Abstract
The E3 ubiquitin-ligases are important for cellular protein homeostasis and their deregulation is implicated in cancer. The E3 ubiquitin-ligase Hakai is involved in tumour progression and metastasis, through the regulation of the tumour suppressor E-cadherin. Hakai is overexpressed in colon cancer, however, the implication in colitis-associated cancer is unknown. Here, we investigated the potential role of Hakai in intestinal inflammation and cancer bowel disease. Several mouse models of colitis and associated cancer were used to analyse Hakai expression by immunohistochemistry. We also analysed Hakai expression in patients with inflamed colon biopsies from ulcerative colitis and Crohn's disease. By Hakai interactome analysis, it was identified Fatty Acid Synthase (FASN) as a novel Hakai-interacting protein. Moreover, we show that Hakai induces FASN ubiquitination and degradation via lysosome, thus regulating FASN-mediated lipid accumulation. An inverse expression of FASN and Hakai was detected in inflammatory AOM/DSS mouse model. In conclusion, Hakai regulates FASN ubiquitination and degradation, resulting in the regulation of FASN-mediated lipid accumulation, which is associated to the development of inflammatory bowel disease. The interaction between Hakai and FASN may be an important mechanism for the homeostasis of intestinal barrier function and in the pathogenesis of this disease.
Subject terms: Cancer, Cell biology, Molecular biology, Diseases
Introduction
Inflammation is an important risk factor for the development of human cancers. Colitis-associated colorectal cancer (CAC) is a specific type of colorectal cancer developed from colitis in inflammatory bowel disease (IBD) patients. IBD, including ulcerative colitis (UC) and Crohn’s disease (CD), is characterized by chronic inflammation in the gastrointestinal tract and can induce pre-neoplastic lesions, being an important risk factor for the onset of colorectal cancer1,2. Ubiquitination is one of the most important translational modifications in eukaryotes that induces substrate degradation, which in consequence controls the “quantity” and “quality” of specific proteins, ensuring cell homeostasis3,4. Ubiquitination process consists in the ubiquitin moiety linkage to a specific substrate that signals for degradation5. Three-enzymes are involved in the ubiquitination cascade: the E1 ubiquitin-activating enzyme that activates the ubiquitin molecule, the E2 ubiquitin-conjugating enzyme that carries the activated ubiquitin molecule as a thioester to the E3 ubiquitin-ligase enzyme that transfers the activated ubiquitin from the E2 to the lysine residue of substrate protein5,6. Among all RING-type E3 ligases, there are a number that require substrate phosphorylation at a tyrosine residue (pTyr) for their recognition, including Cbl family7. Excessive ubiquitin-mediated proteolysis has been observed in different types of cancer as well as in colitis, and its inhibition was shown to improve the colitis in the mouse model8–10.
Hakai protein is an E3 ubiquitin-ligase, identified as the first posttranslational regulator of the E-cadherin stability. Hakai induces E-cadherin ubiquitination and degradation in a Src-dependent manner; which in turn causes the alteration of cell–cell adhesions11,12. E-cadherin is the best-characterized member of adherens junctions, a type of cell–cell contacts that participate in embryogenesis and tissue homeostasis13,14. The loss of E-cadherin at cell–cell contacts induces the epithelial-to-mesenchymal transition (EMT), a process by which cells lose cell polarity and acquire a migratory phenotype, a fundamental program during colon tumor progression and metastasis15–17. In human biopsies from IBD patients, E-cadherin expression is downregulated. Indeed, E-cadherin is the strongest candidate for UC susceptibility and plays a key role in epithelial restitution and repair following mucosal damage. In fact, expression of E-cadherin is significantly reduced in areas of active UC. Of note, E-cadherin has recently been associated with susceptibility to colorectal cancer, which is an established complication of longstanding UC. The new associations suggest that changes in the integrity of the intestinal epithelial barrier may contribute to the pathogenesis of UC18–22. Moreover, In an in vivo mouse model of experimentally induced colitis, it is shown that E-cadherin deficiency was associated with exacerbated acute and chronic inflammation highlighting its role in the pathogenesis of UC23. We have previously demonstrated that Hakai is an important regulator of cell proliferation, epithelial-to-mesenchymal transition, cell invasion, and that it induces tumour progression and metastasis in vivo24–26. We have also shown that Hakai expression is increased in colon and gastric cancer compared to adjacent human colon healthy tissues, with important clinical implications for designing new therapeutic strategies26–28.
Several E3 ubiquitin-ligases were reported to be implicated in the pathogenesis and development of human IBD8. However, whether the E3 ubiquitin-ligase Hakai participates in IBD is still unknown. The main purpose is to explore the implication of Hakai in IBD by analyzing Hakai expression in murine models for CAC. In the present study, we have shown that Hakai is downregulated under pro-inflammatory environment in the mice intestinal tissue regardless of origin of that inflammation. Moreover, by an interactome analysis we have identified Fatty Acid Synthase (FASN) as a novel Hakai-interacting protein. Hakai induces FASN ubiquitination and degradation via lysosome, regulating FASN-mediated lipid accumulation, which is associated to the development of IBD. Importantly, an inverse association of FASN expression with Hakai expression was detected in inflammatory AOM/DSS compared to tumour tissue of CAC and healthy tissues.
Results
Hakai expression in different mouse models of colitis and CAC
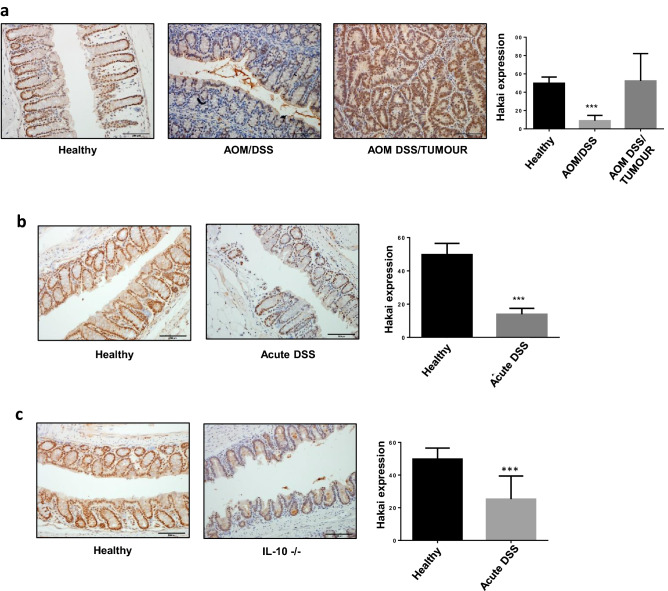
Hakai overexpression in cytoplasm and nucleus was reported in human colon cancer compared to adjacent normal healthy colon tissues24,26,27. In order to compare Hakai expression in the context of intestinal inflammation, three different mouse models that mimic different origins and stages of the IBD disease were used. Firstly, an AOM-DSS model of CAC was used. This model replicates the tumour development process of the human CAC by using the combination of a pro-inflammatory agent (DSS) and a carcinogenic compound (AOM), as previously described29. As shown in Fig. 1a, the immunohistochemistry analysis of the whole intestine samples using Hakai antibody revealed a significant decreased expression of Hakai in inflammatory conditions (AOM/DSS) of the gut epithelium compared to tumour tissue from the CAC (AOM DSS/Tumour) or healthy tissues. The same pattern of expression was observed when analysing an animal model of acute colitis (acute DSS), which resemble the acute phase of the disease or flare-ups by using higher concentrations of pro-inflammatory agent DSS at short times (Fig. 1b). Finally, a third stablished mouse model was used based in a genetically modified mice deficient for the IL-10 gene (IL-10 KO). These mice spontaneously develop a chronic inflammatory bowel disease (IBD) due to the important control of IL-10 on the gut microbiota30. The absence of IL-10 modulates cellular immune response and reduces mice tolerance towards bacterial antigens of enteric bacteria which causes the inflammatory process. In this model, Hakai expression was also statistically reduced in inflamed conditions compared to gut epithelium of healthy mice (Fig. 1c). All these results point out to a regulation of Hakai in different inflammatory conditions, with a great reduction of Hakai expression in the inflamed epithelium. Remarkably, high expression of Hakai in the epithelium of healthy untreated mouse was detected while this observation was not shown in human tumour-adjacent healthy tissue26.
Figure 1.
Expression of Hakai in mouse models of colitis and CAC. (a) Immunohistochemistry staining of Hakai in healthy non-treated mouse colonic mucosa and in gut sections of AOM/DSS-treated mouse (inflamed epithelium and tumours, healthy mice, n = 7; AOM/DSS-treated mice, n = 14). (b) Immunohistochemistry staining of Hakai in healthy non-treated mouse colonic mucosa and in gut sections for DSS-treated mouse (inflamed epithelium, healthy mice, n = 7; DSS-treated mice, n = 3). (c) Immunohistochemistry staining of Hakai in healthy non-treated mouse colonic mucosa and in gut sections for IL-10 deficient mice (inflamed epithelium, healthy mice, n = 7; IL-10 deficient mice, n = 6). Images obtained with ×20 objective. Five pictures of each sample were taken and quantified. Data are represented as scatter plot. Values are means ± SEM of staining intensity signal scoring per area. Quantification and calibration of the images were performed with Immunohistochemistry Image Analysis Toolbox software for Image J. Kruskal–Wallis with Tukey correction test analyses show statistical differences in AOM/DSS-inflamed tissue respect to paired healthy samples (*p < 0.05; **p < 0,01; ***p < 0,001). Scale bar 125 μm.
