Abstract
In recent years, more and more single-cell technologies have been developed. A vast amount of single-cell omics data has been generated by large projects, such as the Human Cell Atlas, the Mouse Cell Atlas, the Mouse RNA Atlas, the Mouse ATAC Atlas, and the Plant Cell Atlas. Based on these single-cell big data, thousands of bioinformatics algorithms for quality control, clustering, cell-type annotation, developmental inference, cell-cell transition, cell-cell interaction, and spatial analysis are developed. With powerful experimental single-cell technology and state-of-the-art big data analysis methods based on artificial intelligence, the molecular landscape at the single-cell level can be revealed. With spatial transcriptomics and single-cell multi-omics, even the spatial dynamic multi-level regulatory mechanisms can be deciphered. Such single-cell technologies have many successful applications in oncology, assisted reproduction, embryonic development, and plant breeding. We not only review the experimental and bioinformatics methods for single-cell research, but also discuss their applications in various fields and forecast the future directions for single-cell technologies. We believe that spatial transcriptomics and single-cell multi-omics will become the next booming business for mechanism research and commercial industry.
Graphical abstract
Public summary
-
•
Single-cell sequencing can reveal molecular characteristics at the cell level
-
•
Spatial omics can reconstruct the organization of cells and their interactions
-
•
Single-cell multi-omics will picture the cell in a comprehensive way
Introduction
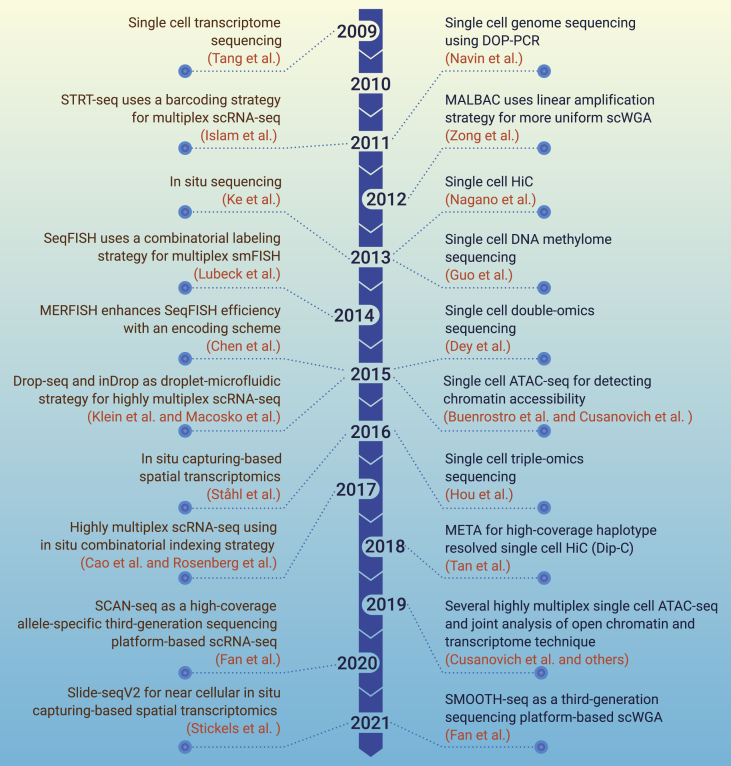
The completion of the Human Genome Project in 2003 was also a beginning to understand the biological meaning of the three billion nucleotides in the human genome.1 A key technological milestone was the release of several massively parallel DNA sequencing (next-generation sequencing) platforms in 2005 and 2007, which allowed for generating sequencing reads of billions of base pairs in a few days at a cost of less than 1,000 dollars. The single-cell omics sequencing research started in 2009 when the single-cell RNA sequencing (scRNA-seq) assay was described by Tang et al.2 Method development has been surging during the subsequent years. Various types of scRNA-seq assays based on different strategies of RNA capture and cDNA amplification were reported. The highly multiplex power of scRNA-seq was realized by the Linnarsson group in 2011 and then was tremendously enhanced by the microfluidics droplet and microwell technologies in 2015.3,4,5,6 The in situ combinatorial indexing strategy allowed for an even higher throughput of thousands or tens of thousands of cells.7,8
A single cell contains multiple omics including genome, epigenome, transcriptome, and others. Single-cell epigenome sequencing technologies were described in 2013, i.e., single-cell HiC and single-cell DNA methylome sequencing assays.9,10 The fastest-growing technique is the scATAC-seq (single-cell sequencing assay of transposase accessible chromatin), which was developed by the Greenleaf group for bulk sample format in 2013 and was quickly adapted to single-cell analysis in 2015.11,12 More recently, combinatorial indexing and droplet-based scATAC-seq assays have been established.13,14,15,16 Single-cell whole genome amplification (scWGA) methods, which were developed many years ago, i.e., degenerative oligonucleotide-primed PCR (DOP-PCR) and multiple displacement amplification, are still applicable for single-cell genome sequencing. In 2011, Navin et al. reported the single-cell genome sequencing study on human cancer cells using DOP-PCR.17 New scWGA techniques, including MALBAC, LIANTI, and META by the Sunney Xie group, aim to increase the coverage and uniformity while reducing the allele dropout, false positive, and chimera problems, for single-nucleotide variation and copy number variation (CNV) detection.18,19,20,21 More recently, third-generation sequencing-based single-cell genome and transcriptome sequencing techniques, SMOOTH-seq and SCAN-seq, have been developed for efficient detection of structure variations and alternative splicing in single cells, respectively.22,23
Single-cell multi-omics sequencing techniques simultaneously measure two or more omics in a single cell. Assays for simultaneously detecting transcriptome and genome were reported in 2015, and those for simultaneously detecting transcriptome and DNA methylome (as well as genome CNVs) were reported in 2016.24,25,26,27,28 More recently, several high-throughput assays were reported for simultaneously detecting chromatin accessibility and transcriptome.29,30,31,32 Another direction is the analysis in a spatial context. Three sets of spatially resolved transcriptomics methods have been reported. The first set, sequential barcoding single-molecule RNA fluorescence in situ hybridization (FISH), was described by the Cai Long group in 2014 (seqFISH), and enhanced by the Zhuang Xiaowei group in 2015 (MERFISH).33,34 The second set, in situ sequencing, was described by the Nilsson group using padlock probes in 2013.35 The third set, in situ capturing, was described by the Lundeberg group in 2016.36 These methods have been developing since their initiation, which will be discussed in detail later in this review. We summarized the milestones of single-cell sequencing technologies in Figure 1.
Figure 1.
The milestones of single-cell sequencing technologies
Single-cell technologies
In recent years, more and more single-cell technologies have been developed, such as single-cell surface functionalization, intracellular electrophysiology, single-cell isolation, and single-cell sequencing. Although single-cell sequencing is most widely used and well known, its development cannot be done without other single-cell technologies, such as single-cell isolation. Therefore, we introduce not only single-cell sequencing but also other essential single-cell technologies.
Single-cell surface functionalization
Inspired by the characteristics of cells in living organisms, single-cell surface functionalization in which single cells are coated by nanostructured materials has been developed to enhance the stability and activity of the cells and endow the cells with abiotic functions that are completely different from their original specializations. Single-cell surface functionalization has features of both biologic and abiotic components.37,38,39 As such, the combination could reach the “one plus one greater than two” results. The integration of the living cells and materials could achieve cell manipulation being more oriented, for the purpose of precise control, on single-cell applications. The surface-functionalized single cell could perform non-natural functions that have significance in creating new functions for multiple applications.40,41,42 These advanced technological targets have become important as a result of the increasing population of the world, which is causing excessive energy and biomass consumption, environmental contamination, and health problems.43,44,45
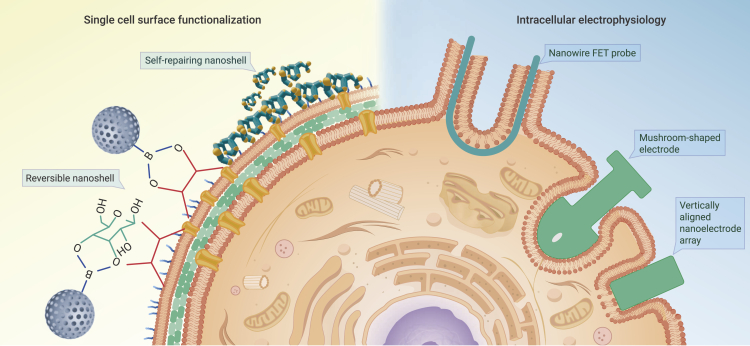
Despite having potential applications, single-cell surface functionalization often limits cell activity and leads to the state of “artificial dormancy” that affects long-term cell functions. In addition, the nanostructured shells formed on a single-cell surface through self-assembly are fragile, leading to a low performance and reduced functions. Dynamic shell technology on a single-cell surface cannot only enhance the cell stability and create abiotic functions but also degrade the shells to activate cell functions in need,46,47 which is known as the future single-cell surface functionalization. Biohybrid nanoshells, which are consisted of gold nanoparticles and amino acid molecules, undergo structure forming deposition on cell surfaces through electrostatic forces.48 This unique system is the first example of a dynamic, self-repairing nanomaterial (Figure 2). Upon growth and division of the entrapped single cell, the biohybrids deposit on the surface of the newly generated cell, forming a supplemental nanoshell. Very recently, a dynamic reversible cell shell technology based on click chemistry has been reported to develop an assembly-dissociation-reassembly smart control of a mesoporous nanoshell on a single-yeast-cell surface by adjusting the additional glucose level in the culture medium (Figure 2). The click chemistry-based method should be highly versatile for introducing protective capsules and controllable assembly-disassembly nano-functionality around single cells as part of biotechnological efforts.49
Figure 2.
Illustration of single-cell detection methods
Single cells in nanoshells, which have sustainable, clean, and green advantages, are already on the market for biomass conversion. To further drive the development of single-cell surface functionalization in industrial and biomedical applications, continuing progress is needed in the design and improvement of these systems. How can the self-repairing dynamic process of nanomaterials evolve from artificial control into autonomic regulation based on natural cells? Materials with autocatalytic proliferation to construct nanoshells for living cells provide a potential alternative, such as micellar with catalysis and autocatalytic proliferation.50 Thus, an autocatalytic proliferation nanoshell for cell functionalization could be obtained that can dynamically function the cells in time.
Intracellular electrophysiology
Recording high-fidelity electrophysiological signals from electrogenic cells, which exhibit good electrical coupling with the cell and provide accurate readout over the entire dynamic range of voltages generated by cells without distorting the readout over time, is critical to deepening our understanding of cellular behaviors at scales ranging from single cells to cellular networks. Taking the neuronal electrophysiological recording as an example, to obtain the most information-rich readings for detailed mapping of brain functions and/or fine control of the neural prosthetics, the electronic electrodes or probes need to provide access to intracellular signals from single to multiple neurons comprising a neuronal circuit. Conventional methodologies utilizing microelectrode arrays residing outside the cellular membrane suffer significantly in signal quality and are blind to subthreshold events.51
The patch-clamp technique is a versatile electrophysiological technique developed in the late 1970s and has been the historical gold standard for studying the function of single or multiple ion channels in living tissue. It has a wide range of applications, from evaluating the functional consequences of ion channel gene mutations to how electrophysiological principles and technologies can be applied to human health. It is particularly useful in the study of electrogenic cells, such as neurons, cardiomyocytes, muscle fibers, and oocytes, and can also be applied to the study of bacterial ion channels in specially prepared giant spheroplasts. Depending on the research interests, different patch-clamp configurations can be applied. For example, the cell-attached configuration can be used to study the behavior of individual ion channels in the section of membrane attached to the electrode, while the whole-cell patch allows the study of the electrical behavior of the entire cell, instead of single channel currents.
The whole-cell patch-clamp technique, on the other hand, is the historical gold standard for intracellular electrophysiological recording52 (Figure 2). A glass capillary filled with an artificial intracellular solution and an AgCl-coated wire inserted on the micro-manipulator is tightly attached to the single-cell membrane, forming a resistance of >1 GW (so-called giga-ohm seal) between the pipette and the membrane. Once the negative pressure is applied, a hole is generated on the cell membrane, leading to the exchange of intrinsic and artificial intracellular solutions. After that, the net dynamics of currents and voltages generated through all ion channels can be measured by the AgCl-coated wire.
Although the patch-clamp technique has served as the primary tool for studying high-fidelity analysis of the electrical properties and functional connectivity of electrogenic cells for many years, several limitations remain: first, it is considered an invasive approach as the whole-cell patch clamp involves destruction of the cell membrane and solution exchange between the intracellular space and the pipette reservoir, which can affect the functioning of the cell. In addition, whole-cell patching requires great skill and is very difficult to use in large-scale parallel recording.
To overcome the above challenges, several strategies have been proposed by reconsidering the design of patch-clamp electrodes or developing new techniques that mimic the nanometer-scale topographical features naturally found in single-cell microenvironments. For example, inspired by the shape of dendritic spines, extracellular micrometer-sized mushroom-shaped gold electrodes (Figure 2) have been developed to enable “intracellular-like” recordings of monophasic action potentials and subthreshold activity, based on enhanced coupling between cells and extracellular electrodes.53 In addition, the development of vertically aligned nanoelectrode arrays and nanowire field-effect-transistor (FET) probes have also been instrumental in enabling high-quality intracellular recordings. Their small tip with nanoscale features can penetrate cell membranes with minimal invasiveness and gain tighter contact with cells, thereby significantly reducing interfacial cracks and contact resistance.
