Abstract
The mechanisms of olfactory dysfunction in COVID-19 are still unclear. In this review, we examine potential mechanisms that may explain why the sense of smell is lost or altered. Among the current hypotheses, the most plausible is that death of infected support cells in the olfactory epithelium causes, besides altered composition of the mucus, retraction of the cilia on olfactory receptor neurons, possibly because of the lack of support cell-derived glucose in the mucus, which powers olfactory signal transduction within the cilia. This mechanism is consistent with the rapid loss of smell with COVID-19, and its rapid recovery after the regeneration of support cells. Host immune responses that cause downregulation of genes involved in olfactory signal transduction occur too late to trigger anosmia, but may contribute to the duration of the olfactory dysfunction.
Keywords: olfactory epithelium, anosmia, SARS-CoV-2, smell loss, parosmia, sustentacular cell
Loss of smell in COVID-19: why?
The frequency of olfactory dysfunction in the coronavirus disease 2019 (COVID-19) pandemic is unprecedented [1,2]. Previous pandemics caused by other viruses have resulted in olfactory loss at a much lower rate [1., 2., 3., 4., 5., 6., 7., 8.], primarily by nasal congestion and obstruction, or by loss of smell and taste as a sequel after the acute infection; regardless, they did not give rise to the sudden and extensive loss of smell on the scale of the hundreds of millions of cases seen in COVID-19. What is different about severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2 virus; see Glossary) compared with previous coronaviruses? Why does this virus have such a devastating impact on the sense of smell? Most of the mechanisms that have been proposed to explain the unique features of anosmia in COVID-19 are not fully consistent with the data that have been reported from humans and animal models.
In this review, we first describe the shortcomings of previous hypotheses attempting to explain anosmia in COVID-19; we then present evidence indicating that largely overlooked properties of support cells may account for all major features of SARS-CoV-2-induced anosmia in both humans and animals. Olfactory receptor neurons depend on two types of support cell: sustentacular cells (Figure 1 ) and Bowman gland cells. This dependence is much more intricate than traditionally appreciated. The olfactory neurons engage in a series of complex interactions with their support cells. When this intimate symbiosis is disrupted by the infection of support cells with SARS-CoV-2, neuronal function is compromised. Given that multiple variants of SARS-CoV-2 have evolved with various mutations, it has been possible to dissect the contributions of viral factors to the effectiveness of host cell entry and tropism, and how these determine the extent of olfactory dysfunction. The COVID-19 pandemic has revealed crucial roles of support cells for neuronal function that were not previously obvious, and this has helped to inform about fundamental workings of the olfactory system. Here, we examine the merits of five hypotheses that may explain the olfactory dysfunction associated with COVID-19. We contend that, among the hypotheses, the most plausible one involves the death of infected support cells in the olfactory epithelium and, as a result, temporary disruption of olfactory receptor neuron function. We also discuss how viral mutations modify the extent of such dysfunction. From a historical perspective, the impact of SARS-CoV-2 on olfaction appears to be unprecedented among the pandemics of recent history.
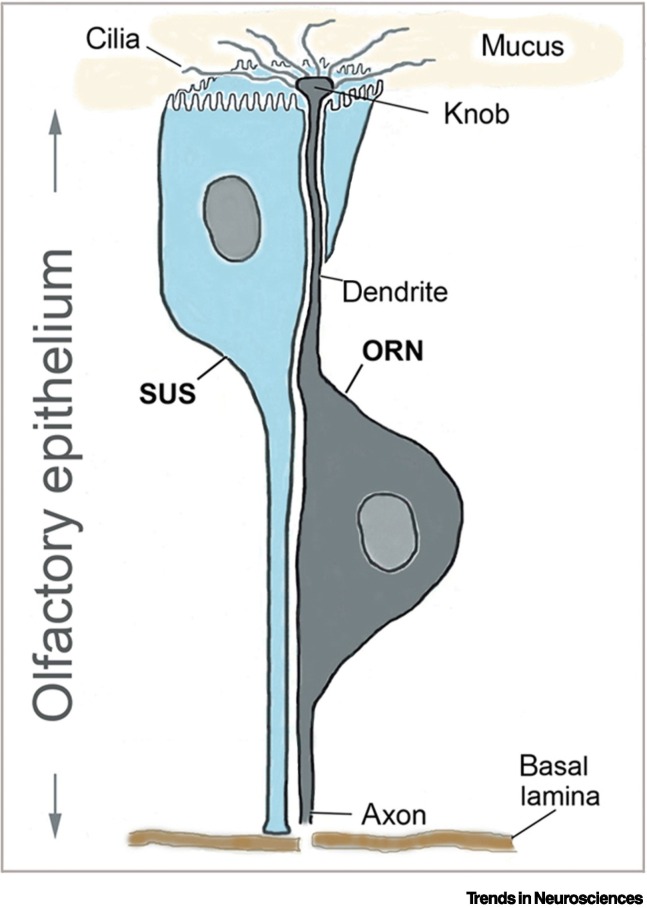
Figure 1.
Intimate relationship between the olfactory receptor neuron (ORN) and its support cell, the sustentacular cell (SUS), in the olfactory epithelium.
The sense of smell in humans depends on 10 million ORNs and a similar number of SUS. The ORN has an apical dendritic process that projects a thickening (knob) into the lumen above the olfactory epithelium (nasal cavity) from which 10–15 cilia extend that bear the odorant receptors. The olfactory epithelium is covered by mucus to protect the dendritic processes. Odorant molecules inhaled in the nasal cavity dissolve in the mucus to bind to odorant receptors on the olfactory cilia. The basal process of the bipolar neuron is an axon that penetrates the basal lamina and the cribriform plate and forms synaptic contacts in a specific glomerulus in the olfactory bulb of the brain. Each mature ORN is tightly wrapped by its SUS [44], as shown. The SUS extends throughout the olfactory epithelium, with its basal process reaching the basal lamina, while the apical surface of the SUS is covered with microvilli, which intermingle with the cilia of the ORNs. The SUS provides structural support to the epithelium, and is thought to have multiple important functions. Notably, the SUS, as well as cells in the Bowman glands (not shown) secrete the mucus, and likely provide energy (glucose) to the cilia [57., 58., 59.] (for details, see Figure 4), so that the cilia can conduct the energy-consuming olfactory signal transduction. Additional important functions include regulation of the ionic composition of the mucus [117], detoxification and odorant clearance [118], and expression of odorant-binding proteins [119].
Previous hypotheses about underlying mechanisms
During the first year of the pandemic, when reports of olfactory dysfunction permeated the literature and the media, a variety of potential mechanisms was considered. These included: congestion of the nasal mucosa due to swelling and obstruction of the olfactory cleft; infection and death of olfactory receptor neurons; viral neuroinvasion along the olfactory nerve; infection and death of neurons in the olfactory bulb and cortex; altered neuronal function due to cytokine release and inflammation; reduced neuronal function due to vascular changes in the olfactory bulb; reduction in mucus that dissolves odorants, due to infection and the death of cells that produce it; immune-mediated downregulation of odorant receptors and other signaling molecules; autoimmune reactions due to the resemblance of viral proteins to odorant receptors; inflammation and damage of the olfactory epithelium; and infection and death of sustentacular support cells [9., 10., 11., 12., 13., 14., 15., 16., 17., 18.].
Scenarios or mechanisms that have been discounted
Any hypothesis about the mechanism of anosmia in COVID-19 must account for the high penetrance, sudden onset, and remarkable transience of the olfactory dysfunction [9,19]. Most of the above-listed hypotheses turned out to be implausible, for various reasons. The olfactory cleft does not become obstructed in most patients with COVID-19 with olfactory dysfunction; thus, congestion cannot explain most cases of anosmia [13,15,20., 21., 22.]. Olfactory receptor neurons do not express the virus entry proteins and, therefore, become infected rarely or not at all [13,22., 23., 24., 25., 26.]. Current evidence indicates that SARS-CoV-2 has very limited neurotropic potential in humans, if any, unlike other viruses that target the olfactory circuits [13,22., 23., 24.]. Since the regeneration of olfactory receptor neurons takes 2–3 weeks, the mostly short-lived loss of smell cannot be caused by death and subsequent regeneration of the olfactory receptor neurons [9,11]. SARS-CoV-2 does not invade the olfactory nerve in patients with COVID-19, and this nerve remains largely intact during anosmia [22., 23., 24.,27]. In animal models and also in humans, many regions of the olfactory epithelium retain half or more of their olfactory receptor neurons after inoculation or infection [22,28], which is thought to be more than sufficient to maintain a basic sense of smell [29., 30., 31.]. Effects of the virus on axonal pathology in the olfactory bulb and cerebral cortex, whether directly by viral infection or indirectly through host immune responses, were not significantly different between patients with COVID-19 with loss of smell and patients without loss of smell [32]. Infection of the olfactory epithelium in animal models causes longer term changes of microglia activation in olfactory targets in the brain [28], apparently mediated through systemic inflammatory responses [33], but the timing of such events is not compatible with the early abrupt onset of complete anosmia and its recovery just days later [21]. In conclusion, since most of the hypotheses fail to explain COVID-19-associated anosmia, which scenarios remain? Before we examine five hypotheses that deserve further scrutiny, we first discuss key features of anosmia in COVID-19 that any mechanism has to explain.
