ABSTRACT
Cervical cancer is the third leading cause of female cancers globally, resulting in more than 300,000 deaths every year. The majority of all cervical cancers are caused by persistent infections with high-risk human papillomaviruses (hrHPV) that can progress to cancer via a series of premalignant lesions. Most women, however, clear this infection within a year, concomitant with disease regression. Both hrHPV clearance and disease regression have been associated with the composition of the cervicovaginal microenvironment, which is defined by the host immune system and the cervicovaginal microbiome (CVM). A healthy microbiome is generally characterized by a high abundance of Lactobacillus species, and a change in the composition may cause bacterial vaginosis (BV), which is associated with an increased susceptibility to persistent hrHPV infections and disease. In this review, the composition of the CVM is discussed, with emphasis on the possible causes that drive changes in the cervicovaginal microbiota in relation to hrHPV infections, disease progression, and disease regression. The literature search focused on the composition of the CVM and its correlation with hrHPV infections and neoplastic lesions as well as the current efforts to adjust the microbiome against adverse viral outcomes.
KEYWORDS: microbiome, cervicovaginal microbiota, microbial communities, CVM, CSTs, human papillomavirus, hrHPV
INTRODUCTION
Cervical cancer is the third leading cause of female cancers globally, with approximately 570,000 cases and 300,000 deaths annually (1). High-risk human papillomaviruses (hrHPV) are the main cause of cervical intraepithelial neoplasia (CIN) and cervical carcinomas (2). hrHPVs are sexually transmitted, nonenveloped, double-stranded DNA viruses from the family Papillomaviridae that infect the basal cells of a variety of epithelial tissues, including the transition zone between the cervix and the endometrium (3). Under conditions of persistence, hrHPV can cause premalignant CIN, which can progress to invasive cancer, a process that can take 5 to 20 years (3). Cervical cancer screening programs aim to detect and remove these CIN lesions in a timely manner. Cervical cancer is highly prevalent in low-income and middle-income countries due to the lack of screening programs and the slow introduction of HPV vaccines (4). Additional risk factors to the disease include smoking, unhealthy lifestyle, and number of sexual partners (5, 6). Around 80% of all sexually active women will experience an HPV infection during their lives (7, 8). In the majority of the cases, the infection remains unnoticed, as the immune system is able to clear it (9–11). However, in some women, hrHPV will persist. A persistent infection with hrHPV is considered the most significant risk factor in the development of cervical carcinomas (1, 12), with integrated hrHPV DNA being present in almost all cervical cancer biopsy specimens (13, 14).
The cervicovaginal mucosa is the intrinsic defense against pathogens entering the vagina. It consists of a stratified squamous epithelium covered by a mucosal layer, which is constantly lubricated by cervicovaginal fluid. The cervicovaginal fluid is composed of many different elements secreted by cells and bacteria that are present in the vagina, and it contains products that are important for the protection of the vagina and the cervix, such as mucins and antimicrobial molecules, including B-defensins, lipocalin, elafin, secretory leukocyte protease inhibitors (SLPI), and immunoglobulins IgA and IgG (15, 16). Moreover, the cervicovaginal fluid helps to maintain the cervicovaginal microbiome (CVM). The CVM is structured into microbial communities that exist in a symbiotic relationship with the host and are often dominated by Lactobacillus species (17, 18). Lactobacillus species secrete lactic acid, maintain a low pH in the vagina, and adhere to epithelial surfaces, thereby preventing the adhesion of pathogenic bacteria to epithelial cells (19). The cervicovaginal ecosystem is also comprised of innate and adaptive immune cells, such as macrophages, neutrophils, NK cells, dendritic cells, Langerhans cells, T cells, and lymphocytes that regulate the immune response in the vagina (20).
The CVM has been overwhelmingly recognized as a candidate biomarker for the progression and regression of hrHPV infections (21). A healthy Lactobacillus-dominated microbiome prevents bacterial vaginosis (BV) and urogenital infections caused by yeasts, fungi, and human immunodeficiency virus (HIV) (22, 23). A highly diverse microbiota is associated with an increased risk of genital tract health problems, including hrHPV and HIV infections (24, 25). In this review, we discuss the composition and function of the lower genital tract microbiome, the CVM, and the effects of changes in the cervicovaginal environment on hrHPV infection outcomes.
HUMAN PAPILLOMAVIRUS INFECTION AND LIFE CYCLE
The human papillomavirus family is divided into five genera (Alpha, Beta, Gamma, Mu, and Nu) based on DNA sequence analysis. They differ in life cycle characteristics and disease association (26, 27). The Beta, Gamma, Mu, and Nu genera are associated with asymptomatic infections that cause cutaneous lesions, while the Alpha type causes asymptomatic infections that result in mucosal and cutaneous lesions. The mucosal Alpha viruses are further divided into high-risk (hrHPV) and low-risk (lrHPV) genotypes based on their association with cancer (27). The lrHPV genotypes are generally associated with warts and do not typically cause neoplasia. In contrast, the hrHPV genotypes are associated with carcinomas, of which genotypes 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68 are defined by the World Health Organization (WHO) as cancer-causing types. Overall, hrHPV genotypes 16 and 18 are responsible for 70% of women’s cervical cancer worldwide and therefore are considered the most oncogenic genotypes (28, 29).
An hrHPV infection is limited to the basal keratinocytes of several stratified epithelial tissues, including the transition zone between the cervix and the uterus in women, where it infects the basement membrane through microwounds and attaches to cells via the viral capsid proteins L1 and L2 (26). After internalization in basal epithelial cells (30) and following uncoating, the viral genome is initially retained as an extrachromosomal circular episome. The expression of the early genes E1 and E2 ensures that the viral DNA is maintained as an episome and facilitates the correct segregation of genomes during cell division (26, 31). The infected cells will go through natural keratinocyte differentiation in the transformation zone while carrying the viral genome. In an uninfected epithelium, basal cells exit the cell cycle soon after migrating into the suprabasal cell layer, where they undergo a process of terminal differentiation. However, when the expression of hrHPV proteins E6 and E7 is induced, cell cycle regulation and differentiation are disrupted (32). The expression of hrHPV E6 and E7 is often induced after the integration of the viral genome into the host genome. Interestingly, integration often leads to the deletion of essential viral genes for the synthesis of new viral particles, as integration mostly occurs in one of the early genes and disrupts its reading frames (33). The loss of E2, a repressor of the viral E6 and E7 gene promoters, will therefore increase the expression of the E6 and E7 genes (33, 34). Via interference with p53 and RB signaling, respectively, the E6 and E7 oncoproteins disrupt cell cycle control and induce genomic instability as a first step to cancer initiation (35, 36).
In productive hrHPV infections, the virus evades local immune responses by minimizing antigen production and by expressing the oncoproteins E6 and E7 (37, 38). In the early phase of infection, hrHPVs express a low abundance of E5, E6, and E7 proteins that are promptly translocated to the cell nucleus, thereby minimizing viral antigen presentation to the host immune cells present in the epithelial layer (37). In the late phase of infection, hrHPV capsid proteins are expressed. These are secreted toward the outer layer of the epithelium, where relatively few antigen-presenting cells are present, allowing immune escape (39). Likewise, the high binding affinity of the oncoproteins E6 and E7 to immune cells blocks immune-related gene expression and immune signaling pathways in infected keratinocytes (40). The impairment of immune responses in infected cells affects their capacity to activate local immune cells, resulting in an overall immunosuppressive environment that allows for carcinogenesis.
COMPOSITION OF THE CERVICOVAGINAL MICROBIOME
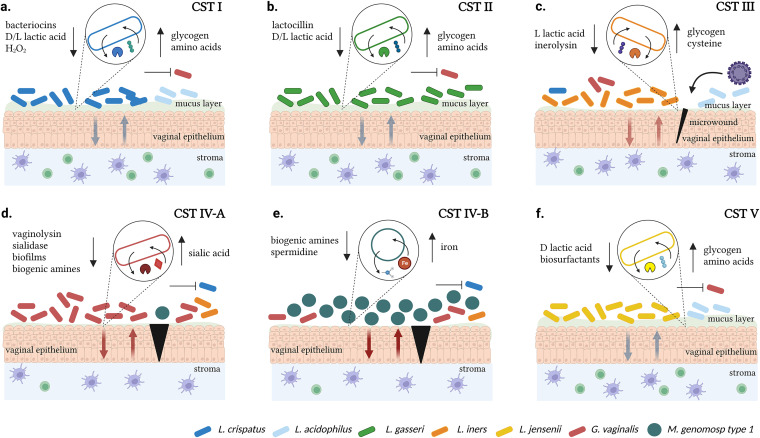
The CVM is typically composed of Lactobacillus species, which are Gram-positive, rod-shaped bacteria. Lactobacillus contribute to women's cervical health by maintaining a low pH (≤4.5) in the vagina through lactic acid production. A stable CVM inhibits colonization by harmful bacterial taxa through the production of biosurfactants, which prevent the attachment of such species to the vaginal epithelium, as well as bacteriocins, which eradicate closely related bacterial species (Fig. 1) (41). Furthermore, the CVM is characterized by microbial communities, which are groups of bacteria that establish symbiotic relationships in the CVM (42–45). These community state types (CSTs) are classified into five major groups: I, II, III, IV, and V, with CSTs I, II, III, and V presenting low diversity and having microbial dominance for Lactobacillus crispatus, Lactobacillus gasseri, Lactobacillus iners, and Lactobacillus jensenii, respectively. Colonization by Lactobacillus acidophilus further subdivides CSTs I and III into A and B subgroups (46, 47). CST IV is characterized by high diversity, Lactobacillus depletion, and a high abundance of BV-associated bacteria, such as Gardnerella vaginalis, Megasphaera genomosp type 1, Prevotella timonensis, and Sneathia amnii (17, 46, 48, 49).
FIG 1.