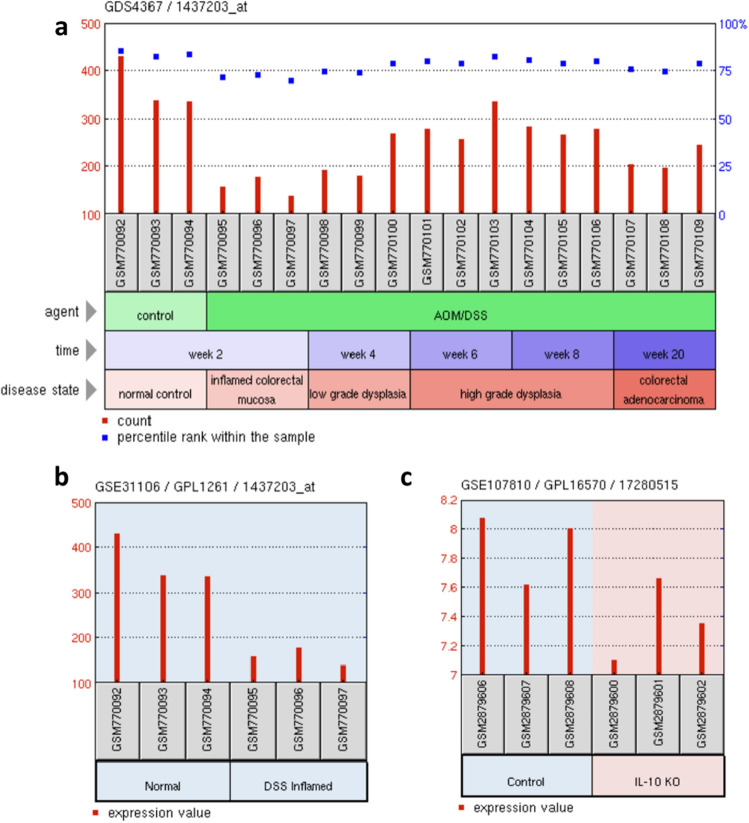
Then we further analysed the Hakai mRNA expression by using microarray data from similar experiments in NCBI’s Gene Expression Omnibus (GEO). Three different mouse models were analysed: the colitis-associated colorectal cancer (AOM/DSS), the acute colitis (acute-DSS), and the IL-10 KO mouse models. As shown, the levels of Hakai mRNA were reduced in the inflamed tissue in AOM/DSS model (Fig. 2a), the DSS mouse model (Fig. 2b) and the IL-10 KO mouse model (Fig. 2c). Moreover, Hakai mRNA expression was recovered during tumour progression (low grade dysplasia, high grade dysplasia and colorectal adenocarcinoma) in the AOM/DSS model (Fig. 2a). Taking together these results suggest that, mRNA and protein expression of Hakai are clearly regulated under a pro-inflammatory environment in the intestinal tissue regardless of origin of that inflammation.
Figure 2.
Gene expression of Hakai in AOM/DSS, acute DSS and IL-10 KO mouse models using microarray data from similar experiments in NCBI’s Gene Expression Omnibus (GEO). (a) Gene expression of Hakai during temporal analysis of mouse colon tissue from AOM/DSS-induced model of UC-associated colorectal cancer. (b) Hakai gene expression differences between normal control and DSS inflamed mucosa in mouse. (c) Hakai gene expression in colon tissue from IL-10 WT and IL-10 KO mice. Data from GEO database accession numbers: GDS4367, GSE31106 and GSE107810.
FASN as a novel Hakai-regulated protein identified by interactome analysis
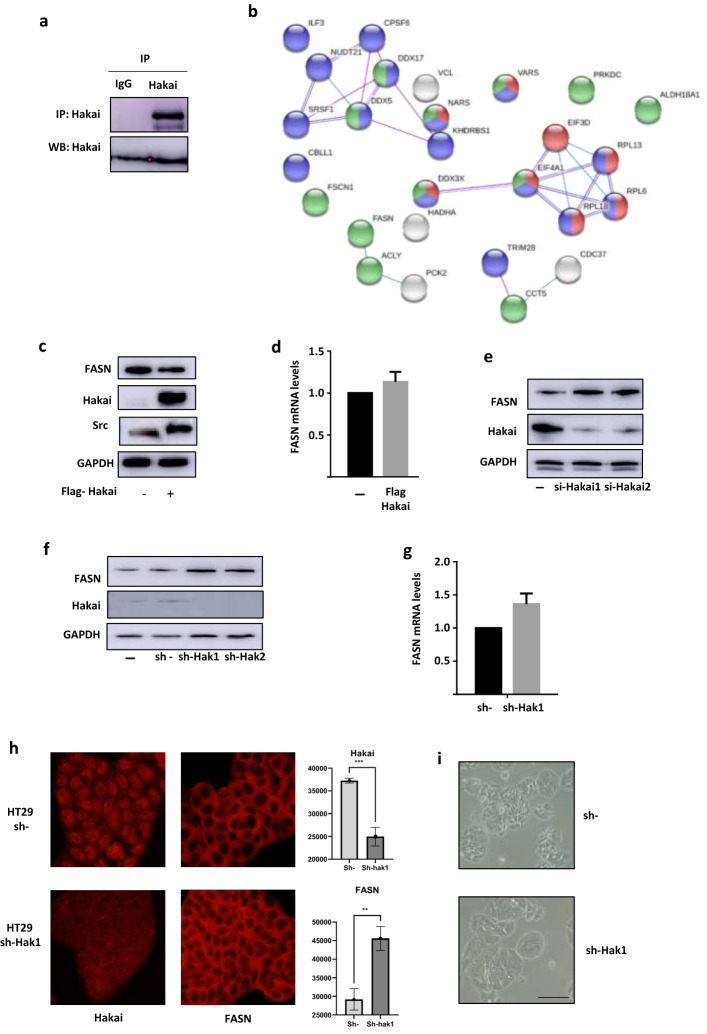
In order to understand the potential molecular mechanism by which Hakai could be involved in inflammation, HCT116 colon cell line was used to perform a large-scale immunoprecipitation of Hakai followed by a mass spectrometric analysis in order to determine novel Hakai-interacting proteins that could be related to inflammatory process. For Hakai interactome, immunoprecipitation of endogenous Hakai protein levels was firstly confirmed (Fig. 3a). A total of 26 proteins with a peptide match ≥ 2 were identified by proteomic analysis in Hakai immunoprecipitated samples compared to non-specific immunoprecipitation using IgG control (Table 1). It should be noted that, Hakai is among the identified proteins and also HSP90 protein, another previously Hakai-interacting protein reported31, ensuring the specificity of the developed technique. A map of Hakai molecular interactions was performed by using the biological database STRING that provide information about both physical and functional properties of proteins (Fig. 3b)32. Interestingly, the Fatty Acid Synthase (FASN) protein was found as a novel Hakai-interacting protein. FASN is a multifunctional enzyme that catalyses the novo synthesis of long chain saturated fatty acids, from acetyl-CoA and malonyl-CoA, in the presence of NADPH33. Long-chain fatty acids considerably contribute to the pathology of intestinal epithelial cells, however, under normal conditions, the synthesis of fatty acids takes place in order to store energy. Abnormalities of lipid metabolism through overexpression of FASN, are associated with the development of inflammatory bowel disease (IBD)34,35. On the other hand, FASN expression has been detected in some benign and preneoplastic lesions of several organs such as colon, prostate, breast, lung, stomach, and skin nevi36,37.
Figure 3.
FASN is a novel identified protein in the Hakai interactome analysis. (a) Immunoprecipitation of endogenous Hakai in HCT116 cells. (b) Network using STRING database of all the identified proteins in Hakai immunoprecipitation in HCT-116. Proteins in green are related to drug binding, red colour, implication in translation and blue colour in RNA processing. (c) Effect of Hakai overexpression reduces FASN protein levels. Transfection with pcDNA-Flag-Hakai (2 µg) and pSG-v-Src (0.5 µg) was performed in HCT116 cells following by western blot with the indicated antibodies. (d) Effect of Hakai overexpression, as described in C, in FASN mRNA levels analysed by RT qPCR using RPL13A and GAPDH as control (e,f) Effect of Hakai silencing on FASN using transient transfection in HCT116 cells (e) or an inducible viral-transduced system with 1 µg/ml doxycycline for Hakai silencing in HT29 (f). Cell lysates were subjected to Western Blotting as described. (g) mRNA levels of FASN determined in the inducible viral-transduced system for Hakai silencing in HT29, analysed by RT qPCR using RPL13A and GAPDH as a housekeeping controls. (h) Effect of Hakai silencing in FASN subcellular localization in the inducible viral-transduced system in HT29 cells. Immunofluorescence images with anti-Hakai and anti-FASN antibodies (left panel) and quantification (right panel) is included. Images taken with 40X magnification. (i) Effect of Hakai on phenotype by using an inducible silencing viral system on HT29 cell. Optical microscopy images taken after 72 h induction. Scale bar 125 μm. The blots images were cropped prior hybridization and original blots are included in Supplementary Information.