The unique features of COVID-19-associated anosmia
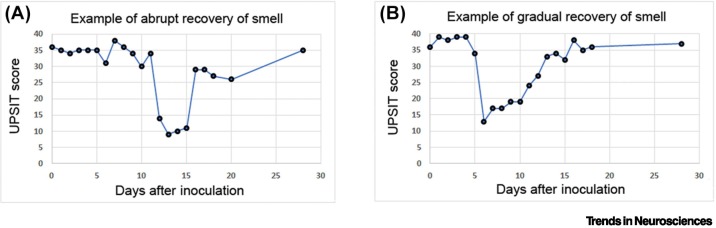
One of the most peculiar characteristics of the olfactory dysfunction in COVID-19 is that it typically starts very abruptly, lasts for only a few days (mean or median ranges: 7–21.6 days [34,35]), and smell can recover just as abruptly as it was lost. The olfactory dysfunction may be quantitative, with reduced function (hyposmia) or complete loss (anosmia), or the dysfunction may be qualitative, with altered smell (parosmia); it may be accompanied by dysfunction of taste; and it may last shorter or longer, and with sudden or gradual recovery. Typical temporal features of anosmia are illustrated in Figure 2 using data from a controlled clinical study on humans [36]. Volunteers underwent objective testing of olfaction daily or every 2 days post inoculation (dpi). As expected from epidemiological meta-analyses of the same virus variant based on subjective recall of patients [37,38], two-thirds of volunteers infected with SARS-CoV-2 lost their sense of smell. The onset of loss of smell varied, but occurred between 5 and 12 dpi. Two main patterns became apparent. In most volunteers with anosmia, smell was lost abruptly (within hours) for an average duration of 8 days, followed by either an abrupt (Figure 2A) or gradual recovery (Figure 2B).
Figure 2.
Key features of anosmia in coronavirus disease 2019 (COVID-19): rapid onset, relatively short duration, and usually rapid recovery.
Examples of two typical cases of anosmia, according to scores obtained with the University of Pennsylvania Smell Identification Test (UPSIT) on volunteers inoculated with the G614 variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) on day 0 [36]. (A) Example of a case with abrupt onset and abrupt recovery of smell. (B) Example of a case with abrupt onset and gradual recovery of the sense of smell. Note the relatively short duration of anosmia (mean of 7.8 days; range of 4–21 days for transient anosmia [36]), which is consistent with a similar time-course reported in meta-analyses that examined epidemiological studies of the same variant and populations in similar geographic regions [37,38]. Modified from [36].
Based on animal studies, it is thought that more than 90% of the olfactory epithelium has to be impaired to lose the sense of smell [29., 30., 31.]. In adult rats, when 30–40% of sustentacular cells were eliminated, with loss of ~25% of olfactory receptor neurons, olfaction was found to be normal [39]. There appears to be redundancy (or a ‘safety buffer’) in that only 10% or less of functional olfactory receptor neurons are needed for a basic sense of smell. Humans have ~10 million olfactory neurons and 10 million support cells in the olfactory epithelium. To lose the sense of smell, as often occurs in COVID-19, it is likely that, by extrapolation from animal studies, at least 90% of the human olfactory receptor neurons (i.e., 9 million cells) must be dysfunctional. Since it is known that, in COVID-19 patients with anosmia, the number of olfactory receptor neurons is not reduced to such an extent [22,24], the neurons appear to become temporarily disabled by a mechanism that differs from those known for other viruses. Any viable hypothesis has to account for these peculiar features of anosmia in COVID-19.
Timing of recovery of smell: implications for the mechanism involved
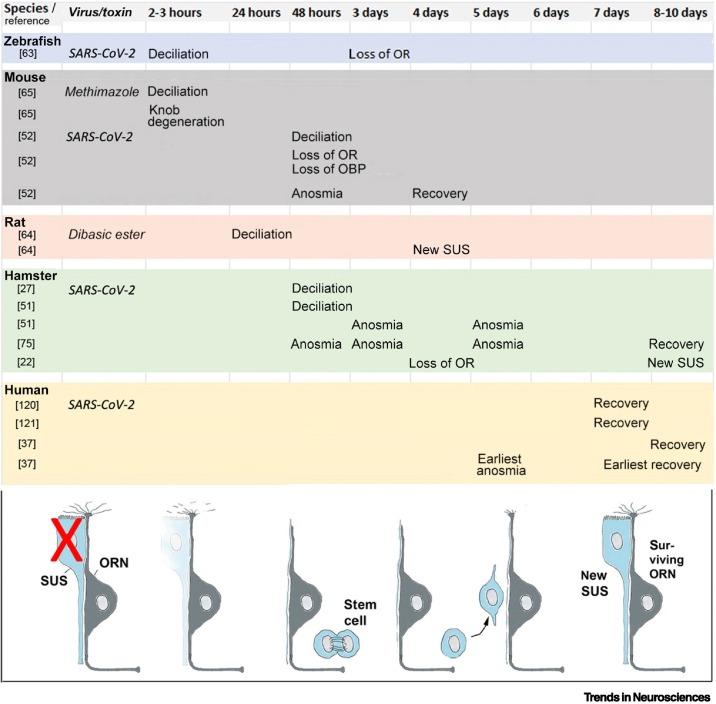
Another important feature of anosmia in COVID-19 is the way in which smell recovers, because this recovery profile rules out several mechanisms. What kind of insult can rapidly disable 90% or more of the olfactory receptor neurons, causing an anosmia that may last only 4 days, on average 9 days [37], and mostly shows an abrupt, not gradual, recovery of the sense of smell (objectively verified in humans [36])? Keeping in mind the ‘safety buffer’, one can conclude that, for subjective or objective recovery of smell, in as few as 4 days, at least 5–10% of olfactory neurons (500 000 to 1 million neurons) must have become functional or have regained their functionality. Under normal conditions, both the adult olfactory receptor neurons and their support cells constantly regenerate with a neuronal turnover of ~30–90 days [40]. Regeneration of these two cell types follows a very different time course. The sustentacular cells rapidly regenerate, within 4–8 days after lesion (Figure 3 ). Regeneration of olfactory receptor neurons is much slower [41., 42., 43.], due to the need for growth of the apical dendritic process toward the nasal cavity, odorant receptor expression and trafficking, and enwrapping of the dendrite by sustentacular support cells [44,45]. The growth of the axon, along the olfactory nerve, through the cribriform plate and into the glomeruli of the olfactory bulb, takes an additional 5–7 days, even in small rodents [43,45., 46., 47.]. These differences in regeneration between cell types provide clues to the plausibility of the proposed mechanisms.
Figure 3.
Timetable of the events that ensue when the sustentacular support cell (SUS) is damaged or eliminated.
This figure summarizes the events when the SUS is damaged by either a toxin or due to virus infection. Notably, damage of the SUS causes within 2–3 h a physical separation of the SUS from the olfactory receptor neuron (ORN), swelling of the knob and its degeneration, and retraction of the cilia [27,51,63., 64., 65.]. Deciliation continues from 2 to 48 h. The ORN resumes an immature stage of its dendritic extension, with focus on growth of its processes rather than on neurotransmission and sensory transduction. Gene expression of odorant receptors (ORs) is downregulated at 2 days (mouse [52]), 3 days (zebrafish [63]), and 4 days (hamster [22]) after inoculation. Odorant-binding proteins (OBPs) and receptor transporting protein 1 (RTP1) are also reduced [52]. In most animal models, loss of smell is evident as early as 2 days after lesion of SUS (mouse) and, depending on the animal model, anosmia lasts from 2 to 8 days. In humans, it lasts from 7 to 10 days (mean values) after infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [37]. The stem cells in the basal layers begin to divide at 3 days after SUS damage [66], and the first newly regenerated SUS appear at 4–8 days [22,64]. Recovery of smell begins at 4 days after SUS lesion in the mouse, at 8 days in the hamster [75], and the olfactory epithelium appears fully intact in the hamster at 7 days [74] or 14 days [51]. Human data on recovery of smell are according to pertinent studies [36,37,120,121].
Focus on elimination of support cells and host immune responses
The currently viable hypotheses aiming to explain anosmia in COVID-19 can be divided into two major categories with five different ‘flavors’ that are not mutually exclusive: (i) elimination of support cells with consequences for neuronal function: (a) reduction or alteration of the mucus covering the olfactory epithelium; (b) loss of energy (glucose) that normally powers olfactory cilia, due to death of sustentacular cells and Bowman gland cells; and (c) retraction of olfactory cilia due to the death of sustentacular cells and loss of a maintenance factor for the cilia; and (ii) host immune responses affecting olfactory receptor neuron function: (a) downregulation of gene expression for odorant receptors (and other signaling molecules) in olfactory receptor neurons; and (b) immune cytokine-caused inflammation and destruction of the olfactory epithelium, including loss or damage of olfactory receptor neurons.
We review these five hypotheses in sequence. The first three envision a central role of the support cells and propose that damage to two types of support cell, sustentacular cells and Bowman gland cells, is responsible for the loss of neuronal function. The immune hypothesis postulates that host-generated immune responses cause neuronal dysfunction, either by changes in gene expression of olfactory receptor neurons or by inflammation of the olfactory epithelium and damage or death of olfactory receptor neurons. Accordingly, these two hypotheses predict fundamentally different mechanisms of how neuronal function may become compromised and result in anosmia.
Mucus reduction
The mucus that covers the olfactory epithelium is produced by sustentacular cells and Bowman gland cells. These cell types abundantly express the viral entry proteins, angiotensin-converting enzyme 2 (ACE2) and TMPRSS2 [19,48., 49., 50.] and, therefore, are the prime target of SARS-CoV-2 within the olfactory system. The virus rapidly and extensively infects and destroys these cells in animal models [25,27,51,52] and humans [22,24]. This could lead to reduction and/or alteration of the mucus, impede diffusion of odorant molecules, and alter processing and signal transduction induced by odorants [53,54]. The amount of mucus after SARS-CoV-2 infection has been quantified in an animal model and was found to be significantly reduced after infection [55], suggesting that changes in the quantity and chemical composition of the mucus contribute to the olfactory dysfunction [51,56].
