Abstract
Neurotransmitter release by Ca2+‐triggered synaptic vesicle exocytosis is essential for information transmission in the nervous system. The soluble N‐ethylmaleimide sensitive factor attachment protein receptors (SNAREs) syntaxin‐1, SNAP‐25, and synaptobrevin‐2 form the SNARE complex to bring synaptic vesicles and the plasma membranes together and to catalyze membrane fusion. Munc18‐1 and Munc13‐1 regulate synaptic vesicle priming via orchestrating neuronal SNARE complex assembly. In this review, we summarize recent advances toward the functions and molecular mechanisms of Munc18‐1 and Munc13‐1 in guiding neuronal SNARE complex assembly, and discuss the functional similarities and differences between Munc18‐1 and Munc13‐1 in neurons and their homologs in other intracellular membrane trafficking systems.
Keywords: Munc13, Munc18, SNARE complex assembly, SNAREs, synaptic exocytosis, synaptic vesicle fusion
Munc18‐1 and Munc13‐1 regulate synaptic vesicle exocytosis via orchestrating neuronal SNARE complex (syntaxin‐1, SNAP‐25, and synaptobrevin‐2) assembly. In this review, we summarized recent advances towards the molecular mechanisms of Munc18‐1 and Munc13‐1 in guiding neuronal SNARE complex assembly and discussed the similarities and differences between Munc18‐1 and Munc13‐1 in neurons and their homologs in other intracellular membrane trafficking systems.
Abbreviations
- CAPS
Ca2+‐dependent activator protein for secretion
- CATCHR
complex associated with tethering containing helical rods
- COG
conserved oligomeric Golgi
- CORVET
class C core vacuolar/endosomal tethering
- Dsl1
dependent on Sly1‐20
- GARP
golgi associated retrograde protein
- HOPS
Homotypic fusion and vacuolar protein sorting
- NSF
N‐ethylmaleimide sensitive factor
- SM
Sec1/Munc18‐1
- SNARE
soluble N‐ethylmaleimide sensitive factor attachment protein receptor
- TRAPP
transport protein particle
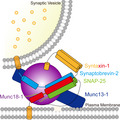
Neurotransmitter release by Ca2+‐triggered synaptic vesicle exocytosis is an exquisitely regulated process essential for information transmission in the nervous system [1, 2, 3]. Under resting conditions, most newly formed and recycled synaptic vesicles are stored in the cytoplasm of the nerve terminal. A subset of synaptic vesicles can be attached to specialized sites at the presynaptic active zones, where a number of multidomain proteins constitute a scaffold platform to mediate vesicle tethering and docking [4, 5]. To achieve fast exocytosis, docked vesicles require maturation into a ‘priming’ state that involves a population of ‘ready‐for‐fusion’ vesicles corresponding to the readily releasable pool (RRP) [6, 7, 8]. When an action potential arrives at the axon terminal, Ca2+ influx triggers primed vesicles to fuse with the presynaptic membrane in the millisecond timescale [9, 10].
The core release machinery governing synaptic vesicle exocytosis consists of components that belong to protein families involved in most types of intracellular membrane trafficking systems and with conserved roles in membrane fusion, including the AAA+ ATPase N‐ethylmaleimide sensitive factor (NSF), soluble NSF adaptor proteins (SNAPs), the SNAP receptors (SNAREs), Sec1/Munc18‐like (SM) protein Munc18‐1, and complex associated with tethering containing helical rods (CATCHR) family protein Munc13s [11, 12, 13]. In addition, the release machinery contains specialized components such as synaptotagmins and complexins, whose functions and mechanisms in regulating Ca2+‐triggered synaptic vesicle fusion have been reviewed previously [14, 15, 16]. The neuronal SNAREs serve as the engine of the release machinery, as their assembly into the SNARE complex provides energy for membrane bridging and fusion [17, 18]. Tight control and regulation of SNARE complex assembly by regulatory components are the prerequisites for synaptic vesicle fusion occurring at the right place, at the right time, and with the right probability.
Priming of synaptic vesicles is believed to be a hallmark of synaptic vesicle exocytosis, which involves activation and regulation of SNARE complex assembly. Recent advances toward the structural insights, intermolecular interactions, and functional properties of the priming components shed new light on understanding the molecular mechanism of synaptic vesicle exocytosis. In this review, we summarize recent progress on crucial priming apparatus that includes the SNAREs, Munc18‐1, and Munc13s—focusing on the functional properties and molecular mechanisms of Munc18‐1 and Munc13‐1 in organizing neuronal SNARE complex assembly—and discuss the functional similarities and differences between Munc18‐1 and Munc13s in neurons and their homologs in other intracellular membrane trafficking systems.
The neuronal SNAREs
The neuronal SNAREs syntaxin‐1, SNAP‐25, and synaptobrevin‐2 contain ~ 65‐residue sequences termed SNARE motifs that are able to form coiled coils [17, 19]. Syntaxin‐1 and synaptobrevin‐2 are anchored at the presynaptic membrane and synaptic vesicles, respectively, via the C‐terminal transmembrane region (TMR) which is connected to the SNARE motif by a short juxtamembrane linker region (JLR). SNAP‐25 lacks a TMR but is attached to the plasma membrane by palmitoyl chains bound to cysteine residues in an extended linker joining its two SNARE motifs. The neuronal SNAREs assemble into an intertwined and parallel four‐helical bundle called the SNARE complex via their SNARE motifs [17]. Crystal structure of the neuronal SNARE complex revealed that the core architecture contains 16 consecutive layers [20, 21], including layers −7 to −1 in the N‐terminal half, layers +1 to +8 in the C‐terminal half, and one central layer 0. These layers contain predominantly hydrophobic residues, except for the central layer 0, which comprises one arginine (R) and three glutamines (Q). The SNARE motifs are accordingly classified into Qa‐ (syntaxin‐1), Qb‐ (the first SNARE motif of SNAP‐25), Qc‐ (the second SNARE motif of SNAP‐25), and R‐SNARE (synaptobrevin‐2) [22, 23] (Fig. 1A). Phylogenetic and structural analysis of the SNARE motifs showed that the four SNARE subfamilies are diverged early during eukaryotic evolution and revealed a ‘QabcR’ pattern for functional SNARE complexes e[23, 24, 25, 26, 27].
Fig. 1.
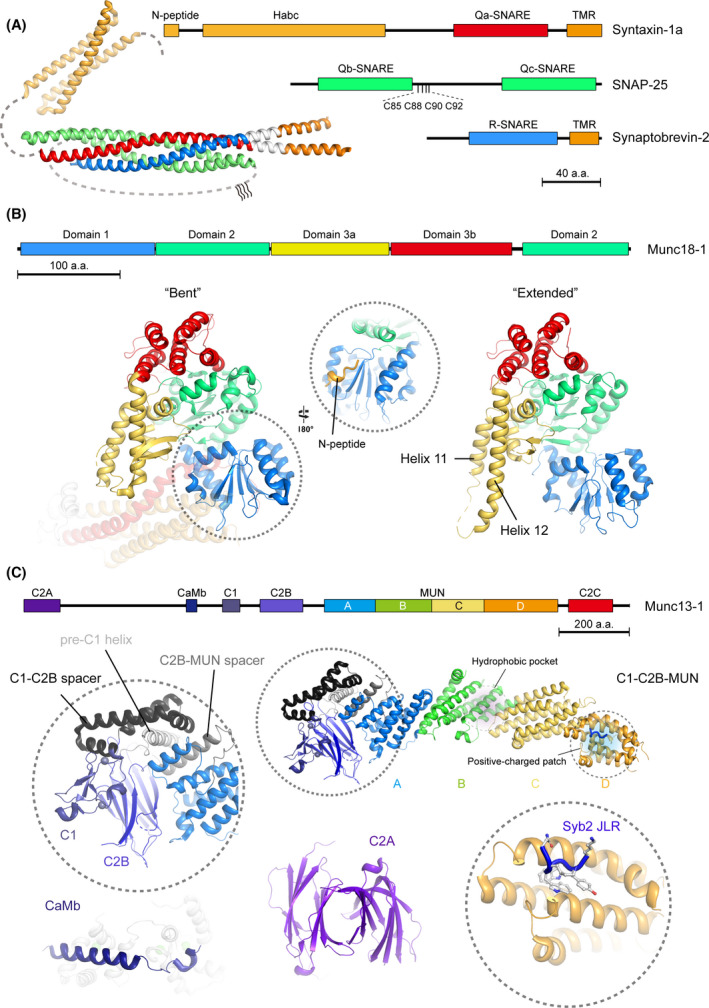
Structure illustration of the neuronal SNAREs, Munc18‐1, and Munc13‐1. (A) Crystal structure and domain illustration of the neuronal SNARE complex. The structure models are fetched from the protein data bank (PDB) by entries of 3HD7 (helical extended neuronal SNARE complex) and 1EZ3 (Habc domain of syntaxin‐1). The SNARE motif of syntaxin‐1 (Qa‐SNARE, residues 191−253), SNAP‐25 (Qb‐ and Qc‐SNARE, residues 19−81 and 140−202), and synaptobrevin‐2 (R‐SNARE, residues 29−87) are colored in red, green, and blue, respectively. The N‐peptide (residues 1−10) and Habc (residues 26−146) of syntaxin‐1 are colored in bright orange. Transmembrane domains (TMRs) of syntaxin‐1 (residues 266−288) and synaptobrevin‐2 (residues 95−114) are colored in orange. The juxtamembrane linker regions (JLRs) of syntaxin‐1 and synaptobrevin‐2 are colored in light gray. The S‐palmitoyl cysteines of SNAP‐25 are indicated as C85, C88, C90, and C92, respectively. Missing densities in the structural model are supplied by dashed lines. (B) Structural illustration of Rattus norvegicus Munc18‐1 with two conformations. The subdomains of Munc18‐1 are colored in blue (domain 1, residues 4−134), green (domain 2, residues 135−245 and 490−592), yellow (domain 3a, residues 246−360), and red (domain 3b, residues 361−479), respectively. Left panel displays the ‘bent’ conformation of Munc18‐1 that binds to syntaxin‐1 (PDB entry: 3C98), where the helix 11 and 12 of domain 3a are folded back. Inset shows the binding between N‐peptide (bright orange) of syntaxin‐1 and domain 1 of Munc18‐1. Right panel displays the ‘extended’ conformation of Munc18‐1 (PDB entry: 3PUJ), in which helix 11 and 12 are outstretched. (C) Structural illustration of Rattus norvegicus Munc13‐1. The domains and subdomains of Munc13‐1 are colored in purple (C2A, residues 1−97), dark blue (calmodulin‐binding domain, CaMb, residues 459−492), navy blue (C1, residues 566−616), purple blue (C2B, residues 683−820), blue (MUN‐A, residues 859−1005), green (MUN‐B, residues 1006−1167), yellow (MUN‐C, residues 1168−1318), bright orange (MUN‐D, residues 1319−1531), and red (C2C, residues 1558−1685), respectively. The structural models of C1‐C2B‐MUN fragment (PDB entry: 5UE8 and 6A30), C2A (dimer form, PDB entry: 2CJT), and CaMb (binding with calmodulin, PDB entry: 2KDU) are fetched from PDB. Two functional regions of the MUN domain, that is, the hydrophobic pocket (binds to syntaxin‐1) and the negative‐charged patch (binds to synaptobrevin‐2), are highlighted by purple and blue shadows, respectively. Inset on the left shows the architecture of the intramolecule contacts of C1, C2B, and MUN domain. Inset on the right shows the interaction between the MUN‐D and synaptobrevin‐2 (Syb2) JLR.
Unlike SNAP‐25 and synaptobrevin‐2, syntaxin‐1 possesses an N‐terminal regulatory sequence that is connected to the SNARE motif by a flexible linker region [28]; this regulatory sequence consists of an N‐terminal short stretch called the N‐peptide followed by an antiparallel three‐helix bundle called the Habc domain. As with the SNARE motif, both the N‐peptide and the Habc domain of syntaxin‐1 play indispensable roles in synaptic vesicle exocytosis [29, 30]. Although the N‐peptide and the Habc domain are widely found in most of the Qa‐SNAREs [31, 32, 33, 34], their regulatory roles and mechanisms differ among different membrane trafficking systems [32, 35, 36, 37, 38].
