Abstract
Aging is accompanied by the decline of organismal functions and a series of prominent hallmarks, including genetic and epigenetic alterations. These aging-associated epigenetic changes include DNA methylation, histone modification, chromatin remodeling, non-coding RNA (ncRNA) regulation, and RNA modification, all of which participate in the regulation of the aging process, and hence contribute to aging-related diseases. Therefore, understanding the epigenetic mechanisms in aging will provide new avenues to develop strategies to delay aging. Indeed, aging interventions based on manipulating epigenetic mechanisms have led to the alleviation of aging or the extension of the lifespan in animal models. Small molecule-based therapies and reprogramming strategies that enable epigenetic rejuvenation have been developed for ameliorating or reversing aging-related conditions. In addition, adopting health-promoting activities, such as caloric restriction, exercise, and calibrating circadian rhythm, has been demonstrated to delay aging. Furthermore, various clinical trials for aging intervention are ongoing, providing more evidence of the safety and efficacy of these therapies. Here, we review recent work on the epigenetic regulation of aging and outline the advances in intervention strategies for aging and age-associated diseases. A better understanding of the critical roles of epigenetics in the aging process will lead to more clinical advances in the prevention of human aging and therapy of aging-related diseases.
Subject terms: Diseases, Senescence
Introduction
Aging is a slow but gradual process that is characterized by a continuous decline in the normal physiological functions of living organisms over their lifespan. With age, the body’s resilience decreases, making it more sensitive to aging-related diseases, such as neurodegenerative diseases and cancer, and increasing the risk of death.1–3 Although most organisms have a similar death curve and a higher mortality rate during aging, dramatic variance in aging rates can be observed within the same species. Taking honey bees as an example, although the queen bee and the worker bees are genetically identical, the queen lives on average ten times longer.4 Intriguingly, there are so-called “non-aging” organisms that have exceptionally long lifespans, exhibit no or late-onset aging-related declines in physiological abilities and are resistant to aging-related diseases, such as hydra and naked mole rats.5–7 These phenomena indicate that aging is a complicated process that may be regulated by a variety of different factors.
Numerous studies have revealed how aging occurs and how it is regulated by complex cellular and molecular mechanisms at different stages of life. Many factors affecting the aging process and longevity have been reported,8–10 including telomere shortening, nutrient sensing, mitochondrial dysfunction and oxidative stress, deterioration of DNA repair and accumulation of DNA damage, changes in protein homeostasis leading to the accumulation and aggregation of misfolded proteins, and changes in epigenetic regulation. The word “epigenetics” is derived from the Greek word “epi” and means “over” or “above” the genome. Epigenetics represents a reversible mechanism in regulating the function of the genome without altering the underlying DNA sequence of the genome; thus, the epigenome links genotype to phenotype, which plays an important role in modulating the aging process in response to environmental stimulation.
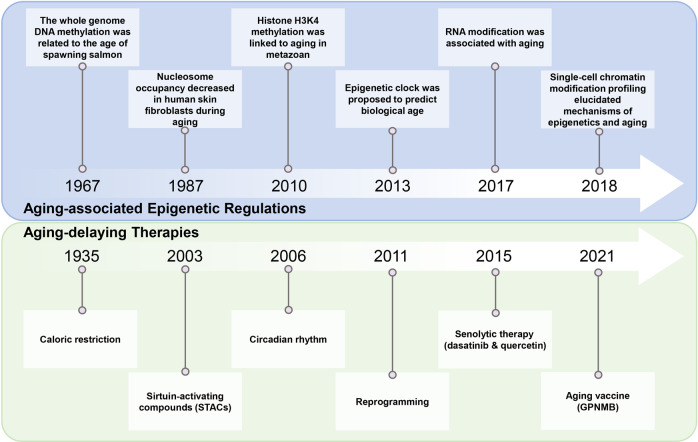
Epigenetic modifications are often reversible with the aid of epigenetic regulators, which lay the theoretical basis for aging modulation and make them promising targets for aging-intervention strategies. However, it was not until recently that a series of important studies have been carried out on epigenetic regulation and interventions for aging. In 1967, whole-genome DNA methylation was found to be related to the age of spawning salmon.11 Subsequent studies revealed that DNA methylation was generally downregulated in a variety of mouse tissues and human fibroblasts during aging.12,13 In 1987, the nucleosome occupancy in human skin fibroblasts was shown to decrease during aging, suggesting that chromatin configuration may change in the aging process.14 In 2010, histone methylation was first linked with life extension, and it was demonstrated that H3K4me3 demethylation in the germline boosted the lifespan of C. elegans.15 With increasing epigenetic evidence related to aging, in 2013, the concept of the “epigenetic clock” was proposed to link DNA methylation with biological age.16 In addition, RNA modifications and ncRNA regulation have recently been demonstrated to be involved in the regulation of aging.17 Furthermore, emerging single-cell chromatin modification profiling may provide molecular information with an unprecedented resolution of the relationship between epigenetics and aging in the future.18–20
Understanding how aging is regulated by epigenetic factors greatly facilitates the development of aging-delaying therapies. Ever since the first report of caloric restriction (CR) to slow down aging in 1935, researchers have been exploring potential approaches to delay aging.21 One of these aging-intervention studies shows that the aging process can be delayed, and the healthy lifespan or healthspan can be extended by CR and lowering the basal metabolic rate.22 Another exemplary study is the discovery of resveratrol, an agonist of the longevity factor SIR2 of the sirtuin family, and its function in extending the lifespan of yeast.23,24 In addition, heterochronic parabiosis (HP) and circadian rhythm models have been found to be effective in identifying factors that delay aging.25,26 In 2011, senescent cells from centenarians or Hutchinson–Gilford progeria syndrome (HGPS) patients can be fully reprogrammed to a pluripotent state with a rejuvenated epigenome, suggesting the potential of reprogramming in the reversal of aging.27,28 In 2015, a combination of dasatinib and quercetin was identified to kill senescent cells selectively; hence they were named senolytic drugs.29 More recently, the concept of aging vaccines was proposed, and glycoprotein nonmetastatic melanoma protein (GPNMB) vaccination has been shown to decrease tissue senescence and alleviate aging-related phenotypes.30 However, how epigenetic mechanisms are involved in these aging-intervention approaches has just begun to be revealed (Fig. 1).31
Fig. 1.
The history of studies on aging-associated epigenetic regulation and interventions
In this review, we will discuss how epigenetic remodeling, including DNA methylation, histone modification, chromatin remodeling, RNA modification, and non-coding RNA regulation, is regulated during aging. We will also introduce current therapeutic strategies to delay aging, including small molecules, reprogramming, active health, and many other epigenetic-associated approaches.
Epigenetic regulation of aging
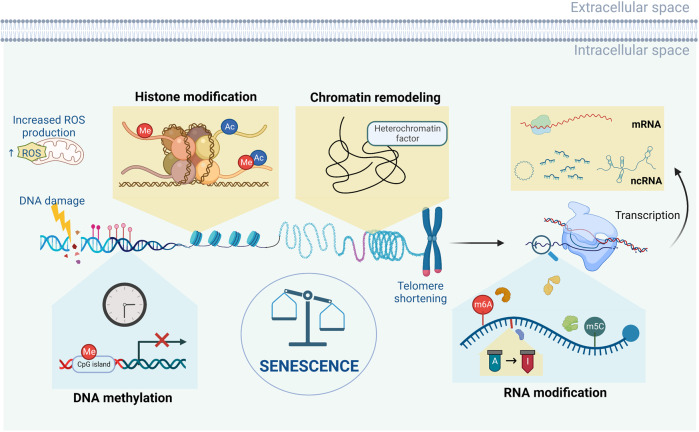
Accumulating evidence from invertebrate and vertebrate organisms, tissues, and in vitro systems links aging with epigenetic mechanisms. In mammals, there are global and local DNA methylation changes in the genome during aging. Additionally, there is a general loss of histones as well as global chromatin remodeling in all aging models. RNA modification and ncRNA regulation also play essential roles in cellular senescence via post-transcriptional regulations. Studies on how these epigenetic mechanisms regulate individual aging can provide targets to delay aging and rejuvenate aging organisms (Fig. 2).
Fig. 2.
An overview of the aging epigenome. During aging and the emergence of cellular senescence, a series of epigenetic changes occur in cells, including alterations in DNA methylation, chromatin remodeling, histone modification, RNA modification, and ncRNA regulation
DNA methylation
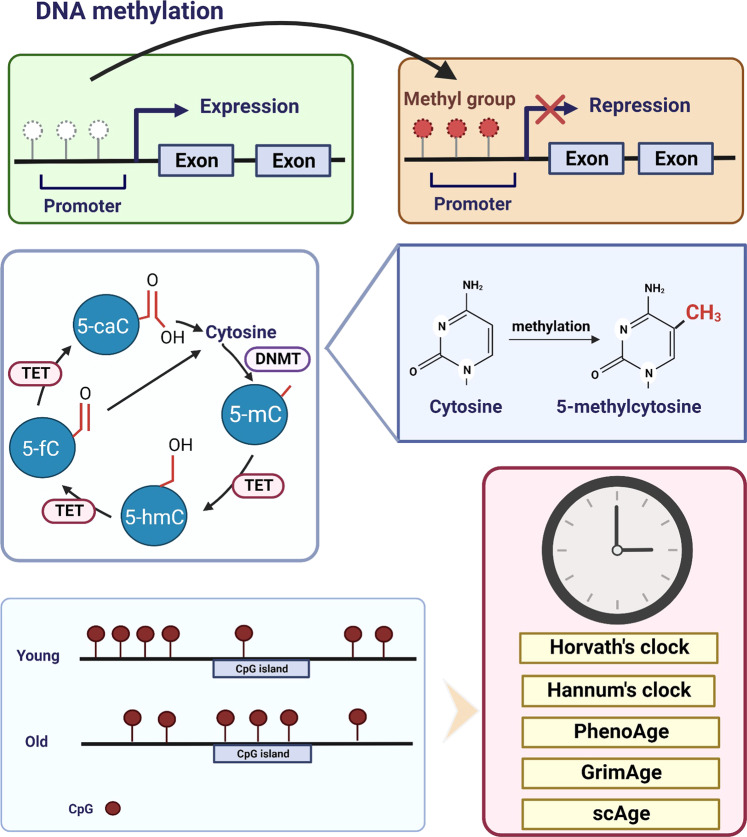
DNA methylation occurs at the cytosines in CpG dinucleotides to form 5-methylcytosine (5-mC), and 60%-90% of CpG sites in the mammalian genome are methylated (Fig. 3). The genome is generally hypomethylated during aging. Consistent with this, genes in energy metabolism and oxidative-stress resistance show higher expression in skeletal muscle of aged individuals.32,33
Fig. 3.
The mechanism of DNA methylation and the epigenetic clock theory of aging. Aging is often marked by global DNA hypomethylation, but hypermethylation also occurs at selective CpG islands. DNA methylation at the promoter of a gene often leads to silencing of that gene. DNA methylation at the 5ʹ cytosine of CpG results in 5-methylcytosine (5-mC). The methylation of DNA is mediated by DNMTs whereas the methyl group on DNA is removed by TET enzymes. TET enzymes oxidize 5-mC to generate 5-mC derivatives, including 5-hydroxymethylcytosine (5-hmC), 5-formylcytosine (5-fC), and 5-carboxylcytosine (5-caC), in mammalian cells. Age estimators, such as Horvath’s clock, Hannum’s clock, PhenoAge, GrimAge and single-cell age clock (scAge) are based on DNA methylation changes in the genome
DNA methyltransferases (DNMTs), namely DNMT1, DNMT3A, and DNMT3B, add methyl groups to nucleotides, resulting in gene silencing.34,35 The expression of DNMT1 decreases with age, resulting in a reduced DNA methylation level. DNMT1 mutants that cause the degeneration of selective central and peripheral neurons have been shown to translocate to the cytoplasm and form aggresomes while failing to bind to heterochromatin.36 In contrast, the expression of DNMT3A and DNMT3B increases with age, and contributes to de novo methylation of CpG islands in mammalian cells, increases with age.37,38 DNA methylation can be removed by ten-eleven translocation (TET) enzymes.39 In clinical research of aged patients, mutations of TET2 or DNMT3A increase the expression of pro-inflammatory cytokines and chronic inflammation, which is associated with conventional cardiovascular disease (CVD).40
DNA methylation shift
DNA methylation generally decreases with age in certain human and mouse tissues or cell cultures.13,41,42,43 Compared with newborns, whole-genome DNA methylation in CD4+ T cells of individuals over 100 years old has been shown to be decreased.41 The decrease of 5-mC from young to old mice is also observed in various organs, such as the brain, liver, and small intestinal mucosa, and the loss of 5-mC impairs the physiological function of cells in old mice.13 However, there is no notable shift in the genome-wide methylation level during aging in other human cell types, such as cells of the epidermis, liver, and heart, or some rat tissues, such as blood and kidney. These differences in DNA methylation may be due to tissue specificity or different detection techniques. On the other hand, many genes tend to be hypermethylated at CpG islands with aging (Fig. 3).44–46 A large meta-analysis of aging-related CpG islands demonstrated that hypermethylation of CpG islands is conserved across 59 tissues, including blood, liver, muscle, skin, brain, and cortex, derived from 128 mammalian species.47
Moreover, there are CpG sites that have increased variability in methylation with age, which are called age-associated variably methylated positions (aVMPs).44,48 Researchers first identified aVMPs in twin studies, in which older monozygotic twins exhibit a higher level of methylation variation in the overall content of 5-mC than younger twins, meaning that the methylation variation increases with age.48 The increased variation in aVMP methylation is associated with the downregulation of the expression of pentose metabolism genes, including PYGL, TALDO1, and PGD.49 Apart from aVMPs, there are also specific CpG sites, named age-associated differentially methylated positions (aDMPs).50 The methylation rate of aDMPs decreases with age in 6 mammalian species, including human beings, mice, dogs, naked mole rats, rhesus macaques, and humpback whales.43 Thus, the DNA methylation shift is associated with different CpG sites, including aVMPs and aDMPs, which can be measured to assess epigenetic age.
Epigenetic clock
The level of CpG site methylation with age is a reliable biomarker to predict chronological age. Researchers have developed age estimators called epigenetic clocks based on these mammalian DNA methylation levels. Epigenetic clocks use machine learning methods and are based on a set of CpG sites, whose DNA methylation states are consistent in multiple cells, tissues, or organs to predict the chronological age.51
The earliest model can estimate the age of a person and predict the risk of aging-related diseases, but shows low precision, only explaining 73% of age variance with a prediction error of 5.2 years.51 Since then, multiple epigenetic clocks have been reported with higher accuracy, precision, and broader application prospects in aging research.52,53 Among them, the first-generation clocks are Horvath’s epigenetic clock and Hannum’s epigenetic clock.54,55 Horvath’s epigenetic clock is a multi-tissue predictor based on 353 CpG sites to estimate the age of most tissues and cell types and is widely used in aging and cancer research.16 Hannum’s epigenetic clock can measure and compare human aging rates and provides a quantitative readout for aging-related diseases using 71 CpG markers from the DNA of blood.52 Based on Horvath’s pan-tissue clock, the DNAge™ algorithm is developed to compare the chronological age of young and aged muscles.56 Later, the second-generation clocks, including PhenoAge and GrimAge, introduced morbidity and mortality into the model, improving accuracy over the first generation.53,55 PhenoAge takes into account the role of multiple clinical biomarkers and can predict 10-year and 20-year mortality.53 GrimAge is based on 12 plasma proteins and smoking pack-years, and is a more predictive epigenetic clock for identifying clinical phenotypes.55 Notably, a recent study built up a single-cell age clock (scAge), which exhibits the epigenetic age using single-cell methylation data.57 ScAge is not only able to epigenetically differentiate “young” and “old” cells in heterogeneous tissues, but also predicts the chronological age of the tissues in mice.57 In summary, different epigenetic clocks have been developed for the prediction of the chronological age, which can be used to assess the efficacy of intervention methods for aging and to advance precision medicine.
Histone modification
Post-translational modifications of histones can activate or silence gene expression and regulate the aging process. The types of histone modifications include methylation, acetylation, phosphorylation, ubiquitination, ADP ribosylation, and others.58 Among these modifications, methylation, and acetylation at lysine residues are the most widely studied and are known to affect the aging process. In vivo and in vitro studies report global changes in H3K9me3, H4K20me3, H3K27me3, and H3K9ac levels during aging.59 Several enzymes are involved in the regulation of histone methylation and acetylation. Histone methyltransferases (HMTs) and histone demethylases (HDMs) play opposite roles in regulating histone methylation, and histone acetyltransferases (HATs) and histone deacetylases (HDACs) antagonistically regulate histone acetylation (Fig. 4).
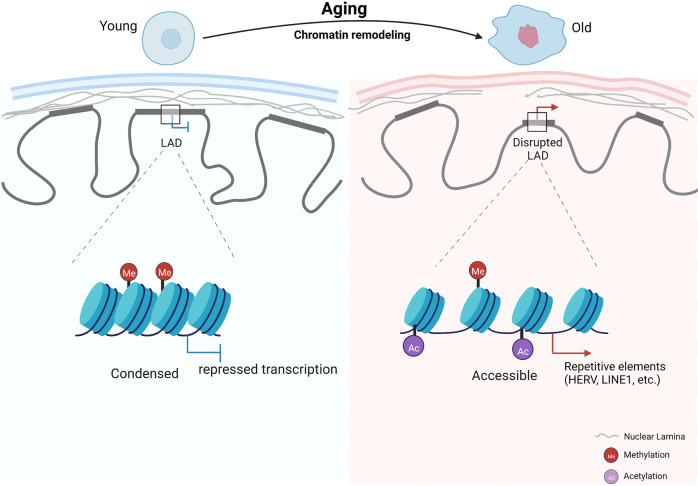
Fig. 4.
Chromatin structural remodeling during aging. A general loss of heterochromatin and detachment of lamina-associated domain (LAD) structures from the nuclear lamina occur during this process. Higher-order chromatin structure alterations during aging are accompanied by the redistribution of various histone modifications, including histone methylation (H3K4me3, H3K27me3, H3K36me3, H3K9me3) and acetylation (H3K9ac, H3K56ac, H4K16ac, H3K18ac). This leads to reactivation of repeating sequences and dysregulated gene expression due to aberrant chromatin accessibility
Histone methylation
Previous studies have shown that H3K4me3, a marker associated with active transcription, plays an important role in determining aging and lifespan by regulating the expression of aging-related genes.60,61 With aging in yeast, H3K4me3 accumulates in non-promoter regions and ribosomal DNA (rDNA), leading to the loss of rDNA heterochromatin along with an increase in genome-wide pervasive transcription.62,63 Studies in C. elegans somatic cells have also shown increased enrichment of H3K4me3 in promoter regions of senescence-related genes, and that this dynamics often occurs in regions with relatively low H3K4me3 markers,64 while down-regulation of the ASH-2 trithorax complex leads to H3K4me3 deficiency and lifespan extension.15 Consistently, ROS stimulation in C. elegans juveniles leads to an overall decrease in the H3K4me3 level and enhances their longevity.65 In a mouse model of Alzheimer’s disease (AD), the level of H3K4me3 and its catalyzing enzymes increases in the prefrontal cortex, a crucial brain region impaired in AD, and treating these mice with an inhibitor of H3K4 HMTs promotes the recovery of prefrontal cortex functions.66 The H3K4me3 level has also been shown to increase with age in mouse hematopoietic stem cells (HSCs).67 In contrast, a recent study using physiologically aged human HSCs demonstrated that aging is associated with reduced H3K4me3, H3K4me1, and H3K27ac.68 Neurons in aged (>60 years) human prefrontal cortex exhibit loss of H3K4me3 at 556 genes and gain of H3K4me3 at 101 genes compared to young (<1 year) neurons.69 Thus, H3K4me3 is related to aging in different species, although its influence on aging is context-dependent and requires further investigation.
