Abstract
Aim
To describe associations of gingival bacterial composition and diversity with self‐reported gingival bleeding and oral hygiene habits in a Norwegian regional‐based population.
Materials and Methods
We examined the microbiome composition of the gingival fluid (16S amplicon sequencing) in 484 adult participants (47% females; median age 28 years) in the Respiratory Health in Northern Europe, Spain and Australia (RHINESSA) study in Bergen, Norway. We explored bacterial diversity and abundance differences by the community periodontal index score, self‐reported frequency of gingival bleeding, and oral hygiene habits.
Results
Gingival bacterial diversity increased with increasing frequency of self‐reported gingival bleeding, with higher Shannon diversity index for “always” β = 0.51 and “often” β = 0.75 (p < .001) compared to “never” gingival bleeding. Frequent gingival bleeding was associated with higher abundance of several bacteria such as Porphyromonas endodontalis, Treponema denticola, and Fretibacterium spp., but lower abundance of bacteria within the gram‐positive phyla Firmicutes and Actinobacteria. Flossing and rinsing with mouthwash twice daily were associated with higher total abundance of bacteria in the Proteobacteria phylum but with lower bacterial diversity compared to those who never flossed or never used mouthwash.
Conclusions
A high frequency of self‐reported gingival bleeding was associated with higher bacterial diversity than found in participants reporting no gingival bleeding and with higher total abundance of known periodontal pathogens such as Porphyromonas spp., Treponema spp., and Bacteroides spp.
Keywords: bacteria, gingival bleeding, microbiome, oral hygiene, periodontal health
Clinical Relevance.
Scientific rationale for study: There is a lack of studies describing the oral microbiome composition in a general adult population and how oral bacterial composition is associated with self‐reported gingival bleeding.
Principal findings: Self‐reported gingival bleeding was associated with higher abundance of “red complex” bacteria and higher diversity, while lower diversity was found in those reporting good dental hygiene.
Practical implications: Our study highlights the importance of oral hygiene habits to prevent the growth of bacteria implicated in poor periodontal health. High bacterial diversity may indicate a deterioration of periodontal health.
1. INTRODUCTION
Gingivitis and periodontitis are considered the most common periodontal diseases worldwide (Frencken et al., 2017). By the age of 30 years, four out of five will have experienced a moderate to severe form of gingivitis. It is usually not painful, rarely causes spontaneous bleeding, and thus often goes unnoticed (Trombelli et al., 2018). Periodontal disease is caused by bacteria‐induced inflammation in the gums. Mild forms (gingivitis) may be treated with optimal oral hygiene, but failure to resolve inflammation may eventually develop into periodontitis (Harvey, 2017).
For oral health surveillance studies, self‐reported tools and self‐reported oral health questionnaires have been discussed as alternatives to clinical measures when the latter are not possible to achieve or difficult to collect (Ramos et al., 2013; Goulão et al., 2021). We have previously reported that self‐reported frequency of gum bleeding and increased Community Periodontal Index (CPI) score were associated with respiratory symptoms and low lung function (Gomez Real et al., 2016; Perez Barrionuevo et al., 2018). It is hypothesized that aspiration of bacteria or bacterial components from the oral cavity can explain the link between poor periodontal health and respiratory disease (Heinrich et al., 2019). But a healthy lung microbiota also has its origin from bacteria in the oral cavity (Dickson et al., 2017). The same types of oral bacteria are often present in both periodontally healthy and diseased subjects, although the abundance of these bacteria may differ considerably (Ximénez‐Fyvie et al., 2000). Perturbation of microbial communities may occur before the full clinical symptoms of the disease become apparent (Liu et al., 2012). To understand the aetiology underlying the microbial shift from the healthy to the diseased state, studies on experimental gingivitis are useful for describing this shift (Kistler et al., 2013; Bamashmous et al., 2021). However, there is a lack of literature describing the composition of the oral bacterial community associated with self‐reported crude markers of periodontal health status, such as gingival bleeding. There is also little information available regarding how oral hygiene habits in the absence of oral hygiene instructions or professional tooth‐cleaning influence the oral bacterial composition in a generally healthy population. Previous studies have found that a high diversity of oral bacteria is associated with periodontal disease (Liu et al., 2012). We hypothesized that, in a general population, self‐reported markers of periodontal health status may be related to differential bacterial composition and diversity and that oral hygiene habits may also influence the oral microbiome composition and diversity.
Therefore, the purpose of this study was to describe the association between gingival bacterial diversity and composition with self‐reported frequency of gingival bleeding and oral hygiene habits in a Norwegian regional‐based population.
2. METHODS
2.1. Study population and sample collection
This analysis is based on the Respiratory Health in Northern Europe, Spain and Australia (RHINESSA) study (www.RHINESSA.net), a community‐based generation study (Svanes et al., 2022). In the present analysis, only participants from the Bergen study centre were included. They were examined in 2014–2015 with questionnaires, interview, and clinical examination. Interview data and samples of gingival fluid were collected concurrently from all participants. This study included all 484 adult participants with gingival fluid samples (excluding 14 participants due to use of antibiotics during the last 4 weeks prior to gingival sample collection). Among these 484 participants, 445 provided information on the frequency of gum bleeding when brushing teeth, whereas the CPI score was available for 399 participants.
Ethical approval was obtained from the Regional Committee for Medical and Health Research Ethics in Western Norway (approval no. #2012/1077). All participants provided written informed consent.
2.2. Markers of periodontal health status
2.2.1. Gingival bleeding
In a self‐reported questionnaire, the study participants were asked how often their gums bled during tooth brushing (“Do your gums bleed when you brush your teeth?”) with the following answer categories: “Never”, “Rarely”, “Sometimes”, “Often”, or “Always”. For the microbial differential abundance analysis, we categorized the gum bleeding frequency into three different groups: “never/rarely”, “sometimes”, and “often/always”, and compared the “never/rarely” (reference groups) with the categories “sometimes” and “often/always”.
2.2.2. Community Periodontal Index
The CPI has been clinically assessed by a modified version of the World Health Organization's (WHO) guidelines “Oral Health Surveys – Basic Methods” (Ainamo et al., 1982), as previously described (Perez Barrionuevo et al., 2018). CPI measurements were performed without saliva control with the patient lying down on an examination bed, with a WHO‐621 periodontal probe. Three indicators (bleeding, calculus, pocket depths) were used to assess the CPI score: healthy gingiva (CPI code = 0); gingival bleeding (CPI code = 1); calculus (CPI code = 2); and periodontal pockets 4–5 mm (CPI code = 3) and ≥6 mm (CPI code = 4). Ten index teeth were examined to screen periodontal status. Using the FDI dental notation, the following teeth were included from the upper jaw: 17, 16, 11, 26, and 27, and from the lower jaw: 47, 46, 31, 36, and 37. Individuals were given the highest score from all 10 teeth.
2.3. General characteristics and oral hygiene habit variables
General characteristics (gender, age, smoking, and body mass index [BMI]), use of antibiotics in the last 4 weeks before gingival fluid sampling, and reported frequency of self‐applied oral hygiene—frequency of toothbrushing (less than once daily, once daily, or twice daily), use of fluoride toothpaste, and frequency of use of dental floss, mouthwash, and toothpicks (never or rarely, once per week, once per day, twice daily, or more)—were obtained from questionnaires and structured interviews (questionnaire forms are available from www.rhinessa.net).
2.4. Oral microbiome samples
The time of the day of gingival fluid sample collection and information regarding fasting before sample collection were recorded. Subgingival fluid was collected with sterile paper points (PROTAPER, Jacobsen Dental) from the gingival crevice at five per‐protocol pre‐determined sites in the lower and upper jaw: the inter‐proximal region of the central incisors; the inter‐proximal region of the central incisor and lateral incisor (left and right side); and inter‐proximal region between second premolar and first molar (left and right side). The gingival fluid sampling was performed with sterile mirror and tweezers, sterile gloves, and surgical face mask. The paper points were frozen directly (−80°C) after collection in 2‐ml microtubes (Biopur Safe‐Lock Tubes) without buffer. Five paper points per individual (lower jaw sites only or mix between upper and lower jaw sites) were pooled and analysed together for Illumina 16S rRNA gene sequencing.