The biology of microbial communities in the cervicovaginal ecosystem. (A) Lactobacillus crispatus dominance (CST I, blue rods) in the cervicovaginal microbiota associates with a lower abundance for pathogenic bacteria (red rods). L. crispatus colonization leads to the production of l-and d-lactic acids, which acidifies the vaginal pH, and other products, such as H2O2 and bacteriocins that inhibit harmful bacteria. d-lactic acid and bacterial proteins modulate the host immune response toward an anti-inflammatory vaginal state. Glycogen, immune peptides, and amino acids found in the mucus layer serve as nutrient sources for the bacterium. (B) Lactobacillus gasseri dominance (CST II, green rods) in the cervicovaginal microbiota associates with a low abundance for pathogenic bacteria (red rods). L. gasseri produces l-and d-lactic acids, which acidify the vaginal pH, and bacteriocins, such as Lactocillin, that inhibit harmful bacteria. d-lactic acid and bacterial proteins modulate the host immune response toward an anti-inflammatory state. Glycogen and immune peptides found in the mucus layer serve as nutrient sources for the bacterium. (C) Lactobacillus iners dominance (CST III, orange rods) in the cervicovaginal microbiota associates with a high abundance of pathogenic bacteria and a low abundance of L. crispatus. L. iners produces only l-lactic acid, which does not properly regulate the vaginal pH, and inerolysin, which promotes bacterial adhesion in the mucus layer. l-lactic acid does not induce an anti-inflammatory vaginal state, and there is a higher susceptibility to a cytotoxic immune response in the vaginal epithelium that results in the disruption of the mucus layer. This imbalance may also lead to microwounds (marked in black) that pose a risk of viral infections. Glycogen and cysteine, an amino acid produced in the mucus layer, serve as nutrient sources for the bacterium. Gardnerella vaginalis dominance (D, red rods) and Megasphaera genomosp type 1 dominance (E, green cocci), along with high diversity (CST IV), in the vaginal cervicovaginal microbiota associate with a high abundance for pathogenic anaerobes and a low abundance for Lactobacillus. G. vaginalis and M. genomosp type 1 produce biogenic amines, which increase the vaginal pH. G. vaginalis also releases vaginolysin and sialidase, which lyse epithelia cells and break mucus sialoglycans, disturbing the mucus layer. Without an anti-inflammatory vaginal state, the bacteria induce a cytotoxic immune response that impairs the vaginal epithelium. This dysbiosis may also lead to microwounds that pose a risk of viral infections. Sialic acid and iron products serve as a nutrient source for G. vaginalis and M. genomosp type 1. (F) Lactobacillus jensenii dominance (CST V, yellow rods) in the cervicovaginal microbiota associates with a lower abundance of pathogenic bacteria (red rods). L. jensenii produces d-lactic acid, which acidifies the vaginal pH, and biosurfactants that inhibit the epithelial adhesion of harmful bacteria. d-lactic acid and bacterial proteins modulate the host immune response toward an anti-inflammatory vaginal state. Glycogen and amino acids found in the mucus layer serve as nutrient sources for the bacterium. An up arrow indicates uptake/consumption and a down arrow indicates release/production.
CST I
CST I microbiomes are stable cervicovaginal environments. L. crispatus dominance maintains a protective environment and mucus layer by producing bacteriocins and peroxidase that kill pathogenic bacteria. The bacterium also utilizes sugars, such as glycogen, to produce d- and l-lactic acids that stimulate beneficial immune responses in the mucus layer, lower the vaginal pH, and prevent the outgrow of anaerobic pathogens, such as G. vaginalis (Fig. 1A) (50–52). L. crispatus promotes an anti-inflammatory state that is characterized by the production of interleukins and proinflammatory cytokines as well as T cell activation (50, 53). The bacterium also uses the immune peptides elafin and S100A7, derived from the mucus layer, as an amino acid source (54). Since colonization by pathogenic species is inhibited in this ecosystem, the crosstalk between the vaginal epithelium and the stromal immune responses creates a balance suitable for Lactobacillus species and the mucus layer (Fig. 1A).
CST II
CST II microbiomes are rarely observed in women and are relatively stable. L. gasseri adheres to the mucus layer through mucus-binding proteins and produces both d- and l-lactic acids by glycogen consumption, thereby maintaining acidic conditions in the cervicovaginal microenvironment (Fig. 1B) (52, 55, 56). L. gasseri also produces the bacteriocins gassericin A and lactocillin, the latter being active against Enterococcus faecalis and G. vaginalis and thus having a protective role in the vagina. Lactocillin is inactive against Lactobacillus species, which suggests a resistance against a compound that they commonly encounter in the CVM (57–59). L. gasseri also promotes an anti-inflammatory state in the mucus layer by modulating tumor necrosis factor-α and interleukin-1β expression and increases vaginal epithelial cell exfoliation (Fig. 1B) (60). CST II has the highest vaginal pH (between 4.5 and 5.0) among the Lactobacillus-dominated CSTs and consequently has been associated with a higher susceptibility to dysbiosis (61, 62).
CST III
CST III microbiomes have a more unstable microenvironment than do CSTs I and II. L. iners is the dominant species in this CST, and it only produces l-lactic acid, which may lead to less control of the vaginal pH (52, 63) than is displayed by species that produces both d- and l-lactic acids and to a lesser protection against the colonization of strict anaerobes and other pathogens (Fig. 1C). L. iners is more capable of surviving and adapting to an environment with a wide pH range and to environmental conditions related to metabolic stress due to its expression of specific stress-associated genes that have not been found in other Lactobacillus species (64, 65). The production of inerolysin by L. iners, an enzyme responsible for adhesion, can result in the impairment of the mucus layer and epithelial cells, which can cause microwounds that are susceptible to viral infections (66). L. iners uses the amino acid cysteine, which is produced in the mucus layer, in its metabolic activities (67). The disturbance of the cervicovaginal ecosystem by CST III microbiomes can promote immune responses that cause cytotoxic effects in the epithelium and can result in a lower number of antigen-presenting cells (Fig. 1C) (66).
CSTs IV-A AND IV-B
CST IV microbiomes are characterized by a high vaginal pH (≥5.0) and a high production of biogenic amines, such as putrescine, cadaverine, tyramine, agmatine, spermine, and trimethylamine that allow pathogenic bacteria to grow, thereby promoting dysbiosis and the inhibition of Lactobacillus species (68, 69). In CST IV-A microbiomes, where G. vaginalis is generally dominant, the bacterium releases vaginolysin and sialidase enzymes that may cause the lysis of epithelial cells and the hydrolysis of sialic acid residue from mucus sialoglycans in the cervicovaginal mucus, respectively, leading to the disruption of the mucus barrier membrane (Fig. 1D) (70, 71). G. vaginalis can also form biofilms that can shelter BV-associated bacteria from adverse conditions, thereby shaping the host immune responses (72). These conditions favor the infiltration of CD4+ T cells and the production of cytokines that are cytotoxic to epithelial cells and consequently promote the outgrowth of pathogenic anaerobes (Fig. 1D). Similarly, in CST IV-B, where M. genomosp type 1 is dominant, the vaginal pH is also basic, and there is production of biogenic amines, including spermidines (Fig. 1E) (46, 73, 74). Megasphaera species also take up iron available in the mucus layer for their metabolic activities (74). Although less investigated so far, M. genomosp type 1 is believed to downregulate the colonization of Lactobacillus species and promote a strong immune response, thereby disrupting the mucus layer and enhancing colonization by pathogenic anaerobes (Fig. 1E).
CST V
CST V microbiomes are characterized by the dominance of L. jensenii. The bacterium produces d-lactic acid with glycogen as a nitrogen source, ensuring a protective acidic cervicovaginal environment (Fig. 1F) (52, 55). L. jensenii also makes biosurfactants that exhibit antimicrobial activity against bacterial pathogens (75–77). Similar to L. crispatus, L. jensenii promotes an anti-inflammatory state and uses the immune peptides elafin and S100A7, derived from the mucus layer, as an amino acid source (Fig. 1F) (54, 78).
DYNAMICS OF COMMUNITY STATE TYPES
The CVM is a dynamic ecosystem, with CSTs transitioning between each other regularly, dependent on women’s cervicovaginal conditions. The most common transition observed is from CST III to CST IV (79). In general, many factors can influence the composition and changes within the CVM. For instance, the menstrual cycle, hygiene, vaginal douching, and hormonal changes all affect the pH balance of the vagina, possibly resulting in BV and CST IV. The prevalence of BV is high among reproductive-aged women globally, ranging from 23% to 29% (80, 81). Other studies have investigated the composition of the CVM and have identified even more CSTs than the classical five groups (46, 82, 83). For instance, the vaginal colonization by L. acidophilus is characteristic of the microbial subgroup B in both CST I and CST III (46, 61). The occurrence of L. acidophilus may promote a decrease in abundance for L. crispatus and L. iners in CSTs I and III, respectively, leading to a transitional state between both communities (46). It must be noted that microbiome clustering analysis is routinely performed with data from different techniques (16S rRNA gene sequencing, metagenomics) making data analyses nonuniform and complicating direct comparisons between different studies. Additionally, microbiome profiles may differ between different populations. Cheng et al. studied the composition of the CVM in young Swedish women and found that the majority of the subjects exhibited a CVM dominated by L. crispatus and L. iners or were Lactobacillus-depleted (32.7%, 30.4%, and 33.5%, respectively) (82). Likewise, Fettweis et al. studied the differences between the CVMs of African American women and Caucasian women (84). They found that in African American women, the most common profiles exhibited dominance for L. iners, followed by G. vaginalis, bacterial vaginosis-associated bacterium-1 (BVAB1), “other”, and L. crispatus. The most common profiles in Caucasian women were L. crispatus, followed by L. iners and G. vaginalis (84). In addition, Serrano et al. also showed that nonpregnant women of African ancestry had CVM profiles with L. iners as the most abundant bacterium, while nonpregnant Caucasian women had CSTs with L. crispatus as the most abundant bacterium (85).
THE MICROBIOME IN hrHPV-POSITIVE WOMEN
Many studies have investigated the correlation between the CVM and hrHPV infection and have reported distinctive microbial signatures in all stages of cervical health and dysplasia. Lee et al. were the first to use 16S rRNA gene sequencing to investigate the impact of hrHPV infections on the CVM composition in a cross-sectional study, using the data of 912 women who participated in a Korean Twin Cohort study (86). They compared the CVMs of 23 hrHPV-positive premenopausal women, 4 of whom had developed CIN, and 27 hrHPV-negative premenopausal women, only one of whom had CIN. The average abundance of Lactobacillus species, such as L. crispatus, L. gasseri, L. iners, and L. jensenii, was much lower in the groups infected with hrHPV compared to the control group (Fig. 2). They also compared the CVMs of 16 premenopausal twin pairs, of which in 9 pairs, one was hrHPV-positive and one was hrHPV-negative. They found that in comparison to their uninfected twins, the hrHPV-positive women had a higher species diversity and a lower abundance for Lactobacillus species. The study identified Sneathia species as a microbiological marker of hrHPV infections, which are three times more frequent among hrHPV-positive women (Table 1).
