Abstract
Pathogenic variants in genes that cause dilated (DCM) and arrhythmogenic cardiomyopathies (ACM) convey high risks for the development of heart failure through unknown mechanisms. Using single nucleus RNA sequencing (snRNAseq), we characterized the transcriptome of 880,000 nuclei from 18 control and 61 failing, non-ischemic human hearts with pathogenic variants in DCM and ACM genes or idiopathic disease. We performed genotype-stratified analyses of the ventricular cell lineages and transcriptional states. The resultant DCM and ACM ventricular cell atlas demonstrated distinct right and left ventricular responses, highlighting genotype-associated pathways, intercellular interactions, and differential gene expression at single cell resolution. Together these data illuminate both shared and distinct cellular and molecular architectures of human heart failure and suggest candidate therapeutic targets.
One-Sentence Summary:
Single nucleus analyses define ventricular responses in human cardiomyopathies with pathogenic variants.
Introduction
Dilated cardiomyopathy (DCM), a prevalent disorder occurring in 1:250 individuals, is characterized by left ventricular (LV) dilatation, cardiomyocyte loss with fibrotic replacement, and impaired contractility (1). Arrhythmogenic cardiomyopathy (ACM) similarly incites ventricular dysfunction, often with more prominent right ventricular (RV) involvement, high arrhythmogenic burden, and fibrofatty accumulations (2). Both disorders can arise from genetic causes (1, 2). DCM genes encode proteins involved in contractility (titin; TTN, troponin T; TNNT2, troponin C; TNNC1, tropomyosin; TPM1, and filamin C; FLNC), that regulate cardiac splicing (RNA-binding motif protein; RBM20) or calcium sequestration (phospholamban; PLN), and maintain cytoskeletal (desmin; DES) or nuclear (lamin A/C; LMNA) integrity. ACM genes often encode desmosome proteins including plakophilin-2 (PKP2) and desmoplakin (DSP). The cardiomyocyte-specific expression and damaging effects of pathogenic variants (PVs) in many DCM and ACM genes propel the development of arrhythmias and heart failure (HF), a highly morbid condition affecting 23 million individuals worldwide (3).
We hypothesized that PVs in different genes would evoke distinct single-cell molecular phenotypes. To address this, we studied the molecular signals underlying HF pathogenesis using snRNAseq of human hearts with advanced DCM and ACM compared to non-failing donor (control) hearts. We revealed differences in the cellular landscape and transcriptional changes between several DCM and ACM genotypes. By leveraging machine learning approaches we illuminated genotype-specific molecular responses, as validated by reconstructing the underlying PVs using snRNAseq data.
Results
Study Cohort
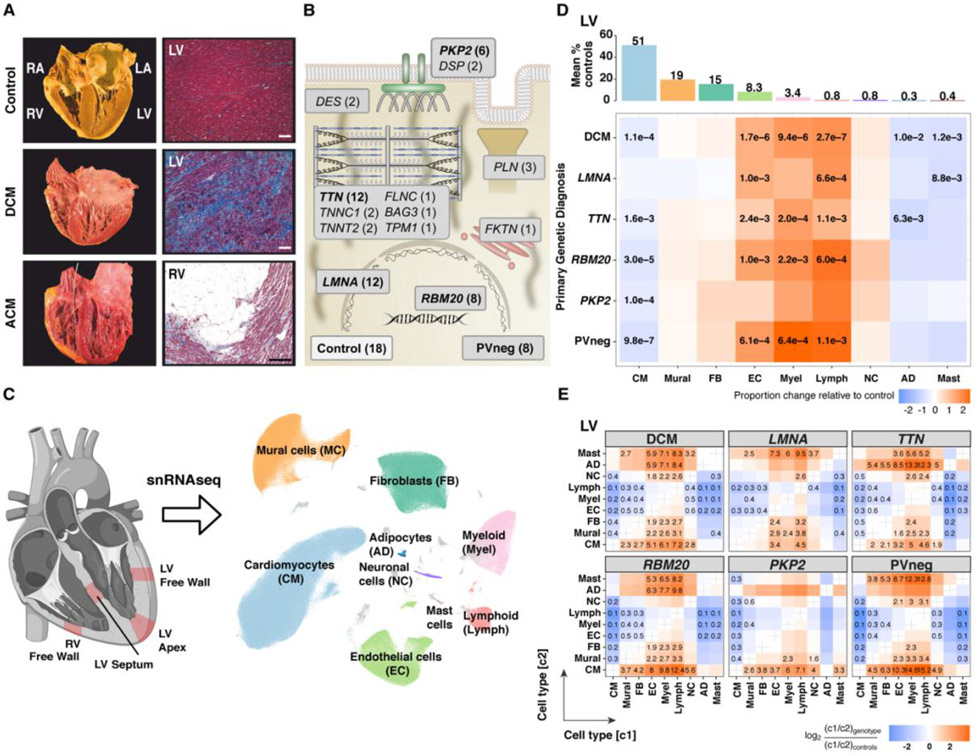
We studied LV and RV tissues (Fig. 1A), obtained prior to any mechanical support in 61 patients and 18 controls (Tables S1, S2) including 12 controls previously reported (4). Thirty-eight independent PVs were identified in three DCM genes (LMNA; n=12, RMB20; n=8, TTN; n=12) and in the ACM gene PKP2 (n=6), while no PVs were detected in eight DCM patients (PVneg) (Fig. 1B). Analyses were performed for these five genotypes individually (n=46), for aggregated DCM genotypes (LMNA, RBM20, TTN, PVneg), ACM (PKP2), and controls. Additionally, we generated data from 15 PVs across PLN, BAG3, DES, FLNC, FKTN, TNNC1, TNNT2, TMP1, and DSP (Table S1, S2). Due to few recurrent PVs, these genes were excluded in downstream analyses, except where indicated.
Figure 1: PVs and unexplained causes of DCM and ACM alter cardiac morphology, histopathology, and cellular compositions.
(A) Comparisons of normal cardiac anatomy and histology to DCM, demonstrating LV dilation with fibrosis, and to ACM, showing RV dilation with fibrofatty degeneration (Masson trichrome staining, magnification 100x, bar 10μm). (B) Schematic depiction of the functions of DCM and ACM genes with PVs (number indicates unique genotypes, bolded denotes ≥6 patients) in studied tissues. (C) Single nuclei isolated from transmural LV (free wall, apex or septum) and RV sections were processed using 10X Chromium 3’ chemistry. UMAP embedding of 881,081 nuclei delineated ten cell types and unassigned populations (gray). (D) Upper panel: Mean abundance (%) of cell types in control LVs. Lower panel: Proportional changes of cell types in specified genotypes or aggregated across DCM genotypes. Proportional changes are scaled by color: increased (red) or decreased (blue) in disease versus control. p-values are indicated for significant proportional changes, FDR≤0.05. (E) Pairwise cell type abundance ratios in specified genotypes or aggregated DCM genotypes in LVs relative. Color scale, FDR, significance depicted as in (D).
Males predominated among patients (60%) and controls (72%) (Fig. S1A). The mean age of patients was 48.4±4.3 years exclusive of RBM20 (mean age 32.9±14.6 years). Clinical manifestations indicated similar LV dysfunction in LMNA, TTN, and PVneg patients, but greater LV dilatation and reduced systolic contraction in RBM20, and preserved LV with reduced RV function in PKP2. LMNA and TTN patients received more pacemaker/resynchronization therapies than those with other genotypes (Table S1).
Disease-associated compositional changes of cell types
Nuclei were isolated as described (4) from full thickness LV free wall (FW), apex (AP), septum (S) and RV free wall (RV) (Fig. 1C). We compared ~500,000 high-quality nuclei from patients’ ventricular tissues and ~380,000 nuclei from controls (Fig. S1B-E). After pre-processing and quality control filtering, data were integrated across samples using Harmony prior to constructing manifolds using Uniform Manifold Approximation and Projections (UMAPs). Clustering identified ten major cardiac cell types encompassing ventricular cardiomyocytes (CM), fibroblasts (FB), adipocytes (AD), pericytes and smooth muscle cells (mural, MC), endothelial (EC), myeloid and lymphoid (immune), neuronal (NC) and mast cells (Figs. 1C) with 71 distinct cell states. States of the same cell type shared a transcriptional profile but also expressed distinct genes, which implied biological differences.
Cell type abundances in sample replicates were highly correlated (Pearson coefficient 0.74 - 0.99) (Figs. S2A, B). Cell composition, states, and transcript counts across the FW, AP, and S showed high similarities, and therefore are reported grouped together as LV (Figs. S2C, D).
Using center log ratio- (CLR) transformed abundance of cell types, we considered the effects of sex on LV and RV cell compositions in DCM (10 female, 29 male) and control (7 female, 11 male) hearts (Fig. S2E). Myeloid cells showed a modest sex-specific difference (FDR=0.016). Only LMNA tissues (from 7 males, 5 females) were sufficient for genotype-specific, sex-associated analyses. Female LV myeloid cells were increased (FDR 0.05), paralleling the findings in DCM vs controls. In addition, RV endothelial cells showed a modest female-specific increase between LMNA patients and controls (FDR 0.048).
The proportions of LV nuclei across the genotypes demonstrated depletion of CMs except in LMNA, and increased ECs and immune cells except in PKP2 (Figs. 1D, S3). In RVs (Fig. S4), CMs were depleted in the DCM subgroup except for TTN, while ECs were increased in LMNA, TTN, and RBM20, and immune cells were not changed. FBs were not increased in LVs and RVs (Figs. 1D), despite histopathological findings of fibrosis, which implied the acquisition of a secretory rather than a proliferative phenotype.
Individual-level abundances for cell types (LV, Table S3; RV, Table S4) and cell states (Table S5) are provided for controls and patients. Different cell type abundances in disease compared to control LVs remained significant using a linear model adjusting for age. Pairwise cell type ratios in disease LVs compared to controls confirmed loss of CMs and showed accompanying increased ECs, lymphoid and myeloid cells, and altered ratios (FB and mural compared to CM) with quantitative, genotype-specific differences (Figs. 1E). RBM20 and PVneg, respectively showed greatest shifts in EC:CM (8- and 10.3-fold), myeloid:CM (9.8- and 14.6-fold), and lymphoid:CM (12.4- and 15.2-fold). By contrast, pairwise ratios of all cell types compared to CM were modest or unchanged in LMNA.
Genotypes diversify cell types and states
Cardiomyocytes
Disease and control ventricles exhibited the previously described (vCM1.0-5; (4)) and four new cardiomyocyte states: vCM1.1, 1.2, 1.3, 3.1 (Fig. 2A-B, Table S6). Across all vCM cell states (Fig. S5), differentially expressed genes (DEGs) in disease connoted increased stress (for example, NPPB) and apoptosis (Tables S7-12, Fig. S6A). While these findings implied late-stage transcriptional convergence, 20-40% of DEGs were genotype-specific (Fig. S5). Only PVneg reduced MYH6 expression, increasing MYH7:MYH6 ratios (Fig. S6B). Conversely, only vCMs with PVs downregulated SMYD1 (Figs. 2C, S6C-E, Table S13), a cardioprotective muscle-specific histone methyltransferase that regulates sarcomere assembly and mitochondrial energetics (5, 6), and ADRB1 (β1-adrenergic receptor; Fig. S6C) indicative of adrenergic activation, which is therapeutically targeted by β-blockade medicines (7, 8). Genotype-selective responses included upregulation of FNIP2 (Fig. 2D), inhibiting AMP-activated protein kinase activity and oxidative metabolism (9-11) in LMNA and PKP2, and CPEB4, an RNA-binding protein that activates glycolysis and stimulates fibrosis (12) in LMNA, RBM20, and PKP2 (Fig. S6F).