2.5. 16S rRNA gene amplicon sequencing
All laboratory procedures were performed at the UNC Microbiome Core Facility of the University of North Carolina in Chapel Hill, NC. The samples were analysed at two time points, one batch (n = 283) in 2016 and the second batch (n = 201) in 2019, with the same procedure and by the same laboratory technician. Bacterial DNA was extracted from the gingival fluid samples. Overall, 12.5 ng of total DNA was amplified using a combination (4:1) of universal and Bifidobacterium‐specific primers targeting the V1–V2 region of the bacterial 16S rRNA gene (Edwards et al., 1989; Fierer et al., 2008). The DNA library pool based on the 16S amplicons was sequenced with Illumina (San Diego, CA) MiSeq, with automated cluster generation and paired‐end sequencing with dual reads, according to the manufacturer's instructions.
Bioinformatic analysis of bacterial 16S amplicon sequencing data was conducted using the Quantitative Insights Into Microbial Ecology (QIIME) software (Caporaso et al., 2010; Bolyen et al., 2019). For more details on the sequencing procedure and quality control and filtering, see Supplementary Files.
Taxonomy was assigned to amplicon sequence variants (ASVs) using the q2‐feature‐classifier (Bokulich et al., 2018) against the expanded Human Oral Microbiome Database (eHOMD), which is a comprehensive database with well‐curated 16S rRNA gene reference sequences from sites along the human aerodigestive tract (version 15.01) (Escapa et al., 2018). Five percent of the ASVs were assigned taxonomy at a lower taxonomic resolution (class, family etc.). The remaining 95% of the ASVs were classified into 188 bacterial genera, of which 21 genera contributed to 79% of the bacteria in the gingival microbiome.
2.6. Statistical analysis
Gingival bleeding and CPI were described according to population characteristics and oral hygiene habits. Statistical significance was defined by p‐values ≤.05. Associations between alpha diversity indices (Faith's phylogenetic diversity index [Faith's PD], Shannon diversity index, and observed species number metrics [at the ASV level]) and general characteristics (independent variables) were fitted by linear regression using robust variance estimates adjusting for clustering within families and adjusted for age, gender, smoking, batch, sample type, and time of sampling using STATA (version IC 14.0; StataCorp LLC, College Station, TX).
Alpha diversity indices (Shannon, Observed, Chao1, and Pileous evenness) were calculated after rarefying, showing the genus‐level difference in bacterial richness (number of bacterial taxa) and bacterial evenness (relative abundance of bacterial taxa) across categories of gum bleeding frequency. Differences in alpha diversity across the frequency of gum bleeding were assessed using a Kruskal–Wallis test.
For beta diversity (at the genus level), principal coordinates analysis (PCoA) was performed to visualize the Bray–Curtis dissimilarity matrices. Permutational multivariate analysis of variance (PERMANOVA) (Anderson & Walsh, 2013) and permutational analysis of multivariate dispersion (PERMDISP) (Anderson, 2006) were applied to compare the microbial community clustering between groups. A significant PERMANOVA test implies that the observed differences are either due to different spatial medians (location effect) or due to the heterogeneity of dispersions (dispersion effect), or a combination of both. Therefore, a follow‐up PERMDISP test was also performed to examine the difference in dispersion between groups.
To test which bacteria differed in abundance between the exposure groups (frequency of gum bleeding, frequency of oral hygiene, and CPI score), we performed a differential abundance analysis using the analysis of compositions of microbiomes with bias correction (ANCOM‐BC) (Lin & Peddada, 2020) at the genus level and at the species level for gum bleeding as outcome. This methodology is aimed at detecting differentially abundant microbes among two or more groups while adjusting for other covariates. The observed microbiome data (shown in operational taxonomic units (OTUs)/ASVs, etc.) represent only relative information of each taxon and thus are compositional data (Gloor et al., 2017). ANCOM‐BC properly adjusts the bias due to sample‐specific sampling fractions and thus transfers the observed abundances (which are compositional and constrained by the library size) to absolute abundances in a unit volume of an ecosystem (e.g., a unit volume of gut) and makes inferences on differentially abundant taxa in the sense of absolute abundances. p‐Values obtained from the ANCOM‐BC model were adjusted by the Holm–Bonferroni method (Holm, 1979) to account for the effect of multiple comparisons. The R packages vegan, phyloseq, and ANCOM‐BC were used for the corresponding analyses.
3. RESULTS
A total of 484 adults (18–47 years of age) were included in the present study (Table 1). Women and men did not differ by smoking status (p = .4), but men had higher BMI (p < .001). Six participants reported to have diabetes (five women and one man). Women reported better oral health practices, with a higher frequency of toothbrushing (p = .001) and more frequent use of dental floss than men (p < .001). Men reported more frequent use of toothpicks than women (p = .04). Self‐reported frequency of gingival bleeding was similar for men and women (p = .8), but a higher proportion of men (17.1%) than women (5.5%) presented with CPI score 2 (overall p < .001). All the participants were native Norwegians, and the study population did not include different ethnicities.
TABLE 1.
Characteristics of the study population (n = 484)
| All (n = 484) | Females (n = 227) | Males (n = 257) | p‐Value for gender diff. | |
|---|---|---|---|---|
| Age (mean, range) | 28.0 (18–47) | 27.2 (18–45) | 28.6 (18–47) | .02 |
| BMIa | ||||
| <20 | 44 (9) | 13.7 | 5.10 | |
| 20–24.99 | 239 (50) | 61.2 | 39.2 | |
| 25–29.99 | 140 (29) | 15.4 | 41.2 | |
| >30 | 59 (12) | 9.7 | 14.5 | <.001 |
| Smoking a | ||||
| Never | 337 (70) | 72.9 | 67.6 | |
| Ex‐smoker | 78 (16) | 13.8 | 18.4 | |
| Current smoker | 66 (14) | 13.3 | 14.1 | .4 |
| Frequency of toothbrushing | ||||
| ≤Once daily | 63 (13) | 8.9 | 16.8 | |
| Twice daily | 389 (81) | 81.8 | 80.1 | |
| >Twice daily | 29 (6) | 9.3 | 3.1 | .001 |
| Use of fluoride toothpaste a | ||||
| ≤Once daily | 75 (16) | 12 | 18.8 | |
| Twice daily | 377 (78) | 79.6 | 77.6 | |
| >Twice daily | 28 (6) | 8.4 | 3.5 | .01 |
| Use of dental floss a | ||||
| Never or rarely | 197 (41) | 30.2 | 50.6 | |
| Once weekly | 130 (27) | 28.4 | 25.9 | |
| Once daily | 125 (26) | 30.2 | 22.4 | |
| ≥Twice daily | 28 (6) | 11.1 | 1.1 | <.001 |
| Use of mouthwash a | ||||
| Never or rarely | 236 (49) | 45.3 | 55.6 | |
| Once weekly | 105 (22) | 22.7 | 21.2 | |
| Once daily | 113 (24) | 24.4 | 22.8 | |
| ≥Twice daily | 26 (5) | 7.6 | 3.5 | .1 |
| Use of toothpicks a | ||||
| Never or rarely | 304 (64) | 69.2 | 58.7 | |
| Once weekly | 120 (25) | 20.1 | 29.5 | |
| ≥Once daily | 54 (11) | 10.7 | 11.8 | .04 |
| Gingival bleeding a | ||||
| Never | 57 (13) | 12.8 | 13.1 | |
| Rarely | 210 (48) | 45.9 | 49.0 | |
| Sometimes | 135 (30) | 29.8 | 30.0 | |
| Often | 32 (7) | 8.3 | 6.3 | |
| Always | 11 (2) | 3.2 | 1.7 | .8 |
| CPI a , b | ||||
| Score 0 | 325 (82) | 89.5 | 73.9 | |
| Score 1 | 17 (4) | 2.5 | 6.0 | |
| Score 2 | 45 (11) | 5.5 | 17.1 | |
| Score 3 | 12 (3) | 3.0 | 3.0 | .001 |
Abbreviations: BMI, body mass index; CPI, community periodontal index.
Missing data for BMI (n = 1); smoking (n = 2), fluoride and flossing and mouthwash (n = 3), toothpicks (n = 5); gingival bleeding (n = 39); CPI (n = 85).
No participants with score above 3.