Development of intracellular electrophysiology tools with recording elements at the nanoscale size could push the limits of spatiotemporal resolution while reducing invasiveness, which could provide a deeper understanding of electrogenic cells at the single-cell to the tissue level, as well as provide novel quantitative parameters in the study of subcellular/molecular electrophysiology. In particular, stable recording and interrogating the same individual neurons in the neural networks and multiple apical dendrites in a single neuron promise to help decipher complex neurological disorders, which develop over time, thus driving the advanced brain-machine interface with higher resolution, and perhaps eventually bringing “cyborgs” to reality.
The intracellular electrophysiology recordings via vertically aligned nanoelectrode arrays (Figure 2) could be achievable by either spontaneous single-cell penetration or assisted single-cell penetration.54 It is worth noting that spontaneous penetration of a single-cell membrane occurs rarely or may only occur immediately after plating and before activation of membrane repair mechanisms, ultimately leading to probe rejection and membrane resealing. Electroporation using one or multiple high-voltage pulses or plasmonics-enabled optoporation is usually applied to facilitate nanoprobe penetration and enhance intracellular recordings.55
Electrical, photothermal, and photoelectric stimulation could also be used to regulate the electrical activity of a single cell. Through proper geometrical and circuit design, as well as the optimization of the physical cell-electrode coupling interface, decent intracellular recordings of action potentials and subthreshold signals can be obtained by vertically aligned nanoelectrode arrays.56 Nevertheless, their small tip size usually results in a high electrode impedance, leading to signal loss in amplitude and shape.
Nanowire FET probes, on the other hand, can detect intracellular voltage independently of electrode impedance, exhibiting higher fidelity of electrophysiological signals of cultured primary neurons and beating cardiomyocytes57,58 (Figure 2). The high-quality recording is due to the nanoscale size of the FET, which enables its insertion into the cytosol, resulting in a negligible access resistance, and the formation of gigaohm seal resistance against the single-cell membrane. The recently developed shape-controlled deterministic nanowire assembly and compressive buckling technique could overcome the scalability issue, which is typically associated with the nanowire FET probes.59,60 Combined with in vivo recording platforms, these advances can enable investigations of dynamics in the brain and drive the development of new brain-machine interfaces with unprecedented resolution and precision.61
High-throughput single-cell isolation
To isolate a certain cell population from a cell mixture at the single-cell level, it is arguably crucial to develop more efficient single-cell isolation techniques. The earliest single-cell isolation was based on manual pipetting to analyze only a handful of cells.2,3 Later, fluorescence-activated cell sorting (FACS) was used as an easy-to-implement alternative to isolating cells into microwell plates.62 A further increase in throughput to about one thousand cells was achieved using an integrated fluidic circuit.63 Then, the application of liquid-handling robotics increased the number of cells that could be measured in a single run to several thousand.64 More recently, nano-droplet and pico-well technologies emerged to isolate tens of thousands of cells at a time, truly realizing “high-throughput cell sorting.”5,65,66,67
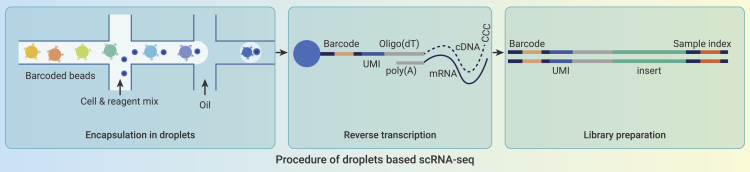
Droplet microfluidic platforms precisely encapsulate a large number of individual cells into nanoliter reaction chambers; such platforms are ideal for high-throughput single-cell isolation and downstream treatments.68,69,70 In particular, droplet microfluidic platforms randomly encapsulate individual cells according to the Poisson distribution.71 The high-throughput characteristics of nanoliter droplets minimize the required reaction reagent volume while improving reaction efficiency within a single droplet (Figure 3). More improvements are needed to accelerate research in the single-cell field, such as more multifunctional single-cell manipulation capabilities and more computational tools to identify single-cell heterogeneity. The introduction of machine learning algorithms would automate single-cell separation to develop more optimized turnkey systems for early diagnosis of diseases, drug screening assessment, and precision medicine.
Figure 3.
Procedure of droplet-based scRNA-seq
High-throughput single-cell sequencing platforms
Based on these single-cell isolation techniques, many commercialized high-throughput single-cell sequencing platforms have been developed. SORT-seq and new VASA-seq platforms offered by Single Cell Discoveries use FACS to sort single cells into the wells of a 384-well cell capture plate.72 Each well in the plate contains barcoded primers and other reagents. After isolation and barcoding, the amplification of the RNA with in vitro transcription is conducted, followed by sequencing analysis. Clontech’s iCell8 single-cell system is using an integrated robotic system to dispense single cells in a 5,184-nanowell chip. It can capture up to 1,800 single cells, and single live cells can be recognized and selected by the automated imaging station.73 BD Rhapsody is also a well-based system, in which individual cells are randomly deposited into an array of picoliter wells under the effect of gravity, followed by loading barcode beads onto the microwell array to saturation. The dimensions of the beads and wells are optimized to prevent double occupancy of beads, and it can capture up to 20,000 cells.74
In addition, commercial microfluidic-based high-throughput single-cell platforms have also been developed. For instance, the Fluidigm C1 platform utilizes the integrated microfluidic chip (IFC) as a reservoir to capture, image, and perform cell lysis, reverse transcription, and initial PCR reactions for single cells,75 which can capture up to 800 cells per run. Chromium from 10X Genomics is a droplet-based system. This system partitions thousands of cells into nanoliter-scale Gel Bead-In-Emulsions generated in a microfluidic device.74 It allows capturing an average of 50% of input cells, approximately 5,000 cells that can be sequenced in a typical Chromium experiment. More recently, based on its all-in-one digital PCR machine MobiGaea, MobiDrop launches the MobiNova single-cell system associated with MobiCube kits and MobiVision software, which combines advanced microfluidic technologies with versatile barcode beads to achieve highly efficient single-cell analysis.76 It can partition tens of thousands of cells with an efficiency up to 60% in a single operation within only 20 min.
All these commercial platforms are very helpful to stimulate the usages of single-cell sequencing in life science research. For example, with the assistance of the MobiNova system, a recent study examined the underlying mechanisms of noise-driven single-cell heterogeneity triggered by volumetric compression, which usually arises from tumor progression.76 In the future, the applications of high-throughput single-cell sequencing will extend into many new fields and will play an even more important role in biological, clinical, and pharmaceutical research.
Bioinformatics analysis of single-cell sequencing data
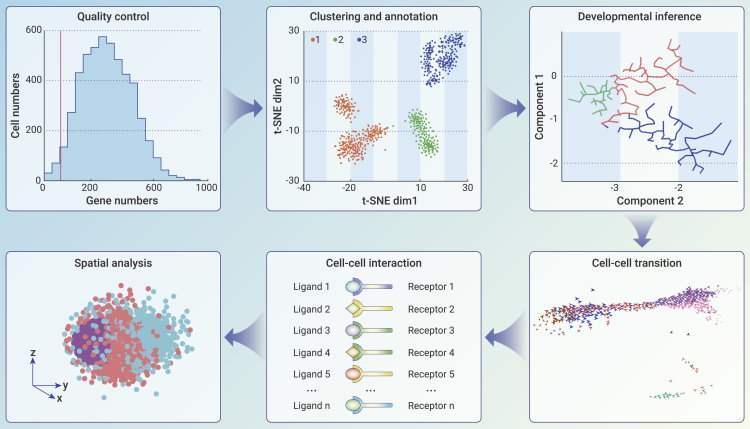
The most mature and commercialized high-throughput single-cell data were scRNA-seq data. Algorithm and software development in bioinformatics are critical to translate scRNA-seq data into biological and medical data and further applications. Thousands of bioinformatics tools are now available for scRNA-seq analysis, which can be found here: https://github.com/seandavi/awesome-single-cell. Guidance to the common practice of scRNA-seq data analysis has been discussed in published reviews and tutorials of multiple scRNA-seq bioinformatics tools, such as Seurat and Scanpy.77,78 However, it is worth noting that none of the available bioinformatics tools fit all conditions because of the wide range of scRNA-seq applications. Therefore, here we provide a brief introduction to those concepts critical to bioinformatics tool selection in this review. We summarize the current bioinformatic analyses of scRNA-seq data in Figure 4 and they are generally composed of two steps: (1) quality control and annotation and (2) biomedical knowledge mining.
Figure 4.
The bioinformatics analysis for scRNA-seq data
Quality control
Because of the limited RNA content of single cells and the stochastic nature of current scRNA-seq techniques, mRNAs within one cell cannot be fully captured and sequenced. The supper and upper gene numbers, together with the percentage of mitochondrial genes, are three commonly applied indices for quality control during scRNA-seq data analysis. The aim of quality control is to control the rates of empty cells, doublets or multiplets, and cells with bad states. It is worth noting that such quality control metrics vary across different biological specimens, and thus no consistent cutoffs exist.
Batch effect
The differences in scRNA-seq data across different experiments, operators, laboratories, or other technical replicates are named as batch effects. In practice, the same specimen cannot be sequenced multiple times by different operators or laboratories, leaving such technical biases mixed with biological signals. A straightforward strategy is adopting batch effect removing algorithms that have been applied in bulk RNA-seq data analysis. While this strategy can mitigate partial batch effects in scRNA-seq, more advanced and customized algorithms have been proposed for scRNA-seq data, including mutual nearest neighbors,79 canonical correlation analysis,80 and optimization model-based methods. It is worth noting that batch effect removal may introduce batch effects if not properly used. Therefore, parameter tuning is critical to balance the performance of existing algorithms.
Clustering and cell-type annotation
Visualization of cell clusters and their mutual relationships is a critical task of scRNA-seq data analysis. Dimension reduction algorithms in the machine learning field are frequently adopted, with principal-component analysis, t-distributed stochastic neighbor embedding, and Uniform Manifold Approximation and Projection as the most frequently used in practice. After clustering, the cell type of each cluster can be predicted with artificial intelligence-based methods,81 such as SingleR,82 or with cell marker-based methods, such as CellAssign.83
Trajectory analysis
Typical algorithms for inferring the developmental relationships among cells include Monocle84 and RNA velocity.85 Monocle represents a large class of algorithms that infer cellular developmental trajectories based on gene expression similarity.84 The assumption underlying such analysis is that cells along a specific developmental trajectory were sampled by scRNA-seq and thus could be reconstructed via examining the similarity of cellular expression profiles. RNA velocity analysis employs the dynamics of the mRNA maturation process and formulates an elegant algorithm to derive the probability and direction of cell-to-cell transitions, which provides informative clues to derive biological conclusions from scRNA-seq data.85 While Monocle and RNA velocity analysis are informative, the results are predictive and validations are needed to confirm the predictions.
Single-cell regulatory network
To identify the critical regulators of individual cells or cell types, we need to construct the single-cell regulatory network based on scRNA-seq data; for this there are tools, such as SCENIC86 and NicheNet.87 SCENIC conducts regulon analysis based on scRNA-seq data and can provide quantitative analysis of the roles of diverse transcription factors in shaping the observed scRNA-seq data.86 Different from SCENIC analyzing the intrinsic factors underlying cellular expression profiles, NicheNet compiles a ligand-target database and tries to prioritize those extrinsic factors critical to the observed gene expression profiles.87 Both algorithms provide insightful clues for understanding scRNA-seq data, but a universal algorithm that could dissect the regulatory mechanisms underlying scRNA-seq data is of pressing need, although it is also challenging.
Cell-cell communication
To interrogate the potential cell-to-cell crosstalks and even spatial relationships, there are two types of algorithms: one is dependent on ligand-receptor interactions and the other is not. Ligand-receptor interactions are the molecular mediators of cell-to-cell crosstalks and thus could be employed together with scRNA-seq to derive cellular interactions. CellphoneDB88 and CSOmap89 are two typical methods of the ligand-receptor interaction-dependent cell-cell communication algorithms. CellphoneDB takes scRNA-seq data and ligand-receptor interactions as inputs and outputs the potential or most significant ligand-receptor interactions among cells or cell types, without considering whether the potential interacting cells were spatially close to or far from each other.88 In comparison, the spatial relationship among cells is the primary aim of CSOmap analysis and cellular interactions are derived from the spatial inference.89 Both CellphoneDB and CSOmap establish statistical tests but the testing hypotheses are different. The statistical tests of CellphoneDB are applied to individual ligand-receptor interactions across different cell pairs while CSOmap examines whether a cell type pair tends to appear in the neighborhood of each other defined by the inferred pseudo-space. NovoSpaRc90 represents a class of algorithms that aim to derive cellular spatial relationships only based on scRNA-seq data without ligand-receptor interactions. The assumption behind NovoSpaRc and its variants is that cells with similar gene expression profiles should have a high probability to share spatial niches. This assumption is well applicable to embryo development where the cellular spatial positions are tightly regulated. However, it may be challenging to study the immune cell compartments where ligand-receptor interactions play a dominant role. All three types of algorithms were developed for scRNA-seq data analysis.
