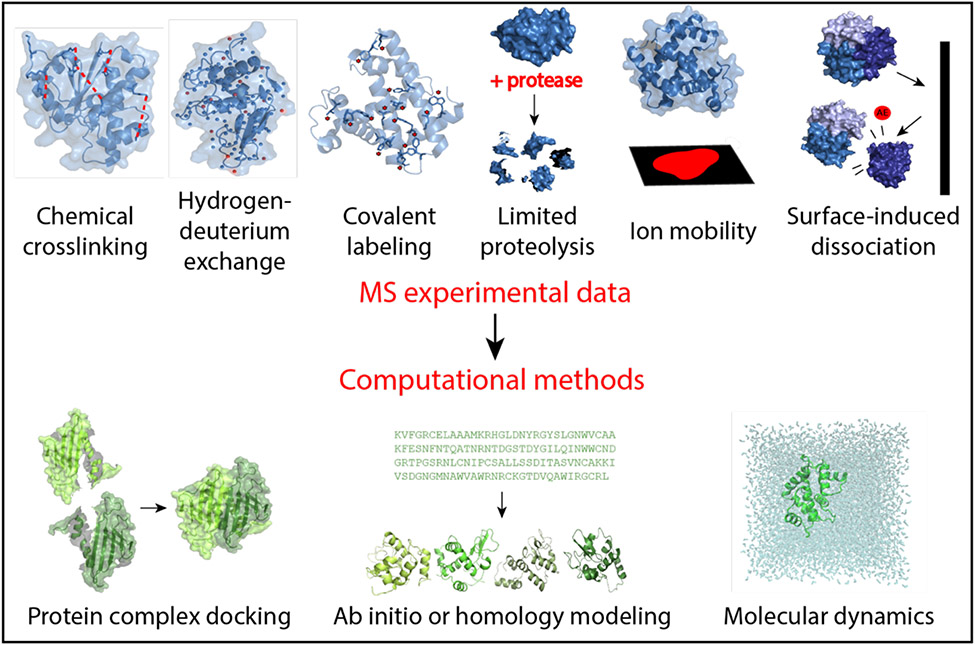
Abstract
Knowledge of protein structure is crucial to our understanding of biological function and is routinely used in drug discovery. High-resolution techniques to determine the three-dimensional atomic coordinates of proteins are available. However, such methods are frequently limited by experimental challenges such as sample quantity, target size, and efficiency. Structural mass spectrometry (MS) is a technique in which structural features of proteins are elucidated quickly and relatively easily. Computational techniques that convert sparse MS data into protein models that demonstrate agreement with the data are needed. This review features cutting-edge computational methods that predict protein structure from MS data such as chemical cross-linking, hydrogen-deuterium exchange, hydroxyl radical protein footprinting, limited proteolysis, ion mobility, and surface-induced dissociation. Additionally, we address future directions for protein structure prediction with sparse MS data.
Keywords: computational methods, mass spectrometry, protein structure prediction, sparse experimental data
Introduction
Proteins are involved in nearly every life process, making them important subjects for studying the molecular basis of disease. Additionally, protein structures can be harnessed for structure-based drug discovery with existing and designed drug-like molecules (1). However, a disparity currently exists between the number of known protein sequences and the number of determined structures. Methodologies to elucidate protein structure are vital to our understanding of molecular biology and for continued use in drug discovery.
Multiple experimental techniques exist to determine high-resolution protein structure. X-ray crystallography is a popular method in which a high concentration of a protein target is crystalized. Then, the crystals are struck with an X-ray beam in order to elucidate a diffraction pattern from which atomic protein coordinates can be determined (2). While powerful, crystallography is rate-limited by the crystallization process, as ascertaining experimental conditions ideal for crystal growth can be a tedious if not impossible process. X-ray crystallography has historically been more successful for ordered and monomeric proteins. Nuclear magnetic resonance (NMR) spectroscopy is another high-resolution technique. It utilizes the chemical shifts of protein atoms for structure determination (3). It is in most cases limited to smaller proteins in order to avoid overlapping peaks. Cryo-electron microscopy (cryo-EM) has recently emerged as a promising structure determination technique that can elucidate larger, more complex proteins while bypassing the need for crystallization, probing the protein more physiological conditions (4). However, further optimization of cryo-EM methodologies is required to consistently determine higher resolution density maps.
Due to the limitations of above techniques, many proteins or protein complexes currently evade high-resolution structure determination. Thus, additional experimental methods are needed to provide insight into structural features. Structural mass spectrometry (MS) is a powerful complementary approach that can overcome limitations of above-mentioned methods with its high sensitivity, theoretically unlimited size constraint, and speed. Although the data provided by MS are too sparse for full high-resolution structure elucidation, structural MS can be used to examine size, solvent accessibility, and topography of proteins (5-7). Several MS techniques exist that can elucidate elements of protein tertiary and quaternary structure, including chemical cross-linking (XL-MS) (8; 9), hydrogen-deuterium exchange (HDX-MS) (10), covalent labeling (CL-MS) (11; 12), limited proteolysis (13), ion mobility (IM-MS) (14), and surface-induced dissociation (SID-MS) (15), reviewed here (Figure 1). In chemical cross-linking, residue modifications provide insight into spatial proximity of modified residues. HDX-MS, CL-MS, and limited proteolysis data are used to infer residue solvent exposure. IM-MS data reveal information about the size and shape of proteins, while SID-MS is used to analyze protein complex connectivity and stoichiometry. Sparse experimental data from structural mass spectrometry generally must be interpreted in combination with computational methods to elucidate protein structure.
Figure 1.
Mass spectrometry-based methods and computational modeling explored in this review. Chemical crosslinking involves the modification of residues, commonly lysine, to provide information regarding spatial proximity. Hydrogen-deuterium exchange examines the exchange rate of amide hydrogens with deuterium solvent to give insight into solvent exposure and residue flexibility. Covalent labeling is reliant upon the irreversible covalent modification of residues, illuminating solvent exposure and topology. Limited proteolysis uses a protease enzyme to cleave proteins into fragments based on solvent exposure. Ion mobility is used to investigate shape and size of proteins based on the collision cross sectional area. Appearance energies (AE) can be deduced from surface-induced dissociation, which is used to study the stoichiometry and connectivity of protein complexes. Data from these techniques is then incorporated into computational modeling techniques such as protein-protein docking to examine complexes, structure prediction via ab initio or homology modeling, and molecular dynamics based on experimental restraints.
Computational methods have increasingly been employed to complement experimental techniques in order to elucidate protein structures (16; 17). As experimental data becomes more readily available, software packages can be employed to combine sparse data with advanced structure sampling and scoring techniques. A number of computational tools currently exist for protein structure modeling, including the Rosetta software suite (17; 18), I-TASSER (19), Phyre2 (20), Integrative Modeling Platform (IMP) (21), HADDOCK (22), and MODELLER (23). Sparse experimental data can be implemented during the computational modeling process or used as a filter during post model generation analysis. Here, we will be highlighting work that combines computational efforts for protein structure examination with sparse experimental data from MS. We will be discussing work that incorporates XL-MS, HDX-MS, CL-MS, limited proteolysis, IM-MS, and SID-MS experimental data into computational modeling.
Chemical cross-linking
Chemical cross-linking (XL) utilizes reagents to chemically link two amino acids, particularly the side chain atoms within lysine residues, in order to assess proximity within a protein or within protein complexes (9). After digestion and separation via liquid chromatography, crosslinks can be identified via tandem MS. XL-MS experiments provide insight into protein structure. Residues that are distant to one another in amino acid sequence can be identified as being within spatial proximity. Interactions between protein complex subunits can also be inferred by residues that are identified as crosslinked. Only residues that are solvent exposed should be modified by a crosslinking reagent. As such, the crosslinking agent can give insight into proximity between surface residues, from which contact information can further be derived with computational methods that utilize XL-MS data. XL-MS efforts have been incorporated into the Critical Assessment of protein Structure Prediction (CASP) challenges to integrate high-density XL-MS data into prediction methods (24).
