Abstract
Purpose:
Propagation of Ewing sarcoma (EwS) requires precise regulation of EWS::FLI1 transcriptional activity. Determining the mechanisms of fusion regulation will advance our understanding of tumor progression. Here we investigated whether HOXD13, a developmental transcription factor that promotes EwS metastatic phenotypes, influences EWS::FLI1 transcriptional activity.
Experimental Design:
Existing tumor and cell line datasets were used to define EWS::FLI1 binding sites and transcriptional targets. Chromatin immunoprecipitation and CRISPR-interference were employed to identify enhancers. CUT&RUN and RNA-seq defined binding sites and transcriptional targets of HOXD13. Transcriptional states were investigated using bulk and single-cell transcriptomic data from cell lines, patient-derived xenografts, and patient tumors. Mesenchymal phenotypes were assessed by geneset enrichment, flow cytometry, and migration assays.
Results:
We found that EWS::FLI1 creates a de novo GGAA microsatellite enhancer in a developmentally conserved regulatory region of the HOXD locus. Knockdown of HOXD13 led to widespread changes in expression of developmental gene programs and EWS::FLI1 targets. HOXD13 binding was enriched at established EWS::FLI1 binding sites where it influenced expression of EWS::FLI1-activated genes. More strikingly, HOXD13 bound and activated EWS::FLI1-repressed genes, leading to adoption of mesenchymal and migratory cell states that are normally suppressed by the fusion. Single-cell analysis confirmed that direct transcriptional antagonism between HOXD13-mediated gene activation and EWS::FLI1-dependent gene repression defines the state of EwS cells along a mesenchymal axis.
Conclusions:
EwS tumors are comprised of tumor cells that exist along a mesenchymal transcriptional continuum. The identity of cells along this continuum is, in large part, determined by the competing activities of EWS::FLI1 and HOXD13.
Keywords: EWS::FLI1, HOXD13, epigenetics, development, enhancer reprogramming, Ewing sarcoma, neuro-mesenchymal, cell state, CUT&RUN, CITE-seq
Statement of Translational Relevance
The capacity of cancer cells to undergo phenotypic state transitions is emerging as a key requirement for tumor progression. Ewing sarcomas (EwS) are highly metastatic tumors that are driven by EWS::FLI1 fusion oncogenes. Paradoxically, high transcriptional activity of EWS::FLI1 inhibits metastasis-promoting mesenchymal gene programs and emergence of metastatic EwS cell states requires moderation of fusion gene activity. We have identified that HOXD13 serves as a rheostat for EWS::FLI1 activity. HOXD13 antagonizes EWS::FLI1 function in individual EwS cells, resulting in derepression of mesenchymal gene programs and induction of migratory cell states. These studies suggest that individual EwS cells possess inherently different metastatic capacities, and that metastasis may be driven by transcriptionally and phenotypically distinct cell subpopulations.
Introduction
Ewing sarcomas (EwS) are highly malignant bone and soft tissue tumors that peak in adolescence (1). They are lethal cancers for over a third of diagnosed patients and for nearly all who develop metastatic disease (1). The tumors are undifferentiated histologically with features of both mesenchymal and neural lineages and are of presumed mesenchymal and/or neural crest stem cell origin. The genetic drivers of EwS arise from chromosomal translocations between a FET family member (FUS/EWSR1/TAF15) and an ETS family transcription factor, most often EWS::FLI1 (1). EWS::ETS proteins largely induce malignant transformation via chromatin remodeling (2). Functioning as a pioneer factor, EWS::FLI1 creates de novo enhancers at GGAA microsatellites throughout the genome, resulting in aberrant activation of heterochromatic regions and widespread transcriptional rewiring (2-6). In addition to its role in transcriptional activation, EWS::ETS fusions also lead to gene repression through incompletely understood mechanisms (2, 7, 8). Importantly, recent studies have suggested that successful propagation and metastatic progression of EwS depends on precise regulation of EWS::FLI1 transcriptional activity. Whereas high levels of fusion activity support a more proliferative and neural tumor phenotype, EwS cells with lower levels of fusion expression or activity display mesenchymal features and have a higher capacity for migration and metastasis (9-15). Elucidation of the mechanisms that control fusion gene expression and activity is critical to advancing knowledge regarding the processes of EWS::FLI1-induced transformation and metastatic progression.
Although EwS harbor few additional mutations (16, 17), they express a unique homeobox (HOX) gene profile that is distinct from other tumors and tissues (18-20). HOX transcription factors are critical for normal embryogenesis and their dysregulation can contribute to malignant transformation, as best exemplified by hijacking of HOXA9 in leukemogenesis (21). In EwS, posterior HOXD genes (HOXD10, HOXD11, HOXD13) are highly expressed and HOXD13 contributes to tumorigenic and metastatic phenotypes (19, 20, 22). In normal development, HOXD13 contributes to limb development and transcriptional regulation of mesenchymal gene programs (23). Germline mutations in human HOXD13 are associated with polydactyly and other defects in distal limb development (24). The molecular mechanisms underlying HOXD13 activation and tumorigenic function in EwS have yet to be elucidated.
In the current work we show that HOXD13 is a direct epigenetic target of EWS::FLI1. In addition, our studies demonstrate that HOXD13 serves as a rheostat for EWS::FLI1 transcriptional activity wherein HOXD13 moderates expression of fusion-dependent target genes, both cooperating with and competing with EWS::FLI1 at cis-regulatory regions of EWS::FLI1-modulated genes. Most strikingly, we find that HOXD13 directly induces mesenchymal gene programs that are normally repressed by EWS::FLI1 and that are implicated in driving metastatic cancer phenotypes. This transcription factor tug-of-war is evident at the level of individual tumor cells and the relative activities of HOXD13 and EWS::FLI1 determine transcriptional and phenotypic EwS cell states along a mesenchymal axis. Together these studies suggest that individual EwS cells may possess inherently different metastatic capacities and that metastatic progression of EwS tumors may be driven by transcriptionally and phenotypically distinct cell subpopulations.
Materials & Methods
Cell Culture
EwS cell lines were obtained and cultured as previously described (25). H7-MSCs (kind gift of Dr. Sweet-Cordero) (26) were maintained in alpha-mem supplemented with 10% FBS, 2mmol/L-glutamine, and 1% Antibiotic-Antimycotic. Cells were cultured at 37°C with 5% CO2. Cells were all confirmed to be mycoplasma free and identities subject to STR-confirmation every 6 months.
Lentivirus Production & Genetic Modification
Virus production and transductions carried out as previously described (25). pLKO.1 shNS and the shFLI1 were used for FLI1 knockdown experiments (Sigma, St. Louis, MO, USA). For HOXD13 knockdown, stable cells lines were created with doxycycline-inducible hairpins: pTripz shNS, shHOXD13 #1 or shHOXD13 #2. To induce the shRNA, 0.5 ug/mL doxycycline was added. EWS::FLI1 overexpression was achieved with pCLS-EGFP empty and EWS::FLI1-V5-2A-EGFP or pLIV empty and EWS::FLI1 (2).
Quantitative PCR
Total RNA extraction, cDNA generation, and qPCR was performed as previously described (25). Primer and probe sequences are in Supplementary Table S1.
Western Blot
Whole cell protein extraction, protein quantification, and western blot analysis was performed as previously described (25). Antibodies and the dilutions are in Supplementary Table S1. Membranes were imaged on the LiCor Odyssey imaging system.
HOXD13 antibody production
Polyclonal anti-HOXD13 antibody was produced through peptide immunization in rabbits (YenZym, Brisbane, CA). The HOXD13 immunizing peptide described previously (23), was modified to contain a 16-amino-acid region with a Cysteine residue added to the N-terminus (C+VGLQQNALKSSPHASL) to facilitate coupling to a carrier protein.
Immunofluorescence
Frozen sections of E13.5 Hoxd13 WT, heterozygous, and knockout embryos (27) were formalin fixed, frozen in OCT and sectioned. Slides were thawed, washed, blocked, and incubated with the HOXD13 primary antibody in blocking buffer overnight (4C). Slides were incubated in donkey anti-rabbit 488 secondary for 1 hour (RT) followed by DAPI addition. Images were taken using an inverted Olympus IX83 microscope with the CellSens Dimensions software. For immunocytochemistry, cells were fixed in 4% paraformaldehyde and permeabilized with 0.5% Triton. Cells were blocked for 1 hour and incubated either HOXD13 or IgG for 1 hour. Following 3 washes, a fluorescent secondary antibody was added and incubated for 1 hour, followed by DAPI (1:10 000) incubation. Images were taken on a Lecia DMi8 microscope using the Lecia software.
Migration assays
XCELLigence real-time cell analysis (RTCA) of cell migration was performed. 50 uL complete media was placed in the upper chambers and 160 uL serum-free media was added to the lower chambers before 1hr equilibration at 37C. 5 x 104 cells/well were plated in the upper chamber (100 uL) and plates were equilibrated for 30 minutes at room temperature. Migration was evaluated up to 30 hours.
