Abstract
Background
Here we release a new version of EchinoDB, EchinoDB v2.0 (https://echinodb.uncc.edu). EchinoDB is a database of genomic and transcriptomic data on echinoderms. The initial database consisted of groups of 749,397 orthologous and paralogous transcripts arranged in orthoclusters by sequence similarity.
Results
The updated version of EchinoDB includes two new major datasets: the RNA-Seq data of the brittle star Ophioderma brevispinum and the high-quality genomic assembly data of the green sea urchin Lytechinus variegatus. In addition, we enabled keyword searches for annotated data and installed an updated version of Sequenceserver to allow Basic Local Alignment Search Tool (BLAST) searches. The data are downloadable in FASTA format. The first version of EchinoDB appeared in 2016 and was implemented in GO on a local server. The new version has been updated using R Shiny to include new features and improvements in the application. Furthermore, EchinoDB now runs entirely in the cloud for increased reliability and scaling.
Conclusion
EchinoDB serves a user base drawn from the fields of phylogenetics, developmental biology, genomics, physiology, neurobiology, and regeneration. As use cases, we illustrate the function of EchinoDB in retrieving components of signaling pathways involved in the tissue regeneration process of different echinoderms, including the emerging model species Ophioderma brevispinum. Moreover, we use EchinoDB to shed light on the conservation of the molecular components involved in two echinoderm-specific phenomena: spicule matrix proteins involved in the formation of stereom endoskeleton and the tensilin protein that contributes to the capacity of the connective tissues to quickly change its mechanical properties. The genes involved in the former had been previously studied in echinoids, while gene sequences involved in the latter had been previously described in holothuroids. Specifically, we ask (a) if the biomineralization-related proteins previously reported only in sea urchins are also present in other, non-echinoid, echinoderms and (b) if tensilin, the protein responsible for the control of stiffness of the mutable collagenous tissue, previously described in sea cucumbers, is conserved across the phylum.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12863-022-01090-6.
Keywords: Database, Echinoderms, Echinoids, Gene family, Genome, Ophiuroids, Orthocluster, Ortholog, Paralog, Transcriptome, Notch, Wnt, Spicule matrix proteins, Mutable collagenous tissue, Tensilin
Background
The phylum Echinodermata is composed of marine invertebrate animals commonly known as echinoderms. It contains five extant classes: Asteroidea, Ophiuroidea, Holothuroidea, Echinoidea, and Crinoidea [1]. Echinoderms share a number of unique characteristics such as: pentaradial body symmetry (or modifications thereof) in adults, a skeleton composed of numerous ossicles formed of stereom (a calcium carbonate material), a water-vascular system, and a mutable collagenous tissue [2–5]. However, the most astonishing feature of echinoderms is their capacity to regenerate complex internal organs following injury or autotomy [6–13]. For instance, sea cucumbers (Echinodermata: Holothuroidea) have the ability to fully regenerate their digestive tube following visceral autotomy (evisceration) [14] and their radial nerve cord following transection [15]. Similarly, brittle stars of the class Ophiuroidea display remarkable regenerative capabilities in arm regeneration post injury or autotomy [16]. Regeneration in these animals involves substantial cell division, but it never goes awry to result in tumor formation [17]. Therefore, EchinoDB provides an opportunity to investigate genes involved in the evolution of echinoderm-specific traits (e.g., stereom skeleton and mutable collagenous tissue) and to deeply study fundamental genomic regulatory mechanisms underlying regeneration.
Researchers motivated by the biomedical potential of echinoderms have assembled a number of resources to study these animals. However current resources are limited to only a small fraction of species that do not represent the diversity within the phylum. Hence, to fill this gap, we have created EchinoDB, a database resource, in which genomic and transcriptomic data on 42 unique echinoderm species, spanning the deepest divergences within the five extant classes, is wrapped in an easy-to-use web-based application [18]. These species and associated raw sequence resources are listed in Table 1 and Additional file 1: Table S1. Our database thus allows for deep phylogenetic sampling within the echinoderm clade to facilitate data retrieval (annotated sequences) for various downstream projects, including regeneration, phylogeny, and gene family studies.
Table 1.
Raw reads from the various echinoderm species that are available in NCBI’s SRA and Zenodo (doi: 10.5281/zenodo.6985492). Each line corresponds to transcriptome or gene expression data. Orthoclusters: number of orthoclusters. Sequences: number of amino acids or coding sequences. Length: sum of base pairs in all sequences. See complete table in Additional file 1: Table S1
| Class: Order: Family | Species | Accession | SRR | Orthoclusters | Sequences | Length |
|---|---|---|---|---|---|---|
| Crinoidea: Comatulida: Zenometridae | Psathryometra fragilis | PRJNA299480 | SRR2846085 | 6651 | 9015 | 3.16E+ 07 |
| Asteroidea: Velatida: Xyloplacidae | Xyloplax sp. Janetae (BJ2) | PRJNA299326 | SRR2846120 | 17,993 | 24,452 | 5.65E+ 07 |
| Asteroidea: Spinulosida: Echinasteridae | Echinaster spinulosus | PRJNA300370 | SRR2844624 | 13,844 | 18,608 | 6.41E+ 07 |
| Ophiuroidea: Ophiocomidea: Ophiocomidae | Ophiocoma wendtii | PRJNA299897 | SRR2845427 | 3662 | 9783 | 8.82E+ 07 |
| Ophiuroidea: Gnathophiuridea: Ophiotrichidae | Ophiothrix spiculata | PRJNA299898 | SRR2845448 | 8118 | 18,816 | 7.34E+ 07 |
| Asteroidea: Velatida: Pterasteridae | Pteraster tesselatus | PRJNA299398 | SRR2846094 | 46,531 | 51,762 | 1.71E+ 08 |
| Holothuroidea: Apodida: Synaptidae | Synapta maculata | PRJNA299890 | SRR2846103 | 5309 | 11,154 | 8.44E+ 07 |
| Echinoidea: Echinoida: Strongylocentrotidae | Strongylocentrotus purpuratus | PRJNA299888 | SRR2846101 | 6885 | 11,368 | 4.15E+ 07 |
| Asteroidea: Forcipulatida: Asteriidae | Pisaster ochraceus | PRJNA299406 | SRR2846074 | 37,807 | 43,479 | 1.68E+ 08 |
| Holothuroidea: Dendrochirotida: Psolidae | Psolus sp. (BJ11) | PRJNA299550 | NA | 24,634 | 35,310 | 1.91E+ 08 |
| Holothuroidea: Aspidochirotida: Stichopodidae | Stichopus chloronotus | PRJNA299896 | SRR2846098 | 17,953 | 24,854 | 1.09E+ 08 |
| Crinoidea: Comatulida: Colobometridae | Oligometra serripinna | PRJNA299464 | SRR2845419 | 55,472 | 70,278 | 2.11E+ 08 |
| Crinoidea: Comatulida: Bourgueticrinidae | Democrinus brevis | PRJNA299465 | SRR2844622 | 6285 | 8287 | 4.72E+ 07 |
| Asteroidea: Velatida: Korethrasteridae | Peribolaster folliculatus (BJ19) | PRJNA299409 | SRR2845673 | 16,927 | 20,462 | 8.32E+ 07 |
| Asteroidea: Paxillosida: Astropectinidae | Psilaster charcoti | PRJNA299410 | SRR2846092 | 24,055 | 28,413 | 9.41E+ 07 |
| Asteroidea: Forcipulatida: Labidiasteridae | Labidiaster annulatus | PRJNA299411 | SRR2845003 | 35,615 | 40,071 | 1.43E+ 08 |
| Asteroidea: Velatida: Korethrasteridae | Remaster gourdoni | PRJNA299412 | SRR2846097 | 18,288 | 22,056 | 8.21E+ 07 |
| Crinoidea: Hyocrinida: Hyocrinidae | Gephyrocrinus messingi | PRJNA300546 | SRR2859800 | 8950 | 12,234 | 4.42E+ 07 |
| Asteroidea: Paxillosida: Luidiidae | Luidia clathrata | PRJNA299414 | SRR2845324 | 36,915 | 77,487 | 9.42E+ 07 |
| Asteroidea: Spinulosida: Echinasteridae | Henricia leviuscula A | PRJNA299415 | SRR2844627 | 47,492 | 76,684 | 9.58E+ 07 |
| Asteroidea: Paxillosida: Astropectinidae | Astropecten duplicatus | PRJNA299417 | SRR2843238 | 42,051 | 73,744 | 9.13E+ 07 |
| Asteroidea: Valvatida: Poraniidae | Glabraster antarctica (BJ28) | PRJNA299418 | SRR2844625 | 28,408 | 54,328 | 7.71E+ 07 |
| Asteroidea: Valvatida: Asteropseidae | Asteropsis carinifera | PRJNA299419 | SRR2843236 | 25,973 | 49,607 | 6.51E+ 07 |
| Asteroidea: Valvatida: Solasteridae | Peribolaster folliculatus (BJ30) | PRJNA299409 | SRR2845673 | 22,319 | 36,551 | 5.25E+ 07 |
| Asteroidea: Notomyotida: Benthopectinidae | Cheiraster hirsutus | PRJNA299420 | SRR2844620 | 325 | 1271 | 6.85E+ 06 |
| Asteroidea: Brisingida: Brisingidae | Odinella nutrix | PRJNA299463 | SRR2845408 | 312 | 1004 | 6.83E+ 06 |
| Crinoidea: Comatulida: Ptilometridae | Ptilometra australis | PRJNA299466 | SRR2846095 | 33,084 | 49,470 | 7.31E+ 07 |
| Crinoidea: Comatulida: Comasteridae | Cenolia new species | PRJNA299468 | SRR2847917 | 11,658 | 18,875 | 3.51E+ 07 |
| Crinoidea: Comatulida: Antedonidae | Isometra vivipara | PRJNA299471 | SRR2844835 | 27,204 | 43,689 | 7.02E+ 07 |
| Crinoidea: Comatulida: Antedonidae | Phrixometra nutrix | PRJNA299469 | SRR2846073 | 4923 | 12,283 | 2.83E+ 07 |
| Crinoidea: Comatulida: Antedonidae | Promachocrinus kerguelensis | PRJNA299478 | SRR2846076 | 8011 | 12,283 | 2.83E+ 07 |
| Echinoidea: Arbacioida: Arbaciidae | Arbacia punctulata | PRJNA299547 | SRR2843235 | 13,324 | 33,220 | 4.86E+ 07 |
| Echinoidea: Cidaroida: Cidaridae | Eucidaris tribuloides | PRJNA299548 | SRR2844624 | 6939 | 16,512 | 2.97E+ 07 |
| Echinoidea: Clypeasteroida: Dendrasteridae | Dendraster excentricus | PRJNA299549 | SRR2844623 | 4619 | 12,561 | 6.57E+ 07 |
| Holothuroidea: Dendrochirotacea: Psolidae | Psolus sp. (BJ41) | PRJNA299550 | NA | 16,398 | 33,062 | 7.32E+ 07 |
| Holothuroidea: Aspidochirotida: Synallactidae | Peniagone sp. (BJ42) | PRJNA299551 | NA | 12,286 | 22,457 | 5.25E+ 07 |
| Holothuroidea: Dendrochirotacea: Cucumariidae | Abyssocucumis sp. (BJ43) | PRJNA299552 | SRR2830762 | 12,309 | 26,171 | 5.47E+ 07 |
| Holothuroidea: Aspidochirotida: Synallactidae | Pseudostichopus sp. (BJ44) | PRJNA299883 | NA | 2464 | 5567 | 1.36E+ 07 |
| Holothuroidea: Molpadida: Molpadidae | Molpadia intermedia | PRJNA299884 | SRR2845419 | 3793 | 6516 | 1.53E+ 07 |
| Holothuroidea: Elasipodida: Laetmogonidae | Pannychia moseleyi | PRJNA299885 | NA | 10,124 | 20,051 | 3.96E+ 07 |
| Ophiuroidea: Euryalida: Gorgonocephalidae | Astrophyton muricatum | PRJNA299886 | SRR2843239 | 11,730 | 26,889 | 7.31E+ 07 |
| Ophiuroidea: Ophiurida: Ophiodermatidae | Ophioderma brevispinum | PRJNA299887 | SRR2845428 | 11,757 | 28,450 | 6.52E+ 07 |
EchinoDB v2.0 is an open-source web-based application (https://echinodb.uncc.edu), designed to provide genomic, transcriptomic and amino acid sequence data on echinoderms. The code for EchinoDB v2.0 is provided in Additional file 5: File S4.