H3K27me3 is generally associated with gene silencing and compacted heterochromatin.70 Earlier studies suggested a global loss of H3K27me3 in aged C. elegans and prematurely aged cells from Hutchinson-Guildford progeroid syndrome (HGPS) patients,71 while in killifish and mouse brains, global H3K27me3 increases with age.72 In C. elegans, the effect of the H3K27me3 demethylase UTX-1 on lifespan seems paradoxical, as both UTX-1 knockdown and overexpression in neurons and the intestine have been shown to extend lifespan.73–75 The conserved histone lysine demethylases jmjd-1.2/PHF8 and jmjd-3.1/JMJD3 could work as positive regulators of lifespan in response to mitochondrial dysfunction across different species, suggesting that increased levels of H3K27me3 in genes involved in the mitochondrial unfolded protein response (UPRmt) are detrimental to lifespan.76 Determining the locus- and cell-type-specific roles of H3K27me3 in lifespan regulation will be the key to unraveling the impact of H3K27me3-modifying enzymes on aging.
H3K36me3 and H3K9me3 also play important roles in the aging process. In both S. cerevisiae and C. elegans, deficiency of H3K36me3 is associated with a shorter lifespan. Consistently, the loss of H3K36me3 demethylase extends the lifespan of S. cerevisiae.77 Similarly, loss of H3K9me3 in the adult Drosophila midgut leads to intestinal stem cell aging.78 Interestingly, in aged somatic tissues of C. elegans, the global H3K9me3 level increases at heterochromatic regions in the distal arms of chromosomes, but decreases in euchromatic central regions of autosomes.79 In aged Drosophila, H3K9me3 and HP1 signals on chromosomes are significantly reduced compared with those in young flies, and overexpression of HP1 extends lifespan.80 Additionally, diminished levels of H3K9me3 and HP1 were identified in mesenchymal stem cells (MSCs) bearing pathogenic mutations of HGPS or Werner Syndrome (WS), another human disease with accelerated aging.81–83 The expression of the H3K9me3 methyltransferase SUV39H1 is decreased during the aging of both human and mouse HSCs,84 leading to a global reduction in H3K9 trimethylation and perturbed heterochromatin function.84 Treatment of Werner syndrome (WS)-specific MSCs with vitamin C, gallic acid (GA), or low-dose chloroquine (CQ) ameliorates a range of senescent phenotypes, promotes cell self-renewal, and upregulates levels of heterochromatin-associated marks, including H3K9me3.85–87 Thus, a reduction or redistribution of H3K9me3 is observed across different species with aging, although this trend is also tissue and cell-type dependent.88,89
Histone acetylation
Unlike histone methylation, the relationship between global histone acetylation and longevity is better understood. Histone acetylation is mediated by lysine acetyltransferases and is increased in active gene regions. HDACs are considered to function as corepressors and, together with HATs, play a critical role in longevity. Sirtuins are class III HDAC that enhance genome stability and regulate the deacetylation of lysine residues in an NAD+ level-dependent manner.90,91 Among the sirtuin family members, SIRT1 has been reported to decrease with age in various tissues of humans and mice, such as the liver, heart, kidney, brain, and lung.92,93 SIRT6 functions as an NAD+-dependent H3K9 deacetylase that modulates telomeric chromatin, and its overexpression contributes to the longevity of rat and human nucleus pulposus cells via inhibiting senescence.94 On the other hand, the HDAC class II family member HDAC4 was reported to be polyubiquitylated and degraded during all types of senescence. HDAC4 selectively binds to and monitors H3K27ac levels at specific enhancers and super-enhancers, such as enhancers of AKR1E2 and VEGFC. Treatment with the inhibitor of the HAT P300 could rescue senescence in HDAC4-depleted cells, suggesting a potential antagonistic effect between HDAC4 and P300.95 In IMR90 cells, P300, which promotes the formation of active enhancer elements in the non-coding genome, significantly increases the levels of H3K122ac and H3K27ac at the proximal senescence-specific gene promoters and is confirmed to be a primary driver of the senescent phenotypes, and depletion of p300 alone is sufficient to downregulate senescence genes and delay replicative senescence.96 CBP-1, the homolog of mammalian acetyltransferase CBP/p300 in C. elegans, is an essential regulator of the UPRmt and mediates H3K27ac and H3K18ac upon mitochondrial stress. Knockdown of CBP-1 decreases the lifespan of the worm in an HSF-1-dependent manner.97,98 Interestingly, abundant H3K4me1 marks are displayed in replicatively senescent IMR90 cells at the senescence-associated secretory phenotype (SASP)-associated super-enhancer loci, which also show substantial H3K27ac marks and BRD4 binding.96,99 Recent studies also demonstrated that aging-mediated changes in H3K27ac and H3K9ac in the human cerebral cortex are associated with AD.100,101
Owing to the antagonistic actions of HATs and HDACs, it is not surprising that HATs have also been implicated in aging. The level of H3K14ac in the brains of aging mice can regulate the expression of aging-related synaptic plasticity genes, and the H3K9me3/H3K14ac bivalent marks are significantly decreased in old mouse hepatocytes.102,103 Inactivation of KAT7 decreases histone H3K14ac and alleviates human mesenchymal precursor cell (hMPC) senescence.104 H3K18ac and H3K56ac are negative markers of senescence in Drosophila and yeast.105,106 Although the mechanism of action is different, knockdown of H3K56ac, Hst3, and Hst4-related HDAC-encoding genes during yeast aging shortens the lifespan.107 In aged yeast, the H3K56ac level decreases while the H4K16ac level increases, leading to the silencing of telomeric repeats.108 Interestingly, H4K16ac may also be involved in brain aging and AD progression. Normal aging leads to H4K16ac enrichment, while H4K16ac in the proximity of genes linked to aging and AD is dramatically reduced in AD.109 Additionally, dysregulation of H4K12ac leads to aging-related memory impairment, suggesting that it may serve as a critical signal of memory formation.110 By administering the HDACi suberoylanilide hydroxamic acid (SAHA) to aged mice, the acetylation deficit of H4K12 could be rescued in neurons.111 Thus, substantial changes in histone acetylation occur during aging and aging-related diseases, and understanding its regulatory mechanisms may provide new insight into the development of aging-intervention strategies.
Histone phosphorylation and ubiquitination
In addition to histone methylation and acetylation, histone phosphorylation and ubiquitination have also been shown to be associated with aging, and in some cases through crosstalk with other histone marks. For example, the effect of histone ubiquitination on DNA damage accumulation can induce premature neuronal aging.112 H3S28A mutants, which depletes H3S28 phosphorylation but also reduces H3K27 methylation to prevent by compromising the activity of its methyltransferase complex in Drosophila, prolong lifespan and improve resistance against starvation and paraquat-induced oxidative stress.113,114 However, the correlation between these modifications and aging is less clear, and more research is needed to refine the mechanisms in the future.
Chromatin remodeling
Chromatin is a flexible and dynamic structure composed of DNA and histones that can exist as heterochromatin or euchromatin. The basic unit of chromatin is the nucleosome core particle, encapsulated in a histone octamer consisting of a central H3-H4 tetramer flanked by two H2A-H2B dimers.115 Chromatin remodeling is defined as a series of genome-wide changes in the nuclear architecture that can be recognized at the level of specific chromosomes or chromosome domains, such as centromeres. Significant chromatin structural remodeling has been identified during cellular senescence, from histone component and modification changes to alterations of the chromatin compartments and topologically associating domains (TADs).83,116,117 Global canonical histone loss is regarded as a common feature of aging from yeast to humans.108,118,119 Overexpression of histone H3/H4 in yeast extends the lifespan, suggesting that an increased pool of free histones promotes survival during aging by facilitating nucleosome exchange and post-transcriptional chromatin repackaging.106 Genome-wide profiling of the core histone H3 occupancy in primary cultures of aging male mouse tissues and neural stem cells (NSCs) reveals local changes in H3 occupancy as tissues and cells age, even though the H3 level remains relatively stable.120 Reversible phosphorylation of serine and threonine residues in the C-terminal tail of H1 histones is responsible for regulating the H1 stacking behavior. Individuals with mutations deleting these residues in one of the histone H1 isoforms show a progeria phenotype, and their fibroblasts exhibit more nucleoid relaxation, less condensed chromosomes, and higher nucleolar instability (Fig. 4).121
In eukaryotes, histone-modifying enzymes and ATP-dependent chromatin-remodeling complexes are the two main factors of the chromatin-remodeling process.122 Modified histones may induce conformational changes in nucleosomes. Restoration of acetyl coenzyme A (acetyl-CoA) production through nutrient supplementation (citrate, acetate, pyruvate, and glucose) could strongly attenuate chromatin reorganization and diminish the extended lifespan of worms under mitochondrial stress conditions.123,124 In mice, aged MSCs show significantly decreased levels of total histone H3-H4 acetylation and an increased abundance of H3K27me3 across the gene body, resulting in a lower transcriptional rate and the loss of chromatin accessibility compared with young MSCs. Restoring cytoplasmic acetyl-CoA levels in aged MSCs can remodel chromatin structure and rejuvenate these cells.125 As H3K9me2 levels decrease, the nuclear peripheral heterochromatin loses its anchor to the nuclear lamina and moves toward the nuclear interior.126 In specific regions during aging, H3K9me2 switches to H3K9me3, another repressive mark but not enriched with direct contacts with the nuclear lamina; this may reflect aging-associated changes in subnuclear location of peripheral chromatin and associate with shortened lifespan in aged C. elegans somatic tissues.79 Interestingly, histone deacetylase or methyltransferase inhibitors alter histone modifications in ways that predominantly increase euchromatin or decrease heterochromatin.127 These results suggest that chromatin remodeling is largely related to the level of histone post-translational modifications. In addition, deletion of autophagy-related 7 (Atg7) leads to disordered nucleosome assembly in mouse CD11b+Ly6G- bone marrow cells, resulting in cellular senescence.128 Promoters of the conserved transcriptional and phenotypic responses to defects in chromatin structure genes and are sensitive to histone dosage. Reducing nucleosome occupancy at these promoters by deleting HHT1-HHF1 allows transcriptional activation induced by the stress-responsive transcription factors Msn2 and Gis1, and thus, responses induced by moderate chromatin architectural defects promote longevity.129
ATP-dependent chromatin-remodeling complexes can be divided into the SWI/SNF, ISWI, CHD, and INO80 families. SWI/SNF is required for the activation of nutrient-responsive genes, and the destruction of this complex impairs the ability of cells to adapt to their environment. SWI/SNF also regulates transcription by remodeling chromatin and promoting a more open chromatin configuration.130 In vitro, the BRM-SWI/SNF complex is required to promote co-expression of the telomere-binding proteins TRF1 and TRF2, which are essential for maintaining telomere length and structure in human fibroblasts and cervical cancer cells, contributing to the development of longevity-related functions.131 SWI plays a role in regulating aging during adulthood, and the absence of SWI shortens the lifespan of nematodes.132 Among CHD chromatin remodelers, the role of NuRD has been widely reported, and disruption of the NuRD complex may compromise the epigenetic composition of histones and the higher-order structure of chromatin, making the NuRD complex more susceptible to the influence of genotoxic stress.133 BAZ1A encodes an accessory subunit of the ATP-dependent chromatin-remodeling complex that regulates cellular senescence in cancer and normal cells.134 Inhibition of the chromatin-remodeling factor SMARCA4 is able to prevent aging-dependent dopaminergic degeneration and shortening of lifespan caused by α-synuclein and LRRK2 in Drosophila PD models.135
Chromatin accessibility states and the expression programs of aging-related genes are positively correlated during aging. Two types of chromatin regions with regular changes in their accessibility during aging are increased accessibility regions (IARs) and decreasing accessibility regions (DARs). IARs mainly exist in genes related to the occurrence and development of aging, whereas DARs mainly exist in genes related to functional decline caused by aging.136,137 The chromatin in human cells is spatially segregated into two compartments, compartment A and compartment B, and chromatin in the same compartment should have more frequent interactions, as revealed by Hi-C analysis.138 The disruption of higher-order chromatin structure and the separation of heterochromatin from the nuclear membrane are observed during cellular senescence and aging.63,83,130,139–141 A hierarchy of integrated structural state changes has been characterized through large-scale epigenomic analyses of isogenic young, senescent, and progeroid hMPCs, manifested as heterochromatin loss in repressive compartments, euchromatin weakening in active compartments, switching in interfacing topological compartments, and increasing epigenetic entropy. Nuclear lamina dysfunction results in the derepression of constitutive heterochromatic regions in repressive LAD structures marked by H3K9 methylations. Diminished histone markers such as H3K27me3 and facultative heterochromatin disruption contributed to an overall increase in epigenetic instability and ectopic expression of lineage restricted genes, SASP genes and repetitive elements.83,142–145 Loss of heterochromatin leads to a global increase in transcription and intercellular transcriptional heterogeneity, which is reported to be associated with cellular senescence and the onset of aging-related diseases.81,137,143,146,147 Higher resolution data show that chromosome compartments are partitioned into TADs, which are more basic domains with high interaction frequency therein and relatively isolated from neighbor regions. Below the scale of TADs, long-range chromatin looping interactions are insulated by TAD boundaries.148 CTCF, a highly conserved architectural protein with 11 zinc fingers, functions as a barrier to inhibit heterochromatin spreading,149 and it has been shown to be reduced during aging in various models and plays an important role in chromatin remodeling.150 Consistent with this, high levels of CTCF in proliferating fibroblasts promote p16INK4a silencing.151
RNA modification
More than 170 types of RNA modifications have been discovered thus far that regulate gene expression at the epitranscriptomic level.152 Although few studies have directly revealed the relationship between RNA modifications and organismal aging, accumulating data show the essential roles of these post-transcriptional regulatory mechanisms in the cellular senescence process, one of the critical causes for aging and aging-related diseases (Fig. 5).
Fig. 5.
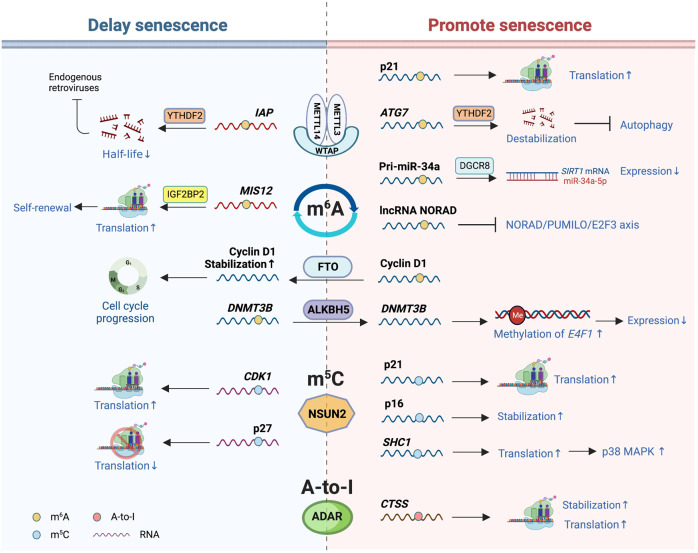
RNA modifications involved in senescence. RNA modifications that have been revealed to be associated with senescence or aging mainly include m6A modification, m5C modification, and adenosine-to-inosine (A-to-I) editing. The m6A modification is regulated by factors including the writer RNA methyltransferases complex, the erasers FTO and ALKBH5, and the reader YTHDF2, whose effects on senescence are complex based on different substrates. Under different stress conditions, m5C modification mediated by NSUN2 plays opposite roles in senescence, retarding replicative senescence and accelerating oxidative-induced senescence. The A-to-I RNA editing catalyzed by the ADAR family mainly exists in the central nervous system, and its relationship to neurodegenerative diseases has been demonstrated
m6A modification
As one of the most extensively studied mRNA modifications in mammalian cells, m6A has been demonstrated to be involved in cellular senescence. m6A is regulated by writer, reader, and eraser proteins.152,153 The multi-subunit writer RNA methyltransferases (MTases) are assembled mainly by methyltransferase like 3 (METTL3), METTL14, and Wilms tumor 1 associating protein (WTAP).154 The first reported m6A modification involved in senescence is methylation at the 3ʹ-UTR of the CDKN1A mRNAs by a METTL3/14 heterodimer, which facilitates p21 translation. Consistently, the expression of METTL3/14 and p21 is enhanced in oxidative-stress-induced senescence.17 Recently, ATG7 mRNAs with METTL3-dependent m6A were found to be destabilized by the reader YTH N6-methyladenosine RNA-binding protein 2 (YTHDF2), which promotes senescence instead of autophagy in fibroblast-like synoviocytes and leads to the progression of osteoarthritis.155 METTL14 also catalyzes the m6A modification affecting miRNAs associated with senescence. For example, TNF-α-induced METTL14 overexpression leads to increased production of miR-34a-5p from m A-modified primary transcript. miR-34a-5p promotes celluar senescence by targeting Sirtuin-1 (SIRT1) in nucleus pulposus cells (NPCs) of patients with intervertebral disc degeneration (IVDD), one of the most prevalent degenerative diseases.156 More recently, the regulator WTAP, which functions to translocate METTL3/14 dimers to nuclear speckles,154 has also been demonstrated to be associated with IVDD. Increased WTAP in senescent NPCs enhances the level of m6A in the lncRNA NORAD, contributing to the disruption of the NORAD/PUMILO/E2F3 axis and accelerating senescence.157
METTL3/14-mediated m6A modification has also been reported to inhibit senescence in some cases. METTL3/14 levels are reduced in LMNA mutant-induced prematurely aged human HGPS cells and senescent fibroblasts, and METTL14 overexpression delays cellular senescence.158 The interaction of Lamin A and METTL3/14 protects the latter from proteasome-mediated degradation to maintain sufficient m6A levels in normal cells.158 Moreover, METTL3-mediated m6A modification of MIS12 mRNAs positively regulates their stabilization by recruiting IGF2BP2, and in young hMSCs, MIS12 facilitates their self-renewal and alleviates cellular senescence. In HGPS and WS hMSCs, cellular models of premature aging, the downregulation of MIS12 is detected at both the mRNA and protein levels.159 Knockdown of DNMT2 in mouse embryonic fibroblasts (MEFs) reduces the m6A level and accelerates senescence.160 Thus, in addition to RNA MTases, DNMT2 participates in senescence regulation by affecting the m6A level. Sulforaphane-mediated cycle arrest and senescence in breast cancer cells are also accompanied by downregulated global m6A levels of mRNAs; however, the underlying mechanism is unclear.161
Similar to these m6A writers, the main erasers of m6A, i.e., fat mass and obesity-associated protein (FTO) and alkB homolog 5 (ALKBH5), are also involved in aging. For example, the expression of FTO declines with ovarian aging, followed by increased m6A levels in old human granulosa cells.162 Furthermore, FTO is crucial for the progression of the G1 phase of the cell cycle by removing m6A from the cyclin D1 mRNAs and stabilizing them.163 The ALKBH5 level is increased during IVDD and NPC senescence, and it removes m6A from the DNMT3B mRNAs, which limits the expression of the transcription factor E4F1 by methylating CpG islands at its promoter region and accelerates NPC senescence.164
To execute the function of m6A, reader proteins are needed, which include YTHDC family members, YTHDF family members, the eukaryotic translation initiation factor eIF3, the insulin-like growth factor 2 mRNA- binding proteins (IGF2BP1/2/3), and the fragile X retardation protein (FMRP).165 Several studies have indicated that the YTHDF family plays an important role in cellular senescence by destabilizing targeted mRNAs. In mouse embryonic stem cells (ESCs), m6A-modified intracisternal A-particle (IAP) mRNAs recruit YTHDFs to shorten their half-life, repressing endogenous retroviruses (ERVs).166 Conversely, the accumulation of IAP mRNAs after deletion of YTHDFs leads to high ERV activity,166 resulting in senescence and diseases.167 The well-known senolytic therapy (discussed in the next chapter) of the combination of dasatinib and quercetin can reduce the lipopolysaccharide (LPS)-induced SASP by upregulating YTHDF2, followed by destabilization of MAP2K4 and MAP4K4 mRNAs in human umbilical vein endothelial cells (HUVECs).168
In addition to cellular senescence, the role of m6A methylation in organs or organismal aging remains elusive. In brain aging and neurodegenerative disease models, dysregulation of m A and related regulatory proteins was indicated but the findings varied. For example, a tendency toward an overall increase in m6A methylation was observed in the cortex and hippocampus of the APP/PS1 transgenic mouse model for AD.169 Yet m6A in the 3ʹ UTR of many AD-associated transcripts in the 5XFAD mouse model of AD is downregulated, which is accompanied by an 8% increase and 4% decrease in FTO and METTL3, respectively, compared to wild-type mice, leading to higher Tau toxicity using the AD fly model.170
m5C modification
The m5C modification of RNAs is also tightly associated with senescence, in which the diverse roles of the tRNA methyltransferase NSUN2 (NOP2/Sun domain family, member 2) depend on the substrates of the m5C modification. For example, the 3ʹ-UTR of cyclin‐dependent kinase 1 (CDK1) mRNAs and the 5ʹ-UTR of the CDK inhibitor p27 mRNAs are targets of NSUN2, and m5C modification facilitates the translation of CDK1 while repressing that of p27, both of which alleviate replicative senescence.171,172 Interestingly, NSUN2 shows the opposite function in senescence under oxidative-stress conditions by targeting other mRNAs. NSUN2-mediated m5C at A988 in the 3ʹ-UTR of cyclin-dependent kinase inhibitor 2A (CDKN2A, i.e., p16) mRNAs stabilizes the mRNAs, contributing to senescence.173 H2O2 treatment leads to upregulation of NSUN2 and Src homology 2 domain-containing (SHC) family proteins in HUVECs with accelerated senescence, whereas knockdown of NSUN2 reduces ROS accumulation and delays senescence. NSUN2 catalyzes m5C modification of SHC1 mRNAs at several sites, which promotes its translation, activates the p38 MAPK pathway, and leads to cell cycle arrest and elevated ROS levels.174 Strikingly, NSUN2 and METTL3/14 have synergistic effects on the p21 mRNA methylation induced by oxidative stress. The m5C modification induced by NSUN2 facilitates METTL3/14 to catalyze the m6A modification at the p21 mRNA 3ʹ-UTR and vice versa.17 Furthermore, the RNA-binding protein human antigen R (HuR) facilitates the m5C modification at the C106 site of TERC to enable telomerase activity and delay cellular senescence.175 In summary, these studies identify RNA m5C as an epitranscriptomic marker for aging, and more investigation at the genome-wide scale will further reveal the dynamics of the m5C landscape and its potential impact on cellular senescence and tissue and organismal aging.