Neuronal SNARE complex assembly
The neuronal SNAREs undergo assembly–disassembly cycles to fulfill constant exocytosis of synaptic vesicles [14, 39, 40]. SNARE complex assembly is assumed to initiate with a contact of the N‐terminal ends of the SNARE motifs and proceeded with the association of the four SNARE motifs into the four‐helical bundle in a trans‐conformation that includes loose and tight intermediates underlying different priming states [41, 42, 43, 44, 45, 46] (Fig. 2A). The N‐ to C‐zippering of the trans‐SNARE complex can generate energy to overcome the repulsion of the opposite membranes thereby bringing the membranes into close proximity [47, 48, 49]. Subsequent assembly proceeding over the JLR and TMR of synaptobrevin‐2 and syntaxin‐1 is believed to transmit the energy into the membrane interface, leading to membrane fusion with the conversion of the SNARE complex from the trans‐ into a cis‐conformation in which the two TMRs are aligned in parallel in the plasma membrane [21, 50, 51, 52, 53, 54] (Fig. 2A). After membrane fusion, the cis‐SNARE complex is disassembled in an ATP‐driven manner by NSF and α‐SNAP [55, 56]. Once disassembled, free SNAREs are recycled for another round of synaptic vesicle fusion [57, 58, 59].
Fig. 2.
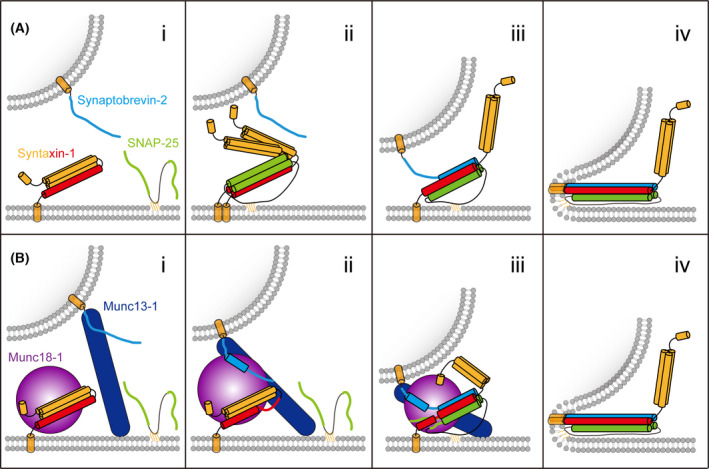
Models of SNARE‐mediated membrane fusion and synaptic exocytosis. (A) The zippering model with merely the three neuronal SNAREs. (i) At rest state, syntaxin‐1 adopts self‐inhibitory conformation; (ii) syntaxin‐1 fluctuates between closed and open conformations and is prone to form a 2 : 1 complex with SNAP‐25; (iii) synaptobrevin‐2 displaces one copy of syntaxin‐1 of the 2 : 1 complex; the N‐termini of the SNARE motifs nucleate together to promote complex assembly; (iv) zippering of the SNARE motifs transfers sufficient energy into the membrane thus catalyzing membrane fusion. (B) Munc18‐1 and Munc13‐1 synergistically organize neuronal SNARE complex assembly and synaptic exocytosis. (i) Munc18‐1 captures syntaxin‐1 into closed conformation; Munc13‐1 bridges the presynaptic membrane and synaptic vesicle to facilitate synaptic vesicle docking; (ii) Munc13‐1 interacts with Munc18‐1−syntaxin‐1 complex to induce conformational changes of the syntaxin‐1 linker region and Munc18‐1 domain 3a, leading to uncaging of the N‐terminus of the syntaxin‐1 SNARE motif and extension of Munc18‐1 domain 3a; in the meantime, Munc18‐1 interacts with the C‐terminal half of synaptobrevin‐2 with the assistance of the binding between Munc13‐1 and synaptobrevin‐2 JLR. This intermediate underlies a potential conformational state, namely the prefusion priming complex; (iii) the N‐termini of the SNARE motifs start to nucleate to produce a half‐zippered SNARE complex, which is organized by Munc18‐1 and Munc13‐1; (iv) full zippering of the SNARE motifs in response to calcium signal is accompanied by the interplay of complexins/synaptotagmin‐1 with the half‐zippered SNARE complex (not shown). The color schemes of the neuronal SNAREs are the same as in Fig. 1. Munc18‐1 and Munc13‐1 are colored in purple and navy blue, respectively.
The SNARE zippering model embodies elegant simplicity and comprehensive unity suitable for most intracellular membrane trafficking systems. However, distinguished from most other membrane trafficking processes, synaptic vesicle exocytosis is extremely fast. Accompanied by large conformational change and high energy release [47, 48, 49], neuronal SNARE complex assembly go through multiple and complicated reactions [60], in which the detailed assembly kinetics and thermodynamics remain elusive. These, therefore, have raised many intriguing conundrums. First, more than 40 SNARE homologs have been identified in mammals, many of which are distributed in different cellular compartments and specific for different intracellular trafficking pathways [17, 27]. This raises an obvious question as to how the cognate SNAREs recognize each other to form a functional SNARE complex. Second, in the zippering model, syntaxin‐1 and SNAP‐25 are able to form heterodimeric complexes in the plasma membrane, which bind vesicle‐bound synaptobrevin‐2 to initiate trans‐SNARE complex formation (Fig. 2A). However, this assembly model faces several challenges: (a) syntaxin‐1 and SNAP‐25 are prone to form a 2:1 ‘dead‐end’ complex in vitro, in which a second copy of syntaxin‐1 occupies the position of synaptobrevin‐2 thus hindering the assembly kinetics of the SNARE complex [61, 62]; (b) isolated syntaxin‐1 prefers to assume a closed conformation in which the Habc domain folds back to the SNARE motif to inhibit SNARE complex assembly [63]; and (b) in vitro lipid mixing driven by the neuronal SNAREs alone is strongly inhibited by NSF and α‐SNAP[64], owing to the disassociation of trans‐SNARE complexes and/or syntaxin‐1−SNAP‐25 heterodimeric complexes. These findings led to crucial questions about how nonproductive side reactions and kinetically trapped intermediates along the assembly pathway are prevented, and how the assembly is protected against disassembly factors. Third, the fusion kinetics obtained from in vitro reconstitution assay using the SNAREs alone is not comparable to that observed in vivo. In addition, previous studies reported that two complementary paired nucleic acid strands linked by the TMRs of syntaxin‐1 and synaptobrevin‐2 could mediate in vitro liposome fusion as well [65, 66]. Another study revealed that artificial coiled‐coil peptides linked by the JLRs and TMRs of the SNAREs could induce liposome fusion[67]. Therefore, it is apparent that liposome fusion catalyzed by the SNAREs alone could not factually reproduce membrane fusion in vivo.
The above considerations indicate a need for crucial factors to prime the SNAREs and organize SNARE complex assembly. A wealth of evidence has revealed that Munc18‐1 and Munc13s play a central function in synaptic vesicle priming via orchestrating SNARE complex assembly.
Munc18‐1
Munc18‐1 is a member of the SM family proteins that have fundamental roles in most types of membrane trafficking processes from fungi to mammals [68, 69]. Loss of Munc18‐1 causes severe defects in neurotransmitter release of Caenorhabditis elegans motor neurons [70, 71, 72, 73] and of mice neocortex and neuromuscular synapse [74, 75, 76], revealing its critical role in synaptic vesicle exocytosis. Munc18‐1 knockout mice die immediately after birth. Analysis of the embryo brain lacking Munc18‐1 displayed massive neurodegeneration after assembly of the neuronal networks [74], indicating that Munc18‐1 is fundamental for the development of the nervous system. Munc18‐1 has been implicated to participate in synaptic vesicle docking, priming, and fusion. The diverse functions of Munc18‐1 depend, at least in part, on its capacity to bind the neuronal SNAREs with multiple conformations [13, 68, 69].
Crystal structures of Munc18‐1 bound to syntaxin‐1 revealed that Munc18‐1 forms an arch‐shaped architecture with three domains. Domain 3 is divided into two subdomains, that is, domain 3a and 3b. Domain 1 and domain 3a jointly create a central cavity to accommodate the Habc domain and the SNARE motif of syntaxin‐1 in a closed conformation, which produces a high binding affinity between the two molecules (K d ~ 1.4 nanomolar) [77, 78] (Fig. 1B). The closed conformation prevents syntaxin‐1 from being prematurely trapped by its cognate or noncognate Qbc‐ and R‐SNAREs during its translocation to the plasma membrane and renders vesicles suspended at the docking stage [76, 79]. Munc18‐1‐clamped syntaxin‐1 requires a transition from the closed to an open conformation to initiate SNARE complex assembly. This ‘closed‐to‐open’ transition appears to be a specialized feature of synaptic vesicle exocytosis, and the underlying mechanism will be discussed in more detail in the following sections.
The N‐peptide of syntaxin‐1 binds to domain 1 on the opposite side of the Munc18‐1 cavity (Fig. 1B), which is crucial for synaptic vesicle exocytosis [29, 30, 80]. This interaction not only assists Munc18‐1 to clamp the closed conformation of syntaxin‐1 [78] but also enhances binding of Munc18‐1 with the SNARE four‐helical bundle [81, 82, 83]. These data suggest that the N‐peptide binding is important for Munc18‐1 to assume diverse conformations available for syntaxin‐1 or the SNARE complex binding.
Moreover, domain 3a of Munc18‐1 can assume both ‘bent’ and ‘extended’ conformations. The bent conformation seen in Munc18‐1 bound to closed syntaxin‐1 is characterized as an inhibitory state of Munc18‐1, whereas the extended conformation represents an active state of Munc18‐1 which is accessible to synaptobrevin‐2 interaction [84, 85, 86, 87, 88, 89, 90] (Fig. 1B). This binding mode is conserved among various SM family proteins (e.g., Vps33 and Vps45) [36, 91]. Later studies showed that the majority of the SNARE binding layers of synaptobrevin‐2 (layer −4 to +6) were captured by the extended domain 3a of Munc18‐1, which is crucial to initiate SNARE complex assembly [92]. Furthermore, emerging evidence has indicated that Munc18‐1 is capable of binding to the SNARE four‐helical bundle. However, there is no consensus on the binding sites between Munc18‐1 and the SNARE four‐helical bundle, as evidence showed that the binding can be mediated by the cavity of Munc18‐1 [82, 84, 93, 94] or by domain 3a at the other side of the cavity [85, 87]. Despite lacking structural evidence, this binding mode is found in a variety of Munc18‐1 homologs and their cognate SNAREs [34, 95, 96, 97, 98], indicating a conserved function of Munc18‐1 in the final step of membrane fusion.
Munc13‐1
As a member of the CATCHR family proteins, Munc13‐1 is a large multidomain protein highly enriched in the presynaptic active zones and conserved from invertebrates to mammals. Deletion of Munc13‐1 causes severe defects in neurotransmitter release, indicating its fundamental role in synaptic transmission [99, 100, 101, 102]. In addition, Munc13‐1 is important for dense‐core vesicle exocytosis in chromaffin cells, pancreatic beta cells, and neurons, demonstrating its essential function in the release of hormones and neuropeptides [103, 104, 105, 106]. Deletion of Munc13‐1 has no significant influence on the ultrastructure of synapse [101]. Loss of a mass of docked vesicles around the presynaptic active zones in Munc13‐1/2 knockout mice indicates that Munc13s are associated with the scaffold matrix formation of the active zones [107]. Similar phenotypes were observed in mice deficient in Rab3‐interacting molecules (RIMs) and RIM‐binding proteins (RIMBPs) [107, 108, 109, 110], and in Ca2+‐dependent activator protein for secretion (CAPS) [107] which belongs to the CATCHR family proteins as well [111].