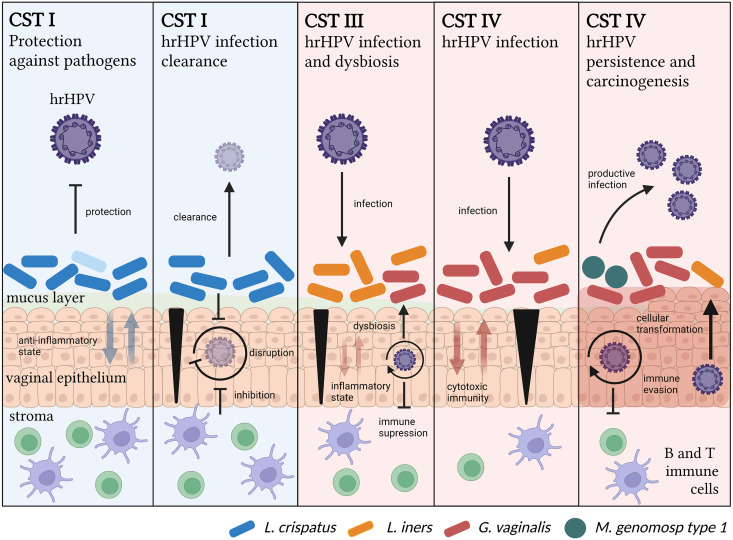
FIG 2.
Temporal relationship between the microbiome and hrHPV. CST I is associated with protection against pathogens, including hrHPV, and hrHPV infection clearance. In this CST, the cervicovaginal microenvironment exhibits an anti-inflammatory state, allowing for effective immune responses against hrHPV and the disruption of its viral life cycle in the case of an infection. Alternatively, CST III is associated with susceptibility to hrHPV infections. In this community, the cervicovaginal microenvironment exhibits an increase in cytotoxic immune responses. Microwounds (marked in black) allow for productive hrHPV infections. The virus suppresses immune responses, which reduces the nutrient sources for Lactobacillus species and causes dysbiosis. CSTs IV are also associated with susceptibility to hrHPV infections, persistence, and carcinogenesis. In this CST, pathogenic bacteria induce the disruption of the mucus layer, which impairs anti-inflammatory immune responses and increases cytotoxic signals that destroy epithelial cells, thus allowing for productive viral infections. Dysbiosis, along with hrHPV persistence, eventually leads to cellular transformation and carcinogenesis.
TABLE 1.
Summary of the microbiome association with hrHPV infections
| Cervical conditions | Microbiome characteristics | References |
|---|---|---|
| Healthy cervix | CSTs I, II, III and V; L. crispatus, L. acidophilus | (47, 83, 104) |
| hrHPV infection | CSTs III, IV, Sneathia species | (86, 93, 94, 140–142) |
| hrHPV persistence | CSTs IV, IV-B | (92, 96) |
| hrHPV progression | CST IV | (91, 93, 94) |
| CIN1+ | CSTs III, IV | (46, 47, 83, 94, 140) |
| Cervical cancer | CST IV, Sneathia and Fusobacterium species | (83, 94) |
| hrHPV clearance | CST II | (92) |
| CIN regression | CSTs I, II, III, and V | (96) |
Aside from correlating with a higher incidence, prevalence, and persistence of hrHPV infections, BV has also been associated with the development of hrHPV-induced CIN (87–91). In a meta-analysis including 12 studies that involved 6,372 women in total, BV was found to be strongly associated with hrHPV infection (89). Similarly, Brotman et al. studied the vaginal microbiome and hrHPV presence in 32 reproductive-aged women, from whom they collected vaginal swabs twice weekly for 16 weeks in total (92). Using 16S rRNA gene sequencing, they clustered the CVM into six CSTs. They found that the largest proportion of hrHPV-positive samples was found with low abundance for Lactobacillus species (L. crispatus, L. jensenii, and L. gasseri), CST IV (71% hrHPV-positive), and CST III (72% hrHPV-positive). They further suggested that CST II was associated with the fastest remission rate of hrHPV infection, whereas CST IV-B was associated with the slowest remission rate (Table 1).
Since hrHPV persistence is necessary but insufficient for the formation of cervical neoplastic lesions, Mitra et al. investigated the structure of the CVM in correlation with CIN disease severity (93). They enrolled 169 women and classified them into 4 groups, depending on the severity of their lesions, and they compared the frequency of the different CSTs (I, II, III, IV, and V) to the CIN disease severity and healthy controls. They found that higher rates of CST IV (Lactobacillus-depleted) were associated with an increase in disease severity, with CST IV being observed twice as frequently in women with low-grade squamous intraepithelial lesions (LSIL), three times as frequently in women with high-grade squamous intraepithelial lesions (HSIL), and four times as frequently in women with invasive cervical cancer (ICC) compared to healthy controls. Furthermore, CST IV was more frequently observed in hrHPV-positive women than in hrHPV-negative women (Table 1) (93).
In a cross-sectional study by Audirac-Chalifour et al., the CVM and the cytokine profile were investigated at various cervical cancer stages (83). The CVM was determined via the high-throughput sequencing of 16S rRNA amplicons and classified in CSTs. In total, they classified eight different CSTs, according to bacterial dominance. Except for CST IV, all of the samples were clustered according to the histopathological diagnosis, and they found that CST I was mainly composed of hrHPV-negative women with noncervical lesions (Fig. 2). CST III was associated with hrHPV-positive women. Sneathia-dominated CSTs were predominantly found in women with squamous intraepithelial lesions (Table 1). A Fusobacterium-dominated CST was comprised of cervical cancer cases and showed higher levels of IL-4 and TGF-β1 mRNA compared to hrHPV-negative women (83).
Laniewski and his colleagues investigated the relationship between hrHPV, CVM composition, the level of genital immune mediators, and the severity of cervical lesions (94). Using data from Hispanic and non-Hispanic women with low-grade and high-grade cervical dysplasia, invasive cervical carcinoma, and healthy controls, they found that vaginal pH is associated with ethnicity and the severity of cervical lesions. This correlates with their finding that Lactobacillus dominance decreased with the severity of cervical lesions. Additionally, they found that hrHPV-positive women and women with LSIL, HSIL, or ICC showed an increase in bacterial taxa associated with BV. The study further reported that Sneathia was the only taxon significantly enriched in women with hrHPV infections as well as women with precancerous lesions and ICC. Interestingly, they also found L. iners to be enriched in hrHPV-positive women and women with LSIL or HSIL. Since L. iners can dominate the CVM of healthy women, and since studies showed that the L. iners-dominated CVM is more likely to transition to a more species-diverse CVM (Table 1) (95), L. iners may contribute to changes in the CVM composition that can eventually lead to disease progression. Similarly, Mitra et al. described the relationship between the CVM and the regression of untreated CIN2 lesions (96). They used vaginal samples from a cohort of 87 subjects with histologically confirmed, untreated CIN2 lesions to determine whether the CVM composition affects regression in a time frame of 2 years. They found that women with a Lactobacillus-dominant microbiome were more likely to have regressive disease after 1 year, whereas Lactobacillus-depleted microbiomes and the presence of pathogenic anerobic bacteria were associated with CIN2 persistence and slower regression. CST IV was also associated with a higher chance of hrHPV persistence after 1 or 2 years compared to women with CST I (Fig. 2).
There are several mechanisms that may explain why hrHPV is able to infect the basal keratinocytes and can lead to the development of cervical cancer in women with CST IV. Increased cell shedding and the reduced proliferation of the vaginal epithelium results in a thinner layer that the virus must get through in order to infect basal keratinocytes. During sexual intercourse, microwounds that facilitate the entry of hrHPV into the basal membrane can develop in the epithelial layer, while the mucus layer that protects the epithelium is reduced due to the depletion of Lactobacillus species (Fig. 2). Likewise, the anti-inflammatory environment that is normally created by lactic acid and the Lactobacillus species is absent in CST IV microbiomes, resulting in a dysregulated immune homeostasis in the vagina. The production of uncontrolled proinflammatory cytokines by vaginal epithelial cells and harmful bacteria results in inflammation, which requires constant cell renewal and, therefore, more cell proliferation, increasing the risk of hrHPV persistence and oncogenesis. This constant cell renewal and proliferation are then able to induce DNA replication stress, a mechanism which has been linked to hrHPV integration (97). The hrHPV then hijacks the host cell’s DNA damage response to replicate, a process which occurs adjacent to the regions in the host DNA that are susceptible to replication stress (14, 98, 99), which are also the preferential integration sites (100). However, this is not enough to facilitate integration into the host genome, as the integration must take place at a site in the DNA that supports viral oncogene expression and epigenetic modulation (101).
The mechanistic biology of CST shifts during hrHPV infections, and cervical disease remains poorly understood. However, Lebeau et al. recently proposed that hrHPV infection promotes a shift to CST IV by downregulating host immune responses, thereby resulting in a decrease of immune peptides, such as SLPI, S100A7, elafin, HβD2, and HβD4, in the mucosal layer (54, 102). Since these peptides are amino acid sources for the Lactobacillus species L. crispatus, L. gasseri, L. jensenii, and L. iners in the CVM and are used to sustain their growth and survival, a reduction of these amino acids will have a negative impact on the lactic bacteria, causing dysbiosis and a higher risk of hrHPV persistence. Likewise, Moscicki et al. observed that hrHPV-positive women who cleared the virus exhibited a higher abundance for G. vaginalis and, thus, CST IV, which they hypothesized occurred due to a switch from antimicrobial surveillance to an antiviral immune response (103, 104). This switch could result in a loss of microbial control, allowing for the expansion of pathogenic bacteria, such as G. vaginalis (103). This overall viral immunopathogenesis implies a timeline in which hrHPV infection precedes BV, which may occur when the virus is able to infect and persist in Lactobacillus-dominated CSTs, such as CST III. The CST III unstable microenvironment has a higher risk of the development of microwounds and detrimental immune responses, which could facilitate productive hrHPV infections and, thus, viral-induced dysbiosis (Fig. 2).
Metabolome analyses on the cervicovaginal environment have also revealed that hrHPV infection leads to a higher concentration of biogenic amines, sialidases (SNA), and phospholipids (105–107). In addition, hrHPV-positive women with CST III exhibit higher concentrations of biogenic amines and glycogen-related metabolites than do hrHPV-negative women with the same community (105). Likewise, hrHPV-positive women with CST IV microbiomes have lower concentrations of glutathione, glycogen, and phospholipid-related metabolites than do hrHPV-negative women with the same CST. In terms of HPV genotypes, hrHPV-infected women had lower concentrations of amino acids, lipids, and peptides compared with lrHPV-infected women (105). Moreover, compared to hrHPV-negative women, hrHPV-positive women with and without cervical dysplasia show a depletion of metabolites associated with taurine, glutamine, and lysine metabolisms, which correlates with the disruption of epithelial cellular growth and dysbiosis (108). Overall, metabolome studies show that there are key signatures on the cervicovaginal ecosystem that are associated with hrHPV infection and with the composition of the CVM, demonstrating that metabolic analyses of the cervicovaginal microenvironment may be a potential tool with which to discriminate hrHPV carcinogenesis and cervical cancer.
