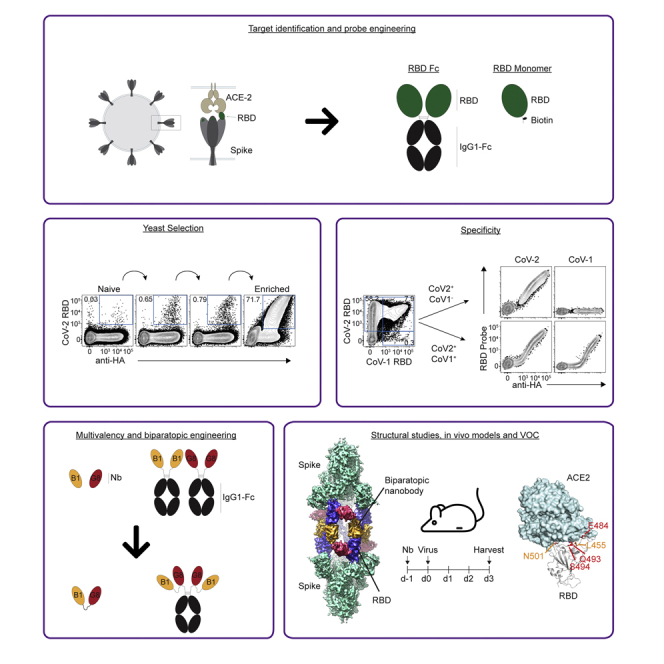
Summary
The development of therapeutics to prevent or treat COVID-19 remains an area of intense focus. Protein biologics, including monoclonal antibodies and nanobodies that neutralize virus, have potential for the treatment of active disease. Here, we have used yeast display of a synthetic nanobody library to isolate nanobodies that bind the receptor-binding domain (RBD) of SARS-CoV-2 and neutralize the virus. We show that combining two clones with distinct binding epitopes within the RBD into a single protein construct to generate biparatopic reagents dramatically enhances their neutralizing capacity. Furthermore, the biparatopic nanobodies exhibit enhanced control over clinically relevant RBD variants that escaped recognition by the individual nanobodies. Structural analysis of biparatopic binding to spike (S) protein revealed a unique binding mode whereby the two nanobody paratopes bridge RBDs encoded by distinct S trimers. Accordingly, biparatopic nanobodies offer a way to rapidly generate powerful viral neutralizers with enhanced ability to control viral escape mutants.
Subject areas: Immunology, microbiology, structural biology
Graphical abstract
Highlights
-
•
Yeast display was used to generate SARS-CoV-2 RBD-specific nanobodies
-
•
Distinct nanobody clones were fused to create biparatopic reagents
-
•
Biparatopic nanobodies protect mice from SARS-CoV-2 infection
-
•
The biparatopics bridge distinct S proteins in a unique “twinned-S” conformation
Immunology; Microbiology; Structural biology.
Introduction
Over two years since coronavirus disease 2019 (COVID-19) was declared a global pandemic (WHO Director-General’s opening remarks at the media briefing on COVID-19 - 11 March 2020, 2020), the spread of the disease continues, causing thousands of deaths daily and widespread social and economic disruption. Despite the fast-tracked development and approval of safe and effective vaccines, major questions remain regarding their long-term efficacy and their ability to provide protection, particularly against the continued emergence of variants of concern (VOC) (Abdool Karim and de Oliveira, 2021; Forni et al., 2021). New therapeutic approaches are thus needed. Recombinant biologics such as monoclonal antibodies (mAb) are one such approach and hold the potential for prophylactic protection, for example by “ring-fencing” of close contacts of infected individuals in quarantine or health care settings, or for reducing risk in immunocompromised individuals (Abraham, 2020). Furthermore, mAb may be used in the treatment of infected individuals to reduce viral burdens and limit the duration and/or severity of disease, as well as to reduce viral transmission (Taylor et al., 2021).
SARS-CoV-2, the causative agent for COVID-19 (Wu et al., 2020; Zhou et al., 2020a; Zhu et al., 2020), is a β-coronavirus and like other coronaviruses, its viral membrane is decorated in membrane-bound Spike (S) proteins (Huang et al., 2020; Li, 2016). S proteins bind surface receptors to facilitate viral entry and this represents a central event during infection. The full S protein exists as a homo-trimer, consisting of three heavily glycosylated protomers (Watanabe et al., 2020; Wrapp et al., 2020b). At the apex of the S trimer, a receptor-binding domain (RBD) of each protomer converges to cap the roof of the trimeric complex (Wrapp et al., 2020b). The SARS-CoV-2 RBD binds the membrane-tethered ectoenzyme angiotensin-converting enzyme 2 (ACE2) (Hoffmann et al., 2020; Lan et al., 2020; Letko et al., 2020; Li et al., 2005; Shang et al., 2020; Walls et al., 2020; Yan et al., 2020), and this binding event triggers membrane fusion with the host cell (Cai et al., 2020). These molecular events are broadly conserved between SARS-CoV-2 and SARS-CoV-1 viruses (Cai et al., 2020; Lan et al., 2020; Li et al., 2005; Song et al., 2018; Wrapp et al., 2020b)—the latter being the causative agent of the 2003 SARS outbreak (Ksiazek et al., 2003; Lee et al., 2003), with the RBDs of both proteins interacting with ACE2 to facilitate viral entry (Hoffmann et al., 2020; Letko et al., 2020; Li et al., 2003; Walls et al., 2020). Given the critical role of S protein and its exposure to the host, it is the dominant target of neutralizing antibodies (nAb) during natural infection and vaccination (Brouwer et al., 2020; Chi et al., 2020b; Ju et al., 2020; Liu et al., 2020; Piccoli et al., 2020; Premkumar et al., 2020; Voss et al., 2021; Wec et al., 2020), and is a key therapeutic target for vaccines and biologics. Of these nAb, the majority bind domains at the apex of the S protein, including the RBD (Brouwer et al., 2020; Ju et al., 2020; Piccoli et al., 2020; Premkumar et al., 2020; Wec et al., 2020) and, to a lesser extent, the N-terminal domain (NTD) (Brouwer et al., 2020; Chi et al., 2020b; Greaney et al., 2021; Liu et al., 2020; Voss et al., 2021), adjacent to the RBD (Wrapp et al., 2020b). Many recombinant, highly potent nAb are now in various stages of clinical development, either as single agents or in antibody cocktails where multiple nAb with distinct target epitopes are combined in an effort to enhance efficacy and reduce the potential of escape mutants (Taylor et al., 2021). Moreover, the field is rapidly evolving, and new approaches that include functional modifications, for example to the Fc domain, are being developed (Garber, 2021).
One approach is the utility of single domain antibodies, also known as nanobodies or VHH. Nanobodies are native to the camelid family including llamas, alpacas and camels and consist of a single variable domain (Hamers-Casterman et al., 1993). The small size, simple architecture, and high stability of nanobodies offers several advantages over traditional mAb including enhanced tissue penetration and a propensity to bind small epitopes with high affinity. Nanobodies typically have high yields and lower costs during recombinant production, and are amenable to covalent linkage to other protein scaffolds to enhance their function, for example Fc domains to increase valency and add Fc-function or to extend their half-life (De Vlieger et al., 2018). Moreover, compared to mAb, nanobodies are more readily adaptable to nebulization, which offers a more direct method of delivery to the airways in the context of respiratory diseases such as COVID-19 (Sasisekharan, 2021).
Here, we used a synthetic nanobody yeast display library (McMahon et al., 2018) to isolate nanobodies that bind and neutralize the SARS-CoV-2 RBD. Multimerization via engineering of a C-terminal human IgG1 Fc domain greatly enhanced the neutralizing capacity of these nanobodies. Moreover, epitope binning identified clones with distinct binding sites, and these were then covalently linked to engineer biparatopic nanobodies which further enhanced neutralizing capacity and resistance to escape from VOC. We subsequently used cryo-electron microscopy to understand the structural basis for biparatopic binding to S proteins, revealing a previously uncharacterized binding mode whereby the biparatopic construct bridged two disparate S proteins. Accordingly, we present evidence that a yeast display library can be used for the generation of anti-SARS-CoV-2 nanobodies with the ability to neutralize VOC that may evade conventional mAb therapy. Furthermore, we show that even when individual nanobodies have relatively low affinity, they can be markedly enhanced through the generation of biparatopic reagents, which may offer some advantages over conventional mAb-based therapies.
Results
Selection of anti-receptor-binding domain nanobodies using yeast-display
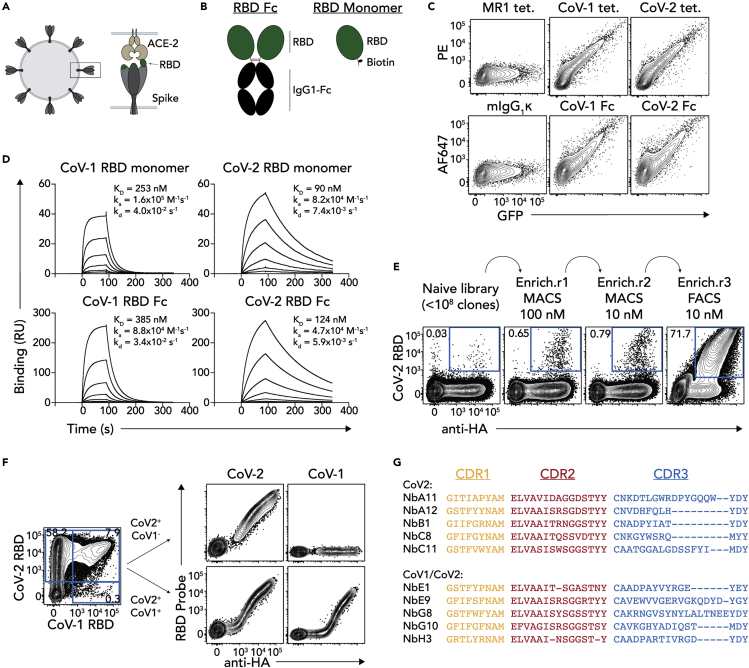
In order to generate nAb, we first engineered and validated a series of tools for use in the selection process. Given the direct binding of the RBD to ACE2 to mediate viral entry, nanobodies directed against the RBD hold obvious therapeutic potential (Figure 1A). RBD proteins were therefore engineered in two formats for both SARS-CoV-1 and SARS-CoV-2 RBDs; monomeric RBD with a C-terminal AVI-tag for site-directed biotinylation, and dimeric RBD-Fc proteins where the RBD was fused to the Fc domain of mouse IgG1 (Figure 1B). To validate these RBD probes, 293T cells were transiently transfected with full-length human ACE2 and stained with tetramerized RBD monomers or AF647-labeled RBD-Fc proteins directly (Figure 1C). Each of the RBD probes, but not an irrelevant MR1-Ag tetramer or IgG1 isotype control, bound 293T cells in an ACE2-dependent manner (as measured by GFP expression which is co-translated with ACE2), validating the functional integrity of the proteins. Surface plasmon resonance (SPR) was also performed, immobilizing RBD ligands and injecting soluble ACE2 analyte (Figure 1D). In line with previous reports (Shang et al., 2020), ACE2 bound the SARS-CoV-2 RBDs with higher affinity than SARS-CoV-1 RBDs (90 nM versus 253 nM for monomeric protein, and 124 nM versus 385 nM for Fc-fusions) as a result of both shorter on-rates and longer off-rates for each of the monomeric and dimeric proteins.
Figure 1.
Selection of anti-RBD nanobodies
(A) Cartoon schematic of RBD. Coronavirus is shown on the left, and a close-up of the S-protein interacting with ACE2 is shown on the right.
(B) Cartoon schematic of RBD-Fc and RBD monomer protein constructs.
(C) FACS plots showing staining of 293T cells transiently transfected to express human ACE2 and stained with SARS-CoV-1 or SARS-CoV-2 RBD tetramer-PE (CoV-1 tet. And CoV-2 tet. respectively), or SARS-CoV-1 or SARS-CoV-2 RBD-Fc conjugated to AF647 (CoV-1 Fc and CoV-2 Fc respectively). GFP is co-expressed with ACE2 thereby representing a surrogate for ACE2 expression. MR1-5-OP-RU tetramers (MR1 tet.) or mIgG1-AF647 were used as negative controls. Data representative of n = 3 separate experiments.
(D) Surface plasmon resonance sensorgrams showing binding kinetics of titrated concentrations of soluble ACE2 to immobilized SARS-CoV-1 or SARS-CoV-2 RBD monomers or Fc dimers. Data representative of n = 3 technical replicates.
(E) FACS plots showing SARS-CoV-2 RBD tetramer staining on yeast library after progressive rounds of enrichment.
(F) left panel shows FACS plot of HA+ yeast cells co-stained with SARS-CoV-1 and SARS-CoV-2 RBD tetramers (CoV-2 RBD and CoV-1 RBD respectively). Right panel shows examples of clones stained with the same tetramers after single-cell sorting for yeast that bound SARS-CoV-2 RBD only (CoV-2+CoV-1-; upper plots) or both SARS-CoV-1 and SARS-CoV-2 RBDs (CoV-2+CoV-1+; lower plots).