Single-cell research projects
In addition to the molecular dimension of omics data in the post-genome era, the rapid development and application of scRNA-seq adds the cellular dimension to omics data. Similar to the Human Genome Project (HGP), which led the leap of biomedical research to an omics era, the Human Cell Atlas ([HCA] https://www.humancellatlas.org/)91 has been initiated to create cellular reference maps for the position, function, and characteristics of every cell type in the human body. While HGP was an international group of researchers whose goals, programs, and organizations were inherently defined by the characteristics of the genome organization in a cell (covalent DNA molecules from limited chromosomes), the goals, programs, and organization of HCA are heavily defined by the characteristics of cells within a body (established research groups from diverse background around the world). The final creation of a human reference cell map across the body will lead to a new leap in biomedical innovation. The National Institutes of Health Common Fund’s Cellular Senescence Network (https://sennetconsortium.org/)92 and the National Cancer Institute-funded Human Tumor Atlas Network (https://humantumoratlas.org/)93 are more specific single-cell projects of cellular senescence and tumor, respectively.
Similar to the HCA, there are also several large single-cell sequencing projects for mouse, such as the Mouse Cell Atlas by Zhejiang University (http://bis.zju.edu.cn/MCA/),94 the Mouse RNA Atlas,8 and the Mouse ATAC Atlas13 by Seattle Organismal Molecular Atlases (https://atlas.gs.washington.edu/). These large-scale high-quality single-cell omics data for human, mouse, and other model organisms (fruit fly, C. elegans) are the basis of many cell-type annotation software, such as SingleR.82 They could be the references for embryo development, tumor genesis, and tissue-specific studies.
Beside animals, the Plant Cell Atlas (https://www.plantcellatlas.org) project was also proposed to create a community resource that incorporates information on nucleic acids, proteins, and metabolites to comprehensively describe the states of various cell types at increasing higher resolutions.95,96,97 To illustrate the mechanisms of plant development and stress response, plant scientists have not stopped their steps to trace the gene expression profiles from the whole plant, different organs, specific cell types, and single cells. At the level of nucleic acids, with the newly developed scATAC-seq and scCUT&Tag approaches,98,99,100 integrated multi-omics analysis would be the next direction for plant single-cell studies. To map proteomics and metabolomics in single plant cells have been tried, but how to realize it at a large scale still needs further efforts.101,102,103,104 Criteria for making data from different labs comparable will facilitate the integrated analysis.
Applications of single-cell sequencing
Although there are many single-cell technologies, the most widely used one is single-cell sequencing. We review its applications in oncology, immunology, plant breeding, reproductive, and developmental studies.
Single-cell sequencing in oncology and immunology
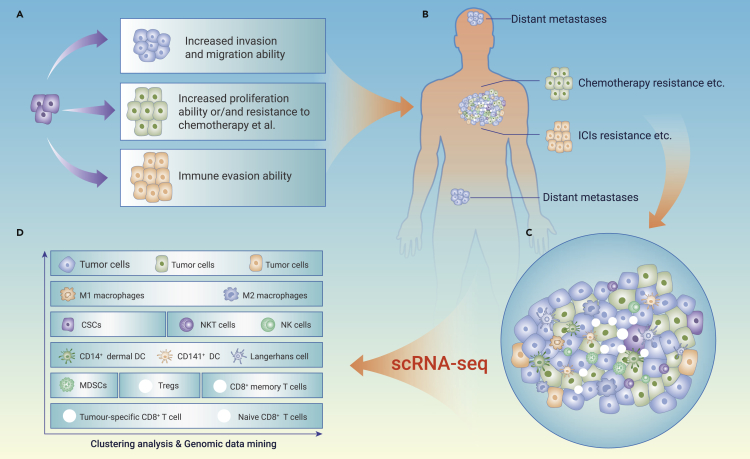
Cancer is always the top threat to human life because of its limitless replicative potential, cell heterogeneity, tissue invasion and metastasis, avoiding immune destruction features, etc.105 In fact, heterogeneity between different malignant cells is one of the fundamental characteristics of almost all human cancers and has been a major obstacle to full recovery from cancers.106,107 The mechanism involves heterogeneity among cancer cells in the same patient including stochastic genetic and epigenetic changes.108,109 With the rapid progress in high-throughput technology, researchers have focused their attention on exhaustive understanding of the molecular mechanisms that influence tumor cells and immune cell heterogeneity during cancer progression. Recently, scRNA-seq has provided an excellent avenue to explore the heterogeneity and characteristics of cancer cells and immune cells (Figure 5).
Figure 5.
The application of single-cell sequencing in oncology
Combined with in vivo lineage tracing, scRNA-seq reveals that the heterogeneity and dynamics of Prom1+ HCC cell have cancer stem cell-like characteristic and contribute to therapeutic resistance and tumor recurrence.110 The heterogeneity of high-grade serous tubo-ovarian cancer (HGSTOC) is characterized by extensive inter-/intratumor and results in therapeutic resistance.111 The scRNA-seq technology provided 43 new promising therapeutic targets and identified 6 cellular phenotypes related to prognosis, which provides an excellent understanding of the tumor microenvironment (TME) of HGSTOC.111 Furthermore, scRNA-seq can be used to identify specific modes of gene expression authorizes for the elucidation of molecular mechanisms underlying tumor migration and invasion.112
Using scRNA-seq, Xu et al. identified 9 cancer cell subclusters from the transcriptome profile of 96,796 single cells from 15 paired samples of primary tumors and axillary lymph nodes. Importantly, the transcriptome data suggested that NECTIN2-TIGIT-mediated interactions between metastatic cancer cells and TME cells promoted lymph node metastasis in breast cancer patients.8 Renal cell carcinoma is characterized by various gene mutations, and the accompanying tumor cell heterogeneity plays a critical role in the promotion of therapeutic resistance, distant metastasis, and poor prognosis.113 Combined with analysis of bulk data and scRNA-seq data, Liu et al. have identified several novel biomarkers for predicting the prognosis and revealed potential molecular mechanisms that participate in tumorigenesis of renal cell carcinoma cells.114
Besides exploring the mechanisms that influence therapeutic resistance, disease relapse, migration, invasion, and gene mutation, advancements in scRNA-seq technology have promoted the progression of gene regulatory networks (GRNs) investigated in several cancers.115,116,117 To construct GRNs based on scRNA-seq data could conduce us to explore the intratumoral heterogeneity and clarify the critical genes involved in the development of cancers. Wouters et al. used single-cell transcriptomics in combination with GRNs and trajectory inference to study 10 melanoma cultures that have mapped the gene regulatory landscape of relapsed melanoma.116 Because of the heterogeneity and complexity of the tumor ecosystem, in-depth analysis of a tumor at single-cell levels using scRNA-seq is indispensable.
Anomalous infiltration of immune cells in the TME of malignant tumors have been verified to play an important role in carcinogenesis, metastasis, and immunotherapy resistance, and predicted a poor clinical prognosis of cancer patients.118,119,120 Therefore, further investigating and identifying critical immune modifiers in tumor immune microenvironments is urgently needed. The heterogeneity and complexity of T cell, B cell, macrophages, and DCs, in TME can be identified by the high-throughput and unbiased gene expression analysis of scRNA-seq.121
Applications of scRNA-seq in dissecting tumor immune microenvironments have made breakthrough progress in the biological function of tumor-infiltrating immune cells, including their heterogeneity, metabolism, dynamics, and potential roles during cancer progression and resistance to immunotherapy.122 Macrophages were characterized by high plasticity and heterogeneity: the M1 type macrophages act as one of the critical immune cells that executes anti-tumor effects in the TME; however, the M2 type macrophages implement a pivotal role during tumor progression.123 Liver metastasis is the leading cause of colorectal cancer-related mortality, and the metastasis demonstrates a significantly heterogeneous and defective immunosurveillance microenvironment. Interestingly, scRNA-seq and spatial transcriptomics (ST) analysis indicate that the colorectal cancer-related liver metastatic microenvironment is infiltrated with increased MRC1+ CCL18+ M2-like macrophages.118
To analyze the plasticity and phenotypes of immune cells in the TME of HBV/HCV-related HCC, Song et al. carried out scRNA-seq on 41,698 immune cells from HBV/HCV-related HCC tumors and non-tumor liver tissues. The results showed 29 immune cell subsets of myeloid cells, NK cells, and lymphocytes with unique transcriptomic profiles in HCC. They also identified a subset of M2 macrophages with forced CCL18 expression and a new subset of activated CD8+ T cells with forced XCL1 expression that were correlated with the prognosis in HCC patients.124 The transcriptomes of ∼17,000 cells from 18 primary or early-relapse HCC cases were analyzed by scRNA-seq. Sun et al. have verified that early-relapse HCC has reduced infiltration of Tregs, and increased dendritic cells and CD8+ T cells. Importantly, CD8+ T cells in recurrent tumor tissues with forced CD161 expression displayed an exhausted phenotype.125
PD-1/PD-L1 antibody-based immunotherapy has revolutionized malignant tumor treatment and accelerated the breakthrough in the field of tumor immunology. Although several types of immunotherapies, including chimeric antigen receptor T cells and immune checkpoint inhibitors, have improved clinical responses, the efficacies varied significantly, and only a small subset of cancer patients can benefit from them.120,126 Exploring all the biological profiles of tumor-infiltrating immune cells in TME is critical to exhibiting tumor immunity and ameliorating cancer immunotherapies. The applications of scRNA-seq to reveal the phenotypic heterogeneity of tumor-infiltrating T cells, macrophages, and DCs in the TMEs have deepened our understanding of the tumor immune evasion regulatory networks. Overall, scRNA-seq showed its superior advantage in the tumor immune microenvironment to discover promising therapeutic targets and optimize treatment strategies of immunotherapy in the management of cancer.
Single-cell sequencing in assisted reproduction
Infertility is a disease of the reproductive system that hinders pregnancy after regular unprotected sexual intercourse.127 Although it is known that it is a heterogeneous pathology with a complex of environmental and genetic factors, nearly 50% of infertility cases are primarily due to genetic defects.128 As a crucial clinical treatment for infertility, assisted reproductive technology (ART) has been widely accepted and revolutionized the lives of millions of families who were previously unable to conceive all over the world.129
Currently, in vitro fertilization (IVF), which involves obtaining eggs, uniting them with sperm in a laboratory setting, growing the embryos, and transferring them directly into the uterus under ultrasound guidance, is the major form of ART and has been used successfully since 1978.130 During IVF treatment, preimplantation genetic screening (PGS) is also required to help identify embryos that are free of inherited mutations, which consequently prevents establishing pregnancy.131 Even though the use of PGS has shown encouraging results, the actual success rate of IVF remains relatively low as it still has a high frequency of implant failure or incomplete pregnancy.132 To improve safety and prevent adverse effects on future generations, every step of the traditional ART procedure, including superovulation, fertilization, preimplantation embryo development, frozen embryo transfer, and PGS, should be examined carefully. Since male and female reproductive tissues and embryos are heterogeneous, only single-cell sequencing techniques can resolve the confounding effects of distinct cells and provide insights into the biological processes that are obscured by bulk measurements. These unprecedented findings will improve the success rate of IVF.
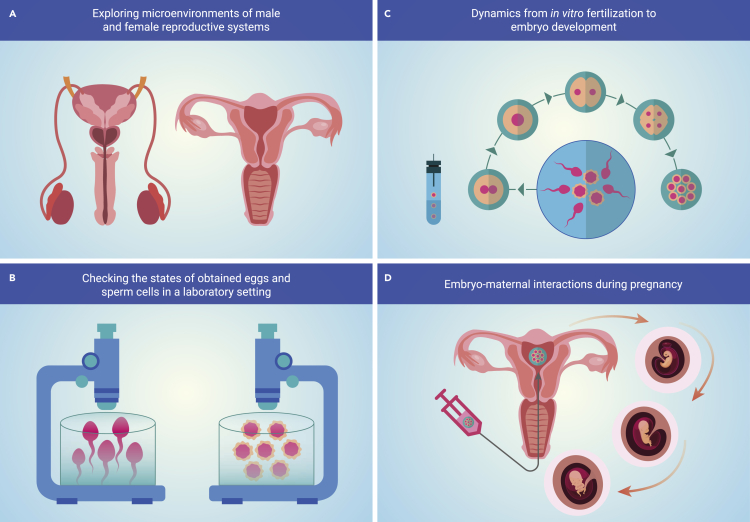
Several ongoing international efforts, including the HCA,91 the Human Cell Landscape,133 and Tabula Sapiens,134 are all aimed at providing the multi-omics data of reproductive organs, such as testis, ovary, and uterus, at the single-cell level. In 2020, Wang et al. obtained the transcriptomic signature of human endometrium at a single-cell resolution.135 In this project, they identified six major endometrial cell types. One of them was a previously uncharacterized epithelial ciliated cell type. Furthermore, they showed that the human window of implantation opens with abrupt and discontinuous transcriptomic activation in the epithelia. Before the opening of the window of implantation, decidualization was found initiated in the stromal compartment by capturing the direct interplay between stromal fibroblasts and lymphocytes during decidualization. All these efforts have a direct impact on the improvement of strategies for ex vivo cultivation in ART (Figure 6A).