Those reporting a high frequency of gingival bleeding (often or always) differed in the reported frequency of toothbrushing from those who never or rarely experienced bleeding from the gums, with the frequent gingival bleeders being more likely to report toothbrushing more than twice daily (11.6% vs. 4.5%) than those who never or rarely have bleeding from the gums. On the other hand, infrequent brushing (once daily or less) was also more commonly reported among the frequent gingival bleeders (16.3%) than the never gingival bleeders (9.4%) (overall p = .009) (Table 2). As expected, a high CPI score was associated with smoking, with 30% of the participants with CPI ≥2 being current smokers compared to those with CPI score of 0 or 1 (9.5% and 5.6%, respectively), p < .001 (Table 2). The reported use of dental floss was similar for participants with CPI = 0 and CPI ≥2, whereas those with CPI = 1 reported more infrequent use of dental floss (p = .05) (Table 2). Among the 18 participants with CPI = 1 (bleeding on probing), 3 reported bleeding from the gums often or always and 7 never or rarely bled from the gums. There was a tendency for higher frequency of self‐reported gingival bleeding among those who scored high on CPI (CPI 2 and 3) compared to those with low CPI (0 and 1), p = .02 (chi‐square test). Of the six participants with diabetes, three had a CPI score of 0 and three had a CPI score of 1, whereas gingival bleeding was reported to be rare (n = 3) or to occur sometimes (n = 3).
TABLE 2.
Participant characteristics and dental hygiene habits according to self‐reported gingival bleeding (n = 445) and community periodontal index (CPI) score (n = 399)
| Oral health outcomes | ||||||||
|---|---|---|---|---|---|---|---|---|
| Frequency of gingival bleeding when brushing teeth | CPI | |||||||
| Never/rarely (n = 267) | Sometimes (n = 135) | Always/often (n = 43) | p‐Value* | CPI = 0 (n = 325) | CPI = 1 (n = 17) | CPI ≥ 2 | p‐Value** | |
| Gender (% female) | 47.2 | 47.4 | 58.1 | .4 | 54.8 | 29.4 | 29.8 | <.001 |
| Age (mean, SD) | 28.4 (6.8) | 27.7 (6.6) | 26.2 (6.5) | .1 | 27.3 (6.8) | 27.2 (6.4) | 31.1 (6.9) | <.001 |
| BMI (mean, SD) | 25.0 (4.6) | 25.2 (4.0) | 24.5 (5.3) | .6 | 24.9 (4.8) | 26.9 (4.5) | 25.5 (4.2) | .2 |
| Smoking (%) | ||||||||
| Never | 74.8 | 69.4 | 55.8 | 74.8 | 52.9 | 56.1 | ||
| Ex‐smoker | 13.5 | 17.9 | 27.9 | 15.7 | 41.2 | 14.0 | ||
| Current smoker | 11.7 | 12.7 | 16.3 | .1 | 9.5 | 5.9 | 29.8 | <.001 |
| Frequency of tooth brushing (%) | ||||||||
| ≤Once daily | 9.4 | 18.7 | 16.3 | 12.9 | 11.8 | 15.8 | ||
| Twice daily | 86.1 | 72.4 | 72.1 | 80.9 | 82.4 | 80.7 | ||
| >Twice daily | 4.5 | 9.0 | 11.6 | .009 | 6.2 | 5.9 | 3.5 | .9 |
| Use of fluoride toothpaste (%) | ||||||||
| ≤Once daily | 12.1 | 20.2 | 20.9 | 15.4 | 23.5 | 17.9 | ||
| Twice daily | 83.4 | 72.4 | 67.4 | 79.1 | 70.6 | 78.6 | ||
| >Twice daily | 4.5 | 7.5 | 11.6 | .03 | 5.5 | 5.9 | 3.6 | .9 |
| Use of dental floss (%) | ||||||||
| Never or rarely | 38.5 | 42.5 | 39.5 | 36.0 | 76.5 | 44.6 | ||
| Once weekly | 27.9 | 28.4 | 20.9 | 28.6 | 11.8 | 21.4 | ||
| Once daily | 26.8 | 23.9 | 32.6 | 28.3 | 11.8 | 28.6 | ||
| ≥Twice daily | 6.8 | 5.2 | 7.0 | .9 | 7.1 | 0 | 5.4 | .05 |
| Use of mouthwash (%) | ||||||||
| Never or rarely | 46.8 | 53.7 | 48.8 | 47.4 | 64.7 | 50.0 | ||
| Once weekly | 20.8 | 23.1 | 16.3 | 21.9 | 17.7 | 23.2 | ||
| Once daily | 26.4 | 19.4 | 27.9 | 24.9 | 17.7 | 21.4 | ||
| ≥Twice daily | 6.0 | 3.7 | 7.0 | .6 | 5.9 | 0 | 5.4 | .8 |
| Use of toothpicks (%) | ||||||||
| Never or rarely | 62.7 | 67.9 | 62.8 | 66.1 | 58.8 | 61.4 | ||
| Once weekly | 25.9 | 24.9 | 20.9 | 24.1 | 29.4 | 22.8 | ||
| ≥Once daily | 11.4 | 8.2 | 16.3 | .6 | 9.9 | 11.8 | 15.8 | .8 |
Abbreviation: BMI, body mass index. * p‐Value for difference between self‐reported frequency of gingival bleeding. ** p‐Value for difference between CPI categories
Alpha diversity assessed by Shannon diversity index and Faith's PD increased with increasing frequency of gingival bleeding (p < .001, Table 3 and Appendix Figure 1). Those reporting frequent bleeding from the gums (often or always) showed statistically significant higher alpha diversity compared to those reporting never or rarely bleed from the gums (p < .05 for Pileou's evenness and p < .001 for Shannon, Observed, Chao1) (Figure 1). There was a statistically significant difference in microbial community clustering (Bray–Curtis dissimilarity) between the gingival‐bleeding frequency groups (Figure 2), with the differences driven by the heterogeneity of both location (PERMANOVA; p = .001) and dispersion (PERMDISP test, p = .024) effects. Shannon diversity index was higher for those with a CPI score of 3 than those with a CPI score 0, but for the overall CPI scores, the trend was not statistically significant (p = .3) (Table 3). Daily or more frequent use of dental floss, mouthwash, and toothpicks was associated with decreased alpha diversities (Table 3); Shannon diversity index was reduced by 0.22 units in those who used toothpicks at least once daily compared to never users.
TABLE 3.
Estimated mean differences for dental hygiene habits and periodontal health status by alpha diversity indices adjusted for age, gender, smoking, batch, sample type time during the day for gingival sample, and clustering by family
| Faith's phylogenetic diversity index | Shannon diversity index (H) | Observed ASVs (count, n) | ||||
|---|---|---|---|---|---|---|
| β (95% CI) | p‐Value | β (95% CI) | p‐Value | β (95% CI) | p‐Value | |
| Frequency of toothbrushing | ||||||
| Twice daily | Reference | Reference | Reference | |||
| >Twice daily | 1.59 (−3.75, 6.92) | 0.18 (−0.01, 0.38) | 21.7 (−8.90, 52.4) | |||
| ≤Once daily | −2.17 (−4.60, 0.25) | .2 | 0.07 (−0.08, 0.22) | .2 | 5.35 (−15.8, 26.5) | .4 |
| Use of fluoride toothpaste | ||||||
| Twice daily | Reference | Reference | Reference | |||
| >Twice daily | 1.62 (−3.89, 7.13) | 0.19 (−0.01, 0.40) | 19.3 (−10.7, 49.3) | |||
| ≤Once daily | −1.19 (−3.48, 1.09) | .5 | 0.07 (−0.07, 0.21) | .2 | 6.29 (−13.4, 25.9) | .4 |
| Use of dental floss | ||||||
| Never or rarely | Reference | Reference | Reference | |||
| Once weekly | −0.50 (−2.87, 1.70) | 0.003 (−0.13, 0.15) | −4.07 (−22.4, 14.3) | |||
| Once daily | −0.39 (−3.07, 2.30) | −0.11 (−0.25, 0.02) | −23.6 (−41.7, −5.42) | |||
| ≥Twice daily | 0.58 (−3.39, 4.55) | .9 | −0.20 (−0.40, 0.01) | .04 | −37.9 (−67.1, −8.66.) | .001 |
| Use of mouthwash | ||||||
| Never or rarely | Reference | Reference | Reference | |||
| Once weekly | 2.85 (−0.13, 5.82) | −0.02 (−0.15, 0.12) | 0.94 (−16.8, 18.7) | |||
| Once daily | 0.60 (−1.48, 2.69) | −0.14 (−0.28, 0.01) | −15.5 (−34.8, 3.82) | |||
| ≥Twice daily | −1.18 (−5.07, 2.70) | .8 | −0.17 (−0.44, 0.09) | .03 | −31.4 (−63.1.7, 0.33) | .0 |
| Use of toothpicks | ||||||
| Never or rarely | Reference | Reference | Reference | |||
| Once weekly | 1.00 (−1.46, 3.46) | −0.03 (−0.16, 0.11) | −0.47 (−17.7, 16.8) | |||
| ≥Once daily | −0.65 (−3.87, 2.57) | .9 | −0.22 (−0.42, −0.04) | .04 | −20.1 (−44.7, 4.46) | .2 |
| Gingival bleeding | ||||||
| Never | Reference | Reference | Reference | |||
| Rarely | −3.37 (−7.00, 0.27) | 0.08 (−0.09, 0.27) | 2.80 (−17.3, 24.9) | |||
| Sometimes | −2.06 (−6.15, 2.03) | 0.14 (−0.06, 0.33) | 13.2 (−10.5, 36.9) | |||
| Often | −0.27 (−5.16, 4.61) | 0.51 (0.27, 0.76) | 71.3 (37.3, 105.2) | |||
| Always | −1.94 (−6.26, 2.39) | .9 | 0.75 (0.45, 1.04) | <.001 | 114.3 (57.5, 171.1) | <.001 |
| CPI | ||||||
| Score 0 | Reference | Reference | Reference | |||
| Score 1 | −4.28 (−7.78, −0.78) | 0.01 (−0.29, 0.31) | 14.1 (−24.1, 52.2) | |||
| Score 2 | −0.78 (−3.98, 2.41) | 0.04 (−0.14, 0.23) | 7.90 (−15.9, 31.7) | |||
| Score 3 | −0.85 (−4.08, 2.39) | .4 | 0.19 (0.00, 0.38) | .3 | 12.7 (−20.8, 46.2) | .3 |
Note: p‐Value for linear trend of the categorical variable.