The objective of EchinoDB is to serve research communities by providing diverse and rich data for a wide diversity of echinoderm species. The previous version of EchinoDB was released in 2016 and consisted of amino acid sequence orthoclusters (orthologous genes) from 42 echinoderm transcriptomes [19]. The new version has now been extended to incorporate new datasets that have been generated since the original release. These new datasets include RNA-Seq data for the brittle star O. brevispinum (Say, 1825) (Echinodermata: Ophiuroidea: Ophiacanthida: Ophiodermatidae) [16], genome assembly data of the green sea urchin Lytechinus variegatus (Lamarck, 1816) (Echinodermata: Echinoidea: Camarodonta: Toxopneustidae) [20], and phylogenomic data for Xyloplax sp. (Echinodermata: Asteroidea) [21]. The RNA-Seq data of the brittle star and the genome assembly data of the green sea urchin form the basis of two newly developed tools, OphiuroidDB [22] and EchinoidDB [23], respectively, integrated within the EchinoDB application.
An effective bioinformatics resource must keep up with new data, advances in software, server architecture, and programming languages. The need to improve reliability and scale well with the increasing amount of data and the number of users warranted an update to EchinoDB. The updated EchinoDB has been rewritten in R Shiny [24] and runs entirely in the cloud environment (AWS) [25]. R Shiny is highly extensible, easy to code and maintain, as compared to the previous implementation built using GO programming language in 2016. R Shiny supports faster development of user interfaces by providing a framework that requires no or little knowledge of scripting languages like HTML, CSS or JavaScript. We have taken advantage of this feature to extend the application’s capabilities to make new data (obtained from collaborations) easily available to the research community, for example, implementing the BLAST [26] search interface for the Lytechinus [20] and Ophioderma [16] sequences via Sequenceserver [27].
To demonstrate the practical utility of the new version of EchinoDB [18] and its associated resources - OphiuroidDB [22] and EchinoidDB [23] – we illustrate how EchinoDB is used in retrieving key components of the Notch and Wnt signaling pathways, that are crucial for tissue regeneration in echinoderms [16, 28–32]. In addition, we describe the use of SequenceServer (BLAST tool) [27, 33, 26] integrated within EchinoDB to find the putative homologs of the skeleton matrix proteins [4, 34–37] and tensilin (a protein that controls tensile strength of mutable collagenous tissues) [5, 38–40, 41, 42], previously reported in sea urchins (Echinodermata:Echinoidea) and sea cucumbers (Echinodermata: Holothuroidea).
Construction and content
EchinoDB is re-factored in R Shiny and currently supports annotated transcriptomic data for 42 echinoderm species (see Table 1 or Additional file 1: Table S1), functional transcriptomic data from a Notch pathway inhibition study in O. brevispinum [16], and protein sequences from a chromosome-level genome assembly of L. variegatus [20]. R Shiny is highly extensible, that is, code developed with R Shiny can be readily integrated with CSS themes, HTML widgets, and scripting languages (e.g. JavaScript). In addition, R Shiny is widely adopted and the code can be modified and tuned at later stages in the development cycle by many developers. EchinoDB v2.0 is hosted using the Nginx web server [43] in Amazon Web Services (AWS) [25]. AWS offers on-demand cloud computing services to build your own web-based applications independent of university information technology bureaus.
EchinoDB contains amino acid sequence clusters of orthologous genes, termed orthoclusters. These orthoclusters were generated by RNA-Seq profiling of adult tissues from 42 echinoderm specimens representing 24 orders and 37 families from all five extant classes [19]. The RNA-Seq data was assembled using Trinity [44] and translated into peptides using Transdecoder [45]. The de novo transcriptome assembly consisted of 1,198,706 amino acid sequences across 42 species. The data was clustered using OrthoMCL, an algorithm for grouping orthologous protein sequences based on sequence similarity [46]. The resulting orthoclusters database consisted of groups of 749,397 orthologous and paralogous transcripts. These orthoclusters were annotated through sequence similarity using the genome of purple sea urchin Strongylocentrotus purpuratus, the best annotated echinoderm genome at the time of the origins of the project [47]. Complete RNA-Seq analysis pipeline (from RNA sampling and isolation to sequencing, de novo transcriptome assembly, translation, orthoclustering and annotation) was described in [19]. These annotated orthoclusters now provide the basis for keyword searches in EchinoDB.
New data resources for ophiuroid and echinoid within the updated EchinoDB
We have added newly generated RNA-Seq data for O. brevispinum [16], a common brittle star found in shallow waters of the western Atlantic Ocean ranging from Canada to Venezuela. This resource can be found in EchinoDB under the name “OphiuroidDB”. We have also added the “EchinoidDB” resource that contains the high-quality genome assembly data of L. variegatus [20], a sea urchin found in shallow waters throughout the western Atlantic Ocean ranging from the United States to Venezuela. The rationale for creating these two new data resources is that there has been a growing use of these two species in recent molecular studies in developmental and regenerative biology [16, 20, 31, 48–52].
OphiuroidDB
We have provided the brittle star, O. brevispinum [22] transcriptome dataset, translated, and annotated using BLASTX [53] against the NCBI collection of predicted proteins of S. purpuratus [54] and protein models from UniProt’s Swiss-Prot [55] and NCBI’s RefSeq [56]. The application can be accessed via “Link to O. brevispinum transcriptome” in EchinoDB and is referred to as “OphiuroidDB”.
The transcriptome data of O. brevispinum were first used to characterize the downstream genes controlled by the Notch signaling pathway, which plays an important role in brittle star arm regeneration [16]. The raw sequencing reads of O. brevispinum transcriptome were submitted to the NCBI as a GEO dataset under the accession number GSE142391 [16, 57], and these sequences can now be also downloaded directly from OphiuroidDB. A total of 30,149 genes were identified, annotated, and included in the application.
EchinoidDB
EchinoidDB facilitates access to a recently published annotated high-quality chromosome-scale genome assembly of L. variegatus [20, 23]. The data (Lvar_3.0) includes 27,232 nucleotide and protein sequences, which were annotated using BLASTP [53] against UniProt Swiss-Prot [55], S. purpuratus [58] and non-S. purpuratus RefSeq invertebrate protein models [56]. These annotations can be downloaded from EchinoidDB.