Mapping mouse enhancers to human
To map mouse enhancer sites in the regulatory HOXD domain the UCSC LIFTOVER tool was used to go from mm9 to hg19.
Chromatin immunoprecipitation qPCR
Chromatin immunoprecipitation and qPCR was performed using the Zymo-Spin ChIP kit (Zymo, D5209) as previously described (25). Antibodies and primer sequences are listed in Supplementary Table S1.
CRISPRi two vector system
Cells transduced with UCOE-SFFV-KRAB-dCas9-P2A-mcherry were FACS sorted twice on mCherry expression. sgRNAs were designed flanking GGAA repeat sites using the Broad institutes GPP sgRNA designer. sgRNAs with high on-target scores were chosen and cloned into the sgOpti vector. Lentiviral sgRNA-containing sgOpti vectors were transduced into stably expressing dCas9-KRAB-mcherry cells and puromycin selected before collection after 8 days. Detailed protocol in Supplementary methods.
Flow cytometry
Cells were washed with PBS + 2% FBS, followed by incubation with antibodies for 30 minutes at 4C in the dark. Cells were washed and analyzed on a BD Accuri C6 machine. 10,000 events were collected, and live cells were gated on the unstained SSC vs FSC. To determine the percentage of positive cells, the isotype IgG control was used to set negative gates. Analyses were performed using FCS express (De Novo Software).
Bru-seq/RNA-seq and analysis
Bru-seq was performed on A673 and CHLA10 cells following HOXD13 knockdown as described previously (28). Poly(A)-capture RNA-seq was performed for TC32 HOXD13 knockdown cells. Libraries were prepared with NEBNext Ultra II RNA Library Prep kit and paired end sequencing was performed on a Novaseq600. Reads were analyzed for quality control, trimmed, aligned to GRCh38, and analyzed for differential analysis (FastQC 0.1.9, Trim Galore, STAR, and DESeq2 v1.18.1. Overrepresentation analysis was performed using the Broad Institute’s Molecular Signatures Database (MSigDB). Heatmaps and volcano plots were made in R using pheatmap and EnhancedVolcano.
Automated CUT&RUN Sequencing and analysis
Automated CUT&RUN protocol was performed as described (29) at the Fred Hutchinson Genomics core. Antibodies listed in Supplementary Table S1. FastQC 0.1.9 was used to examine read quality and paired-end reads were aligned to hg38 using Bowtie2.3.5. Narrow and broad peaks were called and filtered for histone marks using MACS2.2.7. For HOXD13 peaks, both overlapping filtered MACS2 peaks and SEACR peaks (stringent mode) were used. BEDTools 2.30.0 was used to identify overlapping peaks between marks. Peak annotation and motif analysis was performed with HOMER 4.11. The GeneOverlap and ChIPpeakAnno packages were used to calculate gene and genomic site overlap. Number of GGAA/CCTT sites were calculated within 250bp up and downstream the bound peaks. Detailed protocol in Supplementary methods.
Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-seq) processing and analysis
CITE-seq was performed as described (30). 500,000 CHLA10 and A673 cells were resuspended in 25 uL Biolegend staining buffer (420201, San Diego, CA). 2.5 uL human TruStain FcX (422301, San Diego, CA) was added per sample (4C,10min). Hash-Tag Antibodies were incubated for 30 minutes (4C). Samples were pooled 1:1 at 1,000 cells/uL and libraries were generated using the 3’ V3 10X Genomics Chromium Controller (CG000183, Pleasanton, CA). Final library quality was assessed using the Tapestation 4200 (Agilent, Santa Clara, CA) and libraries were quantified by Kapa qPCR (Roche). Pooled libraries were subjected to paired-end sequencing on a NovaSeq 6000 (Illumina). Bcl2fastq2 Conversion Software (Illumina) was used to generate de-multiplexed Fastq files and the CellRanger (3.1) Pipeline (10X Genomics) was used to align reads and generate count matrices. Analysis performed using Seurat and Monocle 3.
DbGAP bulk patient RNA-seq Data analysis
Fastq files (NCBI BioProject PRJNA253759 and PRJNA261990) were downloaded using SRA tools and aligned to the hg38 genome with STAR. Output files were merged into a Summarized Experiment object. RNA counts were then normalized using DESeq and made into a cell_data_set. Using Monocle3, the top 50000 variable genes were selected and used for dimensionality reduction (num_dim = 30). Log sum gene set scores were calculated as previously described (31). The datasets were accessed through the NIH database for Genotypes and Phenotypes (dbGaP) (accession # phs000768.v1.p1).
Statistical analysis
Data were analyzed using GraphPad Prism software version 9.0 (San Diego, CA, USA). All statistical analyses were performed with Student’s t-test / One-way ANOVA followed by Tukey multiple comparison test / or Two-way ANOVA followed by Sidak’s multiple comparison test. Data are expressed as means and SEM from at least three independent experiments. Asterisk denoting p<0.05 (*) or p<0.01 (**).
Data and Code Availability
Public data used in this study reported in Supplementary Table S1. Sequencing data generated in this study have been deposited at GEO (GSE182513). Code available at https://github.com/LawlorLab/HOXD13-Paper.
Results
HOXD13 expression in EwS is dependent on EWS::FLI1.
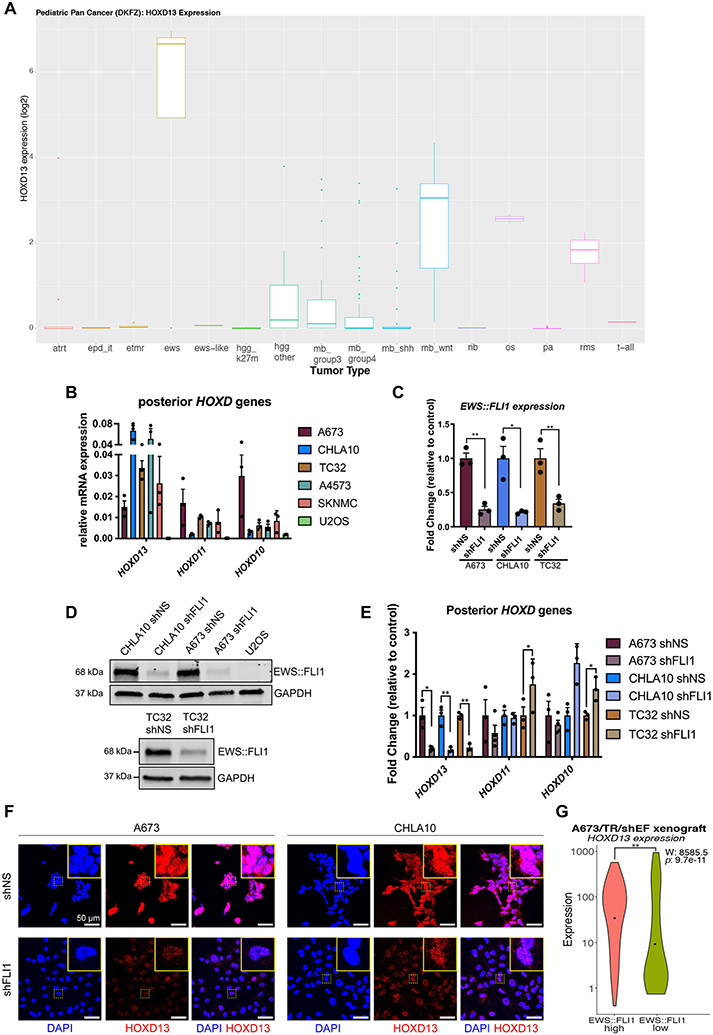
Both cell line encyclopedia (32) and primary pediatric tumor databases (33, 34) show that high HOXD13 expression is a unique and reproducible feature of EwS relative to other cancers (Fig. 1A & B; Supplementary Fig. S1A). Given this, we hypothesized that HOXD13 may be an EWS::FLI1 target and we knocked down EWS::FLI1 in EwS cell lines (Fig. 1C-D, Supplementary Fig. S1B). As shown, loss of the fusion led to downregulation of HOXD13 with no change in either HOXD11 or HOXD10 (Fig. 1E). Loss of HOXD13 protein was confirmed by immunocytochemistry (Fig. 1F and Supplementary Fig. S1G) after authenticating this custom antibody (Supplementary Fig. S1C-H). To test EWS::FLI1-dependent regulation of HOXD13 in an orthogonal system, we interrogated published single-cell data generated from control and EWS::FLI1 knockdown xenografts (35). Consistent with in vitro studies, HOXD13 expression was reduced upon loss of EWS::FLI1 (Fig. 1G). Thus, HOXD13 expression in EwS is, at least partially, dependent on EWS::FLI1.
Figure 1. HOXD13 expression in EwS is dependent on EWS::FLI1.
