Abstract
The spikelet is a unique structure of inflorescence in grasses that generates one to many flowers depending on its determinate or indeterminate meristem activity. The growth patterns and number of spikelets, furthermore, define inflorescence architecture and yield. Therefore, understanding the molecular mechanisms underlying spikelet development and evolution are attractive to both biologists and breeders. Based on the progress in rice and maize, along with increasing numbers of genetic mutants and genome sequences from other grass families, the regulatory networks underpinning spikelet development are becoming clearer. This is particularly evident for domesticated traits in agriculture. This review focuses on recent progress on spikelet initiation, and spikelet and floret fertility, by comparing results from Arabidopsis with that of rice, sorghum, maize, barley, wheat, Brachypodium distachyon, and Setaria viridis. This progress may benefit genetic engineering and molecular breeding to enhance grain yield.
Keywords: Yield improvement, Inflorescence, Spikelet, Fertility, Breeding
Introduction
The family of grasses (Poaceae) contains about 10,000 species, many of which are essential crops, including rice (Oryza stavia), maize (Zea mays), barley (Hordeum vulgare), wheat (Triticum aestivum) and sorghum (Sorghum bicolor). Indeed, the grains produced from these cereals are regarded as staple food and feed for humans and livestock (Kellogg 2001). Considering that the demands for grains will increase due to the projected rise in population, and to changes in our climate, research on grain yield is a pressing scientific challenge (Grierson et al. 2011).
The architecture of the grass inflorescence determines its reproduction and yield, and is, therefore, a key agricultural trait to modify to improve yield and ease of harvesting (Doebley et al. 2006). The architecture of grass inflorescence is complex and diverse, and largely depends on the activity of the inflorescence meristem (IM; see Box 1 for explanations to all acronyms) and axillary meristem (AM) (Kellogg et al. 2013; Koppolu and Schnurbusch 2019; Zhang and Yuan 2014). Based on the lateral organ growth patterns that originate from AMs (branches and spikelets), inflorescence architectures are typically categorized as “racemes” (spikelets are pedicellate in a single central monopodial axis), “spikes” (spikelets lack pedicels, exemplified in wheat, barley and Brachypodium distachyon), and “panicles” (with higher order branching, exemplified in rice and sorghum) (Fig. 1). This inflorescence definition system is borrowed from dicots (Kellogg et al. 2013). However, in contrast to the determinate growth of flowers, a spikelet contains one to many florets depending on whether the spikelet meristem (SpM) is determinate or indeterminate. Therefore, the spikelet is not equivalent to the eudicot flower, and the grass inflorescence is also named as “compound spikes” (Endress 2010). Consequently, the number, growth patterns and morphogenesis of spikelets have profound influence on grain yield potential, with key agricultural potentials in grass breeding selection.
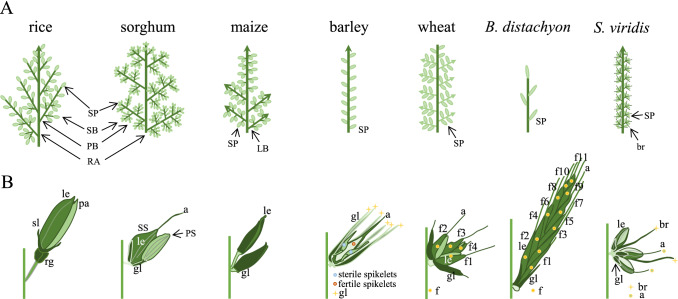
Fig. 1.
Diagrams of the grass inflorescences (A) and spikelets (B). Pictograms of rice (Oryza stavia) and sorghum (Sorghum bicolor) panicle, maize (Zea mays) tassel, barley (Hordeum vulgare), wheat (Triticum aestivum) and Brachypodium distachyon spike, and Setaria viridis inflorescences (A) and spikelets (B). Note that the spikelet is the structural unit of grasses inflorescence, and the diverse growth patterns of the spikelets confers inflorescence complexity (see text for details). a awn, br bristle, le lemma, f flower, gl glume, LB lateral branch, pa palea, PB primary branch, PS pedicellate spikelet, RA rachis, rg rudimentary glume, SB secondary branch, sl sterile lemma, SP spikelet, SS sessile spikelets
The spikelet emerges from SpM, a specialized AM that originates from the IM and branch meristem (BM). The spikelet is enclosed by glumes, or subtended by other subsidiary organs, such as the sterile lemma in rice, and the bristle in Setaria viridis (Fig. 1B). Subsequently, a flower meristem (FM) arises in the spikelet to produce floral organs, which are terminated by seed growth. A spikelet contains one or multiple florets based on the timing of SpM termination. Rice and maize spikelet structures are considered typical determinate spikelets, which generate fixed numbers of florets; one floret in the rice panicle and two florets in the maize tassel (Bommert et al. 2005). Spikelets of wheat and Brachypodium are indeterminate and produce different numbers of florets, largely determined by environmental conditions (Fig. 1B). The grass floret generally consists of non-reproductive (lemma, palea and lodicule) and reproductive (stamen and pistil) organs. Because the spikelet and floret structures are obviously different in dicots and monocots, and even among grass family members, many important and sometimes controversial biological questions remain to be answered. For example, what are the driving forces behind maize and sorghum producing spikelet pairs and not single spikelets as in rice? Why do only the sessile and not the pedicellate spikelets produce perfect flowers in sorghum (Fig. 1B)? Such spikelet structures are also evident in barley, whose inflorescence forms a triplet spikelet, with the two lateral spikelets being sterile in two-rowed barley (Fig. 1B). Other major questions revolve around how we can improve spikelet fertility, and whether the regulatory frameworks of spikelet and floret formation are conserved or developed semi-independently across grass species? Notably, the spikelet of wild-type rice is determinate and produces only one fertile floret, but “two-florets spikelet” and “three-florets spikelet” mutants have been genetically selected (Ren et al. 2019, 2018; Zhang et al. 2017c). This demonstrates that the development of the sterile spikelet or floret is likely to have common genetic grounds in crop inflorescence. Deciphering the molecular regulators that control spikelet and floret fertility will no doubt be of importance for grain number and yield, as recently exemplified in rice, sorghum, barley and wheat (Boden et al. 2015; Dampanaboina et al. 2019; Dixon et al. 2018b; Gladman et al. 2019; Jiao et al. 2018; Ren et al. 2018; Zhang et al. 2017c; Zwirek et al. 2019). There are several recent comprehensive reviews that summarize our knowledge on grass inflorescence branching and flower development (Callens et al. 2018; Gao et al. 2019; Gauley and Boden 2019; Koppolu and Schnurbusch 2019; Kyozuka 2014; Kyozuka et al. 2014; Sakuma and Schnurbusch 2020; Zhang and Yuan 2014; Zhu and Wagner 2020). In this review, we aim to summarize and synthesize current progress on molecular modules that underpin yield improvement, including spikelet initiation and floret fertility, in important grasses.
The florigen pathway decides when to flower and the number of spikelets
Similar to other AMs, the spikelet development typically involves a three-phased transition, including meristem initiation, meristem identity maintenance and termination, which is regulated by environmental and endogenous signals (Fig. 2). Hence, the timing of flowering is adjusted to environmental conditions that in turn determine the initiation of spikelets and florets. Precocious flowering generally causes a determinate inflorescence structure with less axillary organs, e.g. inflorescence branches, spikelets and flowers. In this section, we briefly highlight the mechanisms behind the decision of plants to flower and highlight potential breeding targets.
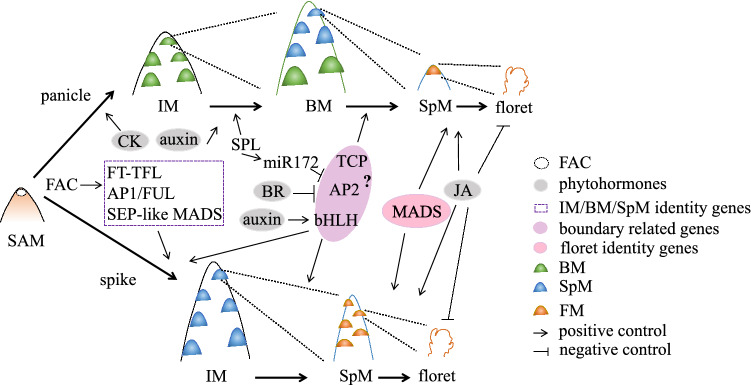
Fig. 2.
Molecular modules for specifying spikelet identity and floret fertility. Spikelet arises from the spikelet meristem, which is derived from inflorescence or branch meristem. The “florigen activation complex” (FAC) complex integrates environmental and genetic signals to promoter flowering and inflorescence development. Phytohormones such as CK and auxin play important roles in maintaining meristem activity and primordium emergence. The FT-TFL, AP1/FUL and SEP-like MADS TFs interact antagonistically or in parallel with each other to establish IM and BM identity, while BR might restrain expression region of boundary genes to establish spikelet identity. Early expression of such boundary genes might confer “unbranch”-spike architecture. The phytohormone JA plays important role in floret fertility, and this pathway could be utilized in increasing floret number. BM branch meristem, BR brassinosteroid, CK cytokinin, FAC florigen activation complex, IM inflorescence meristem, SpM spikelet meristem. See also Table 1 and Box 1 for further explanations
In Arabidopsis, the canonical flowering pathways promote reproductive development by activating floral pathway integrator genes, which respond to environmental conditions, such as light and temperature, and the circadian clock. Here, the FLOWERING LOCUS T (FT) is a key integrator gene of florigen, i.e. a flower-inducing molecule. FT may be induced by increased temperatures (Balasubramanian et al. 2006; Kim et al. 2012), and is activated directly by the photoperiodic timer gene CONSTANS (CO) under long day (LD) conditions in leaves (Imaizumi et al. 2003; Putterill et al. 1995). The FT protein can be transported through the leaf and stem vasculature to the shoot apical meristem (SAM), where it forms the “florigen activation complex” (FAC) with FLOWERING LOCUS D (FD) and 14-3-3 proteins (Andres and Coupland 2012; Song et al. 2013; Turck et al. 2008). The FAC accelerates flowering by activating the expression of the floral integrator gene SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) and FM identity gene APETALA1 (AP1) (Fig. 3) (Abe et al. 2005; Corbesier et al. 2007; Taoka et al. 2011; Wigge et al. 2005). Overexpression of FT may shorten the transition time from IM to AM, as well as antagonize functions of its homologous gene TERMINAL FLOWER1 (TFL1) in IM maintenance. Indeed, increased production of TFL1 results in more branches and flowers in Arabidopsis (Hanano and Goto 2011; Ho and Weigel 2014; Kardailsky et al. 1999; Kobayashi et al. 1999).
