Significance
Androgen receptor (AR) signaling is crucial for driving prostate cancer (PCa), the second leading cause of American male cancer patient death. Castration is a standard therapy for advanced or metastatic PCa. However, patients inevitably develop lethal castration-resistant PCa (CRPC) also resistant to the newest AR inhibitors. GATA2 is a pioneer transcription factor essential for driving AR activation, PCa growth, and castration resistance. However, it is challenging to inhibit GATA2 transcriptional activity. The GATA2 protein is unstable, which makes further enhancement of GATA2 degradation a promising therapeutic strategy. We identified constitutive photomorphogenesis protein 1 (COP1) as the E3 ubiquitin ligase for GATA2 degradation and discovered the striking effects of a COP1-GATA2 axis on suppressing AR activation, PCa growth, and castration resistance.
Keywords: prostate cancer, AR, GATA2, COP1, ubiquitination
Abstract
Androgen receptor (AR) signaling is crucial for driving prostate cancer (PCa), the most diagnosed and the second leading cause of death in male patients with cancer in the United States. Androgen deprivation therapy is initially effective in most instances of AR-positive advanced or metastatic PCa. However, patients inevitably develop lethal castration-resistant PCa (CRPC), which is also resistant to the next-generation AR signaling inhibitors. Most CRPCs maintain AR expression, and blocking AR signaling remains a main therapeutic approach. GATA2 is a pioneer transcription factor emerging as a key therapeutic target for PCa because it promotes AR expression and activation. While directly inhibiting GATA2 transcriptional activity remains challenging, enhancing GATA2 degradation is a plausible therapeutic strategy. How GATA2 protein stability is regulated in PCa remains unknown. Here, we show that constitutive photomorphogenesis protein 1 (COP1), an E3 ubiquitin ligase, drives GATA2 ubiquitination at K419/K424 for degradation. GATA2 lacks a conserved [D/E](x)xxVP[D/E] degron but uses alternate BR1/BR2 motifs to bind COP1. By promoting GATA2 degradation, COP1 inhibits AR expression and activation and represses PCa cell and xenograft growth and castration resistance. Accordingly, GATA2 overexpression or COP1 mutations that disrupt COP1-GATA2 binding block COP1 tumor-suppressing activities. We conclude that GATA2 is a major COP1 substrate in PCa and that COP1 promotion of GATA2 degradation is a direct mechanism for regulating AR expression and activation, PCa growth, and castration resistance.
Prostate cancer (PCa) remains the most diagnosed disease and the second leading cause of death for male patients with cancer in the United States (1). Androgen receptor (AR) signaling is crucial for driving most cases of PCa. Therefore, androgen-ablation (castration) therapy is a standard therapy for advanced and metastatic PCa. However, patients inevitably develop lethal castration-resistant PCa (CRPC), which is also resistant to advanced therapies that use enzalutamide, apalutamide, and abiraterone (2–6). The majority of CRPCs express AR, which maintains activation, and blocking AR signaling remains a main therapeutic approach for CRPC.
GATA2 is a pioneer transcription factor essential for AR expression and activation in PCa, including CRPC (7–9). The expression of GATA2 in human PCa tissues correlates with advanced disease stages and worse prognosis, and GATA2 expression is further increased in metastatic therapy-resistant PCa (9–12). Therefore, GATA2 is emerging as a central driver of lethal CRPC (13). Although it is challenging to inhibit GATA2 transcriptional activity directly, the GATA2 protein is unstable (14, 15). Thus, further increasing GATA2 protein degradation is a promising therapeutic strategy. The molecular mechanisms governing GATA2 protein stability in PCa are currently unknown.
Constitutive photomorphogenesis protein 1 (COP1), also known as ring finger and WD-repeat domain 2 (RFWD2), is a ring finger E3 ubiquitin ligase. COP1 carries both nuclear localization signal and nuclear export signal sequences, which allow it to shuttle between nucleus and cytosol (16). COP1 is a negative modulator of light-regulated development in Arabidopsis thaliana (17). In the dark, A. thaliana COP1 is confined to the nucleus, where it directs the ubiquitination and degradation of several key transcription factors involved in turning on light-activated genes. In contrast, COP1 translocation into cytoplasm in the light stabilizes its nuclear substrates (18, 19). In mammalian cells, COP1 mainly resides in the nucleus, with a small amount present in cytosol (20). However, binding of 14–3-3σ in response to DNA damage promotes its nuclear export (21). Mammalian COP1 primarily functions as a tumor suppressor by targeting degradation of oncogenic transcription factors such as JUN, MTA1, and ETV (22–24). In certain cellular contexts, COP1 may also function as an oncogene by directly targeting p53 degradation. However, it remains unclear whether p53 is a bona fide COP1 target in vivo (25). Studies in COP1 knockout models have confirmed the tumor suppressing, but not tumor promoting, activity of COP1 in vivo (24, 25).
Only two studies have directly addressed COP1 biology in PCa. In 2011, Vitari et al (24) reported that COP1 inhibits the migration of, but not the growth of, human PCa cells by promoting degradation of ETS transcription factors including ETV1, ETV4, and ETV5. Dallavalle et al (26) showed that COP1 inhibits STAT3 to suppress PCa, but without directly assessing AR-positive PCa growth. This apparent lack of direct growth inhibition activity of COP1 in PCa might have decreased interest in COP1 biology in PCa.
Here we report GATA2 as a bona fide COP1 substrate for degradation in PCa and define the detailed molecular mechanism underlying human COP1-GATA2 interaction. By promoting GATA2 protein degradation, COP1 inhibits AR expression and activation and PCa growth, including castration-resistant growth.
Results
COP1 Represses GATA2 Protein Levels in PCa.
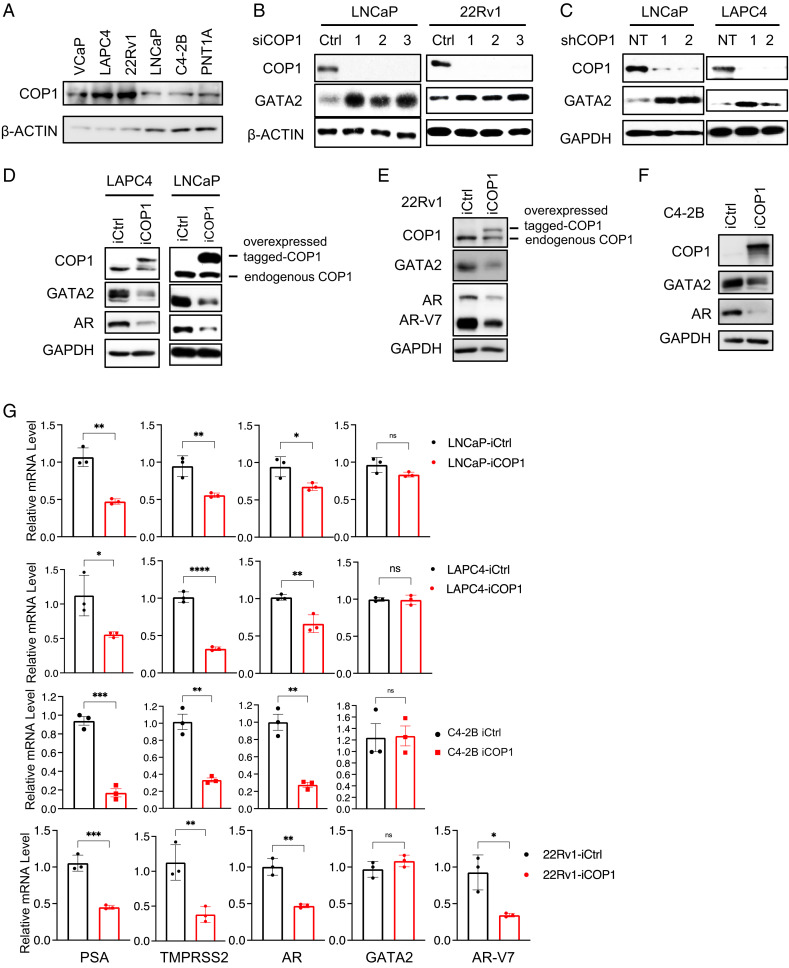
The identity of the E3 ligase for GATA2 degradation in PCa is unknown. A. thaliana COP1 promotes GATA2 degradation in the light-brassinosteroid pathway (27); however, A. thaliana GATA2 protein is only about half the size of human GATA2, and the sequence similarity of both A. thaliana GATA2 and COP1 and human GATA2 and COP1 is very low (SI Appendix, Figs. S1 and S2). It is currently unknown whether human or mammalian COP1 regulates human or mammalian GATA2. To examine whether human COP1 regulates human GATA2 protein levels in PCa, we first confirmed that COP1 is expressed at different levels in commonly used human PCa cell lines and in normal human prostate epithelial PNT1A cells (Fig. 1A). We then performed knockdown of COP1 using three independent short interfering RNAs (siRNAs) against COP1 in LNCaP and 22Rv1 cells. We found that this siRNA-mediated knockdown of COP1 increased GATA2 protein levels (Fig. 1B). Knockdown of COP1 using short hairpin RNA (shRNA) approaches similarly led to enhanced GATA2 levels in the engineered LNCaP and LAPC4 cells (Fig. 1C). Altogether, five different siRNAs and shRNAs were used to knock down COP1, which addressed the potential off-target effects of RNA interference. In accord with these loss-of-function data, doxycycline (Dox)-induced ectopic expression of COP1 tagged by two DYKDDDDK amino acid (aa) sequences (2× Flag-tagged) reduced GATA2 protein levels in LNCaP, LAPC4, 22Rv1, and C4-2B cells (Fig. 1 D–F).