Table 1.
List of identified proteins in Hakai interactome analysis.
| N | Accession number | Gene code | Protein name | Biological role |
|---|---|---|---|---|
| 1 | Q9Z1X4 | ILF3 | Interleukin enhancer-binding factor 3 | Transcription factor (NF45, ILF2) required for T-cell expression of interleukin 2 |
| 2 | Q16630 | CPSF6 | Cleavage and polyadenylation specificity factor subunit 6 | Subunit of a cleavage factor required for 3' RNA cleavage and polyadenylation processing |
| 3 | O43809 | NUDT21 | Cleavage and polyadenylation specificity factor subunit 5 | Subunit of a cleavage factor required for 3' RNA cleavage and polyadenylation processing |
| 4 | Q92841 | DDX17 | ATP-dependent RNA helicase DDX17 | RNA helicases. They are implicated in a number of cellular processes involving alteration of RNA secondary structure |
| 5 | P17844 | DDX5 | ATP-dependent RNA helicase DDX5 | RNA helicases. This protein is involved in pathways that include the alteration of RNA structures |
| 6 | Q07955 | SRSF1 | Serine/arginine-rich splicing factor 1 | Arginine/serine-rich splicing factor protein family |
| 7 | Q16658 | FSCN1 | Fascin | Member of the fascin family of actin-binding proteins |
| 8 | P18206 | VCL | Vinculin | Cytoskeletal protein associated with cell–cell and cell–matrix junctions |
| 9 | Q2KJG3 | NARS | Asparagine tRNA ligase | Asparaginyl-tRNA synthetase |
| 10 | Q07666 | KHDRBS1 | KH domain-containing, RNA-binding, signal transductionassociated protein 1 | Member of the K homology domain-containing, RNA-binding, signal transduction-associated protein family |
| 11 | P26640 | VARS | Valine tRNA ligase | Valyl-tRNA synthetase |
| 12 | O00571 | DDX3X | ATP-dependent RNA helicase DDX3X | ATP-dependent RNA helicase |
| 13 | P40939 | HADHA | Trifunctional enzyme subunit alpha | Catalyzes the last three steps of mitochondrial beta-oxidation of long chain fatty acids |
| 14 | P49327 | FASN | Fatty acid synthase | Catalyze the synthesis of palmitate from acetyl-CoA and malonyl-CoA, in the presence of NADPH, into long-chain saturated fatty acids |
| 15 | P53396 | ACLY | ATP-citrate synthase | Primary enzyme responsible for the synthesis of cytosolic acetyl-CoA in many tissues |
| 16 | Q16822 | PCK2 | Phosphoenolpyruvate carboxykinase | Mitochondrial enzyme that catalyzes the conversion of oxaloacetate to phosphoenolpyruvate in the presence of guanosine triphosphate |
| 17 | Q13263 | TRIM28 | Transcription intermediary factor 1-beta | Gene mediates transcriptional control by interaction with the Kruppel-associated box repression domain found in many transcription factors |
| 18 | P48643 | CCT5 | T-complex protein 1 subunit epsilon | Molecular chaperone that is a member of the chaperonin containing TCP1 complex |
| 19 | Q16543 | CDC37 | Hsp90 co-chaperone | Molecular chaperone with specific function in cell signal transduction. It has been shown to form complex with Hsp90 |
| 20 | P78527 | PRKDC | DNA-dependent protein kinase catalytic subunit | Catalytic subunit of the DNA-dependent protein kinase (DNA-PK). It functions with the Ku70/Ku80 heterodimer protein in DNA double strand break repair and recombination |
| 21 | P54886 | ALDH18A1 | Delta-1-pyrroline-5-carboxylate synthase | Catalyzes the reduction of glutamate to delta1-pyrroline-5-carboxylate, a critical step in the de novo biosynthesis of proline, ornithine and arginine |
| 22 | O15371 | EIF3D | Eukaryotic translation initiation factor 3 subunit D | The complex binds to the 40S ribosome and helps maintain the 40S and 60S ribosomal subunits in a dissociated state |
| 23 | P26373 | RPL13 | 60S ribosomal protein L13 | This gene encodes a ribosomal protein that is a component of the 60S subunit |
| 24 | Q02878 | RPL6 | 60S ribosomal protein L6 | This gene encodes a protein component of the 60S ribosomal subunit |
| 25 | Q07020 | RPL18 | 60S ribosomal protein L18 | Ribosomes, the organelles that catalyze protein synthesis, consist of a small 40S subunit and a large 60S subunit |
| 26 | P60842 | EIF4A1 | Eukaryotic initiation factor 4A-I | Eukaryotic translation initiation factor 4A1 |
In order to determine the possible link between Hakai and FASN, HCT116 cells were transfected with Hakai, in presence of Src, and the effect on FASN expression levels was assessed. Hakai overexpression strongly reduced FASN protein expression in HCT-116 cells, while no effect was detected at mRNA levels (Fig. 3c,d), further suggesting that Hakai may regulate FASN at a post-transcriptional level. Then, we tested the effect of Hakai-silencing on FASN expression. Transient transfections of two different oligos of siRNA Hakai in HCT116 cells and viral transduction of shRNA-Hakai silencing in an inducible system in HT29 were performed. Hakai silencing strongly increased FASN protein expression in both cell lines (Fig. 3e,f), while mRNA expression of FASN was not affected by Hakai silencing (Fig. 3g), further supporting the effect of Hakai on FASN at post-transcriptional level. Then it was analysed the effect of Hakai-silencing in the localization of FASN by immunofluorescence. The strong reduction of Hakai in the cells after inducing the knockdown with doxycycline confirmed the increased of FASN, without affecting FASN cellular localization (Fig. 3h). At the phenotypic level, silencing of Hakai induced a more epithelial phenotype, with cells closer attached between each other accompanied to a more three-dimensional cell sheet, compared to control conditions (Fig. 3i).
Hakai is a post-translational regulator of FASN via ubiquitin-lysosome pathway
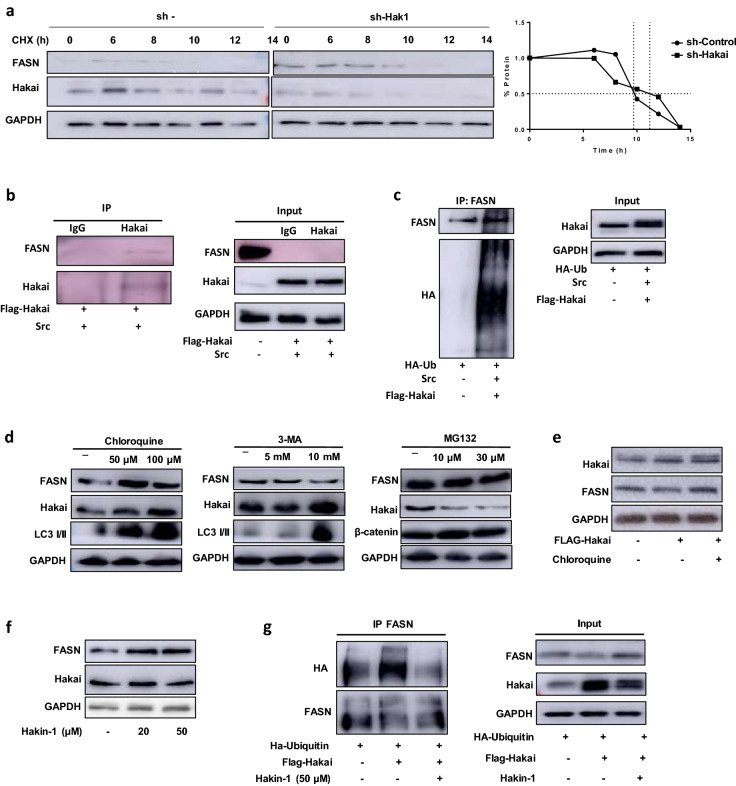
Given that our results suggested that Hakai may control FASN at post-transcriptional level, inhibition of protein synthesis with cycloheximide (CHX) was performed in order to determine the half-life of FASN protein in presence or absence of Hakai (Fig. 4a, left and middle panel). FASN is a rather stable protein in HT29 cell, with an approximate half-life of 9.5 h (Fig. 4a, right panel). FASN half-life was increased to 11.5 h in Hakai-silencing conditions, suggesting Hakai effect on FASN protein stability. We then analysed whether Hakai may act as an E3 ubiquitin-ligase for FASN inducing its ubiquitination and degradation. After Hakai overexpression, an interaction between Hakai and FASN was detected (Fig. 4b), further confirming the results obtained by the interactome analysis (Fig. 3b). Moreover, Hakai induced FASN ubiquitination when transfecting cells in presence of pcDNA-Flag-Hakai, pBSSR-HA-ubiquitin and pSG-v-Src (Fig. 4c). This effect was also detected when Src was not overexpressed (Fig. S1). Taken together these results indicate that Hakai binds to FASN and induces its ubiquitination and degradation in vitro. There are three described pathways by which ubiquitinated substrates can be degraded: proteasome degradation, lysosome degradation and autophagy system. Treatment with the lysosome inhibitor chloroquine did increase FASN protein levels in HCT116 cells, while the autophagy inhibitor 3-methyladenine (3-MA) did not (Fig. 4d). Given that both inhibitors exert their inhibitory function in the autophagy pathway, 3-MA at early stages of autophagosome formation and chloroquine at late-autophagy stage targeting the lysosome38, their eeffectiveness were confirmed by using LC3 I/II levels as positive control. Furthermore, the 26S proteasome inhibitor MG132 did not increase FASN protein (Fig. 4d), suggesting that FASN degradation does occur via lysosome degradation. Besides, FASN levels were recovered after Hakai overexpression when lysosome inhibitor chloroquine was present (Fig. 4e), indicating that Hakai induced FASN degradation is, at least in part, via lysosome. Finally, we used a specific reported Hakai inhibitor, Hakin-1, to inhibit Hakai-mediated ubiquitination without affecting Hakai protein levels28. As shown, Hakin-1 was able to increase FASN protein levels without affecting Hakai protein levels, further supporting Hakai-mediated action on FASN (Fig. 4f). We also observed a reduction of Hakai-dependent ubiquitination of FASN when cells were treated with Hakin-1 (Fig. 4g). Collectively, our data support that Hakai control FASN ubiquitination and degradation via lysosome.