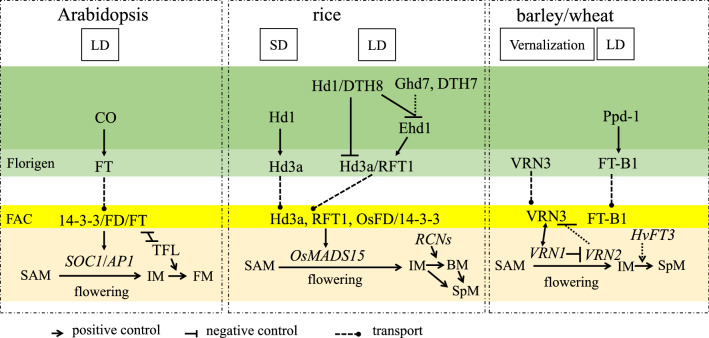
Fig. 3.
Integration of environmental signals into spikelet initiation through florigen pathway. The florigen gene FT plays important role in response to environmental signals, including day length and temperature, which in turn promotes plant flowering by activating the expression of reproductive identity genes, such as MADS TFs SOC1 and AP1 in Arabidopsis, OsMADS15, OsMADS14 and OsMADS18 in rice, VRN1 in wheat and barley. The nucleotide variations of genes involved in florigen pathway, such as Hd1, DTH8, Ghd7 and DTH7 in rice, VRN3 and VRN1 in wheat and barley, as well as duplication of FT family genes contribute to environmental adaptation, and could be targeted in plant breeding
The flowering activation pathway appears to be conserved in cereals, though the functions of FT-like genes are diverse in reproductive development (Fig. 3). Rice is a typical short day (SD) plant and contains two complementary FT-like genes, Heading-date 3a (Hd3a) and RICE FLOWERING LOCUS T1 (RFT1) (Komiya et al. 2008). Hd3a promotes flowering under inductive SD condition (Hayama et al. 2003; Kojima et al. 2002), while RFT1 induces flowering under LD condition (Komiya et al. 2008, 2009). Similar to Arabidopsis, the CO ortholog Heading date 1 (Hd1) activates the expression of Hd3a in leaves under SD condition. Once the FAC complex (Hd3a/RFT1-OsFD-14-3-3) is formed in the rice SAM (Tamaki et al. 2007; Taoka et al. 2011), it induces the TF OsMADS15 by directly binding its promoter, and also alters the expression of two other flowering-promoting AP1/FRUITFULL(FUL)-like TFs, OsMADS14 and OsMADS18 (Tamaki et al. 2015). However, under LD condition, Hd1 typically represses the expression of Hd3a and RFT1, though this depends on the TF DAYS TO HEADING 8 (DTH8) (Du et al. 2017; Zhu et al. 2017). The expression of Hd3a and RFT1 are instead activated by the rice specific gene, EARLY HEADING DATE 1 (Ehd1) under LD condition (Itoh et al. 2010). Furthermore, the expression of Ehd1 is negatively regulated by a group of flowering repressors, including GRAIN, PLANT HEIGHT and HEADING DATE 7 (Ghd7), DTH7 (Ghd7.1/OsPRR37) and DTH8 (Ghd8) (Fig. 3) (Wei et al. 2010; Xue et al. 2008; Yan et al. 2011, 2013). This repressor-Ehd1-florigen pathway, which modulates flowering under LD condition, is vital to adaptation to high-latitude regions (Komiya et al. 2009; Zhao et al. 2015), and natural variations in these genes impact spikelet number and yield (Gao et al. 2014; Yan et al. 2011, 2013). Genetic and molecular studies in rice have repeatedly shown that the heading date is positively correlated with grain yield due to a modified transition from vegetative development to inflorescence differentiation (Liu et al. 2020). Hence, the repressor genes offer interesting breeding targets to change flowering in rice. Indeed, association analysis between Hd1 nucleotide polymorphism and yield/quality variation in 123 major rice varieties, cultivated in China, revealed that haplotypes of Hd1 could be utilized to improve yield of japonica varieties in the southern areas of China by increasing secondary branch number, grain number per plant and grain weight per single panicle (Leng et al. 2020).
Wheat and barley genomes contain at least 12 FT homologs, with multiple roles in plant development (Dixon et al. 2018a; Halliwell et al. 2016). This might indicate that the expansion of the FT-like genes family has a close connection to domestication. Similar to Arabidopsis and rice, the FT ortholog VERNALIZATION3 (VRN3, also referred to as HvFT1 in barley and TaFT in wheat) stimulates flowering by activating the expression of VRN1 (AP1/FUL ortholog) in response to vernalization (Yan et al. 2006). This transition was further enhanced by VRN3 directly repressing the expression of the negative regulator VRN2 (Ghd7 homolog, Fig. 3) (Deng et al. 2015). It is noteworthy that the expression of VRN3 is low before vernalization, independently of the photoperiod, but is induced by LD condition after vernalization (Hemming et al. 2008; Yan et al. 2006). Furthermore, another barley FT homolog, HvFT3, can control spikelet initiation independently of the photoperiod (Mulki et al. 2018). These data indicate sub-functionalization of FT-related genes in barley. In contrast, a wheat FT-like gene, FT-B1, is sensitive to the photoperiod. Mutations, or decreased expression, of this gene extended the time of reproductive developmental transition, resulting in increased numbers of spikelets or paired spikelets (Dixon et al. 2018a; Finnegan et al. 2018). The expression of FT-B1 is regulated by the photo-sensitive gene Photoperiod-1 (Ppd-1), an important regulator of inflorescence architecture and paired spikelet development in wheat (Boden et al. 2015), corroborating that expression of flowering genes could be fine-tuned to increase the number of spikelets and modulate wheat inflorescence architecture.
From the above, it is clear that although the FT pathway differs in different plants, it accelerates flowering in most of them (Putterill and Varkonyi-Gasic 2016). However, there are exceptions to this rule as the FT-like gene BvFT1 represses flowering in sugar beet (Pin et al. 2010). In addition, the FT homolog, MOTHER OF FT AND TFL1 (MFT), plays only a minor role in Arabidopsis flowering (Yoo et al. 2004). A recent study in rice found that the FT-related gene, OsMFT1, may repress the expression of the APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) TF FRIZZY PANICLE (FZP) and SEPALLATA (SEP)-like genes, which in turn regulate heading date and panicle structure. As a consequence, overexpression of OsMFT1 prolonged the transition from BM to SpM to produce more branches and spikelets (Song et al. 2018).
The interactions between florigen FT and the antagonistic “anti-florigen” TFL1-like genes are not well studied in grasses yet, but the TFL1 pathway is conserved among Arabidopsis, rice and maize in regulating inflorescence architecture. Here, reduced expression of TFL1 homologs, RICE CENTRORADIALIS 1–4 (RCN 1–RCN 4), produced small panicles, while overexpression of RCN1, RCN2 or RCN4 led to increased branching due to the delay of transition to the reproductive phase (Liu et al. 2013; Nakagawa et al. 2002). In maize, ectopic expression of the TFL1-like genes can also modify flowering time and inflorescence architecture (Danilevskaya et al. 2010), but the downstream components of the TFL1 pathway in crop inflorescence development and spikelet initiation need to be clarified. Nevertheless, these results indicate that florigen pathway is not only controlling flowering time and its adaption, but also has a prominent role in determining spikelet number and yield selection (Figs. 2, 3).
Optimal seasonal timing of flowering is one of the most important breeding targets as it is essential in adapting cereal crops to temperate climates, and for grain production. Molecular marker-assisted selection has resulted in an increased number of haplotypes and alleles in flowering genes in cereals (Hickey et al. 2019). On the molecular level, CRISPR/Cas9 genome editing systems may further aid in generating many new alleles (Chen et al. 2019; Rodriguez-Leal et al. 2017). For example, such approach may enable fine-tuning the expression of flowering repressors, such as DTH8, Ghd7, DTH7 in rice and VRN2 in wheat and barley, which could boost branching to increase the number of spikelets. On the other hand, enhanced activity of the anti-florigen TFL-like genes might also promote higher-order branching of inflorescence and increase yield, and thus become suitable breeding targets.
Phytohormone gradients determine spikelet initiation and outgrowth
Once an Arabidopsis plant is dedicated to flowering, spatial and temporal distribution of phytohormones, including auxin, cytokinin (CK), brassinosteroids (BRs) and gibberellic acids (GAs), trigger FM initiation and outgrowth (Wils and Kaufmann 2017). In this section, we briefly outline how hormone distributions and components influence inflorescence development.
