyle of the pathogen. The expansion of the WAK and WAK-like gene family in plants suggests that this family of genes is under strong selection for roles in defense responses, much like the expanded intracellular nucleotide-binding, LRR receptor (NLR) family (discussed below). Unlike NLRs however, WAKs seem to maintain diverse roles in cell maintenance and development [100], as well as in abiotic [101] and biotic stress responses.
Intracellular recognition
NLRs are cytosolic proteins that activate immune responses following the detection of an effector. Typically, NLRs are composed of a C-terminal LRR domain, central NB domain, and variable N-terminal domain. The two major classes of NLR are CC-NLRs that have a coiled-coil N-terminal domain and TIR-NLRs that have a toll/interleukin-1 receptor N-terminal domain. In general, the CC and TIR domains are responsible for signalling and the NB and LRR domains regulate NLR activation. The activation of an NLR by a fungal effector typically results in a HR that prevents the growth of biotrophic and hemibiotrophic pathogens [102]. They are the most crucial defense receptor against many adapted pathogens and have been studied and reviewed extensively. For a detailed understanding of how NLRs perceive effectors, and the subsequent immune signaling pathways see [103–106].
Interestingly, necrotrophic pathogens can co-opt NLR effector recognition to promote cell death and enhance disease. LOV1 is an A. thaliana CC-NLR that confers susceptibility to the necrotrophic fungus Cochliobolus victoriae (Table 1) [107]. LOV1 guards a plant thioredoxin TRX-h5 to detect ribosomally produced cyclic hexapeptides (victorin) secreted by C. victoriae during infection, leading to cell death and plant susceptibility [108,109]. Victorin binds directly to the active site of TRX-h5, and in the absence of LOV1 inhibits plant defense [108]. The high conservation of LOV1 across A. thaliana accessions suggests that it plays an important role in plant immunity, likely mediating resistance against a biotrophic or hemibiotrophic pathogen by detecting an effector that interferes with TRX-h5 [110]. Victorin-induced susceptibility to C. victoriae is also controlled by a CC-NLR in common bean and the gene responsible for susceptibility in oats co-segregates with a resistance gene to the biotrophic pathogen Puccinia coronata [111,112]. Collectively, the victorin research indicates that necrotrophic pathogens can promote infection by secreting effectors that mimic recognised effectors from biotrophic or hemibiotrophic pathosystems to activate a HR. An analogous example involves the protein effector ToxA from necrotrophic pathogens Pyrenophora tritici-repentis, Parastagonospora nodorum, and Bipolaris sorokiniana. ToxA is recognized by the intracellular wheat NLR receptor Tsn1, a non-canonical NLR with a kinase domain at the N-terminus (Table 1) [113]. The ToxA protein, in the absence of the pathogen causes plant cell-death in wheat lines carrying Tsn1 [114]. In the context of these necrotrophic pathogens, ToxA is a major determinant in promoting disease in wheat that possess Tsn1 [115–118]. Snn3 is another wheat disease susceptibility gene that encodes an intracellular protein with a kinase domain [119]. The P. nodorum effector Tox3 is detected by Snn3, which results in the upregulation of defense-related transcripts, indicating that Snn3 likely functions in plant defense but is being manipulated by Tox3 to promote infection [120]. Unlike previously characterized pathogen-detecting receptors, Snn3 has a C-terminal major sperm protein (MSP) domain [119]. It is predicted that the MSP domain is responsible for effector perception and the kinase domain then activates defense responses, including the plant cell-death that promotes the necrotrophic growth of P. nodorum. Genes with MSP and protein kinase domains are specific to monocot plants and significant diversity exists in the sequences of the MSP domains, including the presence of uncharacterized integrated domains [119]. It is tempting to speculate that future research efforts will identify more receptors in monocot plants that use an MSP domain to detect effector proteins and activate defense responses. For further discussion on the various receptors hijacked by necrotrophic pathogens see [121].
To defend against biotrophic and hemibiotrophic pathogens intracellular effector-detecting plant receptors often activate immune signaling that culminates in a HR, restricting the growth of pathogens that require living plant tissue. However, the examples presented here demonstrate how necrotrophic pathogens can manipulate intracellular receptors to promote plant-cell death and enable nutrient acquisition.
Receptor cross-talk
We have divided this review into sections based on the structure of the immune receptors. However, regardless of structure, most receptors activate similar, overlapping pathways and recent advances have demonstrated that diverse receptors work co-operatively to promote plant immunity. BAK1, the common LRR-RLP/RLK co-receptor, phosphorylates CERK1 following activation, which promotes rapid activation of CERK1 following chitin perception [122]. A cotton WAK positively regulates resistance to wilt-diseases by promoting the heterodimerization of the LysM-RLKs LYK5 and CERK1 [123]. Likewise, WAK1 from tomato acts to promote apoplastic immunity activated by an LRR-RLK [124].
Extracellular receptors can also enhance the immune responses activated by intracellular receptors, and vice-versa. The Cf-4 LRR receptor complex requires a downstream CC-NLR to activate a HR and restrict the growth of the biotrophic invader [125,126]. Furthermore, two recent publications demonstrated that NLR activation results in the up-regulation of extracellular receptor signaling components, and the HR activated by NLRs requires the activation of extracellular receptors [127,128]. For an in-depth discussion on the cross-over between immunity activated by intracellular and extracellular receptors, we refer readers to recent reviews [129–131].