Fig. 1.
COP1 inhibits GATA2 and AR in PCa cells. (A) COP1 expression in human PCa cell lines. (B) Knockdown of COP1 in LNCaP and 22Rv1 cells using three different COP1 siRNAs (siCOP1, 1–3). Control (Ctrl), siRNA targeting luciferase. (C) Knockdown of COP1 in LNCaP and LAPC4 cells using two independent shRNAs (shCOP1, 1–2). NT, nontargeting control. The engineered (D) LAPC4, LNCaP, (E) 22Rv1, and (F) C4-2B cells with Dox-inducible expression of Flag-tagged COP1 (iCOP1) or control (iCtrl) were treated with 0.5 µg/mL Dox for 5 d before Western blotting analysis. All data shown in A–F are Western blots. (G) qPCR analysis of gene expression of engineered LNCaP, LAPC4, C4-2B, and 22Rv1 iCOP1 cells versus iCtrl cells induced with 0.5 µg/mL Dox for 5 d. Quantification data as means ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. ns, not significant. Data are representative of at least three independent experiments.
COP1 Represses AR Expression and Activation in PCa.
Since COP1 represses GATA2 protein levels (Fig. 1 B–F) and GATA2 plays critical roles in AR expression and activation in PCa (7–9), we examined whether COP1 represses AR expression and activation. Indeed, Dox-induced COP1 overexpression inhibited AR expression and activation as evidenced by inhibition of expression of AR targets prostate-specific antigen (PSA) and TMPRSS2 in LNCaP, LAPC4, C4-2B, and 22Rv1 cells (Fig. 1 D–G). AR-V7 is a splicing variant of AR that exhibits androgen-independent transactivation, thus providing a key mechanism for the development of CRPC (28–33). 22Rv1 cells express high levels of AR-V7 and, at lower levels, full-length AR (30, 31). Dox-induced COP1 overexpression in 22Rv1 cells reduced the expression of both AR-V7 and full-length AR and accordingly repressed AR activation (Fig. 1 E and G). Altogether, these data strongly support that COP1 represses AR expression and activation in PCa.
COP1 is a Major E3 Ubiquitin Ligase for GATA2 in PCa Cells.
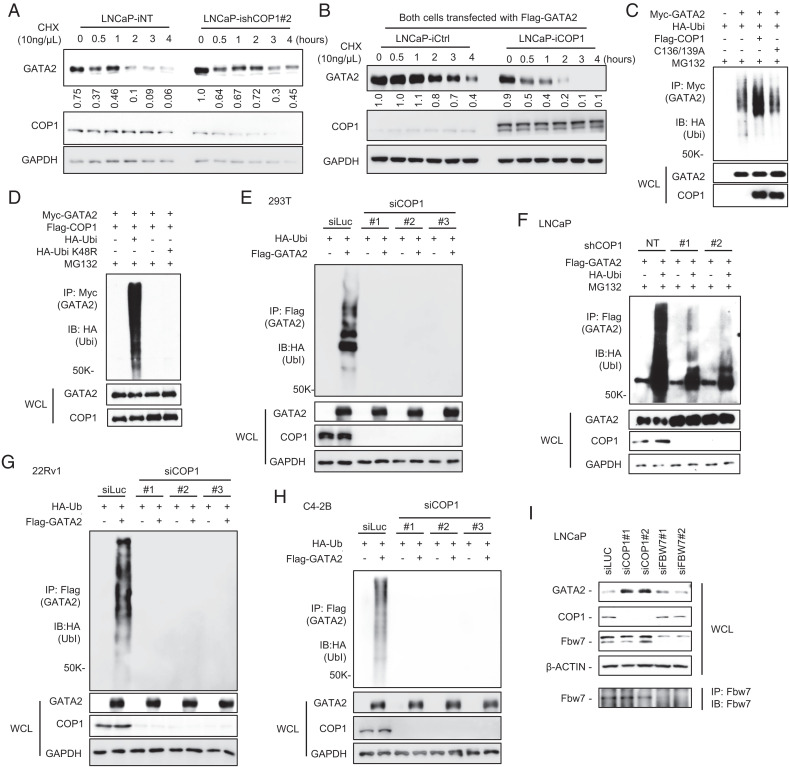
GATA2 messenger RNA (mRNA) levels are comparable among the COP1-overexpressing versus control LNCaP, LAPC4, C4-2B, and 22Rv1 cells (Fig. 1G), indicating that COP1 reduces GATA2 protein but not mRNA levels in PCa. Further analysis revealed that knockdown of COP1 enhances endogenous GATA2 protein stability in LNCaP cells (Fig. 2A), and COP1 overexpression reduces the stability of an ectopically overexpressed GATA2 protein (Fig. 2B). Together, these data suggest that COP1 reduces GATA2 protein levels in PCa by promoting GATA2 degradation.
Fig. 2.
COP1 is an E3 ubiquitin ligase for GATA2. (A) LNCaP cells with 2 µg/mL Dox-induced knockdown of COP1 (ishCOP1#2) versus nontargeting control (iNT) were treated with 10 ng/µL chlorhexidine (CHX) for 0–4 h. (B) Flag-tagged GATA2 was transfected into LNCaP cells with 0.5 µg/mL Dox-induced ectopic expression of COP1 (iCOP1) versus control (iCtrl). After 48 h, the cells were treated with 10 µM MG132 for 2 h. MG132 was then removed, and cells were treated with 10 ng/µL CHX for 0–4 h. Western blots were quantified using Image J. (C) Ubiquitination assay. HEK293T cells were transfected with Myc-tagged GATA2 (Myc-GATA2), HA-tagged ubiquitin (HA-Ubi), and Flag-tagged COP1 (Flag-COP1) versus an E3 ligase-dead COP1 mutant (Flag-C136/139A). After 48 h, the cells were treated with 10 μM MG132 for 6 h before preparing cell lysate for the indicated immunoprecipitation (IP) and Western blot (IB) analyses; WCL, whole cell lysate. (D) HEK293T cells were transfected with Myc-GATA2, Flag-COP1, or HA-Ubi versus HA-Ubi K48R mutant. After 48 h, the cells were treated with 10 μM MG132 for 6 h before preparing cell lysate for IP and IB analysis. (E) HEK293T cells were transfected with Flag-tagged GATA2 (Flag-GATA2), HA-Ubi, and three different siRNAs against COP1 (siCOP1) versus control (siLuc). After 72 h, the cells were treated with 10 μM MG132 for 6 h before preparing cell lysate for IP and IB analysis. (F) LNCaP cells with COP1 knockdown (shCOP1 #1 and #2) versus control (NT) were transfected with Flag-GATA2 and HA-Ubi. After 48 h, the cells were treated with 10 μM MG132 for 4 h before preparing cell lysate for IP and IB analysis. (G) 22Rv1 and (H) C4-2B cells were transfected with three different siRNAs against COP1 (siCOP1) versus control (siLuc). After 24 h, the cells were transfected with FLAG-tagged GATA2 (Flag-GATA2) or HA-Ubi. After 48 h, the cells were treated with 10 μM MG132 for 4 h before preparing cell lysate for IP and IB analysis. (I) LNCaP cells were transfected with siLuc control or two independent siRNAs against COP1 (siCOP1) or FBW7 (siFBW7). After 72 h, the cell lysates were prepared for IB. IP with anti-Fbw7 followed by IB with anti-Fbw7 was used to further confirm knockdown of Fbw7. All data are representative of at least three independent experiments and are presented as Western blots.
