Abstract
The pro‐inflammatory cytokine interleukin 17 (IL‐17), that is mainly produced by Th17 cells, has been recognized as a key regulator in multiple sclerosis (MS) and experimental autoimmune encephalomyelitis (EAE). Reactive astrocytes stimulated by proinflammatory cytokines including IL‐17 are involved in blood brain barrier destruction, inflammatory cells infiltration and spinal cord injury. However, the role of long non‐coding RNAs (lncRNAs) induced by IL‐17 in the pathogenesis of MS and EAE remains unknown. Herein, we found that an IL‐17‐induced lncRNA AK018453 promoted TGF‐β receptor‐associated protein 1 (TRAP1) expression and Smad‐dependent signaling in mouse primary astrocytes. Knockdown of AK018453 significantly suppressed astrocytosis, attenuated the phosphorylation of Smad2/3, reduced NF‐κB p65 and CBP/P300 binding to the TRAP1 promoter, and diminished pro‐inflammatory cytokine production in the IL‐17‐treated astrocytes. AK018453 knockdown in astrocytes by a lentiviral vector in vivo dramatically inhibited inflammation and prevented the mice from demyelination in the spinal cord during the progression of EAE. Together, these results suggest that AK018453 regulates IL‐17‐dependent inflammatory response in reactive astrocytes and potentially promotes the pathogenesis of EAE via the TRAP1/Smad pathway. Targeting this pathway may have a therapeutic potential for intervening inflammatory demyelinating diseases.
Keywords: astrocytes, experimental autoimmune encephalomyelitis, lncRNA AK018453, multiple sclerosis, TGF‐β receptor‐associated protein 1
Main Points
AK018453, an upregulated lncRNA in IL‐17‐activated astrocytes, enhances TRAP1 expression, triggers Smad signaling in the reactive astrocytes and in EAE mice.
AK018453 knockdown in astrocytes dramatically inhibited inflammation and prevented the mice from demyelination during the progression of EAE.
Abbreviations
- CNS
central nervous system
- Ctrl‐shRNA
scrambled control shRNA
- CXCL10
C‐X‐C motif chemokine 10
- EAE
experimental autoimmune encephalomyelitis
- GFAP
glial fibrillary acidic protein
- IL‐17
interleukin‐17
- LFB
luxol fast blue
- LncRNA
long non‐coding RNA
- MCP‐1
monocyte chemoattractant protein‐1
- MOG
myelin oligodendrocyte glycoprotein
- MS
multiple sclerosis
- NC
negative control
- shRNA
short hairpin RNA
- TGF‐β
transforming growth factor‐β
- TNF‐α
tumor necrosis factor‐α
- TRAP1
TGF‐β receptor‐associated protein 1
1. INTRODUCTION
Multiple sclerosis (MS) and its animal model, experimental autoimmune encephalomyelitis (EAE) are a central nervous system (CNS) demyelinating disease accompanied by chronic inflammation and axonal degradation, mainly resulting from an autoimmune attack on the white matter (Faissner et al., 2019; Hafler, 2004). Autoreactive CD4+T effector cells including helper T cell 1 (Th1) and Th17 cell are believed as the major effector cells, which infiltrate into the CNS and initiate MS pathology (Dong & Yong, 2019; Yi et al., 2019). In addition, glial cells are now recognized to be important players contributing to MS (Baecher‐Allan et al., 2018; Dong & Yong, 2019; Yi et al., 2019).
Astrocytes are the most abundant glial cells in the CNS, and play a major role in supporting the neural transmission, maintaining the function and the survival of neurons. During the CNS inflammation, astrocytes secrete pro‐inflammatory cytokines (i.e., IL‐6, TNF‐α, MCP‐1, and CXCL10) and mediate immune cell infiltration into the CNS, resulting in extending further inflammatory reaction (Giovannoni & Quintana, 2020). Furthermore, astrocytes proliferate and enhance the expression of glial fibrillary acidic protein (GFAP) in an activated process termed reactive astrogliosis, a vital cause of the plaque formation of MS (Dong & Benveniste, 2001; Giovannoni & Quintana, 2020; Nair et al., 2008). In response to interleukin‐17 (IL‐17), astrocytes are activated to produce a great number of pro‐inflammatory cytokines, which aggravates the blood–brain barrier (BBB) damage and facilitates autoimmune T cell and inflammatory cell migration into the CNS (Chen, Liu, & Zhong, 2020; Liu et al., 2014, 2019; Tzartos et al., 2008; Yan et al., 2012). Although the accumulating evidence has shown the crucial role of the activated astrocytes in CNS diseases, the mechanisms by which astrocyte activation involved in MS/EAE remains largely unclear.
Transforming growth factor beta (TGF‐β) is a pleiotropic cytokine that has been found to be dysregulated in many neurodegenerative disorders, including MS (Diniz et al., 2019; Lee et al., 2017; Parsa et al., 2016). TGF‐β signaling is mediated mainly by two serine threonine kinase receptors, TGFβRI and TGFβRII. Upon binding to TGFβRII, TGF‐β triggers phosphorylating Smad2 and Smad3 that in turn bind to Smad4, which then translocates to the nucleus and modulates the expression of target genes (Diniz et al., 2019). Recently, emerging data show that TGF‐β signaling is associated with immune‐mediated disease, including MS (Lee et al., 2017). In EAE mice, TGF‐β/Smad pathway in astrocytes is activated to promote the release of inflammatory factors resulting in an inflammatory microenvironment and to exacerbate EAE pathology (Diniz et al., 2019; Parsa et al., 2016; Yoshida et al., 2014). Conversely, blocking TGF‐β decreases the infiltration of inflammatory lymphocytes into the CNS and improves the pathogenesis of EAE (Lee et al., 2017). Of important, experimental evidence reveal that TGF‐β receptor‐associated protein 1 (TRAP1) has been identified as a specific molecular chaperone for interaction of Smad4 that is an essential protein for TGF‐β signaling activation (Wurthner et al., 2001). However, the functions and mechanisms of TRAP1 in modulating inflammatory response in astrocytes remains to be determined.
Long noncoding RNAs (lncRNAs) are transcripts that are more than 200 nucleotides in length but are not protein‐coded. Numerous studies have identified that lncRNAs play key roles in a plethora of cellular processes, including chromatin remodeling, RNA stability, gene transcription, protein translation, and immune response (Kopp & Mendell, 2018; Ransohoff et al., 2018; St Laurent et al., 2015). LncRNAs positively or negatively regulate the expression of some genes by interacting with chromatin‐modifying complexes to modulate the epigenetic landscape of chromatin (Ransohoff et al., 2018). LncRNAs interact with a large number of epigenetic modifier complexes such as PRC2, PRC1, Tip60/P400, ESET, and HDAC1 (Guttman et al., 2011; Khalil et al., 2009). In addition, some research report that a few lncRNAs interact with histone acetyltransferase P300 and nuclear transcription factor kappa B (NF‐κB) to regulate the expression of genes (Chen, Dong, et al., 2020; Li et al., 2018). It is confirmed that lncRNAs contribute to some neurodegenerative diseases, including MS (Wan et al., 2017; Zhang et al., 2016). However, the mechanisms that how lncRNAs regulate the activated astrocytes in MS/EAE process remain largely unknown.
In the present study, we found that TRAP1/Smad pathway was activated both in the spinal cord tissue of EAE mice (in vivo) and in the astrocytes stimulated with IL‐17 (in vitro). We analyzed lncRNA AK018453, an up‐regulated lncRNA in IL‐17‐activated astrocytes, and further determined its function and mechanism in regulating TRAP1/Smad pathway, the production of pro‐inflammatory cytokines and demyelination lesions of EAE mice. Our data indicated that AK018453 promoted TRAP1 expression through interaction with CBP/P300, which was enriched in the promoter of TRAP1 and triggered Smad signaling, resulting in the production and release of pro‐inflammatory cytokines and chemokines in large amounts in the IL‐17‐stimulated astrocytes. Knockdown of both TRAP1 and AK018453 downregulated the production of pro‐inflammatory cytokines in astrocytes and ameliorated the pathogenesis of EAE. Our findings suggest that targeting certain lncRNAs in astrocytes might be a potential strategy for clinical treatment of MS.
2. MATERIALS AND METHODS
2.1. Animal, antibodies and reagents
C57BL/6 mice (Shanghai Experimental Animal Center, Chinese Academy of Sciences) were bred and housed under specific pathogen‐free (SPF) conditions. All experimental procedures were approved by the Institutional Animal Use Committee of Chinese Academy of Sciences. The reagents were used in this study: anti‐GFAP antibody (Rabbit, ab7260, Abcam), anti‐GFAP antibody (Mouse, ab4648, Abcam), anti‐TRAP1 antibody (Rabbit, CL647‐10325, Proteintech), anti‐TGFβRII antibody (Rabbit, ab61213, Abcam), anti‐p‐Smad2/3 antibody (Rabbit, 8828, Cell Signaling Technology), anti‐Smad2/3 antibody (Rabbit, 3102, Cell Signaling Technology), anti‐Smad4 antibody (Rabbit, 9515, Cell Signaling Technology), anti‐NF‐κB p65 antibody (Rabbit, 8242, Cell Signaling Technology), anti‐CBP/P300 antibody (Rabbit, 4771s, Cell Signaling Technology), anti‐RNA Pol II antibody (Mouse, 17‐620, Sigma Aldrich), anti‐H3K27ac antibody (Rabbit, ab177178, Abcam), the secondary antibodies (Sigma Biotechnology), Myelin oligodendrocyte glycoprotein (MOG) amino acids 35–55 (MOG35–55 peptides, MEVGWYRSPFSRVVHLYRNGK) (China Peptides Co. Ltd), recombinant mouse IL‐17 (R&D Systems), and ELISA kits (BD Biosciences).