Utility and discussion
Echinoderms are a phylum of marine invertebrate deuterostomes and thus share a deep common ancestor with vertebrates [59–61]. However, unlike most vertebrates, many echinoderm species can regenerate all their tissue types after injury without developing cancers [17]. The capacity of adult echinoderms to fully regrow lost or damaged parts of their body is among the strongest in the animal kingdom [62]. The highly regenerative body parts include the central nervous system, digestive tube, connective tissue, epidermis, muscles, endoskeleton, and coelomic epithelial structures [2, 7, 10, 63]. However, the genomic and transcriptomic resources currently available today on echinoderms are limited to only a small fraction of species within the phylum. Most importantly, this data availability bias does not reflect the natural diversity in regenerative capacities among echinoderms. For example, the understudied sea cucumbers (class Holothuroidea) regenerate most of their organs [10, 14, 64–68], whereas sea urchins (class Echinoidea), which have been the main focus of the sequencing and annotation efforts so far, are weak in regeneration [49]. The web information systems that are currently available include Echinobase [69], HpBase [70], and SpBase [71]. These databases allow for the querying and exploration of the biological data mostly related to sea urchin and hence, they are not suitable for capturing much of the diversity of the phylum Echinodermata. To illustrate further, the Echinobase information system [69] (https://www.echinobase.org/entry) contains genomic information for eight echinoderm species, five of which are sea urchins – Strongylocentrotus purpuratus (purple sea urchin), Strongylocentrotus fransciscanus (red sea urchin), Allocentrotus fragilis (sea urchin), L. variegatus (green sea urchin), Patiria miniata (bat star), Parastichopus parvimensis (warty sea cucumber), Ophiothrix spiculata (spiny brittle star), and Eucidaris tribuloides (slate pencil urchin). Another commonly used resource, SpBase [71] (https://spbase.org/) is a system of databases that is mostly focused on sea urchin species and contains genomic information of Strongylocentrotus purpuratus, Strongylocentrotus franciscanus, Allocentrotus fragilis, and L. variegatus. Lastly, HpBase [70] contains genomic and transcriptomic information of a single sea urchin species, Hemicentrotus pulcherrimus. In contrast, EchinoDB contains biological data for 42 different echinoderm species representing all five echinoderm classes, in addition to transcriptomic and genomic data for O. brevispinum and L. variegatus. Thus, EchinoDB serves as a valuable information resource to represent the diversity within the phylum and facilitate studies of regenerative phenomenon that varies widely among echinoderms.
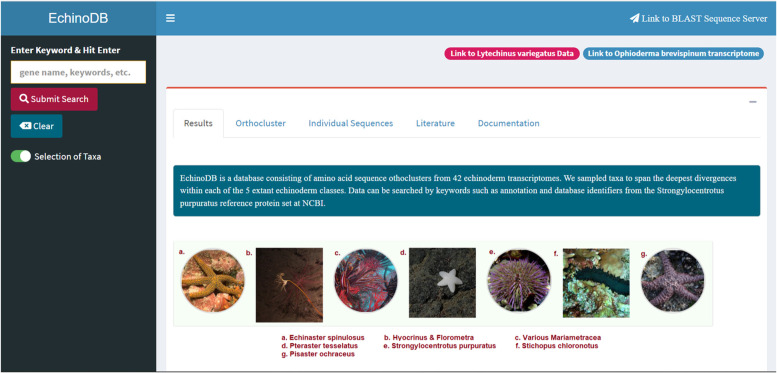
In the latest EchinoDB release, we added a text box that allows users to conduct searches using National Center for Biotechnology Information (NCBI) accession numbers and other keywords with or without the use of wildcard entries. Results include protein sequence(s), annotated description(s), known NCBI GenInfo Identifier (GI ids), and orthocluster(s). The annotations are assigned based on alignment of our sequences to the well-characterized protein sequence dataset of Strongylocentrotus purpuratus (i.e., sequences attributed to taxon 7668 in NCBI’s RefSeq, accessed in August 2012). These results can be further filtered by name or GenInfo Identifier (GI ids) in the search box in the top right corner. Additionally, users are able to expand or narrow their search based on taxonomic class, order, and family via toggle switches. Figure 1 depicts the design created in R Shiny for the EchinoDB application. Each row of the result table represents an orthocluster with the sequence similarity count or total hits. The number of hits is clickable, facilitating the viewing and downloading of related amino acid and nucleotide sequences in FASTA format.
Fig. 1.
Screenshot of the EchinoDB landing page [18], available at https://echinodb.uncc.edu. Users can search against all echinoderm classes, orders, and families or un-toggle to retrieve information for a particular taxon
Use case examples
To demonstrate the utility of EchinoDB v2.0 and associated resources, we used them to retrieve genes associated with the Notch [72] and Wnt [73] signaling pathways. This is a biologically relevant example, as both these pathways are required for regeneration in echinoderms [16, 32]. Knowledge of the Notch and Wnt signaling pathways is important because they are highly conserved in the animal kingdom and regulate a variety of cellular processes, including proliferation, differentiation, fate specification, and cell death [74–77]. Recent studies indicate that inhibiting the Notch signaling pathway prevented the brittle stars from fully regenerating their arms [16, 31]. Furthermore, Wnt signaling pathway is a major regulator of development throughout the animal kingdom. This pathway plays an important role in early regenerative events, including cell division, cell dedifferentiation and apoptosis that contribute to intestinal regeneration in holothurians [62, 78–83]. For example, in sea cucumber Apostichopus japonicus, Wnt6, Wnt7 (Wnt gene family), Fzd7 (Frizzled gene family), and Dvl (Dishevelled gene family) are all significantly upregulated during the early stages of intestinal regeneration [28, 29]. Similarly, in Holothuria glaberrima, Wnt9 is upregulated in early intestinal primordium [30]. Expression knockdown of Wnt7 and Dvl significantly inhibits intestinal regrowth in sea cucumbers, implying that the canonical Wnt signaling is essential for visceral regeneration [29].
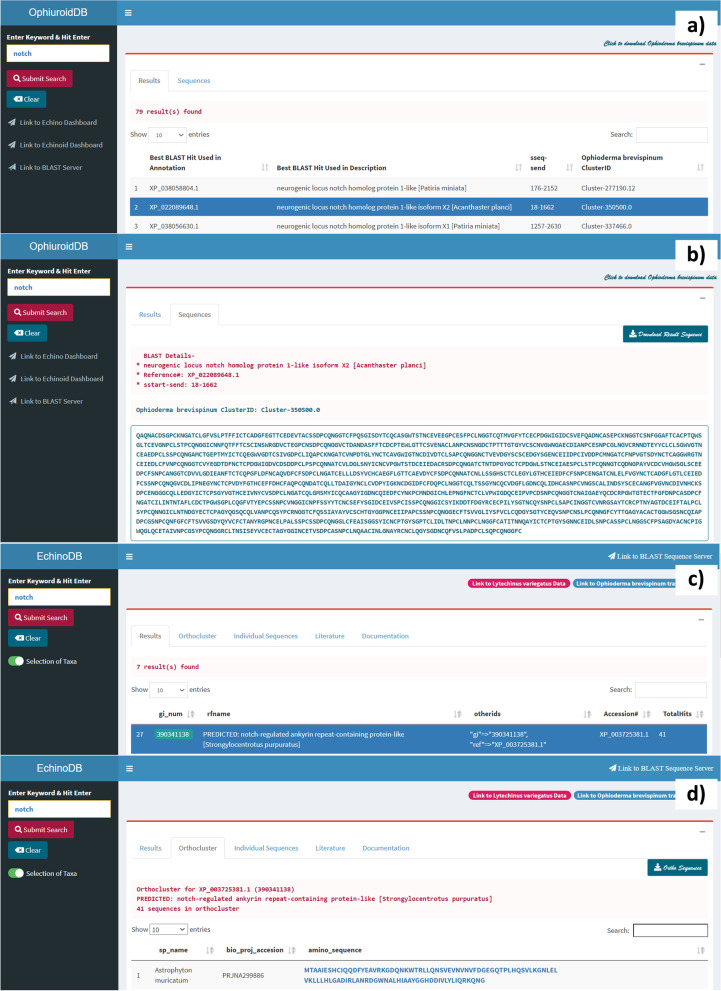
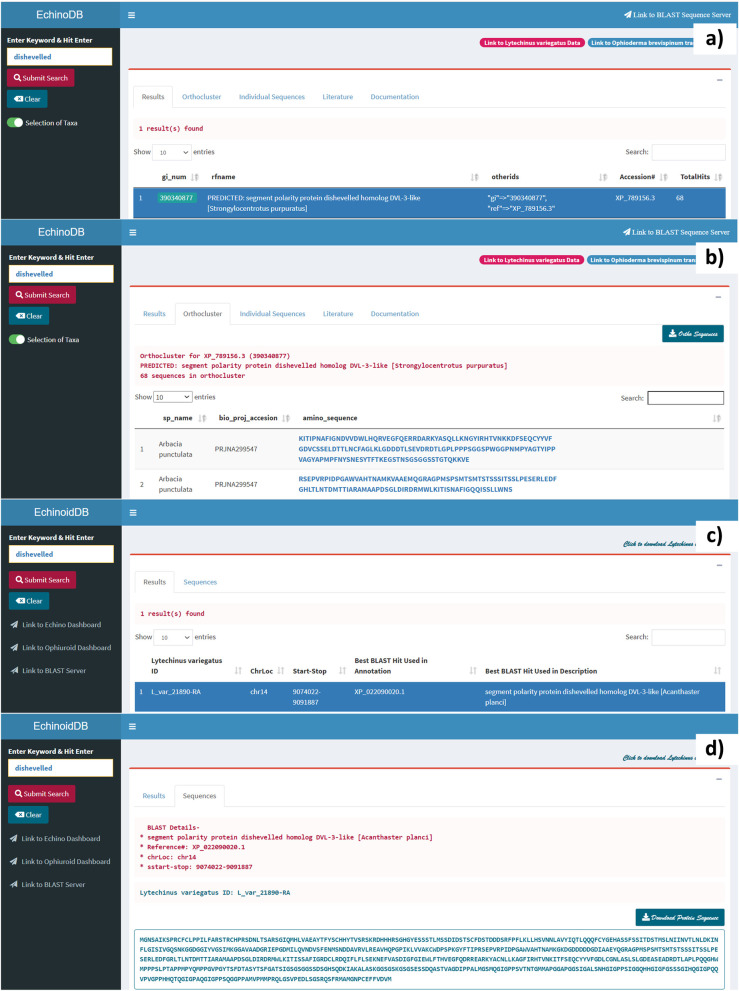
Figure 2 demonstrates the function of EchinoDB v2.0 and some of its outputs. The figure depicts the step-by-step process by locating individual sequences or clusters of Notch-related amino acid sequences in brittle stars and other echinoderms. For example, the user can search EchinoDB for Notch-related genes and obtain the corresponding sequences and metadata from our web resources. To do this, the user can search for the keyword “Notch” in our web resources to locate Notch-related sequences in brittle stars and other echinoderms. The results include NCBI’s accession numbers, other unique identifiers, descriptions of the gene or scaffold, start and end positions of regions of the gene or scaffold, and other details depending on the application used. For the keyword “notch”, a total of 432 amino sequences distributed throughout 7 orthoclusters were found in EchinoDB (amino acid sequence orthoclusters of 42 echinoderm transcriptomes), 54 in OphiuroidDB (transcriptomic data for the brittle star O. brevispinum), and 38 in EchinoidDB (genomic and peptide sequences for the green sea urchin L. variegatus). Similarly, Fig. 3 illustrates the step-by-step process of obtaining the corresponding sequences and metadata for “dishevelled” gene (Dvl) associated with the Wnt signaling pathway from our web resources. A total of 68 amino acid sequences found for “dishevelled” gene, grouped into a single orthocluster (XP_789156.3) in EchinoDB, four sequences were retrieved from OphiuroidDB and one from EchinoidDB. The search results corresponding to canonical Wnt and Notch signaling pathways are summarized in Tables 2 and 3.