(G) CDR1, CDR2 and CDR3 sequence alignments of 10 selected clones. The upper 5 clones bound SARS-CoV-2 RBD tetramers only, while the bottom 5 bound both SARS-CoV-1 and SARS-CoV-2 RBD tetramers.
Traditionally, mAb are generated by producing hybridomas produced from immunized laboratory animals such as mice, cloning B cells from convalescent patients, or by panning phage-display libraries. Yeast display offers several advantages including a flow-cytometry-based selection approach which allows a highly tailored selection strategy to enhance specificity and affinity during the early stages of the selection process. Furthermore, while many mAb targeting the RBD have been described, nanobodies offer several potential advantages as a result of their small size. Accordingly, the RBD tetramers were then used to rapidly generate RBD-specific neutralizing nanobodies from a synthetic nanobody yeast display library (McMahon et al., 2018). The selection strategy consisted of an initial two rounds of magnetic-enrichment of yeast clones that bound to PE-labeled SARS-CoV-2 RBD tetramers, decreasing the concentration of the tetramers between rounds by ten-fold to favour enrichment of high affinity clones. This was followed by a FACS-based enrichment with these tetramers, resulting in an enriched library with ∼72% of yeast clones binding RBD tetramers (Figure 1E). This library was then co-stained with both SARS-CoV-1 and SARS-CoV-2 RBD tetramers in order to detect both SARS-CoV-2 specific nanobodies and SARS-CoV-2 and SARS-CoV-1 cross-reactive nanobodies. The staining pattern revealed two distinct populations; one major population that only bound the SARS-CoV-2 RBD tetramers, and a minor population that co-labeled with both SARS-CoV-1 and SARS-CoV-2 RBD tetramers (Figure 1F, left). These two populations likely contained clones with distinct binding epitopes, and moreover, the SARS-CoV-1/SARS-CoV-2 cross-reactive clones may bind a conserved RBD epitope that could represent an important therapeutic target. Accordingly, clones from both the SARS-CoV-2 only and SARS-CoV-1/CoV-2 cross-reactive populations were single-cell sorted. Screening of individual clonal yeast colonies revealed that the sorted clones maintained their expected binding patterns, with some colonies binding only SARS-CoV-2 tetramers, and others binding both SARS-CoV-1 and SARS-CoV-2 RBD tetramers (Figure 1F, right). The top 10 clones from each population that exhibited the brightest SARS-CoV-2 RBD tetramer staining were then sequenced, yielding five unique clones from each population (Figure 1G).
Characterization of receptor-binding domain-specific nanobodies
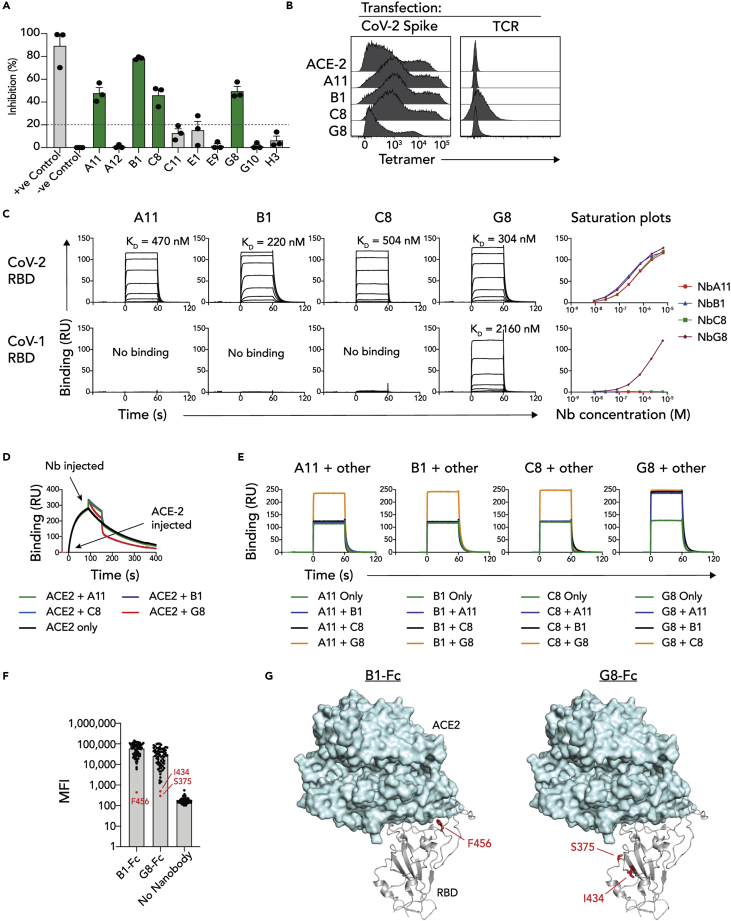
DNA encoding the 10 RBD-specific nanobodies was cloned into mammalian expression vectors and recombinant AVI-tagged nanobodies were expressed and purified. To test whether the isolated nanobodies inhibited the RBD-ACE2 interaction, a surrogate viral neutralization test was utilized (sVNT) (Tan et al., 2020). Here, recombinant HRP-conjugated SARS-CoV-2 RBD was incubated with nanobody before the complex was further incubated with plate-bound ACE2, and RBD binding to ACE2 subsequently detected via ELISA. Incubation with four of the nanobodies, clones A11, B1, C8 (SARS-CoV-2 only) and G8 (SARS-CoV-1/CoV-2 cross-reactive), resulted in a reduced absorbance signal, implying the inhibition of RBD-ACE2 binding by the nanobodies (Figure 2A). Next, these four nanobodies were tested for their ability to bind RBD in the context of full-length S protein. To this end, the nanobodies were biotinylated and tetramerized using streptavidin-PE and used to stain 293T cells transiently transfected to express SARS-CoV-2 S protein at their surface, with biotinylated ACE2 tetramers used as a positive control (Figure 2B). Although these tetramers did not bind 293T cells transfected with an irrelevant T cell receptor protein, they all bound the S-transfected cells, confirming that the nanobodies recognized RBD in its natural context as part of S protein. The RBD-binding capacities of the nanobodies were next characterized using SPR. Here, SARS-CoV-1 and SARS-CoV-2 Fc-fusion proteins were immobilized on SPR chips via their Fc domains, and monomeric nanobody analytes run across these ligands (Figure 2C). All four nanobodies bound SARS-CoV-2 RBD with moderate affinity, exhibiting KD of 470 nM, 220 nM, 504 nM and 304 nM for A11, B1, C8 and G8 respectively. As expected, only G8 exhibited cross-reactivity with SARS-CoV-1 RBD, though this was with lower affinity relative to SARS-CoV-2 RBD-binding (2160 nM). Thus, we had identified four RBD-specific nanobodies that bind and neutralize SARS-CoV-2 S protein, one of which cross-reacted with SARS-CoV-1 RBD.
Figure 2.
Characterization of nanobody RBD-binding
(A) Bar graph showing the percentage inhibition of RBD binding to ACE2 for each nanobody (50 μg/mL) as measured by ELISA-based sVNT assay. Data points represent mean of 2 replicate wells from n = 3 independent experiments.
(B) Flow cytometry histogram overlays showing nanobody or ACE2 tetramers binding to 293T cells transiently transfected to express full-length S protein or control T cell receptor (TCR) protein at their surface.
(C) Top plots: surface plasmon resonance sensorgrams showing binding of soluble nanobody monomers to immobilized SARS-CoV-1 or SARS-CoV-2 RBD Fc dimers. Bottom plots: Saturation plots of nanobody binding to RBD-Fc proteins. Data points represent mean of n = 3 technical replicates.
(D) Overlaid surface plasmon resonance sensorgrams showing ACE-2 or ACE-2/nanobody serial injections over immobilized SARS-CoV-2 RBD-Fc dimers.
(E) Overlaid surface plasmon resonance sensorgrams of simultaneous nanobody injections over immobilized SARS-CoV-2 RBD-Fc dimers.
(F) Bar graph of alanine scan data showing mean fluorescence intensity (MFI) of anti-IgG1 binding to RBD mutant-bound beads. Red data points for B1-Fc and G8-Fc are considered to have no binding.
(G) Structural cartoon representation of the SARS-CoV-2 RBD binding to human ACE2 (PDB: 7KNI (Zhou et al., 2020b)), highlighting residues identified in 2F.
To further interrogate the inhibitory capacity of these nanobodies, SPR-based serial-injection experiments were performed whereby SARS-CoV-2 RBD was immobilized, and two consecutive injections; initially ACE2, followed by individual nanobodies, were conducted (Figure 2D). Although the injection of nanobodies A11, B1 and C8 resulted in additive increases in binding response units which returned to the baseline dissociation of ACE2 upon rapid nanobody-dissociation, the SARS-CoV-1/CoV-2 cross-reactive clone G8, caused a reduction in response units below the ACE2 baseline binding response units. This implied that G8 exhibited a binding mode distinct from the other nanobodies, that potentially resulted in disrupting the interaction between ACE2 and RBD. To further probe the binding modes by the nanobodies, simultaneous-injection SPR experiments were performed whereby each nanobody was injected over immobilized SARS-CoV-2 RBD-Fc protein, either as a single monomeric nanobody, or in an equimolar ratio with one of the other nanobodies (Figure 2E). When A11, B1 or C8 were combined, the binding response units remained unchanged relative to each of them individually, suggesting that they likely compete for overlapping epitopes. In contrast, when any of these nanobodies was injected simultaneously with G8, an additive response was observed, suggesting concurrent binding to distinct epitopes. Collectively, these data suggested that nanobodies A11, B1 and C8 bound a shared epitope, whereas G8 bound a separate epitope that is more conserved within the CoV-1 RBD, and potentially has qualities that allow it to displace ACE2, rather than simply outcompeting it. To map the two distinct RBD-epitopes, an RBD alanine scan based on a previously published multiplex assay was used (Lopez et al., 2021), whereby 68 RBD single alanine mutants (Table S1) were separately conjugated to barcoded beads, incubated with mouse IgG1 Fc-fused G8 and B1 nanobodies (G8-Fc and B1-Fc) and binding of nanobodies to individual beads detected with anti-IgG1 using the Luminex system (Figure 2F). Here, B1-Fc binding was completed abrogated in response to F456A which mapped to the receptor-binding motif (RBM) of the RBD, whereas G8-Fc binding was abrogated in response to S375A and I434A which mapped to a distinct region of the RBD, distal to F456 and underneath the RBM (Figure 2G).
Generation of engineered nanobody protein constructs for enhanced viral neutralization
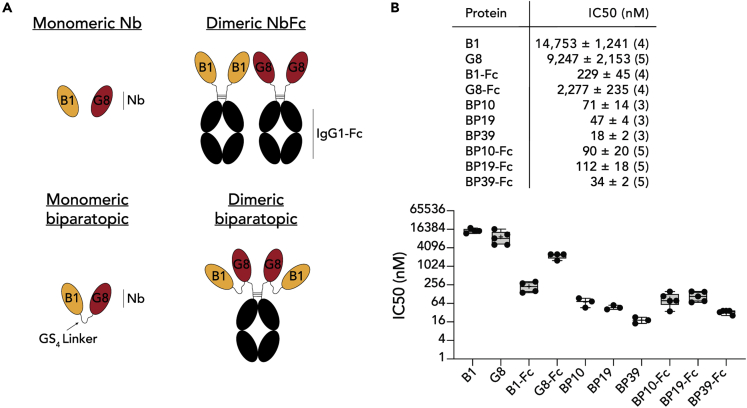
The affinities of the nanobodies, while moderate, were unlikely to be high enough to strongly neutralize viral infection relative to the many high-affinity mAbs that have already been developed. A series of nanobody protein constructs were thus generated that aimed to enhance the neutralizing capacity of the monomeric nanobodies, utilizing clones B1 and G8, as they were the highest affinity binders and also bound distinct RBD epitopes (Figure 3A). Three distinct approaches were taken: First, the nanobodies were fused to the Fc domain of human IgG1 to produce dimeric nanobody-Fc proteins with enhanced avidity for S binding (B1-Fc and G8-Fc). Second, biparatopic nanobodies were generated in which B1 and G8 were fused via a glycine/serine-rich linker, reasoning that fusing these proteins together would provide two points of attachment to each RBD, also increasing the surface area of the RBD bound by the overall nanobody paratope, thus enhancing affinity and avidity, an approach successfully used in the context of anti-influenza hemagglutinin nanobodies (Laursen et al., 2018). Because the precise binding orientations of each nanobody was unknown, three separate constructs were generated with varied linker length, (BP10, BP19 and BP39, each with 10, 19 and 39 amino acid linkers respectively) to test which would be most effective. Additionally, biparatopic Fc fusion proteins using the Fc domain of human IgG1 were generated, thus producing dimeric biparatopics. To test the ability of these proteins to inhibit SARS-CoV-2 infection, a microneutralization assay (MNV) that measures viral cytopathic effect as a measurement of infectivity was employed (Subbarao et al., 2004) (Figure 3B). As monomeric nanobodies, both B1 and G8 showed only modest inhibition, with IC50 scores of 14,750 nM and 9,247 nM respectively. Fc-mediated dimerization of either B1 or G8 clearly enhanced neutralizing activity, registering IC50 scores of 190 nM and 1922 nM respectively. Strikingly, the monomeric biparatopic reagents dramatically enhanced neutralization, to an extent that increased with linker length (71 nM, 47 nM and 18 nM for BP10, BP19 and BP39 respectively). Although Fc-based dimerization of the biparatopics also resulted in potent neutralization, these were moderately less effective than their simple biparatopic counterparts (90 nM, 112 nM and 34 nM for BP10-Fc, BP19-Fc and BP39-Fc respectively). Of note, the BP39 biparatopic which provided the strongest neutralization at 18 nM, was similar to highly potent nanobodies derived from immunized alpacas, as well as mAb cocktails used in clinical trials, when these proteins where assayed in separate experiments performed in the same lab (Pymm et al., 2021). Accordingly, linking distinct nanobodies specific for different epitopes dramatically enhances the neutralizing capacity of RBD-specific nanobodies, while retaining the advantage of small nanobody size and stability.