Abbreviations: ASVs, amplicon sequence variants; CI, confidence interval; CPI, community periodontal index. Numbers in bold indicate the categories that are signficantly different (p < .05) from the reference categories.
FIGURE 1.
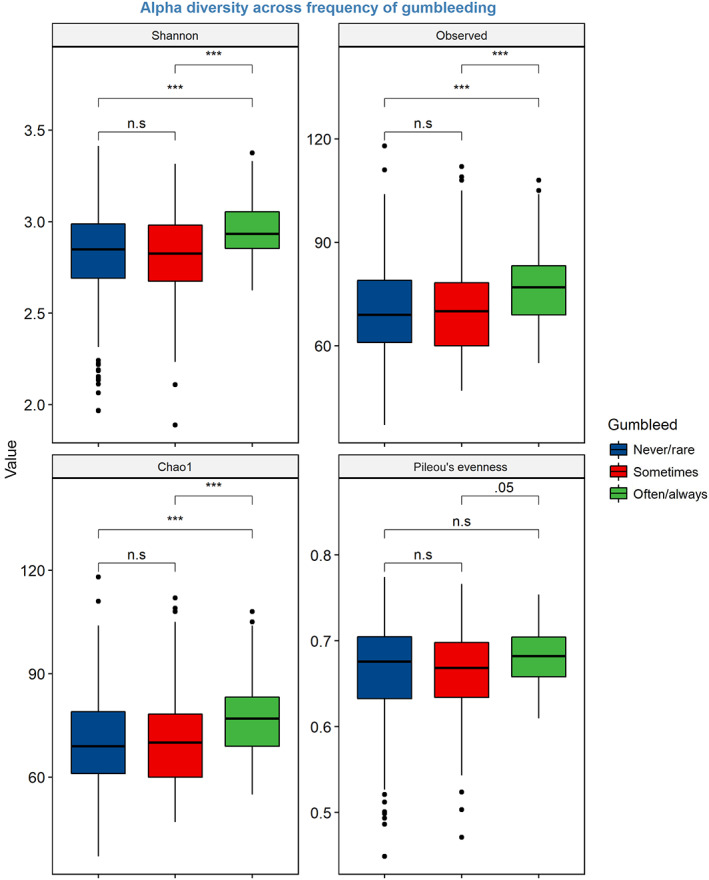
Box plots illustrating alpha‐diversity indices (Shannon, Oserved, Chao1, and Pileous evenness) across self‐reported frequency of gingival bleeding (often/always and sometimes and never/rare). Boxes represent the interquartile range (IQR) between the first and third quartiles (25th and 75th percentiles, respectively), and the horizontal line inside the box defines the median. The whiskers represent the lowest and highest values within 1.5 times the IQR from the first and third quartiles, respectively. Asterisks denote significant differences at Kruskal–Wallis test *p < .05, **p < .01, and ***p < .001; n.s, non‐significant
FIGURE 2.
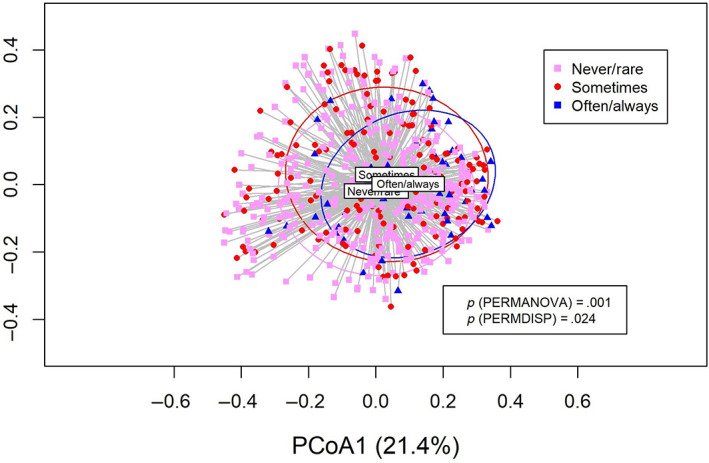
Principal coordinates analysis (PCoA) for self‐reported frequency of gingival bleeding. Permutational multivariate analysis of variance and permutational analysis of multivariate dispersion tests for between‐sample diversity (Bray–Curtis dissimilarity) for the groups: never/rarely, sometimes, and often/always gingival bleeding
3.1. Bacterial composition
A total of 188 bacterial genera were identified, with the most prevalent ones being Fusobacterium spp. (15.3%), Streptococcus spp. (9.4%), Prevotella spp. (8.6%), Capnocytophaga spp. (6.3%), Neisseria spp. (4.5%), Leptotrichia spp. (3.6%), and Porphyromonas spp. (3.4%).
ANCOM‐BC methodology was applied to determine taxa that are differentially abundant in absolute abundance by gingival‐bleeding frequency, CPI score, and oral hygiene habits. Pairwise testing of the groups, while adjusting for age, gender, smoking, and BMI, showed a statistically significant higher absolute abundance for many bacteria taxa among those who reported to always or often to bleed from the gums as compared to the never gum‐bleeders (Table 4, Appendix Table 1, and Figure 3). At the genus level, this included many bacteria from the phylum Bacteroidetes such as Porphyromonas spp., Bacteroidetes [G‐3] and [G‐5] spp., and Tannerella spp. (p‐values from <.001 to .04), and from the Firmicutes phylum genera such as Catonella spp., Filifactor spp., Lachnospiraceae [G‐8] spp., and Peptostreptococcaceae [XI][G‐4] and [G‐7] spp. (all p < .001). A higher absolute abundance of Fretibacterium spp. (phylum Synergistetes, p < .001) and Treponema spp. (phylum Spirochetes, p < .001) was also associated with the highest reported frequency of gingival bleeding when brushing teeth (Figure 3). Four genera showed higher abundance in samples from those reporting “sometimes bleed” from the gums (Dermabacter, Bacteroides, Lysinibacillus, and Novosphingobium) (data not shown). On the species level, frequent gingival bleeding was associated with higher abundance of several bacteria, such as Porphyromonas endodontalis, Treponema denticola, Fretibacterium fastidiosum, and Fretibacterium spp. HMT 360 (all p‐values ≤ .01). Within the gram‐positive bacterial phylum Firmicutes, the species Catonella morbi, Peptostreptococcaceae bacterium HMT 081, and Dialister pneumosintes had higher abundance in those with frequent bleeding from the gingiva, but most bacteria within this phylum showed lower abundance in gingival bleeding (Table 4, all p ≤ .02). Furthermore, a lower abundance of several Prevotella species was also found in the gingival fluid of those reporting the most frequent gum bleeding, such as P. histicola, P. intermedia, and P. salivae (Table 4, all p < .001).
TABLE 4.