A) HOXD13 expression across different pediatric cancers (33, 34). B) qRT-PCR of Posterior HOXD genes in Ewing sarcoma cell lines and U2OS (osteosarcoma) cells. C) qRT-PCR and D) western blot of EWS::FLI1 96 hrs after knockdown. E) qRT-PCR of HOXD13, HOXD11, and HOXD10 expression in control and EWS::FLI1 knockdown cells. F) Fluorescent Immunocytochemistry of HOXD13 (Alexa647) in EWS::FLI1 knockdown cells. Nuclear counterstain was performed with DAPI. Scale bar is 50 um. G) Single-cell gene expression profiles from A673/TR/shEF xenografts (35) quantified by violin plots (Wilcoxon rank sum test). Error bars for qRT-PCR studies are representative of SEM from three independent replicates. Expression levels were determined relative to two housekeeping genes and fold changes expressed relative to control condition. * p<0.05; ** p<0.01; Two-way ANOVA; Sidak’s multiple comparison test; Two-tailed t-test. Atrt: atypical teratoid rhabdoid tumor, epd_it:Ependymoma infratentorial, etmr:embryonal tumor with multilayered rosettes, ews: Ewing sarcoma, ews-like: ewing-like, hgg_k27m: high-grade glioma K27M, hgg_other: high-grade glioma K27WT, mb_group3: medulloblastoma group 3, mb_group4: medulloblastoma group 4, mb_shh: medulloblastoma sonic hedgehog, mb_wnt: medulloblastoma wnt, nb: neuroblastoma, os: osteosarcoma, pa: pilocytic astrocytoma, rms: rhabdomyosarcoma, t-all:T-cell acute lymphoblastic leukemia
EWS::FLI1 creates and activates a de novo HOXD13 enhancer in the developmentally conserved TAD.
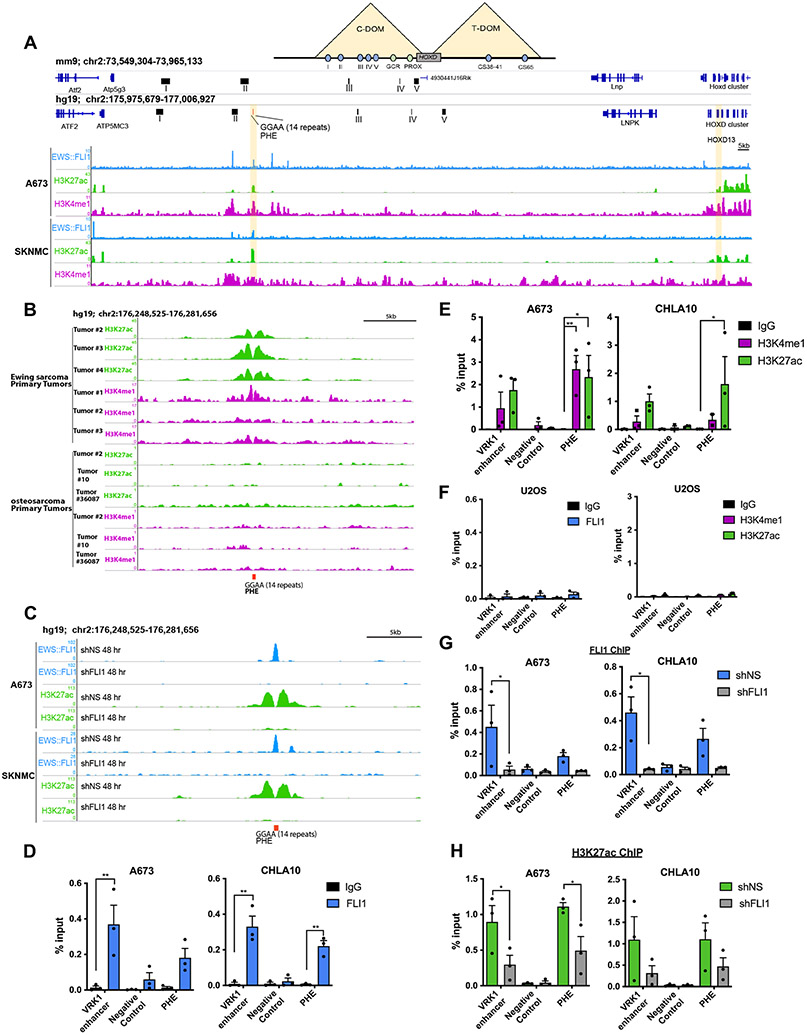
During embryogenesis, expression of HOX genes is tightly orchestrated by epigenetic mechanisms and the HOXD locus is specifically regulated by long-range enhancers in two developmentally conserved topologically associated domains (TADs) (36). In murine development, Hoxd13 is coordinated by five enhancers in a centromeric TAD (C-DOM) (Fig. 2A-Top) and disruption of this region leads to misexpression of Hoxd13, resulting in aberrant limb and posterior skeleton development (36). Given the established role of EWS::FLI1 as a pioneer factor (2, 5), we investigated whether the fusion might influence the chromatin state of this C-DOM region. We mapped the human C-DOM enhancers from their syntenic regions in mice using the UCSC LiftOver tool (Fig. 2A). The C-DOM region in mice, and corresponding region in humans, is a 600kb gene desert that starts ~180kb upstream (5’) of the Hoxd13 promoter (36). We interrogated this region for potential EWS::FLI1 binding sites and identified a 14-consecutive repeat GGAA microsatellite (Fig. 2A). Public chromatin immunoprecipitation (ChIP)-seq data (2, 34, 37) reproducibly show EWS::ETS fusion binding and H3K27ac and H3K4me1 histone modifications at this site in 18 different EWS::FLI1 and EWS::ERG expressing EwS cell lines (Fig. 2A, Supplementary Fig. S2A-D), as well as primary EwS but not osteosarcoma (38) tumors (Fig. 2B). Moreover, the activating histone marks were lost upon EWS::FLI1 knockdown (Fig. 2C).
Figure 2. EWS::FLI1 creates and activates a de novo HOXD13 enhancer in the developmentally conserved TAD.
A) Top- Schematic of the murine HOXD TAD regulatory regions with the associated enhancers for each region (colored circles). Middle- The murine HOXD C-DOM region (mm9) with the highlighted five enhancer (I-V) regions necessary for Hoxd13 expression in limb development. Bottom- The corresponding human syntenic region (hg19) with annotation of human-specific GGAA microsatellite site (posterior HOX enhancer: PHE). ChIP-seq tracks of EWS::FLI1, H3K27ac, and H3K4me1 binding at the PHE region in Ewing sarcoma cells (2). B) ChIP-seq tracks of H3K27ac and H3K4me1 at the PHE of primary Ewing sarcoma (2) and osteosarcoma (38) tumor samples. C) ChIP-seq tracks of EWS::FLI1 and H3K27ac at the PHE region in Ewing cells following EWS::FLI1 knockdown (2). D) ChIP-qPCR for EWS::FLI1, E) H3K27ac, and H3K4me1 in Ewing sarcoma cells. F) ChIP-qPCR for EWS::FLI1, H3K27ac, and H3K4me1 in U2OS cells. ChIP-qPCR for G) EWS::FLI1, H) H3K27ac after EWS::FLI1 knockdown. Negative control: an inert intergenic region in chr2. VRK1 enhancer: positive control GGAA enhancer site (2). C-DOM: centromeric domain; T-DOM: telomeric domain. Error bars representative of SEM from three independent replicates. * p<0.05; ** p<0.01; Two-way ANOVA; Sidak’s multiple comparison test; Two-tailed t-test.
To validate this GGAA microsatellite, which we named the posterior HOXD enhancer (PHE), as an EWS::FLI1-dependent enhancer, we performed ChIP-qPCR using the VRK1 enhancer as a positive control (2, 5). These studies confirmed EWS::FLI1 binding and H3K27ac/H3K4me1 marks at the PHE (Fig. 2D and 2E) in EwS but not U2OS osteosarcoma cells (Fig. 2F). Knockdown of EWS::FLI1 led to loss of fusion-binding (Fig. 2G) and concomitant loss of H3K27ac enrichment (Fig. 2H). Thus, in EwS cells, EWS FLI1 binds and activates a de novo GGAA microsatellite enhancer in the HOXD C-DOM regulatory region.
The PHE controls HOXD13 expression in EwS.