Loss of glucose normally supplied by sustentacular cells and Bowman gland cells
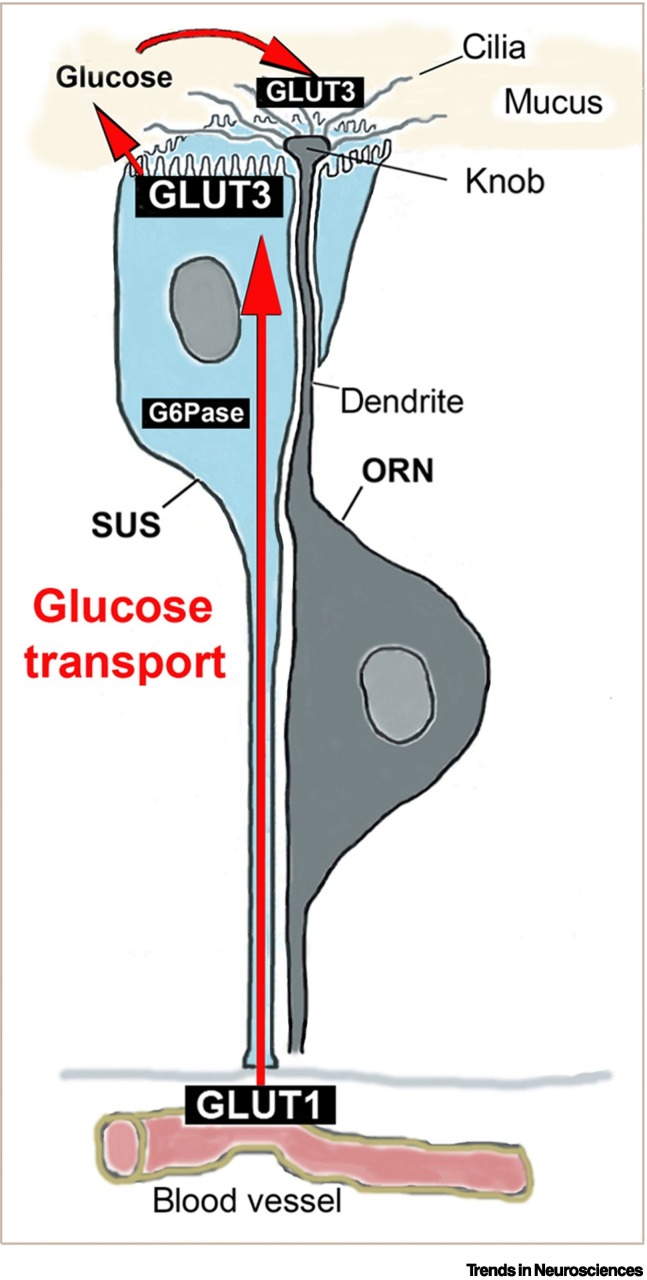
The dendritic knob of olfactory receptor neurons lacks a sufficient number of mitochondria to supply energy for olfactory transduction in the cilia, which extend up to 100 μm from the knob within the mucus [57,58]. Instead, glucose has to be transported from blood vessels below the basal lamina through the sustentacular cells and Bowman gland cells to the mucus. These cells import glucose at their basal domain via glucose transporters (GLUT1) and export (secrete) the glucose at their apical surface via GLUT3 into the mucus [57., 58., 59.] as illustrated for sustentacular cells in Figure 4 . Impairing the glucose transporters and reducing the glucose concentration in the mucus is thought to rapidly abolish the energy-dependent olfactory signal transduction that normally ensues upon binding of odorants to their receptors. When this energy supplementation is disrupted by the infection of the support cells with SARS-CoV-2, the cilia become dysfunctional. Consistent with this hypothesis, genes for glucose transporters are among the earliest downregulated genes according to single-cell RNA-sequencing analyses of support cells after SARS-CoV-2 infection in hamsters [22]. Since ACE2, abundantly expressed in support cells, is part of the renin–angiotensin–aldosterone system (RAAS), RAAS-associated peptidases may be linked to anosmia because of their potential involvement in the regulation of ion/water content and glucose metabolism [60]. SARS-CoV-2 is known to hijack host metabolic pathways to maximize glucose utilization for virus replication [61]. Therefore, it is possible that glucose within infected support cells is diverted even before they die, preventing the glucose from being released and making it unavailable for cilia. The dependence of olfactory signal transduction on energy supplied by the support cells provides a plausible mechanism for COVID-19-induced anosmia [10,11,24,62].
Figure 4.
Illustration of how a sustentacular support cell (SUS) provides the cilia of its olfactory receptor neuron (ORN) with glucose.
The sustentacular cell (SUS) takes up glucose from its basal process, close to the blood vessels, via the glucose transporter 1 (GLUT1). Glucose or glucose-6-phosphate (G6P) diffuses along its gradient from the basal to the apical end, glucose-6 phosphatase (G6Pase) converts the G6P back to glucose, and secretes it through the glucose transporter 3 (GLUT3) into the mucus. Within the mucus, olfactory cilia, which lack mitochondria, uptake glucose via their GLUT3 and generate ATP by glycolysis. Likewise, the SUS-related cells in the Bowman glands also traffic glucose similarly from the blood supply to the mucus (not shown). The glucose-trafficking mechanisms are summarized as previously reported [57., 58., 59.]. The glucose support function may be the most important and acute way by which SUS and Bowman gland cells maintain ORN function.
Retraction of neuronal cilia after damage to sustentacular cells
Several studies of anosmia in COVID-19, in both animal models and humans, have reported rapid deciliation (retraction of cilia from olfactory receptor neurons) after infection of sustentacular cells with SARS-CoV-2 [27,51,52,63] (Figure 3). Such data suggest that the sustentacular cells provide a maintenance factor for olfactory cilia. Surprisingly, deciliation has rarely been discussed as a mechanism for COVID-induced anosmia, with few exceptions [11,27,51,52,60].
Previous studies have shown that toxins that primarily target and destroy the sustentacular cells (e.g., dibasic esters, methimazole, and nickel sulfate) rapidly lead to deciliation of the olfactory receptor neurons [64., 65., 66.] (Figure 3). Olfactory neurons lose contact with their sustentacular cells, dendritic knobs begin to swell and degenerate, losing their cilia within 2 h after treatment with methimazole [65]. Likewise, after destruction of support cells with dibasic esters, cilia disappear within 24 h [64]. Such loss of cilia cannot be an artifact of tissue processing, because cilia are preserved in vehicle control cases, and because specifically those neurons lose their cilia when their adjacent sustentacular cell has died [64], which suggests the existence of a local, contact-mediated maintenance signal between the two cell types [60]. Tight contacts between the two cell types have been described in the rodent [44] as well as in the human olfactory epithelium [67]. SARS-CoV-2 causes deciliation in animals with the same rapid time course as described for the above-listed toxins [27,51,52] (Figure 3). Importantly, the key transcription factor for ciliogenesis, Forkhead box J1 (Foxj1), is already downregulated in olfactory receptor neurons 1 dpi [22]. Although originally proven to be essential only for motile cilia [68], Foxj1 is expressed in some, and possibly most, olfactory receptor neurons, as shown by transcriptome analyses [22,69,70] and examination of tissue sections [69,71]. Rapid deciliation also occurs in the SARS-CoV-2-infected respiratory epithelium [72,73]. Since cilia are required for olfactory signal transduction [45], deciliation after elimination of support cells may be primarily responsible for anosmia in COVID-19.
Can this mechanism also explain the rapid and often abrupt recovery of smell? It is important to consider two possibilities: after deciliation, the recovery of smell may be due to a sufficient number of immature receptor neurons that develop over time to become functional; alternatively, there may be regrowth of cilia on surviving receptor neurons. Regarding the first possibility, assuming a 30–90-day turnover of 10 million olfactory receptor neurons [40], this amounts to 110 000–330 000 new neurons per day. Normally, it would take only 4–10 days to achieve 500 000 to 1 million new functional olfactory receptor neurons with cilia (i.e., 5–10% of the total required for smell [29., 30., 31.]). However, because of the substantial destruction of the olfactory epithelium, numbers of immature neurons in the pipeline may be considerably lower, since some of them become infected or damaged and die during the desquamation of the epithelium [51,52,74] and their maturation may be delayed.
As an alternative, regrowth of cilia may occur on surviving deciliated mature olfactory receptor neurons. Since most of the mature neurons are thought to survive [22,24,25,28], only a small fraction of them (10%) need to regrow their cilia to provide basic olfactory functionality. The abrupt recovery of smell in many cases of anosmia [36] (Figure 2A) is more consistent with a synchronized regeneration of support cells, allowing the regrowth of the cilia on surviving neurons, rather than the gradual increase in regenerating neurons that were already in the pipeline (before infection). Thus, the time course of smell recovery favors the scenario of surviving neurons regrowing their dendrites.
Regardless which of the two scenarios applies (and they are not mutually exclusive), there is strong evidence from animal models that, after having been largely deciliated at 3–5 dpi, the cilia are fully intact at 14 dpi in hamster [51]. Since olfaction returns at 8 dpi in the hamster [75], the time course of cilia recovery (between 5 and 14 dpi) is consistent with the notion that reciliation is involved in the fast recovery of smell in COVID-19.
Downregulation of odorant receptor genes
Increased levels of some cytokines, such as interferons (IFNs), in the olfactory epithelium can reduce the expression of odorant receptors in olfactory receptor neurons [76]. Such downregulation of odorant receptors was proposed as a potential mechanism for anosmia in COVID-19 [9]. This hypothesis was first tested in a mouse model [52], and it was found that not only odorant receptor genes, but also additional molecules involved in olfactory signal transduction were downregulated. However, in humans, normal expression of odorant receptor genes was reported [24], unchanged between infected and non-infected olfactory epithelium. Likewise, no downregulation of odorant receptor genes was found in COVID-19 patients with persistent (12 weeks) loss of smell [56], but ADCY3 (an odorant receptor-related signaling molecule) was reduced. In hamster, downregulation of odorant receptor genes did not occur until 4 dpi [22], and it was concluded that systemic proinflammatory cytokines can induce downregulation of odorant receptor genes as well as genes encoding other signaling molecules, such as ADCY3 [22]. Several studies reported impaired olfactory signal transduction due to a reduction in signaling molecules [22,52,77].
