Abstract
As first demonstrated in budding yeast (Saccharomyces cerevisiae), all eukaryotic cells contain two, distinct multi-component protein kinase complexes that each harbor the TOR (Target Of Rapamycin) polypeptide as the catalytic subunit. These ensembles, dubbed TORC1 and TORC2, function as universal, centrally important sensors, integrators, and controllers of eukaryotic cell growth and homeostasis. TORC1, activated on the cytosolic surface of the lysosome (or, in yeast, on the cytosolic surface of the vacuole), has emerged as a primary nutrient sensor that promotes cellular biosynthesis and suppresses autophagy. TORC2, located primarily at the plasma membrane, plays a major role in maintaining the proper levels and bilayer distribution of all plasma membrane components (sphingolipids, glycerophospholipids, sterols, and integral membrane proteins). This article surveys what we have learned about signaling via the TORC2 complex, largely through studies conducted in S. cerevisiae. In this yeast, conditions that challenge plasma membrane integrity can, depending on the nature of the stress, stimulate or inhibit TORC2, resulting in, respectively, up-regulation or down-regulation of the phosphorylation and thus the activity of its essential downstream effector the AGC family protein kinase Ypk1. Through the ensuing effect on the efficiency with which Ypk1 phosphorylates multiple substrates that control diverse processes, membrane homeostasis is maintained. Thus, the major focus here is on TORC2, Ypk1, and the multifarious targets of Ypk1 and how the functions of these substrates are regulated by their Ypk1-mediated phosphorylation, with emphasis on recent advances in our understanding of these processes.
Keywords: contact sites, lipids, membrane proteins, phosphorylation, plasma membrane, protein kinases
Introduction
For a eukaryotic cell to proliferate, it must be able to grow, i.e. increase in mass—duplicating its genome, augmenting its content of ribosomes and other cytoplasmic constituents, replicating its repertoire of membrane-bounded organelles, and increasing all of the components needed to expand and maintain the function of its boundary with the outside world, the plasma membrane. The decision to grow, often evoked in metazoans via the action of mitogenic growth factors, nonetheless requires that the cell also have mechanisms to assess whether the supply of nutrients available is sufficient to support all of the biosynthetic processes needed. Eukaryotic cells have also evolved additional regulatory circuitry that, on the one hand, impedes such a growth decision when other external and internal cues or stresses indicate conditions for growth are suboptimal and, on the other hand, that elicits compensatory response pathways that restore conditions that then permit growth. The pivotal nexuses for controlling these events are the TOR-containing complexes, TORC1 and TORC2, whether in a unicellular eukaryote, like yeast [1–3], or in mammalian cells [4,5] or in plants [6].
TORC2 core
In both TORC1 and TORC2, the catalytic subunit is a large TOR polypeptide. Metazoan genomes encode a single TOR protein (human mTOR, 2549 residues), whereas budding yeast [7], fission yeast [8], and other fungal species [9] encode two, paralogous TOR proteins, Tor1 and Tor2 (in Saccharomyces cerevisiae, 2470 and 2474 residues, respectively; 70% identity). Tor2 is a plasma membrane-associated protein [10,11]. Yeast cells lacking Tor2 are inviable [12,13] because only Tor2 can serve as the catalytic subunit in TORC2, whereas TORC1 can accommodate either Tor1 or Tor2 as its catalytic subunit [14,15]. Thus, TOR2 is an essential gene, whereas TOR1 is not.
The molecular basis of the preference for incorporation of Tor2 into (and exclusion of Tor1 from) yeast TORC2 has been illuminated, first, by the determination of near-atomic resolution cryo-EM structures for TORC2 from both yeast [16,17] and mammalian cells [18–20]. Second, the construction and analysis of an extensive set of chimeric Tor1–Tor2 proteins [21,22] indicates that Tor2 makes multiple non-contiguous contacts with other constituent subunits of TORC2, but that a handful of single-residue differences between Tor2 and Tor1 within an ∼500-residue segment of their N-terminal HEAT repeat region (425–930 in Tor2), dubbed the ‘major assembly specificity' (MAS) domain (corresponding, in the main, to the so-called Spiral and Bridge structural elements formed by the HEAT repeats in this region [16]), are sufficient to dictate its TORC2-specific assembly.
At the center of TORC2 is a head-to-tail dimer of Tor2 held together by extensive contacts between the fold created by the N-HEAT repeats in one monomer interacting with the fold created by the M-HEAT repeats in the other monomer. Tightly associated with each copy of Tor2 is the small (303 residues) β-propeller (‘WD40 repeat') protein Lst8, which binds tightly to and stabilizes the α-helical lobe of the Tor2 kinase fold. Lst8 does the same with Tor1 in yeast TORC1, and its mammalian ortholog (mLST8) does the same with mTOR in mTORC1 and mTORC2 [16,20,23,24]. Unsurprisingly, given the role of Lst8 in maintaining the structure of the Tor2 catalytic domain, LST8 is also an essential gene.
TORC2 structure and assembly
Aside from Lst8, all of the other known subunits in yeast TORC2 are separate and distinct from those in TORC1 [1,25,26]. Like Lst8, the other subunits are each present in two copies and are organized around the Tor2 dimer in a rhombohedral shape with two-fold rotational symmetry [16,17]. Another key subunit of TORC2 is also encoded by an essential gene, AVO3/TSC11 (mammalian ortholog is RICTOR). Avo3 (1430 residues) comprises a predicted N-terminal antiparallel coiled-coil hairpin (residues 96–132 paired with 145–191) followed by at least 20 Armadillo (ARM) repeats (residues 297–1244) that stack into a pronounced α-solenoid. An apparent C-terminal segment of this α-solenoid is coiled within a cavity created by the junction between the N- and M-HEAT repeats that conjoin each Tor2 molecule in the dimer; thus, two copies of Avo3 serve as extra ‘mortar' to further stabilize TORC2 [27,28]. Our own work has demonstrated that Avo3 is able to associate with the plasma membrane in the absence of either Tor2 or other TORC2 subunits, and that five or so of its more N-terminal ARM repeats (residues 496–710) rich in basic residues is necessary for its plasma membrane recruitment [29], indicating that this portion of Avo3 likely extends away from the body of TORC2. Moreover, our results indicate that, during de novo assembly of TORC2, Avo3 is most likely the first subunit to arrive at the plasma membrane and is responsible for anchoring Tor2 there, which, in turn, then recruits the other associated subunits [29]. Thus, Avo3 is pivotal for TORC2 plasma membrane localization, assembly, and stability in vivo.
The C-terminal-most portion of Avo3, which contains a predicted small 5-helix bundle (residues 1334–1442), appears to interact with the FRB domain in Tor2, in a manner that would sterically block access of the rapamycin-Fpr1 (yeast FKBP12) complex that is inhibitory to TORC1, thereby providing an explanation for the relative resistance of TORC2 to rapamycin inhibition. In agreement with this conclusion, removal of just the last 157 resides of the Avo3 C-terminus renders TORC2 sensitive to rapamycin [30]. Likewise, recent cryo-EM structures unveil how RICTOR sterically occludes mTOR to block inhibition by FKBP12-rapamycin and to prevent recognition of mTORC1 substrates [31].
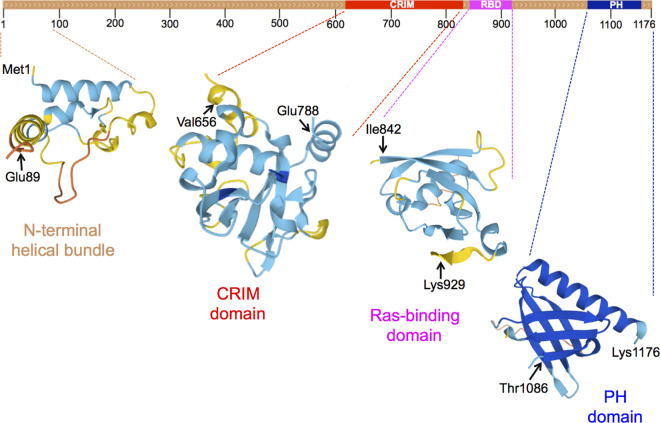
AVO1 (mammalian ortholog is mSIN1) is also an essential gene that encodes another chief subunit of TORC2. Avo1 (1176 residues) is a multi-domain protein (Figure 1) with (i) a predicted N-terminal helical bundle fold, separated by an apparently unstructured segment from (ii) a so-called ‘conserved region in the middle' (CRIM), whose structure has been determined in its fission yeast counterpart [32,33] and whose function is now known to be binding and presentation of substrates to the catalytic center in Tor2 [34–39], (iii) a Ras-binding domain (RBD) whose structure has been determined in mSIN1 [40,41], and (iv) a C-terminal pleckstrin homology (PH) domain specific for binding phosphatidylinositol-4,5-bisphosphate (PtdIns4,5P2), whose structure has been determined for both Avo1 and mSIN1 [42]. In the cryo-EM structure of yeast TORC2, density attributed to Avo1 is rather weak, presumably reflecting its mobility/flexibility, but it appears that the CRIM domain of each Avo1 is situated alongside the rim of the active site cleft of each Tor2 monomer [16,17], consistent with its role in substrate recruitment and presentation to the catalytic center. Likewise, recent cryo-EM structures unveil how mSIN1 contributes contacts important for the selective recruitment of mTORC2 substrates [31].
Figure 1. S. cerevisiae Avo1.
Top, schematized primary structure, highlighting locations of the conserved region in the middle (CRIM, red), Ras-binding domain (RBD, pink) and pleckstrin homology domain (PH, blue). Bottom, depiction of the three-dimensional structures of the indicated sequence segments, as predicted by AlphaFold2 [43]. Actual crystal structures for the C-terminal PH domains of Avo1 and mSIN1 have been determined [42], reflected in the dark blue coloring (the darker the shade of blue the higher the confidence level of the AlphaFold2 prediction).
It was initially posited that the C-terminal PH domain of Avo1 was necessary for both the membrane localization and function of TORC2 [44]. Contrary to that proposal, we demonstrated using three independent genetic methods (including the viability of cells expressing Avo1(Δ1059–1176) as the sole source of this TORC2 subunit) that the PtdIns4,5P2-binding PH domain of Avo1 is not required for the in vivo function of Avo1, or for Avo1 recruitment to the plasma membrane, or for tethering any other components of TORC2 at the plasma membrane [29]. Yet, efficient depletion of plasma membrane PtdIns4,5P2, achieved by chemogenetic degradation of the sole PtdIns4P 5-kinase in S. cerevisiae (Mss4), significantly decreased TORC2 activity, but did so without displacement of any TORC2 component from the PM [29]. The simplest explanation for why the loss of PtdIns4,5P2 markedly reduces yeast TORC2 activity, yet has no effect on the localization or state of assembly of TORC2, is that binding of the PH domain of Avo1 to PtdIns4,5P2 in the plasma membrane induces a conformational change that alleviates a negative constraint that the PH domain exerts, likely intramolecular steric occlusion of the CRIM domain [which would prevent substrate (Ypk1) binding]. In other words, PtdIns4,5P2 serves as a positive allosteric effector of TORC2. This scenario readily explains why deletion of the Avo1 PH domain is well tolerated. Similarly, in mTORC2, the binding of PtdIns3,4,5P3 to the PH domain in mSin1 (the Avo1 ortholog) alleviates an inhibitory constraint that the mSin1 PH domain imposes on mTORC2 activity [45].
However, more needs to be learned about how the dynamic localization, and relative accessibility versus sequestration, of PtdIns4,5P2 influences TORC2 function. In this regard, despite the yeast cell wall, osmotic stresses that change turgor pressure are accompanied by changes in plasma membrane tension (the force per unit length acting on a cross-section of membrane), with ensuing effects on lipid packing and fluidity, which influences the diffusion coefficient of both the lipids and proteins in the membrane [46]. Indeed, it has been reported that the plasma membrane distribution of PtdIns4,5P2 is affected by treatments of yeast cells that alter membrane tension with resulting effects on TORC2 activity [47,48].
Ancillary TORC2 components
When yeast cells are placed in a hypertonic medium, under which condition the cell loses water and the plasma membrane shrinks and invaginates, TORC2 function is very rapidly inactivated [49–51]. Conversely, when yeast cells are placed in a hypotonic medium, under which condition the influx of water subjects the plasma membrane to a stretching force, TORC2 function is stimulated [52,53]. This observed activation was initially attributed to the dynamic movement of two, paralogous, plasma membrane-associated proteins, Slm1 and Slm2 (686 and 656 residues, respectively; 65% identity). Cells lacking both Slm1 and Slm2 are inviable [54].
In unstressed cells, Slm1 and Slm2 are components of eisosomes, furrow-like troughs in the plasma membrane coated with antiparallel stacks of the F-BAR domain-containing proteins Pil1 and Lsp1, which are recruited to the plasma membrane by their binding of PtdIns4,5P2 [55]. Eisosomes, which are scattered around the plasma membrane, also contain a half-dozen small tetraspanins in two families—Sur7 and three relatives (Fmp45, Ynl194c, and Pun1), and Nce10 and its paralog Fhn1—as well as harbor certain nutrient transporters [56,57]. TORC2 also localizes around the cell periphery in multiple plasma membrane-associated puncta [29,44,51]; but, TORC2 does not co-localize with eisosomes and, unlike eisosomes, the TORC2 puncta are quite mobile [29,44,58,59]. Both Slm1 and Slm2 contain an unstructured N-terminus, a central ∼190-residue predicted I-BAR (inverse BAR) domain required for their targeting to eisosomes [60] followed by a structurally characterized ∼110-residue PH domain [61] demonstrated to bind PtdIns4,5P2 specifically [54,62,63] and, at the C-terminus, a high-affinity docking site (PxIxIT motif) for calcineurin (phosphoprotein phosphatase 2B) [62,64,65].
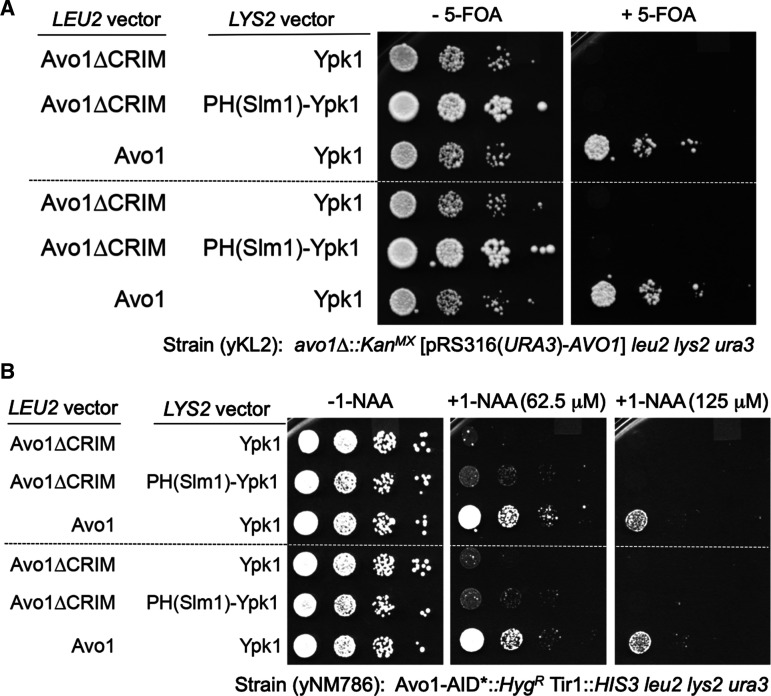
Upon hypotonic stress (or drug-induced sphingolipid depletion), it was observed [50] that Slm1 and Slm2, which can form a heterodimer [62,64,65], dissociate from eisosomes and then co-localize with TORC2. Moreover, it was reported [53,66] that Slm1 and the protein kinase Ypk1, the essential downstream target of TORC2, interact physically, suggesting that the critical function of Slm1 is the delivery of Ypk1 to the plasma membrane for its phosphorylation by both TORC2 (which phosphorylates multiple C-terminal sites in Ypk1 [67]; see further below) and the eisosome-associated protein kinase Pkh1 (which is responsible for phosphorylating the activation loop at Thr504 in Ypk1 [68]; see further below). Consistent with this proposal, targeting Ypk1 to the plasma membrane by fusing the Slm1 PH domain to the Ypk1 N-terminus bypassed the lethality of slm1Δ slm2Δ cells [53,66]. At the time, however, the role of the Avo1 CRIM domain in presenting substrates to the Tor2 active site was not firmly established. Indeed, using two different approaches, we have found that the PHSlm1-Ypk1 construct is unable to support growth, even in otherwise wild-type cells, if Avo1 lacks its CRIM domain (Figure 2).
Figure 2. CRIM domain of Avo1 is necessary for TORC2-mediated Ypk1 activation.