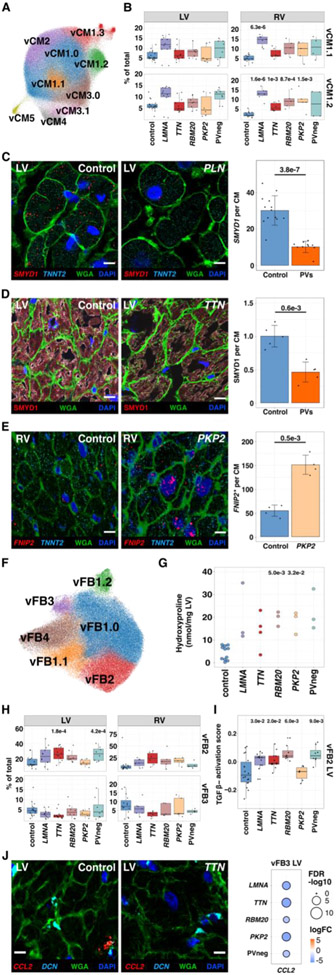
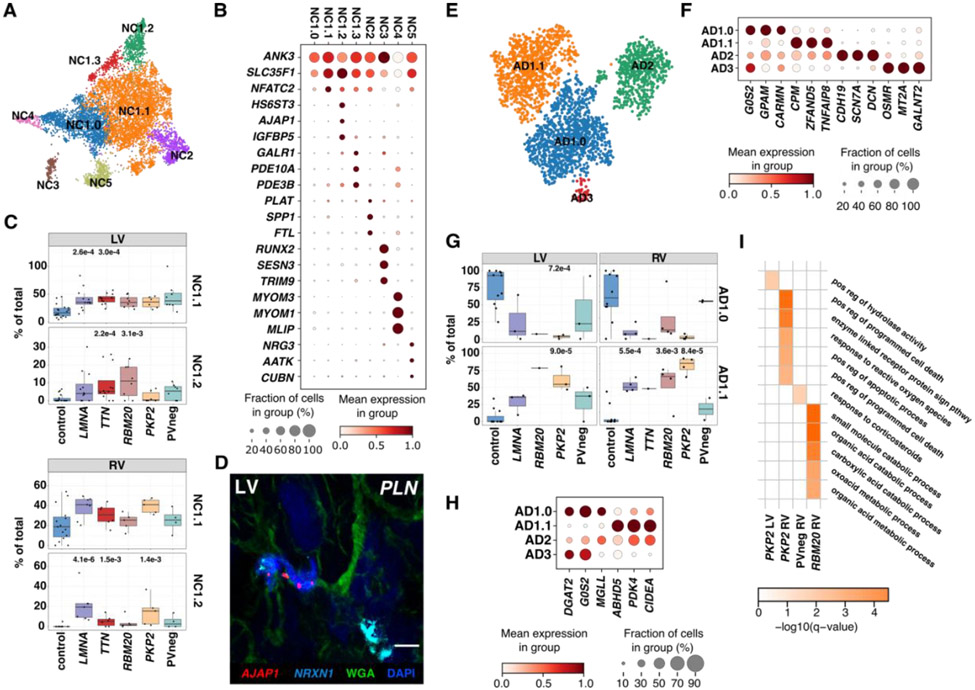
Figure 2: Cardiomyocytes and fibroblast states in control, DCM, and ACM ventricles.
(A) UMAP depicting CM states in all tissues. (B) Control and disease LVs and RVs abundance analyses for vCM1.1 (upper panel) and vCM1.2 (lower panel). (C) Single-molecule RNA fluorescent in situ hybridization exemplifies decreased SMYD1 (red) expression in CMs (identified by TNNT2 transcripts, cyan) within a DCM heart with a PV in PLN (phospholamban). Cell boundaries, WGA-stained (green); nuclei DAPI-stained (blue); bar 10μm. Quantified expression (spots per CM) and p-values from four independent control and disease LVs with PVs were assessed. (D) Immunohistochemistry validated decreased SMYD1 (red) protein in CMs (identified by troponin T staining, Fig. S6E) in TTN LV section. Cell boundaries, WGA-stained (green); nuclei DAPI-stained (blue); bar 10μm. Quantified protein levels (intensity per CM) and p-values were assessed from five independent control and DCM LVs with PVs. (E) Single-molecule RNA fluorescent in situ hybridization demonstrated increased expression of FNIP2 (red). CMs, nuclei, and cell boundaries are labeled as in C; bar 10μm. Quantified expression of FNIP2 (spots per CM and H-score; Methods) and p-values reflect analyses of two independent control and PKP2 samples. (F) UMAP depicting FB states. (G) Hydroxyproline assay (HPA) quantifies cardiac collagen content for each genotype. (H) Control and disease LVs and RVs abundance analyses for vFB2 (upper panel) and vFB3 (lower panel). (I) Pathway score of TGFβ activation in LV vFB2. (J) Single-molecule RNA fluorescent in situ hybridization shows decreased expression of CCL2 (red) in vFB3 (DCN, cyan) in disease compared to controls. WGA-stained (green); Nuclei DAPI-stained (blue); bar 10μm. Dot plot illustrating fold-change (log2FC) and significance (−log10(FDR)) of CCL2 expression in LV vFB3 across genotypes.
Proportional differences in vCM states (Figs. S6G, S7, S8) varied among genotypes and controls and were more prominent in RVs than LVs, perhaps reflecting hemodynamic differences. PVs generally increased vCM1.1 and 1.2, with accompanying decreased vCM3.1 in LMNA and TTN (Figs. 2B, S7A, S8A-D). State-enriched DEGs for vCM1.1 included MYO18B, required for sarcomere formation (13), XPR1, regulating phosphate homeostasis (14), and IGF1R and ROR1, involved in cell survival (8). vCM1.2 had heightened expression of electrophysiologic genes including PCDH7 that enables intercellular contacts (15, 16), but when overexpressed, impedes synaptic currents (17), CAMK2D modulating excitation-contraction coupling (18), and ANK2, harboring arrhythmogenic PVs (19, 20). vCM1.3 was modestly increased in RBM20, associated with the largest numbers of DEGs in all genotypes, and enriched for BMP receptors (BMPR1A, BMPR1B (21, 22); Figs. S7B, D) and GPC5 that regulates BMP signaling (23). vCM3.1, reduced in LMNA and TTN, expressed early-adaptive transcriptional regulators of stress responses (ATF3, ATF4) and cardioprotection (NR4A3; Fig. S7E) (24). Only PVneg depleted vCM2, enriched for SH3RF2, an anti-apoptosis regulator of the JNK pathway (25). As DCM and ACM genes are highly and often selectively expressed in CMs, these disease-associated vCM states and DEGs defined intrinsic responses to PVs as well as cell-cell responses to CM death, depressed contractile performance, and arrhythmogenicity.
Fibroblasts
We identified four previously characterized (4) and two new ventricular FB states (vFB1.1, vFB1.2; Fig. 2E, Table S14). Across all vFB states, upregulated DEGs included genes involved in extracellular matrix (ECM) remodeling (Figs. S9-11, Table S15-21). LMNA, TTN, and PKP2 increased fibrogenic signaling receptor, EGFR and PKP2 also increased AGTR1 (Fig. S10A) that enables EGFR-transactivation (26, 27). The expression of profibrotic TGFβ2 was universally increased (Fig. S10B). DCM hearts were enriched for PCOLCE2, promoting insoluble collagen formation (28), and LMNA, TTN, and PVneg downregulated metalloproteinase inhibitors TIMP1 and 3 (Fig. S10C). Notably, collagen genes showed genotype-specific expression. COL4A1 and COL4A2 were upregulated in LMNA, TTN and PKP2 while COL4A5 and COL24A1 were enriched in PVneg. Thus, although ECM upregulation and collagen deposition (Figs. 2F, S10D-F) was a shared feature, genotype and chamber influenced composition and organization.
FB state abundance was also altered across genotypes (Figs. S11A, S12A-D). vFB1.1 and vFB1.2 expressed canonical vFB1.0 genes but vFB1.1 also up-regulated APOD, APOE and APOO, typifying lipogenic fibroblasts (29) and CST3 (Fig. S13A), involved in matrix remodeling. vFB1.2-enriched genes are related to actin filament assembly (DAAM1) and chondrocyte differentiation (GPC6) (Fig. S13B). vFB2 expressed prominent profibrotic genes including TGFβ-targets (increased in DCM LVs) and fibrogenic IL11 (30) (highest in RBM20 LVs) (Figs. 2H, S14A-C). vFB2 was increased in TTN and PVneg and modestly in other DCM LVs (Figs. 2G-H, S11). vFB3, diminished in hearts with PVs, expressed proinflammatory cytokine genes (CCL2) and genes related to OSM signaling (IL6ST, OSMR, and oncostatin M-target genes) (Figs. 2I, S14D). The resultant increased ratio of vFB2:vFB3 and altered DEGs suggested dysregulation of fibroblast-to-macrophage interactions that would promote deleterious ECM remodeling, particularly in TTN LV and RV (Figs. S12C, D).
Smooth muscle cells and pericytes
Three previously described pericyte (PC1-3) and three smooth muscle cell states (SMC1.1, SMC1.2, SMC2, Fig. 3A, Table S22) were identified. DEGs across all states (Fig. S15, Tables S23-28) showed upregulation of the sodium channel SCN3A, with unknown vascular functions, and the noncoding antisense ADAMTS9-AS2, with concordant downregulation of ADAMTS9, a metalloprotease involved in ECM remodeling (31, 32) (Figs. 3B, S16A). Overall DEGs implied that disease evoked signals to synthesize specific ECM and integrin components.
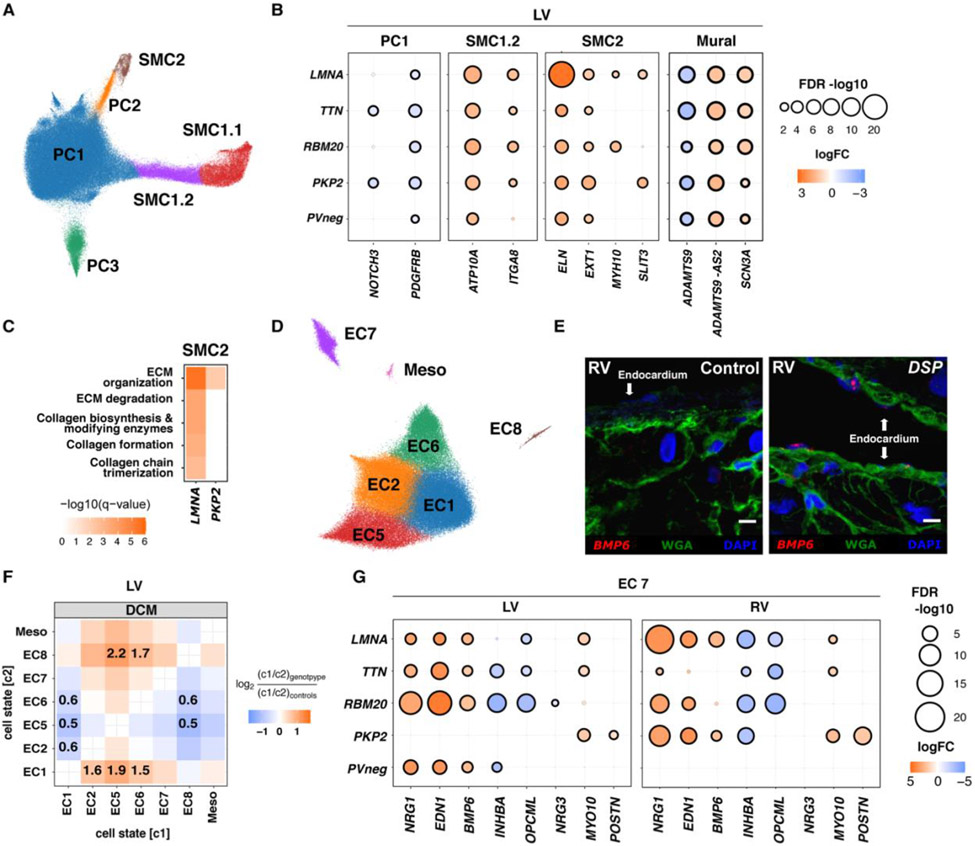
Figure 3: Mural and endothelial cell states in control, DCM, and ACM hearts.
(A) UMAP depicting pericyte (PC) and smooth muscle cell (SMC) states in all tissues. (B) Dot plots illustrate levels (fold-change; logFC) and significance (−log10(FDR)) of selected DEGs in LV PC1, SMC1.2, SMC2, and MC (PC and SMC) across genotypes. (C) KEGG pathway analysis of DEGs in SMC2 among genotypes with ≥1 enriched pathways. Color intensity denotes enrichment significance (−log10(FDR)). (D) UMAP depicting EC states in all tissues. (E) Single-molecule RNA fluorescent in situ hybridization exemplifies BMP6 (red) expression in disease endocardium from a DSP (desmoplakin) compared to control RVs. Cell boundaries, WGA-stained (green); Nuclei DAPI-stained (blue); Bar 10μm. (F) Pairwise cell state abundance ratios in DCM LVs relative to controls. Proportional changes are scaled by color: increased (red) or decreased (blue) in disease versus control. p-values are indicated for significant proportional changes, FDR<0.05. (G) Dot plots illustrate LV and RV levels (fold-change; logFC) and significance (−log10(FDR)) of selected DEGs in EC7 across genotypes. Dot size and color are defined in (B).