Fig. 2.
Usage example illustrating the search for Notch-related sequences in the brittle star O. brevispinum and other echinoderms. a Screenshot of the OphiuroidDB main page (https://echinodb.uncc.edu/BStarApp/) [22]. The image shows the results after searching for the keyword “Notch” against the database of the brittle star O. brevispinum. The interface allows the selection of any record on the results page to view the sequence. b Representative amino acid sequence from one selected Notch-related gene in OphiuroidDB. c Results after searching for the keyword “Notch” in EchinoDB (https://echinodb.uncc.edu) [18]. In this example, the search was conducted against the repository of clusters of orthologous genes discovered from echinoderm transcriptomes. A selected record will be highlighted, and amino acid sequences from the orthocluster repository will be displayed. d Amino acid sequence clusters of the selected orthologous record of the Notch-related gene from the EchinoDB repository
Fig. 3.
A use case illustrating the retrieval of the “dishevelled” gene from EchinoDB that contains orthocluster data from 42 different echinoderm species and EchinoidDB that contains biological data of the green sea urchin L. variegatus. Dishevelled (Dvl) gene functions as a principal component of the Wnt signaling pathway that governs several cellular processes, including cell proliferation, cell differentiation, and apoptosis or cell death. a Results after searching for the keyword “dishevelled” in EchinoDB (https://echinodb.uncc.edu) [18]. In this example, the search was conducted against the repository of clusters of orthologous genes discovered from echinoderm transcriptomes. A selected record will be highlighted, and amino acid sequences from the orthocluster repository will be displayed. b Displays amino acid sequence clusters of the selected orthologous record of the “dishevelled” gene group from the EchinoDB repository. c Screenshot of the EchinoidDB main page (https://echinodb.uncc.edu/SUrchinApp/) [23]. The image shows the results after searching for the keyword “dishevelled” against the database of the green sea urchin L. variegatus. The interface allows the selection of any record on the results page to view the sequence. d Example amino acid sequence from selected record in EchinoidDB
Table 2.
Key components of the Wnt signaling pathway retrieved from the database. For each gene, we list the gene name, the gene group it belongs to, and its role in the pathway. In addition, for each resource – EchinoDB, EchinoidDB, and OphiuroidDB – we show the number of sequences retrieved using keyword and BLAST search. Column search type represents K for keyword search and B for BLAST search. Number 0 indicates that no sequence was found in the database for that gene
| Group | Gene Function | Name | Search Type | EchinoDB | EchinoidDB | OphiuroidDB | Cite |
|---|---|---|---|---|---|---|---|
| Regulator | Negative regulator. Part of the β-catenin destruction complex | APC | B | 32 | 2 | 1 | [73] |
| Regulator | Negative regulator. Part of the β-catenin destruction complex | Axin | K | 54 | 1 | 2 | [73] |
| Regulator | Phosphorylates β-catenin and the cytoplasmic tail of LRP. Part of the β-catenin destruction complex | CK1 | B | 225 | 4 | 2 | [73] |
| Regulator | Negative regulator. Binds to LRP | Dickkopf | B | 2 | 2 | 2 | [73] |
| Regulator | Mediates the recruitment of Axin to the plasmalemma in the ON state of the pathway | Dishevelled | K | 68 | 1 | 4 | [73] |
| Receptor | Wnt receptors | Frizzled | B | 500 | 8 | 7 | [73, 84] |
| Regulator | Negative regulator. Transcriptional co-repressor. Binds to TCF in the OFF state of the pathway | Groucho | K | 330 | 6 | 3 | [73] |
| Regulator | Phosphorylates β-catenin and the cytoplasmic tail of LRP. Part of the β-catenin destruction complex | GSK3 | K | 56 | 1 | 0 | [73] |
| Receptor | Dickkopf receptor. Mediates repression of the Wnt pathway | Kremen | B | 413 | 45 | 49 | [73, 85] |
| Regulator | Pathway enhancer. Receptor for R-spondin | Lgr5 | B | 500 | 1 | 4 | [73] |
| Receptor | Wnt co-receptor | LRP | B | 165 | 11 | 9 | [73] |
| Ligand | Alternative ligand for the Wnt receptors | Norrin | B | 3 | 0 | 0 | [73] |
| Regulator | Negative regulator. Inactivates Wnt in the extracellular space through enzymatic action | Notum (Wingful) | K | 43 | 1 | 0 | [73, 86] |
| Modifier | Palmitoyl transferase, attaches palmitoleic acid to Wnt | Porcupine | K | 23 | 1 | 1 | [73] |
| Regulator | Negative regulator. Wnt target gene | Rnf43 | B | 189 | 7 | 9 | [73] |
| Regulator | Pathway enhancer | R-spondin | K | 12 | 1 | 1 | [73] |
| Regulator | Negative regulator. Binds to LRP | Sclerostin | K | 17 | 1 | 1 | [73] |
| Regulator | Negative regulators. Sequester Wnts in the extracellular space | sFRPs | B | 153 | 4 | 4 | [84] |
| Transcription Factor | Transcriptional factors regulated by the Wnt pathway. Repress the target genes in the OFF state. Acitvate transctiption of the same genes in the ON state | TCF/Lef | K | 48 | 2 | 1 | [73] |
| Receptor | Norrin-specific co-receptor | Tspan12 | B | 146 | 5 | 4 | [73] |
| Ligand | Paracrine/juxtacrine signaling molecules | Wnt | K | 10 | 10 | 7 | [73, 84] |
| Auxiliary protein | Specific intracellular transporter of Wnts | Wntless/Evi (Wls) | K | 44 | 2 | 1 | [73] |
| Regulator | Negative regulator. Wnt target gene | Znrf3 | B | 234 | 1 | 1 | [73] |
| Regulator | Main modulator of the pathway | β-catenin | K | 255 | 3 | 1 | [73] |
| Regulator | Ubiquitinates the phosphorylate β-catenin thus targeting it for proteosomal destruction | β-TrCP | B | 33 | 5 | 2 | [73] |
Table 3.
Key components of the Notch signaling pathway retrieved from the database. Each line corresponds to the gene name, gene group, and its role in the pathway. In addition, we list the number of sequences retrieved from EchinoDB, EchinoidDB and OphiuroidDB using “keyword” and “BLAST” search. Column search type represents K for keyword search and B for BLAST search. Number 0 represents no sequence found in the corresponding database for that gene
| Group | Gene Function | Name | Search Type | EchinoDB | EchinoidDB | OphiuroidDB | Cite |
|---|---|---|---|---|---|---|---|
| Motif | A disintegrin and metalloproteinase with thrombospondin motifs | ADAM 10/17 | K | 344 | 2 | 83 | [72] |
| Receptor | Receptor proteolysis | Presenilin 1 | K | 77 | 2 | 3 | [16, 72] |
| Transcription factor | HES-4-like | HES | K | 46 | 3 | 3 | [16] |
| Auxiliary protein | Mastermind-like protein. Co-activator of RBP-J | Mastermind | B | 122 | 8 | 6 | [72, 87] |
| Enzyme | E3 ubiquitin-protein ligase | Mindbomb | K | 282 | 188 | 367 | [16] |
| Protein coding | Notch Activation Complex Kinase. Co-activator of RBP-J | NACK | K | 68 | 3 | 6 | [16] |
| Transcription factor | CREB-binding protein. Co-activator of RBP-J | p300 | K | 103 | 2 | 2 | [16] |
| Receptor | Neurogenic locus notch | Notch | K | 14 | 35 | 50 | [72, 88] |
| Receptor | Receptor proteolysis | Nicastrin | K | 68 | 1 | 1 | [72, 89] |
| Regulator | Negative regulator of the Notch pathway | Numb | K | 68 | 0 | 1 | [89] |
| Regulator | Context-dependent positive or negative regulator | Notchless | K | 200 | 1 | 10 | [90] |
| Regulator | Neuronal precursor cell-Expressed. Targets Notch and Deltex for degradation | Nedd4 | K | 94 | 2 | 2 | [91] |
| Regulator | E3 ubiquitin-protein ligase/ DTX1. Context-dependent positive or negative regulator. Antagonizes Nedd4 | Deltex | K | 139 | 0 | 0 | [74, 91] |
| Transcription factor | Mesoderm posterior bHLH transcription factor 2. Activates Fringe, induces degradation of Mastermind | Mesp2 | B | 6 | 2 | 1 | [88] |
| Ligand | Ubiquitination of Jagged | Neuralized | K | 159 | 6 | 26 | [16] |
| Receptor | Ligand of the notch receptor | Delta/Serrate (Jagged) | K | 68 | 2 | 2 | [16] |
| Transcription factor | CBF1/ Recombination signal binding protein for immunoglobulin kappa J region. Transcription factor activated by Notch | RBP-J | K | 1 | 1 | 0 | [16, 72] |
| Regulator | Numb-associated kinase. Positive regulator of the Notch pathway | NAK | B | 500 | 39 | 78 | [16, 72] |
| Activator | Acyl-CoA-Binding Domain-Containing Protein 3. Activator of Numb | ACBD3 | B | 132 | 1 | 1 | [16, 72] |
| Ligand | Ligand of Numb Protein 2. Negative regulator of Numb | LNX2 | K | 54 | 1 | 1 | [72, 87] |
| Protein | Hairy/enhancer-of-split related with YRPW motif protein 1. Canonical target gene. | HEY1 | K | 24 | 3 | 1 | [16] |
| Receptor | Paired basic amino acid cleaving enzyme. Receptor proteolysis | Furin | B | 342 | 6 | 11 | [16, 72] |
| Modifier | Protein O-glucosyltransferase. Post-translational maturation of Notch | Poglut | B | 500 | 85 | 46 | [16, 72] |
| Modifier | Protein O-fucosyltransferase 1. Post-translational maturation of Notch | POFUT1 | K | 191 | 5 | 1 | [16, 72] |
| Modifier | beta-1,3-N-acetylglucosaminyltransferase radical fringe/ Lfng (lunatic) or Rfng (Radical). Post-translational maturation of Notch | Fringe | K | 85 | 4 | 3 | [16] |
| Repressor | SHARP/ spen family transcriptional repressor/ Mint/Sharp/SPEN, NCoR/SMRT, KyoT2. Co-repressor of RBP-J | MINT | B | 95 | 1 | 3 | [16, 72] |
| Repressor | Histone deacetylase 1. Co-repressor of RBP-J | HDAC1 | K | 220 | 1 | 0 | [72] |
| Repressor | Nuclear receptor corepressor. Co-repressor of RBP-J | NCoR | B | 77 | 2 | 0 | [16, 72] |
| Repressor | Co-repressor interacting with RBP-J | CIR1 | B | 53 | 2 | 1 | [16, 72] |
In Table 2, we list the components of the canonical Wnt signaling pathway that were searched for in EchinoDB via a “keyword” search function. A number of orthoclusters, genomic, transcriptomic, and peptide sequences were found using this approach. A numerical value of 0 in the table indicates that no hits were returned when a particular gene name was used as a query for a “keyword” search in the database. However, the value 0 immediately raises a question: why are the sequences missing in our databases? For example, no matches are found in EchinoDB, when gene names “Kremen” and “Norrin” were used as keywords. Is it a limitation of the keyword search approach, a failure in annotation, or a true absence of homologs in EchinoDB? To answer this question, we conducted a test study, in which we performed a BLAST search (e-value cutoff 1e-06) [27, 26], instead of keyword search. For all the genes that were not retrieved by keyword search approach, we used reference sequences from the UniProt database [55] as a query in the BLAST search interface of EchinoDB. In all the cases, the genes that were not retrieved by keyword search were retrieved by BLAST search. Hence, in a case study of retrieving components of the Wnt signaling pathway, BLAST search and keyword search turned out to be two complementary strategies, with the former being more sensitive and the latter being faster but dependent on annotation quality of underlying data.