The molecular architecture of Munc13‐1 contains multiple individual domains, including three C2 domains (C2A, C2B and C2C, respectively), a PKC‐like phorbol ester/diacylglycerol (DAG) binding C1 domain, a calmodulin‐binding motif (CaMb), and a central MUN domain (Fig. 1C). C2A resides in the N‐terminal part of Munc13‐1, which is a noncanonical C2 domain insensitive to Ca2+ [112]. C2A can form a homodimer and is able to bind the zinc‐finger (ZF) of RIMs to form a heterodimer [113, 114]. Munc13‐1, RIM, and Rab3 could form a ternary complex crucial for synaptic vesicle docking and priming [115]. Adjacent to C1, C2B binds Ca2+ and is competent for phosphatidylinositol (mainly phosphatidylinositol‐4,5‐diphosphate, PtdIns‐4,5‐P2) binding [116]. C1 and C2B serve as membrane anchors to ensure the binding between Munc13‐1 and the presynaptic membrane. Recent structural evidence and functional data have shown that C1 and C2B are tightly contacted and function in synaptic short‐term plasticity [117, 118, 119, 120]. CaMb is located between C2A and C1, which might modulate synaptic plasticity via calmodulin [121]. C2C is also Ca2+‐insensitive and found as a membrane‐binding module that preferentially interacts with synaptic vesicles. The function of C2C is indispensable, since deletion of C2C causes strong defects in synaptic vesicle exocytosis [122]. The MUN domain between C2B and C2C contains four subdomains (i.e., A, B, C, and D) that mainly consist of stacked α‐helices [123]. Sequence and structural analysis have indicated a remote but significant homology between the MUN domain and the subunits of various tethering complexes, such as the Exocyst complex, Dsl1 complex, COG complex, and GARP complex [123, 124, 125]. Hence, Munc13‐1 and the tethering factors may play a universal role in vesicle tethering and docking. Moreover, recent studies found that Munc13‐1 could form oligomers around the fusion pore in vivo and in vitro [126, 127, 128], indicating that Munc13‐1 serves as a scaffold and tethering factor in exocytosis.
Early studies demonstrated that the MUN domain could partially rescue neurotransmitter release in Munc13‐1 knockout mice, indicating that the MUN domain is the minimal module responsible for Munc13‐1 activity [129, 130]. The finding that syntaxin‐1 bearing the LE mutation that prefers an open conformation [63] can partially rescue release in Caenorhabditis elegans unc13 nulls indicated that Munc13‐1 plays a role in opening syntaxin‐1 [131]. Moreover, the MUN domain has been found to mediate the transition from the closed Munc18‐1−syntaxin‐1 complex to the SNARE complex in vitro [132]. In addition, the crystal structure of a Munc13‐1 C1‐C2B‐MUN fragment indicated an intramolecular synergistic effect in which C1 and C2B may play roles in modulating the activity of the MUN domain [117]. Furthermore, a C1‐C2B‐MUN‐C2C fragment could bridge synaptic vesicles and the presynaptic membrane with at least two different orientations dependent on Ca2+ and presynaptic PtdIns‐4,5‐P2 and DAG levels, which is considered to be relevant to short‐term plasticity [117, 119, 120, 122].
In the past decade, important advances have been made in understanding the structural insights, intermolecular interactions, and functional mechanisms of Munc13‐1 and Munc18‐1 in synaptic vesicle exocytosis. In the next section, we will discuss the synergistic roles of Munc18‐1 and Munc13‐1 in organizing neuronal SNARE complex assembly.
Organizing the SNAREs—synergistic roles of Munc18‐1 and Munc13‐1
Recent reconstitution experiments have illustrated a model in which Munc18‐1 and Munc13‐1 play vital functions in regulating SNARE complex assembly. This model suggests that the starting point of the assembly pathway is the closed Munc18‐1−syntaxin‐1 complex and its transition to the SNARE complex is highly regulated by Munc13‐1 (Fig. 2B) [64, 132]. It was previously observed that syntaxin‐1 together with SNAP‐25 is capable of aggregating in clusters with PtdIns‐4,5‐P2 in the presynaptic membrane under physiological condition [133, 134, 135]. The clusters are expected to involve a large number of syntaxin‐1−SNAP‐25 2:1 complexes unfavorable for synaptobrevin‐2 binding [61]. One of the exciting merits of this model is that the nonproductive intermediate—syntaxin‐1−SNAP‐25 2 : 1 complexes—could be prevented, as Munc18‐1 together with NSF and α‐SNAP effectively displaces SNAP‐25 from its complex with syntaxin‐1 in the membranes [64]. Actually, a 1 : 1 stoichiometry of the syntaxin‐1−SNAP‐25 complex is immensely beneficial for the efficient assembly of the SNARE complex. For instance, it was found that the ∆N‐SNARE complex (i.e., the 1 : 1 syntaxin‐1−SNAP‐25 complex bound to an N‐terminal truncated synaptobrevin‐2) facilitates synaptobrevin‐2 binding thus driving fast SNARE complex assembly and liposome fusion [44, 136].
Another virtue of this model is that Munc18‐1 and Munc13‐1 offer an effective protection mechanism for SNARE complex assembly. NSF and α‐SNAP could abolish fusion between syntaxin‐1−SNAP‐25 liposomes and synaptobrevin‐2 liposomes probably by destructing immature trans‐SNARE complexes and/or syntaxin‐1−SNAP‐25 complexes into individual SNAREs [64]. Intriguingly, fusion between Munc18‐1−syntaxin‐1 liposomes and synaptobrevin‐2 liposomes can robustly proceed in the presence of Munc13‐1, SNAP‐25, NSF, and α‐SNAP [64, 137, 138], suggesting that Munc18‐1 and Munc13‐1 protect SNARE complex assembly against disassembly by NSF and α‐SNAP. In addition, a syntaxin‐binding molecule tomosyn, which possesses an R‐SNARE‐like motif [139, 140, 141], was reported to arrest syntaxin‐1 and SNAP‐25 into a nonfusogenic product that precludes synaptobrevin‐2 entry [142, 143]. Interestingly, in the context of NSF and α‐SNAP, syntaxin‐1 is able to escape from tomosyn arrest and assemble into the Munc18‐1−syntaxin‐1 complex. Remarkably, Munc13‐1 can catalyze the transition from the Munc18‐1−syntaxin‐1 complex to the SNARE complex in a manner specific to synaptobrevin‐2 but resistant to tomosyn [144]. In this process, NSF and α‐SNAP assist Munc13‐1 to promote the conversion from tomosyn‐arrested syntaxin‐1−SNAP‐25 complex to the Munc18‐1−syntaxin‐1 complex, and finally to the functional SNARE complex. These findings illustrate a protection function of Munc18‐1 and Munc13‐1 in SNARE complex assembly and synaptic vesicle priming, consistent with the role of the HOPS tethering complex in protecting SNARE complex assembly and yeast vacuolar fusion against Sec18 and Sec17, the homologs of NSF and α‐SNAP in yeast [145].
As illustrated in the model, the transition from the Munc18‐1−syntaxin‐1 complex to the SNARE complex is catalyzed by Munc13‐1. Initiation of the transition requires activation of the closed Munc18‐1−syntaxin‐1 complex, which involves the opening of syntaxin‐1 and the extension of domain 3a of Munc18‐1. The catalytic site of Munc13‐1 responsible for opening of syntaxin‐1 positions at a hydrophobic pocket located in the middle portion of the Munc13‐1 MUN domain [123] (Fig. 1C). Disruption of the hydrophobic pocket (i.e., NFAA mutation) caused abrogation of MUN‐mediated transition from the closed Munc18‐1−syntaxin‐1 complex to the SNARE complex and led to strong defects in neurotransmitter release of Caenorhabditis elegans neuromuscular junction (NMJ) and mice cortical neurons, indicating that the NF pocket is central for Munc13‐1 catalytic activity [123, 146, 147]. The syntaxin‐1 linker region (bearing the LE sequence) between the Habc domain and the SNARE motif was identified as the binding target for the Munc13‐1 NF pocket [146]. Biochemical and single‐molecule FRET data demonstrated that the Munc13‐1 NF pocket interacts with R151 and I155 in the linker region of syntaxin‐1, and this interaction specifically induces a conformational change of the syntaxin‐1 linker region [146, 148] (Fig. 2B). These data argue against the conventional assumption that syntaxin‐1 needs to be totally escaped from Munc18‐1 clamping to initiate SNARE complex assembly, suggesting that a local conformational change in the syntaxin‐1 linker region is sufficient to initiate SNARE complex assembly. On the other hand, domain 3a of Munc18‐1 is able to undergo ‘bent‐to‐extended’ conformational change during SNARE complex assembly [85, 86, 87, 88, 89]. Recent advances showed that binding of the Munc13‐1 MUN domain to domain 3a is required for MUN activity in opening of syntaxin‐1 [84] (Fig. 1B). Upon the opening of the syntaxin‐1 linker region by the MUN domain, domain 3a of Munc18‐1 is inclined to assume the extended conformation, which allows the N‐terminal end of syntaxin‐1 SNARE motif more accessible to nucleate with SNAP‐25 and synaptobrevin‐2 [84] (Fig. 2B). This cascaded reaction supports the N‐ to C‐zippering model of SNARE complex assembly [44, 45].
Efficient SNARE complex assembly requires N‐terminal nucleation of the SNARE motifs of SNAP‐25, synaptobrevin‐2 and syntaxin‐1 together. An attractive R‐SNARE−SM binding mode has been presented and attracted intense attention [91]. Based on this mode, later studies showed that the majority of the SNARE binding layers of synaptobrevin‐2 (layer −4 to +6) were captured by the extended domain 3a of Munc18‐1, with the N‐terminal layers of the SNARE motif being available for nucleation [92]. It is of note that the interaction between Munc18‐1 and synaptobrevin‐2 is quite weak, compared to their homologs Vps33 and Nyv1 in yeast, respectively [91]. A recent structural study reported a direct binding between the Munc13‐1 MUN domain and the synaptobrevin‐2 JMR [149] (Fig. 1C). This interaction enhances binding of synaptobrevin‐2 to the Munc18‐1−syntaxin‐1 complex [149]. Disruption of the interaction caused abrogation of MUN‐mediated transition from the closed Munc18‐1−syntaxin‐1 complex to the SNARE complex and led to strong defects in neurotransmitter release [149]. In addition, a more recent study identified an interaction between the bottom of domain 3a of Munc18‐1 and the SNARE motif of syntaxin‐1 [84]. It is likely that this binding enables Munc18‐1 to persistently associate with the SNARE motif of syntaxin‐1 during the transit of syntaxin‐1 from the Munc18‐1−syntaxin‐1 complex to the SNARE complex. Hence, Munc18‐1 and Munc13‐1 play crucial roles in SNARE assembly via priming syntaxin‐1 and synaptobrevin‐2.
The above interactions indicate the formation of a quaternary complex comprising Munc13‐1, Munc18‐1, syntaxin‐1, and synaptobrevin‐2 [147, 149] (Fig. 2B), which is available for efficient SNAP‐25 binding. SNAP‐25 is anchored on the presynaptic membrane via palmitoylation, which is naturally advantageous for SNARE nucleation. Notably, this proposed quaternary intermediate fully supports the half‐zippered mechanism of neuronal SNARE complex formation [45, 47], as truncation of the C‐half region of the second SNARE motif of SNAP‐25 (layer +1 to +8, residues 178−206) still supports Munc13‐1‐mediated transition from the closed Munc18‐1−syntaxin‐1 complex to the SNARE complex [149]. These data indicate that the C‐half region of the SNARE complex may be loosely packed along the assembly pathway guided by Munc18‐1 and Munc13‐1 (Fig. 2B), which is available for subsequent binding and regulation by complexins and synaptotagmins that are functionally related to Ca2+‐triggered fast fusion [150, 151, 152].
Moreover, Munc18‐1 and Munc13‐1 are implicated to protect SNARE complex assembly via preventing antiparallel binding of the SNAREs [153], and to promote artificial liposome fusion within 500 milliseconds under single‐vesicle level in vitro [154]. These data reinforce the physiological relevance of Munc18‐1 and Munc13‐1 in synaptic vesicle exocytosis. Altogether, the present data convey a fundamental model whereby Munc18‐1 and Munc13‐1 synergistically organize neuronal SNARE complex assembly and synaptic vesicle exocytosis.
Homologs of Munc18‐1 and Munc13s in different membrane trafficking systems—universality and specificity
SNARE complex assembly is regulated by SM proteins and multisubunit tethering complexes (MTCs) to ensure cargo delivery to proper target organelles or the plasma membrane [124, 155, 156]. Despite being distributed in different subcellular locations across diverse eukaryotic cells, all known SNAREs have their cognate SM proteins (Table 1); and different MTCs also display functional cooperations with specific SM proteins and related SNAREs (Table 1).
Table 1.
