Abstract
Type IV pili are involved in adhesion, twitching motility, aggregation, biofilm formation and virulence in a variety of Gram-negative bacteria. Burkholderia pseudomallei, the causative agent of melioidosis and a Tier 1 biological select agent, is a Gram-negative bacterium with eight type IV pili-associated loci (TFP1 to TFP8). Most have not been fully characterized. In this study, we investigated BPSS2185, an uncharacterized TFP8 gene that encodes a type IVB pilus protein subunit. Using genetic deletion and complementation analysis in B. pseudomallei JW270, we demonstrate that BPSS2185 plays an important role in twitching motility and adhesion to A549 human alveolar epithelial cells. Compared to JW270, the JW270 ΔBPSS2185 mutant failed to display twitching motility and did not adhere to the epithelial cells. These phenotypes were partially reversed by the complementation of BPSS2185 in the mutant strain. The study also shows that BPSS2185 is expressed only during the onset of mature biofilm formation and at the dispersal of a biofilm, suggesting that the motility characteristic is required to form a biofilm. Our study is the first to suggest that the BPSS2185 gene in TFP8 contributes to twitching motility, adhesion and biofilm formation, indicating that the gene may contribute to B. pseudomallei virulence.
Keywords: melioidosis, type IV pili, biofilm formation
Data Summary
Burkholderia pseudomallei is the causative agent of melioidosis and is a Tier 1 biological select agent. A previous study found that the TFP8 genes were important for B. pseudomallei virulence in a murine model of infection, but the function of the encoded pilus was not further evaluated. Our research shows that TFP8 plays an important role in attachment, twitching motility and biofilm formation, all of which can probably affect virulence.
Introduction
Burkholderia pseudomallei is an opportunistic, motile, Gram-negative, environmental saprophyte that carries a wide range of virulence factors and broad antimicrobial drug resistance features [1]. In humans it is the causative agent of melioidosis and is a Tier 1 biological select agent because it poses a severe threat to both human and animal health. The clinical presentation of the disease, which is endemic in Southeast Asia and Australia, can range from an acute febrile illness to a disseminated septicaemia. Long incubation periods before clinical symptoms are possible and recurrence of infection is common despite adequate antimicrobial therapy [1]. The disease is probably under-recognized and under-reported. Based on modelling, the disease may be responsible for up to 165000 cases per year, with 89000 deaths [2]. Recent years have seen an upsurge in B. pseudomallei research because of its potential use as a biological warfare agent and increasing awareness of the disease and its health burden [1–3]. Understanding the mechanisms that contribute to its virulence, allow it to evade the immune system and develop antimicrobial resistance is essential for identifying potential medical countermeasures. Adhesion, twitching motility and biofilm production mediated by type VI pili, which are present in B. pseudomallei and other Gram-negative bacteria, play an important role in their ability to cause disease [4, 5].
Most bacteria have multiple types of pili, which are categorized into different classes based on assembly processes or pathways [6]. Type IV pili (TFP) are long and thin surface filaments that function by repeated extension and retraction [7, 8]. They are often involved in diverse processes such as attachment, twitching motility, DNA uptake, biofilm formation and virulence. There are two distinct subclasses of TFP, termed type IVA and type IVB, that are differentiated by the length of pilin signal peptides and other conserved motifs [8]. B. pseudomallei encodes eight type IV pilus-associated loci, TFP1 to TFP8, and both type IVA and IVB pili genes are represented [9]. The TFP1 locus contains a type IVA gene that is involved in adherence to human epithelial cell lines, and virulence in nematode worms and mice. Strain differences exist with regard to the role of TFP1 in adherence, microcolony formation and expression of the pilus subunit gene pilA [10]. Nandi et al. found that the TFP2, TFP4 and TFP7 loci are positively selected for and may confer an advantage in environmental survival and host adaptation [11]. When the B. pseudomallei TFP4 locus was deleted, the mutant exhibited significantly reduced virulence in the BALB/c mouse intranasal infection assay. The type IVB pilus subunit, PilV, encoded by TFP7 was immunogenic as a vaccine candidate in mice, but was unable to induce protection against B. pseudomallei challenge [12]. Finally, the TFP8 locus was identified as a recombination hotspot in B. pseudomallei and a deletion of the TFP8 genes resulted in a strain that exhibited reduced virulence in a BALB/c mouse intranasal infection assay [13]. The role of TFP8 in other phenotypes, such as adherence, twitching motility and biofilm formation, was not evaluated. These factors probably play a role in colonization of hosts and disease causation.
The adhesion process is critical for a bacterium’s capacity to colonize, infiltrate and cause disease [14]. Gram-negative bacteria can adhere to surfaces using trimeric auto transporters [15], outer membrane proteins [5], filamentous haemagglutinins [16], polysaccharide adhesins [17] and pilus proteins [4]. The pili adhesins are polymeric fibres that are used for biotic and abiotic surface attachment, twitching motility, DNA transfer, biofilm development and transitioning from planktonic to sessile growth [18]. Bacterial adhesion and aggregation are the first stages of biofilm formation, followed by maturation and dispersion to form a new biofilm [19]. Adhesion, aggregation and biofilm formation can be mediated by flagella [20], fimbriae [21], type I pili produced by the CUP pathway [21] or type IV pili [20], or by the presence of various chemical compositions such as extracellular DNA (eDNA) [22, 23] or polysaccharide [24] on the surface of the bacteria. Biofilm production in B. pseudomallei , Staphylococcus aureus and Yersinia pseudotuberculosis has been linked to the higher mortality in both Caenorhabditis elegans and mouse models [25–27], and has been implicated in immune responses [26]. After a matured biofilm is formed, bacteria are released and move to a different location to form a new biofilm [28–30]. Biofilm formation in a B. pseudomallei infected host often results in persistent bacteraemia as cells are continually disseminated in vivo and complete eradication becomes difficult [26].
The role of the TFP8 locus in mediating adherence, twitching motility and biofilm formation has not been described in B. pseudomallei . The objective of our study was to use genetic deletion and complementation analysis in B. pseudomallei JW270 to characterize the contribution of TFP8 to virulence.
Methods
Bacterial strains
Table 1 identifies the strains of bacteria, constructs, plasmids and primers used in this study and their relevant characteristics. B. pseudomallei JW270 was selected because it is an attenuated strain that is exempt from the Select Agent regulations. Escherichia coli strains used for mutant constructs were grown in Lennox LB broth (Sigma-Aldrich) at 37 °C with shaking overnight. E. coli strains containing pBHR2 and pBHR2-BPSS2185 were grown in LB broth supplemented with 50 µg kanamycin ml−1 (Sigma-Aldrich).
Table 1.