Figure 4.
Hakai regulates ubiquitination and degradation of FASN via lysosome. (a) Analysis of FASN degradation by cycloheximide chase. FASN half-life in HT29 cell line was determined by treatment with 10 µg/ml Cycloheximide for the indicated time course and compared with FASN half-life in Hakai-silenced HT29 cells compared to control HT29 cells. Hakai-silencing was performed by viral transduction induced with 1 µg/ml of Doxycycline for 72 h. Quantification of endogenous FASN protein half-life are shown (right panel). (b) Coimmunoprecipitation of Hakai and FASN. Hakai was immunoprecipitated in HCT-116 cells transfected with pcDNA-Flag-Hakai and v-Src. Hakai immunoprecipitation was performed using anti-Hakai antibody. Coimmunoprecipitation was evaluated by western blot as described in materials and methods using the indicated antibodies. GAPDH signal was used as protein loading control. (c) Hakai-dependent ubiquitination of FASN Flag-Hakai, v-Src and HA-ubiquitin were transiently transfected into HCT116 cells. Immunoprecipitation was performed with the anti-FASN antibody before Western blotting, using the indicated antibodies. (d) FASN and Hakai levels in HCT-116 cells treated with the lysosome degradation inhibitor Chloroquine, the autophagy inhibitor 3-Methyladenine and the proteasome inhibitor MG132 at the indicated concentration times described in materials and methods. LC3 I/II levels were analyzed as a positive control of chloroquine and 3-MA treatment and β-catenin for MG132 treatment. (e) HCT116 cells were transiently transfected with pcDNA-Flag-Hakai and the day after transfection cells were treated with chloroquine at 50 µM for 24 h. (f) Effect of Hakin-1 inhibitor on FASN expression. Hakin-1 was used at increasing concentrations for 48 h in HCT-116 cells. (g) Effect of Hakin-1 on Hakai-dependent ubiquitination of FASN. Flag-Hakai and HA-ubiquitin were transiently transfected into HCT116 cells. Immunoprecipitation was performed with the anti-FASN antibody before Western blotting using the indicated antibodies. GAPDH was used as protein loading control. The blots images were cropped prior hybridization and original blots are included in Supplementary Information.
Hakai controls lipid accumulation via FASN
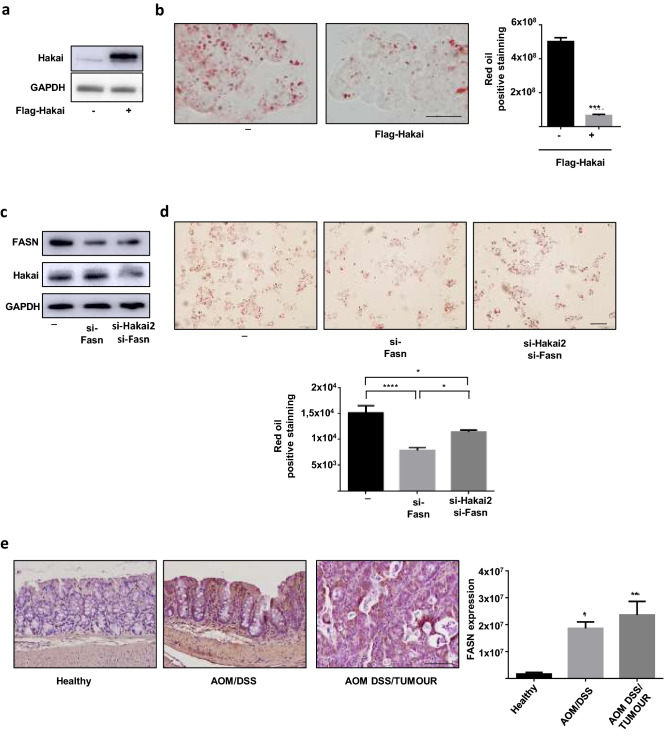
Given the demonstrated role of Hakai controlling FASN protein at posttranslational level, we assessed whether Hakai may have an impact in lipid accumulation. For this purpose, HCT116 cells were first transfected with Flag-Hakai to analyze the effect of its overexpression in lipid accumulation. When Hakai is overexpressed (Fig. 5a), a decrease in lipid accumulation was confirmed by oil red staining (Fig. 5b). Moreover, lower FASN levels decreased lipid accumulation, while the dual knockdown of siRNA-FASN and siRNA-Hakai, did not allow to recover lipid accumulation supporting that Hakai may control lipid accumulation, at least in part, through FASN (Fig. 5c,d). Given the previous results showing that Hakai expression in AOM-DSS model of CAC was decreased in inflammatory conditions compared to tumour tissue of colitis-associated colorectal cancer (AOM DSS/Tumour) and healthy tissues, we extended our results to analyse FASN expression in this mouse model (Fig. 5e). An inverse expression of FASN expression with Hakai expression was detected in inflammatory AOM/DSS compared to tumour tissue of colitis-associated colorectal cancer and healthy tissues (Fig. 5e) further suggesting that Hakai may regulate FASN expression in mouse model of inflammatory bowel disease.
Figure 5.
Hakai controls lipid accumulation via FASN. (a) Hakai expression in HCT116 cells transfected with Flag-Hakai was analyzed by Western blot analysis and (b) the effect on lipid accumulation by staining with Oil Red is shown together with quantification of cell staining (right panel). Scale bar 125 μm. (c) HCT116 cells transfected with siRNA-Hakai2 or siRNA control in presence of absence of siRNA-FASN, analyzed by Western blot analysis and (d) staining with Oil Red together with quantification of cell staining results is. (b and d) Five pictures of each sample were taken and quantified. Values are means ± SD of staining intensity signal scoring per area. Quantification and calibration of the images were performed with Image Analysis Toolbox software for Image J. Kruskal–Wallis with Tukey correction test analyses show statistical differences between overexpression/silencing of Hakai or FASN and controls (*p < 0.05; **p < 0,01; ***p < 0,001; ****P < 0.0001). Scale bar 50 μm. (e) Immunohistochemistry staining (letf panel) and quantification (right panel) of FASN in mouse samples of the AOM/DSS Colitis-associated colorectal cancer model (inflamed epithelium and tumours, healthy mice, n = 7; AOM/DSS-treated mice, n = 14). Immunohistochemistry staining and quantification were performed as indicated in figure legend 1. Scale bar 125 μm. The blots images were cropped prior hybridization and original blots are included in Supplementary Information.