Summary
Pathogenic fungi use biotrophic, hemibiotrophic, and necrotrophic infection strategies to infect plants and reduce crop productivity.
Plants produce a variety of receptor proteins that can detect danger from the cell-surface or inside the cell, leading to defense responses against diverse fungal pathogens.
Recent evidence suggests that pathogenic fungi can interfere with receptor activation via the release of effectors with structural similarity to the recognized effectors.
The success of an immune response can depend on the infection strategy of the invader. A hypersensitive cell-death response is often effective at preventing biotrophic growth but can be manipulated by necrotrophs to promote infection.
Acknowledgements
We thank members of the Williams lab for critically reading the manuscript and providing helpful suggestions during our lab group meeting. We apologize to any colleagues for relevant references that were not included due to space constraints.
Abbreviations
- DAMP
damage-associated molecular pattern
- HR
hypersensitive response
- LRR
leucine-rich repeat
- MAMP
microbe-associated molecular pattern
- MAPK
mitogen-activated protein kinase
- NB
nucleotide binding
- NLR
nucleotide binding leucine-rich repeat receptor
- PAN
plasminogen-apple-nematode
- ROS
reactive oxygen species
- SLG
S-locus glycoprotein
- WAK
Wall-associated kinase
Contributor Information
Peter S. Solomon, Email: peter.solomon@anu.edu.au.
Simon J. Williams, Email: simon.williams@anu.edu.au.
Competing Interests
The authors declare that there are no competing interests associated with this manuscript.
Funding
This work was supported by the Australian Research Council (ARC) [grant numbers DP180102355 to P.S., DP200100388 to S.W.]. C.M. is the recipient of an AINSE Ltd. Postgraduate Research Award (PGRA), and S.W. is supported by an ARC Future fellowship [grant number FT200100135].
Open Access
Open access for this article was enabled by the participation of Australian National University in an all-inclusive Read & Publish agreement with Portland Press and the Biochemical Society under a transformative agreement with CAUL.
Author Contribution
C.L.M. and J.R.G. drafted the manuscript. P.S.S. and S.J.W. conceptualized the manuscript. All authors revised and edited the manuscript.
References
- 1.Fisher M.C., Hawkins N.J., Sanglard D. and Gurr S.J. (2018) Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 360, 739–742 10.1126/science.aap7999 [DOI] [PubMed] [Google Scholar]
- 2.van der Burgh A.M. and Joosten M. (2019) Plant immunity: thinking outside and inside the box. Trends Plant Sci. 24, 587–601 10.1016/j.tplants.2019.04.009 [DOI] [PubMed] [Google Scholar]
- 3.Kanyuka K. and Rudd J.J. (2019) Cell surface immune receptors: the guardians of the plant's extracellular spaces. Curr. Opin. Plant Biol. 50, 1–8 10.1016/j.pbi.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lo Presti L., Lanver D., Schweizer G., Tanaka S., Liang L., Tollot M.et al. (2015) Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 66, 513–545 10.1146/annurev-arplant-043014-114623 [DOI] [PubMed] [Google Scholar]
- 5.Peng Y., van Wersch R. and Zhang Y. (2018) Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol. Plant. Microbe. Interact. 31, 403–409 10.1094/MPMI-06-17-0145-CR [DOI] [PubMed] [Google Scholar]
- 6.Tsuda K. and Katagiri F. (2010) Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465 10.1016/j.pbi.2010.04.006 [DOI] [PubMed] [Google Scholar]
- 7.Macho A.P. and Zipfel C. (2014) Plant PRRs and the activation of innate immune signaling. Mol. Cell. 54, 263–272 10.1016/j.molcel.2014.03.028 [DOI] [PubMed] [Google Scholar]
- 8.Stephens C., Hammond-Kosack K.E. and Kanyuka K. (2022) WAKsing plant immunity, waning diseases. J. Exp. Bot. 73, 22–37 10.1093/jxb/erab422 [DOI] [PubMed] [Google Scholar]
- 9.Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K.et al. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618 10.1073/pnas.0705147104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y.et al. (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214 10.1111/j.1365-313X.2010.04324.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayafune M., Berisio R., Marchetti R., Silipo A., Kayama M., Desaki Y.et al. (2014) Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc. Natl. Acad Sci. U.S.A. 111, E404–E413 10.1073/pnas.1312099111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyata K., Kozaki T., Kouzai Y., Ozawa K., Ishii K., Asamizu E.et al. (2014) The bifunctional plant receptor, OsCERK1, regulates both chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Cell Physiol. 55, 1864–1872 10.1093/pcp/pcu129 [DOI] [PubMed] [Google Scholar]