Auxin is a morphogen that determines almost every aspect of plant growth and development (Zhao 2018). In Arabidopsis inflorescence development, the activity of auxin efflux transporter PIN-FORMED 1 (PIN1) produces an auxin maxima, which determines the site of floral primordium by activating the expression of auxin responsive gene AUXIN RESPONSE FACTOR 5/MONOPTEROS (ARF5/MP) (Okada et al. 1991; Yamaguchi et al. 2013). ARF5/MP then directly activates the expression of FM identity genes, such as LFY and AINTEGUMENTA (ANT), by recruiting SWI/SNF chromatin remodeling complexes RAHMA (BRM) and SPLAYED (SYD) to increase accessibility of the DNA for the induction of key regulators of flower primordium initiation (Wu et al. 2015; Yamaguchi et al. 2013). Then LFY, ANT and other transcription factors (TFs), including AP1-CAULIFLOWER (CAL)-FRUITFULL (FUL) (discussed below), form feed-forward and feed-back loops to establish FM identity (Liu and Mara 2010).
Auxin maxima also determine the site and initiation of spikelet in grasses. In maize, mutations in genes related to auxin biosynthesis or polar auxin transport, such as SPARSE INFLORESCENCE1 (SPI1, an ortholog of YUCCA that regulates auxin biosynthesis), ZmAux1 (an auxin influx transporter) and BARREN INFLORESCENCE2 (BIF2, an ortholog of PINOID), led to barren inflorescence and/or less spikelets (Gallavotti et al. 2008a, b; Huang et al. 2017). Similar phenotypes were also observed in Setaria viridis, where the inflorescence of a SvAUX1 mutant contained less branches than that of wild type (Huang et al. 2017). Although there is no direct genetic evidence for a role of auxin in rice inflorescence development, auxin maxima were observed during IM progression using the auxin biosensor markers DR5rev-VENUS and DII-VENUS (Yang et al. 2017). It is plausible that gene redundancy of certain auxin biosynthesis, transport or response genes might mask the impact of the auxin pathway in rice spikelet initiation.
CK controls many processes in plant growth and development, including cell proliferation and differentiation, shoot and root architecture, light and stress responses and senescence (Hwang et al. 2012). In Arabidopsis, high concentrations of CK promote AM initiation in shoot regeneration and the leaf axils by activating expression of meristem marker gene WUSCHEL (WUS) (Zhang et al. 2017a, d). However, it is unclear if CK also drives WUS expression during FM formation. Nevertheless, AP1 does repress CK accumulation in the axil of sepals to inhibit secondary floret growth by suppressing the cytokinin biosynthetic gene LONELY GUY1 (LOG) and activating the cytokinin degradation gene CYTOKININ OXIDASE/DEHYDROGENASE3 (CKX3) (Han et al. 2014). These results indicate that high content of CK correlates with strong meristem activity in Arabidopsis. In rice, increased levels of CK result in a boost in spikelet numbers and in yield. Indeed, the CK degrading enzyme cytokinin oxidase/dehydrogenase (OsCKX2) has been one of the main yield breeding loci during rice domestication (Ashikari et al. 2005; Kurakawa et al. 2007; Li et al. 2013). In contrast to Arabidopsis, blocking CK signal transduction decreases IM activity in rice (Worthen et al. 2019), implying that distinct pathways might control inflorescence development in grasses. Consistent with this hypothesis, multiple genes involved in CK biosynthesis, degradation and signaling regulate cereal inflorescence development (Chen et al. 2020; Yamburenko et al. 2017). In addition, CK concentration is increasing in an apical-to-basal pattern, which is opposite to the auxin gradient and to the expression pattern of Six-rowed spike 2 (Vrs2), encoding a SHORT INTERNODES (SHI) TF in floral organ patterning (Youssef et al. 2017), during early barley inflorescence development. This indicates that hormone gradients might play a pivotal role in balancing meristem activity and organ outgrowth. However, detailed functions of these distribution patterns in spikelet initiation, fertility and growth duration are still underappreciated.
BRs are a group of steroid hormones known for their function in cell elongation and stress response. The spatial and temporal distribution patterns of BRs affect inflorescence and flower development (Li and He 2020). Several studies in Arabidopsis found that organ boundary formation was altered in BR deficient and constitutive mutants. For example, the BR responsive TFs BRASSINAZOLE RESISTANT 1 (BZR1) and BR1-EMS-SUPPRESSSOR 1 (BES1) could recruit the general repressor TOPLESS (TPL) to repress the expression of boundary identity genes CUP SHAPED COTYLEDON 1 (CUC1), CUC2, CUC3, LATERAL ORGAN FUSION1 and LATERAL ORGAN BOUNDARIES (LOB) (Gendron et al. 2012). Moreover, LOB activated the expression of PHYB ACTIVATION TAGGED SUPPRESSOR1 (BAS1), a cytochrome P450 enzyme that inactivates BRs, to form a negative feed-back loop and limit growth in boundary regions (Bell et al. 2012). Consistent with BRs role as a positional cue, clustered inflorescence and paired spikelets were observed in rice BR-deficient mutant panicle morphology mutant 1 (pmm1) (Li et al. 2018). In addition, the number of spikelets was changed in BR biosynthesis and signaling transduction mutants, such as dwarf 11 (d11) alleles (Wu et al. 2016; Zhou et al. 2017), and bri1-associated receptor kinase (Osbak1)/Ossg2 (Yuan et al. 2017). In some aus rice varieties, two copies of the CGTG motifs, i.e. an OsBZR1 binding motif, are present in the promoter of FZP, which resulted in repression of FZP and in increased number of spikelets due to a longer transition of SpM to FM identity (Bai et al. 2017). Therefore, the numbers of CGTG motifs in the FZP promoter could be targeted in rice breeding efforts. Whether the BZR1-FZP regulatory module is conserved also in other grasses is an interesting question that remains to be addressed.
In Arabidopsis, the FM identity component LFY can suppress the content of GA, a group of tetracyclic diterpenoid hormones that modulate cell division and elongation (Xu et al. 2014). LFY may here activate the expression of EUI-LIKE P450 A1 (ELA1), encoding a P450 enzyme that catabolize bioactive GAs (Yamaguchi et al. 2014). In parallel, the levels of DELLAs (repressors of GA signaling pathway) increase and interact with SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9 (SPL9), which then activate the expression of AP1 to enforce FM identity (Yamaguchi et al. 2014; Yu et al. 2012). Furthermore, studies in Arabidopsis and barley found that accumulation of DELLAs limits IM size (Serrano-Mislata et al. 2017), indicating that the spatio-temporal GA distribution affects the number and onset of flowers that in turn contribute to yield. Indeed, a longitudinal inflorescence GA gradient regulates patterning in barley (Youssef et al. 2017). However, whether such gradients, and the corresponding upstream regulatory loop, i.e. LFY-ELA1-SPL9-AP1 in Arabidopsis, plays similar roles in grass spikelet and floret initiation in other grasses remain elusive.
To summarize, we conclude that auxin plays an important and conserved function in AM initiation, while CK and BR appear to have distinct roles in meristem transition of cereal inflorescence. For breeding applications, it will be crucial to further explore the spatio-temporal distribution patterns of phytohormones, as well as their downstream targets, during the reproductive meristem transition of the inflorescence. Here, it seems that controlling the CK and BR levels/activity before FM initiation may contribute a key factor in increasing the number of spikelets and thus yield.
Multiple transcription factors function synergistically in specifying the identity and development of spikelets
Many TFs regulate inflorescence development. These TFs are typically directed by environmental and hormonal interactions to regulate SpM and FM identity, fertility and determinacy. These TFs typically include members of the MADS, AP2/ERF, SPL, basic helix–loop–helix (bHLH), and Teosinte branched/Cycloidea/PCF (TCP) families (Fig. 2). The TFs may interact with each other to form regulatory complexes that promote or maintain SpM identity by feed-forward or feed-back loops to guarantee the progression of spikelet and floret development (Liu and Mara 2010; Zhu and Wagner 2020). Here, we focus on select recent TF inflorescence studies and discuss how insights from these studies may guide breeding efforts.
The AP1/CAL/FUL TFs respond to FT to promote meristem transition
AP1, CAULIFLOWER (CAL) and FRUITFULL (FUL) are MIKC-type MADS-box TFs that play a critical role in FM identity and are activated by the florigen pathway in Arabidopsis. The inflorescence of ap1 cal ful triple mutant display leafy shoots instead of flowers (Ferrandiz et al. 2000). In rice, the inflorescence of osmads14 osmads15 double mutant plants also produced leaf-like structures (Wu et al. 2017). Furthermore, by suppressing the expression of OsMADS14, OsMADS15 and OsMADS18 in an osmads34/pap2 (panicle phytomer 2, one of SEP-like MADS TFs) mutant background, rice plants formed vegetative tillers on branches other than the primary ones (Kobayashi et al. 2012). Similar phenotypes were observed in the triple SEP-like mutant osmads1-z osmads5-3 osmads34-1 (Wu et al. 2018), indicating that the AP1/FUL and SEP-like MADS box genes have similar roles in maintaining AM identity during reproductive development in Arabidopsis and rice. In wheat, the MADS box TFs VRN1, FUL1 and FUL3 (homologs to the AP1/FUL and SEP-like MADS box TFs above) have redundant roles in promoting spikelet initiation and spike determinacy, as well as in flowering and stem elongation (Li et al. 2019). The number of wheat spikelets increased in both vrn1 and ful2 single mutant, but, perhaps more interestingly, floret numbers increased in ful2 spikelets (Li et al. 2019). Since molecular data show that the AP1/FUL genes can interact with different MADS-box proteins (Li et al. 2019; Wu et al. 2017), it is reasonable to deduce that many of them contribute to IM, SpMs and FMs identity determination.