(A) Cells (Strain yKL2) lacking the AVO1 gene at its normal chromosomal locus, but maintained by a copy of the AVO1 gene on a URA3-marked plasmid (pRS316), were transformed with LEU2- and LYS2-marked vectors carrying the indicated genes, 10-fold serial dilutions of the resulting transformants were spotted in duplicate (top and bottom panels) on plates lacking or containing 5-fluoro-orotic acid, as indicated, to select for loss of the URA3-marked plasmid [69], and the plates were photographed after incubation at 30°C for 2 days. This plasmid shuffle approach revealed that plasma membrane tethering of Ypk1 is not sufficient to bypass the function of the CRIM domain in Avo1. (B) Cells (Strain yNM786) expressing from its chromosomal locus an Avo1 derivative tagged with a degron (AID*) that permits auxin-inducible degradation mediated by the plant Tir1 F-box protein expressed in the same cells [70], were transformed with LEU2- and LYS2-marked vectors carrying the indicated genes, 10-fold serial dilutions of the resulting transformants were spotted in duplicate (top and bottom panels) on plates lacking or containing the synthetic auxin 1-NAA at the indicated concentrations to stimulate Avo1-AID* degradation, and the plates were photographed after incubation at 30°C for 2 days. This chemogenetic approach further confirmed that plasma membrane tethering of Ypk1 is not sufficient to bypass the function of the CRIM domain in Avo1. Strains yKL2 and yNM786 (both derivatives of BY4742) and the LEU2-marked vectors expressing Avo1 and Avo1(ΔCRIM) were described previously [29]. The vectors expressing Ypk1-3xHA and PHSlm1-Ypk1-3xHA are LYS2-marked derivatives of plasmids (pPL433 and pPL495) generously provided by Prof. Ted Powers (Univ. of California, Davis). The results shown were obtained by Dr. Françoise M. Roelants.
Thus, even though tethered to the plasma membrane, PHSlm1-Ypk1 cannot be phosphorylated and activated by TORC2 when the CRIM domain of Avo1 is missing, indicating that the essential function of Slm1 and Slm2 must be something different from simple delivery of Ypk1 to the plasma membrane for encounter with TORC2.
As first detected in global two-hybrid screens [71,72], and confirmed by both coimmunoprecipitation [54] and in vitro binding assays [73], Slm1 and Slm2 also interact specifically with Avo2, another known component of TORC2. Avo2 (426 residues) contains six predicted N-terminal ankyrin (ANK) repeats (residues 1–204) with a ‘tail' of low complexity sequence of about equal length. However, unlike TOR2, LST8, AVO1 and AVO3, AVO2 is not an essential gene. In cryo-EM structures of TORC2, density attributable to the N-terminal ANK domain of Avo2 is nestled between the N-HEAT repeats 18–21 and the FAT domain in Tor2, placing Avo2 in a position to serve some regulatory role on the catalytic subunit. Indeed, we found [51] that Avo2 is a primary target of the stress-activated MAPK Slt2/Mpk1 (mammalian ortholog ERK5/MAPK7), the primary effector of the so-called yeast cell wall integrity pathway [74,75]. Avo2 contains nine -SP- and -TP- sites, eight residing in its C-terminal unstructured region of which five have been detected in global analyses of the S. cerevisiae phosphoproteome [76–80]. Conditions that activate Slt2 caused hyperphosphorylation of Avo2 at all these sites and these modifications impeded the ability of TORC2 to phosphorylate and activate Ypk1 [51]. Correspondingly, a phosphomimetic Avo29E allele and an avo2Δ allele both render cells sensitive to two stresses (myriocin treatment and elevated exogenous acetic acid) that the cell requires Ypk1 activation by TORC2 to survive, indicating that Avo2 promotes optimal TORC2 function and supporting the conclusion that Slt2-mediated phosphorylation of Avo2 down-regulates TORC2 signaling. Likewise, we found that activated Slt2 also phosphorylates Avo3 [51], which contains eleven -SP- and -TP- sites, with seven lying within its first 235 residues of which six have been detected in the yeast phosphoproteome. These Slt2-mediated phosphorylation events appear to dissociate Avo2 from TORC2, but not displace TORC2 from the plasma membrane [51]. In this way, under conditions not amenable to robust cell proliferation, the growth-promoting function of TORC2 is restrained. In general, continued growth and response to stress are incompatible cellular states.
These findings place the role of Slm1 and Slm2 in promoting TORC2 function in a new light, given their documented interaction with Avo2. It seems possible that, when Slm1 and Slm2 are released from eisosomes and encounter any free phosphorylated Avo2, the calcineurin bound to the Slm1 and Slm2 C-termini may act to remove the inhibitory phosphorylations on Avo2 (and perhaps from Avo3 and possibly other TORC2 subunits), thereby alleviating this negative regulation and allowing Avo2 to rejoin the complex. In agreement with this model, when present as the sole source of these proteins, Slm1 or Slm2 mutants with mutated PxIxIT motifs, which were shown to no longer bind calcineurin, displayed elevated sensitivity to the growth inhibitory action of myriocin (a hallmark of impaired TORC2-Ypk1 signaling) [64]. Also consistent with this model, we found that Avo29E, which mimics the phosphorylated state, but is immune to phosphatase action, is inhibitory to TORC2 function in vivo and exhibited reduced association with TORC2 [51]. Moreover, at least one Avo3-derived tryptic peptide containing two of its demonstrated N-terminal Slt2 sites (-LENSAPSPTSPLMAR-) was identified in a phospho-proteomic analysis of calcineurin-deficient cells [81]. Furthermore, the stress-induced release of Slm1 and Slm2 from eisosomes may also contribute to TORC2 function in another way. It has been reported that the absence of Slm1 and Slm2 disrupts eisosome structure [82]. Thus, given that eisosomes represent a protected reservoir of PtdIns,5P2 [83], stresses that cause displacement of Slm1 and Slm2 from eisosomes may result in a certain degree of eisosome disassembly, thereby releasing PtdIns4,5P2 that is more accessible for binding to the PH domain of Avo1, thereby freeing its CRIM domain for more ready presentation of substrate to Tor2.
Two additional characterized components of TORC2 are Bit2 and its paralog Bit61 (545 and 543 residues, respectively; 45% identity), with Bit61 being the more abundant of the two [84]. Unlike a slm1Δ slm2Δ double mutant, a bit2Δ bit61Δ double mutant is viable [85] and, thus, neither protein is essential for the function of TORC2. In keeping with an accessory role for these proteins, in the available cryo-EM structures of TORC2, the density attributed to Bit61 is small, quite weak and located at the solvent-facing periphery of the portion of the Avo3 α-solenoid that is intimately associated with Tor2 [16,17]. Bit61 and Bit2 are considered, by some, to be the equivalent of mammalian PRR5/Protor-1 and PRR5L/Protor-2 [16,25,26] because both sets of proteins purportedly contain a domain (HbrB) found in an Aspergillus nidulans protein involved in filamentous growth [86]; but, there is virtually no detectable sequence similarity between these yeast and mammalian proteins (even in their purported HbrB sequences). Moreover, as generated by AlphaFold2 [43], structural predictions for Bit2 and Bit61 do not well resemble those for Protor-1 and Protor-2. Nonetheless, as observed in yeast for a bit2Δ bit61Δ double mutant, mouse protor-1 (prr5)−/− protor-2 (prr5L)−/− nullizygous animals are viable [87]. Thus, the functions of Bit2 and Bit61 (and of Protor-1 and Protor-2) remain somewhat obscure.
Control of TORC2 by small GTPases
There were reasons to suspect that RAS proteins may play some role in localization, stability and/or function of TORC2 [88]. First, mSIN1 (Avo1 ortholog) was first identified as a partial human cDNA that, when expressed in yeast, inhibited signaling by a constitutively active Ras2 allele, Ras2(G19V), most likely by physical interaction [89]. Second, full-length mSIN1 was shown to be a Ras-binding protein [90]. Third, the slime mold Avo1 ortholog (RIP3) has a sequence similar to a canonical Raf-like RBD [91] and binds the Ras-related protein Rap1, which promotes TORC2 signaling [92]. Likewise, as noted more than two decades ago [14], S. cerevisiae Avo1 also has a predicted RBD (Figure 1). Also, purportedly, Avo3 possesses a RasGEFN-like motif (residues 991 to 1047) [93], but this assignment is very likely a red herring because it is now appreciated that this portion of the sequence of Avo3 is well buried in the stack of ARM repeats that make up its α-solenoid.
Most recently, in mammalian cells, using proximity-dependent biotin labeling (BioID) [94], mSIN1 was identified as one of the targets of oncogenic RAS mutants and the RBD of mSIN1 was required for RAS binding in pull-down assays [95,96]. Moreover, two groups have obtained crystal structures of the mSIN1 RBD in complex with K-RAS (4A isoform) [40] and/or H-RAS [41]. Whereas the former group reported that, in cells expressing as the sole source of mSIN1 a mutant unable to bind RAS, mTORC2 assembly or function were unaffected, the latter group found that lack of Ras-SIN1 association prevents efficient mTORC2-mediated phosphorylation of one of its direct substrates, the AGC family protein kinase SGK1 [97]. Revealingly, the latter group also found that mSIN1 mutants lacking its C-terminal PH domain were able to bind RAS with higher affinity, indicating that the PH domain exerts an inhibitory effect on RAS-mSIN1 binding. It should be noted that the RBD is also immediately adjacent to the CRIM domain in both mSIN1 and Avo1 (Figure 1). Other effects of various splice variants and isoforms of mSIN1 have also been noted [98].
In S. cerevisiae, there are two RAS paralogs, Ras1 and Ras2 (the latter is 5-fold more abundant that the former [84]) and a ras1Δ ras2Δ double mutant is inviable. The sole essential role previously attributed to Ras1 and Ras2 was in binding to and activating adenylate cyclase to generate the 3′,5′-cAMP necessary to stimulate three PKA isoforms (Tpk1, Tpk2, and Tpk3) [99,100]. However, deletion of a 4th kinase (Yak1) permits the growth of otherwise inviable tpk1Δ tpk2Δ tpk3Δ triple mutant cells [101]. Moreover, overexpression of two Ras2 mutants deficient in their ability to stimulate adenylate cyclase, Ras(T42S) and Ras(T42A), were nearly as effective as overexpression of wild-type Ras2 in extending yeast replicative lifespan (which takes prolonged cell growth, it should be noted) [102]. Such findings suggested that essential functions of Ras1 and Ras2 in yeast cell physiology may include more than just the provision of 3′,5′-cAMP.
Given the close juxtaposition of the RBD to the CRIM domain in Avo1, one could argue that the binding of Ras2 may occlude substrate binding and serve to negatively regulate TORC2 function; alternatively, upon exposure of both the RBD and the CRIM domain by binding of the Avo1 PH domain to PtdIns4,5P2 in the membrane, the binding of Ras2 to the RBD may help stabilize the open state and enhance substrate association with the CRIM domain. To distinguish between these possibilities, we constructed functional alleles of Ras2 containing the AID* degron expressed as the sole source of Ras in ras1Δ cells. We found, first, that upon auxin-induced degradation of Ras2-AID*, phosphorylation of the TORC2 sites in Ypk1, the primary downstream substrate, was markedly diminished concomitant with the extent of Ras2-AID* depletion and TORC2 subunits were shunted to the vacuole (Roelants, F.M., Bonnar, J.L., Tiu, R.A.-Y., Thorner, J., unpublished data). Conversely, overexpression of Ras2 stimulated phosphorylation of Ypk1 at its TORC2 sites. These findings suggest that, in yeast, Ras2 is a positive effector of TORC2 and may help stabilize its association with the plasma membrane, given that Ras2 is plasma membrane-anchored via both S-farnesylation [103] and S-palmitoylation [104] of its C-terminal -CCaaX box.
We also uncovered a previously uncharacterized interplay between the Rab5 GTPases and TORC2 signaling through our analysis of Muk1, one of two Rab5-specific guanine nucleotide exchange factors in S. cerevisiae [59]. We demonstrated, first, that Muk1 is a direct physiologically relevant target of TORC2-activated Ypk1 and that Ypk1-mediated phosphorylation of Muk1 activates its guanine nucleotide exchange activity. Second, and unexpectedly, we found that the GTP-bound state of Vps21/Ypt51 (one of the three yeast Rab5 isoforms) physically associates with Tor2 and acts as a direct positive effector required for full TORC2 activity. Our modeling [105] suggests that Rab5 would be accommodated within the same pocket in Tor2 in TORC2 that the small GTPase RHEB occupies in mTOR to activate mTORC1 [106]. This TORC2-Ypk1-Muk1-Rab5-TORC2 circuitry provides, on the one hand, a self-reinforcing mechanism for sustained up-regulation of TORC2-Ypk1 signaling. On the other hand, given the function of Rab5 in promoting the formation of early endosomes [107–109], this regulatory loop also permits the mobilization of TORC2 via its internalization and hence its broader dispersal in the cell and, ultimately, its trafficking to the vacuole and inactivation. Likewise, down-regulation of TORC2 via endocytosis could also explain a report that two protein kinases (Ark1 and Prk1) involved in controlling the assembly and function of plasma membrane-associated so-called actin patches (sites of clathrin-mediated endocytosis) appear to modulate TORC2 signaling [110], as discussed further below.
Quite recently, it has been reported that the RICTOR subunit of mTORC2 physically associates with ARF1 and that ARF1 negatively regulates mTORC2 function [111]. In yeast, however, such an interaction seems unlikely because the bulk of TORC2 in exponentially growing cells under normal nutrient conditions is at the plasma membrane [29,51,59], whereas Arf1 is localized to the Golgi body and never at the PM [112,113]. Moreover, given the dynamics of TORC2 internalization and uptake into the vacuole, any encounter with Arf1 could only be transit, at best.
Ypk1 is the essential downstream effector of TORC2
Stimuli that activate TORC2 include inhibition of sphingolipid biosynthesis [52,114], hypotonic conditions [52,53], heat stress [115], and exposure to a high exogenous concentration of a weak organic acid [116]. The primary direct substrates of TORC2 are the AGC family protein kinase Ypk1 and its paralog Ypk2. Cells lacking both Ypk1 and Ypk2 are inviable under all conditions [68]. Within the human kinome, the kinase domain of Ypk1 shares the greatest homology with that of SGK1 (57% identity) and functional expression of SGK1 cDNA in yeast rescues the lethality of ypk1Δ ypk2Δ double mutant cells, whereas functional expression of cDNAs for other closely related mammalian AGC kinase family members (e.g. AKT1) does not [117]. In fact, the heterologous expression of AKT1 in S. cerevisiae is deleterious [118].
Under normal growth conditions, Ypk1 is the major isoform [84] because YPK2 expression is controlled mainly by three consensus binding sites (TTC(N)2–3GAA) for heat shock transcription factor (Hsf1) at −497, −404, and −104 from its ATG initiator codon [68] and is strongly induced by heat stress [119], whereas YPK1 expression appears to be controlled by the C2H2 zinc finger family transcription factor Com2 [120,121]. Thus, YPK1 ypk2Δ mutant cells manifest no detectably deleterious phenotypes, whereas ypk1Δ YPK2 mutant cells exhibit a cold-sensitive phenotype [68] because YPK2 expression becomes more and more limited the lower the temperature, due to the aforementioned means by which its expression is regulated. There is some indirect evidence that another S. cerevisiae AGC family kinase, Pkc1 (mammalian ortholog is PKN2/PRK2), may also be a substrate for TORC2 [122,123]. However, unlike TORC2-deficient cells, pkc1Δ cells are viable, albeit slow-growing unless provided with an osmotic support in the medium [124]. Most significantly, the expression of a constitutively-active allele of Ypk1, Ypk1(D242A) (see further below), rescues the inviability of tor2ts mutant cells at the restrictive temperature [114], is able to bypass the need for TORC2-mediated phosphorylation at its TORC2 sites [67], and is able to rescue the inviability of loss-of-function mutations in each of the otherwise essential core subunits of TORC2 [29]. Similarly, a corresponding constitutively active allele of Ypk2, Ypk2(D239A), rescues the inviability of avo3ts mutant cells [125]. Collectively, these findings demonstrate that the critical physiological target of TORC2 that is essential for maintaining yeast cell viability is Ypk1.