Bacterial strains, plasmids and primers used for this study
|
Strain/plasmid/primer (5′−3′) |
Relevant characteristics |
Source or reference |
|---|---|---|
|
E. coli INV110 |
For growth and purification of plasmid DNA digested with dam or dcm methylation-sensitive restriction enzymes |
Invitrogen |
|
E. coli TOP10 |
General cloning and blue/white screening |
Life Technologies |
|
E. coli S17-1 |
Mobilizing strain, transfer genes of RP4 integrated in chromosome; Smr |
[51] |
|
pMo130 |
Kmr, Burkholderia gene replacement vector |
[34] |
|
pBHR2 |
Broad-host-range plasmid, Kmr |
[35] |
|
pBHR2-BPSS2185 |
pBHR2 containing a full-length copy of BPSS2185 |
This study |
|
B. pseudomallei JW270 |
Strain excluded from the CDC select agent list as a result of a deletion of the wcb locus encoding the 6-deoxyheptan capsular polysaccharide; ∆(amrR-oprA) rpsL (Smr) |
[52] |
|
B. pseudomallei JW270 ΔBPSS2185 |
JW270 derivative with an in-frame 138 bp deletion of BPSS2185 |
This study |
|
BPSS2185 F1 |
*TTAT CATATG GAATACCGGGGTAA |
This study |
|
BPSS2185 R1 |
GAGGCTGGACATGACT |
This study |
|
BPSS2185 F2 |
AGCCTCACGATCGGCAACGA |
This study |
|
BPSS2185 R2 |
*TGTA GGATCC GAGCGGAAAGCAT |
This study |
|
C-BPSS2185 F |
*TTA CATATG ACGGCAAGGGTTTTA |
This study |
|
C-BPSS2185 R |
*TATA GGATCC ACGCACCCTTACA |
This study |
Kmr, kanamycin resistant; Smr, streptomycin resistant.
*Restriction enzyme sites are bolded, italicized and underlined. CATATG – NdeI, GGATCC – BamHI.
For the adhesion, motility and biofilm experiments, B. pseudomallei JW270 and B. pseudomallei JW270 ΔBPSS2185 were cultured on LB agar, and JW270 ∆BPSS2185 (pBHR2) and JW270 ΔBPSS2185 (pBHR2-BPSS2185) were cultured on LB agar with 50 µg kanamycin ml−1. Overnight colonies were transferred into 1 ml of LB and LB with 50 µg kanamycin ml−1. Unless otherwise stated, these B. pseudomallei strains were grown overnight in a 37 °C incubator with shaking at 250 r.p.m. then transferred to a 24-well plate containing 2 ml of LB broth and grown as a biofilm without agitation for 1–3 days. The strains were subsequently serially diluted, plated on LB agar to determine the bacterial concentration and used for experiments.
RNA-sequencing (RNA-Seq) and transcriptomic analysis
Bacterial aggregation, biofilm formation and extracellular DNA production have been linked to the presence of calcium and divalent cations [31, 32]. Environmental calcium was shown to regulate production of a Type IV Tad pilus responsible for biofilm formation, aggregation and colonization by Vibrio vulnificus [31]. B. pseudomallei JW270 was inoculated in six T175 cell culture flasks, two for each experiment containing either 50 ml LB broth or LB broth supplemented with 1.26 mM CaCl2. Cultures were grown statically for 48 h at 37 °C. Spent medium was removed and the biofilm was washed twice with 20 ml of sterile 1× PBS (Sigma-Aldrich). RNAs from six independent samples of bacteria grown in LB or LB with CaCl2 were extracted using the Direct-zol RNA MiniPrep kit (R2052; Zymo Research), according to the manufacturer’s protocol. The six extracted RNA samples were prepared using the TruSeq Stranded Total RNA Library Prep Kit (Illumina). RNA samples were diluted to 100 ng µl−1 in 10 µl of ultrapure water and prepared as directed by the manufacturer. rRNA was depleted using the RRM G Mix (Illumina). Adapter ligation libraries were quantified using a KAPA Biosystems Library Quantification Kit (Roche Sequencing Systems) with the KAPA SYBR FAST qPCR Master Mix and assessed on a Tapestation 2200 (Agilent) for library size and integrity. All six libraries were pooled at equimolar concentrations yielding a 2 nM pool that was spiked with a 10 % Illumina PhiX loading control. The pooled library was diluted to 14 pM and sequenced using a Paired-End HiSeq Rapid SBS Kit (V2-500 cycles) on a HiSeq 2500 System (Illumina) following the manufacturer’s methods. The sequencing run yielded approximately 300 million reads with a Q30 >83 %. The RNA-Seq data were deposited in Figshare (accession number: https://figshare.com/articles/dataset/Burkholderia_pseudomaelli/17704376).
Statistical and bioinformatics analysis for RNA-Seq data
The raw reads with low-quality ends that had a Phred score of <Q20 were removed from the analysis. Ligated adaptor sequences were trimmed using Trimmomatic [33] needed to check with following filters of a minimum length of 80 bp. Quality reads were further aligned to the B. pseudomallei 1026b reference genome (NC_017831.1 and NC_017832.1) using Spliced Transcripts Alignment to a Reference (STAR) software (GitHub). The total number of raw reads sequenced per sample averaged 47 million and quality filtered reads averaged 14 million. An mRNA read count matrix for each sample was created by identifying the best hits to the genome using featureCounts R (Bioconductor) against the reference gene feature annotation table from the UCSC database. On average, 25 % of the sequence reads were mapped to the Burkholderia genome. The gene-specific library size depths were in the range 3–6 million. Read counts were further filtered with a read count cutoff of <0.5 counts per million across all sample populations to filter out the zero read count mRNAs. Hits for the top genes were confirmed using RT-qPCR. Genes with high expression profiles were selected for further studies.
For the detection of differentially expressed mRNA between bacteria grown in LB media – samples M1, M2 and M3 – to bacteria grown in LB media with CaCl2 – samples C1, C2 and C3 – the read counts were normalized using the Trimmed Mean of M values (TMM) followed by differential expression analysis using the edgeR software package (Bioconductor). Statistically significant differentially expressed mRNAs were identified using empirical Bayes quasi-likelihood F-tests, with the contrast function (samples M1, M2 and M3 compared with samples C1, C2 and C3), after fitting the quasi-likelihood negative binomial generalized log-linear model on counts. Statistically significant differences in mRNA had P-values of <0.05 and false discovery rates (FDRs)<0.05. Differentially expressed genes with P-values of <0.05 were selected for functional gene ontology enrichment analysis using the LIMMA R package (Bioconductor). Based on the results of this analysis, the B. pseudomallei JW270 gene encoding BPSS2185 was selected for use in the following experiments.
Construction of JW270 ΔBPSS2185 and JW270 ΔBPSS2185 (pBHR2-BPSS2185)
To delete the BPSS2185 gene, genomic DNA was extracted from the overnight growth of JW270 cells using the GenElute Bacterial Genomic DNA kit (Sigma-Aldrich), according to the manufacturer’s protocol. PCR primer pairs 2185 F1 and R1 and 2185 F2 and R2 were used to amplify 450 bp upstream and downstream of the BPSS2185 gene. The PCR product was cleaned and purified using the QIAquick PCR Purification Kit (Qiagen) according to the manufacturer’s protocol. Primer pair BPSS2815 F1 and BPSS2185 R2 were used for overlap PCR. The final product of about 900 bp was purified, digested with NdeI and BamHI, and gel-purified using 0.7 % agarose gel. The amplicon was ligated to a pMo130 plasmid [34] previously digested with NdeI and BamHI. The plasmid was transformed in E. coli TOP10 and positive colonies were selected in the presence of 100 µg kanamycin ml−1 and verified using PCR and plasmid sequencing. A positive colony was grown overnight, and the plasmid was purified, electroporated into E. coli S17-1 and incorporated into B. pseudomallei JW270 through conjugation. Transconjugants were selected by plating on LB agar supplemented with 50 µg kanamycin ml−1 and 25 µg polymyxin B ml−1. Colonies were counterselected on agar containing 10 % sucrose for a second crossover event, resulting in the full-length gene being replaced with the truncated version producing B. pseudomallei JW270 ΔBPSS2185. The deletion mutant was confirmed by PCR.