The bHLH TFs respond to auxin to regulate AM initiation
In Arabidopsis, REGULATOR OF AXILLARY MERISTEM FORMATION (ROX) encodes a non-canonical bHLH protein that regulates vegetative AM activity (Yang et al. 2012). In maize, the barren stalk1 (ba1) mutant, which corresponds to a mutation in the maize ROX ortholog, grows unbranched tassels with no spikelet initiation (Gallavotti et al. 2004), indicating that reproductive AM activity may be directed by different regulatory networks in plants. The rice ba1 ortholog, LAX PANICLE1 (LAX1), likewise controls spikelet initiation, and does this by interacting with LAX2 (Tabuchi et al. 2011). This is consistent with data from maize, in which the ortholog of LAX2, BA2, could interact with BA1 to regulate both vegetative and reproductive AM formation (Yao et al. 2019). Bioinformatic analyses further found that LAX2/BA2 has orthologs in Brachypodium and sorghum, perhaps indicating that the LAX1–LAX2/BA1–BA2 pathway is conserved among grasses. Based on the expression pattern of BA2, combined with genetic data of ba1 and other barren inflorescence mutants (Yao et al. 2019), the BA1–BA2 pathway might function downstream of auxin signaling to position boundary regions for AM formation. Therefore, it would be interesting to study the spatio-temporal expression patterns, and interactions, among the AP2 and bHLH boundary marker genes and proteins.
The age pathway drives a phase transition to activate spikelet initiation
In Arabidopsis, the so-called “age pathway” controls, in parallel to environmental and phytohormonal cues, the transition of vegetative-to-reproductive phase and is mediated by the miRNA156-SPL module (Yu et al. 2015). The expression of miRNA156 declines as plant ages and targets for example the TFs SPL3, SPL9 and SPL15 to promote flowering, as they activate AP1, FUL and SOC1 (Wang et al. 2009; Yamaguchi et al. 2009). The SPL family members also play important roles in balancing plant vegetative and reproductive growth (Wang and Wang 2015). In rice, the OsSPL14 expression correlates with the number of spikelets (Jiao et al. 2010; Miura et al. 2010; Wang et al. 2017; Zhang et al. 2017b). In switchgrass (Panicum virgatum L.), PvSPL7 and PvSPL8 induced flowering by directly activating the flower identity genes, PvSEPALLATA3 (PvSEP3) and PvMADS32. Consistent with this observation, down-regulation of PvSPL7 and PvSPL8 induced inflorescence reversion (Gou et al. 2019), indicating that SPL TFs have conserved roles in promoting the transition from vegetative to reproductive growth in grasses. Such role would suggest that they also might engage with LFY; however, such relationships are unknown and will be exciting avenues to explore in the future.
Another aspect of the SPLs in grasses is that PvSPL4 regulates aerial axillary bud formation in switchgrass (Gou et al. 2017). Analogously, OsSPL7 binds directly to the promoter of OsGH3.8, one of the acyl-acid-amido synthetases in auxin catabolism, to regulate tiller number in rice (Dai et al. 2018). A recent study, furthermore, found that TaSPL3 and TaSPL17 interact with the strigolactone (SL) signaling repressor DWARF53 (TaD53) to regulate the expression of TEOSINTE BRANCHED1 (TaTB1) and BARREN STALK1 (TaBA1) to control tillering and spikelet development in wheat (Liu et al. 2017). Hence, the SPL family members control a variety of important inflorescence pathways in grasses.
The miRNA172–euAP2 pathway functions downstream of miRNA156-SPL module in juvenile-to-adult phase transition. In Arabidopsis, the expression level of miRNA172 is activated by SPL9 and the expression increases with age (Wu et al. 2009). Specific alleles of the euAP2 genes, a subfamily of AP2/ERF with a miRNA172-binding site, such as Q in wheat (Zhang et al. 2011) and HvAP2 in barley (Houston et al. 2013), have been selected for high density of spikelets during breeding. These alleles have altered the binding site of miRNA172, rendering elevated levels of euAP2 proteins, which extended the transition duration for spikelet development to increase yield (Houston et al. 2013; Liu et al. 2018).
The AP2/ERF TFs regulate boundary formation and specify spikelet identity
The AP2/ERF family members impact stress responses and plant development; processes that control AM activity and SpM identity via hormone signaling (Chandler 2018; Zhu and Wagner 2020). An increase in floret number is associated with decreased expression of certain members of the AP2 TFs, such as branched silkless1 (bd1) in maize (Chuck et al. 2002), MORE SPIKELETS 1 (MOS1) in Brachypodium (Derbyshire and Byrne 2013), FZP and MULTI-FLORET SPIKELET1 (MFS1) in rice (Bai et al. 2017; Ren et al. 2013), compositum 2 (com2) in barley and branched headt-A1 (bht-A1) in wheat (Poursarebani et al. 2015). Notably, the bd1 genes are expressed specifically in the boundary region between the indeterminate meristem and differentiating lateral organ (Chuck et al. 2002; Komatsu et al. 2003), and OsBZR1 binds directly to the promoter of FZP (Bai et al. 2017). Therefore, as also discussed above, it would be interesting to investigate whether the AP2/ERFs contribute to a conserved boundary-establishment pathway in the inflorescence, perhaps linked to BR signaling. Understanding how AP2/ERF TFs control the fate of the SpM, both at transcriptional and translational levels, is an attractive goal and some of these TFs may be targets for cereal breeding. Potential genetic interactions between AP2/ERF and MADS TFs in grasses are also awaiting to be uncovered.
The TCP TFs promote boundary formation
TEOSINTE BRANCH1 (TB1) encodes TCP protein (named after TB1 in maize, CYC in Antirrhinum majus and the proliferating cell factor DNA-binding proteins of rice), a gene first cloned in maize where it regulates tillering and ear size as one of the genetic loci for maize domestication (Doebley et al. 1997). In Arabidopsis, rice and barely, the role of TB1 orthologs in repressing axillary bud outgrowth is well studied (Wang et al. 2018). However, the TB1s role in reproductive AM development is less well understood. In wheat, TaTB1 interacts with TaFT1 to regulate axillary spikelet development and tiller number in a dosage-dependent manner (Dixon et al. 2018b). But unlike TB1, OsTB1 was apparently not selected for during domestication. Certain alleles of the OsTB1 homolog, OsTB2/RETARDED PALEA1 (REP1), were selected for during upland rice adaptation, and counteract the inhibitory effect of OsTB1 on tillering (Lyu et al. 2020). Here, REP1 is expressed in the adaxial side of the spikelet, a boundary region between IM and SpM, where palea develops. In rep1 mutants, palea development was retarded and cell differentiation was abnormal with less body structure of palea (Yuan et al. 2009). Therefore, sub- and/or neo-functionalization appears to have occurred in the TCP family during evolution. In sorghum, MULTISEEDED 1 (MSD1) belongs to the CYC/TB1 subgroup that promotes JA biosynthesis to repress carpel fertility of pedicellate spikelets (PSs) (Jiao et al. 2018). This function is conserved in barley, where the HvTB1/INTERMEDIUM-C (INT-C) represses carpel fertility of lateral spikelets (Ramsay et al. 2011). The OsTB2/REP1 homologous gene COMPOSITUM 1 (COM1) is expressed specifically in barley inflorescence meristem boundaries, and the com1 mutant grew branch-like structures instead of floret, indicating that COM1 confers SpM identity (Poursarebani et al. 2020). In maize, branch angle defective1 (bad1)/Wavy auricle in blade1 (Wab1), homolog of COM1 and OsTB2/REP1, expresses specifically in the axil of branches, spikelet pair meristems and branch meristems of tassel (Lewis et al. 2014). Furthermore, mutations of SbWab1, TCP homologous in sorghum, caused the plants to grow upright tassel branches and reduced the number of primary inflorescence branches (Poursarebani et al. 2020). Therefore, genes duplicated in the grass CYC/TB1 family might be recruited independently to regulate inflorescence development and contribute to inflorescence branching, SpM identity and carpel fertility, depending on their interactions with other TFs.
Since multiple TFs (Table 1), such as OsSPL14 and FZP in rice, TB1 in maize, Q in wheat, INT-C in barley have been selected for high-yield breeding during domestication (see above), a rational design to create defined ideotypes was proposed as future breeding strategies (Qian et al. 2016). Due to the distinct inflorescence architecture in cereal plants, the basic scheme behind such rational design is to balance the number of branches and spikelets to promote maximum yield. Since the expression dosage and patterns of SPL-AP2 and TCP TFs play essential roles in inflorescence branching and the number of spikelets, their genome duplication and regulatory modules are worth further investigation to optimize yield. As exemplified in tomato breeding, combining different natural alleles in MADS TF SEP4 genes with gene-editing techniques could modulate inflorescence complexity and improve yield (Soyk et al. 2017). Therefore, comparative genomic studies of TFs across different cereals would not only enhance our understanding of inflorescence development but also open a window for rational breeding.
Table 1.