Structure and regulation of Ypk1
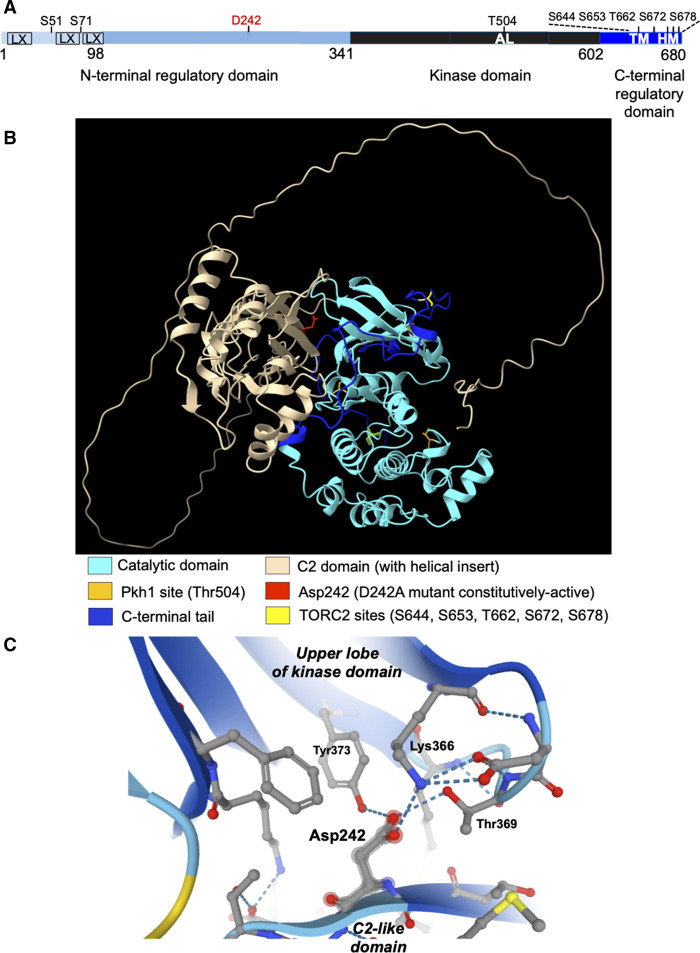
As a prerequisite for its basal activity, Ypk1 requires phosphorylation of its activation loop (T-loop) on T504 (Figure 3A). This phosphorylation is installed by the eisosome-localized protein kinase Pkh1 and, to a lesser extent, by its paralog Pkh2 [68,126]. The kinase domain of Pkh1 most resembles within the human kinome that of 3-phosphoinositide-dependent protein kinase 1 (PDK1) and, correspondingly, the expression of a mammalian PDK1 cDNA rescues the inviability of a pkh1Δ pkh2Δ double mutant [117]. Under stress conditions that activate TORC2, Ypk1 activity is markedly stimulated via TORC2-mediated phosphorylation of five residues at its extreme C-terminal end (Figure 3A) [67,114,126], and this up-regulation is required for the maintenance of cell viability under those same stress conditions. Likewise, corresponding C-terminal residues in Ypk2 are phosphorylated in a TORC2-dependent manner [67,127].
Figure 3. Ypk1 structure and regulation.
(A) Schematic depiction of Ypk1 primary structure. N-terminal regulatory domain (light blue); catalytic domain (black); and, C-terminal regulatory domain (dark blue). LX, low complexity sequence predicted by UniProt [132], SMART [133] and AlphaFold2 [43] databases; AL, Pkh1 site (T504) in activation loop; TORC2 sites: TM, classical turn motif (S644); HM, classical hydrophobic motif (T662); and additional positions (S653, S672, S678); S51 and S71, two, N-terminal sites phosphorylated by Fpk1 and paralog Fpk2. (B) Predicted three-dimensional fold of Ypk1 in the presumed ‘closed’ conformation (AF-P12688-F1-model_v2.pdb); features highlighted as in the color key shown beneath. See also Supplementary Movie S1 for animation. (C) Close-up of predicted contacts made by Asp242.
Ypk1 is located both at the plasma membrane and in the cytosol [68,128]. Based on sequence gazing alone, however, Ypk1 and Ypk2 appeared to lack any identifiable lipid-binding fold that might mediate membrane association. Moreover, it was difficult to explain how a single point mutation, Ypk1(D242A) [29,67,114], and its equivalent (D239A) in Ypk2 [127], is able to confer apparently constitutive and TORC2-independent activity. Remarkably, in the absence of any crystal structure for these enzymes, the AlphaFold2 algorithm strongly predicts in both proteins an elaborately folded domain (in Ypk1, residues G118-to-P341) located in the N-terminal segment (Figure 3B). Most strikingly, comparison of this domain fold to experimentally determined structures using the Dali server [129,130] reveals that its β-sandwich component very strongly resembles C2 domains, a well-characterized class of phospholipid-binding folds [131], with the addition of an α-helical insert or outrigger whose primary sequence is interwoven with the β-strands of the C2-like portion. In contrast, the Dali server was unable to align the α-helical portion with any other known structure; it may simply buttress and stabilize the fold of the C2-like portion. Some C2 domains require Ca2+ for phospholipid head group recognition [131,134], but the residues located in the loops interconnecting the β-strands that are typical of that Ca2+-dependent class appear to be absent from the Ypk1 C2-like fold; however, there is a clear positively-charged pocket, centered around Arg335, that could readily accommodate the head group of an anionic lipid and whose occupancy by lipid would be incompatible with the packing of this N-terminal domain against the kinase domain (Figure 3B).
The first inkling that something in the N-terminal extension upstream of the kinase domain served as a negative regulatory element was that an N-terminal truncation allele, Ypk1(Δ2–336), dubbed Ypk1-ΔN, when overexpressed from the GAL1 promoter, killed cells, whereas full-length Ypk1 overexpressed in the identical fashion and to the same degree was innocuous [68]. These findings suggested that Ypk1-ΔN was hyperactive and phosphorylating inappropriate substrates. In agreement with that conclusion, when a kinase-dead variant (K376A) of Ypk1-ΔN was overexpressed, or Ypk1-ΔN was overexpressed in pkh1Δ cells (to prevent its efficient activation loop phosphorylation and thus squelch its catalytic competence), it was no longer toxic, indicating that indeed it was the hyperactivity of Ypk1-ΔN as a functional protein kinase that was responsible for killing the cells. Similar behavior for Ypk2-ΔN was observed, with Pkh2 mainly responsible for its activation loop phosphorylation [68,127].
The second piece of experimental evidence that defined the N-terminus of Ypk1 as a negative regulatory domain was our isolation of Ypk1(D242A) as an allele that totally bypassed the need for TORC2-mediated phosphorylation [29,67,114]. The fact that Ypk1(D242A) is able to rescue even the inviability of cells deficient in Mss4 [29], the sole PtdIns4P 5-kinase responsible for all PtdIns4,5P2 synthesis in S. cerevisiae [135], suggests that one anionic lipid normally recognized by the C2-like domain in Ypk1 might be PtdIns4,5P2. It is especially noteworthy that D242 is located on a β-strand of the C2-like domain that is packed tightly against the upper lobe of the kinase domain in Ypk1 in a pocket situated between the phosphate anchor loop (-G-x-G-x-x-G359-) and the conserved active-site Lys motif (-ALK376-), where the oxygen atoms of D242 are H-bonded to K366, Thr369, and Y373 (Figure 3C).
Taken together, these observations raise the following speculative model for how TORC2-mediated phosphorylation stimulates Ypk1 function. First, as in other protein kinases, phosphorylation at T504 stabilizes the enzymically competent state if P-T504 is able to form a salt bridge with a conserved Arg in the catalytic loop (-YR469DLKPEN-). However, association of the C2-like fold with the upper lobe of the kinase domain imposes a conformational constraint that precludes the kinase fold from attaining its catalytically competent state. It is also possible that the immediate N-terminus of Ypk1 sterically occludes its active site (Figure 3B), although the prediction for that interaction is rather weak. If so, conformation changes accompanying the activation process must swing the N-terminus out of the way. TORC2 action alleviates the conformational constraint imposed on the kinase domain by the C2-like domain via phosphorylating several sites in the C-terminal tail of Ypk1 because one or more of those negatively charged phospho-sites competes with D242, thereby displacing it, freeing the C2-like fold now to bind instead to the plasma and endoplasmic reticulum (ER) membranes (and likely swinging the immediate N-terminus away, so it no longer blocks the active site). Moreover, release from its previously imposed conformation constraint, concomitantly permits the kinase domain to assume its catalytically active state. Given that the predicted structure indicates that D242 is a linchpin of the interaction between the C2-like fold and the upper lobe of the kinase domain (Figure 3B,C), this scenario provides a ready explanation for how the D242A mutation ablates the inhibitory constraints and allows Ypk1 to access its catalytically-competent conformation without the need for TORC2-mediated phosphorylation.
Recently, based mainly on work analyzing the role of mTORC2 in promoting the function of mammalian PKCβII, a previously uncharacterized mTORC2 phospho-acceptor Thr site located just distal to the kinase fold and fitting the consensus -FD-x-x-FT-, dubbed the ‘TOR interaction motif’ (TIM), was identified [136]. It was further proposed by these investigators that mTORC2-mediated phosphorylation of the TIM acts as the first and rate-limiting step in PKC activation by facilitating subsequent PDK1-mediated activation loop phosphorylation, followed by hydrophobic motif (HM) modification via intramolecular autophosphorylation (rather than by mTORC2 per se), resulting in fully active PKC [137,138]. As pointed out by others [39], this model raises at least some red flags. First, the TIM sequence in the AKT and SGK members of the AGC family kinases is also required for their binding to the CRIM domain of mSIN1 for their presentation to mTORC2. In light of the available structural information about mTORC2 [17,18,20], it is difficult to envision how TIM could be phosphorylated by mTORC2 while remaining bound to mSIN1. In addition, the TIM sequence lies within an α-helix, which, unless somehow unfolded, should not be able to fit into the catalytic cleft in mTOR. Furthermore, it has been observed experimentally for PKC and AKT that the T-loop can still be phosphorylated by PDK1 in mTORC2-deficient cells and, conversely, that the HM can still be phosphorylated in cells lacking PDK1, suggesting that T-loop and HM phosphorylation are independent, rather than sequential and interdependent, events [139–142].
Indeed, in this same regard, a Thr residue in the equivalent location and in the same sequence context as the proposed TIM is present in Ypk1 (and Ypk2). Despite its conservation, however, we had no evidence from either mass spectrometry or Phos-tag™ gel analysis [143] of native Ypk1 and derived phospho-site mutants that the corresponding residue is phosphorylated [67]. Moreover, the C-terminal 78 residues of Ypk1 (603–680) fused to the C-terminus of Escherichia coli maltose-binding protein (MBP), which obviously lacks the catalytic domain of Ypk1, serves as an effective substrate for immuno-purified TORC2 and, thus, phosphorylation of the TM, HM, and other TORC2 sites in Ypk1 need not occur via autophosphorylation. Finally, all of our prior work supports the conclusion that phosphorylation of T504 in Ypk1 by Pkh1 and TORC2-mediated phosphorylation of the C-terminal sites in Ypk1 are independent of each other [2]. Nonetheless, to be fair, the role, if any, of the putative TIM Thr (T637) in Ypk1 has not yet been directly interrogated by mutagenesis or other means.
Functions of Ypk1
The phospho-acceptor site specificities of Ypk1 and SGK1 are virtually identical and have been defined by the use of the purified enzymes and, as in vitro substrates, peptides, unbiased random peptide libraries, protein fragments and, in some cases, full-length proteins [2,59,117,144–146]. Based on those findings and characterization of authentic in vivo substrates, the optimal consensus is -R-x-R-x-x-S > T-(Hpo)-, where (Hpo) at the +1 position indicates a distinct preference for a hydrophobic (or uncharged) residue, and the residues at the −2 and −4 positions are also often basic [2,147], but it is not essential that they be so.
There is also some experimental evidence [148] that Ypk1 may select its highest-affinity substrates in a manner similar to that first observed for PKA, wherein preferred sites bind in the catalytic pocket (via their phospho-acceptor motif) with the residues immediately upstream binding within a prominent groove that extends from the active site across the lower lobe of the kinase domain [149,150] (as opposed, for example, to utilizing for substrate association either joint binding of kinase and substrate to a scaffold protein [151,152], or the presence in the kinase structure of a separate substrate ‘docking groove' distant from the active site, as is the case for recognition of substrates by their cognate MAPKs [153,154]).
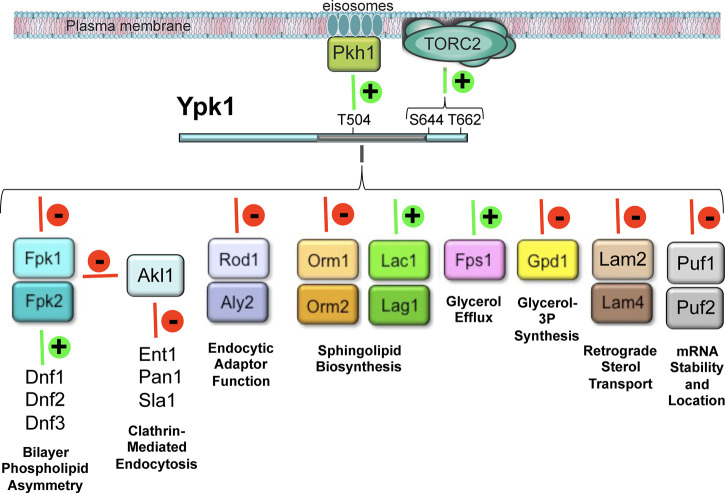
At least for Ypk1, it is noteworthy that substrates for which their Ypk1-mediated phosphorylation has been demonstrated to have a physiological impact are either integral plasma membrane proteins (e.g. aquaglyceroporin Fps1), or peripheral plasma membrane proteins (e.g. protein kinase Fpk1; protein kinase Akl1; α-arrestins Rod1/Art4 and Aly2/Art3), or integral membrane proteins of the cortical ER (e.g. tetraspanins Orm1 and Orm2; ceramide synthase subunits Lac1 and Lag1), or located at ER-plasma membrane contact sites (e.g. retrograde sterol transporters Lam2/Ysp2 and Lam4) (Figure 4). This repertoire of substrates is consistent with the posited role of the C2-like domain in targeting activated Ypk1 to cell membranes by binding to an anionic lipid.
Figure 4. Ypk1 is a master regulator of plasma membrane lipid and protein homeostasis.
Phosphorylation events (lines) that evoke a positive stimulatory effect on the indicated substrate(s) (green) and those that impose a negative inhibitory effect on the indicated substrate(s) (red) are shown. See text for additional details.
Comprehensive reviews are available that describe the various experimental strategies that have been used to identify physiologically relevant substrates of Ypk1 and that discuss what information has been gleaned about how Ypk1-mediated phosphorylation controls their function [2,155]. Hence, those findings will be summarized only briefly here before turning to more recently uncovered and characterized targets of Ypk1.
Control of the leaflet lipid composition of the plasma membrane bilayer
The first bona fide targets of Ypk1 discovered were Fpk1 and Fpk2/Kin82 [156], two paralogous protein kinases that exhibit cell cycle-dependent recruitment to the site of bud emergence and cytokinesis [157]. Fpk1 and Fpk2 phosphorylate and stimulate three, closely related, integral plasma membrane proteins (Dnf1, Dnf2, and Dnf3) [158], which are P-type ATPases (class IV) that function as aminoglycerophospholipid flippases, translocating lipid clients from the outer leaflet to the inner leaflet of the plasma membrane bilayer [159]. Ypk1 phosphorylates three consensus sites within the N-terminal regulatory domain of Fpk1 (893 residues) and one consensus site in the N-terminal segment of the significantly shorter Fpk2 (720 residues) (also, Fpk1 is five times more abundant than Fpk2 [84]). This Ypk1-mediated phosphorylation markedly inhibits Fpk1 (and Fpk2) function [156]. Thus, stimuli that activate TORC2-Ypk1 signaling negatively regulate flippase function, down-regulating the rate of inward aminoglycerophospholipid movement (mainly PtdEth [160], but also PtdCho [161,162] and PtdSer [163]) by inhibiting the ability of Fpk1 (and Fpk2) to phosphorylate and thereby stimulate the flippases.
However, there is an interesting reciprocal relationship between these two classes of protein kinases. Fpk1 phosphorylates Ypk1 at two N-terminal sites (S51 and S71) [156] well upstream if its C2-like domain (118–341) (Figure 3); it is less clear how such modification affects Ypk1 function, but it is likely inhibitory. In Schizosaccharomyces pombe, it has been reported that N-terminal phosphorylation of Gad8 (ortholog of S. cerevisiae Ypk1) by another protein kinase prevents the physical association of Gad8 with TORC2, decreasing TORC2-mediated phosphorylation of its HM (Ser546), thereby reducing TORC2-mediated activation of Gad8 [164]. Hence, it is tempting to speculate that, in the same way, Fpk1-mediated phosphorylation of Ypk1 is a negative feedback mechanism to dampen the TORC2-mediated activation of Ypk1. Alternatively, placing two negative charges at its N-terminus could prevent efficient recruitment of Ypk1 to the membrane, thereby impeding its access to substrates. In any event, this interrelationship means that another outcome of stimuli that evoke TORC2-Ypk1 signaling is that not only will Ypk1-dependent phosphorylation of Fpk1 increase, but, given that Ypk1-mediated phosphorylation of Fpk1 inhibits its function, Fpk1-mediated phosphorylation of Ypk1 will decrease. In this same regard, it is noteworthy that Fpk1 activity in vivo depends, not only on escape from Ypk1-mediated inhibition, but also on the presence of a complex sphingolipid, namely mannosyl-inositolphosphoryl-ceramide (MIPC) [156,157], although whether MIPC is a direct allosteric activator of Fpk1 or acts via a more indirect mechanism has not been elucidated.