Figure 3.
Generation of biparatopic reagents and assessment of viral inhibition
(A) Cartoon schematics of different nanobody protein constructs.
(B) Top: Table showing IC50 minimum neutralizing (MN) concentration from live SARS-CoV-2 virus microneutralization assays for each tested nanobody protein construct. IC50 is shown ±SEM with the number of independent experiments displayed in brackets. Bottom: IC50 values as above, plotted on box and whisker plots. Individual data points are plots. Boxes depict median, upper and lower quartiles, and whiskers show minimum and maximum points. + denotes mean.
The molecular basis for biparatopic nanobody binding
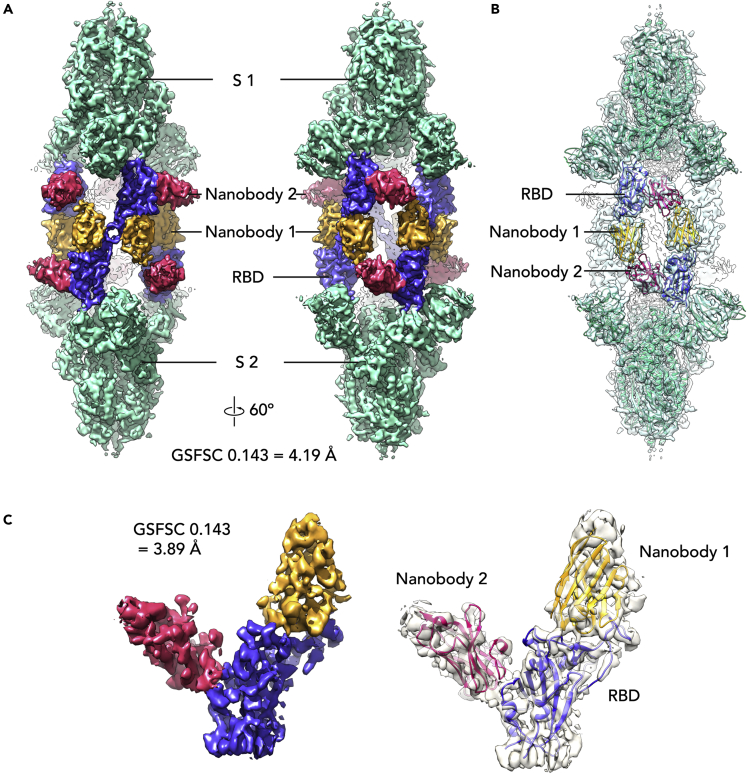
Structural analysis of biparatopic protein binding in the context of the S trimer was carried out using cryo-electron microscopy (cryo-EM). A dataset for the BP10 construct bound to stabilized prefusion S trimer was determined to have a single conformation through the examination of the obtained 2D and 3D classes and was processed to 4.24 Å resolution (Table S2). This allowed the determination of the binding sites of the B1 and G8 nanobodies on the RBD, which make up BP10 (Figure 3A), and assessment of how the 10 amino acid residue linker impacts nanobody occupancy and S conformation.
The cryo-EM map (See Figure S1) revealed a “twinned S″ conformation for the S-BP10 structure, with two copies of the S trimer linked together with six copies of the BP10 protein (Figure 4A). Both S trimers are well resolved (See Figure S1) and each RBD is in the “up” conformation, and fully occupied with two nanobodies (Figure 4B). Although density for the linker is largely absent, given the orientation of the nanobodies on the RBD and the twinning of the S trimers, BP10 most likely binds across two RBDs, bridging two opposing S trimers. Molecular modeling suggests that this observed nanobody orientation could be accommodated by a 10 amino acid linker used in the BP10 construct with only a 15.5 Å distance between these sites (See Figure S2A). Conversely, a longer linker of around 20 amino acids is required to enable BP binding to a single RBD (See Figure S2B) with a distance of 63.9 Å between docked nanobodies. Furthermore, binding adjacent RBD on the same S would require the longest linker of at least 25 amino acids to bridge the 78 Å distance (See Figure S2C), which may explain the enhanced potency of the BP39 in the microneutralization assays.
Figure 4.
Structure of BP10 with the SARS-CoV-2 S-protein
Cryo-EM maps of the BP10-S complex at 4.2 Å resolution with S (green), RBDs (purple-blue) and Nanobodies 1 (gold) and 2 (pink) (A). Refined maps at 60º rotation intervals along the Y axis (B). Homology models for S (PDB: 7VNE (Yang et al., 2021)), RBD (PDB ID: 7LX5 (Pymm et al., 2021)), Nanobody 1 (PDB: 7KKK) and Nanobody 2 (PDB ID: 5IMK (Wen et al., 2017)) docked into the refined S-BP10 complex map (C). Local refinement of density for one copy of the RBD with two nanobodies attached at 3.9 Å resolution with homology models docked into the density. See also Figures S1–S3 and Table S2.
Local refinement of an individual RBD to a global resolution of 3.9 Å revealed clear density corresponding to two nanobodies at separate sites (Figure 4C) and the RBD and RBD-nanobody interfaces were well resolved (See Figure S3). Docking of homology models for the RBD and nanobodies into the map showed the epitope for nanobody 1 (likely nanobody B1 based on the alanine scan data) overlapping the ACE2 binding site and centered approximately on the strand containing RBD residues 491–498. The density for nanobody 2 (likely nanobody G8 based on the alanine scan) is placed on the side of the RBD and has limited overlap with the ACE2 binding site. This also aligns with the cross-reactivity for the RBD of SARS-CoV-1, as there is more sequence diversity at the ACE2 biding site relative to other distal framework regions of the RBD (Wu et al., 2020). Nevertheless, both nanobody B1 and G8 were able to inhibit ACE2 binding to the RBD as monomers (Figure 2A). The positioning of this density on the RBD is similar to several previously published structurally characterized nanobody cocktails including VHH E and VHH U (Koenig et al., 2021) (PDB ID 7KN5) and WNb 2 and WNb 10 (Pymm et al., 2021) (PDB ID 7LX5) and the biparatopic nanobody VHH VE (Koenig et al., 2021) (PDB IDs 7B17, 7B18). However, unlike for BP10, the VHH VE bound to the S trimer did not result in a “twinned” structure, possibly owing to a longer linker length of 15 amino acid residues employed for VHH VE (Koenig et al., 2021).
Biparatopic nanobodies limit escape by variants of concern
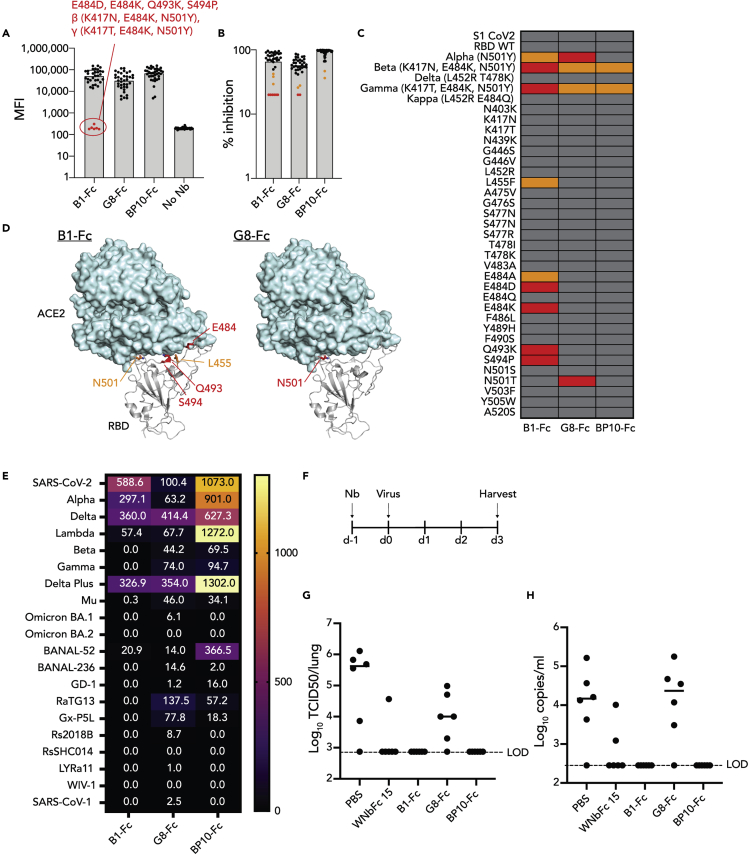
Another advantage of fusing two distinct nanobody paratopes is the potential to overcome viral mutants that escape from any one of the paratopes. Indeed, several VOC have emerged since the beginning of the pandemic, several of which have reduced neutralization from convalescent or post-vaccine plasma and exhibit a complete loss of binding from select mAbs. The ability of the biparatopic nanobody to overcome viral escape was therefore assessed using a multiplex RBD variant array that measures both binding of the nanobodies to RBD variants of concern, as well as their ability to block ACE2 binding to these variants (Lopez et al., 2021). Although G8-Fc exhibited relatively robust binding to all assessed variants, B1-Fc on the other hand had reduced binding to variants with mutations in E484 including E484D, E484K, β (K417N, E484K and N501Y) and (K417N, E484K and N501Y) as well as Q493K and S494P. This mapped to the same region of the RBM identified in the alanine scan in Figure 3. The biparatopic nanobody BP10-Fc however exhibited a similar profile to G8-Fc, but without showing the same loss of binding owing to N501Y and N501T. Thus, the biparatopic conformation apparently compensated for the reduced binding of separate nanobodies B1 and G8 to most of the mutations described, although β and γ represented the two outlying datapoints with moderately reduced MFIs (Figure 5A). Similar results were obtained when assessing the ability of the nanobodies to inhibit ACE2 binding (Figures 5B–5D), whereby the same mutations as identified in direct RBD binding reduced ACE2 inhibition by B1-Fc, with a moderate impact by N501Y, L455F and E484A. G8-Fc showed reduced the inhibition of mutants harboring N501Y or N501T mutations, including the triple mutants β and γ. This is likely influenced by the enhanced binding of N501Y to ACE2. BP10-Fc was not inhibited by either of the E484 and N501 single mutants, Q493K or S494P, and had improved binding to β and γ relative to B1-Fc and G8-Fc. The same nanobody constructs were also tested in a bead-based surrogate virus neutralization test (sVNT) which measures the inhibition of the RBD-ACE2 interaction. A total of 20 distinct sarbecovirus RBDs were used, including some RBDs used above, as well as Omicron BA.1, and BA.2 and a series of bat and pangolin coronavirus RBDs and SARS-CoV-1 (Tan et al., 2021a) (See Figures 5E and S4). The results mirrored but also complemented that of the multiplex array presented above, however neither nanobody demonstrated neutralization against the highly mutated omicron BA.1 and BA.2 RBDs. On the other hand, the biparatopic demonstrated superior neutralization toward Delta Plus RBD as was seen for Delta in this assay and the previous assay, and intriguingly, the biparatopic demonstrated neutralizing activity toward BANAL-52, derived from a highly divergent bat coronavirus (Temmam et al., 2022).
Figure 5.
Biparatopic nanobodies negate escape by RBD variants of concern
(A) Bar graph of variant array data showing MFI of anti-IgG1 binding to RBD variant-bound beads. Red data points for B1-Fc are considered to have no binding.
(B) Bar graph of variant array data showing percentage inhibition of ACE2 binding to RBD variant-bound beads. Red data points are considered to have no binding, whereas orange data points have reduced binding.
(C) A heatmap representation of the data in B.
(D) Structural cartoon representation of the SARS-CoV-2 RBD binding to human ACE2 (PDB: 7KNI (Zhou et al., 2020b)), highlighting residues identified in 2B-C.
(E) A heatmap representation of neutralization titers from multiplex sVNT assay, testing neutralization of RBD-ACE2 binding using RBDs from a range of SARS-CoV-2 variants of concern and sarbecoviruses.
(F) Schematic of mouse challenge model.
(G and H) Bar graphs showing G. viral TCID or H. PCR values from lungs of mice challenged with SARS-CoV-2 after treatment with nanobody proteins. See also Figure S4 and Table S1.