To investigate COP1 as an E3 ligase for GATA2 in mammalian cells, we performed ubiquitination assays of GATA2 by HEK293T cell co-transfection of Myc-tagged GATA2 and hemagglutinin (HA)-tagged ubiquitin along with Flag-tagged wild-type COP1 or the E3 ubiquitin ligase-dead COP1C136/139A mutant (24). After a 6-h treatment with MG132, the cells were processed for immunoprecipitation using an anti-Myc antibody (GATA2) followed by Western blot using an anti-HA antibody (ubiquitin). We confirmed that overexpression of COP1, but not the E3 ligase-dead COP1C136/139A mutant, enhances GATA2 ubiquitination in HEK293T cells (Fig. 2C). We next performed GATA2 ubiquitination assays in HEK293T cells co-transfected with either the wild-type ubiquitin or the ubiquitin K48R mutant that blocks the formation of K48-linked polyubiquitin chains (34). We observed that the K48R mutation totally abolished COP1-induced GATA2 ubiquitination in HEK293T cells (Fig. 2D). This indicates that COP1 promotes the K48-linked polyubiquitination of GATA2, which is consist with the previous reports of K48-linked polyubiquitination as a key pathway for 26S proteasome-mediated degradation (34). Since we observed significant levels of GATA2 ubiquitination in the HEK293T cells without ectopic expression of COP1, we also performed knockdown of COP1 in HEK293T cells using three independent siRNAs and similarly performed ubiquitination assays on the transfected Flag-tagged GATA2. Interestingly, knockdown of endogenous COP1 in HEK293T cells largely abolished GATA2 ubiquitination, indicating that COP1 is the bona fide E3 ubiquitin ligase for GATA2 in these HEK293T cells (Fig. 2E).
To further investigate whether COP1 is the key E3 ligase for GATA2 degradation in PCa cells, we performed ubiquitination assays of GATA2 in LNCaP cells and CRPC 22Rv1 and C4-2B cells. Knockdown of COP1 greatly inhibited GATA2 ubiquitination in all these cells, supporting COP1 as a key E3 ligase for COP1 degradation in PCa, including CRPC (Fig. 2 F–H). Since SKP1–cullin-1–F-box complex containing FBW7 as the F-box protein (SCFFbw7) has been shown to be an E3 ligase for GATA2 in the K562 human leukemia cell line (35), we compared the impact of COP1 and SCFFbw7 in regulating GATA2 in PCa. We confirmed that in accordance with our data in Fig. 1 B and C, COP1 knockdown enhanced GATA2 protein levels in LNCaP cells. In contrast, Fbw7 knockdown had no appreciable effect on GATA2 protein levels (Fig. 2I). Please note that since we observed a higher molecular weight band in the Western blots for Fbw7, we also performed immunoprecipitation using anti-Fbw7 antibody followed by Western blots using the same anti-Fbw7 antibody. We verified the knockdown of Fbw7 in the FBW7 siRNA (SiFbw7)-transfected LNCaP cells (Fig. 2I). Collectively, these data strongly suggest that COP1 is the major E3 ubiquitin ligase for GATA2 degradation in PCa.
COP1 Binds GATA2.
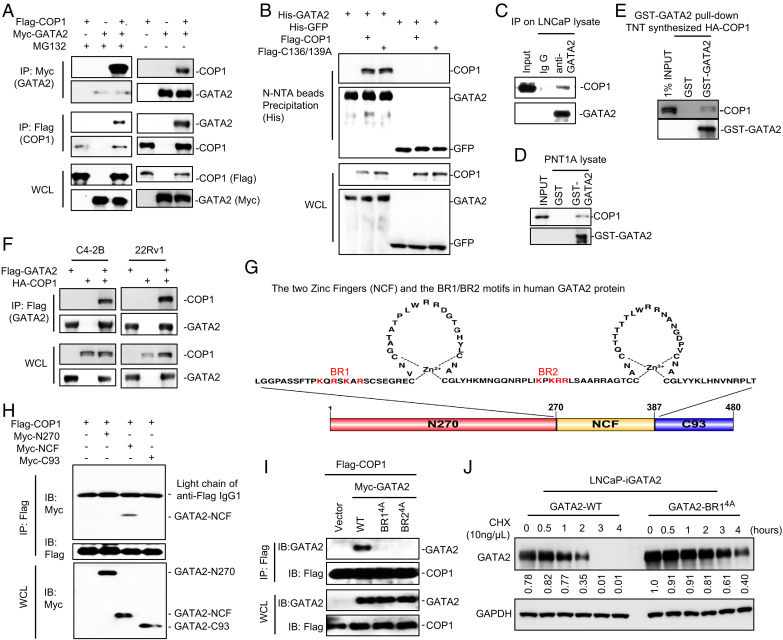
COP1 function as an E3 ligase for GATA2 requires COP1-GATA2 interaction. To investigate this interaction, we expressed Flag-tagged COP1 and Myc-tagged GATA2 in HEK293T cells. After a 6-h MG132 treatment, the cells were processed for immunoprecipitation using anti-Flag antibody-conjugated beads followed by Western blots using an anti-Myc antibody and reciprocal immunoprecipitation using an anti-Myc antibody followed by Western blots using an anti-Flag antibody. In all cases, we detected the co-immunoprecipitation of COP1 and GATA2 (Fig. 3A). We also confirmed this observation in cells without MG132 treatments (Fig. 3A). Next, we similarly expressed histidine (His)-tagged GATA2 (versus His-tagged green fluorescent protein [GFP] as a negative control) together with Flag-tagged COP1 or the Flag-tagged E3 ligase–dead COP1C136/139A mutant in HEK293T cells and used these cells to perform co-immunoprecipitation studies. We demonstrated that the COP1C136/139A mutant retains similar ability to bind GATA2, indicating that the C136 and C139 residues in the ring domain of COP1 are not critical for COP1-GATA2 interaction (Fig. 3B). To further confirm interaction, we prepared cell lysates from LNCaP cells treated with the proteasome inhibitor MG132 and performed immunoprecipitation using anti-GATA2 antibody followed by Western blots using anti-COP1 antibody. We readily detected the co-immunoprecipitation of GATA2 and COP1, confirming that endogenous GATA2 binds with endogenous COP1 in LNCaP cells (Fig. 3C). This direct interaction was further confirmed by the ability of the overexpressed and purified glutathione S-transferase (GST)-GATA2 fusion protein, but not GST protein, to pull down endogenous COP1 from the PNT1A cell lysate (Fig. 3D). GST-GATA2 also pulled down an in vitro synthesized HA-tagged COP1 protein using the TNT Quick Coupled Transcription and Translation System (Fig. 3E). Finally, we confirmed COP1 and GATA2 interaction in CRPC C4-2B and 22Rv1 cells (Fig. 3F).
Fig. 3.
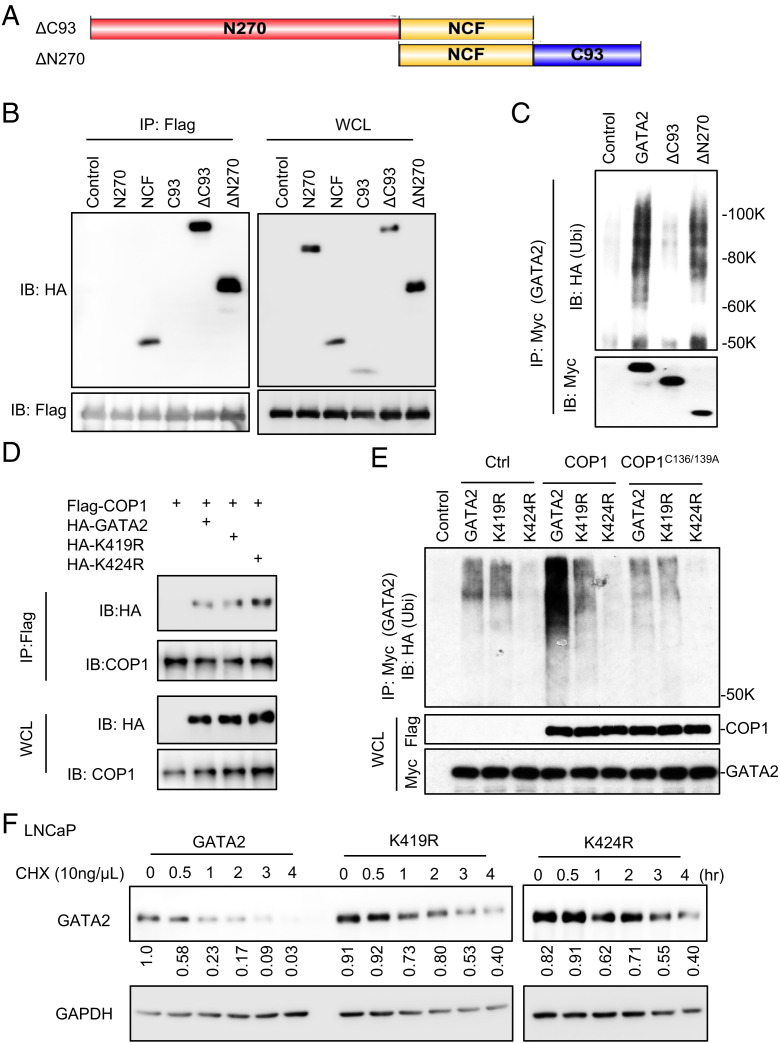