Overall, as a logic circuit, the mutual inhibition exerted by Ypk1 on Fpk1 and, likely, by Fpk1 on Ypk1, makes physiological sense in two ways. First, because one of the primary functions of TORC2-Ypk1 signaling is to up-regulate the production of complex sphingolipids (as discussed further below), when the supply of cellular sphingolipids is adequate, Ypk1 action is no longer needed for that purpose. Given that MIPC promotes optimal Fpk1 function, its phosphorylation of Ypk1 likely serves as a rheostat to adjust the level of Ypk1 activity to meet the demands of the cell for sphingolipids in a finely tuned manner. Second, the interplay between Ypk1 and Fpk1 provides a mechanism to couple the rate of aminoglycerophospholipid removal from the outer leaflet of the plasma membrane bilayer to the rate of synthesis of complex sphingolipids [165], another component of the outer leaflet. The more MIPC available, the faster the internalization of outer leaflet PtdEth, PtdCho and PtdSer. Maintaining this balance in lipid composition and leaflet distribution appears to be crucial for many aspects of membrane function because defects in flippase activity have been reported to cause problems in the vesicle-mediated transport of proteins in both the endocytic and exocytic pathways [166], in the non-vesicular trafficking of sterols [167], and in maintaining the axis of polarized growth through effects on the efficiency with which the small GTPases Cdc42 [168,169] and Rho1 [170] are recruited to the plasma membrane [168,169]. Conversely, it has been claimed recently that PtdSer, Cdc42, and Plc1 (a PtdIns4,5P2-specific phospholipase C) are all involved, by molecular mechanisms not elucidated, in the activation of TORC2 activity upon treatment of cells with non-physiological concentrations of a toxic aldehyde, methylglyoxal [171].
Regulation of integral plasma membrane protein endocytosis
Early observations had implicated Ypk1 action in the control of endocytosis [172]. Indeed, another remarkable consequence of Ypk1-mediated inhibition of Fpk1 is that it affects Fpk1 control of yet another protein kinase, Akl1 (1108 residues). Akl1 phosphorylates and inhibits several proteins necessary for the assembly and function of so-called actin patches [157,173], which are sites of clathrin-mediated endocytosis [174,175]. Fpk1-mediated phosphorylation of two conserved C-terminal sites in Akl1 inhibits its function [157]. Thus, under normal growth conditions, Fpk1-mediated inhibition of Akl1 prevents it from interfering with endocytosis. However, stimuli that elicit TORC2-Ypk1 signaling will down-regulate Fpk1-mediated inhibition of Akl1, thereby promoting Akl1-mediated phosphorylation and inhibition of the actin patch proteins. In this way, under stressful conditions that debilitate the plasma membrane and activate TORC2-Ypk1, endocytosis is impeded, a response that presumably assists in preserving membrane function when its integrity is most at risk.
Strikingly, Ypk1 also controls endocytosis in a manner independently from its regulation of Fpk1. Ypk1 phosphorylates and negatively regulates endocytic adaptors of the α-arrestin family, including well-established Ypk1 targets Rod1/Art4 and Aly2/Art3 [145,176]. The α-arrestins promote the internalization of nutrient permeases and other classes of integral plasma membrane proteins via the recruitment of the ubiquitin ligase (E3) Rsp5 (mammalian ortholog NEDD4L) [177]. In fact, of the 14 recognized α-arrestins encoded in the S. cerevisiae genome, ten of them possess at least one consensus Ypk1 site (Rod1 has two and Aly2 has four). Demonstrated clients of Rod1 include the GPCR Ste2 [178], the lactate transporter Jen1 [179], and glucose transporters Hxt1, Hxt3, and Hxt6 [180,181]. TORC2 also controls glucose transporter endocytosis in fission yeast [182,183]. For Aly2, documented clients include GPCR Ste3 [184], aspartate/glutamate transporter Dip5 [185], general amino acid permease Gap1 [186,187], the proline transporter Put4 [188], the arsenite transporter Acr3 [189], and the glycerophosphoinositol transporter Git1 [190].
So, overall, under stressful conditions (such as sphingolipid limitation, heat shock, or hypotonic conditions) that threaten plasma membrane integrity and activate TORC2-Ypk1 signaling, plasma membrane function will not be further compromised by endocytic removal of integral membrane proteins because Ypk1 action both blocks α-arrestin function and prevents Fpk1 from inhibiting Akl1. Conversely, upon hypertonic shock, where TORC2-Ypk1 signaling is shut off rapidly [49–51], plasma membrane proteins can be removed as part of the clearance of the ‘excess’ membrane created by cell shrinkage because α-arrestins remain fully functional and Fpk1 is able to prevent Akl1 from acting. Thus, by these mechanisms, TORC2-Ypk1 signaling adjusts the rate of endocytosis to meet the needs of the cell.
Control of sphingolipid biosynthesis
Ypk1 also phosphorylates and negatively regulates Orm1 and Orm2 [114,191], which are ER-localized tetraspanins [192,193] that inhibit the heterotrimeric enzyme (Lcb1–Lcb2–Tsc3) that catalyzes the first step unique to sphingolipid biosynthesis, namely L-serine:palmitoyl-CoA acyltransferase [194,195], thereby generally increasing flux into this metabolic pathway. It appears that Ypk1-mediated phosphorylation down-regulates Orm2 (and presumably Orm1) primarily by promoting Orm2 export from the ER, followed by its subsequent endosome- and Golgi-associated degradation [196].
In addition, and equally as important, Ypk1 phosphorylates and stimulates the catalytic subunits of the ER-localized heterotrimeric ceramide synthase (Lac1–Lag1–Lip1) [145,197], thereby ensuring that pathway up-regulation results in the efficient production of its complex sphingolipid end products. In fact, one of the most potent means to stimulate TORC2-mediated phosphorylation and activation of Ypk1 is to treat yeast cells with antibiotics that inactivate enzymes in the sphingolipid biosynthetic pathway, in particular myriocin (a transition state analog that potently blocks L-serine:palmitoyl-CoA acyltransferase [198]) [114] or Aureobasidin A (a compound that efficiently inhibits phosphatidylinositol:ceramide phosphoinositol transferase/IPC synthase [199]) [59].
Recently, a high-content microscopy screen identified Cvm1 as a protein located at sites where the membrane of the yeast vacuole (which, it should be recalled is the equivalent of a mammalian lysosome [200,201]) contacts three other organelles: mitochondria; peroxisomes; and, the ER emanating from the nuclear envelope [202]. Changes in the level of Cvm1 alters the levels of multiple sphingolipid classes, which can reduce the tolerance of yeast cells to various environmental stresses [203], and the contact sites containing Cvm1 are elevated when sphingolipid production is compromised [202]. The central segment of Cvm1 (residues 244–558) has a strongly predicted homology to the α–β hydrolase fold. Members of this protein superfamily include lipases and esterases [204], possibly explaining how Cvm1 influences the spectrum of sphingolipid species [205]. It was also claimed, on the basis of the effect of Cvm1 loss and overproduction on Orm1 phosphorylation assessed by very modest gel mobility shifts [202], that this protein is necessary to elicit the sphingolipid-sensing TORC2-Ypk1 signaling pathway. However, it was not directly tested whether, for example, the absence of Cvm1 prevents the myriocin-induced and Ypk1-dependent phosphorylation of Orm1 or any other well-characterized Ypk1 substrate.
In this same regard, it has also been reported recently that, using a TetOFF strategy to deplete cells of the ceramide synthase regulatory subunit Lip1 by addition of doxycyclin and thereby eventually cripple sphingolipid production, TORC2-Ypk1 signaling was activated. leading to hyperphosphorylation of Lag1, but without concomitant hyperphosphorylation of Orm1, despite the fact that both proteins are integral membrane proteins of the ER membrane [206]. A mechanistic explanation for this differential effect was not provided; however, Lip1 depletion requires a protracted period of time and, in the absence of Lip1, the structure of the complex will be disrupted and, once phosphorylated, perhaps Lag1 phospho-sites are more resistant to counteracting phosphatase action than those in Orm1. In fact, Lag1 seems to be dephosphorylated by calcineurin/PP2B [125,145,197], whereas Orm1 appears to be dephosphorylated by both PP2A [115] and calcineurin [196].
Regulation of intracellular glycerol concentration
Ypk1 phosphorylates and negatively regulates Gpd1 [49], one of two NADH-dependent dehydrogenases that reduce dihydroxyacetone-phosphate to sn-glycerol-3-phosphate (glycerol-3P). Under normal growth conditions, glycerol-3P serves [via its fatty acylation at the sn-1 and sn-2 positions to generate phosphatidic acid (PtdOH)] as the essential precursor to all glycerophospholipids. PtdOH is converted into the various glycerophospholipid species via two main routes, the CDP-diacylglycerol (DAG) pathway and an alternative DAG-dependent route (Kennedy pathway), as described in detail elsewhere [207,208]. Alternatively, DAG can be esterified with a third fatty acyl group on its sn-3 hydroxyl to generate triacylglycerol that is stored in lipid droplets [209,210].
However, when a yeast cell is subjected to the stress of being placed in hypertonic conditions, glycerol-3P has a totally different fate—it is dephosphorylated to glycerol by two phosphatases (Gpp1/Rhr2 and Gpp2/Hor1) whose cognate genes are also up-regulated upon hyperosmotic stress [211]. Generation of high internal glycerol is the strategy S. cerevisiae has evolved to provide a sufficient concentration of an innocuous intracellular osmolyte to combat the loss of water [212,213]. This response is dubbed the HOG (high osmolarity glycerol) pathway and its primary downstream effector is the MAPK Hog1, whose action controls many of the processes needed to cope with hyperosmotic stress, including induction of the expression of appropriate genes [212,213]. The main route for efflux of the accumulated glycerol is through the aquaglyceroporin channel Fps1 [214] and Ypk1 phosphorylation of Fps1 at three sites is necessary to maintain this channel in its open state [50].
The logic of how Ypk1-mediated phosphorylation regulates Gpd1 and Fps1 makes clear physiological sense in light of the need to respond immediately when a yeast cell is suddenly switched from isotonic to hypertonic conditions. Gpd1, which is found in the cytosol and inside peroxisomes, is kept inactive via phosphorylation by Ypk1 at a site just upstream of its catalytic domain. When cells are subjected to hyperosmotic shock, however, TORC2-Ypk1 signaling is dramatically and very rapidly decreased (in less than 1 min) [49–51]. Thus, under conditions were glycerol-3P is needed for glycerol production, Ypk1-mediated inhibition of Gpd1 function is quickly alleviated. This effect is subsequently further potentiated by the fact that, upon hyperosmotic stress, GPD1 mRNA and protein expression are markedly up-regulated in a Hog1-dependent manner [215], allowing the level of glycerol-3P for glycerol production to ramp up. Yet, glycerol will only accumulate inside the cell if the aquaglyceroporin Fps1 closes. Indeed, upon hyperosmotic shock, where TORC2-Ypk1 signaling is rapidly decreased, Ypk1-mediated phosphorylation of Fps1 is rapidly lost, causing channel closure and glycerol accumulation, enhancing survival under hypertonic stress [50]. Another mechanism for channel closure has also been uncovered that involves Hog1-mediated phosphorylation and displacement of two positive channel regulators, the BAR domain- and PH domain-containing proteins Rgc1 and Rgc2/Ask10 [216]. However, given that full Hog1 activation takes significantly longer (at least 5 min) and given that the Fps1 channel still closes in response to hyperosmotic shock even in hog1Δ cells [217,218], the loss of its Ypk1-mediated phosphorylation appears to be the predominant mechanism for blocking Fps1-mediated glycerol efflux.
Thus, overall, inactivation of TORC2-Ypk1 signaling upon hyperosmotic shock has two co-ordinated consequences that work synergistically to facilitate glycerol accumulation and promote cell survival—outcomes that work in conjunction with, but in a much more rapid and mechanistically distinct manner from, the processes evoked by activated Hog1. First, loss of TORC2-Ypk1 signaling alleviates inhibition of Gpd1 [49], which combined with eventual Hog1-stimulated transcriptional induction of GPD1, GPP1, and GPP2, greatly increases the rate of glycerol production. Second, and concomitantly, the loss of TORC2-Ypk1 signaling rapidly closes the Fps1 channel [50], thereby allowing the glycerol produced to be retained by the cell.
Control of retrograde sterol transport
In our combined bioinformatic-genetic-biochemical approach for conducting a proteome-wide screen for potential new high-confidence substrates of Ypk1 [2,145], Ysp2 (also known as Lam2/Ltc4) was identified as a likely physiologically relevant target. Shortly thereafter, Lam2/Ysp2 was shown to be one of a new family of sterol-binding StART-like (also called StARkin) domain-containing proteins located at plasma membrane-ER contact sites [219]. We then documented that Lam2/Ysp2 and its paralog Lam4/Ltc3 are authentic Ypk1 substrates in vivo and demonstrated using both genetic and biochemical criteria that Ypk1-mediated phosphorylation inhibits the ability of Lam2 and Lam4 to promote retrograde transport of sterols (specifically, ergosterol, which performs the equivalent role in fungal membranes that cholesterol does in animal cell membranes [220]) from the PM back to the ER [221]. We further found that, under the membrane-perturbing conditions known to activate TORC2-Ypk1 signaling, retention of sterol in the plasma membrane promotes cell survival, thus defining the underlying molecular basis of a new regulatory mechanism for cellular response to plasma membrane stress [221].
All six of the membrane-tethered sterol-binding Lam/Ltc proteins localize at junctions between the ER membrane and other cellular organelles; however, only Lam2/Ysp2 and Lam4 localize to ER-plasma membrane contact sites. Given our findings [211] that Ypk1-mediated phosphorylation of Lam2 and Lam4 inhibits their function in retrograde sterol transport, we sought to understand the mechanism of this negative regulation. We found [222], in agreement with a prior study [58] that, at ER-plasma membrane contact sites, Lam2 and Lam4 associate with two paralogous β-propeller (WD40 repeat) proteins Laf1/Ymr102c and Dgr2/Ykl121w, one of which was detected as a sterol-binding protein [223]. Using fluorescent tags, we showed that Lam2 and Lam4 remain at ER-plasma membrane contact sites when Laf1 and Dgr2 are absent, whereas neither Laf1 nor Dgr2 remain at ER-plasma membrane contact sites when Lam2 and Lam4 are absent [222]. Furthermore, loss of Laf1 (but not Dgr2) impeded retrograde ergosterol transport, and a laf1Δ mutation did not exacerbate the transport defect of lam2Δ lam4Δ cells, indicating a shared function. We demonstrated that Lam2 and Lam4 bind Laf1 and Dgr2 in vitro in a pull-down assays, and that the PH-like domain in Lam2 hinders its interaction with Laf1 [222]. Most tellingly, Lam2 phosphorylated by Ypk1, or Lam2 with phosphomimetic (Glu) replacements at its Ypk1 phospho-acceptor sites, exhibited a marked reduction in Laf1 binding [222]. Thus, at least one mechanism by which Ypk1-mediated phosphorylation impedes Lam2 function and inhibits retrograde sterol transport is via blocking its interaction with Laf1 at ER-plasma membrane contact sites.
Regulation of stability and localization of mRNAs encoding integral membrane proteins
In our unbiased proteome-wide screen for targets of the TORC2-stimulated protein kinase Ypk1, we also identified the paralogs Puf1/Jsn1 and Puf2 as high-confidence substrates [2,145]. Puf1 and Puf2 are members of the Puf family of RNA-binding proteins, which typically associate via their Pumilio homology domain with specific short motifs in the 3′-UTR of an mRNA and thereby influence the stability, localization and/or efficiency of translation of the bound transcript [224,225]. Remarkably, consistent with all of the studies discussed above documenting that a primary physiological role of TORC2-Ypk1 signaling in yeast is homeostatic maintenance of the components of the plasma membrane, prior work by others had demonstrated that Puf1 and Puf2 exhibit a marked preference for interaction with mRNAs encoding plasma membrane-associated proteins, including integral membrane proteins [226,227]. Indeed, we were able to show, first, that both Puf1 and Puf2 are authentic Ypk1 substrates both in vitro and in vivo [228]. Second, we found that fluorescently tagged Puf1 localizes constitutively in cortical puncta closely apposed to the plasma membrane, whereas Puf2 does so in the absence of its Ypk1 phosphorylation, but is dispersed in the cytosol when phosphorylated [228]. We further demonstrated that Ypk1-mediated phosphorylation of Puf1 and Puf2 up-regulates the production of the protein products of the transcripts to which they bind, with a concomitant increase in the level of the cognate mRNAs [228]. Thus, Ypk1 phosphorylation relieves Puf1- and Puf2-mediated post-transcriptional repression mainly by counteracting their negative effect on transcript stability. Moreover, using a heterologous protein–RNA tethering and fluorescent protein reporter assay, the consequence of Ypk1 phosphorylation in vivo was recapitulated for full-length Puf1 and even for N-terminal fragments (residues 1–340 and 143–295) corresponding to the region upstream of its dimerization domain (an RNA-recognition motif fold) encompassing its two Ypk1 phosphorylation sites (both also conserved in Puf2) [228]. This latter result suggests that alleviation of Puf1-imposed transcript destabilization does not obligatorily require dissociation of Ypk1-phosphorylated Puf1 from a transcript. Overall, these findings have added new insight about yet another level by which the TORC2-Ypk1 signaling axis regulates the content of plasma membrane-associated proteins to promote the maintenance of the integrity of the cell envelope.