Prophylactic treatment of mice with anti-receptor-binding domain nanobodies reduces SARS-CoV-2 viral loads
Biparatopic nanobodies have the potential to overcome escape by VOC, as illustrated by fusion of B1 and G8 nanobodies. The ability of these nanobodies to provide protection from SARS-CoV-2 infection was subsequently tested in a prophylactic setting in which mice were pre-treated with nanobody-Fc proteins and then infected with SARS-CoV-2 virus 24 h later (Figure 5F). Of note, the clinical isolate used in this mouse model harbors both the D614G and N501Y mutations, enabling it to efficiently bind mouse ACE-2 and infect mouse cells as reported previously (Gu et al., 2020). A nanobody previously shown to protect animals from infection in this model, WNbFc 15, was included as a positive control (Pymm et al., 2021). Although G8-Fc moderately reduced viral loads in the lungs of challenged mice, mice treated with B1-Fc and BP10-Fc were fully protected from SARS-CoV-2 challenge as measured by infectious virus titre (Figure 5G) and PCR (Figure 5H). Accordingly, dimeric nanobodies and biparatopics exhibit potential to prevent infection with SARS-CoV-2.
Discussion
The development of biologics for the treatment or prevention of SARS-CoV-2 infection has evolved rapidly, and the first generation of nAbs have entered clinical trials with promising initial results (Garber, 2021). A second generation of biologics are progressing through preclinical development, and these include approaches that enhance the efficacy of natural mAbs, such as optimized Fc-binding, multimerization and nanobody-based biologics (Konwarh, 2020; Ledford, 2020; Sasisekharan, 2021). Here, we isolated a panel of nanobodies with neutralizing capacity from an in vitro yeast display library and used two distinct nanobodies to show that multimerization and the generation of biparatopic nanobodies can greatly enhance the efficacy of these reagents.
Several investigators have now isolated nanobodies targeting the SARS-CoV-2 RBD, either through immunization of camelids (Esparza et al., 2020; Gai et al., 2021; Hanke et al., 2020; Koenig et al., 2021; Ma et al., 2021; Pymm et al., 2021; Valenzuela Nieto et al., 2021; Wagner et al., 2021; Wrapp et al., 2020a; Xiang et al., 2020) or via in vitro library-based approaches (Chi et al., 2020a; Custodio et al., 2020; Huo et al., 2020; Lu et al., 2021; Schoof et al., 2020; Yao et al., 2021). Nanobodies offer several potential advantages over full length mAb, largely through their small and simple structures (Ingram et al., 2018). Despite their size, nanobodies nonetheless bind their targets with affinity comparative to that of antibodies, typically within the sub-low nanomolar range (Ingram et al., 2018). We isolated nanobodies in the mid-nanomolar range, which is in line with another study that used the same yeast-display library, although this study isolated some binders of less than 100 nM affinity (Schoof et al., 2020). Other studies that have utilized camelid immunization to isolate nanobodies have typically achieved higher affinities in the sub-low nanomolar range (Esparza et al., 2020; Gai et al., 2021; Hanke et al., 2020; Ma et al., 2021; Pymm et al., 2021; Wagner et al., 2021; Wrapp et al., 2020a; Xiang et al., 2020), possibly reflecting the limitations imposed by the clone size (approximately 5 × 108) of the yeast-display library (McMahon et al., 2018). Nonetheless, shortcomings in affinity from use of this library can be overcome by in vitro affinity maturation (Schoof et al., 2020). In this study, dimerization of the nanobodies via fusion to the Fc-domain of human IgG1 resulted in enhanced neutralization, likely as a result of increased avidity, in line with other studies that have dimerized anti-RBD nanobodies (Hanke et al., 2020; Huo et al., 2020; Ma et al., 2021; Pymm et al., 2021; Xiang et al., 2020). Similarly, trimeric nanobody fusions have also proven to improve potency over their monomeric counterparts (Dong et al., 2020; Lu et al., 2021; Schoof et al., 2020; Xiang et al., 2020), highlighting the advantage of multimerization and its impact on increasing avidity. Depending on the binding orientation of the nanobodies relative to the trimeric S-associated RBDs, it seems likely that the optimal copy number of a given nanobody within a multimeric scaffold will depend on the RBD epitope and docking mode of individual nanobodies. Indeed, structural analysis of the binding of BP10 to the S trimer revealed a “twinned” S conformation whereby six copies of the BP10 protein “tied” two S trimers together via their RBD domains. There are now over 400 SARS-CoV-2 specific nanobodies on the CoVAbDab database (Raybould et al., 2021) that have been grouped, based on binding epitopes, into three classes (Sun et al., 2021). The density for nanobodies 1 and 2 in the cryo EM structure fall into the class I (nanobody B1) and class II (nanobody G8) families respectively. Combined with data from the multiplex RBD variant array, nanobody 1 is likely to be B1 and nanobody 2 to be G8, this additionally fits well with the previously observed susceptibility of class I nanobodies to the E484K mutation (Sun et al., 2021).
Our cryo-EM results show an S-nanobody conformation which has not been observed before despite the nanobodies binding to previous structurally characterized binding classes (Sun et al., 2021). BP10 has a 10 amino acid linker and we propose that this unique S-nanobody conformation is a result of the shorter linker, and clearly linker length is a critical consideration for design of multivalent nanobody constructs. The BP10 protein showed over 100-fold improvement in neutralizing activity compared with monomeric nanobodies. This is despite the linker being too short to allow binding of both nanobodies to the same RBD or adjacent RBD on the same S protein. Although the physiological relevance of the observed “twinned” S has not been established, it may be that the aggregation of viral particles through binding S in trans on adjacent virions, or distortion of S proteins in cis on individual virions, may contribute to the neutralization profile of multivalent nanobodies.
In this study, fusion of nanobodies with separate RBD epitopes into biparatopic reagents via GS linkers enhanced neutralization to a far greater degree than did the dimerization of a single paratope via Fc-domains. Thus, biparatopic nanobodies offer another potential method of greatly enhancing the efficacy of nanobody-based neutralizers, which is in line with reports from others (Chi et al., 2022; Dong et al., 2020; Koenig et al., 2021; Li et al., 2021; Lu et al., 2021; Ma et al., 2021; Wagner et al., 2021; Xiang et al., 2020; Yao et al., 2021). We tested the linker-length between nanobodies and found that the longest linker length at 39 amino acids was the most effective, although 10 and 19 amino acid linkers still had greatly enhanced neutralization relative to monomeric and Fc-fusion proteins. Optimum linker-length is also likely to depend on the specific paratopes and can be guided by structural studies, as recently reported for other biparatopic nanobodies targeting the SARS-CoV-2 RBD (Koenig et al., 2021) as well as in other viral proteins such as the influenza hemagglutinin stalk (Laursen et al., 2018). Conjoining of two distinct paratopes can also offset the effect of escape mutants including variants of concern (Koenig et al., 2021). Although G8 was sensitive to the N501Y mutation found in several variants of concern or interest, including alpha, beta, and gamma as assessed in this study, B1 was a more sensitive to mutants harboring E484 mutations amongst others. The biparatopic nanobody however was able to overcome these separate sensitivities in in vitro binding/neutralization assays, and indeed provided protection against mice challenged with an N501Y containing virus. Given the extensive mutations in the omicron RBD, including N501Y and E484A, is it perhaps unsurprising that the biparatopic failed to neutralize the omicron variants BA.1 and BA.2 in the sVNT multiplex bead assay. This has been observed for many monoclonal antibodies including those previously used therapeutically, as well as convalescent sera (Takashita et al., 2022; Wilhelm et al., 2022). It was nonetheless intriguing that the biparatopic demonstrated some capacity to neutralize BANAL-52 RBD, derived from a bat coronavirus with marked variation relative to the ancestral SARS-CoV-2 (Temmam et al., 2022). The biparatopic may thus serve as a scaffold for the development of neutralizing reagents against future variants with distinct RBD mutations, for example via yeast display of a mutated B1 and/or G8 nanobody library. Also, while our study did not differentiate between treatment with a cocktail of B1-Fc and G8-Fc versus use of the biparatopic reagent, the fact that the effectiveness of the biparatopic reagents was governed by the linker length suggests that the linked nature of these nanobodies that is important in their increased neutralizing activity.
Interestingly, dimerization of the biparatopic proteins via linkage to the human IgG1 Fc domain failed to enhance their neutralizing capacity—indeed, if anything, it was moderately reduced. This is possibly owing to changes in accessibility of the RBD epitopes when the Fc domain is present. Alternatively, Fc-fusion proteins may have added functionality imparted by the Fc domain, such as Fc-receptor-mediated antibody-dependent cellular cytotoxicity (ADCC), as well as enhanced in vivo half-life as a result of the increased size and reduced renal clearance. Indeed, in the mouse challenge model used here, Fc-bearing biparatopics protected mice. Similarly, in the case of influenza infection, some hemagglutinin stalk-specific mAb require Fc function to elicit protection in vivo (DiLillo et al., 2014), though this is clone-dependent. Fc-function was also shown to be important for in vivo efficacy of Fc-fused biparatopic nanobodies (Laursen et al., 2018).
Supply and distribution are highly limiting factors in the success of biologic-based therapies during a global pandemic. The compact, simple architecture of the single immunoglobulin-like fold of nanobodies results in a protein unit that is highly stable across a wide temperature range (Ingram et al., 2018). These features reduce their susceptibility to temperature fluctuations and decrease cold-chain requirements. Nanobodies are also amenable to lyophilization, which may alleviate challenges with their distribution. Similarly, nanobodies can be nebulized, potentially offering a more direct and non-invasive method of delivery into the airways which may be very effective in the context of treating respiratory illness such as COVID-19 (Detalle et al., 2016; Simoes et al., 2018). In line with this, recent studies showed that nanobodies targeting the SARS-CoV-2 RBD maintained high stability after lyophilization, heat treatment and/or aerosolization (Esparza et al., 2020; Gai et al., 2021; Schoof et al., 2020; Xiang et al., 2020). The simplicity of the nanobody structure also results in high yields from mammalian expression systems as well as other systems such as bacteria and yeast, which are not typically amenable to mAb production (Arezumand et al., 2017). This offers the potential to generate large quantities at lower cost which are desirable in the context of a global pandemic. Because of their small size, however, nanobodies have far shorter serum half-lives relative to mAbs, although this limitation can be overcome by multimerization via Fc domains, or linking them to a larger carrier protein such as human serum albumin (Kontermann, 2011). The speed at which yeast display can be utilized to isolate high-affinity binders specific for SARS-CoV-2 variants of concern, relative to traditional immunization methods also offers great potential to fast-track development of therapeutics in the face of an evolving pandemic. Although the nanobodies from this library are synthetic, they are likely to be minimally immunogenic (Scully et al., 2019) owing to their substantial homology with human variable chains (Klarenbeek et al., 2015), and they are also amenable to further humanization if necessary (Vincke et al., 2009).
In summary, we have developed a series a nanobodies targeting the RBD of SARS-CoV-2 and shown that the multimerization of one or more nanobody paratopes can greatly enhance the neutralizing capacity of individual nanobodies, apparently by cross-linking distinct S proteins to each other. Accordingly, nanobodies offer great potential as therapeutics to target respiratory viruses such as SARS-CoV-2.