The BR1/BR2 motifs of GATA2 are essential for COP1 binding. (A) HEK293T cells were transfected with Myc-tagged GATA2 (Myc-GATA2) and Flag-tagged COP1 (Flag-COP1). After 48 h, cells were treated with 10 μM MG132 versus vehicle control for 6 h before preparing cell lysate for IP using anti-Myc antibody (GATA2) or anti-Flag antibody (COP1) and for IB analysis. (B) HEK293T cells were transfected with His-tagged GATA2 (His-GATA2) versus control His-tagged enhanced green fluorescent protein (eGFP) and Flag-COP1 versus an E3 ligase-dead COP1 mutant (Flag-C136/139A). After 48 h, the cells were treated with 10 μM MG132 for 6 h before preparing cell lysate for His-tagged protein precipitation using Ni-NTA beads and IB analysis. (C) LNCaP cells were treated with 10 μM MG132 for 4 h before preparing cell lysate for the indicated IP and IB analysis. (D) GST-tagged GATA2 (GST-GATA2) protein and GST protein (as control) were overexpressed in E. coli and purified and conjugated onto glutathione Sepharose beads. The beads were then used in the pull-down assay for GATA2 binding with the endogenous COP1 in the PNT1A cell lysate. (E) COP1 protein was synthesized using the TNT In Vitro Kit. GST-GATA2 protein-conjugated beads were used in the pull-down assay for GATA2 binding with the COP1 synthesized in vitro. (F) C4-2B and 22Rv1 cells were transfected with Flag-tagged GATA2 (Flag-GATA2) and HA-tagged COP1 (HA-COP1). After 48 h, cells were treated with 10 μM MG132 for 4 h before preparing cell lysate for IP using anti-FLAG antibody (GATA2) and IB analysis. (G) Illustration of the two zinc finger domains in human GATA2. The basic K/R residues in the BR1 and BR2 motifs are highlighted in red. (H) HEK293T cells were transfected with Flag-COP1 and Myc-tagged truncates of GATA2, including N270 (N-terminal 1–270 aa of GATA2), NCF (N- and C-terminal zinc finger, 271–387 aa), and C93 (C-terminal 388–480 aa, 93 aa long) as shown in E. After 48 h, the cells were treated with 10 μM MG132 for 6 h before preparing cell lysate for IP using anti-Flag affinity gel and IB analysis. (I) HEK293T cells were transfected with Flag-COP1 and Myc-tagged GATA2 versus GATA2 BR1 K/R-4A or GATA2 BR2 K/R-4A mutants. The cells were treated with 10 μM MG132 for 6 h before preparing cell lysate for IP using anti-Flag affinity gel and IB analysis. (J) LNCaP cells with 0.5 µg/mL Dox-induced expression of wild type (WT) versus BR14A-mutated GATA2 were treated with 10 ng/µL chlorhexidine (CHX) for 0–4 h. Data in A–F and H–J are presented as Western blots. Western blot data in (J) were quantified using Image J and are representative of at least three independent experiments. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; WCL, whole cell lysate.
The BR1/BR2 Motifs of GATA2 Are Essential for COP1 Binding.
Most COP1 substrates contain a consensus [D/E](x)xxVP[D/E] degron (20). GATA2 lacks such degrons, raising the question of how COP1 binds GATA2 for degradation. GATA2 contains two zinc finger motifs for binding to DNA and/or interaction with other proteins (Fig. 3G) (36, 37). To assess whether these zinc finger motifs are crucial for GATA2-COP1 interaction, we expressed a Flag-tagged COP1 together with the Myc-tagged GATA2 fragments, including the N270 fragment (containing the N-terminal 270 amino acid [270 aa] of GATA2), the NCF fragment (containing both the N- and C-terminal zinc fingers), and the C93 fragment (containing C-terminal 93 aa) in HEK293T cells. We then performed immunoprecipitation using anti-Flag affinity gel and Western blotting using an anti-Myc antibody. We demonstrated that the NCF domain, but not the N270 or C93 fragments of GATA2, binds to COP1 (Fig. 3H). This revealed that the zinc finger domain of GATA2 is essential for mediating GATA2-COP1 interaction (Fig. 3 G and H).
The NCF domain contains two Lysine or Arginine (K/R) basic aa residue–rich motifs (BR1 and BR2; Fig. 3G), which are essential for mediating GATA2 binding to DNA (36). Each of these BR motifs contains four K/R residues. To investigate the functional significance of these two BR motifs in mediating GATA2 binding to COP1, we mutated all the K and R residues (highlighted in red in Fig. 3G) in the BR1 or BR2 motifs to alanine to generate GATA2-BR14A and GATA2-BR24A mutants. Both mutants exhibited a nucleus-cytoplasm localization pattern similar to that of wild-type GATA2 with primarily nuclear localization (SI Appendix, Fig. S3A). Co-immunoprecipitation studies demonstrated that both mutations effectively block GATA2 binding to COP1 (Fig. 3I). Accordingly, the mutated GATA2 (GATA2-BR14A) protein is more stable than the wild-type GATA2 when it is overexpressed in LNCaP cells (Fig. 3J). Together, these data suggest that the BR1 and BR2 motifs of GATA2 provide essential docking sites for GATA2 binding to COP1.
COP1 Uses a Typical Mechanism to Bind GATA2.
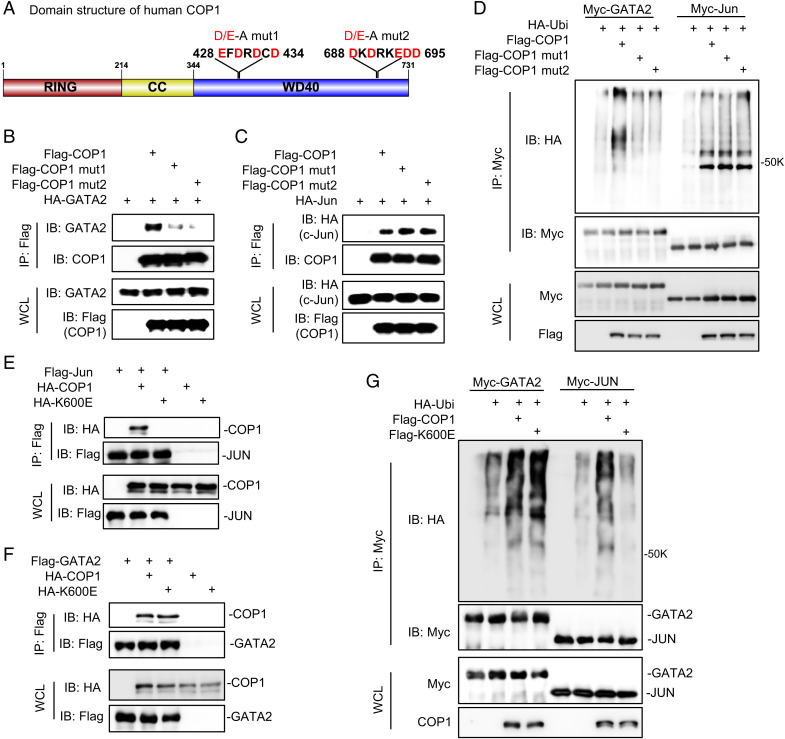
To identify the key COP1 regions for GATA2 binding, we examined COP1 protein sequence and found two D/E-enriched motifs with the potential to bind to the BR1/BR2 motifs in GATA2 through electrostatic interactions (Fig. 4A). Mutations of either 428EFDRDCD434 to AFARACA (COP1mut1) or 688DKDRKEDD695 to AKARKAAA (COP1mut2) inhibited COP1 binding to GATA2 (Fig. 4B), indicating that each of these two D/E-enriched motifs plays a role in COP1-GATA2 interaction. Furthermore, since our data suggests an atypical mechanism for COP1 binding to its substrate GATA2, we predicted that COP1mut1 and COP1mut2 would maintain binding to other known COP1 substrates carrying the consensus [D/E](x)xxVP[D/E] degron. Indeed, both COP1mut1 and COP1mut2 effectively bound to c-Jun (Fig. 4C). Accordingly, COP1mut1 and COP1mut2 could no longer promote GATA2 ubiquitination, but they maintained their ability to enhance c-Jun ubiquitination (Fig. 4D).
Fig. 4.