Summary of the interactions of MTCs, SNAREs, and related SM proteins.
| MTCs a | SNARE interactions b | Related SM protein | SM‐related Qa‐SNARE | SM:Qa‐SNARE binding mode | |||
|---|---|---|---|---|---|---|---|
| N‐peptide | Habc domain | SNARE motif | Four‐helical bundle | ||||
| Dsl1 (3) |
]
|
Sly1 (Sly1p) | Syx18 (Ufe1p) | •[33] | ? | ‐[33] | •[95] |
| TRAPPI (7) |
 [160] [160] |
Sly1 (Sly1p) | Syx5 (Sed5p) | •[161] | ? | ‐[33] | •[34] |
| TRAPPII (10) |
 [160] [160] |
Sly1 (Sly1p)? | ? | ||||
| TRAPPIII (8) | ? | ? | ? | ||||
| COG (8) |
|
Sly1 (Sly1p) | Syx5 (Sed5p) | •[161] | ? | ‐[33] | •[34] |
| GARP (4) | Vps45 (Vps45p) | Syx16 (Tlg2p) | •[36] | •[36] | •[36] | •[96] | |
| CORVET (6) |
|
Vps33 (Vps33p) | ? (Pep12p) | ||||
| HOPS (6) |
|
Vps33 (Vps33p) | Syx7 (Vam3p) | lacking[31] | •[181] | •[91] | •[97] |
| Exocyst (8) |
|
(Sec1p) | (Sso1/2p) | (lacking)[31] | (‐)[98] | (‐)[98] | (•) [98] |
| nCATCHR (2?) |
|
Munc18‐1 | Syx1a/b | •[77] | •[77] | •[77] | •[94] |
‘•’ Indicates direct binding between the motif/domain of Qa‐SNARE and SM protein; ‘‐’ indicates no binding between the motif/domain of Qa‐SNARE and SM protein. Syx, syntaxin; Syb, synaptobrevin.
Values in the parentheses indicate the subunits of MTCs in Homo sapiens and Saccharomyces cerevisiae.
Qa, Qb, Qc, and R‐SNAREs are indicated by red, green, light green, and blue, respectively. Values out of parentheses and in the parentheses indicate mammalian and Saccharomyces cerevisiae homologs, respectively.
In eukaryotic cells, seven MTCs have been identified, namely the Dsl1 complex [190], the TRAPP complex [191], the COG complex [192], the GARP complex [193], the CORVET complex [194], the HOPS complex [195], and the Exocyst complex [196], respectively, all of which participate in a variety of intracellular membrane trafficking pathways [156]. Despite differences in overall architecture and subunit composition, a subset of MTCs (the Dsl1, COG, GARP, and Exocyst complex) was characterized as the members of the CATCHR family proteins, in which the subunits of these complexes are composed of stacked long helical rods [124]. By contrast, the TRAPP, CORVET, and HOPS complex have different assembly and architecture [197, 198]. Different from other MTCs, the CORVET and HOPS complex involve the SM protein Vps33 as a subunit [194, 195]. The architecture of the Munc13‐1 MUN domain is similar to a variety of the CATCHR subunits such as Sec6 (Exocyst), Tip20 (Dsl1), COG5 (COG), and Vps54 (GARP) [124, 125]. Besides, CAPS contains a SNARE binding domain (DAMH), which is structurally similar to the CATCHR subunits and exhibits specific interaction with the Munc13‐1 MUN domain [186]. Therefore, it is expected that Munc13s and CAPSs might constitute a novel MTC—the neuronal CATCHR—to mediate synaptic vesicle exocytosis. It is believed that MTCs are involved in specific recognition of intracellular organelles and membranes by interacting with Rab GTPase [156]. Plenty of literature has reported that MTCs could interact with cognate SNAREs and SM proteins as well (summarized in Table 1). These interactions are crucial for recruiting the fusion components to the fusion sites [124, 156]. In addition, the HOPS complex was found to compete with the disassembly machinery (Sec17 and Sec18) and protect the prefusion SNARE complex from premature disassembly [145]; consistently, Munc13‐1 and Munc18‐1 were found to protect trans‐SNARE complex assembly in the context of Sec17−Sec18 homologs NSF−α‐SNAP [64, 137, 138]. Nevertheless, there still lack clear evidence whether other MTCs retain this function or not. Hence, the convergence of the function of MTCs is still limited among diverse membrane trafficking pathways.
Deletion of SM proteins invariably causes severe defects in membrane fusion (reviewed in [124, 155, 156]). As discussed above, a common characteristic of all SM proteins in membrane fusion is related to their function in templating SNARE complex assembly [34, 36, 84, 91, 92, 199, 200]. In spite of that, it is of note that there is substantial divergence in the binding between SM proteins and the SNAREs. SM proteins directly control the activity of Qa‐SNAREs with diverse binding modes (Table 1). For instance, Sly1p interacts with the N‐peptide to loosen the closed conformation of Sed5p and accelerate SNARE complex formation [201]; Vps45p rescues oligomeric Tlg2p into monomeric open conformation via interaction with the N‐peptide [36]; Vps33p interacts with both the Habc domain and the SNARE motif of Vam3p to template SNARE complex formation [97, 181]; Sec1p binds to the assembled SNARE four‐helical bundle (Sso1p‐Sec9p‐Snc2p) with an unrevealed mechanism that likely involves opening of Sso1p [98]; Munc18‐1 captures syntaxin‐1 into the closed conformation and the opening of syntaxin‐1 requires catalysis by Munc13‐1 [132]. These diverse binding and activation modes indicate multiple mechanisms of SM proteins in regulating Qa‐SNAREs. Essentially, all SM proteins are implicated to interact with the assembled SNARE four‐helical bundle (summarized in Table 1), which not only facilitates SNARE complex assembly but potentially protects partially assembled SNARE complex from disassembly.
The neuronal MTC−SM shares a variety of common features with other ancient MTC−SMs, including the interplay between SM proteins and the N‐peptide of Qa‐SNAREs; the function to template and secure SNARE complex assembly via binding to Qa‐ and R‐SNARE simultaneously. Interestingly, the mode that Munc18‐1 traps syntaxin‐1 into the inactivated conformation has not been found in other membrane trafficking systems. It is conceivable that the neuronal system specifically evolves complementary mechanisms that involve the activation of the closed Munc18‐1−syntaxin‐1 complex by Munc13‐1. Altogether, it is likely that Munc13‐1 and Munc18‐1 inherit some fundamental roles of the ancient MTC−SMs and evolve specialized features that are highly adapted to synaptic vesicle exocytosis.
Future perspective
In this review, we have summarized a variety of important advances toward understanding how Munc18‐1 and Munc13‐1 regulate SNARE complex assembly. However, fundamental questions still remain. First, there still lacks high‐resolution structural models of the whole release machinery that includes Munc18‐1 and Munc13‐1. This is a rather tough task since many interactions among these components are transient and weak, hindering the researchers from obtaining accurate structural models with atomic resolution. Single particle cryo‐electron microscopy (cryo‐EM) [202, 203] and in‐situ cryo‐electron tomography (cryo‐ET) [204] might be powerful tools to solve this issue. In addition, most present reconstitution studies still could not factually reproduce the fast kinetics of Ca2+‐triggered synaptic exocytosis. This may arise because additional components including many tethering and docking factors need to be included in these assays. Moreover, the presynaptic active zone is composed of a large number of macromolecules which create a crowded environment [4, 6]. Macromolecular crowding and liquid‐liquid phase separation (LLPS) might affect multiple reactions such as SNARE complex assembly and Ca2+‐triggered fusion [205, 206, 207, 208]. Hence, improvement of reconstitution methods that can reproduce the fusion events in vitro is necessary to better elucidate the molecular mechanism of synaptic vesicle fusion. Finally, there still exists unrevealed mechanisms of Munc13‐1 and Munc18‐1 in protecting trans‐SNARE complex assembly, in the terminal stage of membrane fusion, and in short‐/long‐term plasticity. Future studies remain challenging in fully resolving the fundamental molecular mechanism of synaptic vesicle exocytosis and the working principle of human brain.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
SW and CM conceived and designed the project, and SW and CM wrote the paper.
Acknowledgements
This work was supported by grant (2020M672326 to S.W.) from the China Postdoctoral Science Foundation, grants (31670846 and 31721002, to C.M.) from the National Science Foundation of China, and grant (2021ZD0202501, to C.M.) from the National Science and Technology Major Project of China. The funders were not involved in study design, data collection, data interpretation, and the decision to submit the manuscript for publication.
Edited by Josep Rizo
References
- 1. Sudhof TC, Rizo J. Synaptic vesicle exocytosis. Cold Spring Harb Perspect Biol. 2011;3:a005637. 10.1101/cshperspect.a005637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sudhof TC. Neurotransmitter release: the last millisecond in the life of a synaptic vesicle. Neuron. 2013;80:675–90. 10.1016/j.neuron.2013.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jahn R, Fasshauer D. Molecular machines governing exocytosis of synaptic vesicles. Nature. 2012;490:201–7. 10.1038/nature11320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Emperador‐Melero J, Kaeser PS. Assembly of the presynaptic active zone. Curr Opin Neurobiol. 2020;63:95–103. 10.1016/j.conb.2020.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rosenmund C, Rettig J, Brose N. Molecular mechanisms of active zone function. Curr Opin Neurobiol. 2003;13:509–19. 10.1016/j.conb.2003.09.011 [DOI] [PubMed] [Google Scholar]
- 6. Sudhof TC. The presynaptic active zone. Neuron. 2012;75:11–25. 10.1016/j.neuron.2012.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Soykan T, Maritzen T, Haucke V. Modes and mechanisms of synaptic vesicle recycling. Curr Opin Neurobiol. 2016;39:17–23. 10.1016/j.conb.2016.03.005 [DOI] [PubMed] [Google Scholar]
- 8. Kaeser PS, Regehr WG. The readily releasable pool of synaptic vesicles. Curr Opin Neurobiol. 2017;43:63–70. 10.1016/j.conb.2016.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sudhof TC. Calcium control of neurotransmitter release. Cold Spring Harb Perspect Biol. 2012;4:a011353. 10.1101/cshperspect.a011353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schneggenburger R, Neher E. Presynaptic calcium and control of vesicle fusion. Curr Opin Neurobiol. 2005;15:266–74. 10.1016/j.conb.2005.05.006 [DOI] [PubMed] [Google Scholar]
- 11. Khan YA, White KI, Brunger AT. The AAA+ superfamily: a review of the structural and mechanistic principles of these molecular machines. Crit Rev Biochem Mol Biol. 2021;1–32. 10.1080/10409238.2021.1979460 [DOI] [PubMed] [Google Scholar]
- 12. Rizo J, Xu J. The synaptic vesicle release machinery. Annu Rev Biophys. 2015;44:339–67. 10.1146/annurev-biophys-060414-034057 [DOI] [PubMed] [Google Scholar]
- 13. Sudhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–7. 10.1126/science.1161748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brunger AT, Choi UB, Lai Y, Leitz J, White KI, Zhou Q. The pre‐synaptic fusion machinery. Curr Opin Struct Biol. 2019;54:179–88. 10.1016/j.sbi.2019.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chapman ER. How does synaptotagmin trigger neurotransmitter release? Annu Rev Biochem. 2008;77:615–41. 10.1146/annurev.biochem.77.062005.101135 [DOI] [PubMed] [Google Scholar]
- 16. Trimbuch T, Rosenmund C. Should I stop or should I go? The role of complexin in neurotransmitter release. Nat Rev Neurosci. 2016;17:118–25. 10.1038/nrn.2015.16 [DOI] [PubMed] [Google Scholar]
- 17. Jahn R, Scheller RH. SNAREs–engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–43. 10.1038/nrm2002 [DOI] [PubMed] [Google Scholar]