2.2. EAE induction and clinical evaluation
EAE mice model was performed as previously described (Liu et al., 2014, 2019). Briefly, C57BL/6 female mice with age of 6–8 weeks were immunized with MOG35–55 (300 μg) emulsified in complete Freund's adjuvant (Sigma Aldrich) containing 5 mg/ml of Mycobacterium tuberculosis (H37Ra strain, Difco). On day 0 and day 2, mice were intraperitoneally injected pertussis toxin (200 ng, Invitrogen), then monitored daily for clinical scores of EAE. The PBS was injected into mice as the negative control (NC).
2.3. Primary astrocyte culture
Primary mouse astrocytes were cultured as described previously (Liu et al., 2014, 2019, 2021). Astrocytes were synchronized with non‐serum culture media for 12–16 h prior to IL‐17 treatment.
2.4. Real‐time PCR assay
The extraction of total RNAs from the spinal cords of mouse or astrocytes and real‐time PCR assay were carried out as described previously (Liu et al., 2019). The levels of gene transcription were calculated with the 2−ΔΔCT method. The primers are listed in the Table S1.
2.5. Detection and analysis of lncRNA profiles
The lncRNA expression profiles were detected as previously described (Liu et al., 2018; Zhou et al., 2018). In brief, Mouse LncRNA Microarray v2.0 (8 × 60K, Arraystar) was performed by Kangchen Bio‐tech (Shanghai, China) to measure the global profiling of mouse LncRNAs in primary mouse astrocytes treated with or without IL‐17. Total 31,423 LncRNAs and 25,376 coding transcripts are detected by second‐generation microarray.
RNAs from astrocytes (50 ng/ml of IL‐17 treatment at 0, 3, and 6 h) were to run microarray analysis as previously described (Liu et al., 2018; Zhou et al., 2018). The acquired raw array images were processed using Agilent Feature Extraction software (version 11.0.1.1), and then normalized and analyzed by the GeneSpring GX v12.0 software package (Agilent Technologies). Differentially expressed lncRNAs were then demonstrated through fold‐change as well as p values calculated with t‐test. The threshold for up‐ and down‐regulation was fold change >2.0 and p value <.05.
2.6. Enzyme‐linked immunosorbent assay (ELISA)
ELISA was applied according to the manufacturer's instructions. Briefly, the serum of mice or the supernatant of cultured astrocytes was collected to detect the secretion level of TGF‐β, TNF‐α, monocyte chemoattractant protein‐1 (MCP‐1) and C‐X‐C motif chemokine 10 (CXCL10).
2.7. Western blot assay
As described previously (Liu et al., 2019), the total protein was extracted from the spinal cords of mouse as well as the cultured astrocytes. The expression level of protein in samples was normalized by GAPDH protein.
2.8. Co‐immunoprecipitation (Co‐IP) assay
Co‐IP assay was performed to assess the interaction between TRAP1 and Smad4 as described (Ma et al., 2019). Briefly, the lysates of cell or tissue in IP lysis buffer (pH 7.4, 0.025 M Tris, 0.15 M NaCl, 0.001 M EDTA, 1% NP40, and 5% glycerol) were incubated with anti‐TRAP1 (2 μg), anti‐Smad4 (2 μg) or IgG (2 μg) overnight at 4°C with shaking. The immune complex was incubated with protein A/G beads for 2 h at room temperature with mixing and then washed to remove the unbound immune complex. Finally, the bound immune complex was dissociated from the beads with 5× loading buffer to perform Western blotting assay.
2.9. Immunofluoresence assay (IFA)
Protein expression was performed by the immunofluoresence assay (IFA) as previously described (Zhou et al., 2019).
2.10. Lentivirus generation
The lentivirus vector system contains the basic element of HIV, 5′ LTR and 3′ LTR and other auxiliary components, such as WPRE (woodchuck hepatitis virus post transcriptional regulatory element). Recombinant lentiviral vectors carrying with EGFP and with astrocyte‐specific promoter of GFAP (Liu et al., 2019, 2021) were constructed by Gene Chem (Shanghai, China), which can cross the blood–brain barrier. To silence the mouse TRAP1 and lncRNA AK018453, shRNA sequences against them were designed. shRNA sequences were as followed: sh‐Ctrl, 5′‐TTC TCC GAA CGT GTC ACG T‐3′; sh‐AK018453, 5′‐GGG CAG TTC CAC TCA CTT CTT‐3′; sh‐TRAP1, 5′‐ACT CTA TAA CCC TGG TTA A‐3′.
2.11. RNA immunoprecipitation (RIP) assay
RIP assay was performed as previously described (Liu et al., 2021; Tsai et al., 2010). Briefly, after stimulating with IL‐17 (50 ng/ml), primary mouse astrocytes were treated with 0.3% formaldehyde in DMEM/F12 medium for 10 min at 37°C. Glycine was dissolved in PBS for 5 min at room temperature. The cell pellets were resuspended in RIPA buffer, incubated on ice with frequent vortex for 30 min, and then the lysate was centrifugated. Antibodies were added and incubated for 4 h at 4°C. The beads were resuspended in RIPA buffer and treated with proteinase K. Then, RNA samples were extracted with Trizol reagent. Co‐precipitated RNAs were purified by RNeasy Mini Kit and detected with qPCR assay. The data of retrieved RNAs is calculated from the subtraction of RT/input ratio and non‐RT/input ratio. The primers for RIP assay were listed in the Table S1.
2.12. Chromatin immunoprecipitation (ChIP) assay
ChIP assay was used as described previously (Liu et al., 2021; Tsai et al., 2010). Briefly, primary mouse astrocytes were stimulated with or without IL‐17 for 6 h, then cells were treated with 1% formaldehyde, and stopped by 1.25 mM of glycine. The antibodies used in ChIP assay were anti‐NF‐κB p65 antibody, anti‐H3K27ac antibody, anti‐Pol II antibody, anti‐CBP/P300 antibody, and normal mouse IgG. All immunoprecipitated chromatin DNA was analyzed by qPCR. The primers for ChIP assay were listed in the Table S1.
2.13. Histopathology assay
Histopathology assay was applied as previously described (Liu et al., 2014, 2019). Hematoxylin and eosin (H&E) and luxol fast blue (LFB) staining on 4 μm paraffin‐embedded spinal cord sections were performed to evaluate inflammation and demyelination, respectively. For immunofluorescent assay (IFA), frozen sections (10 μm) from spinal cord were incubated with anti‐GFAP antibody and anti‐TRAP1 antibody. Furthermore, the ultrastructural of spinal cords were observed under electron microscope (EM).
2.14. Statistical analysis
Statistical analysis was performed by GraphPad Prism version 5.0 software with the two‐tailed unpaired Student's t test or one‐way multiple‐range analysis of variance (ANOVA). A Mann–Whitney test was used for nonparametric data (EAE scoring). Data presented as mean ± standard error of mean (SEM), calculated for all points from at least three independent experiments in triplicates. Values of p < .05 was considered significant.
3. RESULTS
3.1. TRAP1/Smad pathway is activated and inflammatory response is upregulated in EAE mice
To determine whether the TRAP1/Smad pathway is activated in EAE, we firstly analyzed the expression of TRAP1 and the activation of TGF‐β/Smad pathway in spinal cords tissues. As shown in Figure 1a, the mRNA levels of TGF‐β, TRAP1 and TGFβRII were evidently increased by real‐time PCR assay in EAE mice, comparing with negative control (NC) mice. Then, Western blot assay was used to measure the protein expression of TRAP1, TGFβRII, Smad4, p‐Smad2/3, and GFAP. The results indicated that the levels of TRAP1, TGFβRII, and GFAP proteins were obviously elevated in the spinal cords of EAE mice (Figure 1b). Furthermore, the expression of Smad4 and the phosphorylation level of Smad2/3 (p‐Smad2/3) were markedly upregulated in EAE mice (Figure 1b). To further investigate the expression of TRAP1 protein in astrocytes, immunofluorescence assay (IFA) was employed to test TRAP1 co‐expression in GFAP positive cells. The results indicated that the protein expression of TRAP1 was significantly increased in GFAP‐positive cells (Figure 1c). Finally, the levels of mRNA transcription and secretion of pro‐inflammatory cytokines (TNF‐α, CXCL10, and MCP‐1) in the spinal cords and peripheral blood of EAE mice were assessed by real‐time PCR assay and ELISA, respectively. The results showed that mRNA transcription and secretion levels of TNF‐α, CXCL10 and MCP‐1 were dramatically increased along with the EAE process (Figure 1d,e). These results suggest that TRAP1/Smad pathway is activated in the reactive astrocytes, which was associated with the production of pro‐inflammatory cytokines and chemokines in the spinal cords of EAE mice.
FIGURE 1.