Transcription factor functions in spikelet initiation and development
| Family | Gene function | Arabidopsis | Rice | Maize | Barley | Wheat | Sorghum |
|---|---|---|---|---|---|---|---|
| MADS | IM, SMs and FMs identity | APETALA1 (AP1) (Wigge et al. 2005), CAULIFLOWER (CAL) and FRUITFULL (FUL) (Ferrandiz et al. 2000) | OsMADS14, OsMADA15 and OsMADS18 (Wu et al. 2017) | – | – | VRN1, FUL1 and FUL3 (Li et al. 2019) | – |
| OsMADS1, OsMADA5 and OsMADS34 (Wu et al. 2018) | – | – | – | – | |||
| bHLH | AM activity | REGULATOR OF AXILLARY MERISTEM FORMATION (ROX) (Yang et al. 2012) | LAX PANICLE1 (LAX1) (Tabuchi et al. 2011) | Barren stalk1 (ba1) (Gallavotti et al. 2004) | – | – | – |
| – | LAX2 (Tabuchi et al. 2011) | BA2 (Yao et al. 2019) | – | – | – | ||
| SPL | AM activity | SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 (SPL3), SPL9 and SPL15 (Wang et al. 2009; Yamaguchi et al. 2009) | OsSPL14 (Jiao et al. 2010; Miura et al. 2010; Wang et al. 2017; Zhang et al. 2017b) | – | – | TaSPL3, TaSPL17 (Liu et al. 2017) | – |
| euAP2 | Spikelet number | – | – | – | HvAP2 (Houston et al. 2013) | Q (Zhang et al. 2011) | – |
| AP2-ERF | AM activity and SpM identity | – | FRIZZY PANICLE (FZP), MULTI-FLORET SPIKELET1 (MFS1) (Bai et al. 2017; Ren et al. 2013) | Branched silkless1 (bd1) (Chuck et al. 2002) | Compositum 2 (com2) (Poursarebani et al. 2015) | Branched headt-A1 (bht-A1) (Poursarebani et al. 2015) | – |
| TCP | AM activity and boundary formation | – | OsTB1 (Lyu et al. 2020) | TEOSINTE BRANCH1 (TB1) (Doebley et al. 1997) | HvTB1/INTERMEDIUM-C (INT-C) (Ramsay et al. 2011) | TaTB1 (Dixon et al. 2018b) | MULTISEEDED 1 (MSD1) (Jiao et al. 2018) |
| – | OsTB2/RETARDED PALEA1 (REP1) (Lyu et al. 2020) | Branch angle defective1 (bad1)/Wavy auricle in blade1 (Wab1) (Lewis et al. 2014) | COMPOSITUM 1 (COM1) (Poursarebani et al. 2020) | – | SbWab1 (Poursarebani et al. 2020) |
High yield breeding: improving the number and fertility of spikelets and florets
Increasing the number and size of spikelets are main strategies for high-yield breeding. Based on mutant screening and functional genetic analyses of rice long sterile lemma (G1) and LATERAL FLORET 1 (LF1), the three-florets-spikelet model was indicated as a probable ancient rice spikelet structure (Yoshida et al. 2009; Zhang et al. 2017c). This observation led to new breeding strategies for multiple-florets spikelet selection (Ren et al. 2020). With more comparative data from other grass plants, we propose that the grass spikelets could be modified from a spikelet containing one floret to a compound spikelet with multiple spikelets and many florets by modifying different molecular modules that function in releasing space constraint, and improving spikelet and floret fertility (Fig. 4).
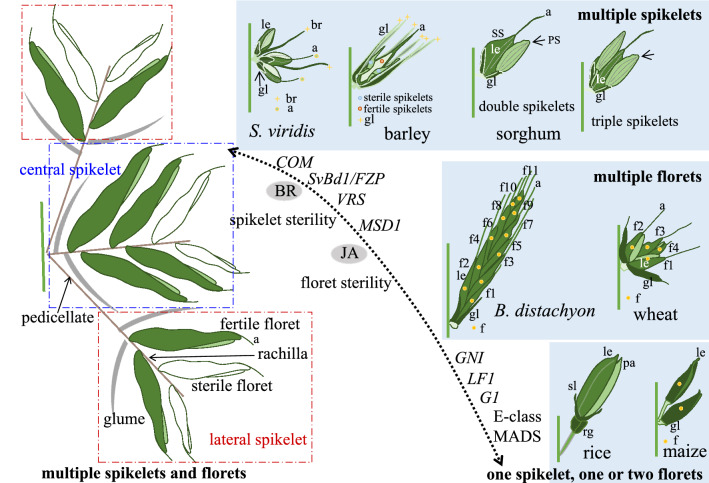
Fig. 4.
A hypothetical model to modify grass spikelet structures. In cereals, the spikelet structure unit contains one to three spikelets, where the lateral one is sterile in barley and sorghum. In Brachypodium and wheat, one spikelet contains many florets, whose fertility can be converted for high yield breeding. Phytohormones (BR, JA), TCP (COM, MSD1), AP2 (SvBd1, FZP), HD-Zip (LF1, VRS1, GNI) TFs are involved in specifying SpM identity and fertility. Manipulation of these regulatory modules to increase spikelet number and floret fertility provides a chance to generate optimal inflorescence and spikelet architecture to improve yield. a awn, br bristle, le lemma, f flower, gl glume, LB lateral branch, pa palea, PB primary branch, PS pedicellate spikelet, RA rachis, rg rudimentary glume, SB secondary branch, sl sterile lemma, SP spikelet, SS sessile spikelets
As indicated above, the structure of the grass spikelets is quite diverse, depending on the fertility of lateral spikelet or floret. Notably, it appears that the spikelet and floret fertility was lost independently several times in different cereal plants during adaptation and domestication (Sakuma and Schnurbusch 2020). The S. viridis, two-rowed barley and sorghum belong to the multiple spikelets group, where three or two spikelets grow in a structural unit. However, the lateral spikelets are sterile in two-rowed barley and sorghum, whereas a bristle structure accompanies spikelet development in S. viridis (Figs. 1B, 4). Recent work in the S. viridis d11 mutant, called bristleless1 (bsl1), found that BR levels specify bristle identity and maintain the SpM activity (Yang et al. 2018). Bsl1 expression was detected at the base of secondary and higher order axillary branches, as well as the initiation sites of lateral spikelet organ. In the bsl1-1 mutant, the boundary gene SvBd1 was ectopically expressed in the developing spikelet (Yang et al. 2018), suggesting that Bd1 class AP2 TFs plays a conserved role in establishing boundary and specifying SpM identity. Therefore, by reducing the BR levels or extending the expression of AP2-type boundary genes during meristem transition, one could generate multiple spikelets in S. viridis. In fact, this strategy was already adopted in 17 accessions in the aus subpopulation of rice, yielding increased spikelets per panicle (Bai et al. 2017). Hence, modulating the BR levels in the boundary region could be a potential way to alter yields.
Studies on mutants with fertile lateral spikelets revealed that a group of Vrs TFs confer lateral spikelet sterility in barley, making the vrs1 a key genetic locus to change lateral spikelet fertility (Zwirek et al. 2019). Vrs1 belongs to the homeodomain leucine zipper I class (HD-Zip I) TFs, and is expressed mainly in the lateral spikelet and inhibits female organ development (Komatsuda et al. 2007; Sakuma et al. 2013). The expression of the Vrs1 ortholog in wheat, Grain Number Increase 1-A (GNI-A1), was detected in the most apical floret primordia, and its expression correlated with floret sterility (Golan et al. 2019; Sakuma et al. 2019). These data indicate that during domestication, Vrs1/GNI-A1 was a key locus for selection in high-yield breeding. Therefore, developing an appropriate number of floret primordia would be helpful to improve grain numbers. Optimizing the functionality of HD-Zip I and AP2 TFs may similarly help improve floret fertility and number, though detailed studies of many of these are lacking.
In the rice lateral floret1 (lf1) mutant, a single T to C substitution in the binding site of miRNA165/166 increased the expression level of LF1, which encodes an HD-Zip III TF, and led to activation of meristem marker gene OSH1 in the axillary side of sterile lemma to produce more florets in a spikelet (Zhang et al. 2017c). Hence, the HD-Zip I and III TFs might play antagonistic roles in maintaining FM activity. How this relationship was established in grasses remains an open question, but may also become a relevant target to change reproductive development.
In sorghum, JA content is correlated with carpel fertility. Inflorescence of sorghum generates two kinds of spikelet, sessile spikelets (SSs) and PSs, and only SSs develop normally to set grain, while growth of PSs is aborted without carpel (Figs. 1B, 4). Mutant screening of fertile PSs identified three genes, MULTISEEDED 1 (MSD1), MSD2 and MSD3 (Dampanaboina et al. 2019; Gladman et al. 2019; Jiao et al. 2018). MSD1 encodes a TCP TF that binds to the promoter of MSD2, which encodes a lipoxygenase (LOX) in the JA biosynthesis pathway (Gladman et al. 2019; Jiao et al. 2018). MSD3 is an ortholog of Arabidopsis FATTY ACID DESATURASE 7 (FAD7), another enzyme involved in JA biosynthesis (Dampanaboina et al. 2019). Although it is still not clear why high JA concentration triggers programmed cell death of SPs, similar to one of the sex-determining pathways reported in maize tassel development (Acosta et al. 2009), the function of JA in FM activity seems conserved in grasses. The JA content also impact organ development and seed numbers in sorghum, maize and rice mutants, i.e. reduced JA content led to less seed setting but new flower organs (Acosta et al. 2009; Cai et al. 2014; Jiao et al. 2018; Li et al. 2009; Ren et al. 2018). Furthermore, genetic and molecular studies revealed that the JA responsive TF OsMYC2 binds to the promoter of FM identity gene OsMADS1 to promote meristem identity transition from SpM to FM during rice inflorescence development (Cai et al. 2014; You et al. 2019). OsMADS1 is one of the SEP-like MADS box TFs that confers floral organ identity and maintains FM activity (Hu et al. 2015). Therefore, the spatiotemporal distribution of JAs plays many roles in regulating plant reproductive organ development. Although the distribution pattern of JAs in spikelet development is unclear, it may be attractive to harness the pathway that modulates JA content to increase floret fertility.
Conclusions
In summary, even though there are some species specific networks that promote flowering, activate spikelet development and increase spikelet fertility, some common regulatory modules certainly exist among cereals. Increasing numbers of studies in the different grass species will improve on similarities and differences in these pathways and modules. Manipulation of the key regulatory modules, such as flowering time, controlled by the FAC-AP1/FUL module, spikelet number, regulated by the SPL-miRNA172-AP2 and BR-FZP modules, and floret fertility, managed by TFs-phytohormone modules, provides many opportunities to enhance inflorescence and spikelet architecture to improve yield.