The cellular source of the cytokines in the host has not yet been identified, and neither have the relevant cytokines. Sustentacular cells may be the only, or the main, source of such cytokines [9], or sustentacular cells may contribute among various other cellular sources [22]. Most studies reported increased levels of IFNs in the olfactory epithelium after SARS-CoV-2 infection [22,51,52], with one exception (to our knowledge) [74]. Some IFN-stimulated genes encode proteins (such as IFITM or LY6E) that inhibit viral entry along the endosomal route [78,79]. Since IFITMs are abundantly expressed in support cells [22], this would reduce endosomal virus entry, but may not prevent virus entry after surface membrane fusion via TMPRSS2 [21,79,80] (Figure 5 ). The importance of viral entry mediated through TMPRSS2 is underscored by the reduced olfactory dysfunction in patients with COVID-19 treated with the TMPRSS2 inhibitor, camostat [81]. Besides IFN, several other cytokines, such as IL6 and TNFα, may be involved in anosmia, although their roles are somewhat controversial and await clarification [76,82., 83., 84., 85.].
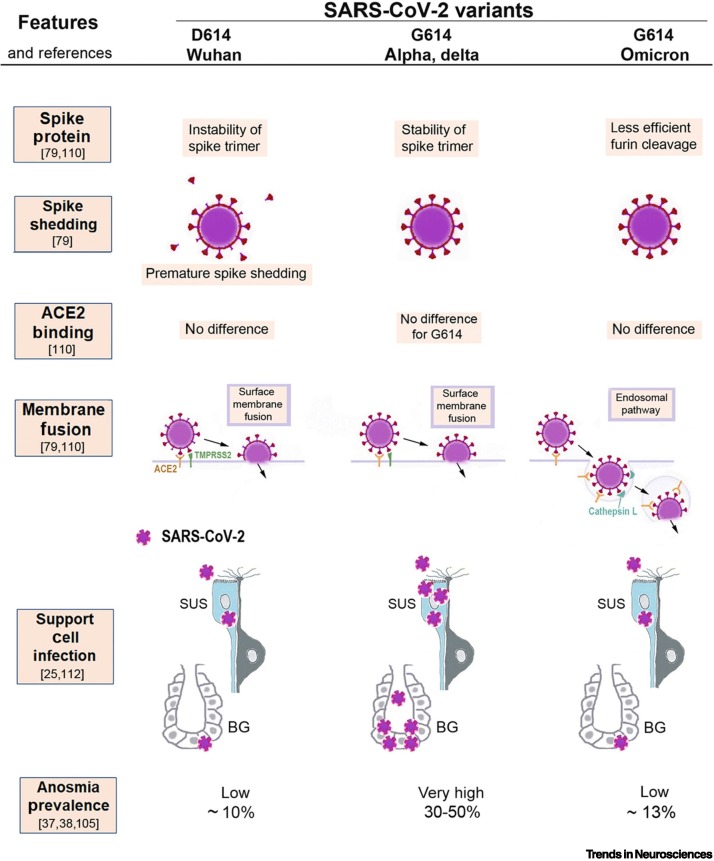
Figure 5.
Illustration of the molecular mechanisms that can explain why severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants cause different amounts of olfactory dysfunction.
This figure summarizes how the properties of three SARS-CoV-2 virus variants (D614, G614, and omicron) differ in ways that likely determine to what extent sustentacular support cells (SUS) in the olfactory epithelium become infected and whether their loss will lead to anosmia. The original D614 (Wuhan) virus results in premature spike shedding, lower spike density, and, therefore, less effective virus entry [79]. This may cause less infection of SUS and, therefore, results in a low prevalence of anosmia (~10%) [37]. The G614 variant has the D614G mutation, which stabilizes the spike trimer and prevents premature spike shedding; the higher spike density allows the G614 variant to infect SUS cells effectively [79], resulting in a high (30–50%) anosmia prevalence [38]. All three variants bind to the virus entry protein angiotensin-converting enzyme 2 (ACE2), expressed by SUS, and with no significant differences in binding affinity to ACE2 [21,79,80]; thus, this cannot explain differences in anosmia. The first two variants, D614 and G614, both enter host cells by using surface membrane fusion mediated by the protease TMPRSS2 [79,110]. The new mutations in the omicron variant cause a less efficient furin cleavage, resulting in reduced surface membrane fusion mediated by TMPRSS2 [79,80,111]. Therefore, omicron prefers an endosomal route that is less efficient for SUS infection, possibly because many host cells have developed defenses for the endosomal entry [78,79,111]. As a result, the omicron variant, despite retaining the D614G mutation, is associated with a lower anosmia prevalence of ~13% [105].
A mechanism involving immune responses is an attractive hypothesis, because it would explain the sudden onset of complete anosmia. However, there are inconsistencies with the timing. If immune-mediated release of cytokines silences olfactory signaling, one would expect that downregulation of odorant receptor genes occurs before the onset of anosmia, and there should be some delay between reduced gene expression and actual depletion of odorant receptor proteins, because of the turnover rate of these proteins [86]. One would also expect that smell does not recover until gene expression has normalized. However, in hamster, the onset of anosmia precedes the gene expression changes by 2 days and extends beyond the time of smell recovery. At 8 dpi, the sense of smell in hamsters has already recovered [75], while odorant receptor genes are still downregulated at 10 dpi [22]. Apparently, smell can recover before normal odorant receptor expression has resumed.
As mentioned previously, the altered regulation of genes enabling olfactory signal transduction is not restricted to odorant receptor genes, but involves additional proteins relevant for olfactory signal transduction, including receptor transporting protein 1 (RTP1 [52]), IFN-controlled G-protein signaling (RGS2 [77]), and ADCY3 [22,56], which is essential for maintenance of olfactory cilia [87] and is also reduced at the protein level [22]. This indicates a broad effect on gene expression in olfactory receptor neurons in response to SARS-CoV-2. Such broad changes in gene expression may be a consequence of deciliation and the regression of the olfactory receptor neurons to a less mature state, with impaired placement of odorant receptors; odorant receptor trafficking to the cilia is thought to be a critical step toward maturation of olfactory neurons [88].
Immune cytokine-induced destruction of the olfactory epithelium
A final possible scenario to consider is that infection of the sustentacular cells by SARS-CoV-2 rapidly causes immune cell infiltration of the olfactory epithelium, which leads to desquamation of the epithelium, with expulsion of epithelial cells into the lumen. Many of these cells die by apoptosis, including some olfactory receptor neurons. This may be part of a host defense mechanism that evolved to protect the brain from pathogens and toxins [89., 90., 91., 92.]. The destruction of the olfactory epithelium may cause the anosmia, and eventual regeneration of the olfactory epithelium may lead to recovery of smell [27,28,51,52,74,93]. Chronic immune responses in the olfactory epithelium appear to delay regeneration of the olfactory epithelium [94,95]. This may explain why an estimated 5% of COVID-19 patients with olfactory dysfunction recover from chemosensory dysfunction late or not at all [96].
While there is substantial, although patchy, damage of the infected olfactory epithelium, a major question is whether such damage indeed leads to the destruction of 90% or more of the olfactory receptor neurons, because of the safety buffer discussed previously [29., 30., 31.]. Studies report that most olfactory receptor neurons do not become infected, and a sufficiently large percentage of them apparently survive [22,24,27,28,51]. This indicates that anosmia in COVID-19 is unlikely to be caused by the death of olfactory receptor neurons.
Parosmias: prevalence, time course and possible mechanisms
The prevalence of qualitative olfactory dysfunction (parosmia) in COVID-19 is not well known. The largest cohort studies have reported 7.5–11% [97,98], and two smaller cohort studies indicate a doubling of the prevalence with COVID-19 compared with the parosmia prevalence without COVID-19 [2,99]. The pre-pandemic level was ~4% [100]. The frequency of parosmia in COVID-19 patients with persistent olfactory dysfunction is higher (11–67%), but survey-type studies likely overestimate prevalence because long-haulers are more motivated to respond [98,101,102]. The more debilitating (persistent) parosmias occur at 1–6 months after diagnosis [97., 98., 99.], suggesting a correlation with smell recovery [98,101]. The mechanism of parosmia is not clear; the most plausible is a peripheral process with aberrant wiring of the olfactory axons into the ‘wrong’ glomerulus in the olfactory bulb [101,103]. A central mechanism at the cerebral level may also contribute [101,103]. Altered mucus may change the effective concentration of odorants, which can transform odorant perception [54], but mucus composition would be expected to normalize after support cell regeneration and, therefore, is not likely to be responsible for parosmias that persist for weeks and months after the infection. The aberrant wiring hypothesis is consistent with our proposal that olfactory receptor neurons become partially ‘immature’ after elimination of their support cells. The dendrite retraction may lead to a disconnect between the innervated glomerulus and the expression of the appropriate odorant receptor. In addition, after extensive destruction of the olfactory epithelium, regenerating olfactory neurons may fail to receive the appropriate signals for innervation of the correct glomerulus, leading to wiring mistakes.
Contributions of virus and host factors to anosmia
Studies from different countries reported widely diverging anosmia prevalences in COVID-19, indicating differences due to ethnicity and/or geographic location [37,60]. It was initially unclear whether such differences in prevalence were due to viral factors or to host factors, or both.
Virus factors: the D614G mutation
When the same population was infected mostly with the G614 variant rather than the D614 virus, olfaction was more often impaired [38]. This constitutes strong evidence that the difference in loss of smell between Western and Asian countries was largely due to infection with different virus variants, with the G614 virus causing a larger anosmia prevalence than the original D614 virus (which affected primarily populations in East and South Asia [38,104]). The D614G mutation in the spike protein appears to enhance entry of this variant into the sustentacular cells and the Bowman gland cells of the olfactory epithelium (Figure 5). It appears that the virus variants differ in how efficiently they infect the support cells, and the population-specific anosmia prevalence reflects such differences.