A-to-I editing
Adenosine to inosine (A-to-I) RNA editing conducted by the adenosine deaminase acting on RNA (ADAR) family occurs most frequently in the central nervous system, and the A-to-I imbalance has been demonstrated to be involved in neurological disorders, metabolic diseases, and other diseases.176,177 Insufficient A-to-I editing influences neurodegenerative processes. The editing level at the GluA2 Q/R site in the hippocampal region is lower in AD, which induces altered Ca2+ influx and neuron death.178 In human endothelial cells under pro-inflammatory conditions, cathepsin S (CTSS) mRNAs, encoding a cysteine protease, can be targeted by ADAR1, thereby recruiting HuR to improve its stability and translation. Consistently, the frequency of A-to-I editing on CTSS mRNAs is much higher in patients with vascular diseases.179
Collectively, these reports indicate that RNA modifications play crucial roles in regulating senescence. However, further studies are needed to uncover more detailed underlying mechanisms and their relationship to organismal aging, which may provide an avenue for developing new treatments for ameliorating senescence.
Non-coding RNA regulation
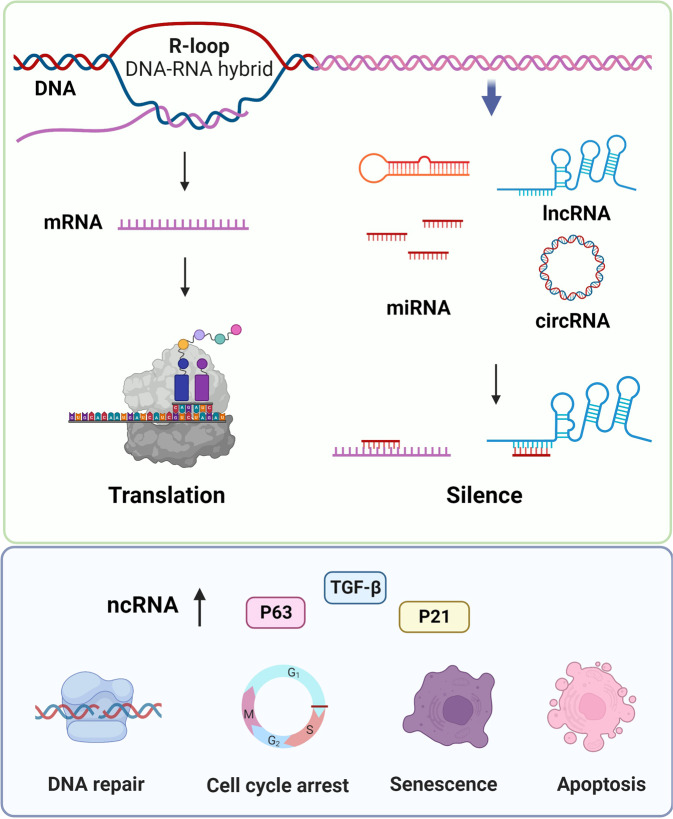
Research on the molecular mechanisms of cellular aging has mainly focused on protein-coding genes. However, accumulating studies have demonstrated that ncRNAs, which widely regulate gene expression in multiple biological processes at the epigenetic, transcriptional, and post-transcriptional levels, also play a critical role in aging. In recent years, studies of ncRNAs in aging have mainly focused on microRNAs (miRNAs),180 long non-coding RNAs (lncRNAs),181 R-loops,182,183 and circular RNAs (circRNAs)184 (Fig. 6).
Fig. 6.
The mechanism of non-coding RNAs regulation during aging. Non-coding RNAs (ncRNAs) include microRNAs (miRNAs), long non-coding RNAs (lncRNAs), R-loop (DNA-RNA hybrids), and circular RNAs (circRNAs). miRNAs bind to mRNAs, lncRNAs or circRNAs to prevent their functions
MiRNAs are small (~22 nucleotides), non-coding and single-stranded RNAs that bind to the 3ʹ-UTR of target mRNAs to degrade these mRNAs or suppress their translation.185,186 Using a microarray containing 863 miRNAs, researchers discovered that 64 miRNAs, such as miR-30d, miR-320d and miR-339-5p, are upregulated and 16 miRNAs, such as miR-103, miR-107, miR-24, and miR-130a, are downregulated in long-lived individuals compared to younger individuals.180 In addition, the miRNA-p53 pathway can maintain the genomic integrity in long-lived individuals during aging.180 The expression of miR-217 increases in late-passage fibroblasts, where miR-217 inhibits DNMT1 expression by targeting its 3′-UTR to induce human skin fibroblast senescence.187 The expression of age-associated miRNAs, including miR-130, miR-138, and miR-181a/b, increases in keratinocytes during cellular senescence, and by binding to p63 and Sirtuin-1 mRNAs, these miRNAs affect cell proliferation pathways.188
LncRNAs are non-protein-coding RNAs longer than 200 nucleotides. LncRNAs bind to DNA, RNA, and proteins to exert their functions as guides, enhancers, or scaffolds in post-transcriptional and post-translational regulations.189 Therefore, lncRNAs have become targets for the treatment of fibrosis in aging. In aged bone marrow mesenchymal stromal cells, the lncRNA NEAT1 promotes CSF1 secretion and enhances osteoclastic differentiation, which may be a therapeutic target for skeletal aging.190 The lncRNA APTR accelerates the cell cycle and cell proliferation of primary hepatic stellate cells in mice.191 Furthermore, targeting the lncRNA Firre by CRISPR/Cas9 delays Ras-induced cellular senescence.192,193
Evidence shows that circRNAs play important roles in the modulation of aging and aging-related diseases, such as cardiovascular disorders, diabetes, and neurodegenerative diseases.194 As circRNAs are relatively stable, aging-related increases in global circRNA levels are potential diagnostic biomarkers for aging.195 R-loops are three-stranded structures composed of a DNA-RNA heteroduplex and a displaced single DNA strand. Although R-loops are often considered as “by-products” of transcription, recent studies have shown that R-loops are important cellular regulators and may contribute to cancer and neurodegeneration.183,196,197 For example, deletion of SPT6 extends lncRNA and increases R-loops associated with DNA damage, which ultimately leads to senescence in HeLa cells.198 In summary, ncRNAs (miRNAs, lncRNAs, and circRNAs) have been proven to serve as biomarkers in regulating cellular senescence.
Strategies to alleviate aging
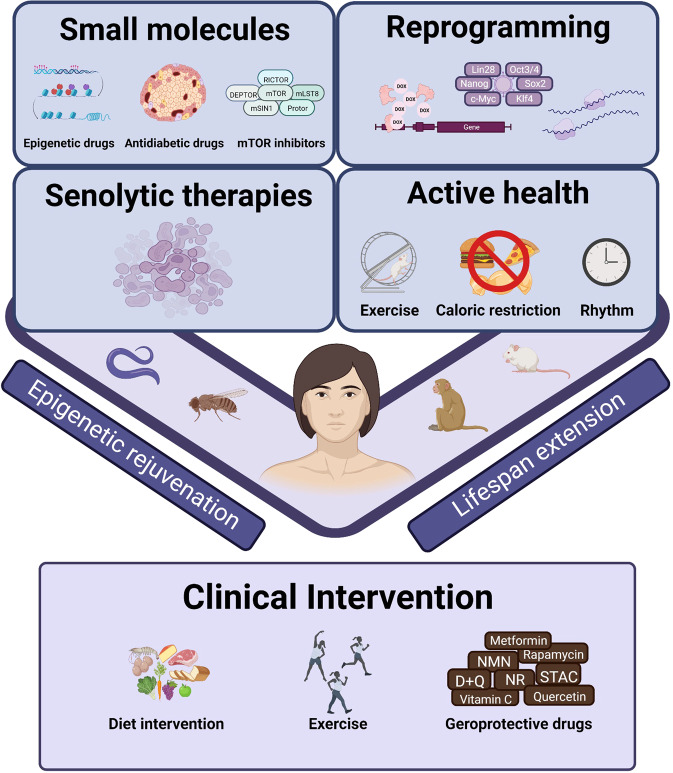
Based on the molecular mechanisms underlying cellular senescence and aging, a series of therapeutic strategies, many of which are closely related to epigenetic regulations, have been proposed (Fig. 7). Reprogramming and geroprotective drugs have been developed to interfere with aging, while senolytics aim to remove senescent cells to delay aging. Active health, such as caloric restriction, exercise, and a healthy circadian rhythm, exerts profound influences on multiple organs, systemic circuitries, and whole-body rejuvenation. Moreover, several advanced intervention methods have entered the clinical trial. Below we will discuss all these aging-intervention strategies and their underlying epigenetic mechanisms.
Fig. 7.
The intervention of aging. Existing intervention strategies aim to alleviate aging in various organisms
Small molecule-based therapy
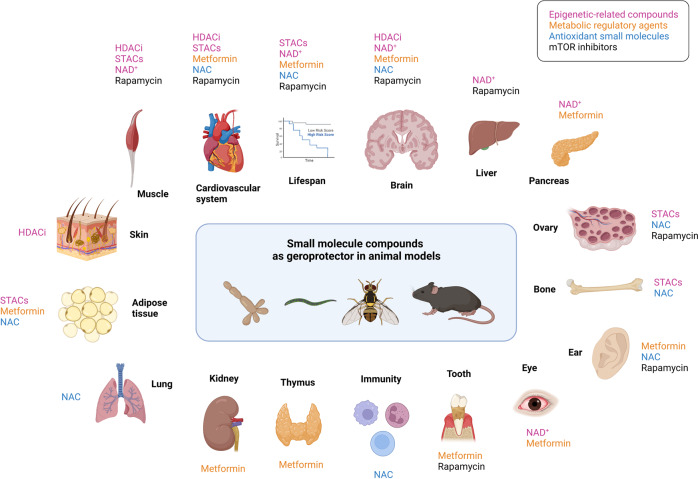
The first class of aging-intervention strategies enumerated here is geroprotective drugs, which include epigenetic-related compounds (e.g., NAD+ precursors, sirtuin-activating compounds, and HDAC inhibitors), small molecules with robust anti-diabetic effects (e.g., metformin), mTOR inhibitors (rapamycin), as well as antioxidant chemicals (N-acetyl-l-cysteine) (Fig. 8).
Fig. 8.
Small molecule compounds as geroprotectors in diverse animal models. A series of small molecule compounds can extend the lifespan or alleviate aging-related phenotypes in different organs. The interventions and corresponding target organs are shown in the diagram
NAD+ precursor
NAD+ is a critical redox coenzyme that plays a unique role in aging through DNA repair and epigenetic regulation.199,200 The effect of sirtuins on histone deacetylation is highly dependent on NAD+, highlighting the indispensable role of NAD+ in the epigenetic regulation of aging.201 Supplementation with NAD+ precursors, such as nicotinamide mononucleotide (NMN), nicotinamide riboside (NR), and nicotinamide (NAM), prevents the decline in NAD+ and exhibits beneficial effects against aging and aging-related diseases. NAD+ repletion extends the lifespan and delays the accelerated aging in C. elegans and Drosophila melanogaster models of Werner syndrome.202 In mammals, NR supplementation increases mitochondrial function, delays the senescence of NSCs, and increases mouse lifespan.203 NR supplementation also increases mitochondrial function and reduces aging-associated amyloidosis in muscle.204 In addition, NAD+ repletion with either NMN or NR ameliorates aging-associated meibomian gland dysfunction in aged mice.205 It also improves cognitive functions in AD mouse models, mainly by rescuing cerebral microvascular endothelial function and neurovascular coupling responses, preventing amyloid-β (Aβ) production in the brain, and reducing DNA damage, neuroinflammation, and apoptosis of hippocampal neurons.206–208 NAM improves glucose homeostasis and reduces hepatic steatosis and inflammation.209 Thus, boosting the NAD+ level appears to be a promising therapeutic strategy to counter aging and aging-associated disorders, although its effects in humans need further clinical studies.
Sirtuin-activating compound
Activators of the sirtuin family of HDACs, also termed sirtuin-activating compounds (STACs), are another class of epigenetic drugs as potential geroprotectors. Since they were found to promote the lifespan of yeast,24 STACs have been demonstrated to extend the longevity of worms, fruit flies, honey bees, and fish.210 In mammals, resveratrol, an activator of sirtuin 2, increases insulin sensitivity and motor function and thus improves the health and survival of mice on a high-calorie diet.211 Resveratrol is also found to attenuate the aging of adipose stem cells via decreasing the levels of 5-mC in DNA and modulating mitochondrial dynamics.212 SRT1720, an activator of sirtuin-1, can attenuate vascular endothelial dysfunction, excessive superoxide production, aging-related metabolic diseases, and inflammation with aging, as well as improve the follicle pool reserve, thereby extending the lifespan and improving the healthspan of mice.213–215 SRT2104, another activator of sirtuin-1, preserves bone and muscle mass and extends the survival of male mice on a standard diet.216
HDAC inhibitor
As discussed earlier, histone acetylation is one of the most important patterns of epigenetic regulation during aging. HDAC inhibitors show geroprotective effects mainly through reversing aging-associated deacetylation of chromatin, acetylation of histones near pro-longevity genes, and activating stress resistance and pro-longevity proteins.217 Administration of the pan-HDAC inhibitor SAHA rescues the skin phenotype, such as loss of subcutaneous fat, inflammation, and fibrosis, in a mouse model of Cockayne syndrome (CS), a hereditary form of premature aging.218 ITF2357 (givinostat) suppresses aging-induced diastolic dysfunction in normotensive mice.219 Another HDAC inhibitor, butyrate protects against aging-related muscle atrophy in mice.220 HDAC inhibitors also exhibit beneficial effects in neurodegenerative disorders by modulating chromatin-mediated neuroplasticity and improving learning consolidation.221,222 Since sirtuins are also a class of HDACs, the mechanism by which both STACs and HDAC inhibitors can delay aging remains to be further investigated.
Metformin
Metformin is an anti-diabetic drug and one of the most attractive geroprotective compounds, and it functions through extensive epigenetic regulation. Metformin retards aging in C. elegans by altering the ratio of S-adenosylmethionine (SAMe)/S-adenosylhomocysteine (SAH), which may affect histone methylation.223 In a spatial restraint stress mouse model, metformin exerts antidepressant effects by increasing the DNA 5-hmC modification level of the Bdnf gene.224 Metformin treatment increases the microRNA-processing protein DICER1 in mice and humans and thus modifies the profile of microRNAs associated with senescence and aging.225 Administration of metformin also alleviates the senescence of dental pulp stem cells through AMPK/mTOR signaling pathway-mediated downregulation of miR-34a-3p and upregulation of CAB39.226 Strikingly, there is evidence that metformin intervention improves the lifespan and healthspan of mice even when the administration starts at middle age (12 months)227 or old age (20–24 months),228–230 and the effect is enhanced when it starts earlier.231 In female SHR mice, however, metformin administration starting at the age of 3 months increases the mean lifespan by 14%, whereas the increase is only 6% when it starts at the age of 9 months, and there is no increase when it starts at the age of 15 months. Consistent with this, the lifespan extension effect of metformin is not seen in male rats232 or aged female mice.233 Nevertheless, metformin relieves many aging-related diseases in rodent models, including cognitive impairment and neurodegeneration,229,234–237 depression,238 chronic kidney disease,239 thymus degeneration,240 aging-related cataract,228 aging-related hearing loss,241 mitochondrial dysfunction in aged hearts,230 adipose tissue senescence and metabolic abnormalities,242,243 and aging-related developmental and metabolic phenotypes.244
Rapamycin
Rapamycin, an approved immunosuppressant in solid organ transplantation, also shows potential to intervene with aging. Rapamycin extends the median and maximum lifespan of both male and female mice in a dose-dependent manner through multiple mechanisms,245–247 including attenuating aging-related DNA methylation changes in the hippocampus to affect brain aging,248 slowing the aging epigenetic signatures in mouse livers, and ameliorating a series of aging-related diseases including cardiovascular dysfunction,249,250 neurodegeneration,251–254 skeletal muscle aging,255,256 ovarian aging,257,258 aging-related hearing loss,259,260 and aging-associated periodontitis.261,262 However, prolonged rapamycin administration is reported to induce muscle insulin resistance in rats, which might increase the incidence of diabetes.263 Considering the immunosuppression and NSC suppression effects of rapamycin,264 its application as a geroprotector should be assessed further.
N-acetyl-l-cysteine
N-acetyl-l-cysteine (NAC) is an antioxidant with a prominent influence on epigenetic regulation. It delays oocyte aging in mice by increasing the expression of sirtuins.265 Similarly, NAC attenuates aging-related oxidative damage and neurodegeneration in rat brains by upregulating sirtuin-1 and downregulating several SASP factors (TNF-α, IL-1β, IL-6).266 In addition, NAC extends the lifespan of mice267 and ameliorates a series of aging-related diseases in rodents, such as AD,268,269 aortic fibrosis,270 immunosenescence,271 oxidative stress and senescence in the lung,272 bone loss in ovariectomized mice,273 adipose tissue senescence and metabolic abnormalities,243 and aging-related hearing loss.274
Other geroprotective drugs
Many other drugs also show geroprotective effects, including anti-diabetic drugs (sodium-glucose cotransporter-2 inhibitors,275 acarbose,276–278) natural compounds (gallic acid,86 quercetin,279,280) antioxidant molecules (vitamin C,85,281 methylene blue,282) antihypertensive drugs (angiotensin-converting enzyme inhibitors and angiotensin receptor blockers),283,284 chloroquine,87,285 aspirin,286 uridine,287 and so on.