- 18. Zhang Y, Hughson FM. Chaperoning SNARE folding and assembly. Annu Rev Biochem. 2021;90:581–603. 10.1146/annurev-biochem-081820-103615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Malsam J, Kreye S, Sollner TH. Membrane fusion: SNAREs and regulation. Cell Mol Life Sci. 2008;65:2814–32. 10.1007/s00018-008-8352-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–53. 10.1038/26412 [DOI] [PubMed] [Google Scholar]
- 21. Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–8. 10.1038/nature08156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q‐ and R‐SNAREs. Proc Natl Acad Sci USA. 1998;95:15781–6. 10.1073/pnas.95.26.15781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bock JB, Matern HT, Peden AA, Scheller RH. A genomic perspective on membrane compartment organization. Nature. 2001;409:839–41. 10.1038/35057024 [DOI] [PubMed] [Google Scholar]
- 24. Kloepper TH, Kienle CN, Fasshauer D. SNAREing the basis of multicellularity: consequences of protein family expansion during evolution. Mol Biol Evol. 2008;25:2055–68. 10.1093/molbev/msn151 [DOI] [PubMed] [Google Scholar]
- 25. Kloepper TH, Kienle CN, Fasshauer D. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol Biol Cell. 2007;18:3463–71. 10.1091/mbc.e07-03-0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kienle N, Kloepper TH, Fasshauer D. Phylogeny of the SNARE vesicle fusion machinery yields insights into the conservation of the secretory pathway in fungi. BMC Evol Biol. 2009;9:19. 10.1186/1471-2148-9-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hong W. SNAREs and traffic. Biochim Biophys Acta. 2005;1744:120–44. 10.1016/j.bbamcr.2005.03.014 [DOI] [PubMed] [Google Scholar]
- 28. Fernandez I, Ubach J, Dulubova I, Zhang X, Sudhof TC, Rizo J. Three‐dimensional structure of an evolutionarily conserved N‐terminal domain of syntaxin 1A. Cell. 1998;94:841–9. 10.1016/s0092-8674(00)81742-0 [DOI] [PubMed] [Google Scholar]
- 29. Zhou P, Pang ZP, Yang X, Zhang Y, Rosenmund C, Bacaj T, et al. Syntaxin‐1 N‐peptide and Habc‐domain perform distinct essential functions in synaptic vesicle fusion. EMBO J. 2013;32:159–71. 10.1038/emboj.2012.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vardar G, Salazar‐Lazaro A, Brockmann M, Weber‐Boyvat M, Zobel S, Kumbol VW, et al. Reexamination of N‐terminal domains of syntaxin‐1 in vesicle fusion from central murine synapses. eLife. 2021;10:e69498. 10.7554/eLife.69498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hu SH, Latham CF, Gee CL, James DE, Martin JL. Structure of the Munc18c/Syntaxin4 N‐peptide complex defines universal features of the N‐peptide binding mode of Sec1/Munc18 proteins. Proc Natl Acad Sci USA. 2007;104:8773–8. 10.1073/pnas.0701124104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Furgason ML, MacDonald C, Shanks SG, Ryder SP, Bryant NJ, Munson M. The N‐terminal peptide of the syntaxin Tlg2p modulates binding of its closed conformation to Vps45p. Proc Natl Acad Sci USA. 2009;106:14303–8. 10.1073/pnas.0902976106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamaguchi T, Dulubova I, Min SW, Chen X, Rizo J, Sudhof TC. Sly1 binds to Golgi and ER syntaxins via a conserved N‐terminal peptide motif. Dev Cell. 2002;2:295–305. 10.1016/s1534-5807(02)00125-9 [DOI] [PubMed] [Google Scholar]
- 34. Peng R, Gallwitz D. Sly1 protein bound to Golgi syntaxin Sed5p allows assembly and contributes to specificity of SNARE fusion complexes. J Cell Biol. 2002;157:645–55. 10.1083/jcb.200202006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nicholson KL, Munson M, Miller RB, Filip TJ, Fairman R, Hughson FM. Regulation of SNARE complex assembly by an N‐terminal domain of the t‐SNARE Sso1p. Nat Struct Biol. 1998;5:793–802. 10.1038/1834 [DOI] [PubMed] [Google Scholar]
- 36. Eisemann TJ, Allen F, Lau K, Shimamura GR, Jeffrey PD, Hughson FM. The Sec1/Munc18 protein Vps45 holds the Qa‐SNARE Tlg2 in an open conformation. eLife. 2020;9:60724. 10.7554/eLife.60724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dulubova I, Yamaguchi T, Wang Y, Sudhof TC, Rizo J. Vam3p structure reveals conserved and divergent properties of syntaxins. Nat Struct Biol. 2001;8:258–64. 10.1038/85012 [DOI] [PubMed] [Google Scholar]
- 38. Dulubova I, Yamaguchi T, Arac D, Li H, Huryeva I, Min SW, et al. Convergence and divergence in the mechanism of SNARE binding by Sec1/Munc18‐like proteins. Proc Natl Acad Sci USA. 2003;100:32–7. 10.1073/pnas.232701299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yoon TY, Munson M. SNARE complex assembly and disassembly. Curr Biol. 2018;28:R397–401. 10.1016/j.cub.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 40. Baker RW, Hughson FM. Chaperoning SNARE assembly and disassembly. Nat Rev Mol Cell Biol. 2016;17:465–79. 10.1038/nrm.2016.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walter AM, Wiederhold K, Bruns D, Fasshauer D, Sorensen JB. Synaptobrevin N‐terminally bound to syntaxin‐SNAP‐25 defines the primed vesicle state in regulated exocytosis. J Cell Biol. 2010;188:401–13. 10.1083/jcb.200907018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sorensen JB, Wiederhold K, Muller EM, Milosevic I, Nagy G, de Groot BL, et al. Sequential N‐ to C‐terminal SNARE complex assembly drives priming and fusion of secretory vesicles. EMBO J. 2006;25:955–66. 10.1038/sj.emboj.7601003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shin J, Lou X, Kweon DH, Shin YK. Multiple conformations of a single SNAREpin between two nanodisc membranes reveal diverse pre‐fusion states. Biochem J. 2014;459:95–102. 10.1042/BJ20131668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pobbati AV, Stein A, Fasshauer D. N‐ to C‐terminal SNARE complex assembly promotes rapid membrane fusion. Science. 2006;313:673–6. 10.1126/science.1129486 [DOI] [PubMed] [Google Scholar]
- 45. Li F, Kummel D, Coleman J, Reinisch KM, Rothman JE, Pincet F. A half‐zippered SNARE complex represents a functional intermediate in membrane fusion. J Am Chem Soc. 2014;136:3456–64. 10.1021/ja410690m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fasshauer D, Margittai M. A transient N‐terminal interaction of SNAP‐25 and syntaxin nucleates SNARE assembly. J Biol Chem. 2004;279:7613–21. 10.1074/jbc.M312064200 [DOI] [PubMed] [Google Scholar]
- 47. Zorman S, Rebane AA, Ma L, Yang G, Molski MA, Coleman J, et al. Common intermediates and kinetics, but different energetics, in the assembly of SNARE proteins. eLife. 2014;3:e03348. 10.7554/eLife.03348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wiederhold K, Fasshauer D. Is assembly of the SNARE complex enough to fuel membrane fusion? J Biol Chem. 2009;284:13143–52. 10.1074/jbc.M900703200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gao Y, Zorman S, Gundersen G, Xi Z, Ma L, Sirinakis G, et al. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–3. 10.1126/science.1224492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kesavan J, Borisovska M, Bruns D. v‐SNARE actions during Ca(2+)‐triggered exocytosis. Cell. 2007;131:351–63. 10.1016/j.cell.2007.09.025 [DOI] [PubMed] [Google Scholar]
- 51. Tarafdar PK, Chakraborty H, Bruno MJ, Lentz BR. Phosphatidylserine‐dependent catalysis of stalk and pore formation by synaptobrevin JMR‐TMD peptide. Biophys J. 2015;109:1863–72. 10.1016/j.bpj.2015.08.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hu Y, Zhu L, Ma C. Structural roles for the juxtamembrane linker region and transmembrane region of synaptobrevin 2 in membrane fusion. Front Cell Dev Biol. 2020;8:609708. 10.3389/fcell.2020.609708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Han J, Pluhackova K, Bruns D, Bockmann RA. Synaptobrevin transmembrane domain determines the structure and dynamics of the SNARE motif and the linker region. Biochim Biophys Acta. 2016;1858:855–65. 10.1016/j.bbamem.2016.01.030 [DOI] [PubMed] [Google Scholar]
- 54. Dhara M, Yarzagaray A, Makke M, Schindeldecker B, Schwarz Y, Shaaban A, et al. v‐SNARE transmembrane domains function as catalysts for vesicle fusion. eLife. 2016;5:e17571. 10.7554/eLife.17571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Otto H, Hanson PI, Jahn R. Assembly and disassembly of a ternary complex of synaptobrevin, syntaxin, and SNAP‐25 in the membrane of synaptic vesicles. Proc Natl Acad Sci USA. 1997;94:6197–201. 10.1073/pnas.94.12.6197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hayashi T, Yamasaki S, Nauenburg S, Binz T, Niemann H. Disassembly of the reconstituted synaptic vesicle membrane fusion complex in vitro. EMBO J. 1995;14:2317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cousin MA. Synaptophysin‐dependent synaptobrevin‐2 trafficking at the presynapse‐Mechanism and function. J Neurochem. 2021;159:78–89. 10.1111/jnc.15499 [DOI] [PubMed] [Google Scholar]
- 58. Littleton JT, Barnard RJ, Titus SA, Slind J, Chapman ER, Ganetzky B. SNARE‐complex disassembly by NSF follows synaptic‐vesicle fusion. Proc Natl Acad Sci USA. 2001;98:12233–8. 10.1073/pnas.221450198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Burgalossi A, Jung S, Meyer G, Jockusch WJ, Jahn O, Taschenberger H, et al. SNARE protein recycling by alphaSNAP and betaSNAP supports synaptic vesicle priming. Neuron. 2010;68:473–87. 10.1016/j.neuron.2010.09.019 [DOI] [PubMed] [Google Scholar]
- 60. Rizo J, Sudhof TC. The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices–guilty as charged? Annu Rev Cell Dev Biol. 2012;28:279–308. 10.1146/annurev-cellbio-101011-155818 [DOI] [PubMed] [Google Scholar]
- 61. Xiao W, Poirier MA, Bennett MK, Shin YK. The neuronal t‐SNARE complex is a parallel four‐helix bundle. Nat Struct Biol. 2001;8:308–11. 10.1038/86174 [DOI] [PubMed] [Google Scholar]
- 62. Margittai M, Fasshauer D, Pabst S, Jahn R, Langen R. Homo‐ and heterooligomeric SNARE complexes studied by site‐directed spin labeling. J Biol Chem. 2001;276:13169–77. 10.1074/jbc.M010653200 [DOI] [PubMed] [Google Scholar]
- 63. Dulubova I, Sugita S, Hill S, Hosaka M, Fernandez I, Sudhof TC, et al. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 1999;18:4372–82. 10.1093/emboj/18.16.4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ma C, Su L, Seven AB, Xu Y, Rizo J. Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science. 2013;339:421–5. 10.1126/science.1230473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sadek M, Berndt D, Milovanovic D, Jahn R, Diederichsen U. Distance regulated vesicle fusion and docking mediated by beta‐peptide nucleic acid SNARE protein analogues. ChemBioChem. 2016;17:479–85. 10.1002/cbic.201500517 [DOI] [PubMed] [Google Scholar]
- 66. Hubrich BE, Kumar P, Neitz H, Grunwald M, Grothe T, Walla PJ, et al. PNA hybrid sequences as recognition units in SNARE‐protein‐mimicking peptides. Angew Chem Int Ed Engl. 2018;57:14932–6. 10.1002/anie.201805752 [DOI] [PubMed] [Google Scholar]
- 67. Meyenberg K, Lygina AS, van den Bogaart G, Jahn R, Diederichsen U. SNARE derived peptide mimic inducing membrane fusion. Chem Commun (Camb). 2011;47:9405–7. 10.1039/c1cc12879e [DOI] [PubMed] [Google Scholar]
- 68. Toonen RF, Verhage M. Vesicle trafficking: pleasure and pain from SM genes. Trends Cell Biol. 2003;13:177–86. 10.1016/s0962-8924(03)00031-x [DOI] [PubMed] [Google Scholar]
- 69. Archbold JK, Whitten AE, Hu SH, Collins BM, Martin JL. SNARE‐ing the structures of Sec1/Munc18 proteins. Curr Opin Struct Biol. 2014;29:44–51. 10.1016/j.sbi.2014.09.003 [DOI] [PubMed] [Google Scholar]