Limitations of the study
Here, we describe a unique molecular interaction whereby the biparatopic nanobodies bridged two distinct S proteins in a “Twinned-S” conformation. The biological implications of this docking mode remain unclear. Although we hypothesize that this may contribute to the enhanced neutralizing capacity of the biparatopic reagent relative to the individual nanobodies, perhaps by aggregating multiple viral particles for example, this has not been formally demonstrated and warrants further investigation in future studies. High resolution crystal structures of the individual nanobodies bound to RBD protein would be helpful to define the precise molecular features and RBD epitopes for each nanobody, which may assist in defining SARS-CoV1/2 cross-reactive epitopes for example. After extensive attempts however, the individual nanobodies failed to crystalize in complex with RBD. Finally, we also note that further animal studies that titrate the dose of prophylactic nanobody treatment would provide helpful insight into how potent the level of protection afforded by the biparatopic reagents is.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-HA-AF647 clone 16B12 | Biolegend | Cat#682404; RRID: AB_2566616 |
| Anti-human IgG-PE | Southern Biotech | Cat#9040–09; RRID: 2796601 |
| AffiniPure sheep anti-mouse IgG | Jackson Research | Cat#515-005-071; RRID: 2340289 |
| Bacterial and virus strains | ||
| DH5α Escherichia coli | University of Melbourne | in-house |
| SARS-CoV-2 isolate CoV/Australia/VIC01/2020 | (Caly et al., 2020) | in-house |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant SARS-CoV-1 RBD-Fc protein | University of Melbourne | in-house |
| Recombinant SARS-CoV-2 RBD-Fc protein | University of Melbourne | in-house |
| Recombinant SARS-CoV-1 RBD-biotin protein | University of Melbourne | in-house |
| Recombinant SARS-CoV-2 RBD-biotin protein | University of Melbourne | in-house |
| Recombinant SARS-CoV-2 RBD-His,AviTag (Ancestral, Alpha, Delta, Beta, Gamma) | Genscript | Custom made |
| Recombinant SARS-CoV-2 RBD-His,AviTag (Delta plus, Mu, Lambda, Omicron BA.2) | Duke-NUS | in-house |
| Recombinant SARS-CoV-2 Omicron BA.1 RBD-His,AviTag | Acrobiosystems | SPD-C82E4 |
| Recombinant SARSr-CoV RBD-His,AviTag | Duke-NUS | In-house |
| PE-labelled human ACE2 | Genscript | Custom made |
| Recombinant SARS-CoV-2 S ‘hexapro’ protein | Walter and Eliza Hall Institute | in-house |
| Recombinant ACE-2 protein | University of Melbourne | in-house |
| Recombinant nanobody monomer proteins | University of Melbourne | in-house |
| Recombinant nanobody-Fc dimer proteins | University of Melbourne | in-house |
| Recombinant biparatopic nanobody proteins | University of Melbourne | in-house |
| Recombinant biparatopic nanobody-Fc proteins | University of Melbourne | in-house |
| Recombinant Bir-A enzyme | University of Melbourne | in-house |
| NheI-HF restriction enzyme | New England Biolabs | Cat#R3131S |
| BamHI-HF restriction enzyme | New England Biolabs | Cat#R3136S |
| Critical commercial assays | ||
| Zymoprep Yeast Plasmid Miniprep II Kits | Zymo Research | Cat#D2004 |
| ZR Plasmid Miniprep - Classic | Zymo Research | Cat#D4054 |
| ZympPURE II Plasmid Maxiprep Kit | Zymo Research | Cat#D4204 |
| ZympPURE II Plasmid Gigaprep Kit | Zymo Research | Cat#D4203 |
| Zymoclean Gel DNA Recovery Kit | Zymo Research | Cat#D4002 |
| HiFi Hotstart ReadyMix PCR Kit | Roche | Cat#7958935001 |
| Expifectamine 293 transfection kits | Thermo Fisher Scientific | Cat#A14525 |
| Nucleospin 96-well plate extraction kit | Machery-Nagel | Cat#740691.4 |
| iTaq Universal Probe Kit | Bio-Rad Inc | Cat#1725140 |
| SARS-CoV-2 Viral Standards kit | Exact Diagnostics | Cat#COV019 |
| sVNT assay | Genscript | Cat#L00847-A |
| Deposited data | ||
| SARS-CoV-2 Spike in complex with biparatopic nanobody BP10 | This paper | EMDB: EMD-28189 |
| SARS-CoV-2 RBD in complex with biparatopic nanobody BP10 local refinement | This paper | EMDB: EMD-28190 |
| Cryo-EM structure of Triple ACE2-bound SARS-CoV-2 Trimer Spike | (Zhou et al., 2020b) | PDB: 7KNI |
| Cryo-EM Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody | (Yang et al., 2021) | PDB: 7VNE |
| Cryo-EM Structure of the SARS-CoV-2 spike protein receptor binding domain bound to neutralizing nanobodies WNb 2 and WNb 10 | (Pymm et al., 2021) | PDB: 7LX5 |
| Crystal structure of a nanobody targeting human Vsig4 in Spacegroup C2 | (Wen et al., 2017) | PDB: 5IMK |
| Experimental models: Cell lines | ||
| Expi293F cells | Thermo Fisher Scientific | Cat#A 14527 |
| HEK293T cells | ATCC | CRL-3216 |
| Experimental models: Organisms/strains | ||
| C57BL/6J Mice | The Walter and Eliza Hall Institute | N/A |
| Oligonucleotides | ||
| 5′-GTTTTGTTCGCTGCTTCTTCTGC-3′ | (McMahon et al., 2018) | pYDS649HM_FWD |
| 5′-GGTAGATTTACTAGGCGATGAGG-3′ | (McMahon et al., 2018) | pYDS649HM_REV |
| 5′-GACCCCAAAATCAGCGAAAT-3′ | (Subbarao et al., 2004) | 2019-nCoV_N1 forward primer |
| 5′-TCTGGTTACTGCCAGTTGAATCTG-3′ | (Subbarao et al., 2004) | 2019-nCoV_N1 reverse primer |
| 5′-FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1-3′ | (Subbarao et al., 2004) | 2019-nCoV_N1 probe |
| Recombinant DNA | ||
| SARS-CoV-2 Hexapro Spike expression plasmid | (Juno et al., 2020) | N/A |
| SARS-CoV-2 RBD monomer expression plasmid | University of Melbourne | In-house |
| SARS-CoV-1 RBD monomer expression plasmid | University of Melbourne | In-house |
| SARS-CoV-2 RBD-Fc dimer expression plasmid | University of Melbourne | In-house |
| SARS-CoV-1 RBD-Fc dimer expression plasmid | University of Melbourne | In-house |
| ACE2-biotin plasmid | University of Melbourne | In-house |
| Nanobody monomer plasmids | University of Melbourne | In-house |
| Nanobody-Fc plasmids | University of Melbourne | In-house |
| Biparatopic Nanobody plasmids | University of Melbourne | In-house |
| Biparatopic Nanobody-Fc plasmids | University of Melbourne | In-house |
| pMIG II plasmid | Addgene | Cat#52107 |
| pHL-sec plasmid | Addgene | Cat#99845 |
| Software and algorithms | ||
| Flowjo v10.8.1 | Beckman Dickinson | https://www.flowjo.com/ |
| PyMOL, opensource | Schrodinger | https://pymol.org/ |
| Prism | Graphpad | v9.3.1 |
| Relion v3.1 | (Zivanov et al., 2018) | https://relion.readthedocs.io/en/latest/ |
| Chimera v1.1.3 | (Pettersen et al., 2004) | https://www.cgl.ucsf.edu/chimera/ |
| CryoSPARC v3.2 | Stuctura Biotechnology | https://structura.bio/ |
| Gautomatch v0.56 | N/A | https://www.mrc-lmb.cam.ac.uk/kzhang/ |
| GCTF | (Zhang, 2016) | N/A |
| Scrubber v2.0 | BioLogic | http://www.biologic.com.au/scrubber.html |
| Other | ||
| AKTA Pure | Cytiva | N/A |
| AKTA Purifier | Cytiva | N/A |
| Flexmap3D | Luminex Corporation | N/A |
| Luminex MagPix Instrument | Luminex Corporation | N/A |
| Titan Krios G4 electron microscope | Thermo Fisher Scientific | N/A |
| LSR Fortessa flow cyometer | BD Biosciences | N/A |
| MoFlo Astrios cell sorter | Beckman Coulter | N/A |
| Quantifoil Cu 300 mesh grids | Quantifoil | R1.2/1.3 |
| Pelco EasiGlow discharge aparatus | Pelco | N/A |
| Vitrobot Mark IV | Thermo Fisher Scientific | N/A |
| Flacon 4 detector | Thermo Fisher Scientific | N/A |
| Mouse inhalation exposure system | Glas-Col | N/A |
| Bullet Blender | Next Advance Inc | N/A |
| DEAE Sepharose | Cytivia | Cat#17070901 |
| Ni-NTA Agarose | Thermo Fisher Scientific | Cat#R90110 |
| Protein A Sepharose | Thermo Fisher Scientific | Cat#101042 |
| MonoQ 10/100 GL anion exchange column | Cytivia | Cat#17517901 |
| HiLoad Superdex 200 16/600 size exclusion column | Cytivia | Cat#28989336 |
| HiLoad Superdex 75 16/600 size exclusion column | Cytivia | Cat#28989333 |
| Superose 6 increase 10/300 column | Cytivia | Cat#29091596 |
| CM5 SPR sensor chip | Cytivia | Cat#BR100012 |
| Streptavidin SPR sensor chip | Cytivia | Cat#BR100032 |
| FITC Streptavidin | Biolegend | Cat#405201 |
| PE Streptavidin | BD Biosciences | Cat#554061 |
| BV421, Streptavidin | BD Bioscience | Cat#563259 |
| R-Phycoerythrin, Biotin-XX conjugate | Thermo Fisher Scientific | Cat#P811 |
| Streptavidin, R-PE | Thermo Fisher Scientific | Cat#SA10041 |
| Live/Dead Fixable Near IR Viability kit | Theromofisher Scientific | Cat#L34994 |
| Streptavidin Dynabeads M-280 | Invitrogen | Cat#112.06D |
| LS magnetic columns | Miltenyi | Cat#130-042-401 |
| LD magnetic columns | Miltenyi | Cat#130-042-901 |
| Anti-PE magnetic micro-beads | Miltenyi | Cat#130-048-801 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Nicholas Gherardin (n.gherardin@unimelb.edu.au).
Materials availability
All requests for resouces or reagents should be directed to and will be fulfilled by the lead contact author. All reagents will be made available upon request after completion of a Materials Transfer Agreement.
Experimental model and subject details
Mice
Animal experiments were performed using 7–10 week old male C57BL/6 mice that were bred and housed at the Walter and Eliza Hall Institute of Medical Research. All animal experiments utilising live SARS-CoV-2 were performed in a Physical Containment Level 3 facility approved by the Office of Gene Technology Regulator at the Walter and Eliza Hall Institute for Medical Research (Cert-3621). All procedures were approved by the Walter and Eliza Hall Institute for Medical Research Animal Ethics Committee.
Cell lines
Expi293F cells (Thermo Fisher Scientific) were used for mammalian expression of recombinant proteins used throughout this study. Cells were maintained in suspension culture at 37°C, 5% CO2 using Expi293F expression media and transfected according to protocols in the ExpiFectamine 293 Transfection Kit (Thermo Fisher Scientific, A14524). The SARS-CoV-2 Spike hexapro construct was produced in ExpiCHO cells (Thermo Fisher Scientific). Cells were maintained in suspension culture at 37°C, 8% CO2 using ExpiCHO expression media and transfected according to the Max titer protocol in the ExpiFectamine CHO Transfection Kit (Thermo Fisher Scientific, A29133). HEK293T cells were obtained from ATCC (CRL-3216) and maintained in RPMI-1640 media (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (JRH Biosciences), Penicillin (100 U/mL), Streptomycin (100 μg/mL), Glutamax (2 mM), sodium pyruvate (1 mM), nonessential amino acids (0.1 mM), HEPES buffer (15 mM), pH7.2–7.5 (all from Invitrogen, Life Technologies) and 2-mercaptoethanol (50 μM, Sigma).
Yeast
Yeast-display nanobody library was provided by the laboratory of Andrew C. Kruse, Harvard University, and yeast culture carried out as per suggested protocol (McMahon et al., 2018). Yeast were maintained at 180 rpm at 25–30°C in tryptophan drop-put media supplemented with glucose during routine culture or galactose during induction of nanobody expression.
Method details
Production of recombinant coronavirus proteins and ACE2
DNA encoding the truncated RBD of SARS-CoV-1 (N321-P513) and SARS-CoV-2 (N334-P527) and the truncated ectodomain of human ACE2 (S19-D615) were codon optimised and synthesised (Thermo Fisher Scientific for SARS-CoV-1 and ACE2, Integrated DNA technologies for SARS-CoV-2). RBD DNA was cloned into pHLSec expression vectors containing a C-terminal AVI-tag and 6xHIS-tag to generate monomeric RBD, as well as a separate pHLSec vector containing the CH2 and CH3 domains of mouse IgG1 such that the C-terminal of the RBD was linked to the N-terminal of the truncated mouse IgG1 with a GS-linker (…CGP-GSGSG-CKPC…), to generate dimeric RBD-Fc protein. ACE2 was cloned into two separate pHLSec plasmids, one with a GSGSGSGTK linker before a C-terminal 6xHIS-tag, and one with the addition of a further GSGSG linker and AVI-tag immediately prior to the 6xHIS-tag (protein referred to as ACE2-AVI). All proteins with the exception of Spike Hexapro were transiently expressed in Expi293F cells using ExpiFectamine 293 Transfection Kits (Thermo Fisher Scientific) and supernatants harvested (cells pelleted and 0.22 μm filtered) on day 6. RBD-Fc proteins were purified using Protein A Sepharose (Thermo Fisher Scientific), eluting protein with 0.1 M citric acid, followed by SEC using a Superdex 200 16/600 column (Cytiva) in PBS. RBD monomers were spiked with 10 mM imidazole and then purified using Ni-NTA (Thermo Fisher Scientific), eluting protein via FPLC with a continuous gradient up to 250 mM imidazole, followed by SEC using a Superdex 75 16/600 column (Cytiva) into PBS. ACE2 protein was dialysed into Tris pH 8.0 and purified by weak anion exchange using DEAE sepharose (Cytiva) via FPLC with a continuous gradient up to 1 M NaCl. Protein was subsequently purified by SEC using a Superdex 200 16/600 column (Cytiva) in 10 mM Tris pH 8.0, 150 mM NaCl, followed by strong anion exchange using a monoQ column (Cytiva) with buffers as per DEAE. Spike hexapro (6P substitutions at 817, 892, 899, 942, 986, 987, C terminal foldon trimerization motif, avitag, 6XHis) was transiently expressed in ExpiCHO cells using the ExpiCHO transfection kit (Thermo Fisher Scientific) using the max titer protocol and supernatants harvested on day 12. Spike was then purified using Ni-NTA (Thermo Fisher Scientific), eluting protein via FPLC with a stepwise gradient up to 400 mM imidazole, followed by SEC using a Superose 6 increase 10/300 column (Cytiva) into HBS.