Figure 6.
The application of single-cell sequencing in assisted reproduction
In addition to exploring reproductive tissues, reproductive cells should also be examined. Primordial germ cells (PGCs) are progenitor cells of mature gametes, oocytes, and sperms, which maintain the continuation of a species through fertilization.136 In 2015, the human PGC transcriptome and DNA methylome along the developmental trajectory were comprehensively analyzed at the single-cell and single-base levels.137 Guo et al. found that human PGCs exhibit unique features compared with mouse PGCs. This research shed light on the safety of ARTs and the clinical assessment of germ cell abnormalities during PGS.
Among germ cells, oocytes are formed already during fetal life, while spermatozoa can be generated throughout adulthood.138 Mature and developmentally competent oocytes are provided by the ovaries, which also can produce hormones to support the female phenotype and pregnancy.139 In 2020, the first single-cell transcriptomic landscape of ovaries from young and aged nonhuman primates was presented.140 Researchers identified seven ovarian cell types with unique gene expression signatures. Meanwhile, somatic cells from the inner part of human ovaries were characterized by single-cell sequencing in 2019.141 In the next year, Wagner et al. reported single-cell transcriptomes and cell surface antigen profiles of over 24,000 cells from the outer lining of the ovary (cortex),142 which confirmed that there are no germline stem cells in adult human ovaries.
Embryo development begins with a unified zygote, failures in zygotic genome activation result in early embryo developmental arrest and implantation failure in mammals (Figure 6B). We cannot perform scRNA-seq directly on the zygote to check the quality and states during IVF because current genome-wide profiling methods would destroy the cells to extract the cellular content. Live-seq143 could be a promising single-cell transcriptome profiling approach since it can keep cells alive after profiling (Figure 6C). Besides, exploring lineage specification during human preimplantation development paves the way to improve ARTs. In 2013, Guo et al. provided a single-cell methylation landscape of mouse embryonic stem cells and early embryos.9 In the same year, a comprehensive set of transcriptome landscapes of human preimplantation embryos and embryonic stem cells were explored using the scRNA-seq technique and published.144 Meanwhile, Xue et al. utilized scRNA-seq to explore the transcriptomes from oocytes to morulae in both human and mouse embryos.145 The methylome of human early embryos from the zygotic stage to post-implantation was published the next year.146 By comparing the single-cell transcriptome of human and mouse preimplantation embryos in early development, Blakeley et al. showed human-specific transcriptional programs.147 In 2016, a comprehensive single-cell transcriptional map of human embryo development from a human preimplantation embryo cohort pool was presented.148 In 2018, Zhu et al. provided a single-cell DNA methylome for human preimplantation embryos.149 With the development of single-cell multi-omics sequencing technology (single-cell COOL-seq), genome-wide maps of DNA methylation and chromatin accessibility at single-cell resolution in preimplantation embryos from mouse150 and human151 were both generated. Then in 2021, Meistermann et al. combined scRNA-seq data time-lapse imaging of annotated embryos to reconstruct early mouse and human embryo development.152 A single-cell multi-omics map of mouse preimplantation development was made available for analysis with the help of a single-cell multi-omics sequencing technology (scNOMeRe-seq).153 Through scRNA-seq, Vento-Tormo et al. analyzed decidual, placental, and maternal blood to explore the interactions at the maternal-embryonic interface during early pregnancy (Figure 6D),154 while Zhang et al. obtained scRNA-seq data from human fetal pituitaries to capture developmental trajectories in human pituitary development.155 Overall, single-cell sequencing techniques provide a deep understanding of the reproductive tissues, embryos, and fetus, which holds the potential to improve the IVF outcome and subsequent embryo and fetal development (Figure 6).
Single-cell sequencing for embryonic development
Early embryonic development, during which an orchestrated cell fate determination occurs in a spatial and temporal context, serves as a paradigm for deciphering the molecular program underlying the tissue architecture and cell organization. Embryonic morphogenesis is accomplished partially owing to the rapid cell behavior changes, which are induced by cell shape change, cell migration, cell proliferation, and programmed cell deaths.1 Over the past decades, transcriptome-wide analyses of differentially expressed genes have gained prevalence, providing overviews of gene expression patterns and more sophisticated molecular changes during development.2 With the discovery of the molecular and functional heterogeneity of cells, scRNA-seq was rapidly developed and optimized with higher throughput, greater seq-depth, lower cost, and long-read coverage.3,4,5,6 scRNA-seq not only establishes more comprehensive molecular atlases,7 but permits the identification of novel cell sub-populations and inference of differentiation trajectories, making it the de facto most widely used technique to screen and resolve preimplantation embryos, gastrulation, early organogenesis, and the development of specific organs over the past decade.2,8,156,157,158 The understanding of individual cell behavior and gene expression regulation was further improved with the explosion of various single-cell multi-omics techniques, covering the proteome, methylome, interactome, chromosome accessibility, chromosome topography, and RNA modifications.159
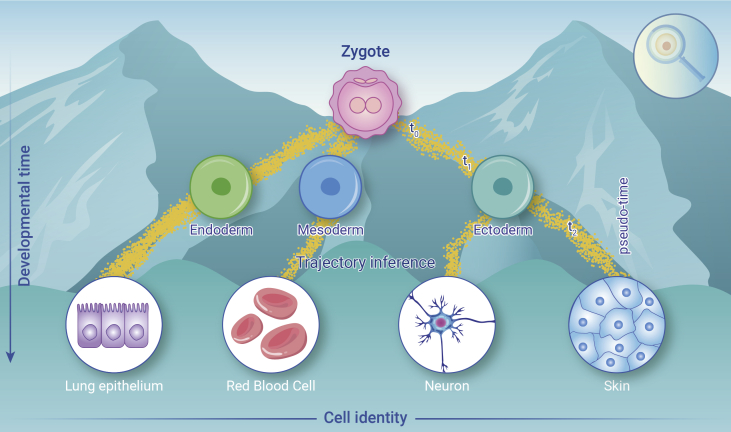
Importantly, besides cataloging cell types and cell states, the understanding of the dynamic process of mammalian cell lineage has been deepened with the development of single-cell transcriptome technology. By connecting cell states in different time series, the trajectory of cell lineage during development can be reconstructed, a long-standing focus and a fundamental goal in stem cell and developmental biology.8 Compared with the traditional study of the lineage process of a certain type of cell through several specific marker genes, single-cell transcriptomics provides a versatile tool for mapping differentiation dynamics and manifolding cell states, facilitating molecular navigation of lineage progress (Figure 7). For example, using Xenopus as a model organism,9 researchers set up "ancestor voting" based on time information among the cell populations obtained by different clusters, mapped the fate of cells over time, and found that many cell fates are determined much earlier than previously thought. The cell fates of endoderm, mesoderm, and ectoderm start to be specialized even after the blastocyst stage. Farrell et al. used the Drop-seq method to obtain a large number of embryonic single-cell transcriptomes with high temporal resolution (from 3.3 h after fertilization to 12 h after fertilization) of embryonic single-cell transcriptomes, revealing that zebrafish changes developmental trajectory during embryogenesis.10 They developed a diffusion-based simulation reconstruction method URD (simulated diffusion based), which uses discrete random walks and graph search to simulate the continuous process of diffusion. This calculation method requires an artificial setting of the start and endpoints. Therefore, it can reflect more biological information and is a powerful visualization method of developmental trajectory. Based on single-cell transcriptome analysis technology, Pijuan-Sala et al. used 10X Genomics to sequence and analyze mouse embryos from 6.5 to 8.5 days after fertilization, and obtained a large number of embryonic single-cell transcriptomes. They found that visceral endoderm cells are mainly involved in the formation of the hindgut.11 Nowotschin et al. conducted single-cell transcription analysis of all endoderm cells from E3.5 to E8.75 in mice and found that the endoderm of embryonic origin and extraembryonic origin in the neonatal intestinal endoderm has a certain spatial pattern.12 Using indexed multiple combinatorial strategies, Cao et al. analyzed 2 million mouse embryonic single-cell transcriptomic landscapes of mammalian organogenesis at a very large scale and combined with Monocle 3 to identify hundreds of cell types and cell lineage trajectories.13
Figure 7.
The application of single-cell sequencing in developmental biology
In addition to the above-mentioned developmental path reconstruction algorithms and studies, there are many other trajectory-inference tools, such as Monocle, Waterfall, Wishbone, TSCAN, Monocle2, Slingshot, and CellRouter.14 Inspired by single-cell transcriptome analysis, these methods offer an unbiased and transcriptome-wide view of dynamic changes in developmental biology and many other fields as well.
It is worth noting that the single-cell transcriptome is inevitably losing spatial information during cell isolation. However, spatial context is one of the key determinants of cellular identity. To resolve the spatial organization and molecular architecture underpinning the cell lineage segregation during embryo development, spatial transcriptome approaches must be properly implemented.15,16,17,18 With the cutting-edge technology of spatially resolved transcriptome, molecular genealogy of germ layers, and cell compositions in spatial resolution can be unraveled.19 We envision that combining spatial and single-cell multi-omics will address fundamental questions and provide new insights into many aspects of development and disease.
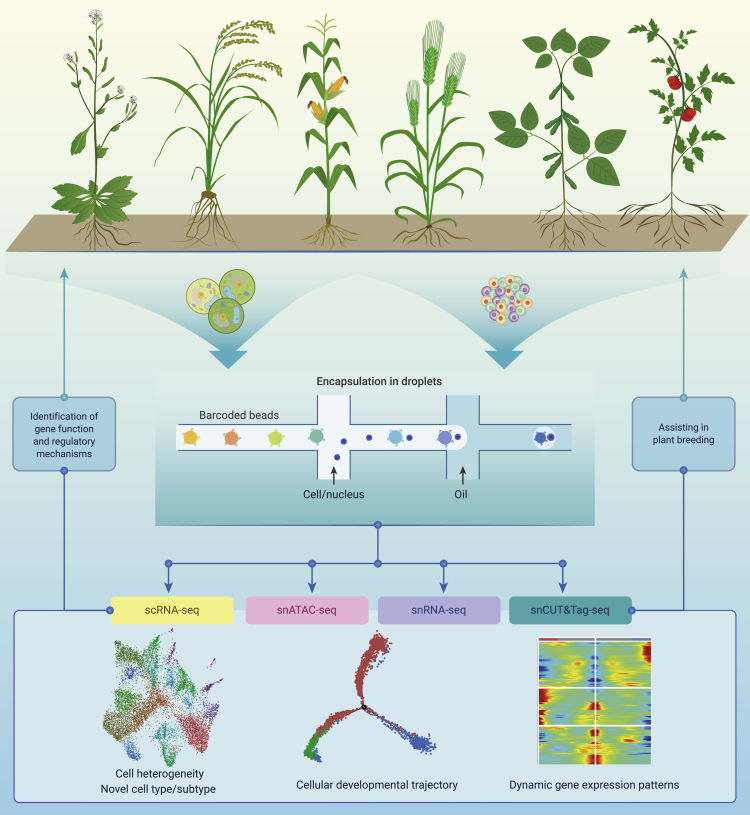
Single-cell sequencing in plant breeding
Single-cell multi-omics is revolutionizing our views from the tissue level to a single-cell level, which allows us to access individual cell modalities, including genomics, transcriptomics, epigenomics, etc.65,160,161 Due to the existence of cell walls out of plant cells, single-cell multi-omics in plants fell behind compared with the explosive development in animals. Microcapillary pick or microscopic isolation of particular cells was first applied to acquire transcript information in one cell,63,162,163,164 and used to try to reconstruct a developmental program in maize male meiosis.165 With the breakthrough of a microfluidic, especially droplet-based high-throughput single-cell sequencing technique, the burst of plant single-cell transcriptomes was in revealed 2019. Papers published successively reported the single-cell transcriptomes of Arabidopsis root, which showed the power of scRNA-seq and opened the door for single-cell multi-omics studies in plants.166,167,168,169,170 Thereafter, single-cell transcriptomes have been applied to other systems, including leaf, vasculature, shoot apex, maize meristem cells, and regeneration callus, highlighting the heterogeneity and developmental dynamics.171,172,173,174,175,176,177,178,179,180,181
Plant tissues and organs commonly have a complex cellular architecture consisting of heterogeneous cell types embedded in cell walls of different compositions. Protoplasting to gain single cells is not generally applicable to all cell types, and disruptive digestion of the cell walls also introduces gene expression responses to stimuli.166,175,182,183 Single-nucleus transcriptomes were developed using isolated nuclei rather than entire cells.98,184,185 This can eliminate the protoplasting-induced transcriptional effects and also the limitation of large cells in encapsulation, which can be used in almost all plant species and organs. Although with a low mRNA capture in a single nucleus than a single cell, studies discovered that snRNA-seq can capture even more cell types than single-cell transcriptomes.98,186 With single nucleus available, scATAC-seq has started its journey in plants.98,187 A recently published paper that profiled histone modification at single-cell resolution means that single-cell epigenomics in plants is accessible.99
The single-cell transcriptome provides an unprecedented perspective on plant development and breeding188; heterogeneity and new regulators have been identified.166,167,168,169,170,173,176 Enrichment and analysis of rare cell types, such as root quiescent center cells, lateral root primordial cells, guard cells, and phloem parenchyma cells, uncovered novel views for the function of these rare cells.171,172,173,174,176,189 Comparing single-cell transcriptomes under different environmental stress, studies found distinct responses among different cell clusters.175 Yang et al. identified key regulators and regulations for the differential response to cytokinin in root and uncovered that root xylem cells act as an organizer by non-autonomous regulation of cytokinin levels.177 Different from animals, plants develop in post-embryonic style with meristemic stem cells dividing and differentiating during the whole lifespan.190 One can capture almost all the cell states in one sample, like those time series studies in animals such as embryogenesis, etc.191,192,193,194,195,196 Tracing the developmental trajectory of specific cell types and illustrating mechanisms of cell fate transition/determination is one of the most outstanding highlights of single-cell studies in plants. With the single-cell transcriptome available, Efroni et al. revealed a rapid identity transition of cells after root tip cutting.197 Using algorithms in the reconstruction of cellular developmental trajectories, studies on root trichoblast differentiation, cortex cell development, and lateral organ initiation, etc., discovered dynamic transcriptional changes and identified regulators involved in the process.166,167,174,180 A cell-by-cell dissection of phloem development conducted by Roszak et al., which combined high-resolution imaging and single-cell omics, illustrated the detailed dynamic maturation gradient for cell specification.198 Spatiotemporal gene expression programs, which correlated with specialized cellular function were encoded by cis-regulatory elements. A cis-regulatory atlas in six maize organs identified cell-type-specific elements as hotspots correlated with selection in modern maize breeding, highlighting the biological meaning of the cis-elements.187 The applications of single-cell omics technology in plant studies are summarized in Figure 8.