COP1 uses a typical mechanism to bind GATA2. (A) Domain structure of human COP1 and the two identified D/E-enriched motifs critical for COP1 binding to GATA2; CC, coiled coils; D/E-A, Mutations of EFDRDCD to AFARACA or DKDRKEDD to AKARKAAA; WD40 domain, a protein domain contains tandem copies of tryptophan–aspartate (WD) repeats of approximately 40 amino acids. (B) HEK293T cells were transfected with HA-tagged GATA2 (HA-GATA2) and Flag-tagged wild-type COP1 (Flag-COP1) versus two COP1 mutants (mut1 and mut2) with D/E-to-A mutations in the two D/E-enriched motifs as illustrated in A. After 48 h, cells were treated with 10 μM MG132 for 6 h before preparing the cell lysate for IP using anti-Flag affinity gel and IB analysis. (C) HEK293T cells were transfected with HA-tagged c-Jun (HA-Jun), Flag-COP1, Flag-COP1mut1, or Flag-COP1mut2. After 48 h, the cells were treated with 10 μM MG132 for 6 h before preparing the cell lysate for IP using anti-Flag affinity gel and IB analysis. (D) Ubiquitination assay. HA-Ubi and Myc-tagged GATA2 (Myc-GATA2) or c-Jun (Myc-Jun), along with Flag-COP1, Flag-COP1mut1, or Flag-COP1mut2 were co-transfected in HEK293T cells. After 48 h, the cells were treated with 10 μM MG132 for 6 h before preparing the cell lysate for IP using anti-Myc antibody and IB analysis. (E, F) HEK293T cells were transfected with HA-tagged COP1 (HA-COP1) or COP1K600E mutant (HA-K600E) together with (E) Flag-tagged c-Jun (Flag-Jun) versus control or (F) Flag-tagged GATA2 (Flag-GATA2) versus control. After 48 h, the cells were treated with 10 μM MG132 for 6 h before preparing the cell lysate for IP using anti-Flag affinity gel and IB analysis. (G) Ubiquitination assay. HA-Ubi and Myc-tagged GATA2 (Myc-GATA2) or c-Jun (Myc-Jun), along with Flag-tagged COP1 (Flag-COP1) or COP1K600E mutant (Flag-K600E) were co-transfected in HEK293T cells. After 48 h, the cells were treated with 10 μM MG132 for 6 h before preparing the cell lysate for IP using anti-Myc antibody and IB analysis. Data in B–G are shown as Western blots and are representative of at least three independent experiments; WCL, whole cell lysate.
Detailed data on how human COP1 binds to its canonical substrates containing the conserved [D/E](x)xxVP[D/E] degron (20) are not yet available, but K550E-mutated A. thaliana COP1 lost binding to its canonical substrates (38). Hence, we tested how mutation of the corresponding K residue in human COP1 (K600E) affects binding to human GATA2. Indeed, K600E mutation totally blocked COP1 binding to its canonical substrate c-Jun but minimally affected binding to GATA2 (Fig. 4 E and F). In accordance with these results, K600E mutation largely blocked COP1-mediated c-Jun ubiquitination but minimally affected GATA2 protein ubiquitination (Fig. 4G).
Together, these data strongly support that COP1 uses an atypical mechanism to bind GATA2, and the COP1mut1, COP1mut2, and COP1K600E mutants provide ideal tools for dissecting GATA2-dependent versus canonical substrate-dependent COP1 biology in PCa.
COP1 Ubiquitinates GATA2 at Lysine 419 and Lysine 424.
As an E3 ubiquitin ligase, COP1 is expected to catalyze the ubiquitination of GATA2 at lysine residues. To assess the key lysine residues in GATA2 for ubiquitination and degradation, we performed ubiquitination assays to assess the ubiquitination status of ectopically overexpressed Myc-tagged wild-type GATA2 and truncated GATA2 with deletion of either C93 (ΔC93) or N270 (ΔN270; Figs. 3G and 5A) in HEK293T cells with endogenous COP1 expression. Interestingly, although ΔC93 and ΔN270 fragments both contain the NCF domain essential for GATA2 binding to COP1 (Figs. 3H and 5B), ubiquitination of ΔC93, but not of ΔN270, was greatly repressed (Fig. 5C). This suggested that the C93 fragment of GATA2 contains the key lysine residues for ubiquitination.
Fig. 5.
COP1 ubiquitinates GATA2 at lysines 419 and 424. (A) Illustration of the ΔC93 (deletion of C93) and ΔN270 (deletion of N270) fragments of GATA2. (B) HEK293T cells were transfected with Flag-COP1 and HA-tagged GATA2 truncated, including N270, NCF, C93, ΔC93, and ΔN270 as shown in A and Fig. 3G. After 48 h, the cells were treated with 10 μM MG132 for 6 h before preparing the cell lysate for IP using anti-Flag affinity gel and IB analysis. (C) Ubiquitination assay. Myc-tagged GATA2 or its truncations ΔC93 or ΔN270 were transfected into HEK293T cells along with HA-Ubi. After 48 h, cells were treated with 10 μM MG132 for 6 h before preparing the cell lysate for IP using anti-Myc antibody and IB analysis. GAPDH, glyceraldehyde 3-phosphate dehydrogenase. (D) HEK293T cells were transfected with Flag-COP1 and HA-tagged GATA2 or GATA2 mutants K419R or K424R. After 48 h, cells were treated with 10 μM MG132 for 6 h before preparing the cell lysate for IP using anti-Flag affinity gel and IB analysis. (E) Ubiquitination assay. Myc-tagged GATA2 versus GATA2 mutants K419R or K424R and COP1 versus Ctrl or COP1C136/139A mutant was transfected into HEK293T cells along with HA-Ubi. After 48 h, cells were treated with 10 μM MG132 for 6 h before preparing the cell lysate for IP using anti-Myc antibody and IB analysis. (F) LNCaP cells with 0.5 µg/mL Dox-induced GATA2 or GATA2 mutant K419R or K424R expression were treated with 10 ng/µL chlorhexidine (CHX) for 0–4 h. GAPDH, glyceraldehyde 3-phosphate dehydrogenase. Data shown are Western blots and are representative of at least three independent experiments. Western blots in panel (F) were also quantified using Image J; WCL, whole cell lysate.
Prediction of GATA2 protein ubiquitination sites using UbPred (39) identified K419 and K424 as candidate sites within the C93 fragment of GATA2. Therefore, we generated mutants that abolish the potential K419- and K424-mediated ubiquitination of GATA2. We first confirmed that both GATA2K419R and GATA2K424R mutants maintain their affinity to COP1 when ectopically expressed in the HEK293T cells (Fig. 5D). We then used these two GATA2 mutants to perform the ubiquitination assays in the HEK293T cells ectopically overexpressing wild-type COP1, the E3 ligase-dead COP1C136/139A mutant, or control. We demonstrated that while overexpression of wild-type COP1 strongly enhanced GATA2 ubiquitination, this effect was greatly inhibited by the K419R mutation and was completely blocked by the K424R mutation (Fig. 5E). In contrast, the COP1C136/139A mutant did not affect the ubiquitination of either wild-type or mutated GATA2. Together, these data suggested that both K419 and K424 are essential sites for COP1-mediated GATA2 ubiquitination. In accordance with this, the GATA2K419R and GATA2K424R mutants were much more stable than the wild-type GATA2 when ectopically expressed in the HEK293T cells (Fig. 5F) in which COP1 is a key E3 ligase for GATA2 (Fig. 2E).
COP1-GATA2 Axis Represses AR Activation in PCa.
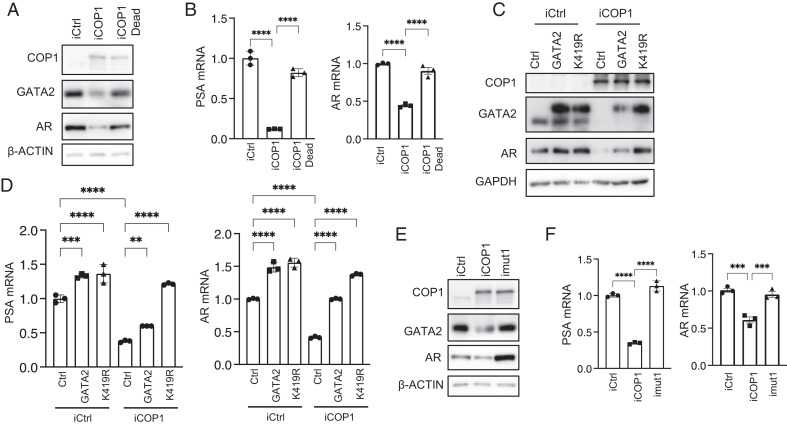
We have shown that COP1 represses AR expression and activation in PCa, including CRPC Fig. 1 D–G. To critically test the functional significance of GATA2 protein degradation in the COP1 repression of AR activation, we first demonstrated that the E3 ligase–dead COP1C136/139A minimally affected GATA2 and AR protein expression and AR activation (PSA expression), supporting that E3-ligase activity is essential for COP1 regulation of AR signaling (Fig. 6 A and B). We then overexpressed the GATA2K419R mutant resistant to COP1-induced ubiquitination and degradation (Fig. 5F) versus wild-type GATA2 in COP1-overexpressing LNCaP cells. This GATA2K419R mutant largely rescued AR expression and activity despite the COP1 overexpression (Fig. 6 C and D). In contrast, despite similar activities in the control LNCaP cells, overexpression of wild-type GATA2 could only partially rescue AR expression and activation in the COP1-overexpressing LNCaP cells, presumably because of their degradation by COP1 (Fig. 6 C and D and SI Appendix, Fig. S3B). To further define the COP1-GATA2-AR signaling axis, we assessed the activity of the COP1mut1, which lacks binding affinity to GATA2 (Fig. 4B) in LNCaP cells. As expected, this COP1mut1 minimally affected AR expression and activation in contrast to the inhibitory effect of wild-type COP1 (Fig. 6 E and F). Together, these data strongly support that COP1 inhibits AR signaling activation in PCa by repressing GATA2.