Reprogramming strategy
Reprogramming somatic cells with Yamanaka factors (Oct3/4, Sox2, Klf4, and c-Myc; OSKM) reverses cell fate and finally generates induced pluripotent stem cells (iPSCs), which possess the characteristics of ESCs.288,289 A classic strategy for combating aging comes from the generation of iPSCs. The durable expression of OSKM leads to widespread chromatin remodeling,290 and interestingly, some aged somatic cells can be reprogrammed to exhibit a youthful state. Ectopic expression of OSK without c-Myc restores the young patterns of DNA methylation and transcriptomes in mouse retinal ganglion cells, which can ameliorate vision problems in glaucomatous in aged mice. The DNA demethylation induced by OSK expression is confirmed to be necessary for the rejuvenation process of retinal ganglion cells.291
Although long-term reprogramming rejuvenates aged cells to varying degrees, some of the aged somatic cells will be fully reversed to iPSCs, which makes it impossible to be used to delay aging in vivo due to the teratoma-forming ability of iPSCs.292 Notably, transient reprogramming, which allows the expression of reprogramming factors in a certain period of time, also exerts a rejuvenating effect on aged somatic cells without altering the original cell identities.293,294 Transfecting aged human fibroblasts, chondrocytes, and endothelial cells with mRNAs expressing OSKMLN (OSKM, LIN28 and NANOG) rejuvenates host cells and significantly reverses the epigenetic clock.295 More recently, a 13-day OSKM reprogramming using the Tet-on expression system significantly reduced the epigenetic age of human fibroblasts without fully changing them into iPSCs, indicating a boundary between the rejuvenation and the pluripotency programs.296 Furthermore, short-term expression of OSKM in vivo significantly expands the lifespan of progeria mice and restores the levels of H3K9me3 and H4K20me3.292,297 In addition, a 2.5-week transient reprogramming in early life (2-month-old mice) is sufficient to extend the lifespan of transgenic progeria mice by 15% and rejuvenates the DNA methylation patterns in skin cells.298 The aging-associated epigenetic and transcriptional changes can also be alleviated by transient reprogramming in naturally aged mice.299 Overall, both long-term and transient reprogramming can achieve the rejuvenation of aged cells, while transient reprogramming also provides a novel method to alleviate aging in vivo in an organism.
Senolytic therapy
Senolytics selectively clear senescent cells in aged individuals and have been studied as a potential therapy for aging intervention. The first proposed senolytic strategy is the combination of dasatinib (D) and quercetin (Q), two pan-tyrosine kinase inhibitors.29 A single dose of D (5 mg/kg) + Q (50 mg/kg) effectively delays the aging phenotypes, such as frailty, cardiovascular diseases, and IVDD in aged mice, and extends the lifespan of Ercc1-/△ mice.29 To date, D + Q has been shown to prolong the healthspan and the physiological or pathological aging process in a variety of tissues or organs, including the cardiovascular system,300,301 skeleton,302–304 brain,305,306 adipose,307,308 lung,309,310 and muscle.311 Most recently, epigenetics regulation has been demonstrated to be an important mechanism by which D + Q eliminates aging cells. D + Q treatment leads to a significant change in epigenetic signatures in the hippocampus and improves the cognitive ability of aged male Wistar rats.312 Moreover, senescent adipose precursor cells exhibit hypomethylation and upregulated expression of the ZMAT3 gene, which is related to type 2 diabetes; 3 days of D + Q treatment is able to increase DNA methylation of ZMAT3 and decrease its expression, and reverse the senescence signature.313 In addition to D + Q, other senolytic drugs such as ABT-263, ABT-737, digoxin, FOXO4-DRI (D-retro inverso), and heat shock protein (HSP) 90 inhibitor 17-DMAG, play their senolytic roles mainly by inducing apoptosis and mitochondrial dysfunction,314–318 but their relationship to epigenetics needs further investigation.
Active health intervention
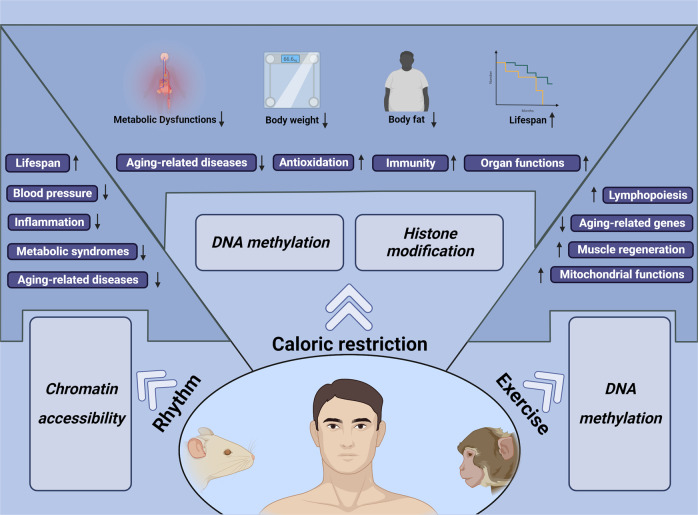
Active health refers to choosing a healthy lifestyle autonomously, such as caloric restriction, regular routine and moderate exercise, which are considered to benefit the quality of life and may exert a rejuvenating effect on the aging process (Fig. 9). With the increasing awareness of active health, various studies have demonstrated that healthy lifestyles ameliorate aging-associated features in different animals and humans.
Fig. 9.
A healthy lifestyle to postpone aging. Active health, including caloric restriction, rhythm control and exercise, improves body function and affects the lifespan of various animals, suggesting that healthy lifestyles exert profound effects on aging intervention
Caloric restriction
CR, which reduces calorie intake ranging from 10 to 40%, has been demonstrated to expand the lifespan of rodents to varying degrees,319–321 attenuating vascular endothelial dysfunction, improving the aerobic function of skeletal muscles, and ameliorating the loss of muscle fibers and turnover of motor neurons.322–325 The effect of CR on lifespan and rejuvenation is at least partially due to the amelioration of aging-related epigenetic changes, such as DNA methylation and histone modification.320,326 It is likely that epitranscriptomic regulation also functions as an effector of CR. For example, CR in rats significantly inhibited the aging-associated down-regulation of the RNA m6A reader protein YBX1, which has been shown to be one of the drivers of stem cell aging.327 According to the epigenetic clock developed in mice, 40% CR treatment slows the molecular changes and reduces the epigenetic age in mouse livers. Notably, a 20-year CR, which reaches a final level at 30%, shows reduced aging-related pathologies and a significant lifespan expansion in adult rhesus monkeys, indicating that moderate CR also exerts a rejuvenating effect on primates.328,329 In humans, moderate CR slows biological aging, improves the function of the liver, and reduces oxidative stress and the incidence of aging-related diseases.330–336 In summary, CR has been widely proven to be an effective method to delay aging, and clinical trials demonstrate the accessibility to applying CR in humans, which will be discussed in the ‘Clinical intervention’ section later.
Circadian rhythm
The circadian rhythm coordinates the behavior with the day/night shift and is also considered to play an essential role in the aging process.337 For example, epidermal and muscle stem cells from aged mice exhibit changed daily rhythms that cope with the stress of aging environments, indicating the continuous change of circadian rhythms along with the aging process.338 Disturbed circadian rhythm is linked to changes in chromatin structure, and it is found that 6 h sleep deprivation affects the chromatin accessibility in the cerebral cortex of mice, which contributes to long-term effects on gene expression.339 Forced circadian change may also accelerate the aging process and impair body function at a systemic level. Light schedule changes significantly affect aged mice, and advanced daytime leads to increased mortality of aged mice.26 In addition, mice with an innate circadian period close to 24 h live 20% longer than those with a shorter or longer innate circadian period.340 The disturbance of the circadian rhythm in rodents indicates that maintaining regular day/night cycles may reduce aging-related mortality and raise the question of whether circadian rhythm affects humans. Notably, a short-term circadian misalignment of 12-h inverted behavioral and environmental cycles for three days increases blood pressure and inflammatory markers in humans.341 However, how regular circadian rhythm benefits the healthspan, especially from the lens of epigenetic mechanisms, still needs further investigation.
Exercise
Exercise may remodel DNA methylation on the promoter of key genes in skeletal muscle342,343 and histone modifications could also be changed by exercise through inhibition of the function of HDACs, thereby influencing the gene expression patterns.344,345 Moreover, exercise can modulate the expression of several miRNAs that mediate the beneficial process.346,347 After voluntary resistance training for 8 weeks, aged mice exhibit nearly 8 weeks of younger epigenetic age in their muscle and a modest lifespan extension.56 In addition, voluntary wheel running benefits aged mice in neurogenesis and learning ability348 and reduces the abnormal changes of the aged synapse.324 Importantly, the rejuvenating effect of exercise is also observed in humans. There is a significant difference in the transcriptional profile between physically active and sedentary aged adults, and endurance exercise improves the function of muscles in aged people.349 In addition, resistance training reduces the level of the mitochondrial methylome in aged human skeletal muscle and partially restores the aging-related change in the nuclear gene methylome in muscle.350,351 Clinical trials aiming to investigate the beneficial effects of exercise will be discussed in the “Clinical intervention” section later.
Current studies have demonstrated that a healthy lifestyle indeed exerts a beneficial effect on aging and therefore raises awareness of vibrant health. However, caloric restriction, circadian control and exercise all need to be moderate during implementation, and the boundary between healthy and unhealthy status requires further investigation. As our understanding of aging deepens, a healthy lifestyle is considered to be the easiest way for humans to interfere with aging, and active health definitively deserves more attention.
Clinical intervention
Although various strategies targeting aging show satisfactory results in animal models, their effects in humans have yet to be demonstrated. Currently, diet intervention and exercise, which are extensively associated with epigenetic regulation, are the mostly studied and accepted strategies to target aging in humans. The clinical trial results demonstrate that CR (11.9%–25%) attenuates aging-related biomarkers, such as decreasing weight, enhancing insulin sensitivity and glucose tolerance, and improving major cardiometabolic risk factors.334,352,353 Time-restricted eating (TRE) in humans also provides benefits to some extent. Under an 8–10 h daily eating window of TRE, reductions in weight, blood pressure, atherogenic lipids, and cardiovascular risks are observed.354,355 A more stringent TRE (6 h window) also shows improvement in insulin sensitivity.356 However, the strategy of TRE is challenging to undertake, especially for cases with longer fasting times. Moreover, skipping breakfast has been found to be associated with an increased risk of mortality from cardiovascular disease.357 Therefore, an 11–12 h daily eating period is suggested to be ideal to avoid the compliance issues and side effects of TRE.358 Considering the difficulty for most subjects to adhere to chronic and extreme diets of CR or TRE, a fasting-mimicking diet (FMD) with low calories, low sugars, and low proteins but high unsaturated fats, provides another choice for a diet intervention. In a randomized phase 2 trial, healthy participants who received 3 monthly 5-day FMD cycles exhibited reduced markers/risk factors for aging, diabetes, cancer, and cardiovascular disease.359 In addition, a comprehensive understanding of the dietary interventions in humans has led to the proposal of the everyday normocaloric longevity diet that includes a mid to high carbohydrate and low but sufficient protein intake that is mostly plant-based but includes regular consumption of pesco-vegetarian-derived proteins.358 Diet intervention has also been found to be associated with epigenetic regulation in clinical trials. For example, CR in healthy and slightly overweight subjects significantly increases plasma concentrations of SIRT1.360 Five days of periodic fasting significantly elevated the expression of SIRT1 and SIRT3 in blood cells.361 Moreover, diet intervention has been shown to slow down the DNA methylation-based biomarkers of aging in several studies.362,363
Exercise has been demonstrated to be an effective geroprotector to improve the lifespan and healthspan in humans. Vigorous exercise, such as running, at middle and older ages, is associated with reduced disability in later life and reduced mortality,364,365 and leisure time physical activity of moderate to vigorous intensity is associated with longer life expectancy.366 In clinical intervention studies, exercise is found to reverse a series of aging-related diseases, including heart failure,367–371 cognitive decline,372,373 atherosclerosis,374 and insulin resistance.375,376 The geroprotective effect of exercise in humans is closely linked to epigenetic regulation. Exercise modifies the DNA methylation patterns in aged human skeletal muscle and reduces stochastic epigenetic mutations in crucial cancer-related pathways.32,363 Endurance exercise upregulates the expression of SIRT3 in the skeletal muscle and upregulates SIRT1, SIRT3, and SIRT6 in the serum.375,377,378 Exercise also modulates the microRNA expression profile (such as miR-423-3p, miR-451a, miR-766-3p, miR-130a, and miRNA-223) in subjects with type 2 diabetes, which may be involved in the improvement of weight loss, blood glucose control, and insulin sensitivity.379–381 In addition, the combination of diet and lifestyle interventions, including exercise, sleep, relaxation guidance, supplemental probiotics and phytonutrients, reverses the epigenetic age in healthy adult males.362
Pharmacological intervention is another major strategy to target natural aging. Epigenetic-related compounds, such as NMN, NR, and STACs, show potential as geroprotectors in clinical trials. Supplementation with NMN increases muscle insulin sensitivity, insulin signaling, and muscle remodeling in prediabetic women.382 NMN prevents aging-related muscle dysfunctions383 and shows benefits in improving aerobic capacity, cardiovascular fitness, sleep quality, fatigue, and physical performance.89,384 As for NR, clinical trials indicate that NR suppresses inflammatory activation of PBMCs in heart failure patients385 and decreases the levels of inflammatory cytokines in the serum and cerebrospinal fluid of Parkinson’s disease (PD) patients.386 However, most of these studies focus on the safety and tolerability of NR in patients.386,387 The effectiveness of NR in preventing or attenuating the progression of aging-related disorders should be verified in further studies, considering that several clinical trials show that NR does not improve insulin resistance.388–391 STACs exhibit inspiring effects in preclinical studies, but the results in clinical trials are not as satisfactory. For example, the natural STAC resveratrol and early synthetic STACs such as SRT1720 have very low bioavailability, potency, and limited target specificity,392 and other STACs, such as SRT2379 and SRT3025, produce no significant clinical responses. Similarly, although SRT2104 shows some benefits on lipid parameters, including cholesterol and triglycerides,393,394 it does not improve glucose or insulin control394,395 and has no significant anti-inflammatory effect in ulcerative colitis patients.396 The poor and variable pharmacokinetics upon oral administration of SRT2104 need to be resolved in the future.
Other geroprotective interventions, such as metformin, rapamycin, and D + Q, have also been explored in clinical trials, and the data show that metformin administration reduces the incidence of diabetes,397 cardiovascular events,398 frailty,399 and cognitive impairment,400 and improves putative longevity effectors in PBMCs.401 Although rapamycin shows exciting effects in preclinical studies, similar results have not been observed in clinical trials. In addition, despite the improved immune function in the elderly after administration of the mTOR inhibitor RAD001,402,403 several studies show that rapamycin does not improve cognitive function or physical performance404 and does not improve frailty.405 The first clinical study of senolytics demonstrated that D + Q improves 6-min walk distance, walking speed, chair raise ability, and short physical performance battery in idiopathic pulmonary fibrosis (IPF) patients.309 D + Q also reduces senescent cell burden in adipose tissue and skin and reduces circulating SASP in people with diabetic kidney disease.406
Collectively, diet intervention and exercise are still the most accepted strategies to intervene in aging and aging-related diseases, mainly because of their effectiveness and safety for humans. Advances in pharmacological interventions such as geroprotectors also show improvement in multiple aging-related conditions; however, the safety concerns and inconsistent results of these strategies demand further clinical evidence. Many other clinical trials related to epigenetic targets and the regulations of aging are still ongoing (Table 1). Together, these findings will provide more candidates for gerotherapeutics and pave the way for fighting aging in the future.
Table 1.
Ongoing clinical trials related to epigenetic targets and regulation of aging
| Interventions | Conditions/diseases | Trial nO. | Phase |
|---|---|---|---|
| Diet intervention | Epigenetic aging | NCT04962464 | 2 |
| Epigenetic aging | NCT05297097 | 2 | |
| Epigenetic aging | NCT05234203 | NA | |
| Aging | NCT05424042 | NA | |
| Exercise | Aging | NCT05232968 | NA |
| Aging; Alzheimer disease | NCT04299308 | NA | |
| Biological aging | NCT03440099 | NA | |
| Aging; Inflammatory response | NCT05042167 | NA | |
| NMN | Glucose metabolism disorders | NCT04571008 | NA |
| Hypertension | NCT04903210 | 4 | |
| Physical activity; Muscle recovery | NCT04664361 | NA | |
| NR | Aging | NCT03818802 | NA |
| Parkinson disease | NCT03568968 | NA | |
| Overweight and obesity; Aging; Type 2 diabetes | NCT04907110 | NA | |
| Sarcopenia; Nicotinamide adenine dinucleotide concentration; Muscle quality and NAD+ content | NCT04691986 | NA | |
| STACs | Vascular resistance; Hypertension | NCT01842399 | 1/2 |
| Healthy | NCT00996229 | 3 | |
| Metformin | Epigenetic aging; Immunosenescence | NCT04375657 | 2 |
| Aging; Insulin sensitivity; Chronic disease; Mitochondria; Insulin resistance | NCT04264897 | 3 | |
| Frailty; Sarcopenic obesity; Aging | NCT04221750 | 3 | |
| Frailty | NCT02570672 | 2 | |
| Mild cognitive impairment | NCT04098666 | 2/3 | |
| Insulin resistance; Obesity | NCT03733132 | 2 | |
| Rapamycin | Epigenetic clock of skin | NCT04608448 | 1 |
| Aging | NCT04488601 | 2 | |
| Aging | NCT04742777 | 2 | |
| Mild cognitive impairment; Alzheimer disease | NCT04629495 | 2 | |
| Mild cognitive impairment; Alzheimer disease | NCT04200911 | 1 | |
| D + Q | Epigenetic aging | NCT04946383 | 2 |
| Diabetic kidney disease | NCT02848131 | 1 | |
| Alzheimer disease | NCT04063124 | 1/2 | |
| Mild cognitive impairment; Alzheimer disease | NCT04785300 | 1/2 | |
| Alzheimer disease; Mild cognitive impairment | NCT04685590 | 2 | |
| Aging-related osteoporosis | NCT04313634 | 2 |
Conclusion and perspective
Studies in C. elegans, Drosophila, and mammals have unraveled the aging-related epigenetic changes in DNA, RNA, and histone modifications and alterations in the more advanced chromatin structure states. Correspondingly, these epigenetic changes have been identified as biomarkers or intervention targets of aging, such as the global decrease in genomic DNA methylation, the global loss of canonical histones, chromatin landscape remodeling caused by heterochromatin loss, and nuclear membrane protein changes in human and mouse tissues during aging. However, the same chromatin modifications (e.g., H3K14ac and H3K27me3) may play opposite roles in regulating aging and longevity across species and even across tissues within the same species, indicating that epigenetic changes need to be interpreted with their context. It is noteworthy that emerging technologies such as single-cell omics sequencing provide a higher resolution for dissecting epigenetic characteristics during aging, and provide new avenues for investigating the heterogeneity of aged cells. In addition, the spatiotemporal transcriptomic atlas across multiple mammalian tissues can provide more information on aging-related interactions between cells or tissues, which may facilitate the design of better and more precise therapeutics for aging and aging-related diseases.
Based on these epigenetic changes in cells during aging, a series of corresponding therapeutic strategies have been developed. Geroprotective drugs targeting longevity-related histone acetylation, including supplementation with NAD+ precursors, STACs have been tested in various species. Metformin, rapamycin, and other drugs have also shown positive effects in alleviating aging-related pathologies and regulating aging-related epigenetic changes in preclinical studies; however, the safety and efficacy of these drugs require more clinical investigation. Currently, a rational diet and exercise are considered the most effective and easiest way to delay aging, but drugs targeting key aging-related molecular and cellular changes are still promising and attractive clinical treatment strategies for intervening in aging and treating aging-related diseases.
Despite all the recent progress, it remains unclear how epigenetic changes interact with other factors, including the genetic background and even the microbiome, to regulate the aging process. It is also unclear how environmental factors, lifestyles, and physiological and psychological states contribute to epigenetic changes in the aging process. Furthermore, as aging is a continuous process that occurs over many years in humans, it would be necessary to track the epigenetic changes in this entire process for a better understanding of what and how epigenetic regulations contribute to each stage of aging.
Acknowledgements
The authors apologize for not citing all important studies in this review due to constraints on manuscript length. This work was supported by the National Key Research and Development Program of China (2020YFA0804000), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16010000), CAS Project for Young Scientists in Basic Research (YSBR-076, YSBR-012), the National Natural Science Foundation of China (81921006, 92149301, 92168201, 92049116, 32121001, 82100140, 31970597), the National Key Research and Development Program of China (2020YFA0803401 2019YFA0802202), the Program of the Beijing Natural Science Foundation (Z190019), the Tencent Foundation (2021–1045), K. C. Wong Education Foundation (GJTD-2019-08), Youth Innovation Promotion Association of CAS (E1CAZW0401), Science & Technology Innovation 2030 of The Ministry of Science and Technology of China (2022ZD0214200). All figures were created with Biorender.com.
Author contributions
G.-H.L., F.Z., J.R., and W.Z. designed and supervised the review, and reviewed the manuscript; K.W., H.L., Q.H., L.W., J.L. wrote the manuscript and drew pictures and tables; Z.Z. performed manuscript reviewing and editing; all authors have read and approved the article.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Kang Wang, Huicong Liu, Qinchao Hu, Lingna Wang, Jiaqing Liu
Contributor Information
Jie Ren, Email: renjie@big.ac.cn.