Figure 8.
The applications of single-cell sequencing in plant biology
Single-cell multi-omics applications are only beginning to unlock their full potential, particularly for plants. Single-cell transcriptome profiling is still in its early stage and has many exciting challenges to overcome. Due to the continuous development of plant cells, how to define a group of cells as a new cell type or subtype is still under discussion.188 The transcripts detected in one single cell account for a low proportion of the total transcripts, which limits the application of lowly expressed cell-type-specific genes. Furthermore, how to combine information from single cells with cellular function in a biological context is still an open question, despite the recent studies that have started to focus on the dissection of molecular mechanisms utilizing single-cell omics.177,178,198
Plants embrace incredible plasticity. Positional signaling plays a pivotal role in determining cell fate specification/transition in plants.199 The adjacent cells can replace the function of damaged stem cells.199,200 Scientists tried to employ a microcapillary manipulation to gain single cell with positional information,201,202 but it cannot be used widely in high throughput. The appearance of spatial transcriptomes paves another way to dissect gene expression patterns with positional information.203,204 Recently, a stereo-seq-based spatial transcriptome was applied in Arabidopsis leaves, discovering the significant transcriptional differences between upper and lower epidermal cells, which started a new era for plant spatial omics.205
Perspectives of single-cell technologies
Single-cell spatial omics
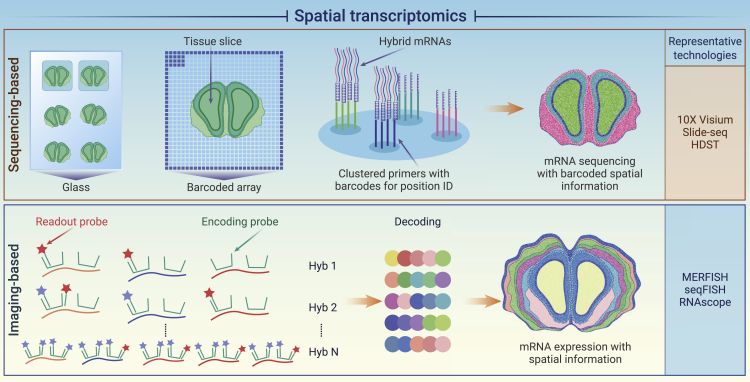
While single-cell omics data are transformative for dissecting the complexities of cell-type composition, they typically start with dissociated cells that lose their physical relationships. Researchers are keen to keep this spatial information to infer how cells are attributed or organized in their native content, which is critical to understanding the fundamental mechanisms underlying embryonic development, neurological science, and tumor heterogeneity. Therefore, spatially resolved single-cell omics technologies are eagerly needed. ST technologies have been developing dramatically over the past years to address these needs.206 Current ST technologies are generally categorized as imaging based and sequencing based, which are often complementary in features, such as spatial resolution and throughput. Here, we briefly summarize major advances in ST technologies and also bring some future perspectives.
The imaging-based approach is originated from FISH technology, which has been used to image selected RNA transcripts in cells, especially with the development of single-molecular FISH to overcome diffraction limitation.207 However, the number of genes that can be simultaneously observed in FISH technology is restricted to the number of distinct fluorescence colors, which presents a major challenge for transcriptome-wide characterizing. We summarize the sequencing-based and imaging-based ST in Figure 9.
Figure 9.
The sequencing-based and imaging-based spatial transcriptomics
One major drawback of imaging-based ST technologies is the throughput, as hybridization and image capturing are all time-consuming. On the other side, high-throughput next-generation sequencing has been applied in numerous biotechnologies. Therefore, it is straightforward to incorporate sequencing methodologies for spatially resolved transcriptomics.
The general principle of sequencing-based ST technologies is to capture and quantify the RNA transcripts in situ by preloading barcoded probes into assigned positions on the surface of glass slides. Tissue slices are then placed on top of the barcoded probes. RNAs in the cells are then released and hybridized in situ with the probes. Sequencing was performed directly on these glass slides. RNA species and the barcodes could be read out from the sequencing reads, and the spatial locations could be decoded with the barcodes.
One major consideration for sequencing-based ST is the resolution that is restricted by the density of the surface probes. In the first generation of the sequencing-based ST developed in 2016,36 the diameter of each spot on the glass slide was around 100 μm, which covered multiple cells to provide averaged transcriptomics of these cells. To push the spatial resolution into the single-cell level, an alternative barcode deposition strategy was developed to reduce the diameter of the probe spots by using densely packed beads. Slide-seq uses 10 μm beads that generally cover one to three cells with decent capture efficiency compared with scRNA-seq.208 In the meanwhile, HDST further improves the resolution by using 2 μm beads and a sequential hybridization strategy, which can provide subcellular resolution.209
ST technologies are continuously evolving in multiple directions to address a broad range of biological questions. To fully take advantage of the spatial information from ST technologies, specialized data analysis algorithms and computational tools should be developed since the ST datasets have unique structures in terms of data complexities, such as imaging, sequencing, and electrophysiology data. Most current ST technologies are based on 2D cross-sectional imaging. By integrating multiple 2D data, we may get the real 3D spatial data. Another direction is to incorporate extra modalities into current transcriptomics-centered spatial technologies, such as genomics, epigenomics, proteomics, and metabolomics. The third one is to consider the dynamics of 3D spatial evolving. Time course 3D spatial single-cell multi-omics data will generate more insights regarding gene regulation, developmental trajectory, and tumor progression.
Single-cell multi-omics
The advancement of scRNA-seq technologies has profoundly altered molecular mechanism research. Proteomics awaits similar progress in protein sequencing techniques, such as single-molecule mass spectrometry, DNA nanotechnologies for protein sequencing, and fingerprinting linearized proteins, which will allow for the analysis of proteins at the single-cell level, even with low-abundance proteins.210 With single-cell multi-omics, a comprehensive view of single cells can be expected (Figure 10). We can validate the cell type identified from one omics with another omics. This will significantly improve the cell-type annotation accuracy and help discover new cell types or new mechanisms.211
Figure 10.
The single-cell multi-omics studies
From the technology perspective of single-cell multi-omics, Guilliams et al. did integrative analysis of single-cell CITE-seq (cellular indexing of transcriptomes and epitopes by sequencing), single-nuclei sequencing, ST, and spatial proteomics to uncover the cellular niches of hepatic macrophages in murine and human liver.212 The single-cell multi-omics and spatial multi-omics data unraveled the cell-cell microenvironmental circuits essential for lipid-associated macrophages (LAMs).212 Perturb-seq has been used in mammalian cells to assist in the screening of individual gene targets, gene signatures, and their genetic interactions.213 Mimitou et al. developed ECCITE-seq (expanded CRISPR-compatible CITE-seq), which can simultaneously detect transcriptome, proteins, clonotypes, and CRISPR perturbations from single cells214 and, later, they developed DOGMA-seq, an adaptation of CITE-seq for measuring gene activity across the central dogma of gene regulation.215 DOGMA-seq showed great values in studying lineage-specific T helper cells.216
From the application perspective of single-cell multi-omics, Bian et al. investigated the complex multi-level dysfunctions in human colorectal cancer using scTrio-seq2 (single-cell triple omics sequencing), which measures CNV, DNA methylation, and transcriptome simultaneously in one cell.217 In immunology, Stephenson et al. revealed the immune response in COVID-19 patients by multi-omics analysis of the single-cell transcriptome, cell-surface protein, and lymphocyte antigen receptor repertoire in peripheral blood.218 In reproductive and developmental medicine, Yan et al. decoded the dynamic epigenetic landscapes in human oocytes using scChaRM-seq (single-cell chromatin accessibility, RNA barcoding, and DNA methylation sequencing), which can measure single-cell ATAC, RNA, and DNA methylation.219
We believe more and more researchers will apply single-cell technologies to their studies. It will be the era of the single cell in which new cell types and new molecular mechanisms will be discovered, just as new species and their evolution paths were discovered in the era of Darwin.
Acknowledgments
This work was supported by the Strategic Priority Research Program of Chinese Academy of Sciences (XDA26040304, XDB38050200), the National Natural Science Foundation of China (82102182, 31961133010, 31970805), Jinfeng Laboratory, Chongqing, China (jfkyjf202203001) and The Youth Innovation Promotion Association of Chinese Academy of Sciences (2017139).
Author contributions
Z.C. and T.H. conceived and designed the research. L.W. wrote the “introduction.” D.Z. and X.R. wrote the “bioinformatics analysis of single-cell sequencing data” section. W.G., H.P., W.S., J.C., P.Z., W.Z., Y.Z., and N.J. wrote the “single-cell technologies” section. P.Z., N.Z., and X.C. wrote the “single-cell sequencing in oncology and immunology” section. J.Y., R.H., and G.T wrote the “single-cell sequencing in assisted reproduction” section. M.Z. and G.P. wrote the “single-cell sequencing for embryonic development” section. X.Z. and C.T. wrote the “single-cell sequencing in plant breeding” section. G.L. wrote the “single-cell spatial omics” section. C.T. and T.H. wrote the “single-cell multi-omics” section.
Declaration of interests
The authors declare no competing interests.
Published Online: October 18, 2022
Contributor Information
Yunlong Zhao, Email: yunlong.zhao@surrey.ac.uk.
Xin Cao, Email: caox@fudan.edu.cn.
Guangdun Peng, Email: peng_guangdun@gibh.ac.cn.
Xianwen Ren, Email: renxwise@pku.edu.cn.
Nan Jiang, Email: jiangnansophia@scu.edu.cn.
Caihuan Tian, Email: tiancaihuan@caas.cn.
Zi-Jiang Chen, Email: chenzijiang@hotmail.com.