Fig. 6.
The COP1-GATA2 axis represses AR expression and activation in PCa cells. (A) Western blot and (B) qPCR assays on AR expression and activation (PSA expression) in the 0.5 µg/mL Dox-induced LNCaP cells expressing COP1 (iCOP1) versus the E3 ligase-dead COP1C136/139A mutant (iCOP1 dead) versus control (iCtrl). (C) Western blot and (D) qPCR assays on 0.5 µg/mL Dox-induced LNCaP cells expressing COP1 (iCOP1) versus control (iCtrl). These cells also overexpressed GATA2 versus GATA2K419R mutant (K419R) versus control (Ctrl). (E) Western blot and (F) qPCR assays on 0.5 µg/mL Dox-induced LNCaP cells expressing control (iCtrl), COP1 (iCOP1), or COP1mut1 with mutated first D/E-enriched motif as shown in Fig. 4A (imut1). Quantification data as means ± SD. **P < 0.01, ***P < 0.001, ****P < 0.0001. Data are representative of at least three independent experiments.
COP1 Represses the Growth of PCa Cells In Vitro.
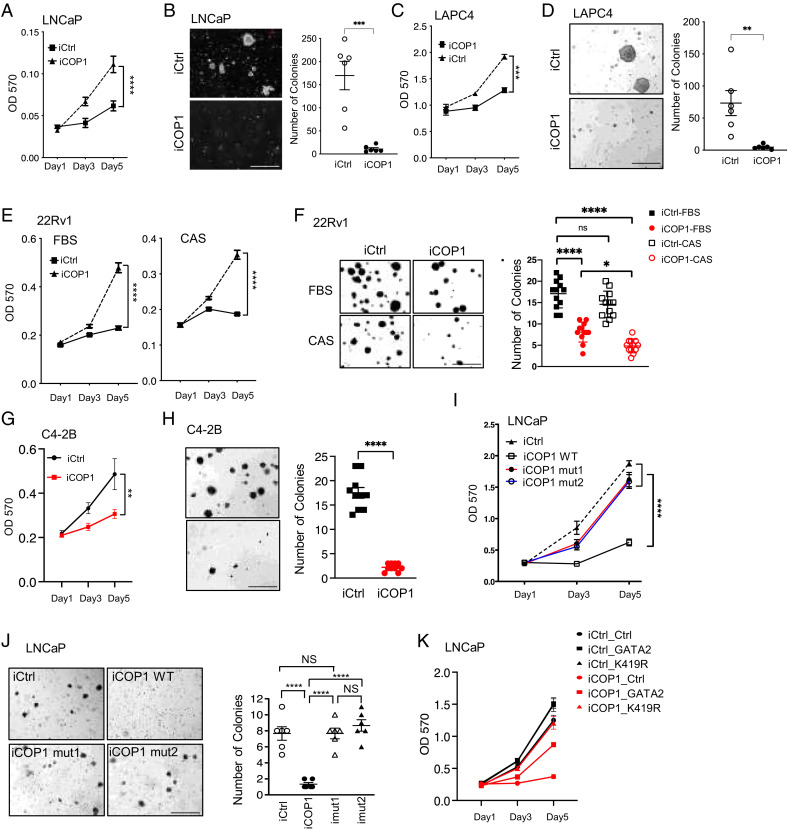
Since AR signaling is crucial for AR-positive PCa growth and since COP1 represses AR expression and activation in PCa and CRPC ( Fig. 1 D–G and 6), we investigated whether COP1 suppresses the growth of castration-naive LNCaP and LAPC4 cells and CRPC C4-2B and 22Rv1 cells. Proliferation and soft-agar assays revealed that COP1 expression greatly repressed the growth, including the anchorage-independent growth, of these PCa and CRPC cells (Fig. 7 A–H). COP1 also robustly repressed the castration-resistant and anchorage-independent growth of 22Rv1 cells cultured under complete castration conditions (10 µM enzalutamide in 5% charcoal-stripped serum, Fig. 7 E and F).
Fig. 7.
COP1 inhibits PCa and CRPC growth. (A, C) Cell proliferation and (B, D) soft-agar assays for 0.5 µg/mL Dox-induced LNCaP-iCOP1 versus LNCaP-iCtrl and LAPC4-iCOP1 versus LAPC4-iCtrl cells. (E) Proliferation and (F) soft-agar assays for 0.5 µg/mL Dox-induced 22Rv1-iCOP1 versus 22Rv1-iCtrl cultured in regular culture condition (10% FBS) versus complete castration condition (CAS:10 µm enzalutamide and 5% charcoal-stripped serum (CSS)). (G) Proliferation and (H) soft-agar assays for 0.5 µg/mL Dox-induced C4-2B-iCOP1 versus C4-2B-iCtrl cells. (I) Proliferation and (J) soft-agar assays for LNCaP cells with 0.5 µg/mL Dox-induced expression of wild-type (WT) COP1 versus COP1 mutants (mut1, mut2) lacking affinity to GATA2. Colony numbers were also quantified. (K) Proliferation assays for LNCaP cells with 0.5 µg/mL Dox-induced expression of COP1 (iCOP1) versus control (iCtrl) that also express GATA2 versus GATA2K419R (K419R) versus control (Ctrl). Quantification data as means ± SEM. *P < 0.05; **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, not significant; OD 570, optical density at 570 nm. (Scale bars: 500 µm.) Data are representative of at least three independent experiments.
Since we identified COP1mut1 and COP1mut2 mutants that could not efficiently bind to GATA2 or promote GATA2 ubiquitination and degradation, we next examined their activities in repressing the growth of PCa cells. Interestingly, both COP1mut1 and COP1mut2 lost their activity in repressing LNCaP cell growth and anchorage-independent growth (Fig. 7 I and J), which supports that the COP1-GATA2 axis is essential for COP1 tumor-inhibiting activities in AR-positive PCa. To further assess the role of GATA2 in mediating COP1 tumor-inhibiting activities in AR-positive PCa, we also compared the growth of COP1-overexpressing LNCaP cells with rescued expression of the GATA2K419R mutant that is resistant to COP1-induced ubiquitination and degradation, wild-type GATA2, or control. As predicted, overexpression of GATA2K419R showed a stronger effect than overexpression of wild-type GATA2 on rescuing the growth of the COP1-overexpressing LNCaP cells (Fig. 7K). Together, these data support that by repressing GATA2, COP1 is a key tumor suppressor in PCa.
COP1 Represses Growth of PCa Xenograft In Vivo.
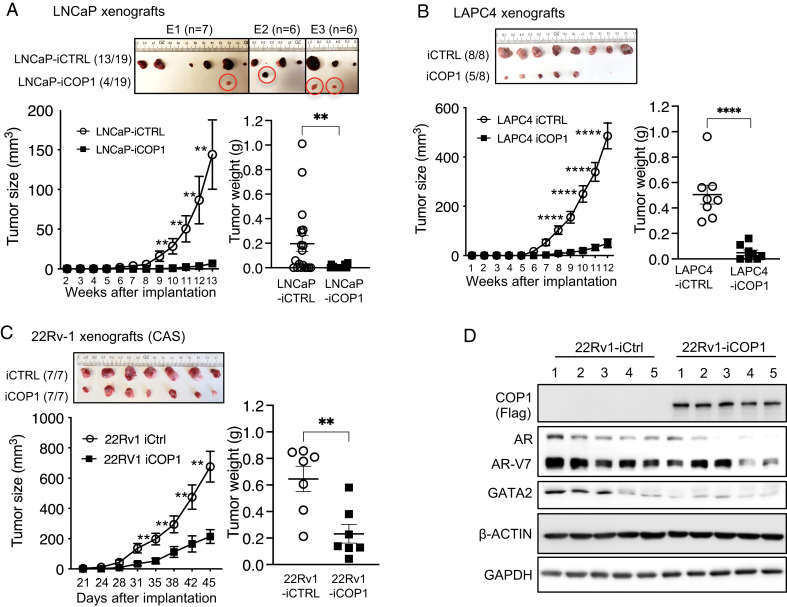
To investigate COP1 biology in vivo, we performed xenograft studies by subcutaneously inoculating control LNCaP-iCtrl cells and inducible COP1-expressing LNCaP-iCOP1 cells into the left and right lateral flanks of male SCID mice at about 8 weeks of age. Before xenograft inoculation, these engineered LNCaP cells were pretreated with Dox for 3 d in vitro for induction of COP1 or control. The injected mice were provided with drinking water containing 0.5 mg/mL of Dox throughout the study for induced expression of COP1 and control in the xenografts. Three months after inoculation, about 70% of the LNCaP-iCtrl inoculations (13 of 19; data are summarized from three independent experiments) formed xenograft tumors; in contrast, only 4 of 19 LNCaP-iCOP1 inoculations developed xenografts that were of minimal size (Fig. 8A). Similarly, Dox-induced COP1 overexpression also greatly reduced engineered LAPC4 xenograft tumor incidence and growth in vivo (Fig. 8B). Finally, we carried out subcutaneous xenograft studies using 22Rv1-iCtrl versus 22Rv1-iCOP1 cells into the left and right lateral flanks of castrated male SCID mice at about 8 weeks of age. Again, COP1 overexpression repressed the growth of these 22Rv1 xenograft tumors in these castrated animals (Fig. 8C). Western blots of representative tumors confirmed COP1 overexpression in the 22Rv1-iCOP1 xenografts. Despite the notable differences in AR/AR-V7 and GATA2 protein expression among these 22Rv1 xenograft tumors, COP1-overexpressing tumors in general expressed less GATA2 and AR/AR-V7 proteins in the paired 22Rv1-iCOP1 versus 22Rv1-iCtrl tumors within the same host (Fig. 8D [i.e., comparing GATA2-AR/AR-V7 protein expression between #1 22Rv1-iCtrl versus #1 22Rv1-iCOP1 tumor in mouse #1]). Collectively, these data support our in vitro observations and further demonstrate that COP1 is a tumor suppressor that strongly inhibits the growth and/or incidence of PCa and CRPC xenograft tumors in vivo.