Fangfang Zhu, Email: zhuff@sjtu.edu.cn.
Guang-Hui Liu, Email: ghliu@ioz.ac.cn.
References
- 1.Catana CS, Atanasov AG, Berindan-Neagoe I. Natural products with anti-aging potential: affected targets and molecular mechanisms. Biotechnol. Adv. 2018;36:1649–1656. doi: 10.1016/j.biotechadv.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Brunet A, Berger SL. Epigenetics of aging and aging-related disease. J. Gerontol. A Biol. Sci. Med. Sci. 2014;69:S17–S20. doi: 10.1093/gerona/glu042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez-Otin C, et al. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson KE, et al. The queen’s gut refines with age: longevity phenotypes in a social insect model. Microbiome. 2018;6:108. doi: 10.1186/s40168-018-0489-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horvath S, et al. DNA methylation clocks tick in naked mole rats but queens age more slowly than nonbreeders. Nat. Aging. 2022;2:46–59. doi: 10.1038/s43587-021-00152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomczyk S, Fischer K, Austad S, Galliot B. Hydra, a powerful model for aging studies. Invertebr. Reprod. Dev. 2015;59:11–16. doi: 10.1080/07924259.2014.927805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montesanto A, et al. Epidemiological, genetic and epigenetic aspects of the research on healthy ageing and longevity. Immun. Ageing. 2012;9:6. doi: 10.1186/1742-4933-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubben N, Misteli T. Shared molecular and cellular mechanisms of premature ageing and ageing-associated diseases. Nat. Rev. Mol. Cell Biol. 2017;18:595–609. doi: 10.1038/nrm.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun, Y., Li, Q. & Kirkland, J. L. Targeting senescent cells for a healthier longevity: the roadmap for an era of global aging. Life Med. lnac030 10.1002/advs.202002611 (2022). [DOI] [PMC free article] [PubMed]
- 10.Cai, Y. et al. The landscape of aging. Sci. China Life Sci. 1–101 10.1007/s11427-022-2161-3 (2022). [DOI] [PMC free article] [PubMed]
- 11.Berdyshev GD, Korotaev GK, Boiarskikh GV, Vaniushin BF. [Nucleotide composition of DNA and RNA from somatic tissues of humpback and its changes during spawning] Biokhimiia. 1967;32:988–993. [PubMed] [Google Scholar]
- 12.Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220:1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- 13.Wilson VL, Smith RA, Ma S, Cutler RG. Genomic 5-methyldeoxycytidine decreases with age. J. Biol. Chem. 1987;262:9948–9951. doi: 10.1016/S0021-9258(18)61057-9. [DOI] [PubMed] [Google Scholar]
- 14.Ishimi Y, et al. Changes in chromatin structure during aging of human skin fibroblasts. Exp. Cell Res. 1987;169:458–467. doi: 10.1016/0014-4827(87)90206-0. [DOI] [PubMed] [Google Scholar]
- 15.Greer EL, et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q, et al. NSUN2-mediated m5C methylation and METTL3/METTL14-mediated m6A methylation cooperatively enhance p21 translation. J. Cell Biochem. 2017;118:2587–2598. doi: 10.1002/jcb.25957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung P, et al. Single-cell chromatin modification profiling reveals increased epigenetic variations with aging. Cell. 2018;173:1385–1397 e1314. doi: 10.1016/j.cell.2018.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou, X. et al. From monkey single-cell atlases into a broader biomedical perspective. Life Med. lnac028 10.1093/lifemedi/lnac028 (2022).
- 20.Aging Atlas C. Aging Atlas: a multi-omics database for aging biology. Nucleic Acids Res. 2021;49:D825–D830. doi: 10.1093/nar/gkaa894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCay CM, Crowell MF, Maynard LA. The effect of retarded growth upon the length of life span and upon the ultimate body size: one figure. J. Nutr. 1935;10:63–79. doi: 10.1093/jn/10.1.63. [DOI] [PubMed] [Google Scholar]
- 22.Acosta-Rodriguez, V. et al. Circadian alignment of early onset caloric restriction promotes longevity in male C57BL/6J mice. Science376, 1192–1202 (2022). [DOI] [PMC free article] [PubMed]
- 23.Kennedy BK, Austriaco NR, Jr, Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- 24.Howitz KT, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 25.Conboy IM, et al. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–764. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- 26.Davidson AJ, et al. Chronic jet-lag increases mortality in aged mice. Curr. Biol. 2006;16:R914–R916. doi: 10.1016/j.cub.2006.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lapasset L, et al. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev. 2011;25:2248–2253. doi: 10.1101/gad.173922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu GH, et al. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 2011;472:221–225. doi: 10.1038/nature09879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Y, et al. The Achilles’ heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell. 2015;14:644–658. doi: 10.1111/acel.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suda M, et al. Senolytic vaccination improves normal and pathological age-related phenotypes and increases lifespan in progeroid mice. Nat. Aging. 2021;1:1117–1126. doi: 10.1038/s43587-021-00151-2. [DOI] [PubMed] [Google Scholar]
- 31.Johnson AA, et al. The role of DNA methylation in aging, rejuvenation, and age-related disease. Rejuvenation Res. 2012;15:483–494. doi: 10.1089/rej.2012.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sailani MR, et al. Lifelong physical activity is associated with promoter hypomethylation of genes involved in metabolism, myogenesis, contractile properties and oxidative stress resistance in aged human skeletal muscle. Sci. Rep. 2019;9:3272. doi: 10.1038/s41598-018-37895-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maegawa S, et al. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20:332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adhikari S, Curtis PD. DNA methyltransferases and epigenetic regulation in bacteria. FEMS Microbiol. Rev. 2016;40:575–591. doi: 10.1093/femsre/fuw023. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, et al. Stella safeguards the oocyte methylome by preventing de novo methylation mediated by DNMT1. Nature. 2018;564:136–140. doi: 10.1038/s41586-018-0751-5. [DOI] [PubMed] [Google Scholar]
- 36.Baets J, et al. Defects of mutant DNMT1 are linked to a spectrum of neurological disorders. Brain. 2015;138:845–861. doi: 10.1093/brain/awv010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z-M, et al. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature. 2018;554:387–391. doi: 10.1038/nature25477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yagi M, et al. Identification of distinct loci for de novo DNA methylation by DNMT3A and DNMT3B during mammalian development. Nat. Commun. 2020;11:3199. doi: 10.1038/s41467-020-16989-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verma N, et al. TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells. Nat. Genet. 2018;50:83–95. doi: 10.1038/s41588-017-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bick AG, et al. Genetic interleukin 6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis. Circulation. 2020;141:124–131. doi: 10.1161/CIRCULATIONAHA.119.044362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heyn H, et al. Distinct DNA methylomes of newborns and centenarians. Proc. Natl Acad. Sci. USA. 2012;109:10522–10527. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, et al. Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation. Science. 2022;375:1053–1058. doi: 10.1126/science.abm0730. [DOI] [PubMed] [Google Scholar]
- 43.Seale, K. et al. Making sense of the ageing methylome. Nat. Rev. Genet. 23, 585–605 (2022). [DOI] [PubMed]
- 44.Ahuja N, et al. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;58:5489–5494. [PubMed] [Google Scholar]
- 45.Waki T, Tamura G, Sato M, Motoyama T. Age-related methylation of tumor suppressor and tumor-related genes: an analysis of autopsy samples. Oncogene. 2003;22:4128–4133. doi: 10.1038/sj.onc.1206651. [DOI] [PubMed] [Google Scholar]
- 46.Kim JY, Siegmund KD, Tavare S, Shibata D. Age-related human small intestine methylation: evidence for stem cell niches. BMC Med. 2005;3:10. doi: 10.1186/1741-7015-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu, A. T. et al. Universal DNA methylation age across mammalian tissues. Preprint at bioRxiv10.1101/2021.01.18.426733 (2021).
- 48.Talens RP, et al. Epigenetic variation during the adult lifespan: cross-sectional and longitudinal data on monozygotic twin pairs. Aging Cell. 2012;11:694–703. doi: 10.1111/j.1474-9726.2012.00835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slieker RC, et al. Age-related accrual of methylomic variability is linked to fundamental ageing mechanisms. Genome Biol. 2016;17:191. doi: 10.1186/s13059-016-1053-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lowe R, et al. Ageing-associated DNA methylation dynamics are a molecular readout of lifespan variation among mammalian species. Genome Biol. 2018;19:22. doi: 10.1186/s13059-018-1397-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bocklandt S, et al. Epigenetic predictor of age. PLoS ONE. 2011;6:e14821. doi: 10.1371/journal.pone.0014821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hannum G, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levine ME, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging. 2018;10:573–591. doi: 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 2018;19:371–384. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- 55.McCrory C, et al. GrimAge outperforms other epigenetic clocks in the prediction of age-related clinical phenotypes and all-cause mortality. J. Gerontol. A Biol. Sci. Med. Sci. 2021;76:741–749. doi: 10.1093/gerona/glaa286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murach KA, et al. Late-life exercise mitigates skeletal muscle epigenetic aging. Aging Cell. 2022;21:e13527. doi: 10.1111/acel.13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trapp A, Kerepesi C, Gladyshev VN. Profiling epigenetic age in single cells. Nat. Aging. 2021;1:1189–1201. doi: 10.1038/s43587-021-00134-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 59.Sidler C, Kovalchuk O, Kovalchuk I. Epigenetic regulation of cellular senescence and aging. Front. Genet. 2017;8:138. doi: 10.3389/fgene.2017.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 61.Bernstein BE, et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl Acad. Sci. USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cruz, C. et al. Tri-methylation of histone H3 lysine 4 facilitates gene expression in ageing cells. Elife. 7, 10.7554/elife.34081 (2018). [DOI] [PMC free article] [PubMed]
- 63.Ren X, et al. Maintenance of nucleolar homeostasis by CBX4 alleviates senescence and osteoarthritis. Cell Rep. 2019;26:3643–3656 e3647. doi: 10.1016/j.celrep.2019.02.088. [DOI] [PubMed] [Google Scholar]
- 64.Pu M, et al. Unique patterns of trimethylation of histone H3 lysine 4 are prone to changes during aging in Caenorhabditis elegans somatic cells. PLoS Genet. 2018;14:e1007466. doi: 10.1371/journal.pgen.1007466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bazopoulou D, et al. Developmental ROS individualizes organismal stress resistance and lifespan. Nature. 2019;576:301–305. doi: 10.1038/s41586-019-1814-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cao, Q. et al. Targeting histone K4 trimethylation for treatment of cognitive and synaptic deficits in mouse models of Alzheimer’s disease. Sci. Adv. 6, eabc8096 (2020). [DOI] [PMC free article] [PubMed]
- 67.Sun D, et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014;14:673–688. doi: 10.1016/j.stem.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adelman ER, et al. Aging human hematopoietic stem cells manifest profound epigenetic reprogramming of enhancers that may predispose to leukemia. Cancer Discov. 2019;9:1080–1101. doi: 10.1158/2159-8290.CD-18-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheung I, et al. Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc. Natl Acad. Sci. USA. 2010;107:8824–8829. doi: 10.1073/pnas.1001702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shumaker DK, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl Acad. Sci. USA. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baumgart M, et al. RNA-seq of the aging brain in the short-lived fish N. furzeri - conserved pathways and novel genes associated with neurogenesis. Aging Cell. 2014;13:965–974. doi: 10.1111/acel.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jin C, et al. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 2011;14:161–172. doi: 10.1016/j.cmet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 74.Guillermo ARR, et al. H3K27 modifiers regulate lifespan in C. elegans in a context-dependent manner. BMC Biol. 2021;19:59. doi: 10.1186/s12915-021-00984-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maures TJ, Greer EL, Hauswirth AG, Brunet A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell. 2011;10:980–990. doi: 10.1111/j.1474-9726.2011.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Merkwirth C, et al. Two conserved histone demethylases regulate mitochondrial stress-induced longevity. Cell. 2016;165:1209–1223. doi: 10.1016/j.cell.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sen P, et al. H3K36 methylation promotes longevity by enhancing transcriptional fidelity. Genes Dev. 2015;29:1362–1376. doi: 10.1101/gad.263707.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jeon HJ, et al. Effect of heterochromatin stability on intestinal stem cell aging in Drosophila. Mech. Ageing Dev. 2018;173:50–60. doi: 10.1016/j.mad.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 79.Li CL, et al. Region-specific H3K9me3 gain in aged somatic tissues in Caenorhabditis elegans. PLoS Genet. 2021;17:e1009432. doi: 10.1371/journal.pgen.1009432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wood JG, et al. Chromatin remodeling in the aging genome of Drosophila. Aging Cell. 2010;9:971–978. doi: 10.1111/j.1474-9726.2010.00624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang W, et al. Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science. 2015;348:1160–1163. doi: 10.1126/science.aaa1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu Z, et al. Differential stem cell aging kinetics in Hutchinson-Gilford progeria syndrome and Werner syndrome. Protein Cell. 2018;9:333–350. doi: 10.1007/s13238-018-0517-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu Z, et al. Large-scale chromatin reorganization reactivates placenta-specific genes that drive cellular aging. Dev. Cell. 2022;57:1347–1368 e1312. doi: 10.1016/j.devcel.2022.05.004. [DOI] [PubMed] [Google Scholar]
- 84.Djeghloul D, et al. Age-associated decrease of the histone methyltransferase SUV39H1 in HSC perturbs heterochromatin and B lymphoid differentiation. Stem Cell Rep. 2016;6:970–984. doi: 10.1016/j.stemcr.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li Y, et al. Vitamin C alleviates aging defects in a stem cell model for Werner syndrome. Protein Cell. 2016;7:478–488. doi: 10.1007/s13238-016-0278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shan, H. et al. Large-scale chemical screen identifies Gallic acid as a geroprotector for human stem cells. Protein Cell13, 532–539 (2021). [DOI] [PMC free article] [PubMed]
- 87.Li W, et al. Low-dose chloroquine treatment extends the lifespan of aged rats. Protein Cell. 2022;13:454–461. doi: 10.1007/s13238-021-00903-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang, Y. et al. Single-nucleus transcriptomics reveals a gatekeeper role for FOXP1 in primate cardiac aging. Protein Cell pwac038, 10.1093/procel/pwac038 (2022). [DOI] [PMC free article] [PubMed]
- 89.Liao B, et al. Nicotinamide mononucleotide supplementation enhances aerobic capacity in amateur runners: a randomized, double-blind study. J. Int. Soc. Sports Nutr. 2021;18:54. doi: 10.1186/s12970-021-00442-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 91.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl Acad. Sci. USA. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gong H, et al. Age-dependent tissue expression patterns of Sirt1 in senescence-accelerated mice. Mol. Med. Rep. 2014;10:3296–3302. doi: 10.3892/mmr.2014.2648. [DOI] [PubMed] [Google Scholar]
- 93.Cho SH, et al. SIRT1 deficiency in microglia contributes to cognitive decline in aging and neurodegeneration via epigenetic regulation of IL-1beta. J. Neurosci. 2015;35:807–818. doi: 10.1523/JNEUROSCI.2939-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang B, et al. Inhibition of histone acetyltransferase GCN5 extends lifespan in both yeast and human cell lines. Aging Cell. 2020;19:e13129. doi: 10.1111/acel.13129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Di Giorgio E, et al. HDAC4 degradation during senescence unleashes an epigenetic program driven by AP-1/p300 at selected enhancers and super-enhancers. Genome Biol. 2021;22:129. doi: 10.1186/s13059-021-02340-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sen P, et al. Histone acetyltransferase p300 induces de novo super-enhancers to drive cellular senescence. Mol. Cell. 2019;73:684–698 e688. doi: 10.1016/j.molcel.2019.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Barrett, L. N. & Westerheide, S. D. The CBP-1/p300 Lysine Acetyltransferase Regulates the Heat Shock Response in C. elegans. Front. Aging. 3, 861761 (2022). [DOI] [PMC free article] [PubMed]
- 98.Li TY, et al. The transcriptional coactivator CBP/p300 is an evolutionarily conserved node that promotes longevity in response to mitochondrial stress. Nat. Aging. 2021;1:165–178. doi: 10.1038/s43587-020-00025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tasdemir N, et al. BRD4 connects enhancer remodeling to senescence immune surveillance. Cancer Discov. 2016;6:612–629. doi: 10.1158/2159-8290.CD-16-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marzi SJ, et al. A histone acetylome-wide association study of Alzheimer’s disease identifies disease-associated H3K27ac differences in the entorhinal cortex. Nat. Neurosci. 2018;21:1618–1627. doi: 10.1038/s41593-018-0253-7. [DOI] [PubMed] [Google Scholar]
- 101.Klein HU, et al. Epigenome-wide study uncovers large-scale changes in histone acetylation driven by tau pathology in aging and Alzheimer’s human brains. Nat. Neurosci. 2019;22:37–46. doi: 10.1038/s41593-018-0291-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Price AJ, et al. Hdac3, Setdb1, and Kap1 mark H3K9me3/H3K14ac bivalent regions in young and aged liver. Aging Cell. 2020;19:e13092. doi: 10.1111/acel.13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Singh P, Thakur MK. Histone deacetylase 2 inhibition attenuates downregulation of hippocampal plasticity gene expression during aging. Mol. Neurobiol. 2018;55:2432–2442. doi: 10.1007/s12035-017-0490-x. [DOI] [PubMed] [Google Scholar]
- 104.Wang, W. et al. A genome-wide CRISPR-based screen identifies KAT7 as a driver of cellular senescence. Sci. Transl. Med. 13, eabd2655 (2021). [DOI] [PubMed]
- 105.Tasselli L, et al. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat. Struct. Mol. Biol. 2016;23:434–440. doi: 10.1038/nsmb.3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Feser J, et al. Elevated histone expression promotes life span extension. Mol. Cell. 2010;39:724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hachinohe M, Hanaoka F, Masumoto H. Hst3 and Hst4 histone deacetylases regulate replicative lifespan by preventing genome instability in Saccharomyces cerevisiae. Genes Cells. 2011;16:467–477. doi: 10.1111/j.1365-2443.2011.01493.x. [DOI] [PubMed] [Google Scholar]
- 108.Dang W, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nativio R, et al. Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nat. Neurosci. 2018;21:497–505. doi: 10.1038/s41593-018-0101-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Peleg S, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 111.Benito E, et al. HDAC inhibitor-dependent transcriptome and memory reinstatement in cognitive decline models. J. Clin. Invest. 2015;125:3572–3584. doi: 10.1172/JCI79942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yang L, et al. Ubiquitylome study identifies increased histone 2A ubiquitylation as an evolutionarily conserved aging biomarker. Nat. Commun. 2019;10:2191. doi: 10.1038/s41467-019-10136-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Joos JP, et al. Ectopic expression of S28A-mutated Histone H3 modulates longevity, stress resistance and cardiac function in Drosophila. Sci. Rep. 2018;8:2940. doi: 10.1038/s41598-018-21372-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yung PYK, et al. Histone H3 serine 28 is essential for efficient polycomb-mediated gene repression in Drosophila. Cell Rep. 2015;11:1437–1445. doi: 10.1016/j.celrep.2015.04.055. [DOI] [PubMed] [Google Scholar]
- 115.Luger K, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 116.Chandra T, et al. Global reorganization of the nuclear landscape in senescent cells. Cell Rep. 2015;10:471–483. doi: 10.1016/j.celrep.2014.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhao, D. & Chen, S. Failures at every level: breakdown of the epigenetic machinery of aging. Life Medicine, lnac016, 10.1093/lifemedi/lnac016 (2022).