- 13.Zhang X., Dong W., Sun J., Feng F., Deng Y., He Z.et al. (2015) The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J. 81, 258–267 10.1111/tpj.12723 [DOI] [PubMed] [Google Scholar]
- 14.Carotenuto G., Chabaud M., Miyata K., Capozzi M., Takeda N., Kaku H.et al. (2017) The rice LysM receptor-like kinase OsCERK1 is required for the perception of short-chain chitin oligomers in arbuscular mycorrhizal signaling. New Phytol. 214, 1440–1446 10.1111/nph.14539 [DOI] [PubMed] [Google Scholar]
- 15.He J., Zhang C., Dai H., Liu H., Zhang X., Yang J.et al. (2019) A LysM receptor heteromer mediates perception of arbuscular mycorrhizal symbiotic signal in rice. Mol. Plant 12, 1561–1576 10.1016/j.molp.2019.10.015 [DOI] [PubMed] [Google Scholar]
- 16.Zhang C., He J., Dai H., Wang G., Zhang X., Wang C.et al. (2021) Discriminating symbiosis and immunity signals by receptor competition in rice. Proc. Natl. Acad Sci. U.S.A. 118, 10.1073/pnas.2023738118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan J., Tanaka K., Zhang X.C., Son G.H., Brechenmacher L., Nguyen T.H.et al. (2012) LYK4, a lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol. 160, 396–406 10.1104/pp.112.201699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao Y., Liang Y., Tanaka K., Nguyen C.T., Jedrzejczak R.P., Joachimiak A.et al. (2014) The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. Elife 3, e03766, 10.7554/eLife.03766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong B.Q., Wang F.Z. and Li J.F. (2020) Hide-and-seek: chitin-triggered plant immunity and fungal counterstrategies. Trends Plant Sci. 25, 805–816 10.1016/j.tplants.2020.03.006 [DOI] [PubMed] [Google Scholar]
- 20.Liu T., Liu Z., Song C., Hu Y., Han Z., She J.et al. (2012) Chitin-induced dimerization activates a plant immune receptor. Science 336, 1160–1164 10.1126/science.1218867 [DOI] [PubMed] [Google Scholar]
- 21.Liu J., Liu B., Chen S., Gong B.Q., Chen L., Zhou Q.et al. (2018) A tyrosine phosphorylation cycle regulates fungal activation of a plant receptor Ser/Thr kinase. Cell Host Microbe 23, 241.e6–253.e6 10.1016/j.chom.2017.12.005 [DOI] [PubMed] [Google Scholar]
- 22.Zhang J., Li W., Xiang T., Liu Z., Laluk K., Ding X.et al. (2010) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7, 290–301 10.1016/j.chom.2010.03.007 [DOI] [PubMed] [Google Scholar]
- 23.Shinya T., Yamaguchi K., Desaki Y., Yamada K., Narisawa T., Kobayashi Y.et al. (2014) Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J. 79, 56–66 10.1111/tpj.12535 [DOI] [PubMed] [Google Scholar]
- 24.Bi G., Zhou Z., Wang W., Li L., Rao S., Wu Y.et al. (2018) Receptor-like cytoplasmic kinases directly link diverse pattern recognition receptors to the activation of mitogen-activated protein kinase cascades in Arabidopsis. Plant Cell. 30, 1543–1561 10.1105/tpc.17.00981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamada K., Yamaguchi K., Yoshimura S., Terauchi A. and Kawasaki T. (2017) Conservation of chitin-induced MAPK signaling pathways in rice and Arabidopsis. Plant Cell Physiol. 58, 993–1002 10.1093/pcp/pcx042 [DOI] [PubMed] [Google Scholar]
- 26.Ao Y., Li Z., Feng D., Xiong F., Liu J., Li J.F.et al. (2014) OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J. 80, 1072–1084 10.1111/tpj.12710 [DOI] [PubMed] [Google Scholar]
- 27.Wang C., Wang G., Zhang C., Zhu P., Dai H., Yu N.et al. (2017) OsCERK1-mediated chitin perception and immune signaling requires receptor-like cytoplasmic kinase 185 to activate an MAPK cascade in Rice. Mol. Plant 10, 619–633 10.1016/j.molp.2017.01.006 [DOI] [PubMed] [Google Scholar]
- 28.Ye W., Munemasa S., Shinya T., Wu W., Ma T., Lu J.et al. (2020) Stomatal immunity against fungal invasion comprises not only chitin-induced stomatal closure but also chitosan-induced guard cell death. Proc. Natl. Acad Sci. U.S.A. 117, 20932–20942 10.1073/pnas.1922319117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y., Maierhofer T., Rybak K., Sklenar J., Breakspear A., Johnston M.G.et al. (2019) Anion channel SLAH3 is a regulatory target of chitin receptor-associated kinase PBL27 in microbial stomatal closure. Elife 8, e44474, 10.7554/eLife.44474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nolan T.M., Vukašinović N., Liu D., Russinova E. and Yin Y. (2020) Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant Cell. 32, 295–318 10.1105/tpc.19.00335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smakowska-Luzan E., Mott G.A., Parys K., Stegmann M., Howton T.C., Layeghifard M.et al. (2018) An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature 553, 342–346 10.1038/nature25184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J., Wen J., Lease K.A., Doke J.T., Tax F.E. and Walker J.C. (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222 10.1016/S0092-8674(02)00812-7 [DOI] [PubMed] [Google Scholar]