Lead contact website
References
- 1.International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 2.Tang F., Barbacioru C., Wang Y., et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 3.Islam S., Kjällquist U., Moliner A., et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein A.M., Mazutis L., Akartuna I., et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macosko E.Z., Basu A., Satija R., et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan H.C., Fu G.K., Fodor S.P.A. Expression profiling. Combinatorial labeling of single cells for gene expression cytometry. Science. 2015;347 doi: 10.1126/science.1258367. [DOI] [PubMed] [Google Scholar]
- 7.Rosenberg A.B., Roco C.M., Muscat R.A., et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018;360:176–182. doi: 10.1126/science.aam8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao J., Spielmann M., Qiu X., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo H., Zhu P., Wu X., et al. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 2013;23:2126–2135. doi: 10.1101/gr.161679.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagano T., Lubling Y., Stevens T.J., et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buenrostro J.D., Wu B., Litzenburger U.M., et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cusanovich D.A., Daza R., Adey A., et al. Epigenetics. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cusanovich D.A., Hill A.J., Aghamirzaie D., et al. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell. 2018;174:1309–1324.e18. doi: 10.1016/j.cell.2018.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Satpathy A.T., Granja J.M., Yost K.E., et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 2019;37:925–936. doi: 10.1038/s41587-019-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preissl S., Fang R., Huang H., et al. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat. Neurosci. 2018;21:432–439. doi: 10.1038/s41593-018-0079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lake B.B., Chen S., Sos B.C., et al. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol. 2018;36:70–80. doi: 10.1038/nbt.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navin N., Kendall J., Troge J., et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zong C., Lu S., Chapman A.R., Xie X.S. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C., Xing D., Tan L., et al. Single-cell whole-genome analyses by linear amplification via transposon insertion (LIANTI) Science. 2017;356:189–194. doi: 10.1126/science.aak9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xing D., Tan L., Chang C.H., et al. Accurate SNV detection in single cells by transposon-based whole-genome amplification of complementary strands. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2013106118. e2013106118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan L., Xing D., Chang C.H., et al. Three-dimensional genome structures of single diploid human cells. Science. 2018;361:924–928. doi: 10.1126/science.aat5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan X., Yang C., Li W., et al. SMOOTH-seq: single-cell genome sequencing of human cells on a third-generation sequencing platform. Genome Biol. 2021;22:195. doi: 10.1186/s13059-021-02406-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan X., Tang D., Liao Y., et al. Single-cell RNA-seq analysis of mouse preimplantation embryos by third-generation sequencing. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dey S.S., Kester L., Spanjaard B., et al. Integrated genome and transcriptome sequencing of the same cell. Nat. Biotechnol. 2015;33:285–289. doi: 10.1038/nbt.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macaulay I.C., Haerty W., Kumar P., et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat. Methods. 2015;12:519–522. doi: 10.1038/nmeth.3370. [DOI] [PubMed] [Google Scholar]
- 26.Angermueller C., Clark S.J., Lee H.J., et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat. Methods. 2016;13:229–232. doi: 10.1038/nmeth.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou Y., Guo H., Cao C., et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016;26:304–319. doi: 10.1038/cr.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y., Huang K., An Q., et al. Simultaneous profiling of transcriptome and DNA methylome from a single cell. Genome Biol. 2016;17:88. doi: 10.1186/s13059-016-0950-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma S., Zhang B., LaFave L.M., et al. Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell. 2020;183:1103–1116.e20. doi: 10.1016/j.cell.2020.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu C., Yu M., Huang H., et al. An ultra high-throughput method for single-cell joint analysis of open chromatin and transcriptome. Nat. Struct. Mol. Biol. 2019;26:1063–1070. doi: 10.1038/s41594-019-0323-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen S., Lake B.B., Zhang K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat. Biotechnol. 2019;37:1452–1457. doi: 10.1038/s41587-019-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao J., Cusanovich D.A., Ramani V., et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science. 2018;361:1380–1385. doi: 10.1126/science.aau0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lubeck E., Coskun A.F., Zhiyentayev T., et al. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen K.H., Boettiger A.N., Moffitt J.R., et al. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348 doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ke R., Mignardi M., Pacureanu A., et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods. 2013;10:857–860. doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- 36.Ståhl P.L., Salmén F., Vickovic S., et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 37.Kim B.J., Cho H., Park J.H., et al. Strategic Advances in Formation of Cell-In-Shell Structures: From Syntheses to Applications. Adv. Mater. 2018;30 doi: 10.1002/adma.201706063. [DOI] [PubMed] [Google Scholar]
- 38.Liu Z., Xu X., Tang R. Improvement of biological organisms using functional. Adv. Funct. Mater. 2016;26:1862–1880. [Google Scholar]
- 39.Park J.H., Hong D., Lee J., Choi I.S. Cell-in-Shell hybrids: chemical nanoencapsulation of individual cells. Acc. Chem. Res. 2016;49:792–800. doi: 10.1021/acs.accounts.6b00087. [DOI] [PubMed] [Google Scholar]
- 40.Zhu W., Guo J., Amini S., et al. SupraCells: living mammalian cells protected within functional modular nanoparticle-based exoskeletons. Adv. Mater. 2019;31 doi: 10.1002/adma.201900545. [DOI] [PubMed] [Google Scholar]
- 41.Yang J., Yang Y., Kawazoe N., Chen G. Encapsulation of individual living cells with enzyme responsive polymer nanoshell. Biomaterials. 2019;197:317–326. doi: 10.1016/j.biomaterials.2019.01.029. [DOI] [PubMed] [Google Scholar]
- 42.Ryu S., Kim H., Kang S., et al. Reversible cell layering for heterogeneous cell assembly mediated by ionic cross-linking of chitosan and a functionalized cell surface membrane. Chem. Mater. 2017;29:5294–5305. [Google Scholar]
- 43.Shi P., Zhao N., Coyne J., Wang Y. DNA-templated synthesis of biomimetic cell wall for nanoencapsulation and protection of mammalian cells. Nat. Commun. 2019;10:2223. doi: 10.1038/s41467-019-10231-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo J., Suástegui M., Sakimoto K.K., et al. Light-driven fine chemical production in yeast biohybrids. Science (New York, N.Y.) 2018;362:813–816. doi: 10.1126/science.aat9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakimoto K.K., Wong A.B., Yang P. Self-photosensitization of nonphotosynthetic bacteria for solar-to-chemical production. Science (New York, N.Y.) 2016;351:74–77. doi: 10.1126/science.aad3317. [DOI] [PubMed] [Google Scholar]
- 46.Li W., Liu Z., Liu C., et al. Manganese dioxide nanozymes as responsive cytoprotective shells for individual living cell encapsulation. Angew. Chem. Int. Ed. Engl. 2017;56:13661–13665. doi: 10.1002/anie.201706910. [DOI] [PubMed] [Google Scholar]
- 47.Liang K., Richardson J.J., Cui J., et al. Metal-organic framework coatings as cytoprotective exoskeletons for living cells. Adv. Mater. 2016;28:7910–7914. doi: 10.1002/adma.201602335. [DOI] [PubMed] [Google Scholar]
- 48.Jiang N., Yang X.Y., Ying G.L., et al. Self-repairing" nanoshell for cell protection. Chem. Sci. 2015;6:486–491. doi: 10.1039/c4sc02638a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geng W., Jiang N., Qing G.Y., et al. Click reaction for reversible encapsulation of single yeast cells. ACS Nano. 2019;13:14459–14467. doi: 10.1021/acsnano.9b08108. [DOI] [PubMed] [Google Scholar]
- 50.Kahana A., Lancet D. Self-reproducing catalytic micelles as nanoscopic protocell precursors. Nat. Rev. Chem. 2021;5:870–878. doi: 10.1038/s41570-021-00329-7. [DOI] [PubMed] [Google Scholar]
- 51.Chen R., Canales A., Anikeeva P. Neural recording and modulation technologies. Nat. Rev. Mater. 2017;2:16093. doi: 10.1038/natrevmats.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamill O.P., Marty A., Neher E., et al. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 53.Shmoel N., Rabieh N., Ojovan S.M., et al. Multisite electrophysiological recordings by self-assembled loose-patch-like junctions between cultured hippocampal neurons and mushroom-shaped microelectrodes. Sci. Rep. 2016;6 doi: 10.1038/srep27110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spira M.E., Hai A. Multi-electrode array technologies for neuroscience and cardiology. Nat. Nanotechnol. 2013;8:83–94. doi: 10.1038/nnano.2012.265. [DOI] [PubMed] [Google Scholar]
- 55.McGuire A.F., Santoro F., Cui B. Interfacing cells with vertical nanoscale devices: applications and characterization. Annu. Rev. Anal. Chem. 2018;11:101–126. doi: 10.1146/annurev-anchem-061417-125705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abbott J., Ye T., Ham D., Park H. Optimizing nanoelectrode arrays for scalable intracellular electrophysiology. Acc. Chem. Res. 2018;51:600–608. doi: 10.1021/acs.accounts.7b00519. [DOI] [PubMed] [Google Scholar]
- 57.Tian B., Cohen-Karni T., Qing Q., et al. Three-dimensional, flexible nanoscale field-effect transistors as localized bioprobes. Science (New York, N.Y.) 2010;329:830–834. doi: 10.1126/science.1192033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao Y., You S.S., Zhang A., et al. Scalable ultrasmall three-dimensional nanowire transistor probes for intracellular recording. Nat. Nanotechnol. 2019;14:783–790. doi: 10.1038/s41565-019-0478-y. [DOI] [PubMed] [Google Scholar]
- 59.Zhao Y., Yao J., Xu L., et al. Shape-controlled deterministic assembly of nanowires. Nano Lett. 2016;16:2644–2650. doi: 10.1021/acs.nanolett.6b00292. [DOI] [PubMed] [Google Scholar]
- 60.Gu Y., Wang C., Kim N., et al. Three-dimensional transistor arrays for intra- and inter-cellular recording. Nat. Nanotechnol. 2022;17:292–300. doi: 10.1038/s41565-021-01040-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang A., Zhao Y., You S.S., Lieber C.M. Nanowire probes could drive high-resolution brain-machine interfaces. Nano Today. 2020;31 [Google Scholar]
- 62.Gierahn T.M., Wadsworth M.H., Hughes T.K., et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods. 2017;14:395–398. doi: 10.1038/nmeth.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brennecke P., Anders S., Kim J.K., et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat. Methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- 64.Jaitin D.A., Kenigsberg E., Keren-Shaul H., et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klein A.M., Mazutis L., Akartuna I., et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bose S., Wan Z., Carr A., et al. Scalable microfluidics for single-cell RNA printing and sequencing. Genome Biol. 2015;16:120. doi: 10.1186/s13059-015-0684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fan H.C., Fu G.K., Fodor S.P.A. Combinatorial labeling of single cells for gene expression cytometry. Science. 2015;347 doi: 10.1126/science.1258367. [DOI] [PubMed] [Google Scholar]
- 68.Guo M.T., Rotem A., Heyman J.A., Weitz D.A. Droplet microfluidics for high-throughput biological assays. Lab Chip. 2012;12:2146–2155. doi: 10.1039/c2lc21147e. [DOI] [PubMed] [Google Scholar]
- 69.Di Carlo D., Wu L.Y., Lee L.P. Dynamic single cell culture array. Lab Chip. 2006;6:1445–1449. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- 70.Xia H.M., Wu J.W., Zheng J.J., et al. Nonlinear microfluidics: device physics, functions, and applications. Lab Chip. 2021;21:1241–1268. doi: 10.1039/d0lc01120g. [DOI] [PubMed] [Google Scholar]
- 71.Brouzes E., Medkova M., Savenelli N., et al. Droplet microfluidic technology for single-cell high-throughput screening. Proc. Natl. Acad. Sci. USA. 2009;106:14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Muraro M.J., Dharmadhikari G., Grün D., et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 2016;3:385–394.e3. doi: 10.1016/j.cels.2016.09.002. e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y.J., Schug J., Lin J., et al. Comparative analysis of commercially available single-cell RNA sequencing platforms for their performance in complex human tissues. bioRxiv. 2019 doi: 10.1101/541433. Preprint at. [DOI] [Google Scholar]
- 74.Gao C., Zhang M., Chen L. The comparison of two single-cell sequencing platforms: BD Rhapsody and 10x Genomics Chromium. Curr. Genomics. 2020;21:602–609. doi: 10.2174/1389202921999200625220812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gong H., Do D., Ramakrishnan R. In: Gene Expression Analysis: Methods and Protocols. Raghavachari N., Garcia-Reyero N., editors. Springer New York; 2018. Single-cell mRNA-seq using the Fluidigm C1 system and integrated fluidics circuits; pp. 193–207. [DOI] [PubMed] [Google Scholar]