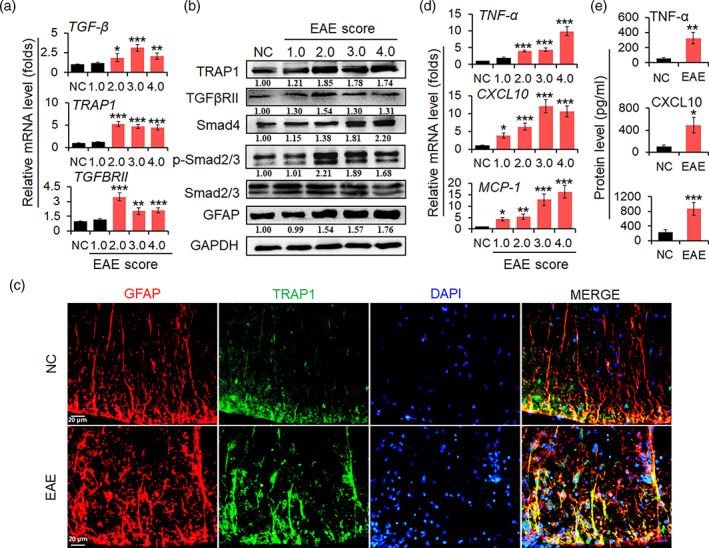
TRAP1/Smad signal pathway is activated in EAE mice. (a) The transcriptional level of TGF‐β, TRAP1, and TGFβRII mRNAs in spinal cords of EAE mice was tested by real‐time PCR assay. (b) The protein level of TRAP1, TGFβRII, Smad4, p‐Smad2/3, and GFAP in spinal cords was measured by Western blot assay. The relative level TRAP1, TGFβRII, Smad4, and GFAP was determined by quantitative densitometry compared to GAPDH; the relative level of phosphorylation Smad2/3 (p‐Smad2/3) was determined by quantitative densitometry after normalized to corresponding non‐phosphorylation ones. The relative value of proteins in the NC group was considered to be 1 for comparison. (c) IFA was used to test the expression of GFAP (red), TRAP1 (green), and nuclear staining of DAPI (blue) in astrocytes from spinal cords of EAE mice. Scale bars, 20 μm. (d) The mRNA levels of TNF‐α, CXCL10, and MCP‐1 in spinal cords were determined by real‐time PCR assay. (e) The secretion levels of TNF‐α, CXCL10, and MCP‐1 in the sera of mice were detected by ELISA. Data are represented as the mean ± SEM. *p < .05, **p < .01, and ***p < .001 versus NC group (n = 10 mice/group)
3.2. IL‐17 activates TRAP1/Smad pathway and promotes the production of pro‐inflammatory cytokines in astrocytes
Our and other groups have documented that IL‐17 is a key mediator in regulating autoimmune responses in MS/EAE through the production of a large number of inflammatory cytokines in astrocytes (Balasa et al., 2020; Kuo et al., 2020; Liu et al., 2019). To investigate the possible roles of IL‐17 in the activation of Smad pathway and the production of pro‐inflammatory cytokines, primary mouse astrocytes were stimulated with IL‐17 (50 ng/ml) in serum‐free media. First, real‐time PCR assay was used to measure the levels of TRAP1 and TGFβRII mRNA transcripts. As shown in Figure 2a, the mRNA level of TRAP1 and TGFβRII was significantly elevated in the astrocytes with IL‐17 stimulation at 3 and 6 h. Then, Western blot assay was used to analyze the protein expression of TRAP1, TGFβRII, Smad4, p‐Smad2/3, and GFAP. The results showed that the protein level of TRAP1, TGFβRII, and GFAP was dramatically increased at 6 h in the astrocytes treated with IL‐17 (Figure 2b). Moreover, the expression of Smad4 and p‐Smad2/3 was increased at 3 h and lasted for 24 h after IL‐17 stimulation (Figure 2b). It is interesting that the interaction of TRAP1 with Smad4 protein was increased in astrocytes by IL‐17 at 6 h (Figure 2c). In addition, the mRNA and protein level of TGF‐β was up‐regulated in the astrocytes with IL‐17 stimulation at 6 and 12 h, respectively (Figure 2d,e). Finally, the mRNA and protein levels of pro‐inflammatory cytokines such as TNF‐α, CXCL10, and MCP‐1 were simultaneously upregulated and peaked at 6 or 12 h in the astrocytes stimulated by IL‐17 (Figure 2d,e), respectively. These data suggest that IL‐17 triggers Smad signal and enhances the production of pro‐inflammatory cytokines in astrocytes.
FIGURE 2.
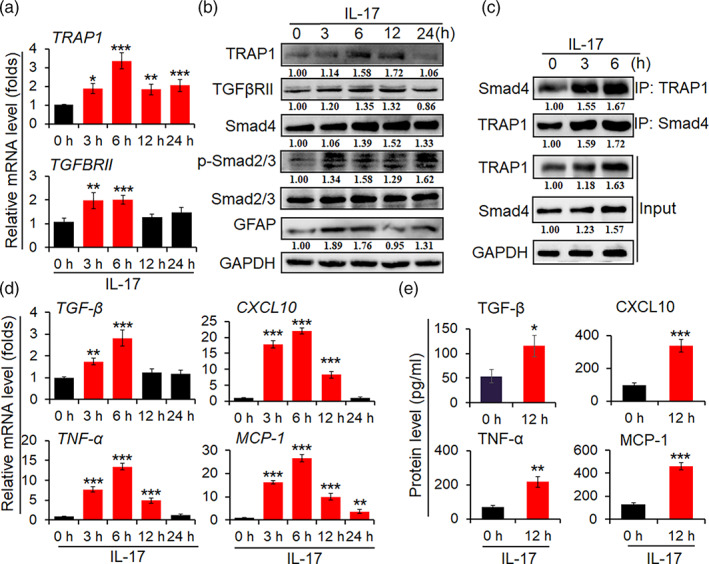
TRAP1/Smad signaling is activated in primary mouse astrocytes by IL‐17. Primary mouse astrocytes were cultured in a serum‐free medium overnight, prior to stimulating with IL‐17 (50 ng/ml) at indicated time point. (a) The mRNA levels of TRAP1 and TGFβRII were determined by real‐time PCR assay. (b) The protein levels of TRAP1, TGFβRII, Smad4, p‐Smad2/3, Smad2/3, and GFAP in astrocytes stimulated with IL‐17 were measured by Western blot assay. (c) Co‐immunoprecipitation (co‐IP) assay was employed to measure the interaction of TRAP1 with Smad4 in astrocytes treated with IL‐17. (d) The mRNA levels of TGF‐β, TNF‐α, CXCL10, and MCP‐1 in astrocytes with IL‐17 were measured by real‐time PCR assay. (e) The secretion levels of TGF‐β, TNF‐α, CXCL10, and MCP‐1 were measured by ELISA in the supernatant from astrocytes treated with IL‐17. The data are from three independent experiments and represented as the mean ± SEM. *p < .05, **p < .01, and ***p < .001 versus 0 h group treated with IL‐17 (n = 3)
3.3. Knockdown TRAP1 inhibits inflammatory response in IL‐17‐activated astrocytes
To further verify whether TRAP1 regulates inflammatory responses in astrocytes by IL‐17, lentivirus of TRAP1‐shRNA (TRAP1‐shRNA) specifically targeting astrocytes was generated, and then infected primary mouse astrocytes. As shown in Figure 3a, TRAP1 protein was significantly decreased in TRAP1‐shRNA group, compared with Ctrl‐shRNA group. Meanwhile, p‐Smad2/3, and Smad4 were dramatically decreased from TRAP1‐shRNA group in astrocytes stimulated with IL‐17 (50 ng/ml) (Figure 3a). Importantly, TRAP1 knockdown significantly inhibited the TRAP1 interaction with Smad4 in astrocytes stimulated with IL‐17 (Figure 3b). Moreover, the production of pro‐inflammatory cytokines of TNF‐α, CXCL10, and MCP‐1 was markedly reduced in TRAP1‐shRNA astrocytes compared to Ctrl‐shRNA ones (Figure 3c,d). These results indicate that IL‐17‐mediated the upregulation of TRAP1 is associated with the production of pro‐inflammatory cytokines in astrocytes.
FIGURE 3.