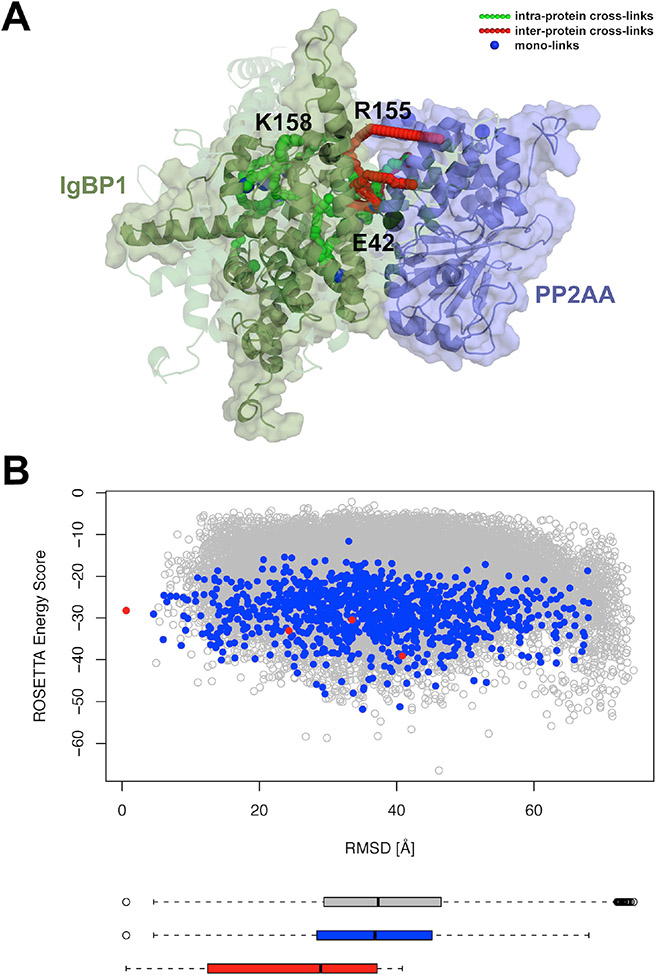
Kahraman and coworkers developed methodologies for applying crosslinking data to homology modeling, de novo modeling, and protein-protein docking (25). A database, XLdb, was also assembled which contained XL-MS data for individual proteins and protein complexes along with the corresponding protein data bank entries, providing a source of accessible data for the mass spectrometry and computational communities. Building on an earlier publication in which X-walk, a program that determines the shortest distance between crosslinked amino acids within solvent accessible regions (26), was established, distance restraints determined from XL-MS data were implemented into the Rosetta scoring function. The Rosetta functionality penalized models that conflicted with the experimental data. For instance, models with residues participating in a cross-link that were spatially farther apart than the spacer length of the crosslinker received a penalty. The distance restraints were also applied as filters to examine existing models. Overall, it was found that usage of the crosslinking distance restraints improved the RMSD of the top scoring models and improved protein-protein docking (Figure 2). Similar methodology was applied in work by Lössl and colleagues in which crosslinking data was used to determine differences in conformational ensembles and interaction modes of singular and interacting proteins (27). Additionally, recent work by Piotrowski and colleagues used XL distance restraints in combination with Rosetta to build models of calmodulin interacting with bMunc13-2 and then subsequently identify a unique binding mode (28).
Figure 2.
Improvement of model prediction and scoring with XL-MS data. A, Best scoring models of IgBP1 (green) complexed with PP2AA (purple), with the opaque cartoon depicting the best scoring model from the largest cluster and the more transparent cartoons depicting the best scoring models from 2-4th largest clusters. Crosslinks are depicted as green, red, and blue spheres, with black spheres representing mutations. B, Rosetta score versus RMSD to the largest cluster plot for models with minimum of six inter-protein XLs (grey), minimum of six inter-protein XLs with binding interface larger than 900 Å2 (blue), and representative models from the four biggest clusters (red). Figure reproduced under the Creative Commons License from Kahraman et al. (2013); copyright 2013 PLOS.
XL-MS data was used in protein structure investigation of human serum albumin protein domains by Belsom and colleagues (29). Instead of traditional XL reagents, this work employed a photo-XL agent that led to increased XL data to probe the protein in isolation and within blood samples. Upon modeling with XL-MS data as restraints and residue contacts predicted with a newly developed software, serum albumin protein models were successfully identified with low RMSD values (3-6 Å) for both the purified and sample models. A similar approach was explored in work from dos Santos et al. in which XL-MS data along with coevolutionary information was applied to protein structure prediction (30). In the work, simplified models containing only alpha carbons were used in combination with restraints from XL-MS and coevolutionary data via direct coupling analysis to elucidate tertiary structure. Models were evaluated by clustering and TM-score for multiple proteins. Quality models were identified from the method, validating the effectiveness of the proposed methodology.
Hauri et al. used computationally determined models for a very large (1.8 MDa) protein complex found in human plasma in order to examine specific peptides from XL-MS, an effort denoted as targeted chemical cross-linking MS (31). Targeted XL-MS used different MS acquisition techniques to discriminate between computational models of the protein complex modeled by Rosetta’s homology modeling protocol. Proteins from the complex were docked together in order to produce a collection of potential models that represented the quaternary structure of the complex. Models of the protein-protein complex that scored well with the crosslinking data were used to identify a short list of potentially crosslinked lysine pairs. Models then underwent a flexible backbone docking workflow with crosslinking data as distance restraints. Overall, the development of targeted XL-MS paved the way for continued improvement of quaternary structure prediction of highly complex systems. Recent work by Khakzad and others sought to elucidate another large protein complex, the membrane attack complex (32). A streamlined protocol for targeted XL-MS was pursued to examine the bacterial protein complex in human plasma. The crosslinking results were utilized to obtain a complete model of the complex that was corroborated with existing models from crystallography and cryo-EM. This work further demonstrated the applicability of XL-MS, particularly to complex targets from bacterial systems relevant to human disease.
XLFF, a force field that relied upon XL-MS restraints was applied to Rosetta’s ab initio protocol by Ferrari and colleagues (33). This was accomplished by determining the probability of identifying residues that could potentially crosslink within a nonredundant set of proteins from the protein data bank. The resulting probability curve was then used to determine a potential energy function reliant on the crosslinker length and the residues involved in linkage. It was observed that usage of the XLFF force field resulted in higher quality, more native-like models occurring within the top scoring model distributions.
In addition to inclusion of crosslinking data within Rosetta, software has been developed outside the Rosetta suite. Degiacomi and coworkers implemented a software tool called DynamXL to consider the implications of dynamics when modeling crosslinking data (34). Contrasting other methods that rely upon the beta carbon for distance measurements, the DynamXL algorithm employed the side chain nitrogen atom of lysine for distance calculations, which was suggested as more experimentally accurate and less computationally expensive. Additionally, the method took the flexibility of residue side chains into account by examining different rotamers and backbone conformations. The work sought to minimize the elimination of reasonable crosslinks, while simultaneously excluding impossible crosslinks, which led to less error when classifying cross linkages. Overall, application of this methodology led to improved RMSD values from protein-protein docking, highlighting the accuracy of the implementation.
Recent work by Mintseris and Gygi explored high density XL-MS efforts in combination with IMP and Rosetta (35). The methodology was used to model carbonic anhydrase proteins and the yeast proteasome. To minimize computational cost, the implemented software reduced sampling of decoy and target peptides to minimize false discovery rates and simplify false discovery rate calculations. Alternative reagents that established crosslinks with additional residue types promoted the crosslinking density, thus providing better results. XL-MS data was applied to modeling of inhibitor-bound carbonic anhydrase via restraints applied during protein-protein docking with Rosetta. High-quality models were identified. Additionally, the work tackled the modeling of the yeast proteasome with both Rosetta and the IMP based on the XL-MS data. Coase-grained models of the complex were elucidated, and regions were verified by existing cryo-EM models.