Virus factors: omicron
The omicron variant causes a lower prevalence of chemosensory dysfunction (Figure 5) [105,106], and this was confirmed by subsequent large-cohort studies [107,108]i. The pooled mean is ~13%, which is a three- to fourfold reduction from the anosmia prevalence caused by the alpha and delta variants (at 35–50%; Figure 5 [107., 108., 109.]). Why does omicron largely spare olfaction? The omicron variant is more hydrophobic and, therefore, may be less soluble in the mucus [105], possibly resulting in fewer virions reaching the support cells. Second, omicron has a lower cell entry efficiency in TMPRSS2-expressing cells (Figure 5), apparently due to less efficient furin cleavage, resulting in lower membrane fusion activity and a shift toward cell entry via the endosomal pathway [79,80,110,111] (Figure 5), a pathway for which support cells have more potent defenses, such as IFITM [22]. Omicron appears to be less efficient in infecting these cells [112], resulting in a lower frequency of anosmia.
Host factors affecting anosmia: ACE2, TMPRSS2, or UGT2A1/A2?
Is there also a contribution of host factors to the anosmia variation between populations? Initially, it was thought that the levels of expression of the virus entry proteins, ACE2 and TMPRSS2, may differ in the frequency of SNPs between ethnicities and this was assumed to contribute to altered binding affinities and enhanced infectivity, and, thus, altered anosmia prevalence [113]. However, more recent studies have concluded that ACE2 expression levels within populations do not correlate with infectivity or chemosensory dysfunction and cannot explain different phenotypes [114,115]. A genome-wide association study (GWAS) on a large number of subjects showed that a different gene locus, the UGT2A1/A2 locus, correlated with differences in anosmia prevalence between populations [116]. The UGT2A1 glucuronosyltransferase metabolizes odorants and other substrates, and genetic variation in this locus differs between ethnicities, with East Asians having the lowest, and populations with European ancestry having the highest levels of expression. This pattern implicates UGT2A1/A2 as the host factor contributing to the differences in anosmia prevalence between populations, rather than ACE2 or TMPRSS2 variants. Since the sustentacular cells are the cell type with most abundant UGT2A1/A2 expression in the olfactory epithelium [24,56], the GWAS [116] further implicates this support cell as the key cell type responsible for COVID-induced anosmia, although the exact role of UGT2A1/A2 awaits clarification.
Concluding remarks
Among the hypotheses attempting to explain anosmia in COVID-19, the strongest evidence favors a lack of support cell-derived cilia-maintenance factors. Immune responses are possibly involved, but appear to occur too late to act as the trigger for anosmia. Broad gene expression changes take place after the olfactory receptor neuron loses its support from sustentacular cells and Bowman gland cells, and the deciliation propels the olfactory neuron back to a less mature state of gene expression, geared toward process growth and away from neurotransmission and signal transduction. When support cells have regenerated, the regrowth of cilia from surviving olfactory neurons enables rapid recovery of smell. SARS-CoV-2 has evolved a novel and unique mechanism of support cell damage, lacking any apparent historic precedent (Box 1 ). While the broader picture of the underlying mechanisms is emerging, many details remain to be clarified (see Outstanding questions). Nevertheless, COVID-19 has revealed a much more intimate relationship between the olfactory neuron and its support cells than previously appreciated.
Box 1. Lack of widespread anosmia in previous pandemics.
Viral infections can cause temporary olfactory dysfunction due to swelling, rhinitis, and obstruction [15,122]. The COVID-19-induced loss of smell differs from common postviral dysfunctions due to its sudden onset, often complete loss of smell, usually short duration, and large numbers of patients affected. Much less olfactory dysfunction was caused by previous flu pandemics (Table I).
It was suggested that COVID-19 has similarities with the 1889 pandemic. The causative agent of the 1889 flu has not been established, unlike the H1N1 influenza pandemic from 1918 (D.A. Pettit, PhD thesis, University of New Hampshire, 1976) [123]. However, circumstantial evidence indicates that the 1889 pandemic may have been caused by a coronavirus. This is based on clinical parallels between COVID-19 and the 1889 pandemic: age risk curve, with older people more affected (unlike the H1N1 pandemic [3]); neurological symptoms; more males affected; pulmonary and cardiac conditions; obesity as a risk factor; multiorgan thrombosis; gastrointestinal symptoms; and long-haulers [123,124]. Based on such similarities, together with genomic similarities between human coronavirus OC43 and bovine coronavirus [125], it was suggested that this pandemic was caused by a coronavirus similar to SARS-CoV-2 [123,124,126].
Among the shared clinical symptoms between COVID-19 and the 1889 pandemic, a frequent loss of smell and taste was emphasized [123,126]. However, the notion that anosmia was frequent in the 1889 pandemic is based on a misunderstanding of one of the transcribed sources ([4], see p. 133): the quoted author [127] in fact refers to a single case ([127], see p. 90). Importantly, in a text providing precise information on the prevalence of loss of smell and taste during the pandemic in Germany [4], it was documented that 20 out of 3042 reports (<1%) mentioned this symptom, prompting the authors to consider chemosensory dysfunction among the ‘more rarely observed nerve symptoms’ ([4], see table in p. 100). Accordingly, one can conclude that loss of smell and taste was not a frequent symptom during the 1889 pandemic. This does not argue against the hypothesis that a coronavirus caused this pandemic, but whatever the virus was, it did not attack the sense of smell and taste to anywhere near the level that SARS-CoV-2 is able to.
In other reports of the 1889 pandemic, chemosensory dysfunction was described to be part of a long-term sequel among long-haulers rather than being an acute symptom during the initial infection [128,129]. We conclude that the widespread olfactory dysfunction caused by SARS-CoV-2 is unique in its intensity and frequency and has no truly comparable precedent during the past 150 years.
Table I.
Comparison of anosmia prevalence in previous pandemicsa
| Pandemic | Years | Virus | Variant | Anosmia (%) | Refs for anosmia prevalence |
|---|---|---|---|---|---|
| Pandemic of 1889 | 1889–1892 | Corona? | <1% | [4] | |
| Pandemic of 1918 | 1918–1920 | Influenza | Not common | [3]b | |
| SARS | 2002 | SARS-CoV-1 | <0.1% | [5,6] | |
| HCoV-NL63 | 2004 | Corona | No reports found | N/A | |
| MERS | 2012 | Corona | Not listed | [7] | |
| COVID-19 | 2019–2020 | SARS-CoV-2 | D614 | ~10% | [37,38] |
| 2020–2021 | G614 | 30–50% | [37,38,105] | ||
| 2021–? | Omicron | ~13% | [105,107,109] |
Abbreviations: D614, original SARS-CoV-2 (Wuhan) strain with an A nucleotide at position 614 of the spike; G614, SARS-CoV-2 variant with a G nucleotide at position 614 of the spike; MERS, Middle East Respiratory Syndrome; N/A, not applicable.
D.A. Pettit, PhD thesis, University of New Hampshire, 1976 (https://scholars.unh.edu/dissertation/1145/).
Alt-text: Box 1
Outstanding questions.
When mucus quantity and composition is assessed in animal models of COVID-induced anosmia, is there a correlation between mucus properties and anosmia?
Is glucose transport/secretion by support cells reduced after virus infection in animal models of COVID-19?
What are the signals that derive from sustentacular cells that cause the deciliation of olfactory receptor neurons?
Can deciliation be prevented by adding glucose to the mucus after SARS-CoV-2 infection?
Does ACE2 as part of the local RAAS have a role in the regulation and chemical composition of the mucus, specifically ion flux, glucose metabolism, and cilia maintenance, and thereby contribute to olfactory dysfunction?
Can deciliated olfactory receptor neurons regrow cilia when they receive signals from regenerated support cells?
The odorant receptor mRNA as well as protein need to be quantified over the course of COVID-19 anosmia; do they correlate with anosmia and with the recovery of smell?
Besides odorant receptors, additional genes and proteins involved in olfactory signal transduction may contribute to anosmia in COVID-19. When such proteins are tracked and manipulated during COVID-19 anosmia, do they correlate and contribute to the cause of anosmia and the recovery of smell?
Can the change of gene expression in olfactory receptor neurons be explained, in part, by an increased fraction of immature neurons following death of mature neurons induced by infection and damage of support cells?
Does the timing of anosmia and smell recovery after anosmia correlate with loss and subsequent regeneration of sustentacular cells and Bowman gland cells?
What is the precise role of the putative host factor UGT2A1/A2 in COVID-19 anosmia?
What are the cytokines, their sources, and their roles in chronic inflammation that may cause the persistent loss of smell in ~5% of COVID-19 patients with olfactory dysfunction?
Alt-text: Outstanding questions
Author contributions
All authors contributed to the writing of this article.
Acknowledgments
Acknowledgments
The authors thank Bing Chen, Nicolas Meunier, and Robert Renden for helpful discussions. Funding was provided by grant 8221 from the Excellence Initiative - Debuts under the Nicolaus Copernicus University Excellence Initiative - Research University programme (to K.B.), and by grant GM103554 from the National Institutes of Health (to C.S.v.B.).
Declaration of interests
The authors declare no competing interests.
Glossary
- ADCY3
this adenylate cyclase participates in olfactory signal transduction and is essential for maintenance of cilia on maturing olfactory receptor neurons.
- Angiotensin-converting enzyme 2 (ACE2)
ACE2 is a host protein that not only increases blood pressure and inflammation, but also allows SARS-CoV-2 to enter host cells.
- Bowman glands
glands located beneath the olfactory epithelium; contribute to the secretion of mucus that covers this epithelium.
- D614G mutation
this aspartic acid to glycine mutation rapidly evolved in the spike protein of SARS-CoV-2; it reduces premature spike shedding and increases infectivity of the virus.