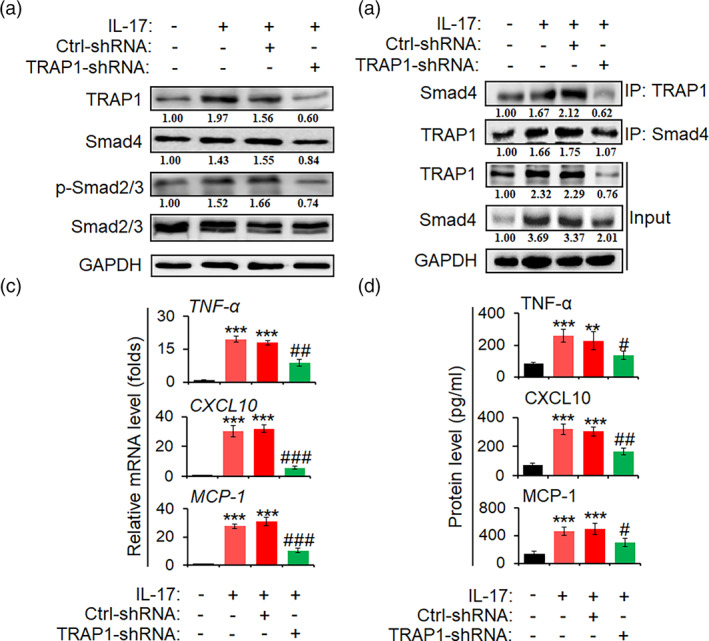
Knockdown of TRAP1 decreases inflammatory reaction in activated astrocytes primary mouse astrocytes were infected with recombinant lentivirus containing TRAP‐shRNA and ctrl‐shRNA sequence, for 72 h, respectively. And then, astrocytes were incubated in a serum‐free medium overnight followed by IL‐17 (50 ng/ml) treatment for 6 h. (a) Western blot assay was utilized to measure the protein level of TRAP1, Smad4, p‐Smad2/3, and Smad2/3. (b) Co‐immunoprecipitation (co‐IP) assay was designed to determine the interaction of TRAP1 with Smad4. (c) The mRNA levels of TNF‐α, CXCL10, and MCP‐1 in astrocytes were detected by real‐time PCR assay. (d) The secretion levels of TNF‐α, MCP‐1, and CXCL10 in the supernatant of astrocytes were tested by ELISA. The data are from three independent experiments and represented as mean ± SEM. **p < .01 and ***p < .001 versus no IL‐17 group; # p < .05, ## p < .01, and ### p < .001 versus ctrl‐shRNA + IL‐17 group (n = 3)
3.4. Knockdown of TRAP1 alleviates EAE pathogenesis in mice
To further explore the role of TRAP1 in the pathogenesis of EAE, 1 × 107 TU of lentivirus specifically targeting astrocytes with TRAP1‐shRNA (TRAP1‐shRNA) was injected into mice via the tail vein, and lentivirus containing Ctrl‐shRNA as shRNA control. After 7 days, EAE mice was immunized by MOG35–55. The clinical score indicated that TRAP1 knockdown not only suppressed the onset of EAE but also relieved illness (Figure 4a). Meanwhile, the protein level of TRAP1, Smad4, p‐Smad2/3, and GFAP was markedly decreased in the spinal cords of EAE mice in TRAP1‐shRNA group compared with Ctrl‐shRNA control mice (Figure 4b). The levels of pro‐inflammatory cytokines of TNF‐α, CXCL10, and MCP‐1 were significantly downregulated in TRAP1‐shRNA group (Figure 4c,d). Furthermore, H&E and LFB staining showed that the EAE mice in TRAP1‐shRNA possessed less inflammatory cell infiltration and demyelination lesion in the spinal cords, while Ctrl‐shRNA mice had more inflammatory cell infiltration and more severer demyelination lesions (Figure 4e,f). Specifically, the myelin sheath of the spinal cords in Ctrl‐shRNA group was disintegrated and damaged under EM, whereas only a slightly loosening of the sheath was shown in TRAP1‐shRNA group (Figure 4g). These data suggest that TRAP1 is involved in the activation and hyperplasia of astrocytes, the infiltration of inflammatory cells and the injury of spinal cords in EAE mice.
FIGURE 4.
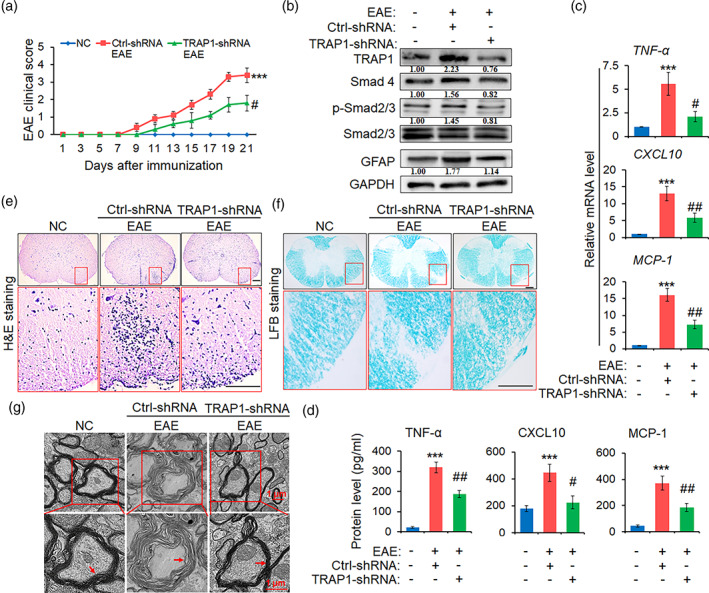
Knockdown of TRAP1 alleviates EAE pathogenesis in mice. Mice were injected to recombinant lentiviruses carrying the promoter of GFAP and TRAP‐shRNA or ctrl‐shRNA sequence, for 7 days, prior to MOG35–55 immunization for 21 days (n = 10 mice per group). (a) The clinical scores of EAE mice by TRAP‐shRNA and ctrl‐shRNA treatment. (b) Western blot assay was employed to detect the protein levels of TRAP1, Smad4, p‐Smad2/3, Smad2/3, and GFAP in the spinal cords. (c and d) The expression level of TNF‐α, CXCL10, and MCP‐1 in the spinal cords and peripheral blood of the TRAP‐shRNA and ctrl‐shRNA mice were measured by real‐time PCR assay and ELISA, respectively. The data are represented as mean ± SEM. ***p < .001 versus NC group; # p < .05 and ## p < .01 versus ctrl‐shRNA group (n = 10 mice/group). (e) Hematoxylin and eosin (H&E) staining was employed to evaluate the infiltrations of inflammatory cells in spinal cords (scale bars, 50 μm). (f) Luxol fast blue (LFB) staining was used to investigate the medullary sheath damages from spinal cords (scale bars, 50 μm). Red box areas in the upper rows are presented enlarged underneath. (g) Electron microscope (EM) was employed to observe the severe disruption or mild loosening of the medullary sheath in spinal cords. Red arrow indicates the changes of the medullary sheath. Scale bars, 1 μm
3.5. The analysis of lncRNA expression profiles in astrocytes treated with IL‐17
To investigate whether lncRNAs is involved in regulating inflammatory astrocytes, mouse lncRNA microarray was performed to analyze the change of lncRNA expression profile. As shown in Figure 5a, the Venn Diagrams exhibited that 222 lncRNAs were differentially co‐expressed in primary mouse astrocytes stimulated with IL‐17 (50 ng/ml) at 3 and 6 h. Meanwhile, a lncRNA named AK018453 was up‐regulated among the differentially‐expressed lncRNAs (Figure 5b). Then, real‐time PCR assay was used to verify the levels of AK018453 in primary mouse astrocytes treated with IL‐17 and in the spinal cord tissues of EAE mice. The results indicated that the level of AK018453 was significantly increased in astrocytes by IL‐17 at 3, 6, 12, and 24 h, and in EAE mice with clinical score 2.0, 3.0, and 4.0 (Figure 5c).
FIGURE 5.
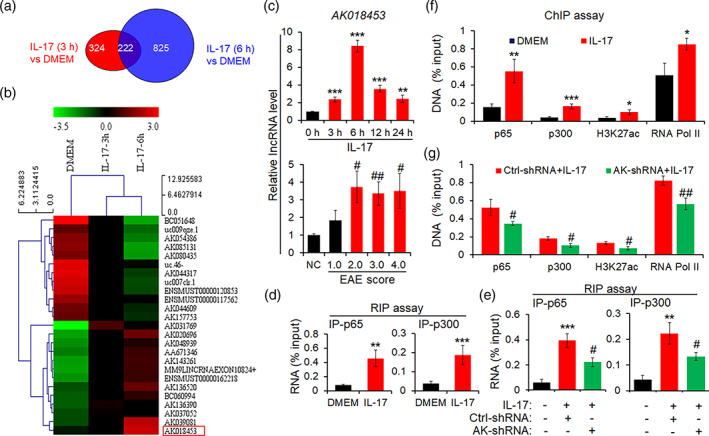
LncRNA AK018453 is upregulated and regulates TRPA1 expression in activated astrocytes by IL‐17. (a) Comparative lncRNA array analysis indicated that differentially upregulated lncRNAs occurred in the primary astrocytes treated with IL‐17 (50 ng/ml) at 3 and 6 h (2.0‐fold higher than DMEM group). (b) Hot map showed some differentially‐expressed lncRNAs in astrocytes stimulated with IL‐17. Red box circled differentially co‐upregulated lncRNA AK018453. (c) Real‐time PCR assay was employed to verify lncRNA AK018453 expression in primary mouse astrocytes treated by IL‐17 and spinal cords from EAE mice. Results were represented as mean ± SEM. **p < .01 and ***p < .001 versus 0 h group. # p < .05 and ## p < .01 versus NC group (n = 3). (d and e) RIP assay was performed to investigate the interaction of AK018453 with NF‐κB p65 and CBP/P300 in astrocytes stimulated with IL‐17 for 6 h, or prior to infection by lentiviruses AK018453‐shRNA (AK‐shRNA) and ctrl‐shRNA for 72 h. The data were from three independent experiments. **p < .01 and ***p < .001 versus DMEM. # p < .05 versus ctrl‐shRNA + IL‐17 group (n = 3). (f and g) ChIP assay analysis of NF‐κB p65, CBP/P300, H3K27ac, and RNA pol II enrichment on the promoter of TRAP1 gene in astrocytes stimulation with IL‐17 for 6 h, or prior to infection by lentiviruses AK‐shRNA and ctrl‐shRNA for 72 h. The data were from three independent experiments and represented as mean ± SEM. *p < .05, **p < .01, ***p < .001 versus DMEM; # p < .05 and ## p < .01 versus ctrl‐shRNA + IL‐17 group (n = 3)
3.6. LncRNA AK018453 regulates the TRAP1 expression
To explore the role of lncRNA in activated astrocytes and EAE mice, we first analyzed the sequence feature of AK018453. We found that the sequence of AK018453 with 1250 bp in length is the antisense sequence of TRAP1 gene at 4788–6035 locus (Figure S1a,b). The results suggested that AK018453 may regulate the TRAP1 expression.