Biotinylation
AVI-tagged proteins were enzymatically biotinylated using BirA enzyme (purified in house). Proteins were buffer exchanged into 10 mM Tris pH 8.0 with 50 mM NaCl and concentrated to 20 μM. Biomix A (0.5 M bicine buffer, pH 8.3) and Biomix B (100 mM ATP, 100 mM MgOAc, 500 μM d-biotin) were each added at 1/8 the starting volume of the protein. 5 μg of BirA enzyme was then added per 1 mg of target protein. Protein was then incubated at 30°C for 4 hrs before proteins were purified from BirA enzyme and buffer components using SEC (Superdex 75 or Superdex 200 as above). Biotinylation was confirmed by streptavidin pull-down assay (Fairhead and Howarth, 2015). In brief, 2 μg of protein was denatured at 95°C for 5 min and then allowed to cool. A molar excess of streptavidin was then added to 1 μg of the denatured protein and incubated for 10 min at RT. Both protein and protein-streptavidin samples were then analysed by SDS-PAGE. The addition of streptavidin typically results in an increase in apparent molecular weight of the biotinylated protein. In all cases, 95–100% of biotinylated protein shifted, suggested almost complete to complete biotinylation. All biotinylated proteins were stored at −80°C.
Selection of nanobodies by yeast display
Yeast were thawed and recovered for 48 h before nanobody expression was induced with galactose for 48 h. Typically, induction resulted in 20–50% of yeast cells staining positive for HA as a surrogate readout of nanobody expression. For the first round of enrichment, 5 billion induced cells were pre-cleared by staining with 2.5 μg/mL of streptavidin-PE (BD) and anti-PE microbeads (Miltenyi) for 30 min at 4°C before passing over Miltenyi LD columns. Cells were then stained with 100 nM (2.5 μg/mL) SARS-CoV-2 RBD tetramer-PE for 1 h at 4°C, washed and incubated with anti-PE microbeads for 30 min at 4°C and then enriched using Miltenyi LS columns. Enriched cells were then recovered in glucose media for 24 h before proceeding to the next round of enrichment. In round 2 of enrichment, the same process was followed, but the concentration of RBD tetramer was dropped to 10 nM and the starting cell number was 100 million. For round 3 of enrichment, 100 million induced cells were stained with Live/Dead Near Infrared fixable viability dye (1:400; Thermo Fisher Scientific), anti-HA-AF647 (clone 16B12; 1:200; Biolegend) and SARS-CoV-2 RBD tetramer-PE (10 nM) for 30 min at 4°C. Cells were washed once and immediately sorted by FACS using a MoFlo Astrios Cell Sorter (Beckman Coulter) for viable, single cells that were HA+, RBDHI. Cells were then recovered and induced as above. For the final sort, 10 million yeast cells were co-stained for 30 min at 4°C with SARS-CoV-2 RBD tetramer-PE and SARS-CoV-1 RBD tetramer-BV421, both at 10 nM. Cells were washed once and then single-cell sorted using a MoFlo Astrios as above. Gating on HA+ cells with the brightest tetramer staining, 2 populations were sorted: SARS-CoV-1 RBD− SARS-CoV-2 RBD+ (CoV-2 only clones) and SARS-CoV-1+ SARS-CoV-2+ (CoV-1/2 cross reactive). Cells were cultured for 4 days before reinduction in galactose. After 48 h of induction, colonies were stained as per pre-sort with the addition of streptavidin-FITC (Biolegend) at 1 μg/mL, and subsequently screened by flow cytometry using an LSR Fortessa (BD). The 10 brightest clones for each of the CoV-2 only and CoV-1/2 cross-reactive clones were selected for sequencing and cloning.
Sequencing, cloning and expression of nanobodies
Colonies of interest were streaked onto agar plates and incubated at 30°C for 72 h. Individual colonies from each plate were then streaked as patches on new agar plates and incubated for a further 48 h. Yeast minipreps were then performed from patches using Zymoprep Yeast Plasmid Miniprep II kits (Zymo). Using miniprep eluate, PCR was performed using KAPA HiFi HotStart ReadyMix PCR kit (Roche) as per manufacturer’s instructions, using pYSD649HM_FWD primer (gttttgttcgctgcttcttctgc) and pYDS649HM_REV primer (ggtagatttactaggcgatgagg). Initial denaturation was performed at 95°C for 3 min followed by 35 cycles of denaturation at 98°C for 20 s, annealing at 65°C for 30 s and extension at 72°C for 1 min, followed by a final extension at 72°C for 1 min. PCR products were then separated on 1.5% agarose gel and purified using DNA gel recovery kit (Zymo). Product was then subject to Sanger sequencing using pYDSM649HM_FWD primer. From the 20 sequenced clones, 11x unique sequences were recovered (6x CoV-2 only and 5x CoV-1/2 cross-reactive), five of each were cloned. PCR product was digested with NheI and BamHI restriction enzymes (NEB) and cloned into 2x separate pHLSec vectors, one with C-terminal AVI-tag and 6xHIS-tag, and the other with the CH2 and CH3 domains of human IgG1 such that the C-terminal of the nanobody was linked to the N-terminal of the truncated human IgG1 with a GS-linker (… VSS-GSGSG-THT …) to generate dimeric nanobody-Fc protein. For biparatopic nanobodies, DNA constructs were synthesised (IDT) and cloned into pHLSec expression vector. B1 was fused to G8 via a (G4S)x linker of variable length (10, 19 or 39 amino acids) and this was either fused to a C-terminal 6xHIS-tag or human the CH2 and CH3 domains of human IgG1 as above. Monomeric nanobody and nanobody-Fc proteins were expressed in Expi293F cells as above. Monomeric nanobody and biparatopic nanobody were purified and biotinylated as per monomeric RBD proteins above, and nanobody-Fc and dimeric biparatopic nanobodies were purified as per RBD-Fc as above.
Transient transfections
DNA encoding full-length human ACE2 and the SARS-CoV-2 S protein was synthesised (Thermo Fisher Scientific) and cloned into pMIGII or pVRC8400 vector respectively. HEK293T cells were then transiently transfected with these plasmids using FUGENE-6 transfection reagent (Promega). After 72 h of transfection, cells were mechanically harvested and used in flow cytometry experiments. To generate RBD or nanobody tetramers, streptavidin-PE (BD) was added to biotinylated RBD or nanobody monomers in 10 additions spaced 10 min apart, to a final molar ratio of 4:1 monomer:streptavidin. Transfected HEK293T cells were stained with LIVE/DEAD NIR viability dye (Thermo Fisher Scientific) and RBD or nanobody tetramers at a concentration of 1 μg/mL. Cells were stained for 30 min at RT, washed twice, then fixed in 2% PFA for 10 min. Cells were then analysed by flow cytometry using a BD LSR Fortessa equipped using a yellow-green laser. Data was analysed using Flowjo software (Treestar). 293T cells were gated based on FSC-A and SSC-A and dead cells and doublets were removed from analysis based on FSC-A versus FSC-H and L/D NIR staining.
Surface plasmon resonance
All SPR experiments were performed at 25°C using Biacore T200 biosensor (Cytiva) with 1x HBS-EP + buffer (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.05% [v/v] Tween 20) as instrument running buffer. Dimeric SARS-CoV-1- and SARS-CoV-2 RBD-Fc constructs were captured onto an SPR chip (CM5, Cytiva) surface containing amine-coupled AffiniPure Sheep α-mouse IgG, Fcγ Fragment Specific (Jackson ImmunoResearch, Code: 515-005-071). Biotinylated monomeric SARS-CoV-1- and SARS-CoV-2-RBD-AVI constructs were immobilized onto an SA sensor chip (Cytiva) containing amine coupled Streptavidin. Kinetic binding experiments were performed by injecting analyte samples (nanobodies and ACE2) over the above described RBD-Fc surfaces for either 60 s (nanobodies) or 90 s (ACE2) at 30 μL/min and then allowed to fully dissociate from the chip surface in the running buffer, typically 60 s for nanobodies and 900 s for ACE2. Injected analyte concentrations were diluted three-fold, ranging from 6561 nM to 9 nM for nanobodies and from 729 nM to 1 nM for ACE2. SPR experimental data were processed and analysed using Scrubber Software (www.biologic.com.au). Flow cell 1 and “zero-buffer-blank” cycle injections were utilized to reference binding signals. Kinetic rate (ka, kd) and affinity (KD) parameters were derived by fitting each set of experimental data to a Langmuir 1:1 binding model. Nanobody epitope binning (binding competition) experiments were performed by simultaneously injecting an equimolar mixture of a pair of nanobodies (both at 6561 nM) over immobilized SARS-CoV-2-RBD-Fc and comparing the resulting SPR binding signal with signals generated when either of the two nanobodies was injected alone (also at 6561 nM). Nanobodies were deemed to be non-competitive when the combined SPR binding signal increased significantly above the signal generated by a single nanobody. SPR competition experiment between ACE2 and nanobodies was performed using a tandem injection approach (Co-inject function in Biacore T200 control software). Thus, a 90 s injection of ACE2 protein (at 729 nM) was immediately followed by a 60 s nanobody injection (at 6561 nM). The nature of the competitive binding between ACE2 and nanobodies was evaluated by comparing SPR binding responses produced during and after the nanobody was injected with the binding signal generated when ACE2 alone was injected.
RBD alanine scans and variant array
The alanine-scan RBD mutant multiplex assays were performed using an established technique (Lopez et al., 2021). In brief, RBD proteins for the alanine scan were produced by Genscript, whereas RBD and ACE2 proteins from the variant arrays were produced in house as above. RBD proteins were coupled to magnetic beads (Bio-Rad). For ACE2 inhibition assay, RBD-coupled beads were pooled and incubated with biotinylated ACE2 in the presence of titrating quantities of nanobody-Fc proteins. ACE2 binding was detected with Streptavidin-PE (Thermo Fisher Scientific), followed by Biotin-PE (Thermo Fisher Scientific). After washing, beads were analysed using FLEXMAP 3D (Luminex corporation). IgG binding was performed similarly; Pooled beads were incubated with titrated quantities of nanobody-Fc proteins and detected using anti-human IgG-PE (Southern Biotech). For analysis, a single dose (non-saturating for WT RBD) of nanobody was chosen to compare for all variants. For the ala-scan, only proteins of high purity (based on SDS-PAGE gel analysis) were used (Table S1) and the full list of variants used in the variant array are depicted in Figure 5C.
Viral microneutralisation assays
Assays were performed using an established technique (Tan et al., 2021b). In brief, SARS-CoV-2 isolate CoV/Australia/VIC01/2020 (Caly et al., 2020) was grown in Vero cells and stored at −80°C. 100 TCID50 of SARS-CoV-2 in MEM/0.5% BSA was added to proteins that were serially diluted 1:4 to 1:2048 and incubated at room temperature for 1 h. Protein/virus mixtures were added in quadruplicate to Vero cells incubated in serum-free media containing 1 μg/mL of TPCK trypsin at 37°C and 5% CO2, and residual viral infectivity was assessed by reading viral cytopathic effect on day 5 (Houser et al., 2016; Subbarao et al., 2004). Neutralising titre was calculated using the Reed-Muench method, and IC50 was calculated by dividing the starting concentration of protein by the neutralising titre.
Surrogate virus neutralisation test (sVNT)
The presence of RBD-directed nanobodies in Figure 2A was assessed using the sVNT (GenScript) according to the manufacturer’s instructions (Tan et al., 2020). Briefly, nanobody samples at a stock concentration of 500 μg/mL were diluted 1:9 and pre-incubated with a horseradish peroxidase-conjugated RBD (HRP-RBD). This mixture was then added to wells of a 96-well plate coated with ACE2. The extent to which the nanobodies inhibited RBD binding to ACE2 was measured by optical density in relation to positive and negative internal controls, with inhibition above 20% being considered positive. For the data presented in Figure 5E, the multiplex sVNT assays were performed using an established technique (Tan et al., 2021a). In brief, Luminex beads coated with RBD proteins were incubated with titrated nanobodies for 15 min prior to addition of ACE conjugated to R-Phycoerytherin and incubated for a further 15 min. Reduction in ACE2 binding was measured as MFI using a Luminex MagPix instrument. Results are expressed as surrogate neutralisation titre 50%.
Cryo-EM sample preparation and data acquisition
Cryo-EM was performed at the Ian Holmes Imaging Centre, Bio21 Institute of Molecular Science and Biotechnology, The University of Melbourne. Prior to EM specimen grid preparation, the pre-fusion stabilised S trimer (Hexapro) was incubated with the BP10 nanobody construct at a 6:1 molar ratio for 30 min at room temperature. The complex was purified over SEC and concentrated to 3 mg/mL. Grids (Quantifoil Cu R1.2/1.3, 300 mesh) were glow discharged in air at 15 mA for 60 s using an EasiGlow glow discharge apparatus (Pelco, USA). The 4 μL samples were applied to the grids at 4°C and 100% humidity, and then plunge frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). Data was collected on a Titan Krios G4 electron microscope (Thermo Fisher Scientific) operated at 300 kV and equipped with a Falcon 4 detector (Thermo Fisher Scientific) and Bioquantum energy filter (Gatan, Pleasanton, USA). The dataset was collected at a pixel size of 0.66 Å with 13,806 movies containing 40 frames each with a total exposure of 50.0 e/Å2. The acquisition was performed at 155Kx indicated magnification using an energy filter slit width of 10 eV to remove contributions from inelastically scattered electrons. All data were collected using EPU software Version 2.9 (Thermo Fisher Scientific) using a 9-hole beam-image shift acquisition scheme with one exposure per hole.