- Forkhead box J1 (Foxj1)
transcription factor that is essential for the development of cilia, especially motile cilia.
- Interferons (IFNs)
cytokines with antiviral properties. Levels of IFN are increased in the olfactory epithelium after infection with SARS-CoV-2.
- Olfactory cilia
extensions protruding from the dendrites of olfactory receptor neurons, which contain the odorant receptors that bind odorants within the mucus covering the olfactory epithelium.
- Olfactory receptor neuron
neuron in the olfactory epithelium that transmits the olfactory signal from the nasal cavity to the brain.
- Omicron variant
variant of SARS-CoV-2 with numerous mutations; compared with previous variants, its preferred route of host cell entry is through endosomal membrane fusion rather than surface membrane fusion.
- Parosmia
dysfunctional sense of smell characterized by the inability of the olfactory system or the brain to correctly identify odors; most often, a pleasant scent is perceived as unpleasant.
- Renin–angiotensin–aldosterone system (RAAS)
not only regulates blood volume and pressure and systemic vascular resistance, but also controls electrolyte balance and is involved in glucose homeostasis.
- Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
coronavirus that causes the respiratory disease called coronavirus disease 2019 (COVID-19).
- Sustentacular cell
non-neuronal cell in the olfactory epithelium provides structural, metabolic, and functional support to olfactory receptor neurons.
- TMPRSS2
transmembrane protease in host cells that cleaves the spike protein of SARS-CoV-2 to allow cell surface membrane fusion and entry of the virus into the host cell.
- UGT2A1/A2 locus
gene encoding a glucuronosyltransferase that metabolizes odorants and thereby enhances odor discrimination in the olfactory system.
Resources
ihttps://doh.wa.gov/sites/default/files/2022-02/420-316-SequencingAndVariantsReport.pdfReferences
- 1.Dhurvey V., et al. Two worst pandemics- Spanish Flu and COVID-19: a review. Magna Sci. Adv. Biol. Pharm. 2021;4:1–12. [Google Scholar]
- 2.Haehner A., et al. SARS-CoV-2 leads to significantly more severe olfactory loss than other seasonal cold viruses. Life (Basel) 2022;12:461. doi: 10.3390/life12030461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edwards S.N. Understanding the present through the past: a comparison of Spanish news coverage of the 1918 flu and COVID-19 pandemics. J. Mass. Commun. Q. 2022;99:12–43. [Google Scholar]
- 4.Leyden E., Guttmann S., editors. Auftrage des Vereins für Innere Medicin in Berlin. Verlag J.F. Bergmann; 1892. [Google Scholar]
- 5.Hwang C.S. Olfactory neuropathy in severe acute respiratory syndrome: report of a case. Acta Neurol. Taiwanica. 2006;15:26–28. [PubMed] [Google Scholar]
- 6.Alshebri M.S., et al. Neurological complications of SARS-CoV, MERS-CoV, and COVID-19. SN Compr. Clin. Med. 2020;2:2037–2047. doi: 10.1007/s42399-020-00589-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zumla A., et al. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flerlage T., et al. Influenza virus and SARS-CoV-2: pathogenesis and host responses in the respiratory tract. Nat. Rev. Microbiol. 2021;19:425–441. doi: 10.1038/s41579-021-00542-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez S., et al. Innate immune signaling in the olfactory epithelium reduces odorant receptor levels: modeling transient smell loss in COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.06.14.20131128. Published online June 16, 2020. [DOI] [Google Scholar]
- 10.Cooper K.W., et al. COVID-19 and the chemical senses: supporting players take center stage. Neuron. 2020;107:219–233. doi: 10.1016/j.neuron.2020.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butowt R., von Bartheld C.S. Anosmia in COVID-19: underlying mechanisms and assessment of an olfactory route to brain infection. Neuroscientist. 2021;27:582–603. doi: 10.1177/1073858420956905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mutiawati E., et al. Anosmia and dysgeusia in SARS-CoV-2 infection: incidence and effects on COVID-19 severity and mortality, and the possible pathobiology mechanisms – a systematic review and meta-analysis. F1000Res. 2021;10:40. doi: 10.12688/f1000research.28393.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang F., Wang Y. COVID-19 anosmia: high prevalence, plural neuropathogenic mechanisms, and scarce neurotropism of SARS-CoV-2? Viruses. 2021;13:2225. doi: 10.3390/v13112225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xydakis M.S., et al. Post-viral effects of COVID-19 in the olfactory system and their implications. Lancet Neurol. 2021;20:753–761. doi: 10.1016/S1474-4422(21)00182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zugaj M., et al. The effect of coronaviruses on olfaction: systematic review. Rhinology. 2021;59:226–235. doi: 10.4193/Rhin20.610. [DOI] [PubMed] [Google Scholar]
- 16.Ojha P., Dixit A. Olfactory training for olfactory dysfunction in COVID-19: a promising mitigation amidst looming neurocognitive sequelae of the pandemic. Clin. Exp. Pharmacol. Physiol. 2022;49:462–473. doi: 10.1111/1440-1681.13626. [DOI] [PubMed] [Google Scholar]
- 17.Karamali K., et al. COVID-19 related olfactory dysfunction. Curr. Opin. Otolaryngol. Head Neck Surg. 2022;30:19–25. doi: 10.1097/MOO.0000000000000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziuzia-Januszewska L., Januszewski M. Pathogenesis of olfactory disorders in COVID-19. Brain Sci. 2022;12:449. doi: 10.3390/brainsci12040449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brann D.H., et al. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci. Adv. 2020;6:eabc5801. doi: 10.1126/sciadv.abc5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keshavarz P., et al. A systematic review of imaging studies in olfactory dysfunction secondary to COVID-19. Acad. Radiol. 2021;28:1530–1540. doi: 10.1016/j.acra.2021.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karimian A., et al. Molecular mechanisms involved in anosmia induced by SARS-CoV-2, with a focus on the transmembrane serine protease TMPRSS2. Arch. Virol. 2022;167:1931–1946. doi: 10.1007/s00705-022-05545-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zazhytska M., et al. Non-cell-autonomous disruption of nuclear architecture as a potential cause of COVID-19-induced anosmia. Cell. 2022;185:1052–1064. doi: 10.1016/j.cell.2022.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butowt R., et al. The olfactory nerve is not a likely route to brain infection in COVID-19: a critical review of data from humans and animal models. Acta Neuropathol. 2021;141:809–822. doi: 10.1007/s00401-021-02314-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan M., et al. Visualizing in deceased COVID-19 patients how SARS-CoV-2 attacks the respiratory and olfactory mucosae but spares the olfactory bulb. Cell. 2021;184:5932–5949. doi: 10.1016/j.cell.2021.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen M., et al. Evolution of nasal and olfactory infection characteristics of SARS-CoV-2 variants. bioRxiv. 2022 doi: 10.1101/2022.04.12.487379. Published online on April 12, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dolgin E. The science behind COVID's assault on smell. Nature. 2022;606:S5–S6. doi: 10.1038/d41586-022-01627-w. [DOI] [PubMed] [Google Scholar]
- 27.Bryche B., et al. Massive transient damage of the olfactory epithelium associated with infection of sustentacular cells by SARS-CoV-2 in golden Syrian hamsters. Brain Behav. Immun. 2020;89:579–586. doi: 10.1016/j.bbi.2020.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kishimoto-Urata M., et al. Prolonged and extended impacts of SARS-CoV-2 on the olfactory neurocircuit. Sci. Rep. 2022;12:5728. doi: 10.1038/s41598-022-09731-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harding J.W., et al. Denervation of the primary olfactory pathway in mice. V. Long-term effect of intranasal ZnSO4 irrigation on behavior, biochemistry and morphology. Brain Res. 1978;140:271–285. doi: 10.1016/0006-8993(78)90460-2. [DOI] [PubMed] [Google Scholar]
- 30.Youngentob S.L., et al. Odorant threshold following methyl bromide-induced lesions of the olfactory epithelium. Physiol. Behav. 1997;62:1241–1252. doi: 10.1016/s0031-9384(97)00301-6. [DOI] [PubMed] [Google Scholar]
- 31.Fleischmann A., et al. Mice with a ‘monoclonal nose’: perturbations in an olfactory map impair odor discrimination. Neuron. 2008;60:1068–1081. doi: 10.1016/j.neuron.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho C.Y., et al. Postmortem assessment of olfactory tissue degeneration and microvasculopathy in patients with COVID-19. JAMA Neurol. 2022;79:544–553. doi: 10.1001/jamaneurol.2022.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spudich S., Nath A. Nervous system consequences of COVID-19. Science. 2022;375:267–269. doi: 10.1126/science.abm2052. [DOI] [PubMed] [Google Scholar]
- 34.Lee Y., et al. Prevalence and duration of acute loss of smell or taste in COVID-19 patients. J. Korean Med. Sci. 2020;35 doi: 10.3346/jkms.2020.35.e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lechien J.R., et al. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): a multicenter European study. Eur. Arch. Otorhinolaryngol. 2020;277:2251–2261. doi: 10.1007/s00405-020-05965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Killingley B., et al. Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults. Nat. Med. 2022;28:1031–1041. doi: 10.1038/s41591-022-01780-9. [DOI] [PubMed] [Google Scholar]
- 37.von Bartheld C.S., et al. Prevalence of chemosensory dysfunction in COVID-19 patients: a systematic review and meta-analysis reveals significant ethnic differences. ACS Chem. Neurosci. 2020;11:2944–2961. doi: 10.1021/acschemneuro.0c00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Bartheld C.S., et al. The D614G virus mutation enhances anosmia in COVID-19 patients: evidence from a systematic review and meta-analysis of studies from South Asia. ACS Chem. Neurosci. 2021;12:3535–3549. doi: 10.1021/acschemneuro.1c00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans J.E., et al. Behavioral, histological, and neurochemical effects of nickel (II) on the rat olfactory system. Toxicol. Appl. Pharmacol. 1995;130:209–220. doi: 10.1006/taap.1995.1026. [DOI] [PubMed] [Google Scholar]