To this end, RIP assay was utilized to measure the interaction of AK018453 with NF‐κB p65 or CBP/P300. As shown in Figure 5d, AK018453 dramatically interacted with NF‐κB p65 and CBP/P300 in activated astrocytes in response to IL‐17. Of importance, knockdown of AK018453 significantly inhibited the interaction with NF‐κB p65 and CBP/P300 (Figure 5e). Moreover, ChIP assay revealed that NF‐κB p65, CBP/P300, H3K27ac, and RNA Pol II were differentially enriched in the TRAP1 gene promoter in astrocytes by IL‐17 (Figure 5f). AK018453 knockdown significantly decreased NF‐κB p65 and CBP/P300 enrichment in the TRAP1 promoter (Figure 5g). Above data suggest that the interaction of AK018453 with CBP/P300 may be involved in epigenetically regulating the expression of TRAP1 gene.
3.7. LncRNA AK018453 is essential for TRAP1/Smad pathway activation and pro‐inflammatory cytokines production in IL‐17‐activated astrocytes
To assess the roles of AK018453 in regulating the activation of TRAP1/Smad signal and the production of pro‐inflammatory cytokines in activated astrocytes by IL‐17, the recombinant lentivirus of AK018453 shRNA (AK‐shRNA) specifically targeting astrocytes were performed to transduce primary mouse astrocytes. As shown in Figure 6a, knockdown of AK018453 suppressed significantly the level of TRAP1 and Smad4 protein, and decreased the phosphorylation of Smad2/3 in astrocytes treated with IL‐17 (50 ng/ml). Meanwhile, AK018453 knockdown reduced the interaction of TRAP1 with Smad4 (Figure 6b). In addition, the production of TNF‐α, CXCL10, and MCP‐1 was reduced in AK‐shRNA group (Figure 6c,d). These results suggest that AK018453 regulates TRAP1 expression and Smad signaling to mediate inflammatory response in astrocytes.
FIGURE 6.
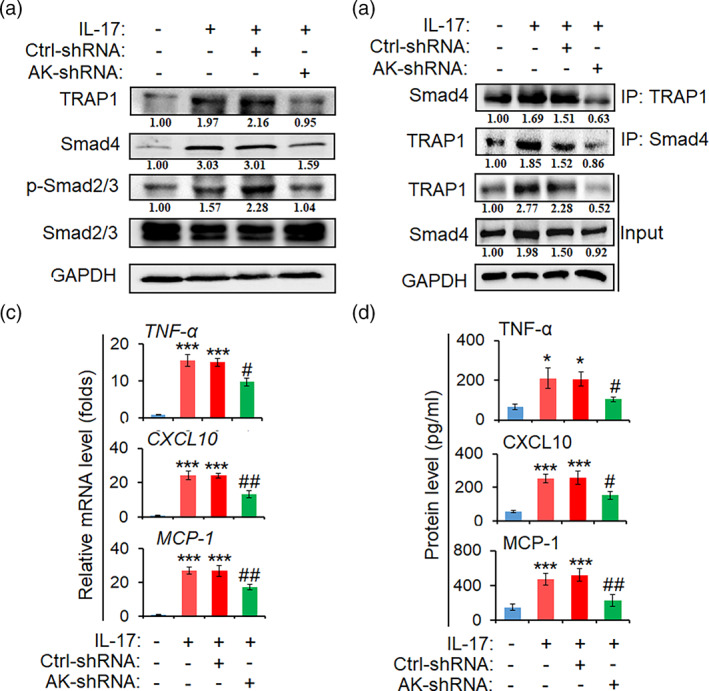
Knockdown of AK018453 decreases the activation of TRAP1/Smad pathway and the production of inflammation cytokines in IL‐17‐activated astrocytes. Primary mouse astrocytes were infected with recombinant lentivirus carrying GFAP promoter and AK‐shRNA or ctrl‐shRNA, for 72 h, respectively, followed by IL‐17 (50 ng/ml) treatment for 6 h. (a) Western blot assay was arranged to detect the protein level of TRAP1, Smad4, p‐Smad2/3, and Smad2/3. (b) Co‐immunoprecipitation (co‐IP) assay was employed to measure the interaction of TRAP1 with Smad4. (c) The mRNA levels of TNF‐α, CXCL10, and MCP‐1 in astrocytes were tested by real‐time PCR assay. (d) The secretion levels of TNF‐α, CXCL10, and MCP‐1 in the supernatant of astrocytes were measured by ELISA. The data are from three independent experiments and represented as mean ± SEM. *p < .05 and ***p < .001 versus no IL‐17 group; # p < .05 and ## p < .01 versus ctrl‐shRNA + IL‐17 group (n = 3)
3.8. AK018453 knockdown in astrocytes ameliorates the pathology of EAE mice
To further investigate whether AK018453 in astrocytes is associated with the pathogenesis of EAE, 1 × 107 lentiviruses of AK018453 shRNA (AK‐shRNA) (Figure 7a) specifically targeting astrocytes was injected to mice via the tail vein, 7 days before EAE induction. As shown in Figure 7b, AK018453 inhibition slowed down significantly EAE progression. Meanwhile, the expression of TRAP1, Smad4, p‐Smad2/3, and GFAP was dramatically down‐regulated in AK‐shRNA mice (Figure 7c). Furthermore, AK018453 knockdown decreased markedly the production of TNF‐α, CXCL10, and MCP‐1 by real‐time PCR assay and ELISA (Figure 7d,e). Additionally, H&E and LFB staining displayed that the inflammatory cell infiltration and demyelination lesion in white matter of the spinal cords were diminished in AK‐shRNA EAE mice, compared with the Ctrl‐shRNA controls (Figure 7f,g). The myelin sheath in Ctrl‐shRNA group was disintegrated under EM, whereas only a light loosening of the sheath was observed in AK‐shRNA group (Figure 7h). Thus, these data suggest that lncRNA AK018453 is involved in regulating TRAP1/Smad signal pathway, which mediates the reactive astrocytosis, inflammatory response and myelin damage.
FIGURE 7.
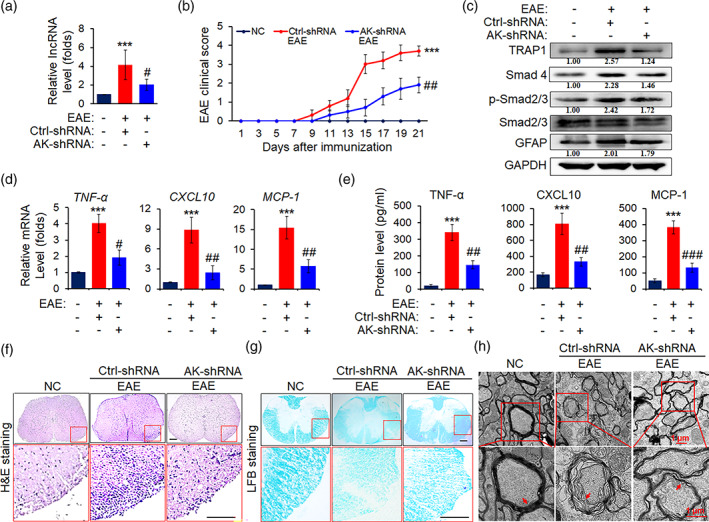
AK018453 knockdown lessens the pathology of EAE mice. On day 7 after administration with recombinant lentivirus carrying GFAP promoter and AK‐shRNA or ctrl‐shRNA sequence, respectively, mice were immunized with or without MOG35–55 for 21 days. (a) The level of lncRNA AK018453 was determined by real‐time PCR assay. ***p < .001 versus NC group; # p < .05 versus ctrl‐shRNA group (n = 10 mice/group). (b) The clinical scores for EAE mice infected with AK‐shRNA or ctrl‐shRNA (n = 10 mice per group). ***p < .001 versus NC group; ## p < .01 versus ctrl‐shRNA group. (c) Western blot assay was used to determine the protein levels of TRAP1, Smad4, p‐Smad2/3, Smad2/3, and GFAP in the spinal cords. (d and e) The production of TNF‐α, CXCL10, and MCP‐1 in the spinal cords and peripheral blood from the AK‐shRNA and ctrl‐shRNA mice was measured by real‐time PCR assay and ELISA assay, respectively. The data are represented as mean ± SEM. ***p < .001 versus NC group; # p < .05, ## p < .01, and ### p < .001 versus ctrl‐shRNA group (n = 10 mice/group). (f) Hematoxylin and eosin (H&E) staining was designed to observe the infiltrations of inflammatory cells in spinal cords (scale bars, 50 μm). (g) Luxol fast blue (LFB) staining was utilized to test the medullary sheath damages from spinal cords (scale bars, 50 μm). Red box areas in the upper rows are presented enlarged underneath. (h) Electron microscope (EM) was employed to investigate the severe disruption or mild loosening of the medullary sheath in spinal cords. Red arrow shows the changes of the medullary sheath. Scale bars, 1 μm
4. DISCUSSION
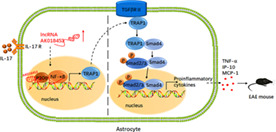
Over the past decades, lncRNAs have been known as key mediators contributing to multiple cellular events associated with neuroinflammation and neurodegeneration diseases. LncRNAs are abundantly expressed in mammalian CNS allowing a quick response to environmental or molecular changes (Riva et al., 2016), which epigenetically regulates the expression of some genes mainly by chromatin remodeling (Khalil et al., 2009; Ransohoff et al., 2018). In this study, we used lncRNA microarray to analyze differentially‐expressed lncRNAs in primary mouse astrocytes by IL‐17, and found that the lncRNA AK018453 was highly expressed. AK018453 is homologous partially to the antisense sequence of TRAP1 gene, but few studies have investigated its function and molecular mechanism. We discovered that downregulating the expression of AK018453 resulted in the reduction of IL‐17‐mediated inflammation and the amelioration of EAE pathogenesis through regulating TRAP1/Smad pathway (Figure 8). Therefore, our findings support the notion that lncRNAs involve in regulating the progression of MS/EAE.