Analysis of compositions of microbiomes with bias correction test for difference in absolute abundance of bacteria (species level) between participants reporting often or always gum bleeding (n = 43) or sometimes (n = 135) compared to those who never or rarely bleed from the gums (n = 267), adjusted for age, gender, smoking, and body mass index
| LFC (SE) | p | |
|---|---|---|
| Bacteria taxa (species [P: phylum]) differences between those reporting often or always to bleed from the gums compared to never/rarely gum bleeding | ||
| Actinomyces graevenitzii (P: Actinobacteria) | −0.07 (0.14) | .00 |
| Actinomyces massiliensis (P: Actinobacteria) | −0.46 (0.29) | .00 |
| Actinomyces odontolyticus (P: Actinobacteria) | −0.32 (0.25) | .00 |
| Actinomyces sp. HMT 169 (P: Actinobacteria) | −2.10 (0.39) | .00 |
| Actinomyces sp. HMT 171 (P: Actinobacteria) | −1.54 (0.37) | .01 |
| Actinomyces sp. HMT 448 (P: Actinobacteria) | −1.25 (0.30) | .01 |
| Scardovia wiggsiae (P: Actinobacteria) | −0.94 (0.12) | .02 |
| Corynebacterium durum (P: Actinobacteria) | −1.55 (0.39) | .00 |
| Dermabacter hominis (P: Actinobacteria) | −0.65 (0.10) | .00 |
| Microbacterium flavescens (P: Actinobacteria) | −0.70 (0.15) | .00 |
| Microbacterium ginsengisoli (P: Actinobacteria) | −0.79 (0.17) | .00 |
| Propionibacteriaceae [G‐2] bacterium HMT 192 (P: Actinobacteria) | −0.75 (0.17) | .00 |
| Atopobium parvulum (P: Actinobacteria) | −1.23 (0.23) | .00 |
| Bacteroidetes [G‐5] bacterium HMT 511 (P: Bacteroidetes) | 1.77 (0.39) | .00 |
| Porphyromonas endodontalis (P: Bacteroidetes) | 2.21 (0.35) | .00 |
| Prevotella histicola (P: Bacteroidetes) | −1.06 (0.21) | .00 |
| Prevotella intermedia (P: Bacteroidetes) | −1.90 (0.58) | .00 |
| Prevotella salivae (P: Bacteroidetes) | −1.26 (0.23) | .00 |
| Prevotella sp. HMT 306 (P: Bacteroidetes) | −1.00 (0.13) | .00 |
| Prevotella sp. HMT 313 (P: Bacteroidetes) | −0.73 (0.15) | .00 |
| Lysinibacillus fusiformis (P: Firmicutes) | −0.69 (0.10) | .00 |
| Streptococcus infantis clade 638 (P: Firmicutes) | −1.03 (0.24) | .01 |
| Streptococcus intermedius (P: Firmicutes) | −1.90 (0.42) | .00 |
| Streptococcus mutans (P: Firmicutes) | −1.23 (0.27) | .00 |
| Streptococcus parasanguinis clade 411 (P: Firmicutes) | −1.11 (0.26) | .01 |
| Streptococcus salivarius (P: Firmicutes) | −1.10 (0.27) | .02 |
| Streptococcus sanguinis (P: Firmicutes) | −1.25 (0.30) | .01 |
| Streptococcus sp. HMT 066 (P: Firmicutes) | −0.93 (0.20) | .00 |
| Catonella morbi (P: Firmicutes) | 0.75 (0.17) | .00 |
| Oribacterium sinus (P: Firmicutes) | −0.83 (0.14) | .00 |
| Stomatobaculum longum (P: Firmicutes) | −0.92 (0.16) | .00 |
| Stomatobaculum sp. HMT 097 (P: Firmicutes) | −0.67 (0.15) | .00 |
| Peptostreptococcaceae bacterium HMT 081 (P: Firmicutes) | 2.10 (0.37) | .00 |
| Dialister pneumosintes (P: Firmicutes) | 1.70 (0.40) | .00 |
| Bosea vestrisii (P: Proteobacteria) | −1.03(0.24) | .01 |
| Novosphingobium panipatense (P: Proteobacteria) | −0.61 (0.11) | .00 |
| Neisseria bacilliformis (P: Proteobacteria) | −1.45 (0.33) | .00 |
| Hemophilus parainfluenzae (P: Proteobacteria) | −1.63 (0.40) | .01 |
| Haemophilus sputorum (P: Proteobacteria) | −0.74 (0.18) | .01 |
| Treponema denticola (P: Spirochaetes) | 1.94 (0.47) | .01 |
| Fretibacterium fastidiosum (P: Synergistetes) | 1.73 (0.38) | .00 |
| Fretibacterium sp. HMT 360 (P: Synergistetes) | 1.94 (0.42) | .00 |
| Bacteria taxa (species [P: phylum]) differences between those reporting sometimes to bleed from the gums compared to never/rarely gum bleeding | ||
| Streptococcus intermedius (P: Firmicutes) | −1.06 (0.25) | .00 |
| Peptostreptococcaceae bacterium HMT 081 (P: Firmicutes) | −0.19 (0.23) | .00 |
| Treponema sp. HMT 270 (P: Spirochaetes) | 0.57 (0.13) | .01 |
Note: Results given by LFC (natural log), SE, and adjusted p‐values.
Abbreviation: LFC, log fold‐change.
FIGURE 3.
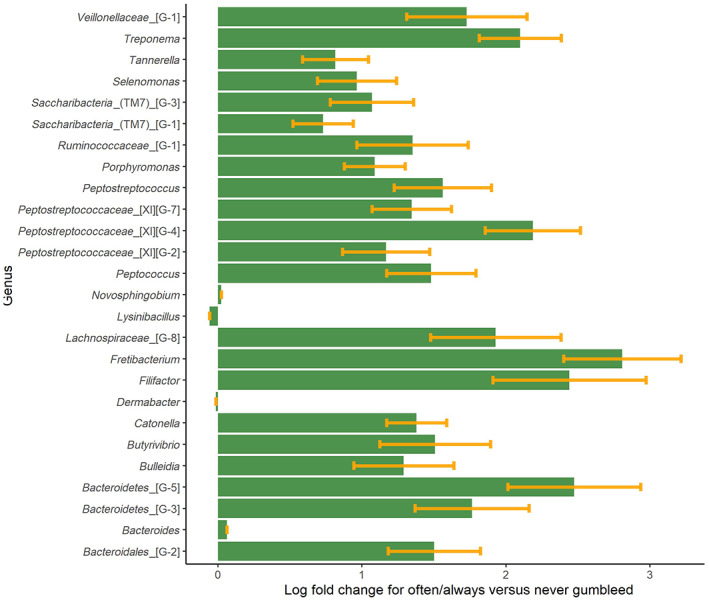
Overview of gingival bacterial genera that differ in abundance between those reported often or always bleeding from the gums compared to those reporting never or rare bleeding from the gums (reference group). The bars represent the log fold‐change and the whiskers represent the SE. The bars displayed on the right‐hand side of 0 indicate a higher abundance of the genus in the often/always category as compared to the reference group, and the bars on the left‐hand side of the 0 is the taxa that appear in lower abundance as compared to the reference group
Regarding oral hygiene habits, using floss twice daily was associated with higher abundance of bacterial genera within phylum Proteobacteria (Novospinogium, Achromobacter, Helicobacter, and Moraxella) (Appendix Table 2, all p < .001). The novel pathogen, Saccharibacteria (TM7), which was more prevalent among the frequent gum bleeders, had lower abundance among those with frequent flossing (Appendix Table 2, p < .001) and among those who used mouthwash twice daily (Appendix Table 3). Also, Bacteroides and Peptostreptococcaceae, which are recognized oral pathogens, were found in lower abundance among those who regularly used dental floss and mouthwash. A higher absolute abundance of Porphyromonas spp. was found in gingival samples for those scoring a CPI of 1 (compared to CPI = 0), p < .01 (Appendix Table 4).
4. DISCUSSION
In this Norwegian general population, we found that participants reporting frequent gingival bleeding had higher bacterial diversity in the gingiva than participants reporting less frequent gingival bleeding. Furthermore, high bacteria diversity in the gingiva was also associated with a low frequency of use of dental flossing, mouthwash, and toothpicks. A higher total abundance of several bacterial genera with known periodontal pathogenic species was present in the gingival fluid from individuals reporting frequent gingival bleeding. Tooth brushing and the use of dental floss twice daily were more likely to be reported by the participants who had a low frequency of gingival bleeding and a low CPI score.