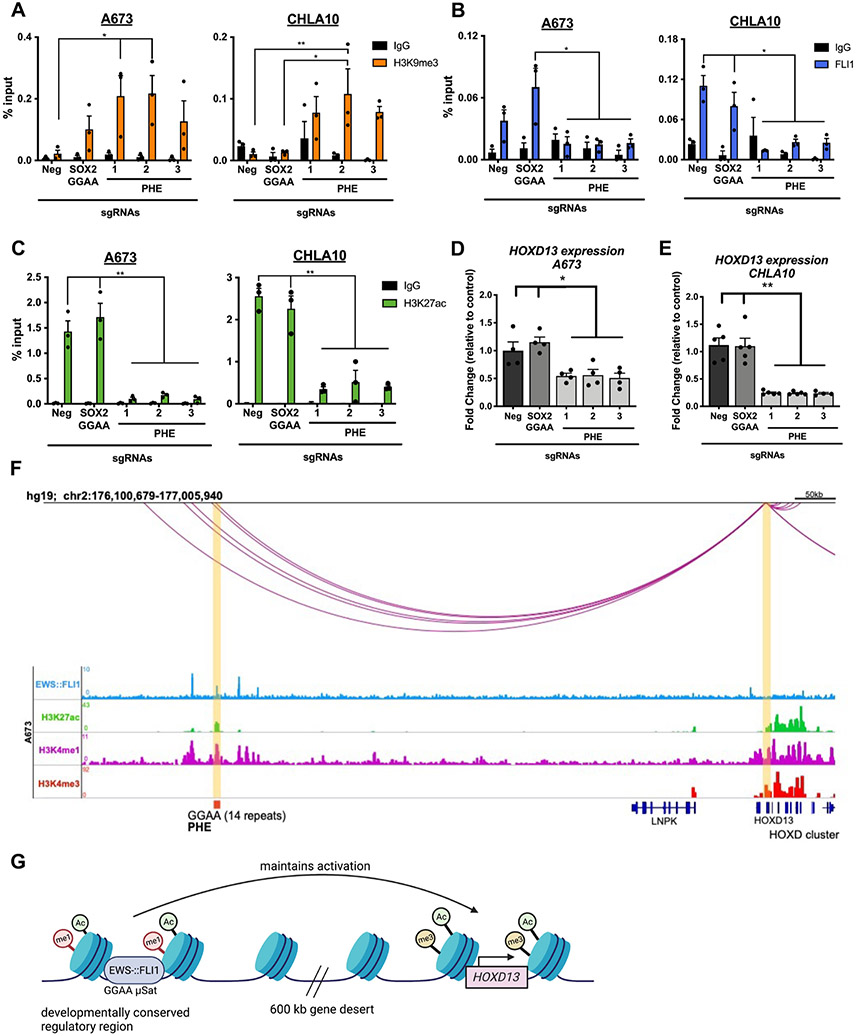
To functionally validate the PHE as an enhancer, we first interrogated public nuclear run-on sequencing (NRO-seq) data from EwS cells (6). These data confirmed expression of an enhancer RNA (eRNA) transcript at the PHE that was lost following EWS::FLI1 knockdown (Supplementary Fig. S3A). This finding, together with enrichment of RNA Polymerase II at the PHE (Supplementary Fig. S3A), provided in silico evidence that the PHE region is actively transcribed in EwS. Next, we used CRISPR interference (CRISPRi) to focally induce the H3K9me3-marked repressive chromatin state at the PHE (39). Since GGAA sites are repetitive and non-specific, we designed three flanking sgRNAs. In parallel to these PHE-targeted sgRNAs, dCas9-KRAB expressing cells were transduced with sgRNAs that target either the SOX2 GGAA enhancer (6) or a non-coding, inert genomic region. Only cells transduced with PHE-targeting sgRNAs acquired H3K9me3 at the PHE locus (Fig. 3A). Similarly, cells transduced with SOX2 GGAA-targeted sgRNA acquired H3K9me3 at the SOX2 enhancer (Supplementary Fig. S3B). SOX2 GGAA-targeted sgRNA had no impact on the chromatin state of the PHE and vice versa (Fig. 3A; Supplementary Fig. S3B).
Figure 3. The PHE uniquely controls HOXD13 expression in Ewing sarcoma.
All cells express dCas9-KRAB-mcherry and sgRNAs targeting either a negative control region (neg), the SOX2 GGAA enhancer, or PHE (1-3). DNA was isolated 8 days after sgRNA transduction. A) ChIP-qPCR for H3K9me3, B) EWS::FLI1, and C) H3K27ac at the PHE region. qRT-PCR of HOXD13 levels in D) A673 and E) CHLA10 cells. F) HiChIP looping data depicting the HOXD13 promoter:enhancer interactions in the C-DOM region in A673 cells (14). G) Model of EWS::FLI1 regulation of HOXD13 in Ewing sarcoma cells (biorender). Error bars are representative of SEM from at least three independent experiments. * p<0.05; ** p<0.01; Two tailed t-test; Two-way ANOVA; Sidak’s multiple comparison test.
Deposition of H3K9me3 at the PHE in EwS cells resulted in site-specific loss of EWS::FLI1 binding (Fig. 3B), loss of H3K27ac (Fig. 3C & Supplementary Fig. S3C-E) and reduced HOXD13 expression (Fig. 3D-E). In contrast, CRISPRi targeting of the PHE had no influence on HOXD13 in 293T cells (Supplementary Fig. S3F-I). Finally, data from high-throughput chromatin conformation capture-ChIP (HiChIP) studies (14) show a looping interaction between the PHE region and the HOXD13 promoter in A673 EwS cells (Fig. 3F, Supplementary Table S2). Together these data strongly support the conclusion that the EWS::FLI1-bound PHE GGAA microsatellite functions as a distal enhancer in EwS, contributing to transcriptional activation of HOXD13 (Fig. 3G).
Significantly, when expressed in human MSCs, EWS::FLI1 binds to the PHE and induces an open and active chromatin state (Supplementary Fig. S4A-B; data from Refs. (2, 5)). Nevertheless, despite successful chromatin remodeling, HOXD13 transcription is not induced in this context (2, 5). Similarly, we did not detect upregulation of HOXD13 expression in human MSCs, U2OS, or SW1353 chondrosarcoma cells following ectopic expression of EWS::FLI1 (Supplementary Fig. S4C-E). Thus, although EWS::FLI1 binding of the PHE leads to epigenetic rewiring, this enhancer hijacking is, by itself, insufficient to induce HOXD13 expression.
HOXD13 regulates mesenchymal gene programs and cell states.
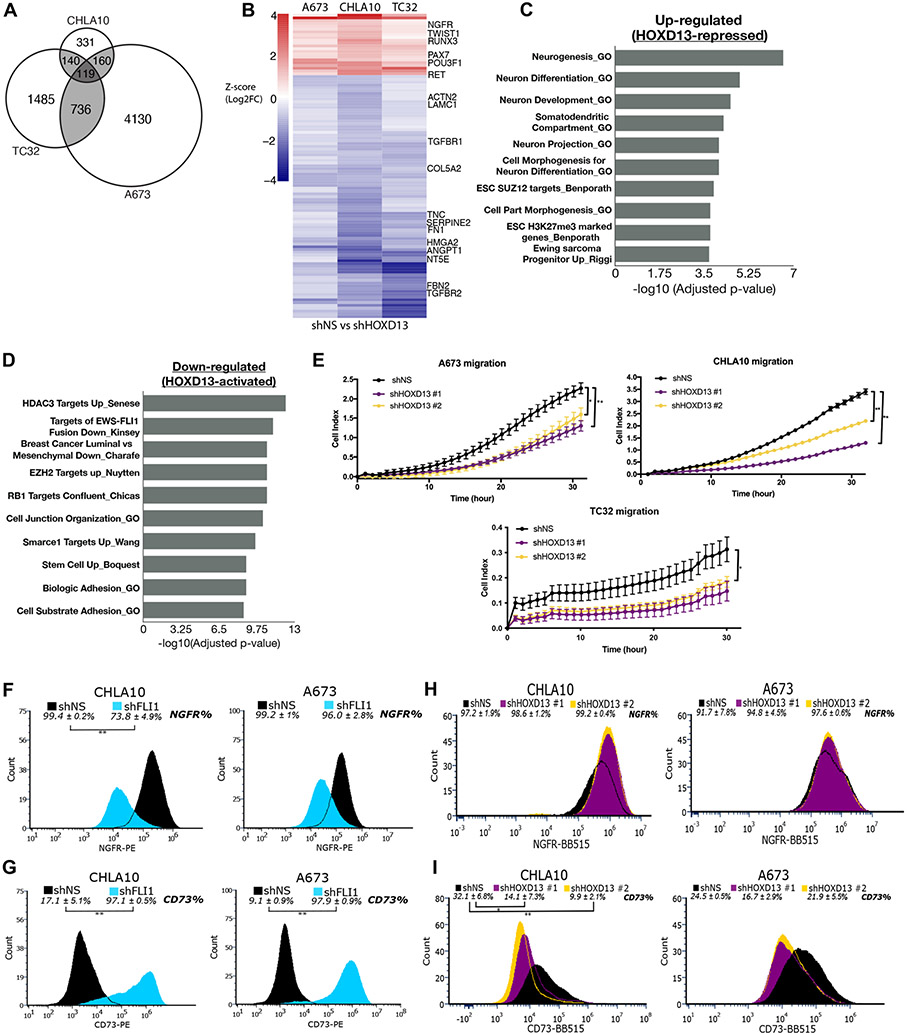
To elucidate how HOXD13 effects its oncogenic function in EwS, we performed RNA-seq on three EwS cell lines following knockdown of HOXD13 (Supplementary Fig. S5A). Significant and widespread changes were observed demonstrating that HOXD13 is highly transcriptionally active in EwS cells (Supplementary Fig. S5B-D, Supplementary Table S3). However, the identity of HOXD13-regulated gene targets was cell line-dependent revealing the importance of cell context. To identify gene programs that are most likely to be relevant for EwS phenotypes we focused on HOXD13-regulated genes among all cell lines (Fig. 4A). Of 119 shared genes, 109 were regulated in the same direction and most (N=87) were downregulated, indicating positive regulation by HOXD13 (Fig. 4B). Gene ontology analysis of significantly altered gene programs showed that neural differentiation and development programs were enriched among upregulated genes, while genes that are commonly associated with mesenchymal programs (i.e. cell adhesion and mesenchymal breast cancer cell signatures) were downregulated (Fig. 4C-D; Supplementary Table S3). Notably, transcription factors and markers of neural and mesenchymal differentiation, as well as genes involved in epithelial mesenchymal transitions (EMT) and metastasis phenotypes, were prominent among HOXD13-regulated transcripts (Fig. 4B).