DEGs in PC states included downregulation of two central signaling receptors, NOTCH3 and PDGFRB, in PKP2 and TTN PC1 (Fig. 3B). NOTCH3 is required for SMC maturation and deficiency causes pericyte dysfunction and arteriovenous malformations. Notch signaling regulates PDGFRB, necessary for angiogenesis and PC recruitment (33-37).
DEGs in SMCs subdivided the previously identified canonical SMC1 (4) into two states. SMC1.1, had higher expression of ACTA2, MYH11, and TAGLN. SMC1.2, strongly expressed ITGA8, required for maintenance of SMC contractile phenotype (38, 39), and ATP10A, suggesting vascular stiffness and increasing diastolic dysfunction (Fig. 3B) Methylation of the ATP10A locus in SMC decreases with age and atherosclerosis (40). SMC1.2 was enriched in LMNA, TTN and PVneg RVs (Figs. S17, S18). SMC2 expressed high levels of genes involved in collagen and elastic fiber formation (ELN, LAMA2) and MYH10, demarcating dedifferentiated SMCs with secretory properties (Figs. S16B, S17C). LMNA and PKP2 SMC2 upregulated SLIT3, a stimulator of fibroblast activity (41), and ECM synthesis and collagen formation genes (Figs. 3B-C). Collectively, disease remodeling of MC indicated modulation of PDGF and NOTCH signaling receptors and synthesis of selective ECM and integrin components.
Endothelial cells
Characterization of ECs identified seven previously described cell states (4) (EC1, 2, 5-8 and Mesothelial) (Figs. 3D, S19-21, Table S29). Shared DEGs occurred across all and between genotypes (Table S30-35) with clear RV and LV differences (Fig. S19). Dysregulation of genes encoding factors involved in EC fate, blood vessel development and angiogenesis (NOTCH4, FLT1, FGFR1, RGCC) (42-44) in disease LVs indicated that vascular remodeling was a common HF feature.
EC composition in DCM RVs and LVs increased EC5 in comparison to controls (Fig. S21A), while genotype-specific cell state ratios in LVs generally decreased EC1 relative to EC2, EC5, and EC6 (Figs. 3F, S21C). Assessment of selective gene enrichment scores for proliferating cells, endothelial-to-mesenchymal transformation (EMT), and cell death showed no difference in disease compared to controls (Fig. S22; Table S36).
EC7 had the most DEGs and highest proportion of unique DEGs across all genotypes. Although initially defined as atrial-enriched (4) EC7 expressed endocardial-enriched genes (SMOC1 (45), NPR3 (46), and POSTN (47) (Figs. S20B-C), which prompted a revised annotation to endocardial cells. Interestingly, upregulated genes in EC7 from DCM LVs and PKP2 RVs (Fig. 3G) encoded secreted proteins involved in myocardial stress-adaptation (NRG1) (48), CM force production (EDN1) (49, 50), and endocardial expansion during development or after cardiac injury (BMP6) (51, 52). Notably, BMP6 was selectively upregulated in the endocardium of DCM LVs and ACM RVs (Fig. 3E). Conversely, INHBA, a TGFB superfamily member involved in atrioventricular canal development (53), and cell adhesion molecule OPCML were downregulated. In PKP2, an unconventional myosin promoting cell adhesion (MYO10), and POSTN, were upregulated in both ventricles. These data highlighted the involvement of the endocardium in chamber-specific remodeling of cardiomyopathies.
Myeloid cells
We classified 14 subclusters of myeloid cells comprising macrophages (MP), monocytes (MO), conventional dendritic (cDC1 and cDC2) and proliferating myeloids (Figs. 4A, S23-27, Tables S37-45). Distinct gene sets were unique to each genotype and were particularly pronounced in PVneg and PKP2 in LVs (Fig. S24).
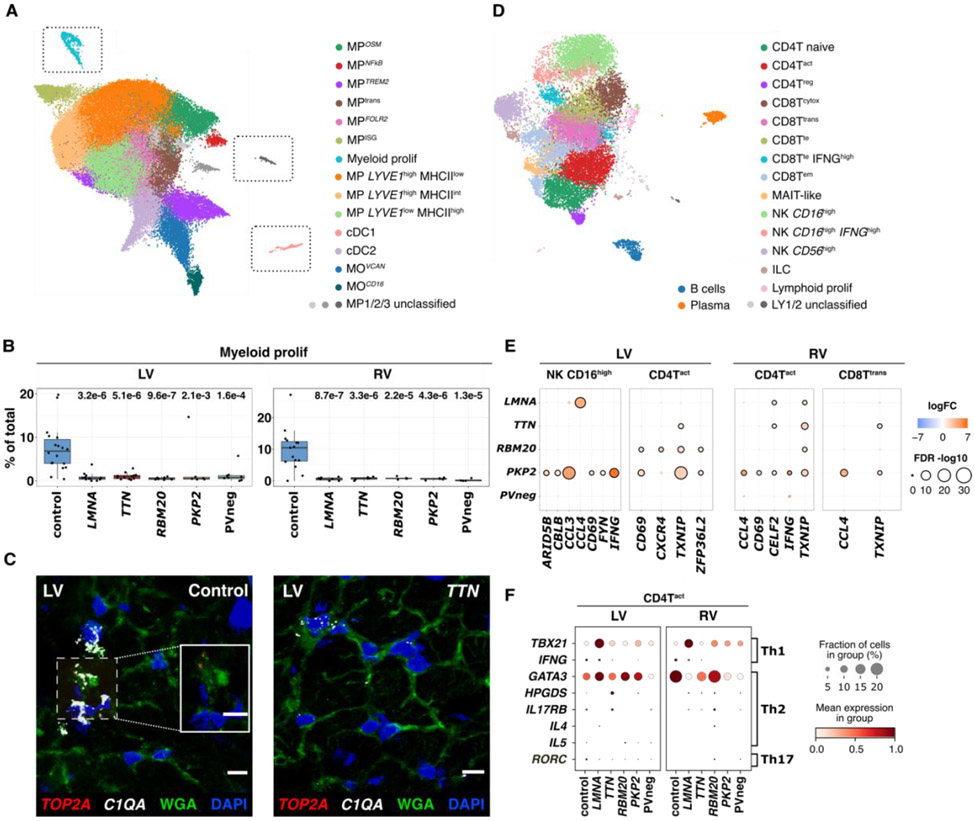
Figure 4: Immune cell states in control, DCM, and ACM hearts.
(A) UMAP depicting myeloid states in all tissues. Unclassified MP1, 2, and 3 require future characterization. Gray boxes enclosing proliferating (prolif) MPs, unclassified MPs, and cDC1s indicate that these were manually juxtaposed toward other states for ease of representation. The unmodified UMAP is in Fig. S23. (B) Myeloid prolif have higher abundance (% total myeloids) in controls versus disease. p-values indicate significant differences in abundances. (C) Single-molecule RNA fluorescent in situ hybridization validated increased expression of TOP2A (red) and C1QA (white) in controls versus disease. Cell boundaries, WGA-stained (green); Nuclei DAPI-stained (blue); Bar 10μm. (D) UMAP depicting lymphoid cell states in all tissues. Unclassified LY1 and 2 require future characterization. (E) Dot plots show the level of fold-change (logFC) and significance (−log10(FDR)) of selected genes in LV NK CD16hi, LV and RV CD4Tact, and RV CD8Ttrans across genotypes. (F) Dot plots highlight gene expression levels of the Th1, 2 and 17 signatures in CD4Tact cells. Dot size, fraction (%) of expressing cells; color, mean expression level.
Although disease increased myeloid cells, the proportions of proliferating myeloids (Figs. 4B-C, S26-27) were consistently lower compared to controls, implying monocyte-infiltration. Tissue resident MPs LYVE1highMHCIIlow and LYVE1lowMHCIIhigh (Fig. S25C) were the most abundant myeloid cells (Figs. S27A-B). MP LYVE1lowMHCIIhigh were increased in the RV of LMNA with similar trends in other genotypes (Fig. S27B). Proportions of MPOSM were modestly decreased with downregulation of OSM in TTN ventricles (Fig. S28A), which would attenuate the MPOSM-vFB3 signaling axis (4), and mirrored the decreased OSM pathway activation score observed in vFB3 (Fig. S14B).
Prominent antigen presenting activities, were evident in MOVCAN, MOCD16, cDC1 and cDC2, also occurred in MPFOLR2, akin to tumor-associated MP (54). Across antigen-presenting MP, RBM20 LVs showed the highest presentation of antigens based on MHCII genes (Figs. S28B-C) and more abundant cDC2 compared to the other genotypes (Fig. S27). PKP2 LVs upregulated MPISG (Fig. S27) with interferon-stimulated genes (ISG) (55), perhaps contributing to inflammatory PKP2 phenotypes (56).
Lymphoids
We classified 15 lymphoid cell states, including T- and natural killer (NK)-cell subsets, innate lymphoid cells (ILC), B-cells, plasma cells plus proliferating lymphoids (Figs. 4D, S29-32, Table S46). Our experimental design did not enrich for immune cells, and few (<40) genotype specific DEGs were identified (Fig. S29). Proliferating lymphoids were rare and abundance was unchanged in disease (Figs. S30, 32A-D, Table S47-52).
The cardiac complexity of CD4+ and CD8+T-cells included naive (CD4Tnaive), activated (CD4Tact), regulatory (CD4Treg) and cytotoxic (CD8Tcytox), transitional (CD8Ttrans), terminal effector (CD8Tte) and effector memory (CD8Tem) cells (Fig. S31B) (57). We detected increased CD4Tact only within DCM samples (Figs. S32A-D). However, DEGs indicated lymphocyte activation (cytokines IFNG, CCL3, CCL4 and signaling molecules CBLB, FYN, TXNIP), and maturation (cell surface receptors CD69, CXCR4), particularly in PKP2 NK−, CD4+T, and CD8+T-cells (Fig. 4E).
CD4+helper T-cells are critical drivers in cardiomyopathy and myocarditis pathogenesis (58, 59). In LMNA CD4Tact we identified upregulation of TBX21, important for Th1 polarization. Conversely, Th2-polarization transcription factor GATA3 was downregulated in PVneg (Fig. 4F).
Neuronal cells
Analysis of cardiac NCs was limited by rarity of this cell type (Figs. 5A, S33, Table S53-57). Across all NC states, DEGs indicated increased NFATC2 in LVs with PVs and PKP2 RVs, and genotype-selective enrichment of LRRK2, an activator of a neurotoxic cascade (60). Other upregulated DEGs function in proteoglycan synthesis for neuronal myelination and axon regeneration (XYLT1 and HS3ST4), and a complement inhibitor (SUSD4) that impacts neural function and morphology (61, 62). Genotype-specific DEGs were highest in PKP2.
Figure 5: Neuronal and Adipocyte cell states in control, DCM, and ACM hearts.
(A) UMAP depicting NC states in all tissues. (B) Dot plot highlights top marker genes for NC states. (C) LV and RV abundance analyses of NC1.1 and 1.2 in controls versus disease. p-values are indicated for significant proportional changes, FDR≤0.05. (D) Single-molecule RNA fluorescent in situ hybridization shows colocalization AJAP1 (red), and NRXN1 (cyan) in disease (exemplified in a DCM LV with a PV in PLN (phospholamban), demarcating the NC1.2 state. Cell boundaries, WGA-stained (green); Nuclei DAPI-stained (blue), Bar 10μm. (E) UMAP depicting AD states in all tissues. (F) Dot plot highlights top marker genes for AD states. (G) LVs and RVs abundance analyses demonstrate decreased AD1.0 and increased AD1.1 in disease versus control. (H) Dot plot of DEGs shows expression differences between AD states. (I) Heatmap of significantly enriched Gene Ontology Biological Processes terms based on significantly upregulated genes in diseased versus control ADs. Dot plots: Dot size, fraction (%) of expressing cells; color, mean expression level. p-values indicate significant differences; FDR≤0.05.
We identified previously described (4) and three new NC states (Figs. 5B-C, S34, S35) that were genotype and chamber-selective: NC1.1 in LMNA and TTN LVs; NC1.2 in TTN and RBM20 LVs and in LMNA, TTN, and PKP2 RVs. NC1.1 was distinguished by the highest upregulation of NFATC2. NC1.2 upregulated genes associated with electrocardiogram intervals (SLC35F1 and AJAP1; Fig. 5D), IGFBP5, involved in neuronal apoptosis and autophagy, and the ion channel and heart rate regulator KCNK1 (63-65). NC1.3 was enriched for the neuromodulator receptor GALR1, and phosphodiesterases PDE10A and PDE3B that participate in neuroprotection and signaling (66-68). Dysregulated expression of genes involved in stress-responses and electrophysiology may account for characteristic life-threatening arrhythmias in DCM and PVs in PKP2 (1, 2).