Other potential Ypk1 targets
Additional candidate substrates for Ypk1 (and Ypk2) have been discussed previously [2,145], but are less well characterized, in that they have not been shown to be directly phosphorylated by Ypk1 either in vitro or in vivo, and/or the sites of phosphorylation have not been mapped, and/or the phenotypic consequences of preventing phosphorylation at demonstrated Ypk1 sites have not been examined. Since that time, Ypk1 action has been implicated in other processes [155], but by what specific mechanism is less clear, including preventing chromosome fragmentation in response to insults that cause double-stranded DNA breaks [229], suggesting that lipids other than PtdIns4,5P2 may regulate TORC2 signaling [197], influencing the expression of the gene encoding Atg8 (mammalian orthologs are GABARAPs) by the transcription factors Msn2 and Msn4 [230], and affecting the function of the transcription factor Swi4 [231]. It has also been noted that Ypk1 is able to phosphorylate a site (S232) very near the C-terminus of the ribosomal 40S subunit protein Rps6, but its physiological impact is unclear [232]. Similarly, it has been claimed that Ypk1 (as well as Pkc1) phosphorylate a site (S223) very near the C-terminus of ribosomal 40S subunit protein Rps5 [233]; however, the context of that site does not match at all the consensus Ypk1 phospho-acceptor site. It has also been reported [234] that TORC2-Ypk1 signaling modulates cell size and growth rate through a rather baroque circuitry, perhaps involving promoting the accumulation of the G1 cyclin Cln3 [235], rather than via all of the other Ypk1-controlled processes that were explicated in the preceding sections and which, collectively, are all necessary for allowing the plasma membrane to expand and to maintain its integrity, thereby permitting cell growth and survival.
Thus, in conclusion, it has emerged from studies in S. cerevisiae that TORC2 plays an crucial role in sensing the status of the plasma membrane and controlling via Ypk1 a multitude of reactions and processes that regulate plasma membrane lipid and protein composition (Figure 4), thereby ensuring maintenance of plasma membrane homeostasis under both balanced growth conditions and in response to stresses that might damage plasma membrane integrity and/or plasma membrane function.
Perspective
Protein kinases controlled by TOR have emerged as universal, centrally important sensors, integrators, and controllers of eukaryotic cell growth.
In human cells, TORC2 dysfunction has been linked to many pathologies, including type 2 diabetes mellitus, hepatic steatosis, obesity, many types of cancer, pulmonary fibrosis, systemic lupus erythematosus, and lifespan.
Analysis of the TORC2-Ypk1 signaling axis in yeast has proven to be a valuable model to extract general principles about the biochemical features and physiological logic of TORC2 signaling.
In S. cerevisiae, there are many potential Ypk1 targets still unexplored and the challenge going forward is to continue to dissect the complexities of the processes and the extent of the networks through which TORC2-Ypk1 signaling affects cell function and viability.
Acknowledgements
The author expresses his profound gratitude to the many generations of postdoctoral associates, graduate students, undergraduate honors students, and visiting student interns from other institutions across the U.S.A., as well as many from other nations—too numerous to mention individually by name here—who contributed to the research that we achieved together over nearly five decades. However, special thanks go to Ms. Riyo Kunisawa and Dr. Françoise M. Roelants, two long-term Research Specialists whose service spanned, respectively, nearly 20 years at the beginning of, and more than 20 years at the end of, the life of the Thorner Lab. Their comradeship, enthusiasm, dedication and skill were exceptional and I will always be deeply indebted to both of them. I also thank my colleague James H. Hurley for his insights about and assistance with protein structure comparisons. Finally, I apologize in advance to any investigators in this field whose work I inadvertently overlooked and failed to cite.
Abbreviations
- ANK
ankyrin
- ARM
armadillo
- CRIM
conserved region in the middle
- DAG
diacylglycerol
- ER
endoplasmic reticulum
- HM
hydrophobic motif
- HOG
high osmolarity glycerol
- MIPC
mannosyl-inositolphosphoryl-ceramide
- RBD
Ras-binding domain
- TIM
TOR interaction motif
Competing Interests
The author declares that there are no competing interests associated with this manuscript.
Funding
All of the work described in this article that was accomplished in the Thorner Lab at UC Berkeley was supported by R01 Research Grant GM021841 from the US National Institute of General Medical Sciences from 1 January 1975 to 30 June 2021.
CRediT Author Contribution
Jeremy Thorner: Conceptualization, Funding acquisition, Writing—original draft, Writing—review and editing.
Supplementary Material
References
- 1.Eltschinger, S. and Loewith, R. (2016) TOR complexes and the maintenance of cellular homeostasis. Trends Cell Biol. 26, 148–159 10.1016/j.tcb.2015.10.003 [DOI] [PubMed] [Google Scholar]
- 2.Roelants, F.M., Leskoske, K.L., Martinez Marshall, M.N., Locke, M.N. and Thorner, J. (2017) The TORC2-dependent signaling network in the yeast Saccharomyces cerevisiae. Biomolecules 7, E66 10.3390/biom7030066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morozumi, Y. and Shiozaki, K. (2021) Conserved and divergent mechanisms that control TORC1 in yeasts and mammals. Genes (Basel) 12, 88 10.3390/genes12010088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.González, A. and Hall, M.N. (2017) Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 36, 397–408 10.15252/embj.201696010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condon, K.J. and Sabatini, D.M. (2019) Nutrient regulation of mTORC1 at a glance. J. Cell Sci. 132, jcs222570 10.1242/jcs.222570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lutt, N. and Brunkard, J.O. (2022) Amino acid signaling for TOR in eukaryotes: sensors, transducers, and a sustainable agricultural fuTORe. Biomolecules 12, 387 10.3390/biom12030387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heitman, J., Movva, N.R. and Hall, M.N. (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253, 905–909 10.1126/science.1715094 [DOI] [PubMed] [Google Scholar]
- 8.Ikai, N., Nakazawa, N., Hayashi, T. and Yanagida, M. (2011) The reverse, but coordinated, roles of Tor2 (TORC1) and Tor1 (TORC2) kinases for growth, cell cycle and separase-mediated mitosis in Schizosaccharomyces pombe. Open Biol. 1, 110007 10.1098/rsob.110007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shertz, C.A., Bastidas, R.J., Li, W., Heitman, J. and Cardenas, M.E. (2010) Conservation, duplication, and loss of the Tor signaling pathway in the fungal kingdom. BMC Genom. 23, 510 10.1186/1471-2164-11-510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunz, J., Schneider, U., Howald, I., Schmidt, A. and Hall, M.N. (2000) HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J. Biol. Chem. 275, 37011–37020 10.1074/jbc.M007296200 [DOI] [PubMed] [Google Scholar]
- 11.Sturgill, T.W., Cohen, A., Diefenbacher, M., Trautwein, M., Martin, D.E. and Hall, M.N. (2008) TOR1 and TOR2 have distinct locations in live cells. Eukaryot. Cell 7, 1819–1830 10.1128/EC.00088-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunz, J., Henriquez, R., Schneider, U., Deuter-Reinhard, M., Movva, N.R. and Hall, M.N. (1993) Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73, 585–596 10.1016/0092-8674(93)90144-f [DOI] [PubMed] [Google Scholar]
- 13.Helliwell, S.B., Wagner, P., Kunz, J., Deuter-Reinhard, M., Henriquez, R. and Hall, M.N. (1994) TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell 5, 105–118 10.1091/mbc.5.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loewith, R., Jacinto, E., Wullschleger, S., Lorberg, A., Crespo, J.L., Bonenfant, D.et al. (2002) Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10, 457–468 10.1016/s1097-2765(02)00636-6 [DOI] [PubMed] [Google Scholar]
- 15.Wedaman, K.P., Reinke, A., Anderson, S., Yates, J., McCaffery, J.M. and Powers, T. (2003) Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 1204–1220 10.1091/mbc.E02-09-0609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karuppasamy, M., Kusmider, B., O, T.M., Gaubitz, C., Prouteau, M., L, R.et al. (2017) Cryo-EM structure of Saccharomyces cerevisiae target of rapamycin complex 2. Nat. Commun. 8, 1729 10.1038/s41467-017-01862-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tafur, L., Kefauver, J. and Loewith, R. (2020) Structural insights into TOR signaling. Genes (Basel) 11, 885 10.3390/genes11080885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, X., Liu, M., Tian, Y., Li, J., Qi, Y., Zhao, D.et al. (2018) Cryo-EM structure of human mTOR complex 2. Cell Res. 28, 518–528 10.1038/s41422-018-0029-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stuttfeld, E., Aylett, C.H., I, S., B, D., Scaiola, A., Sauer, E.et al. (2018) Architecture of the human mTORC2 core complex. eLlife 7, e33101 10.7554/eLife.33101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scaiola, A., Mangia, F., Imseng, S., Boehringer, D., Berneiser, K., Shimobayashi, M.et al. (2020) The 3.2-Å resolution structure of human mTORC2. Sci. Adv. 6, eabc1251. 10.1126/sciadv.abc1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill, A., Niles, B., Cuyegkeng, A. and Powers, T. (2018) Redesigning TOR kinase to explore the structural basis for TORC1 and TORC2 assembly. Biomolecules 8, 36 10.3390/biom8020036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsverov, J., Yegorov, K. and Powers, T. (2022) Identification of defined structural elements within TOR2 kinase required for TOR complex 2 assembly and function in Saccharomyces cerevisiae. Mol. Biol. Cell 33, ar44 10.1091/mbc.E21-12-0611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang, H., Rudge, D.G., Koos, J.D., Vaidialingam, B., Yang, H.J. and Pavletich, N.P. (2013) mTOR kinase structure, mechanism and regulation. Nature 497, 217–223 10.1038/nature12122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baretić, D., Berndt, A., Ohashi, Y., Johnson, C.M. and Williams, R.L. (2016) Tor forms a dimer through an N-terminal helical solenoid with a complex topology. Nat. Commun. 7, 11016 10.1038/ncomms11016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loewith, R. and Hall, M.N. (2011) Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189, 1177–1201 10.1534/genetics.111.133363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaubitz, C., Prouteau, M., Kusmider, B. and Loewith, R. (2016) TORC2 structure and function. Trends Biochem. Sci. 41, 532–545 10.1016/j.tibs.2016.04.001 [DOI] [PubMed] [Google Scholar]
- 27.Wullschleger, S., Loewith, R., Oppliger, W. and Hall, M.N. (2005) Molecular organization of target of rapamycin complex 2. J. Biol. Chem. 280, 30697–30704 10.1074/jbc.M505553200 [DOI] [PubMed] [Google Scholar]
- 28.Ho, H.L., Lee, H.Y., Liao, H.C. and Chen, M.Y. (2008) Involvement of Saccharomyces cerevisiae Avo3p/Tsc11p in maintaining TOR complex 2 integrity and coupling to downstream signaling. Eukaryot. Cell 7, 1328–1343 10.1128/EC.00065-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez Marshall, M. N., Emmerstorfer-Augustin, A., Leskoske, K. L., Zhang, L. H., Li, B. and Thorner, J. (2019) Analysis of the roles of phosphatidylinositol-4,5-bisphosphate and individual subunits in assembly, localization, and function of Saccharomyces cerevisiae target of rapamycin complex 2. Mol. Biol. Cell 30, 1555–1574. 10.1091/mbc.E18-10-0682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaubitz, C., Oliveira, T.M., Prouteau, M., Leitner, A., Karuppasamy, M., Konstantinidou, G.et al. (2015) Molecular basis of the rapamycin insensitivity of target of rapamycin complex 2. Mol. Cell 58, 977–988 10.1016/j.molcel.2015.04.031 [DOI] [PubMed] [Google Scholar]
- 31.Yu, Z., Chen, J., Takagi, E., Wang, F., Saha, B., Liu, X.et al. (2022) Interactions between mTORC2 core subunits Rictor and mSin1 dictate selective and context-dependent phosphorylation of substrate kinases SGK1 and Akt. J. Biol. Chem. 298, 102288 10.1016/j.jbc.2022.102288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furuita, K., Kataoka, S., Sugiki, T., Hattori, Y., Kobayashi, N., Ikegami, T.et al. (2015) Utilization of paramagnetic relaxation enhancements for high- resolution NMR structure determination of a soluble loop-rich protein with sparse NOE distance restraints. J. Biomol. NMR 61, 55–64 10.1007/s10858-014-9882-7 [DOI] [PubMed] [Google Scholar]
- 33.Kataoka, S., Furuita, K., Hattori, Y., Kobayashi, N., Ikegami, T., Shiozaki, K.et al. (2015) 1)H, (15)N and (13)C resonance assignments of the conserved region in the middle domain of S. pombe Sin1 protein. Biomol. NMR Assign. 9, 89–92 10.1007/s12104-014-9550-6 [DOI] [PubMed] [Google Scholar]
- 34.Yang, Q., Inoki, K., Ikenoue, T. and Guan, K.L. (2006) Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 20, 2820–2832 10.1101/gad.1461206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cameron, A.J., Linch, M.D., Saurin, A.T., Escribano, C. and Parker, P.J. (2011) mTORC2 targets AGC kinases through Sin1-dependent recruitment. Biochem. J. 439, 287–297 10.1042/bj20110678 [DOI] [PubMed] [Google Scholar]
- 36.Lu, M., Wang, J., Ives, H.E. and Pearce, D. (2011) mSIN1 protein mediates SGK1 protein interaction with mTORC2 protein complex and is required for selective activation of the epithelial sodium channel. J. Biol. Chem. 286, 30647–30654 10.1074/jbc.M111.257592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao, H.C. and Chen, M.Y. (2012) Target of rapamycin complex 2 signals to downstream effector yeast protein kinase 2 (Ypk2) through adheres-voraciously-to-target-of-rapamycin-2 protein 1 (Avo1) in Saccharomyces cerevisiae. J. Biol. Chem. 287, 6089–6099 10.1074/jbc.M111.303701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tatebe, H., Murayama, S., Yonekura, T., Hatano, T., Richter, D., Furuya, T.et al. (2017) Substrate specificity of TOR complex 2 is determined by a ubiquitin-fold domain of the Sin1 subunit. eLife 6, e19594 10.7554/eLife.19594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Battaglioni, S., Benjamin, D., Wälchli, M., Maier, T. and Hall, M.N. (2022) mTOR substrate phosphorylation in growth control. Cell 185, 1814–1836 10.1016/j.cell.2022.04.013 [DOI] [PubMed] [Google Scholar]
- 40.Castel, P., Dharmaiah, S., Sale, M.J., Messing, S., Rizzuto, G., Cuevas-Navarro, A.et al. (2021) RAS interaction with Sin1 is dispensable for mTORC2 assembly and activity. Proc. Natl Acad. Sci. U.S.A. 118, e2103261118 10.1073/pnas.2103261118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng, Y., Ding, L., Meng, X., Potter, M., Kearney, A.L., Zhang, J.et al. (2022) Structural insights into Ras regulation by SIN1. Proc. Natl Acad. Sci. U.S.A. 119, e2119990119 10.1073/pnas.2119990119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan, D. and Matsuura, Y. (2012) Structures of the pleckstrin homology domain of Saccharomyces cerevisiae Avo1 and its human orthologue Sin1, an essential subunit of TOR complex 2. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68, 386–392 10.1107/s1744309112007178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G.et al. (2022) Alphafold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 10.1093/nar/gkab1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berchtold, D. and Walther, T.C. (2009) TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol. Biol. Cell 20, 1565–1575 10.1091/mbc.E08-10-1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu, P., Gan, W., Chin, Y.R., Ogura, K., Guo, J., Zhang, J.et al. (2015) Ptdins(3,4,5)P3-dependent activation of the mTORC2 kinase complex. Cancer Discov. 5, 1194–1209 10.1158/2159-8290.CD-15-0460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pontes, B., Monzo, P. and Gauthier, N.C. (2017) Membrane tension: a challenging but universal physical parameter in cell biology. Semin. Cell Dev. Biol. 71, 30–41 10.1016/j.semcdb.2017.08.030 [DOI] [PubMed] [Google Scholar]