Cryo-EM data processing
A subset of data, Batch 1(A) (2040 micrographs) were imported into CryoSPARC v3.1.0 (Punjani et al., 2017) and subjected to motion correction. The corrected micrographs were then imported into Relion 3.1.0 (Zivanov et al., 2018) and underwent CTF estimation using GCTF (Zhang, 2016). Template-based particle picking was performed using Gautomatch (http://www.mrc-lmb.cam.ac.uk/kzhang/) using nine projections from EMDB entry 23566 that was low-pass filtered to 20 Å. Particle coordinates were imported into Relion 3.1 (Zivanov et al., 2018). Particle extraction using 6 x binning (96 pixel box size, pixel size 3.96 Å/pix) yielded 279,663 particles. Particles were then imported into CryoSPARC 3.1.0 (Punjani et al., 2017) and subjected to 2 rounds of 2D classification followed by 1 round of heterogeneous refinement (using de novo ab-initio models generated in CryoSPARC) yielding a subset of 28,723 particles. This subset was reextracted using a larger box size (192 pixel box size, pixel size 3.96 Å/pix) and subjected to homogeneous refinement in CryoSPARC yielding an 8.2 Å map. This map was used to generate templates for particle picking of the entire dataset (see below). The full dataset was then imported into CryoSPARC 3.1.0 (Punjani et al., 2017) and processed in two batches, Batch 1 (5,819 movies) and Batch 2 (7,987 movies), each being subjected to the correction of beam-induced motion using Patch Motion, followed by CTF estimation using Patch CTF. Micrographs with CTF fit resolution below 4 Å were selected for particle picking (4,657 and 7,679 movies respectively). Template-based particle picking was performed using projections from the homogeneous refinement output model from Batch 1A, yielding 127,636 and 206,511 particles respectively. Batch 1 then underwent two rounds of heterogeneous refinement using the Batch 1A homogeneous refinement output and three “junk” classes, and a further two with two “junk” classes, yielding a 3D volume at 9.5 Å consisting of 23,213 particles. Particles were then re-extracted using a 2 x binned box size (504 pixels) and underwent a further round of heterogeneous refinement with two identical input models followed by a round of 2D classification. This yielded a dataset of 21,235 particles. Batch 2 underwent a round of 2D classification with 141,988 particles selected and then four rounds of heterogeneous refinement with between one and three “junk” classes, yielding a 14.5 Å 3D volume consisting of 55,726 particles. Particles were then re-extracted using a 2 x binned box size (504 pixels) and were subjected to a further round of 2D classification, 50,480 particles were selected. Batch 1 and Batch 2 were then joined and underwent multiple rounds of heterogeneous and homogeneous refinement to yield a dataset of 37,266 particles. A final 2D classification was performed and a subset of 14,126 particles were selected. This subset was subjected to Non-uniform refinement (Punjani et al., 2020) with D3 symmetry yielding a map at a final resolution of 4.19 Å (“Entire S-Nb map”, EMD-28189). Local refinement of one copy of the RBD and two bound nanobodies was undertaken using a mask generated in Chimera v1.1.3 (Pettersen et al., 2004). Three rounds of local refinement using the prior job output as a mask gave a 3.89 Å map. Model generation: To make the RBD-Nb model (EMD-28190), the map from the last local refinement job was used to rigid body dock the RBD from PDB 7LX5 along with nanobodies from PDBs 5IMK and 7KKK, selected based on the highest scoring BlastP sequence alignments of the G8 and B1 sequences with protein databank sequences. To make the model of the entire S bound to the nanobodies, the RBD-Nb model (described above) and the SARS-CoV-2 S (PDB ID: 7VNE, RBD removed from the model) were rigid body docked into the “Entire S-Nb map”.
Mouse prophylaxis experiments
Animal experiments were performed using an established viral challenge model (Subbarao et al., 2004). In brief, nanobodies were administered at 5 mg/kg in 100 μL PBS by intraperitoneal injection 24 h prior to infection. SARS-CoV-2 infection (clinical isolate hCoV-10/Australia/VIC2089/2020) was performed using an inhalation exposure system (Glas-Col, LLC) for 30 min loaded with 1.5 × 107 SARS CoV-2 TCID50. Three days post-infection, animals were humanely killed and lungs removed and homogenised in a Bullet Blender (Next Advance Inc) in 0.5 mL DME media (Thermo Fisher Scientific) containing steel homogenisation beads (Next Advance Inc). Samples were then diluted with equal volume (0.5 mL) DMEM and clarified by centrifugation before virus quantification by TCID50 and RTqPCR assays. SARS-CoV-2 lung TCID50 was determined by serial dilution of lung tissue homogenate onto confluent Vero cells (clone CCL81). Plates were incubated at 37°C for four days before measuring cytopathic effect under light microscope. TCID50 was calculated using the Spearman and Kärber method. To quantify SARS-CoV2 N1 gene copy number, SARS-CoV-2 RNA was extracted from clarified lung homogenate using a NucleoSpin 96-well plate virus extraction kit (Macherey-Nagel GmbH&Co). Primers and probes specific for the conserved SARS-CoV2 N1 gene were purchased from Integrated DNA Technologies (Iowa, USA). Primer/probe sequences: 2019-nCoV_N1 forward primer – 5′-GAC CCC AAA ATC AGC GAA AT-3′, 2019-nCoV_N1 reverse primer – 5′-TCT GGT TAC TGC CAG TTG AAT CTG-3′, 2019-nCoV_N1 probe – 5′-FAM-ACC CCG CAT TAC GTT TGG TGG ACC-BHQ1-3’. Viral RNA was reverse transcribed and amplified using iTaq Universal Probe Kit (Bio-Rad Inc) and run on a LightCycler 96 machine (Roche). Cycle parameters: RT reaction – 600 s/50°C, pol activation – 120 s/95°C, denaturation – 15 s/95°C, annealing – 60 s/60°C. To determine absolute SARS-CoV-2 N1 gene copy number, standard curves were generated using a commercially validated standard control sample (SKU: COV019, Exact Diagnostics LLC).
Quantification and statistical analysis
No formal statistical tests were performed in this study. In particular, the mouse experiments were not statistically analysed. Given the data is nonparametric with all results from some groups at the lower limit of detection, a parametric test such as ANOVA is not appropriate. We therefore applied a Mann-Whitney test with a Bonferroni correction to account for 4 comparisons to the control group. The lower power of this type of test meant that our results were just outside of the significance threshold (eg p = 0.061) when comparing PBS to B1-Fc and BP10-Fc. Noting that values for the B1-Fc and BP-10-Fc are all below the limit of the detection for the assay, we considered the most appropriate way to present the data was without statistical analysis, instead describing the data in the text accordingly.
Acknowledgments
We thank staff from the flow cytometry facilities at the Department of Microbiology and Immunology at the Peter Doherty Institute. We thank Prof. Andrew C. Kruse (Harvard University, Boston, USA) for provision of the yeast nanobody display library. The authors acknowledge use of the Bio21 Advanced Microscopy Facility, and WEHI Information Technology Services and the WEHI Research Computing Platform for providing facilities and support that contributed to this work. This work was supported by the Jack Ma Foundation, the Ramsay Foundation, The Victorian State Government, Australia, the Medical Research Future Fund (MRFF GNT2002073), the Australian Research Council (ARC; CE140100011) and the National Health and Medical Research Council, Australia (NHMRC; 1113293 and 1145373) and computational resources provided by the Australian Government through MASSIVE HPC facility (www.massive.org.au) under the National Computational Merit Allocation Scheme. N.A.G was supported by an ARC DECRA Fellowship (DE210100705) and D.I.G was supported by an NHMRC Senior Principal Research Fellowship (1117766) and subsequently by an NHMRC investigator grant (2008913). W-H.T. is a Howard Hughes Medical Institute–Wellcome Trust International Research Scholar (208693/Z/17/Z) and is support by an NHMRC Fellowship (1154937). M.P. is supported by an NHMRC Investigator Grant (GNT1175011). A.G. is a CSL Centenary Fellow. K.S. is supported by an NHMRC Investigator Grant. The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health. The work at Duke-NUS is supported by grants from Singapore National Medical Research Council (STPRG-FY19-001, COVID19RF-003, COVID19RF-060 and OFLCG19May-0034).
Author contributions
Conceptualization, N.A.G. and D.I.G.; methodology, N.A.G., P.P., O.D., F.M., E.L., J.P.C., K.C.D., C.W.T, L-F.W, A.L., A.G., M.P., A.W.C., and K.S.; formal analysis, N.A.G.; P.P., S.J.R., O.D., F.M., E.L., J.P.C., E.H., C.W.T., and S.L.G.; investigation, N.A.G.; P.P., S.J.R, A.G., O.D., F.M., E.L., J.P.C., E.H., C.W.T., R.S., and S.L.G.; resources, N.A.G., DIG, P.P., W-H.T., A.P.U., K.S., A.W.C., M.P., A.G., A.L., L-F.W., A.K.W., S.J.K., D.F.J.P., K.C.D., and O.D.; data curation, N.A.G., P.P., A.G., A.L., S.L.G., C.W.T., E.H. J.P.C., E.L., F.M., O.D., and S.J.R.; Writing – Original Draft, N.A.G. P.P., and D.I.G.; writing – review & editing, all authors; visualization, N.A.G. and P.P.; supervision, N.A.G, D.I.G., W-H.T, K.S., A.W.C., M.P., A.G., and D.J.P.; project administration, N.A.G. and D.I.G.; funding acquisition, N.A.G., D.I.G., W.H.T., A.W.C., A.K.W., and S.J.K.
Declaration of interests
Lin-Fa Wang and Chee Wah Tan are co-inventors of the surrogate virus neutralization test commercialized under the tradename cPass by GenScript Biotech.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105259.
Contributor Information
Dale I. Godfrey, Email: godfrey@unimelb.edu.au.
Nicholas A. Gherardin, Email: n.gherardin@unimelb.edu.au.