Hydrogen-deuterium exchange
Hydrogen-deuterium exchange (HDX) is a prevalent non-specific covalent labeling technique in which a protein is exposed to a deuterium-rich solvent (10). Amide hydrogen atoms are able to exchange with deuterium atoms to label the protein backbone. After digestion and separation with liquid chromatography, MS can be used to identify regions of exchange. HDX-MS has also been used in combination with other techniques such as electron capture dissociation to assess hydrogen bonding configurations (36). Regions of the protein are more likely to be modified by HDX if the amide hydrogens are solvent accessible and not actively participating in a hydrogen bond. HDX data is often resolved to fragment level but occasionally residue-specific modifications are reported. From there, data can be expressed as percentage modification, rate constants, or protection factors (PF), all of which are routinely used as input into computational modeling to guide results based upon agreement with HDX data.
HDX-MS data has been used in combination with homology modeling, as seen in work from Zhang and coworkers (37). Homology modeling with MODELLER, Phyre2, and I-TASSER were used to model the tertiary structure of cytochrome C. HDX-MS results were taken into account when examining the models. Additionally, the relationship between HDX modification and SASA was examined to identify the best models. It was determined that the modeling efforts with Phyre2 demonstrated best agreement with the HDX-MS results, and the SASA values from this model led to a better correlation with the percent modification identified from HDX experiments. The results of this work effectively demonstrated that both HDX data and solvent exposure could be used to identify better homology models and to improve upon our previous understanding of the cytochrome C mechanism. While HDX-MS data has not been applied to ab initio modeling, HDX-NMR data has been recently implemented into protein structure prediction (38).
HDX-MS data in combination with molecular dynamics (MD) simulations were employed to examine empirical and fractional population models for G-protein signaling regulator proteins in work from Mohammadiarani et al. (39). Using long timescale MD simulations with AMBER and CHARMM forcefields, PFs were calculated from simulation frames and then compared to experimentally determined percent modification data. It was determined that fractional population models were more accurate and less prone to error than empirical models, arguing that the SASA of amide hydrogens coupled with the distance between the amide hydrogen and first polar atom could be used for accurate predictions. It was also indicated that amide hydrogen atoms could fluctuate in exposure over a sub 100 ps timescale. HDX-MS and MD simulations were also applied to examine interactions between lipids and membrane proteins such as lipid-induced conformational changes in proteins in work from Martens and coworkers (40). The framework developed in the study emphasized a multi-step protocol. After using HDX-MS to evaluate the protein in the presence and absence of lipids, interactions were interpreted via MD simulations in various bilayer conditions. The interactions identified from the simulation were then corroborated by experimental mutagenesis of relevant sites. The methodology presented in this work was suggested as a basis for further study of various lipid-protein interactions in membranes. Beyond this work, size-exclusion chromatography in combination with HDX-MS and circular dichroism were used with computational techniques such as homology modeling and MD simulations to examine the activity of transaminases in work from Makarov and others (41). The study demonstrated that the protocol could be applied to enzyme-directed evolution efforts.
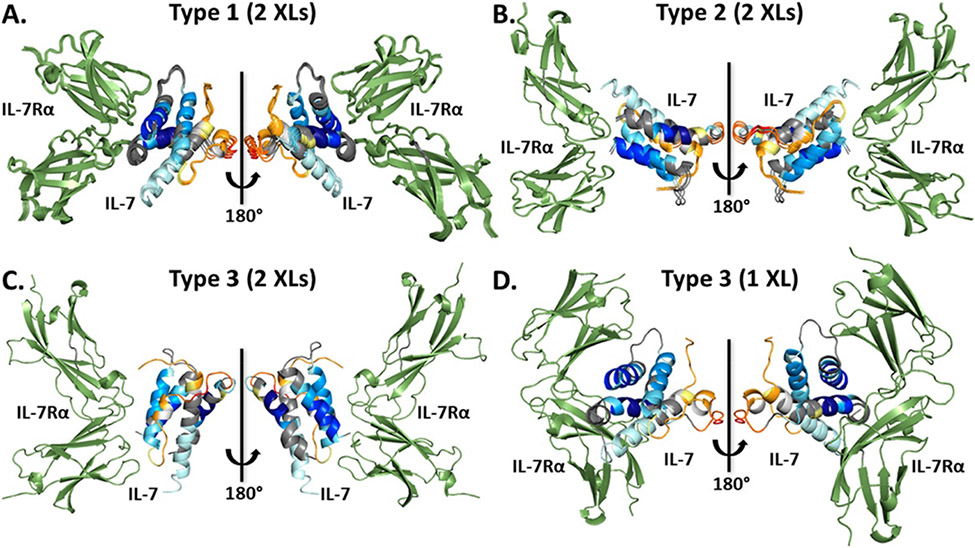
Recently, Zhang and colleagues used both XL-MS and HDX-MS data to evaluate protein-protein docking models of interleukin 7 and its alpha receptor (Figure 3) (42). HDX-MS analysis was performed on free interleukin 7 and when it was bound with its receptor to elucidate changes in exposure. XL-MS was also applied to the system in order to identify residues involved in the receptor binding interface of interleukin 7. Protein-protein docking with RosettaDock produced models of the complex and top-scoring models were subsequently clustered. Clustering data was analyzed for different numbers of crosslinks and subsequently validated by HDX data. When examining the crosslinking data, it was deduced that some crosslinks that suggested an interface at a particular region were undermined by the HDX data that implied protection at the same region, implying that a two-pronged approach was necessary to verify findings. Solvent exposure was additionally examined using SASA for identified models to determine if the models corroborated with regions of protection and exposure identified by HDX. Overall, this methodology elegantly emphasized the importance of more than one structural MS technique being applied to quaternary structure prediction.
Figure 3.
IL-7 (multi-colored, colored with HDX uptake) complexed with IL-7Rα (green) models. Models were docked, clustered, then sorted into types by similarity. Models from each type are depicted in A-C, each utilizing two crosslinking restraints. D, the type 3 model with only one crosslinking restraint. Figure reproduced with permission from Zhang et al. (2019); copyright 2019 American Chemical Society.
HDX-MS data has also been applied to antibody-antigen modeling. Huang et al. used HDX-MS data along with electron-transfer dissociation to examine binding of the mAb1 antibody with a cytokine with implications in autoimmune disease (43). SASA calculations and protein-protein docking provided additional insight into the antibody-antigen binding interface. The study emphasized the importance of HDX-MS data and complementary computational efforts for epitope elucidation. Additionally, recent efforts from Jeliazkov and others were applied to the improvement of Rosetta software for antigen-antibody modeling, RosettaAntibody and SnugDock (44). The SnugDock feature relies upon flexible docking to elucidate the complementarity determining region (CDR) loop, indicated in antigen binding and unique amongst antibody structures, and to configure an adjustment of the heavy and light fragments relevant to antigen-antibody interactions. Restraints from HDX-MS data were used to score antigen-antibody complexes based on agreement with the data. When testing the HDX-MS restraints on an antibody-antigen complex with available labeling data, it was deduced that the HDX-MS restraint-based methodology led to more native-like structure of the CDR loop.
Hydroxyl radical protein footprinting
Hydroxyl radical protein footprinting (HRPF) is a non-specific CL-MS technique in which hydroxyl radicals can covalently modify nineteen of the twenty amino acids types in proteins (11). Synthesized via photolysis or radiolysis of water or hydrogen peroxide, hydroxyl radicals modify residues with varying degrees of reliability and reactivity, as indicated by a broad range of relative intrinsic reactivities (12). Rate constants for labeled peptide fragments and individual residues can be determined and used to calculate protection factor (PF), the relative intrinsic reactivity divided by the labeling rate constant for the particular residue. Because HRPF is more likely to occur in regions that are solvent exposed, residues that are more protected (higher PF) are correlated with lower solvent exposure, and vice versa.