- 70. Park S, Bin NR, Yu B, Wong R, Sitarska E, Sugita K, et al. UNC‐18 and Tomosyn antagonistically control synaptic vesicle priming downstream of UNC‐13 in Caenorhabditis elegans . J Neurosci. 2017;37:8797–815. 10.1523/JNEUROSCI.0338-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hosono R, Hekimi S, Kamiya Y, Sassa T, Murakami S, Nishiwaki K, et al. The unc‐18 gene encodes a novel protein affecting the kinetics of acetylcholine metabolism in the nematode Caenorhabditis elegans . J Neurochem. 1992;58:1517–25. 10.1111/j.1471-4159.1992.tb11373.x [DOI] [PubMed] [Google Scholar]
- 72. Gracheva EO, Maryon EB, Berthelot‐Grosjean M, Richmond JE. Differential regulation of synaptic vesicle tethering and docking by UNC‐18 and TOM‐1. Front Synaptic Neurosci. 2010;2:141. 10.3389/fnsyn.2010.00141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Weimer RM, Richmond JE, Davis WS, Hadwiger G, Nonet ML, Jorgensen EM. Defects in synaptic vesicle docking in unc‐18 mutants. Nat Neurosci. 2003;6:1023–30. 10.1038/nn1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Verhage M, Maia AS, Plomp JJ, Brussaard AB, Heeroma JH, Vermeer H, et al. Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science. 2000;287:864–9. 10.1126/science.287.5454.864 [DOI] [PubMed] [Google Scholar]
- 75. Toonen RF, Wierda K, Sons MS, de Wit H, Cornelisse LN, Brussaard A, et al. Munc18‐1 expression levels control synapse recovery by regulating readily releasable pool size. Proc Natl Acad Sci USA. 2006;103:18332–7. 10.1073/pnas.0608507103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. de Wit H, Walter AM, Milosevic I, Gulyas‐Kovacs A, Riedel D, Sorensen JB, et al. Synaptotagmin‐1 docks secretory vesicles to syntaxin‐1/SNAP‐25 acceptor complexes. Cell. 2009;138:935–46. 10.1016/j.cell.2009.07.027 [DOI] [PubMed] [Google Scholar]
- 77. Misura KM, Scheller RH, Weis WI. Three‐dimensional structure of the neuronal‐Sec1‐syntaxin 1a complex. Nature. 2000;404:355–62. 10.1038/35006120 [DOI] [PubMed] [Google Scholar]
- 78. Burkhardt P, Hattendorf DA, Weis WI, Fasshauer D. Munc18a controls SNARE assembly through its interaction with the syntaxin N‐peptide. EMBO J. 2008;27:923–33. 10.1038/emboj.2008.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Han GA, Malintan NT, Saw NM, Li L, Han L, Meunier FA, et al. Munc18‐1 domain‐1 controls vesicle docking and secretion by interacting with syntaxin‐1 and chaperoning it to the plasma membrane. Mol Biol Cell. 2011;22:4134–49. 10.1091/mbc.E11-02-0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shen C, Liu Y, Yu H, Gulbranson DR, Kogut I, Bilousova G, et al. The N‐peptide‐binding mode is critical to Munc18‐1 function in synaptic exocytosis. J Biol Chem. 2018;293:18309–17. 10.1074/jbc.RA118.005254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rathore SS, Bend EG, Yu H, Hammarlund M, Jorgensen EM, Shen J. Syntaxin N‐terminal peptide motif is an initiation factor for the assembly of the SNARE‐Sec1/Munc18 membrane fusion complex. Proc Natl Acad Sci USA. 2010;107:22399–406. 10.1073/pnas.1012997108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Shen J, Rathore SS, Khandan L, Rothman JE. SNARE bundle and syntaxin N‐peptide constitute a minimal complement for Munc18‐1 activation of membrane fusion. J Cell Biol. 2010;190:55–63. 10.1083/jcb.201003148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Dulubova I, Khvotchev M, Liu S, Huryeva I, Sudhof TC, Rizo J. Munc18‐1 binds directly to the neuronal SNARE complex. Proc Natl Acad Sci USA. 2007;104:2697–702. 10.1073/pnas.0611318104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wang X, Gong J, Zhu L, Wang S, Yang X, Xu Y, et al. Munc13 activates the Munc18‐1/syntaxin‐1 complex and enables Munc18‐1 to prime SNARE assembly. EMBO J. 2020;39:e103631. 10.15252/embj.2019103631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Parisotto D, Pfau M, Scheutzow A, Wild K, Mayer MP, Malsam J, et al. An extended helical conformation in domain 3a of Munc18‐1 provides a template for SNARE (soluble N‐ethylmaleimide‐sensitive factor attachment protein receptor) complex assembly. J Biol Chem. 2014;289:9639–50. 10.1074/jbc.M113.514273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Munch AS, Kedar GH, van Weering JR, Vazquez‐Sanchez S, He E, Andre T, et al. Extension of helix 12 in Munc18‐1 induces vesicle priming. J Neurosci. 2016;36:6881–91. 10.1523/JNEUROSCI.0007-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hu SH, Christie MP, Saez NJ, Latham CF, Jarrott R, Lua LH, et al. Possible roles for Munc18‐1 domain 3a and Syntaxin1 N‐peptide and C‐terminal anchor in SNARE complex formation. Proc Natl Acad Sci USA. 2011;108:1040–5. 10.1073/pnas.0914906108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Han GA, Park S, Bin NR, Jung CH, Kim B, Chandrasegaram P, et al. A pivotal role for pro‐335 in balancing the dual functions of Munc18‐1 domain‐3a in regulated exocytosis. J Biol Chem. 2014;289:33617–28. 10.1074/jbc.M114.584805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Han GA, Bin NR, Kang SY, Han L, Sugita S. Domain 3a of Munc18‐1 plays a crucial role at the priming stage of exocytosis. J Cell Sci. 2013;126:2361–71. 10.1242/jcs.126862 [DOI] [PubMed] [Google Scholar]
- 90. Meijer M, Dorr B, Lammertse HC, Blithikioti C, van Weering JR, Toonen RF, et al. Tyrosine phosphorylation of Munc18‐1 inhibits synaptic transmission by preventing SNARE assembly. EMBO J. 2018;37:300–20. 10.15252/embj.201796484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Baker RW, Jeffrey PD, Zick M, Phillips BP, Wickner WT, Hughson FM. A direct role for the Sec1/Munc18‐family protein Vps33 as a template for SNARE assembly. Science. 2015;349:1111–4. 10.1126/science.aac7906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sitarska E, Xu J, Park S, Liu X, Quade B, Stepien K, et al. Autoinhibition of Munc18‐1 modulates synaptobrevin binding and helps to enable Munc13‐dependent regulation of membrane fusion. eLife. 2017;6:e24278. 10.7554/eLife.24278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Xu Y, Su L, Rizo J. Binding of Munc18‐1 to synaptobrevin and to the SNARE four‐helix bundle. Biochemistry. 2010;49:1568–76. 10.1021/bi9021878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Shen C, Rathore SS, Yu H, Gulbranson DR, Hua R, Zhang C, et al. The trans‐SNARE‐regulating function of Munc18‐1 is essential to synaptic exocytosis. Nat Commun. 2015;6:8852. 10.1038/ncomms9852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Li Y, Gallwitz D, Peng R. Structure‐based functional analysis reveals a role for the SM protein Sly1p in retrograde transport to the endoplasmic reticulum. Mol Biol Cell. 2005;16:3951–62. 10.1091/mbc.e05-02-0114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Carpp LN, Ciufo LF, Shanks SG, Boyd A, Bryant NJ. The Sec1p/Munc18 protein Vps45p binds its cognate SNARE proteins via two distinct modes. J Cell Biol. 2006;173:927–36. 10.1083/jcb.200512024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lobingier BT, Merz AJ. Sec1/Munc18 protein Vps33 binds to SNARE domains and the quaternary SNARE complex. Mol Biol Cell. 2012;23:4611–22. 10.1091/mbc.E12-05-0343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Togneri J, Cheng YS, Munson M, Hughson FM, Carr CM. Specific SNARE complex binding mode of the Sec1/Munc‐18 protein, Sec1p. Proc Natl Acad Sci USA. 2006;103:17730–5. 10.1073/pnas.0605448103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Varoqueaux F, Sigler A, Rhee JS, Brose N, Enk C, Reim K, et al. Total arrest of spontaneous and evoked synaptic transmission but normal synaptogenesis in the absence of Munc13‐mediated vesicle priming. Proc Natl Acad Sci USA. 2002;99:9037–42. 10.1073/pnas.122623799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Richmond JE, Davis WS, Jorgensen EM. UNC‐13 is required for synaptic vesicle fusion in C. elegans . Nat Neurosci. 1999;2:959–64. 10.1038/14755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Augustin I, Rosenmund C, Sudhof TC, Brose N. Munc13‐1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature. 1999;400:457–61. 10.1038/22768 [DOI] [PubMed] [Google Scholar]
- 102. Aravamudan B, Fergestad T, Davis WS, Rodesch CK, Broadie K. Drosophila UNC‐13 is essential for synaptic transmission. Nat Neurosci. 1999;2:965–71. 10.1038/14764 [DOI] [PubMed] [Google Scholar]
- 103. van de Bospoort R, Farina M, Schmitz SK, de Jong A, de Wit H, Verhage M, et al. Munc13 controls the location and efficiency of dense‐core vesicle release in neurons. J Cell Biol. 2012;199:883–91. 10.1083/jcb.201208024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kang L, He Z, Xu P, Fan J, Betz A, Brose N, et al. Munc13‐1 is required for the sustained release of insulin from pancreatic beta cells. Cell Metab. 2006;3:463–8. 10.1016/j.cmet.2006.04.012 [DOI] [PubMed] [Google Scholar]
- 105. Ashery U, Varoqueaux F, Voets T, Betz A, Thakur P, Koch H, et al. Munc13‐1 acts as a priming factor for large dense‐core vesicles in bovine chromaffin cells. EMBO J. 2000;19:3586–96. 10.1093/emboj/19.14.3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Man KN, Imig C, Walter AM, Pinheiro PS, Stevens DR, Rettig J, et al. Identification of a Munc13‐sensitive step in chromaffin cell large dense‐core vesicle exocytosis. eLife. 2015;4:e10635. 10.7554/eLife.10635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Imig C, Min SW, Krinner S, Arancillo M, Rosenmund C, Sudhof TC, et al. The morphological and molecular nature of synaptic vesicle priming at presynaptic active zones. Neuron. 2014;84:416–31. 10.1016/j.neuron.2014.10.009 [DOI] [PubMed] [Google Scholar]
- 108. Brockmann MM, Zarebidaki F, Camacho M, Grauel MK, Trimbuch T, Sudhof TC, et al. A trio of active zone proteins comprised of RIM‐BPs, RIMs, and Munc13s governs neurotransmitter release. Cell Rep. 2020;32:107960. 10.1016/j.celrep.2020.107960 [DOI] [PubMed] [Google Scholar]
- 109. Brockmann MM, Maglione M, Willmes CG, Stumpf A, Bouazza BA, Velasquez LM, et al. RIM‐BP2 primes synaptic vesicles via recruitment of Munc13‐1 at hippocampal mossy fiber synapses. eLife. 2019;8:43243. 10.7554/eLife.43243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Acuna C, Liu X, Sudhof TC. How to make an active zone: unexpected universal functional redundancy between RIMs and RIM‐BPs. Neuron. 2016;91:792–807. 10.1016/j.neuron.2016.07.042 [DOI] [PubMed] [Google Scholar]
- 111. Ann K, Kowalchyk JA, Loyet KM, Martin TF. Novel Ca2+‐binding protein (CAPS) related to UNC‐31 required for Ca2+‐activated exocytosis. J Biol Chem. 1997;272:19637–40. 10.1074/jbc.272.32.19637 [DOI] [PubMed] [Google Scholar]