MODULATING THE MICROBIOME AGAINST hrHPV INFECTIONS
Attempts to establish long-lasting changes in the CVM have been mostly unsuccessful. Current treatments, such as antibiotics against BV (e.g., metronidazole) and estrogen therapy, have only a temporary effect on the CVM (109, 110). Although most of these approaches have been focused on achieving microbiome normalization with biofilm disruptive agents and lactic acid (111, 112), they have provided significant insights for the development of cervicovaginal microbiome-targeted therapies (CVMTs) during hrHPV infections (Fig. 3). Effective CVMTs should consider the protective effect of CST I and the detrimental effect of CST IV in the cervicovaginal environment. It is also important to consider the specific species that dominate the CVM, their metabolism sources, and the genetic material that they carry. This will facilitate the selection of suitable CVMTs that can be produced at large scale to treat hrHPV infections.
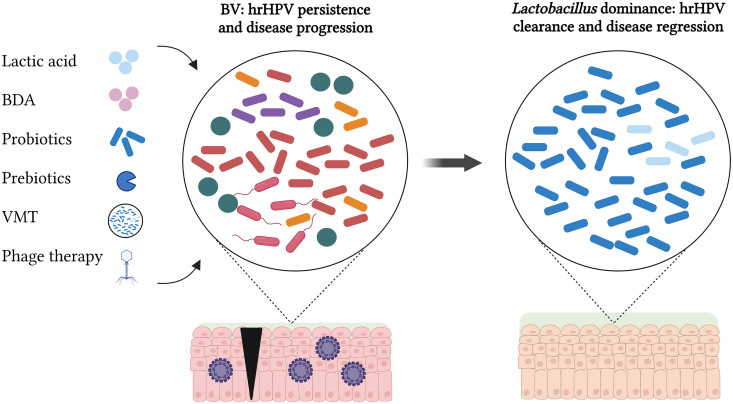
FIG 3.
Therapeutic interventions in the microbiome for hrHPV infections. Current cervicovaginal microbiome-targeted therapies (CVMTs) include agents to lower the vaginal pH, such as probiotics and lactic acid. Potential CVMTs are the use of biofilm disruptive agents (BDA) and prebiotics to counteract harmful activities by BV-associated bacteria. Vaginal microbiome transplants (VMT) and phage therapy are promising therapies that can either replace or modify the vaginal microbiota, respectively. The successful application of these therapies in women with BV, hrHPV persistent infections, and cervical disease should result in a change of the microbial composition into a healthy, Lactobacillus-dominated microbiome, leading to hrHPV clearance and disease regression.
Among the widely studied CVMTs, we find the use of probiotics and prebiotics. Probiotics are live microorganisms that confer a health benefit when consumed in adequate amounts (113), while prebiotics are nutraceutical compounds that induce bacterial growth, the activity of probiotics, or beneficial endogenous microorganisms (Fig. 3). In a clinical trial, Ou et al. tested the use of Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 as oral treatments for hrHPV clearance (114). The probiotic was administered orally (one capsule daily) until hrHPV-negative test results were achieved. However, the study did not find an association of these two probiotics with hrHPV clearance, possibly due to the route of administration, the short-term applications, and the bacterial strains utilized (114). In contrast, a clinical trial by Palma et al. observed long-term restoration of the CVM ecosystem and hrHPV clearance in hrHPV-infected women following a vaginal treatment with Lactobacillus rhamnosus BMX 54 for up to 14 months (115). Lactobacillus crispatus CTV-05 (Lactin-V) has been successfully applied in BV treatment but not for hrHPV infections to date, representing a promising candidate for CVMTs against hrHPV (116). Similarly, there have been significant advances in the application of prebiotics as CVMTs. For instance, lactoferrin, an iron-binding glycoprotein, has been used as a possible treatment for BV and for preterm labor reduction (117–119). Lactoferrin has anti-inflammatory and antimicrobial activities as well as sequestrates iron, thus making it unavailable for the metabolism of harmful bacteria (120). In addition, cysteine inhibitors and the combination of the bacteriocins GasK7B α and β derived from a human intestinal strain of Lactobacillus paragasseri have been recently described as potential CVMTs that could prevent the growth of L. iners and the development of BV, and this could be applied against hrHPV infections, since both have been associated with hrHPV-induced cervical disease (67, 121).
Alternative potential CVMTs consist of either a partial modification or a complete replacement of the cervicovaginal microbiota through genetic therapy by CRISPR-Cas systems and by phages and vaginal microbiome transplants (VMT), respectively (Fig. 3). Microbiome transplants have been extensively studied in the context of the gut microbiome, and they have achieved effective treatment against recurrent Clostridioides difficile infections (122). VMT have encouraging results in animal models and in clinical trials for treating BV and thus could be applied as CVMTs for hrHPV infections (123, 124). Furthermore, Lam et al. reported the use of bacteriophages to deliver a programmable, exogenous CRISPR-Cas9 system to achieve the strain-specific depletion of fluorescently marked, isogenic E. coli strains in the gut microbiomes of mice (125). Phages have also been utilized to target G. vaginalis strains, reaching bacterial clearance in vitro and BV resolution ex vivo (126). Therefore, these genetic tools could be further studied in the genomic edition of the cervicovaginal microbiota during hrHPV infections to prevent viral persistence and carcinogenesis.
INVESTIGATING THE MICROBIOME DURING hrHPV INFECTIONS
Assessing the role of the cervicovaginal microbiota in hrHPV infections and carcinogenesis has its limitations. However, novel approaches may assist in the study of the CVM. Microbiome alterations during cervical disease occur at the community and species levels, and even though amplicon-based techniques have facilitated a breakthrough of most of the CVM observations in the context of hrHPV infections, they generally cannot achieve high-resolution microbiome profiling. In contrast, next generation sequencing (NGS) approaches, such as shotgun metagenomics and circular-probe based RNA sequencing (ciRNAseq), can identify the most relevant microbes in the cervicovaginal microenvironment at the species level, thereby allowing for a better understanding of the CVM composition and hrHPV infections (47, 127). Likewise, in addition to cross-sectional studies, longitudinal studies are required to adequately evaluate the temporal dynamics of microbial communities and species. Microbial communities can naturally change over time, and particular species can induce microbial shifts that have been associated with hrHPV acquisition, viral persistence, and viral-induced dysbiosis. The longitudinal profiling of cervical smears from hrHPV-negative and hrHPV-positive women should elucidate such dynamics. These dynamics should be further validated through suitable in vitro and in vivo models that allow for accurate assessments into the biology of microbial species, CSTs, hosts, and hrHPV (54, 128). Available study models include organotypic raft cultures, 3D models of the human cervix, organoids, and the use of germfree animal models, such as zebrafish, mice, rats, and pigs (129, 130). Integration of the transcriptome, metabolome, and immunobiome into these studies should offer a deeper understanding of the CVM during hrHPV infections and cervical disease (131–133).
Exploring the interplay of the CVM in hrHPV infections also means considering interactions with other microbiomes. Although we focus this review on the lower genital tract microbiome, recent reports suggest the existence of an upper genital microbiome, the endometrial microbiome (EDM), which associates with endometrial diseases, such as endometriosis and cancer, and poor outcomes in assisted reproduction (134–136). Nevertheless, research on the EDM is still ongoing, and there is not clear evidence of established microbial communities or whether there is a relationship with the CVM and hrHPV infections (137). Similarly, the composition of the male genital microbiome has been associated with the protection and risk of sexually transmitted infections (STI) (138, 139), including hrHPV infections. Thus, additional analyses on the dynamics of the male and female genital microbiomes, as well as between the EDM and CVM during hrHPV acquisition and infections, are required to fully understand the role of the CVM in cervical health and disease (138).
CONCLUSION
The composition and dynamics of the CVM have been associated with hrHPV infections and cervical disease behavior. In general, a beneficial cervicovaginal microbiome is characterized by Lactobacillus species, such as Lactobacillus crispatus (CST I), which protects the underlying epithelium and promotes hrHPV clearance and disease regression. In contrast, a disadvantageous microbiome is characterized by Lactobacillus depletion and high microbial diversity (CST IV), which support hrHPV infection, viral persistence, and carcinogenesis and correlates with disease progression and cervical cancer. The anti-inflammatory state promoted by the Lactobacillus species and the inhibition of bacterial species through the regulation of vaginal pH and bacteriocins are key features of CSTs that need to be considered when investigating the relationship between the CVM and hrHPV. High-throughput sequencing methods, as well as longitudinal and omics studies, including in vitro and in vivo experiments, will be essential for this purpose. These research tools, along with clinical trials evaluating potential therapies, will help advance the development of CVMTs and preventive actions against hrHPV-induced neoplasia, which affects thousands of women worldwide.
ACKNOWLEDGMENTS
This work was supported by a research grant obtained from the Ruby and Rose Foundation. BioRender.com was used to design figures for the manuscript. We declare no conflicts of interest.
Contributor Information
Willem J. G. Melchers, Email: Willem.Melchers@radboudumc.nl.
Karl Munger, Tufts University School of Medicine.
Peter Palese, Icahn School of Medicine at Mount Sinai.
REFERENCES
- 1.Bruni L, Albero G, Serrano B, Mena M, Gómez D, Muñoz J, et al. Human papillomavirus and related diseases in the world. Summary report 17 June 2019.