To complement the deleted BPSS2185 gene, primer pairs C-BPSS2185 F and R were used to amplify the 218 nt product of BPSS2185. The amplicon was purified, digested with BamHI and Ndel enzymes, and gel-purified with the PureLink Quick Gel Extraction Kit (Invitrogen). The purified amplicon was ligated into the pBHR2 plasmid using the BamHI and Ndel sites. The plasmid pBHR2 is a medium-copy broad-host-range plasmid that contains a multiple cloning site downstream of a cat promoter that drives the constitutive expression of cloned genes [35]. The plasmid was transformed into chemically competent E. coli TOP10 cells (Life Technologies) and the resulting positive colonies were selected using 50 µg kanamycin ml−1. Colonies were grown in 5 ml of LB media supplemented with 50 µg kanamycin ml−1, and plasmid extraction was done using the PureYield Plasmid Miniprep System (Promega). Sequences were verified by plasmid sequencing and PCR. The plasmid was electroporated into B. pseudomallei JW270 ΔBPSS2185 using a 0.1 cuvette (620; BTX) and pulsed for 4.5 ms and 200 Ω with a constant capacitance of 25 µF. Electroporated cells were incubated in a six-well plate (Thermo Fisher Scientific) containing 1 ml of LB medium for 6 h. Cells were plated on LB agar supplemented with 50 µg kanamycin ml−1 to produce B. pseudomallei JW270 ΔBPSS2185 (pBHR2-BPSS2185).
Adhesion to epithelial cells
To determine if BPSS2185 has an adherence function, we investigated the rate of adhesion to a biotic surface – A549 epithelial cells (American Type Culture Collection) – and an abiotic surface using a 96-well polystyrene plate (Corning). Freshly cultured A549 epithelial cells in high glucose Dulbecco’s modified Eagle Medium (Hyclone Laboratories) supplemented with 10 % heat-inactivated bovine serum (without antibiotics) were grown to 90 % confluence at 37 °C and 5 % CO2. Cells were washed three times with warm Dulbecco’s PBS (DBPS) (Gibco Laboratories) and incubated with 1 : 10 µl of bacteria overnight growth of B. pseudomallei JW270, B. pseudomallei JW270 ΔBPSS2185 and B. pseudomallei JW270 ΔBPSS2185 (pBHR2-BPSS2185) diluted in LB broth. Diluted bacterial samples were plated to determine the initial c.f.u. of inoculum. A549 cells grown without bacteria inoculum were used as a control. Bacterial cells and A549 were incubated for 3 h at 37 °C and 5 % CO2. Spent medium was removed and cells were washed three times with 1 ml of warm DBPS (Gibco Laboratories). Cells were detached and lysed using 100 µl of 1 % Triton X-100 (Sigma-Aldrich), incubated for 10 min at room temperature, and the reaction was then terminated by adding 900 µl of LB medium. One hundred microlitres of serially diluted samples was plated and incubated overnight at 37 °C. Colonies were counted and the percentage adhered was calculated by dividing the number of c.f.u. of adhered bacteria by the number of c.f.u. of the inoculum.
Twitching motility assay
To examine the effect of BPSS2185 on B. pseudomallei twitching motility, JW270, JW270 ΔBPSS2185, JW270 ΔBPSS2185 (pBHR2) and JW270 ΔBPSS2185 (pBHR2-BPSS2185) were inoculated overnight in 1 ml LB medium with corresponding antibiotics. One hundred microlitres of the overnight inoculum was diluted to an OD600 of 0.45 and the bacteria were inoculated with a toothpick onto LB agar plates containing 1 % agar. The plates were incubated at 37 °C for 72 h, and the diameter of the white halo corresponding to the spread of bacteria from the point of inoculation was recorded every 24 h.
Scanning electron microscopy
Samples were grown either on a coverslip or nitrocellulose membrane (Thermo Fisher Scientific). Each coverslip and nitrocellulose membrane was processed at either 3, 24 or 72 h. The samples were washed with 0.1 M sodium cacodylate buffer (EMSciences) and then fixed with 2.5 % glutaraldehyde and 2 % paraformaldehyde in 0.1 M sodium cacodylate buffer for 1 h prior to submission to the electron microscopy facility. Samples were buffer washed, postfixed with 1 % osmium tetroxide and dehydrated through an ethanol series [30, 50, 75, 85, 95%, three changes of 100%, and a single 1 : 1 with 100 % ethanol and hexamethyldisilazane (HMDS)] for 10 min each. Samples were left to dry in 100 % HMDS for 24 h under a chemical fume hood and imaged with a Sigma VPFE microscope (Carl Zeiss Microscopy).
Biofilm assay
Biofilms were monitored using crystal violet (CV) in a 96-well polystyrene plate (Corning) as previously described [36]. Approximately 105 bacterial cells of JW270 and the two JW270 constructs were added to 2 ml of LB broth or LB broth supplemented with 50 µg kanamycin ml−1. Triplicates of 200 µl per well were cultured into three 96-well plates at 37 °C for 1–3 days. The absorbance (OD600nm) of the plates was read daily to determine growth at OD600nm. After 24 h, 0.25 µg of DNase was added. At the end of the experiment, the spent medium was discarded. The wells were gently washed twice with 200 µl of 1× PBS. One hundred microlitres of 0.1 % CV solution (Sigma-Aldrich) was added for 15 min and air-dried overnight in a biosafety cabinet. Biofilm was extracted using 200 µl of 30 % acetic acid and quantified at an optical density of 550 nm (OD550nm) on a SpectraMax M5 (Molecular Devices).
eDNA export
To quantify the amount of eDNA exported, approximately 105 bacteria were added to 3 ml of LB media (MBLE-7030, Molecular Biologicals International) containing 1 µl propidium iodide (PI; Invitrogen). Propidium, an impermeable DNA-binding dye that binds eDNA or impaired cells and emits a red fluorescence, was added to monitor eDNA export [37]. Two hundred microlitres of the mixture was dispensed in six replicates and cultured statically on a 96-well plate (Corning) for 3 days in a SpectraMax M5 sample chamber at 37 °C. Two hundred microlitres of LB plus propidium iodide was used as a blank. eDNA export was measured every 30 min by excitation at 535 nm and measuring the fluorescence at 620 nm. Experiments were repeated three times independently.
Live/dead imaging
Confocal laser scanning microscopy imaging was used to assess the live/dead ratio of B. pseudomallei strains, and the cells were inoculated as described in the biofilm assay protocol. Bacteria were inoculated in 96-well plates, gently washed and exposed to molecular dyes. The Filmtracer LIVE/DEAD Biofilm Viability Kit Film (Invitrogen L10316) was used to stain bacteria in the wells according to the manufacturer’s protocol. The bacterial cells were washed with sterile water and imaged immediately using a Nikon Eclipse 90i microscope.
BPSS2185 RNA expression
To examine the expression of BPSS2185 in a biofilm, B. pseudomallei was cultured in LB media at 37 °C for 3 days on a six-well polystyrene plate (Corning). The supernatant was carefully aspirated to prevent biofilm disruption. The biofilm was gently washed twice with PBS and the RNA was extracted by directly adding Trizol reagent (Invitrogen) to the plate. Ten micrograms of the resulting RNA was treated with Turbo DNase (Invitrogen), and 1 µg was reverse transcribed to cDNA using the iScript cDNA synthesis kit (Bio-Rad Laboratories). qRT‐PCR was performed in a total volume of 25µl, which consisted of 12.5 µl of the 2× Maxima SYBR Green/Fluorescein qPCR kit (Thermo Fisher Scientific), 300 nM of the forward and reverse BPSS2185 primers (C-BPSS2185 f and rev), and 2 µl cDNA. The 50S ribosomal protein L4 (RplD) (P60723; UniProt Consortium) was used as the reference gene for normalization. The reaction conditions were as follows: a single cycle of 95 °C for 3 min, 40 cycles of 95 °C for 10 s and 60 °C for 30 s, followed by 95 °C for 45 s and 55 °C for 1 min. Melt curve analysis was used to confirm that primer dimers were not generated. The comparative Ct method was used to analyse the data.