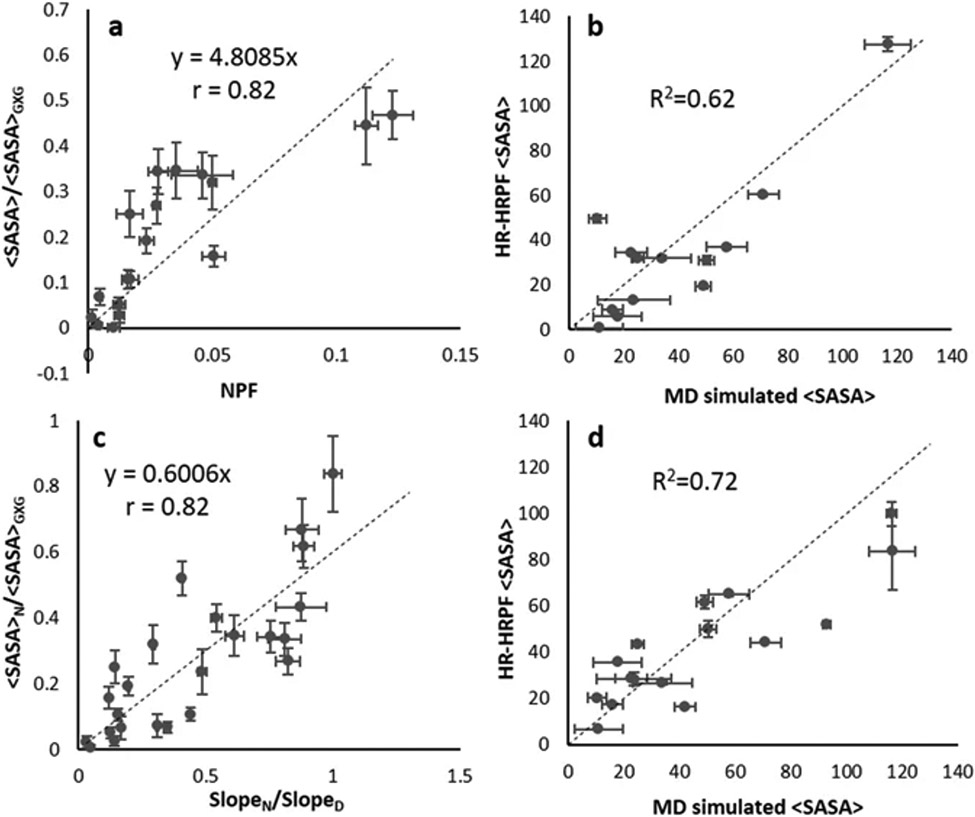
Xie and colleagues recently examined the relationship between residue protection and solvent exposure using MD simulations (45). The work emphasized that normalization of HRPF data should be sequence-dependent, not based on standard values determined from free amino acids. With labeling data for myoglobin and lysozyme, a method was proposed in which accurate side chain SASA values are derived from HRPF data by normalizing labeling data based on sequence context. This was validated by improvements in correlation between labeling data and SASA. When examining the relationship between normalized PF and relative SASA, the correlation was determined to worsen as the relative intrinsic reactivity of the amino acids considered decreased, suggesting that only residues with higher intrinsic reactivity should be used in structural analysis based on PF. When the rate constant of a particular residue in the folded protein was normalized with the rate constant of the same residue in the denatured protein, the correlation improved for all non-sulfur-containing residues (Figure 4). A prediction equation that established a relationship between relative SASA and the normalized rate constant was determined such that relative SASA could be calculated from HRPF data. When the prediction equation was tested with homology models of lysozyme, it was observed that models with backbone RMSD less than 3 Å could be differentiated from models with backbone RMSD greater than 4 Å.
Figure 4.
Comparison of prediction equations using SASA and HRPF data. a, prediction equation between relative SASA (<SASA>/<SASA>GXG) and normalized protection factor (slopeN/relative intrinsic reactivity) using myoglobin data for residue types WYFHLI. b, lysozyme <SASA> calculated using prediction equation derived from (a) versus SASA observed in MD simulations. c, prediction equation between relative SASA of the native (<SASA>N/<SASA>GXG) and rate constant ratio (slopeN/slopeD) for all non-sulfur containing myoglobin residues. d, lysozyme SASA calculated using prediction equation shown in (c) versus SASA observed in MD simulations. Figure reproduced under the Creative Commons License from Xie et al. (2017); copyright 2017 Springer Nature.
Our group has used HRPF labeling data for protein structure prediction. We used the relationship between the natural logarithm of PF (lnPF) and a residue exposure metric, spherical neighbor count, for 15 relaxed crystal structures of calmodulin as a prediction equation. The equation was then implemented in the first available software to use HRPF data for protein structure prediction (46). When tested on ab initio models for four benchmark proteins, the addition of our score term within the Rosetta framework led to improvement in the best scoring model RMSD and funnel-like quality of the score versus RMSD distributions. Results were further validated through use of a confidence metric that assessed the funnel-like quality of the score versus RMSD distribution when RMSD was calculated to the best scoring model. Follow-up work explored the incorporation of labeling data into the ab initio folding algorithm, as opposed to using it for model rescoring.(47)
More recently, we sought to improve the correlation between lnPF and neighbor count, as we hypothesized that accounting for side chain flexibility would improve the relationship (48). We utilized a conical neighbor count for a subset of residue types selected based on intermediate to high intrinsic reactivity and simulated side chain flexibility with MD simulations and with a Rosetta mover ensemble for four benchmark proteins. Upon determining that the normalized root mean square error of lnPF versus conical neighbor count was comparable between MD and the mover ensemble, we developed a new Rosetta score term. 20,000 ab initio models were scored with our term, then a total score was calculated by combining the HRPF score with the Rosetta score. The top 20 scoring models were used as inputs for mover model generation, then scored with both Rosetta and HRPF data. Upon including mover models in our distributions, we found that the best scoring model RMSD was identified at accurate atomic detail for three of the four proteins, indicating that HRPF in combination with a Rosetta mover ensemble can be used to significantly improve model quality.
Other covalent labeling methods and limited proteolysis
Besides the popular HDX and HRPF techniques, other covalent labels have been used to elucidate protein structure. Carbene, another nonspecific covalent labeling reagent, has been used for structural mass spectrometry. Carbene footprinting was applied by Manzi and coworkers to examine the binding sites of lysozyme and a large protease (49). Additional work by Manzi et al. demonstrated that carbene footprinting could be applied to more complex cases by elucidating the interfaces of a trimer membrane protein (50). Radical trifluoromethylation, in which 18 amino acids can be modified, has also been used for covalent labeling structural MS. Myoglobin, beta-lactoglobulin, and membrane protein vitamin K epoxide reductase were explored by radical trifluoromethylation in novel efforts by Cheng and coworkers (51). This work paved the way for an additional study in which trifluoromethyl radicals were produced via synchrotron radiolysis (52). Radical trifluoromethylation is a particularly promising technique for future structure prediction efforts.
In addition to non-specific covalent labeling reagents, other covalent labeling reagents that modify only specific residues have been used to probe protein structure. Diethylpyrocarbonate (DEPC) is a readily available labeling reagent that modifies Cys, Lys, His, Ser, Thr, and Tyr residues along with the N-terminus. It was recently shown that the residue microenvironment played a role in labeling weakly nucleophilic Ser, Thr, and Tyr (STY) residues, as labeled STY residues with lower solvent exposure were found to be in the vicinity of hydrophobic residues (53). Based on this study, we developed a score term within Rosetta to reward models that demonstrated agreement with DEPC labeling data (54). Labeled STY residues with 5-35% relative SASA were rewarded for having more hydrophobic neighbors, while unlabeled STY residues with the same solvent exposure were rewarded for having less hydrophobic neighbors. Additionally, our term rewarded labeled His and Lys residues with higher solvent exposure, as residues that are more exposed are more likely to be covalently labeled. The DEPC score was added to the Rosetta score, and models were ranked by total score. We tested our term with ab initio and homology models for six benchmark proteins and found that the best scoring model RMSD and funnel-like quality of the score versus RMSD distributions improved with use of our term.