- 33.Nam K.H. and Li J. (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212 10.1016/S0092-8674(02)00814-0 [DOI] [PubMed] [Google Scholar]
- 34.Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D.et al. (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500 10.1038/nature05999 [DOI] [PubMed] [Google Scholar]
- 35.Gómez-Gómez L. and Boller T. (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell. 5, 1003–1011 10.1016/S1097-2765(00)80265-8 [DOI] [PubMed] [Google Scholar]
- 36.Li J. and Chory J. (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938 10.1016/S0092-8674(00)80357-8 [DOI] [PubMed] [Google Scholar]
- 37.Stegmann M., Monaghan J., Smakowska-Luzan E., Rovenich H., Lehner A., Holton N.et al. (2017) The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355, 287–289 10.1126/science.aal2541 [DOI] [PubMed] [Google Scholar]
- 38.Jones D.A., Thomas C.M., Hammond-Kosack K.E., Balint-Kurti P.J. and Jones J.D. (1994) Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266, 789–793 10.1126/science.7973631 [DOI] [PubMed] [Google Scholar]
- 39.Dixon M.S., Hatzixanthis K., Jones D.A., Harrison K. and Jones J.D. (1998) The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell. 10, 1915–1925 10.1105/tpc.10.11.1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dixon M.S., Jones D.A., Keddie J.S., Thomas C.M., Harrison K. and Jones J.D. (1996) The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84, 451–459 10.1016/S0092-8674(00)81290-8 [DOI] [PubMed] [Google Scholar]
- 41.Thomas C.M., Jones D.A., Parniske M., Harrison K., Balint-Kurti P.J., Hatzixanthis K.et al. (1997) Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell. 9, 2209–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takken F.L., Thomas C.M., Joosten M.H., Golstein C., Westerink N., Hille J.et al. (1999) A second gene at the tomato Cf-4 locus confers resistance to cladosporium fulvum through recognition of a novel avirulence determinant. Plant J. 20, 279–288 10.1046/j.1365-313X.1999.t01-1-00601.x [DOI] [PubMed] [Google Scholar]
- 43.Liebrand T.W., van den Berg G.C., Zhang Z., Smit P., Cordewener J.H., America A.H.et al. (2013) Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc. Natl. Acad Sci. U.S.A. 110, 10010–10015 10.1073/pnas.1220015110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Postma J., Liebrand T.W., Bi G., Evrard A., Bye R.R., Mbengue M.et al. (2016) Avr4 promotes Cf-4 receptor-like protein association with the BAK1/SERK3 receptor-like kinase to initiate receptor endocytosis and plant immunity. New Phytol. 210, 627–642 10.1111/nph.13802 [DOI] [PubMed] [Google Scholar]
- 45.Cai X., Takken F.L., Joosten M.H. and De Wit P.J. (2001) Specific recognition of AVR4 and AVR9 results in distinct patterns of hypersensitive cell death in tomato, but similar patterns of defence-related gene expression. Mol. Plant Pathol. 2, 77–86 10.1046/j.1364-3703.2001.00053.x [DOI] [PubMed] [Google Scholar]
- 46.de Jonge R., van Esse H.P., Maruthachalam K., Bolton M.D., Santhanam P., Saber M.K.et al. (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad Sci. U.S.A. 109, 5110–5115 10.1073/pnas.1119623109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawchuk L.M., Hachey J., Lynch D.R., Kulcsar F., van Rooijen G., Waterer D.R.et al. (2001) Tomato Ve disease resistance genes encode cell surface-like receptors. Proc. Natl. Acad Sci. U.S.A. 98, 6511–6515 10.1073/pnas.091114198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fradin E.F., Zhang Z., Juarez Ayala J.C., Castroverde C.D., Nazar R.N., Robb J.et al. (2009) Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150, 320–332 10.1104/pp.109.136762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang W., Fraiture M., Kolb D., Löffelhardt B., Desaki Y., Boutrot F.F.et al. (2013) Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell. 25, 4227–4241 10.1105/tpc.113.117010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Böhm H., Albert I., Oome S., Raaymakers T.M., Van den Ackerveken G. and Nürnberger T. (2014) A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis. PLoS Pathog. 10, e1004491 10.1371/journal.ppat.1004491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Albert I., Böhm H., Albert M., Feiler C.E., Imkampe J., Wallmeroth N.et al. (2015) An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plant. 1, 15140 10.1038/nplants.2015.140 [DOI] [PubMed] [Google Scholar]
- 52.Lenarčič T., Albert I., Böhm H., Hodnik V., Pirc K., Zavec A.B.et al. (2017) Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins. Science 358, 1431–1434 10.1126/science.aan6874 [DOI] [PubMed] [Google Scholar]
- 53.Chen J.B., Bao S.W., Fang Y.L., Wei L.Y., Zhu W.S., Peng Y.L.et al. (2021) An LRR-only protein promotes NLP-triggered cell death and disease susceptibility by facilitating oligomerization of NLP in Arabidopsis. New Phytol. 232, 1808–1822 10.1111/nph.17680 [DOI] [PubMed] [Google Scholar]