Statistical analysis for independent experiments
All experiments were independently repeated three times. Unless indicated, graphs represent the mean of three independent experiments. Each biofilm assay and export experiment was conducted in triplicate and mean values were compared between groups using Student’s t-test. Error bars represent the standard error of the mean. Graphpad Prism 9 (GraphPad Software) was used for statistical analysis. Differences were statistically significant at P<0.05.
Results
Five genes encoding outer membrane proteins were upregulated in the presence of CaCl2
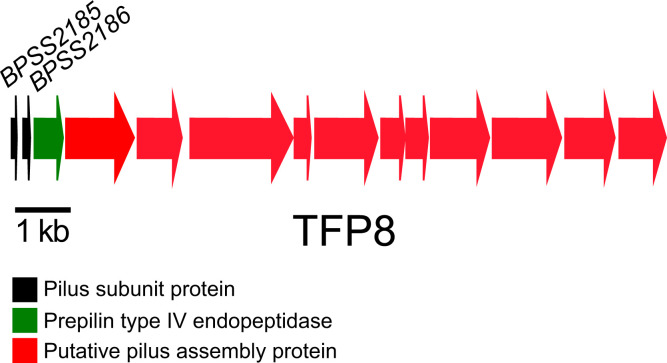
To identify B. pseudomallei genes involved in adhesion, aggregation and biofilm formation, JW270 cells grown in LB +1.26 mM CaCl2 were analysed for transcriptomic analysis using RNA-Seq and compared to JW270 cells grown in LB. The transcriptomic data identified five genes with at least a 1.5 log change that encoded outer membrane proteins (Table 2). BPSS2185 and BPSS2186, two TFP8 genes that encode type IVB subunit proteins, were highly upregulated (Fig. 1). The other three upregulated genes included two that code for fimbriae-related chaperone proteins, BPSS0092 and BPSL1627, and a gene that encodes a transport-related membrane protein (BPSS0019). We focused on type IVB genes, BPSS2185 and BPSS2186, but we were only able to construct an in-frame deletion mutation of BPSS2185. The full-length BPSS2185 gene was cloned into the broad-host-range plasmid pBHR2 and used for complementation studies. JW270, JW270 ΔBPSS2185, JW270 ΔBPSS2185 (pBHR2) and JW270 ΔBPSS2185 (pBHR2-BPSS2185) were used for the experiments described here. There was no growth difference in LB broth for any of the strains (data not shown). To identify the function of this type IVB locus, we looked at adhesion and biofilm formation capabilities.
Table 2.
Transcriptomic data of B. pseudomallei genes upregulated in the presence of CaCl2
Genes represented are those with a P-value <0.05 and at least a 1.5 log fold change (FC).
|
Locus tag |
NCBI ref. seq |
LogFC |
P-value |
Protein function |
|---|---|---|---|---|
|
BPSS2186 |
6.25132 |
0.006138 |
Pilus subunit |
|
|
BPSS2185 |
5.58863 |
0.021329 |
Pilus subunit |
|
|
BPSS0092 |
2.75258 |
0.028668 |
Fimbriae-related chaperone |
|
|
BPSL1627 |
1.68243 |
0.03835 |
Fimbriae assembly chaperone |
|
|
BPSS0019 |
1.60786 |
0.047893 |
Transport related membrane protein |
Fig. 1.
Genetic map of B. pseudomallei TFP8, locus tags BPSS2185–BPSS2198 (K96243) and BP1026B_II2352-BP1026B_II2365 (1026b). The location and direction of transcription of genes are represented by arrows. The putative functions of the proteins encoded by genes are shown in the colour-coded key below. A 1 kb scale is shown below the genetic locus.
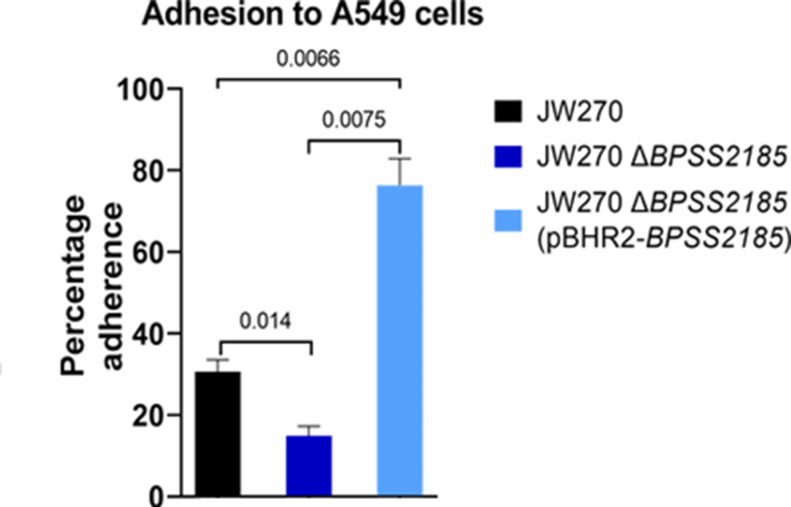
BPSS2185 is required for adhesion to epithelial cells
Adherence is required for cell or organ colonization [38]. To determine if BPSS2185 has an adherence function, we investigated the rate of adhesion to a biotic surface (A549 human epithelial cells) and an abiotic surface (96-well polystyrene plate). When compared to JW270, the ability of JW270 ΔBPSS2185 to attach to A549 alveolar epithelial cells was reduced by about 50 % (Fig. 2, P=0.014). In contrast, JW270 ΔBPSS2185 (pBHR2-BPSS2185) had a considerably higher adherence capability than both JW270 (P=0.0066) and JW270 ΔBPSS2185 (P=0.0075). A control JW270 ΔBPSS2185 (pBHR2) strain did not differ from JW270 ΔBPSS2185, ruling out influence from the empty vector in adhesion to A549 (data not shown). The increased copy number of the pilus subunit gene in the complemented mutant may have mediated the increased adherence phenotype. After 24 h on abiotic surfaces, the numbers of JW270, JW270 ΔBPSS2185 and JW270 ΔBPSS2185 (pBHR2-BPSS2185) cells adhering to 96-well plates were not significantly different (data not shown). While BPSS2185 expression was higher when B. pseudomallei was grown in LB+1.26 mM CaCl2 as compared to LB (Table 2), we found that the expression of BPSS2185 grown in LB media without CaCl2 was sufficient for assessing the phenotype of BPSS2185 in the adherence assay and all subsequent experiments described below.
Fig. 2.
BPSS2185 facilitates adherence to A549 epithelial cells. Graph showing that the deletion of BPSS2185 significantly reduced the ability to bind to A549 epithelial cells. Complementation of the ∆BPSS2185 mutation not only restored, but also enhanced, the adhesive capability of the mutant strain. Graph bars are representative of three independent experiments and error bars represent the standard error of means. A Student’s t-test was conducted to compare groups. P-values are indicated on the graph.