FIGURE 8.
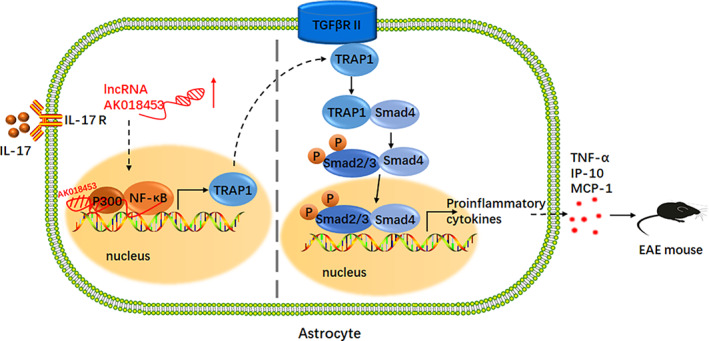
Schematic representation of lncRNA AK018453 contributing to the pathology of EAE mice via regulating TRAP1/Smad pathway. Response to IL‐17 stimulation, AK018453 is upregulated in astrocytes, which regulates epigenetically the expression of TRAP1 through interacting with CBP/P300, in turn promotes the production of pro‐inflammatory cytokines such as TNF‐α, CXCL10, and MCP‐1, thus aggravating the EAE progression
It is now accepted that neuroinflammation is a common feature to almost all neurodegeneration diseases, including MS. In the CNS, reactive astrocytes are vital mediator to construct the detrimental positive‐feedback loop of inflammation, which promotes the inflammatory cascades and accelerates neurological disorders (Baecher‐Allan et al., 2018). Response to inflammatory stress, reactive astrocytes express a great number of gene involved in inflammation, oxidative stress and BBB disruption (Wang et al., 2019). Accumulating evidence has shown that IL‐17 mediates astrocyte activation to produce pro‐inflammatory cytokines and chemokines, in turn facilitating immune cells migration to the CNS to aggravate the disruption of BBB and the damage of myelin sheath in the white matter (Chen, Liu, & Zhong, 2020; Liu et al., 2014, 2019; Tzartos et al., 2008; Yan et al., 2012). Herein, our results exhibited that IL‐17 increased the expression of GFAP and the secretion of pro‐inflammatory cytokines along with the pathological progression of EAE in mice.
TGF‐β/Smad pathway regulates immune response and inflammatory processes (Travis & Sheppard, 2014; Veldhoen et al., 2006; Wu et al., 2021), which are clinically critical in MS. Emerging evidence shows that the Smad signaling regulates the development and function of CD4+ T cells in MS lesions, which is associated with MS pathology (Gorelik & Flavell, 2002; Lee et al., 2017). TGF‐β and IL‐6 have been demonstrated to drive Th17 cell differentiation (Mangan et al., 2006; Veldhoen et al., 2006), and TGF‐β and IL‐4 also have been shown to drive Th9 cell differentiation in vitro (Veldhoen et al., 2008). Importantly, TGF‐β at the side of inflammation, promotes proliferation and cytokine production of Th1 cells to contribute to MS (Lee et al., 2017). However, the understanding of TGF‐β/Smad pathway in astrocytes is still very limited. In this study, our data showed that the high expression of TGF‐β, TRAP1, TGFβRII, p‐Smad2/3, and Smad4 was consistent with GFAP and pro‐inflammatory cytokines production in the spinal cords of EAE mice and in activated astrocytes by IL‐17. Meanwhile, the interaction of TRAP1 with Smad4 occurred in EAE (in vivo) and in IL‐17‐activated astrocytes (in vitro). Of importance, TRAP1 knockdown in astrocytes not only suppressed pro‐inflammatory cytokine production both in vivo and in vitro, but also inhibited inflammatory cell infiltration and demyelination lesion in EAE mice. Nevertheless, the underlying function and mechanisms of TRAP1 in astrocytes are far from known in MS/EAE.
Recent studies indicate lncRNAs are involved in multiple biological processes including immune responses. In CNS, lncRNAs contribute to the brain development, neuron function as well as neuroinflammation (Riva et al., 2016; Tripathi et al., 2021; Wu et al., 2013). Accumulating evidence has shown that lncRNAs as key mediators associated with the onset and progression of MS (Sun et al., 2017; Yang et al., 2018; Zhang et al., 2017), but less data focus on the regulation of astrocytes by lncRNAs. Herein, we assumed that IL‐17‐induced lncRNAs can participate in astrocyte activation, inflammatory response and the pathogeneses of EAE. Our studies indicated that the lncRNA AK018453 is complementary to the sense chain of TRAP1 gene, and was highly‐expressed in astrocytes activated by IL‐17 and in EAE mice. Knockdown AK018453 inhibited the TRAP1 expression and Smad pathway as well as attenuated reactive astrocytosis both in vivo and in vitro, resulted in the alleviation of inflammatory response and demyelination lesion in EAE mice. However, it is still unknown about the mechanisms how lncRNAs regulate TRAP1 in astrocytes. Some studies show that lncRNAs epigenetically regulate gene expression via histone modification. lncRNAs are largely associated with the polycomb repressive complex 2 (PRC2) complex, the CoREST/REST complex and the CBP/p300 complex (Khalil et al., 2009; Tsai et al., 2010; Zhou et al., 2018). In this study, we showed that AK018453 interacted with CBP/P300 and NF‐κB p65 in the promoter of TRAP1gene to promote TRAP1 expression. These results suggest that lncRNA AK018453 might regulate epigenetically TRAP1 expression to trigger Smad signaling in activated astrocytes and EAE.
Taken together, our present data show that IL‐17‐induced lncRNA AK018453 contributes to TRAP1 expression via the interaction with CBP/P300 and NF‐κB p65, and then activates Smad pathway in astrocytes, which is associated with the production pro‐inflammatory cytokines and the pathology of EAE. Therefore, these data support that lncRNAs in astrocytes may be involved in neuroinflammation and pathogenesis of MS.
AUTHOR CONTRIBUTIONS
Xiaomei Liu, Feng Zhou, and Ying Yang designed research; Qingxiu Zhang, Ying Yang, Yingyu Chen, Ruixue Lv, Yifan Wang, Xiangyang Li, and Menglu Zhou performed research; Ying Yang, Yingyu Chen, Xiaomei Liu, Feng Zhou, Qian Yu, and Xiaotian Wang analyzed data; Xiaomei Liu, Feng Zhou, Suping Qin, Hongjuan You, Xiaocui Li, Yuang Wang, and Qingxiu Zhang drafted and revised the paper.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
Supporting information
Figure S1 The feature of lncRNA AK018453. (a) The sequence of AK018453 with 1250 bp in length. (b) AK018453 is complementary to the sense chain of TRAP1 gene
Table S1 The list of primer sequences for the real‐time PCR assay. The table contains all primer sequences for the real‐time PCR assay
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (81971179 to Xiaomei Liu, 82071304 and 81671149 to Qingxiu Zhang), the Natural Science Foundation of Jiangsu Province (BK20191463 to Xiaomei Liu, BK20211347 to Hongjuan You), Jiangsu Commission of Health (Z2019035 to Feng Zhou), Jiangsu Provincial Department of Education (20KJA320004 to Feng Zhou, 21KJA310004 to Hongjuan You), the Jiangsu Graduate Innovation Program (KYCX20_2469 to Menglu Zhou), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (2017 PAPD).
Zhang, Q. , Yang, Y. , Chen, Y. , Wang, Y. , Qin, S. , Lv, R. , Zhou, M. , Yu, Q. , Li, X. , Li, X. , Wang, X. , You, H. , Wang, Y. , Zhou, F. , & Liu, X. (2022). The LncRNA AK018453 regulates TRAP1/Smad signaling in IL‐17‐activated astrocytes: A potential role in EAE pathogenesis. Glia, 70(11), 2079–2092. 10.1002/glia.24239
Qingxiu Zhang, Ying Yang, and Yingyu Chen contributed equally to this work.