Another use case involved retrieving major components of the Notch pathway (i.e., the Notch receptor, the Delta and Serrate ligands, the transcriptional regulator RBPJ, two Notch target genes of the Hes family, and pathway modulators) [16, 72, 88–90]. As above, two complementary approaches were used to find all selected components of the Notch signaling pathway. First, we used a keyword search to retrieve sequences of all those genes of interest from EchinoDB and associated databases. Second, we used SequenceServer (BLAST) functionality in EchinoDB [33] to retrieve putative homologous sequences for the genes that were not retrieved by keyword search. The results of the keyword search and BLAST search are summarized in Table 3. Thus, BLAST search combined with keyword search proved useful in retrieving all major components of the Notch signaling pathway.
EchinoDB can also be used to expand our understanding of the clade-specific biology. For example, biomineralization contributes to the development of the stereome-type endoskeleton unique to echinoderms. Biomineralization is defined as the biologically controlled formation of mineral deposits resulting in structures that function as support, protection, or feeding anatomy [34]. Among echinoderms, biomineralization is best characterized in sea urchins [4]. Hence, we ask if we can use our database to obtain an insight on whether the biomineralization mechanisms described in echinoids are unique to that class or shared across the phylum. To this end, we leveraged the SequenceServer (BLAST search) functionality available within EchinoDB.
Among the proteins involved in biomineralization are spicule matrix proteins. In sea urchins, these secreted proteins are contained within the spicule and closely associated with the mineral component [4]. They have been shown to facilitate all aspects of endoskeleton formation, including nucleation of the crystal formation, as well as control of the orientation, shape and chemical purity of the resulting skeletal structure [35–37]. The spicule matrix protein family consists of nine members, including the most extensively studied SpSM50 and SpSM30B/C [4]. We used SequenceServer (BLAST) integrated in EchinoDB [33] with a cutoff e-value of 1e-06 to compare the amino acid sequences of the echinoid spicule matrix proteins against EchinoDB (42 species), OphiuroidDB (O. brevispinum) and EchinoidDB (L. variegatus). Table 4 lists a number of echinoid and non-echinoid species represented in EchinoDB that had a BLAST match to each of those nine reference echinoid spicule matrix proteins. All nine proteins had a putative ortholog in at least one non-echinoid class, which suggests that the skeletogenesis mechanisms discovered in sea urchins might be also shared by other members of the phylum.
Table 4.
Spicule matrix proteins retrieved from the database. Each line corresponds to individual proteins, for which we list accession numbers of corresponding reference sequences from the NCBI, GenBank or UniProt databases. The numerical values in the table represent the number of species in each class of the phylum that had a BLAST match to the reference sequence
| DataBase (Accession) | Protein | Description | Asteroidea | Ophiuroidea | Echinoidea | Holothuroidea | Crinoidea |
|---|---|---|---|---|---|---|---|
| NCBI (NP_999775.2) | SpSM50 | 50 kDa spicule matrix protein precursor [Strongylocentrotus purpuratus] | 2 | 1 | 4 | 0 | 0 |
| NCBI (NP_999776.1) | SpSM37 | spicule matrix protein SM37 precursor [Strongylocentrotus purpuratus] | 0 | 1 | 3 | 6 | 1 |
| NCBI (NP_999803.1) | SpSM32 | spicule matrix protein SM32 precursor [Strongylocentrotus purpuratus] | 2 | 2 | 4 | 3 | 1 |
| UniProt (P28163/SM30_STRPU) | SpSM30B/C | 30 kDa spicule matrix protein precursor [Strongylocentrotus purpuratus] | 4 | 1 | 4 | 1 | 0 |
| NCBI (NP_999804.1) | SpSM29 | spicule matrix protein SM29 precursor [Strongylocentrotus purpuratus] | 2 | 0 | 4 | 0 | 0 |
| GenBank (CAA42179.1) | LSM34 | spicule matrix 34 kd protein [Lytechinus pictus] | 2 | 2 | 4 | 1 | 0 |
| UniProt (Q25116) | HSM30 | 30 kDa spicule matrix protein [Hemicentrotus pulcherrimus] | 1 | 1 | 4 | 0 | 0 |
| UniProt (Q26264) | HSM41 | 41 kDa spicule matrix protein [Hemicentrotus pulcherrimus] | 2 | 2 | 4 | 0 | 0 |
| UniProt (Q95W96) | PM27 | Primary mesenchyme-specific protein [Heliocidaris erythrogramma] | 1 | 3 | 3 | 3 | 0 |
Another echinoderm-specific phenomenon is the capacity of the connective tissue structures to rapidly change their tensile strength under the control of the central nervous system [5, 41, 42]. A subset of neurosecretory cells is thought to release proteins that can either stiffen or soften the extracellular collagenous matrix. Only one of such effector molecules, the TIMP-like protein tensilin has been characterized so far at the sequence level [92]. Tensilin, upon its release from the neurosecretory cells, stiffens the mutable collagenous tissue [39, 41, 42, 93]. Only three sequences are known thus far, all of them from members of the class Holothuroidea, including sea cucumbers Cucumaria frondosa [39], Apostichopus japonicus [94], and Holothuria forskali [40]. We therefore asked if tensilin, and thus tensilin-induced stiffening mechanisms, are unique to holothurians or are they represented in other classes of the phylum. To this end, we used the published protein and nucleotide sequences of tensilin as a query to perform BLASTP (for amino acid sequence) and BLASTX (for the nucleotide sequences) searches with an e-value threshold of 1e-06 [33]. This allows us to find potential homologs in species from all five echinoderm classes represented in our database, EchinoDB. The BLAST results are summarized in Table 5. They suggest that the tensilin protein, and thus the molecular mechanisms controlling the tensile strength of the mutable collagenous tissue, might be conserved across the phylum. This result is interesting groundwork for further study.
Table 5.
Tensilin proteins. The first row corresponds to protein accession number from UniProt database whereas, second and third row depict nucleotide accession numbers from NCBI databases. The numerical values in the table represent the number of species in each class of the phylum that had a BLAST match to the reference sequence
| DataBase (Accession) | Description | Asteroidea | Ophiuroidea | Echinoidea | Holothuroidea | Crinoidea |
|---|---|---|---|---|---|---|
| UniProt (Q962H0) | Tensilin [Cucumaria frondosa] | 8 | 1 | 1 | 9 | 3 |
| NCBI (KR002726.1) | Apostichopus japonicus tensilin mRNA, complete cds | 5 | 1 | 2 | 9 | 0 |
| NCBI (KY609179.1) | Holothuria forskali tensilin mRNA, complete cds | 9 | 1 | 2 | 9 | 0 |
Finally, the database interface of EchinoDB allows the user to visualize any selected individual sequence or cluster of sequences or download them in FASTA format from the related repository. The downloaded sequences from EchinoDB v2.0 and associated resources can be used in downstream analyses (e.g. BRAKER [95, 96] or BLAST search for gene prediction and annotation in the draft genome of a newly sequenced echinoderm species). Alternatively, the sequences for any specific gene pathway from EchinoDB for example, Notch or Wnt, can be used in NCBI’s Conserved Domain Search (www.ncbi.nlm.nih.gov/Structure/cdd) to identity conserved protein domains in the sequences. The identified conserved domains can facilitate annotation of functionally unknown protein sequences. Hence, the above use cases illustrate how EchinoDB [18] in association with OphiuroidDB [22] and EchinoidDB [23] can be used to retrieve the gene sequences for cell signaling pathways essential in regeneration and facilitate better understanding of genomic underpinnings of phylum-specific biological phenomena. Further, EchinoDB can be used for sequence-similarity-based clustering analysis to get an insight about the conservation of various molecular components across echinoderms.