- 76.Zhao X., Hu J., Li Y., Guo M. Volumetric compression develops noise-driven single-cell heterogeneity. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2110550118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luecken M.D., Theis F.J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 2019;15 doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zappia L., Theis F.J. Over 1000 tools reveal trends in the single-cell RNA-seq analysis landscape. Genome Biol. 2021;22:301. doi: 10.1186/s13059-021-02519-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haghverdi L., Lun A.T.L., Morgan M.D., Marioni J.C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 2018;36:421–427. doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Butler A., Hoffman P., Smibert P., et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu Y., Liu X., Cao X., et al. Artificial intelligence: a powerful paradigm for scientific research. Innovation. 2021;2 doi: 10.1016/j.xinn.2021.100179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aran D., Looney A.P., Liu L., et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang A.W., O'Flanagan C., Chavez E.A., et al. Probabilistic cell-type assignment of single-cell RNA-seq for tumor microenvironment profiling. Nat. Methods. 2019;16:1007–1015. doi: 10.1038/s41592-019-0529-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Qiu X., Mao Q., Tang Y., et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.La Manno G., Soldatov R., Zeisel A., et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van de Sande B., Flerin C., Davie K., et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc. 2020;15:2247–2276. doi: 10.1038/s41596-020-0336-2. [DOI] [PubMed] [Google Scholar]
- 87.Browaeys R., Saelens W., Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods. 2020;17:159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 88.Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 89.Ren X., Zhong G., Zhang Q., et al. Reconstruction of cell spatial organization from single-cell RNA sequencing data based on ligand-receptor mediated self-assembly. Cell Res. 2020;30:763–778. doi: 10.1038/s41422-020-0353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moriel N., Senel E., Friedman N., et al. NovoSpaRc: flexible spatial reconstruction of single-cell gene expression with optimal transport. Nat. Protoc. 2021;16:4177–4200. doi: 10.1038/s41596-021-00573-7. [DOI] [PubMed] [Google Scholar]
- 91.Regev A., Teichmann S.A., Lander E.S., et al. Vol. 6. eLife; 2017. (The Human Cell Atlas). [Google Scholar]
- 92.Roy A.L., Sierra F., Howcroft K., et al. A blueprint for characterizing senescence. Cell. 2020;183:1143–1146. doi: 10.1016/j.cell.2020.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rozenblatt-Rosen O., Regev A., Oberdoerffer P., et al. Human Tumor Atlas Network The human tumor atlas network: charting tumor transitions across space and time at single-cell resolution. Cell. 2020;181:236–249. doi: 10.1016/j.cell.2020.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Han X., Wang R., Zhou Y., et al. Mapping the mouse cell atlas by microwell-seq. Cell. 2018;173:1091–1107. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 95.Rhee S.Y., Birnbaum K.D., Ehrhardt D.W. Towards building a plant cell atlas. Trends Plant Sci. 2019;24:303–310. doi: 10.1016/j.tplants.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jha S.G., Borowsky A.T., Cole B.J., et al. Vision, Challenges and Opportunities for a Plant Cell Atlas. Elife. 2021;10 doi: 10.7554/eLife.66877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Birnbaum K.D., Otegui M.S., Bailey-Serres J., Rhee S.Y. The Plant Cell Atlas: focusing new technologies on the kingdom that nourishes the planet. Plant Physiol. 2022;188:675–679. doi: 10.1093/plphys/kiab584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Farmer A., Thibivilliers S., Ryu K.H., et al. Single-nucleus RNA and ATAC sequencing reveals the impact of chromatin accessibility on gene expression in Arabidopsis roots at the single-cell level. Mol. Plant. 2021;14:372–383. doi: 10.1016/j.molp.2021.01.001. [DOI] [PubMed] [Google Scholar]
- 99.Ouyang W., Luan S., Xiang X., et al. Profiling plant histone modification at single-cell resolution using snCUT&Tag. Plant Biotechnol. J. 2022;20:420–422. doi: 10.1111/pbi.13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thibivilliers S., Libault M. Plant single-cell multiomics: cracking the molecular profiles of plant cells. Trends Plant Sci. 2021;26:662–663. doi: 10.1016/j.tplants.2021.03.001. [DOI] [PubMed] [Google Scholar]
- 101.de Souza L.P., Borghi M., Fernie A. Plant single-cell metabolomics-challenges and perspectives. Int. J. Mol. Sci. 2020;21:E8987. doi: 10.3390/ijms21238987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dai S., Chen S. Single-cell-type proteomics: toward a holistic understanding of plant function. Mol. Cell. Proteomics. 2012;11:1622–1630. doi: 10.1074/mcp.R112.021550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Misra B.B., Assmann S.M., Chen S. Plant single-cell and single-cell-type metabolomics. Trends Plant Sci. 2014;19:637–646. doi: 10.1016/j.tplants.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 104.Fujii T., Matsuda S., Tejedor M.L., et al. Direct metabolomics for plant cells by live single-cell mass spectrometry. Nat. Protoc. 2015;10:1445–1456. doi: 10.1038/nprot.2015.084. [DOI] [PubMed] [Google Scholar]
- 105.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 106.El-Sayes N., Vito A., Mossman K. Tumor heterogeneity: a great barrier in the age of cancer immunotherapy. Cancers. 2021;13:806. doi: 10.3390/cancers13040806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lenz G., Onzi G.R., Lenz L.S., et al. The origins of phenotypic heterogeneity in cancer. Cancer Res. 2022;82:3–11. doi: 10.1158/0008-5472.CAN-21-1940. [DOI] [PubMed] [Google Scholar]
- 108.Magee J.A., Piskounova E., Morrison S.J. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21:283–296. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xie S., Leung A.W.S., Zheng Z., et al. Applications and potentials of nanopore sequencing in the (epi)genome and (epi)transcriptome era. Innovation. 2021;2 doi: 10.1016/j.xinn.2021.100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhou L., Yu K.H., Wong T.L., et al. Lineage tracing and single-cell analysis reveal proliferative Prom1+ tumour-propagating cells and their dynamic cellular transition during liver cancer progression. Gut. 2022;71:1656–1668. doi: 10.1136/gutjnl-2021-324321. [DOI] [PubMed] [Google Scholar]
- 111.Olbrecht S., Busschaert P., Qian J., et al. High-grade serous tubo-ovarian cancer refined with single-cell RNA sequencing: specific cell subtypes influence survival and determine molecular subtype classification. Genome Med. 2021;13:111. doi: 10.1186/s13073-021-00922-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang Y., Wang D., Peng M., et al. Single-cell RNA sequencing in cancer research. J. Exp. Clin. Cancer Res. 2021;40:81. doi: 10.1186/s13046-021-01874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.The Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu K., Gao R., Wu H., et al. Single-cell analysis reveals metastatic cell heterogeneity in clear cell renal cell carcinoma. J. Cell Mol. Med. 2021;25:4260–4274. doi: 10.1111/jcmm.16479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen J., Song Y., Li M., et al. Comprehensive analysis of ceRNA networks reveals prognostic lncRNAs related to immune infiltration in colorectal cancer. BMC Cancer. 2021;21:255. doi: 10.1186/s12885-021-07995-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wouters J., Kalender-Atak Z., Minnoye L., et al. Robust gene expression programs underlie recurrent cell states and phenotype switching in melanoma. Nat. Cell Biol. 2020;22:986–998. doi: 10.1038/s41556-020-0547-3. [DOI] [PubMed] [Google Scholar]
- 117.Zhou S., Huang Y.E., Liu H., et al. Single-cell RNA-seq dissects the intratumoral heterogeneity of triple-negative breast cancer based on gene regulatory networks. Mol. Ther. Nucleic Acids. 2021;23:682–690. doi: 10.1016/j.omtn.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wu Y., Yang S., Ma J., et al. Spatiotemporal immune landscape of colorectal cancer liver metastasis at single-cell level. Cancer Discov. 2022;12:134–153. doi: 10.1158/2159-8290.CD-21-0316. [DOI] [PubMed] [Google Scholar]
- 119.Yuan X., Wang J., Huang Y., et al. Single-cell profiling to explore immunological heterogeneity of tumor microenvironment in breast cancer. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.643692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang Y., Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell. Mol. Immunol. 2020;17:807–821. doi: 10.1038/s41423-020-0488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Suvà M.L., Tirosh I. Single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol. Cell. 2019;75:7–12. doi: 10.1016/j.molcel.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 122.Ren X., Zhang L., Zhang Y., et al. Insights gained from single-cell analysis of immune cells in the tumor microenvironment. Annu. Rev. Immunol. 2021;39:583–609. doi: 10.1146/annurev-immunol-110519-071134. [DOI] [PubMed] [Google Scholar]
- 123.Cassetta L., Pollard J.W. Targeting macrophages: therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018;17:887–904. doi: 10.1038/nrd.2018.169. [DOI] [PubMed] [Google Scholar]
- 124.Song G., Shi Y., Zhang M., et al. Global immune characterization of HBV/HCV-related hepatocellular carcinoma identifies macrophage and T-cell subsets associated with disease progression. Cell Discov. 2020;6:90. doi: 10.1038/s41421-020-00214-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sun Y., Wu L., Zhong Y., et al. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell. 2021;184:404–421.e16. doi: 10.1016/j.cell.2020.11.041. [DOI] [PubMed] [Google Scholar]
- 126.Hong M., Clubb J.D., Chen Y.Y. Engineering CAR-T cells for next-generation cancer therapy. Cancer Cell. 2020;38:473–488. doi: 10.1016/j.ccell.2020.07.005. [DOI] [PubMed] [Google Scholar]
- 127.Carson S.A., Kallen A.N. Diagnosis and management of infertility: a review. JAMA. 2021;326:65–76. doi: 10.1001/jama.2021.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zorrilla M., Yatsenko A.N. The genetics of infertility: current status of the field. Curr. Genet. Med. Rep. 2013;1:247–260. doi: 10.1007/s40142-013-0027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.De Geyter C. Assisted reproductive technology: impact on society and need for surveillance. Best practice & research. Best Pract. Res. Clin. Endocrinol. Metab. 2019;33:3–8. doi: 10.1016/j.beem.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 130.Chen M., Norman R.J., Heilbronn L.K. Does in vitro fertilisation increase type 2 diabetes and cardiovascular risk? Curr. Diabetes Rev. 2011;7:426–432. doi: 10.2174/157339911797579151. [DOI] [PubMed] [Google Scholar]
- 131.Kimelman D., Pavone M.E. Non-invasive prenatal testing in the context of IVF and PGT-A. Best practice & research. Best Pract. Res. Clin. Obstet. Gynaecol. 2021;70:51–62. doi: 10.1016/j.bpobgyn.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 132.Wang Y., Liu Q., Tang F., et al. Epigenetic regulation and risk factors during the development of human gametes and early embryos. Annu. Rev. Genomics Hum. Genet. 2019;20:21–40. doi: 10.1146/annurev-genom-083118-015143. [DOI] [PubMed] [Google Scholar]
- 133.Han X., Zhou Z., Fei L., et al. Construction of a human cell landscape at single-cell level. Nature. 2020;581:303–309. doi: 10.1038/s41586-020-2157-4. [DOI] [PubMed] [Google Scholar]
- 134.Consortium T.T.S., Quake S.R. The Tabula Sapiens: A Multiple Organ Single Cell Transcriptomic Atlas of Humans. bioRxiv. 2021 doi: 10.1101/2021.07.19.452956. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang W., Vilella F., Alama P., et al. Single-cell transcriptomic atlas of the human endometrium during the menstrual cycle. Nat. Med. 2020;26:1644–1653. doi: 10.1038/s41591-020-1040-z. [DOI] [PubMed] [Google Scholar]
- 136.Fujimoto T., Miyayama Y., Fuyuta M. The origin, migration and fine morphology of human primordial germ cells. Anat. Rec. 1977;188:315–330. doi: 10.1002/ar.1091880305. [DOI] [PubMed] [Google Scholar]
- 137.Guo F., Yan L., Guo H., et al. The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell. 2015;161:1437–1452. doi: 10.1016/j.cell.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 138.Jankovičová J., Neuerová Z., Sečová P., et al. Tetraspanins in mammalian reproduction: spermatozoa, oocytes and embryos. Med. Microbiol. Immunol. 2020;209:407–425. doi: 10.1007/s00430-020-00676-0. [DOI] [PubMed] [Google Scholar]
- 139.Ottolenghi C., Uda M., Hamatani T., et al. Aging of oocyte, ovary, and human reproduction. Ann. N. Y. Acad. Sci. 2004;1034:117–131. doi: 10.1196/annals.1335.015. [DOI] [PubMed] [Google Scholar]
- 140.Wang S., Zheng Y., Li J., et al. Single-cell transcriptomic atlas of primate ovarian aging. Cell. 2020;180:585–600.e19. doi: 10.1016/j.cell.2020.01.009. [DOI] [PubMed] [Google Scholar]
- 141.Fan X., Bialecka M., Moustakas I., et al. Single-cell reconstruction of follicular remodeling in the human adult ovary. Nat. Commun. 2019;10:3164. doi: 10.1038/s41467-019-11036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wagner M., Yoshihara M., Douagi I., et al. Single-cell analysis of human ovarian cortex identifies distinct cell populations but no oogonial stem cells. Nat. Commun. 2020;11:1147. doi: 10.1038/s41467-020-14936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen W., Guillaume-Gentil O., Rainer P.Y., et al. Live-seq enables temporal transcriptomic recording of single cells. Nature. 2022;608:733–740. doi: 10.1038/s41586-022-05046-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yan L., Yang M., Guo H., et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 145.Xue Z., Huang K., Cai C., et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500:593–597. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Guo H., Zhu P., Yan L., et al. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- 147.Blakeley P., Fogarty N.M.E., del Valle I., et al. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development (Cambridge, England) 2015;142:3151–3165. doi: 10.1242/dev.123547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Petropoulos S., Edsgärd D., Reinius B., et al. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell. 2016;165:1012–1026. doi: 10.1016/j.cell.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhu P., Guo H., Ren Y., et al. Single-cell DNA methylome sequencing of human preimplantation embryos. Nat. Genet. 2018;50:12–19. doi: 10.1038/s41588-017-0007-6. [DOI] [PubMed] [Google Scholar]
- 150.Guo F., Li L., Li J., et al. Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res. 2017;27:967–988. doi: 10.1038/cr.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li L., Guo F., Gao Y., et al. Single-cell multi-omics sequencing of human early embryos. Nat. Cell Biol. 2018;20:847–858. doi: 10.1038/s41556-018-0123-2. [DOI] [PubMed] [Google Scholar]