- 2.Molijn A, Jenkins D, Chen W, Zhang X, Pirog E, Enqi W, Liu B, Schmidt J, Cui J, Qiao Y, Quint W, on behalf of Chinese HPV Typing Group . 2016. The complex relationship between human papillomavirus and cervical adenocarcinoma. Int J Cancer 138:409–416. doi: 10.1002/ijc.29722. [DOI] [PubMed] [Google Scholar]
- 3.Keller MJ, Burk RD, Xie X, Anastos K, Massad LS, Minkoff H, Xue X, D'Souza G, Watts DH, Levine AM, Castle PE, Colie C, Palefsky JM, Strickler HD. 2012. Risk of cervical precancer and cancer among HIV-infected women with normal cervical cytology and no evidence of oncogenic HPV infection. JAMA 308:362–369. doi: 10.1001/jama.2012.5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Organization WH. Global strategy to accelerate the elimination of cervical cancer as a public health problem. Geneva: World Health Organization; 2020. [Google Scholar]
- 5.Chelimo C, Wouldes TA, Cameron LD, Elwood JM. 2013. Risk factors for and prevention of human papillomaviruses (HPV), genital warts and cervical cancer. J Infect 66:207–217. doi: 10.1016/j.jinf.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 6.Slattery ML, Robison LM, Schuman KL, French TK, Abbott TM, Overall JC, Gardner JW, Jr.. 1989. Cigarette smoking and exposure to passive smoke are risk factors for cervical cancer. JAMA 261:1593–1598. doi: 10.1001/jama.1989.03420110069026. [DOI] [PubMed] [Google Scholar]
- 7.Deleré Y, Schuster M, Vartazarowa E, Hänsel T, Hagemann I, Borchardt S, Perlitz H, Schneider A, Reiter S, Kaufmann AM. 2011. Cervicovaginal self-sampling is a reliable method for determination of prevalence of human papillomavirus genotypes in women aged 20 to 30 years. J Clin Microbiol 49:3519–3522. doi: 10.1128/JCM.01026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chesson HW, Dunne EF, Hariri S, Markowitz LE. 2014. The estimated lifetime probability of acquiring human papillomavirus in the United States. Sexually Transmitted Diseases 41:660–664. doi: 10.1097/OLQ.0000000000000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shannon B, Yi TJ, Perusini S, Gajer P, Ma B, Humphrys MS, Thomas-Pavanel J, Chieza L, Janakiram P, Saunders M, Tharao W, Huibner S, Shahabi K, Ravel J, Rebbapragada A, Kaul R. 2017. Association of HPV infection and clearance with cervicovaginal immunology and the vaginal microbiota. Mucosal Immunol 10:1310–1319. doi: 10.1038/mi.2016.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodríguez AC, Schiffman M, Herrero R, Wacholder S, Hildesheim A, Castle PE, Solomon D, Burk R, Proyecto Epidemiológico Guanacaste Group . 2008. Rapid clearance of human papillomavirus and implications for clinical focus on persistent infections. J Natl Cancer Inst 100:513–517. doi: 10.1093/jnci/djn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho GYF, Bierman R, Beardsley L, Chang CJ, Burk RD. 1998. Natural history of cervicovaginal papillomavirus infection in young women. N Engl J Med 338:423–428. doi: 10.1056/NEJM199802123380703. [DOI] [PubMed] [Google Scholar]
- 12.Bosch FX, Lorincz A, Muñoz N, Meijer CJLM, Shah KV. 2002. The casual relation between human papillomavirus and cervical cancer. J Clin Pathol 55:244–265. doi: 10.1136/jcp.55.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pirami L, Giachè V, Becciolini A. 1997. Analysis of HPV16, 18, 31, and 35 DNA in pre-invasive and invasive lesions of the uterine cervix. J Clin Pathol 50:600–604. doi: 10.1136/jcp.50.7.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cullen AP, Reid R, Campion M, Lörincz AT. 1991. Analysis of the physical state of different human papillomavirus DNAs in intraepithelial and invasive cervical neoplasm. J Virol 65:606–612. doi: 10.1128/JVI.65.2.606-612.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King AE, Wheelhouse N, Cameron S, McDonald SE, Lee K-F, Entrican G, Critchley HO, Horne AW. 2008. Expression of secretory leukocyte protease inhibitor and elafin in human fallopian tube and in an in-vitro model of Chlamydia trachomatis infection. Hum Reprod 24:679–686. doi: 10.1093/humrep/den452. [DOI] [PubMed] [Google Scholar]
- 16.King AE, Critchley HO, Kelly RW. 2003. Innate immune defences in the human endometrium. Reprod Biol Endocrinol 1:116. doi: 10.1186/1477-7827-1-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA 108:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroon SJ, Ravel J, Huston WM. 2018. Cervicovaginal microbiota, women's health, and reproductive outcomes. Fertil Steril 110:327–336. doi: 10.1016/j.fertnstert.2018.06.036. [DOI] [PubMed] [Google Scholar]
- 19.Ghadimi D, de Vrese M, Heller KJ, Schrezenmeir J. 2010. Lactic acid bacteria enhance autophagic ability of mononuclear phagocytes by increasing Th1 autophagy-promoting cytokine (IFN-gamma) and nitric oxide (NO) levels and reducing Th2 autophagy-restraining cytokines (IL-4 and IL-13) in response to Mycobacterium tuberculosis antigen. Int Immunopharmacol 10:694–706. doi: 10.1016/j.intimp.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 20.Lee SK, Kim CJ, Kim DJ, Kang JH. 2015. Immune cells in the female reproductive tract. Immune Netw 15:16–26. doi: 10.4110/in.2015.15.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai W, Du H, Li S, Wu R. 2021. Cervicovaginal microbiome factors in clearance of human papillomavirus infection. Front Oncol 11:722639. doi: 10.3389/fonc.2021.722639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reimers LL, Mehta SD, Massad LS, Burk RD, Xie X, Ravel J, Cohen MH, Palefsky JM, Weber KM, Xue X, Anastos K, Minkoff H, Atrio J, D'Souza G, Ye Q, Colie C, Zolnik CP, Spear GT, Strickler HD. 2016. The cervicovaginal microbiota and its associations with human papillomavirus detection in HIV-infected and HIV-uninfected women. J Infect Dis 214:1361–1369. doi: 10.1093/infdis/jiw374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nunn KL, Wang Y-Y, Harit D, Humphrys MS, Ma B, Cone R, Ravel J, Lai SK. 2015. Enhanced trapping of HIV-1 by human cervicovaginal mucus is associated with Lactobacillus crispatus-dominant microbiota. mBio 6:e01084-15–e01015. doi: 10.1128/mBio.01084-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cone RA. 2014. Vaginal microbiota and sexually transmitted infections that may influence transmission of cell-associated HIV. J Infect Dis 210 Suppl 3:S616–21. doi: 10.1093/infdis/jiu459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atashili J, Poole C, Ndumbe PM, Adimora AA, Smith JS. 2008. Bacterial vaginosis and HIV acquisition: a meta-analysis of published studies. AIDS 22:1493–1501. doi: 10.1097/QAD.0b013e3283021a37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, Stanley MA. 2012. The biology and life-cycle of human papillomaviruses. Vaccine 30 Suppl 5:F55–70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 27.Bernard HU, Burk RD, Chen Z, van Doorslaer K, zur Hausen H, de Villiers EM. 2010. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401:70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.zur Hausen H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 29.Ramakrishnan S, Partricia S, Mathan G. 2015. Overview of high-risk HPV's 16 and 18 infected cervical cancer: pathogenesis to prevention. Biomed Pharmacother 70:103–110. doi: 10.1016/j.biopha.2014.12.041. [DOI] [PubMed] [Google Scholar]
- 30.Horvath CAJ, Boulet GAV, Renoux VM, Delvenne PO, Bogers J-PJ. 2010. Mechanisms of cell entry by human papillomaviruses: an overview. Virol J 7:11. doi: 10.1186/1743-422X-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doorbar J. 2005. The papillomavirus life cycle. J Clin Virol 32 Suppl 1:S7–15. doi: 10.1016/j.jcv.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 32.Sherman L, Jackman A, Itzhaki H, Stöppler MC, Koval D, Schlegel R. 1997. Inhibition of serum- and calcium-induced differentiation of human keratinocytes by HPV16 E6 oncoprotein: role of p53 inactivation. Virology 237:296–306. doi: 10.1006/viro.1997.8778. [DOI] [PubMed] [Google Scholar]
- 33.Choo K-B, Pan C-C, Han S-H. 1987. Integration of human papillomavirus type 16 into cellular DNA of cervical carcinoma: preferential deletion of the E2 gene and invariable retention of the long control region and the E6/E7 open reading frames. Virology 161:259–261. doi: 10.1016/0042-6822(87)90195-4. [DOI] [PubMed] [Google Scholar]
- 34.Francis DA, Schmid SI, Howley PM. 2000. Repression of the integrated papillomavirus E6/E7 promoter is required for growth suppression of cervical cancer cells. J Virol 74:2679–2686. doi: 10.1128/jvi.74.6.2679-2686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas M, Pim D, Banks L. 1999. The role of the E6-p53 interaction in the molecular pathogenesis of HPV. Oncogene 18:7690–7700. doi: 10.1038/sj.onc.1202953. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Sampath A, Raychaudhuri P, Bagchi S. 2001. Both Rb and E7 are regulated by the ubiquitin proteasome pathway in HPV-containing cervical tumor cells. Oncogene 20:4740–4749. doi: 10.1038/sj.onc.1204655. [DOI] [PubMed] [Google Scholar]
- 37.Stoler MH, Rhodes CR, Whitbeck A, Wolinsky SM, Chow LT, Broker TR. 1992. Human papillomavirus type 16 and 18 gene expression in cervical neoplasias. Hum Pathol 23:117–128. doi: 10.1016/0046-8177(92)90232-R. [DOI] [PubMed] [Google Scholar]
- 38.Westrich JA, Warren CJ, Pyeon D. 2017. Evasion of host immune defenses by human papillomavirus. Virus Res 231:21–33. doi: 10.1016/j.virusres.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cason J, Patel D, Naylor J, Lunney D, Shepherd PS, Best JM, McCance DJ. 1989. Identification of immunogenic regions of the major coat protein of human papillomavirus type 16 that contain type-restricted epitopes. J General Virology 70:2973–2987. doi: 10.1099/0022-1317-70-11-2973. [DOI] [PubMed] [Google Scholar]
- 40.Zhou C, Tuong ZK, Frazer IH. 2019. Papillomavirus immune evasion strategies target the infected cell and the local immune system. Front Oncol 9:682. doi: 10.3389/fonc.2019.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson M. Bacteriology of humans: an ecological perspective. Hoboken, NJ: Wiley-Blackwell; 2008. p 360. [Google Scholar]