Figure 4. HOXD13 regulates neuro-mesenchymal gene programs and influences cell states.
A) Venn diagram showing overlap of significantly differentially expressed genes in each cell line. B) Heatmap depicting the differentially expressed genes (padj <0.05) between shHOXD13 and shNS for all cell lines. Scale: Z-score (Log2FC). Overrepresentation analysis of top 10 gene sets C) up- and D) down-regulated following HOXD13 knockdown in all cell lines. E) xCELLigence Real Time Cell Migration assay of control and HOXD13 knockdown cells. Error bars are representative of SEM from at least two independent experiments with 5 technical replicates per condition. * p<0.05; ** p<0.01. Flow cytometry histograms showing the shift in F) NGFR + cells and G) CD73+ cells upon EWS::FLI1 knockdown. Flow cytometry histograms showing the shift in H) NGFR + cells and I) CD73+ cells upon HOXD13 knockdown. Flow Error represents SD from at least three independent experiments. * p<0.05; ** p<0.01; Two-tailed t-test.
EwS tumors display features of both neural and mesenchymal lineages and EWS::FLI1 has been shown to promote neural-, whilst inhibiting mesenchymal-like states (40). Our transcriptomic data suggested to us that HOXD13 might promote transition from a neural to a more mesenchymal cell state. In support of this, HOXD13 knockdown cells were reproducibly less migratory than their respective controls (Fig. 4E). In addition, the MSC marker NT5E (ecto-5’-nucleotidase; CD73) was among HOXD13-induced transcripts and NGFR, a neural marker, was relatively repressed by HOXD13 (Fig. 4B). Thus, we used these cell surface proteins to mark neural-like (NGFR+) and mesenchymal-like (CD73+) EwS cells in control and genetically modified conditions. Most EwS cells express NGFR and knockdown of EWS::FLI1 resulted in loss of NGFR+ cells and gain in the frequency of CD73+ cells (Fig. 4F-G). In direct contrast, HOXD13 knockdown led to diminished numbers of mesenchymal-like CD73+ cells and a modest but still measurable increase in NGFR+ cells (Fig. 4H-I). Thus, HOXD13 promotes mesenchymal gene programs and phenotypes in EwS.
HOXD13 binds active chromatin in EwS cells at intergenic and intronic sites and at EWS::ETS binding sites.
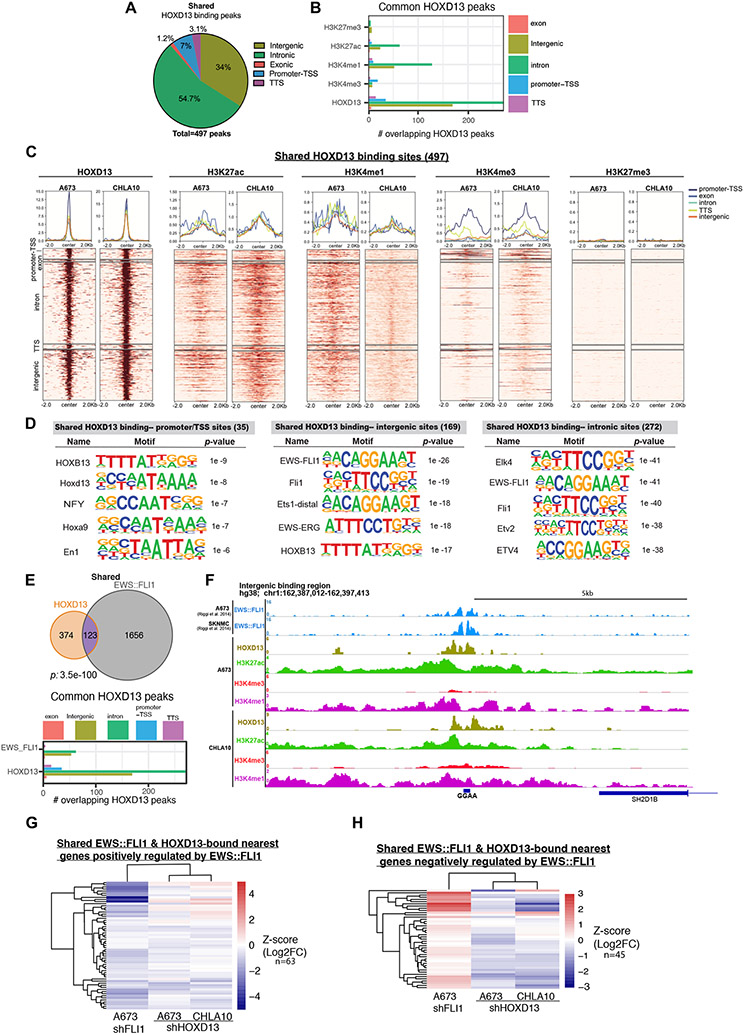
Transcriptional profiling of HOXD13 knockdown cells revealed that numerous genes that are positively regulated by HOXD13 are normally repressed by EWS::FLI1 (Fig. 4D), while EWS::FLI1-induced genes are over-represented among HOXD13-repressed genes (Fig. 4C). This antagonistic effect of HOXD13 on EWS::FLI1-dependent gene regulation was not due to changes in the level of the fusion (Supplemental Fig. S5E-H). To determine how HOXD13 influences EWS::FLI1 transcriptional activity, we performed CUT&RUN-sequencing (41). Binding of HOXD13 protein was detected throughout the genome in both cell lines, with nearly 500 binding sites shared at predominantly intronic and intergenic regions (Fig. 5A; Supplementary Fig. S6A, Supplementary Table S4). Enrichment of H3K27ac and H3K4me1 was evident at these HOXD13 binding sites, identifying them as putative active enhancers (Fig. 5B-C, Supplementary Fig. S6B-C). Likewise, HOXD13-bound promoter/transcription start sites were characterized by enrichment of active chromatin marks, H3K27ac and H3K4me3 (Fig. 5B-C, Supplementary Fig. S6B-C). No enrichment of the repressive H3K27me3 mark was detected (Fig. 5B-C, Supplementary Fig. S6B-C).
Figure 5. HOXD13 binds active chromatin in EwS cells at intergenic and intronic regions and at EWS::ETS binding sites.
A) Pie chart showing the genomic distribution of HOXD13 binding sites shared between CHLA10 and A673 cells. B) Bar chart summarizing shared binding sites and associated histone marks at these sites. C) Tornado plots depicting shared HOXD13 binding and the associated histone marks by genomic location. D) HOMER Motif analysis by genomic location for the shared HOXD13 binding sites. E) Venn diagrams showing the overlap between HOXD13 binding sites and published EWS::FLI1 binding sites in shared sites. Bar graphs depict genomic locations of shared HOXD13 and EWS::FLI1 bound sites. F) Representative CUT&RUN tracks of HOXD13 binding and associated histone marks at a direct target intronic region. G) Heatmaps depicting shared HOXD13 and EWS::FLI1 nearest genes negatively regulated by EWS::FLI1 after HOXD13 knockdown. H) Heatmaps depicting shared HOXD13 and EWS::FLI1 nearest genes negatively regulated by EWS::FLI1 after HOXD13 knockdown.
In normal development, HOX proteins have DNA binding affinities that are determined by their interactions with context-dependent cofactors (42). Therefore, we reasoned that identification of enriched transcription factor binding motifs at HOXD13-bound loci would provide insights into its regulatory partners in EwS. HOMER analysis revealed the expected enrichment of HOX and other early developmental transcription factor motifs at gene promoter/transcription start sites (Fig. 5D, Supplementary Fig. S6D-E, Supplementary Table S5). In contrast, intergenic and intronic peaks showed a striking and reproducible enrichment of ETS family binding motifs, including EWS::ETS sites (Fig. 5D, Supplementary Fig. S6D-E). To directly test whether HOXD13 peaks localized to EWS::ETS binding sites, we compared HOXD13 peaks in A673 and CHLA10 cells to nearly 1800 sites that were identified as EWS::FLI1-bound regions in A673 and SKNMC cells (2). A striking and highly statistically significant overlap exists between HOXD13 and EWS::FLI1-bound sites, especially in intergenic and intronic regions (Fig. 5E, Supplementary Fig. S6F-G, Supplementary Table S6). Although just over half of these shared peaks (67/123) encompass GGAA repeats (e.g. Fig. 5F), the remainder do not. Thus, shared HOXD13/EWS::FLI1 bound loci are not defined by the presence or absence of microsatellites.