In our study population, a higher frequency of women than men reported the use of dental floss and toothbrushing more than twice daily. This is in line with previous studies showing that young women have better oral‐health‐related habits than men (Fernandez de Grado et al., 2018; Virtanen et al., 2019). It has been found that low socio‐economic status is associated with less than optimal oral hygiene habits (Sakki et al., 1998; Astrom & Rise, 2001; Fernandez de Grado et al., 2018). However, our study population is relatively uniform in terms of socio‐economic status, with the majority being highly educated (university level) and 27% not yet having reached the age of maximum attained education. However, for those who had completed education, we did not see differences in oral hygiene habits by the participants' own educational status or by the parental educational status. Nor does our study population include different ethnicities, which could also influence oral health habits (Patino et al., 2018).
We found the highest bacterial community diversity in gingival fluid for those reporting often or always to bleed from the gums when they brushed their teeth. This is in line with studies on experimental gingivitis showing an increase in community diversity during transition from periodontal health to periodontitis (Kistler et al., 2013; Bamashmous et al., 2021). We found that those who reported the highest frequency of gingival bleeding had a lower abundance of the bacteria within the Firmicutes and Actinobacteria phyla. These are typically gram‐positive phyla, which are also found to decrease in relative abundance during the onset of experimental gingivitis (Bamashmous et al., 2021). Although we cannot confirm the presence of gingivitis among our study participants, a recent study found that self‐reported questions regarding bleeding gums performed quite well in sensitivity towards gingival inflammation, as confirmed with bleeding on probing assessed by trained staff (Goulão et al., 2021). Furthermore, in those reporting a high frequency of dental flossing and toothpick use, the microbial alpha diversity was reduced. This might indicate that applying good oral hygiene habits might reduce the risk of having non‐commensal bacteria invading the gingival pockets. In particular, it has been found that using inter‐dental cleaning in combination with toothbrushing may reduce gingivitis or plaque more than toothbrushing alone (Gallie, 2019; Worthington et al., 2019). Brushing teeth twice daily was reported by most of the study participants. However, a higher percentage of those reporting always gingival bleeding brush their teeth more than twice daily. This might indicate that they are aware that their dental health status is not optimal; it is also possible that the gingival bleeding may be unintentionally self‐inflicted due to too hard toothbrushing. Toothbrushing force has been indicated as a risk factor for progression of non‐inflammatory gingival recession (Rajapakse et al., 2007). Thus, we cannot exclude the possibility of reverse causation, with a high frequency of tooth brushing or too forceful toothbrushing leading to damage of the gingiva and thus frequent gingival bleeding.
A higher abundance of suspected periodontal pathogens such as Porphyromonas, Treponema, and Bacteroides spp., which are members of the “red complex” (Socransky et al., 1998), was found in frequent gum bleeders. Bacteria belonging to the “orange complex”, such as Prevotella intermedia and Peptostreptococcus spp., were associated with self‐reported gingival bleeding in our study. These orange complex bacteria are suspected to precede colonization by species of the red complex (Socransky et al., 1998). Since the hallmark paper by Socransky and colleagues, the development of more advanced sequencing techniques and curated databases for assigning sequences to bacteria taxa, such as the Human Oral Microbiome Database, have also opened for identification of “novel” periodontal pathogens. In a review paper by Colombo and Tanner (2019) based on next‐generation sequencing studies, Fretibacterium spp., Saccharibacteria spp., and Dialister spp. were categorized as novel pathogens. Interestingly, these three bacteria were found in the present study in higher abundance in those reporting frequent gingival bleeding. In our study we found that in species‐level analyses, P. endodontalis was present in higher abundance among those who reported the most frequent gingival bleeding compared to the participants reporting no gingival bleeding. Although P. gingivalis has more commonly been reported in gingival inflammation (Kistler et al., 2013; Bamashmous et al., 2021), our findings on P. endodontalis agrees with a previous study, which recognized this as an important periodontal pathogen (Kumar et al., 2003). Also, in line with our results, bacteria in the Synergistetes phylum, in particular F. fastidiosum, have been reported as likely contributors to periodontal disease (Colombo et al., 2009; Pérez‐Chaparro et al., 2014; McCracken & Nathalia, 2021). In the present study, we found that flossing and the use of mouthwash twice daily was associated with a higher abundance of bacteria from the Proteobacteria phylum, which for the most part did not differ in abundance by gingival bleeding. In the present study, we did not have information on periodontitis status. However, only 12 participants had a CPI score of 3 and none had a score of 4, and thus, our study population is likely to include mostly periodontally healthy subjects.
In a study with strict definition of periodontitis (Liu et al., 2012), the oral microbiome community structure was more diverse among the periodontal disease cases than in the healthy samples. In contrast, the healthy microbiome in any individual patient had relatively low taxonomic diversity (Liu et al., 2012). Also, another study comparing subgingival microbial composition in subjects with different periodontal conditions found that both the healthy and the gingivitis groups had higher bacterial diversity than the periodontitis group (Park et al., 2015). As a self‐reported measure for surveillance of periodontitis, it is recommended to include up to five self‐reported questions (e.g., thinking you have gum disease, health of teeth and gums, prior treatment for gum disease, bone loss, and use of dental floss) to correctly predict periodontitis outcomes (Eke et al., 2013). As these self‐reported outcomes are not available in our study, we cannot characterize the participants by periodontitis status. Nevertheless, as discussed above, we may assume that self‐reported gingival bleeding captures gingival inflammation well. We found a higher diversity of bacterial community in the gingival fluid of participants reporting frequent gingival bleeding and in participants with the highest CPI score, despite these being only self‐reported and non‐standard measures of periodontal health status. It is interesting to note that the oral microbiome from periodontally healthy to diseased subjects is different from other parts of the gastrointestinal tract in which disease is often associated with a lower diversity than in a healthy state (Abusleme et al., 2013; Dalal & Chang, 2014).
In line with others (Ximénez‐Fyvie et al., 2000), we found that the so‐called pathogenic bacteria are present not only in those reporting frequent gingival bleeding but also in almost all samples (Porphyromonas in 93% of all samples and Treponema in 94% of all samples). As with periodontal disease, also the transition to caries pathology is caused by the changes in the microbial community structure (Valm, 2019). Thus, changes in the relative abundance of bacteria that are also present in health, transform the bacterial community from healthy to the pathogenic state. This also highlights the importance of using statistical models that can handle the compositional nature of the microbiome, such as ANCOM‐BC, applied in the present study.
4.1. Strengths and limitations
The strength of this study is the large study population with microbiome sequencing data and detailed information on dental hygiene habits. One limitation is the simplified dental protocol and sampling procedure (e.g., gingival fluid sampling without plaque removal). This was needed to enable a uniform collection of samples that can easily be adopted in several study centres and for field workers without dental training. The dental examination was not performed in a dental clinic, but most of the examinations were done by a dentist (Perez Barrionuevo et al., 2018). However, we were not able to distinguish between periodontally healthy, gingivitis, and periodontitis cases, and the CPI score may only provide indications on periodontal pocketing and inflammation. The paper points were pooled before storage, both to minimize the storage volume and maximize the likelihood of having enough bacterial DNA for 16S rRNA sequencing. This limited our possibility to explore the microbiome according to the CPI status of the specific sites. The gingival samples were analysed in two different batches (microbiome sequencing at two different time points), which is unfortunate, but the batch effect is self‐corrected by ANCOM‐BC and adjusted for in the other statistical models. We applied a novel biostatistics model ANCOM‐BC (Lin & Peddada, 2020), which takes into account the compositional nature of the data (Gloor et al., 2017). ANCOM‐BC allows for reporting directionality and effect size for absolute abundances and at the same time controlling for covariates in the models. The model also takes into consideration multiple testing. Finally, species‐level identification may be challenging due to the short sequences obtained from 16S rRNA gene amplicon sequencing, which may give insufficient resolution for accurate identification of species within some predominant genera such as Streptococcus, Veillonella, Neisseria, and Porphyromonas (Wade, 2013).
5. CONCLUSION
To the best of our knowledge, this is the first study describing bacterial community composition associated with dental hygiene habits and markers of periodontal health in a general population of young adults. Self‐reported gingival bleeding was associated with a higher abundance of well‐known and novel periodontal pathogens as compared to those reporting no gingival bleeding when brushing teeth. The study may provide useful information on self‐reported measures in studies where clinically based surveillance is unattainable. Our study highlights the importance of oral hygiene habits by showing reduced microbial diversity and lower abundance of known pathogens such as Peptostreptococcaceae and Bacteroides, as well as newly described pathogens such as Saccharibacteria (TM7) in those reporting good dental hygiene habits. These findings can be used to enhance the already good dental practices in young age.