To improve grain yield, modulating floret fertility by reducing floret abortion are representing a promising breeding strategy in wheat and other plants without branched spikes. Similarly, increasing floret number per spikelet is another strategy in rice and plants whose FM is determinate. However, both these strategies have space constraints (Fig. 4). A ‘compound spikelet’ with multiple spikelets and florets would need more “growth space” as well as nutrient supplements, which depends not only on genetic regulators in spike branching, and on spikelet and floret fertility, but also on developing an appropriate system to balance resistance and growth.
Acknowledgements
The authors would like to thank supporting by the funds from National Natural Science Foundation of China (31671260, 31470397 and 91417311); and China Innovative Research Team, Ministry of Education and the Programme of Introducing Talents of Discipline to Universities (111 Project, B14016), and SMC Morningstar Young Scholarship of Shanghai Jiao Tong University to Z. Y. S. P. is funded by ARC FT and DP Grants (DP190101941; FT160100218), and by a Villum Investigator Grant (Project ID: 25915) and Novo Nordisk Laureate Grant (NNF19OC0056076).
Abbreviations
- AM
Axillary meristem
- AP2/ERF
APETALA2/ETHYLENE RESPONSE FACTOR
- bHLH
Basic helix–loop–helix
- BM
Branch meristem
- BR
Brassinosteroid
- CK
Cytokinin
- FAC
Florigen activation complex
- FM
Flower meristem
- GA
Gibberellic acid
- HD-Zip
Homeodomain leucine zipper
- IM
Inflorescence meristem
- LD
Long day
- PS
Pedicellate spikelet
- SAM
Shoot apical meristem
- SD
Short day
- SpM
Spikelet meristem
- SS
Sessile spikelet
- TCP
Teosinte branched/Cycloidea/PCF
- TF
Transcription factor
- SL
Strigolactone
References
- Abe M, et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- Acosta I, et al. Tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science. 2009;323:262–265. doi: 10.1126/science.1164645. [DOI] [PubMed] [Google Scholar]
- Andres F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- Ashikari M, et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309:741–745. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- Bai X, et al. Duplication of an upstream silencer of FZP increases grain yield in rice. Nat Plants. 2017;3:885–893. doi: 10.1038/s41477-017-0042-4. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Lempe J, Weigel D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2006;2:e106. doi: 10.1371/journal.pgen.0020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell EM, et al. Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc Natl Acad Sci USA. 2012;109:21146–21151. doi: 10.1073/pnas.1210789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden SA, et al. Ppd-1 is a key regulator of inflorescence architecture and paired spikelet development in wheat. Nat Plants. 2015;1:1–6. doi: 10.1038/Nplants.2014.16. [DOI] [PubMed] [Google Scholar]
- Bommert P, Satoh-Nagasawa N, Jackson D, Hirano HY. Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol. 2005;46:69–78. doi: 10.1093/pcp/pci504. [DOI] [PubMed] [Google Scholar]
- Cai Q, et al. Jasmonic acid regulates spikelet development in rice. Nat Commun. 2014;5:3476. doi: 10.1038/ncomms4476. [DOI] [PubMed] [Google Scholar]
- Callens C, Tucker MR, Zhang D, Wilson ZA. Dissecting the role of MADS-box genes in monocot floral development and diversity. J Exp Bot. 2018;69:2435–2459. doi: 10.1093/jxb/ery086. [DOI] [PubMed] [Google Scholar]
- Chandler JW. Class VIIIb APETALA2 ethylene response factors in plant development. Trends Plant Sci. 2018;23:151–162. doi: 10.1016/j.tplants.2017.09.016. [DOI] [PubMed] [Google Scholar]
- Chen K, Wang Y, Zhang R, Zhang H, Gao C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol. 2019;70:667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- Chen L, Zhao J, Song J, Jameson PE. Cytokinin dehydrogenase: a genetic target for yield improvement in wheat. Plant Biotechnol J. 2020;18:614–630. doi: 10.1111/pbi.13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Muszynski M, Kellogg E, Hake S, Schmidt RJ. The control of spikelet meristem identity by the branched silkless1 gene in maize. Science. 2002;298:1238–1241. doi: 10.1126/science.1076920. [DOI] [PubMed] [Google Scholar]
- Corbesier L, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Dai Z, Wang J, Yang X, Lu H, Miao X, Shi Z. Modulation of plant architecture by the miR156f–OsSPL7–OsGH3.8 pathway in rice. J Exp Bot. 2018;69:5117–5130. doi: 10.1093/jxb/ery273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dampanaboina L, et al. Sorghum MSD3 encodes an ω-3 fatty acid desaturase that increases grain number by reducing jasmonic acid levels. Int J Mol Sci. 2019;20:5359. doi: 10.3390/ijms20215359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya ON, Meng X, Ananiev EV. Concerted modification of flowering time and inflorescence architecture by ectopic expression of TFL1-like genes in maize. Plant Physiol. 2010;153:238–251. doi: 10.1104/pp.110.154211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Casao MC, Wang P, Sato K, Hayes PM, Finnegan EJ, Trevaskis B. Direct links between the vernalization response and other key traits of cereal crops. Nat Commun. 2015;6:5882. doi: 10.1038/ncomms6882. [DOI] [PubMed] [Google Scholar]
- Derbyshire P, Byrne ME. MORE SPIKELETS1 is required for spikelet fate in the inflorescence of Brachypodium. Plant Physiol. 2013;161:1291–1302. doi: 10.1104/pp.112.212340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon LE, Farre A, Finnegan EJ, Orford S, Griffiths S, Boden SA. Developmental responses of bread wheat to changes in ambient temperature following deletion of a locus that includes FLOWERING LOCUS T1. Plant Cell Environ. 2018;41:1715–1725. doi: 10.1111/pce.13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon LE, et al. TEOSINTE BRANCHED1 regulates inflorescence architecture and development in bread wheat (Triticum aestivum) Plant Cell. 2018;30:563–581. doi: 10.1105/tpc.17.00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Du A, et al. The DTH8-Hd1 module mediates day-length-dependent regulation of rice flowering. Mol Plant. 2017;10:948–961. doi: 10.1016/j.molp.2017.05.006. [DOI] [PubMed] [Google Scholar]
- Endress PK. Disentangling confusions in inflorescence morphology: patterns and diversity of reproductive shoot ramification in angiosperms. J Syst Evol. 2010;48:225–239. doi: 10.1111/j.1759-6831.2010.00087.x. [DOI] [Google Scholar]
- Ferrandiz C, Gu Q, Martienssen R, Yanofsky MF. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development. 2000;127:725–734. doi: 10.1242/dev.127.4.725. [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, et al. Zebularine treatment is associated with deletion of FT-B1 leading to an increase in spikelet number in bread wheat. Plant Cell Environ. 2018;41:1346–1360. doi: 10.1111/pce.13164. [DOI] [PubMed] [Google Scholar]
- Gallavotti A, et al. The role of barren stalk1 in the architecture of maize. Nature. 2004;432:630–635. doi: 10.1038/nature03148. [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, Schmidt RJ, McSteen P. Sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci USA. 2008;105:15196–15201. doi: 10.1073/pnas.0805596105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Yang Y, Schmidt RJ, Jackson D. The Relationship between auxin transport and maize branching. Plant Physiol. 2008;147:1913–1923. doi: 10.1104/pp.108.121541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, et al. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc Natl Acad Sci. 2014;111:16337–16342. doi: 10.1073/pnas.1418204111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XQ, Wang N, Wang XL, Zhang XS. Architecture of wheat inflorescence: insights from rice. Trends Plant Sci. 2019;24:802–809. doi: 10.1016/j.tplants.2019.06.002. [DOI] [PubMed] [Google Scholar]
- Gauley A, Boden SA. Genetic pathways controlling inflorescence architecture and development in wheat and barley. J Integr Plant Biol. 2019;61:296–309. doi: 10.1111/jipb.12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, et al. Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc Natl Acad Sci USA. 2012;109:21152–21157. doi: 10.1073/pnas.1210799110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladman N, et al. Fertility of pedicellate spikelets in sorghum is controlled by a jasmonic acid regulatory module. Int J Mol Sci. 2019;20:4951. doi: 10.3390/ijms20194951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan G, et al. GNI-A1 mediates trade-off between grain number and grain weight in tetraploid wheat. Theor Appl Genet. 2019;132:2353–2365. doi: 10.1007/s00122-019-03358-5. [DOI] [PubMed] [Google Scholar]
- Gou J, et al. The miR156-SPL4 module predominantly regulates aerial axillary bud formation and controls shoot architecture. New Phytol. 2017;216:829–840. doi: 10.1111/nph.14758. [DOI] [PubMed] [Google Scholar]
- Gou J, et al. SPL7 and SPL8 represent a novel flowering regulation mechanism in switchgrass. New Phytol. 2019;222:1610–1623. doi: 10.1111/nph.15712. [DOI] [PubMed] [Google Scholar]
- Grierson CS, et al. One hundred important questions facing plant science research. New Phytol. 2011;192:6–12. doi: 10.1111/j.1469-8137.2011.03859.x. [DOI] [PubMed] [Google Scholar]
- Halliwell J, et al. Systematic investigation of FLOWERING LOCUS T-like poaceae gene families identifies the short-day expressed flowering pathway gene, TaFT3 in wheat (Triticum aestivum L.) Front Plant Sci. 2016;7:857. doi: 10.3389/fpls.2016.00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Zhang C, Yang H, Jiao Y. Cytokinin pathway mediates APETALA1 function in the establishment of determinate floral meristems in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:6840–6845. doi: 10.1073/pnas.1318532111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano S, Goto K. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell. 2011;23:3172–3184. doi: 10.1105/tpc.111.088641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature. 2003;422:719–722. doi: 10.1038/nature01549. [DOI] [PubMed] [Google Scholar]