Adipocytes
Similar to NCs, limited adipocytes were captured. We identified three adipocyte states previously found in controls: canonical AD1.0, expressing lipid metabolism genes; stromal AD2, expressing ECM genes; and immune AD3, expressing OSMR and cytokine responsive genes (Figs. 5E-F, S36, Table S60-64). While AD3 was not detected in disease, a fourth identified state, AD1.1 was almost exclusive to disease hearts (Figs. 5G, S37). Compositional analysis identified increased proportions of AD1.1 in PKP2 LVs and RVs concurrent with decreased LV proportions of AD1.0. LMNA and RBM20 RVs, unlike LVs, also increased proportions of AD1.1 (Figs. S38A-D).
DEGs between AD1.1 and AD1.0 revealed changes in fatty acid metabolism pathways (Fig. 5H). AD1.1 showed downregulation of DGAT2, encoding a triglyceride forming enzyme (69), and G0S2 and MGLL, encoding lipolysis regulators (70). Conversely ABHD5, a positive lipolysis regulator (70, 71), PDK4, a kinase promoting the shift from glucose to fatty acid metabolism, and CIDEA, a regulator of adipose tissue energy expenditure, were upregulated. PKP2 RVs which typically display fibrofatty replacement also showed an enrichment of Gene Ontology biological processes for apoptosis and cell death (Fig. 5I). These data implied genotype-specific state transitions or replacement of canonical adipocytes in DCM and ACM.
Differential expression of GWAS genes in cardiomyopathies
Genome-wide association studies (GWAS) have identified common genetic variants associated with DCM. We selected candidate genes from 15 previously identified DCM loci (Table S67) and examined expression across cell types and genotypes (Fig. S39). Overall, GWAS genes were more often DEGs in LV and RV than expected by chance in snRNAseq data (LV: OR=7.0, P=0.0007; RV: OR=6.1, P=0.0009, one-sided Fisher’s exact test). Multiple genes showed cell type specific expressions, with the majority highly enriched in CM (ALPK3, BAG3, FHOD3, FLNC, HSPB7, MLIP, SLC6A6, SMARCB1, SVIL and TTN). Among these we observed both genotype- and chamber-specific expression differences. Heat shock protein HSBP7 was reduced in TTN LVs and in LMNA and PKP2 RVs. SLC6A6, a taurine and amino acid transporter with cardioprotective effects, was increased in both LV and RV across all genotypes except for PKP2 and PVneg. CDKN1A, a cell cycle regulator and modulator of apoptosis (72) was increased in LMNA CMs from LVs but not RVs (72).
GWAS genes that were more broadly expressed across cell types included MTSS1, encoding a putative actin-cytoskeletal interactor. MTSS1 showed highest and unchanged expression in myeloids and was widely increased in mural cells as well as in FBs in LMNA, TTN, and PKP2 (Fig. S39), suggesting influences beyond direct effects on CM function (73, 74). SLC39A8, a lowly expressed cardiac solute carrier, was unchanged in CMs but increased in LMNA and TTN LV ECs. We suggest that cell-specific expression changes of GWAS genes may improve interpretation of their biologic effects.
Predicted and altered cell-cell interactions across genotypes
By examining the expression of genes encoding for receptors and ligands, we inferred intercellular signaling and communication (75). We initially quantified the probability of cell-cell interactions and compared signaling between cell states, and then aggregated information to produce cell-specific and across-all-cell-types data, for each genotype relative to controls. This sequential approach accounted for differential abundances of cell states.
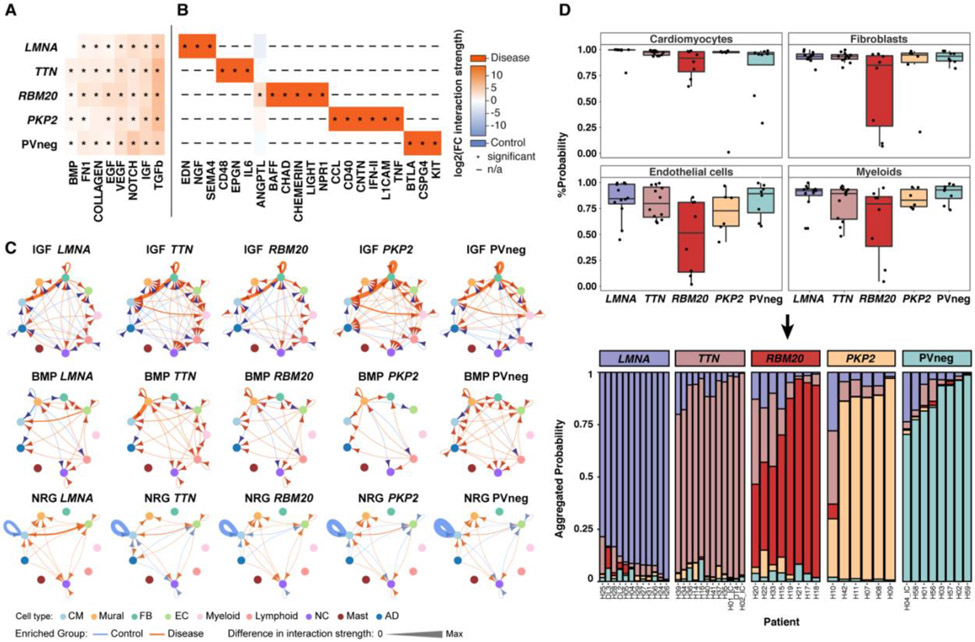
We detected aberrant intercellular signaling across disease (Figs. 6A, S40), including upregulation of BMP, FN1, Collagen, EGF, IGF and TGF pathways that promote fibrosis. Signaling dependent on VEGF, NOTCH and ANGPT was also increased in disease, implying vascular remodeling. Strikingly, genotype-selective increases in intercellular signaling pathways were also identified in LVs (Figs. 6B, S40A, B): EDN in LMNA, the proinflammatory IL6 in TTN, BAFF/LIGHT (denoting TNF signaling) in RBM20, pro-inflammatory CCL and TNF in PKP2, and the immune modulator BTLA in PVneg. Some of these intercellular signaling pathways were similarly dysregulated in RV, but chamber-specific changes were also observed (Figs. S40C, D, S41).
Figure 6: Altered cell-cell interactions and recognition of genotype-specific transcriptional responses.
Heatmaps depict shared (A) and unique (B) signaling pathways in LVs, with significantly different expression in genotypes compared to controls. Signaling pathways are defined in the CellChat database (75). Changes in interaction strength (log2(fold-change)), scaled by color intensity (red, increased; blue, decreased). *denotes significance; adjusted p-values≤0.05; n/a denotes expression not detected in control or disease. (C) Circle plots of significant (adjusted p-value≤0.05) cell-cell communication depict differentially regulated IGF, BMP, and NRG pathways and interactions in disease LVs. The line thickness denotes interaction strength of signals from sending and receiving cell; with color scaled from zero to maximum in disease versus controls (orange, increased; blue, decreased). Arrows indicate directionality. (D) (Top) Genotype prediction probability from graph attention networks (GAT) per cell type. (Bottom) Stacked bar plots represent the likelihood (% aggregated probability) of individual patient genotypes by GAT prediction. The vast majority of established genotypes were predicted with high probability, with lower prediction probability only in H10 (PKP2), H20 (RBM20), H22 (RBM20) and H33 (RBM20).
We also identified genotype-specific differences in the cells sending and receiving signals in disease pathways (Figs. 6C). For example, IGF signaling (Fig. S42), crucial in cell growth and CM hypertrophy, showed increased FB autocrine (highest in PKP2) and paracrine FB to CM signaling, paralleling findings in experimental models (76, 77). In addition, IGF signaling from myeloid cells to CMs only occurred in TTN, PKP2 and PVneg, an interaction that might promote muscle repair (78).
The source of BMP signaling changed in a genotype-specific manner (Figs. 6C, S43). BMP signaling from MC to CM was increased in TTN LVs and RVs but was downregulated in PVneg and LMNA LVs and other RVs (Figs. S41, S43). Notably, BMP signaling in ECs originated solely from EC7, likely depending on BMP6 upregulation (Figs. 3E, G). EDN signaling from EC7 to CM and MC was highly genotype-selective, occurring in LMNA LV and PKP2 RV (Fig. S44).
NRG signaling (comprising NRG1-3 and ERBB receptors) showed multifaceted changes. Disease LVs markedly attenuated autocrine NRG signaling in CMs, while EC and CM signaling was upregulated (highest in LMNA) in all genotypes except RBM20 (Fig. 6C, S45). Additionally, NRG3-ERBB4 interactions identified in controls shifted in disease to NRG1-ERBB4 and NRG1-ERBB3 in a genotype-specific manner, consistent with changes in NRG1/3 expression in EC7 (Figs. 3G, S46) (79). This predicted NRG/ERBB shift may provide compensatory responses to adverse remodeling in cardiomyopathies (48).
Graph attention networks recognized genotype-specific expression patterns
We applied machine learning approaches to snRNAseq data to further advance the recognition of cell and genotype-specific transcriptional patterns. Cell-specific neighborhood graphs showed more connectivities among single-nuclei transcriptomes from PVneg hearts and hearts with PVs in the same gene, compared to PVs from different genes (Fig. S47). Subsequently, we generated a graph attention network (GAT) for multinomial classification of genotypes trained on four major informative LV cell types: CMs, FBs, ECs, and myeloids (Fig. S47A-B). The GAT predicted LMNA, TTN, RBM20, PKP2, and PVneg genotypes with high accuracy. Among LV samples, the genotype prediction accuracy differed by cell types: CMs: 0.93, FBs: 0.92, myeloids: 0.85, ECs: 0.79 (Figs. 6D, S48, Tables S69-70; corresponding RV data Fig. S47C). Aggregation of genotype predictions obtained from these four LV cell types strengthened the correct prediction of genotypes, resulting in a high confidence model (Fig. 6D). Of note, three (H10, H22 and H33) of the four lower prediction probabilities occurred in samples with both a primary and secondary PVs, assigned as such by prior genotype and clinical phenotype review (Table S1). Moreover, as this machine learning model independently confirmed a genotype- and cell type-specific transcriptional signature, we concluded that these snRNAseq datasets accurately described the molecular responses to PVs and unexplained causes of DCM and ACM.
Discussion
Our analyses of snRNAseq of LV and RV samples illuminated the cell types and states, molecular signals, and predicted intercellular communications that characterized DCM and ACM. In comparison to control hearts, we identified differences at multiple levels, including changes in the proportions of cell types and states, additional cell states, and differential gene expression, substantially expanding earlier insights achieved by bulk tissue analyses. Across all genotypes, disease hearts demonstrated some common dissimilarities from control hearts, often with graded differences between LVs and RVs. Despite studying hearts from patients with advanced disease who received diverse therapies, congruent transcriptional signatures emerged for different PVs within the same gene and varied between genotypes.
The differences between genotype groups and controls reflected differences in mean expression and not differences in variance. This was true for each genotype group including PVneg. Transcriptional signatures were complex, diversified both by the proportions of canonical and stressed cell states and by differentially expressed genes within the same states. While interrogation of these datasets provides ongoing opportunities for discovery, our findings provided substantial evidence that genotype influenced pathological remodeling of the heart. These results upend a prevalent dogma that HF results from a common final pathway and can guide the future development of therapies with selective targets to enhance personalized medicine.
Despite anatomical and histopathological differences between DCM and ACM, we identified shared changes in the cellular composition of ventricular tissues, albeit with DCM LV features largely mirrored in ACM RVs. Cardiomyopathies were depleted in CMs while EC and immune cell populations were increased. FBs did not expand, but increased states that augmented ECM gene expression and collagen deposition. Based on these changes and cytokine profiles (TGFβ activation, increased IGF signaling, decreased CCL2 expression; Figs. 2H-I, 6C), we predicted cell-cell interactions and key molecules that are appropriate for mechanistic studies to causally link differential expression with adverse cardiac remodeling.
Disease CMs exhibited loss of the canonical state vCM1.0, and the emergence of genotype-enriched cell states with DEGs. Many of these responses are associated with stress-induced contractile, metabolic, and electrophysiologic properties that are prominent clinical manifestations of some genotypes. For example, attenuated expression of SMYD1, a critical organizer of sarcomere structure, epigenetic and metabolic remodeling, occurred only in CMs with PVs, and was unaltered in PVneg samples. With its global impact on myocardial function, dysregulation of SMYD1 might contribute to earlier presentation and poorer outcomes of cardiomyopathy patients with, versus without, PVs (80). LMNA CMs had the greatest expansion of vCM1.2, enriched for genes encoding Ca2+ regulators and molecules with electrophysiologic functions, while RMB20 and LMNA genotypes had the highest expression of MYL4, a sarcomere protein associated with atrial fibrillation. These data suggested molecular mechanisms whereby particular genotypes convey increased risks for arrhythmias and sudden cardiac death in patients.