Application features within updated EchinoDB
As many “omic” data for echinoderms are not yet well annotated, blast search is an important complement to keyword or accession search.
Using Sequenceserver to run BLAST
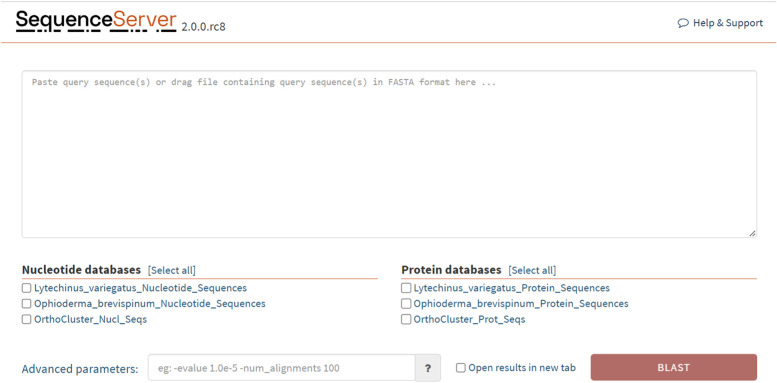
The updated EchinoDB contains an instance of Sequenceserver [27], a web-based BLAST server that supports sequence similarity searches against nucleotide and protein sequence databases. EchinoDB provides nucleotide and protein databases to be queried against user provided sequences to facilitate sequence similarity searches using default or user-selected parameters.
Integration with BLAST allows users of EchinoDB to search data resources with strings of the query sequence. Figure 4 illustrates Sequenceserver for BLAST functionality and can be accessed via “Link to BLAST Sequence Server” in the EchinoDB v2.0 application.
Fig. 4.
Screenshot of our Sequenceserver integrated in EchinoDB [33]. Users can perform BLAST searches against nucleotide and protein sequences of included datasets in the application via https://echinodb.uncc.edu/sequenceserver/
Literature
We provide a repository that contains links to many of the research papers associated with EchinoDB by their title. The literature repository is updated regularly.
Additional data
A link is added in the Literature section to allow users to download data associated with papers. For example, one dataset provides evidence that Xyloplax sp. is a velatid (an order within the class Asteroidea) asteroid rather than a new class [21]. The data included in EchinoDB includes tables and phylogenomic data from large amounts of transcriptome data used in this paper. The additional data repository is updated regularly.
Usage and documentation
EchinoDB, EchinoidDB, and OphiuroidDB user manuals (Additional files 2, 3 and 4: Files S1–3, respectively) are available in a tab named “Documentation” in the EchinoDB website. The user manuals are downloadable and provide instructions with screenshots to assist the user in navigating through the application.
Conclusions
The updated EchinoDB provides, via a cloud-based server, additional tools and data from collaborations and our lab that can be of interest to a variety of scientific communities. One of our focal points in the future is to extend the genomic, transcriptomic, and orthocluster contents of EchinoDB.
Supplementary Information
Additional file 1: Table S1. Raw reads from the various echinoderm species are available in NCBI’s SRA and is also available at Zenodo (doi: https://doi.org/10.5281/zenodo.6985492).
Additional file 2: File S1. EchinoDB user manual contains screenshots of the outputs to assist new users with the features and functionality of the application.
Additional file 3: File S2. EchinoidDB user manual contains instructions to help users with the resources and operations available in the application.
Additional file 4: File S3. OphiuroidDB user manual to describe operations and capabilities of the application.
Additional file 5: File S4. Source code (in R) for EchinoDB, EchinoidDB, and OphiuroidDB. We have also provided three R scripts one for each app.
Acknowledgements
We acknowledge the support of several entities of the University of North Carolina at Charlotte including the College of Computing and Informatics, the Graduate School, the Department of Bioinformatics and Genomics, University Research Computing, and the Bioinformatics Research Center. Additionally, we thank Benjamin Stalcup for issuing a SSL certificate for EchinoDB application and Steven Blanchard for helping us with certificates deployments and instructions for web server setup. We also thank Shantoy Hansel and Jan Kofsky for their testing of the application. Finally, we thank Greg Wray for providing the genomic data of the green sea urchin, L. variegatus [20].
Abbreviations
- BLAST
Basic Local Alignment Search Tool
- AWS
Amazon Web Services
- NCBI
National Center for Biotechnology Information
- GEO
Gene Expression Omnibus
- HTML
HyperText Markup Language
- CSS
Cascading Style Sheets
- RNA-Seq
RNA Sequencing
- RefSeq
NCBI Reference Sequence Database
- FASTA
FAST All
Authors’ contributions
VM, VMA, RR, DJM and DJ: manuscript preparation and revision, data analyses, and annotation. DJ: funding acquisition. VM: BLAST and Sequenceserver implementation, source code, database curation, cloud setup and server maintenance. VMI and DJ: user interface, application design, and usability. VMA provided the transcriptome data for O. brevispinum. All authors read and approved the final manuscript.
Funding
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R15 GM128066–01.
Availability of data and materials
Assembled sequences and orthoclusters are available in EchinoDB (https://echinodb.uncc.edu) [18]. Raw reads from the various echinoderm species are available in NCBI’s SRA (see accession numbers in Additional file 1: Table S1). Additionally, the user manuals and code for EchinoDB v2.0, EchinoidDB, and OphiuroidDB are available as Additional file 2: File S1, Additional file 3: File S2, Additional file 4: File S3, and Additional file 5: File S4, respectively. Additional files are available in Zenodo (doi: 10.5281/zenodo.6985492).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fell HB. Textbook of zoology. London: Palgrave; 1972. Phylum Echinodermata; pp. 776–837. [Google Scholar]
- 2.Mashanov VS, Zueva OR, García-Arrarás JE. Transcriptomic changes during regeneration of the central nervous system in an echinoderm. BMC Genomics. 2014;15(1):1–21. doi: 10.1186/1471-2164-15-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wray GA. Echinodermata. Spiny-skinned animals: sea urchins, starfish, and their allies. Tree of Life. 1999. [Google Scholar]
- 4.Pendola M, Jain G, Evans JS. Skeletal development in the sea urchin relies upon protein families that contain intrinsic disorder, aggregation-prone, and conserved globular interactive domains. PLoS One. 2019;14(10):e0222068. doi: 10.1371/journal.pone.0222068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkie IC, Sugni M, Gupta HS, Carnevali MC, Elphick MR. The mutable collagenous tissue of echinoderms: from biology to biomedical applications. 2021. pp. 1–33. [Google Scholar]
- 6.Bely AE, Nyberg KG. Evolution of animal regeneration: re-emergence of a field. Trends Ecol Evol. 2010;25(3):161–170. doi: 10.1016/j.tree.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Dupont S, Thorndyke M. Bridging the regeneration gap: insights from echinoderm models. Nat Rev Genet. 2007;8(4):320. doi: 10.1038/nrg1923-c1. [DOI] [Google Scholar]
- 8.Ben Khadra Y, Sugni M, Ferrario C, Bonasoro F, Oliveri P, Martinez P, Candia Carnevali MD. Marine organisms as model systems in biology and medicine. Cham: Springer; 2018. Regeneration in stellate echinoderms: Crinoidea, Asteroidea and Ophiuroidea; pp. 285–320. [DOI] [PubMed] [Google Scholar]
- 9.Candia Carnevali MD, Bonasoro F. Introduction to the biology of regeneration in echinoderms. Microsc Res Tech. 2001;55(6):365–368. doi: 10.1002/jemt.1184. [DOI] [PubMed] [Google Scholar]
- 10.Carnevali MC. Regeneration in echinoderms: repair, regrowth, cloning. Invertebr Surviv J. 2006;3(1):64–76. [Google Scholar]
- 11.Drager BJ, Harkey MA, Iwata M, Whiteley AH. The expression of embryonic primary mesenchyme genes of the sea urchin, Strongylocentrotus purpuratus, in the adult skeletogenic tissues of this and other species of echinoderms. Dev Biol. 1989;133(1):14–23. doi: 10.1016/0012-1606(89)90292-3. [DOI] [PubMed] [Google Scholar]
- 12.Dubois P, Ameye L. Regeneration of spines and pedicellariae in echinoderms: a review. Microsc Res Tech. 2001;55(6):427–437. doi: 10.1002/jemt.1188. [DOI] [PubMed] [Google Scholar]
- 13.Heatfield BM, Travis DF. Ultrastructural studies of regenerating spines of the sea urchin Strongylocentrotus purpuratus I. cell types without spherules. J Morphol. 1975;145(1):13–49. doi: 10.1002/jmor.1051450103. [DOI] [PubMed] [Google Scholar]
- 14.García-Arrarás JE, Estrada-Rodgers L, Santiago R, Torres II, Díaz-Miranda L, Torres-Avillán I. Cellular mechanisms of intestine regeneration in the sea cucumber, Holothuria glaberrima Selenka (Holothuroidea: Echinodermata) J Exp Zool. 1998;281(4):288–304. doi: 10.1002/(SICI)1097-010X(19980701)281:4<288::AID-JEZ5>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 15.Miguel-Ruiz S, José E, Maldonado-Soto AR, García-Arrarás JE. Regeneration of the radial nerve cord in the sea cucumber Holothuria glaberrima. BMC Dev Biol. 2009;9(1):1–9. doi: 10.1186/1471-213X-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mashanov V, Akiona J, Khoury M, Ferrier J, Reid R, Machado DJ, Zueva O, Janies D. Active notch signaling is required for arm regeneration in a brittle star. PLoS One. 2020;15(5):e0232981. doi: 10.1371/journal.pone.0232981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medina-Feliciano JG, García-Arrarás JE. Regeneration in echinoderms: molecular advancements. Front Cell Dev Biol. 2021;9. 10.3389/fcell.2021.768641. [DOI] [PMC free article] [PubMed]
- 18.EchinoDB Database. https://echinodb.uncc.edu. Accessed 23 Dec 2021.