AUTHOR CONTRIBUTIONS
Randi Jacobsen Bertelsen, Francisco Gomez Real, and Cecilie Svanes contributed to study conception and study design. Randi Jacobsen Bertelsen, Antonio Manuel Perez Barrionuevo, Francisco Gomez Real, Tamar Ringel‐Kulka, and Cecilie Svanes contributed to data acquisition. Randi Jacobsen Bertelsen, Antonio Manuel Perez Barrionuevo, Rajesh Shigdel, Huang Lin, and Stein Atle Lie contributed to data analyses and interpretation. Randi Jacobsen Bertelsen and Antonio Manuel Perez Barrionuevo drafted the manuscript. Randi Jacobsen Bertelsen, Antonio Manuel Perez Barrionuevo, Rajesh Shigdel, Stein Atle Lie, Huang Lin, Francisco Gomez Real, Tamar Ringel‐Kulka, Anne Nordrehaug Åstrøm, and Cecilie Svanes critically revised the manuscript and approved the final version.
FUNDING INFORMATION
This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (Grant Agreement No. 804199). It was funded also by the Research Council of Norway (Grant Nos 230827 and 273838). The Bergen RHINESSA study is funded by the Research Council of Norway (Grant Nos. 214123 and 228174), the Bergen Medical Research Foundation, the Western Norwegian Regional Health Authorities (Grant Nos 912011, 911892, and 911631), the Norwegian Labour Inspection, and the Norwegian Asthma and Allergy Association.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest for this article.
ETHICS STATEMENT
Ethical approval was obtained from the Regional Committee for Medical and Health Research Ethics in Western Norway (approval # 2012/1077). All participants provided written informed consent.
Supporting information
Appendix S1. Supporting information.
ACKNOWLEDGEMENTS
We thank Andrea Azcarate‐Peril at the UNC Microbiome Core Facility (Chapel Hill, NC) and Jeff Roach at the UNC Research ITS (Chapel Hill, NC) for contributing with laboratory and statistical analyses of the gingival samples.
Bertelsen, R. J. , Barrionuevo, A. M. P. , Shigdel, R. , Lie, S. A. , Lin, H. , Real, F. G. , Ringel‐Kulka, T. , Åstrøm, A. N. , & Svanes, C. (2022). Association of oral bacteria with oral hygiene habits and self‐reported gingival bleeding. Journal of Clinical Periodontology, 49(8), 768–781. 10.1111/jcpe.13644
Funding information Bergen Medical Research Foundation; European Research Council, Grant/Award Number: 804199; Norwegian Asthma and Allergy Association; Norwegian Labour Inspection; Research Council of Norway, Grant/Award Numbers: 214123, 228174; Western Norwegian Regional Health Authorities, Grant/Award Numbers: 912011, 911892, 911631
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in Dryad at https://datadryad.org/stash/share/gNf53i3Vg2tuf6jD8j0CF1K7K2m746Bp6vYllFdkK7U, reference number 10.5061/dryad.r2280gbfh.
REFERENCES
- Abusleme, L. , Dupuy, A. K. , Dutzan, N. , Silva, N. , Burleson, J. A. , Strausbaugh, L. D. , Gamonal, J. , & Diaz, P. I. (2013). The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. The ISME Journal, 7(5), 1016–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainamo, J. , Barmes, D. , Beagrie, G. , Cutress, T. , Martin, J. , & Sardo‐Infirri, J. (1982). Development of the World Health Organization (WHO) community periodontal index of treatment needs (CPITN). International Dental Journal, 32(3), 281–291. [PubMed] [Google Scholar]
- Anderson, M. J. (2006). Distance‐based tests for homogeneity of multivariate dispersions. Biometrics, 62(1), 245–253. [DOI] [PubMed] [Google Scholar]
- Anderson, M. J. , & Walsh, D. C. I. (2013). Permanova, anosim, and the mantel test in the face of heterogeneous dispersions: What null hypothesis are you testing? Ecological Monographs, 83(4), 557–574. [Google Scholar]
- Astrom, A. N. , & Rise, J. (2001). Socio‐economic differences in patterns of health and oral health behaviour in 25 year old norwegians. Clinical Oral Investigations, 5(2), 122–128. [DOI] [PubMed] [Google Scholar]
- Bamashmous, S. , Kotsakis, G. A. , Kerns, K. A. , Leroux, B. G. , Zenobia, C. , Chen, D. , Trivedi, H. M. , McLean, J. S. , & Darveau, R. P. (2021). Human variation in gingival inflammation. Proceedings of the National Academy of Sciences of the United States of America, 118(27), e2012578118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich, N. A. , Kaehler, B. D. , Rideout, J. R. , Dillon, M. , Bolyen, E. , Knight, R. , Huttley, G. A. , & Gregory, C. J. (2018). Optimizing taxonomic classification of marker‐gene amplicon sequences with QIIME 2's q2‐feature‐classifier plugin. Microbiome, 6(1), 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen, E. , Rideout, J. R. , Dillon, M. R. , Bokulich, N. A. , Abnet, C. C. , Al‐Ghalith, G. A. , Alexander, H. , Alm, E. J. , Arumugam, M. , Asnicar, F. , Bai, Y. , Bisanz, J. E. , Bittinger, K. , Brejnrod, A. , Brislawn, C. J. , Brown, C. T. , Callahan, B. J. , Caraballo‐Rodríguez, A. M. , Chase, J. , … Caporaso, J. G. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology, 37(8), 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J. G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F. D. , Costello, E. K. , Fierer, N. , Pena, A. G. , Goodrich, J. K. , Gordon, J. I. , Huttley, G. A. , Kelley, S. T. , Knights, D. , Koenig, J. E. , Ley, R. E. , Lozupone, C. A. , McDonald, D. , Muegge, B. D. , Pirrung, M. , … Knight, R. (2010). QIIME allows analysis of high‐throughput community sequencing data. Nature Methods, 7(5), 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo, A. P. V. , Boches, S. K. , Cotton, S. L. , Goodson, J. M. , Kent, R. , Haffajee, A. D. , Socransky, S. S. , Hasturk, H. , Van Dyke, T. E. , Dewhirst, F. , & Paster, B. J. (2009). Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. Journal of Periodontology, 80(9), 1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo, A. P. V. , & Tanner, A. C. R. (2019). The role of bacterial biofilms in dental caries and periodontal and peri‐implant diseases: A historical perspective. Journal of Dental Research, 98(4), 373–385. [DOI] [PubMed] [Google Scholar]
- Dalal, S. R. , & Chang, E. B. (2014). The microbial basis of inflammatory bowel diseases. The Journal of Clinical Investigation, 124(10), 4190–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, R. P. , Erb‐Downward, J. R. , Freeman, C. M. , McCloskey, L. , Falkowski, N. R. , Huffnagle, G. B. , & Curtis, J. L. (2017). Bacterial topography of the healthy human lower respiratory tract. MBio, 8(1), e02287‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, U. , Rogall, T. , Blocker, H. , Emde, M. , & Bottger, E. C. (1989). Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Research, 17(19), 7843–7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eke, P. I., Dye, B. A., Wei, L., Slade, G. D., Thornton‐Evans, G. O., Beck, J. D., Taylor, G. W., Borgnakke, W. S., Page, R. C., & Genco, R. J. (2013). Self‐reported measures for surveillance of periodontitis. Journal of dental research, 92(11), 1041‐1047. [DOI] [PubMed]
- Escapa, I. F. , Chen, T. , Huang, Y. , Gajare, P. , Dewhirst, F. E. , Lemon, K. P. , & Xu, J. (2018). New insights into human nostril microbiome from the expanded human oral microbiome database (eHOMD): A resource for the microbiome of the human aerodigestive tract. mSystems, 3(6), e00187–e00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez de Grado, G. , Ehlinger, V. , Godeau, E. , Sentenac, M. , Arnaud, C. , Nabet, C. , & Monsarrat, P. (2018). Socioeconomic and behavioral determinants of tooth brushing frequency: Results from the representative French 2010 HBSC cross‐sectional study. Journal of Public Health Dentistry, 78(3), 221–230. [DOI] [PubMed] [Google Scholar]