Shifts in FB states explained the paradoxically increased fibrosis in cardiomyopathies, without expansion of overall FB populations. DCM LVs and ACM RVs showed increased proportions of vFB2, enriched for genes that modulate ECM composition, turnover, stiffness and fibrotic scarring (81) and reciprocally decreased proportions of vFB3 that express transcripts suppressing fibrosis. Notably, fibrogenic genes within vFB2 were highly expressed in RBM20, a genotype within our cohort with the poorest ventricular function and youngest age for HF diagnosis and cardiac transplantation. Strategies to manipulate proteins encoded by these genes may attenuate the prominent myocardial fibrosis that characterizes cardiomyopathies.
Unexpectedly, mural cells (PCs, SMCs) showed no increase compared to ECs across cardiomyopathies. DEG analysis suggested that these cells promoted vascular remodeling and dysfunction. PCs diminished PDGFRB expression and showed aberrant NOTCH signaling in vascular beds, while genotype selective DEGs in SMCs upregulated contractile genes, augmenting fiber formation. Together these molecular signals may contribute to microvascular dysfunction that occurs in cardiomyopathy patients (82) and adversely influences ventricular performance.
Among EC, EC7 demarcated the endocardium and had the most DEGs in disease compared to control. Little is known about this myocardial layer in HF pathogenesis. The endocardium forms via dynamic regulation of NOTCH, neuregulin (83) and BMP (84) signals. Pediatric heart diseases can exhibit pathological expansion of the endocardium (denoted endocardial fibroelastosis) which diminishes cardiac performance and is associated with progression to HF (84). Our data suggested similar dysregulation of these molecular pathways in adult-onset cardiomyopathies with increased BMP6 and NRG1 in EC7 and decreased INHBA (53). These signals may cause pathological changes in endocardium in DCM and ACM hearts and contribute to myocardial dysfunction.
Diseased hearts increased myeloid cells. Expansion of immune cell populations might arise from recruitment of circulating cells or proliferation of resident cells. In our samples proliferating myeloid cells decreased, as previously observed in other DCM studies (85). Surprisingly, myeloid recruitment via the CCR2/CCL2 axis, primarily mediated via vFB3, was decreased in all samples, with particularly dysregulated fibroblast-to-macrophage interactions in TTN. Although distinguishing between proliferation versus recruitment late in disease was problematic, previous studies indicate peak cytokine expression precedes the emergence of HF (86, 87). Analyses of earlier time points during disease progression will be important to discern these key signals.
To confirm and expand the conclusion that genotype-specific signals meaningfully contributed to disease pathogenesis, we employed machine learning strategies. Using GAT to classify patients’ genotypes from each cell type, we found that CMs, FBs, ECs, and myeloid cells provided the highest and most discriminatory information. Harnessing LV and RV data from these cell types, we independently predicted the established genotype of each patient with high accuracy. Moreover, among the four samples with the lowest genotype predictive probability, three samples carried two PVs, indicating that the model detected subtle transcriptional differences with additional influences.
While expanding machine learning models to much larger datasets will undoubtedly improve accuracy, these early analyses supported the conclusion that PVs in different genes evoked cell type and state-specific responses that altered inter-cellular communications and promoted distinct disease pathways. We recognize that pathways may converge, but even in advanced disease, our data indicated that genotypes promoted specific transcriptional signals that likely contributed to distinct as well as common manifestations of genetic cardiomyopathies.
Future studies are needed to comprehensively define the molecular pathophysiology of cardiomyopathies and HF, including assessments of age, sex, and ancestral influences, other DCM and ACM genotypes, additional cardiac regions, and longitudinal analyses to identify initiating and secondary processes. We also expect that the deployment of strategies that upsample conduction system and other rare cell types, and incorporation of techniques to characterize the epigenome, proteome, and spatial relationships between cell types, states, and gene expression will also be highly informative. To promote these initiatives, we freely provide all datasets and an interactive platform (https://cellxgene.cziscience.com/collections/e75342a8-0f3b-4ec5-8ee1-245a23e0f7cb) with cell type and state annotations. We expect that these resources will advance mechanistic studies to improve treatments of cardiomyopathies and enable HF prevention strategies.
Materials and methods
Detailed information on human subject studies, experimental methods, data access, codes, algorithms, and computational programs used in this manuscript is provided in the Supplementary Materials (89).
Human studies were performed using protocols that were reviewed and approved by the ethics boards of participating institutions. DCM and ACM ventricular samples were collected from genotyped patients undergoing mechanical support (n=15) or heart transplantation (n=31) and from deceased donors with non-failing hearts as previously described (4). Nuclei from full-thickness LV and RV regions were isolated and processed for snRNAseq as described (4). Data were mapped to the human genome (GRCh38), processed to remove doublets and identify nuclei that met high quality standards, and harmonized to remove batch effects (105). Manifolds were constructed using Uniform Manifold Approximation and Projections (UMAPs) for all and individual cell types.
Differential abundance analyses of cell types and states were performed using centered log ratio transformation including a linear model. Differential gene expression between disease and control tissues were deduced using a pseudobulk approach and EdgeR (109, 110). Comparative analyses assessed cell type and cell state abundances and differential gene expression between disease samples and controls and between genotypes. Selected genes with differential expression were validated using single molecule fluorescent in situ hybridization or quantitative immunohistochemistry.
We investigated cell-cell communication using CellChat (75). The expression of genes previously identified through genome wide-association studies of DCM was assessed in diseased and control tissues. We interrogated transcriptional datasets with machine learning tools to generate cell type features that distinguished PVs within the same gene from PVs within different genes and used these data to generate a graph attention network (GAT). The accuracy for GAT analyses of randomly selected patient data to assign the correct, clinically assigned genotype was assessed.
Supplementary Material
Acknowledgements:
The authors extend their heartfelt gratitude to the patients and relatives who provided cardiac tissues for research and to the biobanks who supported these efforts (Biobank of the Heart and Diabetes Center NRW, University Hospital of the Ruhr-University Bochum, the HELP Program (Mazankowski Alberta Heart Institute) and HOPE program (University of Alberta Hospital), the Cardiovascular Research Centre Biobank at the Royal Brompton and Harefield hospitals and NHS Blood and Transplant, and the Brigham and Women’s Cardiovascular Tissue Bank. We also thank Hans-Peter Rahn of the Flow Cytometry Technology Platform, the Systems Biology Imaging Platform (both MDC), Tatiana Borodina of the BIH/MDC Genomics Technology Platform, and Sabine Schmidt (MDC) for technical help in single nuclei isolation and library preparation, the Facility for Imaging by Light Microscopy and Stephen Rothery (Imperial College London), the LMS/NIHR Imperial Biomedical Research Centre Flow Cytometry Facility for support, Lorraine Lawrence from the Research Histology Facility at Imperial College, the Biopolymers Laboratory, Paula Montero Llopis of the Microscopy Resources on the North Quad (MicRoN) core (Harvard Medical School), and Greg Plummer at the Faculty of Medicine & Dentistry Cell Imaging Core (University of Alberta) for the technical support. We thank Dr. Dagmar Keinmüller for advice on machine learning and Susanne Bartlett (Brigham and Women’s Hospital and Harvard Medical School) for administrative assistance.
This article is subject to HHMI’s Open Access to Publications policy. HHMI lab heads have previously granted a nonexclusive CC BY 4.0 license to the public and a sublicensable license to HHMI in their research articles. Pursuant to those licenses, the author-accepted manuscript of the article can be made freely available under a CC By 4.0 license immediately upon publication.
Funding:
This project has been made possible in part by the Chan Zuckerberg Foundation (2019-202666) to N.H., M.H., M.N., C.E.S., J.G.S., and S.A.T., the Leducq Foundation (16CVD03) to N.H., C.E.S., and J.G.S.; the British Heart Foundation and Deutsches Zentrum für Herz-Kreislauf-Forschung (BHF/DZHK: SP/19/1/34461) to N. H., M.N., and S.A.T.; the Medical Faculty of the Ruhr-University, FoRUM (F842R-2015) to H.Mi., A.G. & J.G.; the Deutsche Forschungsgemeinschaft to H.Mi. (1164,6-1, and 1146,2-2), to N.H. (SFB-1470 – B03) and to D.R. (Research Fellowship); the Wellcome Trust (107469/Z/15/Z; 200990/A/16/Z) to J.S.W. and to S.A.T. (206194, 108413/A/15/D), the Medical Research Council (UK), the National Institute for Health Research (NIHR) Royal Brompton Cardiovascular Biomedical Research Unit and NIHR Imperial College Biomedical Research Centre to J.S.W. and P.J.B.; the National Institutes of Health (HL080494 and HL084553) to C.E.S. and J.G.S and to Y.K. (KL2-TR002542); the Engineering Research Centers Program of the National Science Foundation (NSF Cooperative Agreement EEC-1647837) to C.E.S. and J.G.S.; the John S. LaDue Fellowship of Harvard Medical School to Y.K. and K.B.; the Wellcome Trust Clinical PhD Fellowship to J.C.; the Overseas Research Fellowship of the Takeda Science Foundation to K.K., the Stanley Sarnoff Foundation Fellowship to E.R.N.; Boehringer Ingelheim Fonds to V.S.; the Brigham and Women’s Hospital Khoury Innovation Award to Y.K.; the Ludwig Institute for Cancer Research grant, National Cancer Institute grant (U54-CA225088) and CCSP Supplemental Award to C.Y.; the British Heart Foundation (RE/18/4/34215) to J.S.W; the ERC-advanced grant under the European Union Horizon 2020 Research and Innovation Program (AdG788970) to N.H.; the University of Alberta Hospital Foundation (UHF), the Canadian Institute of Health Research (CIHR), the Heart and Stroke Foundation (HSF), and the Canada Research Chair in Heart Failure to G.Y.O.; the Choudhrie Family Fund and Howard Hughes Medical Institute to C.E.S.
Footnotes
Competing Interests:
The following authors report competing interests. Consultancy Fees: J.S.W. (MyoKardia Inc, and Foresite Labs); C.E.S. and J.G.S. (Maze, BridgeBio, Bristol Myers Squibb); S.A.T. (Genentech and Roche). Scientific Advisory Boards: S.A.T. (Biogen, GlaxoSmithKline, Qiagen and Foresite Labs); Board of Directors: C.E.S. (Burroughs Wellcome Fund (U.S.) and Merck); Co-founder and equity holder: S.A.T. (Transition Bio). J.S.W. acknowledges research support from MyoKardia, Bristol Myers Squibb, and Pfizer (formerly Array BioPharma). These companies were not involved in any aspect of the study’s experimental design, execution, and analyses or in the preparation of the manuscript.
D.R., E.L.L., H.M., A.M.A.M., A.V., N.S., A.G., E.R.N., M.L., K.K., J.R.-O., V.S., D.M.D., G.P., H.Z., A.W., C.L., Y.K., E.A., J.M.G., S.N.G., K.B., R.J.B., R.A.C., C.C., J.C., L.E.F., H.F., A.G., J.G., M.K., R.L., L.M., B.M., S.S., F.S., C.Y., C.S., P.I.T., F.J.T., A.v.d.B., H.W., C.L.W., P.J.R.B., Y.A.L., H.M., M.N., G.Y.O., M.H., N.H. declare no competing interests.
Data and Materials Availability:
Data
All sequence data generated and analyzed in this study has been deposited as BAM files at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001006374 (https://ega-archive.org/studies/EGAS00001006374). Further information about EGA can be found on https://ega-archive.org, "The European Genome-phenome Archive in 2021" (https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkab1059/6430505).
Processed data can be analyzed using the Cellxgene tool website: https://cellxgene.cziscience.com/collections/e75342a8-0f3b-4ec5-8eel-245a23e0f7cb/private and are available in h5ad-format to download. Metadata sheets and patient information are available in Table S1&2.
Code
All code used to generate the figures in this publication are available on github (https://github.com/heiniglab/DCM_heart_cell_atlas). Code used to reproduce the presented analyses is indexed on Zenodo (88). All scripts run on jupyter notebooks are available as “.ipynb” files, scripts executed in command line are available as .txts or .sh files. R scripts are available as “.R”. Anaconda environments are available as yml files containing information on package versions.