Twitching motility assay
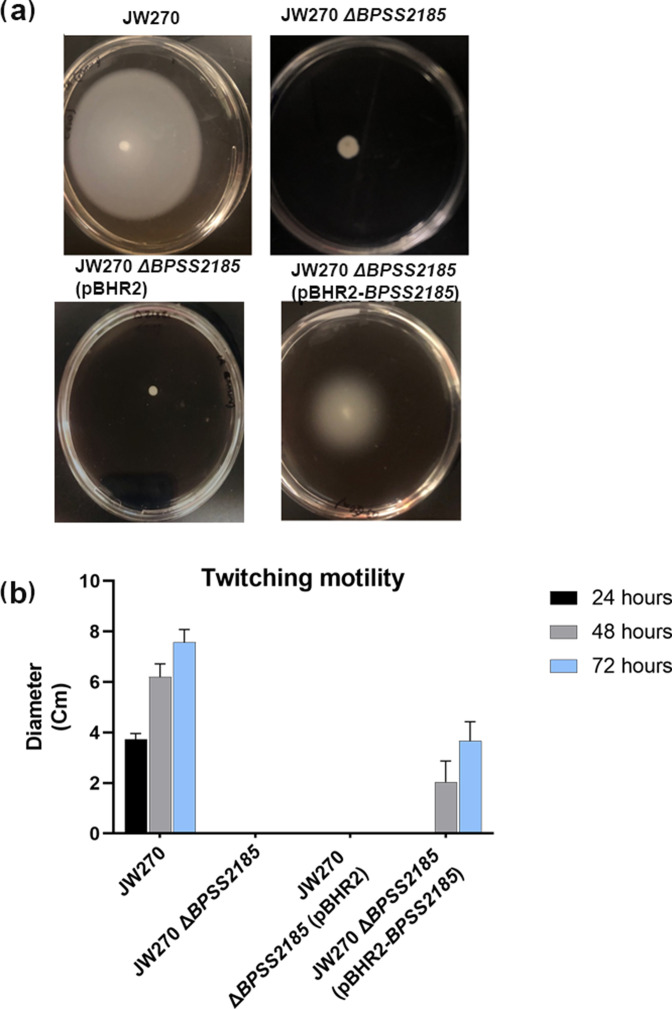
Fig. 3 shows the ability of the bacteria to twitch from an initial point of inoculation and form a circular white halo after 3 days of growth. The diameter of the halo was measured to assess the relative twitching motility of each bacterial strain examined. B. pseudomallei JW270 is a motile strain that formed a twitching halo with a diameter of 4 cm by day 1, 6 cm by day 2 and 8 cm by day 3, but JW270 ΔBPSS2185 does not move from the point of inoculation (Figs. 2 and 3). This loss of function was partially restored in JW270 ΔBPSS2185 (pBHR2-BPSS2185), which formed a twitching halo with a diameter of 2 cm by day 2 and 4 cm by day 3.
Fig. 3.
BPSS2185 is required for B. pseudomallei twitching motility. (a) Agar plate images showing twitching motility of the strains at 48 h. No halo was observed for JW270 ΔBPSS2185. The twitching motility phenotype of JW270 ∆BPSS2185 was partially complemented by pBHR2-BPSS2185, but not pBHR2. (b) Graph showing distance twitched by each strain represented in Fig. 3(a). Twitching motility is measured as the diameter of the halo (cm). Bars are an average of three independent experiments. Error bars are the standard error of means.
BPSS2185 influences biofilm formation
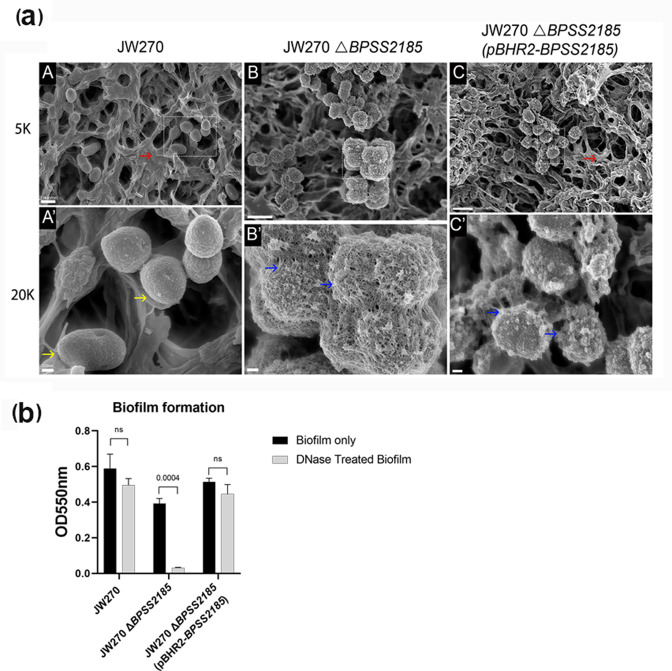
SEM images from a 3 day biofilm grown on nitrocellulose membrane are presented in Fig. 4(a). The top and bottom row images are the same images at different magnifications. In the top row, both JW270 and JW270 ΔBPSS2185 (pBHR2-BPSS2185) were able to form a solid mass of biofilm (red arrows) which covers individual bacterial cells (yellow arrow, bottom row). We observed that the bacterial cells in JW270 ΔBPSS2185 and some JW270 ΔBPSS2185 (pBHR2-BPSS2185) cells were visually coated by an adhesive structure or a weak biofilm (blue arrow, bottom row). A biofilm assay using CV did not show any significant differences in biofilm formation (P=0.586, Fig. S1, available in the online version of this article), but the biofilm formed by JW270 ΔBPSS2185 significantly responded to treatment with DNase (Fig. 4b, P=0.004, JW270 ΔBPSS2185 pre- and post-DNase treatment), indicating that the biofilm observed in Fig. 4(a) by this strain may be mostly eDNA in composition.
Fig. 4.
Biofilm formation by B. pseudomallei . (a) Scanning electron microscope imaging of biofilms formed by B. pseudomallei strains on day 3. The same image for each strain is shown at 5000× (top row) and 20 000× (bottom row). A scale bar for the entire row is shown at the left of each row. Top row: red arrows indicate a robust biofilm. Bottom row: yellow arrows indicate a bare bacterial cell and blue arrows show non-confluent biofilm. (b) Biofilm formed by all three strains on day 3 before and after treatment with DNAse. Each bar is an average of three independent experiments and the standard error of means are represented as error bars. All groups were compared using t-tests.