- Hemming MN, Peacock WJ, Dennis ES, Trevaskis B. Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiol. 2008;147:355–366. doi: 10.1104/pp.108.116418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey LT, et al. Breeding crops to feed 10 billion. Nat Biotechnol. 2019;37:744–754. doi: 10.1038/s41587-019-0152-9. [DOI] [PubMed] [Google Scholar]
- Ho WW, Weigel D. Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell. 2014;26:552–564. doi: 10.1105/tpc.113.115220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston K, et al. Variation in the interaction between alleles of HvAPETALA2 and microRNA172 determines the density of grains on the barley inflorescence. Proc Natl Acad Sci USA. 2013;110:16675–16680. doi: 10.1073/pnas.1311681110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, et al. Interactions of OsMADS1 with floral homeotic genes in rice flower development. Mol Plant. 2015;8:1366–1384. doi: 10.1016/j.molp.2015.04.009. [DOI] [PubMed] [Google Scholar]
- Huang P, et al. Sparse panicle1 is required for inflorescence development in Setaria viridis and maize. Nat Plants. 2017;3:17054. doi: 10.1038/nplants.2017.54. [DOI] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B. Cytokinin signaling networks. Annu Rev Plant Biol. 2012;63:353–380. doi: 10.1146/annurev-arplant-042811-105503. [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature. 2003;426:302–306. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- Itoh H, Nonoue Y, Yano M, Izawa T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat Genet. 2010;42:635–638. doi: 10.1038/ng.606. [DOI] [PubMed] [Google Scholar]
- Jiao Y, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42:541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- Jiao Y, et al. MSD1 regulates pedicellate spikelet fertility in sorghum through the jasmonic acid pathway. Nat Commun. 2018;9:822. doi: 10.1038/s41467-018-03238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, et al. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- Kellogg EA. Evolutionary history of the grasses. Plant Physiol. 2001;125:1198–1205. doi: 10.1104/pp.125.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EA, Camara PE, Rudall PJ, Ladd P, Malcomber ST, Whipple CJ, Doust AN. Early inflorescence development in the grasses (Poaceae) Front Plant Sci. 2013;4:250. doi: 10.3389/fpls.2013.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Lee JH, Kim W, Jung HS, Huijser P, Ahn JH. The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol. 2012;159:461–478. doi: 10.1104/pp.111.192369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, et al. Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell. 2012;24:1848–1859. doi: 10.1105/tpc.112.097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002;43:1096–1105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Chujo A, Nagato Y, Shimamoto K, Kyozuka J. FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development. 2003;130:3841–3850. doi: 10.1242/dev.00564. [DOI] [PubMed] [Google Scholar]
- Komatsuda T, et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci. 2007;104:1424–1429. doi: 10.1073/pnas.0608580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. Hd3a and RFT1 are essential for flowering in rice. Development. 2008;135:767–774. doi: 10.1242/dev.008631. [DOI] [PubMed] [Google Scholar]
- Komiya R, Yokoi S, Shimamoto K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development. 2009;136:3443–3450. doi: 10.1242/dev.040170. [DOI] [PubMed] [Google Scholar]
- Koppolu R, Schnurbusch T. Developmental pathways for shaping spike inflorescence architecture in barley and wheat. J Integr Plant Biol. 2019;61:278–295. doi: 10.1111/jipb.12771. [DOI] [PubMed] [Google Scholar]
- Kurakawa T, et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- Kyozuka J. Chapter seven—grass inflorescence: basic structure and diversity. In: Fornara F, editor. Advances in botanical research. New York: Academic Press; 2014. pp. 191–219. [Google Scholar]
- Kyozuka J, Tokunaga H, Yoshida A. Control of grass inflorescence form by the fine-tuning of meristem phase change. Curr Opin Plant Biol. 2014;17:110–115. doi: 10.1016/j.pbi.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Leng Y, et al. Using Heading date 1 preponderant alleles from indica cultivars to breed high-yield, high-quality japonica rice varieties for cultivation in south China. Plant Biotechnol J. 2020;18:119–128. doi: 10.1111/pbi.13177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MW, Bolduc N, Hake K, Htike Y, Hay A, Candela H, Hake S. Gene regulatory interactions at lateral organ boundaries in maize. Development. 2014;141:4590–4597. doi: 10.1242/dev.111955. [DOI] [PubMed] [Google Scholar]
- Li Z, He Y. Roles of brassinosteroids in plant reproduction. Int J Mol Sci. 2020;21:872. doi: 10.3390/ijms21030872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. A putative lipase gene EXTRA GLUME1 regulates both empty-glume fate and spikelet development in rice. Plant J Cell Mol Biol. 2009;57:593–605. doi: 10.1111/j.1365-313X.2008.03710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, et al. Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc Natl Acad Sci USA. 2013;110:3167–3172. doi: 10.1073/pnas.1300359110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li X, Fu D, Wu C. Panicle morphology mutant 1 (PMM1) determines the inflorescence architecture of rice by controlling brassinosteroid biosynthesis. BMC Plant Biol. 2018;18:348. doi: 10.1186/s12870-018-1577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Lin H, Chen A, Lau M, Jernstedt J, Dubcovsky J. Wheat VRN1, FUL2 and FUL3 play critical and redundant roles in spikelet development and spike determinacy. Development. 2019;146:dev175398. doi: 10.1242/dev.175398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Mara C. Regulatory mechanisms for floral homeotic gene expression. Semin Cell Dev Biol. 2010;21:80–86. doi: 10.1016/j.semcdb.2009.11.012. [DOI] [PubMed] [Google Scholar]
- Liu C, et al. A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Dev Cell. 2013;24:612–622. doi: 10.1016/j.devcel.2013.02.013. [DOI] [PubMed] [Google Scholar]
- Liu J, Cheng X, Liu P, Sun J. miR156-targeted SBP-box transcription factors interact with DWARF53 to regulate TEOSINTE BRANCHED1 and BARREN STALK1 expression in bread wheat. Plant Physiol. 2017;174:1931–1948. doi: 10.1104/pp.17.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Liu J, Dong H, Sun J. Functional regulation of Q by microRNA172 and transcriptional co-repressor TOPLESS in controlling bread wheat spikelet density. Plant Biotechnol J. 2018;16:495–506. doi: 10.1111/pbi.12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhou X, Li Q, Wang L, Xing Y. CCT domain-containing genes in cereal crops: flowering time and beyond. Theor Appl Genet. 2020 doi: 10.1007/s00122-020-03554-8. [DOI] [PubMed] [Google Scholar]
- Lyu J, et al. Neo-functionalization of a Teosinte branched 1 homologue mediates adaptations of upland rice. Nat Commun. 2020;11:725. doi: 10.1038/s41467-019-14264-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, et al. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet. 2010;42:545–549. doi: 10.1038/ng.592. [DOI] [PubMed] [Google Scholar]
- Mulki MA, Bi X, von Korff M. FLOWERING LOCUS T3 controls spikelet initiation but not floral development. Plant Physiol. 2018;178:1170–1186. doi: 10.1104/pp.18.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Shimamoto K, Kyozuka J. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J Cell Mol Biol. 2002;29:743–750. doi: 10.1046/j.1365-313X.2002.01255.x. [DOI] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell. 1991;3:677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin PA, Benlloch R, Bonnet D, Wremerth-Weich E, Kraft T, Gielen JJ, Nilsson O. An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science. 2010;330:1397–1400. doi: 10.1126/science.1197004. [DOI] [PubMed] [Google Scholar]
- Poursarebani N, et al. The genetic basis of composite spike form in barley and ‘miracle-wheat’. Genetics. 2015;201:155–165. doi: 10.1534/genetics.115.176628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poursarebani N, et al. COMPOSITUM 1 (COM1) contributes to the architectural simplification of barley inflorescence via cell wall-mediated and meristem identity signals. bioRxiv. 2020 doi: 10.1101/2020.02.18.952705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Varkonyi-Gasic E. FT and florigen long-distance flowering control in plants. Curr Opin Plant Biol. 2016;33:77–82. doi: 10.1016/j.pbi.2016.06.008. [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- Qian Q, Guo L, Smith SM, Li J. Breeding high-yield superior quality hybrid super rice by rational design. Natl Sci Rev. 2016;3:283–294. doi: 10.1093/nsr/nww006. [DOI] [Google Scholar]
- Ramsay L, et al. INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat Genet. 2011;43:169–172. doi: 10.1038/ng.745. [DOI] [PubMed] [Google Scholar]
- Ren D, et al. MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice. Plant Physiol. 2013;162:872–884. doi: 10.1104/pp.113.216044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, et al. 'Two-floret spikelet' as a novel resource has the potential to increase rice yield. Plant Biotechnol J. 2018;16:351–353. doi: 10.1111/pbi.12849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, et al. FON4 prevents the multi-floret spikelet in rice. Plant Biotechnol J. 2019;17:1007–1009. doi: 10.1111/pbi.13083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Li Y, He G, Qian Q. Multifloret spikelet improves rice yield. New Phytol. 2020;225:2301–2306. doi: 10.1111/nph.16303. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB. Engineering quantitative trait variation for crop improvement by genome editing. Cell. 2017;171(2):470–480.e8. doi: 10.1016/j.cell.2017.08.030. [DOI] [PubMed] [Google Scholar]
- Sakuma S, Schnurbusch T. Of floral fortune: tinkering with the grain yield potential of cereal crops. New Phytol. 2020;225:1873–1882. doi: 10.1111/nph.16189. [DOI] [PubMed] [Google Scholar]
- Sakuma S, et al. Divergence of expression pattern contributed to neofunctionalization of duplicated HD-Zip I transcription factor in barley. New Phytol. 2013;197:939–948. doi: 10.1111/nph.12068. [DOI] [PubMed] [Google Scholar]