- 118.Hu Z, et al. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev. 2014;28:396–408. doi: 10.1101/gad.233221.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.O’Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat. Struct. Mol. Biol. 2010;17:1218–1225. doi: 10.1038/nsmb.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen Y, et al. Remodeling of the H3 nucleosomal landscape during mouse aging. Transl. Med. Aging. 2020;4:22–31. doi: 10.1016/j.tma.2019.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Flex E, et al. Aberrant function of the C-terminal tail of HIST1H1E accelerates cellular senescence and causes premature aging. Am. J. Hum. Genet. 2019;105:493–508. doi: 10.1016/j.ajhg.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu B, Yip R, Zhou Z. Chromatin remodeling, DNA damage repair and aging. Curr. Genomics. 2012;13:533–547. doi: 10.2174/138920212803251373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhu D, et al. NuRD mediates mitochondrial stress-induced longevity via chromatin remodeling in response to acetyl-CoA level. Sci. Adv. 2020;6:eabb2529. doi: 10.1126/sciadv.abb2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhu, D., Li, X. & Tian, Y. Mitochondrial-to-nuclear communication in aging: an epigenetic perspective. Trends Biochem. Sci. 47, 645–659 (2022). [DOI] [PubMed]
- 125.Pouikli A, et al. Chromatin remodeling due to degradation of citrate carrier impairs osteogenesis of aged mesenchymal stem cells. Nat. Aging. 2021;1:810–825. doi: 10.1038/s43587-021-00105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kind J, et al. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 127.Stephens AD, et al. Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol. Biol. Cell. 2018;29:220–233. doi: 10.1091/mbc.E17-06-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fang Y, et al. Loss of Atg7 causes chaotic nucleosome assembly of mouse bone marrow CD11b(+)Ly6G(-) myeloid cells. Aging (Albany NY) 2020;12:25673–25683. doi: 10.18632/aging.104176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yu R, et al. Cellular response to moderate chromatin architectural defects promotes longevity. Sci. Adv. 2019;5:eaav1165. doi: 10.1126/sciadv.aav1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sun, L., Yu, R. & Dang, W. Chromatin architectural changes during cellular senescence and aging. Genes (Basel). 9, 211(2018). [DOI] [PMC free article] [PubMed]
- 131.Wu S, et al. BRM-SWI/SNF chromatin remodeling complex enables functional telomeres by promoting co-expression of TRF2 and TRF1. PLoS Genet. 2020;16:e1008799. doi: 10.1371/journal.pgen.1008799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Riedel CG, et al. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat. Cell Biol. 2013;15:491–501. doi: 10.1038/ncb2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hoffmann A, Spengler D. Chromatin remodeling complex NuRD in neurodevelopment and neurodevelopmental disorders. Front. Genet. 2019;10:682. doi: 10.3389/fgene.2019.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li X, et al. Chromatin remodeling factor BAZ1A regulates cellular senescence in both cancer and normal cells. Life Sci. 2019;229:225–232. doi: 10.1016/j.lfs.2019.05.023. [DOI] [PubMed] [Google Scholar]
- 135.Sun L, et al. Attenuation of epigenetic regulator SMARCA4 and ERK-ETS signaling suppresses aging-related dopaminergic degeneration. Aging Cell. 2020;19:e13210. doi: 10.1111/acel.13210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang C, et al. ATF3 drives senescence by reconstructing accessible chromatin profiles. Aging Cell. 2021;20:e13315. doi: 10.1111/acel.13315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Virk RKA, et al. Disordered chromatin packing regulates phenotypic plasticity. Sci. Adv. 2020;6:eaax6232. doi: 10.1126/sciadv.aax6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Diao Z, et al. SIRT3 consolidates heterochromatin and counteracts senescence. Nucleic Acids Res. 2021;49:4203–4219. doi: 10.1093/nar/gkab161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bi S, et al. SIRT7 antagonizes human stem cell aging as a heterochromatin stabilizer. Protein Cell. 2020;11:483–504. doi: 10.1007/s13238-020-00728-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hu H, et al. ZKSCAN3 counteracts cellular senescence by stabilizing heterochromatin. Nucleic Acids Res. 2020;48:6001–6018. doi: 10.1093/nar/gkaa425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liang C, et al. Stabilization of heterochromatin by CLOCK promotes stem cell rejuvenation and cartilage regeneration. Cell Res. 2021;31:187–205. doi: 10.1038/s41422-020-0385-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu, X. et al. Resurrection of human endogenous retroviruses during aging reinforces senescence. Preprint at bioRxiv10.1101/2021.02.22.432260(2021).
- 144.Zhao H, et al. Destabilizing heterochromatin by APOE mediates senescence. Nat. Aging. 2022;2:303–316. doi: 10.1038/s43587-022-00186-z. [DOI] [PubMed] [Google Scholar]
- 145.Liang C, et al. BMAL1 moonlighting as a gatekeeper for LINE1 repression and cellular senescence in primates. Nucleic Acids Res. 2022;50:3323–3347. doi: 10.1093/nar/gkac146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang, X. et al. The loss of heterochromatin is associated with multiscale three-dimensional genome reorganization and aberrant transcription during cellular senescence. Genome Res.31, 1121–1135 (2021). [DOI] [PMC free article] [PubMed]
- 147.Zhang W, Qu J, Liu GH, Belmonte JCI. The ageing epigenome and its rejuvenation. Nat. Rev. Mol. Cell Biol. 2020;21:137–150. doi: 10.1038/s41580-019-0204-5. [DOI] [PubMed] [Google Scholar]
- 148.Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hou Y, et al. CTCF mediates replicative senescence through POLD1. Front. Cell Dev. Biol. 2021;9:618586. doi: 10.3389/fcell.2021.618586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hirosue A, et al. Quantitative assessment of higher-order chromatin structure of the INK4/ARF locus in human senescent cells. Aging Cell. 2012;11:553–556. doi: 10.1111/j.1474-9726.2012.00809.x. [DOI] [PubMed] [Google Scholar]
- 152.Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science. 2018;361:1346–1349. doi: 10.1126/science.aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sun J, et al. The potential role of m6A RNA methylation in the aging process and aging-associated diseases. Front. Genet. 2022;13:869950. doi: 10.3389/fgene.2022.869950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Oerum S, Meynier V, Catala M, Tisne C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021;49:7239–7255. doi: 10.1093/nar/gkab378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chen X, et al. METTL3-mediated m(6)A modification of ATG7 regulates autophagy-GATA4 axis to promote cellular senescence and osteoarthritis progression. Ann. Rheum. Dis. 2022;81:87–99. doi: 10.1136/annrheumdis-2021-221091. [DOI] [PubMed] [Google Scholar]
- 156.Zhu H, et al. N6-methyladenosine induced miR-34a-5p promotes TNF-alpha-induced nucleus pulposus cell senescence by targeting SIRT1. Front. Cell Dev. Biol. 2021;9:642437. doi: 10.3389/fcell.2021.642437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Li G, et al. WTAP-mediated m(6)A modification of lncRNA NORAD promotes intervertebral disc degeneration. Nat. Commun. 2022;13:1469. doi: 10.1038/s41467-022-28990-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang J, et al. Lamin A safeguards the m(6) A methylase METTL14 nuclear speckle reservoir to prevent cellular senescence. Aging Cell. 2020;19:e13215. doi: 10.1111/acel.13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wu Z, et al. METTL3 counteracts premature aging via m6A-dependent stabilization of MIS12 mRNA. Nucleic Acids Res. 2020;48:11083–11096. doi: 10.1093/nar/gkaa816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lewinska A, Adamczyk-Grochala J, Kwasniewicz E, Wnuk M. Downregulation of methyltransferase Dnmt2 results in condition-dependent telomere shortening and senescence or apoptosis in mouse fibroblasts. J. Cell Physiol. 2017;232:3714–3726. doi: 10.1002/jcp.25848. [DOI] [PubMed] [Google Scholar]
- 161.Lewinska A, Adamczyk-Grochala J, Deregowska A, Wnuk M. Sulforaphane-induced cell cycle arrest and senescence are accompanied by DNA hypomethylation and changes in microRNA profile in breast cancer cells. Theranostics. 2017;7:3461–3477. doi: 10.7150/thno.20657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sun X, et al. Decreased expression of m(6)A demethylase FTO in ovarian aging. Arch. Gynecol. Obstet. 2021;303:1363–1369. doi: 10.1007/s00404-020-05895-7. [DOI] [PubMed] [Google Scholar]
- 163.Hirayama M, et al. FTO demethylates cyclin D1 mRNA and controls cell-cycle progression. Cell Rep. 2020;31:107464. doi: 10.1016/j.celrep.2020.03.028. [DOI] [PubMed] [Google Scholar]
- 164.Li G, et al. m6A hypomethylation of DNMT3B regulated by ALKBH5 promotes intervertebral disc degeneration via E4F1 deficiency. Clin. Transl. Med. 2022;12:e765. doi: 10.1002/ctm2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019;20:608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 166.Chelmicki T, et al. m(6)A RNA methylation regulates the fate of endogenous retroviruses. Nature. 2021;591:312–316. doi: 10.1038/s41586-020-03135-1. [DOI] [PubMed] [Google Scholar]
- 167.Tam OH, Ostrow LW, Gale Hammell M. Diseases of the nERVous system: retrotransposon activity in neurodegenerative disease. Mob. DNA. 2019;10:32. doi: 10.1186/s13100-019-0176-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Fan, T. et al. Senolytics cocktail dasatinib and quercetin alleviate human umbilical vein endothelial cell senescence via the TRAF6-MAPK-NF-kappaB Axis in a YTHDF2-dependent manner. Gerontology68, 920–934 (2022). [DOI] [PubMed]
- 169.Han M, et al. Abnormality of m6A mRNA methylation is involved in Alzheimer’s disease. Front. Neurosci. 2020;14:98. doi: 10.3389/fnins.2020.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Shafik AM, et al. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer’s disease. Genome Biol. 2021;22:17. doi: 10.1186/s13059-020-02249-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Xing J, et al. NSun2 promotes cell growth via elevating cyclin-dependent kinase 1 translation. Mol. Cell Biol. 2015;35:4043–4052. doi: 10.1128/MCB.00742-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tang H, et al. NSun2 delays replicative senescence by repressing p27 (KIP1) translation and elevating CDK1 translation. Aging (Albany NY) 2015;7:1143–1158. doi: 10.18632/aging.100860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhang X, et al. The tRNA methyltransferase NSun2 stabilizes p16INK(4) mRNA by methylating the 3’-untranslated region of p16. Nat. Commun. 2012;3:712. doi: 10.1038/ncomms1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cai X, et al. RNA methyltransferase NSUN2 promotes stress-induced HUVEC senescence. Oncotarget. 2016;7:19099–19110. doi: 10.18632/oncotarget.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tang, H. et al. HuR regulates telomerase activity through TERC methylation. Nat. Commun.9, 2213 (2018). [DOI] [PMC free article] [PubMed]
- 176.Slotkin W, Nishikura K. Adenosine-to-inosine RNA editing and human disease. Genome Med. 2013;5:105. doi: 10.1186/gm508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang, Y. et al. Comparative functional RNA editomes of neural differentiation from human PSCs. Life Med. lnac027, 10.1093/lifemedi/lnac027 (2022).
- 178.Gaisler-Salomon I, et al. Hippocampus-specific deficiency in RNA editing of GluA2 in Alzheimer’s disease. Neurobiol. Aging. 2014;35:1785–1791. doi: 10.1016/j.neurobiolaging.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 179.Stellos K, et al. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat. Med. 2016;22:1140–1150. doi: 10.1038/nm.4172. [DOI] [PubMed] [Google Scholar]
- 180.ElSharawy A, et al. Genome-wide miRNA signatures of human longevity. Aging Cell. 2012;11:607–616. doi: 10.1111/j.1474-9726.2012.00824.x. [DOI] [PubMed] [Google Scholar]
- 181.Leng, S. X. & Pawelec, G. Single-cell immune atlas for human aging and frailty. Life Med. lnac013, 10.1093/lifemedi/lnac013 (2022). [DOI] [PMC free article] [PubMed]
- 182.Lim, Y. W. et al. Genome-wide DNA hypomethylation and RNA:DNA hybrid accumulation in Aicardi-Goutières syndrome. Elife. 4, e08007 (2015). [DOI] [PMC free article] [PubMed]
- 183.Crossley MP, Bocek M, Cimprich KA. R-loops as cellular regulators and genomic threats. Mol. Cell. 2019;73:398–411. doi: 10.1016/j.molcel.2019.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kristensen LS, et al. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 185.Wang S, et al. Corrigendum to: Computational annotation of miRNA transcription start sites. Brief. Bioinform. 2021;22:609. doi: 10.1093/bib/bbaa024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang S, et al. Computational annotation of miRNA transcription start sites. Brief. Bioinform. 2021;22:380–392. doi: 10.1093/bib/bbz178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wang T, et al. Epigenetic aging signatures in mice livers are slowed by dwarfism, calorie restriction and rapamycin treatment. Genome Biol. 2017;18:57. doi: 10.1186/s13059-017-1186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Rivetti di Val Cervo P, et al. p63-microRNA feedback in keratinocyte senescence. Proc. Natl Acad. Sci. USA. 2012;109:1133–1138. doi: 10.1073/pnas.1112257109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Xiang X, et al. Cellular senescence in hepatocellular carcinoma induced by a long non-coding RNA-encoded peptide PINT87aa by blocking FOXM1-mediated. Theranostics. 2021;11:4929–4944. doi: 10.7150/thno.55672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zhang H, et al. LncRNA NEAT1 controls the lineage fates of BMSCs during skeletal aging by impairing mitochondrial function and pluripotency maintenance. Cell Death Differ. 2022;29:351–365. doi: 10.1038/s41418-021-00858-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yu F, et al. Long non-coding RNA APTR promotes the activation of hepatic stellate cells and the progression of liver fibrosis. Biochem. Biophys. Res. Commun. 2015;463:679–685. doi: 10.1016/j.bbrc.2015.05.124. [DOI] [PubMed] [Google Scholar]
- 192.Barutcu AR, et al. A TAD boundary is preserved upon deletion of the CTCF-rich Firre locus. Nat. Commun. 2018;9:1444. doi: 10.1038/s41467-018-03614-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yang, D., Yang, K. & Yang, M. in Aging and Aging-Related Diseases: Mechanisms and Interventions (ed Wang, Z.) 17–35 (Springer Singapore, 2018).
- 195.Inzulza-Tapia A, Alarcon M. Role of non-coding rna of human platelet in cardiovascular disease. Curr. Med. Chem. 2022;29:3420–3444. doi: 10.2174/0929867329666211230104955. [DOI] [PubMed] [Google Scholar]
- 196.Wang, K. et al. Genomic profiling of native R loops with a DNA-RNA hybrid recognition sensor. Sci. Adv. 7, eabe3516 (2021). [DOI] [PMC free article] [PubMed]
- 197.Yan P, et al. Genome-wide R-loop landscapes during cell differentiation and reprogramming. Cell Rep. 2020;32:107870. doi: 10.1016/j.celrep.2020.107870. [DOI] [PubMed] [Google Scholar]
- 198.Nojima, T. et al. Deregulated expression of mammalian lncRNA through loss of SPT6 induces r-loop formation, replication stress, and cellular senescence. Mol Cell. 72, 970–984.e7 (2018). [DOI] [PMC free article] [PubMed]
- 199.Yaku K, Okabe K, Nakagawa T. NAD metabolism: implications in aging and longevity. Ageing Res. Rev. 2018;47:1–17. doi: 10.1016/j.arr.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 200.Gu, X., Yao, H., Kwon, I. & Wang, G. Small-molecule activation of NAMPT as a potential neuroprotective strategy. Life Med. lnac012 10.1093/lifemedi/lnac012 (2022).
- 201.Shen, W. et al. Peroxisome proliferator-activated receptor γ coactivator 1α maintains NAD+ bioavailability protecting against steatohepatitis. Life Med. lnac031 10.1093/lifemedi/lnac031 (2022).