- 47.Kennedy, M.A., Gable, K., Niewola-Staszkowska, K., Abreu, S., Johnston, A., Harris, L.J.et al. (2014) A neurotoxic glycerophosphocholine impacts PtdIns-4, 5-bisphosphate and TORC2 signaling by altering ceramide biosynthesis in yeast. PLoS Genet. 10, e1004010 10.1371/journal.pgen.1004010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riggi, M., Niewola-Staszkowska, K., Chiaruttini, N., Colom, A., Kusmider, B., Mercier, V.et al. (2018) Decrease in plasma membrane tension triggers PtdIns(4,5)P2 phase separation to inactivate TORC2. Nat. Cell Biol. 20, 1043–1051 10.1038/s41556-018-0150-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee, Y.J., Jeschke, G.R., Roelants, F.M., Thorner, J. and Turk, B.E. (2012) Reciprocal phosphorylation of yeast glycerol-3-phosphate dehydrogenases in adaptation to distinct types of stress. Mol. Cell. Biol. 32, 4705–4717 10.1128/mcb.00897-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muir, A., Roelants, F.M., Timmons, G., Leskoske, K.L. and Thorner, J. (2015) Down-regulation of TORC2-Ypk1 signaling promotes MAPK-independent survival under hyperosmotic stress. eLlife 4, e09336 10.7554/eLife.09336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leskoske, K.L., Roelants, F.M., Emmerstorfer-Augustin, A., Augustin, C.M., Si, E.P., Hill, J.M.et al. (2018) Phosphorylation by the stress-activated MAPK Slt2 down-regulates the yeast TOR complex 2. Genes Dev. 32, 1576–1590 10.1101/gad.318709.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berchtold, D., Piccolis, M., Chiaruttini, N., Riezman, I., Riezman, H., Roux, A.et al. (2012) Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat. Cell Biol. 14, 542–547 10.1038/ncb2480 [DOI] [PubMed] [Google Scholar]
- 53.Niles, B.J., Mogri, H., Hill, A., Vlahakis, A. and Powers, T. (2012) Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proc. Natl Acad. Sci. U.S.A. 109, 1536–1541 10.1073/pnas.1117563109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Audhya, A., Loewith, R., Parsons, A.B., Gao, L., Tabuchi, M., Zhou, H.et al. (2004) Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J. 23, 3747–3757 10.1038/sj.emboj.7600384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karotki, L., Huiskonen, J.T., Stefan, C.J., Ziółkowska, N.E., Roth, R., Surma, M.A.et al. (2011) Eisosome proteins assemble into a membrane scaffold. J. Cell Biol. 195, 889–902 10.1083/jcb.201104040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lanze, C.E., Gandra, R.M., Foderaro, J.E., Swenson, K.A., Douglas, L.M. and Konopka, J.B. (2020) Plasma membrane MCC/eisosome domains promote stress resistance in fungi. Microbiol. Mol. Biol. Rev. 84, e00063-19 10.1128/MMBR.00063-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakata, K.T., Hashii, K., Yoshizawa, K., Tahara, Y.O., Yae, K., Tsuda, R.et al. (2022) Coordinated regulation of TORC2 signaling by MCC/eisosome-associated proteins, Pil1 and tetraspan membrane proteins during the stress response. Mol. Microbiol. 117, 1227–1244 10.1111/mmi.14903 [DOI] [PubMed] [Google Scholar]
- 58.Murley, A., Yamada, J., Niles, B.J., Toulmay, A., Prinz, W.A., Powers, T.et al. (2017) Sterol transporters at membrane contact sites regulate TORC1 and TORC2 signaling. J. Cell Biol. 216, 2679–2689 10.1083/jcb.201610032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Locke, M.N. and Thorner, J. (2019) Rab5 GTPases are required for optimal TORC2 function. J. Cell Biol. 218, 961–976 10.1083/jcb.201807154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olivera-Couto, A., Graña, M., Harispe, L. and Aguilar, P.S. (2011) The eisosome core is composed of BAR domain proteins. Mol. Biol. Cell 22, 2360–2372 10.1091/mbc.E10-12-1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anand, K., Maeda, K. and Gavin, A.C. (2012) Structural analyses of the Slm1-PH domain demonstrate ligand binding in the non-canonical site. PLoS One 7, e36526 10.1371/journal.pone.0036526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tabuchi, M., Audhya, A., Parsons, A.B., Boone, C. and Emr, S.D. (2006) The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol. Cell. Biol. 26, 5861–5875 10.1128/MCB.02403-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gallego, O., Betts, M.J., Gvozdenovic-Jeremic, J., Maeda, K., Matetzki, C., Aguilar-Gurrieri, C.et al. (2010) A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae. Mol. Syst. Biol. 6, 430 10.1038/msb.2010.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bultynck, G., Heath, V.L., Majeed, A.P., Galan, J.M., Haguenauer-Tsapis, R. and Cyert, M.S. (2006) Slm1 and Slm2 are novel substrates of the calcineurin phosphatase required for heat stress-induced endocytosis of the yeast uracil permease. Mol. Cell. Biol. 26, 4729–4745 10.1128/MCB.01973-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Daquinag, A., Fadri, M., Jung, S.Y., Qin, J. and Kunz, J. (2007) The yeast PH domain proteins Slm1 and Slm2 are targets of sphingolipid signaling during the response to heat stress. Mol. Cell. Biol. 27, 633–650 10.1128/mcb.00461-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Niles, B.J. and Powers, T. (2012) Plasma membrane proteins Slm1 and Slm2 mediate activation of the AGC kinase Ypk1 by TORC2 and sphingolipids in S. cerevisiae. Cell Cycle 11, 3745–3749 10.4161/cc.21752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leskoske, K.L., Roelants, F.M., Marshall, M.N.M., Hill, J.M. and Thorner, J. (2017) The stress-sensing TORC2 complex activates yeast AGC-family protein kinase Ypk1 at multiple novel sites. Genetics 207, 179–195 10.1534/genetics.117.1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roelants, F.M., Torrance, P.D., Bezman, N. and Thorner, J. (2002) Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol. Biol. Cell 13, 3005–3028 10.1091/mbc.E02-04-0201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boeke, J.D., LaCroute, F. and Fink, G.R. (1984) A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197, 345–346 10.1007/BF00330984 [DOI] [PubMed] [Google Scholar]
- 70.Nishimura, K., Fukagawa, T., Takisawa, H., Kakimoto, T. and Kanemaki, M. (2009) An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- 71.Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R.et al. (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 10.1038/35001009 [DOI] [PubMed] [Google Scholar]
- 72.Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M. and Sakaki, Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. U.S.A. 98, 4569–4574 10.1073/pnas.061034498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fadri, M., Daquinag, A., Wang, S., Xue, T. and Kunz, J. (2005) The pleckstrin homology domain proteins Slm1 and Slm2 are required for actin cytoskeleton organization in yeast and bind phosphatidylinositol-4,5-bisphosphate and TORC2. Mol. Biol. Cell 16, 1883–1900 10.1091/mbc.E04-07-0564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee, J., Liu, L. and Levin, D.E. (2019) Stressing out or stressing in: intracellular pathways for SAPK activation. Curr. Genet. 65, 417–421 10.1007/s00294-018-0898-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.González-Rubio, G., Sellers-Moya, A., Martín, H. and Molina, M. (2021) A walk-through MAPK structure and functionality with the 30-year-old yeast MAPK Slt2. Int. Microbiol. 24, 531–543 10.1007/s10123-021-00183-z [DOI] [PubMed] [Google Scholar]
- 76.Albuquerque, C.P., Smolka, M.B., Payne, S.H., Bafna, V., Eng, J. and Zhou, H. (2008) A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteom. 7, 1389–1396 10.1074/mcp.M700468-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Holt, L.J., Tuch, B.B., Villén, J., Johnson, A.D., Gygi, S.P. and Morgan, D.O. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 10.1126/science.1172867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Swaney, D.L., Beltrao, P., Starita, L., Guo, A., Rush, J., Fields, S.et al. (2013) Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 10, 676–682 10.1038/nmeth.2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.MacGilvray, M.E., Shishkova, E., Place, M., Wagner, E.R., Coon, J.J. and Gasch, A.P. (2020) Phosphoproteome response to dithiothreitol reveals unique versus shared features of Saccharomyces cerevisiae stress responses. J. Proteome Res. 19, 3405–3417 10.1021/acs.jproteome.0c00253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lanz, M.C., Yugandhar, K., Gupta, S., Sanford, E.J., Faça, V.M., Vega, S.et al. (2021) In-depth and 3-dimensional exploration of the budding yeast phosphoproteome. EMBO Rep. 22, e51121 10.15252/embr.202051121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goldman, A., Roy, J., Bodenmiller, B., Wanka, S., Landry, C.R., Aebersold, R.et al. (2014) The calcineurin signaling network evolves via conserved kinase-phosphatase modules that transcend substrate identity. Mol. Cell 55, 422–435 10.1016/j.molcel.2014.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kamble, C., Jain, S., Murphy, E. and Kim, K. (2011) Requirements of Slm proteins for proper eisosome organization, endocytic trafficking and recycling in the yeast Saccharomyces cerevisiae. J. Biosci. 36, 79–96 10.1007/s12038-011-9018-0 [DOI] [PubMed] [Google Scholar]
- 83.Moseley, J.B. (2018) Eisosomes. Curr. Biol. 28, R376–R378 10.1016/j.cub.2017.11.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ho, B., Baryshnikova, A. and Brown, G.W. (2018) Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst. 6, 192–205.e193 10.1016/j.cels.2017.12.004 [DOI] [PubMed] [Google Scholar]
- 85.Shioda, R. (2006) Functional Analysis of TOR Complex 2 and its Control of Sphingolipid Biosynthesis in Saccharomyces cerevisiae. Ph.D. Thesis, Dept. of Biochemistry, University of Basel, Basel, Switzerland, 108pp. https://edoc.unibas.ch/759/1/DissB_8289.pdf
- 86.Gatherar, M., Pollerman, S., Dunn-Coleman, N. and Turner, G. (2004) Identification of a novel gene hbrB required for polarised growth in Aspergillus nidulans. Fungal Genet. Biol. 41, 463–471 10.1016/j.fgb.2003.12.004 [DOI] [PubMed] [Google Scholar]
- 87.Pearce, L.R., Sommer, E.M., Sakamoto, K., Wullschleger, S. and Alessi, D.R. (2011) Protor-1 is required for efficient mTORC2-mediated activation of SGK1 in the kidney. Biochem. J. 436, 169–179 10.1042/bj20102103 [DOI] [PubMed] [Google Scholar]
- 88.An, P., Xu, W., Luo, J. and Luo, Y. (2021) Expanding TOR Complex 2 signaling: emerging regulators and new connections. Front. Cell Dev. Biol. 9, 713806 10.3389/fcell.2021.713806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Colicelli, J., Nicolette, C., Birchmeier, C., Rodgers, L., Riggs, M. and Wigler, M. (1991) Expression of three mammalian cDNAs that interfere with RAS function in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. U.S.A. 88, 2913–2917 10.1073/pnas.88.7.2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schroder, W.A., Buck, M., Cloonan, N., Hancock, J.F., Suhrbier, A., Sculley, T.et al. (2007) Human Sin1 contains Ras-binding and pleckstrin homology domains and suppresses Ras signalling. Cell Signal. 19, 1279–1289 10.1016/j.cellsig.2007.01.013 [DOI] [PubMed] [Google Scholar]
- 91.Lee, S., Parent, C.A., Insall, R. and Firtel, R.A. (1999) A novel Ras-interacting protein required for chemotaxis and cyclic adenosine monophosphate signal relay in Dictyostelium. Mol. Biol. Cell 10, 2829–2845 10.1091/mbc.10.9.2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Khanna, A., Lotfi, P., Chavan, A.J., Montaño, N.M., Bolourani, P., Weeks, G.et al. (2016) The small GTPases Ras and Rap1 bind to and control TORC2 activity. Sci. Rep. 6, 25823 10.1038/srep25823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yaffe, M.B., Leparc, G.G., L, J., Obata, T., Volinia, S. and Cantley, L.C. (2001) A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat. Biotechnol. 19, 348–353 10.1038/86737 [DOI] [PubMed] [Google Scholar]
- 94.Dionne, U. and Gingras, A.C. (2022) Proximity-dependent biotinylation approaches to explore the dynamic compartmentalized proteome. Front. Mol. Biosci. 9, 852911 10.3389/fmolb.2022.852911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kovalski, J.R., Bhaduri, A., Zehnder, A.M., Neela, P.H., Che, Y., Wozniak, G.G.et al. (2019) The functional proximal proteome of oncogenic Ras includes mTORC2. Mol. Cell 73, 830–844.e812 10.1016/j.molcel.2018.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lone, M.U., Miyan, J., Asif, M., Malik, S.A., Dubey, P., Singh, V.et al. (2019) Direct physical interaction of active Ras with mSIN1 regulates mTORC2 signaling. BMC Cancer 19, 1236 10.1186/s12885-019-6422-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.García-Martínez, J.M. and Alessi, D.R. (2008) mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem. J. 416, 375–385 10.1042/BJ20081668 [DOI] [PubMed] [Google Scholar]
- 98.Fu, W. and Hall, M.N. (2020) Regulation of mTORC2 signaling. Genes (Basel) 11, 1045 10.3390/genes11091045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Van Aelst, L., White, M.A. and Wigler, M.H. (1994) Ras partners. Cold Spring Harb. Symp. Quant. Biol. 59, 181–186 10.1101/sqb [DOI] [PubMed] [Google Scholar]
- 100.Broach, J.R. (2012) Nutritional control of growth and development in yeast. Genetics 192, 73–105 10.1534/genetics.111.135731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Garrett, S. and Broach, J. (1989) Loss of Ras activity in Saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAK1, whose product may act downstream of the cAMP-dependent protein kinase. Genes Dev. 3, 1336–1348 10.1101/gad.3.9.1336 [DOI] [PubMed] [Google Scholar]
- 102.Sun, J., Kale, S.P., Childress, A.M., Pinswasdi, C. and Jazwinski, S.M. (1994) Divergent roles of RAS1 and RAS2 in yeast longevity. J. Biol. Chem. 269, 18638–18645 10.1016/S0021-9258(17)32357-8 [DOI] [PubMed] [Google Scholar]
- 103.Fujiyama, A., Tsunasawa, S., Tamanoi, F. and Sakiyama, F. (1991) S-farnesylation and methyl esterification of C-terminal domain of yeast RAS2 protein prior to fatty acid acylation. J. Biol. Chem. 266, 17926–17931 10.1016/S0021-9258(18)55216-9 [DOI] [PubMed] [Google Scholar]
- 104.Linder, M.E. and Deschenes, R.J. (2007) Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74–84 10.1038/nrm2084 [DOI] [PubMed] [Google Scholar]
- 105.Locke, M.N. and Thorner, J. (2019) Regulation of TORC2 function and localization by Rab5 GTPases in Saccharomyces cerevisiae. Cell Cycle 18, 1084–1094 10.1080/15384101.2019.1616999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang, H., Jiang, X., Li, B., Yang, H.J., Miller, M., Yang, A.et al. (2017) Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature 552, 368–373 10.1038/nature25023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Woodman, P.G. (2000) Biogenesis of the sorting endosome: the role of Rab5. Traffic 1, 695–701 10.1034/j.1600-0854.2000.010902.x [DOI] [PubMed] [Google Scholar]
- 108.Carney, D.S., Davies, B.A. and Horazdovsky, B.F. (2006) Vps9 domain-containing proteins: activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 16, 27–35 10.1016/j.tcb.2005.11.001 [DOI] [PubMed] [Google Scholar]