- 40.Brann J.H., Firestein S.J. A lifetime of neurogenesis in the olfactory system. Front. Neurosci. 2014;8:182. doi: 10.3389/fnins.2014.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwob J.E., et al. Reconstitution of the rat olfactory epithelium after methyl bromide-induced lesion. J. Comp. Neurol. 1995;359:15–37. doi: 10.1002/cne.903590103. [DOI] [PubMed] [Google Scholar]
- 42.Schwob J.E. Neural regeneration and the peripheral olfactory system. Anat. Rec. 2002;269:33–49. doi: 10.1002/ar.10047. [DOI] [PubMed] [Google Scholar]
- 43.Schwob J.E., Jang W. In: Neural Development and Stem Cell. 2nd edn. Rao M.S., editor. Humana Press; 2006. Stem cells of the adult olfactory epithelium; pp. 219–233. [Google Scholar]
- 44.Liang F. Sustentacular cell enwrapment of olfactory receptor neuronal dendrites: an update. Genes (Basel) 2020;11:493. doi: 10.3390/genes11050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McClintock T.S., et al. Maturation of the olfactory sensory neuron and its cilia. Chem. Senses. 2020;45:805–822. doi: 10.1093/chemse/bjaa070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kondo K., et al. Age-related changes in cell dynamics of the postnatal mouse olfactory neuroepithelium: cell proliferation, neuronal differentiation, and cell death. J. Comp. Neurol. 2010;518:1962–1975. doi: 10.1002/cne.22316. [DOI] [PubMed] [Google Scholar]
- 47.Liberia T., et al. Sequential maturation of olfactory sensory neurons in the mature olfactory epithelium. eNeuro. 2019;6 doi: 10.1523/ENEURO.0266-19.2019. ENEURO.0266-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bilinska K., et al. Expression of the SARS-CoV-2 entry proteins, ACE2 and TMPRSS2, in cells of the olfactory epithelium: identification of cell types and trends with age. ACS Chem. Neurosci. 2020;11:1555–1562. doi: 10.1021/acschemneuro.0c00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klingenstein M., et al. Evidence of SARS-CoV2 entry protein ACE2 in the human nose and olfactory bulb. Cells Tissues Organs. 2020;209:155–164. doi: 10.1159/000513040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen M., et al. Elevated ACE-2 expression in the olfactory neuroepithelium: implications for anosmia and upper respiratory SARS-CoV-2 entry and replication. Eur. Respir. J. 2020;56:2001948. doi: 10.1183/13993003.01948-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Melo G.D., et al. COVID-19-related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. Sci. Transl. Med. 2021;13:eabf8396. doi: 10.1126/scitranslmed.abf8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye Q., et al. SARS-CoV-2 infection in the mouse olfactory system. Cell Discov. 2021;7:49. doi: 10.1038/s41421-021-00290-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagashima A., Touhara K. Enzymatic conversion of odorants in nasal mucus affects olfactory glomerular activation patterns and odor perception. J. Neurosci. 2010;30:16391–16398. doi: 10.1523/JNEUROSCI.2527-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Breer H. Olfactory receptors: molecular basis for recognition and discrimination of odors. Anal. Bioanal. Chem. 2003;377:427–433. doi: 10.1007/s00216-003-2113-9. [DOI] [PubMed] [Google Scholar]
- 55.Seo J.S., et al. The microvillar and solitary chemosensory cells as the novel targets of infection of SARS-CoV-2 in Syrian golden hamsters. Viruses. 2021;13:1653. doi: 10.3390/v13081653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Finlay J.B., et al. Persistent post-COVID-19 smell loss is associated with inflammatory infiltration and altered olfactory epithelial gene expression. bioRxiv. 2022 doi: 10.1101/2022.04.17.488474. Published online April 18, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Villar P.S., et al. Energy requirements of odor transduction in the chemosensory cilia of olfactory sensory neurons rely on oxidative phosphorylation and glycolytic processing of extracellular glucose. J. Neurosci. 2017;37:5736–5743. doi: 10.1523/JNEUROSCI.2640-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Acevedo C., et al. Possible ATP trafficking by ATP-shuttles in the olfactory cilia and glucose transfer across the olfactory mucosa. FEBS Lett. 2019;593:601–610. doi: 10.1002/1873-3468.13346. [DOI] [PubMed] [Google Scholar]
- 59.Villar P.S., et al. Energy sources that fuel metabolic processes in protruding finger-like organelles. FEBS J. 2021;288:3799–3812. doi: 10.1111/febs.15620. [DOI] [PubMed] [Google Scholar]
- 60.Luchiari H.R., et al. Does the RAAS play a role in loss of taste and smell during COVID-19 infections? Pharmacogenom. J. 2021;21:109–115. doi: 10.1038/s41397-020-00202-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krishnan S., et al. Metabolic perturbation associated with COVID-19 disease severity and SARS-CoV-2 replication. Mol. Cell. Proteomics. 2021;20 doi: 10.1016/j.mcpro.2021.100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baxter B.D., et al. Transcriptional profiling reveals potential involvement of microvillous TRPM5-expressing cells in viral infection of the olfactory epithelium. BMC Genomics. 2021;22:224. doi: 10.1186/s12864-021-07528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kraus A., et al. Intranasal delivery of SARS-CoV-2 spike protein is sufficient to cause olfactory damage, inflammation and olfactory dysfunction in zebrafish. Brain Behav. Immun. 2022;102:341–359. doi: 10.1016/j.bbi.2022.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee K.P., et al. Nasal lesion development and reversibility in rats exposed to aerosols of dibasic esters. Toxicol. Pathol. 1992;20:376–393. doi: 10.1177/019262339202000308. [DOI] [PubMed] [Google Scholar]
- 65.Bergström U., et al. Methimazole-induced damage in the olfactory mucosa: effects on ultrastructure and glutathione levels. Toxicol. Pathol. 2003;31:379–387. doi: 10.1080/01926230390201101. [DOI] [PubMed] [Google Scholar]
- 66.Jia C., et al. Nickel sulfate induces location-dependent atrophy of mouse olfactory epithelium: protective and proliferative role of purinergic receptor activation. Toxicol. Sci. 2010;115:547–556. doi: 10.1093/toxsci/kfq071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morrison E.E., Costanzo R.M. Morphology of the human olfactory epithelium. J. Comp. Neurol. 1990;297:1–13. doi: 10.1002/cne.902970102. [DOI] [PubMed] [Google Scholar]
- 68.Choksi S.P., et al. Switching on cilia: transcriptional networks regulating ciliogenesis. Development. 2014;141:1427–1441. doi: 10.1242/dev.074666. [DOI] [PubMed] [Google Scholar]
- 69.Nickell M.D., et al. Genomics of mature and immature olfactory sensory neurons. J. Comp. Neurol. 2012;520:2608–2629. doi: 10.1002/cne.23052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saraiva L.R., et al. Hierarchical deconstruction of mouse olfactory sensory neurons: from whole mucosa to single-cell RNA-seq. Sci. Rep. 2015;5:18178. doi: 10.1038/srep18178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Larson E.D., et al. A subset of olfactory sensory neurons express forkhead box J1-driven eGFP. Chem. Senses. 2019;44:663–671. doi: 10.1093/chemse/bjz060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robinot R., et al. SARS-CoV-2 infection induces the dedifferentiation of multiciliated cells and impairs mucociliary clearance. Nat. Commun. 2021;12:4354. doi: 10.1038/s41467-021-24521-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schreiner T., et al. SARS-CoV-2 infection dysregulates cilia and basal cell homeostasis in the respiratory epithelium of hamsters. Int. J. Mol. Sci. 2022;23:5124. doi: 10.3390/ijms23095124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang A.J., et al. Severe acute respiratory syndrome coronavirus 2 infects and damages the mature and immature olfactory sensory neurons of hamsters. Clin. Infect. Dis. 2021;73:e503–e512. doi: 10.1093/cid/ciaa995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reyna R.A., et al. Recovery of anosmia in hamsters infected with SARS-CoV-2 is correlated with repair of the olfactory epithelium. Sci. Rep. 2022;12:628. doi: 10.1038/s41598-021-04622-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pozharskaya T., Lane A.P. Interferon gamma causes olfactory dysfunction without concomitant neuroepithelial damage. Int. Forum Allergy Rhinol. 2013;3:861–865. doi: 10.1002/alr.21226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Avnat E., et al. Elevated expression of RGS2 may underlie reduced olfaction in COVID-19 patients. J. Pers. Med. 2022;12:1396. doi: 10.3390/jpm12091396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Majdoul S., Compton A.A. Lessons in self-defence: inhibition of virus entry by intrinsic immunity. Nat. Rev. Immunol. 2021;13:1–14. doi: 10.1038/s41577-021-00626-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jackson C.B., et al. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meng B., et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts tropism and fusogenicity. Nature. 2022;603:706–714. doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chupp G., et al. A phase 2 randomized, double-blind, placebo-controlled trial of oral camostat mesylate for early treatment of COVID-19 outpatients showed shorter illness course and attenuation of loss of smell and taste. medRxiv. 2022 doi: 10.1101/2022.01.28.22270035. Published online January 31, 2022. [DOI] [Google Scholar]
- 82.Henkin R.I., et al. Interleukin 6 in hyposmia. JAMA Otolaryngol. Head Neck Surg. 2013;139:728–734. doi: 10.1001/jamaoto.2013.3392. [DOI] [PubMed] [Google Scholar]