We next mapped the shared binding sites (N=123) to their nearest genes (N=108) as described (2), and assessed how modulation of either transcription factor influenced gene expression. Consistent with the established role of EWS::FLI1 as both a transcriptional activator and repressor, co-bound loci include targets that are positively (63/108, log2FC <0) (Fig. 5G) and negatively (45/108, log2FC >0) regulated by the fusion (Fig. 5H). Conversely, the majority of shared loci are activated by HOXD13 (Fig. 5G & H). To determine if co-bound sites function as enhancers, we again interrogated published A673 CTCF HiChIP data (14) for interactions between promoters of these 108 genes and their CTCF-marked enhancers. Of 108 genes, 22 displayed high confidence promoter: enhancer interactions in the HiChIP data and 9 of these (41%) overlap with HOXD13/EWS::FLI1 co-bound loci (Supplementary Table S2). Thus, binding of HOXD13 at EWS::FLI1-bound loci promotes activation of target genes, likely via enhancer activation, irrespective of whether the gene is positively or negatively regulated by the fusion. Thus, HOXD13 binding can both augment expression of direct EWS::FLI1-induced genes and activate genes that are normally subject to EWS::FLI1-mediated silencing.
Transcriptional antagonism between HOXD13 and EWS::FLI1 is largely indirect and evident at single-cell resolution.
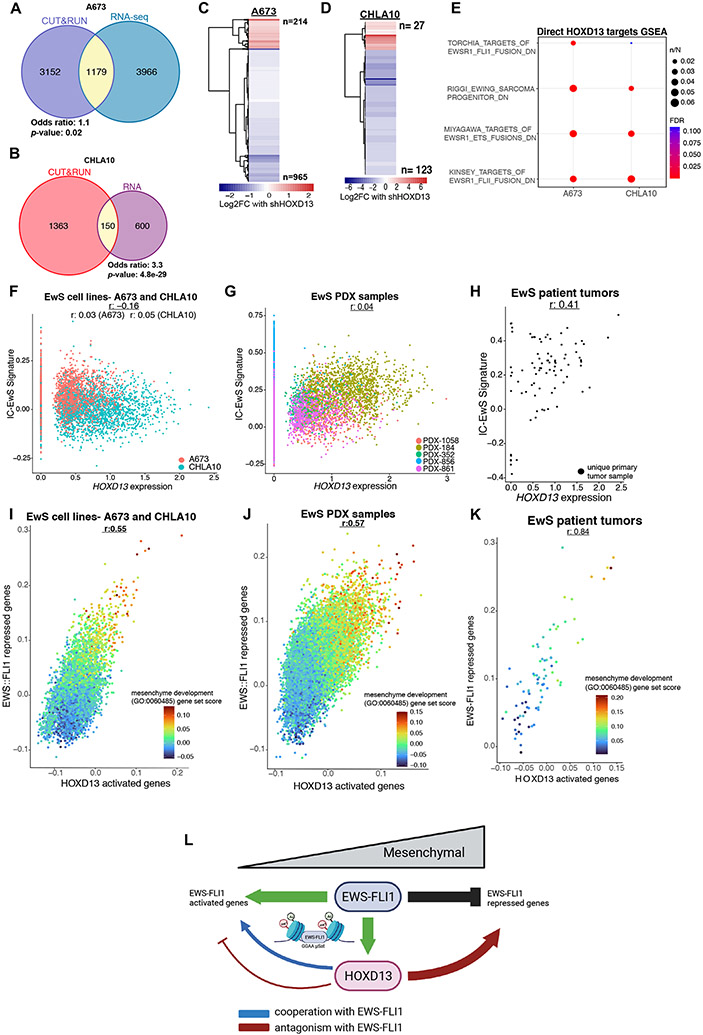
Although a subset of HOXD13 binding sites were found at established EWS::FLI1-bound loci, most were not. We therefore sought to broadly define direct transcriptional targets of HOXD13 in EwS cells, irrespective of fusion binding. Using a nearest gene approach and integration of RNAseq and CUT&RUN data, we identified genes that were both bound and regulated by HOXD13 in A673 (N=1179 genes) and CHLA10 (N=150 genes) cells (Fig. 6A-B). Consistent with its overall distribution, and a putative enhancer function, HOXD13 binding at these loci was in adjacent introns or upstream intergenic regions (Supplementary Fig. S7A-B) and most genes were activated by HOXD13 (Fig. 6C-D). Interrogation of A673 HiChIP data (43) identified high confidence promoter: enhancer interactions for 221 of 1179 HOXD13-regulated genes in A673 cells and 78 (35%) of these enhancer regions overlapped with a HOXD13 binding site (Supplementary Table 6). This supports the conclusion that HOXD13 can directly induce gene expression in EwS cells by binding and activating cis-regulatory regions of target genes.
Figure 6. Transcriptional antagonism between HOXD13 and EWS::FLI1 is evident at single-cell resolution.
A-B) Venn diagrams of the overlap between HOXD13 bound (CUT&RUN) and regulated (RNAseq) genes in each cell line. C-D) Heatmaps show the relative change in expression of these “direct” targets with shHOXD13. E) Overrepresentation analysis of EWS::FLI1 repressed gene sets identified as enriched among directly HOXD13-activated target genes. Data were generated using CITE-seq. F) Scatter plot of HOXD13 expression and the IC-EwS EWS::FLI1 signature by cell line. G) Scatter plot of HOXD13 expression and the IC-EwS EWS::FLI1 signature by PDX. H) Scatter plot of HOXD13 expression and the IC-EwS EWS::FLI1 signature by primary tumor sample. Scatter plot of HOXD13 activated genes and EWS::FLI1 repressed genes colored by the mesenchyme development (GO:0060485) gene set score in I) Ewing cell lines, J) PDX lines, and K) primary tumor samples. J) Summary model of HOXD13 cooperation and antagonism with EWS::FLI1 (Biorender). r: Pearson’s correlation coefficient.
Although there were marked differences between the number and identity of HOXD13 bound and regulated genes in A673 and CHLA10 cells, GSEA of putative direct HOXD13 targets revealed significant and reproducible enrichment of EWS::FLI1-repressed genes (Fig. 6E). These studies of bulk populations indicated that transcriptional antagonism exists between the HOXD13-activated and EWS::FLI1-repressed gene signatures. To address whether this antagonism exists in individual tumor cells, we performed single cell-sequencing of A673 and CHLA10 cells using CITE-seq (30)(Supplementary Fig. S7C). The results showed that HOXD13 expression is highly heterogeneous within and between cell lines (Supplementary Fig. S7D). Similarly, and consistent with prior reports (12, 35), inter- and intra- cell line heterogeneity of EWS::FLI1 is evident, though variability is less than HOXD13 (Supplementary Fig. S7D). Given that direct quantification of the fusion transcript is not feasible using short read sequencing, we used the recently published EWS::FLI1-specific signature (IC-EwS) (35) as a surrogate to infer EWS::FLI1 expression. No correlation was detected between expression of HOXD13 or the IC-EwS signature suggesting that transcriptional antagonism between HOXD13 and EWS::FLI1 cannot be fully explained by differences in the absolute levels of each gene in an individual cell (Fig. 6F). Next, we quantified the relative activity of each transcription factor in individual cells. Transcriptional activity of the fusion was determined by quantifying the relative expression level of established activated (N=1244) and repressed (N=319) EWS::FLI1 target genes (44). The HOXD13 transcriptional activation signature was derived from CHLA10 and A673 cells (N=254 genes). In individual cell transcriptomes, no correlation was detected between HOXD13 activity and expression of EWS::FLI1-activated genes (Supplementary Fig. S7E). In contrast, a significant and reproducible direct correlation was observed between HOXD13 activity and expression of EWS::FLI1-repressed genes (Supplementary Fig. S7F). Moreover, this pattern was also evident in single-cell data generated from five patient derived-xenografts (PDX) (35) (Fig. 6G, Supplementary Fig. S7G-J). Consistent with this observation, bulk RNA-seq data from 84 primary EwS patient tumors shows similar transcriptional antagonism (Fig. 6H, Supplementary Fig. S7K-M). Thus, the direct relationship between HOXD13-activation and upregulated expression of EWS::FLI1-repressed genes is evident in individual tumor cells in both in vitro and in vivo models of EwS and also in fresh tumor biopsies.
Finally, we questioned whether individual cells with differential activity of each of these master transcription factors would harbor differential activation of mesenchymal gene programs. In both cell lines and PDX tumors, single cells with high HOXD13 activity and high expression of the EWS::FLI1-repressed signature are characterized by high level expression of mesenchyme development signature genes (GO:0060485) (Fig. 6I-J). Further, this direct relationship between HOXD13 activity, loss of EWS::FLI1-dependent gene repression, and activation of mesenchymal gene programs is seen in RNA profiles of primary patient tumors (Fig. 6K). Together these data reveal that EwS cells exist along a mesenchymal transcriptional continuum (Fig. 7H, I) and that the relative state of tumor cells along this axis is determined, at least in part, by the competing activities of HOXD13 and EWS::FLI1 (Fig 7H-J).