- 112. Lu J, Machius M, Dulubova I, Dai H, Sudhof TC, Tomchick DR, et al. Structural basis for a Munc13‐1 homodimer to Munc13‐1/RIM heterodimer switch. PLoS Biol. 2006;4:e192. 10.1371/journal.pbio.0040192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Deng L, Kaeser PS, Xu W, Sudhof TC. RIM proteins activate vesicle priming by reversing autoinhibitory homodimerization of Munc13. Neuron. 2011;69:317–31. 10.1016/j.neuron.2011.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Camacho M, Basu J, Trimbuch T, Chang S, Pulido‐Lozano C, Chang SS, et al. Heterodimerization of Munc13 C2A domain with RIM regulates synaptic vesicle docking and priming. Nat Commun. 2017;8:15293. 10.1038/ncomms15293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Dulubova I, Lou X, Lu J, Huryeva I, Alam A, Schneggenburger R, et al. A Munc13/RIM/Rab3 tripartite complex: from priming to plasticity? EMBO J. 2005;24:2839–50. 10.1038/sj.emboj.7600753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Shin OH, Lu J, Rhee JS, Tomchick DR, Pang ZP, Wojcik SM, et al. Munc13 C2B domain is an activity‐dependent Ca2+ regulator of synaptic exocytosis. Nat Struct Mol Biol. 2010;17:280–8. 10.1038/nsmb.1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Xu J, Camacho M, Xu Y, Esser V, Liu X, Trimbuch T, et al. Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13‐1 C1C2BMUN. eLife. 2017;6:e22567. 10.7554/eLife.22567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Michelassi F, Liu H, Hu Z, Dittman JS. A C1–C2 module in Munc13 inhibits calcium‐dependent neurotransmitter release. Neuron. 2017;95:577–590.e5. 10.1016/j.neuron.2017.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Liu X, Seven AB, Camacho M, Esser V, Xu J, Trimbuch T, et al. Functional synergy between the Munc13 C‐terminal C1 and C2 domains. eLife. 2016;5:e13696. 10.7554/eLife.13696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Camacho M, Quade B, Trimbuch T, Xu J, Sari L, Rizo J, et al. Control of neurotransmitter release by two distinct membrane‐binding faces of the Munc13‐1 C1C2B region. eLife. 2021;10:e72030. 10.7554/eLife.72030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Herbst S, Lipstein N, Jahn O, Sinz A. Structural insights into calmodulin/Munc13 interaction. Biol Chem. 2014;395:763–8. 10.1515/hsz-2014-0134 [DOI] [PubMed] [Google Scholar]
- 122. Quade B, Camacho M, Zhao X, Orlando M, Trimbuch T, Xu J, et al. Membrane bridging by Munc13‐1 is crucial for neurotransmitter release. eLife. 2019;8:e42806. 10.7554/eLife.42806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Yang X, Wang S, Sheng Y, Zhang M, Zou W, Wu L, et al. Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic‐vesicle priming. Nat Struct Mol Biol. 2015;22:547–54. 10.1038/nsmb.3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Yu IM, Hughson FM. Tethering factors as organizers of intracellular vesicular traffic. Annu Rev Cell Dev Biol. 2010;26:137–56. 10.1146/annurev.cellbio.042308.113327 [DOI] [PubMed] [Google Scholar]
- 125. Pei J, Ma C, Rizo J, Grishin NV. Remote homology between Munc13 MUN domain and vesicle tethering complexes. J Mol Biol. 2009;391:509–17. 10.1016/j.jmb.2009.06.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Sakamoto H, Ariyoshi T, Kimpara N, Sugao K, Taiko I, Takikawa K, et al. Synaptic weight set by Munc13‐1 supramolecular assemblies. Nat Neurosci. 2018;21:41–9. 10.1038/s41593-017-0041-9 [DOI] [PubMed] [Google Scholar]
- 127. Li F, Kalyana Sundaram RV, Gatta AT, Coleman J, Ramakrishnan S, Krishnakumar SS, et al. Vesicle capture by membrane‐bound Munc13‐1 requires self‐assembly into discrete clusters. FEBS Lett. 2021;595:2185–96. 10.1002/1873-3468.14157 [DOI] [PubMed] [Google Scholar]
- 128. Grushin K, Kalyana Sundaram RV, Sindelar CV, Rothman JE. Munc13 structural transitions and oligomers that may choreograph successive stages in vesicle priming for neurotransmitter release. Proc Natl Acad Sci USA. 2022;119:e2121259119. 10.1073/pnas.2121259119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Stevens DR, Wu ZX, Matti U, Junge HJ, Schirra C, Becherer U, et al. Identification of the minimal protein domain required for priming activity of Munc13‐1. Curr Biol. 2005;15:2243–8. 10.1016/j.cub.2005.10.055 [DOI] [PubMed] [Google Scholar]
- 130. Basu J, Shen N, Dulubova I, Lu J, Guan R, Guryev O, et al. A minimal domain responsible for Munc13 activity. Nat Struct Mol Biol. 2005;12:1017–8. 10.1038/nsmb1001 [DOI] [PubMed] [Google Scholar]
- 131. Richmond JE, Weimer RM, Jorgensen EM. An open form of syntaxin bypasses the requirement for UNC‐13 in vesicle priming. Nature. 2001;412:338–41. 10.1038/35085583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Ma C, Li W, Xu Y, Rizo J. Munc13 mediates the transition from the closed syntaxin‐Munc18 complex to the SNARE complex. Nat Struct Mol Biol. 2011;18:542–9. 10.1038/nsmb.2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. van den Bogaart G, Meyenberg K, Risselada HJ, Amin H, Willig KI, Hubrich BE, et al. Membrane protein sequestering by ionic protein‐lipid interactions. Nature. 2011;479:552–5. 10.1038/nature10545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Honigmann A, van den Bogaart G, Iraheta E, Risselada HJ, Milovanovic D, Mueller V, et al. Phosphatidylinositol 4,5‐bisphosphate clusters act as molecular beacons for vesicle recruitment. Nat Struct Mol Biol. 2013;20:679–86. 10.1038/nsmb.2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Pertsinidis A, Mukherjee K, Sharma M, Pang ZP, Park SR, Zhang Y, et al. Ultrahigh‐resolution imaging reveals formation of neuronal SNARE/Munc18 complexes in situ. Proc Natl Acad Sci USA. 2013;110:E2812–20. 10.1073/pnas.1310654110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Melia TJ, Weber T, McNew JA, Fisher LE, Johnston RJ, Parlati F, et al. Regulation of membrane fusion by the membrane‐proximal coil of the t‐SNARE during zippering of SNAREpins. J Cell Biol. 2002;158:929–40. 10.1083/jcb.200112081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Stepien KP, Prinslow EA, Rizo J. Munc18‐1 is crucial to overcome the inhibition of synaptic vesicle fusion by alphaSNAP. Nat Commun. 2019;10:4326. 10.1038/s41467-019-12188-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Prinslow EA, Stepien KP, Pan YZ, Xu J, Rizo J. Multiple factors maintain assembled trans‐SNARE complexes in the presence of NSF and alphaSNAP. eLife. 2019;8:e38880. 10.7554/eLife.38880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Masuda ES, Huang BC, Fisher JM, Luo Y, Scheller RH. Tomosyn binds t‐SNARE proteins via a VAMP‐like coiled coil. Neuron. 1998;21:479–80. 10.1016/s0896-6273(00)80559-0 [DOI] [PubMed] [Google Scholar]
- 140. Hatsuzawa K, Lang T, Fasshauer D, Bruns D, Jahn R. The R‐SNARE motif of tomosyn forms SNARE core complexes with syntaxin 1 and SNAP‐25 and down‐regulates exocytosis. J Biol Chem. 2003;278:31159–66. 10.1074/jbc.M305500200 [DOI] [PubMed] [Google Scholar]
- 141. Fujita Y, Shirataki H, Sakisaka T, Asakura T, Ohya T, Kotani H, et al. Tomosyn: a syntaxin‐1‐binding protein that forms a novel complex in the neurotransmitter release process. Neuron. 1998;20:905–15. 10.1016/s0896-6273(00)80472-9 [DOI] [PubMed] [Google Scholar]
- 142. Yizhar O, Matti U, Melamed R, Hagalili Y, Bruns D, Rettig J, et al. Tomosyn inhibits priming of large dense‐core vesicles in a calcium‐dependent manner. Proc Natl Acad Sci USA. 2004;101:2578–83. 10.1073/pnas.0308700100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Gracheva EO, Burdina AO, Holgado AM, Berthelot‐Grosjean M, Ackley BD, Hadwiger G, et al. Tomosyn inhibits synaptic vesicle priming in Caenorhabditis elegans. PLoS Biol. 2006;4:e261. 10.1371/journal.pbio.0040261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Li Y, Wang S, Li T, Zhu L, Ma C. Tomosyn guides SNARE complex formation in coordination with Munc18 and Munc13. FEBS Lett. 2018;592:1161–72. 10.1002/1873-3468.13018 [DOI] [PubMed] [Google Scholar]
- 145. Xu H, Jun Y, Thompson J, Yates J, Wickner W. HOPS prevents the disassembly of trans‐SNARE complexes by Sec17p/Sec18p during membrane fusion. EMBO J. 2010;29:1948–60. 10.1038/emboj.2010.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Wang S, Choi UB, Gong J, Yang X, Li Y, Wang AL, et al. Conformational change of syntaxin linker region induced by Munc13s initiates SNARE complex formation in synaptic exocytosis. EMBO J. 2017;36:816–29. 10.15252/embj.201695775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Shu T, Jin H, Rothman JE, Zhang Y. Munc13‐1 MUN domain and Munc18‐1 cooperatively chaperone SNARE assembly through a tetrameric complex. Proc Natl Acad Sci USA. 2020;117:1036–41. 10.1073/pnas.1914361117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Magdziarek M, Bolembach AA, Stepien KP, Quade B, Liu X, Rizo J. Re‐examining how Munc13‐1 facilitates opening of syntaxin‐1. Protein Sci. 2020;29:1440–58. 10.1002/pro.3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Wang S, Li Y, Gong J, Ye S, Yang X, Zhang R, et al. Munc18 and Munc13 serve as a functional template to orchestrate neuronal SNARE complex assembly. Nat Commun. 2019;10:69. 10.1038/s41467-018-08028-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Maximov A, Tang J, Yang X, Pang ZP, Sudhof TC. Complexin controls the force transfer from SNARE complexes to membranes in fusion. Science. 2009;323:516–21. 10.1126/science.1166505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Yang X, Cao P, Sudhof TC. Deconstructing complexin function in activating and clamping Ca2+‐triggered exocytosis by comparing knockout and knockdown phenotypes. Proc Natl Acad Sci USA. 2013;110:20777–82. 10.1073/pnas.1321367110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Zhou Q, Zhou P, Wang AL, Wu D, Zhao M, Sudhof TC, et al. The primed SNARE‐complexin‐synaptotagmin complex for neuronal exocytosis. Nature. 2017;548:420–5. 10.1038/nature23484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Weninger K, Bowen ME, Choi UB, Chu S, Brunger AT. Accessory proteins stabilize the acceptor complex for synaptobrevin, the 1:1 syntaxin/SNAP‐25 complex. Structure. 2008;16:308–20. 10.1016/j.str.2007.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Lai Y, Choi UB, Leitz J, Rhee HJ, Lee C, Altas B, et al. Molecular Mechanisms of Synaptic Vesicle Priming by Munc13 and Munc18. Neuron. 2017;95:591–607.e10. 10.1016/j.neuron.2017.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Mironov AA, Beznoussenko GV. Models of intracellular transport: pros and cons. Front Cell Dev Biol. 2019;7:146. 10.3389/fcell.2019.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Dubuke ML, Munson M. The secret life of tethers: the role of tethering factors in SNARE complex regulation. Front Cell Dev Biol. 2016;4:42. 10.3389/fcell.2016.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Kraynack BA, Chan A, Rosenthal E, Essid M, Umansky B, Waters MG, et al. Dsl1p, Tip20p, and the novel Dsl3(Sec39) protein are required for the stability of the Q/t‐SNARE complex at the endoplasmic reticulum in yeast. Mol Biol Cell. 2005;16:3963–77. 10.1091/mbc.e05-01-0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Hirose H, Arasaki K, Dohmae N, Takio K, Hatsuzawa K, Nagahama M, et al. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J. 2004;23:1267–78. 10.1038/sj.emboj.7600135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Meiringer CT, Rethmeier R, Auffarth K, Wilson J, Perz A, Barlowe C, et al. The Dsl1 protein tethering complex is a resident endoplasmic reticulum complex, which interacts with five soluble NSF (N‐ethylmaleimide‐sensitive factor) attachment protein receptors (SNAREs): implications for fusion and fusion regulation. J Biol Chem. 2011;286:25039–46. 10.1074/jbc.M110.215327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Sacher M, Jiang Y, Barrowman J, Scarpa A, Burston J, Zhang L, et al. TRAPP, a highly conserved novel complex on the cis‐Golgi that mediates vesicle docking and fusion. EMBO J. 1998;17:2494–503. 10.1093/emboj/17.9.2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Bracher A, Weissenhorn W. Structural basis for the Golgi membrane recruitment of Sly1p by Sed5p. EMBO J. 2002;21:6114–24. 10.1093/emboj/cdf608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Willett R, Pokrovskaya I, Kudlyk T, Lupashin V. Multipronged interaction of the COG complex with intracellular membranes. Cell Logist. 2014;4:e27888. 10.4161/cl.27888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Willett R, Kudlyk T, Pokrovskaya I, Schonherr R, Ungar D, Duden R, et al. COG complexes form spatial landmarks for distinct SNARE complexes. Nat Commun. 2013;4:1553. 10.1038/ncomms2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Shestakova A, Suvorova E, Pavliv O, Khaidakova G, Lupashin V. Interaction of the conserved oligomeric Golgi complex with t‐SNARE Syntaxin5a/Sed5 enhances intra‐Golgi SNARE complex stability. J Cell Biol. 2007;179:1179–92. 10.1083/jcb.200705145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Kudlyk T, Willett R, Pokrovskaya ID, Lupashin V. COG6 interacts with a subset of the Golgi SNAREs and is important for the Golgi complex integrity. Traffic. 2013;14:194–204. 10.1111/tra.12020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Zolov SN, Lupashin VV. Cog3p depletion blocks vesicle‐mediated Golgi retrograde trafficking in HeLa cells. J Cell Biol. 2005;168:747–59. 10.1083/jcb.200412003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Sohda M, Misumi Y, Yamamoto A, Nakamura N, Ogata S, Sakisaka S, et al. Interaction of Golgin‐84 with the COG complex mediates the intra‐Golgi retrograde transport. Traffic. 2010;11:1552–66. 10.1111/j.1600-0854.2010.01123.x [DOI] [PubMed] [Google Scholar]