References and Notes
- 1.McNally EM, Mestroni L, Dilated Cardiomyopathy: Genetic Determinants and Mechanisms. Circ. Res 121, 731–748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austin KM, Trembley MA, Chandler SF, Sanders SP, Saffitz JE, Abrams DJ, Pu WT, Molecular mechanisms of arrhythmogenic cardiomyopathy. Nature Reviews Cardiology. 16 (2019), pp. 519–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bui AL, Horwich TB, Fonarow GC, Epidemiology and risk profile of heart failure. Nat. Rev. Cardiol 8, 30–41 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Litviňuková M, Talavera-López C, Maatz H, Reichart D, Worth CL, Lindberg EL, Kanda M, Polanski K, Heinig M, Lee M, Nadelmann ER, Roberts K, Tuck L, Fasouli ES, DeLaughter DM, McDonough B, Wakimoto H, Gorham JM, Samari S, Mahbubani KT, Saeb-Parsy K, Patone G, Boyle JJ, Zhang H, Zhang H, Viveiros A, Oudit GY, Bayraktar OA, Seidman JG, Seidman CE, Noseda M, Hubner N, Teichmann SA, Cells of the adult human heart. Nature. 588, 466–472 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warren JS, Tracy CM, Miller MR, Makaju A, Szulik MW, Oka S-I, Yuzyuk TN, Cox JE, Kumar A, Lozier BK, Wang L, Liana JG, Sabry AD, Cawley KM, Barton DW, Han YH, Boudina S, Fiehn O, Tucker HO, Zaitsev AV, Franklin S, Histone methyltransferase Smyd1 regulates mitochondrial energetics in the heart. Proc. Natl. Acad. Sci. U. S. A 115, E7871–E7880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franklin S, Kimball T, Rasmussen TL, Rosa-Garrido M, Chen H, Tran T, Miller MR, Gray R, Jiang S, Ren S, Wang Y, Tucker HO, Vondriska TM, The chromatin-binding protein Smyd1 restricts adult mammalian heart growth. Am. J. Physiol. Heart Circ. Physiol 311, H1234–H1247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinberg SF, Beta-Adrenergic Receptor Regulation Revisited. Circ. Res 123, 1199–1201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bristow MR, Mechanism of Action of Beta-Blocking Agents in Heart Failure. The American Journal of Cardiology. 80 (1997), p. 26L–40L. [DOI] [PubMed] [Google Scholar]
- 9.Hasumi Y, Baba M, Hasumi H, Huang Y, Lang M, Reindorf R, Oh H-B, Sciarretta S, Nagashima K, Haines DC, Schneider MD, Adelstein RS, Schmidt LS, Sadoshima J, Marston Linehan W, Folliculin (Flcn) inactivation leads to murine cardiac hypertrophy through mTORC1 deregulation. Hum. Mol. Genet 23, 5706–5719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasumi H, Baba M, Hasumi Y, Lang M, Huang Y, Oh HF, Matsuo M, Merino MJ, Yao M, Ito Y, Furuya M, Iribe Y, Kodama T, Southon E, Tessarollo L, Nagashima K, Haines DC, Linehan WM, Schmidt LS, Folliculin-interacting proteins Fnip1 and Fnip2 play critical roles in kidney tumor suppression in cooperation with Flcn. Proc. Natl. Acad. Sci. U. S. A 112, E1624–31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen K, Rogala KB, Chou H-T, Huang RK, Yu Z, Sabatini DM, Cryo-EM Structure of the Human FLCN-FNIP2-Rag-Ragulator Complex. Cell. 179, 1319–1329.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mejias M, Gallego J, Naranjo-Suarez S, Ramirez M, Pell N, Manzano A, Suñer C, Bartrons R, Mendez R, Fernandez M, CPEB4 Increases Expression of PFKFB3 to Induce Glycolysis and Activate Mouse and Human Hepatic Stellate Cells, Promoting Liver Fibrosis. Gastroenterology. 159, 273–288 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Ajima R, Akazawa H, Kodama M, Takeshita F, Otsuka A, Kohno T, Komuro I, Ochiya T, Yokota J, Deficiency of Myo18B in mice results in embryonic lethality with cardiac myofibrillar aberrations. Genes Cells. 13, 987–999 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Hueso M, De Ramon L, Navarro E, Ripoll E, Cruzado JM, Grinyo JM, Torras J, Silencing of CD40 in vivo reduces progression of experimental atherogenesis through an NF-κB/miR-125b axis and reveals new potential mediators in the pathogenesis of atherosclerosis. Atherosclerosis. 255, 80–89 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Kim S-Y, Yasuda S, Tanaka H, Yamagata K, Kim H, Non-clustered protocadherin. Cell Adh. Migr 5, 97–105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Merkel CD, Zeng X, Heier JA, Cantrell PS, Sun M, Stolz DB, Watkins SC, Yates NA, Kwiatkowski AV, The N-cadherin interactome in primary cardiomyocytes as defined using quantitative proximity proteomics. J. Cell Sci 132 (2019), doi: 10.1242/jcs.221606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Kerrisk Campbell M, Tom I, Foreman O, Hanson JE, Sheng M, PCDH7 interacts with GluN1 and regulates dendritic spine morphology and synaptic function. Sci. Rep 10, 10951 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swaminathan PD, Purohit A, Hund TJ, Anderson ME, Calmodulin-dependent protein kinase II: linking heart failure and arrhythmias. Circ. Res 110, 1661–1677 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swayne LA, Murphy NP, Asuri S, Chen L, Xu X, McIntosh S, Wang C, Lancione PJ, Roberts JD, Kerr C, Sanatani S, Sherwin E, Kline CF, Zhang M, Mohler PJ, Arbour LT, Novel Variant in the Membrane-Binding Domain Is Associated With Ankyrin-B Syndrome and Structural Heart Disease in a First Nations Population With a High Rate of Long QT Syndrome. Circ. Cardiovasc. Genet 10 (2017), doi: 10.1161/CIRCGENETICS.116.001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Devalla HD, Gélinas R, Aburawi EH, Beqqali A, Goyette P, Freund C, Chaix M-A, Tadros R, Jiang H, Le Béchec A, Monshouwer-Kloots JJ, Zwetsloot T, Kosmidis G, Latour F, Alikashani A, Hoekstra M, Schlaepfer J, Mummery CL, Stevenson B, Kutalik Z, de Vries AA, Rivard L, Wilde AA, Talajic M, Verkerk AO, Al-Gazali L, Rioux JD, Bhuiyan ZA, Passier R, TECRL, a new life-threatening inherited arrhythmia gene associated with overlapping clinical features of both LQTS and CPVT. EMBO Mol. Med 8, 1390–1408 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaussin V, Morley GE, Cox L, Zwijsen A, Vance KM, Emile L, Tian Y, Liu J, Hong C, Myers D, Conway SJ, Depre C, Mishina Y, Behringer RR, Hanks MC, Schneider MD, Huylebroeck D, Fishman GI, Burch JBE, Vatner SF, Alk3/Bmpr1a receptor is required for development of the atrioventricular canal into valves and annulus fibrosus. Circ. Res 97, 219–226 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrell NW, Bloch DB, ten Dijke P, Goumans M-JTH, Hata A, Smith J, Yu PB, Bloch KD, Targeting BMP signalling in cardiovascular disease and anaemia. Nat. Rev. Cardiol 13, 106–120 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filmus J, Capurro M, The role of glypicans in Hedgehog signaling. Matrix Biol. 35, 248–252 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Schmitz ML, Shaban MS, Albert BV, Gökçen A, Kracht M, The Crosstalk of Endoplasmic Reticulum (ER) Stress Pathways with NF-κB: Complex Mechanisms Relevant for Cancer, Inflammation and Infection. Biomedicines. 6 (2018), doi: 10.3390/biomedicines6020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilhelm M, Kukekov NV, Schmit TL, Biagas KV, Sproul AA, Gire S, Maes ME, Xu Z, Greene LA, Sh3rf2/POSHER protein promotes cell survival by ring-mediated proteasomal degradation of the c-Jun N-terminal kinase scaffold POSH (Plenty of SH3s) protein. J. Biol Chem 287, 2247–2256 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eckenstaler R, Sandori J, Gekle M, Benndorf RA, Angiotensin II receptor type 1 – An update on structure, expression and pathology. Biochem. Pharmacol 192, 114673 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Itabashi H, Maesawa C, Oikawa H, Kotani K, Sakurai E, Kato K, Komatsu H, Nitta H, Kawamura H, Wakabayashi G, Masuda T, Angiotensin II and epidermal growth factor receptor cross-talk mediated by a disintegrin and metalloprotease accelerates tumor cell proliferation of hepatocellular carcinoma cell lines. Hepatol. Res 38, 601–613 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Baicu CF, Zhang Y, Van Laer AO, Renaud L, Zile MR, Bradshaw AD, Effects of the absence of procollagen C-endopeptidase enhancer-2 on myocardial collagen accumulation in chronic pressure overload. Am. J. Physiol. Heart Circ. Physiol 303, H234–40 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, Chang S, Conley SD, Mori Y, Seita J, Berry GJ, Shrager JB, Metzger RJ, Kuo CS, Neff N, Weissman IL, Quake SR, Krasnow MA, A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature. 587, 619–625 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schafer S, Viswanathan S, Widjaja AA, Lim W-W, Moreno-Moral A, DeLaughter DM, Ng B, Patone G, Chow K, Khin E, Tan J, Chothani SP, Ye L, Rackham OJL, Ko NSJ, Sahib NE, Pua CJ, Zhen NTG, Xie C, Wang M, Maatz H, Lim S, Saar K, Blachut S, Petretto E, Schmidt S, Putoczki T, Guimarñes-Camboa N, Wakimoto H, van Heesch S, Sigmundsson K, Lim SL, Soon JL, Chao VTT, Chua YL, Tan TE, Evans SM, Loh YJ, Jamal MH, Ong KK, Chua KC, Ong B-H, Chakaramakkil MJ, Seidman JG, Seidman CE, Hubner N, Sin KYK, Cook SA, IL-11 is a crucial determinant of cardiovascular fibrosis. Nature. 552, 110–115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nandadasa S, Nelson CM, Apte SS, ADAMTS9-Mediated Extracellular Matrix Dynamics Regulates Umbilical Cord Vascular Smooth Muscle Differentiation and Rotation. Cell Reports. 11 (2015), pp. 1519–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koo B-H, Coe DM, Dixon LJ, Somerville RPT, Nelson CM, Wang LW, Young ME, Lindner DJ, Apte SS, ADAMTS9 Is a Cell-Autonomously Acting, Anti-Angiogenic Metalloprotease Expressed by Microvascular Endothelial Cells. The American Journal of Pathology. 176 (2010), pp. 1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nadeem T, Bogue W, Bigit B, Cuervo H, Deficiency of Notch signaling in pericytes results in arteriovenous malformations. JCI Insight. 5 (2020), doi: 10.1172/jci.insight.125940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magnusson PU, Looman C, Ahgren A, Wu Y, Claesson-Welsh L, Heuchel RL, Platelet-derived growth factor receptor-beta constitutive activity promotes angiogenesis in vivo and in vitro. Arterioscler. Thromb. Vasc. Biol 27 (2007), doi: 10.1161/01.ATV.0000282198.60701.94. [DOI] [PubMed] [Google Scholar]
- 35.Jin S, Hansson EM, Tikka S, Lanner F, Sahlgren C, Farnebo F, Baumann M, Kalimo H, Lendahl U, Notch Signaling Regulates Platelet-Derived Growth Factor Receptor-β Expression in Vascular Smooth Muscle Cells. Circulation Research. 102 (2008), pp. 1483–1491. [DOI] [PubMed] [Google Scholar]