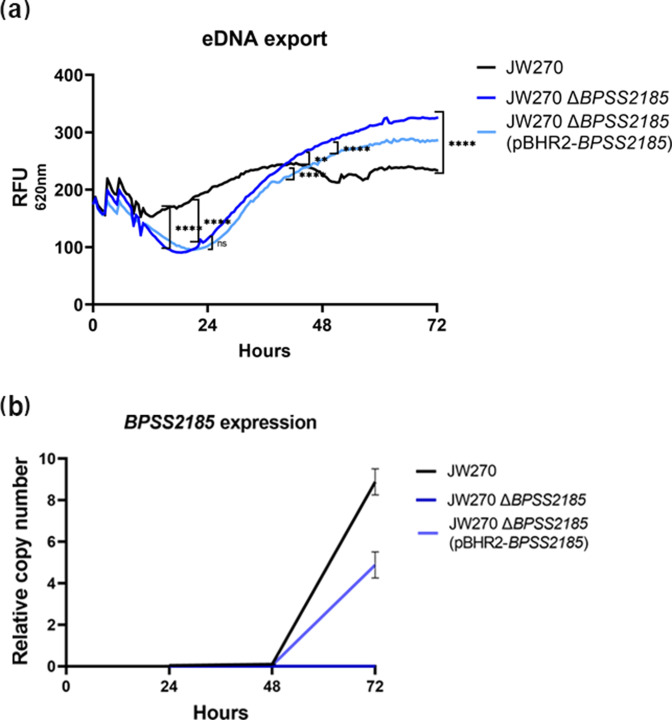
eDNA export increases in JW270 ΔBPSS2185
eDNA is an essential component of B. pseudomallei biofilm [39]. The eDNA export by JW270, JW270 ΔBPSS2185 and JW270 ΔBPSS2185 (pBHR2-BPSS2185) was continuously quantified at 30 min intervals over a 72 h period (Fig. 5a). JW270 exported significantly more eDNA than the other strains by 24 h, but at 72 h both JW270 ΔBPSS2185 and JW270 ΔBPSS2185 (pBHR2-BPSS2185) exported significantly more eDNA than JW270 (P=0.0001). JW270 ΔBPSS2185 (pBHR2-BPSS2185) exported significantly less eDNA than JW270 ΔBPSS2185 at 72 h (P=0.0001), which suggests that BPSS2185 has a negative influence on eDNA export (Fig. 5a). We confirmed that PI fluorescence was from viable cells; a 72 h live/dead microscopic image indicated that all cells were mostly viable, signifying that the increase in eDNA/PI fluorescence was active and not the result of bacterial mortality (Fig. S2).
Fig. 5.
eDNA export and BPSS2185 expression in JW270. (a) A 72 h representation of eDNA export by JW270, JW270 ΔBPSS2185 and JW270 ΔBPSS2185 (pBHR2-BPSS2185) biofilms. Data are the mean of three replicates per strain and readings were taken at 30 min intervals for 72 h. A one-way ANOVA was used to analyse all three groups. The P values for the 24, 48 and 72 h time points are indicated by brackets and asterisks for all strains on the graph.**P=0.0026; ***P≤0.0001; ns, not significant. At 72 h, all strains were significantly different from one another (P=0.0001). (b) RT-qPCR showing expression of JW270 BPSS2185, JW270 ΔBPSS2185 and JW270 ΔBPSS2185 (pBHR2-BPSS2185).
BPSS2185 expression
The JW270 ΔBPSS2185 biofilm scanning electron micrographs show weakened biofilms (Fig. 4a) that were sensitive to DNase treatment (Fig. 4b), and that JW270 ΔBPSS2185 exported more eDNA than the parental and complementation strain (Fig. 5a). We studied the expression of BPSS2185 during eDNA export. On days 1 and 2, BPSS2185 was not expressed in any of the strains. Day 3 of the eDNA export coincides with the formation of a mature biofilm in JW270 and JW270 ∆BPSS2185 (pBHR2-BPSS2185) (Fig. 4a), and BPSS2185 expression increased four- and nine-fold for JW270 and JW270 ΔBPSS2185 (pBHR2-BPSS2185), respectively (Fig. 5b). JW270 ΔBPSS2185 served as a negative control as the gene was deleted in-frame and no expression was expected.
Discussion
Bacterial adhesion and biofilm formation are implicated as major factors for disease progression and persistence [14]. Bacterial adhesion is the first interaction of a microbe and host cell and ultimately leads to biofilm formation and infection. As a result, adhesion interference/anti-adhesion therapy has been suggested as an effective means of preventing or treating bacterial infections [14, 40, 41]. The biofilm-forming strains of B. pseudomallei are highly virulent [26] and B. pseudomallei biofilms facilitate adhesion and intracellular survival in epithelial cells [42]. Adhesion of B. pseudomallei to airway epithelial cells followed by internalization is vital for pathogenesis and bacteria–host interactions [42].
We applied genetic deletion and complementation analysis to this B. pseudomallei JW270 gene to characterize its contribution to biotic and abiotic adherence, twitching motility, and biofilm formation. Using RNA-Seq and transcriptomic analysis, we identified the BPSS2185 TFP8 gene that encodes a type IVB pilin subunit protein. Out results suggest that the TFP8 locus, specifically BPSS2185, plays a role in these phenotypes, which may influence B. pseudomallei virulence and persistence.
The TFP8 locus, BPSS2185–BPSS2198, encodes two pilin subunit proteins, BPSS2185 and BPSS2186 (Fig. 1). In order to study the TFP8 pilus subunit genes, we attempted to construct mutations in both BPSS2185 and BPSS2186. Deletion of BPSS2186 proved unsuccessful after three attempts, but we were able to generate B. pseudomallei JW270 ΔBPSS2185 and JW270 ΔBPSS2185 (pBHR2-BPSS2185), a complementation strain. BPSS2185 is a part of the TFP8 locus in B. pseudomallei and this locus has been suggested as a recombination hotspot and implicated in modulating virulence in B. pseudomallei [13]. Nandi et al. showed that a TFP8 deletion mutant exhibited significantly reduced virulence in a murine intranasal infection assay compared to a parental B. pseudomallei K96243 wild-type control [13]. Likewise, our data suggest that BPSS2185 is required for full epithelial cell adhesion (Fig. 2) and twitching motility (Fig. 3), two characteristics required for full virulence and the formation of a robust biofilm (Fig. 4). BPSS2185 is suppressed during biofilm formation (Fig. 5b), but increased in a mature biofilm to aid with new cell colonization, as noted in the literature with other motile genes [28–30].
Adhesion of B. pseudomallei to airway epithelial cells followed by internalization is vital for pathogenesis and bacteria–host interactions [42]. B. pseudomallei biofilms facilitate adhesion and intracellular survival in epithelial cells, leading to chronic and difficult to eradicate infections [42]. When JW270 ΔBPSS2185 was grown in the presence of A549 epithelial cells, we observed that the deletion mutant failed to adhere efficiently to the cells. There was about a 50 % reduction in the percentage of cells that adhered in comparison to the parent JW270. The complementation strain, B. pseudomallei JW270 ΔBPSS2185 (pBHR2-BPSS2185), presented increased A549 adhesion. This study showed no significant difference in adhesive properties on abiotic surfaces for JW270 and JW270 ΔBPSS2185, suggesting that BPSS2185 is involved with adhesion to cells, but not abiotic surfaces. Therefore, the inability of the JW270 ΔBPSS2185 strain to effectively adhere or form a biofilm may be the mechanism of attenuated virulence demonstrated by Nandi et al. [13].
Twitching motility is a flagella-independent form of bacterial movement over surfaces which occurs through pili extension and retraction and involves a type IV pilus [43, 44]. Twitching motility is also a means of rapid colonization of new surfaces by bacterial communities [45]. Genes implicated in twitching capabilities are required for chronic infection and biofilm formation in Gram-negative bacteria [28, 46]. Chronic infections occur when planktonic bacteria are dispersed from a biofilm, and twitching genes are required for the planktonic bacteria to move to a new location and form a new biofilm [47]. In Fig. 3(a, b), we show that deletion of ΔBPSS2185 attenuated the ability of the bacteria to twitch, a function that was partially restored once the gene was complemented in JW270 ΔBPSS2185 (pBHR2-BPSS2185). Recently, an oligosaccharyltransferase, PglL, was linked to twitching motility in B. pseudomallei [48], but our study is the first to actively implicate BPSS2185 in twitching motility.