- Sakuma S, et al. Unleashing floret fertility in wheat through the mutation of a homeobox gene. Proc Natl Acad Sci. 2019;116:5182–5187. doi: 10.1073/pnas.1815465116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Mislata A, Bencivenga S, Bush M, Schiessl K, Boden S, Sablowski R. DELLA genes restrict inflorescence meristem function independently of plant height. Nat Plants. 2017;3:749–754. doi: 10.1038/s41477-017-0003-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Ito S, Imaizumi T. Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 2013;18:575–583. doi: 10.1016/j.tplants.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Wang G, Hu Y, Liu H, Bai X, Qin R, Xing Y. OsMFT1 increases spikelets per panicle and delays heading date in rice by suppressing Ehd1, FZP and SEPALLATA-like genes. J Exp Bot. 2018;69:4283–4293. doi: 10.1093/jxb/ery232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyk S, et al. Bypassing negative epistasis on yield in tomato imposed by a domestication gene. Cell. 2017;169:1142–1155.e1112. doi: 10.1016/j.cell.2017.04.032. [DOI] [PubMed] [Google Scholar]
- Tabuchi H, et al. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell. 2011;23:3276–3287. doi: 10.1105/tpc.111.088765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Tamaki S, et al. FT-like proteins induce transposon silencing in the shoot apex during floral induction in rice. Proc Natl Acad Sci USA. 2015;112:E901–910. doi: 10.1073/pnas.1417623112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K-i, et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–335. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang H. The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol Plant. 2015;8:677–688. doi: 10.1016/j.molp.2015.01.008. [DOI] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Wang S, et al. Non-canonical regulation of SPL transcription factors by a human OTUB1-like deubiquitinase defines a new plant type rice associated with higher grain yield. Cell Res. 2017;27:1142–1156. doi: 10.1038/cr.2017.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Smith SM, Li J. Genetic regulation of shoot architecture. Annu Rev Plant Biol. 2018;69:437–468. doi: 10.1146/annurev-arplant-042817-040422. [DOI] [PubMed] [Google Scholar]
- Wei X, et al. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010;153:1747–1758. doi: 10.1104/pp.110.156943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- Wils CR, Kaufmann K. Gene-regulatory networks controlling inflorescence and flower development in Arabidopsis thaliana. Biochim Biophys Acta Gene Regul Mech. 2017;1860:95–105. doi: 10.1016/j.bbagrm.2016.07.014. [DOI] [PubMed] [Google Scholar]
- Worthen JM, Yamburenko MV, Lim J, Nimchuk ZL, Kieber JJ, Schaller GE. Type-B response regulators of rice play key roles in growth, development and cytokinin signaling. Development. 2019;146:dev174870. doi: 10.1242/dev.174870. [DOI] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Yamaguchi N, Xiao J, Bargmann B, Estelle M, Sang Y, Wagner D. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife. 2015;4:e09269. doi: 10.7554/eLife.09269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Fu Y, Zhao S, Gu P, Zhu Z, Sun C, Tan L. CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnol J. 2016;14:377–386. doi: 10.1111/pbi.12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Shi X, Lin X, Liu Y, Chong K, Theissen G, Meng Z. The ABCs of flower development: mutational analysis of AP1/FUL-like genes in rice provides evidence for a homeotic (A)-function in grasses. Plant J Cell Mol Biol. 2017;89:310–324. doi: 10.1111/tpj.13386. [DOI] [PubMed] [Google Scholar]
- Wu D, et al. Loss of LOFSEP transcription factor function converts spikelet to leaf-like structures in rice. Plant Physiol. 2018;176:1646–1664. doi: 10.1104/pp.17.00704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Liu Q, Yao T, Fu X. Shedding light on integrative GA signaling. Curr Opin Plant Biol. 2014;21:89–95. doi: 10.1016/j.pbi.2014.06.010. [DOI] [PubMed] [Google Scholar]
- Xue W, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell. 2009;17:268–278. doi: 10.1016/j.devcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, et al. A molecular framework for auxin-mediated initiation of flower primordia. Dev Cell. 2013;24:271–282. doi: 10.1016/j.devcel.2012.12.017. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Winter CM, Wu MF, Kanno Y, Yamaguchi A, Seo M, Wagner D. Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science. 2014;344:638–641. doi: 10.1126/science.1250498. [DOI] [PubMed] [Google Scholar]
- Yamburenko MV, Kieber JJ, Schaller GE. Dynamic patterns of expression for genes regulating cytokinin metabolism and signaling during rice inflorescence development. PLoS ONE. 2017;12:e0176060. doi: 10.1371/journal.pone.0176060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, et al. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA. 2006;103:19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WH, et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol Plant. 2011;4:319–330. doi: 10.1093/mp/ssq070. [DOI] [PubMed] [Google Scholar]
- Yan W, et al. Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res. 2013;23:969–971. doi: 10.1038/cr.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Wang Q, Schmitz G, Müller D, Theres K. The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J. 2012;71:61–70. doi: 10.1111/j.1365-313X.2012.04970.x. [DOI] [PubMed] [Google Scholar]
- Yang J, et al. Dynamic regulation of auxin response during rice development revealed by newly established hormone biosensor markers. Front Plant Sci. 2017;8:256. doi: 10.3389/fpls.2017.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Thames S, Best NB, Jiang H, Huang P, Dilkes BP, Eveland AL. Brassinosteroids modulate meristem fate and differentiation of unique inflorescence morphology in Setaria viridis. Plant Cell. 2018;30:48–66. doi: 10.1105/tpc.17.00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, et al. The barren stalk2 gene is required for axillary meristem development in maize. Mol Plant. 2019;12:374–389. doi: 10.1016/j.molp.2018.12.024. [DOI] [PubMed] [Google Scholar]
- Yoo SY, Kardailsky I, Lee JS, Weigel D, Ahn JH. Acceleration of flowering by overexpression of MFT (MOTHER OF FT AND TFL1) Mol Cells. 2004;17:95–101. [PubMed] [Google Scholar]
- Yoshida A, Suzaki T, Tanaka W, Hirano HY. The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc Natl Acad Sci USA. 2009;106:20103–20108. doi: 10.1073/pnas.0907896106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X, et al. OsPEX5 regulates rice spikelet development through modulating jasmonic acid biosynthesis. New Phytol. 2019;224:712–724. doi: 10.1111/nph.16037. [DOI] [PubMed] [Google Scholar]
- Youssef HM, et al. VRS2 regulates hormone-mediated inflorescence patterning in barley. Nat Genet. 2017;49:157–161. doi: 10.1038/ng.3717. [DOI] [PubMed] [Google Scholar]
- Yu S, et al. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA PROMOTER BINDING–LIKE transcription factors. Plant Cell. 2012;24:3320–3332. doi: 10.1105/tpc.112.101014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Lian H, Wang J-W. Plant developmental transitions: the role of microRNAs and sugars. Curr Opin Plant Biol. 2015;27:1–7. doi: 10.1016/j.pbi.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Yuan Z, et al. RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol. 2009;149:235–244. doi: 10.1104/pp.108.128231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, et al. 08SG2/OsBAK1 regulates grain size and number, and functions differently in Indica and Japonica backgrounds in rice. Rice. 2017;10:25. doi: 10.1186/s12284-017-0165-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Yuan Z. Molecular control of grass inflorescence development. Annu Rev Plant Biol. 2014;65:553–578. doi: 10.1146/annurev-arplant-050213-040104. [DOI] [PubMed] [Google Scholar]
- Zhang Z, et al. Duplication and partitioning in evolution and function of homoeologous Q loci governing domestication characters in polyploid wheat. Proc Natl Acad Sci USA. 2011;108:18737–18742. doi: 10.1073/pnas.1110552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, May A, Irish VF. Type-B ARABIDOPSIS RESPONSE REGULATORs directly activate WUSCHEL. Trends Plant Sci. 2017;22:815–817. doi: 10.1016/j.tplants.2017.08.007. [DOI] [PubMed] [Google Scholar]
- Zhang L, et al. A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat Commun. 2017;8:14789. doi: 10.1038/ncomms14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, et al. LATERAL FLORET 1 induced the three-florets spikelet in rice. Proc Natl Acad Sci USA. 2017;114:9984–9989. doi: 10.1073/pnas.1700504114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TQ, Lian H, Zhou CM, Xu L, Jiao Y, Wang JW. A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell. 2017;29:1073–1087. doi: 10.1105/tpc.16.00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu Rev Plant Biol. 2018;69:417–435. doi: 10.1146/annurev-arplant-042817-040226. [DOI] [PubMed] [Google Scholar]
- Zhao J, et al. Genetic interactions between diverged alleles of early heading date 1 (Ehd1) and heading date 3a (Hd3a)/ RICE FLOWERING LOCUS T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Oryza sativa) New Phytol. 2015;208:936–948. doi: 10.1111/nph.13503. [DOI] [PubMed] [Google Scholar]
- Zhou Y, et al. GNS4, a novel allele of DWARF11, regulates grain number and grain size in a high-yield rice variety. Rice. 2017;10:34. doi: 10.1186/s12284-017-0171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Wagner D. Plant inflorescence architecture: the formation, activity, and fate of axillary meristems. Cold Spring Harbor Perspect Biol. 2020;12:a034652. doi: 10.1101/cshperspect.a034652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, et al. The OsHAPL1–DTH8–Hd1 complex functions as the transcription regulator to repress heading date in rice. J Exp Bot. 2017;68:553–568. doi: 10.1093/jxb/erw468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwirek M, Waugh R, McKim SM. Interaction between row-type genes in barley controls meristem determinacy and reveals novel routes to improved grain. New Phytol. 2019;221:1950–1965. doi: 10.1111/nph.15548. [DOI] [PMC free article] [PubMed] [Google Scholar]