- 36.Domenga V, Fardoux P, Lacombe P, Monet M, Maciazek J, Krebs LT, Klonjkowski B, Berrou E, Mericskay M, Li Z, Tournier-Lasserve E, Gridley T, Joutel A, Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 18 (2004), doi: 10.1101/gad.308904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campos AH, Wang W, Pollman MJ, Gibbons GH, Determinants of Notch-3 receptor expression and signaling in vascular smooth muscle cells: implications in cell-cycle regulation. Circ. Res 91, 999–1006 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Marek I, Lichtneger T, Cordasic N, Hilgers KF, Volkert G, Fahlbusch F, Rascher W, Hartner A, Menendez-Castro C, Alpha8 Integrin (Itga8) Signalling Attenuates Chronic Renal Interstitial Fibrosis by Reducing Fibroblast Activation, Not by Interfering with Regulation of Cell Turnover. PLoS One. 11, e0150471 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hung CF, Wilson CL, Chow Y-H, Schnapp LM, Role of integrin alpha8 in murine model of lung fibrosis. PLOS ONE. 13 (2018), p. e0197937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Connelly JJ, Cherepanova OA, Doss JF, Karaoli T, Lillard TS, Markunas CA, Nelson S, Wang T, Ellis PD, Langford CF, Haynes C, Seo DM, Goldschmidt-Clermont PJ, Shah SH, Kraus WE, Hauser ER, Gregory SG, Epigenetic regulation of COL15A1 in smooth muscle cell replicative aging and atherosclerosis. Hum. Mol. Genet 22, 5107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gong L, Wang S, Shen L, Liu C, Shenouda M, Li B, Liu X, Shaw JA, Wineman AL, Yang Y, Xiong D, Eichmann A, Evans SM, Weiss SJ, Si M-S, SLIT3 deficiency attenuates pressure overload-induced cardiac fibrosis and remodeling. JCI Insight. 5 (2020), doi: 10.1172/jci.insight.136852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sewduth R, Santoro MM, “Decoding” Angiogenesis: New Facets Controlling Endothelial Cell Behavior. Front. Physiol 7, 306 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.House SL, Castro AM, Lupu TS, Weinheimer C, Smith C, Kovacs A, Ornitz DM, Endothelial fibroblast growth factor receptor signaling is required for vascular remodeling following cardiac ischemia-reperfusion injury. Am. J. Physiol. Heart Circ. Physiol 310, H559–71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui X-B, Chen S-Y, Response Gene to Complement 32 in Vascular Diseases. Front Cardiovasc Med. 5, 128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao Y, Hill MC, Zhang M, Martin TJ, Morikawa Y, Wang S, Moise AR, Wythe JD, Martin JF, Hippo Signaling Plays an Essential Role in Cell State Transitions during Cardiac Fibroblast Development. Dev. Cell 45, 153–169.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang J, Zhang H, He L, Huang X, Li Y, Pu W, Yu W, Zhang L, Cai D, Lui KO, Zhou B, Genetic Fate Mapping Defines the Vascular Potential of Endocardial Cells in the Adult Heart. Circ. Res 122, 984–993 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Norris RA, Moreno-Rodriguez RA, Sugi Y, Hoffman S, Amos J, Hart MM, Potts JD, Goodwin RL, Markwald RR, Periostin regulates atrioventricular valve maturation Dev. Biol 316, 200–213 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Keulenaer GW, Feyen E, Dugaucquier L, Shakeri H, Shchendrygina A, Belenkov YN, Brink M, Vermeulen Z, Segers VFM, Mechanisms of the Multitasking Endothelial Protein NRG-1 as a Compensatory Factor During Chronic Heart Failure. Circ. Heart Fail 12, e006288 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Zhao X-S, Pan W, Bekeredjian R, Shohet RV, Endogenous endothelin-1 is required for cardiomyocyte survival in vivo. Circulation. 114, 830–837 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Hathaway CK, Grant R, Hagaman JR, Hiller S, Li F, Xu L, Chang AS, Madden VJ, Bagnell CR, Rojas M, Kim H-S, Wu B, Zhou B, Smithies O, Kakoki M, Endothelin-1 critically influences cardiac function via superoxide-MMP9 cascade. Proc. Natl. Acad. Sci. U. S. A 112, 5141–5146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim RY, Robertson EJ, Solloway MJ, Bmp6 and Bmp7 are required for cushion formation and septation in the developing mouse heart. Dev. Biol 235, 449–466 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Fang Y, Lai KS, She P, Sun J, Tao W, Zhong TP, Tbx20 Induction Promotes Zebrafish Heart Regeneration by Inducing Cardiomyocyte Dedifferentiation and Endocardial Expansion. Front. Cell Dev. Biol 0 (2020), doi: 10.3389/fcell.2020.00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang ACY, Fu Y, Garside VC, Niessen K, Chang L, Fuller M, Setiadi A, Smrz J, Kyle A, Minchinton A, Marra M, Hoodless PA, Karsan A, Notch initiates the endothelial-to-mesenchymal transition in the atrioventricular canal through autocrine activation of soluble guanylyl cyclase. Dev. Cell 21, 288–300 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Puig-Kröger A, Sierra-Filardi E, Domínguez-Soto A, Samaniego R, Corcuera MT, Gómez-Aguado F, Ratnam M, Sánchez-Mateos P, Corbí AL, Folate receptor beta is expressed by tumor-associated macrophages and constitutes a marker for M2 anti-inflammatory/regulatory macrophages. Cancer Res. 69, 9395–9403 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Dick SA, Macklin JA, Nejat S, Momen A, Clemente-Casares X, Althagafi MG, Chen J, Kantores C, Hosseinzadeh S, Aronoff L, Wong A, Zaman R, Barbu I, Besla R, Lavine KJ, Razani B, Ginhoux F, Husain M, Cybulsky MI, Robbins CS, Epelman S, Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat. Immunol 20, 29–39 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pérez-Hernández M, Marrón-Liñares GM, Schlamp F, Heguy A, van Opbergen CJM, Mezzano V, Zhang M, Liang F-X, Cerrone M, Delmar M, Transcriptomic Coupling of PKP2 With Inflammatory and Immune Pathways Endogenous to Adult Cardiac Myocytes. Front. Physiol 0 (2021), doi: 10.3389/fphys.2020.623190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van der Leun AM, Thommen DS, Schumacher TN, CD8 T cell states in human cancer: insights from single-cell analysis. Nat. Rev. Cancer 20, 218–232 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barin JG, Čiháková D, Control of inflammatory heart disease by CD4+ T cells. Ann. N. Y. Acad. Sci 1285, 80–96 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Vdovenko D, Eriksson U, Regulatory Role of CD4 T Cells in Myocarditis. J Immunol Res 2018, 4396351 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim C, Beilina A, Smith N, Li Y, Kim M, Kumaran R, Kaganovich A, Mamais A, Adame A, Iba M, Kwon S, Lee W-J, Shin S-J, Rissman RA, You S, Lee S-J, Singleton AB, Cookson MR, Masliah E, LRRK2 mediates microglial neurotoxicity via NFATc2 in rodent models of synucleinopathies. Sci. Transl. Med 12 (2020), doi: 10.1126/scitranslmed.aay0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwartz NB, Domowicz MS, Proteoglycans in brain development and pathogenesis. FEBS Lett. 592, 3791–3805 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Zhu H, Meissner LE, Byrnes C, Tuymetova G, Tifft CJ, Proia RL, The Complement Regulator Susd4 Influences Nervous-System Function and Neuronal Morphology in Mice. iScience. 23, 100957 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Méndez-Giráldez R, Gogarten SM, Below JE, Yao J, Seyerle AA, Highland HM, Kooperberg C, Soliman EZ, Rotter JI, Kerr KF, Ryckman KK, Taylor KD, Petty LE, Shah SJ, Conomos MP, Sotoodehnia N, Cheng S, Heckbert SR, Sofer T, Guo X, Whitsel EA, Lin HJ, Hanis CL, Laurie CC, Avery CL, GWAS of the electrocardiographic QT interval in Hispanics/Latinos generalizes previously identified loci and identifies population-specific signals. Sci. Rep 7, 17075 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Christensen AH, Chatelain FC, Huttner IG, Olesen MS, Soka M, Feliciangeli S, Horvat C, Santiago CF, Vandenberg JI, Schmitt N, Olesen S-P, Lesage F, Fatkin D, The two-pore domain potassium channel, TWIK-1, has a role in the regulation of heart rate and atrial size. J. Mol. Cell. Cardiol 97, 24–35 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Xu X, Huang E, Luo B, Cai D, Zhao X, Luo Q, Jin Y, Chen L, Wang Q, Liu C, Lin Z, Xie W, Wang H, Methamphetamine exposure triggers apoptosis and autophagy in neuronal cells by activating the C/EBPβ-related signaling pathway. The FASEB Journal. 32 (2018), pp. 6737–6759. [DOI] [PubMed] [Google Scholar]
- 66.Schauwecker PE, Galanin receptor 1 deletion exacerbates hippocampal neuronal loss after systemic kainate administration in mice. PLoS One. 5, e15657 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beaumont V, Zhong S, Lin H, Xu W, Bradaia A, Steidl E, Gleyzes M, Wadel K, Buisson B, Padovan-Neto FE, Chakroborty S, Ward KM, Harms JF, Beltran J, Kwan M, Ghavami A, Häggkvist J, Tóth M, Halldin C, Varrone A, Schaab C, Dybowski JN, Elschenbroich S, Lehtimäki K, Heikkinen T, Park L, Rosinski J, Mrzljak L, Lavery D, West AR, Schmidt CJ, Zaleska MM, Munoz-Sanjuan I, Phosphodiesterase 10A Inhibition Improves Cortico-Basal Ganglia Function in Huntington’s Disease Models. Neuron. 92, 1220–1237 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Mitome-Mishima Y, Miyamoto N, Tanaka R, Oishi H, Arai H, Hattori N, Urabe T, Differences in phosphodiesterase 3A and 3B expression after ischemic insult. Neurosci. Res 75, 340–348 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Chitraju C, Walther TC, Farese RV Jr, The triglyceride synthesis enzymes DGAT1 and DGAT2 have distinct and overlapping functions in adipocytes. J. Lipid Res 60, 1112–1120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schweiger M, Paar M, Eder C, Brandis J, Moser E, Gorkiewicz G, Grond S, Radner FPW, Cerk I, Cornaciu I, Oberer M, Kersten S, Zechner R, Zimmermann R, Lass A, G0/G1 switch gene-2 regulates human adipocyte lipolysis by affecting activity and localization of adipose triglyceride lipase. J. Lipid Res 53, 2307–2317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanders MA, Zhang H, Mladenovic L, Tseng YY, Granneman JG, Molecular Basis of ABHD5 Lipolysis Activation. Sci. Rep 7, 42589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Onoue K, Wakimoto H, Jiang J, Parfenov M, DePalma S, Conner D, Gorham J, McKean D, Seidman JG, Seidman CE, Saito Y, Cardiomyocyte Proliferative Capacity Is Restricted in Mice With Mutation. Front Cardiovasc Med. 8, 639148 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morley MP, Wang X, Hu R, Brandimarto J, Tucker NR, Felix JF, Smith NL, van der Harst P, Ellinor PT, Margulies KB, Musunuru K, Cappola TP, Cardioprotective Effects of MTSS1 Enhancer Variants. Circulation. 139, 2073–2076 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meyer HV, Dawes TJW, Serrani M, Bai W, Tokarczuk P, Cai J, de Marvao A, Henry A, Lumbers RT, Gierten J, Thumberger T, Wittbrodt J, Ware JS, Rueckert D, Matthews PM, Prasad SK, Costantino ML, Cook SA, Birney E, O’Regan DP, Genetic and functional insights into the fractal structure of the heart. Nature. 584, 589–594 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan C-H, Myung P, Plikus MV, Nie Q, Inference and analysis of cell-cell communication using CellChat. Nat. Commun 12, 1088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takeda N, Manabe I, Uchino Y, Eguchi K, Matsumoto S, Nishimura S, Shindo T, Sano M, Otsu K, Snider P, Conway SJ, Nagai R, Cardiac fibroblasts are essential for the adaptive response of the murine heart to pressure overload. J. Clin. Invest 120, 254–265 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Horio T, Maki T, Kishimoto I, Tokudome T, Okumura H, Yoshihara F, Suga S-I, Takeo S, Kawano Y, Kangawa K, Production and autocrine/paracrine effects of endogenous insulin-like growth factor-1 in rat cardiac fibroblasts. Regul. Pept 124, 65–72 (2005). [DOI] [PubMed] [Google Scholar]
- 78.Tonkin J, Temmerman L, Sampson RD, Gallego-Colon E, Barberi L, Bilbao D, Schneider MD, Musarò A, Rosenthal N, Monocyte/Macrophage-derived IGF-1 Orchestrates Murine Skeletal Muscle Regeneration and Modulates Autocrine Polarization. Mol. Ther 23, 1189–1200 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Odiete O, Hill MF, Sawyer DB, Neuregulin in cardiovascular development and disease. Circ. Res 111, 1376–1385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Escobar-Lopez L, Ochoa JP, Mirelis JG, Espinosa MÁ, Navarro M, Gallego-Delgado M, Barriales-Villa R, Robles-Mezcua A, Basurte-Elorz MT, Gutiérrez García-Moreno L, Climent V, Jiménez-Jaimez J, Mogollón-Jiménez MV, Lopez J, Peña-Peña ML, García-Álvarez A, Brion M, Ripoll-Vera T, Palomino-Doza J, Tirón C, Idiazabal U, Brögger MN, García-Hernández S, Restrepo-Córdoba MA, Gonzalez-Lopez E, Méndez I, Sabater M, Villacorta E, Larrañaga-Moreira JM, Abecia A, Fernández AI, García-Pinilla JM, Rodríguez-Palomares JF, Gimeno-Blanes JR, Bayes-Genis A, Lara-Pezzi E, Domínguez F, Garcia-Pavia P, Association of Genetic Variants With Outcomes in Patients With Nonischemic Dilated Cardiomyopathy. J. Am. Coll. Cardiol 78, 1682–1699 (2021). [DOI] [PubMed] [Google Scholar]