- Fierer, N. , Hamady, M. , Lauber, C. L. , & Knight, R. (2008). The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proceedings of the National Academy of Sciences of the United States of America, 105(46), 17994–17999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frencken, J. E. , Sharma, P. , Stenhouse, L. , Green, D. , Laverty, D. , & Dietrich, T. (2017). Global epidemiology of dental caries and severe periodontitis – A comprehensive review. Journal of Clinical Periodontology, 44(Suppl. 18), S94–s105. [DOI] [PubMed] [Google Scholar]
- Gallie, A. (2019). Home use of interdental cleaning devices and toothbrushing and their role in disease prevention. Evidence‐Based Dentistry, 20(4), 103–104. [DOI] [PubMed] [Google Scholar]
- Gloor, G. B. , Macklaim, J. M. , Pawlowsky‐Glahn, V. , & Egozcue, J. J. (2017). Microbiome datasets are compositional: And this is not optional. Frontiers in Microbiology, 8, 2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Real, F. , Perez Barrionuevo, L. , Franklin, K. , Lindberg, E. , Bertelsen, R. J. , Benediktsdottir, B. , Forsberg, B. , Gislason, T. , Jogi, R. , Johannessen, A. , Omenaas, E. , Saure, E. , Schlünssen, V. , Skorge, T. D. , Torén, K. , Saavedra, A. P. , Svanes, Ø. , Åstrøm, A. N. , Janson, C. , & Svanes, C. (2016). The association of gum bleeding with respiratory health in a population based study from Northern Europe. PLoS One, 11(1), e0147518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulão, B. , MacLennan, G. S. , & Ramsay, C. R. (2021). Have you had bleeding from your gums? Self‐report to identify gINGival inflammation (the SING diagnostic accuracy and diagnostic model development study). Journal of Clinical Periodontology, 48(7), 919–928. [DOI] [PubMed] [Google Scholar]
- Harvey, J. D. (2017). Periodontal microbiology. Dental Clinics of North America, 61(2), 253–269. [DOI] [PubMed] [Google Scholar]
- Heinrich, J. , Thiering, E. , Jorres, R. A. , Schulz, H. , Kuhnisch, J. , & Standl, M. (2019). Lung function and oral health in adolescents. The European Respiratory Journal, 53(3), 1801951. [DOI] [PubMed] [Google Scholar]
- Holm, S. (1979). A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics, 6(2), 65–70. [Google Scholar]
- Kistler, J. O. , Booth, V. , Bradshaw, D. J. , & Wade, W. G. (2013). Bacterial community development in experimental gingivitis. PLoS One, 8(8), e71227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, P. S. , Griffen, A. L. , Barton, J. A. , Paster, B. J. , Moeschberger, M. L. , & Leys, E. J. (2003). New bacterial species associated with chronic periodontitis. Journal of Dental Research, 82(5), 338–344. [DOI] [PubMed] [Google Scholar]
- Lin, H. , & Peddada, S. D. (2020). Analysis of compositions of microbiomes with bias correction. Nature Communications, 11(1), 3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B. , Faller, L. L. , Klitgord, N. , Mazumdar, V. , Ghodsi, M. , Sommer, D. D. , Gibbons, T. R. , Treangen, T. J. , Chang, Y. C. , Li, S. , Stine, O. C. , Hasturk, H. , Kasif, S. , Segrè, D. , Pop, M. , & Amar, S. (2012). Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One, 7(6), e37919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken, B. A. , & Nathalia, G. M. (2021). Phylum Synergistetes in the oral cavity: A possible contributor to periodontal disease. Anaerobe, 68, 102250. [DOI] [PubMed] [Google Scholar]
- Park, O. J. , Yi, H. , Jeon, J. H. , Kang, S. S. , Koo, K. T. , Kum, K. Y. , Chun, J. , Yun, C. H. , & Han, S. H. (2015). Pyrosequencing analysis of subgingival microbiota in distinct periodontal conditions. Journal of Dental Research, 94(7), 921–927. [DOI] [PubMed] [Google Scholar]
- Patino, D. , McQuistan, M. R. , Qian, F. , Hernandez, M. , Weber‐Gasparoni, K. , & Macek, M. D. (2018). Oral health knowledge levels of Hispanics in Iowa. Journal of the American Dental Association (1939), 149(12), 1038–1048. [DOI] [PubMed] [Google Scholar]
- Perez Barrionuevo, A. M. , Gomez Real, F. , Igland, J. , Johannessen, A. , Omenaas, E. , Franklin, K. A. , Perez Barrionuevo, L. , Astrom, A. N. , Svanes, C. , & Bertelsen, R. J. (2018). Periodontal health status and lung function in two Norwegian cohorts. PLoS One, 13(1), e0191410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Chaparro, P. J. , Gonçalves, C. , Figueiredo, L. C. , Faveri, M. , Lobão, E. , Tamashiro, N. , Duarte, P. , & Feres, M. (2014). Newly identified pathogens associated with periodontitis: A systematic review. Journal of Dental Research, 93(9), 846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse, P. S. , McCracken, G. I. , Gwynnett, E. , Steen, N. D. , Guentsch, A. , & Heasman, P. A. (2007). Does tooth brushing influence the development and progression of non‐inflammatory gingival recession? A systematic review. Journal of Clinical Periodontology, 34(12), 1046–1061. [DOI] [PubMed] [Google Scholar]
- Ramos, R. Q. , Bastos, J. L. , & Peres, M. A. (2013). Diagnostic validity of self‐reported oral health outcomes in population surveys: Literature review. Revista Brasileira de Epidemiologia, 16(3), 716–728. [DOI] [PubMed] [Google Scholar]
- Sakki, T. K. , Knuuttila, M. L. , & Anttila, S. S. (1998). Lifestyle, gender and occupational status as determinants of dental health behavior. Journal of Clinical Periodontology, 25(7), 566–570. [DOI] [PubMed] [Google Scholar]
- Socransky, S. S. , Haffajee, A. D. , Cugini, M. A. , Smith, C. , & Kent, R. L., Jr. (1998). Microbial complexes in subgingival plaque. Journal of Clinical Periodontology, 25(2), 134–144. [DOI] [PubMed] [Google Scholar]
- Svanes, C. , Johannessen, A. , Bertelsen, R. J. , Dharmage, S. , Benediktsdóttir, B. , Malinovschi, A. , Bråback, L. , Holm, M. , Jogi, N. O. , Martinez‐Moratalla Rovira, J. , Sanchez, J. L. , Janson, C. , Real, F. G. , & Schlünssen, V. (2022). Cohort profile: The multi‐generation respiratory health in northern Europe, Spain and Australia (RHINESSA) cohort. BMJ Open 12:e059434. doi:10.1136/bmjopen‐2021‐059434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombelli, L. , Farina, R. , Silva, C. O. , & Tatakis, D. N. (2018). Plaque‐induced gingivitis: Case definition and diagnostic considerations. Journal of Clinical Periodontology, 45(Suppl. 20), S44–S67. [DOI] [PubMed] [Google Scholar]
- Valm, A. M. (2019). The structure of dental plaque microbial communities in the transition from health to dental caries and periodontal disease. Journal of Molecular Biology, 431(16), 2957–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen, J. I. , Muikku, T. , Simila, T. , Cinar, A. B. , & Pohjola, V. (2019). Physical activity, BMI and oral health behaviour among adolescents: Finnish School Health Promotion Study. European Journal of Public Health, 29(2), 296–302. [DOI] [PubMed] [Google Scholar]
- Wade, W. G. (2013). The oral microbiome in health and disease. Pharmacological Research, 69(1), 137–143. [DOI] [PubMed] [Google Scholar]
- Worthington, H. V. , MacDonald, L. , Poklepovic Pericic, T. , Sambunjak, D. , Johnson, T. M. , Imai, P. , & Clarkson, J. E. (2019). Home use of interdental cleaning devices, in addition to toothbrushing, for preventing and controlling periodontal diseases and dental caries. The Cochrane Database of Systematic Reviews, 4, CD012018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ximénez‐Fyvie, L. A. , Haffajee, A. D. , & Socransky, S. S. (2000). Comparison of the microbiota of supra‐ and subgingival plaque in health and periodontitis. Journal of Clinical Periodontology, 27(9), 648–657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting information.
Data Availability Statement
The data that support the findings of this study are openly available in Dryad at https://datadryad.org/stash/share/gNf53i3Vg2tuf6jD8j0CF1K7K2m746Bp6vYllFdkK7U, reference number 10.5061/dryad.r2280gbfh.