- 42.Pybus V, Onderdonk AB. 1997. Evidence for a commensal, symbiotic relationship between Gardnerella vaginalis and Prevotella bivia involving ammonia: potential significance for bacterial vaginosis. J Infect Dis 175:406–413. doi: 10.1093/infdis/175.2.406. [DOI] [PubMed] [Google Scholar]
- 43.Machado A, Jefferson KK, Cerca N. 2013. Interactions between Lactobacillus crispatus and bacterial vaginosis (BV)-associated bacterial species in initial attachment and biofilm formation. Int J Mol Sci 14:12004–12012. doi: 10.3390/ijms140612004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Witkin SS, Moron AF, Linhares IM, Forney LJ. 2021. Influence of Lactobacillus crispatus, Lactobacillus iners and Gardnerella vaginalis on bacterial vaginal composition in pregnant women. Arch Gynecol Obstet 304:395–400. doi: 10.1007/s00404-021-05978-z. [DOI] [PubMed] [Google Scholar]
- 45.France MT, Mendes-Soares H, Forney LJ, Schloss PD. 2016. Genomic comparisons of Lactobacillus crispatus and Lactobacillus iners reveal potential ecological drivers of community composition in the vagina. Appl Environ Microbiol 82:7063–7073. doi: 10.1128/AEM.02385-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molina MA, Andralojc KM, Huynen MA, Leenders WPJ, Melchers WJG. in press. In-depth insights into cervicovaginal microbial communities and hrHPV infections using high-resolution microbiome profiling. NPJ Biofilms and Microbiomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andralojc KM, Molina MA, Qiu M, Spruijtenburg B, Rasing M, Pater B, Huynen MA, Dutilh BE, Ederveen THA, Elmelik D, Siebers AG, Loopik D, Bekkers RLM, Leenders WPJ, Melchers WJG. 2021. Novel high-resolution targeted sequencing of the cervicovaginal microbiome. BMC Biol 19:267. doi: 10.1186/s12915-021-01204-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou X, Hansmann MA, Davis CC, Suzuki H, Brown CJ, Schütte U, Pierson JD, Forney LJ. 2010. The vaginal bacterial communities of Japanese women resemble those of women in other racial groups. FEMS Immunol Med Microbiol 58:169–181. doi: 10.1111/j.1574-695X.2009.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith BC, Zolnik CP, Usyk M, Chen Z, Kaiser K, Nucci-Sack A, Peake K, Diaz A, Viswanathan S, Strickler HD, Schlecht NF, Burk RD. 2016. Distinct ecological niche of anal, oral, and cervical mucosal microbiomes in adolescent women. Yale J Biol Med 89:277–284. [PMC free article] [PubMed] [Google Scholar]
- 50.Witkin SS, Mendes-Soares H, Linhares IM, Jayaram A, Ledger WJ, Forney LJ. 2013. Influence of vaginal bacteria and D- and L-lactic acid isomers on vaginal extracellular matrix metalloproteinase inducer: implications for protection against upper genital tract infections. mBio 4:e00460-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoyancheva G, Marzotto M, Dellaglio F, Torriani S. 2014. Bacteriocin production and gene sequencing analysis from vaginal Lactobacillus strains. Arch Microbiol 196:645–653. doi: 10.1007/s00203-014-1003-1. [DOI] [PubMed] [Google Scholar]
- 52.Tester R, Al-Ghazzewi FH. 2018. Intrinsic and extrinsic carbohydrates in the vagina: a short review on vaginal glycogen. Int J Biol Macromol 112:203–206. doi: 10.1016/j.ijbiomac.2018.01.166. [DOI] [PubMed] [Google Scholar]
- 53.Mossop H, Linhares IM, Bongiovanni AM, Ledger WJ, Witkin SS. 2011. Influence of lactic acid on endogenous and viral RNA-induced immune mediator production by vaginal epithelial cells. Obstetrics & Gynecology 118. doi: 10.1097/AOG.0b013e31822da9e9. [DOI] [PubMed] [Google Scholar]
- 54.Lebeau A, Bruyere D, Roncarati P, Peixoto P, Hervouet E, Cobraiville G, Taminiau B, Masson M, Gallego C, Mazzucchelli G, Smargiasso N, Fleron M, Baiwir D, Hendrick E, Pilard C, Lerho T, Reynders C, Ancion M, Greimers R, Twizere J-C, Daube G, Schlecht-Louf G, Bachelerie F, Combes J-D, Melin P, Fillet M, Delvenne P, Hubert P, Herfs M. 2022. HPV infection alters vaginal microbiome through down-regulating host mucosal innate peptides used by Lactobacilli as amino acid sources. Nat Commun 13:1076. doi: 10.1038/s41467-022-28724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tachedjian G, Aldunate M, Bradshaw CS, Cone RA. 2017. The role of lactic acid production by probiotic Lactobacillus species in vaginal health. Res Microbiol 168:782–792. doi: 10.1016/j.resmic.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 56.Boekhorst J, Helmer Q, Kleerebezem M, Siezen RJ. 2006. Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology (Reading) 152:273–280. doi: 10.1099/mic.0.28415-0. [DOI] [PubMed] [Google Scholar]
- 57.Pandey N, Malik RK, Kaushik JK, Singroha G. 2013. Gassericin A: a circular bacteriocin produced by lactic acid bacteria Lactobacillus gasseri. World J Microbiol Biotechnol 29:1977–1987. doi: 10.1007/s11274-013-1368-3. [DOI] [PubMed] [Google Scholar]
- 58.Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, Clardy J, Linington RG, Fischbach MA. 2014. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158:1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petrova MI, Lievens E, Malik S, Imholz N, Lebeer S. 2015. Lactobacillus species as biomarkers and agents that can promote various aspects of vaginal health. Front Physiol 6. doi: 10.3389/fphys.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Q, Cheng Q, Cui S, Zhao J, Chen W, Zhang H. 2022. Inhibitory effect of Lactobacillus gasseri CCFM1201 on Gardnerella vaginalis in mice with bacterial vaginosis. Arch Microbiol 204:315. doi: 10.1007/s00203-022-02896-9. [DOI] [PubMed] [Google Scholar]
- 61.France MT, Ma B, Gajer P, Brown S, Humphrys MS, Holm JB, Waetjen LE, Brotman RM, Ravel J. 2020. VALENCIA: a nearest centroid classification method for vaginal microbial communities based on composition. Microbiome 8:166. doi: 10.1186/s40168-020-00934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verstraelen H, Verhelst R, Claeys G, De Backer E, Temmerman M, Vaneechoutte M. 2009. Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L. gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora. BMC Microbiol 9:116. doi: 10.1186/1471-2180-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amabebe E, Anumba DOC. 2018. The vaginal microenvironment: the physiologic role of Lactobacilli. Front Med (Lausanne) 5:181. doi: 10.3389/fmed.2018.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Macklaim JM, Gloor GB, Anukam KC, Cribby S, Reid G. 2011. At the crossroads of vaginal health and disease, the genome sequence of Lactobacillus iners AB-1. Proc Natl Acad Sci USA 108 Suppl 1:4688–4695. doi: 10.1073/pnas.1000086107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Macklaim JM, Fernandes AD, Di Bella JM, Hammond J, Reid G, Gloor GB. 2013. Comparative meta-RNA-seq of the vaginal microbiota and differential expression by Lactobacillus iners in health and dysbiosis. Microbiome 1:12. doi: 10.1186/2049-2618-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng N, Guo R, Wang J, Zhou W, Ling Z. 2021. Contribution of Lactobacillus iners to vaginal health and diseases: a systematic review. Front Cell Infect Microbiol:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bloom SM, Mafunda NA, Woolston BM, Hayward MR, Frempong JF, Abai AB, Xu J, Mitchell AJ, Westergaard X, Hussain FA, Xulu N, Dong M, Dong KL, Gumbi T, Ceasar FX, Rice JK, Choksi N, Ismail N, Ndung'u T, Ghebremichael MS, Relman DA, Balskus EP, Mitchell CM, Kwon DS. 2022. Cysteine dependence of Lactobacillus iners is a potential therapeutic target for vaginal microbiota modulation. Nat Microbiol 7:434–450. doi: 10.1038/s41564-022-01070-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen X, Lu Y, Chen T, Li R. 2021. The female vaginal microbiome in health and bacterial vaginosis. Front Cell Infect Microbiol:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nelson TM, Borgogna J-LC, Brotman RM, Ravel J, Walk ST, Yeoman CJ. 2015. Vaginal biogenic amines: biomarkers of bacterial vaginosis or precursors to vaginal dysbiosis? Front Physiol 6. doi: 10.3389/fphys.2015.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lewis WG, Robinson LS, Gilbert NM, Perry JC, Lewis AL. 2013. Degradation, foraging, and depletion of mucus sialoglycans by the vagina-adapted Actinobacterium Gardnerella vaginalis. J Biological Chemistry 288:12067–12079. doi: 10.1074/jbc.M113.453654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gelber SE, Aguilar JL, Lewis KLT, Ratner AJ. 2008. Functional and phylogenetic characterization of vaginolysin, the human-specific cytolysin from Gardnerella vaginalis. J Bacteriol 190:3896–3903. doi: 10.1128/JB.01965-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swidsinski A, Dörffel Y, Loening-Baucke V, Schilling J, Mendling W. 2011. Response of Gardnerella vaginalis biofilm to 5 days of moxifloxacin treatment. FEMS Immunol Med Microbiol 61:41–46. doi: 10.1111/j.1574-695X.2010.00743.x. [DOI] [PubMed] [Google Scholar]
- 73.Salliss ME, Maarsingh JD, Garza C, Łaniewski P, Herbst-Kralovetz MM. 2021. Veillonellaceae family members uniquely alter the cervical metabolic microenvironment in a human three-dimensional epithelial model. NPJ Biofilms Microbiomes 7:57. doi: 10.1038/s41522-021-00229-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Glascock AL, Jimenez NR, Boundy S, Koparde VN, Brooks JP, Edwards DJ, Strauss JF III, Jefferson KK, Serrano MG, Buck GA, Fettweis JM. 2021. Unique roles of vaginal Megasphaera phylotypes in reproductive health. Microb Genom 7. doi: 10.1099/mgen.0.000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morais IMC, Cordeiro AL, Teixeira GS, Domingues VS, Nardi RMD, Monteiro AS, Alves RJ, Siqueira EP, Santos VL. 2017. Biological and physicochemical properties of biosurfactants produced by Lactobacillus jensenii P6A and Lactobacillus gasseri P65. Microb Cell Fact 16:155. doi: 10.1186/s12934-017-0769-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaewsrichan J, Peeyananjarassri K, Kongprasertkit J. 2006. Selection and identification of anaerobic lactobacilli producing inhibitory compounds against vaginal pathogens. FEMS Immunol Med Microbiol 48:75–83. doi: 10.1111/j.1574-695X.2006.00124.x. [DOI] [PubMed] [Google Scholar]