FASN and Hakai expression in human IBD
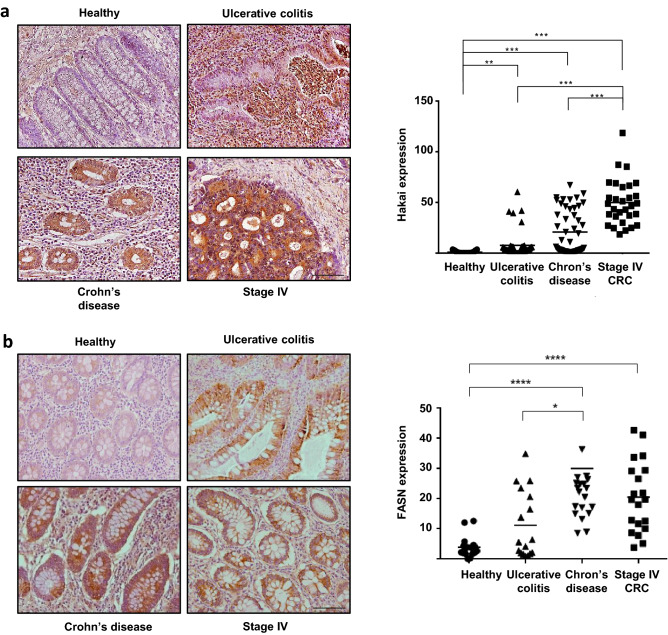
Given that in the mouse model of inflammation-driven colorectal cancer, an inverse expression of Hakai and FASN expression was detected, we decided to analyse the expression of Hakai and FASN in human inflammatory colon disorders, including UC and CD. These two processes are chronic inflammatory conditions of the gastrointestinal tract that increase the risk of developing pre-neoplastic and neoplastic lesions. To compare the expression pattern of Hakai and FASN in the context of intestinal inflammation, human tissue samples from CAC, including UC and CD were analysed by immunohistochemistry. As shown in Fig. 6a, Hakai expression was significantly upregulated in UC and CD compared to normal tissues, and higher expression was detected compared to colorectal adenocarcinoma TNM-stage IV tissues. On the other hand, FASN expression was increased in CD samples compared to healthy tissues, but no significant differences were detected while comparing UC and healthy samples (Fig. 6b). Collectively, these results indicate that Hakai and FASN expression in the IBD mouse models do not mimic in the human IBD.
Figure 6.
Expression of Hakai in human samples of UC, CD and colorectal adenocarcinoma stage IV and its healthy tissue pairs. (a) Immunohistochemistry staining of Hakai (left panel) and quantification (right panel) in healthy colonic mucosa, UC, CD and colorectal adenocarcinoma TNM-stage IV. Images obtained with 20 × objective. Scale bar 125 μm. (b) Staining intensity of FASN (left panel) and quantification (right panel) in healthy colonic mucosa, UC, CD and colorectal adenocarcinoma TNM-stage IV. (Normal colonic mucosa, n = 6, UC, n = 8; CD, n = 10; and colorectal adenocarcinoma, n = 6). Five pictures of each sample were taken and quantified. Data are represented as scatter plot. Values are means ± SEM of staining intensity signal scoring per area. Quantification and calibration of the images were performed with immunohistochemistry Image Analysis Toolbox software for Image J. Kruskal–Wallis with Tukey correction test analyses show statistical differences in colorectal cancer (TNM, SI-IV) respect to paired healthy samples (*p < 0.05; **p < 0,01; ***p < 0,001). Scale bar 125 μm.
Discussion
IBD is a chronic inflammatory disorder in gastrointestinal tract that increases the risk of developing colon cancer39. In the last years, important number of publications highlight the importance of protein ubiquitination in IBD40,41. The E3 ubiquitin-ligase Hakai, highly expressed in colon cancer tissues compared to adjacent normal tissues, plays a critical role during tumor progression42, however, the potential implication in IBD is still unknown. In the current study, we have shown that Hakai expression is downregulated in inflamed intestinal epithelium in different mouse models for IBD (Fig. 1), while higher expression was detected in tumor tissues from AOM-DSS CAC model (Fig. 1a). Furthermore, the Fatty Acid Synthase (FASN) is a novel Hakai-interacting protein identified in the present study. We show that Hakai induces FASN ubiquitination and degradation, thereby resulting in the downregulation of FASN-mediated fatty acid accumulation (Figs. 4, 5). The abnormalities in fatty acid metabolism have been reported in IBD, and are considered as one of the etiologies for the development of this disease43. By immunohistochemistry, we have shown that FASN expression increases in the mucosa in the mice with colitis. This observation agrees with our results and previous reported studies showing high FASN expression in patients with IBD and colorectal neoplasia35,37.
In our in vitro model system, we have demonstrated that Hakai overexpression induces FASN degradation, while Hakai-silencing increases FASN expression and, in consequence, fatty-acid accumulation, which is associated to the development of IBD. In the mouse model, Hakai expression is downregulated in inflamed intestinal epithelium accompanied to an increase expression of FASN. Although, several studies have reported the role of Hakai in different cancers11,44,45, to our knowledge, this is the first study that links Hakai with IBD. Other E3 ubiquitin-ligases were reported to be involved in IBD. For instance, RNF186 controls protein homeostasis in colonic epithelial and regulates intestinal inflammation. Indeed, RNF186 is expressed in colonic epithelial of the mice, and Rnf186−/− mice are reported to lead to an increased risk of intestinal inflammation46. Accordingly, our results open the possibility to elucidate whether, in a similar manner, a decreased expression of Hakai in inflamed intestinal epithelia in mice may increase the risk of IBD. Importantly, in the tumors of the CAC mouse model, a higher expression of FASN and Hakai was detected, further supporting the implication of both proteins in colon cancer. However, in other studies, by gene expression prolife analysis, a decrease of FASN expression was described in patients with UC47. FASN was also described to play a crucial role in maintaining the homeostasis of intestinal barrier function, showing that loss of FASN in mouse intestine can initiate intestinal inflammation48. Intestinal homeostasis depends on multiple factor including the interaction between intestinal epithelium and the host immune system and the microbiota. Several regulatory mechanisms are reported to maintain intestinal homeostasis, and how the alteration of these pathways may precipitate the IBD49. Therefore, further studies are required in order to elucidate in which specific situation Hakai/FASN axis may contribute to intestinal homeostasis or IBD, as well as to reveal additional molecules that may help to orchestrate this regulatory process.
Previous studies have shown an increased ubiquitination mediated proteolysis in colonic biopsy samples from patients with IBD8,9,50. Indeed, other E3 ubiquitin ligases, such as RNF183, are reported to be up-regulated in the inflamed IBD mucosa, including UC and CD. RNF183 induces ubiquitination-mediated degradation of IκBα leading to the activation of NFκB-p65 in intestinal epithelial cells, further suggesting RNF183 role in NF-κB pathway in intestinal inflammation51. Hakai was recently reported to be associated to immune microenvironment regulation of periodontitis, a chronic inflammatory disease occurring in periodontal that involves complex interactions between pathogens and immune reactions52,53. This work demonstrates that Hakai is an important regulator of TNF and cytokine in immune reaction in periodontitis53. Given that TNF-α is a well-known inflammatory mediator that is highly expressed in the inflamed intestines of CD and UC patients54, it opens the possibility to explore whether Hakai may be linked to TNF-α signaling pathway in IBD.
In IBD human biopsies, Hakai expression was significantly upregulated in UC and CD compared to normal tissues, and higher expression was even detected in TNM-stage IV colorectal adenocarcinoma tissues. On the other hand, FASN expression was only increased in CD samples compared to healthy tissues, but no significant differences were detected between IBD and TNM-stage IV colorectal cancer. These results are not according to the ones observed in IBD mice, suggesting that Hakai regulation in mouse models does not accurately mimic human IBD, as observed in UC and CD. Although mouse models have been extensively used to study basic pathophysiological mechanisms, important controversies are reported regarding to how well murine models reflect human inflammatory diseases55,56. Indeed, Seok et at show that the genomic responses to different acute inflammatory stresses are highly similar in humans, but are not reproduced in the mouse models. Several factors can contribute to the differences seen in the molecular response of inflammatory mice and human disease including the evolutional and distance differences in cellular composition between mice and humans, as well as the complexity of the human disease57–60.
Taken together, we found that the E3 ubiquitin-ligase Hakai increases ubiquitination and degradation of FASN, thereby resulting in the regulation of FASN-mediated lipid accumulation, which is associated to the development of IBD. Further studies are needed to deepen into the role of Hakai in IBD in mouse models in order to better understand the regulatory mechanism and pathways that may influence intestinal homeostasis and its breakdown in IBD. Moreover, the role of Hakai/FASN axis in other diseases such as fatty liver disease awaits to be elucidated.
Materials and methods
Human intestinal tissues samples
Human biopsies from patients with UC and CD and colorectal cancer were obtained from the Pathological Anatomy Department from the “Complejo Hospitalario Universitario A Coruña” (CHUAC), under informed consent signed from all patients. Research investigation was approved by the Research Ethics Committee from A Coruña-Ferrol (Ethical Protocol Codes: 2018/257 and 2017/570) following standard ethical procedures of the Spanish regulation (Ley Orgánica de Investigación Biomédica, 14 July 2007) and according to the ethical standards in the 1964 Declaration of Helsinki. Paraffin samples were obtained from CHUAC Biobank integrated in the Spanish Hospital Platform Biobanks Network. Serial 4 µm sections from archived, formalin-fixed and paraffin-embedded intestinal tissue samples from patients (CD, n = 10; UC, n = 8 and colorectal cancer, n = 6) were analysed for Hakai protein expression.