- 54.Kleemann J., Rincon-Rivera L.J., Takahara H., Neumann U., Ver Loren van Themaat E., van der Does H.C.et al. (2012) Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum. PLoS Pathog. 8, e1002643 10.1371/journal.ppat.1002643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gijzen M. and Nürnberger T. (2006) Nep1-like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochemistry 67, 1800–1807 10.1016/j.phytochem.2005.12.008 [DOI] [PubMed] [Google Scholar]
- 56.Ono E., Mise K. and Takano Y. (2020) RLP23 is required for Arabidopsis immunity against the grey mould pathogen Botrytis cinerea. Sci. Rep. 10, 13798 10.1038/s41598-020-70485-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boutrot F. and Zipfel C. (2017) Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 55, 257–286 10.1146/annurev-phyto-080614-120106 [DOI] [PubMed] [Google Scholar]
- 58.Wu J., Reca I.B., Spinelli F., Lironi D., De Lorenzo G., Poltronieri P.et al. (2019) An EFR-Cf-9 chimera confers enhanced resistance to bacterial pathogens by SOBIR1- and BAK1-dependent recognition of elf18. Mol. Plant Pathol. 20, 751–764 10.1111/mpp.12789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perraki A., DeFalco T.A., Derbyshire P., Avila J., Séré D., Sklenar J.et al. (2018) Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling. Nature 561, 248–252 10.1038/s41586-018-0471-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun Y., Qiao Z., Muchero W. and Chen J.G. (2020) Lectin receptor-like kinases: the sensor and mediator at the plant cell surface. Front Plant Sci. 11, 596301 10.3389/fpls.2020.596301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bellande K., Bono J.J., Savelli B., Jamet E. and Canut H. (2017) Plant lectins and lectin receptor-like kinases: how do they sense the outside? Int. J. Mol. Sci. 18, 10.3390/ijms18061164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sherman-Broyles S., Boggs N., Farkas A., Liu P., Vrebalov J., Nasrallah M.E.et al. (2007) S locus genes and the evolution of self-fertility in Arabidopsis thaliana. Plant Cell. 19, 94–106 10.1105/tpc.106.048199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kusaba M., Dwyer K., Hendershot J., Vrebalov J., Nasrallah J.B. and Nasrallah M.E. (2001) Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 13, 627–643 10.1105/tpc.13.3.627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Teixeira M.A., Rajewski A., He J., Castaneda O.G., Litt A. and Kaloshian I. (2018) Classification and phylogenetic analyses of the Arabidopsis and tomato G-type lectin receptor kinases. BMC Genomics 19, 239 10.1186/s12864-018-4606-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kemp B.P. and Doughty J. (2007) S cysteine-rich (SCR) binding domain analysis of the Brassica self-incompatibility S-locus receptor kinase. New Phytol. 175, 619–629 10.1111/j.1469-8137.2007.02126.x [DOI] [PubMed] [Google Scholar]
- 66.Ma R., Han Z., Hu Z., Lin G., Gong X., Zhang H.et al. (2016) Structural basis for specific self-incompatibility response in Brassica. Cell Res. 26, 1320–1329 10.1038/cr.2016.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Giranton J.L., Dumas C., Cock J.M. and Gaude T. (2000) The integral membrane S-locus receptor kinase of Brassica has serine/threonine kinase activity in a membranous environment and spontaneously forms oligomers in planta. Proc. Natl. Acad Sci. U.S.A. 97, 3759–3764 10.1073/pnas.97.7.3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L., Lin Z., Triviño M., Nowack M.K., Franklin-Tong V.E. and Bosch M. (2019) Self-incompatibility in Papaver pollen: programmed cell death in an acidic environment. J. Exp. Bot. 70, 2113–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haque T., Eaves D.J., Lin Z., Zampronio C.G., Cooper H.J., Bosch M.et al. (2020) Self-Incompatibility Triggers Irreversible Oxidative Modification of Proteins in Incompatible Pollen. Plant Physiol. 183, 1391–1404 10.1104/pp.20.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Franklin-Tong V.E., Ride J.P., Read N.D., Trewavas A.J. and Franklin F.C.H. (1993) The self-incompatibility response in Papaver rhoeas is mediated by cytosolic free calcium. Plant J. 4, 163–177 10.1046/j.1365-313X.1993.04010163.x [DOI] [Google Scholar]