- 19.Janies DA, Witter Z, Linchangco GV, Foltz DW, Miller AK, Kerr AM, Jay J, Reid RW, Wray GA. EchinoDB, an application for comparative transcriptomics of deeply-sampled clades of echinoderms. BMC Bioinformatics. 2016;17:48. doi: 10.1186/s12859-016-0883-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davidson PL, Guo H, Wang L, Berrio A, Zhang H, Chang Y, Soborowski AL, McClay DR, Fan G, Wray GA. Chromosomal-level genome assembly of the sea urchin Lytechinus variegatus substantially improves functional genomic analyses. Genome Biol Evol. 2020;12(7):1080–1086. doi: 10.1093/gbe/evaa101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linchangco GV, Jr, Foltz DW, Reid R, Williams J, Nodzak C, Kerr AM, Miller AK, Hunter R, Wilson NG, Nielsen WJ, Mah CL. The phylogeny of extant starfish (Asteroidea: Echinodermata) including Xyloplax, based on comparative transcriptomics. Mol Phylogenet Evol. 2017;115:161–170. doi: 10.1016/j.ympev.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 22.OphiuroidDB Database. https://echinodb.uncc.edu/BStarApp/. Accessed 23 Dec 2021.
- 23.EchinoidDB Database. https://echinodb.uncc.edu/SUrchinApp/. Accessed 23 Dec 2021.
- 24.Shiny from RStudio. https://shiny.rstudio.com/. Accessed 31 Oct 2020.
- 25.Amazon Web Services. https://aws.amazon.com/. Accessed 10 Dec 2020.
- 26.Madden T. The BLAST sequence analysis tool. The NCBI handbook. 2003. [Google Scholar]
- 27.Priyam A, Woodcroft BJ, Rai V, Moghul I, Munagala A, Ter F, Chowdhary H, Pieniak I, Maynard LJ, Gibbins MA, Moon H. Sequenceserver: a modern graphical user interface for custom BLAST databases. Mol Biol Evol. 2019;36(12):2922–2924. doi: 10.1093/molbev/msz185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun LN, Yang HS, Chen MY, Xu DX. Cloning and expression analysis of Wnt6 and Hox6 during intestinal regeneration in the sea cucumber Apostichopus japonicus. Genet Mol Res. 2013;12(4):5321–5334. doi: 10.4238/2013.November.7.7. [DOI] [PubMed] [Google Scholar]
- 29.Yuan J, Gao Y, Sun L, Jin S, Zhang X, Liu C, Li F, Xiang J. Wnt signaling pathway linked to intestinal regeneration via evolutionary patterns and gene expression in the sea cucumber Apostichopus japonicus. Front Genet. 2019;10:112. doi: 10.3389/fgene.2019.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mashanov VS, Zueva OR, Garcia-Arraras JE. Expression of Wnt9, TCTP, and Bmp1/Tll in sea cucumber visceral regeneration. Gene Expr Patterns. 2012;12(1–2):24–35. doi: 10.1016/j.gep.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mashanov V, Machado DJ, Reid R, Brouwer C, Kofsky J, Janies DA. Twinkle twinkle brittle star, how I wonder what your genes are: Ophioderma brevispinum as a genomic resource for regeneration. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alicea-Delgado M, García-Arrarás JE. Wnt/β-catenin signaling pathway regulates cell proliferation but not muscle dedifferentiation nor apoptosis during sea cucumber intestinal regeneration. Dev Biol. 2021;480:105–113. doi: 10.1016/j.ydbio.2021.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.EchinoDB Sequenceserver Integration. https://echinodb.uncc.edu/sequenceserver/. Accessed 23 Dec 2021.
- 34.Livingston BT, Killian CE, Wilt F, Cameron A, Landrum MJ, Ermolaeva O, Sapojnikov V, Maglott DR, Buchanan AM, Ettensohn CA. A genome-wide analysis of biomineralization-related proteins in the sea urchin Strongylocentrotus purpuratus. Dev Biol. 2006;300(1):335–348. doi: 10.1016/j.ydbio.2006.07.047. [DOI] [PubMed] [Google Scholar]
- 35.Evans JS. The biomineralization proteome: protein complexity for a complex bioceramic assembly process. Proteomics. 2019;19(16):1900036. doi: 10.1002/pmic.201900036. [DOI] [PubMed] [Google Scholar]
- 36.Lowenstam HA, Weiner S. On Biomineralization. New York: Oxford University Press; 1989. pp. 1–134. [Google Scholar]
- 37.Mann S, Biomineralization. Principles and concepts in bioinorganic materials chemistry. New York: Oxford University Press; 2001. pp. 6–9. [Google Scholar]
- 38.Tamori M, Yamada A, Nishida N, Motobayashi Y, Oiwa K, Motokawa T. Tensilin-like stiffening protein from Holothuria leucospilota does not induce the stiffest state of catch connective tissue. J Exp Biol. 2006;209(9):1594–1602. doi: 10.1242/jeb.02178. [DOI] [PubMed] [Google Scholar]
- 39.Tipper JP, Lyons-Levy G, Atkinson MA, Trotter JA. Purification, characterization and cloning of tensilin, the collagen-fibril binding and tissue-stiffening factor from Cucumaria frondosa dermis. Matrix Biol. 2002;21(8):625–635. doi: 10.1016/s0945-053x(02)00090-2. [DOI] [PubMed] [Google Scholar]
- 40.Demeuldre M, Hennebert E, Bonneel M, Lengerer B, Van Dyck S, Wattiez R, Ladurner P, Flammang P. Mechanical adaptability of sea cucumber Cuvierian tubules involves a mutable collagenous tissue. J Exp Biol. 2017;220(11):2108–2119. doi: 10.1242/jeb.145706. [DOI] [PubMed] [Google Scholar]
- 41.Inoue M, Birenheide R, Koizumi O, Kobayakawa Y, Muneoka Y, Motokawa T. Localization of the neuropeptide NGIWYamide in the holothurian nervous system and its effects on muscular contraction. Proc R Soc Lond B Biol Sci. 1999;266(1423):993–1000. doi: 10.1098/rspb.1999.0735. [DOI] [Google Scholar]
- 42.Birenheide R, Tamori M, Motokawa T, Ohtani M, Iwakoshi E, Muneoka Y, Fujita T, Minakata H, Nomoto K. Peptides controlling stiffness of connective tissue in sea cucumbers. Biol Bull. 1998;194(3):253–259. doi: 10.2307/1543095. [DOI] [PubMed] [Google Scholar]
- 43.Nginx web server. https://www.nginx.com/resources/wiki/start/. Accessed 21 Jan 2021.
- 44.Henschel R, Lieber M, Wu LS, Nista PM, Haas BJ, LeDuc RD. Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the eXtreme to the campus and beyond. 2012. Trinity RNA-Seq assembler performance optimization; pp. 1–8. [Google Scholar]
- 45.Haas B, Papanicolaou A. Transdecoder. http://transdecoder.github.io/. Accessed 13 July 2022.
- 46.Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.NCBI Protein RefSeqs for taxon 7668. RefSeq assembly accession GCA_000002235.4, Spur_3.1. http://www.ncbi.nlm.nih.gov. Accessed Aug 2012.
- 48.Dupont S, Thorndyke MC. Growth or differentiation? Adaptive regeneration in the brittlestar Amphiura filiformis. J Exp Biol. 2006;209(19):3873–3881. doi: 10.1242/jeb.02445. [DOI] [PubMed] [Google Scholar]
- 49.Reinardy HC, Emerson CE, Manley JM, Bodnar AG. Tissue regeneration and biomineralization in sea urchins: role of notch signaling and presence of stem cell markers. PLoS One. 2015;10(8):e0133860. doi: 10.1371/journal.pone.0133860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bronstein O, Kroh A. The first mitochondrial genome of the model echinoid Lytechinus variegatus and insights into Odontophoran phylogenetics. Genomics. 2019;111(4):710–718. doi: 10.1016/j.ygeno.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 51.Czarkwiani A, Dylus DV, Oliveri P. Expression of skeletogenic genes during arm regeneration in the brittle star Amphiura filiformis. Gene Expr Patterns. 2013;13(8):464–472. doi: 10.1016/j.gep.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Czarkwiani A, Ferrario C, Dylus DV, Sugni M, Oliveri P. Skeletal regeneration in the brittle star Amphiura filiformis. Front Zool. 2016;13(1):1–7. doi: 10.1186/s12983-016-0149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10(1):1–9. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.The NCBI collection of predicted proteins of the sea urchin Stronglocentrotus purpuratus.ftp://ftp.ncbi.nih.gov/genomes/Strongylocentrotus_purpuratus/protein/. Accessed 26 Feb 2020.
- 55.Bateman A. Protein Science. Hoboken: Wiley; 2019. Uniprot: a universal hub of protein knowledge; p. 32. [Google Scholar]
- 56.O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geo Dataset; GSE142391. Active Notch Signaling is Required for Arm Regeneration in a Brittle Star. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142391. Accessed 15 Nov 2021. [DOI] [PMC free article] [PubMed]
- 58.Strongylocentrotus purpuratus Spur_5.0 Protein Models. www.ncbi.nlm.nih.gov/assembly/GCF_000002235.5. Accessed 26 Feb 2020.