- 109.Wandinger-Ness, A. and Zerial, M. (2014) Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb. Perspect. Biol. 6, a022616 10.1101/cshperspect.a022616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alcaide-Gavilán, M., Lucena, R., Banuelos, S. and Kellogg, D.R. (2020) Conserved Ark1-related kinases function in a TORC2 signaling network. Mol. Biol. Cell 31, 2057–2069 10.1091/mbc.E19-12-0685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Luciano, A.K., Korobkina, E., Lyons, S.P., Haley, J.A., Fluharty, S., Jung, S.M.et al. (2022) Proximity labeling of endogenous RICTOR identifies mTOR Complex 2 regulation by ADP ribosylation factor ARF1. J. Biol. Chem. 298. 10.1016/j.jbc.2022.102379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Highland, C.M. and Fromme, J.C. (2021) Arf1 directly recruits the Pik1-Frq1 PI4K complex to regulate the final stages of Golgi maturation. Mol. Biol. Cell 32, 1064–1080 10.1091/mbc.E21-02-0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kolakowski, D., Rzepnikowska, W., Kaniak-Golik, A., Zoladek, T. and Kaminska, J. (2021) The GTPase Arf1 Is a determinant of yeast Vps13 localization to the Golgi apparatus. Int. J. Mol. Sci. 22, 12274 10.3390/ijms222212274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Roelants, F.M., Breslow, D.K., Muir, A., Weissman, J.S. and Thorner, J. (2011) Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. U.S.A. 108, 19222–19227 10.1073/pnas.1116948108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sun, Y., Miao, Y., Yamane, Y., Zhang, C., Shokat, K.M., Takematsu, H.et al. (2012) Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55-PP2A pathways. Mol. Biol. Cell 23, 2388–2398 10.1091/mbc.E12-03-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guerreiro, J.F., Muir, A., Ramachandran, S., Thorner, J. and Sá-Correia, I. (2016) Sphingolipid biosynthesis upregulation by TOR complex 2-Ypk1 signaling during yeast adaptive response to acetic acid stress. Biochem. J. 473, 4311–4325 10.1042/bcj20160565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Casamayor, A., Torrance, P.D., Kobayashi, T., Thorner, J. and Alessi, D.R. (1999) Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr. Biol. 9, 186–197 10.1016/s0960-9822(99)80088-8 [DOI] [PubMed] [Google Scholar]
- 118.Rodríguez-Escudero, I., Fernández-Acero, T., Cid, V.J. and Molina, M. (2018) Heterologous mammalian Akt disrupts plasma membrane homeostasis by taking over TORC2 signaling in Saccharomyces cerevisiae. Sci. Rep. 8, 7732 10.1038/s41598-018-25717-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gasch, A.P., Spellman, P.T., Kao, C.M., Carmel-Harel, O., Eisen, M.B., Storz, G.et al. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257 10.1091/mbc.11.12.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Siggers, T., Reddy, J., Barron, B. and Bulyk, M.L. (2014) Diversification of transcription factor paralogs via noncanonical modularity in C2H2 zinc finger DNA binding. Mol. Cell 55, 640–648 10.1016/j.molcel.2014.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Komatsu, N., Ishino, Y., Shirai, R., Sakata, K.-T., Tani, M., Maeda, T.et al. (2022) Transcriptional regulation of sphingolipid metabolism in budding yeast. bioRxiv 10.1101/2021.11.05.467429 [DOI] [Google Scholar]
- 122.Watanabe, M., Chen, C.Y. and Levin, D.E. (1994) Saccharomyces cerevisiae PKC1 encodes a protein kinase C (PKC) homolog with a substrate specificity similar to that of mammalian PKC. J. Biol. Chem. 269, 16829–16836 10.1016/S0021-9258(19)89466-8 [DOI] [PubMed] [Google Scholar]
- 123.Nomura, W. and Inoue, Y. (2015) Methylglyoxal activates the target of rapamycin complex 2-protein kinase C signaling pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 35, 1269–1280 10.1128/mcb.01118-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Paravicini, G., Cooper, M., Friedli, L., Smith, D.J., Carpentier, J.L., Klig, L.S.et al. (1992) The osmotic integrity of the yeast cell requires a functional PKC1 gene product. Mol. Cell. Biol. 12, 4896–4905 10.1128/mcb.12.11.4896-4905.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Aronova, S., Wedaman, K., Aronov, P.A., Fontes, K., Ramos, K., Hammock, B.D.et al. (2008) Regulation of ceramide biosynthesis by TOR complex 2. Cell Metab. 7, 148–158 10.1016/j.cmet.2007.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Roelants, F.M., Torrance, P.D. and Thorner, J. (2004) Differential roles of PDK1- and PDK2-phosphorylation sites in the yeast AGC kinases Ypk1, Pkc1 and Sch9. Microbiology 150, 3289–3304 10.1099/mic.0.27286-0 [DOI] [PubMed] [Google Scholar]
- 127.Kamada, Y., Fujioka, Y., Suzuki, N.N., Inagaki, F., Wullschleger, S., Loewith, R.et al. (2005) Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol. Cell. Biol. 25, 7239–7248 10.1128/mcb.25.16.7239-7248.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sun, Y., Taniguchi, R., Tanoue, D., Yamaji, T., Takematsu, H., Mori, K.et al. (2000) Sli2 (Ypk1), a homologue of mammalian protein kinase SGK, is a downstream kinase in the sphingolipid-mediated signaling pathway of yeast. Mol. Cell. Biol. 20, 4411–4419 10.1128/MCB.20.12.4411-4419.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Holm, L. (2020) Using Dali for protein structure comparison. Methods Mol. Biol. 2112, 29–42 10.1007/978-1-0716-0270-6_3 [DOI] [PubMed] [Google Scholar]
- 130.Holm, L. (2022) Dali server: structural unification of protein families. Nucleic Acids Res. 50, W210–W215 10.1093/nar/gkac387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Corbalan-Garcia, S. and Gómez-Fernández, J.C. (2014) Signaling through C2 domains: more than one lipid target. Biochim. Biophys. Acta 1838, 1536–1547 10.1016/j.bbamem.2014.01.008 [DOI] [PubMed] [Google Scholar]
- 132.Consortium, U. (2021) Uniprot: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489 10.1093/nar/gkaa1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Letunic, I., Khedkar, S. and Bork, P. (2021) SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460 10.1093/nar/gkaa937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nalefski, E.A. and Falke, J.J. (1996) The C2 domain calcium-binding motif: structural and functional diversity. Protein Sci. 5, 2375–2390 10.1002/pro.5560051201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Strahl, T. and Thorner, J. (2007) Synthesis and function of membrane phosphoinositides in budding yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1771, 353–404 10.1016/j.bbalip.2007.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Baffi, T.R., Lordén, G., Wozniak, J.M., Feichtner, A., Yeung, W., Kornev, A.P.et al. (2021) mTORC2 controls the activity of PKC and Akt by phosphorylating a conserved TOR interaction motif. Sci. Signal. 14, eabe4509 10.1126/scisignal.abe4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Baffi, T.R. and Newton, A.C. (2022) mTOR regulation of AGC kinases: new twist to an old tail. Mol. Pharmacol. 101, 213–218 10.1124/molpharm.121.000310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Baffi, T.R. and Newton, A.C. (2022) Protein kinase C: release from quarantine by mTORC2. Trends Biochem. Sci. 47, 518–530 10.1016/j.tibs.2022.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Williams, M.R., Arthur, J.S., Balendran, A., van der Kaay, J., Poli, V., Cohen, P.et al. (2000) The role of 3-phosphoinositide-dependent protein ki- nase 1 in activating AGC kinases defined in embryonic stem cells. Curr. Biol. 10, 439–448 10.1016/s0960-9822(00)00441-3 [DOI] [PubMed] [Google Scholar]
- 140.Yang, J., Cron, P., Thompson, V., Good, V.M., Hess, D., Hemmings, B.A.et al. (2002) Molecular mechanism for the regulation of protein kinase B/Akt by hydrophobic motif phosphorylation. Mol. Cell 9, 1227–1240 10.1016/s1097-2765(02)00550-6 [DOI] [PubMed] [Google Scholar]
- 141.Jacinto, E., Facchinetti, V., Liu, D., Soto, N., Wei, S., Jung, S.Y.et al. (2006) SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 127, 125–137 10.1016/j.cell.2006.08.033 [DOI] [PubMed] [Google Scholar]
- 142.Facchinetti, V., Ouyang, W., Wei, H., Soto, N., Lazorchak, A., Gould, C.et al. (2008) The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 27, 1932–1943 10.1038/emboj.2008.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kinoshita, E., Kinoshita-Kikuta, E. and Koike, T. (2009) Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat. Protoc. 4, 1513–1521 10.1038/nprot.2009.154 [DOI] [PubMed] [Google Scholar]
- 144.Mok, J., Kim, P.M., Lam, H.Y., Piccirillo, S., Zhou, X., Jeschke, G.R.et al. (2010) Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci. Signal. 3, ra12 10.1126/scisignal.2000482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Muir, A., Ramachandran, S., Roelants, F.M., Timmons, G. and Thorner, J. (2014) TORC2-dependent protein kinase Ypk1 phosphorylates ceramide synthase to stimulate synthesis of complex sphingolipids. eLlife 3, e03779 10.7554/eLife.03779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Johnson, J.L., Yaron, T.M., Huntsman, E.M., Kerelsky, A., Song, J., Regev, A.et al. (2022) A global atlas of substrate specificities for the human serine/threonine kinome. bioRxiv 10.1101/2022.05.22.492882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Plank, M., Perepelkina, M., Müller, M., Vaga, S., Zou, X., Bourgoint, C.et al. (2020) Chemical genetics of AGC-kinases reveals shared targets of Ypk1, protein kinase A and Sch9. Mol. Cell. Proteom. 19, 655–671 10.1074/mcp.RA120.001955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Locke, M.N. (2019) Molecular Mechanisms in Signaling by Target of Rapamycin Complex 2 in Saccharomyces cerevisiae. Ph.D. Thesis, Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, CA, USA, 83pp. https://escholarship.org/uc/item/71q7k9ps
- 149.Knighton, D.R., Zheng, J.H., T, L.F., Xuong, E., Taylor, N.H., and Sowadski, S.S.et al. (1991) Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253, 414–420 10.1126/science.1862343 [DOI] [PubMed] [Google Scholar]
- 150.Taylor, S.S., Wu, J., Bruystens, J.G.H., Del Rio, J.C., Lu, T.W., Kornev, A.P.et al. (2021) From structure to the dynamic regulation of a molecular switch: a journey over 3 decades. J. Biol. Chem. 296, 100746 10.1016/j.jbc.2021.100746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bardwell, A.J., Flatauer, L.J., Matsukuma, K., Thorner, J. and Bardwell, L. (2001) A conserved docking site in MEKs mediates high-affinity binding to MAP kinases and cooperates with a scaffold protein to enhance signal transmission. J. Biol. Chem. 276, 10374–10386 10.1074/jbc.M010271200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Good, M.C., Zalatan, J.G. and Lim, W.A. (2011) Scaffold proteins: hubs for controlling the flow of cellular information. Science 332, 680–686 10.1126/science.1198701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tanoue, T., Maeda, R., Adachi, M. and Nishida, E. (2001) Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J. 20, 466–479 10.1093/emboj/20.3.466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Alexa, A., Ember, O., Szabó, I., Mo'ath, Y., Póti, A.L., Reményi, A.et al. (2021) Peptide-based inhibitors of protein binding to the mitogen-activated protein kinase docking groove. Front. Mol. Biosci. 8, 690429 10.3389/fmolb.2021.690429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Riggi, M., Kusmider, B. and Loewith, R. (2020) The flipside of the TOR coin - TORC2 and plasma membrane homeostasis at a glance. J. Cell Sci. 133, jcs242040 10.1242/jcs.242040 [DOI] [PubMed] [Google Scholar]
- 156.Roelants, F.M., Baltz, A.G., Trott, A.E., Fereres, S. and Thorner, J. (2010) A protein kinase network regulates the function of aminophospholipid flippases. Proc. Natl Acad. Sci. U.S.A. 107, 34–39 10.1073/pnas.0912497106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Roelants, F.M., Leskoske, K.L., Pedersen, R.T., Muir, A., Liu, J.M., Finnigan, G.C.et al. (2017) TOR complex 2-regulated protein kinase Fpk1 stimulates endocytosis via inhibition of Ark1/Prk1-related protein kinase Akl1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 37, e00627-16 10.1128/mcb.00627-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nakano, K., Yamamoto, T., Kishimoto, T., Noji, T. and Tanaka, K. (2008) Protein kinases Fpk1p and Fpk2p are novel regulators of phospholipid asymmetry. Mol. Biol. Cell 19, 1783–1797 10.1091/mbc.e07-07-0646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Best, J.T., Xu, P. and Graham, T.R. (2019) Phospholipid flippases in membrane remodeling and transport carrier biogenesis. Curr. Opin. Cell Biol. 59, 8–15 10.1016/j.ceb.2019.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Roelants, F.M., Su, B.M., von Wulffen, J., Ramachandran, S., Sartorel, E., Trott, A.E.et al. (2015) Protein kinase Gin4 negatively regulates flippase function and controls plasma membrane asymmetry. J. Cell Biol. 208, 299–311 10.1083/jcb.201410076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bai, L., You, Q., Jain, B.K., Duan, H.D., Kovach, A., Graham, T.R.et al. (2020) Transport mechanism of P4 ATPase phosphatidylcholine flippases. eLlife 9, e62163 10.7554/eLife.62163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Xu, J., He, Y., Wu, X. and Li, L. (2022) Conformational changes of a phosphatidylcholine flippase in lipid membranes. Cell Rep. 38, 110518 10.1016/j.celrep.2022.110518 [DOI] [PubMed] [Google Scholar]
- 163.Frøsig, M.M., Costa, S.R., Liesche, J., Østerberg, J., Hanisch, S., Nintemann, S.et al. (2020) Pseudohyphal growth in Saccharomyces cerevisiae involves protein kinase-regulated lipid flippases. J. Cell Sci. 133, jcs235994 10.1242/jcs.235994 [DOI] [PubMed] [Google Scholar]
- 164.Du, W., Forte, G.M., Smith, D. and Petersen, J. (2016) Phosphorylation of the amino-terminus of the AGC kinase Gad8 prevents its interaction with TORC2. Open Biol. 6, 150189 10.1098/rsob.150189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Körner, C. and Fröhlich, F. (2022) Compartmentation and functions of sphingolipids. Curr. Opin. Cell Biol. 74, 104–111 10.1016/j.ceb.2022.01.006 [DOI] [PubMed] [Google Scholar]
- 166.Sebastian, T.T., Baldridge, R.D., Xu, P. and Graham, T.R. (2012) Phospholipid flippases: building asymmetric membranes and transport vesicles. Biochim. Biophys. Acta 1821, 1068–1077 10.1016/j.bbalip.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Muthusamy, B.-P., Raychaudhuri, S., Natarajan, P., Abe, F., Liu, K., Prinz, W.A.et al. (2009) Control of protein and sterol trafficking by antagonistic activities of a type IV P-type ATPase and oxysterol binding protein homologue. Mol. Biol. Cell 20, 2920–2931 10.1091/mbc.e08-10-1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Saito, K., Fujimura-Kamada, K., Hanamatsu, H., Kato, U., Umeda, M., Kozminski, K.G.et al. (2007) Transbilayer phospholipid flipping regulates Cdc42p signaling during polarized cell growth via Rga GTPase-activating proteins. Dev. Cell 13, 743–751 10.1016/j.devcel.2007.09.014 [DOI] [PubMed] [Google Scholar]
- 169.Das, A., Slaughter, B.D., Unruh, J.R., Bradford, W.D., Alexander, R., Rubinstein, B.et al. (2012) Flippase-mediated phospholipid asymmetry promotes fast Cdc42 recycling in dynamic maintenance of cell polarity. Nat. Cell Biol. 14, 304–310 10.1038/ncb2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hatakeyama, R., Kono, K. and Yoshida, S. (2017) Ypk1 and Ypk2 kinases maintain Rho1 at the plasma membrane by flippase-dependent lipid remodeling after membrane stresses. J. Cell Sci. 130, 1169–1178 10.1242/jcs.198382 [DOI] [PubMed] [Google Scholar]
- 171.Nomura, W., Ng, S.P., Takahara, T., Maeda, T., Kawada, T., Goto, T.et al. (2022) Roles of phosphatidylserine and phospholipase C in the activation of TOR complex 2 signaling in Saccharomyces cerevisiae. J. Cell Sci. 135. 10.1242/jcs.259988 [DOI] [PubMed] [Google Scholar]
- 172.deHart,, A.K., Schnell, J.D., Allen, D.A. and Hicke, L. (2002) The conserved Pkh-Ypk kinase cascade is required for endocytosis in yeast. J. Cell Biol. 156, 241–248 10.1083/jcb.200107135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bourgoint, C., Rispal, D., Berti, D., Filipuzzi, I., Helliwell, S.B., Prouteau, M.et al. (2018) Target of rapamycin complex 2-dependent phosphorylation of the coat protein Pan1 by Akl1 controls endocytosis dynamics in Saccharomyces cerevisiae. J. Biol. Chem. 293, 12043–12053 10.1074/jbc.RA117.001615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lu, R., Drubin, D.G. and Sun, Y. (2016) Clathrin-mediated endocytosis in budding yeast at a glance. J. Cell Sci. 129, 1531–1536 10.1242/jcs.182303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lacy, M.M., Ma, R., Ravindra, N.G. and Berro, J. (2018) Molecular mechanisms of force production in clathrin-mediated endocytosis. FEBS Lett. 592, 3586–3605 10.1002/1873-3468.13192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Alvaro, C.G., Aindow, A. and Thorner, J. (2016) Differential phosphorylation provides a switch to control how α-arrestin Rod1 down-regulates mating pheromone response in Saccharomyces cerevisiae. Genetics 203, 299–317 10.1534/genetics.115.186122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kahlhofer, J., Leon, S., Teis, D. and Schmidt, O. (2021) The α-arrestin family of ubiquitin ligase adaptors links metabolism with selective endocytosis. Biol. Cell 113, 183–219 10.1111/boc.202000137 [DOI] [PubMed] [Google Scholar]