- 71.Wilkins K.A., Bosch M., Haque T., Teng N., Poulter N.S. and Franklin-Tong V.E. (2015) Self-incompatibility-induced programmed cell death in field poppy pollen involves dramatic acidification of the incompatible pollen tube cytosol. Plant Physiol. 167, 766–779 10.1104/pp.114.252742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilkins K.A., Bancroft J., Bosch M., Ings J., Smirnoff N. and Franklin-Tong V.E. (2011) Reactive oxygen species and nitric oxide mediate actin reorganization and programmed cell death in the self-incompatibility response of papaver. Plant Physiol. 156, 404–416 10.1104/pp.110.167510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomas S.G. and Franklin-Tong V.E. (2004) Self-incompatibility triggers programmed cell death in Papaver pollen. Nature 429, 305–309 10.1038/nature02540 [DOI] [PubMed] [Google Scholar]
- 74.Catanzariti A.M., Lim G.T.T. and Jones D.A. (2015) The tomato I-3 gene: a novel gene for resistance to Fusarium wilt disease. New Phytol. 207, 106–118 10.1111/nph.13348 [DOI] [PubMed] [Google Scholar]
- 75.Yu D.S., Outram M.A., Smith A., McCombe C.L., Khambalkar P.B., Rima S.A.et al. (2021) The structural repertoire of Fusarium oxysporum f. sp. lycopersici effectors revealed by experimental and computational studies. bioRxiv, 2021.12.14.472499, 10.1016/2021.12.14.472499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rep M., van der Does H.C., Meijer M., van Wijk R., Houterman P.M., Dekker H.L.et al. (2004) A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 53, 1373–1383 10.1111/j.1365-2958.2004.04177.x [DOI] [PubMed] [Google Scholar]
- 77.Catanzariti A.M., Do H.T., Bru P., de Sain M., Thatcher L.F., Rep M.et al. (2017) The tomato I gene for Fusarium wilt resistance encodes an atypical leucine-rich repeat receptor-like protein whose function is nevertheless dependent on SOBIR1 and SERK3/BAK1. Plant J. 89, 1195–1209 10.1111/tpj.13458 [DOI] [PubMed] [Google Scholar]
- 78.Houterman P.M., Cornelissen B.J. and Rep M. (2008) Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog. 4, e1000061 10.1371/journal.ppat.1000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Verica J.A. and He Z.H. (2002) The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol. 129, 455–459 10.1104/pp.011028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Z., Ma W., Ren Z., Wang X., Zhao J., Pei X.et al. (2021) Characterization and expression analysis of wall-associated kinase (WAK) and WAK-like family in cotton. Int. J. Biol. Macromol. 187, 867–879 10.1016/j.ijbiomac.2021.07.163 [DOI] [PubMed] [Google Scholar]
- 81.Tripathi R.K., Aguirre J.A. and Singh J. (2021) Genome-wide analysis of wall associated kinase (WAK) gene family in barley. Genomics 113, 523–530 10.1016/j.ygeno.2020.09.045 [DOI] [PubMed] [Google Scholar]
- 82.Zhang S., Chen C., Li L., Meng L., Singh J., Jiang N.et al. (2005) Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 139, 1107–1124 10.1104/pp.105.069005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Appels R., Eversole K., Feuillet C., Keller B., Rogers J., Stein N.et al. (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, 6403, eaar7191, 10.1126/science.aar7191 [DOI] [PubMed] [Google Scholar]
- 84.He Z.H., Fujiki M. and Kohorn B.D. (1996) A cell wall-associated, receptor-like protein kinase. J. Biol. Chem. 271, 19789–19793 10.1074/jbc.271.33.19789 [DOI] [PubMed] [Google Scholar]
- 85.Wagner T.A. and Kohorn B.D. (2001) Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell. 13, 303–318 10.1105/tpc.13.2.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Decreux A. and Messiaen J. (2005) Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol. 46, 268–278 10.1093/pcp/pci026 [DOI] [PubMed] [Google Scholar]
- 87.Brutus A., Sicilia F., Macone A., Cervone F. and De Lorenzo G. (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad Sci. U.S.A. 107, 9452–9457 10.1073/pnas.1000675107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.De Lorenzo G., Brutus A., Savatin D.V., Sicilia F. and Cervone F. (2011) Engineering plant resistance by constructing chimeric receptors that recognize damage-associated molecular patterns (DAMPs). FEBS Lett. 585, 1521–1528 10.1016/j.febslet.2011.04.043 [DOI] [PubMed] [Google Scholar]
- 89.Saintenac C., Lee W.S., Cambon F., Rudd J.J., King R.C., Marande W.et al. (2018) Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nat. Genet. 50, 368–374 10.1038/s41588-018-0051-x [DOI] [PubMed] [Google Scholar]
- 90.Larkan N.J., Ma L., Haddadi P., Buchwaldt M., Parkin I.A.P., Djavaheri M.et al. (2020) The Brassica napus wall-associated kinase-like (WAKL) gene Rlm9 provides race-specific blackleg resistance. Plant J. 104, 892–900 10.1111/tpj.14966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Haddadi P., Larkan N.J., Van de Wouw A., Zhang Y., Neik T.X., Beynon E.et al. (2021) Brassica napus genes Rlm4 and Rlm7, conferring resistance to Leptosphaeria maculans, are alleles of the Rlm9 wall-associated kinase-like resistance locus. bioRxiv, 2021.12.11.471845, 10.1101/2021.12.11.471845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lazar N., Mesarich C.H., Petit-Houdenot Y., Talbi N., de la Sierra-Gallay I.L., Zélie E.et al. (2021) A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins. bioRxiv, 2020.12.17.423041, 10.1101/2020.12.17.423041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ghanbarnia K., Ma L., Larkan N.J., Haddadi P., Fernando W.G.D. and Borhan M.H. (2018) Leptosphaeria maculans AvrLm9: a new player in the game of hide and seek with AvrLm4-7. Mol. Plant Pathol. 