- 59.Swalla BJ, Smith AB. Deciphering deuterostome phylogeny: molecular, morphological and palaeontological perspectives. Philos T R Soc B: Biol Sci. 2008;363(1496):1557–1568. doi: 10.1098/rstb.2007.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adoutte A, Balavoine G, Lartillot N, Lespinet O, Prud'homme B, De Rosa R. The new animal phylogeny: reliability and implications. Proc Natl Acad Sci U S A. 2000;97(9):4453–4456. doi: 10.1073/pnas.97.9.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blair JE, Hedges SB. Molecular phylogeny and divergence times of deuterostome animals. Mol Biol Evol. 2005;22(11):2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- 62.García-Arrarás JE, Dolmatov IY. Echinoderms: potential model systems for studies on muscle regeneration. Curr Pharm Des. 2010;16(8):942–955. doi: 10.2174/138161210790883426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mashanov VS, Zueva O, García-Arrarás JE. Postembryonic organogenesis of the digestive tube: why does it occur in worms and sea cucumbers but fail in humans? Curr Top Dev Biol. 2014;108:185–216. doi: 10.1016/B978-0-12-391498-9.00006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang X, Sun L, Yuan J, Sun Y, Gao Y, Zhang L, Li S, Dai H, Hamel JF, Liu C, Yu Y. The sea cucumber genome provides insights into morphological evolution and visceral regeneration. PLoS Biol. 2017;15(10):e2003790. doi: 10.1371/journal.pbio.2003790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rojas-Cartagena C, Ortíz-Pineda P, Ramírez-Gómez F, Suárez-Castillo EC, Matos-Cruz V, Rodríguez C, Ortíz-Zuazaga H, García-Arrarás JE. Distinct profiles of expressed sequence tags during intestinal regeneration in the sea cucumber Holothuria glaberrima. Physiol Genomics. 2007;31(2):203–215. doi: 10.1152/physiolgenomics.00228.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun L, Chen M, Yang H, Wang T, Liu B, Shu C, Gardiner DM. Large scale gene expression profiling during intestine and body wall regeneration in the sea cucumber Apostichopus japonicus. Comp Biochem Physiol Part D Genomics Proteomics. 2011;6(2):195–205. doi: 10.1016/j.cbd.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 67.Medina-Feliciano JG, Pirro S, García-Arrarás JE, Mashanov V, Ryan JF. Draft genome of the sea cucumber Holothuria glaberrima, a model for the study of regeneration. Front Mar Sci. 2021;8:603410. doi: 10.3389/fmars.2021.603410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.García-Arrarás JE, Lázaro-Peña MI, Díaz-Balzac CA. Holothurians as a model system to study regeneration. Mar Organ Model Syst Biol Med. 2018:255–83. 10.1007/978-3-319-92486-1_13. [DOI] [PubMed]
- 69.Kudtarkar P, Cameron RA. Echinobase: an expanding resource for echinoderm genomic information. Database. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kinjo S, Kiyomoto M, Yamamoto T, Ikeo K, Yaguchi S. HpBase: a genome database of a sea urchin, Hemicentrotus pulcherrimus. Develop Growth Differ. 2018;60(3):174–182. doi: 10.1111/dgd.12429. [DOI] [PubMed] [Google Scholar]
- 71.Cameron RA, Samanta M, Yuan A, He D, Davidson E. SpBase: the sea urchin genome database and web site. Nucleic Acids Res. 2009;37(suppl_1):D750–D754. doi: 10.1093/nar/gkn887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hurlbut GD, Kankel MW, Lake RJ, Artavanis-Tsakonas S. Crossing paths with Notch in the hyper-network. Curr Opin Cell Biol. 2007;19(2):166–175. doi: 10.1016/j.ceb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 73.Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169(6):985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 74.Gazave E, Lapébie P, Richards GS, Brunet F, Ereskovsky AV, Degnan BM, Borchiellini C, Vervoort M, Renard E. Origin and evolution of the notch signaling pathway: an overview from eukaryotic genomes. BMC Evol Biol. 2009;9:249. doi: 10.1186/1471-2148-9-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marlow H, Roettinger E, Boekhout M, Martindale MQ. Functional roles of notch signaling in the cnidarian Nematostella vectensis. Dev Biol. 2012;362(2):295–308. doi: 10.1016/j.ydbio.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Layden MJ, Martindale MQ. Non-canonical notch signaling represents an ancestral mechanism to regulate neural differentiation. Evodevo. 2014;5:30. doi: 10.1186/2041-9139-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Favarolo MB, López SL. Notch signaling in the division of germ layers in bilaterian embryos. Mech Dev. 2018;154:122–144. doi: 10.1016/j.mod.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 78.Candelaria AG, Murray G, File SK, García-Arrarás JE. Contribution of mesenterial muscle dedifferentiation to intestine regeneration in the sea cucumber Holothuria glaberrima. Cell Tissue Res. 2006;325(1):55–65. doi: 10.1007/s00441-006-0170-z. [DOI] [PubMed] [Google Scholar]
- 79.Dolmatov IY. Molecular aspects of regeneration mechanisms in holothurians. Genes. 2021;12(2):250. doi: 10.3390/genes12020250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dolmatov IY, Ginanova TT. Muscle regeneration in holothurians. Microsc Res Tech. 2001;55(6):452–463. doi: 10.1002/jemt.1190. [DOI] [PubMed] [Google Scholar]
- 81.Ferrario C, Sugni M, Somorjai IM, Ballarin L. Beyond adult stem cells: dedifferentiation as a unifying mechanism underlying regeneration in invertebrate deuterostomes. Front Cell Dev Biol. 2020;8:587320. doi: 10.3389/fcell.2020.587320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mashanov VS, Zueva OR, Rojas-Catagena C, Garcia-Arraras JE. Visceral regeneration in a sea cucumber involves extensive expression of survivin and mortalin homologs in the mesothelium. BMC Dev Biol. 2010;10(1):1–24. doi: 10.1186/1471-213X-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miguel-Ruiz S, José E, García-Arrarás JE. Common cellular events occur during wound healing and organ regeneration in the sea cucumber Holothuria glaberrima. BMC Dev Biol. 2007;7(1):1–9. doi: 10.1186/1471-213X-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Croce JC, Wu SY, Byrum C, Xu R, Duloquin L, Wikramanayake AH, Gache C, McClay DR. A genome-wide survey of the evolutionarily conserved Wnt pathways in the sea urchin Strongylocentrotus purpuratus. Dev Biol. 2006;300(1):121–131. doi: 10.1016/j.ydbio.2006.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler BM, Delius H, Hoppe D, Stannek P, Walter C, Glinka A. Kremen proteins are Dickkopf receptors that regulate Wnt/β-catenin signalling. Nature. 2002;417(6889):664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- 86.Gerlitz O, Basler K. Wingful, an extracellular feedback inhibitor of wingless. Genes Dev. 2002;16(9):1055–1059. doi: 10.1101/gad.991802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ehebauer M, Hayward P, Martinez-Arias A. Notch signaling pathway. Sci STKE. 2006;2006(364):cm7. doi: 10.1126/stke.3642006cm7. [DOI] [PubMed] [Google Scholar]
- 88.Kitagawa M. Notch signalling in the nucleus: roles of mastermind-like (MAML) transcriptional coactivators. J Biochem. 2016;159(3):287–294. doi: 10.1093/jb/mvv123. [DOI] [PubMed] [Google Scholar]
- 89.Kopan R, Ilagan MXG. The canonical notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cormier S, Le Bras S, Souilhol C, Vandormael-Pournin S, Durand B, Babinet C, et al. The murine ortholog of notchless, a direct regulator of the notch pathway in Drosophila melanogaster, is essential for survival of inner cell mass cells. Mol Cell Biol. 2006;26(9):3541–3549. doi: 10.1128/MCB.26.9.3541-3549.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tadashi, Sakata Hiromi, Sakaguchi Leo, Tsuda Atsushi, Higashitani Toshiro, Aigaki Kenji, Matsuno Shigeo, Hayashi. Drosophila Nedd4 Regulates Endocytosis of Notch and Suppresses Its Ligand-Independent Activation. Current Biology. 2004;14(24):2228-36. S0960982204009650. 10.1016/j.cub.2004.12.028. [DOI] [PubMed]
- 92.Clouse RM, Linchangco GV, Jr, Kerr AM, Reid RW, Janies DA. Phylotranscriptomic analysis uncovers a wealth of tissue inhibitor of metalloproteinases variants in echinoderms. R Soc Open Sci. 2015;2(12):150377. doi: 10.1098/rsos.150377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dolmatov IY, Afanasyev SV, Boyko AV. Molecular mechanisms of fission in echinoderms: Transcriptome analysis. PLoS One. 2018;13(4):e0195836. doi: 10.1371/journal.pone.0195836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li L, He C. tensilin [Apostichopus japonicus] GenBank; 2015. [Google Scholar]
- 95.Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M. BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics. 2016;32(5):767–769. doi: 10.1093/bioinformatics/btv661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brůna T, Hoff KJ, Lomsadze A, Stanke M, Borodovsky M. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom Bioinform. 2021;3(1):lqaa108. doi: 10.1093/nargab/lqaa108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Raw reads from the various echinoderm species are available in NCBI’s SRA and is also available at Zenodo (doi: https://doi.org/10.5281/zenodo.6985492).
Additional file 2: File S1. EchinoDB user manual contains screenshots of the outputs to assist new users with the features and functionality of the application.
Additional file 3: File S2. EchinoidDB user manual contains instructions to help users with the resources and operations available in the application.
Additional file 4: File S3. OphiuroidDB user manual to describe operations and capabilities of the application.
Additional file 5: File S4. Source code (in R) for EchinoDB, EchinoidDB, and OphiuroidDB. We have also provided three R scripts one for each app.
Data Availability Statement
Assembled sequences and orthoclusters are available in EchinoDB (https://echinodb.uncc.edu) [18]. Raw reads from the various echinoderm species are available in NCBI’s SRA (see accession numbers in Additional file 1: Table S1). Additionally, the user manuals and code for EchinoDB v2.0, EchinoidDB, and OphiuroidDB are available as Additional file 2: File S1, Additional file 3: File S2, Additional file 4: File S3, and Additional file 5: File S4, respectively. Additional files are available in Zenodo (doi: 10.5281/zenodo.6985492).