BPSS2185 shares 43 % similarity with Flp-1 in Aggregatibacter actinomycetemcomitans [9]. In A. actinomycetemcomitans , Flp-1 is required for non-specific adherence [49] and an flp-1 mutant exhibited reduced surface adherence much like our B. pseudomallei JW270 ∆BPSS2185 data shown in Fig. 2. The BPSS2185-encoded pilin is closely related to Flp-1 of A. actinomycetemcomitans and it is involved with forming a strong biofilm, as indicated by B. pseudomallei JW270 and JW270 ΔBPSS2185 (pBHR2-BPSS2185) [50]. Disruption of A. actinomycetemcomitans flp-1 resulted in a mutant that formed a weakly adherent biofilm [49], similar to what was observed with JW270 ΔBPSS2185 (Fig. 4a). Our biofilm results suggest that the ΔBPSS2185 mutation was only partially complemented by pBHR2-BPSS2185, which supports and is consistent with our finding that complementation partially restored twitching motility.
B. pseudomallei biofilms are made up of proteins, polysaccharides and eDNA, and we assessed eDNA export in strains with and without BPSS2185 over a period of 72 h (Fig. 5a). Surprisingly, JW270 ΔBPSS2185 exported more eDNA than JW270 at 48 and 72 h. We also observed that the amount of eDNA exported by JW270 ΔBPSS2185 (pBHR2-BPSS2185) was decreased relative to JW270 ΔBPSS2185, suggesting that BPSS2185 may impact eDNA export negatively. The mechanism by which eDNA is exported by B. pseudomallei is currently unknown and the inhibition of eDNA export, either directly or indirectly, by type IV pili has not been described in other bacterial species. We speculate that while eDNA export is important during the early stages of biofilm formation, twitching motility is less important and BPSS2185 expression is low. As B. pseudomallei cells are dispersed during biofilm expansion, BPSS2185-mediated twitching motility is required and BPSS2185 expression increases while eDNA export becomes less important and is dampened down. Further studies will be necessary to understand these processes at the molecular level.
In summary, our study is the first to demonstrate a potential mechanism by which TFP8 contributes to B. pseudomallei virulence. By applying genetic deletion and complementation analysis to the B. pseudomallei BPSS2185 pilin gene, we have shown that the gene is necessary for the bacteria to twitch, colonize epithelial cells and form a tenacious biofilm. We propose that TFP8 participates in the dispersal of B. pseudomallei from mature biofilms and contributes to the formation of new biofilms, but future experiments will be necessary to definitively prove this.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper (and its supplementary files). The RNA-Seq data have been deposited in Figshare (accession number: https://figshare.com/articles/dataset/Burkholderia_pseudomaelli/17704376).
Supplementary Data
Funding information
This research was performed while U.O. held an NRC Research Associateship award at the DeShazer Lab at the United States Army Medical Research Institute of Infectious Diseases (USAMRIID), Frederick, Maryland, USA. This work was supported by DTRA/JSTO-CBD under Grant CB10207 (to D.D.). The opinions, interpretations, conclusions and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army.
Author contributions
U.O. conceptualized, performed experiment, curated and analysed data, and wrote and edited the manuscript. S.M. investigated and curated data. G.L. investigated and curated data. J.W. and A.B. performed microscopy imaging and analysis. P.L. and R.K. performed RNA-Seq and analysed data. D.D. conceptualized and supervised the experiment, analysed data, and reviewed and edited the manuscript. J.V. proofread the manuscript for technical writing and grammatical errors.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: CV, crystal violet; eDNA, extracellular DNA; HMDS, hexamethyldisilazane; PI, propidium iodide; TFP, type IV pili.
Two supplementary figures are available with the online version of this article.
References
- 1.Wiersinga WJ, Virk HS, Torres AG, Currie BJ, Peacock SJ, et al. Melioidosis. Nat Rev Dis Primers. 2018;4:17107. doi: 10.1038/nrdp.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Limmathurotsakul D, Golding N, Dance DAB, Messina JP, Pigott DM, et al. Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat Microbiol. 2016;1:15008. doi: 10.1038/nmicrobiol.2015.8. [DOI] [PubMed] [Google Scholar]
- 3.Limmathurotsakul D, Wongratanacheewin S, Teerawattanasook N, Wongsuvan G, Chaisuksant S, et al. Increasing incidence of human melioidosis in Northeast Thailand. Am J Trop Med Hyg. 2010;82:1113–1117. doi: 10.4269/ajtmh.2010.10-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hahn HP. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa-a review. Gene. 1997;192:99–108. doi: 10.1016/s0378-1119(97)00116-9. [DOI] [PubMed] [Google Scholar]
- 5.Pizarro-Cerdá J, Cossart P. Bacterial adhesion and entry into host cells. Cell. 2006;124:715–727. doi: 10.1016/j.cell.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Lukaszczyk M, Pradhan B, Remaut H. The biosynthesis and structures of bacterial pili. Subcell Biochem. 2019;92:369–413. doi: 10.1007/978-3-030-18768-2_12. [DOI] [PubMed] [Google Scholar]
- 7.Craig L, Forest KT, Maier B. Type IV pili: dynamics, biophysics and functional consequences. Nat Rev Microbiol. 2019;17:429–440. doi: 10.1038/s41579-019-0195-4. [DOI] [PubMed] [Google Scholar]
- 8.Strom MS, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 9.Essex-Lopresti AE, Boddey JA, Thomas R, Smith MP, Hartley MG, et al. A type IV pilin, PilA, Contributes To Adherence of Burkholderia pseudomallei and virulence in vivo . Infect Immun. 2005;73:1260–1264. doi: 10.1128/IAI.73.2.1260-1264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boddey JA, Flegg CP, Day CJ, Beacham IR, Peak IR. Temperature-regulated microcolony formation by Burkholderia pseudomallei requires pilA and enhances association with cultured human cells. Infect Immun. 2006;74:5374–5381. doi: 10.1128/IAI.00569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nandi T, Ong C, Singh AP, Boddey J, Atkins T, et al. A genomic survey of positive selection in Burkholderia pseudomallei provides insights into the evolution of accidental virulence. PLoS Pathog. 2010;6:e1000845. doi: 10.1371/journal.ppat.1000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sangdee K, Waropastrakul S, Wongratanachewin S, Homchampa P. Heterologously type IV pilus expressed protein of Burkholderia pseudomallei is immunogenic but fails to induce protective immunity in mice. Southeast Asian J Trop Med Public Health. 2011;42:1190–1196. [PubMed] [Google Scholar]
- 13.Nandi T, Holden MTG, Holden MTG, Didelot X, Mehershahi K, et al. Burkholderia pseudomallei sequencing identifies genomic clades with distinct recombination, accessory, and epigenetic profiles. Genome Res. 2015;25:129–141. doi: 10.1101/gr.177543.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krachler AM, Orth K. Targeting the bacteria-host interface: strategies in anti-adhesion therapy. Virulence. 2013;4:284–294. doi: 10.4161/viru.24606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okaro U, Green R, Mohapatra S, Anderson B. The trimeric autotransporter adhesin BadA is required for in vitro biofilm formation by Bartonella henselae . NPJ Biofilms Microbiomes. 2019;5:10. doi: 10.1038/s41522-019-0083-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Locht C, Bertin P, Menozzi FD, Renauld G. The filamentous haemagglutinin, a multifaceted adhesion produced by virulent Bordetella spp. Mol Microbiol. 1993;9:653–660. doi: 10.1111/j.1365-2958.1993.tb01725.x. [DOI] [PubMed] [Google Scholar]