- 83.Cazzolla A.P., et al. Taste and smell disorders in COVID-19 patients: role of interleukin-6. ACS Chem. Neurosci. 2020;11:2774–2781. doi: 10.1021/acschemneuro.0c00447. [DOI] [PubMed] [Google Scholar]
- 84.Torabi A., et al. Proinflammatory cytokines in the olfactory mucosa result in COVID-19 induced anosmia. ACS Chem. Neurosci. 2020;11:1909–1913. doi: 10.1021/acschemneuro.0c00249. [DOI] [PubMed] [Google Scholar]
- 85.Sanli D.E.T., et al. Relationship between disease severity and serum IL-6 levels in COVID-19 anosmia. Am. J. Otolaryngol. 2021;42 doi: 10.1016/j.amjoto.2020.102796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Francois A., et al. Daily oscillation of odorant detection in rat olfactory epithelium. Eur. J. Neurosci. 2017;45:1613–1622. doi: 10.1111/ejn.13600. [DOI] [PubMed] [Google Scholar]
- 87.Zhang Z., et al. Deletion of type 3 adenylyl cyclase perturbs the postnatal maturation of olfactory sensory neurons and olfactory cilium ultrastructure in mice. Front. Cell. Neurosci. 2017;11:1. doi: 10.3389/fncel.2017.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McClintock T.S., Sammeta N. Trafficking prerogatives of olfactory receptors. Neuroreport. 2003;14:1547–1552. doi: 10.1097/00001756-200308260-00001. [DOI] [PubMed] [Google Scholar]
- 89.Mori I., et al. Olfactory receptor neurons prevent dissemination of neurovirulent influenza A virus into the brain by undergoing virus-induced apoptosis. J. Gen. Virol. 2002;83:2109–2116. doi: 10.1099/0022-1317-83-9-2109. [DOI] [PubMed] [Google Scholar]
- 90.van Riel D., et al. The olfactory nerve: a shortcut for influenza and other viral diseases into the central nervous system. J. Pathol. 2015;235:277–287. doi: 10.1002/path.4461. [DOI] [PubMed] [Google Scholar]
- 91.Tian J., et al. Sendai virus induces persistent olfactory dysfunction in a murine model of PVOD via effects on apoptosis, cell proliferation, and response to odorants. PLoS One. 2016;11 doi: 10.1371/journal.pone.0159033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Le Bon S.D., Horoi M. Is anosmia the price to pay in an immune-induced scorched-earth policy against COVID-19? Med. Hypotheses. 2020;143 doi: 10.1016/j.mehy.2020.109881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bourgon C., et al. Neutrophils initiate the destruction of the olfactory epithelium during SARS-CoV-2 infection in hamsters. bioRxiv. 2022 doi: 10.1101/2022.03.15.484439. Published online March 15, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lane A.P., et al. A genetic model of chronic rhinosinusitis-associated olfactory inflammation reveals reversible functional impairment and dramatic neuroepithelial reorganization. J. Neurosci. 2010;30:2324–2329. doi: 10.1523/JNEUROSCI.4507-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen M., et al. Acute inflammation regulates neuroregeneration through the NF-κB pathway in olfactory epithelium. Proc. Natl. Acad. Sci. U. S. A. 2017;114:8089–8094. doi: 10.1073/pnas.1620664114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tan B.K.J., et al. Prognosis and persistence of smell and taste dysfunction in patients with Covid-19: meta-analysis with parametric cure modelling of recovery curves. BMJ. 2022;378 doi: 10.1136/bmj-2021-069503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Parma V., et al. More than smell-COVID-19 is associated with severe impairment of smell, taste, and chemesthesis. Chem. Senses. 2020;45:609–622. doi: 10.1093/chemse/bjaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Raad N., et al. Parosmia in patients with COVID-19 and olfactory dysfunction. Int. Forum Allergy Rhinol. 2021;11:1497–1500. doi: 10.1002/alr.22818. [DOI] [PubMed] [Google Scholar]
- 99.Gurrola J.G., 2nd, et al. Short-term chemosensory distortions and phantoms in COVID-19. Laryngoscope Investig. Otolaryngol. 2021;6:172–176. doi: 10.1002/lio2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nordin S., et al. Prevalence of parosmia: the Skövde population-based studies. Rhinology. 2007;45:50–53. [PubMed] [Google Scholar]
- 101.Lerner D.K., et al. Clinical features of parosmia associated with COVID-19 infection. Laryngoscope. 2022;132:633–639. doi: 10.1002/lary.29982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ohla K., et al. A follow-up on quantitative and qualitative olfactory dysfunction and other symptoms in patients recovering from COVID-19 smell loss. Rhinology. 2022;60:207–217. doi: 10.4193/Rhin21.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Parker J.K., et al. Insights into the molecular triggers of parosmia based on gas chromatography olfactometry. Commun. Med. (Lond). 2022;2:58. doi: 10.1038/s43856-022-00112-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Korber B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Butowt R., et al. Why does the omicron variant largely spare olfactory function? Implications for the pathogenesis of anosmia in COVID-19. J. Infect. Dis. 2022;226:1304–1308. doi: 10.1093/infdis/jiac113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rodriguez-Sevilla J.J., et al. Is there less alteration of smell sensation in patients with omicron SARS-CoV-2 variant infection? Front. Med. 2022;9 doi: 10.3389/fmed.2022.852998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Menni C., et al. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of omicron and delta variant dominance: a prospective observational study from the ZOE COVID Study. Lancet. 2022;399:1618–1624. doi: 10.1016/S0140-6736(22)00327-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Whitaker M., et al. Variant-specific symptoms of COVID-19 among 1,542,510 people in England. medRxiv. 2022 doi: 10.1101/2022.05.21.22275368. Published online May 23, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vihta K.D., et al. Omicron-associated changes in SARS-CoV-2 symptoms in the United Kingdom. Clin. Infect. Dis. 2022 doi: 10.1093/cid/ciac613. Published online August 3, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang J., et al. Structural and functional impact by SARS-CoV-2 Omicron spike mutations. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Peacock T.P., et al. The altered entry pathway and antigenic distance of the SARS-CoV-2 Omicron variant map to separate domains of spike protein. bioRxiv. 2022 doi: 10.1101/2021.12.31.474653. Published online May 13, 2022. [DOI] [Google Scholar]
- 112.Armando F., et al. SARS-CoV-2 Omicron variant causes mild pathology in the upper and lower respiratory tract of hamsters. Nat. Commun. 2022;13:3519. doi: 10.1038/s41467-022-31200-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Butowt R., et al. Chemosensory dysfunction in COVID-19: integration of genetic and epidemiological data points to D614G spike protein variant as a contributing factor. ACS Chem. Neurosci. 2020;11:3180–3184. doi: 10.1021/acschemneuro.0c00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hashizume M., et al. Population-specific ACE2 single-nucleotide polymorphisms have limited impact on SARS-CoV-2 infectivity in vitro. Viruses. 2021;13:67. doi: 10.3390/v13010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Braga-Paz I., et al. Negative correlation between ACE2 gene expression levels and loss of taste in a cohort of COVID-19 hospitalized patients: new clues to long-term cognitive disorders. Front. Cell. Infect. Microbiol. 2022;12 doi: 10.3389/fcimb.2022.905757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shelton J.F., et al. The UGT2A1/UGT2A2 locus is associated with COVID-19-related loss of smell or taste. Nat. Genet. 2022;54:121–124. doi: 10.1038/s41588-021-00986-w. [DOI] [PubMed] [Google Scholar]
- 117.Getchell T.V., et al. Perireceptor and receptor events in vertebrate olfaction. Prog. Neurobiol. 1984;23:317–345. doi: 10.1016/0301-0082(84)90008-x. [DOI] [PubMed] [Google Scholar]
- 118.Heydel J.M., et al. Odorant-binding proteins and xenobiotic metabolizing enzymes: implications in olfactory perireceptor events. Anat. Rec. (Hoboken) 2013;296:1333–1345. doi: 10.1002/ar.22735. [DOI] [PubMed] [Google Scholar]
- 119.Strotmann J., Breer H. Internalization of odorant-binding proteins into the mouse olfactory epithelium. Histochem. Cell Biol. 2011;136:357–369. doi: 10.1007/s00418-011-0850-y. [DOI] [PubMed] [Google Scholar]
- 120.Al-Ani R.M., Acharya D. Prevalence of anosmia and ageusia in patients with COVID-19 at a primary health center, Doha, Qatar. Indian J. Otolaryngol. Head Neck Surg. 2020 doi: 10.1007/s12070-020-02064-9. Published online August 19, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zayet S., et al. Clinical features of COVID-19 and influenza: a comparative study on Nord Franche-Comte cluster. Microbes Infect. 2020;22:481–488. doi: 10.1016/j.micinf.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hummel T., et al. Position paper on olfactory dysfunction. Rhinology. 2016;56:1–30. doi: 10.4193/Rhino16.248. [DOI] [PubMed] [Google Scholar]
- 123.Brüssow H., Brüssow L. Clinical evidence that the pandemic from 1889 to 1891 commonly called the Russian flu might have been an earlier coronavirus pandemic. Microb. Biotechnol. 2021;14:1860–1870. doi: 10.1111/1751-7915.13889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Erkoreka A., et al. Coronavirus as the possible causative agent of the 1889-1894 pandemic. Infect. Dis. Rep. 2022;14:453–469. doi: 10.3390/idr14030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vijgen L., et al. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ramassy L., et al. Paleoserology points to Coronavirus as possible causative pathogens of the 'Russian flu'. Microb. Biotechnol. 2022;15:1943–1945. doi: 10.1111/1751-7915.14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Habermann J. Zur Erkrankung des Ohres bei Influenza. Prager med. Wochenschr. 1890;15:89–90. [Google Scholar]
- 128.Anonymous The influenza epidemic. Br. Med. J. 1892;1:355–357. [PMC free article] [PubMed] [Google Scholar]
- 129.Dowse T.S. On Brain and Nerve Exhaustion (Neurasthenia); and on the Nervous Sequelae of Influenza. Bailliere, Tindall and Cox; 1894. [Google Scholar]