- 202.Fang EF, et al. NAD(+) augmentation restores mitophagy and limits accelerated aging in Werner syndrome. Nat. Commun. 2019;10:5284. doi: 10.1038/s41467-019-13172-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhang H, et al. NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science. 2016;352:1436–1443. doi: 10.1126/science.aaf2693. [DOI] [PubMed] [Google Scholar]
- 204.Romani M, et al. NAD(+) boosting reduces age-associated amyloidosis and restores mitochondrial homeostasis in muscle. Cell Rep. 2021;34:108660. doi: 10.1016/j.celrep.2020.108660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sasaki L, et al. Intracrine activity involving NAD-dependent circadian steroidogenic activity governs age-associated meibomian gland dysfunction. Nat. Aging. 2022;2:105–114. doi: 10.1038/s43587-021-00167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tarantini S, et al. Nicotinamide mononucleotide (NMN) supplementation rescues cerebromicrovascular endothelial function and neurovascular coupling responses and improves cognitive function in aged mice. Redox Biol. 2019;24:101192. doi: 10.1016/j.redox.2019.101192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Gong B, et al. Nicotinamide riboside restores cognition through an upregulation of proliferator-activated receptor-gamma coactivator 1alpha regulated beta-secretase 1 degradation and mitochondrial gene expression in Alzheimer’s mouse models. Neurobiol. Aging. 2013;34:1581–1588. doi: 10.1016/j.neurobiolaging.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hou Y, et al. NAD(+) supplementation normalizes key Alzheimer’s features and DNA damage responses in a new AD mouse model with introduced DNA repair deficiency. Proc. Natl Acad. Sci. USA. 2018;115:E1876–E1885. doi: 10.1073/pnas.1718819115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Mitchell SJ, et al. Nicotinamide improves aspects of healthspan, but not lifespan, in mice. Cell Metab. 2018;27:667–676 e664. doi: 10.1016/j.cmet.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Bonkowski MS, Sinclair DA. Slowing ageing by design: the rise of NAD(+) and sirtuin-activating compounds. Nat. Rev. Mol. Cell Biol. 2016;17:679–690. doi: 10.1038/nrm.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Baur JA, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kornicka K, Szlapka-Kosarzewska J, Smieszek A, Marycz K. 5-Azacytydine and resveratrol reverse senescence and ageing of adipose stem cells via modulation of mitochondrial dynamics and autophagy. J. Cell Mol. Med. 2019;23:237–259. doi: 10.1111/jcmm.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Zhou XL, et al. SIRT1 activator (SRT1720) improves the follicle reserve and prolongs the ovarian lifespan of diet-induced obesity in female mice via activating SIRT1 and suppressing mTOR signaling. J. Ovarian Res. 2014;7:97. doi: 10.1186/s13048-014-0097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mitchell SJ, et al. The SIRT1 activator SRT1720 extends lifespan and improves health of mice fed a standard diet. Cell Rep. 2014;6:836–843. doi: 10.1016/j.celrep.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Gano LB, et al. The SIRT1 activator SRT1720 reverses vascular endothelial dysfunction, excessive superoxide production, and inflammation with aging in mice. Am. J. Physiol. Heart Circ. Physiol. 2014;307:H1754–H1763. doi: 10.1152/ajpheart.00377.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Mercken EM, et al. SRT2104 extends survival of male mice on a standard diet and preserves bone and muscle mass. Aging Cell. 2014;13:787–796. doi: 10.1111/acel.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.McIntyre RL, et al. From molecular promise to preclinical results: HDAC inhibitors in the race for healthy aging drugs. EMBO Mol. Med. 2019;11:e9854. doi: 10.15252/emmm.201809854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Majora, M. et al. HDAC inhibition improves autophagic and lysosomal function to prevent loss of subcutaneous fat in a mouse model of Cockayne syndrome. Sci. Transl. Med. 10, eaam7510 (2018). [DOI] [PubMed]
- 219.Jeong, M. Y. et al. Histone deacetylase activity governs diastolic dysfunction through a nongenomic mechanism. Sci. Transl. Med. 10, eaao0144 (2018). [DOI] [PMC free article] [PubMed]
- 220.Walsh ME, et al. The histone deacetylase inhibitor butyrate improves metabolism and reduces muscle atrophy during aging. Aging Cell. 2015;14:957–970. doi: 10.1111/acel.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Fass DM, et al. Crebinostat: a novel cognitive enhancer that inhibits histone deacetylase activity and modulates chromatin-mediated neuroplasticity. Neuropharmacology. 2013;64:81–96. doi: 10.1016/j.neuropharm.2012.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Fontan-Lozano A, et al. Histone deacetylase inhibitors improve learning consolidation in young and in KA-induced-neurodegeneration and SAMP-8-mutant mice. Mol. Cell Neurosci. 2008;39:193–201. doi: 10.1016/j.mcn.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 223.Cabreiro F, et al. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell. 2013;153:228–239. doi: 10.1016/j.cell.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wang Y, et al. Metformin exerts antidepressant effects by regulated DNA hydroxymethylation. Epigenomics. 2019;11:655–667. doi: 10.2217/epi-2018-0187. [DOI] [PubMed] [Google Scholar]
- 225.Noren Hooten N, et al. Metformin-mediated increase in DICER1 regulates microRNA expression and cellular senescence. Aging Cell. 2016;15:572–581. doi: 10.1111/acel.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zhang S, et al. Metformin-induced microRNA-34a-3p downregulation alleviates senescence in human dental pulp stem cells by targeting CAB39 through the AMPK/mTOR signaling pathway. Stem Cells Int. 2021;2021:6616240. doi: 10.1155/2021/6616240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Martin-Montalvo A, et al. Metformin improves healthspan and lifespan in mice. Nat. Commun. 2013;4:2192. doi: 10.1038/ncomms3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Chen M, et al. Metformin protects lens epithelial cells against senescence in a naturally aged mouse model. Cell Death Discov. 2022;8:8. doi: 10.1038/s41420-021-00800-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Hong S, et al. A high fat, sugar, and salt Western diet induces motor-muscular and sensory dysfunctions and neurodegeneration in mice during aging: Ameliorative action of metformin. CNS Neurosci. Ther. 2021;27:1458–1471. doi: 10.1111/cns.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Chen Q, Thompson J, Hu Y, Lesnefsky EJ. Chronic metformin treatment decreases cardiac injury during ischemia-reperfusion by attenuating endoplasmic reticulum stress with improved mitochondrial function. Aging (Albany NY) 2021;13:7828–7845. doi: 10.18632/aging.202858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Anisimov VN, et al. If started early in life, metformin treatment increases life span and postpones tumors in female SHR mice. Aging (Albany NY) 2011;3:148–157. doi: 10.18632/aging.100273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Smith DL, Jr., et al. Metformin supplementation and life span in Fischer-344 rats. J. Gerontol. A Biol. Sci. Med. Sci. 2010;65:468–474. doi: 10.1093/gerona/glq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zhu X, et al. Effect of metformin on cardiac metabolism and longevity in aged female mice. Front. Cell Dev. Biol. 2020;8:626011. doi: 10.3389/fcell.2020.626011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Lu, M. et al. Metformin prevents dopaminergic neuron death in MPTP/P-induced mouse model of Parkinson’s disease via autophagy and mitochondrial ROS clearance. Int. J. Neuropsychopharmacol. 19, pyw047 (2016). [DOI] [PMC free article] [PubMed]
- 235.Yan Q, et al. Activation of AMPK/mTORC1-mediated autophagy by metformin reverses Clk1 deficiency-sensitized dopaminergic neuronal death. Mol. Pharm. 2017;92:640–652. doi: 10.1124/mol.117.109512. [DOI] [PubMed] [Google Scholar]
- 236.Wen H, et al. Metformin and cyanidin 3-O-galactoside from Aronia melanocarpa synergistically alleviate cognitive impairment in SAMP8 mice. Food Funct. 2021;12:10994–11008. doi: 10.1039/D1FO02122B. [DOI] [PubMed] [Google Scholar]
- 237.Kodali M, et al. Metformin treatment in late middle age improves cognitive function with alleviation of microglial activation and enhancement of autophagy in the hippocampus. Aging Cell. 2021;20:e13277. doi: 10.1111/acel.13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Lin Y, Dai X, Zhang J, Chen X. Metformin alleviates the depression-like behaviors of elderly apoE4 mice via improving glucose metabolism and mitochondrial biogenesis. Behav. Brain Res. 2022;423:113772. doi: 10.1016/j.bbr.2022.113772. [DOI] [PubMed] [Google Scholar]
- 239.Kim H, et al. Metformin inhibits chronic kidney disease-induced DNA damage and senescence of mesenchymal stem cells. Aging Cell. 2021;20:e13317. doi: 10.1111/acel.13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Yang SP, et al. Metformin ameliorates thymus degeneration of mice by regulating mitochondrial function. Int. Immunopharmacol. 2022;108:108744. doi: 10.1016/j.intimp.2022.108744. [DOI] [PubMed] [Google Scholar]
- 241.Cai H, et al. Metformin attenuates the D-galactose induced aging process via the UPR through the AMPK/ERK1/2 signaling pathways. Int. J. Mol. Med. 2020;45:715–730. doi: 10.3892/ijmm.2020.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Li Q, et al. Obesity and hyperinsulinemia drive adipocytes to activate a cell cycle program and senesce. Nat. Med. 2021;27:1941–1953. doi: 10.1038/s41591-021-01501-8. [DOI] [PubMed] [Google Scholar]
- 243.Chen YW, Harris RA, Hatahet Z, Chou KM. Ablation of XP-V gene causes adipose tissue senescence and metabolic abnormalities. Proc. Natl Acad. Sci. USA. 2015;112:E4556–E4564. doi: 10.1073/pnas.1506954112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zhu Y, et al. Metformin treatment of juvenile mice alters aging-related developmental and metabolic phenotypes. Mech. Ageing Dev. 2022;201:111597. doi: 10.1016/j.mad.2021.111597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Strong R, et al. Rapamycin-mediated mouse lifespan extension: Late-life dosage regimes with sex-specific effects. Aging Cell. 2020;19:e13269. doi: 10.1111/acel.13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Siegmund SE, et al. Low-dose rapamycin extends lifespan in a mouse model of mtDNA depletion syndrome. Hum. Mol. Genet. 2017;26:4588–4605. doi: 10.1093/hmg/ddx341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Bitto, A. et al. Transient rapamycin treatment can increase lifespan and healthspan in middle-aged mice. Elife. 5, e16351 (2016). [DOI] [PMC free article] [PubMed]
- 248.Yin, Z. et al. Dietary restriction and rapamycin affect brain aging in mice by attenuating age-related DNA methylation changes. Genes (Basel). 13, 699 (2022). [DOI] [PMC free article] [PubMed]
- 249.Quarles E, et al. Rapamycin persistently improves cardiac function in aged, male and female mice, even following cessation of treatment. Aging Cell. 2020;19:e13086. doi: 10.1111/acel.13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Lesniewski LA, et al. Dietary rapamycin supplementation reverses age-related vascular dysfunction and oxidative stress, while modulating nutrient-sensing, cell cycle, and senescence pathways. Aging Cell. 2017;16:17–26. doi: 10.1111/acel.12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Van Skike CE, et al. mTOR attenuation with rapamycin reverses neurovascular uncoupling and memory deficits in mice modeling Alzheimer’s disease. J. Neurosci. 2021;41:4305–4320. doi: 10.1523/JNEUROSCI.2144-20.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Towner RA, et al. Rapamycin restores brain vasculature, metabolism, and blood-brain barrier in an inflammaging model. Geroscience. 2021;43:563–578. doi: 10.1007/s11357-021-00363-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Lei H, Wang J, Ladiges W, Jiang Z. Short-term oral rapamycin prevents age-related learning impairment in mice. Aging Pathobiol. Ther. 2020;2:166–167. doi: 10.31491/APT.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Singh AK, et al. Rapamycin confers neuroprotection against aging-induced oxidative stress, mitochondrial dysfunction, and neurodegeneration in old rats through activation of autophagy. Rejuvenation Res. 2019;22:60–70. doi: 10.1089/rej.2018.2070. [DOI] [PubMed] [Google Scholar]
- 255.Ham DJ, et al. Distinct and additive effects of calorie restriction and rapamycin in aging skeletal muscle. Nat. Commun. 2022;13:2025. doi: 10.1038/s41467-022-29714-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Kawakami Y, et al. Rapamycin rescues age-related changes in muscle-derived stem/progenitor cells from progeroid mice. Mol. Ther. Methods Clin. Dev. 2019;14:64–76. doi: 10.1016/j.omtm.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Garcia DN, et al. Effect of caloric restriction and rapamycin on ovarian aging in mice. Geroscience. 2019;41:395–408. doi: 10.1007/s11357-019-00087-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Dou X, et al. Short-term rapamycin treatment increases ovarian lifespan in young and middle-aged female mice. Aging Cell. 2017;16:825–836. doi: 10.1111/acel.12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Altschuler RA, et al. Rapamycin added to diet in late mid-life delays age-related hearing loss in UMHET4 mice. Front. Cell Neurosci. 2021;15:658972. doi: 10.3389/fncel.2021.658972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Altschuler RA, et al. Rapamycin but not acarbose decreases age-related loss of outer hair cells in the mouse Cochlea. Hear Res. 2018;370:11–15. doi: 10.1016/j.heares.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.An, J. Y. et al. Rapamycin rejuvenates oral health in aging mice. Elife. 9, e54318 (2020). [DOI] [PMC free article] [PubMed]
- 262.An JY, et al. Rapamycin treatment attenuates age-associated periodontitis in mice. Geroscience. 2017;39:457–463. doi: 10.1007/s11357-017-9994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Deblon N, et al. Chronic mTOR inhibition by rapamycin induces muscle insulin resistance despite weight loss in rats. Br. J. Pharm. 2012;165:2325–2340. doi: 10.1111/j.1476-5381.2011.01716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Kim Y, Lee JS, Joo YH. Rapamycin increases the incidence of neuropsychiatric illness in kidney transplant patients through the suppression of neural stem cells. Transl. Psychiatry. 2020;10:156. doi: 10.1038/s41398-020-0838-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Liu J, et al. Delay in oocyte aging in mice by the antioxidant N-acetyl-L-cysteine (NAC) Hum. Reprod. 2012;27:1411–1420. doi: 10.1093/humrep/des019. [DOI] [PubMed] [Google Scholar]
- 266.Garg G, Singh S, Singh AK, Rizvi SI. N-acetyl-l-cysteine attenuates oxidative damage and neurodegeneration in rat brain during aging. Can. J. Physiol. Pharm. 2018;96:1189–1196. doi: 10.1139/cjpp-2018-0209. [DOI] [PubMed] [Google Scholar]
- 267.Flurkey K, Astle CM, Harrison DE. Life extension by diet restriction and N-acetyl-L-cysteine in genetically heterogeneous mice. J. Gerontol. A Biol. Sci. Med. Sci. 2010;65:1275–1284. doi: 10.1093/gerona/glq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Costa M, et al. N-acetylcysteine protects memory decline induced by streptozotocin in mice. Chem. Biol. Interact. 2016;253:10–17. doi: 10.1016/j.cbi.2016.04.026. [DOI] [PubMed] [Google Scholar]
- 269.More J, et al. N-acetylcysteine prevents the spatial memory deficits and the redox-dependent RyR2 decrease displayed by an Alzheimer’s disease rat model. Front. Aging Neurosci. 2018;10:399. doi: 10.3389/fnagi.2018.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zhu QY, et al. N-acetyl cysteine ameliorates aortic fibrosis by promoting M2 macrophage polarization in aging mice. Redox Rep. 2021;26:170–175. doi: 10.1080/13510002.2021.1976568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Meryk, A. et al. Antioxidants N-acetylcysteine and vitamin C improve T cell commitment to memory and long-term maintenance of immunological memory in old mice. Antioxidants (Basel). 9, 1152 (2020). [DOI] [PMC free article] [PubMed]
- 272.Kawaguchi K, Hashimoto M, Sugimoto M. An antioxidant suppressed lung cellular senescence and enhanced pulmonary function in aged mice. Biochem. Biophys. Res. Commun. 2021;541:43–49. doi: 10.1016/j.bbrc.2020.12.112. [DOI] [PubMed] [Google Scholar]
- 273.Zhou X, et al. Suppression effect of N-acetylcysteine on bone loss in ovariectomized mice. Am. J. Transl. Res. 2020;12:731–742. [PMC free article] [PubMed] [Google Scholar]
- 274.Marie A, et al. N-acetylcysteine treatment reduces age-related hearing loss and memory impairment in the senescence-accelerated prone 8 (SAMP8) mouse model. Aging Dis. 2018;9:664–673. doi: 10.14336/AD.2017.0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Miller, R. A. et al. Canagliflozin extends life span in genetically heterogeneous male but not female mice. JCI Insight. 5, e140019 (2020). [DOI] [PMC free article] [PubMed]
- 276.Harrison DE, et al. Acarbose improves health and lifespan in aging HET3 mice. Aging Cell. 2019;18:e12898. doi: 10.1111/acel.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Strong R, et al. Longer lifespan in male mice treated with a weakly estrogenic agonist, an antioxidant, an α‐glucosidase inhibitor or a Nrf2‐inducer. Aging Cell. 2016;15:872–884. doi: 10.1111/acel.12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Harrison DE, et al. Acarbose, 17-alpha-estradiol, and nordihydroguaiaretic acid extend mouse lifespan preferentially in males. Aging Cell. 2014;13:273–282. doi: 10.1111/acel.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Geng L, et al. Chemical screen identifies a geroprotective role of quercetin in premature aging. Protein Cell. 2019;10:417–435. doi: 10.1007/s13238-018-0567-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Geng L, et al. Low-dose quercetin positively regulates mouse healthspan. Protein Cell. 2019;10:770–775. doi: 10.1007/s13238-019-0646-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Ma S, et al. Single-cell transcriptomic atlas of primate cardiopulmonary aging. Cell Res. 2021;31:415–432. doi: 10.1038/s41422-020-00412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Zakaria A, Hamdi N, Abdel-Kader RM. Methylene blue improves brain mitochondrial ABAD functions and decreases abeta in a neuroinflammatory Alzheimer’s disease mouse model. Mol. Neurobiol. 2016;53:1220–1228. doi: 10.1007/s12035-014-9088-8. [DOI] [PubMed] [Google Scholar]
- 283.Santos EL, et al. Long term treatment with ACE inhibitor enalapril decreases body weight gain and increases life span in rats. Biochem. Pharm. 2009;78:951–958. doi: 10.1016/j.bcp.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 284.Graus-Nunes F, et al. Differential effects of angiotensin receptor blockers on pancreatic islet remodelling and glucose homeostasis in diet-induced obese mice. Mol. Cell Endocrinol. 2017;439:54–64. doi: 10.1016/j.mce.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 285.Doeppner, T. R. et al. Long-term treatment with chloroquine increases lifespan in middle-aged male mice possibly via autophagy modulation, proteasome inhibition and glycogen metabolism. Aging (Albany NY). 14, 4195–4210 (2022). [DOI] [PMC free article] [PubMed]
- 286.Strong R, et al. Nordihydroguaiaretic acid and aspirin increase lifespan of genetically heterogeneous male mice. Aging Cell. 2008;7:641–650. doi: 10.1111/j.1474-9726.2008.00414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Liu Z, et al. Cross-species metabolomic analysis identifies uridine as a potent regeneration promoting factor. Cell Discov. 2022;8:6. doi: 10.1038/s41421-021-00361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 289.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 290.Koche RP, et al. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Lu Y, et al. Reprogramming to recover youthful epigenetic information and restore vision. Nature. 2020;588:124–129. doi: 10.1038/s41586-020-2975-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Abad M, et al. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature. 2013;502:340–345. doi: 10.1038/nature12586. [DOI] [PubMed] [Google Scholar]
- 293.Manukyan M, Singh PB. Epigenome rejuvenation: HP1β mobility as a measure of pluripotent and senescent chromatin ground states. Sci. Rep. 2014;4:4789. doi: 10.1038/srep04789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Olova N, Simpson DJ, Marioni RE, Chandra T. Partial reprogramming induces a steady decline in epigenetic age before loss of somatic identity. Aging Cell. 2019;18:e12877. doi: 10.1111/acel.12877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Sarkar TJ, et al. Transient non-integrative expression of nuclear reprogramming factors promotes multifaceted amelioration of aging in human cells. Nat. Commun. 2020;11:1545. doi: 10.1038/s41467-020-15174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Gill, D. et al. Multi-omic rejuvenation of human cells by maturation phase transient reprogramming. Elife. 11, e71624 (2022). [DOI] [PMC free article] [PubMed]
- 297.Ocampo A, et al. In vivo amelioration of age-associated hallmarks by partial reprogramming. Cell. 2016;167:1719–1733.e1712. doi: 10.1016/j.cell.2016.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Alle, Q. et al. A single short reprogramming early in life improves fitness and increases lifespan in old age. Preprint at bioRxiv10.1111/acel.13714 (2021).