- 17.Guo H, Yi W, Song JK, Wang PG. Current understanding on biosynthesis of microbial polysaccharides. Curr Top Med Chem. 2008;8:141–151. doi: 10.2174/156802608783378873. [DOI] [PubMed] [Google Scholar]
- 18.Berne C, Ducret A, Hardy GG, Brun YV. Adhesins involved in attachment to abiotic surfaces by gram-negative bacteria. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MB-0018-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 20.Ruhal R, Kataria R. Biofilm patterns in gram-positive and gram-negative bacteria. Microbiol Res. 2021;251:126829. doi: 10.1016/j.micres.2021.126829. [DOI] [PubMed] [Google Scholar]
- 21.Sharma V, von Ossowski I, Krishnan V. Exploiting pilus-mediated bacteria-host interactions for health benefits. Mol Aspects Med. 2021;81:100998. doi: 10.1016/j.mam.2021.100998. [DOI] [PubMed] [Google Scholar]
- 22.Das T, Sehar S, Koop L, Wong YK, Ahmed S, et al. Influence of calcium in extracellular DNA mediated bacterial aggregation and biofilm formation. PLoS One. 2014;9:e91935. doi: 10.1371/journal.pone.0091935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 24.Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol. 2006;188:8213–8221. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Begun J, Gaiani JM, Rohde H, Mack D, Calderwood SB, et al. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 2007;3:e57. doi: 10.1371/journal.ppat.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chin CY, Hara Y, Ghazali AK, Yap SJ, Kong C, et al. Global transcriptional analysis of Burkholderia pseudomallei high and low biofilm producers reveals insights into biofilm production and virulence. BMC Genomics. 2015;16:471. doi: 10.1186/s12864-015-1692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joshua GWP, Karlyshev AV, Smith MP, Isherwood KE, Titball RW, et al. A Caenorhabditis elegans model of Yersinia infection: biofilm formation on a biotic surface. Microbiology (Reading) 2003;149:3221–3229. doi: 10.1099/mic.0.26475-0. [DOI] [PubMed] [Google Scholar]
- 28.O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 29.Otton LM, da Silva Campos M, Meneghetti KL, Corção G. Influence of twitching and swarming motilities on biofilm formation in Pseudomonas strains. Arch Microbiol. 2017;199:677–682. doi: 10.1007/s00203-017-1344-7. [DOI] [PubMed] [Google Scholar]
- 30.Wang T, Guan W, Huang Q, Yang Y, Yan W, et al. Quorum-sensing contributes to virulence, twitching motility, seed attachment and biofilm formation in the wild type strain Aac-5 of Acidovorax citrulli . Microb Pathog. 2016;100:133–140. doi: 10.1016/j.micpath.2016.08.039. [DOI] [PubMed] [Google Scholar]
- 31.Pu M, Storms E, Chodur DM, Rowe-Magnus DA. Calcium-dependent site-switching regulates expression of the atypical iam pilus locus in Vibrio vulnificus . Environ Microbiol. 2020;22:4167–4182. doi: 10.1111/1462-2920.14763. [DOI] [PubMed] [Google Scholar]
- 32.Rose SJ, Bermudez LE. Identification of bicarbonate as a trigger and genes involved with extracellular DNA export in mycobacterial biofilms. mBio. 2016;7:e01597-16. doi: 10.1128/mBio.01597-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamad MA, Zajdowicz SL, Holmes RK, Voskuil MI. An allelic exchange system for compliant genetic manipulation of the select agents Burkholderia pseudomallei and Burkholderia mallei . Gene. 2009;430:123–131. doi: 10.1016/j.gene.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schell MA, Ulrich RL, Ribot WJ, Brueggemann EE, Hines HB, et al. Type VI secretion is a major virulence determinant in Burkholderia mallei . Mol Microbiol. 2007;64:1466–1485. doi: 10.1111/j.1365-2958.2007.05734.x. [DOI] [PubMed] [Google Scholar]
- 36.O’Toole GA. Microtiter dish biofilm formation assay. J Vis Exp. 2011;2011:47. doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenberg M, Azevedo NF, Ivask A. Propidium iodide staining underestimates viability of adherent bacterial cells. Sci Rep. 2019;9:6483. doi: 10.1038/s41598-019-42906-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ribet D, Cossart P. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 2015;17:173–183. doi: 10.1016/j.micinf.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 39.Pakkulnan R, Anutrakunchai C, Kanthawong S, Taweechaisupapong S, Chareonsudjai P, et al. Extracellular DNA facilitates bacterial adhesion during Burkholderia pseudomallei biofilm formation. PLoS One. 2019;14:e0213288. doi: 10.1371/journal.pone.0213288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ofek I, Hasty DL, Sharon N. Anti-adhesion therapy of bacterial diseases: prospects and problems. FEMS Immunol Med Microbiol. 2003;38:181–191. doi: 10.1016/S0928-8244(03)00228-1. [DOI] [PubMed] [Google Scholar]
- 41.Qvortrup K, Hultqvist LD, Nilsson M, Jakobsen TH, Jansen CU, et al. Small molecule anti-biofilm agents developed on the basis of mechanistic understanding of biofilm formation. Front Chem. 2019;7:742. doi: 10.3389/fchem.2019.00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kunyanee C, Kamjumphol W, Taweechaisupapong S, Kanthawong S, Wongwajana S, et al. Burkholderia pseudomallei biofilm promotes adhesion, internalization and stimulates proinflammatory cytokines in human epithelial A549 cells. PLoS One. 2016;11:e0160741. doi: 10.1371/journal.pone.0160741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 44.Rashid MH, Kornberg A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa . Proc Natl Acad Sci U S A. 2000;97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mattingly AE, Weaver AA, Dimkovikj A, Shrout JD. Assessing travel conditions: environmental and host influences on bacterial surface motility. J Bacteriol. 2018;200:e00014–18. doi: 10.1128/JB.00014-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han X, Kennan RM, Davies JK, Reddacliff LA, Dhungyel OP, et al. Twitching motility is essential for virulence in Dichelobacter nodosus . J Bacteriol. 2008;190:3323–3335. doi: 10.1128/JB.01807-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiang P, Burrows LL. Biofilm formation by hyperpiliated mutants of Pseudomonas aeruginosa . J Bacteriol. 2003;185:2374–2378. doi: 10.1128/JB.185.7.2374-2378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willcocks SJ, Denman C, Cia F, McCarthy E, Cuccui J, et al. Virulence of the emerging pathogen, Burkholderia pseudomallei, depends upon the O-linked oligosaccharyltransferase, PglL. Future Microbiol. 2020;15:241–257. doi: 10.2217/fmb-2019-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schreiner HC, Sinatra K, Kaplan JB, Furgang D, Kachlany SC, et al. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc Natl Acad Sci U S A. 2003;100:7295–7300. doi: 10.1073/pnas.1237223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fine DH, Furgang D, Kaplan J, Charlesworth J, Figurski DH. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch Oral Biol. 1999;44:1063–1076. doi: 10.1016/s0003-9969(99)00089-8. [DOI] [PubMed] [Google Scholar]
- 51.Simon R, Priefer U, Pühler A. A Broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol. 1983;1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 52.Warawa JM, Long D, Rosenke R, Gardner D, Gherardini FC. Role for the Burkholderia pseudomallei capsular polysaccharide encoded by the wcb operon in acute disseminated melioidosis. Infect Immun. 2009;77:5252–5261. doi: 10.1128/IAI.00824-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper (and its supplementary files). The RNA-Seq data have been deposited in Figshare (accession number: https://figshare.com/articles/dataset/Burkholderia_pseudomaelli/17704376).