Similar to covalent labeling, limited proteolysis is a technique in which a protein is exposed to a low concentration of protease that cleaves solvent accessible regions of the protein (13; 55). Hennig and coworkers developed a pipeline between MDMDAT, software that analyzes MS data, and HADDOCK, a protein-protein docking algorithm (56). Limited proteolysis data was first analyzed by MDMDAT and then utilized by HADDOCK to dock the protein Rpn13 with ubiquitin. This work demonstrated that limited proteolysis data could be applied to a protocol for protein complex modeling that was easier and quicker than structure determination methods such as NMR. Limited proteolysis was also applied to examine protein complexes in work by Proctor and colleagues (57). Limited proteolysis elucidated by MS guided the modeling of the Cu,Zn superoxide dismutase (SOD1) trimer protein complex. Software was developed to translate locations of proteolysis into restraints that were applied to discrete MD simulations. Such restraints emphasized the importance of regions affected by proteolysis being solvent exposed. After coarse-grained and full atom MD simulations to isolate the lowest energy model, computational mutagenesis was applied to examine interface residues of importance to SOD1 trimer generation.
Ion mobility
Ion mobility (IM) is a structural native mass spectrometry technique in which proteins are subjected to soft ionization in the gas phase and then exposed to a nitrogen or helium gas chamber in which an electric field is applied. Instead of residue or fragment-resolved data as for the previously described techniques, IM-MS provides insight into the shape of the protein. Commonly calculated from IM-MS data is the collision cross section (CCS), which is the rotationally averaged two-dimensional projection area of the protein. Computational methods currently exist to predict CCS from protein structure, including the trajectory method (58; 59), projection superposition approximation (60), and projection approximation (61).
In elegant work by Bleiholder and Liu (62), MD simulations were employed to model ubiquitin at various charge states for ion spectra prediction. The structure relaxation approximation (SRA) method was introduced to examine the similarity of ubiquitin ions to the native protein. SRA operated with input MD simulation frames by removing solvent, adjusting the charge state via charged residues with high exposure, relaxing the structure with a short simulation of the gas-phase protein, calculating average cross sections with the projection superposition approximation, and then determining the ion mobility spectrum based on Gaussian distributions of the averaged cross sections. The method was validated by the agreement of residue interactions between the crystal structure and modeled states, demonstrating that ubiquitin remained native-like during the procedure.
Hall and colleagues examined a modeling method in which coarse-grained models of protein complexes were evaluated with a scoring function based on their agreement with CCS data (63). Complexes from the protein data bank were used to validate the use of coarse-grained models, and it was demonstrated that the CCS of the coarse-grained models were similar to those calculated using all-atom models. The coarse-grained model relied upon spheres to represent individual proteins while a complex was represented by multiple spheres. For the scoring function, volume and CCS restraints were implemented based on the findings from a benchmark set. This method was then applied to influenza B virus neuraminidase, where models were scored based on volume and CCS restraints and then clustered by similarity to other models. The most native-like model was identified within the largest cluster. The method was further applied to tryptophan synthase and nitrobenzene dioxygenase complexes. The case study of nitrobenzene dioxygenase successfully identified high quality models, while the tryptophan synthase uncovered the relevance for symmetry data, which was identified by other experiments. This work confirmed that IM-MS data was able to play a valuable role in protein complex structure investigation.
Eschweiler and coworkers used IM-MS data and computational modeling to elucidate a structural model of the urease activation complex (64). CCS values were determined for the subcomplexes of interest and used to guide coarse-grained model generation with the IMP, representing subunits within the complex as individual spheres. A Monte Carlo algorithm was applied to sample conformational space with the aid of restraints from both CCS data and previous experimental data that established connectivity between particular subunits. IMPACT was applied to determine CCS values for complex models, followed by a clustering and comparison to existing complex structures. This study effectively modeled a very large complex using numerous restraints from experimental and calculated CCS, XL-MS, and SAXS data. A similar methodology was applied in recent work by Wang and others. In order to model apolipoprotein E oligomers relevant to Alzheimer’s disease, IM-MS data was used to identify coarse-grained models using the IMP (65). Additionally, collision-induced unfolding was used to examine the monomer and tetramer of apolipoprotein E. This work deviated from the use of spheres for each individual subunit within the complex. Instead, the monomer was modelled with two domains, or two spheres, within the coarse-grained model, which corroborated the CCS data. A Monte-Carlo algorithm was applied to identify models, which were subsequently clustered by similarity in order to determine a likely complex structure. Intriguingly, electron-capture dissociation was also implemented to validate models based on identification of flexible portions of the complex, demonstrating the capability of IM-MS and IMP modeling coupled with additional experimental techniques.
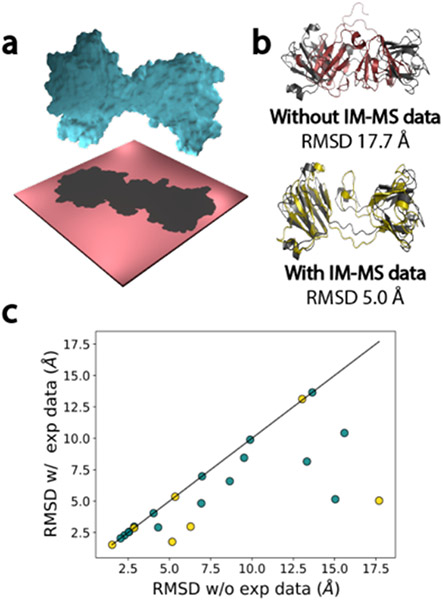
Finally, our group has developed Rosetta functionality to use IM-MS data in protein tertiary structure prediction (66). An algorithm, Projection Approximation using Rough Circular Shapes (PARCS), was implemented to calculate CCS from protein structure. PARCS was shown to perform as accurately and efficiently as the popular IMPACT method. A score term reliant upon IM-MS data was also incorporated into the Rosetta framework based on the PARCS predictions. The score term penalized models with differences in observed and predicted CCS. It was first tested on models for a benchmark set of proteins with PARCS-computed CCS values in which the RMSD of best scoring models was improved for 82 of the 100 proteins examined (Figure 5). The funnel-like quality of the score versus RMSD distributions for model sets also tended to improve upon scoring with IM-MS data. Additionally, the score term was examined with ab initio and homology models for 23 proteins for which experimental IM-MS data was available, with the RMSD improving or exhibiting no change for all 23 instances. This work further solidified the capability of IM-MS methods to elucidate protein structure.
Figure 5.
Incorporation of IM-MS data into Rosetta improved RMSD of best scoring models. a, Depiction of a protein and its projection on a plane upon space-filling measures by the PARCS application. b, Structural alignments of the crystal structure (grey) with the best scoring model when scoring without IM-MS data (burgundy) and with IM-MS data (yellow). c, Comparison of best scoring model RMSDs when scoring with and without IM data. Helium buffer gas conditions are depicted in teal while nitrogen buffer gas conditions are gold. Figure credit: SM Bargeen Alam Turzo.
Surface-induced dissociation
Recently emerging as a structural native MS technique, surface-induced dissociation (SID) relies on the breakage of interfaces within a protein complex when the complex strikes a surface. During SID-MS, protein complexes undergo soft ionization, then are collided with a surface, which can provide insight into the stoichiometry and interfaces within a protein complex. It has been demonstrated that the dissociation observed in SID experiments can be correlated with identified assembly pathways (67-69).
We demonstrated that it is possible to predict SID appearance energy (AE) from protein structure (70). AE, specified as 10% fragmentation, was predicted from quantities such as the number of residues at the interface, number of unsatisfied hydrogen bonds, and rigidity factor, which was determined by intermolecular interactions such as hydrogen bonds, salt bridges, and disulfide bonds. A weighted sum of these terms was used in a prediction equation such that a strong correlation was observed between predicted and experimental AE. The development of this model suggested that the methodology could be applied to structure prediction applications.