Discussion
Successful propagation and metastatic progression of EwS tumors requires tight regulation of EWS::FLI1 expression and transcriptional activity (15). Moreover, the critical level for tumor growth and progression likely differs at different stages of tumor evolution. For example, while local tumor growth is reliant on continued expression of the fusion, migratory and metastatic properties of EwS cells appear to depend on acquisition of an EWS::FLI1-low state (10-12, 40). In addition, too much EWS::FLI1 activity leads to cell death (15). These observations suggest that EwS cells adhere to the “Goldilocks principle”: they require a dose of oncogene activity that is “just right” (15). Our current studies have identified HOXD13 as a key transcription factor that contributes to maintaining “just right” EWS::FLI1 activity, without affecting levels of fusion expression. Specifically, our findings demonstrate that, in addition to cooperating with the fusion at select EWS::FLI1-bound and activated loci to augment gene expression, HOXD13 serves a more striking function as an internal rheostat for the EWS::FLI1-repressed signature, antagonizing EWS::FLI1 at genes that are normally silenced by the fusion. In this manner, EwS cells that express high HOXD13 transcriptional activity display properties of EWS::FLI1-low cells: they activate mesenchymal gene programs, upregulate NT5E transcription and CD73 expression on their cell surface, and show enhanced migratory properties. However, despite clear inhibition of the EWS::FLI1 repressed state, these cells retain much of the EWS::FLI1 activated gene signature. Thus, HOXD13 activity uncouples the activating and repressive functions of the fusion protein. This uncoupling of EWS::FLI1 transcriptional functions enables continued expression of HOXD13, and other positively regulated genes, in cells that have activated the normally silenced mesenchymal gene program. These results provide a molecular explanation for the observation that HOXD13 loss-of-function inhibits EwS metastasis in vivo (20). In addition, they suggest that EwS cells are phenotypically plastic and that transition of tumor cells along a mesenchymal transcriptional axis is determined, in large part, by the competing activities of EWS::FLI1 and HOXD13. These findings are conceptually aligned with recent studies of neuroblastoma (45). Neuroblastoma cell subpopulations with different transcriptional profiles along a neural-mesenchymal axis have been identified and their distinct phenotypic properties may promote different elements of tumor progression. We speculate that cell state transitions along a neural-mesenchymal axis in EwS similarly generate subpopulations of tumor cells that promote metastasis.
We identified an EWS::FLI1-bound GGAA microsatellite in a conserved regulatory region of the posterior HOXD cluster. Targeted silencing of this PHE confirmed that high-level expression of HOXD13 in EwS cells requires EWS::FLI1-mediated activation of this de novo enhancer. However, despite its reproducible capacity for PHE enhancer reprogramming, acute activation of EWS::FLI1 in adult human MSCs or human embryonic stem cell-derived neural crest cells (hNCC) failed to induce HOXD13 (2, 5, 46). HOXD13 mRNA was, however, induced in EWS::FLI1-transduced pediatric MSCs that were cultured in pluripotent stem cell conditions (47) and in fusion-positive hNCC after passaging for several weeks (19). Thus, other yet to be defined, cellular and/or microenvironmental factors are needed for gene activation following epigenetic reprogramming of the PHE. Notably, the syntenic region in the murine HoxD TAD contains only a 4-repeat GGAA, well below the “sweet spot” for EWS::ETS binding and activation (48). Consistent with this, EWS::FLI1 does not bind or activate this region or induce Hoxd13 expression in mouse MSCs (27, 49). Given its function as a rheostat for EWS::FLI1 activity, we speculate HOXD13 may be required for EWS::FLI1-induced transformation. If so, the inability of EWS::ETS proteins to reprogram the murine Hoxd TAD may contribute to the continued absence of a genetically engineered mouse model of Ewing sarcoma.
The precise mechanisms that determine whether and how HOXD13 cooperates with or antagonizes EWS::FLI1 at discrete loci remains to be determined but it is unlikely to be exclusively dependent on co-binding of the transcription factors at enhancers. Although HOXD13 was enriched at GGAA microsatellites and EWS::FLI1-bound enhancers, these shared regions do not control mesenchymal genes such as NT5E that are repressed in EwS cells. Rather, the two transcription factors appear to compete indirectly to modulate mesenchymal programs. Likewise, the mechanism by which HOXD13 dampens select EWS::FLI1-induced neural developmental genes is unknown. Given the marked enrichment for wild type ETS, and other developmental transcription factor motifs at HOXD13 bound loci, we speculate that the impact of HOXD13 on EWS::FLI1 signatures, especially the repressive signature, is likely to involve engagement of other context-dependent transcription factors that govern neural-mesenchymal lineage identity. Wild-type ETS proteins are of particular interest given their proposed role in modulating the EWS::FLI1-repressive signature (2) and the fact that HOX and wild type ETS proteins co-bind at ETS factor motifs in leukemia and hematopoietic cells (50).
In summary, we have identified HOXD13 as a direct target of EWS::FLI1 and shown that the net balance of activity of these two master transcription factors regulates transcriptional cell state along a neural-mesenchymal axis. The mechanism of this “competitive cooperation” is mediated directly by co-binding of the proteins at enhancer elements and indirectly by antagonistic effects on developmental gene programs. Further investigations are required to characterize EwS cell subpopulations and elucidate if and how cells dynamically shift between states. Moreover, it will be critical to determine if cell state transitions, under the control of master transcription factors such as HOXD13 and EWS::FLI1, contribute to metastatic progression.
Supplementary Material
Acknowledgements
The authors thank members of the Lawlor lab for helpful discussion, the staff in the University of Michigan Advanced Genomics and Flow cytometry cores for technical assistance, and the staff at the Fred Hutchinson and Seattle Children’s Research Institute cores for flow cytometry, Bioinformatics and Genomics. We thank Phil Corrin and Dolores Covarrubias in Genomics Shared Resource of Fred Hutchinson Cancer Research Center for performing auto CUT&RUN experiments, NGS library preparation and sequencing. We are thankful to Dr. Brian Parkin, who aided in the design and technical completion of the CITE-seq studies. Financial support for this work was provided by the following sources: National Institute of Health: F31 CA247104 (AA), F31 CA254079 (JJ), T32 CA009676 (AA), K00 CA234810 (AGH), K08 CA208013 (RJHR), R01 CA215981 (ERL), R21 CA215968 (ERL, DW), P30 CA046592 Rogel Cancer Center Support Grant (Flow, genomics, bioinformatics and bioanalytic cores), P30 CA015704 Fred Hutch/University of Washington Cancer Consortium (Flow cytometry, Genomics & Bioinformatics Shared Resource); 1 Million 4 Anna Foundation (ERL, JS, SF); Alex’s Lemonade Stand Innovation Award (ERL, DW); UM Department of Pediatrics Charles Woodson Fund.
Footnotes
Competing interests: The authors declare no potential conflicts of interest.