Mice intestinal tissues samples
Three different mouse models of CAC were used to analyze Hakai protein expression. First, a chronic inflammation-associated carcinogenesis mouse model from which biopsies were taken from C57BL/6J mice, in which the development of colorectal cancer associated with inflammation, was promoted through intraperitoneal administration a chemical carcinogen Azoxymethane (AOM) and the inflammatory agent, dextran sodium sulfate (DSS), was given with the drinking water as reported29. Second, a DSS model that mimics the acute colitis, on which only DSS is used in a single cycle of 8 days and concentration (3%). Animals were sacrificed 4 days after the end of the DSS cycle. Finally, an IL10−/− model61, a genetically modified mice resulting in the enterocolitis appearance in presence of intestinal bacteria and imbalance in the function of the intestinal mucosa. All mouse intestinal tissue samples from C57BL/6J and C57BL/6 IL10 KO mice were kindly provided by Prof. Dr. Christoph Gasche (Medical University of Vienna, Austria).
Microarray data mining in NCBI’s gene expression omnibus (GEO)
Data from Hakai expression obtained from a gene-centric and an experiment-centric perspective was obtained from the GEO Database (https://www.ncbi.nlm.nih.gov/sites/GDSbrowser/). All the data sets available were filtered looking for microarrays of models of colitis associated cancer. Exclusion criteria was common for the 3 searches and included mice models that could interfere in the evaluation of the physiological process, principally studies that evaluated the effect of any drug to prevent the development of the inflammatory process. For the AOM/DSS model subgroups were formed according to the exposure time to the AOM/DSS, which was 2 weeks for the inflamed tissue, 4 weeks for those with low dysplasia, between 6 and 8 weeks for high dysplasia and 20 weeks for CRC with the control group only treated with saline. For the other two models, acute colitis and IL-10 KO, criteria for subgroup formation were only case/control. The final studies selected (GDS4367, GSE31106 and GSE107810) were analysed using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/), in order to identify genes differentially expressed across the experimental conditions. The results of the search for Hakai expression in the created subgroups are presented. Parameters for the GEO2R tool were not customized and were used as default.
Antibodies and inhibitors
Hakai antibody (Invitrogen, Carlsbad, CA, USA) was used for western-blotting, and Hakai-2498 antibody used for immunohistochemistry was kindly provided by Dr. Fujita and Hakai antibody (Bethyl, Montgomery, TX, USA) was used for immunoprecipitation12. The rest of antibodies used for western-blotting are FASN antibody (Santa Cruz, Dallas, TX, USA), LC3 A/B antibody (Cell Signaling, Leiden, The Netherlands), E-cadherin antibody (BD Trans Lab, Franklin Lakes, NJ, USA), β-catenin antibody (Cell Signaling, Leiden, The Netherlands), GAPDH antibody (Invitrogene) and mouse and rabbit secondary antibodies (GE Healthcare, Chicago, IL, USA). Proteasome inhibitor MG132 (Sigma-Aldrich, St. Louis, MO, USA) was added for 6 h using 10 µM and 30 µM. Lysosome degradation inhibitor Chloroquine (Sigma-Aldrich, St. Louis, MO, USA), was added for 24 h at 50 µM and for 6 h at 100 µM. Autophagy inhibitor 3-Methyladenine (Sigma-Aldrich, St. Louis, MO, USA) was added for 24 h at 5 mM and 10 mM. Protein synthesis inhibitor cycloheximide (Sigma-Aldrich) was used at 10 μg/mL for the indicated times.
Immunohistochemistry
For immunohistochemistry, slides containing sections of mice and human intestinal tissues (4 µm) were deparaffinised, rehydrated and processed as previously described31. Incubation with primary antibody was carried out in a wet chamber overnight at 4 °C. Hakai dilution was 1:700 and FASN dilution was 1:500. Commercial kit for immunohistochemistry was Dako EnVision + System, Peroxidase (EnVision + System, HRP) purchased to Agilent Technologies, Inc. Nuclei were counterstained with Gill´s haematoxylin and mounted with DePeX. Pictures were taken with an Olympus BX50 microscope. Quantification of HRP signals was performed with ImageJ software by analysing 5 photographs of each sample, and the represented results are shown as mean ± SEM.
Cell culture, transfection and construction of lentivirus and inducible cell lines
HCT 116 cell line was cultured in Dulbecco’s Modified Eagle Medium (DMEM) and HT29 cell line was cultured in McCoy's 5A medium. All media were supplemented with 1% penicillin/streptomycin and 10% of heat-inactivated fetal bovine serum (FBS). All cell lines were grown at 37 °C in a humidified incubator with 5% of CO2. Cells were regularly tested for mycoplasma. Plasmids used for transfection were pcDNA-Flag-Hakai, pBSSR-HA-Ubiquitin and pSG-v-Src which were kindly provided by Dr. Fujita (University College London, UK). Transient transfection for Hakai silencing was performed by employing two different siRNA oligonucleotides for Hakai: Hakai-1 (5′-CTCGATCGGTCAGTCAGGAAA-3′) and Hakai-2 (5′-CACCGCGAACTCAAAGAACTA-3′) or siRNA FASN oligonucleotide (5′-GCUACAUGGCCCAAGGGAA-3′). Transfection experiments were performed by using Lipofectamine 2000 Transfection Reagent and Opti-MEM (Thermo Fisher Scientific) media following manufacturer’s protocol. Universal Non-coding siRNA (Sigma-Aldrich, St. Louis, MO, USA) was used as a negative control of transfection. Lentiviral vector system for Hakai expression (SMARTvector Inducible Lentiviral shCBLL1) by doxycycline inducible sh-RNAs particles were acquired in Dharmacon (Horizon Perkin Elmer Group). Lentiviruses were propagated using previously described methods62. Doxycycline (Sigma) was added to the medium at final concentration of 1 µg/ml, and cells were incubated for 72 h. Two different clones of HT29 colon cancer cells transduced with viral supernatant, containing sh-Hakai (sh-Hak1 and sh-Hak2) or sh-control (sh-), were selected and Hakai knockdown efficiency was monitored by western blot.
Interactome analyses
HCT116 cells were used for interactome analysis. Cell pellets were incubated with lysis buffer (20 mM Tris–HCl pH 7,5, 150 mM NaCl and 1% Triton X-100, 125 mg/mL N-ethylmaleimide) in rotation at 4 °C for 30 min. Pre-clearing was performed to eliminate unspecific interactions. For large scale immunoprecipitation, cells were used per point of the experiment (IgG and IP). 40 μL of Dynabeads protein A (Thermo Fisher Scientific) were resuspended in filtered PBS-T 0,1% and incubated with 5 μg of Hakai Bethyl antibody or control IgG 2 h at 4 °C on rotation for beads-antibody coupling. Then, beads and protein sample were incubated overnight at 4 °C on rotation. Samples were loaded into SDS-PAGE gel and band was stained with Sypro-Ruby (Lonza) fluorescent and processed to in gel digestion as previously described63. Samples were reduced in 10 mM dithiothreitol and dissolved in 50 mM ammonium bicarbonate (AMBIC) (Sigma-Aldrich). Then samples were alkylated with iodoacetamide 55 mM dissolved in AMBIC 50 mM (Sigma-Aldrich). The gel pieces were rinsed with AMBIC 50 mM in 50% methanol (HPLC grade, Scharlau) and acetonitrile (HPLC grade, Scharlau) was added for dehydration. Finally, they were dried in a SpeedVac (Thermo Fisher Scientific) and digested with porcine trypsin (Promega) to a final concentration of 20 ng/µL in AMBIC 20 mM for a final overnight incubation at 37 °C. For mass spectrometric analysis, samples were processed by the Proteomics Platform of Biomedical Research Institute of Santiago de Compostela (IDIS). The separation of peptides was done by Reverse phase chromatography. The 400 micro nanoLC liquid chromatography system (Eksigent Technologies, ABSciex) combined with a Triple Time-of-flight (TOF) 6600 high speed mass spectrometer (ABSciex) were used for creating a gradient. Analysis of peptides was performed on the C18CL reverse phase column (150 × 0.30 mm, 3 µm, 120 Å) (Eksigent, ABSciex). The peptides were separated using a 90-min gradient ranging from 2 to 90% of mobile phase B (mobile phase A: 0.1% formic acid, 2% acetonitrile; mobile phase B: 0.1% formic acid, 100% acetonitrile). Acquisition of data was performed on a Triple TOF 6600 system (ABSciex). The TF 1.7.1 analyst software was used for instrument operation (ABSciex). A switching criterion was used for ions greater than the mass/charge ratio (m/z) 350 and less than m/z 1400, with a mass tolerance of 250 ppm, a charge state of 2–5 and a threshold of abundance of more than 200 accounts (cps). The computer analysis of the raw data obtained was carried out using ProteinPilotTM 5.0.1 software (ABSciex). Analysis was performed with the Significance Analysis of INTeractome (SAINT) score SAINTexpress64. Preys with SAINT probability score cut-off of 1 detected by at least two exclusive spectral counts were deemed high confidence Hakai-interacting proteins.