Supplemental information
Data and code availability
-
•
CryoEM data have been deposited in the Electron Microscopy Data Bank (EMDB) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
References
- Abdool Karim S.S., de Oliveira T. New SARS-CoV-2 variants - clinical, public health, and vaccine implications. N. Engl. J. Med. 2021;384:1866–1868. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham J. Passive antibody therapy in COVID-19. Nat. Rev. Immunol. 2020;20:401–403. doi: 10.1038/s41577-020-0365-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arezumand R., Alibakhshi A., Ranjbari J., Ramazani A., Muyldermans S. Nanobodies as novel agents for targeting angiogenesis in solid cancers. Front. Immunol. 2017;8:1746. doi: 10.3389/fimmu.2017.01746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly L., Druce J., Roberts J., Bond K., Tran T., Kostecki R., Yoga Y., Naughton W., Taiaroa G., Seemann T., et al. Isolation and rapid sharing of the 2019 novel coronavirus (SARS-CoV-2) from the first patient diagnosed with COVID-19 in Australia. Med. J. Aust. 2020;212:459–462. doi: 10.5694/mja2.50569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Liu X., Wang C., Zhang X., Li X., Hou J., Ren L., Jin Q., Wang J., Yang W. Humanized single domain antibodies neutralize SARS-CoV-2 by targeting the spike receptor binding domain. Nat. Commun. 2020;11:4528. doi: 10.1038/s41467-020-18387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Zhang X., Pan S., Yu Y., Shi Y., Lin T., Duan H., Liu X., Chen W., Yang X., et al. An ultrapotent RBD-targeted biparatopic nanobody neutralizes broad SARS-CoV-2 variants. Signal Transduct. Target. Ther. 2022;7:44. doi: 10.1038/s41392-022-00912-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custódio T.F., Das H., Sheward D.J., Hanke L., Pazicky S., Pieprzyk J., Sorgenfrei M., Schroer M.A., Gruzinov A.Y., Jeffries C.M., et al. Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2. Nat. Commun. 2020;11:5588. doi: 10.1038/s41467-020-19204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vlieger D., Ballegeer M., Rossey I., Schepens B., Saelens X. Single-domain antibodies and their formatting to combat viral infections. Antibodies. 2018;8:E1. doi: 10.3390/antib8010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detalle L., Stohr T., Palomo C., Piedra P.A., Gilbert B.E., Mas V., Millar A., Power U.F., Stortelers C., Allosery K., et al. Generation and characterization of ALX-0171, a potent novel therapeutic nanobody for the treatment of respiratory syncytial virus infection. Antimicrob. Agents Chemother. 2016;60:6–13. doi: 10.1128/AAC.01802-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo D.J., Tan G.S., Palese P., Ravetch J.V. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat. Med. 2014;20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Huang B., Wang B., Titong A., Gallolu Kankanamalage S., Jia Z., Wright M., Parthasarathy P., Liu Y. Development of humanized tri-specific nanobodies with potent neutralization for SARS-CoV-2. Sci. Rep. 2020;10:17806. doi: 10.1038/s41598-020-74761-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esparza T.J., Martin N.P., Anderson G.P., Goldman E.R., Brody D.L. High affinity nanobodies block SARS-CoV-2 spike receptor binding domain interaction with human angiotensin converting enzyme. Sci. Rep. 2020;10:22370. doi: 10.1038/s41598-020-79036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhead M., Howarth M. Site-specific biotinylation of purified proteins using BirA. Methods Mol. Biol. 2015;1266:171–184. doi: 10.1007/978-1-4939-2272-7_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni G., Mantovani A., COVID-19 Commission of Accademia Nazionale dei Lincei, Rome Covid-19 Commission of Accademia Nazionale dei Lincei, R. (2021). COVID-19 vaccines: where we stand and challenges ahead. Cell Death Differ. 2021;28:626–639. doi: 10.1038/s41418-020-00720-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai J., Ma L., Li G., Zhu M., Qiao P., Li X., Zhang H., Zhang Y., Chen Y., Ji W., et al. A potent neutralizing nanobody against SARS-CoV-2 with inhaled delivery potential. MedComm (Beijing) 2021;2:101–113. doi: 10.1002/mco2.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber K. Hunt for improved monoclonals against coronavirus gathers pace. Nat. Biotechnol. 2021;39:9–12. doi: 10.1038/s41587-020-00791-6. [DOI] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Gentles L.E., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Antibodies elicited by mRNA-1273 vaccination bind more broadly to the receptor binding domain than do those from SARS-CoV-2 infection. Sci. Transl. Med. 2021;13:eabi9915. doi: 10.1126/scitranslmed.abi9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.Q., Wang Y., Teng Y., Zhao Z., Cui Y., et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E.B., Bendahman N., Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- Hanke L., Vidakovics Perez L., Sheward D.J., Das H., Schulte T., Moliner-Morro A., Corcoran M., Achour A., Karlsson Hedestam G.B., Hällberg B.M., et al. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 2020;11:4420. doi: 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser K.V., Gretebeck L., Ying T., Wang Y., Vogel L., Lamirande E.W., Bock K.W., Moore I.N., Dimitrov D.S., Subbarao K. Prophylaxis with a Middle East respiratory syndrome coronavirus (MERS-CoV)-Specific human monoclonal antibody protects rabbits from MERS-CoV infection. J. Infect. Dis. 2016;213:1557–1561. doi: 10.1093/infdis/jiw080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N., et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27:846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- Ingram J.R., Schmidt F.I., Ploegh H.L. Exploiting nanobodies' singular traits. Annu. Rev. Immunol. 2018;36:695–715. doi: 10.1146/annurev-immunol-042617-053327. [DOI] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Juno J.A., Tan H.X., Lee W.S., Reynaldi A., Kelly H.G., Wragg K., Esterbauer R., Kent H.E., Batten C.J., Mordant F.L., et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020;26:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- Klarenbeek A., El Mazouari K., Desmyter A., Blanchetot C., Hultberg A., de Jonge N., Roovers R.C., Cambillau C., Spinelli S., Del-Favero J., et al. Camelid Ig V genes reveal significant human homology not seen in therapeutic target genes, providing for a powerful therapeutic antibody platform. mAbs. 2015;7:693–706. doi: 10.1080/19420862.2015.1046648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig P.A., Das H., Liu H., Kümmerer B.M., Gohr F.N., Jenster L.M., Schiffelers L.D.J., Tesfamariam Y.M., Uchima M., Wuerth J.D., et al. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science. 2021;371:eabe6230. doi: 10.1126/science.abe6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontermann R.E. Strategies for extended serum half-life of protein therapeutics. Curr. Opin. Biotechnol. 2011;22:868–876. doi: 10.1016/j.copbio.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Konwarh R. Nanobodies: prospects of expanding the gamut of neutralizing antibodies against the novel coronavirus, SARS-CoV-2. Front. Immunol. 2020;11:1531. doi: 10.3389/fimmu.2020.01531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Laursen N.S., Friesen R.H.E., Zhu X., Jongeneelen M., Blokland S., Vermond J., van Eijgen A., Tang C., van Diepen H., Obmolova G., et al. Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science. 2018;362:598–602. doi: 10.1126/science.aaq0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford H. The race to make covid antibody therapies more potent. Nature. 2020;587:18. doi: 10.1038/d41586-020-02965-3. [DOI] [PubMed] [Google Scholar]
- Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C.B., To K.F., et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li T., Cai H., Yao H., Zhou B., Zhang N., van Vlissingen M.F., Kuiken T., Han W., GeurtsvanKessel C.H., Gong Y., et al. A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection. Nat. Commun. 2021;12:4635. doi: 10.1038/s41467-021-24905-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies directed to multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Lopez E., Haycroft E.R., Adair A., Mordant F.L., O'Neill M.T., Pymm P., Redmond S.J., Lee W.S., Gherardin N.A., Wheatley A.K., et al. Simultaneous evaluation of antibodies that inhibit SARS-CoV-2 variants via multiplex assay. JCI Insight. 2021;6:150012. doi: 10.1172/jci.insight.150012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Zhang Z., Li H., Zhong K., Zhao Q., Wang Z., Wu Z., Yang D., Sun S., Yang N., et al. Development of multivalent nanobodies blocking SARS-CoV-2 infection by targeting RBD of spike protein. J. Nanobiotechnol. 2021;19:33. doi: 10.1186/s12951-021-00768-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Zeng W., Meng X., Huang X., Yang Y., Zhao D., Zhou P., Wang X., Zhao C., Sun Y., et al. Potent neutralization of SARS-CoV-2 by hetero-bivalent alpaca nanobodies targeting the spike receptor-binding domain. J. Virol. 2021;95 doi: 10.1128/JVI.02438-20. JVI.02438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon C., Baier A.S., Pascolutti R., Wegrecki M., Zheng S., Ong J.X., Erlandson S.C., Hilger D., Rasmussen S.G.F., Ring A.M., et al. Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat. Struct. Mol. Biol. 2018;25:289–296. doi: 10.1038/s41594-018-0028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Piccoli L., Park Y.-J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A., Cornaby C., Bartelt L., Weiss S., Park Y., et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5:eabc8413. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Punjani A., Zhang H., Fleet D.J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods. 2020;17:1214–1221. doi: 10.1038/s41592-020-00990-8. [DOI] [PubMed] [Google Scholar]
- Pymm P., Adair A., Chan L.J., Cooney J.P., Mordant F.L., Allison C.C., Lopez E., Haycroft E.R., O'Neill M.T., Tan L.L., et al. Nanobody cocktails potently neutralize SARS-CoV-2 D614G N501Y variant and protect mice. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2101918118. e2101918118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybould M.I.J., Kovaltsuk A., Marks C., Deane C.M. CoV-AbDab: the coronavirus antibody database. Bioinformatics. 2021;37:734–735. doi: 10.1093/bioinformatics/btaa739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasisekharan R. Preparing for the future - nanobodies for covid-19? N. Engl. J. Med. 2021;384:1568–1571. doi: 10.1056/NEJMcibr2101205. [DOI] [PubMed] [Google Scholar]
- Schoof M., Faust B., Saunders R.A., Sangwan S., Rezelj V., Hoppe N., Boone M., Billesbølle C.B., Puchades C., Azumaya C.M., et al. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science. 2020;370:1473–1479. doi: 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully M., Cataland S.R., Peyvandi F., Coppo P., Knöbl P., Kremer Hovinga J.A., Metjian A., de la Rubia J., Pavenski K., Callewaert F., et al. Caplacizumab treatment for acquired thrombotic thrombocytopenic purpura. N. Engl. J. Med. 2019;380:335–346. doi: 10.1056/NEJMoa1806311. [DOI] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões E.A.F., Bont L., Manzoni P., Fauroux B., Paes B., Figueras-Aloy J., Checchia P.A., Carbonell-Estrany X. Past, present and future approaches to the prevention and treatment of respiratory syncytial virus infection in children. Infect. Dis. Ther. 2018;7:87–120. doi: 10.1007/s40121-018-0188-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:e1007236. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K., Packard M., Shieh W.J., Zaki S., Murphy B. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 2004;78:3572–3577. doi: 10.1128/jvi.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Sang Z., Kim Y.J., Xiang Y., Cohen T., Belford A.K., Huet A., Conway J.F., Sun J., Taylor D.J., et al. Potent neutralizing nanobodies resist convergent circulating variants of SARS-CoV-2 by targeting diverse and conserved epitopes. Nat. Commun. 2021;12:4676. doi: 10.1038/s41467-021-24963-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashita E., Kinoshita N., Yamayoshi S., Sakai-Tagawa Y., Fujisaki S., Ito M., Iwatsuki-Horimoto K., Chiba S., Halfmann P., Nagai H., et al. Efficacy of antibodies and antiviral drugs against covid-19 omicron variant. N. Engl. J. Med. 2022;386:995–998. doi: 10.1056/NEJMc2119407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I.C., Tiu C., Hu Z., Chen V.C.W., Young B.E., Sia W.R., et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020;38:1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- Tan C.W., Chia W.N., Young B.E., Zhu F., Lim B.L., Sia W.R., Thein T.L., Chen M.I.C., Leo Y.S., Lye D.C., Wang L.F. Pan-sarbecovirus neutralizing antibodies in BNT162b2-immunized SARS-CoV-1 survivors. N. Engl. J. Med. 2021;385:1401–1406. doi: 10.1056/NEJMoa2108453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H.X., Juno J.A., Lee W.S., Barber-Axthelm I., Kelly H.G., Wragg K.M., Esterbauer R., Amarasena T., Mordant F.L., Subbarao K., et al. Immunogenicity of prime-boost protein subunit vaccine strategies against SARS-CoV-2 in mice and macaques. Nat. Commun. 2021;12:1403. doi: 10.1038/s41467-021-21665-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P.C., Adams A.C., Hufford M.M., de la Torre I., Winthrop K., Gottlieb R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021;21:382–393. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temmam S., Vongphayloth K., Baquero E., Munier S., Bonomi M., Regnault B., Douangboubpha B., Karami Y., Chrétien D., Sanamxay D., et al. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature. 2022;604:330–336. doi: 10.1038/s41586-022-04532-4. [DOI] [PubMed] [Google Scholar]
- Valenzuela Nieto G., Jara R., Watterson D., Modhiran N., Amarilla A.A., Himelreichs J., Khromykh A.A., Salinas-Rebolledo C., Pinto T., Cheuquemilla Y., et al. Potent neutralization of clinical isolates of SARS-CoV-2 D614 and G614 variants by a monomeric, sub-nanomolar affinity nanobody. Sci. Rep. 2021;11:3318. doi: 10.1038/s41598-021-82833-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincke C., Loris R., Saerens D., Martinez-Rodriguez S., Muyldermans S., Conrath K. General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J. Biol. Chem. 2009;284:3273–3284. doi: 10.1074/jbc.M806889200. [DOI] [PubMed] [Google Scholar]
- Voss W.N., Hou Y.J., Johnson N.V., Delidakis G., Kim J.E., Javanmardi K., Horton A.P., Bartzoka F., Paresi C.J., Tanno Y., et al. Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes in COVID-19 convalescent plasma. Science. 2021;372:1108–1112. doi: 10.1126/science.abg5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T.R., Ostertag E., Kaiser P.D., Gramlich M., Ruetalo N., Junker D., Haering J., Traenkle B., Becker M., Dulovic A., et al. NeutrobodyPlex-monitoring SARS-CoV-2 neutralizing immune responses using nanobodies. EMBO Rep. 2021;22:e52325. doi: 10.15252/embr.202052325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y., Ouyang Z., Schoonooghe S., Luo S., De Baetselier P., Lu W., Muyldermans S., Raes G., Zheng F. Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity. Immunobiology. 2017;222:807–813. doi: 10.1016/j.imbio.2016.11.008. [DOI] [PubMed] [Google Scholar]
- WHO Director-General's opening remarks at the media briefing on COVID-19 - 11 March 2020 . 2020. World Health Organisation. [Google Scholar]
- Wilhelm A., Widera M., Grikscheit K., Toptan T., Schenk B., Pallas C., Metzler M., Kohmer N., Hoehl S., Marschalek R., et al. Limited neutralisation of the SARS-CoV-2 Omicron subvariants BA.1 and BA.2 by convalescent and vaccine serum and monoclonal antibodies. EBioMedicine. 2022;82:104158. doi: 10.1016/j.ebiom.2022.104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., VIB-CMB COVID-19 Response Team. Hoffmann M., et al. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1004–1015.e15. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Nambulli S., Xiao Z., Liu H., Sang Z., Duprex W.P., Schneidman-Duhovny D., Zhang C., Shi Y. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science. 2020;370:1479–1484. doi: 10.1126/science.abe4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Wang Y., Jin Y., Zhu Y., Wu Y., Li C., Kong Y., Song W., Tian X., Zhan W., et al. A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants. Signal Transduct. Target. Ther. 2021;6:378. doi: 10.1038/s41392-021-00810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Cai H., Li T., Zhou B., Qin W., Lavillette D., Li D. A high-affinity RBD-targeting nanobody improves fusion partner's potency against SARS-CoV-2. PLoS Pathog. 2021;17:e1009328. doi: 10.1371/journal.ppat.1009328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Tsybovsky Y., Gorman J., Rapp M., Cerutti G., Chuang G.Y., Katsamba P.S., Sampson J.M., Schön A., Bimela J., et al. Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe. 2020;28:867–879.e5. doi: 10.1016/j.chom.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7 doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
CryoEM data have been deposited in the Electron Microscopy Data Bank (EMDB) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.