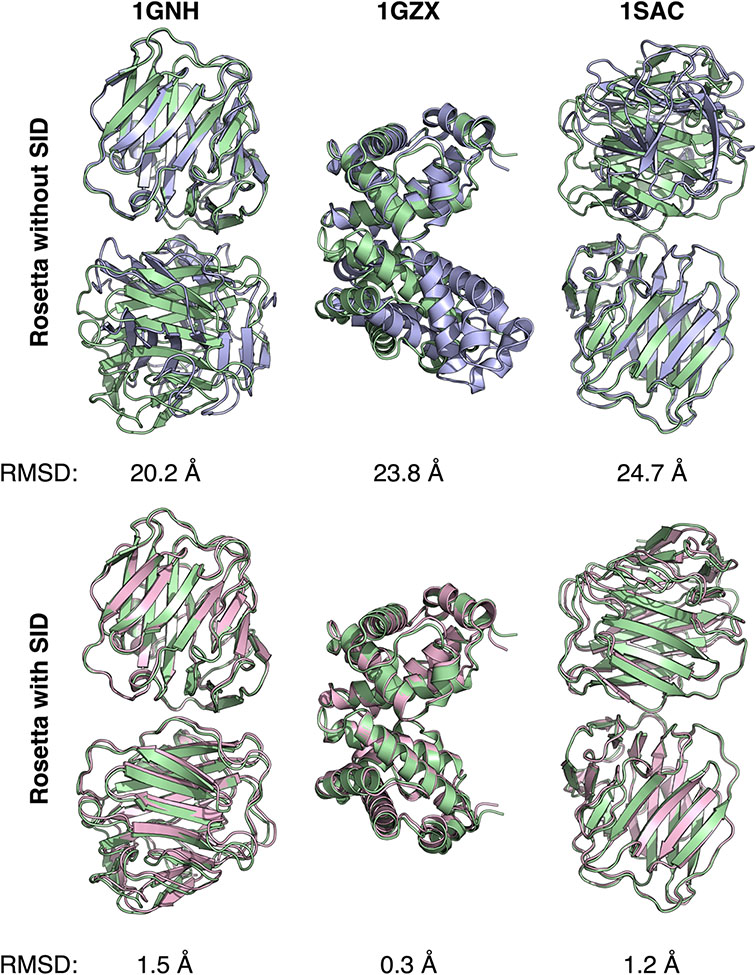
Our group then developed a computational algorithm to use SID-MS data for protein complex structure prediction (71). The number of residues at the interface, rigidity factor, and buried hydrophobic surface area were combined to better predict AE. The new model that combined these three terms was then used in the creation of a Rosetta scoring term that combined SID data with RosettaDock scoring. It was first tested on 57 protein systems using crystal structures to calculate the ‘experimental’ AE, with 54/57 cases demonstrating improvement or no change in best scoring model RMSD. When using experimentally determined AE from SID-MS, it was determined that six of the nine complexes examined demonstrated near-native structures within the top three scoring models (Figure 6). Additionally, a confidence metric was established in this work, using the average score per residue for the best 1,000 models to independently verify the accuracy of scoring. The confidence metric allowed identification of successful predictions, as proteins with more-negative score per residue tended to have improved RMSD values compared to complexes with a higher score per residue. Overall, this work demonstrated that SID data in conjunction with RosettaDock can be used to improve protein complex structure prediction effectively. In follow-up work, it was shown recently that using SID-MS data in combination with cryo-EM data resulted in improved flexible docking results for protein complexes and required less prior knowledge of structures (72).
Figure 6.
Utilization of SID-MS data improved RMSD of best scoring models. Alignment of the crystal structures (green) with one of the top three best scoring models when scoring without SID data (blue, top row) and when including SID-MS data in scoring (pink, bottom row) for three protein complexes. Figure credit: Justin Seffernick.
Future directions of the field
While advances in MS and computational technologies have propelled the field forward in recent years, obstacles still exist and will require provocative solutions to overcome.
As MS data are too sparse to determine protein structure unambiguously, computational techniques will remain relevant to interpret MS data for structure elucidation. One way in which the community can support computational method development is through the establishment of central data repositories. Such databases currently exist for other experimental techniques (73-75). Kahraman and coworkers (25) have started to pave the way for this effort by establishing a crosslinking database. Hopefully, other MS databases will follow suit in the near future. Publicly available datasets can lead to the creation and development of freely accessible, competitive algorithms that can harness sparse experimental data, such as the mass spectrometry data outlined here, to improve structure prediction with machine learning and artificial intelligence methodologies.
Because MS data is sparse, even advanced computational methodologies will inevitably predict false positive structures. Going forward, integrative structural modeling that combines multiple sets of experimental data will be instrumental in reducing the rate at which false positives occur. Further exploration of protein complexes remains a key endeavor for the future of protein structure modeling. Protein complexes have been implicated to have roles in many biological processes, and structural changes to complexes can lead to human disease (76). Elucidation of protein complex structure can provide insight into the mechanisms of such complexes. Structural information can complement efforts to target protein complexes with drugs to alleviate implications in disease. The study of protein complexes benefits greatly from integrative experimental techniques to combat modeling ambiguities. This has been nicely demonstrated in work by Zhang and colleagues that applied both HDX and XL data to quaternary structure investigation (42). The field should continue to emphasize combination of multiple techniques to elucidate structural features of protein complexes.
Recently, the performance of AlphaFold at CASP14 has raised questions about the role of experimental techniques in protein structure determination (77). AlphaFold relies upon artificial intelligence to accomplish protein structure prediction from amino acid sequences. Its impressive global distance test (GDT) median score of 92.4 (78) redefined the field’s expectations of how precise modeling algorithms could be. This inevitably caused speculations about the ability to determine protein structure purely computationally. We believe that this is unlikely to happen in the near future. As AlphaFold is currently not accessible to the academic community, computational researchers should continue to establish techniques that mimic AlphaFold. Callaway indicated in the Nature synopsis of CASP14 (77) that purely computational structure determination is unlikely, but rather that sparse experimental data will soon be sufficient for unambiguous structure elucidation in combination with the new wave of artificial intelligence technologies. As such, we anticipate that MS data will play a continued, if not growing, role alongside tools like AlphaFold.
An additional future avenue of protein structure prediction from MS data is citizen science. FoldIt is one such tool that enlists video game enthusiasts for structure prediction (79). With its colorful graphical user interface and endearing symbols for relevant scientific concepts like steric hinderance and solvent exposure of hydrophobic regions, FoldIt uses the Rosetta software suite to reward user-sampled conformations of proteins. Users can advance through multiple levels of the game while supporting scientific efforts by sampling protein conformations that may be inaccessible to automated protein sampling algorithms. Overall, games such as FoldIt inspire a new generation of scientists while tackling the sampling problem and examining novel protein conformations.
In summary, the future of MS techniques with complementary computational methods appears promising. The combination of MS and computational protocols will, in our opinion, lead to the elucidation of many challenging protein structures.
Conclusion
The field of structural mass spectrometry has significantly benefited from the development of hybrid computational techniques for MS-guided protein structure prediction. Algorithms that use XL-MS, HDX-MS, HRPF-MS, limited proteolysis, IM-MS, and SID-MS data for tertiary and quaternary structure prediction, described here, successfully allow structure elucidation from sparse MS data. The field will continue to thrive with efforts to maintain accessible datasets and software packages, to combine multiple techniques for the purpose of protein complex elucidation, and to pursue out-of-the-box methods like FoldIt that recruit the general public into structure prediction efforts. While it is encouraging to see how far the field has progressed recently, it remains even more exciting to envision where the field will go with continued advances in techniques and technology.
Acknowledgements
The authors are grateful to the members of the Lindert group for insightful discussions. We thank SM Bargeen Alam Turzo for designing and providing Figure 5 and Justin Seffernick for designing and providing Figure 6. MS-guided protein modeling work in the Lindert group was supported by the NSF (Grant No. CHE 1750666), the NIH (Grant No. P41 GM128577), and a Sloan Research Fellowship to S. L.
Footnotes
Competing interest
The authors declare no competing interests.