Western blot analysis, real-time quantitative PCR (RT-qPCR) and immunofluorescence staining
For western blot analysis, whole cell extracts were obtained as described previously31. For RT-qPCR, total RNA was extracted using TriPure isolation reagent (Roche,Basel, Switzerland). mRNA levels were analysed in technical triplicates by quantitative RT-PCR, following specifications of reverse retrotranscriptase kit (NZYTech, Lisbon, Portugal). Amplification was performed in a Light Cycler 480 (Roche, Basel, Switzerland) and data was analysed by qBase + analysis software (Biogazelle, Zwijnaarde, Belgium). Primers used for FASN were F′ TTCTACGGCTCCACGCTCTTCC and R′ GAAGAGTCTTCGTCAGCCAGGA. As housekeeping control HPRT primers F′-TGACCTTGATTTATTTTGCATACC and R′-CGAGCAAGACGTTCAGTCCT or RPL13A F′-CTCAAGGTGTTTGACGGCATCC and R′-TACTTCCAGCCAACCTCGTGAG were used. Immunofluorescence was performed as described31. Briefly, cells were fixed with PFA 4% for 15 min and permeabilized with 0.5% Triton X-100/PBS for 15 min. Primary antibodies were incubated for 2 h, at RT and secondary antibody was incubated at RT for 1 h. Nuclear staining was performed using 1:10,000 Hoechst dilution (Life Technologies, Carlsbad, CA, USA). ProLong Gold Antifade Mountant (Life Technologies, Carlsbad, CA, USA) was employed for coverslips mounting. Images were obtained by using fluorescence microscope “Monitorized reflected Fluorescence System” (Olympus).
Immunoprecipitation and ubiquitination assays
For immunoprecipitation experiments, cells were lysed for 20 min in 1 ml of 1% Triton X-100 lysis buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl and 1% Triton X-100) containing 10 μg/ml leupeptin, 10 μg/ml aprotinin and 1 mM phenylmethanesulphonyl fluoride (PMSF). Supernatants were immunoprecipitated for 2 h with 2 μg of anti-Hakai antibody or anti-HA bound to protein G PLUS-Agarose beads (Santa Cruz Biotechnology, USA), followed by SDS–polyacrylamide gel electrophoresis (PAGE) and western blotting with the indicated antibodies as previously reported. For ubiquitination assays, HCT116 cells were transfected with 0.25 µg Src, 0.75 µg Flag-Hakai, and 0.5 µg HA-ubiquitin using Lipofectamin 2000 (Invitrogen, UK). Cell extracts were obtained in the above lysis buffer used for immunoprecipitation, supplemented with 10 mM N-ethylmaleimide.
Oil-red staining assay
HCT116 and HT29 cells (105) were seeded in 8-well chambers (Millicell EZ SLIDE 8-well glass, Millipore) were washed with 1 × PBS and stained with a freshly prepared Oil Red O (ORO) working solution as described previously33. Staining was observed using light microscope (Olympus BX61). Positively-stained areas were used to quantify lipid accumulation using ImageJ software by analysing 5 photographs. The represented results are shown as mean ± SEM.
Statistical analysis
Shapiro–Wilk test was used to check a normal distribution and Levene test to assess the equality of variances. Statistical significance of data was determined by applying a two-tailed Student t-test or ANOVA depending on the data. Results obtained are expressed as mean ± SD or mean ± SEM. Quantification of human IHQ did not follow a normal distribution therefore we used Kruskal–Wallis with Tukey correction test. Significant differences among the experimental groups indicated in the figures is shown as *P < 0.05, **P < 0.01 ***P < 0.001 and ****P < 0.0001.
Supplementary Information
Acknowledgements
This work was supported by Plan Estatal I + D + I 2013−2016, co-funded by the Instituto de Salud Carlos III (ISCIII, Spain) under grant agreements PI18/00121 and PI21/00238 and by Fondo Europeo de Desarrollo Regional (FEDER) “A way of Making Europe”. The project that gave rise to these results has received funding from ”la Caixa” Foundation and the European Institute of Innovation and Technology, EIT (body of the European Union that receives support from the European Union’s Horizon 2020 research and innovation program), un-der the grant agreements LCF/TR/CI19/52460016 and LCF/TR/CC21/52490003. Also supported by Consolidation of Competitive Research (IN607B2020/14) from GAIN from Xunta de Galicia. DRL was supported by a post-specialization fellowship from Fundación Profesor Novoa Santos and a public grant from Deputación de A Coruña (Spain), ADD by FPU contract (FPU014/02837) from Ministerio de Educación Cultura y Deporte (Spain), MQ was supported by supported by Consolidation of Competitive Research (IN607B2020/14) from GAIN from Xunta de Galicia and AR was supported by a predoctoral contract (PRDLC21591RODR) from Fundación Científica AECC/AECC.
Abbreviations
- CAC
Colitis-associated cancer
- CD
Crohn’s disease
- IBD
Inflammatory bowel diseases
- UC
Ulcerative colitis
Author contributions
Designed the study D.R.L. and A.F., performed the experiments: D.R.L., M.Q., A.D.D., A.R.A. A.B.A. and M.-E.C. contributed to the interactome analysis. A.C. and G.R. contributed to the human I.H.Q. analysis. C.G. and V.K. hosted and supervised D.R.L. at Medical University of Vienna, and reviewed manuscript. Drafted the article: D.R.L. and A.F. All authors read and approved the final version of the manuscript.
Data availability
The datasets generated and/or analysed during the current study are available in the ProteomeXchange repository, accession number PXD029136. Full blots are included in Supplemental Figure file.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Daniel Roca-Lema and Macarena Quiroga.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-22295-w.
References
- 1.Lasry A, Zinger A, Ben-Neriah Y. Inflammatory networks underlying colorectal cancer. Nat. Immunol. 2016;17:230–240. doi: 10.1038/ni.3384. [DOI] [PubMed] [Google Scholar]
- 2.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 3.Rape M. Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 2018;19:59–70. doi: 10.1038/nrm.2017.83. [DOI] [PubMed] [Google Scholar]
- 4.Weissman AM, Shabek N, Ciechanover A. The predator becomes the prey: Regulating the ubiquitin system by ubiquitylation and degradation. Nat. Rev. Mol. Cell Biol. 2011;12:605–620. doi: 10.1038/nrm3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hershko A, Ciechanover A, Varshavsky A. Basic medical research award. The ubiquitin system. Nat. Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 6.Livneh I, Cohen-Kaplan V, Cohen-Rosenzweig C, Avni N, Ciechanover A. The life cycle of the 26S proteasome: From birth, through regulation and function, and onto its death. Cell Res. 2016;26:869–885. doi: 10.1038/cr.2016.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Metzger MB, Hristova VA, Weissman AM. HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 2012;125:531–537. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao Y, Huang Q, Wu Z, Chen W. Roles of protein ubiquitination in inflammatory bowel disease. Immunobiology. 2020;225:152026. doi: 10.1016/j.imbio.2020.152026. [DOI] [PubMed] [Google Scholar]
- 9.Inoue S, et al. The effect of proteasome inhibitor MG132 on experimental inflammatory bowel disease. Clin. Exp. Immunol. 2009;156:172–182. doi: 10.1111/j.1365-2249.2008.03872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng L, Meng T, Chen L, Wei W, Wang P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct Target Ther. 2020;5:11. doi: 10.1038/s41392-020-0107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aparicio LA, Valladares M, Blanco M, Alonso G, Figueroa A. Biological influence of Hakai in cancer: A 10-year review. Cancer Metastasis Rev. 2012;31:375–386. doi: 10.1007/s10555-012-9348-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita Y, et al. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat. Cell Biol. 2002;4:222–231. doi: 10.1038/ncb758. [DOI] [PubMed] [Google Scholar]
- 13.Schneider MR, Kolligs FT. E-cadherin's role in development, tissue homeostasis and disease: Insights from mouse models: Tissue-specific inactivation of the adhesion protein E-cadherin in mice reveals its functions in health and disease. BioEssays. 2015;37:294–304. doi: 10.1002/bies.201400141. [DOI] [PubMed] [Google Scholar]
- 14.Bondow BJ, Faber ML, Wojta KJ, Walker EM, Battle MA. E-cadherin is required for intestinal morphogenesis in the mouse. Dev. Biol. 2012;371:1–12. doi: 10.1016/j.ydbio.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J, et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020 doi: 10.1038/s41580-020-0237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat. Rev. Cancer. 2018;18:128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 17.Aparicio LA, et al. Clinical implications of epithelial cell plasticity in cancer progression. Cancer Lett. 2015;366:1–10. doi: 10.1016/j.canlet.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Jankowski JA, et al. Alterations in classical cadherins associated with progression in ulcerative and Crohn's colitis. Lab Investig. 1998;78:1155–1167. [PubMed] [Google Scholar]