References
- 1.Riggi N, Suva ML, Stamenkovic I. Ewing's Sarcoma. N Engl J Med. 2021;384(2):154–64. [DOI] [PubMed] [Google Scholar]
- 2.Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suva ML, et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell. 2014;26(5):668–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gangwal K, Sankar S, Hollenhorst PC, Kinsey M, Haroldsen SC, Shah AA, et al. Microsatellites as EWS/FLI response elements in Ewing's sarcoma. Proc Natl Acad Sci U S A. 2008;105(29):10149–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomazou EM, Sheffield NC, Schmidl C, Schuster M, Schonegger A, Datlinger P, et al. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell Rep. 2015;10(7):1082–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulay G, Sandoval GJ, Riggi N, Iyer S, Buisson R, Naigles B, et al. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell. 2017;171(1):163–78 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulay G, Volorio A, Iyer S, Broye LC, Stamenkovic I, Riggi N, et al. Epigenome editing of microsatellite repeats defines tumor-specific enhancer functions and dependencies. Genes Dev. 2018;32(15-16):1008–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sankar S, Bell R, Stephens B, Zhuo R, Sharma S, Bearss DJ, et al. Mechanism and relevance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma. Oncogene. 2013;32(42):5089–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sankar S, Theisen ER, Bearss J, Mulvihill T, Hoffman LM, Sorna V, et al. Reversible LSD1 inhibition interferes with global EWS/ETS transcriptional activity and impedes Ewing sarcoma tumor growth. Clin Cancer Res. 2014;20(17):4584–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aryee DN, Niedan S, Kauer M, Schwentner R, Bennani-Baiti IM, Ban J, et al. Hypoxia modulates EWS-FLI1 transcriptional signature and enhances the malignant properties of Ewing's sarcoma cells in vitro. Cancer Res. 2010;70(10):4015–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaturvedi A, Hoffman LM, Welm AL, Lessnick SL, Beckerle MC. The EWS/FLI Oncogene Drives Changes in Cellular Morphology, Adhesion, and Migration in Ewing Sarcoma. Genes Cancer. 2012;3(2):102–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedersen EA, Menon R, Bailey KM, Thomas DG, Van Noord RA, Tran J, et al. Activation of Wnt/beta-Catenin in Ewing Sarcoma Cells Antagonizes EWS/ETS Function and Promotes Phenotypic Transition to More Metastatic Cell States. Cancer Res. 2016;76(17):5040–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franzetti GA, Laud-Duval K, van der Ent W, Brisac A, Irondelle M, Aubert S, et al. Cell-to-cell heterogeneity of EWSR1-FLI1 activity determines proliferation/migration choices in Ewing sarcoma cells. Oncogene. 2017;36(25):3505–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adane B, Alexe G, Seong BKA, Lu D, Hwang EE, Hnisz D, et al. STAG2 loss rewires oncogenic and developmental programs to promote metastasis in Ewing sarcoma. Cancer Cell. 2021;39(6):827–44 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surdez D, Zaidi S, Grossetete S, Laud-Duval K, Ferre AS, Mous L, et al. STAG2 mutations alter CTCF-anchored loop extrusion, reduce cis-regulatory interactions and EWSR1-FLI1 activity in Ewing sarcoma. Cancer Cell. 2021;39(6):810–26 e9. [DOI] [PubMed] [Google Scholar]
- 15.Seong BKA, Dharia NV, Lin S, Donovan KA, Chong S, Robichaud A, et al. TRIM8 modulates the EWS/FLI oncoprotein to promote survival in Ewing sarcoma. Cancer Cell. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014;4(11):1326–41. [DOI] [PubMed] [Google Scholar]
- 17.Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014;4(11):1342–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel M, Simon JM, Iglesia MD, Wu SB, McFadden AW, Lieb JD, et al. Tumor-specific retargeting of an oncogenic transcription factor chimera results in dysregulation of chromatin and transcription. Genome Res. 2012;22(2):259–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Svoboda LK, Harris A, Bailey NJ, Schwentner R, Tomazou E, von Levetzow C, et al. Overexpression of HOX genes is prevalent in Ewing sarcoma and is associated with altered epigenetic regulation of developmental transcription programs. Epigenetics. 2014;9(12):1613–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Heyking K, Roth L, Ertl M, Schmidt O, Calzada-Wack J, Neff F, et al. The posterior HOXD locus: Its contribution to phenotype and malignancy of Ewing sarcoma. Oncotarget. 2016;7(27):41767–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Y, Zhou B, Mao F, Xu J, Miao H, Zou Z, et al. HOXA9 Reprograms the Enhancer Landscape to Promote Leukemogenesis. Cancer Cell. 2018;34(4):643–58 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svoboda LK, Bailey N, Van Noord RA, Krook MA, Harris A, Cramer C, et al. Tumorigenicity of Ewing sarcoma is critically dependent on the trithorax proteins MLL1 and menin. Oncotarget. 2017;8(1):458–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheth R, Barozzi I, Langlais D, Osterwalder M, Nemec S, Carlson HL, et al. Distal Limb Patterning Requires Modulation of cis-Regulatory Activities by HOX13. Cell Rep. 2016;17(11):2913–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao X, Sun M, Zhao J, Leyva JA, Zhu H, Yang W, et al. Mutations in HOXD13 underlie syndactyly type V and a novel brachydactyly-syndactyly syndrome. Am J Hum Genet. 2007;80(2):361–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jimenez JA, Apfelbaum AA, Hawkins AG, Svoboda LK, Kumar A, Ruiz RO, et al. EWS-FLI1 and Menin Converge to Regulate ATF4 Activity in Ewing Sarcoma. Mol Cancer Res. 2021;19(7):1182–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marques Howarth M, Simpson D, Ngok SP, Nieves B, Chen R, Siprashvili Z, et al. Long noncoding RNA EWSAT1-mediated gene repression facilitates Ewing sarcoma oncogenesis. J Clin Invest. 2014;124(12):5275–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfaltzgraff ER, Apfelbaum A, Kassa AP, Song JY, Jiang W, Suhan TK, et al. Anatomic Origin of Osteochondrogenic Progenitors Impacts Sensitivity to EWS-FLI1-Induced Transformation. Cancers (Basel). 2019;11(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulsen MT, Veloso A, Prasad J, Bedi K, Ljungman EA, Magnuson B, et al. Use of Bru-Seq and BruChase-Seq for genome-wide assessment of the synthesis and stability of RNA. Methods. 2014;67(1):45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janssens DH, Wu SJ, Sarthy JF, Meers MP, Myers CH, Olson JM, et al. Automated in situ chromatin profiling efficiently resolves cell types and gene regulatory programs. Epigenetics Chromatin. 2018;11(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14(9):865–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furlan SNSKL C; Tkachev V; Hunt DJ; Hibbard J;Betz KM; Blazar BR; Trapnell C; Kean LS IL-2 enhances ex vivo–expanded regulatory T-cell persistence after adoptive transfer. Blood Advances. 2020;4(8):1594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. Addendum: The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2019;565(7738):E5–E6. [DOI] [PubMed] [Google Scholar]
- 33.Grobner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, et al. The landscape of genomic alterations across childhood cancers. Nature. 2018;555(7696):321–7. [DOI] [PubMed] [Google Scholar]
- 34.Koster J R2 Genomics Analysis and Visualization Platform Department of Oncogenomics in the Academic Medical Center (AMC) Amsterdam, the Netherlands: 2006. [A biologist friendly, web based genomics analysis and visualization application]. Available from: http://r2.amc.nl http://r2platform.com). [Google Scholar]
- 35.Aynaud MM, Mirabeau O, Gruel N, Grossetete S, Boeva V, Durand S, et al. Transcriptional Programs Define Intratumoral Heterogeneity of Ewing Sarcoma at Single-Cell Resolution. Cell Rep. 2020;30(6):1767–79 e6. [DOI] [PubMed] [Google Scholar]
- 36.Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, et al. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147(5):1132–45. [DOI] [PubMed] [Google Scholar]
- 37.Orth MFS D; Marchetto A; Grossetête S; Gerke JS; Zaidi S; Alonso J; Sastre A; Baulande S; Sill M; Cidre-Aranaz F; Ohmura S; Kirchner T; Hauck SM; Reischl E; Gymrek M; Pfister SM; Strauch K; Delattre O; Grünewald TGP Systematic multi-omics cell line profiling uncovers principles of Ewing sarcoma fusion oncogene-mediated gene regulation. bioRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrow JJ, Bayles I, Funnell APW, Miller TE, Saiakhova A, Lizardo MM, et al. Positively selected enhancer elements endow osteosarcoma cells with metastatic competence. Nat Med. 2018;24(2):176–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thakore PI, D'Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015;12(12):1143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11(5):421–9. [DOI] [PubMed] [Google Scholar]
- 41.Skene PJ, Henikoff JG, Henikoff S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc. 2018;13(5):1006–19. [DOI] [PubMed] [Google Scholar]
- 42.Saurin AJ, Delfini MC, Maurel-Zaffran C, Graba Y. The Generic Facet of Hox Protein Function. Trends Genet. 2018;34(12):941–53. [DOI] [PubMed] [Google Scholar]
- 43..!!! INVALID CITATION !!! (14).
- 44.Kinsey M, Smith R, Lessnick SL. NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing's sarcoma. Mol Cancer Res. 2006;4(11):851–9. [DOI] [PubMed] [Google Scholar]
- 45.Gautier M, Thirant C, Delattre O, Janoueix-Lerosey I. Plasticity in Neuroblastoma Cell Identity Defines a Noradrenergic-to-Mesenchymal Transition (NMT). Cancers (Basel). 2021;13(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Levetzow C, Jiang X, Gwye Y, von Levetzow G, Hung L, Cooper A, et al. Modeling initiation of Ewing sarcoma in human neural crest cells. PLoS One. 2011;6(4):e19305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riggi N, Suva ML, De Vito C, Provero P, Stehle JC, Baumer K, et al. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev. 2010;24(9):916–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson KM, Taslim C, Saund RS, Lessnick SL. Identification of two types of GGAA-microsatellites and their roles in EWS/FLI binding and gene regulation in Ewing sarcoma. PLoS One. 2017;12(11):e0186275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimizu R, Tanaka M, Tsutsumi S, Aburatani H, Yamazaki Y, Homme M, et al. EWS-FLI1 regulates a transcriptional program in cooperation with Foxq1 in mouse Ewing sarcoma. Cancer Sci. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamada T, Shimizu T, Sakurai T, Nanashima N, Kihara-Negishi F, Suzuki M, et al. Physical and functional interactions between hematopoietic cell-specific ETS transcription factors and homeodomain proteins. Leuk Res. 2009;33(3):483–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Public data used in this study reported in Supplementary Table S1. Sequencing data generated in this study have been deposited at GEO (GSE182513). Code available at https://github.com/LawlorLab/HOXD13-Paper.