19, 1754–1764 10.1111/mpp.12658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shi G., Zhang Z., Friesen T.L., Raats D., Fahima T., Brueggeman R.S.et al. (2016) The hijacking of a receptor kinase-driven pathway by a wheat fungal pathogen leads to disease. Sci. Adv. 2, e1600822 10.1126/sciadv.1600822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu Z., Zhang Z., Faris J.D., Oliver R.P., Syme R., McDonald M.C.et al. (2012) The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1. PLoS Pathog. 8, e1002467 10.1371/journal.ppat.1002467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McDonald M.C. and Solomon P.S. (2018) Just the surface: advances in the discovery and characterization of necrotrophic wheat effectors. Curr. Opin. Microbiol. 46, 14–18 10.1016/j.mib.2018.01.019 [DOI] [PubMed] [Google Scholar]
- 97.Hurni S., Scheuermann D., Krattinger S.G., Kessel B., Wicker T., Herren G.et al. (2015) The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc. Natl. Acad Sci. U.S.A. 112, 8780–8785 10.1073/pnas.1502522112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang P., Scheuermann D., Kessel B., Koller T., Greenwood J.R., Hurni S.et al. (2021) Alleles of a wall-associated kinase gene account for three of the major northern corn leaf blight resistance loci in maize. Plant J. 106, 526–535 10.1111/tpj.15183 [DOI] [PubMed] [Google Scholar]
- 99.Yang P., Praz C., Li B., Singla J., Robert C.A.M., Kessel B.et al. (2019) Fungal resistance mediated by maize wall-associated kinase ZmWAK-RLK1 correlates with reduced benzoxazinoid content. New Phytol. 221, 976–987 10.1111/nph.15419 [DOI] [PubMed] [Google Scholar]
- 100.Rui Y. and Dinneny J.R. (2020) A wall with integrity: surveillance and maintenance of the plant cell wall under stress. New Phytol. 225, 1428–1439 10.1111/nph.16166 [DOI] [PubMed] [Google Scholar]
- 101.Hu W., Lv Y., Lei W., Li X., Chen Y., Zheng L.et al. (2014) Cloning and characterization of the Oryza sativa wall-associated kinase gene OsWAK11 and its transcriptional response to abiotic stresses. Plant Soil 384, 335–346 10.1007/s11104-014-2204-8 [DOI] [Google Scholar]
- 102.Balint-Kurti P. (2019) The plant hypersensitive response: concepts, control and consequences. Mol. Plant Pathol. 20, 1163–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jubic L.M., Saile S., Furzer O.J., El Kasmi F. and Dangl J.L. (2019) Help wanted: helper NLRs and plant immune responses. Curr. Opin. Plant Biol. 50, 82–94 10.1016/j.pbi.2019.03.013 [DOI] [PubMed] [Google Scholar]
- 104.Bentham A.R., De la Concepcion J.C., Mukhi N., Zdrzałek R., Draeger M., Gorenkin D.et al. (2020) A molecular roadmap to the plant immune system. J. Biol. Chem. 295, 14916–14935 10.1074/jbc.REV120.010852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.van Wersch S., Tian L., Hoy R. and Li X. (2020) Plant NLRs: the whistleblowers of plant immunity. Plant Commun. 1, 100016 10.1016/j.xplc.2019.100016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhou J.M. and Zhang Y. (2020) Plant immunity: danger perception and signaling. Cell 181, 978–989 10.1016/j.cell.2020.04.028 [DOI] [PubMed] [Google Scholar]
- 107.Lorang J.M., Sweat T.A. and Wolpert T.J. (2007) Plant disease susceptibility conferred by a “resistance” gene. Proc. Natl. Acad Sci. U.S.A. 104, 14861–14866 10.1073/pnas.0702572104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lorang J., Kidarsa T., Bradford C.S., Gilbert B., Curtis M., Tzeng S.C.et al. (2012) Tricking the guard: exploiting plant defense for disease susceptibility. Science 338, 659–662 10.1126/science.1226743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kessler S.C., Zhang X., McDonald M.C., Gilchrist C.L.M., Lin Z., Rightmyer A.et al. (2020) Victorin, the host-selective cyclic peptide toxin from the oat pathogen Cochliobolus victoriae, is ribosomally encoded. Proc. Natl. Acad Sci. U.S.A. 117, 24243–24250 10.1073/pnas.2010573117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sweat T.A., Lorang J.M., Bakker E.G. and Wolpert T.J. (2008) Characterization of natural and induced variation in the LOV1 gene, a CC-NB-LRR gene conferring victorin sensitivity and disease susceptibility in Arabidopsis. Mol. Plant. Microbe. Interact. 21, 7–19 10.1094/MPMI-21-1-0007 [DOI] [PubMed] [Google Scholar]
- 111.Litzenberger S. (1949) Nature of susceptibility to Helminthosporium victoriae and resistance to Puccinia coronata in Victoria oats. Phytopathology 39, 300–318 [Google Scholar]