Funding information Jiangsu Commission of Health, Grant/Award Number: Z2019035; Jiangsu Provincial Department of Education, Grant/Award Numbers: 20KJA320004, 21KJA310004; National Natural Science Foundation of China, Grant/Award Numbers: 81671149, 81971179, 82071304; The Jiangsu Graduate Innovation Program, Grant/Award Number: KYCX20_2469; The Natural Science Foundation of Jiangsu Province, Grant/Award Numbers: BK20191463, BK20211347; The Priority Academic Program Development of Jiangsu Higher Education Institutions, Grant/Award Number: 2017 PAPD
Contributor Information
Feng Zhou, Email: zhoufeng_xzhmu@163.com.
Xiaomei Liu, Email: lxmlxm_hi@xzhmu.edu.cn.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
REFERENCES
- Baecher‐Allan, C. , Kaskow, B. J. , & Weiner, H. L. (2018). Multiple sclerosis: Mechanisms and immunotherapy. Neuron, 97(4), 742–768. 10.1016/j.neuron.2018.01.021 [DOI] [PubMed] [Google Scholar]
- Balasa, R. , Barcutean, L. , Balasa, A. , Motataianu, A. , Roman‐Filip, C. , & Manu, D. (2020). The action of TH17 cells on blood brain barrier in multiple sclerosis and experimental autoimmune encephalomyelitis. Human Immunology, 81(5), 237–243. 10.1016/j.humimm.2020.02.009 [DOI] [PubMed] [Google Scholar]
- Chen, J. , Liu, X. , & Zhong, Y. (2020). Interleukin‐17A: The key cytokine in neurodegenerative diseases. Frontiers in Aging Neuroscience, 12, 566922. 10.3389/fnagi.2020.566922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. , Dong, W. H. , Qiu, Z. M. , & Li, Q. G. (2020). The monocyte‐derived exosomal CLMAT3 activates the CtBP2‐p300‐NF‐kappaB transcriptional complex to induce proinflammatory cytokines in ALI. Molecular Therapy Nucleic Acids, 21, 1100. 10.1016/j.omtn.2020.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diniz, L. P. , Matias, I. , Siqueira, M. , Stipursky, J. , & Gomes, F. C. A. (2019). Astrocytes and the TGF‐beta1 pathway in the healthy and diseased brain: A double‐edged sword. Molecular Neurobiology, 56(7), 4653–4679. 10.1007/s12035-018-1396-y [DOI] [PubMed] [Google Scholar]
- Dong, Y. , & Benveniste, E. N. (2001). Immune function of astrocytes. Glia, 36(2), 180–190. 10.1002/glia.1107 [DOI] [PubMed] [Google Scholar]
- Dong, Y. , & Yong, V. W. (2019). When encephalitogenic T cells collaborate with microglia in multiple sclerosis. Nature Reviews. Neurology, 15(12), 704–717. 10.1038/s41582-019-0253-6 [DOI] [PubMed] [Google Scholar]
- Faissner, S. , Plemel, J. R. , Gold, R. , & Yong, V. W. (2019). Progressive multiple sclerosis: From pathophysiology to therapeutic strategies. Nature Reviews. Drug Discovery, 18(12), 905–922. 10.1038/s41573-019-0035-2 [DOI] [PubMed] [Google Scholar]
- Giovannoni, F. , & Quintana, F. J. (2020). The role of astrocytes in CNS inflammation. Trends in Immunology, 41(9), 805–819. 10.1016/j.it.2020.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik, L. , & Flavell, R. A. (2002). Transforming growth factor‐beta in T‐cell biology. Nature Reviews. Immunology, 2(1), 46–53. 10.1038/nri704 [DOI] [PubMed] [Google Scholar]
- Guttman, M. , Donaghey, J. , Carey, B. W. , Garber, M. , Grenier, J. K. , Munson, G. , Young, G. , Lucas, A. B. , Ach, R. , Bruhn, L. , Yang, X. , Amit, I. , Meissner, A. , Regev, A. , Rinn, J. L. , Root, D. E. , & Lander, E. S. (2011). lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature, 477(7364), 295–300. 10.1038/nature10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafler, D. A. (2004). Multiple sclerosis. Journal of Clinical Investigation, 113(6), 788–794. 10.1172/jci200421357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil, A. M. , Guttman, M. , Huarte, M. , Garber, M. , Raj, A. , Rivea Morales, D. , Thomas, K. , Presser, A. , Bernstein, B. E. , van Oudenaarden, A. , Regev, A. , Lander, E. S. , & Rinn, J. L. (2009). Many human large intergenic noncoding RNAs associate with chromatin‐modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America, 106(28), 11667–11672. 10.1073/pnas.0904715106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp, F. , & Mendell, J. T. (2018). Functional classification and experimental dissection of long noncoding RNAs. Cell, 172(3), 393–407. 10.1016/j.cell.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, P. C. , Weng, W. T. , Scofield, B. A. , Paraiso, H. C. , Brown, D. A. , Wang, P. Y. , Yu, I. C. , & Yen, J. H. (2020). Dimethyl itaconate, an itaconate derivative, exhibits immunomodulatory effects on neuroinflammation in experimental autoimmune encephalomyelitis. Journal of Neuroinflammation, 17(1), 138. 10.1186/s12974-020-01768-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, P. W. , Severin, M. E. , & Lovett‐Racke, A. E. (2017). TGF‐beta regulation of encephalitogenic and regulatory T cells in multiple sclerosis. European Journal of Immunology, 47(3), 446–453. 10.1002/eji.201646716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Gong, A. Y. , Zhang, X. T. , Wang, Y. , Mathy, N. W. , Martins, G. A. , Strauss‐Soukup, J. K. , & Chen, X. M. (2018). Induction of a long noncoding RNA transcript, NR_045064, promotes defense gene transcription and facilitates intestinal epithelial cell responses against cryptosporidium infection. Journal of Immunology, 201(12), 3630–3640. 10.4049/jimmunol.1800566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , He, F. , Pang, R. , Zhao, D. , Qiu, W. , Shan, K. , Zhang, J. , Lu, Y. , Li, Y. , & Wang, Y. (2014). Interleukin‐17 (IL‐17)‐induced microRNA 873 (miR‐873) contributes to the pathogenesis of experimental autoimmune encephalomyelitis by targeting A20 ubiquitin‐editing enzyme. The Journal of Biological Chemistry, 289(42), 28971–28986. 10.1074/jbc.M114.577429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Zhang, Q. , Wang, W. , Zuo, D. , Wang, J. , Zhou, F. , Niu, L. , Li, X. , Qin, S. , Kou, Y. , Kong, F. , Pan, W. , Wang, Y. , Gao, D. , Sun, H. , Meves, J. M. , Zheng, K. , & Tang, R. (2018). Analysis of long noncoding RNA and mRNA expression profiles in IL‐9‐activated astrocytes and EAE mice. Cellular Physiology and Biochemistry, 45(5), 1986–1998. 10.1159/000487975 [DOI] [PubMed] [Google Scholar]
- Liu, X. , Zhou, F. , Wang, W. , Chen, G. , Zhang, Q. , Lv, R. , Zhao, Z. , Li, X. , Yu, Q. , Meves, J. M. , Hua, H. , Li, X. , Wang, X. , Sun, H. , & Gao, D. (2021). IL‐9‐triggered lncRNA Gm13568 regulates Notch1 in astrocytes through interaction with CBP/P300: Contribute to the pathogenesis of experimental autoimmune encephalomyelitis. Journal of Neuroinflammation, 18(1), 108. 10.1186/s12974-021-02156-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Zhou, F. , Yang, Y. , Wang, W. , Niu, L. , Zuo, D. , Li, X. , Hua, H. , Zhang, B. , Kou, Y. , Guo, J. , Kong, F. , Pan, W. , Gao, D. , Meves, J. M. , Sun, H. , Xue, M. , Zhang, Q. , Wang, Y. , & Tang, R. (2019). MiR‐409‐3p and MiR‐1896 co‐operatively participate in IL‐17‐induced inflammatory cytokine production in astrocytes and pathogenesis of EAE mice via targeting SOCS3/STAT3 signaling. Glia, 67(1), 101–112. 10.1002/glia.23530 [DOI] [PubMed] [Google Scholar]
- Ma, Z. , Guo, D. , Wang, Q. , Liu, P. , Xiao, Y. , Wu, P. , Wang, Y. , Chen, B. , Liu, Z. , & Liu, Q. (2019). Lgr5‐mediated p53 repression through PDCD5 leads to doxorubicin resistance in hepatocellular carcinoma. Theranostics, 9(10), 2967–2983. 10.7150/thno.30562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan, P. R. , Harrington, L. E. , O'Quinn, D. B. , Helms, W. S. , Bullard, D. C. , Elson, C. O. , Hatton, R. D. , Wahl, S. M. , Schoeb, T. R. , & Weaver, C. T. (2006). Transforming growth factor‐beta induces development of the T(H)17 lineage. Nature, 441(7090), 231–234. 10.1038/nature04754 [DOI] [PubMed] [Google Scholar]