- 77.Matu MN, Orinda GO, Njagi ENM, Cohen CR, Bukusi EA. 2010. In vitro inhibitory activity of human vaginal lactobacilli against pathogenic bacteria associated with bacterial vaginosis in Kenyan women. Anaerobe 16:210–215. doi: 10.1016/j.anaerobe.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 78.Chen H, Min S, Wang L, Zhao L, Luo F, Lei W, Wen Y, Luo L, Zhou Q, Peng L, Li Z. 2022. Lactobacillus modulates chlamydia infectivity and genital tract pathology in vitro and in vivo. Front Microbiol 13:877223. doi: 10.3389/fmicb.2022.877223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Nikita L, Galuppi M, Lamont RF, Chaemsaithong P, Miranda J, Chaiworapongsa T, Ravel J. 2014. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome 2:4. doi: 10.1186/2049-2618-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peebles K, Velloza J, Balkus JE, McClelland RS, Barnabas RV. 2019. High global burden and costs of bacterial vaginosis: a systematic review and meta-analysis. Sexual Trans Dis 46:304–311. doi: 10.1097/OLQ.0000000000000972. [DOI] [PubMed] [Google Scholar]
- 81.Kenyon C, Colebunders R, Crucitti T. 2013. The global epidemiology of bacterial vaginosis: a systematic review. Am J Obstet Gynecol 209:505–523. doi: 10.1016/j.ajog.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 82.Cheng L, Norenhag J, Hu YOO, Brusselaers N, Fransson E, Ährlund-Richter A, Guðnadóttir U, Angelidou P, Zha Y, Hamsten M, Schuppe-Koistinen I, Olovsson M, Engstrand L, Du J. 2020. Vaginal microbiota and human papillomavirus infection among young Swedish women. NPJ Biofilms Microbiomes 6:1–10. doi: 10.1038/s41522-020-00146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Audirac-Chalifour A, Torres-Poveda K, Bahena-Román M, Téllez-Sosa J, Martínez-Barnetche J, Cortina-Ceballos B, López-Estrada G, Delgado-Romero K, Burguete-García AI, Cantú D, García-Carrancá A, Madrid-Marina V. 2016. Cervical microbiome and cytokine profile at various stages of cervical cancer: a pilot study. PLoS One 11:e0153274. doi: 10.1371/journal.pone.0153274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fettweis JM, Brooks JP, Serrano MG, Sheth NU, Girerd PH, Edwards DJ, Strauss JF, Jefferson KK, Buck GA. 2014. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology (Reading) 160:2272–2282. doi: 10.1099/mic.0.081034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Serrano MG, Parikh HI, Brooks JP, Edwards DJ, Arodz TJ, Edupuganti L, Huang B, Girerd PH, Bokhari YA, Bradley SP, Brooks JL, Dickinson MR, Drake JI, Duckworth RA, Fong SS, Glascock AL, Jean S, Jimenez NR, Khoury J, Koparde VN, Lara AM, Lee V, Matveyev AV, Milton SH, Mistry SD, Rozycki SK, Sheth NU, Smirnova E, Vivadelli SC, Wijesooriya NR, Xu J, Xu P, Chaffin DO, Sexton AL, Gravett MG, Rubens CE, Hendricks-Muñoz KD, Jefferson KK, Strauss JF, Fettweis JM, Buck GA. 2019. Racioethnic diversity in the dynamics of the vaginal microbiome during pregnancy. Nat Med 25:1001–1011. doi: 10.1038/s41591-019-0465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee JE, Lee S, Lee H, Song Y-M, Lee K, Han MJ, Sung J, Ko G. 2013. Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS One 8:e63514. doi: 10.1371/journal.pone.0063514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guo YL, You K, Qiao J, Zhao YM, Geng L. 2012. Bacterial vaginosis is conducive to the persistence of HPV infection. Int J STD AIDS 23:581–584. doi: 10.1258/ijsa.2012.011342. [DOI] [PubMed] [Google Scholar]
- 88.King CC, Jamieson DJ, Wiener J, Cu-Uvin S, Klein RS, Rompalo AM, Shah KV, Sobel JD. 2011. Bacterial vaginosis and the natural history of human papillomavirus. Infect Dis Obstet Gynecol 2011:319460. doi: 10.1155/2011/319460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gillet E, Meys JF, Verstraelen H, Bosire C, De Sutter P, Temmerman M, Broeck DV. 2011. Bacterial vaginosis is associated with uterine cervical human papillomavirus infection: a meta-analysis. BMC Infect Dis 11:10. (doi: 10.1186/1471-2334-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gillet E, Meys JFA, Verstraelen H, Verhelst R, De Sutter P, Temmerman M, Broeck DV. 2012. Association between bacterial vaginosis and cervical intraepithelial neoplasia: systematic review and meta-analysis. PLoS One 7:e45201. doi: 10.1371/journal.pone.0045201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oh HY, Kim B-S, Seo S-S, Kong J-S, Lee J-K, Park S-Y, Hong K-M, Kim H-K, Kim MK. 2015. The association of uterine cervical microbiota with an increased risk for cervical intraepithelial neoplasia in Korea. Clin Microbiol Infect 21:674.e1–e9. doi: 10.1016/j.cmi.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 92.Brotman RM, Shardell MD, Gajer P, Tracy JK, Zenilman JM, Ravel J, Gravitt PE. 2014. Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J Infectious Diseases 210:1723–1733. doi: 10.1093/infdis/jiu330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mitra A, MacIntyre DA, Lee YS, Smith A, Marchesi JR, Lehne B, Bhatia R, Lyons D, Paraskevaidis E, Li JV, Holmes E, Nicholson JK, Bennett PR, Kyrgiou M. 2015. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci Rep 5:16865–16811. doi: 10.1038/srep16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Łaniewski P, Barnes D, Goulder A, Cui H, Roe DJ, Chase DM, Herbst-Kralovetz MM. 2018. Linking cervicovaginal immune signatures, HPV and microbiota composition in cervical carcinogenesis in non-Hispanic and Hispanic women. Sci Rep 8:7593. doi: 10.1038/s41598-018-25879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gajer P, Brotman RM, Bai G, Sakamoto J, Schütte UME, Zhong X, Koenig SSK, Fu L, Ma ZS, Zhou X, Abdo Z, Forney LJ, Ravel J. 2012. Temporal dynamics of the human vaginal microbiota. Sci Transl Med 4:132ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mitra A, MacIntyre DA, Ntritsos G, Smith A, Tsilidis KK, Marchesi JR, Bennett PR, Moscicki A-B, Kyrgiou M. 2020. The vaginal microbiota associates with the regression of untreated cervical intraepithelial neoplasia 2 lesions. Nat Commun 11:1999. doi: 10.1038/s41467-020-15856-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McBride AA, Warburton A. 2017. The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog 13:e1006211. doi: 10.1371/journal.ppat.1006211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vojtechova Z, Sabol I, Salakova M, Turek L, Grega M, Smahelova J, Vencalek O, Lukesova E, Klozar J, Tachezy R. 2016. Analysis of the integration of human papillomaviruses in head and neck tumours in relation to patients' prognosis. Int J Cancer 138:386–395. doi: 10.1002/ijc.29712. [DOI] [PubMed] [Google Scholar]
- 99.Jang MK, Shen K, McBride AA. 2014. Papillomavirus genomes associate with BRD4 to replicate at fragile sites in the host genome. PLoS Pathog 10:e1004117. doi: 10.1371/journal.ppat.1004117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gao G, Johnson SH, Vasmatzis G, Pauley CE, Tombers NM, Kasperbauer JL, Smith DI. 2017. Common fragile sites (CFS) and extremely large CFS genes are targets for human papillomavirus integrations and chromosome rearrangements in oropharyngeal squamous cell carcinoma. Genes Chromosomes Cancer 56:59–74. doi: 10.1002/gcc.22415. [DOI] [PubMed] [Google Scholar]
- 101.Chaiwongkot A, Vinokurova S, Pientong C, Ekalaksananan T, Kongyingyoes B, Kleebkaow P, Chumworathayi B, Patarapadungkit N, Reuschenbach M, von Knebel Doeberitz M. 2013. Differential methylation of E2 binding sites in episomal and integrated HPV 16 genomes in preinvasive and invasive cervical lesions. Int J Cancer 132:2087–2094. doi: 10.1002/ijc.27906. [DOI] [PubMed] [Google Scholar]
- 102.Sahin E, Madendag Y, Sahin ME, Madendag IC, Acmaz G, Karakukcu C, Karaman H, Muderris II. 2018. Cervical local immune response for high-risk human papillomavirus infection: involvement with cervical mucus SLPI proteins. Cancer Control 25:107327481879859. 1073274818798598. doi: 10.1177/1073274818798598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moscicki A-B, Shi B, Huang H, Barnard E, Li H. 2020. Cervical-vaginal microbiome and associated cytokine profiles in a prospective study of HPV 16 acquisition, persistence, and clearance. Front Cell Infect Microbiol 10. doi: 10.3389/fcimb.2020.569022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arokiyaraj S, Seo SS, Kwon M, Lee JK, Kim MK. 2018. Association of cervical microbial community with persistence, clearance and negativity of human papillomavirus in Korean women: a longitudinal study. Sci Rep 8:15479. doi: 10.1038/s41598-018-33750-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Borgogna JC, Shardell MD, Santori EK, Nelson TM, Rath JM, Glover ED, Ravel J, Gravitt PE, Yeoman CJ, Brotman RM. 2020. The vaginal metabolome and microbiota of cervical HPV-positive and HPV-negative women: a cross-sectional analysis. BJOG 127:182–192. doi: 10.1111/1471-0528.15981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lin W, Zhang Q, Chen Y, Dong B, Xue H, Lei H, Lu Y, Wei X, Sun P. 2022. Changes of the vaginal microbiota in HPV infection and cervical intraepithelial neoplasia: a cross-sectional analysis. Sci Rep 12:2812. doi: 10.1038/s41598-022-06731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chorna N, Romaguera J, Godoy-Vitorino F. 2020. Cervicovaginal microbiome and urine metabolome paired analysis reveals niche partitioning of the microbiota in patients with human papilloma virus infections. Metabolites 10:36. doi: 10.3390/metabo10010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ilhan ZE, Łaniewski P, Thomas N, Roe DJ, Chase DM, Herbst-Kralovetz MM. 2019. Deciphering the complex interplay between microbiota, HPV, inflammation and cancer through cervicovaginal metabolic profiling. eBioMedicine 44:675–690. doi: 10.1016/j.ebiom.2019.