- 299.Chondronasiou D, et al. Multi-omic rejuvenation of naturally aged tissues by a single cycle of transient reprogramming. Aging Cell. 2022;21:e13578. doi: 10.1111/acel.13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Roos CM, et al. Chronic senolytic treatment alleviates established vasomotor dysfunction in aged or atherosclerotic mice. Aging Cell. 2016;15:973–977. doi: 10.1111/acel.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Lewis-McDougall FC, et al. Aged-senescent cells contribute to impaired heart regeneration. Aging Cell. 2019;18:e12931. doi: 10.1111/acel.12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Zhou Y, et al. Senolytics improve bone forming potential of bone marrow mesenchymal stem cells from aged mice. NPJ Regen. Med. 2021;6:34. doi: 10.1038/s41536-021-00145-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Farr JN, et al. Targeting cellular senescence prevents age-related bone loss in mice. Nat. Med. 2017;23:1072–1079. doi: 10.1038/nm.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Novais EJ, et al. Long-term treatment with senolytic drugs Dasatinib and Quercetin ameliorates age-dependent intervertebral disc degeneration in mice. Nat. Commun. 2021;12:5213. doi: 10.1038/s41467-021-25453-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Zhang P, et al. Senolytic therapy alleviates Abeta-associated oligodendrocyte progenitor cell senescence and cognitive deficits in an Alzheimer’s disease model. Nat. Neurosci. 2019;22:719–728. doi: 10.1038/s41593-019-0372-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Ogrodnik M, et al. Obesity-induced cellular senescence drives anxiety and impairs neurogenesis. Cell Metab. 2019;29:1061–1077 e1068. doi: 10.1016/j.cmet.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Lee G, et al. SREBP1c-PARP1 axis tunes anti-senescence activity of adipocytes and ameliorates metabolic imbalance in obesity. Cell Metab. 2022;34:702–718 e705. doi: 10.1016/j.cmet.2022.03.010. [DOI] [PubMed] [Google Scholar]
- 308.Wang L, et al. Targeting p21(Cip1) highly expressing cells in adipose tissue alleviates insulin resistance in obesity. Cell Metab. 2022;34:75–89 e78. doi: 10.1016/j.cmet.2021.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Justice JN, et al. Senolytics in idiopathic pulmonary fibrosis: Results from a first-in-human, open-label, pilot study. EBioMedicine. 2019;40:554–563. doi: 10.1016/j.ebiom.2018.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Schafer MJ, et al. Cellular senescence mediates fibrotic pulmonary disease. Nat. Commun. 2017;8:14532. doi: 10.1038/ncomms14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Dungan CM, et al. Deletion of SA beta-Gal+ cells using senolytics improves muscle regeneration in old mice. Aging Cell. 2022;21:e13528. doi: 10.1111/acel.13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Krzystyniak A, et al. Combination of dasatinib and quercetin improves cognitive abilities in aged male Wistar rats, alleviates inflammation and changes hippocampal synaptic plasticity and histone H3 methylation profile. Aging (Albany NY) 2022;14:572–595. doi: 10.18632/aging.203835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Spinelli R, et al. ZMAT3 hypomethylation contributes to early senescence of preadipocytes from healthy first-degree relatives of type 2 diabetics. Aging Cell. 2022;21:e13557. doi: 10.1111/acel.13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Kim H, et al. Inhibition of matrix metalloproteinase expression by selective clearing of senescent dermal fibroblasts attenuates ultraviolet-induced photoaging. Biomed. Pharmacother. 2022;150:113034. doi: 10.1016/j.biopha.2022.113034. [DOI] [PubMed] [Google Scholar]
- 315.Chang J, et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 2016;22:78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Baar MP, et al. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell. 2017;169:132–147 e116. doi: 10.1016/j.cell.2017.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Fuhrmann-Stroissnigg, H. et al. Identification of HSP90 inhibitors as a novel class of senolytics. Nat. Commun. 8, 422 (2017). [DOI] [PMC free article] [PubMed]
- 318.Triana-Martinez F, et al. Identification and characterization of cardiac glycosides as senolytic compounds. Nat. Commun. 2019;10:4731. doi: 10.1038/s41467-019-12888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Weindruch R, Walford RL, Fligiel S, Guthrie D. The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. J. Nutr. 1986;116:641–654. doi: 10.1093/jn/116.4.641. [DOI] [PubMed] [Google Scholar]
- 320.Hahn O, et al. Dietary restriction protects from age-associated DNA methylation and induces epigenetic reprogramming of lipid metabolism. Genome Biol. 2017;18:56. doi: 10.1186/s13059-017-1187-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Mitchell SJ, et al. Effects of sex, strain, and energy intake on hallmarks of aging in mice. Cell Metab. 2016;23:1093–1112. doi: 10.1016/j.cmet.2016.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Hepple RT, Baker DJ, Kaczor JJ, Krause DJ. Long-term caloric restriction abrogates the age-related decline in skeletal muscle aerobic function. FASEB J. 2005;19:1320–1322. doi: 10.1096/fj.04-3535fje. [DOI] [PubMed] [Google Scholar]
- 323.Rippe C, et al. Short-term calorie restriction reverses vascular endothelial dysfunction in old mice by increasing nitric oxide and reducing oxidative stress. Aging Cell. 2010;9:304–312. doi: 10.1111/j.1474-9726.2010.00557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Valdez G, et al. Attenuation of age-related changes in mouse neuromuscular synapses by caloric restriction and exercise. Proc. Natl Acad. Sci. USA. 2010;107:14863–14868. doi: 10.1073/pnas.1002220107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Cerletti M, et al. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell Stem Cell. 2012;10:515–519. doi: 10.1016/j.stem.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Cohen HY, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 327.Ma S, et al. Caloric restriction reprograms the single-cell transcriptional landscape of Rattus Norvegicus aging. Cell. 2020;180:984–1001 e1022. doi: 10.1016/j.cell.2020.02.008. [DOI] [PubMed] [Google Scholar]
- 328.Colman RJ, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Maegawa S, et al. Caloric restriction delays age-related methylation drift. Nat. Commun. 2017;8:539. doi: 10.1038/s41467-017-00607-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Rochon J, et al. Design and conduct of the CALERIE study: comprehensive assessment of the long-term effects of reducing intake of energy. J. Gerontol. A Biol. Sci. Med. Sci. 2011;66:97–108. doi: 10.1093/gerona/glq168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Rickman AD, et al. The CALERIE Study: design and methods of an innovative 25% caloric restriction intervention. Contemp. Clin. Trials. 2011;32:874–881. doi: 10.1016/j.cct.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Belsky DW, et al. Change in the rate of biological aging in response to caloric restriction: CALERIE biobank analysis. J. Gerontol. A Biol. Sci. Med. Sci. 2017;73:4–10. doi: 10.1093/gerona/glx096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Dorling JL, et al. Effect of 2 years of calorie restriction on liver biomarkers: results from the CALERIE phase 2 randomized controlled trial. Eur. J. Nutr. 2021;60:1633–1643. doi: 10.1007/s00394-020-02361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Kraus WE, et al. 2 years of calorie restriction and cardiometabolic risk (CALERIE): exploratory outcomes of a multicentre, phase 2, randomised controlled trial. Lancet Diabetes Endocrinol. 2019;7:673–683. doi: 10.1016/S2213-8587(19)30151-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Martin CK, et al. Effect of calorie restriction on mood, quality of life, sleep, and sexual function in healthy nonobese adults: The CALERIE 2 randomized clinical trial. JAMA Intern. Med. 2016;176:743–752. doi: 10.1001/jamainternmed.2016.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Redman LM, et al. Metabolic slowing and reduced oxidative damage with sustained caloric restriction support the rate of living and oxidative damage theories of aging. Cell Metab. 2018;27:805–815.e804. doi: 10.1016/j.cmet.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Acosta-Rodríguez VA, Rijo-Ferreira F, Green CB, Takahashi JS. Importance of circadian timing for aging and longevity. Nat. Commun. 2021;12:2862. doi: 10.1038/s41467-021-22922-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Solanas G, et al. Aged stem cells reprogram their daily rhythmic functions to adapt to stress. Cell. 2017;170:678–692.e620. doi: 10.1016/j.cell.2017.07.035. [DOI] [PubMed] [Google Scholar]
- 339.Hor CN, et al. Sleep-wake-driven and circadian contributions to daily rhythms in gene expression and chromatin accessibility in the murine cortex. Proc. Natl Acad. Sci. USA. 2019;116:25773–25783. doi: 10.1073/pnas.1910590116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Libert S, et al. Deviation of innate circadian period from 24 h reduces longevity in mice. Aging Cell. 2012;11:794–800. doi: 10.1111/j.1474-9726.2012.00846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Morris CJ, Purvis TE, Hu K, Scheer FA. Circadian misalignment increases cardiovascular disease risk factors in humans. Proc. Natl Acad. Sci. USA. 2016;113:E1402–E1411. doi: 10.1073/pnas.1516953113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Barrès R, et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012;15:405–411. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 343.Nitert MD, et al. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes. 2012;61:3322–3332. doi: 10.2337/db11-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Gomez-Pinilla F, et al. Exercise impacts brain-derived neurotrophic factor plasticity by engaging mechanisms of epigenetic regulation. Eur. J. Neurosci. 2011;33:383–390. doi: 10.1111/j.1460-9568.2010.07508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Egan B, et al. Exercise intensity-dependent regulation of peroxisome proliferator-activated receptor coactivator-1 mRNA abundance is associated with differential activation of upstream signalling kinases in human skeletal muscle. J. Physiol. 2010;588:1779–1790. doi: 10.1113/jphysiol.2010.188011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Ogasawara R, et al. MicroRNA expression profiling in skeletal muscle reveals different regulatory patterns in high and low responders to resistance training. Physiol. Genomics. 2016;48:320–324. doi: 10.1152/physiolgenomics.00124.2015. [DOI] [PubMed] [Google Scholar]
- 347.Fyfe JJ, et al. Concurrent exercise incorporating high-intensity interval or continuous training modulates mTORC1 signaling and microRNA expression in human skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016;310:R1297–R1311. doi: 10.1152/ajpregu.00479.2015. [DOI] [PubMed] [Google Scholar]
- 348.van Praag H, Shubert T, Zhao C, Gage FH. Exercise enhances learning and hippocampal neurogenesis in aged mice. J. Neurosci. 2005;25:8680–8685. doi: 10.1523/JNEUROSCI.1731-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Rubenstein AB, et al. Skeletal muscle transcriptome response to a bout of endurance exercise in physically active and sedentary older adults. Am. J. Physiol. Endocrinol. Metab. 2022;322:E260–e277. doi: 10.1152/ajpendo.00378.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Ruple BA, et al. Resistance training rejuvenates the mitochondrial methylome in aged human skeletal muscle. FASEB J. 2021;35:e21864. doi: 10.1096/fj.202100873RR. [DOI] [PubMed] [Google Scholar]
- 351.Blocquiaux S, et al. Recurrent training rejuvenates and enhances transcriptome and methylome responses in young and older human muscle. JCSM Rapid Commun. 2022;5:10–32. doi: 10.1002/rco2.52. [DOI] [Google Scholar]
- 352.Dorling JL, et al. Effects of caloric restriction on human physiological, psychological, and behavioral outcomes: highlights from CALERIE phase 2. Nutr. Rev. 2021;79:98–113. doi: 10.1093/nutrit/nuaa085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Ravussin E, et al. A 2-year randomized controlled trial of human caloric restriction: feasibility and effects on predictors of health span and longevity. J. Gerontol. A Biol. Sci. Med. Sci. 2015;70:1097–1104. doi: 10.1093/gerona/glv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Schroder JD, et al. Effects of time-restricted feeding in weight loss, metabolic syndrome and cardiovascular risk in obese women. J. Transl. Med. 2021;19:3. doi: 10.1186/s12967-020-02687-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Wilkinson MJ, et al. Ten-hour time-restricted eating reduces weight, blood pressure, and atherogenic lipids in patients with metabolic syndrome. Cell Metab. 2020;31:92–104 e105. doi: 10.1016/j.cmet.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Sutton EF, et al. Early time-restricted feeding improves insulin sensitivity, blood pressure, and oxidative stress even without weight loss in men with prediabetes. Cell Metab. 2018;27:1212–1221 e1213. doi: 10.1016/j.cmet.2018.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Rong S, et al. Association of skipping breakfast with cardiovascular and all-cause mortality. J. Am. Coll. Cardiol. 2019;73:2025–2032. doi: 10.1016/j.jacc.2019.01.065. [DOI] [PubMed] [Google Scholar]
- 358.Longo VD, Anderson RM. Nutrition, longevity and disease: from molecular mechanisms to interventions. Cell. 2022;185:1455–1470. doi: 10.1016/j.cell.2022.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Wei, M. et al. Fasting-mimicking diet and markers/risk factors for aging, diabetes, cancer, and cardiovascular disease. Sci. Transl. Med. 9, eaai8700 (2017). [DOI] [PMC free article] [PubMed]
- 360.Mansur AP, et al. Serum concentrations and gene expression of sirtuin 1 in healthy and slightly overweight subjects after caloric restriction or resveratrol supplementation: a randomized trial. Int. J. Cardiol. 2017;227:788–794. doi: 10.1016/j.ijcard.2016.10.058. [DOI] [PubMed] [Google Scholar]
- 361.Lilja, S. et al. Five days periodic fasting elevates levels of longevity related christensenella and sirtuin expression in humans. Int. J. Mol. Sci. 22, 2331 (2021). [DOI] [PMC free article] [PubMed]
- 362.Fitzgerald KN, et al. Potential reversal of epigenetic age using a diet and lifestyle intervention: a pilot randomized clinical trial. Aging (Albany NY) 2021;13:9419–9432. doi: 10.18632/aging.202913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Fiorito G, et al. DNA methylation-based biomarkers of aging were slowed down in a two-year diet and physical activity intervention trial: the DAMA study. Aging Cell. 2021;20:e13439. doi: 10.1111/acel.13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Lee DC, et al. Running as a key lifestyle medicine for longevity. Prog. Cardiovasc. Dis. 2017;60:45–55. doi: 10.1016/j.pcad.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 365.Chakravarty EF, Hubert HB, Lingala VB, Fries JF. Reduced disability and mortality among aging runners: a 21-year longitudinal study. Arch. Intern. Med. 2008;168:1638–1646. doi: 10.1001/archinte.168.15.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Moore SC, et al. Leisure time physical activity of moderate to vigorous intensity and mortality: a large pooled cohort analysis. PLoS Med. 2012;9:e1001335. doi: 10.1371/journal.pmed.1001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Hieda M, et al. One-year committed exercise training reverses abnormal left ventricular myocardial stiffness in patients with stage b heart failure with preserved ejection fraction. Circulation. 2021;144:934–946. doi: 10.1161/CIRCULATIONAHA.121.054117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Kirk B, et al. Leucine-enriched whey protein supplementation, resistance-based exercise, and cardiometabolic health in older adults: a randomized controlled trial. J. Cachexia Sarcopenia Muscle. 2021;12:2022–2033. doi: 10.1002/jcsm.12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Howden EJ, et al. Reversing the cardiac effects of sedentary aging in middle age-a Randomized controlled trial: implications for heart failure prevention. Circulation. 2018;137:1549–1560. doi: 10.1161/CIRCULATIONAHA.117.030617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Sandri M, et al. Age-related effects of exercise training on diastolic function in heart failure with reduced ejection fraction: the Leipzig Exercise Intervention in Chronic Heart Failure and Aging (LEICA) Diastolic Dysfunction Study. Eur. Heart J. 2012;33:1758–1768. doi: 10.1093/eurheartj/ehr469. [DOI] [PubMed] [Google Scholar]
- 371.Kitzman DW, et al. Effect of endurance exercise training on endothelial function and arterial stiffness in older patients with heart failure and preserved ejection fraction: a randomized, controlled, single-blind trial. J. Am. Coll. Cardiol. 2013;62:584–592. doi: 10.1016/j.jacc.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Ngandu T, et al. A 2 year multidomain intervention of diet, exercise, cognitive training, and vascular risk monitoring versus control to prevent cognitive decline in at-risk elderly people (FINGER): a randomised controlled trial. Lancet. 2015;385:2255–2263. doi: 10.1016/S0140-6736(15)60461-5. [DOI] [PubMed] [Google Scholar]
- 373.ten Brinke LF, et al. Aerobic exercise increases hippocampal volume in older women with probable mild cognitive impairment: a 6-month randomised controlled trial. Br. J. Sports Med. 2015;49:248–254. doi: 10.1136/bjsports-2013-093184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Madden KM, et al. Short-term aerobic exercise reduces arterial stiffness in older adults with type 2 diabetes, hypertension, and hypercholesterolemia. Diabetes Care. 2009;32:1531–1535. doi: 10.2337/dc09-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Lanza IR, et al. Endurance exercise as a countermeasure for aging. Diabetes. 2008;57:2933–2942. doi: 10.2337/db08-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Short KR, et al. Impact of aerobic exercise training on age-related changes in insulin sensitivity and muscle oxidative capacity. Diabetes. 2003;52:1888–1896. doi: 10.2337/diabetes.52.8.1888. [DOI] [PubMed] [Google Scholar]
- 377.Johnson ML, et al. Differential effect of endurance training on mitochondrial protein damage, degradation, and acetylation in the context of aging. J. Gerontol. A Biol. Sci. Med. Sci. 2015;70:1386–1393. doi: 10.1093/gerona/glu221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Hooshmand-Moghadam B, et al. The effect of 12-week resistance exercise training on serum levels of cellular aging process parameters in elderly men. Exp. Gerontol. 2020;141:111090. doi: 10.1016/j.exger.2020.111090. [DOI] [PubMed] [Google Scholar]
- 379.Olioso D, et al. Effects of aerobic and resistance training on circulating micro-rna expression profile in subjects with type 2 diabetes. J. Clin. Endocrinol. Metab. 2019;104:1119–1130. doi: 10.1210/jc.2018-01820. [DOI] [PubMed] [Google Scholar]
- 380.Akbarinia A, Kargarfard M, Naderi M. Aerobic training improves platelet function in type 2 diabetic patients: role of microRNA-130a and GPIIb. Acta Diabetol. 2018;55:893–899. doi: 10.1007/s00592-018-1167-2. [DOI] [PubMed] [Google Scholar]
- 381.Taghizadeh M, et al. Long-term aerobic exercise training in type two diabetic patients alters the expression of miRNA-223 and its corresponding target, the P2RY12 receptor, attenuating platelet function. Clin. Hemorheol. Microcirc. 2022;80:107–116. doi: 10.3233/CH-211209. [DOI] [PubMed] [Google Scholar]
- 382.Yoshino M, et al. Nicotinamide mononucleotide increases muscle insulin sensitivity in prediabetic women. Science. 2021;372:1224–1229. doi: 10.1126/science.abe9985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Igarashi M, et al. Chronic nicotinamide mononucleotide supplementation elevates blood nicotinamide adenine dinucleotide levels and alters muscle function in healthy older men. NPJ Aging. 2022;8:5. doi: 10.1038/s41514-022-00084-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Kim, M. et al. Effect of 12-week intake of nicotinamide mononucleotide on sleep quality, fatigue, and physical performance in older Japanese adults: a randomized, double-blind placebo-controlled study. Nutrients14, 755 (2022). [DOI] [PMC free article] [PubMed]
- 385.Zhou B, et al. Boosting NAD level suppresses inflammatory activation of PBMCs in heart failure. J. Clin. Invest. 2020;130:6054–6063. doi: 10.1172/JCI138538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Brakedal B, et al. The NADPARK study: a randomized phase I trial of nicotinamide riboside supplementation in Parkinson’s disease. Cell Metab. 2022;34:396–407 e396. doi: 10.1016/j.cmet.2022.02.001. [DOI] [PubMed] [Google Scholar]
- 387.Martens CR, et al. Chronic nicotinamide riboside supplementation is well-tolerated and elevates NAD(+) in healthy middle-aged and older adults. Nat. Commun. 2018;9:1286. doi: 10.1038/s41467-018-03421-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Remie CME, et al. Nicotinamide riboside supplementation alters body composition and skeletal muscle acetylcarnitine concentrations in healthy obese humans. Am. J. Clin. Nutr. 2020;112:413–426. doi: 10.1093/ajcn/nqaa072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Dollerup OL, et al. Nicotinamide riboside does not alter mitochondrial respiration, content or morphology in skeletal muscle from obese and insulin-resistant men. J. Physiol. 2020;598:731–754. doi: 10.1113/JP278752. [DOI] [PubMed] [Google Scholar]
- 390.Dollerup OL, et al. Effects of nicotinamide riboside on endocrine pancreatic function and incretin hormones in nondiabetic men with obesity. J. Clin. Endocrinol. Metab. 2019;104:5703–5714. doi: 10.1210/jc.2019-01081. [DOI] [PubMed] [Google Scholar]
- 391.Dollerup OL, et al. A randomized placebo-controlled clinical trial of nicotinamide riboside in obese men: safety, insulin-sensitivity, and lipid-mobilizing effects. Am. J. Clin. Nutr. 2018;108:343–353. doi: 10.1093/ajcn/nqy132. [DOI] [PubMed] [Google Scholar]
- 392.Dai H, Sinclair DA, Ellis JL, Steegborn C. Sirtuin activators and inhibitors: promises, achievements, and challenges. Pharm. Ther. 2018;188:140–154. doi: 10.1016/j.pharmthera.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Venkatasubramanian S, et al. Cardiovascular effects of a novel SIRT1 activator, SRT2104, in otherwise healthy cigarette smokers. J. Am. Heart Assoc. 2013;2:e000042. doi: 10.1161/JAHA.113.000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Libri V, et al. A pilot randomized, placebo controlled, double blind phase I trial of the novel SIRT1 activator SRT2104 in elderly volunteers. PLoS ONE. 2012;7:e51395. doi: 10.1371/journal.pone.0051395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Baksi A, et al. A phase II, randomized, placebo-controlled, double-blind, multi-dose study of SRT2104, a SIRT1 activator, in subjects with type 2 diabetes. Br. J. Clin. Pharm. 2014;78:69–77. doi: 10.1111/bcp.12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Sands BE, et al. Assessing colonic exposure, safety, and clinical activity of SRT2104, a novel oral SIRT1 activator, in patients with mild to moderate ulcerative colitis. Inflamm. Bowel Dis. 2016;22:607–614. doi: 10.1097/MIB.0000000000000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Knowler WC, et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N. Engl. J. Med. 2002;346:393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Hong J, et al. Effects of metformin versus glipizide on cardiovascular outcomes in patients with type 2 diabetes and coronary artery disease. Diabetes Care. 2013;36:1304–1311. doi: 10.2337/dc12-0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Wang, C. P., Lorenzo, C. & Espinoza, S. E. Frailty attenuates the impact of metformin on reducing mortality in older adults with type 2 diabetes. J. Endocrinol. Diabetes Obes. 2, 1031 (2014). [PMC free article] [PubMed]
- 400.Luchsinger JA, et al. Metformin in amnestic mild cognitive impairment: results of a pilot randomized placebo controlled clinical trial. J. Alzheimers Dis. 2016;51:501–514. doi: 10.3233/JAD-150493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.de Kreutzenberg SV, et al. Metformin improves putative longevity effectors in peripheral mononuclear cells from subjects with prediabetes. A randomized controlled trial. Nutr. Metab. Cardiovasc. Dis. 2015;25:686–693. doi: 10.1016/j.numecd.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 402.Mannick JB, et al. mTOR inhibition improves immune function in the elderly. Sci. Transl. Med. 2014;6:268ra179. doi: 10.1126/scitranslmed.3009892. [DOI] [PubMed] [Google Scholar]
- 403.Mannick, J. B. et al. TORC1 inhibition enhances immune function and reduces infections in the elderly. Sci. Transl. Med. 10, eaaq1564 (2018). [DOI] [PubMed]
- 404.Kraig E, et al. A randomized control trial to establish the feasibility and safety of rapamycin treatment in an older human cohort: immunological, physical performance, and cognitive effects. Exp. Gerontol. 2018;105:53–69. doi: 10.1016/j.exger.2017.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Singh M, et al. Effect of low-dose rapamycin on senescence markers and physical functioning in older adults with coronary artery disease: results of a pilot study. J. Frailty Aging. 2016;5:204–207. doi: 10.14283/jfa.2016.112. [DOI] [PubMed] [Google Scholar]
- 406.Hickson LJ, et al. Senolytics decrease senescent cells in humans: Preliminary report from a clinical trial of Dasatinib plus Quercetin in individuals with diabetic kidney disease. EBioMedicine. 2019;47:446–456. doi: 10.1016/j.ebiom.2019.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]