- 112.Wolpert T.J. and Lorang J.M. (2016) Victoria Blight, defense turned upside down. Physiol. Mol. Pl. Path. 95, 8–13 10.1016/j.pmpp.2016.03.006 [DOI] [Google Scholar]
- 113.Faris J.D., Zhang Z., Lu H., Lu S., Reddy L., Cloutier S.et al. (2010) A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad Sci. U.S.A. 107, 13544–13549 10.1073/pnas.1004090107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tuori R.P., Wolpert T.J. and Ciuffetti L.M. (1995) Purification and immunological characterization of toxic components from cultures of Pyrenophora tritici-repentis. Mol. Plant. Microbe. Interact. 8, 41–48 10.1094/MPMI-8-0041 [DOI] [PubMed] [Google Scholar]
- 115.Friesen T.L., Holmes D.J., Bowden R.L. and Faris J.D. (2018) ToxA is present in the U.S. bipolaris sorokiniana population and is a significant virulence factor on wheat harboring Tsn1. Plant Dis. 102, 2446–2452 10.1094/PDIS-03-18-0521-RE [DOI] [PubMed] [Google Scholar]
- 116.Ciuffetti L.M., Tuori R.P. and Gaventa J.M. (1997) A single gene encodes a selective toxin causal to the development of tan spot of wheat. Plant Cell. 9, 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McDonald M.C., Ahren D., Simpfendorfer S., Milgate A. and Solomon P.S. (2018) The discovery of the virulence gene ToxA in the wheat and barley pathogen Bipolaris sorokiniana. Mol. Plant Pathol. 19, 432–439 10.1111/mpp.12535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Friesen T.L., Stukenbrock E.H., Liu Z., Meinhardt S., Ling H., Faris J.D.et al. (2006) Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38, 953–956 10.1038/ng1839 [DOI] [PubMed] [Google Scholar]
- 119.Zhang Z., Running K.L.D., Seneviratne S., Peters Haugrud A.R., Szabo-Hever A., Shi G.et al. (2021) A protein kinase-major sperm protein gene hijacked by a necrotrophic fungal pathogen triggers disease susceptibility in wheat. Plant J. 106, 720–732 10.1111/tpj.15194 [DOI] [PubMed] [Google Scholar]
- 120.Winterberg B., Du Fall L.A., Song X., Pascovici D., Care N., Molloy M.et al. (2014) The necrotrophic effector protein SnTox3 re-programs metabolism and elicits a strong defence response in susceptible wheat leaves. BMC Plant Biol. 14, 215 10.1186/s12870-014-0215-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Faris J.D. and Friesen T.L. (2020) Plant genes hijacked by necrotrophic fungal pathogens. Curr. Opin. Plant Biol. 56, 74–80 10.1016/j.pbi.2020.04.003 [DOI] [PubMed] [Google Scholar]
- 122.Gong B.Q., Guo J., Zhang N., Yao X., Wang H.B. and Li J.F. (2019) Cross-microbial protection via priming a conserved immune co-receptor through juxtamembrane phosphorylation in plants. Cell Host Microbe 26, 810.e7–822.e7 10.1016/j.chom.2019.10.010 [DOI] [PubMed] [Google Scholar]
- 123.Wang P., Zhou L., Jamieson P., Zhang L., Zhao Z., Babilonia K.et al. (2020) The cotton wall-associated kinase GhWAK7A mediates responses to fungal wilt pathogens by complexing with the chitin sensory receptors. Plant Cell. 32, 3978–4001 10.1105/tpc.19.00950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang N., Pombo M.A., Rosli H.G. and Martin G.B. (2020) Tomato wall-associated kinase SlWak1 depends on Fls2/Fls3 to promote apoplastic immune responses to pseudomonas syringae. Plant Physiol. 183, 1869–1882 10.1104/pp.20.00144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kourelis J., Contreras M.P., Harant A., Adachi H., Derevnina L., Wu C.-H.et al. (2021) The helper NLR immune protein NRC3 mediates the hypersensitive cell death caused by the cell-surface receptor Cf-4. bioRxiv, 2021.09.28.461843, 10.1101/2021.09.28.461843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gabriëls S.H., Vossen J.H., Ekengren S.K., van Ooijen G., Abd-El-Haliem A.M., van den Berg G.C.et al. (2007) An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins. Plant J. 50, 14–28 10.1111/j.1365-313X.2007.03027.x [DOI] [PubMed] [Google Scholar]
- 127.Yuan M., Jiang Z., Bi G., Nomura K., Liu M., Wang Y.et al. (2021) Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ngou B.P.M., Ahn H.K., Ding P. and Jones J.D.G. (2021) Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115 10.1038/s41586-021-03315-7 [DOI] [PubMed] [Google Scholar]
- 129.Yuan M., Ngou B.P.M., Ding P. and Xin X.F. (2021) PTI-ETI crosstalk: an integrative view of plant immunity. Curr. Opin. Plant Biol. 62, 102030 10.1016/j.pbi.2021.102030 [DOI] [PubMed] [Google Scholar]
- 130.Ngou B.P.M., Jones J.D.G. and Ding P. (2021) Plant immune networks. Trends Plant Sci., 27, 255–273 [DOI] [PubMed] [Google Scholar]
- 131.Lu Y. and Tsuda K. (2021) Intimate association of PRR- and NLR-mediated signaling in plant immunity. Mol. Plant. Microbe. Interact. 34, 3–14 10.1094/MPMI-08-20-0239-IA [DOI] [PubMed] [Google Scholar]