- 19.Gassler N, et al. Inflammatory bowel disease is associated with changes of enterocytic junctions. Am. J. Physiol. Gastrointest. Liver Physiol. 2001;281:G216–228. doi: 10.1152/ajpgi.2001.281.1.G216. [DOI] [PubMed] [Google Scholar]
- 20.Brighenti E, et al. Interleukin 6 downregulates p53 expression and activity by stimulating ribosome biogenesis: A new pathway connecting inflammation to cancer. Oncogene. 2014;33:4396–4406. doi: 10.1038/onc.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett JC, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat. Genet. 2009;41:1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houlston RS, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat. Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grill JI, Neumann J, Hiltwein F, Kolligs FT, Schneider MR. Intestinal E-cadherin deficiency aggravates dextran sodium sulfate-induced colitis. Dig. Dis. Sci. 2015;60:895–902. doi: 10.1007/s10620-015-3551-x. [DOI] [PubMed] [Google Scholar]
- 24.Figueroa A, et al. Novel roles of Hakai in cell proliferation and oncogenesis. Mol. Biol. Cell. 2009;20:3533–3542. doi: 10.1091/mbc.E08-08-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez-Rigueiro T, Valladares-Ayerbes M, Haz-Conde M, Aparicio LA, Figueroa A. Hakai reduces cell-substratum adhesion and increases epithelial cell invasion. BMC Cancer. 2011 doi: 10.1186/1471-2407-11-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castosa R, et al. Hakai overexpression effectively induces tumour progression and metastasis in vivo. Sci. Rep. 2018;8:3466. doi: 10.1038/s41598-018-21808-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abella V, et al. miR-203 regulates cell proliferation through its influence on Hakai expression. PLoS ONE. 2012;7:e52568. doi: 10.1371/journal.pone.0052568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-Iglesias O, et al. Hakin-1, a new specific small-molecule inhibitor for the E3 ubiquitin-ligase Hakai, inhibits carcinoma growth and progression. Cancers. 2020 doi: 10.3390/cancers12051340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dammann K, et al. PAK1 promotes intestinal tumor initiation. Cancer Prev. Res. 2015;8:1093–1101. doi: 10.1158/1940-6207.CAPR-15-0205-T. [DOI] [PubMed] [Google Scholar]
- 30.Sellon RK, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect. Immun. 1998;66:5224–5231. doi: 10.1128/IAI.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Díaz-Díaz A, et al. Heat shock protein 90 chaperone regulates the E3 ubiquitin-ligase Hakai protein stability. Cancers. 2020 doi: 10.3390/cancers12010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franceschini A, et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gang X, et al. Speckle-type POZ protein suppresses lipid accumulation and prostate cancer growth by stabilizing fatty acid synthase. Prostate. 2019;79:864–871. doi: 10.1002/pros.23793. [DOI] [PubMed] [Google Scholar]
- 34.Matsuo S, Yang WL, Aziz M, Kameoka S, Wang P. Fatty acid synthase inhibitor C75 ameliorates experimental colitis. Mol. Med. 2014;20:1–9. doi: 10.2119/molmed.2013.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Consolazio A, et al. Overexpression of fatty acid synthase in ulcerative colitis. Am. J. Clin. Pathol. 2006;126:113–118. doi: 10.1309/pubvqndnvqkjvc8m. [DOI] [PubMed] [Google Scholar]
- 36.Röhrig F, Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer. 2016;16:732–749. doi: 10.1038/nrc.2016.89. [DOI] [PubMed] [Google Scholar]
- 37.Rashid A, et al. Elevated expression of fatty acid synthase and fatty acid synthetic activity in colorectal neoplasia. Am. J. Pathol. 1997;150:201–208. [PMC free article] [PubMed] [Google Scholar]
- 38.Onorati AV, Dyczynski M, Ojha R, Amaravadi RK. Targeting autophagy in cancer. Cancer. 2018;124:3307–3318. doi: 10.1002/cncr.31335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yalchin M, Baker AM, Graham TA, Hart A. Predicting colorectal cancer occurrence in IBD. Cancers. 2021 doi: 10.3390/cancers13122908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cleynen I, et al. Genetic and microbial factors modulating the ubiquitin proteasome system in inflammatory bowel disease. Gut. 2014;63:1265–1274. doi: 10.1136/gutjnl-2012-303205. [DOI] [PubMed] [Google Scholar]
- 41.Ardley HC. Ring finger ubiquitin protein ligases and their implication to the pathogenesis of human diseases. Curr. Pharm. Des. 2009;15:3697–3715. doi: 10.2174/138161209789271807. [DOI] [PubMed] [Google Scholar]
- 42.Rodríguez-Alonso A, et al. Regulation of epithelial-mesenchymal plasticity by the E3 ubiquitin-ligases in cancer. Cancers. 2020 doi: 10.3390/cancers12113093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shores DR, Binion DG, Freeman BA, Baker PR. New insights into the role of fatty acids in the pathogenesis and resolution of inflammatory bowel disease. Inflamm. Bowel Dis. 2011;17:2192–2204. doi: 10.1002/ibd.21560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weng CH, et al. Epithelial-mesenchymal transition (EMT) beyond EGFR mutations per se is a common mechanism for acquired resistance to EGFR TKI. Oncogene. 2019;38:455–468. doi: 10.1038/s41388-018-0454-2. [DOI] [PubMed] [Google Scholar]
- 45.Liu M, et al. Ajuba inhibits hepatocellular carcinoma cell growth via targeting of β-catenin and YAP signaling and is regulated by E3 ligase Hakai through neddylation. J. Exp. Clin. Cancer Res. 2018;37:165. doi: 10.1186/s13046-018-0806-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujimoto K, et al. Regulation of intestinal homeostasis by the ulcerative colitis-associated gene RNF186. Mucosal Immunol. 2017;10:446–459. doi: 10.1038/mi.2016.58. [DOI] [PubMed] [Google Scholar]
- 47.Heimerl S, et al. Alterations in intestinal fatty acid metabolism in inflammatory bowel disease. Biochim. Biophys. Acta. 2006;1762:341–350. doi: 10.1016/j.bbadis.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 48.Wei X, et al. Fatty acid synthase modulates intestinal barrier function through palmitoylation of mucin 2. Cell Host Microbe. 2012;11:140–152. doi: 10.1016/j.chom.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 50.Cenac N, et al. Role for protease activity in visceral pain in irritable bowel syndrome. J. Clin. Investig. 2007;117:636–647. doi: 10.1172/JCI29255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu Q, et al. E3 ubiquitin ligase RNF183 is a novel regulator in inflammatory bowel disease. J. Crohns Colitis. 2016;10:713–725. doi: 10.1093/ecco-jcc/jjw023. [DOI] [PubMed] [Google Scholar]
- 52.Slots J. Periodontitis: Facts, fallacies and the future. Periodontol. 2017;2000(75):7–23. doi: 10.1111/prd.12221. [DOI] [PubMed] [Google Scholar]
- 53.Zhang X, et al. m6A regulator-mediated RNA methylation modification patterns are involved in immune microenvironment regulation of periodontitis. J. Cell Mol. Med. 2021;25:3634–3645. doi: 10.1111/jcmm.16469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Breese EJ, et al. Tumor necrosis factor alpha-producing cells in the intestinal mucosa of children with inflammatory bowel disease. Gastroenterology. 1994;106:1455–1466. doi: 10.1016/0016-5085(94)90398-0. [DOI] [PubMed] [Google Scholar]
- 55.Seok J, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takao K, Miyakawa T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA. 2015;112:1167–1172. doi: 10.1073/pnas.1401965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis MM. A prescription for human immunology. Immunity. 2008;29:835–838. doi: 10.1016/j.immuni.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hayday AC, Peakman M. The habitual, diverse and surmountable obstacles to human immunology research. Nat. Immunol. 2008;9:575–580. doi: 10.1038/ni0608-575. [DOI] [PubMed] [Google Scholar]
- 59.Mestas J, Hughes CC. Of mice and not men: Differences between mouse and human immunology. J. Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 60.Cabrera O, et al. The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. Proc. Natl. Acad. Sci. USA. 2006;103:2334–2339. doi: 10.1073/pnas.0510790103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frick A, et al. Overt increase of oxidative stress and DNA damage in murine and human colitis and colitis-associated neoplasia. Mol. Cancer Res. 2018;16:634–642. doi: 10.1158/1541-7786.MCR-17-0451. [DOI] [PubMed] [Google Scholar]
- 62.Naldini L, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 63.Shevchenko A, et al. Linking genome and proteome by mass spectrometry: Large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. USA. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Teo G, et al. SAINTexpress: Improvements and additional features in Significance Analysis of INTeractome software. J. Proteomics. 2014;100:37–43. doi: 10.1016/j.jprot.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the ProteomeXchange repository, accession number PXD029136. Full blots are included in Supplemental Figure file.