- 81.Kong P, Christia P, Frangogiannis NG, The pathogenesis of cardiac fibrosis. Cell. Mol. Life Sci 71, 549–574 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gulati A, Ismail TF, Ali A, Hsu L-Y, Gonçalves C, Ismail NA, Krishnathasan K, Davendralingam N, Ferreira P, Halliday BP, Jones DA, Wage R, Newsome S, Gatehouse P, Firmin D, Jabbour A, Assomull RG, Mathur A, Pennell DJ, Arai AE, Prasad SK, Microvascular Dysfunction in Dilated Cardiomyopathy: A Quantitative Stress Perfusion Cardiovascular Magnetic Resonance Study. JACC Cardiovasc. Imaging. 12, 1699–1708 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Del Monte-Nieto G, Ramialison M, Adam AAS, Wu B, Aharonov A, D’Uva G, Bourke LM, Pitulescu ME, Chen H, de la Pompa JL, Shou W, Adams RH, Harten SK, Tzahor E, Zhou B, Harvey RP, Control of cardiac jelly dynamics by NOTCH1 and NRG1 defines the building plan for trabeculation. Nature. 557, 439–445 (2018). [DOI] [PubMed] [Google Scholar]
- 84.Xu X, Friehs I, Zhong Hu T, Melnychenko I, Tampe B, Alnour F, Iascone M, Kalluri R, Zeisberg M, Del Nido PJ, Zeisberg EM, Endocardial fibroelastosis is caused by aberrant endothelial to mesenchymal transition. Circ. Res 116, 857–866 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ramirez Flores RO, Lanzer JD, Holland CH, Leuschner F, Most P, Schultz J-H, Levinson RT, Saez-Rodriguez J, Consensus Transcriptional Landscape of Human End-Stage Heart Failure. J. Am. Heart Assoc 10, e019667 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liao X, Shen Y, Zhang R, Sugi K, Vasudevan NT, Amer Alaiti M, Sweet DR, Zhou L, Qing Y, Gerson SL, Fu C, Wynshaw-Boris A, Hu R, Schwartz MA, Fujioka H, Richardson B, Cameron MJ, Hayashi H, Stamler JS, Jain MK, Distinct roles of resident and nonresident macrophages in nonischemic cardiomyopathy. Proc. Natl. Acad. Sci. U. S. A 115, E4661–E4669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dewald O, Zymek P, Winkelmann K, Koerting A, Ren G, Abou-Khamis T, Michael LH, Rollins BJ, Entman ML, Frangogiannis NG, CCL2/Monocyte Chemoattractant Protein-1 regulates inflammatory responses critical to healing myocardial infarcts. Circ. Res 96, 881–889 (2005). [DOI] [PubMed] [Google Scholar]
- 88.Lindberg Eric, Shvetsov Nikolay, Ruiz Jorge, Heinig Matthias, Miranda Antonio, & Kanemaru Kazumasa. (2022). heiniglab/DCM_heart_cell_atlas: v1.0 Release (v1.0). Zenodo. 10.5281/zenodo.6720185 [DOI] [Google Scholar]
- 89.Supplementary Materials [Google Scholar]
- 90.Zhang H, Viveiros A, Nikhanj A, Nguyen Q, Wang K, Wang W, Freed DH, Mullen JC, MacArthur R, Kim DH, Tymchak W, Sergi CM, Kassiri Z, Wang S, Oudit GY, The Human Explanted Heart Program: A translational bridge for cardiovascular medicine. Biochim. Biophys. Acta Mol. Basis Dis 1867 (2021), doi: 10.1016/j.bbadis.2020.165995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, ACMG Laboratory Quality Assurance Committee, Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med 17, 405–424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li H, Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA, The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu X, Jian X, Boerwinkle E, dbNSFP v2.0: A Database of Human Non-synonymous SNVs and Their Functional Predictions and Annotations. Human Mutation. 34 (2013), pp. E2393–E2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, Walters RK, Tashman K, Farjoun Y, Banks E, Poterba T, Wang A, Seed C, Whiffin N, Chong JX, Samocha KE, Pierce-Hoffman E, Zappala Z, O’Donnell-Luria AH, Minikel EV, Weisburd B, Lek M, Ware JS, Vittal C, Armean IM, Bergelson L, Cibulskis K, Connolly KM, Covarrubias M, Donnelly S, Ferriera S, Gabriel S, Gentry J, Gupta N, Jeandet T, Kaplan D, Llanwarne C, Munshi R, Novod S, Petrillo N, Roazen D, Ruano-Rubio V, Saltzman A, Schleicher M, Soto J, Tibbetts K, Tolonen C, Wade G, Talkowski ME, Genome Aggregation Database Consortium, Neale BM, Daly MJ, MacArthur DG, The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Walsh R, Buchan R, Wilk A, John S, Felkin LE, Thomson KL, Chiaw TH, Loong CCW, Pua CJ, Raphael C, Prasad S, Barton PJ, Funke B, Watkins H, Ware JS, Cook SA, Defining the genetic architecture of hypertrophic cardiomyopathy: re-evaluating the role of non-sarcomeric genes. Eur. Heart J 38, 3461–3468 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F, The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Whiffin N, Walsh R, Govind R, Edwards M, Ahmad M, Zhang X, Tayal U, Buchan R, Midwinter W, Wilk AE, Najgebauer H, Francis C, Wilkinson S, Monk T, Brett L, O’Regan DP, Prasad SK, Morris-Rosendahl DJ, Barton PJR, Edwards E, Ware JS, Cook SA, CardioClassifier: disease- and gene-specific computational decision support for clinical genome interpretation. Genet. Med 20, 1246–1254 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nadelmann ER, Gorham JM, Reichart D, Delaughter DM, Wakimoto H, Lindberg EL, Litviňukova M, Maatz H, Curran JJ, Ischiu Gutierrez D, Hübner N, Seidman CE, Seidman JG, Isolation of Nuclei from Mammalian Cells and Tissues for Single-Nucleus Molecular Profiling. Curr Protoc. 1, e132 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, Armean IM, Bennett R, Bhai J, Billis K, Boddu S, Cummins C, Davidson C, Dodiya KJ, Gall A, Girón CG, Gil L, Grego T, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, Kay M, Laird MR, Lavidas I, Liu Z, Loveland JE, Marugán JC, Maurel T, McMahon AC, Moore B, Morales J, Mudge JM, Nuhn M, Ogeh D, Parker A, Parton A, Patricio M, Abdul Salam AI, Schmitt BM, Schuilenburg H, Sheppard D, Sparrow H, Stapleton E, Szuba M, Taylor K, Threadgold G, Thormann A, Vullo A, Walts B, Winterbottom A, Zadissa A, Chakiachvili M, Frankish A, Hunt SE, Kostadima M, Langridge N, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Aken BL, Yates AD, Zerbino DR, Flicek P, Ensembl 2019. Nucleic Acids Res. 47, D745–D751 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lun ATL, Riesenfeld S, Andrews T, Dao TP, Gomes T, participants in the 1st Human Cell Atlas Jamboree, Marioni JC, EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wolf FA, Angerer P, Theis FJ, SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zagar M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LB, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R, Integrated analysis of multimodal single-cell data. Cold Spring Harbor Laboratory (2020), p. 2020.10.12.335331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Solo: Doublet Identification in Single-Cell RNA-Seq via Semi-Supervised Deep Learning. Cell Systems. 11, 95–101.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 105.Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Systems. 8, 281–291.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh P-R, Raychaudhuri S, Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Traag VA, Waltman L, van Eck NJ, From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep 9, 5233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E, Fast unfolding of communities in large networks. Journal of Statistical Mechanics: Theory and Experiment. 2008 (2008), p. P10008. [Google Scholar]
- 109.Gayoso A, Lopez R, Xing G, Boyeau P, Wu K, Jayasuriya M, Melhman E, Langevin M, Liu Y, Samaran J, Misrachi G, Nazaret A, Clivio O, Xu C, Ashuach T, Lotfollahi M, Svensson V, da Veiga Beltrame E, Talavera-López C, Pachter L, Theis FJ, Streets A, Jordan MI, Regier J, Yosef N, scvi-tools: a library for deep probabilistic analysis of single-cell omics data,, doi: 10.1101/2021.04.28.441833. [DOI] [PubMed] [Google Scholar]
- 110.Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McCarthy DJ, Chen Y, Smyth GK, Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lause J, Berens P, Kobak D, Analytic Pearson residuals for normalization of single-cell RNA-seq UMI data. Genome Biol. 22, 258 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hafemeister C, Satija R, Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Martín-Fernández J-A, Hron K, Templ M, Filzmoser P, Palarea-Albaladejo J, Bayesian-multiplicative treatment of count zeros in compositional data sets. Statistical Modelling. 15 (2015), pp. 134–158. [Google Scholar]
- 115.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J, g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yu G, He Q-Y, ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst 12, 477–479 (2016). [DOI] [PubMed] [Google Scholar]
- 117.Díez J, Walter D, Muñoz-Pinedo C, Gabaldón T, DeathBase: a database on structure, evolution and function of proteins involved in apoptosis and other forms of cell death. Cell Death Differ. 17, 735–736 (2010). [DOI] [PubMed] [Google Scholar]
- 118.Shiina T, Hosomichi K, Inoko H, Kulski JK, The HLA genomic loci map: expression, interaction, diversity and disease. J. Hum. Genet 54, 15–39 (2009). [DOI] [PubMed] [Google Scholar]
- 119.Kassner A, Oezpeker C, Gummert J, Zittermann A, Gärtner A, Tiesmeier J, Fox H, Morshuis M, Milting H, Mechanical circulatory support does not reduce advanced myocardial fibrosis in patients with end-stage heart failure. European Journal of Heart Failure. 23 (2021), pp. 324–334. [DOI] [PubMed] [Google Scholar]
- 120.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Preibisch S, Saalfeld S, Tomancak P, Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 25, 1463–1465 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schapiro D, Sokolov A, Yapp C, Muhlich JL, Hess J, Lin J-R, Chen Y-A, Nariya MK, Baker GJ, Ruokonen J, Maliga Z, Jacobson CA, Farhi SL, Abbondanza D, McKinley ET, Betts C, Regev A, Coffey RJ, Coussens LM, Santagata S, Sorger PK, MCMICRO: A scalable, modular image-processing pipeline for multiplexed tissue imaging, doi: 10.1101/2021.03.15.435473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.https://github.com/HMS-IDAC/Cypository, Version 1.1.5
- 124.He K, Gkioxari G, Dollar P, Girshick R, Mask R-CNN (2017), (available at 10.48550/arXiv.1703.06870) [DOI] [PubMed] [Google Scholar]
- 125.Lin Tsung-Yi, Maire Michael, Belongie Serge, Bourdev Lubomir, Girshick Ross, Hays James et al. , Microsoft COCO: Common Objects in Context (2015), (available at 10.48550/arXiv.1405.0312) [DOI] [Google Scholar]
- 126.https://github.com/HMS-IDAC/S3segmenter, Version 1.3.18
- 127.Bronstein MM, Bruna J, LeCun Y, Szlam A, Vandergheynst P, Geometric Deep Learning: Going beyond Euclidean data. IEEE Signal Processing Magazine. 34 (2017), pp. 18–42. [Google Scholar]
- 128.Veličković P, Cucurull G, Casanova A, Romero A, Liò P, Bengio Y, Graph Attention Networks (2017), (available at http://arxiv.org/abs/1710.10903). [Google Scholar]
- 129.Xu C, Lopez R, Mehlman E, Regier J, Jordan MI, Yosef N, Probabilistic harmonization and annotation of single-cell transcriptomics data with deep generative models. Mol. Syst. Biol 17, e9620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data
All sequence data generated and analyzed in this study has been deposited as BAM files at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001006374 (https://ega-archive.org/studies/EGAS00001006374). Further information about EGA can be found on https://ega-archive.org, "The European Genome-phenome Archive in 2021" (https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkab1059/6430505).
Processed data can be analyzed using the Cellxgene tool website: https://cellxgene.cziscience.com/collections/e75342a8-0f3b-4ec5-8eel-245a23e0f7cb/private and are available in h5ad-format to download. Metadata sheets and patient information are available in Table S1&2.
Code
All code used to generate the figures in this publication are available on github (https://github.com/heiniglab/DCM_heart_cell_atlas). Code used to reproduce the presented analyses is indexed on Zenodo (88). All scripts run on jupyter notebooks are available as “.ipynb” files, scripts executed in command line are available as .txts or .sh files. R scripts are available as “.R”. Anaconda environments are available as yml files containing information on package versions.