Literature Cited
- 1.Leelananda SP, Lindert S. 2016. Computational methods in drug discovery. Beilstein journal of organic chemistry 12:2694–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smyth M, Martin J. 2000. x Ray crystallography. Molecular Pathology 53:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavalli A, Salvatella X, Dobson CM, Vendruscolo M. 2007. Protein structure determination from NMR chemical shifts. Proceedings of the National Academy of Sciences 104:9615–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costa TR, Ignatiou A, Orlova EV. 2017. Structural analysis of protein complexes by cryo electron microscopy. Bacterial Protein Secretion Systems:377–413 [DOI] [PubMed] [Google Scholar]
- 5.Limpikirati P, Liu T, Vachet RW. 2018. Covalent labeling-mass spectrometry with non-specific reagents for studying protein structure and interactions. Methods 144:79–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiselar JG, Chance MR. 2010. Future directions of structural mass spectrometry using hydroxyl radical footprinting. Journal of mass spectrometry 45:1373–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu XR, Zhang MM, Gross ML. 2020. Mass Spectrometry-Based Protein Footprinting for Higher-Order Structure Analysis: Fundamentals and Applications. Chemical Reviews 120:4355–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinz A 2006. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein–protein interactions. Mass Spectrometry Reviews 25:663–82 [DOI] [PubMed] [Google Scholar]
- 9.O’Reilly FJ, Rappsilber J. 2018. Cross-linking mass spectrometry: methods and applications in structural, molecular and systems biology. Nature Structural & Molecular Biology 25:1000–8 [DOI] [PubMed] [Google Scholar]
- 10.Konermann L, Pan J, Liu Y-H. 2011. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chemical Society Reviews 40:1224–34 [DOI] [PubMed] [Google Scholar]
- 11.Huang W, Ravikumar KM, Chance MR, Yang S. 2015. Quantitative mapping of protein structure by hydroxyl radical footprinting-mediated structural mass spectrometry: a protection factor analysis. Biophysical Journal 108:107–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu G, Chance MR. 2007. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chemical Reviews 107:3514–43 [DOI] [PubMed] [Google Scholar]
- 13.Hager-Braun C, Tomer KB. 2005. Determination of protein-derived epitopes by mass spectrometry. Expert Review of Proteomics 2:745–56 [DOI] [PubMed] [Google Scholar]
- 14.Jurneczko E, Barran PE. 2011. How useful is ion mobility mass spectrometry for structural biology? The relationship between protein crystal structures and their collision cross sections in the gas phase. Analyst 136:20–8 [DOI] [PubMed] [Google Scholar]
- 15.Zhou M, Wysocki VH. 2014. Surface induced dissociation: dissecting noncovalent protein complexes in the gas phase. Accounts of Chemical Research 47:1010–8 [DOI] [PubMed] [Google Scholar]
- 16.Seffernick JT, Lindert S. 2020. Hybrid methods for combined experimental and computational determination of protein structure. The Journal of Chemical Physics 153:240901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leman JK, Weitzner BD, Lewis SM, Adolf-Bryfogle J, Alam N, et al. 2020. Macromolecular modeling and design in Rosetta: recent methods and frameworks. Nature Methods:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alford RF, Leaver-Fay A, Jeliazkov JR, O’Meara MJ, DiMaio FP, et al. 2017. The Rosetta all-atom energy function for macromolecular modeling and design. Journal of Chemical Theory and Computation 13:3031–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER Suite: protein structure and function prediction. Nature Methods 12:7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols 10:845–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russel D, Lasker K, Webb B, Velázquez-Muriel J, Tjioe E, et al. 2012. Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol 10:e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dominguez C, Boelens R, Bonvin AM. 2003. HADDOCK: a protein–protein docking approach based on biochemical or biophysical information. Journal of the American Chemical Society 125:1731–7 [DOI] [PubMed] [Google Scholar]
- 23.Webb B, Sali A. 2016. Comparative protein structure modeling using MODELLER. Current protocols in bioinformatics 54:5.6. 1–5.6. 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider M, Belsom A, Rappsilber J, Brock O. 2016. Blind testing of cross-linking/mass spectrometry hybrid methods in CASP11. Proteins: Structure, Function, and Bioinformatics 84:152–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kahraman A, Herzog F, Leitner A, Rosenberger G, Aebersold R, Malmström L. 2013. Cross-link guided molecular modeling with ROSETTA. PloS one 8:e73411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kahraman A, Malmström L, Aebersold R. 2011. Xwalk: computing and visualizing distances in cross-linking experiments. Bioinformatics 27:2163–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lössl P, Kölbel K, Tänzler D, Nannemann D, Ihling CH, et al. 2014. Analysis of nidogen-1/laminin γ1 interaction by cross-linking, mass spectrometry, and computational modeling reveals multiple binding modes. PLoS One 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piotrowski C, Moretti R, Ihling CH, Haedicke A, Liepold T, et al. 2020. Delineating the Molecular Basis of the Calmodulin–bMunc13-2 Interaction by Cross-Linking/Mass Spectrometry—Evidence for a Novel CaM Binding Motif in bMunc13-2. Cells 9:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belsom A, Schneider M, Fischer L, Brock O, Rappsilber J. 2016. Serum albumin domain structures in human blood serum by mass spectrometry and computational biology. Molecular & Cellular Proteomics 15:1105–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dos Santos RN, Ferrari AJ, de Jesus HC, Gozzo FC, Morcos F, Martínez L. 2018. Enhancing protein fold determination by exploring the complementary information of chemical cross-linking and coevolutionary signals. Bioinformatics 34:2201–8 [DOI] [PubMed] [Google Scholar]
- 31.Hauri S, Khakzad H, Happonen L, Teleman J, Malmström J, Malmström L. 2019. Rapid determination of quaternary protein structures in complex biological samples. Nature Communications 10:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khakzad H, Happonen L, Van Nhieu GT, Malmström J, Malmström L. 2020. In vivo cross-linking MS of the complement system MAC assembled on live Gram-positive bacteria. Frontiers in Genetics 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrari AJ, Gozzo FC, Martínez L. 2019. Statistical force-field for structural modeling using chemical cross-linking/mass spectrometry distance constraints. Bioinformatics 35:3005–12 [DOI] [PubMed] [Google Scholar]
- 34.Degiacomi MT, Schmidt C, Baldwin AJ, Benesch JL. 2017. Accommodating protein dynamics in the modeling of chemical crosslinks. Structure 25:1751–7. e5 [DOI] [PubMed] [Google Scholar]
- 35.Mintseris J, Gygi SP. 2020. High-density chemical cross-linking for modeling protein interactions. Proceedings of the National Academy of Sciences 117:93–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan J, Han J, Borchers CH, Konermann L. 2009. Hydrogen/deuterium exchange mass spectrometry with top-down electron capture dissociation for characterizing structural transitions of a 17 kDa protein. Journal of the American Chemical Society 131:12801–8 [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Majumder EL-W, Yue H, Blankenship RE, Gross ML. 2014. Structural analysis of diheme cytochrome c by hydrogen–deuterium exchange mass spectrometry and homology modeling. Biochemistry 53:5619–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marzolf DR, Seffernick JT, Lindert S. 2021. Protein Structure Prediction from NMR Hydrogen–Deuterium Exchange Data. Journal of Chemical Theory and Computation 17:2619–29 [DOI] [PubMed] [Google Scholar]
- 39.Mohammadiarani H, Shaw VS, Neubig RR, Vashisth H. 2018. Interpreting hydrogen–deuterium exchange events in proteins using atomistic simulations: Case studies on regulators of G-protein signaling proteins. The Journal of Physical Chemistry B 122:9314–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martens C, Shekhar M, Lau AM, Tajkhorshid E, Politis A. 2019. Integrating hydrogen–deuterium exchange mass spectrometry with molecular dynamics simulations to probe lipid-modulated conformational changes in membrane proteins. Nature Protocols 14:3183–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makarov AA, Iacob RE, Pirrone GF, Rodriguez-Granillo A, Joyce L, et al. 2020. Combination of HDX-MS and in silico modeling to study enzymatic reactivity and stereo-selectivity at different solvent conditions. Journal of Pharmaceutical and Biomedical Analysis 182:113141. [DOI] [PubMed] [Google Scholar]