- 168. Laufman O, Hong W, Lev S. The COG complex interacts with multiple Golgi SNAREs and enhances fusogenic assembly of SNARE complexes. J Cell Sci. 2013;126:1506–16. 10.1242/jcs.122101 [DOI] [PubMed] [Google Scholar]
- 169. Willett R, Blackburn JB, Climer L, Pokrovskaya I, Kudlyk T, Wang W, et al. COG lobe B sub‐complex engages v‐SNARE GS15 and functions via regulated interaction with lobe A sub‐complex. Sci Rep. 2016;6:29139. 10.1038/srep29139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Suvorova ES, Duden R, Lupashin VV. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra‐Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J Cell Biol. 2002;157:631–43. 10.1083/jcb.200111081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Perez‐Victoria FJ, Bonifacino JS. Dual roles of the mammalian GARP complex in tethering and SNARE complex assembly at the trans‐golgi network. Mol Cell Biol. 2009;29:5251–63. 10.1128/MCB.00495-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Siniossoglou S, Pelham HR. An effector of Ypt6p binds the SNARE Tlg1p and mediates selective fusion of vesicles with late Golgi membranes. EMBO J. 2001;20:5991–8. 10.1093/emboj/20.21.5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Dulubova I, Yamaguchi T, Gao Y, Min SW, Huryeva I, Sudhof TC, et al. How Tlg2p/syntaxin 16 'snares' Vps45. EMBO J. 2002;21:3620–31. 10.1093/emboj/cdf381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Liewen H, Meinhold‐Heerlein I, Oliveira V, Schwarzenbacher R, Luo G, Wadle A, et al. Characterization of the human GARP (Golgi associated retrograde protein) complex. Exp Cell Res. 2005;306:24–34. 10.1016/j.yexcr.2005.01.022 [DOI] [PubMed] [Google Scholar]
- 175. Fridmann‐Sirkis Y, Kent HM, Lewis MJ, Evans PR, Pelham HR. Structural analysis of the interaction between the SNARE Tlg1 and Vps51. Traffic. 2006;7:182–90. 10.1111/j.1600-0854.2005.00374.x [DOI] [PubMed] [Google Scholar]
- 176. Abascal‐Palacios G, Schindler C, Rojas AL, Bonifacino JS, Hierro A. Structural basis for the interaction of the Golgi‐Associated Retrograde Protein Complex with the t‐SNARE Syntaxin 6. Structure. 2013;21:1698–706. 10.1016/j.str.2013.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Subramanian S, Woolford CA, Jones EW. The Sec1/Munc18 protein, Vps33p, functions at the endosome and the vacuole of Saccharomyces cerevisiae . Mol Biol Cell. 2004;15:2593–605. 10.1091/mbc.e03-10-0767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Balderhaar HJ, Lachmann J, Yavavli E, Brocker C, Lurick A, Ungermann C. The CORVET complex promotes tethering and fusion of Rab5/Vps21‐positive membranes. Proc Natl Acad Sci USA. 2013;110:3823–8. 10.1073/pnas.1221785110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Collins KM, Thorngren NL, Fratti RA, Wickner WT. Sec17p and HOPS, in distinct SNARE complexes, mediate SNARE complex disruption or assembly for fusion. EMBO J. 2005;24:1775–86. 10.1038/sj.emboj.7600658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Kramer L, Ungermann C. HOPS drives vacuole fusion by binding the vacuolar SNARE complex and the Vam7 PX domain via two distinct sites. Mol Biol Cell. 2011;22:2601–11. 10.1091/mbc.E11-02-0104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Lurick A, Kuhlee A, Brocker C, Kummel D, Raunser S, Ungermann C. The Habc domain of the SNARE Vam3 interacts with the HOPS tethering complex to facilitate vacuole fusion. J Biol Chem. 2015;290:5405–13. 10.1074/jbc.M114.631465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Yue P, Zhang Y, Mei K, Wang S, Lesigang J, Zhu Y, et al. Sec3 promotes the initial binary t‐SNARE complex assembly and membrane fusion. Nat Commun. 2017;8:14236. 10.1038/ncomms14236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Shen D, Yuan H, Hutagalung A, Verma A, Kummel D, Wu X, et al. The synaptobrevin homologue Snc2p recruits the exocyst to secretory vesicles by binding to Sec6p. J Cell Biol. 2013;202:509–26. 10.1083/jcb.201211148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Morgera F, Sallah MR, Dubuke ML, Gandhi P, Brewer DN, Carr CM, et al. Regulation of exocytosis by the exocyst subunit Sec6 and the SM protein Sec1. Mol Biol Cell. 2012;23:337–46. 10.1091/mbc.E11-08-0670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Dubuke ML, Maniatis S, Shaffer SA, Munson M. The exocyst subunit Sec6 interacts with assembled exocytic SNARE complexes. J Biol Chem. 2015;290:28245–56. 10.1074/jbc.M115.673806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Zhou H, Wei Z, Wang S, Yao D, Zhang R, Ma C. Structural and functional analysis of the CAPS SNARE‐binding domain required for SNARE complex formation and exocytosis. Cell Rep. 2019;26:3347–3359.e6. 10.1016/j.celrep.2019.02.064 [DOI] [PubMed] [Google Scholar]
- 187. Parsaud L, Li L, Jung CH, Park S, Saw NM, Park S, et al. Calcium‐dependent activator protein for secretion 1 (CAPS1) binds to syntaxin‐1 in a distinct mode from Munc13‐1. J Biol Chem. 2013;288:23050–63. 10.1074/jbc.M113.494088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. James DJ, Kowalchyk J, Daily N, Petrie M, Martin TF. CAPS drives trans‐SNARE complex formation and membrane fusion through syntaxin interactions. Proc Natl Acad Sci USA. 2009;106:17308–13. 10.1073/pnas.0900755106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Kalyana Sundaram RV, Jin H, Li F, Shu T, Coleman J, Yang J, et al. Munc13 binds and recruits SNAP25 to chaperone SNARE complex assembly. FEBS Lett. 2021;595:297–309. 10.1002/1873-3468.14006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Ren Y, Yip CK, Tripathi A, Huie D, Jeffrey PD, Walz T, et al. A structure‐based mechanism for vesicle capture by the multisubunit tethering complex Dsl1. Cell. 2009;139:1119–29. 10.1016/j.cell.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Kim JJ, Lipatova Z, Segev N. TRAPP complexes in secretion and autophagy. Front Cell Dev Biol. 2016;4:20. 10.3389/fcell.2016.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Willett R, Ungar D, Lupashin V. The Golgi puppet master: COG complex at center stage of membrane trafficking interactions. Histochem Cell Biol. 2013;140:271–83. 10.1007/s00418-013-1117-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Bonifacino JS, Hierro A. Transport according to GARP: receiving retrograde cargo at the trans‐Golgi network. Trends Cell Biol. 2011;21:159–67. 10.1016/j.tcb.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Peplowska K, Markgraf DF, Ostrowicz CW, Bange G, Ungermann C. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo‐lysosomal biogenesis. Dev Cell. 2007;12:739–50. 10.1016/j.devcel.2007.03.006 [DOI] [PubMed] [Google Scholar]
- 195. Brocker C, Kuhlee A, Gatsogiannis C, Balderhaar HJ, Honscher C, Engelbrecht‐Vandre S, et al. Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc Natl Acad Sci USA. 2012;109:1991–6. 10.1073/pnas.1117797109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Heider MR, Munson M. Exorcising the exocyst complex. Traffic. 2012;13:898–907. 10.1111/j.1600-0854.2012.01353.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Yip CK, Berscheminski J, Walz T. Molecular architecture of the TRAPPII complex and implications for vesicle tethering. Nat Struct Mol Biol. 2010;17:1298–304. 10.1038/nsmb.1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. van der Beek J, Jonker C, van der Welle R, Liv N, Klumperman J. CORVET, CHEVI and HOPS ‐ multisubunit tethers of the endo‐lysosomal system in health and disease. J Cell Sci. 2019;132:jcs189134. 10.1242/jcs.189134 [DOI] [PubMed] [Google Scholar]
- 199. Jiao J, He M, Port SA, Baker RW, Xu Y, Qu H, et al. Munc18‐1 catalyzes neuronal SNARE assembly by templating SNARE association. eLife. 2018;7:e41771. 10.7554/eLife.41771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Lobingier BT, Nickerson DP, Lo SY, Merz AJ. SM proteins Sly1 and Vps33 co‐assemble with Sec17 and SNARE complexes to oppose SNARE disassembly by Sec18. eLife. 2014;3:e02272. 10.7554/eLife.02272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Demircioglu FE, Burkhardt P, Fasshauer D. The SM protein Sly1 accelerates assembly of the ER‐Golgi SNARE complex. Proc Natl Acad Sci USA. 2014;111:13828–33. 10.1073/pnas.1408254111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Cheng Y. Single‐particle cryo‐EM‐How did it get here and where will it go. Science. 2018;361:876–80. 10.1126/science.aat4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Bai XC, McMullan G, Scheres SH. How cryo‐EM is revolutionizing structural biology. Trends Biochem Sci. 2015;40:49–57. 10.1016/j.tibs.2014.10.005 [DOI] [PubMed] [Google Scholar]
- 204. Frangakis AS, Forster F. Computational exploration of structural information from cryo‐electron tomograms. Curr Opin Struct Biol. 2004;14:325–31. 10.1016/j.sbi.2004.04.003 [DOI] [PubMed] [Google Scholar]
- 205. Yu H, Rathore SS, Shen C, Liu Y, Ouyang Y, Stowell MH, et al. Reconstituting intracellular vesicle fusion reactions: the essential role of macromolecular crowding. J Am Chem Soc. 2015;137:12873–83. 10.1021/jacs.5b08306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Wu X, Ganzella M, Zhou J, Zhu S, Jahn R, Zhang M. Vesicle tethering on the surface of phase‐separated active zone condensates. Mol Cell. 2021;81:13–24.e7. 10.1016/j.molcel.2020.10.029 [DOI] [PubMed] [Google Scholar]
- 207. Wu X, Cai Q, Shen Z, Chen X, Zeng M, Du S, et al. RIM and RIM‐BP form presynaptic active‐zone‐like condensates via phase separation. Mol Cell. 2019;73:971–984.e5. 10.1016/j.molcel.2018.12.007 [DOI] [PubMed] [Google Scholar]
- 208. Emperador‐Melero J, Wong MY, Wang SSH, de Nola G, Nyitrai H, Kirchhausen T, et al. PKC‐phosphorylation of Liprin‐alpha3 triggers phase separation and controls presynaptic active zone structure. Nat Commun. 2021;12:3057. 10.1038/s41467-021-23116-w [DOI] [PMC free article] [PubMed] [Google Scholar]



 [
[ [
[ [
[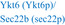 [
[ [
[ [
[ [
[ [
[ [
[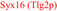 [
[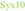 [
[ [
[ [
[ [
[ [
[ [
[ [
[ [
[ [
[ [
[ [
[ [
[ [
[ [
[ [
[ [
[