Fig. 8.
COP1 inhibits PCa and CRPC xenograft tumor growth in vivo. (A) Xenografts were created by inoculating 2 × 106 LNCaP-iCOP1 cells versus LNCaP-iCtrl cells in 100 μL of 1:2 mix of Matrigel into the lateral flanks (subcutaneously, -iCtrl on the left and -iCOP1 on the right) of 8- to 10-wk-old male SCID mice. Mice received 0.5 mg/mL Dox in drinking water throughout the study. Tumor sizes were recorded from 2 to 13 wk after injections. Data are from three independent experiments (E1, E2, E3) using 7, 6, and 6 mice. Thirteen of 19 LNCaP-iCtrl injections versus 4 of 19 LNCaP-iCOP1 injections produced tumors. Xenograft studies were similarly performed using (B) LAPC4-iCOP1 versus LAPC4-iCtrl cells in intact mice and (C) 22Rv1-iCOP1 versus 22Rv1-iCtrl cells in castrated (CAS) mice. (B, C) Representative xenograft data from three independent studies. Quantification data as means ± SEM. **P < 0.01; ****P < 0.0001. (D) Western blots of representative 22Rv1-iCOP1 versus 22Rv1-iCtrl tumors from C. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Discussion
Several previous studies have identified GATA2 as a pioneer transcription factor essential for inducing AR activation in PCa (8). Although it has been known for decades that GATA2 protein is unstable (15), the molecular mechanisms regulating GATA2 stability in PCa have not been elucidated. In this study, we established COP1 as the bona fide E3 ubiquitin ligase for GATA2 in PCa. By targeting GATA2 for ubiquitination and degradation, COP1 reduces GATA2 protein expression, and thus GATA2-mediated AR expression and activation in PCa, and this COP1-GATA2-AR signaling axis seems to be critical for COP1 tumor suppressor activity in AR-positive PCa and CRPC.
Vitari et al (24) reported that COP1 targets ETV1 for degradation to suppress LNCaP cell migration but not proliferation. This is in sharp contrast to our observation that COP1 expression potently inhibits growth of multiple human PCa cell lines in vitro and also decreases xenograft growth in vivo. Currently, the basis of this apparent divergence remains unclear. The potent COP1 repression of AR signaling in PCa represents another distinction between our study and the Vitari et al study (24). We noted that gene expression profiling studies by Vitari et al were performed on LNCaP cells transfected with COP1 siRNAs for 48 h, followed by overnight culture in serum-free medium (24). This overnight culture in serum-free medium is predicted to mask any potential effects of COP1 on AR signaling in these cells.
It has been reported that by directly targeting p53 degradation, COP1 may function as an oncogene in certain cellular contexts. However, it is unclear whether p53 is a bona fide COP1 target in vivo (25). It is well recognized that mutation or loss of p53 is strongly correlated with PCa stages, metastasis, and castration resistance (40, 41), and p53 is the most significantly altered gene in metastatic CRPC (42), which may further reduce the impact of a potential COP1-p53 axis in PCa and CRPC. In contrast, our current data strongly support an overall tumor-suppressing role of the COP1-GATA2 axis in AR-positive PCa and CRPC.
Besides functioning as a bona fide E3 ubiquitin ligase for GATA2 and promoting GATA2 protein degradation in PCa and CRPC, COP1 also regulates GATA2 protein in HEK293T cells (Fig. 2E) and the breast cancer SUM159 cells (SI Appendix, Fig. S3C) beyond the PCa and CRPC context. The biological significance of the COP1-GATA2 signaling axis in regulating the physiology and other types of human diseases remains to be determined.
This study has identified an atypical mechanism for COP1 binding to its noncanonical substrate, GATA2. The two D/E-enriched motifs (D/E1 and D/E2) in COP1 and the two K/R-enriched BR1/BR2 motifs in GATA2 are critical for COP1-GATA2 binding. Interestingly, the D/E1 and D/E2 motifs in COP1 are largely conserved across species (SI Appendix, Figs. S2 and S5). In contrast, although the BR1 and BR2 motifs are largely conserved in mammals, these motifs are less conserved in A. thaliana (SI Appendix, Figs. S1 and S4). It is largely unknown how A. thaliana GATA2 binds A. thaliana COP1. However, if our identified mechanism is conserved, the highlighted basic amino acid–rich regions in A. thaliana GATA2 (SI Appendix, Fig. S1), especially the first two regions, might serve as the potential docking sites for A. thaliana GATA2 binding COP1, potentially through the largely conserved D/E1 and D/E2 motifs.
Materials and Methods
Reagents and Antibodies.
Antibodies against GATA2 (H-116, sc-9008), AR (N-20, sc-816), HA (sc-805), Myc (9E10, sc-40), glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 6C5, sc-32233), Protein G PLUS-Agarose (sc-2002), and Protein A-Agarose (sc-2001) were purchased from Santa Cruz Biotechnology. Antibodies against Flag (M2, F3165), β-actin (A1978), R1881 (R0908), Dihydrotestosterone (DHT) (D073), MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide], MG132 (M7449), and EZview Red Anti-Flag M2 Affinity Gel (F2426) were purchased from Sigma. The anti-COP1/RFWD2 (A300-894A) antibody was from Bethyl Laboratories, anti-GATA2 antibody (#A0677) was from ABclonal Technology, and anti-AR antibody (D6F11, #5153) was from Cell Signaling Technology. Enzalutamide (MDV3100, S1250) was purchased from Selleckchem.
Plasmids.
The pRK5, pInducer20-YF, and pCDH-CMV-MCS-EF1α-RFP+Puro–based GATA2 overexpression vectors were described previously (43). The COP1 complementary DNA (cDNA) was PCR amplified and cloned into the pRK5 vector between EcoRV and XhoI sites. It was also subcloned into the lentiviral pInducer20-YF vector for Dox-inducible overexpression of COP1.
Cell Culture, Transfection, and Lentivirus Infection.
LNCaP, 22Rv1, LAPC4, C4-2B, and VCaP cells were acquired from American Type Culture Collection (ATCC), and PNT1A cells were acquired from the European Collection of Authenticated Cell Cultures. LNCaP, C4-2B, and 22Rv1 cells were cultured in RPMI 1640, and VCaP and PNT1A cells were cultured in Dulbecco’s modified Eagle medium (DMEM), all supplemented with 10% fetal bovine serum (FBS; HyClone or Invitrogen). LAPC4 cells were cultured in Iscove modified Dulbecco medium (IMDM) with 10% FBS and 1 nM R1881. Charcoal-stripped FBS (SH3007103) was from HyClone.
The siRNA and DNA transfections were performed using the GenMute and LipoD293 transfection reagents (SignaGen Laboratories), respectively. Lentivirus packaging, lentivirus-mediated gene delivery, and generation of stable cell lines were performed as previously described (43). We established the LNCaP, LAPC4, 22Rv1, and C4-2B cells with Dox-inducible overexpression of COP1 and established LNCaP and LAPC4 cells with stable knockdown of COP1. At least 3-d induction with up to 0.5 µg/mL Dox was used to obtain COP1 overexpression in these engineered Dox-inducible cells.
Immunoprecipitation and Western Blot.
Cell lysate was prepared, and immunoprecipitation and Western blot assays were performed as previously described (43).