- Nair, A. , Frederick, T. J. , & Miller, S. D. (2008). Astrocytes in multiple sclerosis: A product of their environment. Cellular and Molecular Life Sciences, 65(17), 2702–2720. 10.1007/s00018-008-8059-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsa, R. , Lund, H. , Tosevski, I. , Zhang, X. M. , Malipiero, U. , Beckervordersandforth, J. , Merkler, D. , Prinz, M. , Gyllenberg, A. , James, T. , Warnecke, A. , Hillert, J. , Alfredsson, L. , Kockum, I. , Olsson, T. , Fontana, A. , Suter, T. , & Harris, R. A. (2016). TGFbeta regulates persistent neuroinflammation by controlling Th1 polarization and ROS production via monocyte‐derived dendritic cells. Glia, 64(11), 1925–1937. 10.1002/glia.23033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff, J. D. , Wei, Y. , & Khavari, P. A. (2018). The functions and unique features of long intergenic non‐coding RNA. Nature Reviews. Molecular Cell Biology, 19(3), 143–157. 10.1038/nrm.2017.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva, P. , Ratti, A. , & Venturin, M. (2016). The long non‐coding RNAs in neurodegenerative diseases: Novel mechanisms of pathogenesis. Current Alzheimer Research, 13(11), 1219–1231. 10.2174/1567205013666160622112234 [DOI] [PubMed] [Google Scholar]
- St Laurent, G. , Wahlestedt, C. , & Kapranov, P. (2015). The landscape of long noncoding RNA classification. Trends in Genetics, 31(5), 239–251. 10.1016/j.tig.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, D. , Yu, Z. , Fang, X. , Liu, M. , Pu, Y. , Shao, Q. , Wang, D. , Zhao, X. , Huang, A. , Xiang, Z. , Zhao, C. , Franklin, R. J. , Cao, L. , & He, C. (2017). LncRNA GAS5 inhibits microglial M2 polarization and exacerbates demyelination. EMBO Reports, 18(10), 1801–1816. 10.15252/embr.201643668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis, M. A. , & Sheppard, D. (2014). TGF‐beta activation and function in immunity. Annual Review of Immunology, 32, 51–82. 10.1146/annurev-immunol-032713-120257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi, S. , Shree, B. , Mohapatra, S. , Swati, Basu, A. , & Sharma, V. (2021). The expanding regulatory mechanisms and cellular functions of long non‐coding RNAs (lncRNAs) in neuroinflammation. Molecular Neurobiology, 58(6), 2916–2939. 10.1007/s12035-020-02268-8 [DOI] [PubMed] [Google Scholar]
- Tsai, M. C. , Manor, O. , Wan, Y. , Mosammaparast, N. , Wang, J. K. , Lan, F. , Shi, Y. , Segal, E. , & Chang, H. Y. (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science, 329(5992), 689–693. 10.1126/science.1192002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzartos, J. S. , Friese, M. A. , Craner, M. J. , Palace, J. , Newcombe, J. , Esiri, M. M. , & Fugger, L. (2008). Interleukin‐17 production in central nervous system‐infiltrating T cells and glial cells is associated with active disease in multiple sclerosis. The American Journal of Pathology, 172(1), 146–155. 10.2353/ajpath.2008.070690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen, M. , Hocking, R. J. , Atkins, C. J. , Locksley, R. M. , & Stockinger, B. (2006). TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL‐17‐producing T cells. Immunity, 24(2), 179–189. 10.1016/j.immuni.2006.01.001 [DOI] [PubMed] [Google Scholar]
- Veldhoen, M. , Uyttenhove, C. , van Snick, J. , Helmby, H. , Westendorf, A. , Buer, J. , Martin, B. , Wilhelm, C. , & Stockinger, B. (2008). Transforming growth factor‐beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9‐producing subset. Nature Immunology, 9(12), 1341–1346. 10.1038/ni.1659 [DOI] [PubMed] [Google Scholar]
- Wan, P. , Su, W. , & Zhuo, Y. (2017). The role of long noncoding RNAs in neurodegenerative diseases. Molecular Neurobiology, 54(3), 2012–2021. 10.1007/s12035-016-9793-6 [DOI] [PubMed] [Google Scholar]
- Wang, X. , Mulas, F. , Yi, W. , Brunn, A. , Nishanth, G. , Just, S. , Waisman, A. , Brück, W. , Deckert, M. , & Schlüter, D. (2019). OTUB1 inhibits CNS autoimmunity by preventing IFN‐gamma‐induced hyperactivation of astrocytes. The EMBO Journal, 38(10), e100947. 10.15252/embj.2018100947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, B. , Zhang, S. , Guo, Z. , Bi, Y. , Zhou, M. , Li, P. , Seyedsadr, M. , Xu, X. , Li, J. L. , Markovic‐Plese, S. , & Wan, Y. Y. (2021). The TGF‐beta superfamily cytokine Activin‐a is induced during autoimmune neuroinflammation and drives pathogenic Th17 cell differentiation. Immunity, 54(2), 308–323.e6. 10.1016/j.immuni.2020.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, P. , Zuo, X. , Deng, H. , Liu, X. , Liu, L. , & Ji, A. (2013). Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Research Bulletin, 97, 69–80. 10.1016/j.brainresbull.2013.06.001 [DOI] [PubMed] [Google Scholar]
- Wurthner, J. U. , Frank, D. B. , Felici, A. , Green, H. M. , Cao, Z. , Schneider, M. D. , McNally, J. , Lechleider, R. J. , & Roberts, A. B. (2001). Transforming growth factor‐beta receptor‐associated protein 1 is a Smad4 chaperone. The Journal of Biological Chemistry, 276(22), 19495–19502. 10.1074/jbc.M006473200 [DOI] [PubMed] [Google Scholar]
- Yan, Y. , Ding, X. , Li, K. , Ciric, B. , Wu, S. , Xu, H. , Gran, B. , Rostami, A. , & Zhang, G. X. (2012). CNS‐specific therapy for ongoing EAE by silencing IL‐17 pathway in astrocytes. Molecular Therapy, 20(7), 1338–1348. 10.1038/mt.2012.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Wu, Y. , Zhang, B. , & Ni, B. (2018). Noncoding RNAs in multiple sclerosis. Clinical Epigenetics, 10(1), 149. 10.1186/s13148-018-0586-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, W. , Schluter, D. , & Wang, X. (2019). Astrocytes in multiple sclerosis and experimental autoimmune encephalomyelitis: Star‐shaped cells illuminating the darkness of CNS autoimmunity. Brain, Behavior, and Immunity, 80, 10–24. 10.1016/j.bbi.2019.05.029 [DOI] [PubMed] [Google Scholar]
- Yoshida, Y. , Yoshimi, R. , Yoshii, H. , Kim, D. , Dey, A. , Xiong, H. , Munasinghe, J. , Yazawa, I. , O'Donovan, M. J. , Maximova, O. A. , Sharma, S. , Zhu, J. , Wang, H. , Morse, H. C., 3rd , & Ozato, K. (2014). The transcription factor IRF8 activates integrin‐mediated TGF‐beta signaling and promotes neuroinflammation. Immunity, 40(2), 187–198. 10.1016/j.immuni.2013.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F. , Gao, C. , Ma, X. F. , Peng, X. L. , Zhang, R. X. , Kong, D. X. , Simard, A. R. , & Hao, J. W. (2016). Expression profile of long noncoding RNAs in peripheral blood mononuclear cells from multiple sclerosis patients. CNS Neuroscience & Therapeutics, 22(4), 298–305. 10.1111/cns.12498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F. , Liu, G. , Wei, C. , Gao, C. , & Hao, J. (2017). Linc‐MAF‐4 regulates Th1/Th2 differentiation and is associated with the pathogenesis of multiple sclerosis by targeting MAF. The FASEB Journal, 31(2), 519–525. 10.1096/fj.201600838R [DOI] [PubMed] [Google Scholar]
- Zhou, F. , Liu, X. , Gao, L. , Zhou, X. , Cao, Q. , Niu, L. , Wang, J. , Zuo, D. , Li, X. , Yang, Y. , Hu, M. , Yu, Y. , Tang, R. , Lee, B. H. , Choi, B. W. , Wang, Y. , Izumiya, Y. , Xue, M. , Zheng, K. , & Gao, D. (2019). HIV‐1 tat enhances purinergic P2Y4 receptor signaling to mediate inflammatory cytokine production and neuronal damage via PI3K/Akt and ERK MAPK pathways. Journal of Neuroinflammation, 16(1), 71. 10.1186/s12974-019-1466-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, F. , Liu, X. , Zuo, D. , Xue, M. , Gao, L. , Yang, Y. , Wang, J. , Niu, L. , Cao, Q. , Li, X. , Hua, H. , Zhang, B. , Hu, M. , Gao, D. , Zheng, K. , Izumiya, Y. , & Tang, R. (2018). HIV‐1 Nef‐induced lncRNA AK006025 regulates CXCL9/10/11 cluster gene expression in astrocytes through interaction with CBP/P300. Journal of Neuroinflammation, 15(1), 303. 10.1186/s12974-018-1343-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The feature of lncRNA AK018453. (a) The sequence of AK018453 with 1250 bp in length. (b) AK018453 is complementary to the sense chain of TRAP1 gene
Table S1 The list of primer sequences for the real‐time PCR assay. The table contains all primer sequences for the real‐time PCR assay
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.