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shen J, Song N, Williams CJ, Brown CJ, Yan Z, Xu C, Forney LJ. 2016. Effects of low dose estrogen therapy on the vaginal microbiomes of women with atrophic vaginitis. Sci Rep 6:24380. doi: 10.1038/srep24380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gustin AT, Thurman AR, Chandra N, Schifanella L, Alcaide M, Fichorova R, Doncel GF, Gale M, Klatt NR. 2022. Recurrent bacterial vaginosis following metronidazole treatment is associated with microbiota richness at diagnosis. Am J Obstet Gynecol 226:225.e1–e15. doi: 10.1016/j.ajog.2021.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Marrazzo JM, Dombrowski JC, Wierzbicki MR, Perlowski C, Pontius A, Dithmer D, Schwebke J. 2019. Safety and efficacy of a novel vaginal anti-infective, TOL-463, in the treatment of bacterial vaginosis and vulvovaginal candidiasis: a randomized, single-blind, phase 2, controlled trial. Clin Infect Dis 68:803–809. doi: 10.1093/cid/ciy554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Plummer EL, Bradshaw CS, Doyle M, Fairley CK, Murray GL, Bateson D, Masson L, Slifirski J, Tachedjian G, Vodstrcil LA. 2021. Lactic acid-containing products for bacterial vaginosis and their impact on the vaginal microbiota: a systematic review. PLoS One 16:e0246953. doi: 10.1371/journal.pone.0246953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME. 2014. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 114.Ou Y-C, Fu H-C, Tseng C-W, Wu C-H, Tsai C-C, Lin H. 2019. The influence of probiotics on genital high-risk human papilloma virus clearance and quality of cervical smear: a randomized placebo-controlled trial. BMC Womens Health 19:103. doi: 10.1186/s12905-019-0798-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Palma E, Recine N, Domenici L, Giorgini M, Pierangeli A, Panici PB. 2018. Long-term Lactobacillus rhamnosus BMX 54 application to restore a balanced vaginal ecosystem: a promising solution against HPV-infection. BMC Infect Dis 18:13. doi: 10.1186/s12879-017-2938-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cohen CR, Wierzbicki MR, French AL, Morris S, Newmann S, Reno H, Green L, Miller S, Powell J, Parks T, Hemmerling A. 2020. Randomized trial of lactin-V to prevent recurrence of bacterial vaginosis. N Engl J Med 382:1906–1915. doi: 10.1056/NEJMoa1915254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Giunta G, Giuffrida L, Mangano K, Fagone P, Cianci A. 2012. Influence of lactoferrin in preventing preterm delivery: a pilot study Corrigendum in/mmr/7/4/1366. Mol Med Rep 5:162–166. [DOI] [PubMed] [Google Scholar]
- 118.Russo R, Karadja E, De Seta F. 2019. Evidence-based mixture containing Lactobacillus strains and lactoferrin to prevent recurrent bacterial vaginosis: a double blind, placebo controlled, randomised clinical trial. Benef Microbes 10:19–26. doi: 10.3920/BM2018.0075. [DOI] [PubMed] [Google Scholar]
- 119.Otsuki K, Imai N. 2017. Effects of lactoferrin in 6 patients with refractory bacterial vaginosis. Biochem Cell Biol 95:31–33. doi: 10.1139/bcb-2016-0051. [DOI] [PubMed] [Google Scholar]
- 120.Siqueiros-Cendón T, Arévalo-Gallegos S, Iglesias-Figueroa BF, García-Montoya IA, Salazar-Martínez J, Rascón-Cruz Q. 2014. Immunomodulatory effects of lactoferrin. Acta Pharmacol Sin 35:557–566. doi: 10.1038/aps.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nilsen T, Swedek I, Lagenaur LA, Parks TP, McBain AJ. 2020. Novel selective inhibition of Lactobacillus iners by Lactobacillus-derived bacteriocins. Appl Environ Microbiol 86:e01594-20. doi: 10.1128/AEM.01594-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rohlke F, Stollman N. 2012. Fecal microbiota transplantation in relapsing Clostridium difficile infection. Therap Adv Gastroenterol 5:403–420. doi: 10.1177/1756283X12453637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lev-Sagie A, Goldman-Wohl D, Cohen Y, Dori-Bachash M, Leshem A, Mor U, Strahilevitz J, Moses AE, Shapiro H, Yagel S, Elinav E. 2019. Vaginal microbiome transplantation in women with intractable bacterial vaginosis. Nat Med 25:1500–1504. doi: 10.1038/s41591-019-0600-6. [DOI] [PubMed] [Google Scholar]
- 124.Chen T, Xia C, Hu H, Wang H, Tan B, Tian P, Zhao X, Wang L, Han Y, Deng K-Y, Wei H, Xin H-B. 2021. Dysbiosis of the rat vagina is efficiently rescued by vaginal microbiota transplantation or probiotic combination. Int J Antimicrob Agents 57:106277. doi: 10.1016/j.ijantimicag.2021.106277. [DOI] [PubMed] [Google Scholar]
- 125.Lam KN, Spanogiannopoulos P, Soto-Perez P, Alexander M, Nalley MJ, Bisanz JE, Nayak RR, Weakley AM, Yu FB, Turnbaugh PJ. 2021. Phage-delivered CRISPR-Cas9 for strain-specific depletion and genomic deletions in the gut microbiome. Cell Rep 37:109930. doi: 10.1016/j.celrep.2021.109930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Landlinger C, Tisakova L, Oberbauer V, Schwebs T, Muhammad A, Latka A, Van Simaey L, Vaneechoutte M, Guschin A, Resch G, Swidsinski S, Swidsinski A, Corsini L. 2021. Engineered phage endolysin eliminates Gardnerella biofilm without damaging beneficial bacteria in bacterial vaginosis ex vivo. Pathogens 10:54. doi: 10.3390/pathogens10010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Feehily C, Crosby D, Walsh CJ, Lawton EM, Higgins S, McAuliffe FM, Cotter PD. 2020. Shotgun sequencing of the vaginal microbiome reveals both a species and functional potential signature of preterm birth. NPJ Biofilms Microbiomes 6:50. doi: 10.1038/s41522-020-00162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wolfarth AA, Smith TM, VanInsberghe D, Dunlop AL, Neish AS, Corwin EJ, et al. 2020. A human microbiota-associated murine model for assessing the impact of the vaginal microbiota on pregnancy outcomes. Front Cell Infect Microbiol 10. doi: 10.3389/fcimb.2020.570025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Łaniewski P, Herbst-Kralovetz MM. 2021. Bacterial vaginosis and health-associated bacteria modulate the immunometabolic landscape in 3D model of human cervix. NPJ Biofilms Microbiomes 7:88. doi: 10.1038/s41522-021-00259-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fritz JV, Desai MS, Shah P, Schneider JG, Wilmes P. 2013. From meta-omics to causality: experimental models for human microbiome research. Microbiome 1:14. doi: 10.1186/2049-2618-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Serebrenik J, Wang T, Hunte R, Srinivasan S, McWalters J, Tharp GK, Bosinger SE, Fiedler TL, Atrio JM, Murphy K, Barnett R, Ray LR, Krows ML, Fredricks DN, Irungu E, Ngure K, Mugo N, Marrazzo J, Keller MJ, Herold BC. 2021. Differences in vaginal microbiota, host transcriptome, and proteins in women with bacterial vaginosis are associated with metronidazole treatment response. The J Infectious Diseases 224:2094–2104. doi: 10.1093/infdis/jiab266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li C, Gu Y, He Q, Huang J, Song Y, Wan X, Li Y. 2021. Integrated analysis of microbiome and transcriptome data reveals the interplay between commensal bacteria and fibrin degradation in endometrial cancer. Front Cell Infect Microbiol 11:748558. doi: 10.3389/fcimb.2021.748558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ventolini G, Vieira-Baptista P, De Seta F, Verstraelen H, Lonnee-Hoffmann R, Lev-Sagie A. 2022. The vaginal microbiome: IV. The role of vaginal microbiome in reproduction and in gynecologic cancers. J Low Genit Tract Dis 26:93–98. doi: 10.1097/LGT.0000000000000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Riganelli L, Iebba V, Piccioni M, Illuminati I, Bonfiglio G, Neroni B, et al. 2020. Structural variations of vaginal and endometrial microbiota: hints on female infertility. Front Cell Infect Microbiol 10. doi: 10.3389/fcimb.2020.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Moreno I, Codoñer FM, Vilella F, Valbuena D, Martinez-Blanch JF, Jimenez-Almazán J, Alonso R, Alamá P, Remohí J, Pellicer A, Ramon D, Simon C. 2016. Evidence that the endometrial microbiota has an effect on implantation success or failure. American J Obstetrics & Gynecology 215:684–703. doi: 10.1016/j.ajog.2016.09.075. [DOI] [PubMed] [Google Scholar]
- 136.Wei W, Zhang X, Tang H, Zeng L, Wu R. 2020. Microbiota composition and distribution along the female reproductive tract of women with endometriosis. Ann Clin Microbiol Antimicrob 19:15. doi: 10.1186/s12941-020-00356-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Molina NM, Sola-Leyva A, Haahr T, Aghajanova L, Laudanski P, Castilla JA, Altmäe S. 2021. Analysing endometrial microbiome: methodological considerations and recommendations for good practice. Hum Reprod 36:859–879. doi: 10.1093/humrep/deab009. [DOI] [PubMed] [Google Scholar]
- 138.Tuddenham S, Ravel J, Marrazzo JM. 2021. Protection and risk: male and female genital microbiota and sexually transmitted infections. J Infect Dis 223:S222–S35. doi: 10.1093/infdis/jiaa762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tuominen H, Rautava J, Kero K, Syrjänen S, Collado MC, Rautava S. 2021. HPV infection and bacterial microbiota in the semen from healthy men. BMC Infect Dis 21:373. doi: 10.1186/s12879-021-06029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Norenhag J, Du J, Olovsson M, Verstraelen H, Engstrand L, Brusselaers N. 2020. The vaginal microbiota, human papillomavirus and cervical dysplasia: a systematic review and network meta-analysis. BJOG: Int J Obstet Gy 127:171–180. doi: 10.1111/1471-0528.15854. [DOI] [PubMed] [Google Scholar]
- 141.López-Filloy M, Cortez FJ, Gheit T, Cruz y Cruz O, Cruz-Talonia F, Chávez-Torres M, et al. 2022. Altered vaginal microbiota composition correlates with human papillomavirus and mucosal immune responses in women with symptomatic cervical ectopy. Front Cell Infect Microbiol 12. doi: 10.3389/fcimb.2022.884272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Qingqing B, Jie Z, Songben Q, Juan C, Lei Z, Mu X. 2021. Cervicovaginal microbiota dysbiosis correlates with HPV persistent infection. Microb Pathog 152:104617. doi: 10.1016/j.micpath.2020.104617. [DOI] [PubMed] [Google Scholar]