- 178.Alvaro, C.G., O'Donnell, A.F., Prosser, D.C., Augustine, A.A., Goldman, A., Brodsky, J.L.et al. (2014) Specific α-arrestins negatively regulate Saccharomyces cerevisiae pheromone response by down-modulating the G-protein-coupled receptor Ste2. Mol. Cell. Biol. 34, 2660–2681 10.1128/mcb.00230-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Becuwe, M. and Léon, S. (2014) Integrated control of transporter endocytosis and recycling by the arrestin-related protein Rod1 and the ubiquitin ligase Rsp5. eLife 3, e03307 10.7554/eLife.03307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Nikko, E. and Pelham, H.R. (2009) Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 10, 1856–1867 10.1111/j.1600-0854.2009.00990.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.O'Donnell, A.F., McCartney, R.R., Chandrashekarappa, D.G., Zhang, B.B., Thorner, J. and Schmidt, M.C. (2015) 2-Deoxyglucose impairs Saccharomyces cerevisiae growth by stimulating Snf1-regulated and α-arrestin-mediated trafficking of hexose transporters 1 and 3. Mol. Cell. Biol. 35, 939–955 10.1128/MCB.01183-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Toyoda, Y., Soejima, S., Masuda, F. and Saitoh, S. (2021) TORC2 inhibition of α-arrestin Aly3 mediates cell surface persistence of S. pombe Ght5 glucose transporter in low glucose. J. Cell Sci. 134, jcs257485 10.1242/jcs.257485 [DOI] [PubMed] [Google Scholar]
- 183.Toyoda, Y. and Saitoh, S. (2021) Fission yeast TORC2 signaling pathway ensures cell proliferation under glucose-limited, nitrogen-replete conditions. Biomolecules 11, 1465 10.3390/biom11101465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Prosser, D.C., Pannunzio, A.E., Brodsky, J.L., Thorner, J., Wendland, B. and O'Donnell, A.F. (2015) α-Arrestins participate in cargo selection for both clathrin-independent and clathrin-mediated endocytosis. J. Cell Sci. 128, 4220–4234 10.1242/jcs.175372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hatakeyama, R., Kamiya, M., Takahara, T. and Maeda, T. (2010) Endocytosis of the aspartic acid/glutamic acid transporter Dip5 is triggered by substrate-dependent recruitment of the Rsp5 ubiquitin ligase via the arrestin-like protein Aly2. Mol. Cell. Biol. 30, 5598–5607 10.1128/MCB.00464-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.O'Donnell, A.F., Apffel, A., Gardner, R.G. and Cyert, M.S. (2010) Alpha-arrestins Aly1 and Aly2 regulate intracellular trafficking in response to nutrient signaling. Mol. Biol. Cell 21, 3552–3566 10.1091/mbc.E10-07-0636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Crapeau, M., Merhi, A. and André, B. (2014) Stress conditions promote yeast Gap1 permease ubiquitylation and down-regulation via the arrestin-like Bul and Aly proteins. J. Biol. Chem. 289, 22103–22116 10.1074/jbc.M114.582320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Nishimura, A., Tanahashi, R. and Takagi, H. (2020) The yeast α-arrestin Art3 is a key regulator for arginine-induced endocytosis of the high-affinity proline transporter Put4. Biochem. Biophys. Res. Commun. 531, 416–421 10.1016/j.bbrc.2020.07.117 [DOI] [PubMed] [Google Scholar]
- 189.Wawrzycka, D., Sadlak, J., Maciaszczyk-Dziubinska, E. and Wysocki, R. (2019) Rsp5-dependent endocytosis and degradation of the arsenite transporter Acr3 requires its N-terminal acidic tail as an endocytic sorting signal and arrestin-related ubiquitin-ligase adaptors. Biochim. Biophys. Acta Biomembr. 1861, 916–925 10.1016/j.bbamem.2019.02.004 [DOI] [PubMed] [Google Scholar]
- 190.Robinson, B.P., Hawbaker, S., Chiang, A., Jordahl, E.M., Anaokar, S., Nikiforov, A.et al. (2022) Alpha-arrestins Aly1/Art6 and Aly2/Art3 regulate trafficking of the glycerophosphoinositol. transporter Git1 and impact phospholipid homeostasis. Biol. Cell 114, 3–31 10.1111/boc.202100007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Gururaj, C., Federman, R.S. and Chang, A. (2013) Orm proteins integrate multiple signals to maintain sphingolipid homeostasis. J. Biol. Chem. 288, 20453–20463 10.1074/jbc.M113.472860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Breslow, D.K., Collins, S.R., Bodenmiller, B., Aebersold, R., Simons, K., Shevchenko, A.et al. (2010) Orm family proteins mediate sphingolipid homeostasis. Nature 463, 1048–1053 10.1038/nature08787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Breslow, D.K. and Weissman, J.S. (2010) Membranes in balance: mechanisms of sphingolipid homeostasis. Mol. Cell 40, 267–279 10.1016/j.molcel.2010.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lowther, J., Naismith, J.H., Dunn, T.M. and Campopiano, D.J. (2012) Structural, mechanistic and regulatory studies of serine palmitoyltransferase. Biochem. Soc. Trans. 40, 547–554 10.1042/BST20110769 [DOI] [PubMed] [Google Scholar]
- 195.Harrison, P.J., Dunn, T.M. and Campopiano, D.J. (2018) Sphingolipid biosynthesis in man and microbes. Nat. Prod. Rep. 35, 921–954 10.1039/c8np00019k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Schmidt, O., Weyer, Y., Sprenger, S., Widerin, M.A., Eising, S., Baumann, V.et al. (2020) TOR complex 2 (TORC2) signaling and the ESCRT machinery cooperate in the protection of plasma membrane integrity in yeast. J. Biol. Chem. 295, 12028–12044 10.1074/jbc.RA120.013222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Omnus, D.J., Manford, A.G., Bader, J.M., Emr, S.D. and Stefan, C.J. (2016) Phosphoinositide kinase signaling controls ER-PM cross-talk. Mol. Biol. Cell 27, 1170–1180 10.1091/mbc.E16-01-0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Miyake, Y., Kozutsumi, Y., Nakamura, S., Fujita, T. and Kawasaki, T. (1995) Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem. Biophys. Res. Commun. 211, 396–403 10.1006/bbrc.1995.1827 [DOI] [PubMed] [Google Scholar]
- 199.Sugimoto, Y., Sakoh, H. and Yamada, K. (2004) IPC synthase as a useful target for antifungal drugs. Curr. Drug Targets Infect. Disord. 4, 311–322 10.2174/1568005043340597 [DOI] [PubMed] [Google Scholar]
- 200.Li, S.C. and Kane, P.M. (2008) The yeast lysosome-like vacuole: endpoint and crossroads. Biochim. Biophys. Acta 1793, 650–663 10.1016/j.bbamcr.2008.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Klionsky, D.J. and Eskelinen, E.L. (2014) The vacuole versus the lysosome: when size matters. Autophagy 10, 185–187 10.4161/auto.27367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bisinski, D.D., Gomes Castro, I., Mari, M., Walter, S., Fröhlich, F., Schuldiner, M.et al. (2022) Cvm1 is a component of multiple vacuolar contact sites required for sphingolipid homeostasis. J. Cell Biol. 221, e202103048 10.1083/jcb.202103048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Koga, A., Takayama, C., Ishibashi, Y., Kono, Y., Matsuzaki, M. and Tani, M. (2022) Loss of tolerance to multiple environmental stresses due to limitation of structural diversity of complex sphingolipids. Mol. Biol. Cell 33. 10.1091/mbc.E22-04-0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Marchot, P. and Chatonnet, A. (2012) Enzymatic activity and protein interactions in alpha/beta hydrolase fold proteins: moonlighting versus promiscuity. Protein Pept. Lett. 19, 132–143 10.2174/092986612799080284 [DOI] [PubMed] [Google Scholar]
- 205.Tsuji, T. and Fujimoto, T. (2018) Lipids and lipid domains of the yeast vacuole. Biochem. Soc. Trans. 46, 1047–1054 10.1042/BST20180120 [DOI] [PubMed] [Google Scholar]
- 206.Ishino, Y., Komatsu, N., Sakata, K.-T., Yoshikawa, D., Tani, M., Maeda, T.et al. (2022) Regulation of sphingolipid biosynthesis in the endoplasmic reticulum via signals from the plasma membrane in budding yeast. FEBS J. 289, 457–472 10.1111/febs.16189 [DOI] [PubMed] [Google Scholar]
- 207.Henry, S.A., Kohlwein, S.D. and Carman, G.M. (2012) Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190, 317–349 10.1534/genetics.111.130286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Han, G.-S. and Carman, G.M. (2013) Lipids phospholipid synthesis in yeast. In Encyclopedia of Biological Chemistry, 2nd edn. (Lennarz, W.J. and Lane, M.D., eds.), Vol. 3, pp. 478–481, Elsevier, Inc., New York, NY. 10.1016/B978-0-12-378630-2.00513-2 [DOI] [Google Scholar]
- 209.Graef, M. (2018) Lipid droplet-mediated lipid and protein homeostasis in budding yeast. FEBS Lett. 592, 1291–1303 10.1002/1873-3468.12996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Henne, M., Goodman, J.M. and Hariri, H. (2020) Spatial compartmentalization of lipid droplet biogenesis. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 1865, 158499 10.1016/j.bbalip.2019.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Pahlman, A.K., Granath, K., Ansell, R., Hohmann, S. and Adler, L. (2001) He yeast glycerol 3-phosphatases Gpp1p and Gpp2p are required for glycerol biosynthesis and differentially involved in the cellular responses to osmotic, anaerobic, and oxidative stress. J. Biol. Chem. 276, 3555–3563 10.1074/jbc.M007164200 [DOI] [PubMed] [Google Scholar]
- 212.Blomberg, A. (2022) Yeast osmoregulation - glycerol still in pole position. FEMS Yeast Res. 22, foac035 10.1093/femsyr/foac035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.De Nadal, E. and Posas, F. (2022) The HOG pathway and the regulation of osmoadaptive responses in yeast. FEMS Yeast Res. 22, foac013 10.1093/femsyr/foac013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sabir, F., Loureiro-Dias, M.C., Soveral, G. and Prista, C. (2017) Functional relevance of water and glycerol channels in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 364, fnx080 10.1093/femsle/fnx080 [DOI] [PubMed] [Google Scholar]
- 215.Albertyn, J., Hohmann, S., Thevelein, J.M. and Prior, B.A. (1994) GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 14, 4135–4144 10.1128/mcb.14.6.4135-4144.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Lee, J. and Levin, D.E. (2015) Rgc2 regulator of glycerol channel Fps1 functions as a homo- and heterodimer with Rgc1. Eukaryot. Cell 14, 719–725 10.1128/EC.00073-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Tamás, M.J., Luyten, K., Sutherland, F.C., Hernandez, A., Albertyn, J., Valadi, H.et al. (1999) Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol. Microbiol. 31, 1087–1104 10.1046/j.1365-2958.1999.01248.x [DOI] [PubMed] [Google Scholar]
- 218.Babazadeh, R., Furukawa, T., Hohmann, S. and Furukawa, K. (2014) Rewiring yeast osmostress signalling through the MAPK network reveals essential and non-essential roles of Hog1 in osmoadaptation. Sci. Rep. 4, 4697 10.1038/srep04697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Gatta, A.T., Wong, L.H., Sere, Y.Y., Calderón-Noreña, D.M., Cockcroft, S., Menon, A.K.et al. (2015) A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. eLife 4, e07253 10.7554/eLife.07253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Jordá, T. and Puig, S. (2020) Regulation of ergosterol biosynthesis in Saccharomyces cerevisiae. Genes (Basel) 11, 795 10.3390/genes11070795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Roelants, F.M., Chauhan, N., Muir, A., Davis, J.C., Menon, A.K., Levine, T.P.et al. (2018) TOR complex 2-regulated protein kinase Ypk1 controls sterol distribution by inhibiting StARkin domain-containing proteins located at plasma membrane-endoplasmic reticulum contact sites. Mol. Biol. Cell 29, 2128–2136 10.1091/mbc.E18-04-0229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Topolska, M., Roelants, F.M., Si, E.P. and Thorner, J. (2020) TORC2-dependent Ypk1-mediated phosphorylation of Lam2/Ltc4 disrupts Its association with the β-propeller protein Laf1 at endoplasmic reticulum-plasma membrane contact sites in the yeast Saccharomyces cerevisiae. Biomolecules 10, 1598 10.3390/biom10121598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Chauhan, N., Sere, Y.Y., Sokol, A.M., Graumann, J. and Menon, A.K. (2020) A PhotoClick cholesterol-based quantitative proteomics screen for cytoplasmic sterol-binding proteins in Saccharomyces cerevisiae. Yeast 37, 15–25 10.1002/yea.3448 [DOI] [PubMed] [Google Scholar]
- 224.Wickens, M., Bernstein, D.S., Kimble, J. and Parker, R. (2002) A PUF family portrait: 3′-UTR regulation as a way of life. Trends Genet. 18, 150–157 10.1016/s0168-9525(01)02616-6 [DOI] [PubMed] [Google Scholar]
- 225.Wang, M., Ogé, L., Perez-Garcia, M.-D., Hamama, L. and Sakr, S. (2018) The PUF protein family: overview on PUF RNA targets, biological functions, and post transcriptional regulation. Int. J. Mol. Sci. 19, 410 10.3390/ijms19020410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Gerber, A.P., Herschlag, D. and Brown, P.O. (2004) Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2, E79 10.1371/journal.pbio.0020079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Porter, D.F., Koh, Y.Y., V, B., Raines, R.T. and Wickens, M. (2015) Target selection by natural and redesigned PUF proteins. Proc. Natl Acad. Sci. U.S.A. 112, 15868–15873 10.1073/pnas.1508501112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Galez, H.A., Roelants, F.M., Palm, S.M., Reynaud, K.K., Ingolia, N.T. and Thorner, J. (2021) Phosphorylation of mRNA-binding proteins Puf1 and Puf2 by TORC2-activated protein kinase Ypk1 alleviates their repressive effects. Membranes (Basel) 11, 500 10.3390/membranes11070500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Shimada, K., Filipuzzi, I., Stahl, M., Helliwell, S.B., Studer, C., Hoepfner, D.et al. (2013) TORC2 signaling pathway guarantees genome stability in the face of DNA strand breaks. Mol. Cell 51, 829–839 10.1016/j.molcel.2013.08.019 [DOI] [PubMed] [Google Scholar]
- 230.Vlahakis, A., Lopez Muniozguren, N. and Powers, T. (2016) Calcium channel regulator Mid1 links TORC2-mediated changes in mitochondrial respiration to autophagy. J. Cell Biol. 215, 779–788 10.1083/jcb.201605030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Matos, G.S., Madeira, J.B., Fernandes, C.M., Dasilva, D., Masuda, C.A., Del Poeta, M.et al. (2021) Regulation of sphingolipid synthesis by the G1/S transcription factor Swi4. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 1866, 158983 10.1016/j.bbalip.2021.158983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Yerlikaya, S., Meusburger, M., Kumari, R., Huber, A., Anrather, D., Costanzo, M.et al. (2016) TORC1 and TORC2 work together to regulate ribosomal protein S6 phosphorylation in Saccharomyces cerevisiae. Mol. Biol. Cell 27, 397–409 10.1091/mbc.E15-08-0594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Tomioka, M., Shimobayashi, M., Kitabatake, M., Ohno, M., Kozutsumi, Y., Oka, S.et al. (2018) Ribosomal protein uS7/Rps5 serine-223 in protein kinase-mediated phosphorylation and ribosomal small subunit maturation. Sci. Rep. 8, 1244 10.1038/s41598-018-19652-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Lucena, R., Alcaide-Gavilán, M., Schubert, K., He, M., Domnauer, M.G., Marquer, C.et al. (2018) Cell size and growth rate are modulated by TORC2-dependent signals. Curr. Biol. 28, 196–210.e194 10.1016/j.cub.2017.11.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Sommer, R.A., DeWitt, J.T., Tan, R. and Kellogg, D.R. (2021) Growth-dependent signals drive an increase in early G1 cyclin concentration to link cell cycle entry with cell growth. eLife 10, e64364 10.7554/eLife.64364 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.