Cell Proliferation Assays.
We used two approaches to assess cell proliferation, including crystal violet staining–based cell proliferation assay and MTT assay as previously described (43–46). The first readout was measured at 24 h, followed by measurements every other day as described (43).
Soft-Agar Colony Formation Assay.
Soft-agar colony formation assays were performed as described (43–46). For the engineered LNCaP, LAPC4, 22Rv1, and C4-2B cells with Dox-inducible overexpression of COP1 or control, 0.5 μg/mL Dox was used throughout for induction of COP1 or control.
Xenograft Tumor Models.
Xenograft studies were performed using 8- to 10-wk-old male SCID mice (Envigo) housed in a pathogen-free facility under a protocol approved by the Baylor College of Medicine Institutional Animal Care and Use Committee (IACUC). For LNCaP or LAPC4 xenograft studies, 2 × 106 LNCaP or LAPC4 cells with Dox-inducible overexpression of COP1 (iCOP1) versus control (iCtrl) were subcutaneously injected into the lateral flanks (iCtrl on the left side, iCOP1 on the right side). For 22Rv1 xenograft studies, 2 × 106 22Rv1 cells were subcutaneously injected into the lateral flanks of castrated SCID mice (iCtrl on the left side, and iCOP1 on the right side). Matrigel matrix (1:2 dilution; Corning 354234) was used in all xenograft studies. All cells were pretreated with 0.5 µg/mL Dox for 3 d before tumor inoculation. Mice received 0.5 mg/mL Dox and 10% sucrose in drinking water for inducible overexpression of COP1 or control in the xenografts throughout the studies. Tumors were monitored and measured every 2 d and harvested as indicated and weighed. Tumor sizes were calculated using vol = 0.52 × abc (a, b, c: the maximum length of each tumor dimension).
Study approval.
All animal studies were approved by the Baylor College of Medicine IACUC.
Statistics.
We used the unpaired two-tail Student t test for statistical analysis in our study. Multiple comparisons were performed using one-way or two-way ANOVA followed by Dunnett’s or Sidak’s multiple comparisons test (GraphPad Prism 9.3). P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
This research was supported by grants from the Department of Defense Congressionally Directed Medical Research Programs (W81XWH-17-1-0043 to F.Y.), the Cancer Prevention Research Institute of Texas (RP130651 to F.Y.), and the R.P. Doherty Jr.–Welch Chair in Science (Q-0022 to D.D.M.).
Footnotes
Competing interest statement: F.Y. is the co-founder and CEO of 4Therapeutics Inc., and holds an equity stake there.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2205350119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Siegel R. L., Miller K. D., Fuchs H. E., Jemal A., Cancer statistics, 2022. CA Cancer J. Clin. 72, 7–33 (2022). [DOI] [PubMed] [Google Scholar]
- 2.de Bono J. S., et al. ; COU-AA-301 Investigators, Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 364, 1995–2005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scher H. I., et al. ; AFFIRM Investigators, Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 367, 1187–1197 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Beer T. M., et al. ; PREVAIL Investigators, Enzalutamide in metastatic prostate cancer before chemotherapy. N. Engl. J. Med. 371, 424–433 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan C. J., et al. ; COU-AA-302 Investigators, Abiraterone in metastatic prostate cancer without previous chemotherapy. N. Engl. J. Med. 368, 138–148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith M. R., et al. ; SPARTAN Investigators, Apalutamide treatment and metastasis-free survival in prostate cancer. N. Engl. J. Med. 378, 1408–1418 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Wang Q., et al. , A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol. Cell 27, 380–392 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu D., et al. , Three-tiered role of the pioneer factor GATA2 in promoting androgen-dependent gene expression in prostate cancer. Nucleic Acids Res. 42, 3607–3622 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He B., et al. , GATA2 facilitates steroid receptor coactivator recruitment to the androgen receptor complex. Proc. Natl. Acad. Sci. U.S.A. 111, 18261–18266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Böhm M., Locke W. J., Sutherland R. L., Kench J. G., Henshall S. M., A role for GATA-2 in transition to an aggressive phenotype in prostate cancer through modulation of key androgen-regulated genes. Oncogene 28, 3847–3856 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Chiang Y. T., et al. , GATA2 as a potential metastasis-driving gene in prostate cancer. Oncotarget 5, 451–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vidal S. J., et al. , A targetable GATA2-IGF2 axis confers aggressiveness in lethal prostate cancer. Cancer Cell 27, 223–239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez-Bravo V., et al. , The role of GATA2 in lethal prostate cancer aggressiveness. Nat. Rev. Urol. 14, 38–48 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minegishi N., Suzuki N., Kawatani Y., Shimizu R., Yamamoto M., Rapid turnover of GATA-2 via ubiquitin-proteasome protein degradation pathway. Genes Cells 10, 693–704 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Lurie L. J., Boyer M. E., Grass J. A., Bresnick E. H., Differential GATA factor stabilities: Implications for chromatin occupancy by structurally similar transcription factors. Biochemistry 47, 859–869 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Yi C., Wang H., Wei N., Deng X. W., An initial biochemical and cell biological characterization of the mammalian homologue of a central plant developmental switch, COP1. BMC Cell Biol. 3, 30 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng X. W., Caspar T., Quail P. H., cop1: A regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5, 1172–1182 (1991). [DOI] [PubMed] [Google Scholar]
- 18.Osterlund M. T., Hardtke C. S., Wei N., Deng X. W., Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Yi C., Deng X. W., COP1—From plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15, 618–625 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Marine J. C., Spotlight on the role of COP1 in tumorigenesis. Nat. Rev. Cancer 12, 455–464 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Su C. H., et al. , Nuclear export regulation of COP1 by 14-3-3σ in response to DNA damage. Mol. Cancer 9, 243 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wertz I. E., et al. , Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303, 1371–1374 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Li D. Q., et al. , E3 ubiquitin ligase COP1 regulates the stability and functions of MTA1. Proc. Natl. Acad. Sci. U.S.A. 106, 17493–17498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vitari A. C., et al. , COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature 474, 403–406 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Migliorini D., et al. , Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J. Clin. Invest. 121, 1329–1343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dallavalle C., et al. , MicroRNA-424 impairs ubiquitination to activate STAT3 and promote prostate tumor progression. J. Clin. Invest. 126, 4585–4602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo X. M., et al. , Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell 19, 872–883 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dehm S. M., Schmidt L. J., Heemers H. V., Vessella R. L., Tindall D. J., Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 68, 5469–5477 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dehm S. M., Tindall D. J., Alternatively spliced androgen receptor variants. Endocr. Relat. Cancer 18, R183–R196 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Z., et al. , A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 69, 2305–2313 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu R., et al. , Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 69, 16–22 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun S., et al. , Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J. Clin. Invest. 120, 2715–2730 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antonarakis E. S., et al. , AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 371, 1028–1038 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pickart C. M., Targeting of substrates to the 26S proteasome. FASEB J. 11, 1055–1066 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Nakajima T., et al. , Regulation of GATA-binding protein 2 levels via ubiquitin-dependent degradation by Fbw7: Involvement of cyclin B-cyclin-dependent kinase 1-mediated phosphorylation of THR176 in GATA-binding protein 2. J. Biol. Chem. 290, 10368–10381 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedone P. V., et al. , The N-terminal fingers of chicken GATA-2 and GATA-3 are independent sequence-specific DNA binding domains. EMBO J. 16, 2874–2882 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi X., et al. , Cooperative interaction of Etv2 and Gata2 regulates the development of endothelial and hematopoietic lineages. Dev. Biol. 389, 208–218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holm M., Hardtke C. S., Gaudet R., Deng X. W., Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 20, 118–127 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radivojac P., et al. , Identification, analysis, and prediction of protein ubiquitination sites. Proteins 78, 365–380 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bookstein R., MacGrogan D., Hilsenbeck S. G., Sharkey F., Allred D. C., p53 is mutated in a subset of advanced-stage prostate cancers. Cancer Res. 53, 3369–3373 (1993). [PubMed] [Google Scholar]
- 41.Heidenberg H. B., et al. , Alteration of the tumor suppressor gene p53 in a high fraction of hormone refractory prostate cancer. J. Urol. 154, 414–421 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Robinson D., et al. , Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen T., et al. , MAPK4 promotes prostate cancer by concerted activation of androgen receptor and AKT. J. Clin. Invest. 131, e135465 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang W., et al. , MAPK4 overexpression promotes tumor progression via noncanonical activation of AKT/mTOR signaling. J. Clin. Invest. 129, 1015–1029 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai Q., et al. , MAPK6-AKT signaling promotes tumor growth and resistance to mTOR kinase blockade. Sci. Adv. 7, eabi6439 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang W., et al. , MAPK4 promotes triple negative breast cancer growth and reduces tumor sensitivity to PI3K blockade. Nat. Commun. 13, 245 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.