- 42.Zhang MM, Beno BR, Huang RY-C, Adhikari J, Deyanova EG, et al. 2019. An Integrated Approach for Determining a Protein–Protein Binding Interface in Solution and an Evaluation of Hydrogen–Deuterium Exchange Kinetics for Adjudicating Candidate Docking Models. Analytical Chemistry 91:15709–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang RY-C, Krystek SR Jr, Felix N, Graziano RF, Srinivasan M, et al. Hydrogen/deuterium exchange mass spectrometry and computational modeling reveal a discontinuous epitope of an antibody/TL1A Interaction. Proc. MAbs, 2018, 10:95–103: Taylor & Francis; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeliazkov JR, Frick R, Zhou J, Gray JJ. 2021. Robustification of RosettaAntibody and Rosetta SnugDock. PloS one 16:e0234282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie B, Sood A, Woods RJ, Sharp JS. 2017. Quantitative protein topography measurements by high resolution hydroxyl radical protein footprinting enable accurate molecular model selection. Scientific Reports 7:4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aprahamian ML, Chea EE, Jones LM, Lindert S. 2018. Rosetta protein structure prediction from hydroxyl radical protein footprinting mass spectrometry data. Analytical Chemistry 90:7721–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aprahamian ML, Lindert S. 2019. Utility of Covalent Labeling Mass Spectrometry Data in Protein Structure Prediction with Rosetta. Journal of Chemical Theory and Computation 15:3410–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biehn SE, Lindert S. 2021. Accurate protein structure prediction with hydroxyl radical protein footprinting data. Nature Communications 12:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manzi L, Barrow AS, Scott D, Layfield R, Wright TG, et al. 2016. Carbene footprinting accurately maps binding sites in protein–ligand and protein–protein interactions. Nature Communications 7:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manzi L, Barrow AS, Hopper JT, Kaminska R, Kleanthous C, et al. 2017. Carbene footprinting reveals binding interfaces of a multimeric membrane-spanning protein. Angewandte Chemie 129:15069–73 [DOI] [PubMed] [Google Scholar]
- 51.Cheng M, Zhang B, Cui W, Gross ML. 2017. Laser-initiated radical trifluoromethylation of peptides and proteins: Application to mass-spectrometry-based protein footprinting. Angewandte Chemie International Edition 56:14007–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng M, Asuru A, Kiselar J, Mathai G, Chance MR, Gross ML. 2020. Fast protein footprinting by X-ray mediated radical trifluoromethylation. Journal of the American Society for Mass Spectrometry 31:1019–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Limpikirati P, Pan X, Vachet RW. 2019. Covalent Labeling with Diethylpyrocarbonate: Sensitive to the Residue Microenvironment, Providing Improved Analysis of Protein Higher Order Structure by Mass Spectrometry. Analytical Chemistry 91:8516–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Biehn SE, Limpikirati P, Vachet RW, Lindert S. 2021. Utilization of hydrophobic microenvironment sensitivity in diethylpyrocarbonate labeling for protein structure prediction. Analytical Chemistry [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fontana A, de Laureto PP, Spolaore B, Frare E. 2012. Identifying disordered regions in proteins by limited proteolysis. In Intrinsically disordered protein analysis:297–318: Springer. Number of 297-318 pp. [DOI] [PubMed] [Google Scholar]
- 56.Hennig J, de Vries SJ, Hennig KD, Randles L, Walters KJ, et al. 2012. MTMDAT-HADDOCK: high-throughput, protein complex structure modeling based on limited proteolysis and mass spectrometry. BMC Structural Biology 12:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Proctor EA, Fee L, Tao Y, Redler RL, Fay JM, et al. 2016. Nonnative SOD1 trimer is toxic to motor neurons in a model of amyotrophic lateral sclerosis. Proceedings of the National Academy of Sciences 113:614–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mesleh M, Hunter J, Shvartsburg A, Schatz GC, Jarrold M. 1996. Structural information from ion mobility measurements: effects of the long-range potential. The Journal of Physical Chemistry 100:16082–6 [Google Scholar]
- 59.Ewing SA, Donor MT, Wilson JW, Prell JS. 2017. Collidoscope: an improved tool for computing collisional cross-sections with the trajectory method. Journal of The American Society for Mass Spectrometry 28:587–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bleiholder C, Wyttenbach T, Bowers MT. 2011. A novel projection approximation algorithm for the fast and accurate computation of molecular collision cross sections (I). Method. International Journal of Mass Spectrometry 308:1–10 [Google Scholar]
- 61.Marklund EG, Degiacomi MT, Robinson CV, Baldwin AJ, Benesch JL. 2015. Collision cross sections for structural proteomics. Structure 23:791–9 [DOI] [PubMed] [Google Scholar]
- 62.Bleiholder C, Liu FC. 2019. Structure relaxation approximation (sra) for elucidation of protein structures from ion mobility measurements. The Journal of Physical Chemistry B 123:2756–69 [DOI] [PubMed] [Google Scholar]
- 63.Hall Z, Politis A, Robinson CV. 2012. Structural modeling of heteromeric protein complexes from disassembly pathways and ion mobility-mass spectrometry. Structure 20:1596–609 [DOI] [PubMed] [Google Scholar]
- 64.Eschweiler JD, Farrugia MA, Dixit SM, Hausinger RP, Ruotolo BT. 2018. A structural model of the urease activation complex derived from ion mobility-mass spectrometry and integrative modeling. Structure 26:599–606. e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H, Eschweiler J, Cui W, Zhang H, Frieden C, et al. 2019. Native mass spectrometry, ion mobility, electron-capture dissociation, and modeling provide structural information for gas-phase apolipoprotein E oligomers. Journal of The American Society for Mass Spectrometry 30:876–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Turzo SBA, Seffernick JT, Rolland AD, Donor MT, Heinze S, et al. 2021. Protein shape sampled by ion mobility mass spectrometry consistently improves protein structure prediction. bioRxiv:2021.05.27.445812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Romano CA, Zhou M, Song Y, Wysocki VH, Dohnalkova AC, et al. 2017. Biogenic manganese oxide nanoparticle formation by a multimeric multicopper oxidase Mnx. Nature Communications 8:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song Y, Nelp MT, Bandarian V, Wysocki VH. 2015. Refining the structural model of a heterohexameric protein complex: Surface induced dissociation and ion mobility provide key connectivity and topology information. ACS Central Science 1:477–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quintyn RS, Yan J, Wysocki VH. 2015. Surface-induced dissociation of homotetramers with D2 symmetry yields their assembly pathways and characterizes the effect of ligand binding. Chemistry & Biology 22:583–92 [DOI] [PubMed] [Google Scholar]
- 70.Harvey SR, Seffernick JT, Quintyn RS, Song Y, Ju Y, et al. 2019. Relative interfacial cleavage energetics of protein complexes revealed by surface collisions. Proceedings of the National Academy of Sciences 116:8143–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seffernick JT, Harvey SR, Wysocki VH, Lindert S. 2019. Predicting protein complex structure from surface-induced dissociation mass spectrometry data. ACS Central Science 5:1330–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seffernick JT, Canfield SM, Harvey SR, Wysocki VH, Lindert S. 2021. Prediction of protein complex structure using surface-induced dissociation and cryo-EM. Analytical Chemistry [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berman H, Henrick K, Nakamura H. 2003. Announcing the worldwide protein data bank. Nature Structural & Molecular Biology 10:980- [DOI] [PubMed] [Google Scholar]
- 74.Lawson CL, Patwardhan A, Baker ML, Hryc C, Garcia ES, et al. 2016. EMDataBank unified data resource for 3DEM. Nucleic Acids Research 44:D396–D403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pancsa R, Varadi M, Tompa P, Vranken WF. 2016. Start2Fold: a database of hydrogen/deuterium exchange data on protein folding and stability. Nucleic Acids research 44:D429–D34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bergendahl LT, Gerasimavicius L, Miles J, Macdonald L, Wells JN, et al. 2019. The role of protein complexes in human genetic disease. Protein Science 28:1400–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Callaway E. 2020. 'It will change everything': DeepMind's AI makes gigantic leap in solving protein structures. Nature. [DOI] [PubMed] [Google Scholar]
- 78.Service RF. 2020. ‘The game has changed.’AI triumphs at protein folding. American Association for the Advancement of Science [DOI] [PubMed] [Google Scholar]
- 79.Kleffner R, Flatten J, Leaver-Fay A, Baker D, Siegel JB, et al. 2017. Foldit Standalone: a video game-derived protein structure manipulation interface using Rosetta. Bioinformatics 33:2765–7 [DOI] [PMC free article] [PubMed] [Google Scholar]