- 152.Meistermann D., Bruneau A., Loubersac S., et al. Integrated pseudotime analysis of human pre-implantation embryo single-cell transcriptomes reveals the dynamics of lineage specification. Cell Stem Cell. 2021;28:1625–1640.e6. doi: 10.1016/j.stem.2021.04.027. [DOI] [PubMed] [Google Scholar]
- 153.Wang Y., Yuan P., Yan Z., et al. Single-cell multiomics sequencing reveals the functional regulatory landscape of early embryos. Nat. Commun. 2021;12:1247. doi: 10.1038/s41467-021-21409-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Vento-Tormo R., Efremova M., Botting R.A., et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang S., Cui Y., Ma X., et al. Single-cell transcriptomics identifies divergent developmental lineage trajectories during human pituitary development. Nat. Commun. 2020;11:5275. doi: 10.1038/s41467-020-19012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cheng S., Pei Y., He L., et al. Single-cell RNA-seq reveals cellular heterogeneity of pluripotency transition and X chromosome dynamics during early mouse development. Cell Rep. 2019;26:2593–2607.e3. doi: 10.1016/j.celrep.2019.02.031. [DOI] [PubMed] [Google Scholar]
- 157.Argelaguet R., Clark S.J., Mohammed H., et al. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature. 2019;576:487–491. doi: 10.1038/s41586-019-1825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pijuan-Sala B., Griffiths J.A., Guibentif C., et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature. 2019;566:490–495. doi: 10.1038/s41586-019-0933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hu Y., An Q., Sheu K., et al. Single cell multi-omics technology: methodology and application. Front. Cell Dev. Biol. 2018;6:28. doi: 10.3389/fcell.2018.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jaitin D.A., Kenigsberg E., Keren-Shaul H., et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cusanovich D.A., Daza R., Adey A., et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Han Y., Chu X., Yu H., et al. Single-cell transcriptome analysis reveals widespread monoallelic gene expression in individual rice mesophyll cells. Sci. Bull. 2017;62:1304–1314. doi: 10.1016/j.scib.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 163.Lieckfeldt E., Simon-Rosin U., Kose F., et al. Gene expression profiling of single epidermal, basal and trichome cells of Arabidopsis thaliana. J. Plant Physiol. 2008;165:1530–1544. doi: 10.1016/j.jplph.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 164.Efroni I., Ip P.L., Nawy T., et al. Quantification of cell identity from single-cell gene expression profiles. Genome Biol. 2015;16:9. doi: 10.1186/s13059-015-0580-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nelms B., Walbot V. Defining the developmental program leading to meiosis in maize. Science. 2019;364:52–56. doi: 10.1126/science.aav6428. [DOI] [PubMed] [Google Scholar]
- 166.Denyer T., Ma X., Klesen S., et al. Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev. Cell. 2019;48:840–852.e5. doi: 10.1016/j.devcel.2019.02.022. [DOI] [PubMed] [Google Scholar]
- 167.Jean-Baptiste K., McFaline-Figueroa J.L., Alexandre C.M., et al. Dynamics of gene expression in single root cells of A. thaliana. Plant Cell. 2019;31:993–1011. doi: 10.1105/tpc.18.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhang T.Q., Xu Z.G., Shang G.D., Wang J.W. A single-cell RNA sequencing profiles the developmental landscape of Arabidopsis root. Mol. Plant. 2019;12:648–660. doi: 10.1016/j.molp.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 169.Ryu K.H., Huang L., Kang H.M., Schiefelbein J. Single-cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiol. 2019;179:1444–1456. doi: 10.1104/pp.18.01482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Shulse C.N., Cole B.J., Ciobanu D., et al. High-throughput single-cell transcriptome profiling of plant cell types. Cell Rep. 2019;27:2241–2247.e4. doi: 10.1016/j.celrep.2019.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Satterlee J.W., Strable J., Scanlon M.J. Plant stem-cell organization and differentiation at single-cell resolution. Proc. Natl. Acad. Sci. USA. 2020;117:33689–33699. doi: 10.1073/pnas.2018788117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bezrutczyk M., Zöllner N.R., Kruse C.P.S., et al. Evidence for phloem loading via the abaxial bundle sheath cells in maize leaves. Plant Cell. 2021;33:531–547. doi: 10.1093/plcell/koaa055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kim J.Y., Symeonidi E., Pang T.Y., et al. Distinct identities of leaf phloem cells revealed by single cell transcriptomics. Plant Cell. 2021;33:511–530. doi: 10.1093/plcell/koaa060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lopez-Anido C.B., Vatén A., Smoot N.K., et al. Single-cell resolution of lineage trajectories in the Arabidopsis stomatal lineage and developing leaf. Dev. Cell. 2021;56:1043–1055.e4. doi: 10.1016/j.devcel.2021.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang Y., Huan Q., Li K., Qian W. Single-cell transcriptome atlas of the leaf and root of rice seedlings. J Genet Genomics. 2021;48:881–898. doi: 10.1016/j.jgg.2021.06.001. [DOI] [PubMed] [Google Scholar]
- 176.Xu X., Crow M., Rice B.R., et al. Single-cell RNA sequencing of developing maize ears facilitates functional analysis and trait candidate gene discovery. Dev. Cell. 2021;56:557–568.e6. doi: 10.1016/j.devcel.2020.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yang B., Minne M., Brunoni F., et al. Non-cell autonomous and spatiotemporal signalling from a tissue organizer orchestrates root vascular development. Nat. Plants. 2021;7:1485–1494. doi: 10.1038/s41477-021-01017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhai N., Xu L. Pluripotency acquisition in the middle cell layer of callus is required for organ regeneration. Nat. Plants. 2021;7:1453–1460. doi: 10.1038/s41477-021-01015-8. [DOI] [PubMed] [Google Scholar]
- 179.Zhang T.Q., Chen Y., Liu Y., et al. Single-cell transcriptome atlas and chromatin accessibility landscape reveal differentiation trajectories in the rice root. Nat. Commun. 2021;12:2053. doi: 10.1038/s41467-021-22352-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhang T.Q., Chen Y., Wang J.W. A single-cell analysis of the Arabidopsis vegetative shoot apex. Dev. Cell. 2021;56:1056–1074.e8. doi: 10.1016/j.devcel.2021.02.021. [DOI] [PubMed] [Google Scholar]
- 181.Omary M., Gil-Yarom N., Yahav C., et al. A conserved superlocus regulates above- and belowground root initiation. Science. 2022;375 doi: 10.1126/science.abf4368. [DOI] [PubMed] [Google Scholar]
- 182.Birnbaum K., Shasha D.E., Wang J.Y., et al. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 183.Liu Z., Zhou Y., Guo J., et al. Global dynamic molecular profiling of stomatal lineage cell development by single-cell RNA sequencing. Mol. Plant. 2020;13:1178–1193. doi: 10.1016/j.molp.2020.06.010. [DOI] [PubMed] [Google Scholar]
- 184.Long Y., Liu Z., Jia J., et al. FlsnRNA-seq: protoplasting-free full-length single-nucleus RNA profiling in plants. Genome Biol. 2021;22:66. doi: 10.1186/s13059-021-02288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tian C., Du Q., Xu M., et al. Single-nucleus RNA-seq resolves spatiotemporal developmental trajectories in the tomato shoot apex. bioRxiv. 2020 doi: 10.1101/2020.09.20.305029. Preprint at. [DOI] [Google Scholar]
- 186.Denisenko E., Guo B.B., Jones M., et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 2020;21:130. doi: 10.1186/s13059-020-02048-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Marand A.P., Chen Z., Gallavotti A., Schmitz R.J. A cis-regulatory atlas in maize at single-cell resolution. Cell. 2021;184:3041–3055.e21. doi: 10.1016/j.cell.2021.04.014. [DOI] [PubMed] [Google Scholar]
- 188.Ryu K.H., Zhu Y., Schiefelbein J. Plant cell identity in the era of single-cell transcriptomics. Annu. Rev. Genet. 2021;55:479–496. doi: 10.1146/annurev-genet-071719-020453. [DOI] [PubMed] [Google Scholar]
- 189.Gala H.P., Lanctot A., Jean-Baptiste K., et al. A single cell view of the transcriptome during lateral root initiation in Arabidopsis thaliana. Plant Cell. 2021;33:2197–2220. doi: 10.1093/plcell/koab101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Aichinger E., Kornet N., Friedrich T., Laux T. Plant stem cell niches. Annu. Rev. Plant Biol. 2012;63:615–636. doi: 10.1146/annurev-arplant-042811-105555. [DOI] [PubMed] [Google Scholar]
- 191.Briggs J.A., Weinreb C., Wagner D.E., et al. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science. 2018;360:eaar5780. doi: 10.1126/science.aar5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Farrell J.A., Wang Y., Riesenfeld S.J., et al. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science. 2018;360:eaar3131. doi: 10.1126/science.aar3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Harland R.M. A new view of embryo development and regeneration. Science. 2018;360:967–968. doi: 10.1126/science.aat8413. [DOI] [PubMed] [Google Scholar]
- 194.Plass M., Solana J., Wolf F.A., et al. Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science. 2018;360 doi: 10.1126/science.aaq1723. [DOI] [PubMed] [Google Scholar]
- 195.Zeisel A., Hochgerner H., Lönnerberg P., et al. Molecular architecture of the mouse nervous system. Cell. 2018;174:999–1014.e22. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jordão M.J.C., Sankowski R., Brendecke S.M., et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science. 2019;363:eaat7554. doi: 10.1126/science.aat7554. [DOI] [PubMed] [Google Scholar]
- 197.Efroni I., Mello A., Nawy T., et al. Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell. 2016;165:1721–1733. doi: 10.1016/j.cell.2016.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Roszak P., Heo J.O., Blob B., et al. Cell-by-cell dissection of phloem development links a maturation gradient to cell specialization. Science. 2021;374:eaba5531. doi: 10.1126/science.aba5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kidner C., Sundaresan V., Roberts K., Dolan L. Clonal analysis of the Arabidopsis root confirms that position, not lineage, determines cell fate. Planta. 2000;211:191–199. doi: 10.1007/s004250000284. [DOI] [PubMed] [Google Scholar]
- 200.van den Berg C., Willemsen V., Hage W., et al. Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature. 1995;378:62–65. doi: 10.1038/378062a0. [DOI] [PubMed] [Google Scholar]
- 201.Kubo M., Nishiyama T., Tamada Y., et al. Single-cell transcriptome analysis of Physcomitrella leaf cells during reprogramming using microcapillary manipulation. Nucleic Acids Res. 2019;47:4539–4553. doi: 10.1093/nar/gkz181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Dalal V., Dagan S., Friedlander G., et al. Transcriptome analysis highlights nuclear control of chloroplast development in the shoot apex. Sci. Rep. 2018;8:8881. doi: 10.1038/s41598-018-27305-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Giacomello S., Salmén F., Terebieniec B.K., et al. Spatially resolved transcriptome profiling in model plant species. Nat. Plants. 2017;3 doi: 10.1038/nplants.2017.61. [DOI] [PubMed] [Google Scholar]
- 204.Burgess D.J. Spatial transcriptomics coming of age. Nat. Rev. Genet. 2019;20:317. doi: 10.1038/s41576-019-0129-z. [DOI] [PubMed] [Google Scholar]
- 205.Xia K., Sun H.X., Li J., et al. The single-cell stereo-seq reveals region-specific cell subtypes and transcriptome profiling in Arabidopsis leaves. Dev. Cell. 2022;57:1299–1310.e4. doi: 10.1016/j.devcel.2022.04.011. [DOI] [PubMed] [Google Scholar]
- 206.Marx V. Method of the Year: spatially resolved transcriptomics (vol 85, pg 931, 2020) Nat. Methods. 2021;18:219. doi: 10.1038/s41592-021-01065-y. [DOI] [PubMed] [Google Scholar]
- 207.Femino A.M., Fay F.S., Fogarty K., Singer R.H. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 208.Rodriques S.G., Stickels R.R., Goeva A., et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Vickovic S., Eraslan G., Salmén F., et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods. 2019;16:987–990. doi: 10.1038/s41592-019-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Alfaro J.A., Bohländer P., Dai M., et al. The emerging landscape of single-molecule protein sequencing technologies. Nat. Methods. 2021;18:604–617. doi: 10.1038/s41592-021-01143-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Gaiti F., Hawkins A., Chamely P., et al. Single-cell multi-omics defines the cell-type specific impact of SF3B1 splicing factor mutations on hematopoietic differentiation in human clonal hematopoiesis and myelodysplastic syndromes. Blood. 2021;138:145. [Google Scholar]
- 212.Guilliams M., Bonnardel J., Haest B., et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell. 2022;185:379–396.e38. doi: 10.1016/j.cell.2021.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Dixit A., Parnas O., Li B., et al. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016;167:1853–1866.e17. doi: 10.1016/j.cell.2016.11.038. e1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mimitou E.P., Cheng A., Montalbano A., et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat. Methods. 2019;16:409–412. doi: 10.1038/s41592-019-0392-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Mimitou E.P., Lareau C.A., Chen K.Y., et al. Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells. Nat. Biotechnol. 2021;39:1246–1258. doi: 10.1038/s41587-021-00927-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Xu Z., Heidrich-O’Hare E., Chen W., Duerr R.H. Comprehensive Benchmarking of CITE-Seq versus DOGMA-Seq Single Cell Multimodal Omics. Genome Biol. 2021;23:135. doi: 10.1186/s13059-022-02698-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Bian S., Hou Y., Zhou X., et al. Single-cell multiomics sequencing and analyses of human colorectal cancer. Science. 2018;362:1060–1063. doi: 10.1126/science.aao3791. [DOI] [PubMed] [Google Scholar]
- 218.Stephenson E., Reynolds G., Botting R.A., et al. Cambridge Institute of Therapeutic Immunology and Infectious Disease-National Institute of Health Research CITIID-NIHR COVID-19 BioResource Collaboration Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med. 2021;27:904–916. doi: 10.1038/s41591-021-01329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yan R., Gu C., You D., et al. Decoding dynamic epigenetic landscapes in human oocytes using single-cell multi-omics sequencing. Cell Stem Cell. 2021;28:1641–1656.e7. doi: 10.1016/j.stem.2021.04.012. [DOI] [PubMed] [Google Scholar]